
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol., 05 July 2024
Sec. Cancer Immunity and Immunotherapy
Volume 15 - 2024 | https://doi.org/10.3389/fimmu.2024.1389446
This article is part of the Research TopicMicrobiota in Tumors: Is it a New Hope for Treatment?View all 5 articles
Microbiota has been closely related to human beings, whose role in tumor development has also been widely investigated. However, previous studies have mainly focused on the gut, oral, and/or skin microbiota. In recent years, the study of intratumoral microbiota has become a hot topic in tumor-concerning studies. Intratumoral microbiota plays an important role in the occurrence, development, and response to treatment of malignant tumors. In fact, increasing evidence has suggested that intratumoral microbiota is associated with malignant tumors in various ways, such as promoting the tumor development and affecting the efficacy of chemotherapy and immunotherapy. In this review, the impact of intratumoral microbiota on the immune microenvironment of malignant tumors has been analyzed, as well as its role in tumor immunotherapy, with the hope that it may contribute to the development of diagnostic tools and treatments for related tumors in the future.
Intratumoral microbiota has been confirmed to exist, including bacteria, fungi, viruses, archaea and protist, which has been widley studied as an important part of tumor microenvironment (TME) with tumor specificity (1). There are three main sources of intratumoral microbiota: (1) Microbiota in the oral cavity, intestines, and other areas, which reaches the tumor site through blood circulation; (2) microbiota in the mucosa where the tumor is located, which enters the TME through the mucosal barrier; and (3) those derived from normal tissue adjacent to the tumor (1, 2). With the development of gene sequencing technology, intratumoral microbiota could be well detected, making it possible to study its impact on tumor occurrence, development, and treatment response (3). The tumor immunotherapy refers to enhancing the anti-tumor immune response of the body by using immunological methods to activate immune cells. Due to its specificity in attacking tumor cells and limited side effects, it has been becoming one of the most promising treatment methods (4). The FDA has approved the clinical application of CTLA-4 and PD-1/PD-L1 inhibitors, and other monoclonal antibodies, for the anti-tumor immunotherapy in patients with malignant tumors. The main mechanism underlying these drugs is to activate the T cells to kill tumor cells by blocking the inhibiting signal of T cell activation, which could achieve significant effects in the clinical treatment of melanoma and non-small cell lung cancer. However, the immune checkpoint therapy has no significant effect on tumors with poor T cell infiltration and low tumor antigen mutation (such as pancreatic cancer and ovarian cancer) (5). By comparing the survival time of patients with or without antibiotic treatment, Tucker and Rini et al. have found that immunotherapy was more effective for patients receiving no antibiotic treatment. These findings indicate that the intratumoral microbiota affects the tumor immunotherapy by regulating local or systemic anti-tumor immunity (6). The microbiota and its metabolites can affect the efficacy of immunotherapy by regulating the body’s immune system. Nejman et al. have found that among patients with Melanoma who respond to immune checkpoint inhibitors, Clostridium is more abundant in the tumor, while among those who do not respond, Gardnerella vaginalis is more abundant in the tumor (1). These findings indicate that the intratumoral microbiota promotes the efficacy of anti-tumor therapy by participating in the immune response.
Studies have shown that there are a large number of microorganisms in tumor tissue, some of which may change the TME and participate in the occurrence and development of tumors. Intratumoral microbiota can promote tumor development through three pathways: (1) causing DNA damages with metabolites and secretions, leading to genomic instability; (2) participating in signal transduction in the signaling pathways; and (3) affecting the immune response (7). Escherichia coli could be divided into the A, B1, B2, and D groups, respectively. About 35% of the isolates in Group B2 have a 54kb pks island, which encodes a non-ribosomal peptide synthetase (NRPS)-polyketide synthase (PKS) hybrid gene cluster, encoding colibactin. Colibactin can cause DNA damages in mammals, thereby promoting genomic instability and inducing tumor development (8). In addition to causing DNA damages, intratumor bacteria also participate in cell signal transduction (9). Helicobacter pylori (Hp) produces cytotoxin-associated gene A(Cag A), which activates the β-catenin pathway, enters the cytoplasm of host cells and induces tumor (10). NF-κB is a transcription factor that promotes tumorigenesis (11). Dysbiosis of intratumoral microbiota can activate NF-κB, and the activation of NF-κB affects the immune response, thus promoting the occurrence of tumors (12).
In addition to promoting the occurrence of tumors, intratumoral microbiota can also promote the growth and metastasis of tumors. It has been shown that, some microorganisms can promote the proliferation of Treg cells and myeloid-derived suppressor cells(MDSCs), and promote the tumor growth (13). Ge Zhang et al. have shown that infection with Fusobacterium nucleatum(F. nucleatum) would stimulate tumor cells to release tumor-derived exosomes (TEXs), thereby promoting tumor cell metastasis in patients with colon cancer. Exosomes are extracellular vesicles secreted by cells, which can transmit proteins and genetic information between cells. TEX transfers miR-1246/92b-3p/27a-3p and CXCL16/RhoA/IL-8 from cells infected with F. nucleatum to uninfected cells, promoting tumor cell migration (14). Francesco Curcio et al. have shown that tumor cells can activate Toll-like receptor 4 (TLR4), which belongs to the pattern recognition receptors (PRRs) family. TLR4 can recognize pathogen-associated molecular patterns(PAMP) and damage-associated molecular patterns(DAMP). Some bacterial components would be PAMP, such as lipopolysaccharide (LPS), which is one of the main components of the cell wall of gram-negative bacteria (15). The expression of TLR4 is up-regulated by LPS in the cell wall of E. coli in tumors. After activation, TLR4 can induce inflammatory signals, promote tumor cell proliferation, and facilitate its escape from immune surveillance, thus promoting tumor metastasis (16). Majority of the malignant tumor patients’ death would be related to the tumor metastasis and spread. The key pathway for tumor metastasis is associated with the circulatory system, through which the cells would settle in suitable areas to proliferate. The involved factors mainly include the vascular endothelial integrity or microvascular permeability, hydrodynamic factors, microvascular geometry, cell adhesion molecules and surrounding extracellular matrix (17). Microbiota can break down the gut vascular barrier and promote the tumor cell metastasis (18). Tumor cells undergo fluid shear stress after injected into the circulatory system, which may lead to cellular apoptosis (19). The intratumoral microbiota can inhibit the activation of Actin Cytoskeleton tissue (RhoA and ROCK) in cells and reduce the contractility. The reduction of contractile force induced by bacteria is beneficial for the survival of circulating tumor cells under shear stress in the circulation, thereby enhancing the colonization of tumor cells in other sites (20). (Figure 1) Some microorganisms would promote the recruitment and activation of CD8 T cells, increase IFN-γ, TNF-α and IL-2 in serum, and promote the anti-tumor immune response (13). Compared to drugs, bacteria have motility, which can reach and proliferate at any part of tumor tissue. Most bacteria compete with tumor cells for the necessary nutrients for survival. The presence of bacteria can also activate the body’s immune system, indirectly leading to tumor cell death (21). Based on the above situation, the intratumoral microbiota plays an important role in the occurrence, development, and metastasis of tumors (Table 1). Therefore, studying the impact of intratumoral microbiota on the TME and immunotherapy is of great significance.
