- 1Institute for Maternal and Child Health – IRCCS Burlo Garofolo, Trieste, Italy
- 2Transfusion Medicine Department, Azienda Sanitaria Universitaria Giuliano Isontina, Trieste, Italy
- 3Department of Medical Sciences, University of Trieste, Trieste, Italy
Introduction: Type 1 diabetes is an autoimmune disease with an significant genetic component, played mainly by the HLA class II genes. Although evidence on the role of HLA class I genes in developing type 1 diabetes and its onset have emerged, current HLA screening is limited to determining DR3 and DR4 haplotypes. This study aimed to investigate the role of HLA genes on type 1 diabetes risk and age of onset by extensive typing.
Methods: This study included 115 children and young adults with type 1 diabetes for whom typing of HLA-A, -B, -C, -DRB1, -DRB3/4/5, -DQA1, -DQB1, -DPA1 and -DPB1 genes was conducted using Next Generation Sequencing.
Results: We observed that 13% of type 1 diabetes subjects had non-classical HLA haplotypes that predispose to diabetes. We also found that compared to type 1 diabetes subjects with classical HLA haplotypes, non-classical HLA subjects had a significantly higher frequency of HLA-B*39:06:02 (p-value=0.01) and HLA-C*07:02:01 (p-value=0.03) alleles, known to be involved in activating the immune response. Non-classical HLA subjects also presented peculiar clinical features compared to classical HLA subjects, such as multiple diabetic antibodies and the absence of other autoimmune diseases (i.e., coeliac disease and thyroiditis). We also observed that subjects with early onset had a higher frequency of DQ2/DQ8 genotype than late-onset individuals. Moreover, subjects with late-onset had a higher frequency of alleles HLA-B*27 (p-value=0.003), HLA-C*01:02:01 (p-value=0.027) and C*02:02:02 (p-value=0.01), known to be associated with increased protection against viral infections.
Discussion: This study reveals a broader involvement of the HLA locus in the development and onset of type 1 diabetes, providing insights into new possible disease prevention and management strategies.
1 Introduction
Type 1 diabetes (T1D) is an autoimmune disease characterized by insulin deficiency due to chronic immune-mediated progressive destruction of pancreatic beta cells through the interaction between T lymphocytes and autoantigens (1, 2).
Potential triggers of pancreatic islet autoimmunity are viral infections, toxins, and dietary factors that act on a genetically susceptible background to T1D development (3–5). Over 60 loci across the genome were associated with T1D susceptibility (6, 7). However, the primary susceptibility locus is mapped to the Human Leukocyte Antigen (HLA) class II gene, placed on the short arm of chromosome 6, 6p21.31 (6, 8).
As the HLA system is highly polymorphic, numerous HLA alleles affect the peptides pool, which is recognized as initiating an immune reaction (9).
The strong linkage disequilibrium (LD) in the HLA region leads to the formation of haplotypes consisting of the combination of definite allelic variants. Specific HLA haplotypes are strongly associated with T1D (10) and those associated with the highest risk are HLA-DRB1*04-DQA1*03:01-DQB1*03:02 (or DR4-DQ8) and HLA-DRB1*03:01-DQA1*05:01-DQB1*02:01 (or DR3-DQ2) (11).
On the other hand, other HLA class II haplotypes, such as the DQB1*06:02-DRB1*15:01-DQA1*01:02, which is carried by 20% of the general population compared to 1% of T1D subjects, turn out to have a protective effect against the development of T1D (12).
HLA class II genes do not represent all the observed HLA-related inheritance of T1D (9) and some studies had also revealed the role of HLA class I genes (HLA-A, HLA-B, and HLA-C). The HLA class I molecules are involved in immune response, influencing beta cell destruction, as confirmed by the observation of overexpression of HLA class I molecules on islet cells of deceased T1D subjects (13). For example, HLA-A*24 alleles have been associated with rapid and complete destruction of beta cell function in T1D subjects (14); HLA-B*39:06 allele is known to be the most T1D predisposing HLA class I allele. At the same time, the HLA-B*57:01 has a protective effect (15). Conditional analyses, performed to exclude the effect of LD across the HLA region, also revealed an influence of the HLA-DP alleles on T1D susceptibility. In particular, the study by Varney et al. showed a predisposing effect of the DPA1*01:03 allele in association with the DPB1*03:01, whereas DPA1*01:03 has a protective role when associated with DPB1*04:02 (16).
Th e HLA genes also influence the age at onset of T1D. The high-risk genotype HLA-DRB1*03-DQA1*0501-DQB1*0201/DRB1*0401-DQA1*0301-DQB1*0302 was strongly associated with the onset of diabetes before the age of 5 years and that the frequency of this genotype in T1D subjects decreased with increasing age at onset (17).
Similarly, an association between HLA class I genes and the age at onset of diabetes was observed. For example, HLA-A*24:02, related to the total beta cell destruction, has been associated with an earlier onset of T1D (18). At the same time, diabetic subjects carrying HLA-B*39:06 had an average age at onset 3.7 years lower than subjects without this allele. The same trend was observed for T1D subjects carrying HLA-C*07:02; in contrast, we observed the opposite for carriers of the HLA-B*44:03, who had an age at onset approximately 3.5 years higher than people with diabetes without this allele (19).
Despite this growing body of work, to date, HLA screening in T1D individuals is limited to the determination of DR3 and DR4 haplotypes. Moreover, the previously mentioned studies reporting an involvement of HLA genes in the development of T1D have been conducted on a limited number of subjects and on a limited set of HLA genes.
However, HLA typing is fundamental for studying the risk of T1D and the pathogenesis of the disease, especially with a view to prevention and intervention in high-risk subjects, such as individuals with family history. Moreover, determining the genetic component underlying the early onset of T1D can lead to early intervention and avert greater severity of the disease. Indeed, numerous studies indicate that an early onset of T1D is associated with a poor prognosis of diabetes and an elevated risk of developing diabetes-related diseases (20–23).
Therefore, in this study, DNA samples from 115 children and young adults with T1D were typed for HLA-A, -B, -C, -DRB1, -DRB3/4/5, -DQA1, -DQB1, -DPA1 and -DPB1 by NGS (Next Generation Sequencing), with the aim of investigating the role of these genes on T1D risk and age at onset.
2 Materials and methods
2.1 Study participants
In this observational study, we included 115 T1D European subjects referred to the Endocrinologic Unit of IRCCS Burlo Garofolo of Trieste (Italy). T1D subjects were collected from June 2016 to August 2023 according to the following inclusion criteria: diagnosis of type 1 diabetes for at least one year and age between 6 and 21 years.
For all participants, demographic and anthropometric information (age, sex, height, weight, Body Mass Index (BMI), puberty) at the time of recruitment were collected. BMI standard deviation scores were calculated according to WHO reference charts (24) using the Growth Calculator 4 software (http://www.weboriented.it/gh4/), as recommended by the Italian Society for Pediatric Endocrinology and Diabetology (ISPED).
Puberty was defined as the presence of at least breast budding in girls [B2] and a testicular volume of 4 ml in boys [G2] (25).
