
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol., 02 September 2024
Sec. Microbial Immunology
Volume 15 - 2024 | https://doi.org/10.3389/fimmu.2024.1419782
Bacterial infections remain a significant global health concern, necessitating a comprehensive understanding of the intricate host−pathogen interactions that play a critical role in the outcome of infectious diseases. Recent investigations have revealed that noncoding RNAs (ncRNAs) are key regulators of these complex interactions. Among them, long noncoding RNAs (lncRNAs) have gained significant attention because of their diverse regulatory roles in gene expression, cellular processes and the production of cytokines and chemokines in response to bacterial infections. The host utilizes lncRNAs as a defense mechanism to limit microbial pathogen invasion and replication. On the other hand, some host lncRNAs contribute to the establishment and maintenance of bacterial pathogen reservoirs within the host by promoting bacterial pathogen survival, replication, and dissemination. However, our understanding of host lncRNAs in the context of bacterial infections remains limited. This review focuses on the impact of host lncRNAs in shaping host−pathogen interactions, shedding light on their multifaceted functions in both host defense and bacterial survival, and paving the way for future research aimed at harnessing their regulatory potential for clinical applications.
One of the major breakthroughs in molecular biology over the last few decades has been the discovery and demonstration of long noncoding RNAs (lncRNAs). LncRNAs are a large and relatively novel class of RNAs transcribed from the genome, with a length of >200 nucleotides, and are generally characterized by the lack of protein-coding potential (1). LncRNAs function through a variety of mechanisms via their interactions with DNA, RNA and proteins, regulating various cellular processes in humans and animals (2, 3). Research investigating the engagement of host lncRNAs in host−pathogen interactions during bacterial infections is still emerging. In this review, we briefly discuss the role of host lncRNAs in immunity and then focus on the functional role and mechanisms of host lncRNAs in response to bacterial infections. Finally, we discuss the potential application of host lncRNAs and a perspective on lncRNA research in the context of bacterial infections.
The vast amounts of DNA between protein-coding genes were considered “junk” DNA in the 1970s (4). Two decades later, the discovery of lncRNAs and the identification of their biological functions revolutionized our view of this “junk’ DNA. In the 1980s, differential hybridization screening of eukaryotic cDNA libraries was used to study tissue-specific genes with a temporal pattern of expression. The first lncRNA, H19, was identified by screening a murine fetal liver cDNA library (5, 6). This lncRNA exhibits high sequence conservation between humans and mice and plays an important role in embryonic development (7, 8). The H19 locus and adjacent gene insulin-like growth Factor 2 (Igf2) are reciprocally imprinted and located on chromosome 7 in mice and chromosome 11p15.5 in humans. The expression of H19 and Igf2 is regulated by an intergenic differentially methylated region (DMR) upstream of H19 and several tissue-specific enhancers downstream of the H19 gene. As a result, H19 is exclusively expressed from the maternal chromosome, whereas Igf2 is expressed from the paternal allele (9). H19 was initially thought to be an mRNA until the discovery of another lncRNA, X-inactive-specific transcript (Xist), in the early 1990s. The Xist is critical for X chromosome inactivation in female mammals (8, 10, 11). Unlike H19, Xist has poor primary sequence conservation between humans and mice. Xist triggers gene silencing in cis in the X chromosome through the chromatin-binding region in female cells. This in turn catalyzes a cascade of chromatin changes and global spatial reorganization, ultimately leading to the silencing of the entire chromosome. Since then, H19 and Xist have been widely investigated, and dysregulation of these two lncRNAs has been shown to be involved in various human diseases and conditions, such as tumorigenesis, nervous system diseases, aging and inflammation (12–14).
LncRNAs have attracted increasing attention as a result of the completion of the Human Genome Project (HGP). One of the greatest advantages of HGP is the discovery that the number of protein-coding genes is far less than expected and that less than 2% of the human genome represents protein-coding regions (15–17). This was confirmed by the Encyclopedia of DNA Elements (ENCODE) project, which includes even larger-scale studies conducted in humans and mice (18, 19). Through various techniques, such as microarray hybridization and deep sequencing analyses, it is estimated that 70–90% of the human genome is transcribed, including protein-coding and noncoding RNAs, at some points during development (20–23). The rapid development of high-throughput DNA sequencing technologies and deep RNA sequencing for eukaryotic transcriptomes has led to an explosion in the number of newly identified and uncharacterized lncRNAs (24–26). Although there are arguments regarding poor conservation between humans and mice, shorter lifespans and fewer copies of lncRNAs than of mRNAs (27), it is becoming increasingly clear that lncRNAs have ubiquitous biological functions in translational inhibition, mRNA degradation, RNA decoys, the recruitment of chromatin modifiers, the regulation of protein activity and the availability of microRNAs via a sponging mechanism (28). However, considering lncRNA:microRNA stoichiometry, the extent to which lncRNAs function as sponges for microRNAs is a concern, particularly for those lncRNAs with low abundance. LncRNAs have been detected in various human body fluids and are potential diagnostic biomarkers for human diseases (29, 30). Additionally, an increasing number of studies indicate that lncRNAs are potential drug targets for the development of novel therapies for human diseases (3, 31).
Accumulating evidence suggests that lncRNAs play key roles in the proliferation, differentiation and activation of immune cells, including monocytes, macrophages, dendritic cells (DCs), neutrophils, granulocytes, T cells and B cells. Dysregulated expression of lncRNAs leads to aberrant immune responses in infectious diseases (Figure 1) (32, 33). The engagement of lncRNAs in immunity has been well described in previous reviews (34–36). In the following section, we provide a concise overview of the role of lncRNAs in the host immune response.
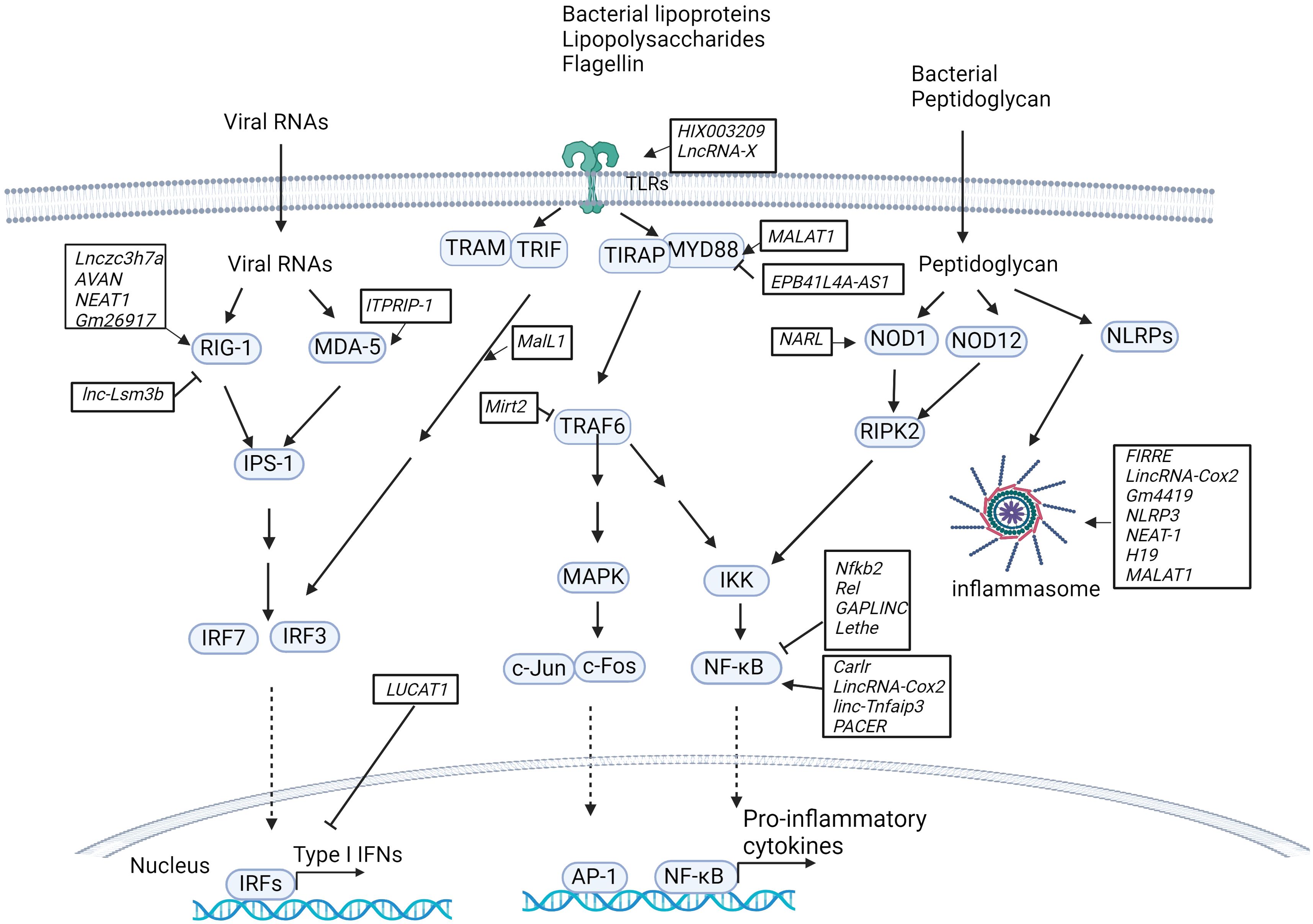
Figure 1. LncRNAs in innate immunity pathways. Host lncRNAs are engaged in various signaling pathways and cellular responses to invading microbial pathogens in immune cells. LncRNAs regulate the RIG-I/MDA-5-dependent nucleic acid-sensing pathways that recognize microbial RNAs in the cytosol of host cells and subsequently activate the expression of type I IFNs. LncRNAs are also important for TLR-dependent pathways in response to microbial infections and activate the expression of proinflammatory cytokines and type I IFNs. Additionally, host lncRNAs also regulate other cellular responses, such as NOD1/NOD12-dependent pathways and NLRP-dependent inflammasome activation.
