
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol., 02 April 2024
Sec. NK and Innate Lymphoid Cell Biology
Volume 15 - 2024 | https://doi.org/10.3389/fimmu.2024.1350470
This article is part of the Research TopicChallenges in NK Cell Therapeutic DevelopmentView all 8 articles
 Johannes Schetelig1,2*
Johannes Schetelig1,2* Henning Baldauf2
Henning Baldauf2 Falk Heidenreich1,2
Falk Heidenreich1,2 Jorinde D. Hoogenboom3
Jorinde D. Hoogenboom3 Stephen R. Spellman4
Stephen R. Spellman4 Alexander Kulagin5
Alexander Kulagin5 Thomas Schroeder6
Thomas Schroeder6 Henrik Sengeloev7
Henrik Sengeloev7 Peter Dreger8
Peter Dreger8 Edouard Forcade9
Edouard Forcade9 Jan Vydra10
Jan Vydra10 Eva Maria Wagner-Drouet11
Eva Maria Wagner-Drouet11 Goda Choi12
Goda Choi12 Shankara Paneesha13
Shankara Paneesha13 Nuno A. A. Miranda14
Nuno A. A. Miranda14 Alina Tanase15
Alina Tanase15 Liesbeth C. de Wreede16
Liesbeth C. de Wreede16 Vinzenz Lange17
Vinzenz Lange17 Alexander H. Schmidt17,18
Alexander H. Schmidt17,18 Jürgen Sauter18
Jürgen Sauter18 Joshua A. Fein19
Joshua A. Fein19 Yung-Tsi Bolon4
Yung-Tsi Bolon4 Meilun He4
Meilun He4 Steven G. E. Marsh20
Steven G. E. Marsh20 Shahinaz M. Gadalla21
Shahinaz M. Gadalla21 Sophie Paczesny22
Sophie Paczesny22 Annalisa Ruggeri23
Annalisa Ruggeri23 Christian Chabannon24
Christian Chabannon24 Katharina Fleischhauer25 on behalf of Cellular Therapy & Immunobiology Working Party of EBMT the Immunobiology Working Committee of the Center for International Blood Marrow Transplant Research (CIBMTR)
Katharina Fleischhauer25 on behalf of Cellular Therapy & Immunobiology Working Party of EBMT the Immunobiology Working Committee of the Center for International Blood Marrow Transplant Research (CIBMTR)Optimizing natural killer (NK) cell alloreactivity could further improve outcome after allogeneic hematopoietic cell transplantation (alloHCT). The donor’s Killer-cell Immunoglobulin-like Receptor (KIR) genotype may provide important information in this regard. In the past decade, different models have been proposed aiming at maximizing NK cell activation by activating KIR-ligand interactions or minimizing inhibitory KIR-ligand interactions. Alternative classifications intended predicting outcome after alloHCT by donor KIR-haplotypes. In the present study, we aimed at validating proposed models and exploring more classification approaches. To this end, we analyzed samples stored at the Collaborative Biobank from HLA-compatible unrelated stem cell donors who had donated for patients with acute myeloid leukemia (AML) or myelodysplastic neoplasm (MDS) and whose outcome data had been reported to EBMT or CIBMTR. The donor KIR genotype was determined by high resolution amplicon-based next generation sequencing. We analyzed data from 5,017 transplants. The median patient age at alloHCT was 56 years. Patients were transplanted for AML between 2013 and 2018. Donor-recipient pairs were matched for HLA-A, -B, -C, -DRB1, and -DQB1 (79%) or had single HLA mismatches. Myeloablative conditioning was given to 56% of patients. Fifty-two percent of patients received anti-thymocyte-globulin-based graft-versus-host disease prophylaxis, 32% calcineurin-inhibitor-based prophylaxis, and 7% post-transplant cyclophosphamide-based prophylaxis. We tested several previously reported classifications in multivariable regression analyses but could not confirm outcome associations. Exploratory analyses in 1,939 patients (39%) who were transplanted from donors with homozygous centromeric (cen) or telomeric (tel) A or B motifs, showed that the donor cen B/B-tel A/A diplotype was associated with a trend to better event-free survival (HR 0.84, p=.08) and reduced risk of non-relapse mortality (NRM) (HR 0.65, p=.01). When we further dissected the contribution of B subtypes, we found that only the cen B01/B01-telA/A diplotype was associated with a reduced risk of relapse (HR 0.40, p=.04) while all subtype combinations contributed to a reduced risk of NRM. This exploratory finding has to be validated in an independent data set. In summary, the existing body of evidence is not (yet) consistent enough to recommend use of donor KIR genotype information for donor selection in routine clinical practice.
Natural Killer (NK) cells have raised great interest as potential mediators of selective graft-versus-leukemia effects after allogeneic hematopoietic cell transplantation (alloHCT) since the first descriptions of a reduced risk of relapse and improved survival of patients transplanted from haploidentical related donors missing the human leukocyte antigen (HLA) ligands for inhibitory Killer-cell Immunoglobulin-like Receptors (KIR) (1). Characterization of the extensive genetic polymorphism of KIR genotypes, including haplotypes with varying numbers of inhibitory and activating KIR genes unravelled the increasing complexity of NK cell mediated alloreactivity in alloHCT.
Clinical evidence for NK cell mediated alloreactivity against cancer cells comes from a series of studies demonstrating activity of NK cell transfusions for patients with relapsed or refractory acute myeloid leukaemia (AML) or myelodysplastic neoplasm (MDS) (2–7). Alloreactivity may be triggered by activating receptors and/or inhibitory receptors on the surface of NK cells which bind to classical and non-classical HLA molecules or to non-HLA ligands. KIR play an important role in NK cytotoxicity. While KIR-expression patterns define the NK-cell repertoire phenotype-wise, KIR genotypes expose remarkable diversity at an individual and population level. The function of this diversity is poorly understood in health and disease.
Evidence towards a potential role of KIR mediating NK- alloreactivity came from a series of retrospective registry studies on patients with HLA-compatible related or unrelated donors, which showed associations between certain KIR genotype patterns of stem cell donors and the risk of relapse after alloHCT (8–13).
The donor’s KIR genotype thus may provide critical information and could be utilized for KIR-informed donor selection. In the past decade, different models have been proposed aiming at maximizing NK cell activation through activating KIR encountering their cognate KIR ligands (KIRL) interactions or minimizing repressive signals through inhibitory KIR-KIRL interactions (12–15). Alternative classifications aimed at predicting outcome after alloHCT according to donor KIR-haplotypes, thereby integrating information of various sets of encoded activating/inhibitory KIR (16, 17). Another classification approach hypothesized that stronger NK-alloreactivity against leukemic blasts could be triggered in the absence of strong KIR-KIRL interactions with a higher risk of relapse among KIR2DL2-positive donors for C1/C1-positive patients, i.e. patients homozygous for the cognate KIR2DL2-ligand in a previous study (18). In the same study, patients heterozygous for the cognate HLA-C ligands transplanted from donors whose genotype did not comprise phylogenetic clade 2 KIR2DL1 allele, had a higher risk of relapse.
