- 1College of Life Sciences and Medicine, Zhejiang Sci-Tech University, Zhejiang Provincial Key Laboratory of Silkworm Bioreactor and Biomedicine, Zhejiang Sci-Tech University, Hangzhou, China
- 2Pediatric Department, The First People’s Hospital of Huzhou, Huzhou, China
In the current preclinical anti-tumor researches, there is a general lack of an in vivo model that can quickly and efficiently screen effective anti-tumor drugs. As a species that is 87% genetically similar to humans, zebrafish have been widely used to model human diseases, and they are considered an alternative economic model for studying cancer development, proliferation, and metastasis. The zebrafish tumor xenograft model has been effectively used for cancer drug development at all levels, including target validation, and high-throughput screening of long non-coding RNAs (lncRNAs) that may be involved in tumor regulation. In this review, we provide a comprehensive overview of zebrafish as an in vivo model for cancer cell growth, migration, anti-tumor immunotherapy, and anti-tumor drug screening. In addition, the regulatory mechanisms of some active lncRNAs have been identified to play a role in the pathogenesis of cancer, but it is still necessary to take advantage of the efficient zebrafish model to screen and learn more about the role of these molecules in tumor development and migration. Current anti-tumor therapies are limited by severe toxicity and multidrug resistance. There is an urgent need for the cost-effective and efficient in vivo research tools to improve our understanding and overcome these problems. This paper reviews the different purposes of anti-tumor research using zebrafish model. We discuss the use of zebrafish in cancer cell proliferation and metastasis, identifying signaling pathways, cancer drug discovery and treatment development, and toxicity studies. Finally, this review highlights the limitations of the field and future directions to effectively utilize zebrafish as a highly efficient model for cancer treatment development.
1 Introduction
Cancer is a disease with complex causes, including genetic mutation, chromosomal translocation and deletion, amplification, and epigenetic alteration (1). While cancer death rates continue to decline, some cancer types are on the rise, such as breast, pancreatic, and uterine cancers, which are increasing in the United States by 0.6% to 1% per year between 2015 and 2019. The incidence of prostate cancer, liver cancer (in women), kidney cancer, human papillomavirus-associated oral cancer, and melanoma is increasing by 2% to 3% per year. The incidence of cervical cancer (30-44 years old) and colorectal cancer (<55 years old) in young people are also increasing at a rate of 1-2% per year (2). Similarly, by 2022, there will be approximately 4,824,700 new cases of malignant tumors and 2,574,100 deaths in China, and the burden of disease is still rising (3). Thus, it remains a major health problem, and effective animal models and therapeutic studies of many types of cancer are lacking. Noncoding RNAs (ncRNAs) differ from mRNAs in that most of them are not translated into proteins and are generally categorized according to the number of bases, of which those larger than 200 bases are called lncRNAs. LncRNAs are important and powerful cis- or trans-regulators of gene activity, acting as scaffolds for chromatin-modifying complexes and nucleosomes, as well as enhancers and mediators of remote chromatin interactions (4–7). A study analyzing the expression profiles of lncRNAs (and other ncRNAs) in samples from patients with a variety of cancers and comparing them with corresponding normal cells showed that many lncRNAs are dysregulated in various types of cancer (8). In prostate cancer, genome-wide RNA-Seq analysis identified many lncRNAs that are up- or down-regulated in prostate cancer, such as PCA3, PCGEM1, and PCAT-1 are highly associated with prostate cancer (9). In breast cancer, lncRNAs involved include HOTAIR, ANRIL, ZFAS1, NEAT1, DANCR, HIF1A-AS, XIST, TOPORS-AS1, LSINCT-5, and LNP1 (10). A number of deregulated lncRNAs have also been identified in other different types of cancers. Tumor research has focused on lncRNAs because of their functional diversity.
Zebrafish (Danio rerio) has become one of essential vertebrate model organisms for biomedical research. Zebrafish xenograft models offer many unique benefits for cancer research, including the optical transparency of zebrafish embryos and juveniles, which allows for in vivo observation and assessment of proliferation and migration at 96 h post-fertilization (hpf) (11). Zebrafish is a reliable model organism for studying tumor growth and metastasis (12, 13), and zebrafish models can be applied to the study of the role of lncRNAs in tumor development. For instance, the roles of lncRNA SNHG4 in the proliferation and migration of colorectal cancer (CRC) (14) and THOR in melanoma (15) have been explored. Zebrafish has emerged as an effective experimental animal model in the transition from in vitro experimental models, such as cells, to mammalian models, such as mice, and is valuable to drug-active molecule screening, antitumor drug development, tumor immunotherapy, and research on the functions of tumor-associated lncRNAs (16–18). In this paper, we mainly review and summarize some zebrafish xenograft tumor models and their uses in the study of tumorigenesis, proliferation, migration, and antitumor mechanism based on lncRNAs.
2 Model organism - zebrafish
2.1 Emergence of zebrafish as a model organism
Zebrafish is a common vertebrate model organism. George Streisinger first recognized the potential of zebrafish as a vertebrate model organism in the early 1980s and published the first paper describing the use of zebrafish as a vertebrate model (19). His research provided a practical basis for the use of zebrafish as a vertebrate model for humans and opened the door to the study of zebrafish. Continuous and in-depth research on zebrafish in the following decades has gradually recognized the potential of zebrafish as a model organism for the study of human diseases. Researchers from the United States and Germany completed the screening of mutated genes in zebrafish in 1996 (20, 21), and the whole genome of zebrafish was sequenced in 2000. Providing insights into the genetic background of zebrafish, several zebrafish models of human diseases have been established. Since then, many zebrafish disease models have been established, demonstrating the importance of zebrafish as a model organism.
2.2 Developmental processes and stages of zebrafish
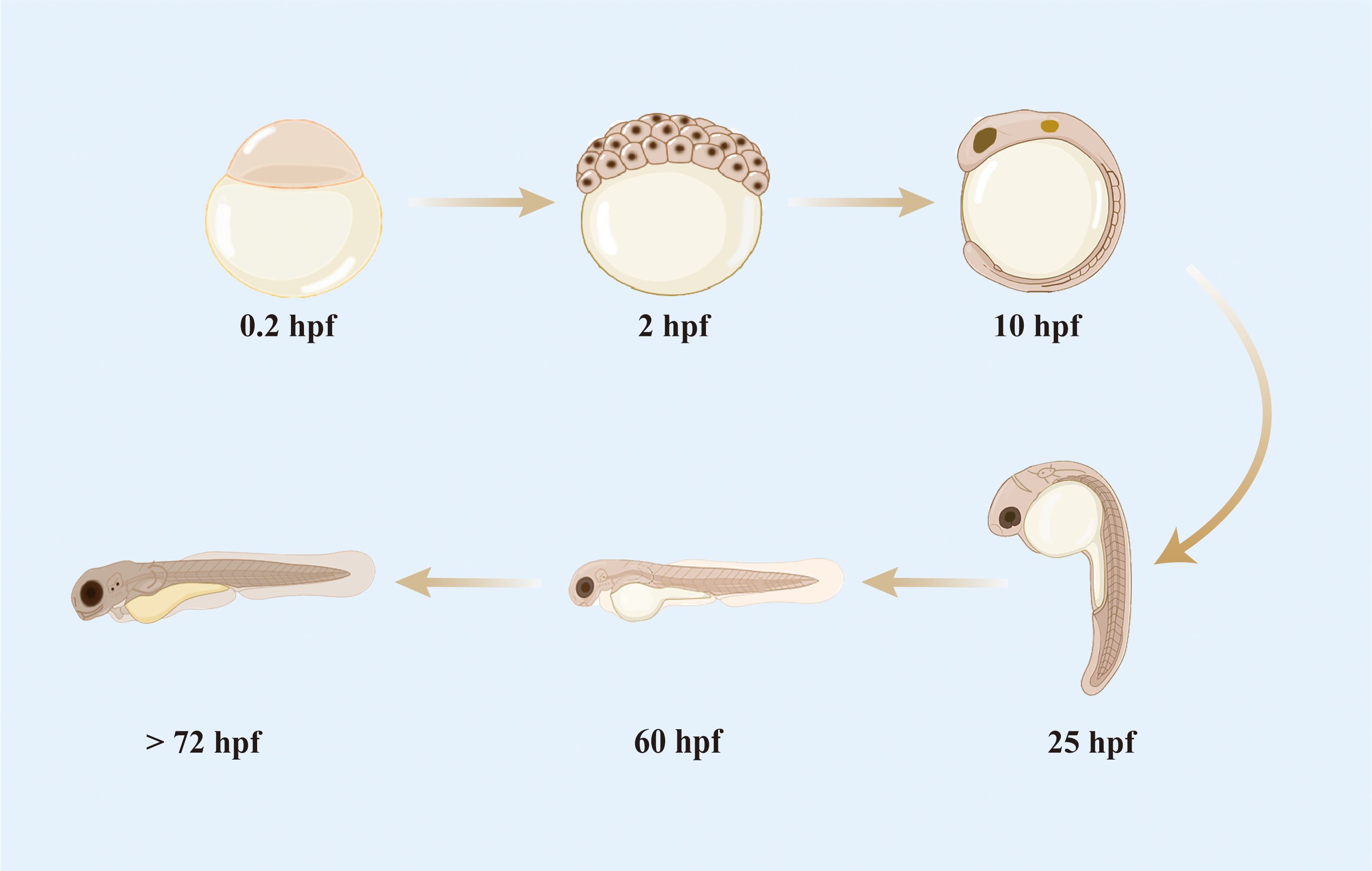
Zebrafish has highly similar development process to the embryos of higher vertebrates, including humans (22–24). Compared with mice, zebrafish are small and easy to manipulate for gene editing and reproduction and can be bred in large numbers at a low cost. The embryonic development of zebrafish is rapid (Figure 1); a single-celled fertilized egg develops into a motile and transparent embryo within 24 h. The majority of morphological changes are completed at 3 days post-fertilization (dpf) (25), and the digestive system and mouth are functional between 5 and 6 dpf. The yolk sacs throughout the embryonic and early larval stages are rapidly depleted and fully absorbed by 7 dpf. Once a zebrafish develops most of its features, it is considered a juvenile; when it is capable of producing living gametes and reproducing, it is considered an adult (25, 26). Under optimal culture conditions, laboratory zebrafish reach sexual maturity in the third month of their development.
