- 1Department of Hepatobiliary Surgery, Affiliated Hospital of North Sichuan Medical College, Nanchong, Sichuan, China
- 2Department of Dermatology, Affiliated Hospital of North Sichuan Medical College, Nanchong, Sichuan, China
- 3Department of General Surgery, Dazhou Central Hospital, Dazhou, China
- 4Department of General Surgery (Hepatopancreatobiliary Surgery), The Affiliated Hospital of Southwest Medical University, Luzhou, Sichuan, China
Hepatocellular carcinoma (HCC) is a high-incidence, poor-prognosis malignancy worldwide, requiring new strategies for treatment. Ubiquitination, especially ubiquitination through E3 ubiquitin ligases, plays an indispensable role in the development and progression of HCC. E3 ubiquitin ligases are crucial enzymes in ubiquitination, controlling the degradation of specific substrate proteins and influencing various cellular functions, such as tumor cell proliferation, apoptosis, migration, and immune evasion. In this review, we systematically summarize the mechanisms of E3 ubiquitin ligases in HCC, with a focus on the significance of RING, HECT, and RBR types in HCC progression. The review also looks at the potential for targeting E3 ligases to modulate the tumor microenvironment (TME) and increase immunotherapy efficacy. Future studies will optimize HCC treatment by formulating specific inhibitors or approaches that will be based on gene therapy targeting E3 ligases in order to overcome resistance issues with present treatments and create optimism in the journey of treatment for HCC patients.
1 Introduction
Hepatocellular carcinoma (HCC) is the sixth most commonly diagnosed cancer worldwide and the third leading cause of cancer-related death (1, 2). And since liver cancer is difficult to diagnose early and has less therapeutic effectiveness, making its prognosis dismal (3–5). Indeed, the application of targeted therapies and immunotherapies strongly increases overall survival in some HCC patients, whereas many remain resistant to these therapies, partly due to TME complexity and heterogeneity (6–11). Recent development in other therapies targeting the liver tumor microenvironment likely means we will need to further characterize the liver cancer microenvironment to design new combination therapies that effectively suppress tumorigenesis or restore the sensitivity of immunotherapy-resistant tumors (9, 12–15).
The ubiquitin-proteasome system (UPS) is the major pathway for proteins to be ubiquitinated and degraded in the cell (16). In fact, ubiquitination represents a dynamic and finely regulated class of PTM; it is realized by a three-enzyme cascade reaction that includes Ub-activating enzymes (E1s), Ub-conjugating enzymes (E2s), and Ub-ligases (E3s) (17, 18). The reaction pathway comprises ATP-dependent activation of Ub by E1, transfer to a cysteine residue of E2, and covalent binding to the amino group of a lysine residue of the substrate protein via E3 (19, 20). (Figure 1A1) E3 ubiquitin ligases are particularly important in this process, as they play a pivotal role in the specific recognition and labeling of substrates. Abnormal expression or malfunction of these ligases may cause signaling pathway disruptions, leading to the build-up of misfolded or dysfunctional proteins and incorrect protein complex assembly, ultimately driving the onset and development of HCC (21, 22).
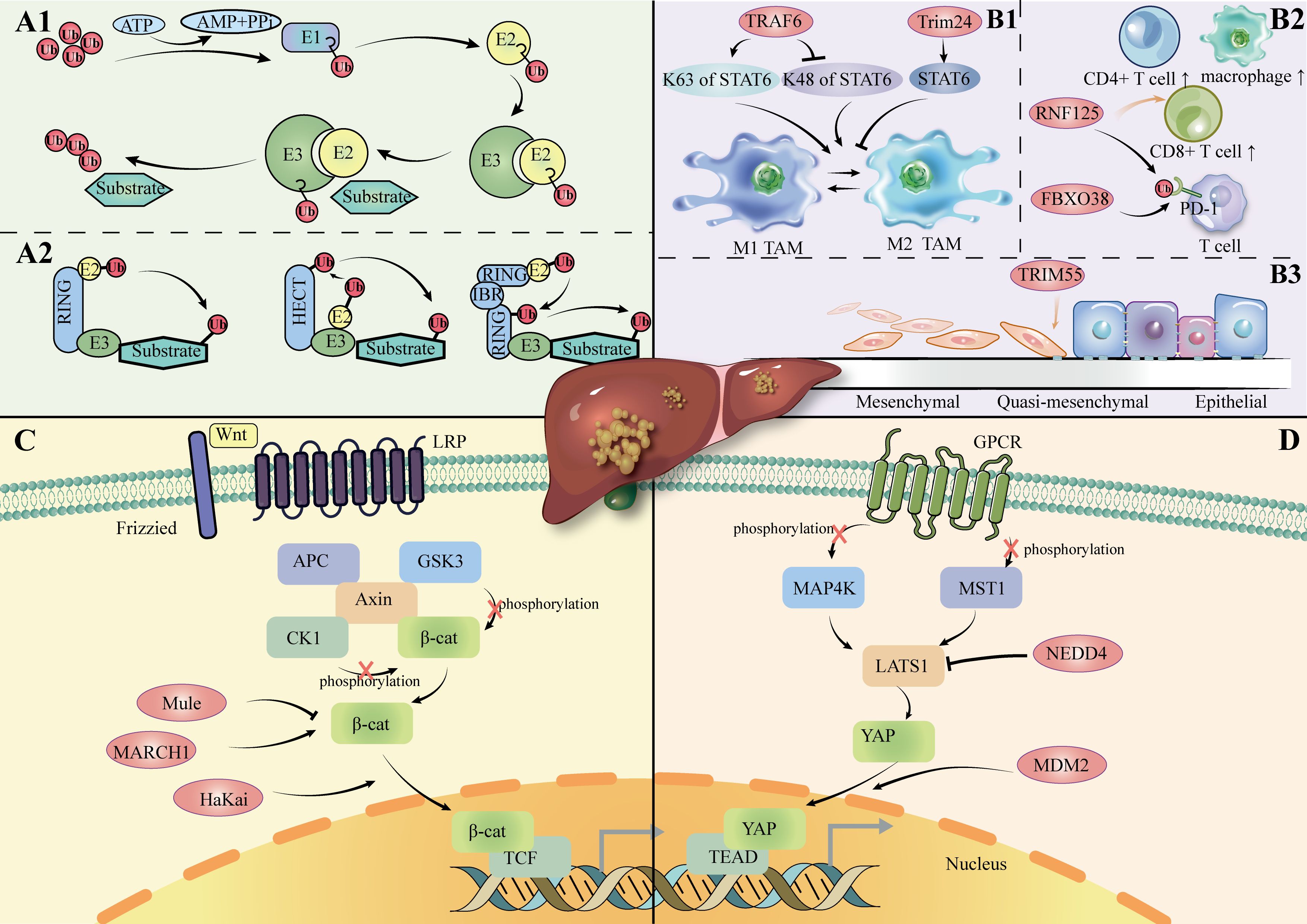
Figure 1. (A1): Ubiquitination process. (A2): Overview of the E3 ligase family. (B1): E3 ligase regulates the polarization of TAM in HCC. (B2): E3 ligase regulates immune cells in HCC. (B3): E3 ligase regulates the EMT process in HCC. (C): Role of E3 ligase in the Wnt/β-catenin channel in HCC. LRP: Low-Density Lipoprotein Receptor-Related; APC: Adenomatous Polyposis Coli; CK1: Casein Kinase 1; GSK3: Glycogen Synthase Kinase 3; TCF: T-cell Factor. (D): Role of E3 ligase in Hippo channels in HCC. MAP4K: Mitogen-Activated Protein Kinase Kinase Kinase Kinase; MST1: Mammalian Sterile 20-like kinase 1; TEAD: TEA Domain Family Member; GPCR: G-Protein-Coupled Receptor; Protein.
