
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Immunol., 06 March 2024
Sec. Molecular Innate Immunity
Volume 15 - 2024 | https://doi.org/10.3389/fimmu.2024.1375143
This article is part of the Research TopicApplication of Multi-omics Analyses in Revealing the Role of Mitochondrial Gene Defects in Disease ProgressionView all 13 articles
 Hao Chi1,2†
Hao Chi1,2† Lanqian Su2†
Lanqian Su2† Yalan Yan2†
Yalan Yan2† Xiang Gu3
Xiang Gu3 Ke Su4
Ke Su4 Han Li2
Han Li2 Lili Yu1
Lili Yu1 Jie Liu5
Jie Liu5 Jue Wang1*
Jue Wang1* Qibiao Wu1*
Qibiao Wu1* Guanhu Yang1,6*
Guanhu Yang1,6*This comprehensive review delves into the complex interplay between mitochondrial gene defects and pancreatic cancer pathogenesis through a multiomics approach. By amalgamating data from genomic, transcriptomic, proteomic, and metabolomic studies, we dissected the mechanisms by which mitochondrial genetic variations dictate cancer progression. Emphasis has been placed on the roles of these genes in altering cellular metabolic processes, signal transduction pathways, and immune system interactions. We further explored how these findings could refine therapeutic interventions, with a particular focus on precision medicine applications. This analysis not only fills pivotal knowledge gaps about mitochondrial anomalies in pancreatic cancer but also paves the way for future investigations into personalized therapy options. This finding underscores the crucial nexus between mitochondrial genetics and oncological immunology, opening new avenues for targeted cancer treatment strategies.
Pancreatic cancer, which is characterized by aggressive progression and a high mortality rate, is one of the most common fatal cancers worldwide (1, 2). Epidemiological studies indicate an increasing incidence of pancreatic cancer in industrialized nations, closely linked with aging demographics and lifestyle factors (3). The treatment landscape of pancreatic cancer is challenging and characterized by late-stage diagnosis and limited effective treatment options (2, 4). The ineffectiveness of conventional treatments is compounded by the prevalence of metastatic disease at diagnosis, coupled with a generally poor response to chemotherapy and radiation therapies (5, 6). Consequently, there is a pressing need for research focused on early detection methods and novel therapeutic approaches (7).
The role of mitochondrial gene defects has emerged as a significant area of interest in pancreatic cancer research (8, 9). These defects can lead to disrupted energy metabolism, increased oxidative stress, and changes in apoptotic pathways, all of which critically affect the survival and proliferation of pancreatic cancer cells (8, 10). Such metabolic and signaling disturbances not only facilitate tumor growth but also may contribute to resistance against standard therapies (11, 12). Furthermore, mitochondrial dysfunction is known to impact the tumor microenvironment, particularly affecting immune cell regulation, a key element in the immune escape mechanisms of pancreatic cancer (13, 14).
Employing a multiomics approach to study mitochondrial gene defects in pancreatic cancer offers significant insights (15). Through the integration of genomics, transcriptomics, and proteomics, we can comprehensively analyze the impact of these genetic defects on tumor cell metabolism, signaling, and immune responses (6, 16). This multifaceted analysis is crucial for understanding how mitochondrial gene defects influence the development of pancreatic cancer at the molecular level and for identifying new therapeutic targets, especially those that regulate immune responses and counteract immune evasion (17–20). As such, deepening our understanding of mitochondrial gene defects in pancreatic cancer is vital not only for deciphering disease mechanisms but also for advancing the development of innovative therapeutic strategies, notably in precision medicine and immunotherapy (18).
This review aims to provide an in-depth overview of the epidemiology, pathological features, and therapeutic challenges of pancreatic cancer, with an emphasis on the pivotal role of mitochondrial gene defects in its development. Additionally, we discuss the potential impact of these findings on the formulation of novel therapeutic strategies, particularly those focused on precision medicine and immunotherapy. Through this comprehensive analysis, we seek to offer new insights and directions for the research and treatment of pancreatic cancer.
Mitochondria, often referred to as cellular powerhouses, are critical for energy production within cells and play a central role in numerous cellular processes (21). They are primarily responsible for generating the bulk of cellular energy in the form of ATP through a process that involves a series of electron transport chain complexes. This process culminates in the activation of ATP synthase, which synthesizes ATP (22).
Furthermore, mitochondria are instrumental in regulating apoptosis, an orderly self-destructive process vital for maintaining tissue health. They exert this control primarily through the release of apoptosis-associated proteins, such as cytochrome. Once released into the cytoplasm, cytochrome c initiates a cascade of reactions culminating in cell death (23, 24).
In cancer cells, mitochondrial function often undergoes significant alterations. A notable example is the Warburg effect, where cancer cells preferentially generate energy through glycolysis rather than oxidative phosphorylation, even in oxygen-rich conditions. This metabolic shift plays a crucial role in the rapid growth and survival of cancer cells (25–28). Specifically, in pancreatic cancer, such mitochondrial metabolic reprogramming is closely linked to increased tumor aggressiveness, resistance to treatment, and a poorer prognosis (29).
The role of mitochondria in tumor progression manifests primarily in three areas: metabolic reprogramming, anti-apoptotic mechanisms, and immune escape, all of which are crucial for cancer progression and resistance to therapy (13, 30–32) (Figure 1A).
