- 1Instituto de Investigación, Desarrollo e Innovación en Biotecnología Sanitaria de Elche (IDiBE), Universidad Miguel Hernández (IDiBE-UMH), Elche, Spain
- 2MISTRAL Beamline Experiments Division, ALBA Synchrotron Light Source, Barcelona, Spain
Introduction: Fish nucleated red blood cells (RBCs), also known as erythrocytes, play a crucial role in maintaining immune system balance by modulating protein expression in response to various stimuli, including viral attack. This study explores the intriguing behavior of rainbow trout RBCs when faced with the viral hemorrhagic septicemia virus (VHSV), focusing on the endoplasmic reticulum (ER) stress and the unfolded protein response (UPR).
Methods: Rainbow trout RBCs were Ficoll-purified and exposed to ultraviolet (UV)-inactivated VHSV or live VHSV at different multiplicities of infection (MOIs). Using cryo-soft X-ray tomography (cryo-SXT), we uncovered structural and cellular modifications in RBCs exposed to UV-inactivated VHSV. Moreover, RBCs were treated with 4-phenylbutyric acid (4-PBA), an ER stress inhibitor, to investigate its effect on viral replication. Quantitative real-time PCR was also used to analyze the expression of genes related to the UPR and other related cellular pathways.
Results and discussion: Beyond their antiviral response, RBCs undergo notable intracellular changes to combat the virus. Cryo-SXT highlighted a significant increase in the ER volume. This increase is associated with ER stress and the activation of the UPR pathway. Interestingly, VHSV replication levels augmented in RBCs under ER-stress inhibition by 4-PBA treatment, suggesting that rainbow trout RBCs tune up ER stress to control viral replication. Therefore, our findings suggested the induction of ER stress and subsequent activation UPR signaling in the antiviral response of RBCs to VHSV. The results open a new line of investigation to uncover additional mechanisms that may become novel cellular targets for the development of RBC-targeted antiviral strategies.
1 Introduction
The main function of red blood cells (RBCs) is to transport oxygen and carbon dioxide, but these cells also play an important role in biological processes related to the immune response in fish (1, 2), such as the recognition of pathogen-associated molecular patterns (PAMPs) (2), the processing and presentation of antigens (3), and the production of effector molecules and cytokines (2, 4). Multiple studies have shown that fish RBCs are also involved in the immune response against viral infections (4–6). Understanding how fish respond to a viral infection is of utmost importance in the search for new preventive treatments, especially for the aquaculture industry.
When exposed to viral hemorrhagic septicemia virus (VHSV), fish RBCs undergo intracellular changes that prevent viral replication (5). For example, autophagy and antigen processing may be involved in minimizing the number of infectious particles in the cell (6–9). On the other hand, other fish rhabdoviruses may manipulate autophagy to evade immunity, promote replication and facilitate their release from infected cells (10–12). Transmission electron microscopy (TEM) has allowed visualization of defense mechanisms in VHSV-exposed RBCs related to the elimination of viral proteins and antigen presentation, which is the formation of vesicles similar to autophagosomes (7). The adaptive immune response of fish RBCs involves the expression of major histocompatibility complex (MHC) class I and II molecules (7, 13). The expression of these molecules implies that RBCs can present antigens and act as atypical APCs, linking RBCs to a vaccine-triggered immune response. Similar to mammalian RBCs, fish RBCs have promise as platforms for immunostimulants or vaccines given that they can express antigens encoded by a DNA vaccine and modulate the expression of interferon-related genes when expressing these antigens (14, 15).
At the intracellular level, the organization of organelles dictates the functional specificity of the cell depending on the environment. This subcellular arrangement may be key to understanding the structure-function trade-off in response to various stimuli. Nevertheless, examination of the morphological organization of a cell in its native state is hindered in part by current analytical platforms and microscopy techniques that lack high resolution and require sample sectioning (16, 17). A new approach, based on full-field cryo-soft X-ray tomography (cryo-SXT), could overcome these limitations given the potential to visualize the three-dimensional nanoscale structure of an intact cryopreserved cell. Moreover, the fast acquisition and large field of view enable rapid collection of tomographic image data. Segmenting this data into usable features is crucial for deriving biologically relevant information from cryo-SXT tomograms (18–20). Using cryo-SXT, Perez-Berna et al. (21) visualized cell structural and topological differences generated by viral exposure and noted a volumetric increase in some cells, especially in the endoplasmic reticulum (ER).
In the present work, we analyzed cryo-SXT images of rainbow trout RBCs exposed to UV-inactivated VHSV (VHSVUV), using the MISTRAL beamline of the ALBA Synchrotron. The purpose of the work was to investigate the structure-function link between the spatial arrangement of organelles and the cellular response after VHSVUV exposure. Notably, virus inactivation by a physical agent, such as ultraviolet (UV) light, preserves virus immunogenicity while destroying infectivity. Therefore, UV-inactivated virus is capable of inducing protective responses as long as the antigen remains intact.
Cryo-SXT images facilitated the identification of RBC organelles that are the core of the response to the VHSVUV, featuring the ER as the main protagonist. Previous transcriptomic and proteomic analysis in RBCs has documented upregulation of molecules related to ER stress, autophagy, and antigen presentation in response to virus and DNA vaccine encoding the viral antigen (7, 14). The ER plays a crucial role in maintaining cellular homeostasis by processing and folding proteins. Alterations in the ER function, such as the accumulation of misfolded proteins or increased demand for protein folding, leads to a state known as ER stress. To resolve this imbalance, cells trigger a series of signaling pathways called the unfolded protein response (UPR) (22, 23). The UPR is controlled by three main ER sensors: inositol-requiring enzyme 1 (IRE1), protein kinase R (PKR)-like ER kinase (PERK), and activating transcription factor 6 (ATF6). Under homeostatic conditions, these transmembrane sensor proteins bind to folding chaperone glucose-regulated protein 78 (GRP78), also known as binding immunoglobulin protein (BiP). Under stress, transmembrane sensor proteins dissociate from GRP78, resulting in the activation of all three pathways (24, 25). ER stress has a major impact on infections, and pathogens can modulate ER stress to their own advantage. Regarding viruses, recent studies have shown that various viral taxa, including Orthomyxoviruses such as influenza virus and Flaviviruses like dengue virus, can significantly modulate the UPR to enhance their replication and evade host immune responses (26–28). However, recent findings suggest a protective role orchestrated by ER stress that the cell activates for its survival (29).
In this work, we investigated the possible correlation between morphological changes and the role of ER stress as a first-aid pathway to invoke an antiviral response in rainbow trout RBCs in response to an inactivated virus. Combining this new knowledge with previous findings will shed light on potential molecular targets for the design of new vaccine approaches, for which RBCs are ideal candidates.
2 Materials and methods
2.1 Animals and ethical statements
Rainbow trout (Oncorhynchus mykiss) individuals approximately 6-7 cm in size were acquired from a certified commercial farm (Piszolla S.L., Cimballa Fish Farm, Zaragoza, Spain). The fish supplier followed strict biosecurity protocols that include the regular monitoring of stock health and compliance with veterinary standards to ensure that the fish are free of pathogens. Fish were maintained at 14°C with a recirculating, dechlorinated water system at the Miguel Hernández University (UMH) animal house facilities. Prior to experimentation, fish were acclimatized to laboratory conditions for two weeks. The number of fish samples used is expressed by an “n” in each experiment’s figure legend.
2.2 Cell culture and virus
The protocol used to obtain and purify rainbow trout RBCs has been previously described (5, 30). In brief, fish were bled from the caudal vein using insulin syringes (NIPRO, Bridgewater). Collected blood samples were placed in a 2 mL eppendorf tube with RPMI-1640 medium (Dutch modification) (Gibco, Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (FBS) gamma irradiated (Cultek), 1 mM pyruvate (Gibco), 2 mM L-glutamine (Gibco), 50 μg/mL gentamicin (Gibco), 2 μg/mL fungizone (Gibco), and 100 U/mL penicillin/streptomycin (Sigma-Aldrich). These samples were layered out onto 2 mL of Histopaque-1077 Ficoll gradient solution (Sigma-Aldrich), a sterile-filtered, ready-to-use solution with a density of 1.077 g/mL. Two consecutive density gradient centrifugations were carried out for purification. Then, RBCs were washed twice with RPMI 2% FBS. Finally, RBCs were cultured with RPMI 10% FBS in 25 cm2 flasks (Nunc Roskilde) at 14°C for 24 hours before experimentation.
The VHSV strain 07.71 (31) was purchased from the ATCC (VR-1388) and cultured in fathead minnow Epithelioma papulosum cyprini (EPC) cell line at 14°C as previously reported (32). VHSV is known to replicate efficiently in EPC cells with a high degree of reproducibility (33).
2.3 Cryo-soft X-ray tomography of RBCs exposed to VHSVUV
VHSV (108 tissue culture infectious dose 50% [TCID50]/mL) was inactivated by two rounds of UV-B irradiation at 1 J/cm2 using a Bio-Link crosslinker BLX E312 (Vilber Lourmat, BLX-E312), following previously validated methods (9). The viral suspension was placed in a shallow multi-well culture plate, to ensure minimal liquid height to maximize UV light penetration and effectiveness. Ficoll-purified rainbow trout RBCs were exposed to VHSVUV in RPMI 2% FBS, at a multiplicity of infection (MOI) of 10. RBCs unexposed to VHSVUV were used as a control. At 12 hours post-exposure (hpe), RBCs were placed on top of Quantifoil holey film copper TEM grids (Au-G200F1) coated with poly-L-lysine (Merck, Germany), with fiducial gold markers (100 nm; BBI Solutions, UK) and vitrified by plunge freezing in liquid ethane in a Leica EM-CPC. We selected the 12 hpe time point based on previous studies indicating that VHSV replication decreases between 6- and 24-hpe (5). This time frame was chosen to capture a critical phase for evaluating cellular responses, while considering the viral replication dynamics.
Vitrified grids were transferred in liquid nitrogen to the cryo-correlative cooling stage (CMS196 stage; Linkam Scientific Instruments, UK) to hold samples stably at 190°C during analysis. The cryo-stage was inserted into a Zeiss Axio Scope (Carl Zeiss, Germany) epifluorescence microscope to visualize the frozen grids. Selected samples were transferred under cryogenic conditions to the MISTRAL beamline (ALBA light source) at ALBA Synchrotron (34, 35). Tomographic data were collected at 520 eV, irradiating the samples for 1-2 s per projection. 520 eV is the energy range in which water is transparent for X-rays and they are mostly absorbed by carbon, allowing the visualization of biological material. In tomographic setup, images obtained at different sample orientations are computationally combined to produce a three-dimensional image, permitting the three-dimensional representation of the sub-cellular ultrastructure of whole, intact cells. A tilt series was acquired for each cell using an angular step of 1° on a 70° angular range with a Fresnel Zone plate (FZP) with a 40 nm outermost zone width and effective pixel size of 13 nm.
Each transmission projection image of the tilt series was normalized using flat-field. This process considers the possibility of different exposure times and the slight decrease of the electron beam current during the acquisition. To increase the image quality, wiener deconvolution considering the experimental impulse response of the optical system was applied to the normalized data (36). The Naperian logarithm was used to reconstruct the linear absorption coefficient (LAC). The resulting stacks were loaded into IMOD software (RRID: SCR_003297), and individual projections were aligned to the common tilt-axis using the internal cellular structures as markers (37). Then, the aligned stacks were reconstructed with algebraic reconstruction techniques (38). The visualization, feature segmentation, and quantification of the LACs and volumes were carried out using Amira 3D software (Thermo Fisher, RRID: SCR_007353). Cellular organelles are distinguished by image contrast, which is due to differential and quantitative absorption of X-ray photons in the ‘water window’, between the absorption edges of carbon (284 eV) and oxygen (543 eV) (39). The interaction of X-rays is element-specific and is due to the relative concentration of organic material and water. We used previous knowledge on the appearance of cellular organelles under cryo-SXT (40) together with representative TEM images (7, 41–45).
The euchromatin/heterochromatin (EU/HET) ratio was calculated taking into account the absorbance of the segmented material that is the corresponding LAC for each voxel of the volume reconstructed with SXT (46). For this purpose, the nuclei were analyzed and compared in terms of volume, absorbance, and shape. As a result, a difference in density can be observed, demonstrating a bimodal distribution. This allows the selection of regions of lower absorption (i.e., light areas) and regions of higher absorption (i.e., dark areas) within each nucleus. Regions with low LAC values correspond to euchromatin regions with low chromatin condensation and more transcriptional machinery. In contrast, regions with high LAC values correspond to heterochromatin, a condensed form of DNA with less transcriptional activity (47).
2.4 Time course of ER stress gene expression in RBCs
Ficoll-purified RBCs from the peripheral blood of rainbow trout individuals (5 x 105 cells/well) were exposed to VHSVUV at MOI 10 and VHSV at MOI 1 and 10, in RPMI 2% FBS at 14°C, and incubated for 6 and 24 hours. Samples were resuspended in TRK lysis buffer (Omega Bio-Tek, Inc.) and stored for RNA extraction and quantitative real-time PCR (qPCR) analysis as previously described, and detailed below (5). For differential expression analyses, we examined a set of genes that are well-known representative components of the UPR (22, 48–50). Evaluated genes and primers used can be found in Table 1.
2.5 4-PBA and niclosamide cytotoxicity evaluation
To examine the cytotoxicity of 4-phenylbutyric acid (4-PBA) (Sigma-Aldrich) or niclosamide (Sigma-Aldrich) on rainbow trout RBCs, propidium-iodide (PI) (Sigma-Aldrich) staining was performed to discriminate between live and dead cells using flow cytometric analysis. Briefly, RBCs were treated with 4-PBA or niclosamide according to the times and conditions used in the assays. For 4-PBA, RBCs were incubated for 24 and 72 hours at the maximum concentration used (8 mM), whereas for niclosamide, RBCs were incubated for 24 hours at 10 µM. As a positive control to determine cell death, 10 μL of 50% hydrogen peroxide (H2O2) (an apoptosis inducer) was added to untreated RBCs for 10 minutes. Afterward, cells were stained with 10 µL PI (1 mg/mL). Possible cell damage was detected by measuring the fluorescence intensity of PI-stained RBCs using a FACS Canto II flow cytometer (BD Biosciences).
