- 1Department of Biology, York University, Toronto, ON, Canada
- 2Young Researchers and Elite Club, Islamic Azad University, Shahrekord, Iran
- 3Department of bioinformatics, School of Advanced Medical Technologies, Isfahan University of Medical Sciences, Isfahan, Iran
- 4Department of Biological Sciences, University of Arkansas, Fayetteville, AR, United States
Cancer treatment has long been fraught with challenges, including drug resistance, metastasis, and recurrence, making it one of the most difficult diseases to treat effectively. Traditional therapeutic approaches often fall short due to their inability to target cancer stem cells and the complex genetic and epigenetic landscape of tumors. In recent years, cancer immunotherapy has revolutionized the field, offering new hope and viable alternatives to conventional treatments. A particularly promising area of research focuses on non-coding RNAs (ncRNAs), especially long non-coding RNAs (lncRNAs), and their role in cancer resistance and the modulation of signaling pathways. To address these challenges, we performed a comprehensive review of recent studies on lncRNAs and their impact on cancer immunotherapy. Our review highlights the crucial roles that lncRNAs play in affecting both innate and adaptive immunity, thereby influencing the outcomes of cancer treatments. Key observations from our review indicate that lncRNAs can modify the tumor immune microenvironment, enhance immune cell infiltration, and regulate cytokine production, all of which contribute to tumor growth and resistance to therapies. These insights suggest that lncRNAs could serve as potential targets for precision medicine, opening up new avenues for developing more effective cancer immunotherapies. By compiling recent research on lncRNAs across various cancers, this review aims to shed light on their mechanisms within the tumor immune microenvironment.
1 Introduction
Cancer is a complex genetic, epigenetic, and environmental disease with an expansive range of tissue, tumor, and cellular levels (1). Additionally, cancer is the second-highest cause of mortality in terms of disability-adjusted life years (DALYs), and it carries the highest clinical, social, and economic burden among human diseases (2). The rate at which cancer progresses depends on a person’s biological, immunological, genetic, and environmental background. Furthermore, the complexity of cancer increases as various gene classes are discovered, particularly tumor-suppressor genes and molecular pathways (3). Cancer cells disregard the rules of cell division by disrupting the order of the body’s cells and resuming their new path. Cancer can stimulate the formation of blood vessels to ensure a continuous supply of nutrients, attack normal tissues, and disrupt their natural processes. Eventually, the cancer cells undergo a process that renders the immune system inoperable (4).
There are many risk factors for developing cancer that fall into two main categories, namely, intrinsic risk factors and non-intrinsic risk factors. Intrinsic risk factors are defined as random errors caused by DNA replication. Then, intrinsic risk factors are classified into two major subgroups: endogenous (biological aging, genetic susceptibility, DNA repair machinery, hormones, growth factors, inflammation, etc.) and exogenous (radiations, chemical carcinogens, tumor-causing viruses, smoking, lack of exercise, nutrient imbalance, etc.) risk factors. Overall, all these risk factors are triggers for cancer (5).
Initially, cancer was considered a genetic disease due to genetic mutations associated with loss of gene function or overexpression, with the assumption that these mutations are the main factor in pathogenesis and disease progression. However, cancer follows accumulative genetic mutations associated with epigenetic changes as well as environmental factors. Many cancer-like pathological conditions can develop and progress with incorrect epigenetic changes, and evidence is emerging that highlights the critical role of epigenetics in carcinogenesis. In this way, epigenetics manages the transcription and posttranscriptional regulation of many genes. These genes control many cellular processes and functions, such as growth, metabolism, immune responses, invasion, and proliferation (6). DNA methylation, histone modification, and regulation by non-coding RNAs (ncRNAs) are the principal epigenetic mechanisms. Furthermore, ncRNA, a functional RNA molecule that transcribes but does not translate into proteins, significantly influences the expression of epigenetic genes.
Long non-coding RNAs (lncRNAs) are notable ncRNA molecules that are major regulators of the epigenetic status of the human genome (6–8). In addition to their involvement in natural physiology, diseases such as cancer are associated with the function and expression of lncRNAs. Studies have demonstrated that numerous disorders in the expression of cancer-specific lncRNAs, such as the misregulation of gene expression that contributes to tumorigenesis, exist. Consequently, the potential role of lncRNAs in regulating multiple biological functions makes them excellent candidates for cancer treatment. To wrap up, controlling the function of lncRNAs in the treatment pathway can effectively contribute to the development of cancer treatments, including cancer immunotherapy (9–13). Recent studies reveal a systematic alteration of RNA levels in cancer. To make these changes happen, the amount of mutations in the genes that code for RNA processing factors, the amount of RNA processing factors, and the type of ncRNAs (like lncRNAs, miRNAs, snRNAs, and circRNAs) all play a role. Indeed, identifying and investigating the mechanisms that are processed by these different RNA subspecies offers opportunities for cancer treatment intervention (9, 10, 13–17).
Different treatments have targeted cancers over the last few decades. Common cancer treatments include surgery, chemotherapy, radiation therapy, hormone therapy, immunotherapy, stem cell therapy, and targeted therapy, each of which varies according to the patient’s clinical parameters. Also, in this study, we discuss cancer immunotherapy in detail. High-potential cancer immunotherapy has caught the attention of the world’s advanced therapies. Then, it aims to create an antitumor effect through the immune system’s response to cancer cells, both preventively and therapeutically (12, 18, 19). Importantly, lncRNAs play a key role in regulating cancer immunity (for example, in the cancer immunity cycle) and immune cells’ transcriptional profiles. The immune system effectively controls the growth of cancer cells through the cancer immunity cycle, which includes seven steps: release of tumor antigens, antigen presentation, immune cell priming and T-cell activation, immune cell migration, immune cell infiltration, recognition, and attack by T cells (10, 11, 13, 17, 19). Then, recent emerging evidence suggests lncRNAs are involved in tumor-stroma overlap and tumor immune microenvironment (TIME) reprogramming. The TIME plays critical roles in cancer development, progression, and control. The TIME has distinct groups of myeloid cells and lymphocytes that influence cancer immune escape, immunotherapy response, and patient survival. On the other hand, understanding the molecular mechanisms and functional roles of lncRNAs in the TIME is helpful for immunotherapy, especially for improving immunotherapy for cancer (11, 12, 18, 20). In this article, we review the advances and challenges in the clinical development of cancer immunotherapy, with a focus on lncRNAs.
In this review, we explore the critical role of lncRNAs in cancer immunotherapy, with a particular focus on their influence on the tumor immune microenvironment and their potential as therapeutic targets. The manuscript is structured to provide a detailed and comprehensive review of the topic. It begins with an overview of cancer immunotherapy, covering various approaches. Following this, the paper delves into the biogenesis and function of lncRNAs, particularly their roles in cancer. It then examines the interaction between lncRNAs and the tumor immune microenvironment, detailing their effects on various immune cells. The discussion then shifts to the potential of lncRNAs as targets for cancer immunotherapy, followed by a discussion of the key findings and their implications. The paper concludes by summarizing the main points and suggesting directions for future research. The primary objective of this paper is to compile and analyze recent research on lncRNAs in various cancers, focusing on their roles within the tumor immune microenvironment, to encourage further exploration and development of precise and effective cancer immunotherapies.
2 Cancer immunotherapy
2.1 Immune checkpoint inhibitors
Immune checkpoints regulate the immune system by initiating a productive immune response against metastatic cancer cells (Table 1). Immune checkpoint inhibitors (ICIs) are possible drugs that stop inhibitory checkpoint molecules from working. This makes the immune system stronger so it can attack a wider range of cells, especially cancer cells that have been hiding from the immune system in the past. One area of intense research is on monoclonal antibodies that target cytotoxic t-lymphocyte-associated protein 4 (CTLA-4), programmed cell death protein 1 (PD-1), and programmed cell death ligand 1 (PD-L1). These antibodies can make T cells more effective at killing cancer cells, which in turn stops the growth of tumors. CTLA-4 is mostly found inside cells and is expressed on CD4+ and CD8+ T cells. It is related to the immunoglobulin-related receptor B7 family. When interacting with CD80 and CD86, CTLA-4 mainly deregulated the immune response of T cells to antigen-presenting cells. The cytoplasm abundantly contains CD28, a homologous receptor that competes with CTLA-4 for affinity binding. CD28 usually enhances the T-cell immune response. The TRIM/LAX/Rab8 pathway is used by CTLA-4 to get out of cells and move to the outer member. This helps T cells mount an immune response (21). PD-1 is another important protein that keeps the T-cell immune response in check upon binding with PD-L1 and PD-L2. PD-1 is a B7 homolog protein encoded by the CD274 gene in humans and expressed in both hematopoietic and non-hematopoietic cells. CD4+ cells express PD-1, a type 1 transmembrane receptor. When PD-1 ligation happened, protein tyrosine phosphates bound to PD-1 phosphorylation motifs. This caused the T-cell signaling molecule of the T-cell receptor (TCR) complex to lose its phosphorylation. This results in inhibition of IL-2 release, one of the major T-cell growth factors. There are several potential immune checkpoints that could serve as potential candidates for immune checkpoint inhibitors (ICIs). NcRNA-RB1 is an lncRNA expressed by the RB1 promotor. It has the potential to impair tumor-inhibiting mechanisms by inhibiting CALR expression in cytotoxic CD8+ T cells. LncRNA ROR is a stress-responsive lncRNA that turns on the TGF-β pathway. This makes more CD-133+ cells proliferate, which makes chemotherapy less effective (22).
2.2 Adoptive cell therapies
Adoptive cell therapy (ACT) is a revolution in immunotherapy, improving the natural T-cell response to tumors (Supplementary Table 1). The T-cell response against tumor-specific antigens is too weak. We surgically remove cells from ACT melanoma and culture them in vitro with IL-2 to herald T cells infiltrating the tumor. This process allows the resident T cells to get the resources against melanoma-specific antigens and differentiate into effector T cells in the culture. We harvest this pure culture of effector T cells and infuse them into the patient’s circulation. The effector T cells actively bind to residual melanoma and selectively diminish their proliferation. ACT immunotherapy employs multiple approaches, including enhancing the T-cell receptor’s affinity for a specific combination of HLA class-1 and genetically allocating peptides to the chimeric antigen receptor (CAR), engineered to exhibit high binding specificity to the defined ligands of tumor cells. In B-cell tumors, CAR-T cells, which express CAR, exhibit high specificity for the CD19 antigen. Finally, we took dendritic cells (monocytes) and cultured them with the recombinant fusion protein sipuleucel-T to treat prostate cancer that has spread (38). Dendritic cells (DC) are the most common antigen-presenting cells. There is a specific marker named lnc-DC. The lnc-DC solely determines the differentiation of DCs. Inhibition of lnc-DC affects the differentiation, proliferation, and growth of the DC and, most importantly, inhibits the DCs from activating T cells. The transcription factor STAT3 regulates lnc-DC function. Lnc-DC plays a crucial role in STAT 3’s movement into the nucleus and the maintenance of transcription activity, with STAT3 setting an example in antigen presentation (39). The balance between T-regulatory cells and T-effector cells is really important for the immunological response; lncRNAs contribute to the differentiation and activation of lymphocytes. The “CECR-7-miR429-CTL4” network system, which is made up of several regulatory genes working together, controls how lymphocytes differentiate. LncRNA CECR7 targets miR-429 to regulate CTLA-4 expression. SGK-1/JunB signaling pathway helps TH2 and Th17 cells differentiate. Other regulatory genes, like linc-serum and glucocorticoid-inducible kinase-1 (Lnc-SGK-1), do the same thing. Linc-MAF is associated with Th1 and Th2 cell differentiation and proliferation by recruiting the repressor protein enhancer of Zesta homolog (EZH2) and lysine-specific demethylase (LSD1), which helps T-cell differentiation toward the Th2 phenotype. It is evident that lncRNAs play a crucial role not only in antigen presentation but also in immune cell differentiation. These contributions range from regulating the STAT3 pathway to inducing over-maturing of decidual DCs, which in turn influences the differentiation of CD8+ cells into Th1 cells, leading to increased inflammation (40).
2.3 Monoclonal antibodies
The most significant advancement in cancer immunotherapy is the development of monoclonal antibodies (Table 2). Monoclonal antibodies aid in the diagnosis of specific tumors as well as precise cancer treatment. Figure 1 illustrates the production process of monoclonal antibodies, from vaccinating mice with cancer-specific antigens to expanding selected hybridomas for antibody generation. CD30 and CD15, expressed by tumor cells, are APC-associated markers. These markers are capable of distinguishing Hodgkin’s lymphoma from other B-cell lymphomas. Conversely, T-cell lymphomas typically display CD30 on their cell surface, serving as a target for therapeutic monoclonal antibodies. The management of Hodgkin’s lymphoma and ALCL involves the use of an anti-CD-30 monoclonal antibody (brentuximab), immunotoxin auristatin (vedotin), and a cleavable linker called cathepsin. On the cell surface, the CD30 conjugate binds to the brentuximab–vedotin conjugate. Cellular cathepsin cleaves off this conjugate upon reaching the cytoplasm. Brentuximab enters the nucleus and inhibits the further division and proliferation of lymphoma cells (41). Human monoclonal antibodies can be administered as “naked antibodies” or as conjugates with cytotoxins or radioisotopes. Naked antibodies reduce tumor growth by adopting a diverse approach, starting with inhibiting the signaling receptor and promoting the opsonization of tumor cells, phagocytosis by NK cells, and most importantly, preventing angiogenesis with antibodies specific to the cell surface of vascular endothelial growth factor (VEGF) (42). Monoclonal antibodies that target inhibitory T-cell response regulators are effective cancer therapies. Advanced melanoma effectively uses ipilimumab, a fully humanized monoclonal anti-CTLA-4 antibody. For this monoclonal antibody, the treatment plan was to stop the checkpoint inhibitor CTLA-4 from working. This stops the normal T-cell response. Ipilimumab stops CTLA-4 from binding to B7 and sending out inhibitory signals that set off the T-cell response by binding to CTLA-4. Nivolumab is an effective treatment for renal cell carcinoma, non-small cell lung cancer, and bladder cancer. Nivolumab is a human IgG4 monoclonal antibody that binds to PD-1 and halts PD-1 from engaging with PD-L1, which attenuates T cells’ antitumor response. T cells, members of the B7 family of co-stimulators, express PD-1 and CD279, which exclusively bind to PD-L1. Engaging T-cell PD-1 with PD-L1 exhausts T cells, triggering an epigenetic program of T-cell exhaustion. Nivolumab inhibits the engagement of PD-L1 in the TIME, thereby enhancing the ability of effector T cells to encounter their tumor antigens (43).
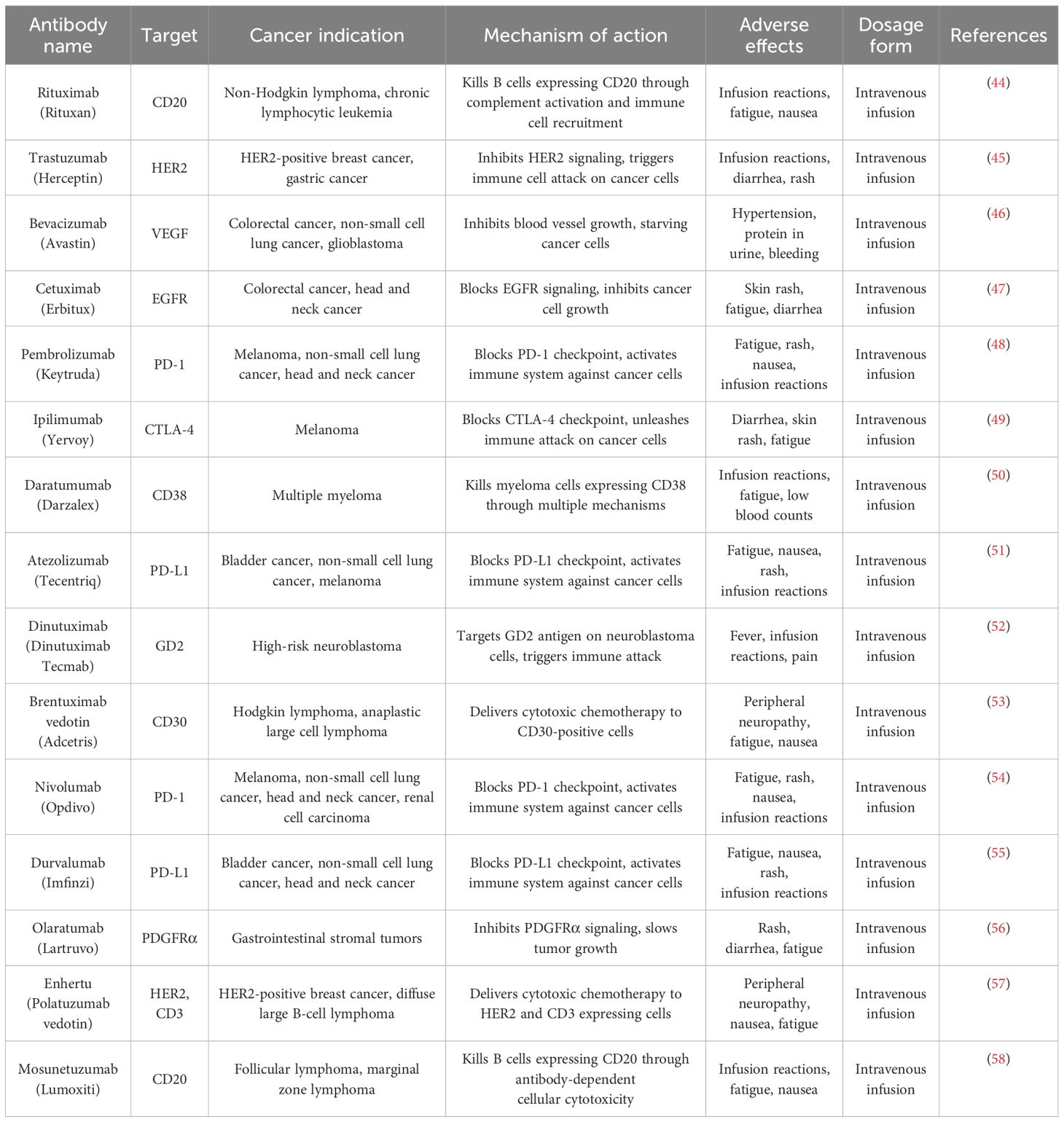
Table 2. Monoclonal antibodies for cancer therapy: targets, indications, and pharmacological profiles.
