- 1Laboratory of Cellular and Molecular Immunology, Gavin Herbert Eye Institute, University of California Irvine, School of Medicine, Irvine, CA, United States
- 2Department of Pathology and Laboratory Medicine, School of Medicine, University of California Irvine, Irvine, CA, United States
- 3Department of Surgery, Divisions of Trauma, Burns and Critical Care, School of Medicine, University of California Irvine, Irvine, CA, United States
- 4Department of Medicine, Division of Infectious Diseases and Hospitalist Program, School of Medicine, University of California Irvine, Irvine, CA, United States
- 5Université Paul Sabatier, Infinity, Inserm, Toulouse, France
- 6Department of Vaccines and Immunotherapies, TechImmune, LLC, University Lab Partners, Irvine, CA, United States
- 7Institute for Immunology, The University of California Irvine, School of Medicine, Irvine, CA, United States
Background: Cross-reactive SARS-CoV-2-specific memory CD4+ and CD8+ T cells are present in up to 50% of unexposed, pre-pandemic, healthy individuals (UPPHIs). However, the characteristics of cross-reactive memory CD4+ and CD8+ T cells associated with subsequent protection of asymptomatic coronavirus disease 2019 (COVID-19) patients (i.e., unvaccinated individuals who never develop any COVID-19 symptoms despite being infected with SARS-CoV-2) remains to be fully elucidated.
Methods: This study compares the antigen specificity, frequency, phenotype, and function of cross-reactive memory CD4+ and CD8+ T cells between common cold coronaviruses (CCCs) and SARS-CoV-2. T-cell responses against genome-wide conserved epitopes were studied early in the disease course in a cohort of 147 unvaccinated COVID-19 patients who were divided into six groups based on the severity of their symptoms.
Results: Compared to severely ill COVID-19 patients and patients with fatal COVID-19 outcomes, the asymptomatic COVID-19 patients displayed significantly: (i) higher rates of co-infection with the 229E alpha species of CCCs (α-CCC-229E); (ii) higher frequencies of cross-reactive functional CD134+CD137+CD4+ and CD134+CD137+CD8+ T cells that cross-recognized conserved epitopes from α-CCCs and SARS-CoV-2 structural, non-structural, and accessory proteins; and (iii) lower frequencies of CCCs/SARS-CoV-2 cross-reactive exhausted PD-1+TIM3+TIGIT+CTLA4+CD4+ and PD-1+TIM3+TIGIT+CTLA4+CD8+ T cells, detected both ex vivo and in vitro.
Conclusions: These findings (i) support a crucial role of functional, poly-antigenic α-CCCs/SARS-CoV-2 cross-reactive memory CD4+ and CD8+ T cells, induced following previous CCCs seasonal exposures, in protection against subsequent severe COVID-19 disease and (ii) provide critical insights into developing broadly protective, multi-antigen, CD4+, and CD8+ T-cell-based, universal pan-Coronavirus vaccines capable of conferring cross-species protection.
Introduction
The coronavirus disease 2019 (COVID-19) pandemic has created one of the largest global health crises in nearly a century (1–3). As of February 2024, the COVID-19 outbreak has affected over 700 million people worldwide, with the number of deaths directly related to severe symptomatic COVID-19 infections reaching 7 million worldwide (1, 2, 4). Some unvaccinated symptomatic COVID-19 patients produce severe symptoms that typically begin with mild upper respiratory syndrome but may further develop into severe respiratory distress and death, particularly in immunocompromised individuals and those with pre-existing co-morbidities (5–8). In contrast, other unvaccinated individuals never develop any COVID-19 symptoms despite being infected with SARS-CoV-2 (5, 9, 10). The underlying mechanisms that lead to protection from symptomatic and fatal SARS-CoV-2 infection in unvaccinated COVID-19 patients remain to be fully elucidated.
There is a growing body of evidence in support of the important role that T-cell responses in protection against COVID-19, as recently reviewed by Wherry and Barouch (11): (i) cross-reactive poly-antigenic CD4+ and CD8+ T-cell responses in COVID-19 patients appear to contribute to the resolution of SARS-CoV-2 infection and reduction in severe symptoms (12–22); (ii) SARS-CoV-2-specific CD4+ and CD8+ T-cell responses reduced viral loads in non-human primates (23, 24); (iii) SARS-CoV-2-infected patients with agammaglobulinemia and B-cell depletion displayed only a small increase in COVID-19 symptoms, indicating that the cross-reactive T cells alone may have protected from severe disease (25–30); and (iv) cancer patients with B-cell deficiencies experience milder COVID-19 disease that correlated with strong SARS-CoV-2-specific CD8+ T-cell responses (22). Conversely, other reports have associated cross-reactive memory CD4+ and CD8+ T cells with poor COVID-19 disease outcomes (16, 31–36). However, the antigen specificity, frequency, phenotype, and function of cross-reactive memory CD4+ and CD8+ T cells that protect against the severity of COVID-19 in unvaccinated asymptomatic patients remain to be determined.
Characterizing the CCCs/SARS-CoV-2 cross-reactive memory CD4+ and CD8+ T cells in unvaccinated COVID-19 patients is a difficult task today because over 85% of adults are currently vaccinated (37–40). Nevertheless, a few studies from our group and others have detected cross-reactive CD4+ and CD8+ T cells, directed toward specific sets of conserved SARS-CoV-2 epitopes, not only from unvaccinated COVID-19 patients but also from a significant proportion (~50%) of unexposed pre-pandemic healthy individuals (UPPHI) who were never exposed to SARS-CoV-2 (1, 16, 18, 20–22, 32, 33, 41–44). Moreover, pre-existing CCCs/SARS-CoV-2 cross-reactive memory CD4+ and CD8+ T cells are also present in unvaccinated UPPHI, suggesting clones of memory T cells induced following previous exposures with seasonal CCCs (1, 16, 21, 31, 41, 43–50). However, it is not yet known whether these cross-reactive memory CD4+ and CD8+ T cells (i) preferentially cross-recognize the alpha CCCs (i.e., α-CCC-229E and α-CCC-NL63) or the beta CCCs (i.e., β-CCC-HKU1 and β-CCC-OC43) and (ii) the antigen specificity, frequency, phenotype, and function of the cross-reactive memory CD4+ and CD8+ T cells associated with protection against COVID-19 severity in unvaccinated asymptomatic patients.
In this study, we hypothesized that different clonal repertoire of CCCs/SARS-CoV-2 cross-reactive memory CD4+ and CD8+ T cells are induced by previous exposures to seasonal alpha CCCs (i.e., α-CCC-229E and α-CCC-NL63) and beta CCCs (i.e., β-CCC-HKU1 and β-CCC-OC43) and that certain clones of T cells are associated with either protective or pathogenic outcomes in SARS-CoV-2 infection. We report that, compared with unvaccinated severely ill COVID-19 patients and unvaccinated patients with fatal COVID-19 outcomes, unvaccinated asymptomatic COVID-19 patients displayed significantly (i) higher rate of the α-CCC species 229E (α-CCC-229E); (ii) higher frequencies of functional memory CD134+CD137+CD4+ and CD134+CD137+CD8+ T cells directed toward cross-reactive α-CCCs/SARS-CoV-2 epitopes from structural, non-structural, and accessory proteins; and (iii) lower frequencies of cross-reactive exhausted PD-1+TIM3+TIGIT+CTLA4+CD4+ and PD-1+TIM3+TIGIT+CTLA4+CD8+ T cells. These findings (i) support the crucial role of functional, poly-antigenic α-CCCs/SARS-CoV-2 cross-reactive memory CD4+ and CD8+ T cells, induced following previous exposures to α-CCC species, in protection against subsequent severe disease caused by SARS-CoV-2 infection and (ii) provides a strong rationale for the development of broadly protective, T-cell-based, multi-antigen universal pan-Coronavirus vaccines.
