- 1Department of Agricultural Sciences, University of Naples Federico II, Portici, Italy
- 2Task Force on Microbiome Studies, University of Naples Federico II, Naples, Italy
- 3Functional Genomics Research Center, Fondazione Human Technopole, Milan, Italy
- 4Department of Chemical Sciences, University of Naples Federico II, Naples, Italy
Salmonella enterica serovar Rissen (S. Rissen) is an emerging causative agent of foodborne diseases. The current emergence of antibiotic resistance makes necessary alternative therapeutic strategies. In this study, we investigated the potential of a phage-resistant strain of S. Rissen (RR) as a tool for developing an effective lipopolysaccharide (LPS)-based vaccine. The LPS O-antigen is known to play critical roles in protective immunity against Salmonella. However, the high toxicity of the LPS lipid A moiety limits its use in vaccines. Here, we demonstrated that the acquisition of bacteriophage resistance by S. Rissen leads to structural modifications in the LPS structure. Using NMR and mass spectrometry, we characterized the LPS from phage-resistant strains as a smooth variant bearing under-acylated Lipid A portions (penta- and tetra-acylated forms). We then combined RT-qPCR and NMR-based metabolomics to explore the effects of phage resistance and LPS modification on bacterial fitness and virulence. Finally, we conducted in vivo studies to determine whether lysogeny-induced remodeling of LPS affects the host immune response. Results revealed that the under-acylated variant of LPS from RR attenuates the inflammatory response in BALB/c mice, while eliciting a specific antibody response that protects against S. Rissen (RW) infection. In conclusion, our findings suggest that phage resistance, through lipid A modification, may offer a novel strategy for reducing LPS toxicity, highlighting its potential as a promising biological approach for developing LPS-based vaccines against Salmonella infections.
Introduction
Salmonellosis is a major enteric infection causing a high rate of mortality worldwide every year (1, 2). The limited treatment options due to antimicrobial resistance and the absence of an effective prophylactic therapy, have contributed to increase the annual morbidity and mortality, positioning salmonellosis as a serious public health concern (3). In the last few years, S. Rissen has emerged as a notable serovar causing salmonellosis in various regions of the world (4), making the development of a successful vaccine a pressing challenge.
To date, only three categories of vaccine are licensed for protection against Salmonella infection, and specifically S. typhi (5). These are the live attenuated vaccine Ty21a, a subunit vaccine based on the Vi capsular polysaccharide (Vi CPS) and the Vi typhoid conjugate vaccine (TCV) (6). Despite their different approaches, both Ty21a and unconjugated Vi CPS vaccines have severe limitations. Neither Ty21a nor unconjugated Vi CPS provide protection for young children, and they offer limited efficacy in adults (7). Additionally, Ty21a contains a mixture of antigens which makes difficult to determine which specific antigen confers protection or increases the risk of adverse effects (8). These difficulties have been successfully overcome by TCV, which promises long-lasting immunity and protection in infants while maintaining a favorable safety profile (9). However, TCV also has a limitation as it confers protection only against a selected subset of Salmonella species expressing the Vi capsular polysaccharide. Consequently, the growing interest in developing Salmonella vaccines has led to the exploration of alternative vaccine strategies. Most of these strategies, including the Generalized Modules for Membrane Antigens (GMMA), focus on targeting the O-antigen of the lipopolysaccharide (LPS) (10, 11).
LPS, also known as endotoxin, is a glycolipid located in the outer membrane of Gram-negative bacteria (12). It consists of a conserved Lipid A, a core oligosaccharide and a variable O-polysaccharide (OPS) (13). For many years, the polysaccharide component of LPS was thought to lack antigenicity. This assumption was definitively disproved by André Boivin, who demonstrated that isolated polysaccharide moieties can act as haptens, retaining antigenic properties without stimulating antibody production (14). Boivin also showed that smooth bacteria, which possess a “complete antigen”, composed of a complete OPS and Lipid A, stimulate robust immune responses by activating the Toll-like receptor 4 (TLR4) and producing polysaccharide-specific antibodies that neutralize pathogenic bacteria (15). As a result, smooth LPS (sLPS) is a virulence factor and a valid target for vaccine development, compared to rough LPS (rLPS) - which lack OPS or contain only a single subunit (16).
sLPS has been proven to be particularly effective in preventing infections caused by various Enterobacteriaceae, such as Klebsiella pneumoniae, Shigella flexneri and Salmonella (17–19). However, due to the diversity of O-antigens among Enterobacteria, sLPS is expected to induce an immune response with restricted specificity. Interestingly, the high similarity of surface molecules among Salmonella strains makes sLPS an attractive candidate for broader-spectrum vaccines capable of cross-reactive immunity and protection against closely related Salmonella serotypes sharing analogous O-antigen profiles (20). Despite this, the high toxicity of sLPS, due to Lipid A component, makes it challenging to develop safe vaccines.
To address this concern and develop safe and immunogenic vaccines, several approaches have been attempted. Among these, genetic manipulation for silencing genes encoding acyl transferases, such as msbB, htrB and pagP, has been the most successful (21, 22). This results in mutant strains that - compared to the parent strain - synthetize LPS molecules with different lengths or numbers of fatty acyl chains in Lipid A, leading to attenuation of the LPS signaling pathway and reduced LPS toxicity (23, 24).
In a previous study, we demonstrated that LPS from the bacteriophage-resistant strain of S. Rissen (RR) induces a reduced inflammatory response in vitro compared to the LPS produced by the bacteriophage-sensitive strain of S. Rissen (RW) (25, 26), and attributed this result to Lipid A modification.
The goal of the present study was to validate our hypothesis, proposing bacteriophage-resistance as an alternative strategy to traditional chemical, enzymatic, or genetic manipulations, to obtain under-acylated variants of Lipid A, useful for producing LPS-based vaccines.
Lysogenic phages play an influential role in shaping various aspects of bacterial lifestyle, significantly contributing to both micro- and macro-diversity observed within bacterial species (27). Of note, prophage Φ1 has been found to introduce variability between RR and RW strains. Unlike RW, the RR strain demonstrated a propensity to spontaneously lose the phage Φ1 without generating phage particle. Yet, the acquisition of phage-resistance has been found to supply the RR strain with additional virulence factors (26). To realize the above transformations, the phage Φ1 provides the host new genetic material which confers several advantages (25).
Morons are prophage genes encoding proteins that enhance bacterial fitness through different mechanisms (28). They can increase bacterial virulence, promote resistance to environmental stressors, and contribute to phage resistance via Superinfection exclusion (Sie) proteins (25, 28). These proteins prevent further infections by the same or closely related phages, inhibiting phage genome entry into bacterial cell (29).
Salmonella phages use LPS to enter host cells and establish a successful infection. Therefore, as part of the Sie mechanism, Salmonella may acquire prophage genes that modify the LPS structure (30).
Based on these considerations, we tested the potential of RR to reduce the LPS reactogenicity through Lipid A modification, proposing it as an alternative approach for developing LPS-based vaccines against Salmonella infections. We found that the acquisition of the phage-resistant phenotype induces metabolic and physiological changes in RR, which alter the host-pathogen interaction by remodeling the LPS acylation pattern. Furthermore, we demonstrated that Lipid A modification in RR-LPS impairs the TLR4 signaling, weakening the innate immune response while mediating early protection in BALB/C mice infected with S. Rissen.
In conclusion, our findings offer a promising strategy for developing effective LPS-based vaccine to prevent Salmonella infection and may pave the way for the development of broad-spectrum vaccines against other Gram-negative bacteria.
Materials and methods
Ethics statement
All mouse experiments were carried out in accordance with the guidelines for the Care and Use of Laboratory Animals of the European Community (Legislative Decree n. 116/92). The Animal Use Protocol (AUP, Number: 86/609/CEE) was approved by the Committee on the Ethics of Animal Experiments of the University of Naples and authorized from the Italian Ministry of Health.
Salmonella Rissen strains and culture conditions
The bacteriophage-sensitive S. enterica ssp. enterica serovar Rissen (RW) was isolated from food products and characterized by Istituto Zooprofilattico Sperimentale del Mezzogiorno (Portici, Naples, Italy), using standard conventional methods (31). The bacteriophage-resistant S. Rissen (RR), instead, was obtained from the RW strain following selection for resistance to phage Ф1, as described by Papaianni et al. (25). Both RR and RW strains were grown in Luria-Bertani (LB) medium (ThermoFisher Scientific, Waltham, Massachusetts, USA) at 37°C, with shaking at 200 rpm.
Phage Ф1 isolation
As previously reported by Papaianni et al. (25), bacteriophage Ф1 was isolated from the RW strain upon induction with mitomycin C. Briefly, the overnight culture of the RW strain was inoculated in 5 mL of fresh LB medium (2 x 108 CFU/5 mL) containing 1 μg/mL mitomycin C (Merck, Darmstadt, Germany) and incubated for 1 hour at 37°C with shaking. After incubation, the induced culture was centrifugated at 5,700 x g for 10 minutes. The supernatant was collected and stored at +4°C, while pellet was resuspended in 5 mL of fresh LB medium and incubated for further 4 hours at 37°C with shaking. After 4 hours, lysate was centrifugated again and the supernatant collected. Supernatants from both the centrifugations were pooled and filtered by 0.22 μm filters (Millipore, Darmstadt, Germany) (25).
The enumeration of phage particles, expressed as plaques forming colony (PFU/mL), was determined by performing the Double Agar Layer (DAL) method (32). Plaques were re-isolated, propagated and stored in SM buffer (NaCl 100 mM, Tris-HCl 50 mM, MgSO4 8 mM; pH 7.5) at + 4°C, or in liquid nitrogen with DMSO 5% (v/v) to avoid loss of phage titer (33).
Bacteriophage-resistant and sensitive Salmonella Rissen genome comparison
Genotyping differences between bacteriophage-resistant and sensitive S. Rissen strains were explored using circular genome visualization methods. Protein Basic Local Alignment Search Tool (BLASTp) was used to match the sequence similarities between the SieB protein of Salmonella Typhimurium phage 22 (GenBank: AF217253, Protein ID: AAF75022) and the Superinfection exclusion protein of S. Rissen phage 29485 (Ф1, GenBank: KY709687, Protein ID: ARB10858). Genome visualization of SieB gene in both S. Rissen strains was performed using BLAST Ring Image Generator (BRIG) software (34) (v0.95). Both RR and RW complete genome sequence were reported by Papaianni et al. (25). S. Rissen phage 29485 was selected as reference genome. The raw sequence data are available via the European Nucleotide Archive (ENA; http://www.ebi.ac.uk/ena) and are accessible through the accession number PRJEB69481. The genome assembly and annotation of the RR strain can be accessed via European Nucleotide Archive under the following accession number GCA_963679805.
Cell culture
The human gastric adenocarcinoma cell line AGS was kindly provided by Prof. Alessandra Tosco from the University of Salerno (Fisciano, SA, Italy). Cells were grown in Dulbecco’s Modified Eagle Medium (DMEM) high glucose (Microtech), supplemented with 10% fetal bovine serum (FBS; Microtech), 1% penicillin/streptomycin (Microtech) and 1% L-glutamine (Microtech) and maintained at 37°C, in a humidified atmosphere of 5% CO2. The cell line was authenticated through the short tandem repeat (STR) analysis and periodically tested for mycoplasma contamination.
Superinfection exclusion gene transcriptional changes
To better investigate genome differences between the bacteriophage-resistant and sensitive Salmonella Rissen strains, due to prophage gene integration, the presence of the SieB gene was analyzed by end-point PCR and electrophoretic run on an automated qiaxcel instrument (Qiagen, Hilden, Germany) as described by Capparelli et al. (26). In addition, transcriptional changes in SieB gene were examined by real-time quantitative PCR experiment (RT-qPCR), using StepOne (Applied Biosystem) Real-Time PCR system. As reported by Capparelli et al., AGS cells were infected with RR or RW strains for 2, 4 and 6 hours and SieB gene expression level was evaluated by qRT-PCR, following the protocol described by Capparelli et al. (26). The primers for detecting SieB gene were: reverse primer 5’-CAAACAAATCCCGAACGACT-3’; forward primer 5’-ATGGTGGCAGGAGTTAATGC-3’.
LPSs isolation and chemical analyses
Dried cells of RR and RW S. Rissen strains were extracted by PCP (phenol:chloroform:petroleum ether 2:5:8) and by water/phenol method as already reported (35).
PolyAcrylamide Gel Electrophoresis (PAGE) was performed using the system of Laemmli (36) with sodium deoxycholate (DOC) as detergent. Purified LPS from S. Rissen RR and RW and the LPS from Escherichia coli O111:B4 were prepared at the concentration of 1 mg/mL, and denaturated with sample buffer (2% DOC and 60 mM Tris/HCl pH 6.8, 25% glycerol, 14.4 mM 2-mercaptoethanol, and 0.1% bromophenol blue). Then, 8 μL of each sample was loaded on a 14% DOC gel with a 5% stacking gel and separated using a mini-PROTEAN electrophoresis instrument (Bio-Rad Laboratories). The electrophoresis was conducted at a constant amperage of 30 mA. The gel was fixed in an aqueous solution of 40% ethanol and 5% acetic acid, and finally visualized after silver nitrate staining (37). Precision Plus Protein Dual Color Standards (4 μL, MW size range 10–250 kDa) were used for molecular weight estimation. Glycosyl and fatty acids composition were obtained after derivatization as acetylated methyl glycosides (AMGs) and fatty acid methyl esters (FAMEs), as already described (38).
The linkage positions of the monosaccharides were determined by GC-MS analysis of the partially methylated alditol acetates (PMAAs). Samples (1 mg) were methylated with CH3I (100 µL) and NaOH powder in DMSO (300 µL) for 20 h (39). The products were totally hydrolyzed with 2 M TFA at 120°C for 2 h, reduced with NaBD4, and acetylated with Ac2O and pyridine (50 µL each, 100°C for 30 min).
All the derivatives were analyzed on Agilent Technologies gas chromatograph 7820A equipped with a mass selective detector 5977B and an HP-5 capillary column (Agilent, 30 m × 0.25 mm i.d.; flow rate, 1 mL/min). AMGs and FAMEs analyses were performed using different temperature programs. In detail, the AMGs analysis started at 140°C for 3 min, followed by an increase from 140°C to 240°C at 3°C/min; while the FAMEs analysis started at 140°C for 3 min, followed by an increase from 140°C to 280°C at 10°C/min. PMAA analysis was performed using a temperature program that began at 90°C for 1 minute, then increased from 90°C to 140°C at 25°C/min, 140°C to 200°C at 5°C/min, and 200°C to 280°C at 10°C/min, with a final hold at 280°C for 10 minutes.
OPSs isolation, purification, and structural characterization
The supernatants obtained after mild acid hydrolysis of the entire LPSs (19.5 mg and 15 mg from RR-LPS and RW-LPS, respectively) were fractionated on a Biogel P-10 column (Biorad, 0.75 x 112 cm) eluted with water. The collected fractions were pooled, freeze-dried, and analyzed by 1H NMR spectroscopy.
The molecular weight determination for OPSs was obtained through a calibration curve by using an Agilent 1100 HPLC system, with a TSK G-5000 PWXL size exclusion column (30 cm × 7.8 mm) equilibrated with NH4HCO3 50 mM as eluent (flow = 0.7 mL/min). The column was calibrated by injecting dextran standards (50 μL of a 1 mg/mL solution) of known molecular weight (12, 50, 150, and 670 kDa). The eluate was monitored with a refractive index detector (40).
