- 1Blizard Institute, Barts and The London Faculty of Medicine and Dentistry, Queen Mary University of London, London, United Kingdom
- 2Department of Gastroenterology, Royal London Hospital, Barts Health NHS Trust, London, United Kingdom
Tissue-resident memory T cells (TRM) are long-lived memory lymphocytes that persist in non-lymphoid tissues and provide the first line of defence against invading pathogens. They adapt to their environment in a tissue-specific manner, exerting effective pathogen control through a diverse T cell receptor (TCR) repertoire and the expression of proinflammatory cytokines and cytolytic proteins. More recently, several studies have indicated that TRM can egress from the tissue into the blood as so-called “ex-TRM”, or “circulating cells with a TRM phenotype”. The numerically small ex-TRM population can re-differentiate in the circulation, giving rise to new memory and effector T cells. Following their egress, ex-TRM in the blood and secondary lymphoid organs can be identified based on their continued expression of the residency marker CD103, alongside other TRM-like features. Currently, it is unclear whether exit is a stochastic process, or is actively triggered in response to unknown factors. Also, it is not known whether a subset or all TRM are able to egress. Ex-TRM may be beneficial in health, as mobilisation of specialised TRM and their recruitment to both their site of origin as well as distant tissues results in an efficient distribution of the immune response. However, there is emerging evidence of a pathogenic role for ex-TRM, with a suggestion that they may perpetuate both local and distant tissue inflammation. Here, we review the evidence for the existence of ex-TRM and examine their potential involvement in disease pathogenesis.
1 Introduction
T cells are crucial for the generation of a successful adaptive immune response against pathogens. Upon entry into barrier tissues, antigen presenting cells (APCs) capture antigen from the invading pathogen and traffic it to the local draining lymph node, where naïve antigen-specific T cells are then activated (1). This is followed by a period of rapid clonal expansion and differentiation, following which newly generated effector T cells migrate to the site of infection and participate in host defence through their production of cytokines and direct cytolytic activity (2–4). Most effector T cells die following pathogen clearance, but a small fraction survives and is capable of recirculating between non-lymphoid tissue and the blood (5). Referred to as memory precursor effector cells (MPECs), this population can differentiate into memory T cells, thus ensuring a potent recall response against invading pathogens (2). Memory T cells can be broadly divided into circulating and resident memory populations, the latter of which was identified a little over a decade ago (2, 6–8). The circulating memory population comprises central memory T cells (TCM), which proliferate extensively upon reactivation, giving rise to expanded effector cell populations, and terminally differentiated effector memory T cells (TEM) which rapidly differentiate into new effector cells upon antigen re-encounter (2, 9). The tissue memory compartment comprises resident memory T cells (TRM), which are localised in non-lymphoid tissues and serve as a “frontline” defence against recurring pathogens at barrier sites (6–8, 10). TRM are phenotypically and transcriptionally distinct from other T cell populations (11) but share features with circulating TEM and effector T cells, including their expression of proinflammatory cytokines and cytolytic proteins, meaning they are primed to rapidly respond to infection. They can proliferate in situ, driving autonomous expansion of their population (10, 12). To date, studies on TRM have focused mostly on CD8+ cells in the mouse, but the importance of CD4+ TRM is increasingly recognized (13) and equivalent populations have been identified in humans. Until recently it was assumed that once residency is established TRM do not exit the tissue. However, several recent studies have suggested that a fraction of TRM are capable of egress and can be found in draining secondary lymphoid organs (SLOs) and in the circulation (Figure 1) (14–19). Here, we will review the evidence for the existence of ex-TRM, discuss their properties, and highlight the emerging evidence for their potential involvement in inflammatory disease.
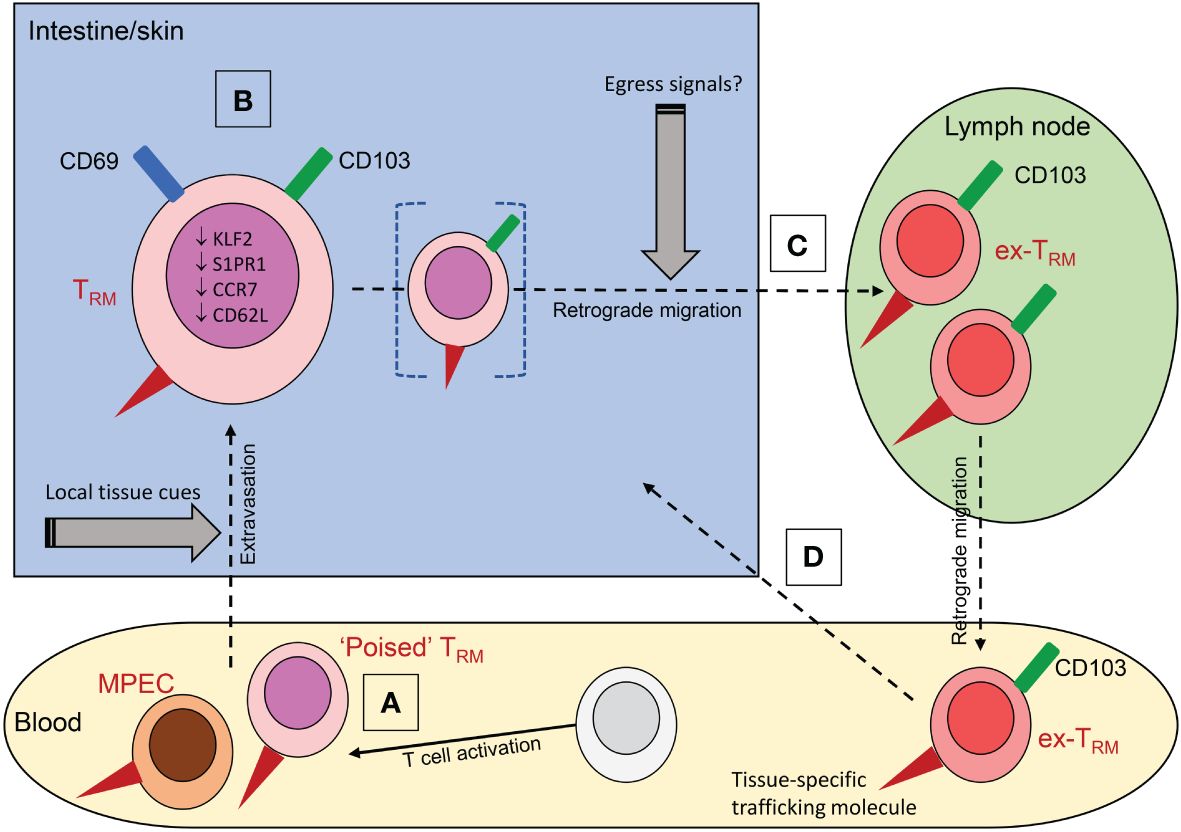