2.6 ER stress inhibition assay
To assess the functional involvement of the ER in viral defense, we used the ER stress inhibitor 4-PBA and tested the susceptibility of cells to VHSV exposure. 4-PBA acts as a chemical chaperone whose hydrophobic regions interact with the hydrophobic segments of the unfolded protein, thereby promoting protein folding and reducing ER stress (51). To evaluate the effect of ER stress inhibition on viral replication, cells were treated with 4-PBA prior to virus exposure as previously described (52). For this purpose, Ficoll-purified rainbow trout RBCs (5 x 105 cells/well) were incubated for 24 hours at 14°C with 2, 4, or 8 mM 4-PBA in RPMI 10% FBS. RBCs were washed and exposed to VHSV at MOI 1 for 6 or 72 hours in RPMI 2% FBS at 14°C. Samples were stored in TRK lysis buffer for RNA extraction and qPCR analysis.
2.7 ER stress inhibition effect on autophagy in RBCs exposed to VHSV
To evaluate the effect of ER stress inhibition on autophagy, in the context of viral infection, Ficoll-purified RBCs (5 x 105 cells/well) were pre-treated with 8 mM of 4-PBA for 24 hours at 14°C and then exposed to VHSV at MOI 10 for 6 hours. In parallel, RBCs were exposed to VHSV for 3 hours and then incubated 24 hours with 10 µM of niclosamide, an autophagic flux blocker. Then, we analyzed the expression of ubiquitin-binding protein p62 to monitor autophagic flux as well as the N-VHSV protein to quantify viral replication, by means of flow cytometry, as explained below.
2.8 Flow cytometry
Flow cytometry analysis was carried out using a FACS Canto II flow cytometer (BD Biosciences). RBCs were fixed with 4% paraformaldehyde (PFA) and 0.008% glutaraldehyde diluted in RPMI medium for 45 minutes. Permeabilization of RBCs was carried out in 0.05% saponin (Sigma-Aldrich) buffer for 15 minutes. We used rabbit anti-p62/SQSTM1 antibody (www.antibodiesonline.com; Ref #ABIN2854836, RRID: AB_3096920) diluted 1/250 in permeabilization buffer to track the autophagy process. We used murine 2C9 anti-NVHSV (53) (RRID: AB_2716276) diluted 1/1000 in permeabilization buffer to track viral replication. Primary antibodies were incubated for 60 minutes. Goat anti-rabbit IgG (H+L) CF647 and goat anti-mouse IgG (H+L) CF488 (Sigma-Aldrich) diluted 1/200 in permeabilization buffer, and incubated for 30 minutes, were used as secondary antibodies. RBCs were stored in 1% PFA in PBS.
2.9 Analysis of genes involved in UPR and related cellular signaling pathways in RBCs from VHSV-challenged rainbow trout
Individual rainbow trout were challenged with 50 µL VHSV (108 TCID50/mL) resuspended in RPMI 2% FBS by intramuscular injection. Negative controls were injected with 50 µL RPMI 2% FBS. Animals were kept at 14°C over the course of the infection. Fish were sacrificed at 2 days post challenge (dpc), and peripheral blood collected from the caudal vein was purified to obtain the RBCs as described above. RBCs were stored in TRK lysis buffer for RNA extraction, and gene expression was analyzed by qPCR. We analyzed the genes involved in UPR and related cellular pathways listed in Table 1.
2.10 RNA isolation, cDNA synthesis, and gene expression by quantitative real-time PCR
Samples were first digested with Proteinase K solution (Omega Bio-Tek) and then RNA was isolated using the E.Z.N.A. Total RNA Kit (Omega Bio-Tek) according to the manufacturer’s instructions. Briefly, samples were applied to the HiBind RNA Mini Column, where total RNA binds to the matrix. Cell debris and contaminants were removed by a series of wash steps. Finally, high-quality RNA was eluted in DEPC-treated water. TURBO DNase (Ambion, Thermo Fisher Scientific Inc.) was used to remove residual genomic DNA as previously described (5). RNA was quantified with a NanoDrop spectrophotometer (Nanodrop Technologies). cDNA was synthesized from RNA using M-MLV reverse transcriptase (Invitrogen, Thermo Fisher Scientific) as previously described (54). qPCR reactions were performed in a total volume of 20 μL containing cDNA, 900 nM of each primer, 10 μL of TaqMan Universal PCR master mix (Thermo Fisher Scientific) with 300 nM probe, or 10 μL of SYBR Green master mix (Thermo Fisher Scientific). qPCR was carried out using the QUANTSTUDIO 3 system (Applied Biosystems, Thermo Fisher Scientific Inc.). Cycling conditions were 50°C for 2 min and 95°C for 10 min, followed by 40 cycles at 95°C for 15 s and 60°C for 1 min (5). Gene expression was analyzed by the 2-ΔCt or 2−ΔΔCt method (55) as indicated in each experiment, and the ef1α gene was used as an endogenous control. Primers and probes sequences are listed in Table 1. The number of samples analyzed in each experiment is indicated by an “n” in the figure legends.
2.11 Confocal microscopy
To visualize the ER changes in RBCs in response to VHSV, the maximum concentration of 4-PBA (8 mM) was chosen. RBCs were incubated with 4-PBA for 24 hours prior to VHSV exposure. Then, RBCs were exposed with VHSV at MOI 1 for 6 hours. Cells were washed with fresh medium and stained with ER-Tracker Red reagent (BODIPY TR Glibenclamide, Thermo Fisher Scientific Inc.) for 30 minutes, CellMask Green Plasma Membrane Stain (Thermo Fisher Scientific Inc.) for 15 minutes, and Hoechst reagent (Thermo Fisher Scientific Inc.) for 5 minutes. Dyes were diluted 1:1000. Images were taken with a Zeiss LSM900 with Airyscan 2 and analyzed with ZEN 3.2 (Blue Edition) software (Carl Zeiss Microscopy, RRID: SCR_013672). This software was also used to measure the fluorescence intensity in the Red Channel (“ER staining”) and to represent and quantify ER volume at multi-depth along the z-axis (Z-stacks). Volume rendering was performed using IMARIS software v9.3 (Bitplane, RRID: SCR_007370).
2.12 Software and statistics
GraphPad Prism 6 software (GraphPad Software Inc., RRID: SCR_002798) was used for graphic creation and statistical analysis. Statistical tests and P values are indicated for each assay. Flowing software 2.5.1 (www.flowingsoftware.com/, RRID: SCR_015781) was used to process and analyze flow cytometry data, and Floreada.io software (RRID: SCR_025286) was applied to generate histograms and dot plots for cytotoxicity assays. ZEN 3.2 (Blue Edition) software and IMARIS v9.3 were used to analyze and produce confocal microscopy images. Amira Thermo Scientific software (Thermo Fisher) was used for the analysis and processing of images obtained by cryo-SXT.
3 Results
3.1 VHSVUV induces ultrastructural alterations in RBCs
Three-dimensional reconstruction of cryo-SXT images of control RBCs and VHSVUV-exposed RBCs was carried out to gain insight into the structural alterations that occur at an intercellular level (Figure 1). The sample preparation and data collection steps carried out for cryo-SXT are outlined in Figure 1A. We also calculated the volume of the organelles of interest and the level of structural packing of the chromatin (Figure 2).
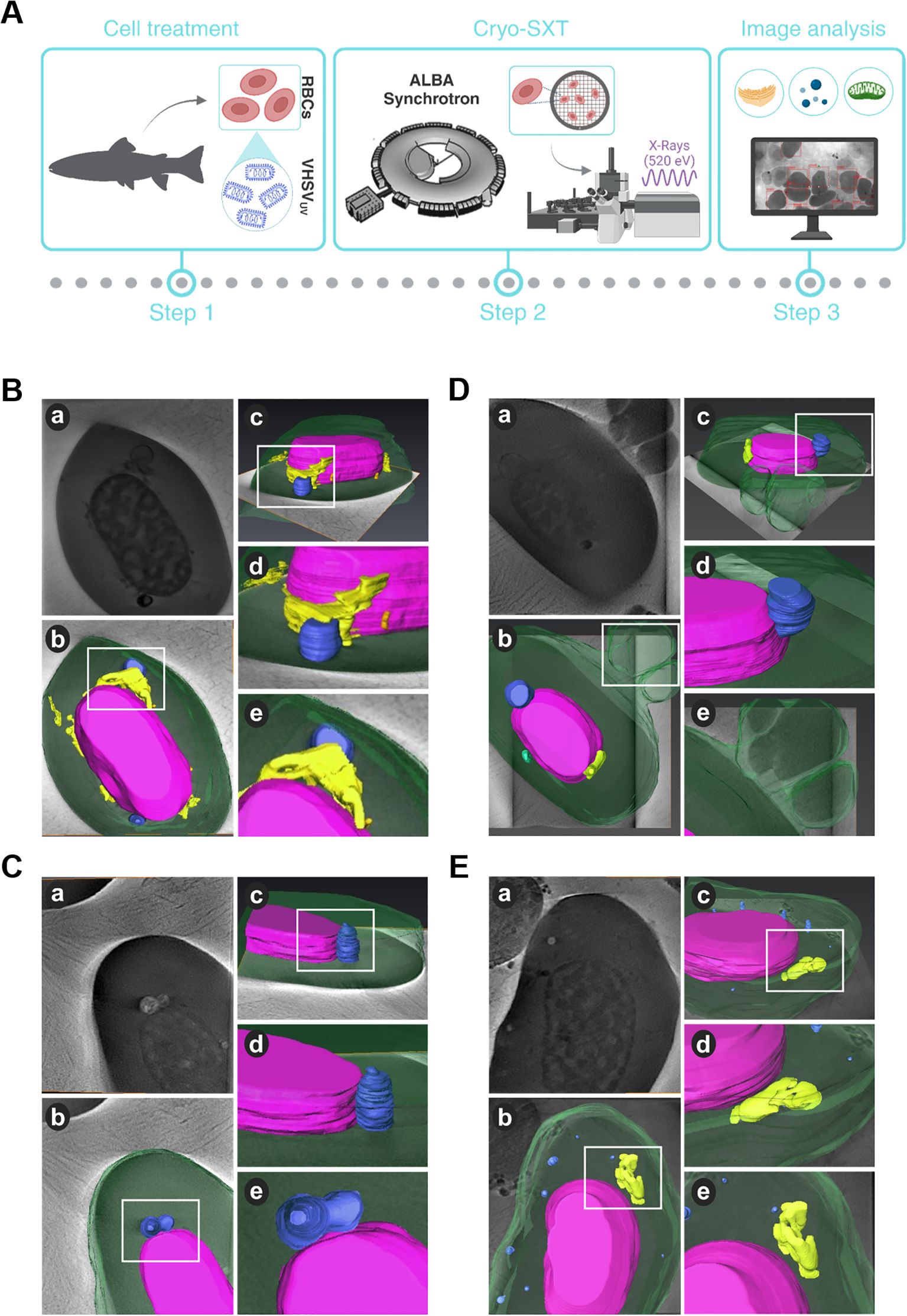
Figure 1. Structural and topological differences in RBCs induced by VHSVUV. (A) Workflow of experimental process. (B–D) Cryo-SXT images of VHSVUV-exposed RBCs. (E) Representative cryo-SXT image of control RBCs (RBCs unexposed to VHSVUV). Within each panel, section (a) is the cell tomography, and sections (b, c) are three-dimensional representations of the RBC in different orientations. Sections (d, e) are magnifications of the highlighted areas (white squares) of the cell. The nucleus is shown in purple, the endoplasmic reticulum in yellow, autophagosome-like vesicles in blue, mitochondria in green, and the cytoplasm and nascent EVs appear in translucent green. Volume rendering was performed with Amira software.
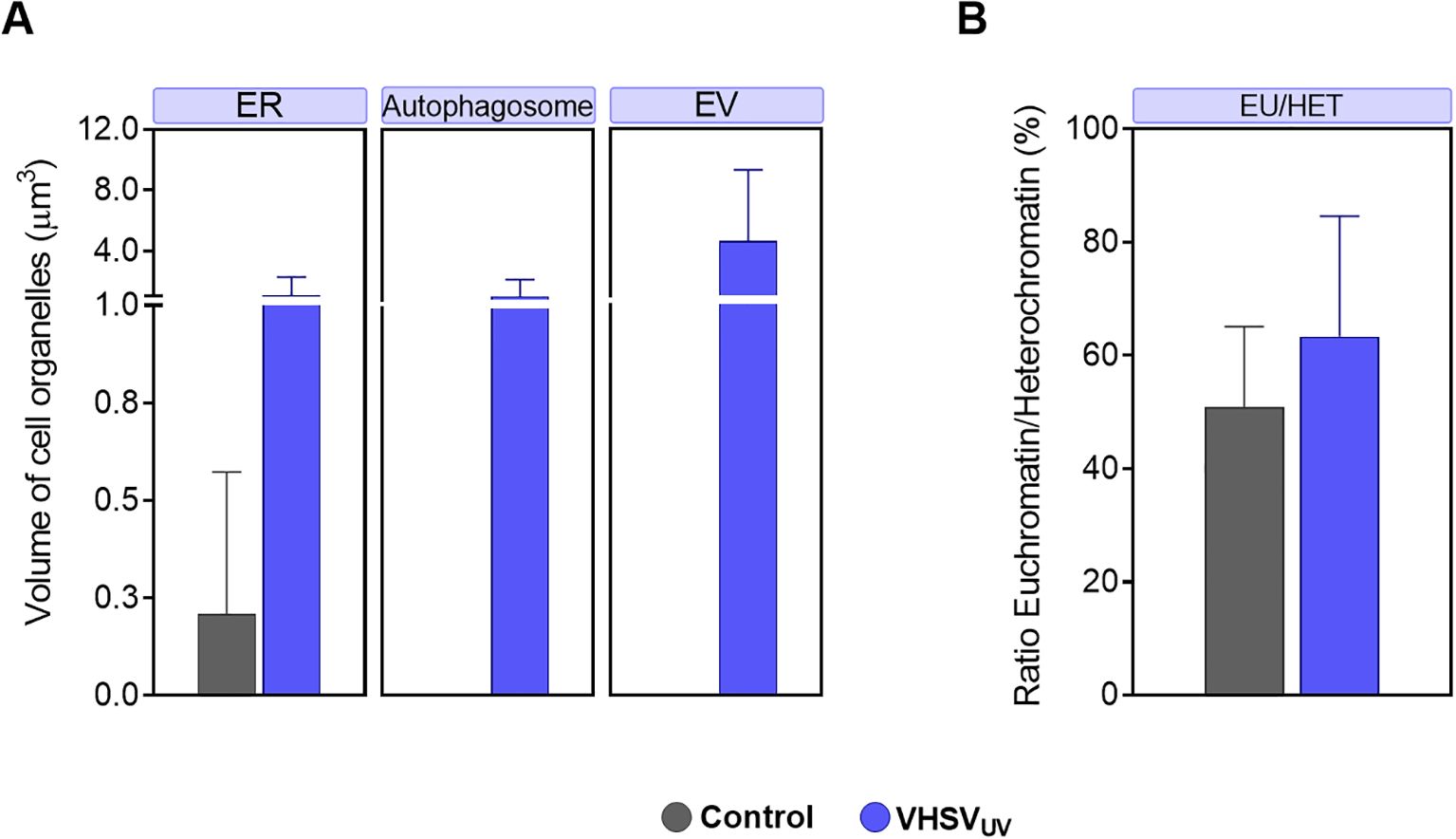
Figure 2. Volume quantification of ultracellular alterations in RBCs induced by VHSVUV. (A) Volume of cellular organelles of rainbow trout RBCs exposed to VHSVUV in comparison to control RBCs, expressed in μm3. ER, endoplasmic reticulum; EV, extracellular vesicle. (B) Euchromatin-heterochromatin (EU/HET) ratio in VHSVUV-exposed rainbow trout RBCs expressed in %. Calculations were made with the Amira software. Data represent the mean ± standard deviation (n=3). A non-parametric Mann-Whitney test was applied for statistical analysis.
