- 1Team Immunity and Cancer, Cancer Research Center of Marseille (CRCM), Inserm U1068, CNRS UMR7258, Paoli-Calmettes Institute, University of Aix-Marseille UM105, Marseille, France
- 2Immunomonitoring Department, Paoli-Calmettes Institute, Marseille, France
- 3Cytometry Platform, Cancer Research Center of Marseille (CRCM), Inserm U1068, CNRS UMR7258, Paoli-Calmettes Institute, University of Aix-Marseille UM105, Marseille, France
- 4Centre for Clinical Investigation in Biotherapy, Paoli-Calmettes Institute, University of Aix-Marseille, Inserm CBT 1409, Marseille, France
- 5Hematology Department, CRCM, Inserm U1068, CNRS UMR7258, Paoli-Calmettes Institute, Marseille, France
- 6Paris Cité University, Saint-Louis Research Institute, Inserm UMRS1160, Paris, France
- 7Immunology and Histocompatibility Laboratory, Assistance Publique-Hôpitaux de Paris, Saint-Louis Hospital, Paris, France
Introduction: Acute myeloid leukemia (AML) is a rare haematological cancer with poor 5-years overall survival (OS) and high relapse rate. Leukemic cells are sensitive to Natural Killer (NK) cell mediated killing. However, NK cells are highly impaired in AML, which promote AML immune escape from NK cell immune surveillance. We made the first report of CD56neg CD16+ NK cells expansion in AML. This unconventional subset has been reported to expand in some chronic viral infections. Although it is unclear whether CD56neg NK cells expansion mechanism is common across diseases, it seems more relevant than ever to further investigate this subset, representing a potential therapeutic target.
Methods: We used PBMCs from AML patients and HV to perform mass cytometry, spectral flow cytometry, bulk RNA-seq and in vitro assays in order to better characterize CD56neg CD16+ NK cells that expand in AML.
Results: We confirmed that CD56neg CD16+ NK cells represent a unique NK cell subset coexpressing Eomes and T-bet. CD56neg CD16+ NK cells could recover CD56 expression in vitro where they displayed unaltered NK cell functions. We previously demonstrated that CD56neg CD16+ NK cells expansion at diagnosis was associated with adverse clinical outcome in AML. Here, we validated our findings in a validation cohort of N=38 AML patients. AML patients with CD56neg CD16+ NK cells expansion at diagnosis had decreased overall survival (HR[CI95]=5.5[1.2-24.5], p=0.0251) and relapse-free survival (HR[CI95]=13.1[1.9-87.5], p=0.0079) compared to AML patients without expansion after 36 months follow-up. RNA-seq unveiled that CD56neg CD16+ NK cells were mature circulating NK cells with functional capacities. Upon expansion, CD56neg CD16+ NK cells from AML patients showed altered proteomic phenotype, with increased frequency of terminally mature CD56neg CD16+ NK cells expressing TIGIT along with decreased frequency of Siglec-7+ CD56neg CD16+ NK cells.
Discussion: Taken together, our results suggest that we could harness CD56neg CD16+ NK cells cytotoxic potential in vitro to restore NK cell anti-tumor response in AML patients with CD56neg CD16+ NK cells expansion and improve patients’ prognosis. To conclude, CD56neg CD16+ NK cells represent a relevant target for future NK-cell-based immunotherapies in AML.
Introduction
NK cells are innate cytotoxic lymphocytes with a pivotal role in the clearance of pathogens and tumor cells (1). They can be divided into three distinct functional subsets based on the surface expressions of CD56 and CD16: CD56dim CD16+, CD56dim CD16- and CD56bright NK cells. Recently, a growing interest in CD56neg NK cells has arisen. Indeed, this subset present in healthy volunteers, can significantly increase in pathological context and is thought to be a hallmark of chronic NK cell activation (2). Their expansion was first identified in human immunodeficiency virus-1 (HIV-1) infected patients (3) and further characterized by Mavilio and al (4). Despite no consensus on their maturation stage and functional capacities, there are convincing evidence of their clinical implications in various diseases including HIV-1 (5), hepatitis C virus infection (6), malaria (7) or AML as described in our previous study (8).
AML is a hematological malignancy developing within the bone marrow and characterized by a maturation arrest of myeloid precursors and an uncontrolled clonal expansion of poorly differentiated cells, called leukemic blasts. AML is sensitive to NK cell-mediated cytotoxicity, however, leukemic blasts can implement escape mechanisms, resulting in impaired NK cells and poor prognosis (9, 10). We demonstrated that an expansion of CD56neg NK cells higher than a threshold of 10% at diagnosis was associated with adverse clinical outcome in AML. Interestingly, it persisted in some patients after complete remission.
Although progress has been made in AML therapies over the past decades, it still suffers from poor prognosis in patients aged ≥ 60 with a 5-years survival rate below 15% (11). To restore NK cell cytotoxicity, NK cell-based immunotherapies are currently under investigation (12). The role of CD56 in NK cell cytotoxicity is controversial, however, two recent studies (13, 14) demonstrated its implication in the establishment of immunological synapse, likely to be compromised in CD56neg NK cells. Molecules like NK-cell engagers (15) could help circumvent the problem and induce synapse formation, triggering NK cells cytotoxicity. Although it is unclear whether CD56neg NK cells expansion mechanism is common across diseases, it seems more relevant than ever to further investigate this subset, representing a potential therapeutic target.
Hereinafter, we presented evidence that CD56neg CD16+ cells define a distinct NK cell subset. CD56neg CD16+ NK cells could recover CD56 expression and exhibit potent NK cell functionalities in vitro similarly to CD56+ NK cells, in a specific culture system. Besides, this subset expressed transcription factors crucial to the maintenance of NK cell identity. Furthermore, we confirmed in a validation cohort of N=38 AML patients that CD56neg CD16+ expansion above 10% of total NK cells at diagnosis is associated with adverse clinical outcome. Next, we established a transcriptomic signature of CD56neg CD16+ NK cells characterized by hallmarks of mature circulating cytotoxic NK cells, although some transcripts were indicative of impaired cytotoxicity. Finally, we showed that expanded CD56neg CD16+ NK cells exhibit a distinct phenotype characterized by the increased frequency of terminally mature CD56neg CD16+ NK cells expressing TIGIT and a decreased frequency of Siglec-7+ CD56neg CD16+ NK cells.
Materials and methods
Study design and patient enrolment
New data generated in this study were obtained from frozen peripheral blood mononuclear cells (PBMCs) of patients (N=38) enrolled in the HEMATOBIO cohort (NCT02320656). Blood collection, PBMCs extraction, and storage were performed by the Paoli-Calmettes Tumor Bank, which operates under authorization #AC-2007-33 granted by the French Ministry of Research. Patients with newly diagnosed AML, except acute promyelocytic leukemia, were eligible for study participation. All patients had received standard chemotherapy. The cohort was divided according to the frequency of CD56neg CD16+ NK cells with a threshold of 10% as previously defined (8). The clinical information of all patients is summarized in Table 1. Healthy volunteers (HV, N = 16) were recruited from the French Blood Agency (EFS). Written informed consent was obtained from all patients in accordance with the Declaration of Helsinki. The study was reviewed and approved by the Institutional Review Boards of Paoli-Calmettes Institute.
Fluorescence-activated cell sorting and RNA extraction
Cryopreserved PBMCs from N=4 AML patients and N=5 age-matched HV were thawed in a 37°C water bath, resuspended in RPMI-1640 (Gibco, Grand Island, NY, USA) supplemented with 10% of heat inactivated fetal bovine serum (FBS) (Gibco), centrifugated for 5 min at 1,500 rpm and incubated in a 37°C, 5% CO2 incubator for 30 min. After resting, cells were filtered on a 30 µm pre-separation filter (Miltenyi), centrifugated and incubated in 1X PBS (Gibco) with the viability marker Live/Dead Fixable Aqua (Thermo Fisher Scientific, Waltham, MA, USA) for 25 min on ice in the dark. After washing in 1X PBS and centrifugation for 5 min at 1,500 rpm twice, cells were incubated with BV605 anti-CD56 (clone NCAM16.2), BV711 anti-CD16 (clone 3G8), BV785 anti-CD45 (cloneHI30), FITC anti-CD13 (clone SJ1D1), FITC anti-CD33 (clone HIM3-4), FITC anti-CD34 (clone 581), PE-CF594 anti-CD3 (clone UCHT1) in Brilliant Stain Buffer (BD Biosciences, Franklin Lakes, NJ, USA) for 30 min on ice in the dark. Before acquisition, cells were passed through a 35 µm cell strainer. Cell sorting of CD56bright, CD56dim CD16-, CD56dim CD16+, and CD56neg CD16+ NK cell subsets was performed on a FACS Aria III (BD Biosciences) following the gating strategy outlined in Supplementary Figure S1. All used antibodies are listed in Supplementary Table S1. After cell sorting, total RNA was immediately isolated with a RNeasy Plus Micro Kit (Qiagen, Hilden, Germany) according to manufacturer’s instructions. High-quality RNA was eluted in 14 µl RNase-free water.