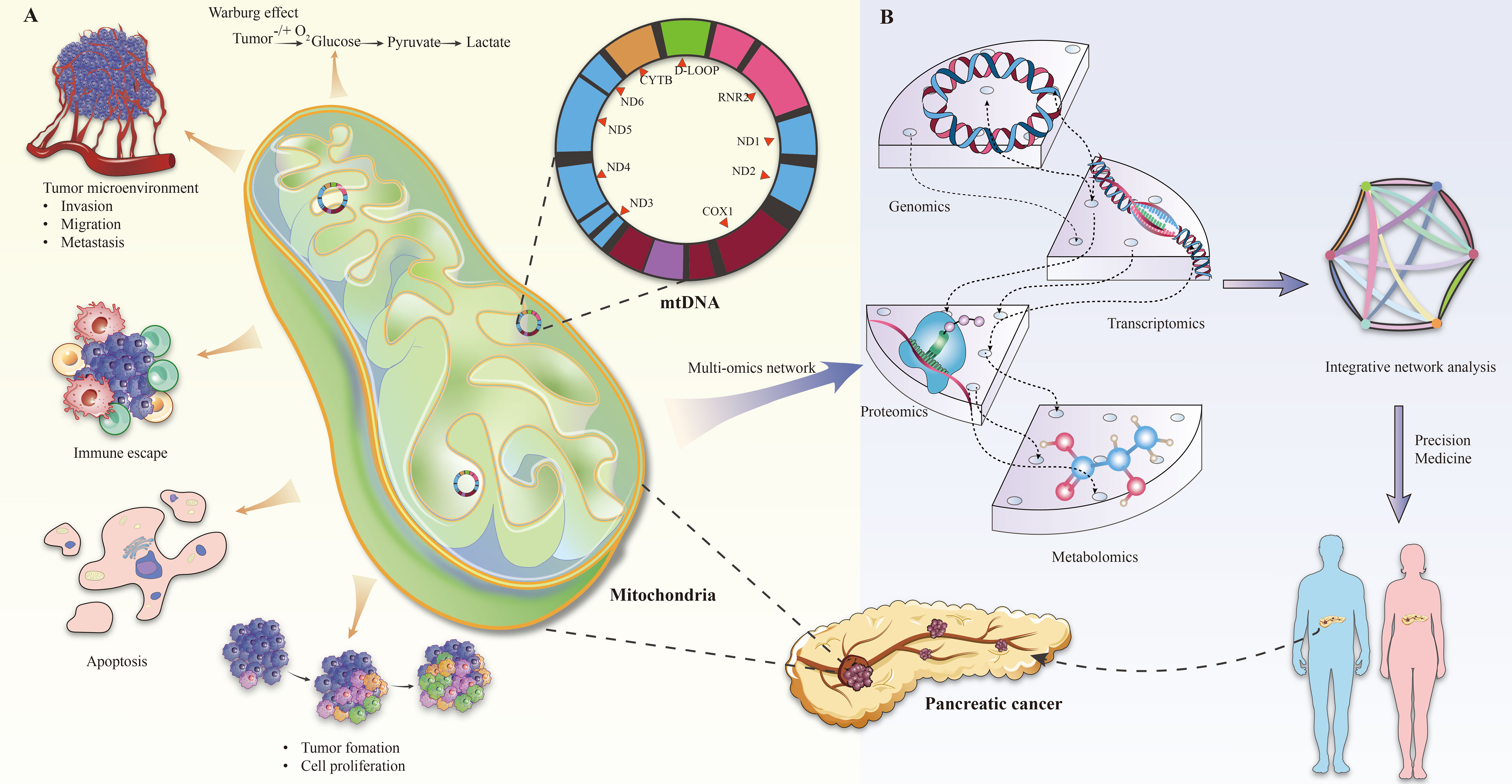
Figure 1 (A) Mitochondrial changes in pancreatic cancer. (B) Multi-omics analysis of mitochondrial gene defects in pancreatic cancer.
The Warburg effect is important for tumor metabolic reprogramming. The (platelet and lymphocyte ratio) PLR describes the propensity of cancer cells to rely on glycolysis for energy production, even in the presence of adequate oxygen (33–35). Although ATP production is less efficient than oxidative phosphorylation, glycolysis supplies rapidly dividing cancer cells with necessary biosynthetic precursors. In pancreatic cancer, for instance, this metabolic shift is facilitated by the upregulation of key glycolytic enzymes such as hexokinase and lactate dehydrogenase, promoting tumor growth and contributing to alterations in the tumor microenvironment. This includes environmental acidification, which subsequently impacts the interactions between tumors and immune cells (29, 36).
The role of mitochondria in antiapoptotic mechanisms is equally significant. Cancer cells often evade apoptosis by modulating mitochondrial pathways, thus ensuring their survival and proliferation (37, 38). This evasion is largely orchestrated through the regulation of the Bcl-2 protein family, which comprises both proapoptotic (e.g., Bax and Bak) and antiapoptotic (e.g., Bcl-2 and Bcl-xl) members (39–44). In various cancers, such as certain breast cancers and leukemias, resistance to apoptosis is achieved by either upregulating antiapoptotic proteins (e.g., Bcl-2) or suppressing proapoptotic proteins (e.g., Bax) (41, 45–47). For instance, overexpression of Bcl-2 in tumor cells has been linked to resistance against certain chemotherapeutic agents (48–50).
In addition, mitochondrial dysfunction is increasingly recognized as a contributing factor to immune escape mechanisms in the tumor microenvironment (30, 51). Mitochondria influence the tumor microenvironment by regulating inflammatory responses and cytokine production, which in turn affects immune cell infiltration and activity (13, 31). Therefore, in some tumors, the anti-tumor immune response is weakened due to impaired mitochondrial function, providing favorable conditions for tumor cell escape and proliferation (30).
In the field of pancreatic cancer research, multiomics analyses have played a pivotal role in elucidating the impact of mitochondrial gene defects. Alterations in the mitochondrial genome (mtDNA), such as mutations or deletions, have been associated with increased chemoresistance and metastatic capabilities in various cancer types. Moreover, recent studies have highlighted the potential of microRNAs that regulate mtDNA-encoded mitochondrial proteins (mitomiRs) and nuclear-encoded mitochondrial proteins as valuable biomarkers for cancer diagnosis and prognosis (9). For example, in the diagnosis of pancreatic cancer, several miRNAs (pancreatic intraepithelial neoplasia) have already been identified by researchers in the PanIN (pancreatic intraepithelial neoplasia) staging of pancreatic cancer, allowing us to study them as potential biomarkers (52).
