- 1Department of Otorhinolaryngology, The Affiliated Fengcheng Hospital of Yichun University, Fengcheng, Jiangxi, China
- 2Department of Urology, The First People's Hospital of Xiushui, Jiujiang, Jiangxi, China
- 3The First Clinical College, Gannan Medical University, Ganzhou, Jiangxi, China
In recent years, the role of gut microbiota (GM) in bladder cancer has attracted significant attention. Research indicates that GM not only contributes to bladder carcinogenesis but also influences the efficacy of adjuvant therapies for bladder cancer. Despite this, interventions targeting GM have not been widely employed in the prevention and treatment of bladder cancer, mainly due to the incomplete understanding of the complex interactions between the host and gut flora. Simultaneously, aberrantly expressed non-coding RNAs (ncRNAs) have been frequently associated with bladder cancer, playing crucial roles in processes such as cell proliferation, invasion, and drug resistance. It is widely known that the regulation of GM-mediated host pathophysiological processes is partly regulated through epigenetic pathways. At the same time, ncRNAs are increasingly regarded as GM signaling molecules involved in GM-mediated epigenetic regulation. Accordingly, this review analyzes the ncRNAs that are closely related to the GM in the context of bladder cancer occurrence and treatment, and summarizes the role of their interaction with the GM in bladder cancer-related phenotypes. The aim is to delineate a regulatory network between GM and ncRNAs and provide a new perspective for the study and prevention of bladder cancer.
1 Introduction
Bladder cancer is one of the most prevalent tumors in urological surgery, exhibiting a high incidence globally. In 2018, there were 430,000 new cases and 165,000 deaths worldwide. By 2020, the number of new cases had increased to 573,278 (1, 2). Urothelial carcinoma is the most common pathological type of bladder cancer. Non-muscle invasive bladder cancer (NMIBC) is the most frequently encountered variant in clinical practice and accounts for approximately three-quarters of cases (3). Fortunately, current treatment strategies for NMIBC result in a 5-year survival rate exceeding 90%. Nevertheless, the management of NMIBC is suboptimal, as indicated by recurrence rates ranging from 15% to 61% at one year and from 31% to 78% at five years (4). As a result, patients with NMIBC who have undergone transurethral resection of bladder tumor (TURBT) are subjected to prolonged cystoscopic surveillance, which significantly increases their physical discomfort and economic burden. The high incidence and recurrence rates of bladder cancer underscore the urgent need to clarify its underlying mechanisms and explore potential therapeutic strategies for both prevention and treatment.
The gut microbiome (GM) can be regarded as an independent system within humans and animals, comprising a highly complex array of components. It plays crucial roles in nutrient absorption, vitamin synthesis, bile acid and sterol metabolism, as well as in immune modulation and the maintenance of intestinal homeostasis (5, 6). Under normal circumstances, the intestinal microflora and the host maintain a mutually beneficial relationship. The composition of intestinal bacteria is closely related to a specific environment, which includes pH, oxygen level/redox state, nutrients, and humidity/temperature. Maintaining this environment is closely tied to the interaction between various intestinal microflora and the host (7). Deviation from its normal composition suggests the occurrence or progression of certain diseases, including bladder cancer. Advances in GM sequencing and the application of Mendelian randomization have led to the identification of numerous intestinal microorganisms associated with the progression of bladder cancer. These will be elaborated on in detail later. Overall, the role of GM in bladder cancer has attracted increasing attention. It is noteworthy that as early as 1984, a study (8) had confirmed that chronic bacterial infection of the bladder can lead to bladder epithelial tumor-like changes. Bladder bacterial infection leads to frequent damage and repair of bladder epithelial cells through immune inflammatory responses, thereby increasing the risk of gene mutation and malignant transformation of cells (9). In addition, bacterial infection can also activate NF-κB and other cancer-promoting signaling pathways, further promoting the growth, survival, and metastasis of tumor cells (10). However, with the progression of research, it has been found that the gut and urine microbiota have a certain correlation. For example, GM can influence the bacterial susceptibility of the bladder through the gut-bladder axis (11, 12). There is also evidence (13) that the gastrointestinal tract forms a connection with the urinary microbiome through bacterial migration and blood circulation. It is proposed that the induction of bladder cancer by urinary microorganisms may also be regarded as one aspect related to GM. Overall, the role of GM in bladder cancer has continued to attract increasing attention.
Non-coding RNA (ncRNA) is a class of RNA that does not encode proteins. Its discovery is regarded as an important breakthrough in life sciences (14). The completion of the Human Genome Project (HGP) and the progress of the ENCODE project have revealed that protein-coding genes account for less than 2% of the genome. Meanwhile, approximately 90% of the human genome is transcribed into ncRNA (15, 16). Increasing evidence shows that ncRNA plays crucial roles in cellular development, physiological functions, and disease progression (16, 17). Based on their length and structure, ncRNAs are classified into small ncRNAs, circular RNAs (CircRNA), and long ncRNAs (lncRNA). Small ncRNAs comprise microRNA (miRNA), ribosomal RNA (rRNA), and transfer RNA (tRNA). These molecules act as key regulators in gene expression processes such as protein translation and post-transcriptional silencing (18, 19). Consequently, ncRNAs have been intensively studied for their significant roles in regulating human diseases, especially cancer development (20). Additionally, ncRNAs exhibit superior histocompatibility and better tissue penetration compared to traditional small molecule drugs, rendering them promising candidates for cancer therapeutics (21). The interaction between the host and GM forms a complex and intricate regulatory network. However, research in this field is still in its early stages. Notably, ncRNAs have emerged as important mediators in this communication. The role of ncRNAs in host-microbiome interactions, as well as their influence on intestinal homeostasis and related cancers, is attracting increasing attention. Accordingly, this paper examines the differential expression of various ncRNAs in bladder cancer and their potential impacts on tumor development. Additionally, we study the interactions between ncRNAs and GM and their implications in tumorigenesis. By clarifying these relationships, we aim to provide insights into the prevention and treatment of bladder cancer and identify potential biomarkers.
2 Role of gut microbiota in bladder cancer
2.1 Gut microbiota and the genesis of bladder cancer
The first intuitive evidence stems from the age of onset of bladder cancer. Data suggest that approximately 75% of newly diagnosed bladder cancer patients are over 65 years old, and around 45% are over 75 years old, classifying it as a disease primarily affecting the elderly (22). Furthermore, the GM undergoes changes with age. Young-derived GM has been demonstrated to restore the health of the elderly by altering the microbial composition of their intestines (23). This leads us to thoroughly question the changes in GM during the occurrence of bladder cancer and its impact on tumors. Several research findings uphold this perspective. A case-control study (24) revealed that bladder cancer patients demonstrated an imbalance in the GM, primarily characterized by a reduction in Clostridium cluster XI and Prevotella, along with decreased butyric acid concentrations and impaired intestinal barrier integrity. In addition, another study (25) found that a soluble high-fiber diet resulted in delayed tumor growth following irradiation compared to other groups. Moreover, rats on the soluble high-fiber diet showed a significantly higher relative abundance of Bacteroides, further corroborating the close relationship between GM and tumor progression. With the continuous progress in GM sequencing and Mendelian randomization research, an increasing number of studies are investigating the differential expression of GM-related genes in bladder cancer patients. A bidirectional Mendelian randomization study (26) identified associations between bladder cancer and several genera, such as Allisonella, Lachnospiraceae, Oscillibacter, Eubacterium coprostanoligenes group, Eubacterium ruminantium group, Ruminococcaceae, and Senegalimassilia. Similarly, Wang et al. (27) employed a comparable method and concluded that higher abundances of Bifidobacterium, Actinobacteria, and Ruminococcus torques in the gut are associated with an elevated risk of bladder cancer. However, another study (28) identified that Lachnospiraceae, Desulfovibrionales (Order), Eubacterium ruminantium group, Olsenella, and Ruminococcaceae have causal impacts on bladder cancer. Conversely, Bacteroidetes (Phylum), Eubacteriumbrachy group, Ruminococcaceae, Rikenellaceae (Family), Lachnospiraceae group, and Adlercreutzia exhibit a protective effect against bladder cancer. The final evidence relates to the preventive effect of sulforaphane on bladder cancer. Research (29) has demonstrated that sulforaphane can restore the disrupted intestinal microbiota in N-butyl-N-(4-hydroxybutyl)-nitrosamine-induced bladder cancer mice, repair physiological damage to the intestinal barrier, reduce inflammation and immune responses, and thus prevent chemical-induced bladder cancer. Overall, various abnormally expressed GMs have been found to be closely related to the development of bladder cancer, potentially serving as both pathogenic and protective factors. Moreover, correcting the GM can significantly reduce the carcinogenic impacts on bladder cancer, indicating that a healthy GM plays a crucial role in reducing the risk of bladder cancer.
2.2 Impact of gut microbiota on bladder cancer treatment
Chemotherapy and immunotherapy constitute key treatment modalities for bladder cancer. As emerging therapeutic approaches, they offer the advantage of specifically targeting tumor cells, alleviating symptoms, and prolonging survival when compared with traditional treatments (30). However, their efficacy varies considerably among individuals, being influenced by gene expression and the body’s immune status. Additionally, chemotherapy drug resistance remains a significant challenge that affects treatment outcomes. The role of the GM in bladder cancer is crucial, not only in the disease’s progression but also in modulating the efficacy of chemotherapy and immunotherapy. Evidence supporting this view is provided by studies investigating the relationship between antibiotic use and the survival rates of patients with urothelial carcinoma undergoing chemotherapy and immunotherapy. Data from Ashley et al. (31) indicated that antibiotic use in patients treated with atezolizumab was associated with decreased survival rates. In contrast, no such association was observed in patients receiving chemotherapy alone. This implies that excessive antibiotic use may disrupt the intestinal microbiota and specifically reduce the efficacy of cancer immunotherapy. Additionally, a multi-institutional prospective cohort study examined differences in the GM between bladder cancer patients and healthy adults, as well as variations in GM composition among patients with different responses to chemotherapy. The study (32) found that patients with higher levels of Bacteroides had a worse response to neoadjuvant chemotherapy. Similarly, Lukas et al. (33) found that Bifidobacterium pseudolongum, Lactobacillus johnsonii, and Olsenella species significantly increased the efficacy of immune checkpoint inhibitors in mouse models of rectal cancer, bladder cancer, and melanoma. Further research has confirmed that this effect is mediated by adenosine, a metabolite of the GM. Another related study (34) found that GM can influence the levels of ketone bodies, such as 3-hydroxybutyrate (3HB), which in turn significantly impacts the anti-tumor efficacy of PD-1 inhibitors. Additionally, fecal microbiota transplantation and single colonization experiments were performed with Parabacteroides distasonis to assess the efficacy of combined treatment with immune checkpoint inhibitors (ICIs). The results indicated that this combination therapy increased the intratumoral density of CD4+ and CD8+ T cells and significantly postponed tumor growth. Transcriptome analysis further revealed that the inclusion of Parabacteroides distasonis significantly upregulated various pathways related to anti-tumor immune responses (35).
The aforementioned evidence indicates that the GM is closely associated with the onset, progression, and treatment of bladder cancer. Bladder cancer development is often accompanied by dysbiosis. Correcting the GM can not only prevent bladder cancer but also improve the effectiveness of immunotherapy. However, understanding how bladder cancer progression influences changes in the GM and the mechanisms by which the GM modulates treatment efficacy remains a critical issue. It is well-known that the interaction between the host and the GM is crucial for maintaining intestinal homeostasis and immune responses. Furthermore, epigenetic regulation is regarded as a significant mechanism through which the microbiota can modify host physiology. Host-intestinal microbiota communication is essential for understanding the relationship between the GM and bladder cancer. Increasing evidence (36, 37) indicates that ncRNAs serve as a significant communication medium between the host and the intestinal microbiota, influencing the host’s pathophysiological processes. The role of ncRNAs in host-microbiome interactions, as well as their influence on intestinal homeostasis and related cancers, is attracting considerable attention. Accordingly, we have examined the differential expression of various non-coding RNAs in bladder cancer and their potential impacts on tumor progression. Additionally, we have explored the interactions between non-coding RNAs and the GM, and their roles in tumor development. Clarifying these relationships offers valuable insights for bladder cancer prevention, treatment, and the identification of potential biomarkers.
3 Another perspective, ncRNA in bladder cancer
3.1 miRNA
MiRNA was discovered in 1993. It is a single-stranded ncRNA with a length of approximately 20–22 nucleotides. MicroRNAs primarily regulate gene expression post-transcriptionally in various organisms, exerting an influence on processes such as cell growth, proliferation, differentiation, and apoptosis (38, 39). It is estimated that microRNAs regulate approximately 60% of the human transcriptome (40). The primary microRNA (pri-miRNA) is processed into precursor microRNA (pre-miRNA) by a microprocessor complex comprising the RNase III enzyme Drosha and the double-stranded RNA-binding protein DGCR8. Subsequently, the pre-miRNA is transported by the exportin XPO5, cleaved by the nuclease Dicer, and integrated into the RNA-induced silencing complex (RISC). When the microRNA is complementary to the 3’ untranslated region (3’ UTR) of the target mRNA, the RNA-induced silencing complex (RISC) inhibits protein translation and promotes mRNA degradation by endonucleases (38, 41, 42). Thus, microRNA plays a crucial role in regulating post-transcriptional gene silencing. As a novel tumor marker and therapeutic target, microRNA has attracted significant attention. Research indicates that microRNAs are crucial in tumor initiation, progression, and metastasis. Certain microRNAs function as tumor suppressors by downregulating oncogene expression, thereby inhibiting tumor growth and proliferation. Conversely, other microRNAs act as oncogenes, promoting tumor cell growth, invasion, and metastasis (43–45). Additionally, microRNAs can influence the tumor microenvironment, modulate interactions between tumor cells and stromal cells, and have an impact on tumor angiogenesis and immune evasion (46, 47). These effects have also been observed in the development of bladder cancer.
