- 1Department of Tumor Biological Treatment, The Third Affiliated Hospital of Soochow University, Changzhou, Jiangsu, China
- 2Jiangsu Engineering Research Center for Tumor Immunotherapy, Changzhou, Jiangsu, China
- 3Institute of Cell Therapy, Soochow University, Changzhou, Jiangsu, China
While biomarkers have been shown to enhance the prognosis of patients with colorectal cancer (CRC) compared to conventional treatments, there is a pressing need to discover novel biomarkers that can assist in assessing the prognostic impact of immunotherapy and in formulating individualized treatment plans. The RUNX family, consisting of RUNX1, RUNX2, and RUNX3, has been recognized as crucial regulators in developmental processes, with dysregulation of these genes also being implicated in tumorigenesis and cancer progression. In our present study, we demonstrated a crucial regulatory role of RUNX in CD8+T and CD103+CD8+T cell-mediated anti-tumor response within the tumor microenvironment (TME) of human CRC. Specifically, RUNXs were significantly differentially expressed between tumor and normal tissues in CRC. Patients with a greater proportion of infiltrating CD8+RUNX1+, CD103+CD8+RUNX1+, CD8+RUNX2+, CD103+CD8+RUNX2+, CD8+RUNX3+, or CD103+CD8+RUNX3+ T cells demonstrated improved outcomes compared to those with lower proportions. Additionally, the proportions of infiltrating CD8+RUNX1+T and CD8+RUNX3+T cells may serve as valuable prognostic predictors for CRC patients, independent of other clinicopathological factors. Moreover, further bioinformatic analysis conducted utilizing the TISIDB and TIMER platforms demonstrated significant associations between the members of the RUNX family and immune-infiltrating cells, specifically diverse subpopulations of CD8+TILs. Our study of human colorectal cancer tissue microarray (TMA) also revealed positive and statistically significant correlations between the expressions of RUNX1, RUNX2, and RUNX3 in both CD8+T cells and CD103+CD8+T cells. Our study comprehensively revealed the varied expressions and prognostic importance of the RUNX family in human colorectal cancer tissues. It underscored their potential as vital biomarkers for prognostic evaluation in colorectal cancer patients and as promising targets for immunotherapy in treating this disease.
Introduction
Colorectal cancer (CRC) is a leading cause of morbidity and mortality worldwide. Traditional treatment including surgery, chemotherapy, and radiotherapy could provide benefits for early-stage patients, but side effects and tumor recurrence due to their non-specificity, cytotoxicity, and incompleteness remain significant challenges in treating CRC (1, 2). Immunotherapy is an emerging treatment that leverages patients’ own immune cells to fight tumor cells. It is regarded as a valuable and essential supplementary approach to conventional standard treatments. Certain therapies, such as immune checkpoint inhibitors targeting PD-1/PD-L1 and CAR-T cell therapy, have shown notably effective outcomes in specific tumor types (3, 4). Due to tumor heterogeneity and the immunosuppressive tumor microenvironment, there is still significant potential for enhancing the responsiveness and overall remission rate of immunotherapy in malignant tumors. The current efficacy of immunotherapy in CRC is closely tied to patient-specific biomarkers, such as microsatellite instability, programmed cell death ligands expression and so on. There is a pressing necessity to discover novel biomarkers that can assist in evaluating the prognostic implications of immunotherapy and in devising individualized immunotherapy protocols. Various subtypes and quantities of infiltrating immune cells, particularly tumor-infiltrating T cells (TILs), are valuable indicators for assessing the effectiveness of immunotherapy and predicting patient outcomes (5, 6). Among these subsets, CD8+ T cells are deemed essential and preferred for their role in anti-tumor immune responses. Despite their importance, the dysfunction and depletion of these cells can result in tumor immunosuppression and tolerance, allowing tumor growth and progression (7). Addressing these challenges is essential for advancing the role of immunotherapy in the treatment of CRC.
The signature profiles of transcription factors, activating receptors or inhibitory receptors expressed by these tumor-infiltrating CD8+T cells play a crucial role in the regulation of their own anti-tumor immune functions. For example, the abundance of intratumorally infiltrating CXCL13+CD8+T in renal clear cell carcinoma correlates with poor clinical prognosis (8), and the abundance of infiltrating PD-1+CD8+T cells in gastric carcinoma correlates with improved prognosis (9). In our prior investigation, it was observed that CD226 exhibited elevated expression levels in CD8+T cells present within gastric cancer tumor infiltrates. Furthermore, the CD226+CD8+T cell subset was identified as a significant prognostic indicator for favorable outcomes in gastric patients (10). Therefore, further identification of more effective biomarkers through well-defined CD8+ T-cell subpopulations will certainly be beneficial in assisting clinical evaluation of tumor patients.
RUNX, a transcription factor family member, plays a crucial role in regulating mammalian cell proliferation, differentiation, lineage development, osteogenesis, and neurogenesis (11–13). The RUNX gene family comprises three members: RUNX1, RUNX2, and RUNX3. The functionality of RUNX proteins varies depending on the cellular context and is influenced by pathways such as transforming growth factor beta (TGF-β), BMP, WNT, hedgehog, Notch, receptor tyrosine kinase 6, and Yes-associated protein 1 (YAP1), which are associated with major developmental pathways (14–17). Dysregulation of RUNX family gene can lead to a variety of diseases, including tumors (18, 19). Knockout studies show that RUNX1 is crucial for the development and upkeep of the blood system. Runx1-deficient mice die before birth from bleeding in the brain and blood system failure, and RUNX1 is closely linked to leukemia and myelodysplasia in human (20). The fusion oncogene RUNX1/RUNX1T1 could reprogram a large transcriptional network to establish and maintain acute myeloid leukemia (AML) via intricate PPI interactions and kinase-driven phosphorylation events in both adult and pediatric patients (21). RUNX2 plays a critical role in the differentiation of osteoblasts and chondrocytes, and mice lacking functional Runx2 alleles perish shortly after birth as a result of impaired ossification and respiratory failure (22). RUNX2 is thought to be associated with human osteosarcoma progression (23), breast cancer-mediated bone metastasis (24) and has also been reported to related with poor prognosis in cervical (25), bladder (26) and pancreatic (27) cancer. RUNX3 plays an important role in neurogenesis and thymogenesis. It is associated with the development of gastric epithelial cells; the majority of Runx3-deficient mice die of gastric epithelial hyperplasia shortly after birth, and those that survive tend to develop spontaneous inflammatory bowel inflammation (28, 29). The inactivation of RUNX3 in a variety of solid tumors has also been identified as an important tumor oncogenic factor.RUNX3 has been reported to act as a tumor suppressor in melanoma (30), gastric cancer (31) and lung cancer (32), inhibited tumor progression and metastasis. The RUNX family has important regulatory roles in both lineage development and differentiation effects of T cells, but there are fewer studies related to how RUNX family members enhance T lymphocyte function in the tumor microenvironment to treat tumors.
In this study, we aimed to examine the clinical correlations and prognostic significance of RUNX family gene expressions in CD8+TILs and CD103+CD8+TILs in human CRC tissues. We conducted a thorough analysis of RUNX genes across various cancers using TCGA data. Additionally, we explored the regulatory role of RUNX genes in CD8+TIL effector function using TIMER and TISIDB databases. We showed that the intensity of CD8+RUNX1+TILs, CD8+RUNX2+TILs and CD8+RUNX3+TILs could be useful prognostic predictors for CRC patients. Additionally, we investigated the interactions between RUNX family genes and immune cells within the colorectal cancer (CRC) tumor microenvironment.
Materials and methods
Patients and tissue specimens
The CRC tissue microarray (TMA, HColA180Su21) was provided by Shanghai Outdo Biotech Co. Ltd and contained tumor tissues and corresponding paracancerous tissues from 94 patients. All patients did not receive preoperative neoadjuvant radiotherapy, chemotherapy and immunotherapy, and were operated from February 2012 to September 2014, and were diagnosed with colorectal carcinoma by pathology after surgery. A total of 89 cases were included in this study excluding factors such as core point detachment during the experiment and incomplete clinical follow-up data. Clinical parameters are detailed in Tables 1–4 and the experiment protocol was approved by the ethics committee of the Third Affiliated Hospital of Soochow University.
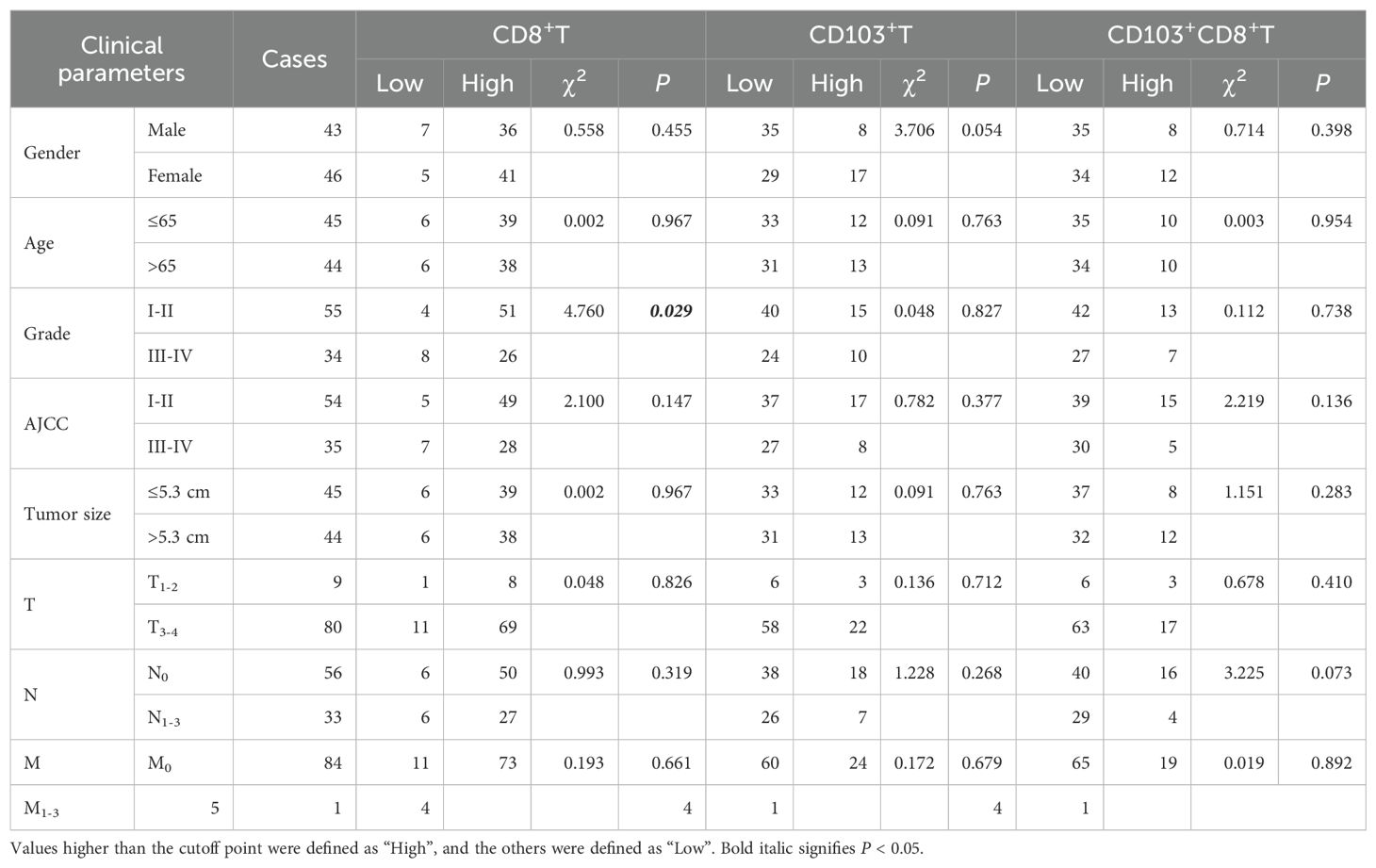
Table 1. Correlation between the infiltrating CD8+T and CD103+CD8+T cells in CRC tissues and clinical parameters of patients.
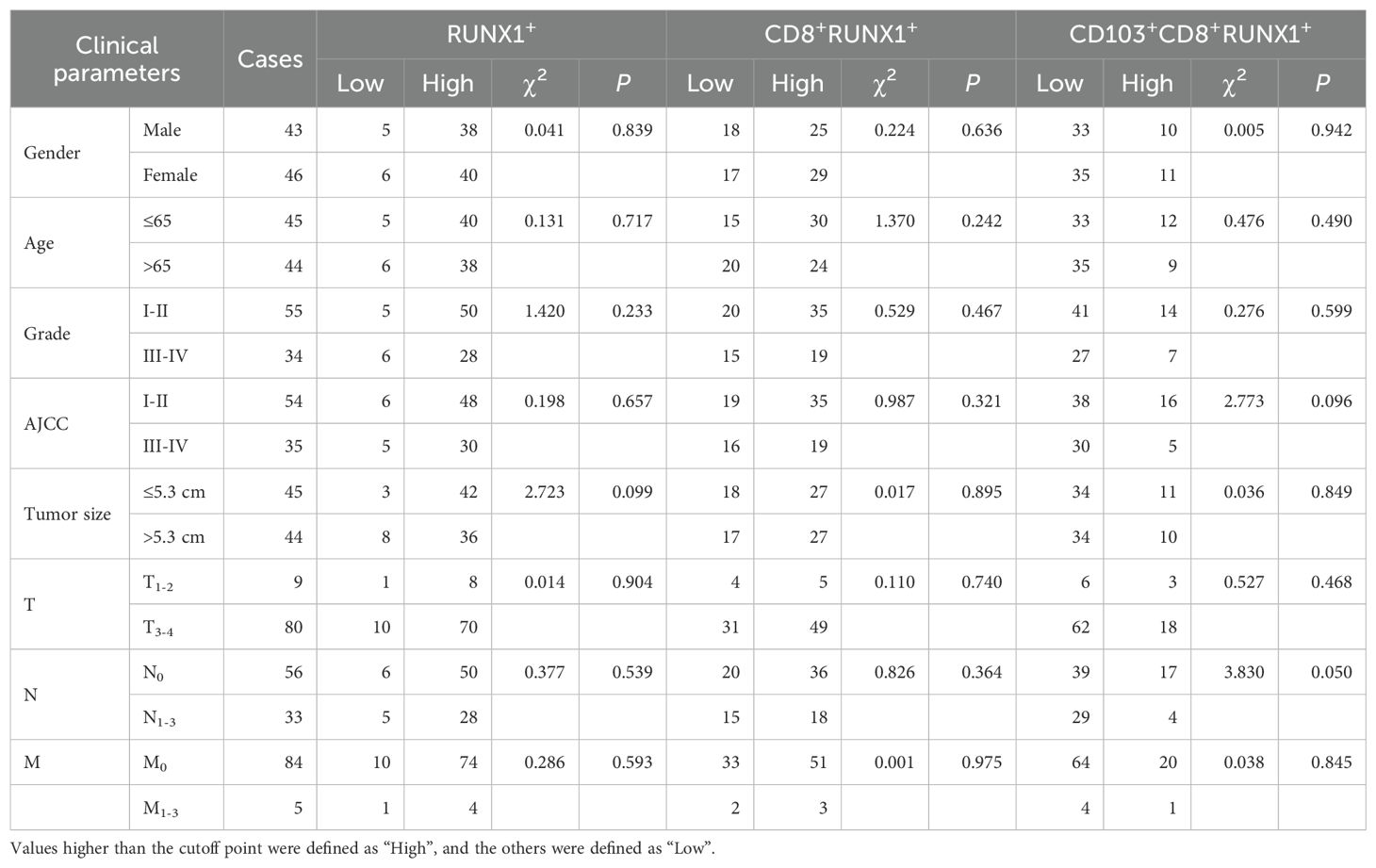
Table 2. Correlation between the infiltrating RUNX1+T, CD8+RUNX1+T and CD103+CD8+RUNX1+T cells in CRC tissues and clinical parameters of patients.