E3 ligase also takes a significant role in the TME of HCC (23). TME comprises a diverse array of cellular components, including immune cells, stromal cells, and blood vessels, along with non-cellular elements such as the extracellular matrix and secreted factors (24, 25). E3 ligases have been shown to influence the invasion and metastasis of tumor cells by regulating key proteins in the TME, such as matrix metalloproteinases (MMPs). Thus, targeting E3 ligases not only holds promise in reducing tumor burden but may also open new avenues for enhancing the efficacy of existing therapies and overcoming drug resistance.
This review aims to provide a comprehensive and updated overview of the role of E3 ubiquitin ligases in HCC, with a particular focus on their impact on the TME and immunotherapy. It offers new insights by emphasizing recent findings on how E3 ligases modulate immune cell activity and therapeutic responses within the TME, distinguishing it from previous reviews in this field.
2 Expression and function of E3 ligase in hepatocellular carcinoma
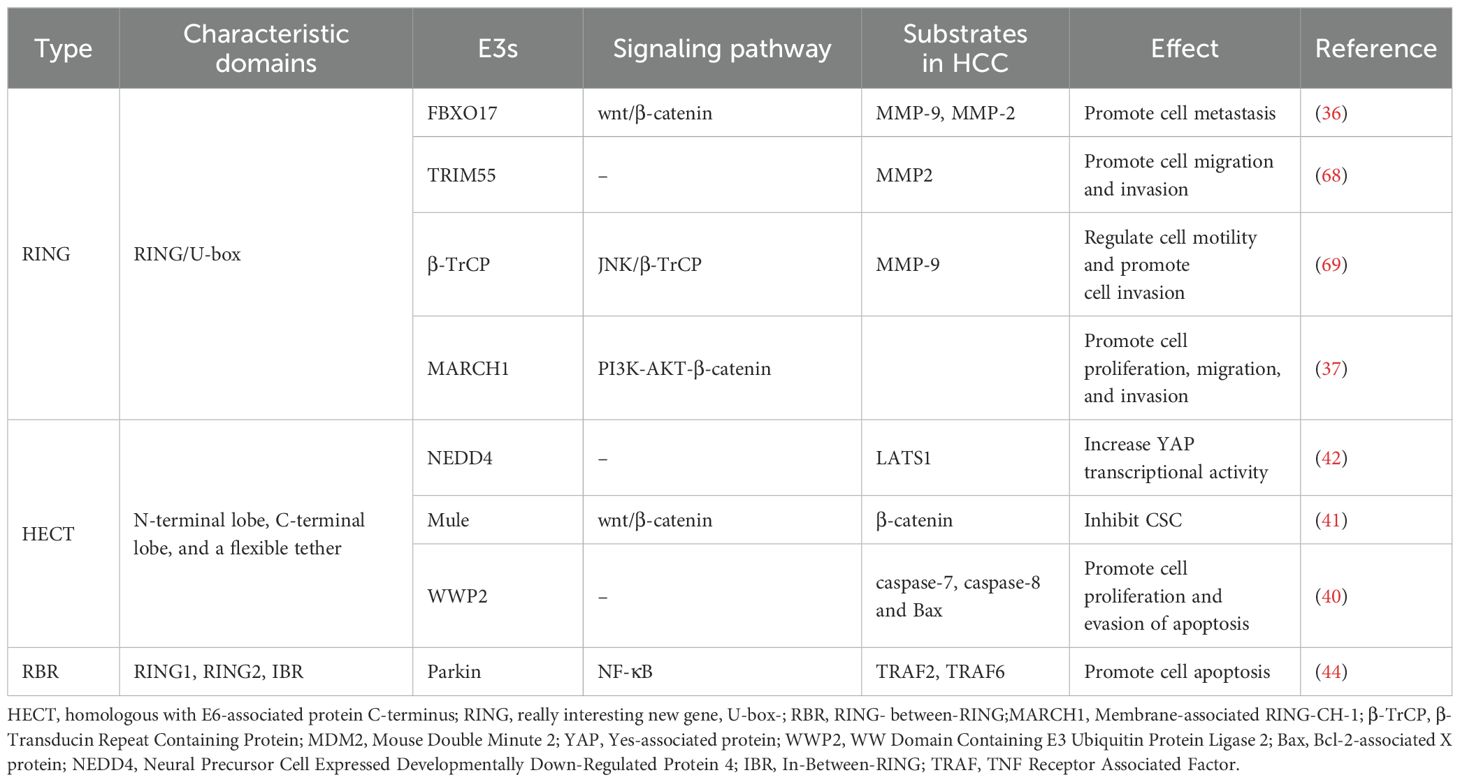
The four identified isoforms of E3 ligase (HECT-, RING-, U-box-, and RBR-type) promotes ubiquitin transfer through different mechanisms (Figure 1A2). The HECT structural domain of HECT E3 ligase transfers ubiquitin to its C-terminal leaflet by binding to the E2 enzyme, first by a process of trans-sulfurylation, followed by further delivery of ubiquitin to the target substrate (26, 27). Upon binding of the ubiquitin-loaded E2 to the RING1 domain of the RBR E3 ligase, ubiquitin is transferred to the RING2 domain through a trans-thioesterification reaction. The RING2 domain then facilitates the transfer of this ubiquitin to the substrate (28). In summary, HECT- and RBR-type E3 ligases transfer ubiquitin (Ub) to substrate proteins after forming a thioester bond between their active site cysteine and Ub. In contrast, RING- and U-box-type E3 ligases directly facilitate the transfer of Ub from E2 enzymes to substrate proteins (29–32).
2.1 RING E3 ligase
The RING-type subgroup represents the primary family of E3 ligases, characterized by two distinct types of RING structural domains: the RING fold structure with a zinc-binding site and a U-box domain. Structures in which both domains can function via monomers, homodimers, heterodimers, or multiple subunits (33, 34). Cullin-RING ligases (CRLs) are a type of multisubunit RING E3 ligases, with F-box proteins serving as an essential part of their structure (35). F-box proteins are categorized into three types: FBXW, FBXL, and FBXO. Studies have shown that the expression of FBXO17 is significantly elevated in the tumor tissues of hepatocellular carcinoma (HCC) patients compared to adjacent normal tissues. FBXO17 may contribute to the malignant progression of HCC by inhibiting the Wnt/β-catenin pathway (36).