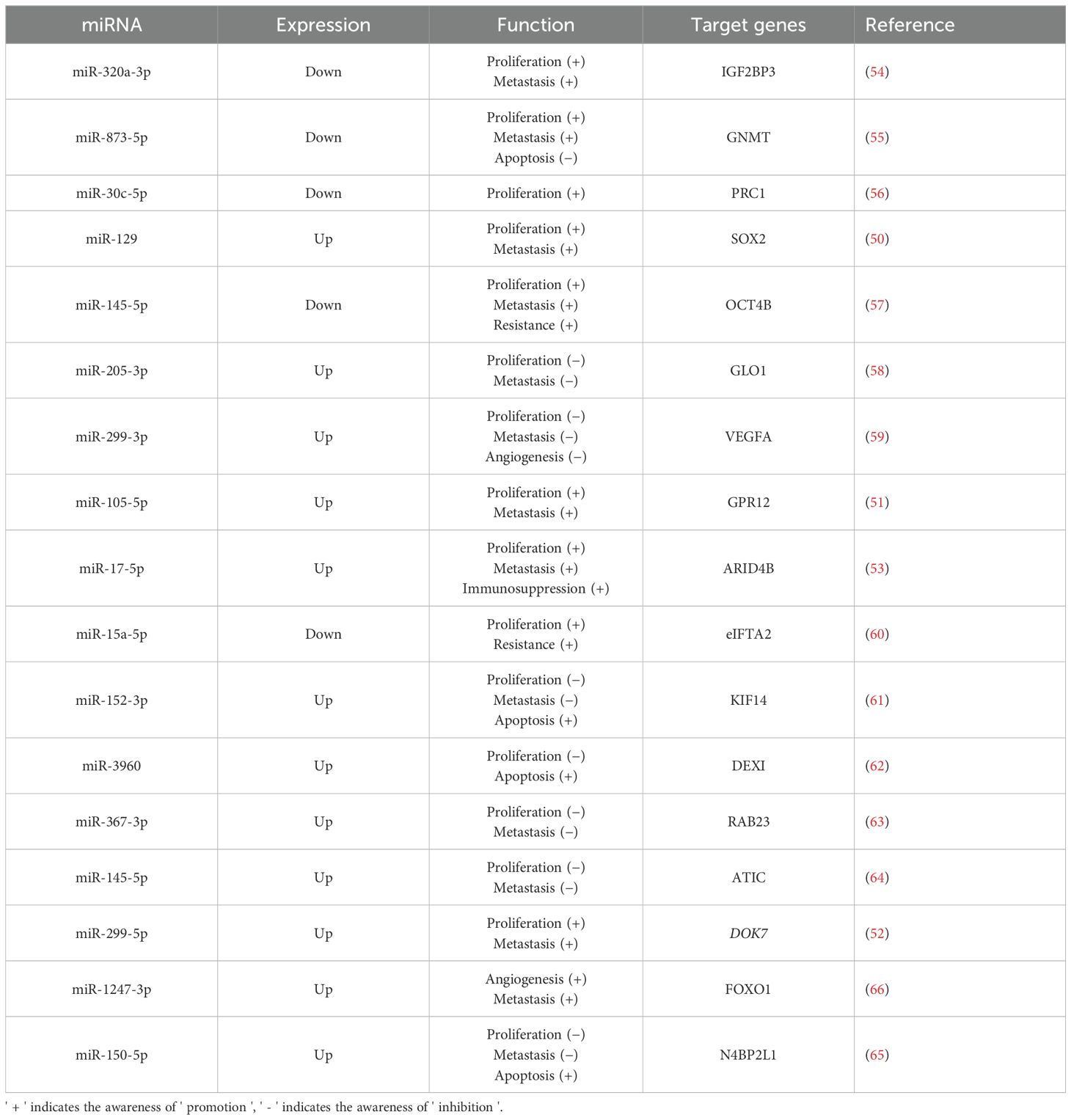
In a case-control study (48), 106 bladder urothelial carcinoma tissues were compared with normal tissues, uncovering a significant difference in the expression level of miRNA-124-3p between bladder cancer and normal samples. This indicates that miR-124-3p is closely related to bladder cancer and has potential as a predictive marker. Similarly, Yang et al. (49) examined the differential expression of urinary exosomal miRNAs in 116 bladder cancer patients compared to 116 healthy volunteers. Their results revealed elevated levels of urinary exosomal miR-146a-5p, miR-93-5p, miR-663b, miR-21, and miR-4454 in bladder cancer patients. The aforementioned studies demonstrate a strong correlation between miRNAs and bladder cancer. Furthermore, additional in vivo and in vitro experiments have examined the complex effects of various miRNAs on bladder cancer (Table 1). Excessive proliferation is a hallmark of tumor cells and a key driver of cancer progression, involving various complex signaling molecules and pathways, such as growth factors and receptors. miRNAs affect cell proliferation by targeting genes related to these signaling pathways and proteins. In bladder cancer development, several miRNAs (such as miR-12, miR-299-5p) have been discovered to promote bladder cancer cell proliferation by targeting specific genes, such as those regulating the cell cycle and promoting glycolysis (50–52). Furthermore, research (53) has demonstrated that miR-17-5p from urinary exosomes can also promote the growth and invasion of bladder cancer cells by influencing the tumor immune microenvironment. Conversely, some studies (54–65) have shown that certain miRNAs can suppress bladder cancer cell proliferation by downregulating tumor-promoting genes. Tissue invasion and metastasis are additional key characteristics of tumor cells. Various miRNAs have been demonstrated to either promote (50–53) or inhibit (54, 55, 57, 61, 63, 64) tumor cell invasion. A study (59) found that miR-299-3p suppresses bladder cancer progression by downregulating VEGFA levels and inhibiting angiogenesis. Conversely, another study (66) shows that miR-1247-3p, which is highly expressed in tumor-derived exosomes, promotes angiogenesis in bladder cancer. At present, bladder cancer treatment mainly involves surgery and adjuvant chemotherapy. The efficacy of these treatments is significantly affected by tumor cell resistance to chemotherapeutic drugs. Encouragingly, miRNAs have demonstrated potential in enhancing drug resistance in bladder cancer cells. For example, Wu and Yang et al. (57, 60) found that miR-145-5p and miR-15a-5p can respectively counteract chemoresistance mediated by OCT4B and eIF5A2, thereby enhancing the effectiveness of chemotherapy. In general, various miRNAs have been demonstrated to affect bladder cancer cell characteristics by regulating the expression of specific target genes, thereby influencing the progression of the disease. Generally, a large number of miRNAs are differentially expressed in bladder cancer, and they are demonstrated to target genes closely related to the proliferation and invasion of bladder cancer. It should be noted that the effect of miRNA on bladder cancer is not always promoting or inhibiting, which depends on the type of miRNA and the stage of tumor development. Moreover, miRNA can also enhance the drug resistance of bladder cancer cells and increase the effectiveness of chemotherapy.
3.2 lncRNA
LncRNA is characterized as RNA with more than 200 nucleotides that does not undergo translation into proteins. Similar to mRNA, the majority of lncRNAs are transcribed by RNA polymerase II (RNA Pol II) and then degraded by exonucleases or processed by exosomes (42, 67). Moreover, certain lncRNAs undergo a maturation process similar to that of most transfer RNAs and are commonly cleaved by the RNase P enzyme (68). In contrast to small RNA molecules, lncRNAs possess longer sequences and complex spatial configurations. Utilizing Watson-Crick base pairing, lncRNAs can adopt secondary or tertiary three-dimensional structures, enabling them to perform RNA-related functions through nucleic acid complementarity and exhibit protein-like functions through their three-dimensional spatial arrangements (69, 70). This complex interplay highlights the diverse and complex role of lncRNAs in regulating gene expression mechanisms. The functional modes of lncRNAs can be broadly classified into four types (signal, decoy, guide, and scaffold) (71, 72). Signal: Some lncRNAs are transcribed in response to specific stimuli and regulate one or more target genes (either in a cis- or trans-acting manner) (73, 74). Guide: lncRNAs can function in a manner similar to molecular chaperones by binding to proteins (usually transcription factors), directing these proteins to specific DNA sequences, and thereby regulating downstream molecular transcription (either in a cis- or trans-acting manner) (75, 76). Decoy: This class of lncRNAs, upon transcription, binds directly to RNAs or transcription factors, thereby suppressing their activity and associated signaling pathways (77, 78). Scaffold: Moreover, lncRNAs can function as a “central platform” to which multiple related transcription factors can attach. Consequently, when multiple signaling pathways are simultaneously activated within a cell or organism, these downstream effector molecules can interact with the same lncRNA. This interaction promotes the convergence and integration of information from different signaling pathways, enabling the cell or organism to rapidly generate feedback and regulate responses to external signals and stimuli (79, 80). Moreover, as research progresses, lncRNAs are being investigated for their increasingly complex roles in transcriptional regulation. This paper also examines their associated functions in bladder cancer.
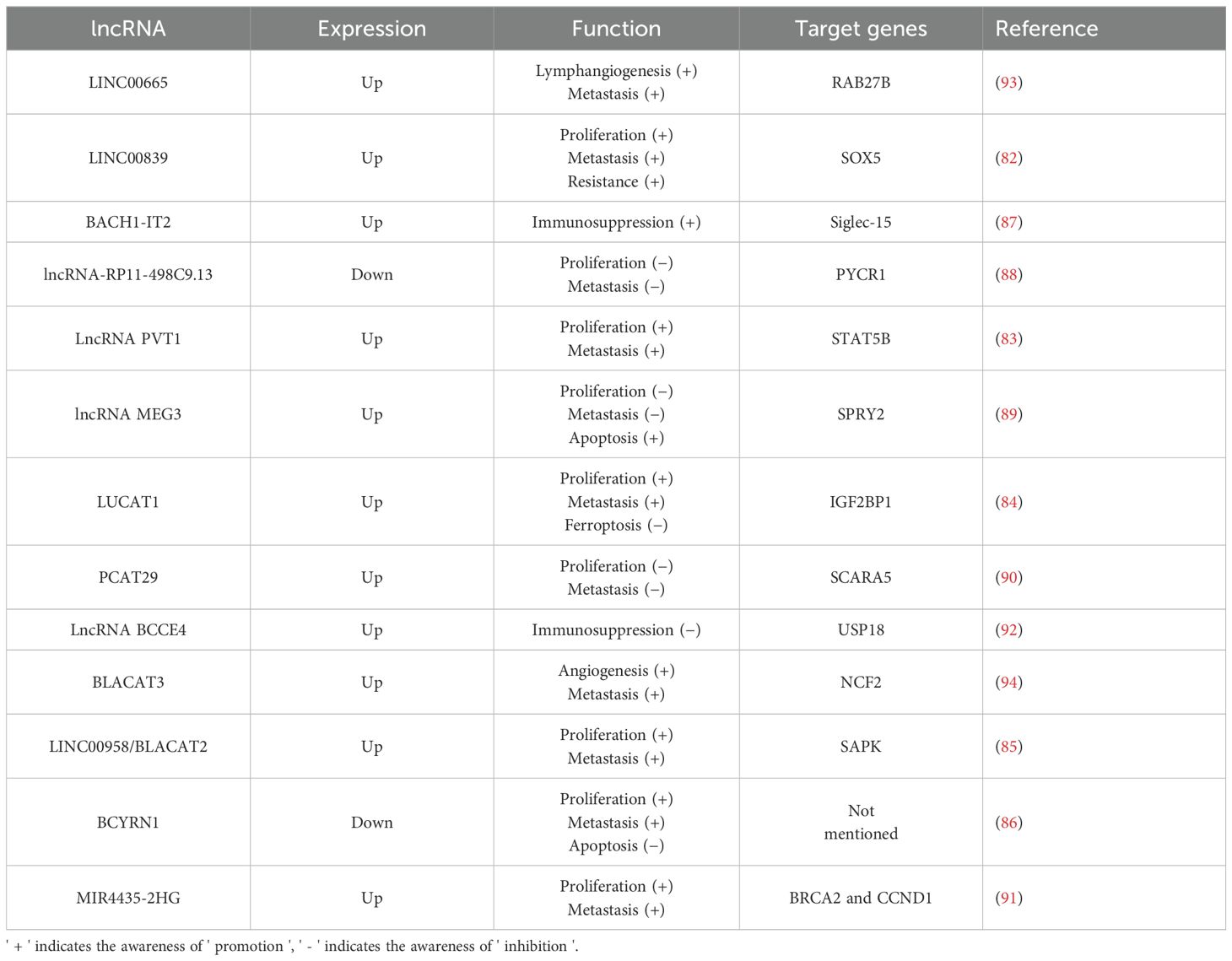
A meta-analysis investigating the relationship between lncRNA MALAT1 expression and prognosis in bladder cancer found that elevated levels of lncRNA MALAT1 are associated with a poor prognosis and an increased risk of lymph node metastasis in these patients (81). Furthermore, numerous studies have elucidated the significant role of lncRNAs in the development of bladder cancer (Table 2). Several lncRNAs, including LINC00839, lncRNA PVT1, LUCAT1, LINC00958, and BCYRN1, exert a positive influence on both the proliferative viability and invasive capabilities of bladder cancer cells. These lncRNAs interact with specific target genes, such as SOX5, enhancing cell viability, reducing apoptosis, and promoting metastasis by facilitating epithelial-mesenchymal transition (EMT) (82–86). Moreover, it has been found that BACH1-IT2, a lncRNA, contributes to the progression of bladder cancer by stabilizing Siglec-15, which in turn suppresses the local tumor immune microenvironment (87). In contrast, some lncRNAs (such as lncRNA-RP11-498C9.13, lncRNA MEG3, PCAT29, MIR4435-2HG) have been demonstrated to significantly inhibit the proliferation and invasion of bladder cancer cells by targeting downstream genes (88–91). Furthermore, lncRNA BCCE4 has been demonstrated to enhance the anti-tumor immune response and suppress bladder tumorigenesis by potentiating the PD-L1/PD-1 interaction in smoking-associated bladder cancer (92). Distant metastasis is a major cause of mortality in bladder cancer patients. Those with high malignancy are particularly susceptible to lymphatic and bloodstream metastases. Li et al. (93) identified a correlation between LINC00665 and both lymph node metastasis and poor prognosis in bladder cancer patients. Subsequent research revealed that LINC00665 enhances RAB27B expression, thereby inducing a RAB27B-HGF-c-Myc positive feedback loop that promotes lymphangiogenesis and lymph node metastasis in bladder cancer. In addition, lncRNAs promoting blood metastasis have also been reported. BLCa-associated transcript 3 (BLACAT3), an abnormally up-regulated lncRNA in bladder cancer, was found to promote blood metastasis by enhancing bladder cancer angiogenesis (94). Similar to miRNAs, lncRNAs have been implicated in inducing drug resistance in bladder cancer cells. Specifically, LINC00839 regulates SOX5 either directly or indirectly through the miR-142 axis, which increases the resistance of bladder cancer cells to gemcitabine by promoting autophagy (82). Similar to miRNA, many differentially expressed lncRNAs have been identified in bladder cancer, which play an important role in regulating tumor proliferation, invasion, and drug resistance. It is noteworthy that in addition to these conventional tumor phenotypes, some differentially expressed lncRNAs have been found to be associated with blood and lymphatic metastasis of bladder cancer, suggesting that these lncRNAs can be not only used as intervention targets to prevent tumor progression but also serve as potential prognostic markers.
