- School of Life Health and Chemical Sciences, The Open University, Milton Keynes, United Kingdom
Invasive fungal diseases have profound effects upon human health and are on increase globally. The World Health Organization (WHO) in 2022 published the fungal priority list calling for improved public health interventions and advance research. Drosophila melanogaster presents an excellent model system to dissect host-pathogen interactions and has been proved valuable to study immunopathogenesis of fungal diseases. In this review we highlight the recent advances in fungal-Drosophila interplay with an emphasis on the recently published WHO’s fungal priority list and we focus on available tools and technologies.
Introduction
Fungal infections
The global impact of opportunistic fungal infections has gone underrecognized for a long time (1). However, with the increase in chronic and immunosuppressive health conditions including HIV/AIDS, cancer, cystic fibrosis and diabetes, antimicrobial therapies and invasive procedures that leave individuals vulnerable to opportunistic infections, the impact of these infections are becoming more apparent (1, 2). Fungi cause disease through direct infection of the host or through their secondary metabolites, mycotoxins, pigments that can contaminate the environment, food products and air (3). The disease burden ranges from superficial to invasive fungal infections and is estimated to be in the 100s of millions of patients per year, resulting in >1.5 million deaths/year (2, 4). These infections are caused by long recognised pathogens such as Aspergillus fumigatus and Candida albicans (5–7), neglected tropical diseases like eumycetoma (8, 9), and newly emerged pathogens, such as Candida auris (10, 11).
With the development of advanced molecular and cellular biology technologies, fungal pathogenicity and virulence factors are being studied in greater detail (12–14). However, the fungal threat continues to grow while the development of novel effective antifungal therapies remains inadequate (15, 16). As a result, in 2022 the WHO published the WHO fungal priority pathogens list, classifying 18 medically relevant fungal species as “Critical”, “High” or “Medium” priority, according to the perceived public health burden (2, 17).
Model organisms
The use of model organisms is one of the technologies that has been developing over time and has become indispensable to investigating the nuances of host-pathogen interactions (18, 19). A cursory search of PubMed using the keywords “Drosophila” AND “fungi” yielded 8,617 results (1948 – 2023), with over a third (36.2%) of the publications having been released in the last decade alone. Seminal proof of concept studies in the 1990s and early 2000s, utilising wild-type and mutant Drosophila strains and fungi, provided a comprehensive framework for employing Drosophila in fungal research (20–25). Over the last decade, more extensive Drosophila-fungi work has taken place, leading to a better understanding of virulence, pathogenicity, and host immune responses (26–30).
Purpose of review
This review sets out to provide a brief update on tools currently being applied to host-fungal interaction studies in Drosophila and highlight examples of research in the last 5 years with a focus on the WHO’s fungal priority pathogens list (17).
Drosophila as the model organism
Drosophila, affectionately dubbed the biology “work horse”, has been used in fundamental biology, inbreeding and heredity studies since the early 1900s (27, 31) and has led to substantial contributions to our understanding of genetics, cellular biology, neurobiology and immunology (31, 32). Of note, the discovery of Drosophila Toll receptor nearly 3 decades ago elucidated the function of the analogous mammalian Toll-like receptor (TLR) pathway, which is indispensable to innate immunity (20; Lemaitre, 21, 26). Drosophila genome can be genetically manipulated, and genome-wide studies performed to determine genes crucial for survival and infection (27, 33). 75% of the genes responsible for human diseases have a homologue identified in Drosophila genome, an observation that highlights Drosophila’s suitability as a model for the study of mammalian disease conditions (34, 35).
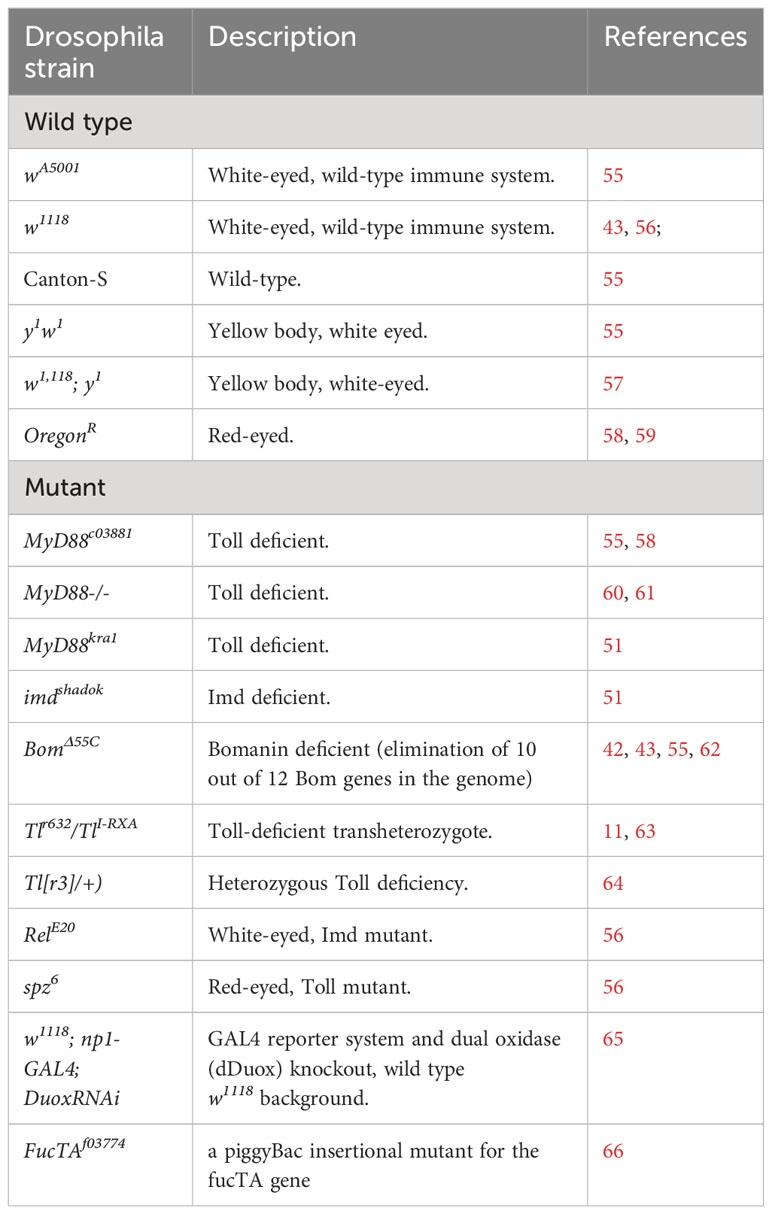
Drosophila immunity relies on the innate immune system, made up of cellular and acellular components and regulatory pathways (Figure 1) (36, 37).These have been traditionally siloed into the humoral and cellular responses, though recent studies have shown that there is considerable crosstalk between the two branches (38, 39). Drosophila shares the following conserved innate immune pathways with vertebrates: the Toll and IMD NF-κB signalling pathways, the JNK pathway and the JAK/STAT pathway (40, 41). The Toll pathway responds to fungi and Gram-positive bacteria, while IMD responds to Gram-negative bacteria (40, 42). These pathways are activated by the recognition of pathogen antigens and host cell damage, and result in the production of effector molecules necessary for eliminating pathogens, autophagy and cellular repair, and immunomodulation as well as other Drosophila-induced Immune Molecules (DIMs) yet to be characterized fully (43, 44). These effectors have not yet been fully identified, but include antimicrobial peptides (AMPs), Boms (encoded by Bomanins), Daisho peptides (39, 45). AMPs are small, positively charged peptides that interact with hydrophobic regions of microbial cells walls and cause cell wall degradation and microbial death and are secreted into the haemolymph by the fat body (45, 46). In addition to AMPs, reactive oxygen species (ROS) are produced by Dual Oxidase (DUOX) and NADPH (Nox) at the epithelial cells (26, 32). The humoral response also provides protection against viral attack through RNAi and autophagy processes (36, 47).
