
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Immunol., 08 July 2024
Sec. Primary Immunodeficiencies
Volume 15 - 2024 | https://doi.org/10.3389/fimmu.2024.1411141
This article is part of the Research TopicAdvancing Research, Clinical Recognition, and Targeted Therapies for WHIM SyndromeView all 4 articles
Warts, Hypogammaglobulinemia, Infections, Myelokathexis (WHIM) syndrome is a rare, combined immunodeficiency disease predominantly caused by gain-of-function variants in the CXCR4 gene that typically results in truncation of the carboxyl terminus of C-X-C chemokine receptor type 4 (CXCR4) leading to impaired leukocyte egress from bone marrow to peripheral blood. Diagnosis of WHIM syndrome continues to be challenging and is often made through clinical observations and/or genetic testing. Detection of a pathogenic CXCR4 variant in an affected individual supports the diagnosis of WHIM syndrome but relies on an appropriate annotation of disease-causing variants. Understanding the genotypic-phenotypic associations in WHIM syndrome has the potential to improve time to diagnosis and guide appropriate clinical management, resulting in a true example of precision medicine. This article provides an overview of the spectrum of CXCR4 variants in WHIM syndrome and summarizes the various lines of clinical and functional evidence that can support interpretation of newly identified variants.
WHIM syndrome is a rare autosomal dominant combined immunodeficiency disease (OMIM #193670). The WHIM acronym refers to a set of typical clinical features of the disease, namely warts, hypogammaglobulinemia, infections, and myelokathexis. However, the full tetrad of symptoms is found only in a minority of individuals (22%-38%) (1–3). The most penetrant clinical and laboratory phenotypes in individuals with WHIM syndrome are severe neutropenia due to impaired release of neutrophils from the bone marrow, lymphopenia, and recurrent bacterial infections (1–3). The disease was first described by Zuelzer in 1964 (4), but the genetic etiology was only discovered 39 years later when variants in the cytoplasmic C-terminal tail of C-X-C chemokine receptor 4 (CXCR4) were linked to the pathology of WHIM syndrome in seven independent kindreds (5). As per European Society for Immunodeficiencies–Pan-American Group for Immunodeficiency diagnostic criteria, identification of a CXCR4 variant in the intracellular C-terminal tail of the receptor or an activating CXCR4 variant supports the definitive diagnosis of WHIM syndrome (6). Owing to the availability of sequencing, the spectrum of CXCR4WHIM variants has grown dramatically since the first discovery of 3 disease-causing variants (5, 7, 8). In this review, we aim to summarize the current knowledge of the landscape of CXCR4 variants in WHIM syndrome, as well as provide an overview of functional assays that can support interpretation of newly discovered CXCR4 variants and their pathogenic role in the disease.
Heterozygous C-terminal CXCR4 variants are found in ≈90% of individuals with clinical diagnosis of WHIM syndrome (2, 3). p.R334* (c.1000C>T), 1 of the 3 originally discovered pathogenic CXCR4 variants (5), is the most frequent one, accounting for 47% to 55% of CXCR4 variants in WHIM syndrome (1–3). p.S338* (c.1013C>G or C>A) is the second most frequent; it is found in 16% to 17% of individuals with CXCR4 variants (1–3). As of March 2023, 33 additional C-terminal variants have been reported, often occurring de novo in a single individual or in several members of 1 family (2, 5, 9–23) (Figure 1A (24), Supplementary Table 1). The total of 36 variants can be divided into 3 subgroups, composed of 1 missense, 8 nonsense, and 27 frameshift variants (Supplementary Table 1). The affected region spans amino acids (aa) 317 to 346, with nonsense variants occurring between aa 332 to 346 and frameshift variants between aa 317 and 346. Both +1 and +2 frameshift variants have been detected in individuals with WHIM syndrome (Figure 1B). The span of CXCR4WHIM variants is very similar to somatic CXCR4 C-terminal variants found in Waldenström macroglobulinemia, a rare indolent B-cell lymphoma (25, 26). Notably, the number of distinct variants is higher in Waldenström macroglobulinemia, and the C-terminal region affected by frameshift variants extends to position T311 (p.T311Ifs*33) (26) and to K327 for nonsense variants (p.K327*) (27). It is therefore likely that additional novel variants will be identified in individuals with WHIM syndrome in the future, especially due to many possible combinations of indels leading to frameshift variants in the C-terminus of CXCR4.