3.3 circRNA
CircRNAs represent a subclass of noncoding RNA molecules lacking a 5’ terminal cap and a 3’ terminal poly(A) tail. They form a covalently bonded ring structure through the variable shearing of a distinctively terminally reverse-complementary precursor mRNA, undergoing head-to-tail cyclization (95). This distinctive loop configuration endows enhanced stability compared to linear RNAs. Despite their typically low expression levels, the remarkable durability of circRNAs enables cellular accumulation to levels exceeding those of corresponding linear RNAs (96–98). Among their diverse functions, circRNAs are prominently acknowledged for their involvement in miRNA sponging. CircRNAs sequester miRNAs by binding to complementary sequences, thereby hindering miRNAs from targeting mRNAs. This liberation of mRNAs from miRNA inhibition enables subsequent ribosomal binding and eventual protein translation (99). Furthermore, circRNAs have been reported to modulate alternative splicing or transcription processes and regulate the gene expression of their parental genes by inhibiting transcription start sites (100). Ongoing investigations are continuously uncovering additional functions of circRNAs. With an expanding set of endogenous circRNAs linked to cancer, nonetheless, many of these molecules remain unexplored with regard to their functional significance (101).
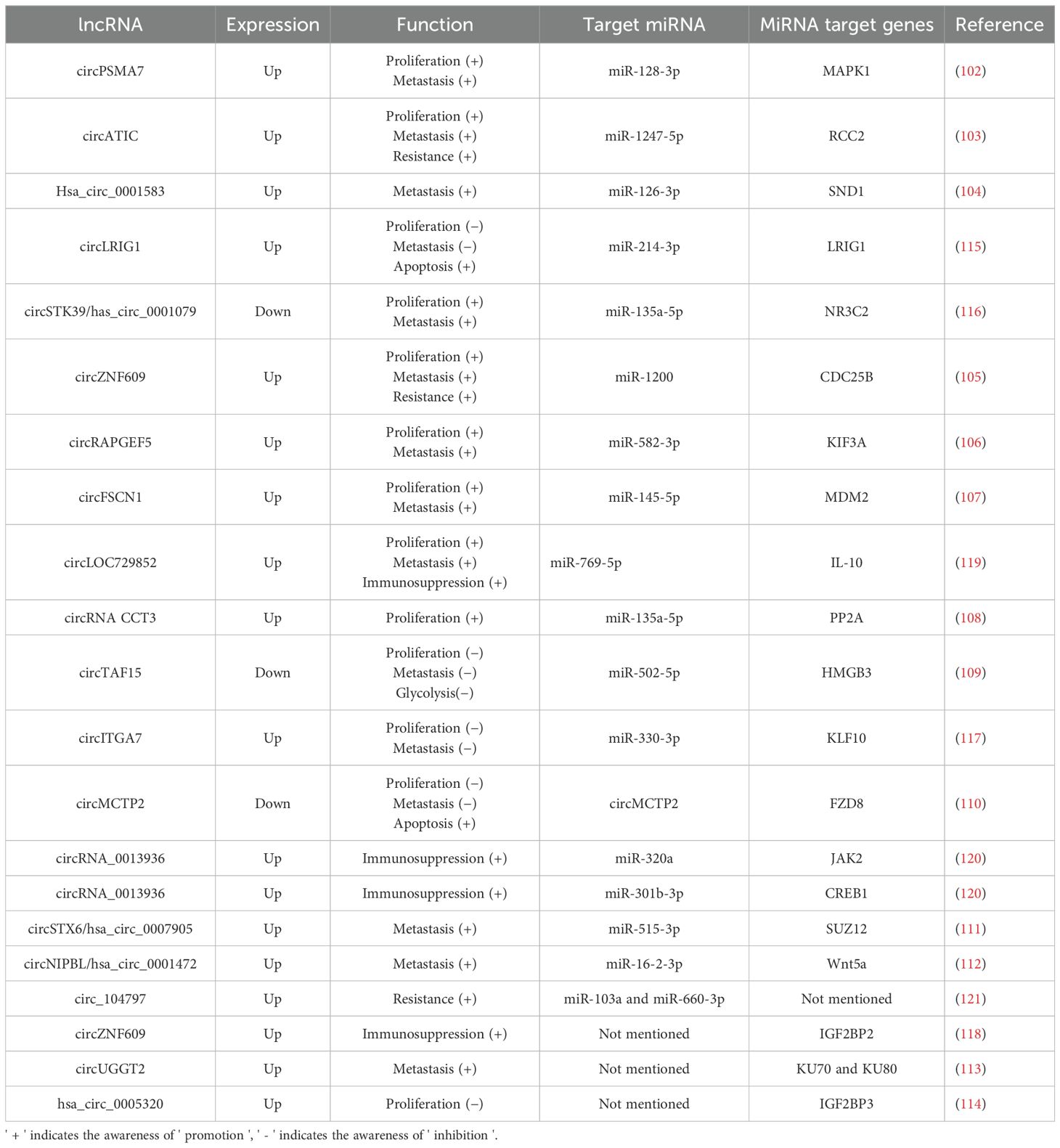
Among studies on the roles of circRNAs in bladder cancer, the most extensive research centers on circRNAs functioning as miRNA sponges (Table 3). Since various miRNAs are known to regulate cell viability and invasive potential in bladder cancer, it is reasonable to deduce that numerous circRNAs also significantly affect the proliferation, apoptosis, and invasion of bladder cancer cells. Generally, abnormally expressed circRNAs contribute to the development of bladder cancer primarily by acting as miRNA sponges, which promotes cell proliferation and invasion through mechanisms such as cell cycle regulation, impacts on cell viability, and epithelial-mesenchymal transition (101–114). Additionally, several circRNAs have been investigated as potential biomarkers for bladder cancer prediction. Notably, some overexpressed circRNAs, including circLRIG1, circSTK39, and circITGA7, have shown a significant ability to inhibit bladder cancer growth and metastasis in vivo and ex vivo, suggesting they may have a protective function (115–117). In the context of tumor immunity regulation, circZNF609 has been identified as a factor that diminishes the efficacy of immunotherapy for bladder cancer. This occurs through the upregulation of IGF2BP2, which facilitates immune escape of bladder cancer cells (118). Similarly, research indicates that circLOC729852 affects autophagy signaling in bladder cancer cells and augments the recruitment and M2 polarization of tumor-associated macrophages, thereby promoting further tumor growth (119). Conversely, increased expression of circRNA_0013936 has been reported to suppress bladder cancer cells by activating JAK2 and CREB1. This activation promotes the production of immunosuppressive cytokines and enhances anti-tumor immunity (120). Additionally, several circRNAs, including circATIC, circZNF609, and circ_104797, have been demonstrated to increase resistance to cisplatin and promote further progression of bladder cancer (103, 105, 121). Unlike the above two types of RNA which directly target bladder cancer-related genes, research on circRNA in bladder cancer mainly centers on its role as a miRNA sponge. Abnormally expressed circRNAs promote the occurrence of bladder cancer through this mechanism, such as promoting cell proliferation and invasion. However, there are also some circRNAs that exhibit inhibitory effects on bladder cancer. Additionally, some circRNAs also demonstrate the ability to regulate the anti-tumor immunity of the host, which exerts an important influence on the regulation of tumor immune resistance and treatment.
4 Bi−direction effects between microbiome and ncRNAs in tumor
As previously elaborated, despite substantial evidence indicating significant differential expression of the GM and ncRNAs in bladder cancer, several questions remain unanswered. These encompass the specific mechanisms by which the GM influences tumor development and the signals that trigger ncRNA alterations during bladder cancer progression. In other words, the mechanisms underlying the interaction between the GM and ncRNAs in bladder cancer necessitate further investigation. Epigenetic regulation is posited as a potential mechanism by which the GM may alter host pathophysiological processes, through various means including non-covalent epigenetic processes that modulate gene expression (122). As previously outlined, a complex and nuanced regulatory network exists between the host and the microbiota, and elucidating this intricate network is a key objective of our future research. Consequently, this review synthesizes the interactions of various non-coding RNAs with the GM and examines the influence of these interactions on tumor development and therapy.
4.1 miRNA-gut microbiota interactions in carcinogenesis
There is accumulating evidence that microRNA plays a crucial role in host-microbiota interactions that govern gut health. MicroRNA is increasingly regarded as a key molecule in microbiota-carcinogenesis interactions (123, 124). Initial evidence of miRNA interactions with gut flora originated from a study on Arabidopsis thaliana’s resistance to Pseudomonas syringae. Navarro et al. (125) discovered that Arabidopsis recognition of a flagellin-derived peptide from Pseudomonas syringae induces miR-393a transcription. This miRNA inhibits the growth hormone receptor, a negative regulator of the plant immune system, ultimately strengthening plant resistance to Pseudomonas syringae infection. Further studies have demonstrated that miRNA and the GM exhibit related changes in the development of human diseases, particularly in tumor-related diseases. For example, a high-starch diet or a high-fat diet can influence the level of some miRNAs while altering the specific microbial composition of the intestine, thereby potentially realizing tumor prevention or promotion (126, 127). With the deepening of research, an increasing amount of evidence (128) shows that miRNA serves as a tool for the GM and plays a role in tumor regulation.
The microbiome has been demonstrated to be associated with various types of cancer, including colorectal, gastric, hepatocellular, and pancreatic cancers (129, 130). It influences carcinogenesis via its impact on metabolism, cell proliferation, inflammation, and immunity (131, 132). Therefore, further elucidation of the interaction between the host microbiome and microRNAs is crucial for regulating the phenotypic role of bladder cancer. Regrettably, although numerous studies have demonstrated that there is a close relationship between the GM, microRNAs, and bladder cancer, there remains a dearth of direct evidence that GM affects bladder cancer through microRNAs. Therefore, we herein summarize the crucial role of the GM in a variety of tumor-related phenotypes via microRNAs. These phenotypes have been demonstrated to be closely related to the occurrence and development of bladder cancer. Consequently, we aim to provide potential evidence of microRNA bridges for the regulation of bladder cancer by the GM, and also hope to attract increased interest from researchers in this research.
Long-term bladder irritants, such as bladder stones and tuberculosis infections, can trigger a localized chronic inflammatory response in the bladder, inducing bladder epithelial cells to release various inflammatory mediators, such as interleukins and tumor necrosis factor, and attracting a variety of immune cells to infiltrate bladder tissue (22). However, these immune cells further release additional inflammatory factors, establishing a persistent chronic inflammatory environment. This inflammatory environment provides favorable conditions for bladder tumorigenesis. On the one hand, inflammatory factors can activate signaling pathways such as NF-κB, resulting in the abnormal expression of cell cycle regulatory proteins and thereby accelerating cell proliferation rates. Simultaneously, they suppress apoptosis, enabling the abnormal proliferation of cells to survive and accumulate (10). On the other hand, chronic inflammation can induce the proliferation and activation of regulatory T cells (Tregs). Tregs induce the formation of an immunosuppressive microenvironment through the secretion of immunosuppressive cytokines, such as IL-10 and TGF-β, thereby facilitating the escape of tumor cells from immune system surveillance and attack and promoting the development of bladder cancer (133). In addition, the inflammatory response can also induce the polarization of tumor-associated macrophages into M2-type macrophages. M2-type macrophages can promote tumor angiogenesis and inhibit the immune response through the secretion of various growth factors and cytokines, thereby offering favorable conditions for the growth and metastasis of tumor cells (134).
Since the discovery of the GM, it has been identified as having a close association with the immune system. This is manifested, on one hand, in its influence on host immune development and, on the other hand, in its modulation of host immunity. Early in life, colonization of the GM can assist the neonatal immune system in differentiating between self and non-self antigens and establishing immune tolerance (135). It can also exert an influence on the development and maturation of systemic immune organs such as the spleen and thymus, further regulating the overall activity of the immune system (136). In regulating host immune function, the GM can enhance intrinsic immune defense by activating Toll-like receptors and the complement system (137). It also regulates adaptive immune responses by influencing the function of intestinal epithelial cells and dendritic cells (DCs), as well as regulating antibody production and immune memory of B cells (138). These effects are crucial for the regulation of the host inflammatory response. Although the importance of the gut microbiome in the immune-inflammatory response is well established, the exact mechanism of action is still being explored.
Recent studies emphasize the significant roles of the GM and miRNAs in regulating immune inflammation (Figure 1). Toll-like receptors (TLRs) that recognize pathogen-associated molecular patterns (PAMPs) from various microorganisms activate numerous microRNAs that modulate inflammation levels, including miR-146a/b, miR-155, miR-21, and let-7. These microRNAs are known to play crucial roles in balancing the host’s inflammatory response (139–141). Pseudomonas aeruginosa infection induces miR-302b expression via a TLR/NF-κB-dependent pathway. miR-302b targets IRAK4, mitigating inflammatory exacerbation by inhibiting TLR-induced NF-κB activation (142). These findings indicate that microRNAs are novel regulators of inflammation, providing negative feedback to TLR-mediated immunity. Additionally, increased miR-301b expression following Pseudomonas aeruginosa infection has been shown to modulate inflammation. miR-301b targets c-Myb, decreasing anti-inflammatory factors IL-4 and TGF-β1, which exacerbates the inflammatory response (143). Neisseria gonorrhoeae also modulates PI3K/AKT signaling by directly targeting phosphatase and tensin homolog (PTEN) via miR-718, suppressing pro-inflammatory factor production (144). Furthermore, Brucella abortus and Mycobacterium tuberculosis inhibit the inflammatory response to enhance their survival by downregulating NF-κB pathway levels via miR-125b-5p and let-7f, respectively (145, 146). Besides influencing inflammatory factors, the GM also affects inflammation progression by altering macrophage polarization through microRNAs. For example, Mycobacterium tuberculosis infection reduces miR-26a-5p, decreasing kruppel—like factor 4 (KLF4). And promoting M2 macrophage polarization (147). While Staphylococcus aureus infection enhances M1 polarization by lowering miR-24, which activates the chi3l1-mediated MAPK pathway (148). Lastly, the GM and microRNAs influence the immune-inflammatory response by modulating inflammatory cell recruitment. Salmonella enterica infection increases miR-128 levels, which inhibits macrophage colony-stimulating factor (M-CSF) aggregation at infection sites by directly targeting it (149). Similarly, during Mycobacterium tuberculosis infection, upregulation of miR-223 reduces the expression of cytokine IL-6 and chemokines C-X-C motif chemokine 2 (CXCL2) and C-C motif chemokine ligand 3 (CCL3), inhibiting proper neutrophil recruitment to infected tissues (150).
