- 1Department of Research and Development, Wecare Probiotics Co., Ltd., Suzhou, China
- 2Department of Food Quality and Safety, Shanghai Business School, Shanghai, China
- 3College of Food and Bioengineering, Henan University of Science and Technology, Luoyang, China
- 4Food Science and Nutrition, University of Leeds, Leeds, United Kingdom
Objective: This study aimed to assess the impact of Lactiplantibacillus plantarum Lp05 (Lp05) on the gastrointestinal microbiome and pathophysiological status of mice infected with Helicobacter pylori (H. pylori), exploring its potential as a probiotic treatment for H. pylori infections.
Methods: In vitro, the interaction between Lp05 and H. pylori was analyzed using laser confocal and scanning electron microscopy. In vivo, C57BL/6 mice infected with H. pylori were treated with Lp05 and divided into six groups: control, model, quadruple therapy, and three dosage levels of Lp05 (2×107, 2×108, 2×109 CFU/mouse/day). Over six weeks, the impact of Lp05 on the gastrointestinal microbiome and physiological markers was assessed. Measurements included digestive enzymes (α-amylase, pepsin, cellulase), inflammatory markers (interleukin-17A, interleukin-23, interleukin-10, interferon-β, interferon-γ, FoxP3, endothelin, IP-10, TGF-β1), oxidative stress markers (catalase, malondialdehyde, superoxide dismutase, myeloperoxidase), and tissue pathology (via modified Warthin-Starry silver and H&E staining). Microbial community structure in the stomach and intestines was evaluated through 16S rRNA gene sequencing.
Results: In vitro studies showed Lp05 and H. pylori formed co-aggregates, with Lp05 potentially disrupting H. pylori cell structure, reducing its stomach colonization. In vivo, Lp05 significantly lowered gastric mucosal urease activity and serum H. pylori-IgG antibody levels in infected mice (p < 0.01). It also mitigated pathological changes in the stomach and duodenum, decreased inflammatory responses (ET, IL-17A, IL-23, TGF-beta1, and IP-10, p < 0.01 for all), and enhanced antioxidant enzyme activities (CAT and SOD, p < 0.01) while reducing MDA and MPO levels (p < 0.01), combating oxidative stress from H. pylori infection. Lp05 treatment significantly modified the intestinal and gastric microbiota, increasing beneficial bacteria like Lactobacillus and Ligilactobacillus, and decreasing harmful bacteria such as Olsenella, linked to pathological conditions.
Conclusion: Lp05 effectively modulates the gastrointestinal microbiome, reduces inflammation and oxidative stress, and suppresses H. pylori, promising for probiotic therapies with further research needed to refine its clinical use.
Introduction
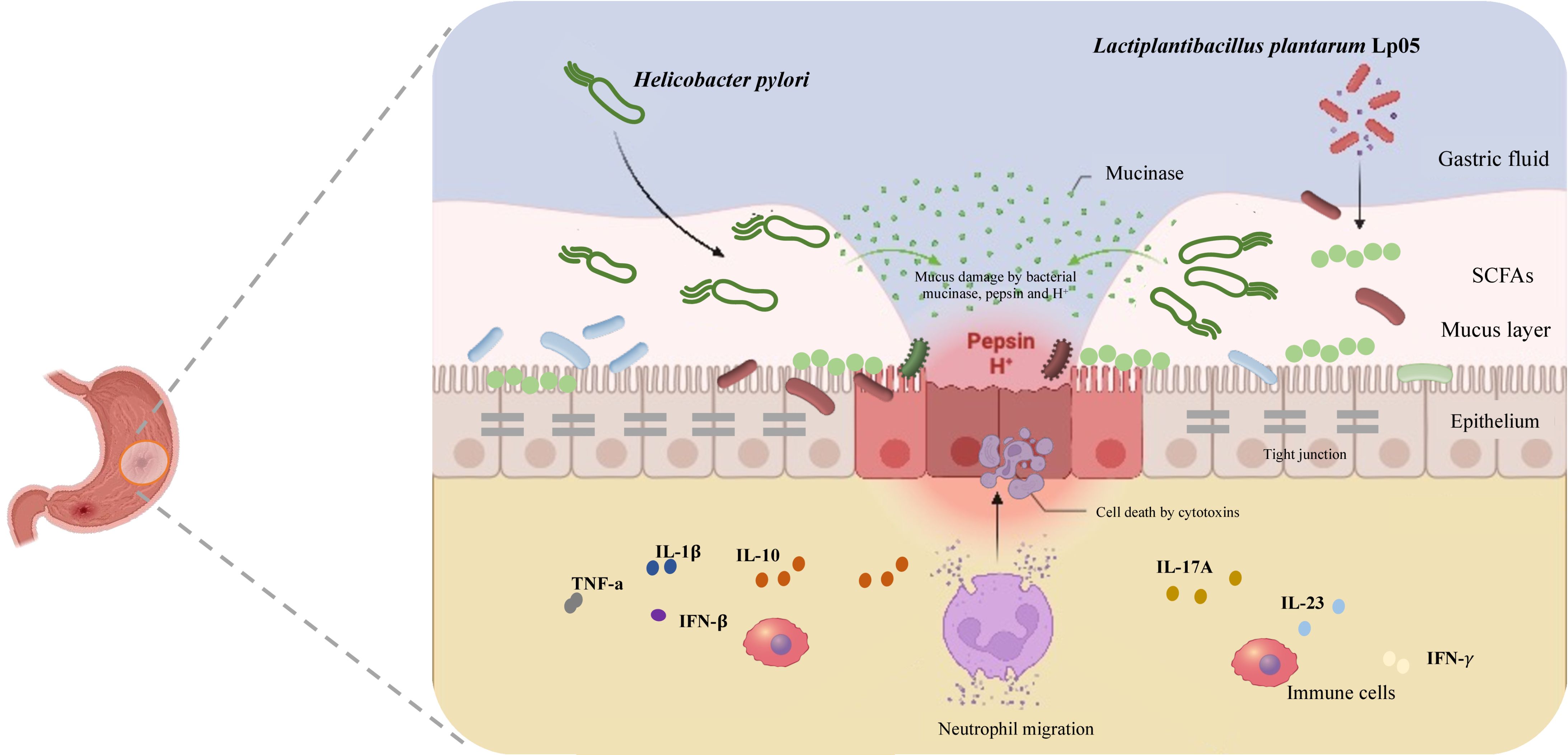
Medically, Helicobacter pylori (H. pylori) is a microaerophilic, spiral-shaped, Gram-negative bacterium that commonly inhabits the human stomach (1). Discovered in 1982, it profoundly influences gastroenterology and infectious diseases by forming persistent, often lifelong infections and adapting to acidic gastric conditions (2). H. pylori adheres to gastric mucosa, forms biofilms, and employs strategies like motility, adhesion, urease activity, and cytotoxin production for its survival and pathogenicity (3, 4). By breaking down urea into ammonia, it neutralizes stomach acidity, maintaining a conducive environment and a stable nitrogen source for itself (5). Additionally, H. pylori triggers chronic inflammation through immune activation, which aids its survival and may lead to gastric damage and cancer (6). It modifies the protective mucosal layer to enhance colonization and evade immune responses. Though often asymptomatic, H. pylori infection correlates with significant gastrointestinal diseases such as peptic ulcers, non-ulcer dyspepsia, and gastric cancer (6, 7), the latter being strongly associated with its classification by the International Agency for Research on Cancer (IARC) as a Group 1 carcinogen (8). Addressing H. pylori infection is essential, particularly in regions with high gastric cancer rates, prompting ongoing research to better understand its pathogenic mechanisms, improve diagnostics, and develop more effective treatments to reduce its health impact.
The standard triple therapy for H. pylori infection, endorsed by the Maastricht IV/Florence Consensus, comprises two antibiotics (usually amoxicillin and clarithromycin) and a proton pump inhibitor (PPI) (9). This regimen faces hurdles like antibiotic resistance, adverse effects, and high costs (10). When clarithromycin resistance occurs, a bismuth-containing quadruple therapy (BQT) is often substituted (11). Although PPIs are crucial for enhancing treatment efficacy and inhibiting bacterial growth, they may increase risks of intestinal infections and acute kidney injury (10). With rising global antibiotic resistance, especially to clarithromycin and levofloxacin, treatment success rates are under strain. The latest Maastricht VI/Florence Consensus advises using triple or bismuth quadruple therapy in areas with low resistance (12). Nevertheless, the broad use of antibiotics has prompted the exploration of alternative treatments (13). Given these circumstances, probiotics have gained significant interest for their safety, immunomodulatory effects, and antimicrobial properties (14). These live microorganisms benefit the host by altering the microbial composition at specific sites (15). Lactobacilli, thriving in the stomach’s acidic conditions (pH 4-6), are particularly suited for treating H. pylori infections (4, 16, 17). Research indicates that certain probiotics can directly inhibit H. pylori growth or modify its lifecycle by enhancing host immune responses, offering a non-antibiotic strategy for managing these infections (4, 16, 17). For example, Lactobacillus johnsonii No. 1088 has shown significant antimicrobial effects in H. pylori-infected mice (18). Moreover, combining probiotics with traditional BQT significantly boosts eradication rates, achieving a 92% cure rate compared to 86.8% without probiotics (19). These insights underline the critical role of gut microbiota in disease progression and support the development of innovative treatment approaches.
While Lactiplantibacillus plantarum is well-studied as a probiotic, its specific anti-H. pylori mechanisms and effects on host microbial communities are not fully understood, especially how it modulates microbial communities and immune responses to diminish H. pylori stomach colonization. In vitro, L. plantarum Lp05 (Lp05) has demonstrated the ability to inhibit H. pylori growth, highlighting its antimicrobial potential. This study focuses on Lp05, analyzing its impact on gastric mucosal urease activity, immune responses, and gastrointestinal microbiota in an H. pylori-infected mouse model. The goal is to explore the mechanisms of its inhibitory effects on H. pylori and assess its potential as a novel treatment approach, providing a foundation for future therapeutic strategies.
Methods and materials
Preparation of strains
Strain Lp05 was provided by Wecare Probiotics Co., Ltd. (Suzhou, China). The strain was cultured in De Man, Rogosa, and Sharpe (MRS) medium at 37°C for 18 h (20). Bacterial cells were harvested by centrifugation at 6000 × g for 8 min and resuspended in sterile water to achieve a final concentration of 1×109 CFU/mL.
H. pylori SS1, obtained from BeNa Culture Collection, was revived from -80°C storage on Columbia agar plates supplemented with 5% sterile defibrinated sheep blood. After being revived for two generations, it was cultured on Columbia blood agar plates for 48 h, washed with BHI broth, and the bacterial suspension adjusted to 1×1010 CFU/mL (21).
Confocal laser scanning microscopy
Cultured H. pylori SS1 was centrifuged and washed twice with artificial gastric juice (22), then resuspended to adjust the OD600 to approximately 0.50. After staining with CFDA SE dye and incubating at 37°C for 30 min, excess dye was removed by centrifuging the bacteria in PBS. The pre-stained H. pylori SS1 was then mixed with Lp05 in equal volumes, vortexed for at least 10 s, and incubated at 37°C for 2 h. A sample of this mixture was placed on a slide and examined under a confocal laser scanning microscope (Ex = 494 nm, Em = 521 nm) (23).
Scanning electron microscopy
Strain Lp05 was centrifuged, washed twice with PBS, and resuspended in sterile PBS. Similarly, H. pylori SS1 was washed and resuspended in artificial gastric juice. Both suspensions were adjusted to an OD600 of approximately 0.50 and mixed equally to induce co-aggregation. After 2 h at 25°C, the aggregates were fixed overnight in 2.5% glutaraldehyde at 4°C, dehydrated in a graded ethanol series, and gold-sputter coated. The samples were then scanned under a scanning electron microscope (24).
