- 1Department of Dermatology, Heji Hospital of Changzhi Medical College, Changzhi, China
- 2Department of Rheumatology, The Second Clinical Medical College of Shanxi Medical University, Taiyuan, China
- 3Shanxi Key Laboratory for immunomicroecology, The Second Hospital of Shanxi Medical University, Taiyuan, China
- 4Department of Rheumatology, The Second Hospital of Shanxi Medical University, Taiyuan, China
- 5Brigham and Women’s Hospital, Harvard Medical School, Boston, MA, United States
Background: Disturbed gut microbiota and associated metabolic dysfunction exist in Psoriasis. Despite the growing use of interleukin-17 inhibitor (anti-IL17) therapy, the effect of anti-IL17 on gut/skin microbiota function is not fully understood in patients with Psoriasis.
Objective: Therefore, we explored whether Psoriasis is associated with alterations in selected gut/skin microbiota in a study cohort, and a longitudinal cohort study to reveal the effects of IL-17A inhibitor treatment on gut microbiota in Psoriasis.
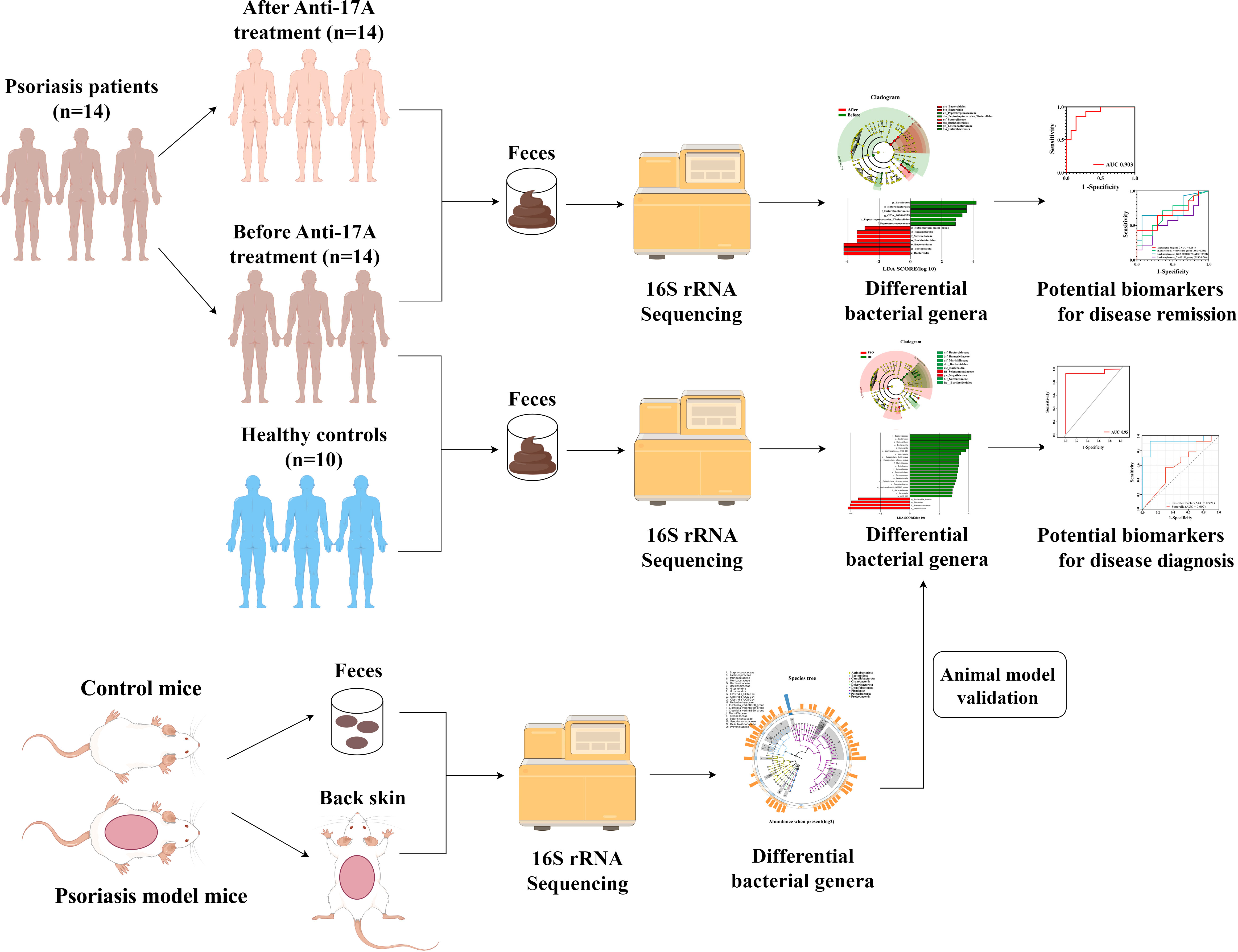
Methods: In a case-control study, 14 patients with Psoriasis and 10 age, sex and body mass index-matched Healthy Controls were recruited. Longitudinal mapping of the gut microbiome was performed using 16S rRNA gene sequencing. Mouse models were used to further study and validate the interrelationship between the skin microbiome and the gut microbiome in Psoriasis. PICRUST2 was applied to predict the function of the bacterial community.
Results: In Psoriasis patients, gut microbiota dysbiosis was present with increased heterogeneity: decreased Bacteroidota and increased Firmicutes as well as Actinobacteriota predominating in Psoriasis. Escherichia-Shigella enrichment was associated with reduction in serum levels of total bile acid and markers in Apoptotic pathways. After IL-17A inhibitor treatment in Psoriasis patients, longitudinal studies observed a trend toward a normal distribution of the gut microbiome and modulation of apoptosis-related metabolic pathways. Results from a mouse model showed dysregulation of the skin microbiota in Psoriasis characterized by Staphylococcus colonization.
Conclusion: The psoriatic gut/skin microbiota exhibits loss of community stability and pathogen enrichment. IL-17A inhibitors restore microbiota homeostasis and metabolic pathways, reduce pro-inflammatory cytokine expression, and alleviate symptoms in patients with Psoriasis.
1 Introduction
Psoriasis (PSO) is a chronic immune-mediated inflammatory disease that affects approximately 2-4% of the population worldwide (1). While it is primarily characterized by skin involvement, it can also cause systemic inflammation in various organs (2). PSO can be highly debilitating (3) and can increase the morbidity and mortality associated with the disease (4). The development of PSO involves a combination of genetic, immunological, and environmental factors (5). An overactive Th17 response can mediate skin inflammation and hyperproliferation of keratinocytes in PSO (6). Although therapies targeting the Th17 pathway can effectively suppress the abnormal immune response and manage symptoms in PSO patients, they do not provide a cure, and the effectiveness of treatment can vary from patient to patient. This emphasizes the need to further understand the factors involved in the development and progression of PSO.
The interaction between the gut microbiome and inflammatory skin diseases is mediated by a dysfunctional gut microbiome, a disrupted gut barrier, increased inflammatory mediators, and metabolites produced by microorganisms (7–9). Gut microecological dysregulation activates a pro-inflammatory state by altering the metabolic environment and activating specific pattern recognition receptors (PRPs) on epithelial cells, which alters the integrity of the tight junctions between epithelial cells, leading to increased permeability of the intestinal, increased release of proinflammatory cytokines, exacerbation of chronic inflammation, and the entry of certain metabolites, toxins, and even bacteria into the systemic circulation (10, 11), and when these microorganisms and their metabolites enter the circulation they can either promote or maintain the proinflammatory state in an organism (12). Polysaccharide A produced by B. fragilis binds to TLR-2 and promotes the differentiation of CD4+ lymphocytes to Treg lymphocytes, along with the production of anti-inflammatory cytokines such as IL-10 (13). Segmented Filamentous Bacteria, as a commensal bacterium, influence the adaptive and innate immune systems by increasing IgA secretion and the development of Th17 lymphocytes (14). Butyrate, produced by intestinal bacteria, promotes the differentiation of Treg cells and has anti-inflammatory properties (15). Elevated claudin 3 has been observed in PSO patients, suggesting increased intestinal permeability (12). Because overexpression of proinflammatory cytokines leading to an imbalance between effector T cells and epithelial permeability, gut microecological dysregulation contributes to PSO-associated inflammatory and autoimmune states (10).
The skin, being the body’s largest organ, serves as a physical barrier against environmental damage and as a dynamical interface for host-skin-microbe interactions. Skin bacteria play a vital role in maintaining skin health by preventing pathogen colonization and regulating T lymphocyte function (16). Keratinocytes can interact with skin bacteria to trigger an immune response in PSO (17). Therefore, disruptions to the skin’s microbiome can potentially affect skin’s immune function. The gut microbiome also influences skin homeostasis by impacting the signaling processes involved in epidermal differentiation (18). The number of Firmicutes, Proteobacteria, and Actinobacteria in PSO has increased. Decreased abundance of Bacteroides in PSO patients indicating that decreased abundance of beneficial microorganisms may disturb skin homeostasis by altering the balance of the immune system (19). Inflammatory mediators, such as IFN-γ, IL-1β, IL-6, IL-12, and TNF, are significantly increased in patients with PSO (20, 21). Additionally, genetic material from intestinal bacteria has been discovered in plasma samples from PSO patients, indicating a potential connection between intestinal microorganisms and psoriatic skin lesions (22).
Current immunologic studies have revealed the important role of immune pathways involving interleukin (IL)-17 in PSO (23). Bacillus hominis (Bu) has been found to attenuate colitis by modulating host Bile Acids (BAs) metabolism, impacting the IL-17 signaling pathway, and inhibiting Th17 cell differentiation (24). Lactobacillus pentosus decrease the suppression of pro-inflammatory cytokines, such as TNF-α, IL-17A, and IL-23 (25). The interaction between the microbiota and the immune system holds clinical significance. The efficacy of Secukinumab, an IL-17 inhibitor, has been demonstrated in the treatment of PSO (26), and studies have suggested that anti-PSO therapy may influence the composition of the microbiota (27).
In this study, we examined and observed the alterations in gut microbiomes in 14 PSO patients compared to 10 healthy subjects using 16S rRNA sequencing. Through a longitudinal observation of Secukinumab treatment, we observed a trend of abnormal changes in the gut microbiomes of PSO patients returning to a more normal state. Additionally, we utilized animal models to identify Staphylococcus as a key factor in PSO skin lesions.
2 Material and methods
2.1 Study design and patients
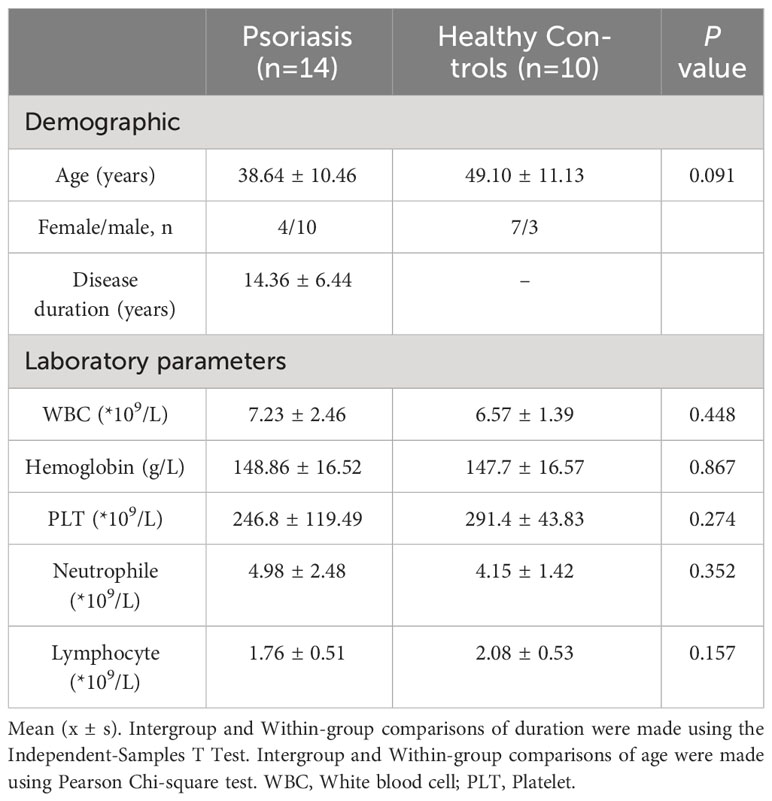
A total of 14 patients diagnosed with Psoriasis (PSO) were included in this study (Figure 1). They were treated with Secukinumab, an interleukin-17 inhibitor, at the Department of Dermatology, Heji Hospital of Changzhi Medical College, from September 2022 to May 2023. All patients had a clear diagnosis of PSO and followed the Guideline for the diagnosis and treatment of psoriasis in China (2018 complete edition) (28). Among them, 2 patients were newly diagnosed, while the other 12 had a previous diagnosis but had not received treatment with hormonal drugs, immunosuppressants, or biologics within the past year. Ten healthy controls (HCs) were also included, matched in terms of age and sex, and had no history of psoriasis, autoimmune diseases, tumors, and gastrointestinal tract disorders, or any other disease that could affect the results of the study were excluded. All included subjects have inclusion criteria: (1) normal range of liver and kidney functional tests; (2) normal blood glucose/lipid, normal urine and stool; (3) free of hepatitis B/C virus antigen; and (4) had no history of antibiotics and probiotics during the past 4 weeks (Supplementary Table 1).
