- 1Institute for Translational Medicine, The Affiliated Hospital of Qingdao University, College of Medicine, Qingdao University, Qingdao, China
- 2Department of Ophthalmology, The Affiliated Hospital of Qingdao University, Qingdao, Shandong, China
- 3Institute of Regenerative Medicine and Laboratory Technology Innovation, Qingdao University, Qingdao, Shandong, China
Hepatocellular carcinoma (HCC) is a highly aggressive cancer with a poor prognosis. The molecular mechanisms underlying its development remain unclear. Recent studies have highlighted the crucial role of RNA modifications in HCC progression, which indicates their potential as therapeutic targets and biomarkers for managing HCC. In this review, we discuss the functional role and molecular mechanisms of RNA modifications in HCC through a review and summary of relevant literature, to explore the potential therapeutic agents and biomarkers for diagnostic and prognostic of HCC. This review indicates that specific RNA modification pathways, such as N6-methyladenosine, 5-methylcytosine, N7-methylguanosine, and N1-methyladenosine, are erroneously regulated and are involved in the proliferation, autophagy, innate immunity, invasion, metastasis, immune cell infiltration, and drug resistance of HCC. These findings provide a new perspective for understanding the molecular mechanisms of HCC, as well as potential targets for the diagnosis and treatment of HCC by targeting specific RNA-modifying enzymes or recognition proteins. More than ten RNA-modifying regulators showed the potential for use for the diagnosis, prognosis and treatment decision utility biomarkers of HCC. Their application value for HCC biomarkers necessitates extensive multi-center sample validation in the future. A growing number of RNA modifier inhibitors are being developed, but the lack of preclinical experiments and clinical studies targeting RNA modification in HCC poses a significant obstacle, and further research is needed to evaluate their application value in HCC treatment. In conclusion, this review provides an in-depth understanding of the complex interplay between RNA modifications and HCC while emphasizing the promising potential of RNA modifications as therapeutic targets and biomarkers for managing HCC.
1 Introduction
Liver cancer as one of the most prevalent malignant tumors worldwide, ranks as the sixth most frequently diagnosed cancer and the third leading cause of cancer-related deaths globally, with 865,000 new cases and 758,000 deaths in 2022, accounting for 4.3% and 7.8% of all malignant tumor morbidities and deaths respectively (1, 2). Hepatocellular carcinoma (HCC), as the most common liver cancer (75%–85% of cases) with the higher incidence and mortality rates, is the top three causes of cancer-related death in many countries (3, 4). HCC development is linked to multiple factors such as hepatitis virus infection, alcohol consumption, non-alcoholic fatty liver disease (NAFLD), and cirrhosis (2, 5). The primary risk factors for the development of HCC are infection with hepatitis B virus (HBV) and hepatitis C virus, with non-alcoholic steatohepatitis linked to metabolic syndrome or diabetes mellitus emerging as a more common risk factor in Western countries (5). HBV is a DNA virus that undergoes a complex life cycle involving reverse transcription. Chronic infection with this virus is a leading cause of liver cancer and cirrhosis on a global scale (6). In addition to environmental factors, certain genetic factors, such as gene mutations of TERT promoter, TP53, CTNNB1, amplifications of VEGFA, etc. also play a role in the occurrence and development of HCC (7, 8). The molecular mechanisms underlying HCC differ based on the specific genotoxic factors and causes (5). Despite advancements in our comprehension of the disease’s pathophysiology and triggers, this information has not yet been sufficient implemented in clinical settings.
Surgical therapies for HCC, including surgical resection and liver transplantation represent potentially curative options for appropriate candidates with tumors detected at earlier stages (4, 9). However, only a minority of patients are eligible for this treatment because of factors such as cirrhosis (10), and local ablation is the preferred method for patients diagnosed with HCC in its early stages who are not candidates for surgery or transplantation (4). For patients with intermediate-stage HCC, chemoembolization is the main treatment strategy. Due to the suboptimal sensitivity of existing HCC surveillance tools and their underutilization in clinical practice, most patients with HCC are diagnosed at an advanced stage, which leaves minimal options for effective treatment (4, 11). Despite the advancements in immune-checkpoint inhibitor-based therapies, the objective response rate for patients with advanced-stage HCC is only around 30%, and the 3-year overall survival rate is still below 50% (4). Challenges faced in the management of HCC include difficulties in early diagnosis, high rates of recurrence, poor prognosis, limited effective treatment options and drug resistance (12).
RNA modification refers to the process of chemically modifying RNA, which can impact RNA stability, translation efficiency, and function (13). Recently, there has been increasing interest in studying RNA modifications in HCC. RNA modification can promote HCC progression by regulating other risk factors for HCC, such as fat metabolism and virus life cycle. RNA modifications have the ability to control viral replication by either modifying the viral genome or altering the expression of genes crucial for viral replication (6). NAFLD stands as a significant risk factor for the development of HCC. Researchers have revealed the impact of RNA modifications on crucial aspects including steatosis, inflammation, fibrosis, and tumorigenesis. RNA modification induces NAFLD by regulating lipid metabolism, ultimately leading to HCC transformation (14). More and more evidences suggest that aberrant modifications of specific RNAs have been correlated with the occurrence, development, metastasis, and prognosis of HCC (15). One common RNA methylation is N6-methyladenine (m6A), which has been found to be associated with tumor proliferation, metastasis, and drug resistance in HCC (16). Additionally, other RNA modifications, such as 5-methylcytosine (m5C), N1-methyladenosine (m1A), and N7-methyladenosine (M7G), have also been reported to play a role in the development and progression of HCC (17). Currently, research on the role of RNA modification in HCC mainly focuses on m6A, m1A, m5C, and m7G. Recent studies have highlighted the crucial role of these RNA modifications in HCC progression, which indicates their potential as therapeutic targets and biomarkers for managing HCC.
In this review, we comprehensively summarize the functional roles, molecular mechanisms, and potential clinical applications of various RNA modifications in HCC. Clarifying the functional mechanism of RNA modifications and identifying new therapeutic targets in HCC will provide novel strategies for treatment. Additionally, RNA modifications also exhibit potential as biomarkers for the early diagnosis and prognostic evaluation of HCC, thereby improving diagnostic accuracy and patient survival rates. Overall, this review enhances our understanding of the role of RNA modifications in HCC and offers new perspectives on its diagnosis, prognosis, and treatment.
2 RNA modifications
As early as the 1950s, scientists discovered that there were some special chemical modifications in RNA molecules (18–22). With advancements in technology and in-depth research, various RNA modification types have been discovered (23). Methylations on RNA nucleotides, such as m6A, were the earliest modification type identified. In 1955, scientists first discovered methylations in RNA molecules (18). Subsequently, there has been continuous identification of new RNA modifications, including m5C, m1A, m7G, and pseudouridine (23). Increasing evidence shows that RNA modifications play complex regulatory roles in the cell and exert important effects on gene expression and cellular functions (24, 25). Recent studies indicate that specific RNA modifications, such as m6A, m5C, m1A, and m7G, through a series of modification regulatory proteins, affect the fate of RNA molecules such as precursor RNA processing, RNA splicing, stability, transport processes and translation, thereby regulating the expression of HCC-associated genes, and are involved in the progression of HCC.
2.1 N6-methyladenosine
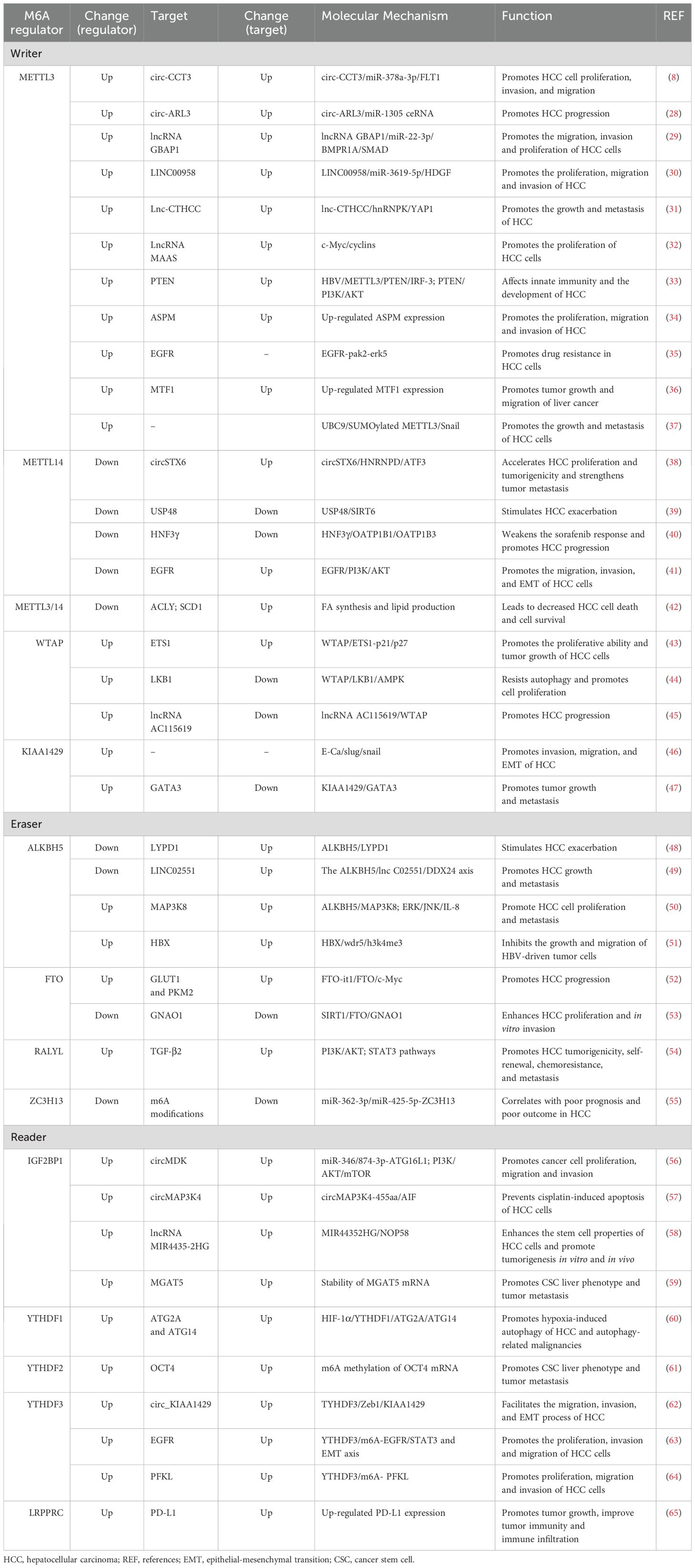
N6-methyladenosine, also referred to as m6A, is an RNA methylation that involves the transfer of a methyl group to adenosine (A) at N6 and is catalyzed by an RNA methyltransferase (Figure 1). It is the most prevalent, abundant, and evolutionarily conserved RNA methylation in eukaryotes (26). The regulation of RNA m6A methylation involves three groups of proteins: m6A methyltransferases (“writers”), m6A demethylases (“erasers”), and m6A methylation recognition proteins (“readers”) (Figure 2) (27). Accumulating evidence suggests that m6A is essential for the progression of HCC. The expression of m6A regulators, including “writer”, “eraser” and “reader” proteins, changed significantly in HCC (Table 1).
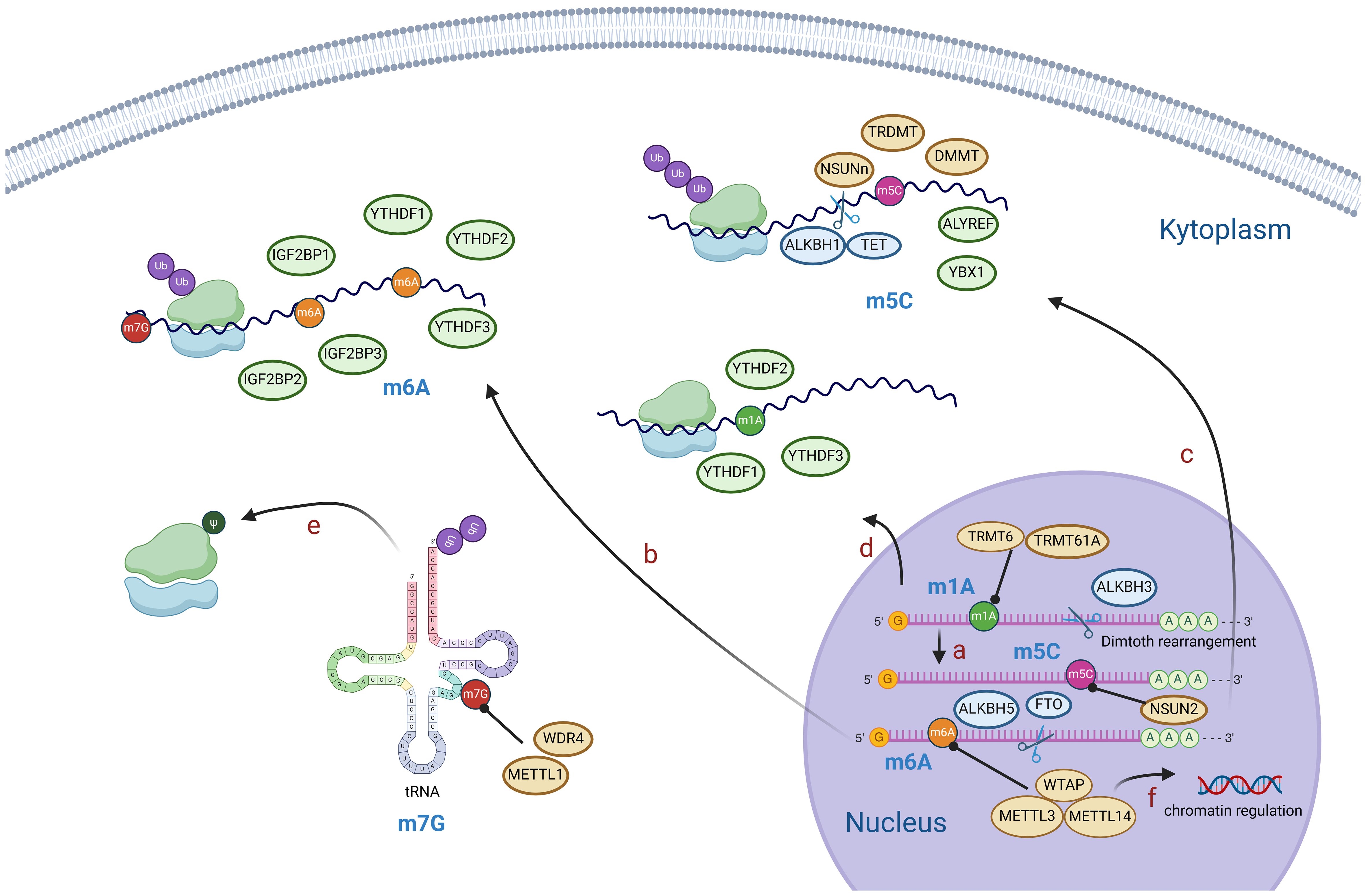
Figure 1. Diagram of RNA modification mechanisms. M1A is generally located in the 5’ end of mRNA and can be converted to m6A by Dimroth rearrangement (A). M6A, m5C, m1A modifications mediate mRNA processing, stability of RNA, post-translational protein modification, etc (B–D). M7G modification mainly affects tRNA and rRNA and mediates post-translational protein modification (E). M6A modification of mRNA reversely regulates chromatin (F). (Created with BioRender.com).
