
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 26 July 2024
Sec. Microbial Immunology
Volume 15 - 2024 | https://doi.org/10.3389/fimmu.2024.1430001
Inflammatory bowel disease (IBD) is an idiopathic and persistent inflammatory illness of the bowels, leading to a substantial burden on both society and patients due to its high incidence and recurrence. The pathogenesis of IBD is multifaceted, partly attributed to the imbalance of immune responses toward the gut microbiota. There is a correlation between the severity of the disease and the imbalance in the oral microbiota, which has been discovered in recent research highlighting the role of oral microbes in the development of IBD. In addition, various oral conditions, such as angular cheilitis and periodontitis, are common extraintestinal manifestations (EIMs) of IBD and are associated with the severity of colonic inflammation. However, it is still unclear exactly how the oral microbiota contributes to the pathogenesis of IBD. This review sheds light on the probable causal involvement of oral microbiota in intestinal inflammation by providing an overview of the evidence, developments, and future directions regarding the relationship between oral microbiota and IBD. Changes in the oral microbiota can serve as markers for IBD, aiding in early diagnosis and predicting disease progression. Promising advances in probiotic-mediated oral microbiome modification and antibiotic-targeted eradication of specific oral pathogens hold potential to prevent IBD recurrence.
Inflammatory bowel disease (IBD) is a type of intestinal disease marked by chronic inflammation with an unclear cause, and it is most commonly associated with ulcerative colitis (UC) and Crohn’s disease (CD). Even though IBD can be diagnosed at any age, from early childhood to old age, most new cases are discovered in adolescence and the early stages of adulthood. It is often clinically manifested as diarrhea, abdominal pain, and even bloody stool, with a tendency to delay and relapse (1). The onset and duration of IBD vary, and the disease’s severity is connected to the scope and degree of the lesions. It is often accompanied by arthritis, iritis, skin lesions, oral ulcers, hepatobiliary diseases, osteoporosis, and other parenteral lesions (2). At present, the primary goals of IBD treatment are symptom relief and quality of life enhancement, but there is no cure (3).
The etiology and pathogenesis of IBD are uncertain, although multiple factors, including disrupted intestinal mucosal immune regulation, persistent intestinal infections, impaired intestinal barrier function, genetic predisposition, and environmental factors (4), are believed to have a role in the onset and development of the disease (5). Of note, substantial research efforts have been directed towards unraveling the connection between the gut microbiome and IBD (6). Numerous studies have demonstrated that patients with IBD have marked dysbiosis in their gut microbiome, including reduced bacterial diversity (7), instability within the bacterial community at inflamed mucosal sites (8), as well as increased bacterial translocation and overgrowth (9).
Oral manifestations are increasingly recognized as extraintestinal manifestations of IBD, including UC and CD. These manifestations can present as various oral lesions, such as aphthous ulcers, mucosal tags, angular cheilitis, gingival inflammation, and periodontal disease (2). Ingesting a significant quantity of disordered oral bacteria, a consequence of periodontitis, could potentially disrupt the balance of gut bacteria, thereby causing changes in bacterial metabolites, compromised gut barrier function, and immune dysfunction (10, 11). The prevalence and severity of these oral manifestations may correlate with the activity and extent of intestinal inflammation. These oral symptoms are crucial for professionals to be aware of since they could have a major influence on the quality of life of patients with IBD. The exact mechanisms underlying the oral manifestations of IBD are not entirely comprehended (12). Patients with IBD had higher levels of microorganisms linked to opportunistic infections than non-IBD patients (13). Furthermore, when compared to non-IBD controls, the gut microbiome of IBD patients is noticeably more comparable to the oral microbiome (14). However, it is believed that systemic inflammation, immune dysregulation, altered oral microbiota, and medication side effects may contribute to their development. Systemic inflammation associated with IBD can lead to increased production of pro-inflammatory cytokines (15), which may directly affect the oral mucosa (16). Immune dysregulation in IBD can result in an abnormal immune response to oral pathogens, leading to oral inflammation. Altered oral microbiota, characterized by an imbalance in the makeup of oral microbial communities, could also influence the pathogenesis of oral manifestations in IBD (17). The purpose of this review is to give a broad overview of the advancements made in the field of oral microbiology and its relationship with IBD.
Extensive research has been conducted on the composition of the oral microbiome. More than 250 types of organisms from the oral cavity have been identified through in vitro isolation and characterization, including a number of key pathogens implicated in the development of dental cavities and periodontal disease, for instance Tannerella forsythia, Streptococcus mutans, Aggregatibacter actinomycetemcomitans and Porphyromonas gingivalis (P. gingivalis) (2). Over time, integrated approaches to understanding oral disease states from a multimicrobial perspective have emerged, attributing disease pathology to co-occurring microbial networks whose aggregate activity contributes to pathogenesis rather than just key pathogens. The constant exposure of oral microbial ecosystems to exogenous foreign substances determines the formation of microorganisms and their ability to survive in this environment, as well as the unique relationship between microbes and hosts that depends on factors of selection (18). The ability to select and bind tongue and cheek cells before teeth emerge and the capacity to compete with other microbial species are key traits of pioneering oral microbial colonists like Streptococcus salivarius, Streptococcus mitis, Streptococcus gordonii, and Streptococcus sanguinis. These traits make them ideal for this specific niche. The gut and the oral cavity are the beginning and the end of the alimentary tract’s microbial aggregation. Both include distinct microbiome linked to human health and illness (19), as well as some shared microbiota. The relationship between gut and mouth microbiota is intricate, erratic, and interwoven (20). They can maintain a precise balance under normal physiological settings, but an imbalance in crosstalk will lead to the onset and progression of illnesses (21). More and more evidence showed connections between the oral microbiome and digestive diseases like esophageal cancer, colorectal cancer, acute appendicitis (22), and IBD. The capacity of numerous oral bacteria to modify the inflammatory microenvironment and obstruct host signaling pathways that regulate cell survival, proliferation, and differentiation may provide a molecular explanation for this connection.
There are more than 1,000 distinct types of bacteria in the human intestinal microbiome, primarily belonging to the Bacteroidetes and Firmicutes phyla, which contain both helpful and harmful microorganisms (23). A condition of homeostasis exists in the gut of healthy people because of ongoing interactions between the microbiome, the human body, and between components of the microbiome, which inhibit the overgrowth of pathogenic organisms (24). The microbiota and the human body are in a reciprocal relationship (25). Gut microbiome mimics an “organ” community that plays a vital role in our bodies, including the biological metabolism of bile acids, the synthesis and utilization of dietary compounds such as vitamins and amino acids, the synthesis of vitamins, increased immune levels, and the protection of intestinal function (26). In exchange, an environment rich in energy sources such as protein and carbohydrates enable gut microorganisms to flourish (27). In recent years, plenty of subjects have studied the significance of the gut microbiome in pathogenesis of IBD and its composition and metabolism (28).
However, changes in the composition of bacteria can disrupt the balance, leading to the proliferation of pathogens that inhibit the growth of beneficial members of the gut microbiome (29). This disruption makes the gut more vulnerable to various pathogenic hazards and has adverse effects on the host. This disruption of the microbiome ecosystem is referred to as an “ecological imbalance”. Ecological imbalance can be categorized into three distinct categories: reduced microbial diversity (30), depletion of symbiotic bacteria, overgrowth of opportunistic pathogens, infection with obligate intestinal pathogens and antibiotic therapy. In fact, ecological imbalance reflects shifts in the constitution of the microbiome, metabolic disturbances, and alterations in the distribution of bacteria that negatively impact the balance and can trigger tumorigenesis (31). One of the most susceptible sites for the extraintestinal symptoms of IBD is the mouth cavity, and multiple studies have revealed that IBD patients are more likely to develop periodontitis than non-IBD patients (32, 33). Furthermore, it was discovered that patients with IBD had more severe and more extensive periodontitis than patients with general periodontitis. Patients with IBD also had significantly higher mean probing depths, plaque and calculus indexes, sulcus hemorrhage indexes, and attachment loss (34). In addition, compared to healthy controls without periodontitis, patients with periodontitis have an increased chance of acquiring IBD (35).
The oral-gut barrier, chemical separation, and physical barriers like bile and stomach acid keep the oral and intestinal microbiomes apart (36). Nevertheless, under certain circumstances, the integrity of the oral-intestinal barrier can be compromised, and acidity of stomach acid may decrease (37), allowing for the translocation and communication of microorganisms between these two organs. For instance, in newborns with immature barrier function, Bifidobacterium that colonize the intestines can migrate into the saliva (38).
The relationship between the oral and intestinal microbiomes exists even in individuals who are considered healthy (39). Research examining microbial strains present in fecal and saliva samples from 310 subjects in five different countries identified around 470 species of microorganisms (40). The study revealed that oral microorganisms can frequently travel from the mouth to the intestinum crassum and then colonize there in healthy individuals. Additionally, most oral microorganisms have the ability to be transferred to the gut (41). Ectopic colonization of oral microbiota in the healthy bowel could help maintain the physiological development of intestinal immunity. For example, intestinal colonization by the oral bacterium Villanella. may regulate host immunity (42). Oral microorganisms can be transferred to the gut through the blood as well as through the digestive tract.
Studies have demonstrated that mechanical actions like tooth brushing or dental procedures can result in the transmission of oral microorganisms to the blood through wounds (43). After oral bacteria enter the bloodstream, they usually colonize the joints, bones, liver, heart and other organs, resulting in disease (44). In addition, oral bacteria can invade dendritic cells and macrophages, colonize them, and then travel through the circulatory system to reach the highly vascularized intestinal tract (45), consequently spreading to the intestinal mucosa (46). However, there are also some oral microorganisms, such as Fusobacterium nucleatum, that express the Fap2 protein, which inhibits the activity of T cells, natural killer (NK) cells, and other immune cells by interacting with T cell immunoglobulin and ITIM domain (TIGIT) receptors on the surface of human immunocytes (47).
The relationship between the oral and intestinal microbiomes is facilitated by saliva, which serves as a medium for transporting various components (48). Saliva can carry effector cytokines and enzymes, both keratin-bound and free-floating bacteria, as well as functional inflammatory cell subpopulations like lymphocytes, neutrophils, and macrophages, to distant positions in the body (49). Additionally, saliva is a mixture of proteins, lipids, water, and mucins that make up mucus, which provides protection for these ingredients against the acidic environment of the stomach (50), allowing them to survive in the gastrointestinal tract (51). On average, an individual produces approximately 1 to 1.5 liters of saliva per day, which includes millions of microorganisms that can colonize the gut (52). In addition to the protective effect of saliva (53), there are also some oral pathogens with strong acid tolerance that can pass through the stomach, such as the well-known Streptococcus anginosus (54) (Figure 1).