Recent studies have revealed that lncRNAs constitute an essential part of the network involved in toll-like receptor (TLR) signaling pathways by acting as competitive endogenous RNAs (ceRNAs) that sponge microRNAs, directly interacting with transcription factors and subsequently activating or inhibiting the expression of downstream genes, regulating protein ubiquitination and activity, or mediating epigenetic chromatin remodeling (Figure 1). For example, by sponging miR-6089, the lncRNA HIX003209 restores the expression of TLR4 and the activation of the downstream signaling molecule NF-κB in human macrophages (37). The lncRNA XIST sponges miR-154-5p, which inhibits the expression of TLR5 in a rat neuropathic pain model (38). The lncRNA MALAT1 upregulates MyD88 by sponging miR-149 to promote LPS-induced inflammatory responses in a lung injury model (39). Lack of lncRNA EPB41L4A-AS1 expression reduces the enrichment of H3K9me3 in the MYD88 promoter region and subsequently promotes activation of the NF-κB pathway in human peripheral blood mononuclear cells (PBMCs) (40). The LPS-inducible lncRNA Mirt2, which serves as a repressor of inflammatory responses, negatively regulates TLR4 signaling. Mirt2 restricts the MyD88-dependent MAPK and NF-κB signaling cascades by interacting with TRAF6 and subsequently inhibiting TRAF6 oligomerization and ubiquitination in mouse macrophages (41).
In addition to the lncRNAs described above, several mammalian lncRNAs have been found to regulate the transcription factor NF-κB, a critical component of TLR-dependent pathways (Figure 1). These lncRNAs can serve as repressors or activators under various conditions. LncRNAs act as repressors: i) LncRNA Nfkb2 and lncRNA Rel regulate the innate immune response upon LPS stimulation in mouse bone marrow-derived macrophages (BMDMs) by binding to the transcription factors NF-κB, IRF3, JunB and c-Jun (42). ii) lncRNA Lethe exerts an anti-inflammatory effect by binding to the p65 subunit of NF-κB and blocking its binding to target promoters and the subsequent expression of downstream genes (43). iii) Gastric adenocarcinoma predictive long intergenic noncoding RNA (lncRNA GAPLINC), a lncRNA that was initially found in cancer progression, is highly expressed during human and mouse macrophage differentiation. A recent study indicated that the lncRNA GAPLINC downregulates innate immunity in response to endotoxic shock induced by bacterial lipopolysaccharide (LPS) (44). LncRNAs act as activators: i) lncRNA Carlr promotes the expression of NF-κB-regulated genes via interactions with the p65 subunit of NF-κB in human THP-1 cells (45). ii) LincRNA-Cox2 (also called Ptgs2os2) is known to regulate immune gene expression in immune cells. RNA-seq analysis revealed that lincRNA-Cox2 is one of the most highly induced lncRNAs following TLR2, TLR4 and TLR7/8 stimulation in mouse BMDMs. LincRNA-Cox2 regulates the expression of inflammatory genes by binding to heterogeneous nuclear ribonucleoproteins hnRNP-A/B and A2/B1 (46). It has also been reported that lincRNA-Cox2 is assembled into the switch/sucrose nonfermentable (SWI/SNF) complex after LPS stimulation in mouse macrophages, where it mediates SWI/SNF-associated chromatin remodeling and NF-κB activation (47). iii) Similarly, p50-associated COX-2 extragenic RNA (PACER) regulates COX-2 expression by interacting with the repressive subunit of NF-κB, p50, and thus activating the NF-κB pathway in human mammary epithelial cells and monocytes/macrophages (48). iv) LincRNA-Tnfaip3 interacts with high-mobility group Box 1 (HMGB1), which facilitates nucleosome remodeling and the accessibility of genomic DNA to the transcription factor NF-κB subunit p50 in mouse macrophages (49).
Additional lncRNAs that are involved in TLR-dependent signaling and the regulation of downstream gene expression have been identified. i) Myristoylated alanine-rich C kinase substrate (MARCKS) cis regulating the lncRNA promoter of cytokines and inflammation (MROCKI), also known as the regulator of cytokines and inflammation (ROCKI), acts as a master regulator of inflammatory responses through reducing MARCKS transcription and inflammatory gene expression in multiple TLR-stimulated human macrophages (48, 50). ii) Macrophage interferon-regulatory lncRNA 1 (MalL1) acts as an architectural RNA component of the TLR4-dependent pathway and stabilizes optineurin, a ubiquitin-adapter platforming TBK1 kinase, thus regulating TLR4–TRIF–IRF3-mediated type I interferon (IFN) expression in human macrophages (51). iii) LncRNA LUCAT1 is an inducible lncRNA that is localized primarily within the nucleus, where it functions as a negative feedback inhibitor of type I IFN and inflammatory gene expression by regulating the splicing and stability of nuclear receptor 4A2 (NR4A2) in human myeloid cells (52, 53). iv) LncRNAIL7–AS is a TLR2/TLR4-responsive gene that positively regulates the expression of inflammatory genes by modulating histone acetylation via interaction with the transcriptional coactivator p300 and assembly of SWI/SNF chromatin remodeling complexes at the promoter regions of these genes (54). v) The miR-155 host gene (MiR155HG) encodes a precursor RNA of microRNA-155 (miR-155) and lncRNA-155 in mice and humans. Previous studies have focused mainly on the role of miR-155 in inflammation and TLR-dependent pathways (55). More recently, the second product of MiR155HG, lncRNA-155, was shown to positively regulate the host response in immune cells and mice in the context of IAV infection (56). In contrast to that of miR-155, the role of lncRNA-155 in immunity remains to be defined. Additionally, determining the crosstalk between miR-155 and lncRNA-155 and understanding their unique roles in the host response to infections would be interesting.
NOD-like receptors (NLRs, also known as nucleotide-binding leucine-rich repeat receptors) play a key role in inflammatory and apoptotic responses by recognizing a variety of pathogen-associated molecular patterns (PAMPs) from microbial pathogens, such as bacterial peptidoglycan, flagellin and viral RNA. Host lncRNAs regulate NLR abundance and the formation and activation of inflammasomes (Figure 1). For example, by sponging miR-217-5p, the lncRNA NARL regulates the abundance of the NOD1 protein and, thus, downstream signaling in teleost fish (57). The lncRNAs FIRRE, lincRNA-Cox2 and Gm4419 bind and activate NF-κB and form a positive feedback loop to promote the activation of the NLRP3 inflammasome in cerebral microglia (58). Suppression of the lncRNA NLRP3 inhibits NLRP3-triggered inflammatory responses in early acute lung injury (59). The lncRNA nuclear paraspeckle assembly transcript 1 (lncRNA NEAT1) enhances NLRP3, NLRC4, and AIM2 inflammasome assembly and promotes caspase-1 activation, cytokine production, and pyroptosis in mouse macrophages (60). The lncRNA H19 significantly promotes NLRP3/6 inflammasome imbalance by sponging miR-21 to facilitate programmed cell death 4 (PDCD4) expression in mouse microglia (61). The lncRNA MALAT1 inhibits Nrf2 by regulating EZH2-mediated epigenetic repression, thereby increasing ROS levels and inflammasome activation in a Parkinson’s disease mouse model (62). The myocardial infarction-associated transcript (lncRNA MIAT) suppresses inflammasome-induced macrophage pyroptosis by inhibiting the expression of IL-1β and TNFα (63).
RIG-I-like receptors (RLRs) are key nucleic acid sensors that detect virus infections in immune cells (64). It has been shown that lncRNAs target the RLR-mediated signaling pathway, regulating the antiviral host response within immune cells, although these studies were performed in the context of virus infection. Some lncRNAs positively modulate the RLR signaling pathway by mediating the expression of type I IFNs (Figure 1). For example, the lncRNA Lnczc3h7a binds to both TRIM25 and RIG-I, serving as a molecular scaffold for stabilizing the RIG-I-TRIM25 complex and promoting RIG-I-mediated antiviral responses in mouse peritoneal macrophages (65). The lncRNA AVAN also binds to TRIM25 and promotes the interaction between TRIM25 and RIG-I. Additionally, the lncRNA AVAN positively regulates FOXO3a expression by remodeling the FOXO3a promoter region, facilitating neutrophil chemotaxis and recruitment in influenza A virus (IAV)-infected patients (66). The lncRNA NEAT1 is induced by virus infection, such as influenza virus, HSV, Hantaan virus (HTNV) and SARS-CoV-2 virus (67–69). NEAT1 interacts with the splicing factor SFPQ and upregulates the expression of RIG-I (70). Gm26917 reduces the degradation of RIG-I mRNA by sponging miR-124-3p in SARS-CoV-infected cells (67). The lncRNA ITPRIP-1 binds to the C-terminus of MDA5, another cytosolic RNA sensor, and promotes MDA5 oligomerization and activation in hepatitis C virus-infected cells (71). Some lncRNAs serve as negative regulators of the RLR-mediated signaling pathway, leading to the inhibition of the antiviral response. For example, lnc-Lsm3b inhibits RIG-I activation by competing with viral RNAs for the binding of RIG-I monomers and therefore attenuates the RIG-I-mediated antiviral response in vesicular stomatitis virus-infected mouse macrophages (72).
As described above, lncRNAs regulate cellular pathways that are involved in various biological processes, including host−pathogen interactions during bacterial infections caused by Mycobacterium tuberculosis, Salmonella typhimurium, Listeria monocytogenes and Pseudomonas aeruginosa. In the following sections, we discuss the role of host lncRNAs in host–bacteria interactions and their potential applications as novel diagnostic biomarkers and in host-directed therapy (Table 1).
Mycobacterium tuberculosis (M. tuberculosis) is an intracellular bacterial pathogen that causes tuberculosis (TB), one of the deadliest infectious diseases in humans. According to a World Health Organization (WHO) report, M. tuberculosis has infected approximately one-third of the world’s population, leading to ~1.5 million active TB cases and ~1.0 million deaths annually (90). TB is transmitted primarily through the inhalation of respiratory droplets containing M. tuberculosis. The transmission of TB occurs when an infected individual expels these droplets into the air through activities such as coughing, sneezing, talking, or even singing. Upon inhalation, M. tuberculosis is engulfed primarily by alveolar macrophages, resulting in three distinct consequences: clearance and active or latent infection (91–93). The engagement of host lncRNAs in each infection stage remains to be defined. However, increasing evidence indicates that host lncRNAs play important roles in host–M. tuberculosis interactions in macrophages, the primary host cells involved in M. tuberculosis intracellular survival. The expression of several host lncRNAs is differentially regulated in human (94, 95) or mouse (96, 97) macrophages following M. tuberculosis infection in cell culture. Differential expression of host lncRNAs was also detected in peripheral blood samples from patients with active pulmonary TB compared with healthy controls (98, 99). The role of the majority of these differentially expressed host lncRNAs in host–Mycobacteria interactions remains unclear.