2.3 Merits of zebrafish as a model in tumor research
A model organism necessarily requires an extremely high degree of conservation of its target genes with respect to the object of a study. Many factors involved in tumor progression are highly conserved between zebrafish and humans. Approximately 71.4% of human genes have at least one zebrafish homolog, and 82% of human disease-related genes have at least one zebrafish homolog (27). In addition, multiple epigenetic marks regulating gene expression are conserved in vertebrates, including zebrafish and humans (28). Despite having cell cycle genes, zebrafish has conserved tumor suppressors and oncogenes, which facilitate studies on oncogenic pathways and targeted tracing in zebrafish models. Moreover, due to the ease of introducing exogenous genes by microinjection at the single-cell stage of zebrafish, as well as the transparent development of the zebrafish embryo, where the expression and distribution of exogenous genes are easy to visualize and detect (29), zebrafish can be easily genetically manipulated through knockouts, overexpression, and transgenesis, allowing for the creation and characterization of cancer models at the genetic level (30). Notably, a high degree of similarity has been observed between the histopathology of human primary tumors and tumors transplanted in zebrafish (31). However, some genes involved in human cancer progression do not have clear homology in zebrafish, including leukemia inhibitory factor, oncostatin M, and breast cancer susceptibility genes (27).
3 Zebrafish tumor xenograft model and its application
3.1 Tumor xenograft models in zebrafish
The zygotes in zebrafish can develop rapidly in vitro and exhibit a certain degree of transparency. These features allow for the direct visualization, microscopic manipulation throughout embryogenesis and larval development, and observation of fluorescently labeled tumor cell lines with a microscope, thereby facilitating studies on the biology of tumor cells in living organisms. These merits establish zebrafish as effective tumor models. In addition, zebrafish shows early embryonic immune system defects at 0–5 dpf, when the adaptive immune system is still underdeveloped; these defect results in the poor development of the thymus in adult zebrafish, enabling it to tolerate exogenous cell implantation (32). Moreover, no rejection reaction occurs when xenogenic tumor transplantation is carried out in this period. A human tumor xenotransplantation model in fish has many benefits.
In 2005, Lee et al. (33) or the first time implanted human melanoma cells into zebrafish through xenotransplantation, providing a theoretical basis and experimental foundation for the construction of zebrafish xenotransplantation model. In 2007, Stoletov et al. (34), constructed a zebrafish juvenile xenotransplantation model by manipulating a xenotransplantation model on a 1 dpf zebrafish embryo, demonstrating that zebrafish larvae possess a window of observation for tumor formation, cell invasion, and tumor-induced angiogenesis. Tumor transplantation in zebrafish has emerged as an important model for oncology research over the past few years (35), and many important genes and pathways involved in cancer formation are highly conserved between the two species due to the high similarity between zebrafish and humans in terms of genetic background (31, 36). Through the study of zebrafish tumor xenograft models, the molecular mechanisms of human tumorigenesis and development, the formation of tumor microenvironment, and the therapeutic effects of antitumor drugs and therapies have been explored in depth.
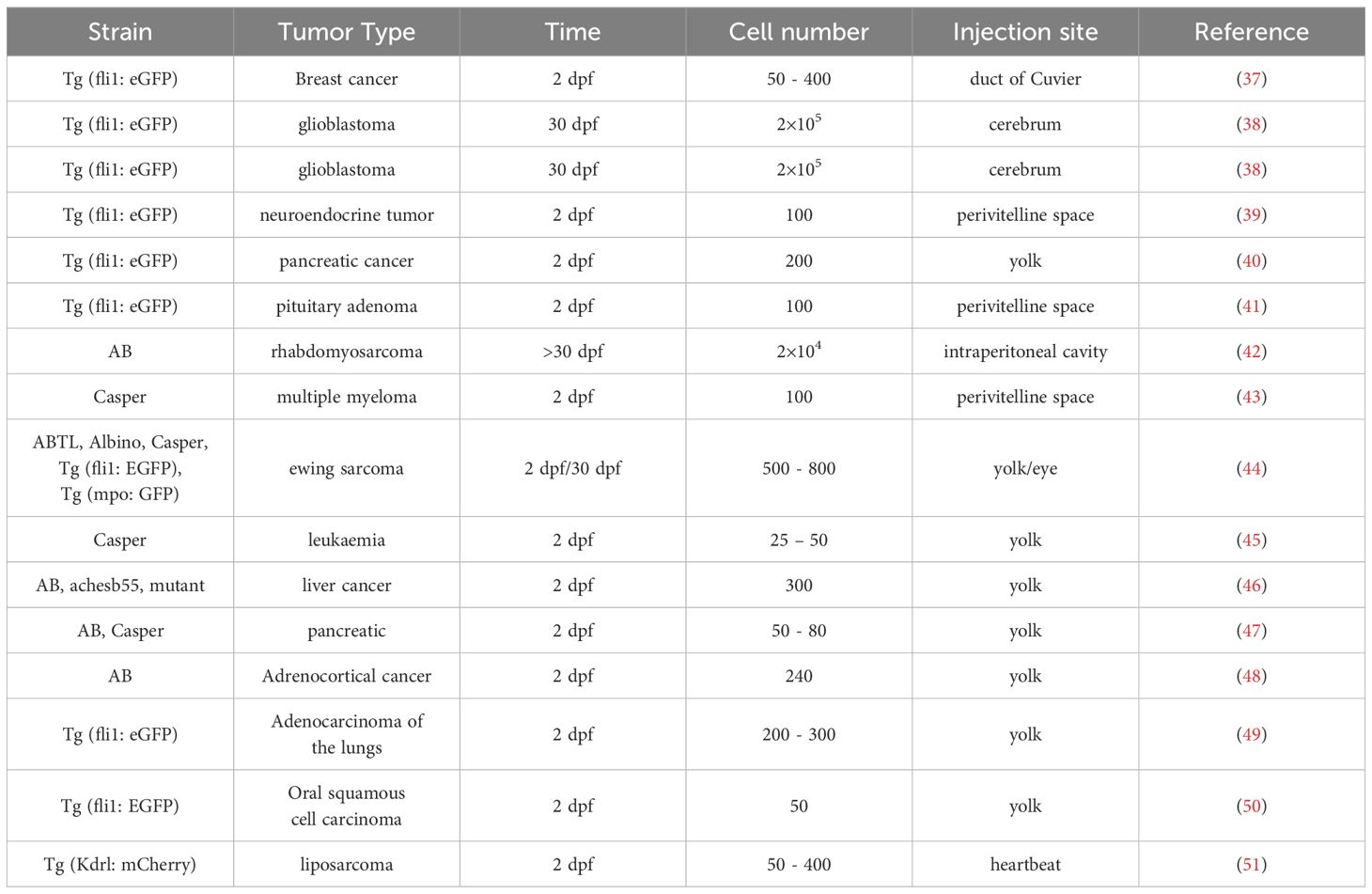
The current application of zebrafish to the construction of various benign and malignant tumor models involves direct oncogenic drug intervention, gene mutation, and tumor cell transplantation, among which there is a rich variety of xenotransplantation models, such as leukemia, melanoma, glioma, and hepatocellular carcinoma models (Table 1). Xenotransplantation is simpler than chemical, transgenic, and genetic mutation. A specific zebrafish tumor model can be established by injecting tumor cells through xenotransplantation. The body of a zebrafish is transparent before adulthood, and thus tumor growth can be directly observed with a body microscope. An adult zebrafish can be observed under a live imager, and the size and position of fluorescently labeled tumor growth can be observed and recorded in real time. In the preparation of zebrafish xenotransplantation models, a large number of model zebrafish can be obtained simultaneously for parallel evaluation and analysis. Tumor models with site and size differences can be constructed by controlling the number and location of injected tumor cell, and tumor models with special properties by modifying tumors. Moreover, the construction of 2–5 day models do not require immunosuppression, thus simplifying experimental procedures and shortening the experimental period.
