
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol., 03 May 2024
Sec. Autoimmune and Autoinflammatory Disorders : Autoimmune Disorders
Volume 15 - 2024 | https://doi.org/10.3389/fimmu.2024.1365554
This article is part of the Research TopicAutoimmunity: novel insights and future perspectivesView all 38 articles
Accumulating studies have indicated that the gut microbiota plays a pivotal role in the onset of autoimmune diseases by engaging in complex interactions with the host. This review aims to provide a comprehensive overview of the existing literatures concerning the relationship between the gut microbiota and autoimmune diseases, shedding light on the complex interplay between the gut microbiota, the host and the immune system. Furthermore, we aim to summarize the impacts and potential mechanisms that underlie the interactions between the gut microbiota and the host in autoimmune diseases, primarily focusing on systemic lupus erythematosus, rheumatoid arthritis, Sjögren’s syndrome, type 1 diabetes mellitus, ulcerative colitis and psoriasis. The present review will emphasize the clinical significance and potential applications of interventions based on the gut microbiota as innovative adjunctive therapies for autoimmune diseases.
Autoimmune diseases represent a group of chronic and systemic disorders characterized by an excessive immune response, abundant inflammation, and the extensive depositions of immune complexes to tissues and organs. Epidemiological investigations have revealed a global incidence of 5~8% for autoimmune diseases (1, 2). The pathogenesis of autoimmune disease is multifaceted involving genetic predisposition, immune dysregulation, and environmental factors like lifestyle, dietary patterns, and medications (3). Despite advancements in diagnosis and treatment, early diagnosis and precise therapeutic strategies of autoimmune diseases remain significant challenges (4). Increasing evidence has highlighted the pivotal role of the gut microbiota in maintaining immune balance and homeostasis in autoimmune diseases, particularly including rheumatoid arthritis (RA), systemic lupus erythematosus (SLE), systemic scleorosis, and type I diabetes mellitus (T1DM). Disturbances in the composition and diversity of the gut microbiota are strongly associated with autoimmune disorders (5–7). This review aims to elucidate the regulatory mechanisms by which the gut microbiota influence autoimmunity and inflammation, consolidating available evidence on their association with autoimmune diseases. Additionally, it seeks to offer novel insights into early diagnosis and precise treatment strategies for autoimmune diseases.
The human gut harbors a diverse array of microbiota, encompassing bacteria, fungi, viruses and assorted microorganisms (8).
With advances in sequencing techniques, such as 16S rRNA and metagenomics, certain intestinal microbiota closely associated with human health have been identified, such as Phyla Firmicutes, Bacteroidetes, Pseudomonadota, and Actinomycetota, while others like Fusobacteria are relatively less studied (9, 10).
Microbiota populations colonize not only the gut but also the skin, respiratory tract, and reproductive system, influencing various physiological processes, including nutrition, tumorigenesis and immune homeostasis (11). It has been well established that the composition and abundance of the gut microbiota can be influenced by various factors, including environmental factors, diet and the host’s immune system (12, 13). The gut microbiota is not static but dynamic throughout human life. The study by Xie et al. has shown that the gut microbiota diversity changes over time, notably in genetically identical twins living apart (14). Dietary habits also significantly impact microbiota; for instance, high-fiber diets positively correlate with increased abundance of Lachnospiraceae (15), while Western diets rich in red meat and low in fiber are associated with Bacteroides spp and Ruminococcus torques dominance (16). Consequently, microbial compositions vary with dietary habits, heredity and other factors.
The gut microbiota, evolved alongside its host, profoundly influences various physiological and pathological processes, including nutrient production, drug effects, resistance to pathogens, and immune regulation (17–19). Gut bacteria ferment indigestible carbohydrates, generating short-chain fatty acids (SCFAs) like acetate, propionate and butyrate. These SCFAs act as biologically active compounds, providing energy for colonic epithelial cells (20, 21). In particular, butyrate acts as a primary energy source for the colonic epithelial cells (22). Furthermore, the gut microbiota significantly impacts medication metabolism. For instance, certain bacteria convert gemcitabine metabolites and irinotecan HCl, affecting therapeutic efficacy and potential side effects (23). It has been demonstrated that Escherichia coli (E. coli), Staphylococcus and Clostridium sporogenes produce an enzyme called beta-glucuronidase, converting the harmless form of the chemotherapeutic drug irinotecan HCl (CPT-11), known as SN-38 glucuronide, to its active form, SN-38 (24). Moreover, the gut microbiota exerts crucial effects on the immune system, including impacts on the proliferation and activation of immune cells, autoantibodies generation, and the onset of autoimmune diseases. Studies comparing germ-free (GF) and specific pathogen-free (SPF) mice highlight the microbiota’s impact on innate immune cell modulation and host defense against bacterial infections (25). Additionally, the microbiota contributes to the production of neurochemicals like gamma-aminobutyric acid (GABA), impacting the central nervous system and the gut through the brain-gut axis, ultimately influencing the immune microenvironment (26). Exploring the makeup, diversity, and structural alterations in the body’s microorganisms, along with interactions with the immune system, holds promise for understanding the fundamental processes of autoimmune diseases. All these findings offer the possibilities of identifying new biomarkers and developing effective therapeutic strategies for various diseases.
The correlation between the gut microbiota and innate immunity has attracted significant attention in academic research. The gut-associated lymphoid tissues (GALT) play a crucial role in protecting the intestinal mucosa, working in coordination with the mucosa-associated lymphoid tissues (MALT). Innate immune cells within these tissues employ non-specific pathogen recognition, innate immune reaction initiation and antigens presentation to activate the adaptive immune system (27, 28). It has been shown that the gut microbiota is important in regulating the physiological functions of GALTs in germ-free (GF) models, aiding in their development and maturation (27). Additionally, metabolic by-products produced by commensal microbiota, such as SCFAs, influence the immune response of GALTs through epigenetic mechanisms, supporting the defensive functions and immune tolerance (27).
Innate lymphoid cells (ILCs) are integral to GALTs. The development of ILCs occurs independently of the gut microbiota, while their specific functions rely on commensal microbiota (29, 30). For instance, ILC3, a prominent ILC class, supports epithelial cells survival, antimicrobial peptides production, and the generation of IL-22 (31). IL-22 is a key cytokine essential for the host’s immune response to Citrobacter (32, 33). It has been well documented that SCFAs promote IL-22 production of ILC3 by activating aromatic hydrocarbon receptor (AHR) through the AKT-STAT3 and ERK-STAT3 signaling pathways (34, 35) (Table 1). Moreover, SCFAs stimulate the proliferation of intestinal ILCs by affecting G protein-coupled receptor (GPCR) activity (45) (Table 1). The gut microbiota also facilitates interactions between ILC3 and other cell types (31), promoting the expression of protective proteins like fucosyltransferase 2 (FUT2) that strengthen the intestinal mucosal barrier (60). As the gut microbiota matures, ILC1 levels increase, indicating their reliance on commensal microbiota for development (30).
Conventional natural killer (NK) cells, the sole cytotoxic cell population within innate immunity, detect pathogens and carry out cytotoxic functions by releasing proteins like granzymes (61). The precise mechanisms regulating NK cell equilibrium between alloreactivity and autotolerance remain unclear. The gut microbiota, containing ligands for NK cell receptors, influences NK cell function and cytotoxicity, as observed in GF mice with the absence of interaction with commensal bacteria.
Macrophages in the gut are crucial for defending against infections and play a pivotal role in maintaining the integrity of the intestinal mucosa. The gut microbiota influences the development of myeloid-derived macrophages and the intestinal inflammation by modulating myeloid cell hematopoiesis, migration, and population sustenance within the gut (37, 38, 62–64) (Table 1).The relationship between the gut microbiota and innate immune cells underscores the profound influence of microbial communities on the development, differentiation, and functionality of these vital immune components, thus significantly contributing to the gut’ s microecology balance.
The intricate interplay between the gut microbiota and the local innate immune system contributes to the shaping of the gut microenvironment. Increasing evidence has demonstrated that the dysregulation of gut microbiota can result in disruptions and imbalance of the innate immune system by regulating the TLR signaling activation, inflammasome response, and ILC alterations, ultimately contributing to the onset of autoimmune diseases, such as RA and SLE (27, 65). Furthermore, fecal transplantation from healthy mice to certain disease-model mice has been shown to alleviate symptoms and ameliorate metabolic irregularities due to impaired activation of innate immune receptors, such as type 1 diabetes mellitus (T1DM) and inflammatory bowel disease (IBD) (31, 66). These findings have suggested the crucial role of gut microbiota in regulating autoimmune disorders through influencing innate immunity.
The gut microbiota plays a pivotal role in establishing and sustaining adaptive immunity in the gut, orchestrating interactions between varieties of immune cell types like T cells and B cells to ensure immunological equilibrium. T cells within the gut, specifically the CD4+T cells known as Th cells, display remarkable diversity and functions shaped by the unique metabolic characteristics of the gut microbiota (67). Notably, the gut microbiota influences the differentiation of naïve CD4+T cells into subsets like Th17 cells and Treg cells. Th17 cells are vital in defending against bacterial and fungal infections in the lamina propria (LP) of the small intestine by producing IL-17A, IL-17F, and IL-22. They also promote intestinal epithelial cells for more production of antimicrobial peptides (AMP), activate endothelial cells (ECs), and aid in neutrophils recruitment (68, 69). Segmented filamentous bacteria (SFB), a relatively low-abundance microbial population in the ileum, can induce the generation of RORγt+Th17 cells in the tissue-associated lymph nodes in the gut, although the excessive activation of RORγt+Th17 cells might lead to autoimmune diseases (70).
In contrast, regulatory T (Treg) cells contribute to immunological tolerance by promoting self-tolerance and suppressing excessive immune activation (71). The forkhead box P3 (Foxp3) is the key transcription factor for CD4+CD25+Treg cells, inhibiting excessive immune reactions. Certain probiotics, like Lactobacillus and Bifidobacterium infantis induce the production of anti-inflammatory CD4+CD25+Foxp3+ Treg cells, while Bacteroides fragilis and its polysaccharide A (PSA) stimulate IL-10 production by CD4+Treg cells depending on IL-2 pathway, suppressing inflammation (72). In addition, some bacterial metabolites, such as adenosine and inosine, can interact with T cells’ adenosine A2A receptor (A2AR), enhancing Treg cell activity while inhibiting Th1 and Th17 inflammatory responses (73).
