- 1The National Key Laboratory of Agricultural Microbiology, College of Veterinary Medicine, Huazhong Agricultural University, Wuhan, China
- 2National Animal Tuberculosis Para-Reference Laboratory (Wuhan) of Ministry of Agriculture and Rural Affairs, Huazhong Agricultural University, Wuhan, China
- 3Department of Biomedical Sciences, College of Veterinary Medicine, Oregon State University, Corvallis, OR, United States
- 4Hubei Hongshan Laboratory, Huazhong Agricultural University, Wuhan, China
The Mycobacterium tuberculosis variant bovis (M. bovis) is a highly pathogenic environmental microorganism that causes bovine tuberculosis (bTB), a significant zoonotic disease. Currently, “test and culling” is the primary measure for controlling bTB, but it has been proven to be inadequate in animals due to their high susceptibility to the pathogen. Selective breeding for increased host resistance to bTB to reduce its prevalence is feasible. In this study, we found a vital host-dependent factor, RBMX2, that can potentially promote M. bovis infection. By knocking RBMX2 out, we investigated its function during M. bovis infection. Through transcriptome sequencing and alternative splicing transcriptome sequencing, we concluded that after M. bovis infection, embryo bovine lung (EBL) cells were significantly enriched in RNA splicing associated with apoptosis compared with wild-type EBL cells. Through protein/molecular docking, molecular dynamics simulations, and real-time quantitative PCR, we demonstrated that RBMX2 promotes the apoptosis of epithelial cells by upregulating and binding to apoptotic peptidase activating factor 1 (APAF-1), resulting in the alternative splicing of APAF-1 as a retention intron. To our knowledge, this is the first report of M. bovis affecting host epithelial cell apoptosis by hijacking RBMX2 to promote the intron splicing of downstream APAF-1. These findings may represent a significant contribution to the development of novel TB prevention and control strategies.
Introduction
Mycobacterium tuberculosis complex (MTBC) is a microorganism that has existed in the environment for thousands of years. Current research indicates that MTBC is indirectly transmitted to humans and animals in the environment, thereby affecting their health and the economics related to its prevention and control (37). In most areas of the world where tuberculosis (TB) exists, Mycobacterium tuberculosis variant bovis (M. bovis) is a member of the MTBC and the main pathogen of bovine TB (bTB), which is designated as a mandatory notifiable disease by the World Organization for Animal Health and classified as a Class II animal disease in China (1). A study demonstrated the existence of dormant MTBC in the environment and, most importantly, its ability to restore metabolic activity when growth conditions are not favorable (38). Therefore, infection with M. bovis from environmental sources may be a severe risk factor for human and animal health (39). In 2012, the Chinese government listed bTB as one of the most attention-seeking diseases for prevention and control in domestic animals (1). It is becoming a significant and challenging disease worldwide, including in China (1). The main control strategy for bTB globally is based on “test and culling” (1). However, it has proven to be insufficient in eradicating the disease due to the high susceptibility of animals to this pathogen and the development by M. bovis of advanced metabolic activities (2). The presence of bTB may be due in part to the higher number of slaughtered animals or the lower accuracy of current diagnostic methods due to high infection rates (2). Quantitative genetic studies have shown genetic variation in cattle resistance to bTB; therefore, selective breeding for increased host resistance to bTB would be a feasible way of reducing its prevalence (3).
Apoptosis, a pivotal biological process, influences the fate of pathogenic M. bovis and M. tuberculosis during host-pathogen interactions (4). It is triggered by M. bovis infection, facilitating bacterial antigen presentation, reducing viable bacteria, and promoting host cell death (5–7). In the process, M. bovis can also hijack the host’s RNA binding proteins (RBPs) and promote the alternative splicing of apoptosis-related RNAs. For example, the Rv3654c protein of M. tuberculosis can bind to polypyrimidine tract-binding protein-associated splicing factor (PSF) and splice it, thereby reducing the splicing of caspase-8 pre mRNA and regulating apoptosis (8). That is why identifying host RBPs and their target RNA splicing events after M. bovis infection may play an important role in the development of drug targets, diagnostic biomarkers, and anti-TB breeding animals.
In this study, using an embryo bovine lung (EBL) cell genome-wide gene knockout library and an M. bovis-infected-EBL cell model, we found the host-dependent factor RBMX2. Knockout of RBMX2 in EBL cells can potentially inhibit cell death after M. bovis infection. Through transcriptomic sequencing, alternative splicing enrichment analysis, molecular docking, and RT-qPCR, it was revealed that RBMX2 facilitates apoptosis in epithelial cells through the alternative splicing of APAF-1 after M. bovis infection. These findings provide a theoretical basis for elucidating the molecular mechanism by which MTBC infection can promote epithelial cell death and are expected to provide new targets and strategies for controlling TB.
Results
RBMX2 is highly expressed and promotes cell death after M. bovis infection
RBMX2 knockout and wild-type EBL cells were infected with M. bovis and M. bovis Bacillus Calmette–Guerin (BCG), respectively, at an MOI of 20:1. RT-qPCR results indicated that the expression level of RBMX2 was significantly upregulated after both M. bovis and BCG infection (Figures 1A, B).
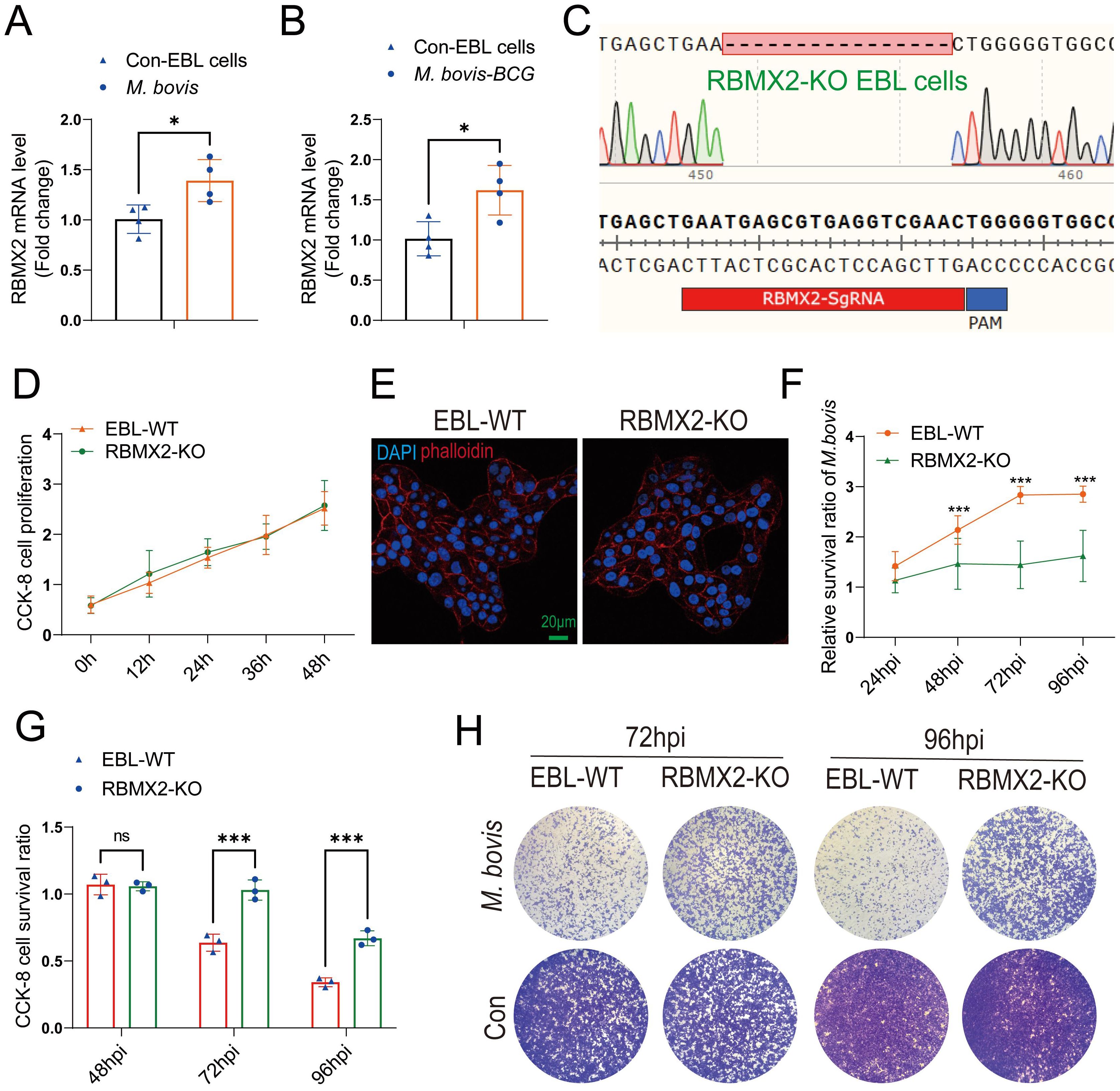
Figure 1. RBMX2 was highly expressed after M. bovis infection and inhibited EBL cell survival. (A, B) The expression of RBMX2 in EBL cells infected by (B) M. bovis (MOI, 20) and (C) M. bovis-BCG (MOI, 20) was analyzed by RT-qPCR. Data are represented as the fold expression relative to uninfected cells. (C) CRISPR-Cas9 technology was used to knockout RBMX2 in EBL cells, and sequencing was used to identify 16 knocked-out bases of RBMX2 in EBL cells. (D) Exploring the effect of RBMX2 on the proliferation of EBL cells through a CCK-8 assay. Data are represented as the absorbance value relative to wild-type (WT) EBL cells. (E) Using phalloidine to stain the EBL cell skeleton, RBMX2 was observed under a high-intensity microscope without affecting cell morphology. DAPI stains the cell nucleus. Scale bar: 20 μm. (F) The impact of M. bovis on the intracellular survival of RBMX2 knocked-out EBL cells through plate counting. Data were relative to wild-type EBL cells after M. bovis infection. (G, H) RBMX2 knockout EBL cells against M. bovis infection by a CCK-8 assay and Crystal Violet staining. Data are presented as the absorbance value relative to wild-type EBL cells after M. bovis infection (MOI 100). A t-test and two-way ANOVA were used to determine the statistical significance of differences between different groups. ns, no significance; *p < 0.05; ***p < 0.001. Data are representative of at least three independent experiments.
