
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol., 20 June 2024
Sec. Cancer Immunity and Immunotherapy
Volume 15 - 2024 | https://doi.org/10.3389/fimmu.2024.1422113
This article is part of the Research TopicDeveloping Biocompatible and Biotechnological Tools against Cancer and Autoimmune DisordersView all 5 articles
Recent breakthroughs in discovering novel immune signaling pathways have revolutionized different disease treatments. SERPINB9 (Sb9), also known as Proteinase Inhibitor 9 (PI-9), is a well-known endogenous inhibitor of Granzyme B (GzmB). GzmB is a potent cytotoxic molecule secreted by cytotoxic T lymphocytes and natural killer cells, which plays a crucial role in inducing apoptosis in target cells during immune responses. Sb9 acts as a protective mechanism against the potentially harmful effects of GzmB within the cells of the immune system itself. On the other hand, overexpression of Sb9 is an important mechanism of immune evasion in diseases like cancers and viral infections. The intricate functions of Sb9 in different cell types represent a fine-tuned regulatory mechanism for preventing immunopathology, protection against autoimmune diseases, and the regulation of cell death, all of which are essential for maintaining health and responding effectively to disease challenges. Dysregulation of the Sb9 will disrupt human normal physiological condition, potentially leading to a range of diseases, including cancers, inflammatory conditions, viral infections or other pathological disorders. Deepening our understanding of the role of Sb9 will aid in the discovery of innovative and effective treatments for various medical conditions. Therefore, the objective of this review is to consolidate current knowledge regarding the biological role of Sb9. It aims to offer insights into its discovery, structure, functions, distribution, its association with various diseases, and the potential of nanoparticle-based therapies targeting Sb9.
Serpinb9 (Sb9), first defined in 1995 (1), is a member of the largest and most widely distributed superfamily of proteinase inhibitors known as serpins, which specifically inhibit serine proteases (2). The serpins superfamily is sub-divided into 16 clades (A-P) based on their phylogenetic relationships (2). These proteins are designated SERPINXy, where ‘X’ signifies the specific clade and ‘y’ denotes the individual number within that clade (2). To date, there have been no less than 37 serpins identified in humans (3). Among these, the extracellular ‘clade A’ serpins and the intracellular ‘clade B’ serpins represent the two largest categories within the human serpin family (4). While the majority of serpins primarily inhibit serine proteases, it’s worth noting that there have been identified cross-class inhibitors that target caspases (5, 6) and papain-like cysteine proteases (7, 8) as well.
Serpins exert their inhibitory effects on proteases, contributing to various physiological processes, including inflammation, complement activation, blood coagulation, tissue remodeling and immune response as well as prevention of inappropriate activity of cytotoxic proteases induced apoptosis (9). Dysfunction within the serpin family is implicated in a wide range of pathophysiological processes and diseases. Several conformational diseases or ‘serpinopathies’ linked to polymerization of serpins within the endoplasmic reticulum have been identified (10). These include emphysema (SERPINA1 mutation) (11), thrombosis (SERPINC1 deficiency) (12), hereditary angioedema (SERPING1 deficiency) (13), cirrhosis (SERPINA1 polymerization) (14) and Alzheimer’s disease (SERPINA3 polymorphism) (15).
All serpin proteins share a conserved tertiary structure characterized by a core domain with three β−sheets, eight-nine α−helices, and a flexible reactive center loop (RCL) that contains seventeen amino acids and is positioned in the protease recognition domain close to the C−terminus of serpin (16). The core structure of serpins is highly conserved, which determines the specificity of serpin inhibitory functions (17). Sb9 is a single-chain protein composed of 376 amino acids with a molecular weight of 42 kDa (18). Consistent with other members in the serpin superfamily, Sb9 possesses a highly preserved tertiary structure. It inhibits GzmB by initially forming a reversible Michaelis complex, followed by the formation of a covalent complex: a covalent ester linkage is formed between the sidechain oxygen atom of GzmB and the carbonyl carbon of Sb9 P1 residue (19–21). In addition, Losasso et al. find that R28K, R201A, and R201K GzmB mutants substantially disrupt the interaction with Sb9. In particular, the activity of R201K GzmB closely resembles that of the wild-type protein, regardless of the presence or absence of Sb9 (22). It is important to note, however, that the structure of human Sb9 has not been fully elucidated. Consequently, additional research is necessary to validate these findings.
Sb9 is extensively expressed in both the cytoplasm and nuclei of various human endothelial, mesothelial and immune cells, particularly in immune-privileged sites such as placenta, testes, ovaries and eyes (23–25). Furthermore, Sb9 is present at high and relatively stable levels in cytotoxic lymphocytes (comprising natural killer cells and CD8+ T cells), as well as in antigen-presenting cells (24, 26).
As previously noted, Sb9 is a potent physiological inhibitor of GzmB through formation of protein complex. GzmB, a caspase-like serine protease, triggers apoptosis of virus-infected cells or tumor cells by activating caspases-3 and -8 (27). Sb9 can keep the GzmB staying in the cytoplasm (28). Subcellular fractionation experiments show that Sb9 can be imported and exported through nuclei to grab any GzmB that evades the nucleocytoplasmic pool of Sb9 (23).
It has been established that Sb9 can protect tumor cells from GzmB mediated destruction, whether GzmB originates from cytotoxic lymphocytes or is produced endogenously (29, 30). Moreover, Sb9 has been revealed to provide cytoprotective effects against self-inflicted injury caused by GzmB in pro-inflammatory leukocytes (cytotoxic T cells (31), dendritic cells and neutrophils) or anti-inflammatory leukocytes (regulatory T cells [Tregs] and myeloid-derived suppressor cells [MDSCs]) (30–33).
It’s worth noting that apart from its role in regulating GzmB, human Sb9 can also influence cytokine production by modulating inflammatory caspase-1, and to a lesser extent caspase-4 and caspase-8, but not caspase-3 (34). Sb9 accounts for the innate inhibitory activity against caspase-1 through protein-protein interaction and prevents processing of the natural substrates of caspase-1, specifically IL-1β and IL-18 precursor in both human vascular smooth muscle cells and monocytes (35, 36). Additionally, there are reports suggesting that Sb9, functioning as a protease inhibitor, impedes tumor necrosis factor- and Fas-induced apoptosis by directly interacting with the pro-apoptotic apical caspases-8 and -10 (37, 38). However, considering the low Kasso (association rate constant) value and lack of data on Sb9 RCL cleavage or caspase-Sb9 complex formation, the physiologically significant interaction between Sb9 and these caspases is limited and unpersuasive (39, 40).
Besides as a protease inhibitor, several studies have indicated murine Sb9 functions in the regulation of antigen presentation and the cell-associated antigen cross-presentation by CD8+ splenic dendritic cells, independently of GzmB (41, 42). In addition, Sb9 has been described to inhibit the activity of subtilisin A and neutrophil elastase rather than human furin via a rapid and single step mechanism (43, 44).
Considering the vital role of Sb9 in numerous physiological functions, gaining a deeper insight into its regulation, encompassing both intracellular factors and extracellular environment, could offer profound implications for clinical practice. Delving into these regulatory mechanisms might pave the way for innovative therapeutic interventions, diagnostics, or preventive measures, especially in those conditions where dysregulation of Sb9 is a key factor. Estrogen is first shown to boost the expression of Sb9 significantly and rapidly in an estrogen receptor-positive human liver cell line. This induction occurs through the Sb9 promoter region by the transcription factor estrogen receptor α (ERα) (45–47). The presence of EGF and the activation of ERK further enhance the estrogen-driven induction of Sb9 (46, 47). These results indicate estrogens regulate the susceptibility of target cells to immune system-induced apoptosis. Another two factors, IL-1β and IL-18, while being downstream targets of Sb9, are also pinpointed as inducers of Sb9 via the transcription factors nuclear factor-κB (NF-κB) and activator protein-1 (AP-1) (48, 49). Notably, mutational ablation of NF-κB and AP-1 binding sites almost entirely suppresses the IL-1β-induced expression of Sb9 (48). Recent studies have uncovered that Sb9 can be modulated by MUC1-C-IFNγ oncogenic signaling via the interferon regulator factor 1 (IRF1) transcription factor and by type 1 interferon in murine tumor cells (50–52). Additionally, cytokines are involved in regulating Sb9 expression in liver cells. Exposure of murine hepatocytes to IFN-α/β/γ induces Sb9. In human hepatoma Huh-7 cells, treatment with IFN-α/γ or TNF-α elevates Sb9 expression, while the related serine proteinase inhibitor, serpinB8, remains unaffected (53). Moreover, the IFN-α-induced upregulation of Sb9 in hepatocytes is specifically triggered by the infiltration of NK cells in the liver, which carry active GzmB and perforin (54). Furthermore, high Sb9 content in breast stem tumorspheres and dendritic cells, expressing high levels of CXCR4 and phospho-p38, might be ascribed to the activation of the CXCR4/p38MAPK proliferative signaling pathway (55, 56). Other factors, including STAT1 and NME1, have been reported to positively correlate with Sb9 expression (57, 58). Granzyme M acts as an endogenous inhibitor of Sb9, with human granzyme M potentially clearing the way for GzmB-mediated target cell damage by inactivating Sb9, a mechanism by which natural killer cells might overcome this escape mechanism (59).
Beyond the endogenous regulators, exogenous compounds have also been shown to affect the expression of Sb9. For example, lipopolysaccharide and 12-O-tetradecanoylphorbol-13-acetate have been shown to induce its expression in HepG2 cell line (48). Pentoxifylline stimulates the expression of Sb9 in K562 cells at the mRNA level, but not protein level (60). Additionally, low concentrations of the soy phytoestrogen genistein have been found to induce Sb9 and subsequently hinder killing of ERα+ breast cancer cells by immune cells (61). This suggests that soy consumption levels may be a relevant consideration for ER+ breast cancer patients (61). Cigarette side stream smoke particulate matter (CSSP), a major component of secondhand smoke, upregulates Sb9 in a dose-dependent manner through the activation of ERα in human lung adenocarcinoma cells (62). In glioma cell lines, Sb9 is also elevated by low pH 6.5, threefold or more, in both 2D tumor cells and 3D glioma stem cells. These findings imply that culturing cancer cells in lower pH conditions, which mimic the endogenous tumor environment, leads to a marked rise in Sb9 and more accurately preserve tumor heterogeneity (63). The upregulation of Sb9 can be mitigated by reactive oxygen species, which inactivate Sb9 by oxidizing a highly conserved cysteine pair in the reactive center loop to form a vicinal disulfide bond (64). Interestingly, we have recently discovered that the expression of Sb9 is significantly induced in both gemcitabine resistant pancreatic cancer cell lines and following transient exposure to gemcitabine. Mechanistic study shows that gemcitabine induces Sb9 expression at transcriptional level through transcription factor ATF3 (unpublished data). Importantly, Sb9 stable knockout (KO) or transient KD via siRNA leads to increased sensitivity to gemcitabine treatment, providing the 1st evidence that Sb9 is also involved in regulation of drug sensitivity in cancer cells (unpublished data).
The primary function of Sb9 is to safeguard somatic cells such as stem cells, immune cells and tumor cells from apoptosis through diverse pathways (65–70). Sb9 plays a dual role in immunity, especially in the regulation of different immune cells and tumor cells. Within immune cells such as cytotoxic T lymphocytes, the primary role of Sb9 is to protect them from apoptosis and cell death mediated by the self-produced GzmB. It ensures that immune cells can utilize GzmB to eliminate target cells without falling prey to their own cytolytic mechanisms. Comprehending the distinct role of Sb9 in modulating various signaling pathways within immune cells is essential for enhancing immune function. However, when co-opted by tumor cells, Sb9 can facilitate immune escape (71), highlighting its intricate role within the immune landscape. Its overexpression in certain cancers and cancer stem cells might be associated with tumor initiation and metastasis, therapy resistance, and epithelial-to-mesenchymal transition (EMT).
Although Sb9 is primarily recognized for its involvement in immunity and cancer, there is evidence Sb9 is involved in other physiological and pathological processes as well. Understanding the differential expression of Sb9 across cell types and tissues is vital for deciphering its physiological and pathological roles in, for example, hepatitis, renal allograft, and autoimmune disorders. It is important to note that while we have general knowledge about Sb9’s roles and expression profiles, the exact levels and functional significance might vary based on the specific tissue or disease context. This knowledge can help devise therapeutic strategies, especially in diseases like cancer where Sb9 expression might influence disease progression and treatment outcomes (Figure 1).
