- 1Institute of Forensic Medicine and Laboratory Medicine, Jining Medical University, Jining, China
- 2College of Fishery, Guangdong Ocean University, Zhanjiang, China
Various types of professional immune cells first emerge in fish and likely represent the primordial form and functions. Recent advancements revealed the direct connection between the central nervous system and the immune system in the mammalian brain. However, the specifics of brain-immune networks in the fish and the underlying mechanisms of teleost’s brain against pathogen infection have not been fully elucidated. In this study, we investigated the distribution of markers representing cerebral cells associated with protection and professional lymphocytes in the seven major components of the Nile tilapia brain through RNA-Seq assay and observed the most dominant abundance in the medulla oblongata. The subsequent challenge test revealed the non-specific cytotoxic cells (NCCs) exhibited the strongest response against streptococcal infection of the brain. The presence of NCCs in the brain was then confirmed using immunofluorescence and the cytotoxic effects usually induced by NCCs under infection were determined as well. Collectively, these findings contribute significantly to comprehending the mechanism of fish neuroimmune interaction and enhancing our understanding of its evolutionary development.
1 Introduction
Since the early 19th century, the brain has been considered a site with limited immune activity and is maintained by the blood-brain barrier (BBB) (1–4). Additionally, various cerebral cells contribute to brain homeostasis and protection (3, 5). For example, microglia function as resident myeloid cells (6), while astrocytes are involved in neuroinflammation and neurodegeneration (7, 8), serving as antigen-presenting cells and participating in inflammatory responses (3, 8). However, recent advancements have expanded our understanding of brain immunity (3, 9–11). This includes the discovery of lymphatic vessels in the brain, the identification of immunological niches, and the recognition of complex brain-immune networks. Additionally, multiple immunological niches have been found in different brain regions (3, 10, 12), such as the choroid plexus (CP), meninges, and cerebrospinal fluid (CSF). These areas contain various types of professional or peripheral immune cells (3, 13), including monocytes (Mo), natural killer cells (NKs), T cells, B cells, and dendritic cells (DCs).
In comparison to the understanding of the evolution of the nervous system in the fish brain, which shows many shared ancestral neural traits among all vertebrates (14), there has been a growing recognition of the critical role of fish immunity due to their position at the intersection of the innate and adaptive immune systems (15–17). The adaptive immune system in fish is notably simpler (16, 18), lacking structures such as bone marrow, lymph nodes, and germinal centers, as well as confirmed class switching of immunoglobulins and memory ability. Despite extensive research on the structure and neurobiological functions of fish brains (14, 19), as well as recent studies on the interactions between the nervous and immune systems in fish (20), the confirmed existence, distribution characteristics, and response patterns of professional immune cells in fish brains against stimulation remain unclear.
Therefore, in this study, we aimed to assess the distribution of immune-related cells in the tilapia brain and identify the cerebral and professional defenders involved in bacterial infection induced by Streptococcus agalactiae, which was well-recorded in the fish neuro-immune study since this bacterial infection usually leads to typical meningitis that was hallmarked by the clinical phenotype such as exophthalmia (21–24). Our findings revealed that cerebral cells associated with protection, as well as professional lymphocytes, were predominantly located in the medulla oblongata of the fish brain. Subsequent evaluation under challenge testing demonstrated that the NCC population exhibited the strongest response, as confirmed by the presence of NCCRP1-positive cells and assessments of cytotoxicity. Collectively, these results indicate that the central antibacterial immunity of fish brains occurs in the medulla oblongata and is mediated by NCCs.
2 Materials and methods
2.1 Fish and brain sample collection
Tilapia specimens weighing approximately 100 ± 10 grams were obtained from Zhanjiang City, China. These specimens were acclimated in recirculating aquaculture systems with appropriate ventilation for 60 days, maintaining a constant temperature of 28°C throughout the acclimation process. Tilapia were fed with commercial feed daily from Guangdong Yuehai Feeds Group Co., Ltd. (Product code 1718). Three fish were collected post the acclimation period (at 61st day), anesthetized using tricaine methanesulfonate (MS-222; Sigma, Darmstadt, Germany), and then euthanized. The entire brain was carefully dissected, and several distinct brain components (25), including the olfactory bulb, cerebrum, optic lobe, cerebellum, hypothalamus, and medulla oblongata, were sampled for RNA extraction. Additionally, the thin medulla oblongata was divided into two segments along the anterior-posterior axis, referred to as the anterior medulla oblongata and posterior medulla oblongata (Supplementary Figure S1).
2.2 RNA extraction, RNA-Seq, and bioinformatics analysis
Total RNA was extracted from each sample using RNAiso Plus from TaKaRa (Dalian, China), and subsequently treated with RNase-free DNase I from TaKaRa (Dalian, China) to eliminate any residual DNA. The quality of the total RNA was confirmed by electrophoresis using 1.2% agarose gels and quantified with NanoDrop 2000 from Thermo Fisher Scientific (Waltham, USA). The mRNA molecules were enriched using oligo(dT) beads (Qiagen, Hilden, Germany), fragmented into short fragments, and subsequently reverse-transcribed into cDNA. The resulting cDNAs were collected, subjected to end repair, ligated with Illumina sequencing adapters, amplified through PCR, and subsequently sequenced using the Illumina NovaSeq 6000 platform (Gene Denovo Biotechnology Co., Guangzhou, China). The raw reads were filtered to obtain high-quality clean reads using fastp (version 0.18.0). Paired-end clean reads were mapped to the reference genome (http://ftp.ensembl.org/pub/release-110/fasta/oreochromis_niloticus/) using HISAT2 (version 2.4.0).
Gene expression abundances were calculated and normalized to fragments per kilobase of transcript per million mapped reads (FPKM). Differentially expressed genes (DEGs) were identified using DESeq2 (version 1.26.0) with the following criteria: |log2(foldchange)| ≥ 1, P value < 0.05, and false discovery rate (FDR, Q value) ≤ 0.05. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were conducted through the KEGG Automatic Annotation Server (KAAS).
In addition, based on our previous study on the characterization of tilapia head kidney leukocytes (HKLs) through RNA-Seq in 2022 (25), the geometric mean of three universal housekeeping genes—β-actin, ef1a, and gapdh (26–28)—were used to normalize the expression abundance of both the HKL and brain cDNA libraries.
2.3 Bacteria and challenge
The preserved S. agalactiae strain ZQ0910 (29) of serotype III was reactivated by incubation in brain-heart infusion broth at 28°C overnight. The bacterial culture was then harvested through centrifugation at 4000 × g for 5 minutes. The bacterial cells were subjected to three successive washes with phosphate-buffered saline (PBS) and were ultimately resuspended in PBS for subsequent experiments.
A total of 20 fish were intraperitoneally injected with 100 μL of S. agalactiae (5 × 107 CFU/mL). Afterward, three parallel individuals were collected and sacrificed at 0, 12, and 24 hours postinfection (hpi), and brain samples were obtained using a previously described protocol for RNA extraction and cDNA synthesis.
2.4 cDNA synthesis and quantitative real-time PCR
cDNAs were synthesized using the PrimeScript™ RT reagent kit with gDNA Eraser from TaKaRa (Dalian, China). Quantitative real-time PCR (qRT−PCR) was performed with TB Green® Premix Ex Taq™ II (Tli RNaseH Plus) from TaKaRa (Dalian, China) and the QuantStudio 6 Flex Real-Time PCR System from Thermo Fisher Scientific (Waltham, USA). Reference genes, including β-actin, ef1a, and gapdh, were utilized for normalization (26–28).
2.5 Polyclonal antibody preparation for On-NCCRP1
The complete sequence of nonspecific cytotoxic cell receptor protein 1 in Nile tilapia Oreochromis niloticus (On-NCCRP1) was previously reported in our 2020 study (30). Subsequently, the ORF sequence of On-NCCRP1 was amplified using PCR with specific primers that incorporated the BamH I and Xho I restriction sites (Supplementary Table S1). The purified DNA fragments were then ligated into the predigested pGEX-6P-1 plasmid (BT Lab, Wuhan, China) and introduced into BL21 (DE3) chemically competent cells (TransGen, Beijing, China). Positive clones were verified by DNA sequencing and cultured in Luria–Bertani broth supplemented with ampicillin sodium (Amp+) at a concentration of 100 µg/mL at 28°C until the OD600 reached 0.6. Isopropyl β-d-thiogalactopyranoside was added to the bacterial culture mixture at a final concentration of 1 μM, and the culture was continued for 8 hours before being harvested by centrifugation. The recombinant protein was then purified using a GST-tag protein purification kit from Beyotime (Shanghai, China), dissolved in sterilized PBS, and confirmed by Coomassie blue staining (Supplementary Figure S2).
