
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol., 07 November 2024
Sec. Alloimmunity and Transplantation
Volume 15 - 2024 | https://doi.org/10.3389/fimmu.2024.1440887
This article is part of the Research TopicAntibody-Mediated Rejection After Solid Organ TransplantationView all 11 articles
 Roberto Littera1,2*
Roberto Littera1,2* Stefano Mocci3,4,5*
Stefano Mocci3,4,5* Davide Argiolas6*
Davide Argiolas6* Letizia Littarru6
Letizia Littarru6 Sara Lai1,2
Sara Lai1,2 Maurizio Melis2
Maurizio Melis2 Celeste Sanna3
Celeste Sanna3 Caterina Mereu3
Caterina Mereu3 Michela Lorrai3
Michela Lorrai3 Alessia Mascia4
Alessia Mascia4 Andrea Angioi6
Andrea Angioi6 Giacomo Mascia6
Giacomo Mascia6 Valeria Matta6
Valeria Matta6 Nicola Lepori7
Nicola Lepori7 Matteo Floris6
Matteo Floris6 Cristina Manieli8
Cristina Manieli8 Paola Bianco8
Paola Bianco8 Daniela Onnis8
Daniela Onnis8 Stefania Rassu1
Stefania Rassu1 Silvia Deidda9
Silvia Deidda9 Mauro Giovanni Carta10
Mauro Giovanni Carta10 Erika Giuressi1
Erika Giuressi1 Andrea Perra2,4
Andrea Perra2,4 Luchino Chessa2,10,11
Luchino Chessa2,10,11 Sabrina Giglio1,3,5*
Sabrina Giglio1,3,5* Antonello Pani6,12*
Antonello Pani6,12*Background: Antibody-mediated rejection is a significant cause of kidney transplant failure. Recent studies have shown that the MHC class I MICA gene influences the transplantation outcome. However, the role of the primary MICA receptor, NKG2D, has yet to be explored.
Aim: We aimed to investigate the correlation between recipient/donor MICA allele matching and NKG2D genotype with the risk of antibody-mediated rejection and their potential clinical effects and implications for organ maintenance therapy.
Methods: Of the 524 patients who underwent transplantation, 387 were eligible for the study. Complete MICA allele and two functional polymorphisms of NKG2D (rs1049174C>G and rs2255336G>A) were analyzed in 148 transplanted patients and 146 controls.
Results: Increased recipient/donor MICA allele mismatches correlate with an elevated risk of antibody-mediated rejection (X2 = 6.95; Log-rank=0.031). Notably, the rs1049174[GG] genotype contributes to a significantly increased risk of antibody-mediated rejection (X2 = 13.44; Log-rank=0.001 and X2 = 0.34; Log-rank=0.84). The combined effect of two MICA allele mismatches and rs1049174[GG] genotype shows the highest risk (X2 = 23.21; Log-rank<0.001). Most importantly, patients with rs1049174[GG] and rs2255336[AA] genotypes may respond less to mTOR inhibitor immunosuppressive therapy than Calcineurin inhibitors (rs1049174[GG]; P=0.035; and rs2255336[AA]; P=0.002).
Conclusion: Recipient/donor MICA allele mismatches and specific NKG2D variants, as well as their combinations, influence kidney transplant outcomes, providing insights for personalized treatment and enhancing graft survival.
Kidney transplantation is the best treatment for kidney function in patients with advanced chronic kidney disease or end-stage kidney disease (ESKD) (1, 2). Transplantation significantly reduces overall mortality and improves the quality of life for patients with renal disease (3, 4). However, kidney transplantation is a complex procedure that relies on several pivotal immunological and non-immunological factors that directly affect the graft’s survival and functionality (5, 6).
Antibody-mediated rejection is undoubtedly the most important factor that adversely affects the survival of the transplanted kidney in the medium and long term (7, 8).
Antibody-mediated rejection (ABMR) constitutes organ injury triggered by circulating donor-specific antibodies (DSA), which can target either human leukocyte antigens (HLAs) or non-HLA antigens (9). Identifying ABMR typically involves assessing the levels of DSAs and performing a kidney biopsy that reveals features such as microvascular inflammation (MVI) and C4d deposition in the endothelium (10–12).
While the compatibility of HLA molecules between donor and recipient has historically been the main focus in kidney transplantation, recent studies suggest that incompatibilities at other loci, such as MICA (major histocompatibility complex class I-related chain A) and KIR genes and NKG2D (natural killer group 2 member D) (KLRK), can influence graft outcome (13–17).
MICA acts as a ligand for NKG2D, an activating receptor expressed on NK cells, NKT cells, γδ T cells, and CD8+ αβ T cells (1, 18, 19). The binding of MICA ligands to NKG2D activates NK cells, enhances their functions, and allows them to function as a bridge between innate and adaptive immunity (20).
The pathogenic role of MICA-specific antibodies in kidney transplantation remains controversial. However, recent studies indicate that patients with MICA mismatches exhibit notably diminished graft survival compared to those with MICA-matched donors, with respective five-year graft survival rates of 88% and 96% (15). Additionally, genetic variability of NKG2D also may influence the receptor’s functional capacity, with at least two haploblocks identified to affect receptor expression activity levels (21).
This study aims to 1) evaluate the impact of MICA polymorphisms on kidney transplantation, focusing on the incidence of antibody-mediated rejection and graft function survival in the Sardinian population 2) Investigate how NKG2D polymorphisms in combination with MICA compatibility may influence immunological activity and assess their immunological significance relative to the classic HLA system.
From July 2012 to July 2022, 524 patients underwent kidney transplantation at the Organ Transplantation Center of the G. Brotzu Hospital in Cagliari, Italy. We excluded 1) cases lacking patient/donor biological material or incomplete clinical data. 2) patients who underwent a second transplant or had pre-transplant donor-specific HLA antibodies (pre-Tx DSA) (Figure 1).
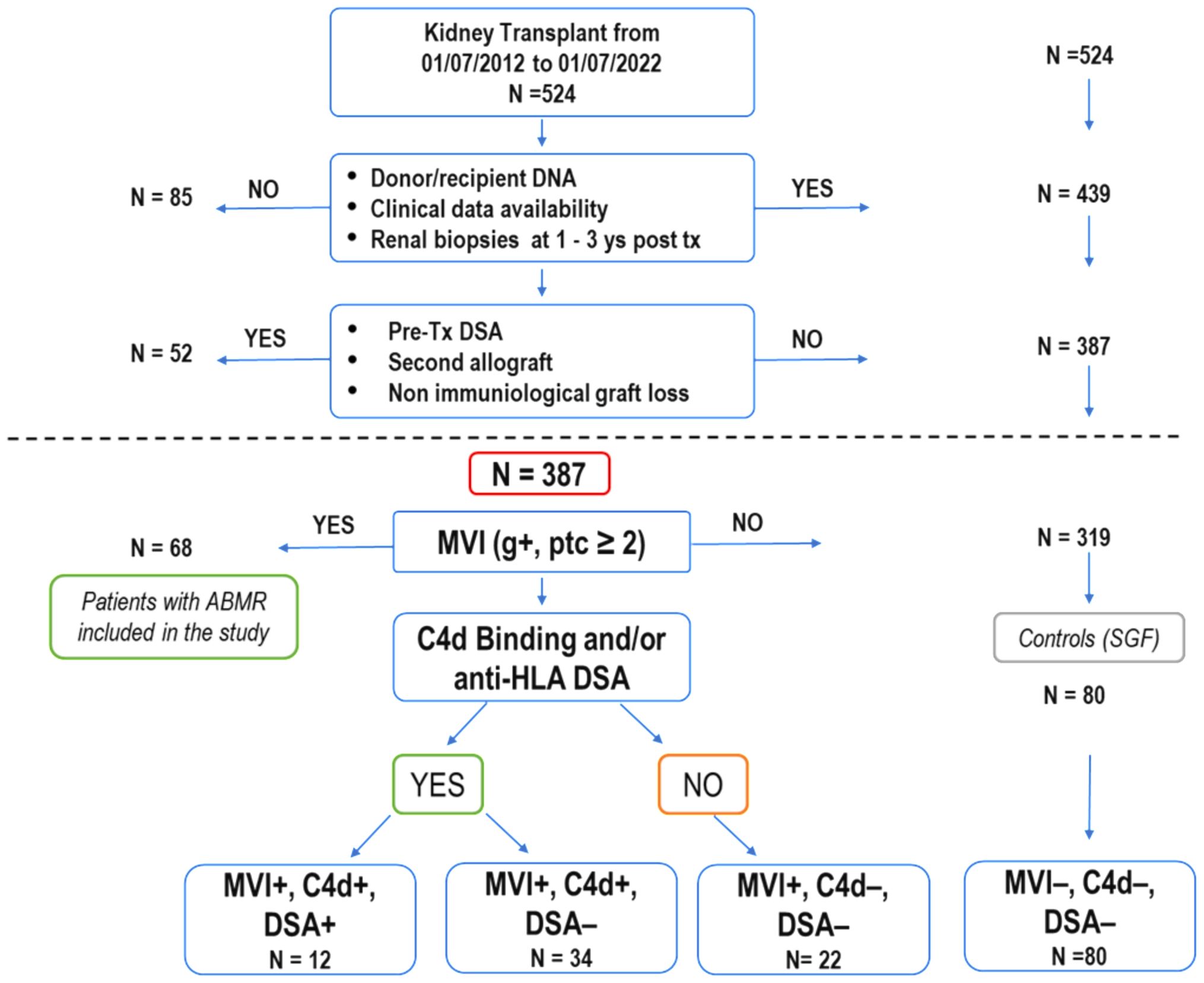
Figure 1. Selection process and characteristics of kidney transplant recipients in the study. Out of 524 patients who underwent kidney transplants over ten years, 387 were enrolled for analysis based on specific criteria, including renal biopsies at 1 and 3 years after transplantation (as required for post-transplant follow-up by the Cagliari transplant center), absence of second transplant and pre-transplant donor-specific HLA antibodies. Among these, 68 patients (21.3%) experienced a progressive decline in graft function attributed to antibody-mediated rejection (ABMR), with histological confirmation through renal biopsies. Some of these patients showed a mixed histological picture of ABMR and TCMR. The 68 patients with ABMR presented with a histological picture of MVI+ (g+, ptc ≥ 2). They were divided into three subgroups based on the presence or absence of C4d and the presence or absence of DSA (MVI+, C4d+, DSA+;MVI+, C4d+, DSA-; MVI+, C4d-, DSA-). The remaining 319 patients never exhibited clinical, histological, or laboratory signs of organ damage. Eighty patients (MVI-, C4d-, DSA-), randomly selected, were used as a control group (SGF). (ABMR, antibody-mediated rejection; DSA, donor-specific HLA antibodies; MVI, microvascular inflammation; TCMR, T-cell-mediated rejection).
Of the remaining 387 eligible patients, 68 manifested ABMR while 319 showed normal renal function. Out of these 319, 80 patients were randomly selected to be used as the stable graft function (SGF) control group.
Moreover, the total number of SGF and ABMR patients (148) was compared with a healthy cohort of 146 individuals of Sardinian descent and underwent a systematic evaluation of clinical and immunological parameters significantly impacting transplantation outcomes. These parameters included HLA class I (0-2 HLA-A, 0-2 HLA-B, 0-2 HLA-C) mismatches (score 0-6), HLA class II (0-4 HLA-DRB1, HLA-DQB1) mismatches (score 0-4), recipient NKG2D (KLRK1) polymorphisms rs1049174 and rs2255336, MICA mismatches (score 0-2). The analysis of MICA alleles matching was performed considering two categories of mismatches: i) mismatches in the host-versus-graft (HvG) direction where the donor but not the recipient is mismatched, and ii) all types of mismatches, independent of their directions. Additional factors examined included panel-reactive antibodies (PRA) exceeding 5-10%, the duration of graft cold ischemia, the type of induction therapy received, and the immunotherapy administered post-transplant.
Kidney biopsies were performed when clinically justified due to suspected graft dysfunction, such as unexplained changes in graft function, new-onset proteinuria, rising serum creatinine, or other deviations from the standard post-transplantation course. These biopsies were carried out to promptly detect and address any potential pathological alterations affecting graft function. Each specimen was assessed according to the standards from the 15th Banff Conference held in 2019 (22). All samples were reviewed by three independent renal pathologists. The analysis included H&E, PAS, AFOG, and silver stain, along with immunofluorescence for IgG, IgA, IgM, kappa, lambda, C3, C4, C1q, and albumin, and immunohistochemistry for C4d, with additional stains as required (e.g., SV40).
The presence of anti-MICA and anti-HLA class I and II antibodies were determined in all pre-transplant patients. The presence of anti-HLA DSA precluded kidney transplantation at our center. Anti-HLA and MICA DSA levels: This assessment was conducted using LAB Screen Single Antigen kits from One Lambda, following the manufacturer’s instructions. Beads with a normalized MFI above 2500 for HLA or 1000 for MICA were considered positive as reported in other work (23). According to the transplant center’s operational standards, antibody testing was performed during the one-year post-transplant follow-up, or earlier if the patient showed clinical signs of rejection (renal injury/graft impairment) within the first year.
HLA and MICA typing. Genomic DNA (gDNA) was isolated from peripheral blood using QIAcube (Qiagen, Hilden, NW, Germany) and the DNA Blood Mini kits according to the manufacturer’s instructions. Isolated gDNA was HLA genotyped using the AlloSeq Tx17 assay (Care Dx, Brisbane, CA) following the recommended protocol. The AlloSeq Tx17 assay incorporates 17 specific probes designed to target HLA-A, HLA-B, HLA-C, HLA-E, HLA-F, HLA-G, HLA-H, HLA-DRB1, HLA-DRB3, HLA-DRB4, HLA-DRB5, HLA-DQA1, HLA-DQB1, HLA-DPA1, HLA-DPB1, MICA, and MICB loci. Library quantification was performed using the Qubit™ dsDNA HS Assay (Life Technologies, Carlsbad, California, USA), and sequencing was performed on the Illumina MiSeq platform (San Diego, CA) with 2x150 sequencing. MICA allele matching/mismatching was analyzed based on the recipient-versus-graft direction, where the donor, but not the recipient, is mismatched. The codon position 129 of the MICA gene was considered to determine the presence of Methionine (Met) or Valine (Val) (rs1051792) and to evaluate recipient/donor MICA mismatches. Allele frequencies of MICA were calculated using direct gene counting.