Several pathways, including B cell receptor (BCR), CD40, Toll-like receptors (TLRs), B cell activating factor receptor (BAFF), and proliferation-inducing ligand (APRIL) receptors, primarily control the activation and differentiation of B cells (74). Activation of TLRs and BAFF/APRIL receptors significantly impacts T cell-dependent B cell antibody production (74). It is firmly established that the gut microbiota modulates B cell function by interacting with BCR through antigenic determinants and activating TLRs and NOD-like receptors (NLRs) via specific metabolites (74). Moreover, the gut microbiota stimulates dendritic cells (DCs) and the intestinal tissue cells to release cytokines like IL-1β and IL-6, which enhances the differentiation of naïve B cells into regulatory B cells (Bregs) within mesenteric lymph nodes (MLN) (41) (Table 1). Additionally, microbiota-derived ATP in the gut is converted to adenosine, activating adenosine receptors on B cells and promoting the production of IgG and IgA antibodies (74). Beyond these direct effects, the gut microbiota indirectly influences B cell differentiation and activation. As gut microbiota-derived metabolites, SCFAs can activate the intestinal epithelial cells via the cell receptors, such as GPR41, GPR43, and FFAR2, which prompts the release of thymic stromal lymphopoietin (TSLP) by epithelial cells and the release of TNF-α and iNOS of dendritic cells. This cascade triggers the expression of APRIL and IL-10, further enhancing IgA antibody production by B cells (75). Additionally, SCFAs can also activate receptors on the intestinal epithelial cells, like FFAR2, FFAR3 or GPR109a, and modulate the nuclear factor-κ-light chain enhancer (NF-κB) pathway involved in B cell activation, thereby suppressing inflammatory responses (Figure 1). Consequently, the gut microbiota and its metabolites are closely intertwined with the development, differentiation, and activation of B cells, exerting a substantial influence on immune responses and inflammation.
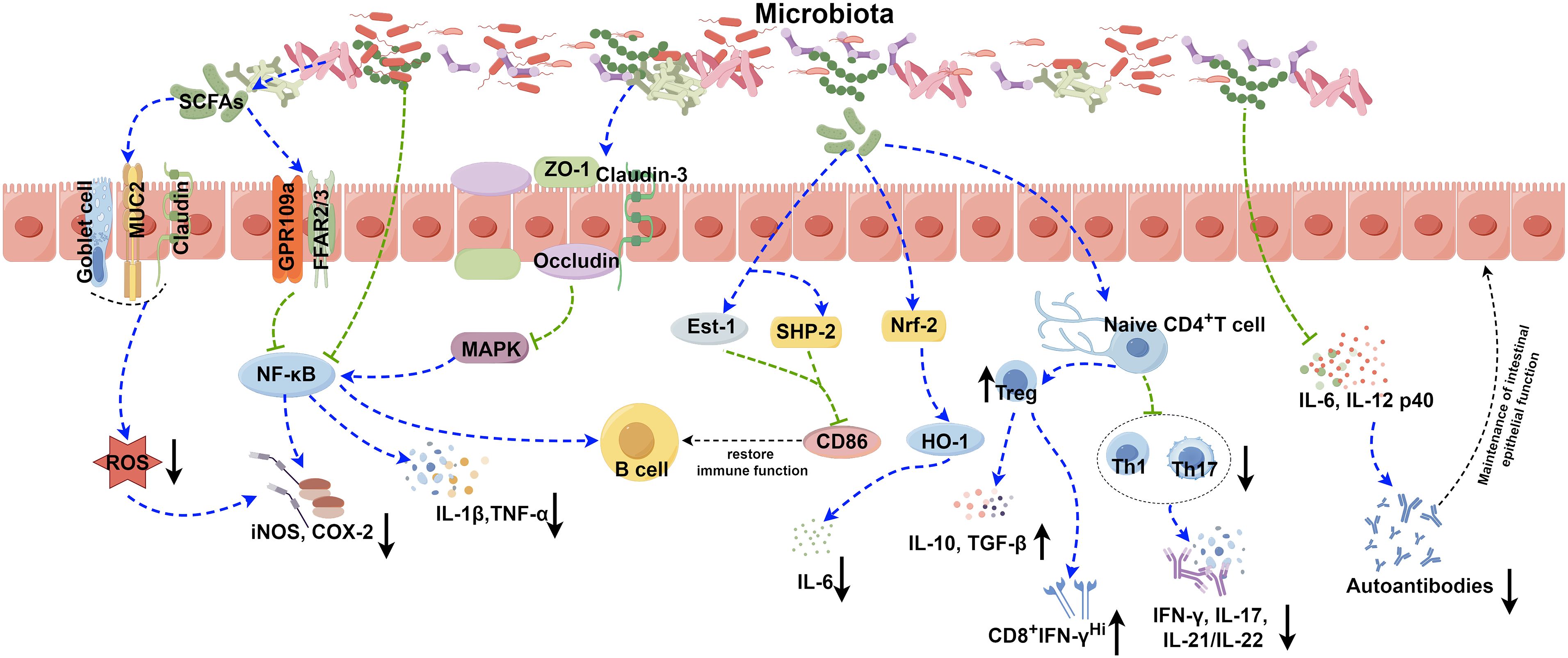
Figure 1 Role of the gut microbiota in autoimmune disease. The gut microbiota contributes to the pathogenesis of autoimmune diseases through various complicated mechanisms, such as the secretion of SCFAs, modulation of NF-κB and Nrf2 signaling pathways, disruption of the balance between Treg, Th1, and Th17 cells, and modulation the release of inflammatory factors. The image was created utilizing the FigDraw online platform (https://www.figdraw.com/#/) and was identified by the in-house image ID: UPORSc4cf4. SCFAs: Short chain fatty acids; MUC2: Recombinant Mucin 2; ROS: Reactive oxygen species; GPR109a: G protein-coupled receptor 109a; FFAR2/3: Recombinant Free Fatty Acid Receptor 2/3; NF-κB: Nuclear factor kappa-B; COX-2: Cyclooxygenase 2; iNOS: Inducible nitric oxide synthase; ZO-1: Zona Occludens 1; MAPK: Mitogen-activated protein kinase; IL-1β: Interleukin-1 beta; TNF-α: Tumor necrosis factor alpha; Est-1: Estrogen sulfotransferase-1; SHP-2: SH2 domain-containing protein-tyrosine phosphatase-2; Nrf-2: NF-E2-related factor 2; HO-1: Recombinant Heme Oxygenase 1; IL-6: Interleukin-6; IL-10: Interleukin-10; TGF-β: Transforming growth factor beta; IFN-γ: Interferon gamma; IL-17: Interleukin-17; IL-21/IL-22: Interleukin-21/Interleukin-22; IL-12 p40: Interleukin-12 p40.
It has been well documented certain gut microorganisms harboring epitopes resembling host proteins, such as the RNA-binding protein Ro60, are capable of activating T and B cells in SLE, thereby triggering abnormal adaptive immune response and inducing production of pathogenic autoantibodies (76). Besides, the roles of Th9 cells and IL-9 have been demonstrated in UC, functioning to impair intestinal barrier function, prevent mucosal wound healing in vivo and compromise tolerance to commensal bacteria by inducing inflammation and adaptive immune disorders (77–79). Therefore, the gut microbiota may contribute to autoimmune diseases by regulating T- or B-cell mediated adaptive immunity. The complex interplay between the gut microbiota and immune cells influences immune responses and homeostasis. Metabolites like SCFAs significantly impact cytokine and immunoglobulin production, affecting the progression of autoimmune diseases. This close interaction between the microbiota and the immune system forms a protective barrier against various threats. Alterations in the microbiota can lead to immune dysfunction and the onset of autoimmune diseases.
The integrity of human gut microbiota has been shown to correlate with susceptibility and outcomes of various diseases, such as metabolic, infectious and autoimmune diseases (80). As suggested in a groundbreaking article in 2002, the incidences of some classic infectious diseases such as tuberculosis and measles as well as intestinal infections have declined significantly but with a higher incidence of autoimmune diseases like T1DM and asthma in western countries in the last 50 years of the 20th century due to the improved sanitation, antibiotics usage and vaccination (81). This trend of change could still be observed in the last few years even until now that the incidence of autoimmune diseases is still steadily increasing, accompanied by a steady decline in the incidence of primary infectious diseases (82). According to “old friend hypothesis”, limited exposure to specific microbes (“old friends”) prevent immune system from forming a tolerogenic microenvironment, especially in early childhood phase (83–86). For children, the delivery mode, diet, and exposure to antibiotics and antacids are common factors for the contact with microbiota. Researches have shown that infants undergoing vaginal delivery and exclusive breastmilk feeding usually have a lower cumulative allergic burden, while those who exposed to antibiotic and antacid have an increased cumulative allergic burden conversely, thereby demonstrating the importance of microbiota in resisting excessive autoimmune response (87). Therefore, investigating the development, management, and prognosis of autoimmune disorders holds paramount importance due to the potential impact of microbiota dysfunction on both immunodeficiency syndromes and autoimmune diseases.
Dysbacteriosis, a commonly recognized term, refers to disruptions in the composition or activity of the microbiota within specific anatomical areas. This disruption encompasses alterations in both α-diversity and β-diversity. α-diversity reflects the variability in types and quantities of microorganisms of the host, while β-diversity delineates differences in microbial community makeup between individuals (88). Recent research has increasingly linked microbiota dysregulation to compromised intestinal barrier integrity, diminished functionality, and enhanced inflammation in autoimmune diseases (89, 90). Therefore, a growing body of evidence has underscored the intimate relationship between microbiota compositions and these conditions.
SLE is one of the most prevalent autoimmune diseases with multisystemic clinical manifestations caused by abundant immune complex depositions to tissues and target organs, leading to long sustained inflammation, immune disorders and ultimately multi-organ damages (91). A previous study has suggested that transferring microbiota from the cecum of lupus-prone mice into healthy mice can induce lupus (92). Similarly, GF mice receiving fecal matter from lupus mice exhibit increased levels of anti-DsDNA antibodies in serum, implicating the crucial role of the gut microbiota in autoantibody formation and immune reactions (93). Consequently, the gut microbiota is recognized as a pivotal factor contributing to SLE (27, 94, 95). The migration of microbial products from the colon and elevated permeability of the intestinal mucosal barrier are well recognized mechanisms implicated in the initiation and progression of SLE. Research in lupus-susceptible (NZB×BXSB) F1 mice has shown the migration of Enterococcus gallinarum from the intestine to mesenteric veins, intestinal draining lymph nodes, liver, and spleen (96). Enterococcus gallinarum has been also detected in liver biopsy specimens from SLE patients (96). Calprotectin, a calcium-containing protein in neutrophils and macrophages, acts as a crucial biomarker for impaired intestinal barrier (97). A close association between damaged intestinal barrier function in SLE patients and elevated levels of fecal calprotectin has been well documented (96, 98). Furthermore, certain gut microorganisms harboring epitopes resembling host proteins are capable of activating T and B cells, thereby triggering abnormal immune responses and abundant production of pathogenic autoantibodies. For instance, the RNA-binding protein Ro60, found in various gut microorganisms, shares homologous sequences with human Ro60 epitopes (76, 99). Anti-Ro60 antibodies can induce the generation of autoantibodies against Ro52, Smith, or U1RNP by spreading their epitopes (76, 99). Therefore, the intestinal microbiota containing sequences akin to human Ro60 epitopes may contribute to SLE by exacerbating pathological damages and dysregulated autoimmune responses in lupus-susceptible individuals.