Figure 1 The origins and properties of ex-TRM. (A) Memory precursor cells (MPEC) recruited into tissues can differentiate into TRM under the influence of local tissue cues including cytokines; a subset of these circulating precursors may be pre-committed to a TRM fate (‘poised TRM’). (B) Residence is associated with expression of CD69 and CD103 and downregulation of the transcriptional profile associated with tissue egress. (C) Under conditions that are poorly defined, TRM can downregulate CD69, while retaining CD103 expression and undergo retrograde migration to the draining lymph node or beyond into the blood. The ability to egress may be a property of a distinct subpopulation of cells (bracketed in figure). Circulating ex-TRM are plastic cells that can undergo further differentiation to yield a variety of effector and memory T cells. (D) They can re-enter tissue, with a propensity to return to the tissue of origin due to retained expression of tissue-specific trafficking molecules (red triangles in figure); these include CLA and α4β7 for skin and intestine respectively.
2 The origin and differentiation of TRM
Cells with the phenotype of TRM are present in most, if not all, human tissues (20, 21), and studies in transplant patients have directly demonstrated long term maintenance of donor-derived T cells in the transplanted tissue (22–24). Despite their prevalence, the origin and development of TRM is not fully understood. TRM are present in both the human fetal and infant intestine, suggesting TRM seeding and development can occur early in life in response to ingested antigens, but the precise nature of the precursor population giving rise to these cells is unclear (25–27).
TRM differentiation is induced by exposure to tissue-derived factors following extravasation of precursors (5, 27, 28). It can be influenced by pre-commitment to this fate prior to arrival in the tissue (28); T cells clones have been identified within circulating mouse T cells that already express TRM associated genes and have an enhanced potential to establish residency in the skin (28). This pre-commitment to a TRM fate occurs early in an immune response and it can occur either within the naïve T cell population (29) or following activation through the TCR, possibly as the result of interaction with specific DC populations (5, 28–32). Regardless of their origin, TRM precursor cells are thought to give rise to TRM by an induced differentiation programme (19, 33–36). The precise signals resulting in the acquisition of a TRM phenotype are complex and likely to be tissue-specific, but there is evidence to suggest that interaction with the extracellular matrix, cytokines including IL-15 and TGFβ, hypoxia, and persistent antigenic stimuli are important for TRM generation in certain contexts (34–38). In response, a number of transcription factors including Runx3, Notch, Hobit, and Blimp-1, function together as a module to silence lymphocyte egress pathways (39–41). These positive, tissue-derived developmental cues result in phenotypic, transcriptional, and metabolic changes to the cell enabling it to attain residency within the tissue.
The conversion of TRM precursor cells to TRM is coupled with the upregulation of tissue retention molecules, including CD69 and CD103 (αEβ7 integrin). CD69 promotes residency by triggering internalisation of sphingosine-1-phosphate receptor 1 (S1PR1) which otherwise mediates migration from the tissue in response to S1P gradients, whereas CD103 binds to E-cadherin and tethers cells to the epithelium (11, 42, 43). TRM are also characterised by reduced expression of the transcription factor KLF2, which promotes S1PR1 expression and a number of additional cell surface markers linked to lymph node-homing and tissue-egress molecules, such as CCR7 and CD62L (11, 44, 45). CD69 and CD103 are often used as phenotypic markers of TRM. CD103 is typically expressed by a subset of CD69+ cells, constituting a greater proportion of CD8+ than of CD4+ TRM. CD103+ and CD103- TRM subsets have distinct functions during infection (46, 47). It should be emphasized that the biology of TRM is complex and caution should be exercised when extrapolating findings to different cell subsets, tissues, or species.
Once resident within the tissue, TRM respond rapidly to infection owing to their diverse tissue-specific TCR repertoire, and close proximity to the cellular targets of specific pathogens (8, 48–52). Upon re-activation, they display immediate cytotoxic potential, and can rapidly release proinflammatory cytokines such as such as IFNγ and TNFα (53, 54). The cytokines released by TRM can render surrounding cells resistant to infection, even with unrelated antigens, and can create tissue-wide anti-pathogen responses via the upregulation of Type I IFN signalling pathway factors and the enhanced recruitment of leucocytes (53, 54). In addition, TRM are able to kill infected cells directly via production of the cytolytic proteins perforin and granzyme B, enabling them to induce the contact-mediated apoptosis of infected cells they encounter (55, 56).
