- 1Murdoch Children’s Research Institute, Melbourne, VIC, Australia
- 2Department of Microbiology and Immunology, University of Melbourne, Melbourne, VIC, Australia
- 3Department of Paediatrics, University of Melbourne, Melbourne, VIC, Australia
- 4Fiona Elsey Cancer Institute, Ballarat, VIC, Australia
- 5Federation University Australia, Ballarat, VIC, Australia
Invariant natural killer T (iNKT) cells are a subset of lipid-reactive, unconventional T cells that have anti-tumor properties that make them a promising target for cancer immunotherapy. Recent studies have deciphered the developmental pathway of human MAIT and Vγ9Vδ2 γδ-T cells as well as murine iNKT cells, yet our understanding of human NKT cell development is limited. Here, we provide an update in our understanding of how NKT cells develop in the human body and how knowledge regarding their development could enhance human treatments by targeting these cells.
Introduction
T cells are highly specialized lymphocytes that undergo a complex maturation and differentiation program that gives rise to distinct T-cell lineages with unique effector functions. Conventional T cells that recognize peptide antigens presented by the major histocompatibility complex (MHC) undergo well-characterized processes of antigen receptor rearrangement followed by positive and negative selection in the thymus. They typically exit the thymus as naive progenitors that can be primed upon primary antigen encounter in the secondary lymphoid organs, which confer functional maturity and effector subset differentiation.
The thymus also supports the development of several populations of unconventional T cells that appear to undergo a decidedly distinct maturation process. Unconventional T cells typically recognize non-peptide antigens presented by distinct antigen-presenting molecules. CD1d molecules present lipid antigens to natural killer T (NKT) cells, MR1 presents microbial riboflavin derivatives to mucosal-associated invariant T (MAIT) cells, and human gamma delta (γδ) T cells are potently activated by a wide range of ligands, e.g., Vδ2+ T cells react to phosphoantigen-bound butyrophilin molecules. NKT cells are defined by their reactivity to CD1d-restricted lipid antigens, and humans contain two main subsets of NKT cells, called type 1 or invariant NKT (iNKT) cells and type 2, or diverse NKT cells (1).
Human iNKT cells express a semi-invariant alpha beta T-cell receptor (αβ-TCR) composed of a Vα24Jα18 alpha chain paired to Vβ11 that recognizes the prototypic ligand alpha-galactosylceramide (αGalCer) presented by CD1d. Diverse NKT cells express diverse TCRs that can also recognize lipid antigens presented by CD1d, but do not recognize αGalCer (1). αGalCer loaded CD1d tetramers and monoclonal antibodies specific to human TCR chains of iNKT cells are widely used tools to stringently identify iNKT cells in health and disease settings (2–6). Diverse NKT cells are considered to be slightly more abundant in humans, but the lack of specific reagents to identify them has hindered their detailed analysis (1). Hence, this review will focus on human iNKT cells and how our knowledge of their development could enhance human immunotherapies that target these cells.
Human iNKT cells
Human iNKT cells display great variability in responding to activation stimuli (7, 8). Remarkably, they exhibit dual functionality via differential secretion of pro- and anti-inflammatory cytokines, and they can also display direct cytotoxicity via the release of lytic granules or via death receptor ligation (9–15). They play important roles in regulating immune responses associated with tumor surveillance, infectious diseases, autoimmunity, and graft versus host disease (GvHD), and are increasingly seen as attractive therapeutic targets to prevent human diseases (16, 17).
In contrast to mouse iNKT cells that represent ~1% of blood and up to 45% of T cells in the liver, human iNKT cells are much rarer. They account for approximately 0.1% of T cells in peripheral blood [reviewed in (1)], and their numbers are only slightly higher in organs such as the liver and spleen, with the exception of the omentum and adipose tissue where they constitute ~10% and 1% of T cells, respectively (1, 18, 19). iNKT cells can be activated in a TCR-dependent manner by recognition of lipid antigens presented by CD1d or in a TCR-independent manner by cytokines such as IL-12 and IL-18 (8). Both pathways are important for their dual functionality.
NKT cell heterogeneity in humans and mice
In humans, iNKT cells can be broadly grouped into three main subsets defined by differential expression of CD4 and CD8 (Figure 1). Whereas mice only have CD4+ and CD4−CD8− double-negative (DN) iNKT subsets, humans contain an additional subset of CD8+ iNKT cells (20–22). Generally, human CD4+ iNKT cells differ functionally from their CD4− counterparts. CD4+ iNKT cells exhibit a T helper, TH0 cytokine signature and can simultaneously secrete IFNγ and IL-4, whereas CD4− iNKT cells lack IL-4 secretion, exhibit an IFNγ-positive TH1 cytokine profile, express several NK receptors, and can also mediate direct cytotoxicity (Figure 1) (23, 24). Minor differences exist between human DN and CD8 iNKT cells in their TH1 cytokine secretion and cytotoxicity (10, 13, 24, 25). Other variations of human iNKT cells have been described, marked by the expression of CD161 or CD62L, although their significance is not well defined (23). Single-cell RNA sequencing of unstimulated and stimulated human iNKT cells revealed additional clusters of functionally diverse iNKT cells, while also corroborating well-established functional differences between CD4+ and CD4− subsets (26).
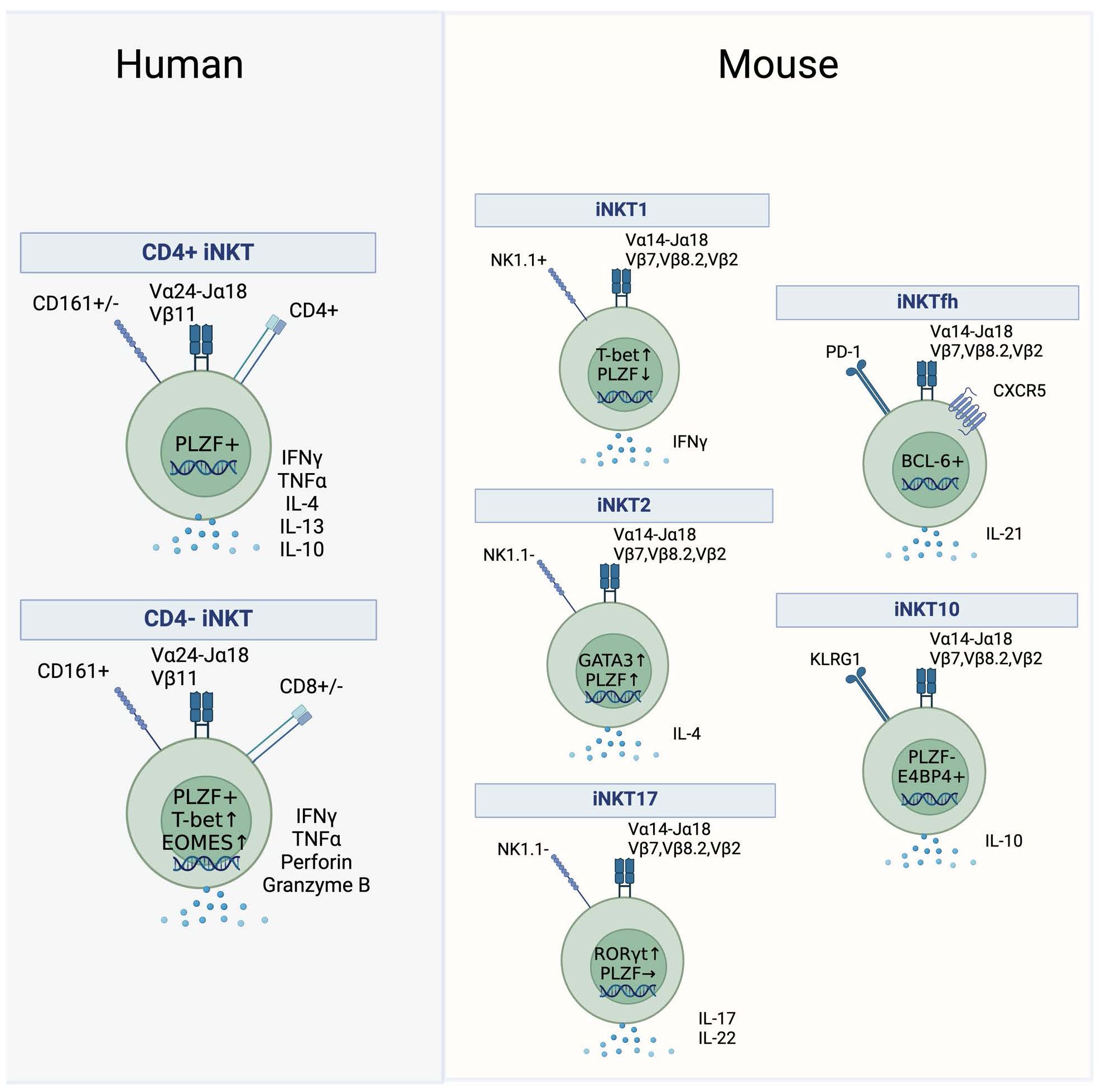
Figure 1. Human and mouse iNKT subsets. Subsets of iNKT cells in humans (left) and mice (right). Key surface markers, cytokines and transcription factors that define the subsets are shown.
