
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Immunol. , 22 July 2024
Sec. Molecular Innate Immunity
Volume 15 - 2024 | https://doi.org/10.3389/fimmu.2024.1375528
This article is part of the Research Topic Enhancing Innate Immunity in Combination Therapy for Solid Tumors View all 7 articles
 Manjusha Biswas1,2*
Manjusha Biswas1,2*Tissue-resident macrophages (TRMs) are an integral part of the innate immune system, but their biology is not well understood in the context of cancer. Distinctive resident macrophage populations are identified in different organs in mice using fate mapping studies. They develop from the yolk sac and self-maintain themselves lifelong in specific tissular niches. Similarly, breast-resident macrophages are part of the mammary gland microenvironment. They reside in the breast adipose tissue stroma and close to the ductal epithelium and help in morphogenesis. In breast cancer, TRMs may promote disease progression and metastasis; however, precise mechanisms have not been elucidated. TRMs interact intimately with recruited macrophages, cytotoxic T cells, and other immune cells along with cancer cells, deciding further immunosuppressive or cytotoxic pathways. Moreover, triple-negative breast cancer (TNBC), which is generally associated with poor outcomes, can harbor specific TRM phenotypes. The influence of TRMs on adipose tissue stroma of the mammary gland also contributes to tumor progression. The complex crosstalk between TRMs with T cells, stroma, and breast cancer cells can establish a cascade of downstream events, understanding which can offer new insight for drug discovery and upcoming treatment choices. This review aims to acknowledge the previous research done in this regard while exploring existing research gaps and the future therapeutic potential of TRMs as a combination or single agent in breast cancer.
Breast cancer is currently the most commonly diagnosed cancer worldwide, surpassing lung cancer in 2020. The International Agency for Research on Cancer (IARC) and partner institutions predict breast cancer cases will rise to 40% by 2040 with an increase of 50% mortality rate worldwide, leading to more than one million deaths per year (1). Various chemotherapy regimens and targeted therapy options are available to treat breast cancer, but immunotherapy is still evasive (2). However, the presence of tumor-infiltrating lymphocytes, identification of different innate immune subsets, and transcriptomics analysis identifying immune gene signatures suggest the potential of combining immunotherapy with standard of care (SOC) (2–4). While the role of immune checkpoint inhibitors, bispecific antibodies, CAR-T cell therapy, etc. have been recognized across various solid tumors including breast cancer (5–7), the innate immune cells, particularly macrophages, are emerging as novel candidates for combination therapy. To exploit macrophage’s potential as a therapeutic target in breast cancer, understanding their ontogeny is crucial as developmental origins dictate their functional commitment in steady state and disease. Single-cell RNA sequencing (scRNA-seq) data indicating existence of macrophage phenotypes beyond the conventional M1 and M2 spectrum (8) inspires a deeper dive into the subject. Ontogenically, macrophages can be either resident or recruited. Tissue-resident macrophages (TRMs) being the guardians of homeostasis garner more attention and also focus of this review. As startling new data have appeared on TRM diversity, their potential mechanistic role in breast cancer, and their influence on varied cellular or stromal partners in breast tumor microenvironment (TME), the complex crosstalk is reviewed and the rising therapeutic scope of resident macrophages in breast cancer as single mechanistic target or a potential candidate for combination therapy is assessed.
The breast cancer microenvironment is intricate (Figure 1). Apart from tumor cells, it has different stromal cells and extracellular matrices (ECMs) (9, 10). Moreover, genome-wide profiling identifies multiple phenotypes within the major cell types of breast TME (10–12). The myeloid population of the breast TME consists of monocytes, dendritic cells, and macrophages, spceially tumor associated macrophages (TAM) (12). The lymphoid components predominantly include T cells, with several internal phenotypes and B cells with predominance of memory B cells (10, 12, 13). The most common mesenchymal cells in breast TME are fibroblasts, followed by other minor groups like pericytes, endothelial cells, adipose-derived stromal cells, and mesenchymal stem cells (14). Myeloid-derived suppressor cells (MDSCs), a distinct state of differentiation within neutrophil and monocyte lineage, also exist in breast cancer patients (15). At the molecular level, the factors involved in pyroptosis pathways, ferroptosis-related genes, and hypoxia-related genes (HRGs), and cuproptosis-related genes (CRGs) have been recently explored to identify overall survival, immunogenicity, and immune checkpoints in breast cancer microenvironment (16–18). Breast microbiota represents another intriguing new member of the TME (19), but the crosstalk of breast microbiome with cellular and stromal partners in breast cancer remains to be elucidated.
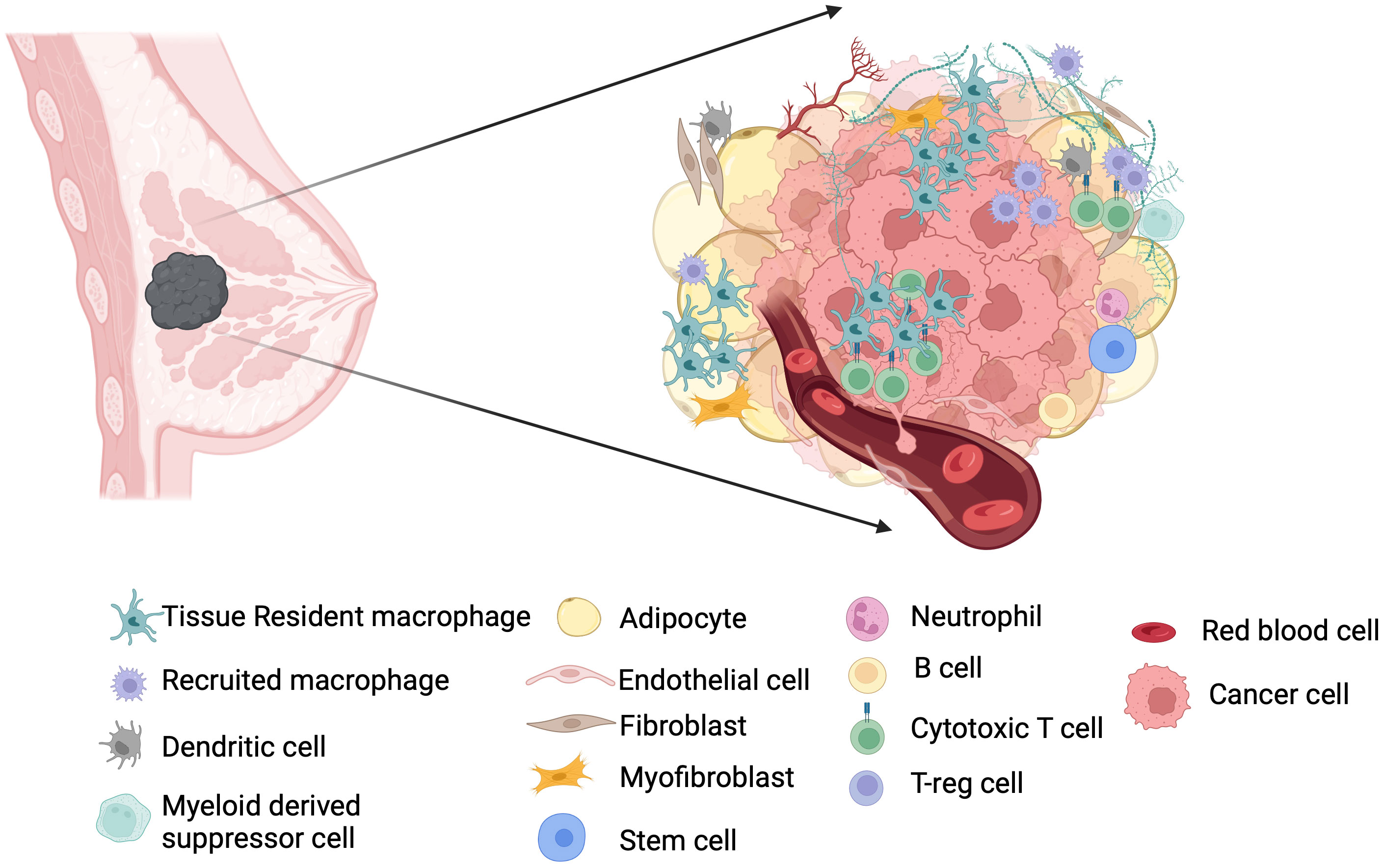
Figure 1 TME in breast cancer. Microenvironment in breast cancer is formed by multiple cell types, and their crosstalk is implicated in the pathogenesis of breast cancer. Resident macrophages predominate the breast TME along with T cells and help in disease progression or attenuation depending on their heterogeneity not only by interacting tumor cells and T cells but also with other stakeholder cells such as fibroblasts and adipocytes. Tumor associated macrophages contain both resident and recruited macrophages.
