
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 13 May 2024
Sec. Viral Immunology
Volume 15 - 2024 | https://doi.org/10.3389/fimmu.2024.1382655
This article is part of the Research Topic Regulation of the Host Immune Activation in Respiratory Virus Infections View all 16 articles
 Cameron R. Bishop1
Cameron R. Bishop1 Kexin Yan1
Kexin Yan1 Wilson Nguyen1
Wilson Nguyen1 Daniel J. Rawle1
Daniel J. Rawle1 Bing Tang1
Bing Tang1 Thibaut Larcher2
Thibaut Larcher2 Andreas Suhrbier1,3*
Andreas Suhrbier1,3*Introduction: Global microplastic (MP) pollution is now well recognized, with humans and animals consuming and inhaling MPs on a daily basis, with a growing body of concern surrounding the potential impacts on human health.
Methods: Using a mouse model of mild COVID-19, we describe herein the effects of azide-free 1 μm polystyrene MP beads, co-delivered into lungs with a SARS-CoV-2 omicron BA.5 inoculum. The effect of MPs on the host response to SARS-CoV-2 infection was analysed using histopathology and RNA-Seq at 2 and 6 days post-infection (dpi).
Results: Although infection reduced clearance of MPs from the lung, virus titres and viral RNA levels were not significantly affected by MPs, and overt MP-associated clinical or histopathological changes were not observed. However, RNA-Seq of infected lungs revealed that MP exposure suppressed innate immune responses at 2 dpi and increased pro-inflammatory signatures at 6 dpi. The cytokine profile at 6 dpi showed a significant correlation with the ‘cytokine release syndrome’ signature observed in some COVID-19 patients.
Discussion: The findings are consistent with the recent finding that MPs can inhibit phagocytosis of apoptotic cells via binding of Tim4. They also add to a growing body of literature suggesting that MPs can dysregulate inflammatory processes in specific disease settings.
Global plastic production has grown exponentially, nearly 475 million tons were produced in 2021, and this is set to climb to ≈ 550 million tons in 2026. The latter part of the Anthropocene can now be referred to as the Plasticene (1), with plastic debris providing a new ecological niche known as the plastisphere (2). Plastics degrade into microplastics (MPs), with MP contamination of our environment recognized with mounting concerns (3, 4), especially as chemical breakdown is very slow and biodegradation pathways are limited (5). Human MP exposure arises from both from use of items manufactured from plastic, and as a result of poor disposal of plastic waste that results in plastic pollution (6–8). Less clear are the human health implications of ingestion and inhalation of MPs by general populations, with considerable speculation available (9–12), but a noteworthy paucity of compelling direct evidence for detrimental human clinical outcomes outside heavily contaminated industrial settings. Nevertheless, some compelling clinical data is emerging. For instance, patients with carotid artery plaques in which MPs were detected had a higher risk of a composite of myocardial infarction, stroke, or death (13). A positive correlation has also been reported between faecal MP concentration and the severity of inflammatory bowel disease (14). More MPs were present in the lungs of pediatric patients with severe community acquired pneumonia when compared with those with non-severe community acquired pneumonia (15). MPs were found in cirrhotic liver tissue, but not in liver samples from individuals without underlying liver disease (16). Unfortunately, whether the different MP levels identified in these studies were causative or a consequence of the disease, often remains unclear. More robust evidence for causation is seen for occupational diseases in workers from synthetic textile, flock and (poly)vinyl chloride industries, following exposure to chronic high doses of airborne MPs (17, 18). For instance, chronic interstitial pneumonitis and breathing difficulties have been associated with workplace exposure to nylon flock (19, 20).
Even outside heavily contaminated industrial settings, humans are exposed to airborne MPs at home, at the office and outdoors (21–23), with widespread reports of MPs found in human lungs (15, 24–27) and sputum (28). Estimates from several studies for MP inhalation in the home were 0.3-2.5 µg/kg/d (29). Using indoor dust measurements from 12 countries, the median MP intake was calculated to be 0.36-150 µg/kg/d, with adult intake 10-fold lower than infants (30). An estimated MP inhalation of 6.5-8.97 µg/kg/day is reported to be derived from human exposure models (31–33), although where this data came from or how it was derived is unclear.
Rodent studies seeking to assess the effects of MP inhalation, introduce MPs into the lungs via intranasal or intratracheal inoculation. Unfortunately, as for many such studies on MP exposure, the doses used were often unrealistically high (34). Doses were, for instance, 40 mg/kg/d for 21 days of carboxy-modified 1-5 and 10-20 µm polystyrene beads (35), 5 mg/kg/d for 2 weeks of three types of MPs (33), 5 mg/kg 3 times per week of 100 nm amino-modified polystyrene beads (36), 4 mg/kg (assuming 25 g mouse) 5 µm and 99 nm polystyrene beads every other day for five weeks (37), or 1.25 and 6.25 mg/kg 3 times per week of 5 µm polystyrene beads for three weeks (38). Such doses are 1 to 2 orders of magnitude higher than even the highest estimates for humans. Another serious confounding issue is the potential presence of highly toxic preservatives such as azide, which are commonly added to commercial polystyrene bead products (39). Such chemicals would provide both acute toxicity if the beads are not washed, and would also slowly leach out of the MPs in vivo, imbuing the MPs with artificial toxicity. Azide inhibits mitochondrial cytochrome C oxidase (the terminal complex of eukaryotic oxidative phosphorylation) and catalase. Catalase protects cells against oxidative damage by reactive oxygen species (ROS). ROS associated toxicity is frequently reported for MP exposure (40, 41) and may thus simply represent an azide artefact.
An important recent observation is that MPs can be recognized by Tim4, a receptor that plays an essential role in binding and phagocytosing apoptotic cells (a process known as efferocytosis), with MPs able to inhibit efferocytosis by macrophages in vitro (42). Azide free, 0.8 µm beads (Sigma-Aldrich, LB8) were used for many of the experiments, with no toxicity or induction of inflammation observed for macrophages treated with MPs in vitro (42). Efferocytosis promotes anti-inflammatory activities and inflammation resolution, and efferocytosis failure or efferocytosis defects can result in apoptotic cells undergoing secondary necrosis, which is generally pro-inflammatory in vivo (43, 44).
We have previously shown that consumption of azide-free 1 µm polystyrene beads had minimal overt effects by themselves, but after infection with the arthritogenic chikungunya virus, the presence of MPs in the digestive track resulted in a significant prolongation of the ensuing viral inflammatory arthritis (45). Given the SARS-CoV-2 pandemic and the aforementioned estimates on human MP inhalation, we examined herein whether the presence of MPs in the lung would affect the outcome of SARS-CoV-2 infection and COVID-19 disease in a mild non-lethal transgenic mouse model. The transgenic mACE2-hACE2 mouse uses the mouse angiotensin converting enzyme 2 (mACE2) promoter to drive expression of the virus receptor, human angiotensin converting enzyme 2 (hACE2) (46). The inflammatory response and the cytokine signatures associated with COVID-19 are well described (47, 48), and are largely recapitulated in mouse models (46, 49, 50), although these studies used earlier variants of concern. Herein we used a recent SARS-CoV-2 variant of concern, omicron BA.5 (51), with omicron variants currently the dominate SARS-CoV-2 viruses infecting human populations (52). Using this mouse model system and RNA-Seq, we illustrate that a single co-inoculation of MPs and virus into the lungs dysregulated the innate inflammatory response to SARS-CoV-2, initially at 2 days post infection (dpi) suppressing the innate inflammatory responses and later at 6 dpi moving the cytokine response profile towards a “cytokine release syndrome” signature.
All mouse work was conducted in accordance with the “Australian code for the care and use of animals for scientific purposes” as defined by the National Health and Medical Research Council of Australia. Mouse work was approved by the QIMR Berghofer Medical Research Institute animal ethics committee (P3600), with infectious SARS-CoV-2 work conducted in a PC3 (Bio-Safety Level 3) facility at the QIMR Berghofer MRI (Australian Department of Agriculture, Water and the Environment certification Q2326 and Office of the Gene Technology Regulator certification 3445). Breeding and use of GM mice was approved under a Notifiable Low Risk Dealing (NLRD) Identifier: NLRD_Suhrbier_Oct2020: NLRD 1.1(a). Mice were euthanized using carbon dioxide.
Collection of nasal swabs from consented COVID-19 patients (to isolate circulating SARS-CoV-2 variants) was approved by the QIMR Berghofer Medical Research Institute Human Research Ethics Committee (P3600).
The omicron BA.5 isolate, SARS-CoV-2QIMR03 (SARS-CoV-2/human/AUS/QIMR03/2022) belongs to the BE.1 sublineage (GenBank: OP604184.1) and was isolated from a nasal swab, voluntarily collected and donated by a de-identified, consented, adult COVID-19 patient with degree level education (53). Virus stocks were propagated in Vero E6 cells, stocks and tissue culture supernatants were checked for endotoxin (54, 55) and mycoplasma (MycoAlert, Lonza) (56). Virus titres were determined by CCID50 assays (57).
mACE2-hACE2 mice (58) were generated as described (46) by Monash Genome Modification Platform (MGMP), Monash University and are freely available as heterozygotes through Phenomics Australia (MGMP code ET26). The strain was initially maintained in-house as heterozygotes by backcrossing to C57BL/6J mice. Heterozygotes were then inter-crossed to generate a homozygous mACE2-hACE2 transgenic mouse line. Genotyping was undertaken by digital droplet PCR (MGMP) to distinguish homozygotes from heterozygotes; hACE2 primers 5′-CCAGATGTACCCTCTGCAAG-3′/5′-TCGTGTTCAGGATGGTGTTC-3′, probe 6-carboxyfluorescein-5′-GCTCCAGCTGCAGGCTCTCCAGCA-3′-ZEN/IowaBlack; RPP30 reference primers CTTTGAACTTGTCTATGGTCCT/GCATCAAATTGAGGGCATTG, probe hexachlorofluorescein-TGTGTACCTTCTCATCGTTGCATC-ZEN/IowaBlack. A Bio-Rad QX200 ddPCR droplet generator was used to generate droplets, amplified products were analysed by QX200 droplet reader, and copy numbers were determined using QuantaSoft Analysis Pro Version 1.0 (Bio-Rad, USA). After 2 inter-crossing of homozygotes, all offspring were homozygotes, and a homozygous line was established.
Mice were infected as described (59), briefly, female mice (≈ 10-20 weeks of age) received intrapulmonary infections delivered via the intranasal route with 5×104 CCID50 of virus in 50 μl RPMI 1640, while under light anaesthesia. Each group of mice within an experiment had a similar age range and distribution, with the mean age for each group not differing by more than 1 week. Mice were weighed and overt disease symptoms scored as described (53). Mice were euthanized using CO2, and tissue titres determined using CCID50 assays and Vero E6 cells (57).
The MPs comprised internally dye loaded, Fluoresbrite® yellow-green polystyrene-based microspheres with a diameter of 1 µm (Cat# 17154-10) purchased from Polysciences as 2.5% w/v in a sterile aqueous suspension without sodium azide. The zeta potential of ≈ 1 µm polystyrene beads has been estimated to be ≈ -20 mV (60). The rationale for choosing these MPs has been described previously (45); briefly, polystyrene MPs are frequently found in the environment, surface labelled microspheres have altered surface characteristics not recapitulate by MPs in the environment (and were thus not used), and the 1 μm size is approximately the size of a bacteria, with bacteria routinely phagocytosed by macrophages. MPs were diluted in PBS and administered alone or together with the viral inoculum in a single dose of 1 µg of MPs per mouse (≈ 40 µg/kg); the total inoculated volume was always 50 µl per mouse.
Lungs were fixed in 10% formalin for 2-3 days, tapped dry and embedded in O.C.T. (Tissue-Tek, Qiagen), with ≈ 7 µm cryosections, under a glass coverslip, viewed by fluorescent microscopy. MPs were counted by eye.
Lungs were weighed, manually chopped using scissors and digested in ammonium sulphate (50 mM), SDS (5 mg/ml) and proteinase K (1 mg/ml) overnight at 37°C as described (61). Digested suspensions were viewed by fluorescent microscopy and a haemocytometer, with fluorescent MPs counted by eye.
Histology was undertaken as described (59, 62), with lungs fixed in formalin, embedded in paraffin, sections stained by H&E and slides scanned by Aperio AT Turbo (Aperio, Vista, CA, USA). Image analysis (nuclear/cytoplasmic staining ratios) was undertaken using Positive Pixel Count v9 algorithm. White space analysis was undertaken using QuPath v0.2.3.
Immunohistochemistry (IHC) was undertaken as described (63) using the macrophage/monocyte monoclonal antibody F4/80 (Abcam, Cambridge, MA) and color developed using NovaRed (Vector Laboratories, Newark, CA, USA).
RNA-Seq and bioinformatic analyses were undertaken as described (46, 59). Briefly, mouse lung tissues were harvested into RNAlater, RNA was extracted using TRIzol (Life Technologies), and RNA concentration and quality measured using TapeStation D1kTapeScreen assay (Agilent). cDNA libraries were generated using Illumina TruSeq Stranded mRNA library prep kit and sequencing performed in-house using Illumina Nextseq 2000 platform (75-base paired end reads). Processed reads were aligned to GRCm39 vM31 (mouse genome) and the BA.5 genome using STAR aligner. Gene expression was calculated using RSEM and EdgeR. For the BA.5+MP vs. BA.5 data sets a term for viral reads was introduced into EdgeR to minimize the effects of viral loads on significance and fold change in the mRNA expression data. A filter was applied to the count matrix of counts per million (cpm) >1 for any given gene in at least 5 samples.
Differentially expressed genes (DEGs) were analysed using Ingenuity Pathway Analysis (IPA) (QIAGEN). Whole gene lists ranked by fold change were interrogated using Gene Set Enrichment Analyses (GSEA v4.0.3) (Broad Institute, UCSanDiego) using the “GSEAPreranked” module. Gene sets were obtained from the complete Molecular Signatures Database (MSigDB) v7.2 (31,120 gene sets) (msigdb.v7.2.symbols.gmt), Blood Transcription modules (BTMs) (64), and Xue at al., 2014 (65). Relative abundance of specific cell types was estimated via cellular deconvolution using the SpatialDecon package in R (66), with cell-type expression matrices obtained from Yoshida et al., 2019 (67) or the NanoString Cell Profile Library, either Mouse/Adult/Lung_MCA (Mouse cell atlas) or Mouse/Adult/ImmuneAtlas_ImmGen_cellfamily (Immune cell family) (available at https://github.com/Nanostring-Biostats/CellProfileLibrary/tree/master/Mouse/Adult).
Kraken metagenomic sequence classification was undertaken as described (68). The Interferome database (69) was used to identify interferon regulated genes among DEG sets.