It has been reported that certain gut microbiota can exert inhibitory effects on SLE through various mechanisms (Figure 1). Lactobacillus has been demonstrated to possess the capability to diminish the number of ILC3 cells and Th17 cells, suppress the pro-inflammatory factor IL-17 production, shift the Treg/Th17 balance in favor of the Treg phenotype, stimulate IL-10 production, and limit the accumulation of IgG-2a in the kidney (94, 100). This cascade ultimately reduces the kidney injures induced by dysregulated autoimmune responses. Moreover, it has been reported that Lactobacillus can significantly enhance the expression of key molecules associated with the intestinal mucosal barrier, such as ZO1, occludin and Cldn1 (100). Lactobacillus helps to enhance the intestinal barrier’s functionality without affecting the expression of Cldn2, responsible for creating lining pores (100) (Table 2). MicroRNAs (miRNAs) play pivotal roles in diverse biological processes, including immune cell maturation, the establishment of central and peripheral tolerance, and the differentiation of T helper (Th) cells. Alterations in miRNA expression can lead to immune system dysfunctions (123). Earlier studies have indicated a positive correlation between elevated expression levels of miR-155 and miR-181a and increased disease severity of SLE patients. Conversely, miR-155 deficiency reduces anti-dsDNA IgG titers and alleviates disease symptoms (124, 125). Interestingly, it has been discovered that Lactobacillus rhamnosus and Lactobacillus delbrueckii can attenuate the activity of miR-155 and miR-181a in peripheral blood mononuclear cells (PBMCs) of SLE patients (126). However, it’s noteworthy that Lactobacillus reuteri can exacerbate SLE by increasing the expression of type I interferon gene in the spleen and ileum of C57/B6 mice, resulting in anemia, increased intestinal permeability, and immune dysfunctions (127). These findings have suggested that different Lactobacillus strains may induce distinct immune responses in varying conditions in mice, leading to diverse outcomes (128). Additionally, the Firmicutes/Bacteroidetes (F/B) ratio is found to be significantly lower in SLE patients compared to non-SLE individuals (129–131), with Firmicutes showing a negative correlation with SLE Disease Activity Index (SLEDAI) scores (132). This suggests that Firmicutes may potentially delay the progression of SLE. Butyric acid and propionic acid, produced by Firmicutes, directly impact B cells by promoting the differentiation and proliferation of extrathymic Treg cells. Additionally, they suppress the expression of LPS-induced inflammatory cytokines such as IL-6, IL-12, and p40, thereby reducing the production of autoantibodies. Furthermore, these acids enhance and sustain the integrity of the intestinal epithelial barrier function in lupus-prone animals (57, 133, 134).
The gut microbiota composition in early RA patients differs significantly from that of healthy individuals, characterized by a significant reduction in Bifidobacterium and Bacteroides families and a notable increase in Prevotella species (135–137). These distinctions suggest a potential contribution of the gut microbiota-host interaction to the onset and progression of RA. However, the precise mechanism remains unclear. Recent studies propose that the influence of the gut microbiota on RA might involve various mechanisms, including the activation of antigen-presenting cells, production of citrullinated peptides through interactions with Toll-like receptors (TLRs) or NOD-like receptors (NLRs), induction of antigen-mimicking cross-reactivity, alterations in the intestinal mucosal permeability, and the promotion of Th17 cell-mediated inflammation in the mucosa (138, 139) (Figure 1).
Citrullinated peptides, formed when arginine residues are converted into citrullinated residues by protein arginine deiminase (PAD), can disrupt immune tolerance and trigger autoimmune reactions in individuals genetically predisposed to RA under specific conditions (140–142). The gut microbiota dysregulation may compromise PAD function, impacting immunological tolerance and contributing to autoimmune diseases (138, 143–145). Additionally, the gut microbiota itself can encode bacterial PAD enzymes, facilitating citrullination (146, 147). Different species of Prevotella play varying roles in RA. Overcolonization of Prevotella copri (P. copri) might intensify mucosal inflammation and induce immune responses, potentially leading to arthritis (138). P. copri can also compromise the intestinal barrier integrity by disrupting the tight junctions (TJs) between intestinal epithelial cells, facilitating disease occurrence (139, 148). Conversely, Marietta et al. have demonstrated that Prevotella histicola (P. histicola) can prevent and treat collagen-induced arthritis (CIA) in HLA-DQ8 transgenic mice by boosting Treg cells, suppressing Th17 responses, enhancing IL-10 release, and stabilizing the intestinal barrier (102) (Table 2, Figure 1).
Various strains of Lactobacillus casei (L. casei) in preclinical studies have shown efficacy in treating RA, including reducing joint swelling, arthritis scores, and serum inflammatory cytokine levels, highlighting probiotics’ potential in RA remission (103, 149, 150). Additionally, the interplay between periodontal disease and RA involves shared pathogenic mechanisms and immunological pathways. RA patients often display elevated antibody levels against Porphyromonas gingivalis (P. gingivalis) and Prevotella, with P. gingivalis antibodies correlating with the levels of RA-specific anti-CCP antibodies (151, 152).
The enzyme PAD from P. gingivalis in periodontal disease can citrullinate human fibrinogen and α-enolase. Antibodies generated against these citrullinated antigens may cross-react with joint antigens, exacerbating RA-related inflammation (153). Researchers are actively exploring strategies to mitigate microbial self-antigen cross-reactivity and curb excessive citrullination, aiming to develop microbiota-based treatments for RA.
SS is one of the most prevalent autoimmune diseases, primarily affecting the lacrimal and salivary glands. The precise pathogenesis of SS remains largely unknown. Dysfunction of T cells and B cells play a critical role in the onset and progression of SS (154, 155). An imbalance, marked by an increase in Th17 cells and a decrease in Treg cells, can prompt lymphocyte infiltration, epithelial cell activation, enhanced proinflammatory cytokines production (e.g., IFN-γ and IL-17), exposure to autoantibodies, and damages to the corneal barrier, contributing to the development of SS (156–158). Furthermore, Th1 cells, known contributors to the pathogenesis of SS, partake in ocular inflammation by secreting pro-inflammatory cytokines like IFN-γ, IL-1β, IL-6 and TNF-α (159). Elevated expression of B cell activating factor (BAFF) is mainly induced by type I and type II interferons (IFN) (160). Significantly elevated levels of BAFF in the peripheral circulation and the salivary gland tissues have been observed in 55% of SS patients, highlighting increased B cell activation in SS (154, 161). A significant reduction in the diversity of the gut microbiota has been reported in SS patients, characterized by diminished symbiotic bacteria and increased potentially pathogenic strains, positively correlated with disease severity (158). Fecal transplantation (FMT) effectively ameliorates ocular symptoms in germ-free (GF) mice with SS, highlighting the strong link between gut microbiota and SS pathogenesis (162–164). The protective role of the gut microbiota in SS operates through two main mechanisms (Figure 1). Firstly, microbiota-produced metabolites like SCFAs exert anti-inflammatory properties, alleviating ocular inflammation (165–167). Secondly, the gut microbiota plays a critical role in regulating the development, differentiation and activation of ocular immune cells, effectively modulating the balance between pro-inflammatory Th17 cells and anti-inflammatory Treg cells (109, 165, 168). As a result, strategies aiming at maintaining microbial balance may hold promise for effective biological prevention approaches for SS.
T1DM emerges from a multifaceted autoimmune process, characterized by the T-cell-mediated destruction of insulin-producing β-cells. It is the most commonly diagnosed diabetes in young population, while the incidence of T1DM in adult has also increased rapidly in the past 15 years (169, 170). More and more evidence provide supports that the composition of the gut microbiota, as an environment factor, is different between diabetes patients and healthy individuals. Bacteroidetes has been recognized as the most common microbial phylum in T1DM patients, while the number of butyrate-producing species from Clostridium clusters IV and XIVa, as well as mucin-degrading bacteria such as Prevotella and Akkermansia in T1DM patients has significantly reduced (171). The importance of butyrate in preserving the integrity of the intestinal mucosal barrier has been highlighted (172) (Figure 1). The reduction in butyrate-producing bacteria leads to increased gut permeability. And it will allow the passage of microbial antigens, products, and even the microorganisms themselves, which may promote the inflammation and the progression of T1DM (173, 174). In addition to this, there are many kinds of microbiota that can have an impact on the development of disease. An increase in the gut microbiota and abundance of Firmicutes was observed in non-obese diabetic (NOD) mice fed with human lacto-oligosaccharide, helping to inhibit islet inflammation and diabetes (175). Although Prevotella has been associated with the development of chronic inflammatory diseases by augmenting mucosal Th17-mediated immune responses, it has also been established that Prevotella play a role in protecting against Bacteroides-induced glucose intolerance, promoting glycogen storage, and enhancing glucose metabolism (176, 177). When transplanting human amniotic mesenchymal stem cells (hAMSC) into T1DM mice, scientists noticed that the therapeutic effect of MSC strongly depends on the modification of the beneficial gut microbiota, including Bifidobacterium, Providencia, Veillonella, and Prevotella, indicating the significant role of the gut microbiota in alleviating symptom and controlling the development of T1DM (178). Except for forming the intestinal mucosal barrier, the metabolic products of the gut microbiota, such as tryptophan derivatives and SCFAs, also regulates intestinal immunity. SCFAs protect NOD mice from insulitis and slow down the development of T1DM by inhibiting inflammatory responses and the accumulation of IFN-γ+T cell in the pancreas (179, 180). As the precursor of SCFAs and an important by-product, intracellular succinic acid could activate intestinal gluconeogenesis positively to regulate gluconeogenesis and blood glucose levels (181, 182). Hence, clinical treatments based on the gut microbiota show promise in maintaining intestinal homeostasis, slow down disease progression, and even reverse T1DM.
UC is a kind of IBD characterized by unpredictable and chronic clinical symptoms, with alternating periods of exacerbation and remission (183). Based on next-generation sequencing technique, scientists have found reduced bacterial diversity and imbalance between beneficial and aggressive bacteria in UC cases (184). Species from the Proteobacteria including E. coli, Enterobacteriaceae, Klebsiella, and Proteus spp., as well as members from Fusobacteria, could enhance inflammatory response and aggravate symptoms, and have been demonstrated to be positively associated with UC (185–188). The microbial metabolites also vary between UC patients and the healthy. SCFAs plays an important role in suppressing intestinal inflammation (34, 189). SCFAs could also induce the production of T-cell-dependent IgA and maintain mucosal homeostasis by regulating the localization of commensal bacteria (190). It has been determined that acetic acid and butyric acid in feces of UC patients were lower than those in the healthy individuals (191). Taken together, all of the changes reveal the close relationships between the gut microbiota and the development of UC (Figure 1). However, given the individual phenotypic differences in the gut microbiota, it is still necessary to further study the exact effect mechanisms between individual microbiota and the host (192).
The pathogenesis of psoriasis is complex and not very clear. For now, Th17/IL-23 axis has been established as a crucial immunological mechanism in the development of psoriasis, which is also the basis of biologics treatment (193–195). The gut microbiota and their metabolites could adjust the balance between immune tolerance and inflammation, such as acting on differentiation of naïve T cells into either regulatory or Th17 lineages, so that affecting the progression of psoriasis (168) (Figure 1). It has been found that the gut microbiota has impact on the manifestation of the psoriatic phenotype through a Th17-mediated T-cell response on imiquimod-induced mouse models; meanwhile, germ-free mice or conventionally housed mice treated with antibiotics showed protective effect on skin (196) (Figure 1). Scientists also noticed an interesting phenomenon that the gut microbiota dysbiosis in psoriasis patients is similar to those of IBD, both with reduced Eubacterium rectale (E. rectale), Alistipes finegoldii (A. finegoldii) and Alistipes shahii (A. shahii) species (197–200). Based on this consistency, using probiotics to restore the gut microbiota homeostasis and reduce inflammation to achieve therapeutic purpose for psoriasis is possible. Probiotic could suppress the expression of TNF−α, IL−6 and proinflammatory cytokines in the IL−23/IL−17 cytokine axis and enhance gut barrier function to prevent further infection (201). In another case, patients with severe pustular psoriasis showed obvious clinical improvement within 2 weeks after applying Lactobacillus sporogenes supplementation 3 times per day, and almost get remission after 4 weeks (202).
