- 1RNA and T cell Biology, St Vincent’s Institute of Medical Research, Fitzroy, VIC, Australia
- 2Department of Medicine, University of Melbourne, Parkville, VIC, Australia
T cells express an enormous repertoire of T cell receptors, enabling them to recognize any potential antigen. This large repertoire undergoes stringent selections in the thymus, where receptors that react to self- or non-danger-associated- antigens are purged. We know that thymic tolerance depends on signals and antigens presented by the thymic antigen presenting cells, but we still do not understand precisely how many of these cells actually contribute to tolerance. This is especially true for thymic dendritic cells (DC), which are composed of diverse subpopulations that are derived from different progenitors. Although the importance of thymic DCs has long been known, the functions of specific DC subsets have been difficult to untangle. There remains insufficient systematic characterization of the ontogeny and phenotype of thymic APCs in general. As a result, validated experimental models for studying thymic DCs are limited. Recent technological advancement, such as multi-omics analyses, has enabled new insights into thymic DC biology. These recent findings indicate a need to re-evaluate the current tools used to study the function of these cells within the thymus. This review will discuss how thymic DC subpopulations can be defined, the models that have been used to assess functions in the thymus, and models developed for other settings that can be potentially used for studying thymic DCs.
Introduction
T cells undergo random somatic recombination of Variable (V), Diversity (D) and Joining (J) gene segments to assemble their T cell receptors (TCR) (1). This rearrangement theoretically allows 1018 possible TCRs to be generated, which must undergo strict selection checkpoints in the thymus to purge non-functional TCRs as well as those than might recognize self-antigens (2). The first checkpoint, positive selection, selects TCRs that recognize major histocompatibility complex (MHC) molecules (3). The second, negative selection, determines the self-reactivity of TCRs by displaying self-peptides on MHC molecules to thymocytes (4). Thymocytes that show strong self-reactivity are deleted or further differentiate into regulatory T cells (Treg) with immunomodulatory functions (2, 5, 6). These two checkpoints are essential for shaping the T cell repertoire. Both checkpoints require antigen presenting cells (APC) in the thymus to interact with TCRs. Therefore, they play pivotal roles in achieving the intricate balance between immunodeficiency and autoimmunity.
APCs in the thymus include the classical hematopoietic APCs, and non-classical thymic epithelial cells (TEC) (7). The role of TECs in supporting T cell development are well characterized relative to hematopoietic APCs. Three lineages of hematopoietic APCs have been identified in the thymus: dendritic cells (DC), macrophages and B cells (8–10). Of these, there is evidence demonstrating that DCs and B cells present antigens and participate in negative selection (8, 11–13). Although it remains unclear if macrophages participate in thymocyte selection, these cells can present antigens ex vivo (14).
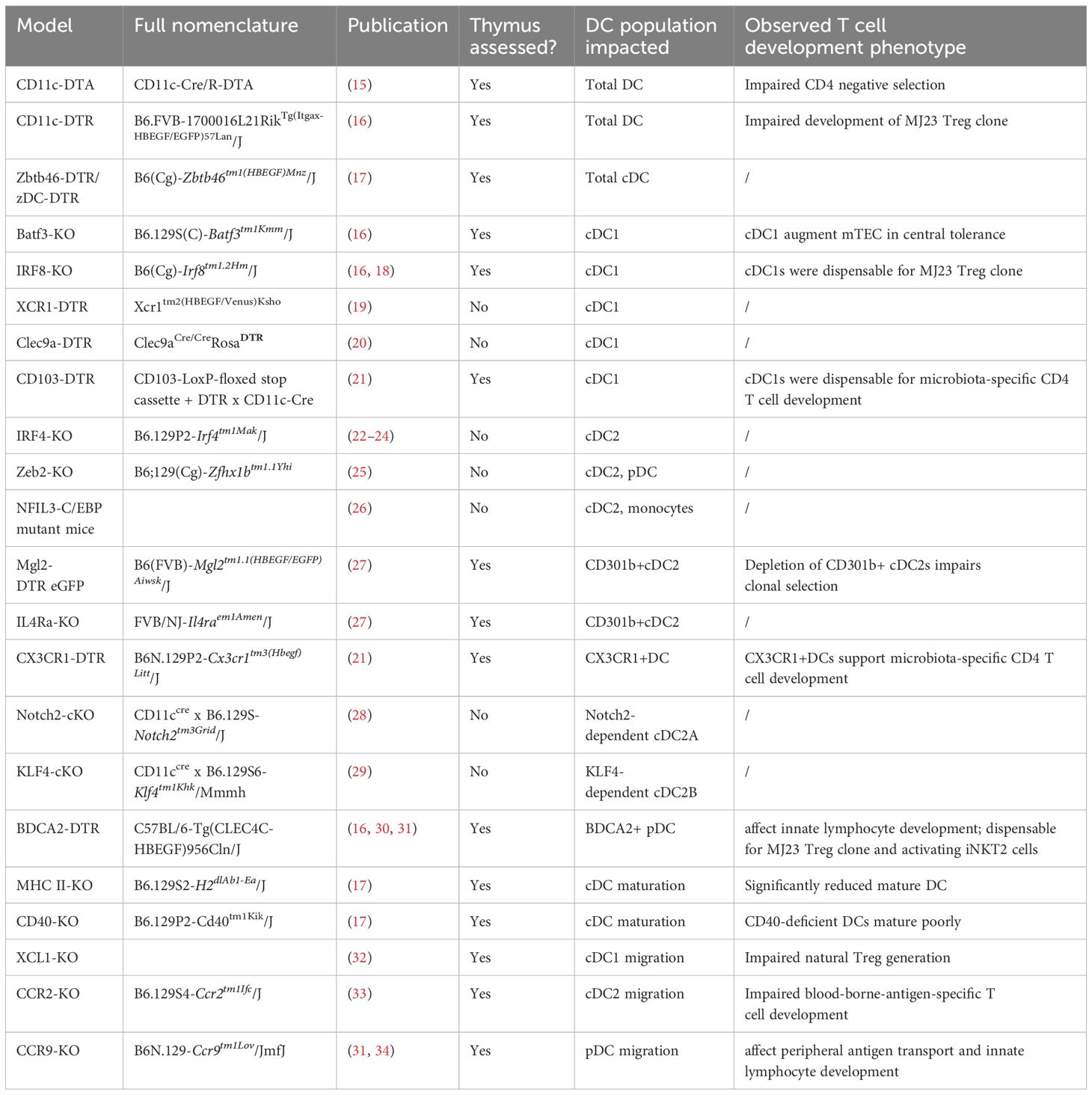
From the study of transgenic mouse models that lack thymic DCs, we know that these cells play non-redundant roles in clonal deletion of self-reactive CD4 T cells and induction of Tregs (15). Despite the importance of thymic DCs, the lineage origins of these cells are not well characterized. As a result, it is difficult to manipulate individual DC subsets to untangle the functions of these subsets. More recent studies that better define DC biology in the thymus and other organs have been enabled by new technologies such as transcriptomic profiling, allowing us to utilize mouse models to manipulate these cells more accurately (Table 1).