The PSO patients in the anti-IL-17 drug group received secukinumab 300 mg at weeks 0, 1, 2, 3, and 4, followed by administration every 4 weeks. Baseline demographic and clinical data, including age, gender, and laboratory markers, were collected. Disease severity was assessed using the Psoriasis Area and Severity Index (PASI) scores (29), with assessments conducted at Weeks 0 and 16 by the same physician. A PASI 75 score indicates a 75% or greater reduction in PASI score from baseline, representing significant disease improvement. This criterion has been traditionally used as a benchmark for treatment efficacy in clinical trials (30, 31). Therefore, respondents are defined as patients who have improved PASI by ≥ 75% after 16 weeks of treatment, whereas non-responders were defined as <75% improvement in PASI. The study received ethical approval from the Ethics Committee of Heji Hospital of Changzhi Medical College (number: 2023-13), and all patients provided written informed consent to participate.
2.2 Construction of Psoriasis mouse model
Male BALB/c mice (SPF grade, aged 5 weeks, weighing 16 ~ 18 g) were obtained from Beijing Spectrum Biotechnology Co. and housed at a temperature of 23°C and a humidity level of 50%. All animal procedures were approved by the Ethics Committee of Changzhi Medical College (number: DW2023051).
After one week of acclimatization, the mice were randomly assigned to two groups: experimental and control, with six mice in each group. Hair on the dorsal area of all mice was removed using depilatory cream. Mice in the experimental group were topically treated with imiquimod cream (IMQ, Sichuan Mingxin Pharmaceutical Co., Ltd., 3 g/tube, [Approval Number] National Drug License H20030129) at a dose of 62.5 mg/day on their dorsal skin for seven consecutive days, while control mice received an equivalent volume of petroleum jelly (Shandong Dezhou Likang Pharmaceutical Science and Technology Co., Ltd., 300 g/box) for the same duration. Throughout the experiment, the experimental and control groups were maintained under identical conditions, with no significant differences in diet, water access, ambient temperature, or humidity. On the 8th day, the mice were euthanized following anesthesia.
2.3 Sample collection
2.3.1 Stool sample collection
Stool specimens were obtained from subjects at baseline (M0) and after 16 weeks of PSO treatment with the anti-IL-17A medication (Secukinumab). The collected specimens were then placed in pre-labeled stool collection tubes containing fecal DNA stabilizer. After blending through shaking, the specimens were immediately stored at -80°C in frozen storage until transportation on ice to the laboratory for further processing.
On the 8th day of mouse modeling, fresh fecal samples were collected from both groups of mice. Two grams of fecal samples were dispensed into sterile freezing tubes immediately after sampling and placed in a -80°C refrigerator for freezing and storage. The collection process and requirements remained unchanged.
2.3.2 Skin tissue sample collection
On the 8th day of conducting mouse modeling, the mice were subjected to anesthesia using a carbon dioxide gas anesthesia machine (PerkinElmer). Skin tissue samples measuring approximately 1cm×1cm×2cm were obtained from the back of the mice and labeled as No. 1 and No. 2 samples. Following the sampling procedure, sample No. 1 was fixed in a sterile formalin solution, while sample No. 2 was placed in a sterile freezing tube and stored in a -80°C refrigerator for freezing. All samples were collected by the same laboratory personnel, and strict adherence to sterility principles was maintained to prevent contamination of the experimental specimens by other microorganisms during the collection process.
2.4 DNA extraction and 16S rRNA gene sequencing
Fecal samples were collected at the baseline and 16 weeks after treatment using the Longsee Fecalpro kit (Longsee Medical Technology Co., Guangzhou, China). DNA extraction was performed using a nucleic acid extraction or purification reagent (Hangzhou Guhe: GHFDE100) according to the manufacturer’s instructions. The quantity and quality of the extracted DNA were assessed using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). The variable regions 3 and 4 (V3-V4) of the 16S rRNA gene, PCR mixture conditions, and the corresponding thermal cycling steps were amplified by PCR using primer sets (Illumina V3 forward 5′-CCTACGGGGNGGCWGCAG-3′ and V4 reverse 5′-GACTACHVGGGTATCTAATCC′). The purified libraries were quantified, normalized, and sequenced using a NovaSeq instrument (Illumina).
2.5 Sequencing data processing and species annotation
The reads from each sample were spliced using Vsearch v2.15.0 in order to obtain the raw Tags data (Raw Tags). Sequence quality was also controlled and filtered. Information was collected from the SILVA database and a pre-fitted QIME2 classifier built with the Scikit Lean package was applied to assign bacterial classifications. Filtered OTU sequences were used in PICRUSt prediction, with default parameters.
2.6 PICRUSt2 functional prediction analysis
Phylogenetic studies of the cohort were performed by reconstructing the unobserved state 2 (PICRUST2) to predict the functional potential of the bacterial community. R (v3.6.1) was utilized for the generation of other graphs.
2.7 Statistical analysis
Alpha diversity and beta diversity were calculated using QIME2. Principal Coordinate Analysis (PCoA) was conducted based on the UniFrac distance metric. Subsequently, Wilcoxon tests, linear discriminant analysis (LDA) effect size (LEfSe) analyses, and metagenomeSeq analyses were performed to identify the differential classification group. All statistical tests were two-tailed, and p-values less than 0.05 were considered statistically significant.
3 Results
3.1 Patient cohort
The study included a cohort of 14 patients diagnosed with PSO and 10 HCs, among which 5 had guttate psoriasis and 9 had plaque psoriasis (Supplementary Table 2). Two groups of subjects, matched for sex and age, were compared (Table 1; Supplementary Table 3). All PSO patients were diagnosed at the Dermatology Department of Changzhi Heji Hospital. The average PASI score for the PSO patients in this study was 22.04, indicating that all enrolled subjects had moderate to severe disease. To minimize treatment-related variability, subjects who had recently undergone antibiotic therapy and/or other biological and systemic therapies were excluded.
3.2 Altered gut microbial diversity in Psoriasis patients
Differences in the overall composition of the microbial community between PSO patients and controls were assessed by measuring α- and β-diversity. Alpha diversity reflects the microbial diversity within a sample, while beta diversity captures differences between samples. It was observed that, although not statistically significant, the gut microbiome in PSO patients exhibited reduced α-diversity (Figures 2A-C). This trend suggests that the diversity of the gut microbial community becomes homogenous as PSO progresses.
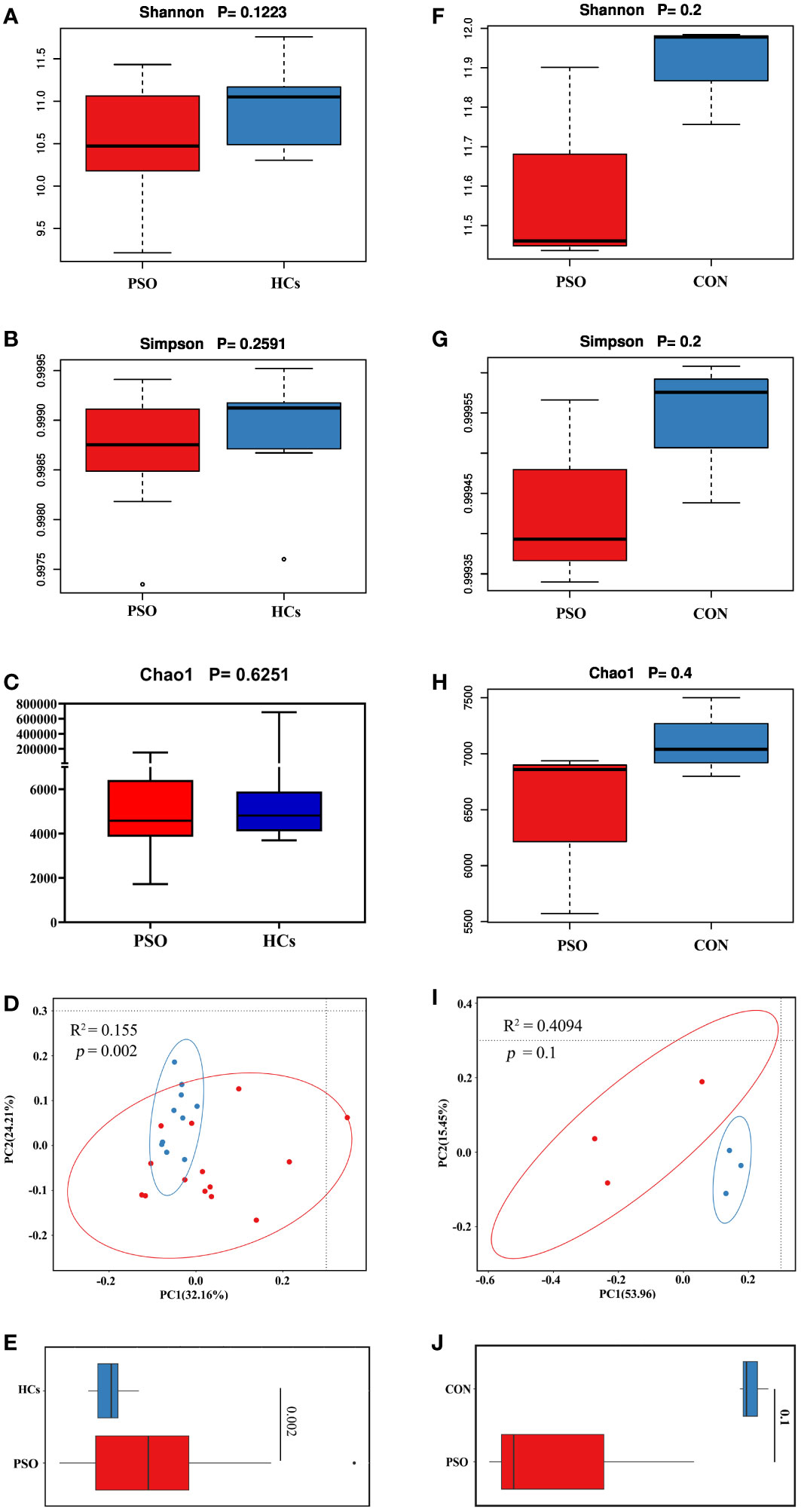
Figure 2 Bacterial community diversity in the healthy and psoriatic gut. Alpha diversity measured according to (A) Shannon index, (B) Simpson’s diversity index, and (C) chao1 index of healthy skin samples. (D) Principal coordinate analysis (PCoA) of the microbial community structures based on weighted UniFrac distance matrix for the first two principal axes. Each point on the PCoA plot represents a fecal microbiome sample (red = HCs, blue = PSO). The first principal coordinate explains 32.16% of variation, and the second principal coordinate explains 24.21% of the variation. (E) The average weighted UniFrac distances among samples within each state are shown in the box plot. (F-H) Alfa diversity of the gut microbiome in PSO model mouse. (I, J) Beta diversity of the gut microbiome in PSO model mouse (the average unweighted UniFrac distances). HCs: Healthy Controls. PSO: Psoriasis patients and psoriasis model mouse, CON, Control Mouse.
Beta diversity was calculated using the weighted UniFrac metric to determine whether the structure of the microbial community differed between psoriatic subjects and HCs. Principal coordinates analysis revealed significant differences in the gut microbiome between PSO patients and HCs, with the former showing greater dispersion (Figure 2D). PSO patients also exhibited higher mean distances in the composition of their gut microbiota compared to HCs (Figure 2E), indicating greater heterogeneity. Consistent results were also observed in a mouse model (Figures 2F-J).
3.3 Changes in gut microbial composition in Psoriasis
Gut microbiome in all states (HCs, PSO) consisted of four dominant phyla: Firmicutes, Proteobacteria, Actinobacteriota, and Bacteroidota (Figures 3A, B). The gut microbiome in PSO patients is characterized by an increase in Firmicutes and Actinobacteriota, while Bacteroidota decreases, in agreement with previous descriptions (12). The dominant genera in the disease state include Faecalibacterium, Megamonas, Prevotella, and Escherichia-Shigella (Figure 3C). We also used LEfSe identified Escherichia-Shigella as an additional discriminatory feature of a healthy gut (Figure 3D).