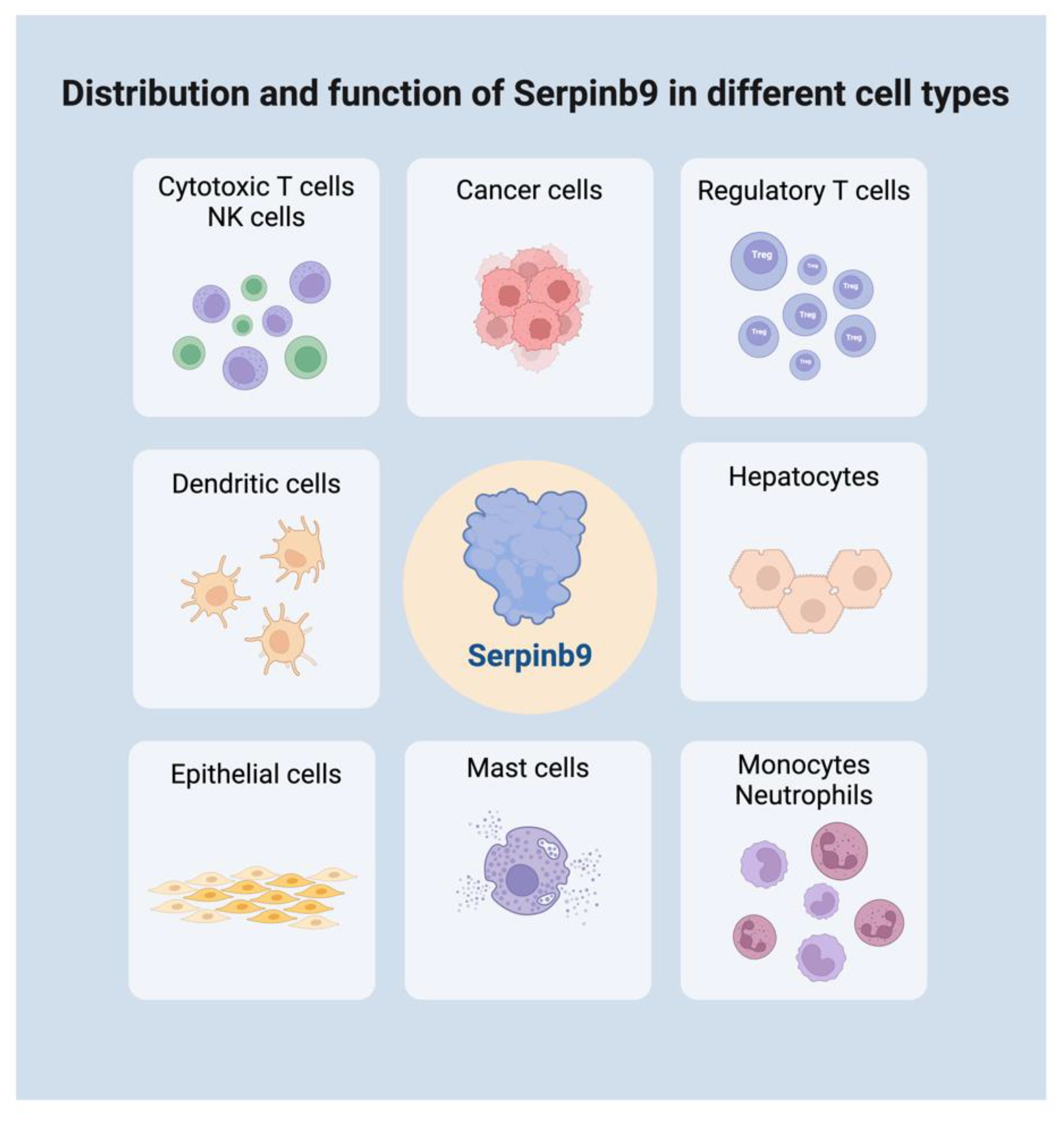
Figure 1 Distribution and function of Serpinb9 in different cell types. Serpinb9 is expressed in various immune cells, somatic cells, and tumor cells. It plays a dual role in immune modulation across different cell types. Created with BioRender.com.
Sb9 emerges as a signature gene of T cells dysfunction from the “statistical interaction test”, which is employed to identify specific genes in several cancer types that could be linked to patients’ abnormal tumor immune microenvironment. The identification of Sb9 as a hallmark of T cell dysfunction implies that the presence or high expression of this gene in a tumor immune microenvironment can serve as a biomarker of tumor immune evasion (50, 72).
It is well established that the appearance of GzmB in the cytoplasm of NK cells and cytotoxic T lymphocytes (CTLs) during activation is responsible for cell death. Restimulation of previously activated T lymphocytes induces lysosomal membrane permeabilization, followed by GzmB release into the cytosol (73). Sb9 acts to neutralize misdirected GzmB following the degranulation of cytotoxic cells. As a result, the suppression of GzmB by Sb9 can enhance the viability of CTLs and NK cells (73–75). This mechanism potentially explains why cytotoxic cells are not damaged by their own GzmB during destruction of abnormal cells (76). The Sb9 is also upregulated during degranulation of effector cells, and its overexpression enhances CTLs potency by improving cell viability (26). Research indicates that the absence of the Sb9 gene renders CTLs more vulnerable to premature death and diminishes their effectiveness in recognizing and eradicating virus-infected cells (31). Invariant natural killer T (iNKT) cells from Sb9 knockout mice exhibit impaired production of IL-4 and IFN-γ, highlighting that Sb9 is required for immunoregulatory function of iNKT cells (77). Similarly, Sb9 plays a critical role in protecting NK cells from GzmB-mediated cell death. Sb9-/- mice are highly susceptible to Ectromelia virus infection, which is attributed to NK cells being more prone to GzmB-mediated death, leading to diminished cytotoxic potential (75, 78).
Sb9 exhibits varied functions across different T cell subtypes from diverse origins. For example, CD8+ effector T cells are categorized into those destined to become long-lived memory CD8+ T cells (memory-precursor-effector-CD8+-cells [MPEC]) and those that are short-lived (short-lived-effector-CD8+-cells [SLEC]). The formation of SLEC is affected by Sb9, but not MPEC, suggesting that Sb9 has a differential impact on the survival of effector and memory alloreactive CD8+ T cells (79). Generally, overexpression of Sb9 in mice leads to an increased frequency and enhanced development of memory T cells (80). Additionally, the expression of Sb9 serves to ensure the survival of metabolically active CD8+ memory T cells, protecting them from the effects from cytotoxic T lymphocytes (80). However, if memory CD8+ T cells originate from the progenitors that are refractory to self-inflicted damage with low levels of Sb9 and GzmB, rather than from fully differentiated CTLs, Sb9 is not required for the development and survival of these subsets of memory CD8+ T cells (81). Intriguingly, TCRαβ+CD3+CD4-CD8-NK1.1- double-negative regulatory T cells have been observed to suppress CD8+ memory T cell but not CD4+ memory T cells, which is attributed to the high expression of Sb9 in CD4+ memory T cells (82).
Regulatory T cells (Tregs) are critical for maintaining immune homeostasis by suppressing the activity of other immune cells, thus preventing autoimmunity and excessive inflammation. Alterations in function or quantity of Tregs can result in immune dysregulation. Interestingly, Sb9 has been found to have a contentious role in Tregs across various diseases, a topic that continues to be at the forefront of intense research and debate. In murine skin graft model, GzmB+ Tregs exhibit an immunosuppressive role, contributing to prolonged graft survival. Overexpression of Sb9 interferes with the function of GzmB in Tregs and results in decreased graft survival compared to tolerized wild-type (WT) mice (83). In contrast, another research shows that Sb9 is critical for maintaining the potency and of survival of activated Tregs in graft versus-host disease, where it exerts immunosuppressive functions (32). Prashant S. Giri et al. identify that Tregs from generalized vitiligo patients express lower level of Sb9 compared to control. The decreased Sb9 lead to the reduced number of Tregs, implicating a role in the pathogenesis of generalized vitiligo (84). In the context of HIV, some infected CD4+ Th1 cells express Sb9, which shields these infected cells from their own GzmB mediated and CD8+ T cells mediated killing. These observations hint at a potential mechanism for HIV-1 RNA+ cells to evade destruction by cytotoxic CD8+ T cells: Sb9 could reside within cytotoxic CD4+ T cells that are inherently resistant to GzmB mediated death. However, further studies involving more extensive single-cell evaluations are essential, given the limited scope of cells analyzed in these studies (85).
Dendritic cells display elevated levels of Sb9 in primary lymphoid organs and inflammatory sites. This presence is noted in thymic DCs (characterized as CD3-, CD4+, CD8-, CD45+), tonsillar DCs, DC subsets from peripheral blood (CD16+ monocytes and CD123+ plasmacytoid DCs), as well as monocyte-derived DCs (24, 26). Furthermore, Sb9 is tightly associated with the maturation of DCs (increased levels of DCs activation marker CD86) induced by TNF-α, LPS, CD40L or ectromelia virus infection (26, 42, 55, 86). The elevated expression of Sb9 is found to be functionally significant for protecting DCs, as LPS-treated DCs become less vulnerable to CTL-mediated killing in vitro (86). Further evidence shows that impaired survival of CD8α DCs is observed in Sb9 knockout mice following infection with lymphocytic choriomeningitis virus (55, 87). However, when these LPS-treated DCs are injected in vivo, they remain susceptible to CTL-mediated killing. This is observed irrespective of whether the CTLs in host mice are elicited via active immunization or passively transferred via the injection of in vitro-activated CTLs. A rapid reduction in Sb9 expression levels in DCs after in vivo injection may explain why DCs are resistant to CTL-mediated killing in vitro but not in vivo (86).
In addition, Sb9 acts as a biomarker for antigen cross-presenting DCs, CD8+DC subsets. Notably, only the subset with high Sb9 expression is capable of cross-presentation (42). On the contrary, DCs from Sb9-deficient mice demonstrate a compromised ability to cross-present cell-associated antigens and cross-prime CTLs, with this deficit being independent of GzmB (41).
Furthermore, a novel subpopulation of IFN-induced DCs with high levels of Sb9 is identified. These cells exhibit direct tumoricidal activity against K562 cells, deserving further study to become a potential regimen for cancer therapy (88).
The recent discovery of Sb9 expression in mast cells, known for their central role in allergic responses and innate immunity, adds to its established presence in T cells and DCs. This finding highlights Sb9’s potentially extensive impact on immune regulation. Activation of mast cell-lines HMC-1 and LAD2, as well as cord blood- and mature skin-derived human mast cells by PMA and calcium ionophore A2318 results in expression and release of GzmB, accompanied by an upsurge in Sb9 expression (89, 90). Similar to cytotoxic lymphocytes, Sb9 is localized in the cytoplasm of mast cells. Furthermore, GzmB has been observed in granules within mast cells that are morphologically akin to the cytotoxic granules seen in CLs, as revealed through immunohistochemical staining and immuno-electron microscopy. This upregulation of Sb9 in activated mast cells strongly suggests that Sb9 serves to protect these cells against apoptosis induced by either autologous misdirected GzmB or GzmB released during the onset of local inflammatory responses, mirroring observations in CLs (89, 90). While the direct link between Sb9 and mast cell activation is yet to be comprehensively elucidated, it is conceivable that Sb9’s modulation of GzmB activity could influence the release of mediators from mast cells, thereby impacting allergic reactions, asthma and inflammatory responses. Incorporating the role of Sb9 in mast cells into diagnostic or therapeutic strategies can provide a holistic approach, especially in conditions influenced by mast cell activity.
Upregulation of Sb9 is observed in lymphocytes and monocytes of patients with acute Epstein-Barr virus infection. Moreover, Sb9 is upregulated by IL-2 and LPS in monocytes, but not lymphocytes. This upregulation can be inhibited by NF-κB inhibitor pyrrolidine dithiocarbamate and dexamethasone, indicating a relevant biological role of Sb9 in the functionality of leucocyte subsets during inflammatory processes (91).
During another inflammatory response triggered by M. tuberculosis infection, a rapid increase in Sb9 levels is observed in both alveolar macrophages and blood monocytes, lasting over seven days. The induction of Sb9 not only disrupts the cell death process but also promotes the survival of both host cells and the intracellular pathogen. These findings suggest that Sb9 might have a multifaceted impact on the progression of human M. tuberculosis infection, due to its roles in reducing inflammation and protecting against GzmB-triggered cell damage (92).
Another function of Sb9 is an intracellular inhibitor of neutrophil elastase, a neutral serine protease released by polymorphonuclear neutrophils during inflammation and in response to Pseudomonas aeruginosa infection (44). Sb9 can also protect neutrophils from self-damage. Ablation of Sb9 in neutrophils can increase the neutrophil elastase activity in the extracellular environment due to elevated neutrophil lysis, therefore boosting neutrophil immunity against Pseudomonas aeruginosa infection (93). Additionally, upregulation of Sb9 is positively correlated with infiltration of neutrophil and negatively correlated with infiltration of CD8+ T cell within tumor microenvironment of uveal melanoma patients, indicating the potential prognostic ability of neutrophils in this context (94).
In 2002, Sb9 was identified for the first time as being expressed in vivo by human tumors, non-Hodgkin and Hodgkin lymphoma (95). This discovery leads to the proposal of a novel mechanism by which Sb9 may enable tumor cells to circumvent immune surveillance (95, 96). However, a subsequent study suggests that primary lymphomas or lymphoma cell lines, despite expressing Sb9, remain sensitive to perforin-dependent cytolysis. This sensitivity is later found to be attributed to the cytolysis caused by the perforin-dependent pathways, rather than the Sb9-targeted protein, GzmB (97). Recent studies have increasingly shown that Sb9 plays a significant role in cancer progression, contributing to increased aggression and treatment failures. For example, there is a robust negative correlation between constitutive Sb9 expression levels and the activity of GzmB in leukemia cells, indicating Sb9 can inhibit the cell apoptosis induced by GzmB (98). Moreover, human cancer cell lines stably expressing Sb9 demonstrate a heightened ability to resist apoptosis triggered by both granzyme A or GzmB and Fas/Fas ligand pathway (37). At the cellular level, overexpression of Sb9 via transfection in leukemia cell line K562 (99) and mastocytoma cell line P815 (100) results in resistance against NK cell-induced apoptosis. Furthermore, Sb9 has been detected in various cancer cell types, including HepG2 hepatocellular carcinoma cells (46, 47), MCF7 breast cancer cells (46, 47), LCLC-103 H and A549 lung cancer cells (101), as well as prostate cancer cells (102), modulating their apoptosis induced by the immune system. Interestingly, in a study involving 8 Ewing sarcoma cell lines, tumor cells remain susceptible to perforin/granzyme-induced apoptosis even in the presence of Sb9, possibly due to alternative pathways (103). Moreover, in vivo studies show that genetic ablation of Sb9 using CRISPR/Cas9 significantly impede tumor growth in various cancer models, including B16 (murine melanoma), 4T1 (murine breast cancer), LLC1 (murine lung cancer), and A375 (human melanoma) (30).