Primer sets targeting specific regions were designed with the assistance of Primer3 version 4.1.0 (24). The annealing temperature was optimized for each primer set. The two SNPs located within the NKG2D (KLRK1) gene rs1049174 and rs2255336 belong to two different haplotype blocks (NKG2D hb-1 and hb-2), each of which generates two major haplotypes associated with low (LNK) and high natural cytotoxic activity (HNK) phenotypes as shown in Table 1 (21). Several studies have demonstrated that high and low natural cytotoxic activity haplotype alleles (HNK1 or LNK1) belonging to NKG2D haplotype blocks 1 (hb-1) may be successfully predicted by only a single SNP (dbSNP: rs1049174) and haplotype blocks 2 (hb-2) by dbSNP rs2255336 (25, 26). Primers are reported in the Supplementary Table S1. The PCR was performed according to the protocol supplied with AmpliTaq Gold™ DNA Polymerase (Applied Biosystems/Thermo Fisher Scientific, Waltham, MA). Sequencing was performed using the BigDye™ Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, USA), with the same primers described previously and cleaned up with CleanSEQ Dye-Terminator Removal Kit (Beckman Coulter, Inc.). Capillary electrophoresis was performed on the ABI 3500 Genetic Analyser (Applied Biosystems), and sequences were analyzed with Sequencher 5.3 (© 2017 Gene Codes Corporation).
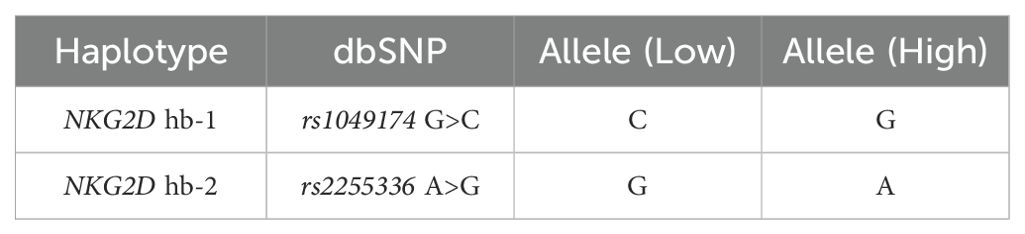
Table 1. Haplotype blocks (NKG2D hb-1 and hb-2) are split into low and high natural cytotoxic activity haplotypes.
Summary statistics were calculated for the clinical and biochemical data of patients diagnosed with or without antibody-mediated rejection: interquartile ranges (IQR), medians, means, standard deviations (SD), and mean differences were calculated on all continuous variables; percentages and odds ratios (OR) were calculated on categorical data. P values and 95% confidence intervals (95% CI) were obtained using Student’s t-test or Fisher’s exact test, as appropriate. Statistical analysis was performed by using R software version 4.3.2 (27). The frequencies of rs1049174 G>C and rs2255336 A>G SNPs in the NKG2D gene and MICA alleles were compared between patients with ABMR, stable graft function (SGF), and an appropriate group of Sardinian healthy controls. The Hardy-Weinberg Equilibrium (HWE) of the SNPs and allele frequencies was examined by computing X2HWE and P values. Deviation from HWE was assessed using HaploView 4.2 software (28). The linkage disequilibrium (LD) between the rs1049174 G/C and rs2255336 A/G haplotypes of the NKG2D gene was evaluated in transplanted patients and healthy group controls. The observed and expected frequencies in each sample were compared using the chi-square test. LD was measured by the parameters D (difference between the observed and expected frequencies) and D′ (i.e., D normalized to one: -1 ≤ D′ ≤ 1). D′ was obtained using the normalization formulas proposed by Lewontin (29) for two-loci haplotypes. We also computed the parameter r2 expressing the correlation between the alleles at two loci. To compare the LD in the control and patient groups or the SGF and ABMR cohorts, we evaluated the P value associated with the chi-square variable (with two degrees of freedom) given by the difference between the chi-square variables in the two groups (with one degree of freedom). Kaplan-Meier curves were used to illustrate the cumulative incidence of antibody-mediated rejection from the date of transplantation to the date of clinical, histopathological, and immunohistochemical detection or the date of the last follow-up or death with a functioning graft. Transplant recipients were stratified into several groups according to genotypes and allele mismatches. The log-rank test was used for comparisons of the different gene profile combinations. Serum creatinine levels and glomerular filtration rate were measured at 1, 6, 12, 36, and 72 months from the date of transplantation. Comparison between groups of stratified patients was performed by computing the area under the curve (AUC) for the corresponding plots (30). AUC was evaluated using the trapezium formula extended to include all times at which serum creatinine and glomerular filtration rate (eGFR) were measured. The Student’s t-test was used to confirm statistical significance. A multivariate analysis was conducted to determine the independence from donor age and gender of the other clinical and genetic variables influencing rejection incidence and graft function. In the multivariate comparison between patients with SGF and ABMR, a logistic regression model was used to compute P values (PM), odds ratios (ORM), and 95% confidence intervals (95% CIM) adjusted accordingly to age and gender and for the potential confounder. The analysis included cold ischemia time, R/D MICA alleles full mismatch (MICA 2MM), allelic mismatches R/D HLA-DRB1, HLA-DQB1 (HLA II Class 1-2MM) and HLA class I (HLA-A, -B, -C > 3MM), genotypes NKG2D rs2255336 AA and NKG2D rs1049174 GG, de novo DSA HLA Class I and II, and the combination of MICA 2MM with the genotype NKG2D rs1049174 GG, which was confirmed to be strongly associated with rejection incidence. The de novo DSA HLA Class I observations in the two groups of patients were too few to yield fully reliable results for ORM and 95% CIM.
Between July 2012 and July 2022, 524 kidney transplant patients (KTPs) were treated at the Brotzu transplant center in Cagliari (Figure 1). Eighty-five patients (16.2%) were excluded due to insufficient patient/donor biological material or incomplete clinical data, including the lack of renal biopsy in the 1st and 3rd year of post-transplant follow-up. To accurately evaluate the immunological impact of allelic MICA mismatch and NKG2D genotype on long-term graft function, we further excluded 52 patients (9.9%) who underwent a second transplant or had pre-transplant donor-specific HLA antibodies (pre-tx DSA). Out of 387 patients enrolled in the study, 68 (17.6%) had clinically manifested a progressive decline in graft function attributed to antibody-mediated rejection (ABMR), characterized by MVI+ (Banff score: g+, ptc ≥ 2) as evidenced in all cases through renal biopsies. They were divided into three subgroups based on the presence or absence of C4d+ and the presence or absence of DSA (12 patients were MVI+, C4d+, DSA+; 34 were MVI+, C4d+, DSA-; and 22 were MVI+, C4d-, DSA-). The presence of calcineurin inhibitor toxicity, hypertensive damage, BK virus, and bacterial infections was ruled out. All patients were compliant with post-transplantation immunosuppressive therapy. Three hundred nineteen patients, who showed no clinical, histological, or laboratory signs of organ damage, were included in the control group (SGF), which consisted of 80 randomly selected patients (MVI-, C4d-, DSA-).
The age, sex, clinical, and demographic characteristics of recipient-donor pairs are detailed in Table 2. No significant differences were observed between the two groups of patients with SGF or ABMR in terms of the age and gender of the recipients. The number of HLA Class I (HLA-A, -B, -C) mismatches (0-6), HLA Class II (HLA-DRB1, HLA-DQB1) mismatches (0-4), and the percentage of sensitized patients (PRA > 5%) (31), showed no substantial differences between the SGF and ABMR groups. Significantly, the cold ischemia time in the ABMR patient group was notably more protracted than in the SGF group (747.5 ± 211.5 vs. 590.3 ± 22.9, OR = 157.2, 95% CI 74.0 – 590.3; P = 3.0 x 10-4).
After the transplant, only 1.3% (1/80) of patients with stable graft function (SGF) developed de novo donor-specific antibodies (DSAs) with a MFI above 2500 for HLA or 1000 for MICA, compared to 17.6% (12/68) of patients with antibody-mediated rejection (ABMR) (OR = 16.9, 95% CI 2.1–134.0; P = 0.001). Most of these DSAs were directed against HLA class II antigens.
Additionally, 17 out of 68 (25.0%) patients with ABMR had MICA DSA alloantibodies (Supplementary Table S3). However, most of these patients had a low MFI level (<1000). Only 4 of the ABMR patients developed MICA DSAs with a mean fluorescence intensity (MFI) > 1000 (ranging from 1100 to 4500). Three of these patients had anti-MICA alloantibodies against the donor MICA-129 Methionine antigen (MICA 18 and MICA 01), while one patient had antibodies against both MICA-129 Methionine and Valine antigens (MICA 01, 08). Overall, 5 out of 6 anti-MICA DSAs with an MFI level > 1000 targeted Methionine at residue 129 (Supplementary Table S3).
Considerable overlap in treatment regimens administered before and after transplantation was observed between the two groups (Table 2). It is noteworthy to observe that the maintenance regimen based on mTOR inhibitors was statistically significantly associated with a higher number of episodes of ABMR compared to therapeutic regimens consisting of CNI [55.8% (48/86) vs. 32.3% (20/62); P = 0.007; OR: 2.6 (1.3 – 5.6)].
MICA alleles were compared in 148 KTPs and 146 healthy controls. The analysis revealed a few substantial differences in allele frequencies between patients and healthy controls (Table 3). The frequency of the MICA*002:01 was significantly lower in the kidney transplant patient group compared to the control group [16.9% (50/296) in KTPs vs. 24.0% (70/292) in controls; p-value: 0.040; OR: 0.65 (0.42 - 0.99)]. The other alleles did not exhibit substantial differences and had comparable frequencies between controls and patients.
Furthermore, when dividing the patients based on transplant outcomes (ABMR or SGF), the most represented alleles in both groups were MICA*001:01 and MICA*002:01 (Table 4). The MICA*010:01 allele was more frequent in SGF patients [0.007% (1/136) in ABMR, 0.05% (8/160) in SGF; P = 0.042; OR: 7.105 (0.877-57.548)]. Furthermore, ABMR and SGF groups were compared based on each allele’s heterozygote and homozygote frequencies. In particular, no significant differences were observed in the frequencies of homozygotes and heterozygotes for the most frequent allele: MICA*001:01, *002:01, *004:01, *008:01, *009:01 and *010:01 (Table 4).

Table 4. MICA alleles their frequencies in the group of antibody-mediated rejection patients and patients with stable graft function.
Figure 2A shows the 95% cumulative incidence curves over 120 months for antibody-mediated rejection (ABMR) in 148 patients, stratified into three groups based on the number of MICA allele mismatches with their donors. Of these, 24 R/D pairs (16.2%) were matched (0 MM), 52 R/D pairs (35.1%) had one mismatch (1 MM), and 72 R/D pairs (48.7%) had two mismatches (2 MM). The median follow-up was 52.9 months for MICA-matched patients and 64.7 months for MICA-mismatched patients. At 5 years post-transplantation, graft survival was 79.2% (19/24) for MICA-matched patients and 64.5% (80/124) for MICA-mismatched patients (1 and 2 MM). Compared to MICA-mismatched patients, those matched for MICA alleles exhibited a significantly reduced risk of antibody-mediated rejection (X² = 6.95; Log-rank = 0.03). Indeed at 120 months post-transplantation, the incidence of ABMR was only 20.8% (5/24) in MICA-matched patients, compared to 49.1% (26/52) and 52.7% (38/72) in patients with 1 and 2 mismatches (1 MM and 2 MM), respectively.
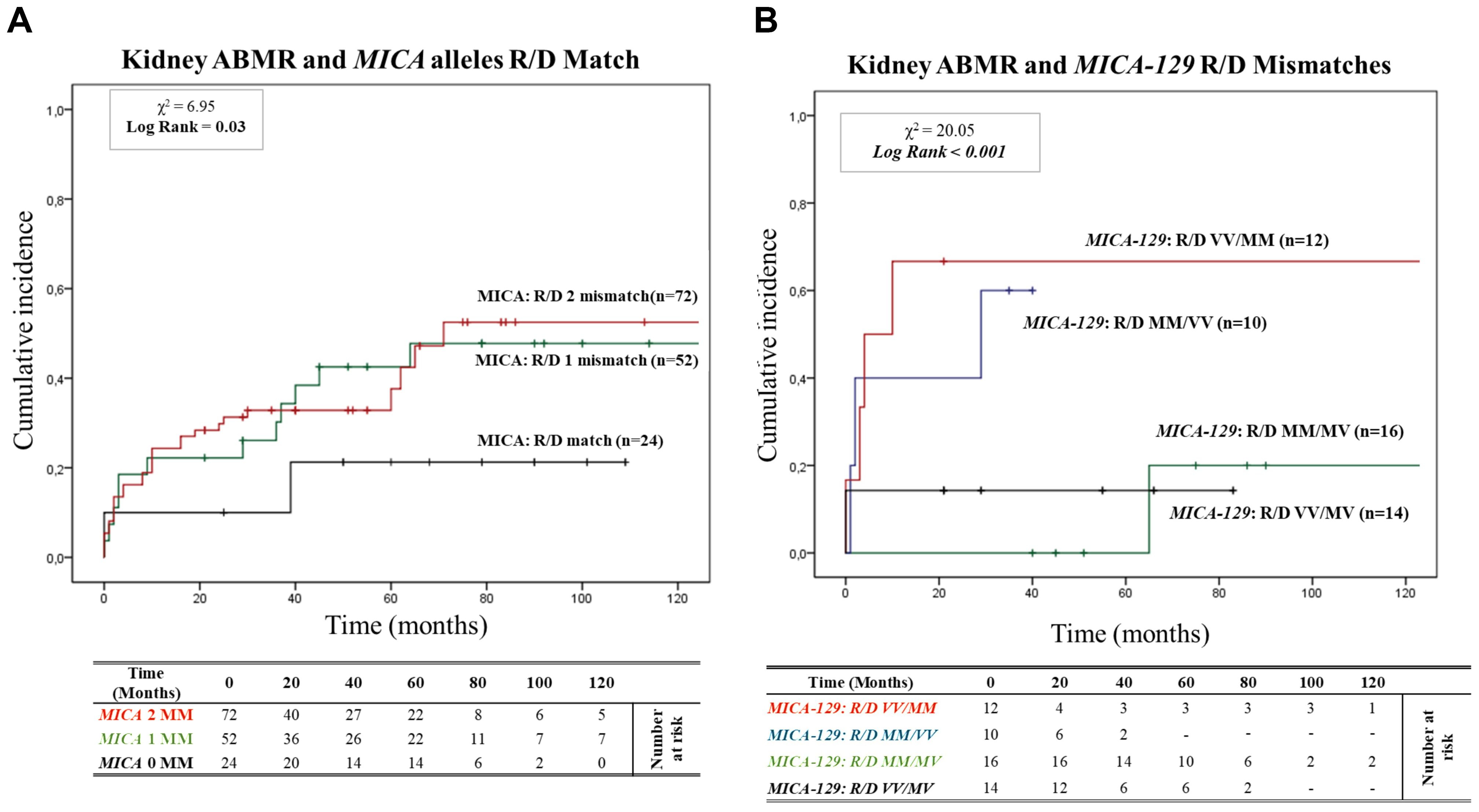
Figure 2. (A) Cumulative incidence for antibody-mediated rejection according to recipient-donor (R/D) MICA allele mismatches. The cumulative incidence of rejection events is graphically presented for a cohort of 148 patients observed over 120 months. Patients were categorized based on three groups of patients stratified according to donor-recipient MICA allele mismatches [0MM (black), 1MM (green), 2 MM (red)]. P-values were calculated using the two-sided Log-rank test without correction. χ2: Chi-square. MM: Mismatches. (B) Cumulative incidence for antibody-mediated rejection according to recipient-donor (R/D) MICA-129 allele mismatches. The cumulative incidence of rejection events is graphically presented for a cohort of 148 patients observed over 120 months. Patients were categorized on four groups of patients stratified according to recipient-donor MICA allele mismatches based on substitution of valine (V) with methionine (M) at position 129 (MICA-129) of the MICA protein: [R/D VV/MM (Red), R/D MM/VV (blue), R/D MM/MV (green) and R/D VV/MV (black)]. P-values were calculated using the two-sided Log-rank test without correction. χ2, Chi-square.
