
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol., 14 June 2024
Sec. Molecular Innate Immunity
Volume 15 - 2024 | https://doi.org/10.3389/fimmu.2024.1425168
This article is part of the Research TopicCommunity Series in C-Reactive Protein in Age-Related Disorders - Volume IIIView all 6 articles
C-reactive protein (CRP) is a plasma protein that is evolutionarily conserved, found in both vertebrates and many invertebrates. It is a member of the pentraxin superfamily, characterized by its pentameric structure and calcium-dependent binding to ligands like phosphocholine (PC). In humans and various other species, the plasma concentration of this protein is markedly elevated during inflammatory conditions, establishing it as a prototypical acute phase protein that plays a role in innate immune responses. This feature can also be used clinically to evaluate the severity of inflammation in the organism. Human CRP (huCRP) can exhibit contrasting biological functions due to conformational transitions, while CRP in various species retains conserved protective functions in vivo. The focus of this review will be on the structural traits of CRP, the regulation of its expression, activate complement, and its function in related diseases in vivo.
Pentraxin (PTX) is a superfamily of multifunctional proteins that are highly conserved in evolution and is named because of its “pentagonal” structure under electron microscope observation. PTX originated early in evolution and is called the “living fossil”. Many forms of PTX in ancient horseshoe crabs about 250 to 300 million years ago (1, 2). As a superfamily protein, PTX has been conserved in phylogeny from arachnids to mammals. Its typical feature is the presence of a 200-amino acid pentraxin domain at its carboxyl end (3, 4), called the pentramerization protein domain. Also, all members of this family share a conservative sequence of 8 amino acids in the pentameric protein domain (His-x-Cys-x-Ser/Thr-Trp-x-Ser, where x can be any amino acid), called the pentameric protein tag, or the “signature sequence” of the pentameric protein (5).
The members of the PTX family identified so far mainly include C-reactive protein (CRP), serum amyloid protein P component (SAP), PTX3, PTX4, and neuronal PTX (NPTX), etc. (3, 5). Tillett and Francis first discovered CRP can react with C-polysaccharides in the pneumococcus cell wall in the presence of calcium ions (6). SAP is calcium-dependent recognition and binding of amyloid deposited in tissues. It is a highly conserved plasma glycoprotein in humans and all other species studied. Almost all amyloid deposits have been detected. SAP is present in both of them, which leads to a very high concentration of SAP in amyloid deposits, which can account for about 15% of the total mass (7). The amino acid sequence homology between SAP and CRP is as high as 51%. The structure observed under the electron microscope is highly similar, both showing a pentameric disc-like structure. SAP is considered to be directly related to CRP and originated from single gene duplication (8, 9). PTX3, as a secreted protein containing a pentameric protein domain, was first identified in fibroblasts, induced by IL-1 in endothelial cells, or produced by TNF stimulation (TSG14) in fibroblasts (10). The long-chain pentameric proteins with the same overall composition identified after PTX3 include guinea pig acrosome pentameric protein, neuronal pentameric protein (NPTX1, NPTX2), PTX4 and neuronal pentameric receptor (NPR). Among them, NPR is a transmembrane molecule with a transmembrane domain at its amino terminus (11, 12). CRP and SAP orthologs in different mammalian species have a high degree of sequence similarity. Still are significant differences in serum basal levels and changes during the acute phase reaction. CRP and SAP are the main acute phases in humans and mice respectively (13). PTX3 is highly conserved in humans and mice, while CRP and SAP, their sequence and expression regulation have evolutionary differences between mice and humans (13).
CRP is a typical short-chain PTX member, mainly synthesized in the liver to respond to inflammation. The most significant is the inflammatory stimulus signal mediated by the cytokine IL-6. Pentraxins recognize a wide range of exogenous pathogenic substances and mutated self-molecules, and behave as acute-phase proteins in a species-specific manner. The high degree of sequence homology and the appearance of the pentameric molecule that can be quickly identified indicate that all the different plasma proteins that bind to the classical pentameric ligands in a calcium-dependent manner belong to the same family member. Although the “long pentamer” contains moderately homologous sequence domains, it does not have the appearance of the pentamer. Also, calcium-dependent binding is necessary for the stability of secondary, tertiary and quaternary structures of most pentamers. It is also an essential element for the binding of pentamers to specific ligands, but “long pentamers” do not have such feature.
CRP is an acute-phase protein primarily synthesized in liver hepatocytes and plays a critical role in inflammation response (14–17). When the body is invaded by pathogens such as bacteria, fungi, parasites or subjected to inflammatory stimuli, as well as after tissue damage caused by trauma and progressive cancer, the plasma concentration of CRP can increase rapidly within 6–8 hours and reach the peak in about 48 hours, rising 1000-fold from the basal level ~ 0.5 μg/ml (18). It will return to baseline once the inflammation subsides (19). The plasma levels of CRP exhibit a positive correlation with the severity of inflammation, making it a commonly employed nonspecific biomarker in clinical applications (20–22).
Furthermore, CRP acting as an innate pattern recognition receptor (23), can precipitate the somatic C-polysaccharide of Streptococcus pneumoniae on microorganisms in the presence of calcium, then triggers the innate immunity through the classical complement pathway by interacting with the global head region of C1q or other cell surface glycoproteins such as FcγR receptors (16, 24, 25). The structural analysis of the CRP binding phosphocholine (PC) ligand and C1 complex using Cryo-Electron Tomography (Cryo-ET) revealed a novel mechanism. Contrary to previous understanding, it was observed that CRP does not form an Fc-mediated hexamer antibody platform to bind and activate the C1 complex. Instead, CRP forms a rectangular platform assembled by tetrameric CRP to effectively bind and activate complement (25).
Human CRP (huCRP) has a molecular weight of approximate 115 kDa and is characterized by a “jelly-like lectin fold”. Generally, CRP is composed of five identical subunits which assemble into a cyclic pentamer around a central pore (26). One side within each promoter contains a calcium-binding pocket, accommodating two calcium irons to stable its structure, which further mediates PC ligand-binding named recognition face (Figure 1). The other side possesses a single α-helix, which binds to ligands such as C1q, therefore named effector face (Figure 1). HuCRP is non-glycosylated and has no inter-subunit covalent bond, while a sole disulfide bond within each subunit formed between Cys36 and Cys97. The formation of disulfide bonds in hCRP protomers should occur in the early spontaneous folding stage driven by the conformational folding of six β strands, which plays a vital role in the subsequent conformational reconstruction of the whole subunit and the pentamer assembly (29).
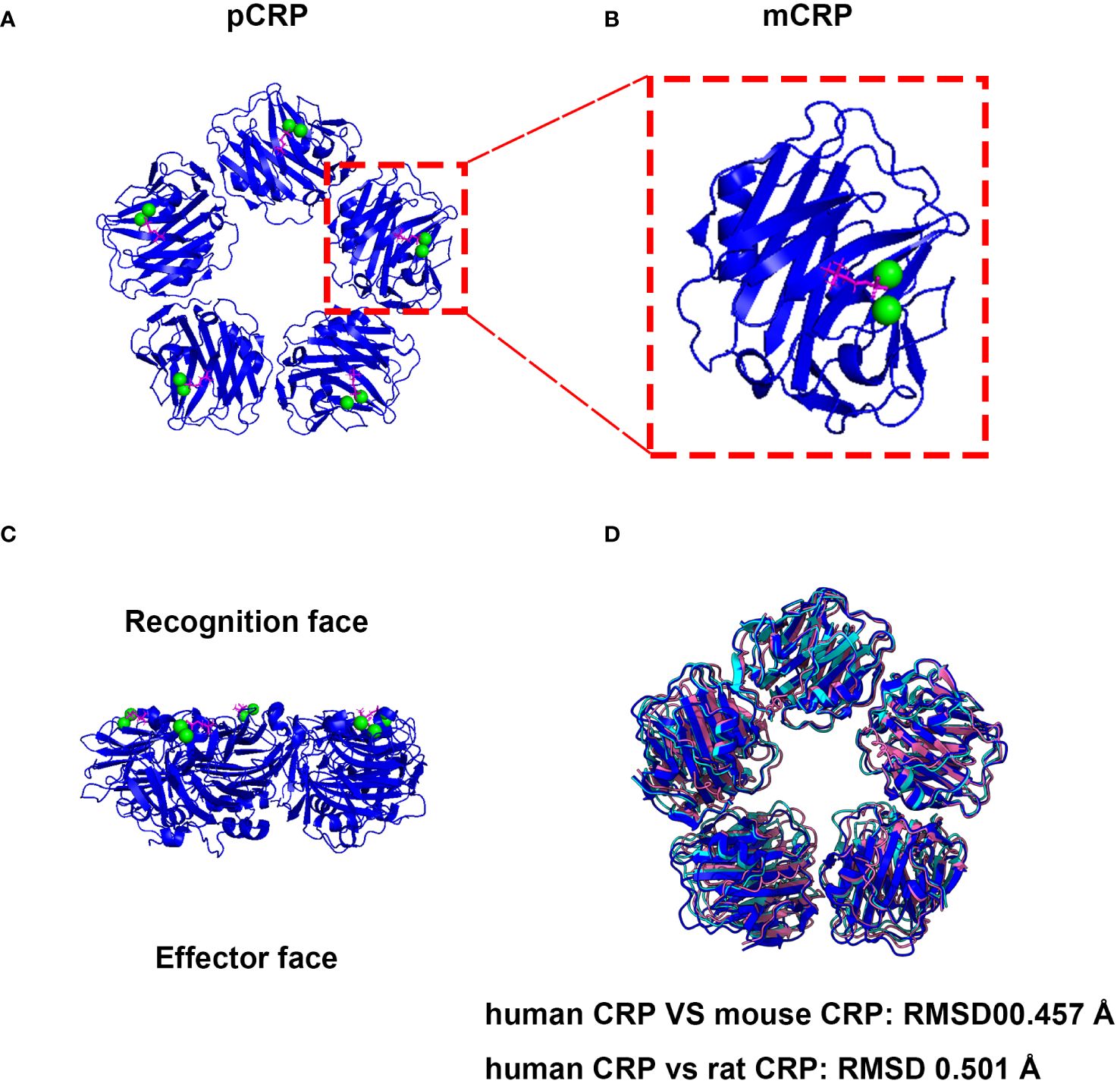
Figure 1 The native structure and associated activities of CRP are evolutionarily conserved. (A) Cryo-EM structure of CRP complexes with PC. The CRP-PC complex was achieved from Protein Data Bank (PDB entry 7PKE) and the ribbon diagram of the CRP Cryo-EM structure of CRP-PC-Ca2+ was generated by ChimeraX software (27). (B) View of the ligand binding face of mCRP. Each protomer contains a binding site, which is shown occupied by 2 calcium (green) and 1 PC molecule (magenta). (C) Side view of the pentameric form of CRP. Which mediates PC ligand-binding named recognition face, and the other side possesses a single α-helix, which binds to ligands such as C1q, therefore named effector face. (D) The pentameric form structures of mouse and rat CRP were generated with AlphaFold 3 using five subunits (28).