Furthermore, we collected data relating to the T1D onset, such as the HbA1c value, the presence or absence of ketoacidosis (DKA) [calculated as a venous pH of <7.3 and/or a bicarbonate (HCO3) level of <15 mmol/L (26)], age at onset and the presence of antibodies: Insulin-directed antibodies (IAA), antibodies directed against tyrosine phosphatase (IA2), antibodies directed against the cytoplasm of pancreatic islet cells (ICA), antibodies directed against glutamic acid decarboxylase (GAD) and zinc transporter 8 autoantibodies (ZnT8A) (27).
We gathered additional clinical data, such as the previous year’s HbA1c of and the presence of diseases concomitant to diabetes (e.g., coeliac and thyroid disease).
All the enrolled participants or their parents, for participants aged <18 years, gave written informed consent. The ethics committee approved the study (CEUR-2018-Em-323-Burlo).
2.2 DNA extraction and HLA typing by next generation sequencing
DNA was extracted from saliva using the EZ1 DNA investigator kit (Qiagen, Milan, Italy) following the manufacturer’s protocols.
HLA typing was performed employing the MIA FORA NGS MFlex HLA typing kit (Immucor, Inc., GA, USA), which uses long-range PCR to capture the clinically relevant class I and II HLA genes. The kit includes each class I genes, HLA-A, -B, -C, and the class II genes, HLA-DRB, -DQA1, -DQB1, -DPA1 and -DPB1.
Sample and sequencing library preparation was performed following the manufacturer’s protocols. NGS sequencing libraries were prepared in sets of 24 samples and were sequenced using iSeq™ 100 Illumina®.
Candidate HLA alleles were computed and the final HLA typing was called and examined using the MIA FORA NGS EXPRESS analysis software (Immucor, Inc., GA, USA).
Using three orthogonal algorithms, including independent mapping and de novo assembly strategies, a probability score was calculated, genotype candidates were ranked, and consensus sequences were created for individual alleles.
Due to the lack of coverage in the intron region, ambiguities in the fourth field could not be ruled out so HLA typings were assigned to the third field.
2.3 Statistical analysis
Demographic, anthropometric and clinical data were reported in contingency tables.
Continuous variables were indicated by mean and standard deviation (mean ± sd), and binary traits by percentages (%).
T1D subjects were grouped into classical HLA (CH) and non-classical HLA (NCH) according to the presence or absence of classical HLA haplotypes predisposing to diabetes.
According to percentile distribution of age at onset, T1D subjects were stratified into three groups: early-onset (EO) (age at onset <5 years old); intermediate-onset (IO) (age at onset ≥5<10 years old); late-onset (LO) (age at onset ≥10 old).
Poor Glycemic Control (PGC) [HbA1c > 7% (53 mmol/mol)] was defined using the mean HbA1c values of the previous year (28).
To test normality, we calculated skewness and kurtosis. We applied for binary variables the Fisher’s exact test to analyzed anthropometric and clinical data among the stratified T1D subjects. Instead, to compare continuous variables between CH and NCH T1D subjects, when the variable was normally or non-normally distributed, we used t-test or Mann-Whitney test, respectively.
Whereas, to compare continuous variables between EO, IO and LO T1D subjects, the ANOVA test for normal data and the Kruskall-Wallis test for non-normal data were used.
Fisher’s exact tests analyzed differences in HLA alleles and haplotypes between CH and NCH, and between EO, IO, and LO.
We set statistical significance at a p-value ≤ 0.05. All statistical analyses were performed with R software (www.r-project.org).
3 Results
3.1 HLA typing
We recruited 115 T1D children and young adults and performed HLA typing.
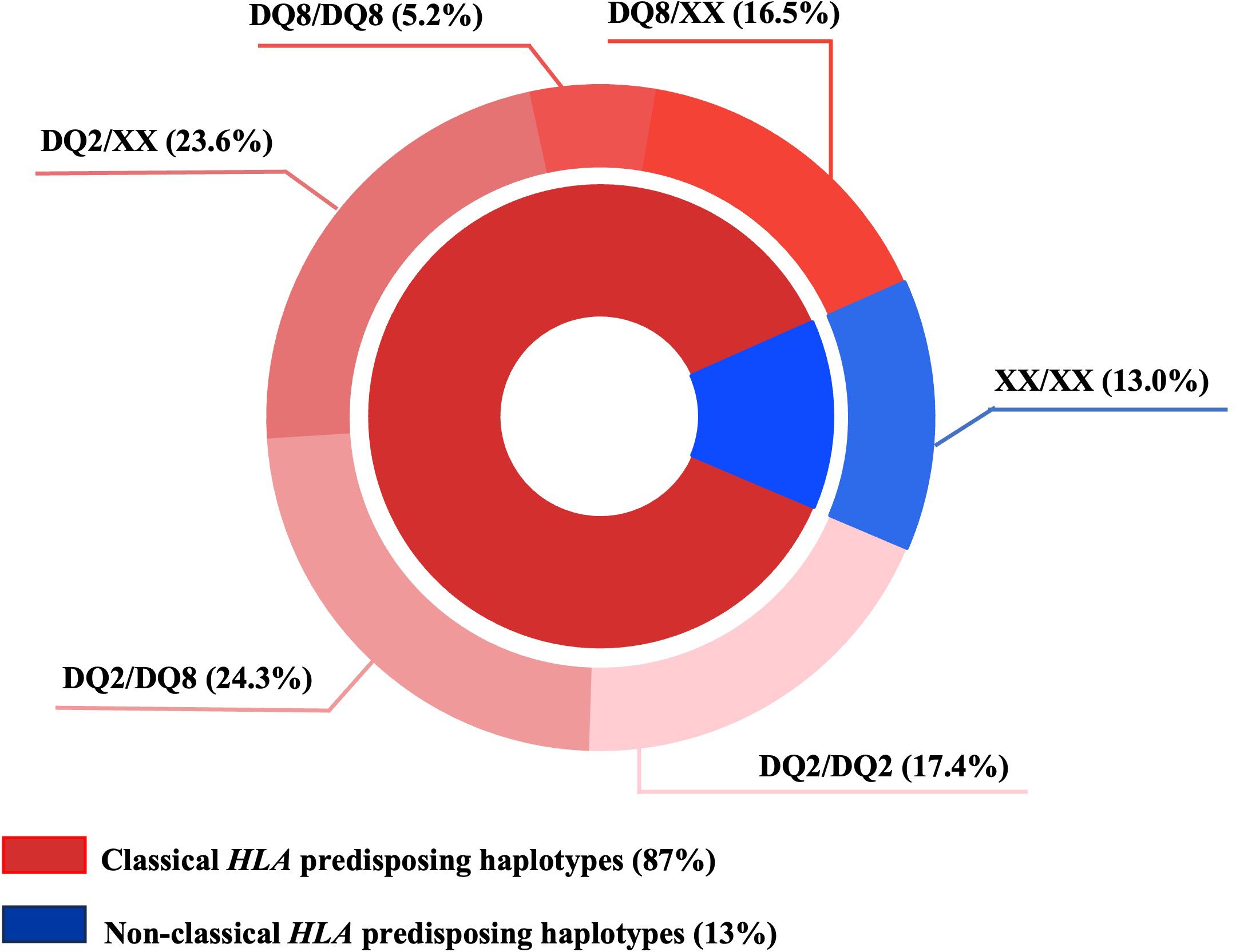
We observed that in 87% of T1D subjects classical HLA predisposing haplotypes were present, represented mainly by the DQ2/DQ8 (24.3%) and the DQ2/XX (23.6%) genotypes (XX indicates a haplotype different from DQ2 and/or DQ8), as shown in the pie chart (Figure 1). Interestingly, HLA typing also showed that 13% of T1D subjects had non-classical HLA haplotypes predisposing to diabetes.
