
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 27 September 2024
Sec. Cancer Immunity and Immunotherapy
Volume 15 - 2024 | https://doi.org/10.3389/fimmu.2024.1457612
This article is part of the Research Topic Emerging Paradigms in Understanding Cancer Metastasis: Focus on Tumor Microenvironment Dynamics View all 3 articles
Esophageal cancer (EC) is among the most aggressive malignancies, ranking as the seventh most prevalent malignant tumor worldwide. Lymph node metastasis (LNM) indicates localized spread of cancer and often correlates with a poorer prognosis, emphasizing the necessity for neoadjuvant systemic therapy before surgery. However, accurate identification of LNM in EC presents challenges due to the lack of satisfactory diagnostic techniques. Imaging techniques, including ultrasound and computerized tomography scans, have low sensitivity and accuracy in assessing LNM. Additionally, the existing serological detection lacks precise biomarkers. The intricate and not fully understood molecular processes involved in LNM of EC contribute to current detective limitations. Recent research has shown potential in using various molecules, circulating tumor cells (CTCs), and changes in the microbiota to identify LNM in individuals with EC. Through summarizing potential biomarkers associated with LNM in EC and organizing the underlying mechanisms involved, this review aims to provide insights that facilitate biomarker development, enhance our understanding of the underlying mechanisms, and ultimately address the diagnostic challenges of LNM in clinical practice.
Esophageal cancer (EC) is the seventh most prevalent cancer and the sixth highest contributor to cancer-related mortality globally (1). Esophageal squamous cell carcinoma (ESCC) accounts for approximately 90% of global EC cases, with high incidence in East Africa, sub-Saharan Africa, and much of Central Asia. In contrast, esophageal adenocarcinoma (EAC) is more prevalent in Europe and high-income North America, where its incidence has quadrupled over the past four decades (2, 3). As immune organs, lymph nodes (LNs) defend against pathogen dissemination and regulate immune responses and homeostasis. Nevertheless, LNs and their lymphatic circulation can serve as conduits for disease progression, contributing to the spread of inflammation and tumors (4). Lymph node metastasis (LNM) is the primary pathway for the spread of most solid tumors. In EAC, LNM increases from less than 5% for intramucosal (pT1a) lesions to 26% for submucosa (pT1b) lesions. For pT1a ESCC, the risk is approximately 4%, while for pT1b, it rises to around 30%, which is higher than in EAC (3).
The esophagus anatomically includes cervical, thoracic, and abdominal segments, with a wide distribution of regional LN drainage and the heterogeneity of LNM patterns in each anatomical region (5). Although tumor cells preferentially metastasize to the corresponding lymphatic drainage area, skip metastasis increases the complexity of LNM in EC (6, 7). The occurrence of LNM in EC is associated with shorter overall survival (OS). A study reported that the 5-year OS rate was over 50% for patients with fewer than one positive LN, and below 30% for those with more than two positive LNs (8). To obtain better clinical outcomes, EC patients with LNM receive lymphatic node dissection and neoadjuvant chemoradiotherapy (NCRT). However, both excessive and insufficient lymph node dissection can harm EC patients because of operative complications, shorter survival, and lower life quality (9, 10). Currently, classification standards of LNM worldwide, clinical practice, and management vary and rely on subjective and personal experience (11, 12). It is essential to accurately determine the presence or absence of LNM and evaluate the extent of metastasis. These assessments facilitate the selection of surgical methods and help evaluate postoperative patient quality and survival.
The imaging modalities include ultrasound, computed tomography (CT), magnetic resonance imaging (MRI), positron emission tomography (PET), and PET/CT, which diagnose LNM based on the size, morphology, and imaging characteristics of the nodes. However, the accuracy of diagnosing cancers and many other diseases relies on the expertise of individual radiologists and pathologists, leading to various comprehension and interpretation of medical images (13). Therefore, current imaging and pathological practices prioritize diagnosing LNM rather than assessing its occurrence, progression, and prognostic value. Biomarkers are measurable and quantifiable characteristics that reflect the physiological and pathological states of the body. Tumor-associated biomarkers are among the essential fields in this area, with extensive research and applications in early diagnosis, progression and prognosis assessment, tumor classification, prediction of treatment responses, and monitoring for recurrence. Traditional tumor markers primarily include proteins secreted by tumor cells and carbohydrate antigens (14, 15). Recent advancements in diagnostic technologies have introduced potential tumor markers, such as genetic mutations, abnormal RNA, epigenetic modifications, circulating tumor cells (CTCs), and microbiome characteristics. These biomarkers help optimize decision-making in clinical practice, particularly in precision oncology, where identifying patients with specific cancer genetic mutations is required for targeted therapy (16, 17).
Consecutive stages of EC progression, from the precancerous, tumor-occurring to premetastatic niche (PMN) stages, are characterized by elements such as hypoxia, acidosis, and changes in the extracellular matrix (ECM), cytokines, inflammatory factors, and tumor-promoting cells that assist in immune avoidance and tolerance. Characteristic products in each stage can be promising biomarkers in LNM of EC. For example, EC cells could secrete IGF2 and elevate VEGF produced by tumor-associated fibroblasts via miR-29c in a p53-dependent manner, activating VEGFR1-positive bone marrow vascular progenitor cells in the tumor microenvironment (TME) and forming PMN (18). After irradiation, the upregulation of MiR-26b-5p in small extracellular vesicles (EVs) derived from dying ESCC cells promotes the proliferation and activation of myeloid-derived suppressor cells and macrophages. These cells suppress the PI3K/AKT pathway by targeting PTEN and eventually facilitate PMN formation (19). Therefore, the primary tumor-derived cells support PMN formation in LNs to aid the spread and colonization of cancer cells. Accordingly, several molecules are promising biomarkers for diagnosing, evaluating, and predicting LNM in EC and other cancer types. The molecular mechanisms underlying LNM are diverse, especially the formation of PMN, which promotes LNM in the esophagus. However, such a microenvironment is complicated, and the specific mechanisms and interactions between molecules and their crosstalk with EC remain insufficient understandings (20). Researchers have focused on the microbiota (21) in the LNM microenvironment, which are essential promoters or regulators of tumor progression.
In this review, we have summarized potential biomarkers that may serve as indicators of LNM in EC and outlined their involvement in the mechanisms, including molecular signaling pathways. This information is expected to guide future biomarker research, deepen researchers’ understanding of the mechanisms underlying LNM in EC, and ultimately offer new tools for clinical practice in the diagnosis and treatment of EC.
NNMT is an important enzyme that controls NAD+ and SAM levels that regulates cellular metabolism, impacting tumor malignancy (22–25). NNMT overexpression has been observed in various cancers (26). Upregulation of NNMT maintains the cancer-associated fibroblast phenotype, activates the PRDM5/COL1A1 axis, and promotes tumor metastasis in gastric cancer (27, 28) and breast cancer (BC) (29). Its inhibitor has been shown to reverse drug resistance in lung cancer cells (30). Therefore, NNMT plays a crucial indicator in cancer progression and offers potential as an effective target for anticancer treatment. NNMT overexpression plays a role in the diagnosis and prognosis of patients with positive LNs through the m6A modification and the EMT pathway (31). SMYD3 expression and the coexpression of histone methyltransferase G9a and MCM7 predict LNM and other adverse outcomes in ESCC (32, 33). Meanwhile, differential methylation of HOXB2 and SEPT9 serves as predictive factors for LNM (34), yet the hypermethylated promoter of FOXF2 only indicates OS in ESCC (35).
MACC1, prevalent in various solid tumors, has emerged as a predictor in tumor stages. Its overexpression is linked to diminished OS in tumors and decreased metastasis-free survival in colorectal cancer (CRC) (36, 37). In CRC and BC, MACC1 overexpression regulates β-catenin, HGF/c-Met, and other signaling pathways to elevate tumor malignancy (37, 38). Its overexpression is closely related to LNM in nasopharyngeal and gastric cancer (39, 40). This understanding guides ongoing research into MACC1-based therapeutic strategies to inhibit tumor growth and metastasis. MACC1 overexpression in EAC has been identified as a predictor of metastasis and other poor survival via the MEK1/ERK signaling pathway both in vitro and in vivo (41). In ESCC, coexpression of MACC1 with Snail and AGR2, along with diminished KAI1, is associated with LNM in ESCC and head and neck squamous cell carcinoma (HNSCC) (42, 43). MACC1 overexpression promotes the LNM via the c-Met/cyclin D1, the PTEN/PI3K/Akt, and the AMPK/ULK1 signaling pathways (44, 45). Moreover, MACC1-AS1, induced by NSD2, mediates cisplatin resistance in ESCC and represents a promising target for enhancing cisplatin-based chemotherapy (46). Therefore, MACC1, with multiple regulatory functions, is identified as a potent indicator and therapeutic target in LNM of EC.
MMPs are enzymes responsible for degrading the collagen and other proteins in the ECM (47), and TWIST1 is an essential EMT-inducing transcription factor (48). The upregulation of crosstalk between MMPs and TWIST1 with other markers, such as HLAG-1 (49) and CD44 (50), indicates the occurrence and development of tumor metastasis. A significant inverse correlation between INPP5A and the overexpression of TWIST1, EGFR, and MMP-2 has been observed, which might predict a poor prognosis in ESCC (51). Meanwhile, upregulation of CD147 and MMP-9 could potentially predict LNM and the prognosis in EAC (52).
TWIST1 also affects lymphatic status by upregulating Notch signaling genes (53). Meanwhile, MAML1 is a transcriptional co-activator in the Notch signaling pathway to promote tumor progression and metastasis. Studies have demonstrated that coexpression of TWIST1 and MAML1 in this pathway promotes a cascade in the aggressiveness and metastasis of HNSCC and ESCC (54, 55). However, in another study, no significant changes in MAML1 expression has been found when TWIST1 overexpresses (53). Further studies are required to explore the molecular mechanisms of TWIST1 and MAML1 and other Notch signaling genes.
ADAMs enhance structural resemblance by exhibiting MMP activity. ADAM17 overexpression is linked to LNM and a poorer prognosis in ESCC, contributing to the formation of lymphatic tubules (56). Cancer-associated fibroblasts stimulate ADAM17 through activation of the ERK1/2 pathway to promote the progression of ESCC (57). Its antibody shows promise as a therapeutic target by inhibiting both EGFR-mediated and non-EGFR-mediated pathways to suppress tumor growth (58). ADAM10 overexpression has been demonstrated as a predictor of LNM in oral SCC (59). CLCA4 involves MMP activity and intracellular calcium-gated chloride channels. It modulates the upregulation of miR-501-5p, which acts as a suppressor in LNM of HNSCC (60, 61).
Essential biomarkers proposed for predicting LNM in EC include the overexpression of cortactin, MLL2, EIF4E, mTOR, and STC1 in ESCC, transcription of CD69, MyD88, and TCR4, and low expression of OLFM4 in EAC (62–64). A study has found that TSTA3 overexpression is associated with advanced LNM and was identified as a predictor in LNM of ESCC (65). Meanwhile, positive WTAP expression is correlated with advanced TNM stages in epithelial-mesenchymal transition (EMT) (66). In stage IIA ESCC after esophagectomy, FPXM1 overexpression predicts a higher 5-year recurrence of LNM (67). The elevated expression of MUC1 is a promising marker to predict advanced LNM and 5-year OS in ESCC after radical resection (68). In addition, the combined analysis of Annexin II, kindlin-2, and myosin-9 is more robust in predicting LNM in EC (69) (Table 1).
Underlying markers (Table 1), for example, Vav1 (70), TACC3 (71), and HMGA2 (72), are positively associated with LNM of EC. Overexpressions of PDIA3 and ITGA5B1 are associated with advanced LNM and pTNM stages in a model for clinic risk stratification (73). KRT15 is overexpressed to promote malignant phenotypes in Barrett’s esophagus and EAC (74, 75). Caprin-1 overexpression indicates a poor prognosis in EC with LNM by regulating the Warburg effect and glycolysis (76–78). Overexpression of STMN1 (79) and NOLC1 (80) in ESCC indicates advanced LNM and poor prognosis by activating the PI3K/AKT pathway. PARK2 also promotes phosphorylation via the Hippo/YAP signaling pathway (81). Upregulation of Rad51 is involved in DNA repair and tumor metastasis (82). Overexpression of ATAD2 (83) and CDCA7 (84) activates TGF-β/Smad signaling to regulate LNM in ESCC, yet conflicting results are observed in RUNX3 (85). In another study, this tumor suppressor overexpressed but showed no significant association with the LNM of EC (86). Through the Wnt/β-catenin signaling pathway, RARα overexpression upregulates MMPs to promote ESCC metastasis (87). Besides, upregulation of FOXC2 and downregulation of ZNF750 are identified as metastatic and prognostic biomarkers, providing therapeutic targets for ESCC. ZNF750 dysfunction promotes tumorigenesis and metastatic ability of ESCC through the DANCR/miR-4707-3p/FOXC2 axis in a ceRNA manner (88). In ESCC, the TNFα/FOXC2/FA2H axis promotes lung metastasis by dysregulating ceramide metabolism via the NF-κB signaling pathways (89). Additionally, the coexpression of AGGF1 and FOXC2 functions in angiogenesis to promote the LNM in ESCC (90). The coexpression of PKM2 and HSP27 promoted the LNM of EC through ubiquitination (91). Additionally, PKM2 elevates STAT3 in TGF-β1-induced EMT to promote LNM (92). Some markers, including SLC38A3 (93), RCAN1.2 mRNA, and P21, are inversely associated with LNM and shorter survival (94, 95). The PTP family members suppress tumor migration and metastasis via RTK signaling pathways, and their silenced epigenetic modulation indicates malignancies (96). PTPRS and PTPRO deficiencies in ESCC are associated with aggressive LNM and other malignant phenotypes (97). PTPROlow/p-METhigh suppresses LNM of ESCC by dephosphorylating p-MET (98). B3GNT3 is suppressed and inversely associated with the LNM of ESCC through the synergistic effect on regulating M2 macrophages (99).