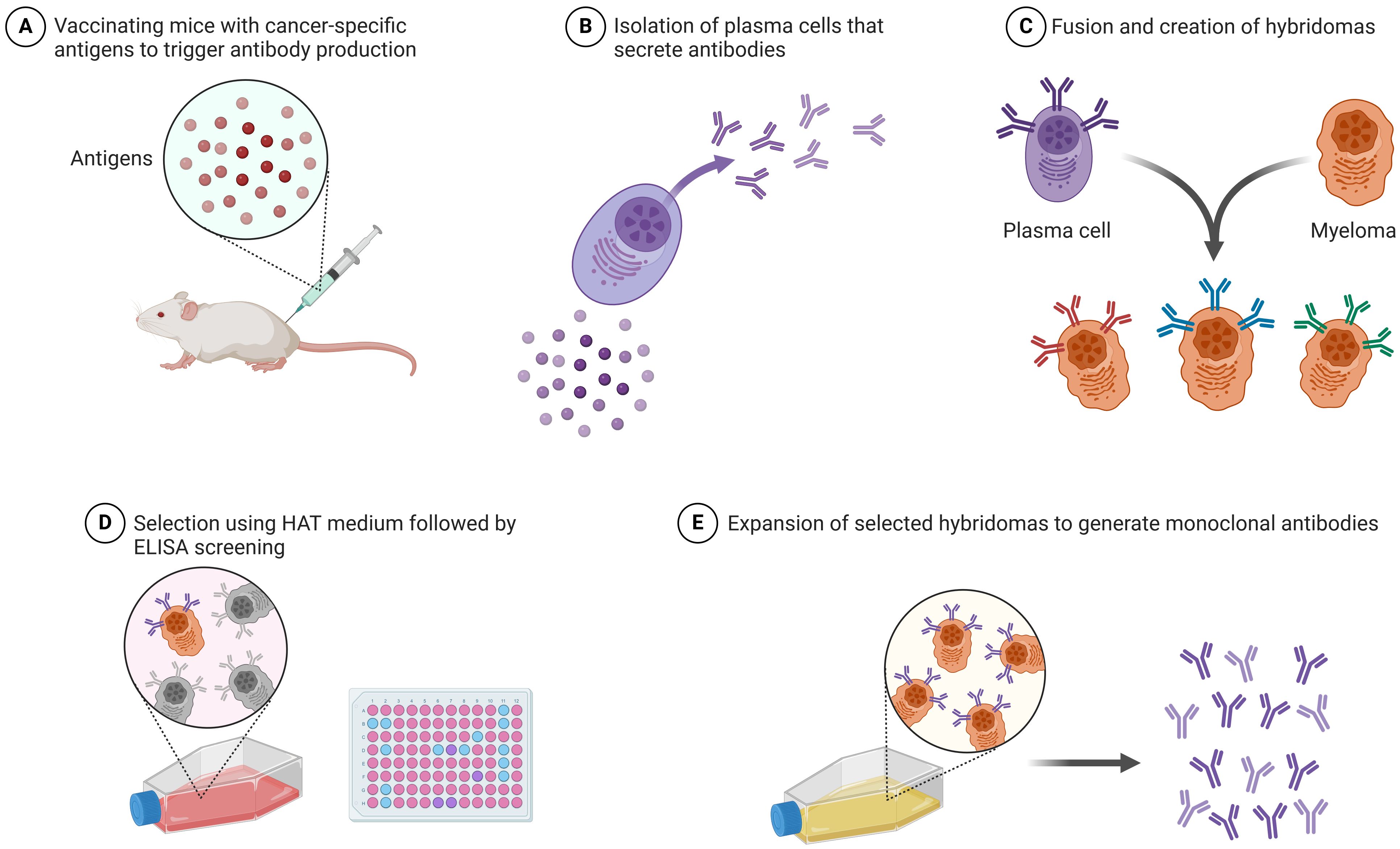
Figure 1. Production of monoclonal antibodies: step-by-step process involved in the production of monoclonal antibodies (this image was created with BioRender).
2.4 Oncolytic virus therapy
Significant progress has been made in cancer diagnosis and prevention. Using oncolytic virus infection on tumor cells has brought attention to the field of immunotherapy. Human endogenous retroviral (HERV) reactivation, oncolytic adenovirus in oral squamous cell carcinoma (OSCC) management, and oncolytic vaccina virus (OVV) are some of the most popular treatment strategies in cancer immunotherapy. LncRNA expression is correlated with HERV reactivation. HERV comprises a significant part of the human genome (approximately 8%). HERV proviral DNA is transcribed into dsRNA, which is recognized by pattern recognition receptors as a danger signal or translated into proteins to detect tumor-specific antigens (TSAs). HERV reactivation can be done either by drugs or by inducing cellular changes in the tumor cells. Reactivation of HERV comes up with some possible targets in cancer immunotherapy, such as the production of viral proteins on tumor cells recognized by our immune system, immunoglobulin viral defense responses against tumors, and the expression of mRNA transcripts, which can be used as biomarkers for prognosis. Inherent DNA hypomethylation activates, and overexpression of HERV, which is recognized by the PRRs, induces a viral mimicry state that synergistically shows an antitumor effect. A novel combinational therapy uses the same antitumor strategy based on histone deacetylase inhibitors (HDACIs) (which induce overexpression of HERV) and TLR 7/8 agonists (which induce apoptosis in susceptible HERV de-repressed tumor cells) in human ovarian cancer cells. In testicular germ cells, overexpression of HERV is usually associated with seminomas DNA hypomethylation, which in turn increases the IFN production and infiltration of CD8+ cells, which may potentiate a viral-like immune response against tumors (59). Researchers are investigating the Lentivirus Display System for monoclonal antibody screening, as well as engineered herpes simplex virus, adenovirus, Newcastle virus, and vaccinia virus as potential oncolytic viruses. The entire life cycle of the Vaccinia virus takes place in the cytoplasm, and it does not integrate with the host genome. OVV replicates fast and spreads through the blood, exerting not only a primary antitumor effect but also a metastatic cancer effect through a remote effect (60). LncRNAs are also associated with SNHG1 expression in OSCC and head and neck cancer. Oncolytic adenovirus H101 shows an antitumor effect with high SNHG1 expression in OSCC. SNHG1 expression acts as a biomarker for OSCC prognosis. Poor prognosis associated with SNHG1 lncRNA expression leads to colon cancer tumorigenesis, hepatocellular carcinoma, and the proliferation of non-small cell lung cancers. Knocking down SNHG1 suppresses cell proliferation both in vivo and in vitro (61).
2.5 Cancer vaccines
In 1980, researchers developed a cancer vaccination based on tumor cells and lysates to treat colorectal cancer using autologous cells. Using human tumor antigens as vaccines against cancer started with the discovery of melanoma-associated antigen-1 in the early 1990s (62). Since 2010, DC vaccination has proven effective in treating prostate cancer, demonstrating the potential of cancer vaccination (63). Cancer vaccines use tumor antigens to promote tumor immunity. By overcoming tumor immunological repression, the most powerful cancer vaccines will stimulate humoral and cellular immunity simultaneously. The effectiveness of any cancer vaccination in the clinic greatly depends on the antigen that is selected as its target. The optimal antigen should have immunological function, be essential for cell viability, and only be present in cancer cells with no expression in normal cells. The two main kinds of cancer antigens are tumor-associated antigens (TAAs) and tumor-specific antigens (TSAs) (64). Supplementary Table 2 presents information on cancer vaccines, distinguishing between preventive and therapeutic vaccines. One possibly beneficial vaccination platform is the nucleic acid vaccine because it elicits robust MHC-I-mediated CD8+ T-cell responses. Several antigens can be delivered concurrently using nucleic acid vaccines, promoting both humoral and cellular protection (65). Researchers primarily use DNA vaccines in clinical studies on cervical, prostate, and breast cancer, whereas melanoma, glioblastoma, and prostate cancer more frequently use mRNA vaccines (18). Small-scale trials investigating mRNA-based immunizations for cancer treatment have been underway for more than a decade, with promising outcomes. In many clinical trials, individuals with melanoma, colorectal cancer, pancreatic cancer, and other malignancies are being investigated for mRNA therapy vaccines (66). In addition, lncRNAs play a role in controlling tumor cell immune evasion and altering the TIME. Researchers can identify and select particular antigens that are crucial for the creation of vaccines using information about lncRNAs. LncRNAs control the activity of immune cells, pathways, and genes. Certain lncRNAs are involved in the regulation of immune response-related genes. Vaccines that elicit a potent and targeted immune response can be developed with the use of knowledge about the epigenetic control of immune-related genes (67). A few of the difficulties encountered in this field are caused by the immunosuppressive properties of cancer, heterogeneity within and between cancer types, inadequate immunogenicity of nucleic acid vaccines, weakened T-cell responses, subpar vaccine formulations and adjuvants, inadequate knowledge of tumor biology and its immune-suppressive TIME, and the selection of suitable tumor-specific antigens. Cancer vaccines have bright future possibilities despite these obstacles. Some of the future prospects comprise mixing different immunotherapies or conventional treatments with cancer vaccines, using personalized mRNA-based cancer vaccines to target neoantigens, combining novel medications with antigen-based therapies, and improving the whole tumor vaccine’s ability to stimulate immune responses that provide protection and improve treatment efficacy against cancers (68).
2.6 Immune system modulators
A type of immunotherapy known as immune system modulators works to strengthen the body’s defenses against cancer. They stimulate the immune system to fight cancer cells more vigorously or intelligently (69). These modulators include a variety of forms, such as BCG, cytokines, and immunomodulatory medications. Cytokines produced by white blood cells have vital roles in fighting the immune system against cancer and regulating the body’s immunological responses. Interferons and interleukins, two kinds of cytokines, are utilized in the treatment of several types of cancer. INFs boost the immune system’s capacity to fight tumor cells by stimulating certain white blood cells, such as DC and natural killer cells. They also inhibit cancer development by inducing apoptosis. The body produces more white blood cells, such as killer T cells and NK cells, when exposed to ILs. These cells can then build an immune response against cancer cells (70). Research is ongoing to better understand the immune response to cancer cells and develop drugs that can enhance the immune system’s natural abilities. This entails identifying and clarifying novel immune response-regulating molecules, researching the biology of regulatory T cells, and learning more about the particular biochemical markers that enable a patient’s T cells to target cancer cells only (71).
2.7 Epigenetics-related cancer immunotherapy
Epigenetic regulation, encompassing DNA methylation, histone modifications, chromatin remodeling, and non-coding RNAs, plays a pivotal role in gene expression and has significant implications for cancer development and treatment. In cancer, epigenetic dysregulation can lead to aberrant gene expression, contributing to tumor initiation, progression, and metastasis. These epigenetic changes critically influence the TIME, impacting both tumor cells and immune cells (72). For example, epigenetic changes in tumor cells can change the expression of tumor-associated antigens, MHC molecules, and immune checkpoint molecules like PD-L1. This can affect how immune cells recognize and kill cancer cells. Additionally, epigenetic mechanisms regulate the function and differentiation of various immune cell types, including T cells, NK cells, and MDSCs (73). Targeting these epigenetic mechanisms has emerged as a promising approach in cancer therapy, with drugs like DNA methyltransferase (DNMT) inhibitors and HDAC inhibitors showing efficacy in treating various malignancies. However, their use as single agents in solid tumors has been limited, prompting exploration of combination strategies, particularly with ICIs (74).
The intersection of epigenetics and cancer immunotherapy has become an area of intense research, as epigenetic modifications can influence the efficacy of immunotherapy by modulating tumor immunogenicity, enhancing T-cell function and persistence, and reprogramming the immunosuppressive TIME. Combining epigenetic drugs with ICIs has shown promising results in preclinical and clinical studies, aiming to overcome resistance to immunotherapy and improve patient outcomes. Emerging strategies include the development of selective HDAC inhibitors, epigenetic editing using CRISPR-Cas9, targeting epigenetic readers like BET proteins, and exploring the synergistic effects of epigenetic drugs with other therapies. Despite the promise, challenges remain, such as improving drug specificity, identifying reliable biomarkers, understanding resistance mechanisms, determining optimal dosing, and assessing long-term effects. Future research should focus on addressing these challenges and further elucidating the complex interplay between epigenetics and cancer immunology, with the goal of integrating epigenetic regulation to enhance the efficacy of existing immunotherapies and develop novel cancer treatment strategies (75, 76).
Despite the promising advances in epigenetics-related cancer immunotherapy, we still need to address several challenges. Improving the specificity of epigenetic drugs to reduce off-target effects and toxicity is a key area of focus. Identifying reliable biomarkers to predict responses to epigenetic therapies and guide patient selection is critical for optimizing treatment outcomes. Understanding and overcoming resistance mechanisms to epigenetic drugs is another important consideration. Determining the most effective treatment regimens, especially in combination therapies, requires further investigation to establish optimal dosing and scheduling. Assessing the potential long-term consequences of epigenetic modulation is also necessary to ensure the safety and efficacy of these treatments. As research in this field progresses, elucidating the complex interplay between epigenetics and cancer immunology may uncover new therapeutic targets and strategies. The integration of epigenetics and cancer immunotherapy represents a promising frontier in cancer treatment, with the potential to enhance the efficacy of existing immunotherapies and develop innovative approaches to combat cancer (77).
3 LncRNAs: biogenesis and function
LncRNAs are a class of RNA molecules that do not encode proteins but play critical roles in regulating gene expression at various levels, including chromatin modification, transcription, and posttranscriptional processing. They are typically longer than 200 nucleotides, distinguishing them from small non-coding RNAs like miRNAs and siRNAs. The biogenesis and function of lncRNAs are complex and multifaceted, involving intricate mechanisms that contribute to their diverse roles in cellular processes and, by extension, in the pathogenesis of diseases, including cancer (15, 78, 79). LncRNAs are defined by their length and lack of an open reading frame (ORF) capable of encoding a protein. Despite their non-coding nature, lncRNAs are involved in crucial regulatory functions within the cell. They are classified based on their proximity to protein-coding genes into several categories: sense, antisense, bidirectional, intronic, and intergenic lncRNAs. Each class has distinct mechanisms of action and functional implications in gene regulation, highlighting the diversity within this group of RNAs (14, 79–83).
The primary components of lncRNAs include regulatory elements that enable their interaction with DNA, RNA, and proteins. These elements facilitate the formation of secondary and tertiary structures that are essential for their function. LncRNAs can act as scaffolds, guides, decoys, or enhancers, depending on their structure and the molecules they interact with. Their ability to fold into specific structures allows them to bind to various targets with high specificity, mediating a wide range of biological processes (14, 84, 85). LncRNAs interact with DNA, RNA, and proteins to exert their regulatory functions. They can bind to DNA to form RNA–DNA hybrids, affecting the accessibility of the DNA to transcription factors and other regulatory proteins. By interacting with RNA molecules, lncRNAs can influence the splicing, stability, and translation of mRNAs. Their interaction with proteins can modulate the activity, localization, and interaction of proteins with other biomolecules. These interactions enable lncRNAs to act as key regulators of gene expression, mediating both activation and repression of gene transcription (86, 87). The impact of lncRNAs on gene expression is profound and varied. They can modulate gene expression at multiple levels, including chromatin remodeling, transcriptional control, and posttranscriptional processing. LncRNAs can recruit chromatin-modifying enzymes to specific genomic loci, leading to alterations in chromatin state and gene expression. At the transcriptional level, they can enhance or inhibit the assembly of transcriptional machinery at gene promoters (88, 89). Posttranscriptionally, lncRNAs can affect mRNA splicing, stability, and translation, further influencing gene expression outcomes. LncRNAs play a pivotal role in the regulation of gene expression, acting through diverse mechanisms to modulate the cellular transcriptome. Their ability to interact with DNA, RNA, and proteins enables them to exert broad regulatory functions, influencing gene expression at multiple levels. The complexity of lncRNA-mediated regulation highlights the intricate network of interactions that govern cellular processes, including those involved in cancer development and progression. In the context of cancer, lncRNAs have been implicated in various aspects of tumor biology, including proliferation, apoptosis, metastasis, and drug resistance. Their involvement in cancer-related pathways underscores the potential of lncRNAs as biomarkers for cancer diagnosis, prognosis, and therapeutic targets (87, 90, 91). LncRNAs are crucial for both innate and adaptive immune responses. The innate immune system serves as the initial barrier against infections, encompassing innate immune molecules and immune cells produced from myeloid cells. The adaptive immune response is a complex system that safeguards the host organism and provides protection against future encounters with the same pathogen. This response involves the activation of B and T cells. Table 3 contains a compilation of research that has reported the presence of both innate and adaptive lncRNAs.
3.1 LncRNAs in cancer
LncRNAs have emerged as key players in the regulation of cancer development and progression. Their diverse roles in modulating gene expression, cellular processes, and signaling pathways have been implicated in various aspects of cancer biology, including proliferation, apoptosis, metastasis, and drug resistance. The involvement of lncRNAs in these processes highlights their potential as biomarkers for cancer diagnosis, prognosis, and therapeutic targets. LncRNAs have been shown to modulate cellular proliferation and apoptosis, two critical processes in cancer development. For example, the lncRNA HOTAIR has been implicated in the regulation of tumor growth and metastasis by modulating the expression of tumor suppressor genes, such as PTEN. Another lncRNA, MALAT1, has been linked to the promotion of cell proliferation and inhibition of apoptosis in various cancer types. These findings underscore the potential of lncRNAs as key regulators of cellular processes in cancer. Researchers have also implicated lncRNAs in the regulation of cancer metastasis and drug resistance. For instance, studies have shown that the lncRNA MEG3 inhibits cancer cell migration and invasion by modulating the expression of genes related to metastasis. Similarly, the lncRNA HOTAIR has been associated with the development of drug resistance in cancer cells by regulating the expression of drug resistance-related genes. These findings suggest that lncRNAs play a crucial role in the progression of cancer and the development of therapy resistance (8, 10, 85, 90, 127). The involvement of lncRNAs in various cancer-related processes has led to their consideration as potential therapeutic targets and biomarkers. For example, the lncRNA HOTAIR has been proposed as a potential therapeutic target for the treatment of breast cancer, as its inhibition has been shown to reduce tumor growth and metastasis in preclinical models. Additionally, lncRNAs have been suggested as biomarkers for cancer diagnosis and prognosis, as their expression levels have been found to correlate with cancer progression and patient outcomes (127, 128). LncRNAs involved in different cancers and their various functions are listed in Supplementary Table 3.
4 LncRNAs and tumor immune microenvironment
The TIME has several constituents, including the extracellular matrix, blood vessels, immune cells, and signaling molecules. Various patterns of immune infiltration and activation are applied in various tumors by the dynamic and varied interactions between these constituents (129). Certain cancers have a high degree of immune cell infiltration and activation, meaning that a large number of immune cells are present to identify and combat the cancer cells. These tumors are referred to as immunological hot or inflammatory tumors, and they typically respond better to immunotherapy. As a result, some types of cancer do not have many or any immune cells that can kill the cancer cells. These tumors are called immunological cold or non-inflammatory tumors, and they do not respond well to immunotherapy. However, they may respond well to other treatments that boost the immune system, such as radiation, chemotherapy, or targeted therapy (129). Numerous elements affect the TIME, including the host immune system, the environment, the kind and location of the tumor, the genetic and epigenetic properties of the cancer cells, and more. Understanding the TIME and its function in tumor advancement and regression is essential for creating efficient and customized cancer therapies (130). LncRNAs have the potential to be both therapeutic targets and prognostic indicators for cancer, and they can also play significant roles in the TIME. Unlike mRNAs, the unique subcellular localizations and activities of lncRNAs are linked to their synthesis. LncRNAs can affect chromatin function, control the stability and translation of cytoplasmic mRNAs, and obstruct signaling pathways, depending on where they are located and how they specifically interact with DNA, RNA, and proteins. LncRNAs play a role in immunological responses, immune cell activation, and antigen presentation, among other immune-related processes (131).
4.1 Myeloid cells
Myeloid cells are a particular type of immune cell that are essential to the cancer TIME (132). They comprise granulocytes, macrophages, dendritic cells, and monocytes that can be produced from either monocytes or neutrophils. Myeloid cells may either stimulate or repress the immune system, which can have a beneficial or negative effect on the spread and metastasis of cancer (133). Myeloid cell accumulation is linked to poor outcomes and resistance to treatments like ICIs and chemotherapy (134). Clinical investigations targeting myeloid cells in cancer are currently in process, despite the little achievement made in huge-scale clinical studies employing myeloid cell modulators. Nonetheless, novel approaches are being developed to target tumor-resident myeloid cells with cancer immunotherapy. In both preclinical and clinical settings, several tactics aimed at the myeloid compartment consist of various strategies (132). One strategy is to modify the myeloid population around a tumor by increasing myeloid cell recruitment, differentiation, and proliferation inside the TIME. The CCL2-CCR2 pathway plays a major role in the activation of myeloid cells, such as TAMs and MDSCs, and is associated with inflammation monocytes (135).
