- 1KU Leuven Department of Microbiology, Immunology and Transplantation, Allergy and Clinical Immunology Research Group, KU Leuven, Leuven, Belgium
- 2Clinical Department of Otorhinolaryngology, Head and Neck Surgery, University Hospitals Leuven, Leuven, Belgium
- 3KU Leuven Department of Chronic Diseases and Metabolism, Translational Research Center for Gastrointestinal Diseases, KU Leuven, Leuven, Belgium
- 4Department of Pediatrics, University Hospitals Leuven, Leuven, Belgium
- 5KU Leuven Department of Cellular and Molecular Medicine, Laboratory of Ion Channel Research Division of Physiology, KU Leuven, Leuven, Belgium
- 6KU Leuven Department of Neurosciences, Experimental Otorhinolaryngology Rhinology Research, KU Leuven, Leuven, Belgium
- 7University of Antwerp (UAntwerp) Department of Bioscience Engineering, Lab of Applied Microbiology and Biotechnology, University of Antwerp (UAntwerp), Antwerp, Belgium
- 8University of Ghent (UGhent) Department of Head and Skin, Upper Airways Research Laboratory, University of Ghent (UGhent), Ghent, Belgium
There is growing evidence that neurogenic inflammation contributes to the pathophysiology of upper airway diseases, with nasal hyperreactivity (NHR) being a key symptom. The rare neuroendocrine cells (NECs) in the epithelium have been linked to the pathophysiology of bronchial and intestinal hyperreactivity, however their presence in the nasal mucosa and their potential role in NHR remains unclear. Therefore, we studied the presence of NECs in the nasal epithelium of controls, allergic rhinitis patients and chronic rhinosinusitis with nasal polyps patients, and their link to NHR. The expression of typical NECs markers, CHGA, ASCL1 and CGRP, were evaluated on gene and protein level in human samples using real-time quantitative PCR (RT-qPCR), western blot, immunohistochemistry fluorescence staining, RNA scope assay, flow cytometry and single cell RNA-sequencing. Furthermore, the change in peak nasal inspiratory flow after cold dry air provocation and visual analogue scale scores were used to evaluate NHR or disease severity, respectively. Limited gene expression of the NECs markers CHGA and ASCL1 was measured in patients with upper airway diseases and controls. Gene expression of these markers did not correlate with NHR severity nor disease severity. In vitro, CHGA and ASCL1 expression was also evaluated in primary nasal epithelial cell cultures from patients with upper airway disease and controls using RT-qPCR and western blot. Both on gene and protein level only limited CHGA and ASCL1 expression was found. Additionally, NECs were studied in nasal biopsies of patients with upper airway diseases and controls using immunohistochemistry fluorescence staining, RNA scope and flow cytometry. Unlike in ileum samples, CHGA could not be detected in nasal biopsies of patients with upper airway diseases and control subjects. Lastly, single cell RNA-sequencing of upper airway tissue could not identify a NEC cluster. In summary, in contrast to the bronchi and gut, there is only limited evidence for the presence of NECs in the nasal mucosa, and without correlation with NHR, thereby questioning the relevance of NECs in upper airway pathology.
Introduction
The airway epithelium represents the first mucosal barrier protecting the host against infiltration of potential harmful airborne stimuli (1–3). Besides the formation of a physical barrier, the epithelium is equipped with specialized cells and receptors that can detect and respond to noxious stimuli within the inhaled air (4–6). Traditionally, the airway epithelium consists of three major cell types, i.e., ciliated cells, mucus-secreting goblet cells and stem cell-like basal cells (2, 7). With the emerging and growing use of advanced sequencing techniques, the traditional view of the airway epithelium as a sole barrier to the environment has changed. Rather, the airway epithelium is a dynamic cellular structure containing several types of rare specialized cells that are able to respond to environmental changes and interact with the immune and neurogenic system (8–11). One of these rare identified epithelial cells in the lower respiratory epithelium are neuroendocrine cells (NECs) which might play a role in the pathophysiology of neurogenic inflammation (8, 12). Neurogenic inflammation is presumed to be triggered by external non-specific environmental airborne stimuli, such as cigarette smoke, temperature or humidity changes and other irritants, leading to sensory nerve activation with the subsequent release of neuropeptides including substance P and calcitonin gene-related peptide (CGRP) (13). These neuropeptides can directly activate inflammatory cells like mast cells, which leads to the release of classic pro-inflammatory mediators causing inflammation (14). One of the pivotal clinical manifestations of neurogenic inflammation in the upper airways is nasal hyperreactivity (NHR) (15). NHR is defined as the induction of one or more nasal symptoms upon encounter of environmental stimuli, such as cigarette smoke, temperature or humidity changes, strong odors or fragrances, and other irritants (15) It is presumed that NHR is triggered by overactivation of the sensory nerves innervating the respiratory mucosa to non-specific environmental airborne stimuli (1, 15, 16). In physiological conditions, the airways are highly sensitive to airborne stimuli since they are continuously exposed to the environment. Upon encounter with potentially harmful airborne stimuli, the epithelium provides a fast protective response to remove potentially harmful substances from the airways by means of sneezing and rhinorrhea via efferent nerves (17). In case of NHR, there presumably is an overactivation of these sensory neurons, also provoking a response to non-specific environmental airborne stimuli (15, 16). In chronic upper airway diseases, like allergic rhinitis (AR) and chronic rhinosinusitis with nasal polyps (CRSwNP), there is growing evidence that neurogenic inflammation also contributes to the pathophysiology aside from the typical type 2 inflammation (1, 13, 18). The reported prevalence of NHR in patients with upper airway diseases is between 50% and 80%, indicating that neurogenic inflammation is indeed part of the pathophysiology of upper airway diseases (15, 19, 20). The cold dry air provocation test has been shown to be a good tool to objectively quantify NHR in patients with a high sensitivity and specificity in the upper airways (21). As NHR develops upon encounter of airborne stimuli, and NECs are embedded in epithelia, NECs form the prime candidate to be involved in the pathophysiology of NHR (13, 22). Indeed, over the last few years NECs have gained more interest given their possible role in sensory neuron hyperactivation (23–27). NECs act as chemosensors to the external environment that, upon activation, release various neuropeptides like substance P and CGRP. These neuropeptides induce further downstream responses, including activation of sensory neurons located at the base of NECs (8). In the lungs, pulmonary NECs have been shown to respond to changes in oxygen, mechanical stretch and chemical stimuli (23, 28–30). Recently, pulmonary NECs were shown to amplify type 2 allergic responses in mice (23). In the gut, enteroendocrine cells, which display large similarities with pulmonary NECs, were similarly identified as chemosensors that regulate a variety of physiological processes, including gastrointestinal motility and visceral hypersensitivity (24, 25). Pulmonary NECs and their intestinal counterpart, enteroendocrine cells are characterized by expression of Chromogranin A (CHGA) and Achaete-Scute Family bHLH transcription factor 1 (ASCL1) as they both have been shown to be constitutively with the top marker genes of this cell type (8, 12, 31). Chromogranin A (CHGA) is a member of the chromogranin family which belongs to the neuroendocrine secretory proteins, while ASCL1 is a known neurogenic transcription factor with a key role in the development of pulmonary NECs (32–35). Unlike the role of NECs in intestinal and bronchial epithelium, the presence and function of NECs in the nasal epithelium is largely uninvestigated. Therefore, the aim of this study was to evaluate the presence of NECs in the nasal mucosa and their potential role in the induction of NHR in upper airway diseases.
Materials and methods
Primary nasal epithelial cell isolation
Primary nasal epithelial cells were isolated from residual surgical waste tissue coming from inferior turbinates of healthy controls and AR patients collected during aesthetic or functional surgery, and from nasal polyp tissue from CRSwNP patients collected during functional endoscopic sinus surgery. Ear-Nose-Throat specialists diagnosed AR patients with symptomatic (≥ 2 nasal symptoms) house dust mite (HDM)-induced AR, and CRSwNP patients based on nasal endoscopy. Control subjects had no clinical signs or history suggestive of AR or CRS. All included subjects were non-smokers (≥ 1 year). Detailed patient’s characteristics can be found in Supplementary Table 1. The study was approved by the Ethical Committee Research of University Hospitals Leuven (S65483). Primary nasal epithelial cells were isolated from residual surgical waste tissue as previously reported (4). Collected cells were expanded in bronchial epithelial basal medium supplemented with the SingleQuot Kit (CC-3170, Lonza) or directly seeded on Thincert inserts (Greiner Bio-One) at a concentration of 100,000 – 120,000 cells per Thincert insert using DMEM/F-12 (11320033, Gibco) supplemented with 100 U/mL penicillin, 100 g/mL streptomycin and 2% Ultroser G (15950-017, Sartorius) in liquid-liquid interface. Once a confluent monolayer was formed (approximately 7-10 days), apical medium was removed to allow cell differentiation in the air-liquid interface for an additional 14 days.
Nasal biopsies
Nasal biopsies were collected from patients with well-document NHR disease status. Patients with HDM-induced AR or CRSwNP, and healthy controls were recruited on a voluntary basis from the outpatient rhinology clinic of the University Hospitals Leuven. All participants were well-characterized by Ear-Nose-Throat specialists and underwent skin prick testing, nasal endoscopy, cold dry air provocation test, and filled in the Visual Analogue Scale (VAS) questionnaire for nasal symptoms. Nasal biopsies were harvested from the inferior turbinates after local anesthesia (cocaine 1%) with a Fokkens forceps and snap frozen in liquid nitrogen. Participants were considered to suffer from NHR using the cold dry air provocation test as previously described (19, 21). Peak Nasal Inspiratory Flow (PNIF) was measured immediately before and after provocation with cold dry air with a PNIF-device (In-Check Nasal Inspiratory Flow Meter, Clement Clarke International, Harlow, UK) and NHR was diagnosed in case of a drop in PNIF of at least 20%. Detailed patient’s characteristics can be found in Supplementary Table 2. The study was approved by the Ethical Committee Research of University Hospitals Leuven (S63139).
Ileum biopsies
Ileum samples were collected from the most proximal unaffected regions of colon rectal cancer and Crohn’s disease patients. Detailed patient’s characteristics can be found in Supplementary Table 3. The study was approved by the Ethical Committee Research of University Hospitals Leuven (S53684, S68720). Biopsies were put in RLT lysis buffer (79216, Qiagen) for RT-qPCR experiments, in PBS containing 5% BSA for western blot experiments or in cold RPMI medium for flow cytometry experiments.
