- 1Laboratory of Genomic and Precision Medicine, Wuxi School of Medicine, Jiangnan University, Wuxi, China
- 2Joint Primate Research Center for Chronic Diseases, Institute of Zoology of Guangdong Academy of Science, Jiangnan University, Wuxi, China
- 3Jiangnan University Brain Institute, Jiangnan University, Wuxi, Jiangsu, China
Background: COVID-19 vaccines are crucial for reducing the threat and burden of the pandemic on global public health, yet the epigenetic, especially RNA editing in response to the vaccines remains unelucidated.
Results: Our current study performed an epitranscriptomic analysis of RNA-Seq data of 260 blood samples from 102 healthy and SARS-CoV-2 naïve individuals receiving different doses of the COVID-19 vaccine and revealed dynamic, transcriptome-wide adenosine to inosine (A-to-I) RNA editing changes in response to COVID-19 vaccines (RNA editing in response to COVID-19 vaccines). 5592 differential RNA editing (DRE) sites in 1820 genes were identified, with most of them showing up-regulated RNA editing and correlated with increased expression of edited genes. These deferentially edited genes were primarily involved in immune- and virus-related gene functions and pathways. Differential ADAR expression probably contributed to RNA editing in response to COVID-19 vaccines. One of the most significant DRE in RNA editing in response to COVID-19 vaccines was in apolipoprotein L6 (APOL6) 3’ UTR, which positively correlated with its up-regulated expression. In addition, recoded key antiviral and immune-related proteins such as IFI30 and GBP1 recoded by missense editing was observed as an essential component of RNA editing in response to COVID-19 vaccines. Furthermore, both RNA editing in response to COVID-19 vaccines and its functions dynamically depended on the number of vaccine doses.
Conclusion: Our results thus underscored the potential impact of blood RNA editing in response to COVID-19 vaccines on the host’s molecular immune system.
Introduction
SARS-CoV-2 emerged in late 2019 and caused a global pandemic of coronavirus disease 2019 (COVID-19), posing serious threats to human health and society (1). COVID-19 vaccines are crucial to prevent severe disease and death caused by SARS-CoV-2. Vaccines could induce a robust immune response, triggering the production of antibodies and memory cells that can recognize and act on the virus (2, 3). However, COVID-19 vaccines could also pose immune-related challenges, such as the risk of allergic reactions (4, 5), vaccine breakthrough infections (6, 7), and waning immunity over time (8, 9). These challenges require further research and monitoring to ensure the safety and efficacy of the vaccines. Additionally, individuals with different immune conditions may respond differently to COVID-19 vaccines. The underlying regulatory mechanism involved in the immune response to COVID-19 vaccines requires further investigation.
Adenines to inosine (A-to-I) RNA editing, mediated by adenosine deaminase acting on RNA (ADARs), is one of the most widespread post-transcriptional epigenetic RNA modifications (10) and is recognized as A-to-G transitions during translation (11, 12). A-to-I RNA editing and ADARs play a key role in host antiviral activities by acting on the negative-strand RNA of SARS-CoV-2 during the viral replication (1, 13–17). Additionally, emerging studies have shown that SARS-CoV-2 infection changes the A-to-I RNA editing level, which could further affect the evolution of SARS-CoV-2 and the immune response (18–20). Nevertheless, it remains to be elucidated whether and how RNA editing plays a role in the immune response to COVID-19 vaccines.
In the current study, we performed a transcriptome-wide analysis of the blood A-to-I RNA editing profiles of individuals vaccinated with COVID-19 vaccines to investigate RNA editing response to COVID-19 vaccines (RNA editing in response to COVID-19 vaccines). By identifying substantial RNA editing in response to COVID-19 vaccines, our results linked A-to-I RNA editing to the immune response to COVID-19 vaccines.
Materials and methods
RNA-Seq reads download and processing
To explore RNA editing in response to COVID-19 vaccines, RNA-Seq raw data was downloaded from the European Nucleotide Archive (ENA) (https://www.ebi.ac.uk/ena). The dataset (PRJNA821445) contained 260 blood samples from 102 adults who received one, two, and three doses of COVID-19 vaccines (2).
By using a pipeline described in our previous study, the FASTQC was used for quality control of sequencing reads (21). Reads that passed quality control were mapped to the human genome (UCSC hg38) using RNA STAR (version 2.7.0e) (22). Then, the output BAM files were filtered by SAMtools (version 1.9) to remove multi-mapped and duplicated reads (23). Finally, GATK (version 4.1.3) was used to conduct base quality score recalibration (24).
RNA editing analysis
Single nucleotide variants (SNVs) were called from the BAM files using VarScan (version 2.4.3) (25), and annotated using the Ensembl Variant Effect Predictor (VEP) (26). Only variants that met the following criteria were retained: base quality ≥ 25, total sequencing depth ≥ 10, alternative allele depth ≥ 2, and alternative allele frequency (AAF) ≥ 1%. SNVs that met the following criteria were removed unless annotated as known RNA editing sites in the REDIportal V2.0 database (27): 1) located in homopolymer runs ≥ 5 nucleotides (nt) or simple repeats; 2) located in mitochondrial DNA; 3) located within 6 nt from splice junctions; 4) located within 1 nt from RNA insertion-deletion (INDEL); 5) within 4% to the ends of reads; 6) annotated as known variants in the dbSNP database Build 142; 7) more than 90% of samples had an AAF equal to 100% or between 40% and 60%; with an editing level < 5% in both the control and COVID-19 vaccines groups.
Gene expression quantification
The Rsubread package of the R language was used to calculate pseudo counts of the RNA expression (28) and normalized gene expression levels (transcript per million, TPM).
Protein structure prediction of missense RNA editing variants
Protein structure prediction was performed using the DDMut online tool (https://biosig.lab.uq.edu.au/ddmut/) to evaluate the impact of the missense changes on the edited protein (29).
Prediction of RNA secondary structure
RNA secondary structure prediction was performed using the RNAfold Web Server online tool (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi) to evaluate the impact of the 3’ UTR DRE changes on the edited gene.
Function enrichment analysis
Gene ontology (GO) and Kyoko Encyclopedia of Genes and Genomes (KEGG) analysis were performed on the DAVID online platform (https://david.ncifcrf.gov/tools.jsp), and Enrichr (https://maayanlab.cloud/Enrichr/) (30). Items with a false discovery rate (FDR) < 0.05 were considered significant.
Statistical analysis
RNA editing levels between V0 and COVID-19 vaccine groups were compared using the general linear model (GLM), and empirical P-values (PGLM) were calculated using the likelihood ratio test. Frequency data were analyzed using Fisher’s exact test. The correlation between RNA editing and gene expression levels was analyzed using the Spearman correlation method, and the correlation coefficient (r) and P-values were calculated accordingly.
Results
A-to-I RNA editing in blood sampled from COVID-19 vaccine recipients
A total of 94573 high-confidence A-to-I RNA editing sites were observed in 5723 genes in all recipients’ blood (Figure 1A). The editing levels of these sites ranged from 1% to 100%, and were widely observed across all chromosomes. Among these RNA editing sites, 92440 (97.7%) were shared by two groups, 158 (0.2%) and 1975 (2.1%) sites were uniquely detected in control (V0) and COVID-19 vaccines groups, respectively (Figure 1B). As for edited genes, 5623 (98.2%) were shared, 15 (0.3%) and 85 (1.5%) genes were uniquely edited in V0 and COVID-19 vaccines groups, respectively (Figure 1C). The functional categories of these RNA editing sites included 73227 (77.4%) intronic variants, 13351 (14.1%) 3’ -untranslated region (3’ UTR) variants, and 1228 (1.3%) missense variants (Figure 1D). Sorts intolerant from tolerant (SIFT) analysis predicted that 361 (29.4%) of these missense variants might have a potential functional impact on the encoded protein (Figure 1E). 61.6% of these edited sites overlapped with Alu repetitive elements (Figure 1F).