Lipid A isolation and characterization
The Lipid A fraction was obtained from crude LPSs of both RR and RW S. Rissen by mild acid hydrolysis, as reported (38). After centrifugation at 3000 rpm for 15 min, the supernatants were removed and the pellet suspended in water and freeze-dried (3 mg and 4mg, for RR and RW respectively). MALDI TOF mass spectrometry was performed in the reflectron negative ions mode using a ABSCIEX TOF/TOF™5800 (AB SCIEX, Darmstadt, Germany) instrument, equipped with an Nd: YLF laser with a λ of 345 nm, and a delayed extraction technology. A solution of 2,4,6-trihydroxyacetophenone (THAP) in MeOH:H2O (v/v 1:1, 10 mg/mL) was used as the matrix. After desalting on a Dowex 50WX8 (H+ form) the samples with a concentration of 2 mg mL-1 were dissolved using CHCl3:MeOH (v/v 2:1). The matrix solution (0.5 µL) and Lipid A solutions (0.5 µL) were spotted on a MALDI plate. Each spectrum was a result of 2000 laser shots. The spectra were calibrated by using an ABSciex calibration mixture, and processed under computer control by using the Data Explorer software (v0.2.0) (41). Fatty acids analyses were obtained as already described (38). In few words, 0.5 mg of each sample was treated with 1 mL of 1.25 M HCl/CH3OH for 20 h at 80°C. The crude mixtures were extracted twice with hexane, and injected into GC-MS. The analyses were performed on Agilent Technologies gas chromatograph 7820A equipped with a mass selective detector 5977B and an HP-5 capillary column (Agilent, 30 m × 0.25 mm i.d.; flow rate, 1 mL/min). A temperature program starting at 140°C for 3 min and followed by an increase from 140°C to 280°C at 10°C/min was used for fatty acid analyses.
LPS purification for biological studies
LPS isolated from both RR and RW (38) were purified according to Stephens et al. (42). Briefly, LPSs were resuspended in endotoxin-free water containing triethylamine (TEA; 0.2%), deoxycholate (0.5%) and water-saturated phenol (500 μL). The obtained solution was vortexed for 5 minutes, incubated on ice for 5 minutes and then centrifugated (10,000 g for 2 minutes at 4°C). The upper phase (aqueous) was transferred into a fresh tube, supplemented with 75% ethanol and 30 mM sodium acetate, precipitated for 1 hour at -20°C and lastly resuspended in TEA.
LPS conjugation for mice immunization
The RR and RW-LPS were coupled to Rat Serum Albumin (RSA; purchased from Sigma Aldrich) using 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC), following the protocol described by Masoud, 2007 (43). Briefly, purified LPSs (10 mg) were dissolved in sterile distilled water (1 mL), and the solution was mixed with 500 μL RSA (10 mg/ml), in the presence of EDC (40 mg). The mixture was stirred on an automatic shaker at room temperature for 3 h. After mixing, the pH was adjusted to 7 and the mixture was centrifuged (1000 rpm for 10 minutes). The supernatants were purified by using a Sephadex G-75 column and eluted with phosphate buffered saline (PBS). Eluates containing both LPS and protein were pooled and dialyzed against distilled water at 48°C, and finally lyophilized.
Bacterial growth culture preparation for metabolomics analysis
RW and RR strains were obtained as indicated above (see the S. Rissen strains and culture conditions paragraph). RWΦ1- and RSΦ1- strains were derived from the RW and RR strains as described by Papaianni et al. (25). All strains were grown in LB medium and centrifugated when bacteria reached the exponential growth phase and the same cell density. After centrifugation, the pellet was discarded, and the supernatant filtered by 0.22 μm filters (Millipore, Darmstadt, Germany). Sterile supernatants were quenched in liquid nitrogen for 10 minutes - in order to stop metabolic activities in the filtrates - and stored at -80°C, until NMR analysis. Ten biological replicates of growth culture of each strain were prepared. Uninoculated growth medium was used as negative control.
NMR sample preparation
To achieve the NMR-based footprinting metabolomics analysis, growth media from RR, RW, RSΦ1- and RWΦ1- were rapidly defrosted and 630 μL of each fluid sample were pipetted into eppendorfs containing 70 µL of D2O, containing sodium 3-(trimethylsilyl)-2,2,3,3-tetradeuteropropionate (TSP) 0.1 mM (for chemical shift reference), and sodium azide 3 mM as antimicrobial agent, reaching 700 μL of total volume.
NMR spectra acquisition
NMR spectra were acquired as described by Papaianni et al. (44). In detail, all NMR experiments were performed on a Bruker Avance III-600 MHz spectrometer (Bruker BioSpin GmbH, Rheinstetten, Germany), equipped with a refrigerated autosampler and a TCI CryoProbe™ fitted with a gradient along the Z-axis, at a probe temperature of 300K (27°C). All spectra were recorded in H2O 90%:D2O 10% with 0.1 mM of TSP. 1H-NMR spectra were acquired with water suppression using excitation sculpting with gradience sequence (45). Spectra were acquired for 32 scans, with an acquisition time of 5.2 s, a relaxation delay of 2 s, a pulse width of 8.000 µs, receiver gain of 101, dwell time of 61 μs, 4 dummy scans and a spectral width of 13.0180 ppm. Two-dimensional homonuclear (1H-1H; TOCSY) spectra were acquired with water suppression using excitation sculpting with gradients, DIPSI-2 sequence (46). Mixing time of 100 ms was applied for TOCSY experiments. TOCSY experiments were acquired with 256 FIDs of 2048 complex points with 80 scans, a relaxation delay of 1 s, a pulse width of 8.000 µs, receiver gain of 101, dwell time of 64 μs, 32 dummy scans and a spectral width of 13.6582 ppm. All data were zero-filled to 4,096 points in both dimensions, and prior to Fourier transformation, a Lorentz- to-Gauss window with different parameters was applied for both t1 and t2 dimensions for all the experiments. The resonances of both 1D and homonuclear 2D spectra were referenced to internal sodium 3-(trimethylsilyl)-2,2,3,3-tetradeuteropropionate (0.1 mM; TSP) which was assumed to resonate at δH = 0.00 ppm. Two-dimensional heteronuclear (1H-13C; HSQC) spectra were recorded on the spectrometer operating at 150.90 MHz for 13C, using pre-saturation for water suppression (47, 48). HSQC resonances spectra were referenced to the lactate doublet (βCH3), assumed to resonate at 1.33 ppm for 1H and 20.76 ppm for 13C.
1H NMR spectra of the O-chains were recorded by using a Bruker 600 MHz spectrometer equipped with a cryo-probe, at 298K. The samples (3 mg) were dissolved in 550 μL of D2O and the spectra were calibrated with external acetone (δH = 2.225 ppm; δC = 31.45 ppm). Spectra were acquired for 32 scans, with an acquisition time of 4.58 s, dwell time of 70 µs, and spectral width of 11.90 ppm.
NMR data analysis
To obtain bacteria growth culture datasets, a spectral area ranging from 9.50 to 0.50 ppm was selected. Prior the data analysis, each proton spectrum was manually phased, and the baseline corrected using Buker Tospin software (v.3.6.3). Successively, they were automatically segmented into integrated regions (buckets) of 0.02 ppm, using the AMIX 3.6 package (Bruker Biospin, Germany). The residual water resonance region (5.00-4.58 ppm) was excluded, and the binned regions were normalized to the total spectrum area. Multivariate statistical data analysis was applied to each dataset, to differentiate culture medium profiles of bacteriophage-sensitive or resistant S. Rissen, before and after lysogenic Ф1 excision (RW or RR and RWΦ1- or RSΦ1-, respectively), through NMR spectra, according to their different metabolic content. Therefore, each integrated dataset – containing information on metabolite concentration - was exported to excel and the derived data matrix imported into SIMCA-P17 package (Umetrics, Umea, Sweden) where unsupervised Principal Component Analysis (PCA), supervised Partial Least Squares Discriminant Analysis (PLS-DA) and Orthogonal Partial Least Squares Discriminant Analysis (OPLS-DA) analyses were performed. PCA was first applied to check outliers and uncover initial trends within the dataset by investigating the systematic variation in the data matrix, in order to identify trends and clusters. Once assessed data homogeneity, PLS-DA or OPLS-DA discriminant analysis was employed to improve group discrimination. The performance of each elaborated model was evaluated via the parameters R2 and Q2 (PCA: R2 = 0.879, Q2 = 0.769; OPLS-DA: R2 = 0.99, Q2 = 0.873), indicating the goodness of fit and the goodness of prediction, respectively. NMR variables with |pcorr| ≥ 0.7, VIP >1 (Variable Importance in the Projection) were then considered for metabolite identification and univariate statistical analysis. Metabolites were identified through the chemical shift, according to both the literature (49) and metabolomics databases, such as the Human Metabolome Database (HMDB). Statistical significance was determined by one-way analysis of variance (ANOVA) followed by Bonferroni post hoc correction. p values < 0.05 were considered statistically significant.
Hemolytic activity
The susceptibility of erythrocytes to RR or RW-LPS was evaluated by performing the hemolysis assay on fresh human blood. Erythrocytes were isolated from blood of healthy donors (n=9), in accordance with the principles of good clinical practices reported in the Declaration of Helsinki and under the approval of the Ethics Committee of Villa Betania Hospital. Here, blood samples were obtained from the arm’s peripheral vein and collected in heparin tubes. An informed consent was obtained from all participants.
As reported by Greco et al. (50), whole blood was centrifuged at 1,500 x g for 5 minutes and the supernatant (plasma) discarded. Cell pellet, instead, was washed with PBS and gently shacked. These operations were repeated 5 times. Aliquots of 5 x 107/mL red blood cells were resuspended in PBS containing different concentrations of RR or RW-LPS (from 1 to 500 μg/mL), incubated at 37°C for 1 hour and centrifuged at 1,500 x g for 5 minutes. The supernatant was removed, and the released hemoglobin measured at 405 nm, using a plate reader. The percentage of hemolysis was calculated as the ratio of the untreated control.
Animal experiments
Animal experiments were carried out on female BALB/c mice aged 8 to 10 weeks - purchased from Charles River Laboratories (Wilmington, Massachusetts, USA) - at the animal facility of the University of Naples “Federico II”. Animal were housed in static microisolator cages, in order to handle them aseptically, and fed with sterile food and beverage. Mice were randomly assigned to the treatment groups. The sample group allocation was not blinded to the investigators.
Six mice per group were infected intraperitoneally with 100 μL of sterile phosphate-buffered saline (PBS), containing grown doses (103, 105, 107 and 109 CFU/animal) of RR or RW strains. Control group, instead, received 100 μL of PBS only. Mice were observed for 21 days, in order to evaluate clinical manifestations and death. Lethal Dose-50 (LD50) was calculated according to the Reed and Muench method (51).
LPS purified from both RR and RW strains was administrated intraperitoneally in a dose responsible for endotoxemia (20 mg/Kg body weight) (52) and mice (6 per group) were observed 2, 8 and 24 hours after the LPS injection, in order to evaluate body temperature and weight. As for viable bacteria, LPS was dissolved in PBS (100 μL per animal) and the control group was injected with PBS only. Mice that lost more than 20% of their starting body weight or decreased their starting body temperature more than 4°C were euthanized.
Cytokine measurement by ELISA assay
Blood samples were collected from mice via tail vein bleeding, after application of a local anesthetic. Blood was collected 2-4-6-8- and 24-hours following RR or RW-derived LPS administration and was immediately centrifugated at 3,000 g for 10 minutes at + 4°C. Blood samples collected from mice injected with sterile PBS were used as control. After centrifugation, serum was separated and frozen at - 80°C, until cytokine measurement was performed. The concentration of IL-6, IL-10, TNF-α, G-CSF and IFN-γ cytokines was determined by multiplex ELISA assay, using the Bio-Plex pro-mouse cytokine 23-plex assay (Bio-Rad), according to the manufacturer’s recommendations. Briefly, samples were diluted 1:4 with sample diluent and assayed as previously described by Verrillo et al. (53). Data were collected using the Bio-Plex 200 System (Bio-Rad) and analyzed by Bio-Plex Manager™ software. In addition, the concentration of IFN-β, was determined by the single target ELISA assay, using the mouse IFN-β ELISA kit (Elabscience), according to the manufacturer’s instructions. Samples were read using the 96-well plate reader (NeoBiotech). Cytokines were measured at different times and their concentration was determined based on the automatically calculated standard curve.
Immunization
Mice were divided into three groups, each group containing 6 mice. The first group was injected intraperitoneally with the RR-entire LPS, while the second one with the RW-entire LPS. Both the antigens were extracted, purified and conjugated with Rat Serum Albumin (RSA) as reported above (“LPS purification for biological studies” and “LPS conjugation for mice immunization”). For the immunization, they were dissolved in sterile PBS (pH=7.4) and administrated twice at two-weeks interval at the final concentration of 10 μg (100 μL). The third group (control group) was injected with sterile PBS only (100 μL). On day 21, a secondary booster was administrated. Blood samples from the tail vein and feces were collected on day 4, 9, 14, 21 and 28 to determine the adaptive immune response and evaluate the cross-protective ability conferred by the RR-LPS immunization, as described below.
Antibody response: IgG, IgA and IgM detection
To investigate the levels of adaptive immune response following animal immunization, the levels of IgG, IgM and IgA were detected. Serum was separated from whole blood by centrifugation (3,000 g, 10 minutes at + 4°C) and used to measure LPS-specific IgG and IgM, while feces were resuspended in protein extraction buffer (100 μL/10 mg), vortexed for 20 minutes and centrifugated 13,000 x g for 10 minutes. Supernatants were collected and used to detect LPS-specific IgA. First, the total antibody level for each Ig was detected using the mouse IgG Elisa kit (Abcam, Cambridge, United Kingdom), IgM ELISA kit (Abcam, Cambridge, United Kingdom) and IgA (Bethyl Laboratories, Montgomery, Texas, USA), according to the manufacturer’s recommendations. Successively, the anti-LPS IgG, IgM or IgA titer was quantified using LPS coated microplates, according to the standard indirect ELISA protocol (54). Briefly, 96-well plate was coated with RR or RW purified LPS (5 µg/mL) in 100 µL/well coating buffer (Na2CO3 0.64 g and NaHCO3 3.7 g) and incubated overnight at + 4°C. The next day, wells were blocked with 200 µL/well of wash-buffer containing 2% bovine serum albumin (BSA) and the plate was incubated for 1h at 37°C. Sera or fecal supernatants were diluted 1:100 in blocking buffer (100 µL/well) and incubated at 37°C for 1 h. Following incubation, 100 µL of the HRP-conjugated goat anti-mouse IgG (1:2,000; Invitrogen), IgA (1:10,000; Abcam) or IgM (1:1,000; Invitrogen) were added to each well and incubated at 37°C for 30 min. Finally, the plate was filled with 100 µL/well of the indicator substrate (TMB; 1 mg/mL to each well) and incubated at room temperature for 10 min in the dark. After incubation the enzymatic reaction was stopped by adding 100 µL/well of stop solution (1M sulphuric acid) and 15-30 minutes later the optical density (OD) was recorded at 450 nm with an automatic ELISA reader (NeoBiotech). The assay was performed in triplicate and each incubation period was followed by multiple washings using PBS containing Tween 20 (0.05%). The standard curve was used to accurately determine the concentration of target LPS-immunoglobulins.
Evaluation of the RR-LPS immunization efficacy
The efficacy of the RR-LPS immunization was evaluated by using a mouse model of salmonellosis producing disseminated infection after intraperitoneal injection of the bacteriophage-sensitive Salmonella Rissen (RW). In detail, mice immunized as detailed above (see the “Immunization” paragraph) were infected with the RW strain (109 CFU/100 μL) two weeks after the last immunization via the intraperitoneal route and the bacterial burden in the spleens was determined. Therefore, mice were sacrificed 3 days post-infection and spleens were aseptically removed. Spleens (1 g) were homogenized in 1 mL of phosphate buffer saline (PBS) solution and serial dilutions were cultured on BHI agar for 24 hours, at 37°C. The next day, colony were counted, and results were expressed as colony forming unit (CFU) per gram of tissue (CFU/spleen).
Additionally, the protective efficacy of the RR-LPS was also evaluated upon a single immunization. Mice (n=13 per group) were immunized intraperitoneally with pure conjugated LPS (10 μg dissolved in PBS 100 μL) isolated from the RR or RW strains or with vehicle (PBS 100 μL; control group). Two weeks post-immunization, each group was infected with the RW strain (109 CFU/100 μL) via the intraperitoneal route and the bacterial burden in the spleens was determined 3 days post-infection.