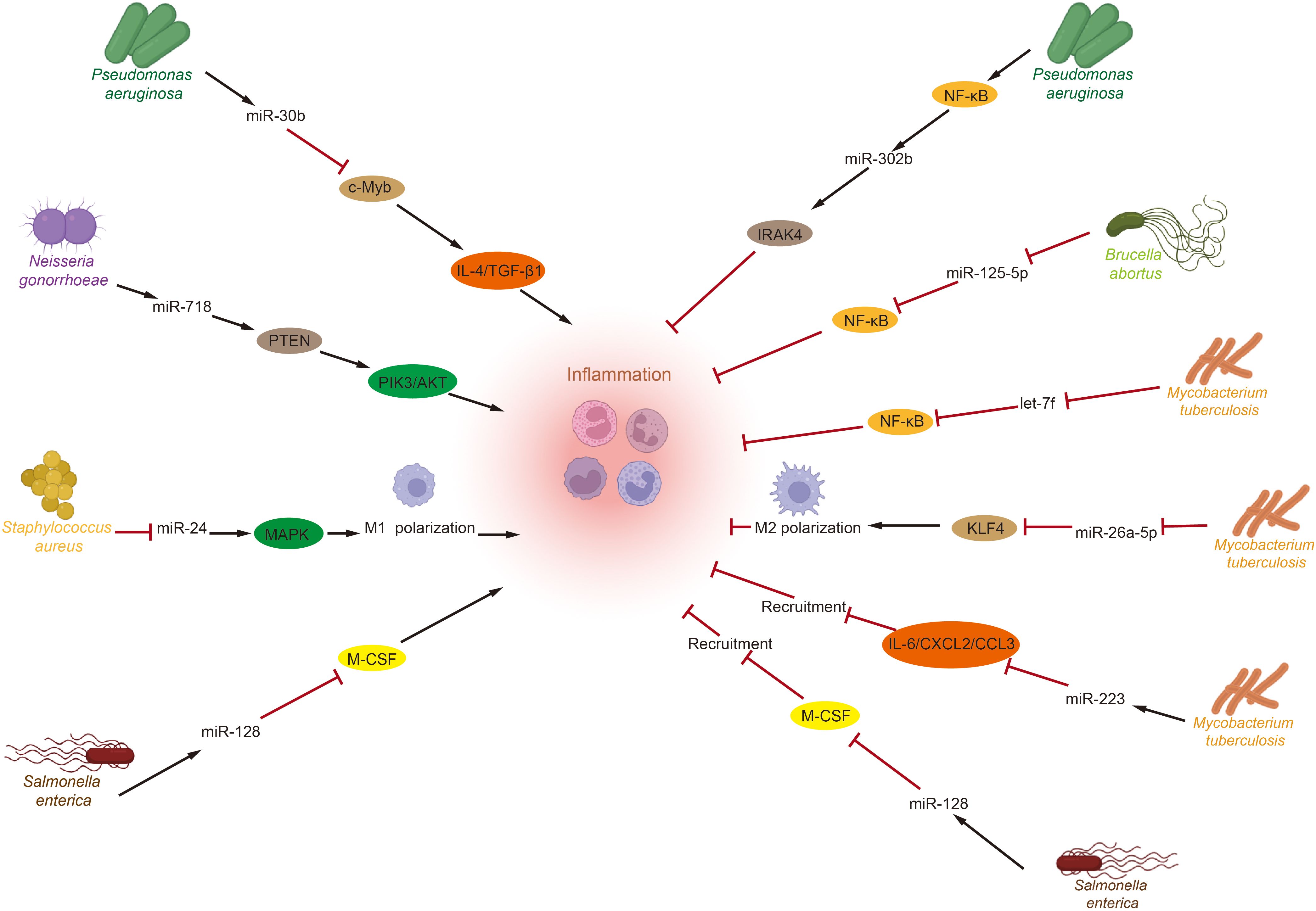
Figure 1. Crosstalk between gut microbiota and miRNAs in inflammation. After Toll-like receptors recognize pathogen-associated molecular patterns from various intestinal microorganisms, several miRNAs related to inflammation regulation are activated. These miRNAs then perform their functions by targeting specific genes, influencing pathways associated with inflammatory responses, regulating macrophage polarization, and modulating the recruitment of inflammatory cells to either promote or inhibit the progression of inflammation. (IL, Interleukin; TGF-β1, transforming growth factor-β1; PTEN, phosphatase and tensin homolog; PI3K, phosphatidylinositide 3-kinase; AKT, protein kinase B; MAPK, mitogen-activated protein kinases; M-CSF, macrophage colony-stimulating factor; IRAK4, interleukin-1 receptor-associated kinase 4; KLF4, kruppel-like factor 4; CXCL2, C-X-C motif chemokine 2; CCL3, C-C motif chemokine ligand 3).
Abnormal levels of cell proliferation and apoptosis are the most crucial factors influencing tumorigenesis. In bladder cancer tissues, abnormal levels of certain growth factors and signaling pathways were detected to be abnormally activated, and dysregulated levels of cell cycle protein expression were observed (151). Simultaneously, the regulation of cell proliferation and apoptosis is also a key target for microbial intervention, with Helicobacter pylori infection serving as a notable example. Several microRNAs, including miR-203, miR-204, miR-212-3p, miR-320, miR-361-3p, miR-375, miR-584, and miR-1290, have been implicated in Helicobacter pylori-dependent cell proliferation and tumorigenesis (152–157). Further research has shown that Helicobacter pylori infection elevates the levels of miR-21 and miR-222 in cells and tissues. These microRNAs target tumor suppressor RECK and protein kinase homeodomain interacting protein kinase 2 (HIPK2), which interacts with the RECK homology domain, thereby promoting tumor cell proliferation and invasion (158–160). This body of research suggests that microRNAs upregulated during Helicobacter pylori infection play a significant role in promoting tumor cell proliferation and development. Beyond Helicobacter pylori, other bacterial microorganisms have also been reported to influence host cell proliferation via microRNAs. For example, Salmonella infection reduces the level of miR-15, thereby inhibiting Cyclin D1 and blocking cell cycle progression (161). Additionally, Citrobacter rodentium infection increases the level of miR-203 and stimulates cell proliferation by counteracting the inhibitory effects of Wnt inhibitory factor-1 (WIF1) on the Wnt/β-catenin pathway (162). Moreover, microRNAs regulated by the GM have been shown to participate in cell death pathways. For instance, miR-582-5p and miR-155, which are upregulated following Mycobacterium tuberculosis infection, reduce apoptosis by decreasing the levels of the transcription factors forkhead box O 1 (FOXO1) and forkhead box O 3 (FOXO3), respectively (163, 164) (Figure 2).
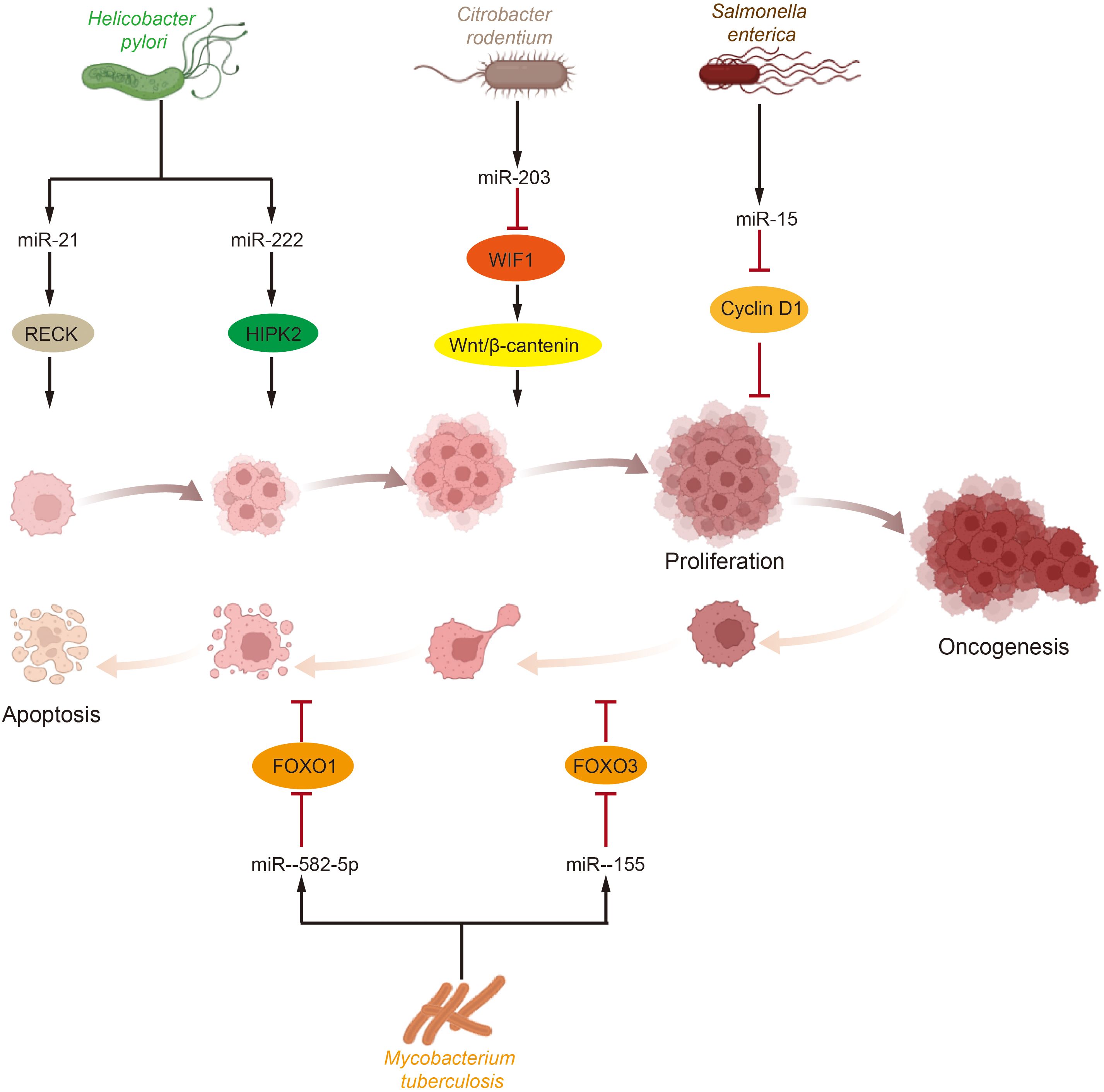
Figure 2. Effects of gut microbiota and miRNA on tumor cell proliferation and apoptosis. Tumor cells proliferation and apoptosis are crucial processes influencing tumorigenesis and development and are significant targets for microbial intervention. Various intestinal microorganisms impact key molecules or pathways involved in tumor survival by modulating miRNA levels, which in turn affect tumor cell proliferation and apoptosis. (HIPK2, homeodomain interacting protein kinase 2; WIF1, Wnt inhibitory factor-1; FOXO, forkhead box O).
Autophagy, regarded as type II programmed cell death, is a cellular digestive process primarily accountable for the degradation of damaged, denatured, or senescent macromolecules and organelles through lysosomes (165). It also plays a vital role in the host’s innate immune response. Like most tumors, the role of autophagy in bladder cancer is often dual. On the one hand, autophagy assists bladder cancer tumor cells in maintaining cell survival by degrading their own cytoplasmic components, such as proteins and organelles, to supply energy and metabolic substrates for the cells. Meanwhile, an elevated autophagy level also augments the invasive and metastatic ability of tumor cells by regulating the EMT process of tumor cells (166). Conversely, autophagy also exerts an inhibitory role in bladder cancer. For instance, autophagy can promptly eliminate damaged organelles and misfolded proteins from tumor cells, reduce intracellular oxidative stress and DNA damage, and thereby prevent genomic instability (167). Meanwhile, autophagy can also facilitate protein degradation and antigen presentation in tumor cells, enhancing the tumor immune response. For instance, autophagy can promote the maturation and antigen-presenting ability of DCs and enhance the activation and proliferation of T cells (168).
Remarkably, bacterial microorganisms have evolved diverse strategies to counter autophagy, including the regulation of host autophagy via microRNAs (Figure 3). For example, different mycobacterial species, such as Mycobacterium tuberculosis and Mycobacterium bovis Bacillus Calmette-Guérin (BCG), modulate host microRNAs to inhibit autophagy. Mycobacterium tuberculosis infection augments bacterial survival by inhibiting the autophagy regulator myeloid cell leukemia 1 (MCL1) via reduced levels of miR-17, thereby diminishing autophagy in host cells (169). Similarly, Mycobacterium tuberculosis can suppress autophagy by targeting the regulator DNA damage regulated autophagy modulator 2 (DRAM2) via upregulation of miR-144-5p. Moreover, Mycobacterium tuberculosis elevates the expression of miR-30a and miR-125a-3p, which disrupt the initial steps of autophagy by inhibiting Beclin-1 and UV radiation resistance associated (UVRAG), respectively (170, 171). Furthermore, Mycobacterium tuberculosis induces miR-33 and miR-33*, which target multiple autophagy and lysosomal effectors (including ATG5, ATG12, LC3B, LAMP1) (172). While microRNAs generally undermine host autophagy to promote mycobacterial survival, Mycobacterium tuberculosis has also been reported to enhance autophagy via miR-26a (147). On the other hand, BCG infection induces miR-17 expression and inhibits the autophagy initiation protein unc-51 like autophagy activating kinase 1 (ULK1) (173). Furthermore, BCG suppresses autophagosome formation by upregulating miR-20a and miR-144-3p, which inhibit ATG7, ATG16L1, and ATG4a (174, 175). The regulation of autophagy by microRNAs has also been observed in other microorganisms. For instance, Helicobacter pylori infection upregulates miR-30b, leading to reduced levels of Beclin-1 and ATG12, thereby inhibiting autophagy and preventing bacterial clearance (176). Similarly, Burkholderia pseudomallei infection in lung epithelial cells elevates the expression of miR-4458, miR-4667-5p, and miR-4668-5p, which inhibit autophagy and promote bacterial survival by suppressing ATG10 expression (177).

Figure 3. Influence of gut microbiota on the process of host autophagy. Autophagy is a crucial process influencing tumor cell survival and drug resistance. Additionally, bacterial microorganisms have evolved various strategies to modulate the host autophagy process through miRNAs. These miRNAs intervene at multiple stages of autophagy, including initiation, elongation, and maturation. (RHEB, ras homology enriched in brain; mTOR, mammalian target of rapamycin; ATG, autophagy-related gene; FIP, focal adhesion kinase family interacting protein; ULK, unc-51 like autophagy activating kinase 1; UVRAG, UV radiation resistance associated; BECN1, Beclin1; DRAM2, DNA damage regulated autophagy modulator 2; MCL1, myeloid cell leukemia 1; VPS, vacuolar protein sorting; LC3, light chain 3).