Western blot
Cells lysates were harvested in cold NP-40 lysis buffer (50 mM Tris.HCl (pH 8.0), 150 mM NaCl, 1% NP-40) containing 1X cOmplete Protease Inhibitor (CO-RO, Roche) and 0.4 mM PMSF protease inhibitor in dry isopropanol (36978, Thermo Fisher Scientific). Protein was extracted and total protein concentration was determined using the Pierce BCA protein assay kit (23227, ThermoFisher Scientific). Protein samples were resuspended in LDS Sample Buffer (1X) (NP007, Invitrogen) and Reducing Agent (1X) (NP0009, Invitrogen), after which denatured proteins were separated by electrophoresis using SDS-PAGE gels (NP0323, Invitrogen). Separated proteins were electroblotted onto an activated PVDF membrane (10600023, VWR International) in transfer buffer (31.2 mM Tris-Base, 0.24 M glycine, 20% methanol, 2% antioxidant). Membranes were blocked with 5% milk powder (MP) (5% milk, 20 mM Tris-HCl, pH 7.6), followed by overnight incubation at 4°C with primary antibodies anti-Chromogranin A antibody (1:1,000; ab45179, abcam), anti-ASCL1 polyclonal antibody (1:1,000; PA5-77868, invitrogen) or anti- Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH) monoclonal antibody (6C5) (1:10,000; AM4300, Invitrogen) in 1% MP. Primary antibodies were detected via enhanced chemiluminescence (NEL105, PerkinElmer) using HRP-linked secondary antibodies (anti-rabbit; 1:5,000; 32460, Thermo Scientific or anti-mouse; 1:5,000; 32430, Thermo Scientific). The signal was visualized with the ImageQuant LAS500 (Bioké, Leiden, The Netherlands).
Real-time quantitative polymerase reaction
Biopsies were homogenized in RLT lysis buffer (79216, Qiagen) using Lysing Matrix D and a FastPrep-24-device (116913100 and 116004500, MP Biomedicals) and RNA was extracted using the RNeasy Mini Kit (74106, Qiagen). cDNA was obtained using a High-Capacity cDNA Reverse Transcription kit (4368814, Thermo Fisher Scientific) starting from 2 µg RNA. RT-qPCR was performed for genes of interest CHGA and ASCL1, and for housekeeping genes actin beta (ACTB) and receptor for activated C kinase 1 (RACK1) (sequences in Supplementary Table 4) with the CFX Connect Real-Time PCR Detection System (Bio-rad, Hercules, California, USA). cDNA plasmid standards of each gene were used to quantify the amount of target genes in unknown samples.
Immunohistochemistry fluorescence staining
Paraffin-embedded tissue slides (5 μm) of nasal and ileum biopsies were subjected to antigen retrieval in citrate buffer (pH 6). Proteins of interest were detected using the following primary antibodies: anti-chromogranin A antibody (1:100, ab45179, abcam), anti-chromogranin A antibody (LK2H10) (1:100, MA5-13096, Invitrogen), anti-CGRP antibody (1:50, ABS 026-04-02, Invitrogen), anti-KRT5-Alexa Fluor 488 antibody (1:100, ab193894, abcam), anti-MUC5AC antibody (45M1) (1:150, MA5-12178, Invitrogen) and anti- acetylated tubulin antibody (clone 6-11B-1) (1:300, T7451, Sigma-aldrich). Secondary antibodies were anti-rabbit AlexaFluor 594 (1:500, A21207, ThermoFisher) and anti-mouse AlexaFluor 488 (1:500, A21202, ThermoFisher). Lastly, nuclei were stained with 4′-6-diamidino-phenylindole dihydrochloride–containing (3nM, D9542, Sigma-Aldrich). After staining, tissues were mounted and images were taken using the ZEISS axioscan 7 (Zeiss, Oberkochen, Germany).
RNA scope
Paraffin-embedded nasal and ileum biopsy slides (5 μm) were prepped and stained using the RNA scope 2.5 reagent Kit (322350, Advanced Cell Diagnostics). Briefly, paraffin-embedded samples were baked for 1h at 60°C and deparaffinized in 100% xylene and 100% ethanol. Airdried slides were pretreated with RNA scope hydrogen peroxide (322330, Advanced Cell Diagnostics) for 10 min, followed by antigen retrieval (322000, Advanced Cell Diagnostics) for 15 min at 99°C. Slides were incubated with Protease plus reagents (322330, Advanced Cell Diagnostics) for 30 min in the humidity and temperature controlled HybEZ oven. The anti-CHGA RNAscope Probe - Hs-CHGA (311111, Advanced Cell Diagnostics) was hybridized for 2h at 40°C. Signal amplification was obtained by sequential hybridization of 6 amplifiers, whereafter signal detection was performed using the Fast RED working solution (322360, Advanced Cell Diagnostics). Slides were counterstained with Hematoxylin, after which mounted slides were evaluated using the ZEISS axioscan 7 (Zeiss, Oberkochen, Germany).
Single cell RNA-sequencing
Single cell suspensions of surgically collected inferior turbinates were obtained by enzymatic digestion using liberase (87.5 µg/ml, 5401127001, Roche) and DNase I (300 µg/ml, 10104159001, Roche) in RPMI with 10% FBS for 30 min with additional mechanical disruption using a 16G needle after both 15 min and 30 min of enzymatic digestion. Digestion was stopped by adding EDTA (20 mM, 15575020, Invitrogen), whereafter red blood cells were lysed using 9.9% NaCl solution. Negative selection for immune cells, myeloid cells and fibroblasts was performed using anti-CD45 (11153D, Invitrogen), anti-CD15 (11137D, Invitrogen) and anti-CD31 (11155D, Invitrogen) dynabeads, respectively. Filtered cells (70 μm, 542070, Greiner) were loaded on the 10X Genomics Chromium 3’ device (3’ V2, 10X Genomics, Pleasanton, California, USA), and afterwards RNA libraries were constructed and sequenced using the NovaSeq 6000 S4 Reagent Kit V1.5 (20028312, Illumina) and using the NovaSeq 6000 device (Illumina, San Diego, California, USA). Raw sequencing files were cleaned, mapped to the human reference genome, filtered, normalized, demultiplexed and counted using fastaq-mcf, fastQC, Cell Ranger and fastMnM, after which data was analyzed using different R tools, such as Seurat, Harmony and Bioconductor in R 4.2.1 (36–41).
Single cell staining and flow cytometry
Single cells solutions of nasal tissues were obtained by overnight enzymatic digestion of surgically collected tissue in 1 mg/ml protease XIV (P5147-1G, Sigma) in Dulbecco’s modified Eagle’s medium (DMEM/F12; Lonza BioWhittaker) supplemented with 100 U/mL penicillin, 100 g/mL streptomycin and 2% Ultroser G (15950-017, Sartorius). Surgically obtained ileum tissue was mechanically disrupted followed by enzymatically digestion for 40 min at 37°C and 5% CO2 in pre-heated HBSS Ca+Mg+ buffer containing 1 mg/ml collagenase D (COLLD-RO, Roche), 20 units/ml DNase I (EN0521, Thermo Fisher Scientific) and 0.02 mM HEPES (15630080, Gibco). 100,000 to 1,000,000 filtered (70 μm, 542070, Greiner) single cells were collected and incubated for 25 min with 1 µl/ml eBioscience Fixable Viability Dye eFluor 780 (65-0865-14, Invitrogen). Cells were blocked using 5 µl/ml human serum for 10 min at 4°C, after which cells were stained with 50 µl/ml Brilliant Violet 650 anti-human CD326 (EpCAM) Antibody (BV650, Biolegend) for 30 min at 4°C. Sequentially, intracellular staining was performed using Flow Cytometry Fixation & permeabilization Buffer Kit I (FC009, Biotechne) with 50 µl/ml PE Anti-Human Chromogranin A Clone S21-537 (RUO) antibody (5564563, BD Pharmingen). Fixated cells were acquired using the BD LSRFortessa Cell Analyzer (BD biosciences, New Jersey, USA) equipped with FACSDiva software. Flow cytometric data was analyzed in FlowJo v10.7.1 (BD Biosciences, New Jersey, USA). Analysis included removal of debris, doublet cells and dead cells.
Statistics
Data was analyzed using GraphPad Prism 10 (GraphPad Software, La Jolla, Calif). Differences between two groups were analyzed by the Mann-Whitney U test. Differences between multiple groups were analyzed by the One-way ANOVA or the Kruskal-Wallis test with post hoc analysis, depending on normality distribution. Categorical values were compared by using the Chi square test and correlations were assessed by using the Spearman r test. Values were considered significantly different at a p value of less than 0.05 adjusted to the appropriate multiple testing correction (Bonferroni) if needed.
Results
Limited expression of NECs markers in the nasal mucosa of patients with upper airway disease and healthy subjects
To investigate whether NECs are present in the nasal mucosa of patients and controls, the expression of NEC markers CHGA and ASCL1 in nasal biopsies and in primary nasal epithelial cell cultures was evaluated. Unlike clear expression of CHGA and ASCL1 in ileum samples, limited CHGA and ASCL1 expression was measured in nasal biopsies and primary nasal epithelial cells (Figures 1A, B). No significant differences in CHGA and ASCL1 expression levels were found between controls, AR patients and CRSwNP patients (Figures 1A, B). The limited mRNA expression of CHGA and ASCL1 in primary nasal epithelial cell cultures was confirmed on protein level, while ileum samples clearly showed more CHGA and ASCL1 protein expression (Figures 1C, D). No significant difference in protein expression was found between controls, AR patients and CRSwNP patients (Figures 1C, D). This observation was confirmed with immunohistochemistry fluorescence staining. A clear double positive (CHGA+/CHGA+ and CHGA+/CGRP+) cell population was observed in ileum samples, while this double positive population was absent in nasal biopsies, irrespective of disease or NHR status (Figure 2). In contrast, immunohistochemistry fluorescence staining of the three main cell types in the upper airway epithelium being KRT5 for basal cells, MUC5AC for goblet cells and acetylated tubulin for ciliated cells, could be detected. These results were further confirmed using the RNA scope assay which detected CHGA+ cells in the ileum, while no CHGA+ cells were detected in controls without NHR, AR with and without NHR and CRSwNP with and without NHR (Figure 3). With flow cytometry, significantly more CHGA+ cells were detected in the epithelial cellular adhesion molecule (EpCAM)-positive epithelial cell population in ileum samples compared to control, AR and CRSwNP samples (Figure 4; Supplementary Figure 1).