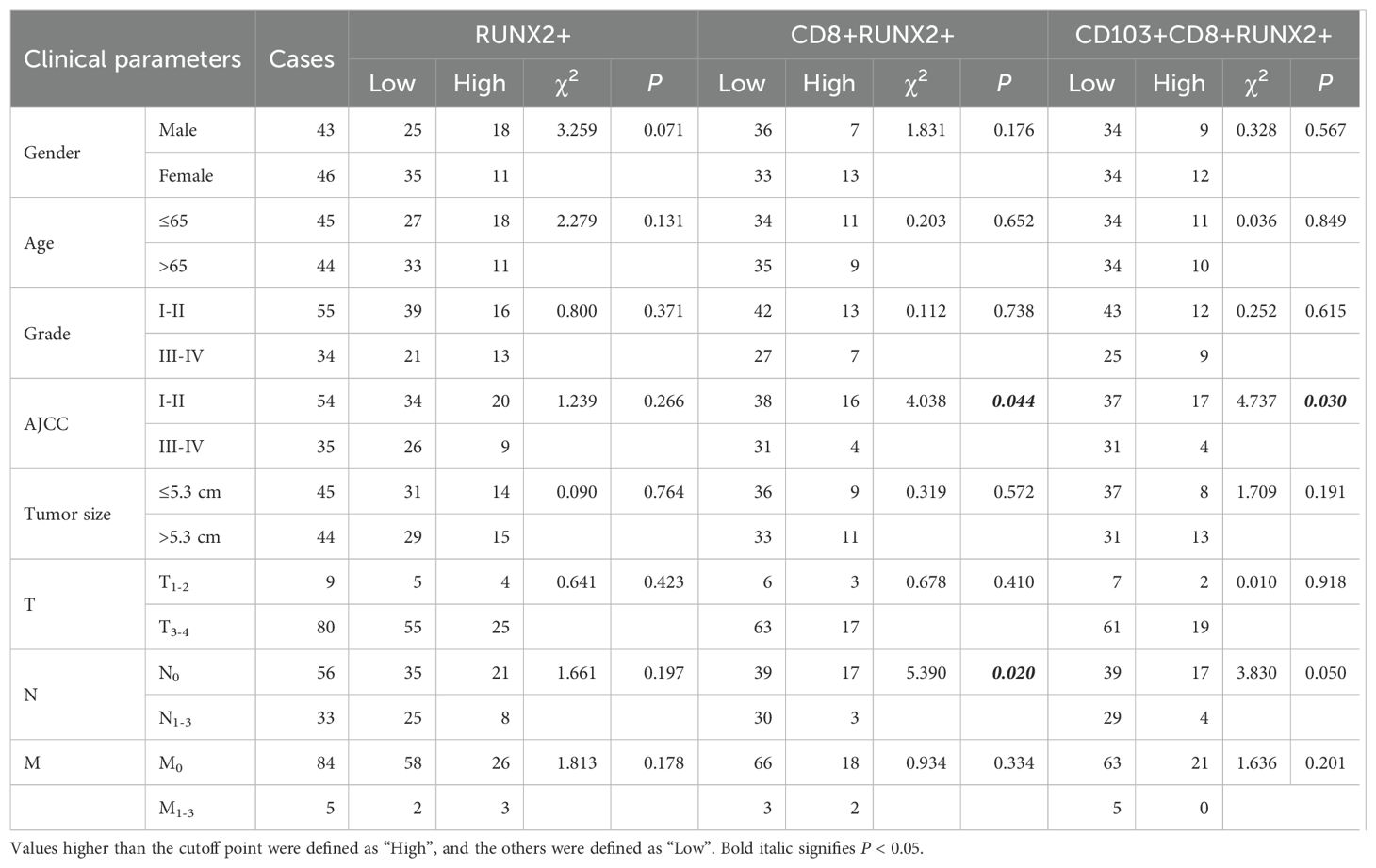
Table 3. Correlation between the infiltrating RUNX2+T, CD8+RUNX2+T and CD103+CD8+RUNX2+T cells in CRC tissues and clinical parameters of patients.
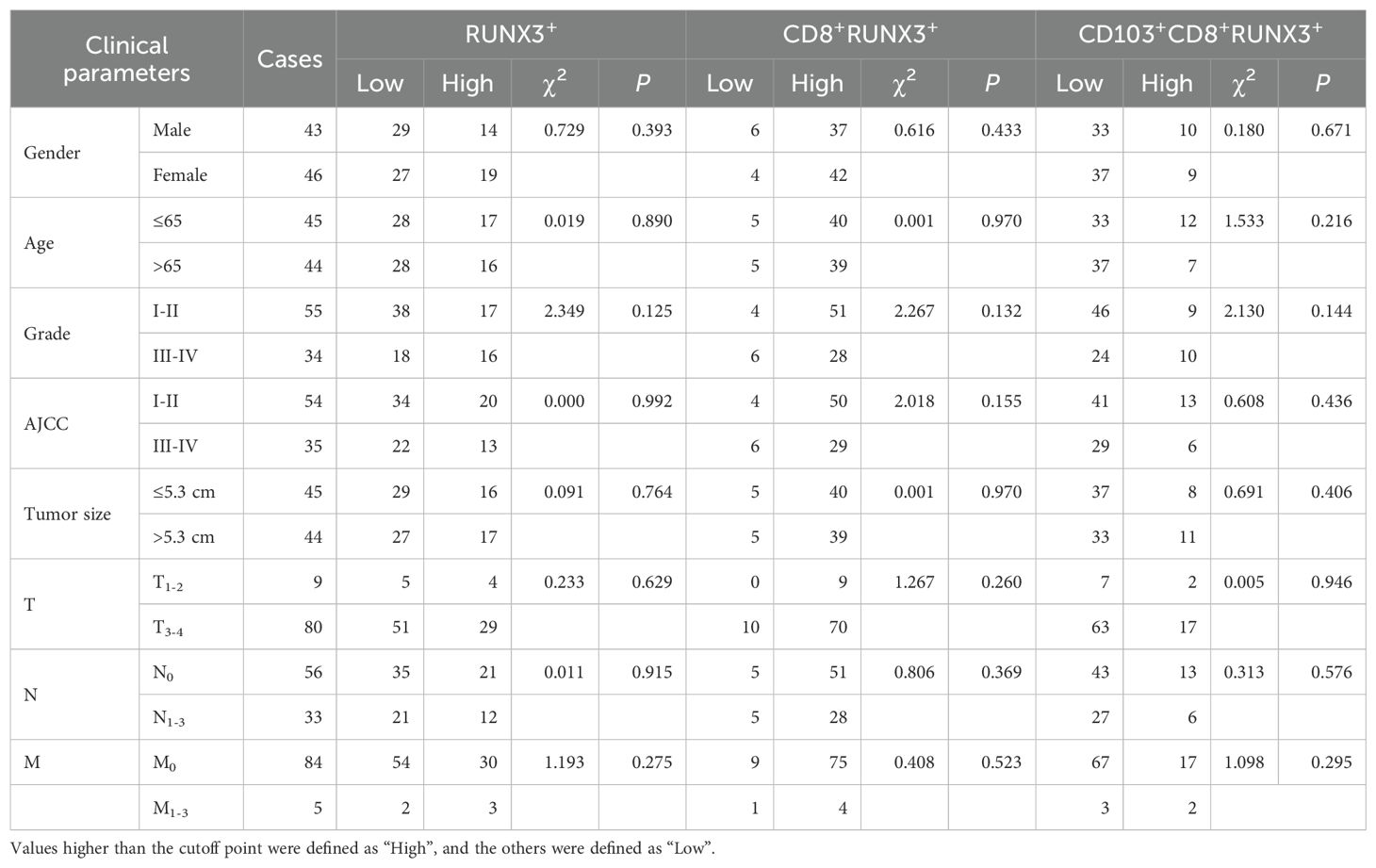
Table 4. Correlation between the infiltrating RUNX3+T, CD8+RUNX3+T and CD103+CD8+RUNX3+T cells in CRC tissues and clinical parameters of patients.
Multi-color immunohistochemically staining
The multi-color immunohistochemically staining (mIHC) was performed using the Opal 5-color fluorescent IHC kit (catalog No. NEL811001KT, PerkinElmer, USA) and automated quantitative analysis (PerkinElmer, USA) to detect CD8, CD103, RUNX1, RUNX2, and RUNX3 in tumor tissues. The 4’, 6-diamidino-2-phenylindole (DAPI) was utilized for nuclear staining. Subsequently, the concentrations of the six antibodies listed above were individually optimized against the markers, and a spectral library was established using single-stained slides. The PC TMA slide underwent dewaxing and rehydration via a sequence of xylene-to-alcohol washes prior to immersion in distilled water. Heat-induced antigen retrieval was done in citric acid solution (PH=6.0), followed by mIHC staining with primary antibodies including anti-CD8A (1:200, CST70306, Cell Signaling Technology), anti-CD103 (1:1000, ab224202, Abcam), anti-RUNX1 (1:1000, ab240639, Abcam), anti-RUNX2 (1:1000, ab192256, Abcam), and anti-RUNX3 (1:500, ab40278, Abcam). The PC TMA slide was then incubated with HRP-conjugated secondary antibodies in Opal working solution (PerkinElmer, USA) and mounted with ProLong Diamond Antifade Reagent with DAPI (Thermofisher, USA).
Imaging analysis
The TissueFAXS system, developed by TissueGnostics Asia Pacific Limited in Austria, was utilized for panoramic multispectral scanning of PC TMA slides. The resulting images were analyzed using StrataQuest software (Version No. 7.0.1.165, TissueGnostics Asia Pacific Limited, Austria), wherein each fluorophore was spectrally unmixed into distinct channels and saved as individual files. DAPI was employed to create a binary mask representing all viable cells within the image. The expressions of RUNX1, RUNX2, and RUNX3 were utilized in conjunction with DAPI to generate binary masks representing cells expressing these biomarkers. Furthermore, binary masks of CD8 and CD103 were employed to assess the infiltration intensity of lymphocytes. Each slide was reviewed independently by two senior pathologists who were unaware of the patients’ clinical characteristics. The immunohistochemical staining was evaluated utilizing the H-score methodology, consistent with our prior publications (33, 34).
Databases applied to analyze RUNX expression in human tumor and immune estimation
The gene expression analyses of the RUNX family in various cancer types were conducted utilizing the publicly available web resources GEPIA (http://gepia.cancer-pku.cn/) and UALCAN (http://ualcan.path.uab.edu/), which analyze cancer transcriptome data from TCGA and MET500 (35). Additionally, the web-based interactive platform TIMER2.0 (https://cistrome.shinyapps.io/timer/) was utilized to systematically analyze immune infiltration in different malignancies (36, 37). The TIMER2.0 database utilizes six sophisticated algorithms to conduct a comprehensive assessment of tumor-infiltrating lymphocyte (TIL) levels in The Cancer Genome Atlas (TCGA) or other tumor-related datasets. Furthermore, the database is capable of accurately estimating tumor purity. Our study examined the expression of RUNXs in different types of cancers and explored the correlation between RUNXs expression and TILs using gene modules. Moreover, an examination of the correlation between the expression of RUNXs and gene markers associated with tumor-infiltrating lymphocytes (TILs), such as CD8+/CD4+ T cells, B cells, monocytes, natural killer (NK) cells, dendritic cells (DCs), tumor-associated macrophages (TAMs), M1 macrophages, M2 macrophages, neutrophils, T cells, and their respective subtypes, has been conducted using correlation modules. The TISIDB database, accessible at http://cis.hku.hk/TISIDB/index.php, serves as an online integrated repository portal that aggregates numerous human cancer datasets from the TCGA database. In the TISIDB database, RUNX gene expression was correlated with immune subtypes or molecular subtypes of different cancer types. Differences with a P value 0.05 were considered statistically significant.
Statistical analyses
Statistical analyses were conducted using Prism 9 software and RStudio 6.3 to compare disease-related factors in patients with varying levels of RUNX1, RUNX2, and RUNX3 expression. The fluorescence intensity of each protein between different patients was truncated using the surv_cutpoint function in the survminer package of the R software package, and the patients were divided into two groups of high and low expression according to the cutoff point truncation value. The survival curve was plotted using the R package survival. Cox model analysis was conducted using the coxph function for both uni-variate and multi-variate analysis. Log-rank survival analysis was used to predict postoperative overall survival (OS) with significance set at P < 0.05.
Result
RUNXs are significantly differentially expressed between tumor and normal tissues in various human cancers
The RUNX gene family exhibited abnormal expression patterns in various human cancers. Analysis of TCGA data through TIMER, UALCAN, and GEPIA databases showed distinct expression profiles for RUNX family members across different tumor types, generally indicating upregulation in most tumors (Figure 1). RUNX1 and RUNX2 had similar cancer expression profiles, with overexpression in most cancer types and downregulation in prostate adenocarcinoma (PRAD). In contrast, RUNX3 was more frequently downregulated in several cancer types, including BLCA, COAD, LIHC, LUAD, THCA, and THYM. Notably, RUNX1 was upregulated, while RUNX3 was downregulated in COAD tumor tissues, with statistical significance. However, RUNX2 showed no significant difference in expression between tumor and normal tissues in COAD (refer to Figures 1A–C). These results suggest a progressive downregulation trend of RUNX1, RUNX2, and RUNX3 in COAD tumor tissues, indicating a potential role for RUNX3 and RUNX2 as tumor suppressors in COAD, in contrast to RUNX1.