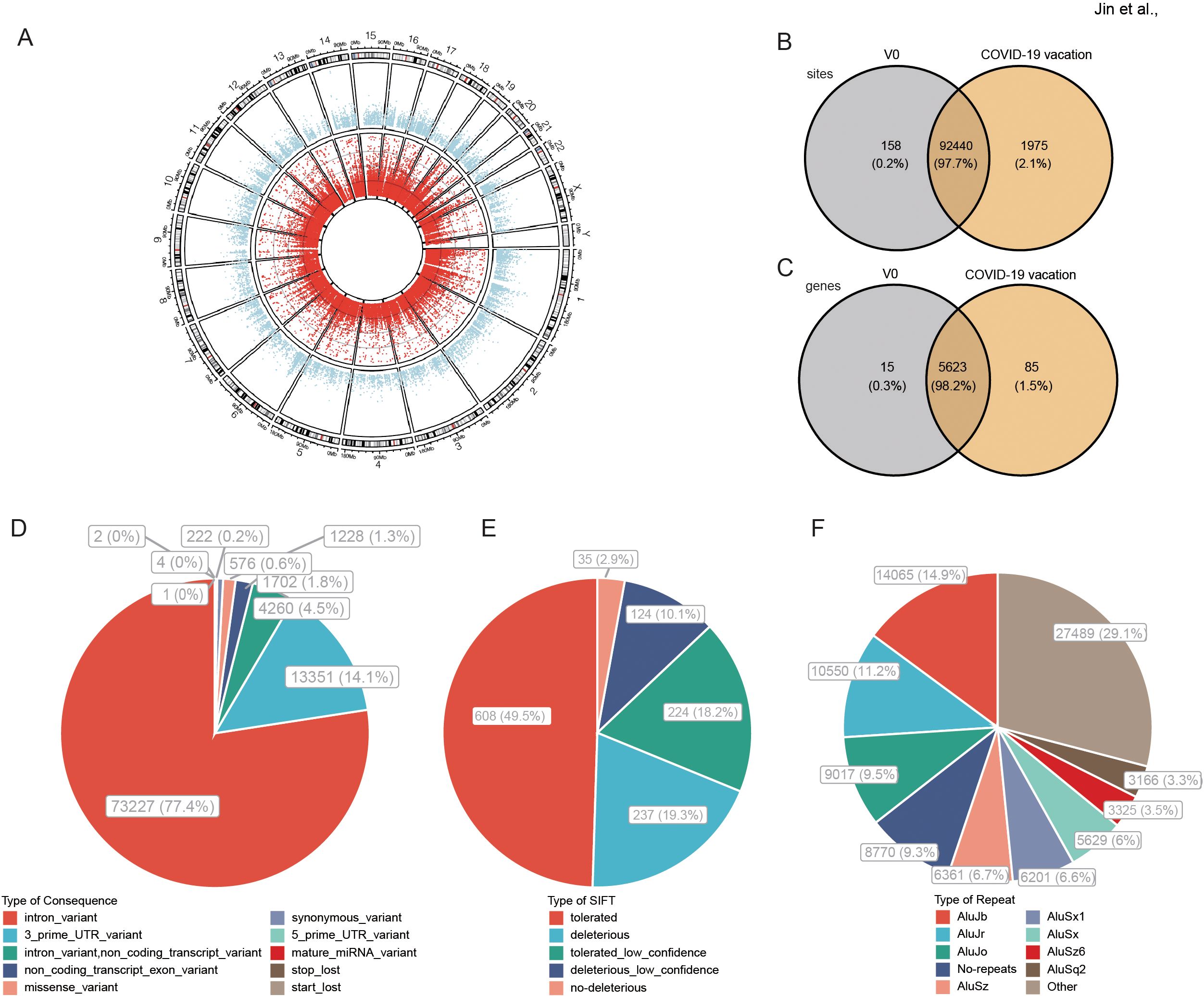
Figure 1. A-to-I RNA editing in response to COVID-19 vaccines identified from blood transcriptome cohort. (A) Circos plot of transcription gene expression (the outer circle) and A-to-I RNA editing sites (the inner circle) in the human brain. (B, C) The A-to-I RNA editing sites (B) and genes (C) of the two groups. (D) Functional types of variants resulted from A-to-I RNA editing. (E) About 29.4% of nonsynonymous variants are predicted by SIFT as possibly deleterious to the encoded proteins. (F) Most RNA editing sites are in Alu repetitive elements.
Temporal dynamics of RNA editing in response to COVID-19 vaccines
RNA editing was compared among the V0 group and vaccination groups that received three different doses of vaccine (V1, V2A, and V3A, as described in the original study). The results showed that ADAR expression and the average editing level, as well as the number of edited genes and editing sites, increased with vaccine doses, especially in V2A and V3A (Figures 2A–D). Among these RNA editing sites, 5592 differential RNA editing (DRE) sites were observed in 1820 genes across all chromosomes (Figure 2E, Supplementary Table S1). The functional categories of these DRE sites included 1828 (32.7%) 3’ UTR variants, 3004 (53.7%) intronic variants, and 219 (3.9%) non-coding transcript exonic variants (Figure 2F). Further analysis identified a subset of these DRE sites with their editing levels positively correlated with ADAR expression (|r| > 0.3, p < 0.05) (Figure 2G). A total of 1785 (98.1%) DRE genes contained two or more edited sites (Supplementary Table S2). The top ten genes ranked by the number of A-to-I RNA editing sites are shown in Supplementary Table S3. The top three genes were nicotinamide phosphoribosyltransferase (NAMPT), ring finger protein 213 (RNF213), and slingshot protein phosphatase 2 (SSH2).
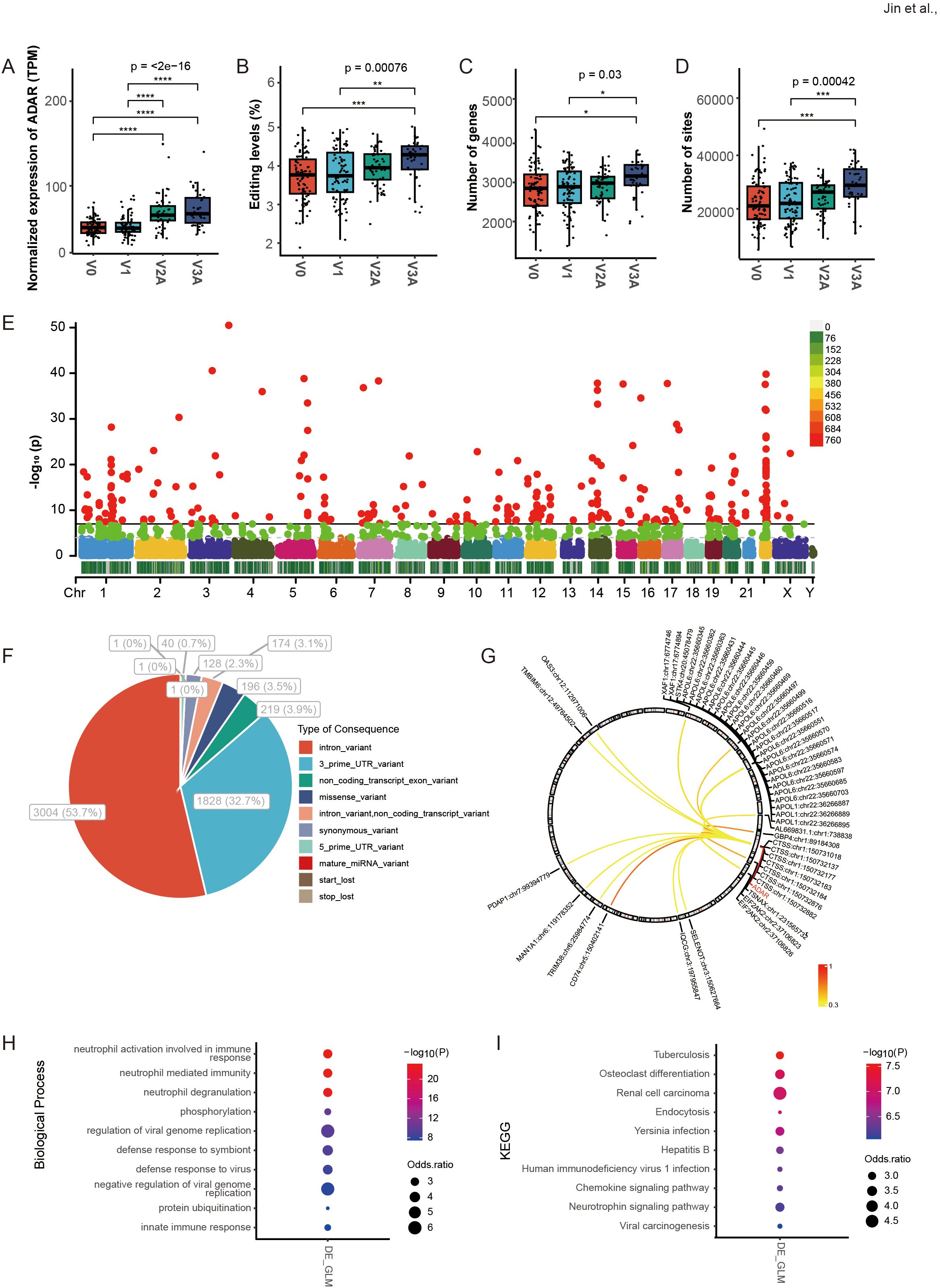
Figure 2. DRE A-to-I RNA editing in response to COVID-19 vaccines identified from blood transcriptome of three different doses of vaccination. (A–D) The ADAR expression and the average A-to-I RNA editing levels, as well as the number of edited genes and sites in four groups of individuals with different doses of vaccination (V0, V1, V2A, and V3A). (E) The Manhattan plot of DRE among the four groups. (F) The functional categories of DRE sites among the four groups. (G) The correlation between ADAR and the editing levels of individual DRE sites. (H, I) The items are shown for (H) biological processes and (I) the KEGG pathway in DRE genes in groups of different doses compared to V0. *P < 0.05; **P < 0.01; ***P < 0.001, ****P < 0.0001.