3.2 Applications in tumor angiogenesis and migration
Given that angiogenesis of tumor tissues plays a key role in tumor growth and metastasis (52), observing and studying the angiogenic process of tumor tissues is of great value in studies on tumor production and migration. In 2001, Sumio Isogai et al. (53) found that the blood vessels of zebrafish and other vertebrates have great similarities, showing that studying the process of tumor angiogenesis in zebrafish is effective and feasible and providing an experimental basis for studying human tumor angiogenesis in zebrafish.
Zebrafish has benefits for evaluating the process of tumor angiogenesis, including its early embryonic transparency, high reproductive efficiency, and fast metabolic efficiency. Thus, it is more suitable for research on tumor angiogenesis than rabbits, mice, and chicken embryos and contributes to the rapid developments in this field (54). In addition, the application of transgenic angiofluorescent strain of zebrafish allows for the direct in vivo real-time evaluation of angiogenesis-inhibiting drugs on the neovascular system of tumor tissues, facilitating the rapid and precise screening and effect analysis of anti-angiogenic drugs (55). The many features of zebrafish facilitate research on the kinetics of microtumor formation and neovascularization through high-resolution imaging. Meanwhile, the interaction between fluorescent tumor cells and GFP-labeled host vasculature can be described in detail by the three-dimensional reconstruction of confocal microscopy images (56). This model system provides a clear window for observing the mechanisms of microtumor formation, cell invasion, and tumor-induced angiogenesis in mature animals. On the other hand, the process of vascular system generation of transplanted tumors in juvenile fish may be close to the process of tumor angiogenesis in cancer patients (57).
3.3 Applications in tumor immunotherapy
Unlike the vascular system that supports circulation, lymphatic vessels form a blind-ended tree-like structure rather than a circulatory loop. Tumors generate not only blood vessels but also lymphatic vessels in the microenvironment. Immune and inflammatory cells in lymphatic vessels enter the tumor microenvironment through infiltration, prompting the body to use autoimmunity against a tumor (58). The molecular mechanisms controlling lymphatic vessel development are largely conserved in zebrafish compared with humans, and lymphatic vessels in both involve key factors such as vegfc, vegfd, vegfr3, ccbe1, and prox1a during development (59). Therefore, the same characteristics of zebrafish embryonic transparency can be utilized in observing the transgenic zebrafish fluorescently labeled with lymphatic vessels in real time under an imager and studying the status of lymphatic vessel generation in the tumor microenvironment, serving as a platform for the observation of metastasis, proliferation, and dissemination of tumor cells via lymphatic vessels.
Based on the generative mechanism of the tumor microenvironmental lymphatic vascular system in zebrafish xenograft models, this model can be used in exploring tumor immunotherapy and provides insights for clinical tumor therapies, such as immune checkpoint inhibitor drugs. Representative drugs in this class include CTLA-4 inhibitors (60) and PD-1 inhibitors (61). Given that zebrafish have similar immune molecular mechanisms to humans, a portion of immune checkpoint inhibitory drugs can be used in zebrafish for studies on relevant drug mechanism of action and efficacy.
CAR-T therapy is one of the methods for tumor immunotherapy. In this method, T-lymphocytes extracted from a tumor lesion site can be tagged labels that recognize tumor markers, expanded, cultured in vitro, and re-infused back to a patient’s body. These cells can specifically kill tumor cells. As zebrafish progressively produce immune-related cells, such as T-cells, B-cells, and natural killer cells, as they develop into larvae, and gradually form a complete innate and acquired immune system, CAR-T therapy can be applied to zebrafish xenograft models for vivo studies. Susana Pascoal et al. (62) demonstrated that CAR-T cell–mediated elimination of target cells can be monitored in zebrafish embryos and CD19-specific CAR-T cells can kill pre-B leukemia cells (Nalm-6) in zebrafish embryos; in addition, they developed a system for quantifying changes in tumor and CAR-T cells over time; they suggest that a zebrafish xenograft model allows for the low-cost and rapid preclinical evaluation of CAR-T cells.
3.4 Application in tumor drug screening
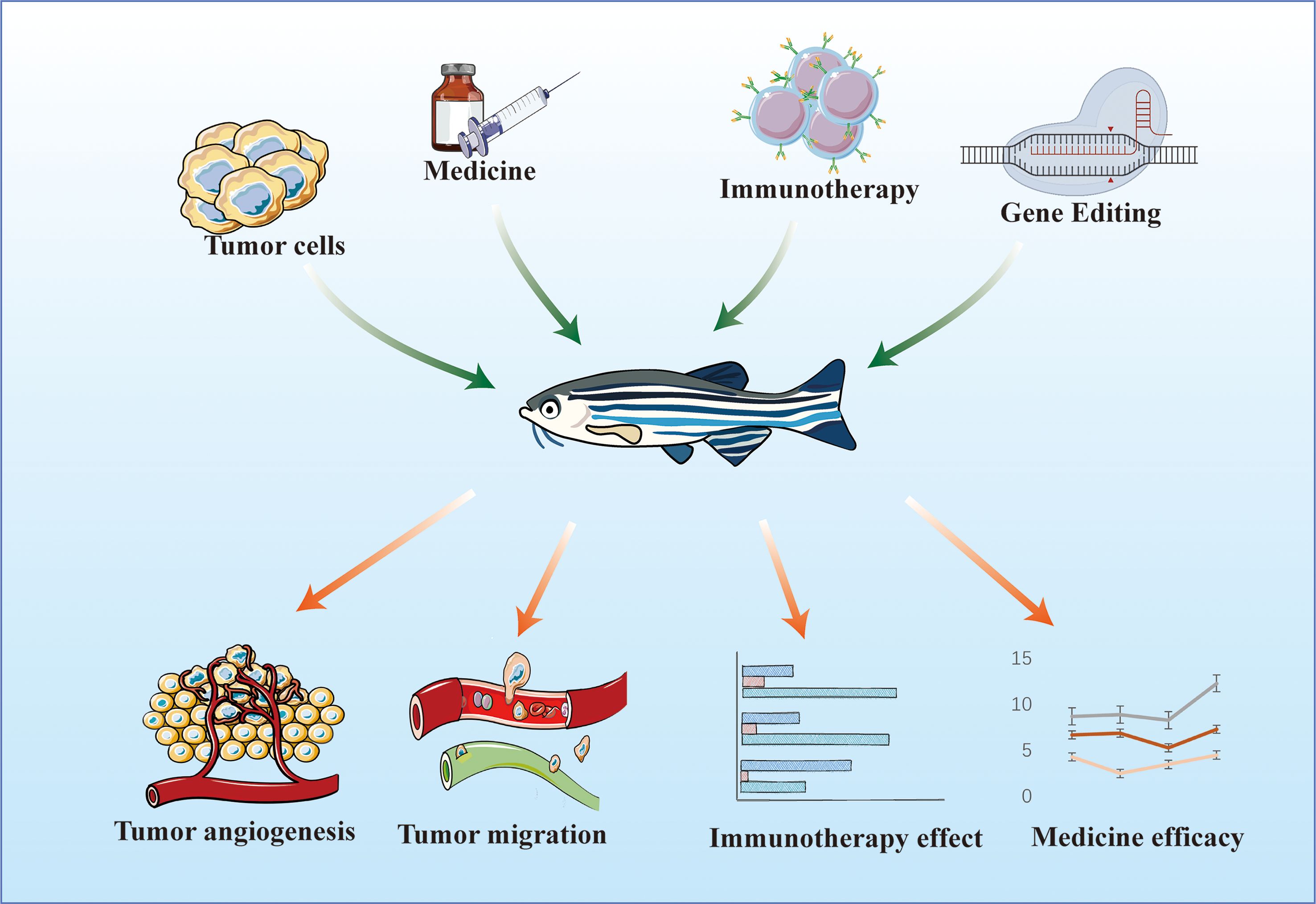
The research and development of novel drugs are long and costly processes. Drugs are first developed through in vitro tests, in which the effects of small-molecule therapies on cells are determined by detecting cell proliferation, cytotoxicity, marker expression, migration, signaling pathway activation, and morphological changes. Then, in vivo tests are conducted, in which the half-life of a drug is evaluated, and final drug screening is performed. Zebrafish are aquatic organisms that can absorb small-molecule compounds directly from water (63), and are thus more suitable for small-molecule drug screening and drug delivery pathway research than mice. In addition, a zebrafish has a short maturation period and produces a large number of offspring, and the cost of manipulating its genetic material is low. Thus, constructing tumor models from zebrafish is less expensive and time consuming than constructing tumor models from nude mice. Owing to the merits of zebrafish, it can be used as an intermediate evaluation model at the cellular and mammalian model levels for the rapid screening of antitumor active drug molecules and evaluation of the efficacy of antitumor therapies. The use of the zebrafish system spans from the discovery phase of high-content and high-throughput screening to the preclinical screening phase (64). We predict that zebrafish models will be increasingly used for drug assessment evaluation in the therapeutic context (Figure 2).