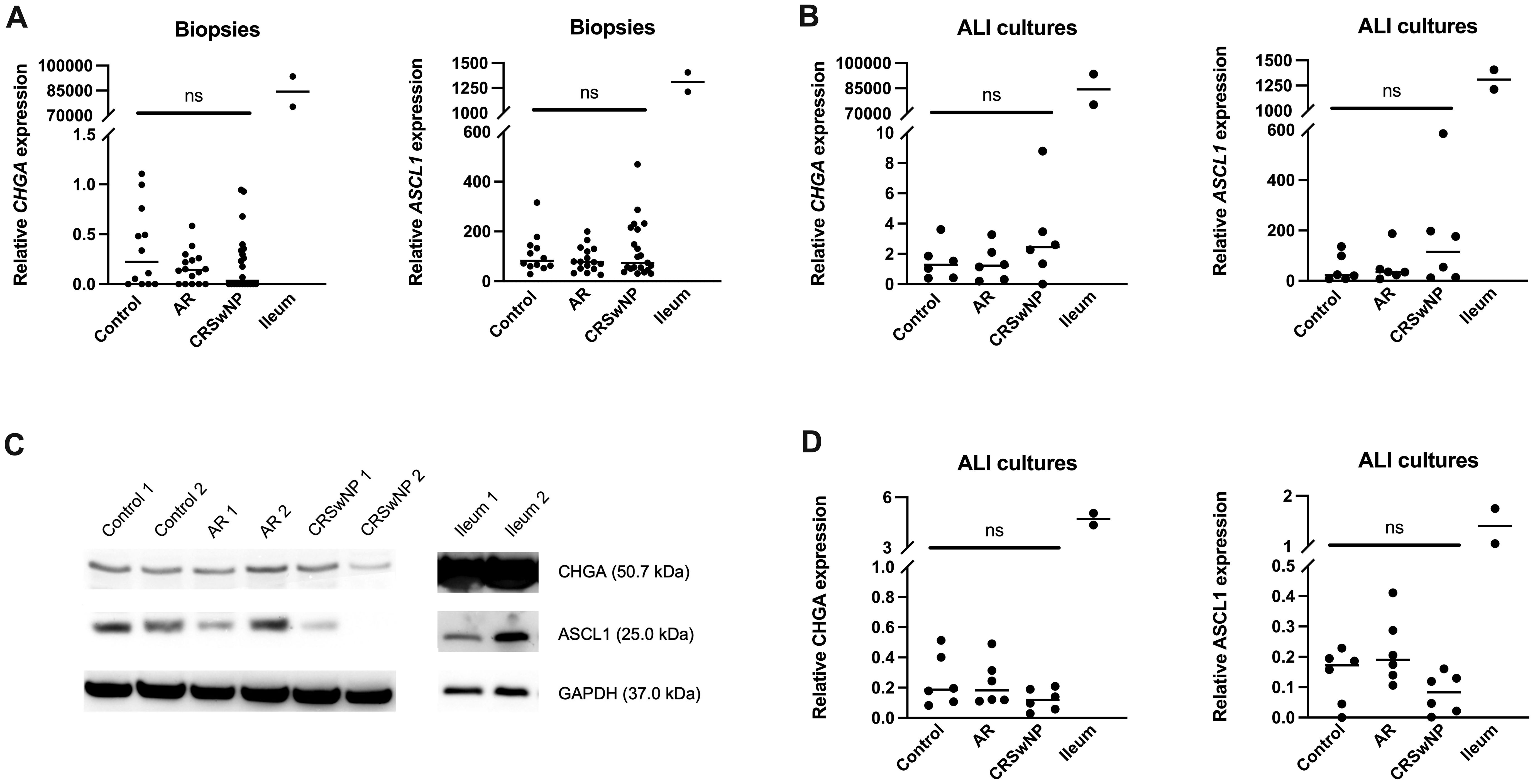
Figure 1 Expression of the NEC markers CHGA and ASCL1 in nasal biopsies and cultured primary nasal epithelial cells. (A) CHGA and ASCL1 mRNA expression levels in nasal biopsies and ileum biopsies relative to the expression of the housekeeping genes ACTB and RACK1. (B) CHGA and ASCL1 mRNA expression levels in air-liquid interface cultured primary nasal epithelial cells and ileum samples relative to the expression of the housekeeping genes β-actin and RACK1. (C) Representative western blot of CHGA, ASCL1 and GAPDH (housekeeping protein) protein expression levels in cultured primary nasal epithelial cells and ileum samples. (D) Quantification of CHGA and ASCL1 protein expression levels relative to GAPDH expression levels based on western blot analysis. AR, allergic rhinithis; CRSwNP, chronic rhinosinusitis with nasal polyps; CHGA, chromogranin A; ASCL1, Achaete-Scute Family bHLH transcription factor 1; data represented as individual values with median, ns = p > 0.05.
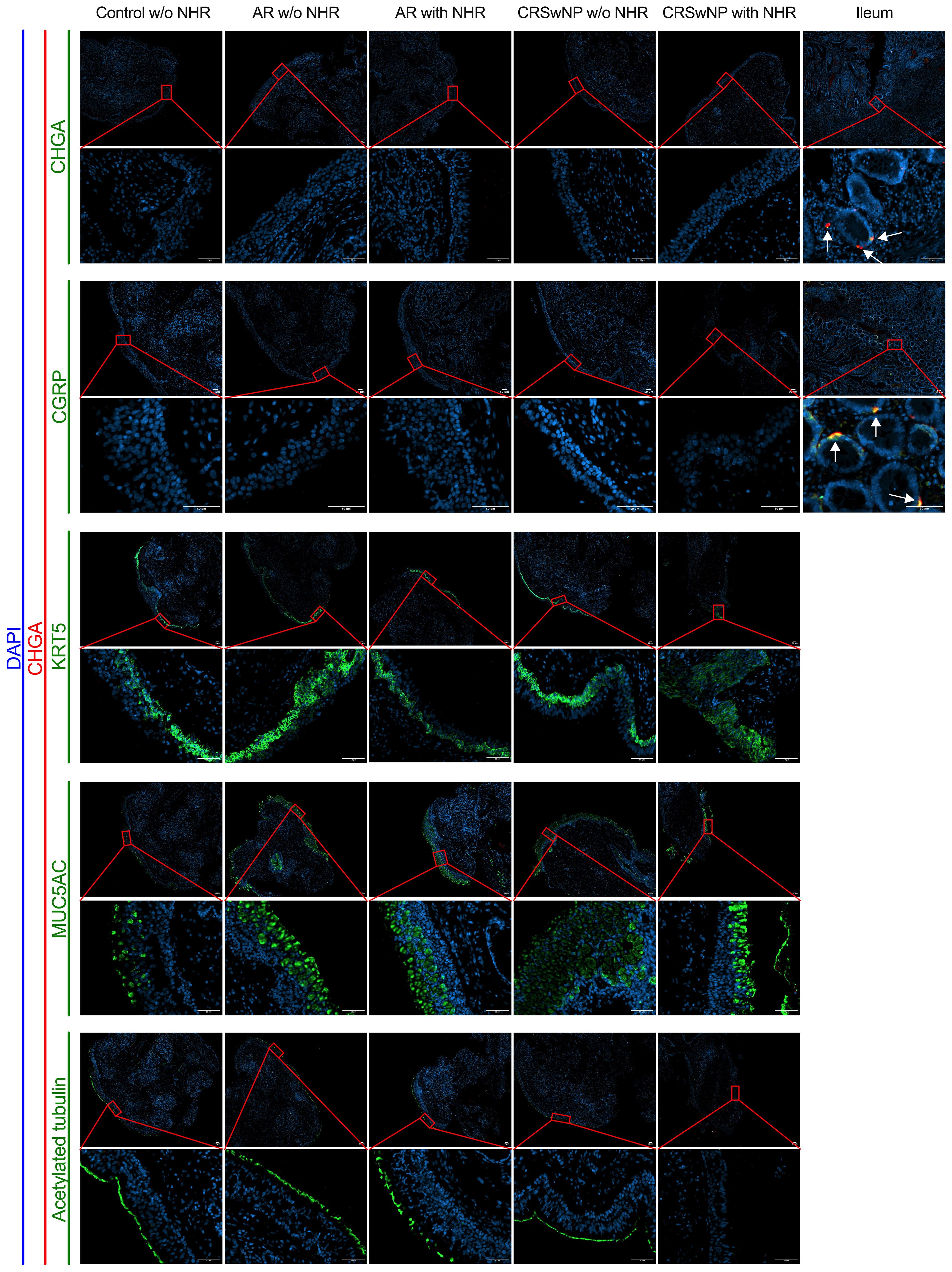
Figure 2 Immunohistochemistry fluorescence staining of neuroendocrine, basal, goblet and ciliated epithelial cells in nasal and ileum biopsies of patients and controls. Representative immunohistochemistry fluorescence staining of the neuroendocrine and entroendocrine cell makers: CHGA (red)/CHGA (green) and CHGA (red)/CGRP (green) in both nasal biopsies from controls without NHR, AR with and without NHR patients and CRSwNP with and without NHR patients and ileum biopsies. Representative immunohistochemistry fluorescence staining of basal cells (KRT5, green), goblet cells (MUC5AC, green) and ciliated cells (acetylated tubulin, green) in combination with CHGA (red) in nasal biopsies from controls without NHR, AR with and without NHR patients and CRSwNP with and without NHR patients. All stainings were costained with DAPI (blue) to visualize the nuclei. NHR = nasal hyperreactivity; AR, allergic rhinithis; CRSwNP, chronic rhinosinusitis with nasal polyps; CHGA, chromogranin A; CGRP, calcitonin gene-related peptide; KRT5, Keratin 5; MUC5AC, Mucin-5AC, DAPI = 4′,6-diamidino-2-phenylindole; scale bar = 50 µm.
