
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 27 March 2024
Sec. Nutritional Immunology
Volume 15 - 2024 | https://doi.org/10.3389/fimmu.2024.1380476
Obesity and chronic low-grade inflammation, often occurring together, significantly contribute to severe metabolic and inflammatory conditions like type 2 diabetes (T2D), cardiovascular disease (CVD), and cancer. A key player is elevated levels of gut dysbiosis-associated lipopolysaccharide (LPS), which disrupts metabolic and immune signaling leading to metabolic endotoxemia, while short-chain fatty acids (SCFAs) beneficially regulate these processes during homeostasis. SCFAs not only safeguard the gut barrier but also exert metabolic and immunomodulatory effects via G protein-coupled receptor binding and epigenetic regulation. SCFAs are emerging as potential agents to counteract dysbiosis-induced epigenetic changes, specifically targeting metabolic and inflammatory genes through DNA methylation, histone acetylation, microRNAs (miRNAs), and long non-coding RNAs (lncRNAs). To assess whether SCFAs can effectively interrupt the detrimental cascade of obesity and inflammation, this review aims to provide a comprehensive overview of the current evidence for their clinical application. The review emphasizes factors influencing SCFA production, the intricate connections between metabolism, the immune system, and the gut microbiome, and the epigenetic mechanisms regulated by SCFAs that impact metabolism and the immune system.
Obesity and inflammation are the two main causes of chronic diseases, which are considered to be the biggest global health issue nowadays (1). The long-term effects of a constant inflammatory state comprise a multitude of chronic inflammatory disorders that significantly increase mortality. These can develop into common conditions like heart disease and cancer, which are all tightly linked to inflammation and obesity (2, 3). Other common chronic disorders are irritable bowel syndrome (IBS), which is characterized by gut dysbiosis and increased activation of immune cells (4) and inflammatory bowel disease (IBD), a group of diseases distinguished by chronic inflammation that includes Crohn’s disease and ulcerative colitis (5). Importantly, dysregulations in the intestinal microbiome and immune system also affect other organs, such as the brain, promoting the development of neurodegenerative disorders (6). Moreover, there is currently no cure for chronic diseases, although medical advances in disease management significantly decreased mortality in the past decade (7). Still, 3 out of 5 people die due to chronic inflammatory diseases worldwide (8). Not only does this constitute a huge health care and economic burden now, but prospects are not very optimistic with the rising trend of prevalence in chronic diseases (9). Hereby, one of the main difficulties of chronic inflammation is the silent progression in which it imperceptibly alters the metabolism and tissue homeostasis of the host, favoring the development of disease (10). One of the consequences can be obesity.
Obesity is considered to be the biggest global epidemic affecting individuals of every age (11). It is also strongly indicated to increase the risk of developing chronic diseases, such as depression, type 2 diabetes (T2D), cardiovascular disease (CVD), and cancer (12). All these are conditions that affect a tremendous amount of the world population and somewhat characterize modern society. The psychosocial costs caused by obesity and difficulties linked to associated comorbidities pose a significant economic burden. Prospects appear to be even worse as childhood obesity results in the same disorders with even earlier onset (13). Weight gain is not only a potential consequence of inflammation but commonly induces obesity-associated inflammation (14).
The most important factors of the vicious cycle between obesity and inflammation are alterations in the adipose tissue, interorgan cross-talk, and most importantly, obesity-induced lipopolysaccharide (LPS) leakage (15). The adipose tissue of obese people is characterized by an increased number or size of adipocytes (16). It is also associated with high infiltration rates of macrophages that are predominantly of the pro-inflammatory M1 phenotype (17). The resulting depletion of interleukin (IL)-10- and TGF-β-producing M2 macrophages does not only have consequences for immunoregulation but also for maintaining insulin sensitivity (18). Moreover, the secretion of proinflammatory cytokines and adipokines affect overall metabolic health, the endocrine and immune system via interorgan crosstalk (19). Thus, pro-inflammatory signals derived from adipose tissue can disrupt metabolic and immunologic homeostasis in other tissues as well, resulting in a systemic chronic inflammatory condition. Ultimately, continuous inflammation, metabolic dysfunction, and tissue damage strongly favor the development of serious diseases.
Although there are many ways in which adiposity and inflammation can be caused and promote the state of one another, gut-derived LPS is accounted to be one of the main causes, if not the main cause, of obesity-linked low-grade inflammation (20). In the case of gut homeostasis, a well-functioning epithelial barrier prevents bacterial components from leaving the gut and entering the bloodstream. Common observations made in obese individuals are alterations in the gut microbiome, increased gut barrier permeability, and higher LPS levels (15). Augmented LPS plasma levels have been linked to obesity-associated low-grade inflammation, as LPS can escape through a weakened gut barrier, travel through the bloodstream to other organs and trigger their local immune cells (21, 22). The pro-inflammatory stimuli in form of LPS prompts residing macrophages to polarize into M1 macrophages and further recruits circulating monocytes, which accelerates the disruptions in tissue homeostasis (15). Thus, serious impairments of the immune system and metabolism are believed to originate from the probably most important influencer of the immune system and systemic metabolism of the human organism – the gut microbiome.
The gut microbiota consists of a large number of microorganisms, which include bacteria, viruses, fungi, and archaea. The number of bacterial genes in the human body exceeds the host genome a hundred-fold, the gut microbiome being the largest and most diverse microbiome of the human body (23). It is dominated by the phyla Firmicutes (now referred to as Bacillota as determined by List of Prokaryotic names with Standing in Nomenclature (LPSN)), Bacteroidetes, Proteobacteria and Actinobacteria (now referred to as Bacteroidota, Pseudomonadota and Actinomycetota, respectively, according to LPSN), with Firmicutes and Bacteroides being the most abundant ones (24). Although it has been long known that bacteria play a crucial role in digestion and vitamin production, their essential role in regulating host metabolism and the immune system has become increasingly evident over the last few decades. For this reason, restoring gut homeostasis shows great potential in combating chronic inflammation and obesity. Gut commensals have a great share in maintaining gut homeostasis, as they impede pathogen growth via inducing colonization resistance and produce metabolites with antimicrobial activity (25). On top of that, bacterial metabolites also regulate host metabolism, such as tryptophan, which binds to the aryl hydrocarbon receptor (AHR), responsible for the transcription of phase I and II metabolizing enzymes (26). To date, certain bacterial residents were found to be crucial in promoting proper gut barrier function and an immunosuppressive environment. SCFA-producing bacteria play an important role in this context.
The three main SCFAs in the intestine are acetate, butyrate and propionate. SCFAs are mainly produced via fiber fermentation, although metabolic pathways of organic and amino acids can also result in SCFA production (27, 28). Although bacterial metabolism is highly strain-specific, generally, strains belonging to the phylum Bacteroidetes mainly produce acetate and propionate while butyrate is mainly produced by members of the Firmicutes phyla (27). Butyrate plays a particularly critical role in maintaining a healthy gut barrier as it is the primary energy source of intestinal epithelial cells. It also increases mucin production, and while it stimulates the proliferation of healthy colonocytes, its effect on cancerous colonocytes is the opposite (29). Thus SCFAs appear to have a sensitive role in regulating gut health. Apart from playing an important role as part of the physical barrier in innate immunity, SCFAs have a great impact on adaptive immunity, as well. For instance, SCFAs induce the differentiation and proliferation of regulatory T cells (Tregs), which are crucial for immunologic homeostasis (30, 31). SCFAs also affect B lymphocyte energy metabolism, shifting antibody production to immunoglobulin (Ig) A and modulating its activity towards commensal bacteria (32, 33). IgA is a key player in humoral mucosal immunity as it poses part of the first line of defense by binding bacterial toxins and fighting pathogens. Importantly, SFCAs also affect innate immune mechanisms, as butyrate and propionate were shown to inhibit mast cell activation (34). Thus, the pleiotropic role of SCFAs modulates both immune defenses and immunological non-reactiveness towards autoantigens, which makes them an essential factor in proper immunoregulation.
Furthermore, SCFAs also regulate host metabolism, as SCFAs can be recognized by several G protein-coupled receptors (GPRs), which are responsible for immune and metabolic regulation (35). This is crucial for limiting food intake and influencing glucose and lipid metabolism, as gut-derived SCFAs act on GPRs resulting in hormone production reaching the brain via the gut-brain axis (36). This way, SCFAs are able to impact appetite and insulin sensitivity. However, gut dysbiosis is not only associated with impairments in these regulations but also with LPS-induced metabolic endotoxemia further exacerbating energy intake and blood glucose levels (37). These mechanisms are also partly induced by epigenetic modifications (38).
Epigenetics modifications affect the transcription of genes without altering the genetic code, which are mainly conveyed by, but not limited to, DNA methylation, posttranslational histone modifications and different types of non-coding RNAs, such as microRNAs (miRNAs) and long non-coding RNAs (lncRNAs) (39). These epigenetic mechanisms globally regulate the expression of genes involved in metabolic and cellular inflammatory responses and are key in maintaining tissue homeostasis. Not only do epigenetics regulate these processes but epigenetic regulators are also strongly affected by metabolites, which is known as ‘metabolo-epigenetics’. SCFAs, particularly butyrate, has been shown to regulate immune cell differentiation and immunological pathways as well as a multitude of metabolic genes associated with obesity (40). This is highly relevant from a clinical point of view since metabolic and immunologic disruptions upon gut dysbiosis might be reversible. SCFA-induced epigenetic modifications might therefore be not merely a reasonable approach for preventing obesity and inflammation-associated pathophysiology but it could potentially reverse harmful developments initially caused by gut dysbiosis. Several murine and human studies show significant correlations between SCFAs and health status, but to translate these striking implications into clinical application, it is important to understand how obesity and inflammation can be practically and controllably modulated via epigenetic manipulation by SCFAs. Since SCFAs are implicated in several health-promoting processes, it would be more than reasonable to exploit their clinical potential by fully embracing their pleiotropic effects on overall health. This will also allow targeting of systemic issues of obesity and inflammation, which is urged by the global health situation.
The aim of this paper, therefore, is to explore the clinical potential of SCFAs, based on the most recent findings and current investigations in this field. To get a realistic picture of possible SCFA manipulation, the first part of this review focuses on the current knowledge about gut-altering factors affecting SCFA production. In the following, the complexity of the bidirectional interactions between metabolism and inflammation and the role of SCFAs in this will be thoroughly discussed. Hereby, both epigenetic-driven and independent activity of SCFAs is reviewed.
The prevalence of gut dysbiosis-associated diseases tremendously increased in the last decades, which can be attributed to the Western diet, sedentary lifestyle, and increased use of antibiotics (41). The importance of early microbial exposure and the danger of increasing sterility in developing an intact immune system has been already stressed in 1989 in Strachan’s hygiene hypothesis (42). Although this notion remains valid, the knowledge about particular influences on the gut microbiota and their impact on overall health becomes continuously deeper and better established. Since SCFA production depends both on the number of bacteria producing SCFAs and the availability of necessary nutritional substrates, it is important to understand the effect of different types of diets and how other factors can cause a shift in the gut microbiota composition. Although age, genetics, environmental and maternal factors are also involved in shaping the gut microbiome (43), this section will merely focus on the most important controllable factors altering SCFA production (Figure 1). This way, the clinical value of SCFA treatment can be realistically assessed.
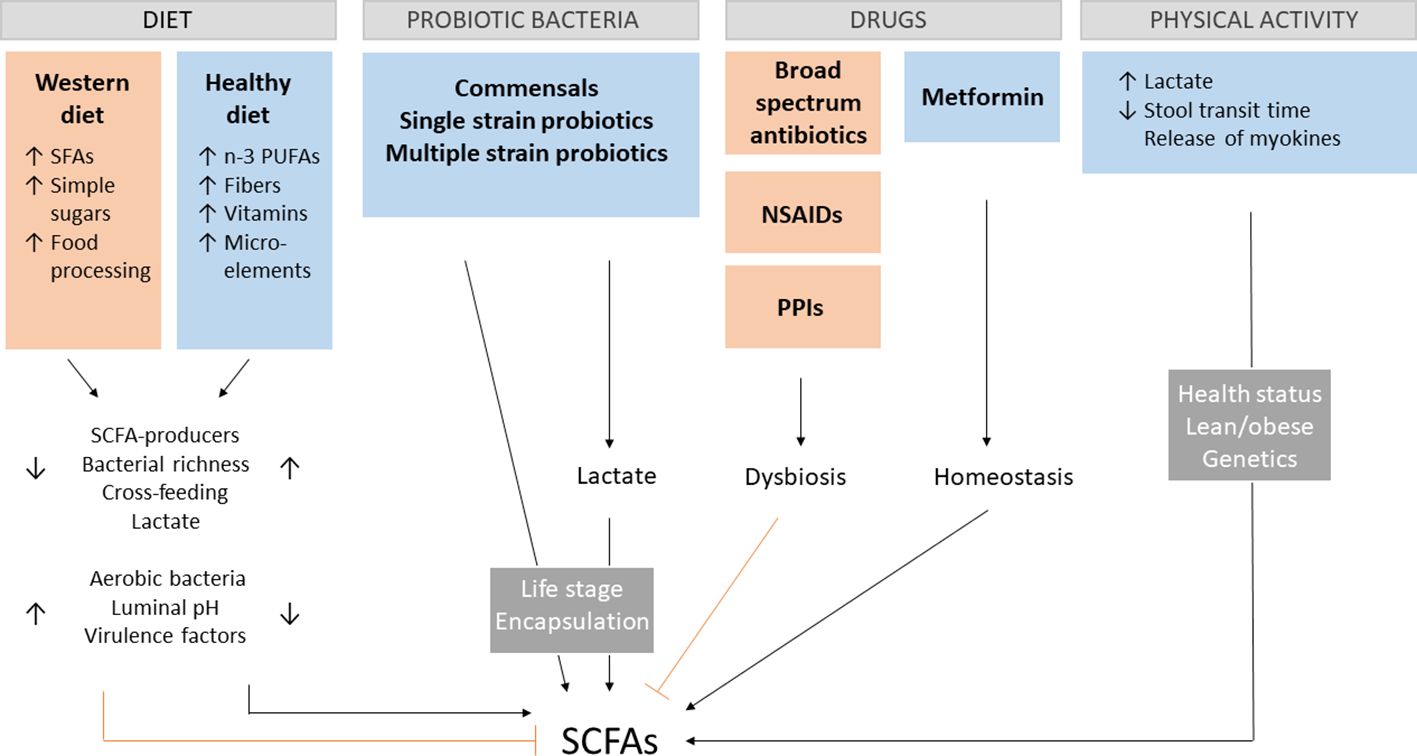
Figure 1 SCFA-influencing factors. Different aspects of the Western diet contribute to dysregulations in the gut environment, leading to the decrease of short-chain fatty acids (SCFAs). A positive impact on SCFA production has a diet rich in n-3 polyunsaturated fatty acids (PUFAs), fibers vitamins and microelements, by increasing the amount of SCFA-producing bacteria, overall bacterial richness, cross-feeding of species, lactate production and decrease the presence of unbeneficial aerobic bacteria, pH and expression of virulence factors. The opposite effect can be observed for the Western diet rich in saturated fatty acids (SFAs), simple sugars and processed food. The presence of key gut commensals and different kinds of probiotics enhance SCFA production directly or indirectly by lactate production with their effectiveness depending on the age of administration and encapsulation. Broad spectrum antibiotics, non-steroidal anti-inflammatory drugs (NSAIDs) and proton pump inhibitors (PPIs) favor gut dysbiosis associated with lower SCFAs levels. The opposite effect is the case for metformin. Physical activity modulates host health by crosstalk between skeletal muscle and the gut via myokines and lactate as well as direct physical stimulation. These effects depend on the health and metabolic state of the host, as well as genetics.
Recent findings emphasize and specify the role of diet, which has a strong influence on the integrity of the intestinal barrier and gut microbiome composition, ultimately affecting SCFA levels. One of the characteristics of the Western Diet is high amounts of dietary fat. A high-fat diet (HFD) is associated with increased imbalances in the gut that also increase the risk for chronic inflammatory diseases (44). In this context, a diet rich in saturated fatty acids (SFAs) is associated with negative health manifestations whereas high amounts of unsaturated fatty acids, especially n-3 polyunsaturated fatty acids (PUFAs), exhibit beneficial health effects (44, 45). SFA interaction with GPRs and Toll-like receptors (TLRs) leads to the activation of pro-inflammatory mediators such as IL-1β and nuclear factor-kappa B (NFκ-B), a process prevented upon PUFA binding (45–47). In fact omega-3 fatty acids were shown to affect lipid metabolism and mediate anti-inflammatory effects via DNA methylation and miRNAs (48, 49), while the conventional Western diet negatively affects gene expression of inflammatory markers and metabolic genes due to epigenetic imprinting that can already take place in utero (50, 51).
Apart from the direct interaction with regulatory receptors, excess SFA intake has negative effects on the gut microbiome, reducing its diversity and richness and promoting the growth of facultative aerobic species which metabolize simple fats and sugars and at the same time decreases the proportion of beneficial anaerobes, including SCFA-producing Akkermansia muciniphila (52). Accordingly, the gut barrier-promoting and pH lowering effects of SCFAs decay as well. Worth noting, SFAs from different sources mediate different effects on gut microbiome diversity (52). Moreover, A. muciniphila and SCFAs were repeatedly shown to protect from HFD-induced obesity and metabolic anomalies, showing the important role of particular microbes in gut homeostasis (53–55), which is promoted by PUFAs (56, 57). Additionally, fibers have been also shown to reverse HFD-induced microbiota changes in diversity (58).
Dietary fibers were shown to counteract the negative effects of a Western Diet directly and by affecting the gut microbiome composition and thus metabolome and mediated epigenetic activity (59). For instance soluble fibers such as psyllium, were shown to hinder glucose and cholesterol absorption and bile salt reabsorption whilst improving the absorption of macro-and micronutrients (60). Fructooligosaccharides (FOS) and galactooligosaccharides (GOS) were shown to remedy gut dysbiosis and the associated gut barrier impairment and systemic inflammation in mice (61). Importantly, the mediated effect also depends on the amount and type of fiber administered and the presence of particular bacteria species. Regarding the amount of fiber, a diet rich in fiber exhibited an immediate effect on the gut microbiome diversity and metabolite production in humans (62), whereas low doses did not affect particular abundances in gut microbes (63, 64). When it comes to the different types of fibers, they can be categorized by fermentability, solubility and viscosity. Due to the fermentability of β-glucans, pectins and inulin, these fibers mainly promote systemic health via gut microbiota modulation (65) whereas non-fermentable fibers like psyllium, which has a high solubility and viscosity, independently improve digestion, glucose homeostasis and cholesterol levels (60). Due to the differently mediated benefits of various kinds of fibers, a plant-based diet comprising different types of dietary fibers is recommended. While complex carbohydrates exhibit various health-promoting effects, the opposite is true for simple carbohydrates.