A rabbit anti-On-NCCRP1 polyclonal antibody was generated using our established protocol beginning in 2023 (31). In summary, two healthy New Zealand White rabbits (~2 kg) were immunized on day 0 with a mixture of On-NCCRP1 protein (400 μg in 750 μL of PBS) and Freund’s complete adjuvant (750 μL) using emulsification. Subsequently, on days 21, 35, and 49, the rabbits were immunized with On-NCCRP1 protein (300 μg in 750 μL of PBS) and Freund’s incomplete adjuvant (750 μL) again. On day 57, the sera of the rabbits were collected, and the antibodies were purified through an affinity chromatography assay using protein A/G agarose beads.
2.6 Head kidney leukocyte preparation
HKL preparation was carried out following previous research from 2020 to 2023 (25, 31, 32). In brief, healthy fish were collected and sacrificed. The head kidney was carefully separated, and the neurochords were removed. The head kidney tissue was then cut and passed through a 40-μm cell strainer (Greiner Bio-One GmbH, Frickenhausen, Germany). The resulting cell suspension was placed in Leibovitz’s L-15 medium (Thermo Fisher Scientific, Waltham, USA). The cells were layered onto a 34%/51% Percoll gradient (Solarbio, Beijing, China) and centrifuged using a swing rotor (400 × g, 40 minutes, 4°C). Subsequently, the cells located at the surface of the 51% Percoll layer were gently aspirated, collected via centrifugation, washed, and resuspended in PBS for use in subsequent experiments.
2.7 Protein extraction and Western blot
Total protein from HKLs and brain components was extracted with a protein extraction kit (BC3710, Solarbio, Beijing, China). Subsequently, 10 μg of protein sample was loaded onto a 12% SDS−PAGE gel and transferred to a PVDF membrane (Merck, Darmstadt, Germany). The membrane was then blocked with a quick blocking buffer (Beyotime, Shanghai, China) for 10 minutes at room temperature, followed by incubation with a primary antibody, rabbit anti-On-NCCRP1, at a dilution ratio of 1:2000 for 1 hour. After the membranes were washed three times with Tris-buffered saline containing 0.1% Tween-20 (TBST), they were incubated with a secondary antibody, HRP-labeled goat anti-rabbit IgG (H+L) (A0208, Beyotime, Shanghai, China), at a dilution ratio of 1:1000 for 30 minutes. After another round of three washes, the antigen−antibody complexes were detected using the DAB Horseradish Peroxidase Color Development Kit (P0203, Beyotime, Shanghai, China). In addition, a rabbit anti-β-actin monoclonal antibody (dilution ratio of 1:20000) (AC026, ABclonal, Wuhan, China) was used to determine the abundance of the reference protein, β-actin, which was used as a loading control. Moreover, the positive bands obtained from the western blot analysis were transformed into gray values by ImageJ (version 1.54g).
2.8 Hematoxylin and eosin staining and immunofluorescence
For the hematoxylin and eosin (H&E) staining and immunofluorescence (IF) assay, the procedures described in our previous work (25, 33) were followed. Briefly, the whole brains of healthy fish were isolated and fixed in Dietrich’s fixative for 24 hours. The brain was then dehydrated in a series of graded alcohol solutions, cleared in xylene, and embedded in paraffin wax. Serial sections (8 μm thick) were rehydrated, stained with an H&E staining kit (C0105S, Beyotime, Shanghai, China), and observed under a microscope.
Selective sections representing typical structures (25) were rehydrated, followed by heat-induced antigen retrieval using a matched solution (P0085, Beyotime, Shanghai, China). The samples were then blocked and incubated with the primary antibody rabbit anti-On-NCCRP1 (dilution ratio of 1:200) for 1.5 hours. After five washes with PBS, the samples were incubated with the secondary antibody Cy3 goat anti-rabbit IgG (H+L) (dilution ratio of 1:500) (AS007, ABclonal, Wuhan, China) for one hour. Finally, the samples were observed and photographed after staining the cell nucleus with 2-(4-amidinophenyl)-6-indolecarbamidine dihydrochloride (DAPI).
2.9 Statistical analysis
All the data are presented as the means ± standard deviations (SDs). Tukey’s HSD test was utilized to analyze significant differences through Prism software (version 8.0), and significant differences (p < 0.05) are indicated by different letters.
2.10 Drawings
TB tools (version 1.108) were used to construct heatmaps. Adobe Photoshop CC (San Jose, CA, USA) and Adobe Illustrator (San Jose, CA, USA) were used to construct and design the final panel.
3 Results
3.1 Overview of the mRNA expression profile of tilapia brain
A study was conducted to examine the mRNA expression profiles of seven brain components in tilapia. An RNA-Seq assay was utilized, resulting in the generation of 21 transcriptome libraries. These libraries generated a total of 1240.4 megabases of clean reads, with an average Q30 score of 95.1% and an average mapping ratio of 95.5% to the tilapia genome. Next, a thorough analysis of the KEGG annotations was performed. The top five significantly enriched items in each component were found to be highly similar, with processes mainly related to RNA processing and endocytosis. However, the olfactory bulb was primarily associated with tRNA synthesis and synapse development (Figure 1A). A comprehensive analysis of 21 pairwise comparisons resulted in the identification of more than 70000 DEGs. The most substantial differences were observed between the cerebellum and posterior medulla oblongata, with a total of 7256 DEGs. Conversely, the fewest differences were detected between the two segments of the medulla oblongata, with a total of 887 DEGs (Figure 1B).
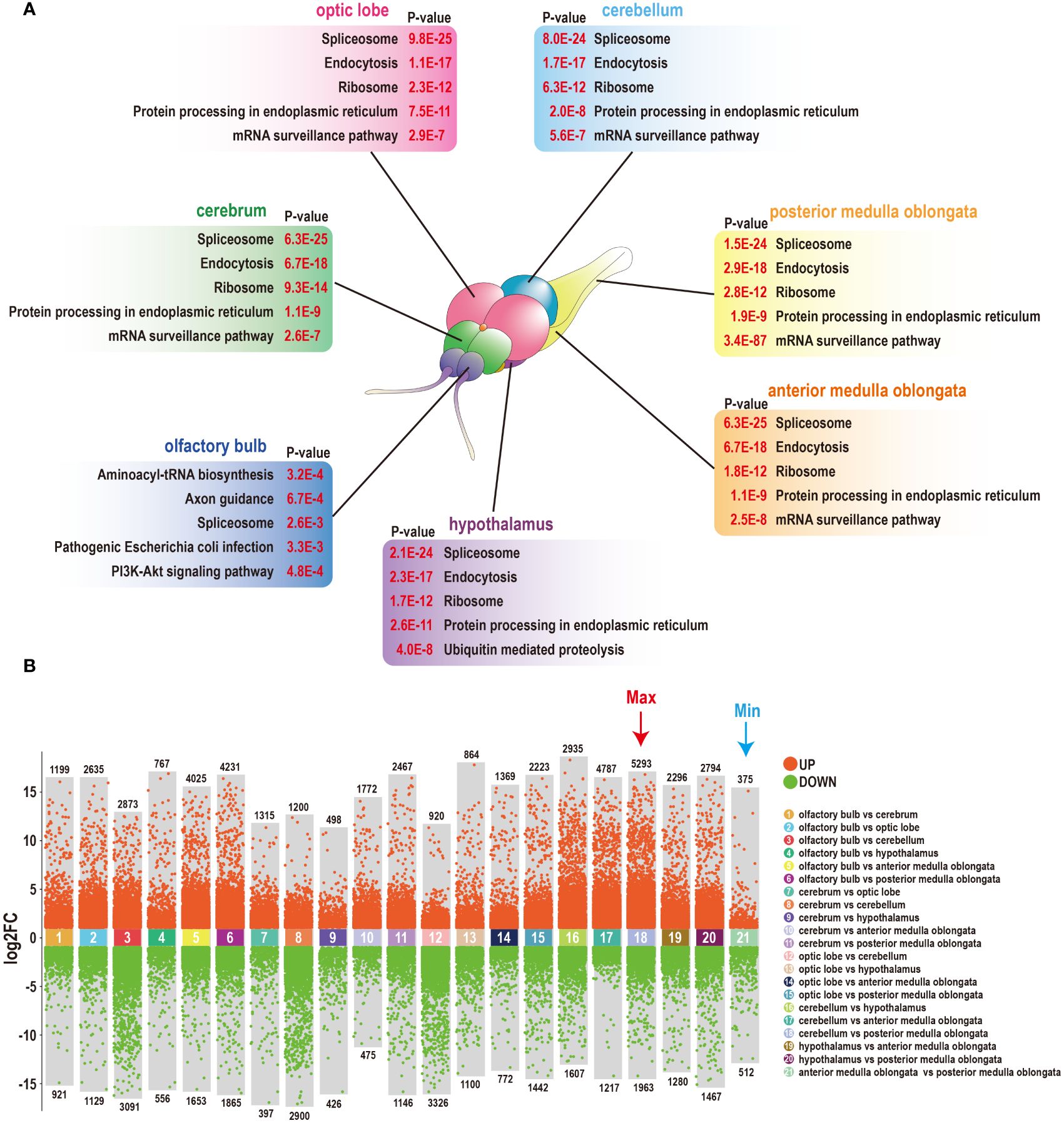
Figure 1 Heterogeneity of seven components of the tilapia brain. (A) Top five KEGG annotations of seven components of the tilapia brain. A diagram of the tilapia brain was generated in our previous study (25). (B) The number of DEGs among seven components of the tilapia brain was obtained through 21 pairwise comparisons. A total of 75579 DEGs were screened, including the most DEGs that existed between the cerebellum and posterior medulla oblongata, and the fewest DEGs were detected between the anterior and posterior medulla oblongata.