Long noncoding RNAs (lncRNAs) are potential predictive, diagnostic, and prognostic biomarkers in LNM of EC (Table 2). LINP1 and LOC440173 are potential indicators and activators of LNM in ESCC (100, 101). Two novel and tumor-specific lncRNAs, ENST00000508406.1 and NR_037652.1, facilitate the prediction of ESCC local invasion and LNM (102). Elevated SNHG6 in ESCC serves as an independent diagnostic biomarker for LNM, distant metastasis, and TNM stages (103). Furthermore, its silencing upregulates miR-186-5p and inhibits tumor migration by targeting HIF1α (104). Lnc-ABCA12-3 overexpression indicates an advanced TNM stage and an unfavorable prognosis, as it competitively binds to miR-200b-3p to upregulate FN1, which is involved in cell adhesion and migration. Moreover, exosome-mediated processes exacerbate metastatic progression in glycolysis through the TLR4/NF-κB signaling pathway (105, 106). LincIN is identified as a potential signature in BC metastasis (107). LincIN overexpression enhances the binding between NF90 and pri-miR-7 and downregulated mature miR-7, a tumor suppressor in various cancers, to elevate and indicate the LNM of ESCC (108, 109). SPRY4-IT1 is considered a diagnostic and predictive biomarker for ESCC surgical procedures and prognosis (110). As a sponge in many miRNAs (111), SPRY4-IT1 improves cellular viability and metastatic ability via ZNF703 overexpression (112) and TFG-β-induced EMT pathways, regulating LNM in ESCC (113). Additionally, lncRNA H19 and MEG promote LNM in EC via the STAT3/EZH2/β-catenin and PSAT1-dependent GSK-3β/Snail signaling pathways, respectively (114, 115). LINC00324 silences miR-493-5p to activate the MAPK1 signaling and enhance LNM in ESCC (116).
MiRNAs are pivotal regulators of tumor growth and development. Expressions of miR-483-5p and miR-625 are positively and negatively correlated with LNM and OS, respectively, making them predictors of LNM and unfavorable prognosis in ESCC (117, 118). Xu Y et al. has demonstrated that miR-17-3p, miR19a-3p, and miR-18b-5p are tumor-specific miRNAs to regulate and predict the regional LNM of ESCC (119). Overexpression of miR-320b promotes the LNM of ESCC depending on m6A modification (120). Besides, exosome-mediated miR-10527-5p suppresses Rab10 by knocking down Wnt/β-catenin signaling to inhibit LNM and other malignant phenotypes of ESCC (121). Meanwhile, miR19a-3p upregulation and the uc.189-EPHA2 axis activates the RAC1/CDC42-PAK1 and the p38/MAPK/VEGF-C pathways to promote the formation of lymphatic vessels and the metastatic ability in ESCC (122, 123). In ESCC, exosomal uc.189 and miR-203 expressions are positively and negatively associated with metastasis and prognosis, respectively (124, 125). The malignancy is activated via the inhibition of KIF5C, the accumulation of Axin, and the sequential or synergistic regulation of miR-203 and miR-21 (124, 126). On the contrary, an anti-tumor miR-133b suppresses TAGLN2 and negatively impacts LNM (127). Additionally, miR-143-5p regulates oncogene expressions, including HMGA2, STMN1, and MET, thereby attenuating migration and invasion in EC cells, similar to the modulator of miR-143-3p in KRT80 (128). Moreover, miR-143-5p is observed to interact with numerous genes involved in LNM of EC, such as the negative regulator AGR2 in HNSCC (42).
Serum markers have consistently attracted attention because of their sample availability. Traditional tumor markers in ESCC patients, such as CEA, CA125, CA199, CA724, and CA242, have been retrospectively analyzed. CA199 and CA125 are positively associated with LNM, and CA125 is a predominant predictor of LNM occurrence (129). Previous studies have demonstrated serum components’ predictive and prognostic role in LNM of EC, including TK1, CYFRA21-1, STMN1, PTEN, and TNFAIP8 (62, 130, 131). A novel homogeneous AlphaLISA improves the sensitivity (81%) and specificity (94%) for detecting serum STMN1, better indicating early-stage LNM in ESCC (132). Besides, the co-detection of autoantibodies against a panel of six tumor-associated antigens (p53, NY-ESO-1, MMP-7, Hsp70, PRDX6, and Bmi-1) is applied to predict early-stage LNM in ESCC (133). The lactate dehydrogenase to albumin ratio (LAR) demonstrates an association with advanced LNM and unfavorable prognosis in patients with resectable EC, regardless of NCRT (134, 135). Similar potential prognostic indices are utilized in CRC and other carcinomas (136, 137). Considering serum metabolites, amino acids are ideal metabolic biomarkers to predict LNM and monitor therapeutic efficacy (138). Combining three serum metabolites, valine, GABA, and pyrrole-2-carboxylic acid, enhances diagnostic efficacy in LNM of EC (139). Additionally, some novel miRNAs (Table 2) in serum, such as miR-21, miR-216a/b, and miR-218, play suppressive and predictive roles in LNM of EC (62, 140–142), while higher concentrations of miR-20b-5p detected in the serum are combined with a nomogram to predict advanced LNM of ESCC accurately (143). CircRNA secreted by ESCC cells, has-circ-0026611, is identified as a novel predictor of tumor prognosis (144). Meanwhile, blood cells may enhance a predictive and prognostic function in LNM of EC. Common blood components, such as platelets and neutrophils, promote the metastatic ability of cancers and interactions with tumor cells (145). Besides, some cell ratios, such as lymphocyte-monocyte, platelet-lymphocyte, and neutrophil-lymphocyte ratios (NLR), are positively associated with predicting and prognosticating LNM of ESCC. NLR is particularly effective in predicting LNM, indicating the need for thorough LND and consideration of preoperative adjuvant therapy for patients with clinical stage I ESCC (146). Meanwhile, NLR and other inflammatory markers, such as red cell width, leukocytosis, and neutrophilia, predict prognosis during NCRT, followed by esophagectomy (147–149). Moreover, the NLR/pre-albumin and NLR/albumin ratios demonstrate potential utility (150, 151).
Thanks to the development of the liquid biopsy, a novel noninvasive method to detect and evaluate changes in body fluids, the concentration of circulating tumor cells (CTCs) in EC, has emerged as a promising marker to assess tumor progression and monitor treatment efficacy (145, 152–154). Specific targets or changes in CTCs, such as a decrease in epithelial cell adhesion molecule (EpCAM) and the presence of cell surface vimentin, are linked to poor prognosis in patients with ESCC, underscoring their significance within the TME (155). Post-NCRT CTCs are applied to evaluate the degree of primary histopathological response and the necessity of esophagectomy (156). In addition, products expressed by CTCs might indicate therapeutic efficacy. For example, the sensitivity to radiotherapy is positively associated with mRNA expression of NRF2 and TP53 in CTCs; meanwhile, CD8+ T-cells accumulate in TME as NRF2 mRNA levels increase (157). Furthermore, CTCs are regarded as promising indicators in LNM of EC. However, this area has a paucity of research (158). The proportion of CTCs with chromosome 7 triploidy is related to distant metastasis and TNM stage (152). Higher levels of EpCAM and CEA in tissues are associated with LNM in EAC and could potentially be applied in tumor-target imaging for EAC (159). Another study has demonstrated that advanced LNM and TNM stages influenced the expression of PTP4A1 in ESCC and efficiently predicted the PTP4A1+ TCTs with an area under the ROC curve of 0.725 (160). Further studies are required to explore the association between CTCs and LNM in EC and other cancers.
Competitive interactions between tumor-infiltrating immune cells and cancer cells create unique lymph metastatic hubs and promote tumor occurrence, development, and metastasis (161). Previous research has demonstrated that innate immune cells in the TME, including tumor-infiltrating lymphocytes (TILs), tumor-associated macrophages (TAMs), and natural killer (NK) cells, play a key role in maintaining esophageal homeostasis and immune defense, providing potential targets for immunotherapy (162). In addition, immune checkpoint inhibitors, such as PD-1/PD-L1 inhibitors, have shown significant efficacy in various malignancies. In ESCC, researches have identified several biomarkers which may influence a patient’s response to immune checkpoint inhibitors (163, 164), yet few have focused on their roles in LNM of EC. For TILs, the silencing of T-cell factor/lymphoid enhancer factor lead to the downregulation of molecules such as SREBP1 (165) and G3BP1 (166) by phosphorylating different targets via Wnt/β-catenin signaling pathways in ESCC. Besides, the combined assessment of reduced CD8+/FOXP3+ TIL density and PD-L1 overexpression is proposed as a potential indicator for tumor differentiation and LN status in ESCC (167). In ESCC patients undergoing neoadjuvant chemotherapy, the PD-L1 status of tumor cells in positive LN tissues and the FOXP3/CD8 ratio in primary tumors are identified as prognostic factors for overall clinical outcomes (168). However, there is insufficient evidence regarding the impact of CD4+ TILs following NCRT for thoracic ESCC (169). The prevalence of PD-L1 immunoreactivity is significantly higher in IL-6-positive EC specimens, and T-cell functions against tumor cells, including proliferation and cytotoxicity, are inhibited (170). Therefore, PD-L1 determines radiation response and potentially serves as a prognostic indicator for patients with ESCC (171). The infiltration of activated cytotoxic T-cells is positively associated with PD-L1 in tumor tissues to predict 3-5-year OS and DFS, as well as TAMs (172). TAMs play a dual role in the TME, where TAM-1 has anti-tumor activity and TAM-2 promotes tumor metastasis. Studies have shown that the higher infiltration of TAM-2, especially CD68-TAMs, is associated with LNM and poor prognosis of EC (130, 173). A polarization tendency to TAM-2 is detected in EAC, and a higher ratio of M2/M1 macrophages serves as a sensitive marker to predict LNM and poor prognosis in EAC without NCRT (174). NK cells exhibit potent cytolytic activity against tumors, while dendritic cells are essential antigen-presenting cells to activate the immune response of TILs. Both cell types are critical regulators in the immune system and potential targets in the immunotherapy combined with PD-1/PD-L1. However, further research is necessary to develop novel markers and verify their efficacy in applying PD-1/PD-L1 inhibitors to target LNM of EC (163, 172).
Alterations in the microbiota have been linked to EC. Compared with direct access to esophageal samples, oral and intestinal flora could be used as noninvasive biomarkers to detect EC and have potential as practical assessment tools (175). In EC, microbiota have shown promise as tumor-associated biomarkers in precancerous lesions and early stages. Dysplasia is associated with bacterial alterations in EC, and Barrett’s esophagus, a precursor of EAC, is categorized into two main types based on Gram-positive or harmful bacterial components (176). Overall, the abundance and diversity of microbiota are lower in EC. Certain bacteria have been enriched or depleted in patients with EC compared to healthy controls, and the abundance of microorganisms is associated with OS and other clinicopathological characteristics (177). A consensus on the dominant bacteria in EC tissues is lacking. However, many studies have acknowledged the predominant position of Fusobacteria in the bacterial composition of EC associated with advanced tumor stages, identifying it as a potentially predictive and prognostic marker (178, 179). Meanwhile, a study has demonstrated the decreasing number of Bacteroidetes, Fusobacteria, and Spirochaetes (177). Another study has demonstrated that a shorter-term survivor has a higher number of Lactobacillus in tumors, while Leptotrichia is the opposite (180). There is an increasing abundance of Prevotella and a decreasing abundance of Streptococcus in the ESCC group, with different proportions in different stages (181). Meanwhile, brush samples from another analysis have revealed that the relative abundance of Streptococcus and Prevotella serve as significant defining characteristics among community types within the esophageal microbiota, and their combined abundance independently emerge as a prognostic predictor for individuals with ESCC (182). The degree of LNM alters microbiota distribution in ESCC (183), and the microbiota also potentially mediates LNM in EC. Besides a higher abundance of Streptococcus and Prevotella associated with the LNM group (182), it has been found that Lactobacillus (180) and Fusobacteria (184) are more abundant in tissues without LNM compared to those with LNM. Zhang et al. have confirmed the positively abundant F. nucleatum associated with LNM in ESCC and the depth of tumor infiltration (178).
Microbiota plays an essential role in tumor metastasis and other malignant phenotypes. Similar functions have been further investigated in other cancers, where metastatic activation occurs through the production of metabolites, bioactive compounds, and the modulation of inflammatory and anti-tumor responses. Genetic and epigenetic changes contribute to increased resistance to specific tumor treatments. Gut and intratumor microbiota can also reconstruct the distal organ microenvironment and form PMN so as to provide better living conditions for the transfer of tumor cells (180, 185). Chronic inflammation is known to be a precursor to EC and many other cancers. Some microbiota-derived metabolites, such as short-chain fatty acids, exert protective effects against inflammation and cancer, while others may promote carcinogenesis pathways (15). Bacteroides are closely associated with EC progression through increased inflammation and the activation of the TLR4/MyD88/NF−κB pathway triggered by lipopolysaccharide (LPS), a surface antigen. This underscores a strong connection between common LPS-induced signaling pathways in both inflammatory responses and the development and progression of EC (186). Induced by Porphyromonas gingivalis LPS in vivo, the NO secretion of macrophages in HNSCC is enhanced to proliferate and invade more aggressively (187). Alterations in microbiome diversity significantly modulate the immune system and impact immune surveillance and anti-tumor responses. The recognition of bacterial flagellin leads to increased nuclear localization of TLRs, especially TLR4 and TLR5. This upregulation is positively associated with malignant behaviors and metastatic abilities of Barrett’s esophagus and EAC (188, 189). When Lactobacillus plays a dominant role in intratumoral-bacterial compositions, it suppresses anti-tumor immune responses and facilitates the progression of EC (180). Robinson W et al. have developed a computational pipeline and suggested that myeloid cells in TME are important origins of microbiota. These cells contribute to inflammation and immunosuppressive responses in tumor tissues, since the abundance of myeloid cells increases with proinflammatory cytokines, IL-1β and CXCL8 upregulation (190). Based on the immune infiltration microenvironment, F. nucleatum invades and persists within different cells to enhance the LNM in ESCC. It facilitates cellular immune escape in Tregs (191). In ESCC cells, F. nucleatum activates the DNA damage response pathways and increases the secretion of cisplatin-induced senescence-associated secretory phenotype, significantly contributing to tumor progression and chemoresistance (178).
EC represents an aggressive malignancy with significant implications for patient survival, particularly when it metastasizes. The lymphatic system is crucial for tumor cell dissemination. Considering thoracic EC, specific regional LNs are particularly susceptible to involvement, including the cervical paraesophageal, recurrent laryngeal nerve, subcarinal, those along the left gastric artery, lesser curvature, and paracardial LNs. A positive thoracic paraesophageal LN status indicates a more advanced disease (5). The length and complexity of the esophageal lymph network results in extensive drainage locations and unexpected “skip metastasis.” Differences in viewpoints and clinical practices regarding LNM in EC lead to disagreements in the locations of LNs between two major TNM staging systems. For example, in the 8th edition of the Union for International Cancer Control (UICC)/American Joint Committee on Cancer (AJCC), supraclavicular LNM is classified as M1 and celiac LNs are regional LNs. Meanwhile, the 12th edition of the Japan Esophageal Society (JES) categorizes a positive supraclavicular LN as M1a due to the possibility of a cure after dissection. However, celiac LNs are not regional for upper thoracic EC. The 2023 Guidelines of the Chinese Society of Clinical Oncology (CSCO) for EC follow the classification of the 8th edition of the UICC/AJCC (192). The incidence of LNM dramatically impacts treatments, including surgical interventions and applications of NCRT, as well as the prognosis and OS of patients.