Figure 1 A simplified schematic overview of Drosophila melanogaster innate immune response to challenges by bacteria, viruses, fungi, or parasites, and to damage induced by stress or wounding. The immune responses are clustered by response type and location. (A) Cellular immunity in the hemocoel is mediated by crystal cells, plasmatocytes and lamellocytes, which are involved in, melanisation, phagocytosis, and encapsulation, respectively. (B) Humoral immunity in the haemolymph is mediated by the activation of signalling cascades in the Toll, Immune deficient (IMD) and JNK, JAK/STAT and mRNA degradation pathways following the recognition of pathogens and their virulence factors. It results in the production of a range of effector molecules including antimicrobial peptides (AMPs), clotting factors, and serine proteases. (C) The gut epithelium functions as an immune organ in response to pathogens and stress damage though the following responses: The JAK/STAT pathway responds to damage to increased proliferation of intestinal stem cells (ISC). The IMD pathway in response to bacteria presence in the gut leads to the production of AMPs. Finally bacterial-derived uracil induces the generation of reactive oxygen species (ROS) through the dual-oxidase (DUOX) and the NADPH oxidase (NOX). Dashed arrows represent additional steps involved in the signalling cascade, transcription, and translation, involved in the immune response.
Drosophila cellular immunity is mediated by the blood cell system which comprises of three differentiated populations. The major class of hemocytes are plasmatocytes which are considered equivalent to vertebrate macrophages. More than 90% of all hemocytes are plasmatocytes in every developmental stage of Drosophila (aside from the early-stage embryo) and they are responsible for the disposal of both microorganism and apoptotic cells. Another class are the crystal cells which are responsible for the melanisation in larvae. They contain the enzyme prophenoloxidase a key enzyme in melanin biosynthesis which is released upon rupture of the crystal cells. The third class refers to the lamellocytes, they are rare, but their number increases following oviposition by parasitoid wasps 48, 49). Haematopoiesis occurs at two different stages of ontogenesis: a first population derives from the head mesoderm during the stage of early embryogenesis, and a following second population that arises from the mesodermal lymph gland at a later stage of development (50).
Drosophila antifungal immune responses rely heavily on the Toll pathway (37, 51). Toll signalling is activated by the binding of the surface antigen β-glucan to Drosophila recognition receptor Gram-negative binding protein 3 (GNBP3) and activates Toll through the activity of the Spätzle ligand and subsequent signalling cascade (51, 52). The Toll signalling cascade is also activated by the cleavage of the haemolymph serine protease Persephone by fungal enzymes contributing to the subsequent downstream activity of the Toll pathway (22, 53). The signalling cascade results in the production of specific AMPs, including Drosomycin, Daisho, Defensin and Metchnikowin, circulated in the hemolymph and the activation of the melanisation cascade to help resist the infection (42, 43, 45, 53).
Application of Drosophila to human fungal pathogens
Tools available for Drosophila-fungal studies
Drosophila is currently being utilised to investigate how medically relevant fungi interact with host immunity, and how they transition from colonization to infection (26–30). Wild-type and genetic mutant strains (e.g., Toll-deficient) are commercially available for distribution across the world from stock centres, such as the Bloomington Drosophila Stock Centre and Kyoto Stock Centre (54, 55). Table 1 summarises Drosophila strains used in fungal research studies included in the current review. The Drosophila Genomics Resource Centre and ATCC are some of the suppliers who distribute Drosophila cell lines, like Schneider’s Drosophila Line 2 (S2) cell line, GFP-tagged cells, and cells from various organs for ex vivo studies (67, 68). These fly strains and cell lines are relatively inexpensive to purchase and maintain, increasing accessibility of the model (68). The model systems are infected or exposed to fungi, fungal secondary metabolites, and antifungal compounds to investigate these interactions (26, 55) via feeding, rolling over, or co-culture and in a standardised manner via needle pricking or microinjection, allowing for rapid inoculation of experimental groups (57, 58). Infection progression can be measured through survival, microbial load, mRNA quantification, melanisation and microscopy assays (55, 64). The efficacy and toxicity of antimicrobial compound screens can be measured in similar ways to determine their efficacy and toxicity (69, 70). Examples of microscopy techniques include confocal microscopy for visualising phagocytosis in fungi-stimulated plasmatocytes (71), electron microscopy for imaging effects of treatment on host cell morphology (35) and fungal burden (72). Immunofluorescence staining and bioluminescence allow visualisation of individual cell types in tissue, larvae or adult flies, and can be done through RNA in situ hybridization (30, 73, 74), intravital 2-d photon microscopy and reporter systems (GFP, lacZ) (62).
