- 1Department of Microbiology, Anatomy, Physiology and Pharmacology, La Trobe University, Bundoora, VIC, Australia
- 2Genomics Research Platform, School of Agriculture, Biomedicine and Environment, La Trobe University, Bundoora, VIC, Australia
- 3School of Animal and Veterinary Sciences, Faculty of Sciences, Engineering and Technology, University of Adelaide, Adelaide, SA, Australia
- 4Infection & Immunity Program, La Trobe Institute for Molecular Science (LIMS), La Trobe University, Bundoora, VIC, Australia
- 5Department of Biochemistry and Chemistry, School of Agriculture, Biomedicine and Environment, La Trobe University, Bundoora, VIC, Australia
- 6Department of Biochemistry and Molecular Biology, Monash University, Clayton, VIC, Australia
Introduction: Long COVID is a debilitating condition that lasts for more than three months post-infection by SARS–CoV–2. On average, one in ten individuals infected with SARS CoV- 2 develops Long COVID worldwide. A knowledge gap exists in our understanding of the mechanisms, genetic risk factors, and biomarkers that could be associated with Long COVID.
Methods: In this pilot study we used RNA-Seq to quantify the transcriptomes of peripheral blood mononuclear cells isolated from COVID-recovered individuals, seven with and seven without Long COVID symptoms (age- and sex-matched individuals), on average 6 months after infection.
Results: Seventy genes were identified as significantly up- or down-regulated in Long COVID samples, and the vast majority were downregulated. The most significantly up- or downregulated genes fell into two main categories, either associated with cell survival or with inflammation. This included genes such as ICOS (FDR p = 0.024) and S1PR1 (FDR p = 0.019) that were both up-regulated, indicating that a pro-inflammatory state is sustained in Long COVID PBMCs compared with COVID recovered PBMCs. Functional enrichment analysis identified that immune-related functions were expectedly predominant among the up- or down-regulated genes. The most frequently downregulated genes in significantly altered functional categories were two leukocyte immunoglobulin like receptors LILRB1 (FDR p = 0.005) and LILRB2 (FDR p = 0.027). PCA analysis demonstrated that LILRB1 and LILRB2 expression discriminated all of the Long COVID samples from COVID recovered samples.
Discussion: Downregulation of these inhibitory receptors similarly indicates a sustained pro-inflammatory state in Long COVID PBMCs. LILRB1 and LILRB2 should be validated as prospective biomarkers of Long COVID in larger cohorts, over time and against clinically overlapping conditions.
1 Introduction
Millions of individuals are suffering from ongoing or new symptoms weeks, months or years after initial infection with SARS-CoV-2 (1). This condition is most often called Long COVID, also referred to as post-acute sequelae of SARS-CoV-2 infection (2). Many studies have now been conducted to measure the prevalence rates of Long COVID, varying widely from 9-83% of people who had been infected with SARS-CoV-2 (3–5). This large variation can be attributed to differences in clinical definition, methodology, vaccination status and the severity of acute infection. Conservative estimates suggest that one in ten people previously infected with SARS-CoV-2 have developed Long COVID (6). Long COVID has been defined by the World Health Organisation (WHO) as the occurrence of new or persistent symptoms three months after a SARS-CoV-2 infection, which persists for at least two months and cannot be explained by an alternative diagnosis (7). There have been over two-hundred symptoms reported which affect most body systems (8). The most common symptoms reported are fatigue (prevalence rate of 21.6%), respiratory problems (14.9%), cognitive impairment (10.1%) and joint/muscle pain (10.6%) (8). The underlying molecular mechanisms responsible for progression to chronicity following the initial infection are not known. Several theories focusing on immune dysfunction have been proposed, relating to viral persistence, reactivation of latent viruses, increased production of autoantibodies and persistent inflammation (6).
Despite the high occurrence rate of Long COVID there are no identified risk loci, diagnostic tests, treatments, or specific clinical biomarkers. This adds a layer of subjectivity and exclusionary process to Long COVID diagnosis that may delay or confound effective clinical management. It also poses a challenge in the recruitment of stringently diagnosed cohorts for research. Considering these issues, many studies have been undertaken which seek biomarkers of Long COVID. These efforts have mostly focused on attempting to identify immunological, neurological, vascular and cardiac signatures specific to the disease. Several studies have reported circulating inflammatory marker proteins to associate with Long COVID disease status, with this body of work strongly indicating a sustained pro-inflammatory state in at least a subset of affected individuals (9–16).
One study examined the immunological profile of Long COVID-affected individuals compared to COVID recovered individuals with varying severity of acute illness. Long COVID-affected individuals were characterized by increased IFN-γ and IL-2, indicative of a pro-inflammatory state and decreased CCL4 production. The authors propose that while the increased IFN-γ and IL-2 results in activation of effector T cells, the concomitantly reduced CCL4 leads to impaired recruitment of them to infected sites (17).
An Australian longitudinal study investigated the transcriptional and immunological blood profile of sixty-nine COVID recovered patients of varying severities, including twenty-one individuals referred to a Long COVID clinic (18). The study found that transcriptional changes which occurred during acute infection were still evident in recovered individuals for at least six months post-infection. This highlights the importance of including COVID recovered samples as controls in new studies. Crucially this study also reported that enriched immune signatures identified in whole blood transcriptomics withstood correction against the differential proportions of cell types in each PBMC sample, thereby demonstrating that transcriptomic assessment of heterogeneous PBMCs in COVID recovered samples is robust against individualized changes in cell type proportions and thus can be applied with confidence in future studies.
Measuring levels of specific mRNA transcripts from blood has high diagnostic potential since blood is readily accessible and the transcripts are measurable by existing Real-Time Quantitative Reverse Transcription PCR (qRT-PCR) infrastructure. This benefit would be accentuated if the number of transcripts required to indicate disease status is few. PBMCs are very easily isolated from whole blood and since, taken together, the prior studies strongly suggest persistent immunological disturbances (including robust changes to PBMCs), PBMC mRNA transcripts as potential discriminators of disease status are a promising avenue.
With this in mind, we undertook a small pilot study to test the effectiveness of using differences in PBMC gene expression to identify candidate biomarkers of Long COVID. We used RNA-Seq to sequence and quantify the transcriptomes of PBMCs isolated from seven Long COVID and seven COVID recovered age and sex matched individuals. The Long COVID participants had been infected with SARS-CoV-2 on average six months prior to sample collection. Functional enrichment analysis identified that immune-related functions were unsurprisingly predominant among the up- and down-regulated genes. The differentially expressed genes supported a persistent or chronic inflammatory state in Long COVID. Using multivariate-based signature discovery, we identified a robust blood-based transcriptomic signature that effectively distinguished patients who have fully recovered from SARS-CoV-2 (COVID recovered) from those experiencing Long COVID using the reduced expression levels of only two genes (LILRB1 and LILRB2). These results if validated in a larger cohort, over time and against similar conditions show promise for development into a simple blood based diagnostic tool for Long COVID.