Effector subsets in mice are not grouped according to the expression of CD4 but are rather defined by the expression of transcription factors and their ability to secrete cytokines. After thymic development in the mouse, functional iNKT subsets diversify into three separate lineages termed iNKT1, iNKT2, and iNKT17, analogous to conventional TH1, TH2 and TH17 subsets (Figure 1). NKT1 cells are low in PLZF and high in Tbet and secrete IFNγ, NKT2 cells express high levels of PLZF and GATA3 and secrete IL-4, whereas NKT17 cells have high expression levels of RORγt coupled with intermediate PLZF expression and secrete IL-17 (Figure 1) (27–29). It is unclear if human equivalents of these subsets exist, as human iNKT cells appear to be polarized towards a TH1 or mixed TH0 functionality while their ability to produce IL-17 ex vivo is more controversial (30–33). Furthermore, even though human iNKT cells express PLZF and the protein levels of Tbet and EOMES vary between the CD4+ and CD4− subset, the classification of human iNKT cells based on the expression of key transcription factors has not been investigated in detail (34–36).
Besides the well-defined murine NKT1, NKT2, and NKT17 and the human CD4+ and CD4− subsets, there are reports of additional iNKT subsets. Immunoregulatory iNKT cells capable of secreting IL-10 were described in mice and in human PBMCs (37). In mice, this subset is termed NKT10 cells, and these cells are enriched in adipose tissue (Figure 1) (38). They are characterized by the absence of PLZF and by the expression of the transcription factor E4BP4 (38). An additional immunoregulatory, suppressive iNKT subset is induced in mice and humans upon stimulation in the presence of TGF-β or rapamycin resulting in the expression of FOXP3 (39, 40). In mice, the emergence of follicular helper NKT (NKTfh) cells was demonstrated following αGalCer administration (Figure 1). This subset closely resembled conventional Tfh cells as it was dependent on the expression of BCL-6, expressed PD-1 and CXCR5, and was able to induce germinal centers via IL-21 signaling (41). An analogous human subset of PD-1 and CXCR5 co-expressing iNKT cells was discovered in human tonsils (41).
Development of mouse iNKT cells
We have previously reviewed the development of mouse iNKT cells, including a detailed description of key signaling molecules, transcription factors, and microRNAs that regulate this process (27). Early studies by us and others suggested that mouse iNKT cells develop in a linear manner, via a 4-stage maturation pathway characterized by the differential expression of CD24, CD44, and NK1.1, with fully mature cells expressing NK1.1 (27, 42, 43). However, subsequent studies revealed that mature subsets of iNKT2 and iNKT17 cells were present within mouse thymus that lacked the expression of NK1.1 (29, 44). Thus, these later studies indicated that thymic iNKT cells developed via lineage diversification, from a common CD24hi, CD44lo, NK1.1− iNKT0 progenitor into distinct mature iNKT1, iNKT2, and iNKT17 subsets. More recently, single-cell sequencing analysis suggested that mouse iNKT2 cells represent a branching point for the generation of iNKT1 and iNKT17 cells, and that iNKT1 cells upregulate NK1.1 upon final maturation (45).
PLZF is described to be the master regulator of unconventional T-cell development, and PLZF is absolutely required for development of functionally mature subsets of mouse iNKT cells and MAIT cells (27, 35, 36, 46, 47). The expression of PLZF in mouse iNKT cells is regulated by the transcription factors Egr2 and Runx1 (48, 49). PLZF expression is first detected in mouse NKT0 progenitors and high levels of PLZF are maintained by iNKT2 cells, whereas PLZF is downregulated in mature subsets of mouse iNKT1 and iNKT17 cells (27, 29, 35). Notably, forced suppression of PLZF results in higher frequencies of mouse iNKT1 cells, and the microRNA, Let7, is required for the downregulation of PLZF in mouse iNKT1 cells (47, 50). Thus, PLZF expression is essential for mouse iNKT cell development.
Development of human iNKT cells
While the developmental pathway of mouse iNKT cells has been well studied (reviewed in (27, 51)), the exploration of human iNKT cell development has proven challenging. The earliest human iNKT cell progenitors emerge in the fetal thymus at 13 weeks of gestation and start to colonize peripheral tissues, especially the gut, but also the spleen and mesenteric lymph nodes in the second trimester (52, 53). In humans, the frequency of fetal thymic iNKT cells negatively correlates with gestational age and early studies failed to detect clear populations of iNKT cells in the postnatal thymus (52, 54). This observation implied that a significant portion of iNKT cell thymic development occurs during early fetal life and led to the initial hypothesis that the postnatal pool of iNKT cells is reconstituted by peripheral expansion of fetal-derived iNKT cells (52, 54). However, subsequent studies identified iNKT cells from human postnatal thymus, conclusively demonstrating that iNKT cells could be generated after fetal development (55, 56). The discrepancy between these studies may be partially attributed to the development of stringent detection reagents that facilitate the detection of these rare thymic progenitor cells (55, 56).
The timing of thymic iNKT cell development in humans and mice is different. While clear evidence of fetal thymic iNKT cells exists for humans, thymic development of iNKT cells in mice appears to be limited to the postnatal thymus (57). Specifically, iNKT cells fail to develop in neonatally thymectimized mice, revealing that the postnatal thymus is essential for mouse iNKT cell development (57). iNKT cells arise from the same lymphoid progenitors as conventional αβ-T cells but diverge at the CD4+CD8+ double-positive (DP) stage (58–60). In mice, selection of iNKT cells depends on CD1d expressed by DP thymocytes (61), and the presence of similar immature iNKT progenitors in both human and mouse thymus suggests that human iNKT cells are selected in a similar manner. Human studies investigating thymic maturation based on phenotypic heterogeneity demonstrate a dominance of CD4+ iNKT cells in the neonatal thymus, with CD4− iNKT cells accumulating in the blood with age (52, 55, 56, 62). Specifically, DN and CD8 iNKT cells do not emerge in peripheral blood until 6 months of age, although their levels in the thymus remain low even in older individuals (55). The scarcity of CD4− iNKT cells among thymic progenitors raises the question of the ontogenic origin of these cells (Figure 2) (55, 56). While the possibility exists that CD4− iNKT cells develop at a distinct extrathymic site, the lower T-cell receptor excision circle (TREC) content in these peripheral cells suggests that small numbers of CD4− iNKT cells exported from the thymus may increase in frequency following peripheral expansion, while CD4+ iNKT cells are maintained by thymic output (56). However, other studies could not find evidence of active proliferation in any peripheral iNKT cell subset outside of the thymus (55). A recent study analyzed iNKT TCR beta clonotypes in longitudinal matched samples and found hints of a linear relationship in which CD4+ cells give rise to CD4− subsets in the periphery but not vice versa (Figure 2) (11). The clear distinction in CD4 expression between thymic and peripheral iNKT cells is unique to humans, because even though the majority of mouse thymic iNKT cells are CD4+, there is a significant proportion of CD4− cells that emerge during development (63, 64).