Manipulating total DCs
The importance of thymic DCs was revealed by exploiting their common expression of CD11c. One of these approaches is diphtheria toxin- (DT) mediated cell knockout models (35, 36). Mice transgenic for CD11c-cre and flox-stop- diphtheria toxin α chain (DTA), which ablates all CD11c-expressing cells, have been used to study the consequence on T cell development (15). In the thymi of these mice, the majority of conventional DCs are depleted and negative selection is severely impaired in the CD4 compartment. Using the same model, another study demonstrated poor development of Treg clones (37). In the CD11c-DTR model, where DCs are depleted upon DT administration, thymic DCs were shown to support the development of a prostate antigen-specific Treg clone (16).
Although CD11c is generally agreed to be a DC-specific marker, a recent study showed that thymic macrophages also express CD11c, as well as class II major histocompatibility complex (MHC II) and SIRPα, which are canonical markers for conventional DC2s (cDC2) (14). This is despite having transcriptional profiles that are clearly distinct from DCs. This suggests that macrophages could be easily mistaken as DCs in phenotypic characterization and the approaches exploiting CD11c expression are likely also affecting macrophages. An alternative marker that is thought to be exclusive to conventional DC (cDC) is the transcription factor Zbtb46 (38). With a Zbtb46-DTR transgene, depletion of close to 80% of thymic cDCs has been reported (17, 39).
Defining DCs by phenotypic subsets
Compared to the other hematopoietic APCs, DCs are relatively well-characterized in the thymus. However, studies on thymic DCs thus far have yet to piece together a clear picture of their composition and functions. The earliest studies identified phenotypic markers to distinguish three DC subsets in mice which are still the canonical markers currently in use: CD8α+ cDC1, SIRPα+CD11b+ cDC2 and PDCA+SiglecH+ plasmacytoid DC (pDC) (Figure 1) (40). cDC1s take up self-antigens expressed by medullary TECs and display them for negative selection (13). cDC2s are also known to participate in negative selection and to generate regulatory T cells (Treg) in vitro (18). It is unclear yet whether one subset contributes towards tolerance more than another, or whether they are functionally redundant.
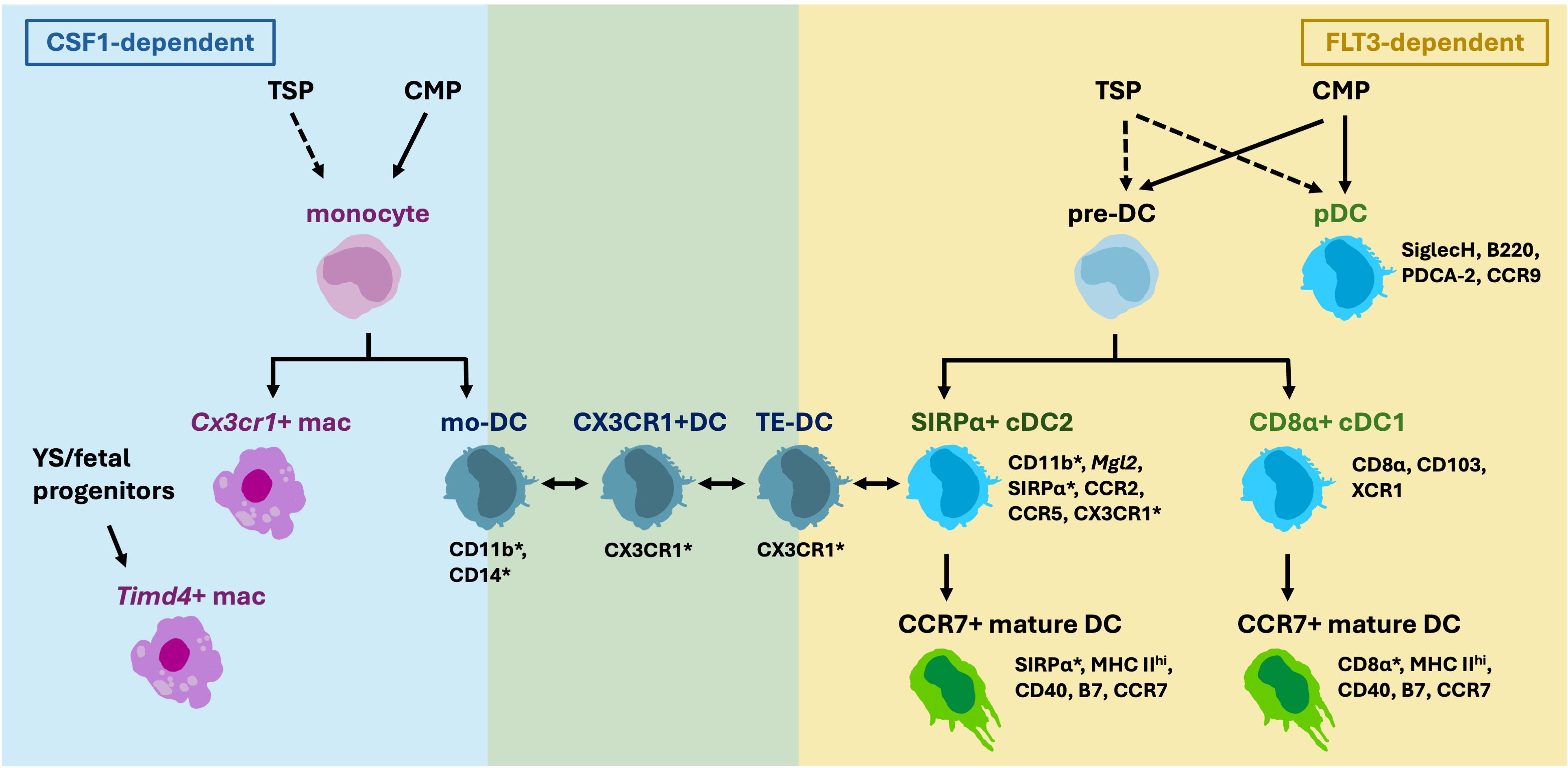
Figure 1 Thymic DCs are composed of various subsets that are derived from different progenitors. Thymic seeding progenitors (TSP) are suggested to give rise to thymic monocytes, pre-DC and pDCs in addition to common myeloid progenitors (CMP). Multiple CX3CR1+ cDC2-like populations have been described, and it is yet unclear whether they are derived from monocytes, or pre-DC, and whether they are CSF1- or FLT3-dependent. Overlapping Markers expressed by DC2s and macrophages (marked by asterisks), such as CD11c, MHC II, SIRPα, CD11b and CX3CR1, increase the difficulty to distinguish the cells apart.