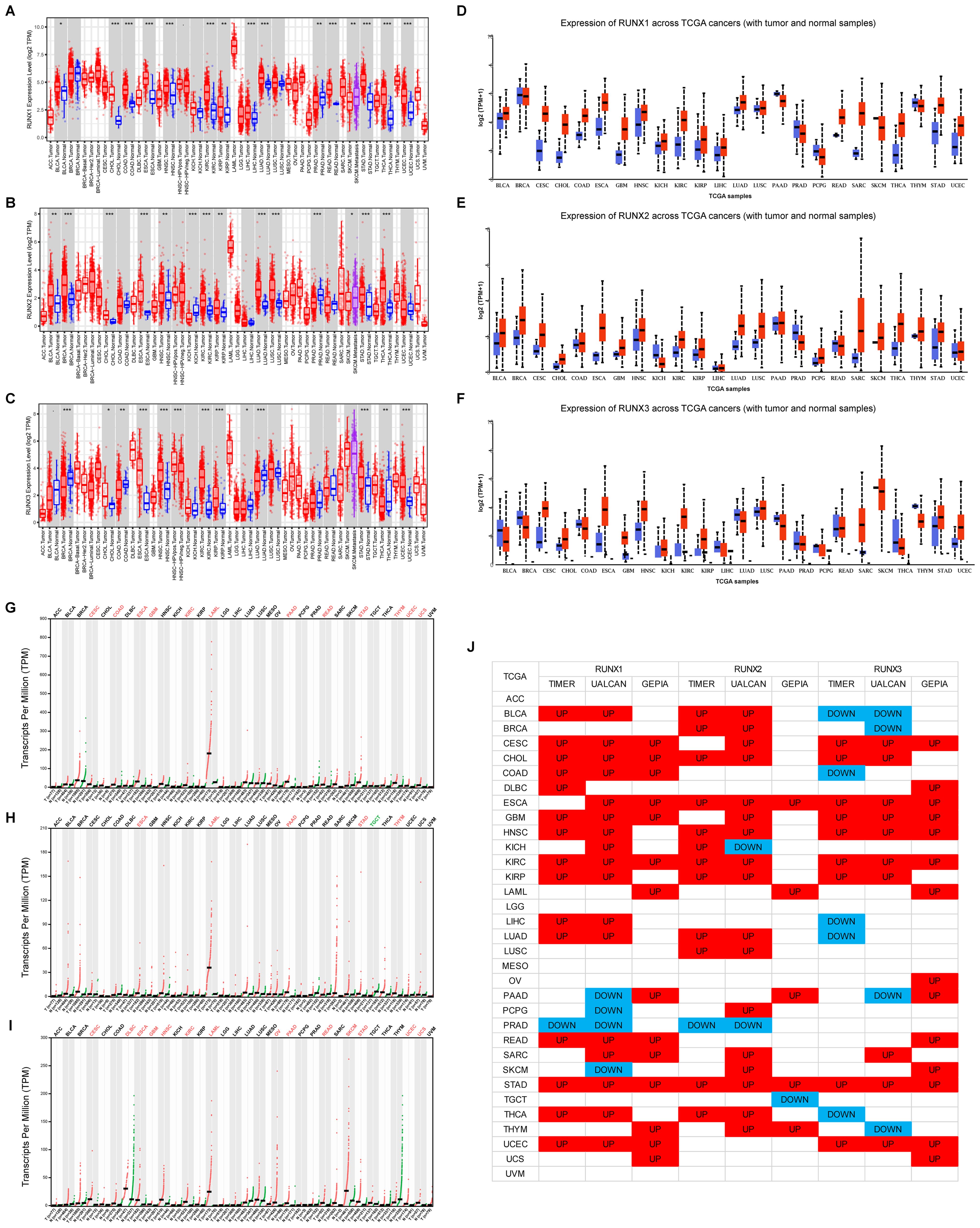
Figure 1. A comparison of expression levels of RUNX genes in human cancers of different types. (A–C) RUNX1, RUNX2 and RUNX3 gene levels in different cancer types (red) and normal tissue (blue) available in TIMER database. (D–F) RUNX1, RUNX2 and RUNX3 gene levels in different cancer types (red) and normal tissue (blue) available in UALCAN database. (G–I) RUNX1, RUNX2 and RUNX3 gene levels in different cancer types (red) and normal tissue (blue) available in GEPIA database. (J) Pan-cancer landscape of differential expression of RUNX1, RUNX2, and RUNX3 across three different TCGA databases. *P< 0.05, **P< 0.01, and ***P< 0.001.
Expressions and localization of CD8, CD103 and RUNX family genes in CRC tissues and normal colorectal tissues
In our previous study, we have already found that CD8+T and tissue-resident CD103+CD8+T cells were prognostic factors for colorectal cancer (38). In this study, we further explored the expression levels of RUNX family members on these two types of tumor-infiltrating lymphoid cells. We examined the localization of CD8, CD103, RUNX1, RUNX2 and RUNX3 in the CRC TMA slide by mIHC. Figure 2A showed that the RUNX1 (green), RUNX2 (red) and RUNX3 (yellow) could be predominantly detected in the nucleus while CD8 (blue) and CD103 (pink) were expressed on the membrane and cytoplasm. Based on the H-score analysis, CRC cancer tissues exhibited a significantly elevated presence of CD8+ cells in comparison to adjacent paracancerous tissues (Figure 2B). Furthermore, the expression levels of RUNX1 were notably higher in cancerous tissues as opposed to paracancerous tissues (Figure 2D), whereas the expression of RUNX3 was comparatively lower in cancerous tissues relative to paracancerous tissues (Figure 2F). Conversely, there was no discernible distinction in the levels of CD103+ cells and RUNX2 expression between cancerous and paracancerous tissues (Figures 2B, E, respectively). These results of RUNX family members between cancer and paracancer were consistent with the previous results in the TCGA database (Figures 1A–C).
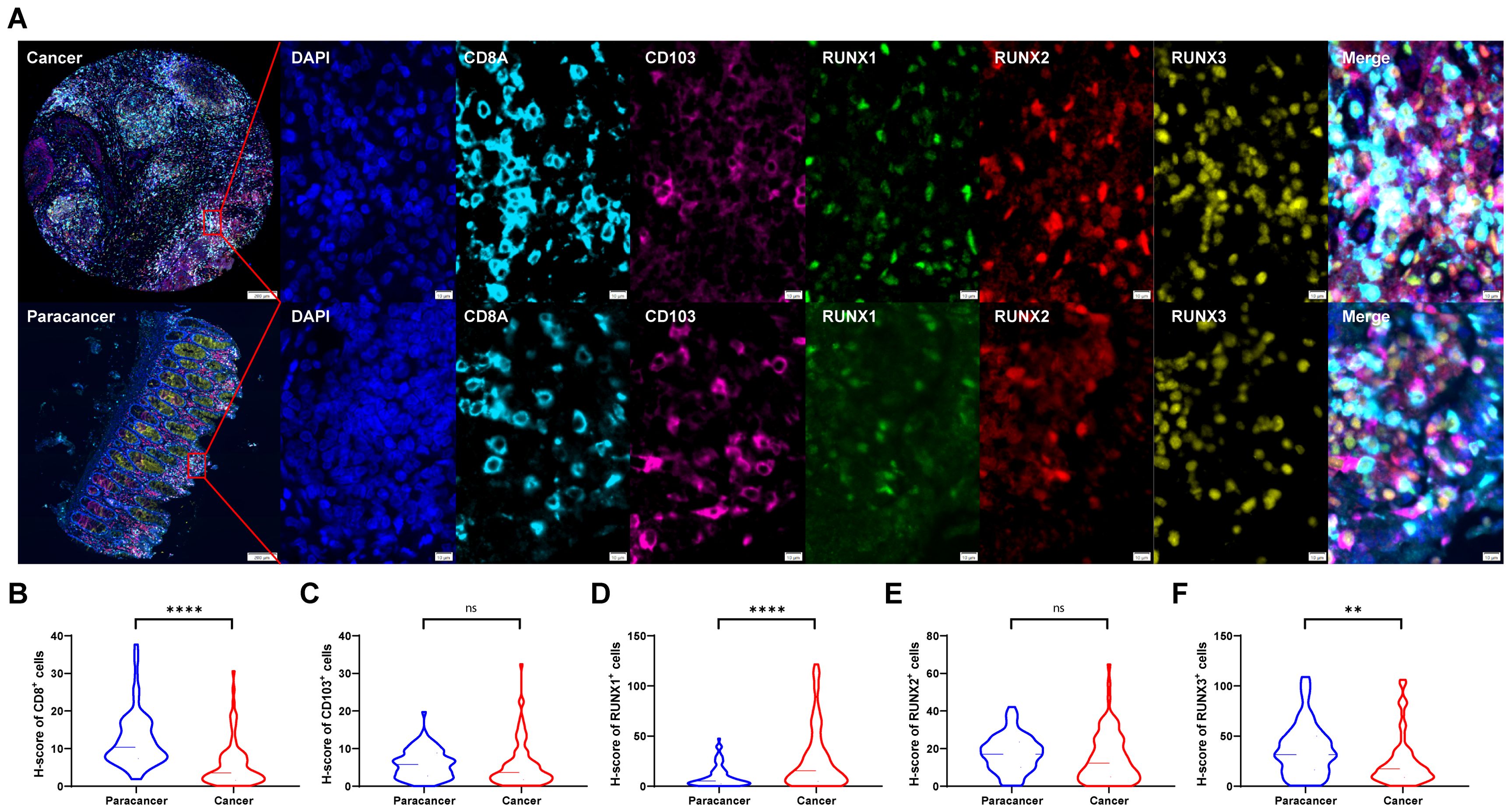
Figure 2. The expressions of CD8, CD103, RUNX1, RUNX2, and RUNX3 in human CRC TMA. (A) Multi-spectral immunohistochemistry (mIHC) was utilized to obtain images from human colorectal cancer tissue and adjacent paracancer tissue. (B–F) H-scores of CD8+ (B), CD103+ (C), RUNX1+ (D), RUNX2+ (E), and RUNX3+ (F) cells. **P < 0.01, ****P < 0.0001 and ns for no significance.
Tumor-infiltrating cytotoxic CD8+ T cells effectively inhibited tumor growth. Based on mIHC and imaging analysis (Figure 2A), membrane staining of CD103 could be found on infiltrating immune cells. The tissues-resident CD8+T cells, which were defined as CD103+CD8+T cells, were found in both adjacent normal and tumor tissues. In the present study, we also found that the proportions of CD8+ T cells and CD103+CD8+ T cells were significantly higher in human CRC tissues than in paraneoplastic tissues (Figures 3A, B). Herein, we also investigated the relationship between the frequency of tumor-infiltrating CD8+T and CD103+CD8+T cells and CRC patients’ survival. Figures 3C, D showed that patients with high frequency of CD8+ TILs (HR=2.917, 95% CI: 0.8615-9.874, P=0.0103, Figure 3C) and CD103+CD8+ TILs (HR=9.093, 95% CI: 3.845-21.51, P=0.0082, Figure 3D) had a better prognosis.
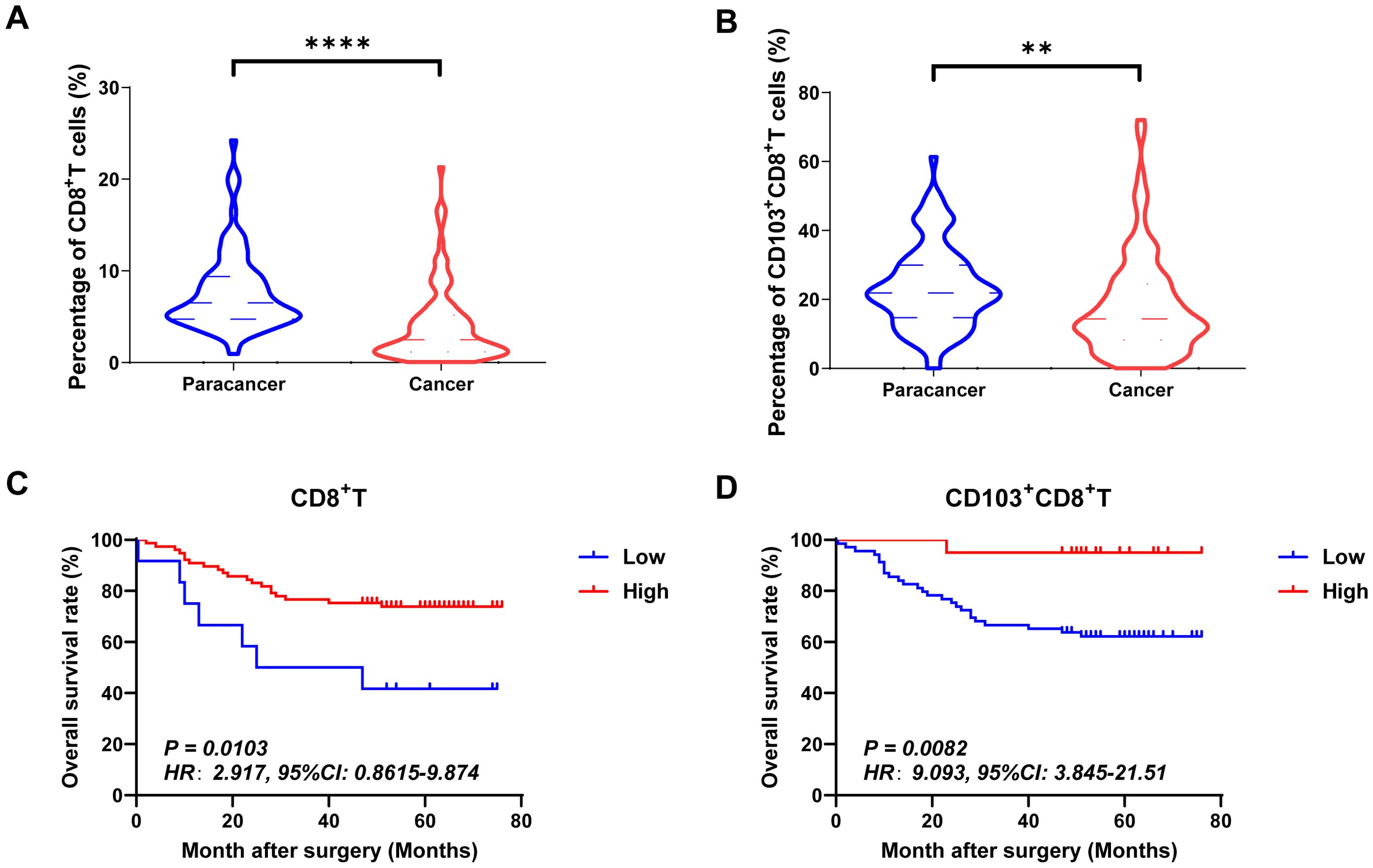
Figure 3. The expressions and prognostic values of CD8+TILs and CD103+CD8+TILs in human CRC TMA. (A, B) The populations of CD8+TILs and CD103+CD8+TILs were compared between CRC tissue and paracancer tissue. (C, D) The prognostic value of CD8+TILs and CD103+CD8+TILs were compared between CRC tissue and paracancer tissue. **P < 0.01, ****P < 0.0001.