TIMER2.0 database can be used to assess the contribution of immune cell infiltration in tumor progression and regression using multiple computational algorithms modules (20). Large-scale cytometry profiling pinpoints that T cells and macrophages are the most abundant immune cells in the breast TME (10). Single-cell RNA sequencing confirms this fact and also shows that compared to normal breast tissue, TME can have more cytotoxic T cells and activated macrophages (11).
Within the macrophage population, two dominant phenotypes M1 and M2 were described earlier by the macrophage polarization model. However sc-RNAseq data hint toward a wider phenotypic diversity (11). Indeed, CITE-Seq highlighted novel PD-L1/PD-L2+ macrophage populations associated with specific disease outcomes (11). Interestingly, Rac/Cdc42i- inhibited macrophages were found to induce an antitumor TME by affecting IL6 secretion and by inhibition of metastatic cancer cells (21). Understandably, macrophage phenotyping in breast TME is not well defined and their mechanism of action in promoting or attenuating the disease is not fully elucidated. It is rather shadowed under the term “tumor associated macrophage” or “TAM”. This allows researchers to explore breast macrophages in light of developmental heterogeneity and functional commitment. Once the full spectrum of macrophage phenotypes is revealed, their interaction with other key components of breast TME will pave the way for new combinatorial approaches.
Macrophage heterogeneity is not yet fully understood (22–24), but their developmental origin certainly plays a role. Tissue-resident macrophages (TRMs or Mϕ) originate from the yolk sac and self-maintain in the peripheral tissue niche. Fate mapping studies show that macrophages develope asynchronously via multiple waves and at different anatomical locations (25, 26). While the first wave can generate primitive microglia, the majority of the tissue-resident macrophages originates in the second wave from erythroid-myeloid progenitor (EMP) through a core macrophage transcriptional program (25, 27). It is important to understand that pMac- and EMP-derived monocytes are two independent progenitors in the developmental trajectory from EMP to long-lived TRMs (25). Hematopoietic stem cells (HSCs) develop as the third wave and migrate via the fetal liver to the bone marrow, where they persist and generate monocytes. These monocytes are constantly recruited in the tissues giving rise to the recruited macrophages (25). The challenge remains due to our limited knowledge about sub-phenotypes of TRM, making their separate anatomical niche within a particular organ, including the breast.
Normal mammary gland (MG) derives from the ectoderm in E10.5, and the F4/80hi and F4/80int macrophage phenotypes are identified in E16.5 (28). Cell fate mapping and antibody depletion studies proved that YS-derived macrophages are F4/80hi in MG, persist lifelong, and express canonical macrophage markers (CD64, MerTK, CD206, C1qa, CSfr1, and Spi) but lack dendritic cell markers (CD11c, Zbtb46, and Itgax) (28, 29). The second population of F4/80int macrophages is fetal liver derived, which contributes to the Mϕ pool postnatally. Mass cytometry data on mammary glands from 3-month-old mice show predominantly F4/80hi CD64hi Siglec-1hi CD206hi TRMs, while CD206lo macrophages were deemed BM derived (28). Another study by Dawson et al., using flow cytometry, showed three Mϕ (CD64+ F4/80+MerTk+) populations, such as CD11clo CD11b+ MHCIIhi (Mϕ1), CD11clo CD11b+ MHCIIlo (Mϕ2), and CD11c+ CD11blo Ly6C− (Mϕ3). These Mϕs express Lyve1 and CD206 to various extents and prefer either nerve or vessel-associated niches (24). These findings support that heterogeneous TRM populations exist within the breast. When compared within the whole breast, Mϕ3 was enriched in the ducts fourfold and was absent in the fat pads cleared of epithelium. Therefore, Mϕ3 represents a unique entity called ductal macrophage (DM) close to the ductal epithelium, enriched for lysosomal genes, matrix metalloproteinase genes, and notch signaling. Their unique expression of Cx3cr1 confirmed their residential nature (24, 27).
The myeloid compartment of breast tumor profiled in transgenic mouse models showed an increase in Mϕ3 (DM) expanding throughout the tumor and a decrease in the adipose-rich stromal TRM (Mϕ1 & Mϕ2). DM-like TAMs suppress cytotoxic T-cell activity and tumor progression (24). Lavrion et al. also demonstrated stromal and ductal TAMs by scRNA-seq and imaging (30). Stromal I Mϕs were (CD11b, CD206, MHC II, and CCR2 positive) located in adipose tissue stroma or adipose islets, and stromal II Mϕs were located in the connective tissue. Ductal Mϕs were elongated, intraepithelial, and parallel to the basement membrane in steady state and surrounded the TME in breast cancer. They showed significant heterogeneity in CD11b and MHCII expression (30). Interestingly, further scRNA-seq from the sorted myeloid components of the tumor identified expression of Trem2, Cadm1, Folr2, and Mrc1, supporting that both TRM and recruited macrophage build the TAM pool (30). These findings were recapitulated in human breast tumors upon analyzing a published sc-RNAseq database (30, 31). TRMs have been associated with BRCA1-associated human breast cancer tissues; however, their significance is not explicated (31). In addition, ER+ cancers are associated with infiltration of TRMs, while HER2+ and triple negative cancers are TRM poor (31), which speculates a possible combination of TRM suppression with hormonal therapy. It is conceivable that in the breast TME, various TRM phenotypes exhibit pro- and anti-tumorigenic activity (32, 33).
In a healthy breast, TRMs play crucial mechanistic roles. They facilitate phagocytosis of apoptotic epithelial cells during puberty and alveolar cells during involution, while also organizing the structure of terminal end bud and ECM (24, 34–36). Locally active TRMs are the major regulator of branching morphogenesis during breast development (24, 37, 38). Conventionally, macrophages in cancer are termed “tumor-associated macrophages” or TAMs, originating from both resident and recruited pools (30, 39–42). It is suggested that tumors reprogram normal epithelium to produce DM-like TAMs (24, 27). Epigenetic reprogramming of TAMs arising from TRMs by DNA methylation results from tumor-directed perturbation, leading to modulation of several ligands and transcription factors, and this is distinct from monocyte-derived macrophage modulation in the TME (43). In a steady state, TRMs maintain breast tissue homeostasis and anti-tumor immunity in a CSF-1-dependent manner. They form a part of the stem cell niche as supported by studies in Csf1op/op mice (34, 44, 45). Indeed, CSF-1 response signatures are found in 25% of breast cancers, which marks the activation of reprogrammed TRMs and is associated with high tumor proliferation and higher grades (45, 46). Furthermore, altered HIF1a signaling can be another reprogramming mechanism, as hypoxia and anaerobic glycolysis induce TRMs to release growth factors and inflammatory cytokines like TGFß, IL-10, TNF-α, and CCL-8 to promote tumor growth and plasticity, tumor cell adherence, angiogenesis, and metastasis (9, 47–51). The tumor-derived exosomes can reprogram TRMs through TLR-2 and activate MYD88 and NF-kB signaling, inducing increased glycolysis and lactic acidosis, which leads to increased PD-L1 expression and immunosuppression (48, 52). This is also supported by HIF1a-mediated lactate-induced arginase expression in macrophages, leading to tumor progression by cell proliferation (50). Resident macrophages promote disease progression by ECM remodeling (41, 42).Tumor nest macrophages are correlated with microvascular density (53), suggesting their role in neo-angiogenesis. TRMs can also increase hormone resistance by activating the PI3K/Akt/mTOR signaling pathway (14, 54). Macrophages can upregulate PD-L1 expression in multiple solid cancers including breast cancer to modulate cytotoxic T-cell activity (55–57). DM-TAMs showed STAT3 expression associated with immunosuppression (30). MϕTAMs are susceptible to chronic inflammation in obesity and upregulate aromatase expression in obese patients in an IL-6-dependent manner, facilitating the development of ER+ breast (58, 59). Interestingly, FOLR2+-resident macrophages locally cohabit with CD8+ T cells and tertiary lymphoid structures and activate T-cell-mediated cytotoxicity instead of immunosuppression (60). Resident macrophages may initiate the recruitment of HSC-derived macrophages for tumor progression by presenting antigens (24, 61). Zeng et al. showed that TAM-secreted CCL18 reprograms breast-resident fibroblast to a CD10+GPR77+cancer-associated fibroblast (CAF), which induces chemoresistance through activated NF-kB signaling (62). However, they did not specify the TAM’s developmental nature.