Materials and methods
Human study population cohort and HLA genotyping
Between July 2020 to November 2022, 600 patients were enrolled at the University of California Irvine Medical Center with various severity of COVID-19 disease under an approved Institutional Review Board–approved protocol (IRB No. 2020-5779). Written informed consent was obtained from participants before inclusion. SARS-CoV-2 positivity was defined by a positive RT-PCR on a respiratory tract sample. None of the patients enrolled in this study received any COVID-19 vaccine.
Patient selection based on HLA-A*02:01 and HLA-DRB1*01:01 alleles
We genotyped all the 600 patients enrolled in our study for class I HLA-A*02:01 and class II HLA-DRB1*01:01 by PCR. Out of the 600 COVID-19 patients, 147 patients were positive for HLA-A*02:01 and/or HLA-DRB1*01:01 and were considered in this study (Supplementary Figure 1). The 147 patients were from mixed ethnicities (Hispanic (28%), Hispanic Latino (22%), Asian (16%), Caucasian (13%), mixed Afro-American and Hispanic (8%), Afro-American (5%), mixed Afro-American and Caucasian (2%), and Native Hawaiian and Other Pacific Islander descent (1%). Six percent of the patients did not reveal their race/ethnicity. The detailed demographic and clinical data for the 147 patients enrolled in this study are shown in Table 1.
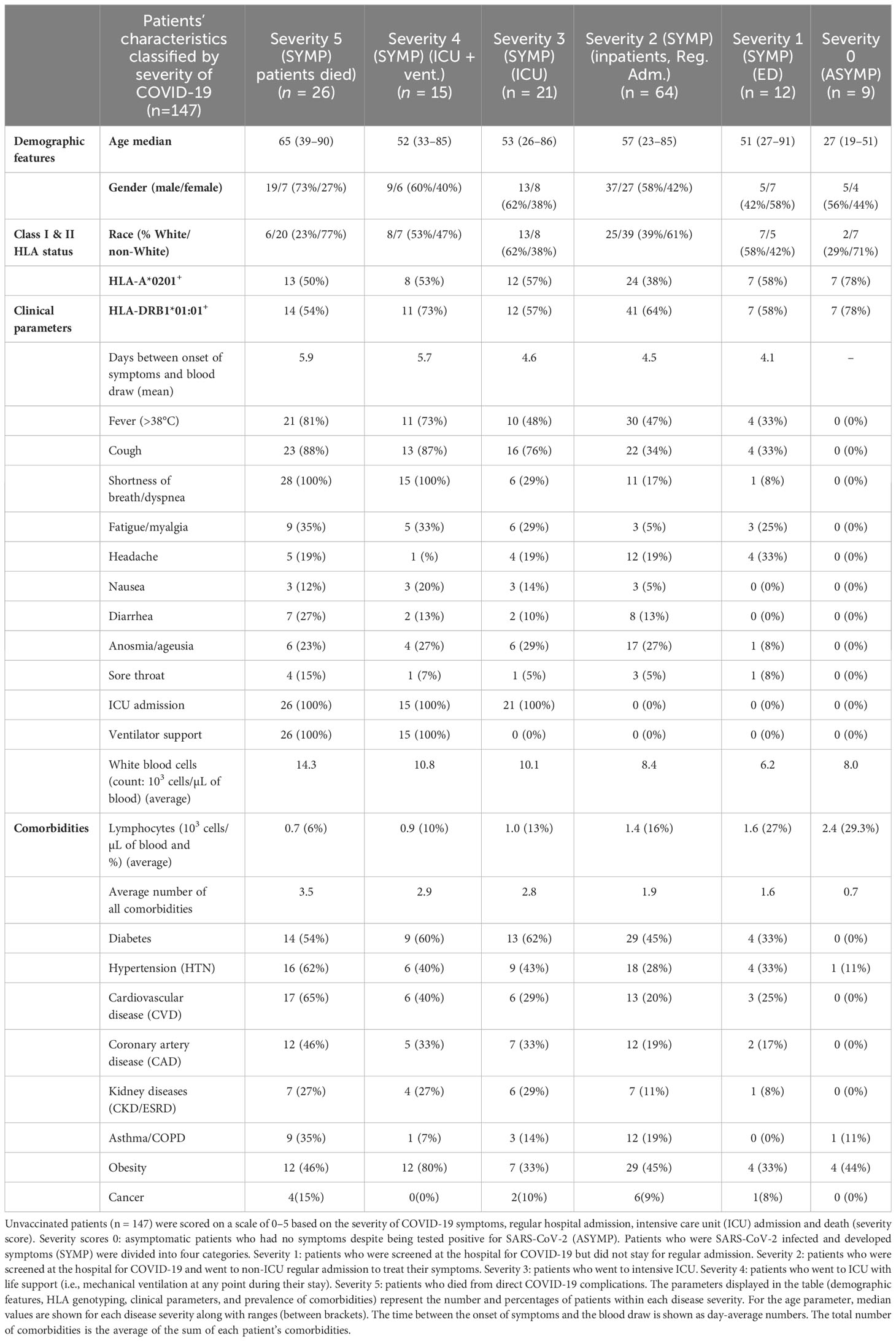
Table 1 Demographic, age, HLA-genotyping, clinical parameters, and prevalence of comorbidities in unvaccinated COVID-19 patients with various degrees of disease severity.
Symptomatic and asymptomatic COVID-19 patient stratification based on disease severity
Following patient discharge, they were divided into six groups depending on the severity of their symptoms and their intensive care unit (ICU) and intubation (mechanical ventilation) status by medical practitioners. The scoring criteria were as follows: severity 5, patients who died from COVID-19 complications; severity 4, infected COVID-19 patients with severe disease who were admitted to the intensive care unit (ICU) and required ventilation support; severity 3, infected COVID-19 patients with severe disease that required enrollment in ICU, but without ventilation support; severity 2, infected COVID-19 patients with moderate symptoms that involved a regular hospital admission; severity 1, infected COVID-19 patients with mild symptoms; and severity 0, infected individuals with no symptoms. Among the 147 COVID-19 patients, subjects with a severity score of 0 were defined as asymptomatic, and subjects with a severity score of 1–5 were defined as symptomatic.
Pre-pandemic healthy controls
Subsequently, we used 15 liquid-nitrogen frozen PBMCs samples (blood collected pre-COVID-19 in 2018) from HLA-A*02:01+/HLA-DRB1*01:01+ unexposed pre-pandemic healthy individuals (UPPHI, 8 men, 7 women; median age, 54 (20–76)] as controls to measure recalled SARS-CoV-2 cross-reactive T-cell responses. The class-II HLA status of each patient was first screened for HLA-DRB1*01:01 by PCR (Supplementary Figure 1A) (51). For class-I HLA, the screening was first performed (two-digit level) by HLA-A*02 flow cytometry staining (data not shown, mAbs clone BB7.2, BioLegend, San Diego, CA). The four-digit class-I HLA-A*02:01 subtype was subsequently screened by PCR (Supplementary Figure 1B) on blood samples (52).
