
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Immunol. , 10 December 2024
Sec. T Cell Biology
Volume 15 - 2024 | https://doi.org/10.3389/fimmu.2024.1445341
The functional specialization of CD4 T lymphocytes into various subtypes, including TH1 and TFH cells, is crucial for effective immune responses. TFH cells facilitate B cell differentiation within germinal centers, while TH1 cells are vital for cell-mediated immunity against intracellular pathogens. Integrin α4, a cell surface adhesion molecule, plays significant roles in cell migration and co-stimulatory signaling. In this study, we investigated the role of Integrin α4 in regulating TFH and TH1 cell populations during acute viral infection using CRISPR-Cas9 gene editing. To effectively delete the Itga4 in primary mouse CD4 T cells, we selected various combinations of crRNAs and generated ribonucleoprotein complexes with fluorochrome-conjugated tracrRNAs and Cas9 proteins. These crRNA pairs enhanced gene deletion by generating deletions in the gene. By analyzing the effects of Itga4 deficiency on TFH and TH1 cell differentiation during acute LCMV infection, we found that optimized crRNA pairs significantly increased the TH1 cell population. Our results highlight the importance of selecting and combining appropriate crRNAs for effective CRISPR-Cas9 gene editing in primary CD4 T cells. Additionally, our study demonstrates the role of Integrin α4 in regulating the differentiation of CD4 T cells, suggesting the potential molecular mechanisms driving T cell subset differentiation through integrin targeting.
The adaptive immune response relies heavily on the functional specialization of CD4 T lymphocytes, which can differentiate into various subtypes, including T helper type 1 (TH1), TH2, TH17, regulatory T (TREG), and follicular helper T (TFH) cells (1). Among these, TFH cells play a crucial role in the formation of germinal centers (GCs) and the differentiation of B cells, which are essential for humoral immunity (2). TFH cells are characterized by the expression of C-X-C chemokine receptor 5 (CXCR5), which facilitates their migration towards the B cell follicles, and B-cell lymphoma 6 (Bcl6), a transcription factor (TF) that upregulates TFH-associated genes, including cytokines and costimulatory molecules, while repressing genes that inhibit TFH cell development (2–4). Conversely, TH1 cells are essential for cell-mediated immunity and are involved in the defense against intracellular pathogens, such as viruses and certain bacteria. Additionally, they play a role in anti-tumor immunity (5, 6). Maintaining a balance between TH1 and TFH cell populations is critical for an effective immune response, ensuring both adequate antibody production and effective cellular immunity.
Integrins, which are cell surface adhesion molecules, are critical for cell-cell interactions, migration, and co-stimulatory signaling (7–9). Integrin α4, in particular, forms heterodimers with Integrin β1 (VLA-4) or Integrin β7 (LPAM-1), and these complexes play significant roles in transendothelial migration and immune cell localization. VLA-4 and LPAM-1 interact with vascular cell adhesion molecule 1 (VCAM-1) and mucosal addressin cell adhesion molecule 1 (MAdCAM-1), respectively, facilitating adhesion and migration signals essential for immune cell trafficking (10–12). Furthermore, Integrin α4β1 (VLA-4) is known to promote TH1 responses by upregulating interferon (IFN)-γ expression and suppressing interleukin (IL)-4, a key cytokine of TH2 cells (13).
CRISPR/Cas9 gene editing has emerged as a powerful tool for investigating gene function and regulation in various biological contexts, including immune cell differentiation and function (14). This technology utilizes guide RNAs to direct the Cas9 nuclease to specific genomic loci, where it induces double-strand breaks. These breaks are typically repaired by the non-homologous end joining (NHEJ) pathway, resulting in insertions or deletions (InDels) that can disrupt gene function (15, 16). The efficiency of gene disruption using CRISPR/Cas9 can be enhanced by employing multiple CRISPR RNAs (crRNAs) targeting different sites within the same gene, thereby increasing the likelihood of generating large deletions and achieving complete gene disruption (17–21).
In the current study, we investigated the role of Integrin α4/CD49d, encoded by Itga4, in regulating the balance between TFH and TH1 cell populations during acute viral infection. We used CRISPR/Cas9 gene editing to delete Itga4 in primary mouse CD4 T cells. We adopted an advanced crRNA/Cas9 ribonucleoprotein (crRNP) complex-mediated gene editing strategy to analyze Itga4 disruption efficiency and nucleotide deletion patterns based on the length of base pairs between target sites using two crRNAs. We then examined the effects of Itga4 deficiency on TFH and TH1 cell differentiation during acute virus infection. Our study provides insights into the molecular mechanisms driving T cell subset differentiation, highlighting the potential role of Integrin α4 in regulating TH1 cell proliferation or differentiation during acute viral infection. Furthermore, we present efficient crRNP-mediated gene deletion methods using Itga4 as an example gene in mouse primary CD4 T cells for their use in in vivo experiments.
C57BL/6 mice (6 weeks old) were acquired from Orient Bio in Korea. CD45.1+ SMARTA mice were generously provided by Yoon Soo Choi at Seoul National University College of Medicine. Specific-pathogen-free male or female donor mice (6-11 weeks old) were used for experiments. All procedures involving animals were conducted in compliance with the protocol approved by the Institutional Animal Care and Use Committee of the College of Medicine at the Catholic University of Korea (CUMC-2023-0268).
Spleens from WT C57BL/6 or CD45.1+ SMARTA mice were collected. CD4 T cells were isolated through a negative selection process according to the manufacturer’s protocol (EasySep™ Mouse CD4 T cell Isolation kit, STEMCELL Technologies). To isolate naive CD4 T cells, biotinylated anti-CD44 (IM7, Biolegend) and anti-CD25 (PC61, Biolegend) were added to the Isolation Cocktail of the Mouse CD4 T cell Isolation kit. After isolation, cells were counted, and the proportion of CD4 T cells was assessed by flow cytometry. The purity of the isolated CD4 T cells exceeded 95%. The isolated WT or SMARTA CD4 T cells were used for in vitro CD4 T cell culture or in vivo adoptive transfer experiments.
The tissue culture plate (24-well) was coated with 330 µl of 8 µg/ml anti-CD3ϵ (145-2C11; BioXCell) and 8 ug/mL anti-CD28 (37.51; BioXCell) in PBS (Corning). Isolated CD4 T cells were plated at 0.5 × 106 cells/well in R10 media (RPMI 1640 with 10% Fetal Bovine Serum (Hyclone), 100 U/ml Penicillin, 100 µg/ml Streptomycin, GlutaMAX™), supplemented with 50 µM 2-Mercaptoethanol (2-ME, Gibco), Non-Essential Amino Acids (NEAA, Gibco) and 2 ng/mL recombinant human IL-7 (rhIL-7, Peprotech), and cultured for 2 days.