Antibody bactericidal activity
To investigate the role of anti-RR-LPS antibodies in imparting immune protection against S. Rissen infection, the bactericidal assay was performed. Mice were immunized as described above and four days after the last immunization, blood samples were collected, and serum separated by centrifugation. Polyclonal antibodies were purified by affinity chromatography, using protein A/G agarose beads as previously described (55, 56). 45 μL of antibody solution, antibody solution plus complement (BioMerieux, France), complement or LB broth (for control group) were mixed with 5 μL of viable RW (final bacterial concentration 1 x 106 CFU/mL) in a 96-well plate and incubated at 37°C for 3 hours. After incubation, the absorbance at 600 nm was measured using a microplate reader. Data are reported as Log10 of CFU/mL.
Analysis of the cross-protective ability of anti-RR-LPS antibodies
To evaluate whether the RR-LPS immunization could confer cross-protection, serum samples from mice immunized with the RR-LPS were tested for antibody reactivity against three different serovars of Salmonella (S. Choleraesuis, S. Infantis, S. Newport) by the agglutination test.
Mice were immunized as described above and four days following the last immunization, blood samples were collected, and the serum was separated by centrifugation (3,000 g, 10 minutes at + 4°C). Serum samples were used to measure IgG levels by the LPS-specific ELISA assay and animals giving the highest level of antibodies were selected for serum harvest. For the agglutination test, colonies of the selected Salmonellae were mixed with 100 μL of serum (undiluted or diluted 1:50 or 1:100 with sterile PBS) or with sterile PBS only (negative control). S. Rissen was used as positive control. The presence of cross-reaction was determined by observing the development of visible clumping of bacterial cells within 3 minutes.
Evaluation of the anti-RR-LPS antibodies target determinant to S. Rissen LPS
To explore the target determinant of the anti-RR-LPS antibodies to S. Rissen LPS, an Indirect ELISA assay was performed. We tested the antibody reactivity against both LPS and O-antigen from the RR and RW strains using serum samples from mice immunized with the RR-LPS, as reported above. Briefly, a 96-well plate was coated with O-antigen (5 µg/mL) or LPS (5 µg/mL) in 100 µL/well coating buffer (Na2CO3 0.64 g and NaHCO3 3.7 g) for 16 h at 4°C. Sera were diluted 1:100 in 100 µL/well of wash-buffer containing 2% bovine serum albumin (BSA) and incubated at 37°C for 1 h. Following incubation, 100 µL of the HRP-conjugated goat anti-mouse IgG diluted 1:2,000 (Invitrogen) was added to each well and incubated at 37°C for 30 min. Finally, the plate was filled with 100 µL/well of the indicator substrate (TMB; 1 mg/mL to each well) and incubated at room temperature for 10 min in the dark. After incubation the enzymatic reaction was stopped by adding 100 µL/well of stop solution (1M sulphuric acid) and 15-30 minutes later the optical density (OD) was recorded at 450 nm with an automatic ELISA reader (NeoBiotech). The assay was performed in triplicate and each incubation period was followed by multiple washings using PBS containing Tween 20 (0.05%).
Statistical analysis
Statistical analysis was conducted using GraphPad Prism 9.0 software (San Diego, CA, USA). One-way ANOVA, followed by either Turkey or Bonferroni post-hoc correction, was utilized to compare two or more experimental groups. For simultaneous comparison of multiple groups with two or more factors, we used two-way ANOVA followed by Bonferroni post-hoc test. The Log-rank test was employed to compare Kaplan–Meier survival curves. A minimum of n = 6 mice per group was included, and no formal randomization procedure was implemented. The reported values represent the means ± SD of two or three biological replicates, as indicated in figure legends. A p-value of < 0.05 was considered statistically significant.
Results
Prophage genes promote the expression of a phage-resistance phenotype
The property of prophages to mediate phenotypic changes in bacterial host is widely known. Our previous studies demonstrated significant differences in virulence profiles between RW and RR (26). Importantly, RR exhibited higher expression levels of prophage-related virulence genes (sspH1, sodC1, gtgE, grvA, and gogB) than RW (26). In the present study, we also observed transcriptional changes in the Superinfection exclusion (SieB) gene, which affects the bacterial membrane integrity and inhibits phage infection (57). As expected, RR expressed SieB at a level 300-fold higher than RW (Figure 1), in a time-dependent manner.
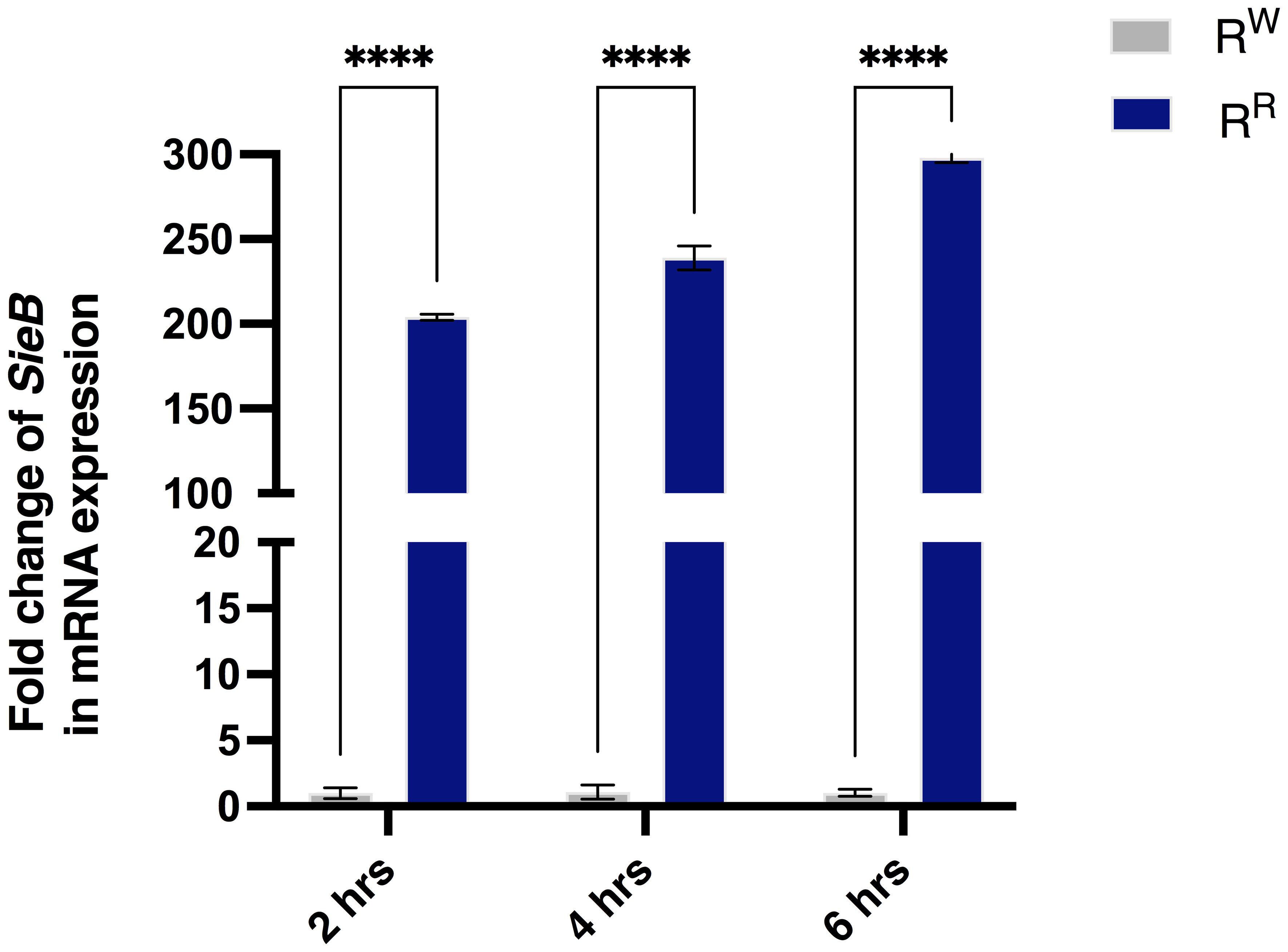
Figure 1. The RR strain expresses the prophage SieB gene. Analysis of mRNA expression levels displayed increased levels of SieB gene in the RR strain - following AGS cell line infection - compared to RW strain. Results are reported as graph bars and represent mean ± SD of three biological replicates from two independent experiments. Two-way ANOVA followed by Turkey post-hoc correction was used to compare the SieB gene expression between RR and/or RW groups, according to different time points. ****p < 0.0001.
To further investigate the genetic diversity between RR and RW, a genome comparison analysis, using the prophage Φ1 genome as a reference (Salmonella phage 29485, GenBank: KY709687), was performed. The circle plot revealed the insertion of the prophage derived SieB gene into the RR genome (Supplementary Figure S1). The evidence that the gene under investigation was SieB, was provided by the Basic Local Alignment Search Toll (BLAST, https://blast.ncbi.nlm.nih.gov/Blast.cgi), showing robust sequence similarity between Salmonella phage 29485 superinfection exclusion protein (Protein Id: ARB10858) and Salmonella phage P22 SieB protein (Protein Id: AAF75022, Supplementary Figure S2).
In conclusion, these results suggest that RR expresses the SieB gene, which confers immunity against secondary phage attack and contributes to the phage-resistance phenotype, likely through modifications of cell membrane elements, such as LPS (28). In addition, these results, along with previous reported results (26), prompted us to investigate whether the prophage-related virulence genes overexpressed by RR confer advantages to the host response against Salmonella.
Phage-resistance alters the LPS structure by inducing the production of under-acylated Lipid A
To investigate whether phage-resistance alters the LPS structure and to assess the effects of these modifications on bacterial virulence and host immune responses, the chemical structure of the Lipid A and O-antigen portions was examined.
Dried cells from both RR and RW strains were extracted as previously described (35). Both RR and RW produced smooth LPS (Supplementary Figure S3A). Additionally, GC-MS chromatograms of the purified O-chain polysaccharides (OPSs) revealed no differences in the sugar content of the O-antigen between the RR and RW strains (Supplementary Figure S3B). Methylation analysis further indicated the presence of 2-substituted mannose units (2-Man), 2,3-disubstituted mannoses (2,3-Man), 3-substituted glucosamine (3-GlcN) and terminal non reducing glucose (t-Glc) in the O-chains of both strains (Supplementary Table S1). Finally, in line with chemical analyses, 1H NMR spectra revealed an identical structure for the purified OPSs (Supplementary Figure S3C). In contrast, differences in the Lipid A portion were detected.
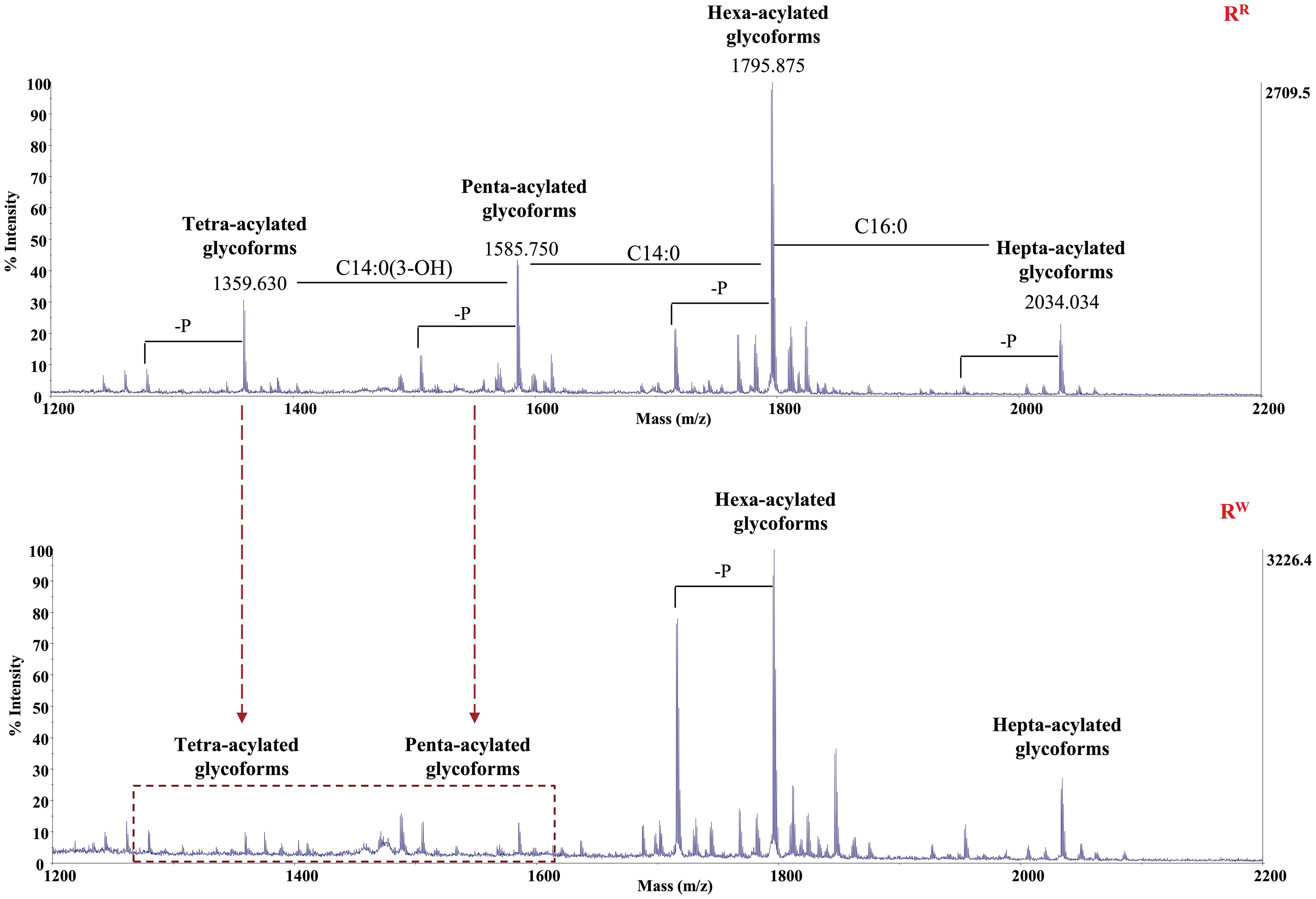
Pure Lipid A was analyzed through chemical techniques and mass spectrometry. GC-MS analysis of the fatty acid methyl esters (FAMEs) disclosed the presence of mainly C12:0, C14:0, C16:0, and C14:0(3-OH), in both the Lipid As. In addition, Lipid A from the RW strain was found to contain C14:0(2-OH). The Lipid A samples were analyzed by negative ions MALDI- TOF in reflectron mode. The acquired spectrum of Lipid A from RR revealed four clusters of signals corresponding to glycoforms with distinct acyl patterns (Figure 2). More in detail, the primary signal at m/z 1795.875 (calculated [M-H]- = 1796.229 Da) was assigned to the di-phosphorylated hexa-acylated glycoforms HexN2P2[C14:0(3-OH)]4(C14:0)(C12:0). The spectrum also showed the presence of a signal at m/z 2034.034 corresponding to the di-phosphorylated hepta-acylated species (calculated [M-H]- =2034.462 Da), which differs from the hexa-acylated species for the additional presence of a C16:0 and, signals at m/z 1585.750 and 1359.630 corresponding to the di-phosphorylated penta- and tetra-acylated glycoforms, respectively. Finally, each cluster exhibited heterogeneity as proven by the occurrence of signals differing in 80 u, corresponding to the phosphate group, and in 14 u (–CH2– unit), which indicates Lipid A species differing in the length of their acyl chains.