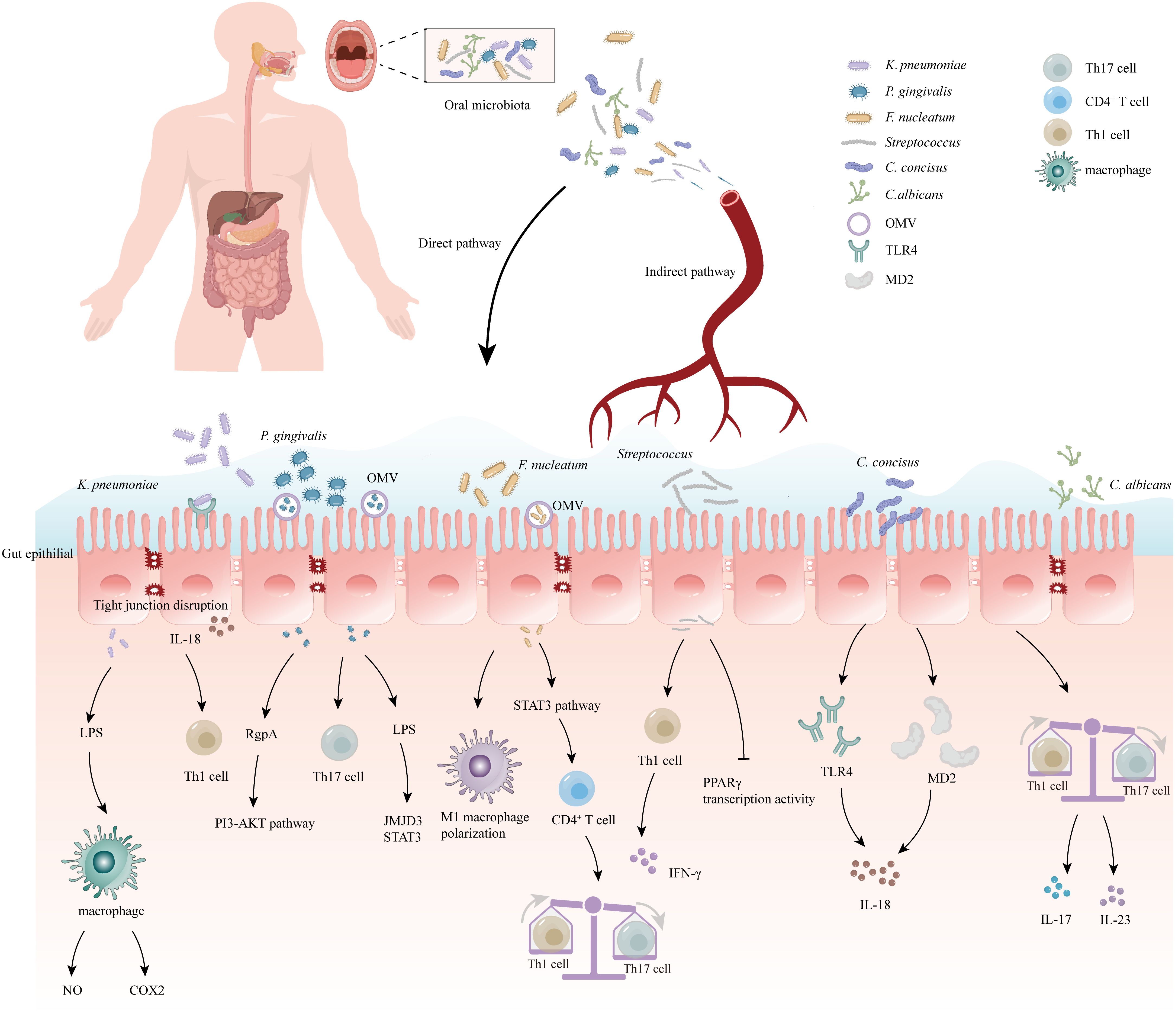
Figure 1 Correlation between oral microbiome and inflammatory bowel disease (IBD). The oral bacteria can induce the development of IBD via several mechanisms: (1) Destruction of the intestinal epithelial barrier: P. gingivalis and K. pneumoniae can downregulate the expressions of tjp-1 and occludin. (2) Release of pro-inflammatory cytokines: F. nucleatum and K.pneumoniae can induce stimulate the pro-inflammatory cytokine LPS. (3) Disruption of the host immune system and induction of immune escape: F. nucleatum and Candida imbalance the Th1/Th17 which induce inflammatory reactions. (4) Migration to the gut and activation of the inflammasome in colonic mononuclear phagocytes: K. pneumoniae and F. nucleatum can migrate to the gut and activate the inflammasome in immune cells, leading to intestinal inflammation. Outer-membrane vesicles (OMV), Toll-like receptor 4 (TLR4), myeloid differentiation protein-2 (MD2), Helper T 17 cell (Th17 cell), Cluster of differentiation 4+ T cell (CD4+ T cell), Helper T 1 cell (Th1 cell), Limited Power Supply (LPS), Nitric Oxide (NO), Phosphoinositide 3-Kinase (PI3K), Cyclooxygenase-2 (COX2), Interleukin-18 (IL-18), Interleukin-17 (IL-17), Interleukin-23 (IL-23), Jumonji domain-containing protein-3 (JMJD3), Signal transducer and activator of transcription (STAT3), Peroxisome proliferator-activated receptorγ (PPARγ), Interferon-γ (IFN-γ).
Extensive research has provided evidence of the involvement of oral microorganisms in various extra-oral diseases, particularly those affecting the digestive system. The associations between these diseases and oral microorganisms are summarized in Table 1. It has been observed that oral microorganisms can reach the intestine through the ingestion of saliva, causing disruptions in the intestinal microecology and eventually contributing to the development of IBD. In the subsequent sections, we will delve into the detailed relationship between six specific types of microorganisms and IBD.
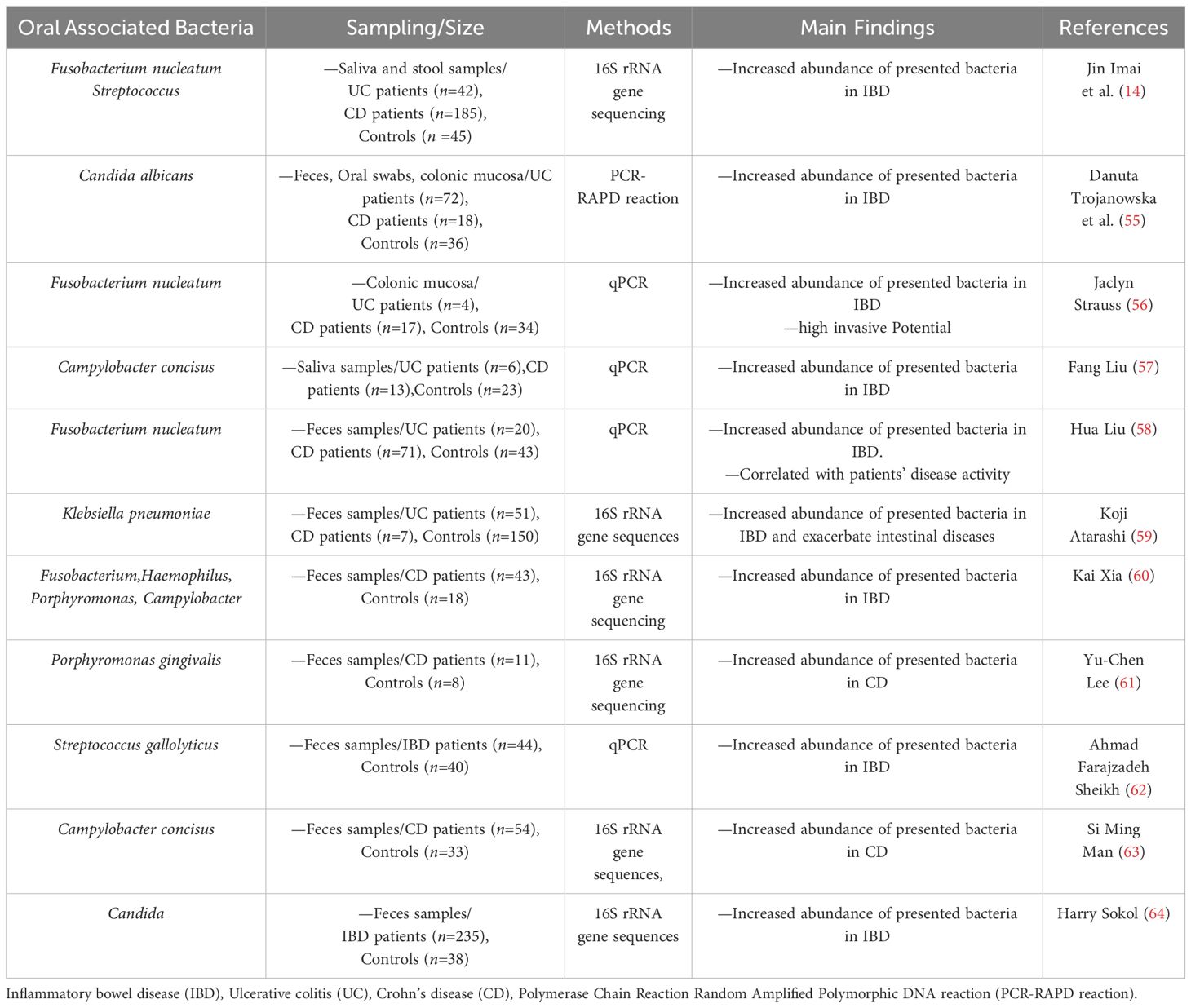
Table 1 Summary of various clinical experimental studies regarding the presence of oral bacteria in Inflammatory bowel disease (IBD).
A well-known keystone pathogen for periodontal illnesses, P. gingivalis can spread from the mouth to the gut in mice, causing a dysbiosis of the gut microbiota characterized by an increased abundance of Bacteroidetes and a decreased abundance of Firmicutes (65). This weakens the function of the intestinal barrier by downregulating the expression of tjp-1 and occluding genes (66). P. gingivalis secretes gingipains that help them selectively inactivate pro-inflammatory substances generated by activated dendritic cells (DCs) and evade innate immune responses. In addition to the effects of metabolites, microbiome dysbiosis can produce abnormal immunological responses. Tim et al. showed that phagocytosis, NETosis, and CAMP activity are the three different ways that PPAD, which is the virulence factor of the oral pathogen P. gingivalis, defuses antibacterial neutrophil assaults (67). This indicates that PPAD plays a significant role in the escape of human natural immunity. Research by Potempa (68) and colleagues demonstrating PPAD-dependent complement system citrullination supports this theory.