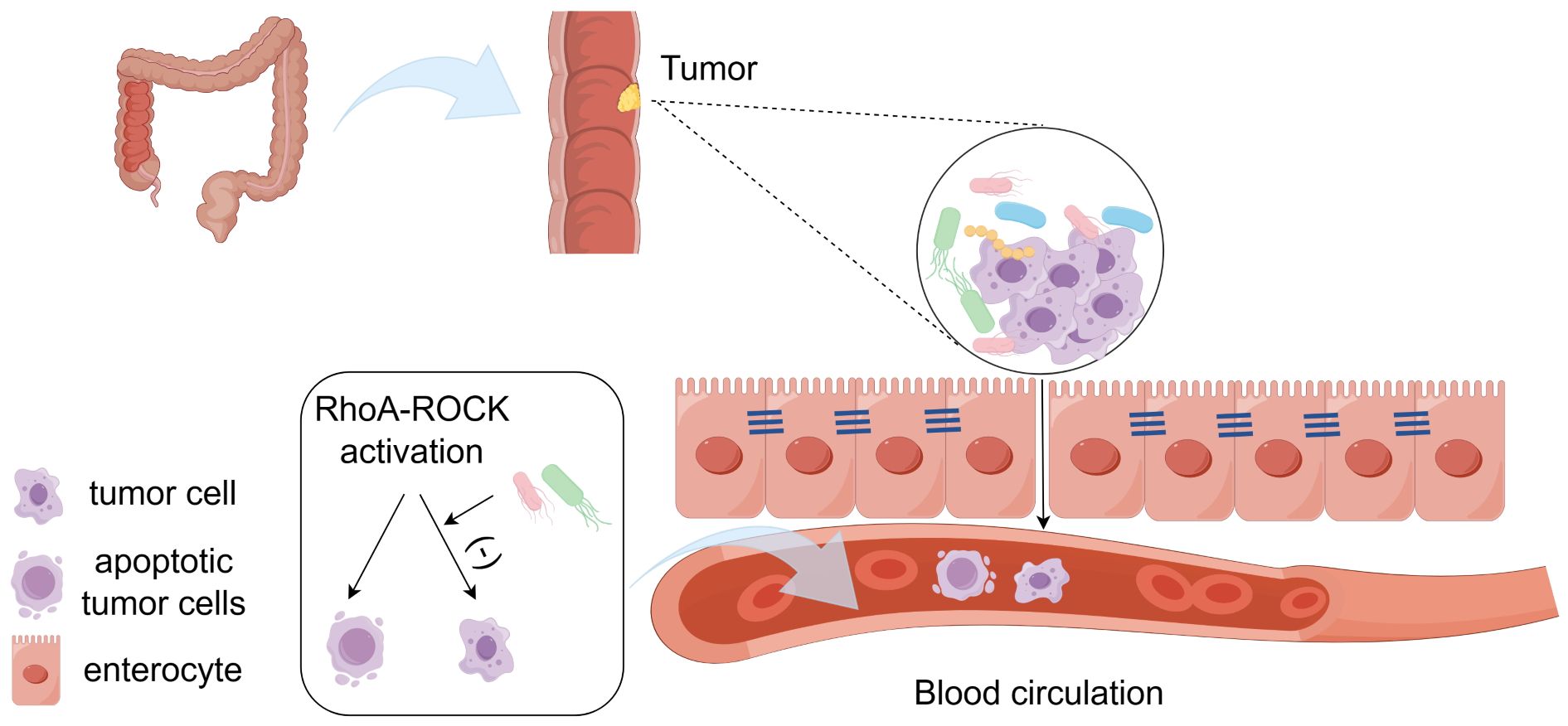
Figure 1 The intratumoral microbiota promotes tumor metastasis. Microbiota can break down the gut vascular barrier and promote the tumor metastasis. Microbiota inhibits the activation of RhoA and ROCK in cells and reduces the contractility, which is beneficial for the survival of circulating tumor cells under shear stress in the circulation. (-), it means inhibition.