2 Materials and methods
2.1 Recruitment, sampling and cohort characteristics
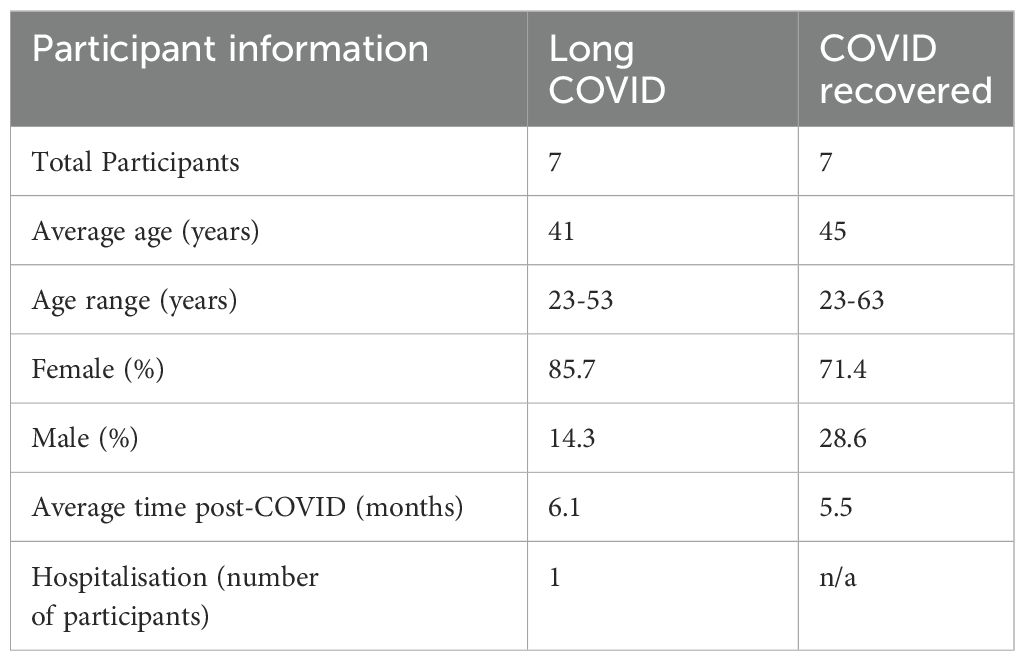
All participants were recruited in accordance with La Trobe University Human Ethics approvals HEC21207 and HEC21907. Seven Long COVID and seven COVID recovered participants were recruited for the study and all participants provided informed consent. Long COVID participants were defined according to the WHO description as exhibiting new or persistent symptoms three months following SARS-CoV-2 infection. Participants were asked to complete a symptom questionnaire and rate their illness severity using a five-point Likert scale. The cohort characteristics are shown in Table 1. COVID recovered participants were age- and sex-matched to the Long COVID cohort and on average all participants in this study were recruited approximately 6 months after an acute SARS-CoV2 infection. To exclude known, measurable alternative explanations for fatigue, blood samples collected from Long COVID participants were subject to pathology analysis conducted by Dorevitch Pathology, Melbourne, Australia. Blood samples for subsequent PBMC isolation were collected in lithium-heparin tubes.
2.2 PBMC isolation
PBMCs were isolated as previously published (19). Briefly, whole blood was separated by centrifugation in SepMate tubes (STEMCELL Technologies) at 1200×G for 10 minutes at RT. Resultant PBMCs were washed with RPMI 1640 to remove residual Ficoll. Aliquots were frozen in fetal bovine serum with 10% DMSO, gradually at -80°C.
2.3 RNA extraction and RNA-seq
RNA was extracted from PBMCs using the Monarch Total RNA Miniprep Kit (NEB, Ipswich, MA, USA). Approximately 5 × 106 PBMCs were harvested by centrifugation at 500 × g for 5 min and resuspended in 300 µL of RNA lysis buffer. The sample was transferred to a gDNA removal column fitted to a microcentrifuge tube and spun at maximum speed for 30 s to collect any contaminating genomic DNA. The RNA in the flow-through was precipitated by the addition of 300 µL ethanol (Chem Supply, Port Adelaide, SA, Australia) and mixed by repeated pipetting. The suspension was transferred to an RNA purification column fitted to a collection tube and again spun at max speed for 30 s and the flow-through discarded. The RNA was washed by the addition of 500 µL RNA wash buffer and centrifugation, as described previously, and the flow-through was discarded. The sample was DNase-treated by the addition of 5 µL DNase I and 75 µL of DNase I reaction buffer to the column matrix and incubated at RT for 15 min. A 500 µL volume of RNA priming buffer was added to the column, centrifuged for 30 s at max speed, and the flow-through was discarded. The RNA in the column was washed twice with 500 µL of RNA wash buffer, followed by centrifugation as described previously. RNA was eluted by the addition of 50 µL nuclease-free water and centrifugation at max speed for 30 s. RNA was then stored at -80°C before being sent to Novogene, Singapore on dry ice for mRNA sequencing and quantification using the Illumina NovaSeq 6000 platform and paired-end 150 bp reads.
2.4 Transcriptomic data analysis, enrichment analysis, and signature discovery
Transcriptomic profiling of PBMC samples from RNA-Seq Long COVID patients (n = 7) and COVID recovered individuals (n = 7) was performed by RNA-Seq (Illumina, paired end reads, 150bp). Quality control of the generated reads, mapping, and selection of genes with significant differential expression was carried out using CLC Genomics Workbench package 22 (QIAGEN) (20), HiSAT2 (21), and edgeR (22). Human genome 38 and its annotations (GRCh38) were downloaded from Ensembl genome browser (https://www.ensembl.org/index.html) and used for mapping and expression profiling. Mapping was carried out with the following parameters: mismatch cost of 2, insertion cost of 3, deletion cost of 3, minimum length fraction of 0.8, and minimum similarity fraction = 0.8. “Reads Per Kilobase of transcript per Million mapped reads” (RPKM) was used as the expression measurement. We used Generalized Linear Models (GLM) based on Negative Binomial distribution for differential expression analysis (22). The p-values were also corrected with false discovery rate (FDR) for multiple testing, and pFDR = 0.05 was used for selection of genes with statistically significant differential expression. The advantage of GLM is fitting the curve to expression values without the assumption that the error is normally distributed. Genes with an average RPKM lower than 4 in both cohorts were removed from the list of significant differentially expressed genes.
To find the key functions that the differentially expressed genes were involved in, enrichment analysis of Gene Ontology (GO) terms was analysed using the STRING web application tool (https://string-db.org/) (23). The significant molecular functions were selected based on pFDR = 0.05 and strength of the function, calculated by STRING (23).
Multivariate analysis, including principal component analysis (PCA) using correlation matrix and hierarchical clustering using average linkage method, was performed on expression values as previously described (24). PCA and clustering were utilized to evaluate the power of the developed transcriptomic signature as well as the functional pathways in distinguishing Long COVID from COVID recovered samples. Minitab Statistical Software 21 was employed for performing multivariate analysis.
3 Results
3.1 Long COVID participant symptoms
Seven Long COVID and seven COVID recovered participants were included in this study (Table 1). All participants had reported a SARS-CoV-2 infection on average six months prior to sample collection and pathology tests revealed no underlying conditions typically associated with fatigue. All pathology results were within the normal range. There were no significant difference in the mean age of each clinical group (p = 0.58, independent t-test) or gender, but both had a higher percentage of women and middle-aged participants. This is in line with the current literature that indicates a high prevalence in this age group and in women (25). Each of the seven Long COVID participants completed the symptom severity questionnaires which used a five-point Likert scale. Eighteen symptoms were included in the questionnaire and encompassed the main symptom clusters of neurocognitive, airway, cardiopulmonary, musculoskeletal and gastrointestinal issues. The most reported symptoms were fatigue, post-exertional malaise (PEM), muscle pain, concentration and memory issues, sore throat, headaches and temperature dysregulation. Fatigue was reported by each of the Long COVID participants and five out of the seven reported PEM. Fatigue and PEM were rated with the greatest overall severity of all symptoms (Figure 1).