The aforementioned studies demonstrate that microRNAs can act as effector molecules of the GM involved in the phenotypic regulation related to bladder cancer. However, it is also shown that the microbial composition in the gut is influenced by microRNAs. Liu et al. (178) identified intestinal epithelial cells and Hopx-positive cells as the primary sources of fecal microRNAs. These microRNAs can regulate bacterial gene expression and affect microbial growth upon entering bacteria. Notably, they discovered that hsa-miR-515-5p and hsa-miR-1226-5p promote the growth of Clostridium nucleatum and Escherichia coli, respectively, thereby contributing to colorectal carcinogenesis. Additionally, their research (179) revealed that exogenous microRNAs can regulate intestinal bacterial abundance. Specifically, orally administered miR-30d modulates lactase expression in Akkermansia muciniphila, further enhancing its abundance in the intestinal tract. Teng et al. (180). demonstrated that exosomal microRNAs from plant sources can affect the intestinal microbiota. Their study showed that exosome-like nanoparticles (ELNs) from edible plants are preferentially absorbed by intestinal bacteria through an ELN lipid-dependent mechanism. Moreover, the extracellular vesicle form of microRNAs can persist in the digestive system, targeting gut microorganisms and strengthening intestinal barrier function. These findings are promising and suggest that therapeutic microRNAs could be extracted from healthy human feces and encapsulated in exosomes to modulate the intestinal microbiota more precisely, potentially preventing disease onset. Additionally, microRNAs might influence bacterial abundance in the tumor microenvironment by regulating glucose metabolism. Yuan et al. (181) observed that bacteria such as cytosolic pseudopods could be regulated by increased polysaccharide production associated with microRNAs, highlighting a potential mechanism by which microRNAs modulate microbial composition through glucose metabolism.
4.2 Interaction of circRNA, lncRNA and gut microbiota in tumors
Several studies have reported a close relationship between lncRNAs and the GM. Zhou et al. (182) compared the expression profiles of lncRNAs in the hippocampus of Specific Pathogen-Free (SPF) and Germ-Free (GF) mice, identifying 2,230 differentially expressed lncRNAs (1,355 upregulated and 875 downregulated). This study was the first to report the influence of the GM on hippocampal lncRNAs. Similarly, Zhang et al. (183) found that intramuscular injection of Esketamine significantly changed the GM in mice, including Adlercreutzia equolifaciens and Akkermansia muciniphila. Additionally, gene expression analysis revealed that 6 lncRNAs were significantly upregulated following Esketamine injection. Several studies have confirmed that the GM can affect host physiological functions by modulating lncRNAs. For example, a recent study demonstrated that the GM could recode lipid metabolism in the intestine via long-chain non-coding RNA Snhg9 (184). Furthermore, another study (185) found that oral administration of Bacillus fragilis enhanced lncRNA-CGB expression and promoted anti-tuberculosis immunity. This research also revealed that lncRNA-CGB interacts with EZH2, negatively regulating the epigenetic programming of H3K27 trimethylation (H3K27Me3) and enhancing IFN-γ expression, thus highlighting the crucial role of the GM in regulating immune responses. Additionally, a study (186) on preeclampsia indicated that dysregulation of the gut microbiome could affect trophoblast cell proliferation, invasion, and migration by modulating the NF-κB pathway through lncRNA BC030099. Similarly, Fusobacterium nucleatum was found to induce IRF5 expression via the lncRNA ENO1-IT1/miR-22-3p axis, exacerbating neonatal necrotizing enterocolitis (187). Additionally, the close relationship between lncRNAs and the GM has been shown to play a similar role in tumors. Stool testing and 16S rRNA sequencing of colorectal cancer patients revealed notable differences in the gut microbiome and lncRNA profiles (188). Further research demonstrated that Fusobacterium nucleatum targets the lncRNA ENO1-IT1 to enhance glycolysis and tumorigenesis in colorectal cancer, thereby elucidating the role of the GM in tumor development through lncRNA (189). Although less common, interactions between circRNAs and the GM have been reported. Chen et al. (190) found that circRNAs can change the composition of the intestinal microbiota and affect intestinal homeostasis and physiology in neonatal mice, thereby linking circRNAs with the intestinal microbiome. Furthermore, an additional study (191) has shown that the gut microbiome can inhibit circRNA expression in tumor stem cells in an interleukin-dependent manner and regulate the levels of corresponding microRNAs, which in turn affects lung cancer metastasis. Although less widespread than microRNAs, this at least indicates that lncRNAs and circRNAs are also potential molecules through which the gut microbiome can influence tumorigenesis. Further considering the molecular sponging role of circRNAs, it is reasonable to hypothesize that there is a complex network of interactions between the gut microbiome and non-coding RNAs in the context of bladder cancer waiting to be further explored.
5 Conclusion
The relationship between the GM and bladder cancer development is significant. Bladder cancer progression is often associated with deviations of the GM from its normal state. Correcting these deviations can alleviate carcinogen-induced bladder cancer and enhance the efficacy of adjuvant therapy. Additionally, ncRNAs also play a crucial role in bladder cancer development by influencing cell proliferation, apoptosis, invasion, metastasis, and vascular lymphangiogenesis. The GM influences the host’s immune-inflammatory response, as well as cell proliferation, apoptosis, and autophagy, through the modulation of ncRNA levels, which are closely related to bladder cancer development and drug resistance. As effector molecules of intestinal microorganisms, ncRNAs contribute to the epigenetic regulation of host genes by the GM, impacting various pathophysiological processes. In addition, the exciting findings (192, 193) that the GM can affect the levels of ncRNAs not only in intestinal epithelial cells but also outside the host intestine, such as in the aorta and hippocampus, further substantiates our conjecture. However, currently, there is no conclusive research on how the GM affects bladder cancer through ncRNAs. Furthermore, dysbiosis has been shown to affect tumorigenesis by altering metabolite levels, such as short-chain fatty acids, tryptophan metabolites, and secondary bile acids (6). NcRNAs can also regulate bacterial gene expression, influencing gut microbial growth and flora abundance. Therefore, supplementing with specific exogenous ncRNAs could potentially correct deviations in gut flora and restore normal host physiology. This suggests a promising new approach: instead of solely relying on fecal transplantation, we could extract and package ncRNAs into exosomes to modulate the GM levels and prevent diseases in a more targeted manner. Further research is needed to explore and validate this approach.
Author contributions
JZ: Data curation, Formal Analysis, Project administration, Resources, Writing – original draft. BX:. PL: Conceptualization, Investigation, Methodology, Validation, Writing – review & editing. TC: Investigation, Methodology, Project administration, Validation, Writing – review & editing. HD: Methodology, Supervision, Validation, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Jiangxi Provincial Health Commission Research Project (No. SKJP220227287).
Acknowledgments
All figures are created with BioRender.com.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Cumberbatch MGK, Jubber I, Black PC, Esperto F, Figueroa JD, Kamat AM, et al. Epidemiology of bladder cancer: A systematic review and contemporary update of risk factors in 2018. Eur Urol. (2018) 74:784–95. doi: 10.1016/j.eururo.2018.09.001
2. Lobo N, Afferi L, Moschini M, Mostafid H, Porten S, Psutka SP, et al. Epidemiology, screening, and prevention of bladder cancer. Eur Urol Oncol. (2022) 5:628–39. doi: 10.1016/j.euo.2022.10.003
3. Dyrskjøt L, Hansel DE, Efstathiou JA, Knowles MA, Galsky MD, Teoh J, et al. Bladder cancer. Nat Rev Dis Primers. (2023) 9:58. doi: 10.1038/s41572-023-00468-9
4. Teoh JY-C, Kamat AM, Black PC, Grivas P, Shariat SF, Babjuk M. Recurrence mechanisms of non-muscle-invasive bladder cancer - a clinical perspective. Nat Rev Urol. (2022) 19:280–94. doi: 10.1038/s41585-022-00578-1
5. Illiano P, Brambilla R, Parolini C. The mutual interplay of gut microbiota, diet and human disease. FEBS J. (2020) 287:833–55. doi: 10.1111/febs.v287.5
6. Luo P, Zheng L, Zou J, Chen T, Zou J, Li W, et al. Insights into vitamin A in bladder cancer, lack of attention to gut microbiota? Front Immunol. (2023) 14:1252616. doi: 10.3389/fimmu.2023.1252616
7. Molinero N, Ruiz L, Sánchez B, Margolles A, Delgado S. Intestinal bacteria interplay with bile and cholesterol metabolism: implications on host physiology. Front Physiol. (2019) 10:185. doi: 10.3389/fphys.2019.00185
8. Davis CP, Cohen MS, Gruber MB, Anderson MD, Warren MM. Urothelial hyperplasia and neoplasia: a response to chronic urinary tract infections in rats. J Urol. (1984) 132:1025–31. doi: 10.1016/S0022-5347(17)49992-7
9. Whiteside SA, Razvi H, Dave S, Reid G, Burton JP. The microbiome of the urinary tract–a role beyond infection. Nat Rev Urol. (2015) 12:81–90. doi: 10.1038/nrurol.2014.361
10. Walter CEJ, Durairajan S, Periyandavan K, C GPD, G DJD, A HRV, et al. Bladder neoplasms and NF-κB: an unfathomed association. Expert Rev Mol Diagn. (2020) 20:497–508. doi: 10.1080/14737159.2020.1743688
11. Worby CJ, Schreiber HL, Straub TJ, van Dijk LR, Bronson RA, Olson BS, et al. Longitudinal multi-omics analyses link gut microbiome dysbiosis with recurrent urinary tract infections in women. Nat Microbiol. (2022) 7:630–9. doi: 10.1038/s41564-022-01107-x
12. Hong L, Huang Y, Han J, Li S, Zhang L, Zhou Q, et al. Pathogen-specific alterations in intestinal microbiota precede urinary tract infections in preterm infants: a longitudinal case-control study. Gut Microbes. (2024) 16:2333413. doi: 10.1080/19490976.2024.2333413
13. Jones-Freeman B, Chonwerawong M, Marcelino VR, Deshpande AV, Forster SC, Starkey MR. The microbiome and host mucosal interactions in urinary tract diseases. Mucosal Immunol. (2021) 14:779–92. doi: 10.1038/s41385-020-00372-5
14. Saw PE, Xu X, Chen J, Song E-W. Non-coding RNAs: the new central dogma of cancer biology. Sci China Life Sci. (2021) 64:22–50. doi: 10.1007/s11427-020-1700-9
15. Palazzo AF, Koonin EV. Functional long non-coding RNAs evolve from junk transcripts. Cell. (2020) 183:1151–61. doi: 10.1016/j.cell.2020.09.047
16. Sullenger BA, Nair S. From the RNA world to the clinic. Science. (2016) 352:1417–20. doi: 10.1126/science.aad8709
17. Cai Z, Cao C, Ji L, Ye R, Wang D, Xia C, et al. RIC-seq for global in situ profiling of RNA-RNA spatial interactions. Nature. (2020) 582:432–7. doi: 10.1038/s41586-020-2249-1
18. Xiao M-S, Ai Y, Wilusz JE. Biogenesis and functions of circular RNAs come into focus. Trends Cell Biol. (2020) 30:226–40. doi: 10.1016/j.tcb.2019.12.004
19. Zong Y, Wang X, Cui B, Xiong X, Wu A, Lin C, et al. Decoding the regulatory roles of non-coding RNAs in cellular metabolism and disease. Mol Ther. (2023) 31:1562–76. doi: 10.1016/j.ymthe.2023.04.012
20. Nemeth K, Bayraktar R, Ferracin M, Calin GA. Non-coding RNAs in disease: from mechanisms to therapeutics. Nat Rev Genet. (2024) 25:211–32. doi: 10.1038/s41576-023-00662-1
21. Li Y, Li G, Guo X, Yao H, Wang G, Li C. Non-coding RNA in bladder cancer. Cancer Lett. (2020) 485:38–44. doi: 10.1016/j.canlet.2020.04.023
22. Martin A, Woolbright BL, Umar S, Ingersoll MA, Taylor JA. Bladder cancer, inflammageing and microbiomes. Nat Rev Urol. (2022) 19:495–509. doi: 10.1038/s41585-022-00611-3
23. Kim KH, Chung Y, Huh JW, Park DJ, Cho Y, Oh Y, et al. Gut microbiota of the young ameliorates physical fitness of the aged in mice. Microbiome. (2022) 10:238. doi: 10.1186/s40168-022-01386-w
24. He C, Li B, Huang L, Teng C, Bao Y, Ren M, et al. Gut microbial composition changes in bladder cancer patients: A case-control study in Harbin, China. Asia Pac J Clin Nutr. (2020) 29(2):395–403. doi: 10.6133/apjcn.202007_29(2).0022