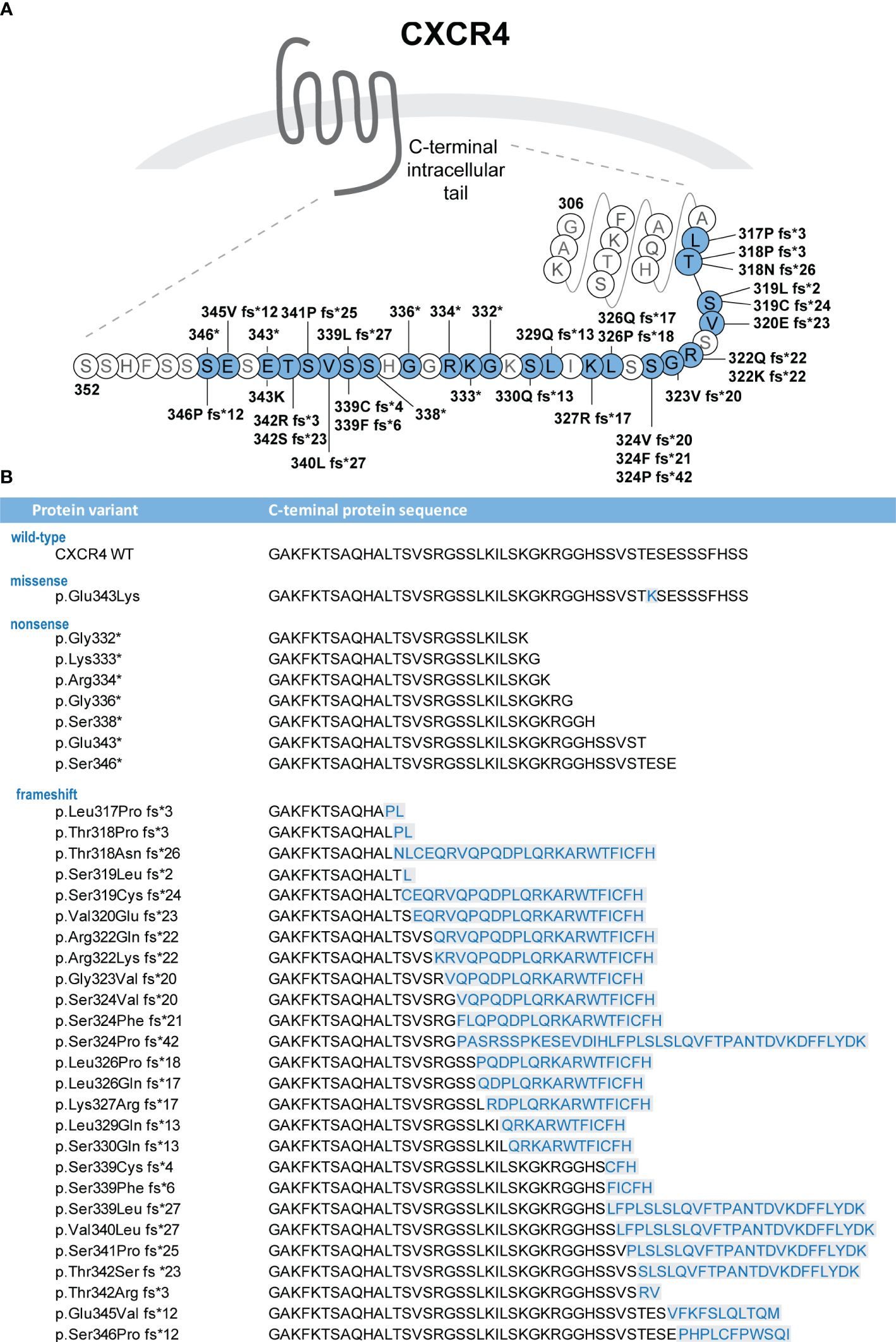
Figure 1 (A) CXCR4 variants identified in individuals with WHIM syndrome are localized in the C-terminal intracellular tail of the receptor (24). The figure indicates protein variants identified to date and positions at which they alter the wild-type sequence of the CXCR4 protein. (B) Protein sequences of the C-terminus variants are shown. Missense variants and de novo sequences resulting from the frameshift variants are highlighted in blue color. *: translation termination codon (stop codon).
Interpretation of variants’ pathogenicity for the underlying condition is an essential aspect of genetic testing. Variant classification is performed according to internationally accepted standards and relies on several lines of clinical and functional types of evidence (28, 29).
One line of clinical evidence in variant classification considers whether the detected variant has been previously observed in clinically affected individuals and families with a condition while being absent in unaffected individuals (28, 29). To access genetic variant data, clinicians and investigators frequently use reference databases such as ClinVar (30), an international public archive of variant-condition interpretations hosted by the National Center for Biotechnology Information. As of December 6, 2023, ClinVar contained entries for 19 of the 36 CXCR4 variants that have been identified in individuals with WHIM syndrome in scientific literature, with 15 of them classified as likely pathogenic or pathogenic, 4 as variant of uncertain significance (Supplementary Table 1) (31). Hence, CXCR4 variant annotation in ClinVar has been outpaced by scientific reports, and to accurately classify the newly identified CXCR4 variants, it is critical to review the recent literature. Of note, a thorough variant interpretation per American College of Medical Genetics and Genomics – Association for Molecular Pathology (ACMG-AMP) guidelines is rarely performed in publications, with a few exceptions (2, 32) (Supplementary Table 1), but such publications still represent a valuable resource for correlating genotype with clinical phenotype.
For a variant to be classified as pathogenic or likely pathogenic, it should segregate with the disease or occur de novo in the affected members of a family, and the allele frequency should be as low as the expected prevalence of WHIM syndrome (29). As of November 29, 2023, a majority of the known CXCR4WHIM variants were absent in gnomAD (formerly ExAC), the largest publicly available aggregated dataset of human variant allele spectrum and frequencies (33). Two variants, c.1006G>T/p.G336*, pathogenic, and c.1013C>A/p.S338*, likely pathogenic, are each found in the gnomAD dataset at allele frequency 1.60e-6 (34).