Based on 3D volume representations, we identified subcellular differences between VHSVUV-exposed and control RBCs. We have been able to clearly identify cell organelles such as ER, mitochondria, autophagosomes and extracellular vesicles (EVs). However, cryo-SXT has limited resolution visualization of some cellular structures, such as the Golgi apparatus.
Regarding ER structures, the most striking result was the thickening of the ER in VHSVUV-exposed RBCs (Figure 1B). Another distinguishing feature was the increased number and size of autophagosome-like vesicles (Figures 1B–D) in contrast to control RBCs (Figure 1E). Interestingly, these structures are often located close to the nucleus where the ER is found. Furthermore, we noted protrusions on the plasma membrane of VHSVUV-exposed RBCs, which could correspond to the secretion of EVs (Figure 1D). Volumetric quantification supported these observations: we detected higher ER volume and more autophagosome and EV formation in VHSVUV-exposed vs control RBCs (Figure 2A).
VHSVUV-exposed RBCs had higher EU/HET ratios than control RBCs (Figure 2B), suggesting increased transcriptional activity in VHSVUV-exposed RBCs. The selection of the LAC regions corresponding to euchromatin and heterochromatin is depicted in Supplementary Figure 1.
3.2 UPR is upregulated in RBCs in response to VHSV or VHSVUV
We evaluated the expression of major UPR genes and their downstream targets in response to ER stress activation by VHSVUV and live VHSV, at two time points. The goal was to identify a differential pattern of gene expression in the experimental conditions of interest: RBCs exposed to VHSVUV (Figure 3B); RBCs exposed to VHSV MOI 1 and MOI 10 (Figures 3C, D, respectively). The experimental workflow is detailed in Figure 3A.
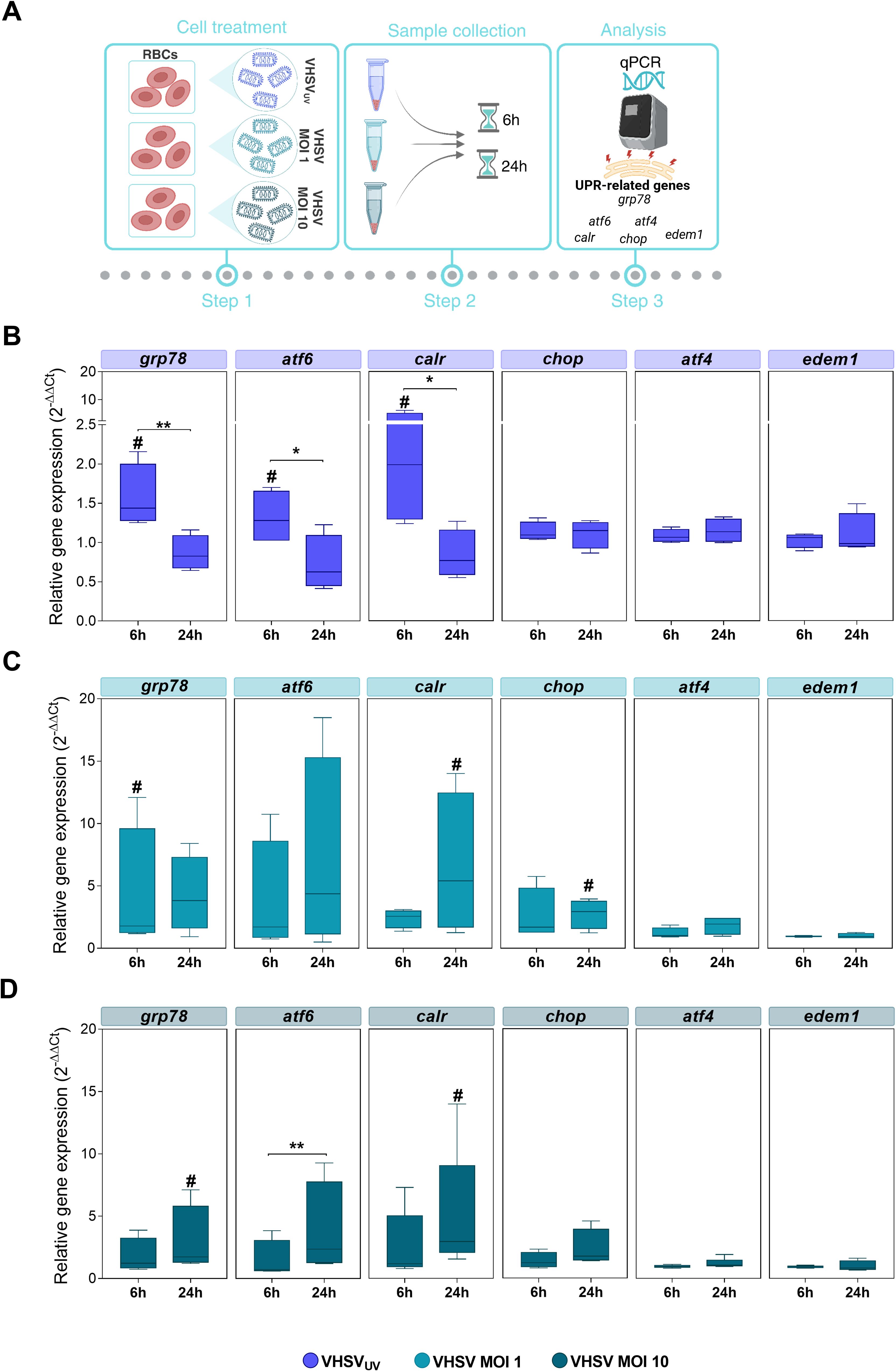
Figure 3. Monitoring UPR-related genes in RBCs exposed to VHSVUV or VHSV, at different time points. (A) Workflow of experimental conditions. Box plots represent grp78, atf6, calr, chop, atf4, and edem1 expression levels, in Ficoll-purified RBCs exposed to VHSVUV (B), VHSV MOI 1 (C), and VHSV MOI 10 (D), at 6 and 24 hpe. Gene expression was relativized to control cells (unexposed RBCs). ef1α was used as an endogenous control gene. Data represent the mean ± standard deviation (n=4). For statistical analysis, the Kruskal-Wallis test with Dunn’s multiple comparisons test was applied between each condition. #P<0.05 with respect to the control. *P<0.05 and **P<0.01 between time point of exposure.
The results showed significant upregulation of the master controller of ER stress response grp78 and ATF6 UPR branch genes, such as atf6 and their target calreticulin (calr), under VHSVUV or live VHSV exposure compared to control RBCs (unexposed RBCs) (Figures 3B–D). In detail, these three genes were overexpressed in RBCs exposed to VHSVUV at 6 hpe (Figure 3B), compared to control RBCs, whereas in RBCs exposed to VHSV MOI 1 and MOI 10, the response was mainly delayed, with significant upregulation observed at 24 hpe (Figures 3C, D), except for grp78 expression in RBCs exposed to VHSV MOI 1 at 6 hpe. Regarding PERK UPR branch, we analyzed the expression of atf4 and chop. In relation to IRE1 UPR branch, we evaluated the ER degradation enhancing alpha-mannosidase like protein 1 (edem1) gene, which is a target gene of XBP1. Neither in RBCs exposed to VHSVUV (Figure 3B) nor in RBCs exposed to VHSV at MOI 1 and MOI 10 (Figures 3C, D, respectively) was the expression of atf4 or edem1 affected, compared to control RBCs. No upregulation of chop was observed after the exposure with VHSVUV, compared to control RBCs, and, likewise, chop expression remained unchanged at both time points (Figure 3B). Nevertheless, chop overexpression was observed in RBCs in response to VHSV MOI 1 and MOI 10, compared to control RBCs, and in particular at MOI 1, at 24 hpe, when it was statistically significant (Figures 3C, D).
Overall, the results indicated that RBCs exposed to VHSVUV or live VHSV triggered an UPR regulation. However, it should be clarified that although VHSVUV confirmed rapid induction of UPR mainly through the ER chaperone GRP78 and the ATF6 pathway, the expression levels of the related genes were higher in RBCs in response to live virus (VHSV MOI 1 and MOI 10).
3.3 ER stress inhibition enhances viral replication in RBCs
Pre-treatment with the ER stress inhibitor 4-PBA at different concentrations (2, 4, and 8 mM) for 24 hours was carried out prior to VHSV exposure to investigate the involvement of ER stress in the response to the virus in rainbow trout RBCs (Figure 4A). Then, viral replication was analyzed by qPCR of the N-VHSV gene in RBCs at 6 and 72 hpe. N-VHSV gene expression increased in a dose-dependent manner after ER stress inhibition with 4-PBA (Figure 4B). It should be highlighted that, as 4-PBA concentration increased, so did N-VHSV gene transcription, even at 72 hpe it was statistically significant for the maximum concentration used. This rise of viral replication at 72 hpe is surprising, since previous reports found that the VHSV replication cycle is halted in RBCs beyond 24 hours (5). The effect of 4-PBA on UPR modulation was also assessed in RBCs (Supplementary Figure 2). grp78 and calr expression levels were significantly lowered in 4-PBA-treated RBCs at 6 hours, relative to their control. In contrast, at this time point, both genes were significantly upregulated in 4-PBA-treated and VHSV-exposed RBCs, when compared to 4-PBA-treated RBCs. Thus, 4-PBA appeared to attenuate UPR in RBCs while VHSV exposure acted as a stress condition that 4-PBA could not resolve. Prior to experiments, the absence of cytotoxicity of 4-PBA on RBCs was confirmed using PI staining method and measured by flow cytometry (Supplementary Figure 3).
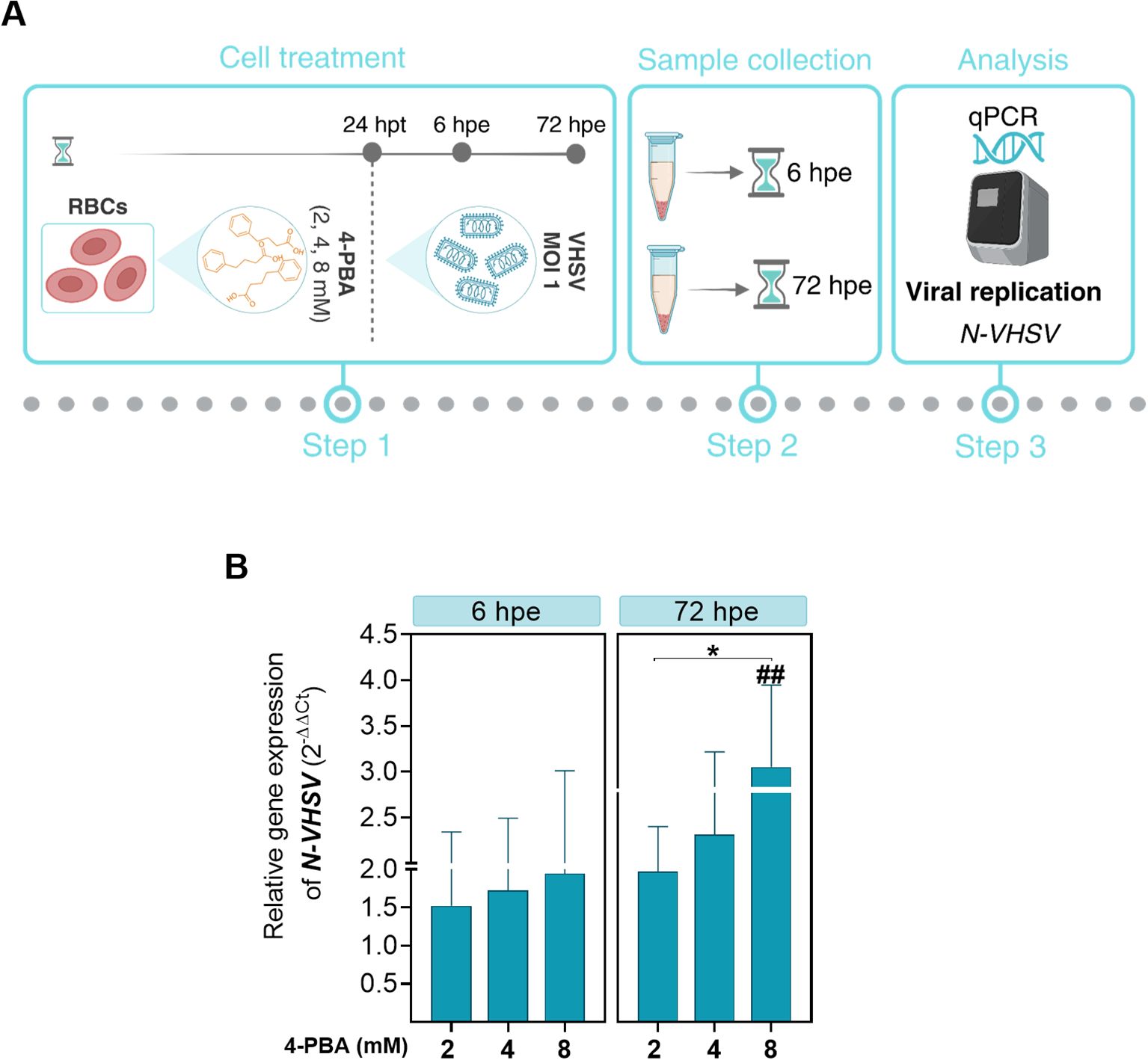
Figure 4. Effect of ER stress inhibition on VHSV replication in RBCs. (A) Workflow of experimental conditions. (B) N-VHSV gene expression in Ficoll-purified RBCs treated with 2, 4, or 8 mM 4-PBA and exposed to VHSV at MOI 1. Gene expression profile was analyzed by qPCR at 6 and 72 hpe. Gene expression was relativized to control cells (RBCs untreated with 4-PBA and exposed to VHSV). The ef1α gene was used as an endogenous control. Data represent mean ± standard deviation (n=6). For statistical analysis, we performed a two-way ANOVA with Tukey’s multiple comparisons test. ##P<0.01 with respect to the control. *P<0.05 between 4-PBA concentrations within each time point.