In this manuscript, our focus was directed towards examining the involvement of certain prevalent autoimmune diseases of the gut microbiota, including SLE, RA, SS, T1DM, UC, and psoriasis. However, diseases such as multiple sclerosis (MS), autoimmune thyroid disease (AITD), celiac disease (CeD), among others, were not addressed. Further research is warranted to explore the underlying mechanisms governing immune disorders.
The interaction between human microbiota and the host plays a crucial role in maintaining health and influencing disease onset. The complex relationship encompasses multiple facets, with microbiota and their metabolites wielding significant influence over host inflammation and immune responses. The gut microbiota participates in regulating immune cell proliferation, differentiation, activation, intestinal permeability, and the integrity of mucosal barriers. Probiotics have emerged as a promising strategy for managing autoimmune diseases, such as SLE and RA. They operate by promoting a healthy gut microbiota and fostering a balanced interaction with the host’s immune system. However, further investigations are warranted to identify specific biomarkers that can accurately distinguish between healthy and compromised microbiota states. Additionally, understanding how microbiota and their metabolites impact normal balanced states versus inflammatory conditions, and discerning potential differences between effects on mucosal surfaces and systemic tissues, remains crucial. In-depth studies investigating the role of microbiota in autoimmune diseases provide insights into the underlying mechanisms of diseases. These insights may reveal prominent diagnostic markers and therapeutic targets, ultimately help us to understand the pathogenesis of autoimmune diseases and explore novel diagnostic and therapeutic strategies.
XW: Writing – original draft. WY: Writing – review & editing. CY: Writing – review & editing. ZW: Writing – review & editing. JZ: Writing – review & editing. DX: Writing – review & editing. XS: Writing – review & editing. WS: Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation, China [82201925 and 82171790] and Natural Science Foundation, Shandong Province, China [ZR2022QH203 and ZR2020KC001].
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Fugger L, Jensen LT, Rossjohn J. Challenges, progress, and prospects of developing therapies to treat autoimmune diseases. Cell. (2020) 181:63–80. doi: 10.1016/j.cell.2020.03.007
2. Zhang Y, Liu J, Wang C, Liu J, Lu W. Toll-like receptors gene polymorphisms in autoimmune disease. Front Immunol. (2021) 12:672346. doi: 10.3389/fimmu.2021.672346
3. De Luca F, Shoenfeld Y. The microbiome in autoimmune diseases. Clin Exp Immunol. (2019) 195:74–85. doi: 10.1111/cei.13158
4. Wang L, Wang FS, Gershwin ME. Human autoimmune diseases: A comprehensive update. J Intern Med. (2015) 278:369–95. doi: 10.1111/joim.12395
5. Shaheen WA, Quraishi MN, Iqbal TH. Gut microbiome and autoimmune disorders. Clin Exp Immunol. (2022) 209:161–74. doi: 10.1093/cei/uxac057
6. Konig MF. The microbiome in autoimmune rheumatic disease. Best Pract Res Clin Rheumatol. (2020) 34:101473. doi: 10.1016/j.berh.2019.101473
7. Miyauchi EA-OX, Shimokawa C, Steimle A, Desai MA-O, Ohno HA-O. The impact of the gut microbiome on extra-intestinal autoimmune diseases. Nat Rev Immunol. (2023) 23:9–23. doi: 10.1038/s41577-022-00727-y
8. de Vos WM, Tilg H, Van Hul M, Cani PD. Gut microbiome and health: mechanistic insights. Gut. (2022) 71:1020–32. doi: 10.1136/gutjnl-2021-326789
9. Seekatz AM, Schnizlein MK, Koenigsknecht MJ, Baker JR, Hasler WL, Bleske BE, et al. Spatial and temporal analysis of the stomach and small-intestinal microbiota in fasted healthy humans. mSphere. (2019) 4:e00126-19. doi: 10.1128/mSphere.00126-19
10. Leite GGS, Weitsman S, Parodi G, Celly S, Sedighi R, Sanchez M, et al. Mapping the segmental microbiomes in the human small bowel in comparison with stool: A reimagine study. Digestive Dis Sci. (2020) 65:2595–604. doi: 10.1007/s10620-020-06173-x
11. Requena T, Velasco M. The human microbiome in sickness and in health. Rev Clin Esp (Barc). (2021) 221:233–40. doi: 10.1016/j.rceng.2019.07.018
12. Berding K, Vlckova K, Marx W, Schellekens H, Stanton C, Clarke G, et al. Diet and the microbiota-gut-brain axis: sowing the seeds of good mental health. Adv Nutr (Bethesda Md). (2021) 12:1239–85. doi: 10.1093/advances/nmaa181
13. Lu Y, Yuan X, Wang M, He Z, Li H, Wang J, et al. Gut microbiota influence immunotherapy responses: mechanisms and therapeutic strategies. J Hematol Oncol. (2022) 15:47. doi: 10.1186/s13045-022-01273-9
14. Xie H, Guo R, Zhong H, Feng Q, Lan Z, Qin B, et al. Shotgun metagenomics of 250 adult twins reveals genetic and environmental impacts on the gut microbiome. Cell Syst. (2016) 3:572–84 e3. doi: 10.1016/j.cels.2016.10.004
15. Wastyk HC, Fragiadakis GK, Perelman D, Dahan D, Merrill BD, Yu FB, et al. Gut-microbiota-targeted diets modulate human immune status. Cell. (2021) 184:4137–53 e14. doi: 10.1016/j.cell.2021.06.019
16. Martinez-Medina M, Denizot J, Dreux N, Robin F, Billard E, Bonnet R, et al. Western diet induces dysbiosis with increased E coli in ceabac10 mice, alters host barrier function favouring aiec colonisation. Gut. (2014) 63:116–24. doi: 10.1136/gutjnl-2012-304119
17. Dominguez-Bello MG, Godoy-Vitorino F, Knight R, Blaser MJ. Role of the microbiome in human development. Gut. (2019) 68:1108–14. doi: 10.1136/gutjnl-2018-317503
18. Adak A, Khan MR. An insight into gut microbiota and its functionalities. Cell Mol Life sciences: CMLS. (2019) 76:473–93. doi: 10.1007/s00018-018-2943-4
19. Lee JS, Wang RX, Alexeev EE, Colgan SP. Intestinal inflammation as a dysbiosis of energy procurement: new insights into an old topic. Gut Microbes. (2021) 13:1–20. doi: 10.1080/19490976.2021.1880241
20. Morrison DJ, Preston T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes. (2016) 7:189–200. doi: 10.1080/19490976.2015.1134082
21. Kimura I, Ichimura A, Ohue-Kitano R, Igarashi M. Free fatty acid receptors in health and disease. Physiol Rev. (2020) 100:171–210. doi: 10.1152/physrev.00041.2018
22. DeMartino P, Cockburn DW. Resistant starch: impact on the gut microbiome and health. Curr Opin Biotechnol. (2020) 61:66–71. doi: 10.1016/j.copbio.2019.10.008
23. Geller LT, Barzily-Rokni M, Danino T, Jonas OH, Shental N, Nejman D, et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science. (2017) 357:1156–60. doi: 10.1126/science.aah5043
24. Yue B, Gao R, Wang Z, Dou W. Microbiota-host-irinotecan axis: A new insight toward irinotecan chemotherapy. Front Cell infection Microbiol. (2021) 11:710945. doi: 10.3389/fcimb.2021.710945
25. Schulz C, Gomez Perdiguero E, Chorro L, Szabo-Rogers H, Cagnard N, Kierdorf K, et al. A lineage of myeloid cells independent of myb and hematopoietic stem cells. Science. (2012) 336:86–90. doi: 10.1126/science.1219179
26. Chen M, Ruan G, Chen L, Ying S, Li G, Xu F, et al. Neurotransmitter and intestinal interactions: focus on the microbiota-gut-brain axis in irritable bowel syndrome. Front Endocrinol. (2022) 13:817100. doi: 10.3389/fendo.2022.817100
27. Jiao Y, Wu L, Huntington ND, Zhang X. Crosstalk between gut microbiota and innate immunity and its implication in autoimmune diseases. Front Immunol. (2020) 11:282. doi: 10.3389/fimmu.2020.00282
28. Luciani C, Hager FT, Cerovic V, Lelouard H. Dendritic cell functions in the inductive and effector sites of intestinal immunity. Mucosal Immunol. (2022) 15:40–50. doi: 10.1038/s41385-021-00448-w
29. Shi Z, Ohno H, Satoh-Takayama N. Dietary derived micronutrients modulate immune responses through innate lymphoid cells. Front Immunol. (2021) 12:670632. doi: 10.3389/fimmu.2021.670632
30. Constantinides MG. Interactions between the microbiota and innate and innate-like lymphocytes. J leukocyte Biol. (2018) 103:409–19. doi: 10.1002/JLB.3RI0917-378R
31. Thaiss CA, Zmora N, Levy M, Elinav E. The microbiome and innate immunity. Nature. (2016) 535:65–74. doi: 10.1038/nature18847
32. Cox CB, Storm EE, Kapoor VN, Chavarria-Smith J, Lin DL, Wang L, et al. Il-1r1-dependent signaling coordinates epithelial regeneration in response to intestinal damage. Sci Immunol. (2021) 6:eabe8856. doi: 10.1126/sciimmunol.abe8856