Because P. gingivalis fimbriae facilitate bacterial adherence to and invasion of specific areas, they are essential for mediating the organism’s contact with host tissues (69). According to the findings from Lagha et al. (70), P. gingivalis penetrated and persisted in the gingival epithelial cells that were cultivated, and it caused the epithelial barrier to be damaged by breaking down the ZO-1 protein. The possible mechanisms are that the three ways that P. gingivalis reduced the amount of ZO-1 protein in vivo were via detaching intestinal mucus, invading epithelial cells of the intestine, and breaking down cytosolic ZO-1. Aleksandra et al. (71) showed that fimbriae isolated from the P. gingivalis mutant strain lacking in PPAD were unable to activate TLR2, suggesting that fimbriae constituents or other proteins related to fimbriae assembly must be citrullinated in order for TLR2 to activate on host cells.
Gingipains (Kgp, RgpA, and RgpB) are complex, lysine- or arginine-specific cysteine proteinases that are secreted to the microbes surface through a type IX secretion system (T9SS) (72) and released into the environment as cargo or as soluble proteins on outer membrane vesicles (OMVs). These proteinases have been identified as the primary pathogenic factors of P. gingivalis. When enzymatically inactivated, RgpA is the gingipain that mediates EGFR-mediated signaling. This is explained by the sequential motif that is specific to the hemagglutinin adhesion domain of RgpA and absent from Kgp (73). Izabela et al. showed that the PI3K-AKT pathway was activated by potent tyrosine residue phosphorylation caused by RgpA in the absence of enzymatic activity in EGFR. The pro-inflammatory response, endocytosis, differentiation, proliferation, and metabolism are all attributed to this route. Phosphorylation of T308 was the primary method by which inactive RgpA activated AKT, indicating the participation of the PDK1 protein, a phosphoinositide-1-dependent kinase (74). This stands in stark contrast to RgpA, which is active enzymatically, which reduces AKT phosphorylation and causes the kinase’s natural form to degrade (75). The mechanism of action of KYT-36 (76), a highly selective inhibitor peptide, against gingipain has been demonstrated in a recent study. This peptide offers potential as a therapeutic agent for the treatment of IBD and periodontitis in clinical settings.
Related studies have shown that the Th17 cell increase in periodontal tissues is stimulated by intestinal priming and oral P. gingivalis infection, which exacerbates periodontitis (77). It was observed that gut-translocated P. gingivalis can enter the Peyer’s patches (PPs) of the small intestine through microfold cells (M cells) in mice and induce a systemic response mediated by P. gingivalis-Th17 cells. However, when P. gingivalis was heat-inactivated and its proteins denatured, M cells were less likely to absorb it, resulting in reduced alveolar bone loss. This indicates that heat-inactivated denatured proteins of P. gingivalis were ineffective as components of the bacterial T cell antigen for uptake by M cells (78). In patients with periodontitis, an upregulation of CCL20, a chemokine involved in cell migration, has been observed in periodontal tissues (79). P. gingivalis-Th17 cell migration aggravates periodontitis progression, indicating that inhibiting the CCL20/CCR6 axis could prevent periodontitis from developing (80). To break the pathogenic gut-oral axis in the inflammatory oral disease process, one possible therapeutic target is to inhibit the invasion of pathobiont-responsive Th17 cells into the dental cavity.
Under normal conditions, the expression of JMJD3, a protein involved in epigenetic regulation, is low. However, different cellular stressors can cause JMJD3 to be expressed (81). Studies have shown that lipopolysaccharide (LPS) stimulation has the ability to activate JMJD3, which in turn controls genes linked to inflammation in peripheral macrophages (82). Along with increased levels of p-STAT3, STAT3, and RORγt, it was also found that P. gingivalis-LPS stimulated the expression of JMJD3 and further improved their interactions. This implies that JMJD3 might be targeting STAT3 and RORγt (83).
The obligate anaerobic bacterium F. nucleatum is spindle-shaped, opportunistic, Gram-negative, and anaerobic (84). It is frequently found in the oral cavity, is a major factor in intrauterine infections linked to pregnancy complications such as stillbirth, neonatal sepsis, and preterm birth, and is present in the dental plague that precedes the beginning of periodontitis (85). The human intestine is not primarily colonized by F. nucleatum, which is actually a protobacterium, which can be both carcinogenic and antigenic. It is a very poor colonizing bacterium found in the gut microbiome of healthy humans, and most significantly, it has a distinct advantage in the microenvironment of colorectal tumors (86). Several symbiotic bacteria, in particular driver bacteria, are protobacteria in the human gut microbiome that are able to directly trigger damage to colon epithelial cells, thereby promoting the development of colorectal cancer. The gastrointestinal pathway is the main pathway for the colonization of oral Fusobacterium tuberculosis colorectal cancer tissue, and its virulent proteins FadA and Fap2 are mainly involved in its adhesion to colorectal cancer cells (87).
The mechanism of F. nucleatum in IBD has been thoroughly studied. F. nucleatum infection aggravates the inflammatory response and damages the integrity of the intestinal mucosal barrier (88). F. nucleatum produces a significant amount of hydrogen sulfide, a very toxic byproduct of cysteine metabolism that prevents colon cells from using butyrate effectively and causes persistent intestinal inflammation (89). According to certain research, the invasion and colonization of F. nucleatum may influence MUC2 production, which could lead to the development of intestinal inflammation (90, 91). Highly invasive F. nucleatum isolates from the lesions of CD patients showed considerably increased expressions of tumor necrosis factor alpha (TNF-α) and MUC2 compared with moderately invasive strains isolated from the healthy intestinal mucosa of control participants (92). By causing intestinal structure to be destroyed, upregulating the expression of TNF-α and IL-1β, and downregulating the expression of IL-10, F. nucleatum accelerates the development of DSS experimental colitis. F. Wei et al. suggest that F. nucleatum-EVs promote colitis by enhancing autophagy through the miR-574-5p/CARD3 axis (93).
The surface of F. nucleatum is home to a number of proteins, including RadD, FadA, Fap2, and FomA. These proteins not only facilitate its copolymerization with other bacteria but also aid in its attachment to host cells and trigger a number of immunological reactions in the host (94). F. nucleatum may cause colitis by controlling the skewing of M1 macrophages (95). Liu et al. found that by increasing the release of cytokines including IL-1β, IL-6, and IL-17 and by triggering the STAT3 signaling pathway, F. nucleatum may worsen intestinal inflammation by promoting the growth of CD4+ T cells and their differentiation into Th1 and Th17 cells (58). Li et al. found that patients with UC had higher levels of FadA genes and F. nucleatum, particularly those with severe pancolitis, indicating a potential role for FadA in the pathophysiology of UC (96). However, further research is required to clarify the precise mechanism of this connection and the functions of FadA genes and F. nucleatum in UC. Yan et al. found that non-surgical periodontal therapy triggers intestinal microbiome regulation and promotes a rehabilitative and healthy microbial environment by using the ApoE−/− mice model (97). Furthermore, studies showed that F. nucleatum could interact with many other microorganisms and showed a synergistic effect of virulence when it was co-infected with other pathogens (98). A down-regulation of the mean relative abundance of probiotics, like Clostridium prai, coincides with an increase in the abundance of opportunistic pathogens, including F. nucleatum (99). The culture supernatant of F. nucleatum has an obvious inhibitory effect on Clostridium prai and Bifidobacterium lactis and can reduce the activity of Lactobacillus rhamnosus in high concentration. F. nucleatum can improve the efficiency of E. coli penetrating vascular endothelial cells (100). In addition, Duan et al. (101) found that Lactobacillus rhamnosus could significantly improve the damage to intestinal epithelial cells caused by infection with F. nucleatum. A variety of probiotics have been proven to help maintain intestinal barrier integrity and relieve or prevent DSS-induced intestinal inflammation in mice with UC, and F. nucleatum may further affect the recurrence of IBD through interaction with these probiotics (102). All the above studies indicated that F. nucleatum could jointly affect the occurrence and development of IBD through interactions with other microorganisms, but the specific effects and mechanisms remain unclear.
Nearly every area of the human body has Streptococcus, which is the predominant species in the mouth (103). Streptococcus is divided into eight different groups, including pyogenic, mitis, bovis, sanguinis, anginosus, downei, mutans, and salivarius (104). At present, the oral cavity contains all groups, with the exception of the pyogenic and bovis groups. The largest group found in the mouth is the mitis group, with 20 species (105). Some researchers have reported increased amounts of Streptococcus in the intestines of patients with irritable bowel syndrome (IBS). These Streptococcus bacteria probably translocated from the mouth (106).
In subsequent investigations, studies found that oral Streptococcus can indeed migrate from the mouth and colonize the gut, even in healthy people (107). In a recent study, patients with IBD had higher concentrations of Streptococcus in their intestines (108). And the researchers confirmed that the Streptococcus originated in the mouth. Therefore, oral streptococci are the only shared genus (109). Streptococcus sanguinis ATCC10556 and TW289 from oral Streptococcus have been shown to aggravate colitis in mice by infecting liver cells and causing interferon gamma secretion, which leads to colitis aggravation. The specific mechanism may be that after infection with Streptococcus TW289 and ATCC10556, the Th1 immune response in the blood is activated, resulting in increased secretion of IFN-γ by T cells in the blood and spleen, which promotes the aggravation of inflammation (110).
In addition, Streptococcus mutans serotype k strain TW295 also has the effect of aggravating colitis. It infects the liver through the blood system and increases the expression of IFN-γ in the liver, thus aggravating colon inflammation (111). While TW295 is difficult to reach the colon through the complex physiological environment of the digestive tract, which is primarily brought on by blood dental surgery (112). However, not all oral streptococci are detrimental. One of the earliest bacteria to colonize the human mouth cavity and digestive system after birth is Streptococcus salivarius, which may be important for the development of immunological homeostasis (113). In mouse models, Streptococcus salivarius JIM8772 strains showed significant palliative effects on colitis. It was found that live Streptococcus salivary strains down-regulated the pro-inflammatory chemokine interleukin-8 (IL-8) secreted by intestinal epithelial cells in vitro and prevented the activation of the nuclear transcription factor-κB (NF-κB) pathway (114). Commensal Streptococcus salivarius can also regulate PPARγ transcription activity in human intestinal epithelial cells and significantly reduce the expression levels of I-FABP and Angptl4, thus contributing to the preservation of the health and equilibrium of the host environment (115).