Establishment of H. pylori infection model in mice
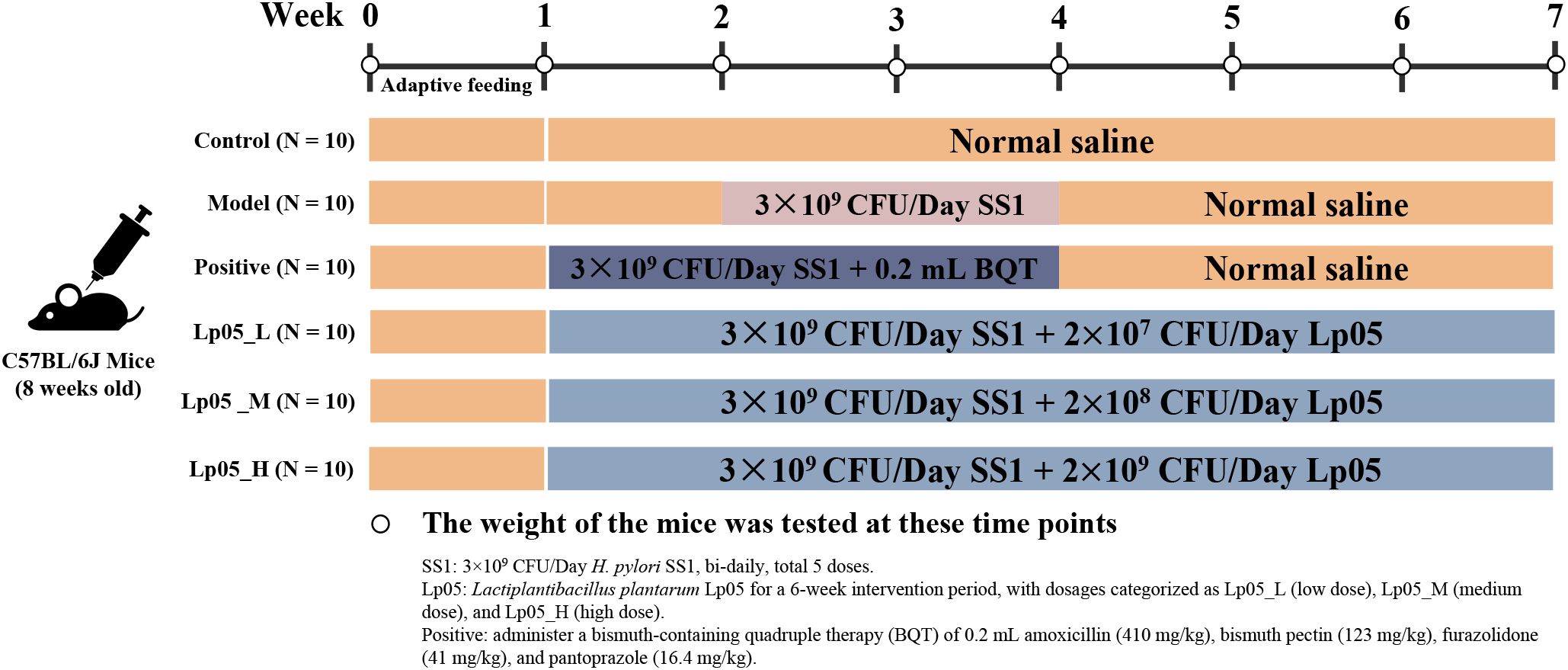
All procedures involving mice were approved by the Experimental Animal Ethics Committee of Zhengzhou University (License No: SYXK(Yu)2021-0011) and followed the ARRIVE guidelines 2.0 (25). To establish the H. pylori infection model, 60 eight-week-old female SPF-grade C57BL/6 mice (SPF Biotechnology Co., Ltd, Beijing, China) were acclimatized for a week and then divided into six groups of ten each for a six-week experiment (26) (Figure 1). The control (CTL) group received daily gavage of 0.2 mL saline throughout the experiment. From the second week, the five treatment groups started modeling H. pylori SS1 infection with bi-daily gavages of a 0.2 mL 0.9% NaCl suspension containing 1×1010 CFU/mL H. pylori for a total of five times. The model control group (MC) followed the same saline regimen and only H. pylori. The quadruple therapy group (PG) received a normal diet in the first week and post-infection, daily gavages of a mix containing amoxicillin (410 mg/kg), bismuth pectin (123 mg/kg), furazolidone (41 mg/kg), and pantoprazole (16.4 mg/kg) during the second and third weeks, followed by 0.2 mL 0.9% NaCl until week six. The Lp05 low, medium, and high dose groups (Lp05_L, Lp05_M, Lp05_H) received daily gavages of 0.2 mL with Lp05 at 1×108, 1×109, and 1×1010 CFU/mL respectively, from the first week and began H. pylori infection the same as MC group from the second week. All mice were kept at 25 ± 2°C with bedding changed two to three times weekly and weighed regularly.
Processing of mice blood and tissue samples
After the final gavage, mice fasted for 12 hours were anesthetized using a 4% chloral hydrate solution intraperitoneally, followed by blood collection via orbital extraction. Blood samples were centrifuged at 3500 rpm for 20 min at 4°C, with serum stored at -80°C for analysis. Post-experiment, mice were euthanized and stomach, duodenum, and cecum tissues were extracted and fixed for histological examination.
Staining of mice stomach and duodenal tissues
H. pylori presence in stomach tissues was detected using a modified Warthin-Starry (W-S) silver stain, effective in revealing H. pylori as black deposits in tissues (27). For structural and pathological assessments, tissues underwent H&E staining after fixing, dehydration, and paraffin embedding. Tissue sections of 4 μm were stained using Dong et al.’s (2022) methods to detail the tissues’ structural and pathological states (28).
Detection of H. pylori colonization in mice stomach tissues
Gastric mucosa samples were incubated in a solution containing 1 g/mL urea and 850 µg/mL phenol red at 37°C for 30 min, observing color changes (29). Additionally, homogenized samples in PBS were diluted, plated on selective media, and cultured under microaerophilic conditions at 37°C for 2-5 days to identify H. pylori based on typical colony characteristics.
Serological testing in mice
Post-experiment, anti-H. pylori IgG antibodies in mouse serum were detected using an ELISA (Yuanju Biotechnology Center, Shanghai, China). Serum samples, diluted 1:50, were added to H. pylori antigen-coated wells, incubated at 37°C for 30 min, washed thrice, and treated with HRP-labeled rabbit anti-mouse IgG for another 30 min. The reaction developed with Tetramethylbenzidine (TMB) was stopped using sulfuric acid and read at 450 nm. Serum was analyzed for gastric enzymes (α-amylase, pepsin, cellulase), inflammatory markers (Interleukin-17A (IL-17A), Interleukin-23 (IL-23), Interleukin-10 (IL-10), Interferon beta (IFN-β), Interferon gamma (IFN-γ), Forkhead Box P3 (FoxP3), endotoxin (ET), interferon–γ–inducible protein 10 (IP-10), transforming growth factor beta-1 (TGF-β1)), and activities of catalase (CAT), malondialdehyde (MDA), superoxide dismutase (SOD), and myeloperoxidase (MPO). All assays were performed according to the manufacturer’s instructions provided with each kit (BIOSCO Biotechnology Co., Ltd, Jiangsu, China).
DNA extraction, 16S rRNA gene amplicon sequencing, and bioinformatics analysis of stomach and cecum microbiomes
We analyzed the microbial communities in stomach and cecum tissues via 16S rRNA gene sequencing. DNA was extracted, and the V3-V4 regions of the 16S rRNA gene were amplified using 341F and 805R primers. The PCR process involved an initial denaturation at 95°C, followed by cycles at 94°C for denaturation, 58°C for annealing, and 72°C for extension, concluding with a final extension at 72°C. Amplified products were indexed, pooled, and sequenced on an Illumina MiSeq platform in a paired-end 2x300 bp format. Raw data were refined using USEARCH to remove low-quality reads and chimeras. Amplicon sequence variants (ASVs) were clustered at 97% similarity with UPARSE (version 7.1) and classified via the RDP classifier (30). Alpha and Beta diversity were analyzed using QIIME, with significant differences between communities determined by linear discriminant analysis effect size (LEfSe), PICRUSt, and statistical analysis of metagenomic profiles (STAMP) analyses (31).
Statistical analysis
All data were presented as mean ± standard deviation (SD). Statistical analyses were performed using R Studio software. The data were first tested for normal distribution using the Kolmogorov-Smirnov test. Multiple group comparisons were made using the non-parametric Kruskal-Wallis test, while differences between two groups were assessed using the Mann-Whitney U test, with a significance level set at p < 0.05. Microbial community analysis was conducted using QIIME software and principal coordinate analysis (PCoA) based on unweighted UniFrac distances (32). Community phylogenetics were investigated using PICRUSt and LEfSe analyses, while Spearman’s rank correlation test was used to explore correlations between bacterial genera in stomach and cecum tissues. All statistical analyses were performed in the R environment version 4.3.2 (33).
Results
In vitro test: CLSM validation of co-aggregation between Lp05 and SS1
This study utilized CLSM to confirm the co-aggregation phenomenon between Lp05 and SS1. CFDA SE fluorescent staining was employed to label SS1, facilitating the identification of its presence within the co-aggregates. Observations under the microscope are presented in Figure 2A (a-c), which shows the distribution of SS1 cultured alone under bright field, dark field, and overlay conditions, with bacteria evenly dispersed and no aggregates formed. In contrast, Figure 2A (d-f) displays the situation after 2 h of co-culturing Lp05 with SS1, where large co-aggregates (indicated by arrows) are visible in all three fields. These aggregates clearly demonstrate SS1 being encapsulated by Lp05, clustering together, which may facilitate its expulsion from the stomach, thus potentially reducing the gastric colonization of H. pylori.
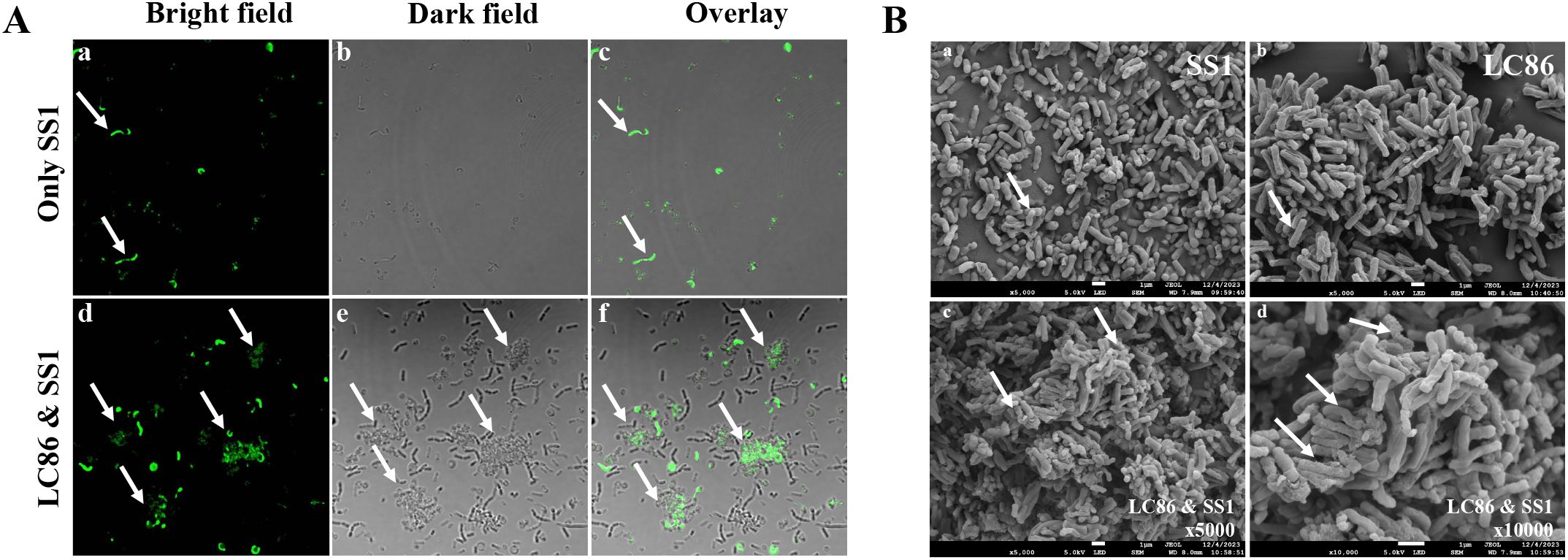
Figure 2. In vitro assessment of Lactiplantibacillus plantarum Lp05 inhibitory effect on Helicobacter pylori SS1. (A) Confocal laser scanning microscopy images after 2 h of co-aggregation between Lp05 and H pylori SS1; (a–c): H pylori SS1 cultured alone; (d–f) Lp05 and H pylori SS1 co-cultured; (a, d) - bright field; (b, e) - dark field; (c, f) - overlay. (B) Scanning electron microscopy images of co-aggregates between Lp05 and H pylori SS1; (a) shows H pylori SS1; (b) shows Lp05; (c, d) show the co-aggregates (c) x5000, (d) x10000.
In vitro test: SEM characterization of the co-aggregation between Lp05 and SS1
The formation of co-aggregates between Lp05 and SS1 was also observed using SEM. After freeze-drying under vacuum, the aggregates were examined under a cold field emission scanning electron microscope to confirm the co-aggregation. Figure 2B (a, b) shows SS1 and Lp05 cultured separately, magnified 5000 times; SS1 appears as curved rods while Lp05 is depicted as short rods, with significant morphological differences between the two. Figure 2B (c, d) display the co-aggregates at magnifications of 5000 and 10000 times, respectively, with arrows pointing to multiple SS1 bodies bonded with Lp05, forming larger aggregates. Notably, the morphology of the Lp05 strain remains stable, whereas the co-cultured SS1 shows signs of edge dispersion, cellular deformation, collapse, and breakage, indicating significant impact on its cell structure. These observations confirm the effective formation of co-aggregates by Lp05 with H. pylori, reducing its gastric colonization.