RNA editing in response to COVID-19 vaccines was mainly immune- and virus-related
Gene function enrichment showed that the differential blood RNA editing in response to COVID-19 vaccines was mainly involved in immune- and virus-related biological processes such as neutrophil activation involved in immune response, neutrophil-mediated immunity, neutrophil degranulation, and regulation of viral genome replication (Figure 2H); and KEGG pathways such as Yersinia infection, Hepatitis B, Human immunodeficiency virus 1 infection and Chemokine signaling pathway (Figure 2I).
Recoded essential immune genes by missense RNA editing in response to COVID-19 vaccines
We then focused on missense RNA editing, which recodes the amino acid sequence of encoded proteins. 196 missense DRE sites in 140 genes were observed, with 27.55% predicted by SIFT to be deleterious or possibly deleterious to the encoded proteins (Figure 3A). Among the edited genes with the top ten missense DRE variants (Supplementary Table S4), two immune-related genes were observed, including IFI30 lysosomal thiol reductase (IFI30) and guanylate-binding protein 1 (GBP1). The missense RNA editing level of IFI30 (IFI30:chr19:18177741, p.T223A) was significantly up-regulated (Figure 3B), whereas the GBP1 missense editing (GBP1:chr1:89057096, p.S305G) was significantly down-regulated (Figure 3C), with the expression of both genes increased during COVID-19 vaccines (Figures 3D, E). The substitution of serine (S) with glycine (G) at codon 305 of GBP1 was predicted to result in profound changes in its protein structure by DDMut (https://biosig.lab.uq.edu.au/ddmut/) (Figure 3F).
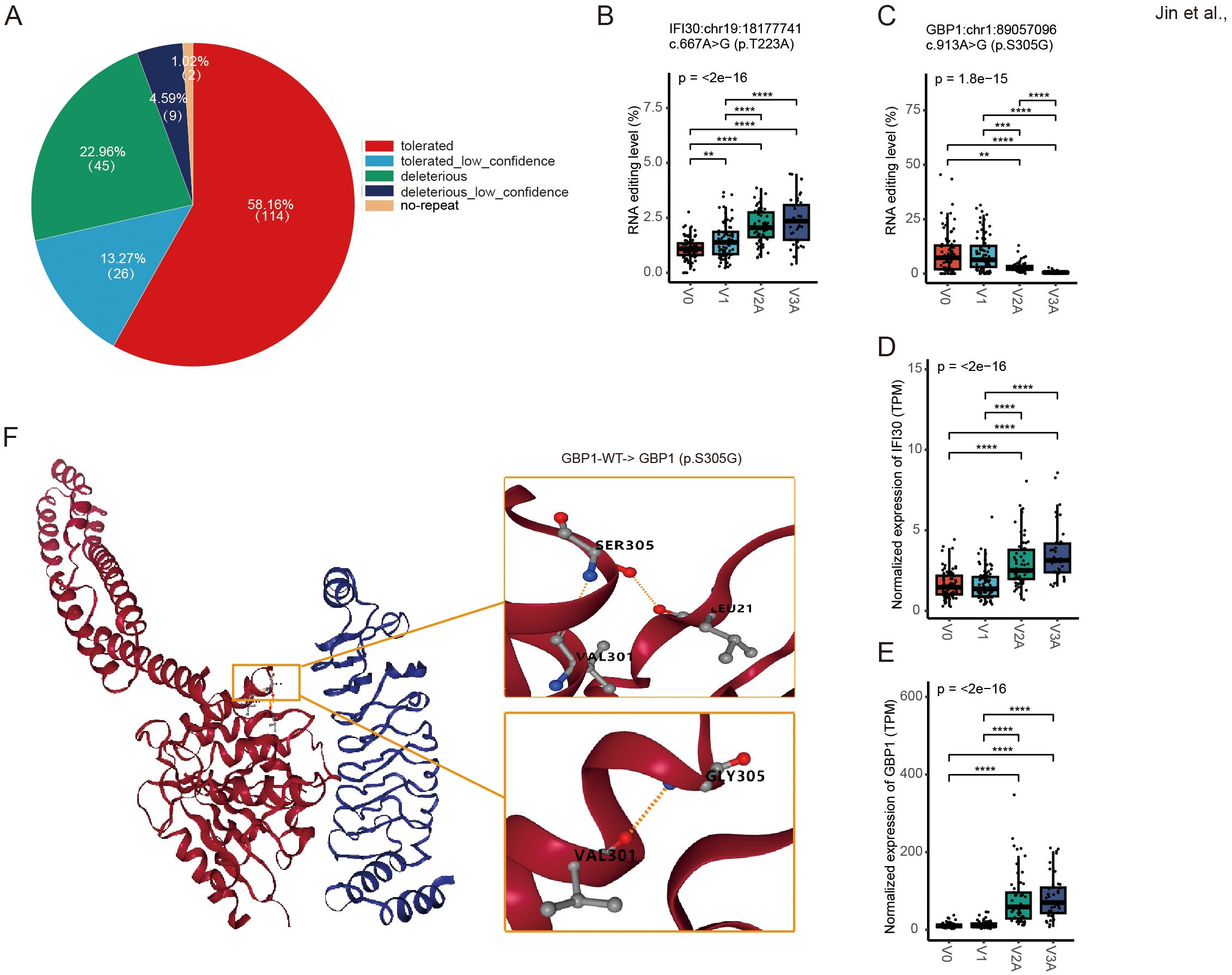
Figure 3. Missense DRE in IFI30 and GBP1. (A) About 27.55% of missense variants are predicted by SIFT as possibly deleterious to the encoded proteins. (B, C) The site editing levels of IFI30:chr19:18177741 (B) and GBP1:chr1:89057096 (C). (D, E) The gene expression levels of IFI30 (D) and GBP1 (E). (F) Prediction of spatial structure of GBP1 protein. **P < 0.01; ***P < 0.001, ****P < 0.0001.
3’ UTR RNA editing regulation in response to COVID-19 vaccines could contribute to gene expression
3’ UTR editing has been reported to potentially be involved in regulating the expression of edited RNA. A total of 1828 3’ UTR DRE sites in 535 genes, with 152 (8.3%) predicted to cis-regulate 92 edited genes (Figure 4A, Supplementary Table S5), and 205 (38.3%) edited genes found differentially expressed (Figure 4B, Supplementary Table S6). The top ten 3’ UTR variants in our results are displayed in Supplementary Table S7. One of the most extensively differential editing was found in the apolipoproteins L6 (APOL6) gene. Both the RNA editing (APOL6:chr22:35660551, and APOL6:chr22:35660499) and gene expression levels of APOL6 significantly increased upon COVID-19 vaccines (Figures 4C–E). APOL6 3’ UTR editing showed a strong positive correlation with its gene expression (Figures 4F, G).