A single polar flagellum, located at either or both sides of the bacterium, enables the majority of gram-negative, spiral- or curved-shaped Campylobacter species to move in a manner akin to a corkscrew (116). In the digestive tracts of different animals, the majority of Campylobacter bacteria exist as regular microbiomes. C. concisus is regarded as a pathogen for intestinal and oral disorders and is found in the digestive tract and oral cavity of IBD patients (117). PCR detection showed that the detection rate of C. concisus in the saliva of patients with UC was 100%, 85% in patients with CD, and only 75% in healthy individuals. Campylobacter has been presented in many studies to raise the chance of IBD (118). Compared with healthy controls, the incidence of Campylobacter in intestinal biopsies and stool samples collected from patients with IBD significantly increases. Especially C. concisus is more likely to colonize the proximal colon. The majority of Campylobacter species are oral bacteria found in humans rather than zoonotic species (119). The innate environmental factor for C. concisus colonization in the intestine is hydrogen gas. H2 gas significantly affects C. concisus growth. In laboratory culture, C. concisus grows very slowly in anaerobic settings but not at all in microaerobic conditions. H2 makes it possible for C. concisus to grow in microaerophilic environments and greatly accelerates its growth in anaerobic environments. The human intestine has microaerobic to anaerobic atmospheric conditions, with hydrogen, carbon dioxide, methane, and other gases making up the majority of colonic gases. In view of this, C. concisus may have the ability to offer a constant amount of available H2 for growth in their intestinal environment (120). Although the relationship between oral Campylobacter and IBD has been proven, the specific mechanism is still not very clear. Some oral C. concisus strains have owned the zonula occludens toxin (Zot) gene from viruses (pre-phages), and C. concisus zot has the same conserved base sequence as the human zonulin receptor binding domain and the cholera vibrio Zot receptor binding domain. The mechanism by which Zot-positive strains of C. concisus may promote the start and recurrence of IBD is primary barrier dysfunction, and Campylobacter can also activate inflammasome signaling in macrophages through Zot, thereby increasing the expression levels of TNF-α and IL-8 (121, 122). C. concisus isolated from the mouth and small intestine of IBD patients is highly invasive to HT-29/Caco2 cell lines, can upregulate the expression of MD-2 and TLR4 on cell surfaces, induce IL-8 production, increase intestinal epithelial permeability, and promote cell apoptosis. This suggests that Campylobacter may repeatedly enter the digestive tract with saliva and colonize the intestine, causing IBD in susceptible populations (123–126).
A German microbiologist named Edwin Klebs made the initial discovery of K. pneumoniae (127). It is a widespread environmental bacterium that can be found on plants, in sewage, surface water, and soil. It belongs to the enterobacteriaceae family and is encapsulated, rod-shaped, gram-negative, and non-motile (128). Moreover, the bacteria are also known to colonize animal mucosa, including those of the gastrointestinal system and oral cavity. However, studies have shown significant differences between the Klebsiella genus identified in healthy human saliva and K. pneumoniae isolated in the saliva of patients with IBD (59, 129, 130). What’s more, recent research has discovered that the ectopic colonization of oral Klebsiella genus can lead to the activation of inflammasomes in resident macrophages, thereby exacerbating colitis (131). Studies have proved that when K. pneumoniae was taken orally, NF-κB activation in the colon, lipid peroxidation, and production of TNF-α, COX-2, IL-1β, and ZO-1 increased, whereas tight junction-associated proteins claudin-1, ZO-1, and occludin decreased (132). In animals with TNBS-induced colitis, K. pneumoniae also worsened the expression of tight junction-associated proteins and inflammatory markers (133). K. pneumoniae generated lipopolysaccharide and β-glucuronidase, which effectively stimulated the production of NO and COX-2 in mice peritoneal macrophages (134). From the colon tissues of colitis patients, they recovered K. pneumoniae J (KLPJ), a strain of KLP. This bacterial strain has the ability to cause and develop colitis, mediated by dextran sodium sulfate. It can also activate caspase-11 inflammasomes and cause mature IL-18 to be produced in colon epithelial cells and intestinal organoids (135). K. pneumoniae isolated from the salivary microbiome of 2 CD patients, specifically Kp-2H7, can activate epithelial cells and DCs through the TLR4 signaling pathway, stimulate IL-18 secretion, and cause recruitment and activation of Th1 cells, thereby leading to colitis (59). K. pneumoniae migrates from the mouth through the digestive tract and colonizes the intestines under harsh conditions. Through fermentation, K. pneumoniae generates hydrogen (H2) to counteract the damaging effects of NO and ROS. These properties keep it active in the mouth. In addition, exopolysaccharide capsules are its most important structure. By blocking immune cell phagocytosis, innate immune response activation, and complement activation-induced lysis avoidance, the capsule provides protection against the host immunological response (136, 137).
Fungi actively participate in affecting health and disease in the intricate and multidimensional relationship that exists between the digestive system and the resident fungal community (the fungal community). Candida is the predominant fungal genus in the digestive tract and mouth of healthy individuals (138). Candida is suitable for colonization in various places in the digestive tract and mouth, increasing the possibility that Candida from the mouth will migrate to the intestine to cause disease. In a study of Candida colonization in the digestive tracts of patients undergoing hematopoietic stem cell transplantation (HSCT), the same candida genotypes were found in oral and intestinal samples. Oral Candida colonization is a risk factor for intestinal Candida colonization (139). In addition, Candida is closely associated with the development of IBD. Research has shown that the abundance of C. albicans in IBD patients, especially those with sudden inflammation, increases significantly. They verified a noteworthy rise in the abundance of C. albicans in IBD patients, especially during flare-ups. In mouse models, it was discovered that Candida tube feeding exacerbated the DSS model without impacting weight loss or diarrhea. This was evidenced by increased intestinal leakage (FITC-glucan assay, serum BG, endotoxemia, and blood bacterial load), a more severe colon histology, and higher mortality. Higher levels of pro-inflammatory cytokines are induced in intestinal and blood tissues by DSS+ Candida (140). And it has been found that colitis can be alleviated by preventing the propagation, pathogenicity, and colonization of Candida (141). Compared to healthy individuals, C. albicans colonized CD patients and their primary healthy relatives (HRs) more frequently and more severely. There was a correlation found between the colonization of C. albicans in the HRs and the antibodies against Saccharomyces cerevisiae antibodies (ASCAs) found in the sera of patients with CD (142). In addition, ASCA is a recognized marker for the clinical distinction between CD and UC (143). CD is associated with genes that are involved in the Candida glycogen response. For nucleotide-binding oligomerization domain-containing proteins (NOD) 1 and NOD2, as well as the NOD-like receptor family (133), genes encoding cytokines or their receptors, like IL-23 or IL-17, and pyrin domain-containing 3 (NLRP3), the direct impact on inflammatory responses is particularly evident. Autophagy is a significant mechanism in the development of IBD. By negatively regulating adaptive immunity and the activation of T cells, it makes patients susceptible to infection by C. albicans (144). Research has demonstrated that in patients with UC, severe cases are associated with the presence of “highly invasive” Candida strains, which can produce Candida to damage macrophages and promote the pro-inflammatory factor (145).
There is increasing evidence in favor of a reciprocal relationship between IBD and the oral microbiota. The oral microbiota is one of the important symbiotic partners in the mouth and the human body. However, periodontal disease can cause systemic disease through the spread of inflammatory mediators and bacterial components through the blood. Current research has found that oral periodontal bacteria passing through the digestive tract can change the intestinal microbiota and cause ectopic colonization of the intestine, thereby exacerbating intestinal inflammation (146). We emphasize the significance of several oral bacteria in IBD via the oral-intestinal axis in this review. These include P. gingivalis, F. nucleatum, C. concisus, K. pneumoniae, and C. albicans.
Microbiota research on IBD has primarily focused on the gut microbiota, but emerging evidence suggests a potential link between dysbiosis of the oral microbiota and the development of IBD. The oral microbiota may have an impact on the inflammatory response of IBD, leading to worsening of the condition in IBD patients with periodontitis (147). Significant dysbiosis in the oral microbiota of IBD patients has been observed, indicating a potential role of the oral microbiota in the pathogenesis of the disease (148). Firstly, alteration in the oral microbiota can serve as markers for IBD, aiding in early diagnosis and predicting disease progression. Some studies have already identified variations in the composition of the oral microbiota between healthy people and IBD patients, suggesting that oral microbiota analysis could be used as an adjunct diagnostic tool for IBD (149, 150). Secondly, the oral microbiota can be a therapeutic target, and modulating the oral microbiota may offer a new approach for treating IBD, for example, reducing the abundance of oral bacteria in the intestine by using mouthwash (151). Future advancements in targeting probiotics for the oral microbiota and eliminating specific oral pathogens with targeted antibiotics hold promise for preventing IBD recurrence. However, this topic of study is still in its infancy, so further research is required. In the future, combining the investigation of oral microbiota with the monitoring of particular microbial products from the oral to the gut may provide fascinating new insights into the oral-gut microbiome axis.