The use of zebrafish in drug discovery not only can improve the success rate of late-stage clinical drug development but also can reduce the cost and time required for screening (65). Owing to the high genetic affinity between zebrafish and humans, low breeding cost, convenient model construction, and easy genetic manipulation, which allows for the simultaneous construction of a large number of xenograft models for high-throughput drug screening, zebrafish is more suitable for high-throughput drug screening than other animal models. The short developmental process and fast metabolic efficiency of zebrafish can greatly reduce the time and increase the rate of drug screening. Thus, zebrafish xenografts have been used for the in vivo screening of drugs for CRC treatment, including cetuximab and regorafenib, and are effective and rapid in vivo model for human tumor drug trials (66).
4 Application of zebrafish modeling in the study of lncRNAs
4.1 Role and mechanisms of lncRNAs in tumor biology
Measurement and analysis of human transcriptional profiles have shown that the human genome is universally transcribed, but only 2% of RNAs encode proteins (67, 68). Transcripts greater than 200 nt in length have been widely categorized as lncRNAs (69–71). LncRNAs play crucial roles in cellular processes, including cell cycle regulation (72), cell differentiation and development (73), metabolism (74), immunity (75), and cancer genesis and development (76) and are potentially involved in viral infections (75, 77).
LncRNAs have various mechanisms of action, can regulate transcriptional and epigenetic modifications by interacting with DNA (78–80), participate in the regulation of translational and post-translational modifications through interactions with proteins (81, 82) and RNAs (83, 84), and can directly interact with signaling receptors (85). Although lncRNAs were previously categorized as noncoding RNAs, multiple lncRNAs contain open reading frames that produce micropeptides involved in a variety of biological processes and associated with many pathophysiological processes, such as carcinogenesis and tumor invasion and metastasis (86, 87) and innate immune responses (88). The discovery of lncRNA-derived peptides has generated a new mode of action by which these peptides interact with proteins and influence biological processes involved in the onset and progression of a disease, particularly cancer and immune diseases (89).
For example, large amounts of lncRNAs promote tumor growth and metastasis.PVT1 is required for high levels of c-Myc protein; PVT1 RNA and c-Myc protein expression are highly correlated in primary human tumors. Inhibition of PVT1 suppresses c-Myc-driven tumorigenic potency (90). MALAT1 was first identified in the lung cancer MALAT1 and is one of the most studied lncRNAs in cancer. For example, MALAT1 can promote tumorigenesis through the Wnt/β-catenin pathway, EMT, PI3K/AKT pathway, ERK/MAPK pathway and angiogenesis (91). HOTAIR alters histone (H3K27) methylation patterns and gene expression, thereby promoting tumor cell invasion. On the other hand, inhibition of HOTAIR decreases cell invasion. This is mainly attributed to the ability of HOTAIR to act as a scaffold for specific histone-modifying enzymes, affecting the expression of specific genes (92). Finally, LINK-A expression and activation of the LINK-Adependent signaling pathway are associated with triple-negative breast cancer (TNBC), and together they promote glycolytic reprogramming and tumorigenesis in breast cancer (93).
A number of lncRNAs have been identified in zebrafish (94, 95) and mice (96, 97); however, their primary sequences are weakly conserved between the species (98, 99). For example, few lncRNAs are conserved in the primary sequence between zebrafish and humans, and the phenotype of functionally inactivated zebrafish with conserved lncRNAs can be recovered using homologous genes from mouse or human (94). These results suggest that the higher-order structures of lncRNAs may be conserved, rather than their primary sequences. Thus, zebrafish may be a suitable model for exploring lncRNAs with function distinct from those indicated by the primary sequences.
4.2 Role of LncRNAs in the regulation of zebrafish tumor models
Since the development of zebrafish as a promising experimental model for human cancer research (100), many researchers have applied zebrafish model to determine the oncogenic role of lncRNAs in tumors. In the study of certain lncRNAs, zebrafish can be easily microinjected with Morpholinos at the embryonic and single-cell stage because zebrafish has homologous lncRNAs (101). Efficient screening of knockout or knockdown lines is ensured by the high number of fast-developing zebrafish zygotes, which facilitates investigations on the function of relevant lncRNAs throughout the course of tumor development and functional roles of relevant lncRNAs during normal development. Similar inferences have been drawn from cultured human and mouse cells (98). For example, in a study of the oncogenic role of the lncRNA THOR in melanoma, Hosono et al. (102), identified an exonic homolog of THOR in the zebrafish genome through bioinformatics prediction and later verified that zebrafish with knocked out THOR gene are resistant to melanoma formation, demonstrating that zebrafish can be used as a model for studying tumor-associated lncRNAs.
LncRNAs that undergo changes in expression in tumor tissues can be explored using a tumor xenograft model of zebrafish. One method is to construct a tumor xenograft model of zebrafish and then subject the zebrafish to different treatments, collect and analyze changes in the expression target lncRNAs, and ultimately validate the function of the relevant lncRNAs in tumor development. For example, in the study of STEAP3-AS1, a zebrafish in situ colorectal cancer model was constructed. Under normal or 8% oxygen condition, STEAP3-AS1 up-regulation may be a key factor in tumor development. AS1 upregulation may be required for hypoxia-mediated tumor metastasis (103). Another way is to treat cells used in tumor xenograft implantation models, such as knocking down, knocking out, and overexpressing relevant genes in tumor cells, before models are established for observation and data related to tumor proliferation and metastasis are collected; this approach allows for the validation of the in vivo functional roles of relevant lncRNAs. For example, Shen et al. (104) constructed a zebrafish xenograft model using lung cancer cells with knocked down PVT1, evaluated cell proliferation by quantifying the yolk fluorescence region of zebrafish larvae, and explored the migration of lung cancer cells with knocked down PVT1 by quantifying the trunk fluorescence region of zebrafish larvae; they verified that the growth and metastasis of lung cancer cells were inhibited when Lnc PVT1 was knocked down.
Furthermore, in the study of LncRNA LETS1, a zebrafish xenograft model was applied to verify that LETS1 deletion attenuated TGF-β-induced EMT and migration of breast and lung cancer cells in vitro (105). Similarly, a zebrafish xenograft model was used to study the functions associated with the lncRNA SNHG4 in studies of colorectal cancer proliferation and migration (14). Although the results of studies applying the zebrafish xenograft model to study the functions associated with LncRNAs in different tumors are not yet abundant, a large number of relevant experiments are being carried out due to the advantages of the zebrafish xenograft model in relevant studies.
The progression of tumor cells can be assessed with a zebrafish xenograft model, and the in vivo function of relevant lncRNAs can be verified 96 h after the model is constructed. By contrast, mouse xenograft models require 3–5 weeks. For studies on endogenous lncRNAs in tumors, shRNA plasmids for gene silencing must be constructed in a mouse model. The zebrafish model only requires the design and synthesis of specific siRNAs. When lncRNAs are examine as indicators of early tumor surveillance, the behavior of early tumor cells can be monitored in vivo according to the characteristics of the immature adaptive immune system of zebrafish larvae and the transparency of the body surface. These procedures are difficult to apply to mouse models. These results suggest that the zebrafish xenograft model is suitable for studying lncRNAs and monitoring tumor progression (Figure 3).