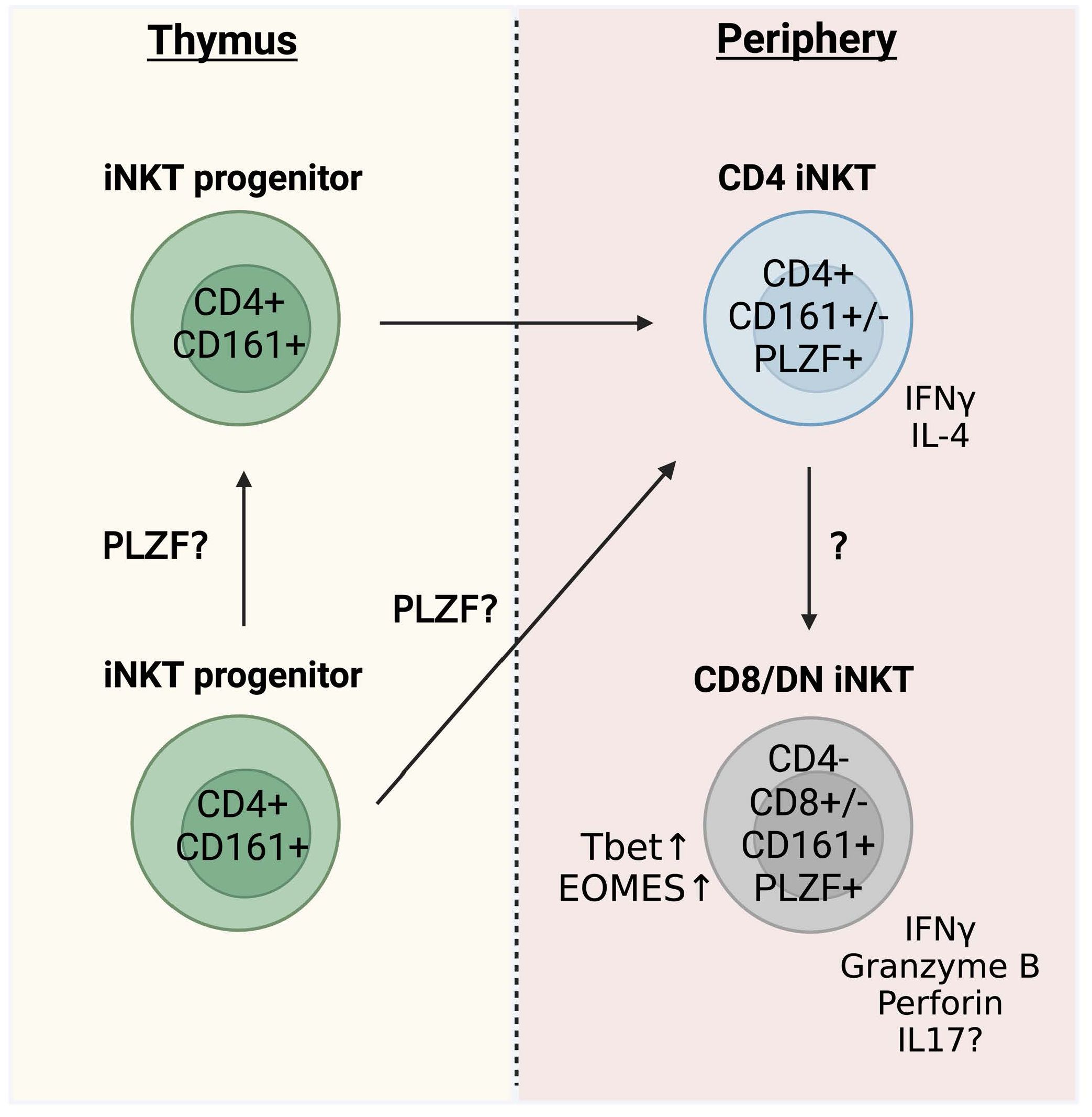
Figure 2. Human thymic iNKT cell development. Proposed pathway of human thymic iNKT cell development.
Like mouse NKT1 cells, the human marker for mouse NK1.1 (CD161) is upregulated on fully mature human NKT cells. Although some CD161+ iNKT cells are already present in the thymus, the majority of human thymic iNKT cells appear to be CD161 negative, and CD161 expression accumulates with age in the periphery (52, 55, 56). Importantly, the increase in CD161+ iNKT cells was restricted to the blood (55). These data indicate that the developmental pathway of iNKT cells in humans is characterized by the upregulation of CD161 (21, 55). In accordance with this, a recent study used single-cell RNA sequencing to reveal the transcriptome of human thymic iNKT cells (65). Although the authors use of cryopreserved thymocytes might skew the ratio of developmental stages due to the preferential death of DP thymocytes, a profound upregulation of CD161 transcripts was identified in the clusters that comprised iNKT cells of later developmental stages (65, 66). Furthermore, a developmental trajectory was proposed that resembled murine iNKT cell development in which human immature “NKT2-like” cells develop into fully matured cells with a NKT1 signature (65).
Development of iNKT cell progenitors appears strongly dependent on IL-7 signaling since thymic and neonatal but also adult peripheral CD4+ iNKT cells express high levels of CD127 and are hyperresponsive to IL-7 (52, 62). CD4− iNKT cells from adult blood on the other hand express higher levels of CD122 and depend on IL-15 signaling (56).
Although the activated memory phenotype that is characteristic of human iNKT cells is imprinted during thymic development and already present in neonatal blood (67, 68), thymic iNKT cell progenitors do not seem to be functionally mature. This was indicated by the absence or comparatively weak cytokine response when stimulated ex vivo, especially when TCR ligation was used instead of PMA and ionomycin (23, 56, 62, 67). The observation that human iNKT cells are subject to substantial peripheral maturation was previously underpinned by a study recently published as a preprint that discovered shared transcriptional states between thymic naïve conventional T cells and iNKT cells using single-cell RNA-sequencing (69). Notably, the paper found no clusters of human iNKT cells that directly corresponded to mouse iNKT1, iNKT2, and iNKT17 subsets, suggesting that human iNKT cells may not fully differentiate into these distinct lineages, but rather acquire a mixed iNKT1/NKT17 effector program in the thymus (69). Similarly, data from Legoux, Lantz, and colleagues also support the idea of human iNKT cells acquiring a mixed Th1/17 effector program akin to the development of MAIT cells (70). Interestingly, only a small subset of thymic iNKT cells exhibited an effector signature, and this signature closely resembled that of iNKT cells in the peripheral blood of adult donors, whereas most thymic iNKT cells appeared to reside in a naïve state (69). It was suggested that the naïve phenotype of human thymic iNKT cells results from positive selection on thymic epithelial cells (TECs) instead of DP thymocytes, since TECs do not provide SLAM signaling, which is critical for the induction of the effector program in iNKT cells (69).
Overall, the cumulative data suggest that iNKT cells in the human thymus are CD4+ and CD161−. Thereby, the expression of CD161 is considered a major control point in the maturation of iNKT cells (21), which mostly occurs extrathymically and increases with age and reaches the highest levels in adult PBMCs. CD4− iNKT cell subsets (DN and CD8+) emerge later and almost exclusively in the periphery. The origin of these cells is still unclear but CD4− cells likely arise from CD4+ cells (Figure 2) (11).
Transcriptional regulation of human iNKT development
PLZF is highly expressed in human peripheral iNKT cells (35, 36), although protein expression has not been examined for human iNKT cells from the thymus. A recent paper used single-cell RNA-sequencing to indicate that PLZF is expressed by mature subsets of human thymic iNKT cells, although gene expression appeared low (70). Interestingly, a patient with a biallelic mutation of PLZF revealed a failure to develop DN and CD161+ iNKT cells, consistent with perturbed thymic NKT cell development (71). These data correspond well with mouse studies (27, 35, 36, 46, 47) and strongly hint towards an equally important role of PLZF in the development of human iNKT cells (Figure 2) (71). It is not known if the dynamics of PLZF expression in humans is similar to the mouse and whether it is differentially expressed during the emergence of distinct functional subsets. Characterizing the expression of PLZF during human iNKT cell thymic development will help to definitively establish a role for PLZF in human iNKT cell development and its impact on lineage commitment.
We know from mouse studies that iNKT cell development is a tightly regulated process that relies crucially on signaling via the SLAM–SAP–Fyn axis, reviewed in (27). Consistent with this, the lack of SAP in patients with X-linked lymphoproliferative syndrome (XLP) correlates with an absence of iNKT cells (72, 73). Multiple other factors exist that are affected in those with primary immunodeficiencies and are associated with defects in iNKT and CD1-restricted T cells, reviewed in (74). Two factors that appear essential for human iNKT development are the Wiskott–Aldrich syndrome protein (WASp) and X-linked inhibitor-of-apoptosis (XIAP). WASp is mutated in Wiskott–Aldrich syndrome patients who exhibit an almost-complete absence of circulating iNKT cells (75). Mutations in XIAP can also cause XLP, and XIAP-deficient patients had low levels of iNKT cell levels, comparable to those seen in SAP-deficient XLP donors (76). The underlying mechanism from a developmental point of view is unexplored, but data from peripheral blood suggest that XIAP counteracts a proneness to increased apoptosis in iNKT cells mediated by PLZF, which may explain the abrogated iNKT cell development in XLP patients with a XIAP deficiency (77).
