- 1Hubei Provincial Key Laboratory of Occurrence and Intervention of Rheumatic Diseases, Minda Hospital of Hubei Minzu University, Hubei Minzu University, Enshi, Hubei, China
- 2Department of Nephrology, Minda Hospital Affiliated to Hubei Minzu University, Hubei Clinical Research Center for Kidney Disease, Hubei Minzu University, Enshi, Hubei, China
- 3Department of Endocrinology, Minda Hospital Affiliated to Hubei Minzu University, Hubei Clinical Research Center for Kidney Disease, Hubei Minzu University, Enshi, Hubei, China
- 4Laboratory of Immunology for Environment and Health, Shandong Analysis and Test Center, Qilu University of Technology (Shandong Academy of Sciences), Jinan, China
Signal transducer and activator of transcription 4 (STAT4) is a member of the STAT family, which is a group of transcription factors that regulate cytokine signaling. Genetic polymorphisms in STAT4 strongly influence immune responses and disease outcomes, especially in cancer and autoimmune diseases. Several studies have indicated that certain STAT4 gene variants are associated with alterations in STAT4 expression and/or activity and that there is a close relationship between STAT4 polymorphisms and drug efficacy. However, the underlying mechanisms are complex, and the roles of these polymorphisms in disease acquisition, progression, and severity are of widespread concern. Therefore, we provide an overview of the clinical significance of polymorphisms in STAT4 and the mechanisms by which these STAT4 variants are involved in various diseases.
1 Introduction
STAT4 plays a central role in signal transduction, particularly in facilitating the production of biomolecules closely associated with autoimmune diseases. Studies have shown that mice lacking STAT4 exhibit a higher resistance to inflammatory conditions such as colitis, arthritis, diabetes, myocarditis, and experimental autoimmune encephalitis (1–6). In the human genetic system, the STAT4 gene is located on chromosome 2q32.2, with its DNA sequence spanning approximately 220 kilobases (kb) and containing 24 exons and 23 introns. The protein encoded by the STAT4 gene has a molecular weight of approximately 84 kDa and consists of 748 amino acids. Among its family members, the STAT4 protein exhibits the highest tissue specificity and is highly expressed in the lymph nodes, myeloid lineage, testicular tissue, and skin. Structurally, STAT4 resembles other STAT proteins in that it is composed of multiple conserved regions in both structure and function, including the N-terminal domain (NTD), the coiled-coil domain (CCD), the DNA-binding domain (DBD), a linker domain (LD), the Src-homology 2 (SH2) domain, and the C-terminal transcription activation domain (TAD) (7). Functionally, the active form of STAT4 is primarily activated through phosphorylation by Janus kinase 2 (JAK2) and tyrosine kinase 2 (TYK2), forming pSTAT4, which promotes STAT4 dimerization, nuclear translocation, and transcriptional activation of its target genes (8, 9). Notably, during this process, the SH2 domains of STAT4 proteins bind to tyrosine-phosphorylation sites (pYs) on receptor complexes as well as pYs on other STAT proteins and work together with the NTD to mediate the formation of homodimers or heterodimers by STAT4. Notably, studies indicate that STATs lacking the NTD cannot form tetramers at DNA-binding sites (10), which potentially impedes the full transcriptional activation of many STAT target genes (11). The CCD and TAD domains allow the STAT4 protein to bind to other transcription factors or co-activators, and depending on the presence of the TAD domain, there can be two spliced variants of STAT4: STAT4α, which contains the TAD, and STAT4β, which lacks it (12).
Although both STAT4α and STAT4β can activate transcription in primary cells and cell lines, studies have demonstrated that the absence of the TAD domain in STAT4β can markedly alter its function (12). In addition, studies have indicated that genetic polymorphisms in STAT4 also influence immune responses and disease susceptibility. For example, unstimulated cells transfected with STAT4 H623Y or A635V variants exhibited greater accumulation of STAT4 in the nucleus (13). Therefore, this review aims to summarize various reports on STAT4 polymorphisms, the effects of these polymorphisms on disease susceptibility and treatment effectiveness, and the mechanisms through which these STAT4 variants are involved in these diseases.
2 Biological effects of STAT4
As shown in Figure 1, STAT4 is mainly activated within the cytoplasm by cytokines such as IL-12, IL-23, and type I interferons, via the JAK2 and TYK2 kinase pathways and/or the p38/MKK6 signaling cascade. This, in turn, leads to the phosphorylation of STAT4 on tyrosine and/or serine residues, thereby promotes STAT4 dimerization, nuclear translocation. Ultimately, this leads to the transcriptional activation of STAT4’s target genes, fulfilling its functional role. Studies have indicated that JAK activation most likely results in the phosphorylation of tyrosine residue 693 on STAT4, which facilitates STAT4 dimerization and nuclear translocation. Mutation at this site completely abrogates IL-12-induced STAT4 transcriptional activity. In contrast, activation of the p38/MKK6 signaling pathway leads to phosphorylation of STAT4 at serine residue 721. Unlike tyrosine phosphorylation, it has been proposed serine phosphorylation of STAT4 is dispensable for nuclear translocation or DNA binding of STAT4, but is required for STAT4 full transcriptional activation (14). Furthermore, it was suggested that serine phosphorylation of STAT4 is partially dependent on precedent tyrosine phosphorylation of STAT4, whereas tyrosine phosphorylation of STAT4 can be seen even in the absence of serine phosphorylation (15), which further implying that the phosphorylation of STAT4 by p38 occurs in the nucleus. Additionally, the phosphorylation of residues serine 733, 743, and 713 is also closely associated with the activation of STAT4. It is speculated that these non-canonical serine phosphorylations may cause the dissociation of STAT4 from the Leukemia Inhibitory Factor Receptor (LIFR), enabling its subsequent nuclear translocation (16).
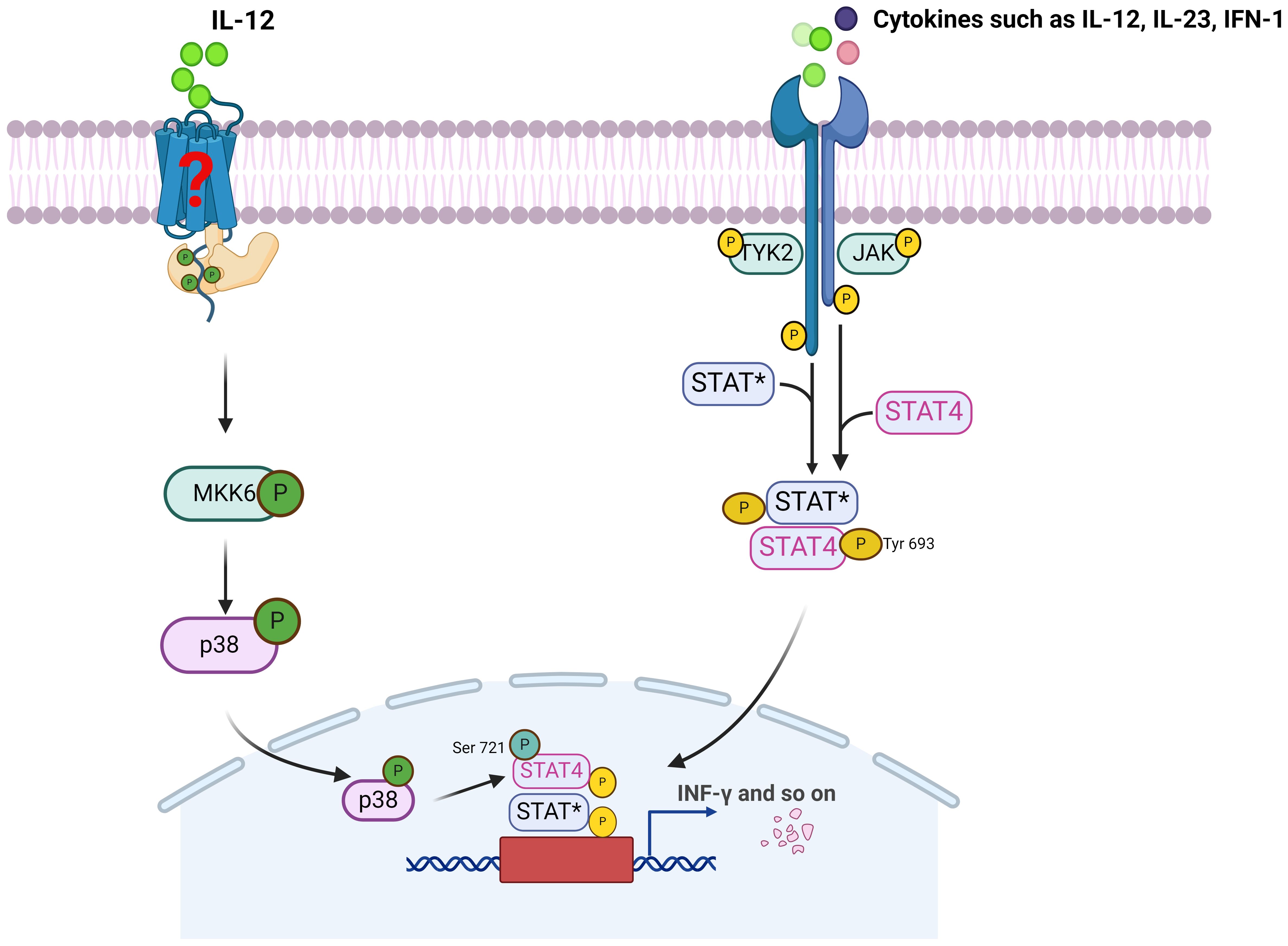
Figure 1. The activation of STAT4 and its mainly signaling pathway. JAK, Janus kinase; TYK2, Tyrosine kinase 2; STAT*, Signal transducer and activator of transcription 4 or other STAT; MKK6, MAP kinase kinase 6.
Although numerous immune cells do not express STAT4 in their basal state (17, 18), the regulation of STAT4 expression appears to play a crucial role in the immune response. For example, STAT4 expression is rarely detected in immature DCs (iDCs) and mucosal mast cells (MMCs), while highly expressed in matured DCs, connective tissue-type mast cells (CTMCs) and activated monocytes (19, 20). Besides, Stat4 expression in T cells is greatly influenced by their state of activation. Human peripheral blood T cells do not have Stat4 in their basal state, but its expression is markedly induced following stimulation (18). It is worth noting that there are differences in the activation mechanisms of STAT4 among different species. For example, in mice, STAT4 can only be activated by IL-12, whereas in humans, STAT4 is not only activated by IL-12 but also phosphorylated by IFN-α (21, 22). Moreover, the activators of STAT4 may vary among different cell types, with monocyte-expressed STAT4 responding to IFN-α but not IL-12 (17).