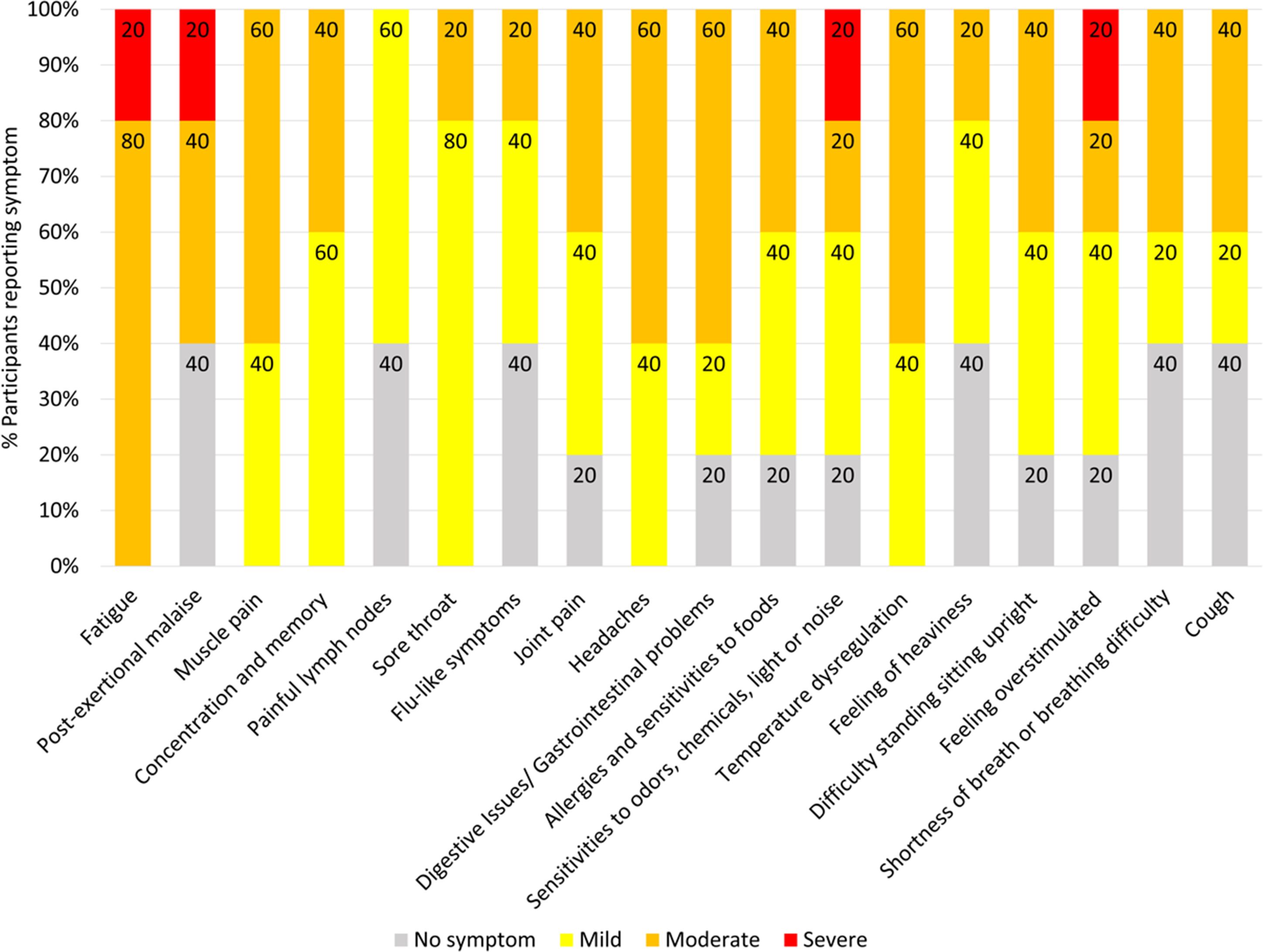
Figure 1. Symptoms affecting Long COVID participants and severity ratings. Seven of the participants with Long COVID completed questionnaires to list and rate the severity of their symptoms. Ratings of the symptoms were classified as 0 = no symptoms, 1-2 = mild, 3-4 = moderate, 5 = severe. Percentages are rounded to the nearest single decimal place.
3.2 Transcriptomic signature of long COVID
Total RNA was isolated from PBMCs of all participants and sent for RNA-Seq analysis (26). Transcriptomic analysis identified 5,144 transcripts, of which seventy were up- or down-regulated in Long COVID compared to COVID recovered controls (Figure 2). Most of the differentially expressed transcripts were downregulated (sixty-six down in total) and four were signficantly up-regulated in Long COVID compared to the COVID recovered controls (FDR p value < 0.05).

Figure 2. Long COVID versus COVID recovered PBMC transcriptomes. (A) Venn diagram depicting the number of gene transcripts significantly up- or down-regulated in PBMCs from people with Long COVID versus PBMCs from COVID recovered individuals. The p-values which determined significance were corrected with FDR for multiple testing, (pFDR < 0.05 was used as the threshold for significance). Genes with an average reads per kilobase per million mapped reads (RPKM) lower than four in both cohorts were removed from the list of significant differentially expressed genes. This was to ensure that any identified genes were present in sufficient amounts to be detected via clinically appropriate methods. (B) Heatmap representation of seventy differentially expressed genes between Long COVID and COVID recovered PBMC samples. Red indicates higher expression while blue indicates lower expression. Each column is a different sample and each row is a different gene.
Principal Component Analysis (PCA) of the seventy up- or down-regulated genes showed separation between Long COVID and COVID recovered samples (Figure 3) where only one COVID recovered sample was clustered in the Long COVID group. The first component was used to classify the samples and explained 81.7% of the variation in the expression data.
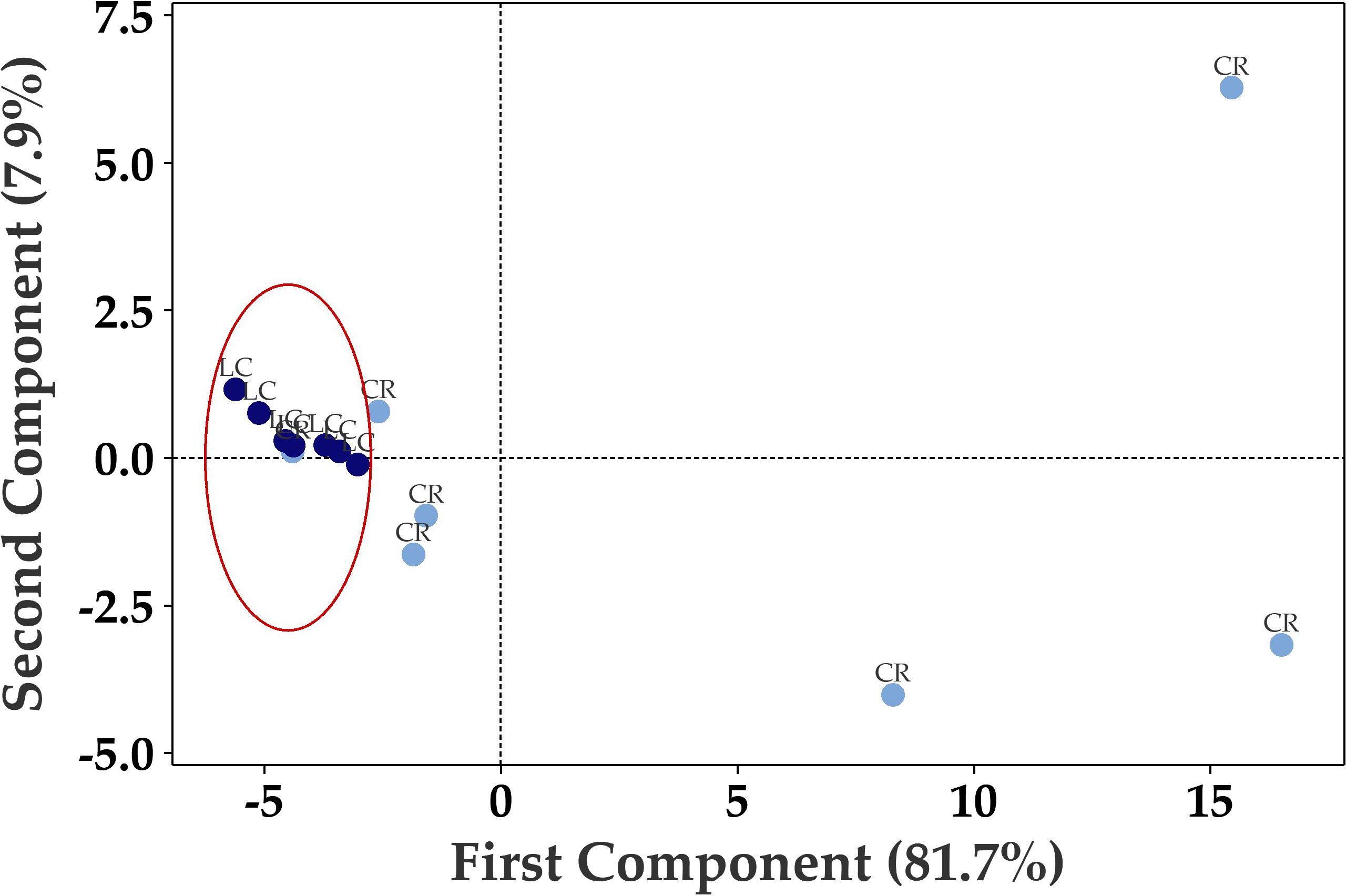
Figure 3. Principal component analysis (PCA) plot based on 70 differentially expressed genes in PBMCs efficiently distinguishes Long COVID (LC, red) PBMCs from most of the COVID recovered PBMCs (CR, blue) with one overlapping COVID recovered sample.
3.3 Dysregulation of immune related pathways
The list of genes with significant (pFDR < 0.05) differential expression between Long COVID and COVID recovered PBMC samples is presented in Table 2. Among the top ten downregulated genes in Long COVID as determined by fold change and FDR corrected p-value were genes associated with cell survival. This included Mesothelin (MSLN), expression of which was reduced more than sixty-fold. Mesothelin is a cell surface glycoprotein and is involved in cell signalling and adhesion. It is associated with various cancers and is thought to promote proliferation through activation of the NF-kB pathway. Downregulation of this gene has been connected with increased cell death via apoptosis (27). Another gene in the top ten which was associated with cell survival is colony stimulating factor receptor (CSF1R). This has been shown to be important for the survival, differentiation and proliferation of myeloid cells. The RNA binding protein, mRNA processing factor 2 (RBPMS2) was also highly downregulated and functions in post-transcriptional regulation of gene expression. Inactivation of this gene via methylation has also been associated with increased apoptosis (28).