3.2 Many immune-related markers are located in the medulla oblongata
To further examine the presence and distribution of lymphocytes marked by specific markers, the abundances of numerous common immune-related markers were gathered and clustered within the brain and tilapia HKL. The analysis revealed that while HKL exhibited a substantial presence of 79 markers, with a mean FPKM value of approximately 64.4, the brain displayed a lower average level of expression, approximately 75% lower, with a mean FPKM value of approximately 16.1 (Figure 2). However, in contrast to other brain components, the medulla oblongata exhibited a greater expression level, with a mean FPKM value of approximately 39.6. Furthermore, the medulla oblongata was clearly distinguished and clustered separately from the other brain components. Notably, a total of 27 immune-related markers were more abundant in the medulla oblongata than in the HKL. Among these markers were several typical T-cell markers, such as CD2, CD6, ZAP70, and LCK (Figure 2).
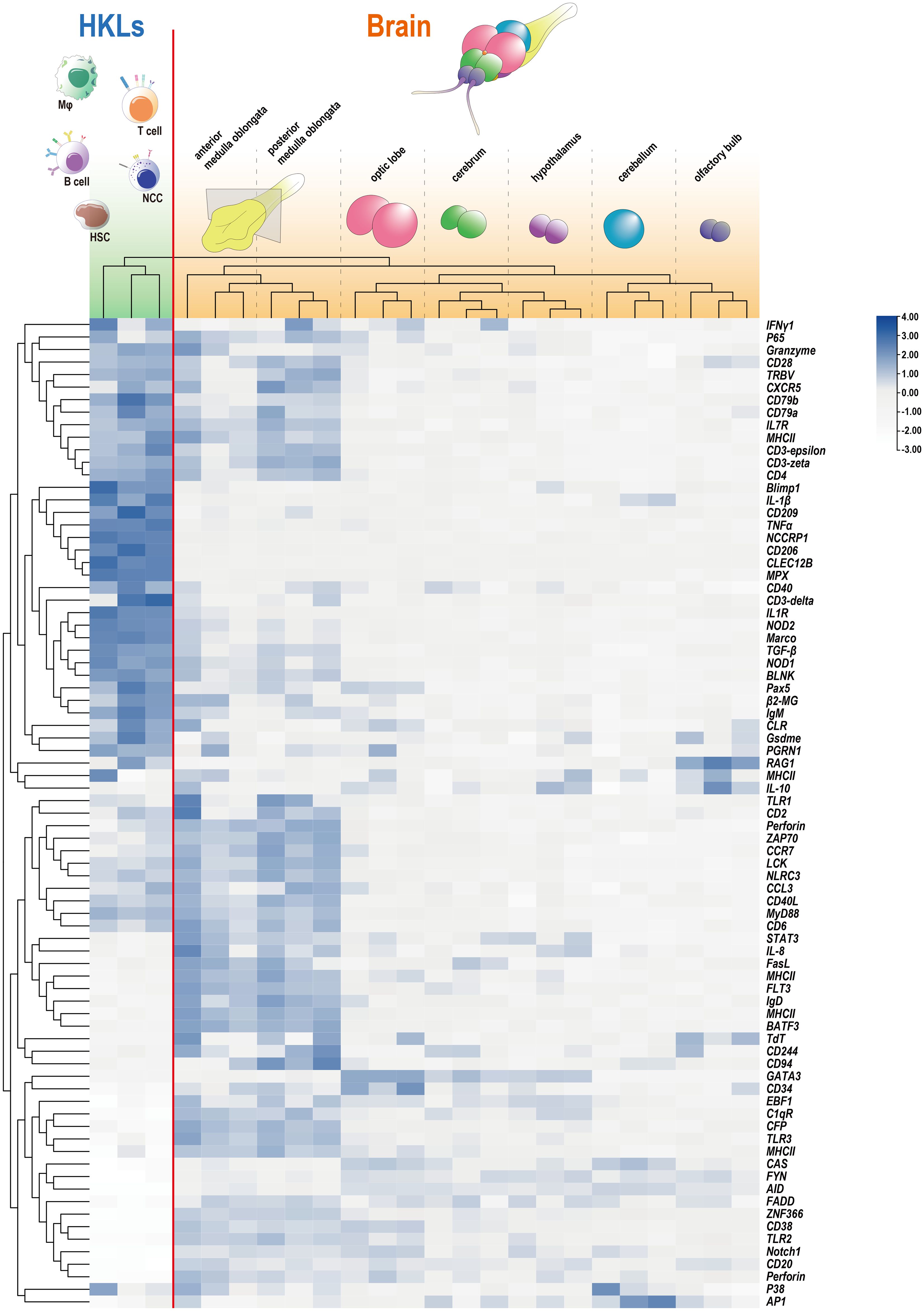
Figure 2 Expression profiles of immune-related genes in HKL and seven components of the tilapia brain detected through RNA-Seq. The RNA-Seq data for tilapia HKL were reported in our previous study (25), and the adjusted FPKM values for HKL and the brain were clustered and presented through a heatmap. The outermost branched sample (HKL) and the remainder (brain) were distinguished by a red line.
The distribution of markers associated with professional lymphocytes in tilapia (25, 32, 34) was initially analyzed. This analysis included hematopoietic stem cells (HSCs) or common lymphoid progenitors (CLPs), B cells, T cells, nonspecific cytotoxic cells (NCCs) or cytotoxic T cells (CTLs), macrophages (Mφ) or granulocytes, and dendritic cells (DCs). The findings indicated that these molecules were predominantly present in the medulla oblongata, with the exception of markers for macrophages or granulocytes, which are also widely distributed in the optic lobe (Figure 3). Additionally, the expression of crucial markers involved in brain surveillance and protection (3, 8, 35, 36), such as GFAP for astrocytes and TEME119 for microglia, was examined, revealing their prominent presence in the medulla oblongata (Figure 3).
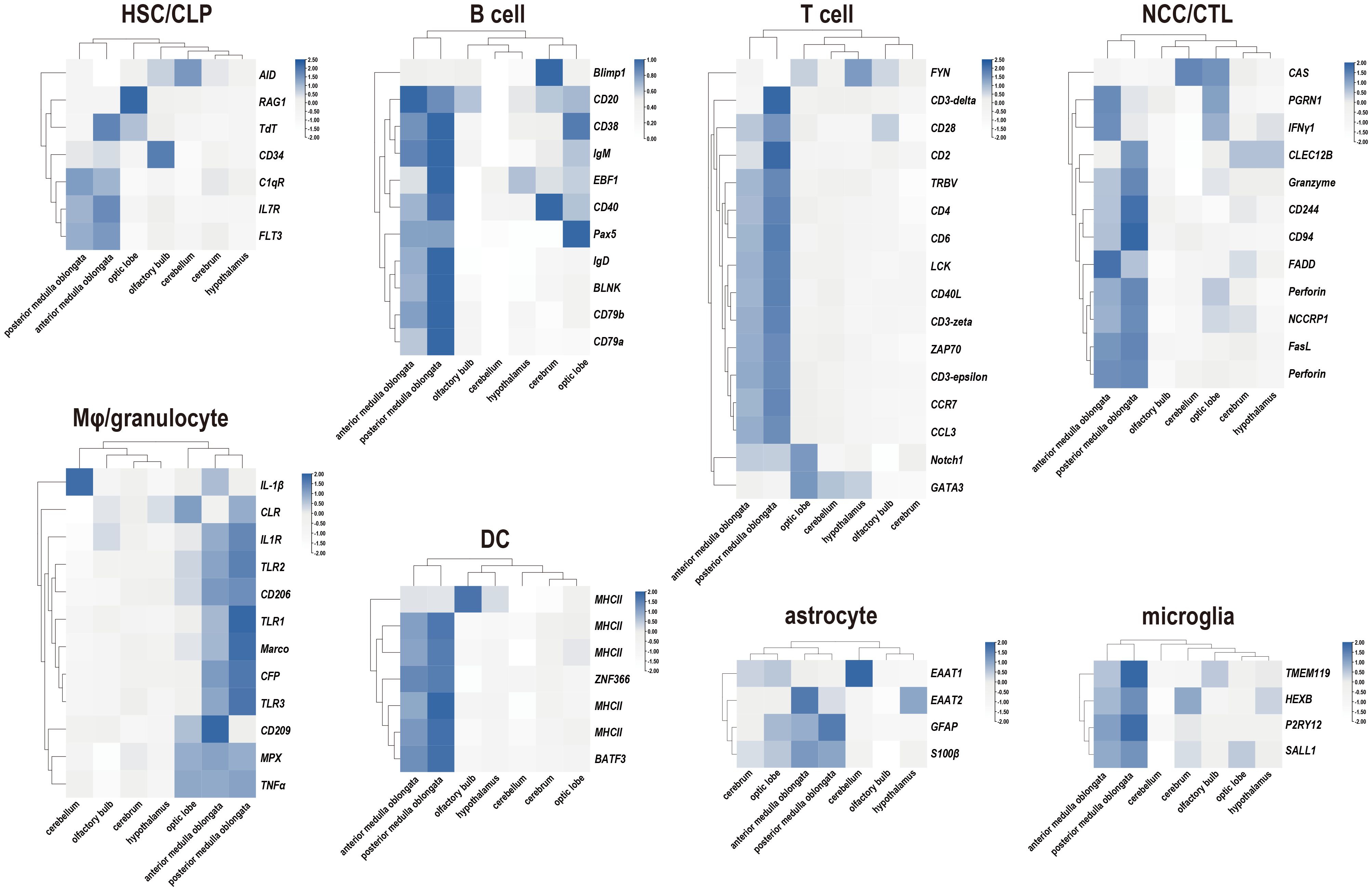
Figure 3 Expression profiles of immune-related markers in seven components of the tilapia brain. Heatmaps showing the clustering of acknowledged marker genes for tilapia lymphocyte subpopulation identification and cerebral defender characteristics, including hematopoietic stem cells (HSCs) or common lymphoid progenitors (CLPs), B cells, T cells, nonspecific cytotoxic cells (NCCs) or cytotoxic T cells (CTLs), macrophages (Mφ) or granulocytes, dendritic cells (DCs), astrocytes, and microglia.
