
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol., 19 April 2024
Sec. Microbial Immunology
Volume 15 - 2024 | https://doi.org/10.3389/fimmu.2024.1360132
Introduction: Considerable evidence has unveiled a potential correlation between gut microbiota and spinal degenerative diseases. However, only limited studies have reported the direct association between gut microbiota and spinal stenosis. Hence, in this study, we aimed to clarify this relationship using a two-sample mendelian randomization (MR) approach.
Materials and Methods: Data for two-sample MR studies was collected and summarized from genome-wide association studies (GWAS) of gut microbiota (MiBioGen, n = 13, 266) and spinal stenosis (FinnGen Biobank, 9, 169 cases and 164, 682 controls). The inverse variance-weighted meta-analysis (IVW), complemented with weighted median, MR-Egger, weighted mode, and simple mode, was used to elucidate the causality between gut microbiota and spinal stenosis. In addition, we employed mendelian randomization pleiotropy residual sum and outlier (MR-PRESSO) and the MR-Egger intercept test to assess horizontal multiplicity. Cochran’s Q test to evaluate heterogeneity, and “leave-one-out” sensitivity analysis to determine the reliability of causality. Finally, an inverse MR analysis was performed to assess the reverse causality.
Results: The IVW results indicated that two gut microbial taxa, the genus Eubacterium fissicatena group and the genus Oxalobacter, have a potential causal relationship with spinal stenosis. Moreover, eight potential associations between genetic liability of the gut microbiota and spinal stenosis were implied. No significant heterogeneity of instrumental variables or horizontal pleiotropy were detected. In addition, “leave-one-out” sensitivity analysis confirmed the reliability of causality. Finally, the reverse MR analysis revealed that no proof to substantiate the discernible causative relationship between spinal stenosis and gut microbiota.
Conclusion: This analysis demonstrated a possible causal relationship between certain particular gut microbiota and the occurrence of spinal stenosis. Further studies focused on the mechanism of gut microbiota-mediated spinal stenosis can lay the groundwork for targeted prevention, monitoring, and treatment of spinal stenosis.
Spinal stenosis, a multifactorial disease, is characterized by the narrowing of the spinal canal, which may occur from exogenous factors like trauma, infections, and tumors, as well as endogenous factors like natural degeneration (1). These factors can change the anatomical structure surrounding the spinal canal (posterior longitudinal ligament, ligamentum flavum, and facet joints) and degenerate the intervertebral discs, leading to symptoms such as ow back pain, leg pain, and intermittent claudication (2). As a result of the aging population, an estimated 100 million individuals worldwide are reported to suffer from spinal stenosis, seriously affecting the quality of life for patients and their families and contributing significantly to the global medical and financial burden (3, 4). Though the function of the existed treatment strategies, to develop high-efficiency and fast prevention, treatment and monitoring strategies is very instant.
As the evolvement of the human health research, studies on the gut microbiota have steadily moved to the center of attention (5). Gut microbiota, including prokaryotes (bacteria and archaea), eukaryotes (fungi, intestinal protozoa, and parasitic helminths), viruses, and phages, forms the largest, most important and diverse micro-ecosystem among the digestive ecosystems (6). Due to the diversity and specificity of its structure, the gut microbiota is essential for maintaining a dynamic balance between the intestinal environment and human systems (7, 8). In addition, gut microbiota and its metabolites are involved in the disease process of various organs inside and outside the intestine (e.g., brain, liver, and colorectum). Especially in some metabolic diseases, changes in the abundance of gut microbiota have become a potential risk factor for inducing inflammatory cytokines (9–11). In recent years, there has been growing evidence with regard to the relationship between gut microbiota and spinal degenerative diseases. Specifically, dysbiosis may affect the health of spinal structures through three mechanisms: (1) nutrition, including calcium, amino acids, and vitamin K; (2) immune regulation, such as estrogen, short-chain fatty acids, and systemic inflammation; and (3) neurotransmitters, including serotonin and leptin, which affect bone metabolism (12). Furthermore, the publication by A et al. serves as an overview of earlier research findings and creatively brings up the concept of the intestinal-spinal axis, thus further illustrating the potential relationship between gut microbiota and spinal degenerative diseases (13).Based on the known interactions between the gut microbiota and diseases, several studies have implicated that the occurrence and development of such diseases can be prevented by precisely and simply regulating the biological abundance of gut microbiota (14), providing a new scheme for spinal stenosis prevention, treatment and monitoring.
Thus, comprehending the connection between the gut microbiota and spinal stenosis is essential. Nevertheless, recent reports have only demonstrated some indirect relationships. It is commonly recognized that spinal stenosis is associated with reduced space available for the neural and vascular elements of the lumbar spine (15). As previously mentioned, dysbiosis of the intestinal flora may affect spinal structures (bone, cartilage, intervertebral discs, ligaments, and muscles) through a range of possible mechanisms, such as immunological and nutritional, leading to the development of vertebral osteophytes, lesser articular eminence hyperplasia and hypertrophy, disc herniation, hypertrophy of the ligamentum flavum, and more. And importantly, when the alterations happen in paravertebral structures, causing the volume of the spinal canal to narrow, it can lead to spinal stenosis (16). Also, the correlation between gut microbiota and disorders associated with chronic inflammation and the validation observed in ligamentum flavum thickening raise the prospect of a relationship between gut microbiota and spinal stenosis (17). However, no direct cause-and-effect association between gut microbiota and spinal stenosis has been reported. Comprehending and clarifying the causal relationship between the two mentioned above can offer novel approaches towards the targeted prevention and management of spinal stenosis.
Employing genetic variants to generate instrumental variables (IVs) for exposure and incorporating collected data from genome-wide association studies (GWAS), mendelian randomization (MR) studies offer a whole new approach to evaluate causal relationships between exposures and outcomes (18). At present, MR studies have been widely used to explore the causal relationship between gut microbiota and diseases, including metabolic diseases, autoimmune diseases, and various types of bone diseases (19). Considering the fact that genotypes are randomly assigned from parents to children, it is unlikely that environmental influences will affect the relationship between genetic variation and outcome (20). Therefore, MR research offers an economical and effective approach to investigating the causal relationship between exposure and the outcome. In this paper, a two-sample MR analysis was performed to assess the causal relationship between gut microbiota and spinal stenosis using the GWAS summary statistics from the MiBioGen and FinnGen consortia.
Based on two-sample MR analyses, we obtained pooled data from published GWAS to evaluate the causal relationship between gut microbiota and spinal stenosis. This study first determined whether gut microbiota contributes to the prevention or promotion of spinal stenosis by selecting gut microbiota as the exposure and spinal stenosis as the outcome. In addition, three crucial presumptions must be met to allow for MR research to proceed: 1. Genetic variation should be significantly linked to exposure; 2. Genetic variation should be independent of confounders associated with selected exposures and outcomes; and 3. Genetic variation can affect outcomes only through exposure and not through other biological pathways (i.e., no horizontal multiplicity of effects) (21). Specific details are shown in Figure 1.