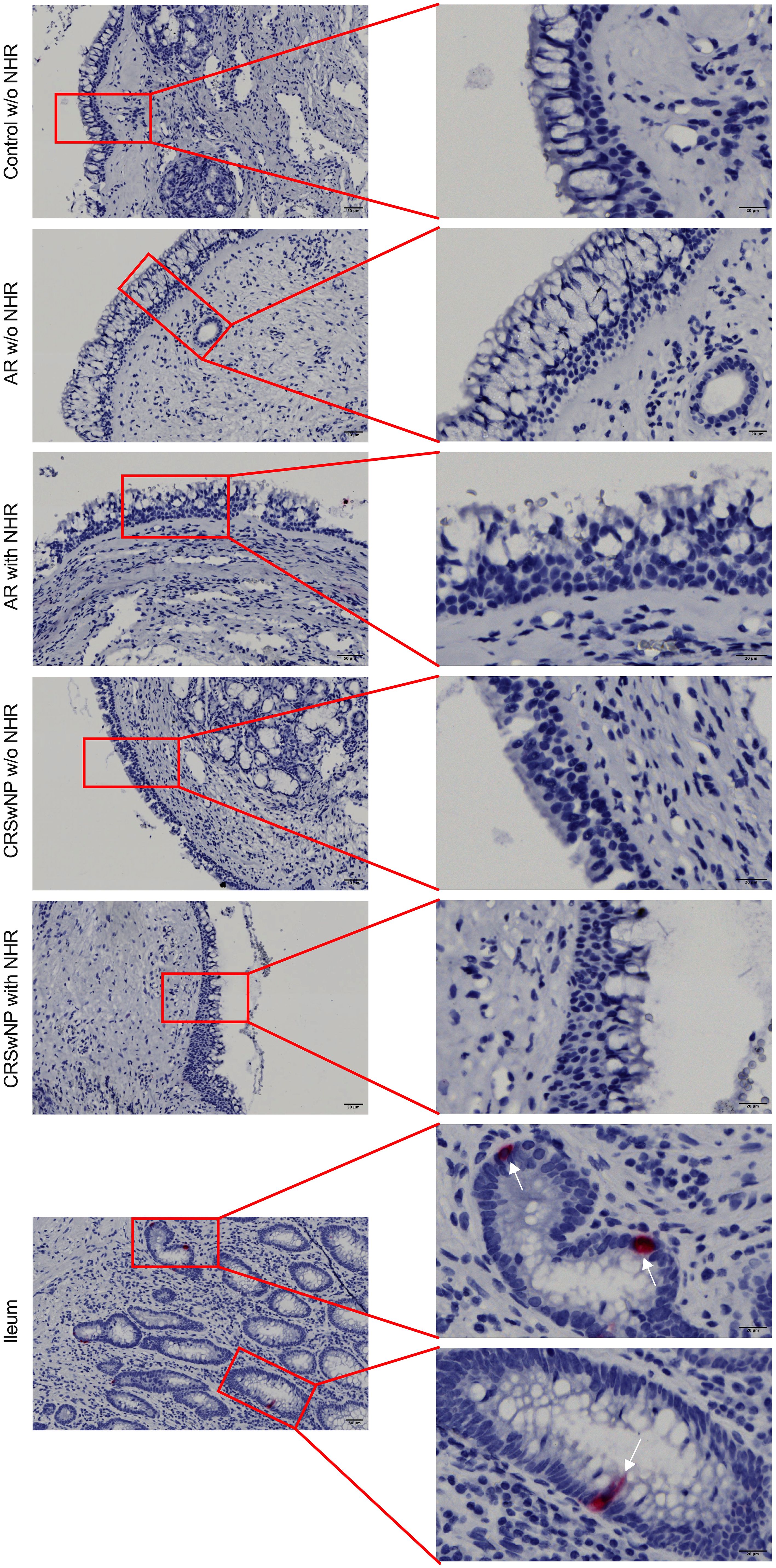
Figure 3 Detection of CHGA RNA molecules in ileum and nasal biopsies using RNA scope RED assay. Representative CHGA RNA detection in nasal biopsies from controls without NHR, AR with and without NHR patients and CRSwNP with and without NHR patients and ileum samples as determined by the RNA scope RED assay. NHR, nasal hyperreactivity; AR, allergic rhinithis; CRSwNP, chronic rhinosinusitis with nasal polyps; CHGA, chromogranin A; scale bar left = 50 µm; scale bar right = 20 µm.
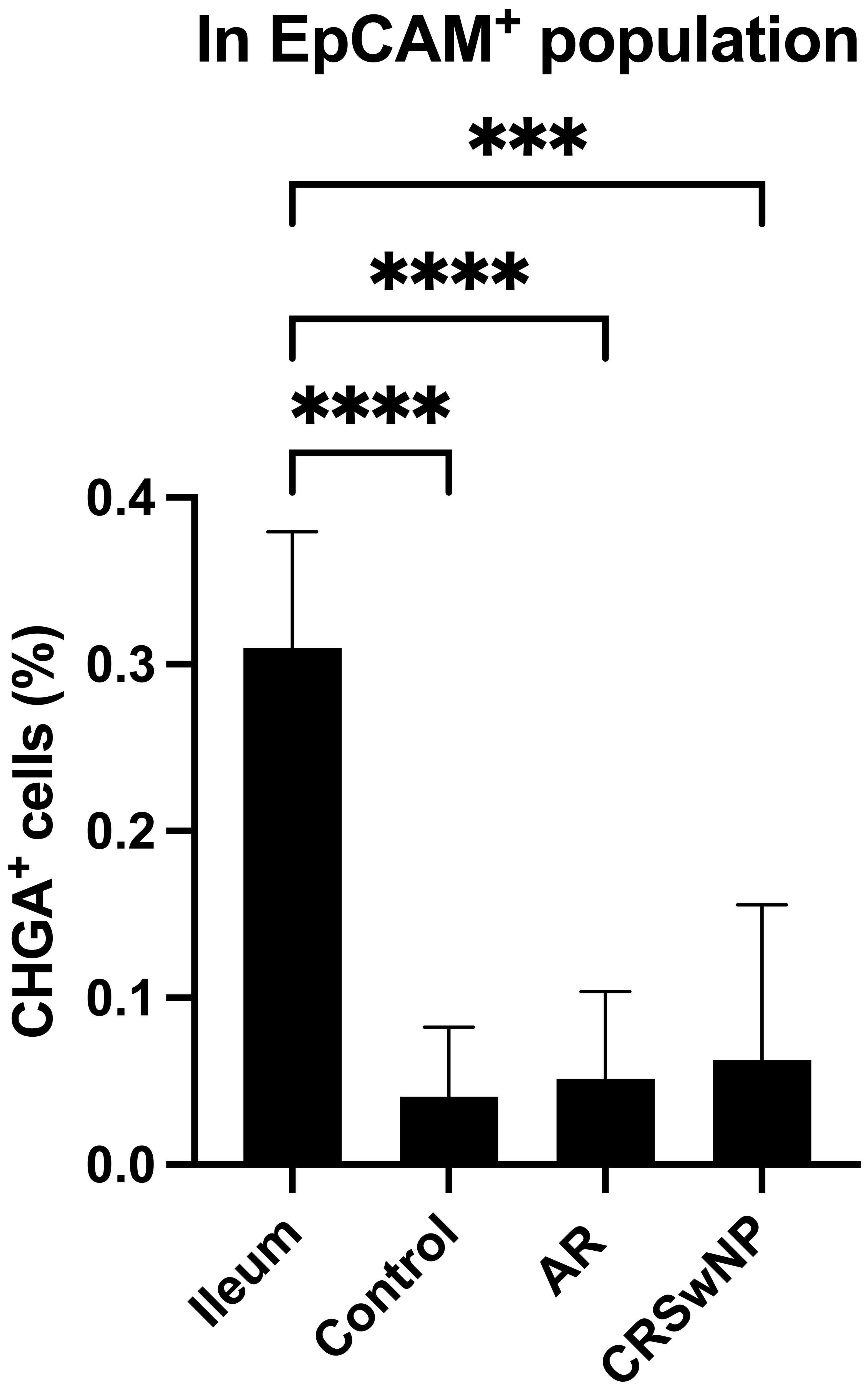
Figure 4 Quantification of CHGA+ cells in ileum and nasal biopsies as determined by flow cytometry. Percentage of CHGA+ cells within the epithelial cell population defined as EpCAM+ expression in ileum tissue and nasal tissue from controls, AR patients and CRSwNP patients (n = 5/group). CHGA, chromogranin A; EpCAM: epithelial cellular adhesion molecule; AR, allergic rhinithis; CRSwNP, chronic rhinosinusitis with nasal polyps; data represented as mean ± SD, *** p < 0.005; **** p < 0.001.
As NECs are a rare cell population (< 0.5%), and potentially are missed with less sensitive methods, we used single cell RNA-sequencing to define the presence of NECs in the nasal mucosa (8, 11, 12). In total 73,487 cells from inferior turbinates of four well characterized subjects of which two controls and two HDM-induced AR patients were sequenced. After quality control and filtering, 61,655 cells remained with a high quality of sequencing data (Supplementary Figure 2). After Harmony integration to remove batch effects, unsupervised graph-based clustering was performed. In total, 20 clusters were defined of which 13 were identified as epithelial cell clusters (blue) after annotation (Figure 5A). Non-epithelial cell clusters included fibroblasts characterized by MXRA8, NBL1, VCAN and LEPR expression and smooth muscle cells characterized by MYH11, CNN1 and ACTA2 expression (Figures 5B–D). Other smaller non-epithelial cell clusters were identified as endothelial cells characterized by PTPRB, PDE2A and ECSCR expression, as well as astrocytes which were identified by expression of CDH19, FOXD3 and MPZ (Figures 5B–D). Lastly, the non-epithelial cell clusters contained two immune cell clusters, being T and B cells, characterized by expression of CD3E, CD2, PTPRC and CD86, CD19, respectively (Figures 5B–D).