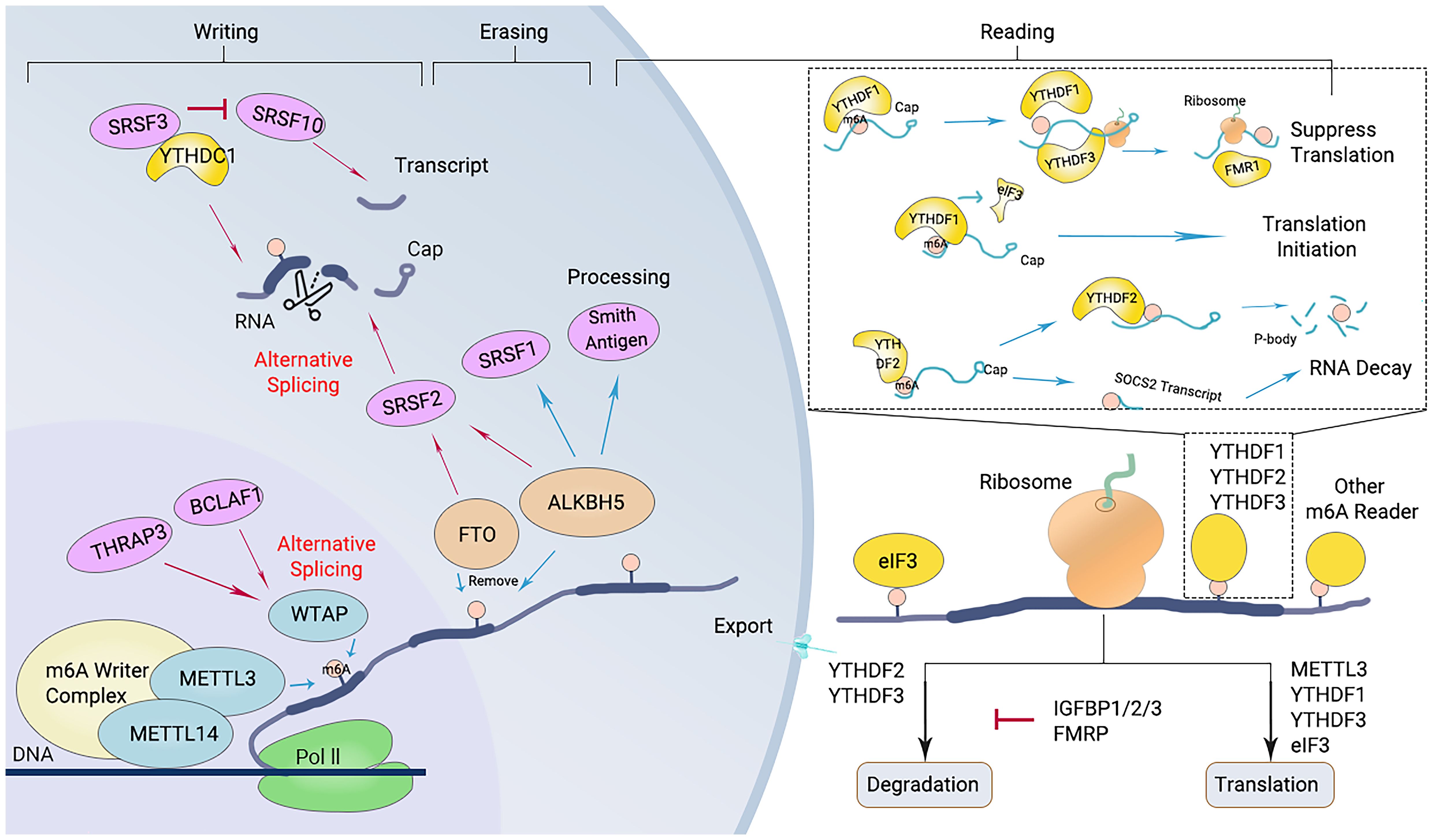
Figure 2. Diagram of the m6A modification mechanism. M6A “writers”, including METTL3/14 and WTAP, catalyze the m6A modification of adenosine on RNA. Removing the methylation of RNA needs the functions of m6A “erasers” that mainly consist of FTO and ALKBH5. M6A “readers” (such as YTHDF1/2/3 and others) recognize m6A modification sites and exert corresponding functions.
The “writers” refer to the m6A methyltransferases, which include methyltransferase-like protein (METTL) 3, METTL14, Wilms’ tumor 1-associating protein (WTAP), and vir like m6A methyltransferase associated (KIAA1429). These enzymes are responsible for catalyzing m6A modification (Figure 2) (27). The methylation of RNA to m6A is primarily carried out by the METTL3/METTL14 complex. Within this complex, METTL3 acts as the catalytic agent, while METTL14 serves as an allosteric activator that aids in binding to target RNA (46, 66).
The m6A modification of RNA is reversible, as it can be removed by “eraser” enzymes such as the m6A demethylases AlkB homolog 5 (ALKBH5) and fat mass and obesity-associated protein (FTO). These enzymes can convert m6A to A and rapidly remove m6A in a dynamic manner (Figure 2) (67). FTO belongs to the Alkb dioxygenase family and is associated with obesity (26). Knockdown of FTO significantly increases the level of RNA m6A modification (26). Another important demethylase, ALKBH5, is responsible for demethylating mRNAs within the nucleus. Knockout of ALKBH5 results in a significant increase in the level of RNA m6A modification (26, 67).
m6A “readers,” such as embryonic tumor-associated RNA-binding protein (IGF2BPs, insulin like growth factor 2 mRNA binding proteins), proteins containing YTH domains (YTH family proteins), and leucine-rich PPR-motif-containing protein (LRPPRC), have the ability to recognize and bind to m6A-modified RNA. Subsequently, they regulate the expression of related genes through various processes (Figure 2) (68). Proteins with YTH domains (YTHDF), such as YTHDF1, YTHDF2, and YTHDF3, can directly bind to m6A and then regulate translation or RNA decay. Specifically, YTHDF1 primarily enhances the translation efficiency of m6A-modified mRNAs, while YTHDF2 accelerates the degradation of m6A-modified mRNAs by recruiting several complexes to promote their degradation (61). The function of YTHDF3 is relatively complex, as it interacts with both YTHDF1 to promote protein synthesis and with YTHDF2 to facilitate mRNA degradation (69). IGF2BPs increase the stability and translational efficiency of m6A-modified mRNAs by recognizing m6A (70). Different members of the IGF2BP family may exhibit differences in regulating mRNA translation and stability; for example, IGF2BP1 and IGF2BP2 may have a wider range of substrate mRNAs in some cell types, whereas IGF2BP3 may focus more on specific mRNA molecules (71).
2.2 5-methylcytosine
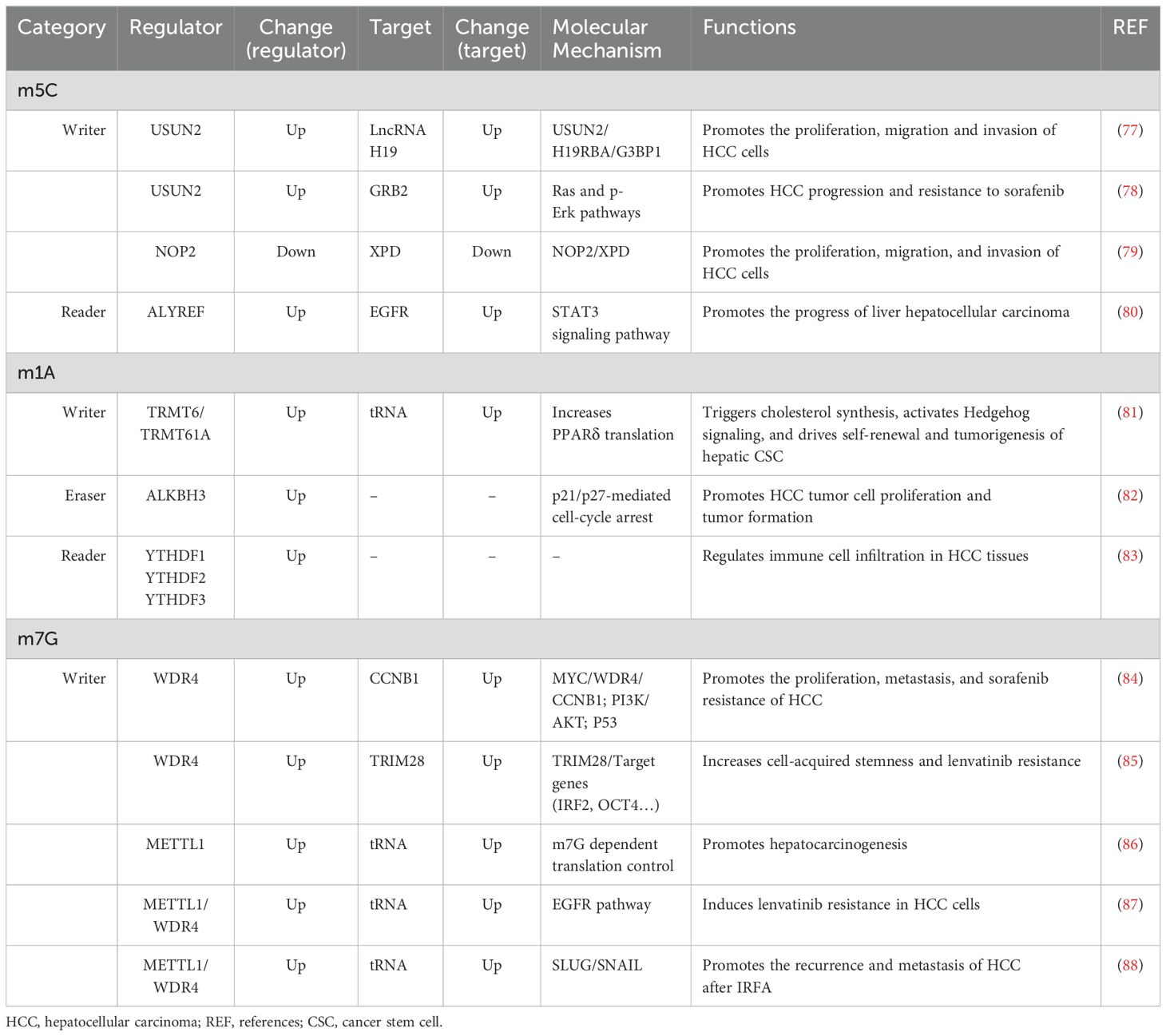
5-Methylcytosine, also known as m5C, plays a pivotal role in the modification of RNA. It is produced during RNA synthesis by converting cytosine to 5-methylcytosine through the action of cytosine deaminase (Figure 1) (72). The methylation of RNA cytosines at the C5 position is facilitated by members of the NOL1/NOP2/SUN structural domain (NSUN) protein family and DNA (cytosine-5)-methyltransferase-like protein 2 (DNMT2). Additionally, this modification can be reversed by the AlkB homolog 1 (ALKBH1) and ten-eleven translocation (TET) demethylases (73). Furthermore, proteins such as Aly/REF export factor (ALYREF) and Y-Box binding protein 1 (YBX1) can recognize and bind to RNA m5C sites, resulting in downstream biological effects (74).m5C modification has been reported to regulate a broad variety of RNA functions, including increasing the stability of RNA molecules and increasing their stability in cells; affecting the translation efficiency of RNA; regulating the expression level of proteins by affecting the translation of mRNA and affecting the splicing process of RNA; and processing and trimming the precursor RNA, thus affecting its maturation and function (72, 75, 76). Via abnormal expression of regulators, m5C modification regulates the expression of HCC-associated genes, and is involved in the progression of HCC (Table 2).
2.3 N1-methyladenosine
N1-methyladenosine is a prevalent RNA modification primarily found on the adenylate residues of mRNAs. This modification is facilitated by an enzyme known as adenylate methyltransferase, which adds a methyl group to the guanine ribose ring of adenylate, resulting in m1A modification (89). The RNA m1A methyltransferases tRNA methyltransferase 10C (TRMT10C), 61B (TRMT61B), 6 (TRMT6), and 61A (TRMT61A) can be reversed by the ALKBH1 and AlkB homolog 3 (ALKBH3) demethylases (90). Additionally, YTHDF1, YTHDF2, YTHDF3, and YTH N6-methyladenosine RNA binding protein C1 (YTHDC1) serve as binding proteins that specifically recognize the m1A site and induce downstream effects (90). m1A modification plays a critical role in enhancing RNA stability, reducing the degradation rate (91), and protecting RNA molecules from damage in the external environment. This modification also extends their lifespan within the cell (92). Furthermore, m1A is involved in the regulation of RNA translation. Research has indicated that it can impact the efficiency and accuracy of RNA translation, thereby controlling protein synthesis levels (89). This may be achieved by influencing the assembly and recognition of translation initiation complexes (93). The abnormal expression of m1A regulators, including “writer”, “eraser” and “reader” proteins, were also found in HCC indicating a pivotal role of m1A in HCC (Table 2).
2.4 N7-methylguanosine
N7-methylguanosine, also referred to as m7G, is an RNA modification that involves the addition of a methyl group (-CH3) to the nitrogen atom at position 7 of the guanine nucleotide within the RNA molecule (88). This process is carried out by methyltransferase enzymes, such as METTL1 and WDR4 (WD repeat domain 4) (71). M7G modifications are primarily found in eukaryotic mRNAs and certain noncoding RNAs, including ribosomal RNA (rRNA) and transfer RNA (tRNA) (Figure 1) (94).
Similar to other modifications, m7G modification is also crucial for regulating gene expression. m7G modifications in RNA can help to stabilize mRNA molecules and reduce their degradation rate, thus extending the lifespan of mRNAs (95). In addition, m7G modification is involved in the translational regulation of mRNAs. The formation of a cap structure and m7G modification can affect the formation and recognition of translation initiation complexes, which in turn affects the rate and precision of protein synthesis (96). Furthermore, m7G modification also affects the transcriptional regulation of mRNAs, including steps such as splicing and translocation (94, 96). The upregulated expressions of m7G “writer” proteins WDR4 and METTL1, are associated with the progression of HCC via regulating the HCC-associated gene expressions (Table 2).
3 Functional mechanism of RNA modifications in HCC
3.1 m6A in HCC
By regulating the RNA m6A modification of HCC-associated genes, m6A regulators are involved in HCC cell proliferation, invasion, migration, epithelial–mesenchymal transition (EMT), autophagy, immune evasion, and drug resistance and play important roles in the progression of HCC (Table 2, Figure 3).
