- Department of Laboratory Medicine, The Third Affiliated Hospital of Zhengzhou University, Zhengzhou, Henan, China
Previous studies have demonstrated that genetic alterations governing epigenetic processes frequently drive tumor development and that modifications in RNA may contribute to these alterations. In the 1970s, researchers discovered that N6-methyladenosine (m6A) is the most prevalent form of RNA modification in advanced eukaryotic messenger RNA (mRNA) and noncoding RNA (ncRNA). This modification is involved in nearly all stages of the RNA life cycle. M6A modification is regulated by enzymes known as m6A methyltransferases (writers) and demethylases (erasers). Numerous studies have indicated that m6A modification can impact cancer progression by regulating cancer-related biological functions. Tumor angiogenesis, an important and unregulated process, plays a pivotal role in tumor initiation, growth, and metastasis. The interaction between m6A and ncRNAs is widely recognized as a significant factor in proliferation and angiogenesis. Therefore, this article provides a comprehensive review of the regulatory mechanisms underlying m6A RNA modifications and ncRNAs in tumor angiogenesis, as well as the latest advancements in molecular targeted therapy. The aim of this study is to offer novel insights for clinical tumor therapy.
1 Introduction
Tumor angiogenesis is an uncontrolled and persistent process that plays a crucial role in tumor growth and metastasis (1). Unlike normal angiogenesis, which is regulated through complex biological processes, tumor angiogenesis has abnormal characteristics, such as irregular morphological structures, disorganized arrangements of endothelial cells, and unstable vascular walls. These abnormalities result in a hypoxic state and the accumulation of metabolic waste within the tumor while also providing nutrients and pathways for tumor cell proliferation and metastasis (2). The regulation of tumor angiogenesis involves multiple signaling pathways and molecular mechanisms (3). Recent studies have highlighted the significant regulatory roles of m6A modifications and noncoding RNAs (ncRNAs) in this process.
In recent years, an increasing number of studies have shown that methylations play crucial roles in the regulation of tumor angiogenesis (4, 5). Methylation is a process that involves the addition of methyl groups to DNA or RNA molecules. This modification can affect gene expression by altering chromatin structure and binding sites for transcription regulatory factors. In tumors, the patterns of methylations on DNA and RNA often undergo changes, which are closely associated with tumor progression, invasiveness, and patient prognosis (6–8).
M6A is a prevalent chemical modification observed in RNA molecules (9). M6A modification involves the addition of a methyl group to the adenine base of RNA molecules. This modification is widely distributed among eukaryotes, including humans, mice, and fruit flies. M6A modification is a dynamic process in which methyl groups are added to RNA molecules by methyltransferases and removed by demethylases (10). This modification can impact various cellular processes, such as RNA stability (11–13), posttranscriptional regulation (14, 15), translation (12, 16), and splicing (17–19) (Figure 1). Importantly, m6A modification can directly or indirectly influence the expression and function of factors related to tumor angiogenesis by regulating the transcription levels, RNA stability, and translation efficiency of key genes. For example, researchers have discovered that m6A modification can enhance or inhibit the regulatory effects of specific ncRNAs on crucial biological processes such as endothelial cell proliferation, migration, and lumen formation. Currently, m6A is increasingly recognized as a promising biomarker for cancer detection and prevention because of its potential clinical value in cancer research.
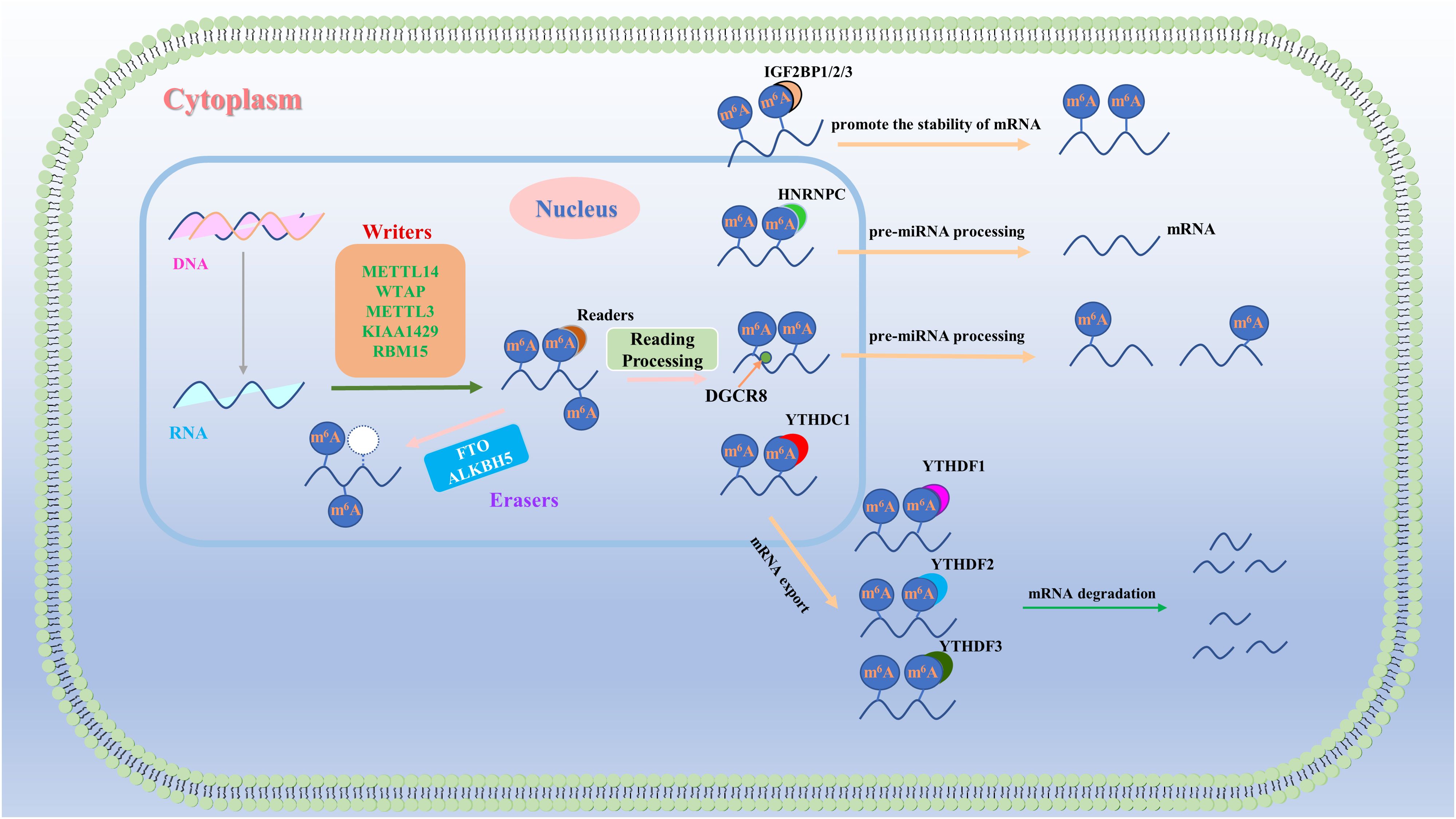
Figure 1. Regulatory mechanism of m6A RNA methylation. M6A RNA methylation is a dynamic process that is governed primarily by three groups of proteins: “writers,” “erasers,” and “readers.” “Writers” and “erasers” are located predominantly in the nucleus, whereas some “readers,” such as HNRNPs and YTHDC1, also function in the nucleus. Other “readers,” including YTHDC2 and IGF2BP1/2/3, are found in both the cytoplasm and the nucleus, whereas “readers”, such as YTHDF1/2/3, are exclusively present in the cytoplasm. These three groups of proteins collaboratively regulate the output, stability, translation, and degradation of RNA.
NcRNAs encompass a group of RNA molecules that do not possess protein-coding capabilities but instead serve as crucial regulators within cells (20). Emerging research has highlighted the significant role of ncRNAs in the regulation of tumor angiogenesis. Specifically, certain ncRNAs interact with the mRNAs of genes associated with angiogenesis, thereby modulating their stability and posttranscriptional modification levels. These interactions ultimately impact the process of tumor angiogenesis.
While studies have examined the regulatory roles of m6A modifications and ncRNAs in tumor angiogenesis, our current knowledge in this area remains limited. Therefore, further investigations into the mechanisms underlying the interaction between m6A modification and ncRNAs in the regulation of tumor angiogenesis are crucial. Therefore, in this review, we assess the regulatory effect of m6A on tumor angiogenesis and provide a theoretical basis for the development of novel targeted therapeutic strategies.
2 M6A, NcRNA and tumor angiogenesis
2.1 M6A
M6A is composed of a ribose with a purine base and a methyl group attached to the sixth nitrogen atom. It was not until 2012 that the genome-wide distribution of m6A was elucidated, and m6A is one of the most abundant modifications in eukaryotic mRNAs (11, 21, 22). M6A is considered the most common, frequent, and conserved internal modification, with an average of 1–2 m6A residues per 1000 nucleotides (23). M6A primarily occurs within the conserved motif RRACH (R = G or A and H = A, C, or U), which tends to be found in stop codons and 3′ untranslated regions (3′ UTRs) (24–27). The basic process of m6A modification involves the installation of methyl groups by “writer” methyltransferases, removal by demethylases known as “erasers,” and recognition by m6A-binding proteins called “readers,” thereby regulating RNA metabolism (28–31). This is a dynamic process (Figure 1). The m6A methyltransferase complex (MTC) consists of methyltransferase-like 3 (METTL3) (32, 33), METTL14 (34), the METTL3 adaptor protein wilms tumor 1-associated protein (WTAP) (35), and other associated proteins, including METTL7A/B (36), METTL5 (37, 38), the METTL5-tRNA MTase subunit 11–2 (TRMT112) complex (39, 40), Vir-like m6A methyltransferase associated protein (VIRMA/KIAA1429) (41, 42), RNA-binding motif protein 15 (RBM15) (43, 44), METTL16 (45), zinc finger CCCH domain-containing protein13 (ZC3H13) (46–48), CBLL1 (Cbl proto-oncogene like 1) (49), and zinc finger CCHC domain-containing protein 4 (ZCCHC4) (50). METTL3 is a key protein in this complex and was first identified as an S-adenosylmethionine-binding protein with methyltransferase activity (33). As a pseudomethyltransferase, METTL14 plays a crucial role in facilitating complex formation and RNA binding. In vitro and in vivo experiments have demonstrated that METTL3 and METTL14 form a heterodimer, and together, they catalyze m6A methylation, while their individual components exhibit lower activity (51). WTAP does not possess catalytic activity but can interact with METTL3 and METTL14, thereby regulating m6A levels during RNA transcription (34). Recently, METTL16 was shown to regulate splicing by targeting pre-mRNAs and various ncRNAs (52). VIRMA is capable of binding m6A with 3’UTRs, whereas ZC3H13 functions to induce the translocation of the writer complex into the nucleus. Recent reports have identified VIRMA and ZC3H13 as new components of the m6A methyltransferase complex that regulate the selectivity of m6A site modifications on mRNAs. These findings provide new insights into the regulatory mechanisms of m6A occurrence (53, 54). Fat mass and obesity-associated protein (FTO) was the first discovered m6A RNA demethylase (55), capable of interacting with various RNA molecules, including mRNA, snRNA, and tRNA. In vitro studies have shown that FTO effectively oxidizes and demethylates m6A and N6,2’-O-dimethyladenosine (m6Am) on RNA (56–58). However, despite its significant demethylation activity on m6A in vitro, its regulatory role on m6A under physiological conditions appears to be relatively limited. Increasing evidence suggests that FTO primarily regulates RNA function through the demethylation of m6Am (59). M6Am is typically situated near the 5’ cap structure of mRNA (60), and its demethylation is more prominent in the cytoplasm than in the nucleus, likely due to the differential subcellular localization of FTO. M6Am plays a critical role in regulating mRNA stability and translation efficiency (61–64). Studies indicate that FTO predominantly acts on m6Am within snRNA in the nucleus (65), while in the cytoplasm, it preferentially demethylates m6A at the 5’ end and within internal regions of mRNA (66). Notably, there is ongoing debate regarding whether FTO’s role in m6A demethylation involves reversible and dynamic regulation, underscoring the need for further in-depth research (59, 67). However, alkB homolog 5 (ALKBH5) exhibits comparable demethylation activity to m6A and shows a preference for consensus sequences (RRACH) that are consistent with m6A (43). Silencing or overexpression of these factors leads to an increase or decrease in m6A on mRNA, respectively. Knockout of FTO or ALKBH5 results in an overall increase in m6A levels in human cells. M6A readers interpret information on RNA methylations and participate in downstream processes such as translation and degradation of RNA. Different readers have distinct physiological functions. The identified readers can be classified into three categories: the first category includes readers with YTH domains (68, 69); the second category consists of HNRNPs (70), which possess the same RNA-binding domains (RBDs); and the third category of readers includes those with KH domains, RNA recognition motifs (RRMs), and arginine/glycine-rich (RGG) domains (71–73). These readers can all bind to m6A-modified mRNAs, including FMR1 and IGF2BP1-3. Interestingly, YTHDF1, YTHDF2, and YTHDF3 are the principal members of the YTHDF protein family (74, 75). Initial studies suggested that these proteins specifically bind to m6A sites, with YTHDF1 enhancing translation, YTHDF2 promoting mRNA degradation, and YTHDF3 both enhancing translation and facilitating mRNA degradation (76, 77). Additionally, YTHDF3 may have a potential role in regulating RNA transport, particularly in neurons or related systems (78). However, recent research has revealed that YTHDF1, YTHDF2, and YTHDF3 exhibit functional redundancy in regulating mRNA degradation (79). The YTHDF protein family collaboratively binds to m6A-modified sites to mediate this process, with all three proteins primarily promoting the degradation of m6A-modified mRNAs rather than directly enhancing translation (80). This finding prompts a reassessment of the roles of YTHDF proteins in RNA metabolism and may challenge our current understanding of m6A regulation (79, 81). The combined actions of writers, erasers, and readers form the m6A modification system that regulates the biological functions and metabolism of RNA (30, 63–65).
2.2 NcRNA
NcRNAs are pivotal regulators of gene expression that function independently of protein translation. These RNAs include small nuclear RNAs (snRNAs), small interfering RNAs (siRNAs), microRNAs (miRNAs), long noncoding RNAs (lncRNAs), and circular RNAs (circRNAs) (82, 83). These ncRNAs exert their regulatory effects through diverse mechanisms, such as posttranscriptional regulation, transcriptional repression, and chromatin remodeling (84). MiRNAs, which are typically 20–24 nucleotides in length, bind to the 3’ untranslated regions of target mRNAs, leading to mRNA degradation or translational inhibition, and are implicated in the pathogenesis of diseases, including cancer and cardiovascular disorders (85, 86). LncRNAs, which are typically longer than 200 nucleotides, regulate gene expression through multiple mechanisms. At the genomic level, lncRNAs modulate gene expression via classical pathways such as transcription, posttranscriptional mRNA processing, turnover, and translation. At the epigenomic level, lncRNAs further fine-tune genomic control by altering chromatin architecture and modifying the chemical properties of DNA and RNA. Additionally, lncRNAs transcend genomic and epigenomic regulation, adding an extra dimension of control by influencing both transcriptional and posttranscriptional processes (87). As a result, lncRNAs play pivotal roles in cellular development, tissue-specific gene expression, and the progression of various diseases (87, 88). CircRNAs, distinguished by their covalently closed loop structures, exhibit high stability and act as miRNA sponges, thereby modulating the activity of miRNAs and potentially regulating transcription factors (89, 90). Dysregulation of these ncRNAs is associated with a range of biological processes and diseases, highlighting their critical roles in gene regulatory networks and their potential as targets for therapeutic intervention.