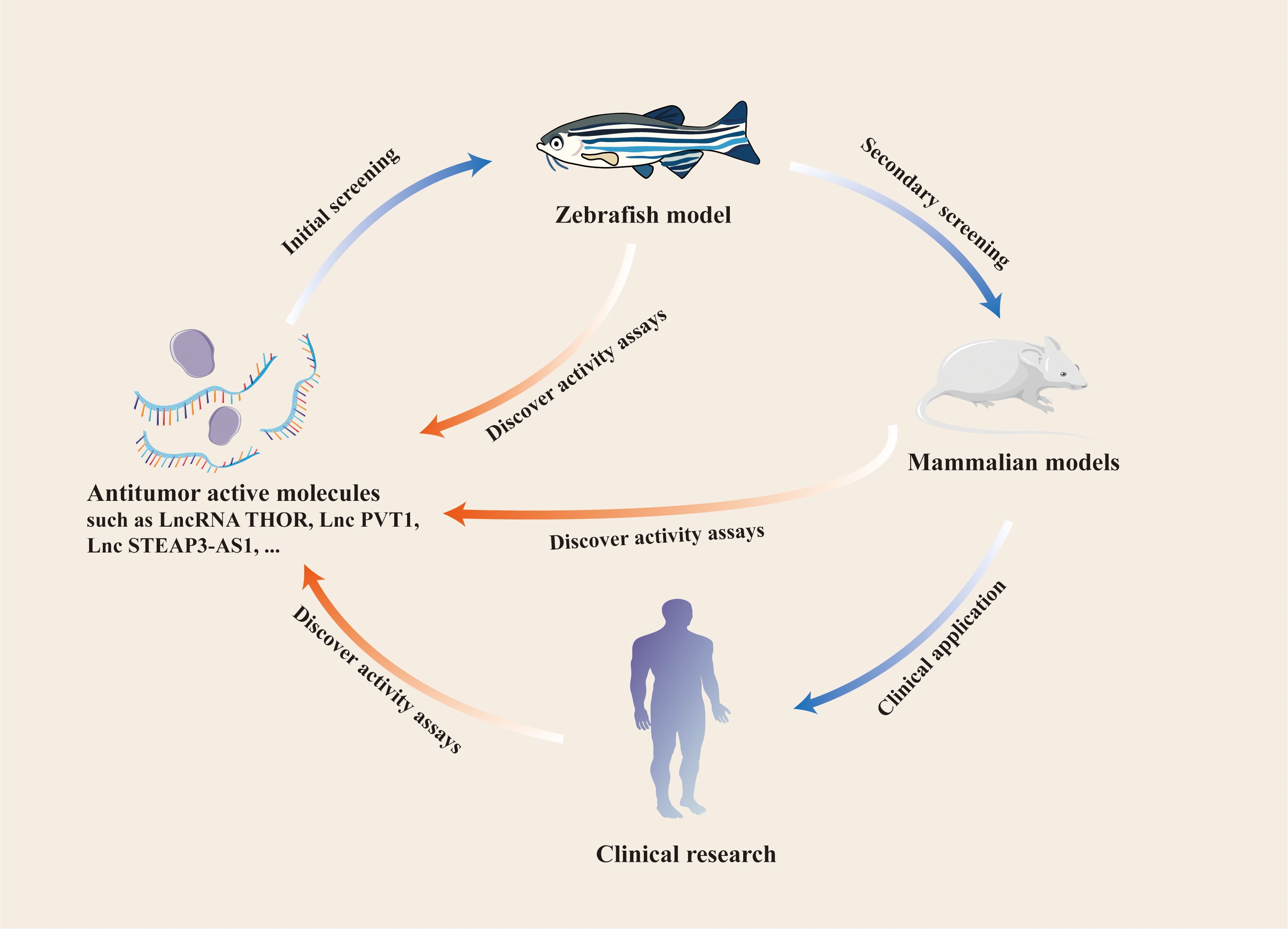
Figure 3. Application of zebrafish model in the study of LncRNAs and other antitumor active molecules.
5 Conclusion
We summarize the development of zebrafish as a model organism, introduce the various growth and development stages of zebrafish, and illustrate the benefits of constructing tumor xenograft models of zebrafish. First, zebrafish tumor xenotransplantation facilitates research on tumor biology, and the proliferation and migration of tumor cells can be evaluated in a relatively short period of time after transplantation; relevant assays can be performed 3 days after drug treatment (49). Second, tumor growth and migration can be continuously monitored in zebrafish larvae in contrast to mouse models, allowing for the real-time observation of tumor cells’ behavior (106). In addition, transparent zebrafish embryos are essential, and single-cell resolution in zebrafish Casper lines exceeds that in mouse transplantation models, making prolonged observation feasible because fish do not need to be sacrificed for analysis (35, 56). These advantages support the use of zebrafish xenograft models in tumor research.
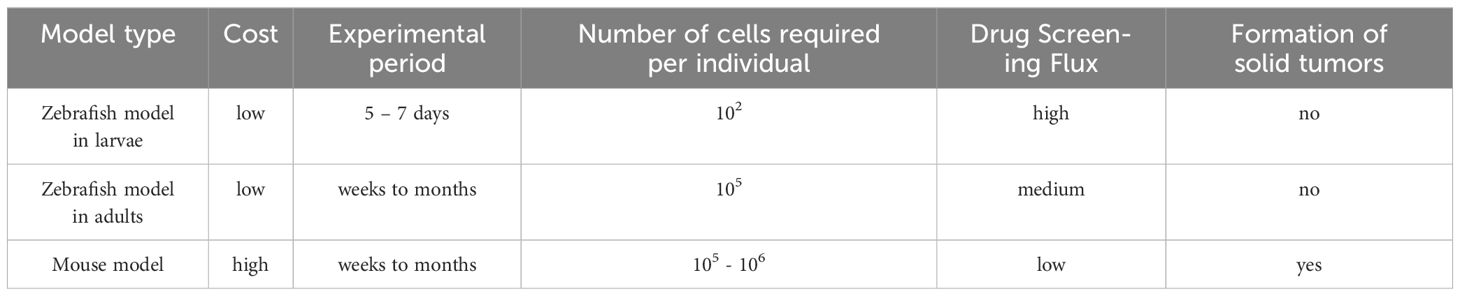
We also summarize and analyze some specific zebrafish xenograft models and their applications to studies on tumor angiogenesis and immunotherapy and drug screening (64, 107, 108). More importantly, we incorporate the application of lncRNAs in zebrafish model studies. Zebrafish xenografts can be used as in vivo tools for studying the role of lncRNAs in tumor cell proliferation and migration in vivo and preliminarily validating the relevant functions of lncRNAs in zebrafish models. Molecular mechanisms by which specific lncRNAs regulate the growth of tumors in vivo can be explored using these models, and some antitumor growth functional lncRNAs can be screened out and validated on mammalian models and in clinical trials (14, 102, 109). The zebrafish model has its own unique advantages over the mouse model, such as simple operation, high efficiency, and low cost. We compare the mouse model, the zebrafish model in more detail (Table 2). These advantages can facilitate the cellular level detection of antitumor active molecules. The in vivo environment of a zebrafish model can mimic the growth characteristics of human tumors and ensures the accurate and efficient identification of effective antitumor active molecules.
Despite the many advantages of the zebrafish tumor xenograft model, there are still some shortcomings. First, it is obvious that zebrafish are devoid of lung, mammary, and prostate organs, and it is not possible for us to directly study primary cancers in these organs on zebrafish models (although relevant xenograft models can be constructed). Second, most transgenic zebrafish models are still based on transgenic expression of human oncogenes rather than knock-in of endogenous or homologous zebrafish loci, and thus may not have physiologic expression levels. Finally, in transplantation models of zebrafish larvae, the small size limits the number of transplanted cells to 100-200 per fish, which often fails to encompass cancer-driving stem cells and accurately recapitulates the genetic heterogeneity and drug-responsive behavior driven by rare subclones in human tumors.
Zebrafish models cannot completely replace mammalian models, such as mice, but can be used as a bridge between the cellular-level and mammalian models in vivo, thus greatly contributing to the progress of related studies and reducing research cost. Overall, the zebrafish xenotransplantation model is a reliable and promising model for human cancer research, and transplantation models are reliable and promising models for human cancer research.