In silico tools that predict the impact of sequence variants can also bring valuable input into the variant classification process (29, 35). Specifically, combined annotation‐dependent depletion (2, 32), MutationTaster and PROVEAN (23) were previously used to assess CXCR4 variants. Additional in silico approaches to predict pathogenicity of missense variants include Polyphen-2 and SIFT, but these tools may be of limited use to evaluate CXCR4WHIM variants that primarily comprise nonsense and frameshift variants (35). Indeed, in silico algorithms are generally not well suited to predicting pathogenicity of nonsense and frameshift variants. Most CXCR4WHIM variants generate a premature stop codon; therefore, it is important to evaluate whether the variant in question will likely escape nonsense-mediated decay and lead to production of pathogenic protein (28, 29).
Per ACMG-AMP guidelines, only variants classified as pathogenic or likely pathogenic should be used to establish a positive molecular diagnosis (29). Identification of a novel variant of unknown significance poses a significant challenge in clinical decision-making (36). In cases where the clinical data, family history, variant frequency, and/or in silico modeling data are not sufficient for a confident variant interpretation, functional biochemical studies can serve as a powerful tool for obtaining additional evidence of pathogenicity (28, 29, 35, 37, 38). The particulars of CXCR4-specific functional tests will be discussed in the next section.
Per ACMG-AMP guidelines, clinical laboratories should implement an internal system to track sequence variants, evidence assertions, and variant classifications (29). Invitae, a certified clinical diagnostic laboratory and the largest submitter of variant data to ClinVar, performed CXCR4 variant interpretation according to Sherloc framework (28), a refined version of the ACMG-AMP criteria (29), considering all applicable lines of evidence. Thirty of the 36 previously observed CXCR4WHIM variants have been classified as pathogenic, and 6 variants as likely pathogenic for WHIM syndrome based on public databases, clinical data at Invitae, published literature at the time of observation, and functional studies. Absence or low frequency in the general population (per gnomAD), segregation with disease, de novo occurrence in affected individual, reports of multiple unrelated cases, variant type (frameshift, nonsense, missense), expected consequence for the gene product (disruption of C-terminus, escape nonsense-mediated decay) and experimental data (impaired internalization) were factors that conferred pathogenic points for CXCR4 variant classification (Supplementary Table 1) (8).
CXCR4 is a 7-transmembrane G protein-coupled receptor that binds cognate ligand C-X-C motif chemokine ligand 12 (CXCL12)/stromal cell-derived factor 1 (39, 40) to regulate leukocyte trafficking and B-cell development (32, 41–43). The cytoplasmic C-terminal tail CXCR4 harbors a set of phosphorylation motifs that regulate downstream signaling, ß-arrestin binding and internalization of the receptor (44, 45). At the molecular level, CXCR4WHIM variants eliminate or dysregulate the C-terminal phosphorylation, resulting in impaired CXCL12-induced receptor internalization (21), leading to hyperactive (gain-of-function) signaling to downstream pathways (5, 21, 46). These altered responses to CXCL12 underlie the clinical manifestations of WHIM syndrome including enhanced leukocyte retention in bone marrow and defects in adaptive immunity (reviewed in Heusinkveld and Majumdar (3, 47)).
These defects in leukocytes from individuals with WHIM syndrome can be utilized as evidence for a deleterious effect associated with newly identified CXCR4 variants (Figure 2). Most previously published studies have used assays to measure CXCR4 internalization in response to CXCL12 stimulation to show the altered function of CXCR4WHIM in comparison with CXCR4WT-expressing cells (2, 13–15, 21, 46, 48, 49). The decrease of internalization is the most consistent defect among the entire spectrum of CXCR4WHIM variants. Cellular chemotaxis in response to CXCL12 (12–14, 21, 46, 48, 49), intracellular calcium mobilization (5, 12–15, 22, 46, 49), and PI3K-Akt/extracellular signal-regulated kinase activation assays (13, 22, 46, 49) have also been frequently used, but the gain-of-function phenotypes in functional assays have not been uniform across all variants, with early frameshift variants lacking the hyperactive phenotype (13, 49). Additional assays used to investigate the molecular pathology downstream of CXCR4WHIM receptor comprise β-arrestin recruitment (impaired in CXCR4WHIM-expressing cells) (13, 22), Gi-protein dissociation (increased) (13), reduction in intracellular cyclic adenosine monophosphate level (increased or equal) (13, 49), and F-actin polymerization (increased) (21, 48). To probe the potential pathogenicity of a newly discovered variant, we recommend assessing CXCR4 internalization in response to a range of CXCL12 concentrations, and chemotaxis or downstream signaling assays to test for gain-of-function phenotype.
