- 1Department of Bone Marrow Transplant, Hebei Yanda Lu Daopei Hospital, Langfang, China
- 2Department of Bone Marrow Transplant, Beijing Lu Daopei Hospital, Beijing, China
- 3Beijing BFR Gene Diagnostics Co., Ltd, Beijing, China
Introduction: The human leukocyte antigen (HLA) evolutionary divergence (HED) reflects immunopeptidome diversity and has been shown to predict the response of tumors to immunotherapy. Its impact on allogeneic hematopoietic stem cell transplantation (HSCT) is controversial in different studies.
Methods: In this study, we retrospectively analyzed the clinical impact of class I and II HED in 225 acute lymphoblastic leukemia patients undergoing HSCT from related haploidentical donors. The HED for recipient, donor, and donor-recipient pair was calculated based on Grantham distance, which accounts for variations in the composition, polarity, and volume of each amino acid within the peptide-binding groove of two HLA alleles. The median value of HED scores was used as a cut-off to stratify patients with high or low HED.
Results: The class I HED for recipient (R_HEDclass I) showed the strongest association with cumulative incidence of relapse (12.2 vs. 25.0%, P = 0.00814) but not with acute graft-versus-host disease. The patients with high class II HED for donor-recipient (D/R_HEDclass II) showed a significantly higher cumulative incidence of severe aGVHD than those with low D/R_HEDclass II (24.0% vs. 6.1%, P = 0.0027). Multivariate analysis indicated that a high D/R_HEDclass II was an independent risk factor for the development of severe aGVHD (P = 0.007), and a high R_HEDclass I had a more than two-fold reduced risk of relapse (P = 0.028). However, there was no discernible difference in overall survival (OS) or disease-free survival (DFS) for patients with high or low HED, which was inconsistent with the previous investigation.
Discussion: While the observation are limited by the presented single center retrospective cohort, the results show that HED has poor prognostic value in OS or DFS, as well as the associations with relapse and aGVHD. In haploidentical setting, class II HED for donor-recipient pair (D/R_HEDclass II) is an independent and novel risk factor for finding the best haploidentical donor, which could potentially influence clinical practice if verified in larger cohorts.
1 Introduction
Allogeneic hematopoietic stem cell transplantation (allo-HSCT) is a curative therapy for many hematopoietic disorders, including acute lymphoblastic leukemia (ALL) (1). The success of allo-HSCT partly depends on the recognition of tumor antigens presented to alloreactive T cells via human leukocyte antigens (HLAs). The importance of HLA matching is currently well established, resulting in a fully HLA-matched sibling or unrelated donor being the preferred source for allo-HSCT to reduce the risk of GVHD through allo-recognition of foreign HLA molecules (2, 3).
The divergence of HLA alleles may lead to an increased functional capability of the immunopeptidome, which would defend against potentially fatal opportunistic infections and leukemia cells causing relapse (4). Heterozygosity was typically used to assess the HLA allelic difference. Recently, HLA evolutionary divergence (HED), a metric reflecting the immunopeptidome diversity, has been utilized to more accurately quantify HLA allele divergence using the Grantham distance, which accounts for variations in the composition, polarity, and volume of each amino acid within the peptide-binding groove of two HLA alleles (5, 6). Previous research has linked the high heterozygosity of HLA class I loci to an improved response to immune checkpoint inhibitors in advanced cancer patients (7). Further, Chowell et al. found that the effect of HED on survival was independent of other clinically relevant variables and that a high HED in class I alleles was strongly related with response to checkpoint inhibitors in advanced cancer patients (8). These findings, however, were subsequently challenged by a study with a large cohort of cancer patients who had undergone anti-PD1 immunotherapy (9).
In the context of liver grafts, Feray et al. discovered that the donor’s HED was an intrinsic feature completely independent of the recipient’s characteristics and that a high class I HED of the donor was strongly related to a poor outcome (10). The influence of class I and II HED in the HSCT setting has primarily been explored in acute myeloid leukemia (AML). In AML patients, a high class I/class II HED ratio was revealed to be an independent factor for improved overall and disease-free survival (11, 12). More recently, HED was utilized to predict the outcome of children and young adults who underwent HSCT from an unrelated donor for a variety of malignant disorders (4). According to this study, patients with a high HED score of the combined HLA-B and -DRB1 loci had significantly increased overall and disease-free survival.
As an alternative donor transplant, HLA-haploidentical transplantation allows patients who do not have fully matched donors to undergo a transplant, and it has been increasingly used globally over the last two decades (13). In the haploidentical HSCT setting, almost all patients have more than one donor. As a result, the search for the best donor is a critical issue because donor selection can considerably affect the incidences of graft-versus-host, relapse, transplant-related mortality, and survival (13). Previous studies have identified a variety of characteristics that influence haploidentical outcomes, including HLA matching, donor age, donor sex, family relationships, and so on. These risk factors should be considered when selecting the best donor. However, the effects of HLA disparity on transplantation outcomes have vanished due to the improved protocols of haploidentical HSCT with anti-thymocyte globulin (ATG) or with post-transplantation cyclophosphamide (PT/Cy). If HLA disparity, either the quantity of HLA-mismatched loci or the mismatch combination of specific sites, is not a risk factor for haploidentical donor selection, it is currently unclear whether HED, which reflects HLA allele spatial epitope information, affects donor selection and clinical outcomes (4). To date, little is known about the impact of HED on outcomes in the HLA-haploidentical HSCT setting. In this study, we scored HED for donors, recipients, and donor-recipient pairs, and assessed the clinical significance of class I and II HED in 225 ALL patients who received HLA-haploidentical HSCT from a related donor. We found that the Grantham distance score of HLA evolutionary divergence was associated with acute GVHD and relapse in ALL patients undergoing HLA-haploidentical HSCT from a related donor, which may be considered a novel risk factor for donor selection in the haploidentical transplant setting.
2 Materials and methods
2.1 Patient characteristics
To investigate the influence of HED on clinical outcomes following HSCT, we conducted a retrospective analysis of consecutive Acute Lymphoblastic Leukemia patients (ALL) receiving allo-HSCT between 2012 and 2017 at Hebei Yanda Lu Daopei Hospital, Langfang City, PR China. HED was calculated using data from all patients. The clinical data collected included graft-versus-host disease (GVHD), relapse, date of the event, survival status, and last follow-up date, etc. All patients were prepared for transplantation using modified myeloablative or reduced intensity conditioning regimens (based on total body irradiation, busulfan, or fludarabine, depending on the patient’s comorbidities) (14). According to Chinese Bone Marrow Transplant Cooperative Group recommendations, GVHD prophylaxis was based on anti-thymoglobulin (ATG), cyclosporin A (CsA), methotrexate (MTX), and mycophenolate mofetil (MMF) (15–17).
This retrospective study was reviewed and approved by the Ethics Committee of Hebei Yanda Lu Daopei Hospital (DEPC-M-2023, No. 20). Before data collection, written informed consent was obtained from the patient or the patient’s parents if the patient was under the age of 18. This study follows the Declaration of Helsinki.
2.2 HED calculation
HLA compatibility was determined at five loci (HLA-A, -B, -C, -DRB1, and -DQB1) using sequencing-based typing (SBT) GenDx excellerator kits (GenDX, Utrecht, Netherlands). The patient and donor two-field resolution typing of these HLA loci served as the input for the HED calculation, and the calculation was performed using a Python script according to the original Grantham distance formula presented in the literature (5).