In addition to a detailed exploration of the consequences of the MICA mismatching model, we closely monitored renal function through assessments of serum creatinine levels and glomerular filtration rate (eGFR) (Supplementary Figures S2, S3). This additional analysis further confirmed the impact of recipient/donor (R/D) MICA allele matching on renal outcomes.
Building on this analysis, we further investigated the specific role of MICA-129 mismatches and their potential interactions with antibody-mediated rejection. We analyzed the cumulative incidence curves over 120 months for ABMR based on the R/D MICA-129 1 or 2 mismatches (1 MM and 2 MM) as R/D: MM/MV, VV/MV, MM/VV, VV/MM (Figure 2B). Two MICA-129 mismatches were observed in 12 R/D pairs VV/MM (8.1%) and 10 R/D pairs MM/VV (6.8%), while 1 MICA-129 mismatch was observed in 16 R/D pairs MM/MV (10.8%) and 14 R/D pairs VV/MV (9.5%).
The median follow-up was 27.9 months for MICA-129 2 mismatched patients (VV/MM and MM/VV) and 60.6 months for patients with 1 MICA-129 mismatch compared to the donor (VV/MV and MM/MV). Patients with 2 MICA-129 mismatches exhibited a significantly higher risk of antibody-mediated rejection (X² = 20.05; Log-rank < 0.001).
At 5 years post-transplantation, graft survival was 100% (16/16) for MICA-129 1 mismatch R/D MM/MV, and 85.7% (12/14) for MICA-129 1 mismatch R/D VV/MV, while in the presence of 2 MICA-129 mismatches, graft survival decreased to 40% (4/10) for R/D MM/VV pairs and to 33.3% (4/12) for R/D VV/MM pairs.
At 120 months post-transplantation, the cumulative incidence of ABMR remained significantly higher in the pairs that presented two MICA-129 mismatches, particularly in R/D VV/MM pairs [66.7% (8/12)].
The two index SNPs, rs1049174 (G>C) and rs2255336 (A>G), located in the RNKG2D gene, were analyzed in 146 healthy control individuals and 148 kidney transplant patients stratified into two groups: SGF and ABMR. The comparison did not reveal significant differences in the frequencies of these SNPs. (Tables 5A, B). The rs1049174 (G>C) variant was found to be in Hardy–Weinberg equilibrium (HWE) within the control group population and the Stable graft function (SGF) group [X2HWE = 0.1222; p = 0.726661 and X2HWE = 0.8765; p = 0.34916 respectively] (Tables 5A, B). Only patients with ABMR had frequencies deviating from HW expectations [X2HWE = 11.0717; p= 0.000877] (Table 5B). Consequently, the entire group of kidney transplant patients lost the HWE [X2HWE = 9.0317; p = 0.002653] (Table 5A). In contrast, rs2255336 (A>G) was in HWE within the control population group and kidney transplant patients [X2HWE = 1.828; p = 0.1763 and X2HWE = 0.0587; p = 0.8080 respectively] (Table 5A) and the due subgroup ABMR and SGF [X2HWE = 0.2369; p = 0.626 and X2HWE = 0.7405; p = 0.3895 respectively] as shown in detail in (Table 5B). Moreover, these two SNPs in the NKG2D gene, despite being a few thousand bases apart (21), do not show strong linkage disequilibrium in all examined groups (Supplementary Tables S2A–C).The only observed association pertains to the SNPs rs1049174G with rs2255336A, which exhibit weak LD in the patient group (D’= 0.70, r2 = 0.32; X2 = 14.87, P =0.0001) including the two subgroups SGF and CR (D’= 0.74, r2 = 0.41; X2 = 9.80, P =0.002 and D’= 0.66, r2 = 0.24; X2 = 4.59, P =0.032, respectively).
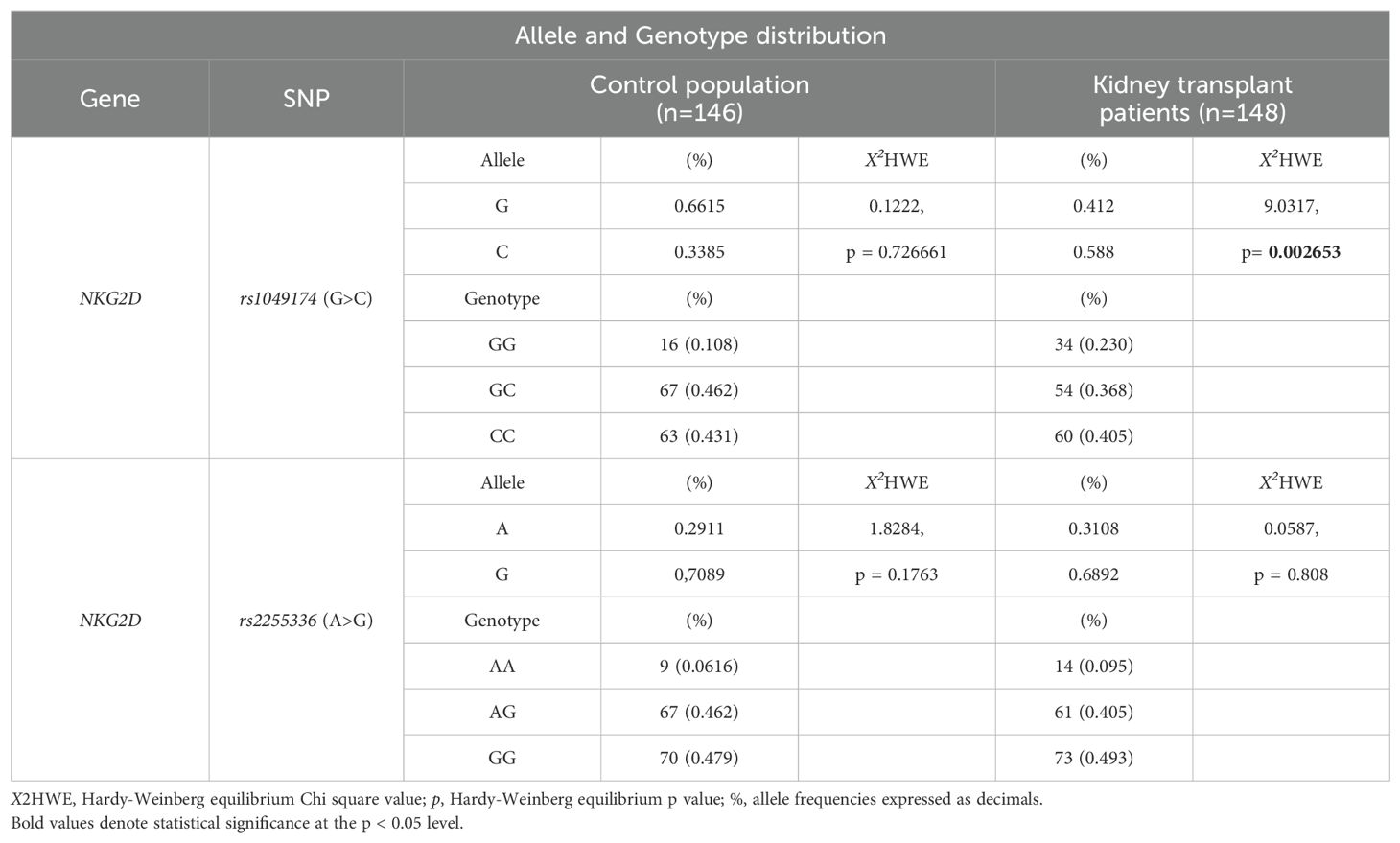
Table 5A. Allele and Genotype distribution of rs1049174 and rs2255336 in the control population and kidney-transplant patients.
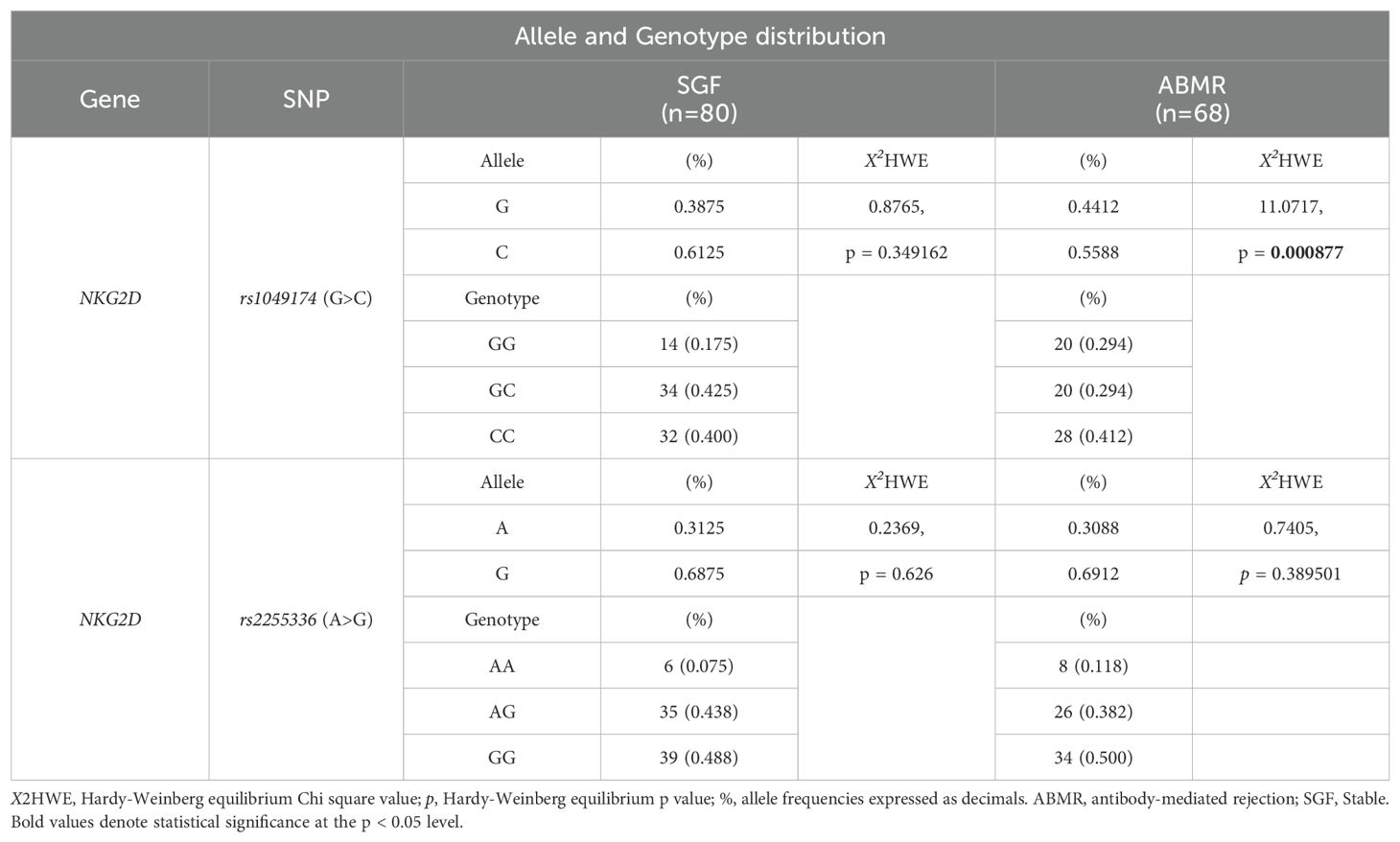
Table 5B. Allele and Genotype distribution of rs1049174 and rs2255336 in the kidney-transplant patients.
Figure 3 depicts the cumulative incidence of ABMR over 120 months in patients categorized based on the three genotypes of rs1049174 (G>C) in the NKG2D gene (GG, CG, and CC). Thirty-four patients (23%) had the rs1049174 [GG] genotype, 54 (36.5%) were heterozygous [CG], and 60 (40.5%) were homozygous [CC]. At 5 years post-transplantation, graft survival was only 38.2% (13/34) for patients with the rs1049174 [GG] genotype, compared to 70.4% (38/54) and 91.7% (49/60) for patients with the rs1049174 [CG] and rs1049174 [CC] genotypes, respectively.