The presence of intratumoral microbiota in liver has been confirmed. It has been found that the most abundant microbial species in the liver microenvironment include the Patesibacteria, Proteobacteria, Bacteroidetes, Firmicutes, and Actinobacteriota (37). The carcinogenic or inhibitory effects of intratumoral microbiota depend on their regulation of the immune system. On one hand, microbiota can lead to chronic inflammation and promote tumor development. Moreover, it can lead to immune activation, which in turn eliminates tumor cells (38). Overexpression of TLR4 in hepatocellular carcinoma contributes to the formation of immunosuppressive microenvironment. Specifically, TLR4 can recognize LPS in the cell wall of Gram-negative bacteria, stimulate the expression of CXCL1 in hepatocytes, activate the JNK/MAPK signal transduction, and promote the epithelial-mesenchymal transition (EMT) of HCC cells, which is conducive to the formation of immunosuppressive TME (39). (Figure 2) Qu et al. have found that there is no significant difference in the microbial diversity between the tumor tissue and the adjacent non-tumor tissue. However, at the family or genus level, due to the anti-tumor effect, Pseudomonas is significantly decreased in tumor tissue, while Rhizobiaceae and Agrobacterium are significantly increased in tumor tissue. Patients with relatively high abundance of Pseudomonas have better prognosis (24). On the contrary, Huang et al. have found that the α diversity of microbiota in HCC is significantly higher than the normal liver. The α diversity (single sample diversity analysis) is used to describe the diversity and abundance of bacterial species in a single sample (37, 40). There may be several reasons for the inconsistent findings from these studies: (1) The non-tumor tissue selected in the experiment are affected by the diseased liver tissue; (2) all the samples are taken from patients with malignant tumor, and there is no contrast between normal liver tissue; (3) it may be contaminated during DNA extraction, PCR amplification and/or sequencing; and (4) the sample size is insufficient. In addition to affecting immune response, intratumoral microbiota can also enhance the biosynthesis of fatty acids and lipids, thus promoting the proliferation of tumor cells (40). Combined immunotherapy based on immune checkpoint inhibitors has become the first-line treatment for advanced hepatocellular carcinoma. Considering that the intratumoral microbiota can regulate the body’s immune response, targeting the microbiota to enhance the effects of immunotherapy is worth further study.
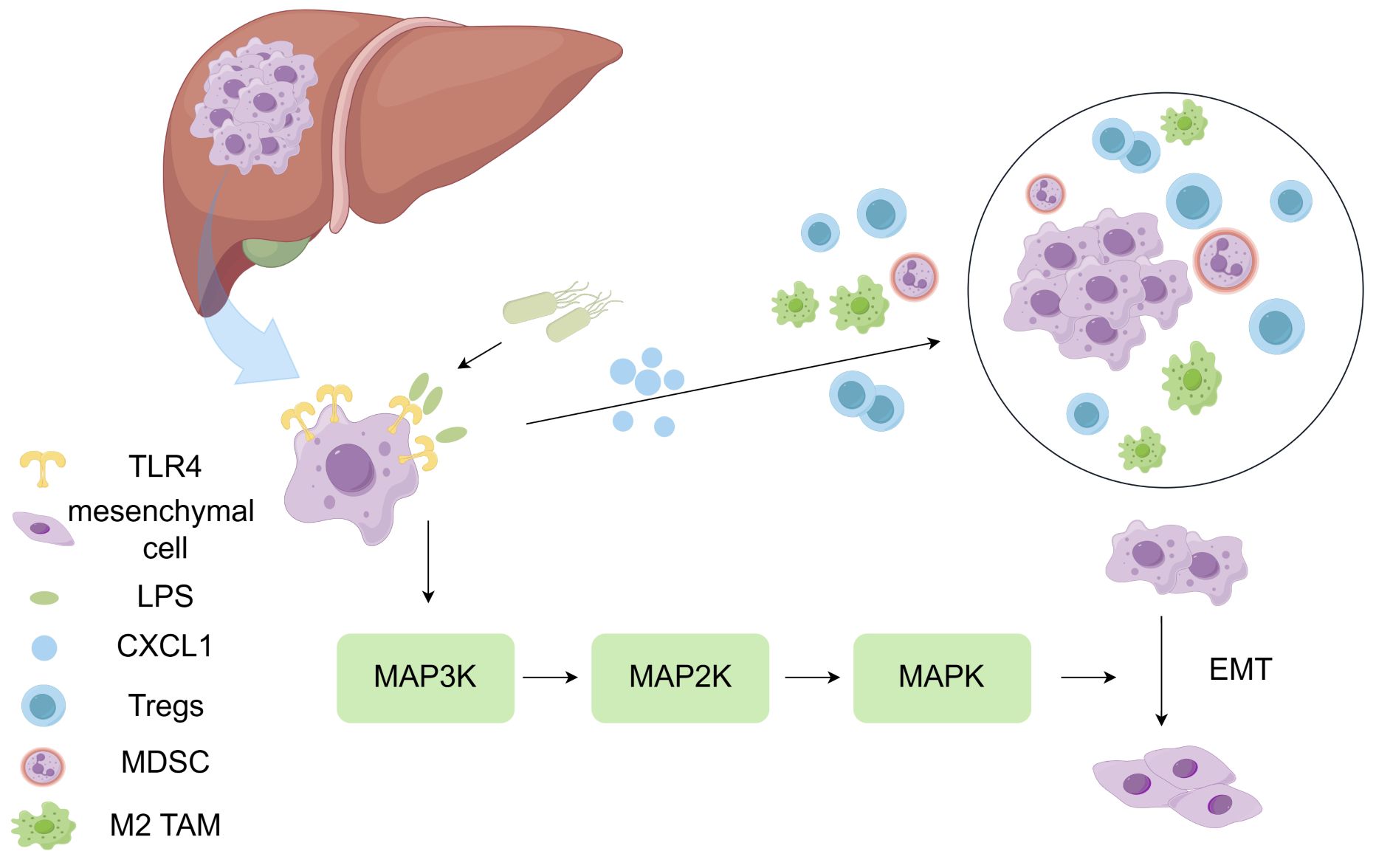
Figure 2 LPS contributes to the formation of immunosuppressive microenvironment. TLR4 can recognize LPS in the cell wall of Gram-negative bacteria, stimulate CXCL1 expression in hepatocytes, activate the JNK/MAPK signal transduction, and promote the epithelial-mesenchymal transition (EMT) of HCC cells. Overexpression of TLR4 in hepatocellular carcinoma contributes to the formation of immunosuppressive microenvironment.