Mechanistic study shows that Sb9 in tumor cells affects NK cell-mediated killing by decelerating the detachment of NK cells from tumor cells, a process recognized as a marker of successful target cell elimination (99). The rate of NK cell detachment from target cell defines the potency of NK cell-mediated killing. Delayed detachment often results from either reduced NK cell cytotoxicity or increased resistance of target cells. Cytokine-stimulated activated NK cells detach earlier than resting NK cells, correlating with their potent cytotoxicity. Overexpression of Sb9 and surface CD107a in K562 decelerates NK cells detachment from target cells, consequently diminishing the effectiveness of NK cell-mediated killing (99).
Recently, the influence of Sb9 on immune cells within the tumor microenvironment has gained attention. Sb9 can be regarded as a tumor antigen, recognized by human leucocytes antigen (HLA) class I-restricted and tumor-reactive CTLs. Sb9-recognizing CTLs can produce IFN-γ, potentially playing a role in the anti-tumor immune response in cancer patients (104). However, in B16-Sb9 KO tumor, the levels of infiltrated Tregs, TAMs, and MDSCs have no difference compared to B16-WT tumor, indicating Sb9 from tumor cells has minimal impact on the proportion of immunosuppressive cell populations in the TME (30). Interestingly, in the Sb9 KO tumors in Sb9 KO mice, there are increased ratios of CD8+, CD44+CD62L-CD8+, TNFα+CD8+, GzmB+CD8+, and IFNγ+CD8+, as compared with those from the WT tumor in WT mice (30). Additionally, the numbers of TAMs, MDSCs and CAFs are significantly lower in TME of Sb9 KO tumors in Sb9 KO mice than in the WT tumors in WT mice. The loss of Sb9 in Tregs, TAMs, and MDSCs drive them more sensitive to GzmB-mediated killing within the TME, ultimately leading to less infiltration of these immunosuppressive cells into the tumor (30). Furthermore, co-administration of siSb9, GzmB and anti-PD1 significantly induces the ratio of mature DCs, CD4+ and CD8+ T cells, as opposed to a pronounced decrease in Tregs in the tumor tissue of murine B16 model, which implies an activated immune system (105).
It should be highlighted that Sb9 is correlated with metastasis formation as well as the generation and maintenance of cancer stem cells. This association is underscored by the observed correlation between Sb9 expression and the epithelial-to-mesenchymal transition, both in vitro and in cancer patients (101). Metastatic melanoma cells have exhibited overexpression of Sb9 (106). Compared to B16-WT tumor, B16-Sb9 KO tumor displays a reduction in metastatic melanoma cells within tumor-draining lymph nodes (TDLNs) (30). Intriguingly, while Sb9 overexpression is evident in metastatic cells, its knockdown results in only a marginal increase in the vulnerability of these cells to CTL-mediated destruction, suggesting that Sb9 expression might not be associated with the resistance of metastatic cells (106). Regarding cancer stem cells, estrogen is recognized for its potential to act as a stimulus in upregulating the Sb9 expression (56). However, in the MCF7 breast cancer tumorspheres, an elevated level of Sb9 was observed despite reduced levels of the ERα66. Treatment with estrogens increases Sb9 level while decreasing ERα66 isoform and promoting cell proliferation, as demonstrated by the increased expression of proliferating cell nuclear antigen as well as increased number and size of tumorspheres (56). Additionally, low pH stress promotes malignancy through the induction of stemness of glioma cells along with increased Sb9 levels (63). This underscores the complex role of Sb9 in cancer progression and its potential as a target for therapeutic intervention.
The expression of Sb9 in hepatocytes is not solely induced by IFN or TNF (53). Other factors, such as the presence of GzmB and perforin in NK cells and CTLs, can also trigger Sb9 expression in hepatocytes (54). However, SPI-8 or other Sb9 homologs (Serpinb9b/9c/9d/9e/9g) in hepatocytes could not be induced significantly following IFN-α stimulation or during viral infection (54). Notably, this increase of Sb9 is absent in mice depleted of NK cells or lacking perforin or GzmB (54). This result indicates a feedback mechanism in hepatocytes where the upregulation of Sb9 during viral infection is correlated with the influx of NK cells and CTLs, and exposure to GzmB and perforin (54). This upregulation of Sb9 protects hepatocytes from NK cells and CTLs-mediated apoptosis. Conversely, inhibition of Sb9 results in increased hepatocellular injury during viral infections, indicating heightened hepatotoxicity (107).
Sb9 is involved in multiple biological pathways and plays a crucial role in regulating the cytotoxic activity of immune cells. Consequently, disruptions in Sb9 can lead to various immune and inflammatory diseases. For example, Sb9 A329S variant contributes to the pathogenesis of autoinflammatory disease due to its impaired ability to inhibit caspase-1 and IL-1β, indicating Sb9 might play an important role in autoimmune diseases (36). Moreover, impaired balance between Sb9 and GzmB is associated with pathophysiological processes of a spectrum of other health issues, including hepatitis (53, 54, 107), pancreatitis (108), non-alcoholic steatohepatitis (109), severe generalized vitiligo (84), villous atrophy in celiac disease (110), chronic obstructive pulmonary disease (111), infections (69), graft injuries (65, 70, 112), cardiovascular diseases (35), Alzheimer disease (113), and polycystic ovary syndrome (114). These associations highlight the broad impact of Sb9 in various inflammatory and immune-related conditions. Furthermore, recent research has also pinpointed Sb9 as a new biomarker and potential target for various cancers (18). These findings collectively underscore the multifaceted roles of Sb9 across various health domains (Figure 2).
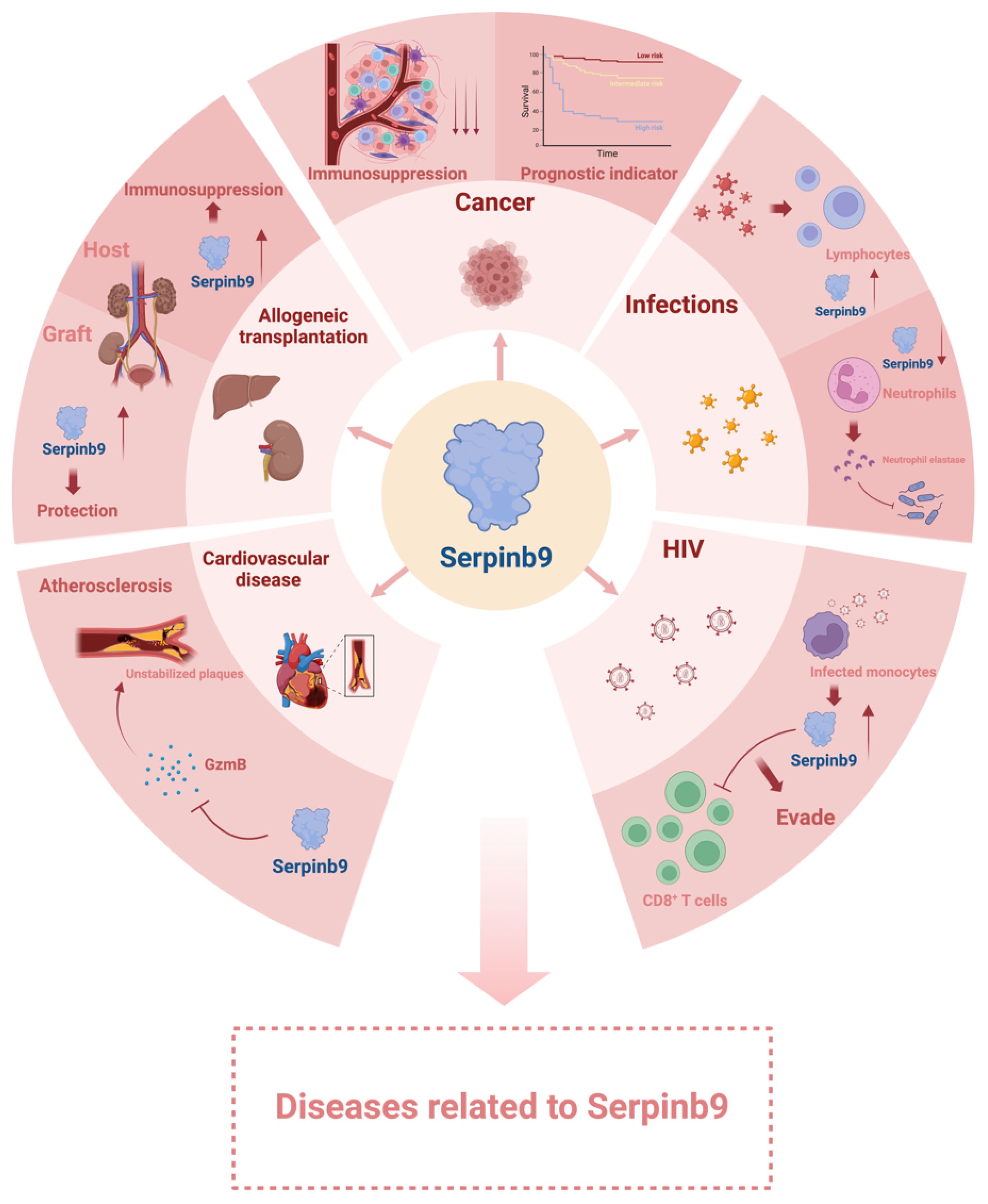
Figure 2 Diseases related to Serpinb9. Serpinb9 is involved in many biological processes. Disruptions in Serpinb9 function may cause various diseases. Created with BioRender.com.
Recent studies have elucidated a strong correlation between Sb9 expression and cancer progression, highlighting its potential role in tumorigenesis. Dysregulation of Sb9 has been found in high grade prostate tumors (102) and acute myeloid leukemia (115). Overexpression of Sb9 has been detected in a variety of cancers, including low grade prostate cancer (102), cervical cancer (58), gastric cancer (49), melanoma (101), nasopharyngeal carcinoma (116), glioma (63), hepatocellular carcinoma (117), non-small cell lung carcinoma (118), breast cancer (119), acute lymphoblastic leukemia (120), Hodgkin lymphomas and non-Hodgkin lymphomas (95, 121). Additionally, Sb9 is found consistently in high grade prostatic intraepithelial neoplasia and atrophic lesions (102).
As mentioned previously, the expression levels of Sb9 in tumors have been explored as a potential biomarker and prognostic indicator in cancer. For instance, in B-cell non-Hodgkin lymphoma, high Sb9 expression is associated with a higher grade of malignancy (95). Sb9 is also identified as the top risk gene for the association with oral squamous cell carcinoma, based on patient databases (122). Interestingly, Sb9 is not typically expressed in normal tissues, but expression can be acquired in oropharyngeal squamous cell carcinoma (123) and CNS-primitive neuroectodermal tumors (124). Likewise, there is significantly increased Sb9 expression in lung cancer tissues compared to non-cancer tissues, with decreased Sb9 expression in airway-derived CD8+ T cells from the cancer group compared to healthy controls (125). Also, Sb9 expression in primary lung cancer cells is significantly correlated with the stage of non-small cell lung cancer (125). However, non-significant differences in Sb9 expression are found between colorectal healthy and tumorous tissues or across different stages (126). The non-significant trend in Sb9 expression is also observed in tumorous tissue and regional lymph node metastases (126). Instead, increased expression of Sb9 in healthy tissue is significantly linked with longer overall survival (OS) in colorectal cancer patients, including those with distant metastases (126). High expression of Sb9 in tumor is negatively associated with prognosis and can be used to predict clinical outcome of ALK-negative anaplastic large cell lymphoma patients (127), hepatocellular carcinoma patients (117) and melanoma patients due to its negative correlation with CD8+ T cells (71, 94). Conversely, in nasal NK/T-cell lymphoma, Sb9 expression by tumor cells is associated with significantly better 5-year OS and 5-year event-free survival (EFS). Absence of Sb9 and low apoptotic index are related to poor outcomes (128).
The expression of Sb9 is widely acknowledged as a critical mechanism in enabling cancer cells to evade immune surveillance. Sb9 has been implicated in the modulation of immunosuppressive environments within the prostate cancer milieu (52) and has exhibited notable upregulation in bortezomib-resistant recurrent and relapsed multiple myeloma, as well as in resistant metastatic melanoma cells (106, 129). These findings highlight its promise as a therapeutic target aimed at overcoming resistance to immunotherapy treatments.
In renal transplants, Sb9 expression in the kidney is increased during infection with cytomegalovirus, Epstein-Barr virus or BK virus, in comparison to stable grafts (69). Similarly, up-regulation of Sb9 is also observed in lymphocytes and monocytes following infection by Epstein-Barr virus, but not in response to bacterial infection (91). Not confined to immune system, serum levels of Sb9 also rise during primary cytomegalovirus infection after transplantation, with a more pronounced increase observed in symptomatic patients than in those who are asymptomatic (130).