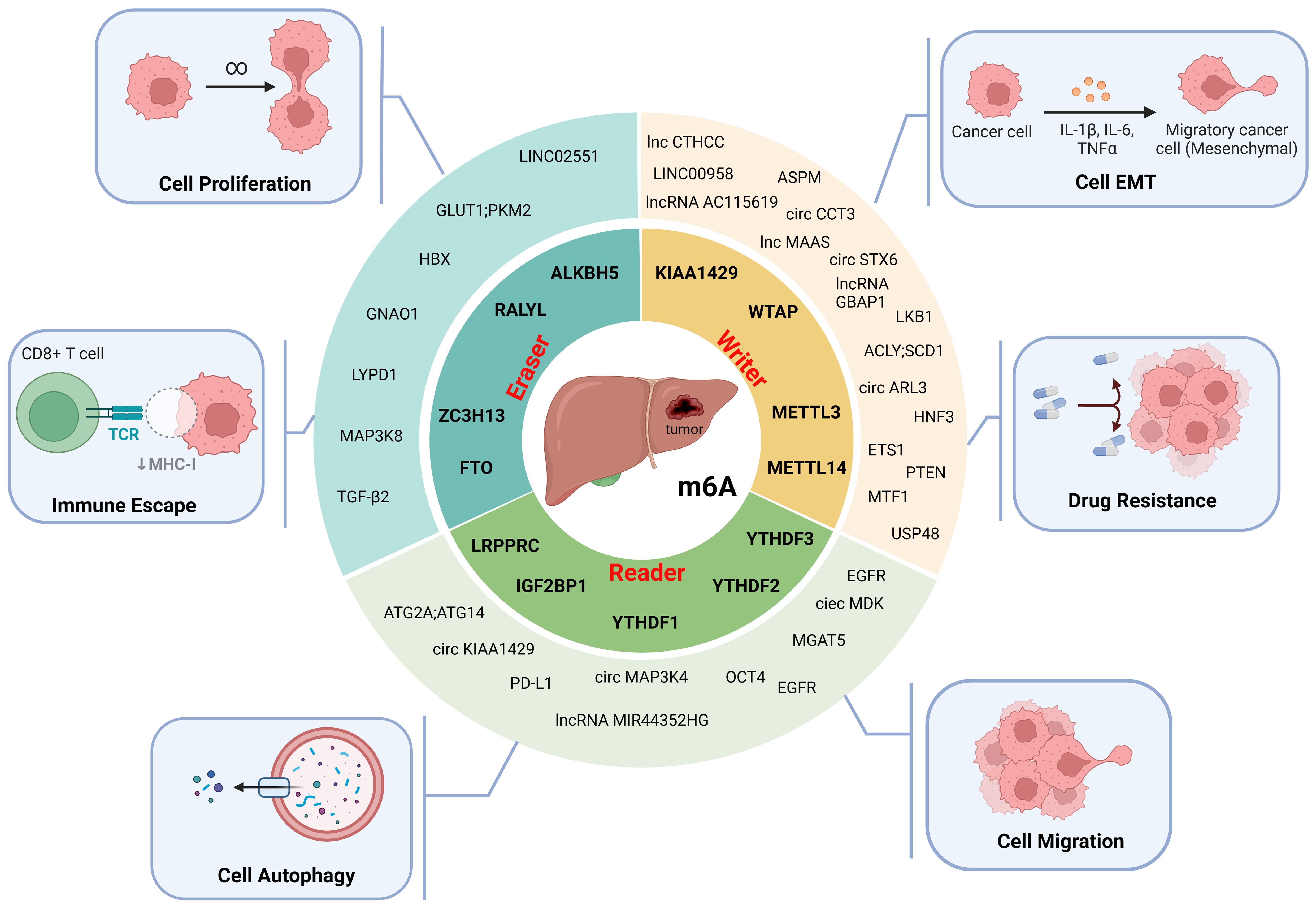
Figure 3. The molecular functions of m6A modification in HCC. M6A modifiers affect the proliferation, epithelial-mesenchymal transition (EMT), migration, autophagy, immune escape and drug resistance of HCC cells. (Created with BioRender.com).
3.1.1 The “writers” and HCC
3.1.1.1 METTL3
METTL3, the first methyltransferase found to be involved in m6A modification, is significantly upregulated in HCC (29). By modifying the methylation of circular RNA (circRNA) (28, 66), long noncoding RNA (lncRNA) (29–32), and transcripts of other cancer-associated genes (33–37), METTL3 is involved in the progression of HCC by regulating the stability of related RNAs (Figure 3, Table 1).
Circ-CCT3 and circ-ARL3 play oncogenic roles in HCC. High expression of circ-CCT3 has been reported to be associated with poor prognosis in HCC patients (66). METTL3 can increase the level of m6A modification of circ-CCT3; promote HCC cell proliferation, invasion, and migration through the circ-CCT3/miR-378a-3p/FLT1 axis; and subsequently promote HCC progression (66). The upregulated expression of METTL3 caused by HBx (an X protein encoded by hepatitis B virus) increases the m6A modification level of circ-ARL3 and then leads to increased stability and enhanced expression of circ-ARL3, causing dysregulation of the circ-ARL3/miR-1305 axis and ultimately facilitating HCC progression (28).
METTL3 regulates m6A modifications of the oncogenes lncRNA GBAP1, LINC00958, Lnc-CTHCC, and MAPKAPK5_AS1 (MAAS) and subsequently promotes the progression of HCC (29–31). The expression of the lncRNA GBAP1 is significantly increased in HCC tissues (29). METTL3 induces the expression and stability of the lncRNA GBAP1 in HCC cells and promotes the migration, invasion, and proliferation of HCC cells through the miR-22-3p/BMPR1A/SMAD pathway (29). Heparin binding growth factor (HDGF) has been identified as an HCC oncogene that affects cellular lipid metabolism (30). METTL3 enhances the stability of LINC00958 through m6A modification and promotes lipogenesis through the miR-3619-5p/HDGF axis, ultimately contributing to HCC proliferation, migration and invasion (30). METTL3-mediated m6A modification in lnc-CTHCC is recognized by (IGF2BP1)/IGF2BP3, which maintains the stability of lnc-CTHCC and promotes HCC growth and metastasis through the lnc-CTHCC/hnRNPK/YAP1 axis (31). MAAS is an oncogene whose expression is upregulated in HCC cancer tissues, and its high expression is closely associated with a low likelihood of patient survival (32). Hepatitis B e antigen secreted by HCC cells upregulates MAAS expression in M2 macrophages by promoting METTL3-mediated m6A modification. MAAS is upregulated in HCC cells via M2 macrophage-derived exosomes and targets the MYC proto-oncogene to promote HCC cell proliferation (32).
Phosphatase and tensin homolog (PTEN) is a tumor suppressor that reduces carcinogenesis by inhibiting the PI3K/AKT pathway (33). HBV increases the m6A methylation of PTEN RNA through the regulation of METTL3, which leads to a decrease in its protein level and promotes HCC (33). Abnormal spindle-like microcephaly (ASPM) has been shown to be involved in tumor progression, and its high expression in HCC predicts a poor prognosis (34). METTL3-mediated m6A modification promotes the expression of ASPM, providing a new therapeutic strategy against HCC (34). Another study suggested that METTL3 may also be associated with lenvatinib resistance in HCC cells, driving cancer cell resistance through the METTL3-M6a/EGFR-pak2-erk5 axis; thus, METTL3 may be a potential therapeutic target for drug resistance (35).
The function of METTL3 is closely associated with its acetylation. When METTL3 undergoes substantial acetylation, its binding to metal-responsive transcription factor 1 (MTF1) mRNA, METTL14, and WTAP weakens, resulting in a decrease in m6A modification induced by the METTL3-METTL14-WTAP methyltransferase complex (36). In HCC, reduced m6A modification of MTF1 mediated by METTL3 acetylation leads to enhanced MTF1 expression, thereby promoting cell proliferation and tumor progression (36). Furthermore, mitogen stimulation leads to an increase in the small ubiquitin-related modifier (SUMO) ylation of METTL3, which is correlated with the upregulation of ubiquitin-conjugating enzyme 9 (UBC9) and is positively associated with the high metastatic potential of liver cancer (37). The UBC9/SUMOylated METTL3/Snail axis represents a novel pathway for SUMO involvement in HCC progression (37).
3.1.1.2 METTL14
As a homolog of METTL3, METTL14 shares some similarities, but METTL14 is downregulated in HCC, is closely associated with tumor metastasis, and plays a regulatory role in the process of HCC tumor metastasis (Figure 3, Table 1) (39).
METTL14 can inhibit circSTX6 expression via m6A modification (38). Downregulation of METTL14 dysregulates the circSTX6/HNRNPD/ATF3 axis, accelerates HCC proliferation and tumorigenicity, and enhances tumor metastasis (38). Notably, the circSTX6-encoded protein circSTX6-144aa also independently promoted HCC progression and is expected to be a potential biomarker and therapeutic target for HCC (38).
Ubiquitin-specific peptidase 48 (USP48) is a member of the ubiquitin-specific protease family and has been identified as an inhibitor of HCC tumorigenesis by stabilizing sirtuin 6 (SIRT6) (39). The downregulation of METTL14 in HCC leads to decreased methylation levels of USP48 mRNA, resulting in reduced expression of USP48. This dysregulation consequently affects glycolysis through the USP48-SIRT6 axis and contributes to the deterioration of HCC (39). These findings indicate that specifically targeting hepatocyte USP48 or the USP48-SIRT6 axis may be a potential therapeutic strategy for future HCC treatment.
Hepatocyte nuclear factor 3γ (HNF3γ) is a hepatocyte nuclear factor that can inhibit HCC growth by transactivating organic anion transporting polypeptide 1B1 (OATP1B1) and 1B3 (OATP1B3) expression, which sensitizes HCC cells to sorafenib-induced growth inhibition and apoptosis (40). METTL14-mediated m6A modification plays an important role in maintaining high HNF3γ expression, and downregulation of METTL14 in HCC cells decreases HNF3γ expression and promotes HCC progression (40). Another study showed that high levels of METTL3/14 enhanced the expression of ATP citrate lyase (ACLY) and stearoyl-CoA desaturase 1 (SCD1) by regulating their mRNA stability, which accelerated fatty acid synthesis and lipogenesis, ultimately leading to lipid peroxidation or endoplasmic reticulum stress, resulting in HCC cell death and a decrease in HCC cell viability (42). Targeting METTL3/14 has proven to be a promising anticancer therapeutic strategy.
3.1.1.3 Other “writers” and HCC
The M6A methylases WTAP and KIAA1429, also known as tumor-associated proteins, are significantly upregulated in HCC (43). WTAP can cause posttranscriptional repression of ETS proto-oncogene 1 (ETS1) through m6A modification and promote the proliferative capacity and tumor growth of HCC cells through the WTAP/ETS1-p21/p27 axis (43). The micropeptide encoded by lncRNA AC115619 inhibited the growth of HCC tumors by binding to WTAP and hindering the assembly of the m6A methyltransferase complex, resulting in a reduction in the overall methylation level (45). In addition, when researchers knocked out the WTAP gene in HCC, the m6A level of liver kinase B1 (LKB1) mRNA decreased, and its stability increased, which in turn promoted autophagy in HCC cells via the WTAP/LKB1/AMPK axis, suggesting that it is a promising target for HCC therapy (44). KIAA1429 was found to be significantly upregulated in HCC tissues. It has been demonstrated to promote the invasion, migration, and EMT of sorafenib-resistant HCC cells (46). Additionally, through its mediation of m6A methylation, KIAA1429 was observed to decrease the expression of the E-Ca protein while increasing the expression of the slug and snail proteins (46).
3.1.2 The “erasers” and HCC
3.1.2.1 ALKBH5
ALKBH5 is a demethylating enzyme with extremely complex regulatory mechanisms in vivo (67). The expression of ALKBH5 was reported to be downregulated in HCC, causing elevated m6A levels and increased stability of Ly6/Plaur domain-containing 1 (LYPD1), leading to dysregulation of the ALKBH5/LYPD1 axis and promoting the progression of HCC (48). DEAD-box helicase 24 (DDX24) is a tumor-associated gene that is often used as a marker to assess tumor risk and treatment efficacy (49). Downregulation of ALKBH5 promotes the expression of LINC02551, which acts as a molecular junction to block the binding of DDX24 to the E3 ligase tripartite motif 27, thereby reducing the ubiquitination and subsequent degradation of DDX24 (49). Moreover, stabilized DDX24 promotes EMT in HCC (49). However, some researchers have reported conflicting results. In contrast to the aforementioned study, they proposed that ALKBH5 is actually upregulated in HCC. They suggested that this upregulation occurs through the activation of the ERK/JNK pathway and the regulation of interleukin-8 (IL-8) expression via the ALKBH5/MAP3K8 axis. Ultimately, this promotes HCC development, metastasis, and macrophage recruitment (50). According to reports, there is a strong positive correlation between HBx and ALKBH5 in HBV-HCC tissues (51). It has been discovered that HBV increases the expression of ALKBH5 through the HBx/wdr5/h3k4me3 pathway, while ALKBH5, in turn, enhances the stability of HBx mRNA by reducing m6A modification (51). Additionally, studies have indicated that the depletion of ALKBH5 significantly suppresses the proliferation and migration of HBV-induced tumor cells both in vitro and in vivo (51).
3.1.2.2 Other “erasers” and HCC
FTO plays key roles in the regulation of adiposity, lipogenesis, and body weight (97). The expression level of FTO is elevated in HCC, which is promoted by the lncRNA FTO-IT1 recruiting interleukin enhancer binding factor 2 (ILF2) and 3 (ILF3) (52). FTO overexpression increases the mRNA expression of the glycolysis-associated genes glucose transporter type 1 (GLUT1), pyruvate kinase M2 (PKM2), and c-Myc by decreasing the expression of m6A modifications, which subsequently promotes HCC progression in a glycolysis-dependent manner (52). Sirtuin 1 (SIRT1) is a known deacetylase silencing information regulator that destabilizes FTO, and its presence is positively correlated with malignancy and metastasis (53). A reduction in FTO by SIRT1 can increase the m6A modification level of guanine nucleotide-binding protein G (o) subunit α (GNAO1) and cause downregulation of its mRNA expression (53). Deletion of GNAO1 significantly enhanced HCC proliferation and invasion in vitro (53).
RALY RNA Binding Protein Like (RALYL) is a recently discovered demethylase that can increase the stability of TGF-β2 mRNA by reducing its m6A modification (54). RALYL promotes the tumorigenicity, self-renewal, chemoresistance, and metastasis of HCC through the TGF-β2/PI3K/AKT and STAT3 pathways (54). Zinc finger CCCH-type containing 13 (ZC3H13) is a methyltransferase whose expression is downregulated in HCC (55). The miR-362-3p/miR-425-5p-ZC3H13 axis leads to downregulation of ZC3H13 and thus a reduction in m6A modifications, which correlates with poor prognosis and poor outcome in HCC patients (55).