Effects of Lp05 on urease activity and H. pylori-IgG antibody levels in H. pylori-infected mice model
The urease activity in gastric mucosa samples was assessed as shown in Figure 3A. Notably, the MC group exhibited significantly increased urease activity, leading to a rise in pH that changed the phenol red indicator from yellow to pink or red. Quantitatively, urease activity in the MC group was markedly higher, with an optical density (OD) measurement at 562 nm reaching 2.0. In comparison, groups treated with various doses of Lp05 and the PG group demonstrated lower urease activities, with minimal change in the color of the phenol red indicator or maintenance of its yellow color. Particularly, the Lp05_H group showed urease activities comparable to those of the PG group (OD562nm = 0.5) and significantly lower than those in the MC group. This suggests that Lp05 can reduce gastric colonization by H. pylori, thereby lowering urease activity induced by H. pylori infection and alleviating tissue inflammation caused by H. pylori colonization and infection. These findings are substantiated by the OD measurements, indicating effective suppression of urease by Lp05_H treatment. Additionally, serum levels of H. pylori-IgG antibodies (as depicted in Figure 3B) were significantly lower (p < 0.01) in mice treated with Lp05 compared to the MC group, approaching levels seen in the CTL group and comparable to those in the PG group. This indicates that Lp05 can effectively reduce the systemic immune response to H. pylori.
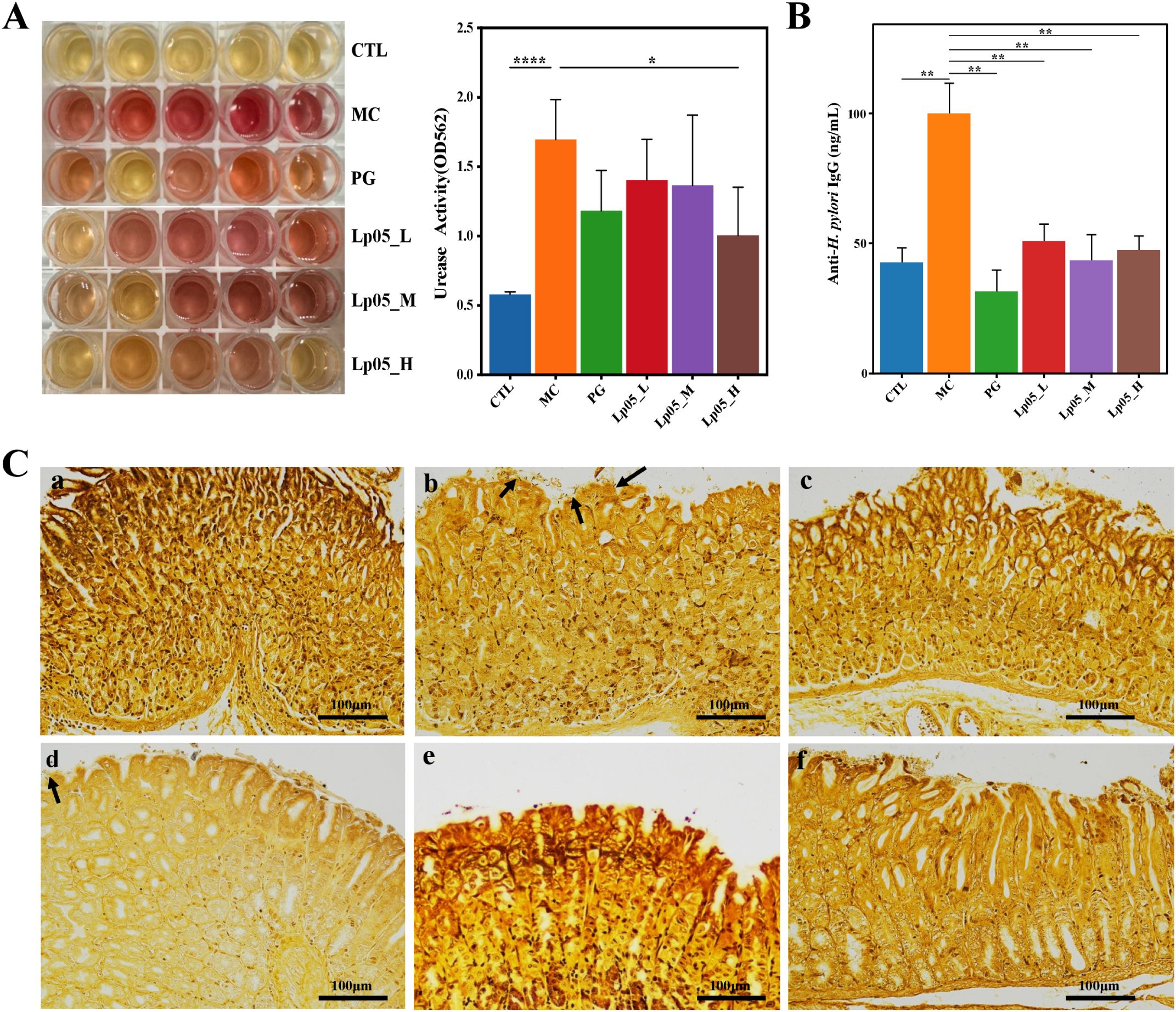
Figure 3. Effects of Lactiplantibacillus plantarum Lp05 on mice infected with H pylori. (A) Impact on gastric mucosal urease activity. (B) Levels of anti-H. pylori IgG antibodies in mouse serum. (C) Gastric tissue sections stained with Warthin-Starry silver staining; subpanels a-f correspond to CTL (control), MC (H. pylori infection model), PG (quadruple therapy group), Lp05_L (low dose, 2×107 CFU), Lp05_M (medium dose, 2×108 CFU), Lp05_H (high dose, 2×109 CFU), respectively. *: p < 0.05, **: p < 0.01, ****: p < 0.0001.
Impact of Lp05 intervention on gastric tissue structure in H. pylori-infected mice model
Figure 3C presents W-S silver-stained sections of mouse gastric tissue. Compared to the CTL group, the MC group displayed dark brown to black rod-shaped or slightly helical bacteria (indicated by arrows) on the mucosal layer, epithelial surface, and intercellular spaces. H. pylori predominantly colonized the gastric pits and gland lumina at the junction of the gastric body and antrum, generally appearing arc-shaped, S-shaped, and occasionally as short rods. After homogenizing gastric tissues and culturing on Brucella agar plates for three days, Gram staining of the colonies (Supplementary Figure S1) revealed typical red S-shaped, slightly helical, or curved rod-shaped morphology of H. pylori, confirming its Gram-negative nature and successful establishment of the H. pylori infection model in the MC group with observable colonization of the gastric mucosa. By contrast, in Lp05_L group, occasional H. pylori colonization was still visible. However, in Lp05_M and Lp05_H groups, the colonization of H. pylori was comparable to that in the CTL and PG groups, indicating that medium and high doses of Lp05 significantly inhibit the colonization of H. pylori on the gastric mucosa.
Impact of Lp05 on gastric and duodenal tissue in H. pylori-infected mice model
HE staining revealed the effects of Lp05 on gastric and duodenal tissues in H. pylori-infected mice. Figures 4A–L show that in the CTL group, gastric structures were intact with well-defined gastric pits, orderly epithelial cells, and no inflammation. Conversely, the MC group exhibited H. pylori-induced pathologies such as epithelial necrosis, disorganized glands, and inflammation, with fewer parietal cells. The PG group had mild inflammation and minor ulcers. The Lp05_L group showed mild to moderate inflammation, whereas the Lp05_M and Lp05_H groups demonstrated notable tissue improvements, especially the Lp05_H group, where the gastric mucosa was clear, intact, and exhibited minimal inflammation, similar to the CTL group. For the duodenum, Figures 4M–R indicates regular mucosal structures in the CTL group, while the MC group showed extensive cellular necrosis and inflammation, disrupting the structure significantly. Despite treatment, the PG group had disordered epithelial cells. Post Lp05 treatment, especially in the Lp05_M and Lp05_H groups, the duodenal villi were intact with regular epithelial cell arrangements, suggesting that medium and high doses of Lp05 effectively alleviate H. pylori-induced duodenal damage.
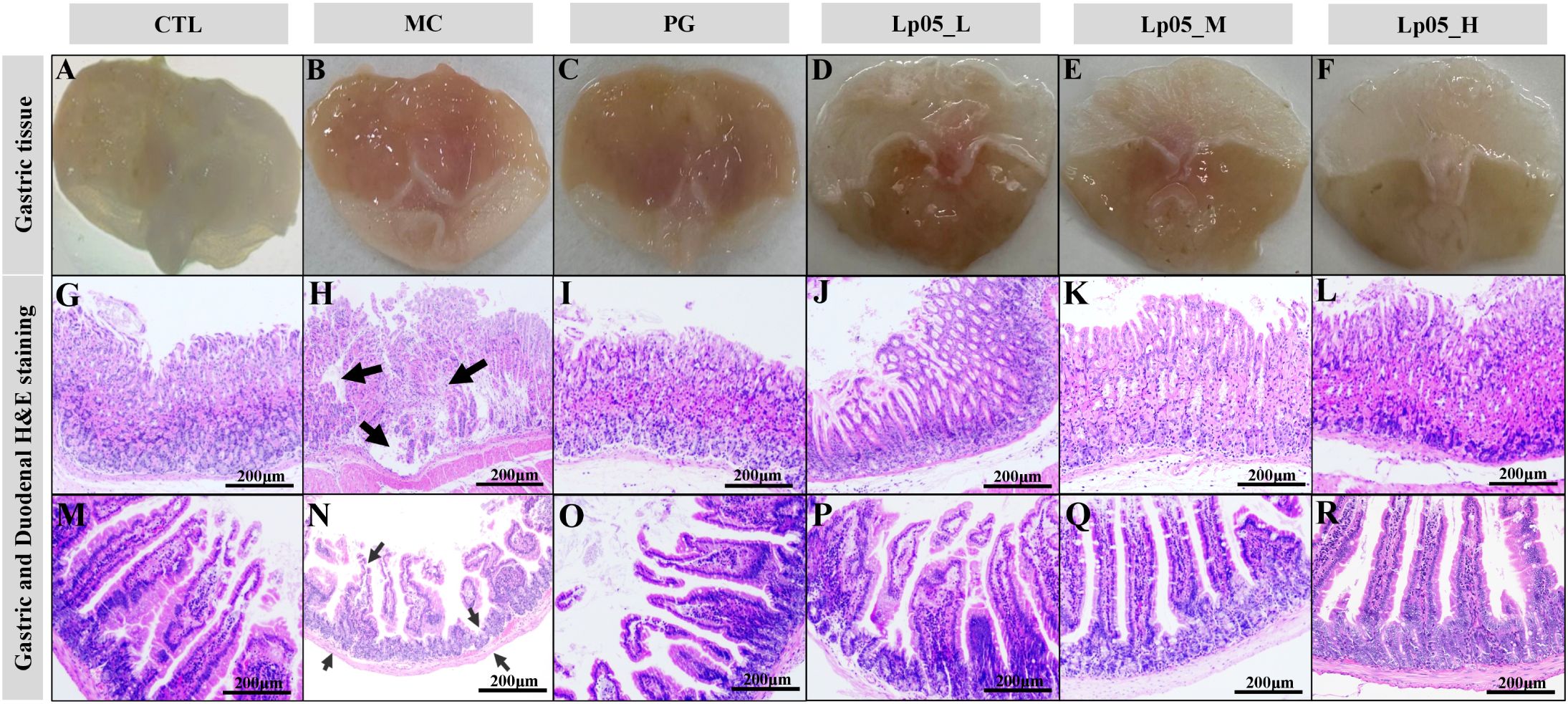
Figure 4. H&E staining of gastric and duodenal tissues in mice infected with H. pylori across Lactiplantibacillus plantarum Lp05 dosages. (A–L) Pathological images of gastric tissue HE stains; (M–R) Pathological images of duodenal tissue HE stains. Groups, CTL (control), MC (H. pylori infection model), PG (quadruple therapy group), Lp05_L (low-dose, 2×107 CFU), Lp05_M (medium-dose, 2×108 CFU), Lp05_H (high-dose, 2×109 CFU).