The development of molecular techniques including DNA and RNA sequencing, RNAi gene silencing and CRISPR/Cas9 has advanced the field in leaps and bounds. Molecular techniques have made it possible to sequence the Drosophila genome (75); sequence coding and non-coding RNA and determine functionality through RNAi-based screening assays and gene silencing or overexpression (66, 76, 77). They facilitate quantification of messenger RNA (mRNA) or transfer RNA (tRNA) through quantitative PCR, reverse transcriptase PCR and modified-induced misincorporation tRNA sequencing (mim-tRNASeq) (78, 79). The gene editing tool CRISPR/Cas9 utilises guide RNA which matches with target gene (CRISPR) and CRISPR-associated protein 9 (Cas9), an endonuclease which helps to break the dsDNA and facilitate editing of the target gene (80) and it can be used for loss-of-function studies (12, 45, 81). Bioinformatics tools have been developed and adapted for genomic studies across microbial, Drosophila and human genomes and these allow for rapid screening of genomes and vast publicly available pathogen and fly data for potential targets for further study (79, 82). These software tools coupled with publicly accessible databases such as Drosophila Evolution over Space and Time (DEST), FlyRNAi (Drosophila RNAi Screening Center and Transgenic RNAi Project (DRSC/TRiP)) and FlyBase form a powerful computational component of the Drosophila tool kit (67, 82, 83).
With a focus on the WHO priority pathogens, we will highlight some examples of how Drosophila has been used to address key questions around host-fungal pathogen interactions, immunology and drug interactions with a focus on developments in the last five years.
Critical priority group
The WHO classified Cryptococcus neoformans, A. fumigatus, C. albicans, and the recently emerged C. auris as “Critical” pathogens. A. fumigatus is a filamentous, airborne pathogen that causes invasive aspergillosis, in vulnerable populations, like cystic fibrosis patients (32, 84). Drosophila has been used to study A. fumigatus pathogenesis since as far back as 2005 (85, 86). In 2010, Chamilos and colleagues showed that pathogenicity of A. fumigatus strains in a Toll-deficient fly model was comparable to that in a mice model (25). Since then, the fly model has been used to study the virulence of A. fumigatus mating types, effects of fungal volatile organic compounds on larval development and comparative pathogenicity of Aspergillus strains collected from diverse sources (environmental, clinical, airborne) (29, 57, 87).
Fungi produce volatile organic compounds (VOCs) which are easily vapourised, carbon-based compounds made up of “alcohols, aldehydes, acids, ethers, esters, ketones, terpenes, thiols and their derivatives” (56, 57, 88). A study of the effects of A. fumigatus VOCs in Drosophila was carried out over a 15-day period, by co-culturing the fungi and fly model. Quantitative measurements of VOCs production showed that greater volumes of VOCs were secreted when A. fumigatus was cultivated at 37°C, than at the fly’s preferred incubation temperature of 25°C (57, 89). In addition, exposure to VOCs resulted in varying levels of toxicity, ranging from mild to severe, including reduced speed and success rate of metamorphosis or death of 3rd instar larvae (57, 90). Gas-chromatography mass spectrometry analysis of A. fumigatus VOCs detected isopentyl alcohol 1-octen-3-ol at the highest volume (57). A Drosophila infection model was subsequently used to show that 1-octen-3-ol caused greater sensitivity in male than female flies, resulting in reduced dehydrogenase activity and nitric oxide production, and increased ROS production (35). The connection to sex may explain the similar sensitivity distribution witnessed in humans postexposure to mould (35, 91). The Drosophila models used to investigate the immune response to mycotoxins and studies have shown that the Toll pathway and secreted Bomanins, specifically neuronal BomS6, mitigate the symptoms of Aspergillus mycotoxin exposure, namely restrictocin and verruculogen (55). This could contribute to our understanding of how mammalian immunity interacts with mycotoxins.
The study by Almaliki (57) investigated the effect of VOCs produced by a single C. neoformans strain and found that the VOCs of this severe pathogen caused more severe morphological effects and higher death rates than all the A. fumigatus strains that were tested (57). C. neoformans is a pathogenic yeast able to establish invasive infections in immunocompromised patients. It has been frequently associated with HIV/AIDS and accounts for as much as 15% of HIV-related deaths (17). A Drosophila S2 protein expression system has been used to produce and purify a recombinant cryptococcal protease, May1. This protease was used for further investigation as a target to identify compounds that could simultaneously inhibit the fungal protease and HIV-1 protease, which would provide dual protection and lower toxicity for HIV/AIDS patients (92). In the study by Almaliki and colleagues (57) regarding the toxicity of VOCs in Drosophila, C. neoformans VOCs cause significant delays in metamorphosis with eclosion rates of 44% compared to 80% for controls.
Contemporaneously with the growing number of studies in Aspergillus, Drosophila has been used to study C. albicans, one of the most common causes of candidiasis and blood stream infections (7, 93). In 2004, Alarco and colleagues published a Toll-deficient Drosophila model through which they demonstrated concordant C. albicans pathogenicity findings with mouse models, giving validity to the use of fly models (94). This was further corroborated by similar study in Candida glabrata mutant libraries (65, 95). Drosophila studies have been used to investigate host adaptation by C. albicans. Liu et al. (59) demonstrated the necessity of phosphate transporter, Pho48, in establishing candidiasis in the wild type OregonR fly via infection with wild type C. albicans and Pho48 null mutants (59). Null mutants were 3.5 times less likely to cause fly death than wild type strains 5 days post-infection (59). Glittenberg and colleagues (66) via a targeted genetic screening of 5698 RNAi lines described the protective impact of fucosylation in immune defence against C. albicans. A recent study in a BomΔ55C fly model, (lacking the ability to produce the full range of Bomanin peptides) highlighted the ability of Candida sp. (including C. albicans and C. auris) to break down proline for energy, which may promote virulence. Moreover C. albicans mutants lacking the Proline UTilization genes put1, put3 or put1/put2 genes) showed reduced virulence compared to control fungal strain in the same fly infection model (62).
Drosophila infection models have been used to investigate the efficacy and toxicity of potential antifungal compounds. These include a Toll heterozygous Drosophila, Tl[r3]/+, used to test the naturally occurring compound, acid ellagic acid, against C. albicans where researchers showed statistically significant survival rates, and no toxicity at the proposed effective doses (64). Raj et al. (96) demonstrated a >70% survival rate of wild-type Drosophila infected with C. albicans when treated with Syzygium samarangense leaf extracted in methanol and dissolved in dimethyl sulfoxide. While the dosage applied to the Drosophila infection model was not specified, 50 mg of the Syzygium samarangense leaf extract was effective at clearing colonisation in an ex vivo porcine tongue and skin model, suggesting it could have utility as part of a topical treatment (96). Drosophila infection models can also be applied to antifungal studies for known compounds with the goal of reintroducing or repurposing old therapies. Clioquinol was administered orally to treat parasitic infections in the mid-1900s, however its use was discouraged due to perceived side effects (63, 97). Researchers investigated the antifungal efficacy and toxicity of Clioquinol in a Toll-deficient Drosophila model infected with C. albicans (63).