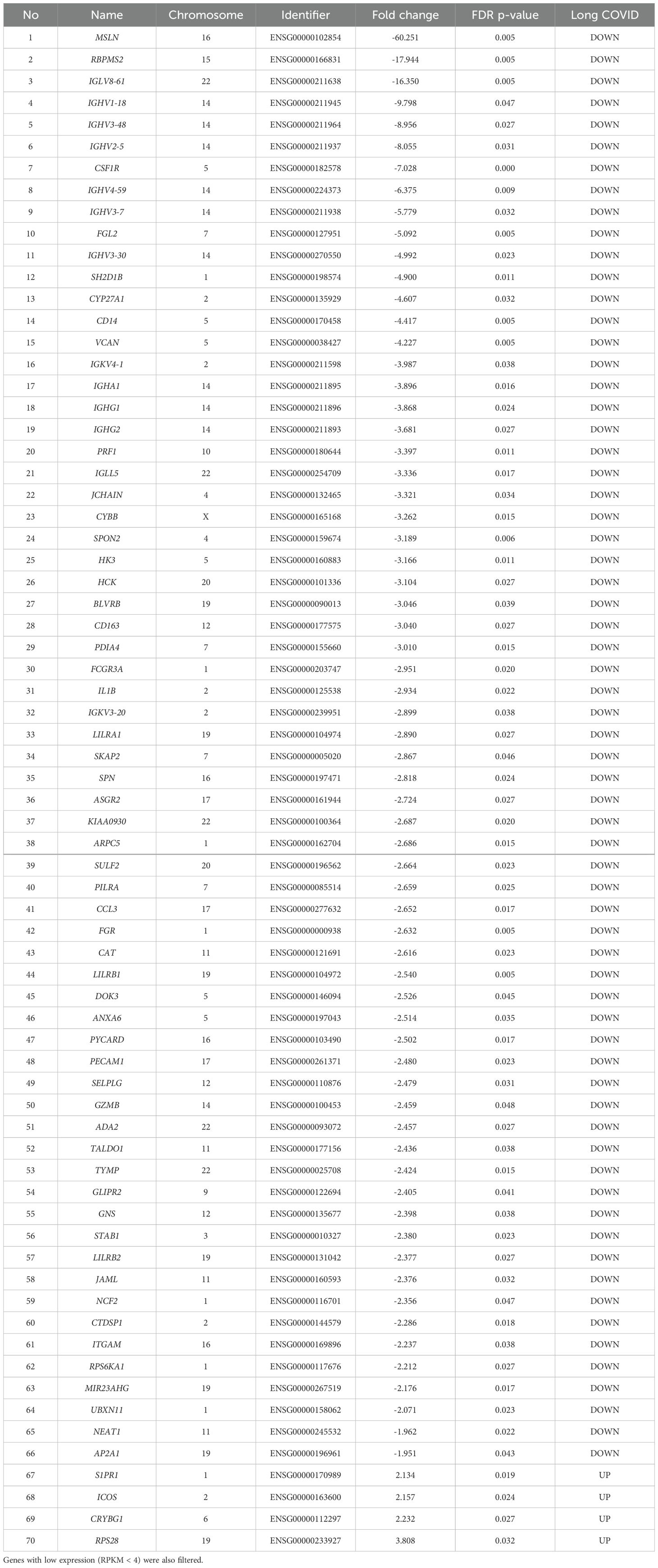
Table 2. The blood-derived PBMC transcriptomic signature of Long COVID compared to COVID recovered individuals The top genes were selected by satisfying a FDR p-value < 0.05 threshold.
Other genes among the top ten downregulated are associated with immune and inflammatory responses. This includes Fibrinogen-like protein 2 (FGL2), which is present in serum as a soluble protein and is a mediator of inflammation (29), and cytochrome b-245 beta chain (CYBB). CYBB encodes a subunit of the NADPH oxidase in phagocytes which is responsible for the microbicidal respiratory burst (30). CYBB is located in chromosome X and so may be related to the greater predisposition of females to Long COVID. Several Immunogloblin Light and Heavy Chain Variable (IGLV or IGHV) region genes were also present in the downregulated genes. All immunogloblin genes are located in chromosome fourteen, and as a group constituted the most frequently downregulated class of proteins in the Long COVID PBMCs. Seven out of the sixty-six downregulated genes belong to this class, namely IGLV8-61, IGHV1-18, IGHV3-48, IGHV2-5, IGHV4-59, IGHV3-7, and IGHV3-30. It is important to note that IGLV8-61 is a secretory protein and is predicted to be active in the extracellular space. Integrated proteomics data analyses, based on the publicly available proteomics data in ProteomicsDB, PaxDb, and MOPED showed that at the protein level, IGLV8-61 is mainly present in urine and skin (https://www.genecards.org/cgi-bin/carddisp.pl?gene=IGLV8-61). Secretory proteins in urine and/or blood are attractive biomarker candidates.
Four genes were significantly up-regulated in the Long COVID PBMC samples and were also associated with survival or immune pathways. Two such genes, Sphingolipid phosphate receptor 1 (S1PR1) and inducible co-stimulator (ICOS) (Figure 4) are associated with inflammation and the differentiation of T cells into T helper cells which produce inflammatory cytokines. Another, the 40S small ribosomal protein 28 (RPS28) has been proposed to play a role in the presentation of Major Histocompatibility Complex (MHC) class I peptides. The final up-regulated gene in Long COVID was crystallin beta-gamma domain containing 1 (CRYBG1) also known as Absent in Melanoma 1 (AIM1). This gene encodes an actin binding protein which acts as a tumour suppressor that is downregulated in various cancers (31).
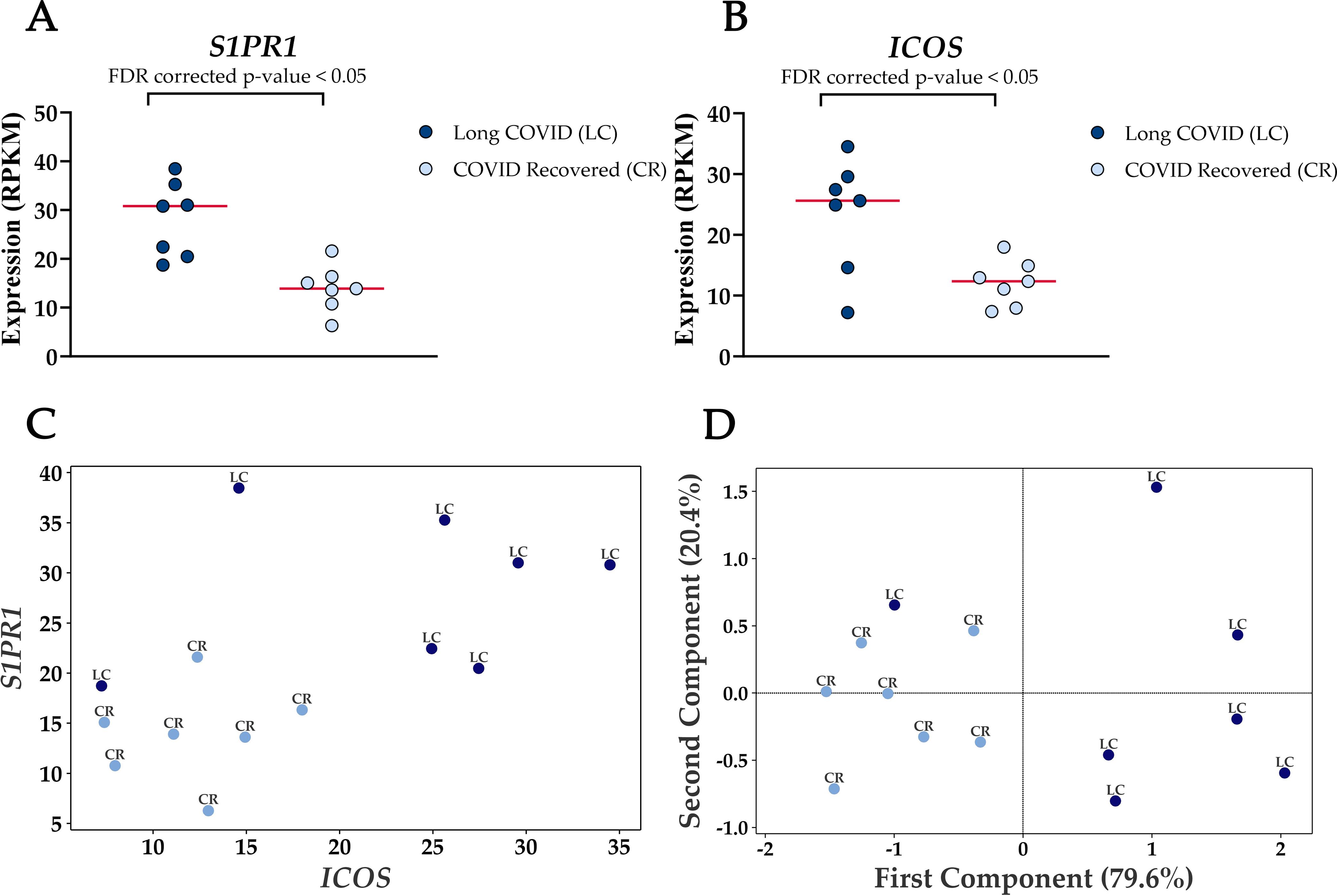
Figure 4. Elevated expression levels of S1PR1 and ICOS in blood-derived PBMCs from Long COVID patients versus COVID recovered individuals. (A) Comparison of S1PR1 expression between Long COVID and COVID recovered PBMCs. (B) Comparison of ICOS expression between Long COVID and COVID recovered PBMCs. (C) Scatter plot demonstrates the elevated expression of S1PR1 and ICOS in Long COVID (LC) samples. (D) PCA analysis demonstrates that S1PR1 and ICOS can discriminate all but one Long COVID (LC) sample from COVID recovered (CR), where the first component described 79.6% of variation in the expression data.