To achieve consistency in our observations on the response of RBCs to the virus under ER stress inhibition, we performed morphological monitoring of the ER under 4-PBA treatment (8mM) and VHSV exposure. For this, we combined live-cell fluorescence microscopy through ER-Tracker with quantification of ER volume based on confocal microscopy Z-stack images. As shown in confocal microscopy images (Figure 5A), VHSV-exposed RBCs displayed a strong fluorescent ER signal followed by RBCs pre-treated with 4-PBA and exposed to VHSV. We calculated the mean fluorescence intensity (MFI) of the ER signal and the results showed that it significantly increased in VHSV-exposed RBCs whereas, in those RBCs treated with 4-PBA (8 mM), ER signal decreased (Figure 5B). Further, as shown in 3D reconstructed images from confocal microscopy, VHSV exposure led to an enlargement of the membranous network of the ER, together with ER expansion (Figure 5C). We next quantified ER volume in the presence or absence of 4-PBA. Analogously with MFI results, a significant increase in ER volume was visible when RBCs were exposed to VHSV but when pre-treated with 4-PBA the ER volume decreased (Figure 5D). It is worth pointing out here that, when considering the results of cryo-SXT, both VHSV and VHSVUV shared a similar pattern in RBCs, expanding the ER in response to the stimulus.
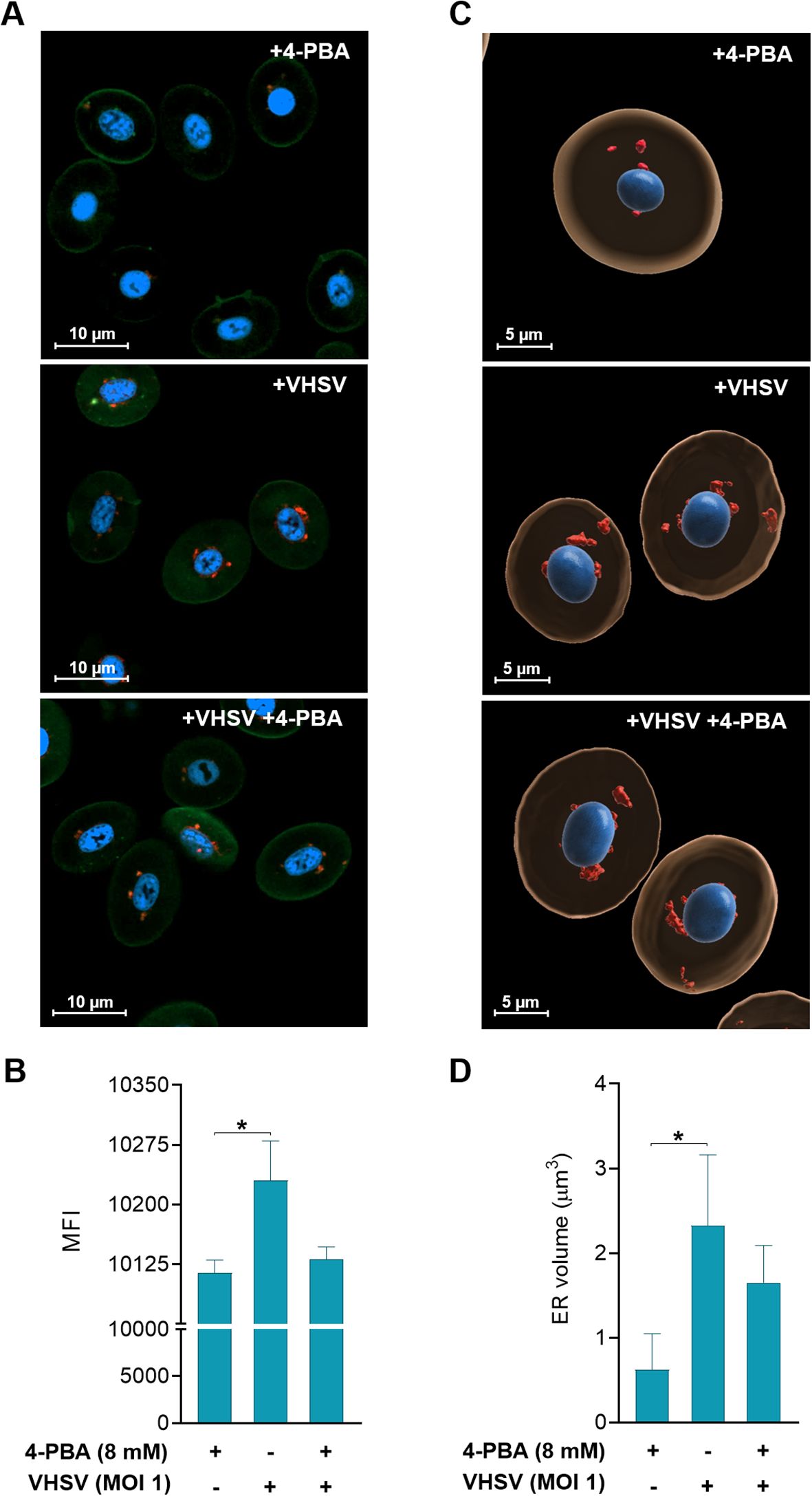
Figure 5. Two- and three-dimensional analysis of ER changes in response to VHSV and 4-PBA in RBCs. (A) Confocal laser scanning microscopy images from RBCs exposed to VHSV in the presence or absence of 4-PBA (8 mM). Images were taken at 63X magnification. The nucleus was stained with Hoechst (blue), the plasma membrane was stained with Cell Mask (green), and the ER was labeled with ER-Tracker (red). The scale bars equal 10 µm. (B) Mean fluorescence intensity (MFI) of the ER-Tracker signal. ZEN (Blue Edition) software was used for image processing and calculation of fluorescence intensity. Data represent the mean ± standard deviation (n=6). (C) Three-dimensional reconstruction of Z-stack confocal datasets. The ER is shown in red, the nucleus in blue, and the cytoplasm in beige. The scale bars equal 5 µm. (D) Volume quantification (µm3) of the ER. Data represent mean ± standard deviation (n=4). Cell volume measurement was performed with IMARIS software. For statistical analysis, the Kruskal-Wallis test with Dunn’s multiple comparisons test was used. *P<0.05.
3.4 ER stress inhibition effect on viral replication and autophagic flux
We decided to investigate whether autophagy may be altered by ER stress inhibition. For this, 4-PBA was used as an ER stress inhibitor, and niclosamide was used to block autophagic flux due to inhibition of autolysosomes formation (56). Previously, we evaluated whether niclosamide treatment had a cytotoxic effect on RBCs and results showed that niclosamide treatment at 10 µM did not induce cell damage (Supplementary Figure 4). As summarized in Figure 6A, for ER stress inhibition, RBCs were preincubated with 4-PBA and then exposed to VHSV. In the case of autophagy blockade, RBCs were first exposed to VHSV and then treated with niclosamide. By means of flow cytometry, we analyzed the ubiquitin-binding protein p62 to monitor autophagic flux as well as the N-VHSV protein to quantify viral replication under these conditions. When VHSV-exposed RBCs were treated with 4-PBA or niclosamide, we found a slight (non-significant) increase in p62 protein levels accompanied by an increase in N-VHSV protein levels, compared to untreated RBCs (Figures 6B, C). Although the differences were not significant, these results suggest that while both ER stress inhibition and autophagy flux blockade appear to affect VHSV replication in RBCs similarly, we cannot say whether ER stress directly affects autophagy or vice versa.
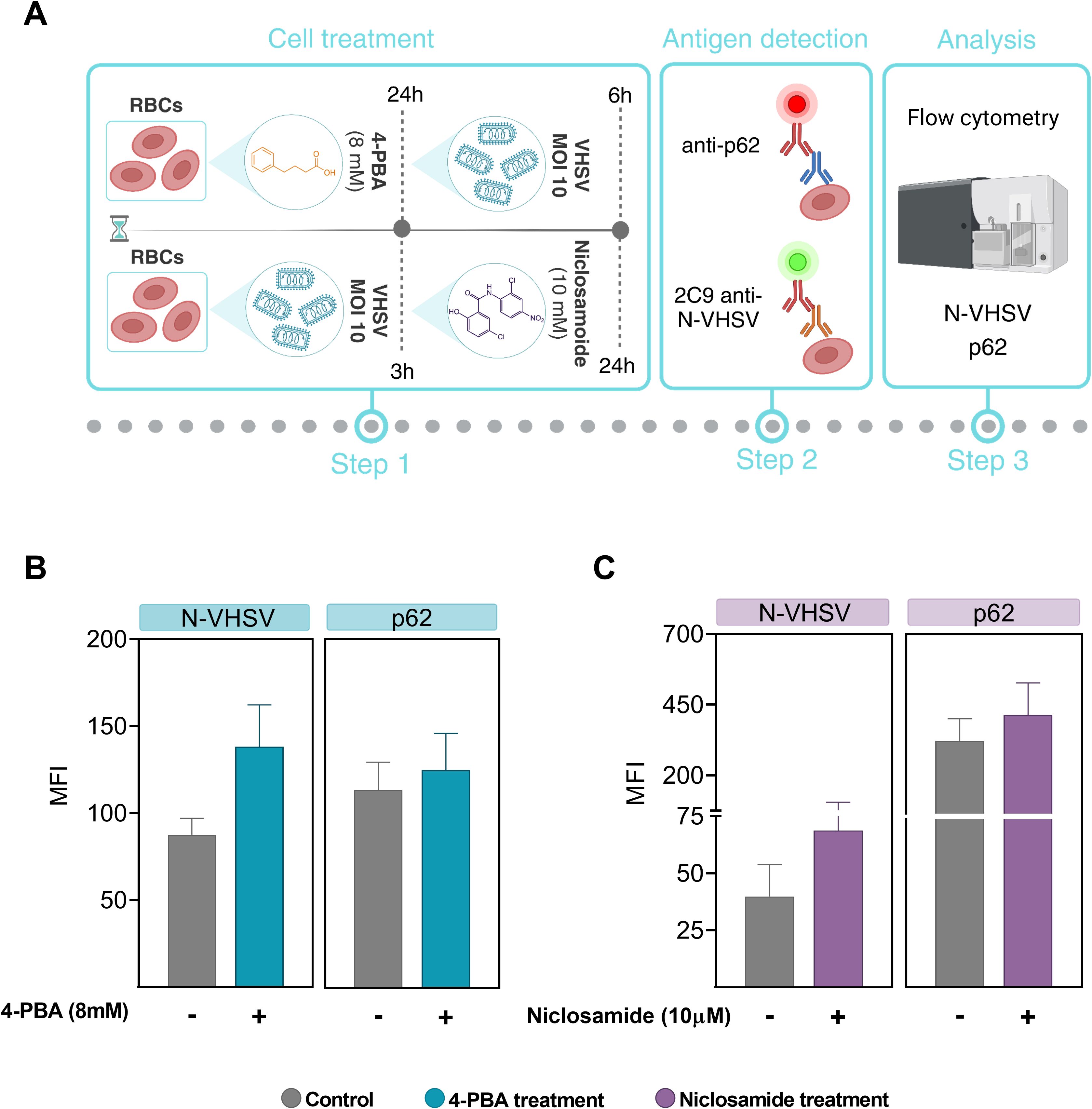
Figure 6. ER stress inhibition effect on viral replication and autophagic flux. (A) Workflow of experimental conditions. (B) MFI of p62 and VHSV levels in RBCs treated with 8 mM 4-PBA (blue bars) and exposed to VHSV at MOI 10, analyzed by flow cytometry at 24 hpe. (C) MFI of p62 and VHSV levels in VHSV-exposed RBCs at MOI 10 and treated with 10 µM niclosamide (purple bars), analyzed by flow cytometry at 24 hpe. Data represent the mean ± standard deviation (n=3). For statistical analysis, a Kruskal-Wallis test with Dunn’s multiple comparisons test was applied.
3.5 Transcriptomic evaluation of UPR connection with antigen presentation and vesicular transport pathways in RBCs from VHSV-challenged rainbow trout
We evaluated the possible correlation between the expression of UPR-associated genes (grp78, chop, atf6, and calr), and genes related to antigen presentation and vesicular transport pathways (mhcI, mhcII, exocyst complex component 1 [exoc1], and SEC13 homolog, nuclear pore and COPII coat complex component [sec13]) in RBCs of VHSV-challenged rainbow trout. Additionally, we evaluated the gene tumor necrosis factor receptor-associated protein 1 (trap1), which is known to be involved in ER stress by modulating the UPR and assisting in protein folding and mitochondrial stress (57, 58) (Figure 7A). At 2 dpc, rainbow trout RBCs showed upregulation of the genes studied, with statistically significant upregulation of grp78, calr, mhcI, and sec13 genes (Figure 7B). The gene heatmap showed a marked differential ER stress, antigen presentation and vesicular transport response in RBCs from VHSV-challenged individuals (Figure 7C). Although we cannot definitively conclude that these pathways are directly interconnected, the observed simultaneous activation suggested a coordinated response to VHSV infection. A more comprehensive analysis is required to explore potential mechanistic links.