2.3 Tumor angiogenesis
In the 1970s, Folkman proposed a hypothesis that was different from traditional theories, the tumor angiogenesis theory (91), which suggests that tumor growth depends on the formation of new blood vessels (92–94). He also first described the potential prospects of antiangiogenic cancer therapy. Tumor angiogenesis refers to the process of forming a new network of blood vessels during tumor growth. Under normal circumstances, angiogenesis is a highly regulated process that maintains the normal physiological function of tissues. However, in tumors, abnormal angiogenesis is a key process involved in tumor growth and metastasis. Tumor cells require a sufficient blood supply to obtain oxygen and nutrients, which are essential for their rapid proliferation and growth. Therefore, as the tumor volume increases, the existing blood vessels in the surrounding tissue are unable to meet its demands (95). To address this issue, tumor cells release a series of proangiogenic factors, such as vascular endothelial growth factor (VEGF) (96) and basic fibroblast growth factor (bFGF) (97), which stimulate the activation of endothelial cells in the surrounding tissue. Activated endothelial cells degrade the surrounding matrix by releasing enzyme substances such as metalloproteinases and proteases and migrating toward the tumor area. This process involves the regulation of multiple growth factors and signaling pathways, such as VEGF/vascular endothelial growth factor receptor (VEGFR) (98, 99), fibroblast growth factor (FGF)/fibroblast growth factor receptor (FGFR), Notch, and phosphatidylinositol 3-kinase (PI3K)/protein kinase B (PKB; also known as AKT) (100). The newly formed vascular structure needs to be stabilized through interactions with the extracellular matrix and other supportive cells in the surrounding tissue. This includes the recruitment of vascular smooth muscle cells and deposition of the extracellular matrix. Tumor cells can also recruit other types of cells, such as endothelial progenitor cells, stromal cells, and immune cells, by releasing chemokines. These cells can participate in the process of angiogenesis and provide support and regulation. During tumor angiogenesis, newly formed blood vessels often exhibit abnormal permeability, leading to the leakage of fluid and proteins into the surrounding tissue. This provides the tumor with increased nutrients and oxygen and creates a pathway for tumor metastasis.
In conclusion, the dynamic regulation of m6A modifications is significantly correlated with gene expression, and disruptions in the balance maintained by writers, erasers, and readers often lead to pathological conditions, ultimately resulting in tumorigenesis (17, 101). M6A is a widely occurring modification of both mRNAs and ncRNAs that participates in RNA splicing, translation, and stability regulation and influences the function of specific ncRNAs through epigenetic mechanisms (102). Research has demonstrated that the regulation of ncRNAs by m6A plays a crucial role in tumor initiation, metastasis, and angiogenesis (103–105). A substantial body of evidence indicates that ncRNAs mediate interactions between RNAs and between RNAs and proteins, regulating specific biological functions and thereby affecting cellular processes and contributing to tumor development and progression. Moreover, m6A and ncRNAs may exhibit synergistic effects in cancer therapy, with their regulatory mechanisms offering significant potential for clinical applications (106–108). In recent years, m6A has emerged as a promising biomarker for cancer detection and prevention, with its clinical potential in oncology becoming increasingly apparent. However, further in-depth research is necessary to elucidate the specific mechanisms and applications of m6A modifications in tumors (109–111). This research is expected to provide novel targeted strategies for cancer treatment and a theoretical foundation for the development of new anticancer therapeutics.
3 Direct and indirect effects of m6A on tumor angiogenesis
3.1 Direct regulation of m6A in tumor angiogenesis
3.1.1 Writers act on tumor angiogenesis
RNA methylation is closely associated with tumor angiogenesis. As one of the core components of m6A modification, METTL3 plays a crucial role in m6A modification and has been identified as an oncogenic target in some hematological malignancies and solid tumors (112, 113). Currently, studies have elucidated the role of METTL3 in promoting cell proliferation and angiogenesis, and its role in promoting tumor cell proliferation and angiogenesis has been confirmed through a series of experiments. METTL3 plays a dual role in normal and leukemic myeloid cells by driving the translation of key genes to maintain acute myeloid leukemia (AML) cell proliferation and undifferentiation. Inhibition of METTL3 induces differentiation and apoptosis, presenting a viable therapeutic strategy (114). Experimental data indicate that in gastric cancer (GC), METTL3 promotes tumor angiogenesis and carcinogenesis by reducing the expression of ADAMTS9 (115). In myeloid leukemia and lung cancer (LC), inhibiting METTL3 has been shown to be a promising therapeutic strategy in the future (114, 116). Recent studies have also demonstrated that METTL3-mediated m6A modification can activate the PI3K/AKT/mammalian target of rapamycin (mTOR) pathway in ovarian cancer (OC) (117) and retinoblastoma (Rb) (118). These pathways play important roles in regulating protein synthesis, cell growth, and protein synthesis related to angiogenesis. They play crucial regulatory roles in tumor angiogenesis. Research has confirmed that in bladder cancer (BCa), METTL3 also regulates the PI3K/AKT pathway, which is involved in tumor angiogenesis (119). Moreover, METTL3-mediated m6A modification is essential for the activation of tie-2 receptor tyrosine kinase (TEK)/vascular endothelial growth Factor A (VEGFA)-mediated BCa progression and angiogenesis. In colorectal cancer (CRC), METTL3 upregulates plasminogen activator urokinase (PLAU) mRNA in a m6A-dependent manner and participates in the mitogen-activated protein kinase (MAPK)/extracellular signal-regulated kinase (ERK) pathway to promote tumor angiogenesis and metastasis (120). In head and neck squamous cell carcinoma and other types of tumor cells, interleukin-8 (IL-8) and VEGF are coexpressed and promote tumor growth, invasion, and angiogenesis (121). The MAPK/ERK pathway can regulate tumor angiogenesis by upregulating or downregulating the synthesis of IL-8 and VEGF. Research on glioblastoma (GBM) has revealed that METTL3 enhances the stability of BUD13 mRNA through m6A methylation (122). BUD13 overexpression enhances the stability of CDK12 mRNA and increases its expression. Subsequently, CDK12 phosphorylates MBNL1, ultimately promoting the proliferation, migration, invasion, and vasculogenic mimicry (VM) of GBM. The METTL3-induced transcription factor interferon regulatory Factor 5 (IRF5) promotes proliferation, migration, invasion, and angiogenesis in cervical cancer (CC) cells by upregulating the protein phosphatase 6 catalytic subunit (PPP6C) (123). In addition, as another important component of the m6A MTC, METTL14 also plays a role in malignant tumors (124–127). Studies have shown that m6A modification of TRAF1, which is dependent on METTL14, promotes sorafenib resistance by regulating apoptosis and angiogenesis pathways (128). Interestingly, in tongue squamous cell carcinoma (TSCC), METTL14 reduces the stability of basic leucine zipper ATF-like transcription Factor 2 (BATF2) through m6A modification, thereby promoting TSCC proliferation, migration, invasion, and angiogenesis (129).
In summary, the m6A writers METTL3 and METTL14 play important roles in the occurrence and development of tumors by regulating specific targets or pathways that affect tumor angiogenesis. These studies may provide new possibilities for the clinical treatment of tumors (Table 1).
3.1.2 Readers act on tumor angiogenesis
Currently, an increasing number of studies have focused on the regulatory roles of m6A readers, such as insulin-like growth Factor 2 mRNA-binding protein 2 (IGF2BP2), IGF2BP3, YTH N6-methyladenosine RNA-binding protein 1 (YTHDF1), YTHDF2, and YTHDF3, in tumor occurrence, development, and angiogenesis. Research has shown that m6A readers play important roles in tumors such as CRC, GC, and BC brain metastasis. In CRC, METTL3 can promote CRC metastasis and progression via m6A-modified IGF2BP2 (134). Another study has shown that in CC, the m6A reader IGF2BP3 recognizes and binds to the m6A modification site on VEGF mRNA, promoting its stability and expression (135). VEGF is a vascular growth factor secreted by tumor cells or lymphocytes and has been shown to be a major factor in tumor angiogenesis. In studies of lung adenocarcinoma (LUAD), IGF2BP2 is transferred from LUAD cells to endothelial cells via exosomes. This transfer enhances the stability of FLT4 RNA, leading to the activation of the PI3K−Akt signaling pathway, which subsequently promotes angiogenesis and metastasis in cancer cells (136). IGF2BP2 is markedly overexpressed in AML and modulates crucial genes such as MYC, GPT2, and SLC1A5 through a m6A-dependent mechanism, thereby playing a pivotal role in glutamine metabolism and contributing to the pathogenesis and progression of AML (137). Therefore, IGF2BP3 can promote angiogenesis in CC cells by regulating VEGF. Experimental data have shown that the m6A reader IGF2BP3 binds to the m6A site on hepatoma-derived growth factor (HDGF) mRNA, maintaining its stability and promoting the secretion of HDGF (138), thereby promoting angiogenesis in GC tumors. IGF2BP3 also interacts with hypoxia-inducible factor-1a (HIF1a) and regulates the migration and angiogenesis of GC cells. YTHDF2 is one of the most efficient m6A readers and recognizes and distributes mRNAs containing m6A to processing bodies, thereby destabilizing the mRNA. Studies have shown that the overexpression of YTHDF2 in hepatocellular carcinoma (HCC) can inhibit tumor cell growth, whereas the knockout of the YTHDF2 gene promotes angiogenic sprouting in human umbilical vein endothelial cells (HUVECs). Silencing YTHDF2 can also promote tumor growth and metastasis in mouse models, and the key targets of YTHDF2 in HCC inflammation have been identified as IL11 and SERPINE2 (139). YTHDF3 expression is increased in BC brain metastases and is directly associated with reduced survival rates in BC patients without brain metastasis (140). It promotes the interaction between BC cells and brain endothelial cells and astrocytes, facilitating extravasation across the blood−brain barrier, angiogenesis, and growth.
In conclusion, the specific mechanisms of action of m6A readers in tumors are receiving increasing attention. Research has shown that m6A readers such as IGF2BP2, IGF2BP3, YTHDF2, and YTHDF3 are involved in tumor angiogenesis, further influencing the occurrence and development of tumors and potentially affecting the prognosis of patients with tumors to some extent (Table 1).
3.1.3 Erasers act on tumor angiogenesis
To date, the demethylases discovered for m6A include mainly FTO and ALKBH5 (141, 142). The dynamic reversibility of RNA methylation is closely related to this process. However, relatively few studies have investigated the regulatory mechanisms of FTO and ALKBH5 in tumor development and angiogenesis. In pancreatic cancer (PC), ALKBH5 can regulate the tumor microenvironment, and its loss reduces the infiltration of CD8+ T cells in PC. Additionally, ALKBH5 can inhibit the motility of PCs by demethylating the lncRNA KCNK15-AS1 (143). In CRC, circ3823 is involved in the regulation of tumor cell growth, metastasis, and angiogenesis through the miR-30c-5p/TCF7 axis. The degradation rate of circ3823 may be regulated by the m6A recognition protein YTHDF3 and the demethylase ALKBH5. Therefore, ALKBH5 can indirectly regulate the expression of circ3823 to control the growth, metastasis, and angiogenesis of CRC (144). Research has shown that METTL14/ALKBH5 affects tumor growth and progression by regulating key cell cycle- and angiogenesis-related transcripts. In addition, the RNA-binding proteins HuR, METTL14/ALKBH5, and their target genes form a feedback loop, regulating each other’s expression in cancer cells and participating in the regulation of tumor occurrence and metabolism. Research has shown that METTL14/ALKBH5 affects tumor growth and progression by regulating key cell cycle- and angiogenesis-related transcripts. In addition, the RNA-binding proteins HuR and METTL14/ALKBH5 and their target genes form a feedback loop, regulating each other’s expression in cancer cells and participating in the regulation of tumor occurrence and metabolism (29). The FTO protein, which is associated with adiposity and obesity, can decrease the concentration of m6A in mRNA transcripts, thereby regulating the expression of target genes such as ASB2 and RARA. It inhibits ATRA-induced differentiation of AML cells and promotes the development of leukemia. Additionally, the FTO/m6A/myelocytomatosis oncogene (MYC)/(CCAAT/enhancer binding protein α) (CEBPA) signaling pathway plays a crucial role in leukemia (145). In patients with intrahepatic cholangiocarcinoma (ICC), the expression level of FTO is lower (146). Patients with low FTO expression are more likely to be CD34 positive (indicating microvessel density), suggesting that the expression of FTO may regulate tumor angiogenesis. However, further research is needed to elucidate the specific mechanism involved.
In summary, m6A demethylases interact with m6A writers and readers, influencing the process of m6A methylation and participating in the regulation of tumor angiogenesis. Therefore, m6A demethylases have the potential to become precise regulatory targets for tumor angiogenesis, playing a role in promoting or inhibiting tumor growth (Table 1).
3.2 Indirect effects of m6A on tumor angiogenesis: interactions with ncRNAs
3.2.1 MiRNAs and m6A in tumor angiogenesis
MiRNAs are a class of evolutionarily conserved noncoding small RNA molecules with lengths of approximately 20–24 nucleotides that play a role in regulating gene expression. Many studies have shown that miRNAs exert their biological functions by regulating the translation process of downstream genes (147). In tumors such as LC, endometrial cancer (EC), and BC, miRNAs regulate the expression of genes related to angiogenesis to influence tumor growth and metastasis. For example, in LC, miR-320b suppresses angiogenesis and tumor growth by downregulating the expression of IGF2BP2 and thymidine kinase 1 (TK1) (148). In LC, METTL3 promotes the maturation of miR-143-3p through methylation and targets vasohibin-1 (VASH1) to inhibit its expression, thereby affecting angiogenesis (149). In breast cancer (BC) brain metastasis, YTHDF3 enriches transcripts associated with metastasis and promotes the interaction between tumor cells and other cells in the tumor microenvironment, facilitating angiogenesis and metastasis (140). In CRC, overexpression of METTL3 leads to the methylation of pri-miR-1246 and promotes its maturation, which is positively correlated with tumor metastasis (150). These findings indicate that miRNAs play crucial roles in tumor angiogenesis and metastasis, further elucidating the regulatory mechanisms of miRNAs in tumor initiation and progression (Figure 2).
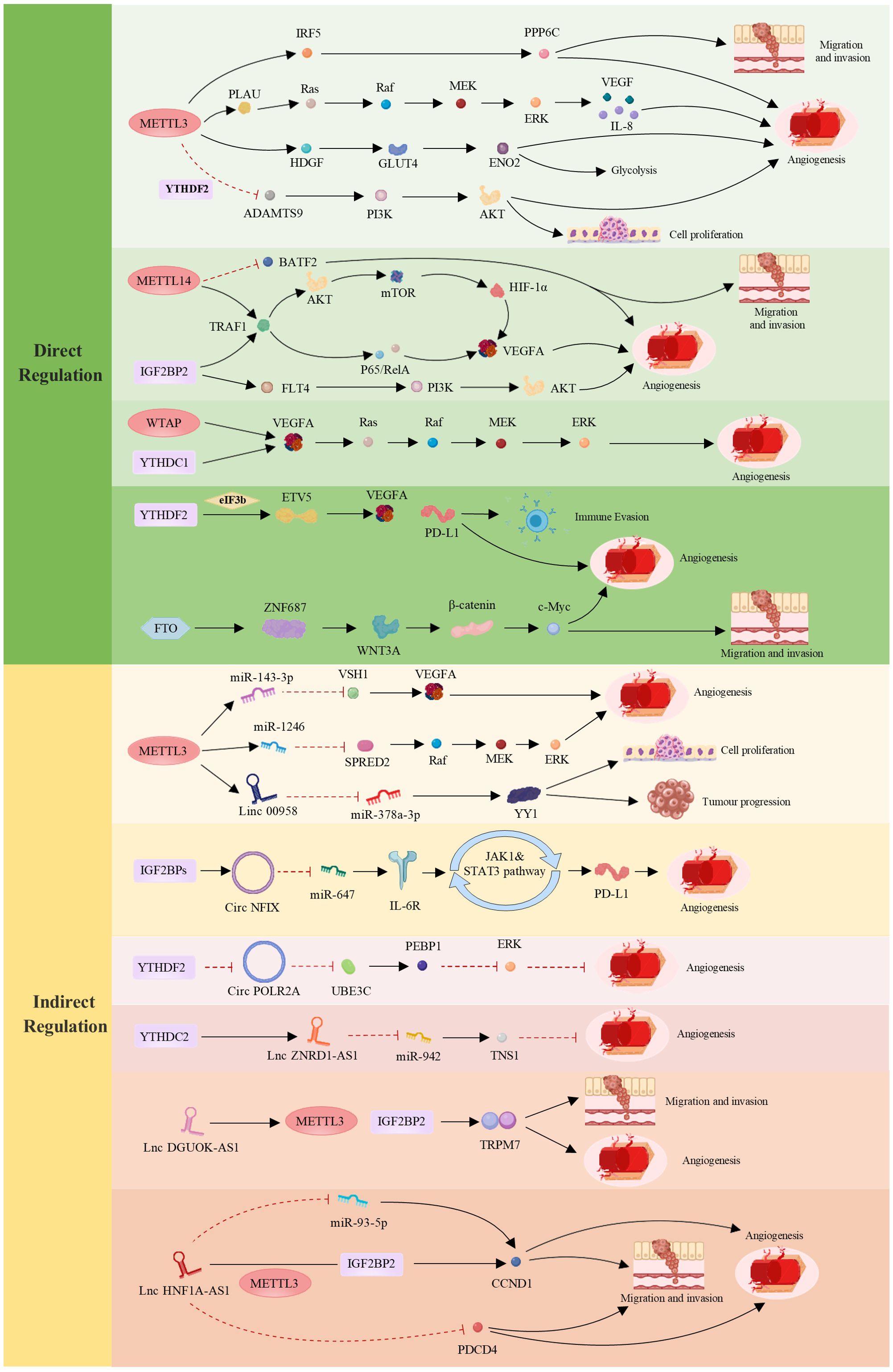
Figure 2. Regulatory roles of m6A in tumor angiogenesis and development—direct and indirect mechanisms. The direct regulation of m6A involves METTLs, ALKBHs, YTHDFs, IGF2BPs, YTHDCs and FTO, which influence specific targets and cellular signaling pathways, thereby modulating tumor angiogenesis and cancer progression. Indirect regulation of m6A occurs through interactions between the three main components of its “life cycle” and ncRNAs (including miRNAs, long noncoding RNAs (lncRNAs), and circular RNAs (circRNAs)), collectively mediating tumor cell proliferation, migration, angiogenesis, and immune evasion. These mechanisms play crucial roles in various tumors, profoundly impacting tumor biology. In the figure, red indicates inhibitory effects, whereas black indicates promoting effects. (Created with MedPeer (www.medpeer.cn)).