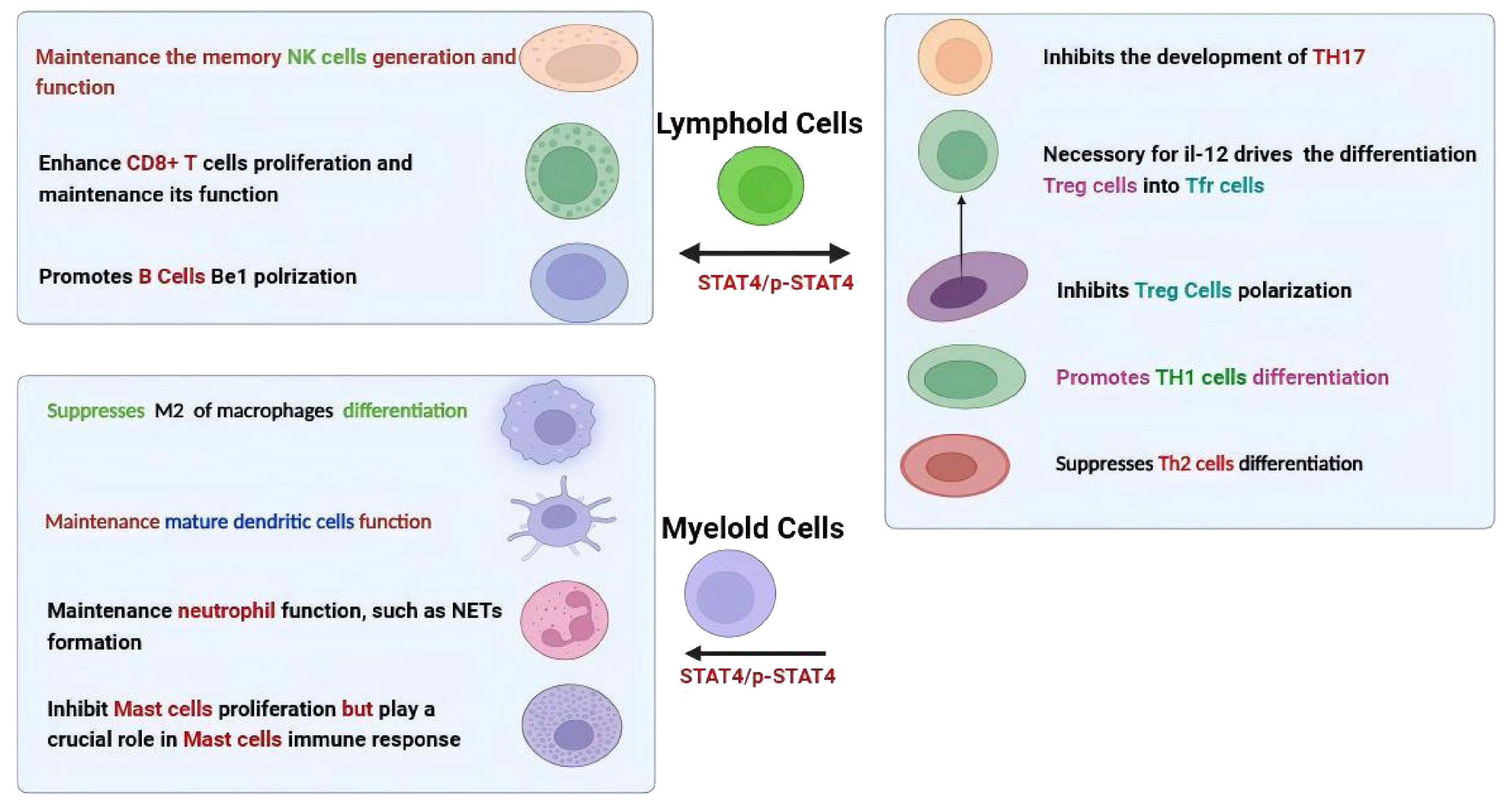
As shown in Figure 2, the function of STAT4 in the immune system mainly encompasses both innate and adaptive immune responses. In particular, for T helper (Th) cells, STAT4 is vital in regulating their development and effector functions. It was suggested STAT4 promotes the differentiation of Th1 cells mainly through upregulating the expression of interleukin-12 receptor β2 (IL-12Rβ2) and enhancing IL-12Rβ2-mediated signaling (23–25). STAT4 is also necessary for follicular helper T (Tfh) cell development, and increased activation of STAT4 in these cells leads to abnormal production of IL-21 and IFN-γ (26, 27). Moreover, STAT4 is involved in the process by which IL-12 drives the differentiation of human regulatory T (Treg) cells into T follicular regulatory (Tfr) cells (28). In contrast, CD4+ T cells lacking STAT4 have a greater tendency to differentiate into Th2 cells (3, 29), and STAT4 also demonstrates a potent ability to inhibit the development of Th17 precursor cells by inducing their conversion into IFN-γ-producing cells (30). In terms of CD8+ T cells, STAT4 plays a vital role in the homeostatic self-renewal of CD8+ T cells and IL-12–induced STAT4 is crucial for the proliferation of effector CD8+ T cells in an mTOR-dependent manner, and upregulated STAT4 expression significantly enhances their survival capacity and infiltration (31–34). Conversely, its absence impairs its effector functions (32, 35). Furthermore, the activation of natural killer (NK) cells also strictly depends on STAT4, which influences NK cell function by regulating the production of IFN-γ and perforin (36–38). High levels of STAT4 contribute to the maintenance of memory NK cell generation, as the number of NK cells in wild-type (WT) mice is greater compared to those in STAT4-deficient mice after infections with mouse cytomegalovirus (MCMV) (39). Meanwhile, pSTAT4 is necessary for the Be1 polarization of human naive B cells (40).
While, STAT4 not only actively participates in the differentiation of lymphocytes but also plays a key role in the activation of myeloid cells such as monocytes and dendritic cells. For instance, studies have shown that the alternative activation (M2) of macrophages is enhanced in diet-induced obese mice due to the lack of STAT4 (41). STAT4 is required for intrinsic signaling in mature dendritic cells (mDCs) function (42), and STAT4-deficient plasmacytoid dendritic cells (pDCs) exhibit defects in the production of IFN-γ (43). Contradictorily, despite research indicating that STAT4 regulates IL-6 through an autocrine mechanism to inhibit the proliferation of CTMCs, it plays a crucial role in mast cell immune response (20, 44, 45). Furthermore, recent research has shown that neutrophil-specific deletion of STAT4 resulted in enhanced susceptibility to methicillin-resistant Staphylococcus aureus (MRSA) (46). This discovery indicates that STAT4 also plays a crucial role in neutrophil function. Specifically, it has been found to exert important effects through multiple aspects such as the production of reactive oxygen species (ROS), chemotaxis, and the formation of neutrophil extracellular traps (NETs).
3 Polymorphisms in the STAT4 gene
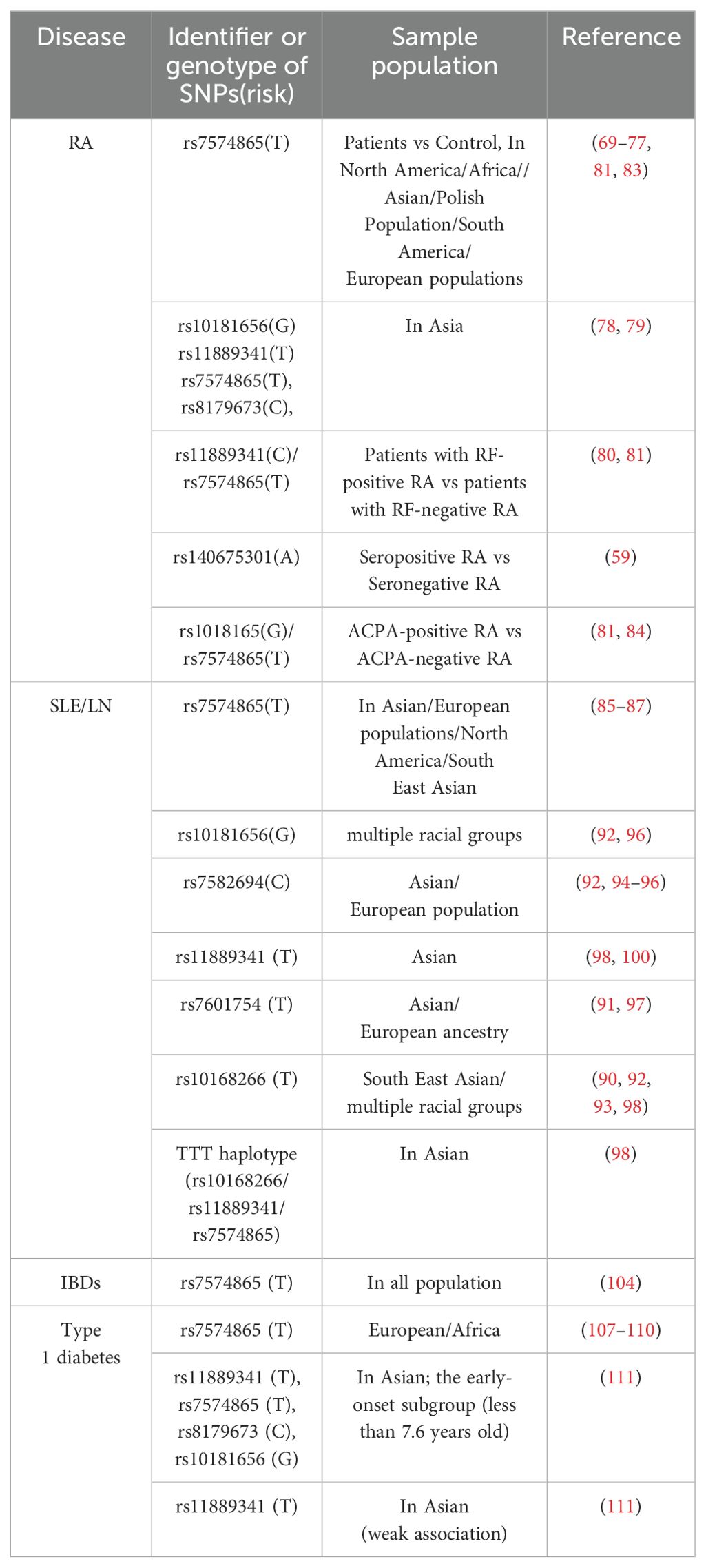
Various polymorphisms within and surrounding the STAT4 gene sequence have been identified. These variations can be broadly categorized into three classes: those in intron regions, those in the 5′-flanking DNA, and those within the open reading frame. Currently, STAT4 polymorphisms and their potential links to diseases have been reported in more than one hundred studies from various countries. These are illustrated in Figure 3.
3.1 Single nucleotide polymorphisms in introns
Presently, the majority of known STAT4 gene variations occur within introns. During RNA processing, the intron sequences of the primary RNA transcripts derived from both normal and variant alleles are excised, resulting in the production of identical STAT4 mRNAs and subsequent synthesis of identical STAT4 proteins. Notably, however, SNPs within the introns of STAT4, particularly the rs7574865 polymorphism in intron 3, play an important role in regulating STAT4 expression. For example, studies have indicated that patients with systemic lupus erythematosus (SLE) carrying the rs7574865 T allele have increased levels of STAT4 mRNA in osteoblasts and peripheral blood mononuclear cells (PBMCs) (47, 48), and when T cells from these patients are stimulated with IL-12 and IFN-α, the levels of STAT4 protein and its phosphorylated form, pSTAT4, are markedly increased (49). Besides, mutations within the STAT4 third intron contain many elements that may alter CD4+ T cell activation induced by IL-12, which may affect the rates of STAT4 transcription and consequently IFNγ production in CD4+ T cells (50). In addition, research has revealed a significant correlation between this risk allele and the methylation status at position -172 of the STAT4 promoter (51). In addition, the SNPs rs3821236, located in intron 16, and rs3024866, located in intron 13, have also been correlated with increased STAT4 expression (48). However, the effects of genetic variants appear to be context-specific, such as significantly elevated STAT4 levels in the serum and peritumoral tissue of hepatocellular carcinoma (HCC) patients with the rs7574865 GG genotype (52). Furthermore, research suggests that rs4853540 located in the STAT4 gene may map to an enhancer of STAT1 (53). And there exists a genotype-dependent repressive element in the DNA surrounding rs11889341. The risk allele rs11889341C enhances HMGA1 binding capability, resulting in reduced repressor activity and consequently higher levels of STAT1 expression (54).