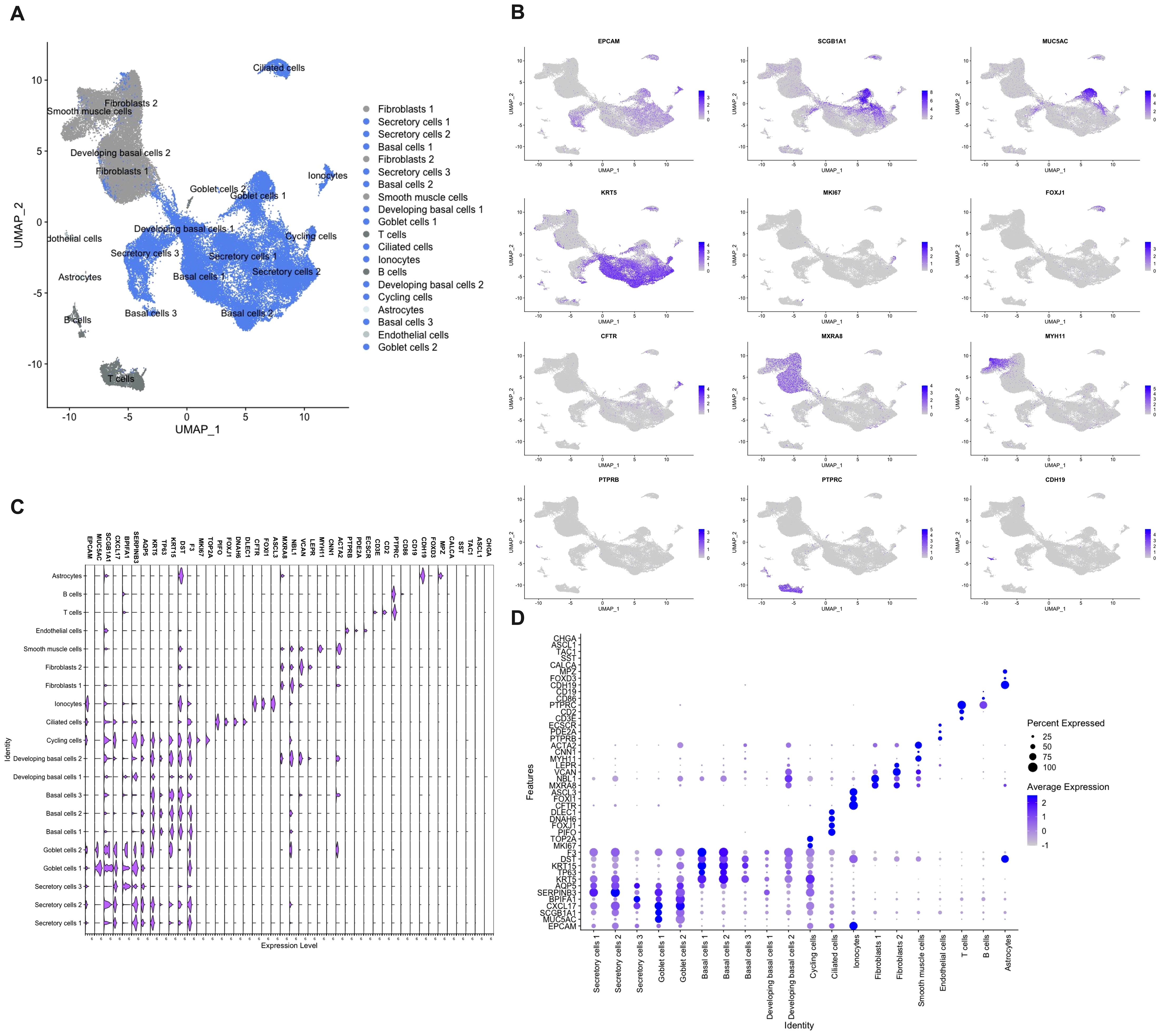
Figure 5 Single cell RNA-sequencing of cells isolated from nasal biopsies from patients and controls. (A) Umap clustering and annotation of single cell RNA-sequencing of allergic rhinitis patients (n=2) and controls (n=2) coloured based on epithelial (blue) and non-epithelial (grey) cells. (B) Feature plots of top marker genes for each cell cluster. (C) Violin plot showing gene expression of cluster and NEC markers. (D) Dot plot displaying mRNA expression levels of cluster and NEC markers.
Within the epithelial cell clusters, most cells were identified as secretory cells, including goblet cells. Secretory and goblet cells were identified by the expression of general secretory markers such as SCGB1A1, CXCL17, BPIFA1, SERPINB3 and AQP5 (Figures 5B–D). Despite strong transcriptional similarity, goblet cells were identified by having higher MUC5AC expression which is the best-known marker for goblet cells in the upper airways (Figures 5B–D). The second largest population that was identified were the basal cells. In total, three basal cell populations and two developing basal cell populations were identified. All basal cell clusters expressed KRT5, TP63, KRT15, DST and F3 to a different extend (Figures 5B–D). More stem cell like basal cell clusters (basal cells 1 and 2) expressed more KRT5 and TP63, while the more developing basal cells expressed less KRT5 and TP63 and started to show low expression of the beforementioned secretory genes (Figures 5B–D). Cycling cells were identified by their mixed expression of both secretory and basal genes, as well as by added expression of genes involved in cell cycling such as MKI67 and TOP2A (Figures 5B–D). In our dataset, a lower amount than expected of ciliated cells, characterized by the expression of PIFO, FOXJ1, DNAH6 and DLEC1, were sequenced. (Figures 5B–D). Furthermore, also a cell cluster of the rare cell type, the ionocytes, defined by CFTR, FOXI1 and ASCL3 expression, was identified (Figures 5B–D). In contrast, known marker genes of NECs, such as CHGA, ASCL1, TAC1, SST and CALCA, could not be found to identify a single cell population of NECs in our dataset (Figures 5C, D). This in contrast with the analysis of the existing CZ CELLxGENE Discover database which shows uniform expression of CHGA in NECs across different human tissues (Supplementary Figure 3) (42). Most pulmonary NECs also express ASCL1 and CALCA, and to a lesser extent SST and TAC1 (Supplementary Figure 3) (42). In conclusion, with the use of several techniques, we were not able to detect the presence of NECs in the nasal mucosa of patients with upper airway disease nor in healthy subjects.
No correlation of NECs marker expression with nasal hyperreactivity in upper airway disease
Since NHR is a hallmark of neurogenic inflammation, we wanted to know if the limited expression levels of CHGA and ASCL1 were correlated with the presence of NHR in patients. Subjects were considered to have NHR if they had a drop of at least 20% in their PNIF after cold dry air exposure as an objective measurement for NHR. No significant differences in CHGA and ASCL1 expression could be found in subjects without NHR compared to patients with NHR (Figure 6A). When the subjects were further classified based on their upper airway disease, being controls, AR patients and CRSwNP patients, as well as based on their NHR diagnosis, we also could not observe a significant difference in CHGA or ASCL1 expression levels (Figure 6B). Furthermore, no significant correlation between the expression of CHGA or ASCL1 with the VAS score of total nasal symptoms, or other measurements of disease severity, could be detected (Figure 6C; Supplementary Figures 4, 5).
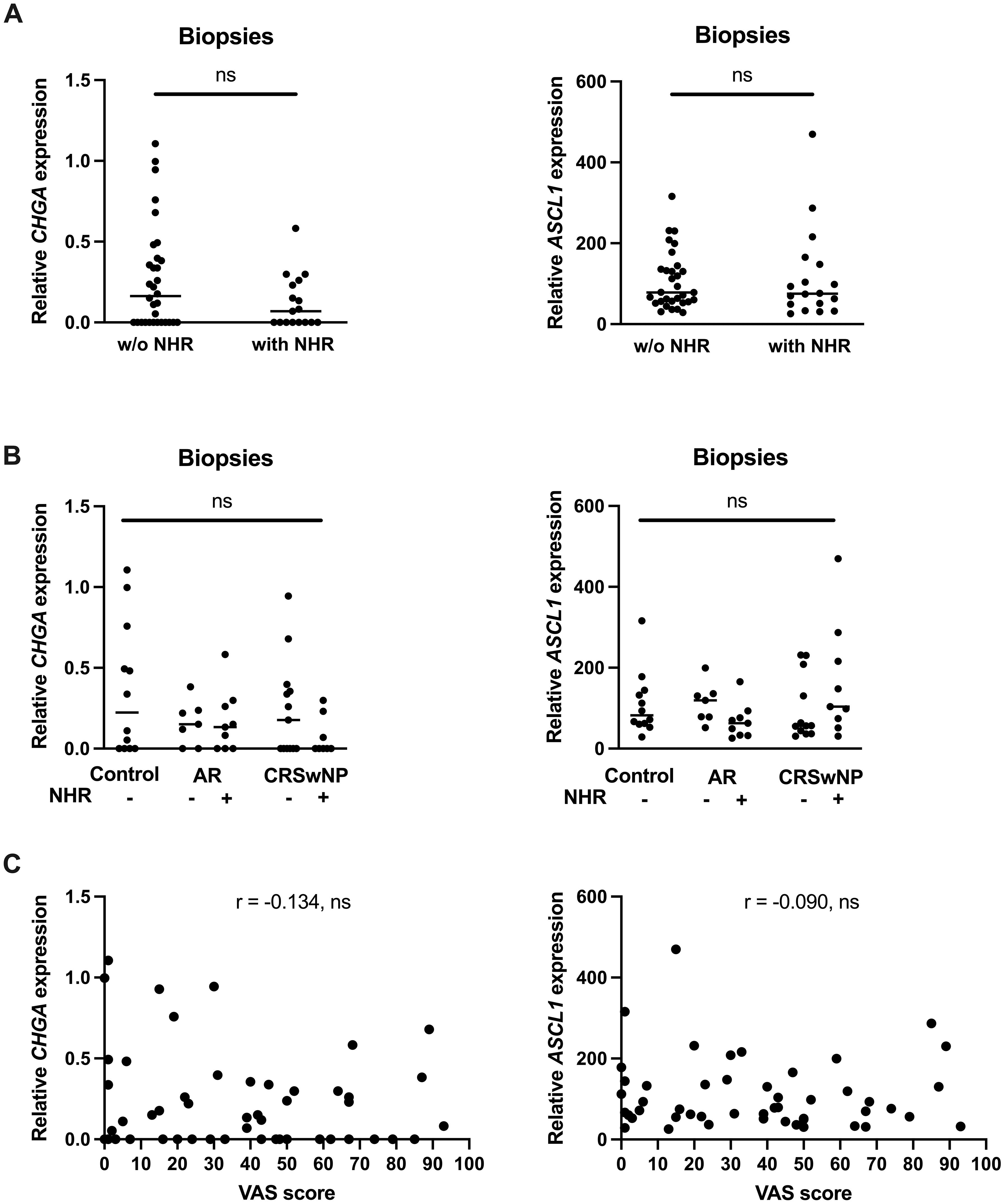
Figure 6 Correlation between the expression of the NEC markers CHGA and ASCL1 and nasal hyperreactivity in upper airway disease. (A) mRNA expression levels of CHGA and ASCL1 in human nasal biopsies with and without NHR. (B) mRNA expression levels of CHGA and ASCL1 in human nasal biopsies in controls, AR with and without NHR patients and CRSwNP with and without NHR patients. (C) Correlation of CHGA and ASCL1 expression with the VAS score of total nasal symptoms as measure of disease severity. NHR, nasal hyperreactivity; AR, allergic rhinithis; CRSwNP, chronic rhinosinusitis with nasal polyps; VAS, visual analogue score; CHGA, chromogranin A; ASCL1, Achaete-Scute Family bHLH transcription factor 1; data represented as individual values with median, ns = p > 0.05.
