- 1Department of Biliary-Pancreatic Surgery, Renji Hospital Affiliated to Shanghai Jiao Tong University, School of Medicine, Shanghai, China
- 2Tilcure Biotherapeutics, Shanghai, China
- 3Department of Training Department, China Medical University Benxi Central Hospital Postgraduate Training Workstation, Shanghai, China
- 4Renal Division, Department of Internal Medicine, Xinhua Hospital Affiliated to Shanghai Jiao Tong University School of Medicine, Shanghai, China
- 5Shanghai Cancer Institute, Shanghai, China
Introduction: The coronavirus disease 2019 (COVID-19) global pandemic has been the most severe public health emergency since 2019. Currently, the Omicron variant of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has been the most dominant. The most prominent symptom of SARS-CoV-2 infection is respiratory. Meanwhile, the fatality of COVID-19 was mainly from pneumonia. However ,in patients with SARS-CoV-2 infection who have pneumonia and those who do not, the differences in the immune repertoire still require further investigation.
Methods: We conducted seven-chain adaptome immune repertoire analyses on patients with SARS-CoV-2 Omicron infection, both with and without pulmonary infiltration.
Results: Patients with pulmonary infiltration exhibit lymphopenia, a decreased proportion of the overall TCR repertoire alongside an increased BCR repertoire, reduced IGHD and IGHM isotype expression, a shorter mean CDR3 length for TRG, and a longer mean length for TRD, as well as diminished clonality and diversity in the TCR/BCR repertoire. Meanwhile, patients with pulmonary infiltration have distinct V-J gene usage and unique CDR3 signature, as well as BCR class switch recombination pattern. Finally, prior vaccination triggered less BCR IGHM/IGHD somatic hypermutation response, preserved the diversity of the entire adaptive immune repertoire, and provided clinical protection against severe or critical conditions following Omicron infection.
Discussion: We report a unique, comprehensive adaptive immune system signature in patients with pulmonary infiltration, which may serve as potential immunological biomarkers and therapeutic targets.
1 Introduction
Coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), had been posing a serious threat to global health (1). As of November 2022, SARS-CoV-2 had infected more than 600 million people and caused more than 6 million deaths worldwide (https://covid19.who.int/). Over the last 5 years of the COVID-19 pandemic, the SARS-CoV-2 virus had undergone a high frequency of mutations and had been evolving swiftly (41). Variants of concern have appeared at regular intervals—Alpha, Beta, Gamma, Delta, and now Omicron. The Omicron variant, first identified in Botswana in November 2021, was rapidly becoming the dominant circulating variant (1). The city of Shanghai, China, encountered a wave of SARS-CoV-2 Omicron spread since March 2022, which caused hundreds of casualties (2).
Although most patients with COVID-19 present with mild illness, even asymptomatic, some patients do develop severe pulmonary, which is an important risk factor for mortality (3–6). The Omicron variant exhibits a higher transmissibility than prior SARS-CoV-2 variants as well as the capability to evade naturally acquired and vaccine-induced immunity (7). As with any other virus infection, the adaptive immune response plays a central role in clearing SARS-CoV-2 (8, 9). In addition to providing host protection, adaptive immune functions may contribute pathologic mediators, including B-cell autoreactivities associated with specific disease-related characteristics in many patients with COVID-19 (10, 11). Severe disease and death caused by SARS-CoV-2 infection appear to be largely due to failures and/or dysregulation of the immune response in vulnerable populations. Currently, there is limited information on the impact of SARS-CoV-2 Omicron variant infection on the adaptive immune responses, especially in patients with pulmonary infiltration.
The immune system comprises innate immunity and adaptive immunity that offer protection against viruses and other pathogens. T cells and B cells are the central mediators of antiviral adaptive immunity (12). Of each T or B cell, there are unique T-cell receptors (TCRs) or B-cell receptors (BCRs), which are expressed on the cell surface. The antigen specificity of each TCR and BCR is primarily determined by the hypervariable complementarity-determining region 3 (CDR3) of the receptor chain, which originates from the recombination of the V (variable), D (diversity), and J (joining) gene segments and the deletion and insertion of nucleotides at the V(D)J junctions (13). Using immune repertoire next-generation sequencing, which offers in-depth quantitative and molecular-level profiling of immune repertoire, helps us understand the dynamics of the antigen-adaptive immune response in humans (14–16).
In humoral immunity, an antibody or immunoglobulin (Ig) can recognize a specific antigen through its N-terminal variable region and activate downstream immune effects through its C-terminal constant region. Upon antigen stimulation, B cells can further diversify the antibody gene by introducing mutations at the Ig variable region exon by somatic hypermutation (SHM) to allow antibody affinity maturation and by switching the antibody class through class switch recombination (CSR) to change the downstream effector functions (17, 18). Various studies have characterized the BCR repertoire feature for SARS-CoV-2 infection and vaccination, thus directing the therapeutic development as well as deepening our understanding of the immunological changes for SARS-CoV-2 infection (19, 20).
An efficacious vaccine is considered essential to prevent further morbidity and mortality from SARS-CoV-2 infection (21). The inactivated SARS-CoV-2 vaccines have shown effective immune responses in eliciting neutralizing antibodies in multiple clinical trials (22, 23). However, with the novel SARS-CoV-2 variants emerging and circulating widely, whether the original vaccines that were designed based on the wild-type SARS-CoV-2 can offer enough protection against emerging variants and the immunological reactions triggered by the vaccines has been under contentious debate (24).
Here, we investigated the peripheral blood immune repertoire of patients infected with the SARS-CoV-2 Omicron variant across different levels of clinical severity. Our goal was to identify a unique immune repertoire signature that could influence the determination of clinical outcomes for SARS-CoV-2 Omicron variant infection.
2 Methods
2.1 Enrollment of patients
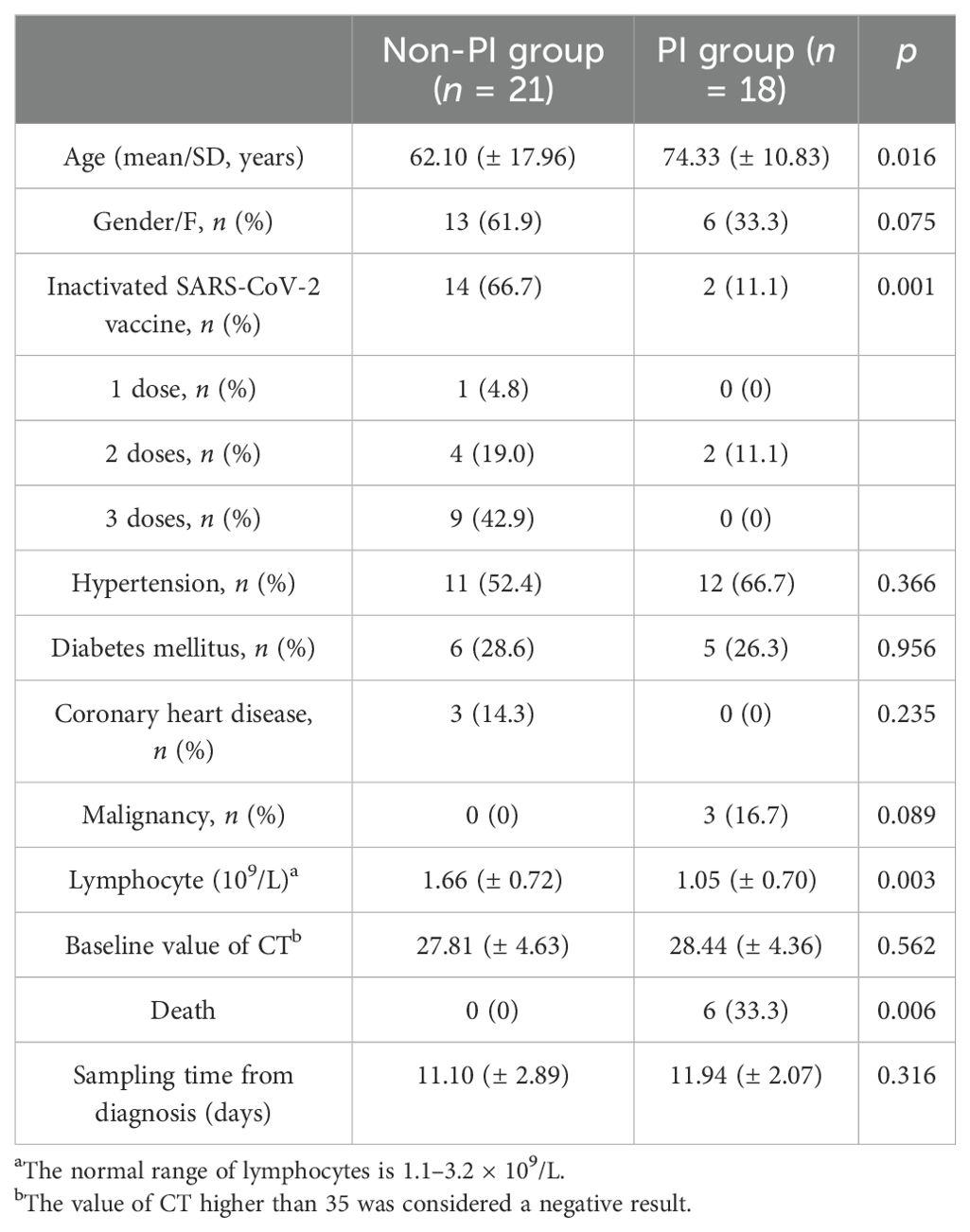
The study protocol was approved by the Ethics Committee of Renji Hospital affiliated with Shanghai Jiao Tong University (KY2022-103-B). A total of 39 patients diagnosed with SARS-CoV-2 Omicron variant infection were enrolled. The classification was performed according to the Diagnosis and Treatment Plan for COVID-19 (trial version 9) recommended by the National Health Commission of the People’s Republic of China as follows: (I) mild type: clinical symptoms are mild, and there are no radiographic signs of pneumonia; (II) common type: the aforementioned clinical symptoms are exhibited, and radiographic imaging shows signs of pneumonia; and (III) severe type: adults meet any of the following criteria: shortness of breath with a respiratory rate (RR) ≥30 breaths/min, oxygen saturation (SpO2) ≤93% while breathing ambient air at rest, and arterial oxygen partial pressure (PaO2)/fraction of inspired oxygen (FiO2) ≤300 mmHg (1 mmHg = 0.133 kPa). In high-altitude areas (altitude over 1,000 m), PaO2/FiO2 should be adjusted using the following formula: PaO2/FiO2 × [760/atmospheric pressure (mmHg)]. In the severe type, there is also progressive worsening of clinical symptoms, with significant lesion progression >50% within 24–48 h as shown by radiographic imaging. (I) pertains to the mild disease group without pulmonary infiltration, and (II) and (III) refer to the severe/critical group with pulmonary infiltration. Data on age, sex, comorbidities, SARS-CoV-2 vaccination status, baseline value of cycle threshold, lymphocyte count, and prognosis were collected.