3.1.3 The “readers” and HCC
3.1.3.1 IGF2BP1
IGF2BP1, a common m6A methylation recognition protein that recognizes a wide range of m6A-modified RNAs, is highly expressed in HCC (59). The expression of autophagy related 16 like 1 (ATG16L1), a member of the autophagy family, is increased in both HCC cells and tissues. This finding suggested that autophagy may play a role in the progression of HCC (56). IGF2BP1 can recognize circMDK via m6A sites, activate the PI3K/AKT/mTOR signaling pathway through the miR-346/874-3p-ATG16L1 axis, and ultimately promote the proliferation, migration and invasion of hepatoma cells (56). IGF2BP1 recognizes the m6A modification of circMAP3K4 and promotes its translation, thereby preventing cisplatin-induced apoptosis in HCC cells by increasing the interaction of circMAP3K4-455 aa with apoptosis inducing factor (AIF) (57). High levels of circMAP3K4 serve as an independent prognostic factor for poor overall survival and disease-free survival in HCC patients (57). Furthermore, IGF2BP1 is capable of recognizing the m6A-modified lncRNA MIR4435-2HG and enhancing its expression in HCC (58). Moreover, overexpression of MIR4435-2HG significantly reduces cell sensitivity to lenvatinib, enhances the stem cell properties of HCC cells, and promotes tumorigenesis in vitro and in vivo (58). Additionally, IGF2BP1 enhances alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase V (MGAT5) mRNA stability by increasing m6A modification, which contributes to the promotion of the HCC stem cell phenotype, self-renewal, chemotherapy resistance, and tumorigenesis in mice (59). In conclusion, IGF2BP1 is highly expressed in HCC and enhances the translational stability of corresponding HCC-associated RNAs by recognizing the m6A site. This finding suggests that it may be an important target for anticancer therapy.
3.1.3.2 YTHDF family
The YTHDF family is classified as m6A recognition proteins and consists of YTHDF1, YTHDF2, and YTHDF3. These proteins have been linked to the progression of HCC (60–63). Under hypoxic conditions, hypoxia-inducible factor-1α (HIF-1α) promotes the expression of YTHDF1 by directly binding to its promoter region. This mechanism facilitates hypoxia-induced autophagy in HCC and autophagy-related malignancies through the HIF-1α/YTHDF1/ATG2A/ATG14 axis (60). Additionally, YTHDF2 regulates the m6A methylation of octamer-binding transcription factor 4 (OCT4) mRNA, thereby promoting a cancer stem cell (CSC) liver phenotype and tumor metastasis (61). Targeting YTHDF2 to eliminate CSCs is a key focus in the development of novel anticancer therapies (61). The m6A modification mediated by YTHDF3 has been shown to promote HCC migration, invasion, and EMT processes, underscoring its potential as a promising therapeutic target for HCC (62, 63). Additionally, Circ_KIAA1429 has been identified as an oncogene that targets zinc finger E-box binding homeobox 1 (Zeb1) downstream of HCC. Its high expression level has been associated with a lower overall survival rate in HCC patients (62). Through M6A-dependent mechanisms, YTHDF3 enhances Zeb1 mRNA stability and promotes HCC progression through the TYHDF3/Zeb1/KIAA1429 axis (62). Furthermore, YTHDF3 plays a role in enhancing the translation and stability of m6A-modified epidermal growth factor receptor (EGFR) mRNA. This stimulation further drives HCC progression through the YTHDF3/m6A EGFR/STAT 3 and EMT pathways (63).
3.1.3.3 LRPPRC
Programmed cell death ligand 1 (PD-L1) is a transmembrane protein expressed on the surface of activated T cells, B cells, and natural killer cells, and acts as a ligand for programmed cell death protein 1 (PD-1) and is commonly found on the surface of tumor cells (98). The interaction between these two proteins can effectively suppress the activation and proliferation of T cells, diminish the cytotoxic capabilities of T-cells towards tumor cells, and promote immune evasion and tumor cell growth (65). PD-L1 plays a crucial role as a coinhibitory immune checkpoint, and the PD1/PD-L1 signaling pathway functions to dampen the cytotoxic T-cell-mediated killing effect within the tumor microenvironment, thereby aiding in immune evasion by tumors (65). Research has indicated that LRPPRC is upregulated in HCC and enhances PD-L1 expression through a m6A-mediated mechanism. This process leads to increased stability of PD-L1 mRNA in cancer cells, ultimately promoting tumor growth and facilitating immune evasion and invasion by tumor cells (65).
3.2 m5C in HCC
NSUNs are a class of m5C methyltransferases in which NSUN2 and NSUN4 are upregulated and NOP2 is downregulated in HCC tissues and cells (77, 78, 99). The expression of NSUN4 varies across different survival rates and grade distributions, indicating its potential as an independent prognostic factor for HCC (99).
NSUN2-mediated gene mRNA and lncRNA H19 m5C modifications regulate the progression of HCC (77, 78). In HCC tissues, the m5C modification level of mRNA is significantly greater than that in normal tissue, and growth factor receptor-bound protein 2 (GRB2), ring finger protein 115 (RNF115) and apoptosis antagonizing transcription factor (AATF) are the top NSUN2-related m5C hypermethylated genes (78). GRB2 is a growth factor receptor-binding protein that is expressed at significantly higher levels in HCC tissues than in normal tissues and is considered a potential therapeutic target for HCC (78). NSUN2 interacts with lin-28B, a protein capable of recognizing m5c, to facilitate the m5c modification of GRB2 and enhance the stability of GRB2 mRNA. This process ultimately contributes to promoting resistance in HCC cells to sorafenib by activating the Ras and p-Erk pathways (78). H19, an important tumor-related lncRNA, is targeted by NSUN2 in HCC (77). The NSUN2-mediated lncRNA H19 m5C modification enhances its stability and increases its ability to bind specifically to Ras-GTPase activating SH3 domain-binding protein 1 (G3BP1) to promote the proliferation, migration and invasion of HCC cells (77).
NOP2, also known as NSUN1, is a member of the NSUN family whose expression is downregulated in HCC (79). NOP2 overexpression enhances the expression of the antioncogene xeroderma pigmentosum D (XPD) via m5C methylation of XPD, thereby inhibiting the proliferation, migration and invasion of HCC cells (79). These findings suggest that XPD may be a potential target for HCC treatment.
ALYREF is a protein that recognizes m5C and is upregulated in HCC (100). The EGFR signaling pathway plays a crucial role in various cellular processes, and abnormal activation of EGFR is observed in a wide range of tumors (100). ALYREF stabilizes EGFR by binding to the m5C site on EGFR mRNA, thereby activating STAT3 signaling and promoting HCC progression (80).
3.3 m1A in HCC
m1A modification is involved in the progression and treatment of HCC (Table 2). In HCC tissues, the m1A modification levels of RNA are aberrantly elevated (81). The expression levels of m1A methyltransferases, such as TRMT6 and TRMT61A, are significantly elevated in HCC tissues and are negatively correlated with patient prognosis (81). The PPARδ protein, also known as peroxisome proliferator-activated receptor, is associated with cholesterol synthesis (81). TRMT6/TRMT61A enhances m1A methylation in tRNA, leading to an increase in PPARδ translation. This subsequently promotes cholesterol synthesis and activates Hedgehog signaling, ultimately driving self-renewal and tumorigenesis of liver CSCs (81).
In addition, the m1A demethylase ALKBH3 is significantly upregulated in HCC tissues compared to nontumor tissues. Furthermore, high expression of ALKBH3 has been associated with poor prognosis (43). The overexpression of ALKBH3 has been found to stimulate the proliferation and tumorigenesis of HCC tumor cells, indicating a functional role for m1A modification in promoting the cell cycle (82).
For binding proteins that specifically recognize the m1A site, members of the YTH domain family, such as YTHDF1, YTHDF2, and YTHDF3, have been reported to be upregulated in HCC tissues compared to normal tissues (83). Additionally, the expression levels of YTHDF1, YTHDF2, and YTHDF3 are positively correlated with immune cell infiltration in HCC tissues. This finding indicates a functional role for m1A regulators in regulating immune cell infiltration (83).
In addition, there have been reports suggesting that patients with smaller tumors and good liver function may benefit from a combined regimen of mitoxantrone, 5-fluorouracil, and cisplatin (90). The clinical use of doxorubicin in patients with elevated levels of m1A modification is more scientifically supported. These findings contribute to mitigating the risk of doxorubicin-induced cardiotoxicity in patients and reducing unnecessary overtreatment (90).
3.4 m7G in HCC
Methylation of m7G has been found to be associated with the progression of HCC and resistance to drugs (Table 2). Both METTL1 and WDR4, which are essential components of the m7G methyltransferase complex, have been reported to be upregulated in HCC tissues and cells (84, 86). In HCC cells, MYC activates the transcription of WDR4, which subsequently enhances the stability and translation of cyclin B1 (CCNB1) mRNA by facilitating binding of eukaryotic initiation factor 2A (EIF2A) to CCNB1 mRNA. This process ultimately promotes proliferation, metastasis, and resistance to sorafenib (84, 86). Furthermore, METTL1-mediated modification of tRNA m7G contributes to hepatocarcinogenesis through translational control of target mRNAs (86).
Long-term drug resistance is a significant challenge in the treatment of HCC. Studies have indicated that METTL1 and WDR4 are upregulated in lenvatinib-resistant cells (87). METTL1/WDR4-mediated tRNA m7G modification enhances the translation of genes within the EGFR pathway, ultimately leading to lenvatinib resistance in HCC cells. This suggests a potential strategy for overcoming drug resistance (87). In glioblastoma, tripartite motif containing 28 (TRIM28) has been identified as a specific marker of stem-like cells, contributing to their invasion (85). Additionally, WDR4 amplifies TRIM28 expression, subsequently affecting the expression of target genes and promoting cell-acquired stemness as well as lenvatinib resistance (85).
Insufficient radiofrequency ablation (IRFA) is a major contributing factor to the high recurrence rate of HCC treatment (88). Studies have shown an association between m7G-tRNA modification and HCC recurrence following IRFA treatment (88). Specifically, after IRFA therapy, there was a significant increase in the level of m7G tRNA modification and its associated methyltransferase complex component METTL1/WDR4. This increase facilitated the translation of SLUG/SNAIL during sublethal heat stress in a manner dependent on codon frequency, ultimately leading to elevated HCC recurrence and metastasis (88).
In summary, RNA modifications, such as m6A, m5C, m1A, and m7G, through a series of modification regulatory proteins regulate the expression of HCC-associated genes and are involved in the progression of HCC via different signal axes. It is important to emphasize that the alterations in the expression of RNA modification regulatory proteins with similar functions in HCC are somewhat inconsistent. Methyltransferases, such as METTL3 and METTL14, as well as USUN2 and NOP2, exhibit opposite expression changes in HCC. Their opposite expression changes play a similar role in the progression of HCC by regulating the expression of different target genes and subsequent signaling pathways through RNA modification. There is no consensus on the expression changes of some RNA modification regulatory proteins, such as ALKBH5 and FTO, in HCC. This suggests that the expression of RNA modification regulatory proteins in HCC may be dynamic and have different mechanisms of action in various stages and types of the HCC. Therefore, more in-depth and detailed classification research is needed to clarify this.
At present, research on the role of RNA modification in HCC mainly focuses on m6A, m1A, m5C, m7G, etc. Their combined interactions form an intricate RNA modification network that significantly influences the physiological and pathological processes of HCC cells. However, whether these RNA modifications interact with each other in HCC has not been studied yet. The interactions between different types of RNA modifications are complex and diverse, and they may affect each other through synergistic effects, competitive relationships, cascade effects (101). Both m6A and inosine occur at the N6 position of the adenosine ring, but they do not compete for the same adenosine (102). m5C and m6A work together to regulate the export of mRNA to the cytoplasm through interactions with ALYREF and ALKBH5 (101). The m6A and m1A modifications on mRNA are recognized by the same reader proteins YTHDF1-3. In addition, m7G plays a role in facilitating the N6, 2-O-dimethyladenosine modification within the cap structure of the extended RNA polymerase II transcript (103). Despite the specific interplay between RNA modifications not being fully elucidated, further research is anticipated to unveil more about the mechanism and function of these modifications, offering new insights and approaches for the treatment and diagnosis of diseases.
4 Potential applications of RNA modifications in HCC
4.1 Potential for use as diagnosis and prognosis biomarkers
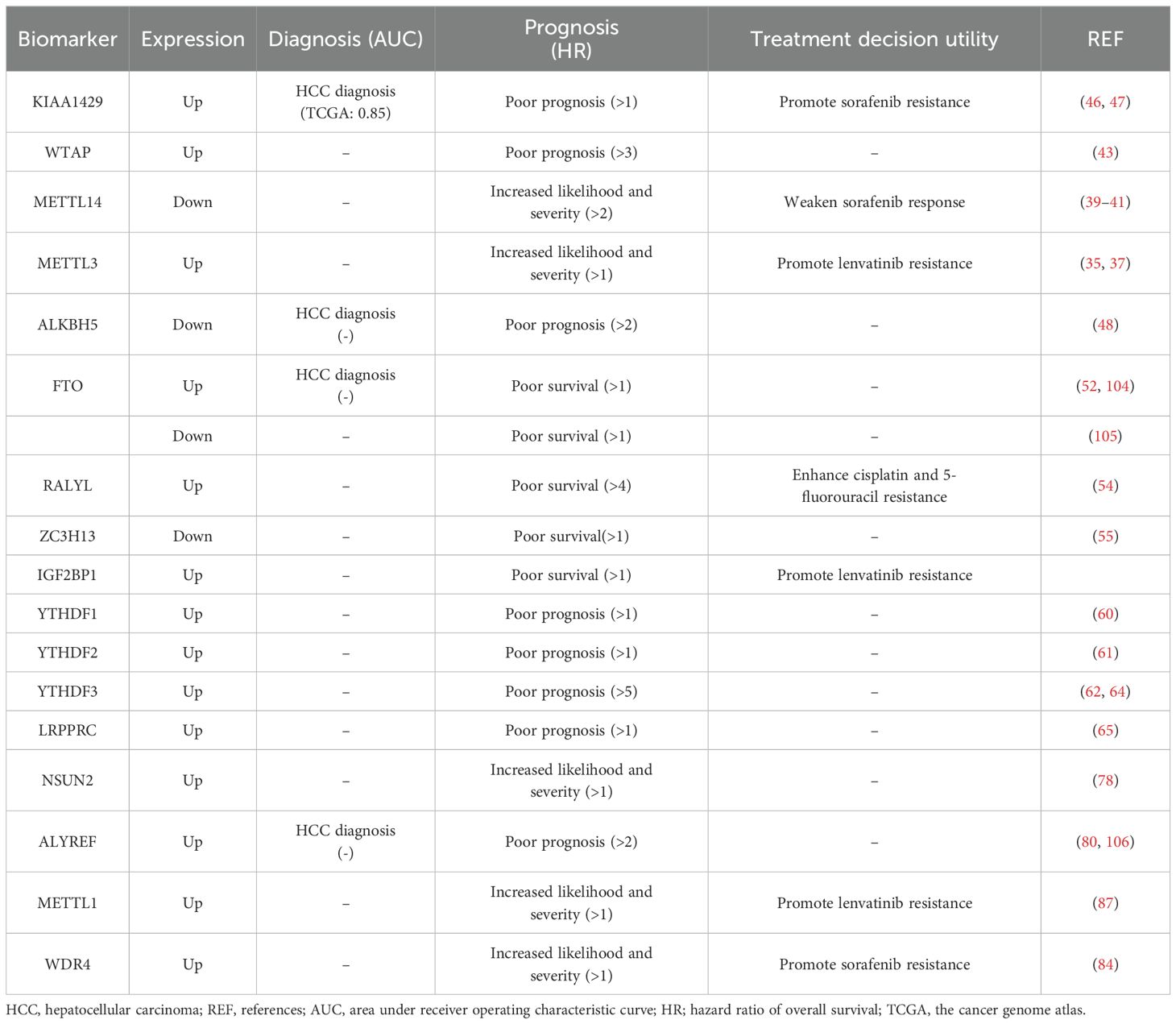
Proteins and genes associated with RNA modifications have the potential to serve as biomarkers for the diagnosis and prognostic evaluation of HCC (Table 3). This capability has the potential to improve diagnostic accuracy and ultimately enhance the survival rate of patients.