In conclusion, with in-depth research on m6A modification and miRNA regulatory mechanisms, we expect to discover more important mechanisms of mutual regulation between m6A modification and miRNAs in cancer initiation and progression. These findings will contribute to a better understanding of the biological characteristics of tumors and provide new targets and insights for the development of novel anticancer treatment strategies (Table 2).
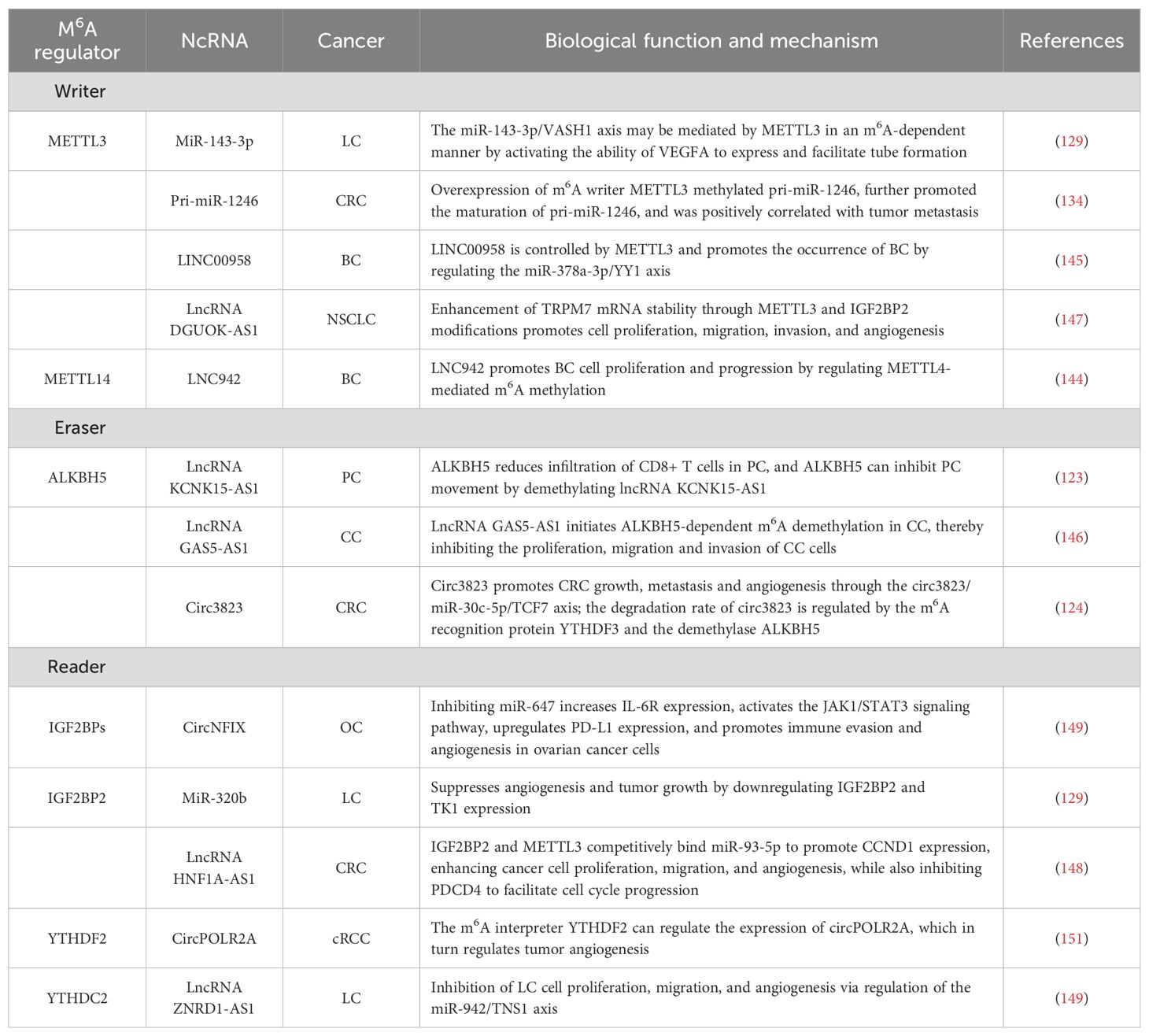
Table 2. NcRNAs and m6A modifications are involved in the regulatory mechanisms of tumor angiogenesis.
3.2.2 LncRNAs and m6A in tumor angiogenesis
LncRNAs are ncRNA molecules that are longer than 200 nucleotides (152). Research has shown that lncRNAs play crucial regulatory roles in various biological processes, including cell proliferation (153, 154), differentiation, apoptosis (155), migration, and invasion (156). LncRNAs can form complementary double-stranded structures with the transcripts of protein-coding genes, interfere with mRNA splicing, regulate protein activity, and serve as precursor molecules for small RNAs such as miRNAs and piRNAs (157). Therefore, lncRNAs play important roles in regulating gene expression (151, 158). In tumors such as BC, CC, and CRC, different lncRNAs are involved in tumor initiation and progression through various mechanisms. For example, in BC, the lncRNA TDRG1 promotes the proliferation, invasion, and metastasis of BC cells through the miR-214-5p/recombinant chloride intracellular channel protein 4 (CLIC4) axis (159). LNC942 regulates METTL14-mediated m6A methylation to promote BC cell proliferation and progression (160). LINC00958, which is controlled by METTL3, promotes the occurrence of BC through the regulation of the miR-378a-3p/YY1 axis (161). Research has confirmed that the lncRNA GAS5-AS1 can initiate ALKBH5-dependent m6A demethylation in CC, thereby inhibiting the proliferation, migration, and invasion of CC cells (162). The evidence indicates that the lncRNA DGUOK-AS1 is markedly overexpressed in non-small cell lung cancer (NSCLC) tissues and cells. It facilitates NSCLC cell proliferation, migration, invasion, and angiogenesis by increasing the stability of transient receptor potential melastatin 7 (TRPM7) mRNA through METTL3-mediated and IGF2BP2-mediated modifications (163). In addition, the m6A reader YTHDF3 promotes the progression of CRC by facilitating the degradation of the m6A-modified lncRNA GAS5-AS1. Notably, the lncRNA HNF1A-AS1 facilitates the progression of the CRC cell cycle by upregulating CyclinD1 (CCND1) and inhibiting programmed cell death 4 (PDCD4). HNF1A-AS1 stabilizes CCND1 mRNA through its interaction with IGF2BP2, a process further enhanced by METTL3-mediated m6A modification. Additionally, HNF1A-AS1 competes with miR-93-5p, leading to increased CCND1 expression. This upregulation promotes CRC cell proliferation, migration, and angiogenesis, thereby driving tumor progression (164). Xiangrui Meng et al. reported that m6A-mediated overexpression of LINC00857 promotes the progression and occurrence of PC through the regulation of the mir-150-5p/E2F3 axis, which is closely associated with PC growth and angiogenesis (143). The m6A modification mediated by YTHDC2 enhances the stability of ZNRD1-AS1. In turn, ZNRD1-AS1 suppresses the proliferation, migration, and angiogenesis of LC cells by modulating the miR-942/tensin 1 (TNS1) axis (165) (Figure 2).
In conclusion, these findings demonstrate the significant role of lncRNAs in tumor initiation and progression, which are closely associated with tumor growth, invasion, and angiogenesis. In recent years, many m6A-modified lncRNAs have been discovered, and they regulate gene expression and function through a complex series of mechanisms. We believe that in the future, more m6A-modified lncRNAs regulated by other mechanisms will be discovered (Table 2).
3.2.3 CircRNAs and m6A in tumor angiogenesis
CircRNAs are a type of ncRNA that exist in organisms. They are characterized by the absence of a 5’ cap and a 3’ poly(A) tail, instead of forming a covalently closed loop structure (166). Most circRNAs are generated through back-splicing of exons, whereas a small portion is derived from introns. Due to their closed circular structure (167), circRNAs are more stable than linear RNAs and are resistant to degradation by exonucleases. Additionally, circRNAs exhibit species-specific, tissue-specific, and time-specific expression patterns (168, 169). It also shares some sequence conservation (170) and can exert regulatory functions at the transcriptional or posttranscriptional level (171). Research has shown that the majority of circRNAs are noncoding, but few can be translated into peptides. Currently, circRNAs have been found to have functions such as acting as miRNA sponges (172, 173), regulating protein binding and gene transcription, and encoding peptides. Many studies have demonstrated that circRNAs play important roles in the growth, development, stress response, and disease progression of organisms. In recent years, interest in the impact of m6A modification on circRNAs in tumor initiation, invasion, and angiogenesis has increased. For example, studies in clear cell kidney cancer (cRCC) have shown that circPOLR2A can downregulate the protein level of recombinant phosphatidylethanolamine binding protein 1 (PEBP1), thereby activating the ERK signaling pathway. Activation of the ERK pathway plays a crucial role in cancer angiogenesis. Therefore, circPOLR2A, an oncogene in cRCC, accelerates the proliferation, migration, invasion, and angiogenesis of cRCC while inhibiting apoptosis (174). In addition, in CRC, circ3823 plays a significant role in tumor growth, metastasis, and angiogenesis (144). Circ3823 can inhibit miR-30c-5p and promote the expression of downstream targets MYC and CCND1, which further promotes CRC growth, metastasis, and angiogenesis through the circ3823/miR-30c-5p/TCF7 axis. We also detected m6A modifications on circ3823. The degradation rate of circ3823 is regulated by the m6A recognition protein YTHDF3 and the demethylase ALKBH5. Therefore, precise regulation of circ3823 by m6A modification is involved in tumor angiogenesis and contributes to tumor initiation and progression. In OC, circNFIX, which is activated by IGF2BPs (IGF2BP1/2/3), inhibits miR-647, resulting in the upregulation of IL-6R expression. This upregulation subsequently triggers the activation of the JAK1/STAT3 signaling pathway, leading to increased PD-L1 expression. These molecular changes facilitate immune evasion and angiogenesis in OC cells (165) (Figure 2).
In summary, precise regulation of circRNAs by m6A modification plays a crucial role in cancer progression and may provide new insights into tumor initiation, development, and precision therapy. However, relatively few examples of the regulation of circRNAs by m6A modification are currently available, and further in-depth research on these modifications is needed. We believe that in the future, more studies on the mutual regulatory mechanisms between circRNAs and m6A modifications will emerge (Table 2).
4 Clinical translational potential of m6A modifications in oncology
4.1 Characterized m6A inhibitors
Given the important role of m6A regulatory proteins in various diseases, small-molecule inhibitors or agonists that target dysregulated m6A regulators may be promising candidates for disease treatment, especially cancer therapy. METTL3, the most extensively studied m6A methyltransferase to date, has attracted considerable attention from researchers. In tumor cells, aberrant expression and activity of METTL3 can lead to changes in m6A modification levels, thereby influencing tumor initiation, progression, and metastasis. Studies have shown that METTL3 inhibitors exhibit significant anticancer effects in various tumor models. For example, STM2457 selectively inhibits METTL3 methyltransferase activity, reducing m6A levels in acute AML cells, inhibiting their proliferation, and inducing apoptosis (175). RSM3 interferes with METTL3’s catalytic activity, reducing m6A modifications and showing antitumor potential in AML cell lines (176). UZH1a, a specific inhibitor, significantly affects viral replication and cell survival in EBV-positive Akata cells by inhibiting METTL3 methyltransferase activity (177). This discovery highlights METTL3 as a potential therapeutic target in EBV-associated diseases such as nasopharyngeal carcinoma (NPC), Hodgkin’s lymphoma, and GC, indicating the clinical application prospects of UZH1a. Quercetin, a natural compound, inhibits METTL3 activity, reduces m6A modifications, and, thus, inhibits tumor cell proliferation and migration, providing new insights into the development of natural product-based anticancer drugs (178).
YTHDF1 recognizes and binds to m6A-modified RNA, regulating the translation of various cancer-related genes and playing crucial roles in tumor initiation, progression, and therapeutic resistance. Studies have shown that RUVBL1/2 interferes with the RNA binding activity of YTHDF1, reducing the translation of cancer-related genes and inhibiting tumor cell proliferation and invasion, demonstrating significant antitumor effects in CRC (179). IGF2BP2, another m6A reader protein, binds to m6A-modified RNA, regulating its stability and translation. In AML, IGF2BP2 regulates key targets in the glutamine metabolic pathway (e.g., MYC, GPT1, and SLC5A6) in a m6A-dependent manner, promoting the development and self-renewal of leukemia stem/initiation cells. The small-molecule compound CWI1-2 inhibits IGF1BP2, showing promising antileukemic effects both in vitro and in vivo (137). In T-cell acute lymphoblastic leukemia (T-ALL), IGF2BP2 is highly expressed. Studies have confirmed that JX5 can inhibit the binding of IGF2BP2 to NOTCH1, thereby inactivating NOTCH1 signaling in T-ALL. However, the off-target effects and toxicity of JX5 require further investigation (180).
ALKBH5 demethylates m6A modifications, maintaining the stability of specific RNAs and regulating specific gene splicing processes, thereby affecting cancer cell proliferation and migration. Studies have shown that the imidazobenzoxazin-5-thione MV1035 inhibits the catalytic activity of ALKBH5 by competitively binding with 2-oxoglutarate (2OG), significantly reducing the migration and invasiveness of the U87 GBM cell line. ALKBH5, as a potential target for cancer therapy, holds significant research value (181). IOX3, a small molecule inhibitor, significantly inhibits the demethylation activity of ALKBH5 in vitro (130). As an ALKBH5 inhibitor, the mechanisms of IOX3, which is related to tumor angiogenesis, need further investigation to provide new drug directions for cancer treatment. ALK-04, an effective ALKBH5-specific inhibitor, has demonstrated good antitumor potential in a B16 melanoma mouse model. Future research should explore the application of ALK-04 in different cancer types and investigate its combined effects with other therapies to increase overall cancer treatment efficacy (182).
Since FTO is one of the most extensively studied regulatory proteins involved in m6A modification, an in-depth understanding of its dysregulation has facilitated the development of small-molecule compounds that target it. Rhein, the first reported FTO inhibitor, competitively blocks the recognition of m6A substrates by FTO, but it lacks selectivity (183, 184). In a mouse GBM model, MA2 binds to the active surface of FTO, inducing m6A methylation, reducing GBM stem cell proliferation in vitro, and exerting good antitumor effects (131, 185). FTO-04 and FTO-43 are effective inhibitors of FTO. FTO-04 has been reported to impair the self-renewal ability of GBM stem cells, thereby inhibiting tumor progression (132). However, FTO-43 is a novel FTO inhibitor with nanomolar potency and high selectivity. Compared with the homologous m6A RNA demethylase ALKBH5, FTO-43 exhibits remarkable selectivity and effectively inhibits the Wnt/PI3K-Akt signaling pathway. Experimental results demonstrate that FTO-43 has potent antiproliferative effects in models of GBM, acute myeloid leukemia, and GC, with efficacy comparable to that of the clinically used chemotherapeutic agent 5-fluorouracil (5-FU) (186). As a novel immunosuppressant, FTO-43 has significant potential to greatly improve therapeutic outcomes in clinical applications. In vitro experiments indicate that the FTO inhibitor CS1 significantly inhibits cell proliferation and induces apoptosis in CRC-related cell lines and that it can downregulate the Akt/mTOR signaling pathway (187). The research team developed two FTO inhibitors, FB23 and FB23-2, and used CRISPR-Cas9 gene editing to create stable FTO knockout (KO) AML cell lines to assess the efficacy of these inhibitors. The results revealed that the antiproliferative effect of FB23-2 was significantly reduced in FTO-KO cells compared with wild-type cells, indicating that the mechanism of FB23-2 activity relies primarily on FTO inhibition. FB23-2 suppresses AML cell proliferation and induces differentiation and apoptosis by increasing m6A levels, demonstrating its potential as a therapeutic strategy for AML. The CRISPR-Cas9-generated FTO-KO model further validated FTO as a viable therapeutic target and provided new insights into its role in AML pathogenesis (188). Dac51 inhibits FTO-mediated tumor cell glycolytic activity, enhancing CD8+ T-cell function and inhibiting solid tumor growth (133). R-2-Hydroxyglutarate (R-2HG) attenuates aerobic glycolysis in leukemia by targeting the FTO/m6A/PFKP/LDHB axis, thereby modulating disease progression. These findings underscore the potential of potent FTO inhibitors as epigenetic modulators and metabolic targets for therapeutic intervention in cancer treatment (189). Recently, a successfully synthesized GSH bioimprinted nanocomposite loaded with an FTO inhibitor (GNPIPP12MA) was proven to inhibit leukemogenesis by targeting the FTO/m6A pathway and synergizing with GSH depletion (190). Notably, in addition to antitumor therapy, FTO inhibitors have also attracted attention for other diseases. For example, entacapone has been reported as a potential FTO inhibitor for the clinical treatment of metabolic syndromes such as obesity and diabetes (191).