Bulk RNA-sequencing and data processing
Samples from patients and HV were respectively pooled, at the same total RNA concentration, according to NK cell subtype prior to sequencing. RNA integrity was assessed on a Fragment Analyzer (Agilent, Santa Clara, CA, USA). Library preparation, RNA Capture Sequencing and were performed by IntegraGen (Evry, France). Briefly, libraries were prepared using NEBNext® Ultra™ II Directional RNA Library Prep Kit for Illumina (NEB, Ipswich, MA, USA) according to supplier recommendations. The RNA was capture with the Twist Human Core Exome (Twist Bioscience, San Francisco, CA, USA) + IntegraGen Custom V2 Enrichment Systems. Sequencing was performed on an Illumina NovaSeq platform and 2 × 100 bp paired-end reads were generated. RNA and sequencing quality control metrics are summarized in Supplementary Table S2.
Spectral flow cytometry, Cell cloning and functional assays, RNA-sequencing analysis, Mass cytometry analysis and Statistical Analysis can be found in Supplementary Materials and Methods.
Results
CD3- CD56neg CD16+ cells represent a distinct NK cell subset
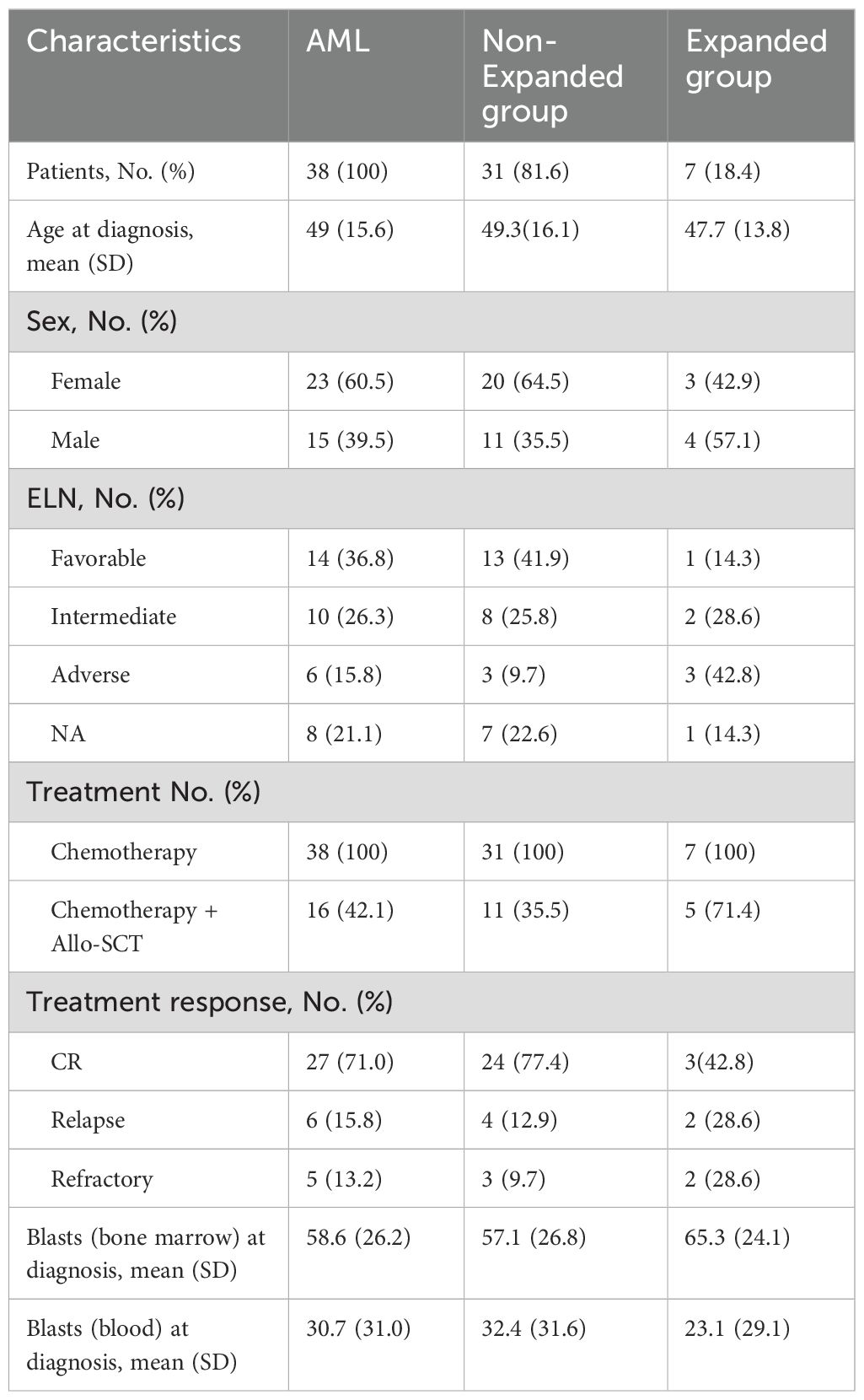
Our team made the first description of CD56neg CD16+ NK cells expansion in AML patients (8). Therefore, we sought to confirm that CD3- CD56neg CD16+ cells identified through mass cytometry belong to NK cell. To this end, we ran Cytosplore+HSNE (16) on our derivation cohort data (8) to perform automatic clustering. We embedded 872 796 CD45+ cells following the gating strategy outlined in Supplementary Figure S2 and isolated from peripheral blood of N=10 HV and N=22 AML patients. Cytosplore+HSNE identified 6 distinct clusters among which NK cells and myeloid/leukemic blasts clusters. Importantly, 88% of CD3- CD56neg CD16+ NK cells clustered among NK cell cluster while 12% clustered among leukemic blasts cluster indicating a strong similarity between CD56neg CD16+ NK cells and conventional NK cells, i.e. CD56dim CD16+,CD56dim CD16- and CD56bright NK cells (p<0.0001) (Figures 1A, B).
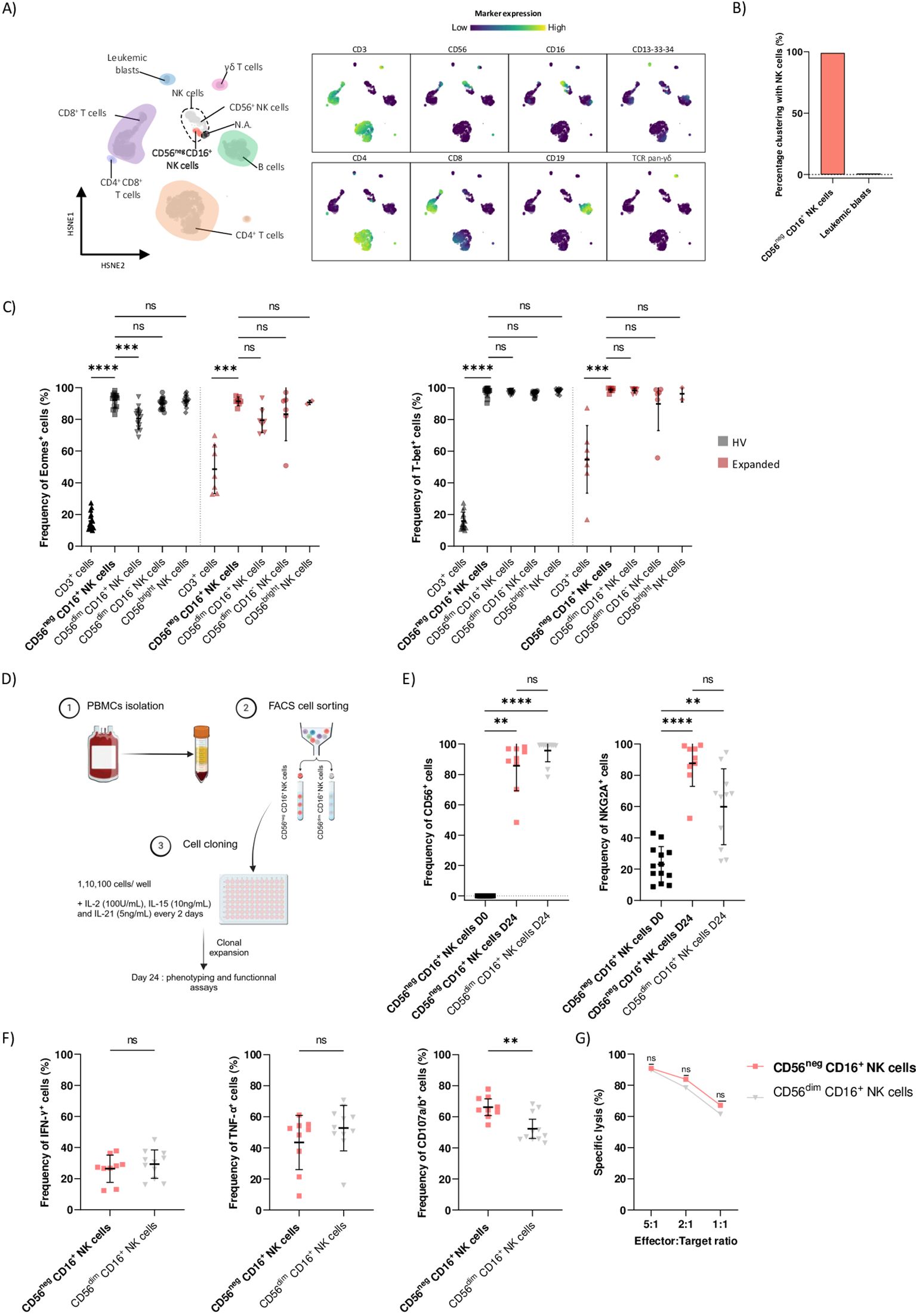
Figure 1. CD3- CD56neg CD16+ cells represent a distinct NK cell subset. (A) HSNE of 416,000 CD45+ cells from peripheral blood of HV and AML patients. Clusters of immune cells are annotated based on the expression of lineage markers. Marker expressions are ArcSinh-5 transformed, blue represents low marker expression and yellow high marker expression. N.A.: not annotated cells. (B) Percentage of CD56neg CD16+ NK cells and leukemic blasts clustering in NK cells cluster. (C) PBMCs from N=16 HV and N=7 AML patients with CD56neg CD16+ expansion (Expanded group) were phenotyped using spectral flow cytometry. Boxplots showing frequencies of Eomes+ and T-bet+ cells among CD3+ cells, CD56neg CD16+, CD56dim CD16+, CD56dim CD16- and CD56bright NK cells were assessed by manual gating. (D) Schematic overview of CD56neg CD16+ NK cells cloning experiment. Briefly, whole blood from HV was used to isolate PBMCs using density gradient centrifugation. CD56neg CD16+ and CD56dim CD16+ NK cells were then FACS sorted from collected PBMCs and plated in a 96-wells plate at a concentration of 1, 10 or 100 cells per well. Phenotyping, cytokine production and cytotoxic assays were performed after expansion on day 24. See Material and Methods section for more details. (E) Boxplots showing frequencies of CD56+ (left) and NKG2A+ (right) cells were assessed on CD56neg CD16+ and CD56dim CD16+ NK cells on days 0 and 24 of culture. (F) On day 24, CD56neg CD16+ and CD56dim CD16+ NK cells were harvested and cocultured with K562 cells at an E:T ratio of 1:1 for 4h at 37°C. Production of IFN-γ and TNF-α and degranulation capacity by CD56neg CD16+ and CD56dim CD16+ NK cells were assessed. (G) Specific lysis by CD56neg CD16+ and CD56dim CD16+ NK cells against K562 cells was assessed. Statistical significance was determined by Kruskal-Wallis and Dunn post-hoc test or Mann-Whitney test. P-values below 0.05 were considered significant. **p<0.01, ***p<0.0001, ****p<0.0001, ns=p not significant. Data is plotted as mean with SD.