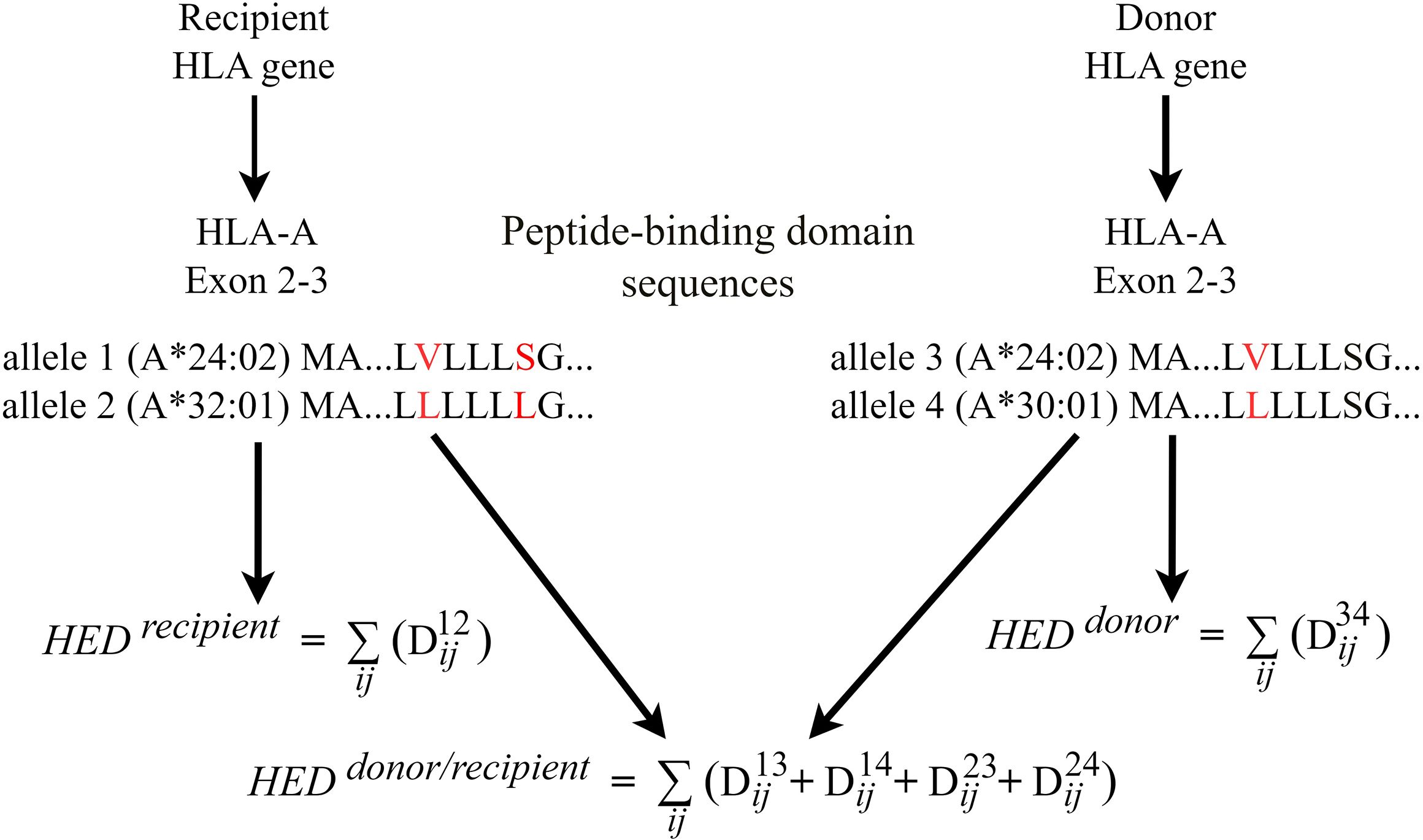
For each donor and recipient, the HED score was determined by calculating the Grantham distance between the peptide-binding domains of the two alleles at the HLA loci (exons 2 and 3 for HLA-A, HLA-B and HLA-C, exons 2 for HLA-DQB1, HLA-DRB1) loci (6, 7). For donor-recipient pair, HED per locus was estimated for pairwise allele combinations between donors and recipients. We take HLA-A as an example to illustrate how HED between donors and recipients was calculated (Figure 1). If recipient has HLA-A allele 1 and 2, donor has HLA-A allele 3 and 4 (Figure 1). Dij is Grantham distance between two alleles and calculated using the original formula (5) as follows:
Where i and j represent paired amino acids of the same position in the sequence of two alleles. c, p and v represent respective composition, polarity and molecular volume of the homologous amino-acids at a given position. α, β and γ are constants. HED between donor and recipient (HED donor/recipient) was calculated by the sum of Grantham distance of four combinations for donor-recipient alleles, given by the formula:
In the context of haploidentical HSCT, where donor and recipient always have one allele shared in any HLA locus, as shown in the diagram allele 1 = allele 3, the formula is:
Furthermore, if HLA-A matched (allele 1 = allele 3, allele 2 = allele 4), the formula is:
The mean HED score of class I HLA (HEDclass I) or class II HLA (HEDclass II) was measured for donor, recipient, and donor-recipient, respectively. HED was denoted by the prefix R (Recipient), D (Donor), or D/R (Donor-Recipient pair). The median HED score was used as the threshold to define a high- or low-HED group.
2.3 Clinical endpoints
The primary objective was to assess the impact of HED on relapse, non-relapse mortality (NRM), and acute and chronic graft-versus-host disease (GVHD). The secondary aim of the study was to assess the effect of HED on prognosis following haploidentical HSCT.
Endpoints of interest included the cumulative incidence of GVHD, relapse and NRM, overall survival (OS), and disease-free survival (DFS). aGVHD incidence was defined as time to first diagnosis of aGVHD (grade 2-4). Because acute GVHD, especially of grade 2 or higher, is probably the most suitable marker of morbidity, an additional sub-analysis for aGVHD (grades 3-4) was performed. Patients who survived more than 14 and 100 days following transplantation were evaluated for acute and chronic GVHD, respectively. The modified Keystone Criteria were used to grade aGVHD (18), while the National Institute of Health Consensus Criteria were used to evaluate cGVHD (19). Relapse incidence was defined as the time to relapse and death without prior recurrence. The NRM event was treated as a competing risk for relapse. NRM was defined as the time to death from any cause other than relapse. OS was defined as the time from transplantation to death, or the last follow-up. DFS was defined as the probability of survival without disease at any period following transplantation, with relapse or death considered events.
At the last follow-up, patients free from the event of interest were censored. The presence of 5% or more leukemic cells in the bone marrow and no indication of extramedullary localization was considered a hematological relapse.
2.4 Statistical analysis
Patient characteristics were summarized using descriptive statistics. Categorical variables are reported as counts (%), while continuous variables are described as the medians. The chi-square test, or Fisher’s exact test, was used to assess differences in categorical variables across two groups. The Mann-Whitney U test was used to compare the intergroup continuous variables.
Cumulative incidences of GVHD, relapse, and NRM were estimated with the methods of Fine and Gray considering the respective competitive risks; comparisons between the high and low HED groups were performed with Gray’s test. The Kaplan-Meier survival curve was used to estimate the probability of OS and DFS, and the significance was determined with a log-rank test. Potential risk factors were identified using the univariate Cox regression method to assess the hazard ratio (HR) for the various factors associated with clinical outcomes. Multivariate Cox regression analysis retained significant HED and other variables that might have been clinically meaningful or statistically significant in univariate analysis (P<0.2). The final multivariate models were built using a backward stepwise model approach.
Variables considered in the multivariate models were donor sex, donor and patient age, donor-recipient HLA disparity, donor-recipient family relationship, disease status at transplant (non-remission vs. complete remission), and donor-recipient sex matching. KIR matching and the HSCT-specific comorbidity index were not included due to insufficient data.
All tests were two-sided, and P<0.05 was considered statistically significant. The date collected is as of December 31, 2017. Statistical analysis was performed using the SPSS 25 package (SPSS Inc., Chicago, USA) and a graphical user interface for R language, EZR version 1.32 (20).