Effects of Lp05 on digestive enzymes in gastric tissue in H. pylori-infected mice model
This test assessed the impact of Lp05 on digestive enzymes in the gastric tissue of mice infected with H. pylori, focusing on α-amylase, cellulase, and pepsin. Specifically, in the CTL group, the mean α-amylase activity was 27.5 mg/min/g. Post-infection in the MC group, activity decreased significantly to 17.2 mg/min/g (p < 0.01), indicating the detrimental effects of H. pylori infection. Following treatment with Lp05, α-amylase activity in the Lp05_H group almost completely recovered to 28 mg/min/g, slightly surpassing the CTL group. The baseline pepsin activity in the CTL group was 1501 nmol/min/g. After H. pylori infection (MC group), activity dropped to 1260 nmol/min/g (p < 0.01). Remarkably, pepsin activity increased to 1614.9 nmol/min/g in the Lp05_H group, significantly higher than the CTL group, which underscores the effectiveness of Lp05 in not only reversing the infection impact but also enhancing enzyme activity. For cellulase, the activity was 454.9 μg/min/g in the CTL group, reduced to 329.5 μg/min/g in the MC group (p < 0.01) due to H. pylori infection. Post-Lp05 treatment, activity in the Lp05_H group reached 481.6 μg/min/g, again exceeding the CTL, indicating significant recovery and enhancement of function. These results (Figures 5A–C) suggest that Lp05 intervention, particularly at higher doses, supports overall gastric function recovery by normalizing and enhancing digestive enzyme activities, which are otherwise compromised by H. pylori infection.
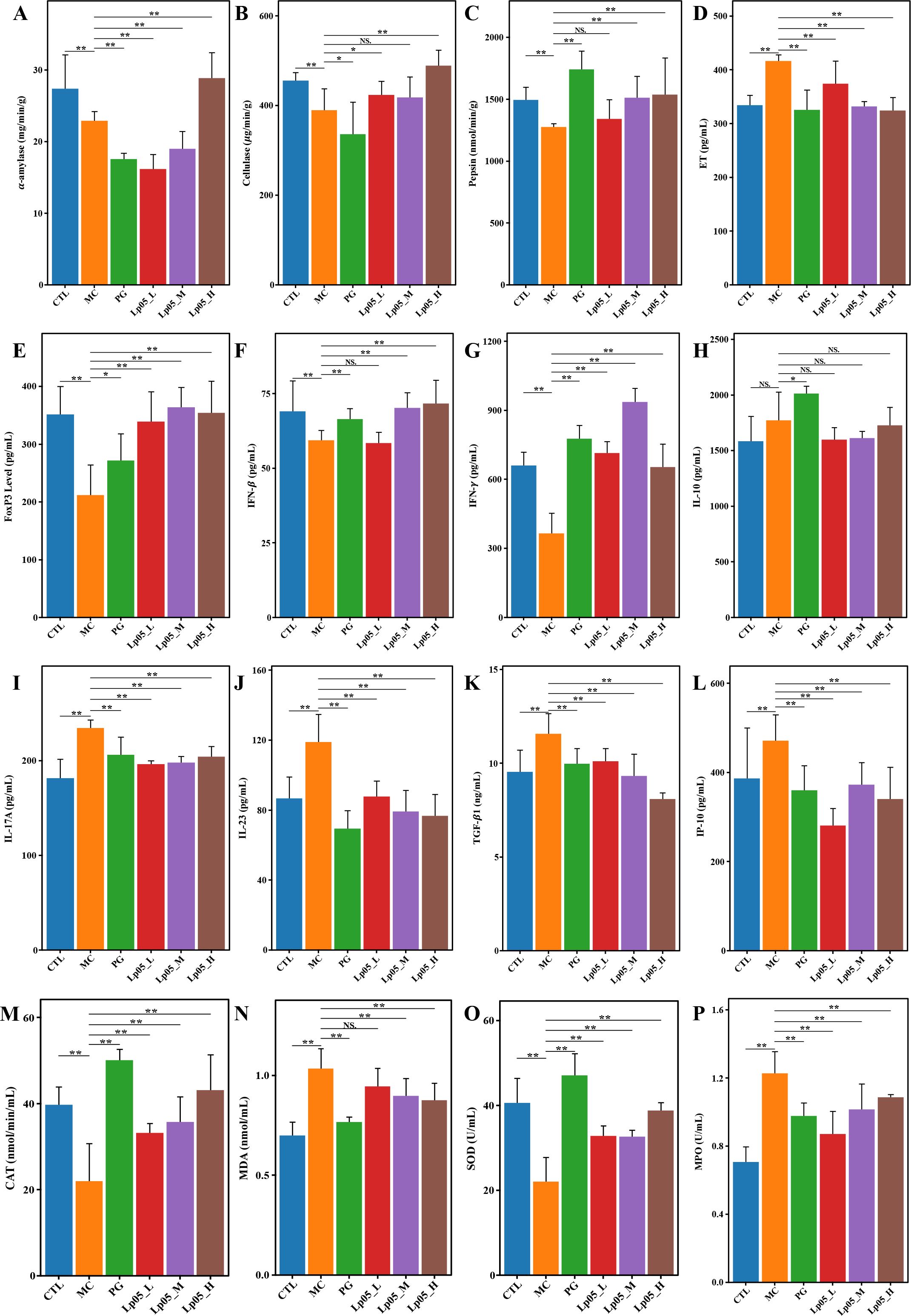
Figure 5. Effects of Lactiplantibacillus plantarum Lp05 on pepsin activity, immune response, and oxidative stress levels in mice infected with Helicobacter pylori. (A–C) Activities of gastric proteases: α-amylase, cellulase, and pepsin; (D–L) Levels of ET, FoxP3, IFN-β, IFN-γ, IL-10, IL-17A, IL-23, TGF- β1, IP-10; (M–P) Levels of CAT, MDA, SOD, and MPO. ** indicates p < 0.01, * indicates p < 0.05. NS indicates p > 0.05.
Impact of Lp05 on serum inflammatory cytokines in H. pylori-infected mice model
As shown in Figures 5D–L, in the MC group, ET levels were measured at 420.1 pg/mL, elevated compared to the CTL group, which had levels of 331 pg/mL. The Lp05_H group effectively reduced ET to 332.24 pg/mL (p < 0.05), closely aligning with the CTL group. IL-17A was higher in MC at 236.15 pg/mL compared to 186.3 pg/mL in CTL, and was reduced to 204 pg/mL in Lp05_H (p < 0.05). IL-23 levels rose to 117.5 pg/mL in MC from 83.9 pg/mL in CTL, but were moderated to 75.70 pg/mL with Lp05 treatment (p < 0.05). IP-10 levels increased to 495.5 pg/mL in MC from 387.1 pg/mL in CTL, with a reduction to 348.7 pg/mL observed in Lp05_H (p < 0.05). TGF-β1 also showed an increase in MC to 11.5 pg/mL from 9.4 pg/mL in CTL (p < 0.05), and was reduced to 8.7 pg/mL in Lp05_H (p < 0.05), indicating a substantial reduction. Conversely, IFN-β and IFN-γ, which decreased to 58.9 pg/mL and 391 pg/mL in MC from CTL levels of 67.7 pg/mL and 646.5 pg/mL respectively, were restored in Lp05_H to 67.8 pg/mL and 672.2 pg/mL (both p < 0.05). Additionally, FoxP3 levels were reduced in MC to 206.76 pg/mL from 343 pg/mL in CTL and were restored in Lp05_H to 341.0 pg/mL (p < 0.05). The effects of Lp05 were similar to those of PG group, particularly in Lp05_H group, where the modulation of inflammatory cytokines was especially pronounced, indicating that Lp05 effectively suppresses the systemic inflammatory response caused by H. pylori infection.
Impact of Lp05 on oxidative stress response in H. pylori-infected mice model
As depicted in Figures 5M–P, oxidative stress markers were quantitatively analyzed to evaluate the impact of H. pylori infection and subsequent treatments. In the CTL group, CAT activity was measured at 38.1 nmol/min/mL, and SOD activity at 40.4 U/mL. Following H. pylori infection, the MC group exhibited significant reductions in these antioxidant enzymes, with CAT decreasing to 21.3 nmol/min/mL and SOD to 23.3 U/mL (all p < 0.05). This decline coincided with increases in MDA and MPO levels, from 0.72 nmol/mL to 1.03 nmol/mL and from 0.75 U/mL to 1.24 U/mL respectively, indicating a marked increase in oxidative stress. Post-treatment analyses showed significant reversals in these oxidative stress markers across all intervention groups (all p < 0.05). After treatment with varying doses of Lp05, CAT levels were increased to 42.2 nmol/min/mL in the Lp05_H group, surpassing CTL levels. Similarly, SOD levels improved to 38.2 U/mL in Lp05_H, nearly reaching CTL values. MDA and MPO levels were significantly reduced to 0.86 nmol/mL and 1.09 U/mL, respectively, in the Lp05_H group. These results suggest that high-dose Lp05 possesses potent antioxidant capabilities, effectively alleviating the oxidative damage caused by H. pylori infection.
Impact of Lp05 on gastric microbiome structure and function in H. pylori-infected mice model
The 16S rRNA gene sequencing assessed Lp05 effect on the gastric microbiome of H. pylori-infected mice. Figure 6A identifies 13,888 ASVs, with 420 common across groups. Unique ASVs were 2366 in the CTL group, 1815 in MC, 3046 in PG, and 2041, 2002, and 2198 in the low, medium, and high dose Lp05 groups respectively, indicating significant microbial diversity changes due to treatment. Observed richness, Chao1, Shannon, and Simpson indices (Figures 6B–E) evaluated microbial abundance, diversity, and evenness, showing a decrease in α-diversity in the MC group compared to CTL. Post-BQT and Lp05 interventions, richness and Chao1 indices increased, albeit not significantly versus the MC group. β-diversity analysis (Figure 6F) showed distinct microbial structures, with significant composition shifts in Lp05, MC, and PG groups compared to CTL. Dominant species and their abundances are detailed in Figures 6G, H. Firmicutes and Bacteroidota were predominant, with significant variations in relative abundance, for instance, 71.18% in MG versus 26.67% in CTL. Lp05 significantly adjusted these proportions, particularly in the low-dose group, aligning more closely with CTL. Dominant genera included Muribaculaceae_unclassified, Lactobacillus, and Muribaculum, with Muribaculaceae_unclassified showing increased abundance in PG and Lp05 groups versus MC, suggesting effective microbial modulation. LEfSe analysis (Figure 7A) highlighted marker species with significant differences across groups, showing beneficial genera like Rikenella were enhanced by Lp05, indicating its role in optimizing the gastric microbiome. STAMP analysis (Figure 7B) noted a significant recovery in Muribaculaceae_unclassified in Lp05-treated groups compared to MC, particularly in low doses. PICRUSt2 analysis (Figure 7C) predicted upregulated metabolic pathways such as amino acid metabolism and polysaccharide biosynthesis in Lp05 groups, essential for microbial community stability and nearing levels in the CTL group.
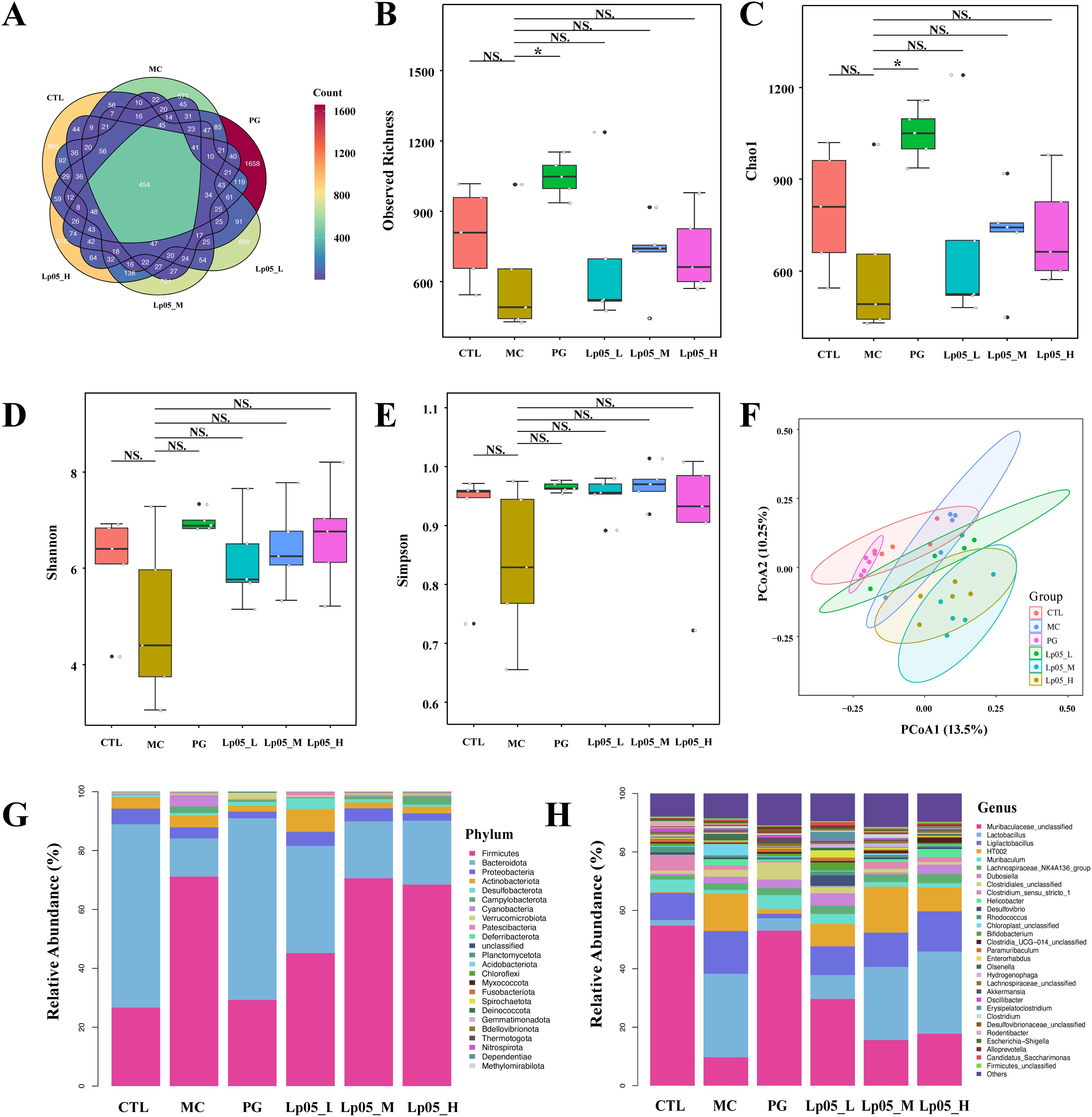
Figure 6. Impact of Lactiplantibacillus plantarum Lp05 doses on gastric microbial composition and structure. (A) Venn diagram analysis of the gastric microbiome sequencing; (B–E) Alpha diversity analysis of the gastric microbiome; (F) Beta diversity analysis of the gastric microbiome through PCoA; (G, H) Relative abundance of species at the phylum and genus levels in the gastric microbiome; CTL, control group; MC, H pylori infection model group; Lp05_L, low-dose group (2×107 CFU); Lp05_M, medium-dose group (2×108 CFU); Lp05_H, high-dose group (2×109 CFU). NS indicates p > 0.05,* indicates p < 0.05.