Various crRNAs targeting Itga4 were selected based on their CHOP-CHOP ranking and the distances between crRNAs (https://chopchop.cbu.uib.no) (Supplementary Table 1). A crRNA targeting Cd8a was used as a control. crRNAs, ATTO-550-conjugated trans-activating CRISPR RNA (tracrRNA), and S.p Cas9 Nuclease V3 were purchased from Integrated DNA Technologies (IDT). crRNA and tracrRNA were duplexed by heating at 95 °C for 5 minutes and incubated at room temperature for 1 hour in the dark. crRNP complexes were generated by mixing crRNA-tracrRNA duplexes (40~240 pmol range) and Cas9 protein (26.6~80 pmol range) for 10 minutes at room temperature. In vitro stimulated CD4 T cells were harvested, resuspended in 20 µl of primary cell nucleofector solution (P4 Primary Cell 4D-Nucleofector™, Lonza), and mixed with the prepared crRNPs complexes. The crRNP-cell mixtures were then transferred to a 16-well Nucleocuvette stirp and electroporated using the CM137 (for in vitro activated CD4 T cells) or DS137 (for naive CD4 T cell) protocol with the 4D-Nucleofector™ (Lonza). The transfected cells were cultured in R10 media supplemented with 50 µM 2-ME, NEAA, and 10 ng/mL recombinant human IL-2 (rhIL-2, Peprotech) without TCR stimulation. Two days later, the transfected CD4 T cells were sorted by FACS Aria fusion (BD Bioscience). The sorted cells were further used for validation of the efficiency of gene delivery by flow cytometry and gene editing by Inference of CRISPR Edits (ICE) analysis or used for adoptive transfer experiments. Gene deletion efficiency, InDel proportion, and InDel contributions by each crRNA from triplicate wells were measured initially, with minimal variations observed between triplicate wells (Supplementary Figures 1A–D). We conducted two to four independent experiments for further analysis.
Genomic DNA of the sorted CD4 T cells was prepared using QIAamp DNA Mini Kit according to the manufacturer’s protocol (Qiagen). PCR was performed for crRNA target site amplification of crRNAs using Q5 polymerase (NEB Biolabs) following the manufacturer’s protocol. Sanger sequencing was performed by Macrogen or Bionics (Korea). ICE analysis was conducted using Synthego online platform (https://ice.synthego.com). The InDel contributions were calculated based on the sum of InDel percentages generated by the crRNA, as provided in the ICE analysis report (Supplementary Figure 1A). For example, the InDel contributions of crItga4 B (g1), crItga4 C (g2), and the deletions in the gene resulting from crItga4 B and crItga4 C (referred to as “fragment” from paired crRNAs) were 20 (11 + 4 + 3 + 1+1), 4 (2 + 1 + 1), and 76 (36 + 34 + 2 + 1+1 + 1+1), respectively, when using the combination of crItga4 B and crItga4 C (Supplementary Figure 1A).
1 × 103 freshly isolated or 3 × 104 sorted crRNP+ CD45.1+ SMARTA CD4 T cells were adoptively transferred into C57BL/6 recipient mice via intravenous injection into the retroorbital sinus. Recipient mice were then injected intraperitoneally with 2 × 105 pfu of LCMVArm in plain DMEM either one day later (for freshly isolated CD4 T cells) or 3 to 4 days later (for crRNP+ CD4 T cells) following the adoptive transfer.
Naive CD4 T cells were electroporated with crRNP and cultured for 2 days in R10 media supplemented with 50 µM 2-ME, NEAA, and 2 ng/mL rhIL-7. For TH1 differentiation, 1.0 × 106 cells/well were cultured in 24-well plates pre-coated anti-CD3 and soluble anti-CD28 antibodies, along with 10 ng/mL recombinant murine IL-12 p70 (rmIL-12, Peprotech) and 10 µg/mL anti-IL-4 (11B11; BioXcell) for 3 days, as previously described (22). For the TH0 control, cells were cultured without rmIL-12 and anti-IL-4 antibody. After 3 days, cells were split at a 1:2 ratio and cultured in fresh rhIL-2 medium for an additional 2 days. For cytokine analysis, cells were stimulated with 50 ng/ml PMA and 1 µM ionomycin for 5 hours. To assess cell proliferation, cells cultured for 3 days under TH0 and TH1 differentiation conditions were labeled with CellTrace Violet (CTV; CellTrace™ Violet Cell Proliferation Kit, Invitrogen), and then cultured in fresh rhIL-2 medium for an additional 2 days. Cytokine expression and CTV levels were measured by flow cytometry.
Surface staining for flow cytometry was performed using monoclonal antibodies against CD8 (53-6.7; APC-Cy7), B220 (RA3-6B2; APC-Cy7), CD4 (RM-4; BV510 or PE-Cy7), CD45.1 (A20; BV605 or APC-Cy7), TCRVα2 (B20.1; FITC), Integrin α4 (R1-2; APC or PE), Integrin β1 (HMb1-1; APC-eF780), Integrin β7 (FIB504; PE.CF594), CD44 (IM7; PE-Cy7), SLAM (TC15-12F12.2; PerCP-Cy5.5 or APC), PD-1 (J43; PE), biotin-conjugated CXCR5 (L138D7) and BV421, PE-Cy7 or BV650-conjugated streptavidin. The antibody cocktail was diluted in FACS buffer (PBS with 0.5% Bovine Serum Albumin). Staining was performed in FACS buffer at 4°C for 30 minutes. For CXCR5 staining, biotinylated anti-CXCR5 was used, followed by a 30-minute incubation at 4°C with BV421, PE-Cy7 or BV650-conjugated streptavidin. Live/dead cell staining was carried out using Fixable Viability Dye eFlour™780 (eBioscience). Intracellular cytokine staining was conducted with monoclonal antibody IFN-γ (XMG1.2; AF700) using the Fixation/Permeabilization kit (BD Biosciences). Intranuclear staining for the transcription factor was performed with a monoclonal antibody with Bcl6 (K112-91; APC), T-bet (4B10; PerCP-Cy5.5), GATA3 (16E10A23; APC), FOXP3 (FJK-16s; PerCP-Cy5.5), Ki-67 (B56; PE-TexasRed) utilizing the Foxp3/Transcription Factor Staining Buffer kit (eBioscience). Stained cells were analyzed using a FACS Canto II or LSRFortessa (BD Biosciences), and data analyses were conducted with FlowJo software v.10.10.0 (FlowJo). The gating strategy used to identify TFH and TH1 populations is shown in Supplementary Figure 2.