Researchers have identified several lncRNAs as regulators of MDSC function and differentiation. MDSCs from lung cancer patients upregulate the lncRNA HOTAIR, which stimulates MDSC expansion and immunosuppressive activity by sponging miR-124. Retinal non-coding RNA3 (RNCR3) is highly expressed in MDSCs from colorectal cancer. It works with miR-185–5p to improve their immune-suppressing effects. In AML-derived MDSCs, LncRNA RUNXOR is overexpressed and contributes to their expansion by regulating RUNX1 expression (136). Key lncRNAs influencing TAM polarization and function include lncRNA NIFK-AS1, which is upregulated in M2-like TAMs and promotes their polarization by sponging miR-146a. In colorectal cancer, LINC00662 enhances M2 polarization of TAMs by regulating the miR-17–5p/STAT3 axis (136). LncRNA GAS5 blocks the miR-222–3p/PTEN axis, which stops M2 macrophage polarization and tumor metastasis (137). Research has also revealed lncRNAs that affect other myeloid cell types within the TIME. LncRNA NEAT1 regulates the differentiation and function of dendritic cells by modulating miR-3076–3p expression. By controlling the expression of CXCL1, HOTAIR affects the migration and activation of neutrophils in hepatocellular carcinoma (136, 137).
A preclinical study indicates that polarized macrophages and elevating the M1 to M2 ratio can reduce myeloid-driven immunosuppression by causing DC stimulation, CD8+ T-cell triggering, and increased phagocytosis when the CD47/SIRPα pathway is inhibited (138). Turning myeloid cells into pro-inflammatory ones by reprogramming them is another way. As an example, TLR9 appears constitutively in the endosomes of pDCs and B cells, but it is also expressed by other myeloid categories in response to immunological stimuli, such as infection. Both innate and adaptive immune cells are highly stimulated by MyD88 signaling when TLR9 identifies unmethylated cytosine-phosphate guanine (CpG) oligodeoxynucleotides (ODNs) on modified or uncommon DNA. Scientists have used this finding to design TLR9 agonists that are based on CpG ODNs. The aforementioned stimulants have shorter half-lives by nature because they are physiologic derivatives; however, by adding a nuclease-resistant phosphonothioate foundation, the half-lives of these agents have been extended from minutes to days (SD-10, CPG 7909, ISS 1018, S-540956, IMO-2055, tilsotolimod, GNKG168, and CpG-28) (139). Another strategy is to manipulate myeloid cells through cytokine activity to enhance antitumor immunity. This tactic affects the function of myeloid cells, which are cells such as TAMs and cells called dendritic cells, by using cytokines. For instance, it has been demonstrated that IL-12 accelerates the growth of dendritic cells and boosts the effectiveness of immunotherapy (140). Utilizing myeloid cells as therapeutic agents to enhance antitumor immunity, referred to as myeloid cell treatments, is also another strategy. For example, vaccines based on DCs have shown promise in enhancing immunity to tumors and enhancing the efficacy of immunotherapy (141). Table 4 provides information on lncRNAs that affect myeloid cells within the TIME.
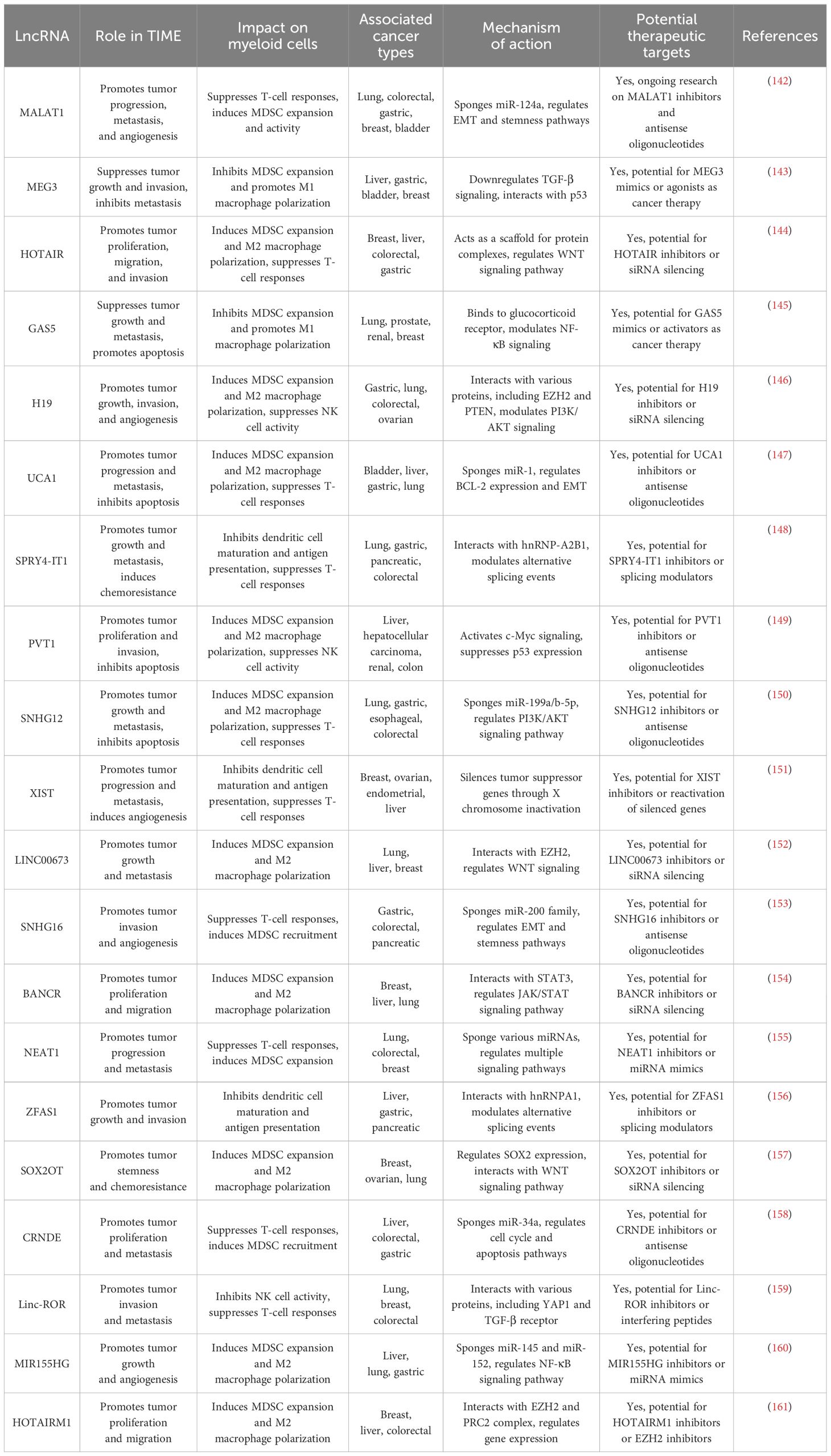
Table 4. Regulatory roles of lncRNAs in modulating myeloid cells within the tumor immune microenvironment.
4.2 Macrophages
One subset of white blood cells that are a component of the body’s innate immune system are macrophages. They are essential for the process of phagocytosis, which involves engulfing and breaking down infections, cancer cells, cellular debris, and foreign objects without proteins unique to healthy body cells on their surface. Macrophages are incredibly malleable cells that may take on a range of activation states in reaction to environmental cues. They play a role in the development of allergy and autoimmune illnesses, non-infectious inflammatory diseases, and the genesis and resolution of sterile inflammation (162). Developing efficient therapies requires an understanding of the function of macrophages in cancer immunotherapy. Macrophages are intricately involved in the immune system’s reaction to malignancies, and a high number of macrophages in tumors are frequently linked to a bad prognosis (163). Macrophages have the ability to affect carcinogenesis in two ways: either by strengthening immune cell responses against tumors or by opposing their cytotoxic effects. Clinical research and experimental models have investigated the targeting of macrophages in cancer, indicating its promise as a therapeutic approach (164). Advances in cancer nano-immunotherapies have also highlighted the function of macrophages within the TIME. An analysis demonstrated the critical function of M1 macrophages in the TIME proinflammatory processes and antitumor immunity. The importance of TAMs in medication resistance, including immunotherapies and cancer prognosis, is emphasized in the study. It also emphasizes how nanoparticles are beginning to be used to target TAMs in order to safely and effectively boost immune responses and sensitize tumors to immunotherapies (165). The distinct functions that M1 and M2 macrophages play in the development of cancer have been extensively studied. These functions are crucial to comprehending the evolution of the TIME. M2 macrophages have a tendency to promote angiogenesis, neovascularization, stromal activation, and remodeling, which can have a favorable or negative influence on cancer development and patient prognosis. M1 and M2 macrophages are both engaged in tumor-related inflammation (166). An investigation explores the functions of M1-like and M2-like macrophages in BC. It highlights that whereas M2-like macrophages are linked to angiogenesis, immunological suppression, and tissue repair, which promote tumor growth, M1-like macrophages exhibit antitumor action by causing inflammation. The study also highlights the M1/M2 macrophage ratio’s clinical significance and the possibility of transforming M2-like macrophages into antitumoral M1-like macrophages, demonstrating the adaptability of macrophages in the TIME (167). The effects of M1 and M2 macrophages on the development of lung cancer were discovered in another investigation. The study comes to the conclusion that M1 and M2 macrophages affect lung cancer cells’ ability to regulate their biological activities and gene expression differently. The M1/M2 macrophage balance in the TIME is important for both patient survival and cancer development, and the study indicates that this balance may be used as a prognostic indication for patients with lung cancer (168). It has been determined that lncRNAs, such as p21 lncRNA, play a crucial role in mammography as they regulate TAMs. This highlights the possibility of using lncRNAs as targets for future cancer immunotherapy biomarkers and therapies. In addition, it emphasizes how lncRNAs influence the TIME and regulate the immune escape of tumor cells, suggesting a bright future for lncRNA-based targeted cancer immunotherapy (169).
4.3 Myeloid-derived suppressor cells
Myeloid-derived suppressor cells (MDSCs) promote immunosuppression, stimulate tumor development, and establish a pre-metastatic niche, all of which are important factors in the course of cancer. These cells are pathologically activated neutrophils and monocytes with strong immunosuppressive activity, and they are produced from hematopoietic stem cells in the bone marrow (170). MDSCs stop T cells from activating and working in a number of ways, such as by making ligands for negative regulators of T-cell activity. It has been noted that they inhibit both innate and adaptive immunity and that those with cancer have higher concentrations of them in their bloodstreams and bone marrow. MDSCs have also been shown to make cancer cells more stem-like by increasing miR-101 and decreasing the activity of the corepressor CtBP2 (171). To develop new approaches for tumor immunotherapy, it is essential to comprehend the mechanisms underlying MDSC generation, expansion, recruitment, and activation (172). Therapeutic interventions targeting MDSCs could be possible, as recent research has confirmed that these cells suppress the immune system and boost cancer through multiple pathways (173). The variety of MDSCs, such as their monocytic (M-MDSCs), granulocytic (PMN-MDSCs), and early forms, emphasizes their distinct position within the myeloid lineage and the intricacy of their function in the advancement of cancer (170). It has also been shown that MDSCs actively take part in immune evasion, angiogenesis, the creation of pre-metastatic niches, the EMT, and additional facets of tumor growth. Their non-immunological actions enhance tumor invasion, and they contribute as well to generating an immunosuppressive environment that affects their own biology and function (174).
The following strategies are the major focus of current clinical therapies for MDSCs (1). Inhibition of MDSCs: This approach, either independently or in conjunction with other treatments, including radiation, chemotherapy, surgery, or immunotherapy, has demonstrated effectiveness in preclinical studies as well as clinical ones. Reversing immunological escape and optimizing immunological-based therapies are the goals of MDSC inactivation (175). One approach that shows promise for improving the effectiveness of cancer immunotherapy is focused on the reduction of MDSCs. In a number of cancer types, including melanoma, breast, colorectal, lung, and hematologic malignancies, high concentrations of MDSCs have been associated with poor prognoses, an increase in malignancy, and decreased therapeutic efficacy of immunotherapies (174, 175). To improve the therapeutic efficiency of cancer immunotherapy, one tactic being investigated is blocking the suppressive actions of MDSCs. It is well recognized that MDSCs suppress antigen-specific CD8+ T-cell activity, which results in a reduction in IFN-γ production (176). Another way to improve the therapeutic efficiency of cancer immunotherapy is to block the suppressive actions of MDSCs. MDSCs obstruct the efficacy of cancer immunotherapies by actively fostering an immune-tolerant TIME (177). MDSCs become stimulated in the TIME, and, through a variety of methods, such as blocking T cells’ ability to eradicate cancer, they encourage immunosuppression and tumor development. The expression of negative immune checkpoint molecules like PD-L1, a reduction of amino acids necessary for the activation of T lymphocytes, the generation of reactive oxygen species and nitric oxide, and the release of immune-suppressive cytokines like TGF-β and IL-10 are the main mechanisms by which MDSCs prevent T cells (178). The function of lncRNAs in the control of MDSCs in the TIME has been the subject of several investigations. In lung cancer patients, MALAT1 lncRNA has been demonstrated to adversely regulate MDSCs (179). In vitro and in vivo MDSC differentiation and function are both inhibited by the knockout of RNCR3lncRNA, which has been shown to be upregulated in MDSCs by both inflammatory and tumor-associated factors (20).
4.4 Neutrophils
White blood cells, known as neutrophils, are vital to the body’s immune system’s reactions. They comprise 50% to 70% of all human white blood cells and are among the most prevalent forms of granulocytes (180). Through a process known as phagocytosis, neutrophils are recognized for their capacity to absorb particles or germs. When bacterial infection, environmental exposure, or some malignancies occur, they are the first inflammatory cells to react and move toward the site of inflammation (181). Neutrophils have a variety of tasks, and their functional variability has been emphasized by a recent study. Numerous subpopulations of neutrophils have been identified so far, and studies have demonstrated that they display morphological and functional variation. This variety shows that neutrophils are a heterogeneous population that responds differently to diverse clinical situations and tissues. Moreover, data indicate the flexibility of mature neutrophils in circulation, suggesting that they can be reprogrammed in response to outside stimuli (182). Recent research has shed light on the role of neutrophils in cancer immunotherapy. Neutrophils accumulate in solid tumors, and their abundance correlates with a poor prognosis.
Neutrophils associated with poor prognosis in solid tumors and PMN-MDSCs are both important players in cancer progression, but they function differently within the tumor microenvironment. Both cell types suppress immune responses and promote tumor growth by infiltrating tumors and secreting growth factors. However, typical neutrophils can switch between tumor-inhibiting and promoting roles, whereas PMN-MDSCs are exclusively immunosuppressive, marked by specific surface molecules that regular neutrophils do not express. The primary difference between these cells is in their development and function. Neutrophils are a standard component of the immune system, potentially adopting antitumor or protumor roles (termed N1 and N2, respectively), influenced by the TIME. In contrast, PMN-MDSCs are aberrantly activated neutrophils with distinct markers like CD11b+, CD14−, CD66b+, and LOX-1, specifically arising under pathological conditions such as cancer, and are uniformly supportive of tumor growth due to their strong immunosuppressive capabilities (183).
According to research, interferon-elicited neutrophils developed an interferon gene signature, which is also observed in human patients. This signature was necessary for therapy to be successful because immunotherapy failed when neutrophils’ interferon-responsive transcription factor (IRF1) was lost. Important elements of antitumor immunity, such as IL-12, IFNγ, and DCs reliant on BATF3, were also necessary for the neutrophil response. Furthermore, they discovered that in lung cancer patients, a systemic neutrophil response induced by medication had a favorable correlation with the course of the illness (184). The treatment and prevention of cancer are being revolutionized by immune-mediated checkpoint blockade-based immunotherapies. Recent research has shown that tumor-associated neutrophils (TANs) play a critical role in regulating the TIME and the immune system’s fight against the tumor. However, these cells were mostly overlooked whereas ligand-1, the cell death program receptor, CTLA-4, and ICIs were being developed as treatment targets. Recent findings about the multifunctional variety of neutrophils in tumors have sparked a lot of debate and may open up new therapeutic avenues when it comes to the emergence of ICIs. Research also showed that a significant change in neutrophil biogenesis and function is linked to the development of cancer and has the potential to both anticipate and disrupt the immune system’s reaction to ICIs. Important characteristics of ICI resistance are also linked to neutrophil infiltration in malignancies. When taken with ICIs, TANs contribute significantly to antiangiogenic drug resistance, decreasing their therapeutic efficacy (185). LncRNAs have been studied recently in relation to cancer immunotherapy and neutrophils. An analysis of BC research, for instance, discovered that an lncRNA profile associated with neutrophil extracellular traps (NETs) might be used to predict prognosis as well as the immunological microenvironment and responsiveness to immunotherapy in BC (186). A further investigation on lung cancer found a lncRNA profile linked to NETs that connected with the TIME and anticipated patient clinical results. SIRLNT, AL365181.3, FAM83A-AS1, and AJ003147.2 are the four NET-associated lncRNAs that were discovered to be highly correlated with patient survival outcomes in the risk model (187). Working with neutrophils in immunotherapy has a number of difficulties and restrictions. Several obstacles and restrictions exist because, due to their immunosuppressive characteristics, neutrophils can be a factor in treatment resistance in a variety of solid tumors. This is a major obstacle to developing potent immunotherapies capable of overcoming this resistance (188). Also, both inflammation and immunological responses depend heavily on neutrophils. In order to effectively manipulate neutrophils for immunotherapy, it is important to strike a careful balance between maximizing their anticancer potential and preventing side effects from exaggerated inflammation or immune response. Additionally, different health and illness situations have been linked to the occurrence of many subpopulations of neutrophils. However, it is difficult to create focused medicines that can successfully modulate neutrophils since the variations in plasma membrane proteins and functional responses of neutrophils under various circumstances are not well known (181).
4.5 Lymphocytes
The production of antigen-specific lymphocytes by host organisms in response to subsequent exposure to comparable antigens or reinfection with the same pathogen is called adaptive immune responses. B cells and T cells constitute the adaptive immune system (189).