However, it has to be noted that the validation of most donor KIR genotype-based prediction models failed so far (19–21). This highlighted the need for independent validation studies with adequate power. Therefore, we set out to validate published models for donor KIR genotype-based outcome prediction in a joint EBMT and Center for International Blood and Marrow Transplant Research (CIBMTR) study. Here, we report results from this dataset of approximately 5,000 patients who had received alloHCT for AML or MDS and whose donors had been typed for KIR genes at high-resolution.
We conducted a joint study of the EBMT and the CIBMTR. For the study DNA samples from stem cell donors stored at the Collaborative Biobank (www.cobi-biobank.com) were genotyped.
Patients were included, if they had a first alloHCT from an unrelated donor between January 2013 and December 2018, a diagnosis of AML or MDS and were aged 18 years or more with an available donor sample in the Collaborative Biobank. Patients receiving cord blood transplantation were excluded.
Information on the genetic risk and disease stage at transplantation was used to calculate Disease Risk Index (DRI). For this purpose, cytogenetic risk was classified according to the rules for the refined DRI (22) except for chromosome 17p abnormalities which were assigned to the adverse risk group. For patients with missing stage, disease or cytogenetic risk information, DRI group was imputed based on largest frequencies reported in the publication of the refined DRI. The intensity of conditioning regimens was classified according to working definitions of EBMT and CIBMTR (23).
Donor information was mapped to the medical data of the patient using the Donor ID as a key. Information on the HLA-genotype was used to cross-check sample identity by comparing the typing result of the study sample with the original typing results for that donor and by checking HLA-compatibility with the corresponding patient information. HLA compatibility between donors and recipients was assessed based on two-field information for HLA-A, -B, -C, -DRB1 and -DQB1. Donor-recipient pairs, whose HLA-compatibility could not be confirmed, were excluded.
Genotyping was performed using a high-resolution short-amplicon-based next generation sequencing workflow. KIR typing at the allele-level was based on sequencing of exons 3, 4, 5, 7, 8, and 9 and subsequent bioinformatic analysis as described previously (24).
Information on KIR3DL1 and KIR2DS1 and their cognate ligands was grouped according to publications by Venstrom et al. (2012) and Boudreau et al. (2017) (12, 13). Further, we classified donors according to A versus B haplotype motifs using definitions for haplotype assignment as provided by Cooley et al (16, 17). Finally, we calculated scores for selected additive models which integrate information on KIR-KIRL combinations of donor-recipient pairs. We calculated the functional inhibitory KIR count by assigning scores for donor KIR2DL1, KIR2DL2, KIR2DL3, and KIR3DL1 when the cognate ligands were encoded by the patient HLA genotype as described in the original paper by Boelen et al (25). The inhibitory score was calculated according to the formula in Supplement 1 of the original paper.
Scores integrating information on inhibitory and activating KIR-KIRL interactions were proposed by Krieger et al (14). We calculated the missing KIR-ligand Score, the inhibitory KIR-ligand Score and the activating KIR-ligand Score according to Supplementary Table 3 in the original publication (26). Another model to integrate KIR-KIRL interactions based on unsupervised, systematic testing was proposed by Fein et al. recently (27).
Exploratory analyses focused on individuals with homozygous KIR haplotypes to better investigate the effects of certain KIR haplotypes. Homozygosity was determined based on haplotypes defined by absence/presence of KIR genes. Centromeric and telomeric motifs were classified according to Pyo et al. and Jiang et al (28, 29). Allele-level KIR typing results were used to further investigate the allelic composition of homozygous diplotypes. KIR2DL1 and KIR2DL3 alleles were assigned to phylogenetic clades according to Hilton et al (30).
HLA-C alleles were grouped into C1 and C2 ligands and HLA-B alleles were grouped into Bw4-80I/Bw4-80T/Bw6 epitope bearing ligands based on information retrieved from https://www.ebi.ac.uk/ipd/kir/ligand.html (31).
Relapse/progression was the primary endpoint. Event-Free Survival (EFS) and Non-Relapse Mortality (NRM) were secondary endpoints. The study was designed to validate the impact of presence/absence of KIR2DL1, KIR2DL2, and KIR2DL3 genes in the donor KIR genotype against combinations of the cognate ligands C1/C2 encoded in the patient HLA genotype on the risk of relapse in a stringent confirmatory statistical setting (18). In addition, we tested alternative published models, explored new classification attempts and tested effects in subgroups and major secondary endpoints. No adjustment of the type I error for multiple testing was made for validation tests and exploratory tests.
EFS probabilities were calculated with the Kaplan-Meier estimator and between-group comparisons were performed with the log-rank test. Relapse/progression and NRM were mutually considered as competing risks. For the calculation of cumulative incidences of acute and chronic graft-versus-host disease (GVHD), relapse or death were considered as competing events. Univariable comparisons for these endpoints were performed with the Gray test. All time-to-event endpoints were evaluated in (cause-specific) multivariable Cox proportional hazards regression models. Risk adjustment in the context of multivariable regression models included information on patients’ performance status, age, sex, cytomegalovirus (CMV) serostatus, disease risk index, conditioning intensity, platform for GVHD prophylaxis, HLA-matching, donor age, donor sex, and donor CMV serostatus. Effect sizes were reported as hazard ratios together with 95%-confidence intervals.
Mapping of data from donors and patients, who met the inclusion criteria, resulted in 5,017 unrelated donor-recipient pairs for whom donor DNA was available for genotyping. The median patient age at alloHCT was 56 years (interquartile range (IQR) 45 years to 64 years). Indications for alloHCT were de novo AML for 81.2% of patients, secondary or therapy-associated AML for 6.3% of patients and MDS for 12.5% of patients. Disease risk according to the Disease Risk Index was assessed as high or very high for 28% of patients. Donors and patients were HLA-A, -B, -C, -DRB1, and -DQB1 matched in 78.6% of pairs, one locus mismatched in 20.1% of pairs, and two locus mismatched in 1.3% of pairs. HLA B mismatches resulted in KIR ligand changes (Bw4-Bw6) in 1.5% of donor-recipient pairs and HLA C mismatches (C1-C2) in 1.6% of donor-recipient pairs. The median donor age was 28 years (IQR, 24 to 36 years). Myeloablative regimens were administered to 57% of the patients and reduced-intensity regimens to 39%. In this cohort of patients with unrelated donors 52% had received ATG for GVHD-prophylaxis and only 7% of patients had received post-transplant cyclophosphamide (PTCY) for GVHD-prophylaxis. Peripheral Blood Stem Cells (PBSC) and Bone Marrow (BM) were used as graft source in 90% and 10% of patients, respectively. Further details on patient characteristics are given in Table 1.