33. Serafini N, Jarade A, Surace L, Goncalves P, Sismeiro O, Varet H, et al. Trained ilc3 responses promote intestinal defense. Sci (New York NY). (2022) 375:859–63. doi: 10.1126/science.aaz8777
34. Yang W, Yu T, Huang X, Bilotta AJ, Xu L, Lu Y, et al. Intestinal microbiota-derived short-chain fatty acids regulation of immune cell il-22 production and gut immunity. Nat Commun. (2020) 11:4457. doi: 10.1038/s41467-020-18262-6
35. Chun E, Lavoie S, Fonseca-Pereira D, Bae S, Michaud M, Hoveyda HR, et al. Metabolite-sensing receptor ffar2 regulates colonic group 3 innate lymphoid cells and gut immunity. Immunity. (2019) 51:871–84 e6. doi: 10.1016/j.immuni.2019.09.014
36. Kim SY, Shin JS, Chung KS, Han HS, Lee HH, Lee JH, et al. Immunostimulatory effects of live lactobacillus sakei K040706 on the cyp-induced immunosuppression mouse model. Nutrients. (2020) 12:3573. doi: 10.3390/nu12113573
37. Khosravi A, Yanez A, Price JG, Chow A, Merad M, Goodridge HS, et al. Gut microbiota promote hematopoiesis to control bacterial infection. Cell Host Microbe. (2014) 15:374–81. doi: 10.1016/j.chom.2014.02.006
38. Lavin Y, Mortha A, Rahman A, Merad M. Regulation of macrophage development and function in peripheral tissues. Nat Rev Immunol. (2015) 15:731–44. doi: 10.1038/nri3920
39. Schnupf P, Gaboriau-Routhiau V, Sansonetti PJ, Cerf-Bensussan N. Segmented filamentous bacteria, th17 inducers and helpers in a hostile world. Curr Opin Microbiol. (2017) 35:100–9. doi: 10.1016/j.mib.2017.03.004
40. Cohen-Poradosu R, McLoughlin RM, Lee JC, Kasper DL. Bacteroides fragilis-stimulated interleukin-10 contains expanding disease. J Infect Dis. (2011) 204:363–71. doi: 10.1093/infdis/jir277
41. Rosser EC, Oleinika K, Tonon S, Doyle R, Bosma A, Carter NA, et al. Regulatory B Cells Are Induced by Gut Microbiota-Driven Interleukin-1β and Interleukin-6 Production. Nat Med. (2014) 20:1334–9. doi: 10.1038/nm.3680
42. Aziz N, Bonavida B. Activation of natural killer cells by probiotics. For Immunopathol Dis Therap. (2016) 7:41–55. doi: 10.1615/ForumImmunDisTher.2016017095
43. Abdi K, Laky K, Abshari M, Hill EM. Dendritic cells trigger ifn-Γ Secretion by nk cells independent of il-12 and il-18. Eur J Immunol. (2022) 52(9):1431–40. doi: 10.1002/eji.202149733
44. Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. (2013) 504:446–50. doi: 10.1038/nature12721
45. Sepahi A, Liu Q, Friesen L, Kim CH. Dietary fiber metabolites regulate innate lymphoid cell responses. Mucosal Immunol. (2021) 14:317–30. doi: 10.1038/s41385-020-0312-8
46. Park J, Kim M, Kang SG, Jannasch AH, Cooper B, Patterson J, et al. Short-chain fatty acids induce both effector and regulatory T cells by suppression of histone deacetylases and regulation of the mtor-S6k pathway. Mucosal Immunol. (2015) 8:80–93. doi: 10.1038/mi.2014.44
47. Sun M, Wu W, Chen L, Yang W, Huang X, Ma C, et al. Microbiota-derived short-chain fatty acids promote th1 cell il-10 production to maintain intestinal homeostasis. Nat Commun. (2018) 9:3555. doi: 10.1038/s41467-018-05901-2
48. Kespohl M, Vachharajani N, Luu M, Harb H, Pautz S, Wolff S, et al. The microbial metabolite butyrate induces expression of th1-associated factors in cd4(+) T cells. Front Immunol. (2017) 8:1036. doi: 10.3389/fimmu.2017.01036
49. Vieira RS, Castoldi A, Basso PJ, Hiyane MI, Camara NOS, Almeida RR. Butyrate attenuates lung inflammation by negatively modulating th9 cells. Front Immunol. (2019) 10:67. doi: 10.3389/fimmu.2019.00067
50. Luu M, Pautz S, Kohl V, Singh R, Romero R, Lucas S, et al. The short-chain fatty acid pentanoate suppresses autoimmunity by modulating the metabolic-epigenetic crosstalk in lymphocytes. Nat Commun. (2019) 10:760. doi: 10.1038/s41467-019-08711-2
51. Bachem A, Makhlouf C, Binger KJ, de Souza DP, Tull D, Hochheiser K, et al. Microbiota-derived short-chain fatty acids promote the memory potential of antigen-activated cd8(+) T cells. Immunity. (2019) 51:285–97. doi: 10.1016/j.immuni.2019.06.002
52. Luu M, Weigand K, Wedi F, Breidenbend C, Leister H, Pautz S, et al. Regulation of the effector function of cd8(+) T cells by gut microbiota-derived metabolite butyrate. Sci Rep. (2018) 8:14430. doi: 10.1038/s41598-018-32860-x
53. Kruglov AA, Grivennikov SI, Kuprash DV, Winsauer C, Prepens S, Seleznik GM, et al. Nonredundant function of soluble ltα3 produced by innate lymphoid cells in intestinal homeostasis. Science. (2013) 342:1243–6. doi: 10.1126/science.1243364
54. Sanchez HN, Moroney JB, Gan H, Shen T, Im JL, Li T, et al. B cell-intrinsic epigenetic modulation of antibody responses by dietary fiber-derived short-chain fatty acids. Nat Commun. (2020) 11:60. doi: 10.1038/s41467-019-13603-6
55. Berndt BE, Zhang M, Owyang SY, Cole TS, Wang TW, Luther J, et al. Butyrate increases il-23 production by stimulated dendritic cells. Am J Physiol Gastrointest Liver Physiol. (2012) 303:G1384–92. doi: 10.1152/ajpgi.00540.2011
56. Liu L, Min J, Wang J, Wu H, Zeng Y, Chen S, et al. Butyrate interferes with the differentiation and function of human monocyte-derived dendritic cells. Cell Immunol. (2012) 277:66–73. doi: 10.1016/j.cellimm.2012.05.011
57. Nastasi C, Candela M, Bonefeld CM, Geisler C, Hansen M, Krejsgaard T, et al. The effect of short-chain fatty acids on human monocyte-derived dendritic cells. Sci Rep. (2015) 5:16148. doi: 10.1038/srep16148
58. Chang PV, Hao L, Offermanns S, Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci U.S.A. (2014) 111:2247–52. doi: 10.1073/pnas.1322269111
59. Liu T, Liu Y, Xiao N, Suo H, Xie K, Yang C, et al. Short-chain fatty acids suppress lipopolysaccharide-induced production of nitric oxide and proinflammatory cytokines through inhibition of nf-Κb pathway in raw264.7 cells. Inflammation. (2012) 35:1676–84. doi: 10.1007/s10753-012-9484-z
60. Stojanović I, Saksida T, Miljković Đ, Pejnović N. Modulation of intestinal ilc3 for the treatment of type 1 diabetes. Front Immunol. (2021) 12:653560. doi: 10.3389/fimmu.2021.653560
61. Wu SY, Fu T, Jiang YZ, Shao ZM. Natural killer cells in cancer biology and therapy. Mol Cancer. (2020) 19:120. doi: 10.1186/s12943-020-01238-x
62. Josefsdottir KS, Baldridge MA-O, Kadmon CS, King KA-O. Antibiotics impair murine hematopoiesis by depleting the intestinal microbiota. Blood. (2017) 129:729–39. doi: 10.1182/blood-2016-03-708594
63. Han H, Yan H, King KY. Broad-spectrum antibiotics deplete bone marrow regulatory T cells. Cells. (2021) 10:277. doi: 10.3390/cells10020277
64. Goenka A, Khan FA-O, Verma B, Sinha P, Dmello CC, Jogalekar MP, et al. Tumor microenvironment signaling and therapeutics in cancer progression. Cancer Commun. (2023) 43:525–61. doi: 10.1002/cac2.12416
65. Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, et al. Nlrp6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. (2011) 145:745–57. doi: 10.1016/j.cell.2011.04.022
66. Carvalho FA, Koren O, Goodrich JK, Johansson MEV, Nalbantoglu I, Aitken JD, et al. Transient inability to manage proteobacteria promotes chronic gut inflammation in tlr5-deficient mice. Cell Host Microbe. (2012) 12:139–52. doi: 10.1016/j.chom.2012.07.004
67. Zhao Q, Elson CO. Adaptive immune education by gut microbiota antigens. Immunology. (2018) 154:28–37. doi: 10.1111/imm.12896
68. Regen T, Isaac S, Amorim A, Núñez NG, Hauptmann J, Shanmugavadivu A, et al. Il-17 controls central nervous system autoimmunity through the intestinal microbiome. Sci Immunol. (2021) 6:eaaz6563. doi: 10.1126/sciimmunol.aaz6563
69. Lee JY, Hall JA, Kroehling L, Wu L, Najar T, Nguyen HH, et al. Serum amyloid a proteins induce pathogenic th17 cells and promote inflammatory disease. Cell. (2020) 180:79–91.e16. doi: 10.1016/j.cell.2019.11.026
70. Sano T, Kageyama T, Fang V, Kedmi R, Martinez CS, Talbot J, et al. Redundant cytokine requirement for intestinal microbiota-induced th17 cell differentiation in draining lymph nodes. Cell Rep. (2021) 36:109608. doi: 10.1016/j.celrep.2021.109608
71. Honda K, Littman DR. The microbiota in adaptive immune homeostasis and disease. Nature. (2016) 535(1476-4687:75–84. doi: 10.1038/nature18848
72. Luckey D, Gomez A, Murray J, White B, Taneja V, Taneja V. Bugs & Us: the role of the gut in autoimmunity. Indian J Med Res. (2013) 138:732–43.