In conclusion, these results suggest that RR LPS contains highly variant Lipid A species, including di-phosphorylated hexa- and hepta-acylated species, along with under-acylated species (di- phosphorylated tetra- and penta-acylated variants). The same pattern was observed in the RW LPS. Nevertheless, RW LPS shows an extremely reduced presence of di-phosphorylated tetra- and penta-acylated species, compared to RR LPS. It is well known that under-acylated variants of Lipid A compete with the hexa-acylated form for TLR4 activation, functioning as weak agonists of TLR4 signaling (58). Specifically, under-acylated Lipid A species destabilize the MD-2–TLR4 complex, leading to a reduced production of pro-inflammatory cytokines (59, 60). Based on these considerations, our findings suggest that RR-LPS is a weak stimulator of the host innate immune response and elicits a mitigated inflammatory response.
Phage-resistance alters host metabolic profiles
In order to investigate the physiology of RR and its capability to adapt to various host conditions and evade host innate immune attack, basic insights into bacterial metabolism are fundamental. Culture supernatants from RR or RW – before and after lysogenic Ф1 excision (RR or RW and RSФ1- or RWФ1, respectively) – were compared by untargeted NMR-based metabolomics analysis. NMR spectra of growth medium from the wild-type RW strain and the selected RR mutant displayed extensive differences. Conversely, growth medium from RW and RR, following phage excision (RWФ1- and RSФ1-, respectively) showed a considerable similarity in the NMR profiles (Supplementary Figure S4). NMR spectra were compared by performing multivariate analysis. The OPLS-DA scores plot revealed a clear separation of the RR group (placed at negative coordinates of the first component t[1]) from RW, RWФ1- and RSФ1- groups (placed at positive coordinates of the first component t[1], Figure 3A). This result might be attributable to metabolic alterations associated with phage-resistance. The second component t[2], instead, revealed a distinct separation of RWФ1- and RSФ1- groups (placed at positive coordinates) from the RW group (placed at negative coordinates), likely due to phage-specific changes in the metabolism of the bacteriophage-sensitive strain (Figure 3A). Based on the first two principal components (t[1] and t[2]), RWФ1- and RSФ1- groups were found closely related each other, forming a single cluster in the hotteling’s T2 plot (Figure 3A). A more evident separation between RWФ1- and RSФ1- groups was detected by the third component t[3], which positioned the RWФ1- group at t[3] positive coordinates and the RSФ1- group at t[3] negative coordinates (Figure 4A).
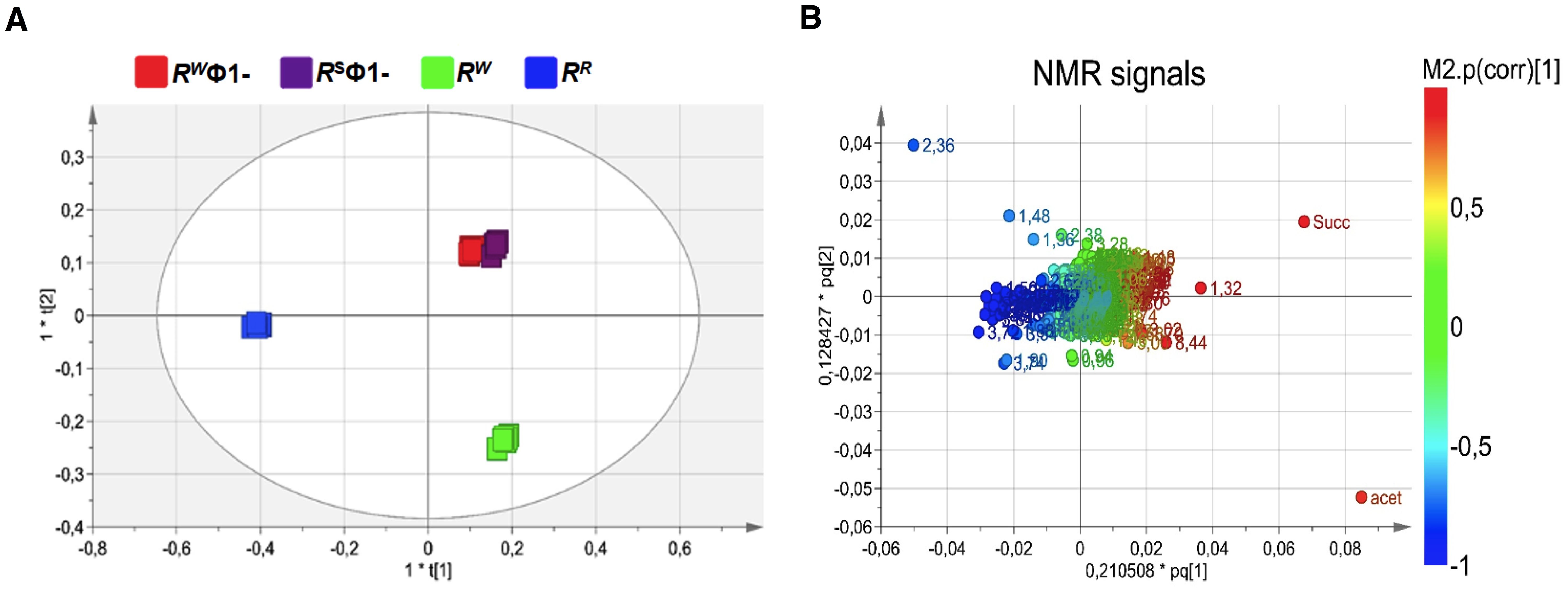
Figure 3. Orthogonal Projection to Latent Structure Discriminant Analysis (OPLS-DA) of bacteriophage-sensitive or resistant S. Rissen medium samples, before or after phage excision (RW; RR; RSФ1-; RWФ1-, respectively). (A) Scores plot showing a distinct separation of RR group (blue squares) from RW, RWФ1- and RSФ1- groups (green, red and purple squares, respectively) and of RWФ1- and RSФ1- groups (red and purple squares, respectively) from the RW group (green squares). (B) Loading plot indicating NMR variables (chemical shift) of metabolites responsible for between-classes separation, characterized by |p(corr)| value > 0.7.
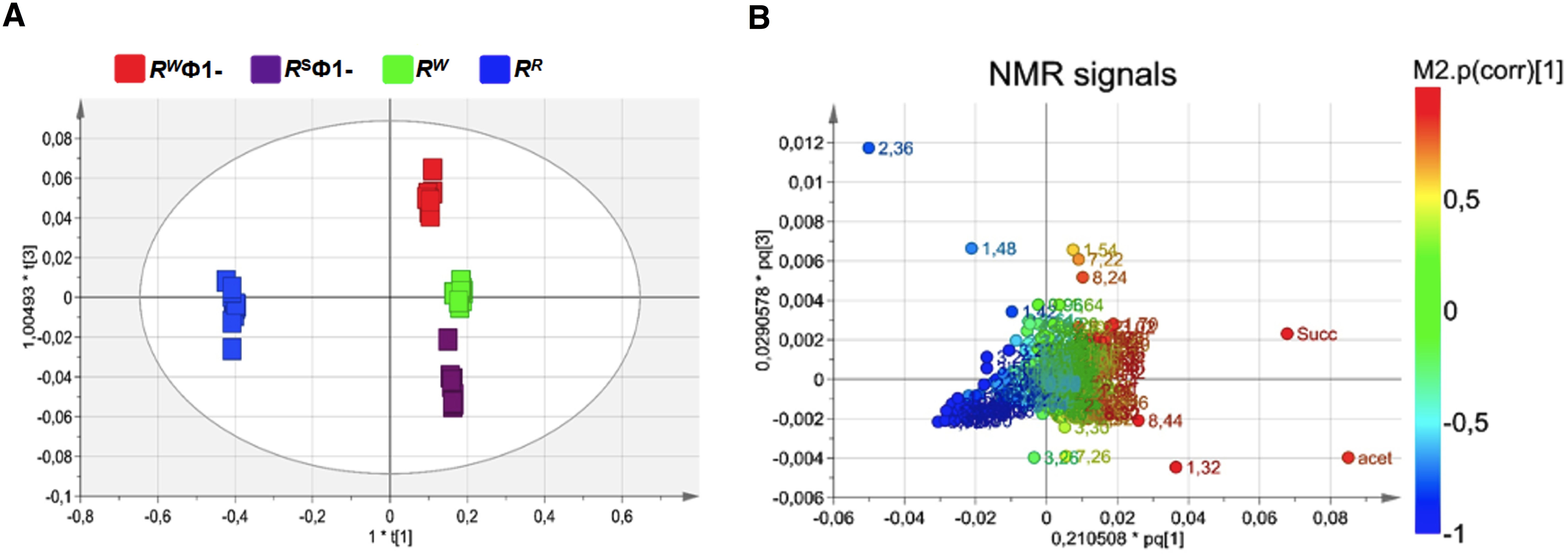
Figure 4. Orthogonal Projection to Latent Structure Discriminant Analysis (OPLS-DA) of bacteriophage-sensitive or resistant S. Rissen medium samples, before or after phage excision (RW; RR; RSФ1-; RWФ1-, respectively). (A) Scores plot showing a distinct separation of RSФ1- group (purple squares) from RWФ1- group (red squares). (B) Loading plot indicating NMR variables (chemical shift) of metabolites responsible for between-classes separation, characterized by |p(corr)| value> 0.7.
A total of 28 metabolites were identified as responsible for the above reported classes separation (Supplementary Table 2). In fact, significant differences in their concentration were detected. In detail, 14 amino acids and derivatives (arginine, lysine, isoleucine, proline, leucine, glutamate, methionine, aspartate, glycine, alanine, phenylalanine, tyrosine, N-acetyl-alanine and γ-aminobutyric acid), 1 sugar (glucose), the nucleic acid derivative hypoxanthine, the short chain fatty acid (SCFA) acetate, and 5 organic acid and derivatives (pyruvate hydrate, 5-aminopentanoic acid, lactate, succinate and formate) discriminated the RW group from the RR (p < 0.05). Interestingly, RSФ1- and RWФ1- groups did not display significant differences (p > 0.05) in the concentration of succinate, lactate, 5-aminopentanoic acid, formate, arginine, lysine, proline and phenylalanine. This result suggests that the different concentration observed in the growth medium from RW and RR is ascribable to metabolic changes induced by the presence of the phage Ф1 and specifically to endogenous phage genome elements responsible for phage-resistance.
Similarly, phage infection altered the expression levels of 3 amino acids and derivatives (betaine, threonine and pyroglutamate), 1 sugar (myo-inositol) and other metabolites (nicotinate and imidazole). These metabolites were present in different concentrations (p < 0.05) in the growth medium from RWФ1- compared to RW. The same result was found in RSФ1- compared to RR. No significant differences (p > 0.05), instead, were detected between RW and RR groups or RSФ1- and RWФ1-, thus addressing the mentioned differences to phage metabolism. On the contrary, RSФ1- and RWФ1- groups were found to show significant differences (p < 0.05) in the concentration of the amino acid tyrosine, the alcohol ethanol and fatty acids, due to the different behavior of Ф1 when excised from the phage-resistant or sensitive strain.
These results suggest that lysogeny affects bacterial metabolism and confers to RR biological advantages, which reflect both an increased fitness and the acquisition of virulence factors, implicated in modulating the host response (Supplementary Figure S5). Thus, taken together, genomics and metabolomics approaches allow for a complete understanding of the RR physiology and its interplay with the host, providing essential information for the development of new candidate vaccines (61).
Under-acylated LPS mitigates bacterial virulence in mice
To evaluate the effects of lysogeny on the virulence profile of S. Rissen, the lethal dose 50 (LD50) of both RR and RW strains was determined in a mouse model of infection. Female BALB/c mice aged 10 weeks were infected intraperitoneally with a dose-range of viable RR or RW bacteria (103, 105, 107, 109 CFU/mouse) and survival was monitored for 21 days (Figure 5).
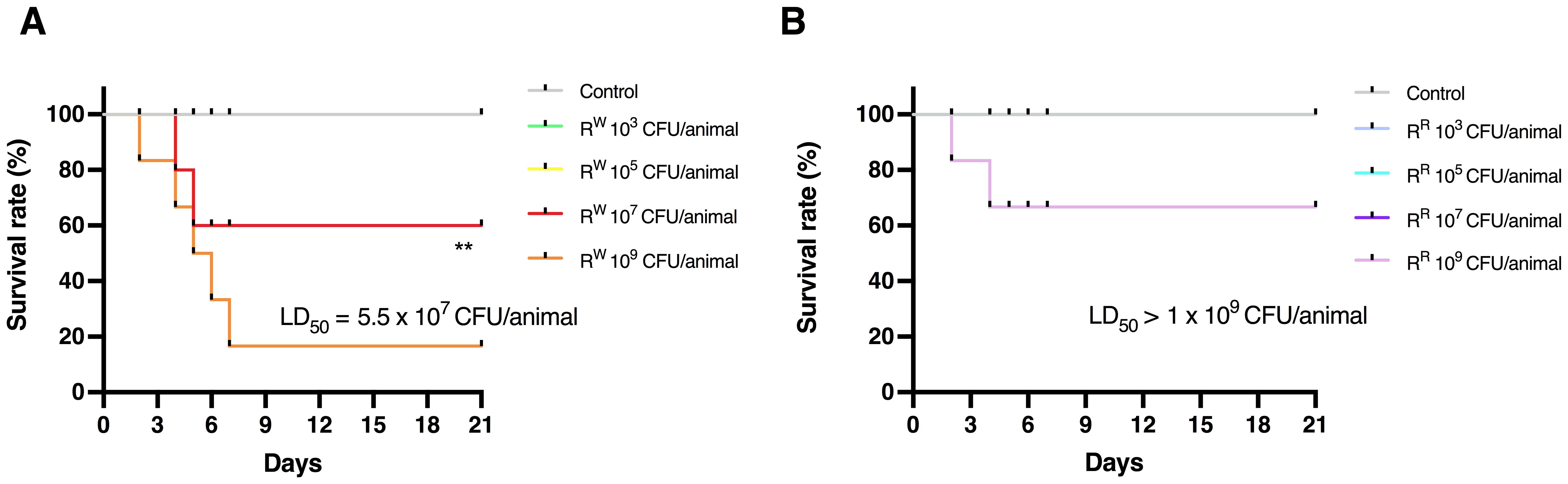
Figure 5. Determination of 50% Lethal Dose (LD50) of RR and RW in BALB/c mice. Survival curves of mice infected with different concentrations of RW strain (A) or RR strain (B). Uninfected control group included mice injected with PBS vehicle (n = 6/group). ** p < 0.01 (Long-rank Mentel-Cox test). Data are representative of two independent experiments. LD50 was calculated according to the Reed and Muench method. LD50 = Log10 end-point bacterial dose = Log10 (bacterial dose showing mortality next below 50%) + {[(50% - mortality at bacterial dose next below 50%)/(mortality at bacterial dose next above 50% - mortality at bacterial dose next below 50%)] x Log10 (differences between bacterial doses used in the assay)]}.
Results displayed that mice infected with lower doses (103 and 105 CFU/animal) of S. Rissen – RR or RW – survived throughout the study (Figure 5; Supplementary Table S3). On the contrary, mice infected with higher doses (107 and 109 CFU/animal) showed different survival rates (Figure 5; Supplementary Table S3). Compared to RR-infected mice, a limited percentage (~30%) of mice infected with RW (107 CFU/animal) began dying 4 days after infection (Figure 5B). Furthermore, while 30% of mice infected with RR (109 CFU/animal) died 6 days after infection, almost all of the mice (~90%) were killed within 7 days following infection with the same amount of RW (Figure 5A). These results show that the RR strain displays a reduced virulence compared to RW, with a LD50 significantly higher (1 x 109 CFU per mouse in RR vs 5.25 x 107 CFU per mouse in RW). This finding could find explanation in differences in the LPS-Lipid A structure between RR and RW (Figure 2). LPS is an endotoxin responsible for the host innate immune response activation, which can result in detrimental effects, such as the endotoxic shock (62). Due to its capacity to synthesize a LPS molecule lacking one or two acyl chains in the Lipid A portion, RR only triggers a mitigated immune response with reduced risk for adverse secondary complications (63). This was confirmed by stimulating mice with a single intraperitoneal injection of LPS (20 mg/Kg body weight) purified from the RR and RW strains (RR-LPS and RW-LPS, respectively) and monitoring clinical signs of endotoxemia (Figure 6A). As expected, mice stimulated with RW-LPS displayed more severe signs of disease (decreased body temperature and weight, diminution of appetite and apathy) compared to the RR one (Figures 6B–E), suggesting that mice are hyporesponsive to RR-LPS stimulation and hence, unable to mount an immediate defensive response. This property may reflect the unique organization of the RR-LPS and its capacity to mitigate the host immune system.