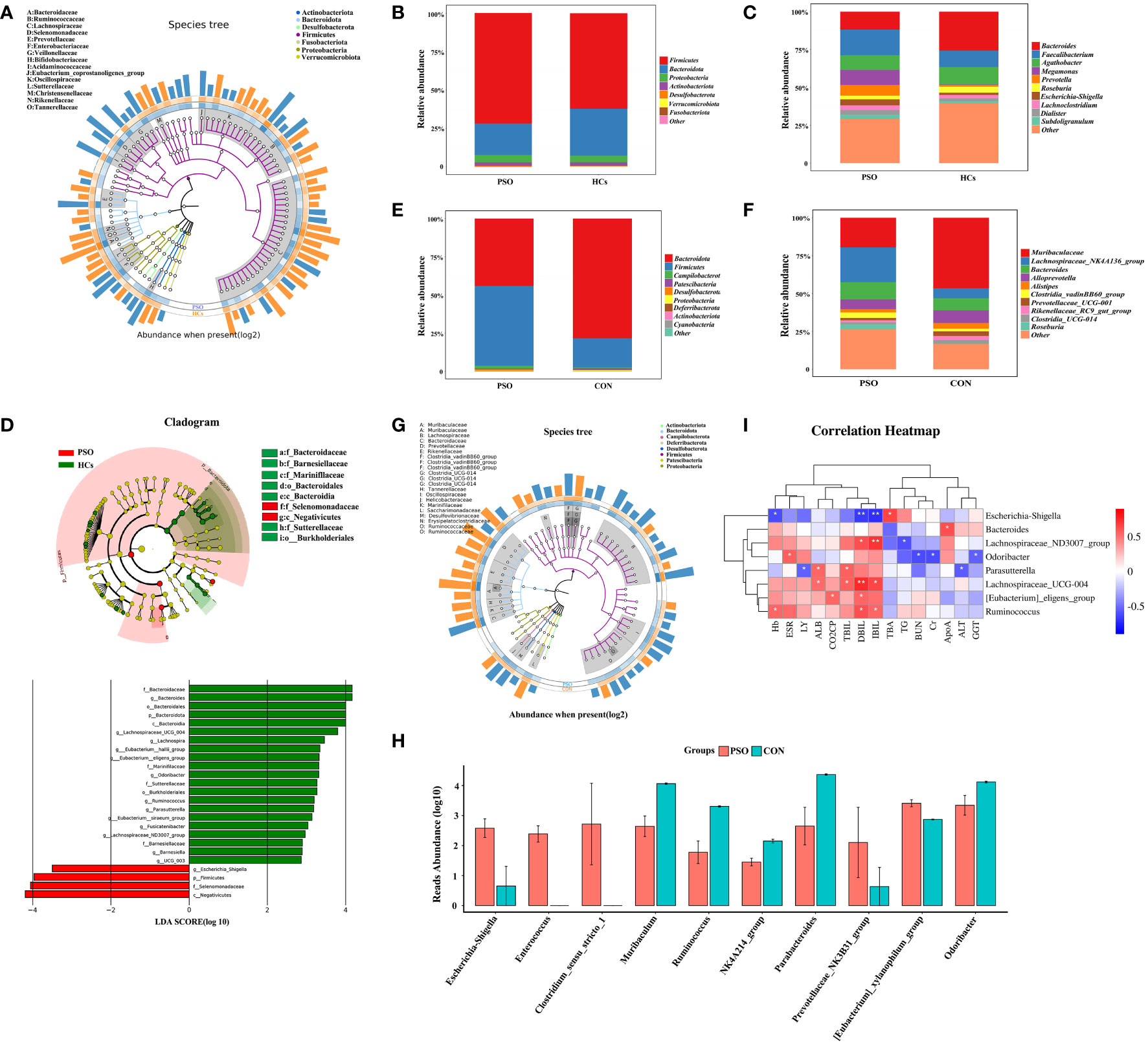
Figure 3 Taxonomic composition and gut microbial characteristics associated with disease states. (A) GraPhlan diagram showing the dominant flora in PSO. (B) Phylum and (C) genus level compositions of gut microbiome in HCs and PSO. Only major taxa are shown. Others represent less abundant taxa and are not plotted. (D) Results of LEfSe analysis of variance. (E, F) represent the composition at the phylum level and genus level in PSO model mouse and CON, respectively. (G) GraPhlan plot showing dominant taxa in PSO model mouse. (H) Results of metagenomeSeq difference analysis in PSO model mouse. (I) Heat map of correlation between different genera of bacteria and laboratory indicators. HCs, Healthy Controls; PSO, Psoriasis patients and psoriasis model mouse; CON, Control Mouse; Hb, Hemoglobin; ALT, Alanine aminotransfease; TBIL, Total bilirubin; DBIL, Direct bilirubin; IBIL, Indirect Bilirubin; ALB, Albumin; GGT, Glutamyltranspeptidase; TBA, Total bile acid; TG, Triglyceride; ApoA, Apolipoprotein A; BUN, Urea nitrogen; Cr, Creatinine; CO2CP, Carbon dioxide-combining power *: p < 0.05, **: p < 0.01.
Similar results were observed in animal experiments using PSO rats, with an increase in Firmicutes and a decrease in Bacteroidota (Figure 3E). The specific genera that increased in the PSO rats were Lachnospiraceae_NK4A136_group, Bacteroides, Clostridia_vadinBB60, and Roseburia, while Alloprevotella, Muribaculaceae, and Alistipes, Prevotellaceae_UCG-001, Rikenellaceae_RC9_gut, and Clostridia_UCG-014 decreased significantly (Figure 3F). It is important to note that the microbiota identified in PSO patients and PSO model mice were not identical (Figure 3G).
Further analysis using metanomeSeq revealed that the relative abundance of Escherichia-Shigella, Enterococcus, Clostridium_sensu_stricto_1 strain, Prevotellaceae_NK3B31_group, and [Eubacterium]_xylanophilum_group increased significantly in PSO model mice, while Muriaculum, Ruminococcus, Oscillospiraceae_NK4A214_group, Parabacteroides and Odoribacter decreased significantly (Figure 3H). These findings suggest that Escherichia-Shigella may play a critical role in the dysregulation of the intestinal microecology in PSO patients.
We next performed a correlation analysis to assess the mechanism of gut microbiota in PSO, which showed that Escherichia-Shigella was positively correlated with serum levels of Total Bile Acids (TBA) and negatively with levels of Hemoglobin (Hb), Direct Bilirubin (DBIL), and Indirect Bilirubin (IBIL) in patients with PSO. Odoribacter was positively correlated with the blood sedimentation rate (ESR). Several other differential genera, including Lachnospiraceae_ND3007_group, Lachnospiraceae_UCG-004, [Eubacterium]_eligens_group, Parabacteroides, and Ruminococcus were found to be positively correlated with Total Bilirubin (TBIL), DBIL and IBIL (Figure 3I). BAs acts as a signaling molecule that differentially affects the gut microbiota and host immunity as well as metabolism. BAs can directly inhibit IL-17A production and block CCL20-mediated transport to ameliorate psoriasiform dermatitis with minimal toxicity (32). Our study found a significant reduction in serum TBA levels in PSO patients (Supplementary Figure 1), therefore, we hypothesized that Escherichia-Shigella may exert pathogenic effects by affecting the BAs metabolic pathway.
3.4 Predictive analysis of faecal microbial function and biomarker screening
KEGG pathway enrichment analysis revealed enhanced pathways in PSO patients, including Lysine degradation, Phosphotransferase system (PTS), Nitrotoluene degradation, Penicillin and cephalosporin biosynthesis, Chlorocyclohexane and chlorobenzene degradation, Bacterial secretion system, Tryptophan metabolism, Propanoate metabolism, and ABC transport proteins. Some pathways, such as Protein digestion and absorption, and Apoptosis pathways, were attenuated (Figure 4A). Additionally, an attenuation of the Apoptosis pathway was observed in a mouse model of PSO (Figure 4B). These findings suggest that dysregulation of the gut microbiome, which leads to an attenuated apoptotic pathway, may play a crucial role in the pathogenesis of PSO.
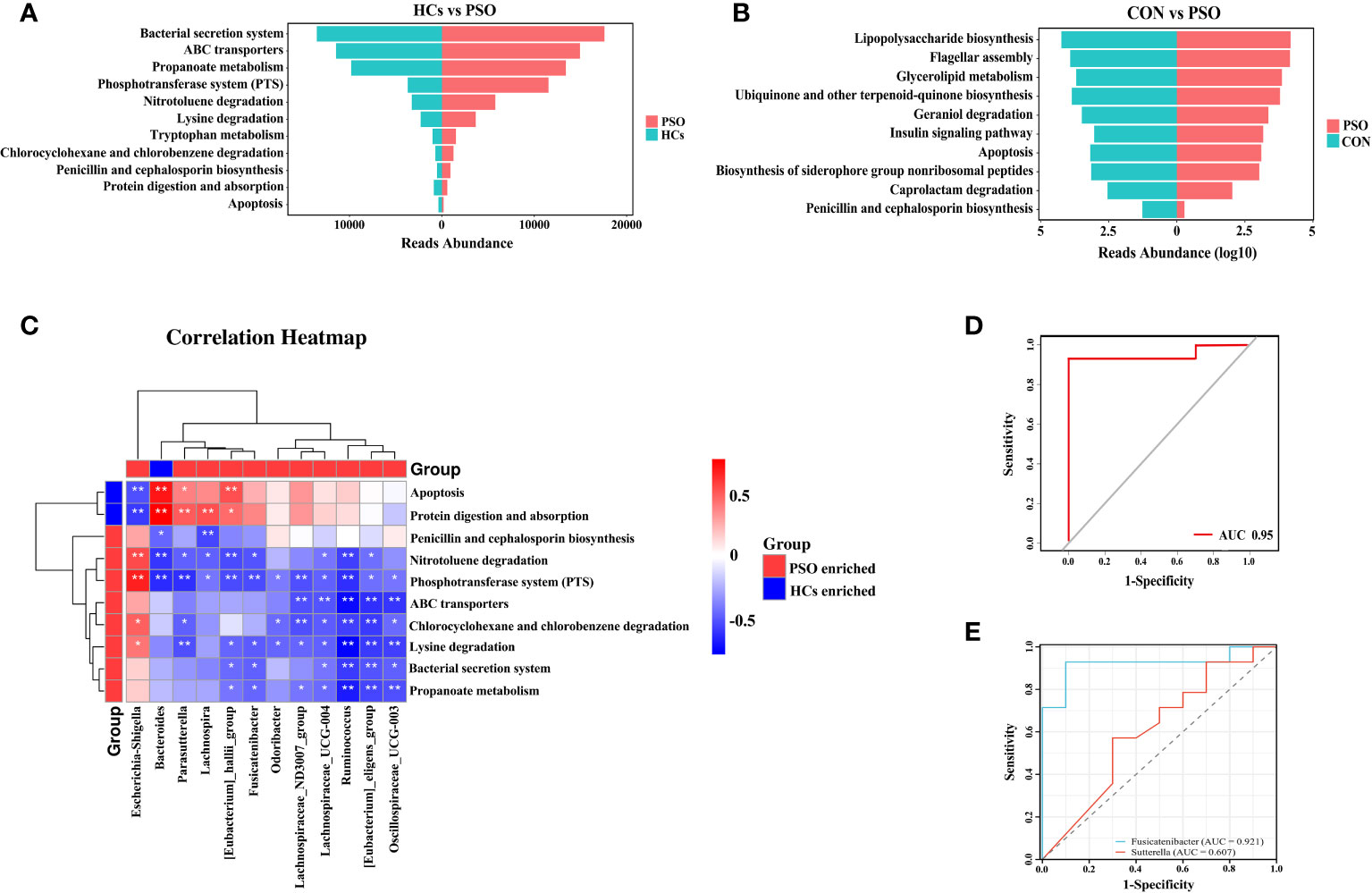
Figure 4 KEGG enrichment analysis and prediction of gut microbiome characteristics with machine learning classification algorithms. (A) KEGG enrichment analysis in PSO. (B) KEGG enrichment analysis in PSO model mice. (C) Correlation heat map revealing the relationship between differential genera and metabolic pathways in the gut of PSO patients. (D, E) Receiver operating characteristic curves using logistic regression for 2 genera. The diagonal line indicates the region under the curve value of 0.5, which is a random decision. HCs, Healthy Controls; PSO, Psoriasis patients and psoriasis model mouse; CON, Control Mouse *: p < 0.05, **: p < 0.01.