Functional enrichment analysis was performed to further define the pathways that were differentially regulated in Long COVID PBMCs (Table 3). The “molecular function” Gene Ontology term was used to identify the molecular processes or activities enriched in the up- or down-regulated genes. Some of the molecular functions identified referred to very broad activities of signalling receptor binding, antigen binding and protein homodimerisation activity. However, the molecular functions with the highest strength were related to MHC class I molecules and immunoglobulin receptor activity. Immunoglobulin and MHC class I are essential for appropriate responses to infection and inflammation. Induction of the MHC class I pathway has been shown to be downregulated by SARS-CoV-2 infection (32) and the immunoglobulin receptor Fc receptor-like 2 has been identified as downregulated in several transcriptomic datasets from SARS-CoV-2 infected individuals (33).
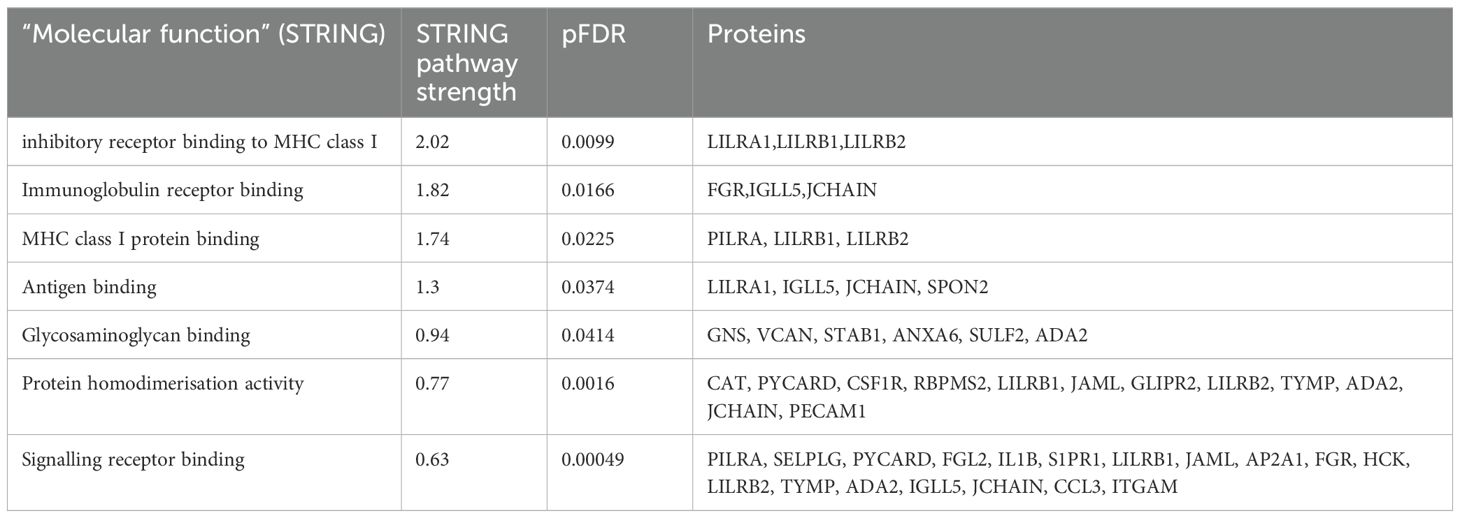
Table 3. Enrichment analysis of genes that were up- or down-regulated in Long COVID compared to COVID recovered individuals in terms of molecular function.
The other molecular function which was enriched in the up- or down-regulated transcripts was glycosaminoglycan binding. Glycosaminoglycans (GAGs) are ubiquitously and abundantly expressed on the surface of cells or in the extracellular matrix and interact with many proteins including chemokines, cytokines and growth factors. Due to the diversity of these interactions, GAGs play important roles in countless biological functions including cell growth and proliferation, resistance to invasion by pathogens and migration of immune cells (34).
3.4 Reduced expression levels of LILRB1 and LILRB2 can effectively differentiate long COVID blood samples from those of recovered individuals
The most frequently occurring genes in the identified molecular functions were leukocyte immunoglobulin-like receptor subfamily B member 1 (LILRB1) and leukocyte immunoglobulin-like receptor subfamily B member 2 (LILRB2) (Table 3) and both genes were significantly downregulated in Long COVID (Figure 5). LILRB1 and LILRB2 are frequently expressed in immune cells and function largely to regulate antigen presenting cells such as macrophages, dendritic cells and B cells, thus are important in a variety of innate and adaptive immune responses (35, 36). Specifically, the gene products are involved in inhibitory responses which suppress downstream pathways to mediate immunosuppression. Downregulation of LILRB1 and LILRB2 as observed here in the Long COVID PBMCs is therefore indicative of a heightened immune response and inflammation.