2.2 RNA extraction
Fresh blood samples (2 ml) were collected from the patients with informed consent, drawn into EDTA tubes, transferred immediately into 15 ml microcentrifuge tubes, and added with 10 ml of TRIzol LS reagent. Total RNAs were extracted according to the manufacturer’s protocol.
2.3 TCR and BCR immune repertoire sequencing
For the current study, we used commercially available iR-RepSeq-plus 7-Chain Cassette (iRepertoire, Inc., USA) to generate NGS libraries covering all TCR and BCR seven chains, namely, TCR alpha (TRA), beta (TRB), delta (TRD), and gamma (TRG) and BCR Ig heavy (IGH), Ig kappa (IGK), and Ig lambda (IGL) from the RNA template. All seven chains were amplified in a single assay using a strategy that allows the incorporation of unique molecular identifiers (UMIs) during the reverse transcription (RT) step. Each disposable cassette is for library preparation for one sample, and all necessary reagents for amplification and purification are preloaded into the cassette. The library size is 500 bp. Quality control was performed with agarose gel to ensure the correct major product, purify the DNA library using SPRI beads, quantify the concentration with a NanoDrop, and ensure equal pooling between samples for sequencing. The RNA (500 ng) of each sample that met the requirements of IR sequencing on concentration and purity was loaded into the cassette. Amplified libraries were multiplexed and pooled for sequencing on the Illumina NovaSeq platform with a 500-cycle kit and sequenced as 250 paired-end reads. The output of the immune receptor sequence covered within the first framework region through the beginning of the constant region including CDR2 and CDR3.
2.4 Data collection and bioinformatics analysis
The raw sequences obtained were collapsed by 10-bp UMI tags into the consensus FASTA format using MiGEC version 1.2.9 (https://migec.readthedocs.io/en/latest/) and were analyzed using the iRmap program (42, 43). Briefly, sequence reads were demultiplexed according to both Illumina dual indices incorporated during the amplification process and barcode sequences at the 5′ end of the reads from the constant region. For immune repertoire gene sequencing, paired-end fastqs were demultiplexed by 6-bp barcode using MiGEC version 1.2.9 and then stitched into a single read using pandaseq version 2.11. The merged reads were mapped using a Smith–Waterman algorithm to germline V, D, J, and C reference sequences using an IMGT reference library. To define the complementarity determining region 3 (CDR3), the position of CDR3 boundaries of reference sequences from the IMGT database was migrated onto reads through mapping results, and the resulting CDR3 regions were extracted and translated into amino acids. The dataset was condensed by the combination of UMIs and CDR3 regions to remove incorrect CDR3s introduced by sequencing and amplification. Reads with the same combination of CDR3 and UMI were condensed into one.
CSR rate:
Where is the accumulation of the number of copies of different immunoglobulins with the same type of CDR3. For example (below)
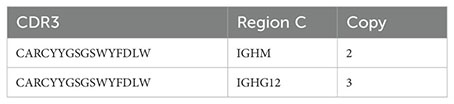
For the sequence “CARCYYGSGSWYFDLW,” the CSR num is 5, and the bottom of the fraction shows the accumulation of the quantity of all copies.
SHM:
The number of mutations from CDR1 -> end of the V region, divided by the alignment length.
VJ usage:
This parameter is also a ratio and tells the type of usage frequency. For example, if the quantity of all copies is 100,000, IGHV1-1 will have 3 occurrences. So, IGHV1-1 usage is 3/100,000.
uCDR3 diversity: For calculating diversity with read, it is more accurate to use uCDR3 for the calculation.
Shannon entropy:
Where , the base of the logarithm, determines the choice of units of the entropy measure. When a repertoire is composed of sequences evenly distributed, the Shannon entropy reaches its maximum, which is the logarithm of the number of unique sequences.
Gini:
Where , refer to the frequency of the respective and jth sequences in the repertoire and is the average of clone frequencies.
Gini is sometimes used to represent the clonal distribution of a repertoire. It is a measure of inequality that is widely used in economics to study wealth distribution.
D50:
Calculated as the percentage of clones that make up the top 50% of reads in the ranked clone distribution.
2.5 Statistical analysis
For continuous variables, data were presented as mean with standard deviation, and differences among groups were compared using Student’s t-test. Categorical variables were described as the number with percentage and compared using the chi-square test or Fisher’s exact test, as appropriate. All p <0.05 on two-sided tests was considered to be statistically significant. All statistical analyses were performed using the statistical software SPSS for Windows, Version 26.0 (IBM Corp., Armonk, NY, USA), GraphPad Prism V 8.0 (GraphPad Software, San Diego, California, USA), and R (ver4.2.0).
3 Results
3.1 Clinical demographics
This study enrolled 39 patients diagnosed with SARS-CoV-2 Omicron variant infection. They were divided into two groups: a mild disease group without pulmonary infiltration (non-PI, n = 21) and a severe/critical group with pulmonary infiltration (PI, n = 18). The PI group was significantly older than the non-PI group (74.33 ± 10.83 years vs. 62.10 ± 17.96 years, p = 0.016), had a lower lymphocyte count (1.05 ± 0.70 × 109/L vs. 1.66 ± 0.72 × 109/L, p = 0.003), a lower vaccination coverage rate (11.1% vs. 66.7%, p < 0.001), and a higher mortality rate (33.3% vs. 0%, p = 0.006) (Table 1). Additionally, there was no significant difference in underlying medical conditions (hypertension, diabetes mellitus, coronary heart disease, and cancer) between the two groups. Various studies (25–28) have reported lymphopenia in COVID-19 patients, characterized by a reduced number of lymphocytes and decreased expression of TCR and BCR. In this study, we also observed a significant reduction in lymphocyte count in PI patients compared to non-PI patients (Table 1).