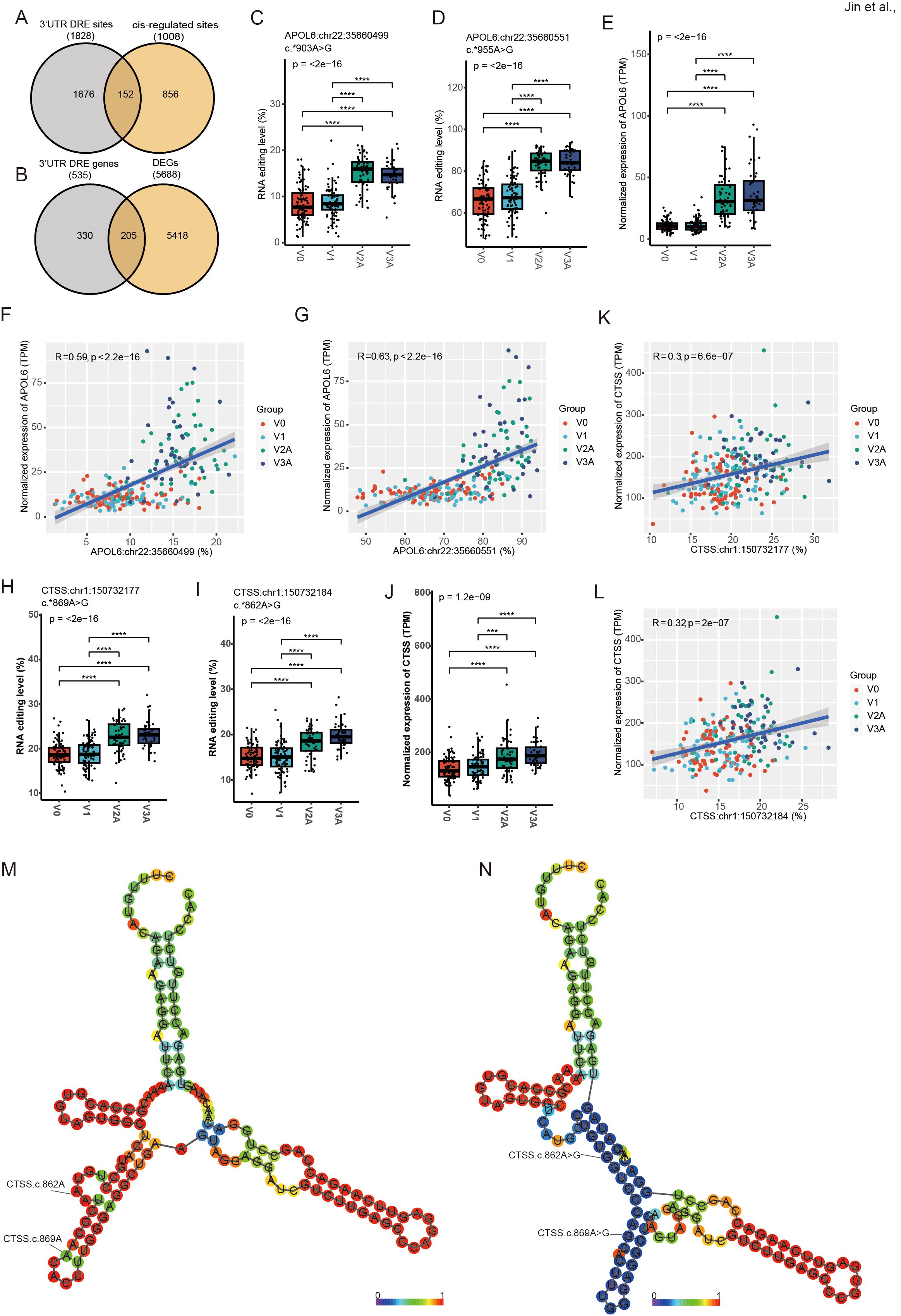
Figure 4. DRE in APOL6 3’ UTR and its potential cis-regulatory effects. (A) The Venn diagram of 3’ UTR DRE genes and cis-regulated genes. (B) The Venn diagram of 3’ UTR DRE genes and DEGs. (C–E) APOL6:chr22:35660499/35660551 editing level (C, D) and gene expression level (E). (F, G) Correlation of cis-regulated APOL6 gene expression level and editing sites (APOL6:chr22:35660499/35660551). (H–J) CTSS:chr1:150732177/150732184 RNA editing level (H, I) and gene expression level (J). (K, L) Correlation of cis-regulated CTSS gene expression level and editing sites (CTSS:chr1:150732177/150732184). (M, N) The RNA secondary structure at the WT (M) and edited (N) of CTSS:chr1:150732177/150732184. ***P < 0.001, ****P < 0.0001.
As shown in Figure 2G, multiple editing sites in the cathepsin S (CTSS) 3’ UTR also showed strong correlations with ADAR expression. The results showed that both the RNA editing (CTSS:chr1:150732177, and CTSS:chr1:150732184) and gene expression levels of CTSS significantly increased upon COVID-19 vaccination (Figures 4H–J). CTSS 3’ UTR editing showed a strong positive correlation with its gene expression (Figures 4K, L) similar to that in APOL6. In addition, prediction of RNA secondary structure showed that both CTSS editing sites could significantly alter their RNA secondary structure (Figures 4M, N), which may contribute to the changes in CTSS expression.
RNA editing in response to COVID-19 vaccines might influence RBP binding activity
To evaluate the potential effect of RNA editing on RBP binding, the RBPmap database was used to predict RBP binding sites that overlapped with DRE sites. The results in Figure 5 showed the top RBPs ranked by their number of predicted overlapping DRE sites, with the top three as RNA-binding protein 45 (RBM45), heterogeneous nuclear ribonucleoprotein A0 (HNRNPA0), and heterogeneous nuclear ribonucleoprotein D-like (HNRNPDL).
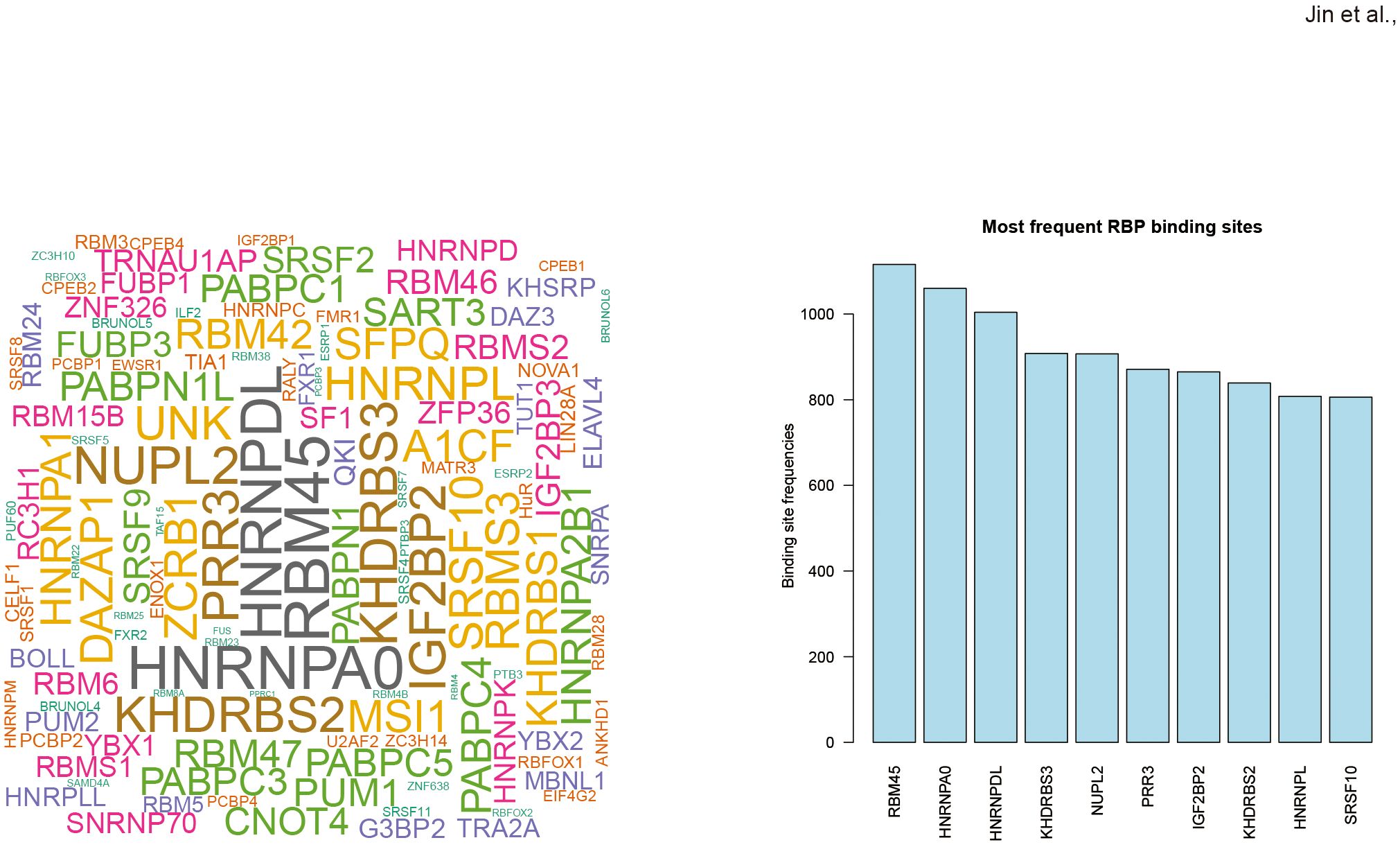
Figure 5. Differential RNA editing in response to COVID-19 vaccines might affect RBP binding sites. (A) Wordcloud plot of RBPs with binding sites overlapped with COVID-19 vaccines-associated DRE. (B) Top ten frequent RBPs with binding sites overlapped with differential RNA editing in response to COVID-19 vaccines.