Drosophila has been utilised to investigate the novel pathogen C. auris. Wurster et al. (11) used a Toll-deficient mutant, Tlr632/TlI-RXA (which shows reduced AMP production and reduced phagocytic ability) to investigate the pathogenicity of C. auris clades identified at the time (Clade I-IV), and to determine the efficacy of azole to treatment (11). Their findings suggested that there was variability among the strains’ pathogenicity, though all strains were more pathogenic than C. albicans (11, 98).
High priority group
Species of non-C. albicans (NCA) have been investigated using Drosophila models. While NCAs have typically accounted for a smaller fraction of candidiasis infections, their prevalence and resistance to azoles and echinocandins is on the rise (17, 53, 99). NCAs in the high priority group include Nakaseomyces glabrata (C. glabrata), Candida tropicalis and Candida parapsilosis.
In 2018, researchers harnessed CRISPR/Cas9 for the targeted deletion of individual Drosophila Bomanin genes to determine their immunoprotective role against C. glabrata (43). Using in vivo and ex vivo infection models, they demonstrated that Bomanin genes do not act in tandem and the short-form Bom peptide was immunoprotective against C. glabrata on its own (43). They showed that flies lacking 10 out of the 12 Bomanin genes (42) were as susceptible to infection as Toll-deficient flies, highlighting the importance of Boms in host immunity (43). Studies in Drosophila cell lines have been used to identify mechanisms by which C. glabrata evades innate immunity strategies, like AMPs and ROS, and potential drug targets. A study by Kounatidis and colleagues showed that C. glabrata ADA2 gene is essential for the pathogen to resist oxidative stress as the ADA2 knockout yeast could only grow in flies with suppressed ROS, while overexpressing ADA2 promoted C. glabrata growth and resulted in lower host survival rates (65). The role of the potassium transporter C. glabrata TRK1 was elucidated through infection of MyD88 and BomΔ55C Drosophila strains with wild type and C. glabrata trk1 knockout (60). Loss of TRK1 gene resulted in cell wall modifications and reduced virulence within the host environment, in a potassium concentration dependent manner (60).
C. parapsilosis is associated with neonatal infections in addition to candidemia and candidiasis in immunosuppressed patients (17, 53). The Toll pathway has been shown to be crucial for Drosophila survival when infected by C. parapsilosis (which was not the case for Persephone protease), by comparing the susceptibility of wild type and mutant MyD88−/− flies to C. parapsilosis (53).
In addition, the high group includes Mucorales, Fusarium sp. Histoplasma sp. and eumycetoma causative agents (17). Mucorales are a large group of ubiquitous, filamentous fungi, frequently found in soil, which can cause infections ranging from mild to invasive (100, 101). The Order includes genera like Rhizopus, Mucor and Lichthiemia (101). Building on previous preexposure studies that showed the utility of Drosophila in Mucorales studies, Wurster and colleagues showed that exposing three Mucorales, Rhizopus arrhizus, R, pusillus, and Mucor circinelloides, to the triazoles isavuconazole and voriconazole, triggered hypervirulence in the fungi, resulting in lower survival rates in a Toll-deficient model (Tlr632/TlI-RXA). This was a significant finding as it could explain infections arising in patients undergoing prophylaxis or treatment with isavuconazole (102). While the number of Mucorales tested was small, this gives some insight into this treatment challenge. This is in contrast with A. fumigatus, which often occupies the same niche and is managed in a similar way, but does not develop isavuconazole-induced hypervirulence (102).
The Fusarium solani species complex, includes F. solani sensu stricto, F. falciforme and F. keratoplasticum, and they are major opportunistic fungal pathogen, capable of causing keratitis (58). A screen of 42 environmental and clinical isolates from South India revealed that all isolates were intrinsically resistant to first-generation azoles and susceptible to imidazole, which contributes to treatment challenges (58). Survival assays comparing Oregon-R wild type and Myd88 mutant flies, infected with 6 Fusarium sp. found that MyD88 is required to mount an effective Toll defence against all Fusarium strains (58). Homa et al. (58) also showed that Fusarium virulence was distinct at strain level (58). Subsequently, Cohen et al. found that survival rates following Daisho peptide knockout also varied among Fusarium species (51).
Medium priority group
The lower priority category has the highest number of pathogens including Scedosporium sp, Lomentospora prolificans, Coccidioides sp, Pichia kudriavzeveii (Candida krusei), Cryptococcus gattii, Talaromyces marneffei, Pneumocystis jirovecii and Paracoccidioides sp (17). These pathogens have the lowest relative global incidence and mortality rates, but still have substantial impacts (2). One of these pathogens, T. marneffei is a thermally dimorphic fungal pathogen localised to South and Southeast Asia (61, 103). It is found in the environment and Bamboo rats and can be inhaled and establish severe invasive infections in humans and animals (61). Its prevalence is not fully known due to limited surveillance and diagnostics and though mortality rates can be as high as 30%, few host-pathogen interaction studies have been performed (103, 104). Qu et al. (61) used the MyD88 −/− fly model to investigate the significance of the T. marneffei mating type on virulence in 107 clinical, Bamboo rat and environmental samples. They demonstrated that the mating type (MAT1-1 or MAT1-2) did not have an impact on flies survival upon infection, despite the fact that MAT1-2 isolates were overabundant across the entire sample population (61).
Perspective and future opportunities
The utility of Drosophila infection models in fungal research has been substantiated through the development of a good range of infection models and relevant findings. In spite of challenges and limitations around selecting the most suitable animal model, the extensive research work carried out in Drosophila over the last decade shows that this model is suitable. Fitting this extensive work within the boundaries of a Mini review article was a key challenge in setting up this review, therefore the WHO fungi prioritisation proved valuable into narrowing down the relevant content. While Drosophila presents a useful and relatively simple tool, subsequent investigations in other animal models are often required and should be considered to further corroborate findings, prior to reaching any general conclusions. Future work could focus on further characterisation of effector molecules (many of them have yet unknown function), on the role of innate immune mechanisms on immune memory adaptions, and on the use of Drosophila as a preclinical model on screening for antimicrobial efficacy against the fungal pathogens highlighted by WHO (17).