To investigate the phenotype and molecular mechanism of RBMX2 regulation in bovine lung epithelial cells infected with M. bovis, we used CRISPR-Cas9 technology to knockout RBMX2 in EBL cells. The sequencing results showed that RBMX2 knockout 16 bases (Figure 1C). Furthermore, a CCK-8 assay demonstrated that RBMX2 does not affect the proliferation of EBL cells (Figure 1D). In addition, to investigate whether the knockout of RBMX2 alters the morphology of EBL cells, we stained the cytoskeleton with phalloidine, and confocal microscopy results showed that RBMX2 knockout does not alter cell morphology in EBL cells (Figure 1E).
A plate-counting assay was used to investigate whether RBMX2 regulates the intracellular survival of M. bovis and found that RBMX2 knockout EBL cells significantly inhibited the intracellular survival ability of M. bovis compared with wild-type EBL cells (Figure 1F). Simultaneously, a CCK-8 assay and Crystal Violet staining uncovered that after M. bovis infection, RBMX2 knockout EBL cells had significantly higher CCK-8 cell proliferation levels than wild-type EBL cells at both 72 and 96 hours post-infection (hpi) (Figures 1G, H). These findings indicated that the mRNA expression level of RBMX2 can be induced by M. bovis and promote cell death.
RNA-seq revealed an association between RBMX2-regulated genes and apoptosis
RNA-seq analysis was conducted to determine how RBMX2 promotes cell death. A total of 173 genes showed significant variation in expression, with 52 upregulated and 121 downregulated in RBMX2 knockout EBL cells compared with WT EBL cells after M. bovis infection (Figures 2A, B) (Supplementary Table 1). The upregulated genes were primarily related to apoptosis-related pathways such as the JAK-STAT signaling, cytokine-receptor interaction, VEGF signaling, and PI3K-Akt signaling pathways (Figure 2C). Conversely, the downregulated genes were primarily associated with innate immune pathways such as the PI3K-Akt signaling and Hippo signaling pathways (Figure 2D).
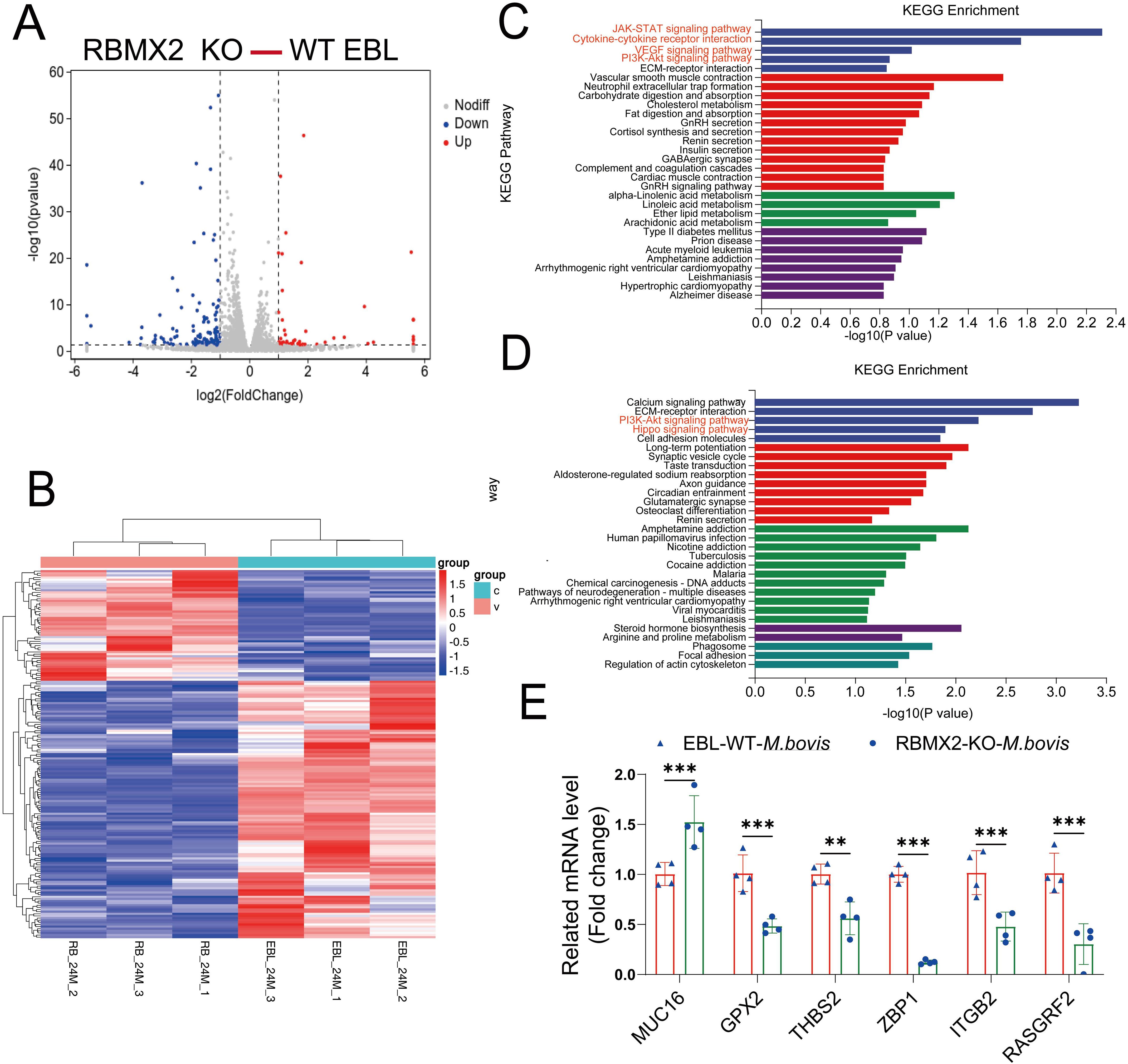
Figure 2. RBMX2-regulated genes associated with apoptosis-related pathways after infection with M. bovis. (A) The volcano plot represents differentially expressed genes in RBMX2 knockout EBL cells and wild-type (WT) EBL cells after M. bovis infection (MOI, 20) 24 hpi. Red represents upregulated genes and blue represents downregulated genes. (B) The heat map illustrates some differentially expressed genes that were all enriched in RBMX2 knockout EBL cells and wild-type EBL cells after M. bovis infection (MOI, 20) 24 hpi. Red represents upregulated genes and blue represents downregulated genes. Each group represents three independent samples. (C, D) KEGG Analysis of upregulated (C) and downregulated (D) genes in 24 hpi. Data representing upregulated and downregulated enriched pathways in RBMX2 knockout EBL cells relative to WT EBL cells after M. bovis-infection (MOI 20). A red-marked region represents the pathways that the research focuses on. (E) Detecting the mRNA expression levels of RNA-seq results using RT-qPCR, including MUC16, GPX2, THBS2, ZBP1, ITGP2, and RASGRF2. Data are represented as the relative mRNA levels in RBMX2 knockout EBL cells relative to wild-type EBL cells with M. bovis infection. A two-way ANOVA was used to determine the statistical significance of differences between different groups. **p < 0.01, ***p < 0.001. Data are representative of at least three independent experiments.
Furthermore, to verify the accuracy of transcriptome sequencing, we randomly selected six genes that regulate apoptosis. The results indicate that the transcriptome sequencing results are consistent with the RT-qPCR results (Figure 2E). Overall, RNA-seq revealed an association between RBMX2-regulated genes and apoptosis.
RBMX2 promotes apoptosis after M. bovis infection
Based on the transcriptome sequencing results, we detected the cell apoptosis level and lactate dehydrogenase (LDH) release in RBMX2 knockout EBL cells and wild-type EBL cells after infection with M. bovis. Under confocal imaging, apoptotic cells were stained red via a terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling assay (TUNEL). There was a stronger red fluorescence in wild-type EBL cells than in RBMX2 knockout EBL cells (Figure 3A). In addition, there was a significant decrease in the release of LDH in RBMX2 knockout EBL cells after infection with M. bovis compared with wild-type EBL cells via an LDH assay (Figure 3B), which meant RBMX2 promotes a high level of apoptosis after M. Bovis infection.