Overlap between NKT cell development and other human unconventional T-cell pathways
We have recently characterized the thymic development of human MAIT cells and Vγ9Vδ2+ γδ T cells (27, 46, 78–80). Both pathways describe a linear three-stage thymic pathway that involves the upregulation of CD161. The earliest stage for human thymic MAIT cells is CD27−CD161− stage 1 cells that upregulate CD27 to become stage 2 CD27+CD161− cells before finally upregulating CD161 to become stage 3 CD27+CD161+ cells (46, 78, 80). For human thymic Vγ9Vδ2+ γδ T cells, stage 1 cells are CD4+CD161− that transition to become CD4−CD161− stage 2 cells and then to CD4−CD161+ stage 3 cells (79). Importantly, the thymic development of both MAIT cells and Vγ9Vδ2+ γδ T cells involves the upregulation of PLZF as cells mature from stage 1 to stage 3. Furthermore, both pathways involve major changes in the expression of transcription factors, surface molecules, chemokines, chemokine receptors, and cytotoxic killing granules as cells mature from stage 1 to stage 3. For example, CD1A, CD4, CCR9, LEF1, TCF7, BACH2, BCL11B, and SOX4 are expressed by stage 1 MAIT and Vγ9Vδ2+ γδ T cells and are downregulated as these cells mature to their stage 3 counterparts (78, 79). These studies highlight clear overlap between the thymic development pathway for human MAIT cells and Vγ9Vδ2+ γδ T cells (78, 79); thus, it will be important to establish if these characteristics of development among these unconventional T-cell subsets extends to the development of human iNKT cells.
Despite the strong overlap in the development of human MAIT cells and Vγ9Vδ2+ γδ T cells, particularly at early stages, some important differences were observed in the degree of maturation of stage 3 subsets from each population. For example, stage 3 thymic MAIT cells produced very little cytokines compared to mature MAIT cells from the blood, suggesting that significant extrathymic development is required for human MAIT cells, and this was supported by RNA-sequencing data (46, 78, 80). In contrast, stage 3 Vγ9Vδ2+ γδ T cells from human thymus produced levels of IFNγ and TNFα upon stimulation similar to their matched blood counterparts (79). Furthermore, transcriptomic analysis revealed very few differences in the genes expressed by CD4−CD161+ stage 3 cells from the thymus and matched blood, suggesting only minor extrathymic development for human Vγ9Vδ2+ γδ T cells (79). As discussed above, very few thymic human iNKT cells express CD161 and acquisition of CD161 occurs predominantly in periphery (52, 55, 56). These data indicate that human MAIT cells and iNKT cells require additional development steps in the periphery that are yet to be fully defined.
Transcriptomic analysis is a powerful approach to dissect the complexities of human unconventional T-cell development (65, 69, 70, 78, 79), although validation of proposed pathways using phenotypic analysis, functional assays, and precursor-product experiments is crucially required for investigating the development of iNKT cells.
Challenges to deciphering a human thymic iNKT developmental pathway
Even though iNKT cells can be detected in the human thymus and there are similarities between mouse and human thymic iNKT development, our knowledge of the developmental pathway in humans is incomplete. A major challenge in defining the pathway for human iNKT cell development is their relative scarcity within thymus tissue. Their frequency in the human thymus is far lower than in mice and approximately 100 times lower than in matched human peripheral blood, which hinders detailed analysis (23). As demonstrated by Berzins et al., detection of an extremely rare cell population in the human thymus requires refined flow cytometry protocols and stringent gating to reliably detect them above background levels (55). The technological advance in high-dimensional full spectrum flow cytometry has significantly expanded and now allows for a more comprehensive phenotyping of rare cell populations (81–84). Furthermore, the use of MACS enrichment of MAIT cells and Vγ9Vδ2+ γδ T cells was critical to our efforts to define the development pathways for these cells from human thymus (46, 78, 79). The integration of MACS technology in combination with advances in flow cytometry and transcriptomics approaches like RNA-seq will likely provide significant new insights into the development of human thymic iNKT cells (85).
How human immunotherapies could benefit from understanding a defined developmental pathway for NKT cells
Since their discovery more than 30 years ago, an ever-growing body of evidence has emerged that human iNKT cells represent a promising therapeutic target for autoimmune disorders, GvHD, and cancer immunotherapy, and as vaccine adjuvants for infectious diseases (86–90). This is based on their potential for off-the-shelf use in immunotherapies (17), and their rapid, multifunctional effector responses. Importantly, iNKT cells express an invariant TCR shared by all individuals that confers common lipid antigen specificity, and moreover, CD1d is monomorphic, which abolishes any concerns caused by highly polymorphic MHC molecules. Hence, a detailed knowledge about iNKT subsets, their function, and especially how these subsets develop will be important to successfully utilize them for distinct therapeutic approaches (91). For example, understanding what genes are switched on and off during the development of cytokine and cytotoxic granule producing iNKT cells could be exploited to develop NKT cell-based therapies with a desired functional output.
Out of several therapeutic options for iNKT cells, iNKT-based tumor immunotherapies are an especially promising field for future therapeutic applications. Reduced numbers of iNKT cells were observed not only in autoimmunity or obesity (18, 92, 93), but also in several human malignancies [reviewed in 94, 95]. Infiltration of IFNγ-producing iNKT cells thereby positively correlated with better survival (96), demonstrating the importance of iNKT cells in the tumor setting. Furthermore, iNKT cells have been shown to mediate potent anti-tumor immunity (14, 97, 98) by a range of different mechanisms like direct cytotoxicity (99, 100), lysis of tumor promoting immune cell subsets (101), or creating a tumor suppressive immune environment by stimulation of NK cells or CD8 T cells via IFNγ release and DC maturation, respectively (96, 102).
Indeed, many clinical trials have been conducted that employed various strategies to stimulate NKT cells to boost their numbers or induce their function with the objective of targeting different cancers (103, 104). While iNKT cell immunotherapy was feasible and well tolerated in most of these trials, there was only partial success in eradicating the tumors. The reasons for this are not clear, but the pan-targeting strategies employed in most of the trials may be a factor. Anti-tumor activity is dependent on a strong type 1 immunity and the release of IFNγ (105). Notably, iNKT cells are not exclusively pro-inflammatory—and thus anti-tumorigenic—but also mediate potent anti-inflammatory immune responses (9) mediated by IL-4 that could promote tumor growth (10, 106). These conflicting iNKT functions are believed to be regulated through the context in which the cells are activated and on the presence of heterogeneous subsets. These factors include the mode of activation (i.e., cytokine- versus ligand-driven activation), the microenvironment (i.e., the presence of cytokines or antigen-presenting cells), and the strength of TCR signaling (8, 107).
As mentioned earlier, iNKT cell subsets elicit heterogeneous immune responses in which anti-tumor functions appear to be primarily mediated by the CD4-negative subset (10, 13, 14). This suggests that trials might need to target specific subsets of iNKT cells, although recent evidence has shown that relying solely on the expression of CD4 to define the anti-tumor potential of iNKT subsets is not enough. Subset differences were demonstrated in a work by Tian et al. that showed superior anti-tumor potential of CD62L+ CAR-iNKT cells, compared with CD62L− CAR-iNKT cells (108). More recently, a circulating NKT cell subpopulation was identified in humans that exhibited pronounced cytotoxic properties that was marked by the expression of CXCR6 and CD244 (109). Investigation of the developmental trajectory of the murine equivalent to these cells showed that they were occupying a separate developmental niche in the thymus. Development of these CXCR6+CD244+ cells was highly dependent on IL-15 in the thymus, whereas “conventional” iNKT cells depend more on IL-15 in the periphery (109). Efforts to generate human iNKT cells for off-the-shelf cancer immunotherapy using human CD34+ stem cells transduced with NKT TCR and cultured in artificial thymic organoids successfully resulted in hematopoietic stem cell (HSC)-engineered iNKT cells of high purity and yield (110). Most HSC-iNKT cells exhibited a CD4+CD8+ DP phenotype after 6 weeks of culture, before further developing into CD8+ or CD4−CD8− DN iNKT cells, but not CD4+ iNKT cells, after 10 weeks of culture (110). Collectively, these studies have important implications for the in vitro manipulation and expansion of specific subsets of iNKT cells for immunotherapy, highlighting how detailed knowledge of iNKT subset development helps to identify subpopulations suitable for therapeutic approaches.