AW: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. ZZ: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. YD: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. JW: Conceptualization, Software, Supervision, Visualization, Writing – review & editing. ZW: Writing – review & editing. HC: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (Grant/Award Number: 82070545), the Tianjin Health Commission of Traditional Chinese and Western medicine integrated research project funding (2023114), and Beijing‐Tianjin‐Hebei Basic Research Cooperation Project (23JCZXJC00200).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Kaplan GG. The global burden of IBD: from 2015 to 2025. Nat Rev Gastroenterol Hepatol. (2015) 12:720–7. doi: 10.1038/nrgastro.2015.150
2. Argollo M, Gilardi D, Peyrin-Biroulet C, Chabot JF, Peyrin-Biroulet L, Danese S. Comorbidities in inflammatory bowel disease: a call for action. Lancet Gastroenterol Hepatol. (2019) 4:643–54. doi: 10.1016/S2468-1253(19)30173-6
3. Rutherford D, Ho GT. Therapeutic potential of human intestinal organoids in tissue repair approaches in inflammatory bowel diseases. Inflammation Bowel Diss. (2023) 29(9):1488–98. doi: 10.1093/ibd/izad044
4. Lu S, Na K, Wei J, Tao T, Zhang L, Fang Y, et al. Alginate oligosaccharide structures differentially affect DSS-induced colitis in mice by modulating gut microbiota. Carbohydr Polym. (2023) 312:120806. doi: 10.1016/j.carbpol.2023.120806
5. Ananthakrishnan AN. Epidemiology and risk factors for IBD. Nat Rev Gastroenterol Hepatol. (2015) 12:205–17. doi: 10.1038/nrgastro.2015.34
6. Cai J, Sun L, Gonzalez FJ. Gut microbiota-derived bile acids in intestinal immunity, inflammation, and tumorigenesis. Cell Host Microbe. (2022) 30:289–300. doi: 10.1016/j.chom.2022.02.004
7. Guo X, Okpara ES, Hu W, Yan C, Wang Y, Liang Q, et al. Interactive relationships between intestinal flora and bile acids. Int J Mol Sci. (2022) 23(15):8343. doi: 10.3390/ijms23158343
8. Liu H, Cai Z, Wang F, Hong L, Deng L, Zhong J, et al. Colon-targeted adhesive hydrogel microsphere for regulation of gut immunity and flora. Adv Sci (Weinh). (2021) 8:e2101619. doi: 10.1002/advs.202101619
9. Liu YJ, Tang B, Wang FC, Tang L, Lei YY, Luo Y, et al. Parthenolide ameliorates colon inflammation through regulating Treg/Th17 balance in a gut microbiota-dependent manner. Theranostics. (2020) 10:5225–41. doi: 10.7150/thno.43716
10. Schroeder BO, Bäckhed F. Signals from the gut microbiota to distant organs in physiology and disease. Nat Med. (2016) 22:1079–89. doi: 10.1038/nm.4185
11. Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, et al. Host-gut microbiota metabolic interactions. Sci (New York NY). (2012) 336:1262–7. doi: 10.1126/science.1223813
12. Saad E, Agab M, Ozcekirdek EC, Awadelkarim A, Idris I. The diagnostic dilemma of acute granulomatous hepatitis in a patient with crohn's disease: A case report and review of literature. J Investig Med High Impact Case Rep. (2022) 10:23247096211069764. doi: 10.1177/23247096211069764
13. Xun Z, Zhang Q, Xu T, Chen N, Chen F. Dysbiosis and ecotypes of the salivary microbiome associated with inflammatory bowel diseases and the assistance in diagnosis of diseases using oral bacterial profiles. Front Microbiol. (2018) 9:1136. doi: 10.3389/fmicb.2018.01136
14. Imai J, Ichikawa H, Kitamoto S, Golob JL, Kaneko M, Nagata J, et al. A potential pathogenic association between periodontal disease and Crohn's disease. JCI Insight. (2021) 6(23):e148543. doi: 10.1172/jci.insight.148543
15. Rogler G, Singh A, Kavanaugh A, Rubin DT. Extraintestinal manifestations of inflammatory bowel disease: current concepts, treatment, and implications for disease management. Gastroenterology. (2021) 161:1118–32. doi: 10.1053/j.gastro.2021.07.042
16. Sun B, Liu B, Gao X, Xing K, Xie L, Guo T. Metagenomic analysis of saliva reveals disease-associated microbiotas in patients with periodontitis and crohn's disease-associated periodontitis. Front Cell Infect Microbiol. (2021) 11:719411. doi: 10.3389/fcimb.2021.719411
17. Iyer N, Williams MA, O'Callaghan AA, Dempsey E, Cabrera-Rubio R, Raverdeau M, et al. Lactobacillus salivarius UCC118 dampens inflammation and promotes microbiota recovery to provide therapeutic benefit in a DSS-induced colitis model. Microorganisms. (2022) 10(7):1383. doi: 10.3390/microorganisms10071383
18. Kawasaki M, Ikeda Y, Ikeda E, Takahashi M, Tanaka D, Nakajima Y, et al. Oral infectious bacteria in dental plaque and saliva as risk factors in patients with esophageal cancer. Cancer. (2021) 127:512–9. doi: 10.1002/cncr.33316
19. Lynch SV, Pedersen O. The human intestinal microbiome in health and disease. New Engl J Med. (2016) 375:2369–79. doi: 10.1056/NEJMra1600266
20. Acharya C, Sahingur SE, Bajaj JS. Microbiota, cirrhosis, and the emerging oral-gut-liver axis. JCI Insight. (2017) 2(19):e94416. doi: 10.1172/jci.insight.94416
21. Albuquerque-Souza E, Sahingur SE. Periodontitis, chronic liver diseases, and the emerging oral-gut-liver axis. Periodontol 2000. (2022) 89:125–41. doi: 10.1111/prd.12427
22. Blohs M, Mahnert A, Brunnader K, Flucher C, Castellani C, Till H, et al. Acute appendicitis manifests as two microbiome state types with oral pathogens influencing severity. Gut Microbes. (2023) 15:2145845. doi: 10.1080/19490976.2022.2145845
23. Zuo T, Ng SC. The gut microbiota in the pathogenesis and therapeutics of inflammatory bowel disease. Front Microbiol. (2018) 9:2247. doi: 10.3389/fmicb.2018.02247
24. Hu Y, Chen Z, Xu C, Kan S, Chen D. Disturbances of the gut microbiota and microbiota-derived metabolites in inflammatory bowel disease. Nutrients. (2022) 14(23):5140. doi: 10.3390/nu14235140
25. Adolph TE, Zhang J. Diet fuelling inflammatory bowel diseases: preclinical and clinical concepts. Gut. (2022) 71:2574–86. doi: 10.1136/gutjnl-2021-326575
26. Taniguchi M, Okumura R, Matsuzaki T, Nakatani A, Sakaki K, Okamoto S, et al. Sialylation shapes mucus architecture inhibiting bacterial invasion in the colon. Mucosal Immunol. (2023) 16:624–41. doi: 10.1016/j.mucimm.2023.06.004
27. Gao Q, Tian W, Yang H, Hu H, Zheng J, Yao X, et al. Shen-Ling-Bai-Zhu-San alleviates the imbalance of intestinal homeostasis in dextran sodium sulfate-induced colitis mice by regulating gut microbiota and inhibiting the NLRP3 inflammasome activation. J Ethnopharmacol. (2024) 319:117136. doi: 10.1016/j.jep.2023.117136
28. Lloyd-Price J, Arze C, Ananthakrishnan AN, Schirmer M, Avila-Pacheco J, Poon TW, et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. (2019) 569:655–62. doi: 10.1038/s41586-019-1237-9
29. Rausch P, Ellul S, Pisani A, Bang C, Tabone T, Marantidis Cordina C, et al. Microbial dynamics in newly diagnosed and treatment naive IBD patients in the mediterranean. Inflammation Bowel Dis. (2023) 29:1118–32. doi: 10.1093/ibd/izad004
30. Fan X, Zhang Z, Gao W, Pan Q, Luo K, He B, et al. An engineered butyrate-derived polymer nanoplatform as a mucosa-healing enhancer potentiates the therapeutic effect of magnolol in inflammatory bowel disease. ACS Nano. (2024) 18(1):229–44. doi: 10.1021/acsnano.3c05732
31. Mehta RS, Abu-Ali GS, Drew DA, Lloyd-Price J, Subramanian A, Lochhead P, et al. Stability of the human faecal microbiome in a cohort of adult men. Nat Microbiol. (2018) 3:347–55. doi: 10.1038/s41564-017-0096-0
32. Papageorgiou SN, Hagner M, Nogueira AV, Franke A, Jäger A, Deschner J. Inflammatory bowel disease and oral health: systematic review and a meta-analysis. J Clin Periodontol. (2017) 44:382–93. doi: 10.1111/jcpe.12698
33. She YY, Kong XB, Ge YP, Liu ZY, Chen JY, Jiang JW, et al. Periodontitis and inflammatory bowel disease: a meta-analysis. BMC Oral Health. (2020) 20:67. doi: 10.1186/s12903-020-1053-5
34. Zhang L, Gao X, Zhou J, Chen S, Zhang J, Zhang Y, et al. Increased risks of dental caries and periodontal disease in Chinese patients with inflammatory bowel disease. Int Dental J. (2020) 70:227–36. doi: 10.1111/idj.12542
35. Kang EA, Chun J, Kim JH, Han K, Soh H, Park S, et al. Periodontitis combined with smoking increases risk of the ulcerative colitis: A national cohort study. World J Gastroenterol. (2020) 26:5661–72. doi: 10.3748/wjg.v26.i37.5661
36. Xu J, Zhang Y, Fang XH, Liu Y, Huang YB, Ke ZL, et al. The oral bacterial microbiota facilitates the stratification for ulcerative colitis patients with oral ulcers. Ann Clin Microbiol Antimicrob. (2023) 22:99. doi: 10.1186/s12941-023-00646-3
37. Kitamoto S, Kamada N. The oral-gut axis: a missing piece in the IBD puzzle. Inflammation Regen. (2023) 43:54. doi: 10.1186/s41232-023-00304-3
38. Park SY, Hwang BO, Lim M, Ok SH, Lee SK, Chun KS, et al. Oral-gut microbiome axis in gastrointestinal disease and cancer. Cancers (Basel). (2021) 13(9):2124. doi: 10.3390/cancers13092124
39. Elghannam MT, Hassanien MH, Ameen YA, Turky EA, GM EL, AA EL, et al. Helicobacter pylori and oral-gut microbiome: clinical implications. Infection. (2024) 52(2):289–300. doi: 10.1007/s15010-023-02115-7