T-cell epitopes screening, selection, and peptide synthesis
CCCs/SARS-CoV-2 cross-reactive peptide epitopes from 12 SARS-CoV-2 proteins, including 27 9-mer long CD8+ T-cell epitopes (ORF1ab84–92, ORF1ab1675–1683, ORF1ab2210–2218, ORF1ab2363–2371, ORF1ab3013–3021, ORF1ab3183–3191, ORF1ab3732–3740, ORF1ab4283–4291, ORF1ab5470–5478, ORF1ab6419–6427, ORF1ab6749–6757, S2–10, S691–699, S958–966, S976–984, S1000–1008, S1220–1228, E20–28, E26–34, M52–60, M89–97, ORF63–11, ORF7b26–34, ORF8a31–39, ORF8a73–81, ORF103–11, and ORF105–13) and 16 13-mer long CD4+ T-cell epitopes (ORF1a1350–1365, ORF1a1801–1815, ORF1ab5019–5033, ORF1ab6088–6102, ORF1ab6420–6434, S1–13, E20–34, E26–40, M176–190, ORF612–26, ORF7a1–15, ORF7a3–17, ORF7a98–112, ORF7b8–22, ORF8b1–15, and N388–403) that we formerly identified were selected as we previously described (1) (Table 2 and Supplementary Table 1). We used the Epitope Conservancy Analysis tool to compute the degree of identity of CD8+ and CD4+ T-cell epitopes within a given protein sequence of SARS-CoV-2 set at 100% identity level (1) (Table 2 and Supplementary Tables 1, 2). Peptides were synthesized (21st Century Biochemicals, Inc., Marlborough, MA), and the purity of peptides determined by both reversed-phase high-performance liquid chromatography and mass spectroscopy was over 95%.
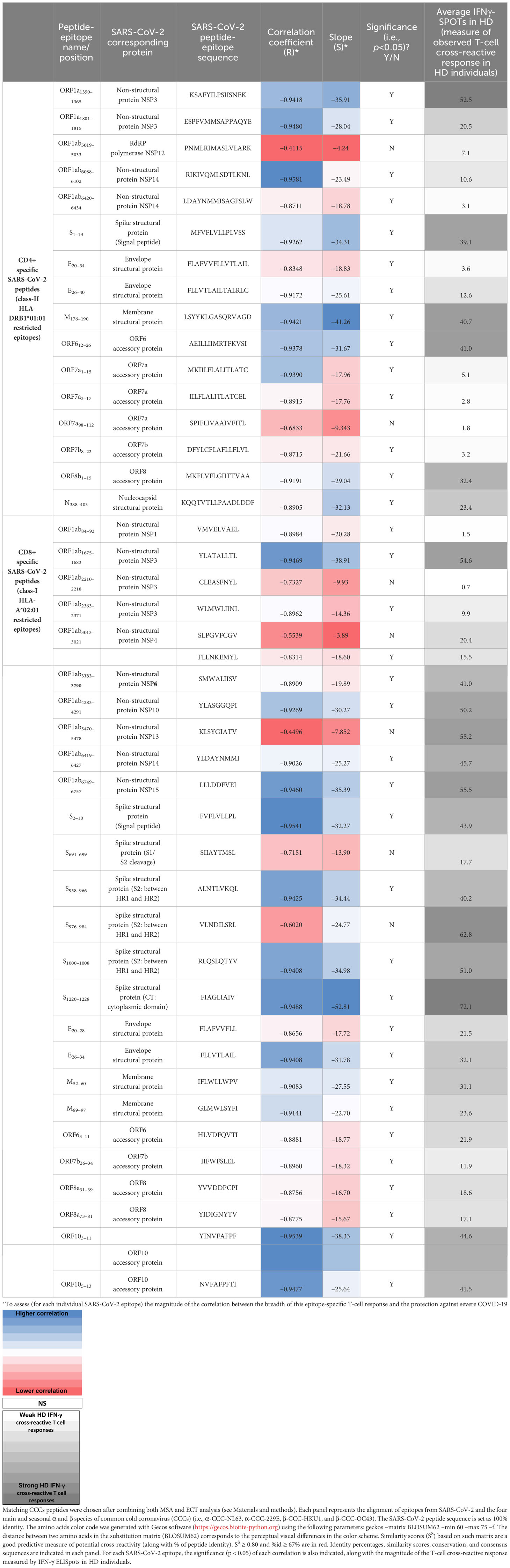
Table 2 Percentages of identity and similarity scores (Ss) between CCCs/SARS-CoV-2 cross-reactive CD4+ and CD8+ T cell epitopes.
Blood differential test
Total white blood cell (WBC) count and lymphocyte count per microliter of blood were performed by the clinicians at the University of California Irvine Medical Center, using a CellaVision™ DM96 automated microscope. Monolayer smears were prepared from anticoagulated blood and stained using the May Grunwald Giemsa (MGG) technique. Subsequently, slides were loaded onto the DM96 magazines and scanned using a ×10 objective focused on nucleated cells to record their exact position. Images were obtained using the ×100 oil objective and analyzed by artificial neural network (ANN).
Peripheral blood mononuclear cells isolation and T-cell stimulation
Peripheral blood mononuclear cells (PBMCs) from COVID-19 patients were isolated from the blood using Ficoll (GE Healthcare) density gradient media and transferred into 96-well plates at a concentration of 2.5 × 106 viable cells per ml in 200 µl (0.5 × 106 cells per well) of RPMI-1640 media (Hyclone) supplemented with 10% (v/v) FBS (HyClone), sodium pyruvate (Lonza), L-glutamine, nonessential amino acids, and antibiotics (Corning). A fraction of the blood was kept separated to perform HLA genotyping of the patients and select only the HLA-A*02:01 and/or DRB1*01:01 positive individuals (Supplementary Figure 1). Fresh peripheral blood mononuclear cells (PBMCs) were used in this study, as they generally have higher viability and functionality compared to frozen PBMCs. Freezing and thawing can lead to cell damage and loss of T-cell functionality, which may affect the accuracy and reliability of experimental results. Frozen PBMCs may exhibit altered activation status compared to fresh cells. Cryopreservation can induce stress responses in cells, leading to changes in their activation state and potentially affecting immune response assays. In the context of COVID-19 research, where precise characterization of immune responses is crucial for understanding disease pathogenesis, vaccine development, and treatment strategies, using fresh PBMCs ensures the accuracy and reliability of experimental results. A side-by-side comparison of frozen and fresh PBMCs and pre-pandemic healthy control PBMCs yielded no significant difference. PBMCs were stimulated with 10 µg/ml of each one of the 43 individual CCCs/SARS-CoV-2 cross-reactive peptide epitopes (27 CD8+ T-cell peptides and 16 CD4+ T-cell peptides) and incubated in a humidified chamber with 5% CO2 at 37°C (Supplementary Figure 2A). Post-incubation, cells were stained by flow cytometry analysis or transferred onto IFN-γ ELISpot plates. The same isolation protocol was followed for HD samples obtained in 2018. Ficoll was kept frozen in liquid nitrogen in FBS DMSO 10%; after thawing, HD PBMCs were stimulated similarly for the IFN-γ ELISpot technique.