Different structures of E3 ligases may be potential tumor promoters in HCC. In vitro and in vivo experiments have demonstrated that monomeric MARCH1 upregulates the PI3K-AKT-β-catenin pathway, thereby promoting the growth and progression of HCC (37). The HaKai heterodimer has been shown to promote the degradation of E-calmodulin, resulting in the nuclear translocation of β-catenin proteins and ultimately driving epithelial-mesenchymal transition (EMT) in HCC (38).However, the homologous structural domain type MDM2 may be a repressor of HCC. MDM2 was shown to diminish YAP’s interaction with other proteins and promote its cytoplasmic translocation and degradation, thereby inhibiting tumorigenesis in HepG2 cells (39).
2.2 HECT E3 ligase
HECTs are second only to human RING E3 ligases in number, and their HECT structural domains consist of an N-terminal lobe, a C-terminal lobe, and a flexible chain (26).Knockdown of WWP2 (HECT-type) significantly elevated the expression levels of apoptosis-related markers in HCC) cell lines, including caspase-7, caspase-8, and Bax, suggesting that inhibition of WWP2 may be a therapeutic tool to negatively regulate HCC overproliferation and escape apoptosis (40). Mule, a member of the HECT E3 ligase family, functions as a tumor suppressor in HCC by inhibiting the Wnt/β-catenin signaling pathway. Specifically, Mule directly targets β-catenin for degradation in HCC, thereby suppressing β-catenin-mediated cancer stem cell (CSC) activity (41).
HECT E3 ligase was shown to mediate the Hippo pathway in HCC cells, including its participation in the Wnt/β-catenin pathway(Figures 1C, D). LATS1 is one of the core components of the Hippo pathway. NEDD4 acts as a direct targeting factor for LATS1, which causes its ubiquitinated degradation and increases the transcriptional activity of YAP. In QGY7703 and SMMC7721 hepatoma cell lines, siRNA-mediated NEDD4 knockdown assays showed that decreased expression of NEDD4 inhibited cell proliferation, invasion, and migration, promoted apoptosis, and further supported the role of the NEDD4-LATS1 pathway in HCC progression (42).
2.3 RBR E3 ligase
The RBR E3 is composed of two RING structural domains (RING1, RING2) and IBR structural domain. Parkin was known to play an oncostatic role in a wide array of tumors including HCC and breast cancer (43). Through direct degradation of TRAF 2 and TRAF6, parkin drives HCC cell apoptosis by inhibition of the NF-κB pathway (44).
3 E3 ligase regulates TME in hepatocellular carcinoma
Given the plasticity of TME and its involvement in the progression of multiple cancers, the modification of TME into an anticancer environment is a promising therapeutic strategy (45–49). Currently, most drugs for TME, such as immunotherapies and antiangiogenic drugs, have limited or unmet efficacy (50, 51).This phenomenon may stem from the complexity of TME and the diversity of its responses to drugs, thus making it difficult to achieve significant clinical results with these therapies in practice. With increasing evidence that ubiquitin signaling cascades modulate immune cell activity and the stability of soluble factors in the TME, a permissive or inhibitory environment for tumor growth can be provided. Moreover, as the first major family of ubiquitinating enzymes, the diversity and specificity of E3 ligases endow them with roles in broadly regulating tumor signaling pathways and biological processes, so making full use of intrinsic E3 ligases to target key mediators seems to be an attractive strategy for anticancer drug development.
3.1 E3 ligase on immune cells
Typically, the immune cells infiltrating the TME CD8+ T cells, CD4+ T helper 1 (Th1), M1 macrophages and NK cells are usually antitumorigenic, whereas the opposite is true for M2 macrophages (52, 53).Regulatory T-cells (Tregs) show these two opposite effects in animal models and clinical trials (54–56) (Figure 1B2).
E3 ligase can regulate the proportion and function of immune cells in the TME by targeting the degradation of tumor suppressors. Analysis of data from The Cancer Genome Atlas (TCGA) public database revealed that RNF125 expression levels are positively correlated with the infiltration of CD4+ and CD8+ T cells, as well as macrophages, within tumors (57).WD repeat 4 (WDR4) has been reported to be a substrate junction for CRL, which can degrade a tumor suppressor, the promyelocytic leukemia (PML) protein.In this process, the expansion of Treg cells, M2 macrophages, and the reduction of CD8+ T cells contribute to the establishment of an immunosuppressive and pro-metastatic TME (58).
In addition, E3 ligases are crucial in immunomodulation by regulating the ubiquitination of key proteins and influencing T cell differentiation. In a study on HCC, In a study on HCC, Jiang et al. found that lncRNA-EGFR binds to EGFR, inhibiting c-CBL-mediated ubiquitination and thus preventing EGFR degradation. This mechanism helps to maintain the continuous activation of the RAS/RAF/MEK/ERK signaling pathway downstream of EGFR, which ultimately promotes the differentiation of Tregs (59).
TME can induce cancer immunosuppression through the upregulation of PD-L1 protein expression. However, E3 ligase plays a role in inhibiting the ubiquitination and degradation of PD-L1, thereby assisting tumor cells in evading T cell-mediated immune surveillance. For example, the RING E3 ligase FBXO38 mediates the ubiquitination of PD-1, thereby regulating antitumor immunity in T cells (60).In hepatocellular carcinoma, RNF125 (RING type) directly ubiquitinates PD-L1 and maintains a stable protein level of PD-L1 (61).
Additionally, E3 ligases are crucial in regulating immune cell differentiation and function. Macrophages, through their M1 and M2 polarization, significantly influence tumor progression and shape the immune environment. Next, we will explore the role of E3 ligases in regulating the polarization of TAMs.
3.2 E3 ligase on TAMs
One of the important processes in which E3 ligases play a role is polarization toward tumor-associated macrophages (TAMs). (Figure 1B1) TAMs are one of the major immune cell types in the tumor microenvironment. When TAMs are exposed to different types of signaling stimuli, they polarize into two contrary functional profiles: differentiation toward M1, with an anti-tumoral effect by response to Th1; differentiation to the M2 type, with pro-tumor effects through Th2 cytokines (62, 63). Specific E3 ligases regulate key signaling pathways such as NF-κB and STAT6 to influence M1 and M2 polarization, further influencing the immune response in the tumor microenvironment.
E3 ligase is one of the mediators that regulate ubiquitination in macrophage polarization. STAT6, for instance, is one of the main transcription factors that drive M2 macrophage polarization (64). TRAF6 is an E3 ligase that is the main activator of K63-linked ubiquitination of STAT6 in M2-polarized macrophages stimulated with IL-4. In contrast, it inhibits the degradation (65).