3 Experimental evidence for the existence of ex-TRM
For years TRM were thought to be permanently resident within the tissue without recirculating. However, it has recently been observed that some TRM are capable of egressing into draining SLOs (57, 58) or the blood (14, 15, 18, 19), a pattern of trafficking that has been termed retrograde migration (15, 58). The cells in the blood, herein termed ex-TRM, can be identified by their continued expression of CD103 and other TRM-like phenotypic features (14, 19, 59). They are plastic cells that contribute to the circulating memory response by re-differentiating into other memory T cell subsets and can enter tissue to re-establish residency (14, 15, 17, 19). In a key study, a fraction of the CD4+CD103+ TRM population present in human skin was found to be capable of downregulating CD69 expression and migrating from the tissue. In an allograft model these cells re-entered the circulation, and migrated to a secondary skin site where they subsequently reassumed their TRM phenotype (14). A corresponding population was detected at low frequency in human blood based on continued expression of CD103 in conjunction with the skin-specific trafficking molecule CLA (cutaneous leucocyte antigen). These cells shared a transcriptional and phenotypic profile with the skin resident population, strengthening the argument that they were related to a bona fide resident population. Importantly, TCRβ sequencing confirmed that both circulating CD4+CD103+ cells and TRM from different skin sites were clonally related, thus highlighting their ability to seed distant tissue locations (14). The continued expression of CLA by the skin derived ex-TRM in the circulation enables them to home preferentially back to their tissue of origin (14). These cells may be related to recirculating T cells previously described in human skin (60). Recent studies of patients undergoing hematopoietic stem cell transplant (HSCT) have also provided evidence for the existence of skin derived ex-TRM (16). In this context the circulating T cell pool is rapidly replaced with donor T cells, but host-derived skin TRM survive conditioning and are maintained for at least ten years, during which time they can egress and be found in the circulation (16, 61). The recipient-derived circulating T cells were identified transcriptionally based on their high expression of ITGAE (encoding CD103) and SEPLG (the gene encoding the protein backbone of CLA). The data discussed above on healthy human skin would suggest that this is not a phenomenon restricted to the unique situation of HSCT.
A mouse study supports the concept that TRM can also exit the intestine and has highlighted the plasticity of TRM during this process. Murine intestinal TRM were shown to undergo retrograde migration (15) and whilst circulating, the ex-TRM population exhibited developmental plasticity and differentiated into TCM or TEM in the circulating pool (15). Interestingly, the circulatory intestinal ex-TRM retained a preference to home back to their tissue of origin, in this case the intestine, and reacquired a stable TRM differentiation programme in response to local cytokine cues (15, 19). The preference of ex-TRM to return to their original tissue was seen even following tertiary reinfection with LCMV, suggesting the maintenance of a stable TRM profile over time (15). Although the phenomenon of ex-TRM is a relatively recent discovery, the presence of these cells may have first been reported several years ago. Jiang et al. (55) first described a population of vaccinia virus-specific CD8+ TRM in mice that could be found at skin sites distant to the site of initial infection. This remote population was highly effective at rapidly eliminating virus and was able to reduce viral loads to those seen at primary sites of infection (55). At the time it was believed this population derived from circulating TEM cells that randomly distributed into skin sites where they differentiated into TRM (55). However, this may be one of the first reports of skin ex-TRM egressing from the initial site of infection and seeding secondary sites to enhance local immunity. This phenomenon was more recently reported following the use of a murine human skin xenograft model (14).
4 Ex-TRM in health and disease
There is considerable evidence that the egress of TRM from tissue occurs in health, generating TCR clonotypes shared between barrier sites, such as the skin or intestine, and the circulation (62, 63). In this context, the ability of TRM to redistribute and contribute to the circulating memory pool is likely to be beneficial. This ‘outside-in’ immune response (15) brings several benefits to the host in the context of infection (Figure 2). Firstly, should an infection overwhelm the local TRM response, there are cells within the circulating compartment which retain a preference to home back to the infected tissue and can re-establish local immunity (15). Secondly, in the case of a large barrier organ such as the gut or skin, infection could occur at any location within the organ and not necessarily at a previous site of infection (19). Thus, the ability of specialised TRM to be mobilised from one site and recruited back from the circulation to other sites results in an efficient distribution of the immune response (19).