73. Liu Y, Alookaran JJ, Rhoads JM. Probiotics in autoimmune and inflammatory disorders. Nutrients. (2018) 10:1537. doi: 10.3390/nu10101537
74. Kim M, Kim CH. Regulation of humoral immunity by gut microbial products. Gut Microbes. (2017) 8:392–9. doi: 10.1080/19490976.2017.1299311
75. Cerdá B, Pérez M, Pérez-Santiago JD, Tornero-Aguilera JF, González-Soltero R, Larrosa M. Gut microbiota modification: another piece in the puzzle of the benefits of physical exercise in health? Front Physiol. (2016) 7:51. doi: 10.3389/fphys.2016.00051
76. Greiling TM, Dehner C, Chen X, Hughes K, Iniguez AJ, Boccitto M, et al. Commensal orthologs of the human autoantigen ro60 as triggers of autoimmunity in lupus. Sci Transl Med. (2018) 10:eaan2306. doi: 10.1126/scitranslmed.aan2306
77. Gerlach K, Hwang Y, Nikolaev A, Atreya R, Dornhoff H, Steiner S, et al. Th9 cells that express the transcription factor pu.1 drive T cell-mediated colitis via il-9 receptor signaling in intestinal epithelial cells. Nat Immunol. (2014) 15:676–86. doi: 10.1038/ni.2920
78. Gerlach K, McKenzie AN, Neurath MF, Weigmann B. Il-9 regulates intestinal barrier function in experimental T cell-mediated colitis. Tissue Barriers. (2015) 3:e983777. doi: 10.4161/21688370.2014.983777
79. Shohan M, Sabzevary-Ghahfarokhi M, Bagheri N, Shirzad H, Rahimian G, Soltani A, et al. Intensified th9 response is associated with the immunopathogenesis of active ulcerative colitis. Immunol Invest. (2018) 47:700–11. doi: 10.1080/08820139.2018.1486411
80. Libertucci J, Young VB. The role of the microbiota in infectious diseases. Nat Microbiol. (2019) 4:35–45. doi: 10.1038/s41564-018-0278-4
81. Bach JF. The effect of infections on susceptibility to autoimmune and allergic diseases. N Engl J Med. (2002) 347:911–20. doi: 10.1056/NEJMra020100
82. Larsen OFA, van der Grint M, Wiegers C, van de Burgwal LHM. The gut microbiota: master of puppets connecting the epidemiology of infectious, autoimmune, and metabolic disease. Front Microbiol. (2022) 13:902106. doi: 10.3389/fmicb.2022.902106
83. Rook GA, Brunet LR. Microbes, immunoregulation, and the gut. Gut. (2005) 54:317–20. doi: 10.1136/gut.2004.053785
84. Kriegel MA, Lohmann T, Gabler C, Blank N, Kalden JR, Lorenz HM. Defective suppressor function of human cd4+ Cd25+ Regulatory T cells in autoimmune polyglandular syndrome type ii. J Exp Med. (2004) 199:1285–91. doi: 10.1084/jem.20032158
85. Viglietta V, Baecher-Allan C, Weiner HL, Hafler DA. Loss of functional suppression by cd4+Cd25+ Regulatory T cells in patients with multiple sclerosis. J Exp Med. (2004) 199:971–9. doi: 10.1084/jem.20031579
86. Badami E, Sorini C, Coccia M, Usuelli V, Molteni L, Bolla AM, et al. Defective differentiation of regulatory foxp3+ T cells by small-intestinal dendritic cells in patients with type 1 diabetes. Diabetes. (2011) 60:2120–4. doi: 10.2337/db10-1201
87. Gabryszewski SJ, Dudley J, Grundmeier RW, Hill DA. Early-life environmental exposures associate with individual and cumulative allergic morbidity. Pediatr Allergy Immunol. (2021) 32:1089–93. doi: 10.1111/pai.13486
88. Moon J, Yoon CH, Choi SH, Kim MK. Can gut microbiota affect dry eye syndrome? Int J Mol Sci. (2020) 21:8443. doi: 10.3390/ijms21228443
89. Christovich A, Luo XM. Gut microbiota, leaky gut, and autoimmune diseases. Front Immunol. (2022) 13:946248. doi: 10.3389/fimmu.2022.946248
90. Kinashi Y. Hase K. Partners in leaky gut syndrome: intestinal dysbiosis and autoimmunity. Front Immunol. (2021) 12:673708. doi: 10.3389/fimmu.2021.673708
91. Owen KA, Grammer AC, Lipsky PE. Deconvoluting the heterogeneity of sle: the contribution of ancestry. J Allergy Clin Immunol. (2022) 149:12–23. doi: 10.1016/j.jaci.2021.11.005
92. Choi SC, Brown J, Gong M, Ge Y, Zadeh M, Li W, et al. Gut microbiota dysbiosis and altered tryptophan catabolism contribute to autoimmunity in lupus-susceptible mice. Sci Transl Med. (2020) 12:eaax2220. doi: 10.1126/scitranslmed.aax2220
93. Ma Y, Xu X, Li M, Cai J, Wei Q, Niu H. Gut microbiota promote the inflammatory response in the pathogenesis of systemic lupus erythematosus. Mol Med. (2019) 25:35. doi: 10.1186/s10020-019-0102-5
94. Chen B, Sun L, Zhang X. Integration of microbiome and epigenome to decipher the pathogenesis of autoimmune diseases. J Autoimmun. (2017) 83:31–42. doi: 10.1016/j.jaut.2017.03.009
95. Li R, Meng X, Chen B, Zhao LA-O, Zhang XA-O. Gut microbiota in lupus: A butterfly effect? Curr Rheumatol Rep. (2021) 1534-6307:27. doi: 10.1007/s11926-021-00986-z
96. Manfredo Vieira S, Hiltensperger M, Kumar V, Zegarra-Ruiz D, Dehner C, Khan N, et al. Translocation of a gut pathobiont drives autoimmunity in mice and humans. Science. (2018) 359:1156–61. doi: 10.1126/science.aar7201
97. Jukic A, Bakiri L, Wagner EF, Tilg H, Adolph TE. Calprotectin: from biomarker to biological function. Gut. (2021) 70:1978–88. doi: 10.1136/gutjnl-2021-324855
98. Azzouz D, Omarbekova A, Heguy A, Schwudke D, Gisch N, Rovin BH, et al. Lupus nephritis is linked to disease-activity associated expansions and immunity to a gut commensal. Ann Rheum Dis. (2019) 78:947–56. doi: 10.1136/annrheumdis-2018-214856
99. Kim JW, Kwok SK, Choe JY, Park SH. Recent advances in our understanding of the link between the intestinal microbiota and systemic lupus erythematosus. Int J Mol Sci. (2019) 20:4871. doi: 10.3390/ijms20194871
100. Mu Q, Zhang H, Liao X, Lin K, Liu H, Edwards MR, et al. Control of lupus nephritis by changes of gut microbiota. Microbiome. (2017) 5:73. doi: 10.1186/s40168-017-0300-8
101. Li D, Pan Y, Xia X, Liang J, Liu F, Dou H, et al. Bacteroides fragilis alleviates the symptoms of lupus nephritis via regulating cd1d and cd86 expressions in B cells. Eur J Pharmacol. (2020) 884:173421. doi: 10.1016/j.ejphar.2020.173421
102. Marietta EV, Murray JA, Luckey DH, Jeraldo PR, Lamba A, Patel R, et al. Suppression of inflammatory arthritis by human gut-derived prevotella histicola in humanized mice. Arthritis Rheumatol. (2016) 68:2878–88. doi: 10.1002/art.39785
103. Fan Z, Yang B, Ross RP, Stanton C, Zhao J, Zhang H, et al. The prophylactic effects of different lactobacilli on collagen-induced arthritis in rats. Food Funct. (2020) 11:3681–94. doi: 10.1039/C9FO02556A
104. Fan Z, Yang B, Ross RP, Stanton C, Shi G, Zhao J, et al. Protective effects of bifidobacterium adolescentis on collagen-induced arthritis in rats depend on timing of administration. Food Funct. (2020) 11:4499–511. doi: 10.1039/D0FO00077A
105. Rossi O, van Berkel LA, Chain F, Tanweer Khan M, Taverne N, Sokol H, et al. Faecalibacterium prausnitzii A2-165 has a high capacity to induce il-10 in human and murine dendritic cells and modulates T cell responses. Sci Rep. (2016) 6:18507. doi: 10.1038/srep18507
106. Kawashima M, Nakamura S, Izuta Y, Inoue S, Tsubota K. Dietary supplementation with a combination of lactoferrin, fish oil, and enterococcus faecium wb2000 for treating dry eye: A rat model and human clinical study. Ocul Surf. (2016) 14:255–63. doi: 10.1016/j.jtos.2015.12.005
107. Chisari G, Chisari EM, Borzi AM, Chisari CG. Aging eye microbiota in dry eye syndrome in patients treated with enterococcus faecium and saccharomyces boulardii. Curr Clin Pharmacol. (2017) 12:99–105. doi: 10.2174/1574884712666170704145046
108. Chisari G, Chisari EM, Francaviglia A, Chisari CG. The mixture of bifidobacterium associated with fructo-oligosaccharides reduces the damage of the ocular surface. Clin Ter. (2017) 168:e181–e5. doi: 10.7417/T.2017.2002
109. Kim J, Choi SH, Kim YJ, Jeong HJ, Ryu JS, Lee HJ, et al. Clinical effect of irt-5 probiotics on immune modulation of autoimmunity or alloimmunity in the eye. Nutrients. (2017) 9:1166. doi: 10.3390/nu9111166
110. Kim SH, Huh CS, Choi ID, Jeong JW, Ku HK, Ra JH, et al. The anti-diabetic activity of bifidobacterium lactis hy8101 in vitro and in vivo. J Appl Microbiol. (2014) 117:834–45. doi: 10.1111/jam.12573
111. Valladares R, Sankar D, Li N, Williams E, Lai KK, Abdelgeliel AS, et al. Lactobacillus johnsonii N6.2 mitigates the development of type 1 diabetes in bb-dp rats. PloS One. (2010) 5:e10507. doi: 10.1371/journal.pone.0010507
112. Teixeira LD, Kling DN, Lorca GL, Gonzalez CF. Lactobacillus johnsonii N6.2 diminishes caspase-1 maturation in the gastrointestinal system of diabetes prone rats. Benef Microbes. (2018) 9:527–39. doi: 10.3920/BM2017.0120
113. Yadav H, Lee JH, Lloyd J, Walter P, Rane SG. Beneficial metabolic effects of a probiotic via butyrate-induced glp-1 hormone secretion. J Biol Chem. (2013) 288:25088–97. doi: 10.1074/jbc.M113.452516
114. Wang CH, Yen HR, Lu WL, Ho HH, Lin WY, Kuo YW, et al. Adjuvant probiotics of lactobacillus salivarius subsp. Salicinius ap-32, L. Johnsonii mh-68, and bifidobacterium animalis subsp. Lactis cp-9 attenuate glycemic levels and inflammatory cytokines in patients with type 1 diabetes mellitus. Front Endocrinol (Lausanne). (2022) 13:754401. doi: 10.3389/fendo.2022.754401
115. Albuquerque R, Brandao ABP, De Abreu I, Ferreira FG, Santos LB, Moreira LN, et al. Saccharomyces boulardii tht 500101 changes gut microbiota and ameliorates hyperglycaemia, dyslipidaemia, and liver inflammation in streptozotocin-diabetic mice. Benef Microbes. (2019) 10:901–12. doi: 10.3920/BM2019.0056
116. Yu F, Hu X, Ren H, Wang X, Shi R, Guo J, et al. Protective effect of synbiotic combination of lactobacillus plantarum sc-5 and olive oil extract tyrosol in a murine model of ulcerative colitis. J Transl Med. (2024) 22:308. doi: 10.1186/s12967-024-05026-9
117. Li H, Li HA-O, Stanton C, Ross RP, Zhao J, Chen WA-O, et al. Exopolysaccharides produced by bifidobacterium longum subsp. Longum ys108r ameliorates dss-induced ulcerative colitis in mice by improving the gut barrier and regulating the gut microbiota. J Agric Food Chem. (2024) 1520-5118:7055–7073. doi: 10.1021/Acs.Jafc.3c06421