25. Then CK, Paillas S, Wang X, Hampson A, Kiltie AE. Association of Bacteroides acidifaciens relative abundance with high-fibre diet-associated radiosensitisation. BMC Biol. (2020) 18:102. doi: 10.1186/s12915-020-00836-x
26. Yin Z, Liu B, Feng S, He Y, Tang C, Chen P, et al. A large genetic causal analysis of the gut microbiota and urological cancers: A bidirectional mendelian randomization study. Nutrients. (2023) 15(18):4086. doi: 10.3390/nu15184086
27. Mingdong W, Xiang G, Yongjun Q, Mingshuai W, Hao P. Causal associations between gut microbiota and urological tumors: a two-sample mendelian randomization study. BMC Cancer. (2023) 23:854. doi: 10.1186/s12885-023-11383-3
28. Zhang F, Yao Z, Zhang B. Genetically proxied intestinal microbiota and risk of bladder cancer. Int J Surg. (2024) 110:1857–9. doi: 10.1097/JS9.0000000000001019
29. He C, Huang L, Lei P, Liu X, Li B, Shan Y. Sulforaphane normalizes intestinal flora and enhances gut barrier in mice with BBN-induced bladder cancer. Mol Nutr Food Res. (2018) 62:e1800427. doi: 10.1002/mnfr.201800427
30. Lenis AT, Lec PM, Chamie K, Mshs MD. Bladder cancer: A review. JAMA. (2020) 324:1980–91. doi: 10.1001/jama.2020.17598
31. Hopkins AM, Kichenadasse G, Karapetis CS, Rowland A, Sorich MJ. Concomitant antibiotic use and survival in urothelial carcinoma treated with atezolizumab. Eur Urol. (2020) 78:540–3. doi: 10.1016/j.eururo.2020.06.061
32. Bukavina L, Ginwala R, Eltoukhi M, Sindhani M, Prunty M, Geynisman DM, et al. Role of gut microbiome in neoadjuvant chemotherapy response in urothelial carcinoma: A multi-institutional prospective cohort evaluation. Cancer Res Commun. (2024) 4:1505–16. doi: 10.1158/2767-9764.CRC-23-0479
33. Mager LF, Burkhard R, Pett N, Cooke NCA, Brown K, Ramay H, et al. Microbiome-derived inosine modulates response to checkpoint inhibitor immunotherapy. Science. (2020) 369:1481–9. doi: 10.1126/science.abc3421
34. Ferrere G, Tidjani Alou M, Liu P, Goubet A-G, Fidelle M, Kepp O, et al. Ketogenic diet and ketone bodies enhance the anticancer effects of PD-1 blockade. JCI Insight. (2021) 6(2):e145207. doi: 10.1172/jci.insight.145207
35. Wang B, Qiu Y, Xie M, Huang P, Yu Y, Sun Q, et al. Gut microbiota Parabacteroides distasonis enchances the efficacy of immunotherapy for bladder cancer by activating anti-tumor immune responses. BMC Microbiol. (2024) 24:237. doi: 10.1186/s12866-024-03372-8
36. Aguilar C, Mano M, Eulalio A. MicroRNAs at the host-bacteria interface: host defense or bacterial offense. Trends Microbiol. (2019) 27:206–18. doi: 10.1016/j.tim.2018.10.011
37. Wang Q, Ding H, Dong G, Xu L, Jiang F, Mao Q. Bi-direction effects between microbiome and MiRNAs in carcinogenesis. J Cancer Res Clin Oncol. (2021) 147:1299–305. doi: 10.1007/s00432-021-03567-w
38. Berindan-Neagoe I, Monroig P, Pasculli B, Calin GA. MicroRNAome genome: a treasure for cancer diagnosis and therapy. CA Cancer J Clin. (2014) 64:311–36. doi: 10.3322/caac.21244
39. Tan W, Liu B, Qu S, Liang G, Luo W, Gong C. MicroRNAs and cancer: Key paradigms in molecular therapy. Oncol Lett. (2018) 15(3):2735–42. doi: 10.3892/ol.2017.7638
40. Friedman RC, Farh KK-H, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. (2009) 19(1):92–105. doi: 10.1101/gr.082701.108
41. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. (2004) 116:281–97. doi: 10.1016/S0092-8674(04)00045-5
42. Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. (2011) 12:861–74. doi: 10.1038/nrg3074
43. Bonci D, Coppola V, Musumeci M, Addario A, Giuffrida R, Memeo L, et al. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med. (2008) 14:1271–7. doi: 10.1038/nm.1880
44. Peng Y, Croce CM. The role of MicroRNAs in human cancer. Signal Transduct Target Ther. (2016) 1:15004. doi: 10.1038/sigtrans.2015.4
45. Tagawa H, Ikeda S, Sawada K. Role of microRNA in the pathogenesis of Malignant lymphoma. Cancer Sci. (2013) 104:801–9. doi: 10.1111/cas.2013.104.issue-7
46. Pencheva N, Tran H, Buss C, Huh D, Drobnjak M, Busam K, et al. Convergent multi-miRNA targeting of ApoE drives LRP1/LRP8-dependent melanoma metastasis and angiogenesis. Cell. (2012) 151:1068–82. doi: 10.1016/j.cell.2012.10.028
47. Zhang Q, Pan J, Xiong D, Wang Y, Miller MS, Sei S, et al. Pulmonary Aerosol Delivery of Let-7b microRNA Confers a Striking Inhibitory Effect on Lung Carcinogenesis through Targeting the Tumor Immune Microenvironment. Adv Sci (Weinh). (2021) 8:e2100629. doi: 10.1002/advs.202100629
48. Fawzy MS, El Faiomy ARM, El Desoky AMZ, Hussein S. The relationship between DNA methyltransferase 3B (DNMT3B) and miR 124-3pa expressions in bladder cancer tissues. Mol Biol Rep. (2023) 50:10005–13. doi: 10.1007/s11033-023-08818-2
49. Yang F-K, Tian C, Zhou L-X, Guan T-Y, Chen G-L, Zheng Y-Y, et al. The value of urinary exosomal microRNA-21 in the early diagnosis and prognosis of bladder cancer. Kaohsiung J Med Sci. (2024) 40:660–70. doi: 10.1002/kjm2.12845
50. Meng H, Yang R, Lin Q, Du W, Chu Z, Cao Y, et al. Isorhapontigenin inhibition of basal muscle-invasive bladder cancer attributed to its downregulation of SNHG1 and DNMT3b. BMC Cancer. (2024) 24:737. doi: 10.1186/s12885-024-12490-5
51. Pan G, Jiang B, Yi Z, Yin J, Liu Y. Exosomal miR-105-5p derived from bladder cancer stem cells targets for GPR12 to promote the Malignancy of bladder cancer. BMC Urol. (2023) 23:155. doi: 10.1186/s12894-023-01326-2
52. Tian X, Liu D, He P, Li L, Wang Y, Qiu M. DOK7, a target of miR-299-5p, suppresses the progression of bladder cancer. Aging (Albany NY). (2023) 15:14306–22. doi: 10.18632/aging.205304
53. Yuan H, Wang T, Peng P, Xu Z, Feng F, Cui Y, et al. Urinary Exosomal miR-17-5p Accelerates Bladder Cancer Invasion by Repressing its Target Gene ARID4B and Regulating the Immune Microenvironment. Clin Genitourin Cancer. (2024) 22(2):569–79.e1. doi: 10.1016/j.clgc.2024.01.012
54. Lv L, Wei Q, Zhang J, Dong Y, Shan Z, Chang N, et al. IGF2BP3 prevent HMGB1 mRNA decay in bladder cancer and development. Cell Mol Biol Lett. (2024) 29:39. doi: 10.1186/s11658-024-00545-1
55. Kishi S, Mori S, Fujiwara-Tani R, Ogata R, Sasaki R, Ikemoto A, et al. ERVK13-1/miR-873-5p/GNMT axis promotes metastatic potential in human bladder cancer though sarcosine production. Int J Mol Sci. (2023) 24(22):16367. doi: 10.3390/ijms242216367
56. Hao Y, Zhu Y, Sun F, Xu D, Wang C. MicroRNA-30c-5p arrests bladder cancer G2/M phase and suppresses its progression by targeting PRC1-mediated blocking of CDK1/Cyclin B1 axis. Cell Signal. (2023) 110:110836. doi: 10.1016/j.cellsig.2023.110836
57. Zhou W, Yang Y, Wang W, Yang C, Cao Z, Lin X, et al. Pseudogene OCT4-pg5 upregulates OCT4B expression to promote bladder cancer progression by competing with miR-145-5p. Cell Cycle. (2024) 23:645–61. doi: 10.1080/15384101.2024.2353554
58. Zhenhai Z, Qi C, Shuchao Z, Zhongqi W, Xue S, Zhijun G, et al. MiR-205-3p suppresses bladder cancer progression via GLO1 mediated P38/ERK activation. BMC Cancer. (2023) 23:956. doi: 10.1186/s12885-023-11175-9
59. Wang C-S, Lee Y-C, Jhan J-H, Li W-M, Chang L-L, Huang AM, et al. MicroRNA-299-3p inhibits cell proliferation, motility, invasion and angiogenesis via VEGFA in upper tract urothelial carcinoma. J Gene Med. (2024) 26:e3616. doi: 10.1002/jgm.v26.1
60. Yang J, Xiang H, Cheng M, Jiang X, Chen Y, Zheng L, et al. microRNA-15a-5p suppresses hypoxia-induced tumor growth and chemoresistance in bladder cancer by binding to eIF5A2. Neoplasma. (2024) 71:60–9. doi: 10.4149/neo_2024_230915N489
61. Meng F, Zhang Z. MicroRNA-152 specifically targets kinesin family member 14 to suppress the advancement of bladder cancer cells via PI3K/AKT pathway. Biochem Biophys Res Commun. (2024) 692:149337. doi: 10.1016/j.bbrc.2023.149337
62. Li W, Wang Z, Jiang Z, Yan Y, Yao X, Pan Z, et al. MiR-3960 inhibits bladder cancer progression via targeting of DEXI. Biochem Biophys Res Commun. (2023) 668:8–18. doi: 10.1016/j.bbrc.2023.05.055
63. Wei X, Jiang Y, Yang G, Chang T, Sun G, Chen S, et al. MicroRNA-367-3p directly targets RAB23 and inhibits proliferation, migration and invasion of bladder cancer cells and increases cisplatin sensitivity. J Cancer Res Clin Oncol. (2023) 149:17807–21. doi: 10.1007/s00432-023-05484-6
64. Luo H-L, Lee Y-C, Chang Y-L, Hsu W-C, Wu Y-T, Jhan J-H, et al. MicroRNA-145-5p suppresses cell proliferation, migration, and invasion in upper tract urothelial carcinoma by targeting 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase. J Cell Biochem. (2023) 124:1324–45. doi: 10.1002/jcb.v124.9
65. Rao P, Li J, Xiong J, Shen S, Zeng J, Zhao H. MicroRNA-150-5p-mediated inhibition of cell proliferation, G1/S transition, and migration in bladder cancer through targeting NEDD4-binding protein 2-like 1 gene. J Physiol Investig. (2024) 67:118–28. doi: 10.4103/ejpi.EJPI-D-24-00009
66. Liu Z, Du D, Zhang S. Tumor-derived exosomal miR-1247-3p promotes angiogenesis in bladder cancer by targeting FOXO1. Cancer Biol Ther. (2024) 25:2290033. doi: 10.1080/15384047.2023.2290033
67. Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. (2016) 17:47–62. doi: 10.1038/nrg.2015.10
68. Marvin MC, Clauder-Münster S, Walker SC, Sarkeshik A, Yates JR, Steinmetz LM, et al. Accumulation of noncoding RNA due to an RNase P defect in Saccharomyces cerevisiae. RNA. (2011) 17:1441–50. doi: 10.1261/rna.2737511
69. Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. (2013) 20:300–7. doi: 10.1038/nsmb.2480
70. Novikova IV, Hennelly SP, Sanbonmatsu KY. Tackling structures of long noncoding RNAs. Int J Mol Sci. (2013) 14:23672–84. doi: 10.3390/ijms141223672
71. Li Y, Egranov SD, Yang L, Lin C. Molecular mechanisms of long noncoding RNAs-mediated cancer metastasis. Genes Chromosomes Cancer. (2019) 58:200–7. doi: 10.1002/gcc.22691
72. Marchese FP, Raimondi I, Huarte M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol. (2017) 18:206. doi: 10.1186/s13059-017-1348-2
73. Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. (2013) 14:699–712. doi: 10.1038/nrm3679
74. Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. (2012) 482:339–46. doi: 10.1038/nature10887
75. Derrien T, Guigó R. Long non-coding RNAs with enhancer-like function in human cells. Med Sci (Paris). (2011) 27:359–61. doi: 10.1051/medsci/2011274009
76. Papaioannou D, Petri A, Dovey OM, Terreri S, Wang E, Collins FA, et al. The long non-coding RNA HOXB-AS3 regulates ribosomal RNA transcription in NPM1-mutated acute myeloid leukemia. Nat Commun. (2019) 10:5351. doi: 10.1038/s41467-019-13259-2
77. Chiu H-S, Somvanshi S, Patel E, Chen T-W, Singh VP, Zorman B, et al. Pan-cancer analysis of lncRNA regulation supports their targeting of cancer genes in each tumor context. Cell Rep. (2018) 23(1):297–312.e12. doi: 10.1016/j.celrep.2018.03.064
78. Xiao H, Tang K, Liu P, Chen K, Hu J, Zeng J, et al. LncRNA MALAT1 functions as a competing endogenous RNA to regulate ZEB2 expression by sponging miR-200s in clear cell kidney carcinoma. Oncotarget. (2015) 6:38005–15. doi: 10.18632/oncotarget.v6i35
79. Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. (2007) 129:1311–23. doi: 10.1016/j.cell.2007.05.022
80. Tsai M-C, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. (2010) 329:689–93. doi: 10.1126/science.1192002
81. Su Y, Chen H, Yao L, Dai H, Liu H. The relationship between the expression of lncRNA MALAT1 and clinical features and prognosis in bladder cancer: A meta-analysis. Cell Mol Biol (Noisy-le-grand). (2023) 69(14):166–71. doi: 10.14715/cmb/2023.69.14.27
82. Wang Z, Wei B, Ma S. EGR1/LINC00839/SOX5 axis modulates migration, invasion and Gemcitabine resistance of bladder cancer cells. Cancer Biol Ther. (2023) 24:2270106. doi: 10.1080/15384047.2023.2270106
83. Li Z, Fu H, Liu J, Song W, Zeng M, Wang J. LncRNA PVT1 promotes bladder cancer progression by forming a positive feedback loop with STAT5B. Pathol Res Pract. (2023) 248:154635. doi: 10.1016/j.prp.2023.154635
84. Cao Y, Zou Z, Wu X, Li W, Lu Z, Hu J, et al. LUCAT1 inhibits ferroptosis in bladder cancer by regulating the mRNA stability of STAT3. Gene. (2024) 894:147974. doi: 10.1016/j.gene.2023.147974
85. Xiao Y, He L, Dong Y, Huang Y, Ma L, Li W. Highly expressed LINC00958 modulates the growth and epithelial-mesenchymal transition of bladder cancer cells through SAPK/JNK signaling pathway. Cancer Biother Radiopharm. (2023) 38:405–14. doi: 10.1089/cbr.2022.0005