4.6 B cells
B cells are essential in the activation of humoral immune responses. B cells are also engaged in initiating T-cell immunological responses, which are required for the control of immune homeostasis (190). Tumor-infiltrating immune cells serve a crucial function in promoting or suppressing tumor growth. Tumor-infiltrating B lymphocytes (TIBs) are found throughout all stages of cancer and contribute to tumor formation as part of the TIME (191). Figure 2 depicts the dual roles of B cells in the cancer tumor microenvironment, highlighting their capacity for both antitumor activity through plasma cell differentiation and tumor cell death, and protumor activity via regulatory B cells and inflammation. TIBs have been linked to tumor immunity in a variety of ways. On the one hand, B cells have been seen invading several malignancies and producing class-switched affinity-matured anticancer antibodies in situ (192). TIBs also operate as antigen-presenting cells, promoting T-cell-mediated immune responses and reinforcing antitumor immunity (193). TIBs, on the other hand, have been identified as regulatory B cells (B-regs) and have been found to enhance cancer progression. B-regs showed their immunosuppressive properties in human and mouse studies by secreting death ligands (TRAIL, FasL) or anti-inflammatory molecules (IL-10, TGF), which can disrupt NK and T-cell responses and promote the protumoral actions of Tregs, MDSCs, and TAMs, reducing antitumor immune responses (193). As measured by the TIME, many factors influence B-cell homeostasis, including intratumoral vascularity, inflammatory degree, cytokines, hypoxia, and cellular infiltration. B cells may be split toward pro- or antitumor orientation once that balance is disrupted. Unfortunately, there have been few studies on lncRNAs and B cells in the tumor environment to date. The majority of them have focused on B-cell lymphomas triggered by lncRNA dysfunction during development and maturation, such as Burkitt lymphoma (BL), classical Hodgkin lymphoma (cHL), and diffuse large B-cell lymphoma (DLBCL) (194). A systematic analysis of the poly-adenylated transcriptome of 116 primary DLBCL specimens identified 2,632 novel lncRNAs in DLBCL, expanding the lymphoma transcriptome (195). This suggests that lncRNAs may play a role in lymphoma development. By modulating the miR-497–5p-mediated expression of the target gene PIM, LncRNASNHG16 promotes lymphoma cell proliferation and inhibits apoptosis (196). TUG1 lncRNA depletion inhibits tumor growth both in vitro and in vivo by inducing MET ubiquitination and subsequent elimination (197). Numerous lncRNAs can influence tumor development by promoting immune evasion. By regulating the PD-1/PD-L1 checkpoint, SNHG14/miR-5590–3p/ZEB1 generates a positive feedback loop that supports immune evasion and lymphoma development (198). MALAT1 promotes lymphoma immune evasion by targeting miR-195. MALAT1 silencing promotes CD8+ T-cell proliferation while inhibiting EMT-like processes via the Ras/ERK signaling pathway (199). MYC, a well-known transcription factor that is considered to be the primary driving force in the development of lymphoma, is activated and regulated by many of them (200). For example, MINCR lncRNA controls the expression of cell cycle genes such as AURKA, AURKB, and CDT1 by regulating the MYC transcriptional pathway (201). The miR-34b-5p/GLI1 axis promotes lymphomagenesis and B-cell proliferation through MYC-induced NEAT1 lncRNA (202). MYC-triggered FIRRE lncRNA promotes lymphoma progression by controlling β-catenin nuclear translocation, which activates the Wnt/-catenin pathway (203). MYC is not the only modulator that interacts with a lncRNA’s promoter region to boost expression. The MAPK/ERK signaling pathway is inactivated by P53-induced PANDA lncRNA, which reduces tumor growth in human lymphoma (204). By activating the Wnt/-catenin signaling pathway, the FOXM1-regulated overexpression of OR3A4 lncRNA controls cell death and proliferation (205). Aside from OR3A4 and FIRRE, the SMAD5-AS1 lncRNA inhibits lymphoma growth via the classic Wnt/-catenin pathway; however, APC expression is required (206). In patients with B-cell lymphoma, a range of lncRNAs are connected with prognosis and have an impact on therapeutic treatment. HULC, HOTAIR, LUNAR1, and PEG10 are lncRNAs that potentially predict a poor clinical outcome, indicate neoplastic activity, and have potential diagnostic value in DLBCL (207). MALAT1 affects DLBCL chemotherapy sensitivity by influencing autophagy-related proteins such as LC3-II/LC3-I, p62, and ATG5 (208). An artificially designed i-lncRNA was described that targeted 13 oncogenic miRNAs based on complementary sequences. In vitro and in vivo, the i-lncRNA inhibited tumor growth by consuming a significant number of oncogenic miRNAs, primarily targeting a set of regulatory genes such as PTEN, TIMP3, p38/MAPK, and c-MYC. Furthermore, the soluble Fas decoy receptor (sFas) can block Fas-ligand-induced apoptosis. FAS-AS1 lncRNA, which is regulated by EZH2-mediated histone methylation, is a new regulator of sFas expression that influences cell apoptosis in non-lymphomas Hodgkin’s by functioning as a decoy for RBM5 (117).
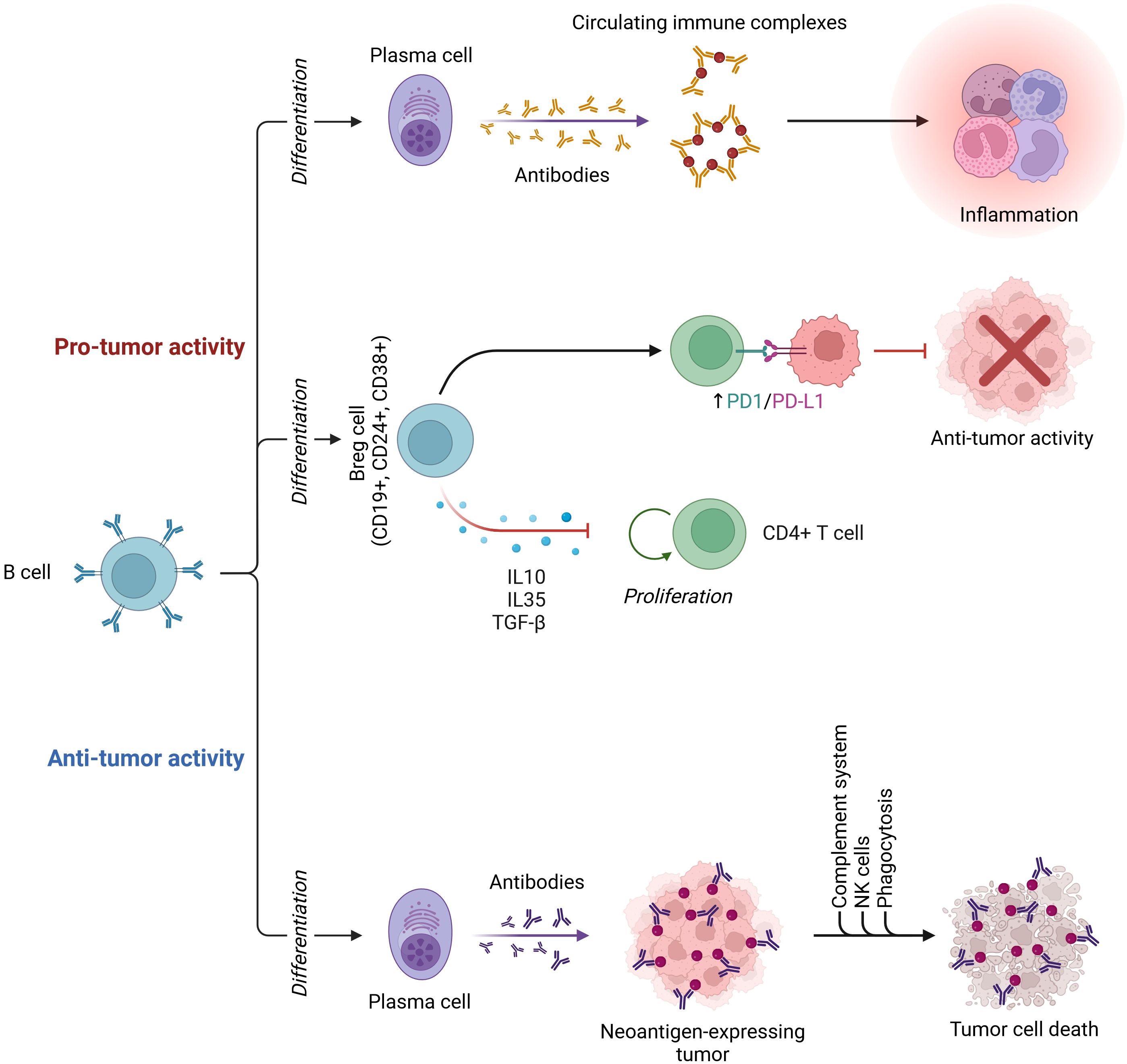
Figure 2. Antitumor and protumor activity of B-cells in the cancer tumor microenvironment: B cells can differentiate into plasma cells, which produce antibodies that target neoantigen-expressing tumors, leading to tumor cell death through the complement system, NK cells, and phagocytosis, thus exhibiting antitumor activity. Conversely, B cells can differentiate into regulatory B cells (Bregs, CD19+, CD24+, CD38+), which secrete IL-10, IL-35, and TGF-β, promoting CD4+ T-cell proliferation and upregulating PD-1/PD-L1, ultimately inhibiting antitumor activity. Additionally, plasma cells can contribute to protumor activity by producing antibodies that form circulating immune complexes, leading to inflammation (this image was created with BioRender).
4.7 T cells
T lymphocytes boost immune cell activation to tackle malignant cells. T cells also signal other immune cells to engage in immunological responses. Numerous types of T cells cause cancer cells to die and stop them from spreading. CTLs are immune cells that have CD8+ surface expression on their surface. These CTLs participate in the apoptosis of specific cells by delivering digestive enzymes or cytotoxic cytokines in granules (TNF and IFN) (209). Cancer cells have been shown to resist the effects of CTLs by rearranging cellular function via the expression of coding RNAs and lncRNAs, resulting in a change in the cellular response to the native immune system. LncRNAs have also been shown to have a role in cancer resistance to CTL activity in recent studies (88, 210). NEAT1 is a nuclear paraspeckle-localized lncRNA found in a variety of cancers (211). In vivo studies revealed that inhibiting NEAT1 reduces CD8+ T-cell death and promotes active cytolytic function via the miR-155/Tim-3 pathway, resulting in increased immunological activity (212). Ji et al. published a paper in 2018 about the involvement of Lnc-Tim-3 in the regulation of Tim-3 protein expression in HCC. According to the researchers, an increase in Lnc-Tim-3 specifically binds to Tim-3 protein and blocks its interaction with Bat-3, suppressing downstream Lck/NFAT1/AP-1 signaling, resulting in nuclear localization of Bat-3 and increased p300-dependent p53 and RelA transcriptional activation of antiapoptotic genes such as MDM2 and Bcl2, allowing exhausted T cells to survive and proliferate (213). This research suggests that Lnc-TIM-3 lncRNA can be exploited as a target for T-cell exhaustion and proliferation. Increased expression of nuclear factor B (κB)-interacting NKILA lncRNA in cytotoxic T cells has also been linked to a poor prognosis in breast and lung cancer. However, knocking down NKILA inhibited tumor growth by boosting CTL-mediated activation-induced cell death, HDAC-mediated P300 nuclear localization, and calmodulin activation after calcium influx (214). The lincRNA MAF-4 is also involved in the development of the Thl response, which is mediated by the epigenetic silencing of the transcriptional factor MAF-4, which is mediated by the recruitment of chromatin modifiers such as polycomb group proteins (PRC1 and PRC2), which facilitates the deposition of H3K27me3 (transcriptional repression) by using the catalytic component partner, primarily EZH2. Lysine-specific demethylase 1 facilitates H3K4me3 (transcriptional activation) and H3K27me3 (gene regulatory regions) in a similar way (LSD1). As a result, lincRNA-MAF-4 interacts with the transcriptional repressors LSD1 and EZH2 to allow the H3K27me3 trimethylation mark at MAF’s promoter, effectively silencing its production (126). One way cancer cells impact immune suppression is by increasing the number of immunosuppressive tumor-infiltrating lymphocytes in the TIME (215). Tregs are one of the most potent and well-studied suppressive characteristics identified in the TIME. Treg cells may potentially play a key role in cancer’s immune evasion strategies, according to mounting data. Furthermore, new data suggest that tumor-derived lncRNA is essential for Treg maintenance and differentiation (216). For example, in HCC, lnc-EGFR plays a role in distinguishing Tregs and suppressing CTLs (217). Lnc-EGFR expression in Tregs was found to be strongly related to EGFR/Foxp3 expression and tumor development but inversely related to IFN-γ expression. Despite the fact that several studies have demonstrated that FOXp3 Tregs play a role in tumor growth, a recent study discovered that the two unique populations of Tregs, FOXp3 low and FOXp3 high, play different roles in tumor promotion (119). Another study discovered the involvement of FLICR lncRNA in controlling FOXp3 expression in Treg. The activation of the conserved noncoding sequence-2 (CNS-2) enhancer by suppressing FLICR has been linked to the enhancement of Foxp3-suppressive actions in Tregs via increased IL-2 production, followed by TCR signaling. Pei et al. (2018) established the interaction of SNHG1 lncRNA with miR-448 in the regulation of IDO expression in TILs to further understand the role lncRNA and miR interplay in IDO synthesis. The study found that miR-448 inhibits IDO expression, particularly in CD4+ TILs, but not in CD4+ cells in circulating peripheral blood (218). In contrast, SNHG1 sponges miR-448, resulting in an elevation of IDO expression in HCC cells. This implies that SNHG1 might be exploited as a possible therapeutic target for improving immunotherapy by decreasing lncRNA expression. Furthermore, it was shown that increased expression of SOX5 and SOX3 in the tongue and colon cancer inhibited T-cell function via upregulating IDO1 (219). A previous study also demonstrated that IDO1-mediated immunosuppression promotes tumor growth by increasing the number of Treg and MDSC regulatory cells.
4.8 Natural killer cells
Natural killer (NK) cells are innate immune lymphocytes that play a vital role in the host’s defense against viral infection and cancer monitoring (220). There is evidence of NK cell cytotoxicity toward adjacent cells displaying oncogenic transformation-associated surface markers (221). Furthermore, NK cells improve antibody response and T-cell activation, demonstrating that they play a significant role in tumor immunotherapy (222). Zhang et al. have discovered the lncRNA expression profile in human primary lymphocyte cells. They revealed that unique NK-specific lncRNAs are involved in the formation and function of NK cells (223). They discovered that the expression of an NK-specific lncRNA, lnc-CD56, correlates with the expression of the NK cell surface marker CD56. Further investigation has revealed that lnc-CD56 may significantly enhance CD56 expression and NK cell development in human CD34+ hematopoietic progenitor cells (223). Another study found that MEG3, GAS5, and several miRNAs influence NK cell killing effectiveness in various cancer types (224). Natural cytotoxicity receptors trigger the production of the IFNG-AS1 lncRNA, and IFNG-AS1 enhances IFN secretion in human NK cells (225). As a result, IFNG-AS1 is a type I immune response modulator in general.
LncRNA expression is tightly linked to the course of NK-related cancers. In cancer patients’ NK cells, the level of GAS5 lncRNA is lower. By sponging miR-544 to target RUNX3, GAS5 enhances NK cell cytotoxicity, IFN production, and the fraction of CD107a+ NK cells, enhancing the killing function of NK cells (224). Furthermore, by modifying miR-18a, GAS5 enhances the secretion of IFN and TNF and cytotoxicity of NK cells against GC (226). Exosomes from tumors include functional lncRNAs that NK cells can pick up and use for intercellular communication. Through miR-4485–5p driven upregulation of NKp46, a crucial natural cytotoxicity receptor, IFNβ-induced exosomal linc-EPHA6–1, strengthens NK cell cytotoxicity against tumor cells and Zika virus-infected tumor cells (227). As a result, the findings from previous research imply that lncRNAs play a significant role in the control of NK cells invading the TIME.
4.9 Dendritic cells
Dendritic cells (DCs) are antigen-presenting cells that help to initiate and modulate both innate and adaptive immune responses. As a result, interest is increasing in controlling DC activity to improve cancer immunotherapy efficacy (228). Tumor-associated typical DCs (cDCs) phagocytose apoptotic cancer cell debris and transport cancer-related antigens to the draining lymph node, exposing them to naive CD4+ or CD8+ T cells, priming and activating T cells (229). Even though DCs have a high potential for anticancer immunity, the TIME causes significant challenges because it disturbs normal DC functions to avoid immune surveillance and therefore obstructs the formation of protective immune responses (230). Neoplastic cells can develop a variety of mechanisms that allow them to thrive in extreme conditions. Numerous cytokines discovered in the tumor environment, for instance, can have a direct impact on the activity of infiltrating DCs, promoting malignant progression. For example, the STAT3-related pathway can be stimulated by IL-6, IL-10, and VEGF, commonly overexpressed in the TIME, causing an immature and tolerogenic phenotype in tumor-associated dendritic cells (TADCs) and thus promoting cancer progression (231). Indeed, immunosuppressive and tolerogenic DC populations are frequently found in the TIME of aggressive malignancies. LncRNAs modulate posttranslational modifications and interact with signaling molecules in the cytoplasm to influence cellular function and differentiation (99). The Lnc-DC lncRNA (gene symbol LOC645638) is only found in human Lin MHC-II+ CD11c+ conventional DCs. DC differentiation is inhibited in vitro and in vivo when Lnc-DC is silent, and DC’s ability to stimulate T-cell activation is reduced. By preventing STAT3 from binding to and dephosphorylating SHP1, Lnc-DC binds to STAT3 in the cytoplasm and stimulates tyrosine-705 phosphorylation (99). Some studies have found that Lnc-DC promotes DC maturation and inhibits trophoblast invasion without the involvement of CD4+ T cells. The p-STAT3 inhibitor can restore lnc-DC function by mediating MMP and TIMP expression (232). In preeclampsia, Lnc-DC causes excessive maturation of decidual DCs and an increase in Th1 cells (233). In addition, the TLR9/STAT3 pathway in DCs modulates the HBV-induced immune response and cellular turnover (234). According to recent research, lncRNAs can delicately regulate the local immune system by regulating DC infiltration, differentiation, and metabolism, as well as influencing other immune cells such as T cells. By using the NLRP3 inflammasome as a molecular decoy for miR-3076–3p, NEAT1 induces a tolerogenic phenotype in DCs (235). MiRNA let-7i modulates NEAT1 expression in the nucleus by interacting with the transcription factor E2F1, which affects the H3K27ac distribution in the NEAT1 promoter. Infusions of NEAT1-depleted DCs reduced the infiltration of inflammatory cells, increased the number of Tregs, and slowed the proliferation of T cells in mouse models (235). Immune tolerance is generally induced as a result of these changes. HOTAIRM1 lncRNA is essential for myeloid development. When monocytes become DCs, their expression of HOTAIRM1 decreases (236). The level of some monocyte differentiation markers, such as B7H2 and CD14, is altered when HOTAIRM1 is knocked out. Furthermore, targeting HOTAIRM1 and HOXA1, a DC differentiation repression gene, miR-3960 acts as a competing endogenous RNA to regulate DC differentiation (236). The chemokine receptor CCR7 induces Lnc-Dpf3, which inhibits DC migration by preventing HIF-1-mediated glycolysis (237). CCR7 stimulation increased lncDpf3 by removing the m6 A modification that prevents RNA degradation. Lnc-Dpf3 knockdown promotes CCR7-mediated DC migration, which worsens inflammation and adaptive immune responses. Lnc-Dpf3 suppresses DC glycolytic metabolism and migration by preventing HIF-1-dependent transcription of the glycolytic gene LDHA (237). As a result, there is a hypothesis that decreasing lnc-Dpf3 in DCs can boost DC recruitment into the hypoxic TIME via a similar mechanism, enhancing adaptive immune responses and improving the TIME. In human colon cancer tissues, tumor-infiltrating DCs release a considerable amount of CCL5 (238). The MALAT1 lncRNA deficiency inhibits CCL5-mediated invasion and migration by lowering Snail levels (238). These results show that MALAT1/Snail signaling is required for TADC-mediated tumor development.