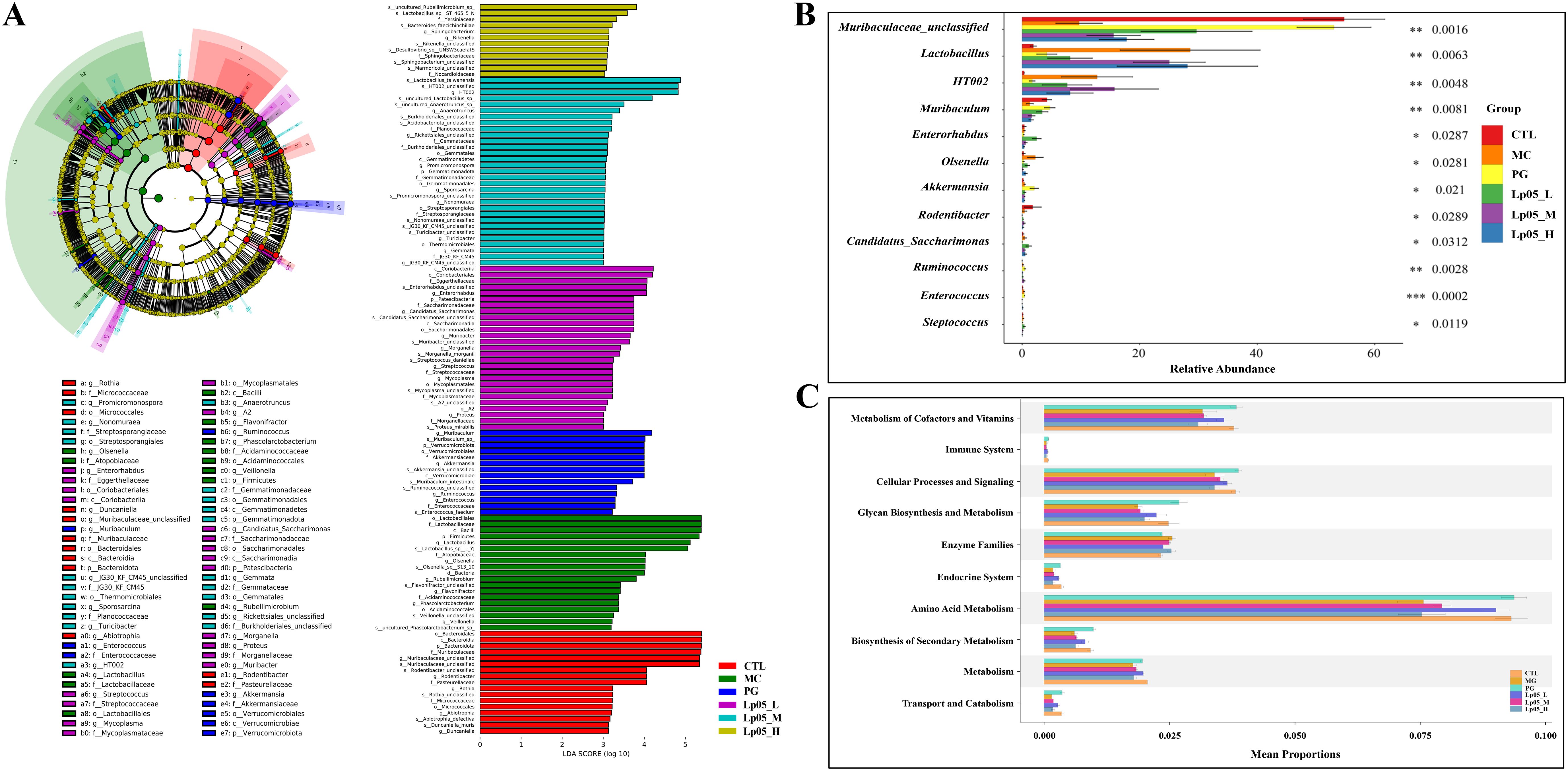
Figure 7. Effects of Lactiplantibacillus plantarum Lp05 doses on gastric microbial species abundance. (A) Phylogenetic cladogram and LefSe analysis LDA score histograms generated from metagenomic data; circle size is proportional to the abundance of taxonomic units. (B) STAMP analysis of differences at the genus level in the gastric microbiome. (C) PICRUSt analysis predicts the functions of gastric microbiome taxa at KEGG level 2.
Impact of Lp05 on the intestinal microbiome structure and function in H. pylori-infected mice model
Venn diagram analysis (Figure 8A) showed 14,342 ASVs, with 340 shared across all six groups. Unique ASVs were 2760 in CTL, 2518 in MC, 1627 in PG, and 2595, 1894, and 2268 in the Lp05 low, medium, and high-dose groups, respectively, indicating significant impacts on intestinal microbiome structure. Alpha diversity metrics, including observed richness, Chao1, Shannon, and Simpson indices (Figures 8B–E), demonstrated a decline in microbial richness and diversity in the MC group compared to CTL. Post-Lp05 treatment, particularly at high doses, significantly enhanced these indices (p < 0.05), suggesting Lp05’s effectiveness in restoring diversity lost due to H. pylori infection. PCoA analysis (Figure 8F) revealed significant microbial composition shifts in the Lp05, MC, and PG groups relative to CTL, indicating effective microbiome rebalancing. Figures 8G, H detailed dominant species at various taxonomic levels, with Firmicutes and Bacteroidota being prevalent. For instance, Firmicutes comprised 60.85% in MC versus 51.91% in CTL. Genus level analysis showed prominence of Lachnospiraceae_NK4A136_group and Muribaculaceae_unclassified, with Lp05 notably enhancing Lachnospiraceae_NK4A136_group, highlighting its potential for supporting intestinal health. LEfSe analysis (Figure 9A) identified significant biomarkers, with dominant species in CTL including Muribaculum and Paramuribaculum. MC showed enrichment in Atopobiaceae and Olsenella, while PG’s biomarkers were primarily Firmicutes and Bifidobacterium. Lp05’s intervention highlighted beneficial biomarkers such as Bacilli and Ligilactobacillus, suggesting a reduction in potential pathogens. STAMP analysis (Figure 9B) at the genus level confirmed significant abundance changes in Ligilactobacillus, Bifidobacterium, and Olsenella, indicating their roles in H. pylori-associated pathologies. PICRUSt2 analysis (Figure 9C) revealed significant enrichment in metabolic pathways, particularly carbohydrate and nucleotide metabolism, essential for cellular growth and repair. This suggests that Lp05 intervention regulates metabolic activities in the gastric microbiome, potentially reducing metabolic disorders and long-term health issues associated with H. pylori infection.
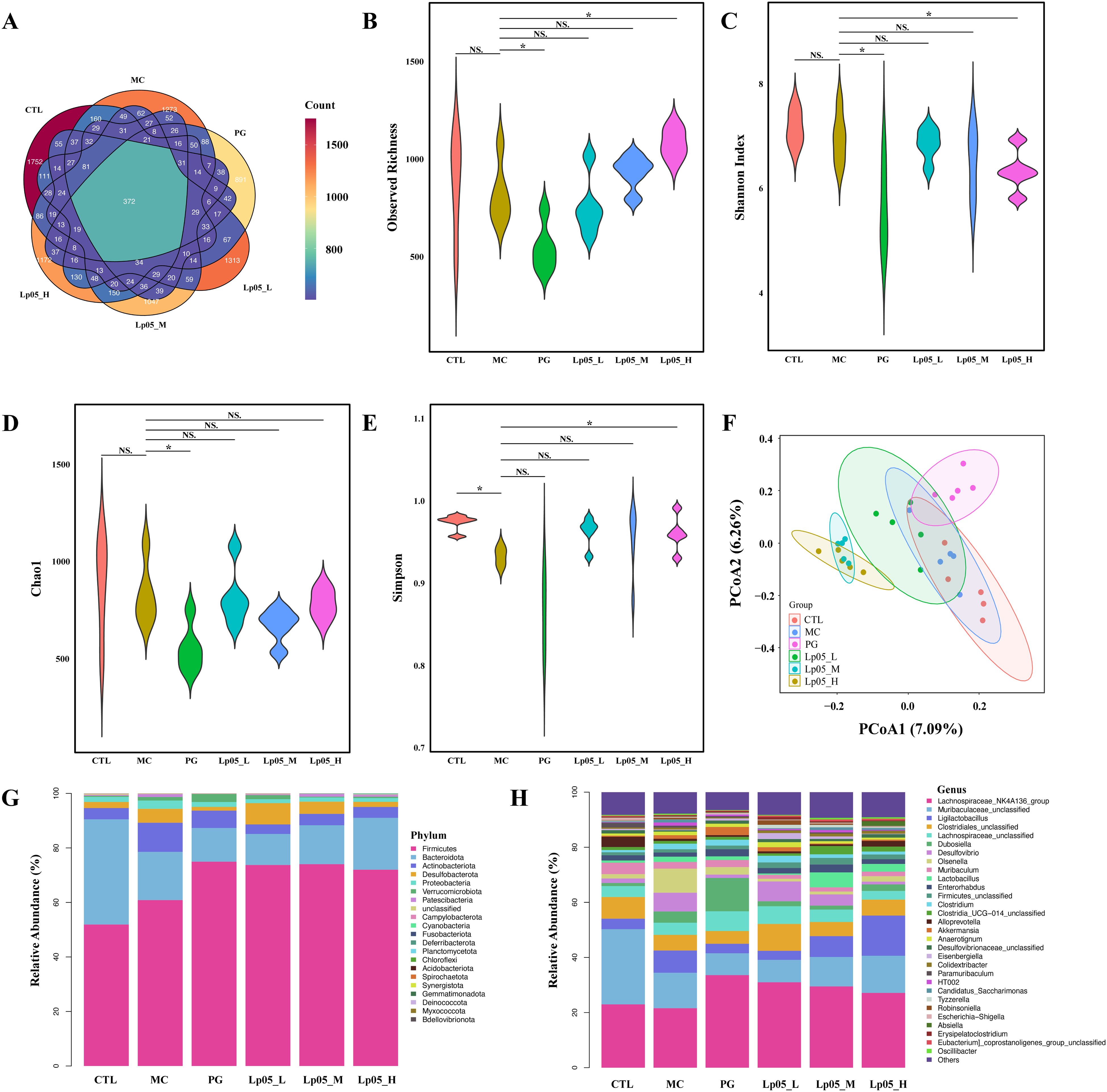
Figure 8. Impact of Lactiplantibacillus plantarum Lp05 doses on cecal microbial composition and structure. (A) Venn diagram analysis of intestinal microbiota sequencing; (B–E) Alpha diversity metrics of the intestinal microbiota; (F) Beta diversity of the intestinal microbiota assessed via PCoA analysis; (G, H) Relative abundance of species at the phylum and genus levels within the intestinal microbiota. Groups, CTL (control), MC (H. pylori infection model), Lp05_L (low dose, 2×107 CFU), Lp05_M (medium dose, 2×108 CFU), Lp05_H (high dose, 2×109 CFU). NS indicates p > 0.05,* indicates p < 0.05.