Controversies surrounding the DC2 lineage
cDCs and pDCs are derived from common DC progenitors that have lost monocyte-potential in the bone marrow (41). There are currently no agreed phenotypic markers that specifically label bona fide cDC2s. Commonly used markers such as SIRPα and CD11b overlap with monocyte-derived cells (14, 42–44). While the current gold standard to define macrophage and DC lineages is by their differential dependence on the growth factors CSF1 and FLT3, respectively, thymic cDC2s are often defined by only phenotypic markers that do not exclusively label cDC2s. In the thymus of Zbtb46-DTR mice, CD8α+cDC1s can be totally depleted with DT, but around 20% of “SIRPα+cDC2s” persist (17).
Recently, “monocyte-derived DCs” (mo-DC) were reported in the thymus (45). These cells are transcriptionally similar to monocytes but express cDC2 markers such as Itgax (CD11c), Itgam (CD11b) and Sirpa. However, whether they are derived from monocytes has not yet been determined. Another study reported the presence of “CX3CR1+DCs” in the thymus that migrate from the gut carrying microbiota antigens (21). In the gut, CX3CR1+ mononuclear phagocytes are derived from monocytes (46). An additional study reported the so called thymic “transendothelial-DCs” (TE-DC) that can present blood-borne antigens also express CX3CR1 (47). It is unknown if these three described populations overlap, or how they are different, other than their common expression of CX3CR1. It will be important to clarify whether these “CX3CR1+DCs” contain DC-progenitor- or monocyte-derived cells, and whether these cells belong to macrophage or DC lineage (14, 48) (Figure 1).
Moreover, heterogeneity among cDC2s has been demonstrated, at least in murine spleen. Splenic cDC2s can be subdivided into Notch2- dependent Tbet+ cDC2A and KLF4-dependent RORγt+ cDC2B subsets (28, 29, 49). No study has directly compared thymic cDC2s to these splenic subsets and thus heterogeneity of thymic cDC2s remain unclear.
Manipulating cDC1s
The transcription factors required for murine cDC1 specification are well defined, and thus there are multiple mouse models that disrupt the development of cDC1s (50). The BATF3- and IRF8-deficient models affect cDC1s (51–53). The thymi of these mice have been characterized and a clear depletion of cDC1s can be seen (16, 18). However, IRF8 deficiency also affects splenic B cells and macrophages (54). It is yet unclear whether thymic B cells and macrophages are impacted, and thus the changes in T cell development observed in IRF8-deficient thymi cannot be solely attributed to the depletion of cDC1s. Alternatively, there are the CD103-DTR, XCR1-DTR or Clec9A-DTR models that utilize cDC1-specific promoters to drive DTR expression to deplete the population (19–21). The integrin CD103 is a common marker used to define intestinal DC1s (55). As this marker is also expressed by thymic DC1s, the conditional CD103-DTR (CD11ccreCD103DTR) model has been shown to efficiently deplete cDC1s in the thymus (21). Thymic cDC1 depletion has not been characterized in XCR1-DTR or Clec9A-DTR mice.
Manipulating cDC2s/mo-DCs
There are currently no mouse models with specific ablation of the SIRPa+ DC population. This is likely due to the possibility that this is a mixed population of cells with a similar phenotype but of different lineages. This is also the case for DC2s in other organs, where their phenotype and lineage origins are still under debate (56).
The transcription factors IRF4 and ZEB2 are implicated in the commitment of the cDC2 lineage (22–24, 57). Due to differences in environmental cues, tissue-specific cDC2s have different dependencies on IRF4. DC-specific IRF4 knockout depletes cDC2s in the lung, but only a proportion of cDC2s in the spleen and small intestinal lamina propria (22, 24). Knockout of Zeb2 depletes cDC2s, but also pDCs (25). To specifically deplete cDC2s, a mouse model with mutations in the Zeb2 enhancer was developed (26). Although B cells and pDCs are not impacted, monocytes are deficient in this model. No assessment of thymic cDC2s in the aforementioned mouse models have so far been reported.
Since the transcriptional regulation of cDC2s is difficult to exploit, other approaches that employ other markers expressed by thymic DC2s in DTR models have been developed, but none achieve total depletion. The Mgl2-DTR model specifically targets Mgl2/CD301b-expressing cells (58). These were found to be 30-60% of the thymic cDC2s (30, 45). The cytokine receptor IL4R regulates the maintenance of CD301b+ cDC2s and Il4Ra-deficient mice exhibit the same degree of depletion as the CD301b-DTR mice (27). CX3CR1-DTR is also a potential tool for depleting cDC2s (21). Using this model, CX3CR1+DCs were shown to be crucial for the development of microbiota-specific CD4 T cells.
The lack of mouse models with effective cDC2 depletion highlights the need to clarify the identity of the SIRPα+ myeloid cells in the thymus. The possibility that there are cDC2s, mo-DCs and macrophages with similar phenotypes implicates that the thymic cDC2s described in previous studies could in fact be comprised of a mixture of populations. The populations of different lineages should be targeted using different approaches. It will also be beneficial to map thymic cDC2s to the well-defined subsets of splenic cDC2s, which could allow existing models such as the Notch2- and Klf4- conditional knockout to be adapted for studying their functions (28, 29).