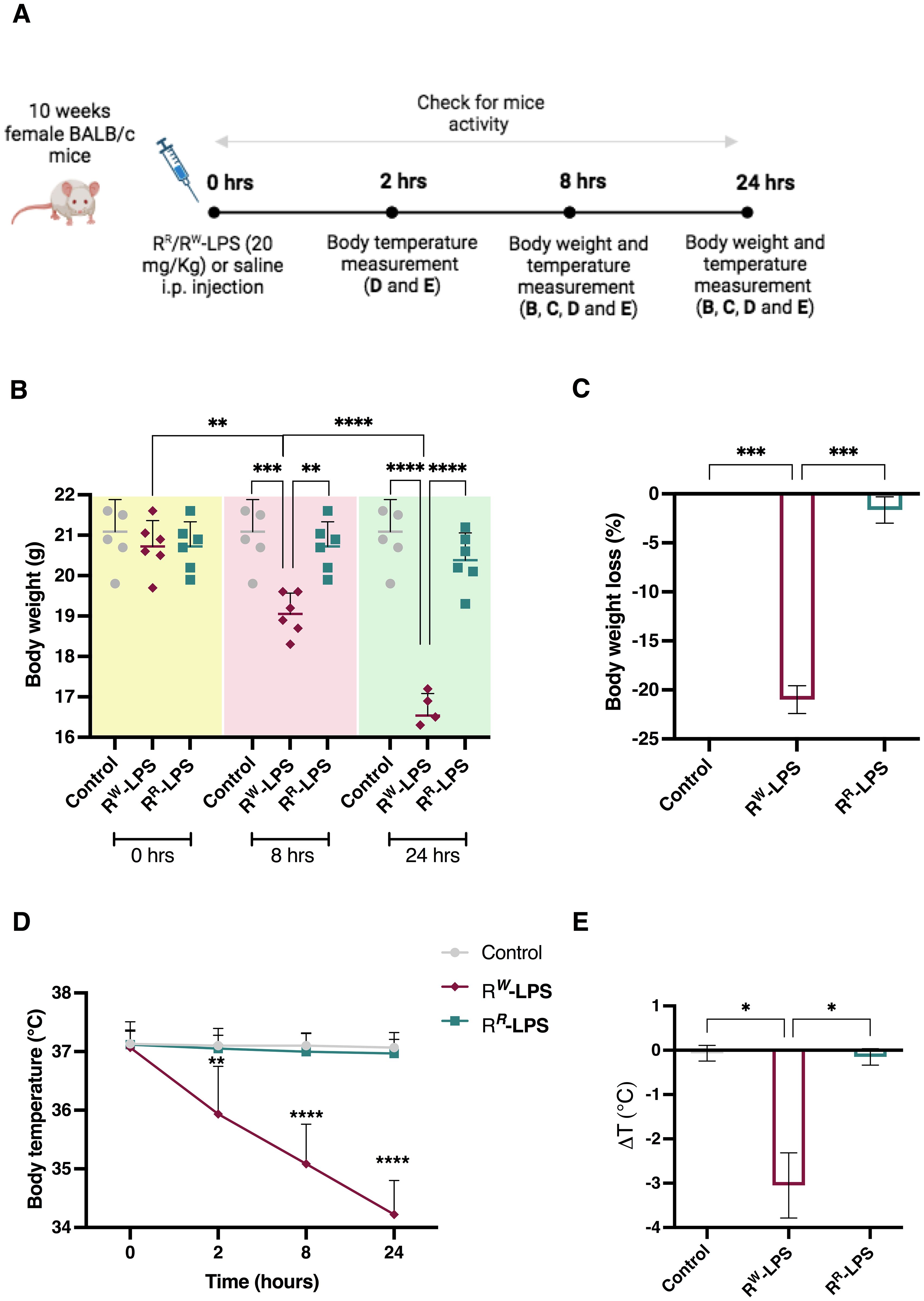
Figure 6. RR-LPS does not induce endotoxemia. (A) Experimental design for the intraperitoneal (i.p.) injection of RR or RW-LPS. Ten-week-old, female BALB/c mice (n = 6 per group) were injected intraperitoneally with 20 mg/Kg of RR or RW-derived LPS (RR-LPS and RW-LPS, respectively) dissolved in PBS solution and then body weight and temperature were evaluated. Mice (n = 6) in the control group (Control) were injected intraperitoneally with the same volume of PBS solution. (B) Body weight was measured 0, 8 and 24 hours after LPS stimulation. Graph represents mean ± SD of values from individual mice (dots). (C) Bar graph represents changes (%) of body weight 24 hours after LPS injection. Data are representative of a single experiment. Two-way ANOVA followed by Turkey post-hoc correction was used to compare body weight among three groups (Control, RR-LPS and RW-LPS) at different times. One-way ANOVA followed by Turkey post-hoc correction was used to compare body weight changes among three groups. **p < 0.01; ***p < 0.001; **** p < 0.0001. (D) Body temperature was measured 0, 2, 8 and 24 hours following LPS injection. (E) Bar graph represents change of temperature 24 hours after LPS injection. Data are representative of a single experiment. Two-way ANOVA followed by Turkey post-hoc correction was used to compare body temperature between RR-LPS and RW-LPS groups at different times following LPS injection. One-way ANOVA followed by Turkey post-hoc correction was used to compare the mean temperature among three groups. *p < 0.05; **p < 0.01; ****p < 0.0001.
Under-acylated LPS triggers a mitigated innate immune response in mice
To assess whether the biochemical diversity in RR and RW-derived LPS affects the host innate immune response, we measured the expression levels of pro-inflammatory cytokine (TNF-α, G-CSF, IL-6, IL-10, IFN-β, RANTES/CCL5 and IFN-γ) using an ELISA assay. LPS (20 mg/Kg body weight) was injected intraperitoneally in female BALB/c mice (n = 6 per group) and blood samples were collected both every 2 hours for a period of 8 hours and at 24 hours post injection. Results showed a much stronger increase in TNF-α, G-CSF and IL-6 levels in blood serum of mice stimulated with RW-LPS compared to those stimulated with RR-LPS (Figures 7A–C).
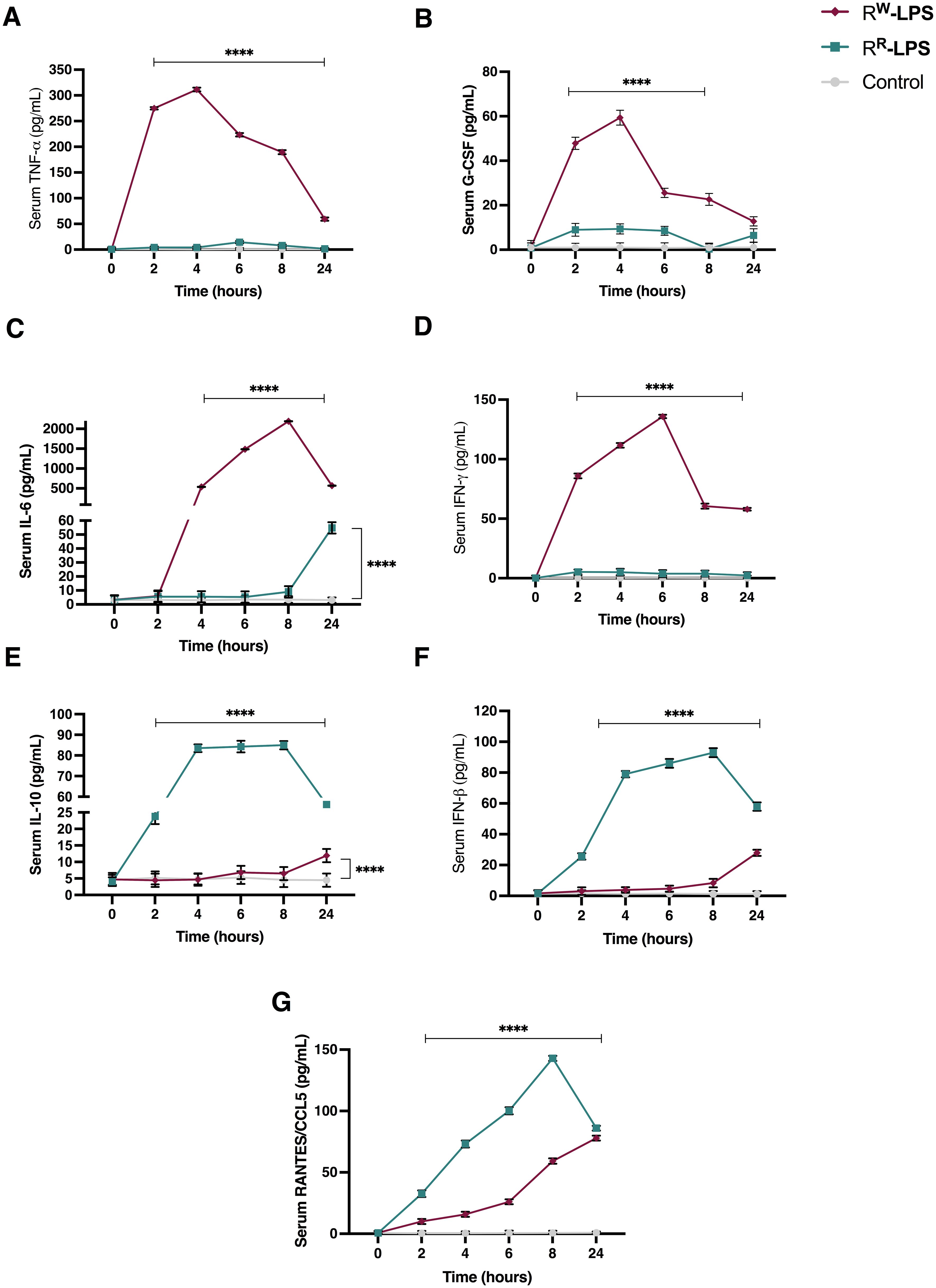
Figure 7. Cytokine levels in serum of BALB/c mice stimulated with RR or RW-LPS. Mice (n = 6 per group) were injected intraperitoneally with 20 mg/Kg of RR or RW-LPS dissolved in PBS solution. Mice in the control group (Control) were injected intraperiteonally with the same volume of PBS solution. Serum was collected every 2 hours for 8 hours and 24 hours following injection. The cytokines: (A) TNF-alpha; (B) G-CSF; (C) IL-6; (D) IFN-gamma; (E) IL-10; (F) IFN-beta and (G) RANTES/CCL5 were analyzed by ELISA. Data are representative of two independent experiments. Two-way ANOVA followed by Turkey post-hoc correction was used to compare cytokine levels among the three groups (control, RR- and RW-LPS groups). ****p < 0.0001.
TNF-α and G-CSF increased rapidly during the first hours of stimulation and reached their highest level 4 hours post-stimulation, then started declining (Figures 7A, B). However, their concentration remained higher than the control group or mice stimulated with RR-LPS (Figures 7A, B). INF-γ and IL-6, instead, were secreted later than TNF-α and G-CSF. The highest levels of these cytokines were observed 6- and 8-hours post-stimulation, respectively and, started decreasing after 24 hours of stimulation (Figures 7C, D). As expected, increased levels of IL-10 were detected at 24 hours, compared to control group (Figure 7E). These results show that RW-LPS initiates sooner an acute inflammatory response via the MyD88-dependent pathway (64), aimed at controlling the injury, as indicated by IL-10 up-regulation at 24 hours after LPS stimulation (Figure 7E). On the contrary, RR-LPS induced a milder inflammatory response. As illustrated in Figure 7, the secretion of the pro-inflammatory cytokines involved in the acute phase response (TNF-α and IL-6) was unaltered during the first hours post-LPS stimulation, compared to the control group (Figures 7A, C), while RANTES, IFN-β and IL-10 were found increased compared to both control group and mice injected with RW-LPS (Figures 7E–G). However, IL-6 showed an increase at 24 hours (Figure 7C). These data indicate the capability of the RR-LPS to selectively engage the TRIF-dependent pathway, while resulting a weaker elicitor of the MyD88-dependent pathway (65–67).
Taken together, these results suggest a different capacity of RR and RW-LPS to stimulate the host immune response and a reduced toxicity of the RR-LPS.
Under-acylated LPS stimulates a more efficient and safer antibody response
As a powerful antigen, LPS plays a pivotal role in immunological processes, stimulating antibody production (68). However, due to the Lipid A reactogenicity, LPS – in its native form - is not suitable as vaccine (69). Instead, the low toxicity of the RR-LPS (Figure 7) – resulting from the deacylation of Lipid A (Figure 2) – could make it safe enough to develop antimicrobial vaccines.
To investigate the protective role of the RR-LPS against S. Rissen infection, the immunization assay was performed. Therefore, we first explored the concentration of the whole LPS structure necessary for mice immunization, by evaluating the hemolytic effect (Supplementary Figure S6) of a wide range of doses (spanning from 1 to 500 μg). Results demonstrated that the minimum concentration of RR-LPS showing hemolytic effect < 8% was 10 μg. Thus, this concentration was selected to assess RR-LPS as a potential vaccine candidate.
To evaluate the level of the antibody response, two groups of BALB/c mice (6 mice per group) were immunized intraperitoneally with RSA-conjugated RR-LPS or RW-LPS (10 μg/100 μL PBS) twice, at an interval of two weeks, while a third group received an intraperitoneal (i.p.) dose of PBS (control group). The secondary booster was administrated seven days after the primary one. Mice immunized with RR-LPS exhibited higher levels of IgG and IgM (IgM > IgG) than those immunized with RW-LPS (Figure 8A). Of note, 7 days after the last immunization, the IgG response prevailed on the IgM one, with a higher extent in mice immunized with RR-LPS compared to those immunized with RW-LPS (Figure 8B). This result reflects the capability of RR-LPS to selectively engage the TLR4-TRIF-dependent pathway compared to RW-LPS (70). We also analyzed the IgA titers, as the primary antibody produced in the intestinal mucosa (71), site of Salmonella infection. Significant LPS-specific IgA titers were detected in feces of mice immunized with RR-LPS compared to both RW-LPS immunized mice and control ones (Figure 8C). Additionally, similar to IgM, IgA levels were found to decrease after the boosting doses (Figure 8D).