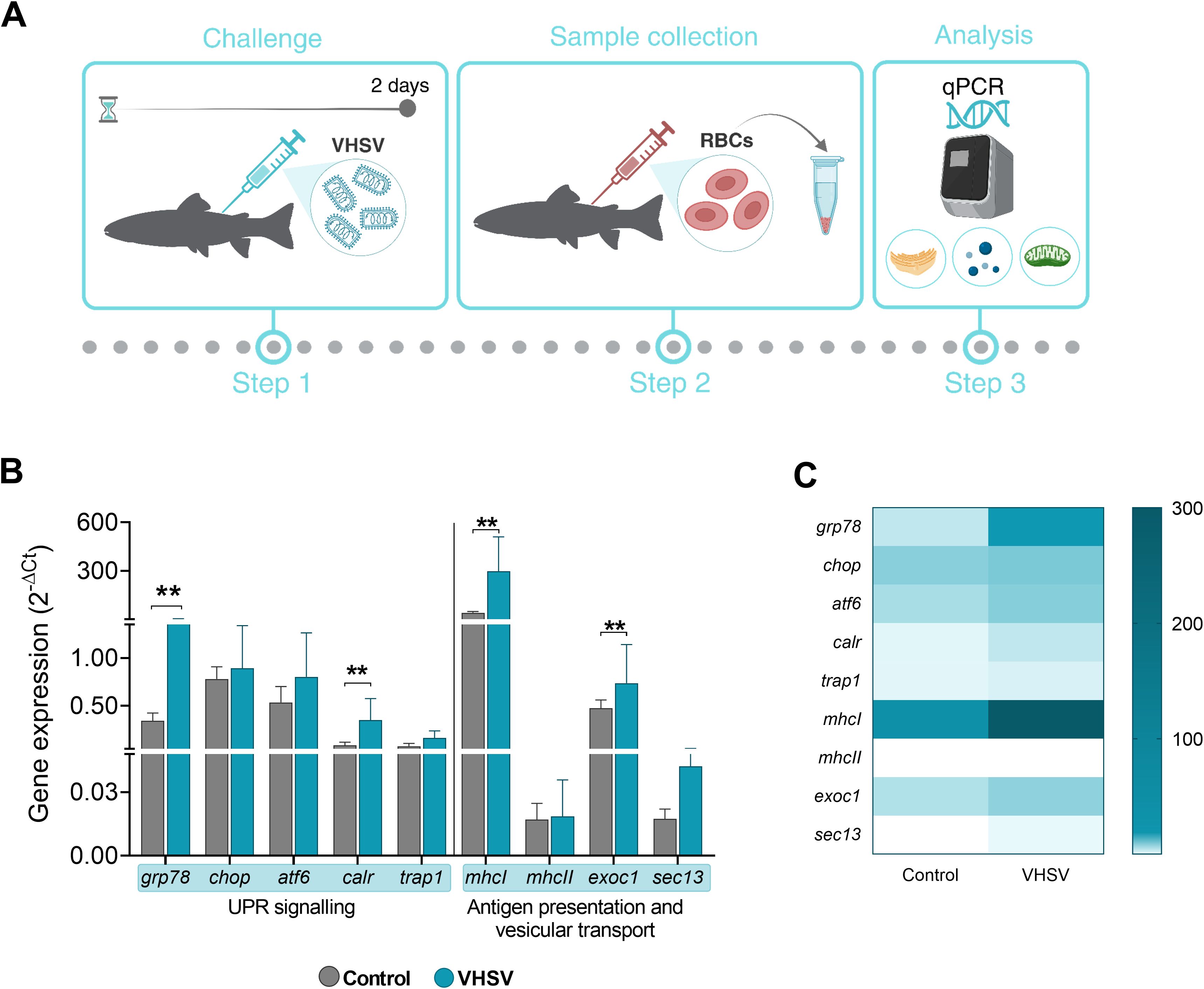
Figure 7. Transcriptomic profile of genes associated with UPR, antigen presentation and vesicular transport in RBCs from rainbow trout challenged with VHSV. (A) Workflow of experimental process. (B) Gene expression relativized to control RBCs (RBCs from non-challenged rainbow trout). (C) Heatmap showing transcription levels of selected genes. Expression levels were analyzed at 2 dpc by qPCR and normalized using ef1α as an endogenous gene. Data show the mean ± standard deviation (n=3). For statistical analysis, the non-parametric Mann-Whitney test was used. **P< 0.01.
4 Discussion
Global aquaculture has faced serious economic problems due to the continuous emergence of infectious diseases precipitated by the high density of farmed fish, the geographical redistribution of aquatic species, and the international expansion of aquaculture (59). Despite efforts to develop therapeutic and prophylactic methods, currently available treatments do not meet certain essential requirements to induce protective immunity (60). Transcriptomic data of RBCs from rainbow trout individuals challenged with VHSV revealed an overrepresentation of pathways related to the ER stress, the UPR, and the signaling pathway between the ER and the nucleus (Supplementary Figure 5) (61). This led us to focus on the potential link between ER stress and immune response in rainbow trout RBCs, with a goal of uncovering novel methods for viral disease prevention through modulation of the UPR.
ER stress is a double-edged sword for the cell and the virus: pathogens promote their own survival and growth by selective modulation of the UPR that affects host immune responses (62–64), but activation of the UPR also exerts a protective role in the innate host defense against the invading pathogen (65, 66). A previous study detailed the link between ER stress and TLR-mediated signaling that allows cells to benefit from the protective functions of the UPR arms while avoiding CHOP-induced apoptosis (67).
In the present study, cryo-SXT revealed enlargement of the ER in RBCs exposed to VHSVUV. When folding capacity of ER is overwhelmed and high levels of misfolded and aggregated proteins accumulate within its lumen, ER stress occurs. This situation is characterized by the expansion or swelling of the ER lumen (68). Outstandingly, some findings have identified an altered 3D ER structure in a human hepatoma cell line during hepatitis C virus (HCV) infection (21, 69). More recently, other research reported that when rainbow trout IgM+ B cells are exposed to a bacterial lipopolysaccharide (LPS), their ER expands. This expansion is important for B cells differentiation process (70). Additionally, multiple studies have investigated the effect of viral infection on the ER (71–73). The stress induced by viral infection alters ER homeostasis, causing malfunction and accumulation of misfolded proteins (48). The UPR mechanism in the ER is activated to maintain equilibrium and promote survival through several mechanisms, including expansion of the ER membrane and the synthesis of key components in protein folding (48, 74), which leads to the enlargement of this cellular organelle observed in our results.
On top of that, tomography images of rainbow trout RBCs exposed to VHSVUV showed autophagosome-like vesicles. Autophagy has been described in relation to ER stress. When proteins cannot be folded correctly, they are degraded by the ER-associated degradation complex (ERAD). If stress persists, the cell activates the autophagy pathway (72, 75, 76). Likewise, autophagosomes can originate in the ER (72), so it is not surprising that these vesicles are observed near the ER in some tomography images. The UPR and autophagy have been reported to play a role in viral pathogenesis, and in recent years there has been a great interest in the regulation of UPR and autophagy as a new therapeutic antiviral strategy (72, 77).
Since VHSVUV causes ER thickening, we analyzed UPR induction as a sign of ER stress in RBCs. The key mediators of UPR, grp78, atf6, and the downstream gene calr, were significantly upregulated at the earliest time after exposure in RBCs treated with VHSVUV. Although gene activation was mild, several recent studies have shown that low levels of ER stress result in the maintenance of an adaptive UPR, which prepares the cell for subsequent stress or attack and is therefore beneficial to the cell (78–80). On the other hand, the expression of grp78, atf6, and calr was much more overexpressed in response to VHSV exposure, MOI 1 and 10, and mainly at the latest time after exposure. ATF6, which stimulates lipid synthesis and increases ER volume (81) was upregulated, a finding that correlated with the results of cryo-SXT. ATF6 is known as an ER stress-regulated transmembrane transcription factor that activates the transcription of ER molecular chaperones. In response to the accumulation of misfolded proteins, ATF6 translocates from the ER to the Golgi where it is processed to its active form. ER stress-induced activation of ATF6 requires a step of dissociation from the regulator GRP78, controlling its transport to the Golgi apparatus (82). Calreticulin is a multifunctional ER luminal protein, which acts as a Ca2+-binding chaperone (83). In addition, calreticulin is a crucial component of the UPR, and participates in protein folding and quality within the ER (84). Also, calreticulin has been connected to ATF6 branch (85). In fact, both genes were found to be simultaneously overexpressed in both VHSVUV- and live VHSV-exposed RBCs. GRP78 is known to function as a pivotal cellular chaperone and its regulation may promote several processes during the viral replicative cycle, demonstrating that it is highly likely that these proteins have virus- and cell-specific mechanisms of action, either antiviral or pro-viral. As a cellular antiviral mechanism, deletion of GRP78 during rotavirus infection was found to reduce infectivity by affecting the maturation of the released virion (86). In addition, some studies have reported that suppression of GRP78 increased hepatitis A virus replication in hepatocytes (87). In the same way, GRP78 can act as an intracellular antiviral factor against hepatitis B virus (88). On the other side of the coin, in the pro-viral scenario, previous reports have shown that GRP78 is positively regulated in dengue virus-infected cells and is essential for viral antigen production (89). Also, knockdown of GRP78 in human cytomegalovirus infection led to intracellular accumulation of viral particles, suggesting that GRP78 is required to accompany virion particle outgrowth (90). Additionally, burgeoning research indicated that, under unfolded protein stress, GRP78 can escape retention in the ER and translocate to the cell surface, where it has binding affinity not only for many endogenous molecules but also for those of exogenous origin, including several viral proteins (91–93). The association of GRP78 with viral proteins has been reported for hepatitis C virus, whose envelope protein E2 has been shown to tightly bind to GRP78 (94), and for coxsackievirus A9, where binding of GRP78 to MHC-I on the cell surface is essential for virus internalization (95) encephalitis virus envelope protein (96). More recently, two independent studies have shown that GRP78 can act as a co-receptor for the SARS-CoV-2 spike protein, facilitating entry into target cells (97, 98). These novel insights point to GRP78 as a multifunctional host factor and a promising line of defense or target against viral infections. Concerning PERK and IRE1 UPR branches, the expression levels of atf4, chop, and edem1 were not affected in either VHSVUV- or live VHSV-exposed RBCs. Accordingly, these two UPR pathways do not seem to play a significant role in the antiviral immune response of RBCs and only the ATF6 pathway seems to be involved in this signaling.
The involvement of ER stress in the antiviral response of RBCs was evaluated at a functional level. VHSV replication levels increased in RBCs pre-treated with 4-PBA, an ER stress inhibitor, establishing a link between ER stress and the immune response triggered by RBCs against the virus. Studies have corroborated that 4-PBA treatment increases viral load in host cells (99), and other studies have shown that an ER stress inducer (e.g., 2-deoxy-d-glucose [2-DG]) inhibits viral infection (99). Similarly, tunicamycin and dithiothreitol, both inducers of ER stress, attenuated Leishmania infantum infection in macrophages, demonstrating a protective role of the host UPR (100). In line with previous reports, our results reveal that disruption in UPR signaling pathways by VHSVUV or VHSV may generate an alarm state that helps the innate immune system to mount an adequate defense by recognizing intracellular infection (101, 102). Viral proteins, such as influenza A virus (IAV) hemagglutinin (HA) glycoproteins, have been shown to induce ER stress, resulting in HA degradation via ERAD and consequent inhibition of IAV replication (103). However, ER stress and UPR pathways have been mainly shown to be modulated in favor of viral replication and propagation. Interestingly, viruses that cause chronic infections have evolved strategies to modulate ER stress signaling, while viruses that cause acute infections use activation of these pathways as a mechanism to induce cell death (104). Rotavirus replication is most efficient under conditions of stress that disrupt host protein synthesis and cellular responses such as the UPR (104). Enterovirus 71 (EV71) induced ER stress to assist viral replication (105). Other work demonstrated that Seneca Valley virus (SVV) infection induced a proviral autophagic process in murine cells, which was regulated by ER stress, via PERK and ATF6 UPR pathways (106). Interestingly, several viral proteins such as SARS coronavirus spike protein (107), dengue 2 virus NS2B-3 protein complex (108), or human cytomegalovirus pUL38 protein (109), were identified as viral components responsible for UPR induction but not directly associated with infection.
ER functions are intrinsically related to autophagy, which is induced downstream of the UPR (110, 111) and both processes are critical for the maintenance of various aspects of cellular homeostasis. Autophagy is a cellular recycling process that can be activated by ER stress to replace unfolded proteins (112, 113). This phenomenon was observed in cryo-SXT, where we observed autophagosomes in RBCs exposed to VHSVUV. Autophagy is a constitutive catabolic degradation pathway that promotes cell survival under stress conditions and has a relevant role in the context of viral infections (114). For example, rainbow trout RBCs exposed to VHSV have increased autophagic activity at both the transcriptional and translational levels (7). Based on this, we investigated the link between ER stress and autophagy. For that, we monitored p62 levels in RBCs treated with an ER stress inhibitor or with an autophagic flux blocker, under VHSV exposure. We observed a slight increment in p62 levels together with an increase in viral load with 4-PBA or niclosamide treatment. These results are not conclusive but suggested that inhibition of ER stress with 4-PBA may affect autophagy, as previously reported (115).
We also observed that in vivo VHSV infection could also stimulate genes related to UPR, antigen presentation, and vesicular transport in RBCs. Our results indicated that these processes may cooperate in the response to the virus, as the expression of grp78 (the main marker of the UPR), atf6, and chop increased simultaneously with the expression of mhcI, mhcII, and calr (antigen presentation pathways) and sec13 and exoc1 (which mediate vesicular transport). The expression of trap1, which is mainly involved in mitochondrial stress, also increased to a lesser extent. Recent reports have found that the UPR plays a role in the regulation of antigen presentation via MHC-I on dendritic cells (116), and forced expression of XBP1 improves vaccine efficacy by stimulating MHC-I/MHC-II pathways (117). Another report described how the association between mitochondria and the ER helps maintain cellular homeostasis under various pathophysiological conditions, with their interaction being mediated by mitochondrial conformational proteins and key chaperones, including calreticulin (118). Furthermore, calreticulin plays a crucial role in ER stress and is involved in the formation and presentation of MHC-I molecules on the cell surface, facilitating antigen recognition (119). This protein may act as a node between the two cellular processes, ER stress response and antigen presentation. Similarly, the secretory pathway has been linked to the UPR and vesicular transport with an active role in the regulation of ER homeostasis (120, 121). Interestingly, we observed protrusions in the cell membrane or EVs in some samples from VHSV-exposed RBCs. RBCs are the major vesicle-secreting cells in the blood, and the mechanism of vesicle biogenesis and production in RBCs has been extensively investigated in mammals (122). EVs have an important role in intercellular communication (123) via the transfer of their contents (e.g., proteins, lipids, and RNA) between cells (124). An example of this is the transfer of micro-RNA from cells infected with the Epstein-Barr virus (EBV) to uninfected cells via exosomes (125), or, in the case of rainbow trout RBCs, the production of cytokines alerting other immune cells in response to the virus (1, 5) or to a DNA vaccine that encodes the VHSV viral antigen (14).