Under hypoxic conditions, Seven in Absentia homologue 2 (SIAH2), an E3 ligase with a RING domain, is the regulator of proteasome degradation of NRF1 (Nuclear Respiratory Factor 1) and, therefore, switches TAM polarization to the tumor-promoting M2 state in breast cancer (66). The underpinning mechanisms of the SIAH2-NRF1 axis are linked to changes in mitochondria-dependent metabolic reprogramming, with an increase in lactate. Another RING E3 ligase, TRIM24, degrades the histone acetyltransferase CBP that acetylates STAT6, which inhibits TAM polarization to M2 (67).
4 E3 ubiquitin ligases target MMPs in TME
4.1 Role of MMPs in TME of hepatocellular carcinoma
MMP is a zinc-dependent endopeptidase and multifunctional enzyme that can be secreted by TAM (68, 69). MMPs are categorized into several groups: collagenase, gelatinase, stromelysin, membrane MMPs, and other unclassified MMPs (70). MMPs have the ability to degrade almost all components of the ECM, leading to structural changes in the cellular and tissue environments. TME is composed of various cellular constituents, along with the biochemical and biophysical elements of ECM, and is defined by their intricate interactions within and surrounding solid tumor masses (71, 72). In TME, MMPs play a crucial role. When MMPs are dysfunctional, they lead to the destruction of the ECM, which promotes cell migration and tumor metastasis (73–75). In tumor stem cells of HCC, MMP remodel the ECM, resulting in tumors that exhibit more aggressive and functional stemness (76). Recent studies have demonstrated that MMP9, secreted by TAMs, is particularly involved in ECM degradation, facilitating tumor invasion and metastasis in HCC. Inhibiting MMP9 activity in TAMs has been shown to reduce ECM breakdown and, consequently, limit the metastatic potential of HCC cells (77).
4.2 Mechanism of regulating the microenvironment of hepatocellular carcinoma by targeting MMPs via E3 ligase
Protein expression of MMP can be controlled by E3 ubiquitin ligases (78, 79). (Table 1) For example, TRIM55 is associated with a decrease in MMP2 (80). TRIM66 reduces MMP9 expression (81). EMT is a process through which epithelial cells acquire mesenchymal traits, facilitating cancer invasion and metastasis (82). Among them, MMP2 is the major MMP in the pathogenesis of EMT in hepatocellular carcinoma (73). It has been demonstrated that overexpression of TRIM55 (RING-type) effectively reduced the migration and invasion ability of HCC cells by modulating epithelial-mesenchymal transition and inhibiting the activity of MMP2 (80). This suggests that E3 ligase can influence the hepatocellular carcinoma microenvironment by affecting MMP protein expression and EMT (epithelial-mesenchymal transition). (Figure 1B3).
5 Potential of E3 ligase as a therapeutic target in hepatocellular carcinoma
A growing body of evidence indicates that abnormal ubiquitination expression is correlated with poor cancer prognosis. Given the critical role of various E3 ligases in the tumorigenesis of HCC, targeting E3 ligase activity is considered a promising therapeutic strategy for cancer treatment. p53 is one of the most important tumor suppressors in vivo, and MDM2 regulates the level of P53. Antagonizing MDM2 seems to be an effective strategy to develop promoter inhibitors for HCC tumors, but further clinical trials are still needed (83–85). For example, in a mouse model, the MDM2 inhibitor APG-115 induced synergistic activity with anti-PD-1 antibody-based immunotherapy (86). Recent studies have shown that Fbxw7 increases the sensitivity of HCC cells to sorafenib (87, 88). This finding implies a potential clinical application of Fbxw7 in enhancing sorafenib efficacy in liver cancer treatment. Multiple compounds can target MMPs via E3 ligase for cancer therapy. Zhang et al. found that the natural agent ALCA upregulated NEDD4L (HECT-type) and caused ubiquitination of β-catenin, which activated Wnt-induced transcription of the MMP9 gene in lung adenocarcinoma cells (89). Gallic acid reduces MMP2 and MMP9 protein levels by inducing β-TrCP in human leukemia cells (90). Notably, MMP9 is the major MMP in the pathogenesis of EMT in hepatocellular carcinoma (73). Therefore, utilizing compounds to regulate the expression of E3 ligases and modulate MMP levels could represent a promising strategy for the treatment of HCC.
6 Conclusion
Hepatocellular carcinoma is heterogeneous at the genetic and epigenetic levels, making the development of therapeutic agents for liver cancer difficult (91, 92). Ubiquitination, a crucial post-translational modification of proteins, has been increasingly recognized on a broader scale. In this intricate environment, E3 ligases, beyond targeting substrates for proteasomal degradation, also regulate various signaling pathways such as PI3K/AKT and Wnt/β-catenin. Moreover, most E3 ligases in HCC are oncoproteins (93, 94). So E3 ligase is an attractive drug target for cancer therapy.
The role and importance of E3 ligases in hepatocellular carcinoma have been widely explored, though numerous questions still persist. As E3 ubiquitin ligases are frequently mutated, their targeting specificity may be insufficient, which leads to less accurate recognition of the target and may trigger off-target effects, ultimately leading to poor therapeutic efficacy. Recent advancements in technologies like CRISPR and PROTAC (Proteolysis Targeting Chimeras) have opened new avenues for more precise targeting of E3 ligases in cancer treatment. In particular, CRISPR can be used to knock out or activate specific genes related to E3 ligases, thereby providing a strategy to mitigate their oncogenic effects in the TME (95, 96). On the other hand, PROTACs (Proteolysis Targeting Chimeras), which are also small molecule inhibitors, degrade POIs (Cullin-RING type) in a substoichiometric manner, leading to more prolonged and potent biological effects on the target compared to SMIs. In addition, PROTAC dBET1 inhibits the pro-inflammatory response by regulating MMP9 in lipopolysaccharide (LPS)-activated microglial cells (97). As a result, PROTACs have emerged as a promising approach for developing new targeted anticancer therapies.
Interestingly, similar to the process of ubiquitination, SUMization (Small Ubiquitin-like Modifier) plays an important role in most organisms, regulating a variety of cellular processes, including DNA replication, transcription, immune response (98, 99). An increasing body of research indicates a strong association between SUMOylation and the progression of hepatocellular carcinoma (100). SUMO E3 ligase may also be a potential target for the treatment of hepatocellular carcinoma.