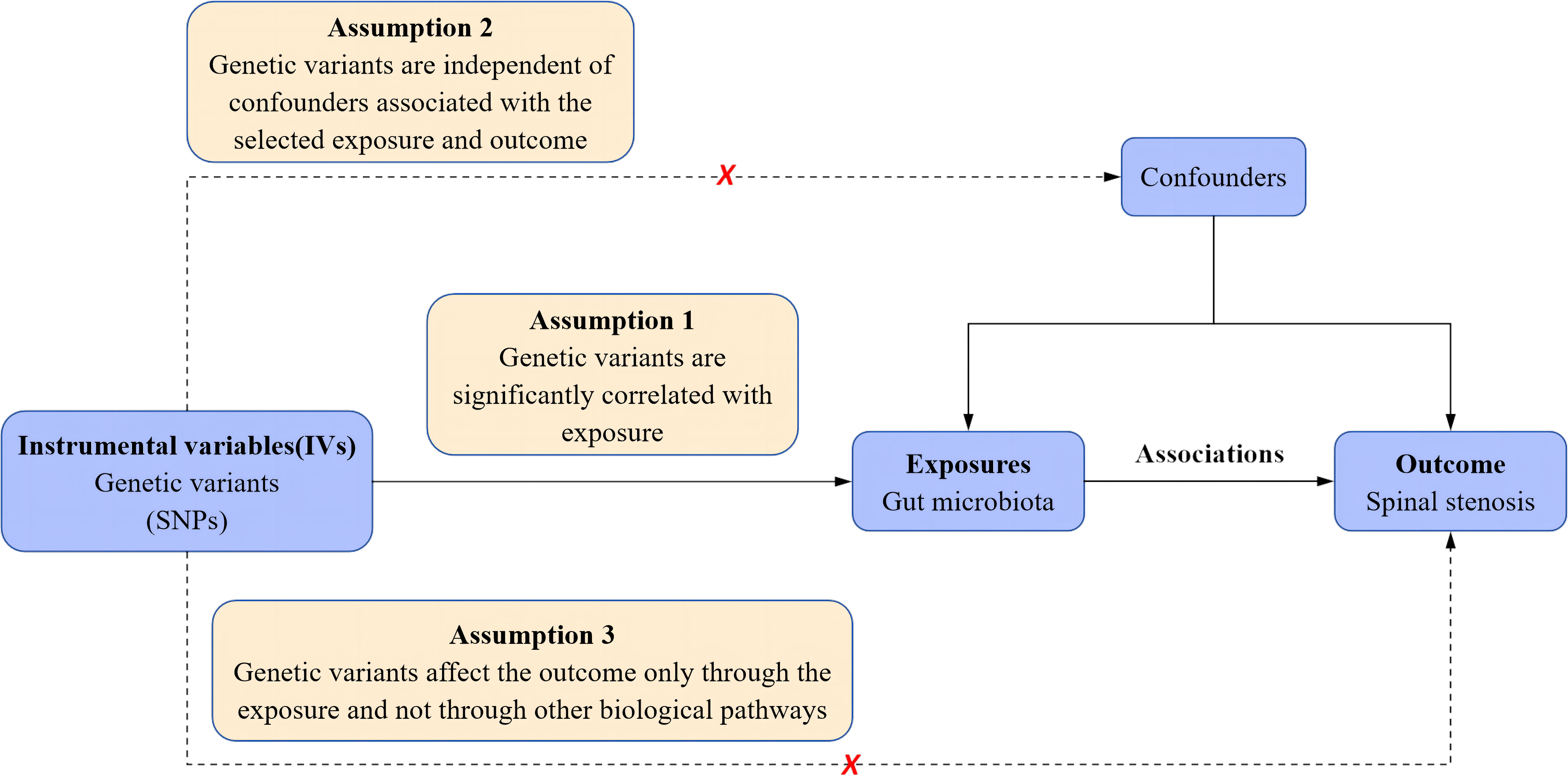
Figure 1 Schematic representation of the a two-sample mendelian randomization analysis. SNPs, single nucleotide polymorphisms.
The MiBioGen consortium, conducting the largest genome-wide meta-analysis on gut microbiota composition, provided genetic variations of gut microbiota in this study. The meta-analysis examined the relationship between human autosomal genetic variation and the gut microbiome by analyzing the microbial composition of 18,340 participants from 24 independent cohorts, the majority of whom were of European ancestry (n = 13,266) (22). A total of 211 taxa (9 phyla, 16 classes, 20 orders, 35 families, and 131 genera) were included in this meta-analysis. The data can be easily accessed from the website (www.mibiogen.org) (23).
GWAS summary statistics for spinal stenosis were obtained from the FinnGen Consortium R8 release (https://r8.risteys.finngen.fi/), which involved 9,169 spinal stenosis cases and 164,682 controls.
To ensure the validity and accuracy of the findings on the causal relationship between gut microbiota and spinal stenosis, we employed a rigorous quality control procedure to screen out acceptable IVs. The flow diagram is shown in Figure 2.
First, referring to previous MR studies and to ensure that a sufficient number of IVs were acquired, we established the significance level at P < 1.0×10-5 to ensure the robustness of the results (24, 25). Second, selected SNPs (R2 < 0.001 and clumping distance = 10,000 kb) were clumped to lessen the offset brought on by linkage disequilibrium to guarantee that the IVs are mutually independent (26). Third, we eliminated palindromic SNPs and SNPs that did not appear in the outcome from the IVs (27). Fourth, the PhenoScanner (http://www.phenoscanner.medschl.cam.ac.uk/phenoscanner) database was employed to detect and exclude IVs associated with confounders (educational attainment, smoking behavior, BMI, length of mobile phone use, and watching TV) to meet the principle of independence assumption in MR analysis (28). Finally, we calculated the F statistic to assess the strength of IVs and removed IVs with F < 10 to satisfy the strong correlation with exposure and attenuate instrumental bias (26, 29). F = R2(n-k-1)/k(1-R2) (n, sample size; k, number of IVs; and R2 variance of the exposure explained by the selected SNPs).
The IVW approach (30) was mostly utilized in this MR analysis to provide unbiased estimates of the causal connection between gut microbiota and spinal stenosis. In addition, to estimate the causal relationship under various circumstances, additional techniques were also employed, including the weighted median approach (31), MR-Egger method (32), simple mode method (33), and weighted mode method (34).
To detect potential horizontal pleiotropy effects, we performed the MR-PRESSO and MR-Egger regression intercept analysis. The intercept term in MR-Egger regression can be utilized for assessing horizontal pleiotropy, which if significant demonstrates the presence of horizontal pleiotropy and vice versa indicates the absence of horizontal pleiotropy (35). By eliminating major outliers, MR-PRESSO analysis identifies and decreases horizontal pleiotropy. However, it is important to note that the MR-PRESSO outlier test requires that at least 50% of the genetic variance be valid instrumental variables and relies on the Instrument Strength Independent of Direct Effect (InSIDE) condition (36). After removing pleiotropy, the remaining SNPs were used for subsequent MR analyses.
Cochran’s Q statistic was used to examine the heterogeneity of the SNPs, with a P-value < 0.05 indicating significant heterogeneity (37). Furthermore, to detect possible heterogeneity, we individually removed each instrumental variable SNP and conducted a “leave-one-out” sensitivity analysis to discover if a single SNP conclusively contributed to the causal signal (38). False discovery rate (FDR) corrections were applied to evaluate the false alarm rate. When P < 0.05 but q-value ≥ 0.1, gut microbiota and spinal stenosis were considered to have a suggestive association.
Finally, we also performed reverse MR analyses to determine whether reverse causality existed. The “TwoSampleMR” package and the “MRPRESSO” package in the R software (version 4.3.1, https://www.r-project.org/) were used for all MR analyses (36, 39).