The biological study of m6A modifications is at a critical stage of translational application, urgently requiring the discovery and development of chemical probes and active lead compounds. The development of highly selective chemical inhibitors targeting m6A writers, readers, and erasers has greatly advanced the understanding of the biological functions of m6A and demonstrated its feasibility as a therapeutic target. These findings lay the foundation for new models in basic research and cancer therapy in the field of RNA epigenetics.
4.2 Prospective m6A inhibitory targets: emerging directions in clinical oncology
4.2.1 Digestive system cancers
4.2.1.1 Gastric cancer
GC is the fifth most common gastrointestinal malignancy and the third leading cause of cancer-related death worldwide (192, 193). Due to the late diagnosis of most GC patients at an advanced stage of malignant spread and metastasis, the prognosis for these patients is generally poor. The dissemination and metastasis of cancer involve various biological processes, such as cell growth, migration, invasion, and angiogenesis. Therefore, there is an urgent need to identify biomarkers for GC diagnosis and prognostic evaluation, as well as therapeutic targets. A study involving in vitro formation and CAM experiments revealed that METTL3 may inhibit the m6A modification of its target gene ADAMTS9 through the ADAMTS9-mediated PI3K/AKT pathway and prevent its transcription in a YTHDF2-dependent manner. These findings suggest that ADAMTS9 could be a novel potential therapeutic target for the treatment of GC carcinogenesis and angiogenesis involving METTL3 (115). This study reveals the pathological role and molecular mechanism of METTL3, further supporting its potential as a prognostic biomarker and therapeutic target for GC. Recently, Jiang et al. reported that knockout of the m6A reader IGF2BP3 can inhibit hypoxia-induced GC cell migration and angiogenesis by regulating HIF1a. Experimental results indicate that HIF1a is a target of IGF2BP3 and that IGF2BP3 positively regulates the expression of HIF1a in GC cells by directly binding to specific m6A sites in the HIF1a mRNA coding sequence (CDS) (138). Under hypoxic conditions, IGF2BP3 promotes angiogenesis in GC cells by upregulating HIF1a. These findings may provide new therapeutic targets for the clinical treatment and prognosis of GC. Wang et al. reported that METTL3-mediated m6A modification maintains the expression of HDGF through IGF2BP3-dependent mRNA stability. Increased secretion of HDGF promotes tumor angiogenesis and glycolysis, thereby accelerating the malignant progression of GC and indicating a poor prognosis. These findings reveal that the METTL3/HDGF/GLUT4/ENO2 axis promotes the occurrence and metastasis of GC by enhancing glycolysis and angiogenesis (194).
In conclusion, the development and progression of GC, as well as angiogenesis, are intricately linked to m6A RNA methylation, which is precisely regulated by m6A-associated proteins. Nonetheless, a deeper investigation into inhibitors targeting m6A-related proteins involved in the mechanistic pathways of GC is warranted. These results will provide significant support for clinical drug development.
4.2.1.2 Colorectal cancer
CRC is one of the most common malignant tumors in the gastrointestinal tract (195). It often lacks obvious early symptoms and is typically diagnosed in the middle to late stages of tumor development. As a result, nearly one million people are diagnosed with CRC each year, with a mortality rate of 33%, ranking it as the second leading cause of cancer-related deaths. Clinical data show that early detection, diagnosis, and treatment can lead to a 5-year survival rate of 90% for CRC patients (196). However, for patients with advanced metastasis, the 5-year survival rate decreases to only 8%. Therefore, it is crucial to study the targets involved in the occurrence, treatment prognosis, and inhibition of angiogenesis in CRC (197). Recent studies have revealed that METTL3 upregulates PLAU mRNA in a manner dependent on m6A modification and is involved in the MAPK/ERK pathway, promoting angiogenesis and metastasis in CRC. Elevated expression of METTL3 in CRC tissues has been associated with lower survival rates during cancer metastasis (134). Furthermore, in describing the epigenetic characteristics of CRC metastasis to the liver and lungs, METTL14 promotes cancer cell proliferation and metastasis by facilitating processes such as epithelial−mesenchymal transition (EMT) and protein phosphorylation through downstream targets such as the lncRNA RP11 and microRNAs (198). This significantly enhances the ability of CRC to metastasize to distant organs, leading to poor patient prognosis. WTAP modifies VEGFA mRNA through m6A, which is mediated by YTHDC1 and activates the MAPK signaling pathway. This process leads to increased VEGFA expression, promoting the progression and angiogenesis of CRC. These findings suggest that m6A inhibitors could be used in clinical treatments (199). In addition, it has been reported that the m6A reader IGF2BP3 can regulate the cell cycle and angiogenesis in CRC cells. Further research data suggest that IGF2BP3 inhibits the expression of VEGF by reading and promoting the decay of m6A-modified mRNAs, thereby suppressing angiogenesis in CC tumor cells and inhibiting tumor growth (135). A research team reported that, in CRC tissues and cell lines, FTO-mediated regulation of ZNF687 promotes tumor growth, metastasis, and angiogenesis through the Wnt/β-catenin pathway. The development of FTO inhibitors offers a new perspective for potential CRC treatment strategies (200).
As a result, in CRC, METTL3, METTL14, WTAP, IGF2BP3, YTHDC1, and FTO orchestrate tumor angiogenesis through diverse pathways and mechanisms, thereby affecting the initiation, progression, and prognosis of CRC. Notably, the writers, erasers, and readers of m6A collaboratively regulate CRC. Hence, the future application of m6A inhibitors in CRC and the equilibrium required for their use merit comprehensive investigation and deeper exploration.
4.2.1.3 Hepatocellular carcinoma
HCC is a highly lethal primary liver cancer and one of the most common malignant tumors worldwide (201). Angiogenesis plays a crucial role in the growth and metastasis of HCC, making the inhibition of angiogenesis an important therapeutic target for HCC (202). Recent studies have shown that the m6A methyltransferase METTL3 is significantly associated with the formation of VM and the expression of VM-related markers and is closely related to poor prognosis in HCC. Additionally, the YAP1 protein promotes VM and malignant progression in HCC through a m6A-dependent mechanism in the Hippo signaling pathway. Therefore, METTL3 and YAP1 may serve as potential targets for the targeted inhibition of VM in the treatment of HCC (203). Recent research has demonstrated that inhibiting YTHDF2 can suppress immune evasion and angiogenesis in HCC through the ETS variant transcription Factor 5 (ETV5)/PD-L1/VEGFA axis. These findings indicate that targeting YTHDF2 may serve as an effective therapeutic strategy for HCC, providing a promising new avenue for combination treatments (204).
In summary, METTL3 and YTHDF2 may act as “oncogenes” in HCC, facilitating tumor angiogenesis and malignant progression. However, further rigorous research is needed to elucidate the precise regulatory mechanisms of m6A modification in HCC. These identified m6A-related proteins are pivotal targets for the development of clinical m6A inhibitors.
4.2.2 Urinary system cancers
4.2.2.1 Renal cell carcinoma
Localized renal cell carcinoma can be treated with surgery, whereas metastatic renal cancer is usually resistant to conventional radiotherapy and chemotherapy (192, 205). In recent years, there has been preliminary progress in inhibiting tumor development and metastasis by targeting the angiogenesis of RCC with drugs (206). A study of RCC revealed that tumor necrosis factor receptor (TNFR)-associated Factor 1 (TRAF1) is closely related to cancer cell angiogenesis and apoptosis. The overexpression of TRAF1 significantly enhances angiogenesis, whereas the downregulation of TRAF1 inhibits angiogenesis. The experimental results indicate that m6A methyltransferase METTL14-mediated m6A modification enhances the stability of TRAF1 mRNA and increases TRAF1 levels through an IGF2BP2-dependent mechanism. The increased expression of TRAF1 subsequently contributes to the activation of downstream antiapoptotic and proangiogenic pathways in sunitinib-resistant cells, thereby promoting angiogenesis and metastasis in RCC. In sunitinib-resistant RCC cells, the expression of TRAF1 can be effectively suppressed by regulating METTL14, thereby inhibiting the angiogenesis signaling pathway and activating the apoptosis signaling pathway (128). This approach can effectively control the drug resistance and metastasis of cancer cells in the clinical treatment of RCC (207).
In summary, current research has demonstrated that m6A modification plays a pivotal role in the clinical treatment and drug resistance of renal cell carcinoma (RCC), providing new hope for patients. We anticipate that future discoveries of relevant regulatory mechanisms and m6A-specific inhibitors will significantly increase the effectiveness of targeted therapies in overcoming drug resistance in RCC.
4.2.2.2 Bladder cancer
Currently, METTL3-catalyzed m6A RNA methylation is widely recognized as a key epigenetic regulatory process for tumorigenic characteristics in various cancer cell lines, including BCa. Kyoto Encyclopedia of Genes and Genomes (KEGG) transcriptome sequencing results revealed a close association between METTL3 and tumor angiogenesis. M6A modification mediated by METTL3 is essential for the activation of tumor progression and angiogenesis mediated by TEK/VEGFA. In BCa, METTL3 regulates the PI3K/AKT pathway associated with tumor angiogenesis and promotes tumor angiogenesis by modulating TEK and VEGFA. METTL3 plays a crucial role in driving the progression of BCa by promoting angiogenesis around tumor cells. It affects bladder malignancies through the METTL3-TEK-VEGFA-CD31/CD34 pathway (119).
Overall, in the clinical treatment of BCa, targeting METTL3 could present novel therapeutic strategies to overcome chemotherapy and immunotherapy resistance driven by tumor angiogenesis, thereby improving the clinical outcomes of patients. The future development of more precise and highly specific METTL3 inhibitors promises to bring new hope to this patient population.
4.2.3 Epidermal cancers
4.2.3.1 Head and neck squamous cell carcinoma
Head and neck squamous cell carcinomas (HNSCCs) are a group of malignant tumors that occur in the head and neck region and account for approximately 90% of all head and neck tumors. They include tumors in the neck, oral and maxillofacial region, and otolaryngology (208, 209). Women are more susceptible to this disease. HNSCCs rank eighth in terms of the global incidence of malignant tumors and twelfth in terms of mortality. The 5-year survival rate is approximately 50%, and there has been no significant improvement in the past 20 years. This may be attributed to the late-stage detection of cancer and a lack of effective therapeutic targets. A recent study revealed that overexpression of METTL3 in HNSCC can promote cell proliferation, migration, invasion, and angiogenesis. METTL3 can mediate m6A modification of CDC25B mRNA, thereby promoting the malignant progression of HNSCC (121).
In conclusion, METTL3 is a promising prognostic biomarker and therapeutic target for HNSCC. This finding has the potential to address the high mortality rate and the current paucity of effective therapeutic targets in HNSCC. Consequently, comprehensive research into the regulatory mechanisms of m6A modification in HNSCC and the development of highly specific clinical inhibitors are crucial.
4.2.4 Female reproductive system cancers
4.2.4.1 Breast cancer
According to the latest data from the International Agency for Research on Cancer (IARC) in 2018, BC has the highest incidence rate among female cancers globally, accounting for 24.2% of all female cancer cases. Among these cases, 52.9% occur in developing countries (210). Although the expression and regulatory patterns of target genes associated with BC have been widely reported, the posttranscriptional regulatory mechanisms of gene expression during BC metastasis remain unclear. This lack of understanding can have implications for tumor treatment and prognosis. Recent studies have shown that the reader protein YTHDF3, which is involved in the epigenetic regulation process, plays a crucial role in BC and its brain metastasis. YTHDF3 enhances the translation of m6A-enriched transcripts such as ST6GALNAC5, gap junction 1 (GJA1), and EGFR, promoting the interaction between BC cells and brain endothelial cells and astrocytes. This interaction leads to extravasation across the blood−brain barrier, angiogenesis, and growth. YTHDF3-mediated m6A modification plays a crucial role in the development of brain metastasis in BC, which relies on the enhanced m6A methylation status and translation efficiency of target transcripts (140).
Consequently, YTHDF3 plays a crucial role in angiogenesis during the brain metastasis of BC. We believe that comprehensive research on m6A modification may provide new evidence and novel strategies for the clinical treatment and drug development of BC metastasis in the future.
Taken together, the m6A methyltransferase METTL3 plays a critical role in regulating tumor angiogenesis in various cancers, including GC, CRC, liver cancer, BCa, and head and neck squamous cell carcinoma. This regulation significantly impacts tumor initiation, progression, invasion, and prognosis. Targeting METTL3 as a novel therapeutic and prognostic marker and developing highly specific inhibitors hold great promise for improving patient prevention, treatment, and survival rates in future clinical applications for these cancers. Notably, in renal cell carcinoma, METTL14 and IGF2BP2 collaboratively regulate tumor angiogenesis and metastasis while also managing drug resistance in cancer cells during clinical treatment. Consequently, we hypothesize that the development of METTL14-targeted inhibitors could have a substantial impact on the clinical management of renal cell carcinoma, paving the way for new therapeutic approaches. IGF2BP3 influences tumor angiogenesis by modulating different targets in various digestive system cancers. For example, in GC, IGF2BP3 enhances tumor angiogenesis and cell migration by promoting HIF1A expression. In contrast, in CRC, IGF2BP3 inhibits VEGF expression, thereby reducing tumor angiogenesis. Thus, IGF2BP3 can be targeted in gastric cancer and CRC cells to control tumor growth by inhibiting angiogenesis, achieving clinical treatment objectives. Notably, YTHDF3 promotes tumor angiogenesis in BC by facilitating the translation of m6A-enriched transcripts such as ST6GALNAC5, GJA1, and EGFR. Therefore, YTHDF3 has emerged as a potential therapeutic target for controlling the development and metastasis of BC (Figure 3).