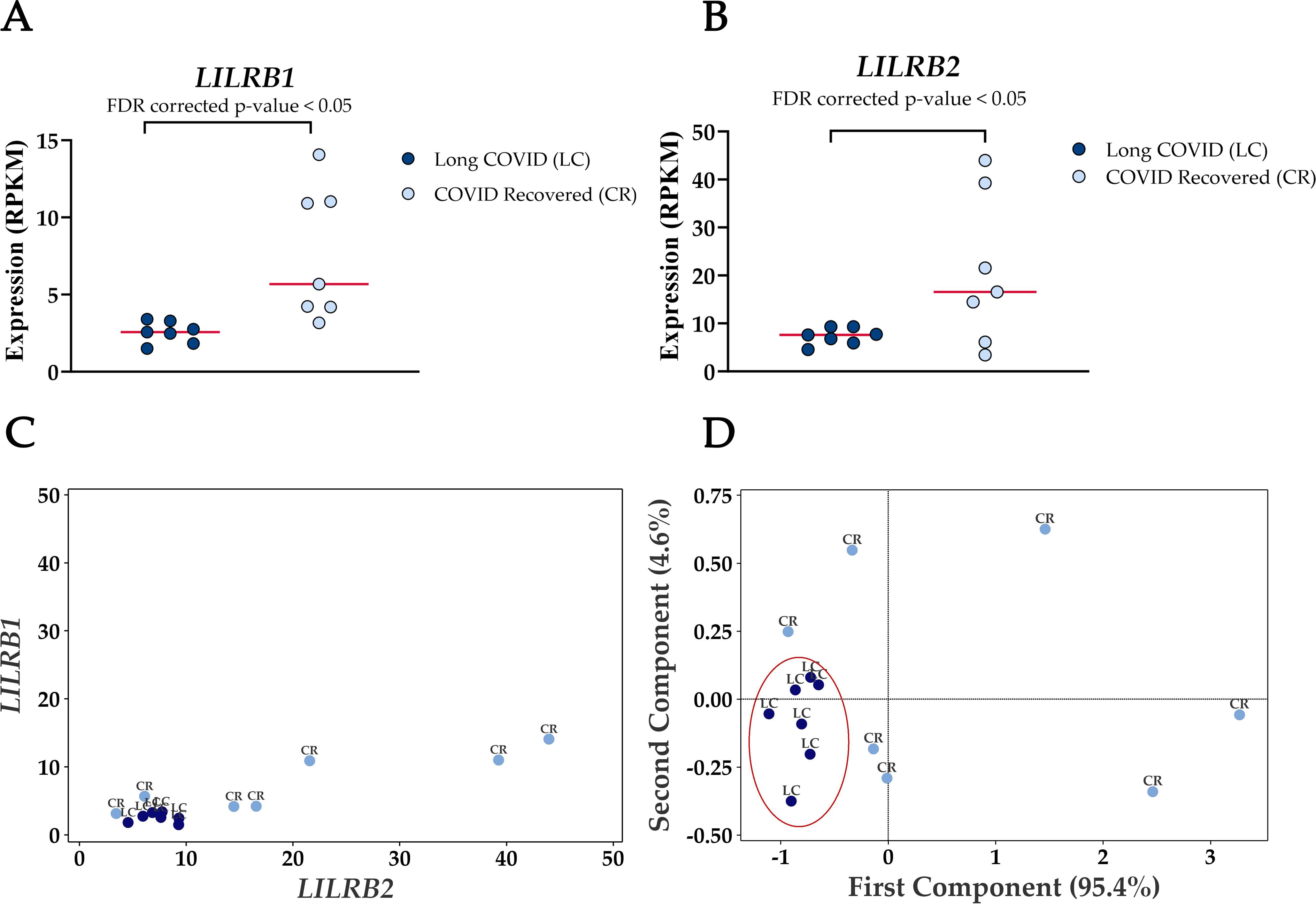
Figure 5. Reduced expression levels of LILRB1 and LILRB2 in blood-derived PBMCs effectively differentiate Long COVID patients from COVID recovered individuals. (A) Comparison of LILRB1 expression between Long COVID and COVID recovered PBMCs. (B) Comparison of LILRB2 expression between Long COVID and COVID recovered PBMCs. (C) Scatter plot demonstrates the reduced expression of LILRB1 and LILRB2 in Long COVID (LC) samples. (D) PCA analysis demonstrates the power of LILRB1 and LILRB2 to discriminate Long COVID (LC) samples from COVID recovered (CR) where the first component described 95.4% of variation in the expression data.
PCA analysis demonstrates the power of LILRB1 and LILRB2 in completely discriminating Long COVID from COVID recovered PBMCs. The first component described 95.4% of variation in the expression data. The ability of only two genes to distinguish the two cohorts with no overlap is a promising avenue for future validation as a prospective biomarker.
4 Discussion
Utilizing PBMC transcriptomic profiling in a small cohort, this study has revealed information about the molecular mechanisms of Long COVID and has identified a prospective transcriptomic signature of the disease in PBMCs for subsequent validation.
Analysis of PBMC transcriptomes from Long COVID individuals compared to COVID recovered controls revealed that the vast majority of differentially expressed genes were downregulated in Long COVID. The most frequently downregulated family of genes in Long COVID PBMCs was Immunoglobulin Heavy Chain Variable Region (IGHV), constituting six of the top ten downregulated genes. This observation in Long COVID samples contrasts with the up-regulation of this family of genes reported in acute SARS-CoV-2 infection (33). An analysis of transcriptomic data from nine different studies that compared SARS-CoV-2 infected cohorts to either healthy controls or other respiratory diseases identified that the expression of IGHV genes was most consistently up-regulated specifically in severe SARS-CoV-2 infections (33). This is noteworthy since severe infections are the most associated with development of Long COVID. A shift from elevated IGHV expression during the acute phase to reduced IGHV expression in Long COVID suggests dysregulation, and perhaps overactivity of the regulatory pathways involved in bringing the immune response back down over time. Interestingly, all of these genes are located in chromosome fourteen which suggests a possible hotspot of shared regulatory control that should be investigated in future studies.
Induction of cell death pathways to decrease the numbers of activated immune cells is an important homeostatic mechanism which can backfire during severe viral infection. This depletion of lymphocytes is termed lymphopenia. The mechanisms of cell death underpinning lymphopenia are diverse. T cell lymphopenia is a feature of many respiratory viral infections and is common in SARS-CoV-2 infection, particularly severe patients (37). T cell lymphopenia has been reported to be more severe in COVID patients and takes longer to resolve than in other viral infections (38). In the current study we did observe a decrease in the expression of genes associated with cell survival. While not measured directly, we also observed that the Long COVID PBMC samples died more quickly than the COVID recovered controls once recovered from frozen storage. Together this is unsurprising given that lymphopenia has been reported to be common in Long COVID (39–41). What these observations do suggest is that the mechanism by which lymphopenia occurs in Long COVID may involve the downregulation of genes associated with cell survival that we observed.
In our data we observed a significant increase in expression of the inducible T cell co-stimulator ICOS. ICOS promotes all fundamental T cell responses to foreign antigen and is crucial in mediating inflammation (42, 43). Among many other functions, ICOS enhances differentiation of T cells into T helper cells which produce pro-inflammatory cytokines. Pro-inflammatory cytokines and persistent low-grade inflammation are ubiquitously reported in Long COVID, although elevated levels of pro-inflammatory cytokines have been reported in recovered individuals too (44). Our observation of elevated ICOS expression, on average six months after infection, confirms that a pro-inflammatory state exists in Long COVID PBMCs until at least six months and that ICOS levels do intuitively wane over time in COVID recovered individuals, as would also be expected of the levels of pro-inflammatory cytokines.
The lipid mediator sphingolipid phosphate (S1P) and the sphingolipid phosphate receptors (S1PR) play critical roles in immune responses. S1PR1 is highly expressed on immune cells and together with S1P has been implicated as a regulator of inflammatory diseases. The S1P/S1PR1 pathway is essential for the trafficking of immune cells and in the differentiation of T helper cells (45). S1PR1 is up-regulated in several autoimmune diseases such as multiple sclerosis, systemic lupus erythematosus or rheumatoid arthritis and agents which inhibit S1PR1 are being investigated for their therapeutic potential (46). It is proposed that, in autoimmune conditions, increased S1PR1 inhibits the number and functions of regulatory T cells leading to increased production of inflammatory cytokines. Elevated ICOS expression is also associated with autoimmune diseases such as rheumatoid arthritis and lupus nephritis (47). Thus, the concomitant up-regulation of ICOS and S1PR1 in our Long COVID PBMCs likely forms a regulatory axis underpinning persistent inflammation and could be related to an autoimmune component based on our knowledge of other diseases.
Regulation of an appropriate inflammatory response is largely dependent on the expression and activation of immunoregulatory receptors, of which the leukocyte immunoglobulin like receptors (LILRBs) are key players. These receptors regulate inflammation via the control of many cellular processes including cell survival, phagocytosis, cell migration, cytokine production and cell death (48). In our data we observed a large downregulation of two LILRBs, LILRB1 and LILRB2, both of which inhibit inflammatory processes. Reduced expression or impaired function of LILRB1 and LILRB2 have been associated with inflammatory autoimmune conditions such as rheumatoid or psoriatic arthritis and systemic lupus erythematosus (49–52). This supports the potential role of reduced expression in promoting a sustained inflammatory state in Long COVID. Future studies characterizing LILRB1 and LILRB2 expression and function in SARS-CoV-2 infection and over time would be useful in determining how these receptors contribute to the severity of disease and susceptibility to development of Long COVID.
In this study we showed that the reduced expression of LILRB1 and LILRB2 alone discriminated a small pilot cohort of Long COVID PBMC samples from COVID recovered controls without overlap. This and the biological roles of these two genes being potentially related to the underlying pathology emphasize their potential as blood-based biomarkers of Long COVID. Given the small sample size of this pilot study this requires validation in larger cohorts, across different disease durations and in different disease groups.
Data availability statement
The data presented in the study are deposited in the Sequence Read Archive NCBI repository, accession number PRJNA1184005.