Eomes and T-bet transcription factors are crucial to sustain NK cell identity and function (17). Using spectral flow cytometry, we assessed the frequencies of Eomes+ and T-bet+ cells among CD3+ cells, CD56neg CD16+ and conventional NK cells from PBMCs of HV (N=16) and AML patients (N=7) with CD56neg CD16+ NK cells expansion at diagnosis (Expanded group). In the HV group, we observed similar frequency of Eomes+ cells among CD56neg CD16+, CD56dim CD16- and CD56bright NK cells whereas CD3+ cells and CD56dim CD16+ NK cells showed lower expression of Eomes compared to CD56neg CD16+, respectively p<0.0001 and p=0.0007. In the Expanded group, frequency of Eomes+ CD56neg CD16+ cells did not differ from the frequencies of conventional NK cells and was significantly increased compared to CD3+ cells, p=0.0004 (Figure 1C, left panel). Besides, frequencies of T-bet+ CD56neg CD16+ NK cells from both HV and Expanded groups matched the frequencies of conventional NK cells and were significantly increased compared to CD3+ cells, respectively p<0.0001 and p=0.0007 (Figure 1C, right panel).
Next, we investigated whether CD56neg CD16+ NK cells express NK cell markers and maintained NK cell killing abilities in vitro. We performed limiting dilution NK cell cloning from whole blood of HV. PBMCs were isolated using density gradient centrifugation and CD56neg CD16+ and CD56dim CD16+ NK cells were FACS-sorted before being plated at a concentration of 1, 10 or 100 cells/well in RPMI supplemented with 10% of heat inactivated FBS. Irradiated K562 cells were used as feeders. IL-2 (100U/mL), IL-15 (10ng/mL) and IL-21 (5ng/mL) were added every two days and medium was changed. After 24 days of culture, cells were harvested, phenotyped and functionality was assessed (Figure 1D). We observed that expression levels of CD56 and NKG2A were significantly increased in CD56neg CD16+ NK cells after 24 days of culture, respectively p=0.0024 and p=0.0003. Importantly, CD56neg CD16+ NK cells and CD56dim CD16+ NK cells expressed similar levels of CD56 and NKG2A after 24 days in culture (Figure 1E). In addition, CD56neg CD16+ and CD56dim CD16+ NK cells exhibited similar production capacities of IFN-γ and TNF-α after 24 days of culture (Figure 1F, left and middle panels). Finally, CD56neg CD16+ NK cells expressed higher levels of CD107a/b, p=0.0057, (Figure 1F, right panel) and showed similar specific lysis against K562 cells than CD56dimCD16+ NK cells after 24 days of culture (Figure 1G). Together these data support that CD3- CD56neg CD16+ cells represent a distinct NK cell subset able to recover CD56 expression in vitro where they display unaltered NK cell functions.
Validation cohort confirmed that CD56neg CD16+ NK cells expansion at diagnosis is associated with adverse clinical outcome in AML patients
We demonstrated that CD56neg CD16+ NK cells expansion at diagnosis was associated with adverse clinical outcome in AML (8). To validate our previous results, we included N=16 HV, N=31 AML patients without CD56negCD16+ NK cells expansion at diagnosis (Non-Expanded group) and N=7 patients in the Expanded group within our validation cohort. Patients’ characteristics are summarized in Table 1. PBMCs at diagnosis were analyzed using spectral flow cytometry and NK cells subsets were gated following the gating strategy outlined in Supplementary Figure S3. We previously defined a threshold of 10% of CD56neg CD16+ NK cells among total NK cells to discriminate between patients. As expected, the threshold could optimally classify samples from the validation cohort in the Non-Expanded or Expanded groups (Figure 2A). Indeed, the Expanded group displayed significant CD56neg CD16+ NK cells expansion compared to the HV and Non-Expanded groups, respectively p=0.0292 and p<0.001 (Figure 2B, left panel). Frequencies of CD56neg CD16+ NK cells in the Expanded group ranged from 11.8% to 75.6% (Supplementary Table S3). Notably, the Expanded group did not show any increase of total NK cells (Figure 2C, left panel). Besides, the Expanded group had significantly less CD56dim CD16+, CD56dim CD16- and CD56bright NK cells than the Non-Expanded (p=0.0150, p=0.0007 and p=0.0074 respectively) and HV (p=0.0237, p=0.0432, p=0.0005 respectively) groups (Figure 2C). Importantly, absolute count of CD56neg CD16+ NK cells was significantly increased in the Expanded group compared to the Non-Expanded group (p<0.0001) (Figure 2B, right panel). Besides, absolute counts of total NK cells and CD56dim CD16+ NK cells were not altered (Figure D, left panel) while absolute counts of CD56bright and CD56dim CD16- NK cells were decreased in the Expanded group compared to the HV group (Figure D, right panel). However, the paucity of these two subsets supports the hypothesis of an expansion from the CD56neg CD16+ NK cells rather than a decrease of the other subsets. On the other hand, we investigated the T cell compartment in the HV, Non-Expanded and Expanded groups and found no difference in the frequencies of CD8+ T cells, CD4+ T cells, TCRVδ2+ T cells and regulatory T cells between groups (Supplementary Figure S4).