Analysis of the allele frequencies of HLA class I and II genes in CH and NCH T1D subjects showed statistically different allele frequencies between T1D CH and NCH groups (Figure 2). Supplementary Table 1 reports all HLA alleles identified in T1D subjects. In particular, for the HLA class I alleles, we found that HLA-A*01:01:01 (19.5% vs. 0%, p-value = 0.002), HLA-B*08:01:01 (24% vs. 0%, p-value = 0.0003) and HLA-C*07:01:01 (29% vs. 7%, p-value = 0.01) were more frequent in CH T1D that in NCH T1D subjects (Figure 2A). Instead, HLA class I alleles more frequent in NCH T1D than CH T1D subjects were HLA-B*39:06:02 (10% vs. 1%, p-value = 0.01), HLA-B*41:01:01 (10% vs. 2%, p-value = 0.04), HLA-C*07:02:01 (17% vs. 3.5%, p-value = 0.03), and HLA-C*17:01:01 (10% vs. 0.5%, p-value = 0.007) (Figure 2A).
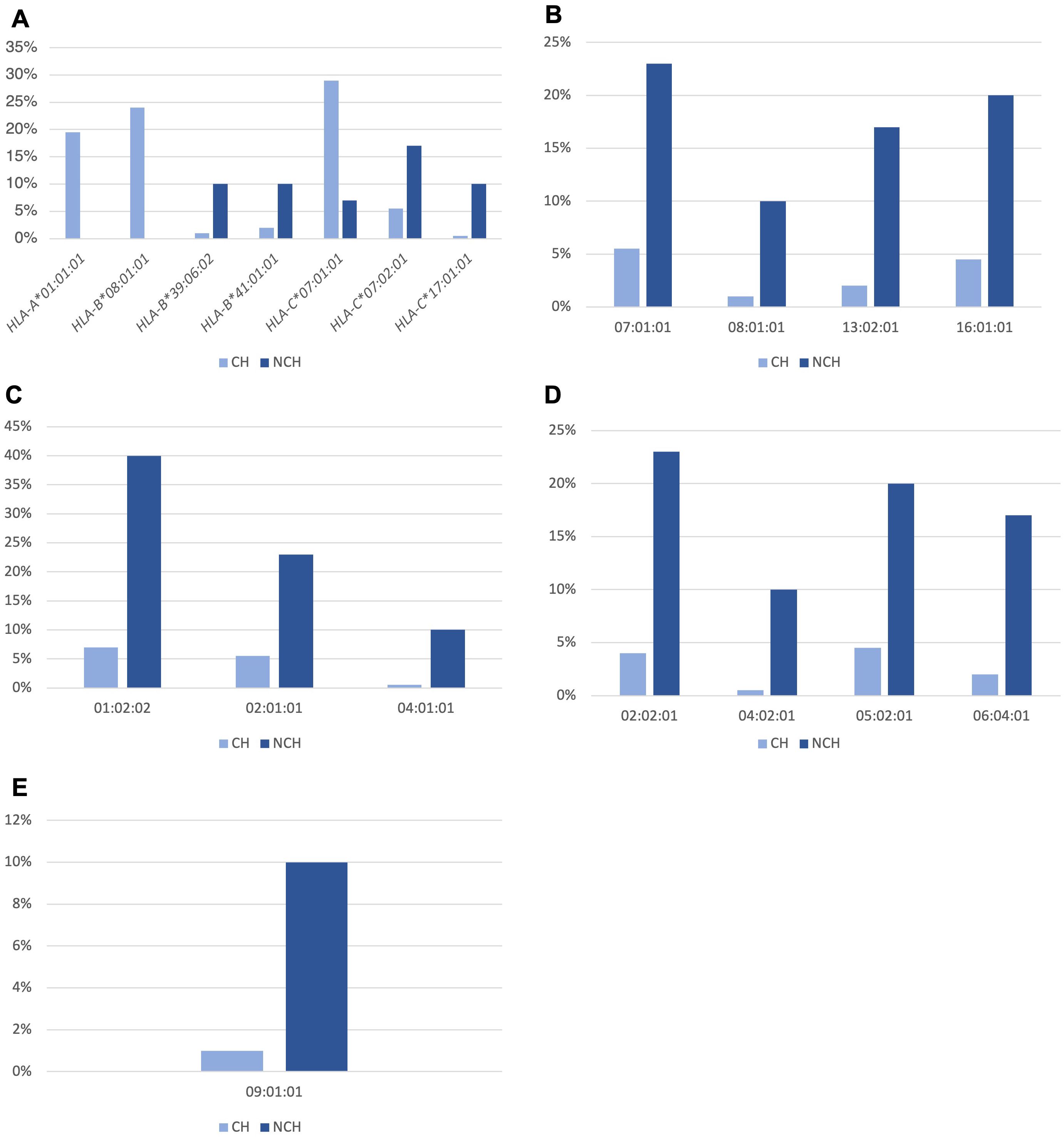
Figure 2. Statistically different allele frequencies in the HLA-A, -B and -C genes of class I (A) and in the HLA-DRB1 (B), -DQA1 (C), -DQB1 (D) and -DPB1 (E) genes of class II, in T1D subjects stratified according to the presence of HLA haplotypes predisposing to T1D, in subjects with classical HLA (CH) and non-classical HLA (NCH).
For HLA class II alleles, those which were part of the HLA DQ2 and DQ8 haplotypes of predisposition to T1D were more frequent in CH T1D subjects than in NCH subjects, i.e., HLA-DRB1*03:01:01 (44.5% vs. 0%, p-value < 0.0001), DRB1*04:01:01 (12.5% vs. 0%, p-value = 0.03); HLA-DQA1*03:01:01 (28% vs. 7%, p-value = 0.01), HLA-DQA1*05:01:01 (45.5% vs. 0%, p-value < 0.0001); HLA-DQB1*02:01:01 (45.5% vs. 7%, p-value < 0.0001) and HLA-DQB1*03:02:01 (30% vs. 3%, p-value = 0.0005).
In NCH T1D subjects, the HLA class II alleles significantly more frequent concerning CH T1D subjects were: HLA-DRB1*07:01:01 (23% vs. 5.5%, p-value = 0.002), HLA-DRB1*08:01:01 (10% vs. 1%, p-value = 0.01), HLA-DRB1*13:02:01 (17% vs. 2%, p-value = 0.009) and HLA-DRB1*16:01:01 (20% vs. 4.5%, p-value = 0.02) (Figure 2B); HLA-DQA1*01:02:02 (40% vs. 7%, p-value = 0.001), HLA-DQA1*02:01:01 (23% vs. 5.5%, p-value = 0.002) and HLA-DQA1*04:01:01 (10% vs. 0.5%, p-value = 0.007) (Figure 2C); HLA-DQB1*02:02:01 (23% vs. 4%, p-value = 0.002), HLA-DQB1*04:02:01 (10% vs. 0.5%, p-value = 0.007), HLA-DQB1*05:02:01 (20% vs. 4.5%, p-value = 0.02) and HLA-DQB1*06:04:01 (17% vs. 2%, p-value = 0.009) (Figure 2D); HLA-DPB1*09:01:01 (10% vs. 1%, p-value = 0.01) (Figure 2E).