ELISpot assay
COVID-19 patients were first screened for their HLA status (DRB1*01:01+ positive = 92 out of 600 tested, HLA-A*02:01+ positive = 71, DRB1*01:01+ and HLA-A*02:01+ positive = 16) (Supplementary Figure 1). The 92 DRB1*01:01 positive individuals were then used to assess the CD4+ T-cell response against CCCs/SARS-CoV-2 cross-reactive class-II restricted epitopes by IFN-γ ELISpot (Supplementary Figure 2). Similarly, we assessed the CD8+ T-cell response against our CCCs/SARS-CoV-2 cross-reactive class-I restricted epitopes in the 71 HLA-A*02:01 positive individuals representing different disease severity categories (Table 1).
All ELISpot reagents were filtered through a 0.22-µm filter. Wells of 96-well Multiscreen HTS Plates (Millipore, Billerica, MA) were pre-wet with 30% ethanol for 60 s and then coated with 100 µl primary anti-IFN-γ antibody solution (10 µg/ml of 1-D1K coating antibody from Mabtech, Cincinnati, OH) OVN at 4°C. After washing, the plate was blocked with 200 µl of RPMI media plus 10% (v/v) FBS for 2 h at room temperature to prevent nonspecific binding. After 24 h, following the blockade, the peptide-stimulated cells from the patient’s PBMCs (0.5 × 106 cells/well) were transferred into the ELISpot-coated plates. PHA-stimulated or non-stimulated cells (DMSO) were used as positive or negative controls of T-cell activation, respectively. Upon incubation in a humidified chamber with 5% CO2 at 37°C for an additional 48 h, cells were next washed using PBS and PBS-Tween 0.02% solution. Next, 100 µl of biotinylated secondary anti-IFN-γ antibody (1 µg/ml, clone 7-B6-1, Mabtech) in blocking buffer (PBS 0.5% FBS) was added to each well. Following a 2-h incubation followed by washing, wells were incubated with 100 µl of HRP-conjugated streptavidin (1:1,000) for 1 h at room temperature. Lastly, wells were incubated for 15–30 min with 100 µl of TMB detection reagent at room temperature, and spots were counted both manually and by an automated ELISpot reader counter (ImmunoSpot Reader, Cellular Technology, Shaker Heights, OH).
Flow cytometry analysis
Surface markers detection and flow cytometry analysis were performed on 147 patients after 72 h of stimulation with each CCCs/SARS-CoV-2 cross-reactive class-I or class-II restricted peptide; PBMCs (0.5 × 106 cells) were stained (Supplementary Figure 2). First, the cells were stained with a live/dead fixable dye (Zombie Red dye, 1/800 dilution—BioLegend, San Diego, CA) for 20 min at room temperature, to exclude dying/apoptotic cells. Subsequently, cells were stained for 45 min at room temperature with five different HLA-A*02*01 restricted tetramers and/or five HLA-DRB1*01:01 restricted tetramers (PE labeled) specific toward the CCCs/SARS-CoV-2 cross-reactive CD8+ T-cell epitopes Orf1ab2210–2218, Orf1ab4283–4291, S976–984, S1220–1228, and ORF103–11 and toward the CCCs/SARS-CoV-2 cross-reactive CD4+ T-cell epitopes ORF1a1350–1365, S1–13, E26–40, M176–190, and ORF612–26, respectively. Cells were alternatively stained with the EBV BMLF-1280–288-specific tetramer for controls (53) (Supplementary Figure 3). We optimized our tetramer staining according to protocol instructions published by Dolton et al. (54). We stained HLA-A*02*01- HLA-DRB1*01:01-negative patients with our 10 tetramers as a negative control aiming to assess tetramers staining specificity. Subsequently, we used anti-human antibodies for surface-marker staining: anti-CD45 (BV785, clone HI30—BioLegend), anti-CD3 (Alexa700, clone OKT3—BioLegend), anti-CD4 (BUV395, clone SK3—BD), anti-CD8 (BV510, clone SK1—BioLegend), anti-TIGIT (PercP-Cy5.5, clone A15153G—BioLegend), anti-TIM-3 (BV 711, clone F38-2E2—BioLegend), anti-PD1 (PE-Cy7, clone EH12.1—BD), anti-CTLA-4 (APC, clone BNI3—BioLegend), anti-CD137 (APC-Cy-7, clone 4B4-1—BioLegend), and anti-CD134 (BV650, clone ACT35—BD). mAbs against these various cell markers were added to the cells, either ex vivo or in vitro, in phosphate-buffered saline (PBS) containing 1% FBS and 0.1% sodium azide [fluorescence-activated cell sorter (FACS) buffer] and incubated for 30 min at 4°C. Cells were then washed twice with FACS buffer and fixed with paraformaldehyde 4% (PFA, Affymetrix, Santa Clara, CA). A total of ∼200,000 lymphocyte-gated PBMCs (140,000 alive CD45+) were acquired by Fortessa X20 (Becton Dickinson, Mountain View, CA) and analyzed using FlowJo software (TreeStar, Ashland, OR). The gating strategy is detailed in Supplementary Figure 2B.
TaqMan quantitative polymerase reaction assay for the detection of CCC species in UPPHI and in COVID-19 patients
To detect common cold coronavirus co-infection in COVID-19 patients, Taqman PCR assays were performed on a total of 85 patients distributed into each different category of disease severity (9 ASYMP, 6 patients of category 1, 32 patients of category 2, 9 patients of category 3, 15 patients of category 4, and 14 patients of category 5). Nucleic acid was first extracted from each nasopharyngeal swab sample using Purelink Viral RNA/DNA mini kit (Thermo Fisher Scientific, Waltham, MA) according to the manufacturer’s instructions. Subsequently, extracted RNA samples were quantified using Qubit and BioAnalyzer. cDNA was synthesized from 10 μL of RNA eluate using random hexamer primers and SuperScript II Reverse Transcriptase (Applied Biosystems, Waltham, MA). The subsequent RT-PCR screening of the enrolled subjects for the four CCCs was performed using specific sets of primers and probes (55).
CCC-229E, CCC-OC43, and CCC-NL63 RT-PCR assays were performed using the following conditions: 50°C for 15 min followed by denaturation at 95°C for 2 min, 40 cycles of PCR performed at 95°C for 8 s, extending and collecting a fluorescence signal at 60°C for 34 s (56). For CCC-HKU1, the amplification conditions were 48°C for 15 min, followed by 40 cycles of 94°C for 15 s and 60°C for 15 s. For each virus, when the Ct-value generated was <35, the specimen was considered positive. When the Ct-value was relatively high (35 ≤ Ct < 40), the specimen was retested twice and considered positive if the Ct-value of any retest was <35 (57).