The lncRNA NEAT1 regulates multiple cellular processes, including gene regulation, the stress response, and immune modulation, by interacting with various proteins and miRNAs in mammalian cells. As described above, NEAT1 is involved in the formation of nuclear bodies called paraspeckles by serving as a scaffold for several proteins, including splicing factor, proline, glutamine rich (SFPQ), non-POU domain-containing cctamer-binding protein (NONO), polypyrimidine tract-binding protein-associated splicing factor (PSF), fused in sarcoma (FUS), RNA-binding fox-1 homolog 2 (RBFOX2) and matrin 3 (MATR3) (100). In addition to interacting with proteins, NEAT1 can also interact with miRNAs, such as miR-377, by acting as a “sponge” or “decoy” (101). The expression of NEAT1 has been reported to be upregulated in human THP-1-derived macrophages after M. tuberculosis infection in cell culture, and this host lncRNA is crucial for controlling M. tuberculosis replication within macrophages (73). The mechanism of action of NEAT1 in macrophages during M. tuberculosis infection remains unclear.
The lncRNA HOX transcript antisense RNA (HOTAIR) is known for its involvement in epigenetic regulation and its impact on gene expression patterns (102, 103). The HOTAIR gene is located within the HOXC gene cluster on chromosome 12 and is transcribed from the opposite strand of the HOXC locus. In a recent study, Subuddhi et al. reported that the expression of the lncRNA HOTAIR is downregulated in virulent M. tuberculosis H37Rv-infected human THP-1-derived macrophages compared with that in cells infected with avirulent M. tuberculosis H37Ra. Furthermore, the lncRNA HOTAIR inhibits the production of dual-specific MAP kinase phosphatase 4 (DUSP4) and specific AT-rich sequence binding protein 1 (SATB1), two host proteins that facilitate the survival of virulent M. tuberculosis in THP-1-derived macrophages, by increasing the trimethylation of histone H3 at lysine 27 (H3K27me3) around the transcription start sites (TSSs) of these two genes (74). Therefore, this study suggests that M. tuberculosis has evolved a mechanism for its intracellular survival by downregulating the lncRNA HOTAIR-mediated antimycobacterial activity in macrophages.
LincRNA-Cox2 is located on chromosome 1 and is transcribed from the antisense strand in mice. The mature sequence of lincRNA-Cox2 comprises 2 exons and spans a length of 1.7 kb. Its closest neighboring protein-coding gene, prostaglandin-endoperoxide synthase 2 (Ptgs2 or Cox2), is located 50 kb upstream and is transcribed from the sense strand (104). Mouse lincRNA-Cox2 is homologous to the human locus that contains p50-associated cyclooxygenase 2 extragenic RNA (PACER) and Cox2-divergent lncRNAs (105). The expression of lincRNA-Cox2 is induced during macrophage activation. Additionally, lincRNA-Cox2 regulates the expression of immune response genes in a cis- and trans-immune-regulatory manner (46). In mouse macrophages, lincRNA-Cox2 has been reported to regulate the host antimycobacterial response and restrict mycobacterial intracellular survival (75). The mechanism of lincRNA-Cox2-mediated antimycobacterial activity in macrophages remains unclear. COX2 is an enzyme involved in the synthesis of prostaglandins that play an important role in host defense against M. tuberculosis infection in macrophages (106). Therefore, one possible mechanism is that lincRNA-Cox2 regulates the expression of the Cox2 gene via inhibition of the transcriptional regulator NF-κB (48). In the context of M. bovis BCG infection, a mycobacterium strain closely related to M. tuberculosis, the expression of lincRNA-Cox2 is induced via a TLR2-dependent pathway in mouse RAW 264.7 cells. Interestingly, the knockdown of lincRNA-Cox2 increases M. bovis BCG-induced apoptosis in mouse macrophages (78).
LncRNA-early secreted parget with a molecular weight of 12 kDa (Lnc-EST12) is a recently identified host lncRNA that facilitates M. tuberculosis survival in mouse macrophages (76). The expression of lnc-EST12 is partially downregulated by the M. tuberculosis protein EST12 via the JAK2-STAT5a signaling pathway in mouse BMDMs and RAW 264.7 cells during mycobacterial infection. However, lnc-EST12 downregulates the expression of several antimycobacterial proinflammatory cytokines, including IL-1β, IL-6, and CCL5/8. Additionally, lnc-EST12 attenuates the activation of the EST12-induced NLRP3 inflammasome and GSDMD pyroptosis-IL-1β immune pathway, which play crucial roles in host immunity in response to M. tuberculosis infection. These regulatory roles of lnc-EST12 in M. tuberculosis-infected macrophages occur when it interacts with the transcription factor far upstream of element-binding protein 3 (FUBP3). Interestingly, lnc-EST12 expression is still partially inhibited by the ΔEST12 M.tb strain compared with the uninfected strain in mouse BMDMs in cell culture, suggesting that additional mycobacterial factors suppress lnc-EST12 expression in addition to EST12 in macrophages (76).
Autophagy and apoptosis are two critical cellular pathways involved in controlling mycobacterial infection in macrophages (107). Several host lncRNAs have been found to regulate these two pathways. These lncRNAs include lncRNA-MIAT (79), PCEED1B-AS1 (77), lncRNA-MEG3 (80) and lncRNA-EPS (81). lncRNA-MIAT is also known as a gene associated with retinoid-IFN-induced mortality 19 (GRIM-19) in mice. It has been studied primarily in the context of cardiovascular diseases, particularly its association with myocardial infarction (heart attack) (108). It was recently reported that the expression of the lncRNA MIAT is highly induced in human THP-1 cells infected with M. bovis BCG. In the same study, it was reported that lncRNA-MIAT represses autophagy and apoptosis by downregulating the miR-665/ULK1 axis in THP-1-derived macrophages in response to M. bovis BCG infection (79). Maternally expressed gene 3 (lncRNA-MEG3) is a well-studied lncRNA that plays an important role in various biological processes. Several studies have demonstrated that lncRNA-MEG3 acts as a tumor suppressor. It inhibits cell proliferation and promotes apoptosis, ultimately suppressing the progression of various tumors, such as glioma and hepatocellular carcinoma (109, 110). Moreover, lncRNA-MEG3 is also expressed in the brain and has been implicated in neurodevelopment and neuronal functions (111, 112). In response to M. bovis BCG infection, the expression of lncRNA-MEG3 was downregulated in human macrophages. Knockdown of lncRNA-MEG3 increases autophagy and M. bovis BCG killing in human THP-1-derived macrophages (80). Compared with the lncRNAs described above, PCED1B antisense RNA 1 (PCED1B-AS1) is a newly discovered and less studied lncRNA. PCED1B-AS1 has been found to facilitate the progression of multiple human tumors, including glioma, pancreatic ductal adenocarcinoma and hepatocellular carcinoma (113). Recently, the expression of PCED1B-AS1 was reported to be attenuated in CD14+ monocytes from patients with active tuberculosis compared with those from healthy individuals. Additionally, PCED1B-AS1 regulates apoptosis and autophagy by binding to miR-155 as a miRNA sponge in human macrophages (77). Similar to PCED1B-AS1, the lncRNA erythroid pro-survival (lncRNA-EPS) is also a less-studied lncRNA. In mouse RAW 264.7 cells, lncRNA-EPS regulates apoptosis and autophagy via the KNK/MAPK-dependent pathway during M. bovis BCG infection (81).
Salmonella typhimurium (S. typhimurium) is a bacterial species belonging to the genus Salmonella. It is a pathogenic bacterium that can cause a type of foodborne disease known as salmonellosis in humans and animals. Like M. tuberculosis, S. typhimurium uses macrophages as primary host cells for their intracellular replication and has evolved various survival mechanisms to evade the host defense system within macrophages, such as inhibiting the fusion of S. typhimurium-containing phagosomes with lysosomes (114). In addition to macrophages, S. typhimurium adheres to epithelial cells lining the gut and can invade epithelial cells and disrupt tight junctions. Therefore, the interaction between S. typhimurium and host cells (such as macrophages and epithelial cells) in the intestine determines the consequences of S. typhimurium infection. Host lncRNAs also play essential roles in host–S. typhimurium interactions.
LncRNA NeST (Nettoie Salmonella pas Theiler’s, also called Tmevpg1) is a lncRNA gene located adjacent to the IFN-γ-encoding gene in both mice (Ifng) and humans (IFNG). The lncRNA NeST has been identified as an important regulator of immune responses, particularly in the context of T-cell activation (82). The lncRNA NeST is expressed primarily in T lymphocytes, including CD4+ and CD8+ T cells, which play a central role in coordinating immune responses (32). The lncRNA NeST has gained attention for its role in regulating the production of IFN-γ, an important cytokine in the immune response to microbial infections. The lncRNA NeST functions as an enhancer RNA, which elevates the expression of nearby genes by interacting with transcription factors. In the context of S. typhimurium infection, the lncRNA NeST interacts with the protein WD repeat domain 5 (WDR5), a component of chromatin-modifying complexes, leading to epigenetic alterations that promote IFN-γ expression in CD8+ T cells in mice. As a result, the lncRNA NeST contributes to host resistance to S. typhimurium infection (82).
The lncRNA NEAT1 variant 2 (NEAT1_2) is a splice variant of the lncRNA NEAT1 gene that we described above in the context of M. tuberculosis infection (100). Similarly, the lncRNA NEAT1_2 is involved in the formation of the subnuclear structure paraspeckles in mammalian cells. S. typhimurium infection significantly induces the expression of the lncRNA NEAT1_2 in HeLa cells via the release of lncRNA degradation by the RNA exosome, aided by the nuclear exosome targeting (NEXT) complex. A decreased level of the NEAT1_2 transcript in S. typhimurium-infected HeLa cells impaired the expression of immune-related genes, including TNF superfamily member 9 (TNFSF9), C-C motif chemokine ligand 2 (CCL2), and colony stimulating factor 1 (CSF1). Additionally, attenuated expression of the lncRNA NEAT1_2 increases host susceptibility to S. typhimurium infection in HeLa cells (83).
Listeria monocytogenes (L. monocytogenes) is a bacterium that can cause a serious foodborne disease called listeriosis, which particularly infects certain populations, such as pregnant women, newborns, elderly individuals, and people with compromised immune systems. L. monocytogenes can be transmitted through the consumption of contaminated food, especially ready-to-eat foods that are not properly cooked or stored. L. monocytogenes has evolved mechanisms to invade host cells, particularly epithelial cells and phagocytic cells such as macrophages. It uses specialized proteins, such as listeriolysin O (LLO), to facilitate entry into cells and replication within the host cell cytoplasm. Like M. tuberculosis and S. typhimurium, L. monocytogenes is capable of replicating within host cells, which allows it to avoid immune detection and contributes to its persistence in the host (115). Host lncRNAs are involved in the intricate crosstalk between host cells and L. monocytogenes, determining the consequences of infection.