3.2 Characteristics of T1D subjects according to the presence of classical and non-classical HLA haplotypes
In Table 1, we reported the characteristics of T1D subjects stratified according to the presence of classical and non-classical HLA haplotypes predisposing to T1D.

Table 1. Characteristics of the T1D subjects stratified according to HLA haplotypes predisposing to T1D in classical HLA (CH) and non-classical HLA (NCH) subjects.
Overall, 54% of the 115 T1D subjects included in the study were female, the mean age of the sample was 15.6 ± 3.7 years, 28% presented DKA at onset, and 65% had a PGC.
Despite the limited number of NCH subjects, comparing CH and NCH subjects, we found that NCH T1D subjects had a shorter disease duration than the CH T1D ones (6.1 vs. 8.4 years old, p-value = 0.03) and that all NCH subjects had more than one antibody compared to 66% of CH T1D subjects (p-value = 0.02).
Furthermore, although differences did not reach statistical significance, higher age at onset was observed in NCH subjects compared to CH T1D subjects (8.5 vs. 7.4 years), as well as a more elevated percentage of DKA at onset in NCH compared to CH (38.5% vs. 26%). Finally, none of the NCH T1D subjects showed coeliac disease or autoimmune thyroid disease, compared to CH T1D subjects (16.5% and 21%, respectively).
3.3 Association between HLA and age at onset
In the present work, we stratified subjects according to their age at onset of T1D and studied its association with HLA genotypes.
In our study population, the age at onset was <5 years old (early-onset, EO) in 31% of participants, between 5 and 10 years old (intermediate-onset, IO) in 46%, and over 10 years old (late-onset, LO) in the remaining 23%. In EO, IO and LO groups, respectively 53%, 45% and 73% were females, while the mean age in the three groups was 13.9 ± 4.1, 15.8 ± 3.5 and 17.5 ± 2.5. Anthropometric and clinical data are reported in Supplementary Table 2.
Table 2 indicates the genotype distribution according to age at onset. The most frequent HLA genotype was DQ2/DQ8, followed by the DQ2/XX genotype, except for LO T1D subjects, for whom the most frequent genotype was DQ2/XX, followed by DQ8/XX.
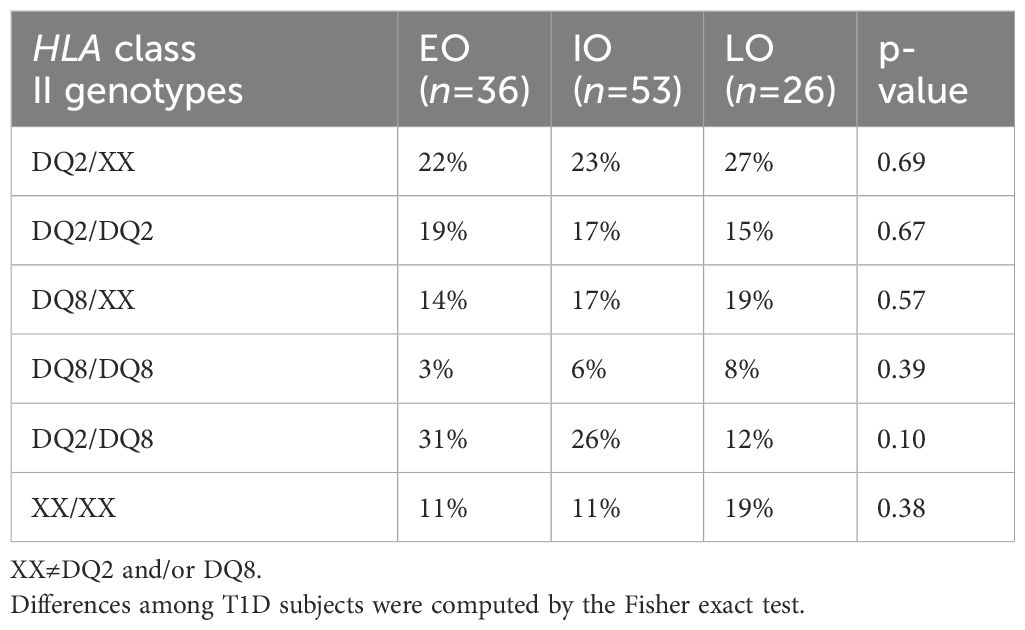
Table 2. Genotype distribution in T1D subjects stratified according to age at onset in early-onset (EO, <5 years old), intermediate-onset (IO, ≥5<10 years old) and late-onset (LO, ≥10 years old).
In T1D subjects, stratified according to age at onset in EO, IO and LO, no significant differences emerged in genotypes distribution. Instead regarding the frequency of HLA alleles, Figure 3 reports significant differences in the three groups. In particular, for the HLA-C locus, HLA-C*01:02:01 and *02:02:02 were more frequent in LO than in EO (7.5% vs. 0%, p-value = 0.027, 10% vs. 0%, p-value = 0.01, respectively), and in IO (7.5% vs. 1% p-value = 0.038, 10% vs. 2%, p-value = 0.036, respectively), while HLA-C*03:04:01 was more frequent in EO (11.5%) compared to IO (2%) and LO (2%) but only the comparison between EO and IO was statistically significant (p-value = 0.01).
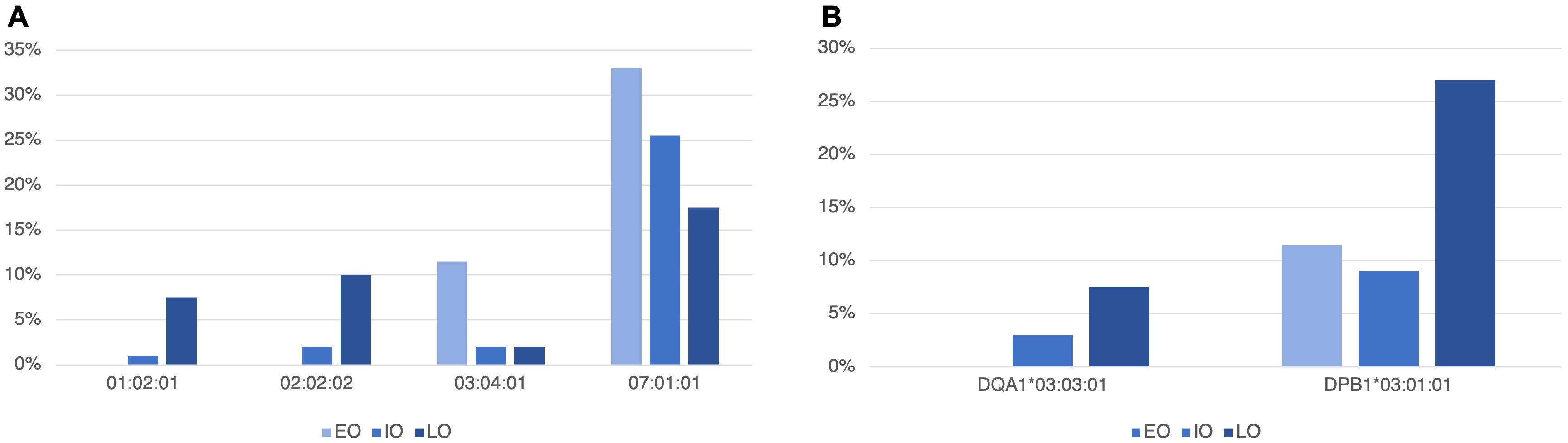
Figure 3. Statistically different allele frequencies in the HLA-C gene of class I (A) and in the HLA-DQA1 and -DPB1 (B) class II genes, in T1D subjects, stratified according to age at onset in early-onset (EO, <5 years old), intermediate-onset (IO, ≥5<10 years old) and late-onset (LO, ≥10 years old).