3 Results
3.1 Patient characteristics
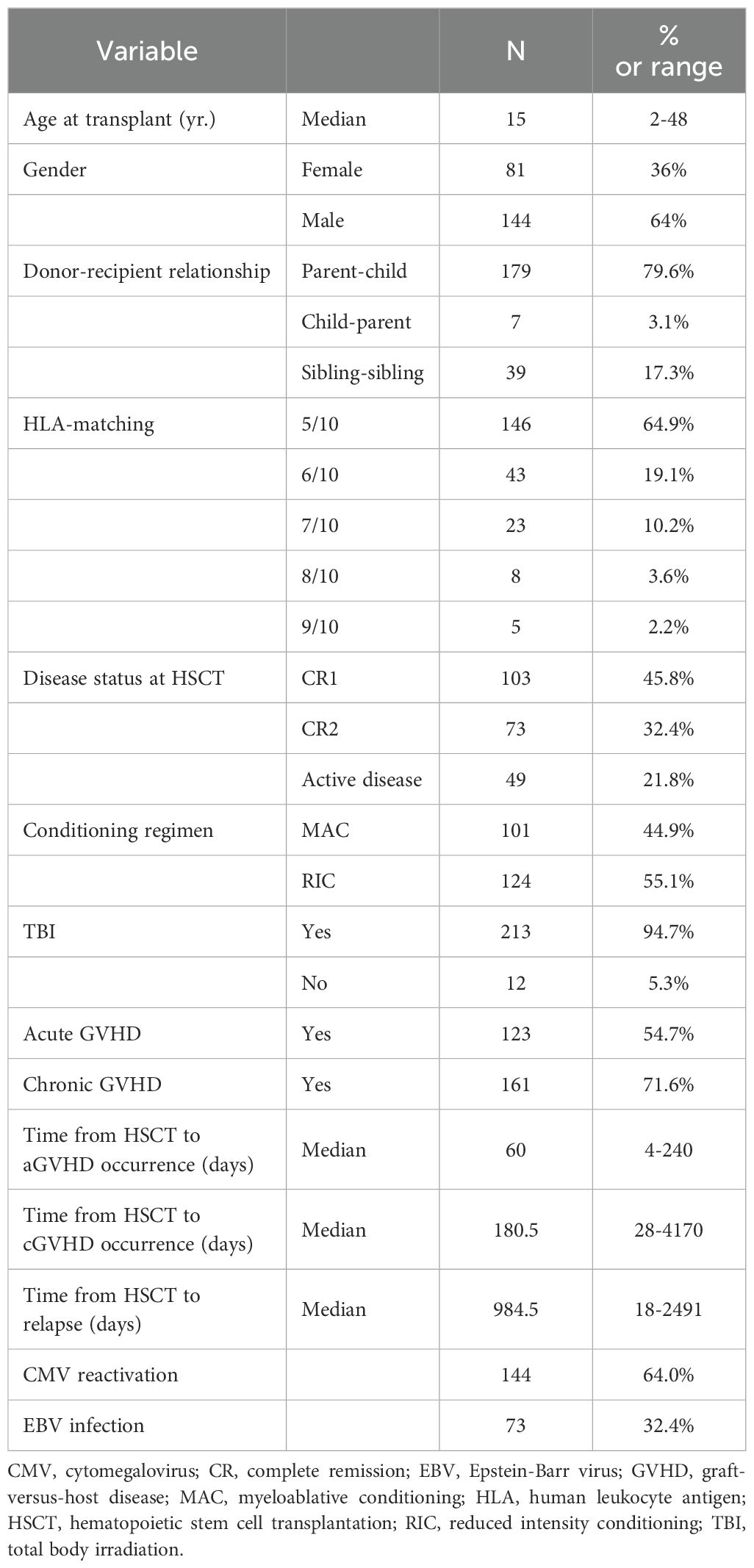
The study comprised 225 ALL patients who had HSCT from a related donor between 2012 and 2017. Most of the transplants (179) were parents as donors. Thirty-nine transplants were siblings as donors. The median age was 15 years, with the range of 2 to 48 years, and the median follow-up time following transplantation was 35.8 months (range, 1-83.9). High-resolution HLA typing revealed that 146 (64.9%) of 225 donor-recipient pairs had five mismatches, 43 (19.1%) had four mismatches, and 36 (16.0%) had three or fewer HLA mismatches. Thirty-one individuals (13.8%) had active disease at the time of transplantation. Table 1 summarizes the patient demographics and characteristics.
3.2 HED scores
We estimated HED strictly following the original formula of Grantham distance. Our HED value for class I was 3.56 times higher than Pierini and Lenz’s (6), and for class II, it was 1.75 times higher (Supplementary Method). This discrepancy is due to differences in data processing, but there is a clear and straightforward relationship between the two calculation methods, thus they can be considered identical in clinical investigations.
For recipients, HLA-B locus showed the highest HED variability (R_HEDB, median 29.7), followed by HLA-A (R_HEDA, median 26.8), HLA-DRB1 and -DQB1 (R_HEDDRB1 and R_HEDDQB1, median 26.5 and 22.5, respectively), and HLA-C locus displayed the lowest HED variation (R_HEDC, median 19.8) (Figure 2A). HLA-B evolutionary divergences were greater than HLA-A and HLA-C, supporting previous findings that HLA-B is the most ancient and diverse of the three HLA-class I loci (6). Class I HLA had a slightly higher mean HED (R_HEDclass I) than class II HLA (R_HEDclass II) (median 23.9, 22.9, respectively) (Figure 2A). The variance and distribution pattern of donor HED were quite comparable to that of the recipient, with HLA-B having the highest value (D_HEDB, median 29.8) and HLA-C having the lowest (D_HEDC, median 20.3) (Figure 2B).
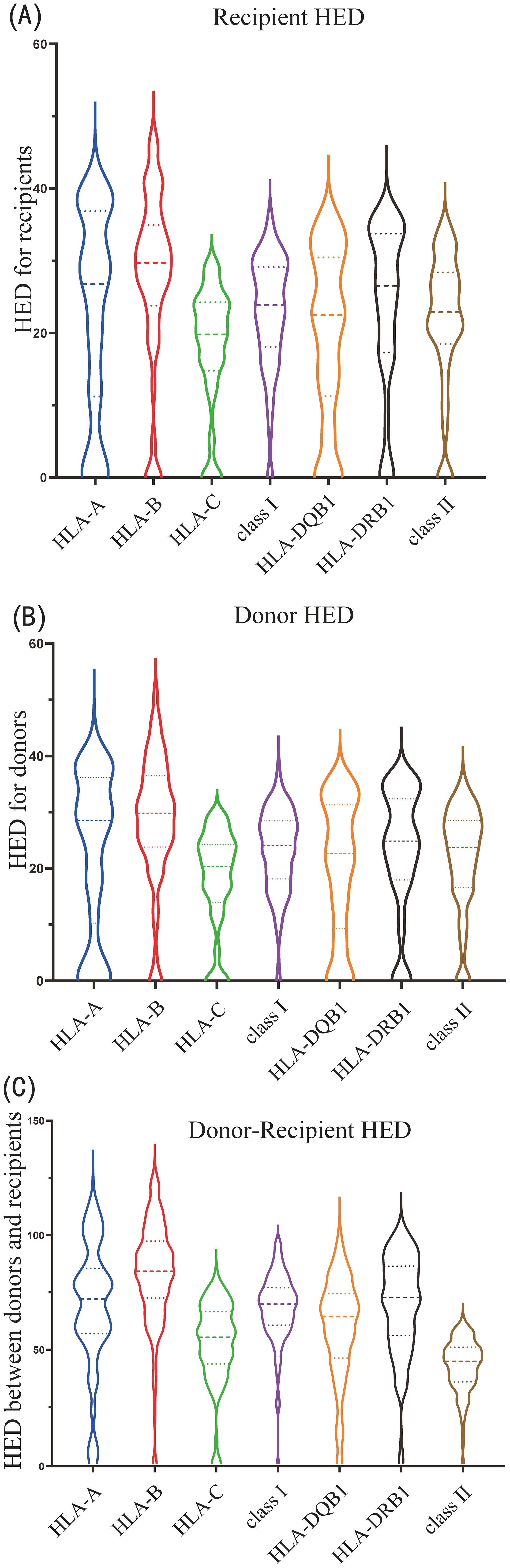
Figure 2. Violin plot of HLA evolutionary divergence (HED) distribution for (A) recipients, (B) donors, and (C) donor-recipient pairs.
Despite the fact that the HED scores for donor-recipient pairs were much higher than those of the donor or recipient due to the computed divergence among the four alleles, the HED distribution and variation patterns of each locus or class were identical to those of the donor or recipient. The highest was D/R_HEDB (median 84.2), followed by D/R_HEDDRB1 (median 72.7), D/R_HEDA (median 72.0), and D/R_HEDDQB1 (median 64.4), while the lowest was D/R_HEDC (median 55.4). D/R_HEDclass I was higher than D/R_HEDclass II (median 69.9 versus 67.4) (Figure 2C).