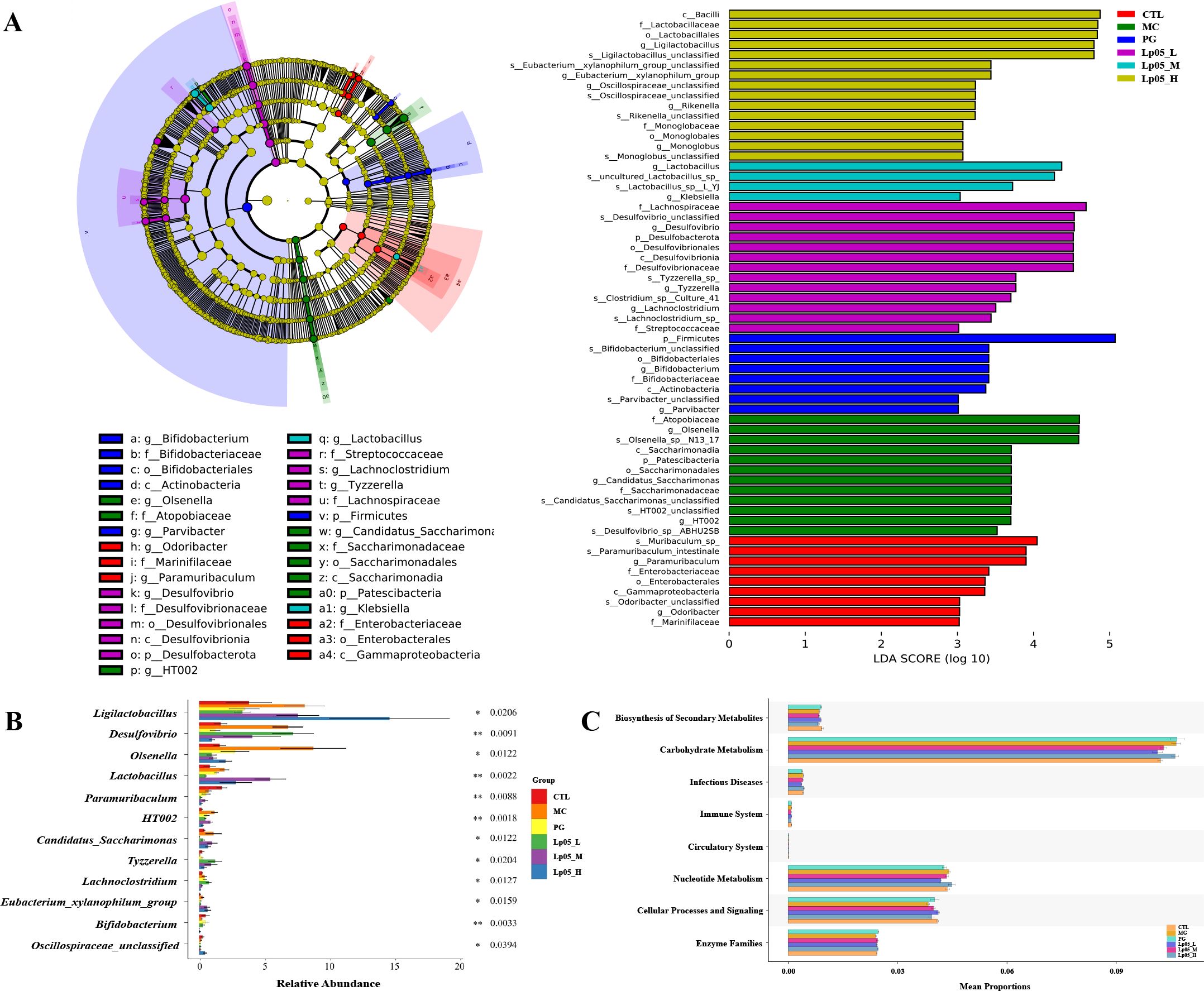
Figure 9. Effects of Lactiplantibacillus plantarum Lp05 doses on cecal microbiome diversity. (A) Cladogram from metagenomic data and LEfSe analysis showing taxa with significant differences, with circle sizes reflecting taxa abundance. (B) STAMP analysis identifies genus-level differences within the intestinal microbiome. (C) PICRUSt predicts functions of KEGG level 2 taxa in the intestinal microbiome. Groups, CTL (control), MC (H. pylori infection model), Lp05_L (low dose, 2×107 CFU), Lp05_M (medium dose, 2×108 CFU), Lp05_H (high dose, 2×109 CFU).
Correlation analysis of gastric and intestinal microbiome in H. pylori-infected mice model
Using Spearman’s correlation, we analyzed the relationship between the relative abundances of microbial genera in the gastric and intestinal microbiomes, as shown in Figure 10. In the gastric microbiome, genera such as Raoultella, Streptomyces, Pseudoxanthomonas, Gemmata, Sporosarcina, and Steroidobacter were found to have significant positive correlations (p < 0.05) with intestinal genera including Actinobacillus, Ureaplasma, Anaerococcus, Oceanivirga, Fusobacterium, Atopostipes, Succinivibrio, Rikenella, Campylobacter, and Anaerovibrio. Conversely, these gastric genera showed negative correlations with Christensenellaceae_R-7_group, Eubacterium_nodatum_group, Bifidobacterium, Mitochondria, Brevundimonas, and Subdoligranulum. Additionally, gastric genera such as Ruminococcus_gnavus_group, Rothia, UBA1819, Subdoligranulum, Blautia, and Phascolarctobacterium demonstrated positive correlations with Christensenellaceae_R-7_group, Eubacterium_nodatum_group, Bifidobacterium, Mitochondria, Brevundimonas, and Subdoligranulum. However, these were significantly negatively correlated (p < 0.05) with the intestinal genera including Actinobacillus, Ureaplasma, Anaerococcus, Oceanivirga, Fusobacterium, Atopostipes, Succinivibrio, Rikenella, Campylobacter, and Anaerovibrio. This complex interplay of correlations between gastric and intestinal microbiomes indicates that Lp05 intervention not only affects specific genera but also influences the broader microbial ecosystem relationships within the host.
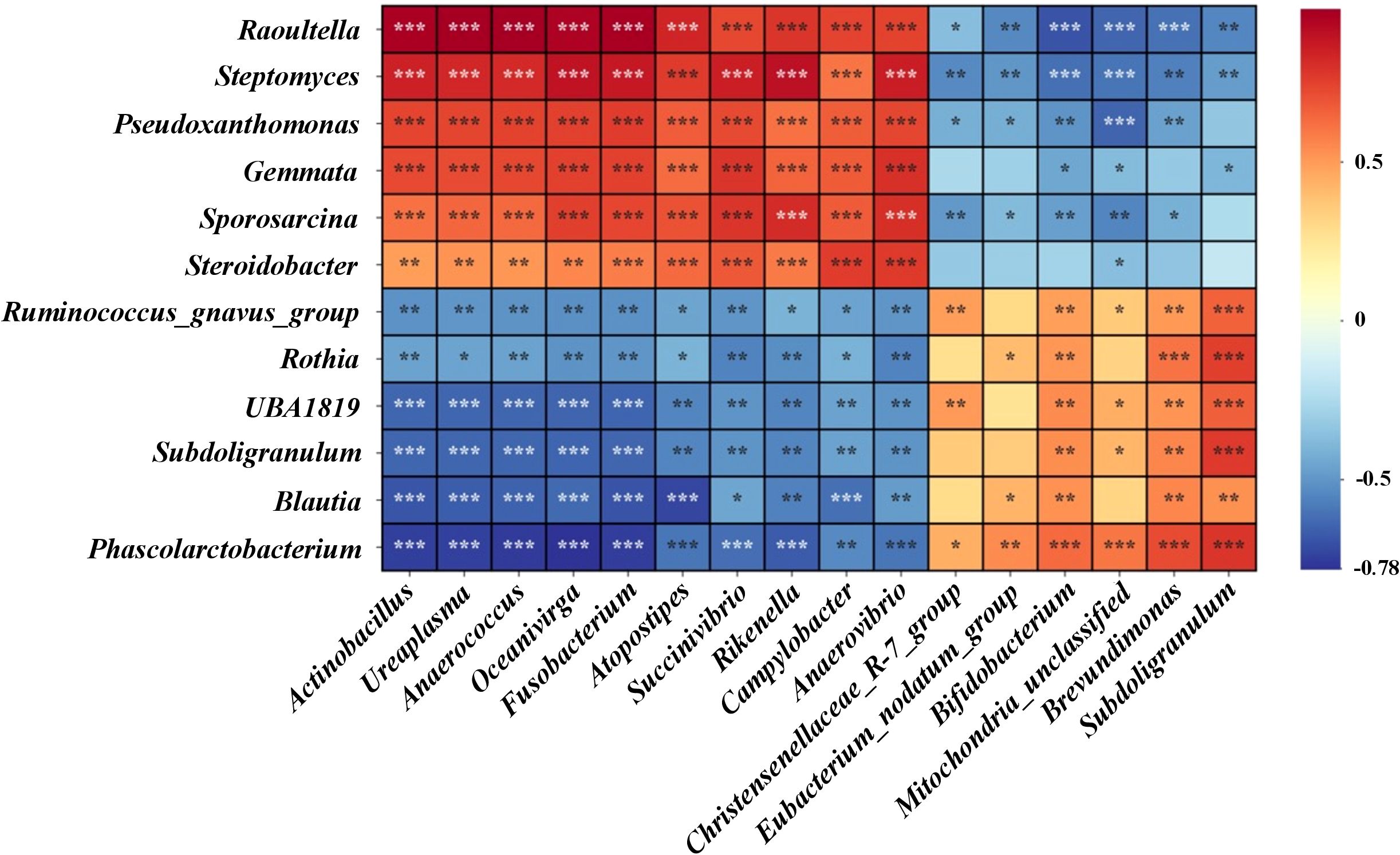
Figure 10. Presents the correlation analysis of differential microbial genera between gastric and intestinal microbiomes. Using Spearman's rank correlation test, the heatmap indicates significance levels with *** for p < 0.001, ** for p < 0.01, and * for p < 0.05.
Using Spearman’s correlation, the study evaluated the relationships between gastric and intestinal microbial genera in H. pylori-infected mice, depicted in Figure 10. Gastric genera like Raoultella, Streptomyces, and Pseudoxanthomonas showed significant positive correlations (p < 0.05) with several intestinal genera such as Actinobacillus and Ureaplasma, indicating potential shared roles in gastrointestinal processes. Conversely, these gastric genera negatively correlated with Christensenellaceae_R-7_group and Bifidobacterium, suggesting differing physiological roles or adaptations to environmental conditions. Additionally, genera like Ruminococcus_gnavus_group displayed both positive and negative correlations with various intestinal genera, reflecting their complex interactions within the gastrointestinal tract. These findings highlight how Lp05 intervention impacts not only specific microbial genera but also the broader inter-genus relationships, affecting overall microbial ecosystem dynamics within the host.
Discussion
This study demonstrates Lp05 potential as a therapeutic agent against H. pylori infection in mice, showcasing its antibacterial and immunomodulatory properties. Lp05 significantly reduces gastric mucosal urease activity, adjusts inflammatory cytokine levels, restores digestive enzyme functions, and alters gastric and intestinal microbiomes to combat H. pylori. These results not only underscore the effectiveness of probiotics in managing H. pylori infections but also highlight their capability to enhance gastrointestinal health. Lp05 may offer advantages over traditional treatments like quadruple therapy, potentially reducing antibiotic-associated gastric damage (34), which is crucial for long-term management.
Our findings are consistent with prior research on probiotics like L. reuteri and L. johnsonii, which have been shown to inhibit H. pylori growth and reduce gastric inflammation (18, 35). Specifically, Lp05’s effectiveness is demonstrated through in vitro studies where its co-aggregates with H. pylori, potentially disrupting its colonization by directly damaging its cellular structure or altering local pH levels (16, 36). In vivo, Lp05 significantly lowers urease activity and H. pylori-IgG levels in infected mice, suggesting reduced colonization and less gastric inflammation (17, 26, 37). Histopathological analysis shows Lp05 markedly improves tissue integrity in the stomach and duodenum, reducing inflammatory infiltration and maintaining epithelial structure, especially at higher doses. Unlike L. reuteri and L. johnsonii, Lp05 not only reduces urease activity and H. pylori colonization but also enhances the gastric environment and immune response, making it a comprehensive treatment option. These multifaceted effects of Lp05 highlight its potential as a holistic approach to managing H. pylori infection and promoting gastrointestinal health.
This study further examined the effects of Lp05 on the gastric microenvironment, focusing on antioxidant enzymes, digestive enzyme restoration, and immune modulation. Lp05 significantly restored the activity of essential digestive enzymes (α-amylase, cellulase, and pepsin) in the gastric tissues of H. pylori-infected mice. Typically, H. pylori infection impairs gastric function through mucosal damage and inflammation, which disrupt enzyme secretion and activity, and through the release of inflammatory mediators affecting enzyme function via complex signaling (38–40). Our findings show that Lp05 markedly enhances these enzymes’ activity, sometimes surpassing baseline levels. This suggests that Lp05 not only counters H. pylori colonization through direct antimicrobial effects but also facilitates recovery of gastric function, essential for combating H. pylori-induced dysfunction (6). Particularly at higher doses, Lp05’s restoration of enzyme activity underscores its multi-mechanistic approach, involving mucosal barrier repair, gastric secretion regulation, and reinstatement of normal gastric functions.