The process of LNM in early EC is intricate and influenced by various factors, including tumor location, type, and depth of invasion. Each stage, from precancerous lesions to tumorigenesis to progression to PMN, involves distinct molecular changes driven by inflammatory, immune, and other responses (193). The risk of LNM increases as early EC invades deeper layers. Notably, when early EC invades the submucosal layer, the risk of LNM significant increases (3). Furthermore, the site of LNM is associated with the location and type of the tumor, with distinct patterns observed in upper, middle, and lower thoracic EC (194). Molecular markers have shown promise in predicting LNM and understanding the biological behavior of EC, informing individualized treatment. Despite the risk of LNM in early EC, not all patients develop it, as seen in patients diagnosed with clinical T1N0 EC who develop LNM postoperatively (195). Thus, comprehensive LN evaluation remains crucial, and further research is needed to explore the mechanisms, interactions, and determinants of tumor metastasis. Understanding the process and mechanism significantly improves the diagnostic accuracy, treatment outcomes, and prognostic management of patients with early EC. Management of early-stage EC differs by tumor stages. The pT1a stage can be managed endoscopically, yet additional interventions are required at the M3 stage. For the pT1b stage, surgical resection or NCRT yields better long-term outcomes (3). EC resection is considered the fundamental treatment for locally advanced cases. However, the 5-year postoperative OS differs significantly based on the presence of LNM. Patients with no LNM or only one LNM exhibit a 5-year OS rate exceeding 50%, while those with two or more LN metastases have a 5-year OS rate of less than 30% (8). A review of 3800 ESCC patients from the nationwide JES registry data suggested that LN dissection range should be based on the location of primary tumors (9). However, excessive LN dissection offers no additional advantages. Compared with standard two-field lymphatic dissection, total two-field lymphatic dissection failed to improve postoperative survival but caused more significant complications (10). Another randomized phase III trial reported that two- and three-field lymphatic dissection showed comparable rates and severities of postoperative complications in EC patients (196). In short, LN evaluation in EC therapy is essential in EC therapy.
Cancer biomarkers objectively measure and evaluate features that indicate normal and pathological processes or pharmacological responses to therapeutic interventions (17). Despite their potential, cancer biomarkers face significant challenges in clinical validation, particularly in detecting, diagnosing, and monitoring early-stage diseases to enhance long-term survival (16). It is estimated that approximately 50% of cancers are diagnosed at an advanced stage. Additionally, early biomarkers are often present in minuscule amounts, making it challenging to isolate accurate signals amidst normal human physiological noise. Potential risks, such as overdiagnosis and overtreatment, must also be addressed (197). Meanwhile, it is imperative to overcome scientific challenges to develop new biomarkers with enhanced sensitivity, specificity, and positive predictive value (198). Besides, combining traditional radiomics and biomarkers has demonstrated heightened predictive capabilities in developing and prognosis EC and other malignancies. This approach surpasses reliance on imaging modalities, such as EpCAM and CEA, novel targets used in tumor-target imaging approaches (159). Notably, signatures exhibit significant variations in radiomics-based imaging parameters. The diagnostic sensitivity and specificity of multislice CT, when coupled with the detection of CA19-9, Bcl-2, and CYFRA21-1, have exhibited superior performance in identifying LNM in EC patients compared to individual indicators (199). Increased Caprin-1 expression in EC samples is accurately anticipated through 18F-FDG PET/CT imaging, exhibiting a sensitivity of 70.8% and a specificity of 77.4%, suggesting the potential of Caprin1 as a robust prognostic biomarker (76). Likewise, elevated eIF6 expression is identified in EC utilizing 18F-FDG PET/CT, with an SUVmax threshold of 18.2, resulting in a predictive accuracy of 0.755 for tumor eIF6 expression (200).
In precision oncology, gene expression signatures related to immunotherapy have shown promise as markers. Analyzing gene expression patterns of immune cells in the TME can provide insights into how tumors evade immune surveillance and guide personalized treatment strategies. Some studies have proposed that genetic evolution is a driving force for tumor metastasis, suggesting that carcinomas might be associated with groups of genes sharing similar characteristics rather than operating independently. Several models with varying sensitivity and specificity levels have been developed for EC, such as ferroptosis (201) and mitochondrial (202) genes. Despite extensive sequencing, there is limited evidence of metastasis-specific genomic alterations, and the underlying mechanisms are still unknown (145). In addition to gene expression products, there are increasingly specific markers. Glycans on the cell surface are structure-variable and tissue-specific, which play a role in tumor occurrence and development. When combined with Lens culinaris lectin, they serve as a predominant predictor in the early detection of LNM in ESCC (203). TMNs significantly modulate tumor development through immune infiltrates and other aspects of tumor progression. Even before metastasis initiation, the primary tumor selectively influences the PMN (204) to enhance the colonization of CTCs (205). Regarding TME and PNM, PD-L1 expression and the abundance and activity of CD8+ T-cells play crucial roles in indicating anti-tumor responses and predicting the efficacy of targeted and immunotherapy (170). The presence of PMN is pivotal for tumor metastasis, as the local microenvironment alone cannot support the dissemination and colonization of CTCs in distant organs and tissues (206). Significant alterations in the microenvironment of LNs and vessels occur before metastasis, creating a PMN that is conducive to the growth of CTCs (207). The formation of this niche is currently debated, with several key factors identified, including changes in the LN vasculature, influence on lymphatic endothelial cells, modulation of the stiffness of the ECM, involvement of fibroblastic reticular cells, and creation of an immunosuppressive microenvironment (4). EVs can establish PMNs and support rapid tumor dissemination to transport cargoes such as nucleic acids and proteins, influencing various signaling pathways, suppressing defensive immune reactions, polarizing tumor-promoting phenotypes, expressing the PD-1 checkpoint protein (208), and targeting EVs represents a promising therapeutic strategy (207). Research and application of these biomarkers demonstrate their potential to enhance the efficacy of cancer treatment and improve patients’ quality of life, particularly in immunotherapy and other targeted therapies. Signaling pathways that regulate LNM in EC are various, such as Wnt/β-catenin, MAPK, and RTK; some rely on EMT signatures (Figures 1, 2). Future studies should focus on underlying therapeutic targets and novel treatments for clinical application.
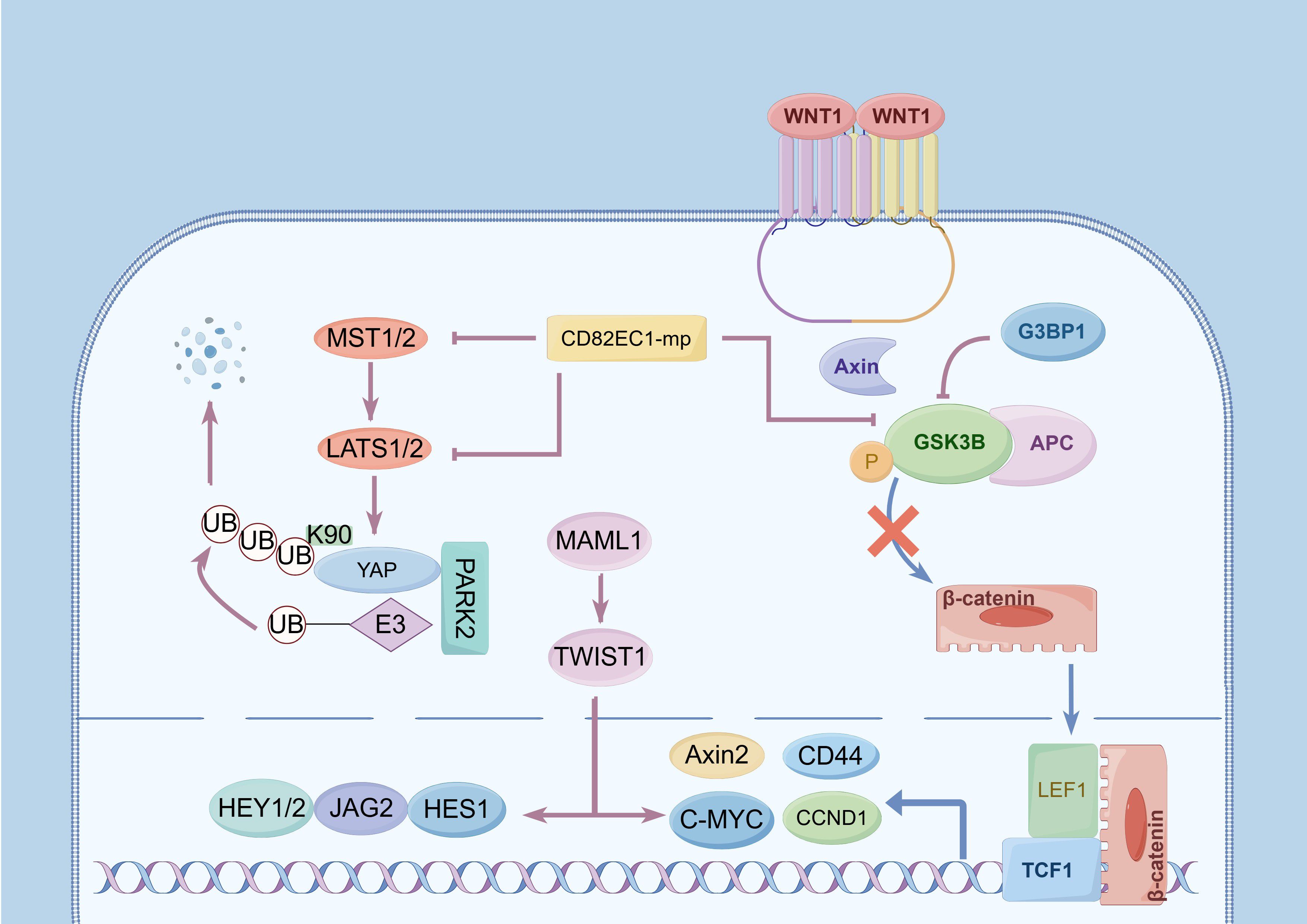
Figure 1. Activation of signaling pathways and upregulation of lymph node metastasis (LNM) of esophageal cancer (EC). (1) The Wnt/β-catenin signaling pathway: The phosphorylation of GSK3B significantly influences this pathway. G3BP1 promotes its phosphorylation, and further promotes the nuclear translocation of β-catenin, activates the expression of TCF/LEF, C-MYC, Axin2,CCND1, and other genes to participate in LNM. (2) The Hippo signaling pathway: MST1/2 and LAST1 are pivotal regulating targets, and YAP encodes a downstream nuclear effector in this pathway. Phosphorylation of MST1/2 and upregulation of LAST1 expression upregulate YAP phosphorylation and activate its proteasome-dependent degradation, and PARK2 facilitates YAP ubiquitination at site K90. CD82EC1-mP can inhibit the above two pathways to inhibit LNM by suppressing the phosphorylation of GSK3β and MIST1. (3) The Notch signaling pathway: MAML1 is an essential transcriptional co-activator in this pathway, and its overexpression induces TWIST1 expression, upregulating downstream targets including C-MYC, JAG2, HEY1, HEY2, and HES1.
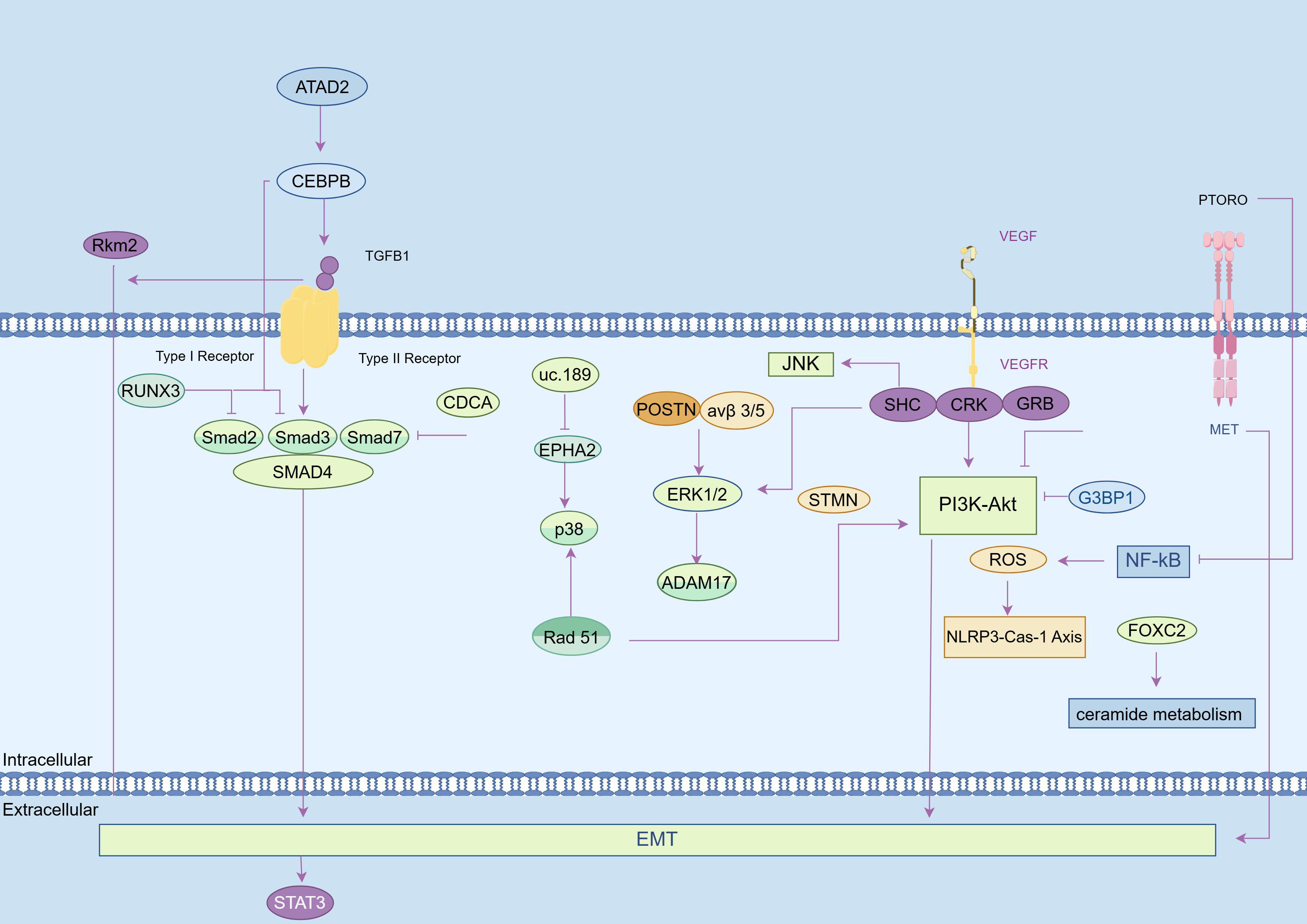
Figure 2. EMT-dependent mechanisms in promoting the lymph node metastasis (LNM) of esophageal cancer (EC). (1) NF-κB signaling pathway: Increased FOXC2 dysregulates ceramide metabolism to activate this pathway and elevate the LNM of EC. Conversely, PTPROt deficiency reduces ROS, inhibiting the NLRP3-Caspase-1 axis and suppressing LNM. (2) RTK signaling pathway: VEGF/VEGFR induces dimerization, phosphorylation, and activation of PTKs, to recruit adaptor proteins such as CRK, SHC, GRB2, thereby promoting LNM of EC. (3) PI3K/AKT signaling pathway: NOLC1 overexpression activates the PI3K/AKT pathway to enhance LNM in EC. G3BP1 knockdown inhibits this pathway by reducing p-PI3K and p-AKT expression levels. Inhibition of the PI3K pathway also suppresses STMN1 expression. Rad51 overexpression upregulates p38 phosphorylation and Snail levels to elevate Akt phosphorylation. (4) MAPK signaling pathway: uc.189 inhibits EPHA2 expression to activate p38 and ultimately promote LNM; POSTN binding to integrin αvβ3/αvβ5 increases ERK1/2 phosphorylation and upregulates ADAM17, leading to tumor aggressiveness. (5) JAK-STAT pathway and TGF-β pathway: Activation of these pathways impedes LNM. PKM2 promotes STAT3 phosphorylation through TGF-β1-induced EMT. ATAD2 interacts with C/EBPβ for nuclear translocation and binding for TGF-β1 promoter activation. TGF-β1 promotes Smad3 phosphorylation and interaction with Smad4 for regulation within the nucleus. Increased RUNX3 reduces Smad2/Smad3 phosphorylation, suppressing TGF-β1-induced EMT. CDCA7 overexpression promotes Smad4 but inhibits Smad7 expression to facilitate EMT in ESCC.