Breast cancer can disseminate even when the tumor is regarded in situ by light microscopy (63, 64). Metastatic breast cancer is a fairly incurable disease with 5 and 10 years survival rates of approximately 27% and 13%, respectively (65, 66). TRMs play an important role in conditioning premetastatic niche to promote breast cancer metastasis and colonization. Wnt/β-catenin signaling pathway is one of the transcriptional regulators of TRMs. Macrophage-derived WNT-7b ligand is implicated for lung metastasis and TNFa-mediated pro-metastatic environment in breast cancer (26, 67). The perivascular macrophages help tumor cell intravasation. Studies on Csf1op/op/PyMT mice show a reduction in circulating tumor cells upon reduction in perivascular macrophage density (68). It is suggested that CD206hi intraepithelial Mϕs (24, 63) produce Wnt-1 causing E-cadherin junction disruption in a CCL2-dependent manner. Subsequently, stromal Mϕs infiltrate into the epithelium leading to early dissemination and subsequent metastasis, especially in HER2+ cancer (63). A study in B6 green fluorescent protein (GFP)-transgenic mice with TNBC shows that FOLR2+ tissue-resident macrophages dominate the TME (42%–49%), and treatment with clodronate liposomes (which induces apoptosis in macrophages) in a local recurrence model prevented lung and liver metastasis in TNBC (69). Breast cancer nodal metastases are associated with the TIE2+ CD31+ breast macrophage subset (70), suggesting their residential origin (71). On the contrary, breast cancer nodal metastasis can also be associated with nodal CD169+-resident macrophages, which often show adjacent PD-L1 expression and better prognosis (72). Intriguingly, metastatic TME of breast cancer is influenced by resident macrophages of the metastatic organ. A study using humanized and genetic mouse models showed that microglia in the brain orchestrate proinflammatory and tumor-suppressive roles in breast cancer brain metastasis. Animals without microglia were susceptible to increased metastasis, poorer survival, and hampered natural killer and T-cell responses (73). On the contrary, osteoclasts confer resistance to breast cancer cells to DNA damage therapy by enhanced glutamine production in bone metastasis (74). ß-Catenin activation in alveolar macrophages leads to a transcriptional programming enriched for inflammatory, vascular development, cytokine, and chemotactic pathways facilitating lung metastasis (67).
TNBC is defined as the absence of ER, PR, and HER2 expression and is associated with a high recurrence rate and poor overall survival. High dimensional single-cell profiling of human BRCA-1-associated TNBC shows that macrophages are the predominant infiltrating immune cells in TME (75). Intriguingly, in early TNBC, F4/80+ Mϕs infiltrate the tumor, with half of them being FOLR2+ and CADM- (69). Recent studies in the 4T1 orthotopic mouse model of TNBC showed the reprogramming of steady-state resident macrophages (referred as MGM). It led to altered cytokine signaling (TGFß, CSF-1, and IFN-gamma) mediated by specific transcription factors such as STAT1, RUNX3, and FOSL2 associated with poor outcome (43).
Adipose tissue stroma (ATS) is part of the breast anatomy and pathology. Large breast volume corresponds to high visceral fat (76) and is associated with worse outcomes in neoadjuvant chemotherapy compared to lean breast, especially in postmenopausal patients (77). In breast cancer, a lipid-associated macrophage (LAM) bearing a TREM2 signature is described, which is usually associated with monocytic origin (78). However, Dawson and colleagues described stromal Mϕs close to ATS (24). Moreover, another study in humans and mice showed two LAM populations by trajectory inference analysis: LAM-STAB1 and LAM-APOC1. Resident LAM-APOC1 was expressed both in the tumor and juxta-tumor area, strongly associated with CD8+ T cells and T-regs, while LAM-STAB1 was mostly expressed in the tumor and had a high level of TREM2 and IL-1B, suggesting their monocytic origin associated with poor prognosis (79). ATS and macrophages respectively release FFA and TNF-α in a paracrine manner to establish a vicious cycle to regulate each other (80). TRMs present antigens from dead adipocytes to attract recruited macrophages to form crown-like structures (CLS) in ATS, which increases aromatase activity, local invasion, and the possibility of metastasis (81).
Targeting macrophages as a potential combination agent in breast cancer treatment is an area of active research. TRMs can be co-targeted based on their ability to interact with other cells, predominantly T cells and fibroblasts. As many available therapies are TAM centric, TRMs being the predominant part of TAMs (28, 69) are also targeted. TRMs inhibit cytotoxic T-cell activity in multiple ways like modulating checkpoints, cytokine release, impaired antigen presentation, and induction of collagen remodeling enzymes (82–84). In human BRCA-1-associated TNBC, PARP inhibitors (PARPi) increase infiltrating CD206+PDL-1+CSF1R+ macrophages, which are immunosuppressive to cytotoxic T cells. Combining CSF-1R inhibitors with PARPi for this group showed improved CD8+ T-cell-mediated survival in mice (75). Another possible combination with PARPi in breast to bone metastasis is zoledronate because it blocks osteoclasts, the local Mϕ in the bone. Osteoclasts reduce the sensitivity of Cisplatin and PARPi by increasing glutamine production (74). Also as half of the HER2+ tumors are immunogenic, targeting macrophage and T cells combined with bispecific antibodies or checkpoint inhibitors can be a therapeutic possibility in those patients (85, 86). Immune checkpoint inhibitors can be useful in targeting TRMs for two different reasons: first, their abundance in breast TME and, second, their ability to modulate CD8+ cytotoxic T cells and immune check points (55–57). Blocking CD47-SIRP-α axis to improve immunosuppression (48) and CD24-Siglec10 in TRMs to evade immune escape of tumor cells are evolving options (87). Blocking immunosuppressive macrophages may also improve the efficacy of immune checkpoint inhibitors. As discussed earlier, TAMs can confer chemoresistance through fibroblasts via CCL18-PITPNM3 signaling. Blocking CCL18–PITPNM3 signaling by inhibiting TAMs can prevent tumor progression, delay metastasis, and prevent immunosuppression (62). TRMs can be targeted by several other mechanisms supplementing SOC (Figure 2). Repolarization of the TRMs using small molecule inhibitors or microbes from pro-tumor TRMs to antitumor ones is one of the proposed models (88). Using macrophage as a drug delivery system such as chimeric antigen receptor macrophage therapy (CAR-M) (89) is gaining more attention recently. As of February 2024, two clinical trials testing CAR-M-based strategy for breast cancer are registered: NCT04660929 on HER2+ patients with refractory or relapsed disease (recruiting) and NCT05007379 (CARMA) on patient derived organoids (90, 91). However, whether resident macrophage as CAR-M has any additional pros is yet to be studied. Two specific mechanistic scenarios can be considered for targeting TRMs. TRMs can present antigen to the monocyte-derived macrophages and T cells, recruiting them to the breast TME. This can be further strategized for drug discovery. Furthermore, metabolic reprogramming of resident macrophages by adipose tissue stroma can be explored, as the crosstalk between the two is the guiding mechanism of metabolic diseases predisposing cancer. Finally, epigenetic reprogramming of resident macrophages such as TMP195 can be an attractive treatment option in the future (92). Current strategies targeting macrophages as single or combination agents are illustrated in Figure 2.