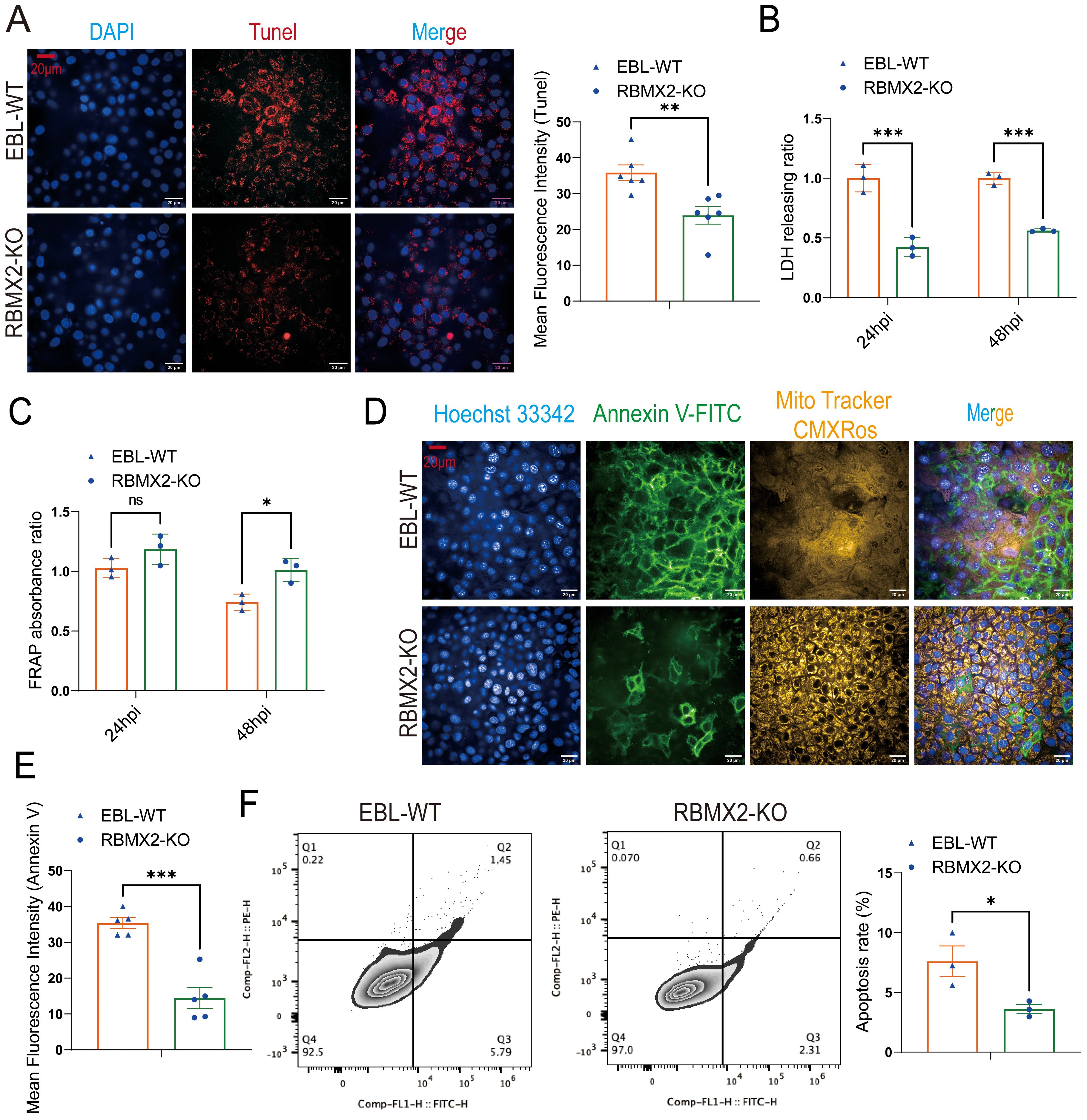
Figure 3. Host factor: RBMX2 promotes apoptosis after M. bovis infection. (A) A TUNEL assay was used to detect the level of cell apoptosis. Scale bars: 20 μm. Data are presented as the red fluorescence intensity in RBMX2 knocked-out EBL cells relative to wild-type EBL cells with M. bovis infection. (B) An LDH assay was used to detect the level of cell apoptosis. Data are presented as the LDH releasing ratio in RBMX2 knocked-out EBL cells relative to wild-type EBL cells with M. bovis infection. (C) A FRAP assay was used to detect the level of antioxidant capacity. Data are presented as the antioxidant capacity in RBMX2 knockout EBL cells relative to wild-type EBL cells with M. bovis infection. (D, E) A high-content imaging assay was used to detect the level of Annexin V-FITC and ROS. Scale Bar: 20μm. Data are presented as the level of mitochondrial membrane potential and ROS relative in RBMX2 knockout EBL cells to wild-type EBL cells with M. bovis infection. (F) Flow cytometry was used to detect the level of Annexin V. Data are presented as the level of the apoptosis rate relative in RBMX2 knockout EBL cells to wild-type EBL cells with M. bovis infection. A t-test and two-way ANOVA were used to determine the statistical significance of differences between different groups. ns, no significance, *p < 0.05; **p < 0.01; ***p < 0.001. Data are representative of at least three independent experiments.
A ferric ion-reducing antioxidant power (FRAP) experiment was conducted to determine whether RBMX2 increases apoptosis by modulating the host’s antioxidant stress capacity following M. bovis infection. Findings showed that RBMX2 knockout EBL cells had a prominent promotion in antioxidant capacity compared with wild-type EBL cells after infection with M. bovis (Figure 3C).
M. bovis BCG is the attenuated strain of M. bovis, and now it is the only official licensed vaccine for human TB. Therefore, we need to find more antigenic markers for controlling TB. To determine whether different virulent strains of M. bovis have different effects on the regulation of apoptosis by RBMX2, we infected RBMX2 knockout EBL cells and wild-type EBL cells with M. bovis BCG and detected the changes in Annexin V-FITC and CMXRos through high-intensity microscopy and flow cytometry. Under the high-content confocal cell imaging analysis system, the membrane of apoptotic cells was stained bright green by Annexin V-FITC, while normal mitochondria were stained bright yellow by Mito Tracker CMXRos. Based on the high mitochondrial membrane potential in normal cells, the results showed green solid and obscure faint yellow signals in wild-type EBL cells, which uncovered that RBMX2 can promote apoptosis after infection with both virulent M. bovis and avirulent M. bovis BCG (Figures 3D–F).
RBMX2 promotes apoptosis through the caspase-3 pathway after M. bovis infection
The above comprehensive assays confirmed that M. bovis infection can promote RBMX2 upregulation and induce EBL cell apoptosis. To determine the specific mechanism, we examined the levels of apoptosis-related genes (BAX, APAF-1, TNF-α, Caspase-9, Caspase-3, and BCL-2) in EBL cells following infection with M. bovis. RT-PCR results showed that RBMX2 knockout cells exhibited a significant downregulation of pro-apoptotic genes (BAX, APAF-1, TNF-a, Caspase-9, and Caspase-3), whereas BCL-2 was notably upregulated (Figure 4A). Furthermore, RBMX2 knockout EBL cells exhibited reduced expression of pro-apoptotic proteins (BAX and cleaved-caspase-3) and an increase in PARP. Collectively, our results underscore the significance of RBMX2 in regulating the apoptotic response to M. bovis infection; its knockout led to the suppression of apoptosis through the inhibition of caspase-3 cleavage (Figure 4B).
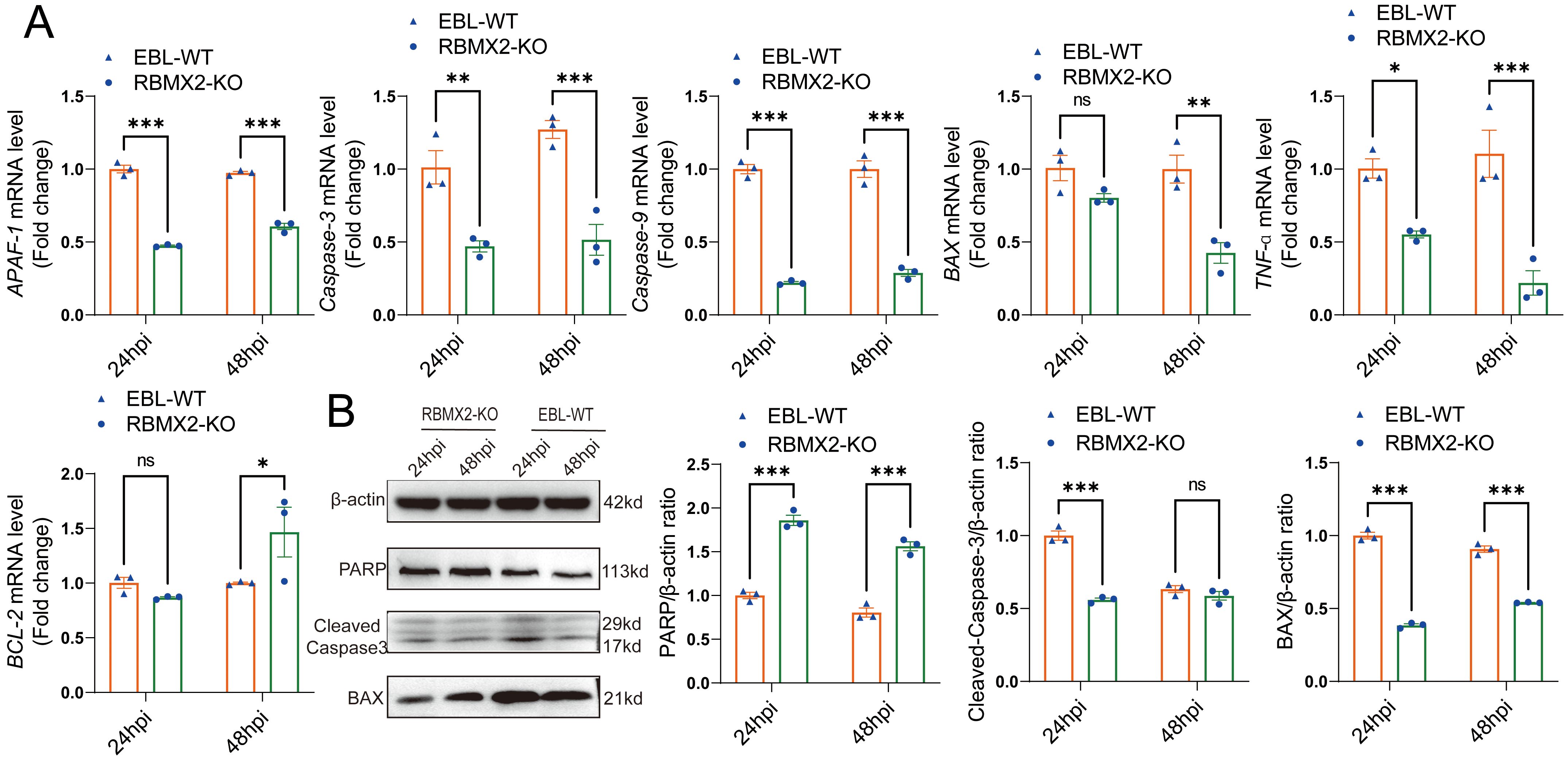
Figure 4. Host factor RBMX2 promotes apoptosis through the caspase-3 pathway after M. bovis infection. (A) Detecting the mRNA expression levels of key apoptotic regulators using RT-qPCR, including BAX, APAF-1, TNF-a, caspase-9, caspase-3, and BCL-2. Data are represented as the relative mRNA level in RBMX2 knockout EBL cells relative to wild-type (WT) EBL cells with M. bovis infection. (B) Assessing the protein expression levels of key apoptotic regulators using western blotting, including BAX, PARP, and cleaved-caspase-3. Data are presented as the relative protein level in RBMX2 knocked-out EBL cells relative to wild-type EBL cells with M. bovis infection. Two-way ANOVA was used to determine the statistical significance of differences between different groups. ns, no significance, *p < 0.05; **p < 0.01; ***p < 0.001. Data are representative of at least three independent experiments.