3.3 GVHD
The overall cumulative incidences of grade 2-4 and 3-4 aGVHD at 100-day were 36.5% (95% confidence interval [CI]: 29.9-43.0%), and 15.1% (95% CI: 10.3-20.7%), respectively. Neither the donor (D_HEDclass I, D_HEDclass II) nor recipient HED values (R_HEDclass I, R_HEDclass II) had any effect on aGVHD. Surprisingly, the HED score of donor-recipient pair (D/R_HEDclass II) was significantly associated with the cumulative incidence of grade 3-4 aGVHD at 100-day. The incidence of grade 3-4 aGVHD was 24.0% (95%CI:15.7-33.3%) in patients with high D/R_HEDclass II compared to 6.1% (95%CI: 2.5-12.2%) in patients with low D/R_HEDclass II (P = 0.0027) (Table 2, Figure 3A). The favorable impact of D/R_HEDclass II appears to be primarily driven by D/R_HEDDRB1. The higher the D/R_HEDDRB1, the higher the incidence of grade 3-4 aGVHD (23.4% [95%CI:15.1-32.8%] vs 7.2% [95%CI: 3.1-13.5%], P = 0.0047).
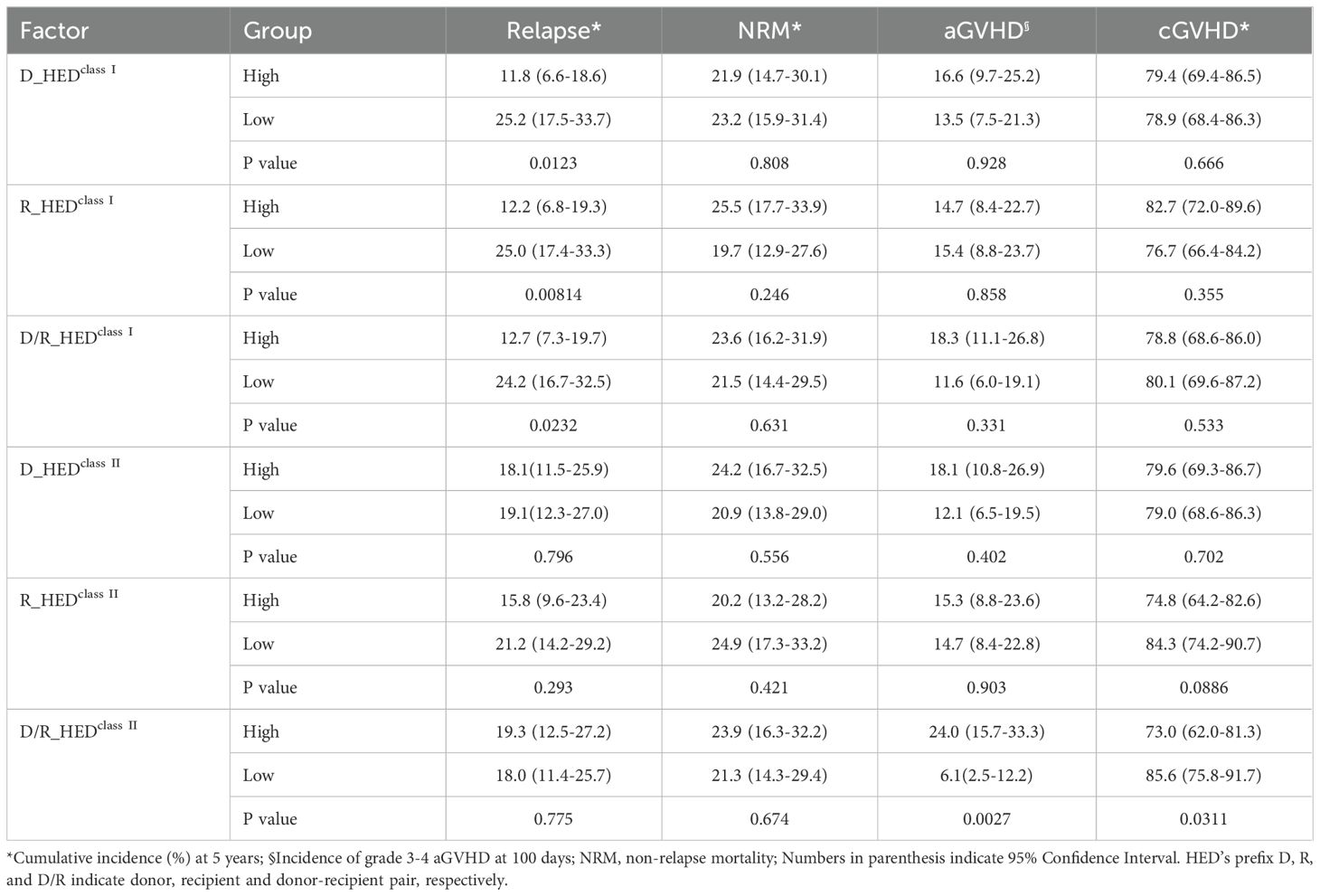
Table 2. Cumulative incidences (%) of Relapse, NRM, cGVHD and aGVHD based on HEDclass I and HEDclass II.
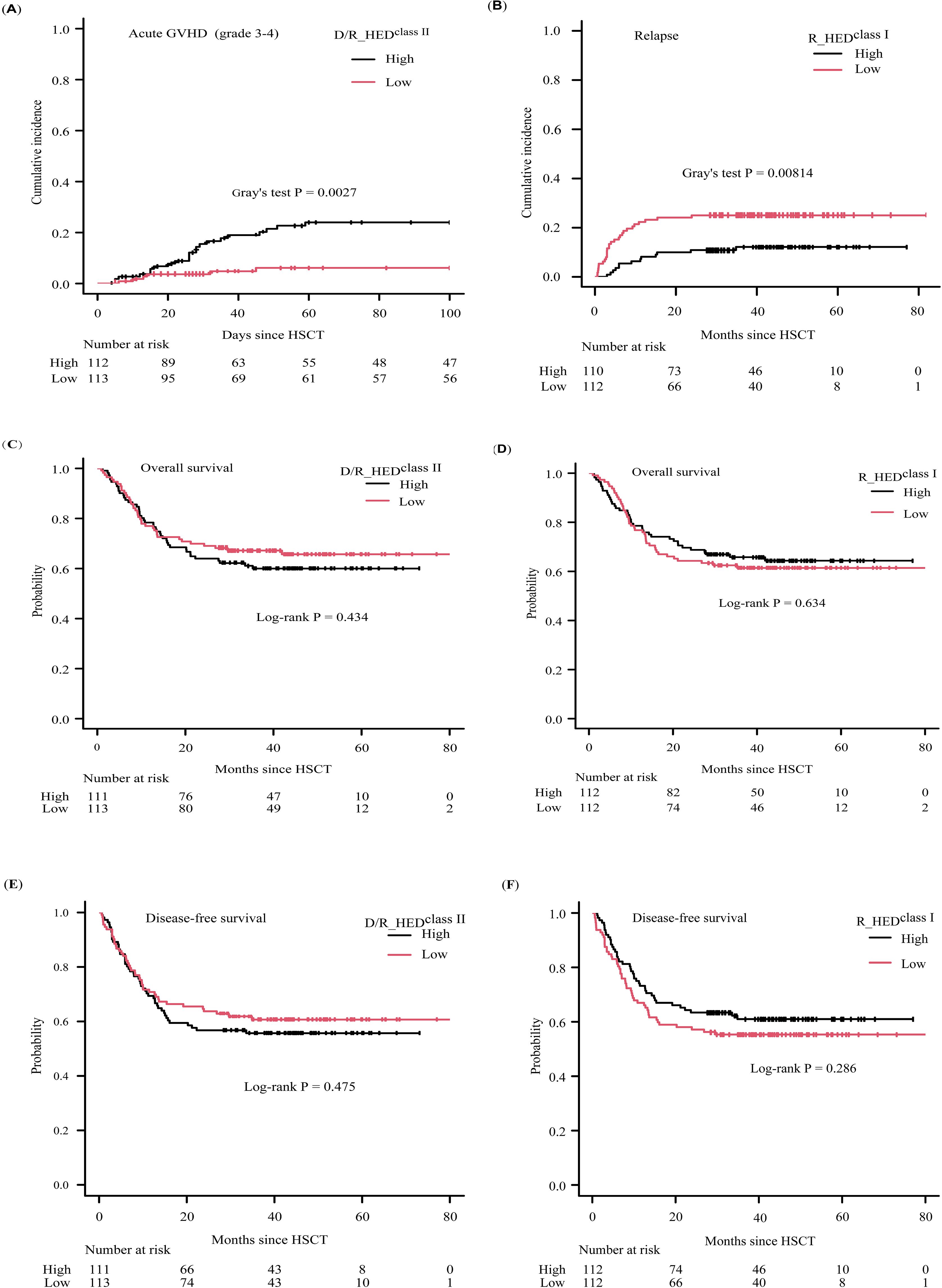
Figure 3. Clinical outcomes according to D/R_HEDclass II and R_HEDclass I (A) cumulative incidence of aGVHD (grade3-4) stratified by D/R_HEDclass II, (B) cumulative incidence of relapse stratified by R_HEDclass I, (C) KM curve of overall survival stratified by D/R_HEDclass II, (D) KM curve of overall survival stratified by R_HEDclass I, (E) KM curve of disease-free survival stratified by D/R_HEDclass II, (F) KM curve of disease-free survival stratified by R_HEDclass I.