The role of Sb9 during infections can be complex and vary depending on the pathogen involved, indicating that its effects on the host’s immune response can be either beneficial or harmful. For example, Sb9 increases in cytotoxic lymphocytes during Ectromelia virus (Mousepox) infection, which is important to protect the host from the infection (78). On the other hand, the elimination of SPI-6 in neutrophils triggers the release of neutrophil elastase, which aids in combating Pseudomonas aeruginosa infections (93). Additionally, mycobacterium tuberculosis (MTB) can stimulate the expression of Sb9 in human mononuclear phagocytes (92, 131). The presence of Sb9 in monocytes is associated with the replication of intracellular MTB, and its inhibition can reduce the replication of MTB within cells, indicating that Sb9 protects these primary targets of infection from apoptosis, thereby enhancing the intracellular survival of the pathogen (92, 131). Mononuclear phagocytes from HIV-infected patients maintain increased activity of intracellular Sb9, which supports higher intracellular replication of MTB (131).
It has been reported that the expression of Sb9 could regulate Treg-mediated immunosuppression in allogeneic transplantation model (32, 83). The functionality of Sb9 within host intestinal epithelial cells can be amplified by IFN-γ originating from donor T cells. This amplification serves as a protective mechanism against donor T cells, presenting a potential strategy for attenuating graft-versus-host disease (132). Conversely, a deficiency in host Sb9 exacerbates graft-versus-host disease, as evidenced by a significant increase in lethality, as well as elevated clinical and histopathological scores (132). In acute renal allograft rejection, the presence of infiltrating cytotoxic T cells within the graft initiates apoptosis of renal tubular epithelial cells. The upregulation of Sb9 within renal allografts serves as a crucial endogenous mechanism, reducing the susceptibility of renal tubular epithelial cells to cytotoxic lymphocyte attacks. Consequently, this process plays a pivotal role in preserving tubular integrity and function (69, 70). The upregulation of Sb9 expression in tubular epithelial cells is induced by proinflammatory cytokines secreted from infiltrated cytotoxic cells (65, 70). This enhanced Sb9 expression provides protection against various forms of inflammatory kidney injury and facilitates long-term allograft survival. (65, 70). In murine studies, Sb9 deficiency is observed to enhance early tubular injury and expedite the rejection of kidney allografts, confirming the pivotal role of Sb9 in inhibiting GzmB-mediated injury to renal tubular cells and promoting renal allograft survival (70). Moreover, levels of Sb9 mRNA in urinary cells vary according to the severity of acute rejection, thereby serving as a diagnostic marker. The Sb9 levels in allograft tubular epithelial cells are higher in patients without rejection compared to those experiencing rejection, accompanied by a lower frequency of apoptosis protein caspase-3 (65). The levels of Sb9 not only predict the histological grade of renal allografts but also anticipate the functional outcomes following acute rejection (65, 112).
Elevated Sb9 DNA methylation, along with reduced Sb9 expression in circulating T cells, has emerged as a novel risk factor for the onset of both initial and subsequent post-transplant cutaneous squamous cell carcinoma (cSCC), a significant complication following organ transplantation (133). Targeting the DNA methylation of Sb9 holds promise as a therapeutic approach for managing cSCC in transplant recipients (133).
GzmB is suggested to play a role in inflammation associated with atherosclerotic coronary artery diseases. Consequently, regulating GzmB through Sb9 could represent a novel therapeutic strategy for preventing cardiovascular diseases, such as atherosclerosis, plaque rupture, and ventricular remodeling after acute myocardial infarction (AMI) (134). In the progression of atherosclerosis, GzmB is prevalently found in unstable atherosclerotic plaques. It is believed to contribute to plaque instability by inducing vascular smooth muscle cells apoptosis and by degrading plaque extracellular matrix (68). In addition, atherosclerotic patients exhibit significantly higher levels of activated caspase-3 and GzmB as well as reduced levels of Sb9 in both peripheral leucocytes and vascular smooth muscle cells compared to control (68, 135). In contrast, the unaffected arterial wall contains abundant and homogeneously distribution of Sb9 (35). Therefore, GzmB and Sb9 could be effective modulators in the development and progression of atherosclerosis (135).
Conversely, retarding the apoptosis of vascular smooth muscle cells by Sb9 is a key process in pathogenesis of abdominal aortic aneurysms (AAA) (136). Hypo-methylated Sb9 gene is positively correlated with AAA patients, along with its elevated mRNA expression (136, 137).
HIV-infected cells with elevated levels of Sb9 can effectively evade detection and destruction by cytotoxic CD8+ T cells, thereby bypassing the immune system’s natural defense mechanisms against infected cells (85). Monocytes isolated from HIV-infected patients exhibit higher expression levels of Sb9 mRNA and protein compared to those from healthy individuals (131). This elevated intracellular Sb9 activity in the monocytes is associated with an increased vulnerability to infections by other pathogens (131).
Given the protective role of Sb9 in regulating cell apoptosis, the modulation of homeostasis between Sb9 and GzmB emerges as a potential therapeutic strategy for various conditions, including cancers (30), HIV, autoimmune disorders (36, 84), renal transplantation (70), and hepatitis (107).
Sb9 serves a dual role in cancer progression: it protects the body against cancer development by maintaining proper immune function, while also promoting cancer progression by assisting malignant cells in evading immune detection and destruction. Given its role in cancer, Sb9 is being studied as a potential therapeutic target, either on its own or in combination with other therapy. Modulating Sb9 could potentially enhance the sensitivity of cancer cells to immune-mediated destruction. Hence, it has become a subject of interest for researchers looking to understand and combat cancer through various therapeutic strategies.
Sb9 is identified as the top immune-resistance gene in a study of somatic gene alterations correlating with immune cytolytic activity in a TCGA cohort of lung cancer (119). Notably, it stands out as the most upregulated gene in immune checkpoint non-responders compared to responders across various cancer types, indicating its potential utility as a predictive biomarker for immune checkpoint blockade (ICB) response and a prognostic marker (50, 71, 72, 101). Sb9 expression is elevated not only in patients who are refractory to ICB therapy but also in those with post- versus pre-treatment cancers (101). High Sb9 expression in melanoma correlates with poor response to ICB (101), leading to lower survival rates following anti-immune checkpoint inhibitor therapy (50, 71). Overexpression of Sb9 reverses the potency of redirected target lysis by AMG-110, a well-characterized EpCAM/CD3-bispecific BiTE® antibody construct currently undergoing clinical trials in late-stage cancer patients with various carcinomas (138). Therefore, pharmacological intervention targeting Sb9 may enhance the efficacy of immunotherapy (71, 129, 138). The resistance to CTLA4 checkpoint inhibition mediated by Sb9 justifies the therapeutic benefit of co-targeting Sb9 and CTLA4 (72).
Immune profile studies demonstrate that high Sb9 levels are closely correlated with significant T cell dysfunction (50, 72), promoting resistance to T cell-mediated killing (50). Combining Sb9 inhibition with tumor-associated antigen-directed T-cell therapy could yield superior efficacy (119). The low expression of Sb9 sensitizes cells to CD20 CAR T cell-mediated killing, indicating that Sb9 plays a crucial role in controlling tumor cell killing by CAR T cells. Combining Sb9 inhibition with CAR T treatment could enhance therapeutic efficacy (139). DNA methylation of Sb9 in circulating T cells represents a persistent risk factor for cutaneous squamous cell carcinoma (cSCC) development, which could be a future target for editing using the CRISPR/Cas9 system. Although this novel technology is far from clinical application, it holds promise for the future (133).
Modifying the expression of Sb9 in dendritic cells (DCs) may offer a novel approach for cancer treatment. Sb9 can prolong the lifespan of DCs by delaying their apoptosis. In one study, genetic modification of dendritic cells to overexpress Sb9 enhanced their ability to prime OVA antigen-specific CTLs in semi-allogeneic recipient mice, presenting a potential cancer prevention method (140). Another research demonstrates that co-administration of Sb9 with human papillomavirus type 16 E7 antigen enhances CD8+ T cell-mediated E7-specific immune responses and antitumor effects more effectively than either strategy alone. These findings suggest that a vaccination strategy combining Sb9 to prolong DC lifespan could enhance vaccine potency and hold promise for future clinical applications (141). Moreover, a novel defined CD56dim IFNα-induced, monocyte-derived DCs exhibit high expression of GzmB, perforin, and Sb9, which contribute to direct tumoricidal activity against K562 cells. Dissecting DCs composed of heterogeneous populations into functional subpopulations defined by molecular markers should enhance our understanding of DC biology and improve the clinical outcomes of DC vaccines (88). KO of Sb9 in mesenchymal stem cells (MSCs) results in diminished activity of downregulating cytotoxic T lymphocyte (CTL)-mediated lysis, suggesting that Sb9 in MSCs could be a potential target due to its immunosuppressive properties (30).
Sb9 may play a multifaceted role in HIV treatment. The expression of Sb9 drives the resistance of HIV-1 RNA+ cells to CD8+ T cell killing, making it a potential target for HIV therapy (85). A novel two-dimensional nanosheet has been developed to enhance Sb9 expression in murine splenocytes. This enhances HIV vaccine-triggered immune responses in vivo by enhancing either antigen presentation or T cell-mediated apoptosis of HIV-infected cells (142).
Sb9 may exert a protective effect in certain somatic cells. For instance, it can protect tubular epithelial cells from apoptosis, thereby increasing the survival rates of renal allograft patients (65, 70). Additionally, the acute induction of GzmB contributes to fulminant hepatitis, a rapidly developing and lethal syndrome (143). Transfection of Sb9 in hepatocytes confers resistance to killing by cytotoxic T cells and hepatotoxicity, suggesting that delivery of Sb9 protein or transgene to liver might be a new strategy for prevention or treatment of GzmB-induced hepatitis (54, 107).
The advancement in our understanding of Sb9-GzmB axis in various physiological and pathophysiological processes has led to several therapeutic approaches for disease treatment, especially in cancer treatment. Development of Sb9 small molecule inhibitors represents an attractive approach due to its simplicity in clinical applications. Compound 3034, a small molecule inhibitor of Sb9, has recently been identified from an in vitro screening, which effectively restrains the growth of melanoma, breast cancer, kidney tumors, and lung cancer in vivo (30). However, further refinement of the structure is needed to improve the specificity and pharmacokinetic (PK) profile. SiRNA-based therapeutics represents another attractive approach to inhibiting Sb9 activity. SiRNAs have the advantage of high selectivity and may have less concern of off-target effects compared to small molecule drugs. However, in vivo delivery of siRNA, especially tissue-specific delivery remains a challenge despite recent progress in siRNA delivery to liver. Another interesting strategy is the use of GzmB variants that are resistant to Sb9-mediated inhibition. However, further in vivo studies with these candidates are necessary to evaluate their activity, as well as potential immunogenicity and nonspecific toxicity (22). All of these approaches remain at the preclinical stage. It is important to emphasize that the development and utilization of Sb9-based therapies would require extensive research and clinical trials to ensure their efficacy and safety.
What is clear from the information outlined thus far is that Sb9-GZMB axis could be regarded as a promising target for different indications, notably in cancer therapy. However, a significant challenge lies in the lack of small molecules effective against Sb9 that also possess desirable pharmacokinetic profiles. To address this, quite a few nanoparticle systems have been developed to enhance the delivery of therapeutic agents targeting Sb9. These nanoparticles are particularly beneficial in targeting cancer cells while minimizing damage to healthy tissues (144–147). This selective targeting reduces potential side effects and toxicity, thereby improving both the bioavailability and antitumor efficacy (148, 149). One of the key advantages of nanoparticle-based drug delivery is the ability to engineer systems that release multiple drugs simultaneously into tumors, at a controlled rate or in response to specific triggers like pH, glutathione or reactive oxygen species (150). This controlled release mechanism ensures the maintenance of optimal drug ratios and therapeutic concentrations in the target tissue over an extended period (151). In summary, nano drug delivery systems offer a way to improve the effectiveness, safety, and patient compliance of various therapeutics, paving the way for more efficient and targeted approaches to healthcare.
One study investigated the application of protocatechuic acid (PCA), a natural phenolic acid known for its inhibitory effects on Sb9. Despite demonstrating excellent antitumor efficacy, PCA’s short half-life limits its widespread use against cancers. To address this limitation, Li and colleagues integrated PCA with isoguanosine (isoG), a natural product with strong self-assembling properties, to create a novel multifunctional supramolecular hydrogel. This PCA-based hydrogel serves as a multifunctional system with significant anticancer effects and an ability to remodel the tumor microenvironment (TME). This offers the potential for localized administration, seamlessly integrating targeted therapy and chemotherapy into a unified approach for treating oral squamous cell carcinoma (OSCC) (152). In another study, Huang et al. combined the reported Sb9 inhibitor 3034 with the immunogenic cell death (ICD)-inducing chemotherapeutic drug doxorubicin and encapsulated them into tumor-targeting peptide (Arg-Gly-Asp, RGD) decorated liposomes for breast cancer chemo-immunotherapy. This nanomedicine effectively accumulates at the tumor site, releasing doxorubicin to directly kill tumor cells and activate the host immune response through ICD induction. Additionally, the locally released 3034 relieves the inhibition of Sb9 on GzmB, facilitating the degradation of matrix components, enhancing immune cell infiltration, and improving nanomedicine penetration, ultimately leading to enhanced therapeutic outcomes (145).