Manipulating pDCs
pDCs express the unique surface marker CLEC4C/BDCA2 and this has been utilized for DTR transgenic models (59). There is efficient depletion of thymic pDCs in this model and they were found to be required for the development of microbiota-specific innate lymphocytes (16, 30, 31).
Defining DCs by activation status
Circulating DCs, or immature DCs are known to undergo maturation upon uptake of an antigen and home to lymph nodes to activate T cells (60). This maturation process refers to the activation of developmentally mature and functional DCs, and not the differentiation of uncommitted progenitors into DCs. Mature DCs upregulate MHC II as well as the costimulation molecules CD80, CD86 and CD40. Mature DC homing to the thymus was first explored in 2006, and it was found that immature DCs are preferentially recruited to the thymus compared to immunogenic LPS-induced mature DCs (61). The authors propose that preferential recruitment of immature and tolerogenic DCs is a safety checkpoint in central tolerance, since DCs that mature with danger signals can lead to undesirable deletion of danger-associated thymocytes.
Using RNA sequencing, the transcriptional changes that take place during maturation of thymic CD8a+ cDC1s were found to resemble maturation of tolerogenic DCs in the periphery (62). Another study reported that both cDC1s and cDC2s undergo maturation in the thymus, and mature cDC1s are transcriptionally very similar to cDC2s (17). Unlike peripheral DCs that become less phagocytic upon maturation, this study suggested that thymic mature DCs are as efficient as immature DCs in the uptake of antigens. Other than antigen presentation, mature DCs have also been found to promote thymic atrophy via the Jagged-Notch2 axis (63). However, it has not been fully resolved whether all DC maturation takes place in the thymus or some mature DCs are recruited (17, 63).
Manipulating DC maturation
Thymic mature DCs are found to depend on cognate MHC II-TCR interactions with CD4 single positive (SP) thymocytes and CD40-mediated costimulation signal (17). Thymic DCs that are deficient for MHC II or CD40 mature poorly. However, CD4 single positive thymocytes are absent in MHC II-deficient mice and it is difficult to study the function of mature DCs with such great impact on the CD4 compartment (64). Further, removing MHC II or CD40 directly affects the antigen presentation capacity of all DCs in the thymus, not just mature DCs. Although there are available approaches to block DC maturation, none of these can be used to address whether mature and immature DCs are functionally distinct, or how mature DC1s are different to DC2s given their transcriptional similarity.
Defining DCs by site of development and source of antigen
The majority of DCs originate from the bone marrow, but early thymic progenitors that enter the thymus can retain multipotency and give rise to DCs (65). This means DCs in the thymus can be either derived from a progenitor in the thymus or enter the thymus as functional DCs. This different origin of DCs can fundamentally impact their function. The unique thymic microenvironment may influence DCs to acquire specialised functions, while a migrated DC can carry antigens that are derived peripherally (66, 67). Therefore, it is desirable to know where a DC has come from when studying their function in the thymus.
The characterization of developmental origin of DCs began with parabiotic mice experiments. The cDC1s were found to be resident, while the other two subsets were found to be migratory (8, 33, 34, 68, 69). These experiments elegantly demonstrate the thymic residency of cDC1s. For the cDC2s and pDCs, such parabiotic experiments only demonstrate that cells from the periphery have migrated into the thymus. It is still possible that a proportion of thymic cDC2s and pDCs develop intrathymically. Recently, single-cell transcriptomic characterization of human thymus-seeding progenitors suggest that they give rise to pDCs and monocytes (70, 71). Further, a rearranged Tcrd locus can be detected in a proportion of human thymic monocytes, pDCs and cDC1s, suggesting an origin via uncommitted thymocytes (70). However, the exact origins of thymic DCs are yet to be demonstrated using in vivo experimental models.
Manipulating DC by migration and antigen
DCs rely on chemokine receptors to migrate throughout lymphoid organs and tissues (72). The unique pattern of chemokine receptor expression determines where subsets migrate and serve as good markers for defining these subsets. cDC1s express XCR1 and they co-localise with XCL1-expressing mTECs in the medulla of the thymus (32). This facilitates the transfer of antigens from mTECs to cDC1s to be efficiently presented or cross-presented to thymocytes (13). cDC2s and monocyte-derived cells express CX3CR1, CCR2 and CCR5, whereas pDCs express CCR9 (21, 33, 34, 47, 73–75). These two subsets are reported to transport antigens from the periphery, including introduced exogenous antigens and microbial antigens (21, 33, 34). All these chemokine receptors can be manipulated to affect their migration into the thymus and localization within the thymus.
In XCL1-deficient mice, which lack the ligand of XCR1, the co-localization of cDC1 and mTEC is impaired, leading to defective generation of thymic Tregs (32). CCR2-deficient mice display a marked reduction in thymic SIRPα+DC2s and leads to impaired negative selection of blood-borne-antigen- and tumor-antigen-specific T cells (33, 74). No decrease in SIRPα+DC2s are seen in CCR5- or CX3CR1-deficient mice (33). When all three chemokine receptors are knocked out or blocked with inhibitors, CX3CR1+DC migration is completely ablated (21). Microbial DNA is present in the thymus as long as mice express one of the three chemokine receptors, suggesting that CX3CR1+DCs can utilize all three for migration.
In CCR9-deficient mice, significantly fewer pDCs are found in the thymus (31, 34). Consequently, peripheral antigen transport is reduced and innate-like thymocyte development is impaired, similar to BDCA2-DTR mice. The downside to manipulating chemokine receptor expression is that thymocytes rely on some of the same receptors, such as CCR9, for migration, their normal development will be affected (76).
Blocking APC migration into the thymus impacts thymic selection partly by removing the antigens they are supposed to present. Therefore, manipulating antigens in peripheral tissues is another approach for studying the functions of the migratory DCs. This can be achieved by introducing non-endogenous antigens at a specific site. For example, by coating the fluorescent protein FITC on the skin, thymus homing DCs have been shown to transport antigens from the skin (61). Otherwise, antigens like ovalbumin (OVA) can be introduced by tissue-specific expression, such as the Cmy promoter in cardiomyocytes, or the insulin promoter in pancreatic β cells (61, 77, 78). Because OVA is not an endogenous antigen, TCR transgenic OT-I CD8+ or OT-II CD4+ T cells that recognise OVA have to be introduced (3, 79). With these approaches, the migratory potential of thymic DCs into different tissues and their roles in the development or deletion of the cognate T cells can be studied.