Total RNA was used to synthesize cDNA using ProtoScript II First Strand cDNA Synthesis Kit (New England Biolabs) and qPCR performed using iTaq Universal SYBR Green Supermix (Bio-Rad) as per manufacturer’s instructions with primers Nr4a1 (Forward 5’-GTTGGGGGAGTGTGCTAGA-3’ and Reverse 5’-AATACAGGGCATCTCCAGCC-3’), Ccn1 (Forward 5’-AAGAGGCTTCCTGTCTTTGGC-3’ and Reverse 5’- AACTCGTGTGGAGATGCCAG-3’), Hspa1a (Forward 5’-TTTGTGTATTGCACGTGGGC-3’ and Reverse 5’-CCAGGGGAGAGTCCAAACAC-3’), Hspa1b (Forward 5’- AATGTTGGGAGCAGCACTGT-3’ and Reverse 5’-TGTCTTCCCAGGCTACTGGA-3’), Oas3 (Forward 5’-TGGCAATCCCATCAAGCCAT-3’ and Reverse 5’- CTGAGGGCTGGTGTCACTTT-3’), Irf7 (Forward 5’-ACCGTGTTTACGAGGAACCC-3’ and Reverse 5’-GTTCTTACTGCTGGGGCCAT-3’), Ccl4 (Forward 5’- GCCAGCTGTGGTATTCCTGA-3’ and Reverse 5’-TGAACGTGAGGAGCAAGGAC-3’). qPCR reactions were performed in duplicate and averaged. Gene expression was normalised using mRPL13a as the house-keeping gene (70). Log2 fold-change was calculated using the 2-ΔΔCt method (71).
Statistical analyses of experimental data were performed using IBM SPSS Statistics for Windows, Version 19.0 (IBM Corp., Armonk, NY, USA). The t-test was used when the difference in variances were <4 fold (determined using Data Analysis ToolPak in Excel), skewness was >-2 and kurtosis was <2 (determined using SPSS). Otherwise, the non-parametric Kolmogorov-Smirnov asymptotic test was used (SPSS).
Experimental delivery of MPs into mouse lungs generally involves delivery of MPs in suspension into the lungs via the intranasal (i.n.) route (33, 35–38), as mimicking MP dust inhalation is currently technically, logistically and ethical difficult in a laboratory setting. Some of the challenges include consistent airborne MP dosing, staff safety considerations, and mouse eye irritation, respectively.
To assess the effects of MPs on uninfected lungs, a single dose of 1 µg of MPs per mouse (≈ 40 µg/kg) of azide-free, 1 µm diameter, fluorescent dye loaded, polystyrene beads (density 0.26 g/ml) was delivered in 50 µl of PBS via the i.n. route into the lungs of lightly anesthetized female C57BL/6J mice. Control mice received 50 µl of PBS. The anaesthesia prevents the mouse sneezing or coughing out the introduced material, but is light enough for the mouse to retain largely normal breathing, promoting deep lung delivery of the material; this is the same method used to infect mice with SARS-CoV-2 (46, 51, 59, 72, 73). The mice were observed daily and no overt clinical signs were observed. Mice were euthanized on day 2 and 6 post MP inoculation, and lungs analysed by RNA-Seq, with bioinformatic treatments comparing +MP day 2 vs. PBS day 2 and +MP day 6 vs. PBS day 6 (Figure 1; Supplementary Tables 1, 2).
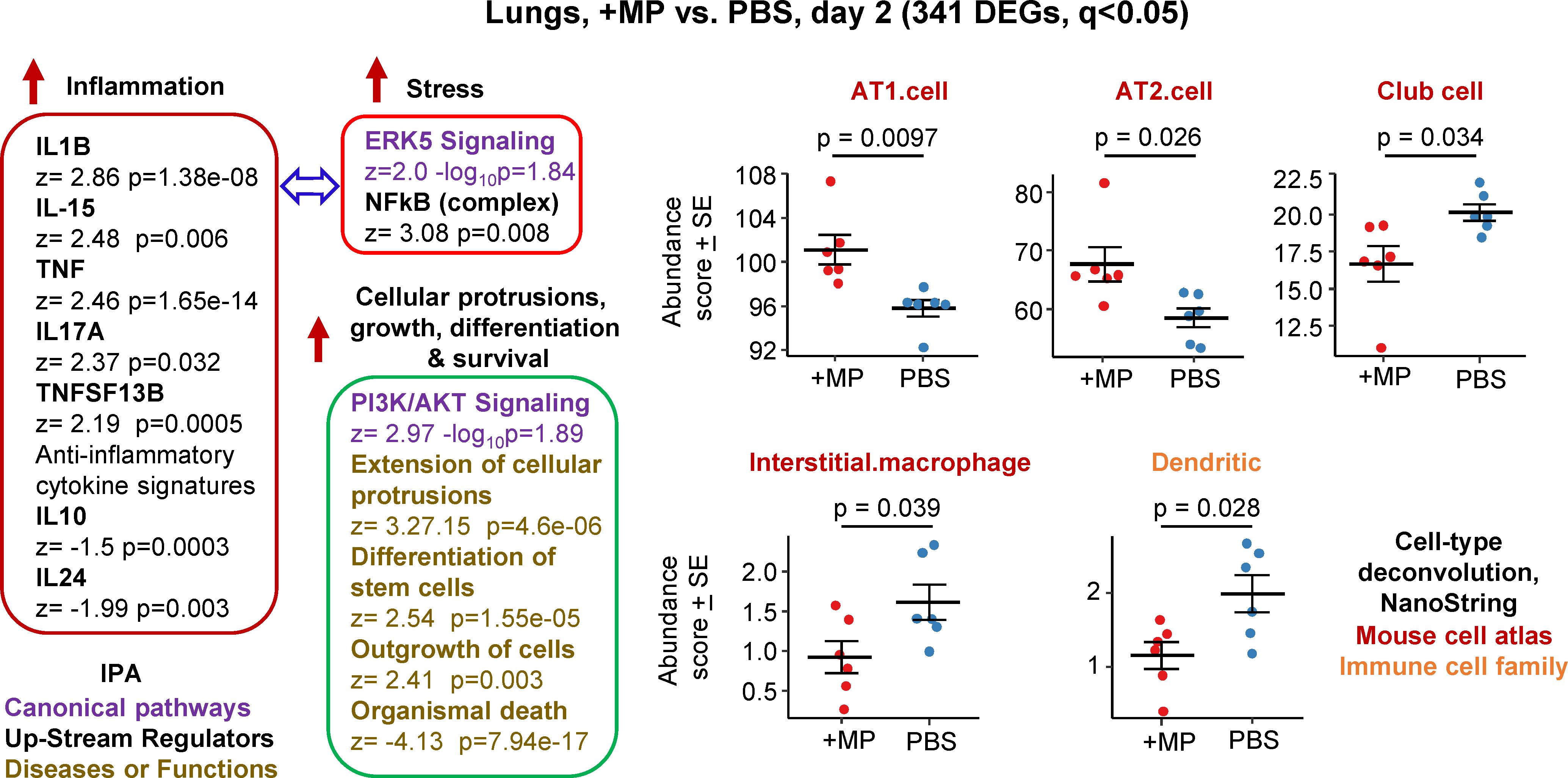
Figure 1 Bioinformatic analyses of RNA-Seq data for lungs inoculated with MPs. C57BL/6J mice were inoculated with MPs or PBS into the lungs via the intranasal route and at 2 days post inoculation lungs were harvested and analysed by RNA-Seq. DEGs (n=341) were identified and analysed by IPA, with selected annotations shown, full data sets are provided in Supplementary Table 1. The whole gene list (Supplementary Table 1) was also analysed by cell type deconvolution using cell-type expression matrices obtained from NanoString; statistics by t tests.
RNA-Seq for +MP vs. PBS day 2 identified 341 differentially expressed genes (DEGs) at q (FDR) <0.05 (Supplementary Table 1). Fold change (FC) was low overall, with only 9 genes showing FC>2 (log2 FC>1) and only 4 of these had a mean normalized counts per million (cpm) >10 across all samples (Supplementary Table 1). Ingenuity Pathway Analysis (IPA) indicated a mild pro-inflammatory response, with IL-1β the highest Cytokine UpStream Regulator (USR) by z score (Figure 1, Inflammation; Supplementary Table 1). Some signatures that can be associated with stress responses were also evident, principally ERK5 (MAPK7) (74, 75) and NFkB (76, 77) (Figure 1, Stress). A series of annotations associated with cellular protrusions, proliferation, differentiation and survival were identified as the top annotations (by z score) by IPA Diseases and Functions (Figure 1), with the largest negative z scores associated with growth failure or death (Supplementary Table 1).
Relative abundance of specific cell types was analysed via cellular deconvolution, SpatialDecon (66), with cell-type expression matrices obtained from Yoshida et al., 2019 (67) or the NanoString Cell Profile Library, either Mouse/Adult/Lung_MCA (Mouse cell atlas) or Mouse/Adult/ImmuneAtlas_ImmGen_cellfamily (Immune cell family) (Supplementary Table 1). These analyses suggested that the cells that were more transcriptionally active (expanding/repairing) in the +MP group were AT1 and AT2 type I and II alveolar epithelial cells (Figure 1, AT1, AT2). The positive z score for “Differentiation of stem cells” (Figure 1, Cellular protrusions, growth, differentiation, survival), may be associated with reduced abundance of Club cells (formally known as Clara cells), which are regional progenitor cells that repair bronchiolar epithelium in response to lung damage. Also identified was a reduced abundance of interstitial macrophages, cells that are associated with nerves and airways and appear to have important immunoregulatory roles (78, 79). A reduction in interstitial macrophages may also be consistent with the reductions in anti-inflammatory cytokines (Figure 1, IL-10 and IL-24) (79). Dendritic cell abundance was also down in the +MP group, perhaps consistent with mobilization to lymph/lymph nodes in response to inflammation (80).
In summary, MP inoculation into lungs generated a transcriptional signature indicating a mild inflammatory response, and mild increases in abundance scores (likely proliferation) for lung epithelial cells, and reductions in abundance scores for interstitial macrophages and dendritic cells.
Harvesting of lungs on day 6 after MP or PBS inoculation and comparing the transcriptome by RNA-Seq (+MP day 6 vs. PBS day 6) identified only 7 DEGs, all with low fold change (Supplementary Table 2). This argues that the lung response to a single exposure to MPs is largely resolved within 6 days. GSEAs using MSigDB gene sets provided a series of significant annotations with high positive Normalised Enrichment Score (NES) associated with epithelial cells (e.g. Epithelial differentiation), suggesting some lung repair activities were still underway on day 6 (Supplementary Table 2).
The best described mouse model for SARS-CoV-2 infection is the K18-hACE2 model which expresses the SARS-CoV-2 receptor, human ACE2 (hACE2), from the keratin 18 promoter (K18). This model is generally lethal within several days, as it usually leads to brain infection and ensuing weight loss that reaches ethically defined criteria for euthanasia (53, 59, 73). A non-lethal, less severe model is the mACE2-hACE2 model, wherein hACE2 is expressed from the mouse ACE2 promoter (mACE2) (58). We generated such a transgenic mouse on a pure C57BL/6J background by microinjection of the mACE2-hACE2 transgene into the pronucleus of C57BL/6J zygotes at the pronuclei stage (46) and generated a homozygous mACE2-hACE2 transgenic mouse line (see Materials and Methods). Given MP-mediated effects were generally mild (Figure 1; Supplementary Table 1), we chose this mouse model so that any MP-mediated perturbations might be more readily detected.
Although infection of mACE2-hACE2 mice with an original strain isolate has been described (46, 58), infection of homozygous mACE2-hACE2 mice with an omicron BA.5 isolate has not. The latter did not result in significant weight loss (data not shown), with lung histology at 6 days post infection (dpi) showing a series of histopathological features that have been described previously in COVID-19 mouse models (49, 58, 72) (Supplementary Figure 1). Lesions were less severe when compared with those seen after infection of K18-hACE2 mice with an original strain isolate (59, 81, 82), although significant loss of white space (unstained air-spaces) in H&E stained lung sections (indicating lung consolidation) (51, 72), was also seen in this model (Supplementary Figure 2).
Infection of mACE2-hACE2 mice with BA.5 (BA.5 vs. PBS) was analysed by RNA-Seq, with lungs harvested on 2 dpi (peak viral load) and 6 dpi (peak lung pathology) and compared with mock infected lungs (PBS) harvested on days 2 and 6, respectively (Supplementary Tables 3, 4). When the lung cytokine response signatures (IPA Cytokine USR z scores) from BA.5-infected mACE2-hACE2 mice, were compared with those from K18-hACE2 mice infected with an original strain isolate (46), a highly significant correlation emerged for 2 dpi. However, at 5/6 dpi a less significant correlation was observed (Supplementary Figure 3), reflecting the lower severity of lung disease in the BA.5 mACE2-hACE2 model.
IPA Diseases and Function annotations for BA.5 infected lungs for mACE2-hACE2 mice (BA.5 vs. PBS) pertinent to the analyses below include, top annotations for leukopoiesis/haematopoiesis, phagocytosis (engulfment of cells) and apoptosis & necroptosis (Supplementary Tables 3, 4).
To evaluate the effects of SARS-CoV-2 infection on MP clearance from lungs, mACE2-hACE2 mice were given a single inoculum of 50 µl containing both BA.5 (5x104 CCID50) and 1 µg (≈40 µg/kg) of azide free MP beads into the lungs via the intranasal route using the same procedure used herein and generally to infect mice with SARS-CoV-2 (59, 72, 83) (BA.5+MP). Control mice were not infected, but received the same 50 µl inoculum containing MP (+MP). Mixing the beads with the viral inoculum prior to infection prevented any complications that might arise from simple liquid occlusion of airways, if for, instance, the SARS-CoV-2 inoculum was given first and MPs were introduced later. Lungs were harvested at ≈ 4 hrs, 24 hrs, 48 hrs (day 2) and 144 hrs (day 6), were fixed in formalin, and the beads (MPs) observed by fluorescent microscopy of cryosections (Figure 2A). Quantitation suggested slower clearance of the MPs in BA.5-infected lungs (Figure 2B), consistent with SARS-CoV-2-mediated disruption of the ciliary layer, which is responsible for mucociliary clearance (59, 84). To provide better quantitation, lungs were digested in proteinase K and dissolved in SDS, and beads counted using a hemocytometer under a fluorescent microscope. The same trend was observed, with significantly more beads present at 24 and 48 hrs in infected mice (Figure 2C). Results from both assays indicated that MPs were largely cleared by day 6 (Figures 2B, C).