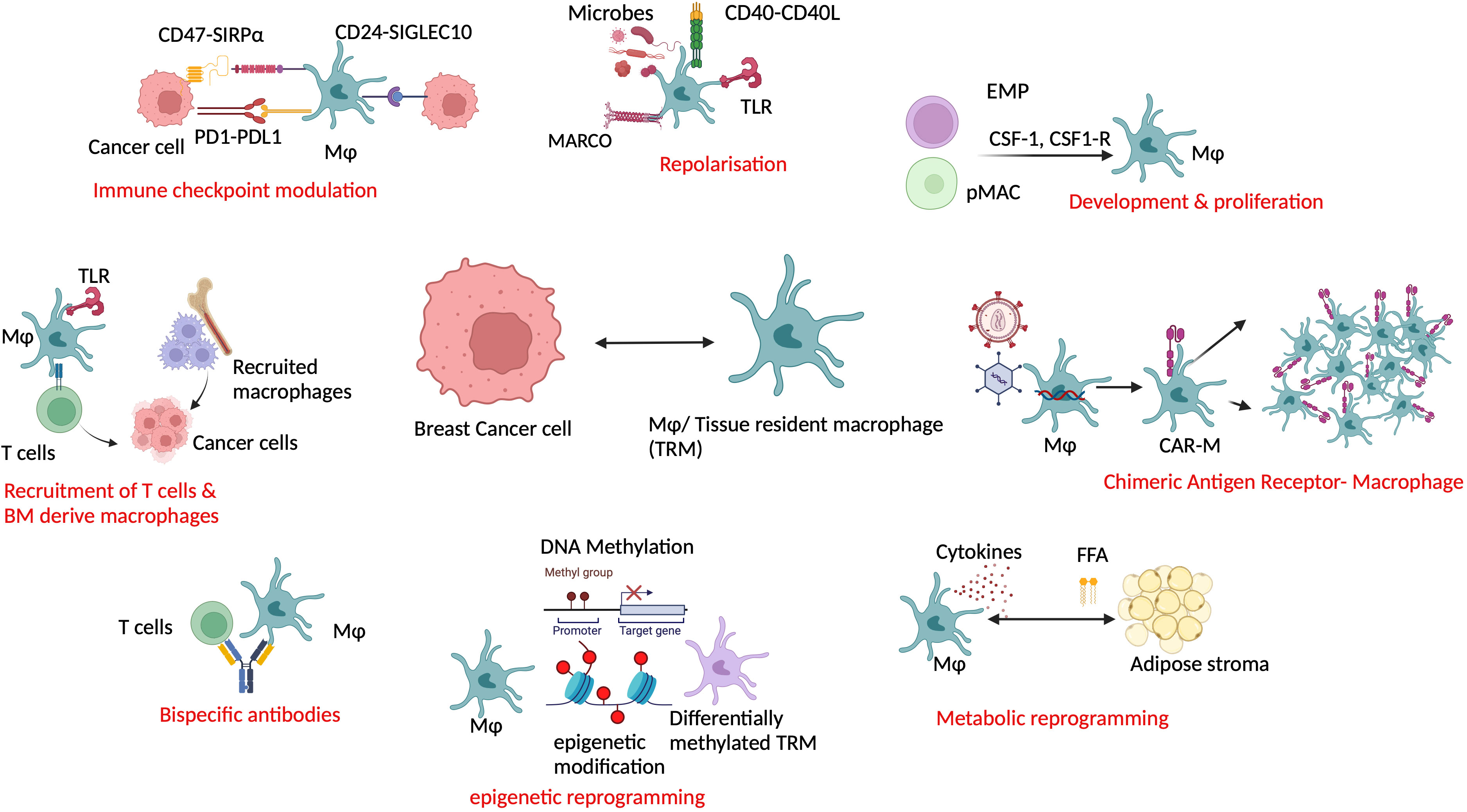
Figure 2 Co-targeting resident macrophage in breast cancer along with the current standard of care can be useful in advanced and refractory cases. Resident macrophages can be exploited in multiple ways such as by immune modulation, repolarization, reprogramming, recruitment, and as a drug delivery system in combination with other modalities. Mφ/TRM, tissue-resident macrophage; TLR, Toll-like receptor; MARCO, macrophage receptor with collagenous structure; BM, bone marrow; EMP, erythroid myeloid progenitor; pMac, pre-macrophage; CSF1, colony-stimulating factor 1; CSF1R, colony stimulating factor 1 receptor; CAR-M, chimeric antigen receptor macrophage; FFA, free fatty acid.
Macrophages are the most abundant and transcriptionally diverse innate immune cells in the breast cancer microenvironment. Their cancer-specific reprogramming facilitates cancer progression and metastasis. However, a major challenge is to separate TRMs from recruited ones within the TAM pool. Such separation is required because they have different and often contradictory functions. For example, TAMs release CSF-1 and CXCL1 helping tumor migration and EMT (93, 94). Furthermore, the hypoxic environment created by TAMs and cancer cell crosstalk triggers NFACTc1-mediated osteoclastogenesis to make a circulating metastatic niche. In addition, TAM causes induction of HIF1a by NF-kB activation, which is implicated in the pathogenesis of breast-to-bone metastasis but by which TAM subpopulation, which is not entirely clear (95–97). One major challenge is the availability of very little imaging information regarding TRMs due to strong overlap of commonly used reporters between myeloid cell subsets. Developing different reporter mice can improve our knowledge of the resident TAMs and further target them (40). Recently, combining scRNA-seq data with spatial mapping using multiple transgenic fluorescent reporter mice revealed a massive increase in intraepithelial CD11b− macrophages in breast TME, interacting with tumor cells at all stages of disease progression (30). These macrophages are ductal and found inside the tumor epithelium passing through the breached basement membrane (30). This is interesting, as these ductal TAMs correspond to the Mϕ3 discussed earlier and their fetal origin was confirmed by fate mapping studies using Ms4a3–Cre/Rosa–tdTomato mice (24). The second CD11b+ TAM population is monocyte derived and expressed in TREM2 and SPP1 (30, 98–100). Another stromal FOLR2+ and LYVE1+ TAM subset found in perivascular tissue niche are part of Mϕ TAMs (30, 101). Intriguingly, in human primary luminal breast cancer, two subsets of APOE+ TAMs are described by scRNA-seq, expressing either TREM2 or FOLR2 (60, 101). TREM2 and FOLR2 expressions determine the functional status and spatial distribution of TAMs. As TREM2+ macrophages are infiltrating in nature during cancer development and transcriptionally proximal to CD14+ CCR2+ monocytes in breast cancer, they are concluded as HSC-derived recruited macrophages (60, 101, 102). LYVE1+ FOLR2+ macrophages found in breast TME are perivascular (101). Studies in mice and the human brain showed that perivascular macrophages, although having a postnatal developmental switch, reside in the CNS without any contribution from HSC-derived precursors (103–105) suggesting their residential nature. Indeed, scRNA-seq analysis of mice macrophages compared with a publicly available database of human macrophages and genetic fate mapping confirms that the FOLR2+ TIMD4+ LYVE1+ macrophages are self-maintaining Mϕs (106). Finally, a SIGLEC-1+TAM is described in human breast cancer associated with aggressive subtypes and shorter survival; however, their developmental origin remains transcriptionally unique (46). Therefore, a clear developmental diversity exists in the breast TAMs. Initial immunotherapy trials for breast cancer were directed toward T cells, but the response was limited, e.g., in JAVELIN (NCT01772004) and KEYNOTE-028 (NCT02054806) (107). However, a bispecific approach with PD1-IL2v to expand stem cells like CD8+ T cells and anti-PD-L1 to reprogram macrophages and vasculature in immunotherapy-resistant pancreatic neuroendocrine tumors in RIP1-Tag5 mouse model showed complete tumor regression (6). Clinical.trial.gov database search using keywords “macrophage” and “breast cancer” showed 81 trials having macrophages in combination with hormonal therapy, chemotherapy, or immunotherapy until January 24. Toward this goal, a recent study isolated five TRM clusters from breast cancer patients by analyzing scRNA-seq data (108). This signature database can help make informed combinatorial treatment decisions by cotargeting TRMs alongside SOC. Moreover, the FOLR2+ TRMs are shown to promote T-cell infiltration in the tumor, thereby increasing immunogenicity and antitumor activity (108). Therefore, combination with immune checkpoint inhibitors and FOLR2+ TRM promoters can help in resistant cases.