3.2 Single nucleotide polymorphisms in 5′-flanking DNA
A second type of allelic variation occurs within the 5’-flanking region of the STAT4 promoter. Shin HJ and his colleagues demonstrated evidence for allelic variation (−149A/G) in noncoding exon 1 of the STAT4 gene, which is located in the 5’-flanking region of the essential promoter. Subsequently, direct sequencing analysis revealed that this noncore promoter region allelic variation frequently appeared in patients with rheumatoid arthritis (RA) and asthma. However, luciferase reporter gene assays of genes transcriptionally controlled by either the wild-type STAT4 promoter or the variant promoter revealed that this polymorphism had no significant effect on promoter activity (55).
In addition, another SNP located at the edge of the 5’ promoter region of STAT4 (rs897200) has been identified (56). Notably, through analysis using the Regulomedb database and predictions made with ALGGEN PROMO software (57), it was discovered that the site at which this SNP occurs could potentially serve as a transcription factor-binding site. In line with this, subjects with the AA genotype presented significantly higher STAT4 mRNA levels in PBMCs and skin cells than did subjects with the GG genotype, and luciferase activity was notably increased in cells harboring the A allele (58).
3.3 Single nucleotide polymorphisms in extrons
A third type of allelic variation has been identified in exons within the STAT4 open reading frame. This type of polymorphism has the potential to affect both protein structure and function, which is different from the effects of reported STAT4 polymorphisms in noncoding sequences. Recently, several SNPs have been identified in exons.
Through a genome-wide association study (GWAS), Saevarsdottir S and colleagues reported that a rare missense variant in STAT4, rs140675301-A, causes a 2.27-fold increase in the risk of seropositive RA. Specifically, this genetic variant causes the replacement of hydrophilic glutamic acid with hydrophobic valine (Glu128Val) in a conserved surface-exposed loop between the N-terminal domain and the helical coiled coil domain (59), although the direct impact of the mutation at this site on protein function remains unknown.
Remarkably, three other heterozygous polymorphisms (STAT4 c.1904 C→T, c.1949C→A, and c.1867C→T), resulting in A635V, A650D, and H623Y amino acid substitutions, respectively, in a specific region of the gene that encodes STAT4 were identified. All of these polymorphisms, occur in the region of the STAT4 gene that encodes the SH2 domain. They also discovered that, in unstimulated cells containing STAT4 variants, there was greater accumulation of STAT4 in the nucleus, and the levels of pSTAT4 were increased, and compared with those in control cells, the levels of pSTAT4 not only increased but also remained elevated for a comparatively longer duration in the interferon-stimulated cells. In addition, they found that mutant STAT4 dimers are more stable than wild-type STAT4 dimers (13), but little is known about how these mutations promote the phosphorylation of STAT4 and maintain the stability of its dimers. Regardless, this experiment further suggested that the SH2 domain is strongly linked to the activity of STAT4, indicating that mutations within this domain could significantly alter the phosphorylation and dimerization of STAT4, which in turn affects its transcriptional activity and potentially contributes to the occurrence of disease.
4 Genetic variation and disease susceptibility
STAT4 can be activated by distinct types of cytokines in multiple cells via the JAK-STAT pathway or p38/MKK6 pathway. Subsequently, it acts as a transcription factor to regulate the expression of various genes. This pivotal function makes STAT4 a central mediator in the induction of inflammation during protective immune responses and immune-mediated diseases. Importantly, numerous studies have identified the association between STAT4 polymorphisms and disease susceptibility.
4.1 Polymorphisms and cancer
Associations between STAT4 SNPs and cancer susceptibility have been reported in different cancer types, but the results have been inconsistent. The STAT4 polymorphism rs7574865 has been extensively studied in the context of hepatocellular carcinoma (HCC) and chronic hepatitis B virus (HBV) infection. Although it is widely accepted that the C allele of rs7574865 increases the risk of HCC and is strongly associated with HBV-related HCC, especially in Asian populations (60–62). However, several studies, such as that of Chen and colleagues, have failed to replicate these findings (63). Additionally, a recent report indicated that this SNP (rs7574865) does not influence the risk of developing HCC in Latin American or European populations (64), suggesting that the association between rs7574865 and HCC risk may be specific to certain populations, and more studies are needed. In addition, a recent study suggested a potential association between STAT4 rs11889341 or rs10174238 and HCC risk among the Chinese Han population (65).
Additionally, the variants of STAT4 rs7574865T and rs1400656G serve as protective alleles against the risk of lung cancer (66). STAT4 rs6738544A was significantly associated with pancreatic cancer risk (67). Furthermore, logistic regression analysis revealed that rs4274624 (STAT4) is linked to an elevated risk of breast cancer, whereas STAT4 rs925847 is associated with a reduced risk of breast cancer (68).
4.2 Polymorphisms and autoimmune diseases
SNPs in STAT4 have been reported as risk factors for the development of autoimmune diseases, including RA, SLE, lupus nephritis (LN), type 1 diabetes (T1D), psoriasis, inflammatory bowel disease (IBD), Behçet’s disease (BD), Sjögren’s syndrome (SS), systemic sclerosis (SSc), primary biliary cirrhosis (PBC), and other diseases (Table 1).
4.2.1 RA
Over the past two decades, the potential effects of STAT4 SNPs on susceptibility to RA have been evaluated. Among these, STAT4 rs7574865 is the most well studied and has been identified as an important risk factor for RA across multiple ethnic group (69–77). Specifically, the T allele of rs7574865 is associated with an increased risk of RA, with the TT genotype predominantly found in RA patient. Besides, studies have also revealed significant associations among the SNPs rs10181656(G), rs11889341(T), rs7574865(T), and rs8179673(C) in the STAT4 gene and RA (78, 79).
Furthermore, some research findings indicate that STAT4 gene polymorphisms may be involved in regulating the seropositive status of RA patients. Specifically, researchers have reported that the frequency of the T allele and TT genotype of rs11889341 is significantly lower in the rheumatoid factor (RF)-positive subgroup of RA patients than in the RF-negative subgroup, but there is a lack of direct evidence linking rs11889341 with RA risk (80). Additionally, the frequency of RF positivity was significantly greater among RA patients with the rs7574865 T allele (81). Furthermore, the rs7574865 and rs10181656 loci in the STAT4 gene were associated with anti-cyclic citrullinated peptide (ACPA)-positive RA (81–84), and a rare missense variant in the exon of STAT4, rs140675301A, has also been shown to increase the risk of seropositive RA (59).
4.2.2 SLE and LN
The association between STAT4 gene polymorphisms and SLE/LN remains controversial according to published studies. However, several meta-analyses have consistently indicated that the T allele or TT genotype of the STAT4 rs7574865 polymorphism serves as a risk factor for SLE across different ethnic populations, despite variations in prevalence rates among different ethnicities (85–87). Moreover, higher levels of IFN-γ (interferon-γ) have been reported in TT allele carriers (88). Interestingly, there are significant differences in the functional manifestations of the STAT4 rs7574865 variant between healthy individuals and patients with SLE, and its function may also be influenced by other mutation sites. Specifically, the STAT4 risk allele rs7574865 T has contrasting effects on cells from healthy individuals compared with those from patients with SLE. In SLE patients carrying the STAT4 risk allele rs7574865[T], T cells exhibit increased STAT4 protein and pSTAT4 lever, and elevated IFN-γ production after PHA/interleukin (IL)-2 activation, whereas in healthy individuals, STAT4 risk allele carriers have reduced pSTAT4 levels in CD8+ and CD4+ T cells (89). However, CD4+ naive T cells from both healthy individuals and SLE patients carrying the non-risk homozygous (NR/NR) genotype (rs7582694G and rs7574865G) exhibit significantly lower levels of STAT4 compared to cells carrying the high-risk homozygous (R/R) genotype (rs7582694C and rs7574865T) during T-cell differentiation, and activation of cells from healthy individuals and SLE patients with the R/R genotype shows increased levels of transcriptionally active STAT4 and production of interferon-γ (50).
In addition, several studies have suggested that the minor allele polymorphisms rs10168266T, rs7601754T, rs7582694C and rs3821236A are also associated with SLE (48, 84, 87, 90–97), and that the TTT haplotype (rs10168266/rs11889341/rs7574865) is also linked to SLE (98). Moreover, SLE-smoking patients with the STAT4 SNP rs11889341T allele have a significantly increased risk of LN (99), and also SLE patients with the rs7582694C and rs7574865T allele exhibit a significantly increased incidence of severe renal insufficiency (100, 101).
4.2.3 IBDs
To date, the associations between STAT4 variants and IBD, specifically ulcerative colitis (UC) and Crohn’s disease (CD), remain uncertain. Previous meta-analyses have suggested that the STAT4 rs7574865 T allele may confer increased susceptibility to UC (102, 103). Subsequently, another study analysis has further clarified that this genetic polymorphism is most strongly associated with UC susceptibility among Caucasians (104). However, a case-control association study conducted in a Korean population revealed that, while the SNP rs925847 polymorphism provides a protective effect against UC, none of the other tested STAT4 SNPs (including rs11889341, rs8179673, rs6752770, rs10168266, rs10181656, and rs11685878) were linked to UC susceptibility (105).
In addition, a protective role for rs7568275T and rs10174238 against the risk of Crohn’s disease (CD) was suggested (106). However, a final meta-analysis found no significant association between STAT4 polymorphisms and CD susceptibility (104).
4.2.4 T1D
Although it has been proven previously that several SNPs in the STAT4 gene contribute to the genetic predisposition to T1D, the role of STAT4 polymorphisms in T1D is poorly understood. This association was first observed in a genetically homogeneous population, where susceptibility to type 1 diabetes was associated with a significant increase in the frequency of the STAT4 rs7574865 T allele (107). Subsequently, several studies on Asian, African and European populations confirmed that rs7574865 plays a role in the incidence of T1D (102, 108, 109), and that carriers of the rs7574865 minor T allele presented earlier T1D onset (110). In addition, analysis of the early-onset subgroup revealed that rs11889341, rs7574865, rs8179673, and rs10181656 were significantly associated with susceptibility to T1D, whereas only a weak association was observed between rs11889341 and T1D in general (111). Over all, these findings suggest that certain STAT4 SNPs may be particularly relevant in the context of early-onset T1D.
4.2.5 SSc
The results of studies on the role of STAT4 SNPs in SSc are somewhat conflicting. Similar to other autoimmune diseases, the T allele of rs7574865 has been identified as a susceptibility factor for SSc (112), and a meta-analysis conducted by Xu et al. encompassing six studies revealed that, depending on the extent of skin involvement, the frequency of the STAT4 (rs7574865) risk T allele was increased in both limited cutaneous (lcSSc) and diffuse cutaneous (dcSSc) SSc patients compared with healthy individuals (113). This finding indicates a potential subtype-specific association, but more experiments are needed to verify this observation. Other variants of STAT4, such as rs11889341A and rs10168266T, have been implicated in systemic sclerosis (SSc) in various studies (114, 115). Moreover, carriers of the rs3821236 A-allele and rs7574865 T-allele, along with rs10168266 T-allele carriers, exhibited an increased prevalence of pulmonary fibrosis in SSc patients, and carriers of rs7574865 and rs10168266 T-alleles were also strongly associated with the presence of anti-topoisomerase I (ATA) (115).