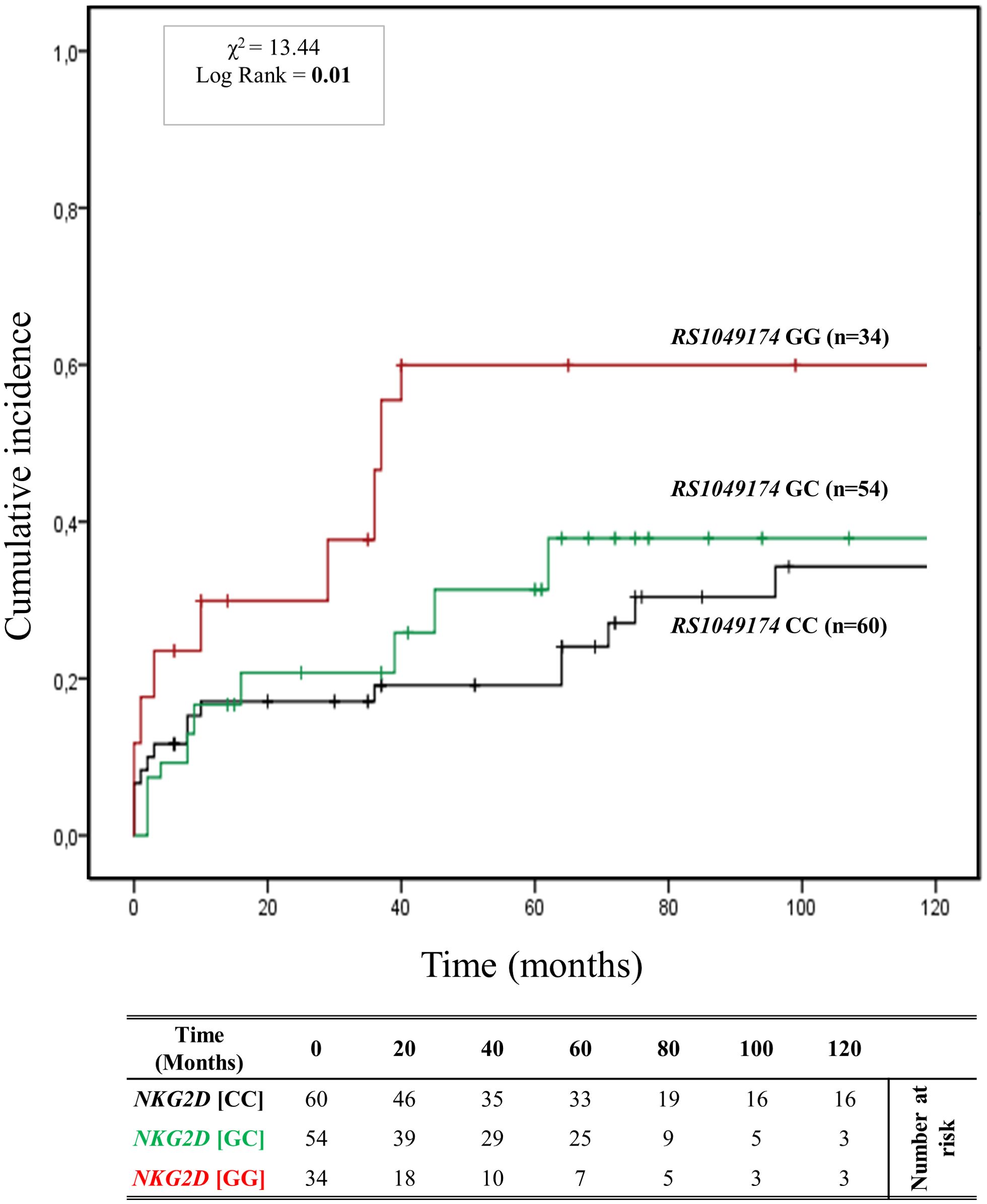
Figure 3. Cumulative incidence for antibody-mediated rejection according to NKG2D rs1049174 (G>C) genotype. The cumulative incidence of rejection events is graphically presented for a cohort of 148 patients observed over 120 months. Patients were categorized into three groups based on their NKG2D genotype for rs1049174 (G>C) [GG (red), GC (green), CC (black)]. This allele is linked to the haplotype blocks NKG2D hb-1, which produces NKG2DR with low (rs1049174 [CC]; LNK) or high (rs1049174 [GG]; HNK) natural cytotoxic activity phenotypes. P-values were calculated using the two-sided Log-rank test without correction. χ2, Chi-square.
Patients with the rs1049174 [GG] genotype exhibited a significantly increased risk of antibody-mediated rejection (X² = 13.44; Log-rank = 0.001). Indeed, at 120 months post-transplantation, the cumulative incidence of ABMR was higher (61.8% (21/34)) in the group of patients with the rs1049174 [GG] genotype compared to 42.6% (23/54) and 40.0% (24/60) in patients with the rs1049174 [CG] and rs1049174 [CC] genotypes, respectively.
This evidence was also confirmed by the trends in renal function, as assessed through serum creatinine levels (SCr) and glomerular filtration rate (eGFR). Difference in eGFR was already evident at 12 months [(57.90 ± 25.98 mL/min/1.73 m2 [GG] vs 63.34 ± 22.74 mL/min/1.73 m2 [CG] vs 71.22 ± 21.93 mL/min/1.73 m2 [CC]; P = 0.085], at 36 months [(52.42 ± 23.22 mL/min/1.73 m2 [GG] vs 58.23 ± 26.92 mL/min/1.73 m2 [CG] vs 77.60 ± 24.74 mL/min/1.73 m2 [CC]; P = 0.007], and continued to rise at 72 months [(42.60 ± 27.43 mL/min/1.73 m2 [GG] vs 63.18 ± 30.48 mL/min/1.73 m2[CG] vs 64.02 ± 31.50 mL/min/1.73 m2 [CC]; P = 0.037] after transplantation. The PAUC was also statistically significant (PAUC = 0.002), (Supplementary Figure S4).
Concurrently, in patients with the rs1049174 [GG] genotype, mean SCr levels were worse than those of patients with the other two genotypes, rs1049174 [CG] and rs1049174 [CC]. In fact, they were significantly higher at 12 months [(128.65 ± 64.52 μmol/L [GG] vs 121.20 ± 73.37 μmol/L [CG] vs 99.28 ± 28.67 μmol/L [CC]; P = 0.046], at 36 months [(140.14 ± 65.93 μmol/L [GG] vs 122.52 ± 57.24 μmol/L [CG] vs 92.55 ± 35.97 μmol/L [CC]; P = 0.009], and continued to increase at 72 months [(195.58 ± 121.70 μmol/L [GG] vs 137.80 ± 168.75 μmol/L [CC] vs 125.87 ± 89.65 μmol/L [CC]; P = 0.042]. Similarly to the eGFR curves, the PAUC for mean SCr levels was also statistically significant (PAUC = 0.023), (Supplementary Figure S5). Moreover, it is noteworthy that the influence exerted by the NKG2D rs1049174 GG polymorphism on the transplant outcome remains independent of other clinical and genetic variables, as elucidated by the multivariate logistic regression analysis (Supplementary Table S4).
The cumulative incidence over 120 months of antibody-mediated rejection (ABMR) in patients divided based on the three genotypes (AA, AG, and GG) of the other haploblock NKG2D identified by the rs2255336 (A>G) as highlighted in Figure 4. Twelve (8.2%) patients had the rs2255336 [AA] genotype, 68 (45.9%) were heterozygous [AG], and 68 (45.9%) were homozygous [GG]. At 5 years post-transplantation, graft survival was only 33.3% (4/12) for patients with the rs2255336 [AA] genotype, compared to 63.9% (43/68) and 69.1% (47/68) for patients with the rs2255336 [AG] and rs2255336 [GG] genotypes, respectively. Patients with the rs2255336 [AA] genotype exhibited a significantly increased risk of antibody-mediated rejection. At 120 months after transplantation, the incidence of antibody-mediated rejection was 66.7% (8/12) in these patients, while in patients with rs2255336 [AG] and rs2255336 [GG] genotypes, it was 41.2% (28/68) and 47.1% (32/68), respectively. However, the Mantel-Cox log-rank test did not reach statistical significance (X2 = 0.34; Log-rank = 0.84). Interestingly, this polymorphism appears to influence the eGFR of transplanted patients over time. Individuals with the rs2255336 [AA] genotype exhibited lower eGFR values compared to patients with the other two genotypes, rs2255336 [AG] and rs2255336 [GG], already at 12 months [(55.60 ± 32.13 mL/min/1.73 m2 [AA] vs 63.40 ± 23.58 mL/min/1.73 m2[AG] vs 69.41 ± 21.31 mL/min/1.73 m2 [GG]; P = 0.194]. This difference reached statistical significance at 36 months [(39.31 ± 20.38 ml/min [AA] vs 60.45 ± 28.05 mL/min/1.73 m2 (AG) vs 73.53 ± 24.29 mL/min/1.73 m2 [GG]; P = 0.047] and continued to rise at 72 months [(26.34 ± 20.32 mL/min/1.73 m2 [GG] vs 62.10 ± 31.89 mL/min/1.73 m2 [AG] vs 61.45 ± 29.28 mL/min/1.73 m2 [GG]; P = 0.015] after transplantation. In this case, the PAUC also reached statistical significance (PAUC = 0.028), (Supplementary Figure S6). SCr levels also appear to be significantly influenced by the three genotypes of RNKG2D rs2255336 (PAUC = 0.030), (Supplementary Figure S7). Higher values are observed in the presence of the rs2255336 [AA] genotype and tend to progressively increase over time: at 12 months [(154.13 ± 89.45 μmol/L [AA] vs 119.19 ± 67.38 μmol/L [AG] vs 100.31 ± 27.70 μmol/L [GG]; P = 0.073], at 36 months [(189.38 ± 68.47 μmol/L [AA] vs 148.80 ± 154.93 μmol/L [AG] vs 95.95 ± 35.32 μmol/L [GG]; P = 0.307] and continued to increase at 72 months [(286.55 ± 154.16 μmol/L [AA] vs 170.28 ± 216.80 μmol/L [AG] vs 152.21 ± 165.73 μmol/L [GG]; P = 0.160].
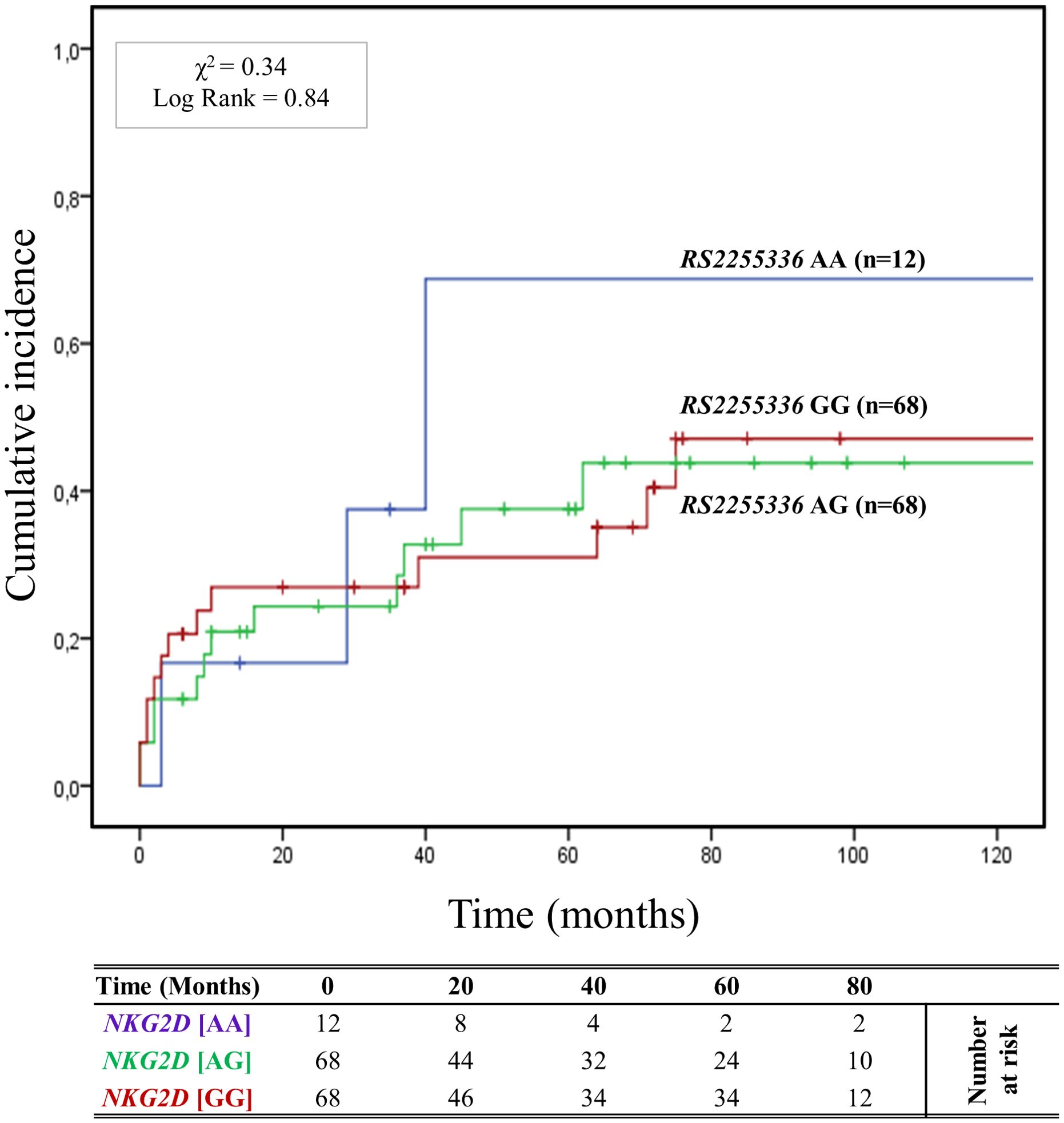
Figure 4. Cumulative incidence for antibody-mediated rejection according to NKG2D rs2255336 (A>G) genotype. The cumulative incidence of rejection events is graphically presented for a cohort of 148 patients observed over 120 months. Patients were categorized into three groups based on their NKG2D genotype for rs2255336 (A>G) [AA (light blue), AG (green), GG (red)]. This allele is linked to the haplotype blocks NKG2D hb-2, which produces RNKG2D with low (rs2255336 [GG]; LNK) or high (rs2255336 [AA]; HNK) natural cytotoxic activity phenotypes. P-values were calculated using the two-sided Log-rank test without correction. χ2, Chi-square.