In conclusion, our results suggested that viral-triggered ER stress contributes to raise a protective antiviral mechanism through the ATF6 UPR branch in RBCs (Figure 8). Moreover, the upregulation of key factors of the UPR appeared to be correlated with the induction of autophagy, antigen presentation, and vesicular transport processes. Nevertheless, it is important to stress that more detailed investigations are warranted to substantiate these findings and to clarify whether UPR signaling arises in an independent manner or is mediated by other cellular mechanisms. This study provides the basis for the identification of novel cellular targets for the development of RBC-targeted antiviral strategies.
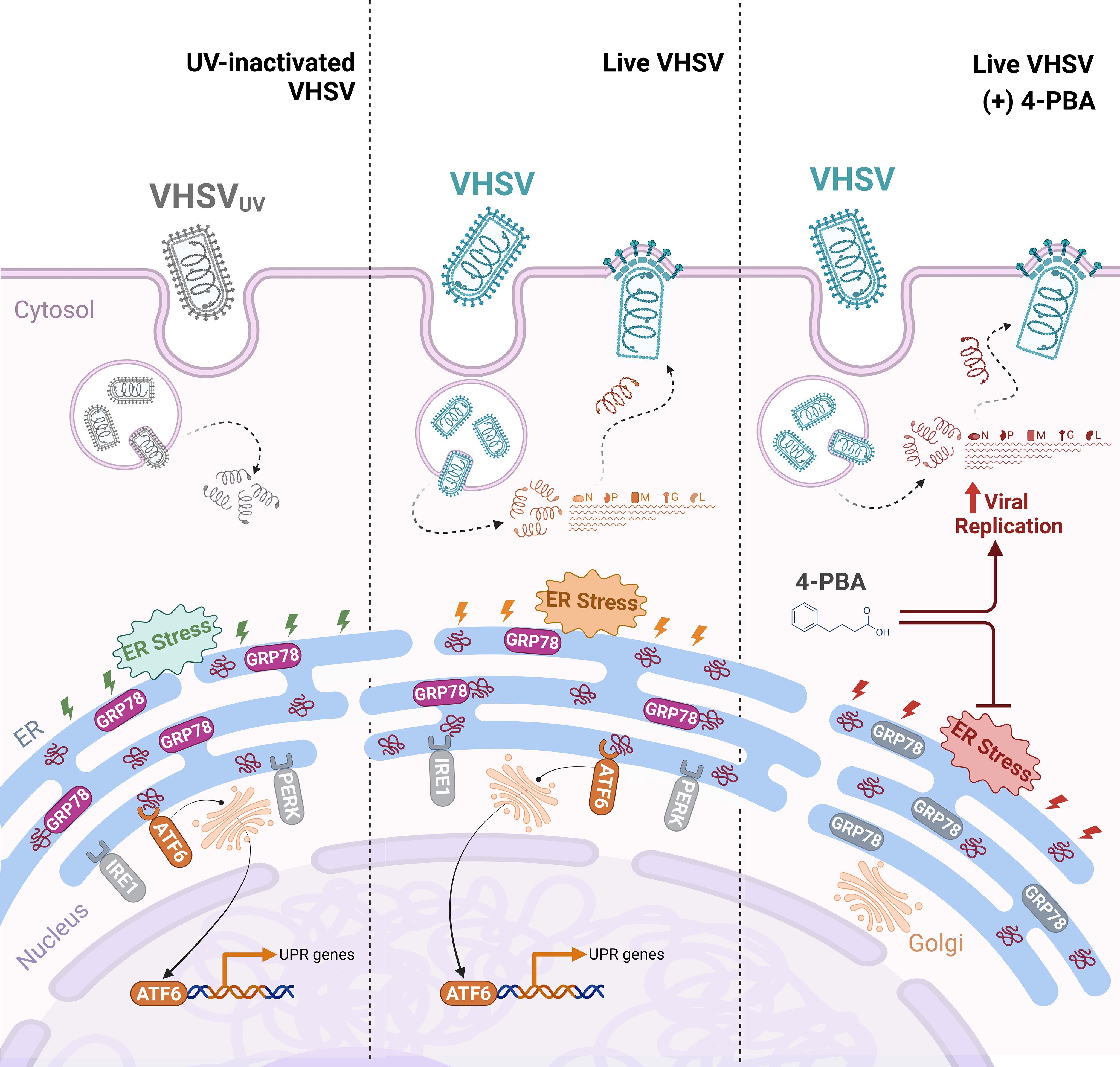
Figure 8. Proposed schematic illustration of the role of UPR regulation in rainbow trout RBCs antiviral response. UV-inactivated VHSV (VHSVUV) or live VHSV enters the RBC cytoplasm via receptor-mediated endocytosis. Then, endosome acidification leads to membrane fusion and release of the nucleoprotein (N) to the cytoplasm, containing all the components necessary for early transcription. Thereafter, ER stress is induced and the UPR is activated. (i) With VHSVUV, GRP78 is released from ER sensors and ATF6 branch is induced while PERK and IRE1 branches remain unaltered. Activation of ATF6 leads to its export to the Golgi apparatus and its translocation to the nucleus to activate the transcription of genes involved in protein folding. (ii) In response to live VHSV, GRP78 is released from ER sensors. ATF6 branch is activated and participates in halting viral replication. (iii) The inhibition of ER stress by 4-PBA is a key trigger of increased VHSV replication in RBCs.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The animal study was approved by The Animal Welfare Body, the Research Ethics Committee at UMH, and the competent authority of the Regional Ministry of Presidency and Agriculture, Fisheries, Food, and Water Supply reviewed and approved all experimental protocols. Animal care and all activities involving animal handling and experiments were done according to Spanish [Real Decreto 1201/2005] regulations. Methodology was performed in accordance with the Spanish [Real Decreto RD 53/2013] and EU [EU Directive 2010/63/EU] regulations and recommendations for animal experimentation and other scientific purposes.
Author contributions
MSM: Conceptualization, Writing – review & editing, Investigation, Methodology, Formal analysis, Validation, Visualization, Writing – original draft. ES: Methodology, Writing – review & editing. MS: Methodology, Writing – review & editing. IN: Methodology, Writing – review & editing. SP: Methodology, Writing – review & editing. VC: Methodology, Writing – review & editing. LP: Writing – review & editing. AP: Writing – review & editing, Methodology. MO: Conceptualization, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by grants from the European Commission (ERC Starting Grant GA639249) to MO-V and the Spanish Ministry of Science (RTI2018-096957-B-C22 MINECO/FEDER) to MO-V and LP. This research includes experiments performed at the MISTRAL beamline at ALBA Synchrotron and in collaboration with ALBA staff (proposal number 2018093095).
Acknowledgments
The authors thank Remedios Torres and Efren Lucas for their excellent technical assistance. Editing was provided by Cecelia Wall (Wall Medical Writing, Wake Forest, NC).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1466870/full#supplementary-material
References
1. Nombela I, Ortega-Villaizan MDM. Nucleated red blood cells: Immune cell mediators of the antiviral response. PloS Pathog. (2018) 14:e1006910. doi: 10.1371/journal.ppat.1006910
2. Morera D, Roher N, Ribas L, Balasch JC, Donate C, Callol A, et al. RNA-Seq reveals an integrated immune response in nucleated erythrocytes. PloS One. (2011) 6:e26998. doi: 10.1371/journal.pone.0026998
3. Passantino L, Altamura M, Cianciotta A, Patruno R, Tafaro A, Jirillo E, et al. Fish immunology. I. Binding and engulfment of Candida albicans by erythrocytes of rainbow trout (Salmo gairdneri Richardson). Immunopharmacol Immunotoxicol. (2002) 24(4):665–78. doi: 10.1081/IPH-120016050
4. Workenhe ST, Kibenge MJ, Wright GM, Wadowska DW, Groman DB, Kibenge FS. Infectious salmon anaemia virus replication and induction of alpha interferon in Atlantic salmon erythrocytes. Virol J. (2008) 5:36. doi: 10.1186/1743-422X-5-36
5. Nombela I, Puente-Marin S, Chico V, Villena AJ, Carracedo B, Ciordia S, et al. Identification of diverse defense mechanisms in rainbow trout red blood cells in response to halted replication of VHS virus. F1000Research. (2018) 6:1958. doi: 10.12688/f1000research
6. Pereiro P, Romero A, Diaz-Rosales P, Estepa A, Figueras A, Novoa B. Nucleated teleost erythrocytes play an nk-lysin- and autophagy-dependent role in antiviral immunity. Front Immunol. (2017) 8:1458. doi: 10.3389/fimmu.2017.01458
7. Nombela I, Requena-Platek R, Morales-Lange B, Chico V, Puente-Marin S, Ciordia S, et al. Rainbow trout red blood cells exposed to viral hemorrhagic septicemia virus up-regulate antigen-processing mechanisms and MHC I&II, CD86, and CD83 antigen-presenting cell markers. Cells. (2019) 8:386. doi: 10.3390/cells8050386
8. Johnstone C, Chaves-Pozo E. Antigen presentation and autophagy in teleost adaptive immunity. Int J Mol Sci. (2022) 23. doi: 10.3390/ijms23094899
9. Garcia-Valtanen P, Ortega-Villaizan Mdel M, Martinez-Lopez A, Medina-Gali R, Perez L, Mackenzie S, et al. Autophagy-inducing peptides from mammalian VSV and fish VHSV rhabdoviral G glycoproteins (G) as models for the development of new therapeutic molecules. Autophagy. (2014) 10:1666–80. doi: 10.4161/auto.29557
10. Jiao X, Y-t Lu, Wang B, Guo Z-y, Qian A-d, Li Y-h. Infection of epithelioma papulosum cyprini (EPC) cells with spring viremia of carp virus (SVCV) induces autophagy and apoptosis through endoplasmic reticulum stress. Microbial Pathogenesis. (2023) 183:106293. doi: 10.1016/j.micpath.2023.106293
11. Liu L, Zhu B, Wu S, Lin L, Liu G, Zhou Y, et al. Spring viraemia of carp virus induces autophagy for necessary viral replication. Cell Microbiol. (2015) 17:595–605. doi: 10.1111/cmi.2015.17.issue-4
12. Bello-Perez M, Pereiro P, Coll J, Novoa B, Perez L, Falco A. Zebrafish C-reactive protein isoforms inhibit SVCV replication by blocking autophagy through interactions with cell membrane cholesterol. Sci Rep. (2020) 10. doi: 10.1038/s41598-020-57501-0
13. Sarder MR, Fischer U, Dijkstra JM, Kiryu I, Yoshiura Y, Azuma T, et al. The MHC class I linkage group is a major determinant in the in vivo rejection of allogeneic erythrocytes in rainbow trout (Oncorhynchus mykiss). Immunogenetics. (2003) 55:315–24. doi: 10.1007/s00251-003-0632-3
14. Puente-Marin S, Nombela I, Chico V, Ciordia S, Mena MC, Coll J, et al. Rainbow Trout Erythrocytes ex vivo Transfection With a DNA Vaccine Encoding VHSV Glycoprotein G Induces an Antiviral Immune Response. Front Immunol. (2018) 9:2477. doi: 10.3389/fimmu.2018.02477
15. Puente-Marin S, Thwaite R, Mercado L, Coll J, Roher N, Ortega-Villaizan MDM. Fish red blood cells modulate immune genes in response to bacterial inclusion bodies made of TNFα and a G-VHSV fragment. Front Immunol. (2019) 10. doi: 10.3389/fimmu.2019.01055
16. Heinrich L, Bennett D, Ackerman D, Park W, Bogovic J, Eckstein N, et al. Whole-cell organelle segmentation in volume electron microscopy. Nature. (2021) 599:141–6. doi: 10.1038/s41586-021-03977-3
17. Xu CS, Hayworth KJ, Lu Z, Grob P, Hassan AM, Garcia-Cerdan JG, et al. Enhanced FIB-SEM systems for large-volume 3D imaging. Elife. (2017) 6. doi: 10.7554/eLife.25916
18. Cossa A, Wien F, Turbant F, Kaczorowski T, Wegrzyn G, Arluison V, et al. Evaluation of the role of bacterial amyloid on nucleoid structure using cryo-soft X-ray tomography. Methods Mol Biol. (2022) 2538:319–33. doi: 10.1007/978-1-0716-2529-3_21
19. McDermott G, Le Gros MA, Knoechel CG, Uchida M, Larabell CA. Soft X-ray tomography and cryogenic light microscopy: the cool combination in cellular imaging. Trends Cell Biol. (2009) 19:587–95. doi: 10.1016/j.tcb.2009.08.005
20. Larabell CA, Le Gros MA. X-ray tomography generates 3-D reconstructions of the yeast, saccharomyces cerevisiae, at 60-nm resolution. Mol Biol Cell. (2004) 15:957–62. doi: 10.1091/mbc.e03-07-0522
21. Perez-Berna AJ, Rodriguez MJ, Chichon FJ, Friesland MF, Sorrentino A, Carrascosa JL, et al. Structural changes in cells imaged by soft X-ray cryo-tomography during hepatitis C virus infection. ACS Nano. (2016) 10:6597–611. doi: 10.1021/acsnano.6b01374
22. Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. (2011) 334:1081–6. doi: 10.1126/science.1209038
23. Zhang K, Kaufman RJ. From endoplasmic-reticulum stress to the inflammatory response. Nature. (2008) 454:455–62. doi: 10.1038/nature07203
24. Hetz C, Zhang K, Kaufman RJ. Mechanisms, regulation and functions of the unfolded protein response. Nat Rev Mol Cell Biol. (2020) 21:421–38. doi: 10.1038/s41580-020-0250-z
25. Almanza A, Carlesso A, Chintha C, Creedican S, Doultsinos D, Leuzzi B, et al. Endoplasmic reticulum stress signalling - from basic mechanisms to clinical applications. FEBS J. (2019) 286:241–78. doi: 10.1111/febs.2019.286.issue-2
26. da Fonseca FG, Serufo ÂV, Leão TL, Lourenço KL. Viral infections and their ability to modulate endoplasmic reticulum stress response pathways. Viruses. (2024) 16:1555. doi: 10.3390/v16101555
27. Macauslane KL, Pegg CL, Short KR, Schulz BL. Modulation of endoplasmic reticulum stress response pathways by respiratory viruses. Crit Rev Microbiol. (2023) 50:750–68. doi: 10.1080/1040841X.2023.2274840
28. Das B, Samal S, Hamdi H, Pal A, Biswas A, Behera J, et al. Role of endoplasmic reticulum stress-related unfolded protein response and its implications in dengue virus infection for biomarker development. Life Sci. (2023) 329:121982. doi: 10.1016/j.lfs.2023.121982
29. Li S, Kong L, Yu X. The expanding roles of endoplasmic reticulum stress in virus replication and pathogenesis. Crit Rev Microbiol. (2015) 41:150–64. doi: 10.3109/1040841X.2013.813899
30. Nombela I, Carrion A, Puente-Marin S, Chico V, Mercado L, Perez L, et al. Infectious pancreatic necrosis virus triggers antiviral immune response in rainbow trout red blood cells, despite not being infective. F1000Res. (2017) 6:1968. doi: 10.12688/f1000research
31. LeBerre M, De Kinkelin P, Metzger A. Identification sérologique des rhabdovirus des salmonidés. Bull Office Int Epizooties. (1977) 87:391–3.