The 5-year cumulative incidence of cGVHD was unexpectedly high, at 80.4% (95%CI: 74.1-86.0%). In contrast to the results for aGVHD, the cumulative incidence of cGVHD at 5-year was significantly associated with higher D/R_HEDclass II (P = 0.0311), with higher D/R_HEDclass II being associated with lower cGVHD risk (73.0% vs. 85.6%), but not with D/R_HEDclass I (P = 0.533) (Table 2). D/R_HEDclass II was therefore included in the subsequent cox regression analysis for GVHD. Regardless of the negative association with D/R_HEDB, there was no significant correlation between cGVHD and D/R_HEDclass I.
3.4 Relapse and NRM
Forty-four of 225 (19.6%) patients relapsed at a median time of 984.5 days (range 18-2491) after transplantation. The 5-year cumulative incidence of relapse (CIR) for all patients after transplantation was 18.6% (95% CI: 13.7-24.0%). The cumulative incidence of NRM at five years was 22.6% (95% CI: 17.3-28.3%), which was greater than the 5-year CIR.
When patients are stratified based on HEDclass I or HEDclass II, all three HEDclass I (D_HEDclass I, R_HEDclass I, and D/R_HEDclass I) scores show an obvious association with CIR (Table 2). Higher D_HEDclass I and D/R_HEDclass I contribute to a lower 5-year CIR (11.8 vs. 25.2%, P = 0.0123; 12.7% vs. 24.2%, P = 0.0232) (Table 2). R_HEDclass I, in particular, exhibited the strongest association with 5-year CIR (12.2 vs. 25.0%, P = 0.00814) (Table 2, Figure 3B). Conversely, neither HEDclass II were correlated with 5-year CIR. Thus, the three HEDclass I(D_HEDclass I, R_HEDclass I, and D/R_HEDclass I) were used as candidate risk factors for subsequent Cox regression analysis. The cumulative incidence of NRM at five years was not associated with any HEDclass I or HEDclass II. These findings suggest that genetic divergence of class I HLA, rather than class II HLA, may be responsible for the differences in CIR, but that genetic differentiation of either class I or II HLA loci has little effect on NRM.
3.5 Multivariate analysis
The impact of HED on GVHD and relapse was further investigated using the Cox proportional hazard regression analysis with consideration of other risk factors in multivariate analysis. The univariate analysis for GVHD, relapse and DFS is shown in Supplementary Table 1.
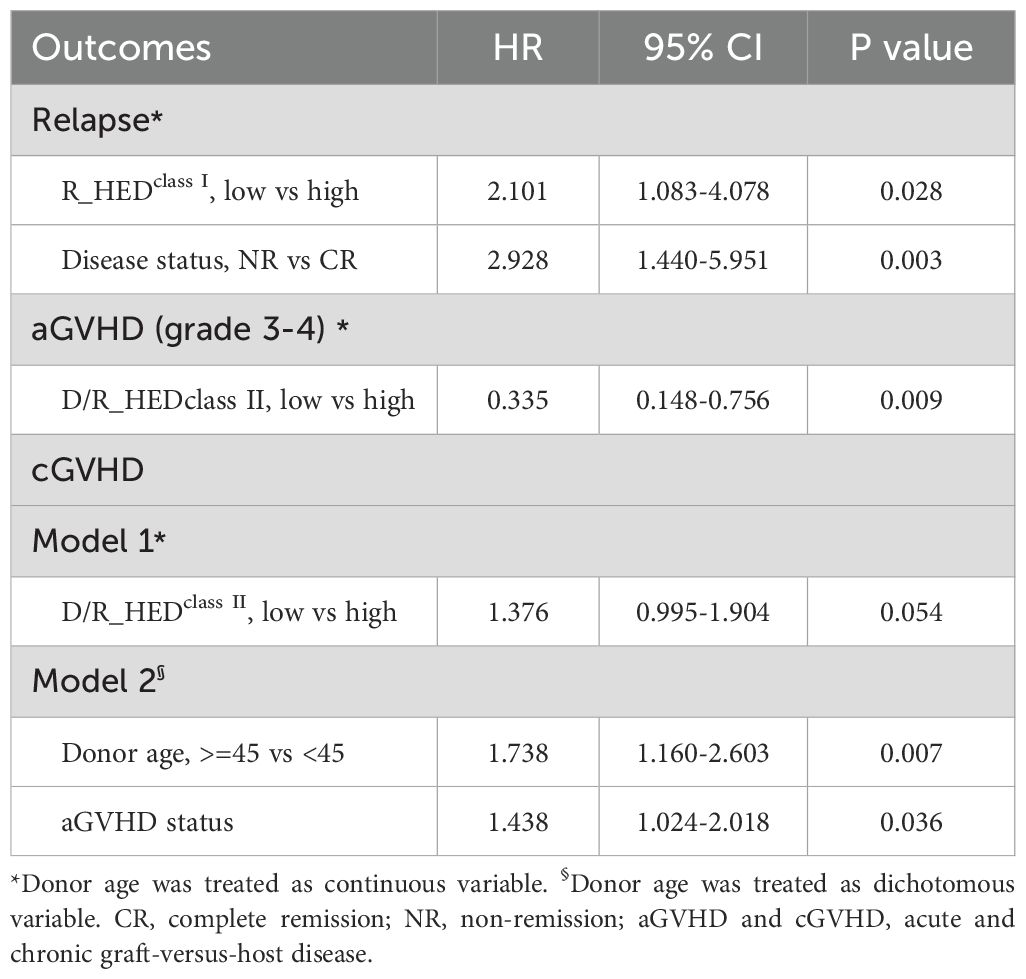
The multivariate regression analysis revealed that the low R_HEDclass I group had a more than two-fold greater risk of relapse (HR = 2.101 [95%CI: 1.083-4.078], P = 0.028) (Table 3, Figure 3B, Supplementary Table 2), whereas non-remission patients exhibited an approximately threefold risk of relapse. Therefore, R_HEDclass I can be considered an independent risk factor for relapse.
In the multivariate model of severe aGVHD, the low D/R_HEDclass II significantly reduced the risk of grade 3-4 aGVHD (HR = 0.335 [95% CI: 0.148-0.756], P = 0.009) as the only protective factor when considering donor age as a continuous variable (Table 3; Supplementary Table 2). However, when donor age was considered a dichotomous variable, it remained in the final model as a risk factor but failed to reach a statistically significant level (HR = 2.153, P = 0.068) (Table 3; Supplementary Table 2).
Regarding cGVHD, Model 1 with donor age as a continuous variable revealed that D/R_HEDclass II was the only independent risk factor, and low D/R_HEDclass II was associated with high risk of cGVHD (HR = 1.376 [95% CI: 0.995-1.904]); however, this association reached marginal statistical significance (P = 0.054, Table 3; Supplementary Table 2). In Model 2, patients with a history of aGVHD or receiving transplantation from donor older than 45 years had a high risk for cGVHD development, while D/R_HEDclass II no longer remained significant.
3.6 Survival
The proportions of 5-year OS and DFS for the entire cohort were 62.9% (95% CI: 56.1-68.9%) and 58.2% (95% CI: 51.4-64.4%), respectively. There was no discernible difference in overall and disease-free survival (OS or DFS) between patients with high- and low-HED (Figures 3C–F; Supplementary Figures 1, 2) except D_HEDclass I associated with DFS, implying that HED was ineffective as a prognostic indicator for ALL patients who underwent HLA-haploidentical HSCT with related donors. Multivariate regression analysis confirmed that HED, including D_HEDclass I, was not associated with survival.