A diet high in simple sugars, like glucose and fructose, is particularly problematic, as it decreases microbial diversity (66). It also increases the amount of aerobic Proteobacteria whilst decreasing Bacteroidetes, amongst which many SCFA-producers can be found (66). In the same study, high glucose and fructose diet ultimately led to gut barrier impairment resulting in metabolic endotoxemia, even without significant weight gain in mice. The consumption of artificial sweeteners was also shown to induce gut dysbiosis with metabolic consequences for the host (67). On the other hand, a diet low in polysaccharides has been shown to negatively affect bacterial abundance and richness (68). SCFA-producing bacteria are particularly affected due to the deprivation of their nutritional source. Moreover, the loss of one bacterial strain may affect the growth of others due to cross-feeding dependence, as is the case with Ruminococcus gnavus and Bifidobacteria, making SCFA-production by other strains possible via prior starch degradation or lactate production (69, 70). Furthermore, enhanced butyrate production upon starch intake was observed in individuals whose microbiota was enriched in R. gnavus and Clostridium chartatabidum (71), in turn eating behavior was shown to be influenced by the presence of SCFA-producing bacteria, as well (72). This suggests, that effective promotion of SCFA production via dietary intervention strongly depends on gut diversity and the presence of key bacteria species. In fact, prebiotic human intervention studies show strong variability amongst individuals, which only emphasized the issue of the complexity of individual genetic and gut microbiome predispositions (73). This highlights the great need for precise interventions, in which discrete dietary fiber structures and arabinoxylan show great potential and consistency among subjects (74, 75).
Apart from a poor-balanced diet, the Western diet is also characterized by various food additives and a high degree of processing. Recent evidence shows significant changes in gut microbiota upon processed food administration, with consequences in bacterial metabolism and weight gain (76). Artificial sweeteners and emulsifiers play an important role in altering host metabolism via the gut microbiota (67, 77). Not only does this affect metabolism, but it also promotes and exacerbates intestinal inflammation and leads to increased expression of virulence factors (78–80). These effects are believed to arise from gut dysbiosis, which favors the growth of pro-inflammatory species as opposed to immunomodulatory commensals. Processed food is not only enriched in chemical additives but certain food processing deprives the product of vitamins and microelements (81), which also affects SCFA production (82, 83).
Thus, the Western diet seems to decrease SCFA production on various levels, by deprivation of nutritional substrates and co-factors, which also play a role in epigenetic regulation (84), as well as by inducing gut dysbiosis and therefore promoting the growth of other species further exacerbating intestinal imbalance in the gut microbiome composition, metabolism and mucosal immune system. As an antidote, continuous intake of unprocessed food, rich in fibers, PUFAs, vitamins and microelements seems the most feasible to promote SCFA production via the diet.
As previously established, the presence and abundance of particular bacteria are crucial in enhancing SCFA production, due to their own ability to produce these compounds but also the effects they exhibit on other gut members. According to the Food and Agriculture Organization of the United Nations (FAO) and the World Health Organization (WHO), probiotics are ‘live microorganisms which when administered in adequate amounts confer a health benefit on the host’ (85). A probiotic may thereby consist of a single bacterial strain or contain multiple.
Regarding the former, several strains including Lacticaseibacillus rhamnosus GG (LGG), Lactobacillus acidophilus CRL 1014, Lactobacillus gasseri PA 16/8, Ligilactobacillus salivarius JCM 1230, Ligilactobacillus agilis JCM 1048, Bifidobacterium longum SP 07/3 and Bifidobacterium bifidum MF 20/5 were shown to produce SCFAs and lactic acid (86). In the study on the effect of oral consumption of Lactiplantibacillus plantarum P-8 (Lp-8) on human intestinal microbiota, and SCFAs of different aged adults (87), the increase in Bifidobacterium and other beneficial bacteria was found, whereas opportunistic pathogens decreased. Furthermore, a statistically significant increase in acetate and propionate levels in all age groups was observed. Thus single-strain-based probiotics seem to be able to competently promote gut homeostasis. Also commensal bacteria such as Faecalibacterium prausnitzii were shown to effectively promote gut diversity and SCFA production and by this mechanism, shows great potential in improving inflammatory and metabolic diseases (88–92). As previously mentioned, A. muciniphila administration can improve metabolic anomalies caused by gut dysbiosis (53, 54, 93). Thus single-strain-based probiotics seem to be able to competently promote gut homeostasis. Regarding multi-strain probiotics, SCFA production can be targeted on multiple levels. Co-cultures of lactic acid bacteria (LAB) were shown to produce SCFAs and stimulate mucus secretion of colonic epithelial cells (94). Moreover, a probiotic supplement (Symprove™), consisting of the four strains L. acidophilus NCIMB 30175, L. plantarum NCIMB 30173, L. rhamnosus NCIMB 30174 and Enterococcus faecium NCIMB 30176) was shown to competently increase SCFA production in vitro, as well (95). Not only were those strains shown to effectively colonize the colon but also to increase lactate concentrations. Lactate was further used by lactate-metabolizing bacteria resulting in enhanced SCFA production, particularly butyrate. In another study, a cocktail of several lactobacilli strains improved SCFA production, which dampened pathogen-induced pro-inflammatory signaling cascades (96). Moreover, another recent study revealed that more effective fiber fermentation occurs with co-cultures of strains from more than one genera, as in this case concerned Lacticaseibacillus and Bifidobacterium strains (97). This shows that the interaction between bacterial species can have a synergistic effect, emphasizing the important role of cross-feeding in the gut.
Nevertheless, less consistency is observed in human intervention studies. While some studies show significant differences in SCFA levels upon probiotic administration (98–100), others do not show any impact (101, 102). The results may differ between individuals due to genetic heterogeneity, differing health conditions, and microbiome composition, as well as due to different probiotic strains applied. The importance to obtain probiotics that are able to survive and colonize the gut can be seen by ex vivo human fecal microbiome culture systems, which are directly exposed to live probiotics and straightforwardly show the desired effects in SCFA production observed in mice (100). On the one hand, a more predictable tool to increase the translatability of mouse studies could be the investigating wild mice which have a more resembling microbial profile to humans (103). On the other hand, the fact that in vivo human studies do not demonstrate such strong translatability of mice studies, suggests that the amounts and viability of bacteria may be affected by several factors of the gastro-intestinal (GI) tract. Effective encapsulation and the ability of the probiotic to adhere to the gut epithelium play also an important role in this context (104–106). Recently, a lot of attention is directed towards bacterial exopolysaccharides (EPSs), which can be used as an effective carrier protecting from GI stressors, increasing muco-adhesion but also a synergistic compound to promote gut homeostasis (107–109). On another note, the robustness of the adult gut microbiome also needs to be acknowledged for effective clinical studies, as short-term interventions might be too weak to exhibit greater and long-lasting changes. The opposite can be observed in young infants, in which probiotic administration demonstrated improved clinical outcomes upon changes in the gut microbiome composition and metabolism (110–112). This can be explained by the critical time window of gut microbiome establishment, which makes it a sensitive target to manipulation (113, 114).
Thus, early prevention of gut dysbiosis, continuous long-term administration in adults and careful strain selection and encapsulation might be the most feasible approach to promote gut homeostasis and SCFA production via probiotics.
Apart from an altered diet, increasing sterility and intake of antibiotics and other pharmaceuticals are additional characteristics of modern Western Society. Narrow-spectrum antibiotics do not impact the gut microbiome composition as greatly as broad-spectrum antibiotics, which can have great consequences for intestinal microbiocenosis (115). These effects vary from the antibiotic administered, as well as from individual to individual. Nevertheless, broad-spectrum antibiotics such as ciprofloxacin and clindamycin revealed immediate decreases in SCFA-producing bacteria, where the restoration time to the initial gut microbiome in humans took up one month up to a year (116), while neomycin and ampicillin-induced gut dysbiosis in mice also affected epigenetic regulation of genes in the gut and liver (117).
SCFAs have an important share in mediating colonization resistance against pathogens and controlling the growth of opportunistic bacteria (118, 119). Their levels were shown to directly correlate with lower levels of the pathosymbiont Escherichia coli in vitro (120). Interestingly, in antibiotic-treated mice, increased levels of Candida albicans directly correlated to lower SCFA levels (121). In another study, similar observations were made and in addition, showed that the decreases in SCFAs also remained after the withdrawal of antibiotics (122). Moreover, long-term antibiotic-mediated SCFAs reduction stands in direct connection with disturbances in intestinal barrier integrity (123). Accordingly, an antibiotic-altered gut is more susceptible to recurring pathogenic infections in humans bearing metabolic consequences that promote obesity (124–126). Thus, antibiotic-induced lack of SCFA-mediated immunomodulatory and pro-homeostatic properties facilitates the colonization of pathogens and overgrowth of pathobionts, which increasingly occupy the niche otherwise dominated by beneficial commensals. The consequences are loss of SCFA-producing bacteria upon death by antibiotic exposure and are further aggravated by disadvantages in the competition of niche colonization by the pathogen-promotive environmental changes in the gut.
Other conventional non-antibiotic drugs such as proton pump inhibitors (PPIs) show similar effects as broad-spectrum antibiotics in altering the gut microbiome composition and immune system (127–129). On the other hand, metformin appears to mediate its beneficial metabolic effects against T2D by modulating the gut microbiome composition in a way that promotes SCFA-producing bacteria (130, 131). Apart from pharmaceuticals prescribed for particular health conditions, non-prescriptive non-steroidal anti-inflammatory drugs (NSAIDs) are not only widely used but also routinely. Routine consumption of aspirin, celecoxib and ibuprofen leaves a drug-specific signature in the gut microbiome, more so with stronger NSAIDs like ketoprofen, naproxen and ketorolac (132). In a recent study, short-term usage of celecoxib did not induce any significant changes in the gut microbiome whereas alterations in the metabolism of commensals decreased the production of butyrate (133).
Therefore it seems that both, the increased intake of antibiotics, PPIs and NSAIDs, with few exceptions, have their own contribution in inducing gut dysbiosis and consequential decrease in SCFAs.
As previously stated, a mainly sedentary lifestyle is a characteristic feature of Western Society. However, the various health benefits associated with physical activity become increasingly evident, as daily exercise decreases the risk of developing chronic metabolic and inflammatory disorders, as well as mental health issues (134, 135). Interestingly, its beneficial effects seem to be to some degree mediated by changes in the gut microbiome and can be also attributed to different epigenetic changes depending on the type and duration of the exercise (136).
Various studies present the benefits of exercise on metabolic health via an increase of SCFA-producing bacteria (137–140). A study comparing the effect of exercise in lean and obese individuals, however, did not show a significant shift in microbial metabolism for the latter (139). It is important to note though, that butyrate- and propionate-regulating genes decreased six weeks after ending the training period for obese individuals. In the same study, within obese individuals, butyrate and the activity of the butyrate-regulating gene BCoAT were associated with less body fat but more lean body mass and higher cardiorespiratory fitness. Thus, exercise-induced changes in the gut microbiota and SCFA-regulating gene expression appear to affect both groups, though differently. Obese individuals might therefore profit from SCFA-associated improvement in cardiorespiratory fitness, which may help in the adaptation process. Nevertheless, in some cases, such as insomnia, excess SCFAs are associated with negative health effects, whereas exercise was shown to beneficially regulate SCFA levels and sleep quality (141). Some studies also show that obesity can be associated with higher amounts of SCFAs as opposed to lean individuals and (142, 143). The same has been shown in mice, which upon receiving microbiota from exercise-trained donors gained more body mass (144). This might suggest that exercise promotes gut homeostasis accordingly to the needs of the individual, including proper balances in SCFA levels, regardless of whether a dysbiotic shift to scarce or excessive SCFA production has previously taken place. This also shows that health status, particularly obesity-related metabolic status, and genetics complicate gut microbiota manipulation via exercise, as it is not equally effective amongst individuals.
Moreover, short-term exercise was not proven to induce sustainable changes, which highlights the need for regular exercise to yield sustainable long-term effects regarding SCFA regulation (139).The individual response to exercise has been repeatedly reported to have a strong genetic base (145, 146). On top of that, different age groups and health conditions of study participants make it difficult to establish a clear relationship between SCFAs and exercise, not to mention the negligence of other confounding factors such as diet, which was shown to synergistically promote SFCA production (147). However, exercise also seems to have a diet-independent mechanism promoting SCFA production (148).
The standardization of SCFA measurements is another issue, as studies do not only differ in study designs (type of physical activity, duration, intensity, time without exercise), SCFA-measuring methods (type of SCFAs measured, gas chromatography, microbial profiling) but also in the sample type (plasma, feces) and sample processing (wet vs dry feces). Other aspects, such as SCFA absorption and interconversion by gut commensals, make it tricky to draw straightforward conclusions from fecal SCFA levels. Future research should therefore consider shedding more light on the underlying mechanism of increased/decreased SCFAs, with regard to the previously mentioned aspects. Thus, identifying SCFA-producing genes and ratios between acetate, propionate and butyrate may give additional insights into SCFA production/absorption (149, 150).
To better understand the varying observations in human studies, it is crucial to look at possible underlying mechanisms of exercise-induced changes in gut microbiota. One of the possible mechanisms is that movement physically stimulates gut microbiome function e.g. by reducing stool transit time (151). Furthermore, the underlying metabolic changes increase lactate production, which can be further metabolized to SCFAs by LAB (152). Another possible explanation could be the form of interorgan crosstalk between the gut and skeletal muscle. One of the mediators involved in this phenomenon are myokines, which are primarily cytokines that are released from skeletal muscle during exercise and exhibit anti-inflammatory properties that seem to have an impact on microbiota function in the intestine as well as systemically (153, 154). In turn, several microbial metabolites have been implicated in skeletal muscle metabolism. SCFAs are able to determine the efficacy of physical exercise by increasing lipolysis, promoting glucose uptake and insulin sensitivity in the skeletal muscle (153). SCFAs are not only an energetic substrate for colonic cells, but they also play an important role in muscle metabolism (155). Increased SCFA production seems to be a major player in mediating exercise-associated health benefits, as the donation of gut microbiomes from exercised mice was able to improve health outcomes in mice with chemically-induced colitis, mediated by the anti-inflammatory properties of SCFAs (144).
Thus, exercise-induced metabolic changes are able to influence the gut microbiome via bowel movement stimulation, myokine and lactate production. However, these effects depend on individual genetics, health status and metabolic profile. Longer-term intervention could possibly lead to more significant results in obese individuals, as increased expression of SCFA-regulating genes is associated with beneficial effects on cardiac and fat tissue metabolism. Thus, future studies should consider the long-term effects of exercise, possible confounding factors and multi-level SCFA analysis that pays special attention to SCFA absorption. More studies are also needed to define a healthy SCFA concentration range, which may differ between different age groups and health and obesity statuses. This also calls for standardizing SCFA-measuring methods.
The unleashing role of SCFAs in overall systemic health also sheds special light to modulating issues directly linked to obesity and inflammation. As there are many mechanisms that particularly affect metabolic and inflammatory processes, it is important to understand the tight relationship between both states and what role gut-derived SCFAs play in it (Figure 2). For this purpose, evidence of the impact of SCFAs on obesity, inflammation, and related chronic conditions will be assessed in the following.
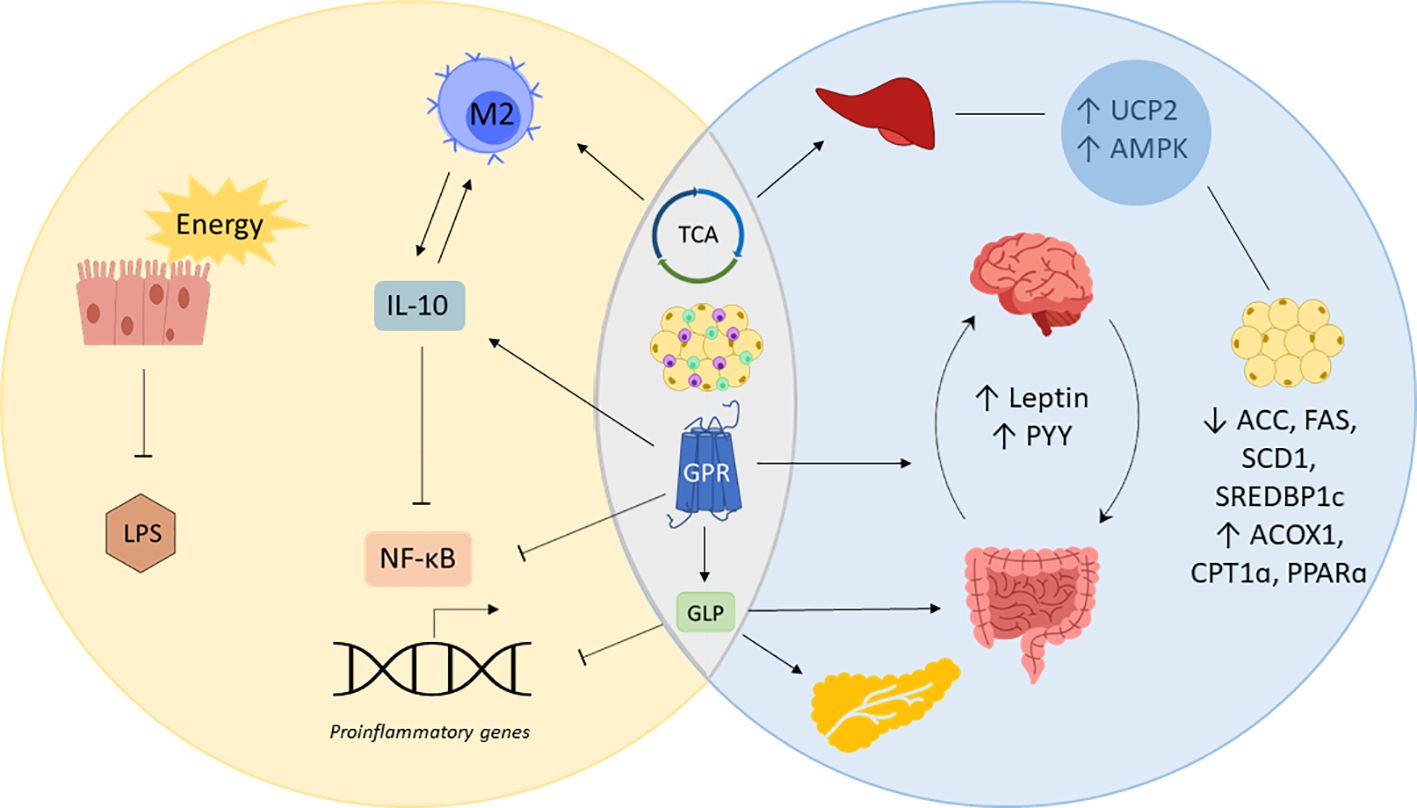
Figure 2 SCFAs in inflammation and obesity. Short-chain fatty acids (SCFAs) prevent inflammation by providing gut epithelial cells with energy enhancing cell renewal and gut barrier integrity, which decreases lipopolysaccharide (LPS) locally and systemically. SCFAs simultaneously regulate obesity and inflammation by acting on G-coupled protein receptors (GPRs), the tricarboxylic acid cycle (TCA) and immune cells of the adipose tissue. Posing a substrate for the TCA cycle and stimulating GPRs to produce interleukin (IL)-10, SCFAs promote the polarization of macrophages into IL-10 producing M2 macrophages, blocking nuclear factor kappa B (NF-κB)-induced expression of proinflammatory genes. Mitochondrial uncoupling protein 2 (UCP2) and AMP-activated protein kinase (AMPK) involved in liver and fat tissue metabolism are also regulated by SCFAs via the TCA. SCFAs decrease the expression of acetyl-CoA carboxylase (ACC), fatty acid synthase (FAS), sterol regulatory element binding protein-1c (SREBDP1c) while upregulating acyl-CoA oxidase 1 (ACOX1), carnitine palmitoyltransferase 1 (CPT1α) and peroxisome proliferator activated receptor α (PPARα). GPR-binding by SCFAs inhibits the transcription of proinflammatory genes and increases the release of appetite regulating hormones leptin and peptide YY (PYY). GPR-binding also causes the release of glucagon like peptide (GLP), which inhibits the expression of proinflammatory genes, promote the gut barrier and insulin sensitivity.