32. Basurco B JMC. Spanish isolates and reference strains of viral haemorrhagic septicaemia virus shown similar protein size patterns. Bull Euro Assoc Fish Pathol. (1989) 9:92–5.
33. Lorenzen E, Carstensen B, Olesen NJ. Inter-laboratory comparison of cell lines for susceptibility to three viruses:VHSV, IHNV and IPNV. Dis Aquat Organisms. (1999) 37:81–8. doi: 10.3354/dao037081
34. Sorrentino A, Nicolas J, Valcarcel R, Chichon FJ, Rosanes M, Avila J, et al. MISTRAL: a transmission soft X-ray microscopy beamline for cryo nano-tomography of biological samples and magnetic domains imaging. J Synchrotron Radiat. (2015) 22:1112–7. doi: 10.1107/S1600577515008632
35. Pereiro E, Nicolas J, Howells MR. A soft X-ray beamline for transmission X-ray microscopy at ALBA. J Synchrotron Radiat. (2009) 16:505–12. doi: 10.1107/S0909049509019396
36. Oton J, Pereiro E, Perez-Berna AJ, Millach L, Sorzano CO, Marabini R, et al. Characterization of transfer function, resolution and depth of field of a soft X-ray microscope applied to tomography enhancement by Wiener deconvolution. BioMed Opt Express. (2016) 7:5092–103. doi: 10.1364/BOE.7.005092
37. Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. (1996) 116:71–6. doi: 10.1006/jsbi.1996.0013
38. Gordon R, Bender R, Herman GT. Algebraic reconstruction techniques (ART) for three-dimensional electron microscopy and x-ray photography. J Theor Biol. (1970) 29:471–81. doi: 10.1016/0022-5193(70)90109-8
39. Schneider G, Guttmann P, Rehbein S, Werner S, Follath R. Cryo X-ray microscope with flat sample geometry for correlative fluorescence and nanoscale tomographic imaging. J Struct Biol. (2012) 177:212–23. doi: 10.1016/j.jsb.2011.12.023
40. Muller WG, Heymann JB, Nagashima K, Guttmann P, Werner S, Rehbein S, et al. Towards an atlas of mammalian cell ultrastructure by cryo soft X-ray tomography. J Struct Biol. (2012) 177:179–92. doi: 10.1016/j.jsb.2011.11.025
41. Aston D, Capel RA, Ford KL, Christian HC, Mirams GR, Rog-Zielinska EA, et al. High resolution structural evidence suggests the Sarcoplasmic Reticulum forms microdomains with Acidic Stores (lysosomes) in the heart. Sci Rep. (2017) 7:40620. doi: 10.1038/srep40620
42. Finstad OW, Dahle MK, Lindholm TH, Nyman IB, Lovoll M, Wallace C, et al. Piscine orthoreovirus (PRV) infects Atlantic salmon erythrocytes. Vet Res. (2014) 45:35. doi: 10.1186/1297-9716-45-35
43. Glomski CA, Tamburlin J, Chainani M. The phylogenetic odyssey of the erythrocyte. III. Fish, the lower vertebrate experience. Histol Histopathol. (1992) 7:501–28.
44. Puente-Marin S, Cazorla D, Chico V, Coll J, Ortega-Villaizan M. Innate immune response of rainbow trout erythrocytes to spinycterins expressing a downsized viral fragment of viral haemorrhagic septicaemia virus. Aquaculture. (2023) 568:739303. doi: 10.1016/j.aquaculture.2023.739303
45. Eskelinen EL. To be or not to be? Examples of incorrect identification of autophagic compartments in conventional transmission electron microscopy of mammalian cells. Autophagy. (2008) 4:257–60. doi: 10.4161/auto.5179
46. Groen J, Conesa JJ, Valcarcel R, Pereiro E. The cellular landscape by cryo soft X-ray tomography. Biophys Rev. (2019) 11:611–9. doi: 10.1007/s12551-019-00567-6
47. Smith EA, McDermott G, Do M, Leung K, Panning B, Le Gros MA, et al. Quantitatively imaging chromosomes by correlated cryo-fluorescence and soft x-ray tomographies. Biophys J. (2014) 107:1988–96. doi: 10.1016/j.bpj.2014.09.011
48. Oslowski CM, Urano F. Measuring ER stress and the unfolded protein response using mammalian tissue culture system. Methods Enzymol. (2011) 490:71–92. doi: 10.1016/B978-0-12-385114-7.00004-0
49. Sicari D, Delaunay-Moisan A, Combettes L, Chevet E, Igbaria A. A guide to assessing endoplasmic reticulum homeostasis and stress in mammalian systems. FEBS J. (2019) 287:27–42. doi: 10.1111/febs.v287.1
50. Park S-M, Kang T-I, So J-S. Roles of XBP1s in transcriptional regulation of target genes. Biomedicines. (2021) 9:791. doi: 10.3390/biomedicines9070791
51. Pao HP, Liao WI, Tang SE, Wu SY, Huang KL, Chu SJ. Suppression of endoplasmic reticulum stress by 4-PBA protects against hyperoxia-induced acute lung injury via up-regulating claudin-4 expression. Front Immunol. (2021) 12:674316. doi: 10.3389/fimmu.2021.674316
52. Wang S, Yang K, Li C, Liu W, Gao T, Yuan F, et al. 4-phenyl-butyric acid inhibits Japanese encephalitis virus replication via inhibiting endoplasmic reticulum stress response. Viruses. (2023) 15. doi: 10.3390/v15020534
53. Sanz F, Basurco B, Babin M, Dominguez J, Coll JM. Monoclonal antibodies against the structural proteins of viral haemorrhagic septicaemia virus isolates. Journal of Fish Diseases. (1993) 16:53–63. doi: 10.1111/j.1365-2761.1993.tb00847.xz
54. Chico V, Gomez N, Estepa A, Perez L. Rapid detection and quantitation of viral hemorrhagic septicemia virus in experimentally challenged rainbow trout by real-time RT-PCR. J Virol Methods. (2006) 132:154–9. doi: 10.1016/j.jviromet.2005.10.005
55. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods. (2001) 25:402–8. doi: 10.1006/meth.2001.1262
56. Li M, Khambu B, Zhang H, Kang JH, Chen X, Chen D, et al. Suppression of lysosome function induces autophagy via a feedback down-regulation of MTOR complex 1 (MTORC1) activity. J Biol Chem. (2013) 288:35769–80. doi: 10.1074/jbc.M113.511212
57. Amoroso MR, Matassa DS, Laudiero G, Egorova AV, Polishchuk RS, Maddalena F, et al. TRAP1 and the proteasome regulatory particle TBP7/Rpt3 interact in the endoplasmic reticulum and control cellular ubiquitination of specific mitochondrial proteins. Cell Death Differ. (2012) 19:592–604. doi: 10.1038/cdd.2011.128
58. Takemoto K, Miyata S, Takamura H, Katayama T, Tohyama M. Mitochondrial TRAP1 regulates the unfolded protein response in the endoplasmic reticulum. Neurochem Int. (2011) 58:880–7. doi: 10.1016/j.neuint.2011.02.015
59. Weidmann M, El-Matbouli M, Zeng W, Bergmann SM. Special issue “Emerging viruses in aquaculture. Viruses. (2021) 13. doi: 10.3390/v13091777
60. Mondal H, Thomas J. A review on the recent advances and application of vaccines against fish pathogens in aquaculture. Aquacult Int. (2022) 30:1971–2000. doi: 10.1007/s10499-022-00884-w
61. Nombela I, Lopez-Lorigados M, Salvador-Mira ME, Puente-Marin S, Chico V, Ciordia S, et al. Integrated transcriptomic and proteomic analysis of red blood cells from rainbow trout challenged with VHSV point towards novel immunomodulant targets. Vaccines (Basel). (2019) 7. doi: 10.3390/vaccines7030063
62. Dubuisson J, Rice CM. Hepatitis C virus glycoprotein folding: disulfide bond formation and association with calnexin. J Virol. (1996) 70:778–86. doi: 10.1128/jvi.70.2.778-786.1996
63. Limjindaporn T, Wongwiwat W, Noisakran S, Srisawat C, Netsawang J, Puttikhunt C, et al. Interaction of dengue virus envelope protein with endoplasmic reticulum-resident chaperones facilitates dengue virus production. Biochem Biophys Res Commun. (2009) 379:196–200. doi: 10.1016/j.bbrc.2008.12.070
64. Huang HL, Wu JL, Chen MH, Hong JR. Aquatic birnavirus-induced ER stress-mediated death signaling contribute to downregulation of Bcl-2 family proteins in salmon embryo cells. PloS One. (2011) 6:e22935. doi: 10.1371/journal.pone.0022935
65. Galluzzi L, Diotallevi A, Magnani M. Endoplasmic reticulum stress and unfolded protein response in infection by intracellular parasites. Future Sci OA. (2017) 3:FSO198. doi: 10.4155/fsoa-2017-0020
66. Smith JA. Regulation of cytokine production by the unfolded protein response; implications for infection and autoimmunity. Front Immunol. (2018) 9:422. doi: 10.3389/fimmu.2018.00422
67. Woo CW, Kutzler L, Kimball SR, Tabas I. Toll-like receptor activation suppresses ER stress factor CHOP and translation inhibition through activation of eIF2B. Nat Cell Biol. (2012) 14:192–200. doi: 10.1038/ncb2408
68. Sriburi R, Bommiasamy H, Buldak GL, Robbins GR, Frank M, Jackowski S, et al. Coordinate regulation of phospholipid biosynthesis and secretory pathway gene expression in XBP-1(S)-induced endoplasmic reticulum biogenesis. J Biol Chem. (2007) 282:7024–34. doi: 10.1074/jbc.M609490200
69. Perez-Berna AJ, Benseny-Cases N, Rodriguez MJ, Valcarcel R, Carrascosa JL, Gastaminza P, et al. Monitoring reversion of hepatitis C virus-induced cellular alterations by direct-acting antivirals using cryo soft X-ray tomography and infrared microscopy. Acta Crystallogr D Struct Biol. (2021) 77:1365–77. doi: 10.1107/S2059798321009955
70. Morel E, Herranz-Jusdado JG, Simón R, Abós B, Perdiguero P, Martín-Martín A, et al. Endoplasmic reticulum expansion throughout the differentiation of teleost B cells to plasmablasts. iScience. (2023) 26:105854. doi: 10.1016/j.isci.2022.105854
71. Grose C, Buckingham EM, Carpenter JE, Kunkel JP. Varicella-zoster virus infectious cycle: ER stress, autophagic flux, and amphisome-mediated trafficking. Pathogens. (2016) 5. doi: 10.3390/pathogens5040067
72. Jheng JR, Ho JY, Horng JT. ER stress, autophagy, and RNA viruses. Front Microbiol. (2014) 5:388. doi: 10.3389/fmicb.2014.00388
73. Choi JA, Song CH. Insights into the role of endoplasmic reticulum stress in infectious diseases. Front Immunol. (2019) 10:3147. doi: 10.3389/fimmu.2019.03147
74. Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol. (2012) 13:89–102. doi: 10.1038/nrm3270
75. B’Chir W, Maurin AC, Carraro V, Averous J, Jousse C, Muranishi Y, et al. The eIF2alpha/ATF4 pathway is essential for stress-induced autophagy gene expression. Nucleic Acids Res. (2013) 41:7683–99. doi: 10.1093/nar/gkt563
76. Senft D, Ronai ZA. UPR. autophagy, and mitochondria crosstalk underlies the ER stress response. Trends Biochem Sci. (2015) 40:141–8. doi: 10.1016/j.tibs.2015.01.002
77. Wu J, Zhang Z, Teng Z, Abdullah SW, Sun S, Guo H. Sec62 regulates endoplasmic reticulum stress and autophagy balance to affect foot-and-mouth disease virus replication. Front Cell Infect Microbiol. (2021) 11:707107. doi: 10.3389/fcimb.2021.707107
78. Mollereau B, Manie S, Napoletano F. Getting the better of ER stress. J Cell Commun Signal. (2014) 8:311–21. doi: 10.1007/s12079-014-0251-9
79. Mendes CS, Levet C, Chatelain G, Dourlen P, Fouillet A, Dichtel-Danjoy ML, et al. ER stress protects from retinal degeneration. EMBO J. (2009) 28:1296–307. doi: 10.1038/emboj.2009.76
80. Taylor RC, Dillin A. XBP-1 is a cell-nonautonomous regulator of stress resistance and longevity. Cell. (2013) 153:1435–47. doi: 10.1016/j.cell.2013.05.042
81. Maiuolo J, Bulotta S, Verderio C, Benfante R, Borgese N. Selective activation of the transcription factor ATF6 mediates endoplasmic reticulum proliferation triggered by a membrane protein. Proc Natl Acad Sci U S A. (2011) 108:7832–7. doi: 10.1073/pnas.1101379108
82. Shen J, Chen X, Hendershot L, Prywes R. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev Cell. (2002) 3:99–111. doi: 10.1016/S1534-5807(02)00203-4
83. Michalak M, Robert Parker JM, Opas M. Ca2+ signaling and calcium binding chaperones of the endoplasmic reticulum. Cell Calcium. (2002) 32:269–78. doi: 10.1016/S0143416002001884