Author contributions
HW: Formal analysis, Writing – original draft, Writing – review & editing. QL: Visualization, Writing – original draft, Writing – review & editing. QT: Data curation, Writing – original draft, Writing – review & editing. GS: Writing – original draft. GW: Writing – original draft. XM: Writing – original draft. CW: Writing – original draft. LZ: Writing – original draft. JL: Conceptualization, Writing – review & editing, Writing – original draft. JDL: Conceptualization, Writing – review & editing, Writing – original draft. BL: Conceptualization, Writing – review & editing, Writing – original draft.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by grants from the Science and Technology Project of Sichuan Province (NO.2024YFHZ0052).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Fares S, Wehrle CJ, Hong H, Sun K, Jiao C, Zhang M, et al. Emerging and clinically accepted biomarkers for hepatocellular carcinoma. Cancers (Basel). (2024) 16:1453. doi: 10.3390/cancers16081453
2. Sim YK, Chong MC, Gandhi M, Pokharkar YM, Zhu Y, Shi L, et al. Real-world data on the diagnosis, treatment, and management of hepatocellular carcinoma in the asia-pacific: the INSIGHT study. Liver Cancer. (2024) 13:298–313. doi: 10.1159/000534513
3. Befeler AS, Di Bisceglie AM. Hepatocellular carcinoma: diagnosis and treatment. Gastroenterology. (2002) 122:1609–19. doi: 10.1053/gast.2002.33411
4. Yang L, He Y, Zhang Z, Wang W. Upregulation of CEP55 predicts dismal prognosis in patients with liver cancer. BioMed Res Int. (2020) 2020:4139320. doi: 10.1155/2020/4139320
5. Wei H, Tang Q, Wang A, Zhang Y, Qin Z, Li W, et al. lncRNA MAGI2-AS3 Exerts Antioncogenic Roles in Hepatocellular Carcinoma via Regulating the miR-519c-3p/TXNIP Axis. J Oncol. (2021) 2021:5547345. doi: 10.1155/2021/5547345
6. Pu Z, Ren Z, Xu Q, Wang X, Chen J, Chen J. Editorial: The role of tumor microenvironment in primary liver cancer therapeutic resistance. Front Oncol. (2022) 12:938557. doi: 10.3389/fonc.2022.938557
7. Deng ZJ, Li L, Teng YX, Zhang YQ, Zhang YX, Liu HT, et al. Treatments of hepatocellular carcinoma with portal vein tumor thrombus: current status and controversy. J Clin Transl Hepatol. (2022) 10:147–58. doi: 10.14218/JCTH.2021.00179
8. Li Q, Dong Y, Pan Y, Tang H, Li D. Case report: clinical responses to tislelizumab as a first-line therapy for primary hepatocellular carcinoma with B-cell indolent lymphoma. Front Immunol. (2021) 12:634559. doi: 10.3389/fimmu.2021.634559
9. Chan T, Wiltrout RH, Weiss JM. Immunotherapeutic modulation of the suppressive liver and tumor microenvironments. Int Immunopharmacol. (2011) 11:879–89. doi: 10.1016/j.intimp.2010.12.024
10. Tang J, Zhang S, Jiang L, Liu J, Xu J, Jiang C, et al. Causal relationship between immune cells and hepatocellular carcinoma: a Mendelian randomisation study. J Cancer. (2024) 15:4219–31. doi: 10.7150/jca.96744
11. Su K, Wang F, Li X, Chi H, Zhang J, He K, et al. Effect of external beam radiation therapy versus transcatheter arterial chemoembolization for non-diffuse hepatocellular carcinoma (≥ 5 cm): a multicenter experience over a ten-year period. Front Immunol. (2023) 14:1265959. doi: 10.3389/fimmu.2023.1265959
12. Zhang J, Huang L, Ge G, Hu K. Emerging epigenetic-based nanotechnology for cancer therapy: modulating the tumor microenvironment. Adv Sci (Weinh). (2023) 10:e2206169. doi: 10.1002/advs.202206169
13. Donne R, Lujambio A. The liver cancer immune microenvironment: Therapeutic implications for hepatocellular carcinoma. Hepatology. (2023) 77:1773–96. doi: 10.1002/hep.32740
14. Chen X, Chi H, Zhao X, Pan R, Wei Y, Han Y. Role of exosomes in immune microenvironment of hepatocellular carcinoma. J Oncol. (2022) 2022:2521025. doi: 10.1155/2022/2521025
15. Su L, Luo H, Yan Y, Yang Z, Lu J, Xu D, et al. Exploiting gender-based biomarkers and drug targets: advancing personalized therapeutic strategies in hepatocellular carcinoma. Front Pharmacol. (2024) 15:1433540. doi: 10.3389/fphar.2024.1433540
16. Farinati F, Sergio A, Baldan A, Giacomin A, Di Nolfo MA, Del Poggio P, et al. Early and very early hepatocellular carcinoma: when and how much do staging and choice of treatment really matter? A multi-center study. BMC Cancer. (2009) 9:33. doi: 10.1186/1471-2407-9-33
17. Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. (2015) 136:E359–86. doi: 10.1002/ijc.v136.5
18. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2018) 68:394–424. doi: 10.3322/caac.21492
19. Hochstrasser M. Ubiquitin, proteasomes, and the regulation of intracellular protein degradation. Curr Opin Cell Biol. (1995) 7:215–23. doi: 10.1016/0955-0674(95)80031-X
20. Sun T, Liu Z, Yang Q. The role of ubiquitination and deubiquitination in cancer metabolism. Mol Cancer. (2020) 19:146. doi: 10.1186/s12943-020-01262-x
21. Hoeller D, Dikic I. Targeting the ubiquitin system in cancer therapy. Nature. (2009) 458:438–44. doi: 10.1038/nature07960
22. Wang W, Yang LY, Yang ZL, Peng JX, Yang JQ. Elevated expression of autocrine motility factor receptor correlates with overexpression of RhoC and indicates poor prognosis in hepatocellular carcinoma. Dig Dis Sci. (2007) 52:770–5. doi: 10.1007/s10620-006-9479-4
23. Deng L, Meng T, Chen L, Wei W, Wang P. The role of ubiquitination in tumorigenesis and targeted drug discovery. Signal Transduct Target Ther. (2020) 5:11. doi: 10.1038/s41392-020-0107-0
24. Zhou H, Wang M, Zhang Y, Su Q, Xie Z, Chen X, et al. Functions and clinical significance of mechanical tumor microenvironment: cancer cell sensing, mechanobiology and metastasis. Cancer Commun (Lond). (2022) 42:374–400. doi: 10.1002/cac2.12294
25. Choi Y, Jung K. Normalization of the tumor microenvironment by harnessing vascular and immune modulation to achieve enhanced cancer therapy. Exp Mol Med. (2023) 55:2308–19. doi: 10.1038/s12276-023-01114-w