3.2 Comprehensive immune repertoire differences between PI and non-PI patients
Previous studies have shown characteristic alterations in the TCR/BCR immune repertoire following SARS-CoV-2 infection (18, 29). However, only a few immune repertoire profiling studies for the SARS-CoV-2 Omicron variant infection have been reported. We collected peripheral blood samples from all 39 patients and amplified the immune repertoire, including all TCR (TRA, TRB, TRD, TRG) and BCR (IGH with its isotypes IGK and IGL), in a single, unbiased PCR reaction. Several studies have revealed the activation patterns and dynamics of the adaptive immune cell response to SARS-CoV-2 infection (20, 30). We observed a pattern of increased IGH and IGK chains and decreased TRA, TRB, TRD, and TRG chains within the total immune repertoire in PI patients compared to non-PI patients (Figures 1A–C), indicating a hyperactivation of humoral immunity over cellular immunity in patients with severe to critical symptoms. We further analyzed the B-cell repertoire composition by calculating unique CDR3 sequences. IGHA was predominant in the BCR repertoire of all patients, while the proportions of IGHD and IGHM were significantly lower in PI patients compared to non-PI patients (Figures 1D–F). Previous studies have reported a dominant IGHA response to viral infections, including SARS-CoV-2 (19, 31), consistent with our findings. Additionally, reduced IGHD and IGHM in PI patients suggest that IGHD and IGHM may offer protection against SARS-CoV-2 Omicron variant lung tissue invasion. Lastly, we measured the mean CDR3 length across all seven immune repertoire chains. The TRD and TRG chains in PI patients had longer and shorter CDR3 lengths, respectively, compared to non-PI patients (Figure 1G), while the other chains showed no significant differences between the groups (Supplementary Figures 1A–E).
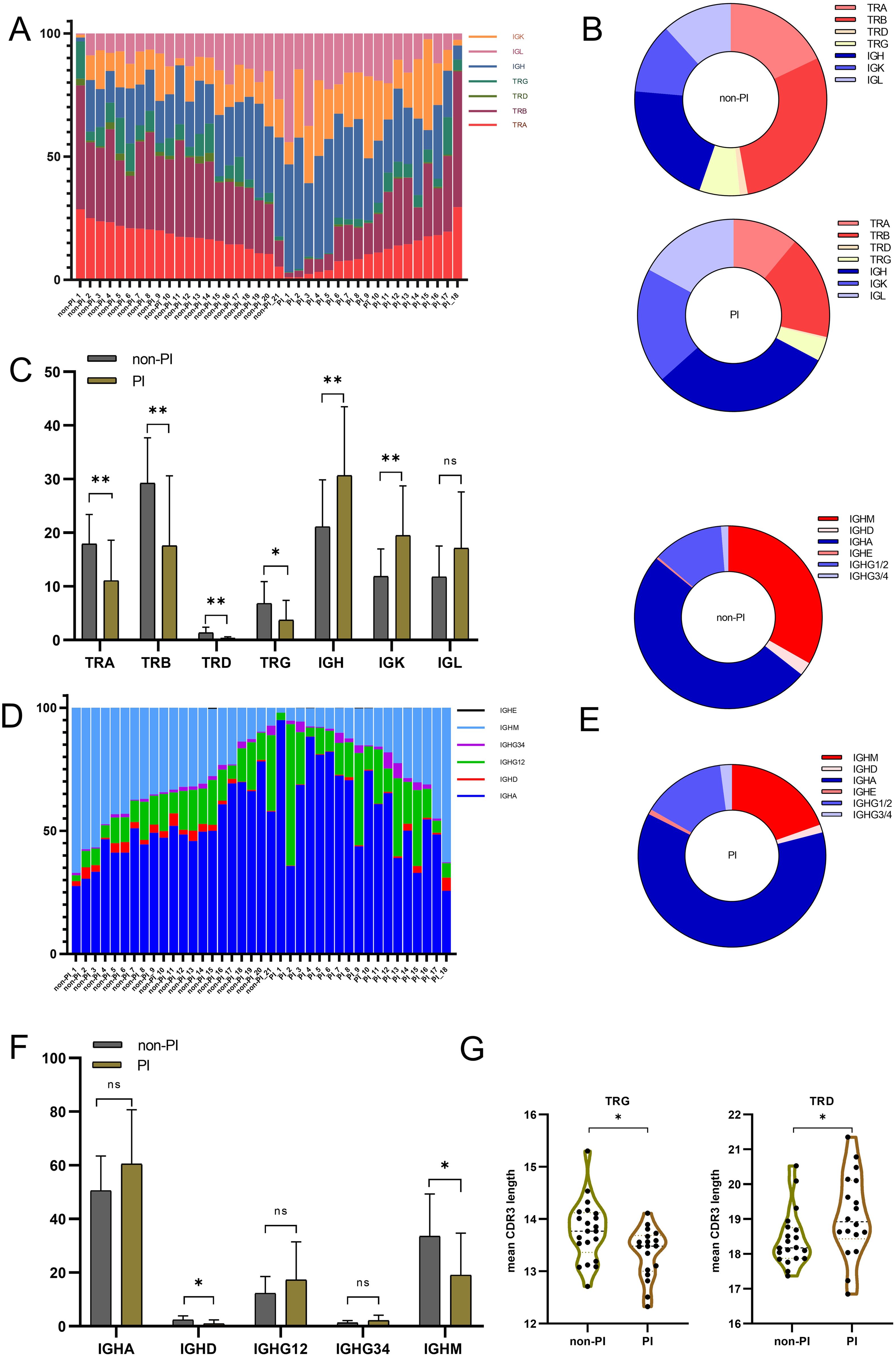
Figure 1. The overall TCR and BCR immune repertoire of patients with SARS−CoV−2 Omicron infection without or with pulmonary infiltration were sequenced, and the reads of each gene were aligned and counted. Data were collected from 39 biological replicated experiments: n = 21 without pulmonary infiltration (non-PI) and n = 18 with pulmonary infiltration (PI). (A) Percentage of the seven-chain repertoire for patients with SARS−CoV−2 Omicron infection without or with pulmonary infiltration. Each bar represents the composition ratio in one sample. (B) The composition ratio of the seven-chain immune repertoire in the two groups by counting unique CDR3. (C) The percentage [details from (A)] for each TCR and BCR chain between the two groups. (D) Comparison of each Ig isotype percentage for patients with SARS−CoV−2 Omicron infection without or with pulmonary infiltration. (E) The composition ratio of Ig isotypes in the two groups by counting unique CDR3. (F) The percentage [details from (D)] for each BCR isotype between the two groups. (G) Mean CDR3 length of the TRG and TRD chains between the two groups. * means P≤ 0.05, ** means P≤ 0.01, ns means No significance.
In summary, patients with pulmonary infiltration exhibited a distinct immune repertoire, characterized by decreased TCR and increased BCR expression. Specifically, IGHD and IGHM showed reduced expression, and the TRD/TRG chains had longer/shorter CDR3 lengths in PI patients, potentially serving as an immune repertoire signature to distinguish patients with or without pulmonary infiltration.
3.3 Altered adaptome clonality and diversity between PI and non-PI patients
A unique CDR3 (uCDR3) sequence, also known as a clonotype, targets specific antigens, thus reflecting the overall adaptive immunity. PI patients showed a reduction in uCDR3 for TRA, TRB, TRD, and TRG, but an increase in uCDR3 for IGH, IGK, and IGL, compared to non-PI patients (Figure 2A). Numerous studies have highlighted a correlation between reduced immune repertoire diversity, an impaired adaptive immune system, and various diseases (28). To determine if PI patients had a more severely damaged repertoire than non-PI patients, we assessed the diversity of the TCR and BCR repertoires in all samples. Diversity, quantified by Shannon entropy, indicates the uniformity of the immune repertoire. PI patients exhibited a significant reduction in Shannon entropy for three immune repertoire chains, TRB, TRD, and IGH, compared to non-PI patients (Figures 2B, C). The clonal diversification index (CDI) analysis was used to assess population unevenness, mitigating potential biases from varying RNA molecule counts per cell. PI patients exhibited a reduced CDI d50 and an increased CDI Gini compared to non-PI patients (Figure 2D). In summary, our findings reveal diminished TCR/BCR clonality and diversity in PI patients, suggesting a further deteriorated adaptome.