Prognostic value of RUNX1, RUNX2 and RUNX3 expression in CD8+TILs and CD103+CD8+TILs in human CRC
Figures 4A–C, 5A–C and 6A–C showed the percentages of RUNX1+ cells, CD8+RUNX1+T/CD8+T cells, CD103+CD8+RUNX1+T/CD8+T cells, RUNX2+ cells, CD8+RUNX2+T/CD8+T cells, CD103+CD8+RUNX2+T/CD8+T cells, RUNX3+ cells, CD8+RUNX3+T/CD8+T cells and CD103+CD8+RUNX3+T/CD8+T cells comparison between paracancer tissues and cancer tissues in CRC. The percentages of RUNX1+ cells, CD8+RUNX1+T/CD8+T cells, and CD103+CD8+RUNX1+T/CD8+T cells were all higher in cancer tissues than in paracancer tissues (Figures 4A–C). The percentage of RUNX2+ cells was lower in cancer tissues than in paracancer tissues (Figure 5A), while the percentage of CD103+CD8+RUNX2+T/CD8+T cells was higher in cancer tissues than in paracancer tissues (Figure 5C). These two differences were both statistically significant. Interestingly, the difference in the percentage of CD8+RUNX2+T among CD8+T cells between cancer or paracancer tissues was not statistically significant (Figure 5B). We also observed that patients with higher density of CD8+RUNX2+T cells or CD103+CD8+RUNX2+T cells proportion tend to have a better OS than lower density (HR=2.629, 95% CI: 1.097-6.302, P=0.0999, Figure 5B; HR=2.827, 95% CI: 1.198-6.672, P=0.0752, Figure 5C). The percentages of RUNX3+ cells (Figure 6A) and CD8+RUNX3+T/CD8+T cells (Figure 6B) were all lower in cancer tissues than in paracancer tissues. These two differences were both statistically significant, while the difference in the percentage of CD103+CD8+RUNX3+T among CD8+T cells between cancer or paracancer tissues was not statistically significant (Figure 6C).
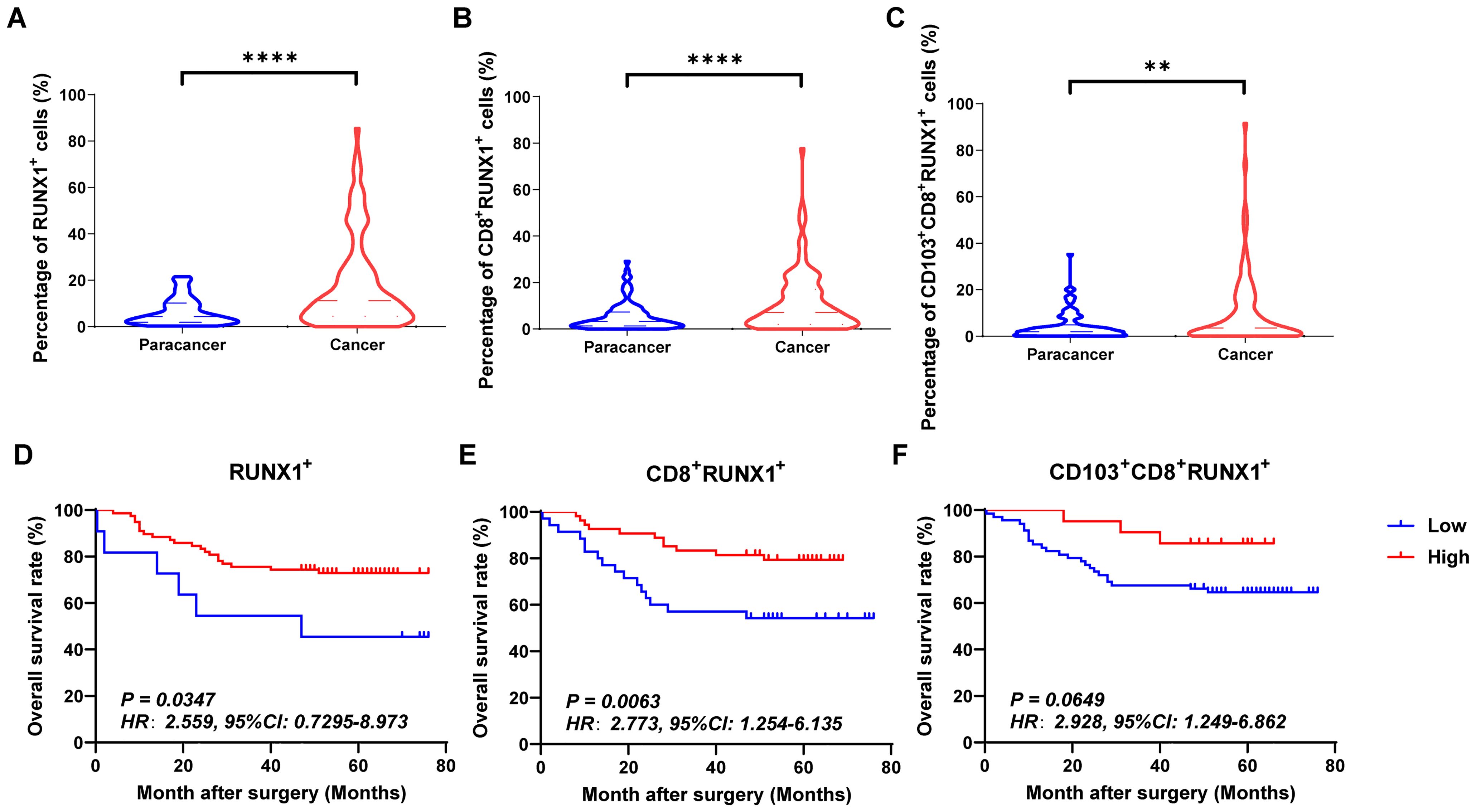
Figure 4. The expressions and prognostic values of RUNX1+TILs, CD8+RUNX1+TILs and CD103+CD8+RUNX1+TILs in human CRC TMA. (A–C) The populations of RUNX1+TILs, CD8+RUNX1+TILs and CD103+CD8+RUNX1+TILs were compared between CRC tissue and paracancer tissue. (D–F) The prognostic significance of RUNX1+TILs, CD8+RUNX1+TILs and CD103+CD8+RUNX1+TILs were assessed between CRC tissue and paracancer tissue. **P < 0.01, ****P < 0.0001.
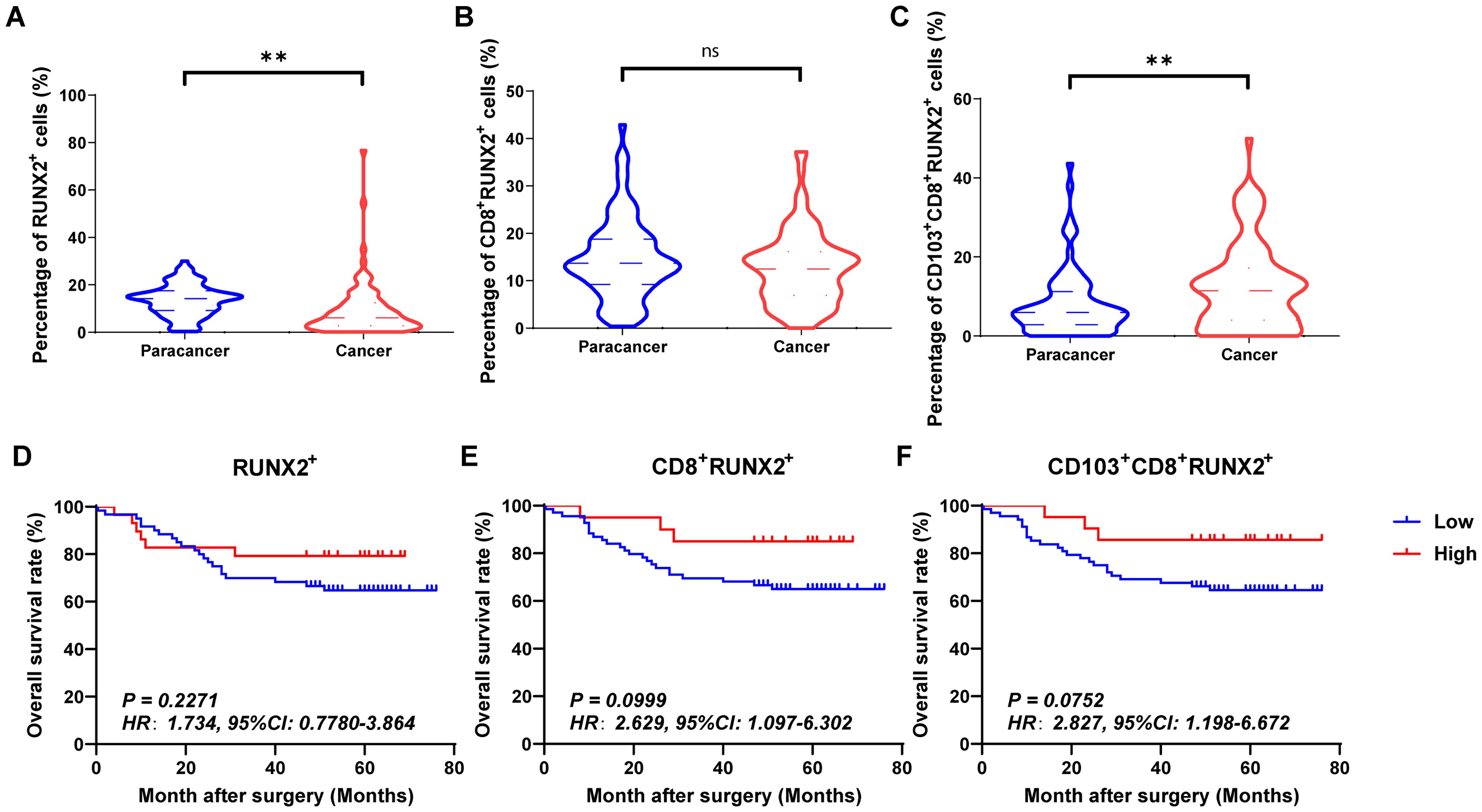
Figure 5. The expressions and prognostic values of RUNX2+TILs, CD8+RUNX2+TILs and CD103+CD8+RUNX2+TILs in human CRC TMA. (A–C) The populations of RUNX2+TILs, CD8+RUNX2+TILs and CD103+CD8+RUNX2+TILs were compared between CRC tissue and paracancer tissue. (D–F) The prognostic value of RUNX2+TILs, CD8+RUNX2+TILs and CD103+CD8+RUNX2+TILs were compared between CRC tissue and paracancer tissue. **P < 0.01, and ns for no significance.
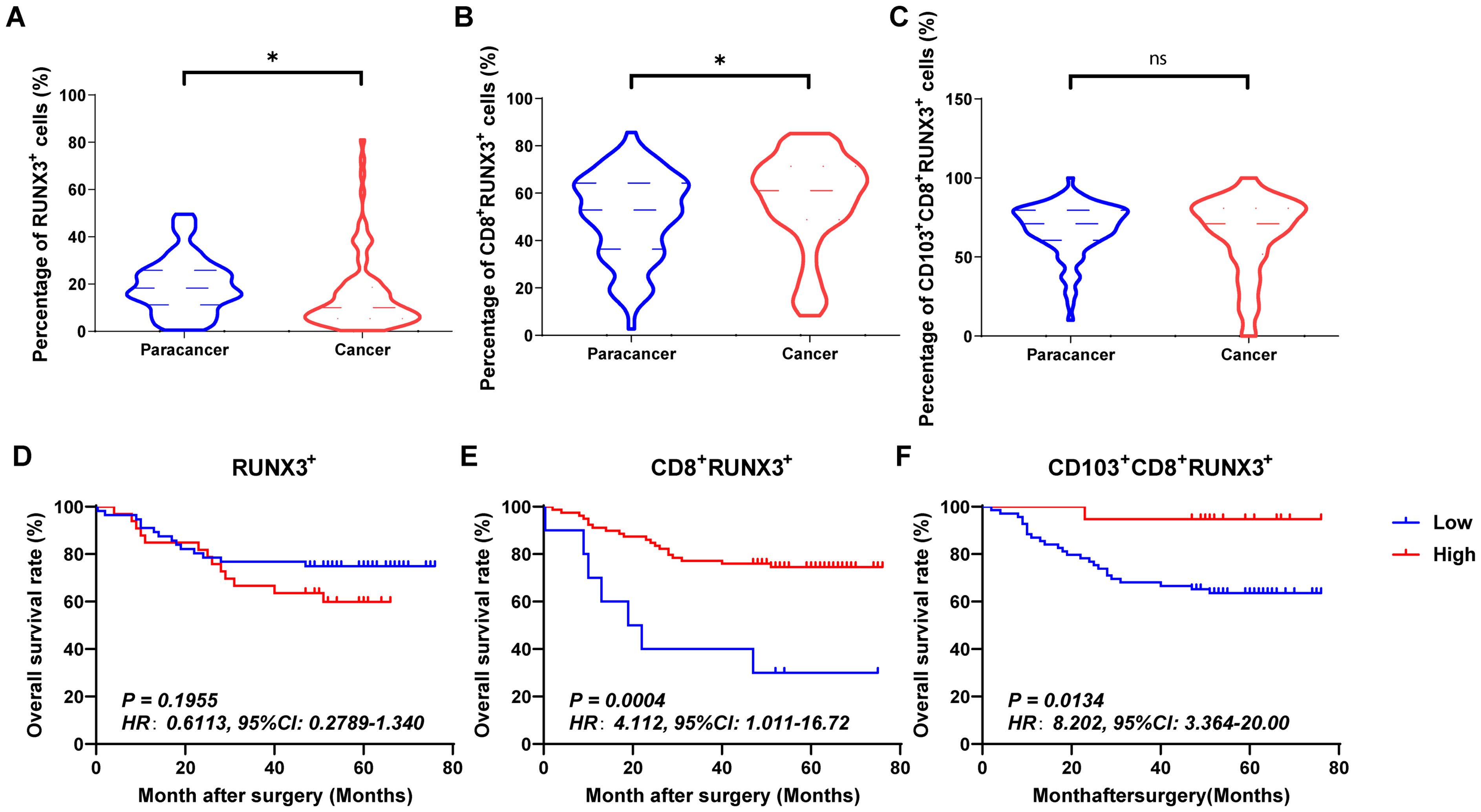
Figure 6. The expressions and prognostic values of RUNX3+TILs, CD8+RUNX3+TILs and CD103+CD8+RUNX3+TILs in human CRC TMA. (A–C) The populations of RUNX3+TILs, CD8+RUNX3+TILs and CD103+CD8+RUNX3+TILs were compared between CRC tissue and paracancer tissue. (D–F) The prognostic value of RUNX3+TILs, CD8+RUNX3+TILs and CD103+CD8+RUNX3+TILs were compared between CRC tissue and paracancer tissue. *P < 0.05, and ns for no significance.