Identity and similarity analysis of CCCs/SARS-CoV-2 cross-reactive epitopes
To assess the % identity (%id) of CCCs/SARS-CoV-2 cross-reactive CD4+ and CD8+ T-cell peptide epitopes, we first identified the best matching CCCs peptide across the CCCs proteomes (Table 2). The full CCCs proteomes sequences were obtained from the National Center for Biotechnology Information (NCBI) GenBank [MH940245.1 (CCC-HUK1), MN306053.1 (CCC-OC43), KX179500.1 (CCC-NL63), and MN306046.1 (CCC-229E)]. We processed this in the following three steps. (1) Corresponding CCCs peptides were determined after protein sequence alignments of all four homologous CCCs proteins plus the SARS-CoV-2 related one using various multiple sequences alignments (MSA) algorithms ran in JALVIEW, MEGA11, and M-coffee software’s (i.e., ClustalO, Kalign3, and M-coffee—the latter computing alignments by combining a collection of multiple alignments from a library constituted with the following algorithms: T-Coffee, PCMA, MAFFT, ClustalW, Dialigntx, POA, MUSCLE, and Probcons). Furthermore, we confirmed our results with global and local pairwise alignments (Needle and Water algorithms ran in Biopython) performed to confirm the results. In case of obtaining different results with the various algorithms, the epitope sequence with the highest BLOSUM62-sum score compared to the SARS-CoV-2 epitope set as reference was selected (Table 2 and Supplementary Tables 1, 2). We calculated the % of identity and similarity score Ss with its related SARS-CoV-2 epitope, for each of these CCCs peptides (Supplementary Tables 1-3). The peptide similarity score Ss calculation is based on the method reported by Sune Frankild et al. (58) and the BLOSUM62 matrix to calculate a BLOSUM62 sum (using the Bio.SubsMat.MatrixInfo package in Biopython) between a pair of peptides (peptide “x” from SARS-CoV-2 and “y” from one CCC) and compared their similarity. 0 ≤ Ss ≤ 1: the closest Ss is to 1, the highest is the potential for T-cell cross-reactivity response toward the related pair of peptides (58). We used a threshold of Ss≥0.8 to discriminate between highly similar and non-similar peptides. (2) Then, we examined if other parts of each of the CCCs proteome (without restricting our search only to peptides present in CCCs homologous proteins) could contain better matching peptides than the CCCs peptides reported in Supplementary Tables 1-3. First, for each one of our 16 CD4+ and 27 CD8+ SARS-CoV-2 epitopes, we spanned the entire proteome of each CCCs using the Epitope Conservancy Tool (ECT: http://tools.iedb.org/conservancy/—with a conservancy threshold of 20%). All the CCCs peptides from the top query (i.e., with the highest % of identity) were reported for every four CCCs in Supplementary Tables 1-3. Second, among these returned top queries (peptides with the same highest % identity), we picked the one with the highest similarity score Ss (bolded in Supplementary Tables 1-3—right column). (3) We compared this peptide with the one previously found in Supplementary Table 1 based on MSA. When both methods returned the same peptide (from the same protein), we kept it (peptides highlighted in beige in Supplementary Tables 1-3). When both matching peptides (using the two different methods) were found to be different, we compared (i) %idMSA with %idECT and (ii) SsMSA with SsECT. If %idMSA ≤ %idECT but SsMSA ≥ SsECT, we kept the CCCs peptide found following the MSA method; however, if %idMSA ≤ %idECT and SsMSA < SsECT, we then picked the CCC peptide found using the ECT instead of the one found using MSA (peptides not highlighted in Supplementary Tables 1-3).
Using the %id and the calculated similarity score with the SARS-CoV-2 epitopes, all related CCCs’ best-matching peptides are reported in Supplementary Tables 1-3. They were then evaluated based on their potential to induce a cross-reactive T-cell response (Supplementary Tables 1-3): (0), CCC best matching peptide with low to no potential to induce a cross-reactive response toward the corresponding SARS-CoV-2 epitope and vice versa (%id with the corresponding SARS-CoV-2 epitope < 67% and similarity score Ss < 0.8); (0.5), CCC best matching peptide that may induce a cross-reactive response (%id with the corresponding SARS-CoV-2 epitope ≥ 67% OR similarity score Ss ≥ 0.8); and (1), CCC best-matching peptide is very likely to induce a cross-reactive response (%id ≥ 67% and Ss ≥ 0.8).
Identification of potential cross-reactive peptides in non-CCC human pathogens and vaccines
We took advantage of the database generated by Pedro A. Reche (59). Queries to find matching peptides with our SARS-CoV-2-derived CD4+ and CD8+ epitopes were performed from the data gathered; only peptides sharing a %id ≥ 67% with our corresponding SARS-CoV-2 epitope were selected. The corresponding similarity score Ss was calculated.
Statistical analyses
To assess the linear negative relationship between COVID-19 severity and the magnitude of each SARS-CoV-2 epitope-specific T-cell response, correlation analysis using GraphPad Prism version 8 (La Jolla, CA) was performed to calculate Pearson correlation coefficients (R), coefficient of determination (R2), and associated p-value (correlation statistically significant for p ≤ 0.05). The slope (S) of the best-fitted line (dotted line) was calculated in Prism by linear regression analysis. The same statistical analysis was performed to compare the cross-reactive pre-existing T-cell response in unexposed pre-pandemic healthy individuals (UPPHI) with the slope S (magnitude of the correlation between this epitope-specific T-cell response in SARS-CoV-2-infected patients and the protection against severe COVID-19). Absolute WBCs and lymphocyte cell numbers (per µL of blood, measured through BDT), corresponding lymphocytes percentages/ratio, flow cytometry data measuring CD3+/CD8+/CD4+ cell percentages and the percentages detailing the magnitude (Tetramer+ T cell %), and the quality (% of PD1+/TIGIT+, CTLA-4+/TIM3+ or AIMs+ cells) of the CD4+ and CD8+ SARS-CoV-2-specific T cells, were compared across groups and categories of disease severity by one-way ANOVA multiple tests. ELISpot SFCs data were compared by Student’s t-tests. Data are expressed as the mean ± SD. Results were considered statistically significant at p ≤ 0.05. To evaluate whether the differences in frequencies of RT-PCR positivity to the four CCCs across categories of disease severity were significant, we used the Chi-squared test or Fisher’s exact test.
Results
Higher magnitudes of common cold coronavirus/SARS-CoV-2 cross-reactive CD4+ T-cell responses detected in unvaccinated asymptomatic COVID-19 patients
We first compared SARS-CoV-2-specific CD4+ T-cell responses in unvaccinated asymptomatic COVID-19 patients (those individuals who never develop any COVID-19 symptoms despite being infected with SARS-CoV-2) to unvaccinated symptomatic (those patients who developed severe to fatal COVID-19 symptoms) (Figure 1). We used 16 recently identified HLA-DR-restricted CD4+ T-cell epitopes that are highly conserved between human SARS-CoVs and CCCs (1). We enrolled 92 unvaccinated HLA-DRB1*01:01+ COVID-19 patients, who were genotyped using PCR (Supplementary Figure 1) and divided into six groups, based on the level of severity of their COVID-19 symptoms (from severity 5 to severity 0, assessed at discharge). Clinical and demographic characteristics of this cohort of COVID-19 patients are detailed in Table 1. Fresh PBMCs were isolated from these COVID-19 patients, on average within 4.8 days after reporting a first COVID-19 symptom or a first PCR-positive test (Table 1). PBMCs were then stimulated in vitro for 72 h using each of the 16 CD4+ T-cell peptide epitopes, as detailed in Materials and methods and illustrated in Supplementary Figure 2. The frequency of responding IFN-γ-producing CD4+ T cells specific to individual epitopes was quantified, in each of the six groups of COVID-19 patients, using ELISpot assay (i.e., number of IFN-γ-spot forming CD4+ T cells or “SFCs”) (Figure 1). A positive IFN-γ-producing CD4+ T-cell responses was determined as the mean SFCs > 50 per 0.5 × 106 PBMCs fixed as threshold.