RNA editing in response to COVID-19 vaccines could be dose-dependent
The three dose groups of COVID-19 vaccines were then compared to the V0 group separately to analyze DRE in individual dose groups (Figure 6A). Among the DRE sites in individual dose groups, 79 were shared by three vaccine dose groups, with 318, 1128, and 2024 exclusively found in V1, V2A, and V3A, respectively. As for genes differentially edited in individual dose groups, 181 were shared by the three vaccine dose groups, whereas 43, 151, and 497 were differentially edited exclusively in V1, V2A, and V3A, respectively (Figure 6B). Notably, such DRE in individual dose groups were found in several anti-viral- and immune-related genes, such as mitochondrial antiviral signaling protein (MAVS:chr20:3868756) and interleukin 1 receptor-associated kinase 4 (IRAK4:chr12:43787946) (Figures 6C, D) in V1, eukaryotic translation initiation factor 2 alpha kinase 2 (EIF2AK2:chr2:37100517/37100873/37100899) (Figures 6E–G) in V2A, and interleukin 6 receptor (IL6R:chr1:154445948) and interferon-stimulated exonuclease gene 20 (ISG20:chr15:88642697) (Figures 6H, I) in V3A.
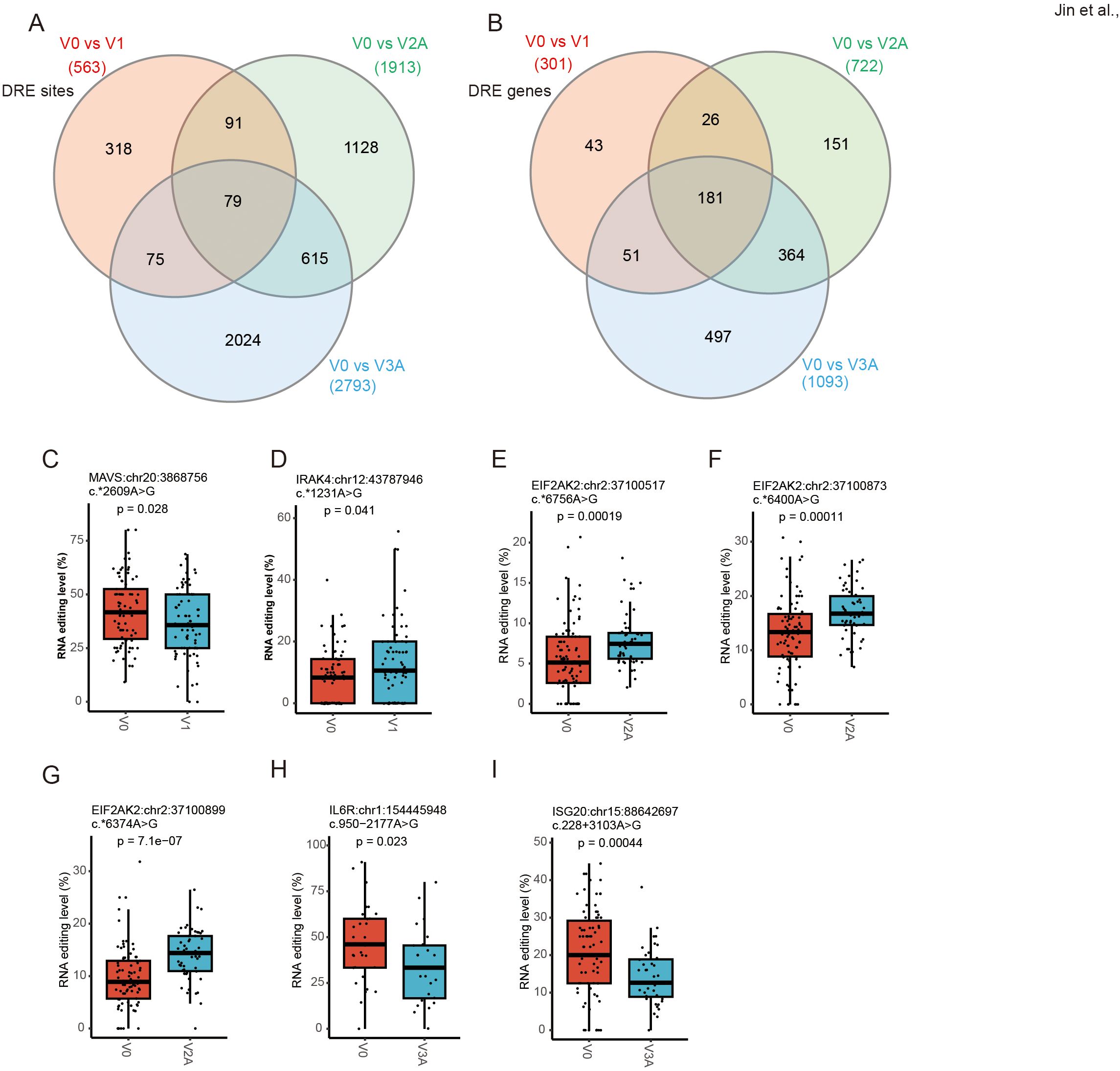
Figure 6. Dose-specific A-to-I RNA editing in response to COVID-19 vaccines. (A, B) The Venn diagram of four groups of DRE genes (A) and sites (B). (C, D) The RNA editing levels of MAVS (chr20:3868756) (C) and IRAK4 (chr12:43787946) (D) in V1. (E, G) The RNA editing levels of EIF2AK2 (chr2:37100517/37100873/37100899) in V2A. (H, I) The RNA editing levels of IL6R:chr1:154445948 (H) and ISG20:chr15:88642697 (I) in V3A.
GO, and KEGG analysis was then performed to explore the biological function of such DRE in individual dose groups. In line with the results of overall DRE across the whole vaccination regime, DRE in different individual dose groups was also found to be mainly related to immune response and viral infections (Figures 7A–F). Notably, a set of gene functions and pathways, such as neutrophil-mediated immunity and defense response to virus, showed enrichment scores increasing with the number of vaccine doses, especially in V2A and V3A.