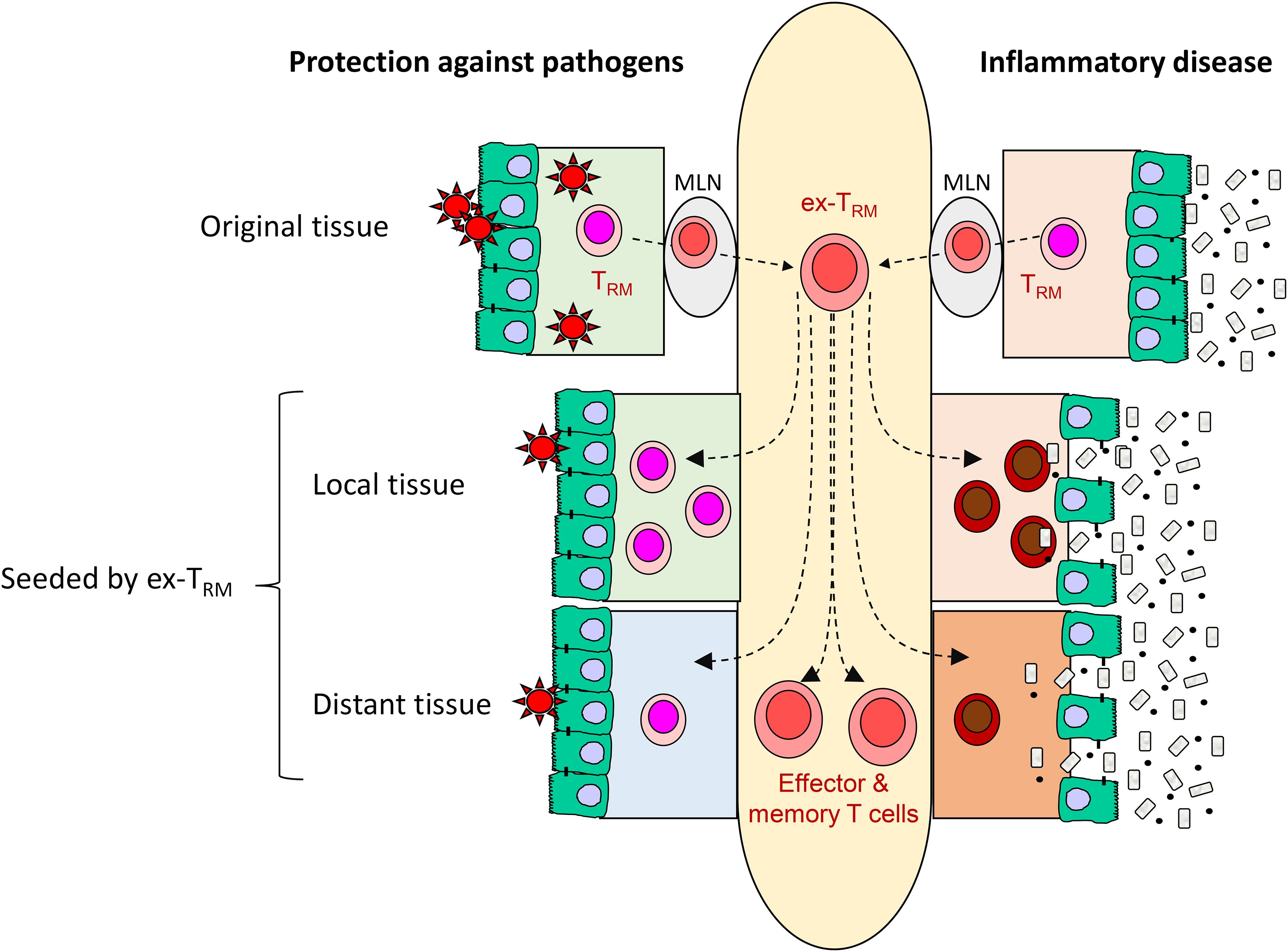
Figure 2 The potential contribution of ex-TRM to protective immunity and inflammatory disease. Here, potentially beneficial and detrimental contributions of ex-TRM are illustrated with refence to the intestine. The plasticity of ex-TRM enables them to generate a variety of effector and memory cells T cells as well as re-establish tissue residency. In the context of infection, these properties may form part of an early warning system responsive in initial stages of colonization (‘outside in’ immunity), provide cellular reinforcements to the site of infection and provide enhanced protection at remote sites that might be vulnerable to the pathogen (left side of figure). However, in chronic infmmatory disease, the same properties may enhance the local deleterious response and spread the inflammation to distant sites within the same tissue or even to other organ systems (right side of figure). MLN: mesenteric lymph node.
However, the presence of ex-TRM may be detrimental in the setting of an immune-mediated disease, and it is conceivable that the redistribution of potentially pathogenic TRM populations may perpetuate both local and distant tissue inflammation. There are currently limited data about ex-TRM in immunological disease but in HSCT, the recipient skin-derived ex-TRM population discussed above induce keratinocyte cell death and tissue damage through their expression of Th2 and Th17-associated cytokines such as IL-13 and IL-17 (16). In addition to skin trafficking molecules, they expressed ITGA4 and ITGB7, encoding the integrin α4β7 which facilitates entry to the intestine, and were localised in gastrointestinal graft versus host disease (GVHD) lesions. These findings not only highlight a potentially pathogenic role for ex-TRM, but also suggest their involvement in the propagation of inflammation to distant organs (16, 61). Potential evidence linking the aberrant trafficking of ex-TRM to distant tissue inflammation has also been found in ankylosing spondylarthritis (AS), a disease characterised by chronic inflammation of the axial skeleton (62). Proteomics and transcriptomic analysis revealed the presence of a specific population of mature CD8+ T cells elevated in both the blood and synovial fluid of AS patients, which were enriched for the cell-surface expression of gut-associated trafficking molecules, including β7 integrin, CD103, and CD49a (64). These cells also exhibited a dual cytotoxic and regulatory profile, with the release of TNFα, perforin, and IL-10 (64). Bulk RNA-sequencing of the CD8+ T cell population revealed a core transcriptional signature reminiscent of TRM, with the upregulation of residency-associated molecules ITGAE (CD103) and CXCR6, and a downregulation of tissue egress factors including SELL (CD62L) and S1PR1 (64). The expression pattern of these molecules was similar to that of intestinal intraepithelial lymphocytes (IELs), leading the authors to hypothesise that the CD8+ T cell population in the blood and synovial fluid may have mucosal origin, potentially having been trafficked from the gut to the joint (64). Interestingly, AS often co-exists with inflammatory bowel disease (IBD), a disease with which it shares many clinical, genetic, and immunological overlaps (64, 65). Given the gut-homing integrin α4β7 is also expressed by immune cells in the synovial tissue, a potential explanation for the co-existence of these conditions may be that inflammation in the gut is transferred to the joint through the aberrant trafficking of ex-TRM cells (64).
There is also recent evidence to suggest that the frequency of ex-TRM may be increased in inflammation, possibly indicating a role for inflammatory signals in the egress of TRM from tissue. In inflammatory bowel disease (IBD; Crohn’s disease and ulcerative colitis), intestinal TRM have been implicated in the inflammatory process (66). Single-cell RNA sequencing (scRNA-seq) analysis of blood and intestinal tissue from ulcerative colitis patients revealed a marked increase in TCR clonotypes shared between a CD8+ T cell cluster derived from intestinal tissue and a cluster derived from peripheral blood, suggesting a migration of clonally related cells between the two compartments (62). Interestingly, the proportion of shared clonotypes was significantly higher in ulcerative colitis patients compared with healthy controls; potentially indicating more frequent or increased TRM egress in a disease setting (62). Further, the presence of a small subset of peripheral blood CD4+ T cells with similar characteristics to synovial CD4+ T cells is correlated with both disease activity and resistance to therapy in juvenile idiopathic arthritis (JIA) (67). Both blood and synovial CD4+ populations exhibited a high degree of clonal overlap, alongside phenotypic similarities including increased IL-17, IFNγ, and TNFα production, thus suggesting the retention of an active TRM proinflammatory profile in the circulation (67). The host-derived ex-TRM population is also increased in the blood of HSCT patients with active GVHD, potentially providing a further link between inflammatory factors and TRM egress (16).