In a straightforward scenario, resident macrophages maintain homeostasis, and recruited macrophages would promote inflammation in breast TME. However, realistically, the reprogramming of resident macrophages confers additional layers of heterogeneity challenging the therapeutic development. Better spatiotemporal delineation of macrophage niche in TME and identifying reprogramming mechanisms may identify dynamic cellular states rather than rigid phenotypes. Understanding the TRM heterogeneity will pave the way for novel targets and potential combinations.
MB: Conceptualization, Data curation, Investigation, Methodology, Project administration, Resources, Software, Visualization, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The author(s) declare financial support was received for the publication (APC) for this article. The open access fee is supported by the University & State Library of Bonn, University of Bonn, Germany.
Figures are partially created using biorender.com. The author is grateful to Professor Alexander Pfeifer and Professor Elvira Mass for their support.
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Arnold M, Morgan E, Rumgay H, Mafra A, Singh D, Laversanne M, et al. Current and future burden of breast cancer: Global statistics for 2020 and 2040. Breast. (2022) 66:15–23. doi: 10.1016/j.breast.2022.08.010
2. Debien V, De Caluwé A, Wang X, Piccart-Gebhart M, Tuohy VK, Romano E, et al. Immunotherapy in breast cancer: an overview of current strategies and perspectives. NPJ Breast Cancer. (2023) 9:7. doi: 10.1038/s41523-023-00508-3
3. Savas P, Salgado R, Denkert C, Sotiriou C, Darcy PK, Smyth MJ, et al. Clinical relevance of host immunity in breast cancer: from TILs to the clinic. Nat Rev Clin Oncol. (2016) 13:228–41. doi: 10.1038/nrclinonc.2015.215
4. Teschendorff AE, Miremadi A, Pinder SE, Ellis IO, Caldas C. An immune response gene expression module identifies a good prognosis subtype in estrogen receptor negative breast cancer. Genome Biol. (2007) 8:R157. doi: 10.1186/gb-2007-8-8-r157
5. Thomas R, Al-Khadairi G, Decock J. Immune checkpoint inhibitors in triple negative breast cancer treatment: promising future prospects. Front Oncol. (2021) 10:600573. doi: 10.3389/fonc.2020.600573
6. Tichet M, Wullschleger S, Chryplewicz A, Fournier N, Marcone R, Kauzlaric A, et al. Bispecific PD1-IL2v and anti-PD-L1 break tumor immunity resistance by enhancing stem-like tumor-reactive CD8+ T cells and reprogramming macrophages. Immunity. (2023) 56:162–79. doi: 10.1016/j.immuni.2022.12.006
7. Dees S, Ganesan R, Singh S, Grewal IS. Bispecific antibodies for triple negative breast cancer. Trends Cancer. (2021) 7:162–73. doi: 10.1016/j.trecan.2020.09.004
8. Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. (2010) 330:841–5. doi: 10.1126/science.1194637
9. Mehraj U, Ganai RA, Macha MA, Hamid A, Zargar MA, Bhat AA, et al. The tumor microenvironment as driver of stemness and therapeutic resistance in breast cancer: New challenges and therapeutic opportunities. Cell Oncol (Dordr). (2021) 44:1209–29. doi: 10.1007/s13402-021-00634-9
10. Wagner J, Rapsomaniki MA, Chevrier S, Anzeneder T, Langwieder C, Dykgers A, et al. A single-cell atlas of the tumor and immune ecosystem of human breast cancer. Cell. (2019) 177:1330–45. doi: 10.1016/j.cell.2019.03.005
11. Azizi E, Carr AJ, Plitas G, Cornish AE, Konopacki C, Prabhakaran S, et al. Single-cell map of diverse immune phenotypes in the breast tumor microenvironment. Cell. (2018) 174:1293–1308.e36. doi: 10.1016/j.cell.2018.05.060
12. Wu SZ, Al-Eryani G, Roden DL, Junankar S, Harvey K, Andersson A, et al. A single-cell and spatially resolved atlas of human breast cancers. Nat Genet. (2021) 53:1334–47. doi: 10.1038/s41588-021-00911-1
13. Tan Z, Kan C, Sun M, Yang F, Wong M, Wang S, et al. Mapping breast cancer microenvironment through single-cell omics. Front Immunol. (2022) 13:868813. doi: 10.3389/fimmu.2022.868813
14. Romano V, Ruocco MR, Carotenuto P, Barbato A, Venuta A, Acampora V, et al. Generation and characterization of a tumor stromal microenvironment and analysis of its interplay with breast cancer cells: an in vitro model to study breast cancer-associated fibroblast inactivation. Int J Mol Sci. (2022) 23:6875. doi: 10.3390/ijms23126875
15. Alshetaiwi H, Pervolarakis N, McIntyre LL, Ma D, Nguyen Q, Rath JA, et al. Defining the emergence of myeloid-derived suppressor cells in breast cancer using single-cell transcriptomics. Sci Immunol. (2020) 5:eaay6017. doi: 10.1126/sciimmunol.aay6017
16. Wu L, Lu H, Pan Y, Liu C, Wang J, Chen B, et al. The role of pyroptosis and its crosstalk with immune therapy in breast cancer. Front Immunol. (2022) 13:973935. doi: 10.3389/fimmu.2022.973935
17. Zhong J, Shen X, Zhou J, Yu H, Wang B, Sun J, et al. Development and validation of a combined hypoxia and ferroptosis prognostic signature for breast cancer. Front Oncol. (2023) 13:1077342. doi: 10.3389/fonc.2023.1077342
18. Song S, Zhang M, Xie P, Wang S, Wang Y. Comprehensive analysis of cuproptosis-related genes and tumor microenvironment infiltration characterization in breast cancer. Front Immunol. (2022) 13:978909. doi: 10.3389/fimmu.2022.978909
19. Thu MS, Chotirosniramit K, Nopsopon T, Hirankarn N, Pongpirul K. Human gut, breast, and oral microbiome in breast cancer: A systematic review and meta-analysis. Front Oncol. (2023) 13:1144021. doi: 10.3389/fonc.2023.1144021
20. Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. (2020) 48:W509–14. doi: 10.1093/nar/gkaa407
21. Torres-Sanchez A, Rivera-Robles M, Castillo-Pichardo L, Martínez-Ferrer M, Dorta-Estremera SM, Dharmawardhane S. Rac and Cdc42 inhibitors reduce macrophage function in breast cancer preclinical models. Front Oncol. (2023) 13:1152458. doi: 10.3389/fonc.2023.1152458
22. Khan F, Pang L, Dunterman M, Lesniak MS, Heimberger AB, Chen P. Macrophages and microglia in glioblastoma: heterogeneity, plasticity, and therapy. J Clin Invest. (2023) 133:e163446. doi: 10.1172/JCI163446
23. Engblom C, Pfirschke C, Pittet MJ. The role of myeloid cells in cancer therapies. Nat Rev Cancer. (2016) 16:447–62. doi: 10.1038/nrc.2016.54
24. Dawson CA, Pal B, Vaillant F, Gandolfo LC, Liu Z, Bleriot C, et al. Tissue-resident ductal macrophages survey the mammary epithelium and facilitate tissue remodelling. Nat Cell Biol. (2020) 22:546–58. doi: 10.1038/s41556-020-0505-0