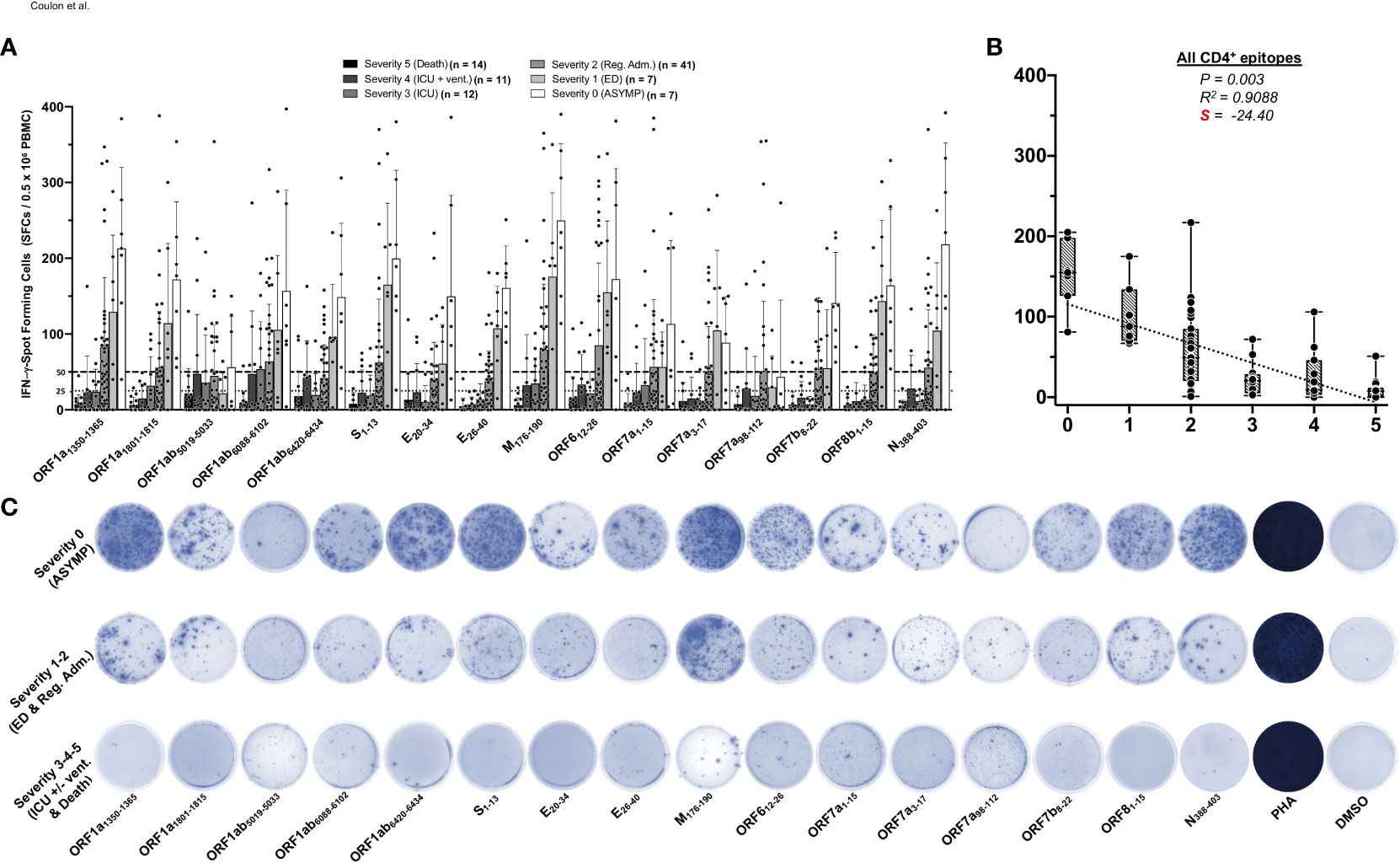
Figure 1 IFN-γ-producing CD4+ T-cell responses to CCCs/SARS-CoV-2 cross-reactive epitopes in unvaccinated COVID-19 patients with various degrees of disease severity. PBMCs from HLA-DRB1*01:01-positive COVID-19 patients (n = 92 are HLA-DRB1*01:01-positive out of 600 tested) were isolated and stimulated for a total of 72 h with 10 µg/ml of each of the previously identified 16 CCCs/SARS-CoV-2 cross-reactive CD4+ T cell epitope peptides. The number of IFN-γ-producing CD4+ T cells was quantified in each of the 92 patients using ELISpot assay. (A) Average/mean numbers (± SD) of IFN-γ-spot forming cells (SFCs) after CD4+ T-cell peptide-stimulation detected in each of the 92 COVID-19 patients divided into six groups based on disease severity scored 0–5, as described in Materials and methods, and as identified by six columns on a grayscale (black columns = severity 5, to white columns = severity 0) is shown. Dotted lines represent an arbitrary threshold set as a cutoff of the positive response. A mean SFC between 25 and 50 SFCs corresponds to a medium/intermediate response, whereas a strong response is defined for mean SFCs > 50 per 0.5 × 106 stimulated PBMCs. (B) Correlation between the overall number of IFN-γ-producing CD4+ T cells induced by each of the 16 CCCs/SARS-CoV-2 cross-reactive CD4+ T-cell epitope peptides in each of the six groups of COVID-19 patients with various disease severity. The coefficient of determination (R2) is calculated from the Pearson correlation coefficients (R). The associated p-value and the slope (S) of the best-fitted line (dotted line) calculated by linear regression analysis are indicated. The gray-hatched boxes in the correlation graphs extend from the 25th to 75th percentiles (hinges of the plots) with the median represented as a horizontal line in each box and the extremity of the vertical bars showing the minimum and maximum values. (C) Representative spots images of the IFN-γ-spot forming cells (SFCs) induced by each of the 16 CCCs/SARS-CoV-2 cross-reactive CD4+ T cell epitope peptides in three representative patients, each falling into one of three groups of disease category: the unvaccinated asymptomatic COVID-19 patients (ASYMP, severity score 0), unvaccinated COVID-19 patients who developed mild to moderate disease (severity scores 1 and 2) and unvaccinated severely ill COVID-19 patients and unvaccinated patients with fatal COVID-19 outcomes (severity scores 3–5). PHA was used as a positive control of T-cell activation. Unstimulated negative control SFCs (DMSO—no peptide stimulation) were subtracted from the SFC counts of peptides-stimulated cells. Results are representative of two independent experiments and were considered statistically significant at p ≤ 0.05.