RBMX2 regulates RNA alternative splicing related to apoptosis after M. bovis infection
M. tuberculosis and its bovine variant strain M. bovis are specialized intracellular bacteria that have evolved complex and sophisticated strategies to evade host surveillance systems (9, 10). It has been found that M. tuberculosis and M. bovis can influence host cell-RBP-mediated RNAs by alternative splicing related to apoptosis. RBMX2, as an RBP, plays an important role in alternative splicing. However, it is still unknown which RNAs are enriched by alternative splicing to promote apoptosis after M. bovis infection. Through transcriptome sequence analysis, we found that 241 RNAs were involved in five different alternative splicing patterns in RBMX2 knockout EBL cells compared with wild-type EBL cells, among which 63 RNAs undergo variable 3’ (A3SS) and 5’ splicing (A5SS), 146 RNAs undergo exon skipping (SE), 10 RNAs undergo intron retention (RI), and 22 RNAs undergo mutually exclusive exon (MXE) splicing (Figure 5A). KEGG pathway analysis revealed that the A3SS-related RNAs were related to the Notch signaling and FoxO signaling pathways (Figure 5B). A5SS-related RNAs were linked to the MAPK signaling and PI3K-Akt signaling pathways (Figure 5C), and RNAs involved in SE were annotated in the FoxO signaling and mTOR signaling pathways (Figure 5D). RI-related RNAs were annotated in the apoptosis and the mTOR signaling pathways (Figure 5E), and the MXE-related RNAs were annotated in the MAPK signaling, TNF signaling, and cGMP-PKG signaling pathways (Figure 5F). As all the pathways mentioned above are potentially related to apoptosis, it was speculated that RBMX2 induces apoptosis through the promotion of alternative splicing after M. bovis infection.
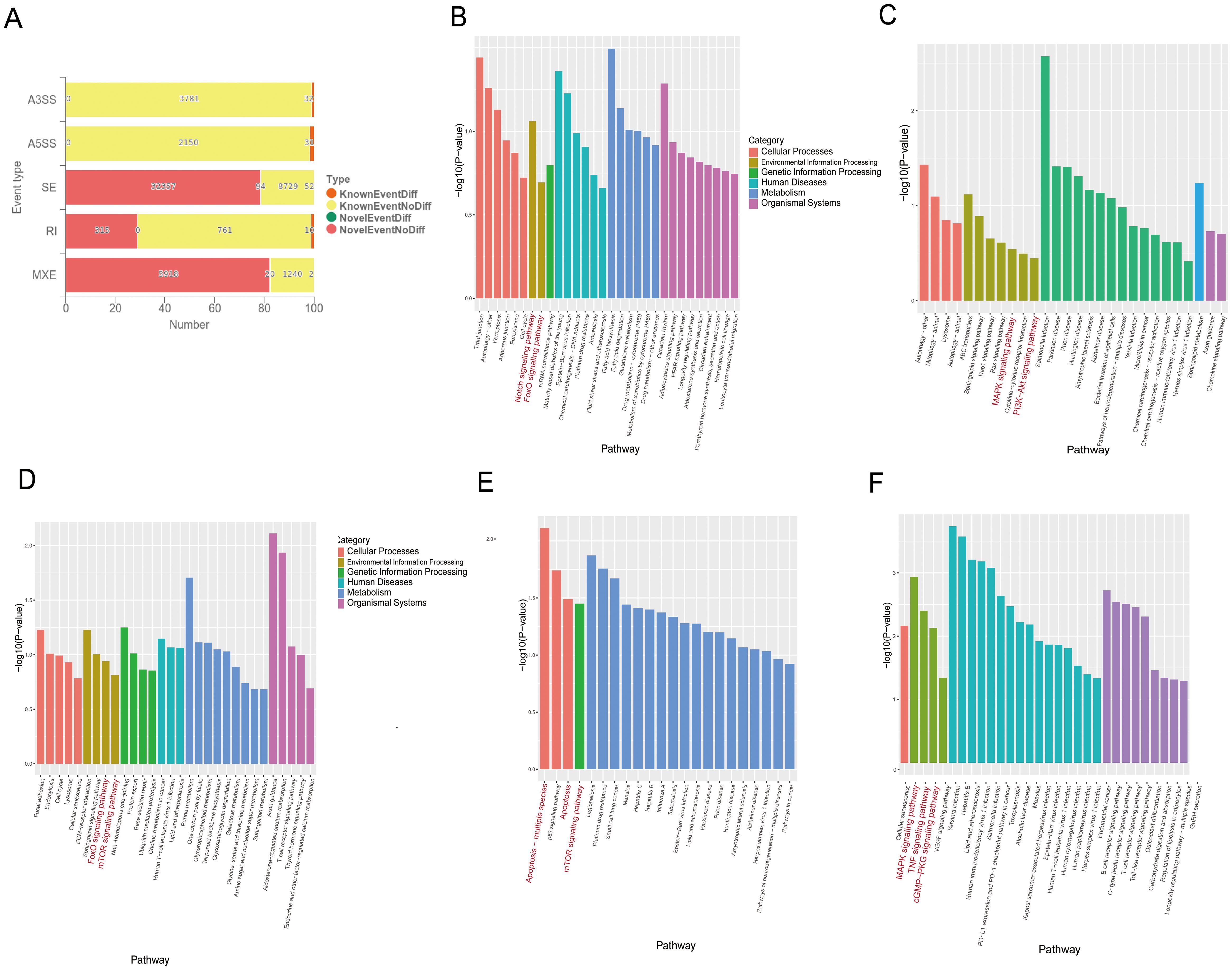
Figure 5. RBMX2 regulates RNA alternative splicing associated with apoptosis after M. bovis infection. (A) Number of different RNAs with five different alternative splicing events occurring after knockout RBMX2 in EBL cells compared with wild-type (WT) EBL cells after M. bovis infection. (B–F) KEGG analysis of RNAs involved in events such as 3′ (B) and 5′ splicing (C), exon skipping (D), intron retention (E), and mutually exclusive exons (F) in RBMX2 knockout EBL cells relative to wild-type EBL cells with M. bovis infection.
RBMX2 promotes apoptosis by regulating the alternative splicing of APAF-1’s RI variant
APAF-1 is a core apoptotic factor that promotes apoptosis through apoptosome formation and by activating caspase-3 (11). RNA-seq shows that APAF-1 is listed among the 241 alternative splicing-related RNAs. Through protein-protein docking, we found that the 73 ASN and 96 SER sites of APAF-1 tightly bind to the 78 GLN and 106 ASP sites of RBMX2 (Figure 6A).
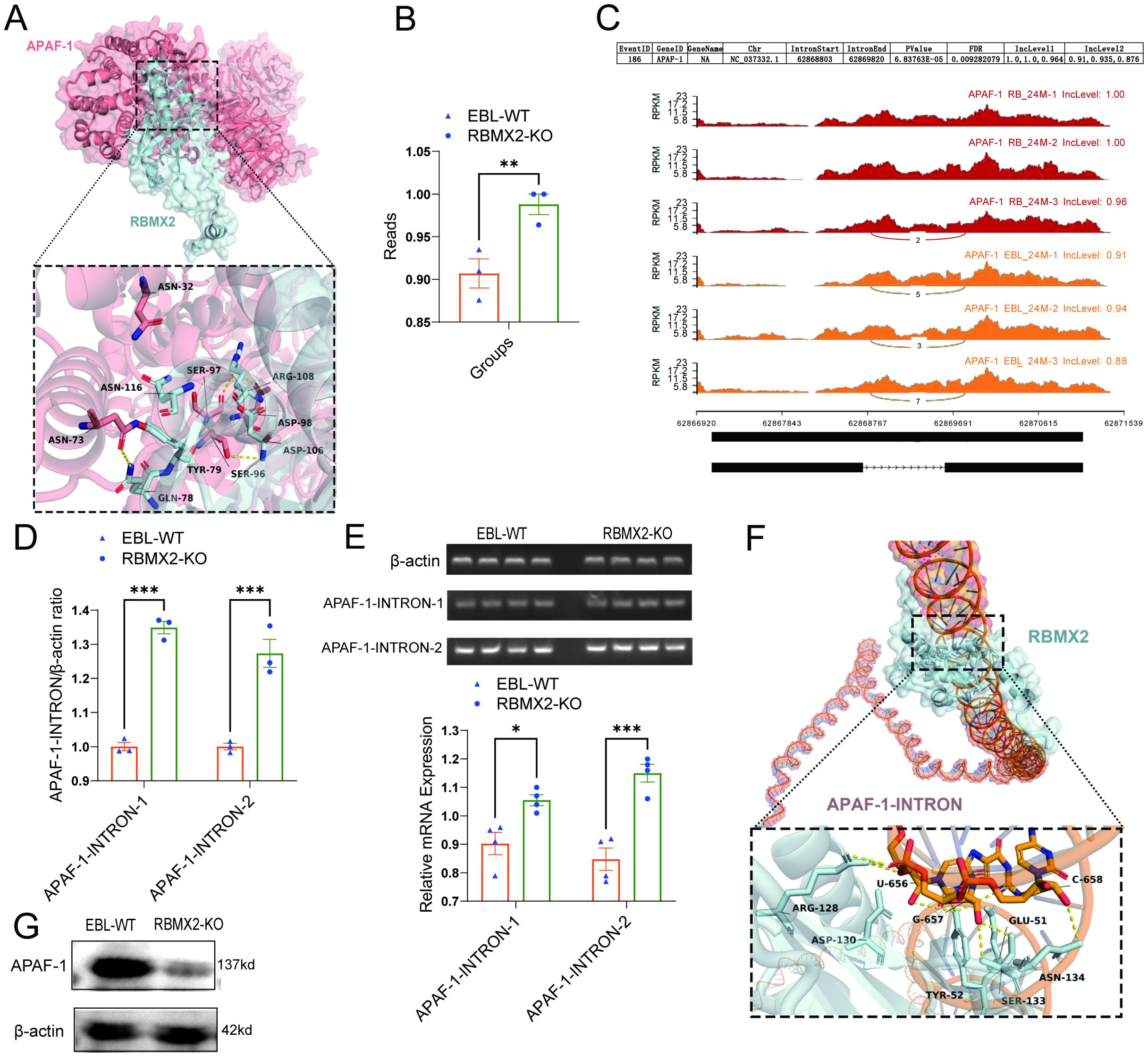
Figure 6. RBMX2 promotes apoptosis in epithelial cells by regulating the alternative splicing of APAF-1’s RI isoform. (A) Conformational diagram of binding sites between RBMX2 protein and APAF-1 protein. (B) The reads of retaining introns in AFAP1 in RBMX2 knockout EBL cells compared with wild-type (WT) EBL cells after M. bovis infection. (C) Sequencing APAF-1 RNA in RBMX2 knocked-out EBL cells compared with wild-type EBL cells after M. bovis infection and visualizing its splicing patterns using software such as rMATS. (D, E) Design of two pairs of primers targeting the intron region (62868803-62869820) of APAF-1 RNA and using RT-qPCR to detect this intron region in RBMX2 knockout EBL cells and wild-type EBL cells after M. bovis infection. (F) The conformational diagram of the binding site between RBMX2 protein and APAF-1 retained introns. (G) Detecting the protein expression of APAF-1 using a western blot assay. Data are presented as the relative protein level in RBMX2 knockout EBL cells relative to wild-type EBL cells with M. bovis infection. A t-test and two-way ANOVA were used to determine the statistical significance of differences between different groups. *p < 0.05; **p < 0.01; ***p < 0.001. Data are representative of at least three independent experiments.