Subsequently, through correlation analysis, we determined that the gut microbiome with high abundance in HCs exhibited a positive correlation with enhanced metabolic pathways in HCs. Specifically, Bacteroides showed a positive correlation with Protein digestion and absorption and Apoptosis pathways. In contrast, Escherichia-Shigella, enriched in PSO, demonstrated a negative correlation with Protein digestion and absorption and Apoptosis pathways, while exhibiting a positive correlation with PTS, Nitrotoluene degradation, Chlorocyclohexane and chlorobenzene degradation, and Lysine degradation (Figure 4C). Previous studies have reported that BAs can exacerbate skin inflammation by activating G-protein bile acid-coupled receptor 5 (TGR5) and sphingosine-1-phosphate (S1P) receptor 2 (S1PR2) in the skin (33). Therefore, we hypothesized that Escherichia-Shigella may facilitate the onset and progression of PSO by inhibiting apoptosis through the BAs metabolic pathway.
The diagnostic efficacy of potential discriminatory biomarkers for PSO was evaluated by ROC curve analysis. By employing Lasso regression analysis, a simplified model consisting of 13 microbial genera successfully discriminated between PSO patients and HCs. To enhance the diagnostic efficacy of PSO, we constructed a classifier based on a logistic regression model. Notably, when the two most important genera were included, the AUC reached 0.95, demonstrating high sensitivity (92.86%) and specificity (100%) (Figure 4D). Ultimately, Fusicatenibacter and Sutterella were identified as robust diagnostic markers for distinguishing PSO from HCs, with their respective areas under the curve being 0.9214 and 0.6071 (Figure 4E).
3.5 Psoriatic skin microbiota enriched with Staphylococcus
PSO is an immune-mediated inflammatory skin disease, so it is particularly important to study the characterization of skin microecology in PSO. In this study, we investigated the composition of the skin microbiota in a PSO mouse model (Figure 5A). Our analysis revealed a significant increase in the phylum Firmicutes and a significant decrease in the phylum Bacteroidota in PSO model mice, in agreement with previous descriptions (34). At the genus level, Staphylococcus was found to dominate the skin microbiome in PSO model mice (Figure 5B). MetagenomeSeq difference analysis further identified Staphylococcus as a discriminatory feature between diseased and healthy skin (Figure 5C). These findings are consistent with previous literature reports, suggesting that Staphylococcus is the main causative genus responsible for the microecological dysbiosis of the skin in PSO patients (35).
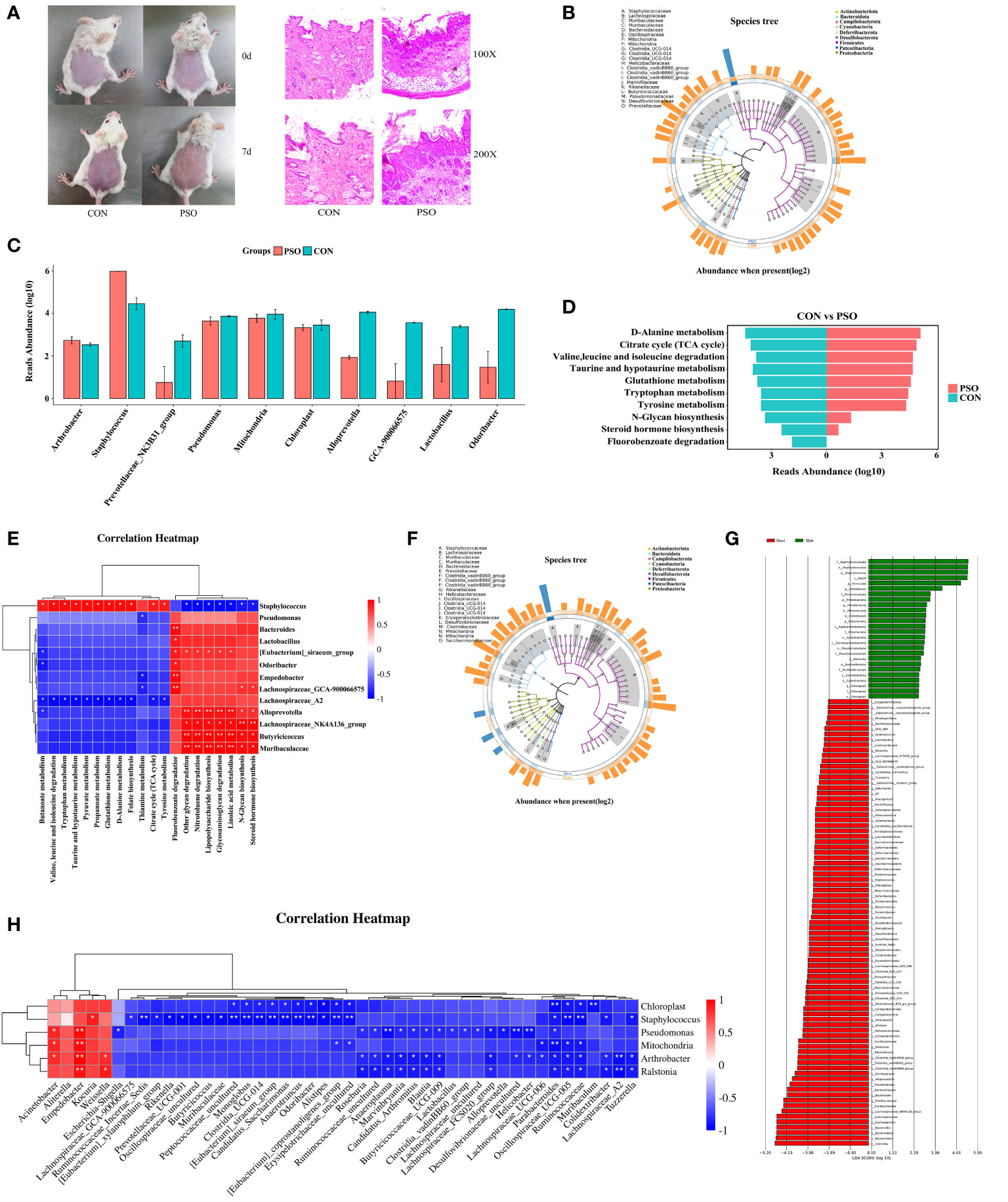
Figure 5 Skin microbiological characteristics of Psoriasis. (A) Diagram of the mouse model and representative micrographs of H&E staining of the skin. (B) GraPhlan plot showing the dominant flora in the skin of PSO model mouse. (C) Results of metagenomeSeq difference analysis showing only the top 10 genera with the most significant differences. (D) Enrichment analysis based on the results of metagenomeSeq difference analysis. The results of the metagenomeSeq difference analysis show only the top 10 genera with the most significant differences. (E) Correlation heatmap revealing the correlation between the differential genera and the enrichment pathway in the skin of PSO model mouse. (F) GraPhlan plot showing the dominant bacterial flora in the gut and in the skin of the PSO model. (G) LEfSe analysis revealing the difference between the gut and skin microbial communities in the PSO model mouse. (H) Correlation heatmap revealing the correlation between gut microbiota and skin microbials in the PSO model mouse. PSO, Psoriasis model mouse; CON, Control Mouse; Skin, Skin tissue of model mouse; Stool, Model mouse feces *: p < 0.05, **: p < 0.01.
Furthermore, KEGG enrichment analysis based on the metagenomeSeq results revealed the degradation of metabolic pathways such as Steroid hormone biosynthesis, Fluorobenzoate degradation, and N-Glycan biosynthesis in PSO model mice. On the other hand, Citrate Cycle (TCA cycle), Valine, leucine and isoleucine degradation, Tyrosine metabolism, Tryptophan metabolism, Taurine and hypotaurine metabolism, D-Alanine metabolism, and Glutathione metabolism were enhanced (Figure 5D). Correlation analysis showed that Staphylococcus is involved in multiple metabolic pathways, suggesting its contribution to skin lesions in PSO patients by affecting these pathways (Figure 5E).
The disruption of gut and skin microecology can have a significant impact on the overall homeostasis of the skin. Imbalances within the microbial community can either positively or negatively affect the integration of the gut and skin (20, 36). To evaluate the effect of an imbalance in the gut microbiome on the skin microecology, we compared the gut and skin microbiomes of mice from the same psoriasis (PSO) model. Our analysis revealed that the gut microbiome was more diverse than the skin microbiome (Supplementary Figure 1A). Additionally, the skin microbiota exhibited a more severe imbalance between Firmicutes and Bacteroidota (Supplementary Figure 2B). At the genus level, we observed that Staphylococcus, Chloroplast, Mitochondria, and Pseudomonas were dominant in the lesional skin of PSO model rats (Supplementary Figure 2C). However, the gut microbiome was dominated by Lachnoclostridium, Rikenellaceae_RC9_gut_group, Clostridia_vadinBB60_group, Muribaculaceae, Butyricoccaceae_UCG-009, Parabacteroides, Alloprevotella, Lachnospiraceae_FCS020_group, Clostridia_UCG-014, Candidatus_Saccharimonas, Bacteroides, Candidatus_Arthromitus, Anaeroplasma, [Eubacterium]_siraeum_ group, and Prevotellaceae_NK3B31_group (Figure 5F). Further analysis using LEfSe identified Staphylococcus, Arthromitus, Pseudomonas, Mitochondria, Ralstonia, and Chloroplast as discriminatory features of diseased skin, which includes many opportunistic pathogens (Figure 5G). This dysregulation of the skin microecology in PSO patients may compromise the skin barrier and lead to increased susceptibility to opportunistic pathogen infections. LEfSe also identified several discriminative features of intestinal microecological dysregulation in PSO, including Lachnospiraceae_NK4A136_group, Muribaculaceae, Bacteroides, Alloprevotella, Clostridia_vadinBB60_group, Roseburia (Figure 5G), which could potentially serve as markers for distinguishing between different tissues affected in PSO.
The understanding of the interplay between gut and skin microbiota can offer valuable insights for further investigations into PSO-related microbial communities, as microbial interactions may serve as the primary driving force behind the imbalance in these communities. By correlating the abundance of microorganisms, we explored the relationships between different microorganisms. Our analysis revealed a positive correlation between Staphylococcus, and Kocuria, Empedobacter, and Weissella at the genus level (Figure 5H). Moreover, we observed that the enrichment of Pseudomonas on the skin was positively associated with Acinetobacter and Empedobacter in the gut, while negatively correlated with various genera in the gut, including Desulfovibrionaceae_uncultured, Helicobacter, Anaeroplasma, Alloprevotella, Lachnospiraceae_FCS020_group, Escherichia-Shigella and Blautia (Figure 5H). Overall, our data suggest that disturbances in the gut microbial community may promote the colocalization of the skin by opportunistic pathogenic microorganisms such as Staphylococcus and Pseudomonas, exacerbating the disruption of skin microbiome in PSO and further contributing to the skin lesions in PSO.
3.6 IL-17A inhibitor modulates the gut microbiome in Psoriasis patients
Th17 cells release IL-17A, which effectively triggers the activation of keratinocytes either alone or in combination with TNF. These keratinocytes subsequently overproduce inflammatory cytokines and chemokines, attracting and activating immune cells. This process establishes a positive feedback loop of sustained inflammation, ultimately leading to skin damage in PSO. Numerous studies have demonstrated the efficacy, rapid action, safety, and long-lasting benefits of Skuticilumab, a monoclonal antibody that inhibits IL-17A. It provides comprehensive advantages for treating moderate-to-severe Psoriasis (26, 37). IL-17 production by γδT cells is inhibited in a histone deacetylase-dependent manner, mediated by Short-chain fatty acids (SCFAs) derived from gut microbiota, particularly propionic acid. These SCFAs exert anti-inflammatory effects (38). The use of IL-17 inhibitors in PSO patients is associated with changes in the composition of gut microbiome (39). Although our findings did not reveal significant alterations in gut microbial diversity before and after Secukinumab treatment (Supplementary Figure 3), further intergroup comparisons demonstrated a decrease in the relative abundance of Firmicutes, Proteobacteria, and Actinobacteriota, while that of Bacteroidota significantly increased (Figure 6A) following Secukinumab treatment. At the genus level, Secukinumab treatment led to an increase in the relative abundance of Bacteroides, Prevotella, Roseburia, Lachnoclostridium, and Phascolarctobacterium, while Faecalibacterium, Agathobacter, Megamonas, Dialister, and Escherichia-Shigella decreased (Figure 6B). These findings suggest that Secukinumab treatment improves gut microbiota dysbiosis in PSO patients (Figure 6C).