The relationship between the microbiota and various diseases has attracted significant attention. Gut microbiota are involved in body homeostasis and influenced by many factors, including diet, smoking, drinking, obesity, and intake of drugs, whose imbalance leads to various diseases (209). Through blood and lymph circulations, gut microbiota reach even distant organs and tissues, where they find and settle in suitable environment, including the TME, becoming one of the essential origins of intratumor microbiota, which are significant components of the cancerous ecosystem (210). Recent studies have demonstrated the existence of intratumor microbiota in several cancers (21, 170), which could increase tumor metastasis by regulating the intrinsic characteristics of tumor cells via EMT regulation, as well as extrinsic characteristics through vessel barrier blocking and modifications in the PMN (211). In BC, intratumor microbiota emerges as a novel and promising biomarker for diagnosis, prognosis, and therapeutic assessments (212–214), prompting further exploration of its role in EC and other cancers. Thanks to next-generation sequencing and non-invasive detection of oral and fecal samples, it is more convenient and practical to explore the pathogenesis and diagnostic or prognostic applications of microbiota in esophageal diseases (215, 216). Studies have suggested that alterations in the gut microbiota composition could be associated with an increased risk of esophageal disorders, such as eosinophilic esophagitis, gastroesophageal reflux disease, and achalasia (217). Recent research on EC has suggested that microbiome may influence its occurrence, development, and early diagnosis or prognosis. Moreover, research is ongoing to explore new avenues for clinical treatments and management, and manipulating the gut microbiome could provide new avenues for clinical treatments and management and enhance the efficacies of chemotherapy and immunotherapy (180, 190). Forsythoside A rejuvenates the diversity of microorganisms to show its efficacy against ESCC and significant implications for clinical implementation (218), and probiotics or dietary modifications aimed at restoring a healthy gut microbiome could complement conventional cancer treatments in ECA (219). The use of antibiotics in EC should be carefully evaluated, as certain species like F. nucleatum could provide a novel targeting strategy to optimize clinical outcomes of cisplatin resistance (178). However, more rigorous research is required to confirm the connection between bacterial biomarkers and LNM in EC, and to translate findings into clinical practice. Current evidence fails to support the view that microbiota is an indicator of EC, particularly EAC. Furthermore, detailed investigations are required to determine whether noninvasive oral or fecal specimens can provide detection as sensitive and specific as biopsy, or even better (175).
Despite ongoing research into biomarkers for evaluating LNM in EC, there is a lack of FDA-approved tests and other commercially available options. A cost-benefit analysis by European surgeons indicated that, among current biomarkers for gastrointestinal cancers, CEA outperformed CA19-9 and CA125 in lower financial cost, higher sensitivity, and better diagnostic accuracy for metastases at presentation (220). Advances in molecular research and clinical trials may eventually lead to reliable biomarkers for routine clinical use in EC management. Our review presents a comprehensive analysis of recent research on potential biomarkers and their underlying mechanisms in LNM of EC. We aim to provide new insights and advancements in predictive approaches by identifying unique biomarker signatures. Utilizing these biomarkers can improve pre- and post-surgical assessments, leading to the development of personalized and effective therapeutic strategies for better patient outcomes and quality of life. Furthermore, the identified biomarkers and their associated pathways may serve as targets for therapeutic interventions in EC patients. However, there is limited research on the specific mechanisms and pathways of biomarkers in LNM among patients with EC. Most biomarkers referring to LNM of EC are prognostic, involving mutations and gene expressions. Only a few, such as serum biomarkers and CTCs, have shown the potential to predict or monitor therapeutic outcomes. In conclusion, future research efforts should concentrate on these areas to enhance our understanding and improve clinical practices.
NW: Resources, Writing – original draft, Writing – review & editing, Formal analysis, Investigation, Visualization. JC: Writing – original draft, Visualization. JJ: Writing – review & editing. YL: Writing – review & editing. XW: Writing – review & editing. WZ: Writing – review & editing. PZ: Formal analysis, Investigation, Supervision, Validation, Visualization, Writing – review & editing. MK: Funding acquisition, Supervision, Visualization, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work has been supported by the National Natural Science Foundation of China (Grant Nos. 82070499 and 82372680) and the Joint Funds for the Innovation of Science and Technology of Fujian Province (Grant No. 2020Y9073).
We thank Lei Ji and Wei Wang for language polishing, and Home for Researchers (www.home-for-researchers.com) for a figure polishing service. We would particularly like to acknowledge the patient support from SandianScience Lab.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
EC, Esophageal cancer; ESCC, esophageal squamous cell carcinoma; EAC, Esophageal adenocarcinoma; LN, Lymph node; LNM, Lymph node metastasis; NCRT, Neoadjuvant chemoradiotherapy; OS, Overall survival; CT, Computed tomography; MRI, Magnetic resonance imaging; PET, Positron emission tomography; PMN, Premetastatic niche; ECM, extracellular matrix; TME, Tumor microenvironment; EVs, Extracellular vesicles; EMT, Epithelial-mesenchymal transition; BC, Breast cancer; HNSCC, Head and neck squamous cell carcinoma; lncRNAs, Long noncoding RNAs; LAR, Lactate dehydrogenase to albumin ratio; NLR, Neutrophil-lymphocyte ratios; CTCs, Circulating tumor cells; EpCAM, Epithelial cell adhesion molecule; TILs, Tumor-infiltrating lymphocytes; TAMs, Tumor-associated macrophages; NK, Natural killer; LPS, Lipopolysaccharide.
1. Fernández-Montes A, Alcaide J, Alsina M, Custodio AB, Franco LF, Gallego Plazas J, et al. SEOM-GEMCAD-TTD Clinical Guideline for the diagnosis and treatment of esophageal cancer (2021). Clin Transl Oncol. (2022) 24:658–69. doi: 10.1007/s12094-022-02801-2
2. Puhr HC, Prager GW, Ilhan-Mutlu A. How we treat esophageal squamous cell carcinoma. ESMO Open. (2023) 8:100789. doi: 10.1016/j.esmoop.2023.100789
3. Deboever N, Jones CM, Yamashita K, Ajani JA, Hofstetter WL. Advances in diagnosis and management of cancer of the esophagus. BMJ. (2024) 385:e074962. doi: 10.1136/bmj-2023-074962
4. Ji H, Hu C, Yang X, Liu Y, Ji G, Ge S, et al. Lymph node metastasis in cancer progression: molecular mechanisms, clinical significance and therapeutic interventions. Signal Transduct Target Ther. (2023) 8:367. doi: 10.1038/s41392-023-01576-4
5. Wang Y, Zhu L, Xia W, Wang F. Anatomy of lymphatic drainage of the esophagus and lymph node metastasis of thoracic esophageal cancer. Cancer Manag Res. (2018) 10:6295–303. doi: 10.2147/CMAR.S182436
6. Shang Q-X, Yang Y-S, Xu L-Y, Yang H, Li Y, Li Y, et al. Prognostic role of nodal skip metastasis in thoracic esophageal squamous cell carcinoma: A large-scale multicenter study. Ann Surg Oncol. (2021) 28:6341–52. doi: 10.1245/s10434-020-09509-z
7. Kumakura Y, Yokobori T, Yoshida T, Hara K, Sakai M, Sohda M, et al. Elucidation of the anatomical mechanism of nodal skip metastasis in superficial thoracic esophageal squamous cell carcinoma. Ann Surg Oncol. (2018) 25:1221–8. doi: 10.1245/s10434-018-6390-0
8. Borggreve AS, Kingma BF, Domrachev SA, Koshkin MA, Ruurda JP, van Hillegersberg R, et al. Surgical treatment of esophageal cancer in the era of multimodality management. Ann N Y Acad Sci. (2018) 1434:192–209. doi: 10.1111/nyas.13677
9. Tachimori Y, Ozawa S, Numasaki H, Matsubara H, Shinoda M, Toh Y, et al. Efficacy of lymph node dissection by node zones according to tumor location for esophageal squamous cell carcinoma. Esophagus. (2016) 13:1–7. doi: 10.1007/s10388-015-0515-3
10. Guo Q, Li H, Wang H, Zhang D, Li Y. Effects of standard and total two-field lymph node dissection on prognosis of patients undergoing Esophagectomy. Pak J Med Sci. (2022) 38:950–4. doi: 10.12669/pjms.38.4.4031
11. Schuring N, Matsuda S, Hagens ERC, Sano J, Mayanagi S, Kawakubo H, et al. A proposal for uniformity in classification of lymph node stations in esophageal cancer. Dis Esophagus. (2021) 34:doab009. doi: 10.1093/dote/doab009
12. Chaudhry SR, Bordoni B. Anatomy, thorax, esophagus, in: StatPearls (2024). Treasure Island (FL: StatPearls Publishing. Available online at: http://www.ncbi.nlm.nih.gov/books/NBK482513/ (Accessed February 2, 2024).