For the whole cohort, 2-year probabilities were 51% (95%-CI: 49% to 52%) for EFS, 29% (95%-CI: 27% to 30%) for the incidence of relapse/progression, and 20% (95%-CI: 19% to 22%) for NRM. In total, 1,485 relapse events and 1,075 cases of death before observation of relapse or progression were recorded.
First, we attempted to validate associations between selected inhibitory donor KIR allele groups, patient KIRL and the risk of relapse which we had observed in an independent previous study (18). Those own findings could not be replicated in this larger dataset. Donor KIR2DL2 positivity for patients with C1/C1 ligands was not associated with a reduced risk of relapse (HR 1.03, 95%-CI 0.87-1.22, p=.8). And in patients with C1/C2 ligands the presence of KIR2DL1 alleles belonging to phylogenetic clade 2 in the donor KIR genotype was not associated with a reduced risk of relapse (HR 1.1, 95%-CI 0.95-1.28, p=.2).
Next, we attempted to validate alternative donor KIR genotype-based prediction models in the current dataset (see Table 2). These models included the concept to optimize NK alloreactivity by increasing activating KIR2DS1 activity and limiting KIR3DL1-mediated inhibition (12, 13) and scores which were designed to capture functional inhibitory and activating KIR (14, 25). Very recently, common combinations of activating and inhibitory KIR genes were investigated in a hypothesis-free approach in a CIBMTR data set of AML patients (27). The authors found several associations between distinct genotypes and patient outcomes. In our independent data set, those findings could not be validated. Moreover, we could not validate the impact of donor KIR haplotype B motifs in the whole cohort and in C1+ recipients (data not shown) (17, 21). Results of donor KIR genotype classifications in subgroups of patients defined by conditioning intensity, use of TBI, and patient KIRL (C2/C2 versus C1+) are shown in Supplementary Tables S2-S7.
Taking advantage of the large number of donor-recipient pairs, we sought to investigate the impact of homozygous genotypes. Homozygous genotypes allow for a more direct analysis of the impact of a certain haplotype or haplotype motif because interactions with a divergent haplotype do not have to be considered and the gene dosage is uniform. A total of 4,166 donors were homozygous for either centromeric (cen) or telomeric (tel) KIR motifs and 1,967 donors (39.5% of all donors) were homozygous for both, centromeric and telomeric KIR motifs. The distribution of homozygous genotypes classified by A or B motifs is shown in Table 3.
After classification of donors with homozygous KIR genotype, we compared outcome of donor-recipient pairs with respect to event-free survival, and cumulative incidences of relapse and non-relapse mortality (see Figure 1). The small group (N=43) of patients with cen B/B tel B/B KIR donors showed a trend towards a lower risk of relapse (multivariable Cox regression, HR 0.45, 95%-CI 0.2-1.01; p=.052) but EFS was not different (multivariable Cox regression, HR 0.78, 95%-CI 0.49-1.24; p=.3) compared to patients with non cen B/B tel B/B KIR donors. Patients who had donors with cen B/B tel A/A KIR genotypes (N=237) had a trend to better event-free survival (multivariable Cox regression, HR 0.84, 95%-CI 0.70-1.02; p=.08) in multivariate Cox regression modelling compared to donors with non cen B/B tel A/A KIR genotypes. This trend was caused by a significantly reduced risk of NRM (multivariable Cox regression, HR 0.65, 95%-CI 0.47-0.90; p=.001). The entire multivariable model is presented in Supplementary Table S1. Based on this observation, we tried to narrow down the putative beneficial haplotypes by segregating the centromeric B motifs of the cen B/B tel A/A diplotypes into B01 and B02 motifs. B01 and B02 motifs differ by absence or presence of KIR2DL5, KIR2DS3, KIR2DP1, and KIR2DL1 genes. Patients whose donors had cen B01/B01-telA/A diplotypes (N=29) showed better EFS (multivariable Cox regression, HR 0.50, 95%-CI 0.27-0.90; p=.02) due to a lower risk for NRM (multivariable Cox regression, HR 0.40, 95%-CI 0.17-0.97; p=.04) compared to non cen B01/B01-telA/A donors. We observed no differences with respect to the risk of NRM for the subgroups of cen B/B tel A/A donors (Figure 2). Up to day +150 after alloHCT the cumulative incidence of acute GVHD II-IV and III-IV was 28% (95%-CI: 22% to 34%) and 9% (95%-CI: 5% to 13%) for patients with cen B/B tel A/A donors compared to 28% (95%-CI: 26% to 29%) and 10% (95%-CI: 9% to 11%) for controls (Figure 3). Also, no difference was found for the cumulative incidences of chronic GVHD.
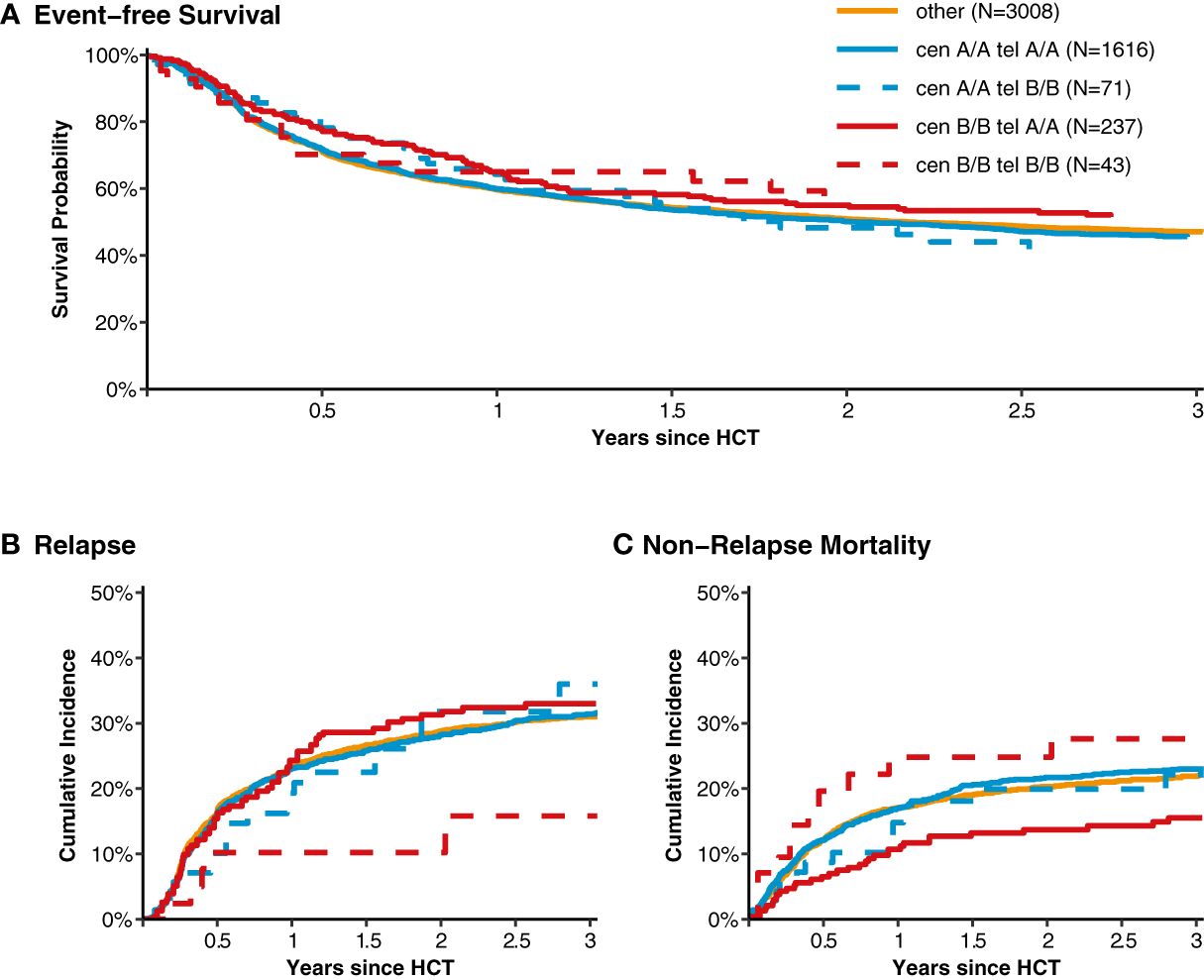
Figure 1 Event-free survival, cumulative incidence of relapse and of non-relapse mortality classified into homozygous versus heterozygous donor KIR diplotypes. Panel (A) shows event-free survival from transplantation for patients with donors whose genotypes were homozygous for the centromeric (cen) and telomeric (tel) KIR gene motifs A/A motifs compared to patients with heterozygous cen & tel KIR gene motifs (displayed in orange). Panels (B, C) show the cumulative incidences of relapse and non-relapse mortality, respectively.