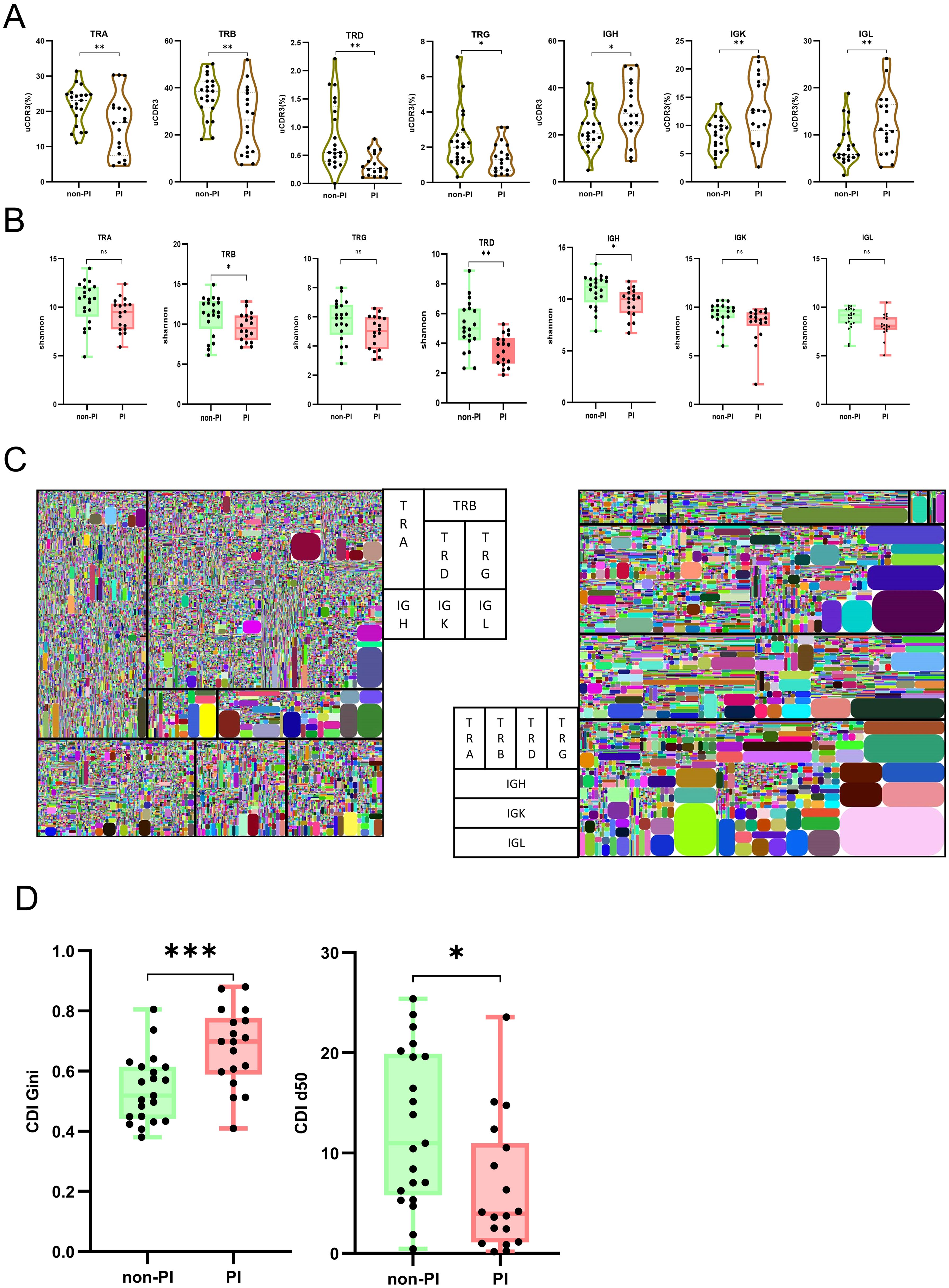
Figure 2. TCR and BCR abundance, clonal expansion, and diversity analysis between patients with SARS−CoV−2 Omicron infection without or with pulmonary infiltration. (A) The abundance of each chain for immune repertoire adaptome is measured by the number of unique CDR3 (uCDR3); the uCDR3 counts of each chain are presented for the two groups as mean value ± SD in the boxplots. (B) The diversity was demonstrated by the Shannon index at the level of unique uCDR3 clones, as shown in TRA, TRB, TRD, TRG, IGH, IGK, and IGL. (C) Seven-chain repertoire tree maps for two representative subjects from each group. Each square represents a chain, with its size being representative of its relative expression out of the seven chains. All tree maps should be read from left to right and then from top to bottom in the following order: TRA, TRB, TRD, TRG, IGH, IGK, and IGL. Each rounded rectangle color block is a clone. (D) The diversity was demonstrated by CDI d50 and CDI Gini. * means P≤ 0.05, ** means P≤ 0.01, *** means P≤ 0.001, ns means No significance.
3.4 Characteristic TRBV/J and IGHV/J gene usage and shared CDR3 sequences between PI and non-PI patients
The TRBV and IGHV families encode the variable segments of the TRB and IGH chains, significantly impacting TCR/BCR diversity (13, 30). We calculated the frequencies of TRBV, TRBJ, IGHV, and IGHJ genes in PI patients and compared them with those in non-PI patients. Siginificant different usage of TRBV5-7, IGHV1-46, IGHV3-30, IGHV3-15, IGHV3-53, and IGHV3-73 was found in PI patients compared to non-PI patients (Figure 3A). Furthermore, significant differences in TRBV_TRBJ and IGHV_IGHJ pairs were observed between the two groups of patients (Figures 3B, C). Uniform manifold approximation and projection (UMAP) analyses were conducted based on the abundance of TRBV (Figure 3B) and IGHV (Figure 3C) segments. Significant differences between PI and non-PI patients were noted, with distinct separation of TRBVJ and IGHVJ clusters (Figures 3D, E). To assess the prevalence of shared CDR3 sequences among the two groups, we combined all sequencing data for analysis. Shared CDR3 sequences across seven chains were expressed at different levels between PI and non-PI patients. The expression levels of uCDR3 and representative sequences were plotted, revealing significant differences between PI and non-PI patients (p < 0.05) (Figures 3F, G). Each column represents a patient, and each cell in a row indicates the expression frequency of a CDR3 sequence, with the color indicating the expression level (blue for low expression and red for high expression). Among the seven chains, shared CDR3 sequences were found in TRA, TRB, TRD, TRG, IGL, and IGK, but not in IGH (Figure 3G), likely due to the high frequency of SHM in the IGH chain. Between PI and non-PI patients, numerous CDR3 sequences showed differential expression, with the most highly expressed shared CDR3 sequences found in non-PI patients (Figures 3F, G), suggesting that these CDR3 sequences may offer protection against pulmonary infiltration. Meanwhile, a small subset of shared CDR3 sequences in the IGK and IGL chains was preferentially expressed in the PI group but absent in the non-PI group, indicating that these clones may be associated with the progression of the infection. In summary, we identified significant differences in V and V-J gene usage between PI and non-PI patients, along with a unique CDR3 signature that was differentially expressed between the two groups. These molecular markers could be further investigated for their roles in the etiology and progression of COVID-19 pulmonary infiltration and their potential as diagnostic biomarkers.
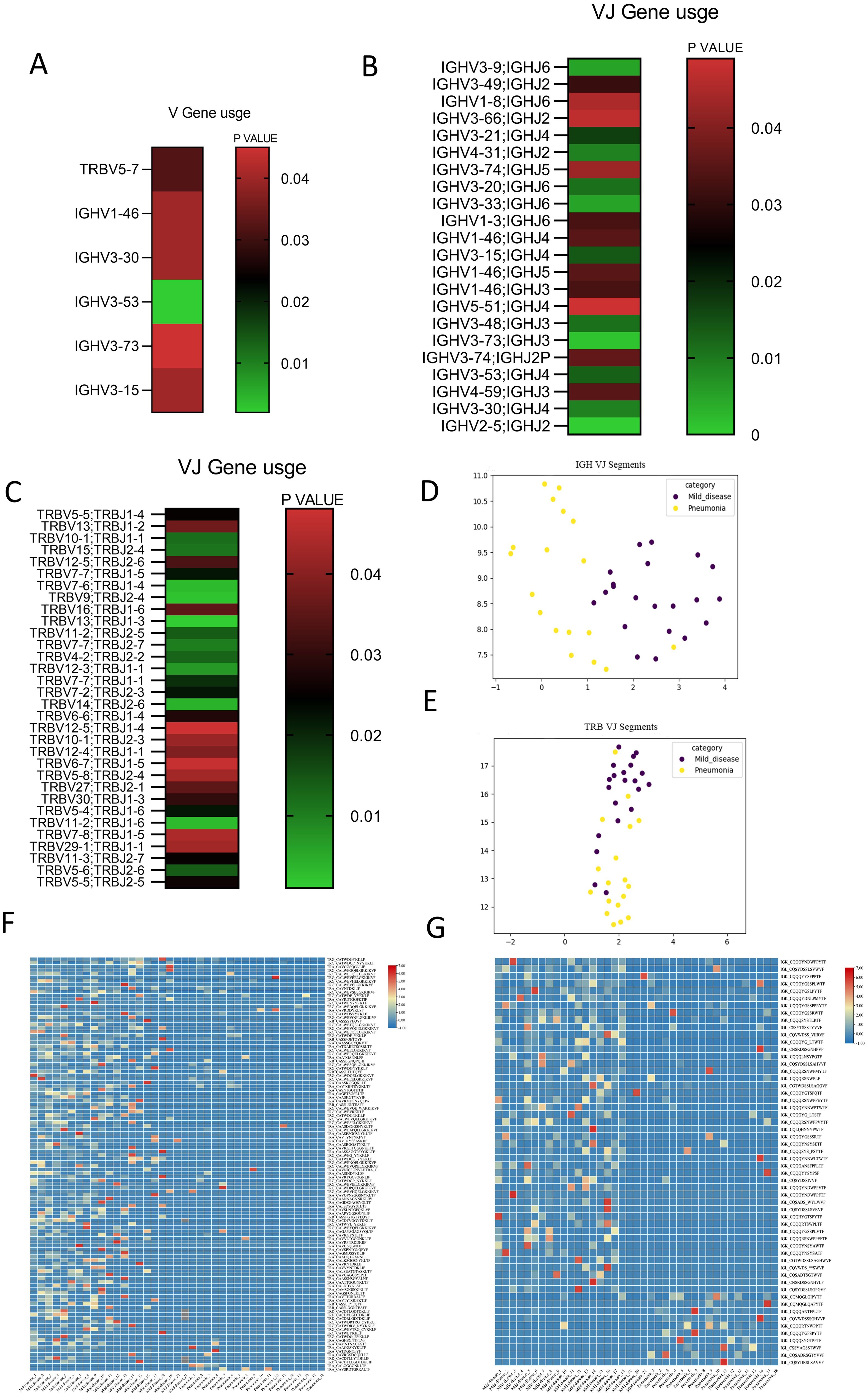
Figure 3. Distinct V/J preference usage and share of CDR3 clones between patients with SARS−CoV−2 Omicron infection without or with pulmonary infiltration. Cluster analysis heat map of the expression level of genes based on the (A) difference of the IGHV gene and the TRBV gene between the two groups. Heat maps of TRBV–TRBJ (B) and IGHV–IGHJ conjunctions (C) between the two groups are presented, showing preferred TRBV–TRBJ and IGHV–IGHJ pairs. Uniform manifold approximation and projection (UMAP) based on the abundance of TRBV (E) and IGHV (D) segments; the distance between the dots indicates the degree of dissimilarity between samples. Cluster analysis heat map for CDR3 between patients with SARS−CoV−2 Omicron infection without or with pulmonary infiltration for TCR (F) and BCR (G).