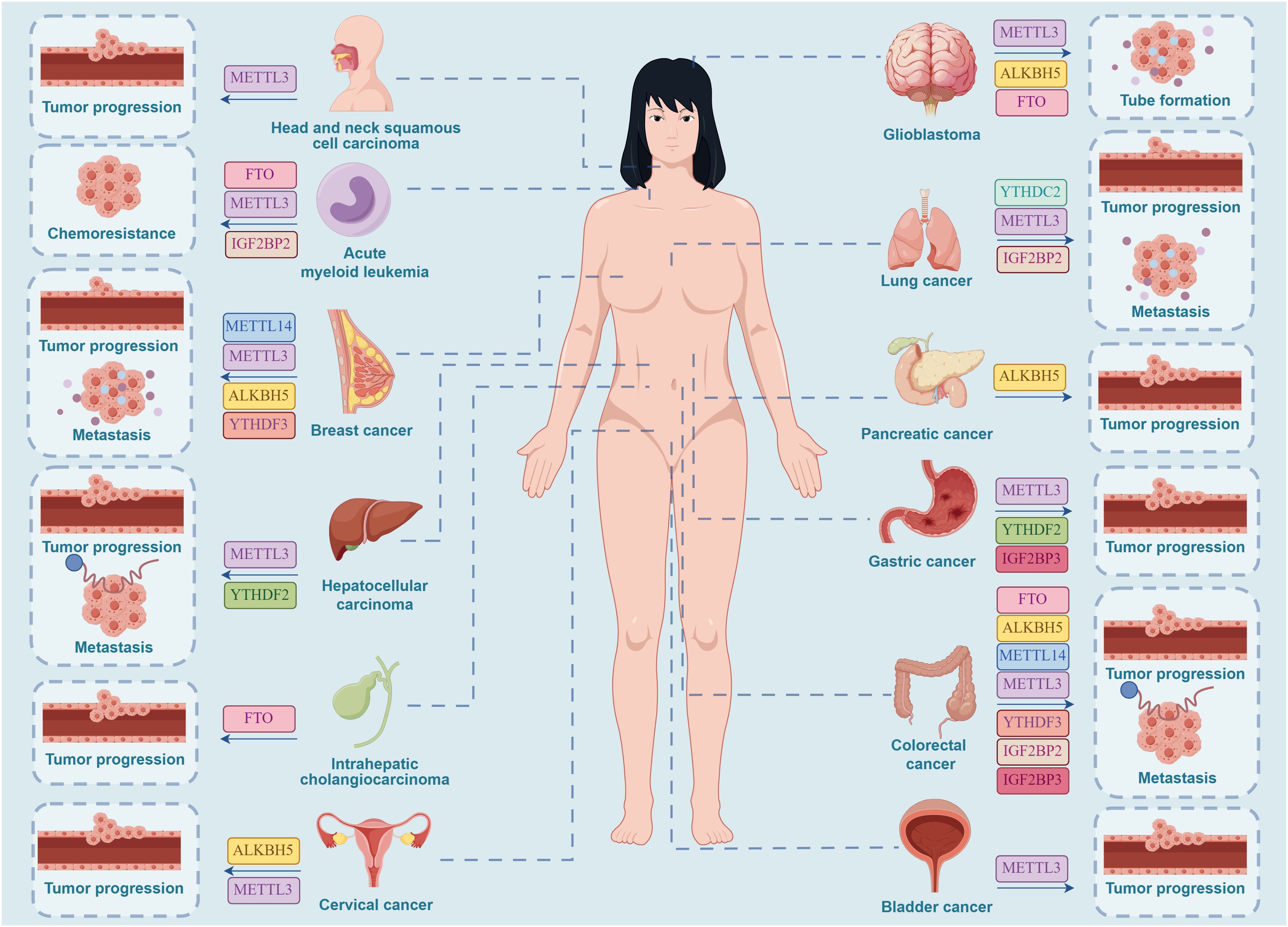
Figure 3. Effective targets of m6A inhibitors in clinical oncology therapy. METTL3 can serve as a clinical inhibitory target for a range of cancers, including HNSCC, AML, GC, BC, CC, HCC, GBM, LC, CRC, and BCa. METTL14 can serve as a clinical inhibitory target for brain metastases of BC and CRC. ALKBH5 can serve as a clinical inhibitory target for CC in females and for GBM, PC, CRC, and brain metastases of BC. FTO can serve as a clinical inhibitory target for AML, GBM, CRC, and ICC. YTHDF2 can serve as a clinical inhibitory target for HCC and GC. YTHDF3 can serve as a clinical inhibitory target for brain metastases of BC and CRC. IGF2BP2 can serve as a clinical inhibitory target for LC, AML, and CRC. IGF2BP3 can serve as a clinical inhibitory target for CRC and GC. YTHDC2 can serve as a clinical inhibitory target for LC. (By Figdraw.).
5 Conclusions and Prospective
M6A modification is pivotal in regulating gene expression and extensively influences various cellular functions and disease processes. By modulating posttranscriptional modifications and RNA metabolism, m6A ultimately governs physiological and pathological processes within the body. Additionally, m6A interacts with histone methylation and acetylation, collaboratively regulating gene transcriptional activity and contributing to heterochromatin formation, thereby affecting chromatin structure and function to regulate gene expression. During tumorigenesis, m6A modification facilitates angiogenesis by regulating the expression of genes associated with tumor vascularization, thus supplying essential nutrients and oxygen to the tumor and accelerating its growth and metastasis. Recent studies suggest that ncRNAs interact with m6A to jointly influence processes such as tumor cell proliferation, migration, invasion, and angiogenesis. Targeting these genes and pathways presents an opportunity for developing m6A inhibitors for various cancers, potentially enabling personalized therapies and enhancing clinical outcomes. This discovery broadens the potential for targeting m6A as a novel therapeutic target in cancer and provides substantial evidence for the development of clinical anticancer drugs.
Despite the rapid progress in m6A research, several challenges persist. Researchers are working to determine the overall abundance of m6A in specific diseases, which will aid in disease diagnosis and prognosis evaluation and reveal the underlying pathological mechanisms involved. This information is vital for developing novel therapeutic strategies and advancing personalized medicine. Current studies have quantified the total m6A abundance in diseases such as abdominal aortic aneurysm (211), diabetic cataracts (212), NSCLC (213), HNSCC (214), and age-related cataracts (215), with the aim of precisely understanding the role of m6A in these conditions. However, there are currently no reports on the quantification of m6A abundance, which is specifically related to angiogenesis in particular tumors. Future research should focus on investigating whether dynamic changes in m6A abundance occur during tumor progression, whether the abundance and modification patterns of m6A vary at different growth stages and treatment phases, and whether these dynamic changes influence tumor angiogenesis and treatment responses.
Numerous studies have demonstrated that m6A methyltransferases and demethylases play critical regulatory roles in the progression of certain tumors. For instance, in AML, the m6A methyltransferase METTL3 is crucial for the myeloid differentiation of both normal and leukemic cells. Compared to healthy hematopoietic stem/progenitor cells, METTL3 expression is significantly elevated in AML cells. Inhibiting METTL3 in AML cells not only induces differentiation but also increases apoptosis and slows the progression of leukemia. This indicates that METTL3 is essential for maintaining the undifferentiated state of leukemic cells (114). Additionally, research by Zejuan Li et al. has shown that the m6A demethylase FTO plays a key oncogenic role in AML. FTO is highly expressed in AML subtypes carrying t(11q23)/MLL rearrangements, t (15, 17)/PML-RARA, FLT3-ITD, and/or NPM1 mutations. FTO enhances leukemogenesis and leukemia oncogene-mediated cell transformation by reducing m6A levels on mRNA transcripts of targets such as ASB2 and RARA, thereby regulating their expression and inhibiting all-trans retinoic acid (ATRA)-induced differentiation of AML cells. Notably, this study also found that FTO expression can be upregulated by certain oncogenic proteins, such as MLL fusion proteins, PML-RARA, FLT3-ITD, and NPM1 mutants, leading to abnormally high levels of FTO in these AML subtypes. This abnormal upregulation typically does not directly rely on m6A regulatory mechanisms (145). Taken together, a noteworthy phenomenon worth deeper exploration is that in the same tumor, m6A methyltransferases and demethylases may jointly regulate tumor development and progression, potentially exerting similar effects (either promoting or inhibiting). This phenomenon could be attributed to multiple factors. On the one hand, tumor development, cancer cell proliferation and migration, and angiogenesis often involve a vast and complex regulatory network, with multiple signaling pathways intertwined. Various components of the m6A machinery may participate in the modification of multiple genes and proteins, and while they may sometimes exhibit consistent functions, their specific target genes and regulatory pathways may differ, leading to this observed phenomenon. On the other hand, during this process, m6A writers and erasers are not solely governed by the m6A regulatory system. The human body is a complex organism with multi-level regulatory mechanisms, where the abnormal expression of certain oncogenes and proteins in the tumor microenvironment can influence the expression of writers and erasers, subsequently participating in regulation in an m6A-dependent manner. Therefore, the precise mechanisms by which m6A influences tumor progression and angiogenesis require more detailed and in-depth research to enable its application in clinical settings, ultimately becoming an effective therapeutic target to improve cancer patient survival rates.
In the future, developing targeted inhibitor treatment strategies for m6A in tumor angiogenesis will be crucial. In combination therapy strategies, m6A modification can affect tumor sensitivity to conventional therapies such as chemotherapy, radiotherapy, or immunotherapy, potentially enhancing therapeutic efficacy and reducing side effects. The levels of m6A modification and the expression of related factors could serve as biomarkers for tumor angiogenesis, aiding in the assessment of angiogenesis status, predicting disease progression, and guiding the formulation of individualized treatment plans. Further exploration of the specific mechanisms of m6A modification in different tumor types, particularly its impact on angiogenesis and the tumor microenvironment, as well as clinical trials of m6A-targeted drugs, is crucial. These trials should evaluate the efficacy, safety, and potential side effects of these therapies in cancer treatment. Personalized treatment strategies based on m6A modification hold promise for improving the precision and effectiveness of therapies, thereby enhancing the clinical outcomes of patients.
Author contributions
YY: Writing – original draft. EY: Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The key project of the Henan Provincial Medical Science and Technology Tackling Plan, jointly established by the province and the ministry, in 2023 (SBGJ202302081).
Acknowledgments
Thanks for every writer. Special thanks to the newsletter author, Professor Yuan, for his guidance.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Glossary
m6A: N6-methyladenosine
mRNA: messenger RNA
NcRNA: Non-coding RNA
MTC: m6A methyltransferase complex
FTO: Fat Mass and Obesity-associated
siRNA: Small interfering RNA
METTL3/5/7A/7B/14/16: Methyltransferase-like 3/5/7A/7B/14/16
TRMT112: tRNA MTase subunit 11–2
WTAP: the METTL3 adaptor protein wilms tumor 1-associated protein
RBM15: RNA-binding motif protein15
ZC3H13: Zinc finger CCCH domain-containing protein13
CBLL1: Cbl proto-oncogene like 1
ZCCHC4: zinc finger CCHC domain-containing protein 4
YTHDC1/2: YTH domain-containing 1/2
YTHDF1-3: YTH N6-methyladenosine RNA-binding protein 1-3
ALKBH5: AlkB homolog 5
VIRMA/KIAA1429: Vir-like m6A methyltransferase associated protein
IGF2BP2/3: Insulin-like growth factor 2 mRNA-binding protein 2/3
miRNA: microRNA
piRNA: PIWI-interacting RNA
circRNA: Circular RNA
LncRNA: long non-coding RNA
m6Am: 2’-O-dimethyladenosine
VEGF: Vascular endothelial growth factor
bFGF: basic fibroblast growth factor
VEGFR: vascular endothelial growth factor receptor
FGF: fibroblast growth factor
FGFR: fibroblast growth factor receptor
PI3K: phosphatidylinositol 3-kinase
PKB: protein kinase B
IRF5: interferon regulatory factor 5
PPP6C: protein phosphatase 6 catalytic subunit
TSCC: tongue squamous cell carcinoma
BATF2: basic leucine zipper ATF-like transcription factor 2
LUAD: lung adenocarcinoma
AML: acute myeloid leukemia
GC: gastric cancer
MTOR: mammalian target of rapamycin
BCa: bladder cancer
TEK: tie-2 receptor tyrosine kinase
VEGFA: vascular endothelial growth factor A
CRC: colorectal cancer
CCND1: CyclinD1
PDCD4: programmed cell death 4
MAPK: mitogen-activated protein kinases
GBM: glioblastoma
ERK: extracellular signal-regulated kinase
IL-8: Interleukin-8
HDGF: hepatoma-derived growth factor
HIF1a: hypoxia-inducible factor-1a
HCC: hepatocellular carcinoma
HUVECs: human umbilical vein endothelial cells
PC: pancreatic cancer
RARA: retinoic acid receptor alpha
MYC: myelocytomatosis oncogene
CEBPA: enhancer binding protein α
ICC: intrahepatic cholangiocarcinoma
OC: ovarian cancer
CC: cervical cancer
LC: lung cancer
TNS1: tensin 1
EC: endometrial cancer
TK1: thymidine kinase 1
VASH1: Vasohibin-1
BC: breast cancer
5-FU: 5-fluorouracil
KO: knockout
R-2HG: R-2-Hydroxyglutarate
CLIC4: recombinant chloride intracellular channel protein 4
NSCLC: non-small cell lung cancer
TRPM7: transient receptor potential melastatin 7
RCC: renal cell carcinoma
cRCC: clear cell kidney cancer
PEBP1: recombinant phosphatidylethanolamine binding protein 1
PLAU: plasminogen activator urokinase
EMT: epithelial-mesenchymal transition
VM: vasculogenic mimicry
ETV5: ETS variant transcription factor 5
TRAF1: Tumor Necrosis Factor Receptor (TNFR)-associated factor 1
GJA1: gap junction 1
IARC: the International Agency for Research on Cancer
References
1. Lugano R, Ramachandran M, Dimberg A. Tumor angiogenesis: causes, consequences, challenges and opportunities. Cell Mol Life Sci. (2020) 77:1745–70. doi: 10.1007/s00018-019-03351-7
2. Lopes-Coelho F, Martins F, Pereira SA, Serpa J. Anti-angiogenic therapy: current challenges and future perspectives. Int J Mol Sci. (2021) 22:3765. doi: 10.3390/ijms22073765
3. Liu Z-L, Chen H-H, Zheng L-L, Sun L-P, Shi L. Angiogenic signaling pathways and anti-angiogenic therapy for cancer. Signal Transduction Targeted Ther. (2023) 8:198. doi: 10.1038/s41392-023-01460-1
4. Han M, Sun H, Zhou Q, Liu J, Hu J, Yuan W, et al. Effects of RNA methylation on Tumor angiogenesis and cancer progression. Mol Cancer. (2023) 22:198. doi: 10.1186/s12943-023-01879-8
5. Pirola L, Ciesielski O, Balcerczyk A. The methylation status of the epigenome: its emerging role in the regulation of tumor angiogenesis and tumor growth, and potential for drug targeting. Cancers (Basel). (2018) 10:268. doi: 10.3390/cancers10080268
6. Liu C, Tang H, Hu N, Li T. Methylomics and cancer: the current state of methylation profiling and marker development for clinical care. Cancer Cell Int. (2023) 23:242. doi: 10.1186/s12935-023-03074-7
7. Liang WW, Lu RJ, Jayasinghe RG, Foltz SM, Porta-Pardo E, Geffen Y, et al. Integrative multi-omic cancer profiling reveals DNA methylation patterns associated with therapeutic vulnerability and cell-of-origin. Cancer Cell. (2023) 41:1567–85.e7. doi: 10.1016/j.ccell.2023.07.013
8. Liu R, Zhao E, Yu H, Yuan C, Abbas MN, Cui H. Methylation across the central dogma in health and diseases: new therapeutic strategies. Signal Transduct Target Ther. (2023) 8:310. doi: 10.1038/s41392-023-01528-y
9. Ries RJ, Zaccara S, Klein P, Olarerin-George A, Namkoong S, Pickering BF, et al. m(6)A enhances the phase separation potential of mRNA. Nature. (2019) 571:424–8. doi: 10.1038/s41586-019-1374-1
10. He PC, He C. m(6) A RNA methylation: from mechanisms to therapeutic potential. EMBO J. (2021) 40:e105977. doi: 10.15252/embj.2020105977
11. Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. (2014) 505:117–20. doi: 10.1038/nature12730
12. Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, et al. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. (2015) 161:1388–99. doi: 10.1016/j.cell.2015.05.014
13. Liu Z, Zhang J. Human C-to-U coding RNA editing is largely nonadaptive. Mol Biol Evol. (2018) 35:963–9. doi: 10.1093/molbev/msy011
14. Barbieri I, Tzelepis K, Pandolfini L, Shi J, Millán-Zambrano G, Robson SC, et al. Promoter-bound METTL3 maintains myeloid leukaemia by m(6)A-dependent translation control. Nature. (2017) 552:126–31. doi: 10.1038/nature24678
15. Su R, Dong L, Li C, Nachtergaele S, Wunderlich M, Qing Y, et al. R-2HG exhibits anti-tumor activity by targeting FTO/m(6)A/MYC/CEBPA signaling. Cell. (2018) 172:90–105.e23. doi: 10.1016/j.cell.2017.11.031
16. Xiang Y, Laurent B, Hsu CH, Nachtergaele S, Lu Z, Sheng W, et al. RNA m(6)A methylation regulates the ultraviolet-induced DNA damage response. Nature. (2017) 543:573–6. doi: 10.1038/nature21671
17. Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. (2012) 485:201–6. doi: 10.1038/nature11112
18. Zhao X, Yang Y, Sun BF, Shi Y, Yang X, Xiao W, et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. (2014) 24:1403–19. doi: 10.1038/cr.2014.151
19. Zaccara S, Ries RJ, Jaffrey SR. Publisher Correction: Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol. (2023) 24:770. doi: 10.1038/s41580-023-00654-3
20. Tian Y, Zhang M, Liu LX, Wang ZC, Liu B, Huang Y, et al. Exploring non-coding RNA mechanisms in hepatocellular carcinoma: implications for therapy and prognosis. Front Immunol. (2024) 15:1400744. doi: 10.3389/fimmu.2024.1400744
21. Jiang X, Liu B, Nie Z, Duan L, Xiong Q, Jin Z, et al. The role of m6A modification in the biological functions and diseases. Signal Transduct Target Ther. (2021) 6:74. doi: 10.1038/s41392-020-00450-x
22. Cun Y, Guo W, Ma B, Okuno Y, Wang J. Decoding the specificity of m(6)A RNA methylation and its implication in cancer therapy. Mol Ther. (2024) 32:2461–9. doi: 10.1016/j.ymthe.2024.05.035
23. Krug RM, Morgan MA, Shatkin AJ. Influenza viral mRNA contains internal N6-methyladenosine and 5'-terminal 7-methylguanosine in cap structures. J Virol. (1976) 20:45–53. doi: 10.1128/jvi.20.1.45-53.1976
24. Kane SE, Beemon K. Precise localization of m6A in Rous sarcoma virus RNA reveals clustering of methylation sites: implications for RNA processing. Mol Cell Biol. (1985) 5:2298–306. doi: 10.1128/mcb.5.9.2298-2306.1985
25. Csepany T, Lin A, Baldick CJ Jr., Beemon K. Sequence specificity of mRNA N6-adenosine methyltransferase. J Biol Chem. (1990) 265:20117–22. doi: 10.1016/S0021-9258(17)30477-5
26. Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell. (2012) 149:1635–46. doi: 10.1016/j.cell.2012.05.003
27. Chen M, Wei L, Law CT, Tsang FHC, Shen J, Cheng CLH, et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. (2018) 67:2254–70. doi: 10.1002/hep.29683
28. Liu ZX, Li LM, Sun HL, Liu SM. Link between m6A modification and cancers. Front Bioeng Biotechnol. (2018) 6:89. doi: 10.3389/fbioe.2018.00089
29. Panneerdoss S, Eedunuri VK, Yadav P, Timilsina S, Rajamanickam S, Viswanadhapalli S, et al. Cross-talk among writers, readers, and erasers of m(6)A regulates cancer growth and progression. Sci Adv. (2018) 4:eaar8263. doi: 10.1126/sciadv.aar8263
30. Chen XY, Zhang J, Zhu JS. The role of m(6)A RNA methylation in human cancer. Mol Cancer. (2019) 18:103. doi: 10.1186/s12943-019-1033-z
31. Patil DP, Pickering BF, Jaffrey SR. Reading m(6)A in the Transcriptome: m(6)A-Binding Proteins. Trends Cell Biol. (2018) 28:113–27. doi: 10.1016/j.tcb.2017.10.001
32. Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M, et al. Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science. (2015) 347:1002–6. doi: 10.1126/science.1261417
33. Bokar JA, Shambaugh ME, Polayes D, Matera AG, Rottman FM. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. Rna. (1997) 3:1233–47.
34. Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. (2014) 10:93–5. doi: 10.1038/nchembio.1432
35. Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. (2014) 24:177–89. doi: 10.1038/cr.2014.3
36. Robey RW, Fitzsimmons CM, Guiblet WM, Frye WJE, González Dalmasy JM, Wang L, et al. The methyltransferases METTL7A and METTL7B confer resistance to thiol-based histone deacetylase inhibitors. Mol Cancer Ther. (2024) 23:464–77. doi: 10.1158/1535-7163.MCT-23-0144
37. Knight HM, Demirbugen Öz M, PerezGrovas-Saltijeral A. Dysregulation of RNA modification systems in clinical populations with neurocognitive disorders. Neural Regener Res. (2024) 19:1256–61. doi: 10.4103/1673-5374.385858
38. Wei Z, Chen Y, Zeng Z, Peng Y, Li L, Hu N, et al. The novel m6A writer METTL5 as prognostic biomarker probably associating with the regulation of immune microenvironment in kidney cancer. Heliyon. (2022) 8:e12078. doi: 10.1016/j.heliyon.2022.e12078
39. van Tran N, Ernst FGM, Hawley BR, Zorbas C, Ulryck N, Hackert P, et al. The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Res. (2019) 47:7719–33. doi: 10.1093/nar/gkz619
40. Oerum S, Meynier V, Catala M, Tisné C. A comprehensive review of m6A/m6Am RNA methyltransferase structures. Nucleic Acids Res. (2021) 49:7239–55. doi: 10.1093/nar/gkab378
41. Schwartz S, Mumbach MR, Jovanovic M, Wang T, Maciag K, Bushkin GG, et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5' sites. Cell Rep. (2014) 8:284–96. doi: 10.1016/j.celrep.2014.05.048
42. Uddin MB, Wang Z, Yang C. The m6A RNA methylation regulates oncogenic signaling pathways driving cell Malignant transformation and carcinogenesis. Mol Cancer. (2021) 20:61. doi: 10.1186/s12943-021-01356-0
43. Melstrom L, Chen J. RNA N(6)-methyladenosine modification in solid tumors: new therapeutic frontiers. Cancer Gene Ther. (2020) 27:625–33. doi: 10.1038/s41417-020-0160-4
44. Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M, et al. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. (2016) 537:369–73. doi: 10.1038/nature19342
45. Koh CWQ, Goh YT, Goh WSS. Atlas of quantitative single-base-resolution N(6)-methyl-adenine methylomes. Nat Commun. (2019) 10:5636. doi: 10.1038/s41467-019-13561-z
46. Wen J, Lv R, Ma H, Shen H, He C, Wang J, et al. Zc3h13 regulates nuclear RNA m(6)A methylation and mouse embryonic stem cell self-renewal. Mol Cell. (2018) 69:1028–38.e6. doi: 10.1016/j.molcel.2018.02.015
47. Bian S, Ni W, Zhu M, Song Q, Zhang J, Ni R, et al. Identification and validation of the N6-methyladenosine RNA methylation regulator YTHDF1 as a novel prognostic marker and potential target for hepatocellular carcinoma. Front Mol Biosci. (2020) 7:604766. doi: 10.3389/fmolb.2020.604766
48. Yang Y, Hsu PJ, Chen YS, Yang YG. Dynamic transcriptomic m(6)A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res. (2018) 28:616–24. doi: 10.1038/s41422-018-0040-8
49. Liang E, Xiao S, Zhao C, Zhang Y, Fu G. M6A modification promotes blood-brain barrier breakdown during cerebral ischemia/reperfusion injury through increasing matrix metalloproteinase 3 expression. Heliyon. (2023) 9:e16905. doi: 10.1016/j.heliyon.2023.e16905
50. Zhu H, Chen K, Chen Y, Liu J, Zhang X, Zhou Y, et al. RNA-binding protein ZCCHC4 promotes human cancer chemoresistance by disrupting DNA-damage-induced apoptosis. Signal Transduct Target Ther. (2022) 7:240. doi: 10.1038/s41392-022-01033-8
51. Wang X, Feng J, Xue Y, Guan Z, Zhang D, Liu Z, et al. Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature. (2016) 534:575–8. doi: 10.1038/nature18298
52. Warda AS, Kretschmer J, Hackert P, Lenz C, Urlaub H, Höbartner C, et al. Human METTL16 is a N(6)-methyladenosine (m(6)A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep. (2017) 18:2004–14. doi: 10.15252/embr.201744940
53. Yue Y, Liu J, Cui X, Cao J, Luo G, Zhang Z, et al. VIRMA mediates preferential m(6)A mRNA methylation in 3'UTR and near stop codon and associates with alternative polyadenylation. Cell Discovery. (2018) 4:10. doi: 10.1038/s41421-018-0019-0
54. Knuckles P, Lence T, Haussmann IU, Jacob D, Kreim N, Carl SH, et al. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m(6)A machinery component Wtap/Fl(2)d. Genes Dev. (2018) 32:415–29. doi: 10.1101/gad.309146.117
55. Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. (2011) 7:885–7. doi: 10.1038/nchembio.687
56. Liu Je, Li K, Cai J, Zhang M, Zhang X, Xiong X, et al. Landscape and Regulation of m6A and m6Am Methylome across Human and Mouse Tissues. Mol Cell. (2020) 77:426–40.e6. doi: 10.1016/j.molcel.2019.09.032
57. Sun H, Li K, Zhang X, Je L, Zhang M, Meng H, et al. m6Am-seq reveals the dynamic m6Am methylation in the human transcriptome. Nat Commun. (2021) 12:4778. doi: 10.1038/s41467-021-25105-5
58. Covelo-Molares H, Obrdlik A, Poštulková I, Dohnálková M, Gregorová P, Ganji R, et al. The comprehensive interactomes of human adenosine RNA methyltransferases and demethylases reveal distinct functional and regulatory features. Nucleic Acids Res. (2021) 49:10895–910. doi: 10.1093/nar/gkab900
59. Mauer J, Jaffrey SR. FTO. m6Am, and the hypothesis of reversible epitranscriptomic mRNA modifications. FEBS Letters. (2018) 592:2012–22. doi: 10.1002/1873-3468.13092
60. Mauer J, Luo X, Blanjoie A, Jiao X, Grozhik AV, Patil DP, et al. Reversible methylation of m6Am in the 5′ cap controls mRNA stability. Nature. (2016) 541:371–5. doi: 10.1038/nature21022
61. Wei J, Liu F, Lu Z, Fei Q, Ai Y, He PC, et al. Differential m(6)A, m(6)A(m), and m(1)A Demethylation Mediated by FTO in the Cell Nucleus and Cytoplasm. Mol Cell. (2018) 71:973–85.e5. doi: 10.1016/j.molcel.2018.08.011
62. Liu Z, Lan P, Liu T, Liu X, Liu T. m6Aminer: Predicting the m6Am Sites on mRNA by Fusing Multiple Sequence-Derived Features into a CatBoost-Based Classifier. Int J Mol Sci. (2023) 24:7878. doi: 10.3390/ijms24097878
63. Luo Z, Su W, Lou L, Qiu W, Xiao X, Xu Z. DLm6Am: A deep-learning-based tool for identifying N6,2′-O-dimethyladenosine sites in RNA sequences. Int J Mol Sci. (2022) 23:11026. doi: 10.3390/ijms231911026
64. Boulias K, Toczydłowska-Socha D, Hawley BR, Liberman N, Takashima K, Zaccara S, et al. Identification of the m(6)Am Methyltransferase PCIF1 Reveals the Location and Functions of m(6)Am in the Transcriptome. Mol Cell. (2019) 75:631–43.e8. doi: 10.1016/j.molcel.2019.06.006
65. Mauer J, Sindelar M, Despic V, Guez T, Hawley BR, Vasseur J-J, et al. FTO controls reversible m6Am RNA methylation during snRNA biogenesis. Nat Chem Biol. (2019) 15:340–7. doi: 10.1038/s41589-019-0231-8
66. Cesaro B, Tarullo M, Fatica A. Regulation of gene expression by m6Am RNA modification. Int J Mol Sci. (2023) 24:2277. doi: 10.3390/ijms24032277
67. Li Y, Su R, Deng X, Chen Y, Chen J. FTO in cancer: functions, molecular mechanisms, and therapeutic implications. Trends Cancer. (2022) 8:598–614. doi: 10.1016/j.trecan.2022.02.010
68. Shi H, Wei J, He C. Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol Cell. (2019) 74:640–50. doi: 10.1016/j.molcel.2019.04.025
69. Haussmann IU, Bodi Z, Sanchez-Moran E, Mongan NP, Archer N, Fray RG, et al. m(6)A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature. (2016) 540:301–4. doi: 10.1038/nature20577
70. Chen JY, Xiong T, Sun YR, Cong J, Gong JS, Peng L, et al. Modification of m6A mediates tissue immune microenvironment in calcific aortic valve disease. Ann Transl Med. (2022) 10:931. doi: 10.21037/atm
71. Bell JL, Wächter K, Mühleck B, Pazaitis N, Köhn M, Lederer M, et al. Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs): post-transcriptional drivers of cancer progression? Cell Mol Life Sci. (2013) 70:2657–75. doi: 10.1007/s00018-012-1186-z
72. Nielsen J, Christiansen J, Lykke-Andersen J, Johnsen AH, Wewer UM, Nielsen FC. A family of insulin-like growth factor II mRNA-binding proteins represses translation in late development. Mol Cell Biol. (1999) 19:1262–70. doi: 10.1128/MCB.19.2.1262
73. Ramesh-Kumar D, Guil S. The IGF2BP family of RNA binding proteins links epitranscriptomics to cancer. Semin Cancer Biol. (2022) 86:18–31. doi: 10.1016/j.semcancer.2022.05.009
74. Chen L, Gao Y, Xu S, Yuan J, Wang M, Li T, et al. N6-methyladenosine reader YTHDF family in biological processes: Structures, roles, and mechanisms. Front Immunol. (2023) 14:1162607. doi: 10.3389/fimmu.2023.1162607
75. Sikorski V, Selberg S, Lalowski M, Karelson M, Kankuri E. The structure and function of YTHDF epitranscriptomic m(6)A readers. Trends Pharmacol Sci. (2023) 44:335–53. doi: 10.1016/j.tips.2023.03.004
76. Chen Y, Wan R, Zou Z, Lao L, Shao G, Zheng Y, et al. O-GlcNAcylation determines the translational regulation and phase separation of YTHDF proteins. Nat Cell Biol. (2023) 25:1676–90. doi: 10.1038/s41556-023-01258-x
77. Ma S, Sun B, Duan S, Han J, Barr T, Zhang J, et al. YTHDF2 orchestrates tumor-associated macrophage reprogramming and controls antitumor immunity through CD8(+) T cells. Nat Immunol. (2023) 24:255–66. doi: 10.1038/s41590-022-01398-6
78. Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, et al. YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. (2017) 27:315–28. doi: 10.1038/cr.2017.15
79. Zaccara S, Jaffrey SR. Understanding the redundant functions of the m(6)A-binding YTHDF proteins. Rna. (2024) 30:468–81. doi: 10.1261/rna.079988.124
80. Zaccara S, Jaffrey SR. A Unified Model for the Function of YTHDF Proteins in Regulating m6A-Modified mRNA. Cell. (2020) 181:1582–95.e18. doi: 10.1016/j.cell.2020.05.012
81. Zou Z, Sepich-Poore C, Zhou X, Wei J, He C. The mechanism underlying redundant functions of the YTHDF proteins. Genome Biol. (2023) 24:17. doi: 10.1186/s13059-023-02862-8
82. Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. (2016) 17:47–62. doi: 10.1038/nrg.2015.10
83. Yan H, Bu P. Non-coding RNA in cancer. Essays Biochem. (2021) 65:625–39. doi: 10.1042/EBC20200032
84. Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. (2012) 482:339–46. doi: 10.1038/nature10887
85. Hill M, Tran N. miRNA interplay: mechanisms and consequences in cancer. Dis Model Mech. (2021) 14:dmm047662. doi: 10.1242/dmm.047662
86. Kousar K, Ahmad T, Abduh MS, Kanwal B, Shah SS, Naseer F, et al. miRNAs in regulation of tumor microenvironment, chemotherapy resistance, immunotherapy modulation and miRNA therapeutics in cancer. Int J Mol Sci. (2022) 23:13822. doi: 10.3390/ijms232213822
87. Herman AB, Tsitsipatis D, Gorospe M. Integrated lncRNA function upon genomic and epigenomic regulation. Mol Cell. (2022) 82:2252–66. doi: 10.1016/j.molcel.2022.05.027
88. Bridges MC, Daulagala AC, Kourtidis A. LNCcation: lncRNA localization and function. J Cell Biol. (2021) 220:e202009045. doi: 10.1083/jcb.202009045
89. Li H, Xu JD, Fang XH, Zhu JN, Yang J, Pan R, et al. Circular RNA circRNA_000203 aggravates cardiac hypertrophy via suppressing miR-26b-5p and miR-140-3p binding to Gata4. Cardiovasc Res. (2020) 116:1323–34. doi: 10.1093/cvr/cvz215
90. Sang Y, Chen B, Song X, Li Y, Liang Y, Han D, et al. circRNA_0025202 Regulates Tamoxifen Sensitivity and Tumor Progression via Regulating the miR-182-5p/FOXO3a Axis in Breast Cancer. Mol Ther. (2021) 29:3525–7. doi: 10.1016/j.ymthe.2021.11.002
91. Folkman J. Tumor angiogenesis: therapeutic implications. N Engl J Med. (1971) 285:1182–6. doi: 10.1056/NEJM197111182852108
92. Ferrara N. From the discovery of vascular endothelial growth factor to the introduction of avastin in clinical trials - an interview with Napoleone Ferrara by Domenico Ribatti. Int J Dev Biol. (2011) 55:383–8. doi: 10.1387/ijdb.103216dr
93. Ribatti D. Judah Folkman, a pioneer in the study of angiogenesis. Angiogenesis. (2008) 11:3–10. doi: 10.1007/s10456-008-9092-6
94. Ribatti D. Napoleone Ferrara and the saga of vascular endothelial growth factor. Endothelium. (2008) 15:1–8. doi: 10.1080/10623320802092377
95. Zhang L, Li C, Song X, Guo R, Zhao W, Liu C, et al. Targeting ONECUT2 inhibits tumor angiogenesis via down-regulating ZKSCAN3/VEGFA. Biochem Pharmacol. (2024) 225:116315. doi: 10.1016/j.bcp.2024.116315
96. Ciccone V, Terzuoli E, Donnini S, Giachetti A, Morbidelli L, Ziche M. Stemness marker ALDH1A1 promotes tumor angiogenesis via retinoic acid/HIF-1α/VEGF signalling in MCF-7 breast cancer cells. J Exp Clin Cancer Res. (2018) 37:311. doi: 10.1186/s13046-018-0975-0
97. Ilhan-Mutlu A, Siehs C, Berghoff AS, Ricken G, Widhalm G, Wagner L, et al. Expression profiling of angiogenesis-related genes in brain metastases of lung cancer and melanoma. Tumour Biol. (2016) 37:1173–82. doi: 10.1007/s13277-015-3790-7
98. Olsson AK, Dimberg A, Kreuger J, Claesson-Welsh L. VEGF receptor signalling - in control of vascular function. Nat Rev Mol Cell Biol. (2006) 7:359–71. doi: 10.1038/nrm1911
99. Takahashi H, Shibuya M. The vascular endothelial growth factor (VEGF)/VEGF receptor system and its role under physiological and pathological conditions. Clin Sci (Lond). (2005) 109:227–41. doi: 10.1042/CS20040370
100. Hu Y, Yu C, Cheng L, Zhong C, An J, Zou M, et al. Flavokawain C inhibits glucose metabolism and tumor angiogenesis in nasopharyngeal carcinoma by targeting the HSP90B1/STAT3/HK2 signaling axis. Cancer Cell Int. (2024) 24:158. doi: 10.1186/s12935-024-03314-4
101. Huang W, Qi CB, Lv SW, Xie M, Feng YQ, Huang WH, et al. Determination of DNA and RNA methylation in circulating tumor cells by mass spectrometry. Anal Chem. (2016) 88:1378–84. doi: 10.1021/acs.analchem.5b03962
102. Ma S, Chen C, Ji X, Liu J, Zhou Q, Wang G, et al. The interplay between m6A RNA methylation and noncoding RNA in cancer. J Hematol Oncol. (2019) 12:121. doi: 10.1186/s13045-019-0805-7