Several host lncRNAs, including lincRNA-Cox2, antisense-interleukin-1 a-subunit (AS-IL1a), Sros1 and lincRNA-EPS, have been studied in the context of L. monocytogenes infection within host cells (32, 46, 84–86). The expression of LincRNA-Cox2 and AS-IL1a is induced by L. monocytogenes infection within host cells both in vitro and in vivo. Within L. monocytogenes-infected mouse BMDMs, AS-IL1α acts as a transcriptional enhancer to recruit RNA polymerase II (RNAPII) to the IL-1α promoter, driving IL-1α expression (46, 84). In contrast to LincRNA-Cox2 and AS-IL1a, the expression of Sros1 and lincRNA-EPS is downregulated during L. monocytogenes infection in mouse BMDMs in cell culture (85, 86). The lncRNA Sros1 can bind to the mRNA of the transcription factor signal transducer and activator of transcription 1 (Stat1) and subsequently prevent its interaction with the RNA-binding protein cytoplasmic activation/proliferation-associated protein-1 (CAPRIN1), which stabilizes the Stat1 mRNA in mouse macrophages. During L. monocytogenes infection, the lncRNA Sros1 is degraded via the miR-1-mediated pathway in mouse BMDMs. It therefore increases the stability of Stat1 mRNA and subsequently elevates the expression of immune-related genes that are critical for host defense against bacterial infection (85). LincRNA-EPS deficiency increases the expression of proinflammatory cytokine genes (including IL-6, TNFα, IL-1β and Ccl5) and inducible nitric oxide synthase (iNOS)), as well as the production of nitric oxide (NO), in mouse BMDMs after L. monocytogenes infection compared with that in wild-type mouse BMMs. These host factors are important antibacterial molecules within host cells against intracellular bacterial pathogens. As expected, lincRNA-EPS-deficient mice are more resistant to L. monocytogenes infection than wild-type mice (86).
Pseudomonas aeruginosa (P. aeruginosa) is an opportunistic bacterial pathogen that can cause serious infections in individuals with compromised immune systems, such as those with cystic fibrosis, burn patients, and chronic wounds. The infection sites include the respiratory tract, urinary tract, blood, skin and soft tissues (116, 117). Host lncRNAs play important roles in host−pathogen interactions during P. aeruginosa infection.
The lncRNA MEG3 transcript 4 (lncRNA MEG3-4) is one of 10 isoforms of the lncRNA MEG3 in mice. Recently, P. aeruginosa lung infection was shown to downregulate the expression of the lncRNA MEG3-4 in the lungs and livers of C57BL/6 mice via the TLR4/NF-κB-mediated pathway (87). Interestingly, the mice receiving engineered MH-S cells that overexpress lncRNA MEG3-4 are more susceptible to P. aeruginosa lung infection than those receiving MH-S cells with only the empty vector. It was further found that lncRNA MEG3-4 acts as a miRNA sponge for miRNA-138, which binds to the mRNA of IL-1β and downregulates the production of the IL-1β protein. In a separate study, Balloy et al. analyzed the lncRNA expression profile in bronchial epithelial cells that were isolated from patients with cystic fibrosis (CF) or healthy donors and then infected with P. aeruginosa in vitro in cell culture for 0, 2, 4 or 6 hr. As a result, they identified 108 unique host lncRNAs that were differentially expressed in P. aeruginosa-infected CF bronchial epithelial cells compared with infected non-CF cells (88). Among them, the expression of 12 lncRNAs was differentially regulated at least at two time points, and the expression of two lncRNAs, LINC00862 and CTD-2619J13, was differentially regulated at all four time points after P. aeruginosa infection. Interestingly, the expression of the lncRNA MEG3 was downregulated in CF bronchial epithelial cells compared with non-CF cells at 4 and 6 hr post-P. aeruginosa infection.
Legionella pneumophila (L. pneumophila) is an intracellular bacterial pathogen that causes legionellosis, a severe and potentially life-threatening pneumonia in humans. L. pneumophila enters the human body primarily through the inhalation of aerosols. Like M. tuberculosis and S. typhimurium, L. pneumophila has evolved the ability to infect and replicate within human cells, particularly within macrophages. After entry into macrophages, L. pneumophila prevents the fusion of Legionella-containing phagosomes with lysosomes, which contain enzymes that can degrade invading pathogens. This prevents bacteria from being destroyed and allows them to establish a specialized compartment called the Legionella-containing vacuole (LCV) as a replication niche for intracellular L. pneumophila (118, 119). Similarly, host lncRNAs play a critical role in the interaction between L. pneumophila and host cells.
The lncRNA MaIL1 (also known as MAILR) is an intergenic lncRNA in humans. The expression of MaIL1 is upregulated via a TLR4-dependent pathway in human blood-derived macrophages in vitro after L. pneumophila infection. Knockdown of the lncRNA MaIL1 significantly increases L. pneumophila survival and replication within human MDMs, which correlates with attenuated production of the type I IFNs IFNα and IFNβ in Legionella-infected macrophages in cell culture (51). Interestingly, MaIL1 deficiency-associated host defense failure in L. pneumophila can be rescued by the addition of exogenous type I IFNs. It was further demonstrated that the lncRNA MaIL1 interacts with optineurin, a ubiquitin-adapter protein that binds and regulates TBK1 kinase activity. Therefore, the lncRNA MaIL1 indirectly upregulates the protein kinase TBK1 and the downstream transcription factor IRF3, ultimately resulting in increased expression of type I IFNs in human macrophages in response to L. pneumophila infection. The same study revealed that the expression of another lncRNA, Linc01215, was also induced in human blood-derived macrophages after L. pneumophila infection. However, the role of linc01215 in the macrophage response to L. pneumophila remains to be defined (51).
Brucella abortus (B. abortus) is a bacterial pathogen that causes brucellosis, a zoonotic infectious disease that primarily affects animals but can also be transmitted to humans. B. abortus enters the host body through mucosal surfaces, such as the respiratory or digestive tract. Once inside, the bacterium is taken up by phagocytic cells such as macrophages. Like M. tuberculosis, B. abortus can evade destruction by inhibiting phagolysosome maturation within macrophages. Additionally, B. abortus induces the formation of a specialized membrane-bound compartment called the “Brucellosome”. This compartment is derived from the endoplasmic reticulum (ER) and provides an environment favorable for bacterial replication (120).
The lncRNA Gm28309 in mice is an ortholog of the human lncRNA P33714. By analyzing differentially expressed lncRNAs in B. abortus-infected human THP-1 cells (vs. uninfected cells) via the human long noncoding RNA V3.0 gene chip, Deng et al. identified 235 differentially expressed lncRNAs (171 upregulated and 64 downregulated) at 24 hr after B. abortus infection (89). Six lncRNAs, namely, P662, P30159, P16218, P3852, P33714 and P12873, were predicted to be involved in the inflammation pathway via KEGG pathway analysis. Among them, the expression of P33714 was significantly downregulated in B. abortus-infected human THP-1 cells compared with that in uninfected cells. Additionally, the knockdown of P33714 increased the expression of the cytokines IL-1β and IL-18 and impaired B. abortus intracellular survival in human THP-1 cells. Similarly, the expression of the mouse ortholog of P33714, Gm28309, was also downregulated in B. abortus-infected mouse macrophage line RAW 264.7 compared with that in uninfected cells. Additionally, overexpression of Gm28309 inhibited the activation of the NLRP3 inflammasome and the expression of IL-1β and IL-18 in B. abortus-infected mouse RAW 264.7 cells. This study further proposed that Gm28309 downregulates NLRP3 inflammasome activation likely via two regulatory mechanisms: i) Gm28309-mediated reduction in TGF-β production attenuates the activation of TAK1 and IKK kinases and subsequent p65 phosphorylation; and ii) Gm28309 acts as a miRNA sponge for miR-3068-5p, which releases kB-Ras2 from NF-κB and promotes p65 phosphorylation. In both cases, the activated NF-κB pathway is required for the activation of the NLRP3 inflammasome (89).
Interferon gamma antisense RNA 1 (lncRNA IFNG-AS1, also known as TMEVPG1 or NeST), as we described above, is a lncRNA that is transcribed from the opposite strand of the gene encoding IFN-γ. The lncRNA IFNG-AS1 regulates immune responses in the host during microbial infections, including enhancing the expression of INF-γ (121, 122). Compared with those in healthy control cells, the expression of the lncRNAs IFNG-AS1 and IFN-γ in peripheral blood mononuclear cells (PBMCs) isolated from patients with brucellosis is highly upregulated. Furthermore, it is correlated with increased production of IFN-γ in the serum of patients with brucellosis, suggesting an antibacterial activity of the lncRNA IFNG-AS1 in response to B. abortus infection (123).
LncRNAs have shown great potential as diagnostic biomarkers for various diseases because of their tissue-specific expression patterns, stability, and dysregulation in different pathological conditions. Moreover, lncRNAs have been detected in various human body fluids, including blood, urine, saliva, cerebrospinal fluid, seminal fluid and breast milk (124–128). Their presence in these fluids makes them attractive candidates for noninvasive diagnostic and prognostic biomarkers for infectious diseases, especially for those that are challenging to diagnose due to atypical symptoms, limited access to health care and/or the lack of appropriate diagnostic tools. For example, active TB patients have nonspecific symptoms such as cough, fever, and weight loss, which can be mistaken for other respiratory diseases. Traditional diagnostic methods include sputum smear microscopy and mycobacterial culture-based tests. However, sputum smear microscopy may have low sensitivity, and culture-based methods take time. Advanced diagnostic techniques, the GeneXpert MTB/RIF test and the interferon-gamma release assay (IGRA) are becoming more accessible but may not be available in all health care settings, especially in high-burden TB developing countries. Additionally, the GeneXpert MTB/RIF test and IGRA have limited accuracy in TB patients coinfected with HIV, children and extrapulmonary TB patients (129). Therefore, novel and cost-effective diagnostic tools are urgently needed to meet the WHO End TB Strategy, which targets a 90% reduction in patients suffering from TB and a 95% reduction in deaths from TB by 2035 (130).