84. Pabst T, Mueller BU, Eyholzer M, Schardt J. The unfolded protein response (UPR) is activated in human acute myeloid leukemia (AML) and suppresses translation of the CCAAT/enhancer binding protein-alpha (CEBPA) by induction of the RNA-binding protein calreticulin. Blood. (2008) 112:2934–. doi: 10.1182/blood.V112.11.2934.2934
85. Hong M, Luo S, Baumeister P, Huang J-M, Gogia RK, Li M, et al. Underglycosylation of ATF6 as a novel sensing mechanism for activation of the unfolded protein response. J Biol Chem. (2004) 279:11354–63. doi: 10.1074/jbc.M309804200
86. Maruri-Avidal L, Lopez S, Arias CF. Endoplasmic reticulum chaperones are involved in the morphogenesis of rotavirus infectious particles. J Virol. (2008) 82:5368–80. doi: 10.1128/JVI.02751-07
87. Jiang X, Kanda T, Haga Y, Sasaki R, Nakamura M, Wu S, et al. Glucose-regulated protein 78 is an antiviral against hepatitis A virus replication. Exp Ther Med. (2017) 13:3305–8. doi: 10.3892/etm.2017.4407
88. Ma Y, Yu J, Chan HL, Chen YC, Wang H, Chen Y, et al. Glucose-regulated protein 78 is an intracellular antiviral factor against hepatitis B virus. Mol Cell Proteomics. (2009) 8:2582–94. doi: 10.1074/mcp.M900180-MCP200
89. Wati S, Soo ML, Zilm P, Li P, Paton AW, Burrell CJ, et al. Dengue virus infection induces upregulation of GRP78, which acts to chaperone viral antigen production. J Virol. (2009) 83:12871–80. doi: 10.1128/JVI.01419-09
90. Buchkovich NJ, Maguire TG, Yu Y, Paton AW, Paton JC, Alwine JC. Human cytomegalovirus specifically controls the levels of the endoplasmic reticulum chaperone BiP/GRP78, which is required for virion assembly. J Virol. (2008) 82:31–9. doi: 10.1128/JVI.01881-07
91. Gonzalez-Gronow M, Gopal U, Austin RC, Pizzo SV. Glucose-regulated protein (GRP78) is an important cell surface receptor for viral invasion, cancers, and neurological disorders. IUBMB Life. (2021) 73:843–54. doi: 10.1002/iub.2502
92. Van Krieken R, Tsai YL, Carlos AJ, Ha DP, Lee AS. ER residential chaperone GRP78 unconventionally relocalizes to the cell surface via endosomal transport. Cell Mol Life Sci. (2021) 78:5179–95. doi: 10.1007/s00018-021-03849-z
93. Honda T, Horie M, Daito T, Ikuta K, Tomonaga K. Molecular chaperone BiP interacts with Borna disease virus glycoprotein at the cell surface. J Virol. (2009) 83:12622–5. doi: 10.1128/JVI.01201-09
94. Liberman E, Fong YL, Selby MJ, Choo QL, Cousens L, Houghton M, et al. Activation of the grp78 and grp94 promoters by hepatitis C virus E2 envelope protein. J Virol. (1999) 73:3718–22. doi: 10.1128/JVI.73.5.3718-3722.1999
95. Triantafilou K, Fradelizi D, Wilson K, Triantafilou M. GRP78, a coreceptor for coxsackievirus A9, interacts with major histocompatibility complex class I molecules which mediate virus internalization. J Virol. (2002) 76:633–43. doi: 10.1128/JVI.76.2.633-643.2002
96. Nain M, Mukherjee S, Karmakar SP, Paton AW, Paton JC, Abdin MZ, et al. GRP78 is an important host factor for Japanese encephalitis virus entry and replication in mammalian cells. J Virol. (2017) 91. doi: 10.1128/JVI.02274-16
97. Carlos AJ, Ha DP, Yeh DW, Van Krieken R, Tseng CC, Zhang P, et al. The chaperone GRP78 is a host auxiliary factor for SARS-CoV-2 and GRP78 depleting antibody blocks viral entry and infection. J Biol Chem. (2021) 296:100759. doi: 10.1016/j.jbc.2021.100759
98. Ibrahim IM, Abdelmalek DH, Elshahat ME, Elfiky AA. COVID-19 spike-host cell receptor GRP78 binding site prediction. J Infect. (2020) 80:554–62. doi: 10.1016/j.jinf.2020.02.026
99. Wang Y, Li JR, Sun MX, Ni B, Huan C, Huang L, et al. Triggering unfolded protein response by 2-Deoxy-D-glucose inhibits porcine epidemic diarrhea virus propagation. Antiviral Res. (2014) 106:33–41. doi: 10.1016/j.antiviral.2014.03.007
100. Galluzzi L, Diotallevi A, De Santi M, Ceccarelli M, Vitale F, Brandi G, et al. Leishmania infantum induces mild unfolded protein response in infected macrophages. PloS One. (2016) 11:e0168339. doi: 10.1371/journal.pone.0168339
101. Carletti T, Zakaria MK, Faoro V, Reale L, Kazungu Y, Licastro D, et al. Viral priming of cell intrinsic innate antiviral signaling by the unfolded protein response. Nat Commun. (2019) 10. doi: 10.1038/s41467-019-11663-2
102. Catanzaro N, Meng X-J. Induction of the unfolded protein response (UPR) suppresses porcine reproductive and respiratory syndrome virus (PRRSV) replication. Virus Res. (2020) 276:197820. doi: 10.1016/j.virusres.2019.197820
103. Frabutt DA, Wang B, Riaz S, Schwartz RC, Zheng Y-H, Schultz-Cherry S. Innate sensing of influenza A virus hemagglutinin glycoproteins by the host endoplasmic reticulum (ER) stress pathway triggers a potent antiviral response via ER-associated protein degradation. J Virol. (2018) 92. doi: 10.1128/JVI.01690-17
104. Trujillo-Alonso V, Maruri-Avidal L, Arias CF, López S. Rotavirus infection induces the unfolded protein response of the cell and controls it through the nonstructural protein NSP3. J Virology. (2011) 85:12594–604. doi: 10.1128/JVI.05620-11
105. Jheng JR, Lau KS, Tang WF, Wu MS, Horng JT. Endoplasmic reticulum stress is induced and modulated by enterovirus 71. Cell Microbiol. (2010) 12:796–813. doi: 10.1111/j.1462-5822.2010.01434.x
106. Hou L, Dong J, Zhu S, Yuan F, Wei L, Wang J, et al. Seneca valley virus activates autophagy through the PERK and ATF6 UPR pathways. Virology. (2019) 537:254–63. doi: 10.1016/j.virol.2019.08.029
107. Chan C-P, Siu K-L, Chin K-T, Yuen K-Y, Zheng B, Jin D-Y. Modulation of the unfolded protein response by the severe acute respiratory syndrome coronavirus spike protein. J Virology. (2006) 80:9279–87. doi: 10.1128/JVI.00659-06
108. Yu C-Y, Hsu Y-W, Liao C-L, Lin Y-L. Flavivirus infection activates the XBP1 pathway of the unfolded protein response to cope with endoplasmic reticulum stress. J Virology. (2006) 80:11868–80. doi: 10.1128/JVI.00879-06
109. Xuan B, Qian Z, Torigoi E, Yu D. Human cytomegalovirus protein pUL38 induces ATF4 expression, inhibits persistent JNK phosphorylation, and suppresses endoplasmic reticulum stress-induced cell death. J Virology. (2009) 83:3463–74. doi: 10.1128/JVI.02307-08
110. Celli J, Tsolis RM. Bacteria, the endoplasmic reticulum and the unfolded protein response: friends or foes? Nat Rev Microbiol. (2015) 13:71–82. doi: 10.1038/nrmicro3393
111. Rashid HO, Yadav RK, Kim HR, Chae HJ. ER stress: Autophagy induction, inhibition and selection. Autophagy. (2015) 11:1956–77. doi: 10.1080/15548627.2015.1091141
112. Bhardwaj M, Leli NM, Koumenis C, Amaravadi RK. Regulation of autophagy by canonical and non-canonical ER stress responses. Semin Cancer Biol. (2020) 66:116–28. doi: 10.1016/j.semcancer.2019.11.007
113. Metcalf MG, Higuchi-Sanabria R, Garcia G, Tsui CK, Dillin A. Beyond the cell factory: Homeostatic regulation of and by the UPR(ER). Sci Adv. (2020) 6:eabb9614. doi: 10.1126/sciadv.abb9614
114. Deretic V. Autophagy in innate and adaptive immunity. Trends Immunol. (2005) 26:523–8. doi: 10.1016/j.it.2005.08.003
115. Wang Z, Zheng S, Gu Y, Zhou L, Lin B, Liu W. 4-PBA Enhances Autophagy by Inhibiting Endoplasmic Reticulum Stress in Recombinant Human Beta Nerve Growth Factor-Induced PC12 cells After Mechanical Injury via PI3K/AKT/mTOR Signaling Pathway. World Neurosurg. (2020) 138:e659–e64. doi: 10.1016/j.wneu.2020.03.038
116. Osorio F, Lambrecht BN, Janssens S. Antigen presentation unfolded: identifying convergence points between the UPR and antigen presentation pathways. Curr Opin Immunol. (2018) 52:100–7. doi: 10.1016/j.coi.2018.04.020
117. Zhang Y, Chen G, Liu Z, Tian S, Zhang J, Carey CD, et al. Genetic vaccines to potentiate the effective CD103+ dendritic cell-mediated cross-priming of antitumor immunity. J Immunol. (2015) 194:5937–47. doi: 10.4049/jimmunol.1500089
118. Malhotra JD, Kaufman RJ. ER stress and its functional link to mitochondria: role in cell survival and death. Cold Spring Harb Perspect Biol. (2011) 3:a004424. doi: 10.1101/cshperspect.a004424
119. Gao B, Adhikari R, Howarth M, Nakamura K, Gold MC, Hill AB, et al. Assembly and antigen-presenting function of MHC class I molecules in cells lacking the ER chaperone calreticulin. Immunity. (2002) 16:99–109. doi: 10.1016/S1074-7613(01)00260-6
120. Tsvetanova NG. The secretory pathway in control of endoplasmic reticulum homeostasis. Small GTPases. (2013) 4:28–33. doi: 10.4161/sgtp.22599
121. Ye J, Liu X. Interactions between endoplasmic reticulum stress and extracellular vesicles in multiple diseases. Front Immunol. (2022) 13:955419. doi: 10.3389/fimmu.2022.955419
122. Yang L, Huang S, Zhang Z, Liu Z, Zhang L. Roles and applications of red blood cell-derived extracellular vesicles in health and diseases. Int J Mol Sci. (2022) 23:5927. doi: 10.3390/ijms23115927
123. Bang C, Thum T. Exosomes: new players in cell-cell communication. Int J Biochem Cell Biol. (2012) 44:2060–4. doi: 10.1016/j.biocel.2012.08.007
124. Wei H, Chen Q, Lin L, Sha C, Li T, Liu Y, et al. Regulation of exosome production and cargo sorting. Int J Biol Sci. (2021) 17:163–77. doi: 10.7150/ijbs.53671
125. Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, van Eijndhoven MA, Hopmans ES, Lindenberg JL, et al. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci U S A. (2010) 107:6328–33. doi: 10.1073/pnas.0914843107
126. Chaves-Pozo E, Montero J, Cuesta A, Tafalla C. Viral hemorrhagic septicemia and infectious pancreatic necrosis viruses replicate differently in rainbow trout gonad and induce different chemokine transcription profiles. Dev Comp Immunol. (2010) 34:648–58. doi: 10.1016/j.dci.2010.01.009
127. Jorgensen TR, Raida MK, Kania PW, Buchmann K. Response of rainbow trout (Oncorhynchus mykiss) in skin and fin tissue during infection with a variant of Gyrodactylus salaris (Monogenea: Gyrodactylidae). Folia Parasitol (Praha). (2009) 56:251–8. doi: 10.14411/fp.2009.029
Keywords: red blood cells, erythrocytes, cryo-soft X-ray tomography, rainbow trout, fish, virus, VHSV, endoplasmic reticulum stress
Citation: Salvador-Mira M, Sanchez-Cordoba E, Solivella M, Nombela I, Puente-Marin S, Chico V, Perez L, Perez-Berna AJ and Ortega-Villaizan MdM (2024) Endoplasmic reticulum stress triggers unfolded protein response as an antiviral strategy of teleost erythrocytes. Front. Immunol. 15:1466870. doi: 10.3389/fimmu.2024.1466870
Received: 18 July 2024; Accepted: 23 October 2024;
Published: 26 November 2024.
Edited by:
Stephanie DeWitte-Orr, Wilfrid Laurier University, CanadaReviewed by:
Alejandro Romero, Spanish National Research Council (CSIC), SpainVikash Kumar, Central Inland Fisheries Research Institute (ICAR), India
Copyright © 2024 Salvador-Mira, Sanchez-Cordoba, Solivella, Nombela, Puente-Marin, Chico, Perez, Perez-Berna and Ortega-Villaizan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maria del Mar Ortega-Villaizan, bW9ydGVnYS12aWxsYWl6YW5AdW1oLmVz
 Maria Salvador-Mira
Maria Salvador-Mira Ester Sanchez-Cordoba1
Ester Sanchez-Cordoba1 Ivan Nombela
Ivan Nombela Sara Puente-Marin
Sara Puente-Marin Veronica Chico
Veronica Chico Luis Perez
Luis Perez Ana Joaquina Perez-Berna
Ana Joaquina Perez-Berna Maria del Mar Ortega-Villaizan
Maria del Mar Ortega-Villaizan