4.2.6 Behçet’s disease/periodic fever, aphthous stomatitis, pharyngitis, and cervical adenitis
GWASs in patients with BD have been performed in Turkish, Japanese, Chinese, and Iranian populations, and STAT4 polymorphisms is considered a common risk factor for BD (58, 116–118). Specifically, the rs7572482 risk allele T, rs7574070 risk allele A, and rs897200 risk allele A have been found to be associated with BD. In addition, studies have explored the relationship between gene polymorphisms at other loci of STAT4 and BD, and revealed that the GG genotype of rs7574865 may be a risk factor for BD patients (119, 120). However, a study conducted in the Korean population revealed no association between SNPs of STAT4 (including rs7574070, rs1031508, rs897200, and rs7572482) and the risk of BD in an analysis of individual polymorphisms (121).
In addition, studies have revealed that PFAPA syndrome shares genetic similarities with BD, and that the rs7574070 risk allele A is also an important susceptibility locus for PFAPA (122).
4.2.7 SS
Several studies have been conducted to determine the associations between STAT4 polymorphisms and susceptibility to SS. Among Europeans, a statistically significant association between SS and STAT4 variants was observed, especially the insertion−deletion polymorphism rs10553577 located in the third intron, which exhibited a particularly strong association (123). Additionally, the T allele of rs7574865 at the genetic locus is significantly more common in SS patients, and carriers of this allele exhibit a higher risk of monoclonal component and leukopenia (124). Consistent with these findings, other GWAS analyses also established associations between SS and the gene regions of STAT4, including rs11889341, rs8179673 (125). Moreover, research indicates that rs7574865, rs7582694, and rs10168266 are significantly associated with primary Sjögren’s syndrome (126–130).
4.2.8 Juvenile idiopathic arthritis
Studies have suggested that the rs7574865 risk allele T plays a role in the development of JIA and is associated with this condition in Han Chinese and American populations (131, 132), but not in Iranian or Greek populations (133, 134). Additionally, in Han Chinese populations, the G allele of rs11893432 was notably associated with an increased risk of oligoarticular JIA, whereas the A allele of rs10931481 and the C allele of rs1018981 were suggested to be associated with higher risk of polyarticular JIA (135).
4.2.9 Optic neuritis/neuromyelitis optica spectrum disorder/idiopathic inflammatory myopathy
Additionally, STAT4 polymorphisms have been reported to be associated with other diseases. For example, a recent study revealed that both the G-G-A-C and C-T-A-T haplotypes of STAT4 (rs10181656, rs7574865, rs7601754, and rs10168266) are associated with the occurrence of optic neuritis (136). Besides, four STAT4 variants, including rs7574865 T, rs10181656 G, rs13426947 A, and rs10168266 T, have been reported to be associated with an increased risk of NMOSD, and similar to SLE, the G allele of rs7601754 also displays a protective effect against NMOSD (137). In addition, studies have revealed an association between STAT4 variants and IIM. Through candidate gene studies, it was discovered that both polymyositis and dermatomyositis are strongly associated with the rs7574865T allele (138). Additionally, by constructing a regional association plot of the STAT4 locus, two other SNPs, rs4853540 and rs6752770, were identified as having a significant association with IIM (53). Although these loci do not overlap, these studies collectively reinforce the association between STAT4 and IIM.
4.3 Infections disease
Several studies have indicated an association between STAT4 polymorphisms and infectious diseases. It was well known the C allele of rs7574865 may enhance susceptibility to hepatitis B virus (HBV) infection (61, 139). In addition, the impact of STAT4 polymorphisms appears to be more pronounced in young patients with pulmonary tuberculosis (PTB). A study revealed an association between STAT4 SNPs (rs6752770, rs3024861, rs7572482, rs1031509, rs1400654 and rs897200) and PTB, particularly highlighting a strong correlation between rs897200 and younger PTB patients (pulmonary tuberculosis onset <25 years) (56). While another study revealed that the rs4853542A allele reduces the risk of tuberculosis in younger adults after applying Bonferroni correction (140). Moreover, the rs7574865 TT genotype was identified as a risk factor for cytomegalovirus (CMV) infection (141).
4.4 Other diseases
In addition, the G allele of rs1031509 is significantly associated with an increased risk of developing doctor-diagnosed asthma when induced by exposure to benzo[α]pyrene in the environment (142), and SLE patients carrying the rs11889341 T risk allele appear to have an increased risk of myocardial infarction (MI) and nephritis (99). Moreover, an association between the rs7574865T allele and type-1 autoimmune hepatitis was observed (143), and an analysis of drug-induced liver injury patients revealed a trend toward an association between a STAT4 variant allele (rs7574865T) and hepatocellular injury, although this association was not significant (144).
Overall, these findings suggest that STAT4 polymorphisms have clinical significance. However, more studies with larger samples are needed to verify the roles of STAT4 polymorphisms in these diseases.
5 Polymorphisms and therapeutic efficacy
Clinical evidence suggests that genetic variations may interfere with the mechanism of drug action. Several studies have reported that SNPs of STAT4 are associated with the clinical efficacy of tumor necrosis factor (TNF) inhibitors in the treatment of RA patients (145), and the STAT4 rs7574865 T allele is associated with the absence of a good/moderate EULAR response at 2 years of treatment in RA patients and ETN-treated patients (146). In addition, carriers of the risk allele exhibit exaggerated CD4+ T-cell activation that, in the context of SLE, contribute to more severe disease, and R/R patients may benefit from blockade of the IL-12/STAT4 pathway (50). Additionally, a prospective cohort study suggested that the effectiveness of peginterferon-α (PEG-IFN) therapy can be altered by STAT4 polymorphisms. Specifically, patients in the GT/TT group presented a notably higher HBeAg seroconversion rate and hepatitis B surface antigen loss rate than those in the GG group (147). Similarly, another study revealed that the SNP rs757486 TT genotype was associated with a greater virological response to PEG-IFN therapy, regardless of baseline HBeAg status (148). However, importantly, this correlation was not observed in nucleoside (acid) analog treatments (149).
In addition, a recent study implicated STAT4 gene polymorphisms as the primary genetic factors that play a role in DPM disease, and the JAK inhibitor ruxolitinib has shown promising results in reducing inflammation in patients with DPM caused by gain-of-function variants in STAT4 by precisely targeting the STAT4 signaling pathway (13). Therefore, STAT4 genetic variation can significantly impact drug treatment effectiveness, emphasizing the importance of understanding these variations for the advancement of personalized medicine and improved patient outcomes.
6 Conclusion
Although STAT4 functions as an immunoregulator contributing to diverse human diseases, it is merely one element within a complex network of both proinflammatory and anti-inflammatory molecules. Its activation is tightly regulated by various cytokines, such as interleukin-12, interferon-gamma, and interleukin-1. Interestingly, the effects of STAT4 depend not only on its absolute expression but also in its functional state. The activity of STAT4 can be modulated by regulatory factors such as suppressor of cytokine signaling protein (SOCS) (150, 151), which inhibits its activity by blocking its receptor binding (152).
The majority of STAT4 gene polymorphisms described here occur within introns, and genetic editing has confirmed that the most significantly associated SNPs linked to autoimmune disease are located in the third intron of the gene (192, 153, 154). Among them, three single nucleotide variations, rs7574865 G/T, rs7582694G/C, and rs10181656C/G, are particularly associated with autoimmune disease, and these three loci are in a state of strong linkage disequilibrium. Specifically, the rs7574865 G > T mutation has been extensively studied, with numerous reports indicating that patients who possess a T allele have an increased risk of autoimmune diseases such as RA, multiple sclerosis, SLE, and primary SS (49, 102, 155). However, this situation is reversed in the context of cancer. Specifically, the G allele of rs7574865 has emerged as a risk factor for cancer development and progression, and STAT4 levels in serum and peritumoral tissue of HCC patients with the GG genotype are significantly higher than those found in TT or TG carriers (52, 139). This highlights the complexity of STAT4 gene variations and their context-dependent effects on different diseases.
Further evidence has indicated associations with autoimmune diseases beyond the SNP variants in intron 3. Specifically, the intronic region spanning from intron 14 to exon 17 (encompassing rs3821236G/A, rs3024886C/T, and rs3024877G/A), as well as the region extending from exon 4 to intron 14 (including rs10168266C/T, rs1517352C/A, rs13017460G/A, and rs16833249T/C), has also demonstrated a significant correlation with autoimmune diseases, especially SLE (48). Specifically, rs3821236A and rs10168266T were significantly more common in patients with SLE than in healthy controls (98), and rs7574865 did not exhibit strong linkage disequilibrium with rs3024866 (R2 = 0.29) or rs3821236 (R2 = 0.42) (48). Furthermore, data from the HapMap project revealed significant differences in the frequency distribution and linkage relationships among STAT4 SNPs between Eastern and Western populations. This finding suggests that different variations may serve as independent risk factors for disease susceptibility, with some having stronger effects in specific populations.
Furthermore, given the intimate connection between STAT4 activity and its protein structure, genetic polymorphisms situated within exon region are likely to exert a more crucial influence on the progression of diseases. However, despite the intricate genetic regulation of STAT4 and its pivotal role in immune responses, the impact of genetic variations on disease susceptibility and outcomes is not absolute, as could be expected. Nonetheless, genetic variations influencing STAT4 activity clearly hold significance. Individuals endowed with a genetically predisposed capacity for heightened STAT4 activation may experience more pronounced inflammatory reactions under certain circumstances. Therefore, more research is needed to elucidate the associations between STAT4 polymorphisms and diseases to further clarify how different polymorphisms affect the function of STAT4 and how these effects are related to the pathogenesis and progression of various diseases.
Author contributions
YX: Conceptualization, Writing – review & editing, Writing – original draft. YNX: Conceptualization, Writing – original draft. HZ: Supervision, Validation, Writing – review & editing. LL: Supervision, Conceptualization, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Natural Science Foundation of China (82401012, 22206092). Scientific Research Project of Education Department of Hubei Province (Q20191901). University and Institute Innovation Team Project of Jinan (Grant No. 202333028).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Deng JC, Zeng X, Newstead M, Moore TA, Tsai WC, Thannickal VJ, et al. STAT4 is a critical mediator of early innate immune responses against pulmonary Klebsiella infection. J Immunol. (2004) 173:4075–83. doi: 10.4049/jimmunol.173.6.4075
2. Hildner KM, Schirmacher P, Atreya I, Dittmayer M, Bartsch B, Galle PR, et al. Targeting of the transcription factor STAT4 by antisense phosphorothioate oligonucleotides suppresses collagen-induced arthritis. J Immunol. (2007) 178:3427–36. doi: 10.4049/jimmunol.178.6.3427
3. Xue YL, Zhang SX, Zheng CF, Li YF, Zhang LH, Hao YF, et al. Silencing of STAT4 Protects Against Autoimmune Myocarditis by Regulating Th1/Th2 Immune Response via Inactivation of the NF-κB Pathway in Rats. Inflammation. (2019) 42:1179–89. doi: 10.1007/s10753-019-00978-3
4. Holz A, Bot A, Coon B, Wolfe T, Grusby MJ, von Herrath MG. Disruption of the STAT4 signaling pathway protects from autoimmune diabetes while retaining antiviral immune competence. J Immunol. (1999) 163:5374–82.