Author contributions
CH: Writing – original draft, Writing – review & editing. LS: Writing – review & editing. JC: Conceptualization, Writing – review & editing. ZL: Conceptualization, Project administration, Resources, Writing – review & editing. CY: Project administration, Resources, Writing – review & editing. XJ: Conceptualization, Project administration, Resources, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by Zhejiang Provincial Public Welfare Technology Application Research Project, China, grant number: LTGY24H100001.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: A new paradigm. Cancer Res. (2017) 77:3965–81. doi: 10.1158/0008-5472.CAN-16-2634
2. Siegel RL, Giaquinto AN, Jemal A. Cancer statistics, 2024. CA Cancer J Clin. (2024) 74:12–49. doi: 10.3322/caac.21820
3. Zheng RS, Chen R, Han BF, Wang SM, Li L, Sun KX, et al. Cancer incidence and mortality in China, 2022. Zhonghua Zhong Liu Za Zhi. (2024) 46:221–31. doi: 10.3760/cma.j.cn112152-20240119-00035
4. Nagano T, Fraser P. No-nonsense functions for long noncoding RNAs. Cell. (2011) 145:178–81. doi: 10.1016/j.cell.2011.03.014
5. Loewer S, Cabili MN, Guttman M, Loh YH, Thomas K, Park IH, et al. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet. (2010) 42:1113–7. doi: 10.1038/ng.710
6. Chaumeil J, Le Baccon P, Wutz A, Heard E. A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes Dev. (2006) 20:2223–37. doi: 10.1101/gad.380906
7. Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. (2010) 142:409–19. doi: 10.1016/j.cell.2010.06.040
8. Bhan A, Mandal SS. Long noncoding RNAs: emerging stars in gene regulation, epigenetics and human disease. ChemMedChem. (2014) 9:1932–56. doi: 10.1002/cmdc.201300534
9. Prensner JR, Iyer MK, Balbin OA, Dhanasekaran SM, Cao Q, Brenner JC, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol. (2011) 29:742–9. doi: 10.1038/nbt.1914
10. Li Y, Wang X. Role of long noncoding RNAs in Malignant disease (Review). Mol Med Rep. (2016) 13:1463–9. doi: 10.3892/mmr.2015.4711
11. Wertman J, Veinotte CJ, Dellaire G, Berman JN. The zebrafish xenograft platform: evolution of a novel cancer model and preclinical screening tool. Adv Exp Med Biol. (2016) 916:289–314. doi: 10.1007/978-3-319-30654-4_13
12. Marques IJ, Weiss FU, Vlecken DH, Nitsche C, Bakkers J, Lagendijk AK, et al. Metastatic behaviour of primary human tumours in a zebrafish xenotransplantation model. BMC Cancer. (2009) 9:128. doi: 10.1186/1471-2407-9-128
13. Haldi M, Ton C, Seng WL, McGrath P. Human melanoma cells transplanted into zebrafish proliferate, migrate, produce melanin, form masses and stimulate angiogenesis in zebrafish. Angiogenesis. (2006) 9:139–51. doi: 10.1007/s10456-006-9040-2
14. Wang J, Zhang XY, Xu DY. Zebrafish xenograft model for studying the function of lncRNA SNHG4 in the proliferation and migration of colorectal cancer. J Gastrointest Oncol. (2022) 13:210–20. doi: 10.21037/jgo
15. Hosono Y, Niknafs YS, Prensner JR, Iyer MK, Dhanasekaran SM, Mehra R, et al. Oncogenic role of THOR, a conserved cancer/testis long non-coding RNA. Cell. (2017) 171:1559–1572.e20. doi: 10.1016/j.cell.2017.11.040
16. Belo MAA, Oliveira MF, Oliveira SL, Aracati MF, Rodrigues LF, Costa CC, et al. Zebrafish as a model to study inflammation: A tool for drug discovery. BioMed Pharmacother. (2021) 144:112310. doi: 10.1016/j.biopha.2021.112310
17. Letrado P, de Miguel I, Lamberto I, Díez-Martínez R, Oyarzabal J. Zebrafish: speeding up the cancer drug discovery process. Cancer Res. (2018) 78:6048–58. doi: 10.1158/0008-5472.CAN-18-1029
18. Zampedri C, Martínez-Flores WA, Melendez-Zajgla J. The use of zebrafish xenotransplant assays to analyze the role of lncRNAs in breast cancer. Front Oncol. (2021) 11:687594. doi: 10.3389/fonc.2021.687594
19. Streisinger G, Walker C, Dower N, Knauber D, Singer F. Production of clones of homozygous diploid zebra fish (Brachydanio rerio). Nature. (1981) 291:293–6. doi: 10.1038/291293a0
20. Driever W, Solnica-Krezel L, Schier AF, Neuhauss SC, Malicki J, Stemple DL, et al. A genetic screen for mutations affecting embryogenesis in zebrafish. Development. (1996) 123:37–46. doi: 10.1242/dev.123.1.37
21. Mullins MC, Hammerschmidt M, Kane DA, Odenthal J, Brand M, van Eeden FJ, et al. Genes establishing dorsoventral pattern formation in the zebrafish embryo: the ventral specifying genes. Development. (1996) 123:81–93. doi: 10.1242/dev.123.1.81
22. Stanton MF. Diethylnitrosamine-induced hepatic degeneration and neoplasia in the aquarium fish, brachydanio rerio. J Natl Cancer Inst. (1965) 34:117–30. doi: 10.1093/jnci/34.1.117
23. Dooley K, Zon LI. Zebrafish: a model system for the study of human disease. Curr Opin Genet Dev. (2000) 10:252–6. doi: 10.1016/S0959-437X(00)00074-5
24. Guyon JR, Steffen LS, Howell MH, Pusack TJ, Lawrence C, Kunkel LM. Modeling human muscle disease in zebrafish. Biochim Biophys Acta. (2007) 1772:205–15. doi: 10.1016/j.bbadis.2006.07.003
25. Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. (1995) 203:253–310. doi: 10.1002/aja.1002030302
26. Parichy DM, Elizondo MR, Mills MG, Gordon TN, Engeszer RE. Normal table of postembryonic zebrafish development: staging by externally visible anatomy of the living fish. Dev Dyn. (2009) 238:2975–3015. doi: 10.1002/dvdy.22113
27. Howe K, Clark MD, Torroja CF, Torrance J, Berthelot C, Muffato M, et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature. (2013) 496:498–503. doi: 10.1038/nature12111
28. Long HK, Sims D, Heger A, Blackledge NP, Kutter C, Wright ML, et al. Epigenetic conservation at gene regulatory elements revealed by non-methylated DNA profiling in seven vertebrates. Elife. (2013) 2:e00348. doi: 10.7554/eLife.00348.016
29. Xiao J, Glasgow E, Agarwal S. Zebrafish xenografts for drug discovery and personalized medicine. Trends Cancer. (2020) 6:569–79. doi: 10.1016/j.trecan.2020.03.012
30. White R, Rose K, Zon L. Zebrafish cancer: the state of the art and the path forward. Nat Rev Cancer. (2013) 13:624–36. doi: 10.1038/nrc3589
31. Amatruda JF, Shepard JL, Stern HM, Zon LI. Zebrafish as a cancer model system. Cancer Cell. (2002) 1:229–31. doi: 10.1016/S1535-6108(02)00052-1
32. Jiang X, Liu G, Hu Z, Chen G, Chen J, Lv Z. cGAMP inhibits tumor growth in colorectal cancer metastasis through the STING/STAT3 axis in a zebrafish xenograft model. Fish Shellfish Immunol. (2019) 95:220–6. doi: 10.1016/j.fsi.2019.09.075
33. Lee LM, Seftor EA, Bonde G, Cornell RA, Hendrix MJ. The fate of human Malignant melanoma cells transplanted into zebrafish embryos: assessment of migration and cell division in the absence of tumor formation. Dev Dyn. (2005) 233:1560–70. doi: 10.1002/dvdy.20471
34. Stoletov K, Montel V, Lester RD, Gonias SL, Klemke R. High-resolution imaging of the dynamic tumor cell vascular interface in transparent zebrafish. Proc Natl Acad Sci U S A. (2007) 104:17406–11. doi: 10.1073/pnas.0703446104
35. Taylor AM, Zon LI. Zebrafish tumor assays: the state of transplantation. Zebrafish. (2009) 6:339–46. doi: 10.1089/zeb.2009.0607
36. Liu S, Leach SD. Zebrafish models for cancer. Annu Rev Pathol. (2011) 6:71–93. doi: 10.1146/annurev-pathol-011110-130330
37. Mercatali L, La Manna F, Groenewoud A, Casadei R, Recine F, Miserocchi G, et al. Development of a patient-derived xenograft (PDX) of breast cancer bone metastasis in a zebrafish model. Int J Mol Sci. (2016) 17:1375. doi: 10.3390/ijms17081375