The curve of cumulative incidence over 120 months of ABMR highlights the effect of different combinations of R/D MICA allele mismatch with the three genotypes of rs1049174 (G>C) in the NKG2D gene (GG, CG, and CC). The six curves (Figure 5) are well distinct and show a gradient of ABMR risk: 2MM/GG+ (91.6%, 11/12) > 1MM/GG+ (62.5%, 10/16) > lMM/GG- (55.5%, 20/36) > 2MM/GG- (38.3%, 23/60) > 0MM/GG+ (33.3%, 2/6) > 0MM/GG- (11.1%, 2/18). Therefore, the highest risk of rejection occurs in the patients with the rs1049174 GG+ genotype transplanted with a donor with complete MICA allele mismatch (2MM/GG+). Conversely, patients with rs1049174 GG- and MICA allele match with the donor (0MM/GG-) present a minimal incidence of ABMR (X2 = 23.21; Log-rank < 0.001).
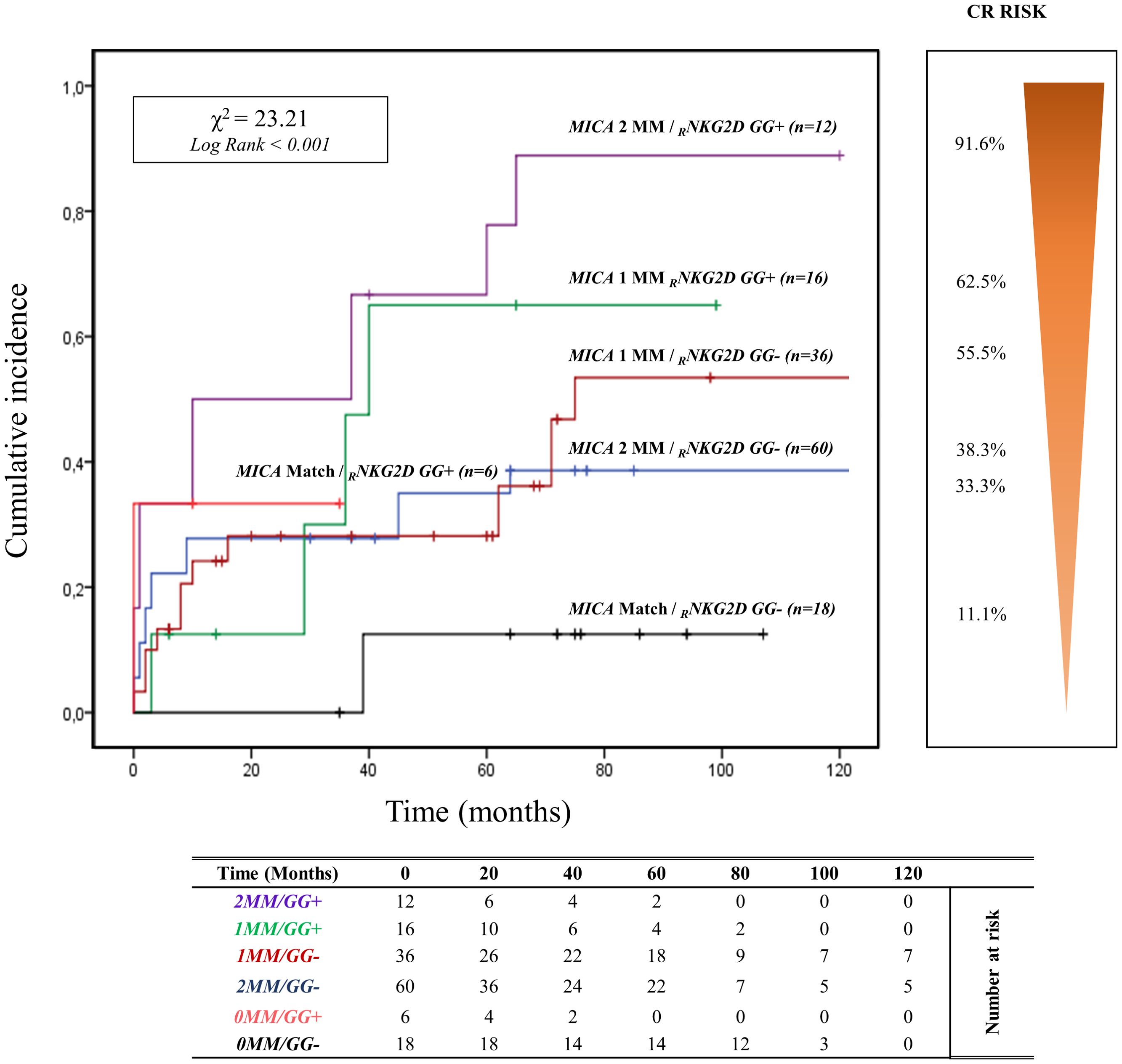
Figure 5. Cumulative incidence for antibody-mediated rejection according to NKG2D rs1049174 [GG] genotype and MICA allele mismatches. The cumulative incidence of rejection events is graphically presented for a cohort of 148 patients observed over 120 months. Patients were categorized based on their NKG2D genotype GG for and rs1049174 in combination with the donor-recipient MICA allele mismatches. The rs1049174 [GG], which produces RNKG2D with high natural cytotoxic activity phenotypes, has been correlated with donor-recipient MICA allele mismatches. Six groups were formed based on the number of MICA allele mismatches and the presence or absence of the GG (rs1049174) genotype: 1. Two MICA R/D allele mismatches and rs1049174 RNKG2D[GG] (purple) 2. One MICA R/D allele mismatches and rs1049174 RNKG2D[GG] (green) 3. One MICA R/D allele mismatches and rs1049174 RNKG2D[GG] (red) 4. Two MICA R/D allele mismatch and rs1049174 RNKG2D[CG] and [CC] (light blue) 5. MICA R/D alleles match and rs1049174 RNKG2D[GG] (orange) 6. MICA R/D alleles match and rs1049174 RNKG2D[CG] and [CC] (black). P-values were calculated using the two-sided Log-rank test without correction. χ2, Chi-square. MM, mismatches.
Renal function, monitored through SCr levels and eGFR, appears to be closely influenced by the combination of R/D MICA allele mismatch with the three genotypes of rs1049174 (G>C) in the NKG2D gene (Supplementary Figures S8, S9 respectively). Moreover, the adverse effect exerted by the 2MM/GG+ combination on the kidney transplant outcome is observed in the subgroup of patients with HLA-DRB1 and HLA-DQB1 match with the donor and those with HLA II class mismatch.
In patients with HLA-DRB1 and HLA-DQB1 full match, the cumulative incidence curve over 120 months illustrates that the highest risk of rejection occurs in individuals with the 2MM/GG+ and 1MM/GG+ combinations (83.3%, 10/12). Conversely, no patients with 0MM/GG+ and 0MM/GG- (0%, 0/8) show episodes of ABMR (X2 = 13.59; Log rank = 0.001; Figure 6). Similar results are observed when analyzing the remaining and more numerous subgroups of patients with 1 or 2 HLA-DRB1 and HLA-DQB1 allele mismatches with the donor (X2 = 14.81; Log-rank = 0.002; Supplementary Figure S10).
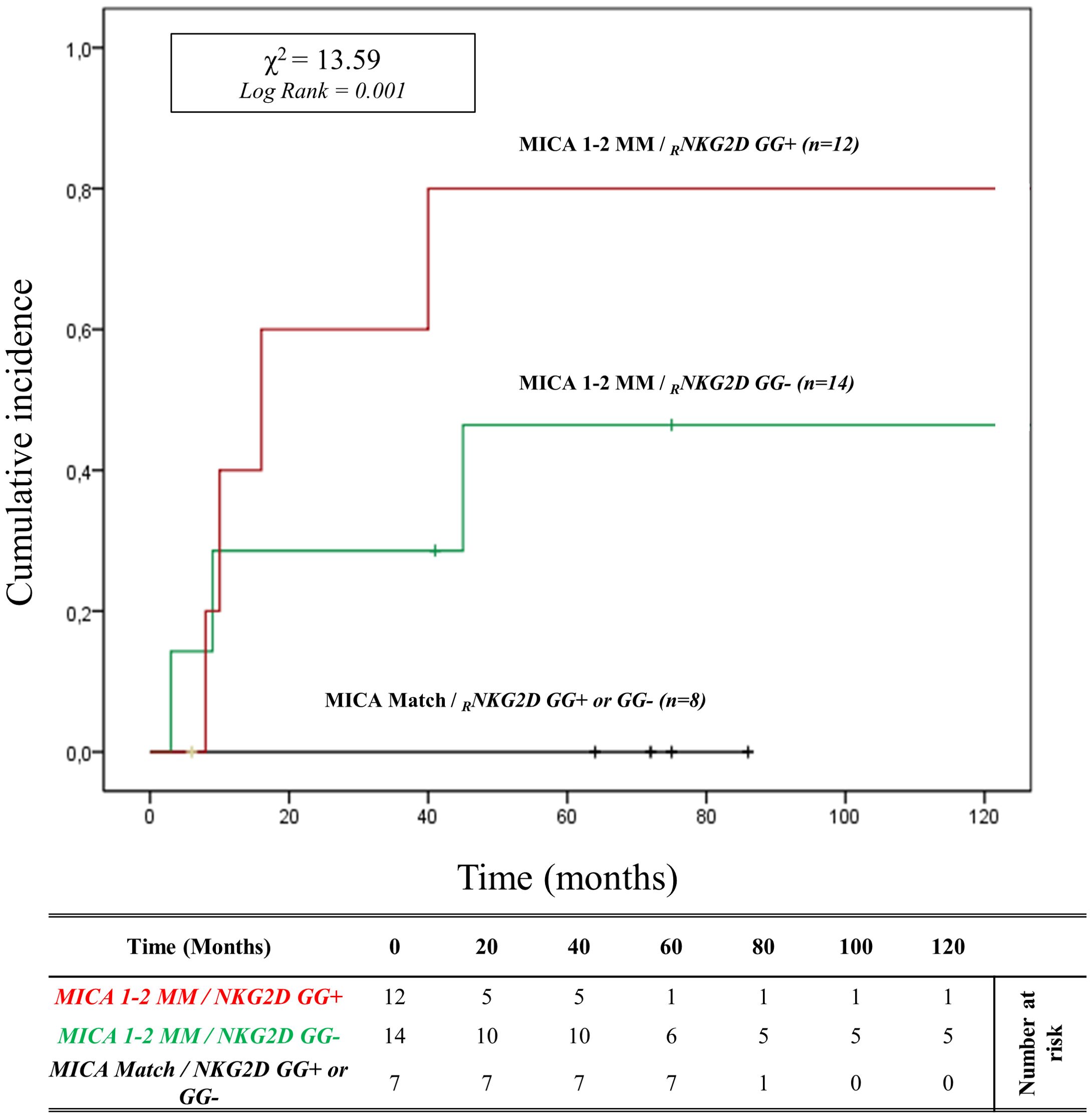
Figure 6. Cumulative incidence for antibody-mediated rejection in patient’s HLA-DRB1 and HLA-DQB1 match R/D according to NKG2D genotype and MICA allele mismatches. The cumulative incidence of rejection events is graphically presented for a cohort of 33 patients with HLA-DRB1 and HLA-DQB1 match R/D observed over 120 months. Patients were categorized into three groups based on the following criteria: 1. R/D MICA alleles match independently of the NKG2D rs1049174 genotype (black). 2. R/D MICA alleles 1-2 mismatches with NKG2D rs1049174 CG or CC genotype (marked as GG-) (green). 3. R/D MICA alleles 1-2 mismatch with NKG2D rs1049174 GG genotype (red). P-values were calculated using the two-sided Log-rank test without correction. χ2, Chi-square. MM, mismatches; R/D, recipient-donor.
It’s well known that there is linkage disequilibrium between MICA and HLA-B due to the proximity of these genes. To better determine the contribution of MICA allele mismatches in the development of ABMR, we analyzed the HLA-B full-match D/R pairs separately. In this small cohort of patients (n=12), the cumulative incidence curve over 120 months illustrates that the highest risk of rejection occurs in individuals with the 1-2MM/GG+ and 1-2MM/GG- combinations (80.0%, 4/5). Conversely, no patients with 0MM/GG+ and 0MM/GG- (0%, 0/7) show episodes of ABMR (X2 = 19.00; Log rank < 0.001; Figure 7).