The physiological function of CRP is related to calcium-dependent ligand affinity (30, 31). Many studies have used calcium-dependent PC affinity to successfully separate CRP from blood, such as horseshoe crab (32), mouse (33), dog (34), cat (35), cow (36), since the calcium-dependent PC affinity is conserved in the species and as a basis for testing the function of multi-species CRP in experiments. The CRP of human, mouse and rat were expressed in different systems and purified based on the calcium-dependent binding properties to PC (37–40). The binding ability of pentamer CRP to pattern recognition ligand PC was no significant difference among different species (38). In addition, CRP not only can bind to various pathogens, including fungi, yeasts, and bacteria, but also has calcium-dependent binding properties to chromatin, histones, and small ribonucleoprotein U1, and glycans (41). In the absence of calcium, CRP can bind to polycations such as poly-L-lysine, poly-L-arginine, and myelin essential protein (42).
Shrive team first obtained the X-ray crystal structure of huCRP in a calcium-binding state (PDB ID: 1GNH) (43). Thompson group further resolved huCRP in PC-binding state in the presence of calcium (PDB ID: 1B09) (44). It reveals that Phe-66 and Glu-81 are two key residues that mediate the binding of PC to CRP. The distance between the point and the calcium ion is only 0.4 nm (45). Among them, Phe-66 provides hydrophobic interaction with the methyl group of PC, while Glu-81 is located at the other end of the pocket and interacts with the positively charged choline nitrogen (46). Ramadan et al. solved huCRP in a calcium-depleted state (PDB ID: 1LJ7) (26), revealing a decamer with two independent pentamers stacked near parallel in a face A-to-face A form. In 2014, Guillon et al. obtained two crystal structures of huCRP, assembled in a staggered decameric form (47). One of them is stabilized by two zinc ions trapped within a cleft on the effector’s face, also in the presence of HIV-1 Tat protein (PDB ID: 3PVN).
With the rapid development of Cryo-Electron Microscopy (Cryo-EM) and artificial intelligence (AI), it is more convenient and fast to analyze and predict the three-dimensional conformation of protein space. Noone et al. used Cryo-EM to analyze the structure of CRP at pH 7.5 or pH 5 and in the presence or absence of the ligand PC, and found that the previous structure obtained from crystallography was an imperfect pentagon with variable angles between each subunit (48). The pentamer CRP obtained by Cryo-EM was found to have C5 symmetry, and the subunit was formed into an isometric regular pentagon (48). Subsequently, they applied the Cryo-ET resolved the structure of CRP binding to PC after binding to C1 complex. Revealed new interfaces, interpreting previously contradictory biochemical data, and clarifying the possible mechanism by which CRP regulates the complement system (25).
Generally, huCRP primarily exists in two conformational forms. One is the native homo-pentameric form, namely pentameric CRP (pCRP) or native CRP (nCRP), the other is the dissociated form, namely monomeric or modified CRP (mCRP) (49–54). Pentameric CRP can undergo continuous structural changes to form mCRP under various conditions, including low pH, absence of calcium, increased temperature, urea chelation, or binding to activated platelets, apoptotic/necrotic cells, and microparticles (55). It is commonly accepted that pCRP responds to tissue damage or infection by rapidly increasing its blood concentration. Accumulating evidence suggests that pCRP dissociates into mCRP and that most of the proinflammatory effects of CRP are expressed only after dissociation of its native pentamer assembly into mCRP.
It has been shown that CRP exhibits a rapid pentamer-decamer equilibrium at 2 mM Ca2+, with the proportion of decamers decreasing as NaCl concentration increases (56). Both pH and the presence of PC appear to affect the ratio of CRP pentamers to decamers, as well as particle orientation within glass ice, with a pH 5 buffer producing more decamers than a pH 7.5 buffer (48). Based on site-directed mutagenesis studies of amino acids at specific sites, Agrawal and Volanakis proposed that the interaction between CRP and C1q is dependent on conformational changes in the CRP pentamer (57). Through the ligand binding ability under simulated acidic conditions, it was found that the ligand binding was significantly increased at pH 5, and the binding to immobilized C1q and immunoglobulin was also increased. That is, subtle changes in charged residues on CRP and immune ligands can significantly increase their association with inflammatory stimulus response, potentially activating the downstream immune effector function of CRP (48).
Monomeric CRP has been considered as a major contributor in local inflammation (54, 55, 58). Not only it can induce pro-inflammatory effects on cells such as monocytes and endothelial cells (59), and trigger the secretion of interleukin-8 (IL-8) by human neutrophils (60), but also promote inflammation of the human coronary artery endothelial cells phenotype through a p38 mitogen-activated protein kinase-dependent mechanism (61) and enhance neutrophil localization and activation at inflamed or injured vascular sites (62, 63). In addition, mCRP can promote neutrophil-endothelial cell adhesion and delay apoptosis of human neutrophils (64). With accumulating in human atherosclerotic lesions, mCRP can induce monocyte chemotaxis, promote neutrophil survival, which may amplify the inflammatory response (52, 65).
The huCRP gene is located in the 1q23–24 region of the chromosome. It encodes 224 amino acids, of which 18 amino acids at the N-terminus are the signal peptide sequence. The whole gene contains only one intron, which separates the region into encoding the signal peptide and the mature protein (66). CRP responds to IL-6 and IL-1β combined stimulation that can produce an acute phase typically. The promoter region contains two acute-phase response elements, including a binding site for the liver-specific transcription factor hepatocyte nuclear factor (HNF1) and two C/EBPβ (CCAAT/enhancer-binding protein β) binding sites for interleukin-6 (IL-6) induced transcription (67, 68).
The expression of CRP is tissue-specific and is mainly produced by hepatocytes. However, it can also be synthesized in monocytes, neuronal cells, lymphocytes, and atherosclerotic plaques, although CRP produced in those cells or tissues does not contribute to the serum level (69). Its serum concentration is significantly increased in inflammatory diseases such as cardiovascular complications (70). CRP plasma base level can be affected by many factors, such as chronic infectious diseases caused by pathogen invasion or smoking, body mass index (BMI), coffee intake, oral contraceptives, and genetic factors, etc. (71).
The synthesis of CRP in hepatocytes is mainly regulated at the transcription level by the stimulator cytokine IL-6 and interleukin-1β (IL-1β) (67, 72). Transcription factors, such as STAT3, Rel p50, c-Rel, and C/EBPβ/δ are mainly involved in the regulation of CRP expression. STAT3 and Rel bind adjacent to the CRP promoter in the non-coding region, allowing closer binding of C/EBP, an important signaling molecule, to the nucleic acid and promoting its efficiency in inducing CRP expression (73). The NF-κB binding site is located at -69 in the proximal CRP promoter, overlapping with the transcription factors OCT-1/HNF-1/HNF-3 binding sites. IL-1β induces CRP transcription through the proximal CRP promoter by activating NF-κB-p50/p65 in synergy with IL-6-activated C/EBPβ (74). In the absence of C/EBPβ, a complex containing C/EBPδ and RBP-Jκ is formed at the C/EBP-p50 site. The synergistic effect of IL-6 and IL-1β in the induction of CRP gene expression is partially mediated through the NF-κB locus (74).
OCT-1 can inhibit CRP transcription induced by IL-6 and IL-1β through the proximal CRP promoter (75). In addition, post-transcriptional level regulation also plays a particular role in CRP expression, especially in the acute phase when CRP is secreted in large amounts. Under normal physiological conditions, CRP expression is at a very low level. Most CRP is bound to carboxylesterase and stored in the endoplasmic reticulum (ER) (76). In the acute phase, the conformation of carboxylesterase changes, which significantly reducing the binding capability with CRP, thus CRP is released from ER (77).