4.1.1 Role of m6A regulators in HCC diagnosis, prognosis and drug resistance
The expression levels of KIAA1429, ALKBH5 and FTO were reported to have the diagnostic potential in HCC (Table 3). A study found that KIAA1429 expression had up to 0.85 of the area under the curve (AUC) in the receiver operating characteristic curve (ROC) analysis, which indicated that it has a relatively high diagnostic value in HCC (47). Although studies have suggested the potential diagnostic potential of ALKBH5 and FTO, their potential and value as diagnostic markers have been reduced due to inconsistent expression in different studies (52, 104, 105) and a lack of ROC analysis data for diagnostic markers (48).
For the upregulated m6A methyltransferases in HCC cells, the higher expression levels of KIAA1429, WTAP, and METTL3 are associated with poorer overall survival outcomes and indicate a poor prognosis and increased likelihood and severity of patients (35, 37, 43, 46). The micropeptide AC115619-22aa encoded by lncRNA AC115619, as an inhibitor of WTAP, is also a potential prognostic indicator for HCC (36). Additionally, the elevated expression of ASPM facilitated by METTL3 via m6A modification is also strongly associated with a poor prognosis of HCC (34). The methyltransferase METTL14 is downregulated in HCC, and lower expression levels of METTL14 are associated with increased likelihood and severity of HCC patients who underwent a poorer overall survival (41). Besides, there are many pathways downstream of METTL14 that increase the likelihood and severity of HCC (107). Reduced HNF3γ expression is associated with the malignant features of HCC and is correlated with poor patient survival (40). CircSTX6 and its encoded proteins are expected to have the potential to serve as biomarkers for the diagnosis, prognosis, and treatment of HCC (38).
Low expressions of demethylases ALKBH5 and ZC3H13 are associated with poor prognosis in HCC patients (48, 50, 55). Additionally, the overexpression of ALKBH5 has been found to cause a decrease in the level of the lincRNA LINC02551, which has been utilized as a prognostic biomarker for HCC (49). Furthermore, the overexpression of the demethylase RALYL has been associated with a poorer prognosis, lower levels of differentiation, and an increased likelihood of metastasis in clinical HCC patients (54). The expression of FTO varies in different studies, with one study showing high expression and another showing low expression, both of which were linked to poorer survival in HCC patients (52, 104, 105). These conflicting findings indicate the multifaceted and functionally complex nature of FTO in HCC, highlighting the need for further research to elucidate its molecular mechanism. The deacetylase SIRT1 reduces the expression of the m6A demethylase FTO, thereby increasing the m6A levels of its downstream target GNAO1 and downregulating its mRNA expression during HCC tumorigenesis (53). The discovery of potential diagnostic biomarkers such as SIRT1, FTO, and GNAO1 offers a promising direction for future research. This area holds significant promise for further investigation and exploration (53).
The significant overexpression of the m6A reading proteins IGF2BP1, YTHDF1, YTHDF2, YTHDF3, and LRPPRC in HCC tissues has been associated with a poorer prognosis for patients (Table 3). IGF2BP1 can promote the progression of HCC through the m6A-mediated upregulation of circMDK and circMAP3K4, which are associated with poor survival in HCC patients and serve as potential tumor biomarkers (56, 57). Furthermore, the results of multivariate Cox regression analysis indicated that the expression of YTHDF1 served as an independent prognostic factor for patients with HCC (60). In addition, scientists have shown a negative correlation between YTHDF2 expression and patient survival (61). Therefore, YTHDF2 plays a significant role in the oncogenesis of HCC and can serve as a valuable biomarker for HCC patients. Additionally, the expression level of YTHDF3 in HCC tissues was found to be significantly greater than that in adjacent liver tissues (62). YTHDF3 plays a crucial role in regulating the progression of HCC, providing a promising new target for HCC treatment (63). In conclusion, the YTHDF family is closely associated with the prognosis of HCC patients and may represent a potential target for the future treatment of HCC. Additionally, LRPPRC is frequently upregulated in HCC tumors, which is linked to advanced disease stages and poor prognosis (65).
Overall, about thirteen m6A regulators are reported to be closely related to HCC prognosis and have a potential for use as prognosis biomarkers, including four methyltransferases (KIAA1429, WTAP, METTL14 and METTL3), four demethylases (ALKBH5, FTO, RALYL and ZC3H13), and five reading proteins (IGF2BP1, YTHDF1-3 and LRPPRC (Table 3). Among them, the high expression of WTAP, RALYL, YTHDF3, etc. showed a higher hazard ratio of overall survival (>3) in survival analysis, indicating a more significant correlation and impact on poor survival rates, suggesting that these markers have relatively higher prognostic value (Table 3). It should be noted that the expression changes of ALKBH5, and FTO in HCC vary in different studies, and these contradictory results need further research and clarification. In addition, the differences in the patient source, quantity, and calculation method of the survival curve analysis of these regulatory proteins indicate strong heterogeneity. Therefore, the confirmation of the prognostic value of each factor still requires multi-center large-scale sample validation. Confirming the prognostic performance of these potential markers through further ROC curve analysis of large sample sizes is an important research direction for the future. In addition, most previous studies have analyzed these factors as independent prognostic factors for patients with HCC, attempting to conduct a joint analysis of these factors for prognosis, which may provide a more comprehensive display of the relationship between m6A regulators and HCC prognosis.
In addition to patient survival rates, some m6A regulators also exhibit a correlation with targeted anti-cancer drugs or chemotherapy drugs, promoting cancer cell drug resistance (Table 3). High expression of KIAA1429 and low expression of METTL14 can promote sorafenib resistance in HCC cells. KIAA1429 is involved in promoting invasion, migration, and EMT in sorafenib-resistant HCC by mediating m6A methylation (21). THNF3γ reduction caused by METTL14 knockdown upregulates the expression of the sorafenib influx transporters OATP1B1 and OATP1B3, thereby rendering sorafenib resistance in HCC, and enforced HNF3γ expression enhances the cellular response to sorafenib in HCC (40). Similarly, high expression of METTL3 and IGF2BP1 can promote lenvatinib resistance in HCC cells, while high expression of RALYL enhances cisplatin and 5-fluorouracil resistance. Targeting METTL3 with the specific inhibitor STM2457 improved sensitivity to lenvatinib in both in vitro and in vivo studies (35). This discovery suggested that METTL3 might serve as a potential therapeutic target for overcoming lenvatinib resistance in HCC. There have been reports suggesting that patients with smaller tumors and good liver function may benefit from a combined regimen of mitoxantrone, 5-fluorouracil, and cisplatin (108). Inhibiting RALYL could potentially improve the effectiveness of a combination therapy involving mitoxantrone, 5-fluorouracil, and cisplatin, providing new strategies for treating HCC. These results not only provide a theoretical basis for personalized medication for patients but also offer new directions for increasing drug sensitivity research. The combination of specific inhibitors or downstream target genes that modify related proteins with chemotherapy drugs may provide new strategies for the treatment of HCC.
4.1.2 Role of m5C regulators in HCC diagnosis, prognosis and drug resistance
Regulators of RNA m5C modification and their target HCC-related oncogenes are potential diagnostic and prognostic biomarkers for HCC (Table 3). A significant increase in the expression of lncRNA H19 was reported to be associated with the development of various types of tumors (77). The RNA m5C methyltransferase NSUN2 regulates the stability of lncRNA H19 through m5C modification and may serve as a potential new target and biomarker for the treatment of HCC (77). In addition, NSUN2 has been found to impact the sensitivity of HCC cells to sorafenib by regulating the activity of the Ras pathway (78). The sensitivity of NSUN2 knockout cell lines to sorafenib was significantly greater than that of control cells (78). ALYREF, functioning as a “reader” for the m5C site in RNA, is a protein that shuttles between the nucleus and cytoplasm, and plays a critical role in facilitating the transportation of RNA from the nucleus to the cytoplasm. The upregulation of ALYREF has been associated with poor prognosis in HCC patients, indicating its potential as a valuable target for diagnosing and predicting prognosis in HCC patients (80, 100). The higher HR value of ALYREF than NSUN2 suggests a priority prognostic value of ALYREF. However, due to the lack of necessary ROC analysis, further analysis of large sample sizes is needed to confirm the prognostic or diagnostic values of NSUN2 and ALYREF.
4.1.3 Role of m7G regulators in HCC prognosis and drug resistance
The upregulation of the m7G methyltransferases METTL1 and WDR4 has been reported to be correlated with advanced tumor stage and unfavorable patient survival outcomes in the literature (86). Investigators have shown that METTL1-mediated tRNA m7G modification plays a critical role in lenvatinib resistance in HCC cells by enhancing the translation of EGFR pathway genes (87). WDR4 overexpression significantly increased the half-maximal inhibitory concentration value of sorafenib in HCC cells (77). Some researchers propose that both METTL1 and WDR4 have the potential to serve as biomarkers for predicting the efficacy of lenvatinib and sorafenib (87). The potential application of m7G regulators in HCC is still in the early stages of research. Further exploration of new m7G regulators involved in the occurrence and progression of HCC may provide new targets for the prognosis and treatment of HCC.
4.1.4 Potential applications of non-coding RNAs regulated by RNA modification
Non-coding RNAs, as important target sequences regulated by RNA modifications, play a crucial role in the pathogenesis of HCC (Figure 3). Upregulated circRNAs dependent on M6A modification, including circ-CCT3, circSTX6, circ-ARL3, circMDK, circMAP3K4, and circ_KIAA1429, are involved in the proliferation, invasion, migration, and apoptosis of HCC cells. Upregulated lncRNAs dependent on m6A modification, such as GBAP1, lnc-CTHCC, LINC00958, MIR4435-2HG, etc., play similar functional roles in the progression of HCC. Certain non-coding RNAs regulated by RNA modification showed significant application potential in the diagnosis, prognosis, and treatment of HCC. The elevated expression of circSTX6 has an AUC of up to 0.8565 in the diagnostic ROC analysis, suggesting a considerable diagnostic significance in HCC (38). The high expression of LINC00958, lnc-CTHCC showed a higher hazard ratio of overall survival (>2) in survival analysis, indicating a more significant correlation and impact on poor survival rates, suggesting that these markers have relatively higher prognostic value (30, 31). Overexpression of MIR4435-2HG significantly reduced cell susceptibility to lenvatinib, suggesting its role in cell resistance to lenvatinib (58). Nevertheless, most research has focused solely on investigating the impact of partial non-coding RNA in m6A modification, leaving a gap in understanding the connection between miRNA and RNA modification, along with the correlation between m5C, m1A, m7G, and RNA modification. This presents a promising avenue for further investigation.
4.2 Potential agents targeting RNA modification
4.2.1 Potential agents targeting m6A
As the link between RNA modifications and cancer continues to be discovered, the demand for inhibitors of the associated proteins is increasing. The role of the m6A methyltransferase METTL3 in various diseases is continuously expanding, and there is also a growing focus on the development of METTL3 inhibitors. This has garnered increasing attention in academic research and scholarly discourse (Figure 4). Adenosine was the first reported METTL3 inhibitor that acts in a SAM-competitive manner to reduce the level of m6A modification (109). UZH2 is a small molecule inhibitor that selectively targets METTL3 and reduces the m6A levels of polyadenylated RNA in the MOLM-13 (acute myeloid leukemia) and PC-3 (prostate cancer) cell lines and has great potential as a therapeutic agent for cancer (110, 111). Curcumin, a polyphenolic pigment derived from turmeric, is yellow in color and has been reported to exhibit anti-inflammatory, anticancer, antioxidant, and antibacterial activities (112). Curcumin can increase the level of m6A modification in piglet liver by affecting the mRNA expression of METTL3, METTL14, ALKBH5, FTO, and YTHDF2 and subsequently attenuate polysaccharide-induced disorders of hepatic lipid metabolism (113).
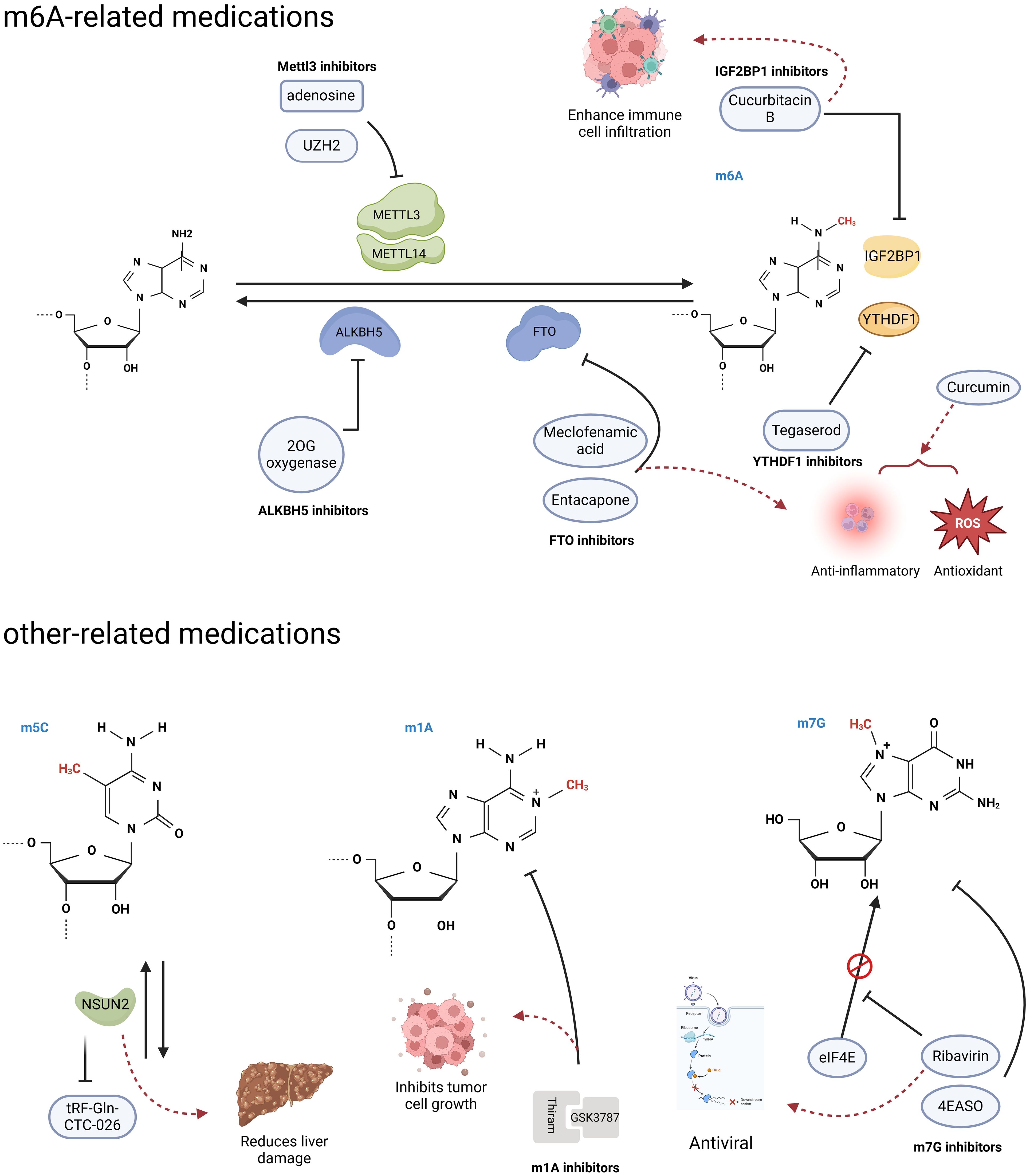
Figure 4. RNA modification-related agents. Research has found that some agents act on RNA modified regulators, and can trigger a series of downstream effects. (Created with BioRender.com).