3.3 Inflammation and immune responses in the brain are induced by S. agalactiae infection
To characterize the immune responses of the tilapia brain, a challenge test was performed through S. agalactiae injection (Figure 4). This bacterium is known to be capable of penetrating the blood−brain barrier and causing severe meningitis (21, 37). Subsequently, the levels of inflammatory factors and key genes involved in various immune pathways were evaluated using qRT−PCR. Notably, robust activation of inflammation was observed, as evidenced by the significant upregulation of the proinflammatory factors IL-1β and TNF-α. Importantly, the medulla oblongata exhibited the most pronounced increase in proinflammatory factors during bacterial infection, with levels reaching several hundred times the baseline. However, in contrast to those of proinflammatory factors, the responses of the anti-inflammatory factors IL-10 and TGF-β were relatively subdued, limited to certain brain components, and less intensive. Next, the expression patterns of six genes involved in the corresponding immune pathways were evaluated, but only limited activation was detected, primarily in the candidate genes STAT3, MyD88, and NOD1 (Figure 4).
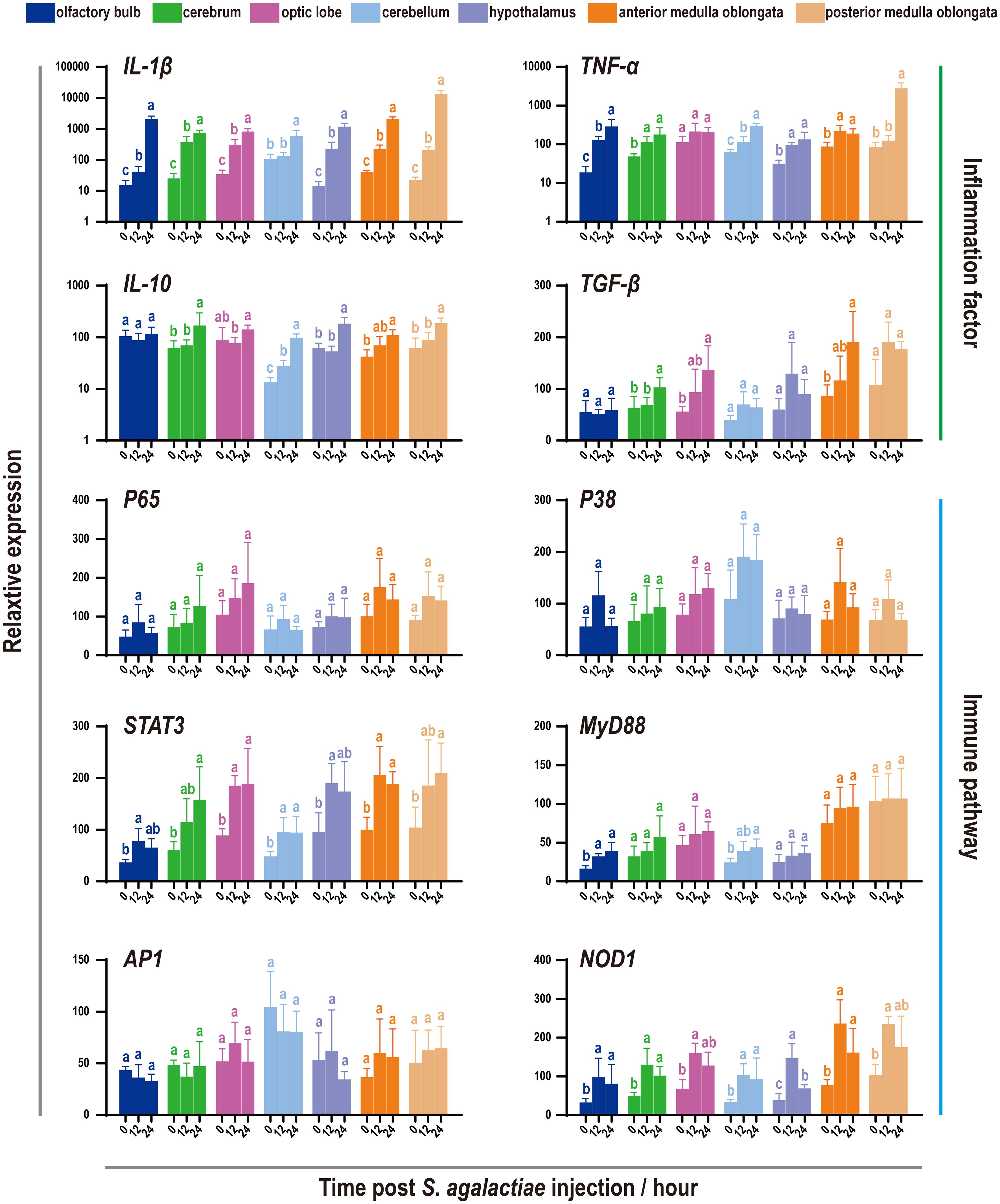
Figure 4 Expression patterns of inflammatory factors and immune pathways associated with seven components of the tilapia brain after S. agalactiae infection. The relative expression of four inflammatory factors and six key genes involved in different immune pathways was detected via qRT−PCR. For each given gene, the expression level of the seven components of the tilapia brain at 0 h with the most abundant transcripts was set as 100 to calculate the relative expression of the remaining samples. Different letters indicate significant differences (p < 0.05).
3.4 Nonspecific cytotoxic cell markers in the tilapia brain were strongly activated under S. agalactiae infection
Given the expression patterns of defense markers previously identified in the tilapia brain, a subsequent investigation was conducted to determine the specific types of cells involved in bacterial infection (Figure 5). The trends of 10 universal markers belonging to five types of tilapia professional lymphocytes were analyzed, revealing widespread promotion following S. agalactiae injection. Notably, NCCRP1, the highest marker in the fish NCC subpopulation, displayed a significant and sharp increase under bacterial infection. A similar phenomenon was observed with CLEC12B, a potential marker of tilapia NCCs (32, 38). However, the candidates commonly used for adaptive immune cell characterization, such as CD79a and IgM for B cells and CD3 and CD4 for T cells, were minimally influenced by bacterial infection. Additionally, little change in the expression of astrocyte and microglial markers was observed, except for a marker of microglia (TMEM119).