The expression pattern of CRP is causally determined by the promoter methylation status tuned by DNMT3A and TET2 (21). The CpG deficient promoter motif of CRP is located at the binding sites of STAT3, C/EBP-B, and NF-κB. These motifs are highly methylated in the resting state but undergo STAT3 and NF-kB-dependent demethylation in response to cytokine stimulation, leading to a significant increase in C/EBP-B, thereby promoting CRP expression (21). Further analysis showed that reversible methylation could also regulate highly induced gene expression with CpG promoters represented by APRs. Thus, these promoters lacking CpG may evolve TF binding sites containing CpG, utilizing dynamic methylation to achieve a rapid and reversible response (21, 78).
Natural conformational CRP is the dominant conformation secreted by most cell types, but some cells also directly release mCRP. When mCRP is injected into circulation can be rapidly redistributed into tissues through a lipid raft mediated mechanism. However, the transport of mCRP locally produced in the inflammatory microenvironment into the circulation remains unclear (79, 80).
Currently, research on the impact of human CRP has predominantly involved introducing human CRP into mouse and rat models either through transgenic methods or direct injection (72, 81, 82). It is recognized that CRP behaves differently in mice and rats compared to humans, particularly in its role as an acute phase protein and its interaction with the complement system (83). This variation has led to the consideration of these animal models as naturally deficient in CRP expression or function, thus rendering their endogenous CRP activity negligible. However, whether CRP can activate auto-complement in rats is still controversial, and there are concerns that serum CRP levels in mice may be inaccurately measured (84). Human CRP recognizes microorganisms and apoptotic cells by binding to PC and promotes phagocytosis of phosphorylated substances by activating the classical pathway of complement (CP) (9). In addition, CRP inhibits complement hyperactivation by binding to complement factor H (CFH), the major inhibitor of the alternative pathway (AP) (85). While human CRP is known to adopt an activated conformation and interact with C1q and CFH to activate or inhibit complement, respectively (86), doubts have been raised about the conservation of these interactions in rodents (86, 87). Our recent findings show that mouse, rat, and human CRPs all exhibit complement activating capacity, binding to their own C1q and activating their respective classical complement pathways alone (38, 86).
Moreover, endogenous CRP knockout in mice and rats, and human CRP complementation (intravenous injection and gene knock-in) showed consistent significant phenotypes in acute liver injury models that contribute significantly to complement activation. CRP has a protective effect on acute liver injury and is able to delay death caused by sepsis, indicating that CRP plays an important protective function and is conserved among species (38, 86). It is further pointed out that endogenous CRP in mouse and rat animal models should not be ignored when studying the function of human CRP. This suggests that the research paradigm and experimental design of relevant animal models need to be reconsidered and optimized.
Until now, no human CRP gene defects have been reported, nor even any sequence polymorphisms in the protein itself. Although their actual function in humans is unknown, gene deletion studies in mice suggest that CRP contribute to innate immunity. CRP is the quintessential human acute-phase protein that is used in clinical practice around the world to monitor disease activity. A growing number of clinical trials have shown that CRP is related to many diseases. In cardiovascular research, CRP has been used to diagnose cardiovascular disease (CVD) and as a marker to indicate disease status and incidence (88). Besides, CRP is associated with atherosclerotic vascular disease (ASVD), systemic lupus erythematosus (SLE), cancer, and other diseases (14, 89–91). Its influence on these diseases occurrence, development, and prognosis should not be underestimated (92, 93). CRP is also related to the epidemic virus COVID-19 and is an independent predictor of mortality from COVID-19 infection (94–97).
CRP participates in the innate immune response as a pattern recognition receptor, and each of five homologous globular subunits has a calcium-regulated binding pocket for ligands expressing the PC moiety. Upon binding to ligands on damaged and apoptotic cells, it undergoes a conformational change that allows binding and activation of C1 and may lead to dissociation of monomeric CRP (81). Although CRP recognition of PC or related molecules on microorganisms plays an important role in our defense, a more important role may be the binding of CRP to PC in damaged membranes. PC are not normally exposed to the cell surface but are exposed due to damage by complement or certain phospholipases (98). CRP binds to apoptotic and necrotic cells, and defective clearance of apoptotic cells is associated with autoimmune diseases (81). The colocalization of CRP with fixed complement in areas of tissue injury suggests that CRP may play a role in clearing cellular debris from tissue. Mark et al. showed that CRP had inherent pro-inflammatory effects, either on human peripheral blood mononuclear cells in vitro or when administered parenterally to mice or healthy human volunteers in vivo (99, 100). Genetic and epigenetic studies of gene targeting mice and humans have shown that CRP plays a crucial non-redundant function in innate immunity, inflammation, and tissue remodeling. The innate immune response is activated when the conserved structure on the pathogen surface, the pathogen-associated molecular pattern (PAMP), is recognized by CRP, a pattern recognition molecule (PRM) (101).
For CRP, the mouse is not an ideal model as its CRP levels do not respond to inflammatory stimuli (102). Several studies have been performed to assess the role of huCRP in transgenic mice overexpressing huCRP. Those studies found that huCRP facilitates the survival of mice infected with S. pneumoniae (103, 104). This effect is mediated mainly by the strong response of CRP to the PC present in the cell walls of these bacteria (105). These data indicate that CRPtg mice infected with S. pneumoniae are resistant to infection, showing longer survival time and lower mortality than non-transgenic littermates (wild type) (106). Similar studies have shown that CRP administration can prevent the invasion and infection of Haemophilus influenzae (107). CRPtg animals can resist infection by the Gram-negative pathogen Salmonella even without CRP binding (108).
CVD is one of the major diseases threatening human health today, mainly caused by atherosclerosis (AS). Cardiovascular risk factors such as obesity, hypertension, diabetes, smoking, and dyslipidemia can lead to intravascular inflammatory response and endothelial cell activation and dysfunction, the earliest event in the development of AS. The most widely studied inflammatory marker related to CVD is CRP. Elevated CRP levels are directly proportional to CVD risk and are an independent risk factor for cardiac death. By measured serum CRP and creatine kinase levels in patients with CVD but without cardiogenic chest pain. The results showed that all individuals with acute myocardial infarction showed elevated CRP secretion levels, and there was a significant correlation between peak CRP concentration and creatine kinase values (109, 110). Mild, 2 - to 5-fold increases in baseline plasma CRP levels in asymptomatic individuals are associated with an increased risk of cardiovascular events such as stroke and myocardial infarction (111, 112). And the use of mildly elevated CRP levels to guide primary prevention has led to a significant reduction in major cardiovascular events in apparently healthy persons (113). Although the exact role of CRP in atherosclerosis and its complications remains unclear, there is now increasing evidence that it may be a direct causative factor (9, 114). The crucial role of CRP in the prevention, treatment, and prognosis of CVD has been agreed upon and is even used as the “gold standard” for CVD risk assessment (115, 116).
CRP is a human acute-phase protein, and its plasma level is associated with cancer risk (117, 118). In clinical practice, the CRP test is widely used to monitor disease severity, the clinical course of the disease, and treatment responses. Recent studies have strongly suggested that CRP acts as a pivotal contributor to the development and progression of tumors. Increased CRP level is reported in colorectal, lung, and gastric cancer cases (50). The host interacts with tumor factors, and these interactions can accelerate tumor progression or regression. Lymphocyte-to-CRP ratio (LCR) has been used as a post-surgical prognostic biomarker in gastric and colorectal cancer (119). Studies have shown that CRP reduction is an early predictor of post-operative complications of gastric cancer and a reliable discharge indicator after gastric cancer (120, 121). Elevated preoperative CRP predicts increased post-operative morbidity in a patient with colorectal neoplasia (122). Anastomotic leakage is associated with higher CRP levels each post-operative day than no anastomotic leakage after colorectal surgery. The cut-off CRP values can be used to predict anastomotic leakage to expedite investigation and treatment (123).
Inflammation in the tumor microenvironment plays a vital role in cancer invasiveness, progression, and metastasis. Preoperative CRP levels help to diagnose differentiated thyroid carcinoma (50, 124). CRP has been reported to be consistently increased in the circulation of patients with body wasting associated with chronic diseases. In addition, CRP as an enhancer of in vitro IgG-mediated erythrocyte and tumor cell destruction (125). Simultaneously, CRP is a highly sensitive marker of inflammation to be considered in diagnosing cancer cachexia (126). Moreover, CRP levels are very responsive to lifestyle and several pathophysiological conditions. Thus, identifying cut-off values for CRP values is needed and should consider the heterogeneity of cancer patients’ clinical profiles.
CRP is associated with the binding and regulation of SLE associated nuclear antigens such as chromatin, histones, small nuclear ribonucleoprotein U1, and apoptotic cells (127). Several studies have shown delayed disease progression and increased survival in NZB/NZW mice carrying huCRP transgene, a protective effect associated with CRP’s ability to limit kidney damage by preventing immune complex deposition (128). SLE patient sera containing high levels of pCRP had an inhibitory effect on type I IFN induction compared with patient sera containing low levels of pCRP (58).