Interestingly, we observed that patients with a higher density of CD8+RUNX1+T cells, CD103+CD8+RUNX1+T cells, CD8+RUNX2+T cells, CD103+CD8+RUNX2+T cells, CD8+RUNX3+T cells, or CD103+CD8+RUNX3+T cells proportion all presented a better OS than those with lower density (HR=2.773, 95% CI: 1.254-6.135, P=0.0063, Figure 4E; HR=2.928, 95% CI: 1.249-6.862, P=0.0649, Figure 4F; HR=2.629, 95% CI: 1.097-6.302, P=0.0999, Figure 5E; HR=2.827, 95% CI: 1.198-6.672, P=0.0752, Figure 5F; HR=4.112, 95% CI: 1.011-16.72, P=0.0134, Figure 6E; HR=8.202, 95% CI: 3.364-20.00, P=0.0134, Figure 6F).
Correlations between patients’ clinical parameters and the intensities of tumor-infiltrating CD8+T cells, CD103+CD8+T cells, CD8+RUNXs+T cells and CD103+CD8+RUNXs+T cells in human CRC tissues
In the current investigation, we sought to examine the associations between clinical parameters of patients and the levels of tumor-infiltrating CD8+T cells, CD103+CD8+T cells, CD8+RUNXs+T cells and CD103+CD8+RUNXs+T cells in human CRC tissues. Table 1 showed the frequency of CD8+T cells was positively associated with tumor grade (P=0.029, Table 1). Additionally, Table 3 demonstrated a significant association between the frequencies of CD8+RUNX2+TILs and CD103+CD8+RUNX2+TILs with AJCC stage (P=0.044, P=0.030, respectively, Table 3), as well as a negative and significant association between the frequency of CD8+RUNX2+TILs and N stage (P=0.020, Table 3). However, based on the data presented in Tables 2, 4, there was no significant correlation observed between the frequencies of various cell types (RUNX1+, CD8+RUNX1+T, CD103+CD8+RUNX1+T, RUNX3+, CD8+RUNX3+T, and CD103+CD8+RUNX3+T cells) and clinical parameters in colorectal cancer tissues.
Moreover, Table 5 revealed that individuals with a pathological grade of III+IV exhibit a markedly higher risk of mortality (multi-variate: HR=2.416, 95% CI: 1.085-5.398, P=0.031) in comparison to those with a pathological grade of I+II, even after adjusting for variables such as gender, age, tumor size, infiltration of CD8+T, infiltration of CD103+CD8+T, and other relevant factors. Patients with an AJCC stage of T3+T4 demonstrated a significantly elevated risk of death (multi-variate: HR=3.566, 95% CI:1.558-8.160, P=0.003) when compared to patients with an AJCC stage of T1+T2. Table 5 illustrated that patients exhibiting elevated levels of infiltrating CD8+T, CD103+CD8+T, RUNX1+T, CD8+RUNX1+T, CD8+RUNX2+T, CD8+RUNX3+T and CD103+CD8+RUNX3+T cells demonstrated a reduced risk of mortality in comparison to patients with lower levels (uni-variate: HR=0.342, 95% CI: 0.144-0.809, P=0.015; HR=0.348, 95% CI: 0.162-0.744, P=0.006; HR=0.390, 95% CI: 0.157-0.968, P=0.0.42; HR=0.359, 95% CI: 0.166-0.775, P=0.009; HR=0.467, 95% CI: 0.168-1.297, P=0.031; HR=0.240, 95% CI: 0.101-0.571, P=0.001; HR=0.362, 95% CI: 0.158-0.828, P=0.016, respectively). Besides, the percentages of infiltrating CD103+CD8+T cells, CD8+RUNX1+T cells and CD8+RUNX3+T cells could also serve as significant prognostic indicators for predicting the survival outcomes of CRC patients, irrespective of other clinicopathological variables including gender, age, tumor size, pathological grade, and AJCC stage (Table 5).
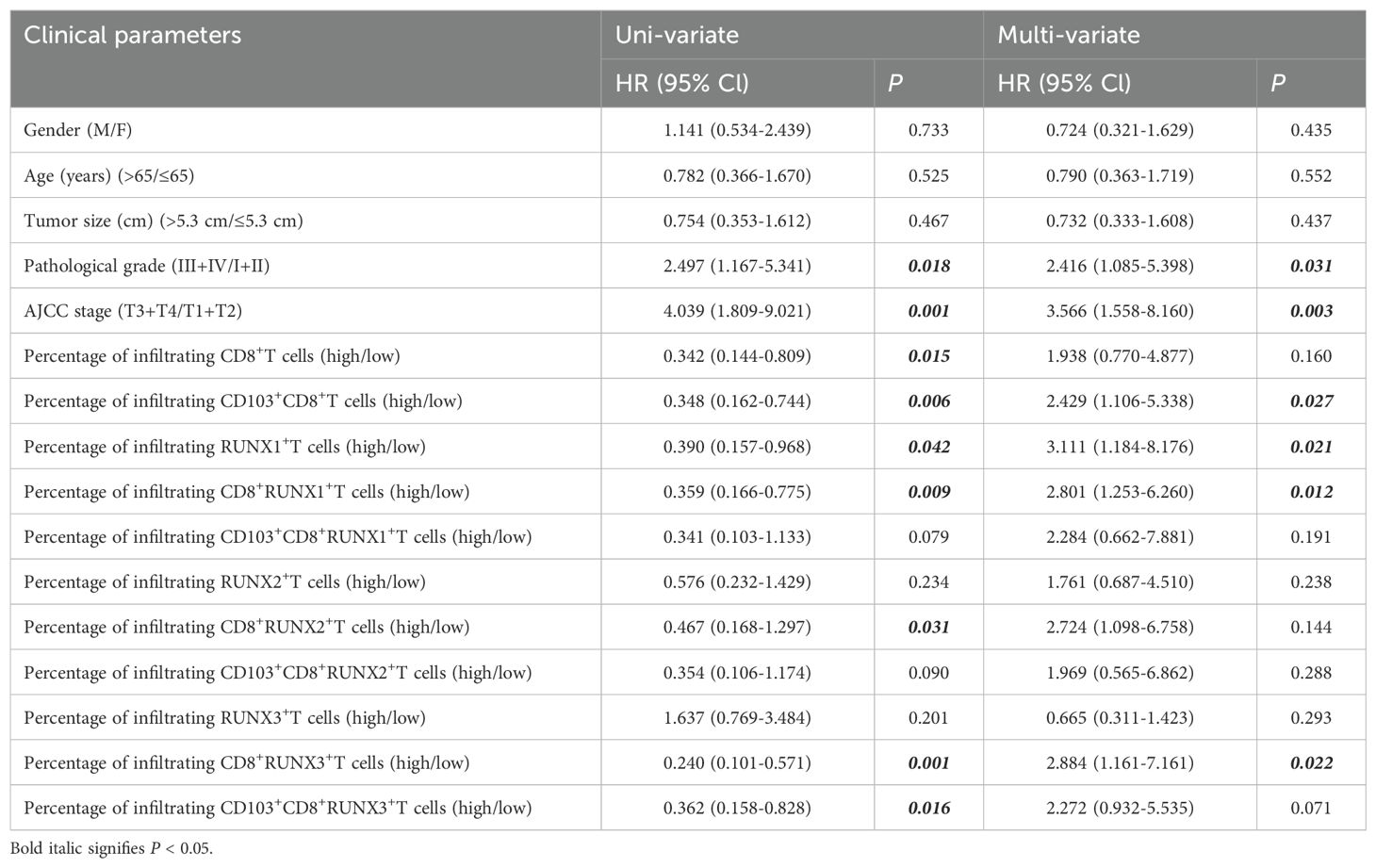
Table 5. Cox model analysis for the correlation between the expression of RUNX, infiltrating CD8+T, CD103+CD8+T cells and clinical parameters of patients.
Correlation between the expression levels of RUNX1, RUNX2, RUNX3 and immune infiltration in colorectal cancer
Immune infiltration plays a vital role in tumor progression. The TIMER and TISIDB platforms were utilized to examine the association between RUNX family genes and immune cell infiltration in COAD. Figure 7 showed a significant correlation between the expression of RUNX1, RUNX2, and RUNX3 and the abundance of TILs in COAD, according to the TIMER database. For instance, RUNX1 expression was positively correlated with infiltrating degree of CD8+T cell (rho=0.24), CD4+T cell (rho=0.467), B cell (rho=0.037), macrophage (rho=1.401), neutrophil (rho=0.36) and dendritic cell (rho=0.384) (Figure 7A). RUNX2 expression was positively correlated with infiltrating degree of CD8+T cell (rho=0.371), CD4+T cell (rho=0.445), B cell (rho=0.22), macrophage (rho=0.553), neutrophil (rho=0.477) and dendritic cell (rho=0.479) (Figure 7B). RUNX3 expression was positively correlated with infiltrating degree of CD8+T cell (rho=0.27), CD4+T cell (rho=0.483), B cell (rho=0.145), macrophage (rho=0.391), neutrophil (rho=0.482) and dendritic cell (rho=0.51) (Figure 7C). All the P-values were significantly less than 0.001. Furthermore, an examination was conducted on the correlation between RUNX family genes and various biomarkers of TILs (including CD8+ T cells, CD4+ T cells, NK cells, B cells, monocytes, DCs, TAMs, M1/M2 macrophages, neutrophils, T cells, and related subtypes) in COAD using data from the GEPIA database (refer to Table 6). The findings suggest that the RUNX1, RUNX2, and RUNX3 genes are correlated with a significant portion of tumor-infiltrating lymphocyte markers in colorectal adenocarcinoma. Various subsets of T cells, such as Th1, Th2, Tfh, Treg, resident, cytotoxic, exhausted, and effector memory T cells, were also analyzed. The RUNX family genes are implicated in the regulation of immune infiltration in colorectal cancer, as shown in Table 6.
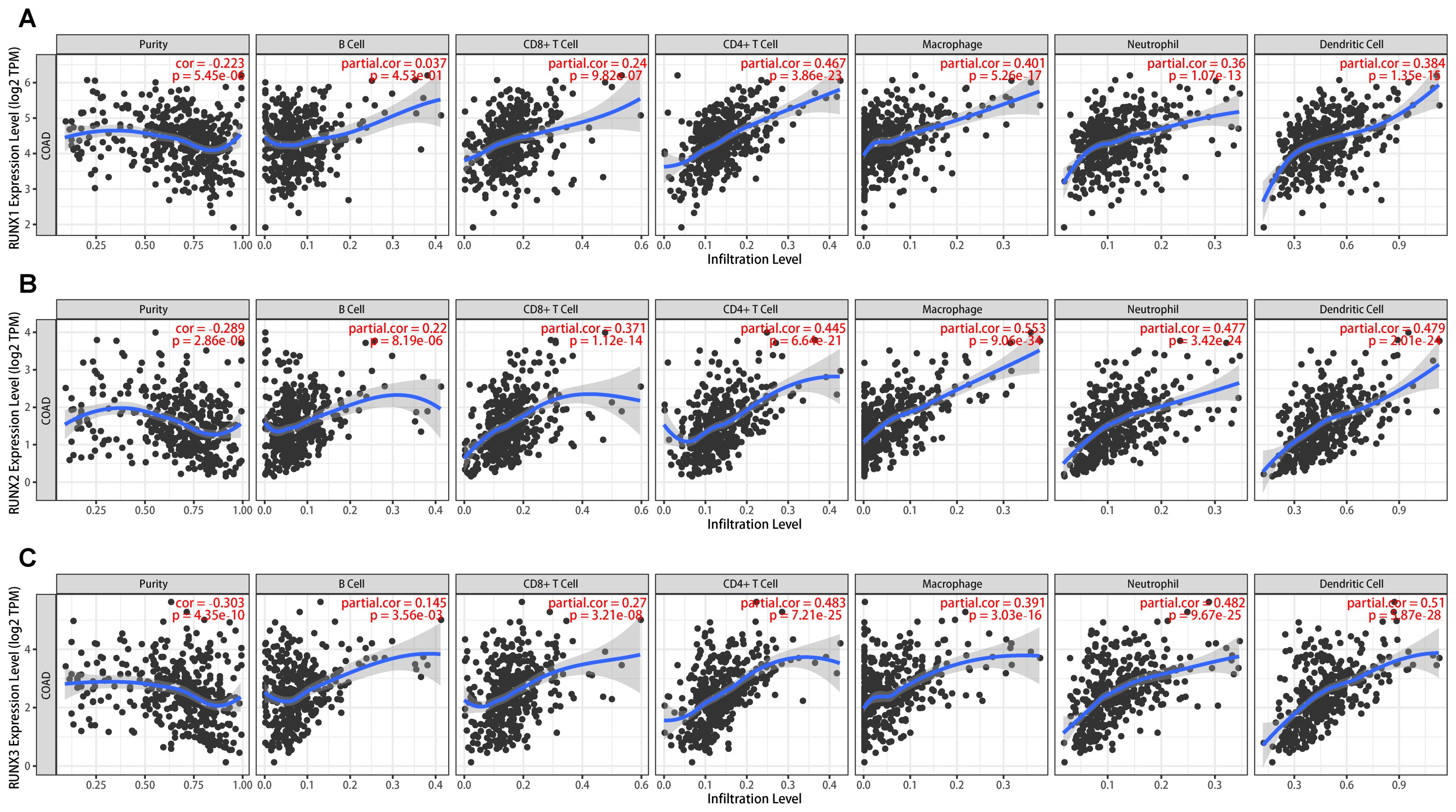
Figure 7. The association between RUNX family gene and cell subsets in human colorectal cancer based on TIMER platform. (A) The scatter plots showed the association between RUNX1 expression and B cell, CD8+T, CD4+T, macrophage, neutrophil, dendritic cell and tumor purity. (B) The scatter plots showed the association between RUNX2 expression and B cell, CD8+T, CD4+T, macrophage, neutrophil, dendritic cell and tumor purity. (C) The scatter plots showed the association between RUNX3 expression and B cell, CD8+T, CD4+T, macrophage, neutrophil, dendritic cell and tumor purity.