According to a series of filtering steps, a total of 102, 180, 216, 380, and 1363 SNPs at the phyla, class, order, family, and genus levels were identified after a series of filtering steps. After clumping and harmonization, the number of SNPs related to spinal stenosis varies from 3 to 20. Furthermore, the genus RuminococcaceaeUCG002 contains the highest number of SNPs (20), whereas the genus with the least number is LachnospiraceaeND3007 (3 SNPs). And no feature, regardless of level, contains a single SNP. Supplementary Table 1 contains comprehensive details regarding the selected SNPs.
Initially, our research examined the cause and effect relationship between the gut microbiota and spinal stenosis. Employing the IVW approach as the primary MR detection method, As shown in Table 1, Supplementary Table 2, and Figure 3, the genus Eubacterium fissicatena group (OR = 1.09, 95% CI = 1.01-1.18, P = 0.02, IVW) and the genus Oxalobacter (OR = 1.12, 95% CI = 1.05-1.20, P = 0.001, IVW) were identified to have a potential causal relationship with spinal stenosis, indicating these two bacterial categories might potentially cause spinal stenosis. Specifically, considering that the above OR values are all greater than 1, the higher genetically predicted Eubacterium fissicatena and Oxalobacter levels were associated with a higher risk of spinal stenosis. When the FDR adjustment was carried out, however, these correlations failed to be significant (q > 0.1). Specific IVs can be found in Table 2.
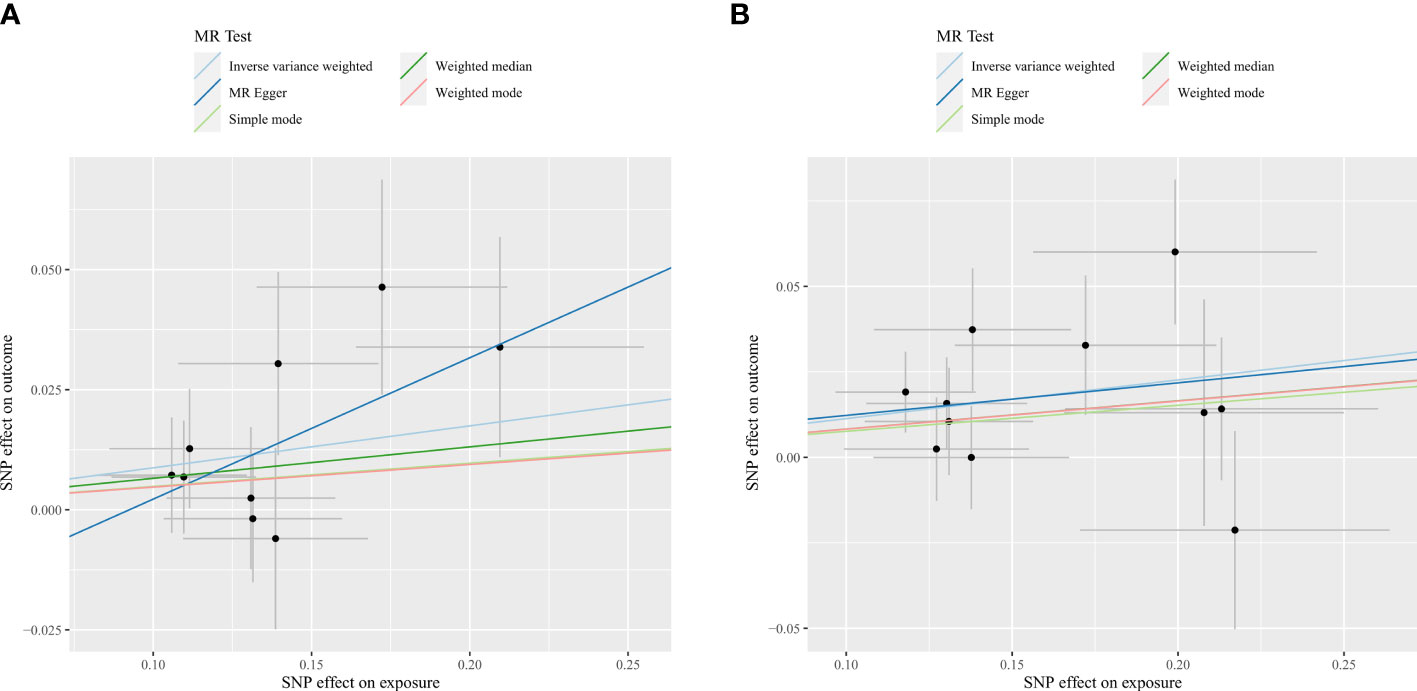
Figure 3 Scatter plots for the causal association between gut microbiota and spinal stenosis. (A) genus Eubacterium fissicatena group; (B) genus Oxalobacter.
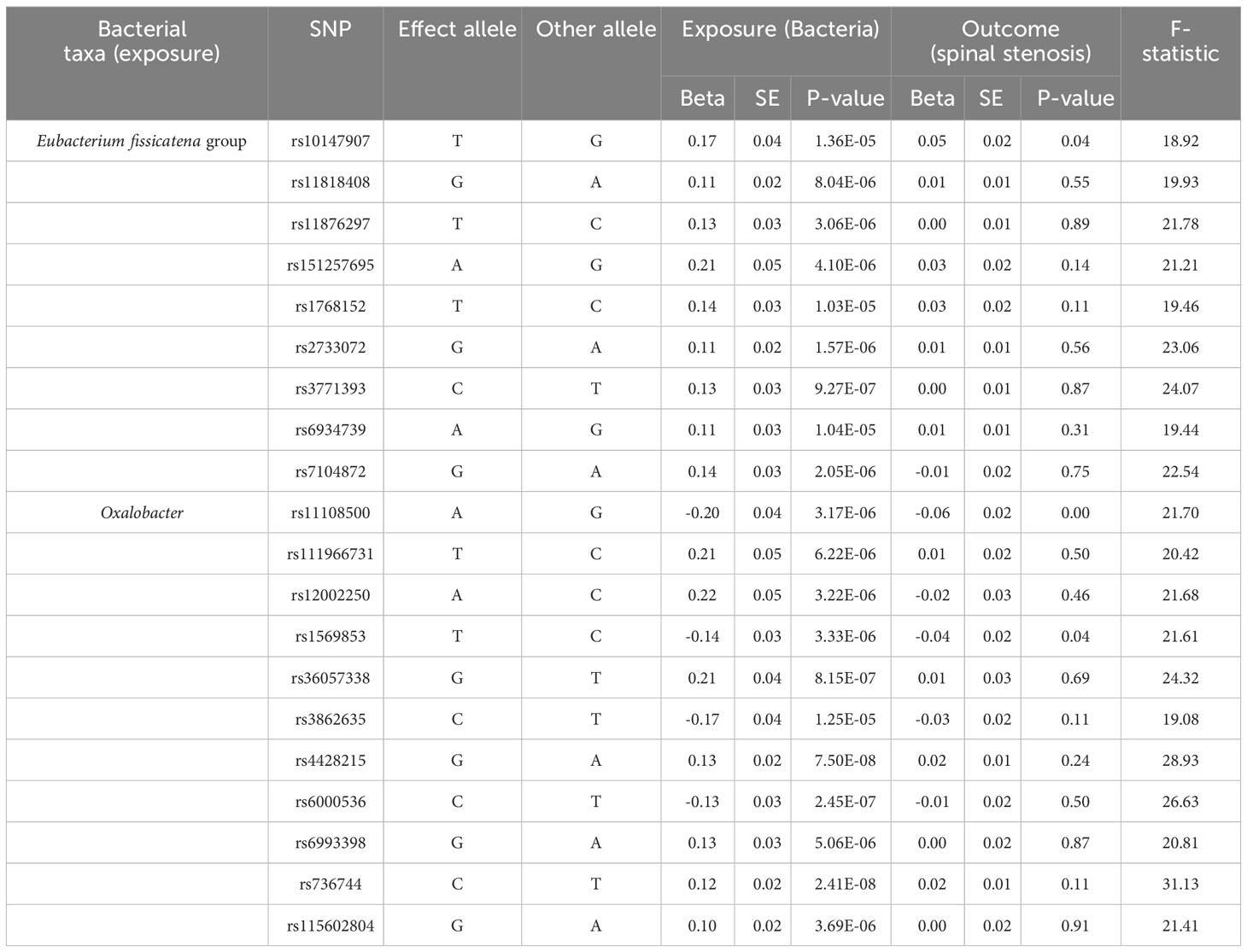
Table 2 SNPs used in MR analysis of the association between the two gut microbiota and spinal stenosis.