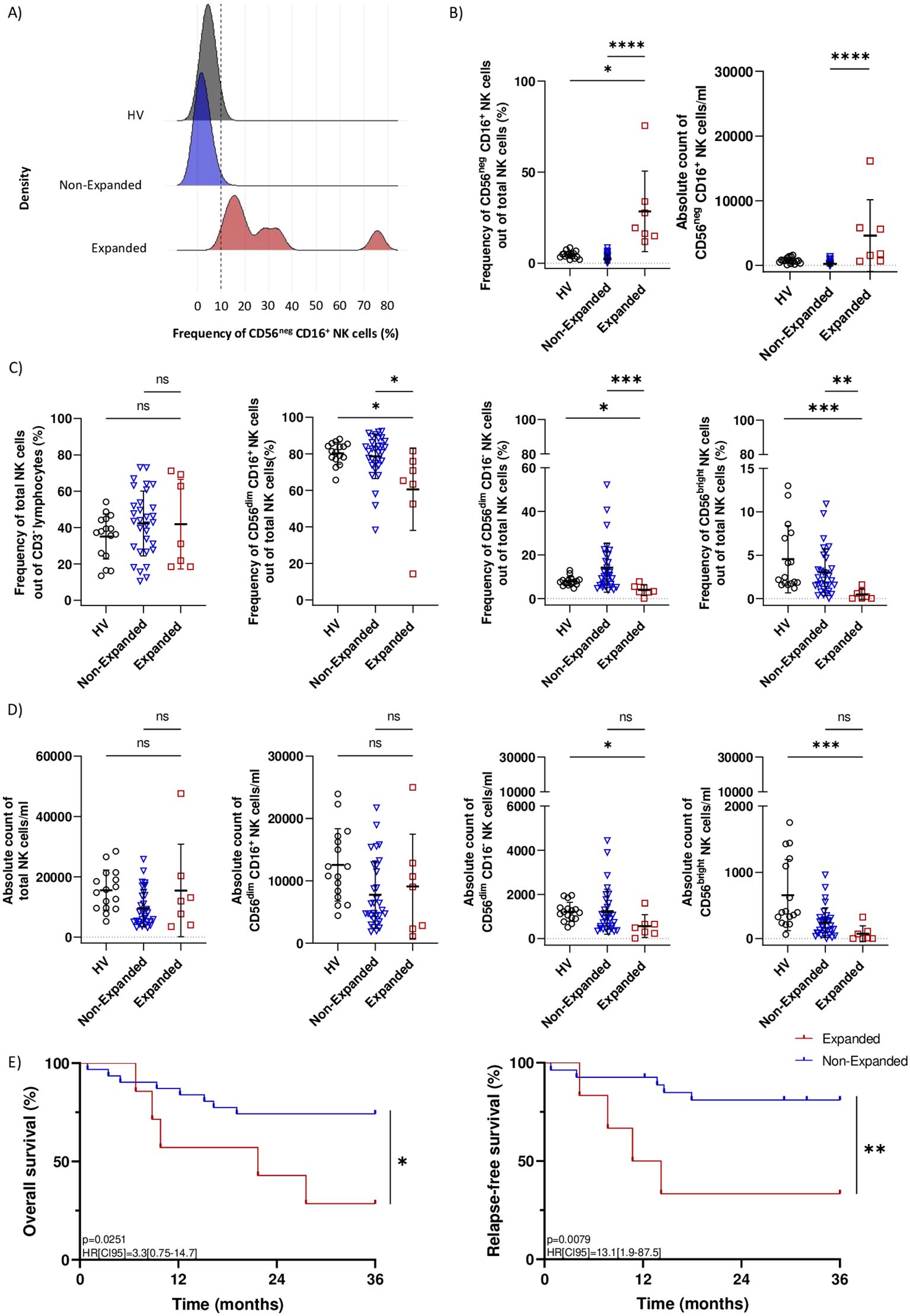
Figure 2. Validation cohort confirmed that CD56neg CD16+ NK cells expansion is associated with adverse clinical outcome in AML patients. (A) Kernel density estimate of CD56neg CD16+ NK cells frequency in N=16 HV, N=31 patients from the Non-Expanded group and N=7 patients from the Expanded group. The dotted line indicates the previously defined 10% threshold used to differentiate Non-Expanded from Expanded patients. (B) Boxplot showing the frequency and absolute count of CD56neg CD16+ NK cells in HV, Non-Expanded and Expanded patients. (C) Boxplots showing frequencies of NK cells, CD56dim CD16+ NK cells, CD56dim CD16- NK cells and CD56bright NK cells in HV, Non-Expanded and Expanded patients. (D) Boxplots showing absolute counts of NK cells, CD56dim CD16+ NK cells, CD56dim CD16- NK cells and CD56bright NK cells in HV, Non-Expanded and Expanded patients. (E) Kaplan-Meier curves for overall survival and relapse-free survival of Non-Expanded and Expanded patients. Statistical significance was determined by Kruskal-Wallis and Dunn post-hoc test and log-rank test. P-values below 0.05 were considered significant. *p<0.05, **p<0.01, ***p<0.0001, ****p<0.0001, ns, p not significant. Data is plotted as mean with SD.
Among patients in the Expanded group, 42.8% achieved sustained complete remission after first induction therapy versus 77.4% in the Non-Expanded group. Besides, 28.6% of the Expanded group relapsed after first complete remission versus 12.9% in the Non-Expanded group. Importantly, patients from the Expanded group had significantly poorer overall survival (HR[CI95]=3.3[0.75-14.7], p=0.0251) and relapse-free survival (HR[CI95]=13.1[1.9-87.5], p=0.0079) after 36 months follow-up (Figures 2D, E). Results from the validation cohort were in line with our previous study (8) and demonstrated that CD56neg CD16+ NK cells expansion at diagnosis results in adverse clinical outcome in AML patients.
CD56neg CD16+ NK cells display a unique transcriptomic profile of mature circulating NK cells and show altered expression of transcripts involved in NK cell function
Considering the clinical implications of CD56neg CD16+ NK cells expansion in AML, we aimed to further characterize this subset. To this end, we performed bulk RNA-seq profiling of CD56neg CD16+, CD56dim CD16+, CD56dim CD16- and CD56bright NK cells isolated from peripheral blood in N=5 HV and N=4 AML patients at diagnosis. NK cell subsets from HV and AML patients were respectively pooled prior to analysis. The purity of CD56neg CD16+ NK cells in our dataset was assessed based on cell type-specific transcriptomic signatures for myeloid DC, non-classical monocytes, B cells and T cells adapted from the human protein atlas (18) (https://www.proteinatlas.org/) and low-density neutrophils signature adapted from (19). We set our minimal abundance threshold log10(TPM+1) to 1 to remove the background noise, as suggested in (20). Sorted CD56neg CD16+ NK cells did not express 10/13 and poorly expressed 3/13 transcripts from the non-classical monocytes signature, and did not express transcripts identifying myeloid DC, B cells, T cells or low-density neutrophils (Supplementary Figure B). In light of these results, we considered that the purity of CD56neg CD16+ NK cells was satisfactory for downstream analysis. CD56neg CD16+ NK cells from HV and AML patients expressed a unique transcriptomic profile with 177 down-regulated and 161 up-regulated transcripts in AML samples and 87 down-regulated and 252 up-regulated transcripts in HV samples (Supplementary Tables S4, S5). As expected, NCAM1, encoding for CD56, was down-regulated in CD56neg CD16+ NK cells from both AML patients and HV.
To assess the impact of AML on CD56neg CD16+ NK cells transcriptomic profile, we focused on differentially expressed transcripts unique to the AML group. Venn diagrams identified 120 down-regulated and 53 up-regulated AML-specific transcripts (Figure 3A, top panel). We first investigated the maturation status of CD56neg CD16+ NK cells from AML patients. This subset down-regulated MYC, involved in NK cell development (21) but up-regulated NFIL3, an essential nuclear factor for NK cell maturation (22). During maturation, NK cells lose CD62L (SELL) and NKG2A (KLRC1) expression and acquire KIRs (23). Consistently, CD56neg CD16+ NK cells down-regulated SELL and KLRC1 and up-regulated KIR2DL1, KIR3DL1, KIR3DL2 and KIR2DS4. Besides, DLL1, which inhibition is involved in Notch-mediated KIRs expression (24), was also down-regulated. Therefore, CD56neg CD16+ NK cells from AML patients seemed to have reached later maturation stage. CD56neg CD16+ NK cells expansion could be a consequence of impaired chemokine or cytokine signaling. We observed that CD56neg CD16+ NK cells from AML patients down-regulated CCR1 and CCR5, involved in NK cell recruitment in inflamed tissues, and up-regulated CCL3 and CCL4, known as CCR5 ligands. Notably, THBS1, shown to take part in late NK cell expansion upon TGF-β stimulation (25), was up-regulated along with TNFRSF1B, involved in TNFα-mediated NK cell proliferation. Besides, KLF4, playing a role in murine NK cell survival and maintenance (26), was also up-regulated. Given the adverse clinical outcome of CD56neg CD16+ NK cells expansion in AML, we also explored relevant transcripts related to NK cell cytotoxicity. Among them, TLR2 and GZMH were up-regulated. However, we also found transcripts that could inhibit NK cell-mediated cytotoxicity. CD56neg CD16+ NK cells down-regulated INF2 described as a key factor of T cells immune synapse (27). In addition, ATF3, a negative regulator of IFN-γ genes in NK cells (28), was up-regulated along with CST7. This molecule is known to be an inhibitor of cathepsins C and H and could promote NK cell split anergy (29) (Figure 3A, bottom panel). Enrichment analysis on AML-specific transcripts mostly unveiled that up-regulated transcripts were enriched in cytokine-related transcripts (TNF, IL-12, and type II IFN) whereas down-regulated transcripts were enriched in cell activation-related transcripts (Figure 3B).
Figure 3. CD56neg CD16+ NK cells transcriptomic signature in AML patients. Differential gene expression analysis was performed between CD56neg CD16+ versus CD56dim CD16+, CD56dim CD16 and CD56bright NK cells in HV and AML patients. (A) Venn diagrams showing down-regulated (left) and up-regulated (right) AML-specific transcripts in CD56neg CD16+ NK cells. Hierarchical clustered heatmap of the top 100 most up and down-regulated AML-specific transcripts in CD56neg CD16+ NK cells. Expression is shown as z-score, lowest z-scores are blue and higher z-scores are red. (B) Gene ontology enrichment analysis of biological processes for up-regulated (top) and down-regulated (bottom) AML-specific transcripts in CD56neg CD16+ NK cells. (C) Venn diagrams showing down-regulated (left) and up-regulated (right) subset-specific transcripts in CD56neg CD16+ NK cells. Hierarchical clustered heatmap of the top 100 most up and down-regulated subset-specific transcripts in CD56neg CD16+ NK cells. Expression is shown as z-scores, lowest z-scores are blue and higher z-scores are red. (D) Pathway enrichment analysis of shared up-regulated (top) and down-regulated (bottom) subset-specific transcripts in CD56neg CD16+ NK cells. DEG threshold: log2FC>2, average expression>1. Enrichment threshold: adjusted p.value < 0.05.