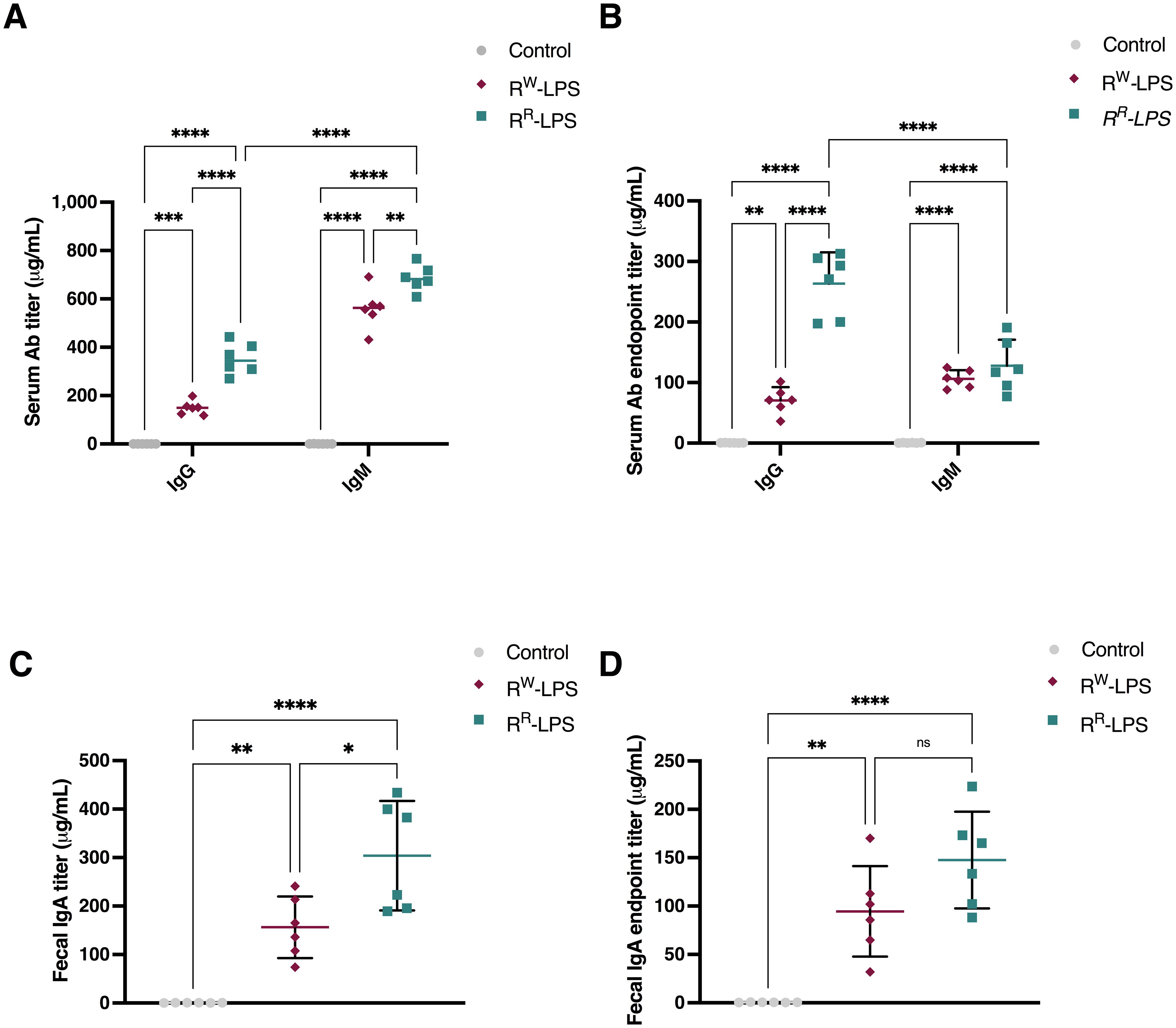
Figure 8. RR-LPS favors antibody response. (A, C) Total IgM, IgG and IgA antibody response elicited after immunization with RR-LPS or RW-LPS. Results are represented as mean titers (μg/mL) in serum or feces ± SD from female BALB/c mice (n = 6 per group) immunized intraperitoneally three times with: 1) RR-LPS (10 μg/100 μL PBS, teal green); 2) RW-LPS (10 μg/100 μL PBS, amaranth red) or 3) vehicle (100 μL PBS, grey). Graph represents mean ± SD of values from individual mice (dots). Data are representative of a single experiment. Two-way ANOVA followed by Turkey post-hoc correction was used to compare total antibody levels among three groups. * p <0.05; **p <0.01; ****p <0.0001. (B, D) IgM, IgG and IgA end-point anti-RR-LPS or RW-LPS antibody titers. Results represent the antibody response elicited on day 7 after last immunization and are represented as mean titers (μg/mL) in serum or feces ± SD. Data are representative of a single experiment. Two-way ANOVA followed by Turkey post-hoc correction was used to compare total antibody levels. ** p <0.01; ****p <0.0001. ns, not statistically significant.
These results clearly suggest RR-LPS as a more effective and safer immunostimulant than the parent RW-LPS, as it retains immunostimulatory properties while lacking most of its toxicity.
Under-acylated LPS protects mice against Salmonella Rissen infection
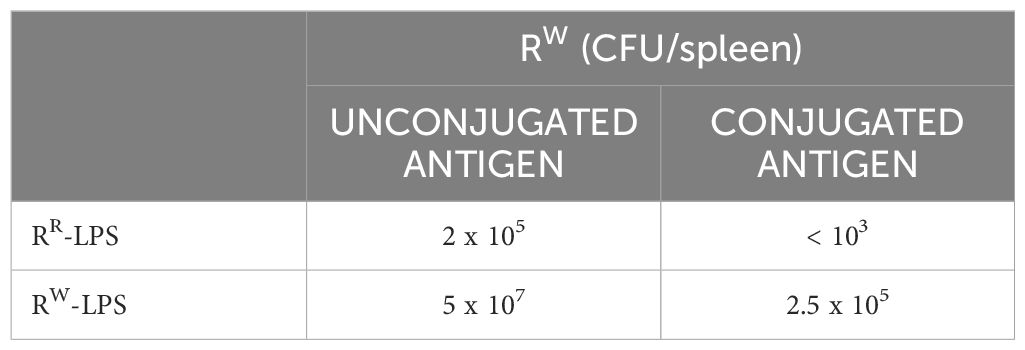
To evaluate the protective efficacy of the RR-LPS induced immunity against S. Rissen infection, all mice were infected with a bacteremia-inducing dose of the RW strain (109 CFU/animal) two weeks after the last immunization. The bacterial count in the splenic tissue was measured on day 4 post infection. Results revealed a significant reduction in bacterial burden in the spleens of mice immunized with RR-LPS, compared to both control mice and mice immunized with RW-LPS (Table 1).
Conversely, mice that received unconjugated RR-LPS exhibited only a modest reduction in bacterial burden. Although bacterial burden was still higher than in those receiving PBS or a single dose of RW-LPS, it was not lethal (Table 1).
Collectively, these findings suggest that LPS-specific antibodies significantly contribute to immune protection against S. Rissen infection, as confirmed by serum bactericidal activity (Figure 9) and highlight RR-LPS as a promising adjuvant for the development of vaccines against Salmonella infections.
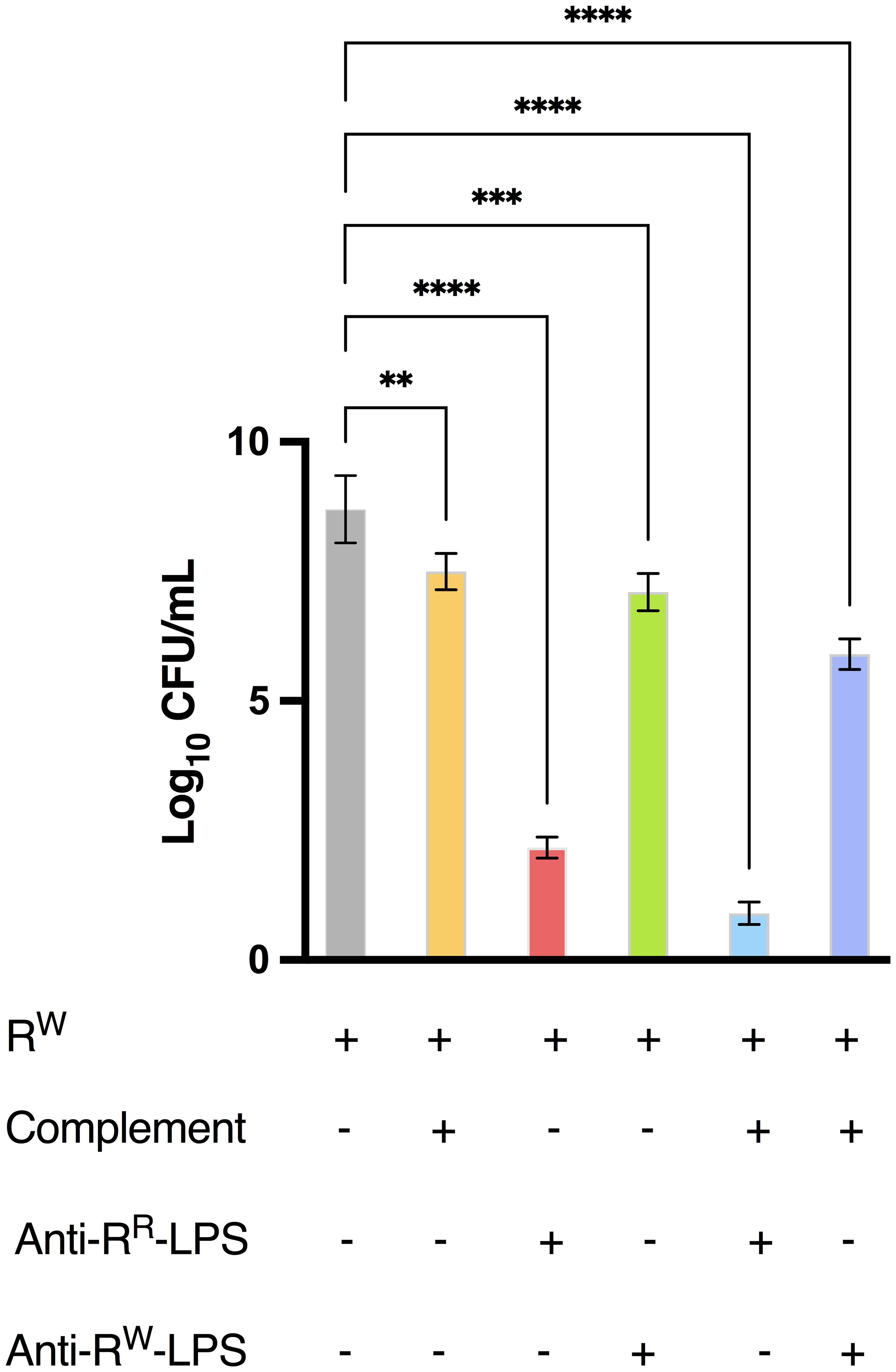
Figure 9. Antibodies from mice immunized with RR-LPS display bactericidal activity against WT S. Rissen. The bacteriophage-sensitive strain of S. Rissen (RW, 1 x 106 CFU/mL) was incubated at 37°C for 3 hours alone, with purified antibodies derived from mice immunized with RR- or RW-LPS, with complement or with both purified antibodies and complement. Data are represented as Log10 of CFU/mL (OD600 = 1 = 5 x 108 CFU/mL) and represent means ± SD of three independent experiments, each performed in triplicate. One-way ANOVA followed by Turkey post-hoc correction was used to compare Log10 CFU/mL among different experimental conditions. **p < 0.01; ***p < 0.001; **** p < 0.0001.
Under-acylated LPS-specific antibodies mediate cross-reactivity
Salmonella species share a high degree of homology in genes encoding cell-surface molecules, including LPS, a major target of antibodies (20). As a result, immunization with LPS from one Salmonella serovar could confer immunity against related serovars.
To explore the potential of anti-RR-LPS antibodies to confer cross-protective immunity to closely related Salmonella serovars, mice were intraperitoneally immunized at two-week intervals with 100 μL of RR-LPS (10 μg) or vehicle (sterile PBS). Four days after the final immunization, blood samples were collected, and serum was separated by centrifugation. Serum samples were then used to test cross-reactivity between anti-RR-LPS antibodies and LPSs from different Salmonella serovars through an agglutination test. As shown in Supplementary Table 4, serum samples (undiluted or diluted 1:50 and 1:100) exhibited variable agglutination with different selected Salmonella strains (S. Infantis, S. Newport and S. Choleresius). Specifically, undiluted serum showed strong agglutination with S. Choleraesuis and S. Infantis, which are both part of the S. Rissen serogroup C1. When diluted 1:50 or 1:100, serum also agglutinated the above Salmonella serovars, although the reaction required more time than the undiluted serum. Remarkably, a late agglutination activity of undiluted serum was observed against S. Newport, expressing O:6,8 antigens (C2 serogroup).
The strong interaction of anti-RR-LPS antibodies with S. Choleraesuis and S. Infantis and, to a lesser extent with S. Newport, suggests the O-antigenic determinants O:6 and 7 as their major target. These results indicate that anti-RR-LPS antibodies may confer cross-protection against related Salmonella serovars by targeting shared O-antigens.
Under-acylated LPS-specific antibodies mediate host protection by targeting the O-antigen
The O-antigen portion of LPS is widely recognized as the key target for immune defense against bacterial infections.
To further determine whether O-antigen is the primary antigenic determinant recognized by anti-RR-LPS antibodies, an indirect ELISA assay was performed. Serum samples from mice immunized with RR-LPS were tested for reactivity against both the O-antigen and LPS from the RR and RW strains. Results revealed that anti-RR-LPS antibodies interacted with both LPS and O-antigen from the two S. Rissen strains (Supplementary Table 5). Specifically, the antibodies showed moderately stronger reactivity with LPS and O-antigen from the RR strain than with those from the RW strain. Notably, LPS exhibited slightly stronger reactivity than the purified O-antigen (p < 0.05, Supplementary Table 5).
These results demonstrate that O-antigen is the primary RR-LPS epitope responsible for modulating the antibody-mediated host protection, supporting our previous findings (Supplementary Table 4). Furthermore, they suggest that other, poorly investigated portions of the LPS molecule may also contribute to the host defense and antibody reactivity.
Discussion
In this study, we investigated the protective effect of an LPS-based vaccine against S. Rissen in BALB/c mice. LPS is a key immunogenic component of Gram-negative bacteria. It stimulates the host humoral immune response and provides protection against pathogens (72, 73). However, LPS also plays an essential role in bacterial virulence through its endotoxic component (Lipid A), which raises concerns about its safety as “subunit” vaccine in its native form.
Several efforts have been made to attenuate the Lipid A toxicity while preserving the LPS immunostimulatory properties. Mutations in the msbB and PagP genes, both involved in the Lipid A acylation pattern, have been shown to reduce the capacity of bacteria to induce a pro-inflammatory response in the host (22, 74). As a result, msbB and PagP mutant strains are considered promising candidates for vaccine development, particularly in the context of outer membrane vesicle (OMV)-based vaccines (75). However, despite this, Salmonella msbB mutants display growth defects and increased sensitivity to environmental stressors such as physiological CO2 levels and low pH (76, 77), limiting their survival and complicating large-scale vaccine production.
In this study, we propose - for the first time - phage resistance as a promising biological tool for producing under-acylated variants of LPS, overcoming the limitations mentioned above.
Phage-resistance, as a naturally occurring selection mechanism, bypasses the need for microbial genetic manipulation, thus avoiding challenging related to the stability of mutations and the maintenance of the desired phenotypic traits. Moreover, by enhancing bacterial fitness, phage-resistance promotes bacterial survival under stress conditions, offering a cost-effective and sustainable alternative to conventional genetic, chemical and enzymatic methods.
To explore the role of prophages in remodeling Lipid A molecules and modulating S. Rissen virulence, we first examined the presence of phage genetic elements in the RR and RW genomes. Both strains contained phage genome elements. However, RR acquired the superinfection exclusion gene (SieB), which confers a competitive advantage increasing bacterial fitness by inhibiting further phage infections (78) (Supplementary Figure S1; Figure 1). This superinfection resistance likely arises from alterations in cell membrane elements, such as the LPS and its endotoxic portion (30). Modifications to the LPS structure may occur at each step of its biosynthetic process, as well as from additional enzymes (79). We recently showed that RR expresses higher levels of the LpxR gene than RW (26). LpxR encodes a deacylase responsible for removing acyl chains from Lipid A. Therefore, we hypothesized the capability of RR of producing deacylated Lipid A species.
In the present study, we validated the above hypothesis. Mass spectrometry analysis revealed that, unlike the RW strain, RR produces a higher abundance of two alternative variants of Lipid A as the penta-acylated and the tetra-acylated form (Figure 2). Both these two structures are less stimulatory for TLR4 and elicit a mitigated innate immune response. These results demonstrate that the acquisition of the SieB gene, which is associated with the distinctive phage-resistant phenotype of RR, improves bacterial fitness and provides virulence factors (LpxR) which attenuate the innate immune response in the eukaryotic host.