Currently, there is no cure for obesity, and only invasive interventions such as bariatric surgery have yielded significant success (156). This treatment is strongly indicated in cases of morbid obesity and metabolic diseases, however, as it is costly and invasive it is not a suitable option for all age groups and people bearing other health issues (157). Therefore, there is a great need for a safer and effective alternative, which brings attention to the clinical potential of SCFAs and their implications for obesity and the associated health conditions.
SCFAs play an important role in obesity and metabolic disease due to their pleiotropic and systemic health-promoting effects. Although some studies claim that SCFAs are increased in obese individuals and are hypothesized to be responsible for increased adiposity through excess energy (158, 159), other studies demonstrate the opposite (160–162). Moreover, recent research links probiotic-induced increased SCFA levels directly to weight loss (163–167), which also seem to exhibit their anti-obesity effect from early on via breastmilk (168, 169). Probiotics enhancing SCFA production decrease the expression of acetyl-CoA carboxylase (ACC), fatty acid synthetase (FAS), stearoyl CoA desaturase 1 (SCD1), and sterol regulatory element binding protein-1c (SREBP1c), all of which are involved in lipogenesis. Acetate, for instance, was shown to induce upregulation of acyl-CoA oxidase 1 (ACOX1), carnitine palmitoyltransferase 1 (CPT1α), and peroxisome proliferator activated receptor α (PPARα) which increased lipid oxidation and reduced adiposity and cholesterol levels (170, 171). Moreover, direct administration of SCFAs was shown to lower triglyceride levels in rats by decreasing ACC expression (172). Importantly, acetate and butyrate directly affect liver metabolism by posing metabolic substrates for the tricarboxylic acid cycle (TCA) (150). Other SCFA-mediated metabolic regulations of liver and adipose tissue concern the expression of mitochondrial uncoupling protein 2 (UCP2) via peroxisome proliferator-activated receptor gamma (PPARγ) (173). This increases the adenosine monophosphate (AMP)/adenosine triphosphate (ATP) ratio and aerobic metabolism in the liver and adipose tissue through AMP-activated protein kinase (AMPK)-signaling. Thus, Besten et al. conclude that SCFAs could serve as efficient PPARγ modulators that could improve metabolic syndrome.
SCFAs are also implicated in various metabolic processes responsible for proper energy expenditure and appetite regulation. They can function as ligands for GPR41 and GPR43 (FFAR3), which are receptors involved in metabolic pathways (171, 174). GPR41 activation causes the release of peptide YY (PYY), a gut hormone that regulates food intake and is involved in energy management. Moreover, leptin is also secreted upon SCFA stimulation of GPR41, which is a key regulator in appetite signaling (175). SCFA-binding to GPR4 triggers the release of glucagon-like peptide (GLP)-1 and GLP-2. GLP-2 promotes the integrity of the intestinal barrier by supporting epithelial cell renewal and tight junction proteins (176). GLP-1 has been shown to improve insulin sensitivity and inhibit glucagon secretion, which is why it is considered crucial in maintaining blood glucose homeostasis (177, 178). Furthermore, it plays an important role in controlling food intake, lipolysis and inflammation (179–181). It, therefore, poses a reasonable target for metabolic diseases and it is no surprise that altered SCFA levels are commonly observed in metabolically ill patients (182, 183).
Low-grade inflammation is a common feature in chronic conditions such as IBS, IBD, T2D, non-alcoholic fatty liver disease (NAFLD), CVD, and cancer. It is thought to originate from the systemic immune response triggered by LPS as a consequence of a leaky gut (20, 184). Since the immune system interacts with the gut microbiome, any dysregulations may lead to an endless loop of gut dysbiosis and systemic low-grade inflammation (185). A leaky gut can be caused by obesity or other factors causing gut dysbiosis, but it can also in turn induce obesity due to the associated systemic metabolic and inflammatory changes. Therefore it is hard to assess the primary cause of the vicious cycle between obesity and inflammation. Moreover, these are not entirely separate arrangements since metabolism determines the immunologic function, as well.
Chronic low-grade inflammation is marked by inflammatory cytokines such as tumor necrosis factor (TNF) α, IL-1, and IL-6 (37, 186). This immune response is activated upon endotoxin binding to lipopolysaccharide-binding protein (LPB). This complex can be bound to the CD14 receptor which activates the myeloid differentiation factor 88 (MyD88) and interleukin-1 receptor-associated kinase (IRAK)-1 signaling cascade upon activation of toll-like receptor (TLR) 4 (187). The fact that obesity is characterized by ongoing low-grade inflammation can be partly explained by the co-regulation of both states via GPRs. Not only are they involved in metabolic homeostasis but they also determine immune response. Since free fatty acids bind to these receptors, the absence of SCFAs necessary for the negative regulation of proinflammatory cytokines leads to the continuous expression of inflammatory mediators. In the presence of SCFAs on the other hand, nuclear factor kappa B (NF-κB)-mediated activation of TNFα, IL-6, and IL-12 is inhibited. Furthermore, SFCAs also increase IL-10 expression, which suppresses TNFα, macrophage inflammatory protein 2 (MIP-2), and IL-1β expression (188–190). Moreover, macrophages are thought to have a great contribution to low-grade inflammation in adipose tissue and systemically, and IL-10 has been shown to protect against endotoxemia and stimulate M2 polarization (191, 192). Moreover, TLR-mediated MyD88 activation and TNF signaling by M1 macrophages have further health consequences by impairing insulin signaling, which shows the tight relationship between inflammation and metabolic health (185). Moreover, the lack of SFCA gut barrier-promoting qualities facilitates the spread of gut-derived inflammatory molecules that can distribute systemically via the bloodstream. This way, LPS leakage through the epithelial barrier can also trigger inflammation in various tissues which might continue until gut homeostasis is reversed (193). If there is no resolution of inflammation, inflammatory mediators might amplify the response by recruitment of leukocytes which can bear systemic consequences in the long run.
Thus, obesity-induced gut dysbiosis that leads to metabolic endotoxemia lays the path for the development of inflammatory chronic disease. This results in aberrant interactions between the gut microbiome, host immune system and metabolism, leading to so-called metaflammation (185). Interestingly, in mice obesity-associated low-grade inflammation was mitigated by probiotics and correlated with SCFA levels (163).
The tight link between immunologic and metabolic function becomes evident in various organelles. For example, nutrient surplus in the endoplasmic reticulum (ER) can disrupt its function and trigger the unfolded protein response (UPR) that ultimately leads to the activation of various inflammatory pathways, such as c-Jun N-terminal kinase (JNK) and iκB kinase (IKK) (194). In consequence, insulin sensitivity and glucose homeostasis is disrupted, which will likely trigger again the UPR. Another organelle affected by obesity is mitochondria. Mitochondrial dysfunction caused by obesity results in increased reactive oxygen species that can further disrupt insulin-signaling and promote M1 polarization of macrophages and activate the inflammasome, which again leads to disruptions in metabolism and promotes obesity (195–198). Disruptions in nutritional or immune system homeostasis therefore further trigger pro-inflammatory responses and metabolic dysfunctions, closing the vicious cycle of promoting both states.
The crucial role of metabolism in the immune response becomes also clear in the case of innate immune cells. Immunological memory is not exclusively part of the adaptive immune system since innate immune cells also exhibit a certain degree of memory function, referred to as ‘trained immunity’. In an aim to elicit a faster and more effective response by the repeated encounter of a certain antigen, innate immune cells become intrinsically wired to increase the response to secondary stimuli, e.g. by LPS. This effect is partly mediated by increased sensitivity of pattern recognition receptors (PRRs) but also by a metabolic shift from oxidative phosphorylation to aerobic glycolysis, called the Warburg effect (199, 200). The latter is responsible for providing, for instance, macrophages with glycolysis- and broken tricarboxylic acid cycle (TCA)-derived substrates necessary to initiate a proinflammatory immune response. These include glycolytic enzymes, succinate, and citrate, but also glutamine and arginine catabolism aid M1 function (201, 202). Interestingly, low levels of LPS have the opposite effect, inducing tolerance (203). Therefore, gut homeostasis or dysbiosis differently impacts innate immune cell metabolism, which in turn directly dictates its function.
Moreover, the mammalian target of the rapamycin (mTOR) pathway in innate immune cells is regulated by AMP kinase (AMPK), which in case of high nutrient availability, becomes deactivated (204). Due to mTOR diverse functions in cellular metabolism and homeostasis, dysregulation of mTOR signaling pathway has been implicated in various diseases (205). The loss of AMPK suppression of proinflammatory pathways can lead to hyperinflammation that can deteriorate underlying conditions such as atherosclerosis (206). Furthermore, it has been shown that the endotoxin-triggered proinflammatory phenotype of macrophages remains a long-lasting feature and that restoration of the anti-inflammatory M2 phenotype is straitened (199). Thus, the series of effects can be primarily triggered by LPS, having its roots in gut dysbiosis (Figure 3).
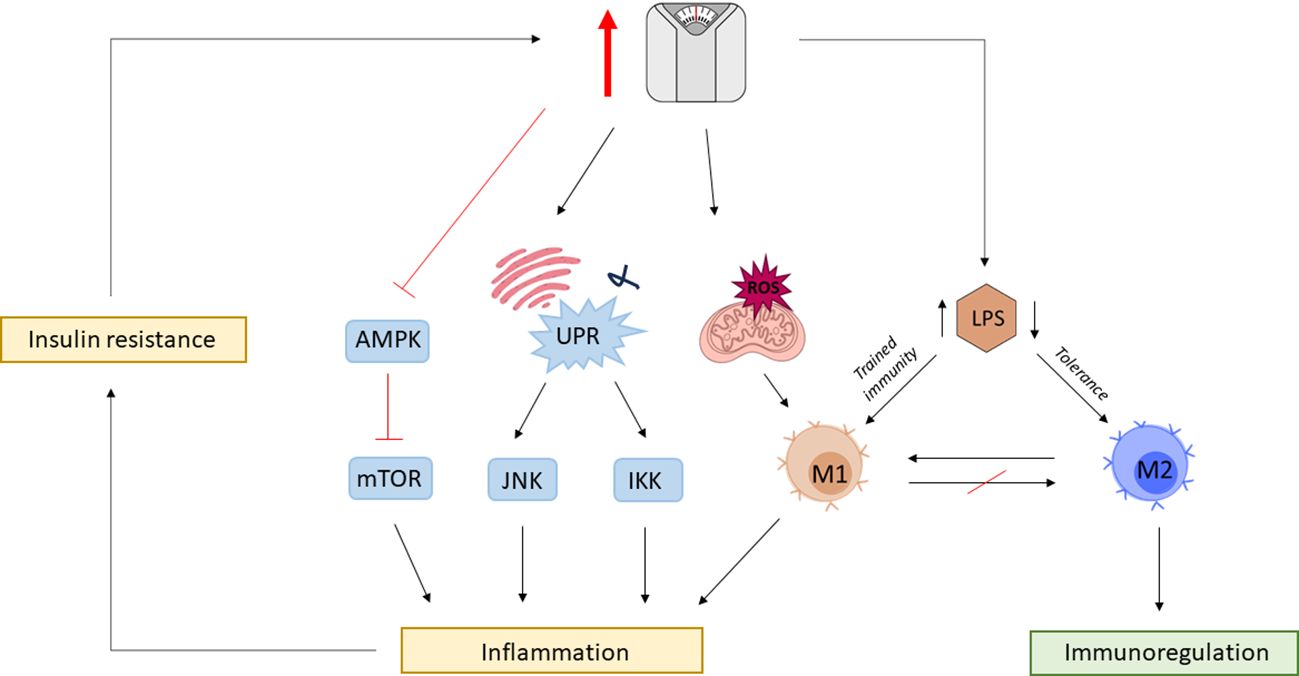
Figure 3 The vicious cycle of obesity and inflammation. Obesity is associated with gut dysbiosis and gut barrier impairment, leading to increased circulating levels of lipopolysaccharide (LPS). Chronic exposure to elevated LPS leaves a permanent phenotypic mark on macrophages, which disables otherwise mediated tissue homeostasis by M2 macrophages. Obesity is associated with mitochondrial dysfunction leading to the production of reactive oxygen species (ROS) promoting M1 polarization. Surplus nutrient intake also triggers mitochondria endoplasmic reticulum (ER) to activate the unfolding protein response (UPR) leading to the activation of c-Jun N-terminal kinase (JNK) and iκB kinase (IKK) proinflammatory signaling cascades. High nutrient availability also impedes the otherwise negative regulation of rapamycin (mTOR) by AMP-activated protein kinase (AMPK). Obesity-associated inflammation leads to metabolic changes resulting in insulin resistance further exacerbating the risk for obesity, closing the vicious cycle between immunologic and metabolic dysregulations.
Interestingly, epigenetic mechanisms are responsible for innate immune memory with metabolic molecules being cofactors for epigenetic enzymes (207). Thus, LPS not only induces metabolic but also epigenetic changes within immune cells that prime them towards a pro-inflammatory phenotype. SCFAs on the other hand, show great potential in reversing those epigenetic dysregulations.
Epigenetic modifications contain a range of different mechanisms that alter DNA or DNA-associated proteins in a post-translational manner. This way the genetic code remains unaffected while protein expression can be regulated. SCFAs have been shown to modulate various pathways involved in inflammatory and metabolic regulations. Strikingly, epigenetic regulations initiated by the gut microbiome are not restricted to the gut but have been proven to impact other tissues as well (208). Epigenetic regulation via the gut microbiome can be mediated by providing cofactors for epigenetic enzymes in form of bacterial metabolites or by influencing the expression and activity of epigenetic enzymes and epigenetic pathways (209, 210). SCFAs play hereby a crucial role, since traveling from the intestinal lumen to other organs via the blood compartment they do not only maintain tissue homeostasis by binding to receptors but also by various epigenetic mechanisms (Figure 4). Under these fall modifications of DNA and chromatin-associated proteins, and non-coding RNAs (ncRNAs) (38).
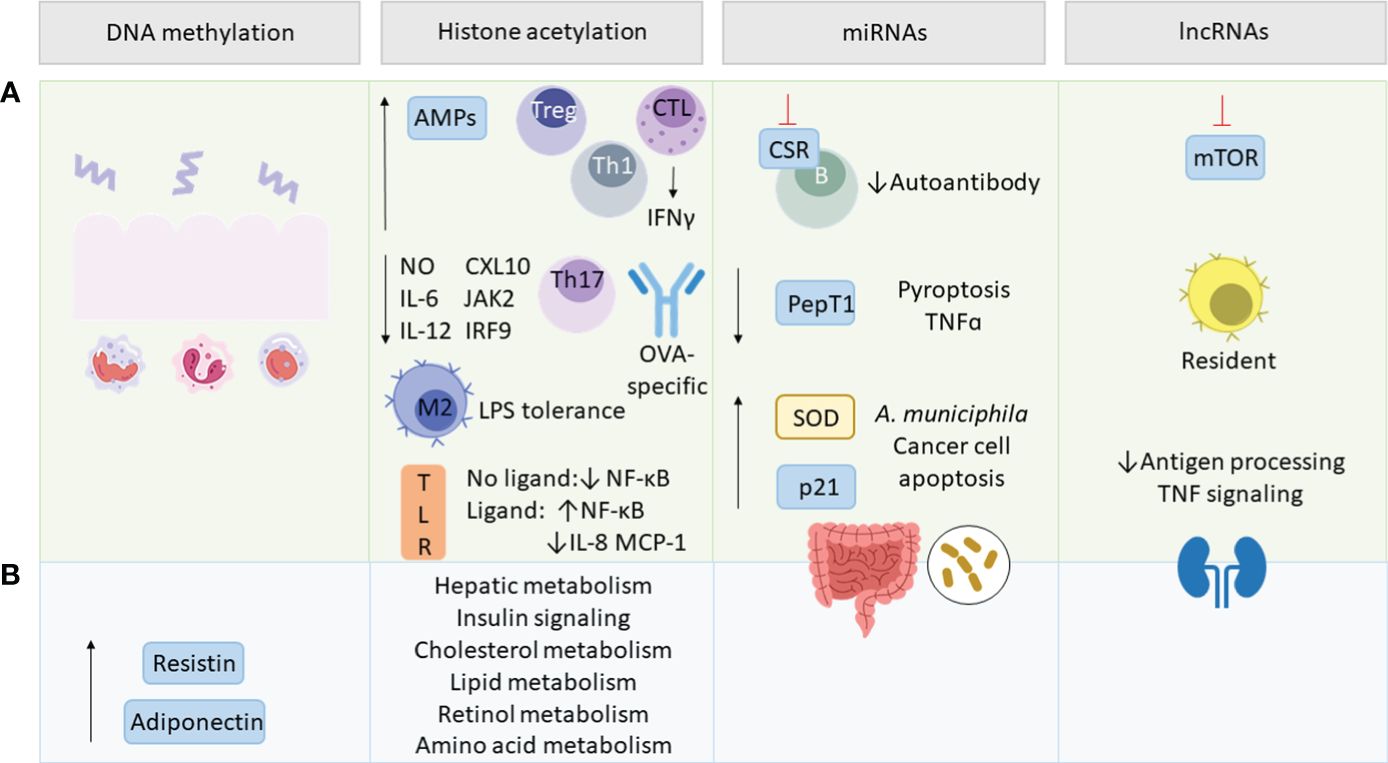
Figure 4 SCFA-mediated epigenetic regulations. Green section (A) addresses epigenetic effects in inflammation and blue section (B) in metabolism. SCFAs regulate intestinal epithelial cell (IEC)-related innate and adaptive immune functions, as well as resistin and adiponectin expression via DNA methylation. By histone deacetylase (HDAC) inhibition, SCFAs upregulate antimicrobial peptides (AMPs), T regulatory (Treg) and T helper (Th) cells and interferon γ (IFNγ) production by cytotoxic T lymphocytes (CTL). SCFAs mediate lipopolysaccharide (LPS) tolerance by M2 macrophage polarization and downregulate nitric oxide (NO), interleukin (IL)-6, IL-12, C-X-C motif chemokine ligand (CXL10), Janus kinase 2 (JAK2), interferon regulatory factor 9 (IRF9), Th17 and ovalbumin (OVA)-specific antibodies. They also modulate ligand-dependent responsiveness of toll-like receptor (TLR) leading to the up- or downregulation of NF-κB, IL-8 and monocyte chemotactic protein 1 (MCP-1) and affect various metabolic pathways. SCFA-modulated micro RNAs (miRNAs) act on class switch recombination (CSR) of B-cells, peptide transporter 1 (PepT1) and tumor necrosis factor α (TNFα) transcription, as well as pyroptosis. miRNAs upregulate superoxide dismutase (SOD), p21, cancer cell apoptosis, Akkermansia muciniphila abundance and impact the gut microbiome composition. SCFAs regulate long non-coding RNAs (lncRNAs) involved in rapamycin (mTOR) signaling, promoting resident macrophages and renal function via decreasing antigen processing and TNF signaling.
Epigenetic modifications of chromatin comprise both modification of DNA and of chromatin-associated proteins. Regarding DNA modification, the main epigenetic mechanism is the methylation or demethylation of genetic material. Methylated DNA is the result of additional methyl group attachment by DNA methyltransferases (DNMTs) to cytosine-phosphate-guanine (CpG) islands. Hereby, increased methylation of DNA is associated with a more compact chromatin structure and therefore decreased transcriptional activity, which can be beneficial in the context of obesity-associated genes and those involved in inflammatory responses (211).