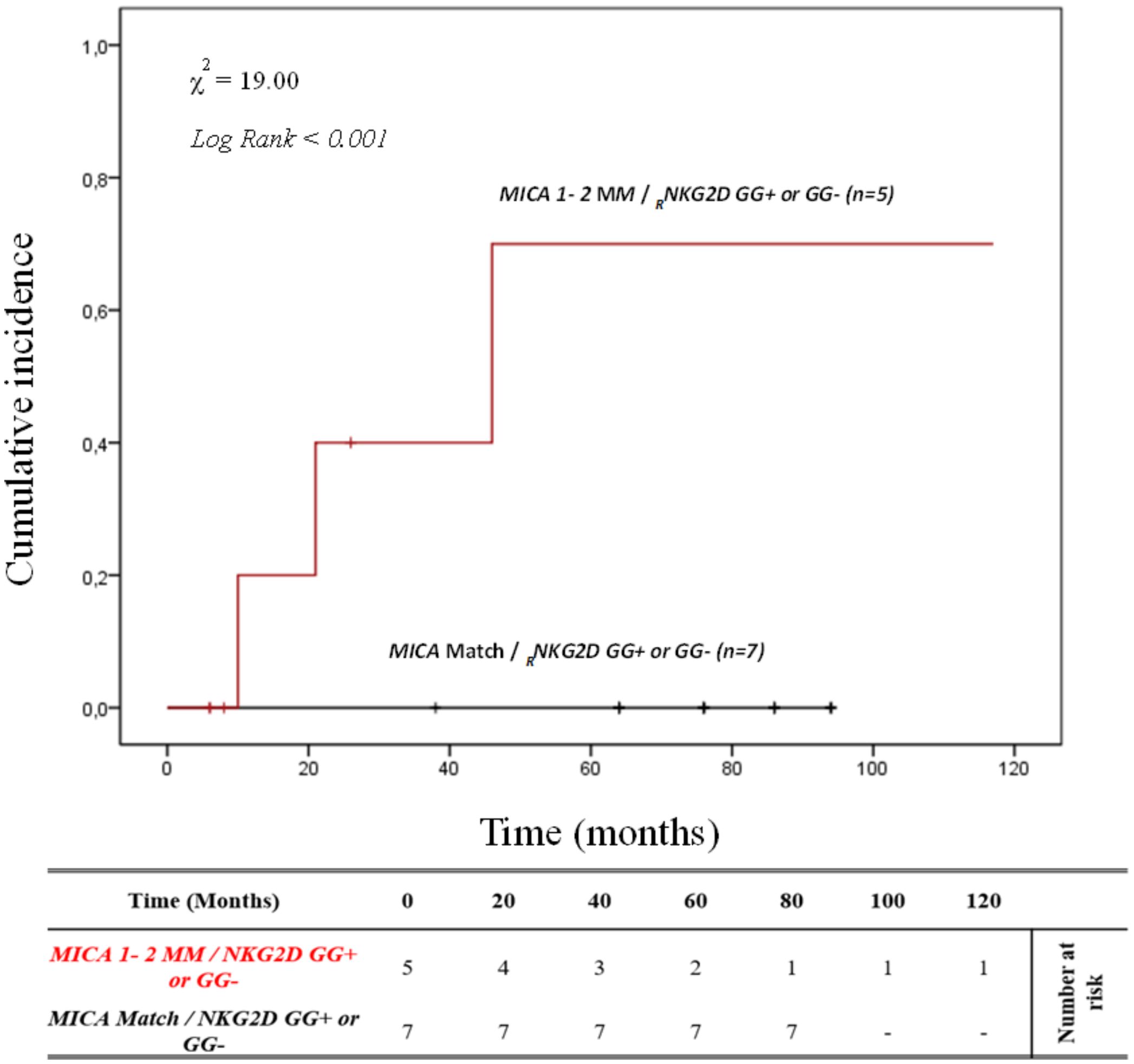
Figure 7. Cumulative incidence for antibody-mediated rejection in patient’s HLA-B match R/D according to NKG2D genotype and MICA allele mismatches. The cumulative incidence of rejection events is graphically presented for a cohort of 12 patients with HLA-B match R/D observed over 120 months. Patients were categorized into two groups based on the following criteria: 1. R/D MICA alleles match independently of the NKG2D rs1049174 genotype (black). 2. R/D MICA alleles 1-2 mismatch independently of the NKG2D rs1049174 genotype (red). P-values were calculated using the two-sided Log-rank test without correction. χ2, Chi-square. MM, mismatches; R/D, recipient-donor.
This data suggests that the effect of the different combinations of these polymorphisms is independent of HLA class I and class II matching.
Furthermore, the effect exerted by the 2MM/GG+ combination on the kidney transplant outcome is also independent of other clinical and genetic variables, as highlighted by the multivariate logistic regression analysis (Supplementary Table S3).
Finally, since the R/D MICA-129 mismatches were significant in the previous analysis, we examined it in combination with the rs1049174 [GG] polymorphism of the NKG2D receptor (Supplementary Figure S1A). The cumulative incidence curves over 120 months for ABMR based on R/D MICA-129 1 or 2 mismatches (1 MM and 2 MM) and the NKG2D rs1049174 [GG] polymorphism, were categorized as R/D: MM/MV, VV/MV, MM/VV, and VV/MM (Supplementary Figure S1A). Two MICA-129 mismatches were identified in 8 R/D pairs with the VV/MM/GG+ genotype (15.3%) and 4 R/D pairs with the MM/VV/GG+ genotype (7.7%), while 10 patients with 2 mismatches did not carry the NKG2D rs1049174 [GG] polymorphism (4 R/D pairs VV/MM/GG- (7.7%) and 6 MM/VV/GG- (11.5%). Among patients with one mismatch, there were 16 R/D pairs MM/MV/GG- (30.8%) and 14 R/D pairs VV/MV/GG- (26.9%). The median follow-up was only 15.3 months for patients with 2 MICA-129 mismatches and the presence of NKG2D rs1049174 [GG] (VV/MM/GG+ and MM/VV/GG+), while it was 35.1 months for those with 2 MICA-129 mismatches and absence of NKG2D rs1049174 [GG] (VV/MM/GG- and MM/VV/GG-). In contrast, in patients with one MICA-129 mismatch the median follow-up was 79.3 months for MM/MV/GG- and 60.6 months for VV/MV/GG-.
At 5 years post-transplantation, for patients with 1 MICA-129 mismatch and rs1049174 [GG-], graft survival was 100% (16/16) for R/D pairs MM/MV/GG- and 85.7% (12/14) for R/D pairs VV/MV/GG-. In the presence of 2 MICA-129 mismatches and rs1049174 [GG+], graft survival was 87.5% (7/8) for R/D pairs VV/MM/GG+ and 0% (0/4) for R/D pairs MM/VV/GG+. When 2 MICA-129 mismatches were associated with rs1049174 [GG-], 5-year graft survival was 50% (2/4) in R/D pairs VV/MM/GG- and 66.7% (4/6) for R/D pairs MM/VV/GG-.
Patients with 2 MICA-129 mismatches and the presence of NKG2D rs1049174 [GG] showed a significantly higher risk of antibody-mediated rejection (X² = 27.33; Log-rank < 0.001).
Notably, at 120 months post-transplantation, the cumulative incidence of ABMR was 100% in patients with 2 MICA-129 mismatches combined with the rs1049174 [GG] genotype (MM/VV/GG+ and VV/MM/GG+). In contrast, in the absence of the rs1049174 [GG] genotype (VV/MM/GG- and MM/VV/GG-), the cumulative incidence of rejection decreased to 50% (2/4) and 33.3% (2/6), respectively (Supplementary Figure S1A).
Lastly, we also analyzed cumulative incidence curves over 120 months for ABMR based on combination R/D MICA-129 1 or 2 mismatches (1 MM and 2 MM) and the other NKG2D polymorphism (rs2255336 [AA]), categorized as R/D: MM/MV, VV/MV, MM/VV, and VV/MM (Supplementary Figure S1B). Two MICA-129 mismatches were identified in 2 R/D pairs with the MM/VV/AA+ genotype (3.8%), while 20 patients with 2 mismatches did not carry the NKG2D rs2255336 [AA] polymorphism (12 R/D pairs VV/MM/AA- (23.1%) and 8 MM/VV/AA- (15.4%)). Among patients with one mismatch, there were 16 R/D pairs MM/MV/AA- (30.8%) and 14 R/D pairs VV/MV/AA- (26.9%). The median follow-up was 29.1 months for patients with 2 MICA-129 mismatches and the presence of rs2255336 [AA] (MM/VV/AA+), while it was 27.8 months for those with 2 MICA-129 mismatches and absence of rs2255336 [AA] (VV/MM/AA- and MM/VV/AA-). Instead, patients with only 1 MICA-129 mismatch and absence of rs2255336 [AA] (VV/MV/AA- and MM/MV/AA-) had a longer follow-up period (60.6 months).
At 5 years post-transplantation, graft survival was 0% (0/2) for R/D pairs with 2 MICA-129 mismatches and rs2255336 [AA+] (MM/VV/AA+), 33.3% (4/12) for R/D pairs with 2 MICA-129 mismatches and rs2255336 [AA-] (VV/MM/AA-), and 50.0% (4/8) for MM/VV/AA-. Instead, patients with just one MICA-129 mismatch the graft survival was 100% (16/16) for MM/MV/AA- and 85.7% (12/14) VV/MV/AA-.
A significantly higher risk of antibody-mediated rejection was associated with the combination of 2 MICA-129 mismatches and the presence of NKG2D rs2255336 [AA] (X² = 20.32; Log-rank < 0.001).
Table 6 highlights how the effectiveness of maintenance regimens in countering graft rejection is correlated with the two genotypes RNKG2D rs1049174 GG and rs2255336 AA. In patients exhibiting these two genotypes, immunosuppressive therapy based on mTOR inhibitors appears to be less effective in preventing ABMR compared to maintenance therapies based on CNIs [rs1049174 GG: 75.0% (15/20) mTOR vs. 35.7% (5/14) CNI; P = 0.035; OR: 0.2 (0.0 – 1.0) and rs2255336 AA: 66.6% (8/14) mTOR vs. 0% (0/4) CNI; P = 0.002; OR: 0.0 (0.0 – 0.3)]. Maintenance therapy does not appear to be influenced by the R/D 2 allele MICA mismatch (2MM), which nonetheless seems to have a synergistic effect in combination with the rs1049174 GG genotype (2MM/GG+), further reducing the efficacy of mTOR inhibitor-based therapy [2MM/GG+: 100.0% (10/10) mTOR vs. 0% (0/2) CNI; P = 0.015; OR: 0.0 (0.0 – 0.7)]. The limited number of patients did not allow for analysis regarding the combination of R/D 2 allele MICA mismatch (2MM) with the rs2255336 AA genotype (2MM/AA+).
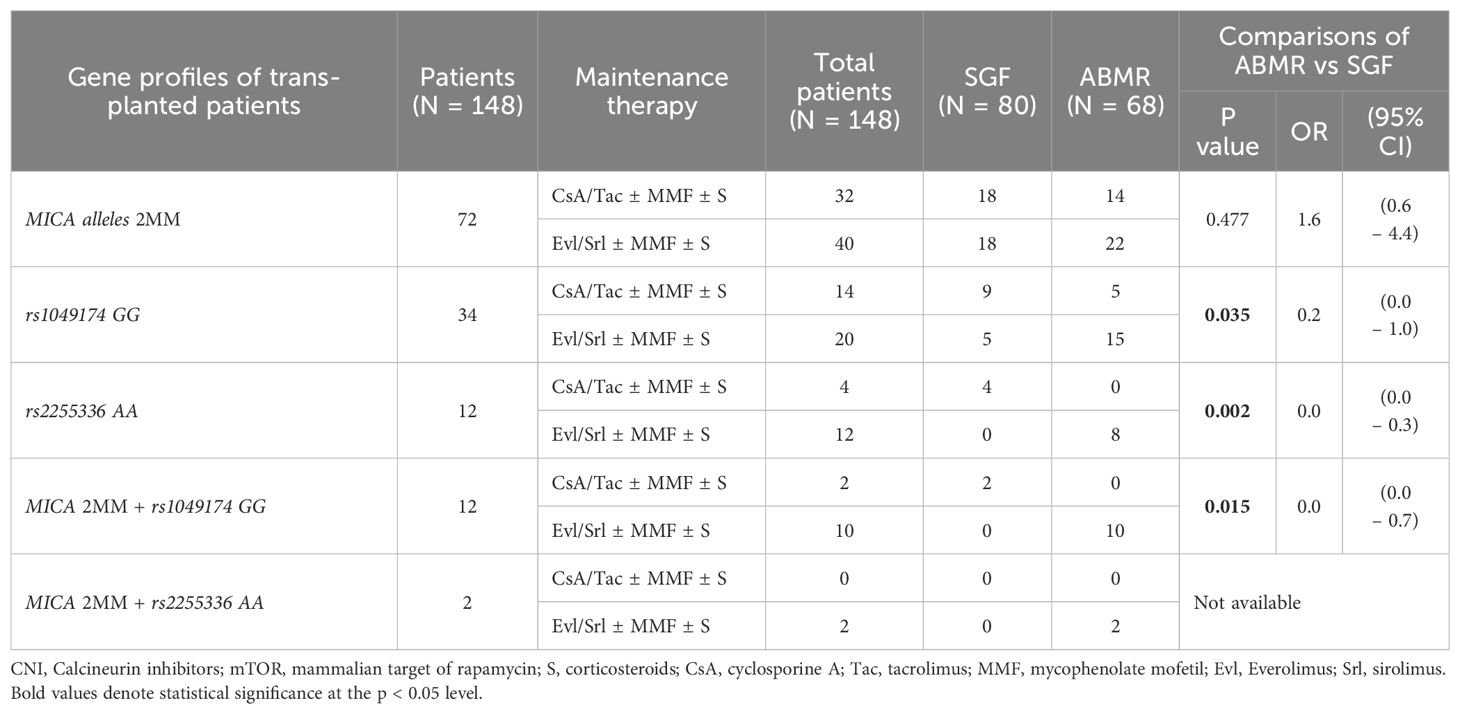
Table 6. Correlation between maintenance therapy and the onset of antibody-mediated rejection (ABMR) based on the genetic profiles of kidney-transplanted patients.
Despite the remarkable advancements in clinical and pharmacological fields in recent years, achieving complete graft tolerance remains challenging in solid organ transplantation. Antibody-mediated rejection stands as the primary immunological barrier, progressively compromising renal function, thus heightening the risk of morbidity and mortality in transplant recipients (32).
Extensive evidence supports that a higher level of compatibility between HLA class I and II in the recipient and donor correlates with improved graft function and survival (33). Moreover, MHC class I chain-related molecules A (MICA) may also play a significant role in kidney transplant outcomes, as recently highlighted by Carapito et al. (15) Our study provides a comprehensive and in-depth analysis of the genetic profile of these molecules in transplant patients, exploring their correlation with one of their main receptors: NKG2D.
An interesting finding that emerges from the study is the substantial overlap in MICA allele frequencies in transplanted patients compared to the control population, except for the MICA*002:01 allele, whose frequency is lower in patients (16% vs 24%). In the Sardinian population, this allele is in strong linkage disequilibrium (LD) (D’ = 1) with HLA-B*58:01, and both are part of the extended haplotype HLA-A*02:05, B*58:01, C*07:01, DRB1*03:01, which has a protective effect against microbiological infections, particularly SARS-CoV-2 infection (34). Furthermore, studies conducted in the Taiwanese population have shown that MICA*002 and MICA*009 play a protective role against the development of psoriasis and rheumatoid arthritis (35, 36). Therefore, the absence of this allele may have implications for the onset and progression of specific types of kidney diseases with an immunological dysregulation background.
Patients experiencing antibody-mediated rejection showed a significantly lower frequency of the MICA*010:01 allele (10 times less) compared to those with SGF. This allele is characterized by a proline-to-arginine substitution at position 6 of the alpha-1 domain, resulting in an unstable protein fold and the absence of cell surface expression (36, 37).
It has been shown that this protein remains trapped intracellularly, preventing the expression of functional MICA, whether in soluble or membrane-bound form (36). One possible explanation for this frequency imbalance between ABMR and SGF is that, due to its lack of expression, the MICA 10 peptide is unable to interact with the NKG2D receptor, thereby limiting immune response activation. This lack of activation may have a protective effect, as observed in Han Chinese populations carrying MICA*010 alleles, who exhibited a lower incidence of systemic lupus erythematosus (SLE) (36).