HLA-C*07:01:01 was also more frequent in EO (33%) compared to IO (25.5%) and LO (17.5%). In this case, only the comparison between EO and LO was statistically significant (p-value = 0.04).
Concerning HLA class II alleles, we found that HLA-DQA1*03:03:01 was more frequent in LO T1D subjects than in EO, in which this allele was absent (7.5% vs. 0%, p-value = 0.027). HLA-DPB1*03:01:01 was also more frequent in LO (27%) compared to EO (11.5%) and IO (9%) (p-value = 0.029 and p-value = 0.016, respectively).
Supplementary Table 3 reports all HLA class I and II allele frequencies in EO, IO and LO. A particular mention should be made about HLA-B*27 alleles (27:02:01, 27:05:02 and 27:07:01), which were not detected in EO subjects, whereas they were present in 2% of IO and 23% of LO, with significant differences in the comparison between EO and LO (p-value = 0.003) and between IO and LO (p-value = 0.004).
4 Discussion
Our findings suggest that haplotypes strongly associated with the development of T1D are not the only HLA genetic component involved in the risk of diabetes and in the age at onset of the disease.
In our cohort, DQ2/DQ8 was the most frequent genotype, in agreement with previous literature data (29, 30). However, in our study HLA high-resolution typing proved that 13% of T1D subjects did not have the classical HLA haplotypes predisposing to the disease. In agreement with our results, approximately the same frequency of T1D subjects without classic HLA predisposition alleles was already found in Polish T1D children and adolescents (31), confirming that HLA class II alleles do not represent the entire genetic component of predisposition to T1D.
In particular, in the present study, analysis of the genetic characteristics of T1D subjects without the classical HLA predisposing haplotypes showed that the HLA class II alleles significantly more frequent in NCH T1D subjects were part of four different haplotypes (HLA-DRB1*07:01:01-DQA1*02:01:01-DQB1*02:02:01; HLA-DRB1*08:01:01-DQA1*04:01:01-DQB1*04:02:01; HLA-DRB1*13:02:01-DQA1*01:02:02-DQB1*06:04:01; and HLA-DRB1*16:01:01-DQA1*01:02:02-DQB1*05:02:01), previously reported to have a neutral or, even, a protective role towards the T1D development risk (29, 32, 33).
These data suggested that NCH T1D subjects probably had other genetic risk factors, which may reside in the HLA class I region. The role of HLA class I genes on the development of T1D had indeed already been assessed, as the encoded molecules bind and present antigens to CD8+ T cells, both by aiding the selection of the T cell repertoire and by initiating antigen-specific cytotoxicity (30).
In our study, one of the more frequent HLA class I alleles in NCH compared to CH T1D subjects was HLA-B*39:06:02. This allele was already known to be the most predisposing HLA class I allele to T1D (15, 34). By developing an HLA-B*39:06 transgenic NOD mouse model, Schloss et al. (35) demonstrated that this allele independently mediates the development of CD8+ T cells that conferred susceptibility to T1D. Some studies reported that HLA-B*39:06 could increase the risk of diabetes in combination with some low-risk haplotypes, containing DRB1*08:01, DRB1*01:01 or DRB1*16:01 alleles (19, 36). The odds ratio values obtained from combination of HLA-B*39:06 with these alleles indicated a significantly raised risk, comparable to that of the DR3/DR4 genotype (36). In our sample, HLA-B*39:06 was indeed found in combination with these alleles, confirming the hypothesis of its involvement in the development of T1D, even in the absence of high-risk haplotypes.
Another HLA class I allele that we have frequently found in NCH T1D subjects was HLA-C*07:02:01. This allele was positively correlated with increased expression of the T cell receptor complex variable region (TCR V) gene, stimulating high immunological activity (37), as suggested by its known association with T1D (34) and psoriasis (37). Some authors hypothesized that the effect of this allele on the development of T1D depended on LD with HLA-B*39:06. However, its strong association with the early onset of diabetes, independently from HLA-B*39:06, supported an autonomous effect on predisposition to T1D (19). In our cohort, we did not find a LD between these two alleles; furthermore, the HLA-C*07:02:01 was always in combination with protective or neutral DR-DQ haplotypes, confirming its autonomous effect on the T1D risk.
Interestingly, the present work suggests that diabetic individuals without the classical high-risk haplotypes present peculiar clinical characteristics. For example, we found that all NCH subjects showed more than one diabetes antibody compared to 66% of the CH group. Our results did not confirm previous studies that reported the role of HLA-DR3 and HLA-DR4 alleles in producing T1D antibodies (38–41). However, the role of other genes has also been proposed (40–42), and future studies should be conducted to investigate their involvement in antibody development better.
In our work, other clinical characteristics were peculiar to NCH T1D subjects, even if the comparison with CH did not result statistically significant, possibly due to the small sample size of our study. For example, we observed that 38% of NCH T1D subjects presented DKA at disease onset, compared to 26% of CH T1D subjects, in agreement with previous studies that had already shown a protective effect of high-risk HLA alleles on the development of DKA (43, 44). Moreover, NCH T1D subjects did not present other autoimmune pathologies, such as coeliac disease and autoimmune thyroid diseases. This observation highlighted the usefulness of carrying out extended HLA typing in helping to identify distinct clinical characteristics and set or avoid specific controls. For example, implementing genetic testing could reduce the number of patients requiring systematic immunological screening.
Finally, the age at onset of the disease in the NCH T1D group was slightly older than in the CH T1D subjects, confirming previous studies on the role of HLA genes in this regard (18, 19, 45).
Infact, investigating the association of HLA genes according to the age at onset, we also observed that T1D subjects with early onset had a higher percentage of DQ2/DQ8 genotype than T1D subjects with late onset. This result suggested that the role of this genotype is not only to increase the risk of developing T1D but also to induce its development at an early age. Although it was difficult to determine whether this high-risk genotype alone was enough to confer a greater risk of developing diabetes early, we showed that the DQ2/DQ8 genotype was associated with an early age at onset (6.37 age at onset) and that the age at onset increased in the absence of DQ2 and in the presence of DQ8 (DQ2/DQ2 = 7.07 age at onset; DQ2/XX = 7.8 age at onset; DQ8/XX = 8.1 age at onset; DQ8/DQ8 = 8.9; XX/XX = 8.6). Interestingly, when we evaluated the mean age at onset associated with both DQ2/DQ8 genotype and HLA-C*03:04:01, that were significantly more frequent in the early onset subjects, we observed that age at onset lowered to 3.9 years. This observation implied that other HLA genetic factors might be involved in early onset besides DQ2 and DQ8.