Then, we sought to identify a specific transcriptomic signature for CD56neg CD16+ NK cells based on the overlapping transcripts between AML and HV samples. A set of 57 down-regulated and 108 up-regulated transcripts were found (Figure 3C, top panel). We observed the differential expression of cytotoxicity-related transcripts. CD56neg CD16+ NK cells up-regulated LYZ, the activating receptor CD86(30) and the inhibitory receptor CSF3R (31). Besides, transcripts from the LILR family (LILRA1, LILRA2 and LILRB2) were also up-regulated. On the other hand, CD56neg CD16+ NK cells differentially expressed transcripts that could hamper NK cell-mediated killing. Namely, TMEM163 implied in NK cell degranulation (32) was down-regulated whereas SIGLEC10, which is thought to decrease NK cell cytotoxicity in hepatocellular carcinoma (33), was up-regulated. Interestingly, complement transcripts C3, C3AR1, which was shown to inhibit NK cell cytotoxicity, as well as C5AR1 with immunoregulatory properties in mice (34) were up-regulated. Next, we investigated transcripts involved in NK cell maturation. CD56neg CD16+ NK cells down-regulated transcripts expressed in immature CD56bright NK cells such as GZMK(35) or KIT(36). They also down-regulated RUNX2, repressed during canonical maturation, along with its target genes ZEB1, BACH2 and PRDM8(37). On the other hand, ZNF683, known to regulated NK cell development (38) but also involved in NK cell exhaustion in multiple myeloma (39), was up-regulated. Then, we assessed the homing abilities of CD56neg CD16+ NK cells. ITGA1, a specific marker of NK cell tissue residency (40), was down-regulated along with CCR7(41) and CXCR3(42). Besides, CXCR2 and its ligands CXCL8 and CXCL16(43), IL3RA and CD4, increasing cytokine production and cell migration in NK cells (44), were up-regulated (Figure 3C, bottom panel). Furthermore, cell activation, cytokine pathways, proliferation and endocytosis were overrepresented among over-expressed transcripts while cell activation, chemotaxis, actin organization and Wnt signaling were overrepresented among under-expressed transcripts as revealed by gene set enrichment analysis from subset-specific transcripts (Figure 3D). Together these results strongly suggested that CD56neg CD16+ NK cells represent a unique subset of mature NK cells with functional capacities relying on different cytotoxic pathways than conventional NK cells. Notably, some transcripts were related to impaired cytotoxicity or exhaustion, which could partly explain the adverse outcome in AML patients with CD56neg CD16+ NK cells.
Altered phenotype of CD56neg CD16+ NK cells in AML patients with CD56neg CD16+ NK cells expansion
After RNA-seq profiling, we attempted to confirm our findings with spectral flow cytometry and investigated whether CD56neg CD16+ NK cells had a distinct protein expression pattern than conventional NK cells in HV, Non-Expanded and Expanded groups.
CD56neg CD16+ NK cells from all three groups had significantly decreased expression of NK cell triggering receptors NKp30 and NKp46 compared to CD56bright NK cells. However, CD56neg CD16+ NK cells from the Expanded group expressed similar levels of NKp30 and NKp46 compared to CD56dim NK cells. CD56neg CD16+ NK cells from the HV group had decreased NKp46 expression compared to CD56dim CD16- NK cells whereas CD56neg CD16+ NK cells from the Non-Expanded group had decreased NKp46 expression compared to CD56dim CD16+ NK cells. Regarding direct cytotoxicity, CD56neg CD16+ NK cells from the HV and Expanded groups significantly expressed higher levels of granzyme B and perforin compared to CD56dim CD16- NK cells. CD56neg CD16+ NK cells from the HV and Non-Expanded groups significantly expressed higher levels of granzyme B and perforin compared to CD56bright NK cells. CD56neg CD16+ NK cells from the Non-Expanded group had decreased expression of granzyme B and perforin compared to CD56dim CD16+ NK cells. On the other hand, CD56neg CD16+ NK cells from the Expanded group had similar expression of granzyme B and perforin than CD56dim CD16+ NK cells. Thus, our results are in line with both our transcriptomic and in vitro data: CD56neg CD16+ NK cells displayed a cytotoxic profile in AML. On the other hand, our results confirmed that CD56neg CD16+ NK cells had reached a mature stage as CD56neg CD16+ NK cells from all three groups expressed significantly lower levels of NKG2A and higher levels of CD158a/b than CD56bright NK cells. Moreover, CD56neg CD16+ NK cells from the HV and Non-Expanded groups had decreased expression of NKG2A compared to CD56dim CD16- NK cells (Figure 4A).
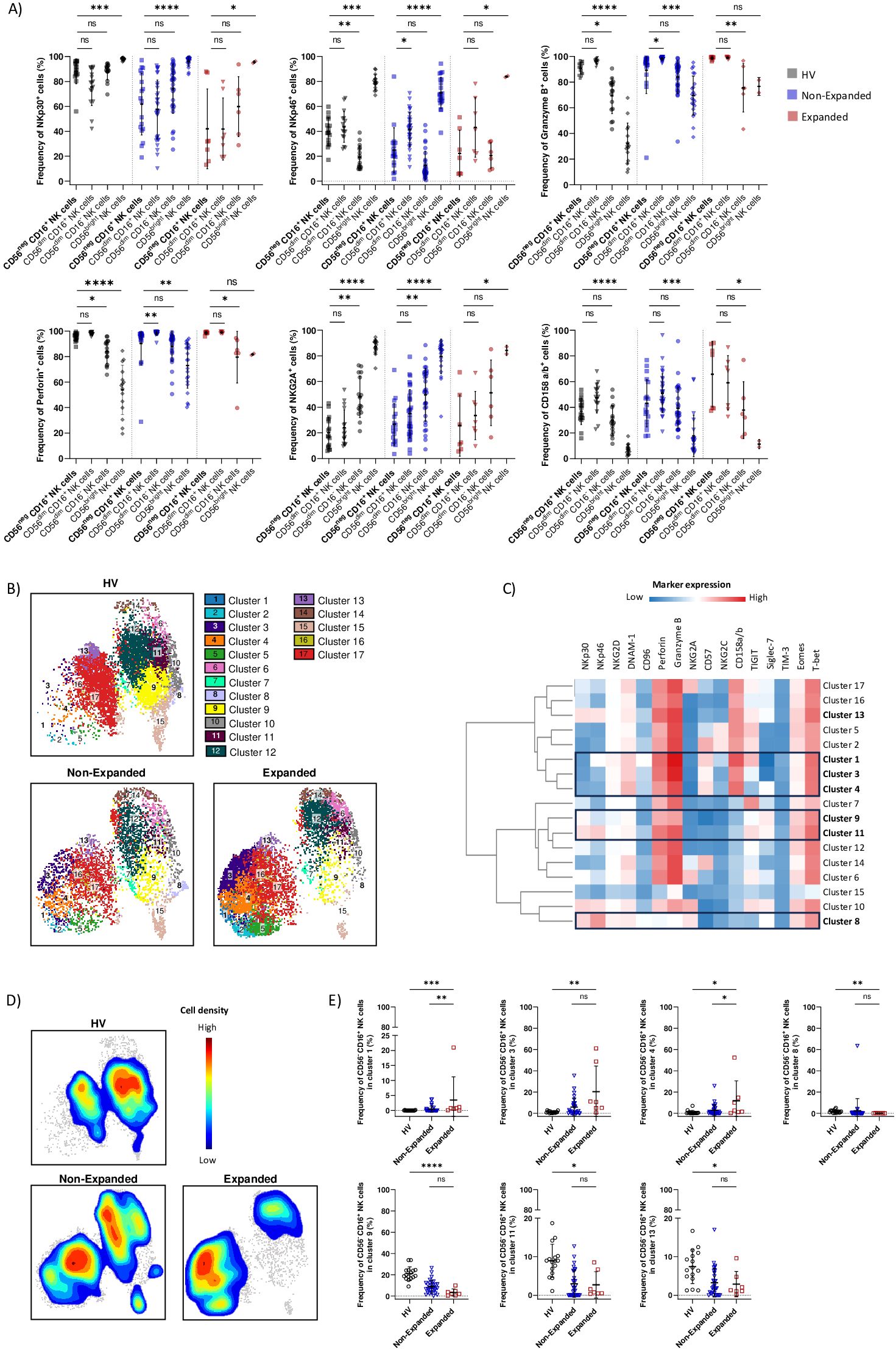
Figure 4. Altered phenotype of CD56neg CD16+ NK cells in AML patients with CD56neg CD16+ NK cells expansion. (A) Boxplots showing frequencies of NK cells markers expression on CD56neg CD16+, CD56dim CD16+, CD56dim CD16- and CD56bright NK cells in HV, Non-Expanded and Expanded groups. (B) UMAP visualization of CD56neg CD16+ NK cell clusters defined with FlowSOM in HV, Non-Expanded and Expanded groups. (C) Marker expression heatmap of CD56neg CD16+ NK cells clusters. Marker expression is normalized, low expression is represented in blue, high expression in red. (D) Cell density plot of CD56neg CD16+ NK cells in HV, Non-Expanded and Expanded groups. Low cell density is represented in blue, high cell density in red. (E) Boxplots showing changes in CD56neg CD16+ NK cells repartition among clusters in HV, Non-Expanded and Expanded groups. Statistical significance was determined by Kruskal-Wallis and Dunn post-hoc test. P-values below 0.05 were considered significant. *p<0.05, **p<0.01, ***p<0.0001, ****p<0.0001, ns, p not significant. Data is plotted as mean with SD.