4 Discussion
In the present investigation, we report the impact of HED scores on clinical outcomes for ALL patients underwent haploidentical HSCT. This represents the first study investigating HED in a pure cohort of haploidentical transplantations with patients affected by only one type of hematologic malignancy. While the observation are limited by the presented single center retrospective cohort, the results find that HED has poor prognostic value in OS/DFS, as well as the associations with relapse and aGVHD. In haploidentical setting, HLA disparity was once considered to have little impact on transplantation benefits, but our results showed that HED is an independent risk factor for selecting the best haploidentical donor.
Previous studies investigated the impact of HED on prognosis in mixed AML patients transplanted from either related or unrelated HLA-matched donors (11, 12). Roerden et al. examined the effect of HED on survival in an AML cohort with an HLA-identical sibling or foreign donor and found that a high class I HED had a favorable impact on OS (12). In AML patients undergoing HSCT, Daul et al. investigated the effect of class I and II HED on survival using four different donor sources: identical siblings, haploidentical donors, matched unrelated donors, and mismatched unrelated donors (11). The authors claimed that the class I/II HED ratio was an independent factor associated with better DFS/OS and could be an additive indication of GVL in addition to the major allogenic effect associated with the mismatched HLA. Recently, various hematological diseases were examined in a study by Merli et al., which supports the use of HED as a predictive marker in young adult and pediatric patients receiving transplantation from unrelated donors (4).
The ability of immune cells to interact with mismatched HLAs, minor histocompatibility antigens, and tumor-associated antigens (TAAs) on the leukemic cells is the foundation of the GVL effect (graft-versus-leukemia) (21). Compared to related patient/donor pairs, the overall genetic divergence for unrelated patient/donor pairs is higher. According to whole exome sequencing of patient-donor pairs undergoing allo-HSCT, an average of 6,445 non-synonymous SNVs were found to be mismatched, offering a sizable pool of possible miHAs (22). Genome-wide SNP array analyses revealed that the average mismatched SNVs in the coding region were 9.4% for sibling donors, rising to 17.3% for unrelated donors (23). To lessen the confounding effect of genetic background divergence, we therefore restricted the analysis to a pure cohort of haploidentical transplantation recipients who received transplantation from the related donor.
Our data showed that high R_HEDclass I was associated with a lower 5-year CIR, confirming the crucial function of CD8+ effective T cells in the GVL immune response and thus directly reflecting the immunological benefit of high HED. Patients with high HED scores potentially exhibit more immunogenic peptides than those with low HED scores, which may be recognized by donor-derived T lymphocytes (24), thereby reducing the likelihood of relapse. This explanation can be supported by similar research conducted recently. It was found that AML patients with high class I HED tended to recover their CD8+ T, B, and NK cells more quickly (11). Recently, Pagliuca et al. found that high recipient class I HED was associated with a higher diversity of TCR repertoire (25). In the first year of HSCT, a higher diversity of TCR repertoire and enhanced immune reconstitution might result in a strong defense against opportunistic infections (4).
However, our studies did not reveal any differences in OS or DFS between high and low R_HEDclass IALL patients, indicating that high R_HEDclass I was not always associated with a good prognosis as seen in AML. Patients with high R_HEDclass I had a relatively high incidence of NRM (25.5%) despite a low relapse rate (12.2%) (Table 2), which in turn offset the survival benefit from high R_HEDclass I, resulting in no significant difference in OS. This possible explanation is related to the Beijing protocol we used. The difference between our results and those of earlier studies may also be due to differences in disease type and ethnicity. Our cohort enrolled ALL patients, which has characteristics that cannot be totally extrapolated from studies of AML patients. For instance, while AML is incredibly sensitive to NK cell alloreactivity, the majority of adult ALL patients are not (26, 27). Furthermore, our homogeneous cohort is limited to Chinese, and distinct HLA alleles and HLA haplotypes are present in each ethnic group (28), emphasizing the significance of studying HED in this particular population. It’s interesting to note that Chhibber et al. (2022) found that genetic diversity of class I or II HLA loci (HED, heterozygosity, genotype) was not associated with clinical outcomes (9), suggesting that this biomarker shouldn’t be used for clinical decision-making for cancer patients receiving pembrolizumab. Similar studies conducted independently have also confirmed Chhibber’s conclusion (29–31). To properly comprehend the overall impact of HED, therefore, more research in larger cohorts and across more centers would be required.
As an alternative donor transplant, haploidentical HSCT offers patients who lack fully matched donors the chance to receive transplant, while donor-derived alloreactive T cells elicit a strong allogeneic response and exert an immense GVL effect (32). Between 2005 and 2015, there was a roughly threefold increase of haplo-HSCT in Europe due to favorable practical aspects of using a haploidentical donor and the accumulation of data of better outcomes achieved with TCR platforms (33). The democratization of using haploidentical donors leads to a fundamental paradigm shift: while donor availability was the key challenge for years, the issue today becomes identifying the best donor among several possible ones when haplo-HSCT (34). In general, the outcome of haploidentical HSCT may be influenced by DSA (donor-specific antibody), donor age, donor sex, KIR (killer immunoglobulin-like receptor), NIMA (noninherited maternal antigen), HLA matching, as well as family relationships (35, 36). Recent studies have confirmed that neither the quantity of HLA loci nor the combination of specific sites would affect the outcome of haploidentical HSCT (35, 37–40). The Beijing protocol showed that 1, 2, or 3 mismatches of 6 HLA loci had no effect on the cumulative incidence of cGVHD or aGVHD. Additionally, the number of HLA mismatches had no influence on the cumulative incidence of relapse, overall survival, and leukemia-free survival (35, 37). The cumulative incidence of GVHD, relapse rate, NRM, and overall survival were not affected by differences in the HLA locus in the T-cell-replete (TCR) haploidentical HSCT with a low dose of anti-T lymphocyte globulin (ATG), according to a prospective multicenter study from Japan (39). In the multivariate analysis, the only significant predictive factor for increased relapse was non-CR status prior to transplantation (P = 0.0424), which tended to be associated with a worse survival rate (P = 0.0524). It was also observed that the degree of HLA mismatching had no effect on post-transplant OS, cumulative incidence of aGVHD, NRM, or 1-year cGVHD in the high-dose PT/Cy haploidentical transplantation protocol, whether in the HVG (host-versus-graft) or GVH (graft-versus-host) settings (40). According to Kasamon et al., survival following nonmyeloablative transplants with posttransplant cyclophosphamide is also not correlated with the degree of HLA disparity (41).
Technique advances in aGVHD prophylaxis, prevention of post-transplant relapse, and treatment strategies have greatly improved the outcome of haploidentical HSCT compared to the past decades. Although the team from the Beijing protocol established the notion of donor selection and the best option of donor selection is to choose youthful, male, and NIMA-incompatible donors (35, 36), the consensus of donor selection, however, is still limited within the TCD and TCR haploidentical systems at this time. New criteria for donor selection may develop as a result of an increase in haploidentical HSCT cases and updated assessments of the factors influencing transplant outcomes (33, 34, 42–44). In this study, we found a strong association between D/R_HEDclass II and aGVHD incidence, with higher D/R_HEDclass II indicating more severe aGVHD. Single locus analysis revealed that the influence of D/R_HEDclass II appears to be predominantly driven by D/R_HEDDRB1,which proves the conclusions that DRB1 has the highest diversity among all HLA class II genes and the highest cell surface expression when compared to other HLA class II antigens (45). A high D/R_HEDclass II implies great spatial structural differences between donors and recipients, as well as more targets from tissue cells being presented. As a result, the greater the effect of T-cells attacking the tissue cells, the more severe the damage to the organ. The number of mismatch loci is obviously a relatively rough indicator, although it also reflects the degree of incompatibility between recipient and donor. Therefore, previous studies and our results suggest that the amount of HLA mismatch should not be used as a criterion for the selection of family haploidentical donors. Instead, D/R_HEDclass II provides more epitope information than mismatch numbers and also indicates donor and recipient mismatches, suggesting that D/R_HEDclass II may be taken into account as a new risk factor for donor selection in related haploidentical HSCT.
There are some limitations to this study. Our research was based on single-center and retrospective data and had a limited number of patients. Independent, prospective, larger, and multicenter investigations would be needed and beneficial to further confirm the impact of HED on outcome and the clinical significance of D/R_HEDclass II in donor selection. Due to the unavailability of data or limitations of the methods themselves, other approaches such as peptide binding motifs (PBM) (46), T-cell epitope (TCE) (47) or KIR-ligand mismatches (34, 45) were not considered.