An increasing number of studies have demonstrated that the expression levels of host lncRNAs are differentially regulated in M. tuberculosis-residing macrophages and the blood of TB patients, indicating the potential application of host lncRNAs as novel biomarkers for TB diagnosis and treatment outcomes. Chen et al. analyzed the lncRNA expression profile in the plasma of 33 TB patients and 11 healthy control subjects via the Arraystar Human LncRNA Microarray V3.0 and identified 511 differentially expressed lncRNAs (163 upregulated and 348 downregulated) in TB patients, such as NR_038221, NR_003142, ENST00000570366 and ENST00000422183. Pathway analysis revealed that these differentially expressed lncRNAs were enriched mainly in the regulation of alpha-beta T-cell activation and the T-cell receptor signaling pathway (131). A similar result was observed in serum exosomes isolated from active TB patients and healthy individuals. By using a public GEO dataset (GSE94907), Fang et al. reported that the abundance of 9 lncRNAs was differentially regulated in serum exosomes isolated from active TB patients compared with exosomes from healthy individuals (132). Among them, four lncRNAs, NONHSAT101518.2, NONHSAT067134.2, NONHSAT148822.1 and NONHSAT078957.2, were verified using plasma samples collected from 69 active TB patients and 69 healthy individuals. The results revealed that the expression of all 4 lncRNAs was significantly lower in the plasma of active TB patients than in that of healthy controls. Taken together, these findings suggest that host lncRNAs represent a potential new strategy for TB diagnosis. However, it is possible that these host lncRNAs are also regulated by other microbial infections and are not specific for mycobacterial infection. Further studies are warranted to explore the specificity of host lncRNAs for TB diagnosis, particularly compared with other microbial infections.
To investigate the potential application of host lncRNAs as biomarkers for pulmonary TB, Hu et al. established a diagnostic TB model in combination with a host lncRNA expression profile and patient electronic health records (EHRs) (98). In this study, the lncRNA expression profiles of PBMCs from 7 clinically diagnosed pulmonary TB patients and 5 healthy donors were analyzed via Affymetrix human transcriptome array 2.0 chips. As a result, they identified a total of 325 lncRNAs that were differentially expressed (287 upregulated and 38 downregulated) between clinically diagnosed pulmonary TB patients and healthy controls. The top five lncRNAs were subsequently selected based on the following criteria: a fold change of 2 between two groups, a P value of < 0.05, a signal intensity of > 25, and unreported lncRNAs in the TB literature. This top 5 list includes three upregulated lncRNAs (n335265, ENST00000518552, and TCONS_00013664) and two downregulated lncRNAs (n333737 and ENST00000497872) in pulmonary TB patients compared with the control group. The expression of these five lncRNAs was further evaluated in samples from 141 clinically diagnosed pulmonary TB patients, 159 nontuberculosis disease controls, and 578 healthy individuals. The expression of n333737, ENST00000497872 and n335265 was differentially regulated in pulmonary TB patients compared with the other two groups, as shown by the microarray results. The remaining two lncRNAs, ENST00000518552 and TCONS_00013664, were excluded from the further modeling step because of their low expression levels. To improve the accuracy of diagnosis, a logistic regression model was trained on three selected host lncRNAs and six EHRs (age, hemoglobin, weight loss, low-grade fever, calcification detected by computed tomography, and IGRA) from the same cohort. The established diagnostic model was validated via a new set of patient data consisting of 97 clinically diagnosed pulmonary TB patients, 140 nontuberculosis disease controls, 392 microbiologically confirmed pulmonary TB patients and 245 healthy individuals. This new diagnostic model works better than the EHR model in discriminating pulmonary TB, including microbiologically confirmed and smear-negative cases (98).
In addition to their application as novel diagnostic biomarkers, host lncRNAs may also be used as criteria for TB treatment outcomes. The host lncRNA NEAT1 (both NEAT1_1 and NEAT1_2) is highly expressed in PBMCs from patients with pulmonary TB compared with those from healthy individuals. Consistently, the expression of both NEAT1_1 and NEAT1_2 is induced by M. tuberculosis infection in THP-1-derived macrophages in cell culture. Interestingly, the expression levels of NEAT1_1 and NEAT1_2 in the PBMCs of pulmonary TB patients correlate with TB treatment and decline over time (73). A high level of the lncRNA LOC152742 was also detected in the plasma of active pulmonary TB patients. However, the abundance of the lncRNA LOC152742 in pulmonary TB patients decreases over the course of TB treatment (133). Similarly, the expression levels of the lncRNAs n333737 and ENST00000497872 are significantly increased in pulmonary TB patients who respond well to TB treatment (98).
RNA-based therapy is a type of medical treatment that utilizes RNA molecules, such as miRNAs, mRNAs and lncRNAs, as therapeutic agents to modulate gene expression, with the goal of treating certain diseases. Currently, 11 RNA-based therapeutics, either small interfering RNAs (siRNAs) or antisense oligonucleotides (ASOs), have been approved by the FDA or the European Medicines Agency (EMA), which target gene expression in the liver, muscle, or central nervous system in human diseases (134). Host lncRNAs have gained significant attention in recent years because of their diverse roles in regulating the cellular response to bacterial infection, as described above. Additionally, many host lncRNAs have structures similar to mRNAs, including poly-A tails and 5’-caps, and both are synthesized by RNA polymerase II. The recent success of mRNA-based vaccines has encouraged the potential application of host lncRNAs as novel therapeutic approaches for infectious diseases (135). In summary, although the therapeutic potential of lncRNAs is still being explored, an increasing number of studies indicate an important role for lncRNAs in the pathophysiology of diseases caused by bacterial pathogens, suggesting their usefulness in host-directed therapies.
The insights gained from this review contribute to a deeper understanding of the mechanisms, challenges, and potential avenues for further research in the field. Our examination of the literature has revealed the complexities surrounding the role of host lncRNAs in host−pathogen interactions during bacterial infections, and our understanding of host lncRNAs in bacterial infections is still at an early stage relative to other research fields of lncRNAs, such as cancers, in mammalian cells. However, emerging discoveries, especially with respect to intracellular bacterial pathogens (such as M. tuberculosis, S. typhimurium and L. monocytogenes), as described in this review, have revealed the importance of host lncRNAs in the complex interactions between bacterial pathogens and their hosts. Research on host lncRNAs will enrich our understanding of immune responses and the survival of bacterial pathogens within host cells. Additionally, this growth holds the potential to drive the advancement of host lncRNAs as novel diagnostic and/or host-directed therapies for combatting bacterial infections. Further efforts are clearly needed to precisely delineate the lncRNA-dependent regulatory mechanisms, including the intracellular signaling pathways that regulate the expression of lncRNAs and/or are regulated by lncRNAs and lncRNA-interacting host molecules (such as genomic sites, miRNAs and proteins) within host cells. Furthermore, additional studies are needed to understand the role of lncRNAs in bacteria−host interactions in animal models such as mice.
YC: Writing – review & editing, Writing – original draft. YL: Writing – review & editing, Writing – original draft. XT: Writing – review & editing, Writing – original draft. LL: Writing – review & editing, Writing – original draft.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Institutes of Health under Award Numbers R21AI166118 (YC and LL), R01AI173180 (YC and LL), P20GM103648 (YC and LL), R01HL157450 (LL), R21AI52004 (LL) and P30GM149368 (LL); the Oklahoma State University Startup Fund (YC); Robert J. Sirny Endowed Professorship Fund (YC); Oklahoma Center for the Advancement of Science & Technology under Award Numbers HR21-050 (YC) and HR-20-050 (LL); Oklahoma Center for Adult Stem Cell Research-A Program of Tobacco Settlement Endowment Trust (TSET) (LL); and Lundberg-Kienlen Endowment fund (LL)”.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. (2007) 316:1484–8. doi: 10.1126/science.1138341
2. Nadhan R, Isidoro C, Song YS, Dhanasekaran DN. Signaling by lncRNAs: structure, cellular homeostasis, and disease pathology. Cells. (2022) 11:2517. doi: 10.3390/cells11162517
3. Pierce JB, Zhou H, Simion V, Feinberg MW. Long noncoding RNAs as therapeutic targets. Adv Exp Med Biol. (2022) 1363:161–75. doi: 10.1007/978-3-030-92034-0_9
4. Palazzo AF, Gregory TR. The case for junk DNA. PloS Genet. (2014) 10:e1004351. doi: 10.1371/journal.pgen.1004351
5. Bartolomei MS, Zemel S, Tilghman SM. Parental imprinting of the mouse H19 gene. Nature. (1991) 351:153–5. doi: 10.1038/351153a0
6. Pachnis V, Belayew A, Tilghman SM. Locus unlinked to alpha-fetoprotein under the control of the murine raf and Rif genes. Proc Natl Acad Sci U.S.A. (1984) 81:5523–7. doi: 10.1073/pnas.81.17.5523
7. Brannan CI, Dees EC, Ingram RS, Tilghman SM. The product of the H19 gene may function as an RNA. Mol Cell Biol. (1990) 10:28–36. doi: 10.1128/mcb.10.1.28-36.1990
8. Brockdorff N, Ashworth A, Kay GF, Cooper P, Smith S, McCabe VM, et al. Conservation of position and exclusive expression of mouse Xist from the inactive X chromosome. Nature. (1991) 351:329–31. doi: 10.1038/351329a0
9. Sasaki H, Ishihara K, Kato R. Mechanisms of Igf2/H19 imprinting: DNA methylation, chromatin and long-distance gene regulation. J Biochem. (2000) 127:711–5. doi: 10.1093/oxfordjournals.jbchem.a022661
10. Borsani G, Tonlorenzi R, Simmler MC, Dandolo L, Arnaud D, Capra V, et al. Characterization of a murine gene expressed from the inactive X chromosome. Nature. (1991) 351:325–9. doi: 10.1038/351325a0
11. Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, et al. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. (1991) 349:38–44. doi: 10.1038/349038a0
12. Ghafouri-Fard S, Khoshbakht T, Hussen BM, Taheri M, Arefian N. Regulatory role of non-coding RNAs on immune responses during sepsis. Front Immunol. (2021) 12:798713. doi: 10.3389/fimmu.2021.798713
13. Lecerf C, Peperstraete E, Le Bourhis X, Adriaenssens E. Propagation and maintenance of cancer stem cells: A major influence of the long non-coding RNA H19. Cells. (2020) 9:2613. doi: 10.3390/cells9122613
14. Wang B, Suen CW, Ma H, Wang Y, Kong L, Qin D, et al. The roles of H19 in regulating inflammation and aging. Front Immunol. (2020) 11:579687. doi: 10.3389/fimmu.2020.579687
15. Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, et al. The transcriptional landscape of the mammalian genome. Science. (2005) 309:1559–63. doi: 10.1126/science.1112014
16. Consortium IHGS. Finishing the euchromatic sequence of the human genome. Nature. (2004) 431:931–45. doi: 10.1038/nature03001
17. Okazaki Y, Furuno M, Kasukawa T, Adachi J, Bono H, Kondo S, et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. (2002) 420:563–73. doi: 10.1038/nature01266
18. Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. (2012) 489:57–74. doi: 10.1038/nature11247
19. Stamatoyannopoulos JA, Snyder M, Hardison R, Ren B, Gingeras T, Gilbert DM, et al. An encyclopedia of mouse DNA elements (Mouse ENCODE). Genome Biol. (2012) 13:418. doi: 10.1186/gb-2012-13-8-418
20. Bertone P, Stolc V, Royce TE, Rozowsky JS, Urban AE, Zhu X, et al. Global identification of human transcribed sequences with genome tiling arrays. Science. (2004) 306:2242–6. doi: 10.1126/science.1103388
21. Birney E, Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. (2007) 447:799–816. doi: 10.1038/nature05874
22. Ota T, Suzuki Y, Nishikawa T, Otsuki T, Sugiyama T, Irie R, et al. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat Genet. (2004) 36:40–5. doi: 10.1038/ng1285
23. Rinn JL, Euskirchen G, Bertone P, Martone R, Luscombe NM, Hartman S, et al. The transcriptional activity of human Chromosome 22. Genes Dev. (2003) 17:529–40. doi: 10.1101/gad.1055203
24. Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, et al. Landscape of transcription in human cells. Nature. (2012) 489:101–8. doi: 10.1038/nature11233
25. Kapranov P, St Laurent G, Raz T, Ozsolak F, Reynolds CP, Sorensen PH, et al. The majority of total nuclear-encoded non-ribosomal RNA in a human cell is ‘dark matter’ un-annotated RNA. BMC Biol. (2010) 8:149. doi: 10.1186/1741-7007-8-149
26. Mercer TR, Gerhardt DJ, Dinger ME, Crawford J, Trapnell C, Jeddeloh JA, et al. Targeted RNA sequencing reveals the deep complexity of the human transcriptome. Nat Biotechnol. (2011) 30:99–104. doi: 10.1038/nbt.2024
27. van Bakel H, Nislow C, Blencowe BJ, Hughes TR. Most “dark matter. transcripts are associated known genes PloS Biol. (2010) 8:e1000371. doi: 10.1371/journal.pbio.1000371
28. Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. (2018) 172:393–407.