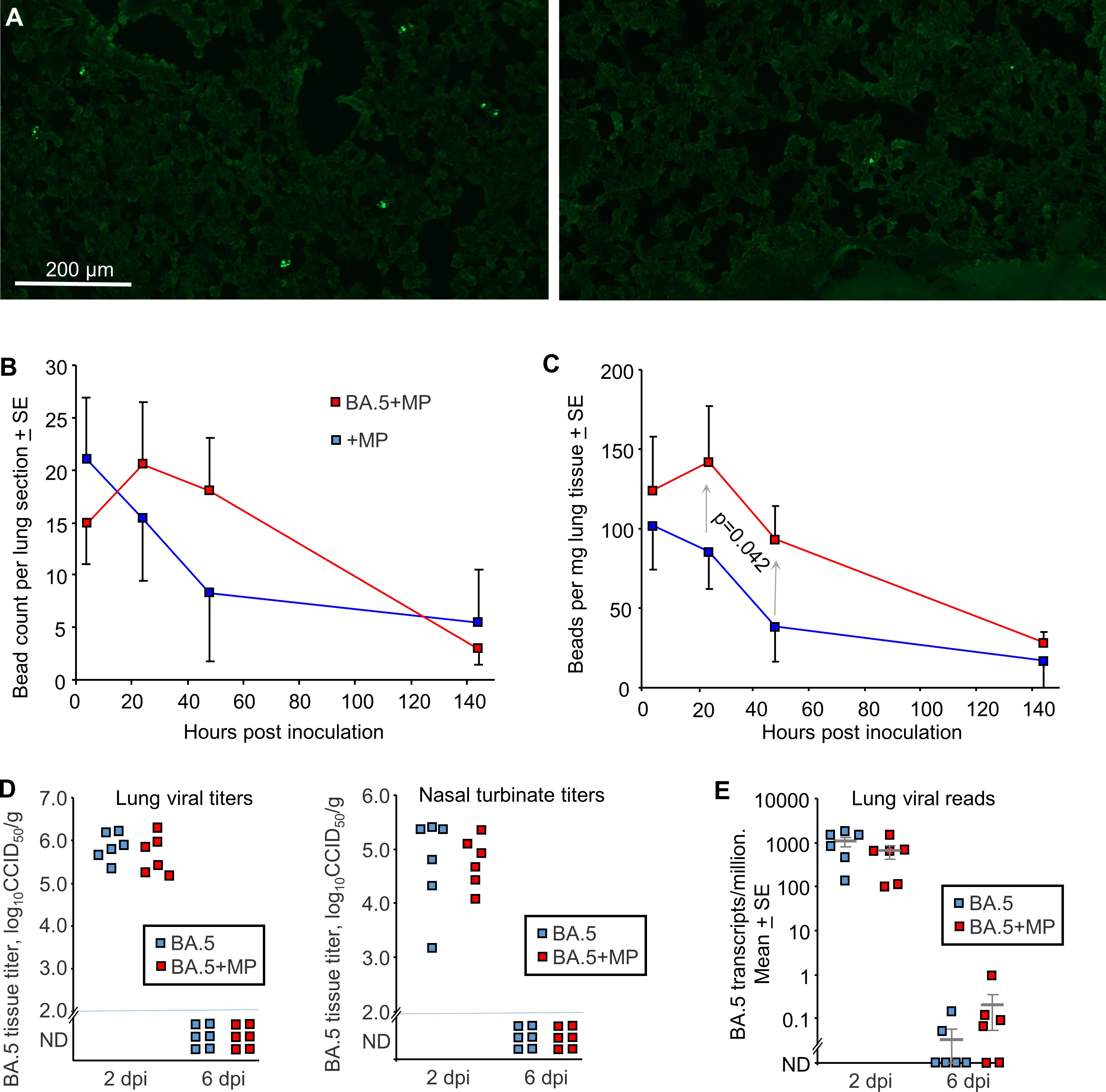
Figure 2 SARS-CoV-2 infection slowed MP clearance, but MPs did not affect viral load. (A) mACE-hACE2 mice were inoculated with BA.5+MP or with just MP (+MP), lungs were harvested at different time points and cryosections of lungs observed by fluorescent microscopy. Examples are shown for the 4 hr (right) and 48 hr (left) time points for the +MP group. (B) Quantitation of MP counts from cryosections (n=6 mice per group for 4 hrs, n=3 for other time points). (C) Quantitation of MP counts using tissue digestion and hemocytometer. The two groups were statistically different by 2 way ANOVA, which included a term for hours post inoculation with data for 24 and 48 hrs included. (D) mACE-hACE2 mice were inoculated with BA.5+MP or with BA.5 (no MP) and lung and nasal turbinate tissue titres determined by CCID50 assays. (E) mACE-hACE2 mice were inoculated with BA.5+MP or with BA.5 and lung analysed by RNA-Seq. Viral read counts are shown as BA.5 counts per million.
To evaluate the effects of MPs on SARS-COV-2 infection, mACE2-hACE2 mice were given a single inoculum of 50 µl containing both BA.5 (5x104 CCID50) and 1 µg (≈40 µg/kg) of beads, delivered into the lungs via the intranasal route (BA.5+MP). Control mice received the same 50 µl inoculum containing BA.5, but no MPs (BA.5). Lungs and nasal turbinates were harvested on 2 and 6 days post infection (dpi) and tissue titres determined by CCID50 assays, with no significant differences in viral titres evident on 2 dpi, and below the level of detection by 6 dpi (Figure 2D).
Lungs were also analysed by RNA-Seq and reads aligned to the viral genome, with the number of viral reads not significantly different for groups with or without MPs at 2 or 6 dpi (Figure 2E; Supplementary Tables 5, 6). Thus overall MP inoculation had no significant effects on viral loads in the respiratory track.
To assess the effects of MPs on the innate immune responses induced by BA.5 infection, the same groups described above (BA.5+MP vs. BA.5, 2 dpi) were compared by RNA-Seq, with reads aligned to the mouse genome. To minimize within-treatment variation in gene expression due to variability in viral load (Figure 2E; Supplementary Table 5), viral loads expressed in counts per-million were included as a term in the linear model used for estimating gene expression in EdgeR. Applying a q<0.05 filter, 596 DEGs were thereby identified (Figure 3; Supplementary Table 5).
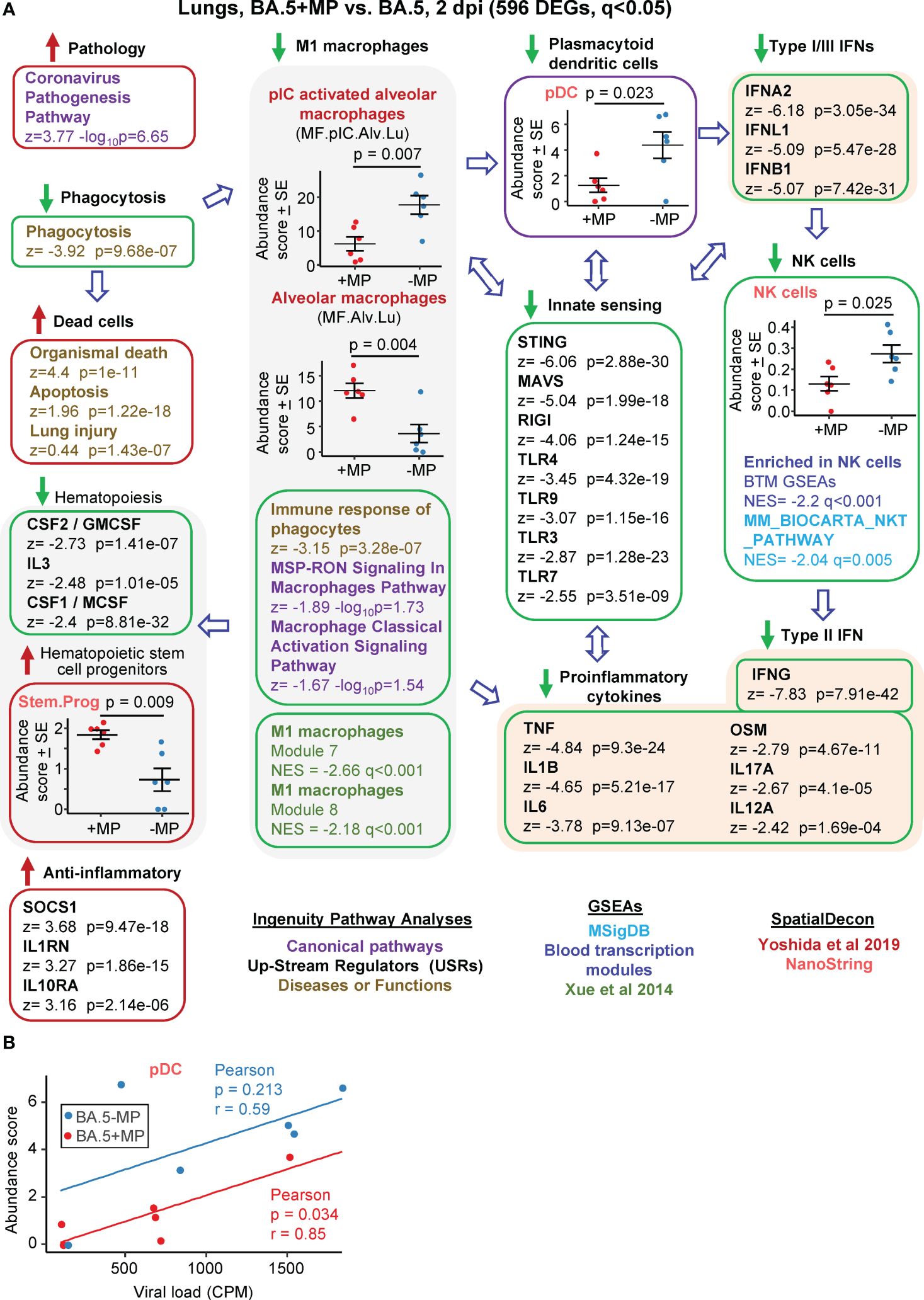
Figure 3 Bioinformatic analyses of RNA-Seq data for BA.5+MP vs. BA.5 at 2 dpi. (A) mACE-hACE2 mice were inoculated with BA.5+MP or with BA.5 and at 2 dpi lungs were harvested and analysed by RNA-Seq. 596 DEGs were identified and analysed by IPA, with selected annotations shown; full data sets are provided in Supplementary Table 5. The whole gene list was also analysed by GSEAs, and cellular deconvolution (SpatialDecon) using cell-type expression matrices obtained from Yoshida et al., 2019 and NanoString (statistics by t tests). (B) Cellular deconvolution data for plasmacytoid dendritic cells shown in ‘a’ plotted against viral reads (counts per million). Pearson correlation significance (p) and correlation coefficient (r) provided.
The DEGs were analysed by IPA as above, and the ‘All gene’ list, ranked by fold change, was used in a series of Gene Set Enrichment Analyses (GSEAs) using gene sets from the Molecular Signatures Data Base (MSigDB), Blood Transcription modules (BTM) and Xue et al., 2014 (65) (Figure 3; Supplementary Table 5). Relative abundance of specific cell types was analysed as above using SpatialDecon (66), using gene expression matrices provided by NanoString Cell Profile Library, and Yoshida et al., 2019 (67) (Figure 3; Supplementary Table 5).
The highest ranked IPA Canonical pathway was the ‘Coronavirus pathogenesis pathway’ (Figure 3A, Pathology; Supplementary Table 5). MPs were recently reported to inhibit phagocytosis of apoptotic cells (known as efferocytosis) (42), and a top IPA Diseases and Functions annotation was ‘Phagocytosis’, with a high negative z score (Figure 3A, Phagocytosis; Supplementary Table 5). During infection, macrophages phagocytose SARS-CoV-2-infected cells and are thereby activated, and in turn mediate strong activation of plasmacytoid dendritic cells (pDC) (85). The reduced phagocytosis is thus consistent with a reduction in activated M1 macrophages identified via a series of annotations (Figure 3, M1 macrophages); specifically (i) reduced abundance of polyinosinic:polycytidylic acid (pIC) stimulated macrophages (a stimulus that mimics viral double stranded RNA), (ii) a series of IPA Canonical pathways and Diseases and Functions macrophage annotations with negative z scores, and (iii) GSEAs with negative NES and q<0.05 using M1 macrophage gene sets from Xue at al., 2014 (65) (Supplementary Table 5).
Significantly reduced numbers of pDC were identified in the BA.5+MP group by cellular deconvolution using NanoString expression matrices (Figure 3, pDC, Supplementary Table 5), consistent with the reduction in M1 macrophages (85). As might be expected, viral load and pDC abundance showed positive correlations, with pDC abundance lower for the BA.5+MP group across all viral loads (Figure 3B). pDC are dominant producers of type I interferons (IFNs) during SARS-CoV-2 infection, with low numbers of pDCs and low type I IFN levels generally associated with increased COVID-19 severity (86, 87). Consistent with reduced abundance of pDC in the BA.5+MP group, was a series of type I IFN IPA USR annotations with highly negative z scores (Figure 3A, Type I IFNs; Supplementary Table 5). Reduction in type I IFN signatures was also evident from GSEAs using MSigDB gene sets (Supplementary Table 5). The general overall reduction in innate sensing signatures (Figure 3A, Innate sensing) was consistent with reduced type I IFN signatures, as well as the signatures associated with reduced phagocytosis, M1 macrophages and pDCs.
The lower type I and type III IFN signatures in the BA.5+MP group were not associated with significantly higher viral titres or higher levels of viral RNA (Figures 2D, E). This might be expected as even elimination of the type I or type III IFN receptor (in IFNAR-/- and IL-28RA-/- mice, respectively) had no significant impact on viral replication in the lungs (49). The ability of multiple SARS-CoV-2 proteins to mediate evasion of innate IFN responses is well described (88, 89), and explains the insensitivity of the virus to these responses. SpatialDecon, and GSEAs using MSigBD and Blood Transcription modules (BTMs), identified significantly reduced Natural Killer (NK) cell signatures in the BA.5+MP group (Figure 3A, NK cells; Supplementary Table 5), with low early type I IFN levels linked to reduced NK activity during viral infections generally (90) and likely also SARS-CoV-2 infections (91). NK cells have the capacity to exert important early innate antiviral activities; however, SARS-CoV-2 shows a remarkable ability to evade this arm of the immune system (92). NK and NKT cells are important sources of early IFNγ (90), thus the high negative z score for the IPA USR annotation for IFNγ (Figure 3A, Type I IFN; Supplementary Table 5) is consistent with reduced abundance of NK and NKT cells.
The general reductions in innate immune signatures for BA.5+MP vs. BA.5 (Figure 3, Innate sensing, Type I IFNs, NK cells) is likely to be responsible for the negative z scores for a series of IPA USR pro-inflammatory cytokine annotations (Figure 3A, Proinflammatory cytokines; Supplementary Table 5). Overall, these data illustrate that MPs can significantly ameliorate SARS-CoV-2-mediated activation of innate immune responses in the lung at 2 dpi.
The Interferome database provides an open-access bioinformatic resource that allows identification of interferon regulated genes (IRGs). The 100 most down-regulated DEGs at 2 dpi (according to EdgeR) were interrogated using the Interferome database, with a total of 45 of these identified as type I IRGs (Supplementary Table 5). Three of these (Ccl4, Irf7, and Oas3) were among the most down-regulated DEGs by RNA-Seq. Their significant differential expression was validated by RT-qPCR (Supplementary Figure 4A).