However, the significance of this finding is limited by the fact that Sardinia is a genetic isolate, and its population exhibits very homogeneous genetic characteristics, which are markedly different from Caucasian and American populations in the allele frequencies of both HLA class I and II, as well as MICA and MICB (38).
In line with the compelling work by Carapito et al. (15), this study underscores the significance of R/D MICA-allele matching to both the occurrence of ABMR episodes and the stable graft function. Graft survival is significantly affected by the number of MICA mismatches, with more mismatches correlating with progressively worse outcomes (0 > 1 > 2 MM). Specifically, the highest cumulative incidence of ABMR (52.7%) was observed in patients with two R/D MICA mismatches (Figure 2A, Log-rank = 0.03). Additionally, graft function deteriorated more rapidly in the presence of two MICA mismatches. This was evidenced by a more pronounced decline in eGFR, which achieved statistical significance at a later stage (P = 0.04 at 72 months post-transplantation) (Supplementary Figures S2, S3).
These data are consistent with the findings in other studies, including multicenter cohorts of kidney transplants, indicating that MICA allelic mismatches are associated with reduced graft survival and increased rejection (15). MICA antigens are not usually expressed in normal cells; however, they are overexpressed in renal, pancreatic, and heart allografts that undergo acute or antibody-mediated rejection (39, 40). The expression of MICA antigens may promote the development of “de novo” anti-MICA DSA, contributing to the onset of antibody-mediated rejection episodes and the progressive loss of graft function. Most likely these antibodies activate NK cells through the interaction with the CD16 receptor (15).
However, the implications of MICA mismatch in solid organ transplantation require further investigation. This necessity arises partly from the high polymorphism of MICA alleles, which can encode both membrane-bound forms and soluble isoforms. Consequently, different MICA alleles can exert varying biological functions by modulating the cytotoxic activity of NK cells and specific subsets of T cells in divergent ways (41). These variations can significantly impact graft rejection.
For example, the MICA*008 allele is characterized by the release of MICA molecules in exosomes, which downregulates the NKG2D receptor in NK cells. In contrast, alleles such as MICA*009 and MICA*002 encode antigens that are released via proteolytic cleavage and act as potent activators of NK cells through the NKG2D receptor (35, 36). Additionally, MICA*010 is not present on the cell membrane, suggesting its direct role in organ rejection may be limited. Indeed, in our study, this allele was found to be 10 times more frequent in patients with SGF compared to those with ABMR.
The study of MICA molecule mechanisms is further complicated by the fact that MICA alleles can encode molecules with methionine at position 129 (instead of valine) in the α2 domain. These methionine-encoded molecules exhibit a stronger binding affinity for the NKG2D receptor, leading to increased NK cell alloreactivity (42, 43).
Indeed, in our study, the highest risk of rejection was observed in R/D pairs with 2 MICA mismatches, particularly when both mismatches were represented by MICA-129 Methionine alleles (R/D: VV/MM; Log Rank < 0.001), (Figure 2B).
In these cases, donor MICA-129 methionine homozygosity appears to lead to high NK cell alloreactivity in the recipient (39, 40), negatively influencing transplant outcomes by increasing the incidence of ABMR (Supplementary Figures S1A, B).
The detrimental effect of the MICA-129 methionine polymorphism on kidney transplantation is further supported by the analysis of anti-MICA DSAs, which were found in 25% of patients who developed ABMR. In fact, the highest anti-MICA DSA titers (expressed as MFI > 1000) were observed for MICA antigens with Methionine at position 129. Specifically, in this study, the antigens that appear to be the most immunogenic are MICA 18 and MICA 01. A similar finding has been reported in previous studies, in which over 80% of patients with acute heart allograft rejection presented anti-MICA antigen-specific antibodies (measured by cytotoxicity) against MICA 01, followed by MICA 04, 11, and 18 (27.3%) (39).
A key finding of this study is the identification of a strong correlation between the high-cytotoxic RNKG2D genotypes (s1049174 [GG] and rs2255336 [AA]) and the worst renal outcomes, including the increase of cumulative incidence of ABMR and detrimental post-transplant renal function (Figures 3, 4). The effect of the rs1049174 [GG] genotype on the incidence of ABMR appears more significant than that of the rs2255336 [AA] genotype (X² = 13.44; Log-rank = 0.001 vs X² = 0.34; Log-rank = 0.84). The rs2255336 [AA] genotype has a frequency of less than 8.1% in transplanted patients, which likely leads to an underestimation of its impact due to the limited number of patients carrying this polymorphism.
Moreover, the detrimental effect on graft function from both polymorphisms (rs1049174 [GG] and rs2255336 [AA]) is markedly evident in the eGFR (PAUC = 0.002 and PAUC= 0.028, respectively) and serum creatinine (SCr) (PAUC = 0.023 and PAUC= 0.030, respectively) curves. Both of these SNPs are associated with high levels of NK cell cytotoxic activity mediated by the NKG2D receptor (43). However, their mechanisms differ: the rs1049174 polymorphism enhances NKG2D mRNA transcription, leading to increased NKG2D expression in vitro (44), while the rs2255336 substitution increases the receptor’s affinity for the DAP10 adaptor molecule when binding to NKG2D ligands (44, 45).
The heightened activation of the NKG2D receptor leads to increased cytotoxic activity by NK cells and specific T cell subsets, including NKT cells, CD8+ TCR-αβ, and CD4+ TCR-γδ T cells. Consequently, it is highly plausible that these specific genotypes play a substantial role in shaping the outcome of kidney transplantation, as indicated by the findings in this study. In hematopoietic stem cell transplantation (HSCT) also, the NKG2D receptor plays a significant role in NK cell cytotoxicity, influencing the transplantation outcome by causing complications such as graft-versus-host disease (GvHD) and post-transplant infections, as well as contributing to the beneficial graft-versus-leukemia (GvL) effect (43, 46, 47).
From a clinical perspective, the most exciting aspect of the study is evident from the analysis of the NKG2D/MICA pathway, which allowed the identification of specific patient categories at a higher risk of antibody-mediated rejection and rapid deterioration of renal function (Figure 5; Supplementary Figures S8, S9).
Indeed, patients at the highest risk of ABMR were those with homozygosity for the RNKG2D rs1049174 [GG] variant, in combination with either 2 MICA mismatches (2MM/GG+), or 1 MICA mismatch (1MM/GG+), (cumulative incidence 91.6% and 62.5% respectively). Conversely, the combination of MICA matching and the absence of the RNKG2D rs1049174 [GG] variant (0MM/GG-) characterized patients with the lowest risk of ABMR (0MM/GG-: cumulative incidence 11.1%) (Figure 5).
It is important to note that the significant cytotoxic effect generated by the 2MM/GG+ combination (two MICA R/D mismatches and homozygosity at rs1049174G variant) manifests independently of R/D HLA II class match, as evidenced by analyzing the subgroup of patients with HLA-DRB1 and HLADQB1 full match (X2 = 13.59; Log-rank = 0.001; Figure 6).
Furthermore, the impact of the 2MM/GG+ combination on the kidney transplant outcome remains independent of other clinical and genetic variables associated with antibody-mediated rejection, as highlighted by the multivariate logistic regression analysis (Supplementary Table S4). The two genetic variants, represented by the presence of two MICA R/D mismatches and homozygosity at rs1049174 [GG] allele, exert such a pronounced synergistic effect on NK cell-mediated alloreactivity that it reaches high statistical significance despite the limited number of examined patients.
In addition, it should be considered that MICA mismatches do not all have the same effect on NK cell activity. The presence of Methionine at codon position 129 (MICA-129) creates mismatches with high affinity for the NKG2D receptor, significantly influencing the alloreactivity of NK cells, including some subsets of T cells. This effect becomes evident when analyzing R/D MICA-129 mismatch combinations with the two high cytotoxic potential NKG2D receptor genotypes (rs1049174 [GG] and rs2255336 [AA]) (Supplementary Figures S1A, B). In fact, graft survival is rapidly compromised when R/D presents 2 MICA-129 mismatches (R/D VV/MM, MM/VV) and the recipient is NKG2D GG+ and/or AA+ (Log-Rank <0.001). These results are highly significant, but they are limited by the small number of patients and need to be validated in larger cohorts.
Although it has been addressed only marginally, one of the most exciting aspects of the study is the high risk of ABMR observed in patients with the genetic profiles rs1049174 [GG] and rs2255336 [AA] of the NKG2D gene (Table 6) when treated with maintenance regimens based on mTOR inhibitors (rapamycin, Everolimus) compared to the use of calcineurin inhibitors (cyclosporine and tacrolimus).
Treatment with mTOR inhibitors appears to be less effective in controlling alloreactivity induced by NKG2D receptors when they are more expressed (rs1049174 [GG] and rs2255336 [AA] genotypes) or have high affinity for their ligands (44, 45). This reduced efficacy in countering antibody-mediated rejection with maintenance therapy based on mTOR inhibitors is even more apparent in the patient subgroup 2MM/GG+ (P = 0.015, Table 6).
Rapamycin and its derivatives, such as Everolimus, are allosteric inhibitors of mTOR, representing one of the main pathways for the proliferation, differentiation, and activation of NK cells (48–50).
Therefore, in our study, we would have expected a more effective immunosuppressive effect from maintenance therapy based on mTOR inhibitors. However, the reduced efficacy of mTOR inhibitors found in the study can be explained by the fact that NKG2D binding with its MICA ligand activates NK cells “via” the DAP10 signaling molecule. This cascade involves several molecular pathways (51), most independent of the mTOR signaling pathways mediated by phosphatidylinositol 3-hydroxy kinase PI3K (51). This hypothesis is supported by in vitro studies indicating that mTOR inhibitors are less effective than CNIs, both in reducing the expression of the C-type lectin receptors (NKG2A and NKG2D) and in the production and secretion of INF-gamma and other pro-inflammatory cytokines (50).
This could result in a progressive and continuous immunological insult to the graft caused by the activation of NK cells and some subsets of T cells such as NKT cells, CD8+ TCR-αβ, and CD4+ TCR-γδ T cells (52). In conclusion, the NKG2D/MICA combination appears to influence the outcome of kidney transplantation strongly. The study of the two polymorphisms, rs1049174 and rs2255336, of the NKG2D gene and the molecular typing of MICA associated with screening for anti-MICA antibodies should be included in pre-transplant assessments. In patients with the NKG2D rs1049174 [GG] genotype at high risk of antibody-mediated rejection, special attention is necessary, and, where possible, efforts should be made to avoid transplantation with donors mismatched for both MICA alleles. Additionally, mTOR therapy seems less effective in limiting rejection onset in patients with this specific genetic profile (2MM/GG+). This observation could open the possibility of tailoring the immunosuppressive therapy scheme to prolong graft survival in the long term. The potential clinical and therapeutic implications are significant, underscoring the importance of confirming these results through multicenter studies conducted on larger and genetically diverse patient cohorts, especially considering the Sardinian population’s limited genetic polymorphism as a genetic isolate.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: PRJNA1077892(SRA).
The study was conducted following the principles outlined in the Declaration of Helsinki, following the approval granted by the relevant local Ethics Committee (Ethics Committee of the G. Brotzu Hospital in Cagliari, Italy; approval date: January 23, 2014; protocol number NP/2014/456). Written informed consent was acquired from all patients and is securely stored in the appropriate Medical Record Office. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin.
RL: Writing – original draft, Writing – review & editing. SM: Writing – original draft, Writing – review & editing. DA: Writing – review & editing. LL: Writing – review & editing. SL: Writing – review & editing. MM: Writing – review & editing. CS: Writing – review & editing. CMe: Writing – review & editing. ML: Writing – review & editing. AM: Writing – review & editing. AA: Writing – review & editing. GM: Writing – review & editing. VM: Writing – review & editing. NL: Writing – review & editing. MF: Writing – review & editing. CMa: Writing – review & editing. PB: Writing – review & editing. DO: Writing – review & editing. SR: Writing – review & editing. SD: Writing – review & editing. MC: Writing – review & editing. EG: Writing – review & editing. APe: Writing – review & editing. LC: Writing – review & editing. SG: Writing – review & editing. APa: Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The research leading to these results has received funding from the European Union - NextGenerationEU through the Italian Ministry of University and Research under PNRR - M4C2-I1.3 Project PE_00000019 "HEAL ITALIA" to Andrea Perra CUP F53C22000750006 University of Cagliari. This study was supported by “Fondazione di Sardegna”, grant #40974 - (2024.0015) to the non-profit organization “Associazione per l’Avanzamento della Ricerca per i Trapianti (AART-ODV).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1440887/full#supplementary-material
ABMR, antibody-mediated rejection; ATG, antithymocyte globulin; AUC, area under the curve; CKTR, Chronic kidney transplant rejection; CNI, Calcineurin inhibitors; CsA, cyclosporine A; DSAs, donor-specific antibodies; eGFR, Estimated glomerular filtration rate; ESKD, end-stage kidney disease; Evl, Everolimus; HWE, Hardy-Weinberg Equilibrium; HvG, host-versus-graft; IRI, ischemia-reperfusion injury; KTPs, kidney transplant patients; LD, linkage disequilibrium; MDRD, Modification of Diet in Renal Disease; MFI, mean fluorescence intensity; MM, mismatches; MMF, mycophenolate mofetil; mTOR, mammalian target of rapamycin; MVI, microvascular inflammation; PRA, Panel-reactive antibody; SCr, serum creatinine; SGF, stable graft function; S, corticosteroids; Srl, sirolimus; Tac, tacrolimus; TCMR, T-cell-mediated rejection.