Metabolic changes in bacteria often influence the production of virulence factors and contribute to bacterial pathogenicity (80). Short chain fatty acids (SCFAs; propionate, acetate and butyrate) regulate the expression of genes in Salmonella pathogenicity island 1 (SPI1, 81). Specifically, acetate promotes SPI1 genes expression, while butyrate suppresses genes like hilD, which plays transcriptional regulatory roles (81). Notably, hilD also activates LpxR (82). In line with these findings, RR uptook from the growth medium more acetate than RW (Supplementary Figure S5), suggesting that phage-resistance enhances the expression of virulence factors like LpxR, which regulate LPS structure and modulate the host innate immune response. This result is confirmed by our previously published data which demonstrate up-regulation of the LpxR gene in AGS cells infected by the RR strain (26). Additionally, RR implemented the extracellular concentration of γ-aminoisobutyric acid (GABA), glucose and pyruvate compared to RW (Supplementary Figure S5). GABA improves bacterial survival in acidic environments and supports colonization of gastro-intestinal tract (83). Similarly, accumulation of glucose in the growth medium, together with pyruvate production from acetate, confer advantages to bacteria, preserving them from acidic environments (84). These findings suggest that phage Ф1 provides RR with an adaptive advantage by enhancing bacterial fitness.
As living organisms, bacteria ordinarily require energy to support their physiological functions. However, under particular conditions - such as the acquisition of phage-resistance - bacteria demand higher levels of energy. In agreement with this requirement, RR uptook more hypoxanthine from the growth medium than RW (Supplementary Figure S5). As an intermediate of the purine biosynthetic pathway, hypoxanthine consumption increases the ATP production to provide energy for the more metabolically active RR cells. RR also uptook more amino acids, including arginine, lysine, isoleucine, leucine, proline and phenylalanine which serve as effective source of carbon and nitrogen (Supplementary Figure S5, 85). Amino acids contribute to ammonium production, which is vital for synthetizing macromolecules through glutamate biosynthesis (86). Consistent with this evidence, RR released glutamate into the growth medium (Supplementary Figure S5), which also participates in glutathione biosynthesis – a key antioxidant molecule that counteracts reactive oxygen species (ROS) produced by the host immune system (87). Higher levels of methionine - the major precursor of glutathione (88) - were detected in the growth medium of RR (Supplementary Figure S5), aligning with our previous results showing increased expression of the SodC1 and gogB genes in cells infected by the RR strain (26). SodC1 is a superoxide dismutase which protects bacteria from the oxidative burst induced by the host immune cells, while gogB attenuates both the inflammatory response and ROS production (28). Taken together, these results suggest that phage-resistance, at least in this case, enhances bacterial fitness and reduces bacterial effectiveness in inducing cell host damage.
These results were confirmed in vivo. RR, in fact, curbs the mortality of mice. The highest tested concentration of RR (109 CFU/animal) resulted in 33% mortality in mice, compared to 100% mortality in those exposed to the same concentration of RW (Figure 5). This reduced mortality may reflect under-acylation of Lipid A and consequently reduced activation of the innate immune response.
Under-acylated Lipid A molecules, in fact, compete with native species of Lipid A (hexa-acylated forms) and inhibit the hexa-acylated Lipid A-MD2 complex functions, thus inducing a deficient TLR4 signaling and a reduced secretion of pro-inflammatory cytokines (58).
As expected, mice stimulated with RR-LPS produced lower levels of pro-inflammatory cytokines, compared to those stimulated with the RW-LPS (Figure 7). They showed lower concentrations of the MyD88-dependent cytokines TNF-α, IL-6 and G-CSF (Figures 7A–C) – all involved in the early stages of infection – thus suggesting the inability of RR in mounting an immediate inflammatory response. Importantly, RR-LPS suppressed the IFN-γ production (Figure 7D), which plays a key role in the pathogenesis of septic shock (89), while increased the production of TRIF-dependent cytokines RANTES and IFN-β (Figures 7F–G), which limit inflammation and promote adaptive immunity (65, 66). Alteration in the stability of TLR4-MD-2, due to under-acylated ligands, in fact, impairs the canonical LPS-induced receptor dimerization and, as a consequence, the traditional signal transduction pathway involving both MyD88 and TRIF-pathways. These results, together with the secretion of the anti-inflammatory cytokine IL-10 (Figure 7E), explain reduced signs of endotoxemia in mice stimulated with the RR-LPS (Figure 6).
To determine the protective effect of the RR-LPS against S. Rissen in vivo, we immunized mice and evaluated the bacterial count in the splenic tissue. A significant reduction in bacterial burden (< 103) was observed in mice immunized with the RR-LPS compared to those immunized with the RW-LPS (Table 1). This protection was attributed to LPS-specific antibodies targeting the O-antigen (Supplementary Table S5; Figure 9). Both RR-LPS and RW-LPS induced an early production of IgA, with a significant higher expression in mice immunized with RR-LPS (Figure 8C).
IgA is the most important class of antibodies released in the intestinal secretion and plays an essential role in protecting the gastro-intestinal tract from Salmonella pathogens (90). Moreover, early IgA production – as in our case – represents a valuable immunological memory to polysaccharidic antigens (91, 92),. In addition, mice immunized with the RR-LPS also showed an immune response profile characterized by higher production of IgG than those immunized with the RW-LPS (Figures 8A, B). As a selective activator of the TRIF-dependent pathway, RR-LPS induces the recruitment of the adaptive immune response through the expression of type I-IFN-encoding genes and IL-10 production (Figure 7), which stimulate both maturation and differentiation of CD80+ and CD86+ dendritic cells through selection and expansion of pathogen-specific T and B cells (93). These events culminate in the T-dependent antibody production – enhanced by the peptide carrier, which enables the antigen presentation to T cell through the MHCII – thus eliciting a protective immunological memory through the antibody switching (93, 94), and explaining the higher expression of IgG in RR-LPS immunized mice. Altogether, these results give reason for attributing the host immune protection to the antibody production (Figure 9) and, hypothesizing that RR-LPS immunization could confer cross-protection against different Salmonella serovars by targeting shared O-antigens (Supplementary Table 4). However, further in-depth investigation is needed to elucidate the exact mechanism of protection and ascertain whether antibodies are the unique contributors to the RR-LPS induced immune protection.
In conclusion, this study demonstrates that under-acylated LPS molecules - derived from the bacteriophage-resistant strain of S. Rissen - induce an attenuated innate immune response, while promoting an effective adaptive immune protection. These properties make RR-LPS a potentially promising candidate for vaccine development. Moreover, this study also highlights the potential use of phages as an alternative to conventional chemical or genetic methods for producing vaccines. Nevertheless, further studies are required to provide further insight into mechanism, length and contribution of additional factors in improving vaccine potency and efficacy, and finally, the possibility to extend this approach to other bacterial infections.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The studies involving humans were approved by the Committee of Villa Betania Hospital, in accordance with the principles of good clinical practices (Declaration of Helsinki). The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study. The animal study was approved by the Italian Ministry of Health. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
PC: Data curation, Formal analysis, Validation, Writing – original draft. CM: Writing – original draft, Data curation, Formal analysis, Validation. AC: Methodology, Writing – review & editing. AG: Validation, Writing – review & editing. CF: Validation, Writing – review & editing. MMC: Methodology, Supervision, Validation, Writing – review & editing. RC: Conceptualization, Supervision, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1450600/full#supplementary-material
References
1. Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O’Brien SJ, et al. International Collaboration on Enteric Disease “Burden of Illness” Studies. The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis. (2010) 50:882–9. doi: 10.1086/650733
2. Radhakrishnan A, Als D, Mintz ED, Crump JA, Stanaway J, Breiman RF, et al. Introductory article on global burden and epidemiology of typhoid fever. Am J Trop Med Hyg. (2018) 99:4–9. doi: 10.4269/ajtmh.18-0032
3. Murugaiyan J, Anand Kumar P, Rao GS, Iskandar K, Hawser S, Hays JP, et al. Progress in alternative strategies to combat antimicrobial resistance: focus on antibiotics. Antibiotics. (2022) 11:200. doi: 10.3390/antibiotics11020200
4. Zhou A, Li J, Xu Z, Ni J, Guo J, Yao YF, et al. Whole-genome comparative and pathogenicity analysis of salmonella enterica subsp. enterica serovar rissen. G3 Genes Genomes Genet. (2020) 10:2159–70. doi: 10.1534/g3.120.401201
5. Sears KT, Galen JE, Tennant SM. Advances in the development of Salmonella-based vaccine strategies for protection against Salmonellosis in humans. J Appl Microbiol. (2021) 131:2640–58. doi: 10.1111/jam.15055
6. Birkhold M, Mwisongo A, Pollard AJ, Neuzil KM. Typhoid conjugate vaccines: advancing the research and public health agendas. J Infect Dis. (2021) 224:S781–7. doi: 10.1093/infdis/jiab449
7. MacLennan CA, Martin LB, Micoli F. Vaccines against invasive Salmonella disease: Current status and future directions. Hum Vaccines Immunother. (2014) 10:1478–93. doi: 10.4161/hv.29054
8. Kantele A, Pakkanen SH, Karttunen R, Kantele JM. Head-to-head comparison of humoral immune responses to vi capsular polysaccharide and salmonella typhi ty21a typhoid vaccines-A randomized trial. PloS One. (2013) 8:e60583. doi: 10.1371/journal.pone.0060583
9. Jossi SE, Arcuri M, Alshayea A, Persaud RR, Marcial-Juárez E, Palmieri E, et al. Vi polysaccharide and conjugated vaccines afford similar early, IgM or IgG-independent control of infection but boosting with conjugated Vi vaccines sustains the efficacy of immune responses. Front Immunol. (2023) 14:1139329. doi: 10.3389/fimmu.2023.1139329
10. De Benedetto G, Alfini R, Cescutti P, Caboni M, Lanzilao L, Necchi F, et al. Characterization of O-antigen delivered by Generalized Modules for Membrane Antigens (GMMA) vaccine candidates against nontyphoidal Salmonella. Vaccine. (2017) 35:419–26. doi: 10.1016/j.vaccine.2016.11.089
11. Mancini F, Micoli F, Necchi F, Pizza M, Berlanda Scorza F, Rossi O. GMMA-based vaccines: the known and the unknown. Front Immunol. (2021) 12:715393. doi: 10.3389/fimmu.2021.715393
12. Wang X, Quinn PJ. Endotoxins: Lipopolysaccharides of gram-negative bacteria. Subcell Biochem. (2010) 53:3–25. doi: 10.1007/978-90-481-9078-2_1
13. Raetz CRH, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. (2002) 71:615–49. doi: 10.1146/annurev.biochem.71.110601.135414
14. Cavaillon JM. André Boivin: A pioneer in endotoxin research and an amazing visionary during the birth of molecular biology. Innate Immun. (2020) 26:165–71. doi: 10.1177/1753425919842307
15. Cross AS. Hit ‘em where it hurts: gram-negative bacterial lipopolysaccharide as a vaccine target. Microbiol Mol Biol Rev. (2023) 87:e0004522. doi: 10.1128/mmbr.00045-22
16. Gorman A, Golovanov AP. Lipopolysaccharide structure and the phenomenon of low endotoxin recovery. Eur J Pharm Biopharm. (2022) 180:289–307. doi: 10.1016/j.ejpb.2022.10.006
17. Ahmad TA, El-Sayed LH, Haroun M, Hussein AA, El Ashry ESH. Development of immunization trials against Klebsiella pneumoniae. Vaccine. (2012) 30:2411–20. doi: 10.1016/j.vaccine.2011.11.027
18. Ledov VA, Golovina ME, Alkhazova BI, Lvov VL, Kovalchuk AL, Aparin PG. A pentavalent shigella flexneri LPS-based vaccine candidate is safe and immunogenic in animal models. Vaccines. (2023) 11:345. doi: 10.3390/vaccines11020345
19. Nagy G, Pál T. Lipopolysaccharide: A tool and target in enterobacterial vaccine development. Biol Chem. (2008) 389:513–20. doi: 10.1515/BC.2008.056/MACHINEREADABLECITATION/RIS
20. Ahmed A, Akhade AS, Qadri A. Accessibility of O antigens shared between salmonella serovars determines antibody-mediated cross-protection. J Immunol. (2020) 205:438–46. doi: 10.4049/jimmunol.1900624
21. Rossi O, Caboni M, Negrea A, Necchi F, Alfini R, Micoli F, et al. Toll-like receptor activation by generalized modules for membrane antigens from lipid A mutants of salmonella enterica serovars typhimurium and enteritidis. Clin Vaccine Immunol. (2016) 23:304–14. doi: 10.1128/CVI.00023-16
22. Watson PR, Benmore A, Khan SA, Jones PW, Maskell DJ, Wallis TS. Mutation of waaN reduces Salmonella enterica serovar typhimurium-induced enteritis and net secretion of type III secretion system 1-dependent proteins. Infect Immun. (2000) 68:3768–71. doi: 10.1128/IAI.68.6.3768-3771.2000
23. Wang X, Quinn PJ. Lipopolysaccharide: Biosynthetic pathway and structure modification. Prog Lipid Res. (2010) 49:97–107. doi: 10.1016/j.plipres.2009.06.002
24. Gerke C, Colucci AM, Giannelli C, Sanzone S, Vitali CG, Sollai L, et al. Production of a Shigella sonnei vaccine based on generalized modules for membrane antigens (GMMA), 1790GAHB. PloS One. (2015) 10. doi: 10.1371/journal.pone.0134478
25. Papaianni M, Contaldi F, Fulgione A, Woo SL, Casillo A, Corsaro MM, et al. Role of phage ϕ1 in two strains of Salmonella Rissen, sensitive and resistant to phage ϕ1. BMC Microbiol 2018 181. (2018) 18:1–10. doi: 10.1186/S12866-018-1360-Z
26. Capparelli R, Cuomo P, Papaianni M, Pagano C, Montone AMI, Ricciardelli A, et al. Bacteriophage-resistant salmonella rissen: an in vitro mitigated inflammatory response. Viruses. (2021) 13:2468. doi: 10.3390/V13122468
27. Castledine M, Buckling A. Critically evaluating the relative importance of phage in shaping microbial community composition. Trends Microbiol. (2024) 32:957–69. doi: 10.1016/j.tim.2024.02.014