Three cytokine signatures associated with haematopoiesis were identified as down-regulated in the IPA USR analysis; CSF1/M-CSF, CSF2/GM-CSF and IL3 (Figure 3, Haematopoiesis; Supplementary Table 5). Cellular deconvolution using SpatialDecon and the Nanostring expression matrices also identified a higher abundance of haematopoietic stem and progenitor cells (HSPC) in the lungs of the BA.5+MP group at 2 dpi (Figure 3A, Stem.Prog; Supplementary Table 5). M-CSF, GM-CSF and IL3 (as well as IFNα, IFNγ and IL1, and TLR) are some of the key cytokine signaling pathways that promote HSPC differentiation (93). These analyses thus suggest an accumulation of undifferentiated HSPC due to reduced differentiation of HSPC into myeloid and/or lymphoid precursors.
Dysregulated emergency myelopoiesis and immature myeloid cells are associated with poor outcomes in COVID-19 patients (94–96), although these are observations from peripheral blood during COVID-19 disease, rather than from lungs early post infection. Nevertheless, these data (Figure 3A, Haematopoiesis & Haematopoietic stem and progenitor cells) suggest MPs in the lung suppress haematopoiesis, likely as a result of the overall reduction in the proinflammatory milieu for BA.5+MP. To clarify the terminology used in these annotations; haematopoiesis includes myelopoiesis, lymphopoiesis and erythropoiesis, whereas leukopoiesis encompasses myelopoiesis and lymphopoiesis.
IPA analysis of BA.5+MP vs. BA.5 indicated some signatures that are usually associated with anti-inflammatory activity (Figure 3A, Anti-inflammatory). The high USR z score for TRIM24 (Supplementary Table 5) argues that, although M1 macrophages are down, M2 macrophages are not increased, given that TRIM24 expression is suppressed in M2 macrophages (97).
Interstitial macrophages can have immunosuppressive properties (79, 98, 99); however, although identified as down in Figure 1 they were not identified as increased for BA.5+MP vs. BA.5. Conceivably, these anti-inflammatory signatures arose from the higher abundance of alveolar macrophages that are not M1 biased.
RNA-Seq was also undertaken for BA.5+MP vs. BA.5 at 6 dpi (the day of peak pathology in this model) with reads again aligned to the mouse genome. Using the same filter (q<0.05) 528 DEGs were identified and were analysed as above (Supplementary Table 6). A number of proinflammatory cytokine IPA USR signatures were up-regulated (Figure 4, Proinflammatory cytokines), all are associated with increased COVID-19 severity; TNF (100, 101), IL1B (102), IL1A (103), OSM (104), IL17A (105), and IL6 (106). Upregulated activity of the transcription factor EPHAS is also associated with pulmonary inflammatory responses in lethal COVID-19 (107). The anti-inflammatory interleukin 1 receptor antagonist (IL1RN) signature was down-regulated, with reduced IL1RN recently associated with increased COVID-19 severity (108).
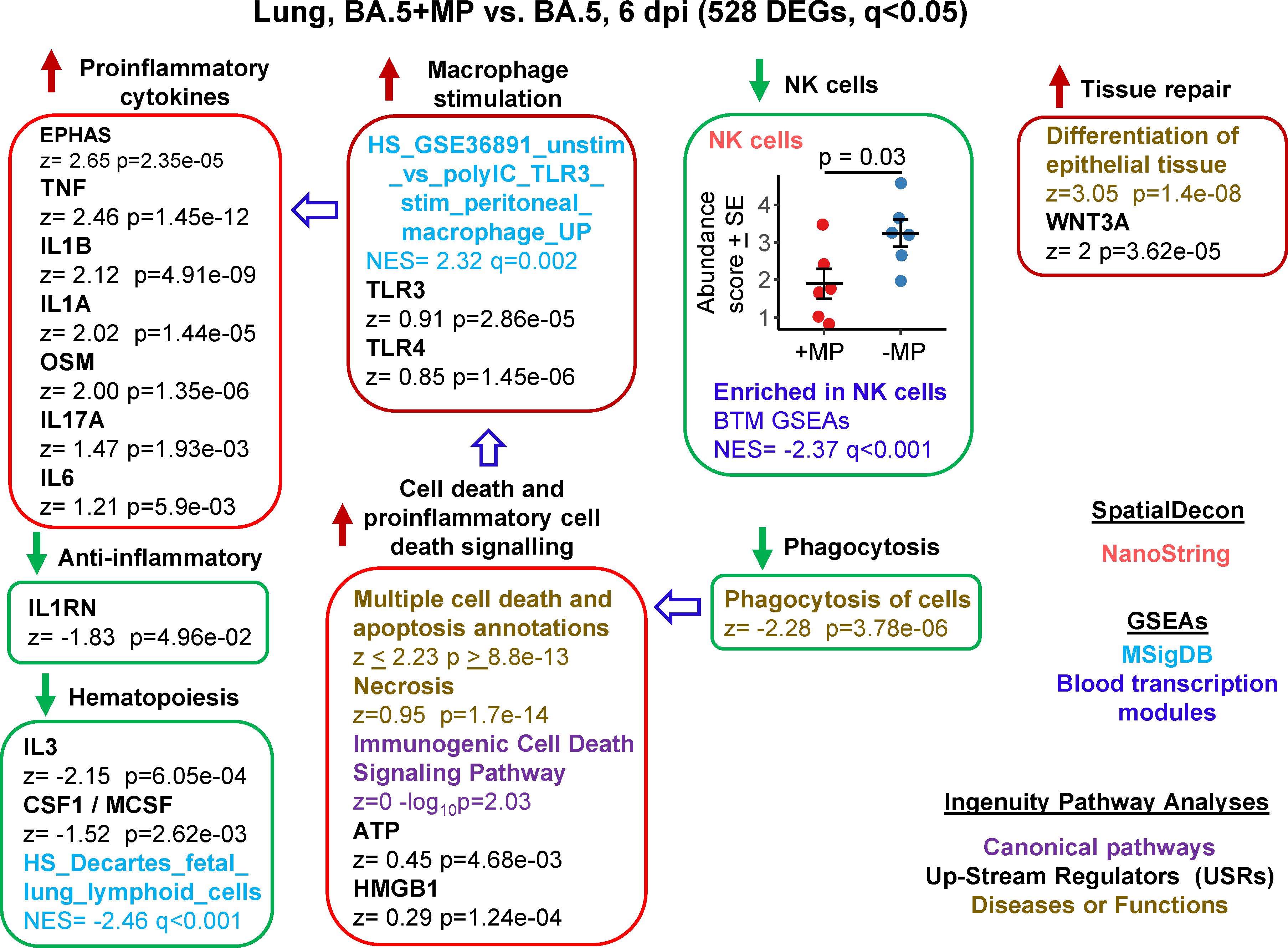
Figure 4 Bioinformatic analyses of RNA-Seq data for BA.5+MP vs. BA.5 at 6 dpi. mACE-hACE2 mice were inoculated with BA.5+MP or with BA.5 and at 6 dpi lungs were harvested and analysed by RNA-Seq; 528 DEGs were identified. Bioinformatic analyses as in Figure 3. Full data sets are provided in Supplementary Table 5.
The increased proinflammatory cytokine signatures (Figure 4) were not associated with reduced viral loads (Figure 2E). Of these responses, TNF is described as having anti-viral activity in some settings (109–111). However, anti-TNF therapy has not been associated with increases in SARS-CoV-2 replication or COVID-19 severity (100, 112), suggesting that TNF has minimal anti-viral activity against SARS-CoV-2 during COVID-19.
The mechanisms responsible for the increase in proinflammatory signatures (Figure 4) remain unclear, with, for instance, reduced type I responses (Figure 3) previously associated with reduced (not increased) lung inflammation (49). However, GSEAs using MSigDB gene sets provided a significant macrophage annotation with high NES for TLR3 stimulation, with an IPA USR annotation for TLR3 signaling also identified, albeit with a relatively low z score (Figure 4, Macrophage stimulation). Viral double-stranded RNA is a probable ligand for TLR3 (113). The reduced phagocytosis (Figure 4, Phagocytosis), and the increased proinflammatory responses (Figure 4, Proinflammatory cytokines), might suggest increased secondary necrosis due to a reduction in the phagocytosis of apoptotic cells (efferocytosis) (114–116). Multiple IPA Disease & Functions annotations suggest an increase in apoptosis signatures (Supplementary Table 6), with secondary necrosis known to be proinflammatory via secretion of a number of mediators such as HMGB1 and ATP (114). These latter mediators were identified as IPA USRs, although again with relatively low z scores (Figure 4, Cell death and proinflammatory cell death signaling).
NK cell and haematopoiesis signatures remained, as at 2 dpi, down-regulated by MPs (Figure 4). Tissue repair signatures were also identified (Figure 4, Tissue repair) as might be expected by 6 dpi, when virus has been largely cleared (Figures 2D, E).
Human lung RNA-Seq data for severe lethal COVID-19 infections was recently provided, with two signatures described, a ‘Classical signature’ and a ‘cytokine release syndrome’ (CRS) signature (117). We re-derived two DEG lists from the fastq files deposited for this study (Supplementary Table 7; PRJNA1036279), with the lists then analysed by IPA. Significant cytokine USR z scores provided by this analysis were compared with the z scores of significant cytokine USRs identified for BA.5+MP vs. BA.5 (Figures 5A, B; Supplementary Table 7). A significant correlation emerged for CRS (Figure 5A). In both human (infected vs. uninfected) and mouse (BA.5+MP vs. BA.5) data sets, TNF, IL1A, IL1B, OSM, IL6 and IL17 signatures were prominently up-regulated, and CSF1, IL3 and EPO were prominently down-regulated (Figure 5A, pink shading). These cytokines and their association with severe COVID-19 are described above for Figure 4. Treatment with EPO (118) and CSF1 (119, 120) have been considered for COVID-19, with low IL-3 levels associated with increased severity (121).
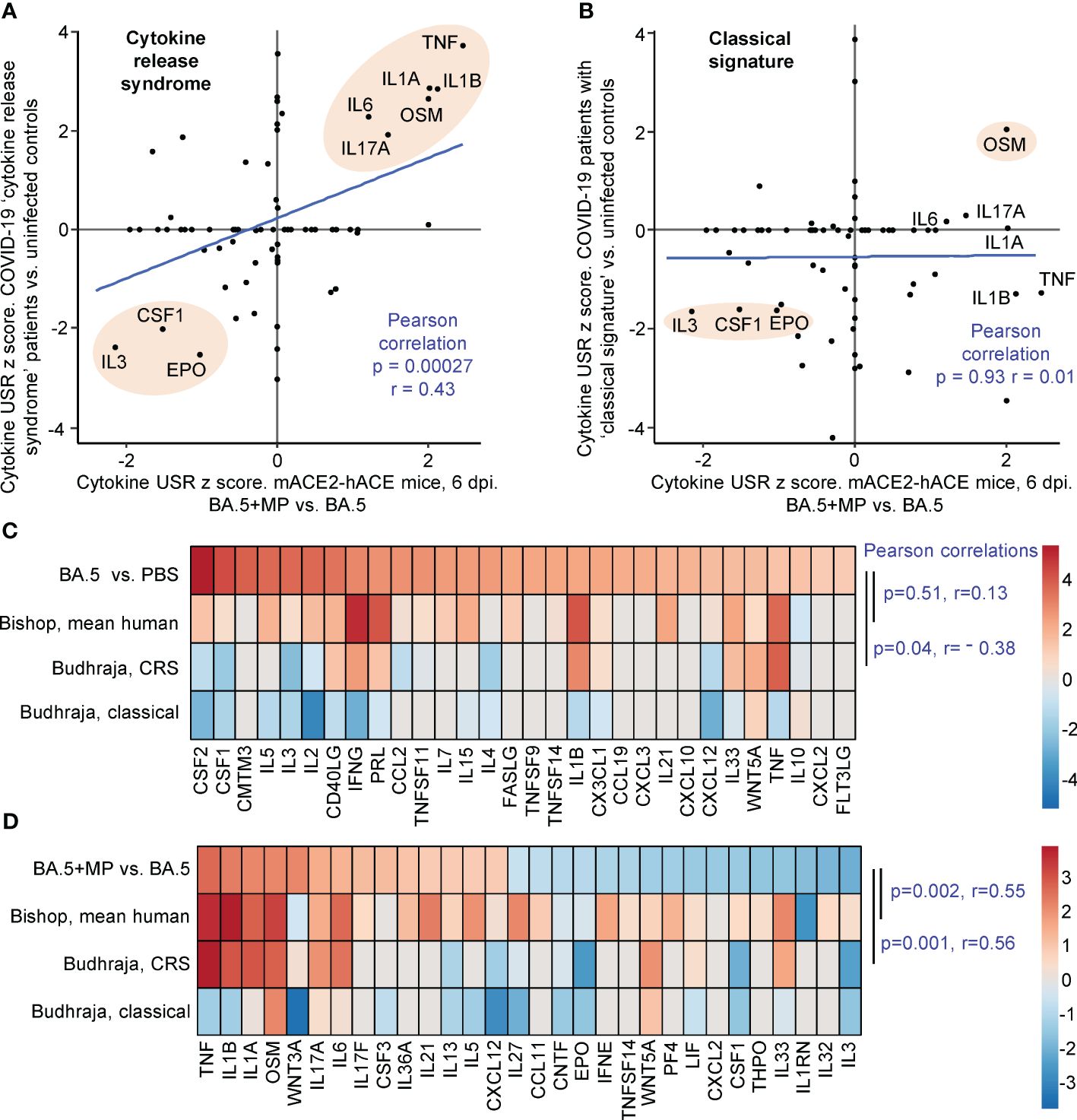
Figure 5 IPA cytokine USR z score correlations between human studies and BA.5+MP vs. BA.5 at 6 dpi. (A, B) IPA Cytokine USR z scores from the IPA analysis of the 528 DEGs described in Figure 4 were plotted against IPA Cytokine USR z scores from the IPA analysis of DEGs generated from fastq files obtained from NCBI SRA Bioproject PRJNA761132 (Budhraja et al., 2022) (Supplementary Table 7). Two distinct patterns were described for severe COVID patients, ‘Cytokine release syndrome’ and ‘Classical signature’; Pearson correlation significance (p) and correlation coefficient (r) are provided for both. Pink shading shows USRs dominant (high z scores) in both human and mouse studies. (C) The top 30 cytokine USRs by absolute z scores for BA.5 vs. PBS are compared to the human lung cytokine USR z scores described in Bishop et al., 2022 (derived from 4 studies of SARS-CoV-2 infected vs. uninfected), and the cytokine USR z scores for ‘Cytokine release syndrome’ and ‘Classical signature’ described in Budhraja et al., 2022 [same as (A, B) above]. (D) The top 30 cytokine USRs by absolute z scores for BA.5+MP vs. BA.5 are compared as for (C). Pearson correlation significance (p) and correlation coefficient (r) are provided.
Budhraja et al., 2022 also described a “Classical signature” as the more common pattern for lethal COVID-19 (117); a similar IPA analysis indicated no correlation when cytokine USR z scores for the ‘Classical signature’ were compared with cytokine USRs z scores identified for BA.5+MP vs. BA.5 (Figure 5B).