Much of the limited data currently available on the role of ex-TRM in disease is associated with skin and gut conditions, perhaps reflecting the large size of these organs and their greater contribution to the ex-TRM pool. Ex-TRM may also contribute to other autoimmune and inflammatory disease, but this possibility largely remains unexplored.
5 Discussion
Whilst the above-mentioned recent work has identified populations of ex-TRM in both murine and human peripheral blood, we are still left with a number of unanswered questions. Firstly, although there is strong evidence that these cells are tissue-derived and share many features with the resident population, the possibility that they are derived from cells distinct from the fully committed TRM population (35) has not been definitively excluded. Further fate mapping in mice or analysis of ex-TRM derived from transplanted human tissue may help resolve this issue. Secondly, it is still unclear whether only a small proportion of the TRM population can egress, or whether all TRM are capable of re-entering the circulation. Given the rarity of ex-TRM in peripheral blood - where they represent less than 1% of the total PBMC compartment – it is perhaps likely that only a small proportion of the vast TRM population are equipped to egress as ex-TRM (16). The ex-TRM so-far identified in blood are from skin and intestine and this may reflect the size of these organs, with the large number of immune cells they contain making rare exit events detectable. Analysis following separation of parabiotic mice show a gradual accumulation of blood ex-TRM over time (18). It is possible that retrograde migration as far as SLOs is a relatively common event but further progress into the circulation occurs only infrequently. Alternatively, the small number of ex-TRM identified in peripheral blood by phenotyping approaches may be explained by a change in phenotype following exit from the tissue. CD103 expression, alongside that of tissue-specific homing markers, is currently the best-defined method by which to identify ex-TRM populations (14–16). Therefore, should ex-TRM rapidly downregulate CD103 following their egress from tissue, they would go largely undetected with significant numbers of cells being missed. Future work should thus aim to identify additional transcriptional or phenotypic characteristics of this population. Finally, studies are also required to address whether the mobilization of resident cells is a stochastic process, or whether it is triggered by as-yet undefined factors. It is possible that there is a consistently low level of TRM egress when the tissue is at steady-state, but this may increase in the presence of inflammation, potentially as a result of increased signaling or antigen re-encounter. In some experimental systems egress of CD103+ TRM has not been seen to occur (47), perhaps suggesting that particular exit signals are lacking in these contexts. Given the potential involvement of ex-TRM in disease pathogenesis, it is important that future studies aim to address these pertinent questions in a range of conditions.
6 Concluding remarks
Recent experimental evidence in mice and humans suggests a fraction of TRM are capable of egressing from the tissue to become ex-TRM which are able support protective immune responses but may also be able to perpetuate local and distant tissue inflammation. Understanding the mechanisms underlying the generation and egress of ex-TRM may provide new therapeutic targets for disease.
Author contributions
BR: Writing – original draft, Writing – review & editing. AS: Conceptualization, Funding acquisition, Supervision, Writing – original draft, Writing – review & editing. JL: Conceptualization, Funding acquisition, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by Bowel Research UK (PhD award).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Sheridan BS, Lefrancois L. Regional and mucosal memory T cells. Nat Immunol. (2011) 12:485–91. doi: 10.1038/ni.2029
2. Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. (1999) 401:708–12. doi: 10.1038/44385
3. Grayson JM, Murali-Krishna K, Altman JD, Ahmed R. Gene expression in antigen-specific CD8+ T cells during viral infection. J Immunol. (2001) 166:795–9. doi: 10.4049/jimmunol.166.2.795
4. Bachmann MF, Barner M, Viola A, Kopf M. Distinct kinetics of cytokine production and cytolysis in effector and memory T cells after viral infection. Eur J Immunol. (1999) 29:291–9. doi: 10.1002/(ISSN)1521-4141
5. Mackay LK, Rahimpour A, Ma JZ, Collins N, Stock AT, Hafon ML, et al. The developmental pathway for CD103(+)CD8+ tissue-resident memory T cells of skin. Nat Immunol. (2013) 14:1294–301. doi: 10.1038/ni.2744
6. Masopust D, Vezys V, Marzo AL, Lefrancois L. Preferential localization of effector memory cells in nonlymphoid tissue. Science. (2001) 291:2413–7. doi: 10.1126/science.1058867