25. Mass E. Delineating the origins, developmental programs and homeostatic functions of tissue-resident macrophages. Int Immunol. (2018) 30:493–501. doi: 10.1093/intimm/dxy044
26. Ginhoux F, Guilliams M. Tissue-resident macrophage ontogeny and homeostasis. Immunity. (2016) 2016:44:439–49. doi: 10.1016/j.immuni.2016.02.024
27. Mass E, Ballesteros I, Farlik M, Halbritter F, Günther P, Crozet L, et al. Specification of tissue-resident macrophages during organogenesis. Science. (2016) 353:aaf4238. doi: 10.1126/science.aaf4238
28. Jäppinen N, Félix I, Lokka E, Tyystjärvi S, Pynttäri A, Lahtela T, et al. Fetal-derived macrophages dominate in adult mammary glands. Nat Commun. (2019) 10:281. doi: 10.1038/s41467-018-08065-1
29. Hoeffel G, Chen J, Lavin Y, Low D, Almeida FF, See P, et al. C-Myb(+) erythro-myeloid progenitor-derived fetal monocytes give rise to adult tissue-resident macrophages. Immunity. (2015) 42:665–78. doi: 10.1016/j.immuni.2015.03.011
30. Laviron M, Petit M, Weber-Delacroix E, Combes AJ, Arkal AR, Barthélémy S, et al. Tumor-associated macrophage heterogeneity is driven by tissue territories in breast cancer. Cell Rep. (2022) 39:110865. doi: 10.1016/j.celrep.2022.110865
31. Pal B, Chen Y, Vaillant F, Capaldo BD, Joyce R, Song X, et al. A single-cell RNA expression atlas of normal, preneoplastic and tumorigenic states in the human breast. EMBO J. (2021) 40:e107333. doi: 10.15252/embj.2020107333
32. Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, Locati M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. (2004) 25:677–86. doi: 10.1016/j.it.2004.09.015
33. Ali HR, Chlon L, Pharoah PD, Markowetz F, Caldas C. Patterns of immune infiltration in breast cancer and their clinical implications: a gene-expression-based retrospective study. PloS Med. (2016) 13:e1002194. doi: 10.1371/journal.pmed.1002194
34. Ingman WV, Wyckoff J, Gouon-Evans V, Condeelis J, Pollard JW. Macrophages promote collagen fibrillogenesis around terminal end buds of the developing mammary gland. Dev Dynam. (2006) 235:3222–9. doi: 10.1002/dvdy.20972
35. O’Brien J, Martinson H, Durand-Rougely C, Schedin P. Macrophages are crucial for epithelial cell death and adipocyte repopulation during mammary gland involution. Development. (2012) 139:269–75. doi: 10.1242/dev.071696
36. Walker N, Bennett R, Kerr J. Cell death by apoptosis during involution of the lactating breast in mice and rats. Am J Anat. (1989) 185:19–32. doi: 10.1002/aja.1001850104
37. Van Nguyen A, Pollard JW. Colony stimulating factor-1 is required to recruit macrophages into the mammary gland to facilitate mammary ductal outgrowth. Dev Biol. (2002) 247:11–25. doi: 10.1006/dbio.2002.0669
38. Gouon-Evans V, Rothenberg ME, Pollard JW. Postnatal mammary gland development requires macrophages and eosinophils. Development. (2000) 127:2269–82. doi: 10.1242/dev.127.11.2269
39. Franklin RA, Liao W, Sarkar A, Kim MV, Bivona MR, Liu K, et al. The cellular and molecular origin of tumor-associated macrophages. Science. (2014) 344:921–25. doi: 10.1126/science.1252510
40. Laviron M, Combadière C, Boissonnas A. Tracking monocytes and macrophages in tumors with live imaging. Front Immunol. (2019) 10:1201. doi: 10.3389/fimmu.2019.01201
41. Loyher P-L, Hamon P, Laviron M, Meghraoui-Kheddar A, Goncalves E, Deng Z, et al. Macrophages of distinct origins contribute to tumor development in the lung. J Exp Med. (2018) 215:2536–53. doi: 10.1084/jem.20180534
42. Zhu Y, Herndon JM, Sojka DK, Kim K-W, Knolhoff BL, Zuo C, et al. Tissue-resident macrophages in pancreatic ductal adenocarcinoma originate from embryonic hematopoiesis and promote tumor progression. Immunity. (2017) 47:323–38. doi: 10.1016/j.immuni.2017.07.014
43. Hey J, Halperin C, Hartmann M, Mayer S, Schönung M, Lipka DB, et al. DNA methylation landscape of tumor-associated macrophages reveals pathways, transcription factors and prognostic value relevant to triple-negative breast cancer patients. Int J Cancer. (2023) 152:1226–42. doi: 10.1002/ijc.34364
44. Gyorki DE, Asselin-Labat ML, van Rooijen N, Lindeman GJ, Visvader JE. Resident macrophages influence stem cell activity in the mammary gland. Breast Cancer Res. (2009) 11:R62. doi: 10.1186/bcr2353
45. Sullivan AR, Pixley FJ. CSF-1R signaling in health and disease: a focus on the mammary gland. J Mammary Gland Biol Neoplasia. (2014) 19:149–59. doi: 10.1007/s10911-014-9320-1
46. Beck AH, Espinosa I, Edris B, Li R, Montgomery K, Zhu S, et al. The macrophage colony stimulating factor 1 response signature in breast carcinoma. Clin Cancer Res. (2009) 15:778–87. doi: 10.1158/1078-0432.CCR-08-1283
47. Cassetta L, Fragkogianni S, Sims AH, Swierczak A, Forrester LM, Zhang H, et al. Human tumor-associated macrophage and monocyte transcriptional landscapes reveal cancer-specific reprogramming, biomarkers, and therapeutic targets. Cancer Cell. (2019) 35:588–602.e10. doi: 10.1016/j.ccell.2019.02.009
48. Wang Y, Johnson KCC, Gatti-Mays ME, Li Z. Emerging strategies in targeting tumor-resident myeloid cells for cancer immunotherapy. J Hematol Oncol. (2022) 15:118. doi: 10.1186/s13045-022-01335-y
49. Guc E, Pollard JW. Redefining macrophage and neutrophil biology in the metastatic cascade. Immunity. (2021) 54:885–902. doi: 10.1016/j.immuni.2021.03.022
50. Colegio OR, Chu NQ, Szabo AL, Chu T, Phebergen AM, Jairam V, et al. Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature. (2014) 513:559–63. doi: 10.1038/nature13490
51. Lewis CE, Pollard JW. Distinct role of macrophages in different tumor microenvironments. Cancer Res. (2006) 66:605–12. doi: 10.1158/0008-5472.CAN-05-4005
52. Morrissey SM, Zhang F, Ding C, Montoya-Durango DE, Hu X, Yang C, et al. Tumor-derived exosomes drive immunosuppressive macrophages in a pre-metastatic niche through glycolytic dominant metabolic reprogramming. Cell Metab. (2021) 33:2040–58.e10. doi: 10.1016/j.cmet.2021.09.002
53. Ch'ng ES, Tuan Sharif SE, Jaafar H. In human invasive breast ductal carcinoma, tumor stromal macrophages and tumor nest macrophages have distinct relationships with clinicopathological parameters and tumor angiogenesis. Virchows Arch. (2013) 462:257–67. doi: 10.1007/s00428-012-1362-4
54. Li D, Ji H, Niu X, Yin L, Wang Y, Gu Y, et al. Tumor-associated macrophages secrete CC-chemokine ligand 2 and induce tamoxifen resistance by activating PI3K/Akt/mTOR in breast cancer. Cancer Sci. (2020) 111:47–58. doi: 10.1111/cas.14230