By using rMATS software, APAF-1 (NC_037332.1) presented a higher RI within the region 62868803-62869820 in RBMX2 knockout EBL cells than in wild-type EBL cells (Figures 6B, C). To verify the bioinformatic results, we employed an RT-qPCR assay to detect the intron region of APAF-1. The results indicated that RBMX2 knockout EBL cells and significantly increased the expression of the 62868803-62869820 intronic sequence compared with wild-type EBL cells (Figures 6D, E). By using the protein-RNA docking technique, we predicted the structural interaction between RBMX2 protein and APAF-1 RNA. We identified multiple spatial sites in the RBMX2 protein and APAF-1 intron regions, such as 656U, 657G, and 658C (Figure 6F). In RBMX2 knockout EBL cells, APAF-1 was significantly decreased after M. bovis infection (Figure 6G). Along with it, RBMX2 can promote epithelial cell apoptosis after M. bovis infection by regulating the alternative splicing of the apoptosis-related RNA of APAF-1.
Discussion
bTB, which is primarily caused by Mycobacterium bovis (12), poses a significant threat to both animal and human health and has serious implications for the environment, agriculture, and economic development worldwide (13). In domestic and wild animals, the transmission of M. bovis may occur through aerosols, the inhalation of infectious droplets, or infected body fluids or tissues (40). M. bovis serves as an important environmental pathogenic microorganism (41); the failure of “test and culling” in many countries, including China, highlights the need for more targeted prevention and control strategies (14).
Host-dependent factors with anti/resistant disease ability play a crucial role in the control of M. bovis. In domestic and wild animals, selective breeding with resistant genes against different pathogens may prove to be a sustainable way of controlling infection. (15, 16). These host-dependent factors can be regulated through genetic improvement and gene editing, contributing to the breeding of animals with higher resistance to specific infectious diseases (17, 18). A previous study reported that double knockout of host-dependent factors CD163 and pAPN in pigs confers resistance to PRRSV and TGEV (19). Transcription activator-like effector (TALE) nickase-mediated SP110 knockout bestows cattle a higher resistance to TB (20). Therefore, the exploration and use of M. bovis host-dependent factors provide crucial theoretical and technical support for TB-resistant breeding, leading to the breeding of animal varieties with increased TB resistance and improving the health and economic benefits of the livestock industry.
RBMX2, an RNA-binding motif, plays an important role in mRNA splicing via spliceosomes; data from past studies showed that it is regarded as an important molecular marker for assessing sperm activity (21). RBMX2 also serves as an X-chromosome-specific gene in telomeric cells (42).
In this study, we found that RBMX2 was increased after M. bovis infection and can lead to the apoptosis of infected EBL cells. To our knowledge, this is the first investigation that reported and highlighted the function of RBMX2 in M. bovis infection.
Apoptosis, or programmed cell death, is a fundamental biological process that plays a dual role in host defense against pathogens (22–24). Apoptosis facilitates the elimination of intracellular pathogens by promoting their engulfment and phagocyte degradation (25). M. bovis/M. tuberculosis infection may promote host cell apoptosis through various mechanisms, such as disrupting the intracellular environment and activating apoptosis-related pathways (26). M. bovis infection can lead to inflammation and tissue damage, increase the activity of immune cells, and promote apoptosis to combat the infection (26–28). Here, we found that RBMX2 was increased after M. bovis infection, leading to significant apoptosis. This indicated that M. bovis induced host cells to express more RBMX2 to induce cell death, through which they combat the infection.
RNA alternative splicing (AS), mediated by RBPs, is a highly regulated process that plays a pivotal role in cellular homeostasis and stress response (29). RBPs can modulate gene expression by regulating AS patterns, thereby influencing diverse biological processes (30). Very few studies have reported how bacteria utilize host RBPs to regulate the alternative splicing (AS) of target RNAs to promote infection and pathogenicity. Limited studies were found to mainly focus on Escherichia coli and Helicobacter pylori infection (31, 32), and reported the enrichment of phosphorylated proteomic RNA and alternative splicing after H. pylori infection in gastric cells (33). However, how M. tuberculosis/M. bovis utilizes host RBPs to regulate the AS of target RNAs to promote infection and pathogenicity has rarely been reported so far (34, 35). In this study, RBMX2 was proven to promote apoptosis through AS and was especially involved with APAF-1, a critical regulator of apoptosis (36).
Furthermore, we observed higher RI within the 62868803-62869820 region of the APAF-1 (NC_037332.1) transcript in RBMX2 knockout EBL cells than in WT EBL cells after M. bovis infection, suggesting a potential molecular mechanism by which RBMX2 modulates to apoptosis through APAF-1 splicing. Protein-protein docking and molecular dynamics simulations further supported the interaction between RBMX2 and APAF-1, highlighting the functional relevance of their association in regulating apoptosis. However, the precise mechanism through which RBMX2 modulates APAF-1 splicing and its impact on M. bovis pathogenesis must be further investigated. However, owing to the fact that M. bovis is a high-grade pathogen, many experiments have not yet been well conducted, such as live-cell imaging and flow cytometry. Therefore, in this article, we partially used attenuated BCG to infect bovine lung epithelial cells to explore the mechanism of RBMX2-mediated apoptosis. In addition, RBMX2, as a global splicing regulatory factor, plays an important role in the host infection of M. bovis. In this article, we only focus on studying and discussing its related mechanisms for mediating apoptosis; other enriched pathways and downstream factors will be the focus of our future work.
In summary, we identified RBMX2 as a key regulator of apoptosis after M. bovis infection and systematically validated that M. bovis hijacks the upregulation of RBMX2 expression and promotes the cleavage of APAF-1 introns, increasing its translation, and then regulates host epithelial cell apoptosis after M. bovis infection. Our findings may provide a novel insight into the molecular mechanisms underlying host-pathogen interactions during M. bovis infection and may point to a new approach to bTB control through selective breeding.
Conclusion
This study identified a novel host factor, RBMX2, implicated in the promotion of cell death after M. bovis infection and the facilitation of apoptosis in epithelial cells through the AS of APAF-1, which activates caspase-3 by forming the apoptosome (Figure 7). These findings provide a new target for the prevention and control of TB based on the genomics of host anti-TB.
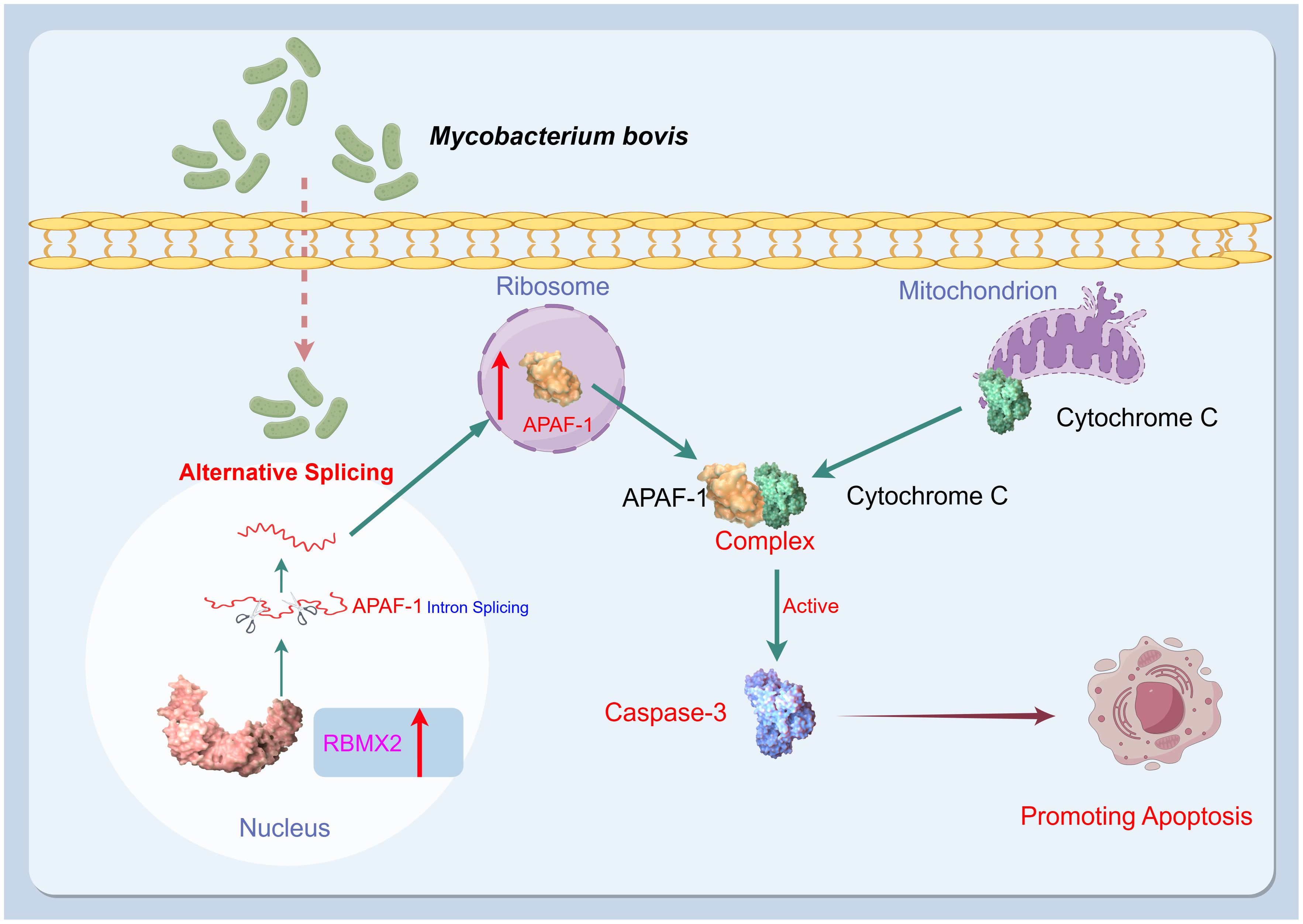
Figure 7. Mechanism diagram. After infection with M. bovis, RBMX2 is upregulated in EBL cells. RBMX2 then increases the translation of APAF-1 RNA and expression of APAF-1 protein by downregulating its retained intron, thereby improving its binding to cytochrome C. Finally, the APAF-1 and cytochrome C complexes activate caspase-3 to promote apoptosis in epithelial cells following M. bovis infection.