The CRP subunit is similar to pCRP and mCRP, but its monomeric structure exposes protein regions hidden in pentameric forms, including hidden protein neoepitopes (129, 130). Autoantibodies against these neoepitopes on mCRP have been demonstrated in SLE patients. The inflammatory environment can induce dissociation of pCRP into mCRP, which may be a way to limit PCRP-induced inflammation. In addition, mCRP has been shown to promote the removal of immune complexes (ICs) (131). Both mCRP and pCRP induced low levels of TNF and IL-1β without the need for immune complexes (ICs) (58). The genetic association between non-coding polymorphisms in CRP and human susceptibility to SLE has not been established. Autoantibodies against CRP are often present in patients with lupus nephritis (LN). Amino acid residues 35–47 constitutes the major epitope recognized by anti-CRP autoantibodies in patients with LN. Anti-a.a.35–47 autoantibodies are closely associated with renal prognosis, suggesting a pivotal role for mCRP in LN (132).
CRP is one of the most frequently tested molecules in clinical medicine. In daily practice, it is used for nonspecific initial diagnosis of viral or bacterial infections and also for monitoring the course of such infections under drug therapy (87). Severe coronavirus disease 2019 (COVID-19) is a public health emergency due to its high infectiousness (133) and high morbidity and mortality rate in critically ill patients. COVID-19 has been associated with inflammation in its induced neurological, cardiovascular, and other end-organ. It is indispensable to explore biomarkers to assess the extent of lung disease and the severity of COVID-19 (134–136). Plasma levels of CRP can be used for the early diagnosis of pneumonia (137), independent of patient age, gender, and physical condition, and correlate with inflammation levels (138), higher in patients with severe pneumonia. Thus, it is an important indicator for diagnosing and evaluating severe pulmonary infectious diseases (139). Some studies have shown that CRP levels in early COVID-19 are positively correlated with lung lesions, and CRP levels can reflect disease severity and should be used as a critical indicator for disease monitoring (140). Based on this, several studies have evaluated CRP levels in COVID-19 patients and their risk of death. Importantly, CRP plasma levels were generally significantly lower in viral infections than in bacterial infections (141). This is particularly important when studying diseases caused by COVID-19. Before COVID - 19 pandemic, as much as 90% of CRP were significantly elevated due to infectious etiology, is one of the most common bacterial pathogens (142). CRP levels are alarmingly high in COVID-19 patients with poor prognosis, despite their viral illness. Plasma concentrations as high as 400 mg/l are usually seen only in severe bacterial infections or sepsis, often in harmful COVID-19 pneumonia without superinfection (143).
Study shows that the magnitude of CRP upregulation exhibited by patients with the severe acute respiratory syndrome (SARS) was associated with their respiratory dysfunction and death (140, 144). Besides, CRP levels were also significantly elevated in patients with the Middle East respiratory syndrome (MERS) and H1N1 influenza (94, 145–147). Based on this, several studies have evaluated CRP levels in COVID-19 patients and their risk of death (148). One of the results showed that CRP levels were significantly higher in deceased patients than surviving patients (95).
Although elevated CRP blood level is associated with death caused by COVID-19, results are inconsistent across populations (149, 150). CRP is closely related to the grade of disease severity caused by the COVID-19, oxygenation rates, radiological evidence of ARDS, and level of respiratory support (134). CRP blood level rise of COVID-19 patients in their first 7 days of hospitalization can predict disease progression and serve as a basis for the need to transfer patients to the ICU at an early stage. The median value of CRP correlates with the severity of COVID-19 and is an independent predictor of mortality caused by COVID-19 (95). However, whether the determined CRP threshold can be used for early risk stratification of patients and to guide intensive management of respiratory support and corticosteroid immunosuppression still needs to be determined in future prospective studies.
At present, the animal models used to study the function of human CRP mainly rely on mice and rats, whose serum CRP baseline levels and inflammatory stress levels are very different from those of humans. The baseline CRP concentrations in mice was trace, ~7.5 μg/ml, which rise only twofold in the acute phase response (151), and have been reported to be a minor acute phase reactant in response to inflammatory stimuli (152). In healthy and pathogen-free rats, CRP ranges from 300–600 µg/ml (153). After injection of casein or croton oil, rising 3- to 4-fold in the acute phase response, it rose only up to about 900 µg/ml, indicating that it is a poor marker of acute inflammation. In humans, the median baseline CRP concentration is 0.8 µg/ml, and can rise to > 500 µg/ml at the peak of the acute phase response.
Nevertheless, the sequence homology of CRP among different species is relatively high, its structural characteristic may vary as indicated by its different in vivo function and the available structure information. Minor alteration in the pentameric assembly and subunit conformation of mouse, rat, and human CRP by electron microscopy visualization, single-particle analysis, and homology modeling (38). These findings suggest that various CRP may have different functions in different species, and therefore a possible functional role of CRP in humans cannot be reliably inferred from experimental animal studies.
Recent findings indicate that pCRP can be separated by platelets in areas of inflammation, leading to the deposition of mCRP. mCRP can exert local pro-inflammatory effects through a variety of mechanisms, which have been studied before. Therefore, the local formation of mCRP in the inflammatory area may represent the “activation signal” of other inflammatory cells, and mCRP may help stimulate the local inflammatory process.
However, the dissociation process of pCRP is still not fully understood, and more experimental models need to be designed to help develop and test potential therapeutic mCRP blockers or pCRP-dissociation blockers. Future research needs to further solve the problem of mCRP receptors and signal transduction. Conclusive animal data on the role of mCRP and the dissociation process of pCRP are still lacking, and it is necessary to establish animal models of acute and chronic inflammation (including AS) suitable for CRP research. This will enable the confirmation of existing in vitro data and therapeutic strategies for the dissociation process of mCRP or pCRP.
Circulating CRP concentration correlates with the severity, degree, and progression of many different pathologies and the prognostic significance of these correlations, which is consistent with CRP as a marker of disease and contributes to the pathogenesis. Understanding the structure and function of CRP, including its 3D structure and its complex with ligand can establish a good platform for drug design. In view of the problem that endogenous CRP in animal models cannot be ignored, the design of animal models using mice and rats as animal models to study the function of human CRP should be reviewed as soon as possible in the field, and the experimental design should be optimized to further clarify the role and function of CRP in diseases.