103. Cesarini V, Silvestris DA, Tassinari V, Tomaselli S, Alon S, Eisenberg E, et al. ADAR2/miR-589-3p axis controls glioblastoma cell migration/invasion. Nucleic Acids Res. (2018) 46:2045–59. doi: 10.1093/nar/gkx1257
104. Li J, Huang C, Zou Y, Ye J, Yu J, Gui Y. CircTLK1 promotes the proliferation and metastasis of renal cell carcinoma by sponging miR-136-5p. Mol Cancer. (2020) 19:103. doi: 10.1186/s12943-020-01225-2
105. Luo H, Zhu G, Xu J, Lai Q, Yan B, Guo Y, et al. HOTTIP lncRNA promotes hematopoietic stem cell self-renewal leading to AML-like disease in mice. Cancer Cell. (2019) 36:645–59.e8. doi: 10.1016/j.ccell.2019.10.011
106. Wang S, Zhu X, Hao Y, Su TT, Shi W. ALKBH5-mediated m6A modification of circFOXP1 promotes gastric cancer progression by regulating SOX4 expression and sponging miR-338-3p. Commun Biol. (2024) 7:565. doi: 10.1038/s42003-024-06274-7
107. Li R, Zhu C, Wang Y, Wang X, Wang Y, Wang J, et al. The relationship between the network of non-coding RNAs-molecular targets and N6-methyladenosine modification in tumors of urinary system. Cell Death Dis. (2024) 15:275. doi: 10.1038/s41419-024-06664-z
108. Rong M, Zhang M, Dong F, Wu K, Cai B, Niu J, et al. LncRNA RASAL2-AS1 promotes METTL14-mediated m6A methylation in the proliferation and progression of head and neck squamous cell carcinoma. Cancer Cell Int. (2024) 24:113. doi: 10.1186/s12935-024-03302-8
109. Chen D, Zhao H, Guo Z, Dong Z, Yu Y, Zheng J, et al. Identification of m6A-related lncRNAs LINC02471 and DOCK9-DT as potential biomarkers for thyroid cancer. Int Immunopharmacol. (2024) 133:112050. doi: 10.1016/j.intimp.2024.112050
110. Tuersun H, Liu L, Zhang J, Maimaitizunong R, Tang X, Li H. m6A reading protein RBMX as a biomarker for prognosis and tumor progression in esophageal cancer. Transl Cancer Res. (2023) 12:2319–35. doi: 10.21037/tcr
111. Zhang J, Zhang Y, Zhang J. N6-Methyladenosine modified circ-NAB1 modulates cell cycle and epithelial-mesenchymal transition via CDKN3 in endometrial cancer. Cell Mol Biol (Noisy-le-grand). (2024) 70:161–9. doi: 10.14715/cmb/2024.70.5.23
112. Zhang G, Wang T, Huang Z, Chen Y, Sun L, Xia X, et al. METTL3 dual regulation of the stability of LINC00662 and VEGFA RNAs promotes colorectal cancer angiogenesis. Discovery Oncol. (2022) 13:89. doi: 10.1007/s12672-022-00557-3
113. Chamorro-Jorganes A, Sweaad WK, Katare R, Besnier M, Anwar M, Beazley-Long N, et al. METTL3 Regulates Angiogenesis by Modulating let-7e-5p and miRNA-18a-5p Expression in Endothelial Cells. Arterioscler Thromb Vasc Biol. (2021) 41:e325–e37. doi: 10.1161/ATVBAHA.121.316180
114. Vu LP, Pickering BF, Cheng Y, Zaccara S, Nguyen D, Minuesa G, et al. The N(6)-methyladenosine (m(6)A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat Med. (2017) 23:1369–76. doi: 10.1038/nm.4416
115. Wang N, Huo X, Zhang B, Chen X, Zhao S, Shi X, et al. METTL3-mediated ADAMTS9 suppression facilitates angiogenesis and carcinogenesis in gastric cancer. Front Oncol. (2022) 12:861807. doi: 10.3389/fonc.2022.861807
116. Zhang H, Zhou J, Li J, Wang Z, Chen Z, Lv Z, et al. N6-methyladenosine promotes translation of VEGFA to accelerate angiogenesis in lung cancer. Cancer Res. (2023) 83:2208–25. doi: 10.1158/0008-5472.CAN-22-2449
117. Bi X, Lv X, Liu D, Guo H, Yao G, Wang L, et al. METTL3-mediated maturation of miR-126-5p promotes ovarian cancer progression via PTEN-mediated PI3K/Akt/mTOR pathway. Cancer Gene Ther. (2021) 28:335–49. doi: 10.1038/s41417-020-00222-3
118. Zhang H, Zhang P, Long C, Ma X, Huang H, Kuang X, et al. m(6) A methyltransferase METTL3 promotes retinoblastoma progression via PI3K/AKT/mTOR pathway. J Cell Mol Med. (2020) 24:12368–78. doi: 10.1111/jcmm.15736
119. Wang G, Dai Y, Li K, Cheng M, Xiong G, Wang X, et al. Deficiency of mettl3 in bladder cancer stem cells inhibits bladder cancer progression and angiogenesis. Front Cell Dev Biol. (2021) 9:627706. doi: 10.3389/fcell.2021.627706
120. Abdul-Sater AA, Edilova MI, Clouthier DL, Mbanwi A, Kremmer E, Watts TH. The signaling adaptor TRAF1 negatively regulates Toll-like receptor signaling and this underlies its role in rheumatic disease. Nat Immunol. (2017) 18:26–35. doi: 10.1038/ni.3618
121. Guo YQ, Wang Q, Wang JG, Gu YJ, Song PP, Wang SY, et al. METTL3 modulates m6A modification of CDC25B and promotes head and neck squamous cell carcinoma Malignant progression. Exp Hematol Oncol. (2022) 11:14. doi: 10.1186/s40164-022-00256-3
122. Liu M, Ruan X, Liu X, Dong W, Wang D, Yang C, et al. The mechanism of BUD13 m6A methylation mediated MBNL1-phosphorylation by CDK12 regulating the vasculogenic mimicry in glioblastoma cells. Cell Death Dis. (2022) 13:1017. doi: 10.1038/s41419-022-05426-z
123. Hou PX, Fan Q, Zhang Q, Liu JJ, Wu Q. M6A-induced transcription factor IRF5 contributes to the progression of cervical cancer by upregulating PPP6C. Clin Exp Pharmacol Physiol. (2024) 51:e13868. doi: 10.1111/1440-1681.13868
124. Liu X, Xiao M, Zhang L, Li L, Zhu G, Shen E, et al. The m6A methyltransferase METTL14 inhibits the proliferation, migration, and invasion of gastric cancer by regulating the PI3K/AKT/mTOR signaling pathway. J Clin Lab Anal. (2021) 35:e23655. doi: 10.1002/jcla.23655
125. Chen XY, Yang YL, Yu Y, Chen ZY, Fan HN, Zhang J, et al. CircUGGT2 downregulation by METTL14-dependent m(6)A modification suppresses gastric cancer progression and cisplatin resistance through interaction with miR-186-3p/MAP3K9 axis. Pharmacol Res. (2024) 204:107206. doi: 10.1016/j.phrs.2024.107206
126. Dong L, Chen C, Zhang Y, Guo P, Wang Z, Li J, et al. The loss of RNA N(6)-adenosine methyltransferase Mettl14 in tumor-associated macrophages promotes CD8(+) T cell dysfunction and tumor growth. Cancer Cell. (2021) 39:945–57.e10. doi: 10.1016/j.ccell.2021.04.016
127. Pomaville M, Chennakesavalu M, Wang P, Jiang Z, Sun HL, Ren P, et al. Small-molecule inhibition of the METTL3/METTL14 complex suppresses neuroblastoma tumor growth and promotes differentiation. Cell Rep. (2024) 43:114165. doi: 10.1016/j.celrep.2024.114165
128. Chen Y, Lu Z, Qi C, Yu C, Li Y, Huan W, et al. N(6)-methyladenosine-modified TRAF1 promotes sunitinib resistance by regulating apoptosis and angiogenesis in a METTL14-dependent manner in renal cell carcinoma. Mol Cancer. (2022) 21:111. doi: 10.1186/s12943-022-01549-1
129. Wen H, Tang J, Cui Y, Hou M, Zhou J. m6A modification-mediated BATF2 suppresses metastasis and angiogenesis of tongue squamous cell carcinoma through inhibiting VEGFA. Cell Cycle. (2022) 22:100–16. doi: 10.1080/15384101.2022.2109897
130. Aik W, Scotti JS, Choi H, Gong L, Demetriades M, Schofield CJ, et al. Structure of human RNA N6-methyladenine demethylase ALKBH5 provides insights into its mechanisms of nucleic acid recognition and demethylation. Nucleic Acids Res. (2014) 42:4741–54. doi: 10.1093/nar/gku085
131. Cui Q, Shi H, Ye P, Li L, Qu Q, Sun G, et al. m(6)A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. (2017) 18:2622–34. doi: 10.1016/j.celrep.2017.02.059
132. Huff S, Tiwari SK, Gonzalez GM, Wang Y, Rana TM. m(6)A-RNA demethylase FTO inhibitors impair self-renewal in glioblastoma stem cells. ACS Chem Biol. (2021) 16:324–33. doi: 10.1021/acschembio.0c00841
133. Liu Y, Liang G, Xu H, Dong W, Dong Z, Qiu Z, et al. Tumors exploit FTO-mediated regulation of glycolytic metabolism to evade immune surveillance. Cell Metab. (2021) 33:1221–33.e11. doi: 10.1016/j.cmet.2021.04.001
134. Yu T, Liu J, Wang Y, Chen W, Liu Z, Zhu L, et al. METTL3 promotes colorectal cancer metastasis by stabilizing PLAU mRNA in an m6A-dependent manner. Biochem Biophys Res Commun. (2022) 614:9–16. doi: 10.1016/j.bbrc.2022.04.141
135. Yang Z, Wang T, Wu D, Min Z, Tan J, Yu B. RNA N6-methyladenosine reader IGF2BP3 regulates cell cycle and angiogenesis in colon cancer. J Exp Clin Cancer Res. (2020) 39:203. doi: 10.1186/s13046-020-01714-8
136. Fang H, Sun Q, Zhou J, Zhang H, Song Q, Zhang H, et al. m6A methylation reader IGF2BP2 activates endothelial cells to promote angiogenesis and metastasis of lung adenocarcinoma. Mol Cancer. (2023) 22:99. doi: 10.1186/s12943-023-01791-1
137. Weng H, Huang F, Yu Z, Chen Z, Prince E, Kang Y, et al. The m(6)A reader IGF2BP2 regulates glutamine metabolism and represents a therapeutic target in acute myeloid leukemia. Cancer Cell. (2022) 40:1566–82.e10. doi: 10.1016/j.ccell.2022.10.004
138. Jiang L, Li Y, He Y, Wei D, Yan L, Wen H. Knockdown of m6A reader IGF2BP3 inhibited hypoxia-induced cell migration and angiogenesis by regulating hypoxia inducible factor-1α in stomach cancer. Front Oncol. (2021) 11:711207. doi: 10.3389/fonc.2021.711207
139. Hou J, Zhang H, Liu J, Zhao Z, Wang J, Lu Z, et al. YTHDF2 reduction fuels inflammation and vascular abnormalization in hepatocellular carcinoma. Mol Cancer. (2019) 18:163. doi: 10.1186/s12943-019-1082-3
140. Chang G, Shi L, Ye Y, Shi H, Zeng L, Tiwary S, et al. YTHDF3 induces the translation of m(6)A-enriched gene transcripts to promote breast cancer brain metastasis. Cancer Cell. (2020) 38:857–71.e7. doi: 10.1016/j.ccell.2020.10.004
141. Alemu A,E, He C, Klungland A. ALKBHs-facilitated RNA modifications and de-modifications. DNA Repair (Amst). (2016) 44:87–91. doi: 10.1016/j.dnarep.2016.05.026
142. Wu G, Yan Y, Cai Y, Peng B, Li J, Huang J, et al. ALKBH1-8 and FTO: potential therapeutic targets and prognostic biomarkers in lung adenocarcinoma pathogenesis. Front Cell Dev Biol. (2021) 9:633927. doi: 10.3389/fcell.2021.633927
143. He Y, Hu H, Wang Y, Yuan H, Lu Z, Wu P, et al. ALKBH5 inhibits pancreatic cancer motility by decreasing long non-coding RNA KCNK15-AS1 methylation. Cell Physiol Biochem. (2018) 48:838–46. doi: 10.1159/000491915
144. Guo Y, Guo Y, Chen C, Fan D, Wu X, Zhao L, et al. Circ3823 contributes to growth, metastasis and angiogenesis of colorectal cancer: involvement of miR-30c-5p/TCF7 axis. Mol Cancer. (2021) 20:93. doi: 10.1186/s12943-021-01372-0
145. Li Z, Weng H, Su R, Weng X, Zuo Z, Li C, et al. FTO plays an oncogenic role in acute myeloid leukemia as a N(6)-methyladenosine RNA demethylase. Cancer Cell. (2017) 31:127–41. doi: 10.1016/j.ccell.2016.11.017
146. Rong Z-X, Li Z, He J-J, Liu L-Y, Ren X-X, Gao J, et al. Downregulation of fat mass and obesity associated (FTO) promotes the progression of intrahepatic cholangiocarcinoma. Front Oncol. (2019) 9. doi: 10.3389/fonc.2019.00369
147. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. (2013) 495:384–8. doi: 10.1038/nature11993
148. Ma YS, Shi BW, Guo JH, Liu JB, Yang XL, Xin R, et al. microRNA-320b suppresses HNF4G and IGF2BP2 expression to inhibit angiogenesis and tumor growth of lung cancer. Carcinogenesis. (2021) 42:762–71. doi: 10.1093/carcin/bgab023
149. Wang H, Deng Q, Lv Z, Ling Y, Hou X, Chen Z, et al. N6-methyladenosine induced miR-143-3p promotes the brain metastasis of lung cancer via regulation of VASH1. Mol Cancer. (2019) 18:181. doi: 10.1186/s12943-019-1108-x
150. Peng W, Li J, Chen R, Gu Q, Yang P, Qian W, et al. Upregulated METTL3 promotes metastasis of colorectal Cancer via miR-1246/SPRED2/MAPK signaling pathway. J Exp Clin Cancer Res. (2019) 38:393. doi: 10.1186/s13046-019-1408-4
151. Tan YT, Lin JF, Li T, Li JJ, Xu RH, Ju HQ. LncRNA-mediated posttranslational modifications and reprogramming of energy metabolism in cancer. Cancer Commun (Lond). (2021) 41:109–20. doi: 10.1002/cac2.12108
152. Longhi MS, Kokkotou E. Lnc-ing RNA expression with disease pathogenesis: MALAT1 and ANRIL in ulcerative colitis. Dig Dis Sci. (2020) 65:3061–3. doi: 10.1007/s10620-020-06216-3
153. Chuang TD, Ton N, Manrique N, Rysling S, Khorram O. Targeting the long non-coding RNA MIAT for the treatment of fibroids in an animal model. Clin Sci (Lond). (2024) 138:699–709. doi: 10.1042/CS20240190
154. Zhang R, Zhou Z, Wang P, He X, Liu Y, Chu M. The SLC19A1-AS/miR-1343/WNT11 axis is a novel positive regulatory ceRNA network governing goat granulosa cell proliferation. Int J Biol Macromolecules. (2024) 264:130658. doi: 10.1016/j.ijbiomac.2024.130658
155. Gao Y, Tong M, Wong TL, Ng KY, Xie YN, Wang Z, et al. Long noncoding RNA URB1-antisense RNA 1 (AS1) suppresses sorafenib-induced ferroptosis in hepatocellular carcinoma by driving ferritin phase separation. ACS Nano. (2023) 17:22240–58. doi: 10.1021/acsnano.3c01199
156. Song Y, Qu H. Identification and validation of a seven m6A-related lncRNAs signature predicting prognosis of ovarian cancer. BMC Cancer. (2022) 22:633. doi: 10.1186/s12885-022-09591-4
157. Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. (2009) 10:155–9. doi: 10.1038/nrg2521
158. Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. (2017) 36:5661–7. doi: 10.1038/onc.2017.184
159. Lv Y, Dong K, Gao H. Long non-coding RNA TDRG1 facilitates cell proliferation, migration and invasion in breast cancer via targeting miR-214-5p/CLIC4 axis. Cancer Biol Ther. (2021) 22:248–56. doi: 10.1080/15384047.2020.1863120
160. Sun T, Wu Z, Wang X, Wang Y, Hu X, Qin W, et al. Correction to: LNC942 promoting METTL14-mediated m 6 A methylation in breast cancer cell proliferation and progression. Oncogene. (2022) 41:1677. doi: 10.1038/s41388-022-02194-0
161. Rong D, Dong Q, Qu H, Deng X, Gao F, Li Q, et al. m(6)A-induced LINC00958 promotes breast cancer tumorigenesis via the miR-378a-3p/YY1 axis. Cell Death Discovery. (2021) 7:27. doi: 10.1038/s41420-020-00382-z
162. Wang X, Zhang J, Wang Y. Long noncoding RNA GAS5-AS1 suppresses growth and metastasis of cervical cancer by increasing GAS5 stability. Am J Transl Res. (2019) 11:4909–21.