26. Goto J, Otaki Y, Watanabe T, Watanabe M. The role of HECT-type E3 ligase in the development of cardiac disease. Int J Mol Sci. (2021) 22:6065. doi: 10.3390/ijms22116065
27. Sosič I, Bricelj A, Steinebach C. E3 ligase ligand chemistries: from building blocks to protein degraders. Chem Soc Rev. (2022) 51:3487–534. doi: 10.1039/D2CS00148A
28. Lorenz S. Structural mechanisms of HECT-type ubiquitin ligases. Biol Chem. (2018) 399:127–45. doi: 10.1515/hsz-2017-0184
29. Jevtić P, Haakonsen DL, Rapé M. An E3 ligase guide to the galaxy of small-molecule-induced protein degradation. Cell Chem Biol. (2021) 28:1000–13. doi: 10.1016/j.chembiol.2021.04.002
30. Ege N, Bouguenina H, Tatari M, Chopra R. Phenotypic screening with target identification and validation in the discovery and development of E3 ligase modulators. Cell Chem Biol. (2021) 28:283–99. doi: 10.1016/j.chembiol.2021.02.011
31. Ramachandran S, Ciulli A. Building ubiquitination machineries: E3 ligase multi-subunit assembly and substrate targeting by PROTACs and molecular glues. Curr Opin Struct Biol. (2021) 67:110–9. doi: 10.1016/j.sbi.2020.10.009
32. Toma-Fukai S, Shimizu T. Structural diversity of ubiquitin E3 ligase. Molecules. (2021) 26:6682. doi: 10.3390/molecules26216682
33. Berndsen CE, Wolberger C. New insights into ubiquitin E3 ligase mechanism. Nat Struct Mol Biol. (2014) 21:301–7. doi: 10.1038/nsmb.2780
34. Leslie PL, Ke H, Zhang Y. The MDM2 RING domain and central acidic domain play distinct roles in MDM2 protein homodimerization and MDM2-MDMX protein heterodimerization. J Biol Chem. (2015) 290:12941–50. doi: 10.1074/jbc.M115.644435
35. Podieh F, Hordijk PL. Regulation of rho GTPases in the vasculature by cullin3-based E3 ligase complexes. Front Cell Dev Biol. (2021) 9:680901. doi: 10.3389/fcell.2021.680901
36. Liu FH, Cui YP, He YK, Shu RH. FBXO17 promotes Malignant progression of hepatocellular carcinoma by activating wnt/β-catenin pathway. Eur Rev Med Pharmacol Sci. (2019) 23:8265–73. doi: 10.26355/eurrev_201910_19137
37. Xie L, Dai H, Li M, Yang W, Yu G, Wang X, et al. MARCH1 encourages tumour progression of hepatocellular carcinoma via regulation of PI3K-AKT-β-catenin pathways. J Cell Mol Med. (2019) 23:3386–401. doi: 10.1111/jcmm.2019.23.issue-5
38. Lu M, Wu J, Hao ZW, Shang YK, Xu J, Nan G, et al. Basolateral CD147 induces hepatocyte polarity loss by E-cadherin ubiquitination and degradation in hepatocellular carcinoma progress. Hepatology. (2018) 68:317–32. doi: 10.1002/hep.29798
39. Jia M, Xiong Y, Li M, Mao Q. Corosolic acid inhibits cancer progress through inactivating YAP in hepatocellular carcinoma. Oncol Res. (2020) 28:371–83. doi: 10.3727/096504020X15853075736554
40. Xu SQ, Qin Y, Pan DB, Ye GX, Wu CJ, Wang S, et al. Inhibition of WWP2 suppresses proliferation, and induces G1 cell cycle arrest and apoptosis in liver cancer cells. Mol Med Rep. (2016) 13:2261–6. doi: 10.3892/mmr.2016.4771
41. Ho NPY, Leung CON, Wong TL, Lau EYT, Lei MML, Mok EHK, et al. The interplay of UBE2T and Mule in regulating Wnt/β-catenin activation to promote hepatocellular carcinoma progression. Cell Death Dis. (2021) 12:148. doi: 10.1038/s41419-021-03403-6
42. Zheng H, Ke X, Li D, Wang Q, Wang J, Liu X, et al. NEDD4 promotes cell growth and motility in hepatocellular carcinoma. Cell Cycle. (2018) 17(6):728–38. doi: 10.1080/15384101.2018.1440879
43. Perwez A, Wahabi K, Rizvi MA. Parkin: A targetable linchpin in human Malignancies. Biochim Biophys Acta Rev Cancer. (2021) 1876:188533. doi: 10.1016/j.bbcan.2021.188533
44. Zhang X, Lin C, Song J, Chen H, Chen X, Ren L, et al. Parkin facilitates proteasome inhibitor-induced apoptosis via suppression of NF-κB activity in hepatocellular carcinoma. Cell Death Dis. (2019) 10:719. doi: 10.1038/s41419-019-1881-x
45. Jin MZ, Jin WL. The updated landscape of tumor microenvironment and drug repurposing. Signal Transduct Target Ther. (2020) 5:166. doi: 10.1038/s41392-020-00280-x
46. Liu F, Qin L, Liao Z, Song J, Yuan C, Liu Y, et al. Microenvironment characterization and multi-omics signatures related to prognosis and immunotherapy response of hepatocellular carcinoma. Exp Hematol Oncol. (2020) 9:10. doi: 10.1186/s40164-020-00165-3
47. Roma-Rodrigues C, Mendes R, Baptista PV, Fernandes AR. Targeting tumor microenvironment for cancer therapy. Int J Mol Sci. (2019) 20:840. doi: 10.3390/ijms20040840
48. Zhang S, Jiang C, Jiang L, Chen H, Huang J, Gao X, et al. Construction of a diagnostic model for hepatitis B-related hepatocellular carcinoma using machine learning and artificial neural networks and revealing the correlation by immunoassay. Tumour Virus Res. (2023) 16:200271. doi: 10.1016/j.tvr.2023.200271
49. Zhang S, Jiang C, Jiang L, Chen H, Huang J, Zhang J, et al. Uncovering the immune microenvironment and molecular subtypes of hepatitis B-related liver cirrhosis and developing stable a diagnostic differential model by machine learning and artificial neural networks. Front Mol Biosci. (2023) 10:1275897. doi: 10.3389/fmolb.2023.1275897
50. Haslam A, Prasad V. Estimation of the percentage of US patients with cancer who are eligible for and respond to checkpoint inhibitor immunotherapy drugs. JAMA Netw Open. (2019) 2:e192535. doi: 10.1001/jamanetworkopen.2019.2535
51. Xiao N, Zhu X, Li K, Chen Y, Liu X, Xu B, et al. Blocking siglec-10(hi) tumor-associated macrophages improves anti-tumor immunity and enhances immunotherapy for hepatocellular carcinoma. Exp Hematol Oncol. (2021) 10:36. doi: 10.1186/s40164-021-00230-5
52. Bhome R, Al Saihati HA, Goh RW, Bullock MD, Primrose JN, Thomas GJ, et al. Translational aspects in targeting the stromal tumour microenvironment: from bench to bedside. New Horiz Transl Med. (2016) 3:9–21. doi: 10.1016/j.nhtm.2016.03.001