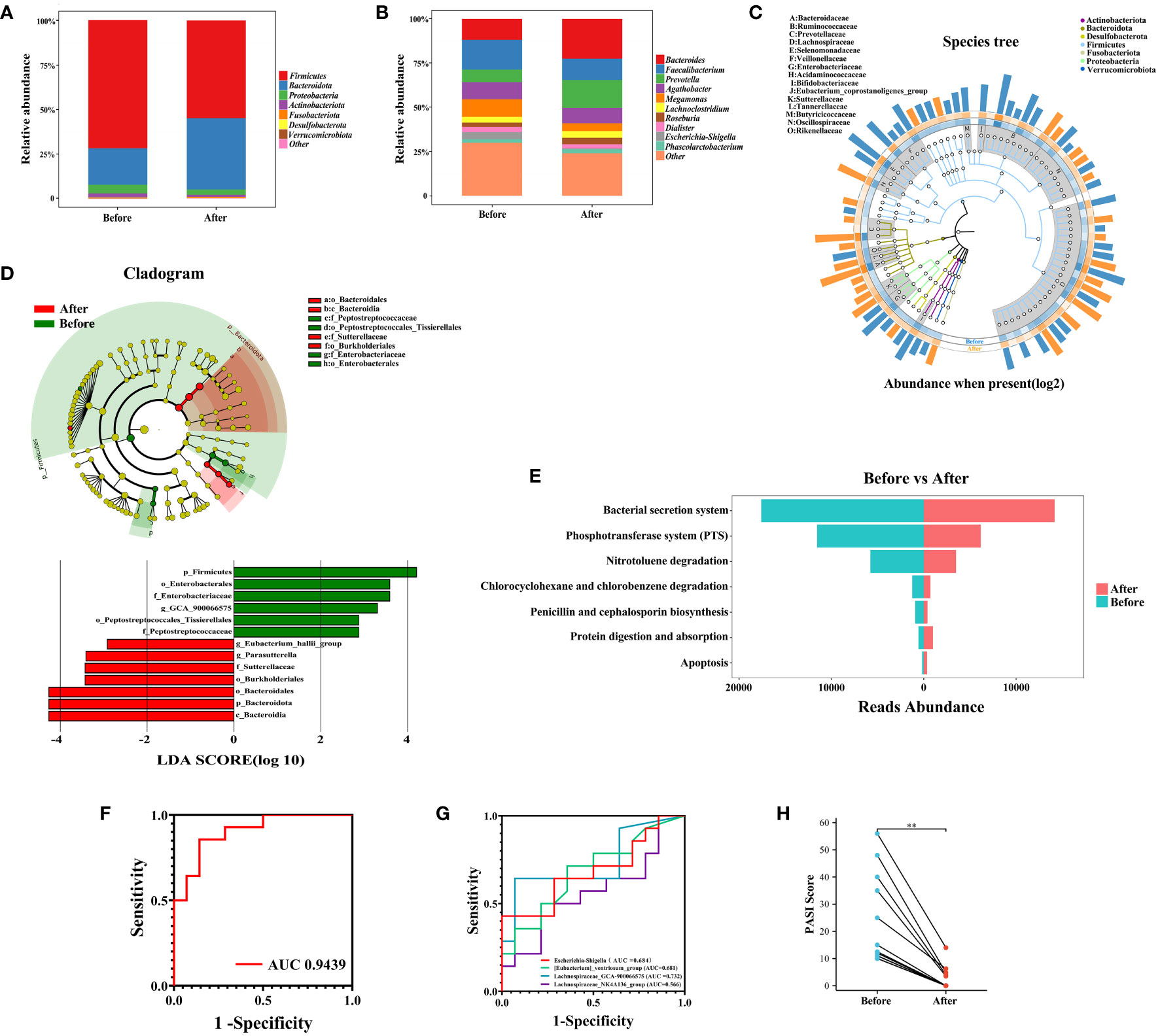
Figure 6 Changes in the gut microbiome before and after IL-17A inhibitor treatment. (A) and (B) Composition of gut microorganisms at the phylum and genus level before and after IL-17A inhibitor treatment. (C) GraPhlan plot showing the dominant flora in the gut before and after IL-17A inhibitor treatment. (D) Results of LEfSe analysis. (E) Results of KEGG enrichment analysis. (F) and (G) Receiver operating characteristic curves using logistic regression for 4 genera. (H) Pairwise plot showing a significant decrease in PASI scores in PSO patients after treatment with IL-17A inhibitor. Before, Before IL-17A inhibitor treatment; After, After IL-17A inhibitor treatment **: p < 0.01.
To investigate the impact of IL-17 inhibitors on the gut microbiome, the LEfSe assay was utilized to identify the key changes in the bacterial composition of PSO patients treated with Secukinumab. After 16 weeks of treatment, the relative abundance of certain bacterial genera in fecal samples showed significant alterations. Specifically, there was a notable increase in Parasutterella and Eubacterium_hallii_group, while the abundance of Lachnospiraceae_GCA_900066575 decreased (Figure 6D). Additionally, through PICRUSt2 functional predictive analysis, it was determined that anti-IL17 intervention in the gut microbiota of PSO patients can lead to changes in various metabolic pathways (40). Notably, Secukinumab treatment had a significant impact on pathways related to bacterial pathogenesis and virulence, such as the Bacterial secretion system, Phosphotransferase system (PTS), and Penicillin and cephalosporin biosynthesis, which were attenuated the most. Conversely, pathways associated with Apoptosis and Protein digestion and absorption showed the strongest increase after Secukinumab treatment (Figure 6E). These findings suggest that IL-17 inhibitor treatment alters the gut microbiota and its metabolites in PSO patients, bringing them closer to the composition seen in healthy individuals.
To identify potential biomarkers for assessing the efficacy of IL-17 inhibitors in patients with psoriasis, we utilized Lasso regression with logistic regression to analyze differential gut microbiota. Our analysis revealed that the inclusion of Lachnospiraceae_GCA-900066575, Escherichia-Shigella, [Eubacterium]_ventriosum_group, and Lachnospiraceae_NK4A136_group resulted in an area under the curve (AUC) of 0.9439 (Figure 6F), with corresponding AUC values of 0.732, 0.684, 0.681, and 0.566 (Figure 6G), respectively. Notably, all patients in our cohort experienced substantial remission of skin lesions upon Secukinumab treatment (Figure 6H). Therefore, we propose that these four gut microbiomes exhibit potential as biomarkers for evaluating the effectiveness of IL-17 inhibitors in PSO patients.
4 Discussion
In this study, we followed a rigorous sampling protocol and combined longitudinal cohort studies and animal experiments to measure the microbiome in the gut and skin by 16S rRNA. Animal experiments were conducted to validate the initial screening results and gain a deeper understanding of the microbiome associated with PSO. Our findings indicate that the gut microbiome in PSO patients exhibits higher heterogeneity. It is well known that impaired intestinal barrier function, increased inflammatory mediators, and release of metabolites can mediate the relationship between inflammatory skin disease and the gut microbiome (9). Most studies have shown no significant differences in α-diversity and β-diversity between HCs and PSO patients (9), which is consistent with our findings. However, some studies have reported severe ecological dysbiosis, lower diversity of the gut microbiota, and changes in the relative abundance of certain bacterial taxa in PSO patients (41). In line with these studies, we observed a significant increase in the phylum of Firmicutes and Actinobacteriota, as well as a significant decrease in the Bacteroidota phylum (41, 42). LEfSe analysis revealed that Escherichia-Shigella was significantly enriched in PSO patients. Correlation analysis showed a positive correlation between Escherichia-Shigella and serum levels of TBAs, as well as a negative correlation with levels of bilirubin. Previous studies have reported fat damage in PSO skin, characterized by lower concentrations of primary and secondary bile acids (43, 44), which aligns with our findings (Supplementary Figure 3). It has also been reported that changes in Escherichia-Shigella after Roux-en-Y gastric bypass (RYGB) surgery correlate with changes in BAs (45). Therefore, we hypothesize that Escherichia-Shigella in the intestine of PSO patients may promote the conversion of BAs into various forms through fecal excretion, leading to reduced BAs levels.
We also found a negative correlation between Escherichia-Shigella enrichment and the attenuated apoptotic pathways in PSO during functional enrichment analysis, suggesting that Escherichia-Shigella enrichment is involved in attenuated apoptotic pathways in PSO patients. Apoptosis is a specific form of cell death that usually prevents inflammation and tissue damage (46). This can result in the inhibition of Extracellular signal regulated kinase (ERK1/2) and phosphoinositide-3 kinase (PI3K)/serine/threonine protein kinase (Akt) signaling pathways by Sphingosine-1-phosphate receptor 2 (S1PR2), leading to the overproliferation of psoriatic keratinocytes and the development of PSO skin lesions (47, 48). Psoriatic keratinocytes in lesions exhibit increased resistance to apoptosis, which disrupts epidermal homeostasis and contributes to the pathogenesis of PSO (49, 50). Microarray transcriptome analysis of psoriatic lesions has revealed the upregulation of anti-apoptotic gene expression and the downregulation of pro-apoptotic gene expression (51). Sphingosine-1-phosphate (S1P), a bioactive sphingolipid, is involved in various cellular processes, including cell proliferation, differentiation, inflammation, and malignant transformation. S1P inhibits macrophage migration to inflammatory sites through the activation of S1PR2 and promotes wound healing (52–54). BAs, as upstream activators of S1PR2, can directly modulate S1PR2 activity and mediate the S1PR2 pathway through the Farnesol X Receptor (FXR) and TGR5 signaling pathways. Conjugated bile acids (CBAs) can activate the ERK1/2 and Akt signaling pathways via S1PR2, exerting an inhibitory effect on apoptosis (55–57). Therefore, the dysregulation of the gut microbiome, specifically the elevation of Escherichia-Shigella, may disrupt BAs metabolism, leading to the inhibition of ERK1/2 and Akt signaling pathways by S1PR2. This dysregulation ultimately promotes the overproliferation of keratinocytes and the development of PSO skin lesions.
Recognition of microbial-associated molecular patterns (MAMP) through specific pattern recognition receptors (PRRs), interactions between symbiotic organisms and hosts (58). Keratinocytes produce the antimicrobial peptide pepsin (LL-37) through contact with symbiotic microorganisms, which bind to nucleic acids in epithelial cells and stimulate the production of type I interferons, while their own RNA interacts with LL-37 to induce nitric oxide synthase (iNOS) to produce TNF-α (59). These cytokines influence the differentiation of primitive T cells into Th17 cells, which produce the interleukins IL-17 and IL-22, leading to the development of psoriatic lesions (60). Furthermore, Malassezia in the skin can metabolize to produce oleic acid (61) and the amount of oleic acid in the lesional skin of patients with psoriasis correlates with a reduction in IL-17 driver signatures following T cell activation (62), suggesting that skin micro ecological dysregulation compromises the skin barrier and alters skin composition, which in turn affects local immune function and promotes the development of psoriatic lesions. In line with previous studies, we noted a decrease in relative abundance of Bacteroides and Proteobacteria associated with PSO skin, while an increase in relative abundance of Firmicutes (63). Multiple studies have reported increased colonization of Staphylococcus in psoriatic skin (64). We found a significant increase in Staphylococcus colonization in both psoriatic lesion and non-lesion sites compared to healthy skin. Additionally, neonatal mice colonized with Staphylococcus exhibit strong Th17 polarization, leading to increased production of IL-17A and exacerbation of skin lesions (35). These findings suggest that the increased presence of Staphylococcus is not simply a result of structural changes in the skin caused by psoriasis, but rather an important trigger for disease progression. Furthermore, our analysis revealed that Staphylococcus colonization is associated with several metabolic pathways that are enhanced in psoriasis, including the TCA cycle, Tryptophan metabolism, Butanoate metabolism, and other Amino Acid metabolic pathways. Staphylococcus may play a regulatory role in energy homeostasis through its involvement in the TCA cycle. Citrate, the first intermediate of the TCA cycle, binds and activates catabolic control protein E (CcpE) (65), a major regulator of Staphylococcus virulence factor expression, central metabolism, iron acquisition, and bacterial virulence. Modulation of TCA cycle activity may provide Staphylococcus with a survival advantage during infection (66, 67). Inactivation of aconitase (CitB) leads to downregulation of the TCA cycle, thereby preventing maximal expression of virulence factors and altering the interaction between Staphylococcus and its host (68–70). Therefore, we hypothesize that Staphylococcus may exert its virulence effects by modulating the activity of the TCA cycle, ultimately contributing to the development of psoriatic skin lesions.