On the other hand, in the group of late onset T1D subjects, the significantly more frequent HLA alleles were HLA-B*27 alleles, HLA-C*01:02:01 and C*02:02:02, all known to be associated with increased protection against viral infections (46–49). Since the peak of early onset observed around the age of 5 was connected to the numerous infections in the early school years (50), protection from viral infections given by these alleles could prevent the early development of the disease. The presence of the alleles mentioned above delayed the age at onset by about 5 years compared to T1D subjects not carrying these alleles (HLA-B*27+, average age at onset 12.48, HLA-B*27- average age at onset 7.21; HLA-C*01:02:01+, average age at onset 11.65, HLA-C*01:02:01-, average age at onset 7.34; HLA-C*02:02:02+, average age at onset 12.14, HLA-C*02:02:02-, average age at onset 7.23), independently from classical HLA risk haplotypes.
Interestingly, some of these HLA alleles were also strongly associated with the development of other diseases, such as seronegative arthritis (HLA-B*27) (51) and psoriasis (HLA-C*01) (52). It means that these subjects, protected from the early development of diabetes, could be more susceptible to the development of other autoimmune diseases. In our study, T1D subjects did not manifest any HLA-related diseases (excluding coeliac and autoimmune thyroid diseases). However, our outcomes suggested that this risk should be considered when screening of diabetic subjects.
Overall, the findings of this study highlight important implications for future clinical practice.
First, our study supports the use of HLA typing by NGS as an accurate and feasible method for HLA-related disease association testing (including T1D), allowing to overcome the detection limitations of low-resolution traditional methods commonly used in clinical practice [i.e., Sequence-Specific Oligonucleotide (SSO) and Sequence-Specific Primer (SSP)] (53). Moreover, considering that the NGS costs continue to decline and are currently similar to or lower than the SSO method, HLA typing by NGS may also become a cost-effective approach, especially in large volume laboratories, thanks to the possibility of analyzing many patients within a single run.
Furthermore, our results underline the possible relevance of HLA extended typing from a prevention perspective. The purpose of screening for T1D is to identify early the risk of developing the disease to prevent a severe onset of diabetes with the presence of DKA, causing both short- and long-term complications. Current T1D screening, in addition to analyzing the presence of antibodies, only assesses the presence of the primary genetic risk factors, DR3 and DR4. In a pediatric population screening context, HLA extended typing would allow the identification of new diabetes predisposition alleles whose identification requires a sample size to counteract the excessive allelic variability of the HLA locus. Moreover, considering their genetic heterogeneity would allow a more accurate genetic risk assessment of different populations. So, if NGS becomes widely used, it could provide a valuable resource for future screening and population studies.
The results of our study show that HLA is associated with clinical characteristics, including age at onset, also emphasize that NGS-based HLA analyses may be relevant for defining clinical phenotypes and predicting the disease course. Moreover, since the HLA region represents the major genetic component for the development of numerous autoimmune diseases, such as coeliac disease, psoriasis, rheumatoid arthritis, and thyroiditis, often concomitant in diabetic subjects, NGS HLA extended typing could also prove useful in the management of T1D subjects (54).
Overall, although low-resolution HLA typing continues to be used in clinical practice, an NGS-based method should be considered in the future since more precise assignments of HLA alleles/haplotypes could help to elucidate the exact role of HLA variation in the etiopathogenesis and progression of T1D, which may result in improved patient care, decreased additional testing, and then reduced costs.
5 Conclusion
The study’s results indicate that the HLA locus’s involvement in the development of T1D and the onset time occurs on a broader spectrum and does not only involve the classical HLA genotypes known to play a predisposing role in diabetes.
However, despite high-resolution genotyping, the extreme allelic variability, in conjunction with the limited number of subjects analyzed, makes these data preliminary. Therefore, it is essential to increase the number of T1D subjects studied to better understand the role of the HLA genes in type 1 diabetes. Moreover, given the small sample size, some associations may have been missed. Furthermore, control group of healthy subjects are not considered in this study.
Overall, despite these limitations, the results of our study suggest that an in-depth understanding of the HLA genetic variations associated with T1D and, in particular, with age at onset, may increase our knowledge of the mechanisms underlying the development of the disease and can also be used in disease prevention and in patient management.
Data availability statement
The original Fastq files obtained from HLA typing are publicly available at https://www.ebi.ac.uk/ena/browser/home with access ID: PRJEB76550. Other information may be available from the corresponding author upon reasonable request.
Ethics statement
The studies involving humans were approved by Comitato Etico Unico Regionale FVG_CEUR-2018-Em-323-Burlo (Italy). The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin.
Author contributions
AR: Writing – review & editing, Writing – original draft, Investigation, Funding acquisition, Formal analysis, Conceptualization. EB: Writing – review & editing, Writing – original draft, Data curation, Conceptualization. LA: Writing – review & editing, Writing – original draft, Data curation. AC: Writing – review & editing, Writing – original draft, Data curation. VB: Writing – review & editing, Writing – original draft, Data curation. GT: Writing – review & editing, Writing – original draft, Conceptualization. EC: Writing – review & editing, Writing – original draft, Supervision, Methodology, Investigation, Funding acquisition, Formal analysis, Data curation, Conceptualization.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Ministry of Health, Roma – Italy, in collaboration with the Institute for Maternal and Child Health IRCCS Burlo Garofolo, Trieste – Italy (GR-2019-12369573 to AR, RC 06/23 to EC).