Transcripts per Million (TPM) values of Bcl6 and Itga4 for WT TH1 and WT TFH cells (GSE140187) were obtained from RNA-seq data previously published (24).
The UCSC tracks and peaks at the ITGA4 locus from BCL6 ChIP–seq data of human tonsillar GC TFH cells (GSE59933) were retrieved from earlier publications (23, 24). Peaks were annotated according to the RefSeq database. Peaks located within ±2 kb of the transcription start site were classified as promoter peaks, those within ±2 kb of the transcription end site were defined as 3′ end peaks, and peaks located more than 2 kb away from genes were categorized as intergenic, following methods described in previous work (24).
We previously showed that TFH cells exhibited lower Itga4 RNA expression compared to TH1 cells during acute lymphocytic choriomeningitis virus (LCMV) infection (24). To investigate whether the protein expression of Integrin α4 correlates with the RNA expression changes in different CD4 T cell populations, including TFH and TH1 cells, we utilized adoptive transfer of antigen-specific CD4 T cells in an acute virus infection mouse model. The mice were adoptively transferred with I-Ab-restricted LCMV glycoprotein (gp)-specific SMARTA CD4 T cells, followed by infection with LCMV Armstrong (LCMVArm) (Figure 1A). Consistent with observations from the previous study (24), LCMVArm infection generated CXCRlo SLAMhi TH1 and CXCR5hi SLAMlo TFH populations in the spleen (Figure 1B) on day 7 post-infection. The expression level of Integrin α4 was measured in each population. Both TH1 and TFH populations expressed higher amounts of Integrin α4 protein than naive CD4 T cells. Consistent with the RNA expression data, TFH cells showed significantly lower expression of Integrin α4 than TH1 cells (Figure 1B).
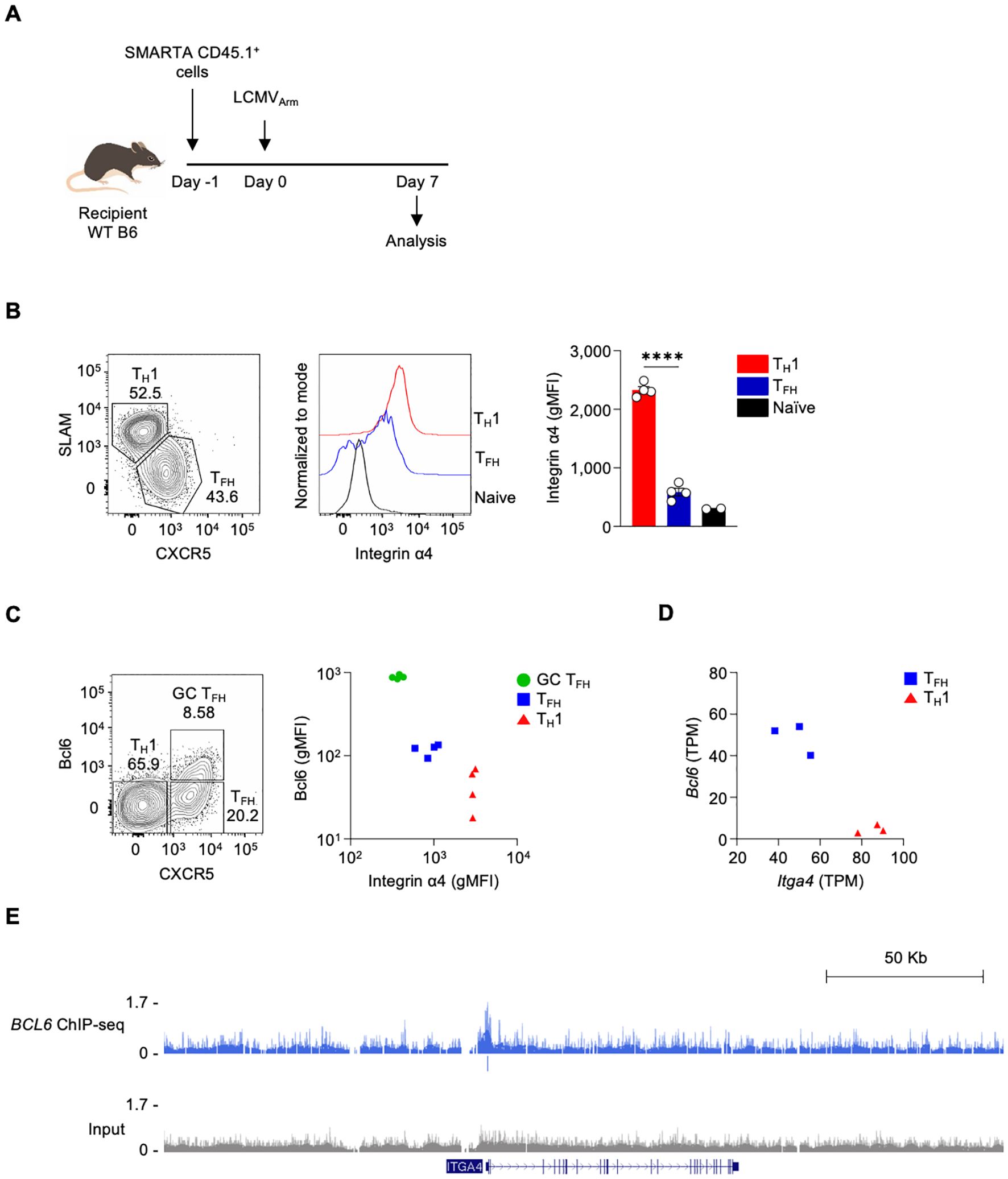
Figure 1. The expression of integrin α4 is downregulated in TFH cells (A) Schematic diagram of the adoptive transfer of SMARTA CD4 T cells in LCMVArm infection model. Isolated splenic SMARTA CD45.1+ CD4 T cells were adoptively transferred into C57BL/6 mice, followed by LCMVArm infection, and analyzed on day 7 post-infection. (B) TFH and TH1 populations were analyzed by flow cytometry. Integrin α4 expression levels in the TFH and TH1 populations are shown. (C) Bcl6 and Integrin α4 expression levels in the GC TFH, TFH, and TH1 populations are shown. A representative of two independent experiments is shown, and each dot represents one mouse (n=4). (D) Gene expression levels of Bcl6 and Itga4 in TFH and TH1 cells from RNA-seq data of LCMVArm-infected mice. (E) Genome browser tracks display a BCL6 ChIP–Seq peak at ITGA4 locus, with peak annotations shown the track. Results are presented as mean ± SEM and were analyzed using an unpaired two-tailed Student’s t-test. ****p < 0.0001.