53. Shen K, Liu T. Comprehensive analysis of the prognostic value and immune function of immune checkpoints in stomach adenocarcinoma. Int J Gen Med. (2021) 14:5807–24. doi: 10.2147/IJGM.S325467
54. Huang J, Zhang L, Wan D, Zhou L, Zheng S, Lin S, et al. Extracellular matrix and its therapeutic potential for cancer treatment. Signal Transduct Target Ther. (2021) 6:153. doi: 10.1038/s41392-021-00544-0
55. Sheth V, Wang L, Bhattacharya R, Mukherjee P, Wilhelm S. Strategies for delivering nanoparticles across tumor blood vessels. Adv Funct Mater. (2021) 31:2007363. doi: 10.1002/adfm.202007363
56. Chi H, Zhao S, Yang J, Gao X, Peng G, Zhang J, et al. T-cell exhaustion signatures characterize the immune landscape and predict HCC prognosis via integrating single-cell RNA-seq and bulk RNA-sequencing. Front Immunol. (2023) 14:1137025. doi: 10.3389/fimmu.2023.1137025
57. Ren X, Wang L, Liu L, Liu J. PTMs of PD-1/PD-L1 and PROTACs application for improving cancer immunotherapy. Front Immunol. (2024) 15:1392546. doi: 10.3389/fimmu.2024.1392546
58. Wang YT, Chen RH. PML degradation fosters an immunosuppressive and pro-metastatic tumor microenvironment. Mol Cell Oncol. (2017) 4:e1364212. doi: 10.1080/23723556.2017.1364212
59. Jiang R, Tang J, Chen Y, Deng L, Ji J, Xie Y, et al. The long noncoding RNA lnc-EGFR stimulates T-regulatory cells differentiation thus promoting hepatocellular carcinoma immune evasion. Nat Commun. (2017) 8:15129. doi: 10.1038/ncomms15129
60. Meng X, Liu X, Guo X, Jiang S, Chen T, Hu Z, et al. FBXO38 mediates PD-1 ubiquitination and regulates anti-tumour immunity of T cells. Nature. (2018) 564:130–5. doi: 10.1038/s41586-018-0756-0
61. Wei M, Mo Y, Liu J, Zhai J, Li H, Xu Y, et al. Ubiquitin ligase RNF125 targets PD-L1 for ubiquitination and degradation. Front Oncol. (2022) 12:835603. doi: 10.3389/fonc.2022.835603
62. Zhang W, Wang M, Ji C, Liu X, Gu B, Dong T. Macrophage polarization in the tumor microenvironment: Emerging roles and therapeutic potentials. BioMed Pharmacother. (2024) 177:116930. doi: 10.1016/j.biopha.2024.116930
63. Recalcati S, Gammella E, Cairo G. Ironing out macrophage immunometabolism. Pharm (Basel). (2019) 12:94. doi: 10.3390/ph12020094
64. Lawrence T, Natoli G. Transcriptional regulation of macrophage polarization: enabling diversity with identity. Nat Rev Immunol. (2011) 11:750–61. doi: 10.1038/nri3088
65. Zhou C, Lu C, Pu H, Li D, Zhang L. TRAF6 promotes IL-4-induced M2 macrophage activation by stabilizing STAT6. Mol Immunol. (2020) 127:223–9. doi: 10.1016/j.molimm.2020.09.001
66. Ma B, Cheng H, Mu C, Geng G, Zhao T, Luo Q, et al. The SIAH2-NRF1 axis spatially regulates tumor microenvironment remodeling for tumor progression. Nat Commun. (2019) 10:1034. doi: 10.1038/s41467-019-08618-y
67. Yu T, Gan S, Zhu Q, Dai D, Li N, Wang H, et al. Modulation of M2 macrophage polarization by the crosstalk between Stat6 and Trim24. Nat Commun. (2019) 10:4353. doi: 10.1038/s41467-019-12384-2
68. Mondal S, Adhikari N, Banerjee S, Amin SA, Jha T. Corrigendum to “Matrix metalloproteinase-9 (MMP-9) and its inhibitors in cancer: A minireview” Eur. J. Med. Chem. 194 (2020) 112260. Eur J Med Chem. (2020) 205:112642. doi: 10.1016/j.ejmech.2020.112260
69. Chanmee T, Ontong P, Konno K, Itano N. Tumor-associated macrophages as major players in the tumor microenvironment. Cancers (Basel). (2014) 6:1670–90. doi: 10.3390/cancers6031670
70. Raeeszadeh-Sarmazdeh M, Do LD, Hritz BG. Metalloproteinases and their inhibitors: potential for the development of new therapeutics. Cells. (2020) 9:9739. doi: 10.3390/cells9051313
71. Popov A, Kozlovskaya E, Rutckova T, Styshova O, Vakhrushev A, Kupera E, et al. Antitumor properties of matrikines of different origins: prospects and problems of their application. Int J Mol Sci. (2023) 24:9502. doi: 10.3390/ijms24119502
72. Baghy K, Ladányi A, Reszegi A, Kovalszky I. Insights into the tumor microenvironment-components, functions and therapeutics. Int J Mol Sci. (2023) 24:17536. doi: 10.3390/ijms242417536
73. Scheau C, Badarau IA, Costache R, Caruntu C, Mihai GL, Didilescu AC, et al. The role of matrix metalloproteinases in the epithelial-mesenchymal transition of hepatocellular carcinoma. Anal Cell Pathol (Amst). (2019) 2019:9423907. doi: 10.1155/2019/9423907
74. Huang CY, Yang JL, Chen JJ, Tai SB, Yeh YH, Liu PF, et al. Fluoroquinolones suppress TGF-β and PMA-induced MMP-9 production in cancer cells: implications in repurposing quinolone antibiotics for cancer treatment. Int J Mol Sci. (2021) 22:11602. doi: 10.3390/ijms222111602
75. Zhang X, Xiao Z, Zhang X, Li N, Sun T, Zhang J, et al. Signature construction and molecular subtype identification based on liver-specific genes for prediction of prognosis, immune activity, and anti-cancer drug sensitivity in hepatocellular carcinoma. Cancer Cell Int. (2024) 24:78. doi: 10.1186/s12935-024-03242-3
76. Ng KY, Shea QT, Wong TL, Luk ST, Tong M, Lo CM, et al. Chemotherapy-enriched THBS2-deficient cancer stem cells drive hepatocarcinogenesis through matrix softness induced histone H3 modifications. Adv Sci (Weinh). (2021) 8:2002483. doi: 10.1002/advs.202002483
77. Frión-Herrera Y, Gabbia D, Cuesta-Rubio O, De Martin S, Carrara M. Nemorosone inhibits the proliferation and migration of hepatocellular carcinoma cells. Life Sci. (2019) 235:116817. doi: 10.1016/j.lfs.2019.116817
78. Yamada S, Yanamoto S, Naruse T, Matsushita Y, Takahashi H, Umeda M, et al. Skp2 regulates the expression of MMP-2 and MMP-9, and enhances the invasion potential of oral squamous cell carcinoma. Pathol Oncol Res. (2016) 22:625–32. doi: 10.1007/s12253-016-0049-6