Genomic analysis plays a crucial role in identifying specific mutations in the mitochondrial DNA of pancreatic cancer cells. An example is the point mutation in the MT-ND4 gene, which is known to disrupt the mitochondrial electron transport chain, leading to functional deficits (53, 54).
Transcriptomic analyses complement these findings by shedding light on the impact of these genetic variations on mitochondrial gene expression. In some cases, pancreatic cancer cells exhibit altered expression of mitochondria-encoded subunits of the oxidative phosphorylation complex, directly influencing cellular energy metabolism (17).
Proteomics studies have delved into the detection of aberrant expression of mitochondrial proteins in cancer cells by mass spectrometry to further confirm that changes in mitochondrial protein function may lead to unexpected mitochondrial dysfunction that can cause disease. This technique, particularly quantitative mitochondrial proteomics, provides a thorough and precise analysis of mitochondrial protein levels, including posttranslational modifications (PTMs), thus offering a comprehensive view of the changes in mitochondrial protein dynamics in cancer cells (55). For example, a study analyzing colorectal cancer tissue revealed significant alterations in the expression of mitochondrial enzymes involved in the Krebs cycle, including a marked increase in the levels of malate dehydrogenase, a key enzyme in this pathway (56). In pancreatic cancer, similar studies have linked changes in the levels of mitochondrial respiratory chain complexes I and II to increased oxidative stress (57).
Metabolomic analyses contribute to elucidating the changes in the metabolic profiles of pancreatic cancer cells caused by mitochondrial gene defects (58, 59). Notably, the increased production of lactate signals a shift toward glycolysis, which is a hallmark of cancer cell metabolism (17, 60).
Overall, the integration of genomic, transcriptomic, proteomic, and metabolomic analyses provides a holistic understanding of the role of mitochondrial gene defects in pancreatic cancer development (Figure 1B). For example, Rae-Anne Hardie and her team performed a multiomics analysis of mitochondrial DNA from 12 pancreatic cancer cell lines (PDCL) and identified 24 somatic mutations in them. The study showed metabolic changes consistent with mitochondrial dysfunction, including decreased oxygen consumption and increased glycolysis (17). Furthermore, individual tumors adapt to increased anabolic demands through different genetic mechanisms, so targeting therapy to the resulting metabolic phenotype may be an effective strategy. This multifaceted approach not only enhances our understanding of disease pathogenesis, but also holds promise in guiding the development of future therapeutic strategies, especially in the context of personalized medicine.
In the context of pancreatic cancer, particular mitochondrial gene defects significantly influence tumor biology (17, 18, 61, 62). Mutations in genes such as MT-ND4 (mitochondrial NADH dehydrogenase subunit 4) have been implicated in compromising the mitochondrial electron transport chain, leading to increased intracellular oxidative stress and destabilized energy metabolism (54, 63). The specific study found that the majority of mtDNA mutations (41.5%) in PDAC-EV were located in respiratory complex I (RCI) (ND1-ND6), followed by the RCIII gene (CYTB; 11.2%).This provides a solid foundation for further research into mtDNA biomarkers for PDAC diagnosis and the development of innovative, clinically feasible EV-based assays. Additionally, variations in mitochondrial DNA copy number, particularly in the MT-CO1 gene (mitochondrial cytochrome c oxidase subunit I), are linked with the proliferative and invasive capacities of pancreatic cancer cells (64, 65). These genetic anomalies not only affect the metabolic and survival pathways of tumor cells but also substantially impact the tumor microenvironment, consequently influencing the progression of cancer and the response to therapy. Thus, understanding these mitochondrial gene defects is vital for comprehending the pathogenesis of pancreatic cancer and for developing new therapeutic strategies (Table 1).
Mitochondrial gene defects profoundly alter the metabolic landscape of the pancreatic cancer tumor microenvironment (13, 18). These defects typically result in enhanced glycolysis and reduced oxidative phosphorylation, culminating in acidification of the tumor microenvironment (66, 69, 70). This acidic milieu not only promotes the survival and proliferation of tumor cells but also hinders the functioning of immune cells, thus facilitating tumor immune escape. Furthermore, mitochondrial dysfunction is associated with elevated reactive oxygen species production, which contributes to DNA damage, gene mutation, and accelerated tumor progression (71–73). Therefore, mitochondrial metabolism is a crucial factor in shaping the tumor microenvironment, impacting both tumor growth and the effectiveness of therapeutic interventions.
Mitochondrial gene defects significantly contribute to immune escape mechanisms in pancreatic cancer (13). Mutations in genes such as MT-CO1 and MT-ND4 can lead to dysfunction in the mitochondrial electron transport chain, affecting oxidative stress levels and altering metabolite concentrations within cells (64, 74, 75). These changes may disrupt antigen processing and presentation on the cell surface, thus impairing the recognition of tumor cells by immune cells. For instance, mitochondrial defects that alter metabolites such as lactate can acidify the tumor microenvironment, which suppresses T-cell activity and supports tumor immune escape (76–79).
Mitochondrial functionality is pivotal in regulating the immune microenvironment in pancreatic cancer (80). Through their influence on metabolic pathways, including ROS production and energy metabolism, mitochondria indirectly regulate the infiltration and activity of immune cells. Increased ROS production triggers the release of inflammatory factors such as IL-6 and TNF-α, altering the tumor microenvironment, which in turn affects the function of T cells and other immune cells (81–83). In addition, mitochondrial adenosine triphosphate (ATP) synthase, which produces most of the ATP required by mammalian cells, has been shown to decrease ATP production when cellular respiration is impaired, potentially affecting autophagy and immune function (84, 85). In certain pancreatic cancer models, the inflammatory milieu resulting from mitochondrial dysfunction has been shown to promote the recruitment of immunosuppressive cells, such as tumor-associated macrophages, further enhancing the tumor’s ability to evade the immune response (18, 86, 87).