Overall, the highest frequencies of CCCs/SARS-CoV-2 cross-reactive epitope-specific IFN-γ-producing CD4+ T cells were detected in the unvaccinated COVID-19 patients with less severe disease (i.e., severity 0, 1, and 2, Figures 1A, B). In contrast, the lowest frequencies of CCCs/SARS-CoV-2 cross-reactive IFN-γ-producing CD4+ T cells were detected in unvaccinated severely ill COVID-19 patients (severity scores 3 and 4) and in unvaccinated COVID-19 patients with fatal outcomes (severity score of 5, Figures 1A, B).
Pearson correlation analysis was performed to determine the linear correlation between the magnitude of CD4+ T-cell responses directed toward each of the 16 highly conserved SARS-CoV-2 epitopes and the severity of COVID-19 symptoms. A negative correlation is usually considered strong when the coefficient R-value is between −0.7 and −1. Except for the ORF1ab5019–5033 and ORF7a98–112 epitopes, we found that a strong positive linear correlation existed between the high magnitude of IFN-γ-producing CD4+ T-cell responses specific to 14 CD4+ T-cell epitopes and the “natural protection” observed in unvaccinated asymptomatic COVID-19 patients (Figures 1A, C). This positive correlation existed regardless of whether CD4+ T cells cross-recognized structural, non-structural, or accessory SARS-CoV-2 antigens. Cross-reactive IFN-γ-producing CD4+ T-cell responses, specific to M176–190, ORF1a1350–1365, S1–13, N388–403, and ORF612–26, and to a slightly lesser extent to ORF8b1–15 and ORF1a1801–1815, were associated with a low COVID-19 severity score (i.e., negatively correlated with an R close to −1) and a very strong negative slope (−41.26 < S < −28.04). Comparatively, the CD4+ T-cell responses against E26–40, ORF1ab6088–6102, ORF7b8–22, E20–34, ORF1ab6420–6434, ORF7a1–15, and ORF7a3–17 were also negatively associated with severe disease in patients, but to a lesser degree (relatively less negative slope: −25.61 < S < −17.76) (Figure 1A and Supplementary Figure 4). In contrast, no significant correlation was found between the magnitude of IFN-γ-producing CD4+ T-cell responses directed toward ORF1ab5019–5033 and ORF7a98–112 epitopes and the disease severity (p > 0.05). For the ORF1ab5019–5033 and ORF7a98–112 epitopes, the slope was comparatively weak: only slightly negative with S > −10 (Figure 1A and Supplementary Figure 4).
Taken together, these results (i) demonstrate an overall higher magnitude of CCCs/SARS-CoV-2 cross-reactive CD4+ T-cell responses present in unvaccinated asymptomatic COVID-19 patients. In contrast, a lower magnitude of CCCs/SARS-CoV-2 cross-reactive CD4+ T-cell responses were detected in unvaccinated severely ill COVID-19 patients and to patients with fatal COVID-19 outcomes and (ii) suggest a crucial role of CCCs/SARS-CoV-2 cross-reactive CD4+ T cells, directed towards structural, non-structural, and accessory protein antigens, in protection from symptomatic and fatal Infections in unvaccinated COVID-19 patients.
Higher magnitudes of CD8+ T-cell responses to common cold coronavirus/SARS-CoV-2 cross-reactive epitopes detected in unvaccinated asymptomatic COVID-19 patients
We next compared the CCCs/SARS-CoV-2 cross-reactive CD8+ T-cell responses in unvaccinated asymptomatic individuals vs. unvaccinated symptomatic COVID-19 patients (Figure 2). We used 27 recently identified HLA-A*0201-restricted CD8+ T-cell epitopes that are highly conserved between human SARS-CoVs and CCCs (1). We enrolled 71 unvaccinated HLA-A*0201+ COVID-19 patients, who were genotyped using PCR (Supplementary Figure 1) and divided into six groups based on the severity of COVID-19 symptoms (i.e., severity 5 to severity 0, Table 1). Fresh PBMCs were isolated from COVID-19 patients on an average of 4.8 days after reporting initial COVID-19 symptoms or a first PCR-positive test (in the case of asymptomatic). Subsequently, PBMCs were stimulated in vitro for 72 h using each of the 27 HLA-A*0201-restricted CD8+ T-cell peptide epitopes (Supplementary Figure 2). The frequency of responding IFN-γ-producing CD8+ T cells specific to individual epitopes was quantified, in each of the six groups of COVID-19 patients, using the ELISpot assay (i.e., number of IFN-γ-spot forming CD8+ T cells or “SFCs”) (Figure 2).
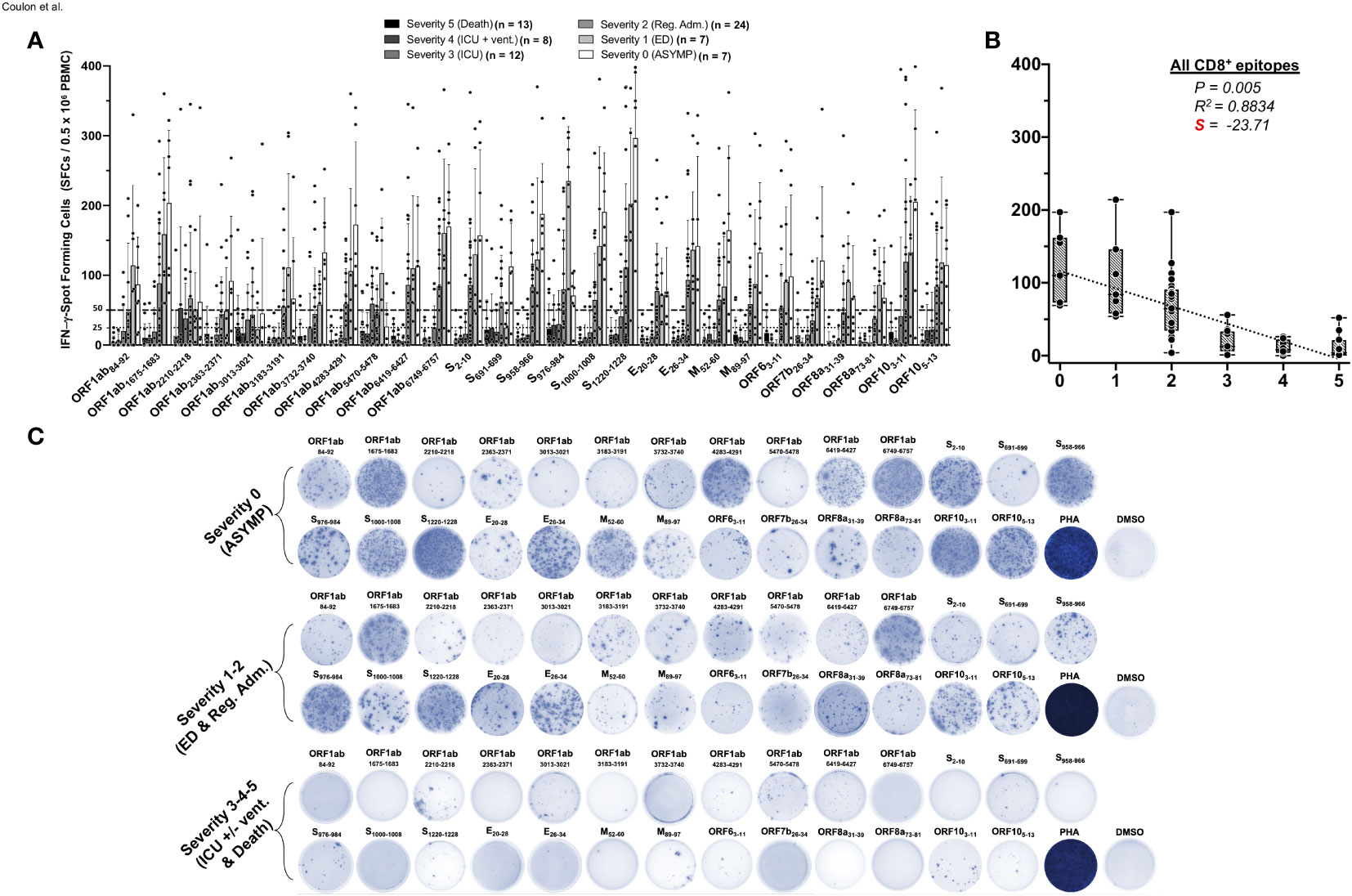
Figure 2 IFN-γ-producing CD8+ T-cell responses to CCCs/SARS-CoV-2 cross-reactive epitopes in unvaccinated COVID-19 patients with various degrees of disease severity. PBMCs from HLA-A*02:01-positive COVID-19 patients (n = 71) were isolated and stimulated for a total of 72 h with 10 µg/ml of each of the previously identified 27 CCCs/SARS-CoV-2 cross-reactive CD8+ T-cell epitope peptides. The number of IFN-γ-producing CD8+ T cells was quantified in each of the 71 patients using ELISpot assay. Panel (A) shows the average/mean numbers (± SD) of IFN-γ-spot forming cells (SFCs) after CD8+ T-cell peptide stimulation detected in each of the 71 COVID-19 patients divided into six groups based on disease severity scored 0–5, as described in Materials and methods, and as identified by six columns on a grayscale (Black columns = severity 5, to white columns = severity 0). Dotted