5 LncRNAs as potential cancer immunotherapeutic targets
Cancer comprises a diverse set of life-threatening diseases caused by abnormal and invasive cell proliferation. Cancer patients around the world are getting treatment with traditional methods, which are surgery, radiation, and cytotoxic drugs. While some patients have achieved cures, a significant number are experiencing remission as a result of either incomplete cancer cell elimination or harmful treatment side effects. The immune response is usually capable of identifying, controlling, and eliminating the cancer cells in the same manner used by the immune system for viral infection, as most of the cancer cells are byproducts of the viral infection. Nk cells and CD8+ cytotoxic T cells play a major role in the elimination of cancer cells (19). A healthy cell needs to go through malignant transformation to become a cancerous cell, which means it needs to accumulate multiple mutations and, in several genes, maintain cell division, proliferation, and survival. Tumor oncogenes encode the protein that prevents the proliferation of mutant cells. p53 is a well-known tumor oncogene that regulates apoptosis in cells with damaged DNA. Loss of p53 or substitution of p53 is the vital reason for compromising the protective function of this protein. In this review paper, we are stressing the contribution of lncRNAs as a target for cancer immunotherapy (239). The p53 protein regulates the lincRNA p-21 locus, which is responsible for gene expression both at the transcriptional and posttranscriptional levels and is associated with the RNA-binding protein HuR. LncRNA also has a role in cell apoptosis, with PANDA being a monoaxenic lncRNA. Targeting lncRNA can be a useful tool for encountering PARP-associated drug resistance, p53 reactivation, and finally tumor suppression. Oncogene suppressor lncRNA is also deemed to inhibit the PD-1/PD-L1 pathway, which is associated with immune invasion of cancer cells, to provide antitumor immunity. Targeting lncRNA could also be associated with regulating other molecular pathways such as STAT, ZEB, and P13k/Akt (19). LncRNA can be targeted in cancer immunotherapy with regard to its specificity in directing the immune response toward the targeted MDC population and function in cancer. LncRNA expression is an important regulator of several biological processes; hence, it is targeted for modulating complications other than cancer (240). TAMs have two classes: proinflammatory, antitumorigenic M1 and anti-inflammatory, pro-tumorigenic M2. LncRNA promotes M2 polarization by regulating TCF-4 and GNAS-AS1/MIR4319/NECAB3. GNAS-AS1 and XIST lncRNAs have a key role in M2 macrophage polarization in NSCLC and clinical tumor cells. In osteosarcoma, tumor progression and proliferation are associated with Lifr-AS1 lncRNA. In contrast, COX2 lncRNA inhibits M2 polarization and thus antagonizes HCC immune escape and tumor growth. In endometrial cancer, M2-macrophage is inhibited by NIFK-AS1 targeting miR-146a. In a nutshell, lncRNA is contributing as a potential biological target for cancer therapy (241). Aptamers are single-stranded oligonucleotides that can bind to a diverse range of molecular targets with high affinity and specificity (239). Designing aptamers for specific upregulated lncRNA is one of the most common strategies for lncRNA-based targeted therapy (242). For instance, knocking out NEAT1 and MALAT1 from the promoter region inhibits cancer metastasis. Aptamers can be used with mAbs or alone, which are capable of inhibiting pro-metastatic miR-214 and simultaneously upregulating anti-metastatic miR-148b, thus inhibiting metastasis (243). LncRNAs play a crucial role in a immunosuppressive TIME. A complex network of fibroblasts, endothelial cells, and adaptive and innate immune cells supports tumor cells by secreting cytokines, chemokines, growth factors, and metabolites that promote tumor growth and proliferation. Cancer-associated fibroblasts (CAFs) induce angiogenesis by upregulating CXCL12 lncRNA and downregulating tumor-suppressor miR-101. Regulatory T cells are CD4+ lymphocytes characterized by the expression of Foxp-3. Flatr lncRNA is the key regulator of Foxp-3, which is responsible for secreting chemokines (CCL1, CCL22, and CCL17), cytokines like IL-10, growth factors TGF-β, and co-inhibitory receptors PD-1 and CTLA-4. To conclude, lncRNA expression is a major player in poor prognosis and malignancies; hence, it could be a novel tool in cancer immunotherapy with high affinity and specificity toward the respective cancer-producing cells (239).
6 Discussion
LncRNAs have been identified as multifunctional regulators that play crucial roles in development, immunological regulation, cellular physiology, and the pathogenesis of various diseases, including cancer (20). Their potential as useful therapeutic targets in the therapy of cancer has been demonstrated by their diverse roles. However, unlike their RNA siblings, mRNA and miRNA, it is clear that the therapeutic targeting of lncRNAs, as well as the effective and safe delivery of therapeutic drugs, have been much neglected. A noteworthy example of the significant gaps that still exist in this developing discipline is the small number of clinical studies employing lncRNAs (20, 244). Numerous studies have previously demonstrated that many lncRNAs can function as oncogenes, and numerous others serve as tumor suppressors. Regarding protein tumor suppressors and oncogenes, this offers new opportunities for targeted cancer treatments by reinstating and boosting lost lncRNA tumor-suppressor behavior and blocking lncRNA oncogenic activity that promotes tumor development along with treatment resistance (244). LncRNAs have been found to be connected with resistance to cancer treatment advancement and oncogenesis. For instance, it has been demonstrated that the GAS5 lncRNA controls cell longevity and development, exhibits tumor suppressor function in a variety of malignancies, and regains sensitivity to chemotherapy medications. Numerous studies, like this one, highlight how oligonucleotide-based treatment may induce extremely targeted control of lncRNA expression, which has obvious applications in cancer therapy (122, 244). Nevertheless, lncRNA-based therapeutics have drawbacks, including how they transmit certain compounds to target cells and safety issues during treatment. Using lncRNA as a therapeutic target is not without its difficulties, such as issues with molecular transport and inappropriate consequences (20). To guarantee that therapeutic drugs are transported precisely and effectively to the targeted lncRNAs, novel delivery mechanisms must be created. Reaching this degree of accuracy is essential to maximizing the effects of therapy (245).
LncRNAs must also be functionally evaluated and validated in vivo in order to be used in therapeutic contexts. Numerous methods have been devised to verify the functional links between lncRNAs using experimental techniques (246–248). These strategies comprise techniques like capture hybridization analysis of RNA targets (CHART) for lncRNA target gene identification, chromatin isolation by RNA purification-sequencing (ChIRP), and RNA antisense purification (RAP) (246). Furthermore, an enormous increase in the quantity of lncRNAs annotated in the human genome has been made possible by advances in transcriptomics; however, only a minor portion of these have been functionally characterized (247). LncRNA expression necessitates the examination of intricate connections between lncRNA molecules and their corresponding target genes and proteins in model organisms. Nevertheless, the conservation of lncRNAs across different species is limited, posing a challenge to the advancement of therapeutic approaches aimed at addressing lncRNAs. Hence, it is imperative to thoroughly investigate the barriers hindering the progress of lncRNA-targeting therapeutics by means of the utilization of bioinformatics, extensive databases, and high-throughput advances in technology. This approach will facilitate the acquisition of a profound comprehension of lncRNA localization, structures, functional motifs, mechanisms of action, and their intricate associations with other molecules in biology (248).
Innovative therapeutic approaches aim to manipulate lncRNAs, either by knocking down oncogenic lncRNAs or by restoring tumor-suppressor lncRNAs. Techniques like RNAi, ASOs, and CRISPR/Cas9-mediated gene editing are employed to modulate the function of lncRNAs specifically. These strategies hold the potential to halt cancer progression, enhance the efficacy of existing treatments, and reduce unwanted side effects by targeting the molecular peculiarities of cancer cells directly (249).
Despite the potential of lncRNA-targeted therapies, the secure and effective delivery of these therapeutic agents poses a significant challenge. Nucleic acid-based drugs, including those targeting lncRNAs, are inherently unstable in the physiological environment and are susceptible to rapid degradation by nucleases. Moreover, they need to overcome various biological barriers to reach their target cells effectively. Researchers are developing advanced delivery systems like lipid nanoparticles (LNPs) and polymer-based carriers to address these issues. These systems protect the therapeutic molecules from enzymatic degradation, improve their half-life in the bloodstream, and facilitate targeted delivery to tumor cells. The development of such delivery vehicles is crucial for the clinical success of lncRNA-targeting therapies, as they ensure that the therapeutic agents are delivered efficiently to the site of action while minimizing off-target effects and toxicity. The targeted delivery of lncRNA therapies might be improved by making nanoparticles better and looking into other new ways to deliver drugs, like exosome-based systems. To make cancer treatments even better, combining lncRNA-targeting strategies with other methods like immunotherapy and chemotherapy could create more complete and effective plans (250). Table 5 outlines the difficulties encountered in immunotherapy directed toward lncRNA.
7 Concluding remarks
Emerging evidence suggests that lncRNAs play an important regulatory role in the immune system and contribute to resistance to cancer immunotherapies. Therefore, lncRNAs are promising cancer immunotherapeutic targets. Many lncRNAs are aberrantly expressed in cancer, and they also affect angiogenesis, metastasis, apoptosis, and tumor growth (257). Recent research has identified immune-related lncRNAs as gene expression regulators specific to immune cells, which mediate immune stimulatory and immunological response repression. This suggests that targeting lncRNAs could enhance the effectiveness of immunotherapy (250). However, there have been a limited number of documented clinical trials using lncRNAs, and little attention has been paid to targeting lncRNAs therapeutically in cancer as well as the effective and secure delivery of the reagents (256).
Author contributions
AA: Writing – original draft, Writing – review & editing. EM: Writing – original draft, Writing – review & editing. FR: Writing – original draft. MD: Writing – original draft. EB: Writing – original draft. YA: Writing – original draft. CP: Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The work was funded in part by the Cancer Research Society and York University Research Chair Program to CP.
Acknowledgments
We would like to thank the Cancer Research Society and York University Research Chair Program for providing financial support for the authorship and publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1446937/full#supplementary-material
References
1. Flavahan WA, Gaskell E, Bernstein BE. Epigenetic plasticity and the hallmarks of cancer. Science. (2017) 357:eaal2380. doi: 10.1126/science.aal2380
2. Fitzmaurice C, Allen C, Barber RM, Barregard L, Bhutta ZA, Brenner H, et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 32 cancer groups, 1990 to 2015: A systematic analysis for the global burden of disease study. JAMA Oncol. (2017) 3:524–48. doi: 10.1001/jamaoncol.2016.5688
3. Hernández Borrero LJ, El-Deiry WS. Tumor suppressor p53: Biology, signaling pathways, and therapeutic targeting. Biochim Biophys Acta Rev Cancer. (2021) 1876:188556. doi: 10.1016/j.bbcan.2021.188556
4. Matthews HK, Bertoli C, de Bruin RAM. Cell cycle control in cancer. Nat Rev Mol Cell Biol. (2022) 23:74–88. doi: 10.1038/s41580-021-00404-3
5. Goding Sauer A, Siegel RL, Jemal A, Fedewa SA. Current prevalence of major cancer risk factors and screening test use in the United States: disparities by education and race/ethnicity. Cancer Epidemiol Biomarkers Prev. (2019) 28:629–42. doi: 10.1158/1055-9965.Epi-18-1169
6. Wei JW, Huang K, Yang C, Kang CS. Non-coding RNAs as regulators in epigenetics (Review). Oncol Rep. (2017) 37:3–9. doi: 10.3892/or.2016.5236
7. Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: A new paradigm. Cancer Res. (2017) 77:3965–81. doi: 10.1158/0008-5472.Can-16-2634
8. Lekka E, Hall J. Noncoding RNAs in disease. FEBS Lett. (2018) 592:2884–900. doi: 10.1002/1873-3468.13182
9. Adams BD, Parsons C, Walker L, Zhang WC, Slack FJ. Targeting noncoding RNAs in disease. J Clin Invest. (2017) 127:761–71. doi: 10.1172/jci84424
10. Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev. (2016) 96:1297–325. doi: 10.1152/physrev.00041.2015
11. Jiang MC, Ni JJ, Cui WY, Wang BY, Zhuo W. Emerging roles of lncRNA in cancer and therapeutic opportunities. Am J Cancer Res. (2019) 9:1354–66.
12. Li Q, Liu Q. Noncoding RNAs in cancer immunology. Adv Exp Med Biol. (2016) 927:243–64. doi: 10.1007/978-981-10-1498-7_9
13. Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. (2021) 22:96–118. doi: 10.1038/s41580-020-00315-9
14. Fang Y, Fullwood MJ. Roles, functions, and mechanisms of long non-coding RNAs in cancer. Genomics Proteomics Bioinf. (2016) 14:42–54. doi: 10.1016/j.gpb.2015.09.006
15. Goodall GJ, Wickramasinghe VO. RNA in cancer. Nat Rev Cancer. (2021) 21:22–36. doi: 10.1038/s41568-020-00306-0
16. Peng C, Li J. Editorial: MicroRNAs in endocrinology and cell signaling. Front Endocrinol (Lausanne). (2022) 13:1118426. doi: 10.3389/fendo.2022.1118426
17. Sullenger BA, Nair S. From the RNA world to the clinic. Science. (2016) 352:1417–20. doi: 10.1126/science.aad8709
18. Waldman AD, Fritz JM, Lenardo MJ. A guide to cancer immunotherapy: from T cell basic science to clinical practice. Nat Rev Immunol. (2020) 20:651–68. doi: 10.1038/s41577-020-0306-5
19. Wu M, Fu P, Qu L, Liu J, Lin A. Long noncoding RNAs, new critical regulators in cancer immunity. Front Oncol. (2020) 10:550987. doi: 10.3389/fonc.2020.550987
20. Pi YN, Qi WC, Xia BR, Lou G, Jin WL. Long non-coding RNAs in the tumor immune microenvironment: biological properties and therapeutic potential. Front Immunol. (2021) 12:697083. doi: 10.3389/fimmu.2021.697083
21. Read S, Malmström V, Powrie F. Cytotoxic T lymphocyte-associated antigen 4 plays an essential role in the function of CD25(+)CD4(+) regulatory cells that control intestinal inflammation. J Exp Med. (2000) 192:295–302. doi: 10.1084/jem.192.2.295
22. Ashrafizadeh M, Zarrabi A, Hushmandi K, Zarrin V, Moghadam ER, Zabolian A, et al. PD-1/PD-L1 axis regulation in cancer therapy: The role of long non-coding RNAs and microRNAs. Life Sci. (2020) 256:117899. doi: 10.1016/j.lfs.2020.117899
23. Prasad V, Kaestner V. Nivolumab and pembrolizumab: Monoclonal antibodies against programmed cell death-1 (PD-1) that are interchangeable. Semin Oncol. (2017) 44:132–5. doi: 10.1053/j.seminoncol.2017.06.007
24. Patnaik A, Kang SP, Rasco D, Papadopoulos KP, Elassaiss-Schaap J, Beeram M, et al. Phase I study of pembrolizumab (MK-3475; anti-PD-1 monoclonal antibody) in patients with advanced solid tumors. Clin Cancer Res. (2015) 21:4286–93. doi: 10.1158/1078-0432.Ccr-14-2607
25. Yang JC, Hughes M, Kammula U, Royal R, Sherry RM, Topalian SL, et al. Ipilimumab (anti-CTLA4 antibody) causes regression of metastatic renal cell cancer associated with enteritis and hypophysitis. J Immunother. (2007) 30:825–30. doi: 10.1097/CJI.0b013e318156e47e
26. Rafae A, Ali S, Sana MK, Rana FK, Ibrahim A, Ahmed Z, et al. Electrolyte abnormalities associated with the use of atezolizumab - A systematic review. J Community Hosp Intern Med Perspect. (2022) 12:35–44. doi: 10.55729/2000-9666.1037
27. Syed YY. Durvalumab: first global approval. Drugs. (2017) 77:1369–76. doi: 10.1007/s40265-017-0782-5
28. Collins JM, Gulley JL. Product review: avelumab, an anti-PD-L1 antibody. Hum Vaccin Immunother. (2019) 15:891–908. doi: 10.1080/21645515.2018.1551671
29. Goodman DT. Cemiplimab and cutaneous squamous cell carcinoma: from bench to bedside. JPRAS Open. (2022) 33:155–60. doi: 10.1016/j.jpra.2022.06.003
30. Qin S, Kudo M, Meyer T, Bai Y, Guo Y, Meng Z, et al. Tislelizumab vs sorafenib as first-line treatment for unresectable hepatocellular carcinoma: A phase 3 randomized clinical trial. JAMA Oncol. (2023) 9:1651–9. doi: 10.1001/jamaoncol.2023.4003
31. Liu J, Yang Y, Liu Z, Fu X, Cai X, Li H, et al. Multicenter, single-arm, phase II trial of camrelizumab and chemotherapy as neoadjuvant treatment for locally advanced esophageal squamous cell carcinoma. J Immunother Cancer. (2022) 10:e004291. doi: 10.1136/jitc-2021-004291
32. Mirza MR, Chase DM, Slomovitz BM, dePont Christensen R, Novák Z, Black D, et al. Dostarlimab for primary advanced or recurrent endometrial cancer. N Engl J Med. (2023) 388:2145–58. doi: 10.1056/NEJMoa2216334
33. Iwasaki T, Hayashi K, Matsushita M, Nonaka D, Matsumoto T, Taniguchi M, et al. Clinical significance of the expression of FOXP3 and TIGIT in Merkel cell carcinoma. Sci Rep. (2023) 13:13114. doi: 10.1038/s41598-023-40050-7
34. Pelaia C, Benfante A, Busceti MT, Caiaffa MF, Campisi R, Carpagnano GE, et al. Real-life effects of dupilumab in patients with severe type 2 asthma, according to atopic trait and presence of chronic rhinosinusitis with nasal polyps. Front Immunol. (2023) 14:1121237. doi: 10.3389/fimmu.2023.1121237
35. Lewis KD, Peris K, Sekulic A, Stratigos AJ, Dunn L, Eroglu Z, et al. Final analysis of phase II results with cemiplimab in metastatic basal cell carcinoma after hedgehog pathway inhibitors. Ann Oncol. (2024) 35:221–8. doi: 10.1016/j.annonc.2023.10.123
36. Herbst RS, Giaccone G, de Marinis F, Reinmuth N, Vergnenegre A, Barrios CH, et al. Atezolizumab for first-line treatment of PD-L1-selected patients with NSCLC. N Engl J Med. (2020) 383:1328–39. doi: 10.1056/NEJMoa1917346
37. Diaz LA Jr., Shiu KK, Kim TW, Jensen BV, Jensen LH, Punt C, et al. Pembrolizumab versus chemotherapy for microsatellite instability-high or mismatch repair-deficient metastatic colorectal cancer (KEYNOTE-177): final analysis of a randomised, open-label, phase 3 study. Lancet Oncol. (2022) 23:659–70. doi: 10.1016/s1470-2045(22)00197-8
38. Vishnubalaji R, Shaath H, Elango R, Alajez NM. Noncoding RNAs as potential mediators of resistance to cancer immunotherapy. Semin Cancer Biol. (2020) 65:65–79. doi: 10.1016/j.semcancer.2019.11.006
39. Aghamiri SS, Puniya BL, Amin R, Helikar T. A multiscale mechanistic model of human dendritic cells for in-silico investigation of immune responses and novel therapeutics discovery. Front Immunol. (2023) 14:1112985. doi: 10.3389/fimmu.2023.1112985
40. Zhu J. T helper 2 (Th2) cell differentiation, type 2 innate lymphoid cell (ILC2) development and regulation of interleukin-4 (IL-4) and IL-13 production. Cytokine. (2015) 75:14–24. doi: 10.1016/j.cyto.2015.05.010
41. Dumitru AV, Ţăpoi DA, Halcu G, Munteanu O, Dumitrascu DI, Ceauşu MC, et al. The polyvalent role of CD30 for cancer diagnosis and treatment. Cells. (2023) 12:1783. doi: 10.3390/cells12131783
42. Tavarozzi R, Zacchi G, Pietrasanta D, Catania G, Castellino A, Monaco F, et al. Changing trends in B-cell non-hodgkin lymphoma treatment: the role of novel monoclonal antibodies in clinical practice. Cancers (Basel). (2023) 15:5397. doi: 10.3390/cancers15225397
43. Savoia P, Astrua C, Fava P. Ipilimumab (Anti-Ctla-4 Mab) in the treatment of metastatic melanoma: Effectiveness and toxicity management. Hum Vaccin Immunother. (2016) 12:1092–101. doi: 10.1080/21645515.2015.1129478
44. Plosker GL, Figgitt DP. Rituximab: a review of its use in non-Hodgkin’s lymphoma and chronic lymphocytic leukaemia. Drugs. (2003) 63:803–43. doi: 10.2165/00003495-200363080-00005
45. Gunturu KS, Woo Y, Beaubier N, Remotti HE, Saif MW. Gastric cancer and trastuzumab: first biologic therapy in gastric cancer. Ther Adv Med Oncol. (2013) 5:143–51. doi: 10.1177/1758834012469429
46. Gerriets V, Kasi A. Bevacizumab. Treasure Island (FL: StatPearls (2024). ineligible companies. Disclosure: Anup Kasi declares no relevant financial relationships with ineligible companies.: StatPearls Publishing Copyright © 2024, StatPearls Publishing LLC.