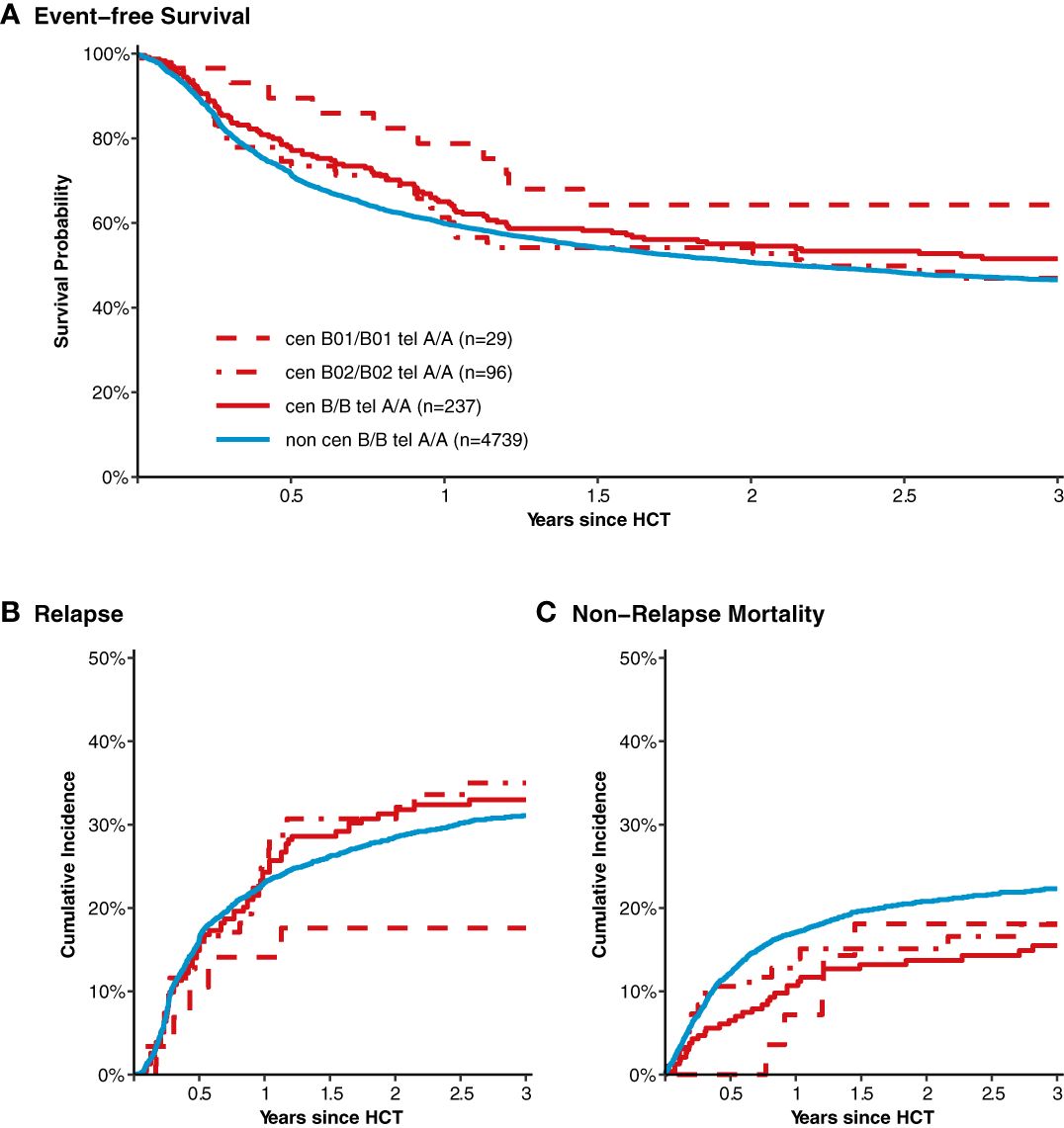
Figure 2 Event-free survival, cumulative incidence of relapse and of non-relapse mortality after transplantation from donors with different homozygous KIR diplotypes. Panel (A) shows event-free survival of patients after transplantation grouped by the donor KIR genotype. Data of patients with segregated homozygous KIR donor genotypes (red curves) are compared to all remaining patients (blue curve). High-resolution donor KIR genotype B motifs were classified into the subgroups B01 and B01. Centromeric B02 motifs differ from Cen B01 by deletion of KIR2DL5, KIR2DS3/5, KIR2DP1, and KIR2DL1 genes. Panels (B, C) show corresponding curves for the cumulative incidences of relapse and non-relapse mortality, respectively.