38. Eden CJ, Ju B, Murugesan M, Phoenix TN, Nimmervoll B, Tong Y, et al. Orthotopic models of pediatric brain tumors in zebrafish. Oncogene. (2015) 34:1736–42. doi: 10.1038/onc.2014.107
39. Gaudenzi G, Albertelli M, Dicitore A, Würth R, Gatto F, Barbieri F, et al. Patient-derived xenograft in zebrafish embryos: a new platform for translational research in neuroendocrine tumors. Endocrine. (2017) 57:214–9. doi: 10.1007/s12020-016-1048-9
40. Weiss FU, Marques IJ, Woltering JM, Vlecken DH, Aghdassi A, Partecke LI, et al. Retinoic acid receptor antagonists inhibit miR-10a expression and block metastatic behavior of pancreatic cancer. Gastroenterology. (2009) 137:2136–45.e1-7. doi: 10.1053/j.gastro.2009.08.065
41. Peverelli E, Giardino E, Treppiedi D, Meregalli M, Belicchi M, Vaira V, et al. Dopamine receptor type 2 (DRD2) and somatostatin receptor type 2 (SSTR2) agonists are effective in inhibiting proliferation of progenitor/stem-like cells isolated from nonfunctioning pituitary tumors. Int J Cancer. (2017) 140:1870–80. doi: 10.1002/ijc.30613
42. Langenau DM, Keefe MD, Storer NY, Guyon JR, Kutok JL, Le X, et al. Effects of RAS on the genesis of embryonal rhabdomyosarcoma. Genes Dev. (2007) 21:1382–95. doi: 10.1101/gad.1545007
43. Lin J, Zhang W, Zhao JJ, Kwart AH, Yang C, Ma D, et al. A clinically relevant in vivo zebrafish model of human multiple myeloma to study preclinical therapeutic efficacy. Blood. (2016) 128:249–52. doi: 10.1182/blood-2016-03-704460
44. van der Ent W, Jochemsen AG, Teunisse AF, Krens SF, Szuhai K, Spaink HP, et al. Ewing sarcoma inhibition by disruption of EWSR1-FLI1 transcriptional activity and reactivation of p53. J Pathol. (2014) 233:415–24. doi: 10.1002/path.4378
45. Corkery DP, Dellaire G, Berman JN. Leukaemia xenotransplantation in zebrafish–chemotherapy response assay in vivo. Br J Haematol. (2011) 153:786–9. doi: 10.1111/j.1365-2141.2011.08661.x
46. Avci ME, Keskus AG, Targen S, Isilak ME, Ozturk M, Atalay RC, et al. Development of a novel zebrafish xenograft model in ache mutants using liver cancer cell lines. Sci Rep. (2018) 8:1570. doi: 10.1038/s41598-018-19817-w
47. Wang L, Chen H, Fei F, He X, Sun S, Lv K, et al. Patient-derived heterogeneous xenograft model of pancreatic cancer using zebrafish larvae as hosts for comparative drug assessment. J Vis Exp. (2019) 146. doi: 10.1167/19.10.14
48. Gianoncelli A, Guarienti M, Fragni M, Bertuzzi M, Rossini E, Abate A, et al. Adrenocortical carcinoma xenograft in zebrafish embryos as a model to study the in vivo cytotoxicity of abiraterone acetate. Endocrinology. (2019) 160:2620–9. doi: 10.1210/en.2019-00152
49. Li XY, Huang LT, Wu JQ, He MF, Zhu SH, Zhan P, et al. Zebrafish xenograft model of human lung cancer for evaluating osimertinib resistance. BioMed Res Int. (2019) 2019:3129748. doi: 10.1155/2019/3129748
50. Yu CI, Chen CY, Liu W, Chang PC, Huang CW, Han KF, et al. Sandensolide induces oxidative stress-mediated apoptosis in oral cancer cells and in zebrafish xenograft model. Mar Drugs. (2018) 16:387. doi: 10.3390/md16100387
51. Liverani C, La Manna F, Groenewoud A, Mercatali L, Van Der Pluijm G, Pieri F, et al. Innovative approaches to establish and characterize primary cultures: an ex vivo 3D system and the zebrafish model. Biol Open. (2017) 6:133–40. doi: 10.1242/bio.023911
52. Carmeliet P, Jain RK. Angiogenesis in cancer and other diseases. Nature. (2000) 407:249–57. doi: 10.1038/35025220
53. Isogai S, Horiguchi M, Weinstein BM. The vascular anatomy of the developing zebrafish: an atlas of embryonic and early larval development. Dev Biol. (2001) 230:278–301. doi: 10.1006/dbio.2000.9995
54. Bahrami N, Childs SJ. Pericyte biology in zebrafish. Adv Exp Med Biol. (2018) 1109:33–51. doi: 10.1007/978-3-030-02601-1_4
55. Cha YR, Weinstein BM. Visualization and experimental analysis of blood vessel formation using transgenic zebrafish. Birth Defects Res C Embryo Today. (2007) 81:286–96. doi: 10.1002/bdrc.20103
56. White RM, Sessa A, Burke C, Bowman T, LeBlanc J, Ceol C, et al. Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell. (2008) 2:183–9. doi: 10.1016/j.stem.2007.11.002
57. Tobia C, De Sena G, Presta M. Zebrafish embryo, a tool to study tumor angiogenesis. Int J Dev Biol. (2011) 55:505–9. doi: 10.1387/ijdb.103238ct
58. Garnier L, Gkountidi AO, Hugues S. Tumor-associated lymphatic vessel features and immunomodulatory functions. Front Immunol. (2019) 10:720. doi: 10.3389/fimmu.2019.00720
59. Okuda KS, Baek S, Hogan BM. Visualization and tools for analysis of zebrafish lymphatic development. Methods Mol Biol. (2018) 1846:55–70. doi: 10.1007/978-1-4939-8712-2_4
60. Leach DR, Krummel MF, Allison JP. Enhancement of antitumor immunity by CTLA-4 blockade. Science. (1996) 271:1734–6. doi: 10.1126/science.271.5256.1734
61. Ishida Y, Agata Y, Shibahara K, Honjo T. Induced expression of PD-1, a novel member of the immunoglobulin gene superfamily, upon programmed cell death. EMBO J. (1992) 11:3887–95. doi: 10.1002/embj.1992.11.issue-11
62. Pascoal S, Salzer B, Scheuringer E, Wenninger-Weinzierl A, Sturtzel C, Holter W, et al. A preclinical embryonic zebrafish xenograft model to investigate CAR T cells in vivo. Cancers (Basel). (2020) 12:567. doi: 10.3390/cancers12030567
63. Peterson RT, Link BA, Dowling JE, Schreiber SL. Small molecule developmental screens reveal the logic and timing of vertebrate development. Proc Natl Acad Sci U S A. (2000) 97:12965–9. doi: 10.1073/pnas.97.24.12965
64. Patton EE, Zon LI, Langenau DM. Zebrafish disease models in drug discovery: from preclinical modelling to clinical trials. Nat Rev Drug Discovery. (2021) 20:611–28. doi: 10.1038/s41573-021-00210-8
65. Lu JW, Yang WY, Tsai SM, Lin YM, Chang PH, Chen JR, et al. Liver-specific expressions of HBx and src in the p53 mutant trigger hepatocarcinogenesis in zebrafish. PLoS One. (2013) 8:e76951. doi: 10.1371/journal.pone.0076951
66. Fior R, Póvoa V, Mendes RV, Carvalho T, Gomes A, Figueiredo N, et al. Single-cell functional and chemosensitive profiling of combinatorial colorectal therapy in zebrafish xenografts. Proc Natl Acad Sci U S A. (2017) 114:E8234–e8243. doi: 10.1073/pnas.1618389114
67. Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, et al. Landscape of transcription in human cells. Nature. (2012) 489:101–8. doi: 10.1038/nature11233
68. Bridges MC, Daulagala AC, Kourtidis A. LNCcation: lncRNA localization and function. J Cell Biol. (2021) 220:e202009045. doi: 10.1083/jcb.202009045
69. Hangauer MJ, Vaughn IW, McManus MT. Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genet. (2013) 9:e1003569. doi: 10.1371/journal.pgen.1003569
70. Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. (2015) 47:199–208. doi: 10.1038/ng.3192
71. Abdi E, Latifi-Navid S. LncRNA polymorphisms and urologic cancer risk. Environ Mol Mutagen. (2022) 63:190–203. doi: 10.1002/em.22472
72. Kitagawa M, Kitagawa K, Kotake Y, Niida H, Ohhata T. Cell cycle regulation by long non-coding RNAs. Cell Mol Life Sci. (2013) 70:4785–94. doi: 10.1007/s00018-013-1423-0
73. Brazão TF, Johnson JS, Müller J, Heger A, Ponting CP, Tybulewicz VL. Long noncoding RNAs in B-cell development and activation. Blood. (2016) 128:e10–9. doi: 10.1182/blood-2015-11-680843
74. Sun X, Wong D. Long non-coding RNA-mediated regulation of glucose homeostasis and diabetes. Am J Cardiovasc Dis. (2016) 6:17–25.