DNA methylation by the microbiota plays an important role in intestinal homeostasis and balanced immune response. Microbiota-dependent DNA demethylation was shown to regulate the function of intestinal epithelial cells (IECs) and immune cells involved in innate and adaptive immune responses (212). Importantly, these epigenetic regulations affect the expression and responsiveness of TLR-4 upon LPS encounter and promote the proliferation of intestinal Tregs and natural killer lymphocytes (213–215). Moreover, IBD-associated mucosal inflammation is associated with aberrant DNA methylation whereas SCFAs exhibit anti-inflammatory effects in IBD via IECs-induced changes in the transcription of inflammatory markers (216–218). On the other hand, gut dysbiosis, the presence of bacterial pathogens or LPS affect epigenetic mechanisms connected to inflammation and metabolism by acting on host DNMTs or by producing the enzyme itself (219–222).
Mouse studies also reveal the direct role of SCFAs in the epigenetic regulation of obesity-associated proteins (223). Supplementation of acetate, propionate, and butyrate in mice was able to reverse the reduced expression of adiponectin and resistin in adipose tissue of HFD-fed mice by lowering the expression of DNA methyltransferases and methyl-CpG-binding domain protein 2 (MBD2). Since adiponectin is also considered to have insulin sensitivity-promoting, anti-inflammatory, and anti-atherogenic properties, there is great interest in increasing its expression in obesity and chronic inflammatory conditions (224). On the other hand, resistin levels above the norm are associated with obesity and T2D, whereas normal levels do not exhibit harmful effects (225, 226). Human studies also confirm disrupted DNA methylation in the adipose tissue of metabolically ill or overweight subjects and point to the probable role of SCFA due to the association of methylation status and abundance of SCFA-producing bacteria (227, 228). However, in yet another mouse study, antibiotic-induced changes in the gut microbiome lead to weight loss and increased adiponectin and resistin in an SCFA-independent manner (229).
Moreover, DNA methylation can be also influenced by other bacterial metabolites serving as cofactors of epigenetic enzymes, such as B group vitamins and polyamines (230–232). Into the bargain, the impact of SCFAs on host health appears to be distinct between lean and obese individuals. While SCFAs seem to prevent lean individuals from obesity and metabolic disturbances, in obese individuals, SCFA-mediated DNA methylation makes them more prone to develop T2D (233). Moreover, epigenetic alterations induced during early gut microbiome development were shown to have a great impact on individual predispositions regarding gut microbiome composition and metabolic and immunological profile (234–236). Importantly, early intervention in gut microbiome-induced epigenetic mechanisms via maternal probiotic intake shows great potential in modulating short- and long-term epigenetic patterns that beneficially regulate health (237, 238).
Amongst all epigenetic modifications, histone acetylation has been most thoroughly studied and the role of SCFAs in this context is also better understood. Histone acetylation is regulated by two catalytic enzymes – histone acetyltransferase (HAT) and histone deacetylase (HDAC). The latter is responsible for deacetylation and poses the main target of SCFAs. As previously mentioned, SCFAs influence the metabolism and increase the availability of metabolic substrates needed for histone acetylation, such as acetyl-CoA (239). Other post-translational histone modifications mediated by different SCFAs are methylation, ethylation, crotonylation, propionilation, butyrylation and hydroxybutyrylation, but these mechanisms just begin to be understood (239–244).
Similarly as DNA methylation, histone methylation of immunoregulatory and antioxidant genes is distinct in IBD patients and modulated by the gut microbiota (245). However, histone acetylation/deacetylation is a better studied process of epigenetic histone modification. Bacterial metabolites were shown to affect the acetylation status of genes from IECs and macrophages, modulating antimicrobial immune response (246, 247). Amongst all SCFAs, butyrate appears to be the strongest HDAC inhibitor and by this was shown to regulate the immune system and metabolism systemically. Mice and in vitro experiments show that the HDAC-inhibiting activity of butyrate increases antimicrobial activity of macrophages while suppressing nitric oxide (NO) expression and thus greater flare up of inflammation (247). In another study, antimicrobial peptides (AMPs) were shown to be upregulated via butyrate-induced HDAC inhibition, enhancing targeted pathogen elimination (248). Importantly, HDAC inhibition by butyrate was shown to induce the anti-inflammatory and resolutive macrophage M2 phenotype and generate tolerance towards LPS by downregulating NO, IL-6 and IL-12 (249, 250). Similarly, other studies show selective histone deacetylation of inflammatory genes during LPS-induced immune response in epithelial cells, suppressing inflammatory pathways induced by C-X-C motif chemokine ligand 10 (CXL10), Janus kinase 2 (JAK2) and interferon regulatory factor 9 (IRF9) (251). Another study points to the regulatory effect of SCFAs on NF-ĸB expression, dependent on the absence or presence of TLR ligands (252). In the latter case, SCFAs enhanced NF-ĸB expression during TLR stimulation, but downregulated the chemotactic cytokines IL-8 and monocyte chemotactic protein 1 (MCP-1). Thus SCFA-mediated histone modifications affecting the immune system enhances defenses against pathogens whilst sparing the intestine from excessive inflammation – relevant in counteracting chronic low-grade inflammation.
Histone modifications also play an important role in the immunoregulation of adaptive immune cells. SCFAs were shown to influence genes responsible for class switching in B cells via HDAC inhibition, reducing affinity to the ovalbumin (OVA) allergen (253). In addition to GPR signaling, HDAC inhibition at the forkhead box P3 (FOXP3) locus promotes differentiation and proliferation of Tregs, a key mediator of tolerance and homeostasis (30, 254–256). Interestingly, SFCAs also exhibit synergistic effects on intestinal immunity via GPR stimulation and HDAC inhibition in T cells and ILCs, resulting in increased expression of the gut barrier-promoting IL-22 (257). Moreover, butyrate was shown to decrease T helper cell (Th) 17 expression and increase Th1 via histone acetylation (258, 259). On top of that, SCFA enhance interferon γ (IFNγ) expression in cytotoxic T lymphocytes (CTLs) via HDAC inhibition, crucial for mediating anti-tumor effects (260).
SCFAs were also shown to affect histone acetylation and methylation beyond the intestine, as they epigenetically regulate gene expression in the liver, kidney and fat tissue (208, 261). Mice fed a Western diet showed decreased levels of SCFA resulting in altered histone modifications ultimately regulating hepatic metabolism via PPAR signaling, genes involved in insulin and cholesterol pathways, as well as genes related to adaptive immunity, lipid, retinol, and amino acid metabolism (208). Interestingly, SCFA supplementation of acetate, propionate, and butyrate was able to reverse the epigenetic phenotype to such a degree that it resembled the one from conventionally raised mice.
Although the sensitive role of SCFAs in modulating the immune system and metabolism becomes clear in the case of histone modification, the effect of SCFAs is not entirely straightforward. Different bacterial metabolites can be antagonistic and can act differently on HDACs. Although it is generally thought that SCFAs act beneficially as HDAC inhibitors, it was shown that butyrate can interfere with IBD-counteracting inositol-1,4,5-triphopshate (INSP3)-mediated histone deacetylation (246). Moreover, Kespohl et al. suggest that the beneficial action of butyrate might be dependent on its concentration and immunological predispositions (262). For safe and effective SCFA-mediated epigenetic regulation, It might be necessary to formulate a personalized approach towards each individual with regard to their metabolic status, immunologic and microbiome profile.
Important regulators of gene expression are ncRNAs, which are RNA molecules that are not coding for a protein and do not undergo translation. Besides the housekeeping ncRNAs like transfer RNA (tRNA) or ribosomal RNA (rRNA), there is a range of regulatory RNAs. Regarding the latter, miRNAs are the most abundant ncRNAs and also the best studied. miRNAs have around 20 nucleotides and regulate gene expression by binding to complementary target messenger RNA (mRNA), which suppresses translation either by degradation or silencing of the bound mRNA fragment.
Studies reveal a correlational relationship between bacteria, SCFAs and miRNAs regulating inflammatory processes in the intestine as well as in other organs (263–267). The gut microbiota was also shown to downregulate proinflammatory cytokines and the transport of proinflammatory bacterial peptides by peptide transporter 1 (PepT1) via miRNA upregulation (268–270). Moreover, probiotic strains producing SCFAs and lactate were shown to modulate miRNA expression resulting in anti-inflammatory and gut barrier-promoting effects (271). What is more, high doses of butyrate and propionate were shown to directly modulate B cell-intrinsic mechanisms of class switch recombination (CSR) by targeting associated genes via the upregulation of miRNAs (253). Not only did this limit autoantibody production in mice but the manner in which miRNAs were upregulated was the result of HDAC inhibition of miRNA-encoding genes and thus was found to be a form of epigenetic crosstalk. The same has been shown for DNA methylation which can affect miRNA expression (272). Interestingly, certain miRNAs can affect DNA methylation by targeting DNMTs and ten-eleven translocation enzymes (TETs) acting as methylation erasers (273–275) and this way can also impact the expression of other miRNAs (276).
In addition, miRNAs also regulate the gut microbiome composition and SCFA receptors showing the bidirectional regulation of SCFAs and miRNAs (277–279). The same goes for diet- and host-derived miRNAs, which were shown to affect gut microbiota (280, 281). Moreover, several epigenetic mechanisms can work in synergism as has been shown for SCFA-mediated regulation of the expression of cyclin-dependent kinase inhibitor p21 in human colonic cancer, which can be enhanced by miRNA degradation and histone acetylation (282, 283). Synergistic regulation was also found for several miRNAs and butyrate regarding colon cancer cell apoptosis (284). Other bacterial metabolites, such as tryptophan, were shown to control adiposity and insulin sensitivity through miR-181 expression (285). Butyrate, on the other hand, was shown to dampen inflammatory dysregulations in T2D patients by decreasing pyroptosis and TNF-α levels whilst increasing superoxide dismutase (SOD) activity and A. muciniphila abundance (286, 287).
lncRNAs are RNA fragments with up to 200 nucleotides and can regulate gene expression by various posttranslational protein modifications such as protein phosphorylation, ubiquitination and acetylation, as well as DNA methylation (288). They can also affect miRNA expression and vice versa (288, 289). The gut microbiome was shown to regulate lncRNAs within and beyond the intestine (290, 291). Hereby, bacterial components such as LPS negatively affect the host transcriptome and proteome, promoting inflammatory processes and cancer development (292–294). On the other hand, SCFAs were shown to suppress mTOR signaling and induce macrophage differentiation into resident macrophages (295, 296). LncRNA also play an important role in obesity, regarding energy expenditure, appetite regulation, insulin sensitivity and other obesity-associated inflammatory conditions (291, 297–300). SCFAs were hereby shown to regulate lncRNAs preventing renal dysfunction of diabetic nephropathic mice by e.g. decreasing antigen-processing and TNF signaling (301).
Recent studies also hint at the involvement of the gut microbiome in regulating other non-coding RNAs, such as small nuclear RNAs (snoRNAs), piwi-interacting RNAs (piRNAs) and circular RNAs (circRNAs) (302–304). However, the underlying mechanisms and possible role of SCFAs in this context remain to be discovered.
Overall, the epigenetic activity of SCFAs shows great potential in reversing the metabolic and immunological defects caused by metabolic endotoxemia and thus break the vicious cycle of obesity and inflammation. It is important to keep in mind, that the treatment in adult, obese and chronically ill individuals requires a long intervention time and personalized approach to overcome the prevailing individual metabolic and immunological predispositions. In order to find an effective strategy in humans, a synbiotic containing fibers and beneficial bacterial key species but also enriched in other SCFA-promoting substances and cofactors such as lactate, B group vitamins and microelements could prove efficacious. Nevertheless, diet and lifestyle should be an additional angle to counteract obesity-associated low-grade inflammation, as not only does it influence SCFA production but also conveys distinct health-promoting effects that can have a synergistic effect, which is desirable for the complex issues of obesity and inflammation. This might give a better chance to reestablish healthy interorgan crosstalk of myokines, cytokines and host miRNAs derived from adipose tissue, skeletal muscle and liver, all of which also influences the gut microbiota and its regulative function. More long-term and standardized research is still needed to better understand how to effectively modulate metabolism and the immune system via SCFA-mediated epigenetic modification. However, in the long run, SCFA treatment could become an accessible standard therapy for both prevention and treatment of obesity and inflammation.
JK: Conceptualization, Visualization, Writing – original draft, Writing – review & editing. MK: Funding acquisition, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. JK and MK were supported by the National Science Center (NCN) [grant number 2021/41/B/NZ9/02236]. Open Access funding enabled by Project [2021/41/B/NZ9/02236] received by MK from NCN.
The authors declare the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Ryan D, Barquera S, Barata Cavalcanti O, Ralston J. “The global pandemic of overweight and obesity.,”. In: Kickbusch I, Ganten D, Moeti M, editors. Handbook of Global Health. Springer International Publishing, Cham (2021). p. 739–73. doi: 10.1007/978-3-030-45009-0_39
2. Divella R, Gadaleta Caldarola G, Mazzocca A. Chronic inflammation in obesity and cancer cachexia. J Clin Med. (2022) 11:2191. doi: 10.3390/jcm11082191
3. Nguyen H-L, Geukens T, Maetens M, Aparicio S, Bassez A, Borg A, et al. Obesity-associated changes in molecular biology of primary breast cancer. Nat Commun. (2023) 14:4418. doi: 10.1038/s41467-023-39996-z
4. Aguilera-Lizarraga J, Hussein H, Boeckxstaens GE. Immune activation in irritable bowel syndrome: what is the evidence? Nat Rev Immunol. (2022) 22:674–86. doi: 10.1038/s41577-022-00700-9
5. McDowell C, Farooq U, Haseeb M. “Inflammatory bowel disease.,”, in: StatPearls (2023). Treasure Island (FL: StatPearls Publishing. Available online at: http://www.ncbi.nlm.nih.gov/books/NBK470312/ (Accessed August 13, 2023).
6. El-Hakim Y, Bake S, Mani KK, Sohrabji F. Impact of intestinal disorders on central and peripheral nervous system diseases. Neurobiol Dis. (2022) 165:105627. doi: 10.1016/j.nbd.2022.105627
7. Shah NN, Wass S, Hajjari J, Heisler AC, Malakooti S, Janus SE, et al. Proportionate cardiovascular mortality in chronic inflammatory disease in adults in the United States from 1999 to 2019. JCR: J Clin Rheumatol. (2022) 28:97. doi: 10.1097/RHU.0000000000001818
8. Pahwa R, Goyal A, Jialal I. “Chronic inflammation.,”, in: StatPearls (2022). Treasure Island (FL: StatPearls Publishing. Available online at: http://www.ncbi.nlm.nih.gov/books/NBK493173/ (Accessed January 11, 2023).
9. Hajat C, Stein E. The global burden of multiple chronic conditions: A narrative review. Prev Med Rep. (2018) 12:284–93. doi: 10.1016/j.pmedr.2018.10.008
10. Lee YS, Olefsky J. Chronic tissue inflammation and metabolic disease. Genes Dev. (2021) 35:307–28. doi: 10.1101/gad.346312.120
11. Obesity and overweight(2021). Available online at: https://www.who.int/news-room/fact-sheets/detail/obesity-and-overweight (Accessed January 22, 2024).
12. Scully T, Ettela A, LeRoith D, Gallagher EJ. Obesity, type 2 diabetes, and cancer risk. Front Oncol. (2021) 10:615375. doi: 10.3389/fonc.2020.615375
13. Kansra AR, Lakkunarajah S, Jay MS. Childhood and adolescent obesity: A review. Front Pediatr. (2020) 8:581461. doi: 10.3389/fped.2020.581461
14. Artemniak-Wojtowicz D, Kucharska AM, Pyrżak B. Obesity and chronic inflammation crosslinking. Cent Eur J Immunol. (2020) 45:461–8. doi: 10.5114/ceji.2020.103418
15. Scheithauer TPM, Rampanelli E, Nieuwdorp M, Vallance BA, Verchere CB, van Raalte DH, et al. Gut microbiota as a trigger for metabolic inflammation in obesity and type 2 diabetes. Front Immunol. (2020) 11:571731. doi: 10.3389/fimmu.2020.571731
16. Russo L, Lumeng CN. Properties and functions of adipose tissue macrophages in obesity. Immunology. (2018) 155:407–17. doi: 10.1111/imm.13002
17. Toubal A, Kiaf B, Beaudoin L, Cagninacci L, Rhimi M, Fruchet B, et al. Mucosal-associated invariant T cells promote inflammation and intestinal dysbiosis leading to metabolic dysfunction during obesity. Nat Commun. (2020) 11:3755. doi: 10.1038/s41467-020-17307-0
18. Suzuki T, Gao J, Ishigaki Y, Kondo K, Sawada S, Izumi T, et al. ER stress protein CHOP mediates insulin resistance by modulating adipose tissue macrophage polarity. Cell Rep. (2017) 18:2045–57. doi: 10.1016/j.celrep.2017.01.076
19. Donato J. Programming of metabolism by adipokines during development. Nat Rev Endocrinol. (2023) 19:385–97. doi: 10.1038/s41574-023-00828-1
20. Cani PD, Amar J, Iglesias MA, Poggi M, Knauf C, Bastelica D, et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. (2007) 56:1761–72. doi: 10.2337/db06-1491
21. Jobe M, Agbla SC, Todorcevic M, Darboe B, Danso E, de Barros J-PP, et al. Possible mediators of metabolic endotoxemia in women with obesity and women with obesity-diabetes in The Gambia. Int J Obes. (2022) 46:1892–900. doi: 10.1038/s41366-022-01193-1
22. Trøseid M, Nestvold TK, Rudi K, Thoresen H, Nielsen EW, Lappegård KT. Plasma lipopolysaccharide is closely associated with glycemic control and abdominal obesity. Diabetes Care. (2013) 36:3627–32. doi: 10.2337/dc13-0451
23. Gilbert JA, Blaser MJ, Caporaso JG, Jansson JK, Lynch SV, Knight R. Current understanding of the human microbiome. Nat Med. (2018) 24:392–400. doi: 10.1038/nm.4517
24. Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett C, Knight R, Gordon JI. The human microbiome project: exploring the microbial part of ourselves in a changing world. Nature. (2007) 449:804–10. doi: 10.1038/nature06244
25. Caballero-Flores G, Pickard JM, Núñez G. Microbiota-mediated colonization resistance: mechanisms and regulation. Nat Rev Microbiol. (2023) 21:347–60. doi: 10.1038/s41579-022-00833-7
26. De Juan A, Segura E. Modulation of immune responses by nutritional ligands of aryl hydrocarbon receptor. Front Immunol. (2021) 12:645168. doi: 10.3389/fimmu.2021.645168
27. Louis P, Flint HJ. Formation of propionate and butyrate by the human colonic microbiota. Environ Microbiol. (2017) 19:29–41. doi: 10.1111/1462-2920.13589
28. Deleu S, Machiels K, Raes J, Verbeke K, Vermeire S. Short chain fatty acids and its producing organisms: An overlooked therapy for IBD? eBioMedicine. (2021) 66. doi: 10.1016/j.ebiom.2021.103293
29. Comalada M, Bailón E, de Haro O, Lara-Villoslada F, Xaus J, Zarzuelo A, et al. The effects of short-chain fatty acids on colon epithelial proliferation and survival depend on the cellular phenotype. J Cancer Res Clin Oncol. (2006) 132:487–97. doi: 10.1007/s00432-006-0092-x
30. Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. (2013) 504:451–5. doi: 10.1038/nature12726
31. Meyer F, Seibert FS, Nienen M, Welzel M, Beisser D, Bauer F, et al. Propionate supplementation promotes the expansion of peripheral regulatory T-Cells in patients with end-stage renal disease. J Nephrol. (2020) 33:817–27. doi: 10.1007/s40620-019-00694-z
32. Takeuchi T, Miyauchi E, Kanaya T, Kato T, Nakanishi Y, Watanabe T, et al. Acetate differentially regulates IgA reactivity to commensal bacteria. Nature. (2021) 595:560–4. doi: 10.1038/s41586-021-03727-5
33. Isobe J, Maeda S, Obata Y, Iizuka K, Nakamura Y, Fujimura Y, et al. Commensal-bacteria-derived butyrate promotes the T-cell-independent IgA response in the colon. Int Immunol. (2020) 32:243–58. doi: 10.1093/intimm/dxz078
34. O’Mahony L. Short-chain fatty acids modulate mast cell activation. Allergy. (2020) 75:1848–9. doi: 10.1111/all.14313
35. Carmody RN, Bisanz JE. Roles of the gut microbiome in weight management. Nat Rev Microbiol. (2023) 21:535–50. doi: 10.1038/s41579-023-00888-0
36. Li Z, Yi C-X, Katiraei S, Kooijman S, Zhou E, Chung CK, et al. Butyrate reduces appetite and activates brown adipose tissue via the gut-brain neural circuit. Gut. (2018) 67:1269–79. doi: 10.1136/gutjnl-2017-314050
37. Violi F, Cammisotto V, Bartimoccia S, Pignatelli P, Carnevale R, Nocella C. Gut-derived low-grade endotoxaemia, atherothrombosis and cardiovascular disease. Nat Rev Cardiol. (2023) 20:24–37. doi: 10.1038/s41569-022-00737-2
38. Amatullah H, Jeffrey KL. Epigenome-metabolome-microbiome axis in health and IBD. Curr Opin Microbiol. (2020) 56:97–108. doi: 10.1016/j.mib.2020.08.005
39. Epigenetics(2024). Available online at: https://www.genome.gov/genetics-glossary/Epigenetics (Accessed January 22, 2024).