FTO, a m6A demethylase, has been reported to play important roles in various diseases, including HCC and diabetes mellitus, and is a potential therapeutic for related diseases (114). Entacapone and meclofenamic acid (MA) are reported to be inhibitors of the m6A demethylase FTO (Figure 4) (114–116). As a chemical inhibitor, entacapone inhibits FTO-mediated metabolic regulation through forkhead box O1 (FOXO1) (114). MA, a nonsteroidal anti-inflammatory drug, has been identified as a highly selective FTO inhibitor that competitively binds to FTO, thereby enhancing overall m6A modification levels (97). In the case of inhibitors targeting another m6A demethylase, ALKBH5, only a small number of nonselective compounds have been discovered (Figure 4). The majority of these compounds are 2OG oxygenase inhibitors, with only a limited few characterized as ALKBH5 inhibitors. Currently, there is a lack of significant selective inhibitors for ALKBH5 (117).
In addition, Hong et al. conducted a structure-based drug screening and identified tegaserod as a potential inhibitor of YTHDF1 (Figure 4) (118). Tegaserod functions by blocking the binding of YTHDF1 to m6A-modified mRNA, thereby inhibiting the YTHDF1-mediated translation of Cyclin E2. This suggests that tegaserod may serve as a promising antitumor agent (119). Modulation of the tumor immune microenvironment has emerged as a novel approach for cancer immunotherapy in HCC, with m6A modification being an ideal target due to its role in the immune response (120). The expression of IGF2BP1, which recognizes m6A modifications, is significantly upregulated in HCC and is associated with immune cell infiltration (121). Cucurbitacin B has been demonstrated to inhibit the binding of IGF2BP1 to m6A-modified mRNA and increase immune cell infiltration, suggesting that it is a potential anti-HCC agent (Figure 4) (121).
4.2.2 Potential agents targeting m5C
The m5C writing protein is a promising target for the treatment of HCC. The upregulation of the m5C methyltransferase NSUN2 in HCC has been observed (77). Knocking down NSUN2 leads to reduced levels of m5C modification, and some unmodified tRNAs undergo complex changes to become tfRNAs (tRF-Gln-CTC-026), which effectively alleviate liver injury by inhibiting global protein synthesis (GPS) (122). Efforts to deplete NSUN2 and provide protective tfRNAs are potential treatments for liver injury and have significant implications for reducing the risk of HCC occurrence.
4.2.3 Potential agents targeting m1A
Thiram, a potent inhibitor of the m1A methyltransferase complex TRMT6/TRMT61A, has been shown to significantly inhibit the self-renewal of hepatic CSCs and hepatic tumor growth (Figure 4) (81). Furthermore, when combined with the PPARS antagonist GSK3787, thiram synergistically inhibits the development of HCC and the growth of tumors that are hypermethylated with m1A (81).
4.2.4 Potential agents targeting m7G
Ribavirin is a widely used antiviral medication that has recently been shown to possess antitumor effects as a m7G cap analog that inhibits cell proliferation (Figure 4) (123). Additionally, the eIF4E antisense oligonucleotide drug (4EASO) is a well-established medication that competitively inhibits eIF4E binding to the m7G cap (124). These medications have shown promising results in halting cancer progression and improving prognosis, offering hopeful prospects for the development of additional m7G-targeted drugs.
5 Challenges and opportunities for RNA modification
Currently, research on the role of RNA modification in HCC mainly focuses on m6A, m1A, m5C, and m7G. The expression of RNA modification regulators, including “writer”, “eraser” and “reader” proteins, changed significantly in HCC, and are involved in the proliferation, autophagy, innate immunity, invasion, metastasis, immune cell infiltration, and drug resistance of HCC via different signal axes. The varying expression of RNA modification regulatory proteins with similar functions in HCC, along with the lack of consensus on the expression changes of some RNA modification regulatory proteins, indicates that the expression of these proteins in HCC may be dynamic and have different mechanisms of action in various stages and types of the disease. Consequently, further comprehensive and detailed classification research is necessary to elucidate this matter. Additionally, additional investigation is required to ascertain the relationships between these various forms of RNA modifications. In addition to the aforementioned modifications, there are still multiple types of RNA modifications involved in regulating post-transcriptional gene expression, including 5-hydroxymethylcytosine, pseudouridine, and 2’-O-methylation. Exploring whether these recently discovered RNA modifications are also involved in the occurrence and development of HCC may be a new research hotspot in the future. As high-throughput sequencing technology and novel biomarkers continue to advance, we will be able to delve deeper into the intricate regulatory network of RNA modifications in HCC, ultimately providing more precise and effective approaches for clinical diagnosis and treatment.
Studying RNA modifications in HCC will contribute to an in-depth understanding of the pathogenesis of HCC, the search for new diagnostic and prognostic markers, the development of new therapeutic strategies, and the assessment of treatment efficacy and the monitoring of recurrence. These findings provide a new perspective for the diagnosis and treatment of HCC by targeting specific RNA-modifying enzymes, recognition proteins, or related RNAs. More than ten RNA-modifying regulators showed the potential for use for the diagnosis, prognosis, and treatment decision utility biomarkers of HCC. KIAA1429 has a relatively high diagnostic value in HCC. For other potential diagnostic markers, further research is needed because of inconsistent expression in different studies and a lack of ROC analysis data for diagnostic markers. WTAP, RALYL, and YTHDF3 exhibit significantly higher prognostic significance in HCC. However, the validation of their prognostic value necessitates extensive multi-center sample validation and ROC curve analysis in the future. On the other hand, prior research has focused on examining these factors separately as prognostic indicators for HCC patients. Conducting a combined analysis of these factors could offer a more comprehensive understanding of the association between RNA-modifying regulators and HCC prognosis. Some RNA-modifying regulators also exhibit an association with targeted anti-cancer drugs or chemotherapy drugs, which provide a theoretical basis for personalized medication for patients. The concurrent administration of drugs and inhibitors of regulatory proteins based on the relationship between regulatory proteins and drug resistance may enhance the sensitivity of drugs and offer novel approaches for managing HCC.
A growing number of RNA modifier inhibitors are being developed, but the lack of preclinical experiments and clinical studies targeting RNA modification in HCC poses a significant obstacle, and further research is needed to evaluate their application value in HCC treatment. Studying RNA modifications in liver cancer necessitates careful consideration of the potential and challenges associated with its translational application. Although RNA modification may present a novel target for HCC treatment, the lack of specific drugs targeting RNA modification poses a significant obstacle. Many RNA modifications occur at low levels, making it difficult to selectively target and manipulate modified RNA molecules without affecting unmodified ones. Additionally, our understanding of the structural domains of certain RNA modifiers, such as those in the NSUN and TRMT families, remains limited. This lack of knowledge makes it even more challenging to design inhibitors that effectively target these proteins. Therefore, further investigation into the mechanism of action of RNA modification is imperative to identify effective strategies for regulating this process and to evaluate its safety and efficacy. The development of effective tools and techniques for targeting and manipulating specific RNA modifications is an ongoing challenge. Overcoming these obstacles will pave the way for the development of targeted therapies and interventions that can harness the potential of RNA modifications in various biological and disease settings.
6 Conclusion
RNA modifications play a crucial role in HCC progression. Specific RNA modification pathways, such as m6A, m5C, m1A, and m7G, are erroneously regulated by a series of modification regulatory proteins, thereby regulating the expression of HCC-associated genes, and are involved in the proliferation, autophagy, innate immunity, invasion, metastasis, immune cell infiltration, and drug resistance of HCC. These functional roles and molecular mechanism advancements of RNA modification offer new perspectives on the pathogenesis, as well as potential new diagnostic and prognostic markers and therapeutic strategies of HCC. At present, over ten RNA-modifying regulators have displayed promise as biomarkers for the diagnosis, prognosis, and treatment decisions related to HCC. However, the practical application of these biomarkers in HCC necessitates thorough validation across multiple centers in the future. Agents targeting RNA modification are potential therapeutic strategies for HCC. The concurrent administration with the inhibitors of RNA modification may enhance the sensitivity of drugs and offer novel approaches for managing HCC. While a growing number of RNA modifier inhibitors are being developed, the absence of preclinical and clinical studies focusing on targeting RNA modification in HCC presents a significant challenge. Further investigation is crucial to assess the efficacy of these inhibitors in the treatment of HCC.
Author contributions
J-XL: Writing – original draft, Writing – review & editing. XZ: Writing – review & editing. W-HX: Funding acquisition, Writing – review & editing. X-DH: Funding acquisition, Investigation, Methodology, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by the Shandong Provincial Natural Science Foundation, China (ZR2020MC059 and RH2200000157).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, et al. Global cancer statistics 2022: globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2024) 74:229–63. doi: 10.3322/caac.21834
2. Huang DQ, Mathurin P, Cortez-Pinto H, Loomba R. Global epidemiology of alcohol-associated cirrhosis and hcc: trends, projections and risk factors. Nat Rev Gastroenterol Hepatol. (2023) 20:37–49. doi: 10.1038/s41575-022-00688-6
3. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: Cancer J Clin. (2021) 71:209–49. doi: 10.3322/caac.21660
4. Singal AG, Kanwal F, Llovet JM. Global trends in hepatocellular carcinoma epidemiology: implications for screening, prevention and therapy. Nat Rev Clin Oncol. (2023) 20:864–84. doi: 10.1038/s41571-023-00825-3
5. Llovet JM, Kelley RK, Villanueva A, Singal AG, Pikarsky E, Roayaie S, et al. Hepatocellular carcinoma. Nat Rev Dis Primers. (2021) 7:6. doi: 10.1038/s41572-021-00245-6
6. Kostyusheva A, Brezgin S, Glebe D, Kostyushev D, Chulanov V. Host-cell interactions in hbv infection and pathogenesis: the emerging role of M6a modification. Emerg Microbes infect. (2021) 10:2264–75. doi: 10.1080/22221751.2021.2006580
7. Wang Y, Deng B. Hepatocellular carcinoma: molecular mechanism, targeted therapy, and biomarkers. Cancer metastasis Rev. (2023) 42:629–52. doi: 10.1007/s10555-023-10084-4
8. Chidambaranathan-Reghupaty S, Fisher PB, Sarkar D. Hepatocellular carcinoma (Hcc): epidemiology, etiology and molecular classification. Adv Cancer Res. (2021) 149:1–61. doi: 10.1016/bs.acr.2020.10.001
9. Yegin EG, Oymaci E, Karatay E, Coker A. Progress in surgical and nonsurgical approaches for hepatocellular carcinoma treatment. Hepatobil Pancreat Dis Int. (2016) 15:234–56. doi: 10.1016/s1499-3872(16)60097-8
10. Delis SG, Dervenis C. Selection criteria for liver resection in patients with hepatocellular carcinoma and chronic liver disease. World J Gastroenterol. (2008) 14:3452–60. doi: 10.3748/wjg.14.3452
11. Jiang C, Cai YQ, Yang JJ, Ma CY, Chen JX, Huang L, et al. Radiomics in the diagnosis and treatment of hepatocellular carcinoma. Hepatobil Pancreat Dis Int. (2023) 22:346–51. doi: 10.1016/j.hbpd.2023.03.010
12. Vogel A, Meyer T, Sapisochin G, Salem R, Saborowski A. Hepatocellular carcinoma. Lancet (London England). (2022) 400:1345–62. doi: 10.1016/s0140-6736(22)01200-4