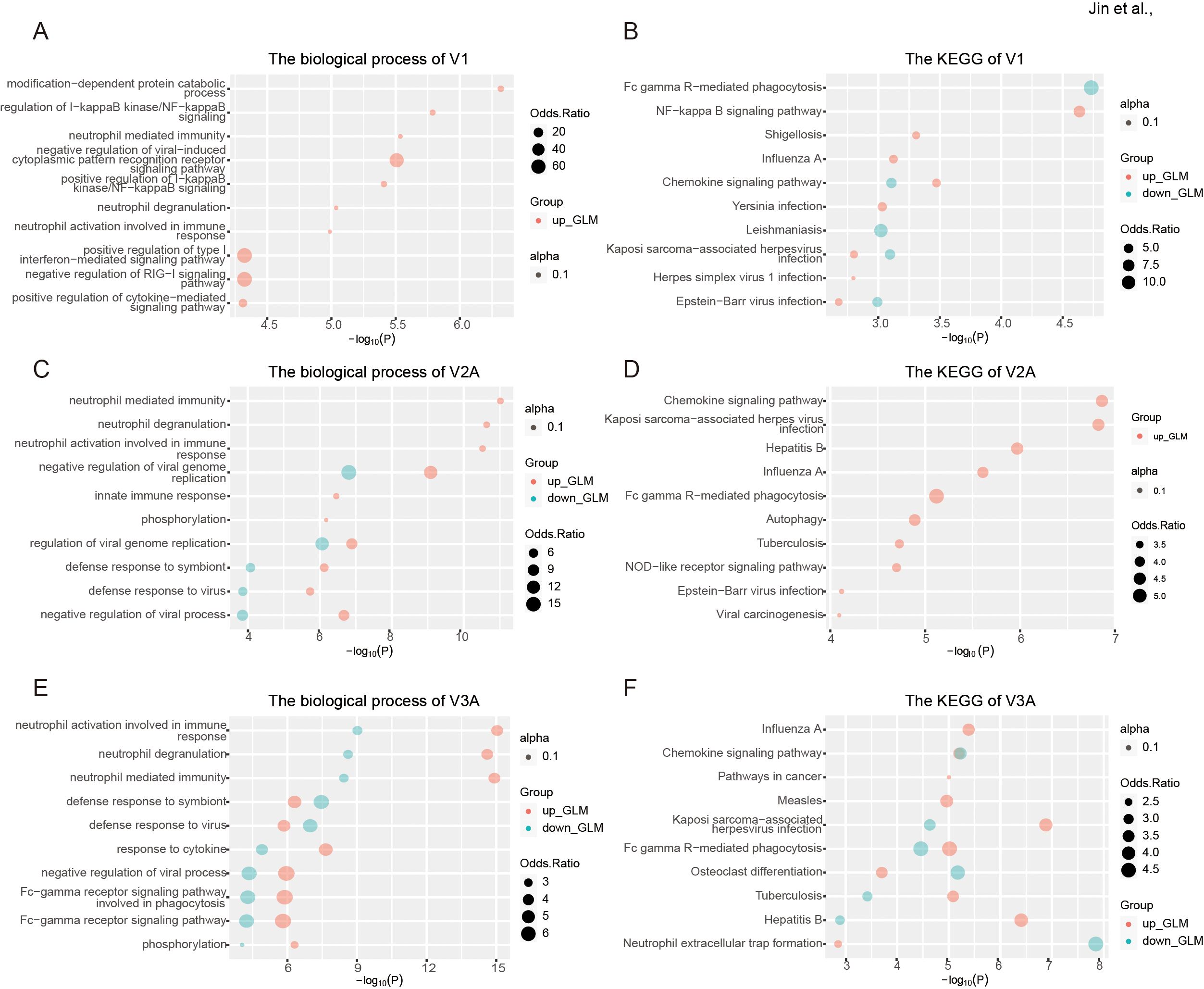
Figure 7. Functional relevance of differential RNA editing in response to COVID-19 vaccines in different individual dose groups. (A, B) The biological process (A) and KEGG pathways (B) of one-dose vaccination in response to one-dose vaccination. (C, D) The biological process (C) and KEGG pathways (D) of two-dose vaccination in response to two-dose vaccination. (E, F) The biological process (E) and KEGG pathways (F) of DRE in response to three-dose vaccination.
Discussion
The underlying epigenetic changes and mechanisms related to RNA editing in response to COVID-19 vaccines remained largely unclear before our studies (31). By systematically investigating blood A-to-I RNA editing in response to COVID-19 vaccines, our results thus suggested a potentially important role of A-to-I RNA editing in regulating the immune response to COVID-19 vaccines.
Compared with V0, our results showed that ADAR expression and the average level of A-to-I RNA editing increased with doses during vaccination (Figure 2), pointing to enhanced RNA editing activity, an essential component of the anti-viral innate immune system. Vaccinated individuals could have enhanced immunity against COVID-19, which could also be in part attributed to the higher level of ADAR and RNA editing activity. Previous studies have shown that ADAR can inhibit hepatitis C virus (HCV) viral RNA replication by editing viral RNA during the virus replication (32). In addition to such direct anti-viral effects through RNA editing, ADAR might also influence the HCV viral cycle in an editing-independent manner by suppressing PKR activation (33, 34) and promoting up-regulated expression of anti-viral microRNAs (35). Emerging studies also reported that ADAR could modulate the immune response by editing host RNA during SARS-CoV-2 infection (20). Therefore, our results of RNA editing in response to COVID-19 vaccines showed a consistent role of ADAR-mediated A-to-I RNA editing in modulating the host’s immunity against SARS-CoV-2 infection. Moreover, individuals receiving more COVID-19 vaccine doses showed a higher immune response, in line with a higher level of RNA editing response that was also mainly related to immune and viral biological processes and pathways. Future research will further explore the underlying mechanism of specific editing events in the host’s antiviral immunity and their potential importance in the treatment and prevention of SARS-CoV-2 infections.
Missense RNA editing alters the amino acid sequences, potentially increasing protein diversity or affecting protein structure, stability, and functions (15). In our study, IFI30 is a very significant missense DRE gene (Supplementary Table S4). IFI30 encodes a crucial enzyme involved in antigen processing and presentation. Its expression could be induced by interferons, signaling molecules released by infected cells to alert neighboring cells of the presence of antigens (36). Additionally, IFI30 is associated with an enhanced immune and inflammatory response mediated by leukocytes and can regulate the IL6-STAT6 pathway (37). Increased IFI30 expression has been shown to enhance the ability of immune cells to eliminate various cancers by promoting the antigen presentation process (38–40). Recent studies have highlighted the therapeutic potential of targeting IFI30 in anti-tumor strategies, suggesting that modulating its expression or activity could regulate the immune response against tumor cells (37, 41). Moreover, IFI30 plays an important role in the initiation of CD4 and CD8 T-cell responses against viral peptides and exerts its antiviral effect by inhibiting cathepsin L activity of SARS-CoV, Ebola virus, and Lassa virus (42). In addition, GBP1 encodes a guanylate-binding protein that plays a key role in inflammatory pyroptosis and is involved in innate immunity against a diverse range of bacterial, viral, and protozoan pathogens (43, 44). Our analysis further suggested that these missense RNA editing events could exert their biological effects by altering the structure and function of the encoded proteins or cis-regulating the edited mRNA expression. Our findings thus pointed to a potentially important role of these missense editing in acting against viral infections by enhancing host immune and proinflammatory responses during COVID-19 vaccines.
Among the 3’ UTR DRE variants, the hyper-edited APOL6 gene was one of the genes that exhibited the most significant differential RNA editing (Supplementary Table S7). Studies have revealed that APOL6 can inhibit the replication of certain viruses, such as coxsackie B virus and poliovirus (45), and its expression was significantly up-regulated in association with HIV-associated neurocognitive disorders (46). Furthermore, APOL6 are strongly upregulated upon inflammation via the Janus kinase (JAK)-signal transducer and activator of transcription (STAT) pathway, which are important signaling pathways involved in immune responses (47). Additionally, studies have shown that APOL6 is upregulated in immunotherapy responders, and enhances the efficacy of anti-tumor immunotherapy by promoting tumor cell apoptosis, necrosis, and pyroptosis pathways (48, 49). Therefore, our findings in APOL6 3’ UTR RNA editing were in line with the role of the gene in both antiviral defense and immune regulation. The CTSS gene was previously reported as a target of ADAR (50), strongly associated with type I IFN signature (51), and pivotal in MHC-II antigen loading and production of autoantibodies (52). Consistent with previous studies, elevated RNA editing levels of specific adenosines within CTSS 3’ UTR Alu elements correlate with increased CTSS expression (51). Moreover, the RNA editing levels of CTSS also significantly increased with vaccine dose (Figures 4H, I), and the changed editing sites also performed noticeable RNA structural alterations (Figures 4M, N). Therefore, the RNA secondary structure changes associated with RNA editing could also increased with vaccine doses. These findings underscore the potential vaccine dose-dependent RNA structure-specific editing and warranted future study on its functional implications.
Our results also indicate the possible involvement of RBP in the biological effects of RNA editing response to COVID-19 vaccines. NUPL2, also known as CG1, is required for the export of mRNAs containing poly(A) tails from the nucleus into the cytoplasm and could participate in the docking of viral Vpr at the nuclear envelope during HIV-1 infection (53). SART3 encoded an RNA-binding nuclear protein, which is found to be an important cellular factor for HIV-1 gene expression and viral replication. It also is transiently associated with U6 and U4/U6 snRNPs during the recycling phase of the spliceosome cycle and is involved in the regulation of mRNA splicing (54). Our findings thus warrant further experimental analysis of the actual biological impact of RBP binding in RNA editing response to COVID-19 vaccines.
While RNA transcription precedes RNA editing, it is well-established that RNA editing is a post-transcriptional modification mechanism that can dynamically modulate RNA stability, alternative splicing patterns, and even translation efficiency (55–57). These processes can significantly impact gene expression profiles in response to external stimuli such as vaccination. Specifically, RNA editing by ADAR enzymes can alter the sequence of transcripts by converting A to I, which are interpreted as G during translation. This editing process is known to affect the functional diversity of RNA molecules and consequently influence the expression of immune-regulatory genes. Furthermore, the temporal relationship between RNA transcription and editing underscores the dynamic nature of cellular responses to vaccine dosage. Changes in editing patterns may reflect adaptive responses of immune cells, where alterations in RNA editing profiles could fine-tune immune gene expression to optimize immune responses.