Nejman et al. have found that the breast tumor tissue is rich in Staphylococcus, Lactobacillus, Enterococcus, and Streptococcus. Compared with the lung cancer, ovarian cancer, pancreatic cancer, melanoma, bone tumor and brain tumor, the diversity of microbiota in the breast cancer is increased (1). Fu et al. have used a mouse spontaneous breast tumor model, i.e., the mouse mammary tumor virus-polyoma middle T-antigen (MMTV-PyMT), which is similar to human breast cancer, to study the intratumoral microbiota. They have found that the bacteria in tumor tissue are similar to those in human beings, and the intracellular bacteria can change the activity of host cells and promote tumor progress (20). F. nucleatum can specifically bind to the abundant Gal-GalNAc on tumor cells of patients with breast cancer through the lectin Fap2 on its surface, so as to colonize breast tumors. F. nucleatum can accelerate the progress of breast cancer and tumor metastasis by inhibiting the aggregation of CD4 T cells and CD8 T cells in TME after colonization. Moreover, F. nucleatum can inhibit the anti-tumor immunity by activating two human immunesuppression checkpoint receptors (the TIGIT and CEACAM1) (25). The abundance of Yanoikuyae is sharply decreased in breast tumor tissue compared with normal breast tissue, indicating that this microorganism may play an anti-tumor role in the breast (41). Yanoikuyae expresses glycosphingolipid ligands, and then activates invariant natural killer T (iNKT) cells. The iNKT cells are CD1d-restricted T lymphocytes, which can recognize specific lipid antigens. The iNKT cells represent important medium for tumor immune monitoring which play important roles in inhibiting the growth and metastasis of breast tumors (42).
With the development of sequencing technology and increasing investigation on pancreatic cancer, the tumor microenvironment, which has previously been considered absolutely sterile, has been found to contain microorganisms (43). It has been found that the α diversity of intratumoral microbiota in long-term survivors of pancreatic cancer is higher than that of short-term survivors, and long-term survivors have characteristic microorganisms (29). Pushalkar et al. have found that Proteobacteria phylum, Bacteroides phylum, and Firmicutes phylum are the most abundant and prevalent bacterial groups in pancreatic cancer. The microbiota can induce the peritumor immunosuppression and promote the progression of pancreatic cancer. Targeting the microbiota would prevent the tumorigenesis and reverse the immune tolerance of tumor cells, thus making the checkpoint-based immunotherapy effective (27). The microbiota can generate tolerant immune response through differential activation of selected Toll-like receptors (TLRs) in monocyte. Ablation of the intratumoral microbiota can inhibit the infiltration of MDSCs, enhance the M1 macrophage differentiation, and promote the activation of CD4 T cells and CD8 T cells. It can also up-regulate the TNF-α and IFN-γ (which have anti-tumor effects), inhibiting the progression of malignant tumors (44, 45). In addition to bacteria, tumor tissue in pancreatic cancer is also rich in fungi. Berk Aykut et al. have found that fungi in gut can migrate to pancreas (46). Compared with normal pancreatic tissue, patients with pancreatic cancer have higher abundance of fungal communities in tumors, the most abundant of which are Malassezia and Alternaria (47, 48). IL-33 is a pro-inflammatory cytokine released by cancer cells. It can combine with the cognate receptor ST2 to recruit Th2 and ILC2 cells. Th2 and ILC2 cells secrete IL-4, IL-5 and IL-13 to promote tumor development. The fungi in the tumor can promote the secretion of IL33. After treatment with antifungal drugs, the fungi in the tumor would be decreased, leading to obvious regression of the tumor, and the infiltration of Th2 and ILC2 would also be decreased (48). Dominant microorganisms within tumors, such as Saccharopolyspora, Pseudoxanthomonas, and Streptomyces, are beneficial to promote the recruitment and activation of CD8 T cells, which can produce plenty of anti-tumor cytokines (such as TNF-α and IFN-γ), thereby enhancing the anti-tumor immune response (29). The intratumoral microbiota in patients with pancreatic cancer would promote the production of the IL-1β. IL-1β derived from tumor cells can activate pancreatic stellate cells (PSCs), which in turn leads to mesenchymal fibrosis, inhibiting T cell infiltration (49, 50). The Megasphaera specifically enriches and produces plenty of short-chain fatty acids in long-term survivors, which promotes the production of effector cytokines and enhances the immune response (51). The immune tolerance may lead to poor effects of immunotherapy drugs on pancreatic ductal adenocarcinoma (PDAC). Therefore, it has a positive impact on the treatment of pancreatic cancer by regulating immunotherapy through microbiota.
There are a large number of microorganisms in human intestine. The dysbiosis of gut microbiota is involved in the occurrence and development of intestinal tumors. Bacteroides fragilis (B. fragilis) has been commonly seen in human intestine. Bacteroides fragilis can be divided into two groups according to the ability to secrete Bacteroides fragilis toxin (BFT), i.e., the enterotoxigenic B. fragilis (ETBF) and nontoxigenic B. fragilis (NTBF) (52). Studies have confirmed that B. fragilis is more common in cancer patients than normal people, mainly ETBF. ETBF can promote chronic inflammation and drive the occurrence of colorectal cancer (53). It has been found that the dominant phyla in tumor tissue of colorectal cancer patients are Firmicutes phylum and Bacteroides phylum, and the secondary dominant phyla include Proteobacteria, Fusobacteria, and Actinobacteriota (54). Escherichia coli in colorectal cancer patients can migrate to the liver when the intestinal vascular barrier is disrupted, which would then activate the liver microenvironment by recruiting innate immune cells, thus promoting tumor metastasis (55). There are abundant F. nucleatum in the colorectal cancer. Bullman et al. have reported that in the patient-derived tumor xenograft (PDX) model containing Fusobacterium, treatment of antibiotics would reduce the load of Fusobacterium in tumor and inhibit the proliferation and growth of tumor cells. Based on these, we infer that F. nucleatum can affect the immune response, thus affecting the efficacy of immunotherapy (31). Microbiota can enhance T cell response in different tumors and improve the therapeutic effect of the anti-PD-L1 therapy (56). Jung Ho Kim et al. have found that among colorectal cancer patients with high microsatellite instability, patients with high content of intratumor F. nucleatum have larger tumors and are more prone to suffer from late invasion beyond the intrinsic muscle layer, compared with those with low content (without F. nucleatum) (57). However, the mechanism of the tumor immune interaction between intratumor F. nucleatum and colorectal cancer has not been fully elucidated, and further investigation is still needed. If this mechanism is clarified, F. nucleatum in TME is expected to become one of the targets of immunotherapy.