40. González-Becerra K, Ramos-Lopez O, Barrón-Cabrera E, Riezu-Boj JI, Milagro FI, Martínez-López E, et al. Fatty acids, epigenetic mechanisms and chronic diseases: a systematic review. Lipids Health Dis. (2019) 18:178. doi: 10.1186/s12944-019-1120-6
41. Martinez JE, Kahana DD, Ghuman S, Wilson HP, Wilson J, Kim SCJ, et al. Unhealthy lifestyle and gut dysbiosis: A better understanding of the effects of poor diet and nicotine on the intestinal microbiome. Front Endocrinol. (2021) 12:667066. doi: 10.3389/fendo.2021.667066
42. Strachan DP. Hay fever, hygiene, and household size. BMJ. (1989) 299:1259–60. doi: 10.1136/bmj.299.6710.1259
43. Hasan N, Yang H. Factors affecting the composition of the gut microbiota, and its modulation. PeerJ. (2019) 7. doi: 10.7717/peerj.7502
44. Andújar-Tenorio N, Prieto I, Cobo A, Martínez-Rodríguez AM, Hidalgo M, Segarra AB, et al. High fat diets induce early changes in gut microbiota that may serve as markers of ulterior altered physiological and biochemical parameters related to metabolic syndrome. Effect of virgin olive oil in comparison to butter. PloS One. (2022) 17:e0271634. doi: 10.1371/journal.pone.0271634
45. Muredda L, Kępczyńska MA, Zaibi MS, Alomar SY, Trayhurn P. IL-1β and TNFα inhibit GPR120 (FFAR4) and stimulate GPR84 (EX33) and GPR41 (FFAR3) fatty acid receptor expression in human adipocytes: implications for the anti-inflammatory action of n-3 fatty acids. Arch Physiol Biochem. (2018) 124:97–108. doi: 10.1080/13813455.2017.1364774
46. Binker-Cosen MJ, Richards D, Oliver B, Gaisano HY, Binker MG, Cosen-Binker LI. Palmitic acid increases invasiveness of pancreatic cancer cells AsPC-1 through TLR4/ROS/NF-κB/MMP-9 signaling pathway. Biochem Biophys Res Commun. (2017) 484:152–8. doi: 10.1016/j.bbrc.2017.01.051
47. Zhou H, Urso C, Jadeja V. Saturated fatty acids in obesity-associated inflammation. J Inflammation Res. (2020) 13:1–14. doi: 10.2147/JIR.S229691
48. Ortega FJ, Cardona-Alvarado MI, Mercader JM, Moreno-Navarrete JM, Moreno M, Sabater M, et al. Circulating profiling reveals the effect of a polyunsaturated fatty acid-enriched diet on common microRNAs. J Nutr Biochem. (2015) 26:1095–101. doi: 10.1016/j.jnutbio.2015.05.001
49. Tremblay BL, Guénard F, Rudkowska I, Lemieux S, Couture P, Vohl M-C. Epigenetic changes in blood leukocytes following an omega-3 fatty acid supplementation. Clin Epigenet. (2017) 9. doi: 10.1186/s13148-017-0345-3
50. Johnson CSC, Shively CA, Michalson KT, Lea AJ, DeBo RJ, Howard TD, et al. Contrasting effects of Western vs Mediterranean diets on monocyte inflammatory gene expression and social behavior in a primate model. Elife. (2021) 10:e68293. doi: 10.7554/eLife.68293
51. Ruegsegger GN, Grigsby KB, Kelty TJ, Zidon TM, Childs TE, Vieira-Potter VJ, et al. Maternal Western diet age-specifically alters female offspring voluntary physical activity and dopamine- and leptin-related gene expression. FASEB J. (2017) 31:5371–83. doi: 10.1096/fj.201700389R
52. An J, Wang Q, Yi S, Liu X, Jin H, Xu J, et al. The source of the fat significantly affects the results of high-fat diet intervention. Sci Rep. (2022) 12:4315. doi: 10.1038/s41598-022-08249-2
53. Wu F, Guo X, Zhang M, Ou Z, Wu D, Deng L, et al. An Akkermansia muciniphila subtype alleviates high-fat diet-induced metabolic disorders and inhibits the neurodegenerative process in mice. Anaerobe. (2020) 61:102138. doi: 10.1016/j.anaerobe.2019.102138
54. Lin X-Q, Chen W, Ma K, Liu Z-Z, Gao Y, Zhang J-G, et al. Akkermansia muciniphila suppresses high-fat diet-induced obesity and related metabolic disorders in beagles. Molecules. (2022) 27. doi: 10.3390/molecules27186074
55. Xu Y, Wang N, Tan H-Y, Li S, Zhang C, Feng Y. Function of akkermansia muciniphila in obesity: interactions with lipid metabolism, immune response and gut systems. Front Microbiol. (2020) 11:219. doi: 10.3389/fmicb.2020.00219
56. Zaloga GP. Narrative Review of n-3 Polyunsaturated Fatty Acid Supplementation upon Immune Functions, Resolution Molecules and Lipid Peroxidation. Nutrients. (2021) 13:662. doi: 10.3390/nu13020662
57. Costantini L, Molinari R, Farinon B, Merendino N. Impact of omega-3 fatty acids on the gut microbiota. Int J Mol Sci. (2017) 18:2645. doi: 10.3390/ijms18122645
58. Wang B, Kong Q, Li X, Zhao J, Zhang H, Chen W, et al. A high-fat diet increases gut microbiota biodiversity and energy expenditure due to nutrient difference. Nutrients. (2020) 12:3197. doi: 10.3390/nu12103197
59. Murga-Garrido SM, Hong Q, Cross T-WL, Hutchison ER, Han J, Thomas SP, et al. Gut microbiome variation modulates the effects of dietary fiber on host metabolism. Microbiome. (2021) 9:117. doi: 10.1186/s40168-021-01061-6
60. Gibb RD, Sloan KJ, McRorie JW. Psyllium is a natural nonfermented gel-forming fiber that is effective for weight loss: A comprehensive review and meta-analysis. J Am Assoc Nurse Pract. (2023) 35:468–76. doi: 10.1097/JXX.0000000000000882
61. Zhang Z, Lin T, Meng Y, Hu M, Shu L, Jiang H, et al. FOS/GOS attenuates high-fat diet induced bone loss via reversing microbiota dysbiosis, high intestinal permeability and systemic inflammation in mice. Metabolism. (2021) 119:154767. doi: 10.1016/j.metabol.2021.154767
62. David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. (2014) 505:559–63. doi: 10.1038/nature12820
63. Davis LMG, Martínez I, Walter J, Goin C, Hutkins RW. Barcoded pyrosequencing reveals that consumption of galactooligosaccharides results in a highly specific bifidogenic response in humans. PloS One. (2011) 6:e25200. doi: 10.1371/journal.pone.0025200
64. Holscher HD, Bauer LL, Gourineni V, Pelkman CL, Fahey GC, Swanson KS. Agave inulin supplementation affects the fecal microbiota of healthy adults participating in a randomized, double-blind, placebo-controlled, crossover trial. J Nutr. (2015) 145:2025–32. doi: 10.3945/jn.115.217331
65. Corrêa RO, Castro PR, Fachi JL, Nirello VD, El-Sahhar S, Imada S, et al. Inulin diet uncovers complex diet-microbiota-immune cell interactions remodeling the gut epithelium. Microbiome. (2023) 11:90. doi: 10.1186/s40168-023-01520-2
66. Do MH, Lee E, Oh M-J, Kim Y, Park H-Y. High-glucose or -fructose diet cause changes of the gut microbiota and metabolic disorders in mice without body weight change. Nutrients. (2018) 10:761. doi: 10.3390/nu10060761
67. Suez J, Korem T, Zeevi D, Zilberman-Schapira G, Thaiss CA, Maza O, et al. Artificial sweeteners induce glucose intolerance by altering the gut microbiota. Nature. (2014) 514:181–6. doi: 10.1038/nature13793
68. Sonnenburg ED, Smits SA, Tikhonov M, Higginbottom SK, Wingreen NS, Sonnenburg JL. Diet-induced extinctions in the gut microbiota compound over generations. Nature. (2016) 529:212–5. doi: 10.1038/nature16504
69. Crost EH, Le Gall G, Laverde-Gomez JA, Mukhopadhya I, Flint HJ, Juge N. Mechanistic Insights Into the Cross-Feeding of Ruminococcus gnavus and Ruminococcus bromii on Host and Dietary Carbohydrates. Front Microbiol. (2018) 9:2558. doi: 10.3389/fmicb.2018.02558
70. Dank A, van Mastrigt O, Boeren S, Lillevang SK, Abee T, Smid EJ. Propionibacterium freudenreichii thrives in microaerobic conditions by complete oxidation of lactate to CO2. Environ Microbiol. (2021) 23:3116–29. doi: 10.1111/1462-2920.15532
71. Baxter NT, Schmidt AW, Venkataraman A, Kim KS, Waldron C, Schmidt TM. Dynamics of human gut microbiota and short-chain fatty acids in response to dietary interventions with three fermentable fibers. mBio. (2019) 10:e02566–18. doi: 10.1128/mBio.02566-18
72. Medawar E, Haange S-B, Rolle-Kampczyk U, Engelmann B, Dietrich A, Thieleking R, et al. Gut microbiota link dietary fiber intake and short-chain fatty acid metabolism with eating behavior. Transl Psychiatry. (2021) 11:1–11. doi: 10.1038/s41398-021-01620-3
73. Holmes ZC, Villa MM, Durand HK, Jiang S, Dallow EP, Petrone BL, et al. Microbiota responses to different prebiotics are conserved within individuals and associated with habitual fiber intake. Microbiome. (2022) 10:114. doi: 10.1186/s40168-022-01307-x
74. Deehan EC, Yang C, Perez-Muñoz ME, Nguyen NK, Cheng CC, Triador L, et al. Precision microbiome modulation with discrete dietary fiber structures directs short-chain fatty acid production. Cell Host Microbe. (2020) 27:389–404.e6. doi: 10.1016/j.chom.2020.01.006
75. Kjølbæk L, Benítez-Páez A, Gómez Del Pulgar EM, Brahe LK, Liebisch G, Matysik S, et al. Arabinoxylan oligosaccharides and polyunsaturated fatty acid effects on gut microbiota and metabolic markers in overweight individuals with signs of metabolic syndrome: A randomized cross-over trial. Clin Nutr. (2020) 39:67–79. doi: 10.1016/j.clnu.2019.01.012
76. Miclotte L, Van de Wiele T. Food processing, gut microbiota and the globesity problem. Crit Rev Food Sci Nutr. (2020) 60:1769–82. doi: 10.1080/10408398.2019.1596878
77. Chassaing B, Koren O, Goodrich JK, Poole AC, Srinivasan S, Ley RE, et al. Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature. (2015) 519:92–6. doi: 10.1038/nature14232
78. Collins J, Robinson C, Danhof H, Knetsch CW, van Leeuwen HC, Lawley TD, et al. Dietary trehalose enhances virulence of epidemic Clostridium difficile. Nature. (2018) 553:291–4. doi: 10.1038/nature25178
79. Furuhashi H, Higashiyama M, Okada Y, Kurihara C, Wada A, Horiuchi K, et al. Dietary emulsifier polysorbate-80-induced small-intestinal vulnerability to indomethacin-induced lesions via dysbiosis. J Gastroenterol Hepatol. (2020) 35:110–7. doi: 10.1111/jgh.14808
80. Harusato A, Chassaing B, Dauriat CJG, Ushiroda C, Seo W, Itoh Y. Dietary emulsifiers exacerbate food allergy and colonic type 2 immune response through microbiota modulation. Nutrients. (2022) 14:4983. doi: 10.3390/nu14234983
81. Martínez Steele E, Popkin BM, Swinburn B, Monteiro CA. The share of ultra-processed foods and the overall nutritional quality of diets in the US: evidence from a nationally representative cross-sectional study. Popul Health Metr. (2017) 15:6. doi: 10.1186/s12963-017-0119-3
82. Dostal A, Lacroix C, Bircher L, Pham VT, Follador R, Zimmermann MB, et al. Iron modulates butyrate production by a child gut microbiota. In Vitro. mBio. (2015) 6:e01453–01415. doi: 10.1128/mBio.01453-15
83. Park J, Hosomi K, Kawashima H, Chen Y-A, Mohsen A, Ohno H, et al. Dietary vitamin B1 intake influences gut microbial community and the consequent production of short-chain fatty acids. Nutrients. (2022) 14:2078. doi: 10.3390/nu14102078
84. Clemente-Suárez VJ, Beltrán-Velasco AI, Redondo-Flórez L, Martín-Rodríguez A, Tornero-Aguilera JF. Global impacts of western diet and its effects on metabolism and health: A narrative review. Nutrients. (2023) 15:2749. doi: 10.3390/nu15122749
85. Food and Agriculture Organization of the United Nations, World Health Organization, Joint FAO/WHO Expert Consultation on Evaluation of Health and Nutritional Properties of Probiotics in Food including Powder Milk with Live Lactic Acid Bacteria, Joint FAO/WHO Working Group on Drafting Guidelines for the Evaluation of Probiotics in Food eds. Probiotics in food: health and nutritional properties and guidelines for evaluation: Report of a Joint FAO/WHO Expert Consultation on Evaluation of Health and Nutritional Properties of Probiotics in Food including Powder Milk with Live Lactic Acid Bacteria, Cordoba, Argentina, 1-4 October 2001 [and] Report of a Joint FAO/WHO Working Group on Drafting Guidelines for the Evaluation of Probiotics in Food, London, Ontario, Canada, 30 April -1 May 2002. Rome [Italy]: Food and Agriculture Organization of the United Nations, World Health Organization (2006). p. 50.