40. Yu Y, Zhao X, Xu X, Cai C, Tang X, Zhang Q, et al. Rational design of orally administered cascade nanozyme for inflammatory bowel disease therapy. Adv Mater. (2023) 35:e2304967. doi: 10.1002/adma.202304967
41. Schmidt TS, Hayward MR, Coelho LP, Li SS, Costea PI, Voigt AY, et al. Extensive transmission of microbes along the gastrointestinal tract. Elife. (2019) 8:e42693. doi: 10.7554/eLife.42693
42. Geva-Zatorsky N, Sefik E, Kua L, Pasman L, Tan TG, Ortiz-Lopez A, et al. Mining the human gut microbiota for immunomodulatory organisms. Cell. (2017) 168:928–43 e11. doi: 10.1016/j.cell.2017.01.022
43. Lockhart PB, Brennan MT, Sasser HC, Fox PC, Paster BJ, Bahrani-Mougeot FK. Bacteremia associated with toothbrushing and dental extraction. Circulation. (2008) 117:3118–25. doi: 10.1161/CIRCULATIONAHA.107.758524
44. Gendron R, Grenier D, Maheu-Robert L. The oral cavity as a reservoir of bacterial pathogens for focal infections. Microbes Infection. (2000) 2:897–906. doi: 10.1016/S1286-4579(00)00391-9
45. Kato S, Okamoto H, Nishino Y, Oyake Y, Nakazato Y, Okuda M, et al. Helicobacter pylori and TT virus prevalence in Japanese children. J Gastroenterol. (2003) 38:1126–30. doi: 10.1007/s00535-003-1218-4
46. Parahitiyawa NB, Jin LJ, Leung WK, Yam WC, Samaranayake LP. Microbiology of odontogenic bacteremia: beyond endocarditis. Clin Microbiol Rev. (2009) 22:46–64. doi: 10.1128/CMR.00028-08
47. Wang N, Fang JY. Fusobacterium nucleatum, a key pathogenic factor and microbial biomarker for colorectal cancer. Trends Microbiol. (2023) 31:159–72. doi: 10.1016/j.tim.2022.08.010
48. Ferretti P, Pasolli E, Tett A, Asnicar F, Gorfer V, Fedi S, et al. Mother-to-infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host Microbe. (2018) 24:133–45 e5. doi: 10.1016/j.chom.2018.06.005
49. Van Daele E, Knol J, Belzer C. Microbial transmission from mother to child: improving infant intestinal microbiota development by identifying the obstacles. Crit Rev Microbiol. (2019) 45:613–48. doi: 10.1080/1040841X.2019.1680601
50. Yang CJ, Chang HC, Sung PC, Ge MC, Tang HY, Cheng ML, et al. Oral fecal transplantation enriches Lachnospiraceae and butyrate to mitigate acute liver injury. Cell Rep. (2023) 43:113591. doi: 10.1016/j.celrep.2023.113591
51. Kageyama S, Sakata S, Ma J, Asakawa M, Takeshita T, Furuta M, et al. High-resolution detection of translocation of oral bacteria to the gut. J Dent Res. (2023) 102:752–8. doi: 10.1177/00220345231160747
52. Byrd KM, Gulati AS. The "Gum-gut" Axis in inflammatory bowel diseases: A hypothesis-driven review of associations and advances. Front Immunol. (2021) 12:620124. doi: 10.3389/fimmu.2021.620124
53. Lu Y, Li Z, Peng X. Regulatory effects of oral microbe on intestinal microbiota and the illness. Front Cell Infect Microbiol. (2023) 13:1093967. doi: 10.3389/fcimb.2023.1093967
54. Fu K, Cheung AHK, Wong CC, Liu W, Zhou Y, Wang F, et al. Streptococcus anginosus promotes gastric inflammation, atrophy, and tumorigenesis in mice. Cell. (2024) 187:882–96.e17. doi: 10.1016/j.cell.2024.01.004
55. Poulopoulos A, Belazi M, Epivatianos A, Velegraki A, Antoniades D. The role of Candida in inflammatory papillary hyperplasia of the palate. J Oral Rehabil. (2007) 34:685–92. doi: 10.1111/j.1365-2842.2007.01758.x
56. Strauss J, Kaplan GG, Beck PL, Rioux K, Panaccione R, Devinney R, et al. Invasive potential of gut mucosa-derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflammation Bowel Dis. (2011) 17:1971–8. doi: 10.1002/ibd.21606
57. Liu F, Ma R, Tay CYA, Octavia S, Lan R, Chung HKL, et al. Genomic analysis of oral Campylobacter concisus strains identified a potential bacterial molecular marker associated with active Crohn's disease. Emerg Microbes Infect. (2018) 7:64. doi: 10.1038/s41426-018-0065-6
58. Liu H, Hong XL, Sun TT, Huang XW, Wang JL, Xiong H. Fusobacterium nucleatum exacerbates colitis by damaging epithelial barriers and inducing aberrant inflammation. J Dig Dis. (2020) 21:385–98. doi: 10.1111/1751-2980.12909
59. Atarashi K, Suda W, Luo C, Kawaguchi T, Motoo I, Narushima S, et al. Ectopic colonization of oral bacteria in the intestine drives T(H)1 cell induction and inflammation. Sci (New York NY). (2017) 358:359–65. doi: 10.1126/science.aan4526
60. Xia K, Gao R, Wu X, Sun J, Wan J, Wu T, et al. Characterization of specific signatures of the oral cavity, sputum, and ileum microbiota in patients with crohn's disease. Front Cell Infect Microbiol. (2022) 12:864944. doi: 10.3389/fcimb.2022.864944
61. Lee YC, Liu CY, Lee CL, Zhang RH, Huang CJ, Yen TL. The periodontopathic pathogen, porphyromonas gingivalis, involves a gut inflammatory response and exacerbates inflammatory bowel disease. Pathogens. (2022) 11(1):84. doi: 10.3390/pathogens11010084
62. Sheikh AF, Masjedi Zadeh AR, Saki M, Khani P, Hashemi SJ, Shahin Zadeh S, et al. Detection of Streptococcus gallolyticus in colorectal cancer and inflammatory bowel disease patients compared to control group in southwest of Iran. Mol Biol Rep. (2020) 47:8361–5. doi: 10.1007/s11033-020-05807-7
63. Man SM, Zhang L, Day AS, Leach ST, Lemberg DA, Mitchell H. Campylobacter concisus and other Campylobacter species in children with newly diagnosed Crohn's disease. Inflammation Bowel Dis. (2010) 16:1008–16. doi: 10.1002/ibd.21157
64. Sokol H, Leducq V, Aschard H, Pham HP, Jegou S, Landman C, et al. Fungal microbiota dysbiosis in IBD. Gut. (2017) 66:1039–48. doi: 10.1136/gutjnl-2015-310746
65. Boyanova L, Markovska R, Yordanov D, Gergova R, Hadzhiyski P. Anaerobes in specific infectious and noninfectious diseases: new developments. Anaerobe. (2023) 81:102714. doi: 10.1016/j.anaerobe.2023.102714
66. Qi Y, Wu HM, Yang Z, Zhou YF, Jin L, Yang MF, et al. New insights into the role of oral microbiota dysbiosis in the pathogenesis of inflammatory bowel disease. Dig Dis Sci. (2022) 67:42–55. doi: 10.1007/s10620-021-06837-2
67. Stobernack T, du Teil Espina M, Mulder LM, Palma Medina LM, Piebenga DR, Gabarrini G, et al. A secreted bacterial peptidylarginine deiminase can neutralize human innate immune defenses. mBio. (2018) 9:e01704–18. doi: 10.1128/mBio.01704-18
68. Bielecka E, Scavenius C, Kantyka T, Jusko M, Mizgalska D, Szmigielski B, et al. Peptidyl arginine deiminase from Porphyromonas gingivalis abolishes anaphylatoxin C5a activity. J Biol Chem. (2014) 289:32481–7. doi: 10.1074/jbc.C114.617142
69. Di Tommaso N, Gasbarrini A, Ponziani FR. Intestinal barrier in human health and disease. Int J Environ Res Public Health. (2021) 18(23):12836. doi: 10.3390/ijerph182312836
70. Lagha AB, Groeger S, Meyle J, Grenier D. Green tea polyphenols enhance gingival keratinocyte integrity and protect against invasion by Porphyromonas gingivalis. Pathog Dis. (2018) 76(4):10.1093/femspd/fty030. doi: 10.1093/femspd/fty030
71. Wielento A, Bereta GP, Lagosz-Cwik KB, Eick S, Lamont RJ, Grabiec AM, et al. TLR2 activation by porphyromonas gingivalis requires both PPAD activity and fimbriae. Front Immunol. (2022) 13:823685. doi: 10.3389/fimmu.2022.823685
72. Veith PD, Glew MD, Gorasia DG, Reynolds EC. Type IX secretion: the generation of bacterial cell surface coatings involved in virulence, gliding motility and the degradation of complex biopolymers. Mol Microbiol. (2017) 106:35–53. doi: 10.1111/mmi.13752
73. de Diego I, Veillard F, Sztukowska MN, Guevara T, Potempa B, Pomowski A, et al. Structure and mechanism of cysteine peptidase gingipain K (Kgp), a major virulence factor of Porphyromonas gingivalis in periodontitis. J Biol Chem. (2014) 289:32291–302. doi: 10.1074/jbc.M114.602052
74. Liu J. P300 increases CSNK2A1 expression which accelerates colorectal cancer progression through activation of the PI3K-AKT-mTOR axis. Exp Cell Res. (2023) 430:113694. doi: 10.1016/j.yexcr.2023.113694
75. Nakayama M, Inoue T, Naito M, Nakayama K, Ohara N. Attenuation of the phosphatidylinositol 3-kinase/Akt signaling pathway by Porphyromonas gingivalis gingipains RgpA, RgpB, and Kgp. J Biol Chem. (2015) 290:5190–202. doi: 10.1074/jbc.M114.591610
76. Guevara T, Rodriguez-Banqueri A, Lasica AM, Ksiazek M, Potempa BA, Potempa J, et al. Structural determinants of inhibition of Porphyromonas gingivalis gingipain K by KYT-36, a potent, selective, and bioavailable peptidase inhibitor. Sci Rep. (2019) 9:4935. doi: 10.1038/s41598-019-41354-3
77. Nagao JI, Kishikawa S, Tanaka H, Toyonaga K, Narita Y, Negoro-Yasumatsu K, et al. Pathobiont-responsive Th17 cells in gut-mouth axis provoke inflammatory oral disease and are modulated by intestinal microbiome. Cell Rep. (2022) 40:111314. doi: 10.1016/j.celrep.2022.111314
78. Soldati KR, Jiang Y, Brandt BW, Exterkate RAM, Buijs MJ, Nazmi K, et al. Differential modulation of saliva-derived microcosm biofilms by antimicrobial peptide LL-31 and D-LL-31. Pathogens. (2023) 12(11):1295. doi: 10.3390/pathogens12111295
79. Williams DW, Greenwell-Wild T, Brenchley L, Dutzan N, Overmiller A, Sawaya AP, et al. Human oral mucosa cell atlas reveals a stromal-neutrophil axis regulating tissue immunity. Cell. (2021) 184:4090–104 e15. doi: 10.1016/j.cell.2021.05.013