Final comments
Despite the known importance of thymic DCs in tolerance, DC defects have yet to be implicated in autoimmune diseases. This presents the conundrum of whether this is because they are not involved, or because there are very limited tools to study them. The biggest roadblock to studying thymic DC functions is our incomplete understanding of their ontogeny and how subsets relate to each other. Due to the unique purpose of the thymus and its specialized microenvironment, assumptions made based on DC phenotypes in other secondary lymphoid organs are often inaccurate. Single-cell transcriptomic analyses of thymic myeloid cells revealed that non-DCs also express CD11c and presented the issue of cDC2 lineage heterogeneity. The phenotype of thymocytes and all hematopoietic APCs should be carefully evaluated in models that are thought to only impact DCs, to better interpret the reported changes in T cell development. This is also important when comparing different models to tease out the function of specific subsets. To further understand the origins and developmental regulation of thymic DCs, experimental approaches such as lineage tracing models are needed. It also remains beneficial to leverage existing understanding of (peripheral) DC biology and study the thymus phenotype in models previously used for assessing DCs in other organs. Having established and validated models for manipulating DC subsets can be used to answer the question of whether these subsets are functionally redundant and how each of them contribute to tolerance under normal and autoimmune settings.
Author contributions
YW: Conceptualization, Writing – original draft, Writing – review & editing. MC: Conceptualization, Funding acquisition, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. Research in the Chong laboratory has been supported by funding from the National Health and Medical Research Council, Breakthrough T1D, mRNA Victoria, Diabetes Australia and The United States Department of Defense.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Chien YH, Gascoigne NR, Kavaler J, Lee NE, Davis MM. Somatic recombination in a murine T-cell receptor gene. Nature. (1984) 309:322–6. doi: 10.1038/309322a0
2. Klein L, Kyewski B, Allen PM, Hogquist KA. Positive and negative selection of the T cell repertoire: what thymocytes see (and don’t see). Nat Rev Immunol. (2014) 14:377–91. doi: 10.1038/nri3667
3. Hogquist KA, Jameson SC, Heath WR, Howard JL, Bevan MJ, Carbone FR. T cell receptor antagonist peptides induce positive selection. Cell. (1994) 76:17–27. doi: 10.1016/0092-8674(94)90169-4
4. Palmer E. Negative selection–clearing out the bad apples from the T-cell repertoire. Nat Rev Immunol. (2003) 3:383–91. doi: 10.1038/nri1085
5. Li MO, Rudensky AY. T cell receptor signalling in the control of regulatory T cell differentiation and function. Nat Rev Immunol. (2016) 16:220–33. doi: 10.1038/nri.2016.26
6. Tai X, Indart A, Rojano M, Guo J, Apenes N, Kadakia T, et al. How autoreactive thymocytes differentiate into regulatory versus effector CD4(+) T cells after avoiding clonal deletion. Nat Immunol. (2023) 24:637–51. doi: 10.1038/s41590-023-01469-2
7. Wang H, Zuniga-Pflucker JC. Thymic microenvironment: interactions between innate immune cells and developing thymocytes. Front Immunol. (2022) 13:885280. doi: 10.3389/fimmu.2022.885280
8. Proietto AI, van Dommelen S, Zhou P, Rizzitelli A, D’Amico A, Steptoe RJ, et al. Dendritic cells in the thymus contribute to T-regulatory cell induction. Proc Natl Acad Sci USA. (2008) 105:19869–74. doi: 10.1073/pnas.0810268105
9. Epstein HD, Mitchell DS, Hunt JS, Wood GW. Ia-positive macrophages bind and internalize viable lymphocytes in murine thymus. Cell Immunol. (1985) 95:15–34. doi: 10.1016/0008-8749(85)90291-6
10. Perera J, Huang H. The development and function of thymic B cells. Cell Mol Life Sci. (2015) 72:2657–63. doi: 10.1007/s00018-015-1895-1
11. Perera J, Meng L, Meng F, Huang H. Autoreactive thymic B cells are efficient antigen-presenting cells of cognate self-antigens for T cell negative selection. Proc Natl Acad Sci USA. (2013) 110:17011–6. doi: 10.1073/pnas.1313001110
12. Perera J, Zheng Z, Li S, Gudjonson H, Kalinina O, Benichou JIC, et al. Self-antigen-driven thymic B cell class switching promotes T cell central tolerance. Cell Rep. (2016) 17:387–98. doi: 10.1016/j.celrep.2016.09.011
13. Hubert FX, Kinkel SA, Davey GM, Phipson B, Mueller SN, Liston A, et al. Aire regulates the transfer of antigen from mTECs to dendritic cells for induction of thymic tolerance. Blood. (2011) 118:2462–72. doi: 10.1182/blood-2010-06-286393
14. Zhou TA, Hsu HP, Tu YH, Cheng HK, Lin CY, Chen NJ, et al. Thymic macrophages consist of two populations with distinct localization and origin. Elife. (2022) 11. doi: 10.7554/eLife.75148.sa2
15. Ohnmacht C, Pullner A, King SB, Drexler I, Meier S, Brocker T, et al. Constitutive ablation of dendritic cells breaks self-tolerance of CD4 T cells and results in spontaneous fatal autoimmunity. J Exp Med. (2009) 206:549–59. doi: 10.1084/jem.20082394
16. Leventhal DS, Gilmore DC, Berger JM, Nishi S, Lee V, Malchow S, et al. Dendritic cells coordinate the development and homeostasis of organ-specific regulatory T cells. Immunity. (2016) 44:847–59. doi: 10.1016/j.immuni.2016.01.025