Acknowledgments
We thank all the children, young people and parents who participated in this study. Thanks to Dr Melissa Frequin, of Immucor Italia S.p.A., for her constant technical support. We thank Martina Bradaschia for the language editing of the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1427349/full#supplementary-material
References
1. Katsarou A, Gudbjörnsdottir S, Rawshani A, Dabelea D, Bonifacio E, Anderson BJ, et al. Type 1 diabetes mellitus. Nat Rev Dis Primers. (2017) 3:17016. doi: 10.1038/nrdp.2017.16
2. Mayer-Davis EJ, Kahkoska AR, Jefferies C, Dabelea D, Balde N, Gong CX, et al. ISPAD Clinical Practice Consensus Guidelines 2018: Definition, epidemiology, and classification of diabetes in children and adolescents. Pediatr Diabetes. (2018) 19:7–19. doi: 10.1111/pedi.2018.19.issue-S27
3. Li Y, Sun F, Yue TT, Wang FX, Yang CL, Luo JH, et al. Revisiting the antigen-presenting function of beta cells in T1D pathogenesis. Front Immunol. (2021) 12:690783. doi: 10.3389/fimmu.2021.690783
4. Krischer JP, Liu X, Lernmark Å, Hagopian WA, Rewers MJ, She JX, et al. Predictors of the initiation of islet autoimmunity and progression to multiple autoantibodies and clinical diabetes: The TEDDY Study. Diabetes Care. (2022) 45:2271–81. doi: 10.2337/dc21-2612
5. Zorena K, Michalska M, Kurpas M, Jaskulak M, Murawska A, Rostami S. Environmental factors and the risk of developing type 1 diabetes-old disease and new data. Biol (Basel). (2022) 11:608. doi: 10.3390/biology11040608
6. Pociot F, Lernmark A. Genetic risk factors for type 1 diabetes. Lancet. (2016) 2387:2331–9. doi: 10.1016/S0140-6736(16)30582-7
7. Diedisheim M, Carcarino E, Vandiedonck C, Roussel R, Gautier J, Venteclef N. Regulation of inflammation in diabetes: from genetics to epigenomics evidence. Mol Metab. (2020) 41:101041. doi: 10.1016/j.molmet.2020.101041
8. Hu X, Deutsch AJ, Lenz TL, Onengut-Gumuscu S, Han B, Chen WM, et al. Additive and interaction effects at three amino acid positions in HLA-DQ and HLA-DR molecules drive type 1 diabetes risk. Nat Gene. (2015) 47:898–905. doi: 10.1038/ng.3353
9. Kennedy AE, Ozbek U, Dorak MT. What has GWAS done for HLA and disease associations? Int J Immunogenet. (2017) 44:195–211. doi: 10.1111/iji.12332
10. Nyaga DM, Vickers MH, Jefferies C, Perry JK, O’Sullivan JM. The genetic architecture of type 1 diabetes mellitus. Mol Cell Endocrinol. (2018) 477:70–80. doi: 10.1016/j.mce.2018.06.002
11. Moheb-Alian A, Forouzesh F, Sadeghi A, Rostami K, Aghamohammadi E, Rostami-Nejad M, et al. Contribution of HLA-DQ2/DQ8 haplotypes in type one diabetes patients with/without celiac disease. J Diabetes Complications. (2019) 33:59–62. doi: 10.1016/j.jdiacomp.2018.10.001
12. Redondo MJ, Kawasaki E, Mulgrew CL, Noble JA, Erlich HA, Freed BM, et al. DR- and DQ-associated protection from type 1A diabetes: comparison of DRB1*1401 and DQA1*0102-DQB1*0602*. J Clin Endocrinol Metab. (2000) 85:3793–7. doi: 10.1210/jcem.85.10.6920
13. Richardson SJ, Rodriguez-Calvo T, Gerling IC, Mathews CE, Kaddis JS, Russell MA, et al. Islet cell hyperexpression of HLA class I antigens: a defining feature in type 1 diabetes. Diabetologia. (2016) 59:2448–58. doi: 10.1007/s00125-016-4067-4
14. Noble JA, Erlich HA. Genetics of type 1 diabetes. Cold Spring Harb Perspect Med. (2012) 2(1):a007732. doi: 10.1101/cshperspect.a007732
15. Nejentsev S, Howson JM, Walker NM, Szeszko J, Field SF, Stevens HE, et al. Localization of type 1 diabetes susceptibility to the MHC class I genes HLA-B and HLA-A. Nature. (2007) 450:887–92. doi: 10.1038/nature06406
16. Varney MD, Valdes AM, Carlson JA, Noble JA, Tait BD, Bonella P, et al. Type 1 Diabetes Genetics Consortium. HLA DPA1, DPB1 alleles and haplotypes contribute to the risk associated with Type 1 Diabetes. Diabetes. (2010) 59:2055–62. doi: 10.2337/db09-0680
17. Lambert AP, Gillespie KM, Thomson G, Cordell HJ, Todd JA, Gale EAM, et al. Absolute risk of childhood-onset type 1 diabetes defined by human leukocyte antigen class II genotype: a population-based study in the United Kingdom. J Clin Endocrinol Metab. (2004) 89:4037–43. doi: 10.1210/jc.2003-032084
18. Valdes AM, Thomson G, Erlich HA, Noble JA. Association between type 1 diabetes age of onset and HLA among sibling pairs. Diabetes. (1999) 48:1658–61. doi: 10.2337/diabetes.48.8.1658
19. Valdes AM, Erlich HA, Noble JA. Human leukocyte antigen class I B and C loci contribute to type 1 diabetes (T1D) susceptibility and age at T1D onset. Hum Immunol. (2005) 66:301–13. doi: 10.1016/j.humimm.2004.12.001
20. Inshaw JRJ, Cutler AJ, Crouch DJ, Wicker LS, Todd JA. Genetic variants predisposing most strongly to type 1 diabetes diagnosed under age 7 years lie near candidate genes that function in the immune system and in pancreatic β-cells. Diabetes Care. (2020) 43:169–77. doi: 10.2337/dc19-0803
21. Leete P, Willcox A, Krogvold L, Dahl-Jørgensen K, Foulis AK, Richardson SJ, et al. Differential insulitic profiles determine the extent of β-cell destruction and the age at onset of type 1 diabetes. Diabetes. (2016) 65:1362–9. doi: 10.2337/db15-1615
22. White NH. Long-term outcomes in youths with diabetes mellitus. Pediatr Clin North Am. (2015) 62:889–909. doi: 10.1016/j.pcl.2015.04.004
23. Hietala K, Harjutsalo V, Forsblom C, Summanen P, Groop PH, FinnDiane Study Group. Age at onset and the risk of proliferative retinopathy in type 1 diabetes. Diabetes Care. (2010) 33:1315–9. doi: 10.2337/dc09-2278
24. World Health Organization. WHO child growth standards: length/height-for-age, weight-for-age, weight-for-length, weight -for-height and body mass index-for-age: methods and development. Geneva, Switzerland: World Health Organization ([amp]]lrm;2006). Available at: https://iris.who.int/handle/10665/43413.