The clinical application and therapeutic prospects of pancreatic cancer treatment are increasingly focused on two main areas: the development of targeted therapeutic strategies and the implementation of precision medicine (88, 89). In regard to therapeutic strategies, a key area of exploration is the creation of drugs that specifically target mitochondrial gene defects in pancreatic cancer. For instance, targeted therapies aimed at identifying mutations within the MT-CO1 gene could inhibit tumor growth by restoring normal mitochondrial electron transport chain function. Additionally, the use of metabolic modulators such as 2-deoxyglucose (2-DG) might prove especially effective in targeting pancreatic cancer cells exhibiting the Warburg effect by impeding their glycolytic pathways (18, 29). In the realm of precision medicine, the integration of multiomics data—including genomics, transcriptomics, and proteomics—enables the development of personalized treatment plans for patients with pancreatic cancer (90–92). This approach involves analyzing the mitochondrial genome of a patient’s tumor to identify and tailor therapies that are best suited to the patient’s unique genetic profile. For instance, selecting small molecule inhibitors that specifically target identified mitochondrial mutations can be a direct outcome of this analysis (93). This method also has the potential to better predict a patient’s response to standard chemotherapy, thereby optimizing treatment regimens, reducing adverse effects, and improving overall treatment efficacy.
Mitochondria are important organelles responsible for many physiological processes, such as energy production, apoptosis, and cellular metabolism. Mitochondrial dysfunction is increasingly recognized as a central mediator of many common diseases, including tumors and cardiovascular diseases, and there is growing evidence that mitochondrial metabolic disorders are involved in cancer development, which explains the significance of the Warburg effect for metabolic reprogramming in tumors (94). Furthermore, specific mitochondrial gene defects substantially affect the tumor microenvironment by influencing metabolic pathways, (including ROS production and energy metabolism), enhancing immune escape of cancer cells; mutations alter cytokine and chemotactic factor release, thereby affecting immune cell infiltration and function in the tumor microenvironment (95). For instance, mitochondrial dysfunction may foster the recruitment of immunosuppressive cells such as tumor-associated macrophages, subsequently dampening T-cell-mediated immune responses (96).
A key future research trajectory is the development of targeted therapies for specific mitochondrial gene mutations. Employing small molecule drugs or gene editing techniques to correct these genetic anomalies could restore mitochondrial function and impede tumor growth. Additionally, the use of metabolic modulators may augment the efficacy of chemotherapy and immunotherapy (97).
The advancement of personalized treatment strategies represents a significant research avenue (98). The integration of multiomics data enables the formulation of optimal treatment plans tailored to the unique genetic and microenvironmental characteristics of each patient’s tumor (99–101). This personalized approach holds the promise of a major breakthrough in pancreatic cancer treatment, potentially enhancing patient prognosis and survival.
Interestingly, we also found that mitochondrial retrograde signaling is also important for metabolic activities (13). Mitochondrial retrograde signaling communicates with the nucleus to maintain mitochondrial homeostasis and respond to stress. This process involves mitochondria-derived molecules (ROS, calcium, exported mtDNA, mitochondrial double-stranded RNA), the mitochondrial unfolded protein response (mtUPR), and the integrative stress response (ISR). Among these, ROS is a by-product of mitochondrial respiration and an important mediator of retrograde mitochondrial signaling. These findings suggest that targeting mitochondrial retrograde signaling may be a potential therapy against cancer progression. In addition, it has been found that mtDNA mutations can be detected in circulating extracellular vesicles (EVs) and may serve as a reliable diagnostic tool for pancreatic ductal adenocarcinoma (PDAC), compensating for the lack of highly sensitive and specific biomarkers for the diagnosis of early-stage pancreatic ductal adenocarcinomas and enriching the choice of multimodal therapy (18).
HC: Conceptualization, Data curation, Validation, Writing – original draft, Writing – review & editing. LS: Data curation, Writing – original draft. YY: Data curation, Writing – original draft. XG: Writing – original draft. KS: Writing – original draft. HL: Writing – original draft. LY: Writing – original draft. JL: Writing – original draft. JW: Writing – original draft, Writing – review & editing. QW: Writing – original draft, Writing – review & editing. GY: Writing – original draft, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Science and Technology Development Fund, Macau SAR (No.: 0098/2021/A2 and 0048/2023/AFJ), the National Natural Science Foundation of China (No.: 82361168663), and Macau University of Science and Technology’s Faculty Research Grant (No: FRG-23-003-FC).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Seufferlein T, Bachet JB, Van Cutsem E, Rougier P. Pancreatic adenocarcinoma: ESMO-ESDO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. (2012) 23 Suppl 7:vii33–40. doi: 10.1093/annonc/mds224