5. Afanasyeva M, Wang Y, Kaya Z, Stafford EA, Dohmen KM, Sadighi Akha AA, et al. Interleukin-12 receptor/STAT4 signaling is required for the development of autoimmune myocarditis in mice by an interferon-gamma-independent pathway. Circulation. (2001) 104:3145–51. doi: 10.1161/hc5001.100629
6. Chitnis T, Najafian N, Benou C, Salama AD, Grusby MJ, Sayegh MH, et al. Effect of targeted disruption of STAT4 and STAT6 on the induction of experimental autoimmune encephalomyelitis. J Clin Invest. (2001) 108:739–47. doi: 10.1172/JCI12563
7. Levy DE, Darnell JE Jr. Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. (2002) 3:651–62. doi: 10.1038/nrm909
8. Watford WT, Hissong BD, Bream JH, Kanno Y, Muul L, O'Shea JJ. Signaling by IL-12 and IL-23 and the immunoregulatory roles of STAT4. Immunol Rev. (2004) 202:139–56. doi: 10.1111/j.0105-2896.2004.00211.x
9. Palmroth M, Kuuliala K, Peltomaa R, Virtanen A, Kuuliala A, Kurttila A, et al. Tofacitinib Suppresses Several JAK-STAT Pathways in Rheumatoid Arthritis In Vivo and Baseline Signaling Profile Associates With Treatment Response. Front Immunol. (2021) 12:738481. doi: 10.3389/fimmu.2021.738481
10. Vinkemeier U, Cohen SL, Moarefi I, Chait BT, Kuriyan J, Darnell JE Jr. DNA binding of in vitro activated Stat1 alpha, Stat1 beta and truncated Stat1: interaction between NH2-terminal domains stabilizes binding of two dimers to tandem DNA sites. EMBO J. (1996) 15:5616–26.
11. Xu X, Sun YL, Hoey T. Cooperative DNA binding and sequence-selective recognition conferred by the STAT amino-terminal domain. Science. (1996) 273:794–7. doi: 10.1126/science.273.5276.794
12. Hoey T, Zhang S, Schmidt N, Yu Q, Ramchandani S, Xu X, et al. Distinct requirements for the naturally occurring splice forms Stat4alpha and Stat4beta in IL-12 responses. EMBO J. (2003) 22:4237–48. doi: 10.1093/emboj/cdg393
13. Baghdassarian H, Blackstone SA, Clay OS, Philips R, Matthiasardottir B, Nehrebecky M, et al. Variant STAT4 and Response to Ruxolitinib in an Autoinflammatory Syndrome. N Engl J Med. (2023) 388:2241–52. doi: 10.1056/NEJMoa2202318
14. Visconti R, Gadina M, Chiariello M, Chen EH, Stancato LF, Gutkind JS, et al. Importance of the MKK6/p38 pathway for interleukin-12-induced STAT4 serine phosphorylation and transcriptional activity. Blood. (2000) 96:1844–52.
15. Morinobu A, Gadina M, Strober W, Visconti R, Fornace A, Montagna C, et al. STAT4 serine phosphorylation is critical for IL-12-induced IFN-gamma production but not for cell proliferation. Proc Natl Acad Sci U S A. (2002) 99:12281–6. doi: 10.1073/pnas.182618999
16. Zhang YS, Xin DE, Wang Z, Song X, Sun Y, Zou QC, et al. STAT4 activation by leukemia inhibitory factor confers a therapeutic effect on intestinal inflammation. EMBO J. (2019) 38:e99595. doi: 10.15252/embj.201899595
17. Frucht DM, Aringer M, Galon J, Danning C, Brown M, Fan S, et al. Stat4 is expressed in activated peripheral blood monocytes, dendritic cells, and macrophages at sites of Th1-mediated inflammation. J Immunol. (2000) 164:4659–64. doi: 10.4049/jimmunol.164.9.4659
18. Bacon CM, Petricoin EF 3rd, Ortaldo JR, Rees RC, Larner AC, Johnston JA, et al. Interleukin 12 induces tyrosine phosphorylation and activation of STAT4 in human lymphocytes. Proc Natl Acad Sci U S A. (1995) 92:7307–11. doi: 10.1073/pnas.92.16.7307
19. Longman RS, Braun D, Pellegrini S, Rice CM, Darnell RB, Albert ML. Dendritic-cell maturation alters intracellular signaling networks, enabling differential effects of IFN-alpha/beta on antigen cross-presentation. Blood. (2007) 109:1113–22. doi: 10.1182/blood-2006-05-023465
20. Kataoka TR, Komazawa N, Morii E, Oboki K, Nakano T. Involvement of connective tissue-type mast cells in Th1 immune responses via Stat4 expression. Blood. (2005) 105:1016–20. doi: 10.1182/blood-2004-07-2811
21. Cho SS, Bacon CM, Sudarshan C, Rees RC, Finbloom D, Pine R, et al. Activation of STAT4 by IL-12 and IFN-alpha: evidence for the involvement of ligand-induced tyrosine and serine phosphorylation. J Immunol. (1996) 157:4781–9.
22. Rogge L, D'Ambrosio D, Biffi M, Penna G, Minetti LJ, Presky DH, et al. The role of Stat4 in species-specific regulation of Th cell development by type I IFNs. J Immunol. (1998) 161:6567–74.
23. Letimier FA, Passini N, Gasparian S, Bianchi E, Rogge L. Chromatin remodeling by the SWI/SNF-like BAF complex and STAT4 activation synergistically induce IL-12Rbeta2 expression during human Th1 cell differentiation. EMBO J. (2007) 26:1292–302. doi: 10.1038/sj.emboj.7601586
24. Chang HC, Han L, Goswami R, Nguyen ET, Pelloso D, Robertson MJ, et al. Impaired development of human Th1 cells in patients with deficient expression of STAT4. Blood. (2009) 113:5887–90. doi: 10.1182/blood-2008-09-179820
25. Tahvanainen J, Kyläniemi MK, Kanduri K, Gupta B, Lähteenmäki H, Kallonen T, et al. Proviral integration site for Moloney murine leukemia virus (PIM) kinases promote human T helper 1 cell differentiation. J Biol Chem. (2013) 288:3048–58. doi: 10.1074/jbc.M112.361709
26. Weinstein JS, Laidlaw BJ, Lu Y, Wang JK, Schulz VP, Li N, et al. STAT4 and T-bet control follicular helper T cell development in viral infections. J Exp Med. (2018) 215:337–55. doi: 10.1084/jem.2017045702062018c
27. Dong X, Antao OQ, Song W, Sanchez GM, Zembrzuski K, Koumpouras F, et al. Type I Interferon-Activated STAT4 Regulation of Follicular Helper T Cell-Dependent Cytokine and Immunoglobulin Production in Lupus. Arthritis Rheumatol (Hoboken NJ). (2021) 73:478–89. doi: 10.1002/art.41532
28. Castaño D, Wang S, Atencio-Garcia S, Shields EJ, Rico MC, Sharpe H, et al. IL-12 drives the differentiation of human T follicular regulatory cells. Sci Immunol. (2024) 9:eadf2047. doi: 10.1126/sciimmunol.adf2047
29. Kaplan MH, Sun YL, Hoey T, Grusby MJ. Impaired IL-12 responses and enhanced development of Th2 cells in Stat4-deficient mice. Nature. (1996) 382:174–7. doi: 10.1038/382174a0
30. Lee YK, Turner H, Maynard CL, Oliver JR, Chen D, Elson CO, et al. Late developmental plasticity in the T helper 17 lineage. Immunity. (2009) 30:92–107. doi: 10.1016/j.immuni.2008.11.005
31. Li Q, Eppolito C, Odunsi K, Shrikant PA. IL-12-programmed long-term CD8+ T cell responses require STAT4. J Immunol. (2006) 177:7618–25. doi: 10.4049/jimmunol.177.11.7618
32. Wang W, Liu S, Lu F, Yang B, Zhuang X, Yin J, et al. STAT4, a potential predictor of prognosis, promotes CD8 T−cell infiltration in ovarian serous carcinoma by inducing CCL5 secretion. Oncol Rep. (2023) 50:140. doi: 10.3892/or.2023.8577
33. Rao RR, Li Q, Odunsi K, Shrikant PA. The mTOR kinase determines effector versus memory CD8+ T cell fate by regulating the expression of transcription factors T-bet and Eomesodermin. Immunity. (2010) 32:67–78. doi: 10.1016/j.immuni.2009.10.010
34. Kye YC, Lee GW, Lee SW, Ju YJ, Kim HO, Yun CH, et al. STAT1 maintains naïve CD8(+) T cell quiescence by suppressing the type I IFN-STAT4-mTORC1 signaling axis. Sci Adv. (2021) 7:eabg8764. doi: 10.1126/sciadv.abg8764
35. Stark R, Hartung A, Zehn D, Frentsch M, Thiel A. IL-12-mediated STAT4 signaling and TCR signal strength cooperate in the induction of CD40L in human and mouse CD8+ T cells. Eur J Immunol. (2013) 43:1511–7. doi: 10.1002/eji.201243218
36. Kim H, Abbasi A, Sharrock J, Santosa EK, Sheppard S, Lau CM, et al. Cutting Edge: STAT4 Promotes Bhlhe40 Induction to Drive Protective IFN-γ from NK Cells during Viral Infection. J Immunol. (2023) 211:1469–74. doi: 10.4049/jimmunol.2300402
37. Jamil KM, Hydes TJ, Cheent KS, Cassidy SA, Traherne JA, Jayaraman J, et al. STAT4-associated natural killer cell tolerance following liver transplantation. Gut. (2017) 66:352–61. doi: 10.1136/gutjnl-2015-309395
38. Guo L, Wu D, Shen J, Gao Y. ERG mediates the inhibition of NK cell cytotoxicity through the HLX/STAT4/Perforin signaling pathway, thereby promoting the progression of myocardial infarction. J Physiol Biochem. (2024) 80:219–33. doi: 10.1007/s13105-023-00999-5
39. Sun JC, Madera S, Bezman NA, Beilke JN, Kaplan MH, Lanier LL. Proinflammatory cytokine signaling required for the generation of natural killer cell memory. J Exp Med. (2012) 209:947–54. doi: 10.1084/jem.20111760
40. Liang Y, Pan HF, Ye DQ. Therapeutic potential of STAT4 in autoimmunity. Expert Opin Ther Targets. (2014) 18:945–60. doi: 10.1517/14728222.2014.920325
41. Dobrian AD, Galkina EV, Ma Q, Hatcher M, Aye SM, Butcher MJ, et al. STAT4 deficiency reduces obesity-induced insulin resistance and adipose tissue inflammation. Diabetes. (2013) 62:4109–21. doi: 10.2337/db12-1275
42. Alakhras NS, Zhang W, Barros N, Sharma A, Ropa J, Priya R, et al. An IL-23-STAT4 pathway is required for the proinflammatory function of classical dendritic cells during CNS inflammation. Proc Natl Acad Sci U S A. (2024) 121:e2400153121. doi: 10.1073/pnas.2400153121
43. Suto A, Nakajima H, Tokumasa N, Takatori H, Kagami S, Suzuki K, et al. Murine plasmacytoid dendritic cells produce IFN-gamma upon IL-4 stimulation. J Immunol. (2005) 175:5681–9. doi: 10.4049/jimmunol.175.9.5681