13. Chen X, Wang X, Zhang K, Fung K-M, Thai TC, Moore K, et al. Recent advances and clinical applications of deep learning in medical image analysis. Med Image Anal. (2022) 79:102444. doi: 10.1016/j.media.2022.102444
14. Tang S, Yuan K, Chen L. Molecular biomarkers, network biomarkers, and dynamic network biomarkers for diagnosis and prediction of rare diseases. Fundam Res. (2022) 2:894–902. doi: 10.1016/j.fmre.2022.07.011
15. Qiu S, Cai Y, Yao H, Lin C, Xie Y, Tang S, et al. Small molecule metabolites: discovery of biomarkers and therapeutic targets. Sig Transduct Target Ther. (2023) 8:1–37. doi: 10.1038/s41392-023-01399-3
16. Sarhadi VK, Armengol G. Molecular biomarkers in cancer. Biomolecules. (2022) 12:1021. doi: 10.3390/biom12081021
17. Mishra A, Verma M. Cancer biomarkers: are we ready for the prime time? Cancers. (2010) 2:190–208. doi: 10.3390/cancers2010190
18. Xu WW, Li B, Guan XY, Chung SK, Wang Y, Yip YL, et al. Cancer cell-secreted IGF2 instigates fibroblasts and bone marrow-derived vascular progenitor cells to promote cancer progression. Nat Commun. (2017) 8:14399. doi: 10.1038/ncomms14399
19. Yin X, Tian M, Zhang J, Tang W, Feng L, Li Z, et al. MiR-26b-5p in small extracellular vesicles derived from dying tumor cells after irradiation enhances the metastasis promoting microenvironment in esophageal squamous cell carcinoma. Cancer Lett. (2022) 541:215746. doi: 10.1016/j.canlet.2022.215746
20. Han P, Cao P, Hu S, Kong K, Deng Y, Zhao B, et al. Esophageal microenvironment: from precursor microenvironment to premetastatic niche. Cancer Manag Res. (2020) 12:5857–79. doi: 10.2147/CMAR.S258215
21. Nejman D, Livyatan I, Fuks G, Gavert N, Zwang Y, Geller LT, et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science. (2020) 368:973–80. doi: 10.1126/science.aay9189
22. Loring HS, Thompson PR. Kinetic mechanism of nicotinamide N-methyltransferase. Biochemistry. (2018) 57:5524–32. doi: 10.1021/acs.biochem.8b00775
23. Yoda M, Mizuno R, Izumi Y, Takahashi M, Bamba T, Kawaoka S. Nicotinamide-N-methyltransferase regulates lipid metabolism via SAM and 1-methylnicotinamide in the AML12 hepatocyte cell line. J Biochem. (2023) 174:89–98. doi: 10.1093/jb/mvad028
24. Novak Kujundžić R, Prpić M, Đaković N, Dabelić N, Tomljanović M, Mojzeš A, et al. Nicotinamide N-methyltransferase in acquisition of stem cell properties and therapy resistance in cancer. Int J Mol Sci. (2021) 22:5681. doi: 10.3390/ijms22115681
25. Yang C, Wang T, Zhu S, Zong Z, Luo C, Zhao Y, et al. Nicotinamide N-methyltransferase remodeled cell metabolism and aggravated proinflammatory responses by activating STAT3/IL1β/PGE2 pathway. ACS Omega. (2022) 7:37509–19. doi: 10.1021/acsomega.2c04286
26. Wang W, Yang C, Wang T, Deng H. Complex roles of nicotinamide N-methyltransferase in cancer progression. Cell Death Dis. (2022) 13:267. doi: 10.1038/s41419-022-04713-z
27. Barrows RD, Jeffries DE, Vishe M, Tukachinsky H, Zheng S-L, Li F, et al. Potent uncompetitive inhibitors of nicotinamide N-methyltransferase (NNMT) as in vivo chemical probes. J Med Chem. (2022) 65:14642–54. doi: 10.1021/acs.jmedchem.2c01166
28. Zhang L, Song M, Zhang F, Yuan H, Chang W, Yu G, et al. Accumulation of nicotinamide N-methyltransferase (NNMT) in cancer-associated fibroblasts: A potential prognostic and predictive biomarker for gastric carcinoma. J Histochem Cytochem. (2021) 69:165–76. doi: 10.1369/0022155420976590
29. Couto JP, Vulin M, Jehanno C, Coissieux M-M, Hamelin B, Schmidt A, et al. Nicotinamide N-methyltransferase sustains a core epigenetic program that promotes metastatic colonization in breast cancer. EMBO J. (2023) 42:e112559. doi: 10.15252/embj.2022112559
30. Bach D-H, Kim D, Bae SY, Kim WK, Hong J-Y, Lee H-J, et al. Targeting nicotinamide N-methyltransferase and miR-449a in EGFR-TKI-resistant non-small-cell lung cancer cells. Mol Ther Nucleic Acids. (2018) 11:455–67. doi: 10.1016/j.omtn.2018.03.011
31. Huang Q, Chen H, Yin D, Wang J, Wang S, Yang F, et al. Multi-omics analysis reveals NNMT as a master metabolic regulator of metastasis in esophageal squamous cell carcinoma. NPJ Precis Oncol. (2024) 8:24. doi: 10.1038/s41698-024-00509-w
32. Zhong X, Chen X, Guan X, Zhang H, Ma Y, Zhang S, et al. Overexpression of G9a and MCM7 in oesophageal squamous cell carcinoma is associated with poor prognosis. Histopathology. (2015) 66:192–200. doi: 10.1111/his.12456
33. Zhu Y, Zhu M-X, Zhang X-D, Xu X-E, Wu Z-Y, Liao L-D, et al. SMYD3 stimulates EZR and LOXL2 transcription to enhance proliferation, migration, and invasion in esophageal squamous cell carcinoma. Hum Pathol. (2016) 52:153–63. doi: 10.1016/j.humpath.2016.01.012
34. Nagata H, Kozaki K-I, Muramatsu T, Hiramoto H, Tanimoto K, Fujiwara N, et al. Genome-wide screening of DNA methylation associated with lymph node metastasis in esophageal squamous cell carcinoma. Oncotarget. (2017) 8:37740–50. doi: 10.18632/oncotarget.17147
35. Chen X, Hu H, Liu J, Yang Y, Liu G, Ying X, et al. FOXF2 promoter methylation is associated with prognosis in esophageal squamous cell carcinoma. Tumour Biol. (2017) 39:1010428317692230. doi: 10.1177/1010428317692230
36. Wang G, Fu Z, Li D. MACC1 overexpression and survival in solid tumors: a meta-analysis. Tumor Biol. (2015) 36:1055–65. doi: 10.1007/s13277-014-2736-9
37. Lemos C, Hardt MS, Juneja M, Voss C, Förster S, Jerchow B, et al. MACC1 induces tumor progression in transgenic mice and colorectal cancer patients via increased pluripotency markers nanog and oct4. Clin Cancer Res. (2016) 22:2812–24. doi: 10.1158/1078-0432.CCR-15-1425
38. Son H-Y, Heo WH, Li SB, Quan M, Her YJ, Lee K-J, et al. Abstract 6241: Macc1 regulates breast cancer progression and metastasis via remodeling tumor immune microenvironment. Cancer Res. (2022) 82:6241. doi: 10.1158/1538-7445.AM2022-6241
39. Meng F, Li H, Shi H, Yang Q, Zhang F, Yang Y, et al. MACC1 down-regulation inhibits proliferation and tumourigenicity of nasopharyngeal carcinoma cells through Akt/β-catenin signaling pathway. PloS One. (2013) 8:e60821. doi: 10.1371/journal.pone.0060821
40. Lin L, Huang H, Liao W, Ma H, Liu J, Wang L, et al. More than metastasis: MACC1 as a glucose metabolic regulator in gastric cancer. JCO. (2014) 32:e15054–4. doi: 10.1200/jco.2014.32.15_suppl.e15054
41. Treese C, Werchan J, von Winterfeld M, Berg E, Hummel M, Timm L, et al. Inhibition of MACC1-induced metastasis in esophageal and gastric adenocarcinomas. Cancers (Basel). (2022) 14:1773. doi: 10.3390/cancers14071773
42. Sun B, Cheng Z, Sun J. Associations of MACC1, AGR2, and KAI1 expression with the metastasis and prognosis in head and neck squamous cell carcinoma. Int J Clin Exp Pathol. (2018) 11:822–30.
43. Song W, Wang X, Yang R, Wu S, Wang D. The expression of metastasis-associated in colon cancer-1, Snail, and KAI1 in esophageal carcinoma and their clinical significance. Int J Clin Exp Pathol. (2019) 12:987–95.
44. Shi Y, Li M-Y, Wang H, Li C, Liu W-Y, Gao Y-M, et al. The relationship between MACC1/c-met/cyclin D1 axis expression and prognosis in ESCC. Anal Cell Pathol (Amst). (2022) 2022:9651503. doi: 10.1155/2022/9651503
45. Wu J, Zhang D, Li J, Deng X, Liang G, Long Y, et al. MACC1 induces autophagy to regulate proliferation, apoptosis, migration and invasion of squamous cell carcinoma. Oncol Rep. (2017) 38:2369–77. doi: 10.3892/or.2017.5889
46. Xue W, Shen Z, Li L, Zheng Y, Yan D, Kan Q, et al. Long non-coding RNAs MACC1-AS1 and FOXD2-AS1 mediate NSD2-induced cisplatin resistance in esophageal squamous cell carcinoma. Mol Ther Nucleic Acids. (2021) 23:592–602. doi: 10.1016/j.omtn.2020.12.007
47. Jabłońska-Trypuć A, Matejczyk M, Rosochacki S. Matrix metalloproteinases (MMPs), the main extracellular matrix (ECM) enzymes in collagen degradation, as a target for anticancer drugs. J Enzyme Inhib Med Chem. (2016) 31:177–83. doi: 10.3109/14756366.2016.1161620
48. Yu X, He T, Tong Z, Liao L, Huang S, Fakhouri WD, et al. Molecular mechanisms of TWIST1-regulated transcription in EMT and cancer metastasis. EMBO Rep. (2023) 24(11):e56902. doi: 10.15252/embr.202356902
49. Golyan FF, Abbaszadegan MR, Forghanifard MM. TWIST1, MMP-21, and HLAG-1 co-overexpression is associated with ESCC aggressiveness. J Cell Biochem. (2019) 120:14838–46. doi: 10.1002/jcb.28745
50. Mahmoudian RA, Gharaie ML, Abbaszadegan MR, Alasti A, Forghanifard MM, Mansouri A, et al. Crosstalk between MMP-13, CD44, and TWIST1 and its role in regulation of EMT in patients with esophageal squamous cell carcinoma. Mol Cell Biochem. (2021) 476:2465–78. doi: 10.1007/s11010-021-04089-2
51. Fardi Golyan F, Forghanifard MM. A new gene panel as a marker for ESCC poor prognosis; INPP5A, TWIST1, MMP2, and EGFR. Adv Med Sci. (2021) 66:231–6. doi: 10.1016/j.advms.2021.03.004
52. Huang L, Xu A-M, Peng Q. CD147 and MMP-9 expressions in type II/III adenocarcinoma of esophagogastric junction and their clinicopathological significances. Int J Clin Exp Pathol. (2015) 8:1929–37.
53. Fahim Y, Yousefi M, Izadpanah MH, Forghanifard MM. TWIST1 correlates with Notch signaling pathway to develop esophageal squamous cell carcinoma. Mol Cell Biochem. (2020) 474:181–8. doi: 10.1007/s11010-020-03843-2
54. Forghanifard MM, Azaraz S, Ardalan Khales S, Morshedi Rad D, Abbaszadegan MR. MAML1 promotes ESCC aggressiveness through upregulation of EMT marker TWIST1. Mol Biol Rep. (2020) 47:2659–68. doi: 10.1007/s11033-020-05356-z
55. Ardalan Khales S, Ebrahimi E, Jahanzad E, Ardalan Khales S, Forghanifard MM. MAML1 and TWIST1 co-overexpression promote invasion of head and neck squamous cell carcinoma. Asia Pac J Clin Oncol. (2018) 14:e434–41. doi: 10.1111/ajco.12843
56. Liu H-B, Yang Q-C, Shen Y, Zhu Y, Zhang X-J, Chen H. A disintegrin and metalloproteinase 17 mRNA and protein expression in esophageal squamous cell carcinoma, as well as its clinicopathological factors and prognosis. Mol Med Rep. (2015) 11:961–7. doi: 10.3892/mmr.2014.2802
57. Ishibashi Y, Mochizuki S, Horiuchi K, Tsujimoto H, Kouzu K, Kishi Y, et al. Periostin derived from cancer-associated fibroblasts promotes esophageal squamous cell carcinoma progression via ADAM17 activation. Biochim Biophys Acta Mol Basis Dis. (2023) 1869:166669. doi: 10.1016/j.bbadis.2023.166669
58. Rios-Doria J, Sabol D, Chesebrough J, Stewart D, Xu L, Tammali R, et al. A monoclonal antibody to ADAM17 inhibits tumor growth by inhibiting EGFR and non-EGFR-mediated pathways. Mol Cancer Ther. (2015) 14:1637–49. doi: 10.1158/1535-7163.MCT-14-1040
59. Jain R, Gosavi S, Sethia D, Trimuke A, Salunke M. Evaluation of expression of ADAM 10 as a predictor of lymph node metastasis in oral squamous cell carcinoma-an immunohistochemical study. Head Neck Pathol. (2022) 16:1055–62. doi: 10.1007/s12105-022-01466-1
60. Li B, Jiang Y-P, Zhu J, Meng L. MiR-501-5p acts as an energetic regulator in head and neck squamous cell carcinoma cells growth and aggressiveness via reducing CLCA4. Mol Biol Rep. (2020) 47:2181–7. doi: 10.1007/s11033-020-05317-6
61. Song X, Zhang S, Li S, Wang Y, Zhang X, Xue F. Expression of the CLCA4 gene in esophageal carcinoma and its impact on the biologic function of esophageal carcinoma cells. J Oncol. (2021) 2021:1649344. doi: 10.1155/2021/1649344
62. Li Y, Wang J-X, Yibi R-H. Prediction of lymph node metastasis in early esophageal cancer. World J Gastrointest Surg. (2023) 15:2294–304. doi: 10.4240/wjgs.v15.i10.2294
63. Suzuki L, Ten Kate FJC, Gotink AW, Stoop H, Doukas M, Nieboer D, et al. Olfactomedin 4 (OLFM4) expression is associated with nodal metastases in esophageal adenocarcinoma. PloS One. (2019) 14:e0219494. doi: 10.1371/journal.pone.0219494
64. Lu J, Pan Y, Lei Y, Gu Y, Zhong F. Clinical significance of the expression of eukaryotic initiation factor 4 E and mammalian target of rapamycin in esophageal squamous carcinoma tissues. Zhonghua Wei Chang Wai Ke Za Zhi. (2015) 18:905–8. doi: 10.3760/cma.j.issn.1671-0274.2015.09.014
65. Yang J, Kong P, Yang J, Jia Z, Hu X, Wang Z, et al. High TSTA3 expression as a candidate biomarker for poor prognosis of patients with ESCC. Technol Cancer Res Treat. (2018) 17:1533033818781405. doi: 10.1177/1533033818781405
66. Luo G, Qi Y, Lei Z, Shen X, Chen M, Du L, et al. A potential biomarker of esophageal squamous cell carcinoma WTAP promotes the proliferation and migration of ESCC. Pathol Res Pract. (2022) 238:154114. doi: 10.1016/j.prp.2022.154114