Among the two causal correlations revealed above, the F-statistics of the IVs were larger than 10, eliminating the bias of weak IVs (Table 2; Supplementary Table 1). Cochran’s IVW Q-test indicated that no significant heterogeneity was observed in the IVs of the genus Eubacterium fissicatena group (P = 0.67) and the genus Oxalobacter (P = 0.45) (Supplementary Table 3). In addition, according to the results of the MR-Egger regression intercept analysis, no significant directional horizontal pleiotropy was identified either in the IVs of the genus Eubacterium fissicatena group (P = 0.33) and the genus Oxalobacter (P = 0.92) (Supplementary Table 4). In addition, none of the above SNPs revealed a potential relationship with confounders (educational attainment, smoking behavior, BMI, length of mobile phone use, and watching TV) associated with spinal stenosis (28).
According to the results of reverse MR analysis, no significant causal association was found between spinal stenosis and the mentioned gut microbiota, the genus Eubacterium fissicatena group (OR = 0.94, 95% CI = 0.87-1.03, P = 0.19, IVW) and the genus Oxalobacter (OR = 0.96, 95% CI = 0.89-1.04, P = 0.31, IVW) (Table 3). Promising results (P > 0.05) were obtained for heterogeneity based on the Cochran’s IVW Q-test and directional horizontal pleiotropy based on the MR-Egger regression intercept analysis. In addition, relevant information on the selected IVs and MR results about reverse MR analysis can be found in Supplementary Table 5.
Moreover, in at least one MR method excluding IVW, eight bacterial taxa, the class Clostridia (OR
= 1.39, 95% CI = 1.14-1.70, p = 0.001, Weighted median; OR = 1.43, 95% CI = 1.09-1.87, P = 0.03,
Weighted mode), the order Clostridiales (OR = 1.37, 95% CI = 1.13-1.65, P = 0.001, Weighted median;
OR = 1.40, 95% CI = 1.08-1.81, P = 0.02, Weighted mode), the order Rhodospirillales (OR = 1.46, 95% CI = 1.05-2.03, P = 0.04, MR Egger), the family Lachnospiraceae (OR = 1.65, 95% CI = 1.06-2.51, P = 0.03, MR Egger), the family Prevotellaceae (OR = 0.86, 95% CI = 0.75-0.98, P = 0.03, Weighted median),
the family Acidaminococcaceae (Causal Estimate = -0.07, SD = 0.03, T = -2.65, P = 0.04, MR-PRESSO), the genus Eisenbergiella (OR = 0.87, 95% CI = 0.78-0.97, P = 0.03, MR Egger), the genus unknowngenus.id.2755 (OR = 1.49, 95% CI = 1.10-2.02, P = 0.03, MR Egger), were revealed to be suggestively related with spinal stenosis (Figure 4; Supplementary Tables 2, 6).
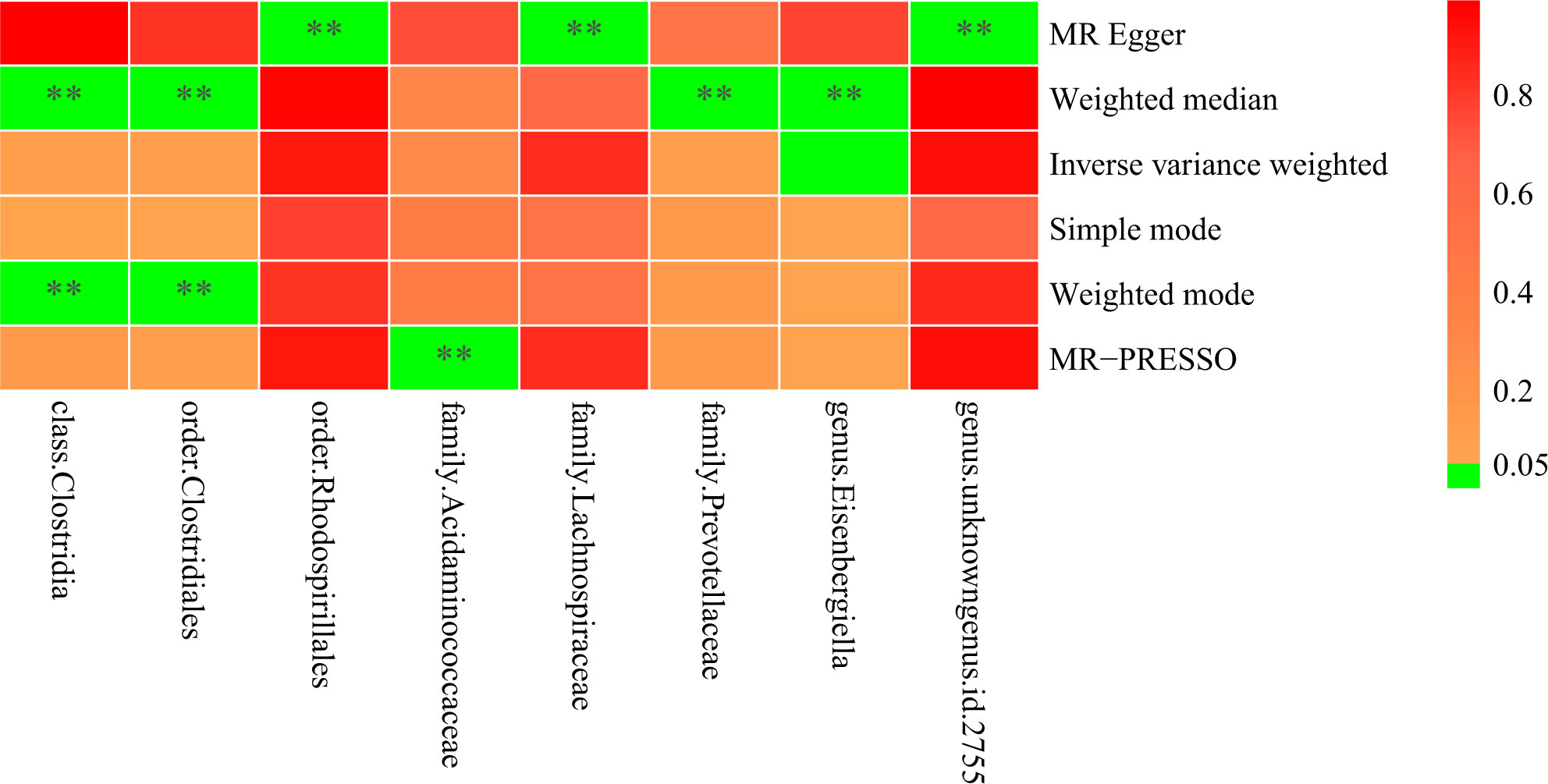
Figure 4 Heatmap illustrating eight bacterial taxa were revealed to be suggestively related with spinal stenosis, with ** indicating P < 0.05.