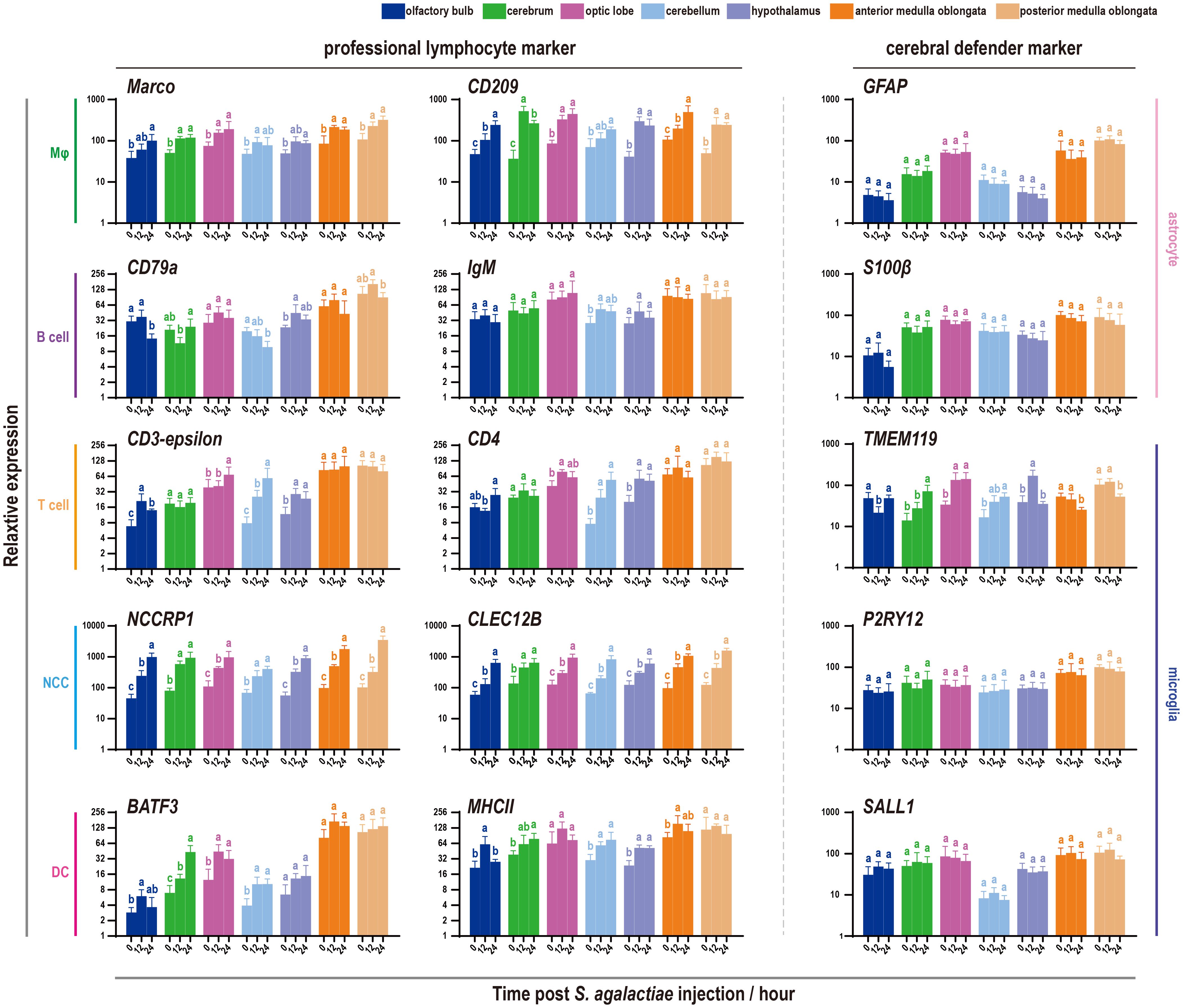
Figure 5 Expression patterns of immune-related markers in seven components of the tilapia brain after S. agalactiae infection. The relative expression of ten markers belonging to five types of lymphocytes and astrocyte and microglial markers was detected via qRT−PCR. For each given gene, the expression level of the seven components of the tilapia brain at 0 h with the most abundant transcripts was set as 100 to calculate the relative expression of the remaining samples. Different letters indicate significant differences (p < 0.05).
3.5 NCCs (NCCRP1-positive cells) are widespread in the tilapia brain
Although previous studies demonstrated the presence of NCCRP1 transcripts in the tilapia brain, with induction evident during bacterial infection (30), the distribution of NCCs (NCCRP1-positive cells) in the tilapia brain remains unclear. Therefore, a prokaryotically expressed recombinant protein, tilapia NCCRP1, was prepared (Supplementary Figure S2), and a corresponding rabbit anti-NCCRP1 polyclonal antibody was obtained. The NCCRP1 protein in the tilapia brain and HKLs was assessed using a Western blot analysis, and the results are shown in Figure 6B. The details of the NCCRP1 transcripts in the tilapia brain and HKLs, as evaluated through RNA-Seq, are displayed in histograms (Figure 6A). Both the quantification of NCCRP1 at the RNA and protein levels indicated that NCCRP1 was highly abundant in tilapia HKL and widely distributed throughout the brain. Subsequently, discernible positive signals for the NCCRP1 protein were observed in the olfactory bulb, cerebellum, and medulla oblongata (Figure 6C). These positive signals were predominantly located on the surface of these tissues, with scarce signals observed in other brain components.
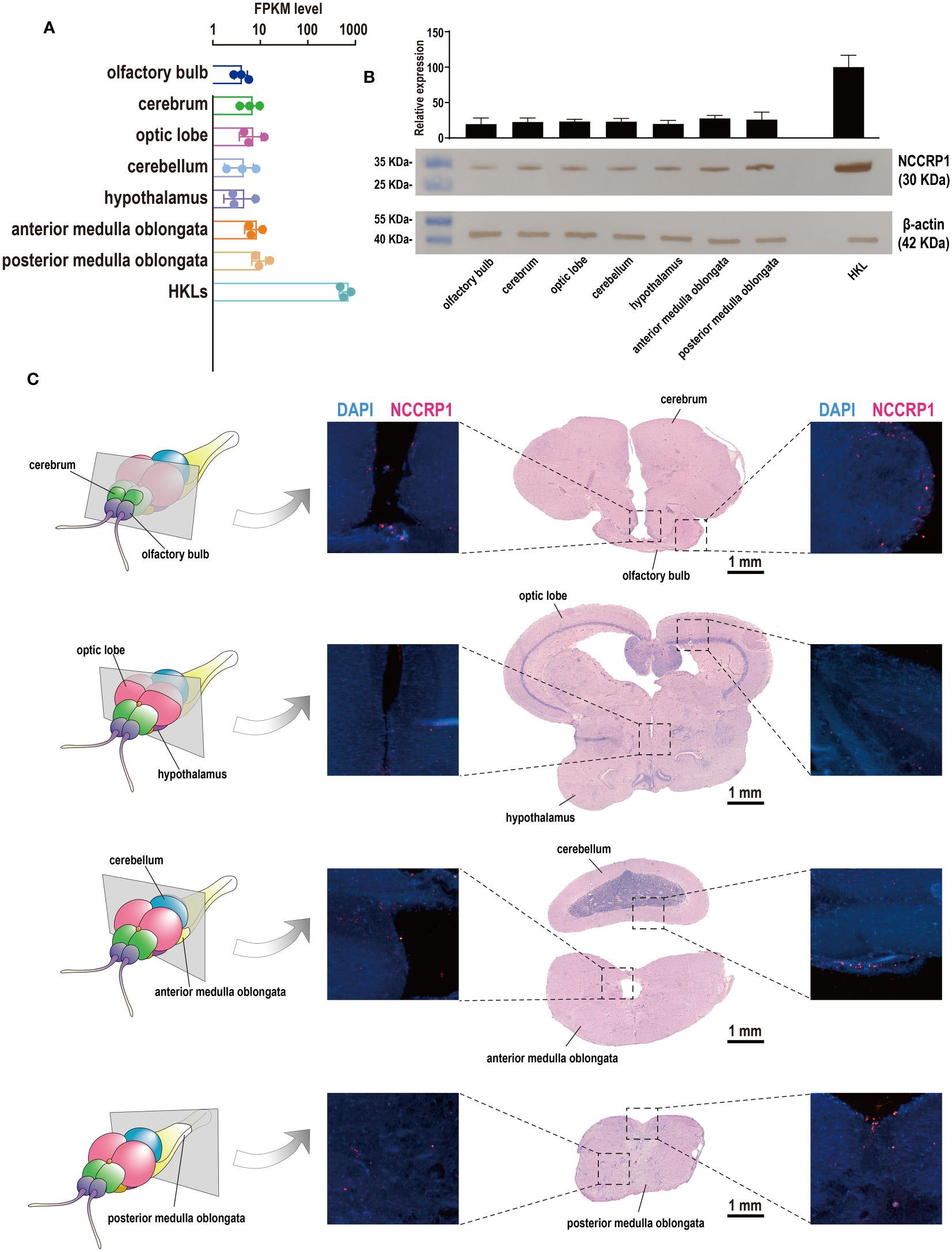
Figure 6 Expression pattern of NCCRP1 in the tilapia brain. (A) Relative expression of NCCRP1 in HKL and seven components of the tilapia brain, as detected through RNA-Seq. (B) The statistical analysis of the corresponding gray values was measured by ImageJ software from three parallel tests (Supplementary Table S2) and the Western blot analysis of NCCRP1 in HKL and seven components of the tilapia brain; β-actin was used as a reference protein. (C) Location of NCCRP1 in the tilapia brain. The left column shows a diagram of sections with typical brain components (25). The corresponding H&E staining results and the presence of NCCRP1 (red) and nuclei (blue) in each section detected by IF are displayed on the right.