In conclusion, our study systematically investigated blood A-to-I RNA editing and revealed its dynamic response to COVID-19 vaccines. Our findings linked RNA editing with the immune response and antiviral effects of COVID-19 vaccines.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
Ethical approval was not required for the study involving humans in accordance with the local legislation and institutional requirements. Written informed consent to participate in this study was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and the institutional requirements.
Author contributions
Y-YJ: Writing – original draft, Writing – review & editing. Y-PL: Formal analysis, Writing – original draft. J-QP: Formal analysis, Writing – original draft. W-HH: Formal analysis, Writing – original draft. Y-MF: Formal analysis, Writing – original draft. W-JS: Software, Writing – review & editing. HY: Software, Writing – review & editing. X-DT: Software, Writing – review & editing. LZ: Writing – review & editing. J-HC: Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported in part by grants from the National Natural Science Foundation of China (No. 82302492, 31671311, 82070987), the “Six Talent Peak” Plan of Jiangsu Province (No. SWYY-127), the Innovative and Entrepreneurial Doctorate of Jiangsu Province (JSSCBS20230197), the Wuxi Science and Technology Development Fund Project (K20231035), the Fundamental Research Funds for the Central Universities (JUSRP123077), Start-Up Research Grants in Medical Colleges (No. 1286010241222110), Jiangsu university students innovation and entrepreneurship project (202410295116Y), and School-Level Innovation and Entrepreneurship Project at Jiangnan University for supporting the study.
Acknowledgments
We thank Ryan et al., for sharing the RNA-Seq data.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1413704/full#supplementary-material
References
1. V'Kovski P, Kratzel A, Steiner S, Stalder H, Thiel V. Coronavirus biology and replication: Implications for Sars-Cov-2. Nat Rev Microbiol. (2021) 19:155–70. doi: 10.1038/s41579-020-00468-6
2. Ryan FJ, Norton TS, McCafferty C, Blake SJ, Stevens NE, James J, et al. A systems immunology study comparing innate and adaptive immune responses in adults to Covid-19 mrna and adenovirus vectored vaccines. Cell Rep Med. (2023) 4:100971. doi: 10.1016/j.xcrm.2023.100971
3. Skelly DT, Harding AC, Gilbert-Jaramillo J, Knight ML, Longet S, Brown A, et al. Two doses of Sars-Cov-2 vaccination induce robust immune responses to emerging Sars-Cov-2 variants of concern. Nat Commun. (2021) 12:5061. doi: 10.1038/s41467-021-25167-5
4. Bian S, Li L, Wang Z, Cui L, Xu Y, Guan K, et al. Allergic reactions after the administration of covid-19 vaccines. Front Public Health. (2022) 10:878081. doi: 10.3389/fpubh.2022.878081
5. Greenhawt M, Abrams EM, Shaker M, Chu DK, Khan D, Akin C, et al. The risk of allergic reaction to Sars-Cov-2 vaccines and recommended evaluation and management: A systematic review, meta-analysis, grade assessment, and international consensus approach. J Allergy Clin Immunol Pract. (2021) 9:3546–67. doi: 10.1016/j.jaip.2021.06.006
6. Gupta RK, Topol EJ. Covid-19 vaccine breakthrough infections. Science. (2021) 374:1561–2. doi: 10.1126/science.abl8487
7. Lipsitch M, Krammer F, Regev-Yochay G, Lustig Y, Balicer RD. Sars-Cov-2 breakthrough infections in vaccinated individuals: measurement, causes and impact. Nat Rev Immunol. (2022) 22:57–65. doi: 10.1038/s41577-021-00662-4
8. Chemaitelly H, Abu-Raddad LJ. Waning effectiveness of Covid-19 vaccines. Lancet. (2022) 399:771–3. doi: 10.1016/s0140-6736(22)00277-x
9. Goldberg Y, Mandel M, Bar-On YM, Bodenheimer O, Freedman L, Haas EJ, et al. Waning immunity after the bnt162b2 vaccine in Israel. N Engl J Med. (2021) 385:e85. doi: 10.1056/NEJMoa2114228
10. Gott JM, Emeson RB. Functions and mechanisms of rna editing. Annu Rev Genet. (2000) 34:499–531. doi: 10.1146/annurev.genet.34.1.499
11. Di Giorgio S, Martignano F, Torcia MG, Mattiuz G, Conticello SG. Evidence for host-dependent rna editing in the transcriptome of Sars-Cov-2. Sci Adv. (2020) 6:eabb5813. doi: 10.1126/sciadv.abb5813
12. Duan Y, Tang X, Lu J. Evolutionary driving forces of a-to-I editing in metazoans. Wiley Interdiscip Rev RNA. (2022) 13:e1666. doi: 10.1002/wrna.1666
13. Emrani J, Ahmed M, Jeffers-Francis L, Teleha JC, Mowa N, Newman RH, et al. Sars-Cov-2, infection, transmission, transcription, translation, proteins, and treatment: A review. Int J Biol Macromol. (2021) 193:1249–73. doi: 10.1016/j.ijbiomac.2021.10.172
14. Heale BS, Keegan LP, McGurk L, Michlewski G, Brindle J, Stanton CM, et al. Editing independent effects of adars on the mirna/sirna pathways. EMBO J. (2009) 28:3145–56. doi: 10.1038/emboj.2009.244
15. Nishikura K. Functions and regulation of rna editing by adar deaminases. Annu Rev Biochem. (2010) 79:321–49. doi: 10.1146/annurev-biochem-060208-105251
16. Picardi E, Mansi L, Pesole G. Detection of a-to-I rna editing in Sars-Cov-2. Genes (Basel). (2021) 13(1):41. doi: 10.3390/genes13010041
17. Song Y, He X, Yang W, Wu Y, Cui J, Tang T, et al. Virus-specific editing identification approach reveals the landscape of a-to-I editing and its impacts on Sars-Cov-2 characteristics and evolution. Nucleic Acids Res. (2022) 50:2509–21. doi: 10.1093/nar/gkac120
18. Crooke PS 3rd, Tossberg JT, Porter KP, Aune TM. Reduced a-to-I Editing of Endogenous Alu Rnas in Lung after Sars-Cov-2 Infection. Curr Res Immunol. (2021) 2:52–9. doi: 10.1016/j.crimmu.2021.04.001
19. Peng X, Luo Y, Li H, Guo X, Chen H, Ji X, et al. Rna editing increases the nucleotide diversity of Sars-Cov-2 in human host cells. PLoS Genet. (2022) 18:e1010130. doi: 10.1371/journal.pgen.1010130
20. Wei ZY, Wang ZX, Li JH, Wen YS, Gao D, Xia SY, et al. Host a-to-I rna editing signatures in intracellular bacterial and single-strand rna viral infections. Front Immunol. (2023) 14:1121096. doi: 10.3389/fimmu.2023.1121096
21. Tao J, Ren CY, Wei ZY, Zhang F, Xu J, Chen JH. Transcriptome-wide identification of G-to-a rna editing in chronic social defeat stress mouse models. Front Genet. (2021) 12:680548. doi: 10.3389/fgene.2021.680548
22. Dobin A, Gingeras TR. Mapping rna-seq reads with star. Curr Protoc Bioinf. (2015) 51:11.4.1–.4.9. doi: 10.1002/0471250953.bi1114s51
23. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and samtools. Bioinformatics. (2009) 25:2078–9. doi: 10.1093/bioinformatics/btp352
24. Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, et al. From fastq data to high confidence variant calls: The genome analysis toolkit best practices pipeline. Curr Protoc Bioinf. (2013) 43:11.0.1–.0.33. doi: 10.1002/0471250953.bi1110s43
25. Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, et al. Varscan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. (2012) 22:568–76. doi: 10.1101/gr.129684.111
26. McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, et al. The ensembl variant effect predictor. Genome Biol. (2016) 17:122. doi: 10.1186/s13059-016-0974-4