Gut microecological dysregulation can negatively impact skin integrity and function (71). Mucosal immune elements and signaling pathways play a crucial role in coordinating epidermal differentiation and intestinal barrier function (72, 73). Additionally, the transmission of intestinal microbes and their metabolites to the skin has been shown to impact skin physiology and the immune system (74). For instance, metabolites like cresol and phenol produced by Clostridium difficile, which are biomarkers of intestinal microbial dysbiosis, enter the bloodstream and accumulate in the skin, leading to compromised skin barrier integrity and impaired epidermal differentiation, thereby affecting keratinization (75). In our study, we observed significant differences in the gut microbiome and skin microbiome of PSO patients. Staphylococcus and Mitochondria were the dominant genera in the skin microbiome, whereas the gut microbiome was characterized by dominant genera such as Lachnospiraceae, Muribaculaceae, Bacteroidaceae, Prevotellaceae, Clostridia vadinBB60 group, Rikenellaceae, and Helicobacteraceae. Correlation analysis revealed that Staphylococcus was correlated with a wide range of genera in the gut, including SCFAs producers like Butyricicoccus, Monoglobus, Clostridia_UCG_014, Prevotellaceae_UCG_001, Parabacteroides, etc. SCFAs can regulate energy metabolism and immunity, suppress inflammatory responses, stabilize the intestinal ecosystem and maintain intestinal integrity. Sodium butyrate significantly down-regulates mRNA expression of biofilm-forming associated gene aggregation factor B (ClfB) and serine aspartate repeat (SdrC) of protein C, thereby inhibiting staphylococcal invasion (76). Butyrate and propionate ameliorate Staphylococcus-induced inflammatory responses by inhibiting NF-κB, IFN-β/STAT1, and HDAC, thereby reducing NO production (77). Based on these findings, we hypothesize that the reduction of SCFAs-producers in the gut of PSO patients may promote Staphylococcus colonization on the skin and accelerate the onset and progression of PSO.
In young individuals, PSO can often be mistaken for Atopic Dermatitis (AD) (78). AD lesional skin typically exhibits low bacterial diversity (79), accompanied by a significant increase in Staphylococcus aureus (80), which aligns with our observations in PSO lesional skin. Recent research has identified dysbiosis of the gut microbiota as a key factor in the pathogenesis of AD. Multiple studies have demonstrated that dysbiosis of the gut microbiota, characterized by a reduction in SCFAs-producing bacteria, plays a pivotal role in the development of AD (81). For instance, Clostridium, Prevotella, Clostridia, Bifidobacteraceae, Bifidobacterium, Bacteroidetes, Christensenellaceae_R7_group, and other bacterial species have been found to be decreased (82–85). Additionally, the “gut-skin” axis has emerged as a novel therapeutic target for the prevention and management of AD.
In mouse models, the skin microbiome, specifically Staphylococcus (35), can enhance Th17 polarization to promote PSO-like skin inflammation (86), while immunotherapy can alter the composition and classification of the gut microbiome and lead to changes in gut metabolism and immune response regulation (87, 88). IL-17 cytokine production effectively activates keratin-forming cells and promotes inflammation in the feed-forward circuit. The IL-17 pathway plays a key pathogenic role in autoimmune and inflammatory diseases and has been the focus of targeted therapies. Secukinumab is a selective IL-17A inhibitor that has been approved for PSO, Psoriatic Arthritis (PsA) in adults and children (26, 37). Our data show that the gut microbiome of PSO patients after Secukinumab treatment tends to change toward the distribution of normal human flora. Parasutterella and Eubacterium_hallii_group were the dominant bacteria found in the intestines of patients with PSO after treatment with the Secukinumab. Parasutterella is a type of Betaproteobacteria and is normally present in the healthy human gastrointestinal tract (89). Previous studies have shown that a high-fat diet (HFD) and Clostridium difficile infection (CDI) can decrease the abundance of Parasutterella in the gut (90, 91). However, an increase in Parasutterella has been found to have a protective effect against Intrahepatic Cholangiocarcinoma (ICC) (92). Parasutterella produces succinic acid and hypoxanthine (93), and is negatively correlated with levels of faecal Deoxycholic Acid (DCA) and Lithocholic Acid (LCA) (94). Hypoxanthine plays a key role in maintaining intestinal barrier function (95). On the other hand, Eubacterium_hallii_group can ameliorate intestinal inflammation and regulate BAs metabolism in the host through the production of SCFAs (96). Based on these findings, it is hypothesized that IL-17A inhibitors may improve intestinal inflammation in PSO patients by increasing the abundance of Eubacterium_hallii_group, which promotes SCFA production while increased Parasutterella promotes hypoxanthine production and regulates BAs metabolism in patients, which in turn improves intestinal barrier function and achieves the relief of clinical symptoms. In addition, the increase in Parasutterella after IL-17A inhibitor therapy may be related to the development of inflammatory bowel disease in some PSO patients (97), as higher levels of Parasutterella are associated with chronic inflammation in Irritable Bowel Syndrome (IBS) (98). Further research is needed to understand the specific mechanism of Parasutterella in IL-17A inhibitor therapy.
KEGG enrichment analyses revealed a significant enhancement of the previously attenuated Protein digestion and absorption and apoptosis pathways. Impaired lipolysis characterized by low concentrations of BAs is present in psoriatic skin. About one-third of patients with PSO eventually convert to PsA, and results from untargeted metabolomics studies have shown that patients with PsA have reduced serum BAs and Butyrate levels and increased levels of inflammatory lipid mediators (43, 99). Blocking the transport mediated by CCL20, a chemokine involved in PSO, can be improved by gut microbiota-derived BAs (32). Gut microbiota-derived SCFAs, which inhibit IL-17 production by intestinal γδ T cells (38), and in vitro supplementation with Bifidobacterium short CCFM683 improved PSO-like dermatitis by restoring the intestinal microbiome of PSO model rats, promoting BAs production, modulating the FXR/NF-κB pathway, reduces the generation of inflammatory cytokines, modulating keratinocytes and maintaining epidermal barrier function to attenuate PSO (100). IL-17 accumulation in lesions is a key factor in PSO pathogenesis, and inhibiting IL-17 production can attenuate abnormal keratinocyte proliferation. Staphylococcus colonization in PSO patients’ epidermis promotes IL-17A expression (35), which delays wound healing and can be abrogated by IL-17-blocking monoclonal antibodies (101–103). Therefore, using IL-17A inhibitors may inhibit Staphylococcus proliferation, reduce IL-17 expression, promote skin lesion healing, and restore the gut microbiome through the “gut-skin” axis. This can increasing Parasutterella and Eubacterium_hallii_group, promoting SCFAs and BAs production, reducing pro-inflammatory cytokine expression, regulating keratinocyte apoptosis, restoring the microbiota and epidermal barrier function, and improving PSO symptoms. In addition, Fusicatenibacter and Sutterella can serve as potential biomarkers for PSO diagnosis, while Lachnospiraceae_GCA-900066575, Escherichia-Shigella, [Eubacterium]_ventriosum_group, and Lachnospiraceae_NK4A136_group have potential value in discriminating PSO disease remission. This provides a rationale for targeting gut microbiome alterations to treat PSO.
5 Conclusion
In this investigation, we discovered that PSO is characterized by alterations in the intrinsic microbial communities of the gut and skin, as well as disruptions in their metabolic pathways. This was determined through longitudinal studies and validation of mouse models, suggesting that the interactions between gut and skin microbes may play a crucial role in the development of PSO. Interestingly, the administration of IL-17A inhibitors restored the gut microbiota of PSO patients, potentially leading to an improvement in their clinical symptoms. This finding highlights the importance of maintaining a healthy gut microbiota for individuals with PSO and supports the connection between the gut-skin axis and PSO. It is noteworthy that this study is the first to observe an increase in Parasutterella in the gut of PSO patients after IL-17A inhibitor treatment. The bidirectional effect of Parasutterella on gut inflammation may serve as a possible mechanism by which IL-17A inhibitors contribute to inflammatory bowel disease. However, further investigation is required to better understand the role of Parasutterella in the gut of PSO patients. It should be noted that this study has limitations, as it involved a small sample size. Therefore, additional research is needed to confirm the causal relationship between gut microbiota and PSO, and these findings should be validated in larger studies.
Data availability statement
The data presented in the study are deposited in the Sequence Read Archive (SRA) repository, accession number PRJNA1078442.
Ethics statement
The studies involving humans were approved by the Ethics Committee of Heji Hospital of Changzhi Medical College (number: 2023-13). The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study. The animal study was approved by the Ethics Committee of Changzhi Medical College (number: DW2023051). The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
HZ: Funding acquisition, Resources, Writing – review & editing, Conceptualization. LS: Formal Analysis, Writing – original draft, Data curation, Writing – review & editing. YZ: Data curation, Writing – original draft. ZL: Investigation, Writing – original draft. NW: Investigation, Writing – original draft. QZ: Resources, Writing – original draft. CG: Methodology, Writing – review & editing. JL: Funding acquisition, Project administration, Supervision, Writing – review & editing, Conceptualization.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Research Project Supported by Shanxi Scholarship Council of China (2020-191), Four “Batches” Innovation Project of Invigorating Medical through Science and Technology of Shanxi Province (2020SYS08), the Project of Central Guides Local Science and Technology Development Funds (YDZJSX2022C031), the Foundation of Shanxi Key Laboratory for immunomicroecology (202104010910012) and Changes of intestinal microflora in patients with Psoriasis before and after treatment with secuchiumab based on 16S rRNA sequencing (202209).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1344963/full#supplementary-material
References
1. Fry L, Baker BS. Triggering Psoriasis: The role of infections and medications. Clin Dermatol. (2007) 25:606–15. doi: 10.1016/j.clindermatol.2007.08.015
2. Korman N. Management of Psoriasis as a systemic disease: What is the evidence? Br J Dermatol. (2020) 182:840–8. doi: 10.1111/bjd.18245
3. Kong HH, Andersson B, Clavel T, Common JE, Jackson SA, Olson ND, et al. Performing skin microbiome research: a method to the madness. J Invest Dermatol. (2017) 137:561–8. doi: 10.1016/j.jid.2016.10.033
4. Loesche MA, Farahi K, Capone K, Fakharzadeh S, Blauvelt A, Duffin KC, et al. Longitudinal study of the Psoriasis-associated skin microbiome during therapy with ustekinumab in a randomized phase 3b clinical trial. J Invest Dermatol. (2018) 138:1973–81. doi: 10.1016/j.jid.2018.03.1501
5. Roberson EDO, Bowcock AM. Psoriasis genetics: breaking the barrier. Trends Genet. (2010) 26:415–23. doi: 10.1016/j.tig.2010.06.006
6. Karczewski J, Dobrowolska A, Rychlewska-Hańczewska A, Adamski Z. New insights into the role of T cells in pathogenesis of Psoriasis and psoriatic arthritis. Autoimmunity. (2016) 49:435–50. doi: 10.3109/08916934.2016.1166214
7. Le S, Toussi A, Maverakis N, Marusina AI, Barton VR, Merleev AA, et al. The cutaneous and intestinal microbiome in psoriatic disease. Clin Immunol. (2020) 218:108537. doi: 10.1016/j.clim.2020.108537
8. Szántó M, Dózsa A, Antal D, Szabó K, Kemény L, Bai P. Targeting the gut-skin axis—Probiotics as new tools for skin disorder management? Exp Dermatol. (2019) 28:1210–8. doi: 10.1111/exd.14016
9. Sikora M, Stec A, Chrabaszcz M, Knot A, Waskiel-Burnat A, Rakowska A, et al. Gut microbiome in Psoriasis: An updated review. Pathogens. (2020) 9:463. doi: 10.3390/pathogens9060463
10. Kosiewicz MM, Dryden GW, Chhabra A, Alard P. Relationship between gut microbiota and development of T cell associated disease. FEBS Lett. (2014) 588:4195–206. doi: 10.1016/j.febslet.2014.03.019
11. Vlachos C, Gaitanis G, Katsanos KH, Christodoulou DK, Tsianos E, Bassukas ID. Psoriasis and inflammatory bowel disease: Links and risks. Psoriasis (Auckl). (2016) 6:73–92. doi: 10.2147/PTT.S85194
12. Visser MJ, Kell DB, Pretorius E. Bacterial dysbiosis and translocation in Psoriasis vulgaris. Front Cell Infect Microbiol. (2019) 9:7. doi: 10.3389/fcimb.2019.00007
13. Bousbaine D, Fisch LI, London M, Bhagchandani P, Rezende de Castro TB, Mimee M, et al. A conserved Bacteroidetes antigen induces anti-inflammatory intestinal T lymphocytes. Science. (2022) 377:660–6. doi: 10.1126/science.abg5645
14. Sano T, Kageyama T, Fang V, Kedmi R, Martinez CS, Talbot J, et al. Redundant cytokine requirement for intestinal microbiota-induced Th17 cell differentiation in draining lymph nodes. Cell Rep. (2021) 36:109608. doi: 10.1016/j.celrep.2021.109608