Author contributions
CM: Writing – review & editing, Investigation, Writing – original draft. IK: Writing – review & editing, Conceptualization, Supervision, Validation.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Almeida F, Rodrigues ML, Coelho C. ‘The still underestimated problem of fungal diseases worldwide’. Front Microbiol. (2019) 10:214. doi: 10.3389/fmicb.2019.00214
2. Rodrigues ML, Nosanchuk JD. Recognition of fungal priority pathogens: What next? PloS Negl Trop Diseases. (2023) 17:e0011136. doi: 10.1371/journal.pntd.0011136
3. Esheli M, Thissera B, El-Seedi HR, Rateb EM. Fungal metabolites in human health and diseases—An overview. Encyclopedia. (2022) 2:1590–601. doi: 10.3390/encyclopedia2030108
5. Glittenberg MT, Kounatidis I, Christensen D, Kostov M, Kimber S, Roberts I, et al. Pathogen and host factors are needed to provoke a systemic host response to gastrointestinal infection of Drosophila larvae by Candida albicans. DMM Dis Models Mech. (2011) 4:515–25. doi: 10.1242/dmm.006627
6. Arastehfar A, et al. Aspergillus fumigatus and aspergillosis: From basics to clinics. Stud Mycol. (2021) 100:100115. doi: 10.1016/j.simyco.2021.100115
7. Lopes JP, Lionakis MS. Pathogenesis and virulence of Candida albicans. Virulence. (2022) 13:89–121. doi: 10.1080/21505594.2021.2019950
8. Zijlstra EE, van de Sande WWJ, Welsh O, Mahgoub ES, Goodfellow M, Fahal AH. Mycetoma: a unique neglected tropical disease. Lancet Infect Dis. (2016) 16:100–12. doi: 10.1016/S1473-3099(15)00359-X
9. Emmanuel P, Dumre SP, John S, Karbwang J, Hirayama K. Mycetoma: A clinical dilemma in resource limited settings. Ann Clin Microbiol Antimicrobials. (2018) 17:35. doi: 10.1186/s12941-018-0287-4
10. Lockhart SR, Etienne KA, Vallabhaneni S, Farooqi J, Chowdhary A, Govender NP, et al. Simultaneous emergence of multidrug-resistant candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis. (2017) 64:134–40. doi: 10.1093/cid/ciw691
11. Wurster S, Bandi A, Beyda ND, Albert ND, Raman NM, Raad II, et al. Drosophila melanogaster as a model to study virulence and azole treatment of the emerging pathogen Candida auris. J Antimicrobial Chemother. (2019) 74:1904–10. doi: 10.1093/jac/dkz100
12. Song R, Zhai Q, Sun L, Huang E, Zhang Y, Zhu Y, et al. CRISPR/Cas9 genome editing technology in filamentous fungi: progress and perspective. Appl Microbiol Biotechnol. (2019) 103:6919–32. doi: 10.1007/s00253-019-10007-w
13. Malavia D, Gow NAR, Usher J. Advances in molecular tools and in vivo models for the study of human fungal pathogenesis. Microorganisms. (2020) 8:803. doi: 10.3390/microorganisms8060803
14. Lorenz A, Papon N. New tools for the new bug Candida auris. Trends Microbiol. (2022) 30:203–5. doi: 10.1016/j.tim.2022.01.010
15. Peyclit L, Yousfi H, Rolain JM, Bittar F. Drug repurposing in medical mycology: identification of compounds as potential antifungals to overcome the emergence of multidrug-resistant fungi. Pharmaceuticals. (2021) 14:488. doi: 10.3390/ph14050488
16. Velazhahan V, McCann BL, Bignell E, Tate CG. Developing novel antifungals: lessons from G protein-coupled receptors. Trends Pharmacol Sci. (2023) 44:162–74. doi: 10.1016/j.tips.2022.12.002
17. World Health Organization. WHO fungal priority pathogens list to guide research, development and public health action(2022). Available online at: https://www.who.int/publications/i/item/9789240060241 (Accessed 29 September 2023).
18. Hedges SB. The origin and evolution of model organisms. Nat Rev Genet. (2002) 3:838–49. doi: 10.1038/nrg929
19. Baldridge D, Wangler MF, Bowman AN, Yamamoto S, Undiagnosed Diseases Network, Schedl T, et al. Model organisms contribute to diagnosis and discovery in the undiagnosed diseases network: Current state and a future vision. Orphanet J Rare Dis. (2021) 16:206. doi: 10.1186/s13023-021-01839-9
20. Belvin MP, Anderson KV. A CONSERVED SIGNALING PATHWAY: the Drosophila toll-dorsal pathway. Annu Rev Cell Dev Biol. (1996) 12:393–416. doi: 10.1146/annurev.cellbio.12.1.393
21. Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. The Dorsoventral Regulatory Gene Cassette spä tzle/Toll/cactus Controls the Potent Antifungal Response in Drosophila Adults. Cell. (1996) 86:973–83. doi: 10.1016/s0092-8674(00)80172-5
22. Ligoxygakis P, Pelte N, Hoffmann JA, Reichhart JM. Activation of Drosophila toll during fungal infection by a blood serine protease. Science. (2002) 297:114–6. doi: 10.1126/science.1072391
23. Lamaris GA, Chamilos G, Lewis RE, Kontoyiannis DP. Virulence studies of Scedosporium and Fusarium species in Drosophila melanogaster. J Infect Dis. (2007) 196:1860–4. doi: 10.1086/523765
24. Levitin A, Marcil A, Tettweiler G, Laforest MJ, Oberholzer U, Alarco AM, et al. Drosophila melanogaster Thor and response to Candida albicans infection. Eukaryotic Cell. (2007) 6:658–63. doi: 10.1128/EC.00346-06
25. Chamilos G, Bignell EM, Schrettl M, Lewis RE, Leventakos K, May GS, et al. Exploring the concordance of aspergillus fumigatus pathogenicity in mice and Toll-deficient flies. Med Mycol. (2010) 48:506–10. doi: 10.3109/13693780903225813
26. Buchon N, Silverman N, Cherry S. Immunity in Drosophila melanogaster — from microbial recognition to whole-organism physiology. Nat Rev Immunol. (2014) 14:796–810. doi: 10.1038/nri3763
27. Harnish JM, Link N, Yamamoto S. Drosophila as a model for infectious diseases. Int J Mol Sci. (2021) 22. doi: 10.3390/ijms22052724
28. Liegeois S, Ferrandon D. Sensing microbial infections in the Drosophila melanogaster genetic model organism. Immunogenetics. (2022) 74:35–62. doi: 10.1007/s00251-021-01239-0