27. Mansi L, Tangaro MA, Lo Giudice C, Flati T, Kopel E, Schaffer AA, et al. Rediportal: millions of novel a-to-I rna editing events from thousands of rnaseq experiments. Nucleic Acids Res. (2021) 49:D1012–d9. doi: 10.1093/nar/gkaa916
28. Liao Y, Smyth GK, Shi W. Featurecounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. (2014) 30:923–30. doi: 10.1093/bioinformatics/btt656
29. Zhou Y, Pan Q, Pires DEV, Rodrigues CHM, Ascher DB. Ddmut: predicting effects of mutations on protein stability using deep learning. Nucleic Acids Res. (2023) 51:W122–w8. doi: 10.1093/nar/gkad472
30. Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. (2016) 44:W90–7. doi: 10.1093/nar/gkw377
31. Zhang Y, Guo X, Li C, Kou Z, Lin L, Yao M, et al. Transcriptome analysis of peripheral blood mononuclear cells in Sars-Cov-2 naïve and recovered individuals vaccinated with inactivated vaccine. Front Cell Infect Microbiol. (2021) 11:821828. doi: 10.3389/fcimb.2021.821828
32. Taylor DR, Puig M, Darnell ME, Mihalik K, Feinstone SM. New antiviral pathway that mediates hepatitis C virus replicon interferon sensitivity through adar1. J Virol. (2005) 79:6291–8. doi: 10.1128/jvi.79.10.6291-6298.2005
33. Garaigorta U, Chisari FV. Hepatitis C virus blocks interferon effector function by inducing protein kinase R phosphorylation. Cell Host Microbe. (2009) 6:513–22. doi: 10.1016/j.chom.2009.11.004
34. Pfaller CK, George CX, Samuel CE. Adenosine deaminases acting on rna (Adars) and viral infections. Annu Rev Virol. (2021) 8:239–64. doi: 10.1146/annurev-virology-091919-065320
35. Liu G, Ma X, Wang Z, Wakae K, Yuan Y, He Z, et al. Adenosine deaminase acting on rna-1 (Adar1) inhibits hepatitis B virus (Hbv) replication by enhancing microrna-122 processing. J Biol Chem. (2019) 294:14043–54. doi: 10.1074/jbc.RA119.007970
36. Wang X, Ge X, Qin Y, Liu D, Chen C. Ifi30 is required for sprouting angiogenesis during caudal vein plexus formation in zebrafish. Front Physiol. (2022) 13:919579. doi: 10.3389/fphys.2022.919579
37. Zhu C, Chen X, Guan G, Zou C, Guo Q, Cheng P, et al. Ifi30 is a novel immune-related target with predicting value of prognosis and treatment response in glioblastoma. Onco Targets Ther. (2020) 13:1129–43. doi: 10.2147/ott.S237162
38. Buetow KH, Meador LR, Menon H, Lu YK, Brill J, Cui H, et al. High gilt expression and an active and intact mhc class ii antigen presentation pathway are associated with improved survival in melanoma. J Immunol. (2019) 203:2577–87. doi: 10.4049/jimmunol.1900476
39. Jiang W, Zheng F, Yao T, Gong F, Zheng W, Yao N. Ifi30 as a prognostic biomarker and correlation with immune infiltrates in glioma. Ann Transl Med. (2021) 9:1686. doi: 10.21037/atm-21-5569
40. Zhou C, Wei Z, Zhang L, Yang Z, Liu Q. Systematically characterizing a-to-I rna editing neoantigens in cancer. Front Oncol. (2020) 10:593989. doi: 10.3389/fonc.2020.593989
41. Fan Y, Wang X, Li Y. Ifi30 expression predicts patient prognosis in breast cancer and dictates breast cancer cells proliferation via regulating autophagy. Int J Med Sci. (2021) 18:3342–52. doi: 10.7150/ijms.62870
42. Majdoul S, Compton AA. Lessons in self-defence: inhibition of virus entry by intrinsic immunity. Nat Rev Immunol. (2022) 22:339–52. doi: 10.1038/s41577-021-00626-8
43. Ghosh A, Praefcke GJ, Renault L, Wittinghofer A, Herrmann C. How guanylate-binding proteins achieve assembly-stimulated processive cleavage of gtp to gmp. Nature. (2006) 440:101–4. doi: 10.1038/nature04510
44. Li P, Jiang W, Yu Q, Liu W, Zhou P, Li J, et al. Ubiquitination and degradation of gbps by a shigella effector to suppress host defence. Nature. (2017) 551:378–83. doi: 10.1038/nature24467
45. Kreit M, Vertommen D, Gillet L, Michiels T. The interferon-inducible mouse apolipoprotein L9 and prohibitins cooperate to restrict theiler's virus replication. PLoS One. (2015) 10:e0133190. doi: 10.1371/journal.pone.0133190
46. Siangphoe U, Archer KJ. Gene expression in hiv-associated neurocognitive disorders: A meta-analysis. J Acquir Immune Defic Syndr. (2015) 70:479–88. doi: 10.1097/qai.0000000000000800
47. Adão R, Guzik TJ. Inside the heart of Covid-19. Cardiovasc Res. (2020) 116:e59–61. doi: 10.1093/cvr/cvaa086
48. Liu K, Chen Y, Li B, Li Y, Liang X, Lin H, et al. Upregulation of apolipoprotein L6 improves tumor immunotherapy by inducing immunogenic cell death. Biomolecules. (2023) 13(3):415. doi: 10.3390/biom13030415
49. Liu Z, Lu H, Jiang Z, Pastuszyn A, Hu CA. Apolipoprotein L6, a novel proapoptotic bcl-2 homology 3-only protein, induces mitochondria-mediated apoptosis in cancer cells. Mol Cancer Res. (2005) 3:21–31.
50. Stellos K, Gatsiou A, Stamatelopoulos K, Perisic Matic L, John D, Lunella FF, et al. Adenosine-to-inosine rna editing controls cathepsin S expression in atherosclerosis by enabling hur-mediated post-transcriptional regulation. Nat Med. (2016) 22:1140–50. doi: 10.1038/nm.4172
51. Vlachogiannis NI, Tual-Chalot S, Zormpas E, Bonini F, Ntouros PA, Pappa M, et al. Adenosine-to-inosine rna editing contributes to type I interferon responses in systemic sclerosis. J Autoimmun. (2021) 125:102755. doi: 10.1016/j.jaut.2021.102755
52. Riese RJ, Wolf PR, Brömme D, Natkin LR, Villadangos JA, Ploegh HL, et al. Essential role for cathepsin S in mhc class ii-associated invariant chain processing and peptide loading. Immunity. (1996) 4:357–66. doi: 10.1016/s1074-7613(00)80249-6
53. Le Rouzic E, Mousnier A, Rustum C, Stutz F, Hallberg E, Dargemont C, et al. Docking of hiv-1 vpr to the nuclear envelope is mediated by the interaction with the nucleoporin hcg1. J Biol Chem. (2002) 277:45091–8. doi: 10.1074/jbc.M207439200
54. Bell M, Schreiner S, Damianov A, Reddy R, Bindereif A. P110, a novel human U6 snrnp protein and U4/U6 snrnp recycling factor. EMBO J. (2002) 21:2724–35. doi: 10.1093/emboj/21.11.2724
55. Hsiao YE, Bahn JH, Yang Y, Lin X, Tran S, Yang EW, et al. Rna editing in nascent rna affects pre-mrna splicing. Genome Res. (2018) 28:812–23. doi: 10.1101/gr.231209.117
56. Rieder LE, Reenan RA. The intricate relationship between rna structure, editing, and splicing. Semin Cell Dev Biol. (2012) 23:281–8. doi: 10.1016/j.semcdb.2011.11.004
Keywords: SARS-CoV-2, A-to-I RNA editing, ADARs, COVID-19 vaccine, immune
Citation: Jin Y-Y, Liang Y-P, Pan J-Q, Huang W-H, Feng Y-M, Sui W-J, Yu H, Tang X-D, Zhu L and Chen J-H (2024) RNA editing in response to COVID-19 vaccines: unveiling dynamic epigenetic regulation of host immunity. Front. Immunol. 15:1413704. doi: 10.3389/fimmu.2024.1413704
Received: 07 April 2024; Accepted: 14 August 2024;
Published: 06 September 2024.
Edited by:
Tanushree Dangi, Northwestern University, United StatesReviewed by:
Yaping Sun, Yale University, United StatesJingzhe Shang, Chinese Academy of Medical Sciences, China
Copyright © 2024 Jin, Liang, Pan, Huang, Feng, Sui, Yu, Tang, Zhu and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian-Huan Chen, Y2poX2Jpb0Bob3RtYWlsLmNvbQ==
 Yun-Yun Jin
Yun-Yun Jin Ya-Ping Liang
Ya-Ping Liang Jia-Qi Pan
Jia-Qi Pan Wen-Hao Huang1,2,3
Wen-Hao Huang1,2,3 Jian-Huan Chen
Jian-Huan Chen