3.5 Elevated CSR in PI patients compared to non-PI patients
The B-cell repertoire is characterized by unique events of CSR and SHM (32, 33). Considering the six main isotypes (IGHD, IGHM, IGHG1/2, IGHG3/4, IGHE, and IGHA), we used the percentage of unique CDR3 sequences observed in more than one isotype to evaluate class switching among multiple isotypes. We observed dramatic differences in CSR rates among different isotypes between PI and non-PI patients (Figure 4A). The CSR rates for IGHE–IGHG12–IGHA, IGHG34–IGHE–IGHA, IGHD–IGHM–IGHA, IGHE–IGHA, IGHG34–IGHA, IGHG34–IGHE, IGHD–IGHA, and IGHD–IGHE were significantly higher in non-PI patients, while the remaining isotypes had higher CSR rates in the PI group. In PI patients, the IGHA isotypes of BCR most frequently undergo class switching. Overall, the cumulative CSR rates in non-PI patients were higher for IGHD, IGHG34, IGHM, and IGHE and lower for IGHG12 and IGHA (Figure 4A). Previous studies have reported changes in SHM among COVID-19 patients (20). In our study, no significant differences in SHM were observed for IGM/IGD, IGHG1/2, IGHG3/4, and IGHA between the two groups (Figure 4B). In summary, PI patients exhibited distinct B-cell repertoire patterns, characterized by lower CSR rates in IGHD, IGHG34, IGHM, and IGHE and higher CSR rates in IGHG12 and IGHA.
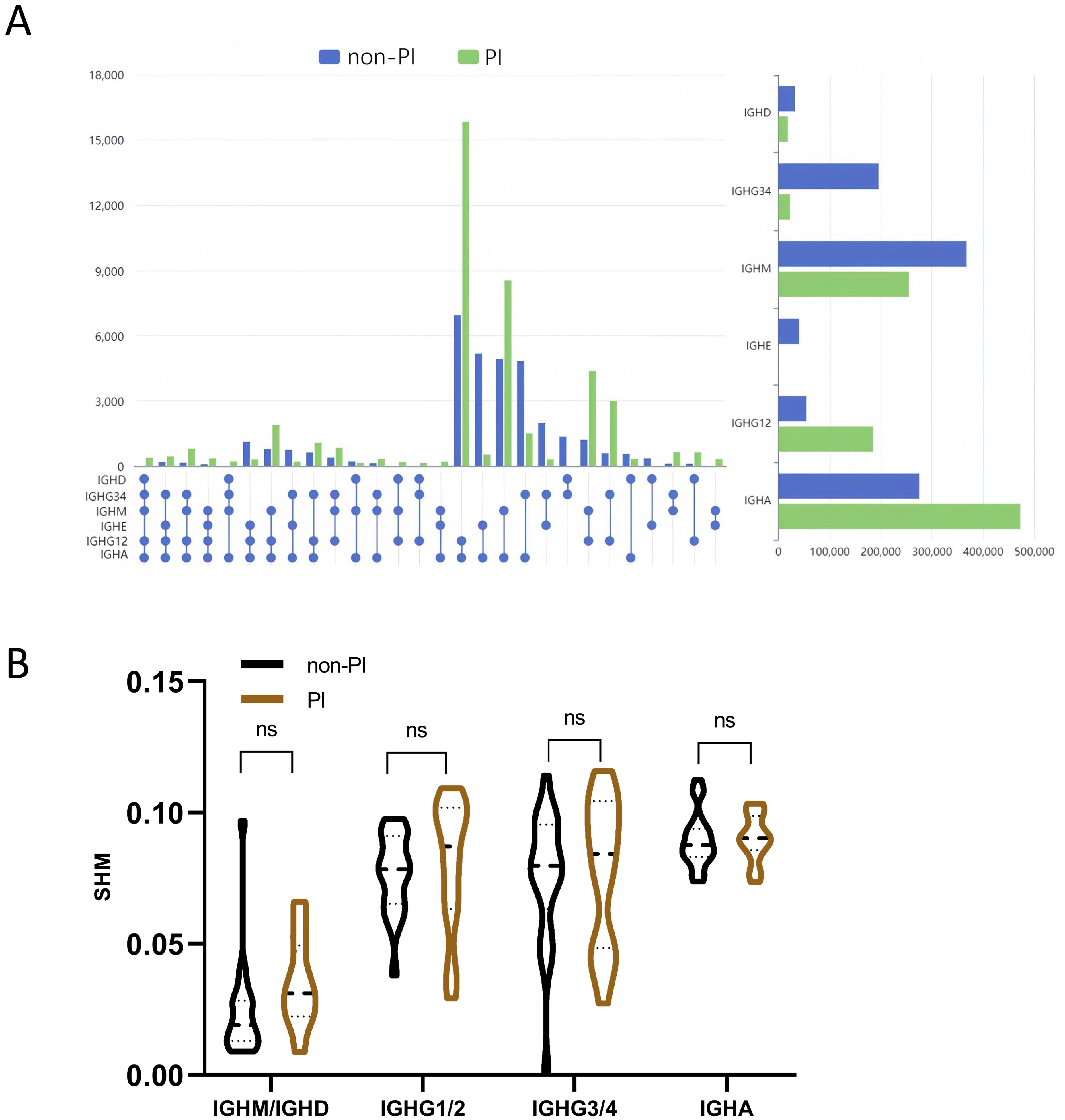
Figure 4. Analysis of class switch recombination (CSR) and somatic hypermutation (SHM) for BCR. (A) The CSR of the six Igs (IGHA, IGHD, IGHE, IGHG12, IGHG34, and IGHM) between the two groups. The spheres represent different immunoglobulin isotypes, and the line in between the dots indicates class switch between these two isotypes. Isotype switch with a significant difference between the two groups (p-value < 0.05) is presented. (B) Somatic hypermutation ratio in the four subtypes of Ig (IGHM and IGHD are shown in the graph as a whole). ns means No significance.