7. Gebhardt T, Wakim LM, Eidsmo L, Reading PC, Heath WR, Carbone FR. Memory T cells in nonlymphoid tissue that provide enhanced local immunity during infection with herpes simplex virus. Nat Immunol. (2009) 10:524–30. doi: 10.1038/ni.1718
8. Gebhardt T, Whitney PG, Zaid A, Mackay LK, Brooks AG, Heath WR, et al. Different patterns of peripheral migration by memory CD4+ and CD8+ T cells. Nature. (2011) 477:216–9. doi: 10.1038/nature10339
9. Butcher EC, Picker LJ. Lymphocyte homing and homeostasis. Science. (1996) 272:60–6. doi: 10.1126/science.272.5258.60
10. Masopust D, Choo D, Vezys V, Wherry EJ, Duraiswamy J, Akondy R, et al. Dynamic T cell migration program provides resident memory within intestinal epithelium. J Exp Med. (2010) 207:553–64. doi: 10.1084/jem.20090858
11. Kumar BV, Ma W, Miron M, Granot T, Guyer RS, Carpenter DJ, et al. Human tissue-resident memory T cells are defined by core transcriptional and functional signatures in lymphoid and mucosal sites. Cell Rep. (2017) 20:2921–34. doi: 10.1016/j.celrep.2017.08.078
12. Masopust D, Vezys V, Wherry EJ, Barber DL, Ahmed R. Cutting edge: gut microenvironment promotes differentiation of a unique memory CD8 T cell population. J Immunol. (2006) 176:2079–83. doi: 10.4049/jimmunol.176.4.2079
13. Oja AE, van Lier RAW, Hombrink P. Two sides of the same coin: Protective versus pathogenic CD4(+) resident memory T cells. Sci Immunol. (2022) 7:eabf9393. doi: 10.1126/sciimmunol.abf9393
14. Klicznik MM, Morawski PA, Hollbacher B, Varkhande SR, Motley SJ, Kuri-Cervantes L, et al. Human CD4(+)CD103(+) cutaneous resident memory T cells are found in the circulation of healthy individuals. Sci Immunol. (2019) 4:eaav8995. doi: 10.1126/sciimmunol.aav8995
15. Fonseca R, Beura LK, Quarnstrom CF, Ghoneim HE, Fan Y, Zebley CC, et al. Developmental plasticity allows outside-in immune responses by resident memory T cells. Nat Immunol. (2020) 21:412–21. doi: 10.1038/s41590-020-0607-7
16. Strobl J, Gail LM, Kleissl L, Pandey RV, Smejkal V, Huber J, et al. Human resident memory T cells exit the skin and mediate systemic Th2-driven inflammation. J Exp Med. (2021) 218(11):e20210417. doi: 10.1084/jem.20210417
17. Behr FM, Parga-Vidal L, Kragten NAM, van Dam TJP, Wesselink TH, Sheridan BS, et al. Tissue-resident memory CD8(+) T cells shape local and systemic secondary T cell responses. Nat Immunol. (2020) 21:1070–81. doi: 10.1038/s41590-020-0723-4
18. Wijeyesinghe S, Beura LK, Pierson MJ, Stolley JM, Adam OA, Ruscher R, et al. Expansible residence decentralizes immune homeostasis. Nature. (2021) 592:457–62. doi: 10.1038/s41586-021-03351-3
19. Rosato PC, Wijeyesinghe S, Stolley JM, Masopust D. Integrating resident memory into T cell differentiation models. Curr Opin Immunol. (2020) 63:35–42. doi: 10.1016/j.coi.2020.01.001
20. Sathaliyawala T, Kubota M, Yudanin N, Turner D, Camp P, Thome JJ, et al. Distribution and compartmentalization of human circulating and tissue-resident memory T cell subsets. Immunity. (2013) 38:187–97. doi: 10.1016/j.immuni.2012.09.020
21. Szabo PA, Miron M, Farber DL. Location, location, location: Tissue resident memory T cells in mice and humans. Sci Immunol. (2019) 4:eaas9673. doi: 10.1126/sciimmunol.aas9673
22. FitzPatrick MEB, Provine NM, Garner LC, Powell K, Amini A, Irwin SL, et al. Human intestinal tissue-resident memory T cells comprise transcriptionally and functionally distinct subsets. Cell Rep. (2021) 34:108661. doi: 10.1016/j.celrep.2020.108661
23. Bartolome-Casado R, Landsverk OJB, Chauhan SK, Saetre F, Hagen KT, Yaqub S, et al. CD4(+) T cells persist for years in the human small intestine and display a TH1 cytokine profile. Mucosal Immunol. (2021) 14:402–10. doi: 10.1038/s41385-020-0315-5
24. Bartolome-Casado R, Landsverk OJB, Chauhan SK, Richter L, Phung D, Greiff V, et al. Resident memory CD8 T cells persist for years in human small intestine. J Exp Med. (2019) 216:2412–26. doi: 10.1084/jem.20190414
25. Zuber J, Shonts B, Lau SP, Obradovic A, Fu J, Yang S, et al. Bidirectional intragraft alloreactivity drives the repopulation of human intestinal allografts and correlates with clinical outcome. Sci Immunol. (2016) 1:eaah3732. doi: 10.1126/sciimmunol.aah3732
26. Schreurs R, Baumdick ME, Sagebiel AF, Kaufmann M, Mokry M, Klarenbeek PL, et al. Human fetal TNF-alpha-cytokine-producing CD4(+) effector memory T cells promote intestinal development and mediate inflammation early in life. Immunity. (2019) 50:462–76.e8. doi: 10.1016/j.immuni.2018.12.010
27. Thome JJ, Bickham KL, Ohmura Y, Kubota M, Matsuoka N, Gordon C, et al. Early-life compartmentalization of human T cell differentiation and regulatory function in mucosal and lymphoid tissues. Nat Med. (2016) 22:72–7. doi: 10.1038/nm.4008
28. Kok L, Dijkgraaf FE, Urbanus J, Bresser K, Vredevoogd DW, Cardoso RF, et al. A committed tissue-resident memory T cell precursor within the circulating CD8+ effector T cell pool. J Exp Med. (2020) 217(10):e20191711. doi: 10.1084/jem.20191711