Concluding remarks
A crucial advance in our efforts to utilize NKT cells in immunotherapies will be to identify functionally distinct subsets and characterize their development pathways. This will help enable subsets with the desired functions to be selectively targeted, ensuring different subsets do not oppose each other’s functions (111). Important unknowns include whether there is direct thymic imprinting of human NKT cell subsets as there is in mice, and determining whether the functions of human iNKT cells are polarized in the thymus or periphery, and understanding what drives this process (i.e., specific stimuli) (112). The frequency of iNKT cells is another important consideration given their low frequency in human blood and tissues. The use of transcriptomics to study the development of unconventional T cells has greatly increased our understanding of the molecular requirements to produce functional subsets of T cells. Therefore, uncovering the origin of mature human iNKT cell subsets and defining a more detailed thymic developmental pathway may then allow the specific targeting of iNKT cell subsets in new-generation immunotherapies that exploit the unique characteristics of iNKT cells.
Author contributions
DP: Funding acquisition, Visualization, Writing – original draft, Writing – review & editing. NT: Writing – original draft. LP: Visualization, Writing – review & editing. SB: Writing – review & editing. CM: Visualization, Writing – original draft.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. DP is funded by a Sylvia & Charles Viertel Fellowship. DP and SB are supported by NHMRC grant 2030186.
Acknowledgments
The figures in this manuscript were generated using BioRender.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Godfrey DI, Uldrich AP, McCluskey J, Rossjohn J, Moody DB. The burgeoning family of unconventional T cells. Nat Immunol. (2015) 16:1114–23. doi: 10.1038/ni.3298
2. Matsuda JL, Naidenko OV, Gapin L, Nakayama T, Taniguchi M, Wang CR, et al. Tracking the response of natural killer T cells to a glycolipid antigen using CD1d tetramers. J Exp Med. (2000) 192:741–54. doi: 10.1084/jem.192.5.741
3. Benlagha K, Weiss A, Beavis A, Teyton L, Bendelac A. In vivo identification of glycolipid antigen-specific T cells using fluorescent CD1d tetramers. J Exp Med. (2000) 191:1895–903. doi: 10.1084/jem.191.11.1895
4. Karadimitris A, Gadola S, Altamirano M, Brown D, Woolfson A, Klenerman P, et al. Human CD1d-glycolipid tetramers generated by in vitro oxidative refolding chromatography. Proc Natl Acad Sci U.S.A. (2001) 98:3294–8. doi: 10.1073/pnas.051604498
5. Exley MA, Wilson SB, Balk SP. Isolation and functional use of human NKT cells. Curr Protoc Immunol. (2017) 119:14111–141120. doi: 10.1002/cpim.33
6. Exley MA, Hou R, Shaulov A, Tonti E, Dellabona P, Casorati G, et al. Selective activation, expansion, and monitoring of human iNKT cells with a monoclonal antibody specific for the TCR alpha-chain CDR3 loop. Eur J Immunol. (2008) 38:1756–66. doi: 10.1002/eji.200737389
7. Sharma A, Lawry SM, Klein BS, Wang X, Sherer NM, Zumwalde NA, et al. LFA-1 ligation by high-density ICAM-1 is sufficient to activate IFN-gamma release by innate T lymphocytes. J Immunol. (2018) 201:2452–61. doi: 10.4049/jimmunol.1800537
8. Brennan PJ, Brigl M, Brenner MB. Invariant natural killer T cells: an innate activation scheme linked to diverse effector functions. Nat Rev Immunol. (2013) 13:101–17. doi: 10.1038/nri3369
9. Bharadwaj NS, Gumperz JE. Harnessing invariant natural killer T cells to control pathological inflammation. Front Immunol. (2022) 13:998378. doi: 10.3389/fimmu.2022.998378
10. Gumperz JE, Miyake S, Yamamura T, Brenner MB. Functionally distinct subsets of CD1d-restricted natural killer T cells revealed by CD1d tetramer staining. J Exp Med. (2002) 195:625–36. doi: 10.1084/jem.20011786
11. Liu J, Hill BJ, Darko S, Song K, Quigley MF, Asher TE, et al. The peripheral differentiation of human natural killer T cells. Immunol Cell Biol. (2019) 97:586–96. doi: 10.1111/imcb.12248
12. Schmid H, Schneidawind C, Jahnke S, Kettemann F, Secker KA, Duerr-Stoerzer S, et al. Culture-expanded human invariant natural killer T cells suppress T-cell alloreactivity and eradicate leukemia. Front Immunol. (2018) 9:1817. doi: 10.3389/fimmu.2018.01817
13. O’Reilly V, Zeng SG, Bricard G, Atzberger A, Hogan AE, Jackson J, et al. Distinct and overlapping effector functions of expanded human CD4+, CD8alpha+ and CD4-CD8alpha- invariant natural killer T cells. PloS One. (2011) 6:e28648. doi: 10.1371/journal.pone.0028648
14. Metelitsa LS, Naidenko OV, Kant A, Wu HW, Loza MJ, PeRussia B, et al. Human NKT cells mediate antitumor cytotoxicity directly by recognizing target cell CD1d with bound ligand or indirectly by producing IL-2 to activate NK cells. J Immunol. (2001) 167:3114–22. doi: 10.4049/jimmunol.167.6.3114
15. Diaz-Basabe A, Burrello C, Lattanzi G, Botti F, Carrara A, Cassinotti E, et al. Human intestinal and circulating invariant natural killer T cells are cytotoxic against colorectal cancer cells via the perforin-granzyme pathway. Mol Oncol. (2021) 15:3385–403. doi: 10.1002/1878-0261.13104
16. Berzins SP, Smyth MJ, Baxter AG. Presumed guilty: natural killer T cell defects and human disease. Nat Rev Immunol. (2011) 11:131–42. doi: 10.1038/nri2904
17. Godfrey DI, Le Nours J, Andrews DM, Uldrich AP, Rossjohn J. Unconventional T cell targets for cancer immunotherapy. Immunity. (2018) 48:453–73. doi: 10.1016/j.immuni.2018.03.009
18. Lynch L, O’Shea D, Winter DC, Geoghegan J, Doherty DG, O’Farrelly C. Invariant NKT cells and CD1d(+) cells amass in human omentum and are depleted in patients with cancer and obesity. Eur J Immunol. (2009) 39:1893–901. doi: 10.1002/eji.200939349
19. Schipper HS, Rakhshandehroo M, Graaf SFvde, Venken K, Koppen A, Stienstra R, et al. Natural killer T cells in adipose tissue prevent insulin resistance. J Clin Invest. (2012) 122:3343–54. doi: 10.1172/JCI62739
20. Gadola SD, Dulphy N, Salio M, Cerundolo V. Valpha24-JalphaQ-independent, CD1d-restricted recognition of alpha-galactosylceramide by human CD4(+) and CD8alphabeta(+) T lymphocytes. J Immunol. (2002) 168:5514–20. doi: 10.4049/jimmunol.168.11.5514
21. Godfrey DI, Berzins SP. Control points in NKT-cell development. Nat Rev Immunol. (2007) 7:505–18. doi: 10.1038/nri2116
22. Montoya CJ, Pollard D, Martinson J, Kumari K, Wasserfall C, Mulder CB, et al. Characterization of human invariant natural killer T subsets in health and disease using a novel invariant natural killer T cell-clonotypic monoclonal antibody, 6B11. Immunology. (2007) 122:1–14. doi: 10.1111/j.1365-2567.2007.02647.x