As Itga4 was observed as a potential direct Bcl6 target gene (24), we analyzed the correlation between the expression of Bcl6 and Integrin α4 in GC TFH, TFH, and TH1 populations. GC TFH, which are the highest Bcl6-expressing cells, express the lowest amount of Integrin α4; in contrast, TH1, which are the lowest Bcl6-expressing cells, express the highest amount of Integrin α4 (Figure 1C). Additionally, RNA-seq analysis revealed a negative correlation between Bcl6 and Itga4 mRNA expression (Figure 1D) (24). To assess whether Bcl6 binds to the regulatory region of Itga4, we analyzed BCL6 ChIP-seq data from human tonsillar GC TFH cells (23, 24). Indeed, BCL6 binding was detected at the ITGA4 promoter region, indicating that Bcl6 may directly repress Itga4 expression (Figure 1E). These results indicate differential expression of Integrin α4 across distinct T cell subsets following LCMVArm infection, with the lowest expression observed in GC TFH cells, demonstrating a negative correlation between Bcl6 and Integrin α4 expression. This differential expression pattern suggests a potential role for Integrin α4 in the differentiation, functional specialization, or migratory behavior of these CD4 T cell subsets during the immune response to LCMVArm infection.
To investigate the role of Integrin α4 in different CD4 T cell populations in LCMV infection, we employed acute gene deletion using CRISPR-Cas9 gene editing (17, 24). Mouse splenic CD4 T cells were isolated and stimulated with anti-CD3/CD28 and IL-7 for two days in vitro, followed by electroporation with the crRNP complex, and the delivery efficiency and deletion efficiency were analyzed two days later (Figure 2A). The tracrRNA+ provides electroporation efficiency as the crRNP complex contains fluorescence-conjugated tracrRNA. The tracrRNA+ population in the crRNP electroporated group was higher than 98%, while it was not observed in no electroporation control (No EP), indicating that the electroporation of the crRNP complex was successful (Figure 2B).
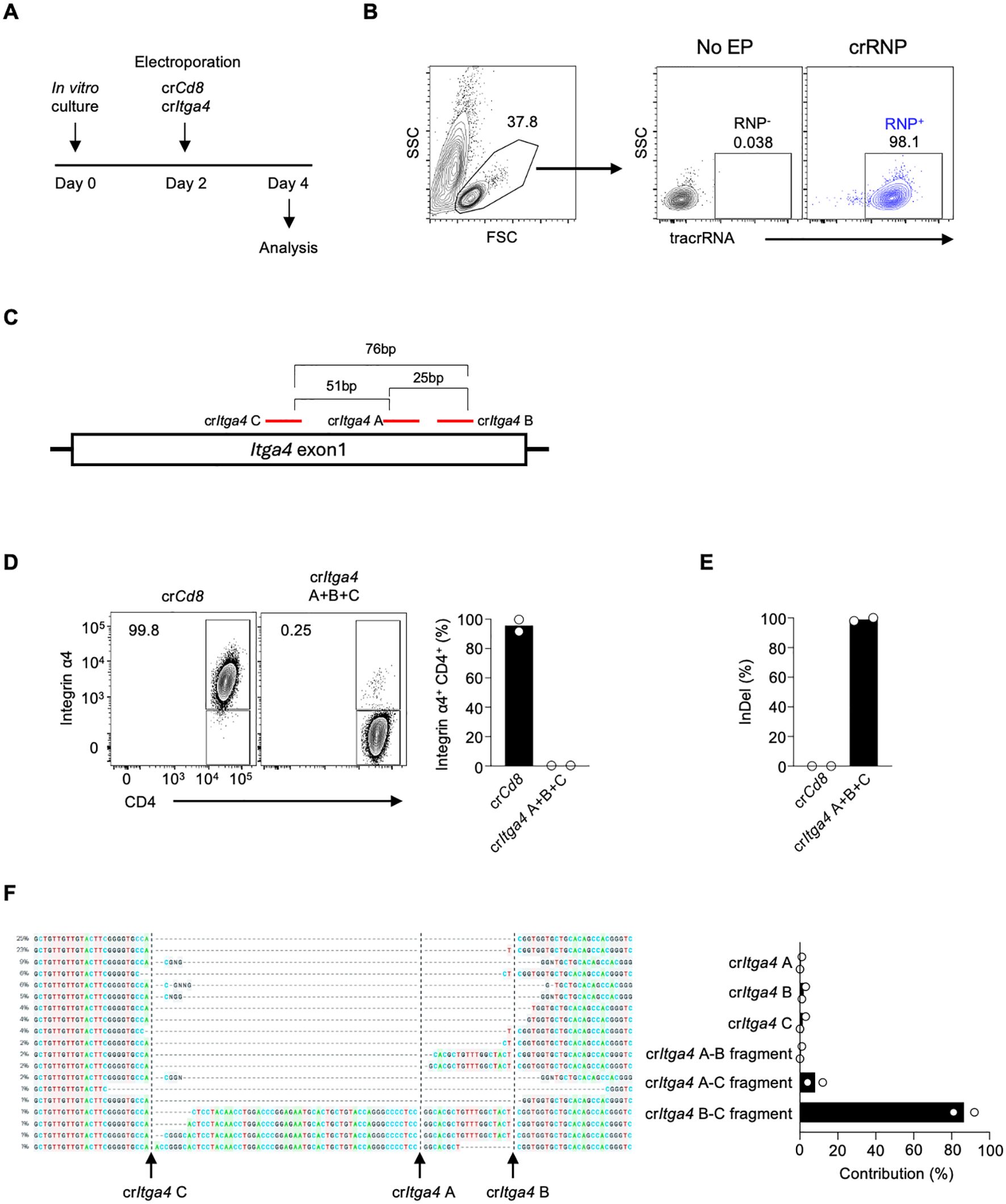
Figure 2. Combining multiple crRNAs efficiently deletes integrin α4 in CD4 T cells (A) Schematic diagram of the in vitro culture and electroporation protocol. Mouse splenic CD4 T cells were isolated and stimulated with anti-CD3/CD28 and IL-7 for two days. On day 2, cells were electroporated with either control crRNP (crCd8) or crRNP targeting Itga4 (crItga4), followed by analysis on day 4. (B) Electroporation efficiency of crRNP delivery was measured by flow cytometry. (C) Diagram of crRNA target sites in Itga4 exon 1. (D) Flow cytometry analysis of Integrin α4 expression in CD4 T cells. A mixture crItga4 A+B+C was generated with 80 pmol each of crItga4 A, B, and C (total 240 pmol) and used for electroporation (E) ICE analysis of InDel percentage. The bar graph shows the percentage of InDels in the crCd8 and crItga4 groups. (F) Sequencing analysis of InDel contributions by each crRNA. The alignment shows the contribution of each crRNA and its combinations to the observed gene editing. The contributions represent the inferred sequences present in the crRNP population and their relative proportions. Actual cut sites are represented by black vertical dotted lines. Arrows mark the double-strand breakage sites by each crRNA. Data shown are representative of two independent experiments.