2. Chi H, Peng G, Wang R, Yang F, Xie X, Zhang J, et al. Cuprotosis programmed-cell-death-related lncRNA signature predicts prognosis and immune landscape in PAAD patients. Cells. (2022) 11(21):3436. doi: 10.3390/cells11213436
3. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2021) 71:209–49. doi: 10.3322/caac.21660
4. Jemal A, Siegel R, Ward E, Murray T, Xu J, Smigal C, et al. Cancer statistics, 2006. CA Cancer J Clin. (2006) 56:106–30. doi: 10.3322/canjclin.56.2.106
5. Boreddy SR, Srivastava SK. Pancreatic cancer chemoprevention by phytochemicals. Cancer Lett. (2013) 334:86–94. doi: 10.1016/j.canlet.2012.10.020
6. Huang X, Chi H, Gou S, Guo X, Li L, Peng G, et al. An aggrephagy-related lncRNA signature for the prognosis of pancreatic adenocarcinoma. Genes (Basel). (2023) 14(1):124. doi: 10.3390/genes14010124
7. Chi H, Chen H, Wang R, Zhang J, Jiang L, Zhang S, et al. Proposing new early detection indicators for pancreatic cancer: Combining machine learning and neural networks for serum miRNA-based diagnostic model. Front Oncol. (2023) 13:1244578. doi: 10.3389/fonc.2023.1244578
9. Moro L, Mitochondrial DNA. and mitomiR variations in pancreatic cancer: potential diagnostic and prognostic biomarkers. Int J Mol Sci. (2021) 22(18):9692. doi: 10.3390/ijms22189692
10. Luo Y, Ma J, Lu W. The significance of mitochondrial dysfunction in cancer. Int J Mol Sci. (2020) 21(16):5598. doi: 10.3390/ijms21165598
11. Jin P, Jiang J, Zhou L, Huang Z, Nice EC, Huang C, et al. Mitochondrial adaptation in cancer drug resistance: prevalence, mechanisms, and management. J Hematol Oncol. (2022) 15:97. doi: 10.1186/s13045-022-01313-4
12. Ghosh P, Vidal C, Dey S, Zhang L. Mitochondria targeting as an effective strategy for cancer therapy. Int J Mol Sci. (2020) 21(9):3363. doi: 10.3390/ijms21093363
13. Wang S-F, Tseng L-M, Lee H-C. Role of mitochondrial alterations in human cancer progression and cancer immunity. J Biomed Sci. (2023) 30:61. doi: 10.1186/s12929-023-00956-w
14. Liu Y, Sun Y, Guo Y, Shi X, Chen X, Feng W, et al. An overview: the diversified role of mitochondria in cancer metabolism. Int J Biol Sci. (2023) 19:897–915. doi: 10.7150/ijbs.81609
15. Steele NG, Hartway KM. Multi-omics analysis of metastatic pancreatic cancer reveals an immunosuppressive landscape. Med. (2023) 4:657–9. doi: 10.1016/j.medj.2023.09.005
16. Khan S, Ince-Dunn G, Suomalainen A, Elo LL. Integrative omics approaches provide biological and clinical insights: examples from mitochondrial diseases. J Clin Invest. (2020) 130:20–8. doi: 10.1172/JCI129202
17. Hardie RA, van Dam E, Cowley M, Han TL, Balaban S, Pajic M, et al. Mitochondrial mutations and metabolic adaptation in pancreatic cancer. Cancer Metab. (2017) 5:2. doi: 10.1186/s40170-017-0164-1
18. Sarwar A, Zhu M, Su Q, Zhu Z, Yang T, Chen Y, et al. Targeting mitochondrial dysfunctions in pancreatic cancer evokes new therapeutic opportunities. Crit Rev Oncol Hematol. (2022) 180:103858. doi: 10.1016/j.critrevonc.2022.103858
19. Hu ZI, O’Reilly EM. Therapeutic developments in pancreatic cancer. Nat Rev Gastroenterol Hepatol. (2024) 21:7–24. doi: 10.1038/s41575-023-00840-w
20. Sainero-Alcolado L, Liaño-Pons J, Ruiz-Pérez MV, Arsenian-Henriksson M. Targeting mitochondrial metabolism for precision medicine in cancer. Cell Death Differentiation. (2022) 29:1304–17. doi: 10.1038/s41418-022-01022-y
21. Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, et al. Sequence and organization of the human mitochondrial genome. Nature. (1981) 290:457–65. doi: 10.1038/290457a0
22. Nolfi-Donegan D, Braganza A, Shiva S. Mitochondrial electron transport chain: Oxidative phosphorylation, oxidant production, and methods of measurement. Redox Biol. (2020) 37:101674. doi: 10.1016/j.redox.2020.101674
23. Newmeyer DD, Ferguson-Miller S. Mitochondria: releasing power for life and unleashing the machineries of death. Cell. (2003) 112:481–90. doi: 10.1016/S0092-8674(03)00116-8
24. Aloysius MM, Zaitoun AM, Bates TE, Ilyas M, Constantin-Teodosiu D, Rowlands BJ, et al. Immunohistochemical expression of mitochondrial membrane complexes (MMCs) I, III, IV and V in Malignant and benign periampullary epithelium: a potential target for drug therapy of periampullary cancer? BMC Cancer. (2010) 10:80. doi: 10.1186/1471-2407-10-80
25. Warburg O, Wind F, Negelein E. THE METABOLISM OF TUMORS IN THE BODY. J Gen Physiol. (1927) 8:519–30. doi: 10.1085/jgp.8.6.519
26. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. (2011) 144:646–74. doi: 10.1016/j.cell.2011.02.013
27. Warburg O. On the origin of cancer cells. Science. (1956) 123:309–14. doi: 10.1126/science.123.3191.309
28. Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. (2009) 324:1029–33. doi: 10.1126/science.1160809