29. Cao M, Luo H, Li D, Wang S, Xuan L, Sun L. Research advances on circulating long noncoding RNAs as biomarkers of cardiovascular diseases. Int J Cardiol. (2022) 353:109–17. doi: 10.1016/j.ijcard.2022.01.070
30. Rashidmayvan M, Sahebi R, Ghayour-Mobarhan M. Long non-coding RNAs: a valuable biomarker for metabolic syndrome. Mol Genet Genomics. (2022) 297:1169–83. doi: 10.1007/s00438-022-01922-1
31. Jiang W, Lv Y, Wang S. Prediction of non-coding RNAs as drug targets. Adv Exp Med Biol. (2018) 1094:109–15. doi: 10.1007/978-981-13-0719-5_11
32. Agliano F, Rathinam VA, Medvedev AE, Vanaja SK, Vella AT. Long noncoding RNAs in host-pathogen interactions. Trends Immunol. (2019) 40:492–510. doi: 10.1016/j.it.2019.04.001
33. Carpenter S. Long noncoding RNA: Novel links between gene expression and innate immunity. Virus Res. (2016) 212:137–45. doi: 10.1016/j.virusres.2015.08.019
34. Atianand MK, Caffrey DR, Fitzgerald KA. Immunobiology of long noncoding RNAs. Annu Rev Immunol. (2017) 35:177–98. doi: 10.1146/annurev-immunol-041015-055459
35. Flores-Concha M, Oñate Á.A. Long non-coding RNAs in the regulation of the immune response and trained immunity. Front Genet. (2020) 11:718. doi: 10.3389/fgene.2020.00718
36. Peltier DC, Roberts A, Reddy P. LNCing RNA to immunity. Trends Immunol. (2022) 43:478–95. doi: 10.1016/j.it.2022.04.002
37. Yan S, Wang P, Wang J, Yang J, Lu H, Jin C, et al. Long Non-coding RNA HIX003209 Promotes Inflammation by Sponging miR-6089 via TLR4/NF-κB Signaling Pathway in Rheumatoid Arthritis. Front Immunol. (2019) 10:2218. doi: 10.3389/fimmu.2019.02218
38. Wei M, Li L, Zhang Y, Zhang ZJ, Liu HL, Bao HG. LncRNA X inactive specific transcript contributes to neuropathic pain development by sponging miR-154-5p via inducing toll-like receptor 5 in CCI rat models. J Cell Biochem. (2019) 120:1271–81. doi: 10.1002/jcb.27088
39. Liang WJ, Zeng XY, Jiang SL, Tan HY, Yan MY, Yang HZ. Long non-coding RNA MALAT1 sponges miR-149 to promote inflammatory responses of LPS-induced acute lung injury by targeting MyD88. Cell Biol Int. (2020) 44:317–26. doi: 10.1002/cbin.11235
40. Wang Z, Liao W, Liu F, Yang T, Xie W, Liao M, et al. Downregulation of lncRNA EPB41L4A-AS1 mediates activation of MYD88-dependent NF-κB pathway in diabetes-related inflammation. Diabetes Metab Syndr Obes. (2021) 14:265–77. doi: 10.2147/DMSO.S280765
41. Du M, Yuan L, Tan X, Huang D, Wang X, Zheng Z, et al. The LPS-inducible lncRNA Mirt2 is a negative regulator of inflammation. Nat Commun. (2017) 8:2049. doi: 10.1038/s41467-017-02229-1
42. Mao AP, Shen J, Zuo Z. Expression and regulation of long noncoding RNAs in TLR4 signaling in mouse macrophages. BMC Genomics. (2015) 16:45. doi: 10.1186/s12864-015-1270-5
43. Rapicavoli NA, Qu K, Zhang J, Mikhail M, Laberge RM, Chang HY. A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. Elife. (2013) 2:e00762. doi: 10.7554/eLife.00762
44. Vollmers AC, Covarrubias S, Kuang D, Shulkin A, Iwuagwu J, Katzman S, et al. A conserved long noncoding RNA, GAPLINC, modulates the immune response during endotoxic shock. Proc Natl Acad Sci. (2021) 118:e2016648118. doi: 10.1073/pnas.2016648118
45. Castellanos-Rubio A, Kratchmarov R, Sebastian M, Garcia-Etxebarria K, Garcia L, Irastorza I, et al. Cytoplasmic Form of Carlr lncRNA Facilitates Inflammatory Gene Expression upon NF-κB Activation. J Immunol. (2017) 199:581–8. doi: 10.4049/jimmunol.1700023
46. Carpenter S, Aiello D, Atianand MK, Ricci EP, Gandhi P, Hall LL, et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science. (2013) 341:789–92. doi: 10.1126/science.1240925
47. Hu G, Gong AY, Wang Y, Ma S, Chen X, Chen J, et al. LincRNA-cox2 promotes late inflammatory gene transcription in macrophages through modulating SWI/SNF-mediated chromatin remodeling. J Immunol. (2016) 196:2799–808. doi: 10.4049/jimmunol.1502146
48. Krawczyk M, Emerson BM. p50-associated COX-2 extragenic RNA (PACER) activates COX-2 gene expression by occluding repressive NF-κB complexes. Elife. (2014) 3:e01776. doi: 10.7554/eLife.01776
49. Ma S, Ming Z, Gong AY, Wang Y, Chen X, Hu G, et al. A long noncoding RNA, lincRNA-Tnfaip3, acts as a coregulator of NF-κB to modulate inflammatory gene transcription in mouse macrophages. FASEB J. (2017) 31:1215–25. doi: 10.1096/fj.201601056R
50. Bastard P, Rosen LB, Zhang Q, Michailidis E, Hoffmann HH, Zhang Y, et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. (2020) 370:eabd4585. doi: 10.1126/science.abd4585
51. Aznaourova M, Janga H, Sefried S, Kaufmann A, Dorna J, Volkers SM, et al. Noncoding RNA MaIL1 is an integral component of the TLR4–TRIF pathway. Proc Natl Acad Sci. (2020) 117:9042–53. doi: 10.1073/pnas.1920393117
52. Agarwal S, Vierbuchen T, Ghosh S, Chan J, Jiang Z, Kandasamy RK, et al. The long non-coding RNA LUCAT1 is a negative feedback regulator of interferon responses in humans. Nat Commun. (2020) 11:6348. doi: 10.1038/s41467-020-20165-5
53. Vierbuchen T, Agarwal S, Johnson JL, Galia L, Lei X, Stein K, et al. The lncRNA LUCAT1 is elevated in inflammatory disease and restrains inflammation by regulating the splicing and stability of NR4A2. Proc Natl Acad Sci U.S.A. (2023) 120:e2213715120. doi: 10.1073/pnas.2213715120
54. Andersen RE, Hong SJ, Lim JJ, Cui M, Harpur BA, Hwang E, et al. The long noncoding RNA pnky is a trans-acting regulator of cortical development in vivo. Dev Cell. (2019) 49:632–642.e637. doi: 10.1016/j.devcel.2019.04.032
55. Bayraktar R, Bertilaccio MTS, Calin GA. The interaction between two worlds: microRNAs and toll-like receptors. Front Immunol. (2019) 10:1053. doi: 10.3389/fimmu.2019.01053
56. Rai KR, Liao Y, Cai M, Qiu H, Wen F, Peng M, et al. MIR155HG plays a bivalent role in regulating innate antiviral immunity by encoding long noncoding RNA-155 and microRNA-155-5p. mBio. (2022) 13:e02510–02522. doi: 10.1128/mbio.02510-22
57. Zheng W, Chu Q, Xu T. The long noncoding RNA NARL regulates immune responses via microRNA-mediated NOD1 downregulation in teleost fish. J Biol Chem. (2021) 296:100414. doi: 10.1016/j.jbc.2021.100414
58. Zang Y, Zhou X, Wang Q, Li X, Huang H. LncRNA FIRRE/NF-kB feedback loop contributes to OGD/R injury of cerebral microglial cells. Biochem Biophys Res Commun. (2018) 501:131–8. doi: 10.1016/j.bbrc.2018.04.194
59. Luo D, Dai W, Feng X, Ding C, Shao Q, Xiao R, et al. Suppression of lncRNA NLRP3 inhibits NLRP3-triggered inflammatory responses in early acute lung injury. Cell Death Dis. (2021) 12:898. doi: 10.1038/s41419-021-04180-y
60. Zhang P, Cao L, Zhou R, Yang X, Wu M. The lncRNA Neat1 promotes activation of inflammasomes in macrophages. Nat Commun. (2019) 10:1495. doi: 10.1038/s41467-019-09482-6
61. Wan P, Su W, Zhang Y, Li Z, Deng C, Li J, et al. LncRNA H19 initiates microglial pyroptosis and neuronal death in retinal ischemia/reperfusion injury. Cell Death Differ. (2020) 27:176–91. doi: 10.1038/s41418-019-0351-4
62. Cai LJ, Tu L, Huang XM, Huang J, Qiu N, Xie GH, et al. LncRNA MALAT1 facilitates inflammasome activation via epigenetic suppression of Nrf2 in Parkinson’s disease. Mol Brain. (2020) 13:130. doi: 10.1186/s13041-020-00656-8
63. Wang Z, Kun Y, Lei Z, Dawei W, Lin P, Jibo W. LncRNA MIAT downregulates IL-1β, TNF-α to suppress macrophage inflammation but is suppressed by ATP-induced NLRP3 inflammasome activation. Cell Cycle. (2021) 20:194–203. doi: 10.1080/15384101.2020.1867788
64. Odendall C, Kagan JC. Host-encoded sensors of bacteria: our windows into the microbial world. Microbiol Spectr. (2019) 7. doi: 10.1128/microbiolspec.BAI-0011-2019