Next, we investigated whether CD56neg CD16+ NK cells from the Expanded group displayed an altered phenotype compared to the HV and Non-Expanded groups. We performed UMAP analysis of CD56neg CD16+ NK cells and automatic clustering using FlowSOM, resulting in 17 clusters, in HV, Non-Expanded and Expanded groups (Figure 4B). Clusters were identified by the co-expression of 16 markers (Figure 4C; Supplementary Figure S6). We plotted CD56neg CD16+ NK cells density for each group and observed significant changes in CD56neg CD16+ NK cells from the Expanded group compared to the HV and Non-Expanded groups (Figure 4D). More CD56neg CD16+ NK cells from the Expanded group were found in mature cytotoxic clusters compared to the HV and Non-Expanded groups. Indeed, cluster 1 (NKp46+ NKG2D+ DNAM-1+ CD96+ Perforin+ Granzyme B+ CD57+ CD158a/b+ TIGIT+) and cluster 4 (NKG2D+ DNAM-1+ Perforin+ Granzyme B+ CD57+ KIR+ Siglec-7+) were significantly increased in the Expanded group compared to the HV and Non-Expanded groups (Figure 4E). Notably, TIGIT was shown to be associated with NK cell exhaustion (45) and TIGIT+ NK cells displayed decreased effector function. Besides, CD56neg CD16+ NK cells from cluster 3 (NKG2D+ DNAM-1+ Perforin+ Granzyme B+ CD57+ KIR+) were more abundant in the Expanded group than in the HV group (Figure 4E). Interestingly, the frequency of terminally mature CD56neg CD16+ NK cells expressing NKG2C present in clusters 2 and 5 was not different between the three groups (Figure 4C). On the other hand, Siglec-7 expressing clusters, cluster 8 (NKp30+ NKp46+ NKG2D+ DNAM-1+ Granzyme B+ NKG2A+ Siglec-7+), cluster 9 (NKp30+ DNAM-1+ Perforin+ Granzyme B+ Siglec-7+), cluster 11 (NKp30+ NKp46+ DNAM-1+ Perforin+ Granzyme B+ TIGIT+ Siglec-7+) and cluster 13 (NKp30+ NKp46+ NKG2D+ DNAM-1+ Perforin+ Granzyme B+ KIR+ TIGIT+ Siglec-7+) were significantly decreased in the Expanded group compared to the HV group. Interestingly, Siglec-7 expression on NK cells were shown to mark fully functional NK cells ligand (46). Therefore, the loss of Siglec-7+ CD56neg CD16+ NK cells could contribute to adverse clinical outcome of patients with CD56neg CD16+ NK cells expansion. Together, our results showed an altered phenotype of CD56neg CD16+ NK cells from the Expanded group, with increased mature cytotoxic NK cell expressing inhibitory receptors such as CD158a/b and TIGIT and decreased Siglec-7+ NK cells.
Discussion
In this study, our aim was to further characterize CD56neg CD16+ NK cells, a subset that expands in AML. We confirmed that CD56neg CD16+ NK cells represent a unique NK cell subset. We used Cytosplore+HSNE, an automated clustering approach, on mass cytometry data and observed that CD56neg CD16+ NK cells located in NK cells cluster. These results suggested a strong similarity between proteomic phenotype of CD56neg CD16+ and conventional NK cells, in line with previous publications (2, 47). Besides, CD56neg CD16+ NK cells co-expressed high levels of Eomes and T-bet, distinguishing them from ILCs (48). CD56neg CD16+ NK cells seem to be circulating NK cells as they down-regulated transcripts involved in homing or tissue residency such as CCR7 and ITGA1. Interestingly, CD56neg CD16+ NK cells from AML patients up-regulated THBS1 and TNFRSF1B involved in TGF-β and TNFα-mediated expansion, respectively. Those two cytokines were reported to be increased in AML patients compared to healthy volunteers (49, 50).
We further unveiled that CD56neg CD16+ NK cells downregulate NCAM1, encoding for CD56, compared to conventional NK cells, suggesting that the loss of CD56 is an intrinsic mechanism. The loss of CD56 was reversible as CD56neg CD16+ NK cells could regain CD56 expression when cultivated with IL-2, IL-15 and IL-21. Importantly, CD56neg CD16+ NK cells displayed unaltered NK cell function in vitro once they regained CD56, known to play a critical role in NK cell cytotoxicity (13).
On the other hand, CD56negCD16+ NK cells expansion has been documented in chronic diseases. In a validation cohort, we confirmed that newly-diagnosed AML patients with CD56neg CD16+ NK cells expansion had significantly poorer clinical outcomes than others. This affords the possibility for adoptive NK cell therapy or the use of NK-cell engagers on CD56neg CD16+ NK cells from AML patients. Importantly, no significant change in CD8+ T cells, CD4+ T cells, TCRVδ2+ T cells and regulatory T cells frequency was found in patients with CD56neg CD16+ NK cell expansion.
Bulk RNA-seq and spectral flow cytometry results showed that CD56neg CD16+ NK cells poorly expressed NKG2A and up-regulated KIRs. Recently, Rebuffet et al. (51) performed high-dimensional single-cell profiling of human NK cells and identified 3 NK cell subsets in peripheral blood. NK1 et NK3 subsets expressed transcripts of NK cell maturity. We applied NK1 and NK3 signatures to our bulk RNA-seq dataset. Interestingly, CD56neg CD16+ NK cells displayed high expression of transcripts from NK3 signature, identifying terminally mature NK cells (Supplementary Figure S7). Therefore, our results strongly suggest that CD56neg CD16+ NK cell have reached a late maturation stage. Importantly, we observed that CD56neg CD16+ NK cells overexpressed ZNF683. This transcription factor was recently identified as a driver of NK cell exhaustion in multiple myeloma (39), another hematological malignancy. The authors performed pseudotime analysis on ZNF683+ NK cells and underlined 5 transcripts indicative of exhaustion state. Among them, 4 were found up-regulated in CD56neg CD16+ NK cells of AML patients, namely ZNF683, LAG3, FGL2 and CST7, involved in NK cell split anergy (29). Furthermore, AML patients with CD56neg CD16+ NK cells expansion displayed increased frequency of terminally mature CD57+ KIR+ CD56neg CD16+ NK cells compared to HV or/and patients without expansion. Interestingly, one cluster contained TIGIT+ NK cells, a marker of NK cell exhaustion. We previously showed that CD56neg CD16+ NK cells from healthy volunteers expressed slightly higher frequencies of PD-1 compared to CD56dim CD16+, CD56dim CD16- and CD56bright NK cells NK cells NK cells (8). However, TIM-3 expression was not increased. Therefore, further investigations are needed to assess CD56neg CD16+ NK cells exhaustion. On the other hand, Siglec-7+ CD56neg CD16+ NK cells were decreased. Siglec-7 was shown to be expressed by terminally differentiated NK cells with high cytotoxic potential. Taken together, our results suggested that AML microenvironment could decrease CD56neg CD16+ NK cells cytotoxicity, which may partly explain the negative impact of CD56neg CD16+ NK cells expansion in AML on survival.
There were some limitations in the study. RNA-seq analysis was performed on pooled samples and without biological replicates. Therefore, we were likely to find high false positive rates in our differential gene expression analysis. Nevertheless, our results corroborated previous studies (2, 5). Besides, CMV status from AML patients were not available and we cannot exclude the possibility that CD56neg CD16+ NK cells expansion is driven by CMV infection (52). However, adaptative CD57+ NKG2C+ NK cells from AML patients with CD56neg CD16+ NK cells expansion were not significantly increased compared to the AML patients without expansion (data not shown).
To conclude, we demonstrated that CD56neg CD16+ NK cells represent a distinct NK cell subset that can recover CD56 expression in vitro and display NK cell functions, which could be leveraged to counteract the adverse clinical outcome of AML patients with CD56neg CD16+ NK cells expansion.
Data availability statement
The original RNA-seq contributions presented in the study are publicly available. This data can be found here: https://www.ebi.ac.uk/biostudies/studies/E-MTAB-14569. Further inquiries can be directed to the corresponding authors.