29. Qin C, Yang G, Yang J, Ren B, Wang H, Chen G, et al. Metabolism of pancreatic cancer: paving the way to better anticancer strategies. Mol Cancer. (2020) 19:50. doi: 10.1186/s12943-020-01169-7
30. Bai R, Cui J. Mitochondrial immune regulation and anti-tumor immunotherapy strategies targeting mitochondria. Cancer Lett. (2023) 564:216223. doi: 10.1016/j.canlet.2023.216223
31. Klein K, He K, Younes AI, Barsoumian HB, Chen D, Ozgen T, et al. Role of mitochondria in cancer immune evasion and potential therapeutic approaches. Front Immunol. (2020) 11:573326. doi: 10.3389/fimmu.2020.573326
32. Moindjie H, Rodrigues-Ferreira S, Nahmias C. Mitochondrial metabolism in carcinogenesis and cancer therapy. Cancers (Basel). (2021) 13(13):3311. doi: 10.3390/cancers13133311
33. Fukushi A, Kim HD, Chang YC, Kim CH. Revisited metabolic control and reprogramming cancers by means of the warburg effect in tumor cells. Int J Mol Sci. (2022) 23(17):10037. doi: 10.3390/ijms231710037
34. Vaupel P, Schmidberger H, Mayer A. The Warburg effect: essential part of metabolic reprogramming and central contributor to cancer progression. Int J Radiat Biol. (2019) 95:912–9. doi: 10.1080/09553002.2019.1589653
35. Lu J, Tan M, Cai Q. The Warburg effect in tumor progression: mitochondrial oxidative metabolism as an anti-metastasis mechanism. Cancer Lett. (2015) 356:156–64. doi: 10.1016/j.canlet.2014.04.001
36. Shiratori R, Furuichi K, Yamaguchi M, Miyazaki N, Aoki H, Chibana H, et al. Glycolytic suppression dramatically changes the intracellular metabolic profile of multiple cancer cell lines in a mitochondrial metabolism-dependent manner. Sci Rep. (2019) 9:18699. doi: 10.1038/s41598-019-55296-3
37. Badrinath N, Yoo SY. Mitochondria in cancer: in the aspects of tumorigenesis and targeted therapy. Carcinogenesis. (2018) 39:1419–30. doi: 10.1093/carcin/bgy148
38. Sharma A, Boise LH, Shanmugam M. Cancer metabolism and the evasion of apoptotic cell death. Cancers (Basel). (2019) 11(8):1144. doi: 10.3390/cancers11081144
39. Qian S, Wei Z, Yang W, Huang J, Yang Y, Wang J. The role of BCL-2 family proteins in regulating apoptosis and cancer therapy. Front Oncol. (2022) 12:985363. doi: 10.3389/fonc.2022.985363
40. Czabotar PE, Garcia-Saez AJ. Mechanisms of BCL-2 family proteins in mitochondrial apoptosis. Nat Rev Mol Cell Biol. (2023) 24:732–48. doi: 10.1038/s41580-023-00629-4
41. Carneiro BA, El-Deiry WS. Targeting apoptosis in cancer therapy. Nat Rev Clin Oncol. (2020) 17:395–417. doi: 10.1038/s41571-020-0341-y
42. Campbell KJ, Tait SWG. Targeting BCL-2 regulated apoptosis in cancer. Open Biol. (2018) 8(5):180002. doi: 10.1098/rsob.180002
43. Brinkmann K, Ng AP, de Graaf CA, Strasser A. What can we learn from mice lacking pro-survival BCL-2 proteins to advance BH3 mimetic drugs for cancer therapy? Cell Death Differentiation. (2022) 29:1079–93. doi: 10.1038/s41418-022-00987-0
44. Kang MH, Reynolds CP. Bcl-2 inhibitors: targeting mitochondrial apoptotic pathways in cancer therapy. Clin Cancer Res. (2009) 15:1126–32. doi: 10.1158/1078-0432.CCR-08-0144
45. Neophytou CM, Trougakos IP, Erin N, Papageorgis P. Apoptosis deregulation and the development of cancer multi-drug resistance. Cancers (Basel). (2021) 13(17):4363. doi: 10.3390/cancers13174363
46. Shahar N, Larisch S. Inhibiting the inhibitors: Targeting anti-apoptotic proteins in cancer and therapy resistance. Drug Resist Update. (2020) 52:100712. doi: 10.1016/j.drup.2020.100712
47. Wilson TR, Johnston PG, Longley DB. Anti-apoptotic mechanisms of drug resistance in cancer. Curr Cancer Drug Targets. (2009) 9:307–19. doi: 10.2174/156800909788166547
48. Kapoor I, Bodo J, Hill BT, Hsi ED, Almasan A. Targeting BCL-2 in B-cell Malignancies and overcoming therapeutic resistance. Cell Death Dis. (2020) 11:941. doi: 10.1038/s41419-020-03144-y
49. Hafezi S, Rahmani M. Targeting BCL-2 in cancer: advances, challenges, and perspectives. Cancers (Basel). (2021) 13(6):1292. doi: 10.3390/cancers13061292
50. Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. Cancer drug resistance: an evolving paradigm. Nat Rev Cancer. (2013) 13:714–26. doi: 10.1038/nrc3599
51. Liu X, Peng G. Mitochondria orchestrate T cell fate and function. Nat Immunol. (2021) 22:276–8. doi: 10.1038/s41590-020-00861-6
52. Sundarbose K, Kartha RV, Subramanian S. MicroRNAs as biomarkers in cancer. Diagnostics (Basel). (2013) 3:84–104. doi: 10.3390/diagnostics3010084
53. Kassauei K, Habbe N, Mullendore ME, Karikari CA, Maitra A, Feldmann G. Mitochondrial DNA mutations in pancreatic cancer. Int J Gastrointest Cancer. (2006) 37:57–64. doi: 10.1007/s12029-007-0008-2
54. van Gisbergen MW, Voets AM, Starmans MH, de Coo IF, Yadak R, Hoffmann RF, et al. How do changes in the mtDNA and mitochondrial dysfunction influence cancer and cancer therapy? Challenges, opportunities and models. Mutat Res Rev Mutat Res. (2015) 764:16–30. doi: 10.1016/j.mrrev.2015.01.001
55. Li N, Zhan X. Mass spectrometry-based mitochondrial proteomics in human ovarian cancers. Mass Spectrom Rev. (2020) 39:471–98. doi: 10.1002/mas.21618