47. Chidharla A, Parsi M, Kasi A. Cetuximab. Treasure Island (FL: StatPearls (2024). ineligible companies. Disclosure: MeGhana Parsi declares no relevant financial relationships with ineligible companies. Disclosure: Anup Kasi declares no relevant financial relationships with ineligible companies.: StatPearls Publishing Copyright © 2024, StatPearls Publishing LLC.
48. Nasser NJ, Gorenberg M, Agbarya A. First line immunotherapy for non-small cell lung cancer. Pharm (Basel). (2020) 13:373. doi: 10.3390/ph13110373
49. Lipson EJ, Drake CG. Ipilimumab: an anti-CTLA-4 antibody for metastatic melanoma. Clin Cancer Res. (2011) 17:6958–62. doi: 10.1158/1078-0432.Ccr-11-1595
50. Gozzetti A, Ciofini S, Simoncelli M, Santoni A, Pacelli P, Raspadori D, et al. Anti CD38 monoclonal antibodies for multiple myeloma treatment. Hum Vaccin Immunother. (2022) 18:2052658. doi: 10.1080/21645515.2022.2052658
51. Moore KF. Use of atezolizumab for bladder and non-small cell lung cancers. J Adv Pract Oncol. (2017) 8:535–8. doi: 10.6004/jadpro.2017.8.5.9
52. Cupit-Link M, Federico SM. Treatment of high-risk neuroblastoma with dinutuximab and chemotherapy administered in all cycles of induction. Cancers (Basel). (2023) 15:4609. doi: 10.3390/cancers15184609
53. Donato EM, Fernández-Zarzoso M, Hueso JA, de la Rubia J. Brentuximab vedotin in Hodgkin lymphoma and anaplastic large-cell lymphoma: an evidence-based review. Onco Targets Ther. (2018) 11:4583–90. doi: 10.2147/ott.S141053
54. Rajan A, Kim C, Heery CR, Guha U, Gulley JL. Nivolumab, anti-programmed death-1 (PD-1) monoclonal antibody immunotherapy: Role in advanced cancers. Hum Vaccin Immunother. (2016) 12:2219–31. doi: 10.1080/21645515.2016.1175694
55. Doki Y, Ueno M, Hsu CH, Oh DY, Park K, Yamamoto N, et al. Tolerability and efficacy of durvalumab, either as monotherapy or in combination with tremelimumab, in patients from Asia with advanced biliary tract, esophageal, or head-and-neck cancer. Cancer Med. (2022) 11:2550–60. doi: 10.1002/cam4.4593
56. Penniman L, Parmar S, Patel K. Olaratumab (Lartruvo): an innovative treatment for soft tissue sarcoma. P T. (2018) 43:267–70.
57. Davis JA, Shockley A, Herbst A, Hendrickson L. Polatuzumab vedotin for the front-line treatment of diffuse large B-cell lymphoma: A new standard of care? J Adv Pract Oncol. (2023) 14:67–72. doi: 10.6004/jadpro.2023.14.1.6
58. Cao Y, Marcucci EC, Budde LE. Mosunetuzumab and lymphoma: latest updates from 2022 ASH annual meeting. J Hematol Oncol. (2023) 16:69. doi: 10.1186/s13045-023-01462-0
59. Petrizzo A, Ragone C, Cavalluzzo B, Mauriello A, Manolio C, Tagliamonte M, et al. Human endogenous retrovirus reactivation: implications for cancer immunotherapy. Cancers (Basel). (2021) 13:1999. doi: 10.3390/cancers13091999
60. Wang X, Yang S, Lv X, Wang L, Li C. Overexpression of lncRNA SNHG1 were suitable for oncolytic adenoviruse H101 therapy in oral squamous-cell carcinoma. Onco Targets Ther. (2020) 13:13033–9. doi: 10.2147/ott.S285536
61. Horita K, Kurosaki H, Nakatake M, Kuwano N, Oishi T, Itamochi H, et al. lncRNA UCA1-mediated cdc42 signaling promotes oncolytic vaccinia virus cell-to-cell spread in ovarian cancer. Mol Ther Oncolytics. (2019) 13:35–48. doi: 10.1016/j.omto.2019.03.003
62. van der Bruggen P, Traversari C, Chomez P, Lurquin C, De Plaen E, Van den Eynde B, et al. A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science. (1991) 254:1643–7. doi: 10.1126/science.1840703
63. Gardner TA, Elzey BD, Hahn NM. Sipuleucel-T (Provenge) autologous vaccine approved for treatment of men with asymptomatic or minimally symptomatic castrate-resistant metastatic prostate cancer. Hum Vaccin Immunother. (2012) 8:534–9. doi: 10.4161/hv.19795
64. Vishweshwaraiah YL, Dokholyan NV. mRNA vaccines for cancer immunotherapy. Front Immunol. (2022) 13:1029069. doi: 10.3389/fimmu.2022.1029069
65. Sadeghi Najafabadi SA, Bolhassani A, Aghasadeghi MR. Tumor cell-based vaccine: an effective strategy for eradication of cancer cells. Immunotherapy. (2022) 14:639–54. doi: 10.2217/imt-2022-0036
66. Bidram M, Zhao Y, Shebardina NG, Baldin AV, Bazhin AV, Ganjalikhany MR, et al. mRNA-based cancer vaccines: A therapeutic strategy for the treatment of melanoma patients. Vaccines (Basel). (2021) 9:1060. doi: 10.3390/vaccines9101060
67. Fonseca-Montaño MA, Vázquez-Santillán KI, Hidalgo-Miranda A. The current advances of lncRNAs in breast cancer immunobiology research. Front Immunol. (2023) 14:1194300. doi: 10.3389/fimmu.2023.1194300
68. Grimmett E, Al-Share B, Alkassab MB, Zhou RW, Desai A, Rahim MMA, et al. Cancer vaccines: past, present and future; a review article. Discovery Oncol. (2022) 13:31. doi: 10.1007/s12672-022-00491-4
69. Naidoo J, Page DB, Wolchok JD. Immune modulation for cancer therapy. Br J Cancer. (2014) 111:2214–9. doi: 10.1038/bjc.2014.348
70. Qiu Y, Su M, Liu L, Tang Y, Pan Y, Sun J. Clinical application of cytokines in cancer immunotherapy. Drug Des Devel Ther. (2021) 15:2269–87. doi: 10.2147/dddt.S308578
71. Sakurai T, Nishida N, Kudo M. Promising anticancer therapy: combination of immune checkpoint inhibitors and molecular-targeted agents. Hepatobiliary Surg Nutr. (2020) 9:777–9. doi: 10.21037/hbsn.2020.03.04
72. Musella M, Manduca N, Maccafeo E, Sistigu A. Epigenetics behind tumor immunology: a mini review. Oncogene. (2023) 42:2932–8. doi: 10.1038/s41388-023-02791-7
73. Yang J, Xu J, Wang W, Zhang B, Yu X, Shi S. Epigenetic regulation in the tumor microenvironment: molecular mechanisms and therapeutic targets. Signal Transduct Target Ther. (2023) 8:210. doi: 10.1038/s41392-023-01480-x
74. Gibney ER, Nolan CM. Epigenetics and gene expression. Heredity. (2010) 105:4–13. doi: 10.1038/hdy.2010.54
75. Shen C, Li M, Duan Y, Jiang X, Hou X, Xue F, et al. HDAC inhibitors enhance the anti-tumor effect of immunotherapies in hepatocellular carcinoma. Front Immunol. (2023) 14:1170207. doi: 10.3389/fimmu.2023.1170207
76. Tai Y, Chen M, Wang F, Fan Y, Zhang J, Cai B, et al. The role of dendritic cells in cancer immunity and therapeutic strategies. Int Immunopharmacol. (2024) 128:111548. doi: 10.1016/j.intimp.2024.111548
77. Hogg SJ, Beavis PA, Dawson MA, Johnstone RW. Targeting the epigenetic regulation of antitumour immunity. Nat Rev Drug Discovery. (2020) 19:776–800. doi: 10.1038/s41573-020-0077-5
78. Arshi A, Sharifi FS, Khorramian Ghahfarokhi M, Faghih Z, Doosti A, Ostovari S, et al. Expression Analysis of MALAT1, GAS5, SRA, and NEAT1 lncRNAs in Breast Cancer Tissues from Young Women and Women over 45 Years of Age. Mol Ther Nucleic Acids. (2018) 12:751–7. doi: 10.1016/j.omtn.2018.07.014
79. Jarroux J, Morillon A, Pinskaya M. History, discovery, and classification of lncRNAs. Adv Exp Med Biol. (2017) 1008:1–46. doi: 10.1007/978-981-10-5203-3_1
80. Aliperti V, Skonieczna J, Cerase A. Long non-coding RNA (lncRNA) roles in cell biology, neurodevelopment and neurological disorders. Noncoding RNA. (2021) 7:36. doi: 10.3390/ncrna7020036
81. Bridges MC, Daulagala AC, Kourtidis A. LNCcation: lncRNA localization and function. J Cell Biol. (2021) 220:e202009045. doi: 10.1083/jcb.202009045
82. Chen LL. Linking long noncoding RNA localization and function. Trends Biochem Sci. (2016) 41:761–72. doi: 10.1016/j.tibs.2016.07.003
83. McDonel P, Guttman M. Approaches for understanding the mechanisms of long noncoding RNA regulation of gene expression. Cold Spring Harb Perspect Biol. (2019) 11:a032151. doi: 10.1101/cshperspect.a032151
84. Dahariya S, Paddibhatla I, Kumar S, Raghuwanshi S, Pallepati A, Gutti RK. Long non-coding RNA: Classification, biogenesis and functions in blood cells. Mol Immunol. (2019) 112:82–92. doi: 10.1016/j.molimm.2019.04.011
85. Jin KT, Yao JY, Fang XL, Di H, Ma YY. Roles of lncRNAs in cancer: Focusing on angiogenesis. Life Sci. (2020) 252:117647. doi: 10.1016/j.lfs.2020.117647
86. Balas MM, Johnson AM. Exploring the mechanisms behind long noncoding RNAs and cancer. Noncoding RNA Res. (2018) 3:108–17. doi: 10.1016/j.ncrna.2018.03.001
87. Chowdhary A, Satagopam V, Schneider R. Long non-coding RNAs: mechanisms, experimental, and computational approaches in identification, characterization, and their biomarker potential in cancer. Front Genet. (2021) 12:649619. doi: 10.3389/fgene.2021.649619
88. Luo Y, Yang J, Yu J, Liu X, Yu C, Hu J, et al. Long non-coding RNAs: emerging roles in the immunosuppressive tumor microenvironment. Front Oncol. (2020) 10:48. doi: 10.3389/fonc.2020.00048
89. Mumtaz PT, Bhat SA, Ahmad SM, Dar MA, Ahmed R, Urwat U, et al. LncRNAs and immunity: watchdogs for host pathogen interactions. Biol Proced Online. (2017) 19:3. doi: 10.1186/s12575-017-0052-7
90. Atianand MK, Caffrey DR, Fitzgerald KA. Immunobiology of long noncoding RNAs. Annu Rev Immunol. (2017) 35:177–98. doi: 10.1146/annurev-immunol-041015-055459
91. Denaro N, Merlano MC, Lo Nigro C. Long noncoding RNAs as regulators of cancer immunity. Mol Oncol. (2019) 13:61–73. doi: 10.1002/1878-0261.12413
92. Chan J, Atianand M, Jiang Z, Carpenter S, Aiello D, Elling R, et al. Cutting edge: A natural antisense transcript, AS-IL1α, controls inducible transcription of the proinflammatory cytokine IL-1α. J Immunol. (2015) 195:1359–63. doi: 10.4049/jimmunol.1500264
93. Atianand MK, Hu W, Satpathy AT, Shen Y, Ricci EP, Alvarez-Dominguez JR, et al. A long noncoding RNA lincRNA-EPS acts as a transcriptional brake to restrain inflammation. Cell. (2016) 165:1672–85. doi: 10.1016/j.cell.2016.05.075
94. Roux BT, Heward JA, Donnelly LE, Jones SW, Lindsay MA. Catalog of differentially expressed long non-coding RNA following activation of human and mouse innate immune response. Front Immunol. (2017) 8:1038. doi: 10.3389/fimmu.2017.01038
95. Zgheib C, Hodges MM, Hu J, Liechty KW, Xu J. Long non-coding RNA Lethe regulates hyperglycemia-induced reactive oxygen species production in macrophages. PloS One. (2017) 12:e0177453. doi: 10.1371/journal.pone.0177453
96. Lu Y, Liu X, Xie M, Liu M, Ye M, Li M, et al. The NF-κB-Responsive Long Noncoding RNA FIRRE Regulates Posttranscriptional Regulation of Inflammatory Gene Expression through Interacting with hnRNPU. J Immunol. (2017) 199:3571–82. doi: 10.4049/jimmunol.1700091
97. Venkatraman A, He XC, Thorvaldsen JL, Sugimura R, Perry JM, Tao F, et al. Maternal imprinting at the H19-Igf2 locus maintains adult haematopoietic stem cell quiescence. Nature. (2013) 500:345–9. doi: 10.1038/nature12303
98. Elling R, Robinson EK, Shapleigh B, Liapis SC, Covarrubias S, Katzman S, et al. Genetic Models Reveal cis and trans Immune-Regulatory Activities for lincRNA-Cox2. Cell Rep. (2018) 25:1511–24.e6. doi: 10.1016/j.celrep.2018.10.027
99. Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S, et al. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science. (2014) 344:310–3. doi: 10.1126/science.1251456
100. Zhu X, Du J, Yu J, Guo R, Feng Y, Qiao L, et al. LncRNA NKILA regulates endothelium inflammation by controlling a NF-κB/KLF4 positive feedback loop. J Mol Cell Cardiol. (2019) 126:60–9. doi: 10.1016/j.yjmcc.2018.11.001
101. Cui H, Xie N, Tan Z, Banerjee S, Thannickal VJ, Abraham E, et al. The human long noncoding RNA lnc-IL7R regulates the inflammatory response. Eur J Immunol. (2014) 44:2085–95. doi: 10.1002/eji.201344126
102. Xie Q, Chen S, Tian R, Huang X, Deng R, Xue B, et al. Long noncoding RNA ITPRIP-1 positively regulates the innate immune response through promotion of oligomerization and activation of MDA5. J Virol. (2018) 92:e00507–18. doi: 10.1128/jvi.00507-18
103. Jiang M, Zhang S, Yang Z, Lin H, Zhu J, Liu L, et al. Self-recognition of an inducible host lncRNA by RIG-I feedback restricts innate immune response. Cell. (2018) 173:906–19.e13. doi: 10.1016/j.cell.2018.03.064
104. Du M, Yuan L, Tan X, Huang D, Wang X, Zheng Z, et al. The LPS-inducible lncRNA Mirt2 is a negative regulator of inflammation. Nat Commun. (2017) 8:2049. doi: 10.1038/s41467-017-02229-1
105. Ma S, Ming Z, Gong AY, Wang Y, Chen X, Hu G, et al. A long noncoding RNA, lincRNA-Tnfaip3, acts as a coregulator of NF-κB to modulate inflammatory gene transcription in mouse macrophages. FASEB J. (2017) 31:1215–25. doi: 10.1096/fj.201601056R
106. Cui H, Banerjee S, Guo S, Xie N, Ge J, Jiang D, et al. Long noncoding RNA Malat1 regulates differential activation of macrophages and response to lung injury. JCI Insight. (2019) 4:e124522. doi: 10.1172/jci.insight.124522
107. Hadjicharalambous MR, Roux BT, Feghali-Bostwick CA, Murray LA, Clarke DL, Lindsay MA. Long non-coding RNAs are central regulators of the IL-1β-induced inflammatory response in normal and idiopathic pulmonary lung fibroblasts. Front Immunol. (2018) 9:2906. doi: 10.3389/fimmu.2018.02906
108. Imamura K, Imamachi N, Akizuki G, Kumakura M, Kawaguchi A, Nagata K, et al. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol Cell. (2014) 53:393–406. doi: 10.1016/j.molcel.2014.01.009
109. Schwerdtfeger M, Desiderio V, Kobold S, Regad T, Zappavigna S, Caraglia M. Long non-coding RNAs in cancer stem cells. Transl Oncol. (2021) 14:101134. doi: 10.1016/j.tranon.2021.101134
110. Mariotti B, Servaas NH, Rossato M, Tamassia N, Cassatella MA, Cossu M, et al. The long non-coding RNA NRIR drives IFN-response in monocytes: implication for systemic sclerosis. Front Immunol. (2019) 10:100. doi: 10.3389/fimmu.2019.00100
111. Krawczyk M, Emerson BM. p50-associated COX-2 extragenic RNA (PACER) activates COX-2 gene expression by occluding repressive NF-κB complexes. Elife. (2014) 3:e01776. doi: 10.7554/eLife.01776
112. Li Z, Chao TC, Chang KY, Lin N, Patil VS, Shimizu C, et al. The long noncoding RNA THRIL regulates TNFα expression through its interaction with hnRNPL. Proc Natl Acad Sci U S A. (2014) 111:1002–7. doi: 10.1073/pnas.1313768111
113. Zhang X, Weissman SM, Newburger PE. Long intergenic non-coding RNA HOTAIRM1 regulates cell cycle progression during myeloid maturation in NB4 human promyelocytic leukemia cells. RNA Biol. (2014) 11:777–87. doi: 10.4161/rna.28828
114. Kambara H, Niazi F, Kostadinova L, Moonka DK, Siegel CT, Post AB, et al. Negative regulation of the interferon response by an interferon-induced long non-coding RNA. Nucleic Acids Res. (2014) 42:10668–80. doi: 10.1093/nar/gku713
115. Kotzin JJ, Spencer SP, McCright SJ, Kumar DBU, Collet MA, Mowel WK, et al. The long non-coding RNA Morrbid regulates Bim and short-lived myeloid cell lifespan. Nature. (2016) 537:239–43. doi: 10.1038/nature19346
116. Wang Y, Zhong H, Xie X, Chen CY, Huang D, Shen L, et al. Long noncoding RNA derived from CD244 signaling epigenetically controls CD8+ T-cell immune responses in tuberculosis infection. Proc Natl Acad Sci U S A. (2015) 112:E3883–92. doi: 10.1073/pnas.1501662112
117. Sehgal L, Mathur R, Braun FK, Wise JF, Berkova Z, Neelapu S, et al. FAS-antisense 1 lncRNA and production of soluble versus membrane Fas in B-cell lymphoma. Leukemia. (2014) 28:2376–87. doi: 10.1038/leu.2014.126
118. Huang W, Thomas B, Flynn RA, Gavzy SJ, Wu L, Kim SV, et al. DDX5 and its associated lncRNA Rmrp modulate TH17 cell effector functions. Nature. (2015) 528:517–22. doi: 10.1038/nature16193