15. Hao F, Tian M, Zhang X, Jin X, Jiang Y, Sun X, et al. Butyrate enhances CPT1A activity to promote fatty acid oxidation and iTreg differentiation. Proc Natl Acad Sci USA. (2021) 118:e2014681118. doi: 10.1073/pnas.2014681118
16. Belkaid Y, Tamoutounour S. The influence of skin microorganisms on cutaneous immunity. Nat Rev Immunol. (2016) 16:353–66. doi: 10.1038/nri.2016.48
17. Lowes MA, Suárez-Fariñas M, Krueger JG. Immunology of psoriasis. Annu Rev Immunol. (2014) 32:227–55. doi: 10.1146/annurev-immunol-032713-120225
18. Alzahrani Y, Alesa D, Alshamrani H, Alamssi D, Alzahrani N, Almohammadi M. The role of gut microbiome in the pathogenesis of Psoriasis and the therapeutic effects of probiotics. J Fami Med Primary Care. (2019) 8:3496. doi: 10.4103/jfmpc.jfmpc_709_19
19. de Francesco MA, Caruso A. The gut microbiome in psoriasis and crohn’s disease: is its perturbation a common denominator for their pathogenesis? Vaccines. (2022) 10:244. doi: 10.3390/vaccines10020244
20. O’Neill CA, Monteleone G, McLaughlin JT, Paus R. The gut-skin axis in health and disease: a paradigm with therapeutic implications. BioEssays. (2016) 38:1167–76. doi: 10.1002/bies.201600008
21. Hsu DK, Fung MA, Chen HL. Role of skin and gut microbiota in the pathogenesis of Psoriasis, an inflammatory skin disease. Med Microecol. (2020) 4:100016. doi: 10.1016/j.medmic.2020.100016
22. Ramírez-Boscá A, Navarro-López V, Martínez-Andrés A, Such J, Francés R, Horga de la Parte J, et al. Identification of bacterial DNA in the peripheral blood of patients with active Psoriasis. JAMA Dermatol. (2015) 151:670. doi: 10.1001/jamadermatol.2014.5585
23. Fletcher JM, Moran B, Petrasca A, Smith CM. IL-17 in inflammatory skin diseases Psoriasis and hidradenitis suppurativa. Clin Exp Immunol. (2020) 201:121–34. doi: 10.1111/cei.13449
24. Yan Y, Lei Y, Qu Y, Fan Z, Zhang T, Xu Y, et al. Bacteroides uniformis-induced perturbations in colonic microbiota and bile acid levels inhibit TH17 differentiation and ameliorate colitis developments. NPJ Biofilms Microbiomes. (2023) 9:56. doi: 10.1038/s41522-023-00420-5
25. Nermes M, Kantele JM, Atosuo TJ, Salminen S, Isolauri E. Interaction of orally administered Lactobacillus rhamnosus GG with skin and gut microbiota and humoral immunity in infants with atopic dermatitis. Clin Exp Allergy. (2011) 41:370–7. doi: 10.1111/j.1365-2222.2010.03657.x
26. Yiu ZZN, Becher G, Kirby B, Laws P, Reynolds NJ, Smith CH, et al. Drug survival associated with effectiveness and safety of treatment with guselkumab, ixekizumab, secukinumab, ustekinumab, and adalimumab in patients with psoriasis. JAMA Dermatol. (2022) 158:1131–41. doi: 10.1001/jamadermatol.2022.2909
27. Yeh N-L, Hsu C-Y, Tsai T-F, Chiu H-Y. Gut microbiome in psoriasis is perturbed differently during secukinumab and ustekinumab therapy and associated with response to treatment. Clin Drug Investig. (2019) 39:1195–203. doi: 10.1007/s40261-019-00849-7
28. Committee on Psoriasis, Chinese Society of Dermatology. Guideline for the diagnosis and treatment of psoriasis in China (2018 complete edition). Chin J Dermatol. (2019) 52:667–710. doi: 10.35541/cjd.20190847
29. de Oca Pedrosa AM, Oakley A, Rogers J, Epiha MR. PASI vs PO-PASI: Patient-Oriented PASI (PO-PASI) is comparable to clinician score PASI. Australas J Dermatol. (2022) 63:e67–70. doi: 10.1111/ajd.13738
30. Gerdes S, Körber A, Biermann M, Karnthaler C, Reinhardt M. Absolute and relative psoriasis area and severity index (PASI) treatment goals and their association with health-related quality of life. J Dermatol Treat. (2020) 31:470–5. doi: 10.1080/09546634.2020.1746734
31. Maravilla-Herrera P, Merino M, Alfonso Zamora S, Filgueiras JB, Carrillo JMC, Delgado Sánchez O, et al. The social value of a PASI 90 or PASI 100 response in patients with moderate-to-severe plaque psoriasis in Spain. Front Public Health. (2023) 11:1000776. doi: 10.3389/fpubh.2023.1000776
32. Shi Z, Wu X, Wu C-Y, Singh SP, Law T, Yamada D, et al. Bile acids improve psoriasiform dermatitis through inhibition of IL-17A expression and CCL20-CCR6-mediated trafficking of T cells. J Invest Dermatol. (2022) 142:1381–1390.e11. doi: 10.1016/j.jid.2021.10.027
33. Prasant Kumar J, Sheng L, Mcneil K, Chau TQ, Yu S, Kiuru M, et al. Long-term Western diet intake leads to dysregulated bile acid signaling and dermatitis with Th2 and Th17 pathway features in mice. J Dermatol Sci. (2019) 95:13–20. doi: 10.1016/j.jdermsci.2019.05.007
34. Mahmud R, Akter S, Tamanna SK, Mazumder L, Esti IZ, Banerjee S, et al. Impact of gut microbiome on skin health: gut-skin axis observed through the lenses of therapeutics and skin diseases. Gut Microbes. (2022) 14:2096995. doi: 10.1080/19490976.2022.2096995
35. Chang H-W, Yan D, Singh R, Liu J, Lu X, Ucmak D, et al. Alteration of the cutaneous microbiome in Psoriasis and potential role in Th17 polarization. Microbiome. (2018) 6:154. doi: 10.1186/s40168-018-0533-1
36. Ellis SR, Nguyen M, Vaughn AR, Notay M, Burney WA, Sandhu S, et al. The skin and gut microbiome and its role in common dermatologic conditions. Microorganisms. (2019) 7:550. doi: 10.3390/microorganisms7110550
37. Gottlieb AB, Deodhar A, Mcinnes IB, Baraliakos X, Reich K, Schreiber S, et al. Long-term safety of secukinumab over five years in patients with moderate-to-severe plaque psoriasis, psoriatic arthritis and ankylosing spondylitis: update on integrated pooled clinical trial and post-marketing surveillance data. Acta Derm Venereol. (2022) 102:adv00698. doi: 10.2340/actadv.v102.563
38. Dupraz L, Magniez A, Rolhion N, Richard ML, Costa GD, Touch S, et al. Gut microbiota-derived short-chain fatty acids regulate IL-17 production by mouse and human intestinal γδ T cells. Cell Rep. (2021) 36:109332. doi: 10.1016/j.celrep.2021.109332
39. Huang Y-H, Chang L-C, Chang Y-C, Chung W-H, Yang S-F, Su S-C. Compositional alteration of gut microbiota in psoriasis treated with IL-23 and IL-17 inhibitors. Int J Mol Sci. (2023) 24:4568. doi: 10.3390/ijms24054568
40. Douglas GM, Maffei VJ, Zaneveld JR, Yurgel SN, Brown JR, Taylor CM, et al. PICRUSt2 for prediction of metagenome functions. Nat Biotechnol. (2020) 38:685–8. doi: 10.1038/s41587-020-0548-6
41. Hidalgo-Cantabrana C, Gomez J, Delgado S, Requena-López S, Queiro-Silva R, Margolles A, et al. Gut microbiota dysbiosis in a cohort of patients with Psoriasis. Br J Dermatol. (2019) 181:1287–95. doi: 10.1111/bjd.17931
42. Shapiro J, Cohen NA, Shalev V, Uzan A, Koren O, Maharshak N. Psoriatic patients have a distinct structural and functional fecal microbiota compared with controls. J Dermatol. (2019) 46:595–603. doi: 10.1111/1346-8138.14933
43. Paine A, Brookes PS, Bhattacharya S, Li D, Garcia-Hernandez MdL, Tausk F, et al. Dysregulation of bile acids, lipids, and nucleotides in psoriatic arthritis revealed by unbiased profiling of serum metabolites. Arthritis Rheumatol. (2023) 75:53–63. doi: 10.1002/art.42288
44. Sorokin AV, Domenichiello AF, Dey AK, Yuan Z-X, Goyal A, Rose SM, et al. Bioactive lipid mediator profiles in human psoriasis skin and blood. J Invest Dermatol. (2018) 138:1518–28. doi: 10.1016/j.jid.2018.02.003
45. Dang JT, Mocanu V, Park H, Laffin M, Tran C, Hotte N, et al. Ileal microbial shifts after Roux-en-Y gastric bypass orchestrate changes in glucose metabolism through modulation of bile acids and L-cell adaptation. Sci Rep. (2021) 11:23813. doi: 10.1038/s41598-021-03396-4
46. Yan Q, Bach DQ, Gatla N, Sun P, Liu J-W, Lu J-Y, et al. Deacetylated GM3 promotes uPAR-associated membrane molecular complex to activate p38 MAPK in metastatic melanoma. Mol Cancer Res. (2013) 11:665–75. doi: 10.1158/1541-7786.MCR-12-0270-T
47. Luo Y, Hara T, Kawashima A, Ishido Y, Suzuki S, Ishii N, et al. Pathological role of excessive DNA as a trigger of keratinocyte proliferation in Psoriasis. Clin Exp Immunol. (2020) 202:1–10. doi: 10.1111/cei.13455
48. Pohla L, Ottas A, Kaldvee B, Abram K, Soomets U, Zilmer M, et al. Hyperproliferation is the main driver of metabolomic changes in Psoriasis lesional skin. Sci Rep. (2020) 10:3081. doi: 10.1038/s41598-020-59996-z
49. Laporte M, Galand P, Fokan D, de Graef C, Heenen M. Apoptosis in established and healing Psoriasis. Dermatology. (2000) 200:314–6. doi: 10.1159/000018394
50. Jia HY, Zhang K, Lu WJ, Xu GW, Zhang JF, Tang ZL. LncRNA MEG3 infuences the proliferation and apoptosis of Psoriasis epidermal cells by targeting miR-21/caspase-8. BMC Mol Cell Biol. (2019) 20:46. doi: 10.1186/s12860-019-0229-9
51. Eding CB, Enerback C. Involved and uninvolved psoriatic keratinocytes display a resistance to apoptosis that may contribute to epidermal thickness. Acta Derm Venereol. (2017) 97:788–96. doi: 10.2340/00015555-2656
52. Maceyka M, Harikumar KB, Milstien S, Spiegel S. Sphingosine-1- phosphate signaling and its role in disease. Trends Cell Biol. (2012) 22:50–60. doi: 10.1016/j.tcb.2011.09.003
53. Michaud J, Im DS, Hla T. Inhibitory role of sphingosine 1-phosphate receptor 2 in macrophage recruitment during inflammation. J Immunol. (2010) 184:1475–83. doi: 10.4049/jimmunol.0901586
54. Serriere-Lanneau V, Teixeira-Clerc F, Li L, Schippers M, de Wries W, Julien B, et al. The sphingosine 1-phosphate receptor S1P2 triggers hepatic wound healing. FASEB J. (2007) 21:2005–13. doi: 10.1096/fj.06-6889com
55. Strub GM, Maceyka M, Hait NC, Milstien S, Spiegel S. Extracellular and intracellular actions of sphingosine-1-phosphate. Adv Exp Med Biol. (2010) 688:141–55. doi: 10.1007/978-1-4419-6741-1_10
56. Studer E, Zhou X, Zhao R, Wang Y, Takabe K, Nagahashi M, et al. Conjugated bile acids activate the sphingosine-1-phosphate receptor 2 in primary rodent hepatocytes. Hepatology. (2012) 55:267–76. doi: 10.1002/hep.24681
57. Cao R, Cronk ZX, Zha W, Sun L, Wang X, Fang Y, et al. Bile acids regulate hepatic gluconeogenic genes and farnesoid X receptor via G (α)i-protein-coupled receptors and the AKT pathway. J Lipid Res. (2010) 51:2234–44. doi: 10.1194/jlr.M004929
58. Chu H, Mazmanian SK. Innate immune recognition of the microbiota promotes host-microbial symbiosis. Nat Immunol. (2013) 14:668–75. doi: 10.1038/ni.2635
59. Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. (2014) 157:121–41. doi: 10.1016/j.cell.2014.03.011
60. Fry L, Baker BS, Powles AV. Is chronic plaque Psoriasis triggered by microbiota in the skin? Br J Dermatol. (2013) 169:47–52. doi: 10.1111/bjd.12322
61. Park M, Park S, Jung WH. Skin commensal fungus malassezia and its lipases. J Microbiol Biotechnol. (2021) 31:637–44. doi: 10.4014/jmb.2012.12048
62. El Mahi Y, Varin A, Vetter M, Dal Zuffo L, Mazzeo L, Pais De Barros J-P, et al. Resolved psoriasis with abundant oleic acid in stratum corneum exhibits lower T-cell-driven IL-17 signature. J Invest Dermatol. (2023) 143:2145–2152.e6. doi: 10.1016/j.jid.2023.04.021
63. Chen YJ, Ho HJ, Tseng CH, Lai ZL, Shieh JJ, Wu CY. Intestinal microbiota profiling and predicted metabolic dysregulation in Psoriasis patients. Exp Dermatol. (2018) 27:1336–43. doi: 10.1111/exd.13786
64. Totté JEE, van der Feltz WT, Bode LGM, van Belkum A, van Zuuren EJ, Pasmans SGMA. A systematic review and meta-analysis on Staphylococcus aureus carriage in Psoriasis, acne and rosacea. Eur J Clin Microbiol Infect Dis. (2016) 35:1069–77. doi: 10.1007/s10096-016-2647-3
65. Hartmann T, Zhang B, Baronian G, Schulthess B, Homerova D, Grubmüller S, et al. Catabolite control protein E (CcpE) is a LysR-type transcriptional regulator of tricarboxylic acid cycle activity in Staphylococcus aureus. J Biol Chem. (2013) 288:36116–28. doi: 10.1074/jbc.M113.516302
66. Hu X, Guo J, Zhao C, Jiang P, Maimai T, Yanyi L, et al. The gut microbiota contributes to the development of Staphylococcus aureus-induced mastitis in mice. ISME J. (2020) 14:1897–910. doi: 10.1038/s41396-020-0651-1
67. Alva-Murillo N, López-Meza JE, Ochoa-Zarzosa A. Nonprofessional phagocytic cell receptors involved in staphylococcus aureus internalization. BioMed Res Int. (2014) 2014:538546. doi: 10.1155/2014/538546
68. Somerville GA, Chaussee MS, Morgan CI, Fitzgerald JR, Dorward DW, Reitzer LJ, et al. Staphylococcus aureus aconitase inactivation unexpectedly inhibits post-exponential-phase growth and enhances stationary-phase survival. Infect Immun. (2002) 70:6373–82. doi: 10.1128/IAI.70.11.6373-6382.2002
69. Ding Y, Liu X, Chen F, Di H, Xu B, Zhou L, et al. Metabolic sensor governing bacterial virulence in Staphylococcus aureus. Proc Natl Acad Sci USA. (2014) 111:E4981–90. doi: 10.1073/pnas.1411077111
70. Akhtar M, Naqvi SU-A, Liu Q, Pan H, Ma Z, Kong N, et al. Short chain fatty acids (SCFAs) are the potential immunomodulatory metabolites in controlling staphylococcus aureus-mediated mastitis. Nutrients. (2022) 14:3687. doi: 10.3390/nu14183687
71. Polkowska-Pruszyńska B, Gerkowicz A, Krasowska D. The gut microbiome alterations in allergic and inflammatory skin diseases–an update. J Eur Acad Dermatol Venereol. (2020) 34:455–64. doi: 10.1111/jdv.15951
72. Sirisinha S. The potential impact of gut microbiota on your health: Current status and future challenges. Asian Pac J Allergy Immunol. (2016) 34:249–64. doi: 10.12932/AP0803
73. Huttenhower C, Gevers D, Knight R, Abubucker S, Badger JH, Chinwalla AT, et al. Structure, function and diversity of the healthy human microbiome. Nature. (2012) 486:207–14. doi: 10.1038/nature11234
74. Purchiaroni F, Tortora A, Gabrielli M, Bertucci F, Gigante G, Ianiro G, et al. The role of intestinal microbiota and the immune system. Eur Rev Med Pharmacol Sci. (2013) 17:323–33.