56. Moro L. The mitochondrial proteome of tumor cells: A snapShot on methodological approaches and new biomarkers. Biol (Basel). (2020) 9(12):479. doi: 10.3390/biology9120479
57. Gonzalez-Franquesa A, Stocks B, Chubanava S, Hattel HB, Moreno-Justicia R, Peijs L, et al. Mass-spectrometry-based proteomics reveals mitochondrial supercomplexome plasticity. Cell Rep. (2021) 35:109180. doi: 10.1016/j.celrep.2021.109180
58. Miyazaki Y, Mori N, Akagi Y, Oda T, Kida YS. Potential metabolite markers for pancreatic cancer identified by metabolomic analysis of induced cancer-associated fibroblasts. Cancers (Basel). (2022) 14(6):1375. doi: 10.20944/preprints202201.0312.v1
59. Gaude E, Frezza C. Defects in mitochondrial metabolism and cancer. Cancer Metab. (2014) 2:10. doi: 10.1186/2049-3002-2-10
60. Kaoutari AE, Fraunhoffer NA, Hoare O, Teyssedou C, Soubeyran P, Gayet O, et al. Metabolomic profiling of pancreatic adenocarcinoma reveals key features driving clinical outcome and drug resistance. EBioMedicine. (2021) 66:103332. doi: 10.1016/j.ebiom.2021.103332
61. Yang H, Cui Y, Zhu Y. Comprehensive analysis reveals signal and molecular mechanism of mitochondrial energy metabolism pathway in pancreatic cancer. Front Genet. (2023) 14:1117145. doi: 10.3389/fgene.2023.1117145
62. Kopinski PK, Singh LN, Zhang S, Lott MT, Wallace DC. Mitochondrial DNA variation and cancer. Nat Rev Cancer. (2021) 21:431–45. doi: 10.1038/s41568-021-00358-w
63. Stoccoro A, Coppedè F. Mitochondrial DNA methylation and human diseases. Int J Mol Sci. (2021) 22(9):4594. doi: 10.3390/ijms22094594
64. Singh RK, Saini SK, Prakasam G, Kalairasan P, Bamezai RNK. Role of ectopically expressed mtDNA encoded cytochrome c oxidase subunit I (MT-COI) in tumorigenesis. Mitochondrion. (2019) 49:56–65. doi: 10.1016/j.mito.2019.07.002
65. Gentiluomo M, Katzke VA, Kaaks R, Tjønneland A, Severi G, Perduca V, et al. Mitochondrial DNA copy-number variation and pancreatic cancer risk in the prospective EPIC cohort. Cancer Epidemiol Biomarkers Prev. (2020) 29:681–6. doi: 10.1158/1055-9965.EPI-19-0868
66. Cruz-Monserrate Z, Roland CL, Deng D, Arumugam T, Moshnikova A, Andreev OA, et al. Targeting pancreatic ductal adenocarcinoma acidic microenvironment. Sci Rep. (2014) 4:4410. doi: 10.1038/srep04410
67. Tuchalska-Czuroń J, Lenart J, Augustyniak J, Durlik M. Is mitochondrial DNA copy number a good prognostic marker in resectable pancreatic cancer? Pancreatology. (2019) 19
68. Hopkins JF, Denroche RE, Aguiar JA, Notta F, Connor AA, Wilson JM, et al. Mutations in mitochondrial DNA from pancreatic ductal adenocarcinomas associate with survival times of patients and accumulate as tumors progress. Gastroenterology. (2018) 154:1620–4.e5.
69. Wang S, Zheng Y, Yang F, Zhu L, Zhu X-Q, Wang Z-F, et al. The molecular biology of pancreatic adenocarcinoma: translational challenges and clinical perspectives. Signal Transduction Targeted Ther. (2021) 6:249. doi: 10.1038/s41392-021-00659-4
70. Fu Y, Ricciardiello F, Yang G, Qiu J, Huang H, Xiao J, et al. The role of mitochondria in the chemoresistance of pancreatic cancer cells. Cells. (2021) 10(3):497. doi: 10.3390/cells10030497
71. Inigo JR, Chandra D. The mitochondrial unfolded protein response (UPRmt): shielding against toxicity to mitochondria in cancer. J Hematol Oncol. (2022) 15:98. doi: 10.1186/s13045-022-01317-0
72. Lin YH, Lim SN, Chen CY, Chi HC, Yeh CT, Lin WR. Functional role of mitochondrial DNA in cancer progression. Int J Mol Sci. (2022) 23(3):1659. doi: 10.3390/ijms23031659
73. Renaudin X. Reactive oxygen species and DNA damage response in cancer. Int Rev Cell Mol Biol. (2021) 364:139–61. doi: 10.1016/bs.ircmb.2021.04.001
74. Mahalaxmi I, Subramaniam MD, Gopalakrishnan AV, Vellingiri B. Dysfunction in mitochondrial electron transport chain complex I, pyruvate dehydrogenase activity, and mutations in ND1 and ND4 gene in autism spectrum disorder subjects from Tamil Nadu population, India. Mol Neurobiol. (2021) 58:5303–11. doi: 10.1007/s12035-021-02492-w
75. Wu S, Akhtari M, Alachkar H. Characterization of mutations in the mitochondrial encoded electron transport chain complexes in acute myeloid leukemia. Sci Rep. (2018) 8:13301. doi: 10.1038/s41598-018-31489-0
76. Cheng H, Qiu Y, Xu Y, Chen L, Ma K, Tao M, et al. Extracellular acidosis restricts one-carbon metabolism and preserves T cell stemness. Nat Metab. (2023) 5:314–30. doi: 10.1038/s42255-022-00730-6
77. Wang K, Zhang Y, Chen Z-N. Metabolic interaction: tumor-derived lactate inhibiting CD8+ T cell cytotoxicity in a novel route. Signal Transduction Targeted Ther. (2023) 8:52. doi: 10.1038/s41392-023-01320-y
78. Pérez-Tomás R, Pérez-Guillén I. Lactate in the tumor microenvironment: an essential molecule in cancer progression and treatment. Cancers (Basel). (2020) 12(11):3244. doi: 10.3390/cancers12113244
79. Gao Y, Zhou H, Liu G, Wu J, Yuan Y, Shang A. Tumor microenvironment: lactic acid promotes tumor development. J Immunol Res. (2022) 2022:3119375. doi: 10.1155/2022/3119375
80. Scharping NE, Menk AV, Moreci RS, Whetstone RD, Dadey RE, Watkins SC, et al. The tumor microenvironment represses T cell mitochondrial biogenesis to drive intratumoral T cell metabolic insufficiency and dysfunction. Immunity. (2016) 45:374–88. doi: 10.1016/j.immuni.2016.07.009