Materials and methods
Cell lines
EBL cells were kindly provided by M. Heller from the Friedrich-Loeffler Institute, Germany. These cells were grown in heat-inactivated 10% fetal bovine serum supplemented with Dulbecco’s modified Eagle medium (Gibco, USA) at 37°C and 5% CO2.
Bacterium culture and infection
M. bovis [American Type Culture Collection (ATCC): 19210] was kindly provided by Dr. Chuanyou Li (Beijing Tuberculosis and Thoracic Tumor Research Institute, Beijing, China). M. bovis BCG-Pasteur (ATCC: 35734) was a gift from Professor Luiz Bermudez (Oregon State University). All strains were cultured in a Middlebrook 7H9 broth (BD, MD, USA) containing 0.5% glycerol (Sigma-Aldrich, MO, USA), 10% oleic acid-albumin-dextrose-catalase (OADC, BD), and 0.05% Tween 80 (Sigma-Aldrich) or on Middlebrook 7H11 agar plates (BD, MD, USA) containing 0.5% glycerol (Sigma-Aldrich) and 10% OADC (BD). Before infection, the optical densities at 600 nm (OD600) of the bacterial culture were adjusted to the required MOI using the standard turbidimetric method. The cultures were then centrifuged at 3,000 × g for 10 min. The precipitated bacteria were resuspended in a medium and dispersed by passage through an insulin syringe. An aliquot of 50 μl of the 10-fold serially diluted bacterial suspension was plated onto Middlebrook 7H11 agar (BD) to count viable bacteria (colony-forming units, CFUs).
For the infection, both M. bovis and M. bovis BCG were used to infect EBL cells at MOIs of 20 (for apoptosis detection) or 100 (for cell viability).
All M. bovis-related experiments in this study were conducted strictly in accordance with the biosafety-related operating procedures at the Animal Biosafety Level 3 Laboratory of the National Key Laboratory of Agricultural Microbiology at Huazhong Agricultural University.
Extraction of total RNA and RT-qPCR
Cold phosphate-buffered saline (PBS, HyClone, China) was used to wash the cells three times, and 1 ml of Trizol (Invitrogen, USA) was added per well. The cells were lysed and collected in Eppendorf tubes. In addition, 200 μl of chloroform was added, followed by vortexing for 30 s and centrifugation at 12,000 rpm at 4°C for 10 min. Afterward, 500 μl of the supernatant was collected in a new Eppendorf tube. Then, 500 μl of isopropanol was added to the collected supernatant, followed by gradually mixing upside down. The mixture was allowed to stand for 10 min at 4°C, followed by centrifugation at 12,000 rpm at 4°C for 15 min. The supernatant was removed, and the RNA pellet was visible. Next, 1 ml of 75% ethanol was added to wash the RNA, followed by centrifugation at 7,500 rpm at 4°C for 5 min. The supernatant was removed, and the RNA was dried for 15 min. Subsequently, 20 μl of DEPC water was added, and the mixture was incubated at 58°C in a water bath for 10 min. Finally, purified RNA was obtained.
As a quality control index of RNA purity, OD260/OD280 values between 1.8 and 2.0 were determined using a NanoDrop ND-1000 instrument (Agilent Inc., USA). Denaturing agarose gel electrophoresis was used to measure RNA integrity and contamination with gDNA. The sample was stored at −80°C for further analysis.
Reverse transcription of the RNA samples was performed using HiScript III RT SuperMix for qPCR (+gDNA wiper, Vazyme, China). A volume of 4 μl of 4 × gDNA wiper mix was added to 1 μg of RNA, which was then added to 16 μl of RNAse-free ddH2O. The mixture was incubated at 42°C for 2 min to remove genomic DNA. Reverse transcription was performed by adding 4 μl of 5 × HiScript III qRT SuperMix. The mixture was incubated at 37°C for 15 min and then at 85°C for 5 s to obtain cDNA.
cDNA expression in different groups of samples was detected using AceQ qPCR SYBR Green Master Mix (Vazyme, China) in a ViiA7 real-time PCR machine (Applied Biosystems Inc, USA). The final volume of the real-time PCR reaction was 20 μl, including 10 μl of 2 × AceQ qPCR SYBR Green Master Mix, 0.4 μl of upstream primer (10 μM), 0.4 μl of downstream primer (10 μM), 0.4 μL of 50 × ROX reference dye 2, 3 μl of cDNA template, and 5.8 μl of ddH2O. The reaction was completed as follows: 95°C for 5 min (1 cycle), 95°C for 10 s (40 cycles), and 60°C for 30 s (40 cycles). Supplementary Table 1 shows the RT-qPCR primer sequences.
Western blot
Radioimmunoprecipitation assay reagent (Sigma-Aldrich, USA) containing protease inhibitors and phosphatase inhibitors (Roche, China) was added to the cell samples, and the cells were lysed for 30 min on ice. The supernatant was collected by centrifugation at 12,000 rpm at 4°C for 10 min. A bicinchoninic acid kit (Beyotime, China) was used to detect the protein concentration of each sample, and the proteins were adjusted to equal concentrations. The protein samples were mixed with 5 × protein loading buffer and boiled for 10 min. Proteins were separated using SDS-PAGE on 10% polyacrylamide gels at 100 V for 90 min and then transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, Germany) at 100 V for 70 min. The PVDF membrane was placed in 5% skimmed milk and sealed for 4 h. The membrane was washed three times with Tris-buffered saline with 0.15% Tween-20 for 5 min. Then, the membrane was incubated with a primary antibody [cleaved-caspase3 (Bioss, China), PARP (Proteintech, China), BAX (Proteintech, China), APAF-1 (Bioss, China), and β-actin (Bioss, China)] at 4°C for 12h followed by a secondary antibody [anti-mouse HRP secondary antibody and anti-rabbit HRP (Invitrogen, USA)] at room temperature for 1 h.
Generation of the RBMX2-KO EBL cells
The small guide RNA (sgRNA) sequence targeting the bovine RBMX2 gene (5′- GAATTGCGGGGTCGAAC-3′) was cloned into pKLV2-U6gRNA5 (BbsI)-PGKpuro2ABFP (# 67991) (gifted by Professor Shuhong Zhao of Huazhong Agricultural University), and a recombinant lentivirus was constructed using a three plasmid helper system (PMD2. G and psPAX2 helper plasmids gifted by Professor Shuhong Zhao of Huazhong Agricultural University). EBL cells were infected with the RBMX2 pKLV2−U6gRNA5(BbsI)−PGKpuro2ABFP (#67991) lentivirus or the empty vector pKLV2−U6gRNA5(BbsI)−PGKpuro2ABFP (#67991) lentivirus (negative control). After 48 h of infection, puromycin (2.0 mg/ml) was added to select positive clone cells. Finally, the monoclonal knockout cells obtained through limited dilution were amplified, and the knockout sites were identified using Sanger sequencing (Supplementary Table S2).
Assay of intracellular bacterial survival
EBL cells were infected with M. bovis at 37°C with a MOI of 20 for 2 h. Then, the cells were washed three times with sterile 1×PBS to remove extracellular bacteria. This time point is considered as 0 h; cells in a complete culture medium containing 100 µg/ml of gentamicin were cultivated for different durations (24 h, 48 h, 72 h, and 96 h). Afterward, the cell lysate was continuously diluted and spread onto a 7H11 agar plate supplemented with 10% OADC, and CFUs were calculated after 3 weeks of cultivation. Each sample was plated three times.
Alternative splicing RNA-Seq
Total RNA was isolated using Trizol Reagent (Invitrogen Life Technologies), and the concentration, quality, and integrity were determined using a NanoDrop spectrophotometer (Thermo Scientific). Three micrograms of RNA were used as input material for RNA sample preparation. Sequencing libraries were generated according to the following steps. mRNA was purified from total RNA by using poly-T oligo-attached magnetic beads. Fragmentation was performed using divalent cations under elevated temperature in an Illumina proprietary fragmentation buffer. First-strand cDNA was synthesized using random oligonucleotides and Super Script II. Second-strand cDNA synthesis was subsequently performed using DNA Polymerase-I and RNase H. The remaining overhangs were converted into blunt ends via exonuclease/polymerase activities, and the enzymes were removed. Illumina PE-adapted oligonucleotides were ligated for hybridization after adenylation of the 3′ ends of the DNA fragments. To select cDNA fragments of the preferred length of 400–500 bp, the library fragments were purified using the AMPure XP system (Beckman Coulter, Beverly, CA, USA). DNA fragments with ligated adaptor molecules at both ends were selectively enriched using the Illumina PCR Primer Cocktail in a 15-cycle PCR reaction. The products were purified (AMPure XP system) and quantified using the Agilent high-sensitivity DNA assay on a Bioanalyzer 2100 system (Agilent Inc., USA). The sequencing library was then sequenced on the NovaSeq 6000 platform (Illumina) by Shanghai Personal Biotechnology Cp. Ltd.