13. Barbieri I, Kouzarides T. Role of rna modifications in cancer. Nat Rev Cancer. (2020) 20:303–22. doi: 10.1038/s41568-020-0253-2
14. Li S, Mehal WZ, Ouyang X. Rna modifications in the progression of liver diseases: from fatty liver to cancer. Sci China Life Sci. (2024). doi: 10.1007/s11427-023-2494-x
15. Feng Q, Wang D, Xue T, Lin C, Gao Y, Sun L, et al. The role of rna modification in hepatocellular carcinoma. Front Pharmacol. (2022) 13:984453. doi: 10.3389/fphar.2022.984453
16. Chen L, Gao Y, Xu S, Yuan J, Wang M, Li T, et al. N6-methyladenosine reader ythdf family in biological processes: structures, roles, and mechanisms. Front Immunol. (2023) 14:1162607. doi: 10.3389/fimmu.2023.1162607
17. Li D, Li K, Zhang W, Yang KW, Mu DA, Jiang GJ, et al. The M6a/M5c/M1a regulated gene signature predicts the prognosis and correlates with the immune status of hepatocellular carcinoma. Front Immunol. (2022) 13:918140. doi: 10.3389/fimmu.2022.918140
18. Dunn DB, Smith JD. Occurrence of a new base in the deoxyribonucleic acid of a strain of bacterium coli. Nature. (1955) 175:336–7. doi: 10.1038/175336a0
19. Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger rna from novikoff hepatoma cells. Proc Natl Acad Sci United States America. (1974) 71:3971–5. doi: 10.1073/pnas.71.10.3971
20. Dubin DT, Taylor RH. The methylation state of poly a-containing messenger rna from cultured hamster cells. Nucleic Acids Res. (1975) 2:1653–68. doi: 10.1093/nar/2.10.1653
21. Adams JM, Cory S. Modified nucleosides and bizarre 5’-termini in mouse myeloma mrna. Nature. (1975) 255:28–33. doi: 10.1038/255028a0
22. Perry RP, Kelley DE, Friderici K, Rottman F. The methylated constituents of L cell messenger rna: evidence for an unusual cluster at the 5’ Terminus. Cell. (1975) 4:387–94. doi: 10.1016/0092-8674(75)90159-2
23. Ontiveros RJ, Stoute J, Liu KF. The chemical diversity of rna modifications. Biochem J. (2019) 476:1227–45. doi: 10.1042/bcj20180445
24. Tang Q, Li L, Wang Y, Wu P, Hou X, Ouyang J, et al. Rna modifications in cancer. Br J Cancer. (2023) 129:204–21. doi: 10.1038/s41416-023-02275-1
25. Yang Z, Zhang S, Xia T, Fan Y, Shan Y, Zhang K, et al. Rna modifications meet tumors. Cancer Manage Res. (2022) 14:3223–43. doi: 10.2147/cmar.S391067
26. Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, et al. N6-methyladenosine in nuclear rna is a major substrate of the obesity-associated fto. Nat Chem Biol. (2011) 7:885–7. doi: 10.1038/nchembio.687
27. Zhu ZM, Huo FC, Pei DS. Function and evolution of rna N6-methyladenosine modification. Int J Biol Sci. (2020) 16:1929–40. doi: 10.7150/ijbs.45231
28. Rao X, Lai L, Li X, Wang L, Li A, Yang Q. N(6) -methyladenosine modification of circular rna circ-arl3 facilitates hepatitis B virus-associated hepatocellular carcinoma via sponging mir-1305. IUBMB Life. (2021) 73:408–17. doi: 10.1002/iub.2438
29. Liu R, Yin G, Tuo H, Guo Y, Zhu Y, Zhang L, et al. Mettl3-induced lncrna gbap1 promotes hepatocellular carcinoma progression by activating bmp/smad pathway. Biol direct. (2023) 18:53. doi: 10.1186/s13062-023-00409-2
30. Zuo X, Chen Z, Gao W, Zhang Y, Wang J, Wang J, et al. M6a-mediated upregulation of linc00958 increases lipogenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. J Hematol Oncol. (2020) 13:5. doi: 10.1186/s13045-019-0839-x
31. Xia A, Yuan W, Wang Q, Xu J, Gu Y, Zhang L, et al. The cancer-testis lncrna lnc-cthcc promotes hepatocellular carcinogenesis by binding hnrnp K and activating yap1 transcription. Nat Cancer. (2022) 3:203–18. doi: 10.1038/s43018-021-00315-4
32. Tao L, Li D, Mu S, Tian G, Yan G. Lncrna mapkapk5_As1 facilitates cell proliferation in hepatitis B virus -related hepatocellular carcinoma. Lab investigation; J Tech Methods Pathol. (2022) 102:494–504. doi: 10.1038/s41374-022-00731-9
33. Kim GW, Imam H, Khan M, Mir SA, Kim SJ, Yoon SK, et al. Hbv-induced increased N6 methyladenosine modification of pten rna affects innate immunity and contributes to hcc. Hepatol (Baltimore Md). (2021) 73:533–47. doi: 10.1002/hep.31313
34. Wang A, Chen X, Li D, Yang L, Jiang J. Mettl3-mediated M6a methylation of aspm drives hepatocellular carcinoma cells growth and metastasis. J Clin Lab Anal. (2021) 35:e23931. doi: 10.1002/jcla.23931
35. Wang L, Yang Q, Zhou Q, Fang F, Lei K, Liu Z, et al. Mettl3-M(6)a-egfr-axis drives lenvatinib resistance in hepatocellular carcinoma. Cancer Lett. (2023) 559:216122. doi: 10.1016/j.canlet.2023.216122
36. Yang Y, Qian Cai Q, Sheng Fu L, Wei Dong Y, Fan F, Zhong Wu X. Reduced N6-methyladenosine mediated by mettl3 acetylation promotes mtf1 expression and hepatocellular carcinoma cell growth. Chem biodivers. (2022) 19:e202200333. doi: 10.1002/cbdv.202200333
37. Xu H, Wang H, Zhao W, Fu S, Li Y, Ni W, et al. Sumo1 modification of methyltransferase-like 3 promotes tumor progression via regulating snail mrna homeostasis in hepatocellular carcinoma. Theranostics. (2020) 10:5671–86. doi: 10.7150/thno.42539
38. Lu J, Ru J, Chen Y, Ling Z, Liu H, Ding B, et al. N(6) -methyladenosine-modified circstx6 promotes hepatocellular carcinoma progression by regulating the hnrnpd/atf3 axis and encoding a 144 amino acid polypeptide. Clin Trans Med. (2023) 13:e1451. doi: 10.1002/ctm2.1451
39. Du L, Li Y, Kang M, Feng M, Ren Y, Dai H, et al. Usp48 is upregulated by mettl14 to attenuate hepatocellular carcinoma via regulating sirt6 stabilization. Cancer Res. (2021) 81:3822–34. doi: 10.1158/0008-5472.Can-20-4163
40. Zhou T, Li S, Xiang D, Liu J, Sun W, Cui X, et al. M6a rna methylation-mediated hnf3γ Reduction renders hepatocellular carcinoma dedifferentiation and sorafenib resistance. Signal transduct target Ther. (2020) 5:296. doi: 10.1038/s41392-020-00299-0
41. Shi Y, Zhuang Y, Zhang J, Chen M, Wu S. Mettl14 inhibits hepatocellular carcinoma metastasis through regulating egfr/pi3k/akt signaling pathway in an M6a-dependent manner. Cancer Manag Res. (2020) 12:13173–84. doi: 10.2147/CMAR.S286275
42. Yang Y, Cai J, Yang X, Wang K, Sun K, Yang Z, et al. Dysregulated M6a modification promotes lipogenesis and development of non-alcoholic fatty liver disease and hepatocellular carcinoma. Mol therapy: J Am Soc Gene Ther. (2022) 30:2342–53. doi: 10.1016/j.ymthe.2022.02.021
43. Chen Y, Peng C, Chen J, Chen D, Yang B, He B, et al. Wtap facilitates progression of hepatocellular carcinoma via M6a-hur-dependent epigenetic silencing of ets1. Mol Cancer. (2019) 18:127. doi: 10.1186/s12943-019-1053-8
44. Li G, Deng L, Huang N, Cui Z, Wu Q, Ma J, et al. M(6)a mrna methylation regulates lkb1 to promote autophagy of hepatoblastoma cells through upregulated phosphorylation of ampk. Genes. (2021) 12:1747. doi: 10.3390/genes12111747
45. Zhang Q, Wei T, Yan L, Zhu S, Jin W, Bai Y, et al. Hypoxia-responsive lncrna ac115619 encodes a micropeptide that suppresses M6a modifications and hepatocellular carcinoma progression. Cancer Res. (2023) 83:2496–512. doi: 10.1158/0008-5472.Can-23-0337
46. Kuang Y, Cheng Y, Wang J, Li H, Cao X, Wang Y. Kiaa1429 mediates epithelial mesenchymal transition in sorafenib-resistant hepatocellular carcinoma through M6a methylation modification. Cancer Med. (2023) 12:7222–33. doi: 10.1002/cam4.5432
47. Lan T, Li H, Zhang D, Xu L, Liu H, Hao X, et al. Kiaa1429 contributes to liver cancer progression through N6-methyladenosine-dependent post-transcriptional modification of gata3. Mol Cancer. (2019) 18:186. doi: 10.1186/s12943-019-1106-z
48. Chen Y, Zhao Y, Chen J, Peng C, Zhang Y, Tong R, et al. Alkbh5 suppresses Malignancy of hepatocellular carcinoma via M(6)a-guided epigenetic inhibition of lypd1. Mol Cancer. (2020) 19:123. doi: 10.1186/s12943-020-01239-w
49. Zhang H, Liu Y, Wang W, Liu F, Wang W, Su C, et al. Alkbh5-mediated M(6)a modification of lincrna linc02551 enhances the stability of ddx24 to promote hepatocellular carcinoma growth and metastasis. Cell Death Dis. (2022) 13:926. doi: 10.1038/s41419-022-05386-4
50. You Y, Wen D, Zeng L, Lu J, Xiao X, Chen Y, et al. Alkbh5/map3k8 axis regulates pd-L1+ Macrophage infiltration and promotes hepatocellular carcinoma progression. Int J Biol Sci. (2022) 18:5001–18. doi: 10.7150/ijbs.70149
51. Qu S, Jin L, Huang H, Lin J, Gao W, Zeng Z. A positive-feedback loop between hbx and alkbh5 promotes hepatocellular carcinogenesis. BMC Cancer. (2021) 21:686. doi: 10.1186/s12885-021-08449-5
52. Wang F, Hu Y, Wang H, Hu P, Xiong H, Zeng Z, et al. Lncrna fto-it1 promotes glycolysis and progression of hepatocellular carcinoma through modulating fto-mediated N6-methyladenosine modification on glut1 and pkm2. J Exp Clin Cancer res: CR. (2023) 42:267. doi: 10.1186/s13046-023-02847-2
53. Liu X, Liu J, Xiao W, Zeng Q, Bo H, Zhu Y, et al. Sirt1 regulates N(6) -methyladenosine rna modification in hepatocarcinogenesis by inducing ranbp2-dependent fto sumoylation. Hepatol (Baltimore Md). (2020) 72:2029–50. doi: 10.1002/hep.31222
54. Wang X, Wang J, Tsui YM, Shi C, Wang Y, Zhang X, et al. Ralyl increases hepatocellular carcinoma stemness by sustaining the mrna stability of tgf-β2. Nat Commun. (2021) 12:1518. doi: 10.1038/s41467-021-21828-7
55. Wu S, Liu S, Cao Y, Chao G, Wang P, Pan H. Downregulation of zc3h13 by mir-362-3p/mir-425-5p is associated with a poor prognosis and adverse outcomes in hepatocellular carcinoma. Aging. (2022) 14:2304–19. doi: 10.18632/aging.203939
56. Du A, Li S, Zhou Y, Disoma C, Liao Y, Zhang Y, et al. M6a-mediated upregulation of circmdk promotes tumorigenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. Mol Cancer. (2022) 21:109. doi: 10.1186/s12943-022-01575-z
57. Duan JL, Chen W, Xie JJ, Zhang ML, Nie RC, Liang H, et al. A novel peptide encoded by N6-methyladenosine modified circmap3k4 prevents apoptosis in hepatocellular carcinoma. Mol Cancer. (2022) 21:93. doi: 10.1186/s12943-022-01537-5
58. Zhu Y, Xiao B, Liu M, Chen M, Xia N, Guo H, et al. N6-methyladenosine-modified oncofetal lncrna mir4435-2hg contributed to stemness features of hepatocellular carcinoma cells by regulating rrna 2’-O methylation. Cell Mol Biol Lett. (2023) 28:89. doi: 10.1186/s11658-023-00493-2
59. Yang Y, Wu J, Liu F, He J, Wu F, Chen J, et al. Igf2bp1 promotes the liver cancer stem cell phenotype by regulating mgat5 mrna stability by M6a rna methylation. Stem Cells Dev. (2021) 30:1115–25. doi: 10.1089/scd.2021.0153
60. Li Q, Ni Y, Zhang L, Jiang R, Xu J, Yang H, et al. Hif-1α-induced expression of M6a reader ythdf1 drives hypoxia-induced autophagy and Malignancy of hepatocellular carcinoma by promoting atg2a and atg14 translation. Signal transduct target Ther. (2021) 6:76. doi: 10.1038/s41392-020-00453-8
61. Zhang C, Huang S, Zhuang H, Ruan S, Zhou Z, Huang K, et al. Ythdf2 promotes the liver cancer stem cell phenotype and cancer metastasis by regulating oct4 expression via M6a rna methylation. Oncogene. (2020) 39:4507–18. doi: 10.1038/s41388-020-1303-7
62. Wang M, Yang Y, Yang J, Yang J, Han S. Circ_Kiaa1429 accelerates hepatocellular carcinoma advancement through the mechanism of M(6)a-ythdf3-zeb1. Life Sci. (2020) 257:118082. doi: 10.1016/j.lfs.2020.118082
63. Hu B, Gao J, Shi J, Wen P, Guo W, Zhang S. M(6) a Reader Ythdf3 Triggers the Progression of Hepatocellular Carcinoma through the Ythdf3/M(6) a-Egfr/Stat3 Axis and Emt. Mol carcinogene. (2023) 62:1599–614. doi: 10.1002/mc.23602
64. Zhou R, Ni W, Qin C, Zhou Y, Li Y, Huo J, et al. A functional loop between yth domain family protein ythdf3 mediated M(6)a modification and phosphofructokinase pfkl in glycolysis of hepatocellular carcinoma. J Exp Clin Cancer Res. (2022) 41:334. doi: 10.1186/s13046-022-02538-4
65. Wang H, Tang A, Cui Y, Gong H, Li H. Lrpprc facilitates tumor progression and immune evasion through upregulation of M(6)a modification of pd-L1 mrna in hepatocellular carcinoma. Front Immunol. (2023) 14:1144774. doi: 10.3389/fimmu.2023.1144774
66. Liu H, Jiang Y, Lu J, Peng C, Ling Z, Chen Y, et al. M(6)a-modification regulated circ-cct3 acts as the sponge of mir-378a-3p to promote hepatocellular carcinoma progression. Epigenetics. (2023) 18:2204772. doi: 10.1080/15592294.2023.2204772
67. Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, et al. Alkbh5 is a mammalian rna demethylase that impacts rna metabolism and mouse fertility. Mol Cell. (2013) 49:18–29. doi: 10.1016/j.molcel.2012.10.015
68. Lence T, Paolantoni C, Worpenberg L, Roignant JY. Mechanistic insights into M(6)a rna enzymes. Biochim Biophys Acta Gene Regul Mech. (2019) 1862:222–9. doi: 10.1016/j.bbagrm.2018.10.014