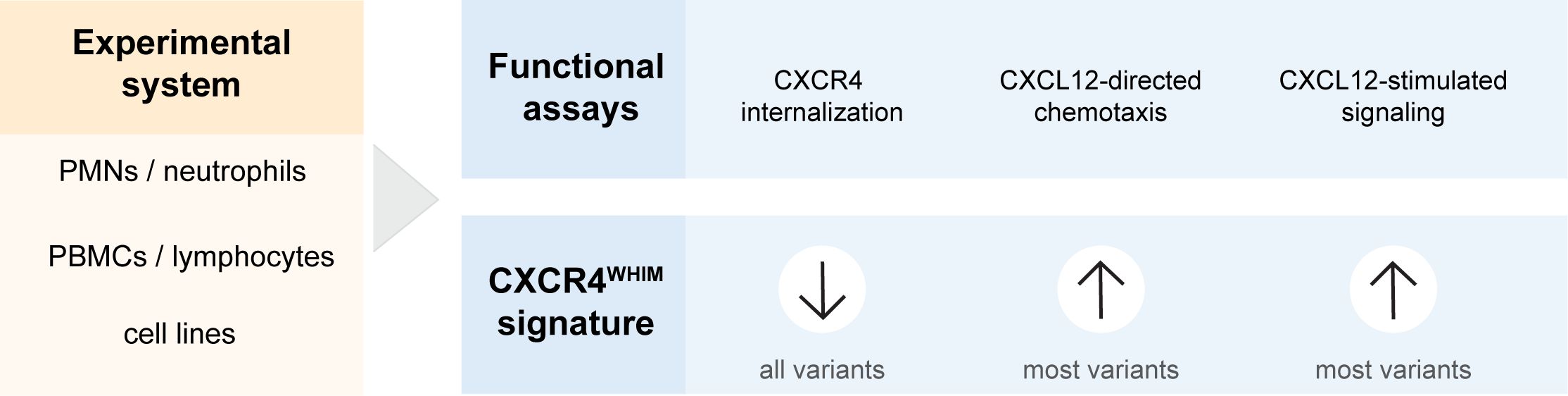
Figure 2 Pipeline of functional tests to support CXCR4 variant interpretation. CXCL12, C-X-C chemokine ligand 12; CXCR4, C-X-C chemokine receptor 4; PBMC, peripheral blood mononuclear cell; PMN, polymorphonuclear leukocyte.
CXCR4 is expressed on the cell surface of mature leukocytes and their progenitors (50), in addition to other cell types such as endothelial cells; therefore, functional experiments can be performed directly with various types of leukocytes isolated from patient blood (Figure 2). Polymorphonuclear leukocytes (12, 14, 46) and peripheral blood mononuclear cells (14, 15, 21, 46) have been used previously to investigate the cellular impairment in CXCR4WHIM variants. However, polymorphonuclear leukocytes may not be preferred due to their short life span and infeasible cryopreservation. Use of peripheral blood mononuclear cells overcomes these drawbacks and additionally enables expansion of T-cell lymphoblasts (12, 13) or generation of immortalized B-cell lines (5, 48) when larger numbers of cells are needed for CXCR4WHIM cellular analysis. Furthermore, cell lines transfected with CXCR4 variants of interest can be used to decipher the pathobiology downstream of CXCR4 receptor in parallel to functional studies with patient peripheral blood mononuclear cells or as an alternative when patient samples are not available (Figure 2). This approach also allows the study of CXCR4 variants in a more standardized and isolated experimental system as they are presented in a homogenous genetic background, allowing direct comparisons of a larger array of variants. Cellular models relying on the overexpression of variant CXCR4 include K562 (2, 14, 15, 46, 49), HEK293 (13, 15, 21, 22), CHO-K1 (21, 46), A0.01 T-cell (21), and Jurkat cell lines (51). In addition, there has been a report of using CRISPR/Cas9 gene editing system to introduce the c.1000C>T p. R334* variant in the endogenous CXCR4 locus in the Jurkat cell line to overcome some of the concerns with the overexpression systems (49).
A small proportion of WHIM syndrome cases were found to be not linked to variants in CXCR4. Two unrelated individuals with a clinical diagnosis of WHIM syndrome, but negative for pathogenic CXCR4 variants, were reported by Balabanian et al. (21). These patients had dysfunction in GRK3, a kinase involved in CXCR4 C-tail phosphorylation (45), resulting in impairment of CXCR4 internalization and thus phenocopying the effects of C-terminal CXCR4 truncation in functional assays (21, 52). Severe congenital neutropenia with myelokathexis and recurrent infections can be also caused by deficiency in CXCR2, a chemokine receptor that mediates neutrophil egression from the bone marrow (53, 54). Mechanistically, CXCR2 loss-of-function recapitulates the pathogenic mechanism of CXCR4 gain-of-function in neutrophils, specifically their excessive retention in bone marrow and impaired egress (55). In terms of functional assays, the 2 published studies on CXCR2 utilized chemotaxis assays showing an impaired chemotactic response to CXCL8 in cells harboring the variant CXCR2 receptors. Although OMIM lists the disease as WHIM syndrome 2 (#619407), it remains to be determined whether these patients fall within the clinical spectrum of WHIM syndrome, as lymphopenia, hypogammaglobulinemia, and warts have not been reported in this patient group thus far (54).