In this study, Cochran ‘s IVW Q-test was utilized to evaluate heterogeneity, and the MR-Egger regression intercept analysis was used to evaluate directional horizontal pleiotropy. Furthermore, we employed MR PRESSO to ensure global directional horizontal pleiotropy and increase the reliability of the study (Supplementary Table 6). At the level of five different strains, MR PRESSO was performed for each intestinal flora except the genus LachnospiraceaeND3007 because there were insufficient SNPs (n ≤ 3) to conduct MR PRESSO. And one of the class levels, one of the order levels, three of the family levels, and eighteen of the genus levels were tested for outlier-corrected MR-PRESSO to ensure the robustness of the MR results (Supplementary Table 6). In addition, the leave-one-out analyses were performed to examine the potential influence of individual SNPs on the observed associations. As shown in Figure 5, when an SNP is eliminated individually, the overall error lines are all positioned on one side of the median line, implying that each SNP affects the results equally and excludes outlier SNPs from affecting the reliability of the results.
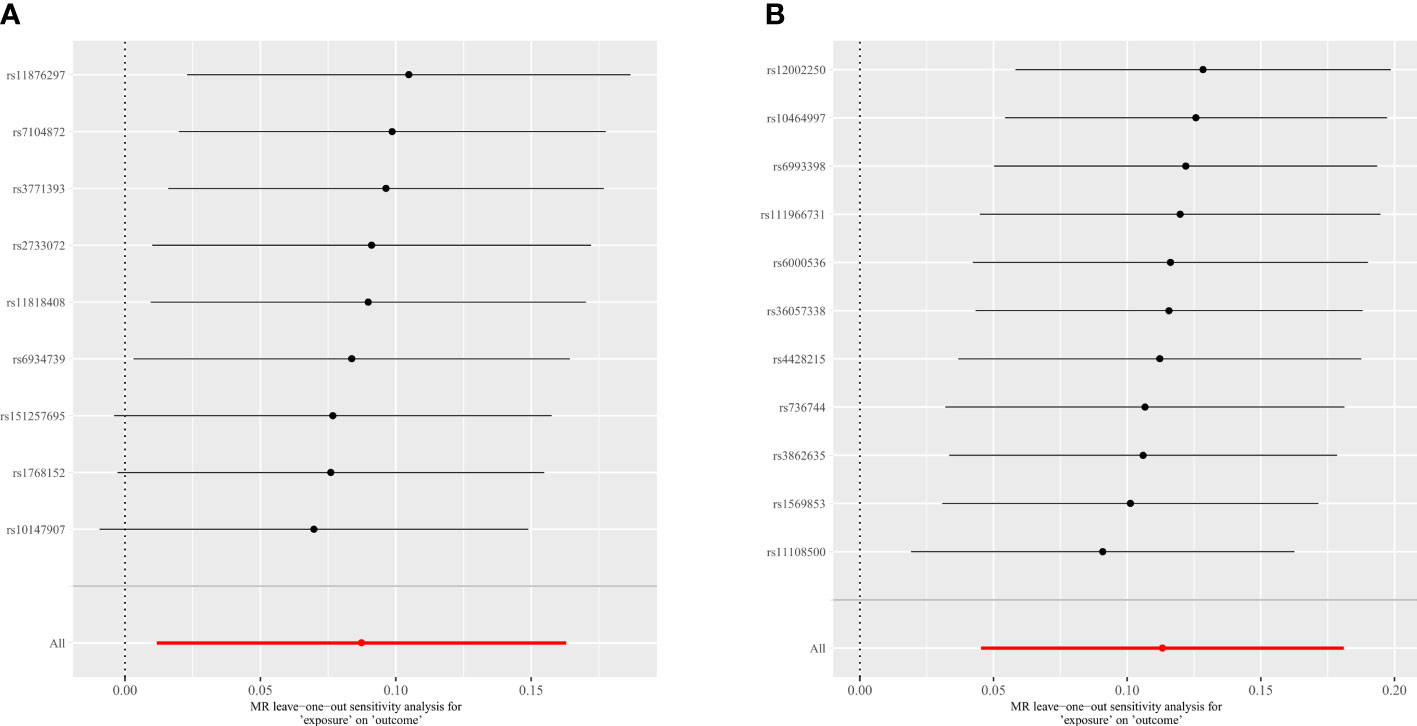
Figure 5 Leave-one-out plots for the causal association between gut microbiota and spinal stenosis. (A) genus Eubacterium fissicatena group; (B) genus Oxalobacter.
Recently, researchers have revealed that the composition and variety of gut microbial components are closely linked to spine-related disorders such as osteoporosis (11), intervertebral disc degeneration (40), spinal sarcopenia (41), adolescent idiopathic scoliosis (42), ankylosing spondylitis (43), and others. An article, based on previous research, innovatively proposes the concept of the gut-spine axis by reviewing articles on gut microbiota and spine-related diseases (13). In terms of the association between gut microbiota and spinal stenosis, Tadatsugu Morimoto et al. demonstrated that a Gut-ligament axis may be present in lumbar spinal stenosis, but the specific connection is unknown (13). In this study, a two-sample MR analysis was performed to assess the causal relationship between gut microbiota and spinal stenosis, employing summary statistics for gut microbiota from the largest GWAS meta-analysis carried out by the MiBioGen consortium and summary statistics for spinal stenosis from the FinnGen consortium R8 release data. With the help of large-scale GWAS data sources, our MR study fills this knowledge gap from a new perspective. To the best of our knowledge, this is the first MR study to investigate the potential causal relationship between gut microbiota and spinal stenosis. Interestingly, the finding indicates that genetically predicted abundance of specific gut microbiota (the genus Eubacterium fissicatena group and genus Oxalobacter) is potentially associated with spinal stenosis. Additionally, we also identified some gut microbiota may be potentially related to spinal stenosis. Importantly, the above results may have implications for public health efforts to minimize the risk of development of spinal stenosis.