In conclusion, we conducted a retrospective analysis to investigate the correlation between HED and outcomes in ALL patients who underwent transplants from related haploidentical donors. Results revealed that only class I HED of the recipient (R_HEDclass I) was associated with 5-year CIR and only D/R_HEDclass II was significantly correlated with severe aGVHD. Multivariate Cox regression analysis did confirm that a high D/R_HEDclass II was an independent risk factor for grade 3-4 aGVHD, and the high R_HEDclass I group had a more than two-fold reduced risk of relapse. KM and multivariate regression analyses confirmed that none of HED was associated with overall or disease-free survival. These results suggest that HEDclass II of donor-recipient pair could be used for donor selection as a novel risk factor for grade 3-4 aGVHD and patient’s HEDclass I for relapse in the setting of related haploidentical HSCT, but not as an independently prognostic factor for predicting OS or DFS.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author/s.
Ethics statement
The studies involving humans were approved by Ethics Committee of Hebei Yanda Lu Daopei Hospital. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin.
Author contributions
H-FZ: Data curation, Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. X-JL: Conceptualization, Investigation, Methodology, Resources, Supervision, Writing – review & editing. X-YC: Data curation, Investigation, Methodology, Project administration, Resources, Writing – review & editing. X-BL: Data curation, Formal analysis, Methodology, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
The authors would like to thank Dr. Ri Xu for her valuable suggestion.
Conflict of interest
Author H-FZ, X-JL and X-BL are employed by Beijing BFR Gene Diagnostics Co., Ltd.
The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1440911/full#supplementary-material
References
1. Kwon M, Bailén R, Díez-Martín JL. Evolution of the role of haploidentical stem cell transplantation: past, present, and future. Expert Rev Hematol. (2020) 13:835–50. doi: 10.1080/17474086.2020.1796621
2. Fleischhauer K, Beelen DW. HLA mismatching as a strategy to reduce relapse after alternative donor transplantation. Semin Hematol. (2016) 53:57–64. doi: 10.1053/j.seminhematol.2016.01.010
3. Tiercy J. How to select the best available related or unrelated donor of hematopoietic stem cells? Haematologica. (2016) 101:680–7. doi: 10.3324/haematol.2015.141119
4. Merli P, Crivello P, Strocchio L, Pinto RM, Algeri M, Del Bufalo F, et al. Human leukocyte antigen evolutionary divergence influences outcomes of paediatric patients and young adults affected by Malignant disorders given allogeneic haematopoietic stem cell transplantation from unrelated donors. Br J Haematol. (2023) 200:622–32. doi: 10.1111/bjh.18561
5. Grantham R. Amino acid difference formula to help explain protein evolution. Science. (1974) 185:862–4. doi: 10.1126/science.185.4154.862
6. Pierini F, Lenz TL. Divergent allele advantage at human MHC genes: signatures of past and ongoing selection. Mol Biol Evol. (2018) 35:2145–58. doi: 10.1093/molbev/msy116
7. Chowell D, Morris LGT, Grigg CM, Weber JK, Samstein RM, Makarov V, et al. Patient HLA class I genotype influences cancer response to checkpoint blockade immunotherapy. Science. (2018) 359:582–7. doi: 10.1126/science.aao4572
8. Chowell D, Krishna C, Pierini F, Makarov V, Rizvi NA, Kuo F, et al. Evolutionary divergence of HLA class I genotype impacts efficacy of cancer immunotherapy. Nat Med. (2019) 25:1715–20. doi: 10.1038/s41591-019-0639-4
9. Chhibber A, Huang L, Zhang H, Xu J, Cristescu R, Liu X, et al. Germline HLA landscape does not predict efficacy of pembrolizumab monotherapy across solid tumor types. Immunity. (2022) 55:56–64. doi: 10.1016/j.immuni.2021.12.006
10. Féray C, Taupin J, Sebagh M, Allain V, Demir Z, Allard M, et al. Donor HLA class 1 evolutionary divergence is a major predictor of liver allograft rejection. Ann Intern Med. (2021) 174:1385–94. doi: 10.7326/M20-7957
11. Daull AM, Dubois V, Labussière-Wallet H, Venet F, Barraco F, Ducastelle-Lepretre S, et al. Class I/Class II HLA evolutionary divergence ratio is an independent marker associated with disease-free and overall survival after allogeneic hematopoietic stem cell transplantation for acute myeloid leukemia. Front Immunol. (2022) 13:841470. doi: 10.3389/fimmu.2022.841470
12. Roerden M, Nelde A, Heitmann JS, Klein R, Rammensee H-G, Bethge WA, et al. HLA Evolutionary divergence as a prognostic marker for AML patients undergoing allogeneic stem cell transplantation. Cancers (Basel). (2020) 12:1835. doi: 10.3390/cancers12071835
13. Sun Y, Chang Y, Huang X. Update on current research into haploidentical hematopoietic stem cell transplantation. Expert Rev Hematol. (2018) 11:273–84. doi: 10.1080/17474086.2018.1447379
14. Stem Cell Application Group, Chinese Society of Hematology, Chinese Medical Association. The consensus of allogeneic hematopoietic transplantation for hematological diseases in China (2014)-indication, conditioning regimen and donor selection. Chin J Hematol. (2014) 35:775–80. doi: 10.3760/cma.j.issn.0253-2727.2014.08.029
15. Wang Y, Wu D-P, Liu Q-F, Xu L-P, Liu K-Y, Zhang X-H, et al. Low-dose post-transplant cyclophosphamide and anti-thymocyte globulin as an effective strategy for GVHD prevention in haploidentical patients. J Hematol Oncol. (2019) 12:88. doi: 10.1186/s13045-019-0781-y
16. Lai Y-R, Chen Y-H, Hu D-M, Jiang M, Liu Q-F, Liu L, et al. Multicenter phase II study of a combination of cyclosporine a, methotrexate and mycophenolate mofetil for GVHD prophylaxis: results of the Chinese Bone Marrow Transplant Cooperative Group (CBMTCG). J Hematol Oncol. (2014) 7:59. doi: 10.1186/s13045-014-0059-3
17. Stem Cell Application Group, Chinese Society of Hematology, Chinese Medical Association. Chinese consensus of allogeneic hematopoietic transplantation for hematological disease (III)-acute graft-versus-host disease. Chin J Hematol. (2020) 41:529–36. doi: 10.3760/cma.j.issn.0253-2727.2020.07.001
18. Przepiorka D, Weisdorf D, Martin P, Klingemann HG, Beatty P, Hows J, et al. 1994 Consensus conference on acute GVHD grading. Bone Marrow Transplant. (1995) 15:825–8.