80. Xia Y, Cheng T, Zhang C, Zhou M, Hu Z, Kang F, et al. Human bone marrow mesenchymal stem cell-derived extracellular vesicles restore Th17/Treg homeostasis in periodontitis via miR-1246. FASEB J. (2023) 37:e23226. doi: 10.1096/fj.202300674RR
81. Zhang X, Liu L, Yuan X, Wei Y, Wei X. JMJD3 in the regulation of human diseases. Protein Cell. (2019) 10:864–82. doi: 10.1007/s13238-019-0653-9
82. De Santa F, Narang V, Yap ZH, Tusi BK, Burgold T, Austenaa L, et al. Jmjd3 contributes to the control of gene expression in LPS-activated macrophages. EMBO J. (2009) 28:3341–52. doi: 10.1038/emboj.2009.271
83. Huang D, Zhang C, Wang P, Li X, Gao L, Zhao C. JMJD3 promotes porphyromonas gingivalis lipopolysaccharide-induced th17-cell differentiation by modulating the STAT3-RORc signaling pathway. DNA Cell Biol. (2022) 41:778–87. doi: 10.1089/dna.2022.0149
84. Zhang X, Xiao H, Zhang H, Jiang Y. Lactobacillus plantarum surface-displayed FomA (Fusobacterium nucleatum) protein generally stimulates protective immune responses in mice. Front Microbiol. (2023) 14:1228857. doi: 10.3389/fmicb.2023.1228857
85. Martin-Gallausiaux C, Salesse L, Garcia-Weber D, Marinelli L, Beguet-Crespel F, Brochard V, et al. Fusobacterium nucleatum promotes inflammatory and anti-apoptotic responses in colorectal cancer cells via ADP-heptose release and ALPK1/TIFA axis activation. Gut Microbes. (2024) 16:2295384. doi: 10.1080/19490976.2023.2295384
86. Bashir A, Miskeen AY, Hazari YM, Asrafuzzaman S, Fazili KM. Fusobacterium nucleatum, inflammation, and immunity: the fire within human gut. Tumour Biol. (2016) 37:2805–10. doi: 10.1007/s13277-015-4724-0
87. Chen L, Zhao R, Kang Z, Cao Z, Liu N, Shen J, et al. Delivery of short chain fatty acid butyrate to overcome Fusobacterium nucleatum-induced chemoresistance. J Control Release. (2023) 363:43–56. doi: 10.1016/j.jconrel.2023.09.028
88. Dharmani P, Strauss J, Ambrose C, Allen-Vercoe E, Chadee K. Fusobacterium nucleatum infection of colonic cells stimulates MUC2 mucin and tumor necrosis factor alpha. Infect Immun. (2011) 79:2597–607. doi: 10.1128/IAI.05118-11
89. Neiers F, Gourrat K, Canon F, Schwartz M. Metabolism of cysteine conjugates and production of flavor sulfur compounds by a carbon-sulfur lyase from the oral anaerobe fusobacterium nucleatum. J Agric Food Chem. (2022) 70:9969–79. doi: 10.1021/acs.jafc.2c01727
90. Tang B, Wang K, Jia YP, Zhu P, Fang Y, Zhang ZJ, et al. Fusobacterium nucleatum-Induced Impairment of Autophagic Flux Enhances the Expression of Proinflammatory Cytokines via ROS in Caco-2 Cells. PloS One. (2016) 11:e0165701. doi: 10.1371/journal.pone.0165701
91. Engevik MA, Danhof HA, Auchtung J, Endres BT, Ruan W, Basseres E, et al. Fusobacteriumnucleatum Adheres to Clostridioides difficile via the RadD Adhesin to Enhance Biofilm Formation in Intestinal Mucus. Gastroenterology. (2021) 160:1301–14 e8. doi: 10.1053/j.gastro.2020.11.034
92. Fan Z, Tang P, Li C, Yang Q, Xu Y, Su C, et al. Fusobacterium nucleatum and its associated systemic diseases: epidemiologic studies and possible mechanisms. J Oral Microbiol. (2023) 15:2145729. doi: 10.1080/20002297.2022.2145729
93. Wei S, Zhang J, Wu X, Chen M, Huang H, Zeng S, et al. Fusobacterium nucleatum Extracellular Vesicles Promote Experimental Colitis by Modulating Autophagy via the miR-574-5p/CARD3 Axis. Inflammation Bowel Dis. (2023) 29:9–26. doi: 10.1093/ibd/izac177
94. Yeoh YK, Chen Z, Wong MCS, Hui M, Yu J, Ng SC, et al. Southern Chinese populations harbour non-nucleatum Fusobacteria possessing homologues of the colorectal cancer-associated FadA virulence factor. Gut. (2020) 69:1998–2007. doi: 10.1136/gutjnl-2019-319635
95. Liu L, Liang L, Liang H, Wang M, Lu B, Xue M, et al. Fusobacterium nucleatum Aggravates the Progression of Colitis by Regulating M1 Macrophage Polarization via AKT2 Pathway. Front Immunol. (2019) 10:1324. doi: 10.3389/fimmu.2019.01324
96. Li DH, Li ZP, Yan Z, Zhou GZ, Ren RR, Zhao HJ, et al. Fecal Fusobacterium nucleatum harbored virulence gene fadA are associated with ulcerative colitis and clinical outcomes. Microb Pathog. (2021) 157:104964. doi: 10.1016/j.micpath.2021.104964
97. Huang Y, Liao Y, Luo B, Li L, Zhang Y, Yan F. Non-surgical periodontal treatment restored the gut microbiota and intestinal barrier in apolipoprotein E(-/-) mice with periodontitis. Front Cell Infect Microbiol. (2020) 10:498. doi: 10.3389/fcimb.2020.00498
98. Brennan CA, Garrett WS. Fusobacterium nucleatum - symbiont, opportunist and oncobacterium. Nat Rev Microbiol. (2019) 17:156–66. doi: 10.1038/s41579-018-0129-6
99. Stange EF, Schroeder BO. Microbiota and mucosal defense in IBD: an update. Expert Rev Gastroenterol Hepatol. (2019) 13:963–76. doi: 10.1080/17474124.2019.1671822
100. Ebbers M, Lubcke PM, Volzke J, Kriebel K, Hieke C, Engelmann R, et al. Interplay between P. gingivalis, F. nucleatum and A. actinomycetemcomitans in murine alveolar bone loss, arthritis onset and progression. Sci Rep. (2018) 8:15129. doi: 10.1038/s41598-018-33129-z
101. Duan C, Tang X, Wang W, Qian W, Fu X, Deng X, et al. Lactobacillus rhamnosus attenuates intestinal inflammation induced by Fusobacterium nucleatum infection by restoring the autophagic flux. Int J Mol Med. (2021) 47:125–36. doi: 10.3892/ijmm
102. Quevrain E, Maubert MA, Michon C, Chain F, Marquant R, Tailhades J, et al. Identification of an anti-inflammatory protein from Faecalibacterium prausnitzii, a commensal bacterium deficient in Crohn's disease. Gut. (2016) 65:415–25. doi: 10.1136/gutjnl-2014-307649
103. Wang L, Liu R, Yan F, Chen W, Zhang M, Lu Q, et al. A newly isolated intestinal bacterium involved in the C-ring cleavage of flavan-3-ol monomers and the antioxidant activity of the metabolites. Food Funct. (2024) 15(2):580–90. doi: 10.1039/D3FO03601D
104. Guo X, Xu J, Huang C, Zhang Y, Zhao H, Zhu M, et al. Rapamycin extenuates experimental colitis by modulating the gut microbiota. J Gastroenterol Hepatol. (2023) 38:2130–41. doi: 10.1111/jgh.16381
105. Abranches J, Zeng L, Kajfasz JK, Palmer SR, Chakraborty B, Wen ZT, et al. Biology of oral streptococci. Microbiol Spectr. (2018) 6(5):10.1128/microbiolspec.GPP3-0042-2018. doi: 10.1128/microbiolspec.GPP3-0042-2018
106. Iribarren C, Nordlander S, Sundin J, Isaksson S, Savolainen O, Tornblom H, et al. Fecal luminal factors from patients with irritable bowel syndrome induce distinct gene expression of colonoids. Neurogastroenterol Motil. (2022) 34:e14390. doi: 10.1111/nmo.14390
107. Lobato-Delgado B, Priego-Torres B, Sanchez-Morillo D. Combining molecular, imaging, and clinical data analysis for predicting cancer prognosis. Cancers (Basel). (2022) 14(13):3215. doi: 10.3390/cancers14133215
108. Gill SK, Rossi M, Bajka B, Whelan K. Dietary fibre in gastrointestinal health and disease. Nat Rev Gastroenterol Hepatol. (2021) 18:101–16. doi: 10.1038/s41575-020-00375-4
109. Abdelbary MMH, Hatting M, Bott A, Dahlhausen A, Keller D, Trautwein C, et al. The oral-gut axis: Salivary and fecal microbiome dysbiosis in patients with inflammatory bowel disease. Front Cell Infect Microbiol. (2022) 12:1010853. doi: 10.3389/fcimb.2022.1010853
110. Gecse KB, Vermeire S. Differential diagnosis of inflammatory bowel disease: imitations and complications. Lancet Gastroenterol Hepatol. (2018) 3:644–53. doi: 10.1016/S2468-1253(18)30159-6
111. Kojima A, Nakano K, Wada K, Takahashi H, Katayama K, Yoneda M, et al. Infection of specific strains of Streptococcus mutans, oral bacteria, confers a risk of ulcerative colitis. Sci Rep. (2012) 2:332. doi: 10.1038/srep00332
112. Ito S, Misaki T, Naka S, Wato K, Nagasawa Y, Nomura R, et al. Specific strains of Streptococcus mutans, a pathogen of dental caries, in the tonsils, are associated with IgA nephropathy. Sci Rep. (2019) 9:20130. doi: 10.1038/s41598-019-56679-2
113. Kaci G, Goudercourt D, Dennin V, Pot B, Dore J, Ehrlich SD, et al. Anti-inflammatory properties of Streptococcus salivarius, a commensal bacterium of the oral cavity and digestive tract. Appl Environ Microbiol. (2014) 80:928–34. doi: 10.1128/AEM.03133-13
114. Kaci G, Lakhdari O, Dore J, Ehrlich SD, Renault P, Blottiere HM, et al. Inhibition of the NF-kappaB pathway in human intestinal epithelial cells by commensal Streptococcus salivarius. Appl Environ Microbiol. (2011) 77:4681–4. doi: 10.1128/AEM.03021-10
115. Couvigny B, de Wouters T, Kaci G, Jacouton E, Delorme C, Dore J, et al. Commensal streptococcus salivarius modulates PPARgamma transcriptional activity in human intestinal epithelial cells. PloS One. (2015) 10:e0125371. doi: 10.1371/journal.pone.0125371
116. Arbour CA, Vuksanovic N, Bernstein HM, Allen KN, Imperiali B. Characterization of pglJ, a glycosyltransferase in the campylobacter concisus N-linked protein glycosylation pathway that expands glycan diversity. Biochemistry. (2024) 63:141–51. doi: 10.1021/acs.biochem.3c00564
117. Cai Z, Zhu T, Liu F, Zhuang Z, Zhao L. Co-pathogens in periodontitis and inflammatory bowel disease. Front Med (Lausanne). (2021) 8:723719. doi: 10.3389/fmed.2021.723719