Eubacterium fissicatena, growing at 20–40 °C (optimum 37 °C) and pH 6.0–8.0 (optimum pH 7.0), was isolated from the alimentary tract of the goat as first reported by M. Taylor in 1972 (44). Eubacterium, regarded as promising targets for microbial therapeutics, forms the core genera of health-associated human gut microbiota (45). Over the past few years, an increasing number of studies have shown that Eubacterium modulates immune and inflammatory responses in humans through the regulation of short-chain fatty acids (SCFAs) which have significant effects on gut health (45, 46). In addition to its anti-inflammatory effects, butyrate, the primary metabolite of SCFAs in the colon, is necessary for the maintenance of intercellular tight junctions, which preserves the normal barrier function of intestinal mucosa (47). Several studies conducted in mice have shown that specific medication components or metabolites can modulate dextran sulfate sodium salt-induced colitis by correcting intestinal flora dysbiosis (decreasing the abundance of pathogenic bacteria, Eubacterium fissicatena) (48–50). Furthermore, in terms of metabolism-related diseases, the reduction of Eubacterium fissicatena abundance is effective in reducing diet-induced obesity, hepatic steatosis, dyslipidaemia, and insulin resistance (51–54). Taken together, the existing research indicates that Eubacterium fissicatena is most often a causative agent in the mediation of inflammation-related diseases and metabolic disorders, including obesity and dyslipidaemia. As mentioned in the background section, there is a correlation between these factors and spinal stenosis, and what seems to serve as an explanation for the existence of a possible causal relationship between Eubacterium fissicatena and spinal stenosis.
In contrast to Eubacterium fissicatena, Oxalobacter, first reported as a novel anaerobic bacterium for degrading oxalic acids in 1985, is a popular species of interest to researchers in our literature search (55). Since the separation of Oxalobacter, numerous studies have concentrated on its role in hyperoxaluria and kidney stone development and attempted to develop associated therapies to limit the formation and advancement of the above-mentioned diseases, owing to its specific function in degrading oxalic acid (56–59). OxlT, an oxalate transporter protein of Oxalobacter, specifically absorbs oxalate from the intestine into bacterial cells, lowering the likelihood that the host animal may develop oxalate-deposition illnesses such as kidney stones (60). Remarkably, when examining the connection between Oxalobacter and spinal stenosis, we discovered a case report by Knight RQ et al. suggesting that lumbar spinal stenosis could potentially be caused by oxalic acid accumulation (61). As a supplement, the correlation between Oxalobacter and spinal stenosis at the genetic level was explored in the present MR study to enhance insights in this field.
Furthermore, we found eight strains of organisms that might be related to spinal stenosis based on five MR statistical approaches other than the IVW method, in addition to the two strains (the genus Eubacterium fissicatena group and the genus Oxalobacter) that were obviously causally linked to the condition. As mentioned above, eight bacterial taxa, 1 class (Clostridia), 2 orders (Clostridiales, Rhodospirillales), 3 families (Lachnospiraceae, Prevotellaceae, Acidaminococcaceae), and 2 genera (Eisenbergiella, unknowngenus.id.2755), may have a potential relationship with spinal stenosis. However, to confirm the aforementioned association between gut microbiota and spinal stenosis, large-scale population sequencing studies need to be performed.
Overall, this study has several strengths. Firstly, employing MR analysis, we established the genetic causal relationship between gut microbiota and spinal stenosis, and importantly identified two gut microbiota (the genus Eubacterium fissicatena group and genus Oxalobacter) that were causally linked to the condition. This work contributes to the body of knowledge in this field and holds the potential to contribute to public prevention strategies for spinal stenosis. Second, to improve the robustness of the study findings, MR-PRESSO and MR-Egger regression intercept terms were utilized to test for horizontal pleiotropy. Furthermore, to avoid bias, the two-sample MR study adopted non-overlapping exposure and outcome summary data (62).
Nevertheless, there are several limitations regarding our study that should be acknowledged. First, as the majority of participants of the aggregated data in this research were of European ancestry, the extent to which findings from this study can be extrapolated to other ethnic groups may be limited. Second, following the relevant research papers, we set the significance level for SNPs to P < 1 × 10-5 to acquire more instrumental variables, which implies that SNPs employed in the analyses in the current study did not satisfy the standard GWAS significance threshold (P < 5 × 10-8) (19, 22). For this, we carefully evaluated the F-value of each SNP to eliminate the potential effect of weak IVs bias. Furthermore, the majority of individuals with spinal stenosis are middle-aged or older, while our current study is unable to divide the intestinal flora based on aging.
In summary, we comprehensively assessed the causal association between the gut microbiota and spinal stenosis. As a result, the genus Eubacterium fissicatena group and genus Oxalobacter are considered to have a potentially genetic causal relationship with spinal stenosis. Undoubtedly, this study might provide new insights into the pathogenesis of gut microbiota-mediated spinal stenosis. However, further large-scale clinical trials are needed to confirm these findings and to develop public prevention strategies for spinal stenosis, as well as to explore the potential of targeted probiotics for the treatment of spinal stenosis.
The original contributions presented in the study are included in the article/Supplementary material. Further inquiries can be directed to the corresponding authors.
JL: Conceptualization, Data curation, Investigation, Methodology, Software, Writing – original draft. JPW: Writing – original draft, Data curation, Formal analysis, Visualization, Software. JNW: Conceptualization, Investigation, Software, Writing – original draft. TX: Conceptualization, Data curation, Writing – original draft. BW: Investigation, Visualization, Writing – original draft. SY: Investigation, Software, Writing – original draft. SJ: Methodology, Writing – original draft. HW: Writing – review & editing. HH: Writing – review & ,editing Supervision.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
The authors appreciate the MiBioGen consortium for providing the gut microbiota GWAS summary statistics, as well as the researchers and participants of the FinnGen project.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1360132/full#supplementary-material
Supplementary Table 1 | Instrumental variables used in MR analysis of the association between gut microbiota and spinal stenosis.
Supplementary Table 2 | Full result of MR estimates for the association between gut microbiota and spinal stenosis.
Supplementary Table 3 | The heterogeneity of gut microbiota instrumental variables.
Supplementary Table 4 | Directional horizontal pleiotropy assessed by intercept term in MR Egger regression of the association between gut microbiota and spinal stenosis.
Supplementary Table 5 | The results of reverse MR analysis between spinal stenosis and gut microbiota.
Supplementary Table 6 | MR-PRESSO analysis for the association between gut microbiota and spinal stenosis.