H-HZ: Conceptualization, Funding acquisition, Investigation, Writing – original draft. Y-LT: Data curation, Investigation, Visualization, Writing – original draft. T-HX: Data curation, Investigation, Visualization, Writing – review & editing. BC: Conceptualization, Data curation, Investigation, Validation, Visualization, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by grants from the National Natural Science Foundation of China (grant numbers: 31960141), Longyuan Youth Innovation and Entrepreneurship Talent Project (grant number: 2022LQGR76).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Shrive AK, Burns I, Chou H-T, Stahlberg H, Armstrong PB, Greenhough TJ. Crystal structures of limulus SAP-like pentraxin reveal two molecular aggregations. J Mol Biol. (2009) 386:1240–54. doi: 10.1016/j.jmb.2009.01.008
2. Shrive AK, Metcalfe AM, Cartwright JR, Greenhough TJ. C-reactive protein and SAP-like pentraxin are both present in limulus polyphemus haemolymph: crystal structure of limulus SAP 1. J Mol Biol. (1999) 290:997–1008. doi: 10.1006/jmbi.1999.2956
3. Garlanda C, Bottazzi B, Bastone A, Mantovani A. Pentraxins at the crossroads between innate immunity, inflammation, matrix deposition, and female fertility. Annu Rev Immunol. (2005) 23:337–66. doi: 10.1146/annurev.immunol.23.021704.115756
4. Volanakis J. Human C-reactive protein: expression, structure, and function. Mol Immunol. (2001) 38:189–97. doi: 10.1016/s0161–5890(01)00042–6
5. Martinez de la Torre Y, Fabbri M, Jaillon S, Bastone A, Nebuloni M, Vecchi A, et al. Evolution of the pentraxin family: the new entry PTX4. J Immunol. (2010) 184:5055–64. doi: 10.4049/jimmunol.0901672
6. Tillett WS, Francis T. Serological reactions in pneumonia with a non-protein somatic fraction of pneumococcus. J Exp Med. (1930) 52:561. doi: 10.1084/jem.52.4.561
8. Chu W, Burns DK, Swerlick RA, Presky DH. Identification and characterization of a novel cytokine-inducible nuclear protein from human endothelial cells. J Biol Chem. (1995) 270:10236–45. doi: 10.1074/jbc.270.17.10236
9. Pepys MB, Hirschfield GM. C-reactive protein: A critical update. J Clin Invest. (2003) 111:1805–12. doi: 10.1172/jci200318921
10. Breviario F, D’Aniello EM, Golay J, Peri G, Bottazzi B, Bairoch A, et al. Interleukin-1-inducible genes in endothelial cells. Cloning of a new gene related to C-reactive protein and serum amyloid P component. J Biol Chem. (1992) 267:22190–7. doi: 10.1016/s0021–9258(18)41653–5
11. Rubio N, Sharp PM, Rits M, Zahedi K, Whitehead AS. Structure, expression, and evolution of Guinea pig serum amyloid P component and C-reactive protein. J Biochem. (1993) 113:277–84. doi: 10.1093/oxfordjournals.jbchem.a124039
12. Xu D, Hopf C, Reddy R, Cho RW, Guo L, Lanahan A, et al. Narp and np1 form heterocomplexes that function in developmental and activity-dependent synaptic plasticity. Neuron. (2003) 39:513–28. doi: 10.1016/S0896-6273(03)00463-X
13. Deban L, Bottazzi B, Garlanda C, de la Torre YM, Mantovani A. Pentraxins: multifunctional proteins at the interface of innate immunity and inflammation. BioFactors. (2009) 35:138–45. doi: 10.1002/biof.21
14. Rajab IM, Majerczyk D, Olson ME, Addams JMB, Choe ML, Nelson MS, et al. C-reactive protein in gallbladder diseases: diagnostic and therapeutic insights. Biophys Rep. (2020) 6:49–67. doi: 10.1007/s41048–020-00108–9
15. Wang Q, Shi Y. Photoacoustic viscoelasticity imaging for the detection of acute hepatitis: A feasibility study. Biophys Rep. (2020) 6:1–8. doi: 10.1007/s41048-020-00104-z
16. Jimenez RV, Wright TT, Jones NR, Wu J, Gibson AW, Szalai AJ. C-reactive protein impairs dendritic cell development, maturation, and function: implications for peripheral tolerance. Front Immunol. (2018) 9:372. doi: 10.3389/fimmu.2018.00372
17. Cottin V, Valenzuela C. C-reactive protein as a candidate biomarker in fibrotic interstitial lung disease. Respirology. (2024) 29:195–8. doi: 10.1111/resp.14666
18. VanDevanter DR, Heltshe SL, Skalland M, West NE, Sanders DB, Goss CH, et al. C-reactive protein (CRP) as a biomarker of pulmonary exacerbation presentation and treatment response. J Cyst Fibros. (2022) 21:588–93. doi: 10.1016/j.jcf.2021.12.003
19. Ji SR, Zhang SH, Chang Y, Li HY, Wang MY, Lv JM, et al. C-reactive protein: the most familiar stranger. J Immunol. (2023) 210:699–707. doi: 10.4049/jimmunol.2200831
20. Pathak A, Agrawal A. Evolution of C-reactive protein. Front Immunol. (2019) 10:943. doi: 10.3389/fimmu.2019.00943
21. Zhang SC, Wang MY, Feng JR, Chang Y, Ji SR, Wu Y. Reversible promoter methylation determines fluctuating expression of acute phase proteins. eLife. (2020) 9:e51317. doi: 10.7554/eLife.51317
22. Plebani M. Why C-reactive protein is one of the most requested tests in clinical laboratories? Clin Chem Lab Med (CCLM). (2023) 61:1540–5. doi: 10.1515/cclm-2023–0086
23. Ngwa DN, Agrawal A. Structurally altered, not wild-type, pentameric C-reactive protein inhibits formation of amyloid-B Fibrils. J Immunol. (2022) 209:1180–8. doi: 10.4049/jimmunol.2200148
24. Jimenez RV, Kuznetsova V, Connelly AN, Hel Z, Szalai AJ. C-reactive protein promotes the expansion of myeloid derived cells with suppressor functions. Front Immunol. (2019) 10:2183. doi: 10.3389/fimmu.2019.02183
25. Noone DP, Isendoorn MME, Hamers SMWR, Keizer ME, Wulffelé J, van der Velden TT, et al. Structural basis for surface activation of the classical complement cascade by the short pentraxin C-reactive protein. bioRxiv [Preprint]. (2024), 585147. doi: 10.1101/2024.03.18.585147
26. Ramadan MAM, Shrive AK, Holden D, Myles DAA, Volanakis JE, DeLucas LJ, et al. The three-dimensional structure of calcium-depleted human C-reactive protein from perfectly twinned crystals. Acta Crystallograph Section D Biol Crystallogr. (2002) 58:992–1001. doi: 10.1107/s0907444902005693
27. Meng EC, Goddard TD, Pettersen EF, Couch GS, Pearson ZJ, Morris JH, et al. Ucsf chimerax: tools for structure building and analysis. Protein Sci. (2023) 32:e4792. doi: 10.1002/pro.4792
28. Abramson J, Adler J, Dunger J, Evans R, Green T, Pritzel A, et al. Accurate structure prediction of biomolecular interactions with alphafold 3. Nature. (2024). doi: 10.1038/s41586-024-07487-w
29. Lv JM, Lü SQ, Liu ZP, Zhang J, Gao BX, Yao ZY, et al. Conformational folding and disulfide bonding drive distinct stages of protein structure formation. Sci Rep. (2018) 8:1494. doi: 10.1038/s41598-018-20014-y
30. Lee RT, Takagahara I, Lee YC. Mapping the binding areas of human C-reactive protein for phosphorylcholine and polycationic compounds. J Biol Chem. (2002) 277:225–32. doi: 10.1074/jbc.M106039200
31. Ngwa DN, Singh SK, Agrawal A. C-reactive protein-based strategy to reduce antibiotic dosing for the treatment of pneumococcal infection. Front Immunol. (2021) 11:620784. doi: 10.3389/fimmu.2020.620784
32. Pathak A, Singh SK, Thirumalai A, Armstrong PB, Agrawal A. Evolution of host-defense function of C-reactive protein from horseshoe crab to humans. J Immunol. (2016) 196:132.5–5. doi: 10.4049/jimmunol.196.Supp.132.5
33. Bodmer B, Siboo R. Isolation of mouse C-reactive protein from liver and serum. J Immunol. (1977) 118:1086–9. doi: 10.4049/jimmunol.118.3.1086
34. Yamamoto S, Tagata K, Nagahata H, Ishikawa Y, Morimatsu M, Naiki M. Isolation of canine C-reactive protein and characterization of its properties. Veterinary Immunol Immunopathol. (1992) 30:329–39. doi: 10.1016/0165-2427(92)90103-W
35. Cerón JJ, Eckersall PD, Martínez SS. Acute phase proteins in dogs and cats: current knowledge and future perspectives. Vet Clin Pathol. (2005) 34:85–99. doi: 10.1111/j.1939-165X.2005.tb00019.x
36. Sarikaputi M, Morimatsu M, Syuto B, Saito M, Naiki M. A new purification procedure for bovine C-reactive protein and serum amyloid P component. Int J Biochem. (1991) 23:1137–42. doi: 10.1016/0020-711X(91)90155-G
37. Cheng B, Wu D, Wu K, Huang XP, Lv JM, Ji SR, et al. Purification of recombinant mouse C-reactive protein from pichia pastorisgs 115 by nickel chelating sepharose fast-flow affinity chromatography and p-aminophenyl phosphoryl choline agarose resin affinity chromatography in tandem. J Chromatograph Sci. (2021) 60:750–9. doi: 10.1093/chromsci/bmab121
38. Cheng B, Lv JM, Liang YL, Zhu L, Huang XP, Li HY, et al. Secretory quality control constrains functional selection-associated protein structure innovation. Commun Biol. (2022) 5:268. doi: 10.1038/s42003–022-03220–3