Prodrug-based nanocarriers combined with photothermal therapy (PTT) represents an effective strategy for achieving a synergistic antitumor effect. Compound 3034 is conjugated to a second near-infrared (NIR-II) semiconducting polymer backbone via a glutathione (GSH)-cleavable linker. Once in the tumor, 3034 can be specifically liberated from the backbone to inhibit Sb9, thereby reactivating the activity of GzmB to enhance cancer immunotherapy. Moreover, the polymer induces photothermal therapy for direct tumor ablation, immune cell death (ICD), and the elicitation of an immune response under NIR-II photoirradiation (153). Furthermore, this well-controlled combination photothermal immunotherapy (PTI) can be synergized with other regimens to improve cancer treatment (105). A photothermal-sensitive liposome fused with M1 macrophage exosomes (M1 Exo) is constructed to efficiently transport protein GzmB and siRNA of Sb9 to the cells. This hybrid nanosystem exhibits pronounced efficacy in suppressing tumor growth in both B16 subcutaneous and lung metastasis models, offering a new strategy of multi-combinational tumor therapy (105).
As discussed before, Sb9 is a double-edged sword with different functions in different cell types especially in most tumor cells and CTLs, achieving cell-type selective delivery is crucial to minimize adverse effects on beneficial immune cells. We have recently developed a novel siRNA delivery system that targets tumor cells specifically, with minimal impact on other healthy immune cells. Furthermore, in a preliminary study, we demonstrate that the combination of siSb9 with gemcitabine (siSb9/GEM) exhibits a better synergistic effect than the combination of gemcitabine with 3034 (unpublished results). This enhanced synergy may be attributed to the siSb9/GEM combination fostering a more favorable tumor immune microenvironment (TIME), which shall bode well with combination with ICB in the future.
Numerous studies on Sb9 have been conducted, particularly in murine models. The murine orthologue of human SERPINB9 is Serpinb9 (SPI6), and at a cellular level, it plays an identical role in inhibiting GzmB. Despite functional similarities, mouse Sb9 and human Sb9 do differ in the in vitro kinetics of interaction with GzmB. Human Sb9 is a direct, highly efficient inhibitor of human GzmB with an SI of 10 and a Kasso above 106 M/s, mainly due to a P1 glutamic acid, which is cleaved only by proteases with a preference for acidic residues (39). In contrast, mouse Sb9 has a nonacidic P1 residue (Cys), with a Kasso outside the defined physiological range with mGzmB (Kasso=5.6 ± 1.4 × 104 M/s and SI=5.8 ± 0.5) (39, 40, 154). These data suggest that while mouse Sb9 can inhibit mGzmB-mediated death, it does not directly interact with mGzmB. Recent reports have indicated that mGzmB is much less cytotoxic than hGzmB and lacks the ability to cleave the key pro-apoptotic substrate Bid due to differences in its catalytic cleft topology (39). These differences suggest that human and mouse Sb9 likely control cell apoptosis through different mechanisms, which can be explained in three non-mutually exclusive ways: other mouse Sb9 homologs are involved in cell death; this process in mice requires some cofactors; mouse Sb9 is also required to control other rodent-specific GzmB paralogues to prevent death (39, 40). Indeed, compared to humans, larger mouse Sb9 homologues are located on chromosome 13, likely reflecting the need to regulate a larger proteinase repertoire arising from differing evolutionary pressures on the reproductive and immune systems. In addition to Sb9, other orthologues of human Sb9 in mice have been characterized, including Serpinb9b/9c/9d/9e/9f/9g (155), but limited information is available regarding the structure and function of other Sb9 homologues except for Serpinb9b. Serpinb9b, which inhibits cell apoptosis mediated by granzyme M, cooperates with Sb9 in protection from effector cell-mediated cytotoxicity (156, 157). Deficiency of Serpinb9b confers leukemia resistance in SPF mice, which is controlled by activation of another kinase, receptor-interacting serine/threonine-protein kinase 2 (RIPK2) (158). These discrepancies must be considered when conducting research on Sb9 in murine models. Further studies are needed to gain a comprehensive understanding of the Sb9 homologues in mice. Moreover, besides GzmB, limited information has been revealed regarding the endogenous inhibitors of Sb9 or the pathways regulated by Sb9. Multi-omics technology is not only a powerful tool for understanding the pathways involving Sb9 but also for facilitating the development of precision medicine for optimal patient treatment (159). Furthermore, advancements in nanotechnology will contribute to improving treatment by regulating Sb9 and its associated upstream or downstream pathways.
HH: Conceptualization, Investigation, Project administration, Validation, Writing – original draft, Writing – review & editing. YM: Writing – review & editing. SL: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the fund from National Institute of Health grants R01CA219399, R01CA223788, RO1CA278608, RO1CA270623 (to SL) and The David and Betty Brenneman Scholar Fund (to SL).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling editor declared a past supervisory role with one of the authors SL.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Sprecher CA, Morgenstern KA, Mathewes S, Dahlen JR, Schrader SK, Foster DC, et al. Molecular cloning, expression, and partial characterization of two novel members of the ovalbumin family of serine proteinase inhibitors. J Biol Chem. (1995) 270:29854–61. doi: 10.1074/jbc.270.50.29854
2. Law RH, Zhang Q, Mcgowan S, Buckle AM, Silverman GA, Wong W, et al. An overview of the serpin superfamily. Genome Biol. (2006) 7:216. doi: 10.1186/gb-2006-7-5-216
3. Maas C, De Maat S. Therapeutic SERPINs: improving on nature. Front Cardiovasc Med. (2021) 8:648349. doi: 10.3389/fcvm.2021.648349
4. Sánchez-Navarro A, González-Soria I, Caldiño-Bohn R, Bobadilla NA. An integrative view of serpins in health and disease: the contribution of SerpinA3. Am J Physiology-Cell Physiol. (2020) 320:C106–18. doi: 10.1152/ajpcell.00366.2020
5. Ray CA, Black RA, Kronheim SR, Greenstreet TA, Sleath PR, Salvesen GS, et al. Viral inhibition of inflammation: cowpox virus encodes an inhibitor of the interleukin-1 beta converting enzyme. Cell. (1992) 69:597–604. doi: 10.1016/0092-8674(92)90223-Y
6. Komiyama T, Ray CA, Pickup DJ, Howard AD, Thornberry NA, Peterson EP, et al. Inhibition of interleukin-1 beta converting enzyme by the cowpox virus serpin CrmA. An example of cross-class inhibition. J Biol Chem. (1994) 269:19331–7. doi: 10.1016/S0021-9258(17)32171-3
7. Schick C, Pemberton PA, Shi GP, Kamachi Y, Cataltepe S, Bartuski AJ, et al. Cross-class inhibition of the cysteine proteinases cathepsins K, L, and S by the serpin squamous cell carcinoma antigen 1: a kinetic analysis. Biochemistry. (1998) 37:5258–66. doi: 10.1021/bi972521d
8. Irving JA, Pike RN, Dai W, Bromme D, Worrall DM, Silverman GA, et al. Evidence that serpin architecture intrinsically supports papain-like cysteine protease inhibition: engineering alpha(1)-antitrypsin to inhibit cathepsin proteases. Biochemistry. (2002) 41:4998–5004. doi: 10.1021/bi0159985
9. Medema JP, Schuurhuis DH, Rea D, Van Tongeren J, De Jong J, Bres SA, et al. Expression of the serpin serine protease inhibitor 6 protects dendritic cells from cytotoxic T lymphocyte-induced apoptosis: differential modulation by T helper type 1 and type 2 cells. J Exp Med. (2001) 194:657–67. doi: 10.1084/jem.194.5.657
10. Lomas DA, Carrell RW. Serpinopathies and the conformational dementias. Nat Rev Genet. (2002) 3:759–68. doi: 10.1038/nrg907
11. Askew DJ, Silverman GA. Intracellular and extracellular serpins modulate lung disease. J Perinatol. (2008) 28 Suppl 3:S127–135. doi: 10.1038/jp.2008.150
12. De La Morena-Barrio ME, Suchon P, Jacobsen EM, Iversen N, Minano A, de la Morena-Barrio B, et al. Two SERPINC1 variants affecting N-glycosylation of Asn224 cause severe thrombophilia not detected by functional assays. Blood. (2022) 140:140–51. doi: 10.1182/blood.2021014708
13. Haslund D, Ryo LB, Seidelin Majidi S, Rose I, Skipper KA, Fryland T, et al. Dominant-negative SERPING1 variants cause intracellular retention of C1 inhibitor in hereditary angioedema. J Clin Invest. (2019) 129:388–405. doi: 10.1172/JCI98869
14. Rabekova Z, Frankova S, Jirsa M, Neroldova M, Lunova M, Fabian O, et al. Alpha-1 antitrypsin and hepatocellular carcinoma in liver cirrhosis: SERPINA1 MZ or MS genotype carriage decreases the risk. Int J Mol Sci. (2021) 22:10560. doi: 10.3390/ijms221910560
15. Kamboh MI, Minster RL, Kenney M, Ozturk A, Desai PP, Kammerer CM, et al. Alpha-1-antichymotrypsin (ACT or SERPINA3) polymorphism may affect age-at-onset and disease duration of Alzheimer’s disease. Neurobiol Aging. (2006) 27:1435–9. doi: 10.1016/j.neurobiolaging.2005.07.015
16. Knaupp AS, Bottomley SP. Serpin polymerization and its role in disease-the molecular basis of α-antitrypsin deficiency. IUBMB Life. (2009) 61:1–5. doi: 10.1002/iub.127
17. Sanrattana W, Maas C, De Maat S. SERPINs-from trap to treatment. Front Med (Lausanne). (2019) 6:25. doi: 10.3389/fmed.2019.00025
18. Wang WJ, Wang J, Ouyang C, Chen C, Xu XF, Ye XQ. Overview of serpin B9 and its roles in cancer (Review). Oncol Rep. (2021) 46:190. doi: 10.3892/or
19. Silverman GA, Bird PI, Carrell RW, Church FC, Coughlin PB, Gettins PG, et al. The serpins are an expanding superfamily of structurally similar but functionally diverse proteins. Evolution, mechanism of inhibition, novel functions, and a revised nomenclature. J Biol Chem. (2001) 276:33293–6. doi: 10.1074/jbc.R100016200
20. Ye S, Cech AL, Belmares R, Bergstrom RC, Tong Y, Corey DR, et al. The structure of a Michaelis serpin-protease complex. Nat Struct Biol. (2001) 8:979–83. doi: 10.1038/nsb1101-979
21. Hedstrom L. Serine protease mechanism and specificity. Chem Rev. (2002) 102:4501–24. doi: 10.1021/cr000033x
22. Losasso V, Schiffer S, Barth S, Carloni P. Design of human granzyme B variants resistant to serpin B9. Proteins. (2012) 80:2514–22. doi: 10.1002/prot.24133
23. Bird CH, Blink EJ, Hirst CE, Buzza MS, Steele PM, Sun J, et al. Nucleocytoplasmic distribution of the ovalbumin serpin PI-9 requires a nonconventional nuclear import pathway and the export factor Crm1. Mol Cell Biol. (2001) 21:5396–407. doi: 10.1128/MCB.21.16.5396-5407.2001
24. Bladergroen BA, Strik MC, Bovenschen N, Van Berkum O, Scheffer GL, Meijer CJ, et al. The granzyme B inhibitor, protease inhibitor 9, is mainly expressed by dendritic cells and at immune-privileged sites. J Immunol. (2001) 166:3218–25. doi: 10.4049/jimmunol.166.5.3218
25. Buzza MS, Hirst CE, Bird CH, Hosking P, Mckendrick J, Bird PI. The granzyme B inhibitor, PI-9, is present in endothelial and mesothelial cells, suggesting that it protects bystander cells during immune responses. Cell Immunol. (2001) 210:21–9. doi: 10.1006/cimm.2001.1806
26. Hirst CE, Buzza MS, Bird CH, Warren HS, Cameron PU, Zhang M, et al. The intracellular granzyme B inhibitor, proteinase inhibitor 9, is up-regulated during accessory cell maturation and effector cell degranulation, and its overexpression enhances CTL potency. J Immunol. (2003) 170:805–15. doi: 10.4049/jimmunol.170.2.805