Previous studies have shown that using combinations of dual or triple crRNAs enhances gene deletion efficiency in human primary T cells by generating insertion and deletion (InDel)-mediated fragment deletions (17, 19, 20). Initially, we designed three different crRNAs targeting Itga4 (crItga4 A, B, and C; Supplementary Table 1). These crRNAs all target exon 1 of the Itga4 gene, with distances between crRNAs being 25 bp (crItga4 A – crItga4 B), 51 bp (crItga4 A – crItga4 C), and 76 bp (crItga4 B – crItga4 C) (Figure 2C). To maximize gene deletion efficiency, we used a mixture of all three crRNAs (crItga4 A+B+C). The Integrin α4 positive population in the control crCd8 group was higher than 95%, but in crItga4 A+B+C group, it decreased to less than 1%, indicating that a mixture of crRNAs targeting Itga4 is highly efficient in disruption of Itga4 gene expression (Figure 2D).
We further analyzed DNA editing efficacy using Inference of CRISPR Edits (ICE) analysis, which can locate the site of InDel and calculate the proportion of InDel by sequencing PCR products that include crRNA target sites. The InDel percentage of crItga4 A+B+C was higher than 90%, indicating crItga4 A+B+C was highly efficient in generating DNA editing (Figure 2E). Interestingly, although a mixture of three different crRNAs was delivered into the cells, each crRNA targeting Itga4 contributed differently to the gene editing. Most of the InDel was contributed by crItga4 B and crItga4 C, which can generate around a 76-bp fragment deletion (Figure 2F). Although crItga4 A was ranked as the top crRNA by CHOP CHOP, the contributions of a crItga4 A and fragments containing crItga4 A (crItga4 A–B or crItga4 A–C fragment) were less than 5% (Figure 2F). It is possible that the effect of crItga4 A could not be measured because crItga4 A targets a site in the middle between crItga4 B and crItga4 C. These findings suggest that using a pair of crRNAs may be more efficient than using a combination of three crRNAs if the pair is sufficient to effectively delete the target gene.
Given that crRNA-induced fragmentation and their locations are important for enhancing gene deletion efficiency, we further tested additional crRNAs targeting the Itga4 gene, including crItga4 E targeting exon 2 to generate fragments of varying lengths (Figure 3A; Supplementary Table 1). We also investigated if a lower amount of crRNP could suffice for effective gene deletion. Mouse splenic CD4 T cells were electroporated with either a single (40 pmol) or a mixture of dual crRNAs (total 80 pmol), which is one-sixth or one-third of the original 240 pmol total crRNA, respectively (Figure 3B). The use of single crItga4 B completely abolished Integrin α4 expression, whereas the efficiencies of crItga4 C, D, or E were similar, leaving 11~18% of Integrin α4+ populations (Figure 3B). Notably, combining two crRNAs, crItga4 C+D or crItga4 C+E, significantly enhanced disruption efficiency, as evidenced by less than 5% of the Integrin α4+ population remaining (Figure 3B).
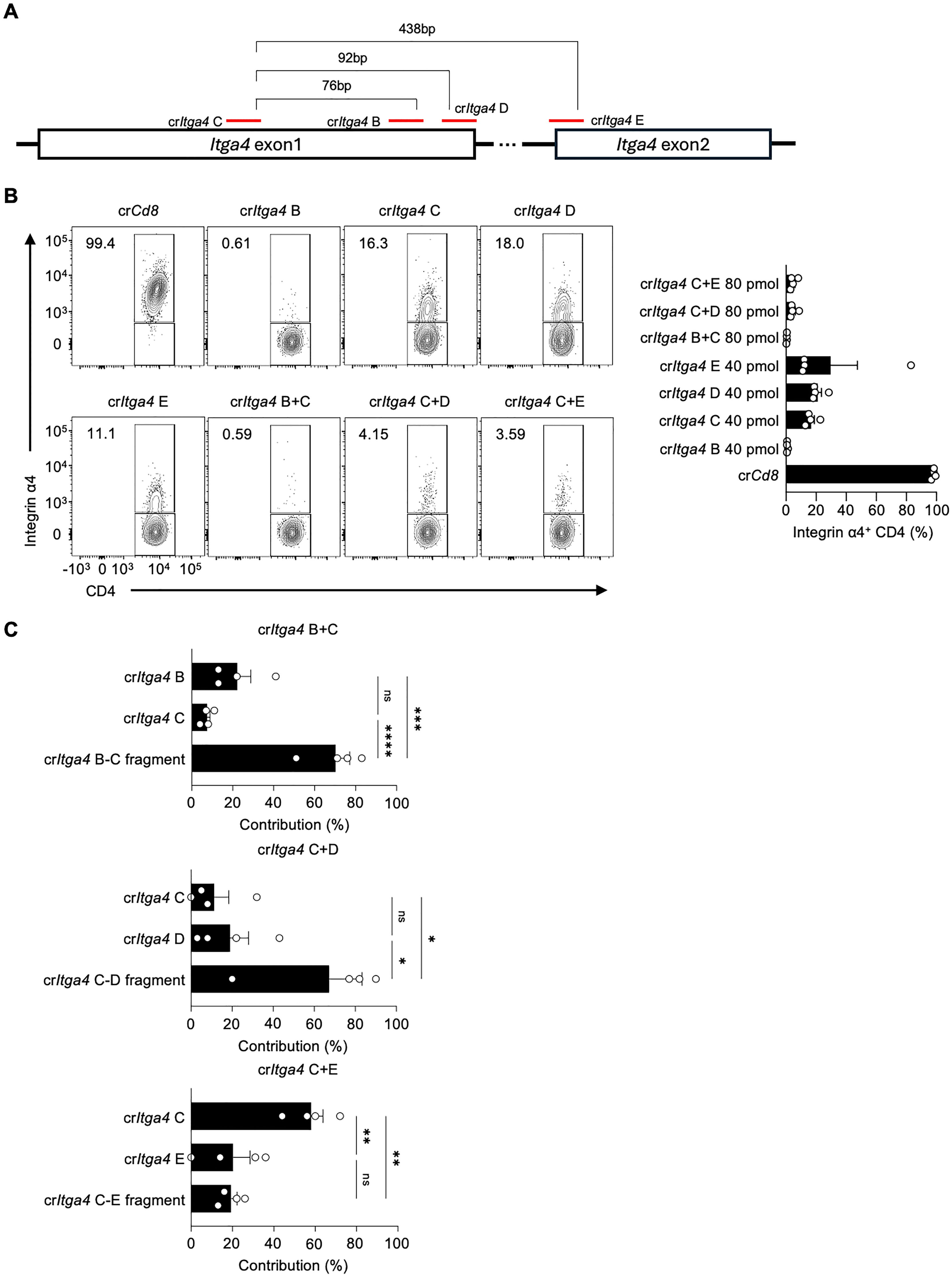
Figure 3. Combinations of proximal crRNA pairs efficiently delete integrin α4 in CD4 T cells (A) Diagram of crRNA target sites in Itga4 exons 1 and 2. (B) Flow cytometry analysis of Integrin α4 expression in CD4 T cells after electroporation with single or paired crRNAs. Single crItga4 was used at 40 pmol, and a mixture crItga4 pair was generated with 40 pmol of each crItga4 (total 80 pmol). The bar graph quantifies the percentage of Integrin α4+ CD4 T cells for each condition. (C) Sequencing analysis of InDel contributions of crRNAs to the overall InDel profile of crRNA pairs. Data shown are representative of four independent experiments. Results are presented as mean ± SEM and were analyzed using one-way ANOVA. ns, non-significant; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
We examined the InDel contributions of crRNAs in the mixture groups in more detail. Intriguingly, although crItga4 C+E, with a distance of 438 bp between the two crRNAs, generated almost complete deletion of Integrin α4 expression, the InDel contribution was primarily from crItga4 C rather than from the crItga4 C–E fragment (Figure 3C). This suggests that the fragmentation effect may be influenced by the distance between the two crRNAs. Among the crRNA mixtures, crItga4 C+D produced the most synergistic effect on gene disruption and fragment generation (Figures 3B, C). These results demonstrate the importance of crRNA selection and combination for efficient gene deletion, providing insights into optimizing CRISPR-Cas9 strategies for targeted gene editing.