44. Iida K, Suzuki K, Yokota M, Nakagomi D, Wakashin H, Iwata A, et al. STAT4 is required for IFN-β-induced MCP-1 mRNA expression in murine mast cells. Int Arch Allergy Immunol. (2011) 155 Suppl 1:71–6. doi: 10.1159/000327300
45. Kataoka TR, Nishizawa Y. Stat4 suppresses the proliferation of connective tissue-type mast cells. Lab Invest. (2008) 88:856–64. doi: 10.1038/labinvest.2008.51
46. Mehrpouya-Bahrami P, Moriarty AK, De Melo P, Keeter WC, Alakhras NS, Nelson AS, et al. STAT4 is expressed in neutrophils and promotes antimicrobial immunity. JCI Insight. (2021) 6:e141326. doi: 10.1172/jci.insight.141326
47. Lamana A, López-Santalla M, Castillo-González R, Ortiz AM, Martín J, García-Vicuña R, et al. The Minor Allele of rs7574865 in the STAT4 Gene Is Associated with Increased mRNA and Protein Expression. PloS One. (2015) 10:e0142683. doi: 10.1371/journal.pone.0142683
48. Abelson AK, Delgado-Vega AM, Kozyrev SV, Sánchez E, Velázquez-Cruz R, Eriksson N, et al. STAT4 associates with systemic lupus erythematosus through two independent effects that correlate with gene expression and act additively with IRF5 to increase risk. Ann Rheum Dis. (2009) 68:1746–53. doi: 10.1136/ard.2008.097642
49. Hagberg N, Joelsson M, Leonard D, Reid S, Eloranta ML, Mo J, et al. The STAT4 SLE risk allele rs7574865[T] is associated with increased IL-12-induced IFN-γ production in T cells from patients with SLE. Ann Rheum Dis. (2018) 77:1070–7. doi: 10.1136/annrheumdis-2017-212794
50. Madera-Salcedo IK, Ramírez-Sánchez AL, Rodríguez-Rodríguez N, García-Quintero R, Rubio RM, Morales-Montes de Oca G, et al. Down-Regulation-Resistant STAT4 Risk Haplotype Contributes to Lupus Nephritis Through CD4+ T Cell Interferon-γ Production. Arthritis Rheumatol (Hoboken NJ). (2023) 75:961–72. doi: 10.1002/art.42435
51. Kim SW, Kim ES, Moon CM, Kim TI, Kim WH, Cheon JH. Abnormal genetic and epigenetic changes in signal transducer and activator of transcription 4 in the pathogenesis of inflammatory bowel diseases. Dig Dis Sci. (2012) 57:2600–7. doi: 10.1007/s10620-012-2199-z
52. Wang C, Gao N, Yang L, Guo Y, Fang Y, Wang T, et al. Stat4 rs7574865 polymorphism promotes the occurrence and progression of hepatocellular carcinoma via the Stat4/CYP2E1/FGL2 pathway. Cell Death Dis. (2022) 13:130. doi: 10.1038/s41419-022-04584-4
53. Rothwell S, Amos CI, Miller FW, Rider LG, Lundberg IE, Gregersen PK, et al. Identification of Novel Associations and Localization of Signals in Idiopathic Inflammatory Myopathies Using Genome-Wide Imputation. Arthritis Rheumatol (Hoboken NJ). (2023) 75:1021–7. doi: 10.1002/art.42434
54. Patel ZH, Lu X, Miller D, Forney CR, Lee J, Lynch A, et al. A plausibly causal functional lupus-associated risk variant in the STAT1-STAT4 locus. Hum Mol Genet. (2018) 27:2392–404. doi: 10.1093/hmg/ddy140
55. Shin HJ, Park HY, Jeong SJ, Park HW, Kim YK, Cho SH, et al. STAT4 expression in human T cells is regulated by DNA methylation but not by promoter polymorphism. J Immunol. (2005) 175:7143–50. doi: 10.4049/jimmunol.175.11.7143
56. Sabri A, Grant AV, Cosker K, El Azbaoui S, Abid A, Abderrahmani Rhorfi I, et al. Association study of genes controlling IL-12-dependent IFN-γ immunity: STAT4 alleles increase risk of pulmonary tuberculosis in Morocco. J Infect diseases. (2014) 210:611–8. doi: 10.1093/infdis/jiu140
57. Messeguer X, Escudero R, Farré D, Núñez O, Martínez J, Albà MM. PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinf (Oxford England). (2002) 18:333–4. doi: 10.1093/bioinformatics/18.2.333
58. Hou S, Yang Z, Du L, Jiang Z, Shu Q, Chen Y, et al. Identification of a susceptibility locus in STAT4 for Behçet's disease in Han Chinese in a genome-wide association study. Arthritis rheumatism. (2012) 64:4104–13. doi: 10.1002/art.37708
59. Saevarsdottir S, Stefansdottir L, Sulem P, Thorleifsson G, Ferkingstad E, Rutsdottir G, et al. Multiomics analysis of rheumatoid arthritis yields sequence variants that have large effects on risk of the seropositive subset. Ann Rheum Dis. (2022) 81:1085–95. doi: 10.1136/annrheumdis-2021-221754
60. Matsuura K, Isogawa M, Tanaka Y. Host genetic variants influencing the clinical course of hepatitis B virus infection. J Med virology. (2016) 88:371–9. doi: 10.1002/jmv.24350
61. Shi H, He H, Ojha SC, Sun C, Fu J, Yan M, et al. Association of STAT3 and STAT4 polymorphisms with susceptibility to chronic hepatitis B virus infection and risk of hepatocellular carcinoma: a meta-analysis. Bioscience Rep. (2019) 39:BSR20190783. doi: 10.1042/BSR20190783
62. Jiang DK, Sun J, Cao G, Liu Y, Lin D, Gao YZ, et al. Genetic variants in STAT4 and HLA-DQ genes confer risk of hepatitis B virus-related hepatocellular carcinoma. Nat Genet. (2013) 45:72–5. doi: 10.1038/ng.2483
63. Chen K, Shi W, Xin Z, Wang H, Zhu X, Wu X, et al. Replication of genome wide association studies on hepatocellular carcinoma susceptibility loci in a Chinese population. PloS One. (2013) 8:e77315. doi: 10.1371/journal.pone.0077315
64. Ayoub A, Anugwom CM, Prieto J, Balderramo D, Ferrer JD, Mattos AZ, et al. Assessment of STAT4 Variants and Risk of Hepatocellular Carcinoma in Latin Americans and Europeans. Cancers. (2023) 15:4530. doi: 10.3390/cancers15184530
65. Chao X, Wu J, Zhang W, Feng X, Zhao L, Huang F, et al. A new discovery of STAT4 single nucleotide polymorphisms associated with hepatocellular carcinoma risk in Chinese Han population: a case-control study. Bioscience Rep. (2021) 41:BSR20210124. doi: 10.1042/BSR20210124
66. Ma Y, Zhou Y, Zhang H, Su X. Immune Response-Related Genes - STAT4, IL8RA and CCR7 Polymorphisms in Lung Cancer: A Case-Control Study in China. Pharmacogenomics personalized Med. (2020) 13:511–9. doi: 10.2147/PGPM.S271983
67. Cotterchio M, Lowcock E, Bider-Canfield Z, Lemire M, Greenwood C, Gallinger S, et al. Association between Variants in Atopy-Related Immunologic Candidate Genes and Pancreatic Cancer Risk. PloS One. (2015) 10:e0125273. doi: 10.1371/journal.pone.0125273
68. Núñez-Marrero A, Arroyo N, Godoy L, Rahman MZ, Matta JL, Dutil J. SNPs in the interleukin-12 signaling pathway are associated with breast cancer risk in Puerto Rican women. Oncotarget. (2020) 11:3420–31. doi: 10.18632/oncotarget.27707
69. Durán-Avelar MJ, Vibanco-Pérez N, Hernández-Pacheco RR, Castro-Zambrano AD, Ortiz-Martínez L, Zambrano-Zaragoza JF. STAT4 rs7574865 G/T polymorphism is associated with rheumatoid arthritis and disease activity, but not with anti-CCP antibody levels in a Mexican population. Clin Rheumatol. (2016) 35:2909–14. doi: 10.1007/s10067-016-3320-z
70. Shen L, Liu R, Zhang H, Huang Y, Sun R, Tang P. Replication study of STAT4 rs7574865 G/T polymorphism and risk of rheumatoid arthritis in a Chinese population. Gene. (2013) 526:259–64. doi: 10.1016/j.gene.2013.05.022
71. Xia Y, Song J, Feng J, Tian R, Zeng Y, Xiang Y, et al. [STAT4 rs7574865 polymorphism is associated with the susceptibility of rheumatoid arthritis in Wuling mountain area]. Xi bao yu fen zi mian yi xue za zhi = Chin J Cell Mol Immunol. (2018) 34:829–33.
72. Budlewski T, Sarnik J, Galita G, Dragan G, Brzezińska O, Popławska M, et al. SNP in PTPN22, PADI4, and STAT4 but Not TRAF1 and CD40 Increase the Risk of Rheumatoid Arthritis in Polish Population. Int J Mol Sci. (2023) 24:7586. doi: 10.3390/ijms24087586
73. Zhao Y, Liu X, Liu X, Su Y, Li Y, Zhang X, et al. Association of STAT4 gene polymorphism with increased susceptibility of rheumatoid arthritis in a northern Chinese Han subpopulation. Int J rheumatic diseases. (2013) 16:178–84. doi: 10.1111/1756-185X.12093
74. Fodil M, Benzaoui A, Zemani-Fodil F, Aberkane M, Boughrara W, Saidi-Mehtar N, et al. Association of PTPN22 (rs2476601) and STAT4 (rs7574865) polymorphisms with Rheumatoid Arthritis in the Western Algerian population. Acta reumatologica portuguesa. (2015) 40:56–62.
75. Orozco G, Alizadeh BZ, Delgado-Vega AM, González-Gay MA, Balsa A, Pascual-Salcedo D, et al. Association of STAT4 with rheumatoid arthritis: a replication study in three European populations. Arthritis rheumatism. (2008) 58:1974–80.
76. Palomino-Morales RJ, Rojas-Villarraga A, González CI, Ramírez G, Anaya JM, Martín J. STAT4 but not TRAF1/C5 variants influence the risk of developing rheumatoid arthritis and systemic lupus erythematosus in Colombians. Genes immunity. (2008) 9:379–82. doi: 10.1038/gene.2008.30
77. Liang YL, Wu H, Li PQ, Xie XD, Shen X, Yang XQ, et al. Signal transducer and activator of transcription 4 gene polymorphisms associated with rheumatoid arthritis in Northwestern Chinese Han population. Life Sci. (2011) 89:171–5. doi: 10.1016/j.lfs.2011.05.012
78. Ghanavati F, Nezhad SRK, Hajjari MR, Akhoond MR. Association of Signal Transducer and Activator of Transcription 4 rs10181656 Polymorphism With Rheumatoid Arthritis and Systemic Sclerosis in Khuzestan Province in Southwestern Iran. Arch Rheumatol. (2019) 34:434–42. doi: 10.5606/ArchRheumatol.2020.7376
79. Lee HS, Remmers EF, Le JM, Kastner DL, Bae SC, Gregersen PK. Association of STAT4 with rheumatoid arthritis in the Korean population. Mol Med (Cambridge Mass). (2007) 13:455–60.