1. Wolfe RA, Ashby VB, Milford EL, Ojo AO, Ettenger RE, Agodoa LY, et al. Comparison of mortality in all patients on dialysis, patients on dialysis awaiting transplantation, and recipients of a first cadaveric transplant. N Engl J Med. (1999) 341:1725–30. doi: 10.1056/NEJM199912023412303
2. Merion RM, Ashby VB, Wolfe RA, Distant DA, Hulbert-Shearon TE, Metzger RA, et al. Deceased-donor characteristics and the survival benefit of kidney transplantation. JAMA. (2005) 294:2726–33. doi: 10.1001/jama.294.21.2726
3. Kostro JZ, Hellmann A, Kobiela J, Skóra I, Lichodziejewska-Niemierko M, Dębska-Ślizień A, et al. Quality of life after kidney transplantation: A prospective study. Transplant Proc. (2016) 48:50–4. doi: 10.1016/j.transproceed.2015.10.058
4. Tucker EL, Smith AR, Daskin MS, Schapiro H, Cottrell SM, Gendron ES, et al. Life and expectations post-kidney transplant: a qualitative analysis of patient responses. BMC Nephrol. (2019) 20:175. doi: 10.1186/s12882-019-1368-0
5. McCaughan JA, Patterson CC, Maxwell AP, Courtney AE. Factors influencing survival after kidney transplant failure. Transplant Res. (2014) 3:18. doi: 10.1186/2047-1440-3-18
6. Boubaker K, Bouabid B, Bardi R, Abderrahim E, Ben Abdallah T, Ayed KH. Immunological factors and renal allograft survival for more than fifteen years: a single center study from Tunisia. Saudi J Kidney Dis Transplant. (2006) 17:70–6.
7. Poggio ED, Augustine JJ, Arrigain S, Brennan DC, Schold JD. Long-term kidney transplant graft survival-Making progress when most needed. Am J Transplant. (2021) 21:2824–32. doi: 10.1111/ajt.16463
8. Lamb KE, Lodhi S, Meier-Kriesche HU. Long-term renal allograft survival in the United States: a critical reappraisal. Am J Transplant. (2011) 11:450–62. doi: 10.1111/j.1600-6143.2010.03283.x
9. Tamargo CL, Kant S. Pathophysiology of rejection in kidney transplantation. J Clin Med. (2023) 12:4130. doi: 10.3390/jcm12124130
10. Cornell LD. Histopathologic features of antibody mediated rejection: the Banff classification and beyond. Front Immunol. (2021) 12:718122. doi: 10.3389/fimmu.2021.718122
11. Bentall A, Herrera LP, Cornell LD, Gonzales MA, Dean PG, Park WD, et al. Differences in chronic intragraft inflammation between positive crossmatch and ABO-incompatible kidney transplantation. Transplantation. (2014) 98:1089–96. doi: 10.1097/TP.0000000000000188
12. Haas M, Rahman MH, Racusen LC, Kraus ES, Bagnasco SM, Segev DL, et al. C4d and C3d staining in biopsies of ABO- and HLA-incompatible renal allografts: correlation with histologic findings. Am J Transplant. (2006) 6:1829–40. doi: 10.1111/j.1600-6143.2006.01356.x
13. Littera R, Piredda G, Argiolas D, Lai S, Congeddu E, Ragatzu P, et al. KIR and their HLA Class I ligands: Two more pieces towards completing the puzzle of antibody-mediated rejection and graft loss in kidney transplantation. PloS One. (2017) 12:e0180831. doi: 10.1371/journal.pone.0180831
14. Choy MK, Phipps ME. MICA polymorphism: biology and importance in immunity and disease. Trends Mol Med. (2010) 16:97–106. doi: 10.1016/j.molmed.2010.01.002
15. Risti M, Bicalho MD. MICA and NKG2D: is there an impact on kidney transplant outcome. Front Immunol. (2017) 8:179. doi: 10.3389/fimmu.2017.00179
16. Nowak I, Magott-Procelewska M, Kowal A, Miazga M, Wagner M, Niepiekło-Miniewska W, et al. Killer immunoglobulin-like receptor (KIR) and HLA genotypes affect the outcome of allogeneic kidney transplantation. PloS One. (2012) 7:e44718. doi: 10.1371/journal.pone.0044718
17. Carapito R, Aouadi I, Verniquet M, Untrau M, Pichot A, Beaudrey T, et al. The MHC class I MICA gene is a histocompatibility antigen in kidney transplantation. Nat Med. (2022) 28:989–98. doi: 10.1038/s41591-022-01725-2
18. Luo L, Li Z, Wu W, Luo G, Xu C, Sun Z, et al. Role of MICA antibodies in solid organ transplantation. Clin Transplant. (2014) 28:152–60. doi: 10.1111/ctr.2014.28.issue-2
19. Luo L, Li Z, Wu W, Luo G, Mei H, Sun Z, et al. The effect of MICA antigens on kidney transplantation outcomes. Immunol Lett. (2013) 156:54–8. doi: 10.1016/j.imlet.2013.08.009
20. Suárez-Alvarez B, López-Vázquez A, Baltar JM, Baltar JM, Ortega F, López-Larrea C, et al. Potential role of NKG2D and its ligands in organ transplantation: new target for immunointervention. Am J Transplant. (2009) 9:251–7. doi: 10.1111/j.1600-6143.2008.02526.x
21. Hayashi T, Imai K, Morishita Y, Hayashi I, Kusunoki Y, Nasaci K. Identification of the NKG2D haplotypes associated with natural cytotoxic activity of peripheral blood lymphocytes and cancer immunosurveillance. Cancer Res. (2006) 66:563–70. doi: 10.1158/0008-5472.CAN-05-2776
22. Loupy A, Haas M, Roufosse C, Naesens M, Adam B, Afrouzian M, et al. The Banff 2019 Kidney Meeting Report (I): Updates on and clarification of criteria for T cell- and antibody-mediated rejection. Am J Transplant. (2020) 20:2318–31. doi: 10.1111/ajt.15898
23. Chowdhry M, Makroo RN, Singh M, Kumar M, Thakur Y, Sharma V. Role of anti-MICA antibodies in graft survival of renal transplant recipients of India. J Immunol Res. (2018) 2018:3434050. doi: 10.1155/2018/3434050
24. Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, et al. Primer3–new capabilities and interfaces. Nucleic Acids Res. (2012) 40:e115. doi: 10.1093/nar/gks596
25. Furue H, Kumimoto H, Matsuo K, Suzuki T, Hasegawa Y, Shinoda M, et al. Opposite impact of NKG2D genotype by lifestyle exposure to risk of aerodigestive tract cancer among Japanese. Int J Cancer. (2008) 123:181–6. doi: 10.1002/ijc.v123:1
26. Vazquez-Gonzalez WG, Martinez-Alvarez JC, Arrazola-Garcia A, Perez-Rodriguez M. Haplotype block 1 variant (HB-1v) of the NKG2 family of receptors. Hum Immunol. (2019) 80:842–7. doi: 10.1016/j.humimm.2019.07.276
27. R core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing (2023). Available at: https://www.R-project.org/.
28. Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. (2005) 21:263–5. doi: 10.1093/bioinformatics/bth457
29. Lewontin RC. The interaction of selection and linkage. I. General considerations; Heterotic models. Genetics. (1964) 49:49–67. doi: 10.1093/genetics/49.1.49
30. Bland M ed. An Introduction to Medical Statistics. 4th ed. Oxford: Oxford University Press; Inc (2015).
31. Jun KW, Kim MH, Hwang JK, Kim SD, Park SC, Won YS, et al. Impact of pretransplant panel-reactive antibody level on renal graft survival in patients with a negative crossmatch and no donor-specific antibody. Transplant Proc. (2016) 48:770–2. doi: 10.1016/j.transproceed.2015.12.099
32. Boesmueller C, Biebl M, Scheidl S, Oellinger R, Margreiter C, Pratschke J, et al. Long-term outcome in kidney transplant recipients over 70 years in the Eurotransplant Senior Kidney Transplant Program: a single center experience. Transplantation. (2011) 92:210–6. doi: 10.1097/TP.0b013e318222ca2f
33. Mohammadhassanzadeh H, Oualkacha K, Zhang W, Klement W, Bourdiec A, Lamsatfi J, et al. On path to informing hierarchy of eplet mismatches as determinants of kidney transplant loss. Kidney Int Rep. (2021) 6:1567–79. doi: 10.1016/j.ekir.2021.03.877
34. Littera R, Campagna M, Deidda S, Angioni G, Cipri S, Melis M, et al. Human leukocyte antigen complex and other immunogenetic and clinical factors influence susceptibility or protection to SARS-CoV-2 infection and severity of the disease course. Sardinian Experience Front Immunol. (2020) 11:605688. doi: 10.3389/fimmu.2020.605688
35. Ashiru O, Boutet P, Fernández-Messina L, Agüera-González S, Skepper JN, Valés-Gómez M, et al. Natural killer cell cytotoxicity is suppressed by exposure to the human NKG2D ligand MICA*008 that is shed by tumor cells in exosomes. Cancer Res. (2010) 70:481–9. doi: 10.1158/0008-5472.CAN-09-1688
36. Wang CM, Tan KP, Wu YJ, Zheng JW, Wu J, Chen JY. Functional MICA variants are differentially associated with immune-mediated inflammatory diseases. Int J Mol Sci. (2024) 25:3036. doi: 10.3390/ijms25053036
37. Li Z, Groh V, Strong RK, Spies T. A single amino acid substitution causes loss of expression of a MICA allele. Immunogenetics. (2000) 51:246–8. doi: 10.1007/s002510050039
38. Birtsas V, Batrinou A, Dinou A, Routsias J, Gennimata V, Iniotaki A, et al. Distribution of MICA alleles and haplotypes associated with HLA-B in Greek population. Hum Immunol. (2021) 82:588–92. doi: 10.1016/j.humimm.2021.04.006
39. Suárez-Alvarez B, López-Vázquez A, Gonzalez MZ, Fdez-Morera JL, Díaz-Molina B, Blanco-Gelaz MA, et al. The relationship of anti-MICA antibodies and MICA expression with heart allograft rejection. Am J Transplant. (2007) 7:1842–8. doi: 10.1111/j.1600-6143.2007.01838.x
40. Hankey KG, Drachenberg CB, Papadimitriou JC, Klassen DK, Philosophe B, Bartlett ST, et al. MIC expression in renal and pancreatic allografts. Transplantation. (2002) 73:304–6. doi: 10.1097/00007890-200201270-00029
41. Baranwal AK, Mehra NK. Major histocompatibility complex class I chain-related A (MICA) molecules: relevance in solid organ transplantation. Front Immunol. (2017) 8:182. doi: 10.3389/fimmu.2017.00182
42. Steinle A, Li P, Morris DL, Groh V, Lanier LL, Strong RK, et al. Interactions of human NKG2D with its ligands MICA, MICB, and homologs of the mouse RAE-1 protein family. Immunogenetics. (2001) 53:279–87. doi: 10.1007/s002510100325
43. Fuerst D, Neuchel C, Niederwieser D, Bunjes D, Gramatzki M, Wagner E, et al. Matching for the MICA-129 polymorphism is beneficial in unrelated hematopoietic stem cell transplantation. Blood. (2016) 128:3169–76. doi: 10.1182/blood-2016-05-716357
44. Espinoza JL, Nguyen VH, Ichimura H, Pham TT, Nguyen CH, Pham TV, et al. A functional polymorphism in the NKG2D gene modulates NK-cell cytotoxicity and is associated with susceptibility to Human Papilloma Virus-related cancers. Sci Rep. (2016) 6:39231. doi: 10.1038/srep39231
45. Mariaselvam CM, Tamouza R, Krishnamoorthy R, Charron D, Misra DP, Jain VK, et al. Association of NKG2D gene variants with susceptibility and severity of rheumatoid arthritis. Clin Exp Immunol. (2017) 187:369–75. doi: 10.1111/cei.12891
46. Marçais A, Cherfils-Vicini J, Viant C, Degouve S, Viel S, Fenis A, et al. The metabolic checkpoint kinase mTOR is essential for IL-15 signaling during the development and activation of NK cells. Nat Immunol. (2014) 15:749–57. doi: 10.1038/ni.2936
47. Pontrelli P, Rascio F, Castellano G, Grandaliano G, Gesualdo L, Stallone G, et al. The role of natural killer cells in the immune response in kidney transplantation. Front Immunol. (2020) 11:1454. doi: 10.3389/fimmu.2020.01454
48. Liu Q, Thoreen C, Wang J, Sabatini D, Gray NS. mTOR mediated anti-cancer drug discovery. Drug Discovery Today Ther Strateg. (2009) 6:47–55. doi: 10.1016/j.ddstr.2009.12.001
49. Siemaszko J, Marzec-Przyszlak A, Bogunia-Kubik K. NKG2D natural killer cell receptor-A short description and potential clinical applications. Cells. (2021) 10:1420. doi: 10.3390/cells10061420
50. Mao B, Zhang Q, Ma L, Zhao DS, Zhao P, Yan P. Overview of Research into mTOR Inhibitors. Molecules. (2022) 27:5295. doi: 10.3390/molecules27165295
51. Pradier A, Papaserafeim M, Li N, Rietveld A, Kaestel C, Gruaz L, et al. Small-molecule immunosuppressive drugs and therapeutic immunoglobulins differentially inhibit NK cell effector functions in vitro. Front Immunol. (2019) 10:556. doi: 10.3389/fimmu.2019.00556
Keywords: kidney transplant, antibody-mediated rejection, MICA, NKG2D, DSA
Citation: Littera R, Mocci S, Argiolas D, Littarru L, Lai S, Melis M, Sanna C, Mereu C, Lorrai M, Mascia A, Angioi A, Mascia G, Matta V, Lepori N, Floris M, Manieli C, Bianco P, Onnis D, Rassu S, Deidda S, Carta MG, Giuressi E, Perra A, Chessa L, Giglio S and Pani A (2024) MICA and NKG2D gene polymorphisms influence graft survival, and response to therapy in kidney transplantation. Front. Immunol. 15:1440887. doi: 10.3389/fimmu.2024.1440887
Received: 30 May 2024; Accepted: 18 October 2024;
Published: 07 November 2024.
Edited by:
Ying Chen, University of Massachusetts Medical School, United StatesReviewed by:
Yadira Palacios, Metropolitan Autonomous University, MexicoCopyright © 2024 Littera, Mocci, Argiolas, Littarru, Lai, Melis, Sanna, Mereu, Lorrai, Mascia, Angioi, Mascia, Matta, Lepori, Floris, Manieli, Bianco, Onnis, Rassu, Deidda, Carta, Giuressi, Perra, Chessa, Giglio and Pani. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Roberto Littera, cm9ieS5saXR0ZXJAZ21haWwuY29t; Stefano Mocci, c3RlZmFuby5tb2NjaS45QGdtYWlsLmNvbQ==; Davide Argiolas, ZGF2aWRlX2FyZ2lvbGFzQHlhaG9vLml0; Antonello Pani, YW50b25lbGxvLnBhbmlAdW5pY2EuaXQ=; Sabrina Giglio, c2FicmluYXIuZ2lnbGlvQHVuaWNhLml0
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.