Another way of representing such data is by heat maps (46). When the top 30 cytokine USRs (by absolute z score) for BA.5 vs. PBS were ranked, and shown next to the aforementioned z scores for CRS and the Classical signature, there were no significant positive correlations (Figure 5C, Pearson correlations). The correlation was also poor for comparisons with the mean cytokine USR z scores previously generated from 4 human studies of SARS-CoV-2 infected lung tissues (46) (Figure 5C, Bishop, mean human, p=0.51). These poor correlations (Figure 5C) likely again reflect the relatively mild disease seen in the BA.5 mACE2-hACE2 mouse model.
When the top 30 cytokine USRs for BA.5+MP vs. BA.5 were ranked and compared with z scores from CRS in COVID-19 infected patients and the Bishop et al., 2022 study, significant positive correlations again emerged (Figure 5D). As in Figure 5B, there was no significant correlation with the classical signature (Figure 5D).
Taken together these analyses suggest that MPs in the SARS-CoV-2-infected lung pushed the cytokine signatures towards a CRS profile. However, MPs did not induce overt clinical disease in the BA.5-infected mice, nor were we able to detect significant histological changes in lung sections (Supplementary Figures 2B, 5).
At the top of the DEG lists for BA.5+MP vs. BA.5 are a number of genes that are associated with stress, and/or genes that have previously been identified as being induced after smoke or diesel particle inhalation, or in other lung diseases/disease models (Table 1). The top DEGs for BA.5+MP vs. BA.5 at 6 dpi where heat shock protein 70 (Hsp70) family members Hspa1a and Hspa1b, with Hsp70 induction well described in the MP literature (see Discussion).
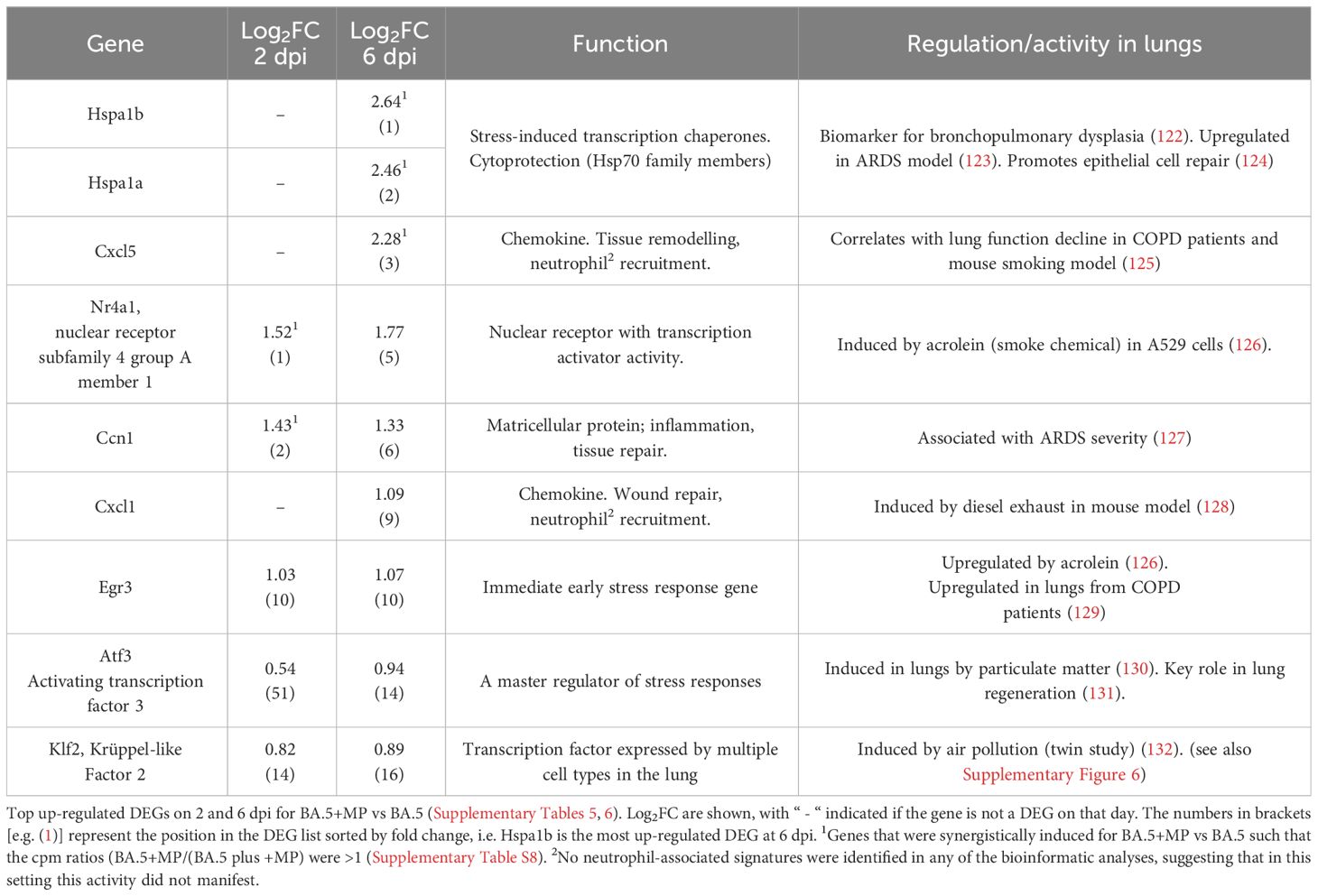
Table 1 Many top up-regulated DEGs are associated with stress responses and are also identified in other lung diseases or disease models.
We also calculated which genes were synergistically induced (i.e. genes for which the cpm for BA.5+MP > +MP alone plus BA.5 alone). Hspa1a and Hspa1b emerged as the most synergistically induced genes, with Cxcl5, Nr4a1 and Ccn1 also identified (Table 1; Supplementary Table 8). RT-qPCR was used to validate differential expression of Nr4a1, Ccn1, Hspa1a and Hspa1b, with log2 fold-changes and statistical significance consistent with the RNA-Seq data (Supplementary Figure 4B).
When the upregulated DEGs for BA.5+MP vs. BA.5 were compared with upregulated DEGs for +MP vs. PBS, only a small number of genes were found to be common to both DEG lists at both time points (n=12 for 2 dpi and n=2 for 6 dpi) (Supplementary Figure 6). Thus MPs on their own induced a largely different set of genes, when compared with MPs in a SARS-CoV-2 infection setting. This observation supports the contention that the detrimental activity of MPs might best be observed in the dysregulation of the inflammatory processes during the course of a disease (45), rather than as an imposition of fixed MP-specific responses.
Despite the overall low level of overlaps, treatment with MPs was consistently associated with upregulated of one DEG, kruppel-like factor 2 (Klf2) (Supplementary Figure 6).
We show herein in a mild disease model of SARS-CoV-2 infection and disease (omicron BA.5 infection of mACE2-hACE2 mice) that MP inoculation into the lungs dysregulated the innate inflammatory responses against the virus. MPs in the SARS-CoV-2 infected lungs lead to depressed innate proinflammatory immune responses at 2 dpi, with an elevated innate proinflammatory profile identified at 6 dpi. The latter profile showed significant correlation with the ‘cytokine release syndrome’, which is a potentially lethal manifestation of severe COVID-19 (117, 133). Thus the influence of MPs might be viewed as moving the inflammatory response away from protective inflammation (134, 135) toward pathological inflammation. However, despite this modulation, MP-mediated influences on SARS-CoV-2-induced disease was clinically inapparent in this model, with no overt clinical or histologically-detectable changes observed.
The ability of MPs to block efferocytosis via binding to the efferocytosis receptor Tim4 (42), may provide a basis for understanding at least some of the transcriptional perturbation described herein. Specifically, the clearly depressed phagocytosis signatures, the identification of multiple annotations associated with modulation of macrophage responses (with macrophages the key mediators of efferocytosis), and the overall lack of detectable changes to adaptive immune responses, might be viewed as consistent with a role for dysregulated phagocytosis/efferocytosis (115, 116). Reduced phagocytosis of SARS-CoV-2 infected cells at 2 dpi might reduce M1 macrophage activation, subsequent pDC activation (85), and the ensuing cytokine responses (86, 136). Less phagocytosis at the peak of infection might also lead to more secondary necrosis and/or necroptosis of SARS-CoV-2 infected cells, thereby promoting inflammation at 6 dpi (137, 138).
Inhibiting phagocytosis/efferocytosis may not be the only mechanism in play, as MPs alone provided some stress-associated, damage and pro-inflammatory signatures (Figure 1), which may also influence the SARS-CoV-2-mediated innate responses in the lung. The top DEGs for BA.5+MP vs. BA.5 support this contention as some of these DEGs have also been identified in studies of smoke or diesel particle inhalation (Table 1). The top DEGs for 6 dpi were heat Hsp70 family members, Hspa1a and Hspa1b, with these genes also synergistically induced (Table 1; Supplementary Table 8). Hsp70 up-regulation is reported in a range of MP exposure settings including mussels (139), goldfish (140) and Daphnia (141). Hsp70 stress responses are involved in a vast range of pathologies (142) and play a role in cytoprotection against environmental challenges (122) including SARS-CoV-2 infection (143). The top cytokine USR by z score for +MP vs. PBS was IL-1β, a cytokine matured and released via inflammasome activation (Figure 1). Our observation thus support a recent speculation that MPs might activate the inflammasome (144), with IL-1β also a dominant signature at 6 dpi for BA.5+MP vs. BA.5.
Nr4a1 (Table 1) was recently identified as a marker of a subset of group 2 innate lymphoid cells (ILCs) (145), with ILCs implicated in our previous study of MPs and a viral arthritis model (45). However, we have been unable to find a compelling signature that implicates ILCs as important players in the current setting. Nr4a1-dependent CD16.2+ monocytes have been implicated as potential precursors of CD206– interstitial macrophages (98, 99, 146). However, changes in interstitial macrophages were identified for +MP vs. PBS (Figure 1), where Nr4a1 was not a DEG, but interstitial macrophages were not identified for BA.5+MP vs. BA.5 (Figures 3, 4) where Nr4a1 was a top DEG (Table 1). Conceivably, the up-regulation of this gene is associated with lung epithelial cells, where it has been identified as a novel allergy-associated gene (147).
Klf2 was the only DEG upregulated by MPs in all settings and time points studied herein (Supplementary Figure 6). Klf2 is upregulated in blood endothelial cells as a result of sheer stress (148), but is ordinarily down-regulated during SARS-CoV-2 infection (149, 150). How these activities might be relevant in the current context is unclear. Klf2 upregulation may be related to the general dysregulation of macrophage activities by MPs seen herein, with Klf2 expression associated with alveolar macrophage self-renewal (151) and inhibition of M1 polarization (152).
Although many reports suggest MPs induce ROS (40, 41), we did not identify an increase in ROS as a major consequence of MP exposure, with the MPs used in this study being free of azide (39). IPA Disease and Functions annotation actually providing a slightly negative z score for “Production of reactive oxygen species” (Supplementary Table 1). MP-induced changes to the lung microbiota (37) might also provide a potential mechanism for the modulation of innate responses; however, our metagenomics analysis failed to identify any changes in the microbiome (Supplementary Figure 7).
This study has a number of limitations; we have not investigated the activity of MPs with different shapes, sizes, compositions and leachates (153, 154). However, such considerations give rise to an unworkably large number of experimental variables that are beyond the scope of this foundational study. We have also not examined longer exposure times or different exposure doses, and we have also only examined the consequences of MPs in one mild mouse model of COVID-19. However, the mild MP-associated transcriptional changes seen herein (Figure 1) might be drowned in a more severe model of COVID-19, where infection-induced gene expression changes are substantially more widespread and robust (46).
In summary, we provide herein evidence that MPs in the lung can dysregulate innate inflammation-associated transcriptional responses to SARS-CoV-2 infection in a mouse model of mild COVID-19. However, the dysregulation did not result in overt changes in disease or histopathology, suggesting MP-mediated changes were generally mild. To what extent MPs might influence COVID-19 disease severity at a population level may warrant investigation. An approach might be to compare matched populations exposed to high (155) and low levels (156) of airborne MP pollution, although separating the influence of MPs from other factors may represent a formidable challenge. Nevertheless, such analyses are conceivable with, for instance, mortality associated with fine particulate matter air pollution from coal powered electricity plants was recently estimated (157).
Considerable speculation surrounds the potential health impacts of MP inhalation by the general public (21, 158–160), with limited compelling in vivo data from human studies and animal models. However, an emerging theme from such studies, including the current report, is that MPs can dysregulate and/or promote inflammatory processes in specific disease settings (15, 45, 161–164). Future animal research in this area would benefit from use of realistic MP doses; although we need (i) better insights into what such doses actually are in different human populations, and (ii) adoption of standard units for MP exposure (165) (e.g. µg/kg/d) so that studies can be compared and dose effects understood. Avoidance of artefacts associated with preservatives such as azide is clearly also critical for future meaningful medical research in the MP space (39). Future animal research might also examine the effect of chronic MP exposure using multiple doses, as well as comparing the effects of different MP sizes, shapes, compositions and leachates.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: PRJNA1036279 (SRA).
The studies involving humans were approved by the QIMR Berghofer Medical Research Institute Human Research Ethics Committee (P3600). The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study. The animal study was approved by QIMR Berghofer Medical Research Institute animal ethics committee -P3600. The study was conducted in accordance with the local legislation and institutional requirements.
CB: Writing – review & editing, Writing – original draft, Visualization, Methodology, Formal analysis, Data curation. KY: Writing – review & editing, Investigation. WN: Writing – review & editing, Formal analysis. DR: Writing – review & editing, Supervision, Methodology, Funding acquisition. BT: Writing – review & editing, Investigation. TL: Writing – review & editing, Investigation, Formal analysis. AS: Writing – review & editing, Writing – original draft, Visualization, Supervision, Project administration, Methodology, Funding acquisition, Data curation, Conceptualization.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was funded by the National Health and Medical Research Council (NHMRC) of Australia (Investigator grant APP1173880 awarded to A.S.). Establishment of the QIMR Berghofer MRI SARS-CoV-2/COVID-19 PC3 research facilities, and research therein, was supported by generous philanthropic donations from the Brazil Family Foundation (and others). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors thank the following QIMRB staff; Dr. I. Anraku for management of the PC3 facility at QIMR Berghofer MRI, Dr. Viviana Lutzky for proof reading, Dr Crystal Chang for histology services, the animal house staff for mouse breeding and agistment, and Dr. Gunter Hartel for assistance with statistics.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1382655/full#supplementary-material
MP, microplastic; COVID-19, Coronavirus disease 2019; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; DEG, differentially expressed gene; ACE2, angiotensin converting enzyme 2; GSEA, Gene Set Enrichment Analyses; IPA, Ingenuity Pathway Analysis; USR, Up-Stream Regulator; FC, Fold Change; CCID50, 50% cell culture infectivity dose; ROS, reactive oxygen species; ILCs, innate lymphoid cells; dpi, days post infection; pDC, plasmacytoid dendritic cell; IFN, interferon; BTM, Blood Transcription Module; MSigDB, Molecular Signatures Data Base; TLR, Toll-like receptor; HSPC, haematopoietic stem and progenitor cell; NK, Natural Killer (cell).