WHIM syndrome is a clinicopathologic diagnosis, and since the initial discovery of the disease, our understanding of its natural history and spectrum of clinical manifestations continues to evolve, and diagnosis of WHIM syndrome remains challenging (2, 3). Increased implementation of genetic testing can expedite and support the clinical diagnosis of WHIM syndrome but relies on annotation of likely pathogenic variants (29). The catalogue of disease-causing CXCR4WHIM variants has grown to 36 (8), and additional novel variants in the C-terminus of the receptor are likely to be detected in the future. The current body of evidence, including patient observations and functional studies, is large enough to make a prediction that any novel truncating variant (nonsense or frameshift) between aa 317 and 346 will likely be a pathogenic variant for WHIM syndrome. A similar prediction for missense variants is not possible, and such variants will have to be assessed for evidence of pathogenicity on an individual basis using cellular functional assays. Of interest is whether activating and/or non-”desensitizable” variants outside of the currently curated C-terminal “hot spot” for CXCR4WHIM variants will be found in individuals with WHIM syndrome. In these instances, a rigorous variant interpretation should be done according to ACMG-AMP guidelines, which will require integrating clinical and functional data. With respect to functional tests, impaired C-X-C chemokine ligand 12−induced CXCR4 internalization has been most consistently associated with pathogenic CXCR4WHIM variants, and it seems to correlate with a decrease in circulating neutrophils in individuals with WHIM syndrome (49). In addition, the genetic landscape of WHIM syndrome may not be restricted to CXCR4; defects in other genes regulating either CXCR4 trafficking or the balance between neutrophil mobilization and retention in bone marrow lead to a spectrum of WHIM-like disease presentations (21, 48, 53, 54, 56). Increased implementation of whole genome/exome sequencing in the diagnostic process of primary immunodeficiencies warrants new discoveries in the future. Understanding the genotypic-phenotypic associations in WHIM syndrome has the potential to improve time to diagnosis and guide appropriate clinical management resulting in a true example of precision medicine. Thus far, there is no standard-of-care treatment addressing the underlying cause of WHIM syndrome resulting in use of therapies focused only on clinical signs and symptoms (57). Therefore, efforts are focused on development of therapies targeting the underlying dysfunction in CXCR4 signaling pathways. Recently, an orally bioavailable small-molecule CXCR4 antagonist, mavorixafor, was approved by the US Food and Drug Administration for the treatment of patients with WHIM syndrome (58).
KZ: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Visualization, Writing – original draft, Writing – review & editing. KN: Writing – original draft, Writing – review & editing. GB: Writing – original draft, Writing – review & editing. MY: Writing – review & editing. JW: Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. X4 Pharmaceuticals funded the development of this article.
Medical writing and editorial assistance were provided by PRECISIONscientia in Yardley, Pennsylvania, USA, which was supported financially by X4 Pharmaceuticals, Inc., in compliance with international Good Publication Practice guidelines.
X4 Pharmaceuticals funded the development of the article. KZ is a current employee and has equity ownership of X4 Pharmaceuticals and was involved in conceptualization, investigation, data curation, methodology, and figure design. KN is a current employee and stockholder of Invitae Corporation. GB reports no conflicts of interest. MY receives funding from X4 Pharmaceuticals for research efforts on WHIM syndrome. JW is a consultant for Takeda, X4 Pharmaceuticals, Grifols, ADMA Biologicals, Enzyvant, Regeneron, and Pharming; receives research funding from Takeda, Janssen, Chiesi, ADMA Biologicals, Octapharma, X4 Pharmaceuticals, Novartis, Regeneron, Bristol-Myers Squibb; and is part of a speaker’s bureau for Pharming.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be constructed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1411141/full#supplementary-material
1. Dotta L, Notarangelo LD, Moratto D, Kumar R, Porta F, Soresina A, et al. Long-term outcome of Whim syndrome in 18 patients: high risk of lung disease and Hpv-related Malignancies. J Allergy Clin Immunol Pract. (2019) 7:1568–77. doi: 10.1016/j.jaip.2019.01.045
2. Geier CB, Ellison M, Cruz R, Pawar S, Leiss-Piller A, Zmajkovicova K, et al. Disease progression of whim syndrome in an international cohort of 66 pediatric and adult patients. J Clin Immunol. (2022) 42:1748–65. doi: 10.1007/s10875–022-01312–7
3. Heusinkveld LE, Majumdar S, Gao JL, McDermott DH, Murphy PM. Whim syndrome: from pathogenesis towards personalized medicine and cure. J Clin Immunol. (2019) 39:532–56. doi: 10.1007/s10875-019-00665-w
4. Zuelzer WW. "Myelokathexis"–a new form of chronic granulocytopenia. Rep Case. N Engl J Med. (1964) 270:699–704. doi: 10.1056/nejm196404022701402
5. Hernandez PA, Gorlin RJ, Lukens JN, Taniuchi S, Bohinjec J, Francois F, et al. Mutations in the chemokine receptor gene Cxcr4 are associated with whim syndrome, a combined immunodeficiency disease. Nat Genet. (2003) 34:70–4. doi: 10.1038/ng1149
6. Esid-Pagid Diagnostic Criteria for Whim (Warts-Hypogammaglobulinemia-Infections-Myelokathexis) Syndrome . Available online at: https://esid.org/Working-Parties/Clinical-Working-Party/Resources/Diagnostic-criteria-for-PID2#Q13.