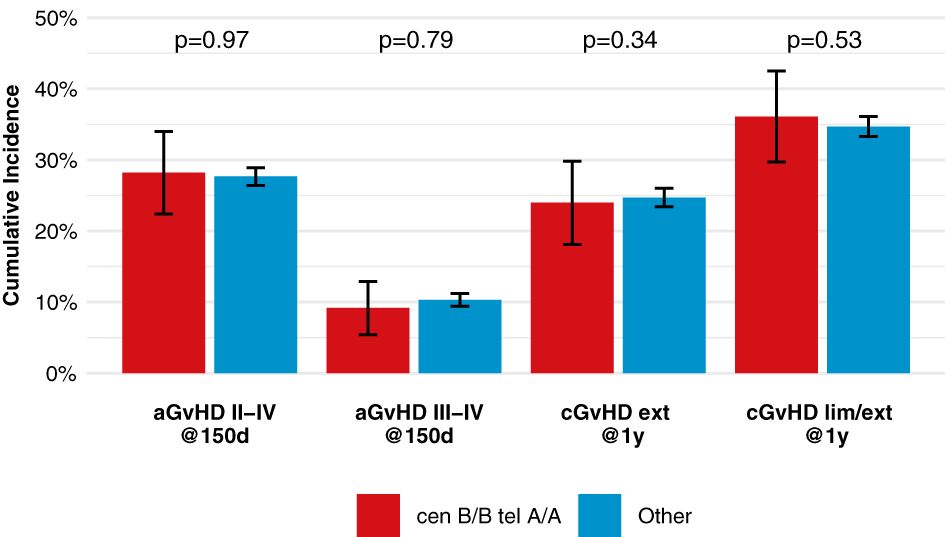
Figure 3 Incidences of acute and chronic GVHD among patients with cen-B/B tel A/A donors, shows point estimates of cumulative incidence curves for acute GVHD on day 150 after transplantation and for chronic GVHD at one year after transplantation for patients with cen-B/B tel A/A donor KIR genotypes compared to all remaining patients.
In order to complement existing association studies and since many scores integrate information on functional inhibitory KIR, we systematically investigated KIR binding to C1 or C2. We classified high-resolution KIR genotypes into phylogenetic clades and tested the impact of the presence versus absence of those KIR with their cognate KIRL. Results of the single KIR – HLA-C-ligand analyses are shown in Table 4. Patients with C2/C2 ligands whose donors were KIR2DL1 phylogenetic clade 3-positive (N=190) showed a lower risk of relapse (HR 0.65, 95%-CI 0.46-0.91, p=.012) and better event-free survival (HR 0.65, 95%-CI 0.50-0.84, p=.001). In contrast, patients with C1/C2 ligands whose donors were KIR2DL1 phylogenetic clade 2-positive (N=1,414) showed a trend toward higher NRM (HR 1.16, 95%-CI 0.97-1.39, p=.096) and worse event-free survival (HR 1.13, 95%-CI 1.00-1.27, p=.042). To provide comprehensive results we present the impact of the presence/absence of all KIR in Supplementary Table S8.
In reference to studies where donor NK-cell transfusions from haploidentical relatives induced remissions of patients with AML, NK alloreactivity is thought to contribute to graft-versus leukemia effects after alloHCT (3, 5–7, 32, 33). Yet, no consistent model exists which allows for the prediction of NK alloreactivity for HLA-matched and –mismatched transplantation. We here present the largest study investigating different classifications for donor KIR-genotype based outcome prediction, so far.
In the confirmatory part of the study, we aimed at validating associations between certain KIR genotypes and patient outcomes, which we had observed in an independent data set (19). The disruption of inhibitory signals for NK cells by down-regulation of KIR ligands from the surface of leukemic blasts is at the core of many predictive models (34, 35). Building on the assumption that phylogenetic clades share different strength of KIR – KIRL binding affinities, we classified KIR2DL1 and KIR2DL3 alleles into their respective clades and showed that certain phylogenetic clades were associated with the risk of relapse in patients with C1/C2-ligand combinations (18, 30). However, the findings of our previous study could not be confirmed in this independent study (see Tables 2, 4). Alternative explanations for the discordant findings could be uncharacterized differences between the patient cohorts resulting in differential impact of NK alloreactivity or that the previous finding was incidental. In an extended set of analyses in the present study (see Table 4), where we investigated systematically all KIR with HLA-C ligands, one association stood out: Patients with C2/C2 ligands whose donors were KIR2DL1 phylogenetic clade 3-positive (N=190) showed a lower risk of relapse (HR 0.65, 95%-CI 0.46-0.91, p=.012). This association should be validated in an independent study.
Further, we investigated homozygous KIR genotypes building on previous work to define KIR haplotypes (36). Our motivation for this approach was that with less complexity in the setting of homozygosity, signals could be more easily detected. Furthermore, this approach appeared as a logical extension of KIR genotype classifications building on KIR haplotype motifs (16, 21, 37). Notably, we did not find significant associations between cen B/B motifs and a reduced risk of relapse. However, patients with donors who were homozygous for the cen B/tel A haplotype had a trend to better EFS (HR 0.84, p=.08) and lower NRM (HR 0.65, p=.01). The lower risk of NRM could not be linked to a reduced risk of GVHD. Further investigating the role of the B01 and B02 subtypes showed a reduced risk of relapse only for patients with homozygous cen B01/tel A genotypes. Since the centromeric B01 motif contains KIR2DL1 clade 3 alleles, this finding partly reflects the association reported above (30, 38). Yet, it has to be noted that the results are not in line with a smaller study of 890 donor-recipient pairs which reported that B02 protects better against relapse than B01 (37). Moreover, Weisdorf et al. reported that cen B motifs were associated with a reduced risk of relapse in C1/C1 or C1/C2 patients but not in C2/C2 patients (21). We did not observe this subset effect in exploratory analyses (Supplementary Tables S6, 7). However, since Weisdorf et al. did not distinguish between B01 and B02 motifs for their analysis, the impact cannot be assigned to specific centromeric B motifs.
We also attempted to validate published models for outcome prediction after mostly matched unrelated donor transplantation (see Table 2). None of the classifications predicted the risk of relapse and EFS significantly as originally published. We found a lower risk of relapse among patients who had received reduced intensity or non-myeloablative conditioning and had donors with two centromeric B motifs (Supplementary Table S3). However, this subgroup effect was not observed in a previous study on 1,140 patients with MDS or secondary AML, who had received reduced-intensity or non-myeloablative conditioning [Supplementary Table S2A of Schetelig et al., 2021 (20)]. So, this finding should be interpreted with caution.