3.6 Cytotoxicity in the brain was induced under S. agalactiae infection
Since the discovery of increasing cytotoxic effects mediated by NCCs or NCCRP1 protein in 2021 and 2022 (39, 40), it has become necessary to examine the cytotoxic effects and cell death levels during S. agalactiae infection. The findings showed that six common cytotoxicity effectors were activated to varying degrees, with the perforin and interferon genes being particularly prominent (Figure 7). However, the activation of apoptosis executors (Caspase3 and Caspase9) and pyroptosis executors (Caspase1 and GsdmE) was minor, while the most significant activation was observed in the cerebellum and medulla oblongata.
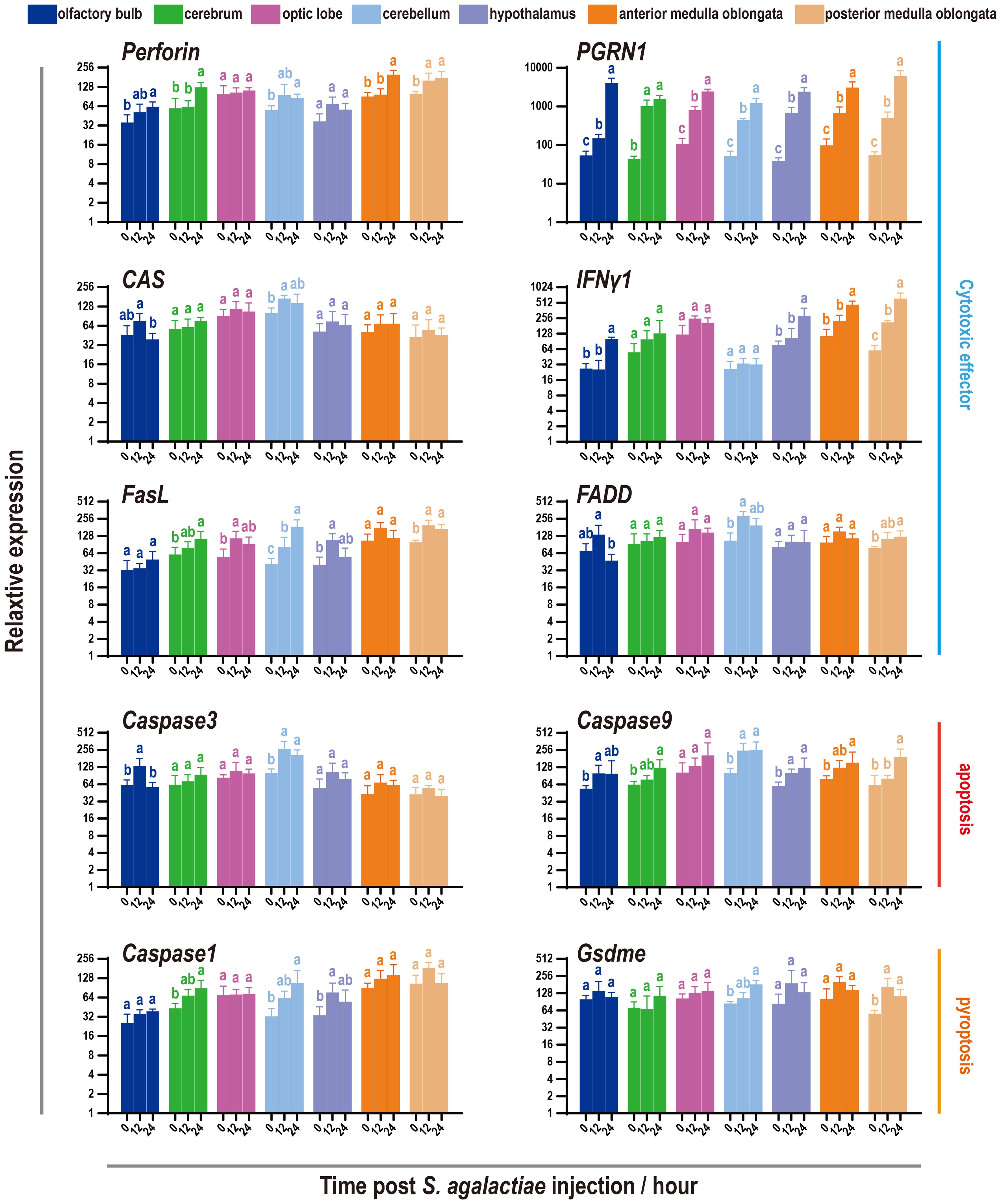
Figure 7 Expression pattern of cytotoxic factors and markers of apoptosis and pyroptosis in seven components of the tilapia brain subjected to S. agalactiae infection. The relative expression of six cytotoxic factors and four markers of the apoptosis/pyroptosis process was detected via qRT−PCR. For each given gene, the expression level of the seven components of the tilapia brain at 0 h with the most abundant transcripts was set as 100 to calculate the relative expression of the remaining samples. Different letters indicate significant differences (p < 0.05).
4 Discussion
Our current study sheds light on the distribution of defenders in the brain and response strategies against S. agalactiae infection. To achieve this goal, we first utilized the classical RNA-Seq assay to assess heterogeneity in the tilapia brain. Although there were few differences in the seven brain components identified through KEGG annotation, which could be attributed to the limited resolution of bulk RNA-seq technology and the dominance of neurons in the brain (41–43), the noticeable discrepancy in the number of DEGs emphasized the significant heterogeneity present.
Consequently, a comparison was made between the abundance of professional or peripheral lymphocyte markers present in both the brain and HKL. Astonishingly, the results revealed exceptionally high expression levels of immune-related markers in the medulla oblongata, surpassing even those in the HKL. This is particularly noteworthy considering that the head kidney of teleosts has traditionally been regarded as the primary hematopoietic tissue and central immune organ (16, 17), and tilapia HKLs were found to be almost exclusively composed of professional immune cells (25, 32, 34). The presence of such a large number of professional lymphocytes in the medulla oblongata strongly suggests frequent migration under steady-state conditions, similar to the situation in mammals (3). However, there are some differences to note, as the main immunological niches in the mammalian brain are located in the CP, meninges, and CSF (3). Although CP has been reported in the zebrafish myelencephalon, the obvious positive signal was limited to the saccus dorsalis zone of adult brains rather than the medulla oblongata (44). In contrast, bony fish have been found to have a high flow of CSF around the medulla oblongata (45), which likely performs the principal transport function and may represent the primordial form. Additionally, in the tilapia medulla oblongata, the majority of cerebral defender microglia and astrocytes were detected. However, conflicting results have been recorded in zebrafish, as more microglia were found in the optic lobe (46, 47). Nevertheless, similar to tilapia, dense astrocytes have also been observed in the medulla oblongata of zebrafish, and these astrocytes are associated with synapses and interact with each other (48). These findings collectively underscore the immunological significance of the medulla oblongata in the tilapia brain.
Furthermore, challenge tests were conducted and evaluated, revealing evident inflammation in the brain occurring no later than 12 hours after injection, which aligns with previous findings (27, 33, 49). Moreover, relatively moderate activation of the immune pathway was also observed, similar to prior reports that the intense response of these candidates mainly occurs in the head kidney and spleen of tilapia (49). This may be associated with the immune tolerance of the brain, which serves to prevent excessive cell death (2, 50).
Notably, the most significant response in the tilapia brain under S. agalactiae infection was observed in the NCCs, followed by Mφ, as indicated by the universal marker genes. In particular, the expression levels of NCC markers during infection were significantly greater than those under normal physiological conditions. NCCs, which are considered potential evolutionary precursors of natural killer (NK) cells, possess spontaneous recognition and binding capabilities against pathogenic molecules and xenogeneic tissues, resulting in the elimination of specific target cells (30). However, the morphology and molecular markers of NCCs in fish differ from those of mammalian NK cells. For instance, the CD56 molecule, which is a core marker of human NK cells (51), is absent in almost all fish species. NCCRP1 was identified as the first recognized marker of NCCs (52). Furthermore, our previous studies reported several potential markers for tilapia NCCs, including CLEC12B, which showed complete overlap with NCCRP1 (25, 32). Similarly, the distribution and response patterns of CLEC12B were found to be similar to those of NCCRP1 (38). In this study, highly consistent profiles of CLEC12B were also observed, highlighting the involvement of NCCs in the defense of the tilapia brain against bacterial infections. Additionally, in mammals, proinflammatory factors are secreted by macrophages, dendritic cells, and CD4+ T cells (53, 54), and similar colocalization phenomena have been reported in tilapia (25, 34), suggesting the activation of inflammation due to the stimulation of macrophages in the brain.