7. Heimall JR, Hagin D, Hajjar J, Henrickson SE, Hernandez-Trujillo HS, Tan Y, et al. Use of genetic testing for primary immunodeficiency patients. J Clin Immunol. (2018) 38:320–9. doi: 10.1007/s10875–018-0489–8
8. Zmajkovicova K, Nykamp K, Badarau A, Maierhofer B, Maier-Munsa S, Mordhorst J, et al. Expanding C-X-C chemokine receptor 4 variant landscape in warts, hypogammaglobulinemia, infections, myelokathexis (Whim) syndrome: integrating clinical and functional data for variant interpretation, in: Poster Presented At: International Primary Immunodeficiencies Congress (Ipic 2023), Rotterdam, the Netherlands, November 8–10, 2023.
9. Abolhassani H, Hammarström L, Cunningham-Rundles C. Current genetic landscape in common variable immune deficiency. Blood. (2020) 135:656–67. doi: 10.1182/blood.2019000929
10. Beaussant Cohen S, Fenneteau O, Plouvier E, Rohrlich P-S, Daltroff G, Plantier I, et al. Description and outcome of a cohort of 8 patients with whim syndrome from the French severe chronic neutropenia registry. Orphanet J Rar. (2012) 7:71. doi: 10.1186/1750–1172-7–71
11. Dale DC, Firkin F, Bolyard AA, Kelley M, Makaryan V, Gorelick KJ, et al. Results of a phase 2 trial of an oral Cxcr4 antagonist, mavorixafor, for treatment of whim syndrome. Blood. (2020) 136:2994–3003. doi: 10.1182/blood.2020007197
12. Gulino AV, Moratto D, Sozzani S, Cavadini P, Otero K, Tassone L, et al. Altered leukocyte response to Cxcl12 in patients with warts hypogammaglobulinemia, infections, myelokathexis (Whim) syndrome. Blood. (2004) 104:444–52. doi: 10.1182/blood-2003–10-3532
13. Kumar R, Milanesi S, Szpakowska M, Dotta L, Di Silvestre D, Trotta AM, et al. Reduced G protein signaling despite impaired internalization and B-arrestin recruitment in patients carrying a Cxcr4leu317fsx3 mutation causing whim syndrome. JCI Insight. (2023) 8. doi: 10.1172/jci.insight.145688
14. Liu Q, Chen H, Ojode T, Gao X, Anaya-O'Brien S, Turner NA, et al. Whim syndrome caused by a single amino acid substitution in the carboxy-tail of chemokine receptor Cxcr4. Blood. (2012) 120:181–9. doi: 10.1182/blood-2011–12-395608
15. Liu Q, Pan C, Lopez L, Gao J, Velez D, Anaya-O’Brien S, et al. Whim syndrome caused by Waldenström’s macroglobulinemia-associated mutation Cxcr4 L329fs. J Clin Immunol. (2016) 36:397–405. doi: 10.1007/s10875–016-0276–3
16. Moens L, Frans G, Bosch B, Bossuyt X, Verbinnen B, Poppe W, et al. Successful hematopoietic stem cell transplantation for myelofibrosis in an adult with warts-hypogammaglobulinemia-immunodeficiency-myelokathexis syndrome. J Allergy Clin Immunol. (2016) 138:1485–9.e2. doi: 10.1016/j.jaci.2016.04.057
17. Nijman IJ, van Montfrans JM, Hoogstraat M, Boes ML, van de Corput L, Renner ED, et al. Targeted next-generation sequencing: A novel diagnostic tool for primary immunodeficiencies. J Allergy Clin Immunol. (2014) 133:529–34. doi: 10.1016/j.jaci.2013.08.032
18. Shin DW, Park SN, Kim SM, Im K, Kim JA, Hong KT, et al. Whim syndrome with a novel Cxcr4 variant in a Korean child. Ann Lab Med. (2017) 37:446–9. doi: 10.3343/alm.2017.37.5.446
19. Tassone L, Notarangelo LD, Bonomi V, Savoldi G, Sensi A, Soresina A, et al. Clinical and genetic diagnosis of warts, hypogammaglobulinemia, infections, and Myelokathexis syndrome in 10 patients. J Allergy Clin Immunol. (2009) 123:1170–3.E3. doi: 10.1016/j.jaci.2008.12.1133
20. Thaventhiran JED, Lango Allen H, Burren OS, Rae W, Greene D, Staples E, et al. Whole-genome sequencing of a sporadic primary immunodeficiency cohort. Nature. (2020) 583:90–5. doi: 10.1038/s41586–020-2265–1
21. Balabanian K, Lagane B, Pablos JL, Laurent L, Planchenault T, Verola O, et al. Whim syndromes with different genetic anomalies are accounted for by impaired Cxcr4 desensitization to Cxcl12. Blood. (2005) 105:2449–57. doi: 10.1182/blood-2004–06-2289
22. Luo J, De Pascali F, Richmond GW, Khojah AM, Benovic JL. Characterization of a new whim syndrome mutant reveals mechanistic differences in regulation of the chemokine receptor Cxcr4. J Biol Chem. (2021) 298(2):101551. doi: 10.1016/j.jbc.2021.101551
23. Ma X, Wang Y, Wu P, Kang M, Hong Y, Xue Y, et al. Case report: A novel Cxcr4 mutation in a Chinese child with Kawasaki disease causing whim syndrome. Front Immunol. (2022) 13:857527. doi: 10.3389/fimmu.2022.857527
24. Cxcr4 (Cxcr4_Human) . Gpcrdb. Available online at: https://Gpcrdb.Org/Protein/Cxcr4_Human/ (Accessed December 21, 2023).