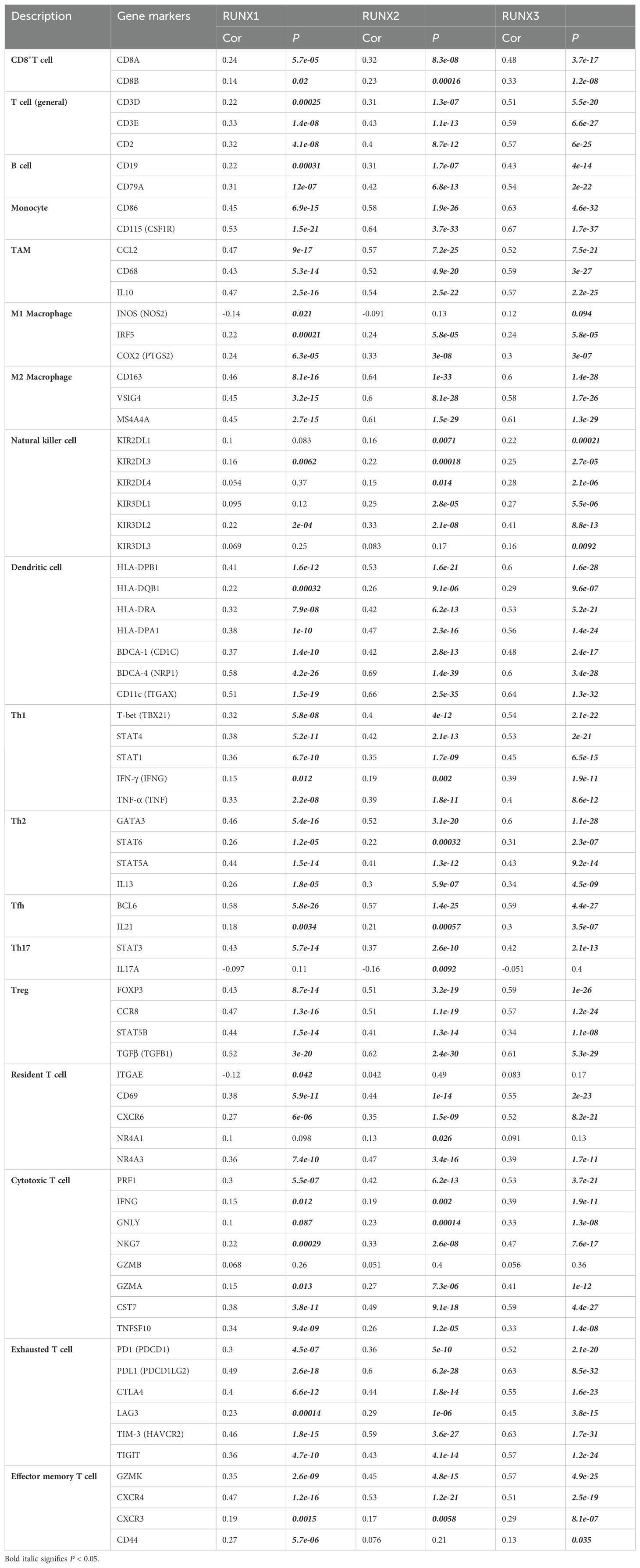
Table 6. Correlation analysis between RUNX family genes and related gene markers of monocyte, macrophages, and T-cell exhaustion in Gene Expression Profiling Interaction Analysis (GEPIA).
Correlation between the expression levels of RUNX1, RUNX2, RUNX3 and tumor infiltrating CD8+T cells in colorectal cancer
Upon closer examination of the relationship between the RUNX family gene and infiltrating CD8+T cells, it was observed that the values of Spearman’s correlation test rho exhibited a gradual increase from RUNX1 to RUNX3 on the TISIDB platform (Figures 8A, E, I). For instance, the spearman correlation rho was -0.133 between RUNX1 and active CD8+T (Act_CD8), data statistically significant, revealed a negative correlation between RUNX1 and active CD8+T cells (Figure 8B). While the rho value was 0.039 between RUNX2 and active CD8+T with a 0.401 P value, while revealed a none significant statistically correlation (Figure 8F). And the rho value became 0.309 between RUNX3 and active CD8+T, with statistically significant data again (Figure 8J). Similarly, the Spearman correlation coefficient rho between RUNX1, RUNX2 to RUNX3 and central memory CD8+T (Tcm_CD8) cells showed a stepwise increase (Figures 8C, G, K), so did the effector memory CD8+T (Tem_CD8) (Figures 8D, H, L). Besides, the Spearman’s correlation test rho value also presented that the correlation between RUNX genes with active CD8+T, central memory CD8+T and effector memory CD8+T were progressively increased, respectively (Figures 8B–D, F–H, J–L). These results indicated that RUNX family genes might be more relevant to effector CD8+ T than to central memory CD8+ T cells and activated CD8+ T cells and RUNX3 showed more relevant to effector CD8+ T than to central memory CD8+ T cells and activated CD8+ T cells than RUNX2 and RUNX1.
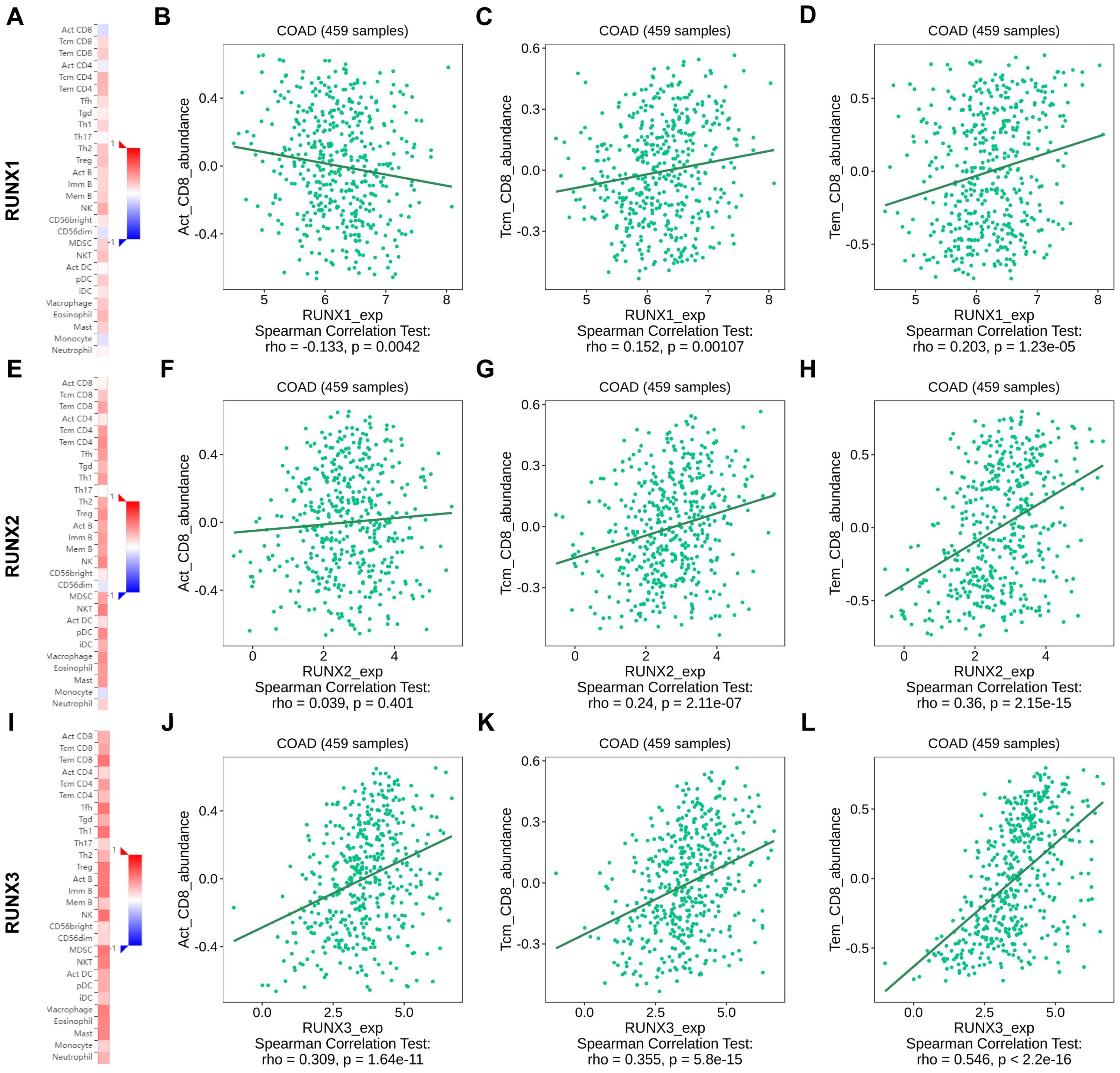
Figure 8. The association between RUNX family gene and immune infiltrating CD8+T cell subsets in human colorectal cancer based on TISIDB platform. (A, E, I) Heatmap showed the correlation of RUNX1 (A), RUNX2 (E) and RUNX3 (I) expression and immune infiltration in pan-cancer. (B–D) Correlation between RUNX1 and active CD8+T, central memory CD8+T and effector memory CD8+T. (F–H) Correlation between RUNX1 and active CD8+T, central memory CD8+T and effector memory CD8+T. (J–L) Correlation between RUNX1 and active CD8+T, central memory CD8+T and effector memory CD8+T.
Figure 9 illustrates significant positive correlations between the expressions of RUNX1 and RUNX2, RUNX1 and RUNX3, as well as RUNX2 and RUNX3 in both CD8+T cells and CD103+CD8+T cells within the context of human CRC TMA in our mIHC findings. The correlation coefficients between CD8+RUNX2+T and CD8+RUNX3+T, as well as CD103+CD8+RUNX2+T and CD103+CD8+RUNX3+T, were notably higher compared to other comparison groups (R=0.8548, P<0.0001, Figure 9C, R=0.7783, P<0.0001, Figure 9F, respectively).
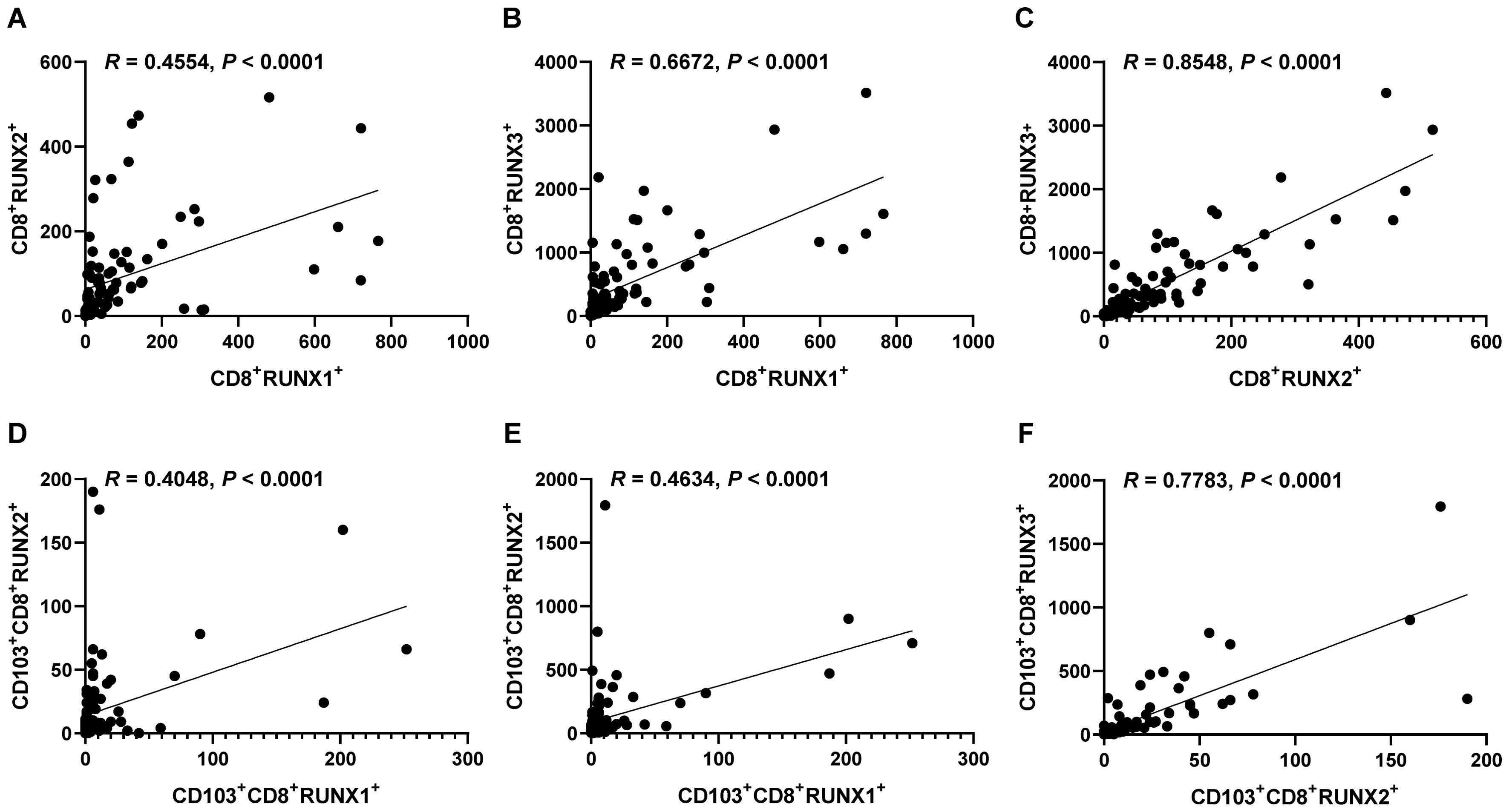
Figure 9. Correlations of the expressions of RUNX1, RUNX2 and RUNX3 in CD8+TILs and CD103+CD8+TILs in human CRC TMA. (A) The dot plot showed the correlation between CD8+RUNX2+T and CD8+RUNX1+T. (B) The dot plot showed the correlation between CD8+RUNX3+T and CD8+RUNX1+T. (C) The dot plot showed the correlation between CD8+RUNX3+T and CD8+RUNX2+T. (D) The dot plot showed the correlation between CD103+CD8+RUNX2+T and CD103+CD8+RUNX1+T. (E) The dot plot showed the correlation between CD103+CD8+RUNX3+T and CD103+CD8+RUNX1+T. (F) The dot plot showed the correlation between CD103+CD8+RUNX3+T and CD103+CD8+RUNX2+T.