The RNA Sequencing service was provided by Personal Biotechnology Co., Ltd., Shanghai, China. The data were analyzed using the free online platform Personalbio GenesCloud (https://www.genescloud.cn). Furthermore, we randomly selected genes related to apoptosis for validation. Supplementary Table S2 shows the RT-qPCR primer sequences.
TUNEL assay
The TUNEL assay was performed to detect apoptotic cells after infection. Cell samples were prepared and subjected to deoxyribonucleotide triphosphates (dNTPs) blocking to minimize the background signals. The TUNEL reaction mixture, consisting of terminal deoxynucleotidyl transferase (TdT) and labeled deoxyuridine triphosphate (dUTP), was applied to the samples and incubated under humid conditions for 30 min to 1 h. Subsequently, samples were washed with PBS to remove unincorporated labeled dUTP. Optionally, selective nuclear staining was performed to visualize nuclear morphology. Finally, samples were observed using a fluorescence microscope to detect TUNEL-positive cells. Experimental procedures were carried out strictly according to the manufacturer’s instructions provided with the TUNEL assay kit (Beyotime, China).
Apoptosis detection assay via a confocal high-content cell imaging system
After infection, a confocal high-content cell imaging analysis system was used to determine the mitochondrial membrane potential according to the instructions provided with the mitochondrial membrane potential and apoptosis detection kit (Beyotime, China). Cells were analyzed using a confocal high-content cell imaging analysis system (PerkinElmer Life And Analytical Sciences Ltd, UK).
Apoptosis detection assay using a flow cytometer
Cold PBS was used to wash the cell samples three times after infection. Cells were digested by trypsin, and the digestion was terminated with DME/F-12 medium containing 10% FBS. Cells were collected by centrifugation at 1,500 rpm for 5 min. The samples were washed three times in PBS by centrifugation at 1,500 rpm for 5 min. The supernatant was discarded. An Annexin V-FITC Apoptosis Detection Kit (Beyotime, China) was used to detect apoptosis. An aliquot of 100 μl of binding buffer was added to resuspend the cell. EBL cells were stained with Annexin V-FITC and propidium iodide for 10 min before being detected by CytoFLEX-LX (Becton Dickinson, USA).
LDH assay
LDH activity was determined using a commercially available assay kit (Beyotime, China). Cell samples were homogenized in ice-cold lysis buffer after infection, followed by centrifugation to collect the supernatant. LDH activity was measured by monitoring the decrease in absorbance at 340 nm over time, reflecting the rate of NADH oxidation. Protein concentrations were quantified using the Bradford assay, and LDH activity was normalized to protein content.
Cell viability assay
EBL cells were infected with M. bovis at an MOI of 100. Cell viability was then detected at different hours post-infection using CCK-8 (Dojindo, Kumamoto, Japan). Briefly, at 48, 72, and 96 hpi, fresh medium containing 10% CCK-8 was added to a 96-well cell plate and incubated for 60 min at 37°C. The cell viability was then determined as the absorbance at 450 nm.
Protein-protein docking assay
We obtained the protein structures of APAF-1 and RBMX2 from the UniProt database (https://www.uniprot.org/) and evaluated the interaction pattern between APAF-1 and RBMX2 using Z-dock. Pymol 2.3.0 (https://pymol.org/) was used to analyze the interaction mode of the docking results.
Bioinformatics analysis of RBMX2 protein and APAF-1 RNA interaction
Sequence Retrieval and Alignment: the amino acid sequence of the RBMX2 protein and the RNA sequences of APAF-1 were retrieved from the National Center for Biotechnology Information database (https://www.ncbi.nlm.nih.gov/). Multiple sequence alignments of RBMX2 protein sequences and APAF-1 RNA sequences from various species were performed using Clustal Omega software (https://www.ebi.ac.uk/Tools/msa/clustalo/).
Structural Prediction: the secondary structure of APAF-1 RNA was predicted using the RNAfold web server (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi). The tertiary structure of the RBMX2 protein was predicted using I-TASSER (https://zhanglab.ccmb.med.umich.edu/I-TASSER/).
Statistical analysis
Statistical analysis involved conducting all assays in triplicate and expressing the data as the mean ± standard error with three independent repetitions of all experiments. Statistical analyses were performed using GraphPad Prism 7.0, with t-tests to compare two groups and two-way ANOVA to compare multiple variables.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA1105205.
Author contributions
CW: Writing – review & editing, Writing – original draft, Methodology, Data curation, Conceptualization. YJ: Writing – original draft, Investigation, Data curation, Conceptualization. ZY: Writing – original draft, Validation, Investigation, Data curation, Conceptualization. HX: Writing – review & editing, Methodology, Investigation, Data curation, Conceptualization. AK: Writing – review & editing, Methodology, Investigation, Data curation, Conceptualization. TI: Writing – review & editing, Methodology, Investigation, Data curation, Conceptualization. YP: Writing – original draft, Investigation, Conceptualization. LL: Writing – review & editing, Methodology, Investigation, Data curation, Conceptualization. LZ: Writing – review & editing, Project administration, Methodology, Investigation, Formal Analysis, Data curation, Conceptualization. LB: Writing – review & editing, Investigation, Data curation, Conceptualization. AG: Writing – review & editing, Supervision, Investigation, Funding acquisition, Conceptualization. YC: Writing – review & editing, Supervision, Software, Methodology, Investigation, Funding acquisition, Data curation, Conceptualization.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Key Research and Development Program of China (2021YFD1800403), the National Natural Science Foundation of China (32072942), and the China Agriculture Research System of MOF.
Acknowledgments
We would like to thank the National Key Laboratory of Agricultural Microbiology Core Facility for assistance in high-throughput microscopy, and we are grateful to Zhe Hu for his support in the sample preparation. At the same time, I would like to thank Dr. Taufik Walhidayah and Dr. Shirani Ihsanullah for their help in modifying the language and formatting of the article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1431207/full#supplementary-material
Supplementary Table 1 | RT-qPCR primer sequences.
Supplementary Table 2 | RBMX2 sgRNA sequence.
Supplementary Table 3 | RT-qPCR primer sequences.
Abbreviations
M. bovis, Mycobacterium tuberculosis variant bovis; TB, Tuberculosis; MTBC, Mycobacterium tuberculosis complex; WOAH, World Organization for Animal Health; RBMX2, RNA-binding motif protein X-linked 2; RBPs, RNA binding proteins; EBL, embryo bovine lung cells; RT-qPCR, Real-time quantitative polymerase chain reaction; LDH, lactate dehydrogenase; Tunel Terminal Deoxynucleotidyl Transferase mediated dUTP Nick-End Labeling assay; FRAP, Ferric ion reducing antioxidant power; PSF, Polypyrimidine tract binding Protein-associated Splicing Factor; AS, Alternative Splicing; APAF-1, Apoptotic peptidase activating factor 1; RI, Retention Intron; A3SS and A5SS Variable 3’ and 5’ splicing; SE, Skipping Exon; MXE, Mutually Exclusive Exons; CFUs, colony forming units; TALE, Transcription activator-like effector.