In normal lungs, the most abundant microorganisms include Firmicutes, Actinobacteriota, Bacteroidetes, and Proteobacteria, and the core microorganisms include Veillonella, Haemophilus, Neisseria, Streptococcus, Fusobacterium, and Prevotella (58). Lee et al. have found that the microorganisms significantly enriched in lung cancers are TM7 and Firmicutes at the phylum level, and Megasphaera, Selenomonas, Atopobium, and Veillonella at the genus level (59). In 2020, Lindsay et al. have grouped patients with lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) based on age and sex, and then they have studied whether there are differences in the composition of microbiota in each cohort. In LUSC, Pseudomonas putida str. KT2440 is common in young male patients, which can participate in immune regulation. Specifically, patients with higher abundance of Pseudomonas putida str. KT2440 have more neutrophils and monocytes, while CD8 T cells and macrophages are fewer. In LUAD, Escherichia coli str. K-12 substr. W3110 is related to the survival rate and genome changes of elderly females and males (60). The intratumoral microbiota of lung cancer can produce TLR ligands (such as lipopolysaccharide and peptidoglycan), stimulate myeloid cells to produce Myd88-dependent IL-1b and IL-23, induce the proliferation and activation of gamma delta (γδ) T cells, and then produce other pro-inflammatory cytokines, such as IL-17, which can promote the occurrence of inflammation and the growth of tumor cells (61).
Intratumoral microbiota not only participates in the immunomodulation of tumors mentioned above, but also plays a role in the immunotherapy of other tumors. Mackenzie J. Bender et al. have found that Lactobacillus reuteri can colonize melanoma continuously. When mice are fed on food rich in tryptophan, Lactobacillus reuteri can release indole-3-aldehyde (I3A), which can activate the aryl hydrocarbon receptor (AhR) within CD8 T cells, and enhance the therapeutic effect of immune checkpoint inhibitors (62). Bifidobacterium derived from the gastrointestinal tract can accumulate within tumors and enhance immunotherapy efficacy through STING signaling (63). To sum up, the intratumoral microbiota can regulate the immune response and affect the efficacy of immunotherapy.
Engineering bacteria refer to the microorganisms that are modified based on natural bacteria using modern biotechnology to deliver drugs, induce apoptosis of tumor cells, and cause anti-tumor immunity. Engineering bacteria can target tumors and accumulate in TME. Microorganisms have better permeability in tumor tissue than drugs, which can reach the depth of tumor tissue and kill tumor cells comprehensively (21, 64). In the absence of external force, some anaerobic or facultative anaerobic bacteria can invade or colonize solid tumors, and enter the tumors that cannot be completely penetrated by passive treatment, thereby inhibiting the tumor growth or eliminating the tumors. Some bacteria are toxic and can cause immune response in the body. After entering the solid tumor, the engineering bacteria are less susceptible to be attacked by the body’s immune system, and therefore can continue to release the anti-tumor drugs and inhibit the growth of tumor cells (65). Due to the heterogeneity of intratumoral microbiota, engineering bacteria can avoid killing healthy tissue and reduce the side effects compared with other drugs. Engineering bacteria can also alter the tumor microenvironment and enhance the host’s anti-tumor immune response by regulating immune response and/or metabolism. With the development of modern biotechnology, many engineering bacteria have been studied for the treatment of tumors. Attenuated Salmonella typhimurium has the characteristic of selective growth in tumors. Yoon et al. have genetically modified the bacterium to express IFN-γ, which has a greater cytotoxic effect and can inhibit tumor growth and prolong survival in mouse model (66). Geng et al. have bound aptamer to the surface of bacteria, and compared with unmodified bacteria, aptamer conjugated Salmonella has a stronger ability to activate tumor immune response and exert better anti-tumor efficacy (67). Sreyan Chowdhury et al. have designed a non-pathogenic Escherichia coli strain, which can colonize tumors and locally release an encoded nanobody antagonist of CD47, increasing the activation of tumor-infiltrating T cells, and generating anti-tumor immunity (68). Fernando P Canale et al. have developed an engineering probiotic Escherichia coli Nissle 1917 strain, which can colonize tumor and convert ammonia into L-arginine. The increase of L-arginine concentration in the tumor can increase the number of tumor-infiltrating T cells, thus improving the efficacy of immunotherapy. In addition, this strain can synergistically enhance the therapeutic effect of PD-L1 blockade (69, 70).
At present, the researches only characterize the intratumoral microbiota and their effects on the occurrence, development and treatment of tumors in some tumors. We made a table to summarize clinical trials related to microbiota (Table 2). In the future, the researchers need to study more types of tumors and more researches are needed to elucidate the mechanism by which intratumoral microbiota affects the efficacy of treatment, particularly the relationship between intratumoral microbiota and immune response, as this may be a key factor in regulating the efficacy of immunotherapy. In addition, it is of great significance to balance the relationship between the therapeutic effect and the inflammation caused by microbiota. Some of the intratumoral microbiota has been used in the clinical treatment without safety assessment. It is necessary to conduct more clinical researches in the future. And the composition of intratumoral microbiota in some tumors has not yet been determined. In the future, more advanced and accurate technology is needed to be invented and used for microbiota sequencing, so as to find and develop new treatments for malignant tumors by using the intratumoral microbiota.
In this paper, the role of intratumoral microbiota in the occurrence and development of tumors and their influence on the immune microenvironment of five different tumors and the immunotherapy of malignant tumors have been reviewed. Intratumoral microbiota and their products in tumors have both tumor-promoting and -inhibiting effects, depending on different cases and situations. Intratumoral microbiota can affect the immune response and serve as an emerging biomarker in immunotherapy. Antibiotic therapy that precisely targets on the intratumoral microbiota seems to be a promising treatment option, but its clinical safety and efficacy are still needed to be evaluated. The mechanisms of immune response and immunotherapy between the intratumoral microbiota and the host are still not fully understood, and more relevant researches are still needed in the future to gain in-depth understanding. Exploring changes in the composition of intratumoral microbiota to regulate its impact on immune status can help to develop new treatment methods for cancers in clinic.
JX: Writing – original draft. MCh: Writing – original draft. JLiu: Writing – original draft. MCu: Writing – original draft. BY: Writing – review & editing. JLia: Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the following projects: Projects ZR202111300475 and ZR2023MH333 supported by Shandong Provincial Natural Science Foundation.