Discussion
There is increasing evidence that NECs play a possible role in sensory neuron hyperactivation (23–26). The role of NEC-like cells in the pathology of upper airways diseases has not been thoroughly studied yet. An extensive analysis of the presence and potential involvement of NECs in NHR in the upper airways of patients with upper airway disease and control subjects was performed. Surprisingly, only limited expression of NECs markers on both RNA and protein level in the upper airway epithelium was found, irrespective of disease status, using pooled methods in which we could not specify from which cells these signals were derived from. In single cell methods, no evidence of NECs in the upper airways was found. Western blotting detects pooled protein signals from all cells in a sample but cannot specify the source cells, while flow cytometry quantifies protein expression in individual cells, providing cell-specific analysis. The strong CHGA response in the ileum by western blotting, compared to only 0.5% detection of CHGA+ cells by flow cytometry, highlights the differences between these techniques in sensitivity and specificity. Flow cytometry clearly detects single CHGA+ cells in the ileum, however, in the nasal EpCAM+ population, the CHGA+ cell population and expression are so minimal that it is challenging to definitively confirm the presence of CHGA+ cells within the nasal epithelial cell population, thereby questioning the presence of neuroendocrine cells in the nasal epithelium.
Chromogranins, such as CHGA, belong to the family of proteins that constitute a major component of the secretory granules of various endocrine and neuroendocrine cells and is therefore considered as a general marker for NECs (43). In daily clinical setting, CHGA is a well-established biomarker to diagnose neuroendocrine tumors (44). Furthermore, CHGA was consistently identified as a top marker gene for NECs in both the lungs and gut (8, 12, 45, 46). Surprisingly, CHGA and other NEC markers could not be observed in our dataset. Only four subjects were sequenced, which might be too limited to identify NECs. Nevertheless, we have sequenced up to 20,000 cells per subject, which, based on the estimated prevalence, should have resulted in 200 to 1,000 sequenced NECs. It might be plausible that NECs in the upper airways are characterized by other unidentified marker genes. For this reason, CHGA detection was complemented with the additional marker ASCL1. Ascl1 knock out studies demonstrated that Ascl1 is not only an early expressed marker of pulmonary NECs, but is also necessary but not sufficient for pulmonary NECs cell fate specification (23, 47, 48). In addition, CHGA detection was further complemented with the detection of neuropeptides associated with neurogenic inflammation like CGRP, TAC1 and SST (11, 12, 23). In mice, CGRP was shown to be expressed in 95% of pulmonary NECs, and was shown to be released by pulmonary NECs upon allergen challenge, thereby highlighting the role of CGRP as a classical NEC neuropeptide (23, 49). Despite the combination of CHGA detection with multiple other known NEC-associated markers, no relevant expression in the nasal mucosa could be found. Although CHGA, ASCL1 and CALCA (encoding CGRP) are all considered as canonical NECs marker, it is still plausible that nasal NECs are characterized by other markers such as GRP, PIEZO2 or NMU (42, 49, 50).
Pulmonary NECs first differentiate as scattered solitary cells, whereafter they migrate to form clusters called neuroepithelial bodies (NEB) (30, 51). NEB are frequently observed at the bifurcation points of branching airways (52, 53). This specific location at the airway branch points is also the location were inhaled aerosolized particles congregate, therefore positioning the pulmonary NECs at the ideal location to detect and respond to inhaled environmental particles (54). Additionally, in the lower airways, pulmonary NECs have been described as intrapulmonary chemosensors to detect several environmental stimuli such as oxygen, nicotine, mechanical changes, and succinate (55–64). In the intestine, enteroendocrine cells respond to irritants, metabolites, norepinephrine or interleukin (IL)-33 via activation of TRPA1, olfactory receptor 558, alpha-2A adrenergic receptor and IL-33 receptor, respectively (24, 65). Activation of the enteroendocrine cells through one of the aforementioned receptors results in the release of 5-HT leading to activation of the nearby serotonergic afferent nerve endings (24, 65). As mentioned earlier, a variety of intestinal physiological processes, including gastrointestinal motility and visceral hypersensitivity have been shown to be regulated via 5-HT release (24, 25, 65). Given the accumulation of pulmonary NECs at airway branch points and the chemosensory function of NECs in the lower airways and intestine, it can be assumed that if NECs are present in the nasal epithelium, they would be localized at regions with the highest possibility to encounter environmental triggers. In the nasal cavity, the inferior turbinate represents the best hypothetical location for NECs (66). All samples used in this study were therefore coming from inferior turbinates of controls and AR patients. However, we cannot fully exclude that NECs are localized at a different location in the nasal cavity.
Pulmonary NECs and their intestinal counterpart are known to be extremely rare cell types, with their prevalence being estimated to be less than 0.5% of the epithelium (45–47). It is possible that, even with very sensitive techniques, NECs could not be detected in the upper airway epithelium due to their scarcity. Unlike previous publications which successfully identified NEC clusters in intestines and in the lower airways, we could not detect any NECs in the nasal mucosa using single cell RNA-sequencing (12, 42, 45, 67). When applying all the other assays to ileum samples, enteroendocrine cells were successfully detected, emphasizing that NECs are unlikely to be present in nasal mucosa. The lack of NECs being present in the upper airways was unexpected, given the implication of pulmonary NECs in several pulmonary diseases. Increased numbers of pulmonary NECs were observed in lungs of asthmatic patients (23). Pulmonary NECs play a role in amplifying type 2 inflammation and goblet cell hyperplasia in an allergen-induced mouse model via CGRP and GABA release, respectively (23). Also, chronic obstructive pulmonary disease (COPD) patients have a higher number of pulmonary NECs and pulmonary NECs products in their airways compared to healthy controls (68). Both in COPD patients and in smokers, the major risk factor of COPD, higher levels of bombesin-like peptide were found, suggesting an association with pulmonary NECs hyperplasia and increased sensitivity to chemical stimuli possibly leading to COPD development (69). In neuroendocrine cell hyperplasia of infant syndrome it was shown that patients showed NEC hyperplasia mainly in the distal respiratory airways (70, 71). Furthermore, NECs have also been thought to be the originating cancer cells in neuroendocrine small cell lung cancer (72, 73).
Despite the association of NECs in lower airway diseases, no correlation was found between NEC marker expression and disease or NHR severity in the upper airways. Therefore, it is unlikely that increased numbers of NECs in the upper airways are responsible for upper airway diseases or NHR. As mentioned, the extremely rare nature of NECs, as well as their scattered location can hamper the detection of a relevant correlation. Furthermore, the functional capacity of NECs was not investigated. Different studies have already used specific ASCL1-cre-mediated knock out or labeling of NECs in the murine lower airways to investigate the functional role of NECs (23, 49, 64, 74, 75). An analogue upper airway epithelial specific NEC knock in or knock out mouse model could be used to elucidate the potential functional role of NECs in the upper airways.
In conclusion, only limited expression of NEC markers on both gene and protein level was found in the human upper airway epithelium using multiple methods in which we could not specify from which cells these signals were derived from. Furthermore, no correlation between NECs and NHR could be detected. In contrast, enteroendocrine cells were detected in the ileum using the same protocols. In our single cell RNA-sequencing dataset, no NECs cluster or any type of cells in the nasal mucosa expressing NEC-associated markers was detected. Therefore, the implication of NECs in upper respiratory diseases remains questionable. This in contrast with the bronchial and intestinal epithelium where NECs have been described to be present, to act as chemosensor to environmental stimuli and are associated with various pathophysiology’s.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of therepository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/geo/, GSE261706.
Ethics statement
The studies involving humans were approved by Ethical Committee Research of University Hospitals Leuven. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
TW: Conceptualization, Data curation, Funding acquisition, Investigation, Methodology, Visualization, Writing – original draft, Writing – review & editing. WB: Data curation, Methodology, Writing – review & editing. IJ: Methodology, Writing – review & editing. ER: Methodology, Writing – review & editing. JC: Methodology, Writing – review & editing. ED: Methodology, Writing – review & editing. DB: Supervision, Writing – review & editing. KT: Supervision, Writing – review & editing. BS: Conceptualization, Data curation, Methodology, Supervision, Writing – review & editing. LV: Methodology, Supervision, Writing – review & editing. KM: Methodology, Supervision, Writing – review & editing. PH: Supervision, Writing – review & editing, Funding acquisition, Investigation, Methodology, Validation, Conceptualization.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The author’s laboratories are supported by grants from the research council of the KU Leuven (C14/18/086). TW is supported by a predoctoral grant of the Fund for Scientific Research Flanders (FWO), Belgium (11L8922N). KM received a junior postdoctoral fellowship from the Fund for Scientific Research Flanders (FWO), Belgium (12Z0622N). BS received a postdoctoral fellowship from the Fund for Scientific Research Flanders (FWO), Belgium (12U6721N). LG is supported by a grant of the Fund for Scientific Research Flanders (FWO), Belgium (18B2222N). DB is a recipient of a senior clinical investigator grant of the Fund for Scientific Research Flanders (FWO), Belgium (1801014N) and from the Bijzonder Onderzoeksfonds – fundamenteel klinisch mandaat (BOF-FKM) of the KU Leuven (FKO CM23-FKO-02).
Acknowledgments
The authors would like to thank Professor Dr. M. Jorissen and the personnel of the Otorhinolaryngology, head and neck surgery department of UZ Leuven, as well as Professor Dr. G. Boeckxstaens, Professor Dr. S. Vermeire, Professor Dr. B. Verstockt and the personnel of the Gastroenterology and hepatology department of UZ Leuven for their contribution in harvesting nasal and ileum tissue, respectively. We thank the UZ Leuven Genomics Core for their help and expertise in the sequencing experiment. Images were recorded on the Zeiss Axioscan 7 thanks to the department of Woman and Child, UZ Leuven. The authors would also express their gratitude to Dr. M. Khan and the Max Planck Research Unit for Neurogenetics for their advice on the RNA scope assay.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1394539/full#supplementary-material
Abbreviations
AR, Allergic rhinitis; ASCL1, Achaete-Scute Family bHLH transcription factor 1; CGRP, Calcitonin gene-related peptide; CHGA, Chromogranin A; CRSwNP, Chronic rhinosinusitis with nasal polyps; EpCAM, Epithelial cellular adhesion molecule; HDM, House dust mite; IL, interleukin; KRT5, Keratin 5; MP, Milk powder; MUC5AC, Mucin-5AC; NECs, Neuroendocrine cells; NHR, Nasal hyperreactivity; PNIF, Peak nasal inspiratory flow; VAS, Visual Analogue Scale.