80. Li H, Zou Q, Xie Z, Liu Y, Zhong B, Yang S, et al. A haplotype in STAT4 gene associated with rheumatoid arthritis in Caucasians is not associated in the Han Chinese population, but with the presence of rheumatoid factor. Rheumatol (Oxford England). (2009) 48:1363–8. doi: 10.1093/rheumatology/kep207
81. El-Lebedy D, Raslan H, Ibrahim A, Ashmawy I, El-Aziz SA, Mohammed AM. Association of STAT4 rs7574865 and PTPN22 rs2476601 polymorphisms with rheumatoid arthritis and non-systemically reacting antibodies in Egyptian patients. Clin Rheumatol. (2017) 36:1981–7. doi: 10.1007/s10067-017-3632-7
82. Seddighzadeh M, Gonzalez A, Ding B, Ferreiro-Iglesias A, Gomez-Reino JJ, Klareskog L, et al. Variants within STAT genes reveal association with anticitrullinated protein antibody-negative rheumatoid arthritis in 2 European populations. J Rheumatol. (2012) 39:1509–16. doi: 10.3899/jrheum.111284
83. Bravo-Villagra KM, Muñoz-Valle JF, Baños-Hernández CJ, Cerpa-Cruz S, Navarro-Zarza JE, Parra-Rojas I, et al. STAT4 Gene Variant rs7574865 Is Associated with Rheumatoid Arthritis Activity and Anti-CCP Levels in the Western but Not in the Southern Population of Mexico. Genes (Basel). (2024) 15:241. doi: 10.3390/genes15020241
84. Tarakji I, Habbal W, Monem F. Association Between STAT4 rs7574865 Polymorphism and Rheumatoid Arthritis: Debate Unresolved. Open Rheumatol J. (2018) 12:172–8. doi: 10.2174/1874312901812010172
85. Shancui Z, Jinping Z, Guoyuan L, Liu L, Zhiyong D. Polymorphism in STAT4 Increase the Risk of Systemic Lupus Erythematosus: An Updated Meta-Analysis. Int J Rheumatol. (2022) 2022:5565057. doi: 10.1155/2022/5565057
86. Ji JD, Lee WJ, Kong KA, Woo JH, Choi SJ, Lee YH, et al. Association of STAT4 polymorphism with rheumatoid arthritis and systemic lupus erythematosus: a meta-analysis. Mol Biol Rep. (2010) 37:141–7. doi: 10.1007/s11033-009-9553-z
87. Wang JM, Xu WD, Huang AF. Association of STAT4 Gene Rs7574865, Rs10168266 Polymorphisms and Systemic Lupus Erythematosus Susceptibility: A Meta-analysis. Immunol investigations. (2021) 50:282–94. doi: 10.1080/08820139.2020.1752712
88. Esparza Guerrero Y, Vazquez Villegas ML, Nava Valdivia CA, Ponce Guarneros JM, Perez Guerrero EE, Gomez Ramirez EE, et al. Association of the STAT4 Gene rs7574865 Polymorphism with IFN-γ Levels in Patients with Systemic Lupus Erythematosus. Genes (Basel). (2023) 14:537. doi: 10.3390/genes14030537
89. Hagberg N, Rönnblom L. Interferon-α enhances the IL-12-induced STAT4 activation selectively in carriers of the STAT4 SLE risk allele rs7574865[T]. Ann Rheum Dis. (2019) 78:429–31. doi: 10.1136/annrheumdis-2018-213836
90. Chai HC, Chua KH, Lim SK, Phipps ME. Insight into gene polymorphisms involved in toll-like receptor/interferon signalling pathways for systemic lupus erythematosus in South East Asia. J Immunol Res. (2014) 2014:529167. doi: 10.1155/2014/529167
91. Yang W, Ng P, Zhao M, Hirankarn N, Lau CS, Mok CC, et al. Population differences in SLE susceptibility genes: STAT4 and BLK, but not PXK, are associated with systemic lupus erythematosus in Hong Kong Chinese. Genes immunity. (2009) 10:219–26. doi: 10.1038/gene.2009.1
92. Namjou B, Sestak AL, Armstrong DL, Zidovetzki R, Kelly JA, Jacob N, et al. High-density genotyping of STAT4 reveals multiple haplotypic associations with systemic lupus erythematosus in different racial groups. Arthritis rheumatism. (2009) 60:1085–95. doi: 10.1002/art.24387
93. Yang W, Shen N, Ye DQ, Liu Q, Zhang Y, Qian XX, et al. Genome-wide association study in Asian populations identifies variants in ETS1 and WDFY4 associated with systemic lupus erythematosus. PloS Genet. (2010) 6:e1000841. doi: 10.1371/journal.pgen.1000841
94. Luan H, Li P, Cao C, Li C, Hu C, Zhang S, et al. A single-nucleotide polymorphism of the STAT4 gene is associated with systemic lupus erythematosus (SLE) in female Chinese population. Rheumatol Int. (2012) 32:1251–5. doi: 10.1007/s00296-010-1767-9
95. Piotrowski P, Lianeri M, Wudarski M, Olesińska M, Jagodziński PP. Contribution of STAT4 gene single-nucleotide polymorphism to systemic lupus erythematosus in the Polish population. Mol Biol Rep. (2012) 39:8861–6. doi: 10.1007/s11033-012-1752-3
96. Hellquist A, Sandling JK, Zucchelli M, Koskenmies S, Julkunen H, D'Amato M, et al. Variation in STAT4 is associated with systemic lupus erythematosus in a Finnish family cohort. Ann Rheum Dis. (2010) 69:883–6. doi: 10.1136/ard.2009.112284
97. Harley JB, Alarcón-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP, Moser KL, et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. (2008) 40:204–10. doi: 10.1038/ng.81
98. Kawasaki A, Ito I, Hikami K, Ohashi J, Hayashi T, Goto D, et al. Role of STAT4 polymorphisms in systemic lupus erythematosus in a Japanese population: a case-control association study of the STAT1-STAT4 region. Arthritis Res Ther. (2008) 10:R113. doi: 10.1186/ar2516
99. Reid S, Hagberg N, Sandling JK, Alexsson A, Pucholt P, Sjöwall C, et al. Interaction between the STAT4 rs11889341(T) risk allele and smoking confers increased risk of myocardial infarction and nephritis in patients with systemic lupus erythematosus. Ann Rheum Dis. (2021) 80:1183–9. doi: 10.1136/annrheumdis-2020-219727
100. Bolin K, Sandling JK, Zickert A, Jönsen A, Sjöwall C, Svenungsson E, et al. Association of STAT4 polymorphism with severe renal insufficiency in lupus nephritis. PloS One. (2013) 8:e84450. doi: 10.1371/journal.pone.0084450
101. Taylor KE, Remmers EF, Lee AT, Ortmann WA, Plenge RM, Tian C, et al. Specificity of the STAT4 genetic association for severe disease manifestations of systemic lupus erythematosus. PloS Genet. (2008) 4:e1000084. doi: 10.1371/journal.pgen.1000084
102. Liang YL, Wu H, Shen X, Li PQ, Yang XQ, Liang L, et al. Association of STAT4 rs7574865 polymorphism with autoimmune diseases: a meta-analysis. Mol Biol Rep. (2012) 39:8873–82. doi: 10.1007/s11033-012-1754-1
103. Diaz-Gallo LM, Palomino-Morales RJ, Gómez-García M, Cardeña C, Rodrigo L, Nieto A, et al. STAT4 gene influences genetic predisposition to ulcerative colitis but not Crohn's disease in the Spanish population: a replication study. Hum Immunol. (2010) 71:515–9. doi: 10.1016/j.humimm.2010.02.005
104. Liu QF, Li Y, Zhao QH, Wang ZY, Hu S, Yang CQ, et al. Association of STAT4 rs7574865 polymorphism with susceptibility to inflammatory bowel disease: A systematic review and meta-analysis. Clin Res Hepatol Gastroenterol. (2015) 39:627–36. doi: 10.1016/j.clinre.2015.04.002
105. Moon CM, Cheon JH, Kim SW, Shin DJ, Kim ES, Shin ES, et al. Association of signal transducer and activator of transcription 4 genetic variants with extra-intestinal manifestations in inflammatory bowel disease. Life Sci. (2010) 86:661–7. doi: 10.1016/j.lfs.2010.02.016
106. Glas J, Seiderer J, Nagy M, Fries C, Beigel F, Weidinger M, et al. Evidence for STAT4 as a common autoimmune gene: rs7574865 is associated with colonic Crohn's disease and early disease onset. PloS One. (2010) 5:e10373. doi: 10.1371/journal.pone.0010373
107. Zervou MI, Mamoulakis D, Panierakis C, Boumpas DT, Goulielmos GN. STAT4: a risk factor for type 1 diabetes? Hum Immunol. (2008) 69:647–50. doi: 10.1016/j.humimm.2008.07.004
108. Bi C, Li B, Cheng Z, Hu Y, Fang Z, Zhai A. Association study of STAT4 polymorphisms and type 1 diabetes in Northeastern Chinese Han population. Tissue Antigens. (2013) 81:137–40. doi: 10.1111/tan.12057
109. Abdelmajed SS, El-Dessouky MA, SalahElDin DS, Hassan NA, Zaki ME, Ismail S. Assessing the association of rs7574865 STAT4 gene variant and type 1 diabetes mellitus among Egyptian patients. Journal Genet Eng Biotechnol. (2021) 19:112. doi: 10.1186/s43141-021-00214-2
110. Fichna M, Żurawek M, Bogusz-Górna K, Małecki PP, Niechciał E, Sidoruk A, et al. STAT4 sequence variant and elevated gene expression are associated with type 1 diabetes in Polish children. Central-European J Immunol. (2020) 45:22–8. doi: 10.5114/ceji.2019.92492
111. Park Y, Lee HS, Park Y, Min D, Yang S, Kim D, et al. Evidence for the role of STAT4 as a general autoimmunity locus in the Korean population. Diabetes/metabolism Res Rev. (2011) 27:867–71. doi: 10.1002/dmrr.1263
112. Rueda B, Broen J, Simeon C, Hesselstrand R, Diaz B, Suárez H, et al. The STAT4 gene influences the genetic predisposition to systemic sclerosis phenotype. Hum Mol Genet. (2009) 18:2071–7. doi: 10.1093/hmg/ddp119
113. Xu Y, Wang W, Tian Y, Liu J, Yang R. Polymorphisms in STAT4 and IRF5 increase the risk of systemic sclerosis: a meta-analysis. Int J Dermatol. (2016) 55:408–16. doi: 10.1111/ijd.12839
114. Gourh P, Agarwal SK, Divecha D, Assassi S, Paz G, Arora-Singh RK, et al. Polymorphisms in TBX21 and STAT4 increase the risk of systemic sclerosis: evidence of possible gene-gene interaction and alterations in Th1/Th2 cytokines. Arthritis rheumatism. (2009) 60:3794–806. doi: 10.1002/art.24958
115. Yi L, Wang JC, Guo XJ, Gu YH, Tu WZ, Guo G, et al. STAT4 is a genetic risk factor for systemic sclerosis in a Chinese population. Int J immunopathology Pharmacol. (2013) 26:473–8. doi: 10.1177/039463201302600220
116. Deng Y, Zhu W, Zhou X. Immune Regulatory Genes Are Major Genetic Factors to Behcet Disease: Systematic Review. Open Rheumatol J. (2018) 12:70–85. doi: 10.2174/1874312901812010070
117. Sousa I, Shahram F, Francisco D, Davatchi F, Abdollahi BS, Ghaderibarmi F, et al. Brief report: association of CCR1, KLRC4, IL12A-AS1, STAT4, and ERAP1 With Behçet's disease in Iranians. Arthritis Rheumatol (Hoboken NJ). (2015) 67:2742–8. doi: 10.1002/art.39240
118. Kirino Y, Bertsias G, Ishigatsubo Y, Mizuki N, Tugal-Tutkun I, Seyahi E, et al. Genome-wide association analysis identifies new susceptibility loci for Behçet's disease and epistasis between HLA-B*51 and ERAP1. Nat Genet. (2013) 45:202–7. doi: 10.1038/ng.2520