17. Oh J, Wu N, Barczak AJ, Barbeau R, Erle DJ, Shin JS. CD40 mediates maturation of thymic dendritic cells driven by self-reactive CD4(+) thymocytes and supports development of natural regulatory T cells. J Immunol. (2018) 200:1399–412. doi: 10.4049/jimmunol.1700768
18. Herbin O, Bonito AJ, Jeong S, Weinstein EG, Rahman AH, Xiong H, et al. Medullary thymic epithelial cells and CD8alpha(+) dendritic cells coordinately regulate central tolerance but CD8alpha(+) cells are dispensable for thymic regulatory T cell production. J Autoimmun. (2016) 75:141–9. doi: 10.1016/j.jaut.2016.08.002
19. Yamazaki C, Sugiyama M, Ohta T, Hemmi H, Hamada E, Sasaki I, et al. Critical roles of a dendritic cell subset expressing a chemokine receptor, XCR1. J Immunol. (2013) 190:6071–82. doi: 10.4049/jimmunol.1202798
20. Piva L, Tetlak P, Claser C, Karjalainen K, Renia L, Ruedl C. Cutting edge: Clec9A+ dendritic cells mediate the development of experimental cerebral malaria. J Immunol. (2012) 189:1128–32. doi: 10.4049/jimmunol.1201171
21. Zegarra-Ruiz DF, Kim DV, Norwood K, Kim M, Wu WH, Saldana-Morales FB, et al. Thymic development of gut-microbiota-specific T cells. Nature. (2021) 594:413–7. doi: 10.1038/s41586-021-03531-1
22. Bajana S, Turner S, Paul J, Ainsua-Enrich E, Kovats S. IRF4 and IRF8 act in CD11c+ Cells to regulate terminal differentiation of lung tissue dendritic cells. J Immunol. (2016) 196:1666–77. doi: 10.4049/jimmunol.1501870
23. Schlitzer A, McGovern N, Teo P, Zelante T, Atarashi K, Low D, et al. IRF4 transcription factor-dependent CD11b+ dendritic cells in human and mouse control mucosal IL-17 cytokine responses. Immunity. (2013) 38:970–83. doi: 10.1016/j.immuni.2013.04.011
24. Persson EK, Uronen-Hansson H, Semmrich M, Rivollier A, Hagerbrand K, Marsal J, et al. IRF4 transcription-factor-dependent CD103(+)CD11b(+) dendritic cells drive mucosal T helper 17 cell differentiation. Immunity. (2013) 38:958–69. doi: 10.1016/j.immuni.2013.03.009
25. Wu X, Briseno CG, Grajales-Reyes GE, Haldar M, Iwata A, Kretzer NM, et al. Transcription factor Zeb2 regulates commitment to plasmacytoid dendritic cell and monocyte fate. Proc Natl Acad Sci USA. (2016) 113:14775–80. doi: 10.1073/pnas.1611408114
26. Liu TT, Kim S, Desai P, Kim DH, Huang X, Ferris ST, et al. Ablation of cDC2 development by triple mutations within the Zeb2 enhancer. Nature. (2022) 607:142–8. doi: 10.1038/s41586-022-04866-z
27. Breed ER, Voboril M, Ashby KM, Martinez RJ, Qian L, Wang H, et al. Type 2 cytokines in the thymus activate Sirpalpha(+) dendritic cells to promote clonal deletion. Nat Immunol. (2022) 23:1042–51. doi: 10.1038/s41590-022-01218-x
28. Lewis KL, Caton ML, Bogunovic M, Greter M, Grajkowska LT, Ng D, et al. Notch2 receptor signaling controls functional differentiation of dendritic cells in the spleen and intestine. Immunity. (2011) 35:780–91. doi: 10.1016/j.immuni.2011.08.013
29. Tussiwand R, Everts B, Grajales-Reyes GE, Kretzer NM, Iwata A, Bagaitkar J, et al. Klf4 expression in conventional dendritic cells is required for T helper 2 cell responses. Immunity. (2015) 42:916–28. doi: 10.1016/j.immuni.2015.04.017
30. Wang H, Breed ER, Lee YJ, Qian LJ, Jameson SC, Hogquist KA. Myeloid cells activate iNKT cells to produce IL-4 in the thymic medulla. Proc Natl Acad Sci USA. (2019) 116:22262–8. doi: 10.1073/pnas.1910412116
31. Ennamorati M, Vasudevan C, Clerkin K, Halvorsen S, Verma S, Ibrahim S, et al. Intestinal microbes influence development of thymic lymphocytes in early life. Proc Natl Acad Sci USA. (2020) 117(5):2570–8. doi: 10.1073/pnas.1915047117
32. Lei Y, Ripen AM, Ishimaru N, Ohigashi I, Nagasawa T, Jeker LT, et al. Aire-dependent production of XCL1 mediates medullary accumulation of thymic dendritic cells and contributes to regulatory T cell development. J Exp Med. (2011) 208:383–94. doi: 10.1084/jem.20102327
33. Baba T, Nakamoto Y, Mukaida N. Crucial contribution of thymic Sirp alpha+ conventional dendritic cells to central tolerance against blood-borne antigens in a CCR2-dependent manner. J Immunol. (2009) 183:3053–63. doi: 10.4049/jimmunol.0900438
34. Hadeiba H, Lahl K, Edalati A, Oderup C, Habtezion A, Pachynski R, et al. Plasmacytoid dendritic cells transport peripheral antigens to the thymus to promote central tolerance. Immunity. (2012) 36:438–50. doi: 10.1016/j.immuni.2012.01.017
35. Saito M, Iwawaki T, Taya C, Yonekawa H, Noda M, Inui Y, et al. Diphtheria toxin receptor-mediated conditional and targeted cell ablation in transgenic mice. Nat Biotechnol. (2001) 19:746–50. doi: 10.1038/90795
36. Pappenheimer AM Jr., Harper AA, Moynihan M, Brockes JP. Diphtheria toxin and related proteins: effect of route of injection on toxicity and the determination of cytotoxicity for various cultured cells. J Infect Dis. (1982) 145:94–102. doi: 10.1093/infdis/145.1.94
37. Perry JSA, Lio CJ, Kau AL, Nutsch K, Yang Z, Gordon JI, et al. Distinct contributions of Aire and antigen-presenting-cell subsets to the generation of self-tolerance in the thymus. Immunity. (2014) 41:414–26. doi: 10.1016/j.immuni.2014.08.007