Additionally, Lp05 treatment significantly reduced the systemic inflammatory responses caused by H. pylori infection, which triggers both local gastric inflammation and systemic reactions, worsening the condition (41, 42). Following Lp05 intervention, inflammatory markers such as ET, IL-17A, IL-23, TGF-β1, and IP-10 were notably decreased, while regulatory markers like FoxP3, IFN-β, and IFN-γ were increased, indicating Lp05’s role in both reducing H. pylori colonization and modulating the host’s immune response. The high-dose group showed particularly strong improvements in inflammatory markers, underscoring Lp05’s effectiveness in immune modulation. Besides, oxidative stress, a key pathological response induced by H. pylori, involves reduced activity of antioxidant enzymes like CAT and SOD (43, 44), and increased oxidative markers like MDA and MPO (45, 46). Oxidative stress not only causes further damage to gastric tissues but may also trigger a range of downstream pathological responses, including cellular apoptosis and mutations (44, 47). Lp05 intervention significantly enhanced antioxidant enzyme activity and reduced oxidative stress markers, especially at higher doses, demonstrating its potential to mitigate oxidative stress and protect gastric tissues from H. pylori-induced damage. This suggests that probiotics could play a role in managing oxidative stress linked to infections.
We conducted further analyses of the gastric and cecal microbiota to explore how Lp05 mitigates H. pylori infection. The MC group displayed a marked decrease in alpha diversity, showing the pathogen’s impact on the host’s microbiome. Lp05 intervention significantly increased the observed richness and Chao1 indices in the gastric microbiome, suggesting it helps restore microbial diversity crucial for maintaining a healthy gastric environment and reducing H. pylori recurrence (48). Lp05 particularly raised the abundance of Firmicutes, known for their anti-inflammatory and protective functions (49), which may explain its effects on reducing inflammation. The recovery of Bacteroidota could relate to positive effects on intestinal barrier functions, aligning with improvements in intestinal structures (48, 50). Lp05 also significantly reduced urease activity, a crucial factor for H. pylori’s survival in acidic conditions (48). This suggests that Lp05’s modification of Firmicutes and Bacteroidota levels may alter gastric pH and microbial interactions, indirectly inhibiting urease activity (51, 52). LEfSe analysis showed that Lp05 enhanced bacterial taxa with anti-inflammatory and immunoregulatory functions like Rikenella (53, 54), correlating with decreases in inflammatory cytokines such as IL-10 and TGF-β1, indicating enhanced immune modulation. PICRUSt2 analysis revealed that Lp05 enriched pathways related to amino acid metabolism and polysaccharide biosynthesis, important for antioxidant defenses (55, 56). This corresponds with boosts in antioxidant enzyme activities such as CAT and SOD, suggesting that Lp05 promotes host antioxidant capabilities by modulating microbial communities and metabolic pathways, thus alleviating oxidative damage from H. pylori infection.
H. pylori infections disrupt gastric and intestinal microbial balance, evident from significant changes in biomarkers and microbial communities. Lp05 intervention notably restores diversity and richness in the intestinal microbiota, suggesting its role in rebalancing the microbial ecosystem to counteract H. pylori adverse effects. The observed enhancement in digestive enzyme activities, such as α-amylase, cellulase, and pepsin under Lp05 treatment, may directly relate to improved microbial health, as certain gut microbes influence the expression and activity of host enzymes, enhancing metabolic pathways and nutrient absorption (57, 58). LEfSe analysis revealed that Lp05 significantly alters microbial abundance related to health, particularly increasing the proportions of Lachnospiraceae_NK4A136_group and Muribaculaceae_unclassified, which could directly correlate with decreased levels of inflammatory factors like IL-17A, IL-23, and TGF-β1 (59). This increase in beneficial microbes might directly impact the host’s immune modulation by producing anti-inflammatory metabolites such as butyrate, thereby reducing inflammation (60). Additionally, Lp05 intervention reduces the abundance of Olsenella, reflecting its potential anti-inflammatory and gut health-promoting capabilities (61, 62). PICRUSt2 analysis of predicted functional changes shows that Lp05 influences microbial metabolic pathways, particularly enhancing carbohydrate and nucleotide metabolism, essential for cell growth and repair (63–65). This suggests that Lp05 not only affects the composition of the microbial community but also potentially improves metabolic health, which could help alleviate metabolic disturbances caused by H. pylori infections, reducing the risk of long-term health issues such as gastric cancer. Lp05’s intervention shows significant effects in reducing inflammation induced by H. pylori, adjusting the abundance of inflammation-related microbes and increasing anti-inflammatory microbial groups, which may help modulate the host’s immune response and reduce inflammation.
Further analysis of the interactions between gastric and intestinal microbiomes shows significant correlations between certain genera, such as Raoultella and Streptomyces in the stomach, and Actinobacillus and Fusobacterium in the intestines. These positive correlations suggest joint participation in gastrointestinal processes and emphasize the microbial communities’ synergistic role in overall health and stress response. Conversely, negative correlations like that between the Ruminococcus_gnavus_group in the stomach and Actinobacillus in the intestines highlight their differing physiological roles or unique environmental adaptations. For example, the Ruminococcus_gnavus_group may support mucosal barrier integrity and anti-inflammatory functions in the stomach, whereas it may be linked to inflammation or pathogen defense in the intestines (66). This analysis underlines the importance of microbial communities in systemic health management, influencing nutrient absorption, immune regulation, and pathogen defense. Notably, the negative correlation of Bifidobacterium suggests its supportive role in intestinal health might decrease in the stomach due to the stomach’s acidic environment (67). These insights reveal the intricate interactions between gastric and intestinal microbiomes and highlight the importance of considering the systemic impact of microbial communities in managing gastrointestinal diseases, suggesting a holistic approach could lead to better microbial health and host disease management strategies.
Overall, this study has unveiled the potential of Lp05 in strategies against H. pylori, demonstrating that its intervention not only influences the microbial communities in the stomach and intestines but also exerts positive effects on host physiological functions such as digestive enzyme activities and immune responses. However, the study’s limitations include the use of a specific animal model that may not fully reflect the complex physiological and microbiotic environments of humans, and the inability to completely explore the causal relationships between changes in microbial communities and host pathology. These factors underscore the necessity for future research to further validate these findings under diverse host models and broader environmental conditions.
Conclusion
This study has confirmed the efficacy of Lp05 in treating mice infected with H. pylori, illustrating its ability to modulate the gastrointestinal microbiota, alleviate inflammation and oxidative stress, and optimize host health. These results highlight the potential of Lp05 as a probiotic and provide a scientific basis for further research into its clinical applications. Future studies should delve deeper into its specific mechanisms of action to expand its use in the prevention and treatment of gastrointestinal diseases.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA1138072 https://www.ncbi.nlm.nih.gov/, PRJNA1138073.
Ethics statement
The animal studies were approved by Experimental Animal Ethics Committee of Zhengzhou University. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
YD: Writing – original draft. MH: Writing – review & editing. YQ: Writing – original draft. YW: Writing – original draft. ZZ: Writing – original draft. DJ: Conceptualization, Investigation, Methodology, Writing – original draft. ZG: Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (grant number 32102007) to MH.
Conflict of interest
Authors YD, YQ and ZG were employed by Wecare Probiotics Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1469885/full#supplementary-material
References
1. Crowe SE. Helicobacter pylori infection. New Engl J Med. (2019) 380:1158–65. doi: 10.1056/NEJMcp1710945
2. Hooi JKY, Lai WY, Ng WK, Suen MMY, Underwood FE, Tanyingoh D, et al. Global prevalence of helicobacter pylori infection: systematic review and meta-analysis. Gastroenterology. (2017) 153:420–9. doi: 10.1053/j.gastro.2017.04.022
3. FitzGerald R, Smith SM. An overview of helicobacter pylori infection. In: Smith SM, editor. Helicobacter pylori. Springer US, New York, NY (2021). p. 1–14.
4. FitzGerald R, Smith SM. An overview of helicobacter pylori infection. Methods Mol Biol. (2021) 2283:1–14. doi: 10.1007/978-1-0716-1302-3_1
5. Kao C-Y, Sheu B-S, Wu J-J. Helicobacter pylori infection: An overview of bacterial virulence factors and pathogenesis. Biomed J. (2016) 39:14–23. doi: 10.1016/j.bj.2015.06.002
6. Fong I, Fong I. Helicobacter pylori infection: when should it be treated? Curr Trends Concerns Infect Dis. (2020), 81–102. doi: 10.1007/978-3-030-36966-8_4
7. Ishikawa E, Nakamura M, Satou A, Shimada K, Nakamura S. Mucosa-associated lymphoid tissue (MALT) lymphoma in the gastrointestinal tract in the modern era. Cancers. (2022) 14:446. doi: 10.3390/cancers14020446
8. I.A.R.C. Schistosomes, liver flukes and Helicobacter pylori Vol. 61. Lyon: IARC Working Group on the Evaluation of Carcinogenic Risks to Humans (1994) p. 7–14 IARC Monogr. Eval. Carcinog. Risks Hum.
9. Guevara B, Cogdill AG. Helicobacter pylori: a review of current diagnostic and management strategies. Digestive Dis Sci. (2020) 65:1917–31. doi: 10.1007/s10620-020-06193-7
10. Malfertheiner P, Megraud F, O'Morain CA, Atherton J, Axon AT, Bazzoli F, et al. Management of Helicobacter pylori infection–the Maastricht IV/ Florence Consensus Report. Gut. (2012) 61:646–64. doi: 10.1136/gutjnl-2012-302084
11. Peng C, Sang S, Shen X, Zhang W, Yan J, Chen P, et al. In vitro anti-Helicobacter pylori activity of Syzygium aromaticum and the preliminary mechanism of action. J Ethnopharmacology. (2022) 288:114995. doi: 10.1016/j.jep.2022.114995
12. Malfertheiner P, Megraud F, Rokkas T, Gisbert JP, Liou JM, Schulz C, et al. Management of Helicobacter pylori infection: the Maastricht VI/Florence consensus report. Gut. (2022) 71(9):1724–62. doi: 10.1136/gutjnl-2022-327745
13. Saniee P, HosseinI F, Kadkhodaei S, Siavoshi F, Khalili-Saman S. Helicobacter pylori multidrug resistance due to misuse of antibiotics in Iran. Arch Iranian Med. (2018) 21:283–8.
14. Wang Z-J, Chen X-F, Zhang Z-X, Li Y-C, Deng J, Tu J, et al. Effects of anti-Helicobacter pylori concomitant therapy and probiotic supplementation on the throat and gut microbiota in humans. Microbial pathogenesis. (2017) 109:156–61. doi: 10.1016/j.micpath.2017.05.035
15. Zendeboodi F, Khorshidian N, Mortazavian AM, da Cruz AG. Probiotic: conceptualization from a new approach. Curr Opin Food Sci. (2020) 32:103–23. doi: 10.1016/j.cofs.2020.03.009
16. Wu S, Xu Y, Chen Z, Chen Y, Wei F, Xia C, et al. Lactiplantibacillus plantarum ZJ316 reduces Helicobacter pylori adhesion and inflammation by inhibiting the expression of adhesin and urease genes. Mol Nutr Food Res. (2023) 67:2300241. doi: 10.1002/mnfr.202300241
17. Sun L, Zhao H, Liu L, Wu X, Gao Q, Zhao Y. Effects of Lactobacillus on the inhibition of Helicobacter pylori growth. Biotechnol Biotechnol Equip. (2018) 32:1533–40. doi: 10.1080/13102818.2018.1515599
18. Aiba Y, Ishikawa H, Tokunaga M, Komatsu Y. Anti-Helicobacter pylori activity of non-living, heat-killed form of lactobacilli including Lactobacillus johnsonii No. 1088. FEMS Microbiol Lett. (2017) 364:fnx102. doi: 10.1093/femsle/fnx102
19. Sun X, Zhang S, Ren J, Udenigwe CC. Sialic acid-based strategies for the prevention and treatment of Helicobacter pylori infection: emerging trends in food industry. Crit Rev Food Sci Nutr. (2022) 62:1713–24. doi: 10.1080/10408398.2020.1846157
20. Cai Y, Dong Y, Han M, Jin M, Liu H, Gai Z, et al. Lacticaseibacillus paracasei LC86 mitigates age-related muscle wasting and cognitive impairment in SAMP8 mice through gut microbiota modulation and the regulation of serum inflammatory factors. Front Nutr. (2024) 11:1390433. doi: 10.3389/fnut.2024.1390433
21. Day AS, Jones NL, Policova Z, Jennings HA, Yau EK, Shannon P, et al. Characterization of virulence factors of mouse-adapted helicobacter pylori strain SS1 and effects on gastric hydrophobicity. Digestive Dis Sci. (2001) 46:1943–51. doi: 10.1023/A:1010691216207
22. Zhang L, Ma H, Kulyar M-e, Pan H, Li K, Li A, et al. Complete genome analysis of Lactobacillus fermentum YLF016 and its probiotic characteristics. Microbial Pathogenesis. (2022) 162:105212. doi: 10.1016/j.micpath.2021.105212
23. Auty MAE, Gardiner GE, McBrearty SJ, O'Sullivan EO, Mulvihill DM, Collins JK, et al. Direct in situ viability assessment of bacteria in probiotic dairy products using viability staining in conjunction with confocal scanning laser microscopy. Appl Environ Microbiol. (2001) 67:420–5. doi: 10.1128/AEM.67.1.420-425.2001
24. Kumar A, Kumar D. Characterization of Lactobacillus isolated from dairy samples for probiotic properties. Anaerobe. (2015) 33:117–23. doi: 10.1016/j.anaerobe.2015.03.004
25. Percie du Sert N, Ahluwalia A, Alam S, Avey MT, Baker M, Browne WJ, et al. Reporting animal research: Explanation and elaboration for the ARRIVE guidelines 2.0. PloS Biol. (2020) 18:e3000411. doi: 10.1371/journal.pbio.3000411
26. Chen Z, Tang Z, Li W, Deng X, Yu L, Yang J, et al. Weizmannia coagulans BCF-01: a novel gastrogenic probiotic for Helicobacter pylori infection control. Gut Microbes. (2024) 16:2313770. doi: 10.1080/19490976.2024.2313770
27. Warthin AS, Starry A. A more rapid and improved method of demonstrating spirochetes in tissues. Am J Syph. (1920) 4:97–103.