39. Dortay H, Schmöckel SM, Fettke J, Mueller-Roeber B. Expression of human C-reactive protein in different systems and its purification from leishmania tarentolae. Protein Expression Purificat. (2011) 78:55–60. doi: 10.1016/j.pep.2011.03.010
40. Thirumalai A, Singh SK, Hammond DJ, Gang TB, Ngwa DN, Pathak A, et al. Purification of recombinant C-reactive protein mutants. J Immunol Methods. (2017) 443:26–32. doi: 10.1016/j.jim.2017.01.011
41. Ngwa DN, Pathak A, Agrawal A. Il-6 regulates induction of C-reactive protein gene expression by activating stat3 isoforms. Mol Immunol. (2022) 146:50–6. doi: 10.1016/j.molimm.2022.04.003
42. Du Clos TW. Pentraxins: structure, function, and role in inflammation. ISRN Inflammation. (2013) 2013:1–22. doi: 10.1155/2013/379040
43. Shrive AK, Gheetham GMT, Holden D, Myles DAA, Turnell WG, Volanakis JE, et al. Three dimensional structure of human C-reactive protein. Nat Struct Mol Biol. (1996) 3:346–54. doi: 10.1038/nsb0496–346
44. Thompson D, Pepys MB, Wood SP. The physiological structure of human C-reactive protein and its complex with phosphocholine. Structure. (1999) 7:169–77. doi: 10.1016/s0969–2126(99)80023–9
45. Black S, Kushner I, Samols D. C-reactive protein. J Biol Chem. (2004) 279:48487–90. doi: 10.1074/jbc.R400025200
46. Peng YN, Ho YL, Wu CY, Liu MY. Investigation of C-reactive protein binding to phosphatidyl choline by CZE and ESI-mass analysis. Electrophoresis. (2009) 30:1564–71. doi: 10.1002/elps.200800608
47. Guillon C, Bigouagou U, Folio C, Jeannin P, Delneste Y, Gouet P. A staggered decameric assembly of human C-reactive protein stabilized by zinc ions revealed by X-ray crystallography. Protein Pept Lett. (2015) 22:248–55. doi: 10.2174/0929866522666141231111226
48. Noone DP, van der Velden TT, Sharp TH. Cryo-Electron Microscopy and Biochemical Analysis Offer Insights into the Effects of Acidic pH, Such as Occur During Acidosis, on the Complement Binding Properties of C-Reactive Protein. Front Immunol. (2021) 12:757633. doi: 10.3389/fimmu.2021.757633
49. Hammond DJ, Singh SK, Thompson JA, Beeler BW, Rusiñol AE, Pangburn MK, et al. Identification of acidic pH-dependent ligands of pentameric C-reactive protein. J Biol Chem. (2010) 285:36235–44. doi: 10.1074/jbc.M110.142026
50. Potempa LA, Rajab IM, Olson ME, Hart PC. C-reactive protein and cancer: interpreting the differential bioactivities of its pentameric and monomeric, modified isoforms. Front Immunol. (2021) 12:744129. doi: 10.3389/fimmu.2021.744129
51. Rajab IM, Hart PC, Potempa LA. How C-reactive protein structural isoforms with distinctive bioactivities affect disease progression. Front Immunol. (2020) 11:2126. doi: 10.3389/fimmu.2020.02126
52. Pathak A, Singh SK, Thewke DP, Agrawal A. Conformationally altered C-reactive protein capable of binding to atherogenic lipoproteins reduces atherosclerosis. Front Immunol. (2020) 11:1780. doi: 10.3389/fimmu.2020.01780
53. McFadyen JD, Kiefer J, Braig D, Loseff-Silver J, Potempa LA, Eisenhardt SU, et al. Dissociation of C-reactive protein localizes and amplifies inflammation: evidence for a direct biological role of C-reactive protein and its conformational changes. Front Immunol. (2018) 9:1351. doi: 10.3389/fimmu.2018.01351
54. Wu Y, Potempa LA, El Kebir D, Filep JG. C-reactive protein and inflammation: conformational changes affect function. Biol Chem. (2015) 396:1181–97. doi: 10.1515/hsz-2015–0149
55. Yao Z, Zhang Y, Wu H. Regulation of C-reactive protein conformation in inflammation. Inflammation Res. (2019) 68:815–23. doi: 10.1007/s00011–019-01269–1
56. Okemefuna AI, Stach L, Rana S, Ziai Buetas AJ, Gor J, Perkins SJ. C-reactive protein exists in an naCl concentration-dependent pentamer-decamer equilibrium in physiological buffer. J Biol Chem. (2010) 285:1041–52. doi: 10.1074/jbc.M109.044495
57. Agrawal A, Volanakis JE. Probing the C1q-binding site on human C-reactive protein by site-directed mutagenesis. J Immunol. (1994) 152:5404–10. doi: 10.4049/jimmunol.152.11.5404
58. Svanberg C, Enocsson H, Govender M, Martinsson K, Potempa LA, Rajab IM, et al. Conformational state of C-reactive protein is critical for reducing immune complex-triggered type I interferon response: implications for pathogenic mechanisms in autoimmune diseases imprinted by type I interferon gene dysregulation. J Autoimmun. (2023) 135:102998. doi: 10.1016/j.jaut.2023.102998
59. Crawford JR, Trial J, Nambi V, Hoogeveen RC, Taffet GE, Entman ML. Plasma levels of endothelial microparticles bearing monomeric C-reactive protein are increased in peripheral artery disease. J Cardiovasc Trans Res. (2016) 9:184–93. doi: 10.1007/s12265–016-9678–0
60. Khreiss T, József L, Potempa LA, Filep JG. Loss of pentameric symmetry in C-reactive protein induces interleukin-8 secretion through peroxynitrite signaling in human neutrophils. Circ Res. (2005) 97:690–7. doi: 10.1161/01.RES.0000183881.11739.CB
61. Khreiss T, József L, Potempa LA, Filep JG. Conformational rearrangement in C-reactive protein is required for proinflammatory actions on human endothelial cells. Circulation. (2004) 109:2016–22. doi: 10.1161/01.CIR.0000125527.41598.68
62. Zhang Z, Na H, Gan Q, Tao Q, Alekseyev Y, Hu J, et al. Monomeric C-reactive protein via endothelial CD31 for neurovascular inflammation in an apoE genotype-dependent pattern: A risk factor for alzheimer’s disease? Aging Cell. (2021) 20:e13501. doi: 10.1111/acel.13501
63. Jimenez RV, Szalai AJ. Therapeutic lowering of C-reactive protein. Front Immunol. (2021) 11:619564. doi: 10.3389/fimmu.2020.619564
64. Khreiss T, József L, Hossain S, Chan JS, Potempa LA, Filep JG. Loss of pentameric symmetry of C-reactive protein is associated with delayed apoptosis of human neutrophils. J Biol Chem. (2002) 277:40775–81. doi: 10.1074/jbc.M205378200
65. Hornick MG, Potempa LA. Monomeric C-reactive protein as a biomarker for major depressive disorder. Front Psychiatry. (2024) 14:1325220. doi: 10.3389/fpsyt.2023.1325220
66. Sheriff A, Kayser S, Brunner P, Vogt B. C-reactive protein triggers cell death in ischemic cells. Front Immunol. (2021) 12:630430. doi: 10.3389/fimmu.2021.630430
67. Wang MY, Zhang CM, Zhou HH, Ge ZB, Su CC, Lou ZH, et al. Identification of a distal enhancer that determines the expression pattern of acute phase marker C-reactive protein. J Biol Chem. (2022) 298:102160. doi: 10.1016/j.jbc.2022.102160
68. Zeng G, Chen D, Zhou R, Zhao X, Ye C, Tao H, et al. Combination of C-reactive protein, procalcitonin, IL-6, IL-8, and IL-10 for early diagnosis of hyperinflammatory state and organ dysfunction in pediatric sepsis. J Clin Lab Anal. (2022) 36:e24505. doi: 10.1002/jcla.24505
69. Pandey A. C-reactive protein (CRP) and its association with periodontal disease: A brief review. J Clin Diagn Res. (2014) 8:ZE21–24. doi: 10.7860/jcdr/2014/8355.4646
70. Ridker PM. C-reactive protein: eighty years from discovery to emergence as a major risk marker for cardiovascular disease. Clin Chem. (2009) 55:209–15. doi: 10.1373/clinchem.2008.119214
71. Peisajovich A, Marnell L, Mold C, Du CT. C-reactive protein at the interface between innate immunity and inflammation. Expert Rev Clin Immunol. (2008) 4:379. doi: 10.1586/1744666X.4.3.379
72. Singh SK, Prislovsky A, Ngwa DN, Munkhsaikhan U, Abidi AH, Brand DD, et al. C-reactive protein lowers the serum level of IL-17, but not TNF-α, and decreases the incidence of collagen-induced arthritis in mice. Front Immunol. (2024) 15:1385085. doi: 10.3389/fimmu.2024.1385085
73. Wang MY, Zhou HH, Zhang SC, Hui F, Zhu W, Su HX, et al. Recurrent mutations at C-reactive protein gene promoter SNP position –286 in human cancers. Cell Res. (2014) 24:505–8. doi: 10.1038/cr.2014.7
74. Jia ZK, Li HY, Liang YL, Potempa LA, Ji SR, Wu Y. Corrigendum: monomeric C-reactive protein binds and neutralizes receptor activator of NF-κb ligand-induced osteoclast differentiation. Front Immunol. (2020) 11:619847. doi: 10.3389/fimmu.2020.619847
75. Voleti B. Mechanism of transcriptional regulation of C-reactive protein gene expression. [electronic theses and dissertations]. (2007). p. 2058. Available at: https://dc.etsu.edu/etd/2058.