86. Markowiak-Kopeć P, Śliżewska K. The effect of probiotics on the production of short-chain fatty acids by human intestinal microbiome. Nutrients. (2020) 12:1107. doi: 10.3390/nu12041107
87. Wang L, Zhang J, Guo Z, Kwok L, Ma C, Zhang W, et al. Effect of oral consumption of probiotic Lactobacillus planatarum P-8 on fecal microbiota, SIgA, SCFAs, and TBAs of adults of different ages. Nutrition. (2014) 30:776–783.e1. doi: 10.1016/j.nut.2013.11.018
88. Zhou Y, Xu H, Xu J, Guo X, Zhao H, Chen Y, et al. prausnitzii and its supernatant increase SCFAs-producing bacteria to restore gut dysbiosis in TNBS-induced colitis. AMB Express. (2021) 11:33. doi: 10.1186/s13568-021-01197-6
89. Demirci M. Could faecalibacterium prausnitzii amount in the gut microbiota be used to monitor the COVID-19 severity? Turk J Gastroenterol. (2022) 33:899–900. doi: 10.5152/tjg.2022.22100
90. Maioli TU, Borras-Nogues E, Torres L, Barbosa SC, Martins VD, Langella P, et al. Possible benefits of faecalibacterium prausnitzii for obesity-associated gut disorders. Front Pharmacol. (2021) 12:740636. doi: 10.3389/fphar.2021.740636
91. Hu W, Gao W, Liu Z, Fang Z, Wang H, Zhao J, et al. Specific strains of faecalibacterium prausnitzii ameliorate nonalcoholic fatty liver disease in mice in association with gut microbiota regulation. Nutrients. (2022) 14:2945. doi: 10.3390/nu14142945
92. Lopez-Siles M, Duncan SH, Garcia-Gil LJ, Martinez-Medina M. Faecalibacterium prausnitzii: from microbiology to diagnostics and prognostics. ISME J. (2017) 11:841–52. doi: 10.1038/ismej.2016.176
93. Zhao S, Liu W, Wang J, Shi J, Sun Y, Wang W, et al. Akkermansia muciniphila improves metabolic profiles by reducing inflammation in chow diet-fed mice. J Mol Endocrinol. (2017) 58:1–14. doi: 10.1530/JME-16-0054
94. Chang YH, Jeong CH, Cheng WN, Choi Y, Shin DM, Lee S, et al. Quality characteristics of yogurts fermented with short-chain fatty acid-producing probiotics and their effects on mucin production and probiotic adhesion onto human colon epithelial cells. J Dairy Sci. (2021) 104:7415–25. doi: 10.3168/jds.2020-19820
95. Moens F, Van den Abbeele P, Basit AW, Dodoo C, Chatterjee R, Smith B, et al. A four-strain probiotic exerts positive immunomodulatory effects by enhancing colonic butyrate production in vitro. Int J Pharm. (2019) 555:1–10. doi: 10.1016/j.ijpharm.2018.11.020
96. Thananimit S, Pahumunto N, Teanpaisan R. Characterization of short chain fatty acids produced by selected potential probiotic lactobacillus strains. Biomolecules. (2022) 12:1829. doi: 10.3390/biom12121829
97. Farooq U, Liu X, Zhang H. Enhancement of short chain fatty acid production by co-cultures of probiotics fermentation with pearl millet (Pennisetum glaucum) fibre fractions. J PURE Appl MICROBIO. (2017) 11:2031–8. doi: 10.22207/JPAM.11.4.47
98. McMurdie PJ, Stoeva MK, Justice N, Nemchek M, Sieber CMK, Tyagi S, et al. Increased circulating butyrate and ursodeoxycholate during probiotic intervention in humans with type 2 diabetes. BMC Microbiol. (2022) 22:19. doi: 10.1186/s12866-021-02415-8
99. Zhu H, Cao C, Wu Z, Zhang H, Sun Z, Wang M, et al. The probiotic L. casei Zhang slows the progression of acute and chronic kidney disease. Cell Metab. (2021) 33:1926–1942.e8. doi: 10.1016/j.cmet.2021.06.014
100. Nagpal R, Wang S, Ahmadi S, Hayes J, Gagliano J, Subashchandrabose S, et al. Human-origin probiotic cocktail increases short-chain fatty acid production via modulation of mice and human gut microbiome. Sci Rep. (2018) 8:12649. doi: 10.1038/s41598-018-30114-4
101. Łagowska K, Drzymała-Czyż S. A low glycemic index, energy-restricted diet but not Lactobacillus rhamnosus supplementation changes fecal short-chain fatty acid and serum lipid concentrations in women with overweight or obesity and polycystic ovary syndrome. Eur Rev Med Pharmacol Sci. (2022) 26:917–26. doi: 10.26355/eurrev_202202_28001
102. Adams JB, Johansen LJ, Powell LD, Quig D, Rubin RA. Gastrointestinal flora and gastrointestinal status in children with autism–comparisons to typical children and correlation with autism severity. BMC Gastroenterol. (2011) 11:22. doi: 10.1186/1471-230X-11-22
103. Rosshart SP, Vassallo BG, Angeletti D, Hutchinson DS, Morgan AP, Takeda K, et al. Wild mouse gut microbiota promotes host fitness and improves disease resistance. Cell. (2017) 171:1015–1028.e13. doi: 10.1016/j.cell.2017.09.016
104. Gorreja F, Walker WA. The potential role of adherence factors in probiotic function in the gastrointestinal tract of adults and pediatrics: a narrative review of experimental and human studies. Gut Microbes. (2022) 14:2149214. doi: 10.1080/19490976.2022.2149214
105. Pupa P, Apiwatsiri P, Sirichokchatchawan W, Pirarat N, Muangsin N, Shah AA, et al. The efficacy of three double-microencapsulation methods for preservation of probiotic bacteria. Sci Rep. (2021) 11:13753. doi: 10.1038/s41598-021-93263-z
106. Yoha KS, Nida S, Dutta S, Moses JA, Anandharamakrishnan C. Targeted delivery of probiotics: perspectives on research and commercialization. Probiotics Antimicrob Proteins. (2022) 14:15–48. doi: 10.1007/s12602-021-09791-7
107. Gao J, Sadiq FA, Zheng Y, Zhao J, He G, Sang Y. Biofilm-based delivery approaches and specific enrichment strategies of probiotics in the human gut. Gut Microbes. (2022) 14:2126274. doi: 10.1080/19490976.2022.2126274
108. Lu Y, Han S, Zhang S, Wang K, Lv L, McClements DJ, et al. The role of probiotic exopolysaccharides in adhesion to mucin in different gastrointestinal conditions. Curr Res Food Sci. (2022) 5:581–9. doi: 10.1016/j.crfs.2022.02.015
109. Miyamoto J, Shimizu H, Hisa K, Matsuzaki C, Inuki S, Ando Y, et al. Host metabolic benefits of prebiotic exopolysaccharides produced by Leuconostoc mesenteroides. Gut Microbes. (2023) 15:2161271. doi: 10.1080/19490976.2022.2161271
110. Athalye-Jape G, Esvaran M, Patole S, Simmer K, Nathan E, Doherty D, et al. Effect of single versus multistrain probiotic in extremely preterm infants: a randomised trial. BMJ Open Gastroenterol. (2022) 9:e000811. doi: 10.1136/bmjgast-2021-000811
111. Patole S, Keil AD, Chang A, Nathan E, Doherty D, Simmer K, et al. Effect of Bifidobacterium breve M-16V supplementation on fecal bifidobacteria in preterm neonates–a randomised double blind placebo controlled trial. PloS One. (2014) 9:e89511. doi: 10.1371/journal.pone.0089511
112. Alcon-Giner C, Dalby MJ, Caim S, Ketskemety J, Shaw A, Sim K, et al. Microbiota supplementation with bifidobacterium and lactobacillus modifies the preterm infant gut microbiota and metabolome: an observational study. Cell Rep Med. (2020) 1:100077. doi: 10.1016/j.xcrm.2020.100077
113. Dogra SK, Chung CK, Wang D, Sakwinska O, Colombo Mottaz S, Sprenger N. Nurturing the early life gut microbiome and immune maturation for long term health. Microorganisms. (2021) 9:2110. doi: 10.3390/microorganisms9102110
114. Lynch CMK, Cowan CSM, Bastiaanssen TFS, Moloney GM, Theune N, van de Wouw M, et al. Critical windows of early-life microbiota disruption on behaviour, neuroimmune function, and neurodevelopment. Brain Behavior Immun. (2023) 108:309–27. doi: 10.1016/j.bbi.2022.12.008
115. Maier L, Goemans CV, Wirbel J, Kuhn M, Eberl C, Pruteanu M, et al. Unravelling the collateral damage of antibiotics on gut bacteria. Nature. (2021) 599:120–4. doi: 10.1038/s41586-021-03986-2
116. Rashid M-U, Zaura E, Buijs MJ, Keijser BJF, Crielaard W, Nord CE, et al. Determining the long-term effect of antibiotic administration on the human normal intestinal microbiota using culture and pyrosequencing methods. Clin Infect Dis. (2015) 60:S77–84. doi: 10.1093/cid/civ137
117. Liu P, Zhang Y, Zhang Z, Huang X, Su X, Yang S, et al. Antibiotic-induced dysbiosis of the gut microbiota impairs gene expression in gut-liver axis of mice. Genes. (2023) 14:1423. doi: 10.3390/genes14071423
118. Stecher B, Hardt W-D. Mechanisms controlling pathogen colonization of the gut. Curr Opin Microbiol. (2011) 14:82–91. doi: 10.1016/j.mib.2010.10.003
119. Kadry AA, El-Antrawy MA, El-Ganiny AM. Impact of short chain fatty acids (SCFAs) on antimicrobial activity of new β-lactam/β-lactamase inhibitor combinations and on virulence of Escherichia coli isolates. J Antibiot. (2023) 76:225–35. doi: 10.1038/s41429-023-00595-1
120. Zhang S, Dogan B, Guo C, Herlekar D, Stewart K, Scherl EJ, et al. Short chain fatty acids modulate the growth and virulence of pathosymbiont escherichia coli and host response. Antibiotics. (2020) 9:462. doi: 10.3390/antibiotics9080462
121. Guinan J, Wang S, Hazbun TR, Yadav H, Thangamani S. Antibiotic-induced decreases in the levels of microbial-derived short-chain fatty acids correlate with increased gastrointestinal colonization of Candida albicans. Sci Rep. (2019) 9:8872. doi: 10.1038/s41598-019-45467-7
122. Zhao X, Jiang Z, Yang F, Wang Y, Gao X, Wang Y, et al. Sensitive and simplified detection of antibiotic influence on the dynamic and versatile changes of fecal short-chain fatty acids. PloS One. (2016) 11:e0167032. doi: 10.1371/journal.pone.0167032
123. Holota Y, Dovbynchuk T, Kaji I, Vareniuk I, Dzyubenko N, Chervinska T, et al. The long-term consequences of antibiotic therapy: Role of colonic short-chain fatty acids (SCFA) system and intestinal barrier integrity. PloS One. (2019) 14:e0220642. doi: 10.1371/journal.pone.0220642
124. van Staa TP, Palin V, Li Y, Welfare W, Felton TW, Dark P, et al. The effectiveness of frequent antibiotic use in reducing the risk of infection-related hospital admissions: results from two large population-based cohorts. BMC Med. (2020) 18:40. doi: 10.1186/s12916-020-1504-5
125. Shekhar S, Petersen FC. The dark side of antibiotics: adverse effects on the infant immune defense against infection. Front Pediatr. (2020) 8:544460. doi: 10.3389/fped.2020.544460
126. Vallianou N, Dalamaga M, Stratigou T, Karampela I, Tsigalou C. Do antibiotics cause obesity through long-term alterations in the gut microbiome? A review of current evidence. Curr Obes Rep. (2021) 10:244–62. doi: 10.1007/s13679-021-00438-w
127. Imhann F, Bonder MJ, Vich Vila A, Fu J, Mujagic Z, Vork L, et al. Proton pump inhibitors affect the gut microbiome. Gut. (2016) 65:740–8. doi: 10.1136/gutjnl-2015-310376
128. McDonald EG, Milligan J, Frenette C, Lee TC. Continuous proton pump inhibitor therapy and the associated risk of recurrent clostridium difficile infection. JAMA Internal Med. (2015) 175:784–91. doi: 10.1001/jamainternmed.2015.42
129. Vich Vila A, Collij V, Sanna S, Sinha T, Imhann F, Bourgonje AR, et al. Impact of commonly used drugs on the composition and metabolic function of the gut microbiota. Nat Commun. (2020) 11:362. doi: 10.1038/s41467-019-14177-z
130. de la Cuesta-Zuluaga J, Mueller NT, Corrales-Agudelo V, Velásquez-Mejía EP, Carmona JA, Abad JM, et al. Metformin is associated with higher relative abundance of mucin-degrading akkermansia muciniphila and several short-chain fatty acid-producing microbiota in the gut. Diabetes Care. (2017) 40:54–62. doi: 10.2337/dc16-1324
131. Wu H, Esteve E, Tremaroli V, Khan MT, Caesar R, Mannerås-Holm L, et al. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat Med. (2017) 23:850–8. doi: 10.1038/nm.4345
132. Rogers M a. M, Aronoff DM. The influence of non-steroidal anti-inflammatory drugs on the gut microbiome. Clin Microbiol Infect. (2016) 22:178.e1–9. doi: 10.1016/j.cmi.2015.10.003
133. Hernandez-Sanabria E, Heiremans E, Calatayud Arroyo M, Props R, Leclercq L, Snoeys J, et al. Short-term supplementation of celecoxib-shifted butyrate production on a simulated model of the gut microbial ecosystem and ameliorated in vitro inflammation. NPJ Biofilms Microbiomes. (2020) 6:1–12. doi: 10.1038/s41522-020-0119-0
134. Wang Y, Ashokan K. Physical exercise: an overview of benefits from psychological level to genetics and beyond. Front Physiol. (2021) 12:731858. doi: 10.3389/fphys.2021.731858
135. Giles A, Nasstasia Y, Baker A, Kelly B, Dascombe B, Halpin S, et al. Exercise as treatment for youth with major depression: the healthy body healthy mind feasibility study. J Psychiatr Pract. (2020) 26:444–60. doi: 10.1097/PRA.0000000000000516
136. Barrón-Cabrera E, Ramos-Lopez O, González-Becerra K, Riezu-Boj JI, Milagro FI, Martínez-López E, et al. Epigenetic modifications as outcomes of exercise interventions related to specific metabolic alterations: A systematic review. Lifestyle Genom. (2019) 12:25–44. doi: 10.1159/000503289
137. Valder S, Brinkmann C. Exercise for the diabetic gut—Potential health effects and underlying mechanisms. Nutrients. (2022) 14:813. doi: 10.3390/nu14040813
138. Carey RA, Montag D. Exploring the relationship between gut microbiota and exercise: short-chain fatty acids and their role in metabolism. BMJ Open Sport Exercise Med. (2021) 7:e000930. doi: 10.1136/bmjsem-2020-000930
139. Allen JM, Mailing LJ, Niemiro GM, Moore R, Cook MD, White BA, et al. Exercise alters gut microbiota composition and function in lean and obese humans. Med Sci Sports Exerc. (2018) 50:747–57. doi: 10.1249/MSS.0000000000001495
140. Hintikka JE, Ahtiainen JP, Permi P, Jalkanen S, Lehtonen M, Pekkala S. Aerobic exercise training and gut microbiome-associated metabolic shifts in women with overweight: a multi-omic study. Sci Rep. (2023) 13:11228. doi: 10.1038/s41598-023-38357-6
141. Magzal F, Shochat T, Haimov I, Tamir S, Asraf K, Tuchner-Arieli M, et al. Increased physical activity improves gut microbiota composition and reduces short-chain fatty acid concentrations in older adults with insomnia. Sci Rep. (2022) 12:2265. doi: 10.1038/s41598-022-05099-w
142. Wang Y, Wang H, Howard AG, Meyer KA, Tsilimigras MCB, Avery CL, et al. Circulating short-chain fatty acids are positively associated with adiposity measures in Chinese adults. Nutrients. (2020) 12:2127. doi: 10.3390/nu12072127
143. Martínez-Cuesta MC, Del Campo R, Garriga-García M, Peláez C, Requena T. Taxonomic characterization and short-chain fatty acids production of the obese microbiota. Front Cell Infect Microbiol. (2021) 11:598093. doi: 10.3389/fcimb.2021.598093
144. Allen JM, Mailing LJ, Cohrs J, Salmonson C, Fryer JD, Nehra V, et al. Exercise training-induced modification of the gut microbiota persists after microbiota colonization and attenuates the response to chemically-induced colitis in gnotobiotic mice. Gut Microbes. (2017) 9:115–30. doi: 10.1080/19490976.2017.1372077
145. de Geus EJC. Genetic pathways underlying individual differences in regular physical activity. Exerc Sport Sci Rev. (2023) 51:2–18. doi: 10.1249/JES.0000000000000305
146. Barh D, Ahmetov II. Sports, Exercise, and Nutritional Genomics: Current Status and Future Directions. Elsevier, Amsterdam: Academic Press (2019). p. 610.
147. Stavsky J, Maitra R. The synergistic role of diet and exercise in the prevention, pathogenesis, and management of ulcerative colitis: an underlying metabolic mechanism. Nutr Metab Insights. (2019) 12:1178638819834526. doi: 10.1177/1178638819834526
148. Nagano T, Yano H. Effect of dietary cellulose nanofiber and exercise on obesity and gut microbiota in mice fed a high-fat-diet. Bioscience Biotechnology Biochem. (2020) 84:613–20. doi: 10.1080/09168451.2019.1690975
149. Hippe B, Zwielehner J, Liszt K, Lassl C, Unger F, Haslberger AG. Quantification of butyryl CoA:acetate CoA-transferase genes reveals different butyrate production capacity in individuals according to diet and age. FEMS Microbiol Lett. (2011) 316:130–5. doi: 10.1111/fml.2011.316.issue-2
150. Boets E, Gomand SV, Deroover L, Preston T, Vermeulen K, De Preter V, et al. Systemic availability and metabolism of colonic-derived short-chain fatty acids in healthy subjects: a stable isotope study. J Physiol. (2017) 595:541–55. doi: 10.1113/JP272613
151. Kim J-H. The physical activity level in female affects colon transit time. J Neurogastroenterol Motil. (2012) 18:4–5. doi: 10.5056/jnm.2012.18.1.4
152. Scheiman J, Luber JM, Chavkin TA, MacDonald T, Tung A, Pham L-D, et al. Meta-omics analysis of elite athletes identifies a performance-enhancing microbe that functions via lactate metabolism. Nat Med. (2019) 25:1104–9. doi: 10.1038/s41591-019-0485-4
153. Ticinesi A, Lauretani F, Tana C, Nouvenne A, Ridolo E, Meschi T. Exercise and immune system as modulators of intestinal microbiome: implications for the gut-muscle axis hypothesis. Exerc Immunol Rev. (2019) 25:84–95.
154. Severinsen MCK, Pedersen BK. Muscle-organ crosstalk: the emerging roles of myokines. Endocr Rev. (2020) 41:594–609. doi: 10.1210/endrev/bnaa016
155. Frampton J, Murphy KG, Frost G, Chambers ES. Short-chain fatty acids as potential regulators of skeletal muscle metabolism and function. Nat Metab. (2020) 2:840–8. doi: 10.1038/s42255-020-0188-7
156. De A, Da T, Rf K, Ap C. Benefits and risks of bariatric surgery in adults: A review. JAMA. (2020) 324. doi: 10.1001/jama.2020.12567
157. Stahl JM, Malhotra S. “Obesity surgery indications and contraindications.,”, in: StatPearls (2023). Treasure Island (FL: StatPearls Publishing. Available online at: http://www.ncbi.nlm.nih.gov/books/NBK513285/ (Accessed November 16, 2023).
158. Yamamura R, Nakamura K, Ukawa S, Okada E, Nakagawa T, Imae A, et al. Fecal short-chain fatty acids and obesity in a community-based Japanese population: The DOSANCO Health Study. Obes Res Clin Pract. (2021) 15:345–50. doi: 10.1016/j.orcp.2021.06.003
159. Farup PG, Valeur J. Changes in faecal short-chain fatty acids after weight-loss interventions in subjects with morbid obesity. Nutrients. (2020) 12:802. doi: 10.3390/nu12030802
160. Ecklu-Mensah G, Choo-Kang C, Maseng MG, Donato S, Bovet P, Viswanathan B, et al. Gut microbiota and fecal short chain fatty acids differ with adiposity and country of origin: the METS-microbiome study. Nat Commun. (2023) 14:5160. doi: 10.1038/s41467-023-40874-x
161. Barczyńska R, Litwin M, Sliżewska K, Szalecki M, Berdowska A, Bandurska K, et al. Bacterial microbiota and fatty acids in the faeces of overweight and obese children. Polish J Microbiol. (2018) 67:339–45. doi: 10.21307/pjm-2018-041
162. Fang W, Xue H, Chen X, Chen K, Ling W. Supplementation with sodium butyrate modulates the composition of the gut microbiota and ameliorates high-fat diet-induced obesity in mice. J Nutr. (2019) 149:747–54. doi: 10.1093/jn/nxy324
163. Joung H, Chu J, Kim B-K, Choi I-S, Kim W, Park T-S. Probiotics ameliorate chronic low-grade inflammation and fat accumulation with gut microbiota composition change in diet-induced obese mice models. Appl Microbiol Biotechnol. (2021) 105:1203–13. doi: 10.1007/s00253-020-11060-6
164. Wang T, Yan H, Lu Y, Li X, Wang X, Shan Y, et al. Anti-obesity effect of Lactobacillus rhamnosus LS-8 and Lactobacillus crustorum MN047 on high-fat and high-fructose diet mice base on inflammatory response alleviation and gut microbiota regulation. Eur J Nutr. (2020) 59:2709–28. doi: 10.1007/s00394-019-02117-y
165. Nguyen HT, Gu M, Werlinger P, Cho J-H, Cheng J, Suh J-W. Lactobacillus sakei MJM60958 as a potential probiotic alleviated non-alcoholic fatty liver disease in mice fed a high-fat diet by modulating lipid metabolism, inflammation, and gut microbiota. Int J Mol Sci. (2022) 23:13436. doi: 10.3390/ijms232113436
166. Liang Y, Liang S, Zhang Y, Deng Y, He Y, Chen Y, et al. Oral administration of compound probiotics ameliorates HFD-induced gut microbe dysbiosis and chronic metabolic inflammation via the G protein-coupled receptor 43 in non-alcoholic fatty liver disease rats. Probiotics Antimicrob Proteins. (2019) 11:175–85. doi: 10.1007/s12602-017-9378-3
167. Kim HR, Seo E, Oh S, Seo M, Byun K, Kim B-Y. Anti-obesity effects of multi-strain probiotics in mice with high-carbohydrate diet-induced obesity and the underlying molecular mechanisms. Nutrients. (2022) 14:5173. doi: 10.3390/nu14235173
168. Prentice PM, Schoemaker MH, Vervoort J, Hettinga K, Lambers TT, van Tol EAF, et al. Human milk short-chain fatty acid composition is associated with adiposity outcomes in infants. J Nutr. (2019) 149:716–22. doi: 10.1093/jn/nxy320
169. Olga L, van Diepen JA, Chichlowski M, Petry CJ, Vervoort J, Dunger DB, et al. Butyrate in human milk: associations with milk microbiota, milk intake volume, and infant growth. Nutrients. (2023) 15:916. doi: 10.3390/nu15040916
170. Kondo T, Kishi M, Fushimi T, Kaga T. Acetic acid upregulates the expression of genes for fatty acid oxidation enzymes in liver to suppress body fat accumulation. J Agric Food Chem. (2009) 57:5982–6. doi: 10.1021/jf900470c
171. Liu L, Fu C, Li F. Acetate affects the process of lipid metabolism in rabbit liver, skeletal muscle and adipose tissue. Anim (Basel). (2019) 9:799. doi: 10.3390/ani9100799
172. Shah S, Fillier T, Pham TH, Thomas R, Cheema SK. Intraperitoneal administration of short-chain fatty acids improves lipid metabolism of long–evans rats in a sex-specific manner. Nutrients. (2021) 13:892. doi: 10.3390/nu13030892
173. den Besten G, Bleeker A, Gerding A, van Eunen K, Havinga R, van Dijk TH, et al. Short-chain fatty acids protect against high-fat diet-induced obesity via a PPARγ-dependent switch from lipogenesis to fat oxidation. Diabetes. (2015) 64:2398–408. doi: 10.2337/db14-1213
174. Shimizu H, Masujima Y, Ushiroda C, Mizushima R, Ohue-Kitano R, Kimura I. Dietary short-chain fatty acid intake improves the hepatic metabolic condition via FFAR3. Sci Rep. (2019) 9:16574. doi: 10.1038/s41598-019-53242-x
175. Samuel BS, Shaito A, Motoike T, Rey FE, Backhed F, Manchester JK, et al. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proc Natl Acad Sci U.S.A. (2008) 105:16767–72. doi: 10.1073/pnas.0808567105
176. Cani PD, Possemiers S, Van de Wiele T, Guiot Y, Everard A, Rottier O, et al. Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP-2-driven improvement of gut permeability. Gut. (2009) 58:1091–103. doi: 10.1136/gut.2008.165886
177. Collins L, Costello RA. “Glucagon-like peptide-1 receptor agonists.,”, in: StatPearls (2022). Treasure Island (FL: StatPearls Publishing. Available online at: http://www.ncbi.nlm.nih.gov/books/NBK551568/ (Accessed March 6, 2023).