118. Zhang L, Budiman V, Day AS, Mitchell H, Lemberg DA, Riordan SM, et al. Isolation and detection of Campylobacter concisus from saliva of healthy individuals and patients with inflammatory bowel disease. J Clin Microbiol. (2010) 48:2965–7. doi: 10.1128/JCM.02391-09
119. Castano-Rodriguez N, Kaakoush NO, Lee WS, Mitchell HM. Dual role of Helicobacter and Campylobacter species in IBD: a systematic review and meta-analysis. Gut. (2017) 66:235–49. doi: 10.1136/gutjnl-2015-310545
120. Lee H, Ma R, Grimm MC, Riordan SM, Lan R, Zhong L, et al. Examination of the anaerobic growth of campylobacter concisus strains. Int J Microbiol. (2014) 2014:476047. doi: 10.1155/2014/476047
121. Zhang L, Lee H, Grimm MC, Riordan SM, Day AS, Lemberg DA. Campylobacter concisus and inflammatory bowel disease. World J Gastroenterol. (2014) 20:1259–67. doi: 10.3748/wjg.v20.i5.1259
122. Mahendran V, Liu F, Riordan SM, Grimm MC, Tanaka MM, Zhang L. Examination of the effects of Campylobacter concisus zonula occludens toxin on intestinal epithelial cells and macrophages. Gut Pathog. (2016) 8:18. doi: 10.1186/s13099-016-0101-9
123. Man SM, Kaakoush NO, Leach ST, Nahidi L, Lu HK, Norman J, et al. Host attachment, invasion, and stimulation of proinflammatory cytokines by Campylobacter concisus and other non-Campylobacter jejuni Campylobacter species. J Infect Dis. (2010) 202:1855–65. doi: 10.1086/657316
124. Nielsen HL, Nielsen H, Ejlertsen T, Engberg J, Gunzel D, Zeitz M, et al. Oral and fecal Campylobacter concisus strains perturb barrier function by apoptosis induction in HT-29/B6 intestinal epithelial cells. PloS One. (2011) 6:e23858. doi: 10.1371/journal.pone.0023858
125. Heimesaat MM, Ismail Y, Lee H, Riordan SM, Grimm MC, Zhang L. The effects of oral and enteric campylobacter concisus strains on expression of TLR4, MD-2, TLR2, TLR5 and COX-2 in HT-29 cells. PloS One. (2013) 8(2):e56888. doi: 10.1371/journal.pone.0056888
126. Ismail Y, Mahendran V, Octavia S, Day AS, Riordan SM, Grimm MC, et al. Investigation of the enteric pathogenic potential of oral Campylobacter concisus strains isolated from patients with inflammatory bowel disease. PloS One. (2012) 7:e38217. doi: 10.1371/journal.pone.0038217
127. Kharaghani AA, Harzandi N, Khorsand B, Rajabnia M, Kharaghani AA, Houri H. High prevalence of Mucosa-Associated extended-spectrum beta-Lactamase-producing Escherichia coli and Klebsiella pneumoniae among Iranain patients with inflammatory bowel disease (IBD). Ann Clin Microbiol Antimicrob. (2023) 22:86. doi: 10.1186/s12941-023-00630-x
128. Golob J, Rao K, Berinstein J, Chey W, Owyang C, Kamada N, et al. The fecal microbiome in quiescent crohn's disease with persistent gastrointestinal symptoms show enrichment of oral microbes but depletion of butyrate and indole producers. medRxiv. (2023) 2023.05.16.23290065. doi: 10.1101/2023.05.16.23290065
129. Baker JL, Hendrickson EL, Tang X, Lux R, He X, Edlund A, et al. Klebsiella and Providencia emerge as lone survivors following long-term starvation of oral microbiota. Proc Natl Acad Sci United States America. (2019) 116:8499–504. doi: 10.1073/pnas.1820594116
130. Guo Y, Kitamoto S, Caballero-Flores G, Kim Y, Watanabe D, Sugihara K, et al. Oral pathobiont Klebsiella chaperon usher pili provide site-specific adaptation for the inflamed gut mucosa. Gut Microbes. (2024) 16(1):2333463. doi: 10.1080/19490976.2024.2333463
131. Guo Y, Kitamoto S, Caballero-Flores G, Kim Y, Watanabe D, Sugihara K, et al. Oral pathobiont Klebsiella chaperon usher pili provide site-specific adaptation for the inflamed gut mucosa. Gut Microbes. (2024) 16:2333463. doi: 10.1080/19490976.2024.2333463
132. Li K, Mei X, Xu K, Jia L, Zhao P, Tian Y, et al. Comparative study of cigarette smoke, Klebsiella pneumoniae, and their combination on airway epithelial barrier function in mice. Environ Toxicol. (2023) 38:1133–42. doi: 10.1002/tox.23753
133. Panpetch W, Phuengmaung P, Hiengrach P, Issara-Amphorn J, Cheibchalard T, Somboonna N, et al. Candida Worsens Klebsiella pneumoniae Induced-Sepsis in a Mouse Model with Low Dose Dextran Sulfate Solution through Gut Dysbiosis and Enhanced Inflammation. Int J Mol Sci. (2022) 23(13):7050. doi: 10.3390/ijms23137050
134. Lee IA, Kim DH. Klebsiella pneumoniae increases the risk of inflammation and colitis in a murine model of intestinal bowel disease. Scand J Gastroenterol. (2011) 46:684–93. doi: 10.3109/00365521.2011.560678
135. Zhang Q, Su X, Zhang C, Chen W, Wang Y, Yang X, et al. Klebsiella pneumoniae induces inflammatory bowel disease through caspase-11-mediated IL18 in the gut epithelial cells. Cell Mol Gastroenterol Hepatol. (2023) 15:613–32. doi: 10.1016/j.jcmgh.2022.11.005
136. Kanazuru T, Sato EF, Nagata K, Matsui H, Watanabe K, Kasahara E, et al. Role of hydrogen generation by Klebsiella pneumoniae in the oral cavity. J Microbiol. (2010) 48:778–83. doi: 10.1007/s12275-010-0149-z
137. Garrett WS, Gallini CA, Yatsunenko T, Michaud M, DuBois A, Delaney ML, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. (2010) 8:292–300. doi: 10.1016/j.chom.2010.08.004
138. Mukherjee PK, Sendid B, Hoarau G, Colombel JF, Poulain D, Ghannoum MA. Mycobiota in gastrointestinal diseases. Nat Rev Gastroenterol Hepatol. (2015) 12:77–87. doi: 10.1038/nrgastro.2014.188
139. Zollner-Schwetz I, Auner HW, Paulitsch A, Buzina W, Staber PB, Ofner-Kopeinig P, et al. Oral and intestinal Candida colonization in patients undergoing hematopoietic stem-cell transplantation. J Infect Dis. (2008) 198:150–3.
140. Panpetch W, Hiengrach P, Nilgate S, Tumwasorn S, Somboonna N, Wilantho A, et al. Additional Candida albicans administration enhances the severity of dextran sulfate solution induced colitis mouse model through leaky gut-enhanced systemic inflammation and gut-dysbiosis but attenuated by Lactobacillus rhamnosus L34. Gut Microbes. (2020) 11:465–80. doi: 10.1080/19490976.2019.1662712
141. Mao X, Ma J, Jiao C, Tang N, Zhao X, Wang D, et al. Faecalibacterium prausnitzii Attenuates DSS-Induced Colitis by Inhibiting the Colonization and Pathogenicity of Candida albicans. Mol Nutr Food Res. (2021) 65:e2100433. doi: 10.1002/mnfr.202100433
142. Standaert-Vitse A, Sendid B, Joossens M, Francois N, Vandewalle-El Khoury P, Branche J, et al. Candida albicans colonization and ASCA in familial Crohn's disease. Am J Gastroenterol. (2009) 104:1745–53. doi: 10.1038/ajg.2009.225
143. Ferrante M, Henckaerts L, Joossens M, Pierik M, Joossens S, Dotan N, et al. New serological markers in inflammatory bowel disease are associated with complicated disease behaviour. Gut. (2007) 56:1394–403. doi: 10.1136/gut.2006.108043
144. Gerard R, Sendid B, Colombel JF, Poulain D, Jouault T. An immunological link between Candida albicans colonization and Crohn's disease. Crit Rev Microbiol. (2015) 41:135–9. doi: 10.3109/1040841X.2013.810587
145. Li XV, Leonardi I, Putzel GG, Semon A, Fiers WD, Kusakabe T, et al. Immune regulation by fungal strain diversity in inflammatory bowel disease. Nature. (2022) 603:672–8. doi: 10.1038/s41586-022-04502-w
146. Yamazaki K. Oral-gut axis as a novel biological mechanism linking periodontal disease and systemic diseases: A review. Jpn Dent Sci Rev. (2023) 59:273–80. doi: 10.1016/j.jdsr.2023.08.003
147. Madsen GR, Bertl K, Pandis N, Stavropoulos A, Burisch J. The impact of periodontitis on inflammatory bowel disease activity. Inflammatory Bowel Dis. (2023) 29:396–404. doi: 10.1093/ibd/izac090
148. Zhou T, Xu W, Wang Q, Jiang C, Li H, Chao Y, et al. The effect of the "Oral-Gut" axis on periodontitis in inflammatory bowel disease: A review of microbe and immune mechanism associations. Front Cell Infect Microbiol. (2023) 13:1132420. doi: 10.3389/fcimb.2023.1132420
149. Elmaghrawy K, Fleming P, Fitzgerald K, Cooper S, Dominik A, Hussey S, et al. The oral microbiome in treatment-naïve paediatric IBD patients exhibits dysbiosis related to disease severity that resolves following therapy. J Crohn's Colitis. (2023) 17:553–64. doi: 10.1093/ecco-jcc/jjac155
150. Hu S, Mok J, Gowans M, Ong DEH, Hartono JL, Lee JWJ. Oral microbiome of crohn's disease patients with and without oral manifestations. J Crohn's Colitis. (2022) 16:1628–36. doi: 10.1093/ecco-jcc/jjac063
Keywords: inflammatory bowel disease, ulcerative colitis, Crohn’s disease, oral microorganism, oral-gut microbiome axis
Citation: Wang A, Zhai Z, Ding Y, Wei J, Wei Z and Cao H (2024) The oral-gut microbiome axis in inflammatory bowel disease: from inside to insight. Front. Immunol. 15:1430001. doi: 10.3389/fimmu.2024.1430001
Received: 09 May 2024; Accepted: 09 July 2024;
Published: 26 July 2024.
Edited by:
Jiajia Song, Southwest University, ChinaReviewed by:
Catherine Chaput, NEC Laboratories Europe, GermanyCopyright © 2024 Wang, Zhai, Ding, Wei, Wei and Cao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hailong Cao, Y2FvaGFpbG9uZ0B0bXUuZWR1LmNu; Zhiqiang Wei, d2VpemhpcWlhbmdva0AxMjYuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.