163. Feng Y, Wu F, Wu Y, Guo Z, Ji X. LncRNA DGUOK-AS1 facilitates non-small cell lung cancer growth and metastasis through increasing TRPM7 stability via m6A modification. Trans Oncol. (2023) 32:101661. doi: 10.1016/j.tranon.2023.101661
164. Bian Y, Wang Y, Xu S, Gao Z, Li C, Fan Z, et al. m6A Modification of Long Non-Coding RNA HNF1A-AS1 Facilitates Cell Cycle Progression in Colorectal Cancer via IGF2BP2-Mediated CCND1 mRNA Stabilization. Cells. (2022) 11:3008. doi: 10.3390/cells11193008
165. Wang R, Ye H, Yang B, Ao M, Yu X, Wu Y, et al. m6A-modified circNFIX promotes ovarian cancer progression and immune escape via activating IL-6R/JAK1/STAT3 signaling by sponging miR-647. Int Immunopharmacol. (2023) 124:110879. doi: 10.1016/j.intimp.2023.110879
166. Shen T, Han M, Wei G, Ni T. An intriguing RNA species–perspectives of circularized RNA. Protein Cell. (2015) 6:871–80. doi: 10.1007/s13238-015-0202-0
167. Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. (2015) 12:381–8. doi: 10.1080/15476286.2015.1020271
168. Cui W, Dang Q, Chen C, Yuan W, Sun Z. Roles of circRNAs on tumor autophagy. Mol Ther Nucleic Acids. (2021) 23:918–29. doi: 10.1016/j.omtn.2021.01.002
169. Kohansal M, Alghanimi YK, Banoon SR, Ghasemian A, Afkhami H, Daraei A, et al. CircRNA-associated ceRNA regulatory networks as emerging mechanisms governing the development and biophysiopathology of epilepsy. CNS Neurosci Ther. (2024) 30:e14735. doi: 10.1111/cns.14735
170. Hentze MW, Preiss T. Circular RNAs: splicing's enigma variations. EMBO J. (2013) 32:923–5. doi: 10.1038/emboj.2013.53
171. Zhou Y, Mao X, Peng R, Bai D. CircRNAs in hepatocellular carcinoma: characteristic, functions and clinical significance. Int J Med Sci. (2022) 19:2033–43. doi: 10.7150/ijms.74713
172. Han D, Li J, Wang H, Su X, Hou J, Gu Y, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. (2017) 66:1151–64. doi: 10.1002/hep.29270
173. Chen X, Chen RX, Wei WS, Li YH, Feng ZH, Tan L, et al. PRMT5 Circular RNA Promotes Metastasis of Urothelial Carcinoma of the Bladder through Sponging miR-30c to Induce Epithelial-Mesenchymal Transition. Clin Cancer Res. (2018) 24:6319–30. doi: 10.1158/1078-0432.CCR-18-1270
174. Xu Z, Chen S, Liu R, Chen H, Xu B, Xu W, et al. Circular RNA circPOLR2A promotes clear cell renal cell carcinoma progression by facilitating the UBE3C-induced ubiquitination of PEBP1 and, thereby, activating the ERK signaling pathway. Mol Cancer. (2022) 21:146. doi: 10.1186/s12943-022-01607-8
175. Yankova E, Blackaby W, Albertella M, Rak J, De Braekeleer E, Tsagkogeorga G, et al. Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature. (2021) 593:597–601. doi: 10.1038/s41586-021-03536-w
176. Li Z, Feng Y, Han H, Jiang X, Chen W, Ma X, et al. A stapled peptide inhibitor targeting the binding interface of N6-adenosine-methyltransferase subunits METTL3 and METTL14 for cancer therapy. Angewandte Chemie Int Edition. (2024) 63:e202402611. doi: 10.1002/anie.202402611
177. Yanagi Y, Watanabe T, Hara Y, Sato Y, Kimura H, Murata T. EBV exploits RNA m6A modification to promote cell survival and progeny virus production during lytic cycle. Front Microbiol. (2022) 13. doi: 10.3389/fmicb.2022.870816
178. Deng L-J, Deng W-Q, Fan S-R, Chen M-F, Qi M, Lyu W-Y, et al. m6A modification: recent advances, anticancer targeted drug discovery and beyond. Mol Cancer. (2022) 21:52. doi: 10.1186/s12943-022-01510-2
179. Chen D, Ji F, Zhou Q, Cheung H, Pan Y, Lau HC, et al. RUVBL1/2 blockade targets YTHDF1 activity to suppress m6A-dependent oncogenic translation and colorectal tumorigenesis. Cancer Res. (2024), CAN-23-2081. doi: 10.1158/0008-5472.CAN-23-2081
180. Feng P, Chen D, Wang X, Li Y, Li Z, Li B, et al. Inhibition of the m6A reader IGF2BP2 as a strategy against T-cell acute lymphoblastic leukemia. Leukemia. (2022) 36:2180–8. doi: 10.1038/s41375-022-01651-9
181. Malacrida A, Rivara M, Di Domizio A, Cislaghi G, Miloso M, Zuliani V, et al. 3D proteome-wide scale screening and activity evaluation of a new ALKBH5 inhibitor in U87 glioblastoma cell line. Bioorganic Medicinal Chem. (2020) 28:115300. doi: 10.1016/j.bmc.2019.115300
182. Li N, Kang Y, Wang L, Huff S, Tang R, Hui H, et al. ALKBH5 regulates anti–PD-1 therapy response by modulating lactate and suppressive immune cell accumulation in tumor microenvironment. Proc Natl Acad Sci. (2020) 117:20159–70. doi: 10.1073/pnas.1918986117
183. Zannella C, Rinaldi L, Boccia G, Chianese A, Sasso FC, De Caro F, et al. Regulation of m6A Methylation as a New Therapeutic Option against COVID-19. Pharmaceuticals. (2021) 14:1135. doi: 10.3390/ph14111135
184. Azzam SK, Alsafar H, Sajini AA. FTO m6A demethylase in obesity and cancer: implications and underlying molecular mechanisms. Int J Mol Sci. (2022) 23:3800. doi: 10.3390/ijms23073800
185. Huang Y, Yan J, Li Q, Li J, Gong S, Zhou H, et al. Meclofenamic acid selectively inhibits FTO demethylation of m6A over ALKBH5. Nucleic Acids Res. (2015) 43:373–84. doi: 10.1093/nar/gku1276
186. Huff S, Kummetha IR, Zhang L, Wang L, Bray W, Yin J, et al. Rational design and optimization of m(6)A-RNA demethylase FTO inhibitors as anticancer agents. J Med Chem. (2022) 65:10920–37. doi: 10.1021/acs.jmedchem.1c02075
187. Phan T, Nguyen VH, Su R, Li Y, Qing Y, Qin H, et al. Targeting fat mass and obesity-associated protein mitigates human colorectal cancer growth in vitro and in a murine model. Front Oncol. (2023) 13:1087644. doi: 10.3389/fonc.2023.1087644
188. Huang Y, Su R, Sheng Y, Dong L, Dong Z, Xu H, et al. Small-molecule targeting of oncogenic FTO demethylase in acute myeloid leukemia. Cancer Cell. (2019) 35:677–91.e10. doi: 10.1016/j.ccell.2019.03.006
189. Qing Y, Dong L, Gao L, Li C, Li Y, Han L, et al. R-2-hydroxyglutarate attenuates aerobic glycolysis in leukemia by targeting the FTO/m6A/PFKP/LDHB axis. Mol Cell. (2021) 81:922–39.e9. doi: 10.1016/j.molcel.2020.12.026
190. Cao K, Du Y, Bao X, Han M, Su R, Pang J, et al. Glutathione-bioimprinted nanoparticles targeting of N6-methyladenosine FTO demethylase as a strategy against leukemic stem cells. Small. (2022) 18:e2106558. doi: 10.1002/smll.202106558
191. Peng S, Xiao W, Ju D, Sun B, Hou N, Liu Q, et al. Identification of entacapone as a chemical inhibitor of FTO mediating metabolic regulation through FOXO1. Sci Transl Med. (2019) 11:eaau7116. doi: 10.1126/scitranslmed.aau7116
192. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. (2021) 71:7–33. doi: 10.3322/caac.21654
193. Rawla P, Barsouk A. Epidemiology of gastric cancer: global trends, risk factors and prevention. Prz Gastroenterol. (2019) 14:26–38. doi: 10.5114/pg.2018.80001
194. Wang Q, Chen C, Ding Q, Zhao Y, Wang Z, Chen J, et al. METTL3-mediated m(6)A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut. (2020) 69:1193–205. doi: 10.1136/gutjnl-2019-319639
195. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2021) 71:209–49. doi: 10.3322/caac.21660
196. O'Connell JB, Maggard MA, Ko CY. Colon cancer survival rates with the new American Joint Committee on Cancer sixth edition staging. J Natl Cancer Inst. (2004) 96:1420–5. doi: 10.1093/jnci/djh275
197. GBD 2017 Colorectal Cancer Collaborators. The global, regional, and national burden of colorectal cancer and its attributable risk factors in 195 countries and territories, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol Hepatol. (2019) 4:913–33. doi: 10.1016/S2468-1253(19)30345-0
198. Yang X, Zhang S, He C, Xue P, Zhang L, He Z, et al. METTL14 suppresses proliferation and metastasis of colorectal cancer by down-regulating oncogenic long non-coding RNA XIST. Mol Cancer. (2020) 19:46. doi: 10.1186/s12943-020-1146-4
199. Ye M, Chen J, Yu P, Hu C, Wang B, Bao J, et al. WTAP activates MAPK signaling through m6A methylation in VEGFA mRNA-mediated by YTHDC1 to promote colorectal cancer development. FASEB J. (2023) 37:e23090. doi: 10.1096/fj.202300344RRR
200. Li J, Li S, Xing X, Liu N, Lai S, Liao D, et al. FTO-mediated ZNF687 accelerates tumor growth, metastasis, and angiogenesis in colorectal cancer through the Wnt/β-catenin pathway. Biotechnol Appl Biochem. (2023) 71:245–55. doi: 10.1002/bab.2536
201. Petrick JL, Braunlin M, Laversanne M, Valery PC, Bray F, McGlynn KA. International trends in liver cancer incidence, overall and by histologic subtype, 1978-2007. Int J Cancer. (2016) 139:1534–45. doi: 10.1002/ijc.30211
202. Morse MA, Sun W, Kim R, He AR, Abada PB, Mynderse M, et al. The role of angiogenesis in hepatocellular carcinoma. Clin Cancer Res. (2019) 25:912–20. doi: 10.1158/1078-0432.CCR-18-1254
203. Qiao K, Liu Y, Xu Z, Zhang H, Zhang H, Zhang C, et al. RNA m6A methylation promotes the formation of vasculogenic mimicry in hepatocellular carcinoma via Hippo pathway. Angiogenesis. (2021) 24:83–96. doi: 10.1007/s10456-020-09744-8
204. Wen J, Xue L, Wei Y, Liang J, Jia W, Yong T, et al. YTHDF2 is a therapeutic target for HCC by suppressing immune evasion and angiogenesis through ETV5/PD-L1/VEGFA axis. Advanced Sci. (2024) 11:e2307242. doi: 10.1002/advs.202307242
205. Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. (2013) 499:43–9. doi: 10.1038/nature12222
206. Bhatt JR, Finelli A. Landmarks in the diagnosis and treatment of renal cell carcinoma. Nat Rev Urol. (2014) 11:517–25. doi: 10.1038/nrurol.2014.194
207. Zarrabi K, Fang C, Wu S. New treatment options for metastatic renal cell carcinoma with prior anti-angiogenesis therapy. J Hematol Oncol. (2017) 10:38. doi: 10.1186/s13045-016-0374-y
208. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. (2019) 69:7–34. doi: 10.3322/caac.21551
209. Gupta B, Johnson NW, Kumar N. Global epidemiology of head and neck cancers: A continuing challenge. Oncology. (2016) 91:13–23. doi: 10.1159/000446117
210. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2018) 68:394–424. doi: 10.3322/caac.21492
211. Fu C, Feng L, Zhang J, Sun D. Bioinformatic analyses of the role of m6A RNA methylation regulators in abdominal aortic aneurysm. Ann Trans Med. (2022) 10:547–. doi: 10.21037/atm
212. Cai L, Han X-Y, Li D, Ma D-M, Shi Y-M, Lu Y, et al. Analysis of N6-methyladenosine-modified mRNAs in diabetic cataract. World J Diabetes. (2023) 14:1077–90. doi: 10.4239/wjd.v14.i7.1077
213. Xiao P, Y-k L, Han W, Hu Y, Zhang B-y, Liu W-l. Exosomal Delivery of FTO Confers Gefitinib Resistance to Recipient Cells through ABCC10 Regulation in an m6A-dependent Manner. Mol Cancer Res. (2021) 19:726–38. doi: 10.1158/1541-7786.MCR-20-0541
214. Chen X, Liu Y, Sun D, Sun R, Wang X, Li M, et al. Long noncoding RNA lnc-H2AFV-1 promotes cell growth by regulating aberrant m6A RNA modification in head and neck squamous cell carcinoma. Cancer Science. (2022) 113:2071–84. doi: 10.1111/cas.15366
Keywords: N6-methyladenosine, tumor, noncoding RNA, angiogenesis, regulatory role, anti-tumor therapy
Citation: YuYan and Yuan E (2024) Regulatory effect of N6-methyladenosine on tumor angiogenesis. Front. Immunol. 15:1453774. doi: 10.3389/fimmu.2024.1453774
Received: 24 June 2024; Accepted: 19 August 2024;
Published: 04 September 2024.
Edited by:
Xiangpeng Dai, Jilin University, ChinaCopyright © 2024 YuYan and Yuan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Enwu Yuan, eXVhbmVud3VAMTI2LmNvbQ==
 YuYan
YuYan Enwu Yuan
Enwu Yuan