178. Senior M. After GLP-1, what’s next for weight loss? Nat Biotechnol. (2023) 41:740–3. doi: 10.1038/s41587-023-01818-4
179. Li H, Donelan W, Wang F, Zhang P, Yang L, Ding Y, et al. GLP-1 induces the expression of FNDC5 derivatives that execute lipolytic actions. Front Cell Dev Biol. (2021) 9:777026. doi: 10.3389/fcell.2021.777026
180. Bendotti G, Montefusco L, Lunati ME, Usuelli V, Pastore I, Lazzaroni E, et al. The anti-inflammatory and immunological properties of GLP-1 Receptor Agonists. Pharmacol Res. (2022) 182:106320. doi: 10.1016/j.phrs.2022.106320
181. Li Z, Li S, Wang N, Xue P, Li Y. Liraglutide, a glucagon-like peptide-1 receptor agonist, suppresses osteoclastogenesis through the inhibition of NF-κB and MAPK pathways via GLP-1R. Biomedicine Pharmacotherapy. (2020) 130:110523. doi: 10.1016/j.biopha.2020.110523
182. Vitale M, Giacco R, Laiola M, Della Pepa G, Luongo D, Mangione A, et al. Acute and chronic improvement in postprandial glucose metabolism by a diet resembling the traditional Mediterranean dietary pattern: Can SCFAs play a role? Clin Nutr. (2021) 40:428–37. doi: 10.1016/j.clnu.2020.05.025
183. Vetrani C, Costabile G, Luongo D, Naviglio D, Rivellese AA, Riccardi G, et al. Effects of whole-grain cereal foods on plasma short chain fatty acid concentrations in individuals with the metabolic syndrome. Nutrition. (2016) 32:217–21. doi: 10.1016/j.nut.2015.08.006
184. Zhao L. The gut microbiota and obesity: from correlation to causality. Nat Rev Microbiol. (2013) 11:639–47. doi: 10.1038/nrmicro3089
185. Hotamisligil GS. Inflammation, metaflammation and immunometabolic disorders. Nature. (2017) 542:177–85. doi: 10.1038/nature21363
186. Amabebe E, Robert FO, Agbalalah T, Orubu ESF. Microbial dysbiosis-induced obesity: role of gut microbiota in homoeostasis of energy metabolism. Br J Nutr. (2020) 123:1127–37. doi: 10.1017/S0007114520000380
187. Pereira M, Durso DF, Bryant CE, Kurt-Jones EA, Silverman N, Golenbock DT, et al. The IRAK4 scaffold integrates TLR4-driven TRIF and MYD88 signaling pathways. Cell Rep. (2022) 40:111225. doi: 10.1016/j.celrep.2022.111225
188. Föh B, Buhre JS, Lunding HB, Moreno-Fernandez ME, König P, Sina C, et al. Microbial metabolite butyrate promotes induction of IL-10+IgM+ plasma cells. PloS One. (2022) 17:e0266071. doi: 10.1371/journal.pone.0266071
189. Ipseiz N, Pickering RJ, Rosas M, Tyrrell VJ, Davies LC, Orr SJ, et al. Tissue-resident macrophages actively suppress IL-1beta release via a reactive prostanoid/IL-10 pathway. EMBO J. (2020) 39:e103454. doi: 10.15252/embj.2019103454
190. Sun M, Wu W, Chen L, Yang W, Huang X, Ma C, et al. Microbiota-derived short-chain fatty acids promote Th1 cell IL-10 production to maintain intestinal homeostasis. Nat Commun. (2018) 9:3555. doi: 10.1038/s41467-018-05901-2
191. Lopes RL, Borges TJ, Zanin RF, Bonorino C. IL-10 is required for polarization of macrophages to M2-like phenotype by mycobacterial DnaK (heat shock protein 70). Cytokine. (2016) 85:123–9. doi: 10.1016/j.cyto.2016.06.018
192. Chen S, Saeed AFUH, Liu Q, Jiang Q, Xu H, Xiao GG, et al. Macrophages in immunoregulation and therapeutics. Sig Transduct Target Ther. (2023) 8:1–35. doi: 10.1038/s41392-023-01452-1
193. van den Brink W, van Bilsen J, Salic K, Hoevenaars FPM, Verschuren L, Kleemann R, et al. Current and future nutritional strategies to modulate inflammatory dynamics in metabolic disorders. Front Nutr. (2019) 6:129. doi: 10.3389/fnut.2019.00129
194. Lemmer IL, Willemsen N, Hilal N, Bartelt A. A guide to understanding endoplasmic reticulum stress in metabolic disorders. Mol Metab. (2021) 47:101169. doi: 10.1016/j.molmet.2021.101169
195. Xu L, Yan X, Zhao Y, Wang J, Liu B, Yu S, et al. Macrophage polarization mediated by mitochondrial dysfunction induces adipose tissue inflammation in obesity. Int J Mol Sci. (2022) 23:9252. doi: 10.3390/ijms23169252
196. Miao H, Ou J, Ma Y, Guo F, Yang Z, Wiggins MD, et al. Macrophage CGI-58 deficiency activates ROS-inflammasome pathway to promote insulin resistance in mice. Cell Rep. (2014) 7:223–35. doi: 10.1016/j.celrep.2014.02.047
197. Stienstra R, van Diepen JA, Tack CJ, Zaki M, van de Veerdonk FL, Perera D, et al. Inflammasome is a central player in the induction of obesity and insulin resistance. Proc Natl Acad Sci. (2011) 108:15324–9. doi: 10.1073/pnas.1100255108
198. Sokolova M, Yang K, Hansen SH, Louwe MC, Kummen M, Hov JER, et al. NLRP3 inflammasome deficiency attenuates metabolic disturbances involving alterations in the gut microbial profile in mice exposed to high fat diet. Sci Rep. (2020) 10:21006. doi: 10.1038/s41598-020-76497-1
199. Netea MG, Joosten LAB, Latz E, Mills KHG, Natoli G, Stunnenberg HG, et al. Trained immunity: A program of innate immune memory in health and disease. Science. (2016) 352:aaf1098. doi: 10.1126/science.aaf1098
200. Riksen NP, Netea MG. Immunometabolic control of trained immunity. Mol Aspects Med. (2021) 77:100897. doi: 10.1016/j.mam.2020.100897
201. Kelly B, O’Neill LA. Metabolic reprogramming in macrophages and dendritic cells in innate immunity. Cell Res. (2015) 25:771–84. doi: 10.1038/cr.2015.68
202. Soto-Heredero G, Gómez de las Heras MM, Gabandé-Rodríguez E, Oller J, Mittelbrunn M. Glycolysis – a key player in the inflammatory response. FEBS J. (2020) 287:3350–69. doi: 10.1111/febs.15327
203. Zubair K, You C, Kwon G, Kang K. Two faces of macrophages: training and tolerance. Biomedicines. (2021) 9:1596. doi: 10.3390/biomedicines9111596
204. Leprivier G, Rotblat B. How does mTOR sense glucose starvation? AMPK is the usual suspect. Cell Death Discovery. (2020) 6:1–5. doi: 10.1038/s41420-020-0260-9
205. Kostiuchenko O, Lushnikova I, Kowalczyk M, Skibo G. mTOR/α-ketoglutarate-mediated signaling pathways in the context of brain neurodegeneration and neuroprotection. BBA Adv. (2022) 2:100066. doi: 10.1016/j.bbadva.2022.100066
206. Poznyak AV, Sukhorukov VN, Zhuravlev A, Orekhov NA, Kalmykov V, Orekhov AN. Modulating mTOR signaling as a promising therapeutic strategy for atherosclerosis. Int J Mol Sci. (2022) 23:1153. doi: 10.3390/ijms23031153
207. Netea MG, Domínguez-Andrés J, Barreiro LB, Chavakis T, Divangahi M, Fuchs E, et al. Defining trained immunity and its role in health and disease. Nat Rev Immunol. (2020) 20:375–88. doi: 10.1038/s41577-020-0285-6
208. Krautkramer KA, Kreznar JH, Romano KA, Vivas EI, Barrett-Wilt GA, Rabaglia ME, et al. Diet-microbiota interactions mediate global epigenetic programming in multiple host tissues. Mol Cell. (2016) 64:982–92. doi: 10.1016/j.molcel.2016.10.025
209. Kumar H, Lund R, Laiho A, Lundelin K, Ley RE, Isolauri E, et al. Gut microbiota as an epigenetic regulator: pilot study based on whole-genome methylation analysis. mBio. (2014) 5:e02113–14. doi: 10.1128/mBio.02113-14
210. Woo V, Alenghat T. Epigenetic regulation by gut microbiota. Gut Microbes. (2022) 14:2022407. doi: 10.1080/19490976.2021.2022407
211. Williamson AK, Zhu Z, Yuan Z-M. Epigenetic mechanisms behind cellular sensitivity to DNA damage. Cell Stress. (2018) 2:176–80. doi: 10.15698/cst2018.07.145
212. Ansari I, Raddatz G, Gutekunst J, Ridnik M, Cohen D, Abu-Remaileh M, et al. The microbiota programs DNA methylation to control intestinal homeostasis and inflammation. Nat Microbiol. (2020) 5:610–9. doi: 10.1038/s41564-019-0659-3
213. Takahashi K, Sugi Y, Hosono A, Kaminogawa S. Epigenetic regulation of TLR4 gene expression in intestinal epithelial cells for the maintenance of intestinal homeostasis. J Immunol. (2009) 183:6522–9. doi: 10.4049/jimmunol.0901271
214. Obata Y, Furusawa Y, Endo TA, Sharif J, Takahashi D, Atarashi K, et al. The epigenetic regulator Uhrf1 facilitates the proliferation and maturation of colonic regulatory T cells. Nat Immunol. (2014) 15:571–9. doi: 10.1038/ni.2886
215. Poupeau A, Garde C, Sulek K, Citirikkaya K, Treebak JT, Arumugam M, et al. Genes controlling the activation of natural killer lymphocytes are epigenetically remodeled in intestinal cells from germ-free mice. FASEB J. (2019) 33:2719–31. doi: 10.1096/fj.201800787R
216. Ryan FJ, Ahern AM, Fitzgerald RS, Laserna-Mendieta EJ, Power EM, Clooney AG, et al. Colonic microbiota is associated with inflammation and host epigenomic alterations in inflammatory bowel disease. Nat Commun. (2020) 11:1512. doi: 10.1038/s41467-020-15342-5
217. Fofanova TY, Petrosino JF, Kellermayer R. Microbiome–epigenome interactions and the environmental origins of inflammatory bowel diseases. J Pediatr Gastroenterol Nutr. (2016) 62:208. doi: 10.1097/MPG.0000000000000950
218. Alsharairi NA. The therapeutic role of short-chain fatty acids mediated very low-calorie ketogenic diet–gut microbiota relationships in paediatric inflammatory bowel diseases. Nutrients. (2022) 14:4113. doi: 10.3390/nu14194113
219. Zhao C, Yang Q, Tang R, Li W, Wang J, Yang F, et al. DNA methyltransferase 1 deficiency improves macrophage motility and wound healing by ameliorating cholesterol accumulation. NPJ Regener Med. (2023) 8:1–17. doi: 10.1038/s41536-023-00306-2
220. Tolg C, Sabha N, Cortese R, Panchal T, Ahsan A, Soliman A, et al. Uropathogenic E. coli infection provokes epigenetic downregulation of CDKN2A (p16INK4A) in uroepithelial cells. Lab Invest. (2011) 91:825–36. doi: 10.1038/labinvest.2010.197
221. Tahara T, Hirata I, Nakano N, Tahara S, Horiguchi N, Kawamura T, et al. Potential link between Fusobacterium enrichment and DNA methylation accumulation in the inflammatory colonic mucosa in ulcerative colitis. Oncotarget. (2017) 8:61917–26. doi: 10.18632/oncotarget.18716
222. Chernov AV, Reyes L, Xu Z, Gonzalez B, Golovko G, Peterson S, et al. Mycoplasma CG- and GATC-specific DNA methyltransferases selectively and efficiently methylate the host genome and alter the epigenetic landscape in human cells. Epigenetics. (2015) 10:303–18. doi: 10.1080/15592294.2015.1020000
223. Lu Y, Fan C, Liang A, Fan X, Wang R, Li P, et al. Effects of SCFA on the DNA methylation pattern of adiponectin and resistin in high-fat-diet-induced obese male mice. Br J Nutr. (2018) 120:385–92. doi: 10.1017/S0007114518001526
224. Nguyen TMD. Adiponectin: role in physiology and pathophysiology. Int J Prev Med. (2020) 11:136. doi: 10.4103/ijpvm.IJPVM_193_20
225. Su K-Z, Li Y-R, Zhang D, Yuan J-H, Zhang C-S, Liu Y, et al. Relation of circulating resistin to insulin resistance in type 2 diabetes and obesity: A systematic review and meta-analysis. Front Physiol. (2019) 10:1399. doi: 10.3389/fphys.2019.01399
226. Giandalia A, Alibrandi A, Giorgianni L, Lo Piano F, Consolo F, Longo Elia G, et al. Resistin levels and inflammatory and endothelial dysfunction markers in obese postmenopausal women with type 2 diabetes mellitus. Diabetol Metab Syndrome. (2021) 13:98. doi: 10.1186/s13098-021-00715-7
227. Castellano-Castillo D, Moreno-Indias I, Sanchez-Alcoholado L, Ramos-Molina B, Alcaide-Torres J, Morcillo S, et al. Altered adipose tissue DNA methylation status in metabolic syndrome: relationships between global DNA methylation and specific methylation at adipogenic, lipid metabolism and inflammatory candidate genes and metabolic variables. J Clin Med. (2019) 8:87. doi: 10.3390/jcm8010087
228. Ramos-Molina B, Sánchez-Alcoholado L, Cabrera-Mulero A, Lopez-Dominguez R, Carmona-Saez P, Garcia-Fuentes E, et al. Gut microbiota composition is associated with the global DNA methylation pattern in obesity. Front Genet. (2019) 10:613. doi: 10.3389/fgene.2019.00613
229. Yao H, Fan C, Lu Y, Fan X, Xia L, Li P, et al. Alteration of gut microbiota affects expression of adiponectin and resistin through modifying DNA methylation in high-fat diet-induced obese mice. Genes Nutr. (2020) 15:12. doi: 10.1186/s12263-020-00671-3
230. Kok DEG, Dhonukshe-Rutten RAM, Lute C, Heil SG, Uitterlinden AG, van der Velde N, et al. The effects of long-term daily folic acid and vitamin B12 supplementation on genome-wide DNA methylation in elderly subjects. Clin Epigenet. (2015) 7:121. doi: 10.1186/s13148-015-0154-5
231. Soda K. Overview of polyamines as nutrients for human healthy long life and effect of increased polyamine intake on DNA methylation. Cells. (2022) 11:164. doi: 10.3390/cells11010164
232. Li X, Wang T, Zhao M, Huang T, Sun D, Han L, et al. DNA methylation variant, B-vitamins intake and longitudinal change in body mass index. Int J Obes. (2019) 43:468–74. doi: 10.1038/s41366-018-0106-1
233. Guo W, Zhang Z, Li L, Liang X, Wu Y, Wang X, et al. Gut microbiota induces DNA methylation via SCFAs predisposing obesity-prone individuals to diabetes. Pharmacol Res. (2022) 182:106355. doi: 10.1016/j.phrs.2022.106355
234. Cortese R, Lu L, Yu Y, Ruden D, Claud EC. Epigenome-Microbiome crosstalk: A potential new paradigm influencing neonatal susceptibility to disease. Epigenetics. (2016) 11:205–15. doi: 10.1080/15592294.2016.1155011
235. Wandro S, Osborne S, Enriquez C, Bixby C, Arrieta A, Whiteson K. The microbiome and metabolome of preterm infant stool are personalized and not driven by health outcomes, including necrotizing enterocolitis and late-onset sepsis. mSphere. (2018) 3:e00104–18. doi: 10.1128/mSphere.00104-18
236. Pan W-H, Sommer F, Falk-Paulsen M, Ulas T, Best P, Fazio A, et al. Exposure to the gut microbiota drives distinct methylome and transcriptome changes in intestinal epithelial cells during postnatal development. Genome Med. (2018) 10:27. doi: 10.1186/s13073-018-0534-5
237. Cuinat C, Stinson SE, Ward WE, Comelli EM. Maternal intake of probiotics to program offspring health. Curr Nutr Rep. (2022) 11:537–62. doi: 10.1007/s13668-022-00429-w
238. Teoh CM, Cooper A, Renteria KM, Lane M, Zhu J, Koh GY. Supplementation of methyl-donor nutrients to a high-fat, high-sucrose diet during pregnancy and lactation normalizes circulating 25-dihydroxycholecalciferol levels and alleviates inflammation in offspring. Metabolites. (2022) 12:1252. doi: 10.3390/metabo12121252
239. Luu M, Monning H, Visekruna A. Exploring the molecular mechanisms underlying the protective effects of microbial SCFAs on intestinal tolerance and food allergy. Front Immunol. (2020) 11:1225. doi: 10.3389/fimmu.2020.01225
240. Fellows R, Denizot J, Stellato C, Cuomo A, Jain P, Stoyanova E, et al. Microbiota derived short chain fatty acids promote histone crotonylation in the colon through histone deacetylases. Nat Commun. (2018) 9:105. doi: 10.1038/s41467-017-02651-5
241. Röth D, Chiang AJ, Hu W, Gugiu GB, Morra CN, Versalovic J, et al. Two-carbon folate cycle of commensal Lactobacillus reuteri 6475 gives rise to immunomodulatory ethionine, a source for histone ethylation. FASEB J. (2019) 33:3536–48. doi: 10.1096/fj.201801848R
242. Tan M, Luo H, Lee S, Jin F, Yang JS, Montellier E, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. (2011) 146:1016–28. doi: 10.1016/j.cell.2011.08.008
243. Goudarzi A, Zhang D, Huang H, Barral S, Kwon OK, Qi S, et al. Dynamic competing histone H4 K5K8 acetylation and butyrylation are hallmarks of highly active gene promoters. Mol Cell. (2016) 62:169–80. doi: 10.1016/j.molcel.2016.03.014