25. Tanner JM, Whitehouse RH. Clinical longitudinal standards for height, weight, height velocity, weight velocity, and stages of puberty. Arch Dis Child. (1976) 51:170–9. doi: 10.1136/adc.51.3.170
26. Raghupathy P. Diabetic ketoacidosis in children and adolescents. Indian J Endocrinol Metab. (2015) 19:S55–7. doi: 10.4103/2230-8210.155403
27. Kawasaki E. Anti-islet autoantibodies in type 1 diabetes. Int J Mol Sci. (2023) 24:10012. doi: 10.3390/ijms241210012
28. De Bock M, Codner E, Craig ME, Huynh T, Maahs DM, Mahmud FH, et al. ISPAD Clinical Practice Consensus Guidelines 2022: Glycemic targets and glucose monitoring for children, adolescents, and young people with diabetes. Pediatr Diabetes. (2022) 23:1270–6. doi: 10.1111/pedi.13455
29. Erlich H, Valdes AM, Noble J, Carlson JA, Varney M, Concannon P, et al. HLA DR-DQ haplotypes and genotypes and type 1 diabetes risk: analysis of the type 1 diabetes genetics consortium families. Diabetes. (2008) 57:1084–92. doi: 10.2337/db07-1331
30. Noble JA, Valdes AM. Genetics of the HLA region in the prediction of type 1 diabetes. Curr Diabetes Rep. (2011) 11:533–42. doi: 10.1007/s11892-011-0223-x
31. Zubkiewicz-Kucharska A, Jamer T, Chrzanowska J, Akutko K, Pytrus T, Stawarski A, et al. Prevalence of haplotype DQ2/DQ8 and celiac disease in children with type 1 diabetes. Diabetol Metab Syndr. (2022) 14:128. doi: 10.1186/s13098-022-00897-8
32. Kieleväinen V, Turtinen M, Luopajärvi K, Härkönen T, Ilonen J, Knip M, et al. Increased HLA class II risk is associated with a more aggressive presentation of clinical type 1 diabetes. Acta Pediatr. (2023) 112:522–8. doi: 10.1111/apa.16621
33. Anand V, Li Y, Liu B, Ghalwash M, Koski E, Ng K, et al. Isles autoimmunity and HLA markers of presymptomatic and clinical type 1 diabetes: joint analyses of prospective cohort studies in Finland, Germany, Sweden and the U. S Diabetes Care. (2021) 44:2269–76. doi: 10.2337/dc20-1836
34. Noble JA, Valdes AM, Varney MD, Carlson JA, Moonsamy P, Fear AL, et al. HLA class I and genetic susceptibility to type 1 diabetes: results from the Type 1 Diabetes Genetics Consortium. Diabetes. (2010) 59:2972–9. doi: 10.2337/db10-0699
35. Schloss J, Ali R, Racine JJ, Chapman HD, Serreze DV, DiLorenzo TP. HLA-B*39:06 efficiently mediates type 1 diabetes in a mouse model incorporating reduced thymic insulin expression. J Immunol. (2018) 200:3353–63. doi: 10.4049/jimmunol.1701652
36. Baschal EE, Baker PR, Eyring KR, Siebert JC, Janinski JM, Eisenbarth GS. The HLA-B 3906 allele imparts a high risk of diabetes only on specific HLA-DR/DQ haplotypes. Diabetologia. (2011) 54:1702–9. doi: 10.1007/s00125-011-2161-1
37. Li J, Li X, He F, Zhao X, Hou R, Lin H, et al. Cross-sectional study reveals that HLA-C*07:02 is a potential biomarker of early onset/lesion severity of psoriasis. Exp Dermatol. (2020) 29:639–46. doi: 10.1111/exd.14127
38. Triolo TM, Pyle L, Broncucia H, Armstrong T, Yu L, Gottlieb PA, et al. Association of high-affinity autoantibodies with type 1 diabetes high-risk HLA haplotypes. J Clin Endocrinol Metab. (2022) 107:e1510–7. doi: 10.1210/clinem/dgab853
39. Lempainen J, Laine AP, Hammais A, Toppari J, Simell O, Veijola R, et al. Non-HLA gene effects on the disease process of type 1 diabetes: From HLA susceptibility to overt disease. J Autoimmun. (2015) 61:45–53. doi: 10.1016/j.jaut.2015.05.005
40. Steck AK, Zhang W, Bugawan TL, Barriga KJ, Blair A, Erlich HA, et al. Do non-HLA genes influence development of persistent islet autoimmunity and type 1 diabetes in children with high-risk HLA-DR, DQ genotypes? Diabetes. (2009) 58:1028–33. doi: 10.2337/db08-1179
41. Steck AK, Wong R, Wagner B, Johnson K, Liu E, Romanos J, et al. Effects of non-HLA gene polymorphisms on development of islet autoimmunity and type 1 diabetes in a population with high-risk HLA-DR, DQ genotypes. Diabetes. (2012) 61:753–8. doi: 10.2337/db11-1228
42. Törn C, Hadley D, Lee HS, Hagopian W, Lernmark A, Simell O, et al. Role of type 1 diabetes associated snps on risk of autoantibody positivity in the TEDDY study. Diabetes. (2015) 64:1818–29. doi: 10.2337/db14-1497
43. Hekkala A, Ilonen J, Knip M, Veijola R, Finnish Pediatric Diabetes Register. Family history of diabetes and distribution of class II HLA genotypes in children with newly diagnosed T1D: effect on diabetic ketoacidosis. Eur J Endocrinol. (2011) 165:813–7. doi: 10.1530/EJE-11-0376
44. Usher-Smith JA, Thompson MJ, Sharp SJ, Walter FM. Factors associated with the presence of diabetic ketoacidosis at diagnosis of diabetes in children and young adults: a systematic review. BMJ. (2011) 343:d4092. doi: 10.1136/bmj.d4092
45. Jiang Z, Ren W, Liang H, Yan J, Yang D, Luo S, et al. HLA class I genes modulate disease risk and age at onset together with DR-DQ in Chinese patients with insulin-requiring type 1 diabetes. Diabetologia. (2021) 64:2016–36. doi: 10.1007/s00125-021-05476-6
46. Goulder PJ, Watkins DI. Impact of MHC class I diversity on immune control of immunodeficiency virus replication. Nat Rev Immunol. (2008) 8:619–30. doi: 10.1038/nri2357
47. McKiernan SM, Hagan R, Curry M, McDonald GS, Kelly A, Nolan N, et al. Distinct MHC class I and II alleles are associated with hepatitis C viral clearance, originating from a single source. Hepatology. (2004) 40:108–14. doi: 10.1002/(ISSN)1527-3350
48. Velastegui E, Vera E, Vanden Berghe W, Muñoz MS, Orellana-Manzano A. HLA-C: evolution, epigenetics, and pathological implications in the major histocompatibility complex. Front Genet. (2023) 14:1206034. doi: 10.3389/fgene.2023.1206034
49. Mori M, Leitman E, Walker B, Ndung’u T, Carrington M, Goulder P. Impact of HLA allele-KIR pairs on HIV clinical outcome in South Africa. J Infect Dis. (2019) 219:1456–63. doi: 10.1093/infdis/jiy692
50. Maahs DM, West NA, Lawrence JM, Mayer-Davis EJ. Epidemiology of type 1 diabetes. Endocrinol Metab Clin North Am. (2010) 39:481–97. doi: 10.1016/j.ecl.2010.05.011
51. Kavadichanda CG, Geng J, Nethra Bulusu S, Singh Negi V, Raghavan M. Spondyloarthritis and the human leukocyte antigen (HLA)-B*27 connection. Front Immunol. (2021) 12:601518. doi: 10.3389/fimmu.2021.601518
52. Huang YW, Tsai TF. HLA-Cw1 and psoriasis. Am J Clin Dermatol. (2021) 22:339–47. doi: 10.1007/s40257-020-00585-1
53. Profaizer T, Pole A, Monds C, Delgado JC, Lázár-Molnár E. Clinical utility of next generation sequencing based HLA typing for disease association and pharmacogenetic testing. Hum Immunol. (2020) 81:354–60. doi: 10.1016/j.humimm.2020.05.001
Keywords: age at onset, human leukocyte antigen, next generation sequencing, pediatric, type 1 diabetes
Citation: Robino A, Bevilacqua E, Aldegheri L, Conti A, Bazzo V, Tornese G and Catamo E (2024) Next-generation sequencing reveals additional HLA class I and class II alleles associated with type 1 diabetes and age at onset. Front. Immunol. 15:1427349. doi: 10.3389/fimmu.2024.1427349
Received: 03 May 2024; Accepted: 22 July 2024;
Published: 09 August 2024.
Edited by:
Bernadete Liphaus, University of São Paulo, BrazilReviewed by:
Howard Davidson, University of Colorado, United StatesVasile Valeriu Lupu, Grigore T. Popa University of Medicine and Pharmacy, Romania
Copyright © 2024 Robino, Bevilacqua, Aldegheri, Conti, Bazzo, Tornese and Catamo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Eulalia Catamo, ZXVsYWxpYS5jYXRhbW9AYnVybG8udHJpZXN0ZS5pdA==
 Antonietta Robino
Antonietta Robino Elena Bevilacqua2
Elena Bevilacqua2 Andrea Conti
Andrea Conti Gianluca Tornese
Gianluca Tornese Eulalia Catamo
Eulalia Catamo