1. Rangel-Buitrago N, Neal W, Williams A. The plasticene: time and rocks. Mar pollut Bull. (2022) 185:114358. doi: 10.1016/j.marpolbul.2022.114358
2. Du Toit A. Plastic communities. Nat Rev Microbiol. (2022) 20:575. doi: 10.1038/s41579-022-00790-1
3. Kozlov M. Landmark study links microplastics to serious health problems. Nature. (2024). doi: 10.1038/d41586-024-00650-3
4. United Nations Development Programme. Microplastics on Human Health: How much do they harm us? (2024). Available online at: https://www.undp.org/kosovo/blog/microplastics-human-health-how-much-do-they-harm-us.
5. Bertocchini F, Arias CF. Why have we not yet solved the challenge of plastic degradation by biological means? PloS Biol. (2023) 21:e3001979. doi: 10.1371/journal.pbio.3001979
6. Kutralam-Muniasamy G, Shruti VC, Pérez-Guevara F, Roy PD. Microplastic diagnostics in humans: “The 3Ps” Progress, problems, and prospects. Sci Total Environ. (2023) 856:159164. doi: 10.1016/j.scitotenv.2022.159164
7. Le VG, Nguyen MK, Nguyen HL, Lin C, Hadi M, Hung NTQ, et al. A comprehensive review of micro- and nano-plastics in the atmosphere: Occurrence, fate, toxicity, and strategies for risk reduction. Sci Total Environ. (2023) 904:166649. doi: 10.1016/j.scitotenv.2023.166649
8. Priya AK, Muruganandam M, Imran M, Gill R, Vasudeva Reddy MR, Shkir M, et al. A study on managing plastic waste to tackle the worldwide plastic contamination and environmental remediation. Chemosphere. (2023) 341:139979. doi: 10.1016/j.chemosphere.2023.139979
9. Zheng Y, Xu S, Liu J, Liu Z. The effects of micro- and nanoplastic on the central nervous system: A New Threat to Humanity? Toxicology. (2024) 504:153799. doi: 10.1016/j.tox.2024.153799
10. Jones LR, Wright SJ, Gant TW. A critical review of microplastics toxicity and potential adverse outcome pathway in human gastrointestinal tract following oral exposure. Toxicol Lett. (2023) 385:51–60. doi: 10.1016/j.toxlet.2023.08.011
11. Koelmans AA, Redondo-Hasselerharm PE, Nor NHM, de Ruijter VN, Mintenig SM, Kooi M. Risk assessment of microplastic particles. Nat Rev Materials. (2022) 7:138–52. doi: 10.1038/s41578-021-00411-y
12. Blackburn K, Green D. The potential effects of microplastics on human health: What is known and what is unknown. Ambio. (2022) 51:518–30. doi: 10.1007/s13280-021-01589-9
13. Marfella R, Prattichizzo F, Sardu C, Fulgenzi G, Graciotti L, Spadoni T, et al. Microplastics and nanoplastics in atheromas and cardiovascular events. New Engl J Med. (2024) 390:900–10. doi: 10.1056/NEJMoa2309822
14. Yan Z, Liu Y, Zhang T, Zhang F, Ren H, Zhang Y. Analysis of microplastics in human feces reveals a correlation between fecal microplastics and inflammatory bowel disease status. Environ Sci Technol. (2022) 56:414–21. doi: 10.1021/acs.est.1c03924
15. Chen C, Liu F, Quan S, Chen L, Shen A, Jiao A, et al. Microplastics in the bronchoalveolar lavage fluid of chinese children: associations with age, city development, and disease features. Environ Sci Technol. (2023) 57:12594–601. doi: 10.1021/acs.est.3c01771
16. Horvatits T, Tamminga M, Liu B, Sebode M, Carambia A, Fischer L, et al. Microplastics detected in cirrhotic liver tissue. EBioMedicine. (2022) 82:104147. doi: 10.1016/j.ebiom.2022.104147
17. Prata JC. Airborne microplastics: Consequences to human health? Environ pollut. (2018) 234:115–26. doi: 10.1016/j.envpol.2017.11.043
18. Zuri G, Karanasiou A, Lacorte S. Microplastics: Human exposure assessment through air, water, and food. Environ Int. (2023) 179:108150. doi: 10.1016/j.envint.2023.108150
19. Eschenbacher WL, Kreiss K, Lougheed MD, Pransky GS, Day B, Castellan RM. Nylon flock-associated interstitial lung disease. Am J Respir Crit Care Med. (1999) 159:2003–8. doi: 10.1164/ajrccm.159.6.9808002
20. Washko RM, Day B, Parker JE, Castellan RM, Kreiss K. Epidemiologic investigation of respiratory morbidity at a nylon flock plant. Am J Ind Med. (2000) 38:628–38. doi: 10.1002/1097-0274(200012)38:6<628::aid-ajim3>3.0.co;2-u
21. Perera K, Ziajahromi S, Nash SB, Leusch FDL. Microplastics in Australian indoor air: Abundance, characteristics, and implications for human exposure. Sci Total Environ. (2023) 889:164292. doi: 10.1016/j.scitotenv.2023.164292
22. Romarate RA 2nd, Ancla SMB, Patilan DMM, Inocente SAT, Pacilan CJM, Sinco AL, et al. Breathing plastics in Metro Manila, Philippines: presence of suspended atmospheric microplastics in ambient air. Environ Sci pollut Res Int. (2023) 30:53662–73. doi: 10.1007/s11356-023-26117-y
23. Liu C, Li J, Zhang Y, Wang L, Deng J, Gao Y, et al. Widespread distribution of PET and PC microplastics in dust in urban China and their estimated human exposure. Environ Int. (2019) 128:116–24. doi: 10.1016/j.envint.2019.04.024
24. Jenner LC, Rotchell JM, Bennett RT, Cowen M, Tentzeris V, Sadofsky LR. Detection of microplastics in human lung tissue using muFTIR spectroscopy. Sci Total Environ. (2022) 831:154907. doi: 10.1016/j.scitotenv.2022.154907
25. Baeza-Martinez C, Olmos S, Gonzalez-Pleiter M, Lopez-Castellanos J, Garcia-Pachon E, Masia-Canuto M, et al. First evidence of microplastics isolated in European citizens' lower airway. J Hazard Mater. (2022) 438:129439. doi: 10.1016/j.jhazmat.2022.129439
26. Jiang Y, Han J, Na J, Fang J, Qi C, Lu J, et al. Exposure to microplastics in the upper respiratory tract of indoor and outdoor workers. Chemosphere. (2022) 307:136067. doi: 10.1016/j.chemosphere.2022.136067
27. Qiu L, Lu W, Tu C, Li X, Zhang H, Wang S, et al. Evidence of microplastics in bronchoalveolar lavage fluid among never-smokers: A prospective case series. Environ Sci Technol. (2023) 57:2435–44. doi: 10.1021/acs.est.2c06880
28. Huang S, Huang X, Bi R, Guo Q, Yu X, Zeng Q, et al. Detection and analysis of microplastics in human sputum. Environ Sci Technol. (2022) 56:2476–86. doi: 10.1021/acs.est.1c03859
29. Soltani NS, Taylor MP, Wilson SP. Quantification and exposure assessment of microplastics in Australian indoor house dust. Environ pollut. (2021) 283:117064. doi: 10.1016/j.envpol.2021.117064
30. Zhang J, Wang L, Kannan K. Microplastics in house dust from 12 countries and associated human exposure. Environ Int. (2020) 134:105314. doi: 10.1016/j.envint.2019.105314
31. Wang Y, Huang J, Zhu F, Zhou S. Airborne microplastics: A review on the occurrence, migration and risks to humans. Bull Environ Contam Toxicol. (2021) 107:657–64. doi: 10.1007/s00128-021-03180-0
32. Kannan K, Vimalkumar K. A review of human exposure to microplastics and insights into microplastics as obesogens. Front Endocrinol (Lausanne). (2021) 12:724989. doi: 10.3389/fendo.2021.724989
33. Danso IK, Woo JH, Lee K. Pulmonary toxicity of polystyrene, polypropylene, and polyvinyl chloride microplastics in mice. Molecules. (2022) 27:7926. doi: 10.3390/molecules27227926
34. Zolotova N, Kosyreva A, Dzhalilova D, Fokichev N, Makarova O. Harmful effects of the microplastic pollution on animal health: a literature review. PeerJ. (2022) 10:e13503. doi: 10.7717/peerj.13503
35. Cao J, Xu R, Geng Y, Xu S, Guo M. Exposure to polystyrene microplastics triggers lung injury via targeting toll-like receptor 2 and activation of the NF-kappaB signal in mice. Environ pollut. (2023) 320:121068. doi: 10.1016/j.envpol.2023.121068
36. Wu Y, Yao Y, Bai H, Shimizu K, Li R, Zhang C. Investigation of pulmonary toxicity evaluation on mice exposed to polystyrene nanoplastics: The potential protective role of the antioxidant N-acetylcysteine. Sci Total Environ. (2023) 855:158851. doi: 10.1016/j.scitotenv.2022.158851
37. Zha H, Xia J, Li S, Lv J, Zhuge A, Tang R, et al. Airborne polystyrene microplastics and nanoplastics induce nasal and lung microbial dysbiosis in mice. Chemosphere. (2023) 310:136764. doi: 10.1016/j.chemosphere.2022.136764
38. Li X, Zhang T, Lv W, Wang H, Chen H, Xu Q, et al. Intratracheal administration of polystyrene microplastics induces pulmonary fibrosis by activating oxidative stress and Wnt/beta-catenin signaling pathway in mice. Ecotoxicol Environ Saf. (2022) 232:113238. doi: 10.1016/j.ecoenv.2022.113238
39. Pikuda O, Xu EG, Berk D, Tufenkji N. Toxicity assessments of micro- and nanoplastics can be confounded by preservatives in commercial formulations. Environ Sci Technol Lett. (2019) 6:21–5. doi: 10.1021/acs.estlett.8b00614
40. Hu M, Palic D. Micro- and nano-plastics activation of oxidative and inflammatory adverse outcome pathways. Redox Biol. (2020) 37:101620. doi: 10.1016/j.redox.2020.101620
41. Subaramaniyam U, Allimuthu RS, Vappu S, Ramalingam D, Balan R, Paital B, et al. Effects of microplastics, pesticides and nano-materials on fish health, oxidative stress and antioxidant defense mechanism. Front Physiol. (2023) 14:1217666. doi: 10.3389/fphys.2023.1217666
42. Kuroiwa M, Yamaguchi SI, Kato Y, Hori A, Toyoura S, Nakahara M, et al. Tim4, a macrophage receptor for apoptotic cells, binds polystyrene microplastics via aromatic-aromatic interactions. Sci Total Environ. (2023) 875:162586. doi: 10.1016/j.scitotenv.2023.162586
43. Mehrotra P, Ravichandran KS. Drugging the efferocytosis process: concepts and opportunities. Nat Rev Drug Discovery. (2022) 21:601–20. doi: 10.1038/s41573-022-00470-y
44. Schilperoort M, Ngai D, Sukka SR, Avrampou K, Shi H, Tabas I. The role of efferocytosis-fueled macrophage metabolism in the resolution of inflammation. Immunol Rev. (2023) 319:65–80. doi: 10.1111/imr.13214
45. Rawle DJ, Dumenil T, Tang B, Bishop CR, Yan K, Le TT, et al. Microplastic consumption induces inflammatory signatures in the colon and prolongs a viral arthritis. Sci Total Environ. (2022) 809:152212. doi: 10.1016/j.scitotenv.2021.152212
46. Bishop CR, Dumenil T, Rawle DJ, Le TT, Yan K, Tang B, et al. Mouse models of COVID-19 recapitulate inflammatory pathways rather than gene expression. PloS Pathog. (2022) 18:e1010867. doi: 10.1371/journal.ppat.1010867
47. Al-Nesf MAY, Abdesselem HB, Bensmail I, Ibrahim S, Saeed WAH, Mohammed SSI, et al. Prognostic tools and candidate drugs based on plasma proteomics of patients with severe COVID-19 complications. Nat Commun. (2022) 13:946. doi: 10.1038/s41467-022-28639-4
48. Maity S, Santra A, Hebbani AV, Pulakuntla S, Chatterjee A, Badri KR, et al. Targeting cytokine storm as the potential anti-viral therapy: Implications in regulating SARS-CoV-2 pathogenicity. Gene. (2023) 881:147612. doi: 10.1016/j.gene.2023.147612
49. Rawle DJ, Le TT, Dumenil T, Yan K, Tang B, Nguyen W, et al. ACE2-lentiviral transduction enables mouse SARS-CoV-2 infection and mapping of receptor interactions. PloS Pathog. (2021) 17:e1009723. doi: 10.1371/journal.ppat.1009723
50. Liu K, Tang M, Xu W, Meng X, Jin H, Han M, et al. An inducible hACE2 transgenic mouse model recapitulates SARS-CoV-2 infection and pathogenesis in vivo. Proc Natl Acad Sci. (2023) 120:e2207210120. doi: 10.1073/pnas.2207210120
51. Chen Q, Gao J, Yu H, Su H, Yang Y, Cao Y, et al. An emerging role of microplastics in the etiology of lung ground glass nodules. Environ Sci Europe. (2022) 34:25. doi: 10.1186/s12302-022-00605-3
52. Our_World_in_Data. SARS-CoV-2 sequences by variant (2023). Available online at: https://ourworldindata.org/grapher/covid-variants-bar.