67. Xiao Z, Jia Y, Jiang W, Wang Z, Zhang Z, Gao Y. FOXM1: A potential indicator to predict lymphatic metastatic recurrence in stage IIA esophageal squamous cell carcinoma. Thorac Cancer. (2018) 9:997–1004. doi: 10.1111/1759-7714.12776
68. Sun Z-G, Yu L, Gao W, Wang Z, Zhu L-M. Clinical and prognostic significance of MUC1 expression in patients with esophageal squamous cell carcinoma after radical resection. Saudi J Gastroenterol. (2018) 24:165–70. doi: 10.4103/sjg.SJG_420_17
69. Cao H-H, Zhang S-Y, Shen J-H, Wu Z-Y, Wu J-Y, Wang S-H, et al. A three-protein signature and clinical outcome in esophageal squamous cell carcinoma. Oncotarget. (2015) 6:5435–48. doi: 10.18632/oncotarget.3102
70. Zhu X, Jin H, Xia Z, Wu X, Yang M, Zhang H, et al. Vav1 expression is increased in esophageal squamous cell carcinoma and indicates poor prognosis. Biochem Biophys Res Commun. (2017) 486:571–6. doi: 10.1016/j.bbrc.2017.03.091
71. Huang Z-L, Lin Z-R, Xiao Y-R, Cao X, Zhu L-C, Zeng M-S, et al. High expression of TACC3 in esophageal squamous cell carcinoma correlates with poor prognosis. Oncotarget. (2015) 6:6850–61. doi: 10.18632/oncotarget.3190
72. Wei R, Shang Z, Leng J, Cui L. Increased expression of high-mobility group A2: A novel independent indicator of poor prognosis in patients with esophageal squamous cell carcinoma. J Cancer Res Ther. (2016) 12:1291–7. doi: 10.4103/0973-1482.180616
73. Zeng F-M, He J-Z, Wang S-H, Liu D-K, Xu X-E, Wu J-Y, et al. A novel three-gene model predicts prognosis and therapeutic sensitivity in esophageal squamous cell carcinoma. BioMed Res Int. (2019) 2019:9828637. doi: 10.1155/2019/9828637
74. Lin J-B, Feng Z, Qiu M-L, Luo R-G, Li X, Liu B. KRT 15 as a prognostic biomarker is highly expressed in esophageal carcinoma. Future Oncol. (2020) 16:1903–9. doi: 10.2217/fon-2019-0603
75. Stavniichuk R, DeLaForest A, Thompson CA, Miller J, Souza RF, Battle MA. GATA4 blocks squamous epithelial cell gene expression in human esophageal squamous cells. Sci Rep. (2021) 11:3206. doi: 10.1038/s41598-021-82557-x
76. Gao Y, Yuan L, Ke C, Pei Z, Liu X, Wu R, et al. Caprin-1 plays a role in cell proliferation and Warburg metabolism of esophageal carcinoma by regulating METTL3 and WTAP. J Transl Med. (2023) 21:159. doi: 10.1186/s12967-023-04001-0
77. Jiang M, Wu Y, Qi L, Li L, Song D, Gan J, et al. Dihydroartemisinin mediating PKM2-caspase-8/3-GSDME axis for pyroptosis in esophageal squamous cell carcinoma. Chem Biol Interact. (2021) 350:109704. doi: 10.1016/j.cbi.2021.109704
78. Chai J, Lei Y, Xiang X, Ye J, Zhao H, Yi L. High expression of caspase-8 as a predictive factor of poor prognosis in patients with esophageal cancer. Cancer Med. (2023) 12:7651–66. doi: 10.1002/cam4.5496
79. Jiang W, Huang S, Song L, Wang Z. STMN1, a prognostic predictor of esophageal squamous cell carcinoma, is a marker of the activation of the PI3K pathway. Oncol Rep. (2018) 39:834–42. doi: 10.3892/or.2017.6145
80. Kong F, Shang Y, Diao X, Huang J, Liu H. Knockdown of NOLC1 inhibits PI3K-AKT pathway to improve the poor prognosis of esophageal carcinoma. J Oncol. (2021) 2021:9944132. doi: 10.1155/2021/9944132
81. Zhou X, Li Y, Wang W, Wang S, Hou J, Zhang A, et al. Regulation of Hippo/YAP signaling and Esophageal Squamous Carcinoma progression by an E3 ubiquitin ligase PARK2. Theranostics. (2020) 10:9443–57. doi: 10.7150/thno.46078
82. Chiu W-C, Fang P-T, Lee Y-C, Wang Y-Y, Su Y-H, Hu SC-S, et al. DNA Repair Protein Rad51 Induces Tumor Growth and Metastasis in Esophageal Squamous Cell Carcinoma via a p38/Akt-Dependent Pathway. Ann Surg Oncol. (2020) 27:2090–101. doi: 10.1245/s10434-019-08043-x
83. Cao L-J, Zhang Y-J, Dong S-Q, Li X-Z, Tong X-T, Chen D, et al. ATAD2 interacts with C/EBPβ to promote esophageal squamous cell carcinoma metastasis via TGF-β1/Smad3 signaling. J Exp Clin Cancer Res. (2021) 40:109. doi: 10.1186/s13046-021-01905-x
84. Li H, Wang S, Li X, Weng Y, Guo D, Kong P, et al. CDCA7 promotes TGF-β-induced epithelial-mesenchymal transition via transcriptionally regulating Smad4/Smad7 in ESCC. Cancer Sci. (2023) 114:91–104. doi: 10.1111/cas.15560
85. Xiao Z, Tian Y, Jia Y, Shen Q, Jiang W, Chen G, et al. RUNX3 inhibits the invasion and migration of esophageal squamous cell carcinoma by reversing the epithelial−mesenchymal transition through TGF−β/Smad signaling. Oncol Rep. (2020) 43:1289–99. doi: 10.3892/or.2020.7508
86. Rehman AU, Iqbal MA, Sattar RSA, Saikia S, Kashif M, Ali WM, et al. Elevated expression of RUNX3 co-expressing with EZH2 in esophageal cancer patients from India. Cancer Cell Int. (2020) 20:445. doi: 10.1186/s12935-020-01534-y
87. Mao X-M, Li H, Zhang X-Y, Zhou P, Fu Q-R, Chen Q-E, et al. Retinoic acid receptor α Knockdown suppresses the tumorigenicity of esophageal carcinoma via wnt/β-catenin pathway. Dig Dis Sci. (2018) 63:3348–58. doi: 10.1007/s10620-018-5254-6
88. Bi Y, Guo S, Xu X, Kong P, Cui H, Yan T, et al. Decreased ZNF750 promotes angiogenesis in a paracrine manner via activating DANCR/miR-4707-3p/FOXC2 axis in esophageal squamous cell carcinoma. Cell Death Dis. (2020) 11:296. doi: 10.1038/s41419-020-2492-2
89. Zhou X, Huang F, Ma G, Wei W, Wu N, Liu Z. Dysregulated ceramides metabolism by fatty acid 2-hydroxylase exposes a metabolic vulnerability to target cancer metastasis. Signal Transduct Target Ther. (2022) 7:370. doi: 10.1038/s41392-022-01199-1
90. Ma L, Yang R, Gu J, Jiang H, Li H. The expression of AGGF1, FOXC2, and E-cadherin in esophageal carcinoma and their clinical significance. Med (Baltimore). (2020) 99:e22173. doi: 10.1097/MD.0000000000022173
91. Zhang X, Liu T, Zheng S, Liu Q, Shen T, Han X, et al. SUMOylation of HSP27 regulates PKM2 to promote esophageal squamous cell carcinoma progression. Oncol Rep. (2020) 44:1355–64. doi: 10.3892/or.2020.7711
92. Ma R, Liu Q, Zheng S, Liu T, Tan D, Lu X. PKM2-regulated STAT3 promotes esophageal squamous cell carcinoma progression via TGF-β1-induced EMT. J Cell Biochem. (2019) 120:11539–50. doi: 10.1002/jcb.28434
93. Liu R, Hong R, Wang Y, Gong Y, Yeerken D, Yang D, et al. Defect of SLC38A3 promotes epithelial-mesenchymal transition and predicts poor prognosis in esophageal squamous cell carcinoma. Chin J Cancer Res. (2020) 32:547–63. doi: 10.21147/j.issn.1000-9604.2020.05.01
94. Yang H, Zhou J, He K, Li J, Zhao F, Dai N, et al. Low RCAN1.2 mRNA expression is associated with poor prognosis of patients with esophageal squamous cell carcinoma. J Cancer. (2023) 14:2361–72. doi: 10.7150/jca.84307
95. Wu J, Liu L, Wu F, Qiu L, Luo M, Ke Q, et al. Clinical and prognostic implications of P21 (WAF1/CIP1) expression in patients with esophageal cancer: A systematic review and meta-analysis. Dis Markers. (2020) 2020:6520259. doi: 10.1155/2020/6520259
96. You Y-J, Chen Y-P, Zheng X-X, Meltzer SJ, Zhang H. Aberrant methylation of the PTPRO gene in peripheral blood as a potential biomarker in esophageal squamous cell carcinoma patients. Cancer Lett. (2012) 315:138–44. doi: 10.1016/j.canlet.2011.08.032
97. Zhang G, Liu X, Wu J, Zhu Q, Zeng H, Wang T, et al. Expression and clinical relations of protein tyrosine phosphatase receptor type S in esophageal squamous cell carcinoma. Histol Histopathol. (2018) 33:1181–8. doi: 10.14670/HH-18-002
98. Dong H, Lin W, Du L, Yao Z, Li F, Chen S, et al. PTPRO suppresses lymph node metastasis of esophageal carcinoma by dephosphorylating MET. Cancer Lett. (2023) 567:216283. doi: 10.1016/j.canlet.2023.216283
99. Yang H, Chen Y, Huang X, Gu Y, Chen Z, Mao W. Bioinformatics analysis reveals a novel prognostic model for esophageal squamous cell carcinoma. Int J Med Sci. (2024) 21:1213–26. doi: 10.7150/ijms.93423
100. Lu T, Ma K, Zhan C, Yang X, Shi Y, Jiang W, et al. Downregulation of long non-coding RNA LINP1 inhibits the Malignant progression of esophageal squamous cell carcinoma. Ann Transl Med. (2020) 8:675. doi: 10.21037/atm-20-1009
101. Wang G, Feng B, Niu Y, Wu J, Yang Y, Shen S, et al. A novel long noncoding RNA, LOC440173, promotes the progression of esophageal squamous cell carcinoma by modulating the miR-30d-5p/HDAC9 axis and the epithelial-mesenchymal transition. Mol Carcinog. (2020) 59:1392–408. doi: 10.1002/mc.23264
102. Wang J, Sun D, Wu K, Liu J, Zhao M, Li X, et al. Genome-wide analysis of long non-coding RNAs in esophageal squamous cell carcinoma reveals their potential role in invasion and metastasis. Thorac Cancer. (2019) 10:78–89. doi: 10.1111/1759-7714.12904
103. Zhang Y, Li R, Ding X, Zhang K, Qin W. Upregulation of long non-coding RNA SNHG6 promote esophageal squamous cell carcinoma cell Malignancy and its diagnostic value. Am J Transl Res. (2019) 11:1084–91.
104. Du F, Guo T, Cao C. Silencing of Long Noncoding RNA SNHG6 Inhibits Esophageal Squamous Cell Carcinoma Progression via miR-186-5p/HIF1α Axis. Dig Dis Sci. (2020) 65:2844–52. doi: 10.1007/s10620-019-06012-8
105. Ma J, Xiao Y, Tian B, Chen S, Zhang B, Wu J, et al. Long noncoding RNA lnc-ABCA12-3 promotes cell migration, invasion, and proliferation by regulating fibronectin 1 in esophageal squamous cell carcinoma. J Cell Biochem. (2020) 121:1374–87. doi: 10.1002/jcb.29373
106. Ma J, Luo Y, Liu Y, Chen C, Chen A, Liang L, et al. Exosome-mediated lnc-ABCA12-3 promotes proliferation and glycolysis but inhibits apoptosis by regulating the toll-like receptor 4/nuclear factor kappa-B signaling pathway in esophageal squamous cell carcinoma. Korean J Physiol Pharmacol. (2023) 27:61–73. doi: 10.4196/kjpp.2023.27.1.61
107. Wasson M-CD, Brown JM, Venkatesh J, Fernando W, Marcato P. Datasets exploring putative lncRNA-miRNA-mRNA axes in breast cancer cell lines. Data Brief. (2021) 37:107241. doi: 10.1016/j.dib.2021.107241
108. Tan Z, Zhou P, Zhu Z, Wang Y, Guo Z, Shen M, et al. Upregulated long non−coding RNA LincIN promotes tumor progression via the regulation of nuclear factor 90/microRNA−7/HOXB13 in esophageal squamous cell carcinoma. Int J Mol Med. (2021) 47:78. doi: 10.3892/ijmm.2021.4911
109. Li R-C, Ke S, Meng F-K, Lu J, Zou X-J, He Z-G, et al. CiRS-7 promotes growth and metastasis of esophageal squamous cell carcinoma via regulation of miR-7/HOXB13. Cell Death Dis. (2018) 9:838. doi: 10.1038/s41419-018-0852-y
110. Qie P, Yin Q, Xun X, Song Y, Zhou S, Liu H, et al. Long non-coding RNA SPRY4-IT1 as a promising indicator for three field lymph-node dissection of thoracic esophageal carcinoma. J Cardiothorac Surg. (2021) 16:48. doi: 10.1186/s13019-021-01433-x
111. Ghafouri-Fard S, Khoshbakht T, Taheri M, Shojaei S. A review on the role of SPRY4-IT1 in the carcinogenesis. Front Oncol. (2021) 11:779483. doi: 10.3389/fonc.2021.779483
112. Xue-Liang J, Ming-Dong W, Ya-Bi Z, Wang-Yue W. Upregulated long noncoding RNA SPRY4-IT1 contributes to increased cell viability by activating zinc finger 703 expression in esophageal squamous cell carcinoma. Indian J Cancer. (2015) 52 Suppl 3:E164–167. doi: 10.4103/0019-509X.186566
113. Zhang C-Y, Li R-K, Qi Y, Li X-N, Yang Y, Liu D-L, et al. Upregulation of long noncoding RNA SPRY4-IT1 promotes metastasis of esophageal squamous cell carcinoma via induction of epithelial-mesenchymal transition. Cell Biol Toxicol. (2016) 32:391–401. doi: 10.1007/s10565-016-9341-1
114. Chen M-J, Deng J, Chen C, Hu W, Yuan Y-C, Xia Z-K. LncRNA H19 promotes epithelial mesenchymal transition and metastasis of esophageal cancer via STAT3/EZH2 axis. Int J Biochem Cell Biol. (2019) 113:27–36. doi: 10.1016/j.biocel.2019.05.011
115. Li M-K, Liu L-X, Zhang W-Y, Zhan H-L, Chen R-P, Feng J-L, et al. Long non−coding RNA MEG3 suppresses epithelial−to−mesenchymal transition by inhibiting the PSAT1−dependent GSK−3β/Snail signaling pathway in esophageal squamous cell carcinoma. Oncol Rep. (2020) 44:2130–42. doi: 10.3892/or.2020.7754
116. Sharma U, Kaur Rana M, Singh K, Jain A. LINC00324 promotes cell proliferation and metastasis of esophageal squamous cell carcinoma through sponging miR-493-5p via MAPK signaling pathway. Biochem Pharmacol. (2023) 207:115372. doi: 10.1016/j.bcp.2022.115372
117. Xue L, Nan J, Dong L, Zhang C, Li H, Na R, et al. Upregulated miR-483-5p expression as a prognostic biomarker for esophageal squamous cell carcinoma. Cancer biomark. (2017) 19:193–7. doi: 10.3233/CBM-160506
118. Li C, Li D-C, Che S-S, Ma K, Wang Y-J, Xia L-H, et al. The decreased expression of miR-625 predicts poor prognosis of esophageal squamous cell carcinoma. Int J Clin Exp Med. (2015) 8:9560–4.