75. Miyazaki K, Masuoka N, Kano M, Iizuka R. Bifidobacterium fermented milk and galacto-oligosaccharides lead to improved skin health by decreasing phenols production by gut microbiota. Benef Microbes. (2014) 5:121–8. doi: 10.3920/BM2012.0066
76. Frutis-Murillo M, Sandoval-Carrillo MA, Alva-Murillo N, Ochoa-Zarzosa A, López-Meza JE. Immunomodulatory molecules regulate adhesin gene expression in Staphylococcus aureus: Effect on bacterial internalization into bovine mammary epithelial cells. Microb Pathog. (2019) 131:15–21. doi: 10.1016/j.micpath.2019.03.030
77. Park JW, Kim HY, Kim MG, Jeong S, Yun C-H, Han SH. Short-chain fatty acids inhibit staphylococcal lipoprotein-induced nitric oxide production in murine macrophages. Immune Netw. (2019) 19:e9. doi: 10.4110/in.2019.19.e9
78. Kapila S, Hong E, Fischer G. A comparative study of childhood psoriasis and atopic dermatitis and greater understanding of the overlapping condition, psoriasis-dermatitis. Australas J Dermatol. (2012) 53:98–105. doi: 10.1111/j.1440-0960.2012.00878.x
79. Baurecht H, Rühlemann MC, Rodríguez E, Thielking F, Harder I, Erkens A-S, et al. Epidermal lipid composition, barrier integrity, and eczematous inflammation are associated with skin microbiome configuration. J Allergy Clin Immunol. (2018) 141:1668–1676.e16. doi: 10.1016/j.jaci.2018.01.019
80. Bjerre RD, Bandier J, Skov L, Engstrand L, Johansen JD. The role of the skin microbiome in atopic dermatitis: A systematic review. Br J Dermatol. (2017) 177:1272–8. doi: 10.1111/bjd.15390
81. Nylund L, Nermes M, Isolauri E, Salminen S, de Vos WM, Satokari R. Severity of atopic disease inversely correlates with intestinal microbiota diversity and butyrate-producing bacteria. Allergy. (2015) 70:241–4. doi: 10.1111/all.12549
82. Xue Y, Zhang L, Chen Y, Wang H, Xie J. Gut microbiota and atopic dermatitis: a two-sample Mendelian randomization study. Front Med (Lausanne). (2023) 10:1174331. doi: 10.3389/fmed.2023.1174331
83. Pessôa R, Clissa PB, Sanabani SS. The interaction between the host genome, epigenome, and the gut-skin axis microbiome in atopic dermatitis. Int J Mol Sci. (2023) 24:14322. doi: 10.3390/ijms241814322
84. Ye S, Yan F, Wang H, Mo X, Liu J, Zhang Y, et al. Diversity analysis of gut microbiota between healthy controls and those with atopic dermatitis in a Chinese population. J Dermatol. (2021) 48:158–67. doi: 10.1111/1346-8138.15530
85. Wang Y, Hou J, Tsui JC-C, Wang L, Zhou J, Chan UK, et al. Unique gut microbiome signatures among adult patients with moderate to severe atopic dermatitis in southern Chinese. Int J Mol Sci. (2023) 24:12856. doi: 10.3390/ijms241612856
86. Zakostelska Z, Malkova J, Klimesova K, Rossmann P, Hornova M, Novosadova I, et al. Intestinal microbiota promotes Psoriasis-like skin infammation by enhancing Th17 response. PloS One. (2016) 11:e0159539. doi: 10.1371/journal.pone.0159539
87. Zhang Z, Liu L, Tang H, Jiao W, Zeng S, Xu Y, et al. Immunosuppressive efect of the gut microbiome altered by high-dose tacrolimus in mice. Am J Transplant. (2018) 18:1646–56. doi: 10.1111/ajt.14661
88. Huang EY, Inoue T, Leone VA, Dalal S, Touw K, Wang Y, et al. Using corticosteroids to reshape the gut microbiome: implications for inflammatory bowel diseases. Inflammation Bowel Dis. (2015) 21:963–72. doi: 10.1097/MIB.0000000000000332
89. Willing BP, Dicksved J, Halfvarson J, Andersson AF, Lucio M, Zheng Z, et al. A pyrosequencing study in twins shows that GI microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology. (2010) 139:1844–1854.e1. doi: 10.1053/j.gastro.2010.08.049
90. Danneskiold-Samsøe NB, Andersen D, Radulescu ID, Normann-Hansen A, Brejnrod A, Kragh M, et al. A safflower oil based highfat/high-sucrose diet modulates the gut microbiota and liver phospholipid profiles associated with early glucose intolerance in the absence of tissue inflammation. Mol Nutr Food Res. (2017) 61 (5):1–19. doi: 10.1002/mnfr.201600528
91. Zhang L, Dong D, Jiang C, Li Z, Wang X, Peng Y. Insight into alteration of gut microbiota in Clostridium difficile infection and asymptomatic C. difficile colonization. Anaerobe. (2015) 34:1–7. doi: 10.1016/j.anaerobe.2015.03.008
92. Chen Z, Shi W, Chen K, Lu C, Li X, Li Q. Elucidating the causal association between gut microbiota and intrahepatic cholangiocarcinoma through Mendelian randomization analysis. Front Microbiol. (2023) 14:1288525. doi: 10.3389/fmicb.2023.1288525
93. Ju T, Kong JY, Stothard P, Willing BP. Defining the role of Parasutterella, a previously uncharacterized member of the core gut microbiota. ISME J. (2019) 13:1520–34. doi: 10.1038/s41396-019-0364-5
94. Acharya C, Betrapally NS, Gillevet PM, Sterling RK, Akbarali H, White MB, et al. Chronic opioid use is associated with altered gut microbiota and predicts readmissions in patients with cirrhosis. Aliment Pharmacol Ther. (2017) 45:319–31. doi: 10.1111/apt.13858
95. Scott Lee J, Wang RX, Alexeev EE, Lanis JM, Battista KD, Glover LE, et al. Hypoxanthine is a checkpoint stress metabolite in colonic epithelial energy modulation and barrier function. J Biol Chem. (2018) 293:6039–51. doi: 10.1074/jbc.RA117.000269
96. Mukherjee A, Lordan C, Ross RP, Cotter PD. Gut microbes from the phylogenetically diverse genus Eubacterium and their various contributions to gut health. Gut Microbes. (2020) 12:1802866. doi: 10.1080/19490976.2020.1802866
97. Orzan OA, Ţieranu CG, Olteanu AO, Dorobanţu AM, Cojocaru A, Mihai MM, et al. An insight on the possible association between inflammatory bowel disease and biologic therapy with IL-17 inhibitors in psoriasis patients. Pharmaceutics. (2023) 15:2171. doi: 10.3390/pharmaceutics15082171
98. Chen Y-J, Wu H, Wu S-D, Lu N, Wang Y-T, Liu H-N, et al. Parasutterella, in association with irritable bowel syndrome and intestinal chronic inflammation. J Gastroenterol Hepatol. (2018) 33:1844–52. doi: 10.1111/jgh.14281
99. Wang N, Yang L, Shang L, Liang Z, Wang Y, Feng M, et al. Altered fecal metabolomics and potential biomarkers of psoriatic arthritis differing from rheumatoid arthritis. Front Immunol. (2022) 13:812996. doi: 10.3389/fimmu.2022.812996
100. Chen X, Chen Y, Stanton C, Ross RP, Zhao J, Chen W, et al. Dose-response efficacy and mechanisms of orally administered bifidobacterium breve CCFM683 on IMQ-induced psoriasis in mice. Nutrients. (2023) 15:1952. doi: 10.3390/nu15081952
101. Lecron J-C, Charreau S, Jégou J-F, Salhi N, Petit-Paris I, Guignouard E, et al. IL-17 and IL-22 are pivotal cytokines to delay wound healing of S. aureus and P. aeruginosa infected skin. Front Immunol. (2022) 13:984016. doi: 10.3389/fimmu.2022.984016
102. Takagi N, Kawakami K, Kanno E, Tanno H, Takeda A, Ishii K, et al. IL-17A promotes neutrophilic inflammation and disturbs acute wound healing in skin. Exp Dermatol. (2017) 26:137–44. doi: 10.1111/exd.13115
Keywords: psoriasis, gut microbiome, skin microbiome, gut-skin axis, IL-17A inhibitor
Citation: Zhao H, Shang L, Zhang Y, Liang Z, Wang N, Zhang Q, Gao C and Luo J (2024) IL-17A inhibitors alleviate Psoriasis with concomitant restoration of intestinal/skin microbiota homeostasis and altered microbiota function. Front. Immunol. 15:1344963. doi: 10.3389/fimmu.2024.1344963
Received: 27 November 2023; Accepted: 12 February 2024;
Published: 28 February 2024.
Edited by:
Philippe Saas, Etablissement Français du Sang AuRA, FranceReviewed by:
Makoto Kondo, Mie University, JapanXiaocang Cao, Tianjin Medical University General Hospital, China
Copyright © 2024 Zhao, Shang, Zhang, Liang, Wang, Zhang, Gao and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jing Luo, bGp0eTk2NkBob3RtYWlsLmNvbQ==
†These authors have contributed equally to this work and share first authorship
 Huixia Zhao1†
Huixia Zhao1† Lili Shang
Lili Shang Yuting Zhang
Yuting Zhang Zhaojun Liang
Zhaojun Liang Nan Wang
Nan Wang Jing Luo
Jing Luo