29. Gaide O, Emerson RO, Jiang X, Gulati N, Nizza S, Desmarais C, et al. Common clonal origin of central and resident memory T cells following skin immunization. Nat Med. (2015) 21:647–53. doi: 10.1038/nm.3860
30. Bourdely P, Anselmi G, Vaivode K, Ramos RN, Missolo-Koussou Y, Hidalgo S, et al. Transcriptional and functional analysis of CD1c(+) human dendritic cells identifies a CD163(+) subset priming CD8(+)CD103(+) T cells. Immunity. (2020) 53:335–52.e8. doi: 10.1016/j.immuni.2020.06.002
31. Milner JJ, Toma C, Yu B, Zhang K, Omilusik K, Phan AT, et al. Runx3 programs CD8(+) T cell residency in non-lymphoid tissues and tumours. Nature. (2017) 552:253–7. doi: 10.1038/nature24993
32. Herndler-Brandstetter D, Ishigame H, Shinnakasu R, Plajer V, Stecher C, Zhao J, et al. KLRG1(+) effector CD8(+) T cells lose KLRG1, differentiate into all memory T cell lineages, and convey enhanced protective immunity. Immunity. (2018) 48:716–29.e8. doi: 10.1016/j.immuni.2018.03.015
33. Iijima N, Iwasaki A. Tissue instruction for migration and retention of TRM cells. Trends Immunol. (2015) 36:556–64. doi: 10.1016/j.it.2015.07.002
34. Schenkel JM, Fraser KA, Casey KA, Beura LK, Pauken KE, Vezys V, et al. IL-15-independent maintenance of tissue-resident and boosted effector memory CD8 T cells. J Immunol. (2016) 196:3920–6. doi: 10.4049/jimmunol.1502337
35. Hirai T. [Competition between T cells for a finite epidermal niche promotes selective retention of antigen-specific memory T cells in the epidermis]. Yakugaku Zasshi. (2022) 142:1327–32. doi: 10.1248/yakushi.22-00155
36. Crowl JT, Heeg M, Ferry A, Milner JJ, Omilusik KD, Toma C, et al. Tissue-resident memory CD8(+) T cells possess unique transcriptional, epigenetic and functional adaptations to different tissue environments. Nat Immunol. (2022) 23:1121–31. doi: 10.1038/s41590-022-01229-8
37. Wienke J, Veldkamp SR, Struijf EM, Yousef Yengej FA, van der Wal MM, van Royen-Kerkhof A, et al. T cell interaction with activated endothelial cells primes for tissue-residency. Front Immunol. (2022) 13:827786. doi: 10.3389/fimmu.2022.827786
38. Hasan MH, Beura LK. Cellular interactions in resident memory T cell establishment and function. Curr Opin Immunol. (2022) 74:68–75. doi: 10.1016/j.coi.2021.10.005
39. Mackay LK, Minnich M, Kragten NA, Liao Y, Nota B, Seillet C, et al. Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science. (2016) 352:459–63. doi: 10.1126/science.aad2035
40. Parga-Vidal L, Taggenbrock R, Beumer-Chuwonpad A, Aglmous H, Kragten NAM, Behr FM, et al. Hobit and Blimp-1 regulate T(RM) abundance after LCMV infection by suppressing tissue exit pathways of T(RM) precursors. Eur J Immunol. (2022) 52:1095–111. doi: 10.1002/eji.202149665
41. Parga-Vidal L, Behr FM, Kragten NAM, Nota B, Wesselink TH, Kavazovic I, et al. Hobit identifies tissue-resident memory T cell precursors that are regulated by Eomes. Sci Immunol. (2021) 6:eabg3533. doi: 10.1126/sciimmunol.abg3533
42. Chen H, Qin Y, Chou M, Cyster JG, Li X. Transmembrane protein CD69 acts as an S1PR1 agonist. Elife. (2023) 12:e88204. doi: 10.7554/eLife.88204
43. Schon MP, Arya A, Murphy EA, Adams CM, Strauch UG, Agace WW, et al. Mucosal T lymphocyte numbers are selectively reduced in integrin alpha E (CD103)-deficient mice. J Immunol. (1999) 162:6641–9. doi: 10.4049/jimmunol.162.11.6641
44. Skon CN, Lee JY, Anderson KG, Masopust D, Hogquist KA, Jameson SC. Transcriptional downregulation of S1pr1 is required for the establishment of resident memory CD8+ T cells. Nat Immunol. (2013) 14:1285–93. doi: 10.1038/ni.2745
45. Preston GC, Feijoo-Carnero C, Schurch N, Cowling VH, Cantrell DA. The impact of KLF2 modulation on the transcriptional program and function of CD8 T cells. PloS One. (2013) 8:e77537. doi: 10.1371/journal.pone.0077537
46. Fung HY, Teryek M, Lemenze AD, Bergsbaken T. CD103 fate mapping reveals that intestinal CD103(-) tissue-resident memory T cells are the primary responders to secondary infection. Sci Immunol. (2022) 7:eabl9925. doi: 10.1126/sciimmunol.abl9925
47. von Hoesslin M, Kuhlmann M, de Almeida GP, Kanev K, Wurmser C, Gerullis AK, et al. Secondary infections rejuvenate the intestinal CD103(+) tissue-resident memory T cell pool. Sci Immunol. (2022) 7:eabp9553. doi: 10.1126/sciimmunol.abp9553
48. Schoettler N, Hrusch CL, Blaine KM, Sperling AI, Ober C. Transcriptional programming and T cell receptor repertoires distinguish human lung and lymph node memory T cells. Commun Biol. (2019) 2:411. doi: 10.1038/s42003-019-0657-2
49. Miron M, Meng W, Rosenfeld AM, Dvorkin S, Poon MML, Lam N, et al. Maintenance of the human memory T cell repertoire by subset and tissue site. Genome Med. (2021) 13:100. doi: 10.1186/s13073-021-00918-7
50. Poon MML, Caron DP, Wang Z, Wells SB, Chen D, Meng W, et al. Tissue adaptation and clonal segregation of human memory T cells in barrier sites. Nat Immunol. (2023) 24:309–19. doi: 10.1038/s41590-022-01395-9