27. Adrain C, Murphy BM, Martin SJ. Molecular ordering of the caspase activation cascade initiated by the cytotoxic T lymphocyte/natural killer (CTL/NK) protease granzyme B. J Biol Chem. (2005) 280:4663–73. doi: 10.1074/jbc.M410915200
28. Blink EJ, Jiansheng Z, Hu W, Calanni ST, Trapani JA, Bird PI, et al. Interaction of the nuclear localizing cytolytic granule serine protease granzyme B with importin alpha or beta: modulation by the serpin inhibitor PI-9. J Cell Biochem. (2005) 95:598–610. doi: 10.1002/(ISSN)1097-4644
29. Medema JP, De Jong J, Peltenburg LT, Verdegaal EM, Gorter A, Bres SA, et al. Blockade of the granzyme B/perforin pathway through overexpression of the serine protease inhibitor PI-9/SPI-6 constitutes a mechanism for immune escape by tumors. Proc Natl Acad Sci U.S.A. (2001) 98:11515–20. doi: 10.1073/pnas.201398198
30. Jiang L, Wang YJ, Zhao J, Uehara M, Hou Q, Kasinath V, et al. Direct tumor killing and immunotherapy through anti-serpinB9 therapy. Cell. (2020) 183:1219–33:e1218. doi: 10.1016/j.cell.2020.10.045
31. Zhang M, Park SM, Wang Y, Shah R, Liu N, Murmann AE, et al. Serine protease inhibitor 6 protects cytotoxic T cells from self-inflicted injury by ensuring the integrity of cytotoxic granules. Immunity. (2006) 24:451–61. doi: 10.1016/j.immuni.2006.02.002
32. Azzi J, Skartsis N, Mounayar M, Magee CN, Batal I, Ting C, et al. Serine protease inhibitor 6 plays a critical role in protecting murine granzyme B-producing regulatory T cells. J Immunol. (2013) 191:2319–27. doi: 10.4049/jimmunol.1300851
33. Kumar V, Patel S, Tcyganov E, Gabrilovich DI. The nature of myeloid-derived suppressor cells in the tumor microenvironment. Trends Immunol. (2016) 37:208–20. doi: 10.1016/j.it.2016.01.004
34. Annand RR, Dahlen JR, Sprecher CA, De Dreu P, Foster DC, Mankovich JA, et al. Caspase-1 (interleukin-1beta-converting enzyme) is inhibited by the human serpin analogue proteinase inhibitor 9. Biochem J. (1999) 342 Pt 3:655–65. doi: 10.1042/bj3420655
35. Young JL, Sukhova GK, Foster D, Kisiel W, Libby P, Schonbeck U. The serpin proteinase inhibitor 9 is an endogenous inhibitor of interleukin 1beta-converting enzyme (caspase-1) activity in human vascular smooth muscle cells. J Exp Med. (2000) 191:1535–44. doi: 10.1084/jem.191.9.1535
36. Van Der Burgh R, Meeldijk J, Jongeneel L, Frenkel J, Bovenschen N, Van Gijn M, et al. Reduced serpinB9-mediated caspase-1 inhibition can contribute to autoinflammatory disease. Oncotarget. (2016) 7:19265–71. doi: 10.18632/oncotarget.v7i15
37. Cunningham TD, Jiang X, Shapiro DJ. Expression of high levels of human proteinase inhibitor 9 blocks both perforin/granzyme and Fas/Fas ligand-mediated cytotoxicity. Cell Immunol. (2007) 245:32–41. doi: 10.1016/j.cellimm.2007.03.004
38. Kummer JA, Micheau O, Schneider P, Bovenschen N, Broekhuizen R, Quadir R, et al. Ectopic expression of the serine protease inhibitor PI9 modulates death receptor-mediated apoptosis. Cell Death Differ. (2007) 14:1486–96. doi: 10.1038/sj.cdd.4402152
39. Mangan MS, Kaiserman D, Bird PI. The role of serpins in vertebrate immunity. Tissue Antigens. (2008) 72:1–10. doi: 10.1111/j.1399-0039.2008.01059.x
40. Kaiserman D, Bird PI. Control of granzymes by serpins. Cell Death Differ. (2010) 17:586–95. doi: 10.1038/cdd.2009.169
41. Rizzitelli A, Meuter S, Vega Ramos J, Bird CH, Mintern JD, Mangan MS, et al. Serpinb9 (Spi6)-deficient mice are impaired in dendritic cell-mediated antigen cross-presentation. Immunol Cell Biol. (2012) 90:841–51. doi: 10.1038/icb.2012.29
42. Mangan MS, Vega-Ramos J, Joeckel LT, Mitchell AJ, Rizzitelli A, Roediger B, et al. Serpinb9 is a marker of antigen cross-presenting dendritic cells. Mol Immunol. (2017) 82:50–6. doi: 10.1016/j.molimm.2016.12.011
43. Dahlen JR, Foster DC, Kisiel W. Human proteinase inhibitor 9 (PI9) is a potent inhibitor of subtilisin A. Biochem Biophys Res Commun. (1997) 238:329–33. doi: 10.1006/bbrc.1997.7282
44. Dahlen JR, Foster DC, Kisiel W. Inhibition of neutrophil elastase by recombinant human proteinase inhibitor 9. Biochim Biophys Acta. (1999) 1451:233–41. doi: 10.1016/S0167-4889(99)00095-6
45. Kanamori H, Krieg S, Mao C, Di Pippo VA, Wang S, Zajchowski DA, et al. Proteinase inhibitor 9, an inhibitor of granzyme B-mediated apoptosis, is a primary estrogen-inducible gene in human liver cells. J Biol Chem. (2000) 275:5867–73. doi: 10.1074/jbc.275.8.5867
46. Jiang X, Orr BA, Kranz DM, Shapiro DJ. Estrogen induction of the granzyme B inhibitor, proteinase inhibitor 9, protects cells against apoptosis mediated by cytotoxic T lymphocytes and natural killer cells. Endocrinology. (2006) 147:1419–26. doi: 10.1210/en.2005-0996
47. Jiang X, Ellison SJ, Alarid ET, Shapiro DJ. Interplay between the levels of estrogen and estrogen receptor controls the level of the granzyme inhibitor, proteinase inhibitor 9 and susceptibility to immune surveillance by natural killer cells. Oncogene. (2007) 26:4106–14. doi: 10.1038/sj.onc.1210197
48. Kannan-Thulasiraman P, Shapiro DJ. Modulators of inflammation use nuclear factor-kappa B and activator protein-1 sites to induce the caspase-1 and granzyme B inhibitor, proteinase inhibitor 9. J Biol Chem. (2002) 277:41230–9. doi: 10.1074/jbc.M200379200
49. Majima T, Ichikura T, Chochi K, Kawabata T, Tsujimoto H, Sugasawa H, et al. Exploitation of interleukin-18 by gastric cancers for their growth and evasion of host immunity. Int J Cancer. (2006) 118:388–95. doi: 10.1002/(ISSN)1097-0215
50. Jiang P, Gu S, Pan D, Fu J, Sahu A, Hu X, et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med. (2018) 24:1550–8. doi: 10.1038/s41591-018-0136-1
51. Chen J, Cao Y, Markelc B, Kaeppler J, Vermeer JA, Muschel RJ. Type I IFN protects cancer cells from CD8+ T cell-mediated cytotoxicity after radiation. J Clin Invest. (2019) 129:4224–38. doi: 10.1172/JCI127458
52. Hagiwara M, Fushimi A, Bhattacharya A, Yamashita N, Morimoto Y, Oya M, et al. MUC1-C integrates type II interferon and chromatin remodeling pathways in immunosuppression of prostate cancer. Oncoimmunology. (2022) 11:2029298. doi: 10.1080/2162402X.2022.2029298
53. Barrie MB, Stout HW, Abougergi MS, Miller BC, Thiele DL. Antiviral cytokines induce hepatic expression of the granzyme B inhibitors, proteinase inhibitor 9 and serine proteinase inhibitor 6. J Immunol. (2004) 172:6453–9. doi: 10.4049/jimmunol.172.10.6453
54. Stout-Delgado HW, Getachew Y, Miller BC, Thiele DL. Intrahepatic lymphocyte expression of dipeptidyl peptidase I-processed granzyme B and perforin induces hepatocyte expression of serine proteinase inhibitor 6 (Serpinb9/SPI-6). J Immunol. (2007) 179:6561–7. doi: 10.4049/jimmunol.179.10.6561
55. Bots M, De Bruin E, Rademaker-Koot MT, Medema JP. Proteinase inhibitor-9 expression is induced by maturation in dendritic cells via p38 MAP kinase. Hum Immunol. (2007) 68:959–64. doi: 10.1016/j.humimm.2007.10.011
56. Lauricella M, Carlisi D, Giuliano M, Calvaruso G, Cernigliaro C, Vento R, et al. The analysis of estrogen receptor-alpha positive breast cancer stem-like cells unveils a high expression of the serpin proteinase inhibitor PI-9: Possible regulatory mechanisms. Int J Oncol. (2016) 49:352–60. doi: 10.3892/ijo.2016.3495
57. Mccorkle JR, Leonard MK, Kraner SD, Blalock EM, Ma D, Zimmer SG, et al. The metastasis suppressor NME1 regulates expression of genes linked to metastasis and patient outcome in melanoma and breast carcinoma. Cancer Genomics Proteomics. (2014) 11:175–94.
58. Chen J, Chen C, Zhan Y, Zhou L, Chen J, Cai Q, et al. Heterogeneity of IFN-mediated responses and tumor immunogenicity in patients with cervical cancer receiving concurrent chemoradiotherapy. Clin Cancer Res. (2021) 27:3990–4002. doi: 10.1158/1078-0432.CCR-20-4521
59. Mahrus S, Kisiel W, Craik CS. Granzyme M is a regulatory protease that inactivates proteinase inhibitor 9, an endogenous inhibitor of granzyme B. J Biol Chem. (2004) 279:54275–82. doi: 10.1074/jbc.M411482200
60. Lazarczyk M, Dziunycz P, Niderla J, Milewski L, Lazarczyk MA, Boszczyk A, et al. Influence of pentoxifylline on natural cytotoxicity and expression of granzymes and PI-9, a specific granzyme B inhibitor. Int J Mol Med. (2006) 17:135–9. doi: 10.3892/ijmm
61. Jiang X, Patterson NM, Ling Y, Xie J, Helferich WG, Shapiro DJ. Low concentrations of the soy phytoestrogen genistein induce proteinase inhibitor 9 and block killing of breast cancer cells by immune cells. Endocrinology. (2008) 149:5366–73. doi: 10.1210/en.2008-0857
62. Kuo LC, Cheng LC, Lee CH, Lin CJ, Chen PY, Li LA. Estrogen and cigarette sidestream smoke particulate matter exhibit ERalpha-dependent tumor-promoting effects in lung adenocarcinoma cells. Am J Physiol Lung Cell Mol Physiol. (2017) 313:L477–90. doi: 10.1152/ajplung.00322.2016
63. Hjelmeland AB, Wu Q, Heddleston JM, Choudhary GS, Macswords J, Lathia JD, et al. Acidic stress promotes a glioma stem cell phenotype. Cell Death Differ. (2011) 18:829–40. doi: 10.1038/cdd.2010.150
64. Mangan MS, Bird CH, Kaiserman D, Matthews AY, Hitchen C, Steer DL, et al. A novel serpin regulatory mechanism: serpinB9 IS REVERSIBLY INHIBITED BY VICINAL DISULFIDE BOND FORMATION IN THE REACTIVE CENTER LOOP. J Biol Chem. (2016) 291:3626–38. doi: 10.1074/jbc.M115.699298
65. Rowshani AT, Florquin S, Bemelman F, Kummer JA, Hack CE, Ten Berge IJ. Hyperexpression of the granzyme B inhibitor PI-9 in human renal allografts: a potential mechanism for stable renal function in patients with subclinical rejection. Kidney Int. (2004) 66:1417–22. doi: 10.1111/j.1523-1755.2004.00903.x
66. El Haddad N, Heathcote D, Moore R, Yang S, Azzi J, Mfarrej B, et al. Mesenchymal stem cells express serine protease inhibitor to evade the host immune response. Blood. (2011) 117:1176–83. doi: 10.1182/blood-2010-06-287979
67. El Haddad N, Moore R, Heathcote D, Mounayar M, Azzi J, Mfarrej B, et al. The novel role of SERPINB9 in cytotoxic protection of human mesenchymal stem cells. J Immunol. (2011) 187:2252–60. doi: 10.4049/jimmunol.1003981
68. Hendel A, Cooper D, Abraham T, Zhao H, Allard MF, Granville DJ. Proteinase inhibitor 9 is reduced in human atherosclerotic lesion development. Cardiovasc Pathol. (2012) 21:28–38. doi: 10.1016/j.carpath.2010.12.002
69. Heutinck KM, Kassies J, Florquin S, Ten Berge IJ, Hamann J, Rowshani AT. SerpinB9 expression in human renal tubular epithelial cells is induced by triggering of the viral dsRNA sensors TLR3, MDA5 and RIG-I. Nephrol Dial Transplant. (2012) 27:2746–54. doi: 10.1093/ndt/gfr690
70. Lau A, Khan K, Pavlosky A, Yin Z, Huang X, Haig A, et al. Serine protease inhibitor-6 inhibits granzyme B-mediated injury of renal tubular cells and promotes renal allograft survival. Transplantation. (2014) 98:402–10. doi: 10.1097/TP.0000000000000237
71. Van Houdt IS, Oudejans JJ, Van Den Eertwegh AJ, Baars A, Vos W, Bladergroen BA, et al. Expression of the apoptosis inhibitor protease inhibitor 9 predicts clinical outcome in vaccinated patients with stage III and IV melanoma. Clin Cancer Res. (2005) 11:6400–7. doi: 10.1158/1078-0432.CCR-05-0306
72. Bozorgui B, Kong EK, Luna A, Korkut A. Mapping the functional interactions at the tumor-immune checkpoint interface. Commun Biol. (2023) 6:462. doi: 10.1038/s42003-023-04777-3