23. Chan AC, Leeansyah E, Cochrane A, d’Udekem d’Acoz Y, Mittag D, Harrison LC, et al. Ex-vivo analysis of human natural killer T cells demonstrates heterogeneity between tissues and within established CD4(+) and CD4(-) subsets. Clin Exp Immunol. (2013) 172:129–37. doi: 10.1111/cei.12045
24. Lee PT, Benlagha K, Teyton L, Bendelac A. Distinct functional lineages of human V(alpha)24 natural killer T cells. J Exp Med. (2002) 195:637–41. doi: 10.1084/jem.20011908
25. Lin H, Nieda M, Hutton JF, Rozenkov V, Nicol AJ. Comparative gene expression analysis of NKT cell subpopulations. J Leukoc Biol. (2006) 80:164–73. doi: 10.1189/jlb.0705421
26. Zhou L, Adrianto I, Wang J, Wu X, Datta I, Mi QS. Single-cell RNA-seq analysis uncovers distinct functional human NKT cell sub-populations in peripheral blood. Front Cell Dev Biol. (2020) 8:384. doi: 10.3389/fcell.2020.00384
27. Pellicci DG, Koay HF, Berzins SP. Thymic development of unconventional T cells: how NKT cells, MAIT cells and gammadelta T cells emerge. Nat Rev Immunol. (2020) 20:756–70. doi: 10.1038/s41577-020-0345-y
28. Wang H, Hogquist KA. How lipid-specific T cells become effectors: the differentiation of iNKT subsets. Front Immunol. (2018) 9:1450. doi: 10.3389/fimmu.2018.01450
29. Lee YJ, Holzapfel KL, Zhu J, Jameson SC, Hogquist KA. Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nat Immunol. (2013) 14:1146–54. doi: 10.1038/ni.2731
30. Moreira-Teixeira L, Resende M, Coffre M, Devergne O, Herbeuval JP, Hermine O, et al. Proinflammatory environment dictates the IL-17-producing capacity of human invariant NKT cells. J Immunol. (2011) 186:5758–65. doi: 10.4049/jimmunol.1003043
31. Snyder-Cappione JE, Tincati C, Eccles-James IG, Cappione AJ, Ndhlovu LC, Koth LL, et al. A comprehensive ex vivo functional analysis of human NKT cells reveals production of MIP1-alpha and MIP1-beta, a lack of IL-17, and a Th1-bias in males. PloS One. (2010) 5:e15412. doi: 10.1371/journal.pone.0015412
32. Li S, Joseph C, Becourt C, Klibi J, Luce S, Dubois-Laforgue D, et al. Potential role of IL-17-producing iNKT cells in type 1 diabetes. PloS One. (2014) 9:e96151. doi: 10.1371/journal.pone.0096151
33. Venken K, Jacques P, Mortier C, Labadia ME, Decruy T, Coudenys J, et al. RORgammat inhibition selectively targets IL-17 producing iNKT and gammadelta-T cells enriched in Spondyloarthritis patients. Nat Commun. (2019) 10:9. doi: 10.1038/s41467-018-07911-6
34. Knox JJ, Cosma GL, Betts MR, McLane LM. Characterization of T-bet and eomes in peripheral human immune cells. Front Immunol. (2014) 5:217. doi: 10.3389/fimmu.2014.00217
35. Savage AK, Constantinides MG, Han J, Picard D, Martin E, Li B, et al. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. (2008) 29:391–403. doi: 10.1016/j.immuni.2008.07.011
36. Kovalovsky D, Uche OU, Eladad S, Hobbs RM, Yi W, Alonzo E, et al. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat Immunol. (2008) 9:1055–64. doi: 10.1038/ni.1641
37. Sag D, Krause P, Hedrick CC, Kronenberg M, Wingender G. IL-10-producing NKT10 cells are a distinct regulatory invariant NKT cell subset. J Clin Invest. (2014) 124:3725–40. doi: 10.1172/JCI72308
38. Lynch L, Michelet X, Zhang S, Brennan PJ, Moseman A, Lester C, et al. Regulatory iNKT cells lack expression of the transcription factor PLZF and control the homeostasis of T(reg) cells and macrophages in adipose tissue. Nat Immunol. (2015) 16:85–95. doi: 10.1038/ni.3047
39. Monteiro M, Almeida CF, Caridade M, Ribot JC, Duarte J, Agua-Doce A, et al. Identification of regulatory Foxp3+ invariant NKT cells induced by TGF-beta. J Immunol. (2010) 185:2157–63. doi: 10.4049/jimmunol.1000359
40. Moreira-Teixeira L, Resende M, Devergne O, Herbeuval JP, Hermine O, Schneider E, et al. Rapamycin combined with TGF-beta converts human invariant NKT cells into suppressive Foxp3+ regulatory cells. J Immunol. (2012) 188:624–31. doi: 10.4049/jimmunol.1102281
41. Chang PP, Barral P, Fitch J, Pratama A, Ma CS, Kallies A, et al. Identification of Bcl-6-dependent follicular helper NKT cells that provide cognate help for B cell responses. Nat Immunol. (2011) 13:35–43. doi: 10.1038/ni.2166
42. Benlagha K, Kyin T, Beavis A, Teyton L, Bendelac A. A thymic precursor to the NK T cell lineage. Science. (2002) 296:553–5. doi: 10.1126/science.1069017
43. Pellicci DG, Hammond KJ, Uldrich AP, Baxter AG, Smyth MJ, Godfrey DI. A natural killer T (NKT) cell developmental pathway iInvolving a thymus-dependent NK1.1(-)CD4(+) CD1d-dependent precursor stage. J Exp Med. (2002) 195:835–44. doi: 10.1084/jem.20011544
44. Engel I, Seumois G, Chavez L, Samaniego-Castruita D, White B, Chawla A, et al. Innate-like functions of natural killer T cell subsets result from highly divergent gene programs. Nat Immunol. (2016) 17:728–39. doi: 10.1038/ni.3437
45. Baranek T, Lebrigand K, de Amat Herbozo C, Gonzalez L, Bogard G, Dietrich C, et al. High dimensional single-cell analysis reveals iNKT cell developmental trajectories and effector fate decision. Cell Rep. (2020) 32:108116. doi: 10.1016/j.celrep.2020.108116
46. Koay HF, Gherardin NA, Enders A, Loh L, Mackay LK, Almeida CF, et al. A three-stage intrathymic development pathway for the mucosal-associated invariant T cell lineage. Nat Immunol. (2016) 17:1300–11. doi: 10.1038/ni.3565
47. Park JY, DiPalma DT, Kwon J, Fink J, Park JH. Quantitative difference in PLZF protein expression determines iNKT lineage fate and controls innate CD8 T cell generation. Cell Rep. (2019) 27:2548–2557.e4. doi: 10.1016/j.celrep.2019.05.012
48. Seiler MP, Mathew R, Liszewski MK, Spooner CJ, Barr K, Meng F, et al. Elevated and sustained expression of the transcription factors Egr1 and Egr2 controls NKT lineage differentiation in response to TCR signaling. Nat Immunol. (2012) 13:264–71. doi: 10.1038/ni.2230
49. Mao AP, Ishizuka IE, Kasal DN, Mandal M, Bendelac A. A shared Runx1-bound Zbtb16 enhancer directs innate and innate-like lymphoid lineage development. Nat Commun. (2017) 8:863. doi: 10.1038/s41467-017-00882-0
50. Pobezinsky LA, Etzensperger R, Jeurling S, Alag A, Kadakia T, McCaughtry TM, et al. Let-7 microRNAs target the lineage-specific transcription factor PLZF to regulate terminal NKT cell differentiation and effector function. Nat Immunol. (2015) 16:517–24. doi: 10.1038/ni.3146
51. Hogquist K, Georgiev H. Recent advances in iNKT cell development. F1000Res. (2020) 9. doi: 10.12688/f1000research
52. Sandberg JK, Stoddart CA, Brilot F, Jordan KA, Nixon DF. Development of innate CD4+ alpha-chain variable gene segment 24 (Valpha24) natural killer T cells in the early human fetal thymus is regulated by IL-7. Proc Natl Acad Sci USA. (2004) 101:7058–63. doi: 10.1073/pnas.0305986101
53. Loh L, Ivarsson MA, Michaelsson J, Sandberg JK, Nixon DF. Invariant natural killer T cells developing in the human fetus accumulate and mature in the small intestine. Mucosal Immunol. (2014) 7:1233–43. doi: 10.1038/mi.2014.13