53. Stewart R, Ellis SA, Yan K, Dumenil T, Tang B, Nguyen W, et al. SARS-CoV-2 omicron BA.5 and XBB variants have increased neurotropic potential over BA.1 in K18-hACE2 mice and human brain organoids. Front Microbiol. (2023) 14:1320856. doi: 10.3389/fmicb.2023.1320856
54. Johnson BJ, Le TT, Dobbin CA, Banovic T, Howard CB, Flores Fde M, et al. Heat shock protein 10 inhibits lipopolysaccharide-induced inflammatory mediator production. J Biol Chem. (2005) 280:4037–47. doi: 10.1074/jbc.M411569200
55. Hirata TD, Nakaya H, Le TT, Poo YS, Suhrbier A. Early pregnancy factor, chaperonin 10 and rheumatoid arthritis; the story unravels. J Trans Sci. (2018) 4:1–3. doi: 10.15761/JTS
56. La Linn M, Bellett AJ, Parsons PG, Suhrbier A. Complete removal of mycoplasma from viral preparations using solvent extraction. J Virol Methods. (1995) 52:51–4. doi: 10.1016/0166-0934(94)00136-5
57. Yan K, Rawle DJ, Le TT, Suhrbier A. Simple rapid in vitro screening method for SARS-CoV-2 anti-virals that identifies potential cytomorbidity-associated false positives. Virol J. (2021) 18:123. doi: 10.1186/s12985-021-01587-z
58. Bao L, Deng W, Huang B, Gao H, Liu J, Ren L, et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. (2020) 583:830–3. doi: 10.1038/s41586-020-2312-y
59. Dumenil T, Le TT, Rawle DJ, Yan K, Tang B, Nguyen W, et al. Warmer ambient air temperatures reduce nasal turbinate and brain infection, but increase lung inflammation in the K18-hACE2 mouse model of COVID-19. Sci Total Environ. (2023) 859:160163. doi: 10.1016/j.scitotenv.2022.160163
60. Delgado A, González-Caballero F, Salcedo J. On the zeta potential of spherical polystyrene particles from electrophoresis theories. Acta Polymerica. (1986) 37:361–4. doi: 10.1002/actp.1986.010370608
61. Walczak AP, Hendriksen PJM, Woutersen RA, van der Zande M, Undas AK, Helsdingen R, et al. Bioavailability and biodistribution of differently charged polystyrene nanoparticles upon oral exposure in rats. J Nanoparticle Res. (2015) 17:231. doi: 10.1007/s11051-015-3029-y
62. Yan K, Dumenil T, Tang B, Le TT, Bishop CR, Suhrbier A, et al. Evolution of ACE2-independent SARS-CoV-2 infection and mouse adaption after passage in cells expressing human and mouse ACE2. Virus Evol. (2022) 8:veac063. doi: 10.1093/ve/veac063
63. Morgan MS, Yan K, Le TT, Johnston RA, Amarilla AA, Muller DA, et al. Monoclonal antibodies specific for SARS-CoV-2 spike protein suitable for multiple applications for current variants of concern. Viruses. (2022) 15:139. doi: 10.3390/v15010139
64. Hagan T, Gerritsen B, Tomalin LE, Fourati S, Mulè MP, Chawla DG, et al. Transcriptional atlas of the human immune response to 13 vaccines reveals a common predictor of vaccine-induced antibody responses. Nat Immunol. (2022) 23:1788–98. doi: 10.1038/s41590-022-01328-6
65. Xue J, Schmidt SV, Sander J, Draffehn A, Krebs W, Quester I, et al. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity. (2014) 40:274–88. doi: 10.1016/j.immuni.2014.01.006
66. Danaher P, Kim Y, Nelson B, Griswold M, Yang Z, Piazza E, et al. Advances in mixed cell deconvolution enable quantification of cell types in spatial transcriptomic data. Nat Commun. (2022) 13:385. doi: 10.1038/s41467-022-28020-5
67. Yoshida H, Lareau CA, Ramirez RN, Rose SA, Maier B, Wroblewska A, et al. The cis-regulatory atlas of the mouse immune system. Cell. (2019) 176:897–912.e20. doi: 10.1016/j.cell.2018.12.036
68. Hazlewood JE, Dumenil T, Le TT, Slonchak A, Kazakoff SH, Patch AM, et al. Injection site vaccinology of a recombinant vaccinia-based vector reveals diverse innate immune signatures. PloS Pathog. (2021) 17:e1009215. doi: 10.1371/journal.ppat.1009215
69. Rusinova I, Forster S, Yu S, Kannan A, Masse M, Cumming H, et al. INTERFEROME v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res. (2013) 41:D1040–D6. doi: 10.1093/nar/gks1215
70. Schroder WA, Le TTT, Major L, Street S, Gardner J, Lambley E, et al. A physiological function of inflammation-associated serpinB2 is regulation of adaptive immunity. J Immunol. (2010) 184:2663–70. doi: 10.4049/jimmunol.0902187
71. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods. (2001) 25:402–8. doi: 10.1006/meth.2001.1262
72. Amarilla AA, Sng JDJ, Parry R, Deerain JM, Potter JR, Setoh YX, et al. A versatile reverse genetics platform for SARS-CoV-2 and other positive-strand RNA viruses. Nat Commun. (2021) 12:3431. doi: 10.1038/s41467-021-23779-5
73. Mills RJ, Humphrey SJ, Fortuna PRJ, Lor M, Foster SR, Quaife-Ryan GA, et al. BET inhibition blocks inflammation-induced cardiac dysfunction and SARS-CoV-2 infection. Cell. (2021) 184:2167–82.e22. doi: 10.1016/j.cell.2021.03.026
74. Paudel R, Fusi L, Schmidt M. The MEK5/ERK5 pathway in health and disease. Int J Mol Sci. (2021) 22:7594. doi: 10.3390/ijms22147594
75. Gomez N, Erazo T, Lizcano JM. ERK5 and cell proliferation: nuclear localization is what matters. Front Cell Dev Biol. (2016) 4:105. doi: 10.3389/fcell.2016.00105
76. Wang T, Zhang X, Li JJ. The role of NF-κB in the regulation of cell stress responses. Int Immunopharmacol. (2002) 2:1509–20. doi: 10.1016/S1567-5769(02)00058-9
77. Bartolini D, Stabile AM, Vacca C, Pistilli A, Rende M, Gioiello A, et al. Endoplasmic reticulum stress and NF-kB activation in SARS-CoV-2 infected cells and their response to antiviral therapy. IUBMB Life. (2022) 74:93–100. doi: 10.1002/iub.2537
78. Aegerter H, Lambrecht BN, Jakubzick CV. Biology of lung macrophages in health and disease. Immunity. (2022) 55:1564–80. doi: 10.1016/j.immuni.2022.08.010
79. Ural BB, Yeung ST, Damani-Yokota P, Devlin JC, de Vries M, Vera-Licona P, et al. Identification of a nerve-associated, lung-resident interstitial macrophage subset with distinct localization and immunoregulatory properties. Sci Immunol. (2020) 5:eaax8756. doi: 10.1126/sciimmunol.aax8756
80. Prow TW, Chen X, Prow NA, Fernando GJP, Tan CSE, Raphael AP, et al. Nanopatch-targeted skin vaccination against west nile virus and chikungunya virus in mice. Small. (2010) 6:1776–84. doi: 10.1002/smll.201000331
81. Arce VM, Costoya JA. SARS-CoV-2 infection in K18-ACE2 transgenic mice replicates human pulmonary disease in COVID-19. Cell Mol Immunol. (2021) 18:513–4. doi: 10.1038/s41423-020-00616-1
82. Winkler ES, Bailey AL, Kafai NM, Nair S, McCune BT, Yu J, et al. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat Immunol. (2020) 21:1327–35. doi: 10.1038/s41590-020-0778-2
83. Guimond SE, Mycroft-West CJ, Gandhi NS, Tree JA, Le TT, Spalluto CM, et al. Synthetic heparan sulfate mimetic pixatimod (PG545) potently inhibits SARS-coV-2 by disrupting the spike-ACE2 interaction. ACS Cent Sci. (2022) 8:527–45. doi: 10.1021/acscentsci.1c01293
84. Robinot R, Hubert M, de Melo GD, Lazarini F, Bruel T, Smith N, et al. SARS-CoV-2 infection induces the dedifferentiation of multiciliated cells and impairs mucociliary clearance. Nat Commun. (2021) 12:4354. doi: 10.1038/s41467-021-24521-x
85. Garcia-Nicolas O, Godel A, Zimmer G, Summerfield A. Macrophage phagocytosis of SARS-CoV-2-infected cells mediates potent plasmacytoid dendritic cell activation. Cell Mol Immunol. (2023) 20:835–49. doi: 10.1038/s41423-023-01039-4
86. Van der Sluis RM, Holm CK, Jakobsen MR. Plasmacytoid dendritic cells during COVID-19: Ally or adversary? Cell Rep. (2022) 40:111148. doi: 10.1016/j.celrep.2022.111148
87. Contoli M, Papi A, Tomassetti L, Rizzo P, Vieceli Dalla Sega F, Fortini F, et al. Blood interferon-alpha levels and severity, outcomes, and inflammatory profiles in hospitalized COVID-19 patients. Front Immunol. (2021) 12:648004. doi: 10.3389/fimmu.2021.648004
88. Minkoff JM, tenOever B. Innate immune evasion strategies of SARS-CoV-2. Nat Rev Microbiol. (2023) 21:178–94. doi: 10.1038/s41579-022-00839-1
89. Bastard P, Zhang Q, Zhang SY, Jouanguy E, Casanova JL. Type I interferons and SARS-CoV-2: from cells to organisms. Curr Opin Immunol. (2022) 74:172–82. doi: 10.1016/j.coi.2022.01.003
90. Paolini R, Bernardini G, Molfetta R, Santoni A. NK cells and interferons. Cytokine Growth Factor Rev. (2015) 26:113–20. doi: 10.1016/j.cytogfr.2014.11.003
91. Acharya D, Liu G, Gack MU. Dysregulation of type I interferon responses in COVID-19. Nat Rev Immunol. (2020) 20:397–8. doi: 10.1038/s41577-020-0346-x
92. Lee MJ, Blish CA. Defining the role of natural killer cells in COVID-19. Nat Immunol. (2023) 24:1628–38. doi: 10.1038/s41590-023-01560-8
93. Chavakis T, Mitroulis I, Hajishengallis G. Hematopoietic progenitor cells as integrative hubs for adaptation to and fine-tuning of inflammation. Nat Immunol. (2019) 20:802–11. doi: 10.1038/s41590-019-0402-5
94. Schulte-Schrepping J, Reusch N, Paclik D, Bassler K, Schlickeiser S, Zhang B, et al. Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell. (2020) 182:1419–40. doi: 10.1016/j.cell.2020.08.001
95. Romo-Rodriguez R, Gutierrez-de Anda K, Lopez-Blanco JA, Zamora-Herrera G, Cortes-Hernandez P, Santos-Lopez G, et al. Chronic comorbidities in middle aged patients contribute to ineffective emergency hematopoiesis in covid-19 fatal outcomes. Arch Med Res. (2023) 54:197–210. doi: 10.1016/j.arcmed.2023.03.003
96. Townsend L, Dyer AH, Naughton A, Imangaliyev S, Dunne J, Kiersey R, et al. Severe COVID-19 is characterised by inflammation and immature myeloid cells early in disease progression. Heliyon. (2022) 8:e09230. doi: 10.1016/j.heliyon.2022.e09230
97. Yu T, Gan S, Zhu Q, Dai D, Li N, Wang H, et al. Modulation of M2 macrophage polarization by the crosstalk between Stat6 and Trim24. Nat Commun. (2019) 10:4353. doi: 10.1038/s41467-019-12384-2
98. Vanneste D, Bai Q, Hasan S, Peng W, Pirottin D, Schyns J, et al. MafB-restricted local monocyte proliferation precedes lung interstitial macrophage differentiation. Nat Immunol. (2023) 24:827–40. doi: 10.1038/s41590-023-01468-3
99. Schyns J, Bai Q, Ruscitti C, Radermecker C, De Schepper S, Chakarov S, et al. Non-classical tissue monocytes and two functionally distinct populations of interstitial macrophages populate the mouse lung. Nat Commun. (2019) 10:3964. doi: 10.1038/s41467-019-11843-0
100. Kokkotis G, Kitsou K, Xynogalas I, Spoulou V, Magiorkinis G, Trontzas I, et al. Systematic review with meta-analysis: COVID-19 outcomes in patients receiving anti-TNF treatments. Aliment Pharmacol Ther. (2022) 55:154–67. doi: 10.1111/apt.16717
101. Song H, Lei N, Zeng L, Li X, Jiang C, Feng Q, et al. Mendelian randomization analysis identified tumor necrosis factor as being associated with severe COVID-19. Front Pharmacol. (2023) 14:1171404. doi: 10.3389/fphar.2023.1171404
102. Labzin LI, Schroder K. Stoking inflammasome fires in the COVID-19 neighborhood. Cell Host Microbe. (2023) 31:168–70. doi: 10.1016/j.chom.2023.01.008
103. Sanchez-de Prada L, Gorgojo-Galindo O, Fierro I, Martinez-Garcia AM, de Quintana GS, Gutierrez-Bustillo R, et al. Time evolution of cytokine profiles associated with mortality in COVID-19 hospitalized patients. Front Immunol. (2022) 13:946730. doi: 10.3389/fimmu.2022.946730
104. Arunachalam PS, Wimmers F, Mok CKP, Perera R, Scott M, Hagan T, et al. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science. (2020) 369:1210–20. doi: 10.1126/science.abc6261
105. Yin J-X, Agbana YL, Sun Z-S, Fei S-W, Zhao H-Q, Zhou X-N, et al. Increased interleukin-6 is associated with long COVID-19: a systematic review and meta-analysis. Infect Dis Poverty. (2023) 12:43. doi: 10.1186/s40249-023-01086-z
106. Avdeev SN, Trushenko NV, Tsareva NA, Yaroshetskiy AI, Merzhoeva ZM, Nuralieva GS, et al. Anti-IL-17 monoclonal antibodies in hospitalized patients with severe COVID-19: A pilot study. Cytokine. (2021) 146:155627. doi: 10.1016/j.cyto.2021.155627
107. Lopez-Cortes A, Guerrero S, Ortiz-Prado E, Yumiceba V, Vera-Guapi A, Leon Caceres A, et al. Pulmonary inflammatory response in lethal COVID-19 reveals potential therapeutic targets and drugs in phases III/IV clinical trials. Front Pharmacol. (2022) 13:833174. doi: 10.3389/fphar.2022.833174
108. Mukundan A, Christopher P, Samrachana A, Eduardo I, Xiyue L, Stephanie T, et al. Interleukin-1 receptor antagonist gene (IL1RN) variants modulate the cytokine release syndrome and mortality of SARS-CoV-2. J Infect Dis. (2024) jiae031. doi: 10.1093/infdis/jiae031
109. Alejo A, Ruiz-Argüello MB, Pontejo SM, Fernández de Marco M, Saraiva M, Hernáez B, et al. Chemokines cooperate with TNF to provide protective anti-viral immunity and to enhance inflammation. Nat Commun. (2018) 9:1790. doi: 10.1038/s41467-018-04098-8
110. Ramshaw IA, Ramsay AJ, Karupiah G, Rolph MS, Mahalingam S, Ruby JC. Cytokines and immunity to viral infections. Immunol Rev. (1997) 159:119–35. doi: 10.1111/j.1600-065X.1997.tb01011.x