73. Ida H, Nakashima T, Kedersha NL, Yamasaki S, Huang M, Izumi Y, et al. Granzyme B leakage-induced cell death: a new type of activation-induced natural killer cell death. Eur J Immunol. (2003) 33:3284–92. doi: 10.1002/eji.200324376
74. Laforge M, Bidere N, Carmona S, Devocelle A, Charpentier B, Senik A. Apoptotic death concurrent with CD3 stimulation in primary human CD8+ T lymphocytes: a role for endogenous granzyme B. J Immunol. (2006) 176:3966–77. doi: 10.4049/jimmunol.176.7.3966
75. Bird CH, Christensen ME, Mangan MS, Prakash MD, Sedelies KA, Smyth MJ, et al. The granzyme B-Serpinb9 axis controls the fate of lymphocytes after lysosomal stress. Cell Death Differ. (2014) 21:876–87. doi: 10.1038/cdd.2014.7
76. Sun J, Bird CH, Sutton V, Mcdonald L, Coughlin PB, De Jong TA, et al. A cytosolic granzyme B inhibitor related to the viral apoptotic regulator cytokine response modifier A is present in cytotoxic lymphocytes. J Biol Chem. (1996) 271:27802–9. doi: 10.1074/jbc.271.44.27802
77. Ansari AW, Temblay JN, Alyahya SH, Ashton-Rickardt PG. Serine protease inhibitor 6 protects iNKT cells from self-inflicted damage. J Immunol. (2010) 185:877–83. doi: 10.4049/jimmunol.1000651
78. Mangan MS, Melo-Silva CR, Luu J, Bird CH, Koskinen A, Rizzitelli A, et al. A pro-survival role for the intracellular granzyme B inhibitor Serpinb9 in natural killer cells during poxvirus infection. Immunol Cell Biol. (2017) 95:884–94. doi: 10.1038/icb.2017.59
79. Azzi J, Ohori S, Ting C, Uehara M, Abdoli R, Smith BD, et al. Serine protease inhibitor-6 differentially affects the survival of effector and memory alloreactive CD8-T cells. Am J Transplant. (2015) 15:234–41. doi: 10.1111/ajt.13051
80. Phillips T, Opferman JT, Shah R, Liu N, Froelich CJ, Ashton-Rickardt PG. A role for the granzyme B inhibitor serine protease inhibitor 6 in CD8+ memory cell homeostasis. J Immunol. (2004) 173:3801–9. doi: 10.4049/jimmunol.173.6.3801
81. Zhang M, Byrne S, Liu N, Wang Y, Oxenius A, Ashton-Rickardt PG. Differential survival of cytotoxic T cells and memory cell precursors. J Immunol. (2007) 178:3483–91. doi: 10.4049/jimmunol.178.6.3483
82. Su Y, Jevnikar AM, Huang X, Lian D, Zhang Z. Spi6 protects alloreactive CD4(+) but not CD8 (+) memory T cell from granzyme B attack by double-negative T regulatory cell. Am J Transplant. (2014) 14:580–93. doi: 10.1111/ajt.12614
83. Gondek DC, Devries V, Nowak EC, Lu LF, Bennett KA, Scott ZA, et al. Transplantation survival is maintained by granzyme B+ regulatory cells and adaptive regulatory T cells. J Immunol. (2008) 181:4752–60. doi: 10.4049/jimmunol.181.7.4752
84. Giri PS, Bharti AH, Dwivedi M. Decreased GZMB, NRP1, ITPR1, and SERPINB9 transcripts lead to reduced regulatory T cells suppressive capacity in generalized vitiligo patients. J Immunol Res. (2022) 2022:3426717. doi: 10.1155/2022/3426717
85. Collora JA, Liu R, Pinto-Santini D, Ravindra N, Ganoza C, Lama JR, et al. Single-cell multiomics reveals persistence of HIV-1 in expanded cytotoxic T cell clones. Immunity. (2022) 55:1013–31:e1017. doi: 10.1016/j.immuni.2022.03.004
86. Andrew KA, Simkins HM, Witzel S, Perret R, Hudson J, Hermans IF, et al. Dendritic cells treated with lipopolysaccharide up-regulate serine protease inhibitor 6 and remain sensitive to killing by cytotoxic T lymphocytes in vivo. J Immunol. (2008) 181:8356–62. doi: 10.4049/jimmunol.181.12.8356
87. Lovo E, Zhang M, Wang L, Ashton-Rickardt PG. Serine protease inhibitor 6 is required to protect dendritic cells from the kiss of death. J Immunol. (2012) 188:1057–63. doi: 10.4049/jimmunol.1102667
88. Watanabe A, Togi M, Koya T, Taniguchi M, Sakamoto T, Iwabuchi K, et al. Identification of CD56(dim) subpopulation marked with high expression of GZMB/PRF1/PI-9 in CD56(+) interferon-alpha-induced dendritic cells. Genes Cells. (2021) 26:313–27. doi: 10.1111/gtc.12844
89. Bladergroen BA, Strik MC, Wolbink AM, Wouters D, Broekhuizen R, Kummer JA, et al. The granzyme B inhibitor proteinase inhibitor 9 (PI9) is expressed by human mast cells. Eur J Immunol. (2005) 35:1175–83. doi: 10.1002/eji.200425949
90. Strik MC, De Koning PJ, Kleijmeer MJ, Bladergroen BA, Wolbink AM, Griffith JM, et al. Human mast cells produce and release the cytotoxic lymphocyte associated protease granzyme B upon activation. Mol Immunol. (2007) 44:3462–72. doi: 10.1016/j.molimm.2007.03.024
91. Classen CF, Bird PI, Debatin KM. Modulation of the granzyme B inhibitor proteinase inhibitor 9 (PI-9) by activation of lymphocytes and monocytes in vitro and by Epstein-Barr virus and bacterial infection. Clin Exp Immunol. (2006) 143:534–42. doi: 10.1111/j.1365-2249.2006.03006.x
92. Toossi Z, Wu M, Rojas R, Kalsdorf B, Aung H, Hirsch CS, et al. Induction of serine protease inhibitor 9 by Mycobacterium tuberculosis inhibits apoptosis and promotes survival of infected macrophages. J Infect Dis. (2012) 205:144–51. doi: 10.1093/infdis/jir697
93. Zhang M, Liu N, Park SM, Wang Y, Byrne S, Murmann AE, et al. Serine protease inhibitor 6-deficient mice have increased neutrophil immunity to Pseudomonas aeruginosa. J Immunol. (2007) 179:4390–6. doi: 10.4049/jimmunol.179.7.4390
94. Luo H, Ma C. Identification of prognostic genes in uveal melanoma microenvironment. PloS One. (2020) 15:e0242263. doi: 10.1371/journal.pone.0242263
95. Bladergroen BA, Meijer CJ, Ten Berge RL, Hack CE, Muris JJ, Dukers DF, et al. Expression of the granzyme B inhibitor, protease inhibitor 9, by tumor cells in patients with non-Hodgkin and Hodgkin lymphoma: a novel protective mechanism for tumor cells to circumvent the immune system? Blood. (2002) 99:232–7. doi: 10.1182/blood.V99.1.232
96. Bots M, Offringa R, Medema JP. Does the serpin PI-9 protect tumor cells? Blood. (2006) 107:4974–5. doi: 10.1182/blood-2006-01-0291
97. Godal R, Keilholz U, Uharek L, Letsch A, Asemissen AM, Busse A, et al. Lymphomas are sensitive to perforin-dependent cytotoxic pathways despite expression of PI-9 and overexpression of bcl-2. Blood. (2006) 107:3205–11. doi: 10.1182/blood-2005-07-2880
98. Fritsch K, Finke J, Grullich C. Suppression of granzyme B activity and caspase-3 activation in leukaemia cells constitutively expressing the protease inhibitor 9. Ann Hematol. (2013) 92:1603–9. doi: 10.1007/s00277-013-1846-6
99. Anft M, Netter P, Urlaub D, Prager I, Schaffner S, Watzl C. NK cell detachment from target cells is regulated by successful cytotoxicity and influences cytokine production. Cell Mol Immunol. (2020) 17:347–55. doi: 10.1038/s41423-019-0277-2
100. Vermijlen D, Luo D, Froelich CJ, Medema JP, Kummer JA, Willems E, et al. Hepatic natural killer cells exclusively kill splenic/blood natural killer-resistant tumor cells by the perforin/granzyme pathway. J Leukoc Biol. (2002) 72:668–76. doi: 10.1189/jlb.72.4.668
101. Ibanez-Molero S, Van Vliet A, Pozniak J, Hummelink K, Terry AM, Monkhorst K, et al. SERPINB9 is commonly amplified and high expression in cancer cells correlates with poor immune checkpoint blockade response. Oncoimmunology. (2022) 11:2139074. doi: 10.1080/2162402X.2022.2139074
102. Ray M, Hostetter DR, Loeb CR, Simko J, Craik CS. Inhibition of Granzyme B by PI-9 protects prostate cancer cells from apoptosis. Prostate. (2012) 72:846–55. doi: 10.1002/pros.21486
103. De Hooge AS, Berghuis D, Santos SJ, Mooiman E, Romeo S, Kummer JA, et al. Expression of cellular FLICE inhibitory protein, caspase-8, and protease inhibitor-9 in Ewing sarcoma and implications for susceptibility to cytotoxic pathways. Clin Cancer Res. (2007) 13:206–14. doi: 10.1158/1078-0432.CCR-06-1457
104. Tanaka K, Harashima N, Niiya F, Miyagi Y, Hida N, Ochi M, et al. Serine proteinase inhibitor 9 can be recognized by cytotoxic T lymphocytes of epithelial cancer patients. Jpn J Cancer Res. (2002) 93:198–208. doi: 10.1111/j.1349-7006.2002.tb01259.x
105. Han R, Yu L, Zhao C, Li Y, Ma Y, Zhai Y, et al. Inhibition of SerpinB9 to enhance granzyme B-based tumor therapy by using a modified biomimetic nanoplatform with a cascade strategy. Biomaterials. (2022) 288:121723. doi: 10.1016/j.biomaterials.2022.121723
106. Hamai A, Meslin F, Benlalam H, Jalil A, Mehrpour M, Faure F, et al. ICAM-1 has a critical role in the regulation of metastatic melanoma tumor susceptibility to CTL lysis by interfering with PI3K/AKT pathway. Cancer Res. (2008) 68:9854–64. doi: 10.1158/0008-5472.CAN-08-0719
107. Stout-Delgado HW, Getachew Y, Rogers TE, Miller BC, Thiele DL. The role of serpinb9/serine protease inhibitor 6 in preventing granzyme B-dependent hepatotoxicity. Hepatology. (2007) 46:1530–40. doi: 10.1002/hep.21820
108. Nakada S, Tsuneyama K, Kato I, Tabuchi Y, Takasaki I, Furusawa Y, et al. Identification of candidate genes involved in endogenous protection mechanisms against acute pancreatitis in mice. Biochem Biophys Res Commun. (2010) 391:1342–7. doi: 10.1016/j.bbrc.2009.12.047
109. Skat-Rordam J, Ipsen DH, Seemann SE, Latta M, Lykkesfeldt J, Tveden-Nyborg P. Modelling nonalcoholic steatohepatitis in vivo-A close transcriptomic similarity supports the Guinea pig disease model. Biomedicines. (2021) 9:1198. doi: 10.3390/biomedicines9091198
110. Pohjanen VM, Kokkonen TS, Arvonen M, Augustin MA, Patankar M, Turunen S, et al. Decreased expression of protease inhibitor 9, a granzyme B inhibitor, in celiac disease: a potential mechanism in enterocyte destruction and villous atrophy. Int J Immunopathol Pharmacol. (2013) 26:897–905. doi: 10.1177/039463201302600408
111. Kelly-Robinson GA, Reihill JA, Lundy FT, Mcgarvey LP, Lockhart JC, Litherland GJ, et al. The serpin superfamily and their role in the regulation and dysfunction of serine protease activity in COPD and other chronic lung diseases. Int J Mol Sci. (2021) 22:6351. doi: 10.3390/ijms22126351
112. Muthukumar T, Ding R, Dadhania D, Medeiros M, Li B, Sharma VK, et al. Serine proteinase inhibitor-9, an endogenous blocker of granzyme B/perforin lytic pathway, is hyperexpressed during acute rejection of renal allografts. Transplantation. (2003) 75:1565–70. doi: 10.1097/01.TP.0000058230.91518.2F
113. Deming Y, Dumitrescu L, Barnes LL, Thambisetty M, Kunkle B, Gifford KA, et al. Sex-specific genetic predictors of Alzheimer’s disease biomarkers. Acta Neuropathol. (2018) 136:857–72. doi: 10.1007/s00401-018-1881-4
114. Liao M, Li X, Zhang H, Zhou L, Shi L, Li W, et al. Effects and plasma proteomic analysis of GLP-1RA versus CPA/EE, in combination with metformin, on overweight PCOS women: a randomized controlled trial. Endocrine. (2023) 83:227–41. doi: 10.1007/s12020-023-03487-4
115. Stirewalt DL, Meshinchi S, Kopecky KJ, Fan W, Pogosova-Agadjanyan EL, Engel JH, et al. Identification of genes with abnormal expression changes in acute myeloid leukemia. Genes Chromosomes Cancer. (2008) 47:8–20. doi: 10.1002/gcc.20500
116. Oudejans JJ, Harijadi H, Kummer JA, Tan IB, Bloemena E, Middeldorp JM, et al. High numbers of granzyme B/CD8-positive tumour-infiltrating lymphocytes in nasopharyngeal carcinoma biopsies predict rapid fatal outcome in patients treated with curative intent. J Pathol. (2002) 198:468–75. doi: 10.1002/path.1236
117. Zhou B, Chen E, Chen J, Zhang J, Zhang N, Chen Z. Overexpression of proteinase inhibitor 9 is associated with poor prognosis in human hepatocellular carcinoma and with proliferation and apoptosis in HepG2 cells in vitro. Int J Clin Exp Pathol. (2019) 12:3719–27.