Discussion
Recent studies have shown that each member of the RUNX family may play a role in various stages of tumor development, such as cell proliferation, apoptosis, and metastasis, indicating their potential as targets for diagnostic and therapeutic interventions in cancer. For instance, RUNX1, including its mutations, has been reported to serve as a prognostic factor in tumors including leukemia, myeloid neoplasia, and renal cancer (39–41). RUNX2 has been reported to be a novel prognostic signature and correlate with immune infiltrates in bladder cancer, cervical cancer and gastric cancer (25, 26, 42). RUNX3 has been found elevated in renal cancer and is associated with shorter progression-free survival, and high RUNX3 expression predicts a greater benefit of IO/TKI (immune checkpoint inhibitor combined with a lorosine kinase inhibitor) therapy compared to TKI monotherapy (43). In our current study, we initially examined the expression of all the three RUNX genes across different tumors using the TCGA database (Figure 1). By analyzing data from GEPIA, UALCAN, and TIMER, three widely utilized TCGA database platforms, we observed that the expression patterns of RUNX1 and RUNX2 were quite similar across most tumors, both showing increased expression levels, whereas RUNX3 was more frequently down-regulated in a larger number of tumors (Figure 1J). These findings align with previous reports on RUNX expression in pan-cancer contexts (44, 45), suggesting that abnormal RUNX expression is linked to prognosis in various cancers.
In the next section, we explored the relationship between RUNX family and prognosis. According to our mIHC results from Figure 2, especially Figures 2D–F, there was an up-regulation of RUNX1 expression and a down-regulation of RUNX3 expression in CRC TMA tumor tissues. The result was also consistent with our previous findings in the UALCAN database (Figures 1A–C). The different expression status of RUNX family members in the same one COAD tumor type, made us further explore their correlation with immune cells in the surrounding immune microenvironment. It has been shown that tumor-infiltrating lymphocytes (TILs) within the TME are a reliable determinant of prognosis and immunity to immunotherapy (46–48). Our study demonstrated that RUNX family gene were all strongly associated with TILs and played a key role in the development of TMEs (Figure 7, Table 6).
The RUNX family is essential for lineage characterization of various types of hematopoietic cells, including T lymphocytes (11, 12, 49). Current research has shown that the RUNX gene family plays a critical role in regulating the development, function, and immune response of CD8+T cells. RUNX1 and RUNX3 play important regulatory roles in promoting the development of immature CD4+CD8+ double-positive (DP) thymocytes into mature CD4+Th and CD8+CTL cells. RUNX1 binds to CD4 to silence and inhibit transcription in immature double-negative thymocytes while activating CD8 progression from double-negative to double-positive thymocytes. RUNX3, on the other hand can bind the core sequence of CD4 silencing and establish epigenetic silencing in CD4-CD8+ cytotoxic T cells (12). One of the major targets of tumor immunotherapy is to rescue and/or maintain optimal effector CD8+ T-cell function by minimizing tumor-induced negative factors. RUNX family proteins establish a core transcriptional program in CD8+CTL cells. T cells acquire specific effector functions by preparing chromatin landscapes early in development, a process that involves the sequential cooperation of transcription factors such as RUNX1, PU.1, and BCL11B, which sequentially and synergistically anchor mSWI/SNF with RUNX1 to balance the T cell effector landscape (50). RUNX3 acts as an upstream transcription factor that binds directly to cis-regulatory regions of Prf1 (which encodes perforin) and Eomes (a type of T-box protein) regulatory region, with T-box driving the gene expression program in activated CD8+CTL cells (13). RUNX3 is also recognized as a central regulator of tissue-resident memory CD8+T (TRM) cell development (51). The integrin CD103 on TRM cells is a target gene of RUNX3. In CD8+ T cells stimulated by antigen receptors, RUNX3 is essential for establishing the signature cytotoxic effector function of CTLs to promote their clearance from tumors in the microenvironment. In addition to RUNX1 and RUNX3, RUNX2 also affects the development of memory CD8+T cells (52). In a murine model of acute lymphocytic choroidal meningitis virus infection, RUNX2 has been identified as a crucial factor for the maintenance of long-term memory CD8+ T-cell persistence and for influencing the development of memory CD8+ T cells (52). In our previous studies, we found that tumor infiltration CD8+T and CD103+CD8+T could serve as good clinical prognostic indicators in CRC (38). We also confirmed this conclusion at the beginning of this study (Figure 3). So, in our present study, we further analyzed the prognostic values of RUNX1, RUNX2 and RUNX3 expressed on CD8+TILs and CD103+CD8+TILs in human CRC. According to the results from Figures 4–6, Table 5 in this study, we found that CD8+RUNX1+T, CD8+RUNX2+T, CD8+RUNX3+T and CD103+CD8+RUNX3+T cells could be important prognostic predictors for the survival prediction of CRC patients. Some previous researches have mentioned that CROX (Cluster Regulation of RUNX) could serve as a potential novel therapeutic approach for tumors (53, 54). In this study, our analysis of correlation between the expression levels of RUNX1, RUNX2 and RUNX3 in infiltrating CD8+T and CD103+CD8+T cells in colorectal cancer, implicated that the transcription factors RUNX1, RUNX2, and RUNX3 might exhibit a synergistic interaction effect within clusters and mutually influence each other (Figure 9). Although the comprehensive RUNX family cluster regulation has yet to be applied in the foundational and clinical fields, manipulating the RUNX gene family may have potential applications in immunotherapy, further study in RUNX gene family cluster will provide new insights and strategies for tumor immunotherapy.
The following defects and limitations may exist in this paper: 1) Since we used commercial tissue microarrays, insufficient sample size and insufficient sample resource may be limited. In subsequent research, more intraoperative clinical fresh tissue samples, in addition to the patient’s tumor and adjacent normal tissue, peripheral blood samples could also be included, to further validate the exploratory conclusions drawn in this paper. 2) This article does not provide a detailed study on how the RUNX family members, including RUNX1, RUNX2, and RUNX3, specifically regulate the mechanisms of tumor-infiltrating CD8+ T cells and CD103+CD8+ T cells in colorectal cancer, as well as the functional roles they play in the anti-tumor immune response within the CRC microenvironment. It has been reported that in antigen-stimulated CD8+ T cells, RUNX is crucial for developing CTL’s cytotoxic function and enhancing tumor clearance in the TME (13). The RUNX family members also appear to play a critical role in regulating the effector differentiation of effector T cells (Teff) and memory T cells (Tmem) in vivo. For instance, within an in vivo T-cell CRISPR screening platform, the ETS family transcription factor Fli1 was identified as a repressor of Teff cell biological function. The deletion of Fli1 led to increased chromatin accessibility at RUNX motifs, thereby enhancing the biological efficacy of RUNX-driven Teff cells, which demonstrated increased resistance to infection and tumorigenesis (55). Furthermore, existing literature indicates that the reversal of RUNX methylation facilitates CD8+ TILs infiltration, mitigates CD8+ T cell exhaustion, and augments the anti-tumor immune response of CD8+ T cells (56). Studies in pancreatic ductal adenocarcinoma and hepatocellular carcinoma have found that RUNX gene-edited CAR-T cells showed enhanced persistence, cytotoxic potential and tumor-resident capacity, and could enhance anti-tumor effects (57, 58). These studies indicate that the RUNX family may play a significant role in the functional differentiation of T effector cells, thereby establishing a theoretical foundation for considering RUNX as a potential target for predicting response rates to immunotherapy. The mechanism through which the RUNX family augments anti-tumor efficacy in CRC by modulating the effector functions of tumor-infiltrating CD8+ T cells and CD103+CD8+ T cells deserve further elucidation.
To sum up, our recent research highlights the prognostic importance of the infiltration intensity of RUNX family genes in CD8+T and resident CD103+CD8+T cells in human colorectal cancer. The RUNX family could be a vital and promising prognostic biomarker for forecasting disease progression and immune evasion in colorectal cancer patients.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethics statement
All participants provided informed consent for their involvement in the study, which was approved by the ethics committee of the Third Affiliated Hospital of Soochow University. The research involving human subjects was also reviewed and approved by Shanghai Outdo Biotech Co., Ltd., Shanghai, China (YB M-05-02). Written consent was obtained from all patients prior to their participation. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
YL: Data curation, Formal analysis, Investigation, Methodology, Writing – original draft. JC: Formal analysis, Visualization, Writing – original draft. AL: Formal analysis, Methodology, Writing – original draft. YW: Software, Visualization, Writing – original draft. JG: Investigation, Methodology, Writing – original draft. MY: Resources, Writing – original draft. BX: Software, Visualization, Writing – original draft. XZ: Formal analysis, Project administration, Writing – original draft. LC: Conceptualization, Project administration, Writing – original draft. JJ: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by grants from the National Natural Science Foundation of China (32270955, 81972869, 82172689, 82473269), Natural Science Foundation of Jiangsu Province (BK20211065), China Postdoctoral Science Foundation (2021M700547), Key Medical Disciplines in Jiangsu Province (YXZDXK202236), Postgraduate Research & Practice Innovation Program of Jiangsu Province (KYCX23_3265), Major Program of Science and Technology Project of Changzhou Health Commission (ZD202329), Young Talent Development Plan of Changzhou Health Commission (CZQM2020044), the Science and Technology Support Plan (Social Development) Project of Changzhou (CE20235058), Changzhou Sci&Tech Program (CZ20241101), Changzhou Clinical Medical Center (CZZX202201), Outstanding Talent of Changzhou ‘The 14th Five-Year Plan’ High-Level Health Talents Training Project, and Prospective Research Program of Changzhou Xitaihu Development Foundation For Frontier Cell-Therapeutic Technology
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Glossary
RUNX: runt-related transcription factor
CRC: colorectal cancer
TME: tumor microenvironment
PD-1: programmed death 1
PD-L1: programmed cell death 1 ligand 1
CAR-T: chimeric antigen receptor T-cell immunotherapy
TIL: tumor-infiltrating lymphocyte
CXCL13: chemokine C-X-C motif ligand 13
TGF-β: transforming growth factor beta
BMP: bone morphogenetic protein
YAP1: Yes-associated protein 1
mIHC: multi-color immunohistochemically staining
DAPI: Diamidino-2-phenylindole
DC: dendritic cell
TAM: tumor-associated macrophage
TCGA: The Cancer Genome Atlas
BLCA: bladder urothelial carcinoma
BRCA: breast carcinoma
CESC: cervical squamous cell carcinoma and endocervical adenocarcinoma
COAD: colon adenocarcinoma
DLBC: lymphoid neoplasm diffuse large B-cell lymphoma
ESCA: esophageal carcinoma
GBM: glioblastoma multiforme
HNSC: head and neck squamous cell carcinoma
KICH: kidney chromophobe
KIRC: kidney renal clear cell carcinoma
KIRP: kidney renal papillary cell carcinoma
LAML: acute myeloid leukemia
LGG: lower-grade glioma
LIHC: liver hepatocellular carcinoma
LUAD: lung adenocarcinoma
LUSC: lung squamous cell carcinoma
PAAD: pancreatic adenocarcinoma
PRAD: prostate adenocarcinoma
SKCM: skin cutaneous melanoma
STAD: stomach adenocarcinoma
TGCT: testicular germ cell tumors
THCA: thyroid carcinoma
THYM: thymoma
UCEC: uterine corpus endometrial carcinoma
UVM: uveal melanoma
References
1. Johdi NA, Sukor NF. Colorectal cancer immunotherapy: Options and strategies. Front Immunol. (2020) 11:1624. doi: 10.3389/fimmu.2020.01624
2. Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA: Cancer J Clin. (2023) 73:17–48. doi: 10.3322/caac.21763
3. Dagar G, Gupta A, Masoodi T, Nisar S, Merhi M, Hashem S, et al. Harnessing the potential of CAR-T cell therapy: Progress, challenges, and future directions in hematological and solid tumor treatments. J Transl Med. (2023) 21:449. doi: 10.1186/s12967-023-04292-3