86. Arima J, Yoshino H, Fukumoto W, Kawahara I, Saito S, Li G, et al. LncRNA BCYRN1 as a potential therapeutic target and diagnostic marker in serum exosomes in bladder cancer. Int J Mol Sci. (2024) 25(11):5955. doi: 10.3390/ijms25115955
87. Li X, Liang Z, Pan J, Zhang M, Liu J, Hu R, et al. Identification of BACH1-IT2-miR-4786-Siglec-15 immune suppressive axis in bladder cancer. BMC Cancer. (2024) 24:328. doi: 10.1186/s12885-024-12061-8
88. Song W, Li Z, Yang K, Gao Z, Zhou Q, Li P. Antisense lncRNA-RP11-498C9.13 promotes bladder cancer progression by enhancing reactive oxygen species-induced mitophagy. J Gene Med. (2023) 25(9):e3527. doi: 10.1002/jgm.3527
89. Hong Y, Li Z, Su Y, Pu H, Zhang X. The ceRNA Mechanism of lncRNA MEG3/miR-21-5p/SPRY2 in Cell Proliferation and Apoptosis in Bladder Cancer. Crit Rev Eukaryot Gene Expr. (2024) 34:55–68. doi: 10.1615/CritRevEukaryotGeneExpr.2023048011
90. Lu X-S, Huang M-L, Chen L-B, Liu S-C, Huang Z-X, Liu S-M. SCARA5 as a downstream factor of PCAT29, inhibits proliferation, migration, and invasion of bladder cancer. Genomics. (2023) 115:110667. doi: 10.1016/j.ygeno.2023.110667
91. Pei L, Yan D, He Q, Kong J, Yang M, Ruan H, et al. LncRNA MIR4435-2HG drives cancer progression by modulating cell cycle regulators and mTOR signaling in stroma-enriched subtypes of urothelial carcinoma of the bladder. Cell Oncol (Dordr). (2023) 46:1509–27. doi: 10.1007/s13402-023-00826-5
92. Zheng R, Gao F, Mao Z, Xiao Y, Yuan L, Huang Z, et al. LncRNA BCCE4 genetically enhances the PD-L1/PD-1 interaction in smoking-related bladder cancer by modulating miR-328-3p-USP18 signaling. Adv Sci (Weinh). (2023) 10:e2303473. doi: 10.1002/advs.202303473
93. Li Y, Zheng H, Luo Y, Lin Y, An M, Kong Y, et al. An HGF-dependent positive feedback loop between bladder cancer cells and fibroblasts mediates lymphangiogenesis and lymphatic metastasis. Cancer Commun (Lond). (2023) 43:1289–311. doi: 10.1002/cac2.v43.12
94. Xie J, Zhang H, Wang K, Ni J, Ma X, Khoury CJ, et al. M6A-mediated-upregulation of lncRNA BLACAT3 promotes bladder cancer angiogenesis and hematogenous metastasis through YBX3 nuclear shuttling and enhancing NCF2 transcription. Oncogene. (2023) 42:2956–70. doi: 10.1038/s41388-023-02814-3
95. Barrett SP, Salzman J. Circular RNAs: analysis, expression and potential functions. Development. (2016) 143:1838–47. doi: 10.1242/dev.128074
96. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. (2013) 19:141–57. doi: 10.1261/rna.035667.112
97. Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PloS Genet. (2013) 9:e1003777. doi: 10.1371/journal.pgen.1003777
98. Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PloS One. (2012) 7:e30733. doi: 10.1371/journal.pone.0030733
99. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. (2019) 20:675–91. doi: 10.1038/s41576-019-0158-7
100. Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, et al. A new star of noncoding RNAs. Cancer Lett. (2015) 365:141–8. doi: 10.1016/j.canlet.2015.06.003
101. Kristensen LS, Hansen TB, Venø MT, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. (2018) 37:555–65. doi: 10.1038/onc.2017.361
102. Yi J, Ma X, Ying Y, Liu Z, Tang Y, Shu X, et al. N6-methyladenosine-modified CircPSMA7 enhances bladder cancer Malignancy through the miR-128-3p/MAPK1 axis. Cancer Lett. (2024) 585:216613. doi: 10.1016/j.canlet.2024.216613
103. Huang C, Yang Y, Wang X, Chen S, Liu Z, Li Z, et al. PTBP1-mediated biogenesis of circATIC promotes progression and cisplatin resistance of bladder cancer. Int J Biol Sci. (2024) 20:3570–89. doi: 10.7150/ijbs.96671
104. Liu C, Cong Y, Chen L, Lv F, Cheng L, Song Y, et al. Hsa_circ_0001583 fuels bladder cancer metastasis by promoting staphylococcal nuclease and tudor domain containing 1-mediated MicroRNA decay. Neoplasia. (2024) 47:100963. doi: 10.1016/j.neo.2023.100963
105. Feng D, Lv J, Li K, Cao Q, Han J, Yu H, et al. CircZNF609 promotes bladder cancer progression and inhibits cisplatin sensitivity via miR-1200/CDC25B pathway. Cell Biol Toxicol. (2023) 39(5):1–18. doi: 10.1007/s10565-022-09715-3
106. Wang C, Yang X. CircRAPGEF5 sponges miR-582-3p and targets KIF3A to regulate bladder cancer cell proliferation, migration and invasion. Int Immunopharmacol. (2024) 131:111613. doi: 10.1016/j.intimp.2024.111613
107. Deng W, Chen R, Xiong S, Nie J, Yang H, Jiang M, et al. CircFSCN1 induces tumor progression and triggers epithelial-mesenchymal transition in bladder cancer through augmentation of MDM2-mediated p53 silencing. Cell Signal. (2024) 114:110982. doi: 10.1016/j.cellsig.2023.110982
108. Luo L, Xie Q, Wu Y, Li P, Qin F, Liao D, et al. Circular RNA CCT3 is a unique molecular marker in bladder cancer. BMC Cancer. (2023) 23:977. doi: 10.1186/s12885-023-11510-0
109. Yang H, He P, Luo W, Liu S, Yang Y. circRNA TATA-box binding protein associated factor 15 acts as an oncogene to facilitate bladder cancer progression through targeting miR-502-5p/high mobility group box 3. Mol Carcinog. (2024) 63:629–46. doi: 10.1002/mc.23677
110. Liu Y, Zhang K, Yang X. CircMCTP2 enhances the progression of bladder cancer by regulating the miR-99a-5p/FZD8 axis. J Egypt Natl Canc Inst. (2024) 36:8. doi: 10.1186/s43046-024-00206-6
111. Wei W, Liu K, Huang X, Tian S, Wang H, Zhang C, et al. EIF4A3-mediated biogenesis of circSTX6 promotes bladder cancer metastasis and cisplatin resistance. J Exp Clin Cancer Res. (2024) 43:2. doi: 10.1186/s13046-023-02932-6
112. Li Y, Kong Y, An M, Luo Y, Zheng H, Lin Y, et al. ZEB1-mediated biogenesis of circNIPBL sustains the metastasis of bladder cancer via Wnt/β-catenin pathway. J Exp Clin Cancer Res. (2023) 42:191. doi: 10.1186/s13046-023-02757-3
113. Lyu F, Huang S, Yan Z, He Q, Liu C, Cheng L, et al. CircUGGT2 facilitates progression and cisplatin resistance of bladder cancer through nonhomologous end-joining pathway. Cell Signal. (2024) 119:111164. doi: 10.1016/j.cellsig.2024.111164
114. Ji Q, Ma F, Zhang X, Liu Y, Wang P, Li M. Hsa_circ_0005320 affects cell proliferation and the cell cycle via the IGF2BP3/CDK2 axis in bladder cancer. Cell Signal. (2024) 119:111154. doi: 10.1016/j.cellsig.2024.111154
115. Cheng S, Li C, Liu L, Liu X, Li M, Zhuo J, et al. Dysregulation and antimetastatic function of circLRIG1 modulated by miR-214-3p/LRIG1 axis in bladder carcinoma. Biol Direct. (2024) 19:20. doi: 10.1186/s13062-023-00446-x
116. Li Z, Wang Z, Yang S, Shen C, Zhang Y, Jiang R, et al. CircSTK39 suppresses the proliferation and invasion of bladder cancer by regulating the miR-135a-5p/NR3C2-mediated epithelial-mesenchymal transition signaling pathway. Cell Biol Toxicol. (2023) 39:1815–34. doi: 10.1007/s10565-022-09785-3
117. Yang X-X, Wang C. CircITGA7 regulates Malignant phenotypes in bladder cancer cells via targeting miR-330-3p/KLF10 axis. Kaohsiung J Med Sci. (2024) 40:324–34. doi: 10.1002/kjm2.12821
118. Li K, Lv J, Wang J, Wei Y, Zhang Y, Lin J, et al. CircZNF609 inhibited bladder cancer immunotherapy sensitivity via enhancing fatty acid uptake through IGF2BP2/CD36 pathway. Int Immunopharmacol. (2024) 137:112485. doi: 10.1016/j.intimp.2024.112485
119. Dong C, Hui P, Wu Z, Li J, Man X. CircRNA LOC729852 promotes bladder cancer progression by regulating macrophage polarization and recruitment via the miR-769-5p/IL-10 axis. J Cell Mol Med. (2024) 28:e18225. doi: 10.1111/jcmm.18225
120. Shi X, Pang S, Zhou J, Yan G, Gao R, Wu H, et al. Bladder-cancer-derived exosomal circRNA_0013936 promotes suppressive immunity by up-regulating fatty acid transporter protein 2 and down-regulating receptor-interacting protein kinase 3 in PMN-MDSCs. Mol Cancer. (2024) 23:52. doi: 10.1186/s12943-024-01968-2
121. Xu C, Zhou J, Zhang X, Kang X, Liu S, Song M, et al. N6-methyladenosine-modified circ_104797 sustains cisplatin resistance in bladder cancer through acting as RNA sponges. Cell Mol Biol Lett. (2024) 29:28. doi: 10.1186/s11658-024-00543-3
122. Malmuthuge N, Guan LL. Noncoding RNAs: regulatory molecules of host-microbiome crosstalk. Trends Microbiol. (2021) 29:713–24. doi: 10.1016/j.tim.2020.12.003
123. Creemers EE, Tijsen AJ, Pinto YM. Circulating microRNAs: novel biomarkers and extracellular communicators in cardiovascular disease? Circ Res. (2012) 110:483–95. doi: 10.1161/CIRCRESAHA.111.247452
124. Tomkovich S, Gharaibeh RZ, Dejea CM, Pope JL, Jiang J, Winglee K, et al. Human Colon Mucosal Biofilms and Murine Host Communicate via Altered mRNA and microRNA Expression during Cancer. mSystems. (2020) 5(1):e00451–19. doi: 10.1128/msystems.00451-19
125. Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, et al. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science. (2006) 312:436–9. doi: 10.1126/science.1126088
126. Wang T, Cai G, Qiu Y, Fei N, Zhang M, Pang X, et al. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. (2012) 6:320–9. doi: 10.1038/ismej.2011.109
127. Nielsen TS, Bendiks Z, Thomsen B, Wright ME, Theil PK, Scherer BL, et al. High-amylose maize, potato, and butyrylated starch modulate large intestinal fermentation, microbial composition, and oncogenic miRNA expression in rats fed A high-protein meat diet. Int J Mol Sci. (2019) 20(9):2137. doi: 10.3390/ijms20092137
128. Peck BCE, Mah AT, Pitman WA, Ding S, Lund PK, Sethupathy P. Functional transcriptomics in diverse intestinal epithelial cell types reveals robust microRNA sensitivity in intestinal stem cells to microbial status. J Biol Chem. (2017) 292:2586–600. doi: 10.1074/jbc.M116.770099
129. Mima K, Nakagawa S, Sawayama H, Ishimoto T, Imai K, Iwatsuki M, et al. The microbiome and hepatobiliary-pancreatic cancers. Cancer Lett. (2017) 402:9–15. doi: 10.1016/j.canlet.2017.05.001
130. Vogtmann E, Goedert JJ. Epidemiologic studies of the human microbiome and cancer. Br J Cancer. (2016) 114:237–42. doi: 10.1038/bjc.2015.465
131. Burns MB, Montassier E, Abrahante J, Priya S, Niccum DE, Khoruts A, et al. Colorectal cancer mutational profiles correlate with defined microbial communities in the tumor microenvironment. PloS Genet. (2018) 14:e1007376. doi: 10.1371/journal.pgen.1007376
132. Dzutsev A, Badger JH, Perez-Chanona E, Roy S, Salcedo R, Smith CK, et al. Microbes and cancer. Annu Rev Immunol. (2017) 35:199–228. doi: 10.1146/annurev-immunol-051116-052133
133. Kübler H, Gschwend JE. Active immunotherapy of urologic Malignancies and tumor-mediated immunosuppression. Urologe A. (2008) 47(9):1122–7. doi: 10.1007/s00120-008-1822-2
134. Bevers RFM, Kurth KH, Schamhart DHJ. Role of urothelial cells in BCG immunotherapy for superficial bladder cancer. Br J Cancer. (2004) 91:607–12. doi: 10.1038/sj.bjc.6602026
135. Méndez CS, Bueno SM, Kalergis AM. Contribution of gut microbiota to immune tolerance in infants. J Immunol Res. (2021) 2021:7823316. doi: 10.1155/2021/7823316
136. Kayisoglu Ö, Schlegel N, Bartfeld S. Gastrointestinal epithelial innate immunity-regionalization and organoids as new model. J Mol Med (Berl). (2021) 99:517–30. doi: 10.1007/s00109-021-02043-9
137. Cristofori F, Dargenio VN, Dargenio C, Miniello VL, Barone M, Francavilla R. Anti-inflammatory and immunomodulatory effects of probiotics in gut inflammation: A door to the body. Front Immunol. (2021) 12:578386. doi: 10.3389/fimmu.2021.578386
138. Kim M, Qie Y, Park J, Kim CH. Gut microbial metabolites fuel host antibody responses. Cell Host Microbe. (2016) 20:202–14. doi: 10.1016/j.chom.2016.07.001
139. Das K, Garnica O, Dhandayuthapani S. Modulation of host miRNAs by intracellular bacterial pathogens. Front Cell Infect Microbiol. (2016) 6:79. doi: 10.3389/fcimb.2016.00079
140. Maudet C, Mano M, Eulalio A. MicroRNAs in the interaction between host and bacterial pathogens. FEBS Lett. (2014) 588:4140–7. doi: 10.1016/j.febslet.2014.08.002
141. Sheedy FJ. Turning 21: induction of miR-21 as a key switch in the inflammatory response. Front Immunol. (2015) 6:19. doi: 10.3389/fimmu.2015.00019
142. Zhou X, Li X, Ye Y, Zhao K, Zhuang Y, Li Y, et al. MicroRNA-302b augments host defense to bacteria by regulating inflammatory responses via feedback to TLR/IRAK4 circuits. Nat Commun. (2014) 5:3619. doi: 10.1038/ncomms4619
143. Li X, He S, Li R, Zhou X, Zhang S, Yu M, et al. Pseudomonas aeruginosa infection augments inflammation through miR-301b repression of c-Myb-mediated immune activation and infiltration. Nat Microbiol. (2016) 1:16132. doi: 10.1038/nmicrobiol.2016.132