Ethics statement
The studies involving humans were approved by Review Boards of Paoli-Calmettes Institute. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
JW: Conceptualization, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. FO: Investigation, Methodology, Writing – review & editing. MR: Investigation, Methodology, Writing – review & editing. CD: Investigation, Methodology, Writing – review & editing. AA: Investigation, Methodology, Writing – review & editing. MSR: Investigation, Methodology, Writing – review & editing. PL: Investigation, Methodology, Writing – review & editing. LG: Formal analysis, Writing – review & editing. MH: Writing – review & editing. ND: Writing – review & editing. RD: Writing – review & editing. NV: Writing – review & editing. DO: Conceptualization, Funding acquisition, Project administration, Supervision, Validation, Writing – review & editing. AC: Conceptualization, Project administration, Supervision, Validation, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This article is supported by grants from Assistance Publique-Hôpitaux de Marseille (AP-HM) and by the Association Laurette Fugain (grant ALF2020/05 for DO).
Acknowledgments
We would like to thank all patients that participated in this study. We thank the Gladstone Bioinformatics Core for their interactive enrichment analysis tools, the CRCM cytometry facility core, the Paoli-Calmettes Tumor Bank as well as the reviewers for their time and insightful comments. The team “Immunity and Cancer” was labelled Equipe Fondation pour la Recherche Médicale (FRM) (DEQ20180339209 for DO). Figures were generated with BioRender.com.
Conflict of interest
DO is a cofounder and shareholder of ImCheck Therapeutics, Alderaan Biotechnology and Emergence Therapeutics and has research funds from ImCheck Therapeutics, Alderaan Biotechnology, Cellectis and Emergence Therapeutics.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1487792/full#supplementary-material
References
1. Boudreau JE, Hsu KC. Natural killer cell education and the response to infection and cancer therapy: stay tuned. Trends Immunol. (2018) 39:222–39. doi: 10.1016/j.it.2017.12.001
2. Forconi CS, Oduor CI, Oluoch PO, Ong’echa JM, Münz C, Bailey JA, et al. A new hope for CD56negCD16pos NK cells as unconventional cytotoxic mediators: an adaptation to chronic diseases. Front Cell Infect Microbiol. (2020) 10:162. doi: 10.3389/fcimb.2020.00162
3. Hu PF, Hultin LE, Hultin P, Hausner MA, Hirji K, Jewett A, et al. Natural killer cell immunodeficiency in HIV disease is manifest by profoundly decreased numbers of CD16+CD56+ cells and expansion of a population of CD16dimCD56- cells with low lytic activity. J Acquir Immune Defic Syndr Hum Retrovirol. (1995) 10:331–40. doi: 10.1097/00042560-199511000-00005
4. Mavilio D, Lombardo G, Benjamin J, Kim D, Follman D, Marcenaro E, et al. Characterization of CD56–/CD16+ natural killer (NK) cells: A highly dysfunctional NK subset expanded in HIV-infected viremic individuals. Proc Natl Acad Sci. (2005) 102:2886–91. doi: 10.1073/pnas.0409872102
5. Cao WJ, Zhang XC, Wan LY, Li QY, Mu XY, Guo AL, et al. Immune dysfunctions of CD56neg NK cells are associated with HIV-1 disease progression. Front Immunol. (2021) 12:811091. doi: 10.3389/fimmu.2021.811091
6. Gonzalez VD, Falconer K, Björkström NK, Blom KG, Weiland O, Ljunggren HG, et al. Expansion of functionally skewed CD56-negative NK cells in chronic hepatitis C virus infection: correlation with outcome of pegylated IFN-alpha and ribavirin treatment. J Immunol. (2009) 183:6612–8. doi: 10.4049/jimmunol.0901437
7. Ty M, Sun S, Callaway PC, Rek J, Press KD, van der Ploeg K, et al. Malaria-driven expansion of adaptive-like functional CD56-negative NK cells correlates with clinical immunity to malaria. Sci Transl Med. (2023) 15:eadd9012. doi: 10.1126/scitranslmed.add9012
8. Chretien AS, Devillier R, Granjeaud S, Cordier C, Demerle C, Salem N, et al. High-dimensional mass cytometry analysis of NK cell alterations in AML identifies a subgroup with adverse clinical outcome. Proc Natl Acad Sci. (2021) 118:e2020459118. doi: 10.1073/pnas.2020459118
9. D’Silva SZ, Singh M, Pinto AS. NK cell defects: implication in acute myeloid leukemia. Front Immunol. (2023) 14:1112059. doi: 10.3389/fimmu.2023.1112059
10. Lion E, Willemen Y, Berneman ZN, Van Tendeloo VFI, Smits ELJ. Natural killer cell immune escape in acute myeloid leukemia. Leukemia. (2012) 26:2019–26. doi: 10.1038/leu.2012.87
11. Kantarjian H, Kadia T, DiNardo C, Daver N, Borthakur G, Jabbour E, et al. Acute myeloid leukemia: current progress and future directions. Blood Cancer J. (2021) 11:1–25. doi: 10.1038/s41408-021-00425-3
12. Xu J, Niu T. Natural killer cell-based immunotherapy for acute myeloid leukemia. J Hematol Oncol. (2020) 13:167. doi: 10.1186/s13045-020-00996-x
13. Gunesch JT, Dixon AL, Ebrahim TA, Berrien-Elliott MM, Tatineni S, Kumar T, et al. CD56 regulates human NK cell cytotoxicity through Pyk2. Elife. (2020) 9:e57346. doi: 10.7554/eLife.57346
14. Taouk G, Hussein O, Zekak M, Abouelghar A, Al-Sarraj Y, Abdelalim EM, et al. CD56 expression in breast cancer induces sensitivity to natural killer-mediated cytotoxicity by enhancing the formation of cytotoxic immunological synapse. Sci Rep. (2019) 9:8756. doi: 10.1038/s41598-019-45377-8
15. Gauthier L, Virone-Oddos A, Beninga J, Rossi B, Nicolazzi C, Amara C, et al. Control of acute myeloid leukemia by a trifunctional NKp46-CD16a-NK cell engager targeting CD123. Nat Biotechnol. (2023) 41:1296–306. doi: 10.1038/s41587-022-01626-2
16. van Unen V, Höllt T, Pezzotti N, Li N, Reinders MJT, Eisemann E, et al. Visual analysis of mass cytometry data by hierarchical stochastic neighbor embedding reveals rare cell types. Nat Commun. (2017) 8:1740. doi: 10.1038/s41467-017-01689-9
17. Wong P, Foltz JA, Chang L, Neal CC, Yao T, Cubitt CC, et al. T-BET and EOMES sustain mature human NK cell identity and antitumor function(2023). Available online at: https://www.jci.org/articles/view/162530 (Accessed 2024 May 7).