The design of the figure was completed by Figdraw.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Nejman D, Livyatan I, Fuks G, Gavert N, Zwang Y, Geller LT, et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science. (2020) 368:973–80. doi: 10.1126/science.aay9189
2. Xie Y, Xie F, Zhou X, Zhang L, Yang B, Huang J, et al. Microbiota in tumors: from understanding to application. Adv Sci (Weinh). (2022) 9:e2200470. doi: 10.1002/advs.202200470
3. An Y, Zhang W, Liu T, Wang B, Cao H. The intratumoural microbiota in cancer: new insights from inside. Biochim Biophys Acta Rev Cancer. (2021) 1876:188626. doi: 10.1016/j.bbcan.2021.188626
4. Yang L, Ning Q, Tang SS. Recent advances and next breakthrough in immunotherapy for cancer treatment. J Immunol Res. (2022) 2022:8052212. doi: 10.1155/2022/8052212
5. Guo Y, Chen Y, Liu X, Min JJ, Tan W, Zheng JH. Targeted cancer immunotherapy with genetically engineered oncolytic Salmonella typhimurium. Cancer Lett. (2020) 469:102–10. doi: 10.1016/j.canlet.2019.10.033
6. Tucker MD, Rini BI. Predicting response to immunotherapy in metastatic renal cell carcinoma. Cancers (Basel). (2020) 12:2662. doi: 10.3390/cancers12092662
7. Chen J, Li T, Liang J, Huang Q, Huang JD, Ke Y, et al. Current status of intratumour microbiome in cancer and engineered exogenous microbiota as a promising therapeutic strategy. BioMed Pharmacother. (2022) 145:112443. doi: 10.1016/j.biopha.2021.112443
8. Barrett M, Hand CK, Shanahan F, Murphy T, O’Toole PW. Mutagenesis by microbe: the role of the microbiota in shaping the cancer genome. Trends Cancer. (2020) 6:277–87. doi: 10.1016/j.trecan.2020.01.019
9. Wong-Rolle A, Wei HK, Zhao C, Jin C. Unexpected guests in the tumor microenvironment: microbiome in cancer. Protein Cell. (2021) 12:426–35. doi: 10.1007/s13238-020-00813-8
10. Liu J, Zhang Y. Intratumor microbiome in cancer progression: current developments, challenges and future trends. biomark Res. (2022) 10:37. doi: 10.1186/s40364-022-00381-5
11. Hoesel B, Schmid JA. The complexity of NF-kappaB signaling in inflammation and cancer. Mol Cancer. (2013) 12:86. doi: 10.1186/1476-4598-12-86
12. Elinav E, Nowarski R, Thaiss CA, Hu B, Jin C, Flavell RA. Inflammation-induced cancer: crosstalk between tumours, immune cells and microorganisms. Nat Rev Cancer. (2013) 13:759–71. doi: 10.1038/nrc3611
13. Chen Y, Liu B, Wei Y, Kuang DM. Influence of gut and intratumoral microbiota on the immune microenvironment and anti-cancer therapy. Pharmacol Res. (2021) 174:105966. doi: 10.1016/j.phrs.2021.105966
14. Guo S, Chen J, Chen F, Zeng Q, Liu WL, Zhang G. Exosomes derived from Fusobacterium nucleatum-infected colorectal cancer cells facilitate tumour metastasis by selectively carrying miR-1246/92b-3p/27a-3p and CXCL16. Gut. (2020) 71(2):e1–3. doi: 10.1136/gutjnl-2020-321187
15. Zindel J, Kubes P. DAMPs, PAMPs, and LAMPs in immunity and sterile inflammation. Annu Rev Pathol. (2020) 15:493–518. doi: 10.1146/annurev-pathmechdis-012419-032847
16. Domenis R, Cifu A, Marino D, Fabris M, Niazi KR, Soon-Shiong P, et al. Toll-like receptor-4 activation boosts the immunosuppressive properties of tumor cells-derived exosomes. Sci Rep. (2019) 9:8457. doi: 10.1038/s41598-019-44949-y
17. Fu BM. Tumor metastasis in the microcirculation. Adv Exp Med Biol. (2018) 1097:201–18. doi: 10.1007/978-3-319-96445-4
18. Murota Y, Jobin C. Bacteria break barrier to promote metastasis. Cancer Cell. (2021) 39:598–600. doi: 10.1016/j.ccell.2021.03.009
19. Mitchell MJ, King MR. Fluid shear stress sensitizes cancer cells to receptor-mediated apoptosis via trimeric death receptors. New J Phys. (2013) 15:015008. doi: 10.1088/1367-2630/15/1/015008
20. Fu A, Yao B, Dong T, Chen Y, Yao J, Liu Y, et al. Tumor-resident intracellular microbiota promotes metastatic colonization in breast cancer. Cell. (2022) 185:1356–1372 e1326. doi: 10.1016/j.cell.2022.02.027
21. Zheng JH, Min JJ. Targeted cancer therapy using engineered salmonella typhimurium. Chonnam Med J. (2016) 52:173–84. doi: 10.4068/cmj.2016.52.3.173
22. Sun L, Ke X, Guan A, Jin B, Qu J, Wang Y, et al. Intratumoural microbiome can predict the prognosis of hepatocellular carcinoma after surgery. Clin Transl Med. (2023) 13:e1331. doi: 10.1002/ctm2.1331
23. Xue C, Gu X, Shi Q, Ma X, Jia J, Su Y, et al. The interaction between intratumoral bacteria and metabolic distortion in hepatocellular carcinoma. J Transl Med. (2024) 22:237. doi: 10.1186/s12967-024-05036-7
24. Qu D, Wang Y, Xia Q, Chang J, Jiang X, Zhang H. Intratumoral microbiome of human primary liver cancer. Hepatol Commun. (2022) 6:1741–52. doi: 10.1002/hep4.1908
25. Parhi L, Alon-Maimon T, Sol A, Nejman D, Shhadeh A, Fainsod-Levi T, et al. Breast cancer colonization by Fusobacterium nucleatum accelerates tumor growth and metastatic progression. Nat Commun. (2020) 11:3259. doi: 10.1038/s41467-020-16967-2