3.6 The inactivated COVID-19 vaccine triggered a specific adaptome response and protected patients from severe or critical conditions
The inactivated vaccine has been proven to be an effective strategy for preventing the COVID-19 pandemic (34). However, the rapid evolution of the virus has led to numerous variants and mutations, compromising the efficacy of the original vaccines. Since November 2021, the SARS-CoV-2 Omicron variant has become the most dominant strain globally, as also found in the sample cohort of this study. In this study, of the 21 patients with mild cases, 14 were vaccinated, whereas only 2 of the 18 severe/critical cases had been vaccinated. This validates that the inactivated vaccine provides significant protection against severe or critical conditions in patients with SARS-CoV-2 Omicron infection (Table 1). To further explore the adaptome features of vaccinated patients, we compared the immune repertoire features in mild cases with and without prior vaccination. The overall composition of the seven-chain immune repertoire and the proportion of B-cell isotypes showed no difference between vaccinated and unvaccinated subjects (Figures 5A, B). Interestingly, the diversity of all TRA, TRB, TRD, TRG, IGH, IGK, and IGL chains, as measured by the Shannon index, was significantly lower in unvaccinated compared to vaccinated patients (Figures 5C), indicating more severe damage to the immune system of unvaccinated patients upon infection. Finally, we observed a higher SHM rate in non-vaccinated compared to vaccinated patients (Figures 5D–F). The SHM rate has been reported to be related to the efficacy of vaccination (35), reflecting the preparedness of B cells against infection. Therefore, the inactivated COVID-19 vaccine triggered a lesser B-cell IGHM/IGHD SHM response upon SARS-CoV-2 Omicron infection, preserved the diversity of the entire adaptome immune repertoire, and provided clinical protection against severe or critical conditions caused by SARS-CoV-2 Omicron infection.
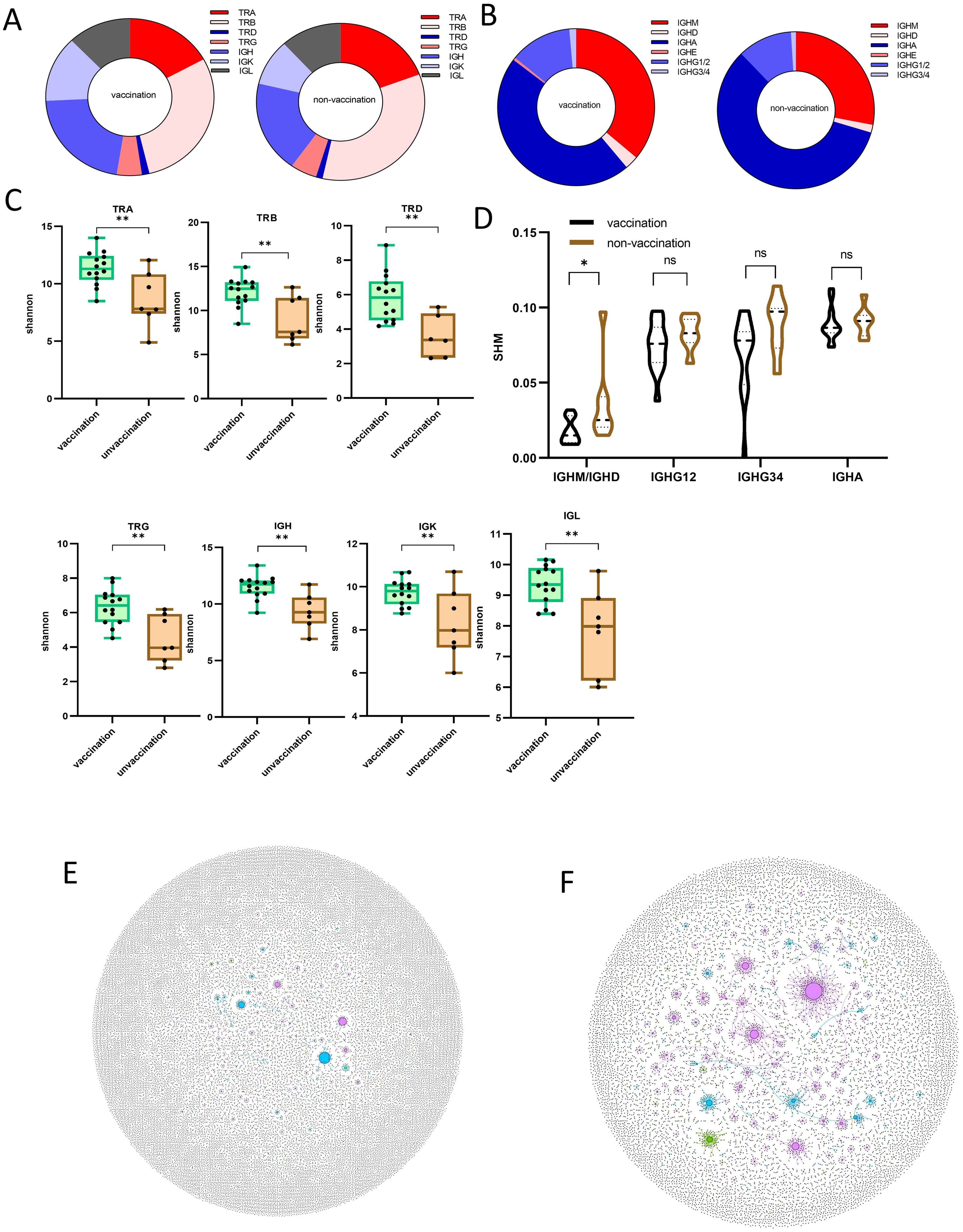
Figure 5. Immune repertoire pattern difference between vaccinated and unvaccinated patients after being infected with SARS−CoV−2 Omicron. (A) The composition ratio of the seven-chain immune repertoire in vaccinated and unvaccinated patients. (B) Comparison of each Ig isotype percentage for patients with SARS−CoV−2 Omicron infection with or without prior vaccination. (C) The diversity was demonstrated by the Shannon index at the level of unique uCDR3 clones, as shown in TRA, TRB, TCD, TRG, IGH, IGK, and IGL for patients with SARS−CoV−2 Omicron infection with or without prior vaccination. (D) Somatic hypermutation ratio in the four subtypes of Ig (IGHM and IGHD are shown in the graph as a whole) of patients with SARS−CoV−2 Omicron infection with or without prior vaccination. (E) B-cell hypermutation is shown in two representative individual samples (F), in which each dot represents a unique B-cell uCDR3. The size of each dot denotes the relative frequency of that uCDR3 clone, and the dots connected by a line represent hypermutation. * means P≤ 0.05, ** means P≤ 0.01, ns means No significance.
4 Discussion
Since December 2019, the COVID-19 pandemic has had a devastating impact on the global economy and health. Furthermore, SARS-CoV-2 continues to mutate, generating new variants, with Omicron becoming the dominant variant so far (36, 37). The host’s immune response against SARS-CoV-2 infection not only plays an antiviral role but also leads to simultaneous pathogenic injury of the organs and tissues, especially in the lungs of patients with COVID-19, which determines the disease severity and outcome (38, 39). Therefore, it is of great importance to understand immune responses to SARS-CoV-2 infection, especially the SARS-CoV-2 Omicron variant. To address this, we profiled the immune repertoire landscape, characterizing the composition and diversity using iR-RepSeq-plus 7-Chain Cassette in patients with SARS-CoV-2 Omicron infection, both with and without pulmonary infiltration (PI and non-PI patients).
This study delineated the disrupted immune repertoire patterns in patients with SARS-CoV-2 Omicron infection, particularly those with PI. Our results revealed reduced diversity in TRB, TRD, and IGH chains in PI patients compared to non-PI patients, potentially constituting a risk factor for disease progression. Several studies found that SARS-CoV-2 infection caused the TRB chain or IGH chain diversity to decline (16, 29, 30). By definition, an exaggerated focus would limit immune cell diversity that may be particularly important antibody-escape variants of concern and/or other coinfections. We also observed preferential V(D)J gene segment usage of IGH and TRB between the two groups of patients. The skewed usage of the V/J segments may be associated with immune dysfunction and the pathogenesis of the disease. Upon infection, SARS-CoV-2 antigens can lead to the targeted rearrangement and excessive abnormal cloning of one or a few V subfamilies, and the cloning of other immune cells may be suppressed by the dominant immune cell clones, which may result in impaired immune function and decreased ability to clear the virus (30).
The distribution of CDR3 sequence lengths is another key feature that provides an integrative view of repertoire composition. Biases in CDR3 length are often observed in epitope-specific repertoire (40). In our study, we observed differences in the mean CDR3 length of the TRD and TRG chains between the two groups. We found a cluster of shared TCR and BCR CDR3 clones, most of which were highly shared among non-PI patients. Therefore, these TCR and BCR clonotypes shared between individuals are likely stimulated against common antigens and are thought to play a significant role in the efficacy of pathogen-specific responses and the control of infection. Thus, if linked to certain infections, such TCRs or BCRs could become invaluable tools for immune diagnosis of human disease and vaccine development. Furthermore, in our cohort, IGHA presented the highest proportion in total BCR repertoire in the two groups. Meanwhile, compared with the other isotypes, the IGHA isotypes of BCR frequently undergo most class switches in PI patients. This discovery thus highlights the importance of developing and using inhaled vaccine. Meanwhile, for SARS-CoV-2, neutralizing antibodies primarily target five structural regions of the spike protein, especially the receptor-binding domain (RBD), and can block the virus from infecting host cells. Previous studies have shown that most neutralizing antibodies in certain populations are encoded by the IGHV1-2 gene family. However, our research found an increased usage of IGHV3 in pneumonia patients. Additionally, human neutralizing antibodies against COVID-19 are predominantly of the IgA class, and our results indicate a higher rate of CSR to IGHA in pneumonia patients. This may suggest increased production of non-neutralizing antibodies, potentially leading to immune-related lung damage.