References
1. MOARA. National medium and long term animal disease prevention and control plan, (2012–2020). (2012).
2. Zhu X, Wang J, Zhao Y, Zhang Z, Yan L, Xue Y, et al. Prevalence, distribution, and risk factors of bovine tuberculosis in dairy cattle in central China. Prev Vet Med. (2023) 213:105887. doi: 10.1016/j.prevetmed.2023.105887
3. Raphaka K, Sánchez-Molano E, Tsairidou S, Anacleto O, Glass EJ, Woolliams JA, et al. Impact of genetic selection for increased cattle resistance to bovine tuberculosis on disease transmission dynamics. Front Vet Sci. (2018) 5:237. doi: 10.3389/fvets.2018.00237
4. Perskvist N, Long M, Stendahl O, Zheng L. Mycobacterium tuberculosis promotes apoptosis in human neutrophils by activating caspase-3 and altering expression of Bax/Bcl-xL via an oxygen-dependent pathway. J Immunol. (2002) 168:6358–65. doi: 10.4049/jimmunol.168.12.6358
5. Escobar-Chavarría O, Benitez-Guzman A, Jiménez-Vázquez I, Carrisoza-Urbina J, Arriaga-Pizano L, Huerta-Yépez S, et al. Necrotic cell death and inflammasome NLRP3 activity in mycobacterium bovis-infected bovine macrophages. Cells-Basel. (2023) 12(16):2079. doi: 10.3390/cells12162079
6. Grace PS, Ernst JD. Suboptimal antigen presentation contributes to virulence of mycobacterium tuberculosis in vivo. J Immunol. (2016) 196:357–64. doi: 10.4049/jimmunol.1501494
7. Smith VL, Cheng Y, Bryant BR, Schorey JS. Exosomes function in antigen presentation during an in vivo Mycobacterium tuberculosis infection. Sci Rep. (2017) 7:43578. doi: 10.1038/srep43578
8. Danelishvili L, Yamazaki Y, Selker J, Bermudez LE. Secreted Mycobacterium tuberculosis Rv3654c and Rv3655c proteins participate in the suppression of macrophage apoptosis. PloS One. (2010) 5:e10474. doi: 10.1371/journal.pone.0010474
9. Blanco FC, Gravisaco MJ, Bigi MM, García EA, Marquez C, McNeil M, et al. Identifying bacterial and host factors involved in the interaction of mycobacterium bovis with the bovine innate immune cells. Front Immunol. (2021) 12:674643. doi: 10.3389/fimmu.2021.674643
10. Li J, Chai QY, Zhang Y, Li BX, Wang J, Qiu XB, et al. Mycobacterium tuberculosis Mce3E suppresses host innate immune responses by targeting ERK1/2 signaling. J Immunol. (2015) 194:3756–67. doi: 10.4049/jimmunol.1402679
11. De Zio D, Ferraro E, D’Amelio M, Simoni V, Bordi M, Soroldoni D, et al. Faf1 is expressed during neurodevelopment and is involved in Apaf1-dependent caspase-3 activation in proneural cells. Cell Mol Life Sci. (2008) 65:1780–90. doi: 10.1007/s00018-008-8075-5
13. Palmer MV, Thacker TC, Waters WR, Gortázar C, Corner LA. Mycobacterium bovis: A Model Pathogen at the Interface of Livestock, Wildlife, and Humans. Vet Med Int. (2012) 2012:236205. doi: 10.1155/2012/236205
14. Schiller I, Oesch B, Vordermeier HM, Palmer MV, Harris BN, Orloski KA, et al. Bovine tuberculosis: a review of current and emerging diagnostic techniques in view of their relevance for disease control and eradication. Transbound Emerg Dis. (2010) 57:205–20. doi: 10.1111/j.1865-1682.2010.01148.x
15. Jasin M, Moynahan ME, Richardson C. Targeted transgenesis. Proc Natl Acad Sci U.S.A. (1996) 93:8804–8. doi: 10.1073/pnas.93.17.8804
16. Whitworth KM, Lee K, Benne JA, Beaton BP, Spate LD, Murphy SL, et al. Use of the CRISPR/Cas9 system to produce genetically engineered pigs from in vitro-derived oocytes and embryos. Biol Reprod. (2014) 91:78. doi: 10.1095/biolreprod.114.121723
17. Whitworth KM, Rowland RR, Ewen CL, Trible BR, Kerrigan MA, Cino-Ozuna AG, et al. Gene-edited pigs are protected from porcine reproductive and respiratory syndrome virus. Nat Biotechnol. (2016) 34:20–2. doi: 10.1038/nbt.3434
18. You X, Li G, Lei Y, Xu Z, Zhang P, Yang Y. Role of genetic factors in different swine breeds exhibiting varying levels of resistance/susceptibility to PRRSV. Virus Res. (2023) 326:199057. doi: 10.1016/j.virusres.2023.199057
19. Xu K, Zhou Y, Mu Y, Liu Z, Hou S, Xiong Y, et al. CD163 and pAPN double-knockout pigs are resistant to PRRSV and TGEV and exhibit decreased susceptibility to PDCoV while maintaining normal production performance. Elife. (2020) 9:e57132. doi: 10.7554/eLife.57132
20. Wu H, Wang Y, Zhang Y, Yang M, Lv J, Liu J, et al. TALE nickase-mediated SP110 knockin endows cattle with increased resistance to tuberculosis. Proc Natl Acad Sci U.S.A. (2015) 112:E1530–1539. doi: 10.1073/pnas.1421587112
21. Ahmadi Rastegar D, Sharifi Tabar M, Alikhani M, Parsamatin P, Sahraneshin Samani F, Sabbaghian M, et al. Isoform-level gene expression profiles of human Y chromosome azoospermia factor genes and their X chromosome paralogs in the testicular tissue of non-obstructive azoospermia patients. J Proteome Res. (2015) 14:3595–605. doi: 10.1021/acs.jproteome.5b00520
22. Gao J, Wang Q, Tang YD, Zhai J, Hu W, Zheng C. When ferroptosis meets pathogenic infections. Trends Microbiol. (2023) 31:468–79. doi: 10.1016/j.tim.2022.11.006
23. Hasnain SE, Begum R, Ramaiah KV, Sahdev S, Shajil EM, Taneja TK, et al. Host-pathogen interactions during apoptosis. J Biosci. (2003) 28:349–58. doi: 10.1007/BF02970153
24. Rodríguez-González J, Gutiérrez-Kobeh L. Apoptosis and its pathways as targets for intracellular pathogens to persist in cells. Parasitol Res. (2023) 123:60. doi: 10.1007/s00436-023-08031-x.
25. Behar SM, Briken V. Apoptosis inhibition by intracellular bacteria and its consequence on host immunity. Curr Opin Immunol. (2019) 60:103–10. doi: 10.1016/j.coi.2019.05.007
26. Zhai WJ, Wu FJ, Zhang YY, Fu YR, Liu ZJ. The immune escape mechanisms of. Int J Mol Sci. (2019) 20(2):340. doi: 10.3390/ijms20020340.
27. Dudani R, Chapdelaine Y, van Faassen H, Smith DK, Shen H, Krishnan L, et al. Preexisting inflammation due to Mycobacterium bovis BCG infection differentially modulates T-cell priming against a replicating or nonreplicating immunogen. Infect Immun. (2002) 70:1957–64. doi: 10.1128/IAI.70.4.1957-1964.2002
28. Ji N, Long M, Garcia-Vilanova A, Ault R, Moliva JI, Yusoof KA, et al. Selective delipidation of Mycobacterium bovis BCG retains antitumor efficacy against non-muscle invasive bladder cancer. Cancer Immunol Immunother. (2023) 72:125–36. doi: 10.1007/s00262-022-03236-y
29. Ule J, Darnell RB. RNA binding proteins and the regulation of neuronal synaptic plasticity. Curr Opin Neurobiol. (2006) 16:102–10. doi: 10.1016/j.conb.2006.01.003
30. Kang D, Lee Y, Lee JS. RNA-binding proteins in cancer: functional and therapeutic perspectives. Cancers (Basel). (2020) 12(9):2699. doi: 10.3390/cancers12092699
31. Stenum TS, Kumar AD, Sandbaumhüter FA, Kjellin J, Jerlström-Hultqvist J, Andrén PE, et al. RNA interactome capture in Escherichia coli globally identifies RNA-binding proteins. Nucleic Acids Res. (2023) 51:4572–87. doi: 10.1093/nar/gkad216
32. Yang J, Ding S. Chimeric RNA-binding protein-based killing switch targeting hepatocellular carcinoma cells. Mol Ther Nucleic Acids. (2021) 25:683–95. doi: 10.1016/j.omtn.2021.08.012
33. Holland C, Schmid M, Zimny-Arndt U, Rohloff J, Stein R, Jungblut PR, et al. Quantitative phosphoproteomics reveals a link between Helicobacter pylori infection and RNA splicing modulation in host cells. Proteomics. (2011) 11:2798–811. doi: 10.1002/pmic.201000793
34. Kalam H, Fontana MF, Kumar D. Alternate splicing of transcripts shape macrophage response to Mycobacterium tuberculosis infection. PloS Pathog. (2017) 13:e1006236. doi: 10.1371/journal.ppat.1006236
35. Zhang W, Niu C, Fu RY, Peng ZY. Mycobacterium tuberculosis H37Rv infection regulates alternative splicing in Macrophages. Bioengineered. (2018) 9:203–8. doi: 10.1080/21655979.2017.1387692
36. Cecconi F, Gruss P. Apaf1 in developmental apoptosis and cancer: how many ways to die? Cell Mol Life Sci. (2001) 58:1688–97. doi: 10.1007/pl00000806
37. Zhang H, Liu M, Fan W, Sun S, Fan X. The impact of Mycobacterium tuberculosis complex in the environment on one health approach. Front Public Health. (2022) 10:994745. doi: 10.3389/fpubh.2022.994745
38. Pereira AC, Pinto D, Cunha MV. Unlocking environmental contamination of animal tuberculosis hotspots with viable mycobacteria at the intersection of flow cytometry, PCR, and ecological modelling. Sci Total Environ. (2023) 891:164366. doi: 10.1016/j.scitotenv.2023.164366
39. Allen AR, Ford T, Skuced RA. Does Mycobacterium tuberculosis var. bovis survival in the Environment Confound Bovine Tuberculosis Control and Eradication? A literature review. Vet Med Int. (2021) 2021:8812898. doi: 10.1155/2021/8812898
40. Devi KR, Lee LJ, Yan LT, Syafinaz AN, Rosnah I, Chin VK. Occupational exposure and challenges in tackling M. bovis at human-animal interface: a narrative review. Int Arch Occup Environ Health. (2021) 94(6):1147–71. doi: 10.1007/s00420-021-01677-z
41. Mardare C, Delahay RJ, Dale JW. Environmental amoebae do not support the long-term survival of virulent mycobacteria. J Appl Microbiol. (2013) 114(5):1388–94. doi: 10.1111/jam.12166
Keywords: RBMX2, Mycobacterium tuberculosis variant bovis, embryo bovine lung cells (EBL), apoptosis, molecular docking, APAF-1, RNA alternative splicing
Citation: Wang C, Jiang Y, Yang Z, Xu H, Khalid AK, Iftakhar T, Peng Y, Lu L, Zhang L, Bermudez L, Guo A and Chen Y (2024) Host factor RBMX2 promotes epithelial cell apoptosis by downregulating APAF-1’s Retention Intron after Mycobacterium bovis infection. Front. Immunol. 15:1431207. doi: 10.3389/fimmu.2024.1431207
Received: 11 May 2024; Accepted: 15 August 2024;
Published: 06 September 2024.
Edited by:
Bart Tummers, King’s College London, United KingdomReviewed by:
Abhinav Joshi, National Cancer Institute (NIH), United StatesXiaomin Wang, Shenzhen Third People’s Hospital, China
Aruna Pal, West Bengal University of Animal and Fishery Sciences, India
Copyright © 2024 Wang, Jiang, Yang, Xu, Khalid, Iftakhar, Peng, Lu, Zhang, Bermudez, Guo and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yingyu Chen, Y2hlbnlpbmd5dUBtYWlsLmh6YXUuZWR1LmNu; Aizhen Guo, YWl6aGVuQG1haWwuaHphdS5lZHUuY24=
 Chao Wang
Chao Wang Yanzhu Jiang
Yanzhu Jiang Zhiming Yang
Zhiming Yang Haojun Xu
Haojun Xu Abdul Karim Khalid
Abdul Karim Khalid Tahira Iftakhar
Tahira Iftakhar Yongchong Peng
Yongchong Peng Lu Lu
Lu Lu Lei Zhang
Lei Zhang Luiz Bermudez
Luiz Bermudez Aizhen Guo
Aizhen Guo Yingyu Chen
Yingyu Chen