29. Ramírez-Camejo LA, Bayman P, Mejía LC. Drosophila melanogaster as an emerging model host for entomopathogenic fungi. Fungal Biol Rev. (2022) 42:85–97. doi: 10.1016/j.fbr.2022.09.001
30. White JS, Lafever KS, Page-McCaw A. Dissecting, fixing, and visualizing the Drosophila pupal notum. J Visualized Experiments. (2022) 2022. doi: 10.3791/63682
31. Jennings BH. Drosophila – a versatile model in biology and medicine. Mater Today. (2011) 14:190–5. doi: 10.1016/S1369-7021(11)70113-4
32. Touré H, Herrmann J-L, Szuplewski S, Girard-Misguich F. ‘Drosophila melanogaster as an organism model for studying cystic fibrosis and its major associated microbial infections’. Infect Immun. (2023) 91:e0024023. doi: 10.1128/iai.00240-23
33. Hamilos G, Samonis G, Kontoyiannis DP. Recent advances in the use of Drosophila melanogaster as a model to study immunopathogenesis of medically important filamentous fungi. Int J Microbiol. (2012) 2012:1–10:583792. doi: 10.1155/2012/583792
34. Cheng L, Baonza A, Grifoni D. Drosophila models of human disease. BioMed Res Int. (2018), 7214974. doi: 10.1155/2018/7214974
35. Macedo GE, Vieira PB, Rodrigues NR, Gomes KK, Rodrigues JF, Franco JL, et al. Effect of fungal indoor air pollutant 1-octen-3-ol on levels of reactive oxygen species and nitric oxide as well as dehydrogenases activities in drosophila melanogaster males. J Toxicol Environ Health - Part A: Curr Issues. (2022) 85:573–85. doi: 10.1080/15287394.2022.2054887
36. Yu S, Luo F, Xu Y, Zhang Y, Jin LH. ‘Drosophila innate immunity involves multiple signaling pathways and coordinated communication between different tissues’. Front Immunol. (2022) 13:905370. doi: 10.3389/fimmu.2022.905370
37. Qin B, Yu S, Chen Q, Jin LH. Atg2 regulates cellular and humoral immunity in Drosophila. Insects. (2023) 14:706. doi: 10.3390/insects14080706
38. Tafesh-Edwards G, Eleftherianos I. Drosophila immunity against natural and nonnatural viral pathogens. Virology. (2020) 540:165–71. doi: 10.1016/j.virol.2019.12.001
39. Arch M, Vidal M, Koiffman R, Melkie ST, Cardona PJ. Drosophila melanogaster as a model to study innate immune memory. Front Microbiol. (2022) 10:991678. doi: 10.3389/fmicb.2022.991678
40. Kounatidis I, Ligoxygakis P. Drosophila as a model system to unravel the layers of innate immunity to infection. Open Biol. (2012) 2:120075. doi: 10.1098/rsob.120075
41. Tafesh-Edwards G, Eleftherianos I. JNK signaling in Drosophila immunity and homeostasis. Immunol Lett. (2020) 226:7–11. doi: 10.1016/j.imlet.2020.06.017
42. Clemmons AW, Lindsay SA, Wasserman SA. An effector peptide family required for Drosophila toll-mediated immunity. PloS Pathog. (2015) 11:e1004876. doi: 10.1371/journal.ppat.1004876
43. Lindsay SA, Lin SJH, Wasserman SA. Short-form bomanins mediate humoral immunity in Drosophila. J Innate Immun. (2018) 10:306–14. doi: 10.1159/000489831
44. Huang C, Xu R, Liégeois S, Chen D, Li Z, Ferrandon D. Differential requirements for mediator complex subunits in Drosophila melanogaster host defense against fungal and bacterial pathogens. Front Immunol. (2021) 11:478958. doi: 10.3389/fimmu.2020.478958
45. Lin SJH, Cohen LB, Wasserman SA. Effector specificity and function in Drosophila innate immunity: Getting AMPed and dropping Boms. PloS Pathog. (2020) 16:e1008480. doi: 10.1371/journal.ppat.1008480
46. Huang J, Lou Y, Liu J, Bulet P, Cai C, Ma K, et al. A Toll pathway effector protects Drosophila specifically from distinct toxins secreted by a fungus or a bacterium. Proc Natl Acad Sci United States America. (2023) 120:e2205140120. doi: 10.1073/pnas.2205140120
47. Hanson MA, Lemaitre B. New insights on Drosophila antimicrobial peptide function in host defense and beyond. Curr Opin Immunol. (2020) 62:22–30. doi: 10.1016/j.coi.2019.11.008
48. Meister M, Laguex M. Drosophila blood cells. Cell Microbiol. (2003) 5:573–80. doi: 10.1046/j.1462-5822.2003.00302.x
49. Vlisidou I, Wood W. Drosophila blood cells and their role in immune responses. FEBS J. (2015) 282:1368–82. doi: 10.1111/febs.13235
50. Williams MJ. Drosophila hemopoiesis and cellular immunity. J Immunol. (2007) 178:4711–6. doi: 10.4049/jimmunol.178.8.4711
51. Cohen LB, Lindsay SA, Xu Y, Lin SJH, Wasserman SA. The daisho peptides mediate Drosophila defense against a subset of filamentous fungi. Front Immunol. (2020) 11:9. doi: 10.3389/fimmu.2020.00009
52. Mishima Y, Quintin J, Aimanianda V, Kellenberger C, Coste F, Clavaud C, et al. The N-terminal domain of Drosophila gram-negative binding protein 3 (GNBP3) defines a novel family of fungal pattern recognition receptors. J Biol Chem. (2009) 284:28687–97. doi: 10.1074/jbc.M109.034587
53. Csonka K, Tasi Z, Vedelek V, Vágvölgyi C, Sinka R, Gácser A. Deciphering of Candida parapsilosis induced immune response in Drosophila melanogaster. Virulence. (2021) 12:2571–82. doi: 10.1080/21505594.2021.1980989
54. Troha K, Buchon N. Methods for the study of innate immunity in Drosophila melanogaster. WIREs Dev Biol. (2019) 8:e344. doi: 10.1002/wdev.344
55. Xu R, Lou Y, Tidu A, Bulet P, Heinekamp T, Martin F, et al. The Toll pathway mediates Drosophila resilience to Aspergillus mycotoxins through specific Bomanins. EMBO Rep. (2023) 24:e56036. doi: 10.15252/embr.202256036
56. Almaliki HS, Drazanin E, Sadovnik R, Yin G, Bennett WJ. Eight-carbon volatiles are more toxic to wild-type Drosophila melanogaster than to flies with blocked immune system mutations. Entomologia Experimentalis Applicata. (2021) 169:1092–102. doi: 10.1111/eea.13108