144. Kalantari P, Harandi OF, Agarwal S, Rus F, Kurt-Jones EA, Fitzgerald KA, et al. miR-718 represses proinflammatory cytokine production through targeting phosphatase and tensin homolog (PTEN). J Biol Chem. (2017) 292:5634–44. doi: 10.1074/jbc.M116.749325
145. Kumar M, Sahu SK, Kumar R, Subuddhi A, Maji RK, Jana K, et al. MicroRNA let-7 modulates the immune response to Mycobacterium tuberculosis infection via control of A20, an inhibitor of the NF-κB pathway. Cell Host Microbe. (2015) 17:345–56. doi: 10.1016/j.chom.2015.01.007
146. Liu N, Wang L, Sun C, Yang L, Sun W, Peng Q. MicroRNA-125b-5p suppresses Brucella abortus intracellular survival via control of A20 expression. BMC Microbiol. (2016) 16:171. doi: 10.1186/s12866-016-0788-2
147. Sahu SK, Kumar M, Chakraborty S, Banerjee SK, Kumar R, Gupta P, et al. MicroRNA 26a (miR-26a)/KLF4 and CREB-C/EBPβ regulate innate immune signaling, the polarization of macrophages and the trafficking of Mycobacterium tuberculosis to lysosomes during infection. PloS Pathog. (2017) 13:e1006410. doi: 10.1371/journal.ppat.1006410
148. Jingjing Z, Nan Z, Wei W, Qinghe G, Weijuan W, Peng W, et al. MicroRNA-24 modulates staphylococcus aureus-induced macrophage polarization by suppressing CHI3L1. Inflammation. (2017) 40(3):995–1005. doi: 10.1007/s10753-017-0543-3
149. Zhang T, Yu J, Zhang Y, Li L, Chen Y, Li D, et al. Salmonella enterica serovar enteritidis modulates intestinal epithelial miR-128 levels to decrease macrophage recruitment via macrophage colony-stimulating factor. J Infect Dis. (2014) 209:2000–11. doi: 10.1093/infdis/jiu006
150. Dorhoi A, Iannaccone M, Farinacci M, Faé KC, Schreiber J, Moura-Alves P, et al. MicroRNA-223 controls susceptibility to tuberculosis by regulating lung neutrophil recruitment. J Clin Invest. (2013) 123:4836–48. doi: 10.1172/JCI67604
151. Stein JP, Grossfeld GD, Ginsberg DA, Esrig D, Freeman JA, Figueroa AJ, et al. Prognostic markers in bladder cancer: a contemporary review of the literature. J Urol. (1998) 160:645–59. doi: 10.1016/S0022-5347(01)62747-2
152. Miao L, Liu K, Xie M, Xing Y, Xi T. miR-375 inhibits Helicobacter pylori-induced gastric carcinogenesis by blocking JAK2-STAT3 signaling. Cancer Immunol Immunother. (2014) 63:699–711. doi: 10.1007/s00262-014-1550-y
153. Noto JM, Piazuelo MB, Chaturvedi R, Bartel CA, Thatcher EJ, Delgado A, et al. Strain-specific suppression of microRNA-320 by carcinogenic Helicobacter pylori promotes expression of the antiapoptotic protein Mcl-1. Am J Physiol Gastrointest Liver Physiol. (2013) 305:G786–96. doi: 10.1152/ajpgi.00279.2013
154. Teng G, Dai Y, Chu Y, Li J, Zhang H, Wu T, et al. Helicobacter pylori induces caudal-type homeobox protein 2 and cyclooxygenase 2 expression by modulating microRNAs in esophageal epithelial cells. Cancer Sci. (2018) 109:297–307. doi: 10.1111/cas.2018.109.issue-2
155. Zhou X, Li L, Su J, Zhang G. Decreased miR-204 in H. pylori-associated gastric cancer promotes cancer cell proliferation and invasion by targeting SOX4. PloS One. (2014) 9(7):e101457. doi: 10.1371/journal.pone.0101457
156. Zhou X, Xu G, Yin C, Jin W, Zhang G. Down-regulation of miR-203 induced by Helicobacter pylori infection promotes the proliferation and invasion of gastric cancer by targeting CASK. Oncotarget. (2014) 5:11631–40. doi: 10.18632/oncotarget.v5i22
157. Zhu Y, Jiang Q, Lou X, Ji X, Wen Z, Wu J, et al. MicroRNAs up-regulated by CagA of Helicobacter pylori induce intestinal metaplasia of gastric epithelial cells. PloS One. (2012) 7:e35147. doi: 10.1371/journal.pone.0035147
158. Li N, Tang B, Zhu E-D, Li B-S, Zhuang Y, Yu S, et al. Increased miR-222 in H. pylori-associated gastric cancer correlated with tumor progression by promoting cancer cell proliferation and targeting RECK. FEBS Lett. (2012) 586:722–8. doi: 10.1016/j.febslet.2012.01.025
159. Tan X, Tang H, Bi J, Li N, Jia Y. MicroRNA-222-3p associated with Helicobacter pylori targets HIPK2 to promote cell proliferation, invasion, and inhibits apoptosis in gastric cancer. J Cell Biochem. (2018) 119:5153–62. doi: 10.1002/jcb.v119.7
160. Zhang Z, Li Z, Gao C, Chen P, Chen J, Liu W, et al. miR-21 plays a pivotal role in gastric cancer pathogenesis and progression. Lab Invest. (2008) 88:1358–66. doi: 10.1038/labinvest.2008.94
161. Maudet C, Mano M, Sunkavalli U, Sharan M, Giacca M, Förstner KU, et al. Functional high-throughput screening identifies the miR-15 microRNA family as cellular restriction factors for Salmonella infection. Nat Commun. (2014) 5:4718. doi: 10.1038/ncomms5718
162. Roy BC, Subramaniam D, Ahmed I, Jala VR, Hester CM, Greiner KA, et al. Role of bacterial infection in the epigenetic regulation of Wnt antagonist WIF1 by PRC2 protein EZH2. Oncogene. (2015) 34:4519–30. doi: 10.1038/onc.2014.386
163. Huang J, Jiao J, Xu W, Zhao H, Zhang C, Shi Y, et al. MiR-155 is upregulated in patients with active tuberculosis and inhibits apoptosis of monocytes by targeting FOXO3. Mol Med Rep. (2015) 12:7102–8. doi: 10.3892/mmr.2015.4250
164. Liu Y, Jiang J, Wang X, Zhai F, Cheng X. miR-582-5p is upregulated in patients with active tuberculosis and inhibits apoptosis of monocytes by targeting FOXO1. PloS One. (2013) 8:e78381. doi: 10.1371/journal.pone.0078381
165. Luo P-Y, Zou J-R, Chen T, Zou J, Li W, Chen Q, et al. Autophagy in erectile dysfunction: focusing on apoptosis and fibrosis. Asian J Androl. (2024). doi: 10.4103/aja202433
166. Liu K, Chen H, Li Y, Wang B, Li Q, Zhang L, et al. Autophagy flux in bladder cancer: Cell death crosstalk, drug and nanotherapeutics. Cancer Lett. (2024) 591:216867. doi: 10.1016/j.canlet.2024.216867
167. Nassour J, Radford R, Correia A, Fusté JM, Schoell B, Jauch A, et al. Autophagic cell death restricts chromosomal instability during replicative crisis. Nature. (2019) 565:659–63. doi: 10.1038/s41586-019-0885-0
168. Jagannath C, Lindsey DR, Dhandayuthapani S, Xu Y, Hunter RL, Eissa NT. Autophagy enhances the efficacy of BCG vaccine by increasing peptide presentation in mouse dendritic cells. Nat Med. (2009) 15:267–76. doi: 10.1038/nm.1928
169. Kumar R, Sahu SK, Kumar M, Jana K, Gupta P, Gupta UD, et al. MicroRNA 17-5p regulates autophagy in Mycobacterium tuberculosis-infected macrophages by targeting Mcl-1 and STAT3. Cell Microbiol. (2016) 18:679–91. doi: 10.1111/cmi.12540
170. Chen Z, Wang T, Liu Z, Zhang G, Wang J, Feng S, et al. Inhibition of autophagy by miR-30A induced by mycobacteria tuberculosis as a possible mechanism of immune escape in human macrophages. Jpn J Infect Dis. (2015) 68:420–4. doi: 10.7883/yoken.JJID.2014.466
171. Kim JK, Yuk J-M, Kim SY, Kim TS, Jin HS, Yang C-S, et al. MicroRNA-125a inhibits autophagy activation and antimicrobial responses during mycobacterial infection. J Immunol. (2015) 194:5355–65. doi: 10.4049/jimmunol.1402557
172. Ouimet M, Koster S, Sakowski E, Ramkhelawon B, van Solingen C, Oldebeken S, et al. Mycobacterium tuberculosis induces the miR-33 locus to reprogram autophagy and host lipid metabolism. Nat Immunol. (2016) 17:677–86. doi: 10.1038/ni.3434
173. Duan X, Zhang T, Ding S, Wei J, Su C, Liu H, et al. microRNA-17-5p modulates bacille calmette-guerin growth in RAW264.7 cells by targeting ULK1. PloS One. (2015) 10(9):e0138011. doi: 10.1371/journal.pone.0138011
174. Guo L, Zhao J, Qu Y, Yin R, Gao Q, Ding S, et al. microRNA-20a inhibits autophagic process by targeting ATG7 and ATG16L1 and favors mycobacterial survival in macrophage cells. Front Cell Infect Microbiol. (2016) 6:134. doi: 10.3389/fcimb.2016.00134
175. Guo L, Zhou L, Gao Q, Zhang A, Wei J, Hong D, et al. MicroRNA-144-3p inhibits autophagy activation and enhances Bacillus Calmette-Guérin infection by targeting ATG4a in RAW264.7 macrophage cells. PloS One. (2017) 12(6):e0179772. doi: 10.1371/journal.pone.0179772
176. Yang X-J, Si R-H, Liang Y-H, Ma B-Q, Jiang Z-B, Wang B, et al. Mir-30d increases intracellular survival of Helicobacter pylori through inhibition of autophagy pathway. World J Gastroenterol. (2016) 22:3978–91. doi: 10.3748/wjg.v22.i15.3978
177. Li Q, Fang Y, Zhu P, Ren C-Y, Chen H, Gu J, et al. Burkholderia pseudomallei survival in lung epithelial cells benefits from miRNA-mediated suppression of ATG10. Autophagy. (2015) 11:1293–307. doi: 10.1080/15548627.2015.1058474
178. Liu S, da Cunha AP, Rezende RM, Cialic R, Wei Z, Bry L, et al. The host shapes the gut microbiota via fecal microRNA. Cell Host Microbe. (2016) 19:32–43. doi: 10.1016/j.chom.2015.12.005
179. Liu S, Rezende RM, Moreira TG, Tankou SK, Cox LM, Wu M, et al. Oral Administration of miR-30d from Feces of MS Patients Suppresses MS-like Symptoms in Mice by Expanding Akkermansia muciniphila. Cell Host Microbe. (2019) 26(6):779–94.e8. doi: 10.1016/j.chom.2019.10.008
180. Teng Y, Ren Y, Sayed M, Hu X, Lei C, Kumar A, et al. Plant-derived exosomal microRNAs shape the gut microbiota. Cell Host Microbe. (2018) 24(5):637–52.e8. doi: 10.1016/j.chom.2018.10.001
181. Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell. (2017) 170(3):548–63.e16. doi: 10.1016/j.cell.2017.07.008
182. Zhou C, Rao X, Wang H, Zeng B, Yu Y, Chen J, et al. Hippocampus-specific regulation of long non-coding RNA and mRNA expression in germ-free mice. Funct Integr Genomics. (2020) 20:355–65. doi: 10.1007/s10142-019-00716-w
183. Zhang Y, Ma W, Lin H, Gu X, Xie H. The effects of esketamine on the intestinal microenvironment and intestinal microbiota in mice. Hum Exp Toxicol. (2023) 42:9603271231211894. doi: 10.1177/09603271231211894
184. Wang Y, Wang M, Chen J, Li Y, Kuang Z, Dende C, et al. The gut microbiota reprograms intestinal lipid metabolism through long noncoding RNA Snhg9. Science. (2023) 381:851–7. doi: 10.1126/science.ade0522
185. Yang F, Yang Y, Chen L, Zhang Z, Liu L, Zhang C, et al. The gut microbiota mediates protective immunity against tuberculosis via modulation of lncRNA. Gut Microbes. (2022) 14:2029997. doi: 10.1080/19490976.2022.2029997
186. Tang R, Xiao G, Jian Y, Yuan Q, Jiang C, Wang W. The Gut Microbiota Dysbiosis in Preeclampsia Contributed to Trophoblast Cell Proliferation, Invasion, and Migration via lncRNA BC030099/NF-κB Pathway. Mediators Inflammation. (2022) 2022:6367264. doi: 10.1155/2022/6367264
187. Lin JC, Ma XY, Liu JY, Luo YZ, Lin L, Zhang LY. One gut microbiota, Fusobacterium nucleatum aggravates Neonatal necrotizing enterocolitis by induction of IRF5 expression through lncRNA ENO1-IT1/miR-22-3p axis. Eur Rev Med Pharmacol Sci. (2021) 25(20):4714–28. doi: 10.26355/eurrev_202110_26984
188. Xi Y, Yuefen P, Wei W, Quan Q, Jing Z, Jiamin X, et al. Analysis of prognosis, genome, microbiome, and microbial metabolome in different sites of colorectal cancer. J Transl Med. (2019) 17:353. doi: 10.1186/s12967-019-2102-1
189. Hong J, Guo F, Lu S-Y, Shen C, Ma D, Zhang X, et al. F. nucleatum targets lncRNA ENO1-IT1 to promote glycolysis and oncogenesis in colorectal cancer. Gut. (2021) 70:2123–37. doi: 10.1136/gutjnl-2020-322780
190. Diling C, Longkai Q, Yinrui G, Yadi L, Xiaocui T, Xiangxiang Z, et al. CircNF1-419 improves the gut microbiome structure and function in AD-like mice. Aging (Albany NY). (2020) 12:260–87. doi: 10.18632/aging.102614
191. Zhu Z, Huang J, Li X, Xing J, Chen Q, Liu R, et al. Gut microbiota regulate tumor metastasis via circRNA/miRNA networks. Gut Microbes. (2020) 12:1788891. doi: 10.1080/19490976.2020.1788891
192. Moloney GM, O'Leary OF, Salvo-Romero E, Desbonnet L, Shanahan F, Dinan TG, et al. Microbial regulation of hippocampal miRNA expression: Implications for transcription of kynurenine pathway enzymes. Behav Brain Res. (2017) 334:50–4. doi: 10.1016/j.bbr.2017.07.026
Keywords: non-coding RNA, gut microbiota, bladder cancer, inflammation, autophagy
Citation: Zou J, Xu B, Luo P, Chen T and Duan H (2024) Non-coding RNAs in bladder cancer, a bridge between gut microbiota and host? Front. Immunol. 15:1482765. doi: 10.3389/fimmu.2024.1482765
Received: 18 August 2024; Accepted: 30 October 2024;
Published: 19 November 2024.
Edited by:
Khashayarsha Khazaie, Mayo Clinic Arizona, United StatesReviewed by:
Kulbhushan Thakur, University of Delhi, IndiaShirin Eyvazi Khasraghi, Mayo Clinic Arizona, United States
Copyright © 2024 Zou, Xu, Luo, Chen and Duan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huanglin Duan, MTU3Nzk4MzExNzlAMTYzLmNvbQ==
†These authors have contributed equally to this work and share first authorship
 Jun Zou
Jun Zou Baisheng Xu2†
Baisheng Xu2† Peiyue Luo
Peiyue Luo Tao Chen
Tao Chen Huanglin Duan
Huanglin Duan