111. Zaid A, Rulli NE, Rolph MS, Suhrbier A, Mahalingam S. Disease exacerbation by etanercept in a mouse model of alphaviral arthritis and myositis. Arthritis Rheum. (2011) 63:488–91. doi: 10.1002/art.30112
112. Long MD, Parlett L, Lewis JD, Haynes K, Adimadhyam S, Hou L, et al. Corticosteroids but not Anti-TNF Are Associated With Increased COVID-19 Complications in Patients With Inflammatory Bowel Disease. Inflammation Bowel Dis. (2023). doi: 10.1093/ibd/izad176
113. Ullah TR, Johansen MD, Balka KR, Ambrose RL, Gearing LJ, Roest J, et al. Pharmacological inhibition of TBK1/IKKepsilon blunts immunopathology in a murine model of SARS-CoV-2 infection. Nat Commun. (2023) 14:5666. doi: 10.1038/s41467-023-41381-9
114. Sachet M, Liang YY, Oehler R. The immune response to secondary necrotic cells. Apoptosis. (2017) 22:1189–204. doi: 10.1007/s10495-017-1413-z
115. Ge Y, Huang M, Yao Y-m. Efferocytosis and its role in inflammatory disorders. Front Cell Dev Biol. (2022) 10:839248. doi: 10.3389/fcell.2022.839248
116. Razi S, Yaghmoorian Khojini J, Kargarijam F, Panahi S, Tahershamsi Z, Tajbakhsh A, et al. Macrophage efferocytosis in health and disease. Cell Biochem Funct. (2023) 41:152–65. doi: 10.1002/cbf.3780
117. Budhraja A, Basu A, Gheware A, Abhilash D, Rajagopala S, Pakala S, et al. Molecular signature of postmortem lung tissue from COVID-19 patients suggests distinct trajectories driving mortality. Dis Model Mech. (2022) 15:dmm049572. doi: 10.1242/dmm.049572
118. Maufak MM, Khan G, Purushothaman A, Ahamed F, Jallo MK, Rizwan M, et al. Ameliorating effect of erythropoietin in a severe case of COVID-19: case report. Pan Afr Med J. (2022) 43:129. doi: 10.11604/pamj.2022.43.129.35014
119. De Ponti FF, Scott CL. A breath of fresh macrophages ameliorates inflammation in the hypoxic lung. Nat Immunol. (2022) 23:820–2. doi: 10.1038/s41590-022-01214-1
120. Mirchandani AS, Jenkins SJ, Bain CC, Sanchez-Garcia MA, Lawson H, Coelho P, et al. Hypoxia shapes the immune landscape in lung injury and promotes the persistence of inflammation. Nat Immunol. (2022) 23:927–39. doi: 10.1038/s41590-022-01216-z
121. Bénard A, Jacobsen A, Brunner M, Krautz C, Klösch B, Swierzy I, et al. Interleukin-3 is a predictive marker for severity and outcome during SARS-CoV-2 infections. Nat Commun. (2021) 12:1112. doi: 10.1038/s41467-021-21310-4
122. Evgen'ev MB, Onikienko SB, Chuvakova LN, Garbuz DG, Zatsepina OG. The role of Hsp70 in adaptation to adverse conditions and its possible medical application. Front Biosci. (2023) 28:25. doi: 10.31083/j.fbl2802025
123. Rico-Llanos G, Porras-Perales Ó, Escalante S, Vázquez-Calero DB, Valiente L, Castillo MI, et al. Cellular stress modulates severity of the inflammatory response in lungs via cell surface BiP. Front Immunol. (2022) 13:1054962. doi: 10.3389/fimmu.2022.1054962
124. Duan Y, Huang S, Yang J, Niu P, Gong Z, Liu X, et al. HspA1A facilitates DNA repair in human bronchial epithelial cells exposed to Benzo[a]pyrene and interacts with casein kinase 2. Cell Stress Chaperones. (2014) 19:271–9. doi: 10.1007/s12192-013-0454-7
125. Chen J, Dai L, Wang T, He J, Wang Y, Wen F. The elevated CXCL5 levels in circulation are associated with lung function decline in COPD patients and cigarette smoking-induced mouse model of COPD. Ann Med. (2019) 51:314–29. doi: 10.1080/07853890.2019.1639809
126. Thompson CA, Burcham PC. Genome-wide transcriptional responses to acrolein. Chem Res Toxicol. (2008) 21:2245–56. doi: 10.1021/tx8001934
127. Morrell ED, Grazioli S, Hung C, Kajikawa O, Kosamo S, Stapleton RD, et al. Alveolar CCN1 is associated with mechanical stretch and acute respiratory distress syndrome severity. Am J Physiology-Lung Cell Mol Physiol. (2020) 319:L825–L32. doi: 10.1152/ajplung.00073.2020
128. Kim DI, Song MK, Kim HI, Han KM, Lee K. Diesel exhaust particulates induce neutrophilic lung inflammation by modulating endoplasmic reticulum stress-mediated CXCL1/KC expression in alveolar macrophages. Molecules. (2020) 25:6046. doi: 10.3390/molecules25246046
129. Chen P, Jiang P, Chen J, Yang Y, Guo X. XIST promotes apoptosis and the inflammatory response in CSE-stimulated cells via the miR-200c-3p/EGR3 axis. BMC Pulm Med. (2021) 21:215. doi: 10.1186/s12890-021-01582-8
130. Yan F, Wu Y, Liu H, Wu Y, Shen H, Li W. ATF3 is positively involved in particulate matter-induced airway inflammation in vitro and in vivo. Toxicol Lett. (2018) 287:113–21. doi: 10.1016/j.toxlet.2018.01.022
131. Niethamer TK, Levin LI, Morley MP, Babu A, Zhou S, Morrisey EE. Atf3 defines a population of pulmonary endothelial cells essential for lung regeneration. Elife. (2023) 12:e83835. doi: 10.7554/eLife.83835
132. Madaniyazi L, Li S, Li S, Guo Y. Candidate gene expression in response to low-level air pollution. Environ Int. (2020) 140:105610. doi: 10.1016/j.envint.2020.105610
133. Ghosh S, Durgvanshi S, Han SS, Bhaskar R, Sinha JK. Therapeutics for the management of cytokine release syndrome in COVID-19. Curr Top Med Chem. (2023) 23:128–42. doi: 10.2174/1568026622666220707114121
134. Mangalmurti N, Hunter CA. Cytokine storms: understanding COVID-19. Immunity. (2020) 53:19–25. doi: 10.1016/j.immuni.2020.06.017
135. Carolin A, Yan K, Bishop CR, Tang B, Nguyen W, Rawle DJ, et al. Tracking inflammation resolution signatures in lungs after SARS-CoV-2 omicron BA.1 infection of K18-hACE2 mice. bioRxiv. (2024). doi: 10.1101/2024.03.12.584739
136. Severa M, Diotti RA, Etna MP, Rizzo F, Fiore S, Ricci D, et al. Differential plasmacytoid dendritic cell phenotype and type I Interferon response in asymptomatic and severe COVID-19 infection. PloS Pathog. (2021) 17:e1009878. doi: 10.1371/journal.ppat.1009878
137. Li S, Zhang Y, Guan Z, Li H, Ye M, Chen X, et al. SARS-CoV-2 triggers inflammatory responses and cell death through caspase-8 activation. Signal Transduction Targeted Ther. (2020) 5:235. doi: 10.1038/s41392-020-00334-0
138. Fredman G, Khan S. Specialized pro-resolving mediators enhance the clearance of dead cells. Immunol Rev. (2023) 319:151–7. doi: 10.1111/imr.13278
139. Detree C, Gallardo-Escarate C. Single and repetitive microplastics exposures induce immune system modulation and homeostasis alteration in the edible mussel Mytilus galloprovincialis. Fish Shellfish Immunol. (2018) 83:52–60. doi: 10.1016/j.fsi.2018.09.018
140. Abarghouei S, Hedayati A, Raeisi M, Hadavand BS, Rezaei H, Abed-Elmdoust A. Size-dependent effects of microplastic on uptake, immune system, related gene expression and histopathology of goldfish (Carassius auratus). Chemosphere. (2021) 276:129977. doi: 10.1016/j.chemosphere.2021.129977
141. Yin J, Long Y, Xiao W, Liu D, Tian Q, Li Y, et al. Ecotoxicology of microplastics in Daphnia: A review focusing on microplastic properties and multiscale attributes of Daphnia. Ecotoxicology Environ Saf. (2023) 249:114433. doi: 10.1016/j.ecoenv.2022.114433
142. Nunes KP, de Oliveira AA. HSP70: from signaling mechanisms to therapeutics. Biomolecules. (2023) 13:1141. doi: 10.3390/biom13071141
143. Rébé C, Ghiringhelli F, Garrido C. Can the hyperthermia-mediated heat shock factor/heat shock protein 70 pathway dampen the cytokine storm during SARS-CoV-2 infection? Br J Pharmacol. (2022) 179:4910–6. doi: 10.1111/bph.15343
144. Alijagic A, Hedbrant A, Persson A, Larsson M, Engwall M, Särndahl E. NLRP3 inflammasome as a sensor of micro- and nanoplastics immunotoxicity. Front Immunol. (2023) 14:1178434. doi: 10.3389/fimmu.2023.1178434
145. Xu S, Zhang Y, Liu X, Liu H, Zou X, Zhang L, et al. Nr4a1 marks a distinctive ILC2 activation subset in the mouse inflammatory lung. BMC Biol. (2023) 21:218. doi: 10.1186/s12915-023-01690-3
146. Strickland AB, Chen Y, Sun D, Shi M. Alternatively activated lung alveolar and interstitial macrophages promote fungal growth. iScience. (2023) 26:106717. doi: 10.1016/j.isci.2023.106717
147. Zhu T, Brown AP, Cai LP, Quon G, Ji H. Single-cell RNA-seq analysis reveals lung epithelial cell type-specific responses to HDM and regulation by tet1. Genes (Basel). (2022) 13:880. doi: 10.3390/genes13050880
148. Zheng Q, Zou Y, Teng P, Chen Z, Wu Y, Dai X, et al. Mechanosensitive channel PIEZO1 senses shear force to induce KLF2/4 expression via caMKII/MEKK3/ERK5 axis in endothelial cells. Cells. (2022) 11:2191. doi: 10.3390/cells11142191
149. Chrysanthopoulou A, Antoniadou C, Natsi AM, Gavriilidis E, Papadopoulos V, Xingi E, et al. Down-regulation of KLF2 in lung fibroblasts is linked with COVID-19 immunofibrosis and restored by combined inhibition of NETs, JAK-1/2 and IL-6 signaling. Clin Immunol. (2023) 247:109240. doi: 10.1016/j.clim.2023.109240
150. Xu S, Liu Y, Ding Y, Luo S, Zheng X, Wu X, et al. The zinc finger transcription factor, KLF2, protects against COVID-19 associated endothelial dysfunction. Signal Transduct Target Ther. (2021) 6:266. doi: 10.1038/s41392-021-00690-5
151. Pang J, Koh TJ. Proliferation of monocytes and macrophages in homeostasis, infection, injury and disease. J Leukoc Biol. (2023) 114:532–46. doi: 10.1093/jleuko/qiad093
152. Sun Q, Xia Y, Qin H, Zhang W, Wang J, Ning Y, et al. MEF2 intervened LPS-induced acute lung injury by binding to KLF2 promoter and modulating macrophage phenotype. Int Immunopharmacol. (2022) 108:108873. doi: 10.1016/j.intimp.2022.108873
153. Rozman U, Kalčíková G. Seeking for a perfect (non-spherical) microplastic particle – The most comprehensive review on microplastic laboratory research. J Hazardous Materials. (2022) 424:127529. doi: 10.1016/j.jhazmat.2021.127529
154. WT J. Health effects of fossil fuel–derived endocrine disruptors. New Engl J Med. (2024) 390:922–33. doi: 10.1056/NEJMra2300476
155. Liu K, Wang X, Fang T, Xu P, Zhu L, Li D. Source and potential risk assessment of suspended atmospheric microplastics in Shanghai. Sci Total Environ. (2019) 675:462–71. doi: 10.1016/j.scitotenv.2019.04.110
156. GGGI. Rwanda: lessons learnt from a pioneer in the fight against plastic pollution (2022). Available online at: https://gggi.org/rwanda-lessons-learnt-from-a-pioneer-in-the-fight-against-plastic-pollution/ (Accessed Nov 2023).
157. Gutiérrez-Avila I, Riojas-Rodríguez H, Colicino E, Rush J, Tamayo-Ortiz M, Borja-Aburto VH, et al. Short-term exposure to PM2.5 and 1.5 million deaths: a time-stratified case-crossover analysis in the Mexico City Metropolitan Area. Environ Health. (2023) 22:70. doi: 10.1186/s12940-023-01024-4
158. Yang W, Jannatun N, Zeng Y, Liu T, Zhang G, Chen C, et al. Impacts of microplastics on immunity. Front Toxicol. (2022) 4:956885. doi: 10.3389/ftox.2022.956885
159. Liaquat A, Kashif A, Rathi S, Raza A. Microplastics in freshly fallen snow: How may it adversely impact human health and exacerbate the COVID-19 crisis? Ann Med Surg (Lond). (2022) 80:104276. doi: 10.1016/j.amsu.2022.104276
160. Fiorito S, Soligo M, Gao Y, Ogulur I, Akdis Cezmi A, Bonini S. Is the epithelial barrier hypothesis the key to understanding the higher incidence and excess mortality during COVID-19 pandemic? The case of Northern Italy. Allergy. (2022) 77:1408–17. doi: 10.1111/all.15239
161. Luo T, Wang D, Zhao Y, Li X, Yang G, Jin Y. Polystyrene microplastics exacerbate experimental colitis in mice tightly associated with the occurrence of hepatic inflammation. Sci Total Environ. (2022) 844:156884. doi: 10.1016/j.scitotenv.2022.156884
162. Yan Z, Liu Y, Zhang T, Zhang F, Ren H, Zhang Y. Response to comment on “Analysis of microplastics in human feces reveals a correlation between fecal microplastics and inflammatory bowel disease status”. Environ Sci Technol. (2022) 56:12779–80. doi: 10.1021/acs.est.2c05327
163. Liu S, Li H, Wang J, Wu B, Guo X. Polystyrene microplastics aggravate inflammatory damage in mice with intestinal immune imbalance. Sci Total Environ. (2022) 833:155198. doi: 10.1016/j.scitotenv.2022.155198
164. Taş BM, Tuna A, Başaran Kankılıç G, Koçak FM, Şencan Z, Cömert E, et al. Role of microplastics in chronic rhinosinusitis without nasal polyps. Laryngoscope. (2023) 134:1077. doi: 10.1002/lary.30926
Keywords: microplastics, SARS-CoV-2, COVID-19, RNA-Seq, inflammation, mouse
Citation: Bishop CR, Yan K, Nguyen W, Rawle DJ, Tang B, Larcher T and Suhrbier A (2024) Microplastics dysregulate innate immunity in the SARS-CoV-2 infected lung. Front. Immunol. 15:1382655. doi: 10.3389/fimmu.2024.1382655
Received: 06 February 2024; Accepted: 24 April 2024;
Published: 13 May 2024.
Edited by:
Avijit Dutta, Chang Gung University, TaiwanReviewed by:
Shetty Ravi Dyavar, Adicet Bio Inc, United StatesCopyright © 2024 Bishop, Yan, Nguyen, Rawle, Tang, Larcher and Suhrbier. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andreas Suhrbier, Andreas.Suhrbier@qimrberghofer.edu.au
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.