Given the disappointing validation studies, it may help to reflect on the specific challenges in this research field. First, small animal models to explore human NK alloreactivity after transplantation, do not exist and cytotoxicity assays do not mimic the complex process of NK cell education. Yet, the plasticity of NK cells is remarkable and averts autoimmunity reliably. As an example, until now, it is not clear if in the setting of alloHCT functional inhibitory KIR, i.e. KIR who encounter their cognate ligand, exert more powerful anti-leukemic effects than non-functional KIR. Further, still not all KIR-ligands are known. Second, the diversity of KIR genotypes is substantial. With the availability of high-resolution KIR genotyping the number of alleles increased substantially and, as of now, more than 1,600 distinct KIR alleles have been described (24, 31). When combined with compound KIR-ligands (C1, C2, Bw4 and Bw6), hundreds of possibilities may be tested. Ambiguity in some genotype calls further hampers optimal statistical analysis of the effect of KIR haplotypes on outcomes. Recently, an expectation maximization-based algorithm has been proposed to take these ambiguous values properly into account in the analysis, leading to improved estimates of haplotype frequencies. However, even then the complexity of the KIR infrastructure makes obtaining unbiased effect estimates very difficult (39). This problem is further aggravated since it is unclear, if NK alloreactivity interacts with conditioning intensity, the use of certain drugs or total-body irradiation, the type of GVHD-prophylaxis, or the graft source (21). The poor understanding of human NK cell biology spawned multiple classifications and hypotheses on patient subsets more susceptible to NK allo reactivity. This setting constitutes a multiplicity problem which is hard to control. Finally, the extent to which peptides displayed in the HLA groove modify KIR–binding has been elicited only rudimentarily, but could be a major force which determines graft-versus-leukemia effects (40, 41). As a consequence, we believe that prediction models have to build on rigorous confirmatory testing in independent, adequately powered studies. Whether artificial intelligence could help resolving the problem, is unknown. While artificial intelligence is a powerful tool to create complex classifications when abundant data is available it does not resolve the multiplicity problem when data are scarce.
In three independent studies, we analysed data on 8,943 patients with AML or MDS whose donors had been typed for KIR genes at the allele-level (19, 20). These data can be accessed for validation studies of other research groups. However, future large studies will be critical. The current shift to post transplant cyclophosphamide (PTCY)-based GVHD-prophylaxis might have an important impact on NK alloreactivity, since cyclophosphamide eliminates most mature donor NK cells infused with the graft (42). Further studies are thus warranted to unravel PTCY-associated changes of NK alloreactivity after alloHCT.
In summary, despite the availability of KIR genotype information for more than 3 million stem cell donors, no donor KIR-genotype based algorithm for unrelated donor selection can be recommended for clinical practice. It is still not possible to predict patient outcome based on donor KIR genotype information at present.
The dataset of this study may be accessed by academic research groups beginning 12 months and ending 48 months following article publication. Access is conditional on approval by the responsible registries and signed data transfer agreements. Three organizations are responsible for the data, the Cellular Therapy & Immunobiology Working Party (CTIWP) of EBMT, the Immunobiology Working Committee of the Center for International Blood and Marrow Transplant Research (CIBMTR), and the Collaborative Biobank (www.cobi-biobank.com).
All stem cell donors had provided written informed consent when they contributed a sample to the biobank. Donor information was linked to patient outcome data with a unique donor identifier. The study was approved by the Ethical Committee at the Technische Universtität (TU) Dresden, Germany, the Cellular Therapy and Immunobiology Working Party of EBMT, and the Institutional Review Board of the National Marrow Donor Program (NMDP; on behalf of CIBMTR).
JSc: Writing – original draft, Conceptualization, Investigation, Methodology, Project administration, Writing – review & editing. HB: Writing – original draft, Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Software, Visualization, Writing – review & editing. FH: Writing – review & editing, Conceptualization, Investigation, Methodology, Supervision. JH: Writing – review & editing, Data curation. SS: Writing – review & editing, Conceptualization, Data curation, Investigation, Methodology, Project administration, Supervision. AK: Writing – review & editing, Data curation, Investigation. TS: Writing – review & editing, Data curation, Investigation. HS: Writing – review & editing, Data curation, Investigation. PD: Writing – review & editing, Data curation, Investigation. EF: Writing – review & editing, Data curation, Investigation. JV: Writing – review & editing, Data curation, Investigation. EW-D: Writing – review & editing, Data curation, Investigation. GC: Writing – review & editing, Data curation, Investigation. ShP: Writing – review & editing, Data curation, Investigation. NM: Writing – review & editing, Data curation, Investigation. AT: Writing – review & editing, Data curation, Investigation. LW: Writing – review & editing, Data curation, Investigation, Methodology, Supervision. VL: Writing – review & editing, Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Project administration, Supervision. AS: Writing – review & editing, Conceptualization, Investigation, Project administration, Resources, Supervision. JSa: Writing – review & editing, Conceptualization, Data curation, Investigation, Methodology, Supervision. JF: Writing – review & editing, Conceptualization, Investigation, Methodology. Y-TB: Writing – review & editing, Data curation, Investigation. MH: Writing – review & editing, Data curation, Investigation. SM: Writing – review & editing, Conceptualization, Investigation, Supervision. SG: Writing – review & editing, Conceptualization, Investigation, Supervision. SoP: Writing – review & editing, Conceptualization, Investigation, Supervision. AR: Writing – review & editing, Conceptualization, Investigation, Supervision. CC: Writing – review & editing, Conceptualization, Investigation, Supervision. KF: Writing – review & editing, Conceptualization, Data curation, Investigation, Methodology, Supervision.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was enabled by DKMS Group who financed donor KIR typing. The CIBMTR is supported primarily by Public Health Service U24CA076518 from the National Cancer Institute (NCI), the National Heart, Lung and Blood Institute (NHLBI) and the National Institute of Allergy and Infectious Diseases (NIAID); 75R60222C00011 from the Health Resources and Services Administration (HRSA); and N00014-21-1-2954 and N00014-23-1-2057 from the Office of Naval Research. The views expressed in this article do not reflect the official policy or position of the National Institute of Health, the Department of the Navy, the Department of Defense, Health Resources and Services Administration (HRSA) or any other agency of the U.S. Government.
We are grateful to the patients and their families for contributing data on their clinical course. Also, we would like to thank all center data managers at EBMT and CIBMTR for facilitating analyses of that kind.
All authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer TT declared shared affiliation with the author’s JSc and FH to the handling editor at the time of review.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1350470/full#supplementary-material
1. Ruggeri L, Capanni M, Urbani E, Perruccio K, Shlomchik WD, Tosti A, et al. Effectiveness of donor natural killer cell alloreactivity in mismatched hematopoietic transplants. Science. (2002) 295:2097–100. doi: 10.1126/science.1068440
2. Miller JS, Soignier Y, Panoskaltsis-Mortari A, McNearney SA, Yun GH, Fautsch SK, et al. Successful adoptive transfer and in vivo expansion of human haploidentical NK cells in patients with cancer. Blood. (2005) 105:3051–7. doi: 10.1182/blood-2004-07-2974
3. Shaffer BC, Le Luduec JB, Forlenza C, Jakubowski AA, Perales MA, Young JW, et al. Phase ii study of haploidentical natural killer cell infusion for treatment of relapsed or persistent myeloid Malignancies following allogeneic hematopoietic cell transplantation. Biol Blood Marrow Transplant. (2016) 22:705–9. doi: 10.1016/j.bbmt.2015.12.028
4. Romee R, Rosario M, Berrien-Elliott MM, Wagner JA, Jewell BA, Schappe T, et al. Cytokine-induced memory-like natural killer cells exhibit enhanced responses against myeloid leukemia. Sci Transl Med. (2016) 8:357ra123. doi: 10.1126/scitranslmed.aaf2341
5. Bjorklund AT, Carlsten M, Sohlberg E, Liu LL, Clancy T, Karimi M, et al. Complete remission with reduction of high-risk clones following haploidentical NK-cell therapy against MDS and AML. Clin Cancer Res. (2018) 24:1834–44. doi: 10.1158/1078-0432.CCR-17-3196
6. Parisi S, Ruggeri L, Dan E, Rizzi S, Sinigaglia B, Ocadlikova D, et al. Long-term outcome after adoptive immunotherapy with natural killer cells: alloreactive NK cell dose still matters. Front Immunol. (2021) 12:804988. doi: 10.3389/fimmu.2021.804988
7. Shapiro RM, Birch GC, Hu G, Vergara Cadavid J, Nikiforow S, Baginska J, et al. Expansion, persistence, and efficacy of donor memory-like NK cells infused for posttransplant relapse. J Clin Invest. (2022) 132. doi: 10.1172/jci154334
8. Giebel S, Locatelli F, Lamparelli T, Velardi A, Davies S, Frumento G, et al. Survival advantage with KIR ligand incompatibility in hematopoietic stem cell transplantation from unrelated donors. Blood. (2003) 102:814–9. doi: 10.1182/blood-2003-01-0091
9. Hsu KC, Keever-Taylor CA, Wilton A, Pinto C, Heller G, Arkun K, et al. Improved outcome in HLA-identical sibling hematopoietic stem-cell transplantation for acute myelogenous leukemia predicted by KIR and HLA genotypes. Blood. (2005) 105:4878–84. doi: 10.1182/blood-2004-12-4825
10. Miller JS, Cooley S, Parham P, Farag SS, Verneris MR, McQueen KL, et al. Missing KIR ligands are associated with less relapse and increased graft-versus-host disease (GVHD) following unrelated donor allogeneic HCT. Blood. (2007) 109:5058–61. doi: 10.1182/blood-2007-01-065383
11. Cooley S, Trachtenberg E, Bergemann TL, Saeteurn K, Klein J, Le CT, et al. Donors with group B KIR haplotypes improve relapse-free survival after unrelated hematopoietic cell transplantation for acute myelogenous leukemia. Blood. (2009) 113:726–32. doi: 10.1182/blood-2008-07-171926
12. Venstrom JM, Pittari G, Gooley TA, Chewning JH, Spellman S, Haagenson M, et al. HLA-C-dependent prevention of leukemia relapse by donor activating KIR2DS1. N Engl J Med. (2012) 367:805–16. doi: 10.1056/NEJMoa1200503
13. Boudreau JE, Giglio F, Gooley TA, Stevenson PA, Le Luduec JB, Shaffer BC, et al. KIR3DL1/Hl a-B Subtypes Govern Acute Myelogenous Leukemia Relapse after Hematopoietic Cell Transplantation. J Clin Oncol. (2017) 35:2268–78. doi: 10.1200/JCO.2016.70.7059
14. Krieger E, Sabo R, Moezzi S, Cain C, Roberts C, Kimball P, et al. Killer immunoglobulin-like receptor-ligand interactions predict clinical outcomes following unrelated donor transplantations. Biol Blood Marrow Transplant. (2020) 26:672–82. doi: 10.1016/j.bbmt.2019.10.016
15. Krieger E, Qayyum R, Keating A, Toor A. Increased donor inhibitory KIR with known HLA interactions provide protection from relapse following HLA matched unrelated donor HCT for aml. Bone Marrow Transplant. (2021) 56:2714–22. doi: 10.1038/s41409-021-01393-9
16. Cooley S, Weisdorf DJ, Guethlein LA, Klein JP, Wang T, Le CT, et al. Donor selection for natural killer cell receptor genes leads to superior survival after unrelated transplantation for acute myelogenous leukemia. Blood. (2010) 116:2411–9. doi: 10.1182/blood-2010-05-283051
17. Cooley S, Weisdorf DJ, Guethlein LA, Klein JP, Wang T, Marsh SG, et al. Donor killer cell ig-like receptor B haplotypes, recipient HLA-C1, and HLA-C mismatch enhance the clinical benefit of unrelated transplantation for acute myelogenous leukemia. J Immunol. (2014) 192:4592–600. doi: 10.4049/jimmunol.1302517
18. Schetelig J, Baldauf H, Schaefer G, Frank S, Sauter J, Stelljes M, et al eds. The Impact of Donor KIR2DL Allele Combinations on Outcome after Unrelated Donor Transplantation in Patients with Acute Myeloid Leukemia. In: The 18th meeting of the Society for Natural Immunity. Luxembourg.