References
1. Doulaptsi M, Steelant B, Prokopakis E, Ierodiakonou D, Tsinaslanidou Z, Cools L, et al. Prevalence and impact of nasal hyperreactivity in chronic rhinosinusitis. Allergy. (2020) 75:1768–71. doi: 10.1111/all.14199
2. Bankova LG, Barrett NA. Epithelial cell function and remodeling in nasal polyposis. Ann Allergy Asthma Immunol. (2020) 124:333–41. doi: 10.1016/j.anai.2020.01.018
3. Lambrecht BN, Hammad H. The airway epithelium in asthma. Nat Med. (2012) 18:684–92. doi: 10.1038/nm.2737
4. Steelant B, Farré R, Wawrzyniak P, Belmans J, Dekimpe E, Vanheel H, et al. Impaired barrier function in patients with house dust mite–induced allergic rhinitis is accompanied by decreased occludin and zonula occludens-1 expression. J Allergy Clin Immunol. (2016) 137:1043–1053.e5. doi: 10.1016/j.jaci.2015.10.050
5. Xiao C, Puddicombe SM, Field S, Haywood J, Broughton-Head V, Puxeddu I, et al. Defective epithelial barrier function in asthma. J Allergy Clin Immunol. (2011) 128:549–456. doi: 10.1016/j.jaci.2011.05.038
6. Soyka M, Holzmann D, Basinski T, Wawryniak M, Bannert C, Burgler S. The induction of IL-33 in the sinus epithelium and its influence on T-helper cell responses. PloS One. (2012) 10:e0123163. doi: 10.1371/journal.pone.0123163
7. Hellings PW, Steelant B. Epithelial barriers in allergy and asthma. J Allergy Clin Immunol. (2020) 145:1499–509. doi: 10.1016/j.jaci.2020.04.010
8. Vieira Braga FA, Kar G, Berg M, Carpaij OA, Polanski K, Simon LM, et al. A cellular census of human lungs identifies novel cell states in health and in asthma. Nat Med. (2019) 25:1153–63. doi: 10.1038/s41591-019-0468-5
9. Plasschaert LW, Žilionis R, Choo-Wing R, Savova V, Knehr J, Roma G, et al. A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte. Nature. (2018) 560:377–81. doi: 10.1038/s41586-018-0394-6
10. Ordovas-Montanes J, Dwyer DF, Nyquist SK, Buchheit KM, Vukovic M, Deb C, et al. Allergic inflammatory memory in human respiratory epithelial progenitor cells. Nature. (2018) 560:649–54. doi: 10.1038/s41586-018-0449-8
11. Travaglini KJ, Nabhan AN, Penland L, Sinha R, Gillich A, Sit RV, et al. A molecular cell atlas of the human lung from single-cell RNA sequencing. Nature. (2020) 587:619–25. doi: 10.1038/s41586-020-2922-4
12. Deprez M, Zaragosi LE, Truchi M, Becavin C, Ruiz Garcia S, Arguel MJ, et al. A single-cell atlas of the human healthy airways. Am J Respir Crit Care Med. (2020) 202:1636–45. doi: 10.1164/rccm.201911-2199OC
13. De Greve G, Hellings PW, Pugin B, Seys SF. Endotype-driven treatment in chronic upper airway diseases. Clin Transl Allergy. (2017) 7:4–12. doi: 10.1186/s13601-017-0157-8
14. Callebaut I, De Vries A, Steelant B, Hox V, Bobic S, Van Gerven L, et al. Nasal allergen deposition leads to conjunctival mast cell degranulation in allergic rhinoconjunctivitis. Am J Rhinol Allergy. (2014) 28:290–6. doi: 10.2500/ajra.2014.28.4052
15. Van Gerven L, Steelant B, Hellings PW. Nasal hyperreactivity in rhinitis: A diagnostic and therapeutic challenge. Allergy. (2018) 73:1784–91. doi: 10.1111/all.13453
16. Bernstein JA, Singh U. Neural abnormalities in nonallergic rhinitis. Curr Allergy Asthma Rep. (2015) 15:18. doi: 10.1007/s11882-015-0511-7
17. Mutolo D. Brainstem mechanisms underlying the cough reflex and its regulation. Respir Physiol Neurobiol. (2017) 243:60–76. doi: 10.1016/j.resp.2017.05.008
18. Van Gerven L, Alpizar YA, Wouters MM, Hox V, Hauben E, Jorissen M, et al. Capsaicin treatment reduces nasal hyperreactivity and transient receptor potential cation channel subfamily V, receptor 1 (TRPV1) overexpression in patients with idiopathic rhinitis. J Allergy Clin Immunol. (2014) 133:1332–9. doi: 10.1016/j.jaci.2013.08.026
19. Backaert W, Steelant B, Jorissen M, Van Oudenhove L, Talavera K, Hellings PW, et al. Self-reported nasal hyperreactivity is common in all chronic upper airway inflammatory phenotypes and not related to general well-being. Allergy. (2021) 76:3806–9. doi: 10.1111/all.15060
20. Feijen J, Seys SF, Steelant B, Bullens DMA, Dupont LJ, García-Cruz M, et al. Prevalence and triggers of self-reported nasal hyperreactivity in adults with asthma. World Allergy Organ J. (2020) 13:100132. doi: 10.1016/j.waojou.2020.100132
21. Van Gerven L, Boeckxstaens G, Jorissen M, Fokkens WJ, Hellings PW. Short-time cold dry air exposure: A useful diagnostic tool for nasal hyperresponsiveness. Laryngoscope. (2012) 122:2615–20. doi: 10.1002/lary.23495
22. Xu J, Yu H, Sun X. Less is more: rare pulmonary neuroendocrine cells function as critical sensors in lung. Dev Cell. (2020) 55:123–32. doi: 10.1016/j.devcel.2020.09.024
23. Sui P, Wiesner DL, Xu J, Zhang Y, Lee J, Van Dyken S, et al. Pulmonary neuroendocrine cells amplify allergic asthma responses. Science. (2018) 360:eaan8546. doi: 10.1126/science.aan8546
24. Bellono NW, Bayrer JR, Leitch DB, Castro J, Zhang C, O’Donnell TA, et al. Enterochromaffin cells are gut chemosensors that couple to sensory neural pathways. Cell. (2017) 170:185–98. doi: 10.1016/j.cell.2017.05.034
25. Linan-Rico A, Ochoa-Cortes F, Beyder A, Soghomonyan S, Zuleta-Alarcon A, Coppola V, et al. Mechanosensory signaling in enterochromaffin cells and 5-HT release: potential implications for gut inflammation. Front Neurosci. (2016) 10. doi: 10.3389/fnins.2016.00564
26. Devos FC, Boonen B, Alpizar YA, Maes T, Hox V, Seys S, et al. Neuro-immune interactions in chemical-induced airway hyperreactivity. Eur Respir J. (2016) 48:380–92. doi: 10.1183/13993003.01778-2015
27. López-Requena A, Boonen B, Van Gerven L, Hellings P, Alpizar Y, Talavera K. Roles of neuronal TRP channels in neuroimmune interactions. In: Neurobiology of TRP Channels 2nd ed. CRC Press/Taylor & Francis, Boca Raton (FL), USA (2017). p. 277–94.
28. Lommel AV. Pulmonary neuroendocrine cells (PNEC) and neuroepithelial bodies (NEB): chemoreceptors and regulators of lung development. Paediatr Respir Rev. (2001) 2:171–6. doi: 10.1053/prrv.2000.0126
29. Cutz E, Jackson A. Neuroepithelial bodies as airway oxygen sensors. Respir Physiol. (1999) 115:201–14. doi: 10.1016/S0034-5687(99)00018-3
30. Branchfield K, Nantie L, Verheyden JM, Sui P, Wienhold MD, Sun X. Pulmonary neuroendocrine cells function as airway sensors to control lung immune response. Science. (2016) 351:707–10. doi: 10.1126/science.aad7969
31. Gehart H, van Es JH, Hamer K, Beumer J, Kretzschmar K, Dekkers JF, et al. Identification of enteroendocrine regulators by real-time single-cell differentiation mapping. Cell. (2019) 176:1158–73. doi: 10.1016/j.cell.2018.12.029
32. Laguerre F, Anouar Y, Montero-Hadjadje M. Chromogranin A in the early steps of the neurosecretory pathway. Int Union Biochem Mol Biol Life. (2020) 72:524–32. doi: 10.1002/iub.2218
33. Borromeo MD, Savage TK, Kollipara RK, He M, Augustyn A, Osborne JK, et al. ASCL1 and NEUROD1 reveal heterogeneity in pulmonary neuroendocrine tumors and regulate distinct genetic programs. Cell Rep. (2016) 16:1259–72. doi: 10.1016/j.celrep.2016.06.081
34. Mou H, Yang Y, Riehs MA, Fine A, Wellington CV, Ai X. Airway basal stem cells generate distinct subpopulations of PNECs. Cell Rep. (2021) 35:109011. doi: 10.1016/j.celrep.2021.109011
35. Hor P, Punj V, Calvert BA, Castaldi A, Miller AJ, Carraro G, et al. Efficient generation and transcriptomic profiling of human iPSC-derived pulmonary neuroendocrine cells. iScience. (2020) 23:101083. doi: 10.1016/j.isci.2020.101083
37. Aronesty E. Comparison of sequencing utility programs. Ea-Utils. (2013). doi: 10.2174/1875036201307010001
38. Andrews S. FASTQC. A quality control tool for high throughput sequence data. Cambridge: Babraham Bioinformatics (2010).