81. Hatinguais R, Pradhan A, Brown GD, Brown AJP, Warris A, Shekhova E. Mitochondrial reactive oxygen species regulate immune responses of macrophages to aspergillus fumigatus. Front Immunol. (2021) 12:641495. doi: 10.3389/fimmu.2021.641495
82. Mills EL, Kelly B, O'Neill LAJ. Mitochondria are the powerhouses of immunity. Nat Immunol. (2017) 18:488–98. doi: 10.1038/ni.3704
83. Kuo C-L, Ponneri Babuharisankar A, Lin Y-C, Lien H-W, Lo YK, Chou H-Y, et al. Mitochondrial oxidative stress in the tumor microenvironment and cancer immunoescape: foe or friend? J Biomed Sci. (2022) 29:74. doi: 10.1186/s12929-022-00859-2
84. Su WL, Wu CC, Wu SV, Lee MC, Liao MT, Lu KC, et al. A review of the potential effects of melatonin in compromised mitochondrial redox activities in elderly patients with COVID-19. Front Nutr. (2022) 9:865321. doi: 10.3389/fnut.2022.865321
85. Pei DD, Sun JL, Zhu CH, Tian FC, Jiao K, Anderson MR, et al. Contribution of mitophagy to cell-mediated mineralization: revisiting a 50-year-old conundrum. Adv Sci (Weinh). (2018) 5:1800873. doi: 10.1002/advs.201800873
86. Li Y, Wang J, Wang H, Zhang S, Wei Y, Liu S. The interplay between inflammation and stromal components in pancreatic cancer. Front Immunol. (2022) 13:850093. doi: 10.3389/fimmu.2022.850093
87. Yang J, Li Y, Sun Z, Zhan H. Macrophages in pancreatic cancer: An immunometabolic perspective. Cancer Lett. (2021) 498:188–200. doi: 10.1016/j.canlet.2020.10.029
88. Neoptolemos JP, Kleeff J, Michl P, Costello E, Greenhalf W, Palmer DH. Therapeutic developments in pancreatic cancer: current and future perspectives. Nat Rev Gastroenterol Hepatol. (2018) 15:333–48. doi: 10.1038/s41575-018-0005-x
89. Stower H. Precision-medicine success in pancreatic cancer. Nat Med. (2020) 26:649–9. doi: 10.1038/s41591-020-0903-7
90. Nassar SF, Raddassi K, Ubhi B, Doktorski J, Abulaban A. Precision medicine: steps along the road to combat human cancer. Cells. (2020) 9(9):2056. doi: 10.3390/cells9092056
91. Olivier M, Asmis R, Hawkins GA, Howard TD, Cox LA. The need for multi-omics biomarker signatures in precision medicine. Int J Mol Sci. (2019) 20(19):4781. doi: 10.3390/ijms20194781
92. Raufaste-Cazavieille V, Santiago R, Droit A. Multi-omics analysis: Paving the path toward achieving precision medicine in cancer treatment and immuno-oncology. Front Mol Biosci. (2022) 9:962743. doi: 10.3389/fmolb.2022.962743
93. Aldea M, Friboulet L, Apcher S, Jaulin F, Mosele F, Sourisseau T, et al. Precision medicine in the era of multi-omics: can the data tsunami guide rational treatment decision? ESMO Open. (2023) 8:101642. doi: 10.1016/j.esmoop.2023.101642
94. Ayer A, Fazakerley DJ, Suarna C, Maghzal GJ, Sheipouri D, Lee KJ, et al. Genetic screening reveals phospholipid metabolism as a key regulator of the biosynthesis of the redox-active lipid coenzyme Q. Redox Biol. (2021) 46:102127. doi: 10.1016/j.redox.2021.102127
95. Kang S, Xie J, Miao J, Li R, Liao W, Luo R. A knockdown of Maml1 that results in melanoma cell senescence promotes an innate and adaptive immune response. Cancer Immunol Immunother. (2013) 62:183–90. doi: 10.1007/s00262-012-1318-1
96. Zhang L, Zhang W, Li Z, Lin S, Zheng T, Hao B, et al. Mitochondria dysfunction in CD8+ T cells as an important contributing factor for cancer development and a potential target for cancer treatment: a review. J Exp Clin Cancer Res. (2022) 41:227. doi: 10.1186/s13046-022-02439-6
97. Morgan MA, Lange L, Schambach A. Prime time for base editing in the mitochondria. Signal Transduction Targeted Ther. (2022) 7:213. doi: 10.1038/s41392-022-01068-x
98. Fraunhoffer NA, Abuelafia AM, Bigonnet M, Gayet O, Roques J, Nicolle R, et al. Multi-omics data integration and modeling unravels new mechanisms for pancreatic cancer and improves prognostic prediction. NPJ Precis Oncol. (2022) 6:57. doi: 10.1038/s41698-022-00299-z
99. Huang X, Zhang G, Tang T-Y, Gao X, Liang T-B. Personalized pancreatic cancer therapy: from the perspective of mRNA vaccine. Military Med Res. (2022) 9:53. doi: 10.1186/s40779-022-00416-w
100. Connor AA, Gallinger S. Pancreatic cancer evolution and heterogeneity: integrating omics and clinical data. Nat Rev Cancer. (2022) 22:131–42. doi: 10.1038/s41568-021-00418-1
Keywords: mitochondrial gene defects, pancreatic cancer, multiomics analysis, immune escape, disease progression, clinical application prospects
Citation: Chi H, Su L, Yan Y, Gu X, Su K, Li H, Yu L, Liu J, Wang J, Wu Q and Yang G (2024) Illuminating the immunological landscape: mitochondrial gene defects in pancreatic cancer through a multiomics lens. Front. Immunol. 15:1375143. doi: 10.3389/fimmu.2024.1375143
Received: 23 January 2024; Accepted: 16 February 2024;
Published: 06 March 2024.
Edited by:
Gang Ye, Sichuan Agricultural University, ChinaReviewed by:
Wanli Ge, Nanjing Medical University, ChinaCopyright © 2024 Chi, Su, Yan, Gu, Su, Li, Yu, Liu, Wang, Wu and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jue Wang, d2FuZ2p1ZUBtdXN0LmVkdS5tbw==; Qibiao Wu, cWJ3dUBtdXN0LmVkdS5tbw==; Guanhu Yang, Z3Vhbmh1eWFuZ0BnbWFpbC5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.