79. Guo F, Liu J, Han X, Zhang X, Lin T, Wang Y, et al. Erratum: FBXO22 suppresses metastasis in human renal cell carcinoma via inhibiting MMP-9-mediated migration and invasion and VEGF-mediated angiogenesis. Int J Biol Sci. (2022) 18:1052. doi: 10.7150/ijbs.69330
80. Li X, Huang L, Gao W. Overexpression of tripartite motif conaining 55 (TRIM55) inhibits migration and invasion of hepatocellular carcinoma (HCC) cells via epithelial-mesenchymal transition and matrix metalloproteinase-2 (MMP2). Med Sci Monit. (2019) 25:771–7. doi: 10.12659/MSM.910984
81. Zhang HG, Pan YW, Feng J, Zeng CT, Zhao XQ, Liang B, et al. TRIM66 promotes Malignant progression of hepatocellular carcinoma by inhibiting E-cadherin expression through the EMT pathway. Eur Rev Med Pharmacol Sci. (2019) 23:2003–12. doi: 10.26355/eurrev_201903_17239
82. Pastushenko I, Blanpain C, Transition States during Tumor Progression EMT. and metastasis. Trends Cell Biol. (2019) 29:212–26. doi: 10.1016/j.tcb.2018.12.001
83. Cheok CF, Verma CS, Baselga J, Lane DP. Translating p53 into the clinic. Nat Rev Clin Oncol. (2011) 8:25–37. doi: 10.1038/nrclinonc.2010.174
84. Tuval A, Strandgren C, Heldin A, Palomar-Siles M, Wiman KG. Pharmacological reactivation of p53 in the era of precision anticancer medicine. Nat Rev Clin Oncol. (2024) 21:106–20. doi: 10.1038/s41571-023-00842-2
85. Zhu H, Gao H, Ji Y, Zhou Q, Du Z, Tian L, et al. Targeting p53-MDM2 interaction by small-molecule inhibitors: learning from MDM2 inhibitors in clinical trials. J Hematol Oncol. (2022) 15:91. doi: 10.1186/s13045-022-01314-3
86. Fang DD, Tang Q, Kong Y, Wang Q, Gu J, Fang X, et al. MDM2 inhibitor APG-115 synergizes with PD-1 blockade through enhancing antitumor immunity in the tumor microenvironment. J Immunother Cancer. (2019) 7:327. doi: 10.1186/s40425-019-0750-6
87. Tang X, Yang W, Shu Z, Shen X, Zhang W, Cen C, et al. MicroRNA−223 promotes hepatocellular carcinoma cell resistance to sorafenib by targeting FBW7. Oncol Rep. (2019) 41:1231–7. doi: 10.3892/or.2018.6908
88. Zhang Z, Guo M, Li Y, Shen M, Kong D, Shao J, et al. RNA-binding protein ZFP36/TTP protects against ferroptosis by regulating autophagy signaling pathway in hepatic stellate cells. Autophagy. (2020) 16:1482–505. doi: 10.1080/15548627.2019.1687985
89. Zhang W, Zhang R, Zeng Y, Li Y, Chen Y, Zhou J, et al. ALCAP2 inhibits lung adenocarcinoma cell proliferation, migration and invasion via the ubiquitination of β-catenin by upregulating the E3 ligase NEDD4L. Cell Death Dis. (2021) 12:755. doi: 10.1038/s41419-021-04043-6
90. Chen YJ, Chang LS. Gallic acid downregulates matrix metalloproteinase-2 (MMP-2) and MMP-9 in human leukemia cells with expressed Bcr/Abl. Mol Nutr Food Res. (2012) 56:1398–412. doi: 10.1002/mnfr.201200167
91. Li H, Wu Z, Chen J, Su K, Guo L, Xu K, et al. External radiotherapy combined with sorafenib has better efficacy in unresectable hepatocellular carcinoma: a systematic review and meta-analysis. Clin Exp Med. (2023) 23:1537–49. doi: 10.1007/s10238-022-00972-4
92. Su K, Liu Y, Wang P, He K, Wang F, Chi H, et al. Heat-shock protein 90α is a potential prognostic and predictive biomarker in hepatocellular carcinoma: a large-scale and multicenter study. Hepatol Int. (2022) 16:1208–19. doi: 10.1007/s12072-022-10391-y
93. Barik GK, Sahay O, Islam S, Ghate NB, Kalita B, Alam A. Ubiquitination in cancer metastasis: emerging functions, underlying mechanisms, and clinical implications. Technol Cancer Res Treat. (2023) 22:15330338231210720. doi: 10.1177/15330338231210720
94. Dong R, Wang Z, Cao D, Li Y, Fei Y, Gao P, et al. The ‘Other’ subfamily of HECT E3 ubiquitin ligases evaluate the tumour immune microenvironment and prognosis in patients with hepatocellular carcinoma. IET Syst Biol. (2024) 18:23–39. doi: 10.1049/syb2.12086
95. Zeng Z, Gu SS, Wong CJ, Yang L, Ouardaoui N, Li D, et al. Machine learning on syngeneic mouse tumor profiles to model clinical immunotherapy response. Sci Adv. (2022) 8:eabm8564. doi: 10.1126/sciadv.abm8564
96. Su K, Shen Q, Tong J, Gu T, Xu K, Li H, et al. Construction and validation of a nomogram for HBV-related hepatocellular carcinoma: A large, multicenter study. Ann Hepatol. (2023) 28:101109. doi: 10.1016/j.aohep.2023.101109
97. DeMars KM, Yang C, Castro-Rivera CI, Candelario-Jalil E. Selective degradation of BET proteins with dBET1, a proteolysis-targeting chimera, potently reduces pro-inflammatory responses in lipopolysaccharide-activated microglia. Biochem Biophys Res Commun. (2018) 497:410–5. doi: 10.1016/j.bbrc.2018.02.096
98. Eifler K, Vertegaal ACO. SUMOylation-mediated regulation of cell cycle progression and cancer. Trends Biochem Sci. (2015) 40:779–93. doi: 10.1016/j.tibs.2015.09.006
99. Flotho A, Melchior F. Sumoylation: a regulatory protein modification in health and disease. Annu Rev Biochem. (2013) 82:357–85. doi: 10.1146/annurev-biochem-061909-093311
Keywords: HCC, ubiquitination, E3 ubiquitin ligase, tumor microenvironment, MMP
Citation: Wang H, Li Q, Tang Q, Shi G, Wu G, Mao X, Wu C, Zhang L, Liu J, Li J and Li B (2024) Role and therapeutic potential of E3s in the tumor microenvironment of hepatocellular carcinoma. Front. Immunol. 15:1483721. doi: 10.3389/fimmu.2024.1483721
Received: 20 August 2024; Accepted: 14 October 2024;
Published: 31 October 2024.
Edited by:
Wenyi Jin, City University of Hong Kong, Hong Kong SAR, ChinaReviewed by:
Qihang Yuan, Dalian Medical University, ChinaKaige Chen, Wake Forest University, United States
Copyright © 2024 Wang, Li, Tang, Shi, Wu, Mao, Wu, Zhang, Liu, Li and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jie Liu, MTIzNTc0NTE0QHFxLmNvbQ==; Jingdong Li, TGlqaW5nZG9uZzM1OEAxMjYuY29t; Bo Li, bGlib2VyMjAwMkAxMjYuY29t
†These authors have contributed equally to this work
 Hailin Wang
Hailin Wang Qiang Li1†
Qiang Li1† Jie Liu
Jie Liu Jingdong Li
Jingdong Li