1. Parenteau CS, Lau EC, Campbell IC, Courtney A. Prevalence of spine degeneration diagnosis by type, age, gender, and obesity using Medicare data. Sci Rep. (2021) 11:5389. doi: 10.1038/s41598-021-84724-6
2. Katz JN, Zimmerman ZE, Mass H, Makhni MC. Diagnosis and management of lumbar spinal stenosis: A review. JAMA. (2022) 327:1688–99. doi: 10.1001/jama.2022.5921
3. Ravindra VM, Senglaub SS, Rattani A, Dewan MC, Hartl R, Bisson E, et al. Degenerative lumbar spine disease: estimating global incidence and worldwide volume. Global Spine J. (2018) 8:784–94. doi: 10.1177/2192568218770769
4. Fang EF, Xie C, Schenkel JA, Wu C, Long Q, Cui H, et al. A research agenda for ageing in China in the 21st century (2nd edition): Focusing on basic and translational research, long-term care, policy and social networks. Ageing Res Rev. (2020) 64:101174. doi: 10.1016/j.arr.2020.101174
5. Afzaal M, Saeed F, Shah YA, Hussain M, Rabail R, Socol CT, et al. Human gut microbiota in health and disease: Unveiling the relationship. Front Microbiol. (2022) 13:999001. doi: 10.3389/fmicb.2022.999001
6. Sebastian Domingo JJ, Sanchez Sanchez C. From the intestinal flora to the microbiome. Rev Esp Enferm Dig. (2018) 110:51–6. doi: 10.17235/reed.2017.4947/2017
7. Wang L, Lilburn M, Yu Z. Intestinal microbiota of broiler chickens as affected by litter management regimens. Front Microbiol. (2016) 7:593. doi: 10.3389/fmicb.2016.00593
8. Gao B, Duan Y, Lang S, Barupal D, Wu TC, Valdiviez L, et al. Functional microbiomics reveals alterations of the gut microbiome and host co-metabolism in patients with alcoholic hepatitis. Hepatol Commun. (2020) 4:1168–82. doi: 10.1002/hep4.1537
9. Yachida S, Mizutani S, Shiroma H, Shiba S, Nakajima T, Sakamoto T, et al. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat Med. (2019) 25:968–76. doi: 10.1038/s41591-019-0458-7
10. Azzini GOM, Santos GS, Visoni SBC, Azzini VOM, Santos RGD, Huber SC, et al. Metabolic syndrome and subchondral bone alterations: The rise of osteoarthritis - A review. J Clin Orthop Trauma. (2020) 11:S849–55. doi: 10.1016/j.jcot.2020.06.021
11. Wei M, Li C, Dai Y, Zhou H, Cui Y, Zeng Y, et al. High-throughput absolute quantification sequencing revealed osteoporosis-related gut microbiota alterations in han chinese elderly. Front Cell Infect Microbiol. (2021) 11:630372. doi: 10.3389/fcimb.2021.630372
12. Chen Y, Wang X, Zhang C, Liu Z, Li C, Ren Z. Gut microbiota and bone diseases: A growing partnership. Front Microbiol. (2022) 13:877776. doi: 10.3389/fmicb.2022.877776
13. Morimoto T, Kobayashi T, Kakiuchi T, Esaki M, Tsukamoto M, Yoshihara T, et al. Gut-spine axis: a possible correlation between gut microbiota and spinal degenerative diseases. Front Microbiol. (2023) 14:1290858. doi: 10.3389/fmicb.2023.1290858
14. Tanabe K, Nakamura S, Moriyama-Hashiguchi M, Kitajima M, Ejima H, Imori C, et al. Dietary fructooligosaccharide and glucomannan alter gut microbiota and improve bone metabolism in senescence-accelerated mouse. J Agric Food Chem. (2019) 67:867–74. doi: 10.1021/acs.jafc.8b05164
15. Lurie J, Tomkins-Lane C. Management of lumbar spinal stenosis. Bmj. (2016) 352:h6234. doi: 10.1136/bmj.h6234
16. Byvaltsev VA, Kalinin AA, Hernandez PA, Shepelev VV, Pestryakov YY, Aliyev MA, et al. Molecular and genetic mechanisms of spinal stenosis formation: systematic review. Int J Mol Sci. (2022) 23(21):13479. doi: 10.3390/ijms232113479
17. Sairyo K, Biyani A, Goel VK, Leaman DW, Booth R Jr., Thomas J, et al. Lumbar ligamentum flavum hypertrophy is due to accumulation of inflammation-related scar tissue. Spine (Phila Pa 1976). (2007) 32:E340–347. doi: 10.1097/01.brs.0000263407.25009.6e
18. Burgess S, Timpson NJ, Ebrahim S, Davey Smith G. Mendelian randomization: where are we now and where are we going? Int J Epidemiol. (2015) 44:379–88. doi: 10.1093/ije/dyv108
19. Li P, Wang H, Guo L, Gou X, Chen G, Lin D, et al. Association between gut microbiota and preeclampsia-eclampsia: a two-sample Mendelian randomization study. BMC Med. (2022) 20:443. doi: 10.1186/s12916-022-02657-x
20. Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet. (2014) 23:R89–98. doi: 10.1093/hmg/ddu328
21. Davies NM, Holmes MV, Davey Smith G. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ. (2018) 362:k601. doi: 10.1136/bmj.k601
22. Kurilshikov A, Medina-Gomez C, Bacigalupe R, Radjabzadeh D, Wang J, Demirkan A, et al. Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat Genet. (2021) 53:156–65. doi: 10.1038/s41588-020-00763-1
23. consortium, M. MiBioGen (2021). Available online at: https://mibiogen.gcc.rug.nl/.
24. Sanna S, van Zuydam NR, Mahajan A, Kurilshikov A, Vich Vila A, Vosa U, et al. Causal relationships among the gut microbiome, short-chain fatty acids and metabolic diseases. Nat Genet. (2019) 51:600–5. doi: 10.1038/s41588-019-0350-x
25. Yu XH, Yang YQ, Cao RR, Bo L, Lei SF. The causal role of gut microbiota in development of osteoarthritis. Osteoarthritis Cartilage. (2021) 29:1741–50. doi: 10.1016/j.joca.2021.08.003
26. Genomes Project C, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. A global reference for human genetic variation. Nature. (2015) 526:68–74. doi: 10.1038/nature15393
27. Cao Z, Wu Y, Li Q, Li Y, Wu J. A causal relationship between childhood obesity and risk of osteoarthritis: results from a two-sample Mendelian randomization analysis. Ann Med. (2022) 54:1636–45. doi: 10.1080/07853890.2022.2085883
28. Wan B, Ma N, Lu W. Evaluating the causal relationship between five modifiable factors and the risk of spinal stenosis: a multivariable Mendelian randomization analysis. PeerJ. (2023) 11:e15087. doi: 10.7717/peerj.15087
29. Burgess S, Thompson SG. Bias in causal estimates from Mendelian randomization studies with weak instruments. Stat Med. (2011) 30:1312–23. doi: 10.1002/sim.4197
30. Larsson SC, Burgess S. Appraising the causal role of smoking in multiple diseases: A systematic review and meta-analysis of Mendelian randomization studies. EBioMedicine. (2022) 82:104154. doi: 10.1016/j.ebiom.2022.104154
31. Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. (2016) 40:304–14. doi: 10.1002/gepi.21965
32. Zheng C, He MH, Huang JR, He Y. Causal relationships between social isolation and osteoarthritis: A mendelian randomization study in european population. Int J Gen Med. (2021) 14:6777–86. doi: 10.2147/IJGM.S331864
33. Li C, Niu M, Guo Z, Liu P, Zheng Y, Liu D, et al. A mild causal relationship between tea consumption and obesity in general population: A two-sample mendelian randomization study. Front Genet. (2022) 13:795049. doi: 10.3389/fgene.2022.795049
34. Hartwig FP, Davey Smith G, Bowden J. Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int J Epidemiol. (2017) 46:1985–98. doi: 10.1093/ije/dyx102
35. Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. (2015) 44:512–25. doi: 10.1093/ije/dyv080
36. Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. (2018) 50:693–8. doi: 10.1038/s41588-018-0099-7
37. Bowden J, Del Greco MF, Minelli C, Zhao Q, Lawlor DA, Sheehan NA, et al. Improving the accuracy of two-sample summary-data Mendelian randomization: moving beyond the NOME assumption. Int J Epidemiol. (2019) 48:728–42. doi: 10.1093/ije/dyy258
38. Hemani G, Tilling K, Davey Smith G. Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PloS Genet. (2017) 13:e1007081. doi: 10.1371/journal.pgen.1007081
39. Davies NM, Hill WD, Anderson EL, Sanderson E, Deary IJ, Davey Smith G. Multivariable two-sample Mendelian randomization estimates of the effects of intelligence and education on health. Elife. (2019) 8:e43990. doi: 10.7554/eLife.43990
40. Geng Z, Wang J, Chen G, Liu J, Lan J, Zhang Z, et al. Gut microbiota and intervertebral disc degeneration: a bidirectional two-sample Mendelian randomization study. J Orthop Surg Res. (2023) 18:601. doi: 10.1186/s13018-023-04081-0
41. Zhang T, Cheng JK, Hu YM. Gut microbiota as a promising therapeutic target for age-related sarcopenia. Ageing Res Rev. (2022) 81:101739. doi: 10.1016/j.arr.2022.101739
42. Shen N, Chen N, Zhou X, Zhao B, Huang R, Liang J, et al. Alterations of the gut microbiome and plasma proteome in Chinese patients with adolescent idiopathic scoliosis. Bone. (2019) 120:364–70. doi: 10.1016/j.bone.2018.11.017
43. Song ZY, Yuan D, Zhang SX. Role of the microbiome and its metabolites in ankylosing spondylitis. Front Immunol. (2022) 13:1010572. doi: 10.3389/fimmu.2022.1010572
44. Taylor MM. Eubacterium fissicatena sp.nov. isolated from the alimentary tract of the goat. J Gen Microbiol. (1972) 71:457–63. doi: 10.1099/00221287-71-3-457
45. Mukherjee A, Lordan C, Ross RP, Cotter PD. Gut microbes from the phylogenetically diverse genus Eubacterium and their various contributions to gut health. Gut Microbes. (2020) 12:1802866. doi: 10.1080/19490976.2020.1802866
46. Chambers ES, Preston T, Frost G, Morrison DJ. Role of gut microbiota-generated short-chain fatty acids in metabolic and cardiovascular health. Curr Nutr Rep. (2018) 7:198–206. doi: 10.1007/s13668-018-0248-8
47. Silva YP, Bernardi A, Frozza RL. The role of short-chain fatty acids from gut microbiota in gut-brain communication. Front Endocrinol (Lausanne). (2020) 11:25. doi: 10.3389/fendo.2020.00025
48. Liu Y, Huang W, Ji S, Wang J, Luo J, Lu B. Sophora japonica flowers and their main phytochemical, rutin, regulate chemically induced murine colitis in association with targeting the NF-κB signaling pathway and gut microbiota. Food Chem. (2022) 393:133395. doi: 10.1016/j.foodchem.2022.133395
49. Li H, Wang Y, Shao S, Yu H, Wang D, Li C, et al. Rabdosia serra alleviates dextran sulfate sodium salt-induced colitis in mice through anti-inflammation, regulating Th17/Treg balance, maintaining intestinal barrier integrity, and modulating gut microbiota. J Pharm Anal. (2022) 12:824–38. doi: 10.1016/j.jpha.2022.08.001
50. Li Y, Li N, Liu J, Wang T, Dong R, Ge D, et al. Gegen qinlian decoction alleviates experimental colitis and concurrent lung inflammation by inhibiting the recruitment of inflammatory myeloid cells and restoring microbial balance. J Inflammation Res. (2022) 15:1273–91. doi: 10.2147/jir.S352706
51. Yue C, Li M, Li J, Han X, Zhu H, Yu G, et al. Medium-, long- and medium-chain-type structured lipids ameliorate high-fat diet-induced atherosclerosis by regulating inflammation, adipogenesis, and gut microbiota in ApoE(-/-) mice. Food Funct. (2020) 11:5142–55. doi: 10.1039/D0FO01006E
52. Song Y, Shen H, Liu T, Pan B, De Alwis S, Zhang W, et al. Effects of three different mannans on obesity and gut microbiota in high-fat diet-fed C57BL/6J mice. Food Funct. (2021) 12:4606–20. doi: 10.1039/D0FO03331F
53. Mohr AE, Jasbi P, Bowes DA, Dirks B, Whisner CM, Arciero KM, et al. Exploratory analysis of one versus two-day intermittent fasting protocols on the gut microbiome and plasma metabolome in adults with overweight/obesity. Front Nutr. (2022) 9:1036080. doi: 10.3389/fnut.2022.1036080
54. Wang J, Hao J, Miao D, Xiao P, Jiang X, E. Hu L. Compound chenpi tea consumption reduces obesity-related metabolic disorders by modulating gut microbiota and serum metabolites in mice. J Sci Food Agric. (2024) 104:431–42. doi: 10.1002/jsfa.12940
55. Allison MJ, Dawson KA, Mayberry WR, Foss JG. Oxalobacter formigenes gen. nov., sp. nov.: oxalate-degrading anaerobes that inhabit the gastrointestinal tract. Arch Microbiol. (1985) 141:1–7. doi: 10.1007/bf00446731
56. Kaufman DW, Kelly JP, Curhan GC, Anderson TE, Dretler SP, Preminger GM, et al. Oxalobacter formigenes may reduce the risk of calcium oxalate kidney stones. J Am Soc Nephrol. (2008) 19:1197–203. doi: 10.1681/asn.2007101058
57. Mehta M, Goldfarb DS, Nazzal L. The role of the microbiome in kidney stone formation. Int J Surg. (2016) 36:607–12. doi: 10.1016/j.ijsu.2016.11.024
58. Ticinesi A, Nouvenne A, Meschi T. Gut microbiome and kidney stone disease: not just an Oxalobacter story. Kidney Int. (2019) 96:25–7. doi: 10.1016/j.kint.2019.03.020
59. Chmiel JA, Carr C, Stuivenberg GA, Venema R, Chanyi RM, Al KF, et al. New perspectives on an old grouping: The genomic and phenotypic variability of Oxalobacter formigenes and the implications for calcium oxalate stone prevention. Front Microbiol. (2022) 13:1011102. doi: 10.3389/fmicb.2022.1011102
60. Jaunet-Lahary T, Shimamura T, Hayashi M, Nomura N, Hirasawa K, Shimizu T, et al. Structure and mechanism of oxalate transporter OxlT in an oxalate-degrading bacterium in the gut microbiota. Nat Commun. (2023) 14:1730. doi: 10.1038/s41467-023-36883-5
61. Knight RQ, Taddonio RF, Smith FB, Weseley S, Pecker HM. Oxalosis: cause of degenerative spinal stenosis. A case report and review of the literature. Orthopedics. (1988) 11:955–8. doi: 10.3928/0147-7447-19880601-18
Keywords: two-sample mendelian randomization, gut microbiota, spinal stenosis, causal inference, single nucleotide polymorphism
Citation: Li J, Wei J, Wang J, Xu T, Wu B, Yang S, Jing S, Wu H and Hao H (2024) Association between gut microbiota and spinal stenosis: a two-sample mendelian randomization study. Front. Immunol. 15:1360132. doi: 10.3389/fimmu.2024.1360132
Received: 22 December 2023; Accepted: 09 April 2024;
Published: 19 April 2024.
Edited by:
Xiaoqian Dang, The Second Affiliated Hospital of Xi’an Jiaotong University, ChinaReviewed by:
Lijin Guo, South China Agricultural University, ChinaCopyright © 2024 Li, Wei, Wang, Xu, Wu, Yang, Jing, Wu and Hao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haihu Hao, aGhoMzMwMUAxMjYuY29t; Hua Wu, d3VodWFAaHVzdC5lZHUuY24=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.