38. Satpathy AT, Kc W, Albring JC, Edelson BT, Kretzer NM, Bhattacharya D, et al. Zbtb46 expression distinguishes classical dendritic cells and their committed progenitors from other immune lineages. J Exp Med. (2012) 209:1135–52. doi: 10.1084/jem.20120030
39. Meredith MM, Liu K, Darrasse-Jeze G, Kamphorst AO, Schreiber HA, Guermonprez P, et al. Expression of the zinc finger transcription factor zDC (Zbtb46, Btbd4) defines the classical dendritic cell lineage. J Exp Med. (2012) 209:1153–65. doi: 10.1084/jem.20112675
40. Wu L, Shortman K. Heterogeneity of thymic dendritic cells. Semin Immunol. (2005) 17:304–12. doi: 10.1016/j.smim.2005.05.001
41. Schlitzer A, Sivakamasundari V, Chen J, Sumatoh HR, Schreuder J, Lum J, et al. Identification of cDC1- and cDC2-committed DC progenitors reveals early lineage priming at the common DC progenitor stage in the bone marrow. Nat Immunol. (2015) 16:718–28. doi: 10.1038/ni.3200
42. Leon B, Lopez-Bravo M, Ardavin C. Monocyte-derived dendritic cells formed at the infection site control the induction of protective T helper 1 responses against Leishmania. Immunity. (2007) 26:519–31. doi: 10.1016/j.immuni.2007.01.017
43. Plantinga M, Guilliams M, Vanheerswynghels M, Deswarte K, Branco-Madeira F, Toussaint W, et al. Conventional and monocyte-derived CD11b(+) dendritic cells initiate and maintain T helper 2 cell-mediated immunity to house dust mite allergen. Immunity. (2013) 38:322–35. doi: 10.1016/j.immuni.2012.10.016
44. Menezes S, Melandri D, Anselmi G, Perchet T, Loschko J, Dubrot J, et al. The Heterogeneity of Ly6C(hi) Monocytes Controls Their Differentiation into iNOS(+) Macrophages or Monocyte-Derived Dendritic Cells. Immunity. (2016) 45:1205–18. doi: 10.1016/j.immuni.2016.12.001
45. Voboril M, Brabec T, Dobes J, Splichalova I, Brezina J, Cepkova A, et al. Toll-like receptor signaling in thymic epithelium controls monocyte-derived dendritic cell recruitment and Treg generation. Nat Commun. (2020) 11:2361. doi: 10.1038/s41467-020-16081-3
46. Diehl GE, Longman RS, Zhang JX, Breart B, Galan C, Cuesta A, et al. Microbiota restricts trafficking of bacteria to mesenteric lymph nodes by CX(3)CR1(hi) cells. Nature. (2013) 494:116–20. doi: 10.1038/nature11809
47. Vollmann EH, Rattay K, Barreiro O, Thiriot A, Fuhlbrigge RA, Vrbanac V, et al. Specialized transendothelial dendritic cells mediate thymic T-cell selection against blood-borne macromolecules. Nat Commun. (2021) 12:6230. doi: 10.1038/s41467-021-26446-x
48. Liu Z, Wang H, Li Z, Dress RJ, Zhu Y, Zhang S, et al. Dendritic cell type 3 arises from Ly6C(+) monocyte-dendritic cell progenitors. Immunity. (2023) 56:1761–77 e6. doi: 10.1016/j.immuni.2023.07.001
49. Brown CC, Gudjonson H, Pritykin Y, Deep D, Lavallee VP, Mendoza A, et al. Transcriptional basis of mouse and human dendritic cell heterogeneity. Cell. (2019) 179:846–63 e24. doi: 10.1016/j.cell.2019.09.035
50. Schlitzer A, Zhang W, Song M, Ma X. Recent advances in understanding dendritic cell development, classification, and phenotype. F1000Res. (2018) 7:F1000 Faculty Rev–1558. doi: 10.12688/f1000research.14793.1
51. Hildner K, Edelson BT, Purtha WE, Diamond M, Matsushita H, Kohyama M, et al. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science. (2008) 322:1097–100. doi: 10.1126/science.1164206
52. Holtschke T, Lohler J, Kanno Y, Fehr T, Giese N, Rosenbauer F, et al. Immunodeficiency and chronic myelogenous leukemia-like syndrome in mice with a targeted mutation of the ICSBP gene. Cell. (1996) 87:307–17. doi: 10.1016/S0092-8674(00)81348-3
53. Lanca T, Ungerback J, Da Silva C, Joeris T, Ahmadi F, Vandamme J, et al. IRF8 deficiency induces the transcriptional, functional, and epigenetic reprogramming of cDC1 into the cDC2 lineage. Immunity. (2022) 55(8):1431–47 e11. doi: 10.1016/j.immuni.2022.06.006
54. Qi CF, Li Z, Raffeld M, Wang H, Kovalchuk AL, Morse HC 3rd. Differential expression of IRF8 in subsets of macrophages and dendritic cells and effects of IRF8 deficiency on splenic B cell and macrophage compartments. Immunol Res. (2009) 45:62–74. doi: 10.1007/s12026-008-8032-2
55. Scott CL, Aumeunier AM, Mowat AM. Intestinal CD103+ dendritic cells: master regulators of tolerance? Trends Immunol. (2011) 32(9):412–9. doi: 10.1016/j.it.2011.06.003
56. Backer RA, Probst HC, Clausen BE. Classical DC2 subsets and monocyte-derived DC: Delineating the developmental and functional relationship. Eur J Immunol. (2023) 53:e2149548. doi: 10.1002/eji.202149548
57. Scott CL, Soen B, Martens L, Skrypek N, Saelens W, Taminau J, et al. The transcription factor Zeb2 regulates development of conventional and plasmacytoid DCs by repressing Id2. J Exp Med. (2016) 213:897–911. doi: 10.1084/jem.20151715
58. Kumamoto Y, Linehan M, Weinstein JS, Laidlaw BJ, Craft JE, Iwasaki A. CD301b(+) dermal dendritic cells drive T helper 2 cell-mediated immunity. Immunity. (2013) 39:733–43. doi: 10.1016/j.immuni.2013.08.029
59. Swiecki M, Gilfillan S, Vermi W, Wang Y, Colonna M. Plasmacytoid dendritic cell ablation impacts early interferon responses and antiviral NK and CD8(+) T cell accrual. Immunity. (2010) 33:955–66. doi: 10.1016/j.immuni.2010.11.020
60. Dalod M, Chelbi R, Malissen B, Lawrence T. Dendritic cell maturation: functional specialization through signaling specificity and transcriptional programming. EMBO J. (2014) 33:1104–16. doi: 10.1002/embj.201488027