Therefore, an antibody against tilapia NCCRP1 was prepared, and the NCCs (NCCRP1+ cells) in tilapia were examined. These results indicated that NCCRP1 is widely distributed in the tilapia brain, although its abundance remains lower than that in the HKL. However, RNA-Seq data revealed that the rank of NCCRP1 transcripts more than 60% of all genes in the tilapia brain. The visually poor content may be attributed to the extreme abundance of NCCRP1 in HKLs, where it exhibited a positive rate of greater than 25% (25). Furthermore, the mean FPKM of NCCRP1 in HKLs (approximately 3000) was comparable to that of common reference genes such as GAPDH and EF1a. Subsequently, abundant NCCRP1+ cells were detected in the tilapia brain, particularly on the surfaces of the olfactory bulb, cerebellum, and medulla oblongata, suggesting that the meninges in fish might have similar immunological functions to those in mammalian brains, acting as neuroimmune interface (3, 13). Finally, cytotoxicity and cell death were assessed and confirmed, and the results indicated that these effects were likely induced by NCCs (39, 40), further supporting the involvement of NCCs in dominant antibacterial immunity in the tilapia brain.
In summary, our study provides insight into the distribution of professional and autochthonous defenders in the tilapia brain and identifies the primary immune cells involved in combating streptococcal infection. These findings suggest that the complex neuroimmune connection observed in mammals was already emerging in the tilapia brain, with NCCs playing a central role. This research significantly contributes to our understanding of the evolutionary development of neuroimmune mechanisms from fish to humans.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA1084852.
Ethics statement
The animal study was approved by Ethics Committee of Guangdong Ocean University. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
XH: Conceptualization, Funding acquisition, Writing – original draft, Writing – review & editing. QL: Conceptualization, Data curation, Investigation, Methodology, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by Shandong Provincial Natural Science Foundation (Grant ZR2022QC194 to XH), National Natural Science Foundation of China (Grant 32303065 to XH), Research Fund for Lin He’s Academician Workstation of New Medicine and Clinical Translation in Jining Medical University (Grant JYHL2021MS21 to XH), and College Students’ Innovation Training Program (Grant cx2023085z to XH).
Acknowledgments
We would like to thank the reviewers for their kind and helpful comments on the original manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1442906/full#supplementary-material
Abbreviations
BBB, Blood-brain barrier; CLP, Common lymphoid progenitor; CP, Choroid plexus; CTL, Cytotoxic T-cell; CSF, Cerebrospinal fluid; DC, Dendritic cell; DEG, Differentially expressed gene; FPKM, Fragments per kilobase of transcript per million mapped reads; HKL, Head kidney leukocyte; HSC, Hematopoietic stem cell; H&E, Hematoxylin and eosin; IF, Immunofluorescence; Mo, Monocyte; Mφ, Macrophage; NCC, Nonspecific cytotoxic cell; NCCRP, Nonspecific cytotoxic cell receptor protein 1; NK, Natural killer cell; PBS, Phosphate-buffered saline; qRT−PCR, Quantitative real-time PCR.
References
1. Louveau A, Harris TH, Kipnis J. Revisiting the mechanisms of CNS immune privilege. Trends Immunol. (2015) 36:569–77. doi: 10.1016/j.it.2015.08.006
2. Forrester JV, McMenamin PG, Dando SJ. CNS infection and immune privilege. Nat Rev Neurosci. (2018) 19:655–71. doi: 10.1038/s41583-018-0070-8
3. Croese T, Castellani G, Schwartz M. Immune cell compartmentalization for brain surveillance and protection. Nat Immunol. (2021) 22:1083–92. doi: 10.1038/s41590-021-00994-2
4. Wu D, Chen Q, Chen X, Han F, Chen Z, Wang Y. The blood-brain barrier: structure, regulation, and drug delivery. Signal Transduct Target Ther. (2023) 8:217. doi: 10.1038/s41392-023-01481-w
5. Kipnis J, Filiano AJ. Neuroimmunology in 2017: The central nervous system: privileged by immune connections. Nat Rev Immunol. (2018) 18:83–4. doi: 10.1038/nri.2017.152
6. Deczkowska A, Amit I, Schwartz M. Microglial immune checkpoint mechanisms. Nat Neurosci. (2018) 21:779–86. doi: 10.1038/s41593-018-0145-x
7. Patani R, Hardingham GE, Liddelow SA. Functional roles of reactive astrocytes in neuroinflammation and neurodegeneration. Nat Rev Neurol. (2023) 19:395–409. doi: 10.1038/s41582-023-00822-1
8. Geyer S, Jacobs M, Hsu NJ. Immunity against bacterial infection of the central nervous system: An astrocyte perspective. Front Mol Neurosci. (2019) 12:57. doi: 10.3389/fnmol.2019.00057
9. Castellani G, Croese T, Peralta Ramos JM, Schwartz M. Transforming the understanding of brain immunity. Science. (2023) 380:eabo7649. doi: 10.1126/science.abo7649
10. Prinz M, Priller J. The role of peripheral immune cells in the CNS in steady state and disease. Nat Neurosci. (2017) 20:136–44. doi: 10.1038/nn.4475
11. Louveau A, Smirnov I, Keyes TJ, Eccles JD, Rouhani SJ, Peske JD, et al. Structural and functional features of central nervous system lymphatic vessels. Nature. (2015) 523:337–41. doi: 10.1038/nature14432
12. Noe FM, Marchi N. Central nervous system lymphatic unit, immunity, and epilepsy: Is there a link? Epilepsia Open. (2019) 4:30–9. doi: 10.1002/epi4.12302
13. Rustenhoven J, Drieu A, Mamuladze T, de Lima KA, Dykstra T, Wall M, et al. Functional characterization of the dural sinuses as a neuroimmune interface. Cell. (2021) 184:1000–16. doi: 10.1016/j.cell.2020.12.040
14. Wullimann MF, Vernier P. Evolution of the brain in fishes. In: Encyclopedia of Neuroscience, 1st ed. Springer Press, Berlin, Heidelberg (2009). p. 1318–26. doi: 10.1007/978-3-540-29678-2_3151
15. Rauta PR, Nayak B, Das S. Immune system and immune responses in fish and their role in comparative immunity study: a model for higher organisms. Immunol Lett. (2012) 148:23–33. doi: 10.1016/j.imlet.2012.08.003
16. Mokhtar DM, Zaccone G, Alesci A, Kuciel M, Hussein MT, Sayed RK. Main components of fish immunity: An overview of the fish immune system. Fishes. (2023) 8:93. doi: 10.3390/fishes8020093
17. Lieschke GJ, Trede NS. Fish immunology. Curr Biol. (2009) 19:678–82. doi: 10.1016/j.cub.2009.06.068
18. Stosik M, Tokarz-Deptula B, Deptula W. Immunological memory in teleost fish. Fish Shellfish Immunol. (2021) 115:95–103. doi: 10.1016/j.fsi.2021.05.022
19. Bshary R, Gingins S, Vail AL. Social cognition in fishes. Trends Cogn. Sci. (2014) 18:465–71. doi: 10.1016/j.tics.2014.04.005
20. Levraud JP, Rawls JF, Clatworthy AE. Using zebrafish to understand reciprocal interactions between the nervous and immune systems and the microbial world. J Neuroinflamm. (2022) 19:170. doi: 10.1186/s12974-022-02506-x
21. Fan B, Chen F, Li Y, Wang Z, Wang Z, Lu Y, et al. A comprehensive profile of the tilapia (Oreochromis niloticus) circular RNA and circRNA-miRNA network in the pathogenesis of meningoencephalitis of teleosts. Mol Omics. (2019) 15:233–46. doi: 10.1039/c9mo00025a
22. Palang I, Withyachumnarnkul B, Senapin S, Sirimanapong W, Vanichviriyakit R. Brain histopathology in red tilapia Oreochromis sp. experimentally infected with Streptococcus agalactiae serotype III. Microsc. Res Tech. (2020) 83:877–88. doi: 10.1002/jemt.23481
23. Eto SF, Fernandes DC, Moraes AC, Alecrim J, Souza PG, Carvalho FCA, et al. Meningitis caused by Streptococcus agalactiae in Nile tilapia (Oreochromis niloticus): Infection and inflammatory response. Animals. (2020) 10:2166. doi: 10.3390/ani10112166