28. Dong Y, Liao W, Tang J, Fei T, Gai Z, Han M. Bifidobacterium BLa80 mitigates colitis by altering gut microbiota and alleviating inflammation. AMB Express. (2022) 12:67. doi: 10.1186/s13568-022-01411-z
29. Uotani T, Graham DY. Diagnosis of Helicobacter pylori using the rapid urease test. Ann Trans Med. (2015) 3(1). doi: 10.3978/j.issn.2305-5839.2014.12.04
30. Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. (2013) 10:996–8. doi: 10.1038/nmeth.2604
31. Chen T, Liu YX, Huang L. ImageGP: An easy-to-use data visualization web server for scientific researchers. Imeta. (2022) 1:e5. doi: 10.1002/imt2.v1.1
32. Oksanen J, Blanchet F, Friendly M, Kindt R, Legendre P, McGlinn D, et al. vegan: Community Ecology Package. R package version 2.5-6, Vol. 2019. CRAN (The Comprehensive R Archive Network). (2019). Available online at: https://CRAN.R-project.org/package=vegan.
33. Gómez-Rubio V. ggplot2 - elegant graphics for data analysis (2nd edition). J Stat Software. (2017) 77:1-3.
34. Rimbara E, Fischbach LA, Graham DY. Optimal therapy for Helicobacter pylori infections. Nat Rev Gastroenterol Hepatol. (2011) 8:79–88. doi: 10.1038/nrgastro.2010.210
35. Dore MP, Cuccu M, Pes GM, Manca A, Graham DY. Lactobacillus reuteri in the treatment of Helicobacter pylori infection. Internal Emergency Med. (2014) 9:649–54. doi: 10.1007/s11739-013-1013-z
36. Neshani A, Zare H, Akbari Eidgahi MR, Hooshyar Chichaklu A, Movaqar A, Ghazvini K. Review of antimicrobial peptides with anti-Helicobacter pylori activity. Helicobacter. (2019) 24:e12555. doi: 10.1111/hel.2019.24.issue-1
37. Crowe Sheila E. Helicobacter pylori infection. New Engl J Med. (2019) 380:1158–65. doi: 10.1056/NEJMcp1710945
38. Zhang Q-Q, Xie M, Guo R-X, Liu X-H, Ma S-J, Na L, et al. The effects of Helicobacter pylori eradication therapy on salivary pepsin concentration in patients with laryngopharyngeal reflux. Eur Arch Oto-Rhino-Laryngology. (2022) 279:5289–97. doi: 10.1007/s00405-022-07439-y
39. Nguyen SPH, Do TCMV, Tran TTT, Pham TKA, De Tran V, Nguyen KT. Characteristics of basal gastric juice in Helicobacter pylori-associated gastritis before and after eradication therapy. Trop J Pharm Res. (2022) 21:2241–6. doi: 10.4314/tjpr.v21i10.27
40. Yang H, Hu B. Immunological perspective: Helicobacter pylori infection and gastritis. Mediators Inflammation. (2022) 2022:2944156. doi: 10.1155/2022/2944156
41. He J, Liu Y, Ouyang Q, Li R, Li J, Chen W, et al. Helicobacter pylori and unignorable extragastric diseases: Mechanism and implications. Front Microbiol. (2022) 13:972777. doi: 10.3389/fmicb.2022.972777
42. Toyoshima O, Nishizawa T, Koike K. Endoscopic Kyoto classification of Helicobacter pylori infection and gastric cancer risk diagnosis. World J Gastroenterol. (2020) 26:466. doi: 10.3748/wjg.v26.i5.466
43. Hosoda K, Wanibuchi K, Amgalanbaatar A, Shoji M, Hayashi S, Shimomura H. A novel role of catalase in cholesterol uptake of Helicobacter pylori. Steroids. (2023) 191:109158. doi: 10.1016/j.steroids.2022.109158
44. Han L, Shu X, Wang J. Helicobacter pylori-mediated oxidative stress and gastric diseases: a review. Front Microbiol. (2022) 13:811258. doi: 10.3389/fmicb.2022.811258
45. Ndrepepa G. Myeloperoxidase - A bridge linking inflammation and oxidative stress with cardiovascular disease. Clin Chim Acta. (2019) 493:36–51. doi: 10.1016/j.cca.2019.02.022
46. Wang Y-K, Chiang W-C, Kuo F-C, Wu M-C, Shih H-Y, Wang SSW, et al. Levels of malondialdehyde in the gastric juice: Its association with Helicobacter pylori infection and stomach diseases. Helicobacter. (2018) 23:e12460. doi: 10.1111/hel.2018.23.issue-2
47. Toh JW, Wilson RB. Pathways of gastric carcinogenesis, Helicobacter pylori virulence and interactions with antioxidant systems, vitamin C and phytochemicals. Int J Mol Sci. (2020) 21:6451. doi: 10.3390/ijms21176451
48. Baj J, Forma A, Sitarz M, Portincasa P, Garruti G, Krasowska D, et al. Helicobacter pylori virulence factors—mechanisms of bacterial pathogenicity in the gastric microenvironment. Cells. (2020) 10:27. doi: 10.3390/cells10010027
49. Stojanov S, Berlec A, Štrukelj B. The influence of probiotics on the firmicutes/bacteroidetes ratio in the treatment of obesity and inflammatory bowel disease. Microorganisms. (2020) 8(11):1715. doi: 10.3390/microorganisms8111715
50. Manilla V, Di Tommaso N, Santopaolo F, Gasbarrini A, Ponziani FR. Endotoxemia and gastrointestinal cancers: insight into the mechanisms underlying a dangerous relationship. Microorganisms. (2023) 11:267. doi: 10.3390/microorganisms11020267
51. Schuchmann K, Müller V. Autotrophy at the thermodynamic limit of life: a model for energy conservation in acetogenic bacteria. Nat Rev Microbiol. (2014) 12:809–21. doi: 10.1038/nrmicro3365
52. Vesper BJ, Jawdi A, Altman KW, Haines GK 3rd, Tao L, Radosevich JA. The effect of proton pump inhibitors on the human microbiota. Curr Drug Metab. (2009) 10:84–9. doi: 10.2174/138920009787048392
53. Ma R-X, Hu J-Q, Fu W, Zhong J, Cao C, Wang C-C, et al. Intermittent fasting protects against food allergy in a murine model via regulating gut microbiota. Front Immunol. (2023) 14:1167562. doi: 10.3389/fimmu.2023.1167562
54. Graf J. The family rikenellaceae. prokaryotes. (2014), 857–9. doi: 10.1007/978-3-642-38954-2_134
55. Wu G. Amino acids: metabolism, functions, and nutrition. Amino Acids. (2009) 37:1–17. doi: 10.1007/s00726-009-0269-0
56. Gong F, Zhou X, Cao K, Xu H, Zhou X. Analyses of the metabolism and assessment of antioxidant activity in Rhododendron chrysanthum Pall. after UV-B Irradiation. Plant Growth Regul. (2024), 1–13. doi: 10.21203/rs.3.rs-3084712/v1
57. Sewaka M, Trullas C, Chotiko A, Rodkhum C, Chansue N, Boonanuntanasarn S, et al. Efficacy of synbiotic Jerusalem artichoke and Lactobacillus rhamnosus GG-supplemented diets on growth performance, serum biochemical parameters, intestinal morphology, immune parameters and protection against Aeromonas veronii in juvenile red tilapia (Oreochromis spp.). Fish shellfish Immunol. (2019) 86:260–8. doi: 10.1016/j.fsi.2018.11.026
58. Falcinelli S, Rodiles A, Hatef A, Picchietti S, Cossignani L, Merrifield DL, et al. Influence of probiotics administration on gut microbiota core: A review on the effects on appetite control, glucose, and lipid metabolism. J Clin Gastroenterol. (2018) 52:S50–6. doi: 10.1097/MCG.0000000000001064
59. Wu M-R, Chou T-S, Huang C-Y, Hsiao J-K. A potential probiotic-Lachnospiraceae NK4A136 group: Evidence from the restoration of the dietary pattern from a high-fat diet. (2020). doi: 10.21203/rs.3.rs-48913/v1
60. Siddiqui MT, Cresci GA. The immunomodulatory functions of butyrate. J Inflammation Res. (2021), 6025–41. doi: 10.2147/JIR.S300989
61. Wang Q, Zhan X, Wang B, Wang F, Zhou Y, Xu S, et al. Modified montmorillonite improved growth performance of broilers by modulating intestinal microbiota and enhancing intestinal barriers. Anti-Inflammatory Response Antioxidative Capacity Antioxidants. (2022) 11:1799. doi: 10.3390/antiox11091799
62. Obata M, Ohtsuji M, Iida Y, Shirai T, Hirose S, Nishimura H. Genome-wide genetic study in autoimmune disease-prone mice. Arthritis Research: Methods Protoc. (2014), 111–41. doi: 10.1007/978-1-4939-0404-4_13
63. Mason EF, Rathmell JC. Cell metabolism: An essential link between cell growth and apoptosis. Biochim Biophys Acta (BBA) - Mol Cell Res. (2011) 1813:645–54. doi: 10.1016/j.bbamcr.2010.08.011
64. Kaneko JJ. ”Carbohydrate metabolism and its diseases“. In Clinical Biochemistry of Domestic Animals. San Diego, CA: Academic Press. (1997) p. 45–81.
65. Zhao H, Raines LN, Huang SC-C. Carbohydrate and amino acid metabolism as hallmarks for innate immune cell activation and function. Cells. (2020) 9:562. doi: 10.3390/cells9030562
66. Mao A, Zhao W, Zhu Y, Kong F, Chen D, Si H, et al. Gut bacterial community determines the therapeutic effect of ginsenoside on canine inflammatory bowel disease by modulating the colonic mucosal barrier. Microorganisms. (2023) 11:2616. doi: 10.3390/microorganisms11112616
Keywords: Helicobacter pylori, Lactiplantibacillus plantarum, gut microbiota, gastric microenvironment, inflammatory markers
Citation: Dong Y, Han M, Qi Y, Wu Y, Zhou Z, Jiang D and Gai Z (2025) Enhancement of host defense against Helicobacter pylori infection through modulation of the gastrointestinal microenvironment by Lactiplantibacillus plantarum Lp05. Front. Immunol. 15:1469885. doi: 10.3389/fimmu.2024.1469885
Received: 24 July 2024; Accepted: 17 December 2024;
Published: 17 January 2025.
Edited by:
Esaki M. Shankar, Central University of Tamil Nadu, IndiaReviewed by:
Vanitha Mariappan, National University of Malaysia, MalaysiaRavi Verma, Jawaharlal Nehru University, India
Jawaria Mansoor, University of Veterinary and Animal Sciences, Pakistan
Copyright © 2025 Dong, Han, Qi, Wu, Zhou, Jiang and Gai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhonghui Gai, emhnYWlAYWxpeXVuLmNvbQ==
 Yao Dong
Yao Dong Mei Han
Mei Han Yongmei Qi1
Yongmei Qi1 Ying Wu
Ying Wu Zhonghui Gai
Zhonghui Gai