54. Gurney KB, Yang OO, Wilson SB, Uittenbogaart CH. TCR gamma delta+ and CD161+ thymocytes express HIV-1 in the SCID-hu mouse, potentially contributing to immune dysfunction in HIV infection. J Immunol. (2002) 169:5338–46. doi: 10.4049/jimmunol.169.9.5338
55. Berzins SP, Cochrane AD, Pellicci DG, Smyth MJ, Godfrey DI. Limited correlation between human thymus and blood NKT cell content revealed by an ontogeny study of paired tissue samples. Eur J Immunol. (2005) 35:1399–407. doi: 10.1002/eji.200425958
56. Baev DV, Peng XH, Song L, Barnhart JR, Crooks GM, Weinberg KI, et al. Distinct homeostatic requirements of CD4+ and CD4- subsets of Valpha24-invariant natural killer T cells in humans. Blood. (2004) 104:4150–6. doi: 10.1182/blood-2004-04-1629
57. Hammond K, Cain W, van Driel I, Godfrey D. Three day neonatal thymectomy selectively depletes NK1.1+ T cells. Int Immunol. (1998) 10:1491–9. doi: 10.1093/intimm/10.10.1491
58. Gapin L, Matsuda JL, Surh CD, Kronenberg M. NKT cells derive from double-positive thymocytes that are positively selected by CD1d. Nat Immunol. (2001) 2:971–8. doi: 10.1038/ni710
59. Egawa T, Eberl G, Taniuchi I, Benlagha K, Geissmann F, Hennighausen L, et al. Genetic evidence supporting selection of the Valpha14i NKT cell lineage from double-positive thymocyte precursors. Immunity. (2005) 22:705–16. doi: 10.1016/j.immuni.2005.03.011
60. Bezbradica JS, Hill T, Stanic AK, Van Kaer L, Joyce S. Commitment toward the natural T (iNKT) cell lineage occurs at the CD4 + 8+ stage of thymic ontogeny. Proc Natl Acad Sci United States America. (2005) 102:5114–9. doi: 10.1073/pnas.0408449102
61. Bendelac A. Positive selection of mouse NK1+ T cells by CD1-expressing cortical thymocytes. J Exp Med. (1995) 182:2091–6. doi: 10.1084/jem.182.6.2091
62. de Lalla C, Festuccia N, Albrecht I, Chang HD, Andolfi G, Benninghoff U, et al. Innate-like effector differentiation of human invariant NKT cells driven by IL-7. J Immunol. (2008) 180:4415–24. doi: 10.4049/jimmunol.180.7.4415
63. Godfrey DI, Stankovic S, Baxter AG. Raising the NKT cell family. Nat Immunol. (2010) 11:197–206. doi: 10.1038/ni.1841
64. Coquet JM, Chakravarti S, Kyparissoudis K, McNab FW, Pitt LA, McKenzie BS, et al. Diverse cytokine production by NKT cell subsets and identification of an IL-17-producing CD4-NK1.1- NKT cell population. Proc Natl Acad Sci United States America. (2008) 105:11287–92. doi: 10.1073/pnas.0801631105
65. Maas-Bauer K, Kohler N, Stell AV, Zwick M, Acharya S, Rensing-Ehl A, et al. Single-cell transcriptomics reveal different maturation stages and sublineage commitment of human thymic invariant natural killer T cells. J Leukoc Biol. (2024) 115:401–9. doi: 10.1093/jleuko/qiad113
66. Hagen RR, Xu C, Koay HF, Konstantinov IE, Berzins SP, Kedzierska K, et al. Methodological optimisation of thymocyte isolation and cryopreservation of human thymus samples. J Immunol Methods. (2024) 528:113651. doi: 10.1016/j.jim.2024.113651
67. D’Andrea A, Goux D, De Lalla C, Koezuka Y, Montagna D, Moretta A, et al. Neonatal invariant Valpha24+ NKT lymphocytes are activated memory cells. Eur J Immunol. (2000) 30:1544–50. doi: 10.1002/1521-4141(200006)30:6<1544::AID-IMMU1544>3.0.CO;2-I
68. van Der Vliet HJ, Nishi N, de Gruijl TD, von Blomberg BM, Eertwegh AJvd, Pinedo HM, et al. Human natural killer T cells acquire a memory-activated phenotype before birth. Blood. (2000) 95:2440–2. doi: 10.1182/blood.V95.7.2440
69. Loh L, Carcy S, Krovi HS, Domenico J, Spengler A, Lin Y, et al. Unraveling the phenotypic states of human innate-like T cells: comparative insights with conventional T cells and mouse models. bioRxiv. (2023). doi: 10.1101/2023.12.07.570707
70. Bugaut H, El Morr Y, Mestdagh M, Darbois A, Paiva RA, Salou M, et al. A conserved transcriptional program for MAIT cells across mammalian evolution. J Exp Med. (2024) 221(2):e20231487. doi: 10.1084/jem.20231487
71. Eidson M, Wahlstrom J, Beaulieu AM, Zaidi B, Carsons SE, Crow PK, et al. Altered development of NKT cells, gammadelta T cells, CD8 T cells and NK cells in a PLZF deficient patient. PloS One. (2011) 6:e24441. doi: 10.1371/journal.pone.0024441
72. Pasquier B, Yin L, Fondaneche MC, Relouzat F, Bloch-Queyrat C, Lambert N, et al. Defective NKT cell development in mice and humans lacking the adapter SAP, the X-linked lymphoproliferative syndrome gene product. J Exp Med. (2005) 201:695–701. doi: 10.1084/jem.20042432
73. Nichols KE, Hom J, Gong SY, Ganguly A, Ma CS, Cannons JL, et al. Regulation of NKT cell development by SAP, the protein defective in XLP. Nat Med. (2005) 11:340–5. doi: 10.1038/nm1189
74. Zeissig S, Blumberg RS. Primary immunodeficiency associated with defects in CD1 and CD1-restricted T cells. Ann N Y Acad Sci. (2012) 1250:14–24. doi: 10.1111/j.1749-6632.2011.06380.x
75. Locci M, Draghici E, Marangoni F, Bosticardo M, Catucci M, Aiuti A, et al. The Wiskott-Aldrich syndrome protein is required for iNKT cell maturation and function. J Exp Med. (2009) 206:735–42. doi: 10.1084/jem.20081773
76. Rigaud S, Fondaneche MC, Lambert N, Pasquier B, Mateo V, Soulas P, et al. XIAP deficiency in humans causes an X-linked lymphoproliferative syndrome. Nature. (2006) 444:110–4. doi: 10.1038/nature05257
77. Gerart S, Siberil S, Martin E, Lenoir C, Aguilar C, Picard C, et al. Human iNKT and MAIT cells exhibit a PLZF-dependent proapoptotic propensity that is counterbalanced by XIAP. Blood. (2013) 121:614–23. doi: 10.1182/blood-2012-09-456095
78. Koay HF, Su S, Amann-Zalcenstein D, Daley SR, Comerford I, Miosge L, et al. A divergent transcriptional landscape underpins the development and functional branching of MAIT cells. Sci Immunol. (2019) 4(41):eaay6039. doi: 10.1126/sciimmunol.aay6039
79. Perriman L, Tavakolinia N, Jalali S, Li S, Hickey PF, Amann-Zalcenstein D, et al. A three-stage developmental pathway for human Vgamma9Vdelta2 T cells within the postnatal thymus. Sci Immunol. (2023) 8:eabo4365. doi: 10.1126/sciimmunol.abo4365
80. Koay HF, Godfrey DI, Pellicci DG. Development of mucosal-associated invariant T cells. Immunol Cell Biol. (2018) 96:598–606. doi: 10.1111/imcb.12039
81. Robinson JP, Ostafe R, Iyengar SN, Rajwa B, Fischer R. Flow cytometry: the next revolution. Cells. (2023) 12:1875. doi: 10.3390/cells12141875
82. Jalali S, Harpur CM, Piers AT, Auladell M, Perriman L, Li S, et al. A high-dimensional cytometry atlas of peripheral blood over the human life span. Immunol Cell Biol. (2022) 100:805–21. doi: 10.1111/imcb.12594
83. Park LM, Lannigan J, Jaimes MC. OMIP-069: forty-color full spectrum flow cytometry panel for deep immunophenotyping of major cell subsets in human peripheral blood. Cytometry A. (2020) 97:1044–51. doi: 10.1002/cyto.a.24213