51. Hegazy AN, West NR, Stubbington MJT, Wendt E, Suijker KIM, Datsi A, et al. Circulating and tissue-resident CD4(+) T cells with reactivity to intestinal microbiota are abundant in healthy individuals and function is altered during inflammation. Gastroenterology. (2017) 153:1320–37 e16. doi: 10.1053/j.gastro.2017.07.047
52. Turner DL, Bickham KL, Thome JJ, Kim CY, D’Ovidio F, Wherry EJ, et al. Lung niches for the generation and maintenance of tissue-resident memory T cells. Mucosal Immunol. (2014) 7:501–10. doi: 10.1038/mi.2013.67
53. Ariotti S, Beltman JB, Chodaczek G, Hoekstra ME, van Beek AE, Gomez-Eerland R, et al. Tissue-resident memory CD8+ T cells continuously patrol skin epithelia to quickly recognize local antigen. Proc Natl Acad Sci U S A. (2012) 109:19739–44. doi: 10.1073/pnas.1208927109
54. Roychoudhury P, Swan DA, Duke E, Corey L, Zhu J, Dave V, et al. Tissue-resident T cell-derived cytokines eliminate herpes simplex virus-2-infected cells. J Clin Invest. (2020) 130:2903–19. doi: 10.1172/JCI132583
55. Jiang X, Clark RA, Liu L, Wagers AJ, Fuhlbrigge RC, Kupper TS. Skin infection generates non-migratory memory CD8+ T(RM) cells providing global skin immunity. Nature. (2012) 483:227–31. doi: 10.1038/nature10851
56. Hombrink P, Helbig C, Backer RA, Piet B, Oja AE, Stark R, et al. Programs for the persistence, vigilance and control of human CD8(+) lung-resident memory T cells. Nat Immunol. (2016) 17:1467–78. doi: 10.1038/ni.3589
57. Beura LK, Mitchell JS, Thompson EA, Schenkel JM, Mohammed J, Wijeyesinghe S, et al. Intravital mucosal imaging of CD8(+) resident memory T cells shows tissue-autonomous recall responses that amplify secondary memory. Nat Immunol. (2018) 19:173–82. doi: 10.1038/s41590-017-0029-3
58. Stolley JM, Johnston TS, Soerens AG, Beura LK, Rosato PC, Joag V, et al. Retrograde migration supplies resident memory T cells to lung-draining LN after influenza infection. J Exp Med. (2020) 217(8):e20192197. doi: 10.1084/jem.20192197
59. Fonseca R, Burn TN, Gandolfo LC, Devi S, Park SL, Obers A, et al. Runx3 drives a CD8(+) T cell tissue residency program that is absent in CD4(+) T cells. Nat Immunol. (2022) 23:1236–45. doi: 10.1038/s41590-022-01273-4
60. Watanabe R, Gehad A, Yang C, Scott LL, Teague JE, Schlapbach C, et al. Human skin is protected by four functionally and phenotypically discrete populations of resident and recirculating memory T cells. Sci Transl Med. (2015) 7:279ra39. doi: 10.1126/scitranslmed.3010302
61. Strobl J, Pandey RV, Krausgruber T, Bayer N, Kleissl L, Reininger B, et al. Long-term skin-resident memory T cells proliferate in situ and are involved in human graft-versus-host disease. Sci Transl Med. (2020) 12:eabb7028. doi: 10.1126/scitranslmed.abb7028
62. Bishu S, El Zaatari M, Hayashi A, Hou G, Bowers N, Kinnucan J, et al. CD4+ Tissue-resident memory T cells expand and are a major source of mucosal tumour necrosis factor alpha in active crohn’s disease. J Crohns Colitis. (2019) 13:905–15. doi: 10.1093/ecco-jcc/jjz010
63. Risnes LF, Eggesbo LM, Zuhlke S, Dahal-Koirala S, Neumann RS, Lundin KEA, et al. Circulating CD103(+) gammadelta and CD8(+) T cells are clonally shared with tissue-resident intraepithelial lymphocytes in celiac disease. Mucosal Immunol. (2021) 14:842–51. doi: 10.1038/s41385-021-00385-8
64. Qaiyum Z, Gracey E, Yao Y, Inman RD. Integrin and transcriptomic profiles identify a distinctive synovial CD8+ T cell subpopulation in spondyloarthritis. Ann Rheum Dis. (2019) 78:1566–75. doi: 10.1136/annrheumdis-2019-215349
65. Braun J, Sieper J. Ankylosing spondylitis. Lancet. (2007) 369:1379–90. doi: 10.1016/S0140-6736(07)60635-7
66. Yokoi T, Murakami M, Kihara T, Seno S, Arase M, Wing JB, et al. Identification of a unique subset of tissue-resident memory CD4(+) T cells in Crohn’s disease. Proc Natl Acad Sci U S A. (2023) 120:e2204269120. doi: 10.1073/pnas.2204269120
67. Spreafico R, Rossetti M, van Loosdregt J, Wallace CA, Massa M, Magni-Manzoni S, et al. A circulating reservoir of pathogenic-like CD4+ T cells shares a genetic and phenotypic signature with the inflamed synovial micro-environment. Ann Rheum Dis. (2016) 75:459–65. doi: 10.1136/annrheumdis-2014-206226
Keywords: tissue-resident memory T cells, ex-TRM, tissue egress, recirculation, inflammatory disease
Citation: Rodger B, Stagg AJ and Lindsay JO (2024) The role of circulating T cells with a tissue resident phenotype (ex-TRM) in health and disease. Front. Immunol. 15:1415914. doi: 10.3389/fimmu.2024.1415914
Received: 11 April 2024; Accepted: 26 April 2024;
Published: 16 May 2024.
Edited by:
Tara Marlene Strutt, University of Central Florida, United StatesReviewed by:
Nidhi Kedia-Mehta, Maynooth University, IrelandCopyright © 2024 Rodger, Stagg and Lindsay. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andrew J. Stagg, YS5zdGFnZ0BxbXVsLmFjLnVr
 Beverley Rodger
Beverley Rodger Andrew J. Stagg
Andrew J. Stagg James O. Lindsay1,2
James O. Lindsay1,2