119. Zemmour D, Pratama A, Loughhead SM, Mathis D, Benoist C. Flicr, a long noncoding RNA, modulates Foxp3 expression and autoimmunity. Proc Natl Acad Sci U S A. (2017) 114:E3472–E80. doi: 10.1073/pnas.1700946114
120. Sharma S, Findlay GM, Bandukwala HS, Oberdoerffer S, Baust B, Li Z, et al. Dephosphorylation of the nuclear factor of activated T cells (NFAT) transcription factor is regulated by an RNA-protein scaffold complex. Proc Natl Acad Sci U S A. (2011) 108:11381–6. doi: 10.1073/pnas.1019711108
121. Yang CA, Li JP, Yen JC, Lai IL, Ho YC, Chen YC, et al. lncRNA NTT/PBOV1 axis promotes monocyte differentiation and is elevated in rheumatoid arthritis. Int J Mol Sci. (2018) 19:2806. doi: 10.3390/ijms19092806
122. Mourtada-Maarabouni M, Hedge VL, Kirkham L, Farzaneh F, Williams GT. Growth arrest in human T-cells is controlled by the non-coding RNA growth-arrest-specific transcript 5 (GAS5). J Cell Sci. (2008) 121:939–46. doi: 10.1242/jcs.024646
123. Hu G, Tang Q, Sharma S, Yu F, Escobar TM, Muljo SA, et al. Expression and regulation of intergenic long noncoding RNAs during T cell development and differentiation. Nat Immunol. (2013) 14:1190–8. doi: 10.1038/ni.2712
124. Collier SP, Henderson MA, Tossberg JT, Aune TM. Regulation of the Th1 genomic locus from Ifng through Tmevpg1 by T-bet. J Immunol. (2014) 193:3959–65. doi: 10.4049/jimmunol.1401099
125. Spurlock CF 3rd, Tossberg JT, Guo Y, Collier SP, Crooke PS 3rd, Aune TM. Expression and functions of long noncoding RNAs during human T helper cell differentiation. Nat Commun. (2015) 6:6932. doi: 10.1038/ncomms7932
126. Ranzani V, Rossetti G, Panzeri I, Arrigoni A, Bonnal RJ, Curti S, et al. The long intergenic noncoding RNA landscape of human lymphocytes highlights the regulation of T cell differentiation by linc-MAF-4. Nat Immunol. (2015) 16:318–25. doi: 10.1038/ni.3093
127. Bolha L, Ravnik-Glavač M, Glavač D. Long noncoding RNAs as biomarkers in cancer. Dis Markers. (2017) 2017:7243968. doi: 10.1155/2017/7243968
128. Shi T, Gao G, Cao Y. Long noncoding RNAs as novel biomarkers have a promising future in cancer diagnostics. Dis Markers. (2016) 2016:9085195. doi: 10.1155/2016/9085195
129. Rajbhandary S, Dhakal H, Shrestha S. Tumor immune microenvironment (TIME) to enhance antitumor immunity. Eur J Med Res. (2023) 28:169. doi: 10.1186/s40001-023-01125-3
130. Asadi M, Zarredar H, Zafari V, Soleimani Z, Saeedi H, Caner A, et al. Immune features of tumor microenvironment: A genetic spotlight. Cell Biochem Biophys. (2023) 82:107–18. doi: 10.1007/s12013-023-01192-7
131. Guo Y, Xie Y, Luo Y. The role of long non-coding RNAs in the tumor immune microenvironment. Front Immunol. (2022) 13:851004. doi: 10.3389/fimmu.2022.851004
132. Barry ST, Gabrilovich DI, Sansom OJ, Campbell AD, Morton JP. Therapeutic targeting of tumour myeloid cells. Nat Rev Cancer. (2023) 23:216–37. doi: 10.1038/s41568-022-00546-2
133. Deng J, Fleming JB. Inflammation and myeloid cells in cancer progression and metastasis. Front Cell Dev Biol. (2021) 9:759691. doi: 10.3389/fcell.2021.759691
134. Arpita B, Mohini V, Shweta D. Role of myeloid cells in the tumor microenvironment. J Cancer Metastasis Treat. (2022) 8:27. doi: 10.20517/2394-4722.2022.33
135. Grossman JG, Nywening TM, Belt BA, Panni RZ, Krasnick BA, DeNardo DG, et al. Recruitment of CCR2(+) tumor associated macrophage to sites of liver metastasis confers a poor prognosis in human colorectal cancer. Oncoimmunology. (2018) 7:e1470729. doi: 10.1080/2162402x.2018.1470729
136. Liu Y, Han Y, Zhang Y, Lv T, Peng X, Huang J. LncRNAs has been identified as regulators of Myeloid-derived suppressor cells in lung cancer. Front Immunol. (2023) 14:1067520. doi: 10.3389/fimmu.2023.1067520
137. Tian X, Tian J, Tang X, Ma J, Wang S. Long non-coding RNAs in the regulation of myeloid cells. J Hematol Oncol. (2016) 9:99. doi: 10.1186/s13045-016-0333-7
138. Pengam S, Durand J, Usal C, Gauttier V, Dilek N, Martinet B, et al. SIRPα/CD47 axis controls the maintenance of transplant tolerance sustained by myeloid-derived suppressor cells. Am J Transplant. (2019) 19:3263–75. doi: 10.1111/ajt.15497
139. Li TT, Ogino S, Qian ZR. Toll-like receptor signaling in colorectal cancer: carcinogenesis to cancer therapy. World J Gastroenterol. (2014) 20:17699–708. doi: 10.3748/wjg.v20.i47.17699
140. Brunda MJ, Luistro L, Rumennik L, Wright RB, Dvorozniak M, Aglione A, et al. Antitumor activity of interleukin 12 in preclinical models. Cancer Chemother Pharmacol. (1996) 38 Suppl:S16–21. doi: 10.1007/s002800051031
141. Wang Y, Johnson KCC, Gatti-Mays ME, Li Z. Emerging strategies in targeting tumor-resident myeloid cells for cancer immunotherapy. J Hematol Oncol. (2022) 15:118. doi: 10.1186/s13045-022-01335-y
142. Guo L, Zhang X, Pan H, Li Y, Wang J, Li L, et al. Prognostic and immunological significance of metastasis associated lung adenocarcinoma transcript 1 among different kinds of cancers. Bioengineered. (2021) 12:4247–58. doi: 10.1080/21655979.2021.1955511
143. Xu X, Zhong Z, Shao Y, Yi Y. Prognostic value of MEG3 and its correlation with immune infiltrates in gliomas. Front Genet. (2021) 12:679097. doi: 10.3389/fgene.2021.679097
144. Botti G, Scognamiglio G, Aquino G, Liguori G, Cantile M. LncRNA HOTAIR in tumor microenvironment: what role? Int J Mol Sci. (2019) 20:2279. doi: 10.3390/ijms20092279
145. Kaur J, Salehen N, Norazit A, Rahman AA, Murad NAA, Rahman N, et al. Tumor suppressive effects of GAS5 in cancer cells. Noncoding RNA. (2022) 8:39. doi: 10.3390/ncrna8030039
146. Batista S, Gregório AC, Hanada Otake A, Couto N, Costa-Silva B. The gastrointestinal tumor microenvironment: an updated biological and clinical perspective. J Oncol. (2019) 2019:6240505. doi: 10.1155/2019/6240505
147. Zhou L, Zhu Y, Sun D, Zhang Q. Emerging roles of long non-coding RNAs in the tumor microenvironment. Int J Biol Sci. (2020) 16:2094–103. doi: 10.7150/ijbs.44420
148. Ghafouri-Fard S, Khoshbakht T, Taheri M, Shojaei S. A review on the role of SPRY4-IT1 in the carcinogenesis. Front Oncol. (2021) 11:779483. doi: 10.3389/fonc.2021.779483
149. Park EG, Pyo SJ, Cui Y, Yoon SH, Nam JW. Tumor immune microenvironment lncRNAs. Brief Bioinform. (2022) 23:bbab504. doi: 10.1093/bib/bbab504
150. Tamang S, Acharya V, Roy D, Sharma R, Aryaa A, Sharma U, et al. SNHG12: an lncRNA as a potential therapeutic target and biomarker for human cancer. Front Oncol. (2019) 9:901. doi: 10.3389/fonc.2019.00901
151. Han W, Shi CT, Ma J, Chen H, Shao QX, Gao XJ, et al. Pan-cancer analysis of LncRNA XIST and its potential mechanisms in human cancers. Heliyon. (2022) 8:e10786. doi: 10.1016/j.heliyon.2022.e10786
152. Zhu K, Gong Z, Li P, Jiang X, Zeng Z, Xiong W, et al. A review of linc00673 as a novel lncRNA for tumor regulation. Int J Med Sci. (2021) 18:398–405. doi: 10.7150/ijms.48134
153. Tan P, Xu M, Nie J, Qin J, Liu X, Sun H, et al. LncRNA SNHG16 promotes colorectal cancer proliferation by regulating ABCB1 expression through sponging miR-214-3p. J BioMed Res. (2022) 36:231–41. doi: 10.7555/jbr.36.20220049
154. Hussen BM, Azimi T, Abak A, Hidayat HJ, Taheri M, Ghafouri-Fard S. Role of lncRNA BANCR in human cancers: an updated review. Front Cell Dev Biol. (2021) 9:689992. doi: 10.3389/fcell.2021.689992
155. Gu J, Zhang B, An R, Qian W, Han L, Duan W, et al. Molecular interactions of the long noncoding RNA NEAT1 in cancer. Cancers (Basel). (2022) 14:4009. doi: 10.3390/cancers14164009
156. Liu J, Zhu Y, Ge C. LncRNA ZFAS1 promotes pancreatic adenocarcinoma metastasis via the RHOA/ROCK2 pathway by sponging miR-3924. Cancer Cell Int. (2020) 20:249. doi: 10.1186/s12935-020-01322-8
157. Zhan Y, Chen Z, He S, Gong Y, He A, Li Y, et al. Long non-coding RNA SOX2OT promotes the stemness phenotype of bladder cancer cells by modulating SOX2. Mol Cancer. (2020) 19:25. doi: 10.1186/s12943-020-1143-7
158. Li X, Liang S, Fei M, Ma K, Sun L, Liu Y, et al. LncRNA CRNDE drives the progression of hepatocellular carcinoma by inducing the immunosuppressive niche. Int J Biol Sci. (2024) 20:718–32. doi: 10.7150/ijbs.85471
159. Wen X, Wu Y, Lou Y, Xia Y, Yu X. The roles of Linc-ROR in the regulation of cancer stem cells. Transl Oncol. (2023) 28:101602. doi: 10.1016/j.tranon.2022.101602
160. Peng L, Chen Z, Chen Y, Wang X, Tang N. MIR155HG is a prognostic biomarker and associated with immune infiltration and immune checkpoint molecules expression in multiple cancers. Cancer Med. (2019) 8:7161–73. doi: 10.1002/cam4.2583
161. Zhan DT, Xian HC. Exploring the regulatory role of lncRNA in cancer immunity. Front Oncol. (2023) 13:1191913. doi: 10.3389/fonc.2023.1191913
162. Lendeckel U, Venz S, Wolke C. Macrophages: shapes and functions. ChemTexts. (2022) 8:12. doi: 10.1007/s40828-022-00163-4
163. Li C, Xu X, Wei S, Jiang P, Xue L, Wang J. Tumor-associated macrophages: potential therapeutic strategies and future prospects in cancer. J Immunother Cancer. (2021) 9:e001341. doi: 10.1136/jitc-2020-001341
164. Poh AR, Ernst M. Targeting macrophages in cancer: from bench to bedside. Front Oncol. (2018) 8:49. doi: 10.3389/fonc.2018.00049
165. Kumari N, Choi SH. Tumor-associated macrophages in cancer: recent advancements in cancer nanoimmunotherapies. J Exp Clin Cancer Res. (2022) 41:68. doi: 10.1186/s13046-022-02272-x
166. Liu J, Geng X, Hou J, Wu G. New insights into M1/M2 macrophages: key modulators in cancer progression. Cancer Cell Int. (2021) 21:389. doi: 10.1186/s12935-021-02089-2
167. Oshi M, Tokumaru Y, Asaoka M, Yan L, Satyananda V, Matsuyama R, et al. M1 Macrophage and M1/M2 ratio defined by transcriptomic signatures resemble only part of their conventional clinical characteristics in breast cancer. Sci Rep. (2020) 10:16554. doi: 10.1038/s41598-020-73624-w
168. Yuan A, Hsiao YJ, Chen HY, Chen HW, Ho CC, Chen YY, et al. Opposite effects of M1 and M2 macrophage subtypes on lung cancer progression. Sci Rep. (2015) 5:14273. doi: 10.1038/srep14273
169. Mofed D, Omran JI, Sabet S, Baiomy AA, Emara M, Salem TZ. The regulatory role of long non- coding RNAs as a novel controller of immune response against cancer cells. Mol Biol Rep. (2022) 49:11775–93. doi: 10.1007/s11033-022-07947-4
170. Veglia F, Sanseviero E, Gabrilovich DI. Myeloid-derived suppressor cells in the era of increasing myeloid cell diversity. Nat Rev Immunol. (2021) 21:485–98. doi: 10.1038/s41577-020-00490-y
171. Cheng JN, Yuan YX, Zhu B, Jia Q. Myeloid-derived suppressor cells: A multifaceted accomplice in tumor progression. Front Cell Dev Biol. (2021) 9:740827. doi: 10.3389/fcell.2021.740827
172. Umansky V, Blattner C, Gebhardt C, Utikal J. The role of myeloid-derived suppressor cells (MDSC) in cancer progression. Vaccines (Basel). (2016) 4:36. doi: 10.3390/vaccines4040036
173. Ai L, Mu S, Wang Y, Wang H, Cai L, Li W, et al. Prognostic role of myeloid-derived suppressor cells in cancers: a systematic review and meta-analysis. BMC Cancer. (2018) 18:1220. doi: 10.1186/s12885-018-5086-y
174. Yang Y, Li C, Liu T, Dai X, Bazhin AV. Myeloid-derived suppressor cells in tumors: from mechanisms to antigen specificity and microenvironmental regulation. Front Immunol. (2020) 11:1371. doi: 10.3389/fimmu.2020.01371
175. De Cicco P, Ercolano G, Ianaro A. The new era of cancer immunotherapy: targeting myeloid-derived suppressor cells to overcome immune evasion. Front Immunol. (2020) 11:1680. doi: 10.3389/fimmu.2020.01680
176. Wu Y, Yi M, Niu M, Mei Q, Wu K. Myeloid-derived suppressor cells: an emerging target for anticancer immunotherapy. Mol Cancer. (2022) 21:184. doi: 10.1186/s12943-022-01657-y
177. Law AMK, Valdes-Mora F, Gallego-Ortega D. Myeloid-derived suppressor cells as a therapeutic target for cancer. Cells. (2020) 9:561. doi: 10.3390/cells9030561
178. Ozbay Kurt FG, Lasser S, Arkhypov I, Utikal J, Umansky V. Enhancing immunotherapy response in melanoma: myeloid-derived suppressor cells as a therapeutic target. J Clin Invest. (2023) 133:e170762. doi: 10.1172/jci170762
179. Hu Q, Egranov SD, Lin C, Yang L. Long noncoding RNA loss in immune suppression in cancer. Pharmacol Ther. (2020) 213:107591. doi: 10.1016/j.pharmthera.2020.107591
180. Rosales C. Neutrophils at the crossroads of innate and adaptive immunity. J Leukoc Biol. (2020) 108:377–96. doi: 10.1002/jlb.4mir0220-574rr
181. Rosales C. Neutrophil: A cell with many roles in inflammation or several cell types? Front Physiol. (2018) 9:113. doi: 10.3389/fphys.2018.00113
182. Hedrick CC, Malanchi I. Neutrophils in cancer: heterogeneous and multifaceted. Nat Rev Immunol. (2022) 22:173–87. doi: 10.1038/s41577-021-00571-6
183. Raskov H, Orhan A, Gaggar S, Gögenur I. Neutrophils and polymorphonuclear myeloid-derived suppressor cells: an emerging battleground in cancer therapy. Oncogenesis. (2022) 11:22. doi: 10.1038/s41389-022-00398-3
184. Gungabeesoon J, Gort-Freitas NA, Kiss M, Bolli E, Messemaker M, Siwicki M, et al. A neutrophil response linked to tumor control in immunotherapy. Cell. (2023) 186:1448–64.e20. doi: 10.1016/j.cell.2023.02.032
185. Faget J, Peters S, Quantin X, Meylan E, Bonnefoy N. Neutrophils in the era of immune checkpoint blockade. J Immunother Cancer. (2021) 9:e002242. doi: 10.1136/jitc-2020-002242
186. Jiang T, Wang Y, Chen X, Xia W, Xue S, Gu L, et al. Neutrophil extracellular traps (NETs)-related lncRNAs signature for predicting prognosis and the immune microenvironment in breast cancer. Front Cell Dev Biol. (2023) 11:1117637. doi: 10.3389/fcell.2023.1117637
187. Ding W, Li B, Zhang Y, He L, Su J. A neutrophil extracellular traps-associated lncRNA signature predicts the clinical outcomes in patients with lung adenocarcinoma. Front Genet. (2022) 13:1047231. doi: 10.3389/fgene.2022.1047231
188. Zahid KR, Raza U, Tumbath S, Jiang L, Xu W, Huang X. Neutrophils: Musketeers against immunotherapy. Front Oncol. (2022) 12:975981. doi: 10.3389/fonc.2022.975981
189. Chen Z, Li JL, Lin S, Cao C, Gimbrone NT, Yang R, et al. cAMP/CREB-regulated LINC00473 marks LKB1-inactivated lung cancer and mediates tumor growth. J Clin Invest. (2016) 126:2267–79. doi: 10.1172/jci85250
190. Ron Y, De Baetselier P, Gordon J, Feldman M, Segal S. Defective induction of antigen-reactive proliferating T cells in B cell-deprived mice. Eur J Immunol. (1981) 11:964–8. doi: 10.1002/eji.1830111203
191. Wang SS, Liu W, Ly D, Xu H, Qu L, Zhang L. Tumor-infiltrating B cells: their role and application in anti-tumor immunity in lung cancer. Cell Mol Immunol. (2019) 16:6–18. doi: 10.1038/s41423-018-0027-x
192. Zirakzadeh AA, Marits P, Sherif A, Winqvist O. Multiplex B cell characterization in blood, lymph nodes, and tumors from patients with Malignancies. J Immunol. (2013) 190:5847–55. doi: 10.4049/jimmunol.1203279
193. Sarvaria A, Madrigal JA, Saudemont A. B cell regulation in cancer and anti-tumor immunity. Cell Mol Immunol. (2017) 14:662–74. doi: 10.1038/cmi.2017.35
194. Huang L, Li X, Ye H, Liu Y, Liang X, Yang C, et al. Long non-coding RNA NCK1-AS1 promotes the tumorigenesis of glioma through sponging microRNA-138-2-3p and activating the TRIM24/Wnt/β-catenin axis. J Exp Clin Cancer Res. (2020) 39:63. doi: 10.1186/s13046-020-01567-1