28. Taylor VL, Fitzpatrick AD, Islam Z, Maxwell KL. The diverse impacts of phage morons on bacterial fitness and virulence. Adv Virus Res. (2019) 103:1–31. doi: 10.1016/bs.aivir.2018.08.001
29. Folimonova SY. Superinfection exclusion is an active virus-controlled function that requires a specific viral protein. J Virol. (2012) 5554:61. doi: 10.1128/jvi.00310-12
30. Wahl A, Battesti A, Ansaldi M. Prophages in Salmonella enterica: a driving force in reshaping the genome and physiology of their bacterial host? Mol Microbiol. (2019) 111:303–16. doi: 10.1111/MMI.14167
31. Lennette EH, Spaulding EH, Truant JP. Manual of clinical microbiology. Amsocmicrobiol,Washington,Dc. (1974) 970p. doi: 10.4269/ajtmh.1971.20.3.tm0200030508a
32. Glonti T PJ. In vitro techniques and measurements of phage characteristics that are important for phage therapy success. Viruses. (2022) 14:1490. doi: 10.3390/v14071490
33. Bonilla N, Rojas MI, Cruz GNF, Hung SH, Rohwer F, Barr JJ. Phage on tap-a quick and efficient protocol for the preparation of bacteriophage laboratory stocks. PeerJ. (2016) 4:e2261. doi: 10.7717/peerj.2261
34. Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genomics. (2011) 12:402. doi: 10.1186/1471-2164-12-402
35. Galanos C, Lüderitz O, Westphal O. A new method for the extraction of R lipopolysaccharides. Eur J Biochem. (1969) 9:245–9. doi: 10.1111/j.1432-1033.1969.tb00601.x
36. Laemmli UK FM. Maturation of the head of bacteriophage T4. I. DNA packaging events.e. J Mol Biol. (1973) 80:575–99. doi: 10.1016/0022-2836(73)90198-8
37. Tsai CM FC. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal Biochem. (1982) 119:115–9. doi: 10.1016/0003-2697(82)90673-X
38. Casillo A, Parrilli E, Filomena S, Lindner B, Lanzetta R, Parrilli M, et al. Structural investigation of the oligosaccharide portion isolated from the lipooligosaccharide of the permafrost psychrophile Psychrobacter arcticus 273-4. Mar Drugs. (2015) 13:4539–55. doi: 10.3390/md13074539
39. Ionel Ciucanu FK. A simple and rapid method for the permethylation of carbohydrates. Carbohydr Res. (1984) 131:209–17. doi: 10.1016/0008-6215(84)85242-8
40. Abid Y, Casillo A, Gharsallah H, Joulak I, Lanzetta R, Corsaro MM, et al. Production and structural characterization of exopolysaccharides from newly isolated probiotic lactic acid bacteria. Int J Biol Macromol. (2018) 108:719–28. doi: 10.1016/j.ijbiomac.2017.10.155
41. Carillo S, Pieretti G, Parrilli E, Tutino ML, Gemma S, Molteni M, et al. Structural investigation and biological activity of the lipooligosaccharide from the psychrophilic bacterium pseudoalteromonas haloplanktis TAB 23. Chem - A Eur J. (2011) 17:7053–60. doi: 10.1002/chem.201100579
42. Stephens M, Liao S, von der Weid PY. Ultra-purification of Lipopolysaccharides reveals species-specific signalling bias of TLR4: importance in macrophage function. Sci Rep. (2021) 11:1335. doi: 10.1038/s41598-020-79145-w
43. Masoud H. LPS-based conjugate vaccines composed of saccharide antigens of smooth-type Salmonella enteritidis and rough-type S. gallinarum 9R bound to bovine serum albumin. Scand J Infect Dis. (2007) 39:315–22. doi: 10.1080/00365540601034782
44. Papaianni M, Cuomo P, Fulgione A, Albanese D, Gallo M, Paris D, et al. Bacteriophages promote metabolic changes in bacteria biofilm. Microorganisms. (2020) 8:480. doi: 10.3390/microorganisms8040480
45. Hwang TL, Shaka AJ. Water suppression that works. Excitation sculpting using arbitrary wave-forms and pulsed-field gradients. J Magn Reson - Ser A. (1995) 112. doi: 10.1006/jmra.1995.1047
46. Bax A, Davis DG. MLEV-17-based two-dimensional homonuclear magnetization transfer spectroscopy. J Magn Reson. (1985) 65. doi: 10.1016/0022-2364(85)90018-6
47. Kay LE, Keifer P, Saarinen T. Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J Am Chem Soc. (1992) 114. doi: 10.1021/ja00052a088
48. Schleucher J, Schwendinger M, Sattler M, Schmidt P, Schedletzky O, Glaser SJ, et al. A general enhancement scheme in heteronuclear multidimensional NMR employing pulsed field gradients. J Biomol NMR. (1994) 4:301–6. doi: 10.1007/BF00175254
49. Fan TWM. Metabolite profiling by one- and two-dimensional NMR analysis of complex mixtures. Prog Nucl Magn Reson Spectrosc. (1996) 28:161–219. doi: 10.1016/0079-6565(95)01017-3
50. Greco I, Molchanova N, Holmedal E, Jenssen H, Hummel BD, Watts JL, et al. Correlation between hemolytic activity, cytotoxicity and systemic in vivo toxicity of synthetic antimicrobial peptides. Sci Rep. (2020) 10:13206. doi: 10.1038/s41598-020-69995-9
51. Ramakrishnan MA. Determination of 50% endpoint titer using a simple formula. World J Virol. (2016) 5:85–6. doi: 10.5501/wjv.v5.i2.85
52. Soromou LW, Jiang L, Wei M, Chen N, Huo M, Chu X, et al. Protection of mice against lipopolysaccharide-induced endotoxic shock by pinocembrin is correlated with regulation of cytokine secretion. J Immunotoxicol. (2014) 11:56–61. doi: 10.3109/1547691X.2013.792886
53. Verrillo M, Cuomo P, Montone AMI, Savy D, Spaccini R, Capparelli R PA. Humic substances from composted fennel residues control the inflammation induced by Helicobacter pylori infection in AGS cells. PloS One. (2023) 18:e0281631. doi: 10.1371/journal.pone.0281631
55. Capparelli R, Ventimiglia I, Longobardo L, Iannelli D. Quantification of gliadin levels to the picogram level by flow cytometry. Cytom Part A. (2005) 63:108–13. doi: 10.1002/cyto.a.20109
56. Fishman JB, Berg EA. Protein A and protein G purification of antibodies. Cold Spring Harb Protoc. (2019) 2019. doi: 10.1101/pdb.prot099143
57. Ranade K, Poteete AR. Superinfection exclusion (sieB) genes of bacteriophages P22 and lambda. J Bacteriol. (1993) 175:4712. doi: 10.1128/JB.175.15.4712-4718.1993
58. Teghanemt A, Zhang D, Levis EN, Weiss JP, Gioannini TL. Molecular basis of reduced potency of underacylated endotoxins. J Immunol. (2005) 175:4669–76. doi: 10.4049/jimmunol.175.7.4669
59. Park BS, Lee JO. Recognition of lipopolysaccharide pattern by TLR4 complexes. Exp Mol Med. (2013) 45:e66. doi: 10.1038/emm.2013.97
60. Kim HM, Park BS, Kim JI, Kim SE, Lee J, Oh SC, et al. Crystal structure of the TLR4-MD-2 complex with bound endotoxin antagonist eritoran. Cell. (2007) 130:906–17. doi: 10.1016/j.cell.2007.08.002
61. Schulz C, Hammerschmidt S. Exploitation of physiology and metabolomics to identify pneumococcal vaccine candidates. Expert Rev Vaccines. (2013) 12:1061–75. doi: 10.1586/14760584.2013.824708
62. Caroff M, Karibian D, Cavaillon JM, Haeffner-Cavaillon N. Structural and functional analyses of bacterial lipopolysaccharides. Microbes Infect. (2002) 4:915–26. doi: 10.1016/S1286-4579(02)01612-X
63. Ogawa R, Yen H, Kawasaki K, Tobe T. Activation of lpxR gene through enterohaemorrhagic Escherichia coli virulence regulators mediates lipid A modification to attenuate innate immune response. Cell Microbiol. (2018) 20. doi: 10.1111/cmi.12806
64. Watanabe S, Kumazawa Y, Inoue J. Liposomal lipopolysaccharide initiates TRIF-dependent signaling pathway independent of CD14. PloS One. (2013) 8:2–8. doi: 10.1371/journal.pone.0060078
65. Ciesielska A, Matyjek M, Kwiatkowska K. TLR4 and CD14 trafficking and its influence on LPS-induced pro-inflammatory signaling. Cell Mol Life Sci. (2021) 78:1233–61. doi: 10.1007/s00018-020-03656-y
66. Chang EY, Guo B, Doyle SE, Cheng G. Cutting edge: involvement of the type I IFN production and signaling pathway in lipopolysaccharide-induced IL-10 production. J Immunol. (2007) 178:6705–9. doi: 10.4049/jimmunol.178.11.6705
67. Lin TL, Shu CC, Chen YM, Lu JJ, Wu TS, Lai WF, et al. Like cures like: pharmacological activity of anti-inflammatory lipopolysaccharides from gut microbiome. Front Pharmacol. (2020) 11:554. doi: 10.3389/fphar.2020.00554
68. Zarri A, van der L. Biosynthetically engineered lipopolysaccharide as vaccine adjuvant. Expert Rev Vaccines. (2015) 14:861–76. doi: 10.1586/14760584.2015.1026808
69. Arenas J, Pupo E, Phielix C, David D, Zariri A, Zamyatina A, et al. Shortening the lipid A acyl chains of Bordetella pertussis enables depletion of lipopolysaccharide endotoxic activity. Vaccines. (2020) 8:594. doi: 10.3390/vaccines8040594
70. Yanagibashi T, Nagai Y, Watanabe Y, Ikutani M, Hirai Y, Takatsu K. Differential requirements of MyD88 and TRIF pathways in TLR4-mediated immune responses in murine B cells. Immunol Lett. (2015) 163:22–31. doi: 10.1016/j.imlet.2014.11.012
71. Lin R, Chen H, Shu W, Sun M, Fang L, Shi Y, et al. Clinical significance of soluble immunoglobulins A and G and their coated bacteria in feces of patients with inflammatory bowel disease. J Transl Med. (2018) 16:359. doi: 10.1186/s12967-018-1723-0
72. Singh SP, Williams YU, Klebba PE, Macchia P, Miller S. Immune recognition of porin and lipopolysaccharide epitopes of Salmonella typhimurium in mice. Microb Pathog. (2000) 28:157–67. doi: 10.1006/mpat.1999.0332
73. Zhao Y, Arce-Gorvel V, Conde-Álvarez R, Moriyon I, Gorvel JP. Vaccine development targeting lipopolysaccharide structure modification. Microbes Infect. (2018) 20:455–60. doi: 10.1016/j.micinf.2017.11.006
74. d’Hauteville H, Khan S, Maskell DJ, Kussak A, Weintraub A, Mathison J, et al. Two msbB Genes Encoding Maximal Acylation of Lipid A Are Required for Invasive Shigella flexneri to Mediate Inflammatory Rupture and Destruction of the Intestinal Epithelium. J Immunol. (2002) 168:5240–51. doi: 10.4049/jimmunol.168.10.5240
75. Micoli F, Rondini S, Alfini R, Lanzilao L, Necchi F, Negrea A, et al. Comparative immunogenicity and efficacy of equivalent outer membrane vesicle and glycoconjugate vaccines against nontyphoidal Salmonella. Proc Natl Acad Sci U.S.A. (2018) 115:10428–33. doi: 10.1073/pnas.1807655115
76. Murray SR, Bermudes D, De Felipe KS, Low KB. Extragenic suppressors of growth defects in msbB Salmonella. J Bacteriol. (2001) 183:5554–61. doi: 10.1128/JB.183.19.5554-5561.2001
77. Karsten V, Murray SR, Pike J, Troy K, Ittensohn M, Kondradzhyan M, et al. MsbB deletion confers acute sensitivity to CO2in Salmonella enterica serovar Typhimurium that can be suppressed by a loss-of-function mutation in zwf. BMC Microbiol. (2009) 9:170. doi: 10.1186/1471-2180-9-170
78. Bondy-Denomy J, Qian J, Westra ER, Buckling A, Guttman DS, Davidson AR, et al. Prophages mediate defense against phage infection through diverse mechanisms. ISME J. (2016) 10:2854–66. doi: 10.1038/ismej.2016.79
79. Simpson BW, Trent MS. Pushing the envelope: LPS modifications and their consequences. Nat Rev Microbiol. (2019) 17:403–16. doi: 10.1038/s41579-019-0201-x
80. Rudra P, Boyd JM. Metabolic control of virulence factor production in Staphylococcus aureus. Curr Opin Microbiol. (2020) 55:81–7. doi: 10.1016/j.mib.2020.03.004
81. Gantois I, Ducatelle R, Pasmans F, Haesebrouck F, Hautefort I, Thompson A, et al. Butyrate specifically down-regulates Salmonella pathogenicity island 1 gene expression. Appl Environ Microbiol. (2006) 72:946–9. doi: 10.1128/AEM.72.1.946-949.2006
82. Martínez-Flores I, Pérez-Morales D, Sánchez-Pérez M, Paredes CC, Collado-Vides J, Salgado H, et al. In silico clustering of Salmonella global gene expression data reveals novel genes co-regulated with the SPI-1 virulence genes through HilD. Sci Rep. (2016) 6:37858. doi: 10.1038/srep37858
83. De Biase D, Pennacchietti E. Glutamate decarboxylase-dependent acid resistance in orally acquired bacteria: Function, distribution and biomedical implications of the gadBC operon. Mol Microbiol. (2012) 86:779–86. doi: 10.1111/mmi.12020
84. Wu J, Li Y, Cai Z, Jin Y. Pyruvate-associated acid resistance in bacteria. Appl Environ Microbiol. (2014) 80:4108–13. doi: 10.1128/AEM.01001-14
85. Liu YK, Kuo HC, Lai CH, Chou CC. Single amino acid utilization for bacterial categorization. Sci Rep. (2020) 10:12686. doi: 10.1038/s41598-020-69686-5
86. Krysenko S WW. Polyamine and ethanolamine metabolism in bacteria as an important component of nitrogen assimilation for survival and pathogenicity. Med Sci. (2022) 10:40. doi: 10.3390/medsci10030040
87. Fu RY, Chen J, Li Y. The function of the glutathione/glutathione peroxidase system in the oxidative stress resistance systems of microbial cells. Chin J Biotechnol. (2007) 23:770–5. doi: 10.1016/S1872-2075(07)60048-X
88. Mohany NAM, Totti A, Naylor KR, Janovjak H. Microbial methionine transporters and biotechnological applications. Appl Microbiol Biotechnol. (2021) 105:3919–29. doi: 10.1007/s00253-021-11307-w
89. Varma TK, Lin CY, Toliver-Kinsky TE, Sherwood ER. Endotoxin-induced gamma interferon production: Contributing cell types and key regulatory factors. Clin Diagn Lab Immunol. (2002) 9:530–43. doi: 10.1128/CDLI.9.3.530-543.2002
90. Costello CM, Willsey GG, Richards AF, Kim J, Pizzuto MS, Jaconi S, et al. Transcytosis of igA attenuates salmonella invasion in human enteroids and intestinal organoids. Infect Immun. (2022) 90:e0004122. doi: 10.1128/iai.00041-22
91. Ledov VA, Golovina ME, Markina AA, Knirel YA, L’vov VL, Kovalchuk AL, et al. Highly homogenous tri-acylated S-LPS acts as a novel clinically applicable vaccine against Shigella flexneri 2a infection. Vaccine. (2019) 37:1062–72. doi: 10.1016/j.vaccine.2018.12.067
92. Pauli NT, Henry Dunand CJ, Wilson PC. Exploiting human memory B cell heterogeneity for improved vaccine efficacy. Front Immunol. (2011) 2:77. doi: 10.3389/fimmu.2011.00077
93. Gandhapudi SK, Chilton PM, Mitchell TC. TRIF is required for TLR4 mediated adjuvant effects on T cell clonal expansion. PloS One. (2013) 8:e56855. doi: 10.1371/journal.pone.0056855
Keywords: bacteriophages, host immune response, lipid A, salmonella infection, vaccine
Citation: Cuomo P, Medaglia C, Casillo A, Gentile A, Fruggiero C, Corsaro MM and Capparelli R (2024) Phage-resistance alters Lipid A reactogenicity: a new strategy for LPS-based conjugate vaccines against Salmonella Rissen. Front. Immunol. 15:1450600. doi: 10.3389/fimmu.2024.1450600
Received: 17 June 2024; Accepted: 22 November 2024;
Published: 11 December 2024.
Edited by:
Jiae Kim, Henry M Jackson Foundation for the Advancement of Military Medicine (HJF), United StatesReviewed by:
Ho Seong Seo, Korea Atomic Energy Research Institute (KAERI), Republic of KoreaDaniela Eletto, University of Salerno, Italy
Copyright © 2024 Cuomo, Medaglia, Casillo, Gentile, Fruggiero, Corsaro and Capparelli. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rosanna Capparelli, Y2FwcGFyZWxAdW5pbmEuaXQ=
†These authors have contributed equally to this work and share first authorship
 Paola Cuomo
Paola Cuomo Chiara Medaglia
Chiara Medaglia Angela Casillo
Angela Casillo Antonio Gentile
Antonio Gentile Carmine Fruggiero
Carmine Fruggiero Maria Michela Corsaro
Maria Michela Corsaro Rosanna Capparelli
Rosanna Capparelli