To assess the role of Itga4 deficiency in CD4 T cells during acute viral infection, we adoptively transferred SMARTA CD4 T cells, either transfected with crItga4 C+D or control crCd8, into WT B6 recipient mice. Following LCMVArm infection, we analyzed the cells on day 6 post-infection (Figure 4A). Most cells from the crItga4 group were Integrin α4-negative during viral infection (Supplementary Figure 3A). Interestingly, the lack of Integrin α4 also led to a decrease in expression of its heterodimeric partners, Integrin β1/CD29 and Integrin β7 (Supplementary Figure 3B). This outcome likely results from the process by which integrins are translated into proteins, paired with their partner integrin subunits to form a heterodimer before being expressed on the cell surface (8).

Figure 4. Deficiency of Itga4 in CD4 T cells increases the accumulation of TH1 cells in the spleen during acute viral infection (A) SMARTA CD4 T cells were transfected with either crCd8 or crItga4 C+D, adoptively transferred into C57BL/6 recipient mice, followed by LCMVArm infection, and analyzed on day 6 post-infection. A mixture of crItga4 C+D was generated with 40 pmol of each crItga4 (total 80 pmol). (B) TFH and TH1 populations were analyzed by flow cytometry. (C) Expression levels of CXCR5 were calculated in TFH and TH1 cells. (D) Cell numbers of TFH and TH1 populations were calculated. (E) Comparative cell numbers of TFH and TH1 populations within the same mouse were calculated. The graph shows the cell numbers of TFH and TH1 populations in individual mice receiving either control crCd8 or crItga4 C+D transfected SMARTA CD4 T cells. A representative of two independent experiments is shown, and each dot represents one mouse (n=7). Results are presented as mean ± SEM and were analyzed by using unpaired (B–D, F) or paired (E) two-tailed Student’s t-test. **p < 0.01; ***p < 0.001.
We initially hypothesized that the deficiency of Itga4 might increase the TFH population since its expression level is downregulated in TFH cells (Figure 1). In stark contrast, a significant decrease in the proportion of CXCR5hi SLAMlo TFH cells was observed in the crItga4 group compared to the crCd8 control group, whereas the proportion of CXCR5lo SLAMhi TH1 cells was increased (Figure 4B). Furthermore, CXCR5 expression was significantly reduced in both TFH and TH1 cells in the crItga4 group, while PD-1 levels remained unchanged (Figure 4C; Supplementary Figure 4A). Notably, Bcl6 expression was also reduced in TFH (Supplementary Figure 4B), whereas the levels of T-bet (the lineage defining TF of TH1), GATA-3 (TH2), and Foxp3 (TREG) remained unchanged in the TH1 population in LCMV-infected mice (Supplementary Figures 4B, C). These findings suggest that Integrin α4 signaling may play a role in inducing Bcl6 and CXCR5, indicating that Itga4 deficiency may alter TFH phenotypic characteristics.
To further investigate changes in the proportions of TFH and TH1 cells, we quantified the cell numbers in each population. There was no significant difference in the total number of TFH cells between the two groups. However, the crItga4 group showed an apparent increase in the number of TH1 cells (Figure 4D). Consistently, there were more TH1 cells than TFH cells in the crItga4 group, whereas no significant difference was found between these subsets in the crCd8 group (Figure 4E). To better understand this increase TH1 population, we tested whether the accumulation was due to enhanced proliferation following Integrin α4 disruption. We evaluated Ki-67 levels in TH1 cells in vivo, but found no significant differences between the crCd8 and crItga4 groups in LCMV-infected mice (Supplementary Figure 4D). We further examined whether Itga4 disruption affected TH1 proliferation in vitro, as Ki-67 expression in in vivo TH1 cells might reflect the steady-state level. Itga4 was ablated, followed by in vitro TH1 differentiation. Interestingly, Itga4 disruption significantly increased the proliferation of TH1 cells while reducing the proliferation of TH0 cells (Supplementary Figure 4E). This suggests that Integrin α4 may regulate CD4 T cell proliferation differently, depending on TCR and IL-12 cytokine signaling pathways. We also investigated whether Itga4 disruption affected TH1 differentiation. In contrast to the findings for TH1 cell proliferation, the frequency of IFN-γ-producing TH1 cells decreased in the crItga4 group (Supplementary Figure 4F). Therefore, the accumulation of TH1 cells in the crItga4 group may result from the combined effects of both proliferation and differentiation.