18. Uhlen M, Karlsson MJ, Zhong W, Tebani A, Pou C, Mikes J, et al. A genome-wide transcriptomic analysis of protein-coding genes in human blood cells. Science. (2019) 366:eaax9198. doi: 10.1126/science.aax9198
19. Montaldo E, Lusito E, Bianchessi V, Caronni N, Scala S, Basso-Ricci L, et al. Cellular and transcriptional dynamics of human neutrophils at steady state and upon stress. Nat Immunol. (2022) 23:1470–83. doi: 10.1038/s41590-022-01311-1
20. Cockrum C, Kaneshiro KR, Rechtsteiner A, Tabuchi TM, Strome S. A primer for generating and using transcriptome data and gene sets. Dev (Cambridge England). (2020) 147:dev193854. doi: 10.1242/dev.193854
21. Khameneh HJ, Fonta N, Zenobi A, Niogret C, Ventura P, Guerra C, et al. Myc controls NK cell development, IL-15-driven expansion, and translational machinery. Life Sci Alliance. (2023) 6:e202302069. doi: 10.26508/lsa.202302069
22. Kamizono S, Duncan GS, Seidel MG, Morimoto A, Hamada K, Grosveld G, et al. Nfil3/E4bp4 is required for the development and maturation of NK cells in vivo. J Exp Med. (2009) 206:2977–86. doi: 10.1084/jem.20092176
23. Björkström NK, Riese P, Heuts F, Andersson S, Fauriat C, Ivarsson MA, et al. Expression patterns of NKG2A, KIR, and CD57 define a process of CD56dim NK-cell differentiation uncoupled from NK-cell education. Blood. (2010) 116:3853–64. doi: 10.1182/blood-2010-04-281675
24. Felices M, Ankarlo DEM, Lenvik TR, Nelson HH, Blazar BR, Verneris MR, et al. Notch signaling at later stages of Natural Killer cell development enhances KIR expression and functional maturation. J Immunol. (2014) 193:3344–54. doi: 10.4049/jimmunol.1400534
25. Pierson BA, Gupta K, Hu WS, Miller JS. Human natural killer cell expansion is regulated by thrombospondin-mediated activation of transforming growth factor-β1 and independent accessory cell-derived contact and soluble factors. Blood. (1996) 87:180–9. doi: 10.1182/blood.V87.1.180.180
26. Shik Park C, Lee PH, Yamada T, Burns A, Shen Y, Puppi M, et al. Krüppel-like factor 4 (KLF4) promotes the survival of natural killer cells and maintains the number of conventional dendritic cells in the spleen. J Leukoc Biol. (2012) 91:739–50. doi: 10.1189/jlb.0811413
27. Andrés-Delgado L, Antón OM, Bartolini F, Ruiz-Sáenz A, Correas I, Gundersen GG, et al. INF2 promotes the formation of detyrosinated microtubules necessary for centrosome reorientation in T cells. J Cell Biol. (2012) 198:1025–37. doi: 10.1083/jcb.201202137
28. Rosenberger CM, Clark AE, Treuting PM, Johnson CD, Aderem A. ATF3 regulates MCMV infection in mice by modulating IFN-γ expression in natural killer cells. Proc Natl Acad Sci U S A. (2008) 105:2544–9. doi: 10.1073/pnas.0712182105
29. Perišić Nanut M, Sabotič J, Švajger U, Jewett A, Kos J. Cystatin F affects natural killer cell cytotoxicity. Front Immunol. (2017) 8:1459. doi: 10.3389/fimmu.2017.01459
30. Peng Y, Luo G, Zhou J, Wang X, Hu J, Cui Y, et al. CD86 is an activation receptor for NK cell cytotoxicity against tumor cells. PloS One. (2013) 8:e83913. doi: 10.1371/journal.pone.0083913
31. Cao X, Han TT, Zhao M, Juan X, Chang YJ, Huang XJ, et al. G-CSFR is a checkpoint of natural killer cells against tumor. Blood. (2023) 142:3923. doi: 10.1182/blood-2023-182511
32. Gilchrist JJ, Makino S, Naranbhai V, Sharma PK, Koturan S, Tong O, et al. Natural Killer cells demonstrate distinct eQTL and transcriptome-wide disease associations, highlighting their role in autoimmunity. Nat Commun. (2022) 13:4073. doi: 10.1038/s41467-022-31626-4
33. Zhang P, Lu X, Tao K, Shi L, Li W, Wang G, et al. Siglec-10 is associated with survival and natural killer cell dysfunction in hepatocellular carcinoma. J Surg Res. (2015) 194:107–13. doi: 10.1016/j.jss.2014.09.035
34. Min X, Liu C, Wei Y, Wang N, Yuan G, Liu D, et al. Expression and regulation of complement receptors by human natural killer cells. Immunobiology. (2014) 219:671–9. doi: 10.1016/j.imbio.2014.03.018
35. Cheng S, Li Z, Gao R, Xing B, Gao Y, Yang Y, et al. A pan-cancer single-cell transcriptional atlas of tumor infiltrating myeloid cells. Cell. (2021) 184:792–809.e23. doi: 10.1016/j.cell.2021.01.010
36. Matos ME, Schnier GS, Beecher MS, Ashman LK, William DE, Caligiuri MA. Expression of a functional c-kit receptor on a subset of natural killer cells. J Exp Med. (1993) 178:1079–84. doi: 10.1084/jem.178.3.1079
37. Holmes TD, Pandey RV, Helm EY, Schlums H, Han H, Campbell TM, et al. The transcription factor Bcl11b promotes both canonical and adaptive NK cell differentiation. Sci Immunol. (2021) 6:eabc9801. doi: 10.1126/sciimmunol.abc9801
38. Post M, Cuapio A, Osl M, Lehmann D, Resch U, Davies DM, et al. The transcription factor ZNF683/HOBIT regulates human NK-cell development. Front Immunol. (2017) 8:535. doi: 10.3389/fimmu.2017.00535
39. Li X, Chen M, Wan Y, Zhong L, Han X, Chen X, et al. Single-cell transcriptome profiling reveals the key role of ZNF683 in natural killer cell exhaustion in multiple myeloma. Clin Trans Med. (2022) 12:e1065. doi: 10.1002/ctm2.v12.10
40. Hegewisch-Solloa E, Seo S, Mundy-Bosse BL, Mishra A, Waldman EH, Maurrasse S, et al. Differential integrin adhesome expression defines human natural killer cell residency and developmental stage. J Immunol. (2021) 207:950–65. doi: 10.4049/jimmunol.2100162
41. Pesce S, Moretta L, Moretta A, Marcenaro E. Human NK cell subsets redistribution in pathological conditions: A role for CCR7 receptor. Front Immunol. (2016) 7:414. doi: 10.3389/fimmu.2016.00414
42. Ali A, Canaday LM, Feldman HA, Cevik H, Moran MT, Rajaram S, et al. Natural killer cell immunosuppressive function requires CXCR3-dependent redistribution within lymphoid tissues. J Clin Invest. 131(18):e146686. doi: 10.1172/JCI146686
43. Susek KH, Karvouni M, Alici E, Lundqvist A. The role of CXC chemokine receptors 1–4 on immune cells in the tumor microenvironment. Front Immunol. (2018) 9:2159/full. doi: 10.3389/fimmu.2018.02159/full
44. Bernstein HB, Plasterer MC, Schiff SE, Kitchen CMR, Kitchen S, Zack JA. CD4 expression on activated NK cells: ligation of CD4 induces cytokine expression and cell migration. J Immunol. (2006) 177:3669–76. doi: 10.4049/jimmunol.177.6.3669
45. Zhang Q, Bi J, Zheng X, Chen Y, Wang H, Wu W, et al. Blockade of the checkpoint receptor TIGIT prevents NK cell exhaustion and elicits potent anti-tumor immunity. Nat Immunol. (2018) 19:723–32. doi: 10.1038/s41590-018-0132-0
46. Shao JY, Yin WW, Zhang QF, Liu Q, Peng ML, Hu HD, et al. Siglec-7 defines a highly functional natural killer cell subset and inhibits cell-mediated activities. Scandinavian J Immunol. (2016) 84:182–90. doi: 10.1111/sji.2016.84.issue-3
47. Voigt J, Malone DFG, Dias J, Leeansyah E, Björkström NK, Ljunggren HG, et al. Proteome analysis of human CD56neg NK cells reveals a homogeneous phenotype surprisingly similar to CD56dim NK cells. Eur J Immunol. (2018) 48:1456–69. doi: 10.1002/eji.201747450
48. Zhang J, Marotel M, Fauteux-Daniel S, Mathieu AL, Viel S, Marçais A, et al. T-bet and Eomes govern differentiation and function of mouse and human NK cells and ILC1. Eur J Immunol. (2018) 48:738–50. doi: 10.1002/eji.201747299
49. Wang CZ, Zhang ZQ, Zhang Y, Zheng LF, Liu Y, Yan AT, et al. Comprehensive characterization of TGFB1 across hematological Malignancies. Sci Rep. (2023) 13:19107. doi: 10.1038/s41598-023-46552-8
50. Sanchez-Correa B, Bergua JM, Campos C, Gayoso I, Arcos MJ, Bañas H, et al. Cytokine profiles in acute myeloid leukemia patients at diagnosis: Survival is inversely correlated with IL-6 and directly correlated with IL-10 levels. Cytokine. (2013) 61:885–91. doi: 10.1016/j.cyto.2012.12.023
51. Rebuffet L, Melsen JE, Escalière B, Basurto-Lozada D, Bhandoola A, Björkström NK, et al. High-dimensional single-cell analysis of human natural killer cell heterogeneity. Nat Immunol. (2024) 25:1474–88. doi: 10.1038/s41590-024-01883-0
Keywords: CD56 neg NK cells, AML, high-dimensional cytometry, RNA-sequencing, cancer
Citation: Wlosik J, Orlanducci F, Richaud M, Demerle C, Amara AB, Rouviere M-S, Livrati P, Gorvel L, Hospital M-A, Dulphy N, Devillier R, Vey N, Olive D and Chretien A-S (2025) CD56neg CD16+ cells represent a distinct mature NK cell subset with altered phenotype and are associated with adverse clinical outcome upon expansion in AML. Front. Immunol. 15:1487792. doi: 10.3389/fimmu.2024.1487792
Received: 28 August 2024; Accepted: 02 December 2024;
Published: 10 January 2025.
Edited by:
Jin S. Im, University of Texas MD Anderson Cancer Center, United StatesReviewed by:
Jules Russick, Commissariat à l’Energie Atomique et aux Energies Alternatives (CEA), FranceSilvia Guglietta, Medical University of South Carolina, United States
Emilie Narni-Mancinelli, INSERM U1104 Centre d’immunologie de Marseille-Luminy (CIML), France
Copyright © 2025 Wlosik, Orlanducci, Richaud, Demerle, Amara, Rouviere, Livrati, Gorvel, Hospital, Dulphy, Devillier, Vey, Olive and Chretien. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Julia Wlosik, anVsaWEud2xvc2lrQGluc2VybS5mcg==; Anne-Sophie Chretien, YW5uZS1zb3BoaWUuY2hyZXRpZW5AaW5zZXJtLmZy
†These authors have contributed equally to this work
 Julia Wlosik
Julia Wlosik Florence Orlanducci
Florence Orlanducci Manon Richaud3
Manon Richaud3 Clemence Demerle
Clemence Demerle Marie-Sarah Rouviere
Marie-Sarah Rouviere Laurent Gorvel
Laurent Gorvel Nicolas Dulphy
Nicolas Dulphy Raynier Devillier
Raynier Devillier Norbert Vey
Norbert Vey Daniel Olive
Daniel Olive Anne-Sophie Chretien
Anne-Sophie Chretien