The above results indicated that a disruptive immune repertoire may contribute to COVID-19 progress. Vaccine remains the most effective approach against the COVID-19 global pandemic. Meanwhile, we found a higher vaccination coverage in the mild disease group. By immune repertoire sequencing, compared to unvaccinated patients, upon SARS-CoV-2 infection, vaccinated patients possess increased IGH chain and TRB chain diversity, and more IGHM and IGHD SHMs were observed in unvaccinated patients. To some extent, vaccination increases the immune system’s buffering capacity to ameliorate the SARS-CoV-2 Omicron infection. A previous study (35) had a different observation, and we believe the contradictory conclusion is due to the different mechanisms of the vaccine. Their study focused on mRNA vaccines, targeting specific immunity against parts of the S protein, while our study encompassed subjects vaccinated with the inactivated vaccine, eliciting a broader immune response due to a wider range of epitopes. Our findings still have shortcomings, including the lack of strict negative controls (patients with no infection) and a relatively small number of cases included, which may lead to instability in the results. However, our findings still aid in understanding the immune repertoire in patients infected with the SARS-CoV-2 Omicron variant.
In summary, this study provides a comprehensive overview of the immune repertoire in patients infected with the SARS-CoV-2 Omicron variant. We also found a correlation between the impaired immune repertoire and the clinical severity of COVID-19. Finally, our study suggests that the TCR/BCR repertoire could serve as a biomarker for predicting COVID-19 severity, offering diagnostic value and guiding more precise therapeutic strategies.
Data availability statement
The data presented in the study are deposited in the National Genomics Data Center repository, accession number HRA009242.
Ethics statement
The studies involving humans were approved by Renji Hospital affiliated Shanghai Jiao Tong University. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
XL: Conceptualization, Data curation, Formal Analysis, Writing – original draft, Writing – review & editing. HZ: Writing – original draft. PX: Writing – original draft. JZ: Writing – review & editing. ZW: Writing – review & editing. HH: Writing – review & editing. FS: Writing – review & editing. YJ: Writing – review & editing. LS: Writing – review & editing. JX: Writing – review & editing. LY: Writing – review & editing. CY: Writing – review & editing. HJ: Writing – review & editing. GG: Writing – review & editing. JJ: Writing – review & editing. HS: Writing – review & editing. YW: Writing – review & editing. LW: Writing – review & editing. CQ: Writing – review & editing. DL: Writing – review & editing. WQ: Writing – review & editing. QL: Writing – review & editing. YC: Writing – review & editing. FL: Supervision, Writing – review & editing. YL: Investigation, Methodology, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by a research grant from the Chinese PLA General Hospital Chronic Kidney Disease Clinical Medicine Research Centre (kfkt202015).
Acknowledgments
The authors are grateful for the assistance provided by the Shanghai Cancer Institute and Biliary-Pancreatic Surgery, Renji Hospital Affiliated with Shanghai Jiao Tong University School of Medicine. The authors also thank the following individuals from Renji Hospital, School of Medicine, Shanghai Jiao Tong University for their contributions to this study: Drs. Min Li from the Department of Laboratory Medicine, Yuan Gao from the Department of Critical Care Medicine, Shuang Ye from the Department of Rheumatology, Jing Ma from the Department of Endocrinology, Yi Chen from the Department of Emergency, and Hua Zhong from the Department of Hematology.
Conflict of interest
The authors ZW and HH were employed by Tilcure Biotherapeutics.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1486352/full#supplementary-material
Supplementary Figure 1 | (A-E) Mean CDR3 length of TRA, TRB, IGK, IGH and IGL chains between two groups.
References
1. Vitiello A, Ferrara F, Auti AM, Di Domenico M, Boccellino M. Advances in the Omicron variant development. J Intern Med. (2022) 292:81–90. doi: 10.1111/joim.13478
2. Sun F, Lin Y, Wang X, Gao Y, Ye S. Paxlovid in patients who are immunocompromised and hospitalised with SARS-CoV-2 infection. Lancet Infect Dis. (2022) 22(9):1279. doi: 10.1016/s1473-3099(22)00430-3
3. Huang C, Huang L, Wang Y, Li X, Ren L, Gu X, et al. 6-month consequences of COVID-19 in patients discharged from hospital: a cohort study. Lancet. (2021) 397:220–32. doi: 10.1016/s0140-6736(20)32656-8
4. Shi L, Huang H, Lu X, Yan X, Jiang X, Xu R, et al. Effect of human umbilical cord-derived mesenchymal stem cells on lung damage in severe COVID-19 patients: a randomized, double-blind, placebo-controlled phase 2 trial. Signal transduction targeted Ther. (2021) 6:58. doi: 10.1038/s41392-021-00488-5
5. Shi L, Yuan X, Yao W, Wang S, Zhang C, Zhang B, et al. Human mesenchymal stem cells treatment for severe COVID-19: 1-year follow-up results of a randomized, double-blind, placebo-controlled trial. EBioMedicine. (2022) 75:103789. doi: 10.1016/j.ebiom.2021.103789
6. Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. A novel coronavirus from patients with pneumonia in China 2019. New Engl J Med. (2020) 382:727–33. doi: 10.1056/NEJMoa2001017
7. Thakur V, Ratho RK. OMICRON (B.1.1.529): A new SARS-CoV-2 variant of concern mounting worldwide fear. J Med Virol. (2022) 94:1821–4. doi: 10.1002/jmv.27541
8. Guo YR, Cao QD, Hong ZS, Tan YY, Chen SD, Jin HJ, et al. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak - an update on the status. Military Med Res. (2020) 7:11. doi: 10.1186/s40779-020-00240-0
9. Yu S, Di C, Chen S, Guo M, Yan J, Zhu Z, et al. Distinct immune signatures discriminate between asymptomatic and presymptomatic SARS-CoV-2(pos) subjects. Cell Res. (2021) 31:1148–62. doi: 10.1038/s41422-021-00562-1
10. Wang EY, Mao T, Klein J, Dai Y, Huck JD, Jaycox JR, et al. Diverse functional autoantibodies in patients with COVID-19. Nature. (2021) 595:283–8. doi: 10.1038/s41586-021-03631-y
11. Zuo Y, Estes SK, Ali RA, Gandhi AA, Yalavarthi S, Shi H, et al. Prothrombotic autoantibodies in serum from patients hospitalized with COVID-19. Sci Trans Med. (2020) 12:570–80. doi: 10.1126/scitranslmed.abd3876
12. Tian W, Zhang N, Jin R, Feng Y, Wang S, Gao S, et al. Immune suppression in the early stage of COVID-19 disease. Nat Commun. (2020) 11:5859. doi: 10.1038/s41467-020-19706-9
13. Bashford-Rogers RJM, Bergamaschi L, McKinney EF, Pombal DC, Mescia F, Lee JC, et al. Analysis of the B cell receptor repertoire in six immune-mediated diseases. Nature. (2019) 574:122–6. doi: 10.1038/s41586-019-1595-3
14. Li F, Luo M, Zhou W, Li J, Jin X, Xu Z, et al. Single cell RNA and immune repertoire profiling of COVID-19 patients reveal novel neutralizing antibody. Protein Cell. (2021) 12:751–5. doi: 10.1007/s13238-020-00807-6
15. Niu X, Li S, Li P, Pan W, Wang Q, Feng Y, et al. Longitudinal analysis of T and B cell receptor repertoire transcripts reveal dynamic immune response in COVID-19 patients. Front Immunol. (2020) 11:582010. doi: 10.3389/fimmu.2020.582010