4. Banchereau R, Leng N, Zill O, Sokol E, Liu G, Pavlick D, et al. Molecular determinants of response to PD-L1 blockade across tumor types. Nat Commun. (2021) 12:3969. doi: 10.1038/s41467-021-24112-w
5. Shang S, Yang Y-W, Chen F, Yu L, Shen S-H, Li K, et al. TRIB3 reduces CD8+ T cell infiltration and induces immune evasion by repressing the STAT1-CXCL10 axis in colorectal cancer. Sci Transl Med. (2022) 14:eabf0992. doi: 10.1126/scitranslmed.abf0992
6. McAuliffe J, Chan HF, Noblecourt L, Ramirez-Valdez RA, Pereira-Almeida V, Zhou Y, et al. Heterologous prime-boost vaccination targeting MAGE-type antigens promotes tumor T-cell infiltration and improves checkpoint blockade therapy. J Immunotherapy Cancer. (2021) 9:e003218. doi: 10.1136/jitc-2021-003218
7. Sun Q, Cai D, Liu D, Zhao X, Li R, Xu W, et al. BCL6 promotes a stem-like CD8+ T cell program in cancer via antagonizing BLIMP1. Sci Immunol. (2023) 8:eadh1306. doi: 10.1126/sciimmunol.adh1306
8. Dai S, Zeng H, Liu Z, Jin K, Jiang W, Wang Z, et al. Intratumoral CXCL13 + CD8 + T cell infiltration determines poor clinical outcomes and immunoevasive contexture in patients with clear cell renal cell carcinoma. J Immunotherapy Cancer. (2021) 9:e001823. doi: 10.1136/jitc-2020-001823
9. Choo J, Kua LF, Soe MY, Asuncion BR, Tan BKJ, Teo CB, et al. Clinical relevance of PD-1 positive CD8 T-cells in gastric cancer. Gastric Cancer. (2023) 26:393–404. doi: 10.1007/s10120-023-01364-7
10. Huang H, Huang Z, Ge J, Yang J, Chen J, Xu B, et al. CD226 identifies functional CD8+T cells in the tumor microenvironment and predicts a better outcome for human gastric cancer. Front Immunol. (2023) 14:1150803. doi: 10.3389/fimmu.2023.1150803
11. Seo W, Taniuchi I. The roles of RUNX family proteins in development of immune cells. Mol Cells. (2020) 43:107–13. doi: 10.14348/molcells.2019.0291
12. Korinfskaya S, Parameswaran S, Weirauch MT, Barski A. Runx transcription factors in T cells—what is beyond thymic development? Front Immunol. (2021) 12:701924. doi: 10.3389/fimmu.2021.701924
13. Pipkin ME. Runx proteins and transcriptional mechanisms that govern memory CD8 T cell development. Immunol Rev. (2021) 300:100–24. doi: 10.1111/imr.12954
14. Hong D, Knelson EH, Li Y, Durmaz YT, Gao W, Walton E, et al. Plasticity in the absence of NOTCH uncovers a RUNX2-dependent pathway in small cell lung cancer. Cancer Res. (2022) 82:248–63. doi: 10.1158/0008-5472.CAN-21-1991
15. Kim BR, Na YJ, Kim JL, Jeong YA, Park SH, Jo MJ, et al. RUNX3 suppresses metastasis and stemness by inhibiting hedgehog signaling in colorectal cancer. Cell Death Differentiation. (2020) 27:676–94. doi: 10.1038/s41418-019-0379-5
16. Sweeney K, Cameron ER, Blyth K. Complex interplay between the RUNX transcription factors and wnt/β-catenin pathway in cancer: A tango in the night. Mol Cells. (2020) 43:188–97. doi: 10.14348/molcells.2019.0310
17. Meng G, Wei J, Wang Y, Qu D, Zhang J. miR-21 regulates immunosuppression mediated by myeloid-derived suppressor cells by impairing RUNX1-YAP interaction in lung cancer. Cancer Cell Int. (2020) 20:495. doi: 10.1186/s12935-020-01555-7
18. Dutta B, Osato M. The RUNX family, a novel multifaceted guardian of the genome. Cells. (2023) 12:255. doi: 10.3390/cells12020255
19. Krishnan V. The RUNX family of proteins, DNA repair, and cancer. Cells. (2023) 12:1106. doi: 10.3390/cells12081106
20. Simon L, Spinella J-F, Yao C-Y, Lavallée V-P, Boivin I, Boucher G, et al. High frequency of germline RUNX1 mutations in patients with RUNX1-mutated AML. Blood. (2020) 135:1882–6. doi: 10.1182/blood.2019003357
21. Kanellou P, Georgakopoulos-Soares I, Zaravinos A. Deregulated gene expression profiles and regulatory networks in adult and pediatric RUNX1/RUNX1T1-positive AML patients. Cancers (Basel). (2023) 15:1795. doi: 10.3390/cancers15061795
22. Xu X, Zhang C, Trotter TN, Gowda PS, Lu Y, Ponnazhagan S, et al. Runx2 deficiency in osteoblasts promotes myeloma progression by altering the bone microenvironment at new bone sites. Cancer Res. (2020) 80:1036–48. doi: 10.1158/0008-5472.CAN-19-0284
23. Wang Y, Qin D, Gao Y, Zhang Y, Liu Y, Huang L. Identification of therapeutic targets for osteosarcoma by integrating single-cell RNA sequencing and network pharmacology. Front Pharmacol. (2022) 13:1098800. doi: 10.3389/fphar.2022.1098800
24. Vishal M, Swetha R, Thejaswini G, Arumugam B, Selvamurugan N. Role of Runx2 in breast cancer-mediated bone metastasis. Int J Biol Macromol. (2017) 99:608–14. doi: 10.1016/j.ijbiomac.2017.03.021
25. Cruz-De la Rosa MI, Jiménez-Wences H, Alarcón-Millán J, Romero-López MJ, Castañón-Sánchez CA, Salmerón-Bárcenas EG, et al. miR-218-5p/RUNX2 axis positively regulates proliferation and is associated with poor prognosis in cervical cancer. Int J Mol Sci. (2022) 23:6993. doi: 10.3390/ijms23136993
26. Jin Y, Huang S, Wang Z. Identify and validate RUNX2 and LAMA2 as novel prognostic signatures and correlate with immune infiltrates in bladder cancer. Front Oncol. (2023) 13:1191398. doi: 10.3389/fonc.2023.1191398
27. Jin G, Ruan Q, Shangguan F, Lan L. RUNX2 and LAMC2: Promising pancreatic cancer biomarkers identified by an integrative data mining of pancreatic adenocarcinoma tissues. Aging (milano). (2021) 13:22963–84. doi: 10.18632/aging.203589
28. Wu K, Nie Y, Guo C, Chen Y, Ding J, Fan D. Molecular basis of therapeutic approaches to gastric cancer. J Gastroenterol Hepatol. (2009) 24:37–41. doi: 10.1111/j.1440-1746.2008.05753.x
29. Yano T, Ito K, Fukamachi H, Chi X-Z, Wee H-J, Inoue K, et al. The RUNX3 tumor suppressor upregulates bim in gastric epithelial cells undergoing transforming growth factor beta-induced apoptosis. Mol Cell Biol. (2006) 26:4474–88. doi: 10.1128/MCB.01926-05
30. Zhang Z, Chen G, Cheng Y, Martinka M, Li G. Prognostic significance of RUNX3 expression in human melanoma. Cancer. (2011) 117:2719–27. doi: 10.1002/cncr.25838
31. Yu J, Tian X, Chang J, Liu P, Zhang R. RUNX3 inhibits the proliferation and metastasis of gastric cancer through regulating miR-182/HOXA9. Biomedicine Pharmacotherapy = Biomedecine Pharmacotherapie. (2017) 96:782–91. doi: 10.1016/j.biopha.2017.08.144
32. Lee Y-S, Lee J-Y, Song S-H, Kim D-M, Lee J-W, Chi X-Z, et al. K-ras-activated cells can develop into lung tumors when Runx3-mediated tumor suppressor pathways are abrogated. Mol Cells. (2020) 43:889–97. doi: 10.14348/molcells.2020.0182
33. Chen L, Sun R, Xu J, Zhai W, Zhang D, Yang M, et al. Tumor-derived IL33 promotes tissue-resident CD8+ T cells and is required for checkpoint blockade tumor immunotherapy. Cancer Immunol Res. (2020) 8:1381–92. doi: 10.1158/2326-6066.CIR-19-1024
34. Liu Y, Xing Z, Yuan M, Xu B, Chen L, Zhang D, et al. IL1R2 promotes tumor progression via JAK2/STAT3 pathway in human clear cell renal cell carcinoma. Pathol - Res Pract. (2022) 238:154069. doi: 10.1016/j.prp.2022.154069
35. Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK, et al. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia (New York NY). (2017) 19:649–58. doi: 10.1016/j.neo.2017.05.002
36. Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. (2020) 48:W509–14. doi: 10.1093/nar/gkaa407
37. Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, et al. TIMER: A web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. (2017) 77:e108–10. doi: 10.1158/0008-5472.CAN-17-0307
38. Hu W, Sun R, Chen L, Zheng X, Jiang J. Prognostic significance of resident CD103+CD8+T cells in human colorectal cancer tissues. Acta Histochem. (2019) 121:657–63. doi: 10.1016/j.acthis.2019.05.009
39. Lew-Derivry L, Marceau-Renaut A, Fenwarth L, Cuccuini W, Ballerini P, Ferreboeuf M, et al. Prognostic impact of RUNX1 mutations and deletions in pediatric acute myeloid leukemia: Results from the french ELAM02 study group. Leukemia. (2023) 37:1723–6. doi: 10.1038/s41375-023-01931-y
40. Ma J, He S, Li M, Peng Y, Yang X, Chen L, et al. RUNX1 predicts poor prognosis and correlates with tumor progression in clear cell renal carcinoma. Pathology Res Pract. (2023) 251:154886. doi: 10.1016/j.prp.2023.154886
41. Kubota Y, Gu X, Terkawi L, Bodo J, Przychodzen BP, Awada H, et al. Molecular and clinical analyses of PHF6 mutant myeloid neoplasia provide their pathogenesis and therapeutic targeting. Nat Commun. (2024) 15:1832. doi: 10.1038/s41467-024-46134-w
42. Guo Z, Zhou K, Wang Q, Huang Y, Ji J, Peng Y, et al. The transcription factor RUNX2 fuels YAP1 signaling and gastric cancer tumorigenesis. Cancer Sci. (2021) 112:3533–44. doi: 10.1111/cas.15045
43. Wang J, Zhang S, Wang Y, Zhu Y, Xu X, Guo J. RUNX3 pathway signature predicts clinical benefits of immune checkpoint inhibition plus tyrosine kinase inhibition in advanced renal cell carcinoma. BMC Urol. (2024) 24:8. doi: 10.1186/s12894-023-01356-w
44. Pan S, Sun S, Liu B, Hou Y. Pan-cancer landscape of the RUNX protein family reveals their potential as carcinogenic biomarkers and the mechanisms underlying their action. J Trans Internal Med. (2022) 10:156–74. doi: 10.2478/jtim-2022-0013
45. Zhao H, Chen Y, Shen P, Gong L. Prognostic value and immune characteristics of RUNX gene family in human cancers: A pan-cancer analysis. Aging (milano). (2022) 14:4014–35. doi: 10.18632/aging.204065
46. Hu C, Shu L, Chen C, Fan S, Liang Q, Zheng H, et al. A prediction model integrated genomic alterations and immune signatures of tumor immune microenvironment for early recurrence of stage I NSCLC after curative resection. Trans Lung Cancer Res. (2022) 11:24–42. doi: 10.21037/tlcr-21-751
47. Qin M, Chen G, Hou J, Wang L, Wang Q, Wang L, et al. Tumor-infiltrating lymphocyte: Features and prognosis of lymphocytes infiltration on colorectal cancer. Bioengineered. (2022) 13:14872–88. doi: 10.1080/21655979.2022.2162660
48. Bie F, Tian H, Sun N, Zang R, Zhang M, Song P, et al. Comprehensive analysis of PD-L1 expression, tumor-infiltrating lymphocytes, and tumor microenvironment in LUAD: Differences between asians and caucasians. Clin Epigenet. (2021) 13:229. doi: 10.1186/s13148-021-01221-3
49. Shin B, Zhou W, Wang J, Gao F, Rothenberg EV. Runx factors launch T cell and innate lymphoid programs via direct and gene network-based mechanisms. Nat Immunol. (2023) 24:1458–72. doi: 10.1038/s41590-023-01585-z
50. Gamble N, Bradu A, Caldwell JA, McKeever J, Bolonduro O, Ermis E, et al. PU.1 and BCL11B sequentially cooperate with RUNX1 to anchor mSWI/SNF to poise the T cell effector landscape. Nat Immunol. (2024) 25:860–72. doi: 10.1038/s41590-024-01807-y
51. Milner JJ, Toma C, Yu B, Zhang K, Omilusik K, Phan AT, et al. Runx3 programs CD8+ T cell residency in non-lymphoid tissues and tumours. Nature. (2017) 552:253–7. doi: 10.1038/nature24993
52. Olesin E, Nayar R, Saikumar-Lakshmi P, Berg LJ. The transcription factor Runx2 is required for long-term persistence of antiviral CD8+ memory T cells. ImmunoHorizons. (2018) 2:251–61. doi: 10.4049/immunohorizons.1800046
53. Kamikubo Y. CROX (cluster regulation of RUNX) as a potential novel therapeutic approach. Mol Cells. (2020) 43:198–202. doi: 10.14348/molcells.2019.0268
54. Kamikubo Y. Genetic compensation of RUNX family transcription factors in leukemia. Cancer Sci. (2018) 109:2358–63. doi: 10.1111/cas.13664
55. Chen Z, Arai E, Khan O, Zhang Z, Ngiow SF, He Y, et al. In vivo CD8+ T cell CRISPR screening reveals control by Fli1 in infection and cancer. Cell. (2021) 184:1262–80.e22. doi: 10.1016/j.cell.2021.02.019
56. Liu Z, Li X, Gao Y, Liu J, Feng Y, Liu Y, et al. Epigenetic reprogramming of Runx3 reinforces CD8 + T-cell function and improves the clinical response to immunotherapy. Mol Cancer. (2023) 22:84. doi: 10.1186/s12943-023-01768-0
57. Tang J, Sheng J, Zhang Q, Ji Y, Wang X, Zhang J, et al. Runx3-overexpression cooperates with ex vivo AKT inhibition to generate receptor-engineered T cells with better persistence, tumor-residency, and antitumor ability. J Immunotherapy Cancer. (2023) 11:e006119. doi: 10.1136/jitc-2022-006119
Keywords: RUNX, tumor-infiltrating CD8+T cells, multi-color immunohistochemical staining, colorectal cancer, prognosis
Citation: Liu Y, Chen J, Li A, Wu Y, Ge J, Yuan M, Xu B, Zheng X, Chen L and Jiang J (2024) Novel biomarkers: the RUNX family as prognostic predictors in colorectal cancer. Front. Immunol. 15:1430136. doi: 10.3389/fimmu.2024.1430136
Received: 09 May 2024; Accepted: 21 November 2024;
Published: 09 December 2024.
Edited by:
Valentyn Oksenych, University of Bergen, NorwayReviewed by:
Apostolos Zaravinos, European University Cyprus, CyprusKanagaraj Palaniyandi, SRM Institute of Science and Technology, India
Copyright © 2024 Liu, Chen, Li, Wu, Ge, Yuan, Xu, Zheng, Chen and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jingting Jiang, amlhbmdqaW5ndGluZ0BzdWRhLmVkdS5jbg==
 Yingting Liu
Yingting Liu Junjun Chen
Junjun Chen An Li
An Li Yue Wu1,2,3
Yue Wu1,2,3 Junwei Ge
Junwei Ge Maoling Yuan
Maoling Yuan Bin Xu
Bin Xu Xiao Zheng
Xiao Zheng Lujun Chen
Lujun Chen Jingting Jiang
Jingting Jiang