118. Rousalova I, Krepela E, Prochazka J, Cermak J, Benkova K. Expression of proteinase inhibitor-9/serpinB9 in non-small cell lung carcinoma cells and tissues. Int J Oncol. (2010) 36:275–83. doi: 10.3892/ijo_00000498
119. Dervovic D, Malik AA, Chen ELY, Narimatsu M, Adler N, Afiuni-Zadeh S, et al. In vivo CRISPR screens reveal Serpinb9 and Adam2 as regulators of immune therapy response in lung cancer. Nat Commun. (2023) 14:3150. doi: 10.1038/s41467-023-41255-0
120. Juric D, Lacayo NJ, Ramsey MC, Racevskis J, Wiernik PH, Rowe JM, et al. Differential gene expression patterns and interaction networks in BCR-ABL-positive and -negative adult acute lymphoblastic leukemias. J Clin Oncol. (2007) 25:1341–9. doi: 10.1200/JCO.2006.09.3534
121. Ten Berge RL, Oudejans JJ, Ossenkoppele GJ, Meijer CJ. ALK-negative systemic anaplastic large cell lymphoma: differential diagnostic and prognostic aspects–a review. J Pathol. (2003) 200:4–15. doi: 10.1002/path.1331
122. Sun W, Lv W, Lv H, Zhang R, Jiang Y. Genome-wide haplotype association analysis identifies SERPINB9, SERPINE2, GAK, and HSP90B1 as novel risk genes for oral squamous cell carcinoma. Tumour Biol. (2016) 37:1845–51. doi: 10.1007/s13277-015-3965-2
123. Van Kempen PM, Noorlag R, Swartz JE, Bovenschen N, Braunius WW, Vermeulen JF, et al. Oropharyngeal squamous cell carcinomas differentially express granzyme inhibitors. Cancer Immunol Immunother. (2016) 65:575–85. doi: 10.1007/s00262-016-1819-4
124. Vermeulen JF, Van Hecke W, Spliet WG, Villacorta Hidalgo J, Fisch P, Broekhuizen R, et al. Pediatric primitive neuroectodermal tumors of the central nervous system differentially express granzyme inhibitors. PloS One. (2016) 11:e0151465. doi: 10.1371/journal.pone.0151465
125. Soriano C, Mukaro V, Hodge G, Ahern J, Holmes M, Jersmann H, et al. Increased proteinase inhibitor-9 (PI-9) and reduced granzyme B in lung cancer: mechanism for immune evasion? Lung Cancer. (2012) 77:38–45. doi: 10.1016/j.lungcan.2012.01.017
126. Vycital O, Pitule P, Hosek P, Kriz T, Treska V, Liska V. Expression of serpin B9 as a prognostic factor of colorectal cancer. Anticancer Res. (2019) 39:6063–6. doi: 10.21873/anticanres.13813
127. Ten Berge RL, Meijer CJ, Dukers DF, Kummer JA, Bladergroen BA, Vos W, et al. Expression levels of apoptosis-related proteins predict clinical outcome in anaplastic large cell lymphoma. Blood. (2002) 99:4540–6. doi: 10.1182/blood.V99.12.4540
128. Bossard C, Belhadj K, Reyes F, Martin-Garcia N, Berger F, Kummer JA, et al. Expression of the granzyme B inhibitor PI9 predicts outcome in nasal NK/T-cell lymphoma: results of a Western series of 48 patients treated with first-line polychemotherapy within the Groupe d’Etude des Lymphomes de l’Adulte (GELA) trials. Blood. (2007) 109:2183–9. doi: 10.1182/blood-2006-07-033142
129. Chen Y, Quan L, Jia C, Guo Y, Wang X, Zhang Y, et al. Proteomics-based approach reveals the involvement of SERPINB9 in recurrent and relapsed multiple myeloma. J Proteome Res. (2021) 20:2673–86. doi: 10.1021/acs.jproteome.1c00007
130. Rowshani AT, Strik MC, Molenaar R, Yong SL, Wolbink AM, Bemelman FJ, et al. The granzyme B inhibitor SERPINB9 (protease inhibitor 9) circulates in blood and increases on primary cytomegalovirus infection after renal transplantation. J Infect Dis. (2005) 192:1908–11. doi: 10.1086/497606
131. Toossi Z, Wu M, Liu S, Hirsch CS, Walrath J, Van Ham M, et al. Role of protease inhibitor 9 in survival and replication of Mycobacterium tuberculosis in mononuclear phagocytes from HIV-1-infected patients. AIDS. (2014) 28:679–87. doi: 10.1097/QAD.0000000000000192
132. Mohammadpour H, Du W, O’neill R, Khalili S, Qiu J, Repasky EA, et al. Host-derived serine protease inhibitor 6 provides granzyme B-independent protection of intestinal epithelial cells in murine graft-versus-host disease. Biol Blood Marrow Transplant. (2018) 24:2397–408. doi: 10.1016/j.bbmt.2018.07.003
133. Peters FS, Peeters AMA, Van Den Bosch TPP, Mooyaart AL, Van De Wetering J, Betjes MGH, et al. Disrupted regulation of serpinB9 in circulating T cells is associated with an increased risk for post-transplant skin cancer. Clin Exp Immunol. (2019) 197:341–51. doi: 10.1111/cei.13309
134. Saito Y, Kondo H, Hojo Y. Granzyme B as a novel factor involved in cardiovascular diseases. J Cardiol. (2011) 57:141–7. doi: 10.1016/j.jjcc.2010.10.001
135. Sanad EF, Hamdy NM, El-Etriby AK, Sebak SA, El-Mesallamy HO. Peripheral leucocytes and tissue gene expression of granzyme B/perforin system and serpinB9: Impact on inflammation and insulin resistance in coronary atherosclerosis. Diabetes Res Clin Pract. (2017) 131:132–41. doi: 10.1016/j.diabres.2017.07.013
136. Ryer EJ, Ronning KE, Erdman R, Schworer CM, Elmore JR, Peeler TC, et al. The potential role of DNA methylation in abdominal aortic aneurysms. Int J Mol Sci. (2015) 16:11259–75. doi: 10.3390/ijms160511259
137. Toghill BJ, Saratzis A, Freeman PJ, Sylvius N, Collaborators U, Bown MJ. SMYD2 promoter DNA methylation is associated with abdominal aortic aneurysm (AAA) and SMYD2 expression in vascular smooth muscle cells. Clin Epigenet. (2018) 10:29. doi: 10.1186/s13148-018-0460-9
138. Deisting W, Raum T, Kufer P, Baeuerle PA, Munz M. Impact of diverse immune evasion mechanisms of cancer cells on T cells engaged by epCAM/CD3-bispecific antibody construct AMG 110. PloS One. (2015) 10:e0141669. doi: 10.1371/journal.pone.0141669
139. Kimman T, Slomp A, Martens A, Grabherr S, Li S, Van Diest E, et al. Serpin B9 controls tumor cell killing by CAR T cells. J Immunother Cancer. (2023) 11:e006364. doi: 10.1136/jitc-2022-006364
140. Fukuma D, Matsuyoshi H, Hirata S, Kurisaki A, Motomura Y, Yoshitake Y, et al. Cancer prevention with semi-allogeneic ES cell-derived dendritic cells. Biochem Biophys Res Commun. (2005) 335:5–13. doi: 10.1016/j.bbrc.2005.06.096
141. Kim TW, Hung CF, Boyd DA, He L, Lin CT, Kaiserman D, et al. Enhancement of DNA vaccine potency by coadministration of a tumor antigen gene and DNA encoding serine protease inhibitor-6. Cancer Res. (2004) 64:400–5. doi: 10.1158/0008-5472.CAN-03-1475
142. Liu Y, Balachandran YL, Li Z, Cong Y, Shao Y, Jiang X. Two dimensional nanosheets as immunoregulator improve HIV vaccine efficacy. Chem Sci. (2021) 13:178–87. doi: 10.1039/D1SC04044H
143. Welz M, Eickhoff S, Abdullah Z, Trebicka J, Gartlan KH, Spicer JA, et al. Perforin inhibition protects from lethal endothelial damage during fulminant viral hepatitis. Nat Commun. (2018) 9:4805. doi: 10.1038/s41467-018-07213-x
144. Huang H, Huang Y, Chen Y, Luo Z, Zhang Z, Sun R, et al. A novel immunochemotherapy based on targeting of cyclooxygenase and induction of immunogenic cell death. Biomaterials. (2021) 270:120708. doi: 10.1016/j.biomaterials.2021.120708
145. Huang T, Sun X, Qi Y, Yang X, Fan L, Chen M, et al. Immunogenic cell death effects induced by doxorubicin improved chemo-immunotherapy via restoration of granzyme B activity. Nano Res. (2023) 16:13250–8. doi: 10.1007/s12274-023-5581-6
146. Chen Y, Huang Y, Li Q, Luo Z, Zhang Z, Huang H, et al. Targeting Xkr8 via nanoparticle-mediated in situ co-delivery of siRNA and chemotherapy drugs for cancer immunochemotherapy. Nat Nanotechnol. (2023) 18:193–204. doi: 10.1038/s41565-022-01266-2
147. Luo Z, Huang Y, Batra N, Chen Y, Huang H, Wang Y, et al. Inhibition of iRhom1 by CD44-targeting nanocarrier for improved cancer immunochemotherapy. Nat Commun. (2024) 15:255. doi: 10.1038/s41467-023-44572-6
148. Wan Z, Huang H, West RE 3rd, Zhang M, Zhang B, Cai X, et al. Overcoming pancreatic cancer immune resistance by codelivery of CCR2 antagonist using a STING-activating gemcitabine-based nanocarrier. Mater Today (Kidlington). (2023) 62:33–50. doi: 10.1016/j.mattod.2022.11.008
149. Luo Z, Wan Z, Ren P, Zhang B, Huang Y, West RE 3rd, et al. In situ formation of fibronectin-enriched protein corona on epigenetic nanocarrier for enhanced synthetic lethal therapy. Adv Sci (Weinh). (2024) 11:e2307940. doi: 10.1002/advs.202307940
150. Sun J, Wan Z, Chen Y, Xu J, Luo Z, Parise RA, et al. Triple drugs co-delivered by a small gemcitabine-based carrier for pancreatic cancer immunochemotherapy. Acta Biomater. (2020) 106:289–300. doi: 10.1016/j.actbio.2020.01.039
151. Sun J, Wan Z, Xu J, Luo Z, Ren P, Zhang B, et al. Tumor size-dependent abscopal effect of polydopamine-coated all-in-one nanoparticles for immunochemo-photothermal therapy of early- and late-stage metastatic cancer. Biomaterials. (2021) 269:120629. doi: 10.1016/j.biomaterials.2020.120629
152. Li Z, Cao L, Yang C, Liu T, Zhao H, Luo X, et al. Protocatechuic acid-based supramolecular hydrogel targets serpinB9 to achieve local chemotherapy for OSCC. ACS Appl Mater Interfaces. (2022) 14:36379–94. doi: 10.1021/acsami.2c07534
153. Xu M, Zhang C, He S, Xu C, Wei X, Pu K. Activatable immunoprotease nanorestimulator for second near-infrared photothermal immunotherapy of cancer. ACS Nano. (2023) 17:8183–94. doi: 10.1021/acsnano.2c12066
154. Kaiserman D, Bird CH, Sun J, Matthews A, Ung K, Whisstock JC, et al. The major human and mouse granzymes are structurally and functionally divergent. J Cell Biol. (2006) 175:619–30. doi: 10.1083/jcb.200606073
155. Kaiserman D, Knaggs S, Scarff KL, Gillard A, Mirza G, Cadman M, et al. Comparison of human chromosome 6p25 with mouse chromosome 13 reveals a greatly expanded ov-serpin gene repertoire in the mouse. Genomics. (2002) 79:349–62. doi: 10.1006/geno.2002.6716
156. Bots M, Kolfschoten IG, Bres SA, Rademaker MT, De Roo GM, Kruse M, et al. SPI-CI and SPI-6 cooperate in the protection from effector cell-mediated cytotoxicity. Blood. (2005) 105:1153–61. doi: 10.1182/blood-2004-03-0791
157. Bots M, Bostelen LV, Rademaker MT, Offringa R, Medema JP. Serpins prevent granzyme-induced death in a species-specific manner. Immunol Cell Biol. (2006) 84:79–86. doi: 10.1111/j.1440-1711.2005.01417.x
158. Spring J, Khan AA, Lara S, O’grady K, Wilks J, Gurbuxani S, et al. Gut commensal bacteria enhance pathogenesis of a tumorigenic murine retrovirus. Cell Rep. (2022) 40:111341. doi: 10.1016/j.celrep.2022.111341
Keywords: SerpinB9, granzyme B, immunotherapy, immune cell, nanotechnology
Citation: Huang H, Mu Y and Li S (2024) The biological function of Serpinb9 and Serpinb9-based therapy. Front. Immunol. 15:1422113. doi: 10.3389/fimmu.2024.1422113
Received: 23 April 2024; Accepted: 10 June 2024;
Published: 20 June 2024.
Edited by:
Zhuoya Wan, AbbVie, United StatesReviewed by:
Peiqiang Yan, Harvard Medical School, United StatesCopyright © 2024 Huang, Mu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Song Li, c29sNEBwaXR0LmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.