25. Paludo J, Ansell SM. Waldenström macroglobulinemia: biology, genetics, and therapy. Blood Lymphat Cancer. (2016) 6:49–58. doi: 10.2147/blctt.S84157
26. Treon SP, Cao Y, Xu L, Yang G, Liu X, Hunter ZR. Somatic mutations in myd88 and cxcr4 are determinants of clinical presentation and overall survival in Waldenstrom macroglobulinemia. Blood. (2014) 123:2791–6. doi: 10.1182/blood-2014–01-550905
27. Shin DW, Kim SM, Kim JA, Park HS, Hwang SM, Im K, et al. Characteristics of Waldenström macroglobulinemia in Korean patients according to mutational status of myd88 and Cxcr4: analysis using ultra-deep sequencing. Clin Lymphoma Myeloma Leuk. (2019) 19:e496–505. doi: 10.1016/j.clml.2019.03.009
28. Nykamp K, Anderson M, Powers M, Garcia J, Herrera B, Ho YY, et al. Sherloc: A comprehensive refinement of the Acmg-Amp variant classification criteria. Genet Med. (2017) 19:1105–17. doi: 10.1038/gim.2017.37
29. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American college of medical genetics and genomics and the association for molecular pathology. Genet Med. (2015) 17:405–24. doi: 10.1038/gim.2015.30
30. What Is Clinvar? National Library of Medicine . Available online at: https://www.Ncbi.Nlm.Nih.Gov/Clinvar/Intro/ (Accessed November 1, 2023).
31. Clinvar. Cxcr4[Gene]. National Library of Medicine . Available online at: https://www.Ncbi.Nlm.Nih.Gov/Clinvar/?Term=Cxcr4%5bgene%5d (Accessed December 6, 2023).
32. Ma Q, Jones D, Borghesani PR, Segal RA, Nagasawa T, Kishimoto T, et al. Impaired B-lymphopoiesis, myelopoiesis, and derailed cerebellar neuron migration in Cxcr4- and Sdf-1-deficient mice. Proc Natl Acad Sci U.S.A. (1998) 95:9448–53. doi: 10.1073/pnas.95.16.9448
33. Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. (2020) 581:434–43. doi: 10.1038/s41586–020-2308–7
34. C-X-C Motif Chemokine Receptor 4. Gnomad . Available online at: https://Gnomad.Broadinstitute.Org/Gene/Ensg00000121966?Dataset=Gnomad_R4 (Accessed November 29, 2023).
35. Anderson CL, Munawar S, Reilly L, Kamp TJ, January CT, Delisle BP, et al. How functional genomics can keep pace with Vus identification. Front Cardiovasc Med. (2022) 9:900431. doi: 10.3389/fcvm.2022.900431
36. Hoffman-Andrews L. The known unknown: the challenges of genetic variants of uncertain significance in clinical practice. J Law Biosci. (2017) 4:648–57. doi: 10.1093/jlb/lsx038
37. Brnich SE, Abou Tayoun AN, Couch FJ, Cutting GR, Greenblatt MS, Heinen CD, et al. Recommendations for application of the functional evidence Ps3/Bs3 criterion using the Acmg/Amp sequence variant interpretation framework. Genome Med. (2019) 12:3. doi: 10.1186/s13073–019-0690–2
38. Woods NT, Baskin R, Golubeva V, Jhuraney A, De-Gregoriis G, Vaclova T, et al. Functional assays provide a robust tool for the clinical annotation of genetic variants of uncertain significance. NPJ Genom Med. (2016) 1:16001–. doi: 10.1038/npjgenmed.2016.1
39. Loetscher M, Geiser T, O'Reilly T, Zwahlen R, Baggiolini M, Moser B. Cloning of a human seven-transmembrane domain receptor, Lestr, that is highly expressed in leukocytes. J Biol Chem. (1994) 269:232–7. doi: 10.1016/S0021-9258(17)42339-8
40. Oberlin E, Amara A, Bachelerie F, Bessia C, Virelizier JL, Arenzana-Seisdedos F, et al. The Cxc chemokine Sdf-1 is the ligand for lestr/fusin and prevents infection by T-cell-line-adapted Hiv-1. Nature. (1996) 382:833–5. doi: 10.1038/382833a0
41. Balabanian K, Brotin E, Biajoux V, Bouchet-Delbos L, Lainey E, Fenneteau O, et al. Proper desensitization of cxcr4 is required for lymphocyte development and peripheral compartmentalization in mice. Blood. (2012) 119:5722–30. doi: 10.1182/blood-2012–01-403378
42. De Filippo K, Rankin SM. Cxcr4, the master regulator of neutrophil trafficking in homeostasis and disease. Eur J Clin Invest. (2018) 48 Suppl 2:e12949. doi: 10.1111/eci.12949
43. Levy E, Reger R, Segerberg F, Lambert M, Leijonhufvud C, Baumer Y, et al. Enhanced bone marrow homing of natural killer cells following Mrna transfection with gain-of-function variant Cxcr4(R334x). Front Immunol. (2019) 10:1262. doi: 10.3389/fimmu.2019.01262