84. Anderson J, Quah L, Mangano K, Pellicci DG, Mazarakis N, Licciardi PV. A 38-colour high dimensional immunophenotyping panel for human peripheral blood mononuclear cells. J Immunol Methods. (2024) 532:113726. doi: 10.1016/j.jim.2024.113726
85. Zhou Y, Xu X, Tian Z, Wei H. Multi-omics” Analyses of the development and function of natural killer cells. Front Immunol. (2017) 8:1095. doi: 10.3389/fimmu.2017.01095
86. Van Kaer L, Wu L. Therapeutic potential of invariant natural killer T cells in autoimmunity. Front Immunol. (2018) 9:519. doi: 10.3389/fimmu.2018.00519
87. Mavers M, Maas-Bauer K, Negrin RS. Invariant natural killer T cells as suppressors of graft-versus-host disease in allogeneic hematopoietic stem cell transplantation. Front Immunol. (2017) 8:900. doi: 10.3389/fimmu.2017.00900
88. Vogt S, Mattner J. NKT cells contribute to the control of microbial infections. Front Cell Infect Microbiol. (2021) 11:718350. doi: 10.3389/fcimb.2021.718350.5
89. Cerundolo V, Silk JD, Masri SH, Salio M. Harnessing invariant NKT cells in vaccination strategies. Nat Rev Immunol. (2009) 9:28–38. doi: 10.1038/nri2451
90. Nelson A, Lukacs JD, Johnston B. The current landscape of NKT cell immunotherapy and the hills ahead. Cancers (Basel). (2021) 13:5174. doi: 10.3390/cancers13205174
91. Berzins SP, Ritchie DS. Natural killer T cells: drivers or passengers in preventing human disease? Nat Rev Immunol. (2014) 14:640–6. doi: 10.1038/nri3725
92. Cho YN, Kee SJ, Lee SJ, Seo SR, Kim TJ, Lee SS, et al. Numerical and functional deficiencies of natural killer T cells in systemic lupus erythematosus: their deficiency related to disease activity. Rheumatol (Oxford). (2011) 50:1054–63. doi: 10.1093/rheumatology/keq457
93. van der Vliet HJ, von Blomberg BM, Nishi N, Reijm M, Voskuyl AE, van Bodegraven AA, et al. Circulating V(alpha24+) Vbeta11+ NKT cell numbers are decreased in a wide variety of diseases that are characterized by autoreactive tissue damage. Clin Immunol. (2001) 100:144–8. doi: 10.1006/clim.2001.5060
94. Delfanti G, Dellabona P, Casorati G, Fedeli M. Adoptive immunotherapy with engineered iNKT cells to target cancer cells and the suppressive microenvironment. Front Med (Lausanne). (2022) 9:897750. doi: 10.3389/fmed.2022.897750
95. Wolf BJ, Choi JE, Exley MA. Novel approaches to exploiting invariant NKT cells in cancer immunotherapy. Front Immunol. (2018) 9:384. doi: 10.3389/fimmu.2018.00384
96. Nair S, Dhodapkar MV. Natural killer T cells in cancer immunotherapy. Front Immunol. (2017) 8:1178. doi: 10.3389/fimmu.2017.01178
97. Metelitsa LS, Weinberg KI, Emanuel PD, Seeger RC. Expression of CD1d by myelomonocytic leukemias provides a target for cytotoxic NKT cells. Leukemia. (2003) 17:1068–77. doi: 10.1038/sj.leu.2402943
98. Jahnke S, Schmid H, Secker KA, Einhaus J, Duerr-Stoerzer S, Keppeler H, et al. Invariant NKT Cells From Donor Lymphocyte Infusions (DLI-iNKTs) Promote ex vivo Lysis of Leukemic Blasts in a CD1d-Dependent Manner. Front Immunol. (2019) 10:1542. doi: 10.3389/fimmu.2019.01542
99. Diaz-Basabe A, Strati F, Facciotti F. License to kill: when iNKT cells are granted the use of lethal cytotoxicity. Int J Mol Sci. (2020) 21(11):3909. doi: 10.3390/ijms21113909
100. Kawano T, Nakayama T, Kamada N, Kaneko Y, Harada M, Ogura N, et al. Antitumor cytotoxicity mediated by ligand-activated human V alpha24 NKT cells. Cancer Res. (1999) 59:5102–5.
101. Song L, Asgharzadeh S, Salo J, Engell K, Wu HW, Sposto R, et al. Valpha24-invariant NKT cells mediate antitumor activity via killing of tumor-associated macrophages. J Clin Invest. (2009) 119:1524–36. doi: 10.1172/JCI37869
102. Look A, Burns D, Tews I, Roghanian A, Mansour S. Towards a better understanding of human iNKT cell subpopulations for improved clinical outcomes. Front Immunol. (2023) 14:1176724. doi: 10.3389/fimmu.2023.1176724
103. Waldowska M, Bojarska-Junak A, Rolinski J. A brief review of clinical trials involving manipulation of invariant NKT cells as a promising approach in future cancer therapies. Cent Eur J Immunol. (2017) 42:181–95. doi: 10.5114/ceji.2017.69361
104. Courtney AN, Tian G, Metelitsa LS. Natural killer T cells and other innate-like T lymphocytes as emerging platforms for allogeneic cancer cell therapy. Blood. (2023) 141:869–76. doi: 10.1182/blood.2022016201
105. Terabe M, Berzofsky JA. The role of NKT cells in tumor immunity. Adv Cancer Res. (2008) 101:277–348. doi: 10.1016/S0065-230X(08)00408-9
106. LaMarche NM, Hegde S, Park MD, Maier BB, Troncoso L, Le Berichel J, et al. An IL-4 signalling axis in bone marrow drives pro-tumorigenic myelopoiesis. Nature. (2024) 625:166–74. doi: 10.1038/s41586-023-06797-9
107. Matulis G, Sanderson JP, Lissin NM, Asparuhova MB, Bommineni GR, Schumperli D, et al. Innate-like control of human iNKT cell autoreactivity via the hypervariable CDR3beta loop. PloS Biol. (2010) 8:e1000402. doi: 10.1371/journal.pbio.1000402
108. Tian G, Courtney AN, Jena B, Heczey A, Liu D, Marinova E, et al. CD62L+ NKT cells have prolonged persistence and antitumor activity in vivo. J Clin Invest. (2016) 126:2341–55. doi: 10.1172/JCI83476
109. Cui G, Shimba A, Jin J, Ogawa T, Muramoto Y, Miyachi H, et al. A circulating subset of iNKT cells mediates antitumor and antiviral immunity. Sci Immunol. (2022) 7:eabj8760. doi: 10.1126/sciimmunol.abj8760
110. Li YR, Zhou Y, Kim YJ, Zhu Y, Ma F, Yu J, et al. Development of allogeneic HSC-engineered iNKT cells for off-the-shelf cancer immunotherapy. Cell Rep Med. (2021) 2:100449. doi: 10.1016/j.xcrm.2021.100449
111. Cui G, Abe S, Kato R, Ikuta K. Insights into the heterogeneity of iNKT cells: tissue-resident and circulating subsets shaped by local microenvironmental cues. Front Immunol. (2024) 15:1349184. doi: 10.3389/fimmu.2024.1349184
Keywords: unconventional T cells, unconventional T cell development, cytokines, cytotoxic granules, immunotherapy
Citation: Pellicci DG, Tavakolinia N, Perriman L, Berzins SP and Menne C (2024) Thymic development of human natural killer T cells: recent advances and implications for immunotherapy. Front. Immunol. 15:1441634. doi: 10.3389/fimmu.2024.1441634
Received: 31 May 2024; Accepted: 05 August 2024;
Published: 29 August 2024.
Edited by:
Laurent Gapin, University of Colorado Denver, United StatesReviewed by:
Mihalis Verykokakis, Alexander Fleming Biomedical Sciences Research Center, GreeceChristophe Paget, Institut National de la Santé et de la Recherche Médicale (INSERM), France
Mariolina Salio, Immunocore (United Kingdom), United Kingdom
Copyright © 2024 Pellicci, Tavakolinia, Perriman, Berzins and Menne. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daniel G. Pellicci, ZGFuLnBlbGxpY2NpQG1jcmkuZWR1LmF1
 Daniel G. Pellicci
Daniel G. Pellicci Naeimeh Tavakolinia
Naeimeh Tavakolinia Louis Perriman
Louis Perriman Stuart P. Berzins
Stuart P. Berzins Christopher Menne
Christopher Menne