119. Hu K, Yang P, Jiang Z, Hou S, Du L, Li F. STAT4 polymorphism in a Chinese Han population with Vogt-Koyanagi-Harada syndrome and Behçet's disease. Hum Immunol. (2010) 71:723–6. doi: 10.1016/j.humimm.2010.04.007
120. Kim ES, Kim SW, Moon CM, Park JJ, Kim TI, Kim WH, et al. , vol. 90. Life Sci (2012). p. 740–6. doi: 10.1016/j.lfs.2012.03.017
121. Kang EH, Kim S, Park MY, Choi JY, Choi IA, Kim MJ, et al. Behçet's disease risk association fine-mapped on the IL23R-IL12RB2 intergenic region in Koreans. Arthritis Res Ther. (2017) 19:227. doi: 10.1186/s13075-017-1435-5
122. Manthiram K, Preite S, Dedeoglu F, Demir S, Ozen S, Edwards KM, et al. Common genetic susceptibility loci link PFAPA syndrome, Behçet's disease, and recurrent aphthous stomatitis. Proc Natl Acad Sci U S A. (2020) 117:14405–11. doi: 10.1073/pnas.2002051117
123. Lessard CJ, Li H, Adrianto I, Ice JA, Rasmussen A, Grundahl KM, et al. Variants at multiple loci implicated in both innate and adaptive immune responses are associated with Sjögren's syndrome. Nat Genet. (2013) 45:1284–92. doi: 10.1038/ng.2792
124. Colafrancesco S, Ciccacci C, Priori R, Latini A, Picarelli G, Arienzo F, et al. STAT4, TRAF3IP2, IL10, and HCP5 Polymorphisms in Sjögren's Syndrome: Association with Disease Susceptibility and Clinical Aspects. J Immunol Res. (2019) 2019:7682827. doi: 10.1155/2019/7682827
125. Taylor KE, Wong Q, Levine DM, McHugh C, Laurie C, Doheny K, et al. Genome-Wide Association Analysis Reveals Genetic Heterogeneity of Sjögren's Syndrome According to Ancestry. Arthritis Rheumatol (Hoboken NJ). (2017) 69:1294–305. doi: 10.1002/art.40040
126. Gestermann N, Mekinian A, Comets E, Loiseau P, Puechal X, Hachulla E, et al. STAT4 is a confirmed genetic risk factor for Sjögren's syndrome and could be involved in type 1 interferon pathway signaling. Genes immunity. (2010) 11:432–8. doi: 10.1038/gene.2010.29
127. Li Y, Zhang K, Chen H, Sun F, Xu J, Wu Z, et al. A genome-wide association study in Han Chinese identifies a susceptibility locus for primary Sjögren's syndrome at 7q11. 23. Nat Genet. (2013) 45:1361–5. doi: 10.1038/ng.2779
128. Korman BD, Alba MI, Le JM, Alevizos I, Smith JA, Nikolov NP, et al. Variant form of STAT4 is associated with primary Sjögren's syndrome. Genes immunity. (2008) 9:267–70. doi: 10.1038/gene.2008.1
129. Palomino-Morales RJ, Diaz-Gallo LM, Witte T, Anaya JM, Martín J. Influence of STAT4 polymorphism in primary Sjögren's syndrome. J Rheumatol. (2010) 37:1016–9. doi: 10.3899/jrheum.091007
130. Nordmark G, Kristjansdottir G, Theander E, Eriksson P, Brun JG, Wang C, et al. Additive effects of the major risk alleles of IRF5 and STAT4 in primary Sjögren's syndrome. Genes immunity. (2009) 10:68–76. doi: 10.1038/gene.2008.94
131. Fan ZD, Wang FF, Huang H, Huang N, Ma HH, Guo YH, et al. STAT4 rs7574865 G/T and PTPN22 rs2488457 G/C polymorphisms influence the risk of developing juvenile idiopathic arthritis in Han Chinese patients. PloS One. (2015) 10:e0117389. doi: 10.1371/journal.pone.0117389
132. Prahalad S, Hansen S, Whiting A, Guthery SL, Clifford B, McNally B, et al. Variants in TNFAIP3, STAT4, and C12orf30 loci associated with multiple autoimmune diseases are also associated with juvenile idiopathic arthritis. Arthritis rheumatism. (2009) 60:2124–30. doi: 10.1002/art.24618
133. Dimopoulou DG, Zervou MI, Trachana M, Myrthianou E, Pratsidou-Gertsi P, Kardassis D, et al. Investigation of juvenile idiopathic arthritis susceptibility loci: results from a Greek population. Hum Immunol. (2013) 74:1194–8. doi: 10.1016/j.humimm.2013.06.018
134. Aslani S, Mahmoudi M, Salmaninejad A, Poursani S, Ziaee V, Rezaei N. Lack of Association between STAT4 Single Nucleotide Polymorphisms and Iranian Juvenile Rheumatoid Arthritis Patients. Fetal Pediatr pathology. (2017) 36:177–83. doi: 10.1080/15513815.2016.1253809
135. Huang X, Wang Z, Jia N, Shangguan S, Lai J, Cui X, et al. Association between STAT4 polymorphisms and the risk of juvenile idiopathic arthritis in Han Chinese populations. Clin Exp Rheumatol. (2019) 37:333–7.
136. Gedvilaite G, Duseikaitė M, Dubinskaite G, Kriauciuniene L, Zemaitiene R, Liutkevicienė R. Optic Neuritis: The Influence of Gene Polymorphisms and Serum Levels of STAT4 (rs10181656, rs7574865, rs7601754, rs10168266). J Clin Med. (2023) 13:10. doi: 10.3390/jcm13010010
137. Shi Z, Zhang Q, Chen H, Lian Z, Liu J, Feng H, et al. STAT4 Polymorphisms are Associated with Neuromyelitis Optica Spectrum Disorders. Neuromolecular Med. (2017) 19:493–500. doi: 10.1007/s12017-017-8463-9
138. Sugiura T, Kawaguchi Y, Goto K, Hayashi Y, Tsuburaya R, Furuya T, et al. Positive association between STAT4 polymorphisms and polymyositis/dermatomyositis in a Japanese population. Ann Rheum Dis. (2012) 71:1646–50. doi: 10.1136/annrheumdis-2011-200839
139. Yang C, Chen H, Zhou B, Yin J, Cao G, Hou J, et al. The effects of the interactions of STAT4 rs7574865 with HBV mutations on the risk of hepatocellular carcinoma. Mol Carcinog. (2022) 61:933–40. doi: 10.1002/mc.23449
140. Wu S, Wang M, Wang Y, Zhang M, He JQ. Polymorphisms in the STAT4 gene and tuberculosis susceptibility in a Chinese Han population. Microbial pathogenesis. (2019) 128:288–93. doi: 10.1016/j.micpath.2019.01.024
141. Wun CM, Piao Z, Hong KT, Choi JY, Hong CR, Park JD, et al. Effect of donor STAT4 polymorphism rs7574865 on clinical outcomes of pediatric acute leukemia patients after hematopoietic stem cell transplant. Int immunopharmacology. (2017) 43:62–9. doi: 10.1016/j.intimp.2016.12.007
142. Choi H, Tabashidze N, Rossner P Jr., Dostal M, Pastorkova A, Kong SW, et al. Altered vulnerability to asthma at various levels of ambient Benzo[a]Pyrene by CTLA4, STAT4 and CYP2E1 polymorphisms. Environ pollut. (2017) 231. doi: 10.1016/j.envpol.2017.07.057
143. Migita K, Nakamura M, Abiru S, Jiuchi Y, Nagaoka S, Komori A, et al. Association of STAT4 polymorphisms with susceptibility to type-1 autoimmune hepatitis in the Japanese population. PloS One. (2013) 8:e71382. doi: 10.1371/journal.pone.0071382
144. Urban TJ, Shen Y, Stolz A, Chalasani N, Fontana RJ, Rochon J, et al. Limited contribution of common genetic variants to risk for liver injury due to a variety of drugs. Pharmacogenetics Genomics. (2012) 22:784–95. doi: 10.1097/FPC.0b013e3283589a76
145. Yang MJ, Hou YL, Yang XL, Wang CX, Zhi LX, You CG. Development and application of a PCR-HRM molecular diagnostic method of SNPs linked with TNF inhibitor efficacy. Diagnosis (Berlin Germany). (2019) 6:277–86. doi: 10.1515/dx-2018-0062
146. Conigliaro P, Ciccacci C, Politi C, Triggianese P, Rufini S, Kroegler B, et al. Polymorphisms in STAT4, PTPN2, PSORS1C1 and TRAF3IP2 Genes Are Associated with the Response to TNF Inhibitors in Patients with Rheumatoid Arthritis. PloS One. (2017) 12:e0169956. doi: 10.1371/journal.pone.0169956
147. Qi X, Li F, Zhang Y, Zhu H, Yang F, Li X, et al. STAT4 genetic polymorphism significantly affected HBeAg seroconversion in HBeAg-positive chronic hepatitis B patients receiving Peginterferon-α therapy: A prospective cohort study in China. J Med virology. (2022) 94:4449–58. doi: 10.1002/jmv.27880
148. Limothai U, Chuaypen N, Poovorawan K, Poovorawan Y, Tangkijvanich P. Genetic variation in STAT4 is associated with treatment response to pegylated interferon in patients with chronic hepatitis B. Asian Pacific J Allergy Immunol. (2022) 40:87–93. doi: 10.12932/AP-020419-0533
149. Chen H, Sun J, Zhou B, Xie Q, Liang X, Fan R, et al. Variants in STAT4 Associated With Cure of Chronic HBV Infection in HBeAg-positive Patients Treated With Pegylated Interferon-alpha. Clin Gastroenterol Hepatol. (2020) 18:196–204.e8. doi: 10.1016/j.cgh.2019.04.044
150. Egwuagu CE, Yu CR, Zhang M, Mahdi RM, Kim SJ, Gery I. Suppressors of cytokine signaling proteins are differentially expressed in Th1 and Th2 cells: implications for Th cell lineage commitment and maintenance. J Immunol. (2002) 168:3181–7. doi: 10.4049/jimmunol.168.7.3181
151. Yoon YH, Hwang HJ, Sung HJ, Heo SH, Kim DS, Hong SH, et al. Upregulation of Complement Factor H by SOCS-1/3⁻STAT4 in Lung Cancer. Cancers. (2019) 11:471. doi: 10.3390/cancers11040471
152. Yamamoto K, Yamaguchi M, Miyasaka N, Miura O. SOCS-3 inhibits IL-12-induced STAT4 activation by binding through its SH2 domain to the STAT4 docking site in the IL-12 receptor beta2 subunit. Biochem Biophys Res Commun. (2003) 310:1188–93. doi: 10.1016/j.bbrc.2003.09.140
153. Remmers EF, Plenge RM, Lee AT, Graham RR, Hom G, Behrens TW, et al. STAT4 and the risk of rheumatoid arthritis and systemic lupus erythematosus. N Engl J Med. (2007) 357:977–86. doi: 10.1056/NEJMoa073003
154. Sigurdsson S, Nordmark G, Garnier S, Grundberg E, Kwan T, Nilsson O, et al. A risk haplotype of STAT4 for systemic lupus erythematosus is over-expressed, correlates with anti-dsDNA and shows additive effects with two risk alleles of IRF5. Hum Mol Genet. (2008) 17:2868–76. doi: 10.1093/hmg/ddn184
Keywords: STAT4, single nucleotide polymorphism, clinical significance, polymorphisms and therapeutic efficacy, autoimmune disease
Citation: Xia Y, Xie Y, Zhang H and Liu L (2024) STAT4 gene polymorphisms in human diseases. Front. Immunol. 15:1479418. doi: 10.3389/fimmu.2024.1479418
Received: 19 August 2024; Accepted: 11 October 2024;
Published: 07 November 2024.
Edited by:
Florencia Rosetti, National Institute of Medical Sciences and Nutrition Salvador Zubirán, MexicoReviewed by:
Duy Pham, University of Alabama at Birmingham, United StatesCopyright © 2024 Xia, Xie, Zhang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lunzhi Liu, MTk5MzAyNEBoYm16dS5lZHUuY24=
†These authors have contributed equally to this work
 Yan Xia1,2†
Yan Xia1,2† Hao Zhang
Hao Zhang Lunzhi Liu
Lunzhi Liu