Ethics statement
The studies involving humans were approved by La Trobe University Human Ethics HEC21207 and HEC21907. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
DM: Writing – original draft, Writing – review & editing. EE: Data curation, Formal analysis, Investigation, Methodology, Writing – review & editing. MM: Data curation, Formal analysis, Methodology, Writing – review & editing. CA: Data curation, Formal analysis, Writing – review & editing. OS: Methodology, Writing – review & editing. PF: Conceptualization, Writing – review & editing. SG: Resources, Writing – review & editing. SA: Conceptualization, Formal analysis, Funding acquisition, Investigation, Project administration, Resources, Supervision, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by La Trobe University ABC internal investment scheme (awarded to SA), Tracey Banivanua Mar Research Fellowship (awarded to SA), the Medical Research Future Fund (MRFF, awarded to SG grant number MRF2005654), an NHMRC SRF (awarded to SG grant number #11592720).
Acknowledgments
We would like to thank all participants who kindly donated biosamples to this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Vehar S, Boushra M, Ntiamoah P, Biehl M. Post-acute sequelae of SARS-CoV-2 infection: Caring for the 'long-haulers'. Cleve Clin J Med. (2021) 88:267–72. doi: 10.3949/ccjm.88a.21010
2. Proal AD, VanElzakker MB, Aleman S, Bach K, Boribong BP, Buggert M, et al. SARS-CoV-2 reservoir in post-acute sequelae of COVID-19 (PASC). Nat Immunol. (2023) 24:1616–27. doi: 10.1038/s41590-023-01601-2
3. Byambasuren O, Stehlik P, Clark J, Alcorn K, Glasziou P. Effect of covid-19 vaccination on long covid: systematic review. BMJ Med. (2023) 2:e000385. doi: 10.1136/bmjmed-2022-000385
4. Notarte KI, Catahay JA, Velasco JV, Pastrana A, Ver AT, Pangilinan FC, et al. Impact of COVID-19 vaccination on the risk of developing long-COVID and on existing long-COVID symptoms: A systematic review. EClinicalMedicine. (2022) 53:101624. doi: 10.1016/j.eclinm.2022.101624
5. De Domenico M. Prevalence of long COVID decreases for increasing COVID-19 vaccine uptake. PloS Glob Public Health. (2023) 3:e0001917. doi: 10.1371/journal.pgph.0001917
6. Davis HE, McCorkell L, Vogel JM, Topol EJ. Long COVID: major findings, mechanisms and recommendations. Nat Rev Microbiol. (2023) 21:133–46. doi: 10.1038/s41579-022-00846-2
7. WHO. A clinical case definition of post COVID-19 condition by a Delphi consensus, 6 October 2021. Lancet Infect Dis. (2021). doi: 10.1016/S1473-3099(21)00703-9
8. Woodrow M, Carey C, Ziauddeen N, Thomas R, Akrami A, Lutje V, et al. Systematic review of the prevalence of long COVID. Open Forum Infect Dis. (2023) 10. doi: 10.1093/ofid/ofad233
9. Magdy R, Eid RA, Fathy W, Abdel-Aziz MM, Ibrahim RE, Yehia A, et al. Characteristics and risk factors of persistent neuropathic pain in recovered COVID-19 patients. Pain Med. (2022) 23:774–81. doi: 10.1093/pm/pnab341
10. Colarusso C, Maglio A, Terlizzi M, Vitale C, Molino A, Pinto A, et al. Post-COVID-19 patients who develop lung fibrotic-like changes have lower circulating levels of IFN-β but higher levels of IL-1α and TGF-β. Biomedicines. (2021) 9. doi: 10.3390/biomedicines9121931
11. Littlefield KM, Watson RO, Schneider JM, Neff CP, Yamada E, Zhang M, et al. SARS-CoV-2-specific T cells associate with inflammation and reduced lung function in pulmonary post-acute sequalae of SARS-CoV-2. PloS Pathog. (2022) 18:e1010359. doi: 10.1371/journal.ppat.1010359
12. Maamar M, Artime A, Pariente E, Fierro P, Ruiz Y, Gutiérrez S, et al. Post-COVID-19 syndrome, low-grade inflammation and inflammatory markers: a cross-sectional study. Curr Med Res Opin. (2022) 38:901–9. doi: 10.1080/03007995.2022.2042991
13. Pasini E, Corsetti G, Romano C, Scarabelli TM, Chen-Scarabelli C, Saravolatz L, et al. Serum metabolic profile in patients with long-covid (PASC) syndrome: clinical implications. Front Med (Lausanne). (2021) 8:714426. doi: 10.3389/fmed.2021.714426
14. Martone AM, Tosato M, Ciciarello F, Galluzzo V, Zazzara MB, Pais C, et al. Sarcopenia as potential biological substrate of long COVID-19 syndrome: prevalence, clinical features, and risk factors. J Cachexia Sarcopenia Muscle. (2022) 13:1974–82. doi: 10.1002/jcsm.12931
15. Zhao J, Schank M, Wang L, Dang X, Cao D, Khanal S, et al. Plasma biomarkers for systemic inflammation in COVID-19 survivors. Proteomics Clin Appl. (2022) 16:e2200031. doi: 10.1002/prca.202200031
16. Zhang JY, Whalley JP, Knight JC, Wicker LS, Todd JA, Ferreira RC. SARS-CoV-2 infection induces a long-lived pro-inflammatory transcriptional profile. Genome Med. (2023) 15:69. doi: 10.1186/s13073-023-01227-x
17. Patterson BK, Guevara-Coto J, Yogendra R, Francisco EB, Long E, Pise A, et al. Immune-based prediction of COVID-19 severity and chronicity decoded using machine learning. Front Immunol. (2021) 12:700782. doi: 10.3389/fimmu.2021.700782
18. Ryan FJ, Hope CM, Masavuli MG, Lynn MA, Mekonnen ZA, Yeow AEL, et al. Long-term perturbation of the peripheral immune system months after SARS-CoV-2 infection. BMC Med. (2022) 20:26. doi: 10.1186/s12916-021-02228-6
19. Lineburg KE, Grant EJ, Swaminathan S, Chatzileontiadou DSM, Szeto C, Sloane H, et al. CD8(+) T cells specific for an immunodominant SARS-CoV-2 nucleocapsid epitope cross-react with selective seasonal coronaviruses. Immunity. (2021) 54:1055–1065.e5. doi: 10.1016/j.immuni.2021.04.006
20. Govic A, Nasser H, Levay EA, Zelko M, Ebrahimie E, Mohammadi Dehcheshmeh M, et al. Long-term calorie restriction alters anxiety-like behaviour and the brain and adrenal gland transcriptomes of the ageing male rat. Nutrients. (2022) 14. doi: 10.3390/nu14214670
21. Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. (2016) 11:1650–67. doi: 10.1038/nprot.2016.095
22. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. (2010) 26:139–40. doi: 10.1093/bioinformatics/btp616
23. Szklarczyk D, Kirsch R, Koutrouli M, Nastou K, Mehryary F, Hachilif R, et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. (2023) 51:D638–d646. doi: 10.1093/nar/gkac1000
24. Ebrahimie E, Zamansani F, Alanazi IO, Sabi EM, Khazandi M, Ebrahimi F, et al. Advances in understanding the specificity function of transporters by machine learning. Comput Biol Med. (2021) 138:104893. doi: 10.1016/j.compbiomed.2021.104893
25. Chen C, Haupert SR, Zimmermann L, Shi X, Fritsche LG, Mukherjee B. Global prevalence of post-coronavirus disease 2019 (COVID-19) condition or long COVID: A meta-analysis and systematic review. J Infect Dis. (2022) 226:1593–607. doi: 10.1093/infdis/jiac136
26. Annesley SJ. Long COVID versus COVID recovered pilot study. In: Sequence Read Archive, ID 1184005 - BioProject - NCBI (2024).
27. Qualiotto AN, Baldavira CM, Balancin M, Ab'Saber A, Takagaki T, Capelozzi VL. Mesothelin expression remodeled the immune-matrix tumor microenvironment predicting the risk of death in patients with Malignant pleural mesothelioma. Front Immunol. (2023) 14:1268927. doi: 10.3389/fimmu.2023.1268927
28. Cheng M, Zhan X, Xu Y, Wang S, Zhang H, Fang L, et al. DNA methylation of RNA-binding protein for multiple splicing 2 functions as diagnosis biomarker in gastric cancer pathogenesis and its potential clinical significance. Bioengineered. (2022) 13:4347–60. doi: 10.1080/21655979.2022.2032965
29. Fu L, Liu Z, Liu Y. Fibrinogen-like protein 2 in inflammatory diseases: A future therapeutic target. Int Immunopharmacol. (2023) 116:109799. doi: 10.1016/j.intimp.2023.109799
30. Chanock SJ, J. el Benna RM. Smith and B. M. Babior: The respiratory burst oxidase. J Biol Chem. (1994) 269:24519–22. doi: 10.1016/S0021-9258(17)31418-7
31. Ray ME, Su YA, Meltzer PS, Trent JM. Isolation and characterization of genes associated with chromosome-6 mediated tumor suppression in human Malignant melanoma. Oncogene. (1996) 12:2527–33.