69. Xiong M, Ou C, Yu C, Qiu J, Lu J, Fu C, et al. Qi-shen-tang alleviates retinitis pigmentosa by inhibiting ferroptotic features via the nrf2/gpx4 signaling pathway. Heliyon. (2023) 9:e22443. doi: 10.1016/j.heliyon.2023.e22443
70. Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, et al. Recognition of rna N(6)-methyladenosine by igf2bp proteins enhances mrna stability and translation. Nat Cell Biol. (2018) 20:285–95. doi: 10.1038/s41556-018-0045-z
71. Ramesh-Kumar D, Guil S. The igf2bp family of rna binding proteins links epitranscriptomics to cancer. Semin Cancer Biol. (2022) 86:18–31. doi: 10.1016/j.semcancer.2022.05.009
72. Courtney DG, Tsai K, Bogerd HP, Kennedy EM, Law BA, Emery A, et al. Epitranscriptomic addition of M(5)C to hiv-1 transcripts regulates viral gene expression. Cell Host Microbe. (2019) 26:217–27.e6. doi: 10.1016/j.chom.2019.07.005
73. Reid R, Greene PJ, Santi DV. Exposition of a family of rna M(5)C methyltransferases from searching genomic and proteomic sequences. Nucleic Acids Res. (1999) 27:3138–45. doi: 10.1093/nar/27.15.3138
74. Song H, Zhang J, Liu B, Xu J, Cai B, Yang H, et al. Biological roles of rna M(5)C modification and its implications in cancer immunotherapy. biomark Res. (2022) 10:15. doi: 10.1186/s40364-022-00362-8
75. Kong W, Biswas A, Zhou D, Fiches G, Fujinaga K, Santoso N, et al. Nucleolar protein nop2/nsun1 suppresses hiv-1 transcription and promotes viral latency by competing with tat for tar binding and methylation. PloS Pathog. (2020) 16:e1008430. doi: 10.1371/journal.ppat.1008430
76. Zhou Y, Kong Y, Fan W, Tao T, Xiao Q, Li N, et al. Principles of rna methylation and their implications for biology and medicine. Biomed pharmacother = Biomed pharmacother. (2020) 131:110731. doi: 10.1016/j.biopha.2020.110731
77. Sun Z, Xue S, Zhang M, Xu H, Hu X, Chen S, et al. Aberrant nsun2-mediated M(5)C modification of H19 lncrna is associated with poor differentiation of hepatocellular carcinoma. Oncogene. (2020) 39:6906–19. doi: 10.1038/s41388-020-01475-w
78. Song D, An K, Zhai W, Feng L, Xu Y, Sun R, et al. Nsun2-mediated mrna M(5)C modification regulates the progression of hepatocellular carcinoma. Genom Proteomics Bioinf. (2023) 21:823–33. doi: 10.1016/j.gpb.2022.09.007
79. Sun GF, Ding H. Nop2-mediated M5c methylation of xpd is associated with hepatocellular carcinoma progression. Neoplasma. (2023) 70:340–9. doi: 10.4149/neo_2023_230110N17
80. Nulali J, Zhang K, Long M, Wan Y, Liu Y, Zhang Q, et al. Alyref-mediated rna 5-methylcytosine modification promotes hepatocellular carcinoma progression via stabilizing egfr mrna and pstat3 activation. Int J Biol Sci. (2024) 20:331–46. doi: 10.7150/ijbs.82316
81. Wang Y, Wang J, Li X, Xiong X, Wang J, Zhou Z, et al. N(1)-methyladenosine methylation in trna drives liver tumourigenesis by regulating cholesterol metabolism. Nat Commun. (2021) 12:6314. doi: 10.1038/s41467-021-26718-6
82. Wang Q, Wang G, Wang Y, Liu C, He X. Association of alkb homolog 3 expression with tumor recurrence and unfavorable prognosis in hepatocellular carcinoma. J Gastroenterol Hepatol. (2018) 33:1617–25 doi: 10.1111/jgh.14117
83. Liu M, Zhao Z, Cai Y, Bi P, Liang Q, Yan Y, et al. Yth domain family: potential prognostic targets and immune-associated biomarkers in hepatocellular carcinoma. Aging. (2021) 13:24205–18. doi: 10.18632/aging.203674
84. Xia P, Zhang H, Xu K, Jiang X, Gao M, Wang G, et al. Myc-targeted wdr4 promotes proliferation, metastasis, and sorafenib resistance by inducing ccnb1 translation in hepatocellular carcinoma. Cell Death Dis. (2021) 12:691. doi: 10.1038/s41419-021-03973-5
85. Han WY, Wang J, Zhao J, Zheng YM, Chai XQ, Gao C, et al. Wdr4/trim28 is a novel molecular target linked to lenvatinib resistance that helps retain the stem characteristics in hepatocellular carcinomas. Cancer Lett. (2023) 568:216259. doi: 10.1016/j.canlet.2023.216259
86. Chen Z, Zhu W, Zhu S, Sun K, Liao J, Liu H, et al. Mettl1 promotes hepatocarcinogenesis via M(7) G trna modification-dependent translation control. Clin Trans Med. (2021) 11:e661. doi: 10.1002/ctm2.661
87. Huang M, Long J, Yao Z, Zhao Y, Zhao Y, Liao J, et al. Mettl1-mediated M7g trna modification promotes lenvatinib resistance in hepatocellular carcinoma. Cancer Res. (2023) 83:89–102. doi: 10.1158/0008-5472.Can-22-0963
88. Zhu S, Wu Y, Zhang X, Peng S, Xiao H, Chen S, et al. Targeting N(7)-methylguanosine trna modification blocks hepatocellular carcinoma metastasis after insufficient radiofrequency ablation. Mol therapy: J Am Soc Gene Ther. (2023) 31:1596–614. doi: 10.1016/j.ymthe.2022.08.004
89. Li J, Zhang H, Wang H. N(1)-methyladenosine modification in cancer biology: current status and future perspectives. Comput Struct Biotechnol J. (2022) 20:6578–85. doi: 10.1016/j.csbj.2022.11.045
90. Tong C, Wang W, He C. M1a methylation modification patterns and metabolic characteristics in hepatocellular carcinoma. BMC Gastroenterol. (2022) 22:93. doi: 10.1186/s12876-022-02160-w
91. Saikia M, Fu Y, Pavon-Eternod M, He C, Pan T. Genome-wide analysis of N1-methyl-adenosine modification in human trnas. RNA (New York NY). (2010) 16:1317–27. doi: 10.1261/rna.2057810
92. Helm M, Brulé H, Degoul F, Cepanec C, Leroux JP, Giegé R, et al. The presence of modified nucleotides is required for cloverleaf folding of a human mitochondrial trna. Nucleic Acids Res. (1998) 26:1636–43. doi: 10.1093/nar/26.7.1636
93. Kawarada L, Suzuki T, Ohira T, Hirata S, Miyauchi K, Suzuki T. Alkbh1 is an rna dioxygenase responsible for cytoplasmic and mitochondrial trna modifications. Nucleic Acids Res. (2017) 45:7401–15. doi: 10.1093/nar/gkx354
94. Roundtree IA, Evans ME, Pan T, He C. Dynamic rna modifications in gene expression regulation. Cell. (2017) 169:1187–200. doi: 10.1016/j.cell.2017.05.045
95. Zhang LS, Liu C, Ma H, Dai Q, Sun HL, Luo G, et al. Transcriptome-wide mapping of internal N(7)-methylguanosine methylome in mammalian mrna. Mol Cell. (2019) 74:1304–16.e8. doi: 10.1016/j.molcel.2019.03.036
96. Malbec L, Zhang T, Chen YS, Zhang Y, Sun BF, Shi BY, et al. Dynamic methylome of internal mrna N(7)-methylguanosine and its regulatory role in translation. Cell Res. (2019) 29:927–41. doi: 10.1038/s41422-019-0230-z
97. Azzam SK, Alsafar H, Sajini AA. Fto M6a demethylase in obesity and cancer: implications and underlying molecular mechanisms. Int J Mol Sci. (2022) 23:3800. doi: 10.3390/ijms23073800
98. Han Y, Liu D, Li L. Pd-1/pd-L1 pathway: current researches in cancer. Am J Cancer Res. (2020) 10:727–42.
99. Cui M, Qu F, Wang L, Liu X, Yu J, Tang Z, et al. M5c rna methyltransferase-related gene nsun4 stimulates Malignant progression of hepatocellular carcinoma and can be a prognostic marker. Cancer biomark: sect A Dis Markers. (2022) 33:389–400. doi: 10.3233/cbm-210154
100. Xue C, Gu X, Zheng Q, Shi Q, Yuan X, Su Y, et al. Alyref mediates rna M(5)C modification to promote hepatocellular carcinoma progression. Signal transduct target Ther. (2023) 8:130. doi: 10.1038/s41392-023-01395-7
101. Rengaraj P, Obrdlik A, Vukic D, Varadarajan NM, Keegan LP, Vanacova S, et al. Interplays of different types of epitranscriptomic mrna modifications. RNA Biol. (2021) 18:19–30. doi: 10.1080/15476286.2021.1969113
102. Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible M(6)a rna methylation. Nat Rev Genet. (2014) 15:293–306. doi: 10.1038/nrg3724
103. Akichika S, Hirano S, Shichino Y, Suzuki T, Nishimasu H, Ishitani R, et al. Cap-specific terminal N (6)-methylation of rna by an rna polymerase ii-associated methyltransferase. Science. (2019) 363:eaav0080. doi: 10.1126/science.aav0080
104. Li J, Zhu L, Shi Y, Liu J, Lin L, Chen X. M6a demethylase fto promotes hepatocellular carcinoma tumorigenesis via mediating pkm2 demethylation. Am J Transl Res. (2019) 11:6084–92.
105. Liu L, Gu M, Ma J, Wang Y, Li M, Wang H, et al. Circgpr137b/mir-4739/fto feedback loop suppresses tumorigenesis and metastasis of hepatocellular carcinoma. Mol Cancer. (2022) 21:149. doi: 10.1186/s12943-022-01619-4
106. Xue C, Zhao Y, Li G, Li L. Multi-omic analyses of the M(5)C regulator alyref reveal its essential roles in hepatocellular carcinoma. Front Oncol. (2021) 11:633415. doi: 10.3389/fonc.2021.633415
107. Liu X, Qin J, Gao T, Li C, Chen X, Zeng K, et al. Analysis of mettl3 and mettl14 in hepatocellular carcinoma. Aging. (2020) 12:21638–59. doi: 10.18632/aging.103959
108. Yang TS, Chang HK, Chen JS, Lin YC, Liau CT, Chang WC. Chemotherapy using 5-fluorouracil, mitoxantrone, and cisplatin for patients with advanced hepatocellular carcinoma: an analysis of 63 cases. J Gastroenterol. (2004) 39:362–9. doi: 10.1007/s00535-003-1303-8
109. Fiorentino F, Menna M, Rotili D, Valente S, Mai A. Mettl3 from target validation to the first small-molecule inhibitors: A medicinal chemistry journey. J medicinal Chem. (2023) 66:1654–77. doi: 10.1021/acs.jmedchem.2c01601
110. Dolbois A, Bedi RK, Bochenkova E, Müller A, Moroz-Omori EV, Huang D, et al. 1,4,9-triazaspiro[5.5]Undecan-2-one derivatives as potent and selective mettl3 inhibitors. J medicinal Chem. (2021) 64:12738–60. doi: 10.1021/acs.jmedchem.1c00773
111. Xu P, Ge R. Roles and drug development of mettl3 (Methyltransferase-like 3) in anti-tumor therapy. Eur J medicinal Chem. (2022) 230:114118. doi: 10.1016/j.ejmech.2022.114118
112. Giordano A, Tommonaro G. Curcumin and cancer. Nutrients. (2019) 11:2376. doi: 10.3390/nu11102376
113. Lu N, Li X, Yu J, Li Y, Wang C, Zhang L, et al. Curcumin attenuates lipopolysaccharide-induced hepatic lipid metabolism disorder by modification of M(6) a rna methylation in piglets. Lipids. (2018) 53:53–63. doi: 10.1002/lipd.12023
114. Peng S, Xiao W, Ju D, Sun B, Hou N, Liu Q, et al. Identification of entacapone as a chemical inhibitor of fto mediating metabolic regulation through foxo1. Sci Trans Med. (2019) 11:eaau7116. doi: 10.1126/scitranslmed.aau7116
115. Huang Y, Yan J, Li Q, Li J, Gong S, Zhou H, et al. Meclofenamic acid selectively inhibits fto demethylation of M6a over alkbh5. Nucleic Acids Res. (2015) 43:373–84. doi: 10.1093/nar/gku1276
116. Su R, Dong L, Li Y, Gao M, Han L, Wunderlich M, et al. Targeting fto suppresses cancer stem cell maintenance and immune evasion. Cancer Cell. (2020) 38:79–96.e11. doi: 10.1016/j.ccell.2020.04.017
117. Shen D, Wang B, Gao Y, Zhao L, Bi Y, Zhang J, et al. Detailed resume of rna M(6)a demethylases. Acta Pharm Sin B. (2022) 12:2193–205. doi: 10.1016/j.apsb.2022.01.003
118. Hong YG, Yang Z, Chen Y, Liu T, Zheng Y, Zhou C, et al. The rna M6a reader ythdf1 is required for acute myeloid leukemia progression. Cancer Res. (2023) 83:845–60. doi: 10.1158/0008-5472.Can-21-4249
119. Micaelli M, Dalle Vedove A, Cerofolini L, Vigna J, Sighel D, Zaccara S, et al. Small-molecule ebselen binds to ythdf proteins interfering with the recognition of N (6)-methyladenosine-modified rnas. ACS Pharmacol Trans Sci. (2022) 5:872–91. doi: 10.1021/acsptsci.2c00008
120. Cao X, Geng Q, Fan D, Wang Q, Wang X, Zhang M, et al. M(6)a methylation: A process reshaping the tumour immune microenvironment and regulating immune evasion. Mol Cancer. (2023) 22:42. doi: 10.1186/s12943-022-01704-8
121. Liu Y, Guo Q, Yang H, Zhang XW, Feng N, Wang JK, et al. Allosteric regulation of igf2bp1 as a novel strategy for the activation of tumor immune microenvironment. ACS Cent Sci. (2022) 8:1102–15. doi: 10.1021/acscentsci.2c00107
122. Ying S, Li P, Wang J, Chen K, Zou Y, Dai M, et al. Trf-gln-ctg-026 ameliorates liver injury by alleviating global protein synthesis. Signal transduct target Ther. (2023) 8:144. doi: 10.1038/s41392-023-01351-5
123. Kentsis A, Volpon L, Topisirovic I, Soll CE, Culjkovic B, Shao L, et al. Further evidence that ribavirin interacts with eif4e. RNA (New York NY). (2005) 11:1762–6. doi: 10.1261/rna.2238705
Keywords: hepatocellular carcinoma, N6-methyladenosine, 5-methylcytosine, N1-methyladenosine, N7-methylguanosine, biomarkers, therapeutic targets
Citation: Liu J-X, Zhang X, Xu W-H and Hao X-D (2024) The role of RNA modifications in hepatocellular carcinoma: functional mechanism and potential applications. Front. Immunol. 15:1439485. doi: 10.3389/fimmu.2024.1439485
Received: 28 May 2024; Accepted: 05 August 2024;
Published: 20 August 2024.
Edited by:
Zhihao Wang, Wuhan University, ChinaReviewed by:
Zhen Wang, National Institutes of Health (NIH), United StatesShengjian Huang, Chengdu Chipscreen Pharmaceutical Ltd, China
Dilara Akbulut, National Institutes of Health (NIH), United States
Copyright © 2024 Liu, Zhang, Xu and Hao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiao-Dan Hao, aGFveGlhb2RhbjE5ODdAMTYzLmNvbQ==
 Jin-Xiu Liu
Jin-Xiu Liu Xiaoping Zhang2
Xiaoping Zhang2 Wen-Hua Xu
Wen-Hua Xu Xiao-Dan Hao
Xiao-Dan Hao