57. Almaliki HS, Angela A, Goraya NJ, Yin G, Bennett JW. Volatile organic compounds produced by human pathogenic fungi are toxic to Drosophila melanogaster. Front Fungal Biol. (2020) 1:629510. doi: 10.3389/ffunb.2020.629510
58. Homa M, Galgóczy L, Manikandan P, Narendran V, Sinka R, Csernetics Á, et al. South Indian Isolates of the Fusarium solani species complex from clinical and environmental samples: Identification, antifungal susceptibilities, and virulence. Front Microbiol. (2018) 9:1052. doi: 10.3389/fmicb.2018.01052
59. Liu NN, Uppuluri P, Broggi A, Besold A, Ryman K, Kambara H, et al. Intersection of phosphate transport, oxidative stress and TOR signalling in Candida albicans virulence. PloS Pathog. (2018) 14:e1007076. doi: 10.1371/journal.ppat.1007076
60. Llopis-Torregrosa V, Vaz C, Monteoliva L, Ryman K, Engstrom Y, Gacser A, et al. Trk1-mediated potassium uptake contributes to cell-surface properties and virulence of Candida glabrata. Sci Rep. (2019) 9:7529. doi: 10.1038/s41598-019-43912-1
61. Qu Q, Lu S, Li Z, Zhang J, Wang X, Zheng H, et al. The relationship between the preference of mating type (MAT) and source in the opportunistic pathogen Talaromyces marneffei. Med Mycol. (2023) 61:myad027. doi: 10.1093/mmy/myad027
62. Silao FGS, Jiang T, Bereczky-Veress B, Kühbacher A, Ryman K, Uwamohoro N, et al. Proline catabolism is a key factor facilitating Candida albicans pathogenicity. PloS Pathog. (2023) 19:e1011677. doi: 10.1371/journal.ppat.1011677
63. Pippi B, Merkel S, Staudt KJ, Teixeira ML, de Araújo BV, Zanette RA, et al. Oral clioquinol is effective in the treatment of a fly model of Candida systemic infection. Mycoses. (2019) 62:475–81. doi: 10.1111/myc.12888
64. Sampaio ADG, Gontijo AVL, Araujo HM, Koga-Ito CY. In vivo efficacy of ellagic acid against Candida albicans in a drosophila melanogaster infection model. Antimicrobial Agents Chemother. (2018) 62:e01716-18. doi: 10.1128/AAC.01716-18
65. Kounatidis I, Ames L, Mistry R, Ho HL, Haynes K, Ligoxygakis P. A host-pathogen interaction screen identifies ada2 as a mediator of candida glabrata defenses against reactive oxygen species. G3: Genes Genomes Genet. (2018) 8:1637–47. doi: 10.1534/g3.118.200182
66. Glittenberg MT, Kounatidis I, Atilano M, Ligoxygakis P. A genetic screen in Drosophila reveals the role of fucosylation in host susceptibility to Candida infection. DMM Dis Models Mech. (2022) 15:dmm049218. doi: 10.1242/dmm.049218
68. Kúthy-Sutus E, Kharrat B, Gábor E, Csordás G, Sinka R, Honti V. A novel method for primary blood cell culturing and selection in Drosophila melanogaster. Cells. (2023) 12:24. doi: 10.3390/cells12010024
69. Pippi B, Joaquim AR, Merkel S, Zanette RA, Nunes MEM, da Costa Silva DG, et al. Antifungal activity and toxicological parameters of 8-hydroxyquinoline-5-sulfonamides using alternative animal models. J Appl Microbiol. (2021) 130:1925–34. doi: 10.1111/jam.14915
70. Lorenz A. Candida auris . In: Lorenz A, editor. Methods in Molecular Biology. Springer US, New York, NY (2022). doi: 10.1007/978-1-0716-2417-3
71. Weaver CM, Makdissi S, Di Cara F. Modified protocol for culturing Drosophila S2 R+ cells and adult plasmatocytes to study actin cytoskeleton dynamics. STAR Protoc. (2022) 3:101588. doi: 10.1016/j.xpro.2022.101588
72. Flores Dalla Lana D, Neiva Lavorato S, Minussi Giuliani L, Cruz L, Lopes W, Henning Vainstein M. Discovery of a novel and selective fungicide that targets fungal cell wall to treat dermatomycoses: 1,3-bis(3,4-dichlorophenoxy)propan-2-aminium chloride. Mycoses. (2020) 63:197–211. doi: 10.1111/myc.13027
73. Chen J, Johnston DS. Dissection, fixation, and immunostaining of the Drosophila midgut. In: Chang C, Wang J, editors. Cell Polarity Signalling, Methods and Protocols. Humana Press, Springer Protocols, New York, NY (2022). p. 309–21. doi: 10.1007/978-1-0716-2035-9_20
74. Amin S, Basu M, Buzinova V, Delgado A, Mahadevan T, Mishra S. Glyoxal-based fixation of Drosophila embryos for immunofluorescence staining and RNA in situ hybridization. STAR Protoc. (2023) 4:102385. doi: 10.1016/j.xpro.2023.102385
75. Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, et al. The genome sequence of Drosophila melanogaster. Science. (2000) 287:2185–95. doi: 10.1126/science.287.5461.2185
76. Kanoh H, Kato H, Suda Y, Hori A, Kurata S, Kuraishi T. Dual comprehensive approach to decipher the Drosophila Toll pathway, ex vivo RNAi screenings and immunoprecipitation-mass spectrometry. Biochem Biophys Res Commun. (2019) 508:332–7. doi: 10.1016/j.bbrc.2018.11.007
77. Kim ES, Rajan A, Chang K, Govindarajan S, Gulick C, English E, et al. Generation of LexA enhancer-trap lines in Drosophila by an international scholastic network. G3: Genes Genomes Genet. (2023) 13:jkad124. doi: 10.1093/g3journal/jkad124
78. Behrens A, Rodschinka G, Nedialkova DD. High-resolution quantitative profiling of tRNA abundance and modification status in eukaryotes by mim-tRNAseq. Mol Cell. (2021) 81:1802–1815.e7. doi: 10.1016/j.molcel.2021.01.028
79. Zhou H, Li S, Pan W, Wu S, Ma F, Jin P. Interaction of lncRNA-CR33942 with dif/dorsal facilitates antimicrobial peptide transcriptions and enhances Drosophila toll immune responses. J Immunol. (2022) 208:1978–88. doi: 10.4049/jimmunol.2100658
80. Redman M, King A, Watson C, King D. What is CRISPR/cas9? Arch Dis Childhood - Educ Pract Ed. (2016) 101:213–5. doi: 10.1136/archdischild-2016-310459