195. Verma A, Jiang Y, Du W, Fairchild L, Melnick A, Elemento O. Transcriptome sequencing reveals thousands of novel long non-coding RNAs in B cell lymphoma. Genome Med. (2015) 7:110. doi: 10.1186/s13073-015-0230-7
196. Zhu Q, Li Y, Guo Y, Hu L, Xiao Z, Liu X, et al. Long non-coding RNA SNHG16 promotes proliferation and inhibits apoptosis of diffuse large B-cell lymphoma cells by targeting miR-497-5p/PIM1 axis. J Cell Mol Med. (2019) 23:7395–405. doi: 10.1111/jcmm.14601
197. Hu L, Zhao J, Liu Y, Liu X, Lu Q, Zeng Z, et al. Geniposide inhibits proliferation and induces apoptosis of diffuse large B-cell lymphoma cells by inactivating the HCP5/miR-27b-3p/MET axis. Int J Med Sci. (2020) 17:2735–43. doi: 10.7150/ijms.51329
198. Zhao L, Liu Y, Zhang J, Liu Y, Qi Q. LncRNA SNHG14/miR-5590-3p/ZEB1 positive feedback loop promoted diffuse large B cell lymphoma progression and immune evasion through regulating PD-1/PD-L1 checkpoint. Cell Death Dis. (2019) 10:731. doi: 10.1038/s41419-019-1886-5
199. Wang QM, Lian GY, Song Y, Huang YF, Gong Y. LncRNA MALAT1 promotes tumorigenesis and immune escape of diffuse large B cell lymphoma by sponging miR-195. Life Sci. (2019) 231:116335. doi: 10.1016/j.lfs.2019.03.040
200. Boxer LM, Dang CV. Translocations involving c-myc and c-myc function. Oncogene. (2001) 20:5595–610. doi: 10.1038/sj.onc.1204595
201. Doose G, Haake A, Bernhart SH, López C, Duggimpudi S, Wojciech F, et al. MINCR is a MYC-induced lncRNA able to modulate MYC’s transcriptional network in Burkitt lymphoma cells. Proc Natl Acad Sci U S A. (2015) 112:E5261–70. doi: 10.1073/pnas.1505753112
202. Qian CS, Li LJ, Huang HW, Yang HF, Wu DP. MYC-regulated lncRNA NEAT1 promotes B cell proliferation and lymphomagenesis via the miR-34b-5p-GLI1 pathway in diffuse large B-cell lymphoma. Cancer Cell Int. (2020) 20:87. doi: 10.1186/s12935-020-1158-6
203. Shi X, Cui Z, Liu X, Wu S, Wu Y, Fang F, et al. LncRNA FIRRE is activated by MYC and promotes the development of diffuse large B-cell lymphoma via Wnt/β-catenin signaling pathway. Biochem Biophys Res Commun. (2019) 510:594–600. doi: 10.1016/j.bbrc.2019.01.105
204. Wang Y, Zhang M, Xu H, Wang Y, Li Z, Chang Y, et al. Discovery and validation of the tumor-suppressive function of long noncoding RNA PANDA in human diffuse large B-cell lymphoma through the inactivation of MAPK/ERK signaling pathway. Oncotarget. (2017) 8:72182–96. doi: 10.18632/oncotarget.20053
205. Meng H, Zhao B, Wang Y. FOXM1-induced upregulation of lncRNA OR3A4 promotes the progression of diffuse large B-cell lymphoma via Wnt/β-catenin signaling pathway. Exp Mol Pathol. (2020) 115:104451. doi: 10.1016/j.yexmp.2020.104451
206. Zhao CC, Jiao Y, Zhang YY, Ning J, Zhang YR, Xu J, et al. Lnc SMAD5-AS1 as ceRNA inhibit proliferation of diffuse large B cell lymphoma via Wnt/β-catenin pathway by sponging miR-135b-5p to elevate expression of APC. Cell Death Dis. (2019) 10:252. doi: 10.1038/s41419-019-1479-3
207. Peng W, Feng J. Long noncoding RNA LUNAR1 associates with cell proliferation and predicts a poor prognosis in diffuse large B-cell lymphoma. BioMed Pharmacother. (2016) 77:65–71. doi: 10.1016/j.biopha.2015.12.001
208. Tao S, Chen Y, Hu M, Xu L, Fu CB, Hao XB. LncRNA PVT1 facilitates DLBCL development via miR-34b-5p/Foxp1 pathway. Mol Cell Biochem. (2022) 477:951–63. doi: 10.1007/s11010-021-04335-7
209. Luheshi N, Coates-Ulrichsen J, Harper J, Davies G, Legg J, Wilkinson R. The combination of CD40 agonism and PD-L1 blockade enhances anti-tumor immunity in a mouse syngeneic orthotopic pancreatic tumor model. Cancer Res. (2015) 75:289. doi: 10.1158/1538-7445.AM2015-289
210. Zhang L, Xu X, Su X. Noncoding RNAs in cancer immunity: functions, regulatory mechanisms, and clinical application. Mol Cancer. (2020) 19:48. doi: 10.1186/s12943-020-01154-0
211. Peng W, Wang Z, Fan H. LncRNA NEAT1 impacts cell proliferation and apoptosis of colorectal cancer via regulation of akt signaling. Pathol Oncol Res. (2017) 23:651–6. doi: 10.1007/s12253-016-0172-4
212. Yan K, Fu Y, Zhu N, Wang Z, Hong JL, Li Y, et al. Repression of lncRNA NEAT1 enhances the antitumor activity of CD8(+)T cells against hepatocellular carcinoma via regulating miR-155/Tim-3. Int J Biochem Cell Biol. (2019) 110:1–8. doi: 10.1016/j.biocel.2019.01.019
213. Ji J, Yin Y, Ju H, Xu X, Liu W, Fu Q, et al. Long non-coding RNA Lnc-Tim3 exacerbates CD8 T cell exhaustion via binding to Tim-3 and inducing nuclear translocation of Bat3 in HCC. Cell Death Dis. (2018) 9:478. doi: 10.1038/s41419-018-0528-7
214. Vigneau S, Rohrlich PS, Brahic M, Bureau JF. Tmevpg1, a candidate gene for the control of Theiler’s virus persistence, could be implicated in the regulation of gamma interferon. J Virol. (2003) 77:5632–8. doi: 10.1128/jvi.77.10.5632-5638.2003
215. Vickers NJ. Animal communication: when I’m calling you, will you answer too? Curr Biol. (2017) 27:R713–r5. doi: 10.1016/j.cub.2017.05.064
216. Vinay DS, Ryan EP, Pawelec G, Talib WH, Stagg J, Elkord E, et al. Immune evasion in cancer: Mechanistic basis and therapeutic strategies. Semin Cancer Biol. (2015) 35 Suppl:S185–s98. doi: 10.1016/j.semcancer.2015.03.004
217. Jiang R, Tang J, Chen Y, Deng L, Ji J, Xie Y, et al. The long noncoding RNA lnc-EGFR stimulates T-regulatory cells differentiation thus promoting hepatocellular carcinoma immune evasion. Nat Commun. (2017) 8:15129. doi: 10.1038/ncomms15129
218. Pei X, Wang X, Li H. LncRNA SNHG1 regulates the differentiation of Treg cells and affects the immune escape of breast cancer via regulating miR-448/IDO. Int J Biol Macromol. (2018) 118:24–30. doi: 10.1016/j.ijbiomac.2018.06.033
219. Wang L, Ye S, Wang J, Gu Z, Zhang Y, Zhang C, et al. HuR Stabilizes lnc-Sox5 mRNA to Promote Tongue Carcinogenesis. Biochem (Mosc). (2017) 82:438–45. doi: 10.1134/s0006297917040046
220. Chiossone L, Dumas P-Y, Vienne M, Vivier E. Natural killer cells and other innate lymphoid cells in cancer. Nat Rev Immunol. (2018) 18:671–88. doi: 10.1038/s41577-018-0061-z
221. Shin MH, Kim J, Lim SA, Kim J, Kim SJ, Lee KM. NK cell-based immunotherapies in cancer. Immune Netw. (2020) 20:e14. doi: 10.4110/in.2020.20.e14
222. Souza-Fonseca-Guimaraes F, Cursons J, Huntington ND. The emergence of natural killer cells as a major target in cancer immunotherapy. Trends Immunol. (2019) 40:142–58. doi: 10.1016/j.it.2018.12.003
223. Cichocki F, Cooley S, Davis Z, DeFor TE, Schlums H, Zhang B, et al. CD56dimCD57+NKG2C+ NK cell expansion is associated with reduced leukemia relapse after reduced intensity HCT. Leukemia. (2016) 30:456–63. doi: 10.1038/leu.2015.260
224. Fang P, Xiang L, Chen W, Li S, Huang S, Li J, et al. LncRNA GAS5 enhanced the killing effect of NK cell on liver cancer through regulating miR-544/RUNX3. Innate Immun. (2019) 25:99–109. doi: 10.1177/1753425919827632
225. Stein N, Berhani O, Schmiedel D, Duev-Cohen A, Seidel E, Kol I, et al. IFNG-AS1 enhances interferon gamma production in human natural killer cells. iScience. (2019) 11:466–73. doi: 10.1016/j.isci.2018.12.034
226. Wei MF, Gu ZS, Zheng LL, Zhao MX, Wang XJ. Long non-coding RNA GAS5 promotes natural killer cell cytotoxicity against gastric cancer by regulating miR-18a. Neoplasma. (2020) 67:1085–93. doi: 10.4149/neo_2020_191014N1034
227. Li S, Zhu A, Ren K, Li S, Chen L. IFNβ-induced exosomal linc-EPHA6-1 promotes cytotoxicity of NK cells by acting as a ceRNA for hsa-miR-4485-5p to up-regulate NKp46 expression. Life Sci. (2020) 257:118064. doi: 10.1016/j.lfs.2020.118064
228. Böttcher JP, Reis e Sousa C. The role of type 1 conventional dendritic cells in cancer immunity. Trends Cancer. (2018) 4:784–92. doi: 10.1016/j.trecan.2018.09.001
229. Gardner A, Ruffell B. Dendritic cells and cancer immunity. Trends Immunol. (2016) 37:855–65. doi: 10.1016/j.it.2016.09.006
230. Giovanelli P, Sandoval TA, Cubillos-Ruiz JR. Dendritic cell metabolism and function in tumors. Trends Immunol. (2019) 40:699–718. doi: 10.1016/j.it.2019.06.004
231. Nefedova Y, Huang M, Kusmartsev S, Bhattacharya R, Cheng P, Salup R, et al. Hyperactivation of STAT3 is involved in abnormal differentiation of dendritic cells in cancer. J Immunol. (2004) 172:464–74. doi: 10.4049/jimmunol.172.1.464
232. Zhang W, Yang M, Yu L, Hu Y, Deng Y, Liu Y, et al. Long non-coding RNA lnc-DC in dendritic cells regulates trophoblast invasion via p-STAT3-mediated TIMP/MMP expression. Am J Reprod Immunol. (2020) 83:e13239. doi: 10.1111/aji.13239
233. Zhang W, Zhou Y, Ding Y. Lnc-DC mediates the over-maturation of decidual dendritic cells and induces the increase in Th1 cells in preeclampsia. Am J Reprod Immunol. (2017) 77:e12647. doi: 10.1111/aji.12647
234. Zhuang L, Tian J, Zhang X, Wang H, Huang C. Lnc-DC regulates cellular turnover and the HBV-induced immune response by TLR9/STAT3 signaling in dendritic cells. Cell Mol Biol Lett. (2018) 23:43. doi: 10.1186/s11658-018-0108-y
235. Zhang M, Zheng Y, Sun Y, Li S, Chen L, Jin X, et al. Knockdown of NEAT1 induces tolerogenic phenotype in dendritic cells by inhibiting activation of NLRP3 inflammasome. Theranostics. (2019) 9:3425–42. doi: 10.7150/thno.33178
236. Xin J, Li J, Feng Y, Wang L, Zhang Y, Yang R. Downregulation of long noncoding RNA HOTAIRM1 promotes monocyte/dendritic cell differentiation through competitively binding to endogenous miR-3960. Onco Targets Ther. (2017) 10:1307–15. doi: 10.2147/ott.S124201
237. Liu J, Zhang X, Chen K, Cheng Y, Liu S, Xia M, et al. CCR7 chemokine receptor-inducible lnc-dpf3 restrains dendritic cell migration by inhibiting HIF-1α-mediated glycolysis. Immunity. (2019) 50:600–15.e15. doi: 10.1016/j.immuni.2019.01.021
238. Kan JY, Wu DC, Yu FJ, Wu CY, Ho YW, Chiu YJ, et al. Chemokine (C-C motif) ligand 5 is involved in tumor-associated dendritic cell-mediated colon cancer progression through non-coding RNA MALAT-1. J Cell Physiol. (2015) 230:1883–94. doi: 10.1002/jcp.24918
239. Di Martino MT, Riillo C, Scionti F, Grillone K, Polerà N, Caracciolo D, et al. miRNAs and lncRNAs as novel therapeutic targets to improve cancer immunotherapy. Cancers (Basel). (2021) 13:1587. doi: 10.3390/cancers13071587
240. Daveri E, Vergani E, Shahaj E, Bergamaschi L, Magra S, Dosi M, et al. microRNAs shape myeloid cell-mediated resistance to cancer immunotherapy. Front Immunol. (2020) 11:1214. doi: 10.3389/fimmu.2020.01214
241. Li Z, Feng C, Guo J, Hu X, Xie D. GNAS-AS1/miR-4319/NECAB3 axis promotes migration and invasion of non-small cell lung cancer cells by altering macrophage polarization. Funct Integr Genomics. (2020) 20:17–28. doi: 10.1007/s10142-019-00696-x
242. Shabgah A, Salmaninejad A, Thangavelu L, Alexander M, Yumashev A, Goleij P, et al. The role of non-coding genome in the behavior of infiltrated myeloid- derived suppressor cells in tumor microenvironment; a perspective and state-of-the-art in cancer targeted therapy. Prog Biophys Mol Biol. (2020) 161:17–26. doi: 10.1016/j.pbiomolbio.2020.11.006
243. Quirico L, Orso F, Esposito CL, Bertone S, Coppo R, Conti L, et al. Axl-148b chimeric aptamers inhibit breast cancer and melanoma progression. Int J Biol Sci. (2020) 16:1238–51. doi: 10.7150/ijbs.39768
244. Williams G, Pickard M. Long non-coding RNAs: new opportunities and old challenges in cancer therapy. Transl Cancer Res. (2016) 5:S564–S6. doi: 10.21037/tcr.2016.09.04
245. Martinez-Castillo M, ME A, López-Berestein G, Amero P, Rodríguez-Aguayo C. An overview of the immune modulatory properties of long non-coding RNAs and their potential use as therapeutic targets in cancer. Noncoding RNA. (2023) 9:70. doi: 10.3390/ncrna9060070
246. Su H, Hailin Z, Dongdong L, Jiang Y, Shuncheng H, Shun Z, et al. Long non-coding RNA LINC01018 inhibits human glioma cell proliferation and metastasis by directly targeting miRNA-182-5p. J Neurooncol. (2022) 160:67–78. doi: 10.1007/s11060-022-04113-5
247. Wei C, Wu W, Hou X, Yuan S. Study on oil distribution and oil content of oil bath lubrication bearings based on MPS method. TRIBOL T. (2022) 65:942–51. doi: 10.1080/10402004.2022.2113193
248. Xu J, Wang J, Zhao M, Li C, Hong S, Zhang J. LncRNA LINC01018/miR-942-5p/KNG1 axis regulates the Malignant development of glioma in vitro and in vivo. CNS Neurosci Ther. (2023) 29:691–711. doi: 10.1111/cns.14053
249. Wang T, Tang Y, Tao Y, Zhou H, Ding D. Nucleic acid drug and delivery techniques for disease therapy: Present situation and future prospect. Interdiscip Med. (2024) 2:e20230041. doi: 10.1002/INMD.20230041
250. Varier KM, Dhandapani H, Liu W, Song J, Wang C, Hu A, et al. An immunotherapeutic approach to decipher the role of long non-coding RNAs in cancer progression, resistance and epigenetic regulation of immune cells. J Exp Clin Cancer Res. (2021) 40:242. doi: 10.1186/s13046-021-01997-5
251. Alzhrani R, Alsaab HO, Petrovici A, Bhise K, Vanamala K, Sau S, et al. Improving the therapeutic efficiency of noncoding RNAs in cancers using targeted drug delivery systems. Drug Discovery Today. (2020) 25:718–30. doi: 10.1016/j.drudis.2019.11.006
252. Xin X, Li Q, Fang J, Zhao T. LncRNA HOTAIR: A potential prognostic factor and therapeutic target in human cancers. Front Oncol. (2021) 11:679244. doi: 10.3389/fonc.2021.679244
253. Cheng X, Li Z, Shan R, Li Z, Wang S, Zhao W, et al. Modeling CRISPR-Cas13d on-target and off-target effects using machine learning approaches. Nat Commun. (2023) 14:752. doi: 10.1038/s41467-023-36316-3
254. Sangeeth A, Malleswarapu M, Mishra A, Gutti RK. Long non-coding RNA therapeutics: recent advances and challenges. Curr Drug Targets. (2022) 23:1457–64. doi: 10.2174/1389450123666220919122520
255. Toden S, Zumwalt TJ, Goel A. Non-coding RNAs and potential therapeutic targeting in cancer. Biochim Biophys Acta Rev Cancer. (2021) 1875:188491. doi: 10.1016/j.bbcan.2020.188491
256. Fathi Dizaji B. Strategies to target long non-coding RNAs in cancer treatment: progress and challenges. Egypt J Med Hum Genet. (2020) 21:41. doi: 10.1186/s43042-020-00074-4
Keywords: cancer immunotherapy, lncRNA, therapeutic target, cancer biomarkers, immune response
Citation: Arshi A, Mahmoudi E, Raeisi F, Dehghan Tezerjani M, Bahramian E, Ahmed Y and Peng C (2024) Exploring potential roles of long non-coding RNAs in cancer immunotherapy: a comprehensive review. Front. Immunol. 15:1446937. doi: 10.3389/fimmu.2024.1446937
Received: 10 June 2024; Accepted: 05 August 2024;
Published: 27 August 2024.
Edited by:
Simona Kranjc Brezar, Institute of Oncology Ljubljana, SloveniaReviewed by:
Yao Luo, Sichuan University, ChinaPavel Loskot, The Zhejiang University-University of Illinois at Urbana-Champaign Institute, United States
Copyright © 2024 Arshi, Mahmoudi, Raeisi, Dehghan Tezerjani, Bahramian, Ahmed and Peng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chun Peng, Y3BlbmdAeW9ya3UuY2E=
†These authors have contributed equally to this work
 Asghar Arshi
Asghar Arshi Esmaeil Mahmoudi2†
Esmaeil Mahmoudi2† Masoud Dehghan Tezerjani
Masoud Dehghan Tezerjani Elham Bahramian
Elham Bahramian