61. Bonasio R, Scimone ML, Schaerli P, Grabie N, Lichtman AH, von Andrian UH. Clonal deletion of thymocytes by circulating dendritic cells homing to the thymus. Nat Immunol. (2006) 7:1092–100. doi: 10.1038/ni1385
62. Ardouin L, Luche H, Chelbi R, Carpentier S, Shawket A, Montanana Sanchis F, et al. Broad and largely concordant molecular changes characterize tolerogenic and immunogenic dendritic cell maturation in thymus and periphery. Immunity. (2016) 45:305–18. doi: 10.1016/j.immuni.2016.07.019
63. Wu H, Li X, Zhou C, Yu Q, Ge S, Pan Z, et al. Circulating mature dendritic cells homing to the thymus promote thymic epithelial cells involution via the Jagged1/Notch3 axis. Cell Death Discov. (2021) 7:225. doi: 10.1038/s41420-021-00619-5
64. Huss R, Deeg HJ. Intrathymic maturation of CD4+ T-lymphocytes in an MHC class II deficient transplant model. Tissue Antigens. (1997) 49:70–3. doi: 10.1111/j.1399-0039.1997.tb02714.x
65. Luche H, Ardouin L, Teo P, See P, Henri S, Merad M, et al. The earliest intrathymic precursors of CD8alpha(+) thymic dendritic cells correspond to myeloid-type double-negative 1c cells. Eur J Immunol. (2011) 41:2165–75. doi: 10.1002/eji.201141728
66. Garg G, Nikolouli E, Hardtke-Wolenski M, Toker A, Ohkura N, Beckstette M, et al. Unique properties of thymic antigen-presenting cells promote epigenetic imprinting of alloantigen-specific regulatory T cells. Oncotarget. (2017) 8:35542–57. doi: 10.18632/oncotarget.v8i22
67. Herppich S, Beckstette M, Huehn J. The thymic microenvironment gradually modulates the phenotype of thymus-homing peripheral conventional dendritic cells. Immun Inflamm Dis. (2022) 10(2):175–88. doi: 10.1002/iid3.559
68. Li J, Park J, Foss D, Goldschneider I. Thymus-homing peripheral dendritic cells constitute two of the three major subsets of dendritic cells in the steady-state thymus. J Exp Med. (2009) 206:607–22. doi: 10.1084/jem.20082232
69. Donskoy E, Goldschneider I. Two developmentally distinct populations of dendritic cells inhabit the adult mouse thymus: demonstration by differential importation of hematogenous precursors under steady state conditions. J Immunol. (2003) 170:3514–21. doi: 10.4049/jimmunol.170.7.3514
70. Cordes M, Cante-Barrett K, van den Akker EB, Moretti FA, Kielbasa SM, Vloemans SA, et al. Single-cell immune profiling reveals thymus-seeding populations, T cell commitment, and multilineage development in the human thymus. Sci Immunol. (2022) 7:eade0182. doi: 10.1126/sciimmunol.ade0182
71. Lavaert M, Liang KL, Vandamme N, Park JE, Roels J, Kowalczyk MS, et al. Integrated scRNA-seq identifies human postnatal thymus seeding progenitors and regulatory dynamics of differentiating immature thymocytes. Immunity. (2020) 52:1088–104 e6. doi: 10.1016/j.immuni.2020.03.019
72. Tiberio L, Del Prete A, Schioppa T, Sozio F, Bosisio D, Sozzani S. Chemokine and chemotactic signals in dendritic cell migration. Cell Mol Immunol. (2018) 15:346–52. doi: 10.1038/s41423-018-0005-3
73. Cosway EJ, Ohigashi I, Schauble K, Parnell SM, Jenkinson WE, Luther S, et al. Formation of the intrathymic dendritic cell pool requires CCL21-mediated recruitment of CCR7(+) progenitors to the thymus. J Immunol. (2018) 201:516–23. doi: 10.4049/jimmunol.1800348
74. Baba T, Badr Mel S, Tomaru U, Ishizu A, Mukaida N. Novel process of intrathymic tumor-immune tolerance through CCR2-mediated recruitment of Sirpalpha+ dendritic cells: a murine model. PloS One. (2012) 7(7):e41154. doi: 10.1371/journal.pone.0041154
75. Wendland M, Czeloth N, Mach N, Malissen B, Kremmer E, Pabst O, et al. CCR9 is a homing receptor for plasmacytoid dendritic cells to the small intestine. Proc Natl Acad Sci USA. (2007) 104(15):6347–52. doi: 10.1371/journal.pone.0041154
76. Lee HS, Kim HR, Lee EH, Jang MH, Kim SB, Park JW, et al. Characterization of CCR9 expression and thymus-expressed chemokine responsiveness of the murine thymus, spleen and mesenteric lymph node. Immunobiology. (2012) 217:402–11. doi: 10.1016/j.imbio.2011.10.014
77. Kurts C, Heath WR, Carbone FR, Allison J, Miller JF, Kosaka H. Constitutive class I-restricted exogenous presentation of self antigens in vivo. J Exp Med. (1996) 184:923–30. doi: 10.1084/jem.184.3.923
78. Shibaki A, Sato A, Vogel JC, Miyagawa F, Katz SI. Induction of GVHD-like skin disease by passively transferred CD8(+) T-cell receptor transgenic T cells into keratin 14-ovalbumin transgenic mice. J Invest Dermatol. (2004) 123:109–15. doi: 10.1111/j.0022-202X.2004.22701.x
Keywords: thymus, T cell development, central tolerance, dendritic cells, mouse models
Citation: Wang Y and Chong MMW (2024) Evaluating in vivo approaches for studying the roles of thymic DCs in T cell development in mice. Front. Immunol. 15:1451974. doi: 10.3389/fimmu.2024.1451974
Received: 20 June 2024; Accepted: 23 July 2024;
Published: 06 August 2024.
Edited by:
Takuya Tada, New York University, United StatesReviewed by:
Ivan Dzhagalov, National Yang Ming Chiao Tung University, TaiwanCopyright © 2024 Wang and Chong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mark M. W. Chong, bWNob25nQHN2aS5lZHUuYXU=
 Yi Wang
Yi Wang Mark M. W. Chong
Mark M. W. Chong