118. Li H, Li H, Stanton C, Ross RP, Zhao J, Chen W, et al. Alleviative effects of exopolysaccharides from limosilactobacillus mucosae ccfm1273 against ulcerative colitis via modulation of gut microbiota and inhibition of fas/fasl and tlr4/nf-Κb pathways. Int J Biol Macromol. (2024) 260:129346. doi: 10.1016/j.ijbiomac.2024.129346
119. Bu L, Li Y, Wang C, Jiang Y, Suo H. Preventive effect of lacticaseibacillus rhamnosus 2016swu.05.0601 and its postbiotic elements on dextran sodium sulfate-induced colitis in mice. Front Microbiol. (2024) 15:1342705. doi: 10.3389/fmicb.2024.1342705
120. Groeger D, O'Mahony L, Murphy EF, Bourke JF, Dinan TG, Kiely B, et al. Bifidobacterium infantis 35624 modulates host inflammatory processes beyond the gut. Gut Microbes. (2013) 4:325–39. doi: 10.4161/gmic.25487
121. Moludi J, Khedmatgozar H, Saiedi S, Razmi H, Alizadeh M, Ebrahimi B. Probiotic supplementation improves clinical outcomes and quality of life indicators in patients with plaque psoriasis: A randomized double-blind clinical trial. Clin Nutr ESPEN. (2021) 46:33–9. doi: 10.1016/j.clnesp.2021.09.004
122. Buhas MC, Candrea R, Gavrilas LI, Miere D, Tataru A, Boca A, et al. Transforming psoriasis care: probiotics and prebiotics as novel therapeutic approaches. Int J Mol Sci. (2023) 24:11225. doi: 10.3390/ijms241311225
123. Cho S, Dong J, Lu LF. Cell-intrinsic and -extrinsic roles of mirnas in regulating T cell immunity. Immunol Rev. (2021) 304:126–40. doi: 10.1111/imr.13029
124. Abdul-Maksoud RS, Rashad NM, Elsayed WSH, Ali MA, Kamal NM, Zidan HE. Circulating mir-181a and mir-223 expression with the potential value of biomarkers for the diagnosis of systemic lupus erythematosus and predicting lupus nephritis. J Gene Med. (2021) 23:e3326. doi: 10.1002/jgm.3326
125. Li X, Tian F, Wang F. Rheumatoid arthritis-associated microrna-155 targets socs1 and upregulates tnf-Α and il-1β in pbmcs. Int J Mol Sci. (2013) 14:23910–21. doi: 10.3390/ijms141223910
126. Vahidi Z, Samadi M, Mahmoudi M, RezaieYazdi Z, Sahebari M, Tabasi N, et al. Lactobacillus rhamnosus and lactobacillus delbrueckii ameliorate the expression of mir-155 and mir-181a in sle patients. J Funct Foods. (2018) 48:228–33. doi: 10.1016/j.jff.2018.07.025
127. Zegarra-Ruiz DF, El Beidaq A, Iniguez AJ, Lubrano Di Ricco M, Manfredo Vieira S, Ruff WE, et al. A diet-sensitive commensal lactobacillus strain mediates tlr7-dependent systemic autoimmunity. Cell Host Microbe. (2019) 25:113–27 e6. doi: 10.1016/j.chom.2018.11.009
128. Johnson BM, Gaudreau MC, Gudi R, Brown R, Gilkeson G, Vasu C. Gut microbiota differently contributes to intestinal immune phenotype and systemic autoimmune progression in female and male lupus-prone mice. J Autoimmun. (2020) 108:102420. doi: 10.1016/j.jaut.2020.102420
129. Hevia A, Milani C, Lopez P, Cuervo A, Arboleya S, Duranti S, et al. Intestinal dysbiosis associated with systemic lupus erythematosus. mBio. (2014) 5:e01548–14. doi: 10.1128/mBio.01548-14
130. Li Y, Wang HF, Li X, Li HX, Zhang Q, Zhou HW, et al. Disordered intestinal microbes are associated with the activity of systemic lupus erythematosus. Clin Sci (Lond). (2019) 133:821–38. doi: 10.1042/CS20180841
131. Guo M, Wang H, Xu S, Zhuang Y, An J, Su C, et al. Alteration in gut microbiota is associated with dysregulation of cytokines and glucocorticoid therapy in systemic lupus erythematosus. Gut Microbes. (2020) 11:1758–73. doi: 10.1080/19490976.2020.1768644
132. He J, Chan T, Hong X, Zheng F, Zhu C, Yin L, et al. Microbiome and metabolome analyses reveal the disruption of lipid metabolism in systemic lupus erythematosus. Front Immunol. (2020) 11:1703. doi: 10.3389/fimmu.2020.01703
133. Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. (2013) 504:451–5. doi: 10.1038/nature12726
134. Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic treg cell homeostasis. Science. (2013) 341:569–73. doi: 10.1126/science.1241165
135. Lee JY, Mannaa M. Comparative analysis of fecal microbiota composition between rheumatoid arthritis and osteoarthritis patients. Genes (Basel). (2019) 10:748. doi: 10.3390/genes10100748
136. Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C, et al. Expansion of intestinal prevotella copri correlates with enhanced susceptibility to arthritis. Elife. (2013) 2:e01202. doi: 10.7554/eLife.01202
137. Jiang L, Shang M, Yu S, Liu Y, Zhang H, Zhou Y, et al. A high-fiber diet synergizes with prevotella copri and exacerbates rheumatoid arthritis. Cell Mol Immunol. (2022) 19:1414–24. doi: 10.1038/s41423-022-00934-6
138. Horta-Baas GA-O, Romero-Figueroa MA-OX, Montiel-Jarquín AJ, Pizano-Zárate ML, García-Mena JA-O, Ramírez-Durán NA-OX. Intestinal dysbiosis and rheumatoid arthritis: A link between gut microbiota and the pathogenesis of rheumatoid arthritis. J Immunol Res. (2017) 2017:4835189. doi: 10.1155/2017/4835189
139. Guerreiro CS, Calado A, Sousa J, Fonseca JE. Diet, microbiota, and gut permeability-the unknown triad in rheumatoid arthritis. Front Med (Lausanne). (2018) 5:349. doi: 10.3389/fmed.2018.00349
140. Alghamdi M, Alasmari D, Assiri A, Mattar E, Aljaddawi AA, Alattas SG, et al. An overview of the intrinsic role of citrullination in autoimmune disorders. J Immunol Res. (2019) 2019:7592851. doi: 10.1155/2019/7592851
141. Sugawara E, Kato M, Kudo Y, Lee W, Hisada R, Fujieda Y, et al. Autophagy promotes citrullination of vim (Vimentin) and its interaction with major histocompatibility complex class ii in synovial fibroblasts. Autophagy. (2020) 16:946–55. doi: 10.1080/15548627.2019.1664144
142. Wu CY, Yang HY, Lai JH. Anti-citrullinated protein antibodies in patients with rheumatoid arthritis: biological effects and mechanisms of immunopathogenesis. Int J Mol Sci. (2020) 21:4015. doi: 10.3390/ijms21114015
143. Holers VM, Demoruelle MK, Kuhn KA, Buckner JH, Robinson WH, Okamoto Y, et al. Rheumatoid arthritis and the mucosal origins hypothesis: protection turns to destruction. Nat Rev Rheumatol. (2018) 14:542–57. doi: 10.1038/s41584-018-0070-0
144. Bennike TB, Ellingsen T, Glerup H, Bonderup OK, Carlsen TG, Meyer MK, et al. Proteome analysis of rheumatoid arthritis gut mucosa. J Proteome Res. (2017) 16:346–54. doi: 10.1021/acs.jproteome.6b00598
145. Alghamdi M, Al Ghamdi KA, Khan RH, Uversky VN, Redwan EM. An interplay of structure and intrinsic disorder in the functionality of peptidylarginine deiminases. Family Key Autoimmunity-Related Enzymes. (2019) 76:4635–62. doi: 10.1007/s00018-019-03237-8
146. El Shikh MEM, El Sayed R, Nerviani A, Goldmann K, John CR, Hands R, et al. Extracellular traps and pad4 released by macrophages induce citrullination and auto-antibody production in autoimmune arthritis. J Autoimmun. (2019) 105(1095-9157:102297. doi: 10.1016/j.jaut.2019.06.008
147. Lerner A, Matthias T. Rheumatoid arthritis-celiac disease relationship: joints get that gut feeling. Autoimmun Rev. (2015) 14:1038–47. doi: 10.1016/j.autrev.2015.07.007
148. Makki K, Deehan EC, Walter J, Backhed F. The impact of dietary fiber on gut microbiota in host health and disease. Cell Host Microbe. (2018) 23:705–15. doi: 10.1016/j.chom.2018.05.012
149. Thalayasingam N, Baldwin K, Judd C, Ng WF. New developments in sjogren's syndrome. Rheumatol (Oxford England). (2021) 60:vi53–61. doi: 10.1093/rheumatology/keab466
150. Fan Z, Ross RP, Stanton C, Hou B, Zhao J, Zhang H, et al. Lactobacillus casei ccfm1074 alleviates collagen-induced arthritis in rats via balancing treg/th17 and modulating the metabolites and gut microbiota. Front Immunol. (2021) 12:680073. doi: 10.3389/fimmu.2021.680073
151. Wells PM, Williams FMK, Matey-Hernandez ML, Menni C, Steves CJ. ‘Ra and the microbiome: do host genetic factors provide the link? J Autoimmun. (2019) 99:104–15. doi: 10.1016/j.jaut.2019.02.004
152. Hamamoto Y, Ouhara K. Effect of porphyromonas gingivalis infection on gut dysbiosis and resultant arthritis exacerbation in mouse model. Arthritis Res Ther. (2020) 22(1):249. doi: 10.1186/s13075-020-02348-z
153. Floudas A, Canavan M, McGarry T, Mullan R, Nagpal S, Veale DJ, et al. Acpa status correlates with differential immune profile in patients with rheumatoid arthritis. Cells. (2021) 10:647. doi: 10.3390/cells10030647
154. Both T, Dalm VA, van Hagen PM, van Daele PL. Reviewing primary sjogren’s syndrome: beyond the dryness - from pathophysiology to diagnosis and treatment. Int J Med Sci. (2017) 14:191–200. doi: 10.7150/ijms.17718
155. Psianou K, Panagoulias I, Papanastasiou AD, de Lastic AL, Rodi M, Spantidea PI, et al. Clinical and immunological parameters of sjögren's syndrome. Autoimmun Rev. (2018) 17:1053–64. doi: 10.1016/j.autrev.2018.05.005
156. Wang C, Zaheer M, Bian F, Quach D, Swennes AG, Britton RA, et al. Sjogren-like lacrimal keratoconjunctivitis in germ-free mice. Int J Mol Sci. (2018) 19:565. doi: 10.3390/ijms19020565
157. Coursey TG, Bian F, Zaheer M, Pflugfelder SC, Volpe EA, de Paiva CS. Age-related spontaneous lacrimal keratoconjunctivitis is accompanied by dysfunctional T regulatory cells. Mucosal Immunol. (2017) 10:743–56. doi: 10.1038/mi.2016.83
158. Schaefer L, Trujillo-Vargas CM, Midani FS, Pflugfelder SC, Britton RA, de Paiva CS. Gut microbiota from sjogren syndrome patients causes decreased T regulatory cells in the lymphoid organs and desiccation-induced corneal barrier disruption in mice. Front Med (Lausanne). (2022) 9:852918. doi: 10.3389/fmed.2022.852918
159. Simon Q, Grasseau A, Boudigou M, Le Pottier L, Bettachioli E, Cornec D. A proinflammatory cytokine network profile in th1/type 1 effector B cells delineates a common group of patients in four systemic autoimmune diseases. Arthritis Rheumatol. (2021) 73(8):1550–61. doi: 10.1002/art.41697