76. Kadokura H, Dazai Y, Fukuda Y, Hirai N, Nakamura O, Inaba K. Observing the nonvectorial yet cotranslational folding of a multidomain protein, LDL receptor, in the ER of mammalian cells. Proc Natl Acad Sci. (2020) 117:16401–8. doi: 10.1073/pnas.2004606117
77. Kim Y, Noren HN, Dluzen DF, Martindale JL, Gorospe M, Evans MK. Post-transcriptional regulation of the inflammatory marker C-reactive protein by the RNA-binding protein huR and miR-637. Mol Cell Biol. (2015) 35:4212–21. doi: 10.1128/MCB.00645–15
78. Horvath S, Raj K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat Rev Genet. (2018) 19:371–84. doi: 10.1038/s41576–018-0004–3
79. Li HY, Liu XL, Liu YT, Jia ZK, Filep JG, Potempa LA, et al. Matrix sieving-enforced retrograde transcytosis regulates tissue accumulation of C-reactive protein. Cardiovasc Res. (2019) 115:440–52. doi: 10.1093/cvr/cvy181
80. Dix C, Zeller J, Stevens H, Eisenhardt SU, Shing KSCT, Nero TL, et al. C-reactive protein, immunothrombosis and venous thromboembolism. Front Immunol. (2022) 13:1002652. doi: 10.3389/fimmu.2022.1002652
81. Singh SK, Ngwa DN, Agrawal A. Complement activation by C-reactive protein is critical for protection of mice against pneumococcal infection. Front Immunol. (2020) 11:1812. doi: 10.3389/fimmu.2020.01812
82. Ngwa DN, Singh SK, Gang TB, Agrawal A. Treatment of pneumococcal infection by using engineered human C-reactive protein in a mouse model. Front Immunol. (2020) 11:586669. doi: 10.3389/fimmu.2020.586669
83. Zhang L, Li W, Gong M, Zhang Z, Xue X, Mao J, et al. C-reactive protein inhibits C3a/C3aR-dependent podocyte autophagy in favor of diabetic kidney disease. FASEB J. (2022) 36:e22332. doi: 10.1096/fj.202200198R
84. Torzewski M, Waqar AB, Fan J. Animal models of C-reactive protein. Mediators Inflammation. (2014) 2014:1–7. doi: 10.1155/2014/683598
85. Mold C, Gewurz H, Du Clos TW. Regulation of complement activation by C-reactive protein. Immunopharmacology. (1999) 42:23–30. doi: 10.1016/s0162–3109(99)00007–7
86. Li HY, Tang ZM, Wang Z, Lv JM, Liu XL, Liang YL, et al. C-reactive protein protects against acetaminophen-induced liver injury by preventing complement overactivation. Cell Mol Gastroenterol Hepatol. (2022) 13:289–307. doi: 10.1016/j.jcmgh.2021.09.003
87. Torzewski J, Brunner P, Ries W, Garlichs CD, Kayser S, Heigl F, et al. Targeting C-reactive protein by selective apheresis in humans: pros and cons. J Clin Med. (2022) 11:1771. doi: 10.3390/jcm11071771
88. Gan Q, Wong A, Zhang Z, Na H, Tian H, Tao Q, et al. Monomeric C-reactive protein induces the cellular pathology of alzheimer’s disease. Alzheimer’s Dementia: Trans Res Clin Interventions. (2022) 8:e12319. doi: 10.1002/trc2.12319
89. Bienstock S, Lee SE, Lin FY, Blankstein R, Leipsic J, Patel K, et al. Systemic inflammation with highsensitivity C-reactive protein and atherosclerotic plaque progression. JACC-Cardiovasc Imag. (2024) 17:212–3. doi: 10.1016/j.jcmg.2023.08.019of
90. Singh SK, Agrawal A. Functionality of C-reactive protein for atheroprotection. Front Immunol. (2019) 10:1655. doi: 10.3389/fimmu.2019.01655
91. Pathak A, Singh SK, Agrawal A. C-reactive protein is an atheroprotective molecule. J Immunol. (2018) 200:170.116–170.116. doi: 10.4049/jimmunol.200.Supp.170.16
92. Gheita TA, El-Gazzar II, Azkalany G, El-Fishawy HS, El-Faramawy A. High-sensitivity C-reactive protein (Hs-CRP) in systemic lupus erythematosus patients without cardiac involvement; relation to disease activity, damage and intima-media thickness. Egyptian Rheumatologist. (2012) 34:147–52. doi: 10.1016/j.ejr.2012.08.002
93. Candido J, Hagemann T. Cancer-related inflammation. J Clin Immunol. (2012) 33:79–84. doi: 10.1007/s10875–012-9847–0
94. Ruan Q, Yang K, Wang W, Jiang L, Song J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from wuhan, China. Intensive Care Med. (2020) 46:846–8. doi: 10.1007/s00134-020-05991-x
95. Loukides S, Sharifpour M, Rangaraju S, Liu M, Alabyad D, Nahab FB, et al. C-reactive protein as a prognostic indicator in hospitalized patients with COVID-19. PloS One. (2020) 15(11):E0242400-E0242400. doi: 10.1371/journal.pone.0242400
96. Zheng Z, Peng F, Xu B, Zhao J, Liu H, Peng J, et al. Risk factors of critical & Mortal COVID-19 cases: A systematic literature review and meta-analysis. J Infection. (2020) 81:e16–25. doi: 10.1016/j.jinf.2020.04.021
97. Li HY, Wang JW, Xu LW, Zhao XL, Feng JX, Xu YZ. Clinical analysis of 132 cases COVID-19 from wuhan. Medicine. (2020) 99:e22847. doi: 10.1097/md.0000000000022847
98. Alberts A, Klingberg A, Wessig AK, Combes C, Witte T, Brand K, et al. C-reactive protein (CRP) recognizes uric acid crystals and recruits proteases C1 and masp1. Sci Rep. (2020) 10:6391. doi: 10.1038/s41598–020-63318–8
99. Pepys MB, Gallimore JR, Lloyd J, Li Z, Graham D, Taylor GW, et al. Isolation and characterization of pharmaceutical grade human pentraxins, serum amyloid P component and C-reactive protein, for clinical use. J Immunol Methods. (2012) 384:92–102. doi: 10.1016/j.jim.2012.07.013
100. Pepys MB. C-reactive protein predicts outcome in COVID-19: is it also a therapeutic target? Eur Heart J. (2021) 42:2280–3. doi: 10.1093/eurheartj/ehab169
101. Magrini E, Mantovani A, Garlanda C. The dual complexity of ptx3 in health and disease: A balancing act? Trends Mol Med. (2016) 22:497–510. doi: 10.1016/j.molmed.2016.04.007
102. Suresh MV, Singh SK, Ferguson DA, Agrawal A. Role of the property of C-reactive protein to activate the classical pathway of complement in protecting mice from pneumococcal infection. J Immunol. (2006) 176:4369–74. doi: 10.4049/jimmunol.176.7.4369
103. Agrawal A, Suresh M, Singh S, Ferguson D. The protective function of human C-reactive protein in mouse models of streptococcus pneumoniae infection. Endocrine, Metab Immune Disorders-Drug Targets. (2008) 8:231–7. doi: 10.2174/187153008786848321
104. Ngwa DN, Agrawal A. Structure-function relationships of C-reactive protein in bacterial infection. Front Immunol. (2019) 10:166. doi: 10.3389/fimmu.2019.00166
105. Gang TB, Hanley GA, Agrawal A. C-reactive protein protects mice against pneumococcal infection via both phosphocholine-dependent and phosphocholine-independent mechanisms. Infect Immun. (2015) 83:1845–52. doi: 10.1128/IAI.03058–14
106. Yother J, Volanakis JE, Briles DE. Human C-reactive protein is protective against fatal streptococcus pneumoniae infection in mice. J Immunol. (1982) 128:2374–6. doi: 10.4049/jimmunol.155.5.2557
107. Lysenko E, Richards JC, Cox AD, Stewart A, Martin A, Kapoor M, et al. The position of phosphorylcholine on the lipopolysaccharide of haemophilus influenzae affects binding and sensitivity to C-reactive protein-mediated killing. Mol Microbiol. (2000) 35:234–45. doi: 10.1046/j.1365-2958.2000.01707.x
108. Szalai AJ, Vancott JL, Mcghee JR, Volanakis JE. Human C-reactive protein is protective against fatal salmonella enterica serovar typhimurium infection in transgenic mice. J Immunol. (1995) 155:2557–63. doi: 10.1128/IAI.68.10.5652–5656.2000
109. Isaacs AW, Macaluso F, Smith C, Myburgh KH. C-reactive protein is elevated only in high creatine kinase responders to muscle damaging exercise. Front Physiol. (2019) 10:86. doi: 10.3389/fphys.2019.00086
110. Franklin ME, Chamness MS, Smith LL, Chenier TC, Sizemore CS, Rogers M, et al. Effects of isokinetic soreness-inducing exercise on blood levels of C-reactive protein and creatine kinase. J Orthop Sports Phys Ther. (1992) 16:208–14. doi: 10.2519/jospt.1992.16.5.208
111. Ridker PM, Rifai N, Rose L, Buring JE, Cook NR. Comparison of C-reactive protein and low-density lipoprotein cholesterol levels in the prediction of first cardiovascular events. New Engl J Med. (2002) 347:1557–65. doi: 10.1056/NEJMoa021993
112. Eisenhardt SU, Habersberger J, Murphy A, Chen YC, Woollard KJ, Bassler N, et al. Dissociation of pentameric to monomeric C-reactive protein on activated platelets localizes inflammation to atherosclerotic plaques. Circ Res. (2009) 105:128–37. doi: 10.1161/circresaha.108.190611
113. Ridker PM, Danielson E, Fonseca FAH, Genest J, Gotto AM, Kastelein JJP, et al. Rosuvastatin to prevent vascular events in men and women with elevated C-reactive protein. New Engl J Med. (2008) 359:2195–207. doi: 10.1056/NEJMoa0807646
114. Jialal I, Devaraj S, Venugopal SK. C-reactive protein: risk marker or mediator in atherothrombosis? Hypertension. (2004) 44:6–11. doi: 10.1161/01.HYP.0000130484.20501.df
115. Ueland T, Gullestad L, Nymo SH, Yndestad A, Aukrust P, Askevold ET. Inflammatory cytokines as biomarkers in heart failure. Clinica Chimica Acta; Int J Clin Chem. (2015) 443:71–7. doi: 10.1016/j.cca.2014.09.001
116. Pilely K, Fumagalli S, Rosbjerg A, Genster N, Skjoedt M-O, Perego C, et al. C-reactive protein binds to cholesterol crystals and co-localizes with the terminal complement complex in human atherosclerotic plaques. Front Immunol. (2017) 8:1040. doi: 10.3389/fimmu.2017.01040