65. Lin H, Jiang M, Liu L, Yang Z, Ma Z, Liu S, et al. The long noncoding RNA Lnczc3h7a promotes a TRIM25-mediated RIG-I antiviral innate immune response. Nat Immunol. (2019) 20:812–23. doi: 10.1038/s41590-019-0379-0
66. Lai C, Liu L, Liu Q, Wang K, Cheng S, Zhao L, et al. Long noncoding RNA AVAN promotes antiviral innate immunity by interacting with TRIM25 and enhancing the transcription of FOXO3a. Cell Death Differ. (2021) 28:2900–15. doi: 10.1038/s41418-021-00791-2
67. Arora S, Singh P, Dohare R, Jha R, Ali Syed M. Unravelling host-pathogen interactions: ceRNA network in SARS-CoV-2 infection (COVID-19). Gene. (2020) 762:145057. doi: 10.1016/j.gene.2020.145057
68. Friedl MS, Djakovic L, Kluge M, Hennig T, Whisnant AW, Backes S, et al. HSV-1 and influenza infection induce linear and circular splicing of the long NEAT1 isoform. PloS One. (2022) 17:e0276467. doi: 10.1371/journal.pone.0276467
69. Imamura K, Imamachi N, Akizuki G, Kumakura M, Kawaguchi A, Nagata K, et al. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol Cell. (2014) 53:393–406. doi: 10.1016/j.molcel.2014.01.009
70. Ma H, Han P, Ye W, Chen H, Zheng X, Cheng L, et al. The long noncoding RNA NEAT1 exerts antihantaviral effects by acting as positive feedback for RIG-I signaling. J Virol. (2017) 91:e02250-16. doi: 10.1128/JVI.02250-16
71. Xie Q, Chen S, Tian R, Huang X, Deng R, Xue B, et al. Long noncoding RNA ITPRIP-1 positively regulates the innate immune response through promotion of oligomerization and activation of MDA5. J Virol. (2018) 92:e00507-18. doi: 10.1128/JVI.00507-18
72. Jiang M, Zhang S, Yang Z, Lin H, Zhu J, Liu L, et al. Self-recognition of an inducible host lncRNA by RIG-I feedback restricts innate immune response. Cell. (2018) 173:906–919.e913. doi: 10.1016/j.cell.2018.03.064
73. Huang S, Huang Z, Luo Q, Qing C. The expression of lncRNA NEAT1 in human tuberculosis and its antituberculosis effect. BioMed Res Int. (2018) 2018:9529072. doi: 10.1155/2018/9529072
74. Subuddhi A, Kumar M, Majumder D, Sarkar A, Ghosh Z, Vasudevan M, et al. Unraveling the role of H3K4 trimethylation and lncRNA HOTAIR in SATB1 and DUSP4-dependent survival of virulent Mycobacterium tuberculosis in macrophages. Tuberculosis (Edinburgh Scotland). (2020) 120:101897. doi: 10.1016/j.tube.2019.101897
75. Chen W, Yang X, Zhao Y-L, Xu S, Wang D. lncRNA-cox 2 enhance the intracellular killing ability against mycobacterial tuberculosis via up-regulating macrophage M 1 polarization/nitric oxide production. Int J Clin Exp Med. (2019) 12:2402–10.
76. Yao Q, Xie Y, Xu D, Qu Z, Wu J, Zhou Y, et al. Lnc-EST12, which is negatively regulated by mycobacterial EST12, suppresses antimycobacterial innate immunity through its interaction with FUBP3. Cell Mol Immunol. (2022) 19:883–97. doi: 10.1038/s41423-022-00878-x
77. Li M, Cui J, Niu W, Huang J, Feng T, Sun B, et al. Long non-coding PCED1B-AS1 regulates macrophage apoptosis and autophagy by sponging miR-155 in active tuberculosis. Biochem Biophys Res Commun. (2019) 509:803–9. doi: 10.1016/j.bbrc.2019.01.005
78. Xu Y, Yu J, Ma C, Gong Z, Wu X, Deng G. Impact of knockdown LincRNA-Cox2 on apoptosis of macrophage infected with Bacillus Calmette-Guérin. Mol Immunol. (2021) 130:85–95. doi: 10.1016/j.molimm.2020.11.008
79. Jiang F, Lou J, Zheng XM, Yang XY. LncRNA MIAT regulates autophagy and apoptosis of macrophage infected by Mycobacterium tuberculosis through the miR-665/ULK1 signaling axis. Mol Immunol. (2021) 139:42–9. doi: 10.1016/j.molimm.2021.07.023
80. Pawar K, Hanisch C, Palma Vera SE, Einspanier R, Sharbati S. Down regulated lncRNA MEG3 eliminates mycobacteria in macrophages via autophagy. Sci Rep. (2016) 6:19416. doi: 10.1038/srep19416
81. Ke Z, Lu J, Zhu J, Yang Z, Jin Z, Yuan L. Down-regulation of lincRNA-EPS regulates apoptosis and autophagy in BCG-infected RAW264.7 macrophages via JNK/MAPK signaling pathway. Infect Genet Evol. (2020) 77:104077. doi: 10.1016/j.meegid.2019.104077
82. Gomez JA, Wapinski OL, Yang YW, Bureau JF, Gopinath S, Monack DM, et al. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-γ locus. Cell. (2013) 152:743–54. doi: 10.1016/j.cell.2013.01.015
83. Imamura K, Takaya A, Ishida YI, Fukuoka Y, Taya T, Nakaki R, et al. Diminished nuclear RNA decay upon Salmonella infection upregulates antibacterial noncoding RNAs. EMBO J. (2018) 37:e97723. doi: 10.15252/embj.201797723
84. Chan J, Atianand M, Jiang Z, Carpenter S, Aiello D, Elling R, et al. Cutting edge: A natural antisense transcript, AS-IL1α, controls inducible transcription of the proinflammatory cytokine IL-1α. J Immunol. (2015) 195:1359–63. doi: 10.4049/jimmunol.1500264
85. Xu H, Jiang Y, Xu X, Su X, Liu Y, Ma Y, et al. Inducible degradation of lncRNA Sros1 promotes IFN-γ-mediated activation of innate immune responses by stabilizing Stat1 mRNA. Nat Immunol. (2019) 20:1621–30. doi: 10.1038/s41590-019-0542-7
86. Agliano F, Fitzgerald KA, Vella AT, Rathinam VA, Medvedev AE. Long non-coding RNA lincRNA-EPS inhibits host defense against listeria monocytogenes infection. Front Cell Infect Microbiol. (2019) 9:481. doi: 10.3389/fcimb.2019.00481
87. Li R, Fang L, Pu Q, Bu H, Zhu P, Chen Z, et al. MEG3-4 is a miRNA decoy that regulates IL-1β abundance to initiate and then limit inflammation to prevent sepsis during lung infection. Sci Signal. (2018) 11:eaao2387. doi: 10.1126/scisignal.aao2387
88. Balloy V, Koshy R, Perra L, Corvol H, Chignard M, Guillot L, et al. Bronchial Epithelial Cells from Cystic Fibrosis Patients Express a Specific Long Non-coding RNA Signature upon Pseudomonas aeruginosa Infection. Front Cell Infect Microbiol. (2017) 7:218. doi: 10.3389/fcimb.2017.00218
89. Deng X, Guo J, Sun Z, Liu L, Zhao T, Li J, et al. Brucella-Induced Downregulation of lncRNA Gm28309 Triggers Macrophages Inflammatory Response Through the miR-3068-5p/NF-κB Pathway. Front Immunol. (2020) 11:581517. doi: 10.3389/fimmu.2020.581517
91. Pai M, Behr MA, Dowdy D, Dheda K, Divangahi M, Boehme CC, et al. Tuberculosis. Nat Rev Dis Primers. (2016) 2:1–23. doi: 10.1038/nrdp.2016.76
92. Philips JA, Ernst JD. Tuberculosis pathogenesis and immunity. Annu Rev Pathol. (2012) 7:353–84. doi: 10.1146/annurev-pathol-011811-132458
93. Simmons JD, Stein CM, Seshadri C, Campo M, Alter G, Fortune S, et al. Immunological mechanisms of human resistance to persistent Mycobacterium tuberculosis infection. Nat Rev Immunol. (2018) 18:575–89. doi: 10.1038/s41577-018-0025-3
94. Yan H, Xu R, Zhang X, Wang Q, Pang J, Zhang X, et al. Identifying differentially expressed long non-coding RNAs in PBMCs in response to the infection of multidrug-resistant tuberculosis. Infection Drug Resistance. (2018) 11:945–59. doi: 10.2147/IDR
95. Yang X, Yang J, Wang J, Wen Q, Wang H, He J, et al. Microarray analysis of long noncoding RNA and mRNA expression profiles in human macrophages infected with Mycobacterium tuberculosis. Sci Rep. (2016) 6:38963. doi: 10.1038/srep38963
96. Huang Z, Liu J, Li L, Guo Y, Luo Q, Li J. Long non-coding RNA expression profiling of macrophage line RAW264.7 infected by Mycobacterium tuberculosis. Biotechnic Histochemistry: Off Publ Biol Stain Commission. (2020) 95:403–10. doi: 10.1080/10520295.2019.1707874
97. Roy S, Schmeier S, Kaczkowski B, Arner E, Alam T, Ozturk M, et al. Transcriptional landscape of Mycobacterium tuberculosis infection in macrophages. Sci Rep. (2018) 8:6758. doi: 10.1038/s41598-018-24509-6
98. Hu X, Liao S, Bai H, Gupta S, Zhou Y, Zhou J, et al. Long noncoding RNA and predictive model to improve diagnosis of clinically diagnosed pulmonary tuberculosis. J Clin Microbiol. (2020) 58:e01973-19. doi: 10.1128/JCM.01973-19