19. Schetelig J, Baldauf H, Heidenreich F, Massalski C, Frank S, Sauter J, et al. External validation of models for KIR2DS1/KIR3DL1-informed selection of hematopoietic cell donors fails. Blood. (2020) 135:1386–95. doi: 10.1182/blood.2019002887
20. Schetelig J, Baldauf H, Koster L, Kuxhausen M, Heidenreich F, de Wreede LC, et al. Haplotype motif-based models for KIR-genotype informed selection of hematopoietic cell donors fail to predict outcome of patients with myelodysplastic syndromes or secondary acute myeloid leukemia. Front Immunol. (2020) 11:584520. doi: 10.3389/fimmu.2020.584520
21. Weisdorf D, Cooley S, Wang T, Trachtenberg E, Vierra-Green C, Spellman S, et al. KIR B donors improve the outcome for aml patients given reduced intensity conditioning and unrelated donor transplantation. Blood Adv. (2020) 4:740–54. doi: 10.1182/bloodadvances.2019001053
22. Armand P, Kim HT, Logan BR, Wang Z, Alyea EP, Kalaycio ME, et al. Validation and refinement of the disease risk index for allogeneic stem cell transplantation. Blood. (2014) 123:3664–71. doi: 10.1182/blood-2014-01-552984
23. Bacigalupo A, Ballen K, Rizzo D, Giralt S, Lazarus H, Ho V, et al. Defining the intensity of conditioning regimens: working definitions. Biol Blood Marrow Transplant. (2009) 15:1628–33. doi: 10.1016/j.bbmt.2009.07.004
24. Wagner I, Schefzyk D, Pruschke J, Schofl G, Schone B, Gruber N, et al. Allele-level KIR genotyping of more than a million samples: workflow, algorithm, and observations. Front Immunol. (2018) 9:2843. doi: 10.3389/fimmu.2018.02843
25. Boelen L, Debebe B, Silveira M, Salam A, Makinde J, Roberts CH, et al. Inhibitory killer cell immunoglobulin-like receptors strengthen Cd8(+) T cell-mediated control of HIV-1, HCV, and HTLV-1. Sci Immunol. (2018) 3. doi: 10.1126/sciimmunol.aao2892
26. Krieger E, Sabo R, Moezzi S, Cain C, Roberts C, Kimball P, et al. Killer immunoglobulin-like receptor-ligand interactions predict clinical outcomes following unrelated donor transplantations. Biol Blood Marrow Transplant. (2019) 26(4):672–82. doi: 10.1016/j.bbmt.2019.10.016
27. Fein JA, Shouval R, Krieger E, Spellman SR, Wang T, Baldauf H, et al. Systematic evaluation of donor-KIR/recipient-HLA interactions in HLA-matched hematopoietic cell transplantation for AML. Blood Adv. (2024) 8:581–90. doi: 10.1182/bloodadvances.2023011622
28. Pyo CW, Guethlein LA, Vu Q, Wang R, Abi-Rached L, Norman PJ, et al. Different patterns of evolution in the centromeric and telomeric regions of group a and B haplotypes of the human killer cell ig-like receptor locus. PloS One. (2010) 5:e15115. doi: 10.1371/journal.pone.0015115
29. Jiang W, Johnson C, Jayaraman J, Simecek N, Noble J, Moffatt MF, et al. Copy number variation leads to considerable diversity for B but not a haplotypes of the human KIR genes encoding NK cell receptors. Genome Res. (2012) 22:1845–54. doi: 10.1101/gr.137976.112
30. Hilton HG, Guethlein LA, Goyos A, Nemat-Gorgani N, Bushnell DA, Norman PJ, et al. Polymorphic HLA-C receptors balance the functional characteristics of KIR haplotypes. J Immunol. (2015) 195:3160–70. doi: 10.4049/jimmunol.1501358
31. Robinson J, Halliwell JA, McWilliam H, Lopez R, Marsh SG. Ipd–the immuno polymorphism database. Nucleic Acids Res. (2013) 41:D1234–40. doi: 10.1093/nar/gks1140
32. Ciurea SO, Kongtim P, Soebbing D, Trikha P, Behbehani G, Rondon G, et al. Decrease post-transplant relapse using donor-derived expanded NK-cells. Leukemia. (2022) 36:155–64. doi: 10.1038/s41375-021-01349-4
33. Bednarski JJ, Zimmerman C, Berrien-Elliott MM, Foltz JA, Becker-Hapak M, Neal CC, et al. Donor memory-like NK cells persist and induce remissions in pediatric patients with relapsed aml after transplant. Blood. (2022) 139:1670–83. doi: 10.1182/blood.2021013972
34. Verheyden S, Ferrone S, Mulder A, Claas FH, Schots R, De Moerloose B, et al. Role of the inhibitory KIR ligand HLA-bw4 and HLA-C expression levels in the recognition of leukemic cells by natural killer cells. Cancer Immunol Immunother. (2009) 58:855–65. doi: 10.1007/s00262-008-0601-7
35. Mastaglio S, Wong E, Perera T, Ripley J, Blombery P, Smyth MJ, et al. Natural killer receptor ligand expression on acute myeloid leukemia impacts survival and relapse after chemotherapy. Blood Adv. (2018) 2:335–46. doi: 10.1182/bloodadvances.2017015230
36. Solloch UV, Schefzyk D, Schafer G, Massalski C, Kohler M, Pruschke J, et al. Estimation of german KIR allele group haplotype frequencies. Front Immunol. (2020) 11:429. doi: 10.3389/fimmu.2020.00429
37. Guethlein LA, Beyzaie N, Nemat-Gorgani N, Wang T, Ramesh V, Marin WM, et al. Following transplantation for acute myelogenous leukemia, donor KIR cen B02 better protects against relapse than KIR cen B01. J Immunol. (2021) 206:3064–72. doi: 10.4049/jimmunol.2100119
38. Vierra-Green C, Roe D, Hou L, Hurley CK, Rajalingam R, Reed E, et al. Allele-level haplotype frequencies and pairwise linkage disequilibrium for 14 kir loci in 506 european-american individuals. PloS One. (2012) 7:e47491. doi: 10.1371/journal.pone.0047491
39. van der Burg LLJ, de Wreede LC, Baldauf H, Sauter J, Schetelig J, Putter H, et al. Haplotype reconstruction for genetically complex regions with ambiguous genotype calls: illustration by the KIR gene region. Genet Epidemiol. (2024) 48:3–26. doi: 10.1002/gepi.22538
40. Sim MJ, Malaker SA, Khan A, Stowell JM, Shabanowitz J, Peterson ME, et al. Canonical and cross-reactive binding of NK cell inhibitory receptors to HLA-C allotypes is dictated by peptides bound to HLA-C. Front Immunol. (2017) 8:193. doi: 10.3389/fimmu.2017.00193
41. Hilton HG, Parham P. Missing or altered self: human NK cell receptors that recognize HLA-C. Immunogenetics. (2017) 69:567–79. doi: 10.1007/s00251-017-1001-y
Keywords: killer-cell immunoglobulin-like receptor (KIR), allogeneic hematopoietic cell transplantation (alloHCT), risk of relapse, donor selection, prediction model
Citation: Schetelig J, Baldauf H, Heidenreich F, Hoogenboom JD, Spellman SR, Kulagin A, Schroeder T, Sengeloev H, Dreger P, Forcade E, Vydra J, Wagner-Drouet EM, Choi G, Paneesha S, Miranda NAA, Tanase A, de Wreede LC, Lange V, Schmidt AH, Sauter J, Fein JA, Bolon Y-T, He M, Marsh SGE, Gadalla SM, Paczesny S, Ruggeri A, Chabannon C and Fleischhauer K (2024) Donor KIR genotype based outcome prediction after allogeneic stem cell transplantation: no land in sight. Front. Immunol. 15:1350470. doi: 10.3389/fimmu.2024.1350470
Received: 05 December 2023; Accepted: 04 March 2024;
Published: 02 April 2024.
Edited by:
David Wald, Case Western Reserve University, United StatesReviewed by:
Torsten Tonn, Technical University Dresden, GermanyCopyright © 2024 Schetelig, Baldauf, Heidenreich, Hoogenboom, Spellman, Kulagin, Schroeder, Sengeloev, Dreger, Forcade, Vydra, Wagner-Drouet, Choi, Paneesha, Miranda, Tanase, de Wreede, Lange, Schmidt, Sauter, Fein, Bolon, He, Marsh, Gadalla, Paczesny, Ruggeri, Chabannon and Fleischhauer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Johannes Schetelig, Sm9oYW5uZXMuU2NoZXRlbGlnQHVrZGQuZGU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.