117. Motevalli SM, Eltahan AS, Liu L, Magrini A, Rosato N, Guo W, et al. Co-encapsulation of curcumin and doxorubicin in albumin nanoparticles blocks the adaptive treatment tolerance of cancer cells. Biophys Rep. (2019) 5:19–30. doi: 10.1007/s41048–018-0079–6
118. Hart PC, Rajab IM, Alebraheem M, Potempa LA. C-reactive protein and cancer-diagnostic and therapeutic insights. Front Immunol. (2020) 11:595835. doi: 10.3389/fimmu.2020.595835
119. Yildirim M, Koca B. Lymphocyte C-reactive protein ratio: A new biomarker to predict early complications after gastrointestinal oncologic surgery. Cancer biomark. (2021) 31:409–17. doi: 10.3233/cbm-210251
120. Lee SH, Kim KH, Choi CW, Kim SJ, Kim DH, Choi CI, et al. Reduction rate of C-reactive protein as an early predictor of postoperative complications and a reliable discharge indicator after gastrectomy for gastric cancer. Ann Surg Treat Res. (2019) 97:65–73. doi: 10.4174/astr.2019.97.2.65
121. Yu Q, Li KZ, Fu YJ, Tang Y, Liang XQ, Liang ZQ, et al. Clinical significance and prognostic value of C-reactive protein/albumin ratio in gastric cancer. Ann Surg Treat Res. (2021) 100:338–46. doi: 10.4174/astr.2021.100.6.338
122. Alsaif SH, Rogers AC, Pua P, Casey PT, Aherne GG, Brannigan AE, et al. Preoperative C-reactive protein and other inflammatory markers as predictors of postoperative complications in patients with colorectal neoplasia. World J Surg Oncol. (2021) 19:74. doi: 10.1186/s12957–021-02142–4
123. Yeung DE, Peterknecht E, Hajibandeh S, Hajibandeh S, Torrance AW. C-reactive protein can predict anastomotic leak in colorectal surgery: A systematic review and meta-analysis. Int J Colorectal Dis. (2021) 36:1147–62. doi: 10.1007/s00384–021-03854–5
124. Zhang X, Li S, Wang J, Liu F, Zhao Y. Relationship between serum inflammatory factor levels and differentiated thyroid carcinoma. Technol Cancer Res Treat. (2021) 20:1533033821990055. doi: 10.1177/1533033821990055
125. Temming AR, Tammes Buirs M, Bentlage AEH, Treffers LW, Feringa H, de Taeye SW, et al. C-reactive protein enhances igg-mediated cellular destruction through igG-fc receptors in vitro. Front Immunol. (2021) 12:594773. doi: 10.3389/fimmu.2021.594773
126. Tavares P, Gonçalves DM, Santos LL, Ferreira R. Revisiting the clinical usefulness of C-reactive protein in the set of cancer cachexia. Porto Biomed J. (2021) 6:e123. doi: 10.1097/j.pbj.0000000000000123
127. Ballarano CA, Frishman WH. Cardiovascular disease in patients with systemic lupus erythematosus: potential for improved primary prevention with statins. Cardiol Rev. (2021) 29:323–7. doi: 10.1097/CRD.0000000000000383
128. Enocsson H, Karlsson J, Li HY, Wu Y, Kushner I, Wetterö J, et al. The complex role of C-reactive protein in systemic lupus erythematosus. J Clin Med. (2021) 10:5837. doi: 10.3390/jcm10245837
129. Ji SR, Wu Y, Zhu L, Potempa LA, Sheng FL, Lu W, et al. Cell membranes and liposomes dissociate C-reactive protein (CRP) to form a new, biologically active structural intermediate: mCRPm. FASEB J. (2006) 21:284–94. doi: 10.1096/fj.06-6722com
130. Bell SA, Du Clos TW, Khursigara G, Picazo JJ, Rubin RL. Autoantibodies to cryptic epitopes of C-reactive protein and other acute phase proteins in the toxic oil syndrome. J Autoimmun. (1995) 8:293–303. doi: 10.1006/jaut.1995.0022
131. Motie M, Brockmeier S, Potempa LA. Binding of model soluble immune complexes to modified C-reactive protein. J Immunol. (1996) 156:4435–41. doi: 10.4049/jimmunol.156.11.4435
132. Li QY, Li HY, Fu G, Yu F, Wu Y, Zhao MH. Autoantibodies against C-reactive protein influence complement activation and clinical course in lupus nephritis. J Am Soc Nephrol. (2017) 28:3044–54. doi: 10.1681/asn.2016070735
133. Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, et al. Clinical characteristics of coronavirus disease 2019 in China. New Engl J Med. (2020) 382:1708–20. doi: 10.1056/NEJMoa2002032
134. Keddie S, Ziff O, Chou MKL, Taylor RL, Heslegrave A, Garr E, et al. Laboratory biomarkers associated with COVID-19 severity and management. Clin Immunol. (2020) 221:108614. doi: 10.1016/j.clim.2020.108614
135. Tahery N, Khodadost M, Jahani SS, Rezaei TM, Ahmadi N, Montazer F, et al. C-reactive protein as a possible marker for severity and mortality of COVID-19 infection. Gastroenterol Hepatol Bed Bench. (2021) 14:S118–22. doi: 10.22037/ghfbb.vi.2388
136. Singh K, Mittal S, Gollapudi S, Butzmann A, Kumar J, Ohgami RS. A meta-analysis of SARS-cov-2 patients identifies the combinatorial significance of D-dimer, C-reactive protein, lymphocyte, and neutrophil values as a predictor of disease severity. Int J Lab Hematol. (2020) 43:324–8. doi: 10.1111/ijlh.13354
137. Zhong Y, Wu G, Wu C, Zhang S, Li T, Li Y, et al. C-reactive protein level may predict the risk of COVID-19 aggravation. Open Forum Infect Dis. (2020) 7:ofaa153. doi: 10.1093/ofid/ofaa153
138. Gándara E, Warusevitane A, Karunatilake D, Sim J, Smith C, Roffe C. Early diagnosis of pneumonia in severe stroke: clinical features and the diagnostic role of C-reactive protein. PloS One. (2016) 11:e0150269. doi: 10.1371/journal.pone.0150269
139. Muñoz-Bermúdez R, Abella E, Zuccarino F, Masclans JR, Nolla-Salas J. Successfully non-surgical management of flail chest as first manifestation of multiple myeloma: A case report. World J Crit Care Med. (2019) 8:59–63. doi: 10.5492/wjccm.v8.i5.59
140. Wang L. C-reactive protein levels in the early stage of COVID-19. Med Mal Infect. (2020) 50:332–4. doi: 10.1016/j.medmal.2020.03.007
141. Torzewski J, Torzewski M, Bowyer DE, Froühlich M, Koenig W, Waltenberger J, et al. C-reactive protein frequently colocalizes with the terminal complement complex in the intima of early atherosclerotic lesions of human coronary arteries. Arteriosclerosis Thrombosis Vasc Biol. (1998) 18:1386–92. doi: 10.1161/01.Atv.18.9.1386
142. Landry A, Docherty P, Ouellette S, Cartier LJ. Causes and outcomes of markedly elevated C-reactive protein levels. Can Fam Physician. (2017) 63:e316–23.
143. Smilowitz NR, Kunichoff D, Garshick M, Shah B, Pillinger M, Hochman JS, et al. C-reactive protein and clinical outcomes in patients with COVID-19. Eur Heart J. (2021) 42:2270–9. doi: 10.1093/eurheartj/ehaa1103
144. Cherubini F, Cristiano A, Valentini A, Bernardini S, Nuccetelli M. Circulating calprotectin as a supporting inflammatory marker in discriminating SARS-cov-2 infection: an observational study. Inflammation Res. (2021) 70:687–94. doi: 10.1007/s00011-021-01465-y
145. Roxby AC, Greninger AL, Hatfield KM, Lynch JB, Dellit TH, James A, et al. Outbreak investigation of COVID-19 among residents and staff of an independent and assisted living community for older adults in seattle, washington. JAMA Internal Med. (2020) 180:1101–5. doi: 10.1001/jamainternmed.2020.2233
146. Sproston NR, Ashworth JJ. Role of C-reactive protein at sites of inflammation and infection. Front Immunol. (2018) 9:754. doi: 10.3389/fimmu.2018.00754
147. Vasileva D, Badawi A. C-reactive protein as a biomarker of severe H1N1 influenza. Inflammation Res. (2018) 68:39–46. doi: 10.1007/s00011–018-1188-x
148. Dai Z, Zeng D, Cui D, Wang D, Feng Y, Shi Y, et al. Prediction of COVID-19 patients at high risk of progression to severe disease. Front Public Health. (2020) 8:574915. doi: 10.3389/fpubh.2020.574915
149. Sahu BR, Kampa RK, Padhi A, Panda AK. C-reactive protein: A promising biomarker for poor prognosis in COVID-19 infection. Clin Chim Acta. (2020) 509:91–4. doi: 10.1016/j.cca.2020.06.013
150. Amezcua-Guerra LM, Audelo K, Guzmán J, Santiago D, González-Flores J, García-Ávila C, et al. A simple and readily available inflammation-based risk scoring system on admission predicts the need for mechanical ventilation in patients with COVID-19. Inflammation Res. (2021) 70:731–42. doi: 10.1007/s00011-021-01466-x
151. Teupser D, Weber O, Rao TN, Sass K, Thiery J, Fehling HJ. No reduction of atherosclerosis in C-reactive protein (CRP)-deficient mice. J Biol Chem. (2011) 286:6272–9. doi: 10.1074/jbc.M110.161414
152. Lin CS, Xia D, Yun JS, Wagner T, Magnuson T, Mold C, et al. Expression of rabbit C-reactive protein in transgenic mice. Immunol Cell Biol. (1995) 73:521–31. doi: 10.1038/icb.1995.82
Keywords: C-reactive protein, acute phase, inflammation, structure, pentamer
Citation: Zhou H-H, Tang Y-L, Xu T-H and Cheng B (2024) C-reactive protein: structure, function, regulation, and role in clinical diseases. Front. Immunol. 15:1425168. doi: 10.3389/fimmu.2024.1425168
Received: 29 April 2024; Accepted: 03 June 2024;
Published: 14 June 2024.
Edited by:
Alok Agrawal, East Tennessee State University, United StatesReviewed by:
Lawrence Albert Potempa, Roosevelt University, United StatesCopyright © 2024 Zhou, Tang, Xu and Cheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bin Cheng, Y2hlbmdiaW5nQGx6dS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.