75. Liu W, Wang Z, Liu L, Yang Z, Liu S, Ma Z, et al. LncRNA Malat1 inhibition of TDP43 cleavage suppresses IRF3-initiated antiviral innate immunity. Proc Natl Acad Sci U S A. (2020) 117:23695–706. doi: 10.1073/pnas.2003932117
76. Tan YT, Lin JF, Li T, Li JJ, Xu RH, Ju HQ. LncRNA-mediated posttranslational modifications and reprogramming of energy metabolism in cancer. Cancer Commun (Lond). (2021) 41:109–20. doi: 10.1002/cac2.12108
77. Wang Y, Huang L, Wang Y, Luo W, Li F, Xiao J, et al. Single-cell RNA-sequencing analysis identifies host long noncoding RNA MAMDC2-AS1 as a co-factor for HSV-1 nuclear transport. Int J Biol Sci. (2020) 16:1586–603. doi: 10.7150/ijbs.42556
78. Arora R, Lee Y, Wischnewski H, Brun CM, Schwarz T, Azzalin CM. RNaseH1 regulates TERRA-telomeric DNA hybrids and telomere maintenance in ALT tumour cells. Nat Commun. (2014) 5:5220. doi: 10.1038/ncomms6220
79. Clemson CM, McNeil JA, Willard HF, Lawrence JB. XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. J Cell Biol. (1996) 132:259–75. doi: 10.1083/jcb.132.3.259
80. Postepska-Igielska A, Giwojna A, Gasri-Plotnitsky L, Schmitt N, Dold A, Ginsberg D, et al. LncRNA khps1 regulates expression of the proto-oncogene SPHK1 via triplex-mediated changes in chromatin structure. Mol Cell. (2015) 60:626–36. doi: 10.1016/j.molcel.2015.10.001
81. Ahn JH, Lee HS, Lee JS, Lee YS, Park JL, Kim SY, et al. nc886 is induced by TGF-β and suppresses the microRNA pathway in ovarian cancer. Nat Commun. (2018) 9:1166. doi: 10.1038/s41467-018-03556-7
82. Jiang L, Shao C, Wu QJ, Chen G, Zhou J, Yang B, et al. NEAT1 scaffolds RNA-binding proteins and the Microprocessor to globally enhance pri-miRNA processing. Nat Struct Mol Biol. (2017) 24:816–24. doi: 10.1038/nsmb.3455
83. Kleaveland B, Shi CY, Stefano J, Bartel DP. A network of noncoding regulatory RNAs acts in the mammalian brain. Cell. (2018) 174:350–362.e17. doi: 10.1016/j.cell.2018.05.022
84. Zealy RW, Fomin M, Davila S, Makowsky D, Thigpen H, McDowell CH, et al. Long noncoding RNA complementarity and target transcripts abundance. Biochim Biophys Acta Gene Regul Mech. (2018) 1861:224–34. doi: 10.1016/j.bbagrm.2018.02.001
85. Schmidt K, Weidmann CA, Hilimire TA, Yee E, Hatfield BM, Schneekloth JS, et al. Targeting the oncogenic long non-coding RNA SLNCR1 by blocking its sequence-specific binding to the androgen receptor. Cell Rep. (2020) 30:541–554.e5. doi: 10.1016/j.celrep.2019.12.011
86. Zhou B, Yang H, Yang C, Bao YL, Yang SM, Liu J, et al. Translation of noncoding RNAs and cancer. Cancer Lett. (2021) 497:89–99. doi: 10.1016/j.canlet.2020.10.002
87. Wu P, Mo Y, Peng M, Tang T, Zhong Y, Deng X, et al. Emerging role of tumor-related functional peptides encoded by lncRNA and circRNA. Mol Cancer. (2020) 19:22. doi: 10.1186/s12943-020-1147-3
88. Jackson R, Kroehling L, Khitun A, Bailis W, Jarret A, York AG, et al. The translation of non-canonical open reading frames controls mucosal immunity. Nature. (2018) 564:434–8. doi: 10.1038/s41586-018-0794-7
89. Zhang Y, Wang X, Hu C, Yi H. Shiny transcriptional junk: lncRNA-derived peptides in cancers and immune responses. Life Sci. (2023) 316:121434. doi: 10.1016/j.lfs.2023.121434
90. Tseng YY, Moriarity BS, Gong W, Akiyama R, Tiwari A, Kawakami H, et al. PVT1 dependence in cancer with MYC copy-number increase. Nature. (2014) 512:82–6. doi: 10.1038/nature13311
91. Liu J, Peng WX, Mo YY, Luo D. MALAT1-mediated tumorigenesis. Front Biosci (Landmark Ed). (2017) 22:66–80. doi: 10.2741/4472
92. Tsai MC, Manor O.Wan Y, Mosammaparast N, Wang JK, Lan F, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. (2010) 329:689–93. doi: 10.1126/science.1192002
93. Lin A, Li C, Xing Z, Hu Q, Liang K, Han L, et al. The LINK-A lncRNA activates normoxic HIF1α signalling in triple-negative breast cancer. Nat Cell Biol. (2016) 18:213–24. doi: 10.1038/ncb3295
94. Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. (2011) 147:1537–50. doi: 10.1016/j.cell.2011.11.055
95. Pauli A, Valen E, Lin MF, Garber M, Vastenhouw NL, Levin JZ, et al. Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res. (2012) 22:577–91. doi: 10.1101/gr.133009.111
96. Okazaki Y, Furuno M, Kasukawa T, Adachi J, Bono H, Kondo S, et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. (2002) 420:563–73. doi: 10.1038/nature01266
97. Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. (2009) 458:223–7. doi: 10.1038/nature07672
98. Feyder M, Goff LA. Investigating long noncoding RNAs using animal models. J Clin Invest. (2016) 126:2783–91. doi: 10.1172/JCI84422
99. Washietl S, Kellis M, Garber M. Evolutionary dynamics and tissue specificity of human long noncoding RNAs in six mammals. Genome Res. (2014) 24:616–28. doi: 10.1101/gr.165035.113
100. Hason M, Bartůněk P. Zebrafish models of cancer-new insights on modeling human cancer in a non-mammalian vertebrate. Genes (Basel). (2019) 10:935. doi: 10.3390/genes10110935
101. Nasevicius A, Ekker SC. Effective targeted gene ‘knockdown’ in zebrafish. Nat Genet. (2000) 26:216–20. doi: 10.1038/79951
102. Hosono Y, Niknafs YS, Prensner JR, Iyer MK, Dhanasekaran SM, Mehra , et al. Oncogenic role of THOR, a conserved cancer/testis long non-coding RNA. Cell. (2023) 186:4254–5. doi: 10.1016/j.cell.2023.08.025
103. Zhou L, Jiang J, Huang Z, Jin P, Peng L, Luo M, et al. Hypoxia-induced lncRNA STEAP3-AS1 activates Wnt/β-catenin signaling to promote colorectal cancer progression by preventing m(6)A-mediated degradation of STEAP3 mRNA. Mol Cancer. (2022) 21:168. doi: 10.1186/s12943-022-01638-1
104. Shen W, Pu J, Zuo Z, Gu S, Sun J, Tan B, et al. The RNA demethylase ALKBH5 promotes the progression and angiogenesis of lung cancer by regulating the stability of the LncRNA PVT1. Cancer Cell Int. (2022) 22:353. doi: 10.1186/s12935-022-02770-0
105. Fan C, González-Prieto R, Kuipers TB, Vertegaal ACO, van Veelen PA, Mei H, et al. The lncRNA LETS1 promotes TGF-β-induced EMT and cancer cell migration by transcriptionally activating a TβR1-stabilizing mechanism. Sci Signal. (2023) 16:eadf1947. doi: 10.1126/scisignal.adf1947
106. Kwiatkowska I, Hermanowicz JM, Iwinska Z, Kowalczuk K, Iwanowska J, Pawlak D. Zebrafish-an optimal model in experimental oncology. Molecules. (2022) 27:4223. doi: 10.3390/molecules27134223
107. Cho YS, Jung HJ, Seok SH, Payumo AY, Chen JK, Kwon HJ. Functional inhibition of UQCRB suppresses angiogenesis in zebrafish. Biochem Biophys Res Commun. (2013) 433:396–400. doi: 10.1016/j.bbrc.2013.02.082
108. Miao KZ, Kim GY, Meara GK, Qin X, Feng H. Tipping the scales with zebrafish to understand adaptive tumor immunity. Front Cell Dev Biol. (2021) 9:660969. doi: 10.3389/fcell.2021.660969
Keywords: zebrafish model, anti-tumor research, cancer therapy, lncRNAs, immunotherapy
Citation: Hu C, Sun L, Chen J, Lyu Z, Yuan C and Jiang X (2024) Advantages of the zebrafish tumor xenograft model: the evaluation of efficacy in cancer therapy and the application to the study of lncRNAs. Front. Immunol. 15:1483192. doi: 10.3389/fimmu.2024.1483192
Received: 19 August 2024; Accepted: 11 September 2024;
Published: 30 September 2024.
Edited by:
Xudong Zhu, University of Kentucky, United StatesReviewed by:
Jieyi Li, Shanghai Yunying Medical Technology Co., Ltd, ChinaJiawen Bu, China Medical University, China
Copyright © 2024 Hu, Sun, Chen, Lyu, Yuan and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaofeng Jiang, eGZqaWFuZ0B6c3R1LmVkdS5jbg==; Chen Yuan, MTI0MzY2MDM2M0BxcS5jb20=
 Chengwu Hu1
Chengwu Hu1 Xiaofeng Jiang
Xiaofeng Jiang