160. Itotagawa E, Tomofuji Y, Kato Y, Konaka H, Tsujimoto K, Park J, et al. Sle stratification based on baff and ifn-I bioactivity for biologics and implications of baff produced by glomeruli in lupus nephritis. Rheumatol (Oxford England). (2022):1988–1997. doi: 10.1093/rheumatology/keac528
161. Nocturne G, Mariette X. B cells in the pathogenesis of primary sjögren syndrome. Nat Rev Rheumatol. (2018) 14:133–45. doi: 10.1038/nrrheum.2018.1
162. Mandl T, Marsal J, Olsson P, Ohlsson B, Andreasson K. Severe intestinal dysbiosis is prevalent in primary sjogren's syndrome and is associated with systemic disease activity. Arthritis Res Ther. (2017) 19:237. doi: 10.1186/s13075-017-1446-2
163. Moon J, Choi SH, Yoon CH, Kim MK. Gut dysbiosis is prevailing in sjogren’s syndrome and is related to dry eye severity. PloS One. (2020) 15:e0229029. doi: 10.1371/journal.pone.0229029
164. van der Meulen TA, Harmsen HJM, Vila AV, Kurilshikov A, Liefers SC, Zhernakova A, et al. Shared gut, but distinct oral microbiota composition in primary sjogren’s syndrome and systemic lupus erythematosus. J Autoimmun. (2019) 97:77–87. doi: 10.1016/j.jaut.2018.10.009
165. Chen X, Su W, Wan T, Yu J, Zhu W, Tang F, et al. Sodium butyrate regulates th17/treg cell balance to ameliorate uveitis via the nrf2/ho-1 pathway. Biochem Pharmacol. (2017) 142:111–9. doi: 10.1016/j.bcp.2017.06.136
166. Takahashi D, Hoshina N, Kabumoto Y, Maeda Y, Suzuki A, Tanabe H, et al. Microbiota-derived butyrate limits the autoimmune response by promoting the differentiation of follicular regulatory T cells. EBioMedicine. (2020) 58:102913. doi: 10.1016/j.ebiom.2020.102913
167. Jiang T, Lu W, Fang Z, Wang H, Zhu J, Zhang H, et al. Bifidobacterium treated by electrostatic spray drying relieved constipation by changing the relative abundance of bacteria associated with gastrointestinal regulatory peptides. Front Cell infection Microbiol. (2022) 12:894216. doi: 10.3389/fcimb.2022.894216
168. Omenetti S, Pizarro TT. The treg/th17 axis: A dynamic balance regulated by the gut microbiome. Front Immunol. (2015) 6:639. doi: 10.3389/fimmu.2015.00639
169. Liu C, Yuan YC, Guo MN, Xin Z, Chen GJ, Bentley AR, et al. Incidence of type 1 diabetes may be underestimated in the chinese population: evidence from 21.7 million people between 2007 and 2017. Diabetes Care. (2021) 44:2503–9. doi: 10.2337/dc21-0342
170. Maahs DM, West NA, Lawrence JM, Mayer-Davis EJ. Epidemiology of type 1 diabetes. Endocrinol Metab Clin North Am. (2010) 39:481–97. doi: 10.1016/j.ecl.2010.05.011
171. Knip M, Siljander H. The role of the intestinal microbiota in type 1 diabetes mellitus. Nat Rev Endocrinol. (2016) 12:154–67. doi: 10.1038/nrendo.2015.218
172. Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, Brummer RJ. Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther. (2008) 27:104–19. doi: 10.1111/j.1365-2036.2007.03562.x
173. Bosi E, Molteni L, Radaelli MG, Folini L, Fermo I, Bazzigaluppi E, et al. Increased intestinal permeability precedes clinical onset of type 1 diabetes. Diabetologia. (2006) 49:2824–7. doi: 10.1007/s00125-006-0465-3
174. Monsted MO, Falck ND, Pedersen K, Buschard K, Holm LJ, Haupt-Jorgensen M. Intestinal permeability in type 1 diabetes: an updated comprehensive overview. J Autoimmun. (2021) 122:102674. doi: 10.1016/j.jaut.2021.102674
175. Xiao L, Van't Land B, Engen PA, Naqib A, Green SJ, Nato A, et al. Human milk oligosaccharides protect against the development of autoimmune diabetes in nod-mice. Sci Rep. (2018) 8:3829. doi: 10.1038/s41598-018-22052-y
176. Brown CT, Davis-Richardson AG, Giongo A, Gano KA, Crabb DB, Mukherjee N, et al. Gut microbiome metagenomics analysis suggests a functional model for the development of autoimmunity for type 1 diabetes. PloS One. (2011) 6:e25792. doi: 10.1371/journal.pone.0025792
177. Kovatcheva-Datchary P, Nilsson A, Akrami R, Lee Ying S, De Vadder F, Arora T, et al. Dietary fiber-induced improvement in glucose metabolism is associated with increased abundance of prevotella. Cell Metab. (2015) 22:971–82. doi: 10.1016/j.cmet.2015.10.001
178. Zheng SJ, Luo Y, Wang JB, Chen XM, Xu Y, Xiao JH. Regulated intestinal microbiota and gut immunity to ameliorate type 1 diabetes mellitus: A novel mechanism for stem cell-based therapy. BioMed Pharmacother. (2024) 170(1950-6007:116033. doi: 10.1016/j.biopha.2023.116033
179. Wen L, Wong FS. Dietary short-chain fatty acids protect against type 1 diabetes. Nat Immunol. (2017) 18:484–6. doi: 10.1038/ni.3730
180. Jia L, Cao M, Chen H, Zhang M, Dong X, Ren Z, et al. Butyrate ameliorates antibiotic-driven type 1 diabetes in the female offspring of nonobese diabetic mice. J Agric Food Chem. (2020) 68:3112–20. doi: 10.1021/acs.jafc.9b07701
181. Fernandez-Veledo S, Vendrell J. Gut microbiota-derived succinate: friend or foe in human metabolic diseases? Rev Endocr Metab Disord. (2019) 20:439–47. doi: 10.1007/s11154-019-09513-z
182. De Vadder F, Kovatcheva-Datchary P, Zitoun C, Duchampt A, Backhed F, Mithieux G. Microbiota-produced succinate improves glucose homeostasis via intestinal gluconeogenesis. Cell Metab. (2016) 24:151–7. doi: 10.1016/j.cmet.2016.06.013
183. Kobayashi T, Siegmund B, Le Berre C, Wei SC, Ferrante M, Shen B, et al. Ulcerative colitis. Nat Rev Dis Primers. (2020) 6:74. doi: 10.1038/s41572-020-0205-x
184. Schirmer M, Denson L, Vlamakis H, Franzosa EA, Thomas S, Gotman NM, et al. Compositional and temporal changes in the gut microbiome of pediatric ulcerative colitis patients are linked to disease course. Cell Host Microbe. (2018) 24:600–10 e4. doi: 10.1016/j.chom.2018.09.009
185. Mukhopadhya I, Hansen R, El-Omar EM, Hold GL. Ibd-what role do proteobacteria play? Nat Rev Gastroenterol Hepatol. (2012) 9:219–30. doi: 10.1038/nrgastro.2012.14
186. Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of enterobacteriaceae. Cell Host Microbe. (2007) 2:119–29. doi: 10.1016/j.chom.2007.06.010
187. Zhang J, Hoedt EC, Liu Q, Berendsen E, Teh JJ, Hamilton A, et al. Elucidation of proteus mirabilis as a key bacterium in crohn’s disease inflammation. Gastroenterology. (2021) 160:317–30 e11. doi: 10.1053/j.gastro.2020.09.036
188. Strauss J, Kaplan GG, Beck PL, Rioux K, Panaccione R, Devinney R, et al. Invasive potential of gut mucosa-derived fusobacterium nucleatum positively correlates with ibd status of the host. Inflammation Bowel Dis. (2011) 17:1971–8. doi: 10.1002/ibd.21606
189. Koh A, De Vadder F, Kovatcheva-Datchary P, Backhed F. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell. (2016) 165:1332–45. doi: 10.1016/j.cell.2016.05.041
190. Takeuchi T, Miyauchi E, Kanaya T, Kato T, Nakanishi Y, Watanabe T, et al. Acetate differentially regulates iga reactivity to commensal bacteria. Nature. (2021) 595:560–4. doi: 10.1038/s41586-021-03727-5
191. Marchesi JR, Holmes E, Khan F, Kochhar S, Scanlan P, Shanahan F, et al. Rapid and noninvasive metabonomic characterization of inflammatory bowel disease. J Proteome Res. (2007) 6:546–51. doi: 10.1021/pr060470d
192. Zeevi D, Korem T, Godneva A, Bar N, Kurilshikov A, Lotan-Pompan M, et al. Structural variation in the gut microbiome associates with host health. Nature. (2019) 568:43–8. doi: 10.1038/s41586-019-1065-y
193. Nakajima K. Critical role of the interleukin-23/T-helper 17 cell axis in the pathogenesis of psoriasis. J Dermatol. (2012) 39:219–24. doi: 10.1111/j.1346-8138.2011.01458.x
194. Alesa DI, Alshamrani HM, Alzahrani YA, Alamssi DN, Alzahrani NS, Almohammadi ME. The role of gut microbiome in the pathogenesis of psoriasis and the therapeutic effects of probiotics. J Family Med Prim Care. (2019) 8:3496–503. doi: 10.4103/jfmpc.jfmpc_709_19
196. Zakostelska Z, Malkova J, Klimesova K, Rossmann P, Hornova M, Novosadova I, et al. Intestinal microbiota promotes psoriasis-like skin inflammation by enhancing th17 response. PloS One. (2016) 11:e0159539. doi: 10.1371/journal.pone.0159539
197. Xiao YA-O, Wang Y, Tong B, Gu Y, Zhou X, Zhu N, et al. Eubacterium rectale is a potential marker of altered gut microbiota in psoriasis and psoriatic arthritis. Microbiol Spectr. (2024) 2165-0497:e0115423. doi: 10.1128/spectrum.01154-23
198. Scher JU, Ubeda C, Artacho A, Attur M, Isaac S, Reddy SM, et al. Decreased bacterial diversity characterizes the altered gut microbiota in patients with psoriatic arthritis, resembling dysbiosis in inflammatory bowel disease. Arthritis Rheumatol. (2014) 67:128–39. doi: 10.1002/art.38892
199. Hidalgo-Cantabrana CA-O, Gómez J, Delgado S, Requena-López S, Queiro-Silva R, Margolles A, et al. Gut microbiota dysbiosis in a cohort of patients with psoriasis. Br J Dermatol. (2019) 181:1287–95. doi: 10.1111/bjd.17931
200. Willing BP, Dicksved J, Halfvarson J, Andersson AF, Lucio M, Zheng Z, et al. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology. (2010) 139:1844–54 e1. doi: 10.1053/j.gastro.2010.08.049
201. Nermes M, Kantele JM, Atosuo TJ, Salminen S, Isolauri E. Interaction of orally administered lactobacillus rhamnosus gg with skin and gut microbiota and humoral immunity in infants with atopic dermatitis. Clin Exp Allergy. (2011) 41:370–7. doi: 10.1111/cea.2011.41.issue-3
Keywords: autoimmune disease, homeostasis, gut microbiota, probiotics, microbiome
Citation: Wang X, Yuan W, Yang C, Wang Z, Zhang J, Xu D, Sun X and Sun W (2024) Emerging role of gut microbiota in autoimmune diseases. Front. Immunol. 15:1365554. doi: 10.3389/fimmu.2024.1365554
Received: 04 January 2024; Accepted: 22 April 2024;
Published: 03 May 2024.
Edited by:
Monica Neagu, Victor Babes National Institute of Pathology (INCDVB), RomaniaReviewed by:
Mihaela Surcel, Victor Babes National Institute of Pathology (INCDVB), RomaniaCopyright © 2024 Wang, Yuan, Yang, Wang, Zhang, Xu, Sun and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenchang Sun, c3Vud2NoQHdmbWMuZWR1LmNu; Xicai Sun, cm15eXN4Yzk5MjJAd2ZtYy5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.