55. Lin H, Wei S, Hurt EM, Green MD, Zhao L, Vatan L, et al. Host expression of PD-L1 determines efficacy of PD-L1 pathway blockade-mediated tumor regression. J Clin Invest. (2018) 128:805–15. doi: 10.1172/JCI96113
56. Petty AJ, Dai R, Lapalombella R, Baiocchi RA, Benson DM, Li Z, et al. Hedgehog-induced PD-L1 on tumor-associated macrophages is critical for suppression of tumor-infiltrating CD8+ T cell function. JCI Insight. (2021) 6:e146707. doi: 10.1172/jci.insight.146707
57. Kuang DM, Zhao Q, Peng C, Xu J, Zhang JP, Wu C, et al. Activated monocytes in peritumoral stroma of hepatocellular carcinoma foster immune privilege and disease progression through PD-L1. J Exp Med. (2009) 206:1327–37. doi: 10.1084/jem.20082173
58. Sun XZ, Casbas-Hernandez P, Bigelow C, Makowski L, Joseph Jerry D, Smith Schneider S, et al. Normal breast tissue of obese women is enriched for macrophage markers and macrophage-associated gene expression. Breast Cancer Res Treat. (2012) 131:1003–12. doi: 10.1007/s10549-011-1789-3
59. Brady NJ, Chuntova P, Schwertfeger KL. Macrophages: regulators of the inflammatory microenvironment during mammary gland development and breast cancer. Mediators Inflamm. (2016) 2016:4549676. doi: 10.1155/2016/4549676
60. Nalio Ramos R, Missolo-Koussou Y, Gerber-Ferder Y, Bromley CP, Bugatti M, Núñez NG, et al. Tissue-resident FOLR2+ macrophages associate with CD8+ T cell infiltration in human breast cancer. Cell. (2022) 185:1189–1207.e25. doi: 10.1016/j.cell.2022.02.021
61. Wang Y, Chaffee TS, LaRue RS, Huggins DN, Witschen PM, Ibrahim AM, et al. Tissue-resident macrophages promote extracellular matrix homeostasis in the mammary gland stroma of nulliparous mice. Elife. (2020) 9:e57438. doi: 10.7554/eLife.57438.sa2
62. Zeng W, Xiong L, Wu W, Li S, Liu J, Yang L, et al. CCL18 signaling from tumor-associated macrophages activates fibroblasts to adopt a chemoresistance-inducing phenotype. Oncogene. (2023) 42:224–37. doi: 10.1038/s41388-022-02540-2
63. Linde N, Casanova-Acebes M, Sosa MS, Mortha A, Rahman A, Farias E, et al. Macrophages orchestrate breast cancer early dissemination and metastasis. Nat Commun. (2018) 9:21. doi: 10.1038/s41467-017-02481-5
64. Husemann Y, Geigl JB, Schubert F, Musiani P, Meyer M, Burghart E, et al. Systemic spread is an early step in breast cancer. Cancer Cell. (2008) 13:58–68. doi: 10.1016/j.ccr.2007.12.003
65. AlSendi M, O'Reilly D, Zeidan YH, Kelly CM. Oligometastatic breast cancer: Are we there yet? Int J Cancer. (2021) 149:1520–8. doi: 10.1002/ijc.33693
66. Eng LG, Dawood S, Sopik V, Haaland B, Tan PS, Bhoo-Pathy N, et al. Ten-year survival in women with primary stage IV breast cancer. Breast Cancer Res Treat. (2016) 160:145–52. doi: 10.1007/s10549-016-3974-x
67. Kramer ED, Tzetzo SL, Colligan SH, Hensen ML, Brackett CM, Clausen BE, et al. β-Catenin signaling in alveolar macrophages enhances lung metastasis through a TNF-dependent mechanism. JCI Insight. (2023) 8:e160978. doi: 10.1172/jci.insight.160978
68. Wyckoff JB, Wang Y, Lin EY, Li JF, Goswami S, Stanley ER, et al. Direct visualization of macrophage-assisted tumor cell intravasation in mammary tumors. Cancer Res. (2007) 67:2649–56. doi: 10.1158/0008-5472.CAN-06-1823
69. Hirano R, Okamoto K, Shinke M, Sato M, Watanabe S, Watanabe H, et al. Tissue-resident macrophages are major tumor-associated macrophage resources, contributing to early TNBC development, recurrence, and metastases. Commun Biol. (2023) 6:144. doi: 10.1038/s42003-023-04525-7
70. Kim OH, Kang GH, Noh H, Cha JY, Lee HJ, Yoon JH, et al. Proangiogenic TIE2(+)/CD31 (+) macrophages are the predominant population of tumor associated macrophages infiltrating metastatic lymph nodes. Molecules Cells. (2013) 36:432–8. doi: 10.1007/s10059-013-0194-7
71. Gomez Perdiguero E, Klapproth K, Schulz C, Busch K, Azzoni E, Crozet L, et al. Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature. (2015) 518:547–51. doi: 10.1038/nature13989
72. Björk Gunnarsdottir F, Auoja N, Bendahl PO, Rydén L, Fernö M, Leandersson K. Co-localization of CD169+ macrophages and cancer cells in lymph node metastases of breast cancer patients is linked to improved prognosis and PDL1 expression. Oncoimmunology. (2020) 9:1848067. doi: 10.1080/2162402X.2020.1848067
73. Evans KT, Blake K, Longworth A, Coburn MA, Insua-Rodríguez J, McMullen TP, et al. Microglia promote anti-tumour immunity and suppress breast cancer brain metastasis. Nat Cell Biol. (2023) 25:1848–59. doi: 10.1038/s41556-023-01273-y
74. Fan H, Xu Z, Yao K, Zheng B, Zhang Y, Wang X, et al. Osteoclast cancer cell metabolic cross-talk confers PARP inhibitor resistance in bone metastatic breast cancer. Cancer Res. (2024) 84:449–67. doi: 10.1158/0008-5472.CAN-23-1443
75. Mehta AK, Cheney EM, Hartl CA, Pantelidou C, Oliwa M, Castrillon JA, et al. Targeting immunosuppressive macrophages overcomes PARP inhibitor resistance in BRCA1-associated triple-negative breast cancer. Nat Cancer. (2021) 2:66–82. doi: 10.1038/s43018-020-00148-7
76. Janiszewski PM, Saunders TJ, Ross R. Breast volume is an independent predictor of visceral and ectopic fat in premenopausal women. Obes (Silver Spring). (2010) 18:1183–7. doi: 10.1038/oby.2009.336
77. Iwase T, Sangai T, Nagashima T, Sakakibara M, Sakakibara J, Hayama S, et al. Impact of body fat distribution on neoadjuvant chemotherapy outcomes in advanced breast cancer patients. Cancer Med. (2016) 5:41–8. doi: 10.1002/cam4.571
78. Liu Z, Gao Z, Li B, Li J, Ou Y, Yu X, et al. Lipid-associated macrophages in the tumor-adipose microenvironment facilitate breast cancer progression. Oncoimmunology. (2022) 11:2085432. doi: 10.1080/2162402X.2022.2085432
79. Timperi E, Gueguen P, Molgora M, Magagna I, Kieffer Y, Lopez-Lastra S, et al. Lipid-associated macrophages are induced by cancer-associated fibroblasts and mediate immune suppression in breast cancer. Cancer Res. (2022) 82:3291–306. doi: 10.1158/0008-5472.CAN-22-1427
80. Suganami T, Nishida J, Ogawa Y. A paracrine loop between adipocytes and macrophages aggravates inflammatory changes: role of free fatty acids and tumor necrosis factor alpha. Arterioscler Thromb Vasc Biol. (2005) 25:2062–8. doi: 10.1161/01.ATV.0000183883.72263.13
81. Engin AB, Engin A, Gonul II. The effect of adipocyte-macrophage crosstalk in obesity-related breast cancer. J Mol Endocrinol. (2019) 62:R201–22. doi: 10.1530/JME-18-0252