26. Parida S, Wu S, Siddharth S, Wang G, Muniraj N, Nagalingam A, et al. A procarcinogenic colon microbe promotes breast tumorigenesis and metastatic progression and concomitantly activates notch and beta-catenin axes. Cancer Discovery. (2021) 11:1138–57. doi: 10.1158/2159-8290.CD-20-0537
27. Pushalkar S, Hundeyin M, Daley D, Zambirinis CP, Kurz E, Mishra A, et al. The pancreatic cancer microbiome promotes oncogenesis by induction of innate and adaptive immune suppression. Cancer Discovery. (2018) 8:403–16. doi: 10.1158/2159-8290.CD-17-1134
28. Guo W, Zhang Y, Guo S, Mei Z, Liao H, Dong H, et al. Tumor microbiome contributes to an aggressive phenotype in the basal-like subtype of pancreatic cancer. Commun Biol. (2021) 4:1019. doi: 10.1038/s42003-021-02557-5
29. Riquelme E, Zhang Y, Zhang L, Montiel M, Zoltan M, Dong W, et al. Tumor microbiome diversity and composition influence pancreatic cancer outcomes. Cell. (2019) 178:795–806 e712. doi: 10.1016/j.cell.2019.07.008
30. Geller LT, Barzily-Rokni M, Danino T, Jonas OH, Shental N, Nejman D, et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science. (2017) 357:1156–60. doi: 10.1126/science.aah5043
31. Bullman S, Pedamallu CS, Sicinska E, Clancy TE, Zhang X, Cai D, et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science. (2017) 358:1443–8. doi: 10.1126/science.aal5240
32. Zhang J, Wang P, Wang J, Wei X, Wang M. Unveiling intratumoral microbiota: An emerging force for colorectal cancer diagnosis and therapy. Pharmacol Res. (2024) 203:107185. doi: 10.1016/j.phrs.2024.107185
33. Sikavi DR, Nguyen LH, Haruki K, Ugai T, Ma W, Wang DD, et al. The sulfur microbial diet and risk of colorectal cancer by molecular subtypes and intratumoral microbial species in adult men. Clin Transl Gastroenterol. (2021) 12:e00338. doi: 10.14309/ctg.0000000000000338
34. Liu NN, Yi CX, Wei LQ, Zhou JA, Jiang T, Hu CC, et al. The intratumor mycobiome promotes lung cancer progression via myeloid-derived suppressor cells. Cancer Cell. (2023) 41:1927–1944 e1929. doi: 10.1016/j.ccell.2023.08.012
35. Ma Y, Chen H, Li H, Zheng M, Zuo X, Wang W, et al. Intratumor microbiome-derived butyrate promotes lung cancer metastasis. Cell Rep Med. (2024) 5:101488. doi: 10.1016/j.xcrm.2024.101488
36. Peters BA, Hayes RB, Goparaju C, Reid C, Pass HI, Ahn J. The microbiome in lung cancer tissue and recurrence-free survival. Cancer Epidemiol Biomarkers Prev. (2019) 28:731–40. doi: 10.1158/1055-9965.EPI-18-0966
37. Huang JH, Wang J, Chai XQ, Li ZC, Jiang YH, Li J, et al. The intratumoral bacterial metataxonomic signature of hepatocellular carcinoma. Microbiol Spectr. (2022) 10:e0098322. doi: 10.1128/spectrum.00983-22
38. Chakladar J, Wong LM, Kuo SZ, Li WT, Yu MA, Chang EY, et al. The liver microbiome is implicated in cancer prognosis and modulated by alcohol and hepatitis B. Cancers (Basel). (2020) 12:1632. doi: 10.3390/cancers12061642
39. Chen C, Wang Z, Ding Y, Qin Y. Tumor microenvironment-mediated immune evasion in hepatocellular carcinoma. Front Immunol. (2023) 14:1133308. doi: 10.3389/fimmu.2023.1133308
40. He Y, Zhang Q, Yu X, Zhang S, Guo W. Overview of microbial profiles in human hepatocellular carcinoma and adjacent nontumor tissues. J Transl Med. (2023) 21:68. doi: 10.1186/s12967-023-03938-6
41. Gong Y, Huang X, Wang M, Liang X. Intratumor microbiota: a novel tumor component. J Cancer Res Clin Oncol. (2023) 149:6675–91. doi: 10.1007/s00432-023-04576-7
42. Terabe M, Berzofsky JA. NKT cells in immunoregulation of tumor immunity: a new immunoregulatory axis. Trends Immunol. (2007) 28:491–6. doi: 10.1016/j.it.2007.05.008
43. Guan SW, Lin Q, Yu HB. Intratumour microbiome of pancreatic cancer. World J Gastrointest Oncol. (2023) 15:713–30. doi: 10.4251/wjgo.v15.i5.713
44. Yu ZK, Xie RL, You R, Liu YP, Chen XY, Chen MY, et al. The role of the bacterial microbiome in the treatment of cancer. BMC Cancer. (2021) 21:934. doi: 10.1186/s12885-021-08664-0
45. Wei X, Mei C, Li X, Xie Y. The unique microbiome and immunity in pancreatic cancer. Pancreas. (2021) 50:119–29. doi: 10.1097/MPA.0000000000001744
46. Aykut B, Pushalkar S, Chen R, Li Q, Abengozar R, Kim JI, et al. The fungal mycobiome promotes pancreatic oncogenesis via activation of MBL. Nature. (2019) 574:264–7. doi: 10.1038/s41586-019-1608-2
47. Xue C, Chu Q, Zheng Q, Yuan X, Su Y, Bao Z, et al. Current understanding of the intratumoral microbiome in various tumors. Cell Rep Med. (2023) 4:100884. doi: 10.1016/j.xcrm.2022.100884
48. Li X, Saxena D. The mycobiome-immune axis: The next frontier in pancreatic cancer. Cancer Cell. (2022) 40:120–2. doi: 10.1016/j.ccell.2022.01.009
49. Zhang Z, Liao Y, Tang D. Intratumoral microbiota: new frontiers in tumor immunity. Carcinogenesis. (2022) 43:719–27. doi: 10.1093/carcin/bgac063
50. Das S, Shapiro B, Vucic EA, Vogt S, Bar-Sagi D. Tumor cell-derived IL1beta promotes desmoplasia and immune suppression in pancreatic cancer. Cancer Res. (2020) 80:1088–101. doi: 10.1158/0008-5472.CAN-19-2080