16. Xiang H, Zhao Y, Li X, Liu P, Wang L, Wang M, et al. Landscapes and dynamic diversifications of B-cell receptor repertoires in COVID-19 patients. Hum Immunol. (2022) 83:119–29. doi: 10.1016/j.humimm.2021.10.007
17. Alt FW, Zhang Y, Meng FL, Guo C, Schwer B. Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell. (2013) 152:417–29. doi: 10.1016/j.cell.2013.01.007
18. Lin K, Zhou Y, Ai J, Wang YA, Zhang S, Qiu C, et al. B cell receptor signatures associated with strong and poor SARS-CoV-2 vaccine responses. Emerg Microbes Infect. (2022) 11:452–64. doi: 10.1080/22221751.2022.2030197
19. He B, Liu S, Xu M, Hu Y, Lv K, Wang Y, et al. Comparative global B cell receptor repertoire difference induced by SARS-CoV-2 infection or vaccination via single-cell V(D)J sequencing. Emerg Microbes Infect. (2022) 11:2007–20. doi: 10.1080/22221751.2022.2105261
20. Jin X, Zhou W, Luo M, Wang P, Xu Z, Ma K, et al. Global characterization of B cell receptor repertoire in COVID-19 patients by single-cell V(D)J sequencing. Briefings Bioinf. (2021) 22. doi: 10.1093/bib/bbab192
21. Hodgson SH, Mansatta K, Mallett G, Harris V, Emary KRW, Pollard AJ. What defines an efficacious COVID-19 vaccine? A review of the challenges assessing the clinical efficacy of vaccines against SARS-CoV-2. Lancet Infect Dis. (2021) 21:e26–35. doi: 10.1016/s1473-3099(20)30773-8
22. Al Kaabi N, Zhang Y, Xia S, Yang Y, Al Qahtani MM, Abdulrazzaq N, et al. Effect of 2 inactivated SARS-CoV-2 vaccines on symptomatic COVID-19 infection in adults: A randomized clinical trial. JAMA. (2021) 326:35–45. doi: 10.1001/jama.2021.8565
23. Xia S, Duan K, Zhang Y, Zhao D, Zhang H, Xie Z, et al. Effect of an inactivated vaccine against SARS-CoV-2 on safety and immunogenicity outcomes: interim analysis of 2 randomized clinical trials. JAMA. (2020) 324:951–60. doi: 10.1001/jama.2020.15543
24. Dong Y, Dai T, Wang B, Zhang L, Zeng LH, Huang J, et al. The way of SARS-CoV-2 vaccine development: success and challenges. Signal transduction targeted Ther. (2021) 6:387. doi: 10.1038/s41392-021-00796-w
25. Tang G, Huang M, Luo Y, Liu W, Lin Q, Mao L, et al. The dynamic immunological parameter landscape in coronavirus disease 2019 patients with different outcomes. Front Immunol. (2021) 12:697622. doi: 10.3389/fimmu.2021.697622
26. Wu H, Zhu H, Yuan C, Yao C, Luo W, Shen X, et al. Clinical and immune features of hospitalized pediatric patients with coronavirus disease 2019 (COVID-19) in Wuhan, China. JAMA network Open. (2020) 3:e2010895. doi: 10.1001/jamanetworkopen.2020.10895
27. Wu M, Pan W, Jia C, He Z, Zhao M, Tang C, et al. Systemic lupus erythematosus patients contain B-cell receptor repertoires sensitive to immunosuppressive drugs. Eur J Immunol. (2022) 52:669–80. doi: 10.1002/eji.202149596
28. Zhou Y, Zhang J, Wang D, Wang D, Guan W, Qin J, et al. Profiling of the immune repertoire in COVID-19 patients with mild, severe, convalescent, or retesting-positive status. J Autoimmun. (2021) 118:102596. doi: 10.1016/j.jaut.2021.102596
29. Joseph M, Wu Y, Dannebaum R, Rubelt F, Zlatareva I, Lorenc A, et al. Global patterns of antigen receptor repertoire disruption across adaptive immune compartments in COVID-19. Proc Natl Acad Sci United States America. (2022) 119:e2201541119. doi: 10.1073/pnas.2201541119
30. Hou X, Wang G, Fan W, Chen X, Mo C, Wang Y, et al. T-cell receptor repertoires as potential diagnostic markers for patients with COVID-19. Int J Infect Dis. (2021) 113:308–17. doi: 10.1016/j.ijid.2021.10.033
31. Stacey HD, Golubeva D, Posca A, Ang JC, Novakowski KE, Zahoor MA, et al. IgA potentiates NETosis in response to viral infection. Proc Natl Acad Sci United States America. (2021) 118. doi: 10.1073/pnas.2101497118
32. De Silva NS, Klein U. Dynamics of B cells in germinal centres. Nat Rev Immunol. (2015) 15:137–48. doi: 10.1038/nri3804
33. Stavnezer J, Guikema JE, Schrader CE. Mechanism and regulation of class switch recombination. Annu Rev Immunol. (2008) 26:261–92. doi: 10.1146/annurev.immunol.26.021607.090248
34. Muhsen K, Maimon N, Mizrahi AY, Boltyansky B, Bodenheimer O, Diamant ZH, et al. Association of receipt of the fourth BNT162b2 dose with omicron infection and COVID-19 hospitalizations among residents of long-term care facilities. JAMA Internal Med. (2022) 182:859–67. doi: 10.1001/jamainternmed.2022.2658
35. Fraley ER, Khanal S, Pierce SH, LeMaster CA, McLennan R, Pastinen T, et al. Effects of prior infection with SARS-CoV-2 on B cell receptor repertoire response during vaccination. Vaccines. (2022) 10:1477–82. doi: 10.3390/vaccines10091477
36. Huang M, Wu L, Zheng A, Xie Y, He Q, Rong X, et al. Atlas of currently available human neutralizing antibodies against SARS-CoV-2 and escape by Omicron sub-variants BA.1/BA.1.1/BA.2/BA.3. Immunity. (2022) 55:1501–14.e1503. doi: 10.1016/j.immuni.2022.06.005
37. Pulliam JRC, van Schalkwyk C, Govender N, von Gottberg A, Cohen C, Groome MJ, et al. Increased risk of SARS-CoV-2 reinfection associated with emergence of Omicron in South Africa. Sci (New York NY). (2022) 376:eabn4947. doi: 10.1126/science.abn4947
38. Qin C, Zhou L, Hu Z, Zhang S, Yang S, Tao Y, et al. Dysregulation of immune response in patients with coronavirus 2019 (COVID-19) in Wuhan, China. Clin Infect Dis. (2020) 71:762–8. doi: 10.1093/cid/ciaa248
39. Zhang JY, Wang XM, Xing X, Xu Z, Zhang C, Song JW, et al. Single-cell landscape of immunological responses in patients with COVID-19. Nat Immunol. (2020) 21:1107–18. doi: 10.1038/s41590-020-0762-x
40. Huang C, Li X, Wu J, Zhang W, Sun S, Lin L, et al. The landscape and diagnostic potential of T and B cell repertoire in Immunoglobulin A Nephropathy. J Autoimmun. (2019) 97:100–7. doi: 10.1016/j.jaut.2018.10.018
41. CDC COVID-19 Response Team. SARS-CoV-2 B.1.1.529 (Omicron) variant - United States, December 1-8, 2021. MMWR Morbidity mortality weekly Rep. (2021) 70:1731–4. doi: 10.15585/mmwr.mm7050e1
42. Wang C, Sanders CM, Yang Q, Schroeder HW Jr, Wang E, Babrzadeh F, et al. High throughput sequencing reveals a complex pattern of dynamic interrelationships among human T cell subsets. Proc Natl Acad Sci USA. (2010) 107(4):1518–23. doi: 10.1073/pnas.0913939107
Keywords: COVID-19, SARS-CoV-2 Omicron variant, immune repertoire, vaccine, TCR
Citation: Li X, Zhu H, Xu P, Zhang J, Wang Z, He H, Shen F, Jiang Y, Shen L, Xiang J, Yang L, Yang C, Jiang H, Gao G, Jin J, Shen H, Wang Y, Wu L, Qian C, Liu D, Qiu W, Li Q, Chen Y, Lin F and Liu Y (2024) A comprehensive immune repertoire signature distinguishes pulmonary infiltration in SARS-CoV-2 Omicron variant infection. Front. Immunol. 15:1486352. doi: 10.3389/fimmu.2024.1486352
Received: 26 August 2024; Accepted: 27 November 2024;
Published: 17 December 2024.
Edited by:
Yunlong Cao, Peking University, ChinaReviewed by:
Liangzhi Xie, Sinocelltech Ltd., ChinaMarcos Jessé Abrahão Silva, Evandro Chagas Institute, Brazil
Copyright © 2024 Li, Zhu, Xu, Zhang, Wang, He, Shen, Jiang, Shen, Xiang, Yang, Yang, Jiang, Gao, Jin, Shen, Wang, Wu, Qian, Liu, Qiu, Li, Chen, Lin and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yun Liu, bGl1eXVuMDUwNTA2NTBAcmVuamkuY29t; Fujun Lin, bGluZnVqdW5AeGluaHVhbWVkLmNvbS5jbg==
†These authors have contributed equally to this work and share first authorship
 Xuechuan Li1†
Xuechuan Li1† Peipei Xu
Peipei Xu Zhe Wang
Zhe Wang Changlin Qian
Changlin Qian Qiwei Li
Qiwei Li Fujun Lin
Fujun Lin Yun Liu
Yun Liu