39. 10x Genomics Cell Ranger 4.0.0 using 10x Genomics Cloud Analysis. Pleasanton, California, USA: 10X genomics (2020)
40. Love M, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. (2015) 15:550. doi: 10.1186/s13059-014-0550-8
41. Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, et al. Comprehensive integration of single-cell data. Cell. (2019) 177:1888–902. doi: 10.1016/j.cell.2019.05.031
42. CZI Single-Cell Biology, et al. CZ CELLxGENE Discover: A single-cell data platform for scalable exploration, analysis and modeling of aggregated data. bioRxiv (2023). 10.30. doi: 10.1101/2023.10.30.563174
43. Gut P, Czarnywojtek A, Fischbach J, Baczyk M, Ziemnicka K, Wrotkowska E, et al. Chromogranin A – unspecific neuroendocrine marker. Clinical utility and potential diagnostic pitfalls. Arch Med Sci. (2016) 12:1–9. doi: 10.5114/aoms.2016.57577
44. Rindi G, Mete O, Uccella S, Basturk O, La Rosa S, Brosens LAA, et al. Overview of the 2022 WHO classification of neuroendocrine neoplasms. Endocr Pathol. (2022) 33:115–54. doi: 10.1007/s12022-022-09708-2
45. Harber AL, Biton M, Rogel N, Herbst RH, Shekhar K, Smillie C, et al. A single-cell survey of the small intestinal epithelium. Nature. (2017) 551:333–9. doi: 10.1038/nature24489
46. Zeve D, Stas E, de Sousa Casal J, Mannam P, Wanshu Q, Carlone DL, et al. Robust differentiation of human enteroendocrine cells from intestinal stem cells. Nat Commun. (2022) 13:261. doi: 10.1038/s41467-021-27901-5
47. Linnoila RI, Zhao B, DeMayo JL, Nelkin BD, Baylin SB, DeMayo FJ, et al. Constitutive achaete-scute homologue-1 promotes airway dysplasia and lung neuroendocrine tumors in transgenic mice. Cancer Res. (2000) 60:4005–9.
48. Ito T, Udaka N, Yazawa T, Okudela K, Hayashi H, Sudo T, et al. Basic helix-loop-helix transcription factors regulate the neuroendocrine differentiation of fetal mouse pulmonary epithelium. Development. (2000) 127:3913–21. doi: 10.1242/dev.127.18.3913
49. Kuo CS, Darmanis S, Diaz de Arce A, Liu Y, Almanzar N, Wu TTH, et al. Neuroendocrinology of the lung revealed by single-cell RNA sequencing. Noble PW, Shen M, editors. eLife. (2022) 11:e78216. doi: 10.7554/eLife.78216
50. Klose CSN, Mahlakõiv T, Moeller JB, Rankin LC, Flamar AL, Kabata H, et al. The neuropeptide neuromedin U stimulates innate lymphoid cells and type 2 inflammation. Nature. (2017) 549:282–6. doi: 10.1038/nature23676
51. Van Lommel A, Lauweryns JM. Postnatal development of the pulmonary neuroepithelial bodies in various animal species. J Auton Nerv Syst. (1997) 65:17–24. doi: 10.1016/S0165-1838(97)00030-1
52. Avadhanam KP, Plopper CG, Pinkerton KE. Mapping the distribution of neuroepithelial bodies of the rat lung. A whole-mount immunohistochemical approach. Am J Pathol. (1997) 150:851–9.
53. Noguchi M, Sumiyama K, Morimoto M. Directed migration of pulmonary neuroendocrine cells toward airway branches organizes the stereotypic location of neuroepithelial bodies. Cell Rep. (2015) 13:2679–86. doi: 10.1016/j.celrep.2015.11.058
54. Peake JL, Reynolds SD, Stripp BR, Stephens KE, Pinkerton KE. Alteration of Pulmonary Neuroendocrine Cells during Epithelial Repair of Naphthalene-Induced Airway Injury Author links open overlay panel. Am J Pathol. (2000) 156:279–86. doi: 10.1016/S0002-9440(10)64728-1
55. Shivaraju M, Chitta UK, Grange RMH, Jain IH, Capen D, Liao L, et al. Airway stem cells sense hypoxia and differentiate into protective solitary neuroendocrine cells. Science. (2021) 371:52–7. doi: 10.1126/science.aba0629
56. Cherniack NS. Oxygen sensing: applications in humans. J Appl Physiol. (1985) 96:352–8. doi: 10.1152/japplphysiol.00755.2003
57. Youngson C, Nurse C, Yeger H, Cutz E. Oxygen sensing in airway chemoreceptors. Nature. (1993) 365:153–5. doi: 10.1038/365153a0
58. Semenza GL. Regulation of oxygen homeostasis by hypoxia-inducible factor 1. Physiol Bethesda. (2009) 24:97–106. doi: 10.1152/physiol.00045.2008
59. Pan J, Yeger H, Ratcliffe P, Bishop T, Cutz E. Hyperplasia of pulmonary neuroepithelial bodies (NEB) in lungs of prolyl hydroxylase -1(PHD-1) deficient mice. Adv Exp Med Biol. (2012) 758:149–55. doi: 10.1007/978-94-007-4584-1_21
60. Sartelet H, Maouche K, Laurent TJ, Petit J, Burlet H, Monteau M, et al. Expression of nicotinic receptors in normal and tumoral pulmonary neuroendocrine cells (PNEC). Pathol Res Pract. (2008) 204:891–8. doi: 10.1016/j.prp.2008.05.006
61. Nonomura K, Woo SH, Chang RB, Gillich A, Qiu Z, Francisco AG, et al. Piezo2 senses airway stretch and mediates lung inflation-induced apnoea. Nature. (2017) 541:176–81. doi: 10.1038/nature20793
62. Lembrechts R, Brouns I, Schnorbusch K, Pintelon I, Timmermans JP, Adriaensen D. Neuroepithelial bodies as mechanotransducers in the intrapulmonary airway epithelium involvement of TRPC5. Am J Respir Cell Mol Biol. (2012) 47:315–23. doi: 10.1165/rcmb.2012-0068OC
63. Pan J, Copland I, Post M, Yeger H, Cutz E. Mechanical stretch-induced serotonin release from pulmonary neuroendocrine cells: implications for lung development. Am J Physiol-Lung Cell Mol Physiol. (2006) 290:185–93. doi: 10.1152/ajplung.00167.2005
64. Yu W, Moninger TO, Rector MV, Stoltz DA, Welsh MJ. Pulmonary neuroendocrine cells sense succinate to stimulate myoepithelial cell contraction. Dev Cell. (2022) 57:2221–36. doi: 10.1016/j.devcel.2022.08.010
65. Chen Z, Luo J, Li J, Kim G, Stewart A, Urban JF Jr, et al. Interleukin-33 promotes serotonin release from enterochromaffin cells for intestinal homeostasis. Immunity. (2021) 54:151–62. doi: 10.1016/j.immuni.2020.10.014
66. Xiong GX, Zhan JM, Jiang HY, Li JF, Rong LW, Xu G. Computational fluid dynamics simulation of airflow in the normal nasal cavity and paranasal sinuses. Am J Rhinol Allergy. (2008) 22:477–82. doi: 10.2500/ajr.2008.22.3211
67. Goldfarbmuren KC, Jackson ND, Sajuthi SP, Dyjack N, Li KS, Rios CL, et al. Dissecting the cellular specificity of smoking effects and reconstructing lineages in the human airway epithelium. Nat Commun. (2020) 11:2485. doi: 10.1038/s41467-020-16239-z
68. Gu X, Karp PK, Brody SL, Pierce RA, Welsh MJ, Holtzman M, et al. Chemosensory functions for pulmonary neuroendocrine cells. Am J Respir Cell Mol Biol. (2014) 50:637–46. doi: 10.1165/rcmb.2013-0199OC
69. Cutz E, Yeger H, Pan J, Ito T. Pulmonary neuroendocrine cell system in health and disease. Curr Respir Med Rev. (2008) 4:174–86. doi: 10.2174/157339808785161314
70. Young LR, Brody AS, Inge TH, Acton JD, Bokulic RE, Langston C, et al. Neuroendocrine cell distribution and frequency distinguish neuroendocrine cell hyperplasia of infancy from other pulmonary disorders. Chest. (2011) 139:1060–71. doi: 10.1378/chest.10-1304
71. Deterding RR, Pye C, Fan LL, Landgston C. Persistent tachypnea of infancy is associated with neuroendocrine cell hyperplasia. Pediatr Pulmonol. (2005) 40:157–65. doi: 10.1002/ppul.20243
72. Petty JW, Paz-Ares L. Emerging strategies for the treatment of small cell lung cancer: A review. JAMA Oncol. (2023) 9:419–29. doi: 10.1001/jamaoncol.2022.5631
73. Raso MG, Bota-Rabassedas N, Wistuba II. Pathology and classification of SCLC. Cancers. (2021) 13:820. doi: 10.3390/cancers13040820
74. Pan J, Bishop T, Ratcliffe P, Yeger H, Cutz E. Hyperplasia and hypertrophy of pulmonary neuroepithelial bodies, presumed airway hypoxia sensors, in hypoxia inducible factor prolyl hydroxylase deficient mice. Hypoxia. (2016) 4:69–80. doi: 10.2147/HP.S103957
Keywords: neuroendocrine cells, nasal hyperreactivity, nasal mucosa, upper airway epithelium, neurogenic inflammation
Citation: Wils T, Backaert W, Jacobs I, Ruysseveldt E, Cremer J, Dilissen E, Bullens DM, Talavera K, Steelant B, Van Gerven L, Martens K and Hellings PW (2024) Rare presence and function of neuroendocrine cells in the nasal mucosa. Front. Immunol. 15:1394539. doi: 10.3389/fimmu.2024.1394539
Received: 01 March 2024; Accepted: 24 July 2024;
Published: 07 August 2024.
Edited by:
Hiroshi Wakao, Dokkyo Medical University, JapanReviewed by:
Abdullah Al Tarique, The University of Queensland, AustraliaShradha Wali, McGill University Health Centre, Canada
Anna Ermund, University of Gothenburg, Sweden
Copyright © 2024 Wils, Backaert, Jacobs, Ruysseveldt, Cremer, Dilissen, Bullens, Talavera, Steelant, Van Gerven, Martens and Hellings. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peter W. Hellings, cGV0ZXIuaGVsbGluZ3NAa3VsZXV2ZW4uYmU=
 Tine Wils
Tine Wils Wout Backaert1,2
Wout Backaert1,2 Inge Jacobs
Inge Jacobs Emma Ruysseveldt
Emma Ruysseveldt Ellen Dilissen
Ellen Dilissen Dominique M. Bullens
Dominique M. Bullens Karel Talavera
Karel Talavera Brecht Steelant
Brecht Steelant Katleen Martens
Katleen Martens Peter W. Hellings
Peter W. Hellings