24. Wang B, Thompson KD, Wangkahart E, Yamkasem J, Bondad-Reantaso MG, Tattiyapong P, et al. Strategies to enhance tilapia immunity to improve their health in aquaculture. Rev Aquac. (2022) 15:41–56. doi: 10.1111/raq.12731
25. Li Q, Jiang B, Zhang Z, Huang Y, Xu Z, Chen X, et al. Serotonin system is partially involved in immunomodulation of Nile tilapia (Oreochromis niloticus) immune cells. Front Immunol. (2022) 13:944388. doi: 10.1111/raq.12731
26. Jiang B, Li Q, Zhang Z, Huang Y, Wu Y, Li X, et al. Selection and evaluation of stable reference genes for quantitative real-time PCR in the head kidney leukocyte of. Oreochromis niloticus. Aquac. Rep. (2023) 31:101660. doi: 10.1016/j.aqrep.2023.101660
27. Li Q, Liu R, Ma R, Huang Y, Zhang Z, Zhang L, et al. Brain transcriptome response to Streptococcus agalactiae infection and the heterogeneous regulation of neuropeptides on immune response in tilapia, Oreochromis niloticus. Aquaculture. (2022) 555:738222. doi: 10.1016/j.aquaculture.2022.738222
28. Yang CG, Wang XL, Tian J, Liu W, Wu F, Jiang M, et al. Evaluation of reference genes for quantitative real-time RT-PCR analysis of gene expression in Nile tilapia (Oreochromis niloticus). Gene. (2013) 527:183–92. doi: 10.1016/j.gene.2013.06.013
29. Wang B, Jian J, Lu Y, Cai S, Huang Y, Tang J, et al. Complete genome sequence of Streptococcus agalactiae ZQ0910, a pathogen causing meningoencephalitis in the GIFT strain of Nile tilapia (Oreochromis niloticus). J Bacteriol. (2012) 194:5132–3. doi: 10.1128/JB.01080-12
30. Huang Y, Liu X, Cai J, Tang J, Cai S, Lu Y, et al. Biological characterisation, expression and functional analysis of non-specific cytotoxic cell receptor protein 1 in Nile tilapia (Oreochromis niloticus). Fish Shellfish Immunol. (2020) 104:579–86. doi: 10.1016/j.fsi.2020.05.048
31. Jiang B, Li Q, Zhang Z, Huang Y, Wu Y, Li X, et al. Involvement of CD27 in innate and adaptive immunities of Nile tilapia (Oreochromis niloticus). Fish Shellfish Immunol. (2023) 139:108923. doi: 10.1016/j.fsi.2023.108923
32. Niu J, Huang Y, Liu X, Zhang Z, Tang J, Wang B, et al. Single-cell RNA-seq reveals different subsets of non-specific cytotoxic cells in teleost. Genomics. (2020) 112:5170–9. doi: 10.1016/j.ygeno.2020.09.031
33. Zhang Z, Li Q, Huang Y, Xu Z, Chen X, Jiang B, et al. Vasoactive intestinal peptide (VIP) protects Nile tilapia (Oreochromis niloticus) against Streptococcus agalatiae infection. Int J Mol Sci. (2022) 23:14895. doi: 10.3390/ijms232314895
34. Wu L, Gao A, Li L, Chen J, Li J, Ye J. A single-cell transcriptome profiling of anterior kidney leukocytes from Nile tilapia (Oreochromis niloticus). Front Immunol. (2021) 12:783196. doi: 10.3389/fimmu.2021.783196
35. Ruan C, Elyaman W. A new understanding of TMEM119 as a marker of microglia. Front Cell Neurosci. (2022) 16:902372. doi: 10.3389/fncel.2022.902372
36. Abdelhak A, Foschi M, Abu-Rumeileh S, Yue JK, D’Anna L, Huss A, et al. Blood GFAP as an emerging biomarker in brain and spinal cord disorders. Nat Rev Neurol. (2022) 18:158–72. doi: 10.1038/s41582-021-00616-3
37. Li Y, Huang Y, Cai J, Jiang D, Jian J, Lu Y, et al. Establishment of an astrocyte-like cell line from the brain of tilapia (Oreochromis niloticus) for virus pathogenesis and a vitro model of the blood–brain barrier. J Fish. Dis. (2022) 45:1451–62. doi: 10.1111/jfd.13674
38. Zhang Z, Niu J, Li Q, Huang Y, Jiang B, Li X, et al. A novel C-type lectin (CLEC12B) from Nile tilapia (Oreochromis niloticus) is involved in host defense against bacterial infection. Fish Shellfish Immunol. (2022) 131:218–28. doi: 10.1016/j.fsi.2022.09.073
39. Huang Y, Chen Z, Xie R, Wang P, Zhang Z, Cai J, et al. Transferrin mediated NCC killing activity through NCCRP-1 in Nile tilapia (Oreochromis niloticus). Fishes. (2022) 7:253. doi: 10.1016/j.fsi.2024.109594
40. Huang Y, Zheng Q, Wang Z, Tang J, Lu Y, Qin Q, et al. Fish natural killer enhancing factor-A (NKEF-A) enhance cytotoxicity of nonspecific cytotoxic cells against bacterial infection. Mol Immunol. (2021) 133:184–93. doi: 10.1016/j.molimm.2021.02.017
41. Shainer I, Kuehn E, Laurell E, Kassar M, Mokayes N, Sherman S, et al. A single-cell resolution gene expression atlas of the larval zebrafish brain. Sci Adv. (2023) 9:eade9909. doi: 10.1126/sciadv.ade9909
42. Wang Q, Peng C, Yang M, Huang F, Duan X, Wang S, et al. Single-cell RNA-seq landscape midbrain cell responses to red spotted grouper nervous necrosis virus infection. PloS Pathog. (2021) 17:e1009665. doi: 10.1371/journal.ppat.1009665
43. Tibi M, Hayun SB, Hochgerner H, Lin Z, Givon S, Ophir O, et al. A telencephalon cell type atlas for goldfish reveals diversity in the evolution of spatial structure and cell types. Sci Adv. (2023) 9:eadh7693. doi: 10.1126/sciadv.adh7693
44. Henson HE, Parupalli C, Ju B, Taylor MR. Functional and genetic analysis of choroid plexus development in zebrafish. Front Neurosci. (2014) 8:364. doi: 10.3389/fnins.2014.00364
45. Olstad EW, Ringers C, Hansen JN, Wens A, Brandt C, Wachten D, et al. Ciliary beating compartmentalizes cerebrospinal fluid flow in the brain and regulates ventricular development. Curr Biol. (2019) 29:229–41. doi: 10.1016/j.cub.2018.11.059
46. Wu S, Nguyen LT, Pan H, Hassan S, Dai Y, Xu J, et al. Two phenotypically and functionally distinct microglial populations in adult zebrafish. Sci Adv. (2020) 6:eabd1160. doi: 10.1126/sciadv.abd1160
47. Silva NJ, Dorman LC, Vainchtein ID, Horneck NC, Molofsky AV. In situ and transcriptomic identification of microglia in synapse-rich regions of the developing zebrafish brain. Nat Commun. (2021) 12:5916. doi: 10.1038/s41467-021-26206-x
48. Chen J, Poskanzer KE, Freeman MR, Monk KR. Live-imaging of astrocyte morphogenesis and function in zebrafish neural circuits. Nat Neurosci. (2020) 23:1297–306. doi: 10.1038/s41593-020-0703-x
49. Li Q, Jiang B, Zhang Z, Huang Y, Xu Z, Chen X, et al. SP protects Nile tilapia (Oreochromis niloticus) against acute Streptococcus agalatiae infection. Fish Shellfish Immunol. (2022) 123:218–28. doi: 10.1016/j.fsi.2022.03.002
50. Mohammad MG, Tsai VW, Ruitenberg MJ, Hassanpour M, Li H, Hart PH, et al. Immune cell trafficking from the brain maintains CNS immune tolerance. J Clin Invest. (2014) 124:1228–41. doi: 10.1172/JCI71544
51. Shi FD, Ljunggren HG, La Cava A, Van Kaer L. Organ-specific features of natural killer cells. Nat Rev. Immunol. (2011) 11:658–71. doi: 10.1038/nri3065
52. Evans DL, Jaso-Friedmann L. Nonspecific cytotoxic cells as effectors of immunity in fish. Annu Rev Fish Dis. (1992) 2:109–21. doi: 10.1016/0959-8030(92)90059-7
53. Kany S, Vollrath JT, Relja B. Cytokines in inflammatory disease. Int J Mol Sci. (2019) 20:6008. doi: 10.3390/ijms20236008
Keywords: Nile tilapia, Streptococcus agalactiae, medulla oblongata, NCCs, immune response
Citation: Hou X and Li Q (2024) Medulla oblongata and NCCs are central defenders against Streptococcus agalactiae infection of the tilapia brain. Front. Immunol. 15:1442906. doi: 10.3389/fimmu.2024.1442906
Received: 03 June 2024; Accepted: 18 June 2024;
Published: 01 July 2024.
Edited by:
Ming Xian Chang, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Guan-Jun Yang, Ningbo University, ChinaVikash Kumar, Central Inland Fisheries Research Institute (ICAR), India
Copyright © 2024 Hou and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xitan Hou, aG91eGl0YW5AMTI2LmNvbQ==; Qi Li, Vmlja3lxaTEwMTVAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Xitan Hou
Xitan Hou Qi Li
Qi Li