Since Integrin α4 is known to regulate CD4 T cell trafficking and homing, we examined the distribution of TH1 cells across various lymphoid tissues, including the spleen, mesenteric lymph nodes (mLNs), inguinal lymph nodes (iLNs), and blood. No significant differences in TH1 populations were observed between the crCd8 and crItga4 groups in mLN, iLN, and blood. However, the TH1 population in the spleen of the crItga4 group was significantly higher than in the crCd8 group (Figure 4F), suggesting that TH1 cells may preferentially accumulate in the spleen over other lymphoid organs. In conclusion, Itga4 deficiency in CD4 T cells may skew the immune response towards a TH1 phenotype by specifically controlling TH1 cell accumulation in the spleen and regulating CD4 T cell proliferation during acute LCMV infection. This highlights a potential role for Integrin α4 in balancing TFH and TH1 cell populations during antiviral immune responses.
In the current study, we demonstrate the potentially important role of Integrin α4 in regulating the balance between TFH and TH1 cell populations during acute viral infection. The downregulation of Integrin α4 in TFH cells, compared to its upregulation in TH1 cells, aligns with our previous analysis of its expression at the RNA level (24). This differential expression suggests that Integrin α4 may be involved in the functional specialization and migratory behavior of these T cell subsets and also serve as a useful marker for TFH and TH1 cells in viral infection or vaccination.
Integrins are known to be involved in cell-cell interaction, localization, migration, and co-stimulatory functions (7–9). For instance, VLA-4 (Integrin α4β1) plays a role in TCR-mediated CD4 T cell activation by facilitating CD3-dependent downstream signaling (9). Moreover, stimulation of VLA-4 with monoclonal antibodies against Integrin α4 and Integrin β1 induces T cell proliferation (25). While a high-affinity form of the integrin LFA-1 is known to regulate the development and maintenance of TFH cells (26), the role of VLA-4 in the regulation of TFH and TH1 subsets has remained unclear. The significant reduction in Integrin α4 expression in TFH cells suggests a possible mechanism that helps these cells to be retained in specific microenvironments, such as germinal centers, whereas higher Integrin α4 expression in TH1 cells aids their distribution to inflammatory sites. Indeed, the conjugation of VLA-4 is known to mediate the polarization of TH1 cells (13). Interestingly, the deficiency of Itga4 in CD4 T cells led to a notable shift in T cell populations, with an increased accumulation of TH1 cells in the spleen, contrary to our initial hypothesis that Itga4 deficiency might favor TFH cell development. This indicates that Integrin α4 plays a potential role in maintaining the balance between these subsets by regulating TH1 accumulation. It is possible that Integrin α4 may influence activation signaling for the differentiation, proliferation, or migration of TH1 cells. Itga4 deficiency showed increased in vitro TH1 cell proliferation, which may ultimately affect TH1 accumulation. Future studies are needed to further explore how Integrin α4 regulates spleen-specific accumulation, proliferation, or migration of TH1 cells. Moreover, the altered expression of CXCR5 in TFH cells from the Itga4-deficient group suggests that signaling pathways downstream of VLA-4 may impact the optimal localization to the B cell follicle and stable maintenance of TFH cell features despite its low expression in TFH cells. Further research is warranted to investigate the localization and functional impact of Itga4-deficient TFH and TH1 populations.
The utilization of CRISPR-Cas9 gene editing allowed us to effectively disrupt Itga4 expression in CD4 T cells. Combinations of multiple crRNA mixtures have been tried in human primary T cells and mouse CD8 T cells with varied gene deletion efficiency (17, 19–21). Our results also showed that combinations of proximal crRNA pairs achieved high deletion efficiency, as evidenced by the substantial reduction of Integrin α4 expression. Interestingly, the analysis of InDel contributions revealed that certain crRNA pairs within the combination were more effective in deletions in the gene. For crItga4 C, D, and E, the gene deletion efficacy of single use of these crRNA was similar. The distance between the crRNAs may be an unignorable factor for efficient fragmentation and gene deletion. Indeed, crRNA pairs with 32 ~ 51 bp interval targeting TET2, DOT1L, and PRDM1 in human primary T cells generated highly effective gene deletion, while a mixture of three different crRNAs targeting Itgav/CD51 with distance longer than 10 ~ 75 kb in mouse CD8 T cells left 25 ~ 75% of CD51 positive populations (19, 21). Our research suggests that the distance between crRNA pairs, between approximately 70 and 100 bp and shorter than 400 bp, might be effective for deletions in the gene. Furthermore, reducing the amounts of crRNA through optimized crRNA paring and Cas9 protein usage could enhance cost-effectiveness. These findings emphasize the importance of strategic crRNA selection to optimize gene editing outcomes, suggesting a robust approach for gene deletion in primary T cells.
Our study elucidates an important role of Integrin α4 in regulating TFH and TH1 cell dynamics during acute viral infection. The use of CRISPR-Cas9 gene editing to disrupt Itga4 expression in CD4 T cells provided valuable insights into the molecular mechanisms driving T cell subset differentiation. Future studies should explore the detailed mechanism of Integrin α4 on TH1 regulation, broader implications of integrin-mediated T cell regulation, and investigate potential therapeutic applications for modulating immune responses through integrin targeting.
The original contributions presented in the study are included in the article/Supplementary Material. The data presented in the study are deposited in the Sequence Read Archive (SRA) repository, BioProject accession number PRJNA1183801. Further inquiries can be directed to the corresponding author.
The animal study was approved by Institutional Animal Care and Use Committee of the College of Medicine at The Catholic University of Korea. The study was conducted in accordance with the local legislation and institutional requirements.
TW: Data curation, Formal analysis, Investigation, Visualization, Writing – original draft, Writing – review & editing. YC: Investigation, Writing – review & editing. JK: Investigation, Writing – review & editing. YSC: Resources, Writing – review & editing. MP: Funding acquisition, Supervision, Writing – review & editing. JC: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Resources, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) No. 2021R1F1A1060347, 2023R1A2C1007319, and RS-2023-00258956 to JC), NIH P01AI145815 (sub-award to JC), and by the Catholic Medical Center Research Foundation made in the program years of 2022 and 2023 (JC).