19. Jagasia M, Greinix H, Arora M, Williams K, Wolff D, Cowen E, et al. National Institutes of Health consensus development project on criteria for clinical trials in chronic graft-versus-host disease: I. The 2014 diagnosis and staging working group report. Biol Blood Marrow Transplant. (2015) 21:389–401.e381. doi: 10.1016/j.bbmt.2014.12.001
20. Kanda Y. Investigation of the freely available easy-to-use software 'EZR' for medical statistics. Bone Marrow Transplant. (2013) 48:452–8. doi: 10.1038/bmt.2012.244
21. Janelle V, Rulleau C, Del Testa S, Carli C, Delisle JS. T-Cell immunotherapies targeting histocompatibility and tumor antigens in hematological Malignancies. Front Immunol. (2020) 11:276. doi: 10.3389/fimmu.2020.00276
22. Jameson-Lee M, Koparde V, Griffith P, Scalora A, Sampson J, Khalid H, et al. In silico derivation of HLA-specific alloreactivity potential from whole exome sequencing of stem-cell transplant donors and recipients: Understanding the quantitative immunobiology of allogeneic transplantation. Front Immunol. (2014) 5:529. doi: 10.3389/fimmu.2014.00529
23. Martin P, Levine D, Storer B, Warren E, Zheng X, Nelson S, et al. Genome-wide minor histocompatibility matching as related to the risk of graft-versus-host disease. Blood. (2017) 129:791–8. doi: 10.1182/blood-2016-09-737700
24. MacDonald KP, Hill GR, Blazar BR. Chronic graft-versus-host disease: biological insights from preclinical and clinical studies. Blood. (2017) 129:13–21. doi: 10.1182/blood-2016-06-686618
25. Pagliuca S, Gurnari C, Hong S, Zhao R, Kongkiatkamon S, Terkawi L, et al. Clinical and basic implications of dynamic T cell receptor clonotyping in hematopoietic cell transplantation. JCI Insight. (2021) 6:e149080. doi: 10.1172/jci.insight.149080
26. Locatelli F, Pende D, Falco M, Della Chiesa M, Moretta A, Moretta L. NK cells mediate a crucial graft-versus-leukemia effect in haploidentical-HSCT to cure high-risk acute leukemia. Trends Immunol. (2018) 39:577–90. doi: 10.1016/j.it.2018.04.009
27. Ruggeri L, Vago L, Eikema D-J, de Wreede LC, Ciceri F, Diaz MA, et al. Natural killer cell alloreactivity in HLA-haploidentical hematopoietic transplantation: a study on behalf of the CTIWP of the EBMT. Bone Marrow Transplant. (2021) 56:1900–7. doi: 10.1038/s41409-021-01259-0
28. Pidala J, Kim J, Schell M, Lee SJ, Hillgruber R, Nye V, et al. Race/ethnicity affects the probability of finding an HLA-A, -B, -C and -DRB1 allele-matched unrelated donor and likelihood of subsequent transplant utilization. Bone Marrow Transplant. (2013) 48:346–50. doi: 10.1038/bmt.2012.150
29. Negrao MV, Lam VK, Reuben A, Rubin ML, Landry LL, Roarty EB, et al. PD-L1 expression, tumor mutational burden, and cancer gene mutations are stronger predictors of benefit from immune checkpoint blockade than HLA class I genotype in non-small cell lung cancer. J Thorac Oncol. (2019) 14:1021–31. doi: 10.1016/j.jtho.2019.02.008
30. Anagnostou V, Niknafs N, Marrone K, Bruhm DC, White JR, Naidoo J, et al. Multimodal genomic features predict outcome of immune checkpoint blockade in non-small-cell lung cancer. Nat Cancer. (2020) 1:99–111. doi: 10.1038/s43018-019-0008-8
31. Litchfield K, Reading JL, Puttick C, Thakkar K, Abbosh C, Bentham R, et al. Meta-analysis of tumor- and T cell-intrinsic mechanisms of sensitization to checkpoint inhibition. Cell. (2021) 184:596–614.e514. doi: 10.1016/j.cell.2021.01.002
32. Wu H, Shi J, Luo Y, Yu J, Lai X, Liu L, et al. Assessment of patient-specific human leukocyte antigen genomic loss at relapse after antithymocyte globulin-based T-cell-replete haploidentical hematopoietic stem cell transplant. JAMA Network Open. (2022) 5:e226114. doi: 10.1001/jamanetworkopen.2022.6114
33. Passweg JR, Baldomero H, Bader P, Bonini C, Duarte RF, Dufour C, et al. Use of haploidentical stem cell transplantation continues to increase: The 2015 European Society for Blood and Marrow Transplant Activity Survey Report. Bone Marrow Transplant. (2017) 52:811–7. doi: 10.1038/bmt.2017.34
34. Dhuyser A, Aarnink A, Pérès M, Jayaraman J, Nemat-Gorgani N, Rubio MT, et al. KIR in allogeneic hematopoietic stem cell transplantation: need for a unified paradigm for donor selection. Front Immunol. (2022) 13:821533. doi: 10.3389/fimmu.2022.821533
35. Wang Y, Chang Y-J, Xu L-P, Liu K-Y, Liu D-H, Zhang X-H, et al. Who is the best donor for a related HLA haplotype-mismatched transplant? Blood. (2014) 124:843–50. doi: 10.1182/blood-2014-03-563130
36. Wang Y, Wu DP, Liu QF, Xu LP, Liu KY, Zhang XH, et al. Donor and recipient age, gender and ABO incompatibility regardless of donor source: validated criteria for donor selection for haematopoietic transplants. Leukemia. (2018) 32:492–8. doi: 10.1038/leu.2017.199
37. Fuchs EJ. Human leukocyte antigen-haploidentical stem cell transplantation using T-cell-replete bone marrow grafts. Curr Opin Hematol. (2012) 19:440–7. doi: 10.1097/MOH.0b013e32835822dc
38. Wang Y, Liu D-H, Liu K-Y, Xu L-P, Zhang X-H, Han W, et al. Long-term follow-up of haploidentical hematopoietic stem cell transplantation without in vitro T cell depletion for the treatment of leukemia. Cancer. (2013) 119:978–85. doi: 10.1002/cncr.27761
39. Ikegame K, Yoshida T, Yoshihara S, Daimon T, Shimizu H, Maeda Y, et al. Unmanipulated haploidentical reduced-intensity stem cell transplantation using fludarabine, busulfan, low-dose antithymocyte globulin, and steroids for patients in non-complete remission or at high risk of relapse: A prospective multicenter phase I/II study in Japan. Biol Blood Marrow Transplant. (2015) 21:1495–505. doi: 10.1016/j.bbmt.2015.04.012
40. Raiola A, Risitano A, Sacchi N, Giannoni L, Signori A, Aquino S, et al. Impact of HLA disparity in haploidentical bone marrow transplantation followed by high-dose cyclophosphamide. Biol Blood Marrow Transplant. (2018) 24:119–26. doi: 10.1016/j.bbmt.2017.10.002
41. Kasamon Y, Luznik L, Leffell M, Kowalski J, Tsai H, Bolaños-Meade J, et al. Nonmyeloablative HLA-haploidentical bone marrow transplantation with high-dose posttransplantation cyclophosphamide: effect of HLA disparity on outcome. Biol Blood Marrow Transplant. (2010) 16:482–9. doi: 10.1016/j.bbmt.2009.11.011
42. Shook DR, Triplett BM, Eldridge PW, Kang G, Srinivasan A, Leung W. Haploidentical stem cell transplantation augmented by CD45RA negative lymphocytes provides rapid engraftment and excellent tolerability. Pediatr Blood Cancer. (2015) 62:666–73. doi: 10.1002/pbc.25352
43. Bleakley M, Heimfeld S, Loeb KR, Jones LA, Chaney C, Seropian S, et al. Outcomes of acute leukemia patients transplanted with naive T cell-depleted stem cell grafts. J Clin Invest. (2015) 125:2677–89. doi: 10.1172/jci81229
44. Chang YJ, Luznik L, Fuchs EJ, Huang XJ. How do we choose the best donor for T-cell-replete, HLA-haploidentical transplantation? J Hematol Oncol. (2016) 9:35. doi: 10.1186/s13045-016-0265-2
45. Fürst D, Neuchel C, Tsamadou C, Schrezenmeier H, Mytilineos J. HLA Matching in unrelated stem cell transplantation up to date. Transfus Med Hemoth. (2019) 46:326–36. doi: 10.1159/000502263
46. Crivello P, Arrieta-Bolaños E, He M, Wang T, Fingerson S, Gadalla SM, et al. Impact of the HLA immunopeptidome on survival of leukemia patients after unrelated donor transplantation. J Clin Oncol. (2023) 41:2416–27. doi: 10.1200/JCO.22.01229
Keywords: human leukocyte antigen (HLA) evolutionary divergence (HED), acute lymphoblastic leukemia, haploidentical hematopoietic stem cell transplantation, donor selection, risk factor
Citation: Cao X-Y, Zhou H-F, Liu X-J and Li X-B (2024) Human leukocyte antigen evolutionary divergence as a novel risk factor for donor selection in acute lymphoblastic leukemia patients undergoing haploidentical hematopoietic stem cell transplantation. Front. Immunol. 15:1440911. doi: 10.3389/fimmu.2024.1440911
Received: 30 May 2024; Accepted: 01 August 2024;
Published: 19 August 2024.
Edited by:
Rémy Dulery, Hôpital Saint-Antoine, FranceReviewed by:
Marco Andreani, Bambino Gesù Children’s Hospital (IRCCS), ItalyPietro Crivello, Essen University Hospital, Germany
Copyright © 2024 Cao, Zhou, Liu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xing-Yu Cao, Y2FveGluZ3l1MjAyMEBzaW5hLmNvbQ==; Xiang-Jun Liu, eGpsaXVAYmZyYmlvdGVjaC5jb20uY24=
†These authors have contributed equally to this work
 Xing-Yu Cao
Xing-Yu Cao Hai-Fei Zhou
Hai-Fei Zhou Xiang-Jun Liu
Xiang-Jun Liu Xiao-Bo Li
Xiao-Bo Li