32. Yoo JS, Sasaki M, Cho SX, Kasuga Y, Zhu B, Ouda R, et al. SARS-CoV-2 inhibits induction of the MHC class I pathway by targeting the STAT1-IRF1-NLRC5 axis. Nat Commun. (2021) 12:6602. doi: 10.1038/s41467-021-26910-8
33. Välikangas T, Junttila S, Rytkönen KT, Kukkonen-Macchi A, Suomi T, Elo LL. COVID-19-specific transcriptomic signature detectable in blood across multiple cohorts. Front Genet. (2022) 13:929887. doi: 10.3389/fgene.2022.929887
34. Casale J, Crane JS. Biochemistry, Glycosaminoglycans. In: StatPearls. StatPearls Publishing LLC., Treasure Island (FL (2024).
35. Jan RM, Al-Numan HH, Al-Twaty NH, Alrayes N, Alsufyani HA, Alaifan MA, et al. Rare variant burden analysis from exomes of three consanguineous families reveals LILRB1 and PRSS3 as potential key proteins in inflammatory bowel disease pathogenesis. Front Med (Lausanne). (2023) 10:1164305. doi: 10.3389/fmed.2023.1164305
36. Barkal AA, Weiskopf K, Kao KS, Gordon SR, Rosental B, Yiu YY, et al. Engagement of MHC class I by the inhibitory receptor LILRB1 suppresses macrophages and is a target of cancer immunotherapy. Nat Immunol. (2018) 19:76–84. doi: 10.1038/s41590-017-0004-z
37. André S, Picard M, Cezar R, Roux-Dalvai F, Alleaume-Butaux A, Soundaramourty C, et al. T cell apoptosis characterizes severe Covid-19 disease. Cell Death Differ. (2022) 29:1486–99. doi: 10.1038/s41418-022-00936-x
38. Chen Z, John Wherry E. T cell responses in patients with COVID-19. Nat Rev Immunol. (2020) 20:529–36. doi: 10.1038/s41577-020-0402-6
39. Alfadda AA, Rafiullah M, Alkhowaiter M, Alotaibi N, Alzahrani M, Binkhamis K, et al. Clinical and biochemical characteristics of people experiencing post-coronavirus disease 2019-related symptoms: A prospective follow-up investigation. Front Med (Lausanne). (2022) 9:1067082. doi: 10.3389/fmed.2022.1067082
40. Mandal S, Barnett J, Brill SE, Brown JS, Denneny EK, Hare SS, et al. 'Long-COVID': a cross-sectional study of persisting symptoms, biomarker and imaging abnormalities following hospitalisation for COVID-19. Thorax. (2021) 76:396–8. doi: 10.1136/thoraxjnl-2020-215818
41. Moreno-Pérez O, Merino E, Leon-Ramirez JM, Andres M, Ramos JM, Arenas-Jiménez J, et al. Post-acute COVID-19 syndrome. Incidence and risk factors: A Mediterranean cohort study. J Infect. (2021) 82:378–83. doi: 10.1016/j.jinf.2021.01.004
42. Beier KC, Hutloff A, Dittrich AM, Heuck C, Rauch A, Büchner K, et al. Induction, binding specificity and function of human ICOS. Eur J Immunol. (2000) 30:3707–17. doi: 10.1002/1521-4141(200012)30:12<3707::Aid-immu3707>3.0.Co;2-q
43. Hutloff A, Dittrich AM, Beier KC, Eljaschewitsch B, Kraft R, Anagnostopoulos I, et al. ICOS is an inducible T-cell co-stimulator structurally and functionally related to CD28. Nature. (1999) 397:263–6. doi: 10.1038/16717
44. Gomes SMR, Brito ACS, Manfro WFP, Ribeiro-Alves M, Ribeiro RSA, da Cal MS, et al. High levels of pro-inflammatory SARS-CoV-2-specific biomarkers revealed by in vitro whole blood cytokine release assay (CRA) in recovered and long-COVID-19 patients. PloS One. (2023) 18:e0283983. doi: 10.1371/journal.pone.0283983
45. Garris CS, Blaho VA, Hla T, Han MH. Sphingosine-1-phosphate receptor 1 signalling in T cells: trafficking and beyond. Immunology. (2014) 142:347–53. doi: 10.1111/imm.12272
46. Li J, Huang Y, Zhang Y, Liu P, Liu M, Zhang M, et al. S1P/S1PR signaling pathway advancements in autoimmune diseases. Biomol BioMed. (2023) 23:922–35. doi: 10.17305/bb.2023.9082
47. Hu YL, Metz DP, Chung J, Siu G, Zhang M. B7RP-1 blockade ameliorates autoimmunity through regulation of follicular helper T cells. J Immunol. (2009) 182:1421–8. doi: 10.4049/jimmunol.182.3.1421
48. Abdallah F, Coindre S, Gardet M, Meurisse F, Naji A, Suganuma N, et al. Leukocyte immunoglobulin-like receptors in regulating the immune response in infectious diseases: A window of opportunity to pathogen persistence and a sound target in therapeutics. Front Immunol. (2021) 12:717998. doi: 10.3389/fimmu.2021.717998
49. Monsiváis-Urenda A, Niño-Moreno P, Abud-Mendoza C, Baranda L, Layseca-Espinosa E, López-Botet M, et al. Analysis of expression and function of the inhibitory receptor ILT2 (CD85j/LILRB1/LIR-1) in peripheral blood mononuclear cells from patients with systemic lupus erythematosus (SLE). J Autoimmun. (2007) 29:97–105. doi: 10.1016/j.jaut.2007.05.003
50. Kuroki K, Tsuchiya N, Shiroishi M, Rasubala L, Yamashita Y, Matsuta K, et al. Extensive polymorphisms of LILRB1 (ILT2, LIR1) and their association with HLA-DRB1 shared epitope negative rheumatoid arthritis. Hum Mol Genet. (2005) 14:2469–80. doi: 10.1093/hmg/ddi247
51. Guerra-de Blas Pdel C, Villaseñor-Talavera YS, Cruz-González Dde J, Baranda L, Doníz-Padilla L, Abud-Mendoza C, et al. Analysis of the expression and function of immunoglobulin-like transcript 4 (ILT4, LILRB2) in dendritic cells from patients with systemic lupus erythematosus. J Immunol Res. (2016) 2016:4163094. doi: 10.1155/2016/4163094
Keywords: COVID-19, Long COVID, biomarker, transcriptomics, inflammation, LILRB1, LILRB2
Citation: Missailidis D, Ebrahimie E, Dehcheshmeh MM, Allan C, Sanislav O, Fisher P, Gras S and Annesley SJ (2024) A blood-based mRNA signature distinguishes people with Long COVID from recovered individuals. Front. Immunol. 15:1450853. doi: 10.3389/fimmu.2024.1450853
Received: 18 June 2024; Accepted: 13 November 2024;
Published: 03 December 2024.
Edited by:
Fatemeh Saheb Sharif-Askari, University of Sharjah, United Arab EmiratesReviewed by:
Stelvio Tonello, University of Eastern Piedmont, ItalyJuliana Gonçalves, Human Immunobiology and Pathogenesis Laboratory, Portugal
Copyright © 2024 Missailidis, Ebrahimie, Dehcheshmeh, Allan, Sanislav, Fisher, Gras and Annesley. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sarah J. Annesley, Uy5Bbm5lc2xleUBsYXRyb2JlLmVkdS5hdQ==
†These authors have contributed equally to this work and share first authorship
 Daniel Missailidis
Daniel Missailidis Esmaeil Ebrahimie
Esmaeil Ebrahimie Manijeh Mohammadi Dehcheshmeh
Manijeh Mohammadi Dehcheshmeh Claire Allan
Claire Allan Oana Sanislav
Oana Sanislav Paul Fisher
Paul Fisher Stephanie Gras
Stephanie Gras Sarah J. Annesley
Sarah J. Annesley