44. Busillo JM, Benovic JL. Regulation of Cxcr4 signaling. Biochim Biophys Acta (BBA) - Biomembranes. (2007) 1768:952–63. doi: 10.1016/j.bbamem.2006.11.002
45. Mueller W, Schütz D, Nagel F, Schulz S, Stumm R. Hierarchical organization of multi-site phosphorylation at the Cxcr4 C terminus. PloS One. (2013) 8:e64975. doi: 10.1371/journal.pone.0064975
46. McDermott DH, Lopez J, Deng F, Liu Q, Ojode T, Chen H, et al. Amd3100 is a potent antagonist at Cxcr4 R334x, a hyperfunctional mutant chemokine receptor and cause of whim syndrome. J Cell Mol Med. (2011) 15:2071–81. doi: 10.1111/j.1582-4934.2010.01210.x
47. Majumdar S, Murphy PM. Adaptive immunodeficiency in whim syndrome. Int J Mol Sci. (2018) 20. doi: 10.3390/ijms20010003
48. Balabanian K, Levoye A, Klemm L, Lagane B, Hermine O, Harriague J, et al. Leukocyte analysis from whim syndrome patients reveals a pivotal role for Grk3 in Cxcr4 signaling. J Clin Invest. (2008) 118:1074–84. doi: 10.1172/jci33187
49. Zmajkovicova K, Pawar S, Maier-Munsa S, Maierhofer B, Wiest I, Skerlj R, et al. Genotype-phenotype correlations in whim syndrome: A systematic characterization of Cxcr4(Whim) variants. Genes Immun. (2022) 23:196–204. doi: 10.1038/s41435–022-00181–9
50. Murphy PM, Heusinkveld L. Multisystem multitasking by Cxcl12 and its receptors Cxcr4 and Ackr3. Cytokine. (2018) 109:2–10. doi: 10.1016/j.cyto.2017.12.022
51. García-Cuesta EM, Rodríguez-Frade JM, Gardeta SR, D'Agostino G, Martínez P, Soler Palacios B, et al. Altered Cxcr4 dynamics at the cell membrane impairs directed cell migration in whim syndrome patients. Proc Natl Acad Sci U.S.A. (2022) 119:e2119483119. doi: 10.1073/pnas.2119483119
52. Lagane B, Chow KY, Balabanian K, Levoye A, Harriague J, Planchenault T, et al. Cxcr4 dimerization and beta-arrestin-mediated signaling account for the enhanced chemotaxis to Cxcl12 in whim syndrome. Blood. (2008) 112:34–44. doi: 10.1182/blood-2007–07-102103
53. Auer PL, Teumer A, Schick U, O'Shaughnessy A, Lo KS, Chami N, et al. Rare and low-frequency coding variants in Cxcr2 and other genes are associated with hematological traits. Nat Genet. (2014) 46:629–34. doi: 10.1038/ng.2962
54. Marin-Esteban V, Youn J, Beaupain B, Jaracz-Ros A, Barlogis V, Fenneteau O, et al. Biallelic Cxcr2 loss-of-function mutations define a distinct congenital neutropenia entity. Haematologica. (2022) 107:765–9. doi: 10.3324/haematol.2021.279254
55. Eash KJ, Greenbaum AM, Gopalan PK, Link DC. Cxcr2 and Cxcr4 antagonistically regulate neutrophil trafficking from murine bone marrow. J Clin Invest. (2010) 120:2423–31. doi: 10.1172/jci41649
56. Tarrant TK, Billard MJ, Timoshchenko RG, McGinnis MW, Serafin DS, Foreman O, et al. G protein-coupled receptor kinase-3-deficient mice exhibit whim syndrome features and attenuated inflammatory responses. J Leukoc Biol. (2013) 94:1243–51. doi: 10.1189/jlb.0213097
57. Badolato R, Donadieu J. How I treat warts, hypogammaglobulinemia, infections, and Myelokathexis syndrome. Blood. (2017) 130:2491–8. doi: 10.1182/blood-2017–02-708552
Keywords: WHIM syndrome, congenital neutropenia, primary immunodeficiency disease, CXCR4, genetic testing, functional assays
Citation: Zmajkovicova K, Nykamp K, Blair G, Yilmaz M and Walter JE (2024) Expanding CXCR4 variant landscape in WHIM syndrome: integrating clinical and functional data for variant interpretation. Front. Immunol. 15:1411141. doi: 10.3389/fimmu.2024.1411141
Received: 02 April 2024; Accepted: 24 June 2024;
Published: 08 July 2024.
Edited by:
Teresa Kathleen Tarrant, Duke University, United StatesReviewed by:
María Bravo García-Morato, University Hospital La Paz, SpainCopyright © 2024 Zmajkovicova, Nykamp, Blair, Yilmaz and Walter. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Katarina Zmajkovicova, a2F0YXJpbmEuem1hamtvdmljb3ZhQHg0cGhhcm1hLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.