244. Xie Z, Zhang D, Chung D, Tang Z, Huang H, Dai L, et al. Metabolic regulation of gene expression by histone lysine β-hydroxybutyrylation. Mol Cell. (2016) 62:194–206. doi: 10.1016/j.molcel.2016.03.036
245. Kelly D, Kotliar M, Woo V, Jagannathan S, Whitt J, Moncivaiz J, et al. Microbiota-sensitive epigenetic signature predicts inflammation in Crohn’s disease. JCI Insight. (2018) 3. doi: 10.1172/jci.insight.122104
246. Wu S, Hashimoto-Hill S, Woo V, Eshleman EM, Whitt J, Engleman L, et al. Microbiota-derived metabolite promotes HDAC3 activity in the gut. Nature. (2020) 586:108–12. doi: 10.1038/s41586-020-2604-2
247. Fernando MR, Saxena A, Reyes J-L, McKay DM. Butyrate enhances antibacterial effects while suppressing other features of alternative activation in IL-4-induced macrophages. Am J Physiol Gastrointest Liver Physiol. (2016) 310:G822–831. doi: 10.1152/ajpgi.00440.2015
248. Schulthess J, Pandey S, Capitani M, Rue-Albrecht KC, Arnold I, Franchini F, et al. The short chain fatty acid butyrate imprints an antimicrobial program in macrophages. Immunity. (2019) 50:432–445.e7. doi: 10.1016/j.immuni.2018.12.018
249. Ji J, Shu D, Zheng M, Wang J, Luo C, Wang Y, et al. Microbial metabolite butyrate facilitates M2 macrophage polarization and function. Sci Rep. (2016) 6:24838. doi: 10.1038/srep24838
250. Chang PV, Hao L, Offermanns S, Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci. (2014) 111:2247–52. doi: 10.1073/pnas.1322269111
251. Korsten SGPJ, Peracic L, van Groeningen LMB, Diks MAP, Vromans H, Garssen J, et al. Butyrate prevents induction of CXCL10 and non-canonical IRF9 expression by activated human intestinal epithelial cells via HDAC inhibition. Int J Mol Sci. (2022) 23:3980. doi: 10.3390/ijms23073980
252. Lin MY, de Zoete MR, van Putten JPM, Strijbis K. Redirection of epithelial immune responses by short-chain fatty acids through inhibition of histone deacetylases. Front Immunol. (2015) 6:554. doi: 10.3389/fimmu.2015.00554
253. Sanchez HN, Moroney JB, Gan H, Shen T, Im JL, Li T, et al. B cell-intrinsic epigenetic modulation of antibody responses by dietary fiber-derived short-chain fatty acids. Nat Commun. (2020) 11:60. doi: 10.1038/s41467-019-13603-6
254. Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. (2013) 504:446–50. doi: 10.1038/nature12721
255. Kaisar MMM, Pelgrom LR, van der Ham AJ, Yazdanbakhsh M, Everts B. Butyrate Conditions Human Dendritic Cells to Prime Type 1 Regulatory T Cells via both Histone Deacetylase Inhibition and G Protein-Coupled Receptor 109A Signaling. Front Immunol. (2017) 8:1429. doi: 10.3389/fimmu.2017.01429
256. Zhang M, Zhou L, Wang Y, Dorfman RG, Tang D, Xu L, et al. Faecalibacterium prausnitzii produces butyrate to decrease c-Myc-related metabolism and Th17 differentiation by inhibiting histone deacetylase 3. Int Immunol. (2019) 31:499–514. doi: 10.1093/intimm/dxz022
257. Yang W, Yu T, Huang X, Bilotta AJ, Xu L, Lu Y, et al. Intestinal microbiota-derived short-chain fatty acids regulation of immune cell IL-22 production and gut immunity. Nat Commun. (2020) 11:4457. doi: 10.1038/s41467-020-18262-6
258. Luu M, Pautz S, Kohl V, Singh R, Romero R, Lucas S, et al. The short-chain fatty acid pentanoate suppresses autoimmunity by modulating the metabolic-epigenetic crosstalk in lymphocytes. Nat Commun. (2019) 10:760. doi: 10.1038/s41467-019-08711-2
259. Chen L, Sun M, Wu W, Yang W, Huang X, Xiao Y, et al. Microbiota metabolite butyrate differentially regulates th1 and th17 cells’ Differentiation and function in induction of colitis. Inflammation Bowel Dis. (2019) 25:1450–61. doi: 10.1093/ibd/izz046
260. Luu M, Weigand K, Wedi F, Breidenbend C, Leister H, Pautz S, et al. Regulation of the effector function of CD8+ T cells by gut microbiota-derived metabolite butyrate. Sci Rep. (2018) 8:14430. doi: 10.1038/s41598-018-32860-x
261. Li L, Zhao S, Xiang T, Feng H, Ma L, Fu P. Epigenetic connection between gut microbiota-derived short-chain fatty acids and chromatin histone modification in kidney diseases. Chin Med J (Engl). (2022) 135:1692–4. doi: 10.1097/CM9.0000000000002295
262. Kespohl M, Vachharajani N, Luu M, Harb H, Pautz S, Wolff S, et al. The microbial metabolite butyrate induces expression of th1-associated factors in CD4+ T cells. Front Immunol. (2017) 8:1036. doi: 10.3389/fimmu.2017.01036
263. Ambrozkiewicz F, Karczmarski J, Kulecka M, Paziewska A, Niemira M, Zeber-Lubecka N, et al. In search for interplay between stool microRNAs, microbiota and short chain fatty acids in Crohn’s disease - a preliminary study. BMC Gastroenterol. (2020) 20:307. doi: 10.1186/s12876-020-01444-3
264. Viennois E, Chassaing B, Tahsin A, Pujada A, Wang L, Gewirtz AT, et al. Host-derived fecal microRNAs can indicate gut microbiota healthiness and ability to induce inflammation. Theranostics. (2019) 9:4542–57. doi: 10.7150/thno.35282
265. Golonka RM, Cooper JK, Issa R, Devarasetty PP, Gokula V, Busken J, et al. Impact of nutritional epigenetics in essential hypertension: targeting microRNAs in the gut-liver axis. Curr Hypertens Rep. (2021) 23:28. doi: 10.1007/s11906-021-01142-9
266. Yan X-Y, Yao J-P, Li Y-Q, Zhang W, Xi M-H, Chen M, et al. Global trends in research on miRNA–microbiome interaction from 2011 to 2021: A bibliometric analysis. Front Pharmacol. (2022) 13:974741. doi: 10.3389/fphar.2022.974741
267. Dunislawska A, Pietrzak E, Wishna Kadawarage R, Siwek M. MicroRNA expression in immune tissues of adult chickens after embryo stimulation with bioactive substances. Sci Rep. (2023) 13:3076. doi: 10.1038/s41598-023-30299-3
268. Dai X, Chen X, Chen Q, Shi L, Liang H, Zhou Z, et al. MicroRNA-193a-3p reduces intestinal inflammation in response to microbiota via down-regulation of colonic pepT1. J Biol Chem. (2015) 290:16099–115. doi: 10.1074/jbc.M115.659318
269. Xue X, Feng T, Yao S, Wolf KJ, Liu C-G, Liu X, et al. Microbiota downregulates dendritic cell expression of miR-10a, which targets IL-12/IL-23p40. J Immunol. (2011) 187:5879–86. doi: 10.4049/jimmunol.1100535
270. Xue X, Cao AT, Cao X, Yao S, Carlsen ED, Soong L, et al. Downregulation of microRNA-107 in intestinal CD11c(+) myeloid cells in response to microbiota and proinflammatory cytokines increases IL-23p19 expression. Eur J Immunol. (2014) 44:673–82. doi: 10.1002/eji.201343717
271. Rodríguez-Nogales A, Algieri F, Garrido-Mesa J, Vezza T, Utrilla MP, Chueca N, et al. Differential intestinal anti-inflammatory effects of Lactobacillus fermentum and Lactobacillus salivarius in DSS mouse colitis: impact on microRNAs expression and microbiota composition. Mol Nutr Food Res. (2017) 61:1700144. doi: 10.1002/mnfr.201700144
272. Glaich O, Parikh S, Bell RE, Mekahel K, Donyo M, Leader Y, et al. DNA methylation directs microRNA biogenesis in mammalian cells. Nat Commun. (2019) 10:5657. doi: 10.1038/s41467-019-13527-1
273. Cheng J, Guo S, Chen S, Mastriano SJ, Liu C, D’Alessio AC, et al. An extensive network of TET2-targeting MicroRNAs regulates Malignant hematopoiesis. Cell Rep. (2013) 5:471–81. doi: 10.1016/j.celrep.2013.08.050
274. Lin LL, Wang W, Hu Z, Wang LW, Chang J, Qian H. Negative feedback of miR-29 family TET1 involves in hepatocellular cancer. Med Oncol. (2014) 31:291. doi: 10.1007/s12032-014-0291-2
275. Wang H, An X, Yu H, Zhang S, Tang B, Zhang X, et al. MiR-29b/TET1/ZEB2 signaling axis regulates metastatic properties and epithelial-mesenchymal transition in breast cancer cells. Oncotarget. (2017) 8:102119–33. doi: 10.18632/oncotarget.22183
276. Niu M, Gao D, Wen Q, Wei P, Pan S, Shuai C, et al. MiR-29c regulates the expression of miR-34c and miR-449a by targeting DNA methyltransferase 3a and 3b in nasopharyngeal carcinoma. BMC Cancer. (2016) 16:218. doi: 10.1186/s12885-016-2253-x
277. Weber GJ, Foster J, Pushpakumar SB, Sen U. Altered microRNA regulation of short chain fatty acid receptors in the hypertensive kidney is normalized with hydrogen sulfide supplementation. Pharmacol Res. (2018) 134:157–65. doi: 10.1016/j.phrs.2018.06.012
278. Johnston DGW, Williams MA, Thaiss CA, Cabrera-Rubio R, Raverdeau M, McEntee C, et al. Loss of microRNA-21 influences the gut microbiota, causing reduced susceptibility in a murine model of colitis. J Crohns Colitis. (2018) 12:835–48. doi: 10.1093/ecco-jcc/jjy038
279. Feng Q, Li Y, Zhang H, Wang Z, Nie X, Yao D, et al. Deficiency of miRNA-149-3p shaped gut microbiota and enhanced dextran sulfate sodium-induced colitis. Mol Ther Nucleic Acids. (2022) 30:208–25. doi: 10.1016/j.omtn.2022.09.018
280. Teng Y, Ren Y, Sayed M, Hu X, Lei C, Kumar A, et al. Plant-derived exosomal microRNAs shape the gut microbiota. Cell Host Microbe. (2018) 24:637–652.e8. doi: 10.1016/j.chom.2018.10.001
281. Liu S, da Cunha AP, Rezende RM, Cialic R, Wei Z, Bry L, et al. The Host Shapes the Gut Microbiota via Fecal microRNA. Cell Host Microbe. (2016) 19:32–43. doi: 10.1016/j.chom.2015.12.005
282. Hu S, Dong TS, Dalal SR, Wu F, Bissonnette M, Kwon JH, et al. The Microbe-Derived Short Chain Fatty Acid Butyrate Targets miRNA-Dependent p21 Gene Expression in Human Colon Cancer. PloS One. (2011) 6. doi: 10.1371/journal.pone.0016221
283. Hinnebusch BF, Meng S, Wu JT, Archer SY, Hodin RA. The effects of short-chain fatty acids on human colon cancer cell phenotype are associated with histone hyperacetylation. J Nutr. (2002) 132:1012–7. doi: 10.1093/jn/132.5.1012
284. Ali SR, Humphreys KJ, Simpson KJ, McKinnon RA, Meech R, Michael MZ. Functional high-throughput screen identifies microRNAs that promote butyrate-induced death in colorectal cancer cells. Mol Ther Nucleic Acids. (2022) 30:30–47. doi: 10.1016/j.omtn.2022.08.037
285. Virtue AT, McCright SJ, Wright JM, Jimenez MT, Mowel WK, Kotzin JJ, et al. The gut microbiota regulates white adipose tissue inflammation and obesity via a family of microRNAs. Sci Transl Med. (2019) 11:eaav1892. doi: 10.1126/scitranslmed.aav1892
286. Roshanravan N, Alamdari NM, Jafarabadi MA, Mohammadi A, Shabestari BR, Nasirzadeh N, et al. Effects of oral butyrate and inulin supplementation on inflammation-induced pyroptosis pathway in type 2 diabetes: A randomized, double-blind, placebo-controlled trial. Cytokine. (2020) 131:155101. doi: 10.1016/j.cyto.2020.155101
287. Roshanravan N, Mahdavi R, Jafarabadi MA, Alizadeh E, Ghavami A, Saadat YR, et al. The effects of sodium butyrate and high-performance inulin supplementation on the promotion of gut bacterium Akkermansia muciniphila growth and alterations in miR-375 and KLF5 expression in type 2 diabetic patients: A randomized, double-blind, placebo-controlled trial. Eur J Integr Med. (2018) 18:1–7. doi: 10.1016/j.eujim.2017.12.011
288. Zhang X, Wang W, Zhu W, Dong J, Cheng Y, Yin Z, et al. Mechanisms and functions of long non-coding RNAs at multiple regulatory levels. Int J Mol Sci. (2019) 20:5573. doi: 10.3390/ijms20225573
289. Heydarzadeh S, Ranjbar M, Karimi F, Seif F, Alivand MR. Overview of host miRNA properties and their association with epigenetics, long non-coding RNAs, and Xeno-infectious factors. Cell Bioscience. (2021) 11:43. doi: 10.1186/s13578-021-00552-1
290. Liang L, Ai L, Qian J, Fang J-Y, Xu J. Long noncoding RNA expression profiles in gut tissues constitute molecular signatures that reflect the types of microbes. Sci Rep. (2015) 5:11763. doi: 10.1038/srep11763
291. Dempsey J, Zhang A, Cui JY. Coordinate regulation of long non-coding RNAs and protein-coding genes in germ-free mice. BMC Genomics. (2018) 19. doi: 10.1186/s12864-018-5235-3
292. Chowdhury IH, Narra HP, Sahni A, Khanipov K, Schroeder CLC, Patel J, et al. Expression profiling of long noncoding RNA splice variants in human microvascular endothelial cells: lipopolysaccharide effects. In Vitro. Mediators Inflammation. (2017) 2017:3427461. doi: 10.1155/2017/3427461
293. Singh KK, Matkar PN, Muhammad S, Quan A, Gupta V, Teoh H, et al. Investigation of novel LPS-induced differentially expressed long non-coding RNAs in endothelial cells. Mol Cell Biochem. (2016) 421:157–68. doi: 10.1007/s11010-016-2797-8
294. Wang J, Liu Z, Xu Y, Wang Y, Wang F, Zhang Q, et al. Enterobacterial LPS-inducible LINC00152 is regulated by histone lactylation and promotes cancer cells invasion and migration. Front Cell Infect Microbiol. (2022) 12:913815. doi: 10.3389/fcimb.2022.913815
295. Wang J, Nie D. Abstract B19: Short chain fatty acids suppress mTOR activation in colon cancer cells via the long noncoding RNA rhabdomyosarcoma 2 associated transcript. Mol Cancer Ther. (2015) 14:B19. doi: 10.1158/1538-8514.PI3K14-B19
296. Gao Y, Zhou J, Qi H, Wei J, Yang Y, Yue J, et al. LncRNA lncLy6C induced by microbiota metabolite butyrate promotes differentiation of Ly6Chigh to Ly6Cint/neg macrophages through lncLy6C/C/EBPβ/Nr4A1 axis. Cell Discovery. (2020) 6:1–14. doi: 10.1038/s41421-020-00211-8
297. Bai Z, Chai X, Yoon MJ, Kim H-J, Lo KA, Zhang Z, et al. Dynamic transcriptome changes during adipose tissue energy expenditure reveal critical roles for long noncoding RNA regulators. PloS Biol. (2017) 15:e2002176. doi: 10.1371/journal.pbio.2002176
298. Wijesinghe SN, Nicholson T, Tsintzas K, Jones SW. Involvements of long noncoding RNAs in obesity-associated inflammatory diseases. Obes Rev. (2021) 22:e13156. doi: 10.1111/obr.13156
299. Zhang F, Yang Y, Chen X, Liu Y, Hu Q, Huang B, et al. The long non-coding RNA βFaar regulates islet β-cell function and survival during obesity in mice. Nat Commun. (2021) 12:3997. doi: 10.1038/s41467-021-24302-6
300. Dallner OS, Marinis JM, Lu Y-H, Birsoy K, Werner E, Fayzikhodjaeva G, et al. Dysregulation of a long noncoding RNA reduces leptin leading to a leptin-responsive form of obesity. Nat Med. (2019) 25:507–16. doi: 10.1038/s41591-019-0370-1
301. Yang H, Zhang Z, Peng R, Zhang L, Liu H, Wang X, et al. RNA-Seq analysis reveals critical transcriptome changes caused by sodium butyrate in DN mouse models. Biosci Rep. (2021) 41:BSR20203005. doi: 10.1042/BSR20203005
302. Chiappori F, Cupaioli FA, Consiglio A, Di Nanni N, Mosca E, Licciulli VF, et al. Analysis of Faecal Microbiota and Small ncRNAs in Autism: Detection of miRNAs and piRNAs with Possible Implications in Host–Gut Microbiota Cross-Talk. Nutrients. (2022) 14:1340. doi: 10.3390/nu14071340
303. Zhou H, Yang L, Ding J, Xu K, Liu J, Zhu W, et al. Dynamics of small non-coding RNA profiles and the intestinal microbiome of high and low weight chickens. Front Microbiol. (2022) 13:916280. doi: 10.3389/fmicb.2022.916280
Keywords: short-chain fatty acids, obesity, low-grade inflammation, lipopolysaccharide, epigenetics
Citation: Kopczyńska J and Kowalczyk M (2024) The potential of short-chain fatty acid epigenetic regulation in chronic low-grade inflammation and obesity. Front. Immunol. 15:1380476. doi: 10.3389/fimmu.2024.1380476
Received: 01 February 2024; Accepted: 18 March 2024;
Published: 27 March 2024.
Edited by:
Ronan Lordan, University of Pennsylvania, United StatesReviewed by:
Robin Joshi, University of Pennsylvania, United StatesCopyright © 2024 Kopczyńska and Kowalczyk. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Julia Kopczyńska, amtvcGN6eW5za2FAaWJiLndhdy5wbA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.