119. Xu Y, Sun D, Zhang X, Liu J, Wang J, Cao Q, et al. Microarray analysis of miRNA based on the regional lymph node metastasis status of esophageal squamous cell carcinoma. Transl Cancer Res. (2021) 10:273–87. doi: 10.21037/tcr-20-1969
120. Liu T, Li P, Li J, Qi Q, Sun Z, Shi S, et al. Exosomal and intracellular miR-320b promotes lymphatic metastasis in esophageal squamous cell carcinoma. Mol Ther Oncolytics. (2021) 23:163–80. doi: 10.1016/j.omto.2021.09.003
121. Xiao Z, Feng X, Zhou Y, Li P, Luo J, Zhang W, et al. Exosomal miR-10527-5p Inhibits Migration, Invasion, Lymphangiogenesis and Lymphatic Metastasis by Affecting Wnt/β-Catenin Signaling via Rab10 in Esophageal Squamous Cell Carcinoma. Int J Nanomedicine. (2023) 18:95–114. doi: 10.2147/IJN.S391173
122. Zhong H, Xu Y, Wang J, Cao Q, Hu L, Sun D. Overexpression of microRNA-19a-3p promotes lymph node metastasis of esophageal squamous cell carcinoma via the RAC1/CDC42-PAK1 pathway. Transl Cancer Res. (2021) 10:2694–706. doi: 10.21037/tcr-21-254
123. Ding Z, Yan Y, Guo YL, Wang C. Esophageal carcinoma cell-excreted exosomal uc.189 promotes lymphatic metastasis. Aging (Albany NY). (2021) 13:13846–58. doi: 10.18632/aging.202979
124. He R, Wang J, Ye K, Du J, Chen J, Liu W. Reduced miR-203 predicts metastasis and poor survival in esophageal carcinoma. Aging (Albany NY). (2019) 11:12114–30. doi: 10.18632/aging.102543
125. Guo Y, Wang C, Miao X, Chen S, Qian Y, Li G, et al. Upregulation of uc.189 in patients with esophageal squamous cell carcinoma and its clinicopathologic value. Pathol Res Pract. (2017) 213:1400–3. doi: 10.1016/j.prp.2017.09.005
126. Xia Y, Wang Y, Wang Q, Ghaffar M, Wang Y, Sheng W, et al. Increased miR-203-3p and reduced miR-21-5p synergistically inhibit proliferation, migration, and invasion in esophageal cancer cells. Anticancer Drugs. (2019) 30:38–45. doi: 10.1097/CAD.0000000000000687
127. Tang Y, Liu JH, Shi ZX, Li Z, Liu HT, Lu P. MicroRNA-133b suppresses cell proliferation and invasion of esophageal squamous cell carcinoma via downregulating TAGLN2 expression. Zhonghua Zhong Liu Za Zhi. (2019) 41:91–6. doi: 10.3760/cma.j.issn.0253-3766.2019.02.003
128. Wada M, Goto Y, Tanaka T, Okada R, Moriya S, Idichi T, et al. RNA sequencing-based microRNA expression signature in esophageal squamous cell carcinoma: oncogenic targets by antitumor miR-143-5p and miR-143-3p regulation. J Hum Genet. (2020) 65:1019–34. doi: 10.1038/s10038-020-0795-x
129. Zhao H, Chen W, Wu J, Wang L, Mao W. Clinical significance of preoperative serum tumor markers in esophageal squamous cell carcinoma. J Cancer Res Ther. (2014) 10 Suppl:C179–85. doi: 10.4103/0973-1482.145863
130. Li J, Qi Z, Hu Y-P, Wang Y-X. Possible biomarkers for predicting lymph node metastasis of esophageal squamous cell carcinoma: a review. J Int Med Res. (2019) 47:544–56. doi: 10.1177/0300060518819606
131. Sun Z, Ji N, Bi M, Zhang Z, Liu X, Wang Z. Negative expression of PTEN identifies high risk for lymphatic-related metastasis in human esophageal squamous cell carcinoma. Oncol Rep. (2015) 33:3024–32. doi: 10.3892/or.2015.3928
132. Yan L, Dong X, Gao J, Liu F, Zhou L, Sun Y, et al. A novel rapid quantitative method reveals stathmin-1 as a promising marker for esophageal squamous cell carcinoma. Cancer Med. (2018) 7:1802–13. doi: 10.1002/cam4.1449
133. Xu Y-W, Chen H, Guo H-P, Yang S-H, Luo Y-H, Liu C-T, et al. Combined detection of serum autoantibodies as diagnostic biomarkers in esophagogastric junction adenocarcinoma. Gastric Cancer. (2019) 22:546–57. doi: 10.1007/s10120-018-0894-y
134. Feng J-F, Wang L, Yang X, Jiang Y-H. Prognostic value of lactate dehydrogenase to albumin ratio (LAR) in patients with resectable esophageal squamous cell carcinoma. Cancer Manag Res. (2019) 11:7243–51. doi: 10.2147/CMAR.S208320
135. Shiratori F, Suzuki T, Yajima S, Oshima Y, Nanami T, Funahashi K, et al. Is high score of preoperative lactate dehydrogenase to albumin ratio predicting poor survivals in esophageal carcinoma patients? Ann Thorac Cardiovasc Surg. (2023) 29:215–22. doi: 10.5761/atcs.oa.23-00004
136. Xu H, Lin T, Ai J, Zhang J, Zhang S, Li Y, et al. Utilizing the lactate dehydrogenase-to-albumin ratio for survival prediction in patients with bladder cancer after radical cystectomy. J Inflammation Res. (2023) 16:1733–44. doi: 10.2147/JIR.S384338
137. Xie Z, Zhou H, Wang L, Wu Y. The Significance of the preoperative lactate dehydrogenase/albumin Ratio in the Prognosis of Colon Cancer: a retrospective study. PeerJ. (2022) 10:e13091. doi: 10.7717/peerj.13091
138. Karno B, Edwards DN, Chen J. Metabolic control of cancer metastasis: role of amino acids at secondary organ sites. Oncogene. (2023) 42:3447–56. doi: 10.1038/s41388-023-02868-3
139. Jin H, Qiao F, Chen L, Lu C, Xu L, Gao X. Serum metabolomic signatures of lymph node metastasis of esophageal squamous cell carcinoma. J Proteome Res. (2014) 13:4091–103. doi: 10.1021/pr500483z
140. Fong LY, Huebner K, Jing R, Smalley KJ, Brydges CR, Fiehn O, et al. Zinc treatment reverses and anti-Zn-regulated miRs suppress esophageal carcinomas in vivo. Proc Natl Acad Sci U.S.A. (2023) 120:e2220334120. doi: 10.1073/pnas.2220334120
141. Jiang Z, Song Q, Yang S, Zeng R, Li X, Jiang C, et al. Serum microRNA-218 is a potential biomarker for esophageal cancer. Cancer biomark. (2015) 15:381–9. doi: 10.3233/CBM-150480
142. Dong S, Yin H, Dong C, Sun K, Lv P, Meng W, et al. Predictive value of plasma microRNA-216a/b in the diagnosis of esophageal squamous cell carcinoma. Dis Markers. (2016) 2016:1857067. doi: 10.1155/2016/1857067
143. Xue L, Zhao Z, Wang M, Ma L, Lin H, Wang S, et al. A liquid biopsy signature predicts lymph node metastases in T1 oesophageal squamous cell carcinoma: implications for precision treatment strategy. Br J Cancer. (2022) 127(11):2052–9. doi: 10.1038/s41416-022-01997-y
144. Fan L, Cao Q, Liu J, Zhang J, Li B. Circular RNA profiling and its potential for esophageal squamous cell cancer diagnosis and prognosis. Mol Cancer. (2019) 18:16. doi: 10.1186/s12943-018-0936-4
145. Lambert AW, Pattabiraman DR, Weinberg RA. Emerging biological principles of metastasis. Cell. (2017) 168:670–91. doi: 10.1016/j.cell.2016.11.037
146. Ohsawa M, Hamai Y, Emi M, Ibuki Y, Kurokawa T, Yoshikawa T, et al. Blood biomarkers as predictors of pathological lymph node metastasis in clinical stage T1N0 esophageal squamous cell carcinoma. Dis Esophagus. (2022) 36:doac042. doi: 10.1093/dote/doac042
147. Tustumi F, Takeda FR, Viyuela MS, da Cruz Junior JB, Brandão AAGS, Sallum RAA, et al. The value of cellular components of blood in the setting of trimodal therapy for esophageal cancer. J Surg Oncol. (2020) 121:784–94. doi: 10.1002/jso.25778
148. Hirahara N, Tajima Y, Fujii Y, Hyakudomi R, Yamamoto T, Ishitobi K, et al. Prognostic significance of red cell distribution width in esophageal squamous cell carcinoma. J Surg Res. (2018) 230:53–60. doi: 10.1016/j.jss.2018.04.051
149. Schernberg A, Moureau-Zabotto L, Rivin Del Campo E, Escande A, Ducreux M, Nguyen F, et al. Leukocytosis and neutrophilia predict outcome in locally advanced esophageal cancer treated with definitive chemoradiation. Oncotarget. (2017) 8:11579–88. doi: 10.18632/oncotarget.14584
150. Lv Y, Zhang J, Liu Z, Tian Y, Liu F. A novel inflammation-based prognostic index for patients with esophageal squamous cell carcinoma: Neutrophil lymphocyte ratio/prealbumin ratio. Med (Baltimore). (2019) 98:e14562. doi: 10.1097/MD.0000000000014562
151. Zhao Q, Chen S, Feng J-F. A novel inflammation-based prognostic index for patients with esophageal squamous cell carcinoma: neutrophil lymphocyte ratio/albumin ratio. Oncotarget. (2017) 8:103535–42. doi: 10.18632/oncotarget.21989
152. Li Y, Wang Z, Fu R, Wang S, Zhang T, Tian X, et al. Clinical utility of circulating tumor cells in patients with esophageal cancer. Front Oncol. (2022) 12:828368. doi: 10.3389/fonc.2022.828368
153. Xu H-T, Miao J, Liu J-W, Zhang L-G, Zhang Q-G. Prognostic value of circulating tumor cells in esophageal cancer. World J Gastroenterol. (2017) 23:1310–8. doi: 10.3748/wjg.v23.i7.1310
154. Zhang Y, Deng H, Chen G, Tang Z, Mao J, Mi Y, et al. Clinicopathological and prognostic value of circulating tumor cells in esophageal carcinoma: a meta-analysis. Ann Palliat Med. (2020) 9:4271–82. doi: 10.21037/apm-20-590
155. Gao Y, Fan W-H, Song Z, Lou H, Kang X. Comparison of circulating tumor cell (CTC) detection rates with epithelial cell adhesion molecule (EpCAM) and cell surface vimentin (CSV) antibodies in different solid tumors: a retrospective study. PeerJ. (2021) 9:e10777. doi: 10.7717/peerj.10777
156. Gao X, Leow OQ-Y, Chiu C-H, Hou M-M, Hsieh JC-H, Chao Y-K. Clinical utility of circulating tumor cells for predicting major histopathological response after neoadjuvant chemoradiotherapy in patients with esophageal cancer. J Pers Med. (2022) 12:1440. doi: 10.3390/jpm12091440
157. Xu H, Wu J, Zhang L, Li Y, Gao L, Cheng Y. The measurement of NRF2 and TP53 in blood expects radiotherapeutic sensitivity in patients with esophageal cancer. Mol Cell Probes. (2022) 66:101860. doi: 10.1016/j.mcp.2022.101860
158. Lin D, Shen L, Luo M, Zhang K, Li J, Yang Q, et al. Circulating tumor cells: biology and clinical significance. Signal Transduct Target Ther. (2021) 6:404. doi: 10.1038/s41392-021-00817-8
159. de Gouw DJJM, Rijpkema M, de Bitter TJJ, Baart VM, Sier CFM, Hernot S, et al. Identifying biomarkers in lymph node metastases of esophageal adenocarcinoma for tumor-targeted imaging. Mol Diagn Ther. (2020) 24:191–200. doi: 10.1007/s40291-020-00448-9
160. Wu J, Wang W-X, Zhou Y, Yang D-S, Wu Z-N, Li X, et al. Expression of PTP4A1 in circulating tumor cells and its efficacy evaluation in patients with early- and intermediate-stage esophageal cancer. Med (Baltimore). (2023) 102:e36603. doi: 10.1097/MD.0000000000036603
161. Li H-X, Wang S-Q, Lian Z-X, Deng S-L, Yu K. Relationship between tumor infiltrating immune cells and tumor metastasis and its prognostic value in cancer. Cells. (2022) 12:64. doi: 10.3390/cells12010064
162. Cui K, Hu S, Mei X, Cheng M. Innate immune cells in the esophageal tumor microenvironment. Front Immunol. (2021) 12:654731. doi: 10.3389/fimmu.2021.654731
163. Baba Y, Nomoto D, Okadome K, Ishimoto T, Iwatsuki M, Miyamoto Y, et al. Tumor immune microenvironment and immune checkpoint inhibitors in esophageal squamous cell carcinoma. Cancer Sci. (2020) 111:3132–41. doi: 10.1111/cas.14541
164. Zhou X, Ren T, Zan H, Hua C, Guo X. Novel immune checkpoints in esophageal cancer: from biomarkers to therapeutic targets. Front Immunol. (2022) 13:864202. doi: 10.3389/fimmu.2022.864202
165. Wang J, Ling R, Zhou Y, Gao X, Yang Y, Mao C, et al. SREBP1 silencing inhibits the proliferation and motility of human esophageal squamous carcinoma cells via the Wnt/β-catenin signaling pathway. Oncol Lett. (2020) 20:2855–69. doi: 10.3892/ol.2020.11853
166. Zhang L-N, Zhao L, Yan X-L, Huang Y-H. Loss of G3BP1 suppresses proliferation, migration, and invasion of esophageal cancer cells via Wnt/β-catenin and PI3K/AKT signaling pathways. J Cell Physiol. (2019) 234:20469–84. doi: 10.1002/jcp.28648
167. Zhu Y, Li M, Mu D, Kong L, Zhang J, Zhao F, et al. CD8+/FOXP3+ ratio and PD-L1 expression associated with survival in pT3N0M0 stage esophageal squamous cell cancer. Oncotarget. (2016) 7:71455–65. doi: 10.18632/oncotarget.12213
168. Konno-Kumagai T, Fujishima F, Nakamura Y, Nakano T, Nagai T, Kamei T, et al. Programmed death-1 ligands and tumor infiltrating T lymphocytes in primary and lymph node metastasis of esophageal cancer patients. Dis Esophagus. (2019) 32:doy063. doi: 10.1093/dote/doy063
169. Fassan M, Cavallin F, Guzzardo V, Kotsafti A, Scarpa M, Cagol M, et al. PD-L1 expression, CD8+ and CD4+ lymphocyte rate are predictive of pathological complete response after neoadjuvant chemoradiotherapy for squamous cell cancer of the thoracic esophagus. Cancer Med. (2019) 8:6036–48. doi: 10.1002/cam4.2359
170. Otoshi T, Nagano T, Tachihara M, Nishimura Y. Possible biomarkers for cancer immunotherapy. Cancers (Basel). (2019) 11:935. doi: 10.3390/cancers11070935
171. Chen M-F, Chen P-T, Chen W-C, Lu M-S, Lin P-Y, Lee KD. The role of PD-L1 in the radiation response and prognosis for esophageal squamous cell carcinoma related to IL-6 and T-cell immunosuppression. Oncotarget. (2016) 7:7913–24. doi: 10.18632/oncotarget.6861
172. Pan C, Wang Y, Liu Q, Hu Y, Fu J, Xie X, et al. Phenotypic profiling and prognostic significance of immune infiltrates in esophageal squamous cell carcinoma. Oncoimmunology. (2021) 10:1883890. doi: 10.1080/2162402X.2021.1883890
173. Wang XL, Liu K, Liu JH, Jiang XL, Qi LW, Xie YF, et al. High infiltration of CD68-tumor associated macrophages, predict poor prognosis in Kazakh esophageal cancer patients. Int J Clin Exp Pathol. (2017) 10:10282–92.