99. Zhang X, Liang Z, Zhang Y, Dai K, Zhu M, Wang J, et al. Comprehensive analysis of long non-coding RNAs expression pattern in the pathogenesis of pulmonary tuberculosis. Genomics. (2020) 112:1970–7. doi: 10.1016/j.ygeno.2019.11.009
100. Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. (2021) 22:96–118. doi: 10.1038/s41580-020-00315-9
101. Geng F, Jia WC, Li T, Li N, Wei W. Knockdown of lncRNA NEAT1 suppresses proliferation and migration, and induces apoptosis of cervical cancer cells by regulating the miR−377/FGFR1 axis. Mol Med Rep. (2022) 25:10. doi: 10.3892/mmr.2021.12526
102. Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. (2010) 464:1071–6. doi: 10.1038/nature08975
103. Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. (2007) 129:1311–23. doi: 10.1016/j.cell.2007.05.022
104. Elling R, Robinson EK, Shapleigh B, Liapis SC, Covarrubias S, Katzman S, et al. Genetic Models Reveal cis and trans Immune-Regulatory Activities for lincRNA-Cox2. Cell Rep. (2018) 25:1511–1524.e1516. doi: 10.1016/j.celrep.2018.10.027
105. Murphy MB, Medvedev AE. Long noncoding RNAs as regulators of Toll-like receptor signaling and innate immunity. J Leukoc Biol. (2016) 99:839–50. doi: 10.1189/jlb.2RU1215-575R
106. Kaushal D. Eicosanoids, prostaglandins, and the progression of tuberculosis. J Infect Dis. (2012) 206:1803–5. doi: 10.1093/infdis/jis611
107. Lam A, Prabhu R, Gross CM, Riesenberg LA, Singh V, Aggarwal S. Role of apoptosis and autophagy in tuberculosis. Am J Physiol Lung Cell Mol Physiol. (2017) 313:L218–l229. doi: 10.1152/ajplung.00162.2017
108. Liao J, He Q, Li M, Chen Y, Liu Y, Wang J. LncRNA MIAT: Myocardial infarction associated and more. Gene. (2016) 578:158–61. doi: 10.1016/j.gene.2015.12.032
109. Moradi MT, Fallahi H, Rahimi Z. Interaction of long noncoding RNA MEG3 with miRNAs: A reciprocal regulation. J Cell Biochem. (2019) 120:3339–52. doi: 10.1002/jcb.27604
110. Zhang Z, Shi S, Li J, Costa M. Long non-coding RNA MEG3 in metal carcinogenesis. Toxics. (2023) 11:157. doi: 10.3390/toxics11020157
111. Tan MC, Widagdo J, Chau YQ, Zhu T, Wong JJ, Cheung A, et al. The activity-induced long non-coding RNA meg3 modulates AMPA receptor surface expression in primary cortical neurons. Front Cell Neurosci. (2017) 11:124. doi: 10.3389/fncel.2017.00124
112. Vangoor VR, Gomes-Duarte A, Pasterkamp RJ. Long non-coding RNAs in motor neuron development and disease. J Neurochem. (2021) 156:777–801. doi: 10.1111/jnc.15198
113. Wang B, Yao L, Dong Y, Liu J, Wu J. LncRNA PCED1B-AS1 knockdown inhibits osteosarcoma via methylation-mediated miR-10a downregulation. J Orthop Surg Res. (2022) 17:464. doi: 10.1186/s13018-022-03284-1
114. Galán JE. Salmonella Typhimurium and inflammation: a pathogen-centric affair. Nat Rev Microbiol. (2021) 19:716–25. doi: 10.1038/s41579-021-00561-4
115. Radoshevich L, Cossart P. Listeria monocytogenes: towards a complete picture of its physiology and pathogenesis. Nat Rev Microbiol. (2018) 16:32–46. doi: 10.1038/nrmicro.2017.126
116. Ma LZ, Wang D, Liu Y, Zhang Z, Wozniak DJ. Regulation of biofilm exopolysaccharide biosynthesis and degradation in pseudomonas aeruginosa. Annu Rev Microbiol. (2022) 76:413–33. doi: 10.1146/annurev-micro-041320-111355
117. Muggeo A, Coraux C, Guillard T. Current concepts on Pseudomonas aeruginosa interaction with human airway epithelium. PloS Pathog. (2023) 19:e1011221. doi: 10.1371/journal.ppat.1011221
118. Gonçalves IG, Simões LC, Simões M. Legionella pneumophila. Trends Microbiol. (2021) 29:860–1. doi: 10.1016/j.tim.2021.04.005
119. Mraz AL, Weir MH. Knowledge to Predict Pathogens: Legionella pneumophila Lifecycle Systematic Review Part II Growth within and Egress from a Host Cell. Microorganisms. (2022) 10:141. doi: 10.3390/microorganisms10010141
120. de Figueiredo P, Ficht TA, Rice-Ficht A, Rossetti CA, Adams LG. Pathogenesis and immunobiology of brucellosis: review of Brucella-host interactions. Am J Pathol. (2015) 185:1505–17. doi: 10.1016/j.ajpath.2015.03.003
121. Peng H, Liu Y, Tian J, Ma J, Tang X, Rui K, et al. The long noncoding RNA IFNG-AS1 promotes T helper type 1 cells response in patients with hashimoto’s thyroiditis. Sci Rep. (2015) 5:17702. doi: 10.1038/srep17702
122. Stein N, Berhani O, Schmiedel D, Duev-Cohen A, Seidel E, Kol I, et al. IFNG-AS1 enhances interferon gamma production in human natural killer cells. iScience. (2019) 11:466–73. doi: 10.1016/j.isci.2018.12.034
123. Gheitasi R, Jourghasemi S, Pakzad I, Hosseinpour Sarmadi V, Samieipour Y, Sekawi Z, et al. A potential marker in brucellosis, long non coding RNA IFNG-AS1. Mol Biol Rep. (2019) 46:6495–500. doi: 10.1007/s11033-019-05095-w
124. Tang H, Wu Z, Zhang J, Su B. Salivary lncRNA as a potential marker for oral squamous cell carcinoma diagnosis. Mol Med Rep. (2013) 7:761–6. doi: 10.3892/mmr.2012.1254
125. Tingö L, Ahlberg E, Johansson L, Pedersen SA, Chawla K, Sætrom P, et al. Non-coding RNAs in human breast milk: A systematic review. Front Immunol. (2021) 12:725323. doi: 10.3389/fimmu.2021.725323
126. Xie Y, Yao J, Zhang X, Chen J, Gao Y, Zhang C, et al. A panel of extracellular vesicle long noncoding RNAs in seminal plasma for predicting testicular spermatozoa in nonobstructive azoospermia patients. Hum Reprod. (2020) 35:2413–27. doi: 10.1093/humrep/deaa184
127. Xu K, Jiang X, Ariston Gabriel AN, Li X, Wang Y, Xu S. Evolving landscape of long non-coding RNAs in cerebrospinal fluid: A key role from diagnosis to therapy in brain tumors. Front Cell Dev Biol. (2021) 9:737670. doi: 10.3389/fcell.2021.737670
128. Zhan Y, Du L, Wang L, Jiang X, Zhang S, Li J, et al. Expression signatures of exosomal long non-coding RNAs in urine serve as novel non-invasive biomarkers for diagnosis and recurrence prediction of bladder cancer. Mol Cancer. (2018) 17:142. doi: 10.1186/s12943-018-0893-y
129. Nogueira BMF, Krishnan S, Barreto-Duarte B, Araújo-Pereira M, Queiroz ATL, Ellner JJ, et al. Diagnostic biomarkers for active tuberculosis: progress and challenges. EMBO Mol Med. (2022) 14:e14088. doi: 10.15252/emmm.202114088
131. Chen ZL, Wei LL, Shi LY, Li M, Jiang TT, Chen J, et al. Screening and identification of lncRNAs as potential biomarkers for pulmonary tuberculosis. Sci Rep. (2017) 7:16751. doi: 10.1038/s41598-017-17146-y
132. Fang Y, Zhao J, Wang X, Wang X, Wang L, Liu L, et al. Identification of differentially expressed lncRNAs as potential plasma biomarkers for active tuberculosis. Tuberculosis (Edinb). (2021) 128:102065. doi: 10.1016/j.tube.2021.102065
133. Wang L, Xie B, Zhang P, Ge Y, Wang Y, Zhang D. LOC152742 as a biomarker in the diagnosis of pulmonary tuberculosis infection. J Cell Biochem. (2019) 120:8949–55. doi: 10.1002/jcb.27452
134. Winkle M, El-Daly SM, Fabbri M, Calin GA. Noncoding RNA therapeutics - challenges and potential solutions. Nat Rev Drug Discovery. (2021) 20:629–51. doi: 10.1038/s41573-021-00219-z
Keywords: long noncoding RNAs, host-pathogen interactions, bacterial infections, immunity, host cells
Citation: Cheng Y, Liang Y, Tan X and Liu L (2024) Host long noncoding RNAs in bacterial infections. Front. Immunol. 15:1419782. doi: 10.3389/fimmu.2024.1419782
Received: 18 April 2024; Accepted: 15 August 2024;
Published: 02 September 2024.
Edited by:
Daniel P. Potaczek, University of Marburg, GermanyReviewed by:
Leon Nicolas Schulte, University of Marburg, GermanyCopyright © 2024 Cheng, Liang, Tan and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yong Cheng, eWNoZW5nQG9rc3RhdGUuZWR1; Lin Liu, bGluLmxpdUBva3N0YXRlLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.