82. Matlack R, Yeh K, Rosini L, Gonzalez D, Taylor J, Silberman D, et al. Peritoneal macrophages suppress T-cell activation by amino acid catabolism. Immunology. (2006) 117:386–95. doi: 10.1111/j.1365-2567.2005.02312.x
83. Blumenthal RL, Campbell DE, Hwang P, DeKruyff RH, Frankel LR, Umetsu DT. Human alveolar macrophages induce functional inactivation in antigen-specific CD4 T cells. J Allergy Clin Immunol. (2001) 107:258–64. doi: 10.1067/mai.2001.112845
84. Hamilton MJ, Antignano F, von Rossum A, Boucher JL, Bennewith KL, Krystal G. TLR agonists that induce IFN-beta abrogate resident macrophage suppression of T cells. J Immunol. (2010) 185:4545–53. doi: 10.4049/jimmunol.1002045
85. Kim A, Lee SJ, Kim YK, Park WY, Park DY, Kim JY, et al. Programmed death-ligand 1 (PD-L1) expression in tumour cell and tumour infiltrating lymphocytes of HER2-positive breast cancer and its prognostic value. Sci Rep. (2017) 7:11671. doi: 10.1038/s41598-017-11905-7
86. Kurozumi S, Inoue K, Matsumoto H, Fujii T, Horiguchi J, Oyama T, et al. Clinicopathological values of PD-L1 expression in HER2-positive breast cancer. Sci Rep. (2019) 9:16662. doi: 10.1038/s41598-019-52944-6
87. Barkal AA, Brewer RE, Markovic M, Kowarsky M, Barkal SA, Zaro BW, et al. CD24 signalling through macrophage Siglec-10 is a target for cancer immunotherapy. Nature. (2019) 572:392–6. doi: 10.1038/s41586-019-1456-0
88. Cotechini T, Atallah A, Grossman A. Tissue-resident and recruited macrophages in primary tumor and metastatic microenvironments: potential targets in cancer therapy. Cells. (2021) 10:960. doi: 10.3390/cells10040960
89. Zhang W, Liu L, Su HF, Liu Q, Shen J, Dai H, et al. Chimeric antigen receptor macrophage therapy for breast tumours mediated by targeting the tumour extracellular matrix. Br J Cancer. (2019) 121:837–45. doi: 10.1038/s41416-019-0578-3
90. Chen Y, Yu Z, Tan X, Jiang H, Xu Z, Fang Y, et al. CAR-macrophage: A new immunotherapy candidate against solid tumors. Biomed Pharmacother. (2021) 139:111605. doi: 10.1016/j.biopha.2021.111605
91. Schepisi G, Gianni C, Palleschi M, Bleve S, Casadei C, Lolli C, et al. The new frontier of immunotherapy: chimeric antigen receptor T (CAR-T) cell and macrophage (CAR-M) therapy against breast cancer. Cancers (Basel). (2023) 15:1597. doi: 10.3390/cancers15051597
92. Guerriero JL, Sotayo A, Ponichtera HE, Castrillon JA, Pourzia AL, SChad S, et al. Class IIa HDAC inhibition reduces breast tumours and metastases through anti-tumour macrophages. Nature. (2017) 543:428–32. doi: 10.1038/nature21409
93. Coffelt SB, Hughes R, Lewis CE. Tumor-associated macrophages: Effectors of angiogenesis and tumor progression. Biochim Biophys Acta. (2009) 1796:11–8. doi: 10.1016/j.bbcan.2009.02.004
94. Wang N, Liu W, Zheng Y, Wang S, Yang B, Li M, et al. CXCL1 derived from tumor-associated macrophages promotes breast cancer metastasis via activating NF-kappaB/SOX4 signaling. Cell Death Dis. (2018) 9:880. doi: 10.1038/s41419-018-0876-3
95. Mehta AK, Kadel S, Townsend MG, Oliwa M and Guerriero JL. Macrophage biology and mechanisms of immune suppression in breast cancer. Front Immunol. (2021) 12:643771. doi: 10.3389/fimmu.2021.643771
96. Johnson RW, Schipani E, Giaccia AJ. HIF targets in bone remodeling and metastatic disease. Pharmacol Ther. (2015) 150:169–77. doi: 10.1016/j.pharmthera.2015.02.002
97. Lee CH, Wu CL, Shiau AL. Toll-like receptor 4 signaling promotes tumor growth. J Immunother. (2010) 33:73–82. doi: 10.1097/CJI.0b013e3181b7a0a4
98. Cheng S, Li Z, Gao R, Xing B, Gao Y, Yang Y, et al. A pan-cancer single-cell transcriptional atlas of tumor infiltrating myeloid cells. Cell. (2021) 184:792–809.e23. doi: 10.1016/j.cell.2021.01.010
99. Katzenelenbogen Y, Sheban F, Yalin A, Yofe I, Svetlichnyy D, Jaitin DA, et al. Coupled scRNA-seq and intracellular protein activity reveal an immunosuppressive role of TREM2 in cancer. Cell. (2020) 182:872–885.e19. doi: 10.1016/j.cell.2020.06.032
100. Mulder K, Patel AA, Kong WT, Piot C, Halitzki E, Dunsmore G, et al. Cross-tissue single-cell landscape of human monocytes and macrophages in health and disease. Immunity. (2021) 54:1883–900.e5. doi: 10.1016/j.immuni.2021.07.007
101. Matusiak M, Hickey JW, Luca B, Lu G, Kidziński L, Zhu S, et al. A spatial map of human macrophage niches reveals context-dependent macrophage functions in colon and breast cancer. Res Sq [Preprint]. (2023) 10:rs.3.rs-2393443. doi: 10.21203/rs.3.rs-2393443/v1
102. Molgora M, Esaulova E, Vermi W, Hou J, Chen Y, Luo J, et al. TREM2 modulation remodels the tumor myeloid landscape enhancing anti-PD-1 immunotherapy. Cell. (2020) 182:886–900.e17. doi: 10.1016/j.cell.2020.07.013
103. Masuda T, Amann L, Monaco G, Sankowski R, Staszewski O, Krueger M, et al. Specification of CNS macrophage subsets occurs postnatally in defined niches. Nature. (2022) 604:740–8. doi: 10.1038/s41586-022-04596-2
104. Goldmann T, Wieghofer P, Jordão MJ, Prutek F, Hagemeyer N, Frenzel K, et al. Origin, fate and dynamics of macrophages at central nervous system interfaces. Nat Immunol. (2016) 17:797–805. doi: 10.1038/ni.3423
105. Mass E, Nimmerjahn F, Kierdorf K, Schlitzer A. Tissue-specific macrophages: how they develop and choreograph tissue biology. Nat Rev Immunol. (2023) 15:1–17. doi: 10.1038/s41577-023-00848-y
106. Dick SA, Wong A, Hamidzada H, Nejat S, Nechanitzky R, Vohra S, et al. Three tissue resident macrophage subsets coexist across organs with conserved origins and life cycles. Sci Immunol. (2022) 7:eabf7777. doi: 10.1126/sciimmunol.abf7777
107. Onkar S, Cui J, Zou J, Cardello C, Cillo AR, Uddin MR, et al. Publisher Correction: Immune landscape in invasive ductal and lobular breast cancer reveals a divergent macrophage-driven microenvironment. Nat Cancer. (2023) 4(4):582. doi: 10.1038/s43018-023-00549-4
Keywords: breast cancer, tissue resident macrophage, macrophage ontogeny, tumor microenvironment, immunotherapy
Citation: Biswas M (2024) Understanding tissue-resident macrophages unlocks the potential for novel combinatorial strategies in breast cancer. Front. Immunol. 15:1375528. doi: 10.3389/fimmu.2024.1375528
Received: 24 January 2024; Accepted: 14 June 2024;
Published: 22 July 2024.
Edited by:
David Pejoski, Adoram Therapeutics SA, SwitzerlandReviewed by:
Guangchuan Wang, Jinzhou Medical University, ChinaCopyright © 2024 Biswas. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Manjusha Biswas, bWFuanVzaGFAdW5pLWJvbm4uZGU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.