We thank the staff of the animal facility and flow cytometry facility for their support.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1445341/full#supplementary-material
1. Luckheeram RV, Zhou R, Verma AD, Xia B. CD4+T cells: differentiation and functions. Clin Dev Immunol. (2012) 2012:925135. doi: 10.1155/2012/925135
2. Crotty S. T follicular helper cell biology: A decade of discovery and diseases. Immunity. (2019) 50:1132–48. doi: 10.1016/j.immuni.2019.04.011
3. Choi J, Crotty S. Bcl6-mediated transcriptional regulation of follicular helper T cells (TFH). Trends Immunol. (2021) 42:336–49. doi: 10.1016/j.it.2021.02.002
4. Choi J, Crotty S, Choi YS. Cytokines in follicular helper T cell biology in physiologic and pathologic conditions. Immune Netw. (2024) 24:e8. doi: 10.4110/in.2024.24.e8
5. Dong C, Flavell RA. Cell fate decision: T-helper 1 and 2 subsets in immune responses. Arthritis Res Ther. (2000) 2:179. doi: 10.1186/ar85
6. Topchyan P, Lin S, Cui W. The role of CD4 T cell help in CD8 T cell differentiation and function during chronic infection and cancer. Immune Netw. (2023) 23:e41. doi: 10.4110/in.2023.23.e41
7. Bouvard D, Pouwels J, Franceschi ND, Ivaska J. Integrin inactivators: balancing cellular functions in vitro and in vivo. Nat Rev Mol Cell Biol. (2013) 14:430–42. doi: 10.1038/nrm3599
8. Kechagia JZ, Ivaska J, Roca-Cusachs P. Integrins as biomechanical sensors of the microenvironment. Nat Rev Mol Cell Bio. (2019) 20:457–73. doi: 10.1038/s41580-019-0134-2
9. Nojima Y, Humphries MJ, Mould AP, Komoriya A, Yamada KM, Schlossman SF, et al. Vla-4 mediates cd3-dependent cd4+ T-cell activation via the cs1 alternatively spliced domain of fibronectin. J Exp Med. (1990) 172:1185–92. doi: 10.1084/jem.172.4.1185
10. Yang Y, Harrison JE, Print CG, Lehnert K, Sammar M, Lazarovits A, et al. Interaction of monocytoid cells with the mucosal addressin MAdCAM-1 via the integrins VLA-4 and LPAM-1. Immunol Cell Biol. (1996) 74:383–93. doi: 10.1038/icb.1996.67
11. Rüegg C, Postigo A, Sikorski E, Butcher E, Pytela R, Erle D. Role of integrin alpha 4 beta 7/alpha 4 beta P in lymphocyte adherence to fibronectin and VCAM-1 and in homotypic cell clustering. J Cell Biol. (1992) 117:179–89. doi: 10.1083/jcb.117.1.179
12. DeNucci CC, Pagán AJ, Mitchell JS, Shimizu Y. Control of α4β7 integrin expression and CD4 T cell homing by the β1 integrin subunit. J Immunol. (2010) 184:2458–67. doi: 10.4049/jimmunol.0902407
13. Mittelbrunn M, Molina A, Escribese MM, Yáñez-Mó M, Escudero E, Ursa Á, et al. VLA-4 integrin concentrates at the peripheral supramolecular activation complex of the immune synapse and drives T helper 1 responses. Proc Natl Acad Sci. (2004) 101:11058–63. doi: 10.1073/pnas.0307927101
14. Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. (2013) 8:2281–308. doi: 10.1038/nprot.2013.143
15. Ma Y, Zhang L, Huang X. Genome modification by CRISPR/cas9. FEBS J. (2014) 281:5186–93. doi: 10.1111/febs.2014.281.issue-23
16. Pickar-Oliver A, Gersbach CA. The next generation of CRISPR–Cas technologies and applications. Nat Rev Mol Cell Bio. (2019) 20:490–507. doi: 10.1038/s41580-019-0131-5
17. Seki A, Rutz S. Optimized RNP transfection for highly efficient CRISPR/Cas9-mediated gene knockout in primary T cells. J Exp Med. (2018) 215:985–97. doi: 10.1084/jem.20171626
18. Zou K, Wang F, Zhang Z, Zhou Y, Li P, Wang D, et al. Optimized CRISPR/Cas9 system for gene knockout in chicken DF1 cells. Poult Sci. (2023) 102:102970. doi: 10.1016/j.psj.2023.102970
19. Ito Y, Inoue S, Nakashima T, Zhang H, Li Y, Kasuya H, et al. Epigenetic profiles guide improved CRISPR/Cas9-mediated gene knockout in human T cells. Nucleic Acids Res. (2023) 52:141–53. doi: 10.1093/nar/gkad1076
20. Sandner L, Alteneder M, Rica R, Woller B, Sala E, Frey T, et al. The guanine nucleotide exchange factor Rin-like controls Tfh cell differentiation via CD28 signaling. J Exp Med. (2023) 220:e20221466. doi: 10.1084/jem.20221466
21. Lee S, Ma J, Im SJ. Expression and function of CD51 on CD8 T cells as an immunomodulatory target. Biochem Biophys Res Commun. (2023) 661:56–63. doi: 10.1016/j.bbrc.2023.04.040
22. Choi J, Pease DR, Chen S, Zhang B, Phee H. P21-activated kinase 2 is essential in maintenance of peripheral Foxp3+ regulatory T cells. Immunology. (2018) 154:309–21. doi: 10.1111/imm.2018.154.issue-2
23. Hatzi K, Nance JP, Kroenke MA, Bothwell M, Haddad EK, Melnick A, et al. BCL6 orchestrates Tfh cell differentiation via multiple distinct mechanisms. J Exp Med. (2015) 212:539–53. doi: 10.1084/jem.20141380
24. Choi J, Diao H, Faliti CE, Truong J, Rossi M, Bélanger S, et al. Bcl-6 is the nexus transcription factor of T follicular helper cells via repressor-of-repressor circuits. Nat Immunol. (2020) 21:777–89. doi: 10.1038/s41590-020-0706-5
25. Bednarczyk JL, Teague TK, Wygant JN, Davis LS, Lipsky PE, McLntyre BW. Regulation of T cell proliferation by anti-CD49d and anti-CD29 monoclonal antibodies. J Leukoc Biol. (1992) 52:456–62. doi: 10.1002/jlb.1992.52.issue-4
Keywords: integrin α4, CRISPR-Cas9 gene editing, TFH, TH1, viral infection
Citation: Wi T, Choi Y, Kim J, Choi YS, Pipkin ME and Choi J (2024) Efficient gene deletion of Integrin alpha 4 in primary mouse CD4 T cells using CRISPR RNA pair-mediated fragmentation. Front. Immunol. 15:1445341. doi: 10.3389/fimmu.2024.1445341
Received: 07 June 2024; Accepted: 18 November 2024;
Published: 10 December 2024.
Edited by:
Noah Isakov, Ben-Gurion University of the Negev, IsraelReviewed by:
Vasco Barreto, New University of Lisbon, PortugalCopyright © 2024 Wi, Choi, Kim, Choi, Pipkin and Choi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinyong Choi, amNob2lAY2F0aG9saWMuYWMua3I=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.