51. Huang Y, Zhu N, Zheng X, Liu Y, Lu H, Yin X, et al. Intratumor microbiome analysis identifies positive association between megasphaera and survival of chinese patients with pancreatic ductal adenocarcinomas. Front Immunol. (2022) 13:785422. doi: 10.3389/fimmu.2022.785422
52. Sears CL, Geis AL, Housseau F. Bacteroides fragilis subverts mucosal biology: from symbiont to colon carcinogenesis. J Clin Invest. (2014) 124:4166–72. doi: 10.1172/JCI72334
53. Gagniere J, Raisch J, Veziant J, Barnich N, Bonnet R, Buc E, et al. Gut microbiota imbalance and colorectal cancer. World J Gastroenterol. (2016) 22:501–18. doi: 10.3748/wjg.v22.i2.501
54. Gao Z, Guo B, Gao R, Zhu Q, Qin H. Microbiota disbiosis is associated with colorectal cancer. Front Microbiol. (2015) 6:20. doi: 10.3389/fmicb.2015.00020
55. Bertocchi A, Carloni S, Ravenda PS, Bertalot G, Spadoni I, Lo Cascio A, et al. Gut vascular barrier impairment leads to intestinal bacteria dissemination and colorectal cancer metastasis to liver. Cancer Cell. (2021) 39:708–724 e711. doi: 10.1016/j.ccell.2021.03.004
56. Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre ML, et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science. (2018) 359:104–8. doi: 10.1126/science.aao3290
57. Lee JA, Yoo SY, Oh HJ, Jeong S, Cho NY, Kang GH, et al. Differential immune microenvironmental features of microsatellite-unstable colorectal cancers according to Fusobacterium nucleatum status. Cancer Immunol Immunother. (2021) 70:47–59. doi: 10.1007/s00262-020-02657-x
58. Gao F, Yu B, Rao B, Sun Y, Yu J, Wang D, et al. The effect of the intratumoral microbiome on tumor occurrence, progression, prognosis and treatment. Front Immunol. (2022) 13:1051987. doi: 10.3389/fimmu.2022.1051987
59. Lee SH, Sung JY, Yong D, Chun J, Kim SY, Song JH, et al. Characterization of microbiome in bronchoalveolar lavage fluid of patients with lung cancer comparing with benign mass like lesions. Lung Cancer. (2016) 102:89–95. doi: 10.1016/j.lungcan.2016.10.016
60. Wong LM, Shende N, Li WT, Castaneda G, Apostol L, Chang EY, et al. Comparative analysis of age- and gender-associated microbiome in lung adenocarcinoma and lung squamous cell carcinoma. Cancers (Basel). (2020) 12:1447. doi: 10.3390/cancers12061447
61. Jin C, Lagoudas GK, Zhao C, Bullman S, Bhutkar A, Hu B, et al. Commensal Microbiota Promote Lung Cancer Development via gammadelta T Cells. Cell. (2019) 176:998–1013 e1016. doi: 10.1016/j.cell.2018.12.040
62. Bender MJ, McPherson AC, Phelps CM, Pandey SP, Laughlin CR, Shapira JH, et al. Dietary tryptophan metabolite released by intratumoral Lactobacillus reuteri facilitates immune checkpoint inhibitor treatment. Cell. (2023) 186:1846–1862 e1826. doi: 10.1016/j.cell.2023.03.011
63. Shi Y, Zheng W, Yang K, Harris KG, Ni K, Xue L, et al. Intratumoral accumulation of gut microbiota facilitates CD47-based immunotherapy via STING signaling. J Exp Med. (2020) 217(5):e20192282. doi: 10.1084/jem.20192282
64. Gurbatri CR, Lia I, Vincent R, Coker C, Castro S, Treuting PM, et al. Engineered probiotics for local tumor delivery of checkpoint blockade nanobodies. Sci Transl Med. (2020) 12(530):eaax0876. doi: 10.1126/scitranslmed.aax0876
65. Yang M, Yang F, Chen W, Liu S, Qiu L, Chen J. Bacteria-mediated cancer therapies: opportunities and challenges. Biomater Sci. (2021) 9:5732–44. doi: 10.1039/D1BM00634G
66. Yoon W, Park YC, Kim J, Chae YS, Byeon JH, Min SH, et al. Application of genetically engineered Salmonella typhimurium for interferon-gamma-induced therapy against melanoma. Eur J Cancer. (2017) 70:48–61. doi: 10.1016/j.ejca.2016.10.010
67. Geng Z, Cao Z, Liu R, Liu K, Liu J, Tan W. Aptamer-assisted tumor localization of bacteria for enhanced biotherapy. Nat Commun. (2021) 12:6584. doi: 10.1038/s41467-021-26956-8
68. Chowdhury S, Castro S, Coker C, Hinchliffe TE, Arpaia N, Danino T. Programmable bacteria induce durable tumor regression and systemic antitumor immunity. Nat Med. (2019) 25:1057–63. doi: 10.1038/s41591-019-0498-z
69. Canale FP, Basso C, Antonini G, Perotti M, Li N, Sokolovska A, et al. et al: Metabolic modulation of tumours with engineered bacteria for immunotherapy. Nature. (2021) 598:662–6. doi: 10.1038/s41586-021-04003-2
Keywords: intratumoral microbiota, tumor immune microenvironment, immunosuppressive microenvironment, immunotherapy, engineering bacteria
Citation: Xu J, Cheng M, Liu J, Cui M, Yin B and Liang J (2024) Research progress on the impact of intratumoral microbiota on the immune microenvironment of malignant tumors and its role in immunotherapy. Front. Immunol. 15:1389446. doi: 10.3389/fimmu.2024.1389446
Received: 22 February 2024; Accepted: 21 June 2024;
Published: 05 July 2024.
Edited by:
Elena Niccolai, University of Florence, ItalyCopyright © 2024 Xu, Cheng, Liu, Cui, Yin and Liang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Beibei Yin, Y3J5c3RhbHJlZDgzQDE2My5jb20=; Jing Liang, bGlhbmdqaW5nMDUzMUAxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.