174. Cao W, Peters JH, Nieman D, Sharma M, Watson T, Yu J. Macrophage subtype predicts lymph node metastasis in oesophageal adenocarcinoma and promotes cancer cell invasion in vitro. Br J Cancer. (2015) 113:738–46. doi: 10.1038/bjc.2015.292
175. Moreira C, Figueiredo C, Ferreira RM. The role of the microbiota in esophageal cancer. Cancers (Basel). (2023) 15:2576. doi: 10.3390/cancers15092576
176. Lopetuso LR, Severgnini M, Pecere S, Ponziani FR, Boskoski I, Larghi A, et al. Esophageal microbiome signature in patients with Barrett’s esophagus and esophageal adenocarcinoma. PloS One. (2020) 15:e0231789. doi: 10.1371/journal.pone.0231789
177. Yang W, Chen C-H, Jia M, Xing X, Gao L, Tsai H-T, et al. Tumor-associated microbiota in esophageal squamous cell carcinoma. Front Cell Dev Biol. (2021) 9:641270. doi: 10.3389/fcell.2021.641270
178. Zhang J-W, Zhang D, Yin H-S, Zhang H, Hong K-Q, Yuan J-P, et al. Fusobacterium nucleatum promotes esophageal squamous cell carcinoma progression and chemoresistance by enhancing the secretion of chemotherapy-induced senescence-associated secretory phenotype via activation of DNA damage response pathway. Gut Microbes. (2023) 15:2197836. doi: 10.1080/19490976.2023.2197836
179. Shao D, Vogtmann E, Liu A, Qin J, Chen W, Abnet CC, et al. Microbial characterization of esophageal squamous cell carcinoma and gastric cardia adenocarcinoma from a high-risk region of China. Cancer. (2019) 125:3993–4002. doi: 10.1002/cncr.32403
180. Zhang S, Zhang S, Ma X, Zhan J, Pan C, Zhang H, et al. Intratumoral microbiome impacts immune infiltrates in tumor microenvironment and predicts prognosis in esophageal squamous cell carcinoma patients. Front Cell Infect Microbiol. (2023) 13:1165790. doi: 10.3389/fcimb.2023.1165790
181. Li M, Shao D, Fan Z, Qin J, Xu J, Huang Q, et al. Non-invasive early detection on esophageal squamous cell carcinoma and precancerous lesions by microbial biomarkers combining epidemiological factors in China. J Gastroenterol. (2024) 59:531–42. doi: 10.1007/s00535-024-02117-8
182. Liu Y, Lin Z, Lin Y, Chen Y, Peng X-E, He F, et al. Streptococcus and Prevotella are associated with the prognosis of oesophageal squamous cell carcinoma. J Med Microbiol. (2018) 67:1058–68. doi: 10.1099/jmm.0.000754
183. Hu M, Bai W, Zhao C, Wang J. Distribution of esophagus flora in esophageal squamous cell carcinoma and its correlation with clinicopathological characteristics. Transl Cancer Res. (2020) 9:3973–85. doi: 10.21037/tcr-20-1954
184. Li Z, Shi C, Zheng J, Guo Y, Fan T, Zhao H, et al. Fusobacterium nucleatum predicts a high risk of metastasis for esophageal squamous cell carcinoma. BMC Microbiol. (2021) 21:301. doi: 10.1186/s12866-021-02352-6
185. Chen Z, Huang L. Fusobacterium nucleatum carcinogenesis and drug delivery interventions. Adv Drug Delivery Rev. (2024) 209:115319. doi: 10.1016/j.addr.2024.115319
186. Wu Z, Guo J, Zhang Z, Gao S, Huang M, Wang Y, et al. Bacteroidetes promotes esophageal squamous carcinoma invasion and metastasis through LPS-mediated TLR4/Myd88/NF-κB pathway and inflammatory changes. Sci Rep. (2024) 14:12827. doi: 10.1038/s41598-024-63774-6
187. Utispan K, Pugdee K, Koontongkaew S. Porphyromonas gingivalis lipopolysaccharide-induced macrophages modulate proliferation and invasion of head and neck cancer cell lines. BioMed Pharmacother. (2018) 101:988–95. doi: 10.1016/j.biopha.2018.03.033
188. Helminen O, Huhta H, Leppänen J, Kauppila JH, Takala H, Lehenkari PP, et al. Nuclear localization of Toll-like receptor 5 in Barrett’s esophagus and esophageal adenocarcinoma is associated with metastatic behavior. Virchows Arch. (2016) 469:465–70. doi: 10.1007/s00428-016-1989-7
189. Huhta H, Helminen O, Lehenkari PP, Saarnio J, Karttunen TJ, Kauppila JH. Toll-like receptors 1, 2, 4 and 6 in esophageal epithelium, Barrett’s esophagus, dysplasia and adenocarcinoma. Oncotarget. (2016) 7:23658–67. doi: 10.18632/oncotarget.8151
190. Robinson W, Stone JK, Schischlik F, Gasmi B, Kelly MC, Seibert C, et al. Identification of intracellular bacteria from multiple single-cell RNA-seq platforms using CSI-Microbes. Sci Adv. (2024) 10:eadj7402. doi: 10.1126/sciadv.adj7402
191. Zhang N, Liu Y, Yang H, Liang M, Wang X, Wang M, et al. Clinical significance of fusobacterium nucleatum infection and regulatory T cell enrichment in esophageal squamous cell carcinoma. Pathol Oncol Res. (2021) 27:1609846. doi: 10.3389/pore.2021.1609846
192. Chinese Society of Clinical Oncology. Guidelines of Chinese Society of Clinical Oncology (CSCO) Esophageal Cancer. Beijing: People's Medical Publishing House. (2023).
193. Otto B, Koenig AM, Tolstonog GV, Jeschke A, Klaetschke K, Vashist YK, et al. Molecular changes in pre-metastatic lymph nodes of esophageal cancer patients. PloS One. (2014) 9:e102552. doi: 10.1371/journal.pone.0102552
194. Chen J, Cai W, Lin Y, Chen Y, Zheng Q, Pan J, et al. Patterns and rates of abdominal lymphatic metastasis following esophageal carcinoma. PloS One. (2017) 12:e0185424. doi: 10.1371/journal.pone.0185424
195. Akutsu Y, Kato K, Igaki H, Ito Y, Nozaki I, Daiko H, et al. The prevalence of overall and initial lymph node metastases in clinical T1N0 thoracic esophageal cancer: from the results of JCOG0502, a prospective multicenter study. Ann Surg. (2016) 264:1009–15. doi: 10.1097/SLA.0000000000001557
196. Li B, Hu H, Zhang Y, Zhang J, Miao L, Ma L, et al. Three-field versus two-field lymphadenectomy in transthoracic oesophagectomy for oesophageal squamous cell carcinoma: short-term outcomes of a randomized clinical trial. Br J Surg. (2020) 107:647–54. doi: 10.1002/bjs.11497
197. Crosby D, Bhatia S, Brindle KM, Coussens LM, Dive C, Emberton M, et al. Early detection of cancer. Science. (2022) 375:eaay9040. doi: 10.1126/science.aay9040
198. Cohen JD, Li L, Wang Y, Thoburn C, Afsari B, Danilova L, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. (2018) 359:926–30. doi: 10.1126/science.aar3247
199. Hao J, Liu W, Zhao C, Xia T. The diagnostic significance of 64-slice spiral CT combined with serological CA19-9, Bcl-2, CYFRA21-1 detection in thoracic esophageal carcinoma. Transl Cancer Res. (2021) 10:5383–9. doi: 10.21037/tcr-21-2522
200. Gao Y, Yuan L, Zeng J, Li F, Li X, Tan F, et al. eIF6 is potential diagnostic and prognostic biomarker that associated with 18F-FDG PET/CT features and immune signatures in esophageal carcinoma. J Transl Med. (2022) 20:303. doi: 10.1186/s12967-022-03503-7
201. Wang J, Guo Z, Sun F, Xu T, Wang J, Yu J. A novel ferroptosis-related gene signature to predict prognosis of esophageal carcinoma. J Oncol. (2022) 2022:7485435. doi: 10.1155/2022/7485435
202. Zhang X, Wu H, Niu J, Hu Y, Zhang W, Chang J, et al. A novel mitochondria-related gene signature in esophageal carcinoma: prognostic, immune, and therapeutic features. Funct Integr Genomics. (2023) 23:109. doi: 10.1007/s10142-023-01030-2
203. Xia M, Shao J, Qiao M, Luo Z, Deng X, Ke Q, et al. Identification of LCA-binding glycans as a novel biomarker for esophageal cancer metastasis using a lectin array-based strategy. J Cancer. (2020) 11:4736–45. doi: 10.7150/jca.43806
204. Peinado H, Zhang H, Matei IR, Costa-Silva B, Hoshino A, Rodrigues G, et al. Pre-metastatic niches: organ-specific homes for metastases. Nat Rev Cancer. (2017) 17:302–17. doi: 10.1038/nrc.2017.6
205. Fares J, Fares MY, Khachfe HH, Salhab HA, Fares Y. Molecular principles of metastasis: a hallmark of cancer revisited. Sig Transduct Target Ther. (2020) 5:1–17. doi: 10.1038/s41392-020-0134-x
206. Doglioni G, Parik S, Fendt S-M. Interactions in the (Pre)metastatic niche support metastasis formation. Front Oncol. (2019) 9:219. doi: 10.3389/fonc.2019.00219
207. Kiya Y, Yoshioka Y, Nagakawa Y, Ochiya T. Extracellular vesicles are important mediators that regulate tumor lymph node metastasis via the immune system. Int J Mol Sci. (2023) 24:1362. doi: 10.3390/ijms24021362
208. Chen G, Huang AC, Zhang W, Zhang G, Wu M, Xu W, et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature. (2018) 560:382–6. doi: 10.1038/s41586-018-0392-8
209. Hitch TCA, Hall LJ, Walsh SK, Leventhal GE, Slack E, de Wouters T, et al. Microbiome-based interventions to modulate gut ecology and the immune system. Mucosal Immunol. (2022) 15:1095–113. doi: 10.1038/s41385-022-00564-1
210. Panebianco C, Ciardiello D, Villani A, Maiorano BA, Latiano TP, Maiello E, et al. Insights into the role of gut and intratumor microbiota in pancreatic ductal adenocarcinoma as new key players in preventive, diagnostic and therapeutic perspective. Semin Cancer Biol. (2022) 86:997–1007. doi: 10.1016/j.semcancer.2021.11.007
211. Fu A, Yao B, Dong T, Cai S. Emerging roles of intratumor microbiota in cancer metastasis. Trends Cell Biol. (2023) 33:583–93. doi: 10.1016/j.tcb.2022.11.007
212. Parida S, Wu S, Siddharth S, Wang G, Muniraj N, Nagalingam A, et al. A procarcinogenic colon microbe promotes breast tumorigenesis and metastatic progression and concomitantly activates notch and β-catenin axes. Cancer Discovery. (2021) 11:1138–57. doi: 10.1158/2159-8290.CD-20-0537
213. Chiba A, Bawaneh A, Velazquez C, Clear KYJ, Wilson AS, Howard-McNatt M, et al. Neoadjuvant chemotherapy shifts breast tumor microbiota populations to regulate drug responsiveness and the development of metastasis. Mol Cancer Res. (2020) 18:130–9. doi: 10.1158/1541-7786.MCR-19-0451
214. Fu A, Yao B, Dong T, Chen Y, Yao J, Liu Y, et al. Tumor-resident intracellular microbiota promotes metastatic colonization in breast cancer. Cell. (2022) 185:1356–1372.e26. doi: 10.1016/j.cell.2022.02.027
215. Peters BA, Wu J, Pei Z, Yang L, Purdue MP, Freedman ND, et al. Oral microbiome composition reflects prospective risk for esophageal cancers. Cancer Res. (2017) 77:6777–87. doi: 10.1158/0008-5472.CAN-17-1296
216. Gilbert JA, Blaser MJ, Caporaso JG, Jansson JK, Lynch SV, Knight R. Current understanding of the human microbiome. Nat Med. (2018) 24:392–400. doi: 10.1038/nm.4517
217. Busing JD, Buendia M, Choksi Y, Hiremath G, Das SR. Microbiome in eosinophilic esophagitis-metagenomic, metatranscriptomic, and metabolomic changes: A systematic review. Front Physiol. (2021) 12:731034. doi: 10.3389/fphys.2021.731034
218. Yang Y, Shen J, Deng P, Chen P. Mechanism investigation of Forsythoside A against esophageal squamous cell carcinoma in vitro and in vivo. Cancer Biol Ther. (2024) 25:2380023. doi: 10.1080/15384047.2024.2380023
219. Weh KM, Howard CL, Zhang Y, Tripp BA, Clarke JL, Howell AB, et al. Prebiotic proanthocyanidins inhibit bile reflux-induced esophageal adenocarcinoma through reshaping the gut microbiome and esophageal metabolome. JCI Insight. (2024) 9:e168112. doi: 10.1172/jci.insight.168112
Keywords: lymph node metastasis, esophageal cancer, molecular mechanism, biomarker, diagnosis
Citation: Wu N, Cai J, Jiang J, Lin Y, Wang X, Zhang W, Kang M and Zhang P (2024) Biomarkers of lymph node metastasis in esophageal cancer. Front. Immunol. 15:1457612. doi: 10.3389/fimmu.2024.1457612
Received: 01 July 2024; Accepted: 12 September 2024;
Published: 27 September 2024.
Edited by:
Imran Khan, University of Nebraska Medical Center, United StatesReviewed by:
Asad Ur Rehman, University of Nebraska Medical Center, United StatesCopyright © 2024 Wu, Cai, Jiang, Lin, Wang, Zhang, Kang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mingqiang Kang, bWluZ3FpYW5nX2thbmdAMTYzLmNvbQ==; Peipei Zhang, emhhbmdwZWlwZWkwNTkxQDE2My5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.