81. Szebenyi C, Gu Y, Gebremariam T, Kocsubé S, Kiss-Vetráb S, Jáger O, et al. cotH genes are necessary for normal spore formation and virulence in mucor lusitanicus. mBio. (2023) 14:e0338622. doi: 10.1128/mbio.03386-22
82. Kapun M, Nunez JCB, Bogaerts-Márquez M, Murga-Moreno J, Paris M, Outten J, et al. Drosophila evolution over space and time (DEST): A new population genomics resource. Mol Biol Evol. (2021) 38:5782–805. doi: 10.1093/molbev/msab259
83. Hu Y, Comjean A, Rodiger J, Liu Y, Gao Y, Chung V, et al. FlyRNAi.org—the database of the Drosophila RNAi screening center and transgenic RNAi project: 2021 update. Nucleic Acids Res. (2021) 49:D908–15. doi: 10.1093/nar/gkaa936
84. Van De Veerdonk FL, Gresnigt MS, Romani L, Netea MG, Latgé JP. Aspergillus fumigatus morphology and dynamic host interactions. Nat Rev Microbiol. (2017) 15:661–74. doi: 10.1038/nrmicro.2017.90
85. Lionakis MS, Kontoyiannis DP. Fruit flies as a minihost model for studying drug activity and virulence in Aspergillus. Med Mycol. (2005) 43:S111–4. doi: 10.1080/13693780400020030
86. Spikes S, Xu R, Nguyen CK, Chamilos G, Kontoyiannis DP, Jacobson RH, et al. Gliotoxin production in Aspergillus fumigatus contributes to host-specific differences in virulence. J Infect Dis. (2008) 197:479–86. doi: 10.1086/525044
87. Nafis MMH, Quach ZM, Al-Shaarani AAQA, Muafa MHM, Pecoraro L. Pathogenicity of aspergillus airborne fungal species collected from indoor and outdoor public areas in Tianjin, China. Pathogens. (2023) 12:1154. doi: 10.3390/pathogens12091154
88. Bennett JW, Inamdar AA. Are some fungal volatile organic compounds (VOCs) mycotoxins? Toxins. (2015) 7:3785–804. doi: 10.3390/toxins7093785
89. Dillon ME, Wang G, Garrity PA, Huey RB. Thermal preference in Drosophila. J Thermal Biol. (2009) 34:109–19. doi: 10.1016/j.jtherbio.2008.11.007
90. Almaliki HS, Niu M, Keller NP, Yin G, Bennett JW. Mutational analysis of Aspergillus fumigatus volatile oxylipins in a Drosophila eclosion assay. J Fungi. (2023) 9:402. doi: 10.3390/jof9040402
91. Belmonte RL, Corbally MK, Duneau DF, Regan JC. Sexual dimorphisms in innate immunity and responses to infection in Drosophila melanogaster. Front Immunol. (2020) 10:3075. doi: 10.3389/fimmu.2019.03075
92. Kryštůfek R, Šácha P, Starková J, Brynda J, Hradilek M, Tloušt'ová E, et al. Re-emerging aspartic protease targets: examining cryptococcus neoformans major aspartyl peptidase 1 as a target for antifungal drug discovery. J Medicinal Chem. (2021) 64:6706–19. doi: 10.1021/acs.jmedchem.0c02177
93. Mroczyńska M, Brillowska-Dabrowska A. Virulence of clinical candida isolates. Pathogens. (2021) 10:466. doi: 10.3390/pathogens10040466
94. Alarco A-M, et al. Immune-Deficient Drosophila melanogaster: A Model for the Innate Immune Response to Human Fungal Pathogens 1,2, The Journal of Immunology(2004). Available online at: http://journals.aai.org/jimmunol/article-pdf/172/9/5622/1186248/5622.pdf.
95. Brunke S, Quintin J, Kasper L, Jacobsen ID, Richter ME, Hiller E, et al. Of mice, flies - and men? Comparing fungal infection models for large-scale screening efforts. DMM Dis Models Mech. (2015) 8:473–86. doi: 10.1242/dmm.019901
96. Raj S, Vinod V, Jayakumar J, Suresh P, Kumar A, Biswas R, et al. Antifungal activity of Syzygium samarangense leaf extracts against Candida. Lett Appl Microbiol. (2021) 73:31–8. doi: 10.1111/lam.13471
97. Perez DR, Sklar LA, Chigaev A. Clioquinol: To harm or heal. Pharmacol Ther. (2019) 199:155–63. doi: 10.1016/j.pharmthera.2019.03.009
98. Wurster S, Albert ND, Kontoyiannis DP. Drosophila melanogaster as a Rapid and Reliable In Vivo Infection Model to Study the Emerging Yeast Pathogen Candida auris. In: Lorenz A, editor. Candida auris, Methods and Protocols. Springer Press, New York, NY (2022). p. 375–405. doi: 10.1007/978-1-0716-2417-3_24
99. Pristov KE, Ghannoum MA. Resistance of Candida to azoles and echinocandins worldwide. Clin Microbiol Infect. (2019) 25:792–8. doi: 10.1016/j.cmi.2019.03.028
100. Lecointe K, Cornu M, Leroy J, Coulon P, Sendid B. Polysaccharides cell wall architecture of mucorales. Front Microbiol. (2019) 10:469. doi: 10.3389/fmicb.2019.00469
101. Tahiri G, Lax C, Cánovas-Márquez JT, Carrillo-Marín P, Sanchis M, Navarro E, et al. Mucorales and mucormycosis: recent insights and future prospects. J Fungi. (2023) 9:335. doi: 10.3390/jof9030335
102. Wurster S, Lewis RE, Albert ND, Kontoyiannis DP. Preexposure to Isavuconazole Increases the Virulence of Mucorales but Not Aspergillus fumigatus in a Drosophila melanogaster Infection Model. Antimicrobial Agents Chemother. (2019) 63:e01896-18. doi: 10.1128/AAC.01896-18
103. Singulani JL, Scorzoni L, de Oliveira HC, Marcos CM, Assato PA, Fusco-Almeida AM, et al. Applications of invertebrate animal models to dimorphic fungal infections. J Fungi. (2018) 4:118. doi: 10.3390/jof4040118
Keywords: Drosophila, model organisms, fungal diseases, WHO, FFPL, infection models
Citation: Mpamhanga CD and Kounatidis I (2024) The utility of Drosophila melanogaster as a fungal infection model. Front. Immunol. 15:1349027. doi: 10.3389/fimmu.2024.1349027
Received: 04 December 2023; Accepted: 27 February 2024;
Published: 14 March 2024.
Edited by:
Susanna Valanne, Tampere University, FinlandReviewed by:
Yeon Soo Han, Chonnam National University, Republic of KoreaIoannis Eleftherianos, George Washington University, United States
Copyright © 2024 Mpamhanga and Kounatidis. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ilias Kounatidis, aWxpYXMua291bmF0aWRpc0BvcGVuLmFjLnVr
 Chengetai D. Mpamhanga
Chengetai D. Mpamhanga Ilias Kounatidis
Ilias Kounatidis