lines represent an arbitrary threshold set as a cutoff of the positive response. A mean SFCs between 25 and 50 SFCs corresponds to a medium/intermediate response, whereas a strong response is defined for mean SFCs > 50 per 0.5 × 106 stimulated PBMCs. (B) Correlation between the overall number of IFN-γ-producing CD8+ T cells induced by each of the 27 CCCs/SARS-CoV-2 cross-reactive CD8+ T-cell epitope peptides in each of the six groups of COVID-19 patients with various disease severity. The coefficient of determination (R2) is calculated from the Pearson correlation coefficients (R). The associated p-value and the slope (S) of the best-fitted line (dotted line) calculated by linear regression analysis are indicated. The gray-hatched boxes in the correlation graphs extend from the 25th to 75th percentiles (hinges of the plots) with the median represented as a horizontal line in each box and the extremity of the vertical bars showing the minimum and maximum values. (C) Representative spots images of the IFN-γ-spot forming cells (SFCs) induced by each of the 27 CCCs/SARS-CoV-2 cross-reactive CD8+ epitope peptides in three representative patients, each falling into one of three groups of disease category: the unvaccinated asymptomatic COVID-19 patients (ASYMP, severity score 0), unvaccinated COVID-19 patients who developed mild to moderate disease (severity scores 1 and 2), and unvaccinated severely ill COVID-19 patients and unvaccinated patients with fatal COVID-19 outcomes, (severity scores 3–5). PHA was used as a positive control of T-cell activation. Unstimulated negative control SFCs (DMSO—no peptide stimulation) were subtracted from the SFC counts of peptides-stimulated cells. Results are representative of two independent experiments and were considered statistically significant at p ≤ 0.05.
Overall, the highest frequencies of CCCs/SARS-CoV-2 cross-reactive epitope-specific functional IFN-γ-producing CD8+ T cells (mean SFCs > 50 per 0.5 × 106 PBMCs) were detected in the three groups of unvaccinated COVID-19 patients who presented little to no severe COVID-19 symptoms (i.e., severity 0, 1, and 2, Figures 2A, B). In contrast, significantly lower frequencies of CCCs/SARS-CoV-2 cross-reactive functional IFN-γ-producing CD8+ T cells were detected in the two groups of unvaccinated severely ill symptomatic COVID-19 patients (i.e., severity 3 and 4, mean SFCs < 50) and the unvaccinated COVID-19 patients with fatal outcomes (i.e., severity 5, mean SFCs < 25).
Out of the 27 CD8+ T-cell epitopes, there was a significant positive linear correlation between CD8+ T-cell responses specific to 22 epitopes and little to no severe COVID-19 disease (Figures 2A, B). For these 22 epitopes, the Pearson correlation coefficients (R) ranged from −0.8314 to −0.9541, and slopes (S) of the best-fitted lines comprised between −14.36 and −52.81. For the remaining five epitopes (ORF1ab2210–2218, ORF1ab3013–3021, ORF1ab5470–5478, S691–699, and S976–984), no significant linear correlation was observed. Nonetheless, among these five epitopes, the slope for ORF1ab2210–2218, ORF1ab3013–3021, and ORF1ab5470–5478 was comparatively less negative (S > −10) (Figures 2A, C and Supplementary Figure 5). Additionally, although we could not establish any significant linear correlation between S691–699 and S976–984 epitope-specific CD8+ T-cell responses and disease severity, more complex (non-linear) associations might exist. For example, the magnitude of the S976–984-specific IFN-γ-producing CD8+ T-cell response followed a clear downside trend, as the disease severity increased in severely ill symptomatic COVID-19 patients and patients with fatal outcomes (i.e., severity 3–5) (Figures 2A, C and Supplementary Figure 5).
Taken together, these results demonstrate that, like SARS-CoV-2-specific CD4+ T cells, an overall higher magnitude of CCCs/SARS-CoV-2 cross-reactive CD8+ T-cell responses were present in asymptomatic COVID-19 patients who never presented any COVID-19 symptoms, despite being infected. In contrast, a lower magnitude of CCCs/SARS-CoV-2 cross-reactive CD8+ T-cell responses was detected in severely ill COVID-19 patients and patients with fatal COVID-19 outcomes. These observations also highlight the importance of rapidly mounting strong CCCs/SARS-CoV-2 cross-reactive CD8+ T-cell responses, directed toward structural, non-structural, and accessory protein antigens, for protection against symptomatic and fatal Infections in unvaccinated COVID-19 patients.
A broad lymphopenia, leukocytosis, and low frequencies of CD4+ and CD8+ T cells specific to highly conserved CCCs/SARS-CoV-2 cross-reactive epitopes are present in unvaccinated severely ill symptomatic COVID-19 patients
We next determined whether the low magnitudes of CCCs/SARS-CoV-2 cross-reactive CD4+ and CD8+ T-cell responses detected in unvaccinated severely ill and fatal COVID-19 patients was a result of an overall deficit in the frequencies of total CD4+ and CD8+ T cells. Using a blood differential test (BDT), we compared the absolute numbers of white blood cells (WBCs) and blood-derived lymphocytes, ex vivo, in the unvaccinated COVID-19 patients (Figure 3A).
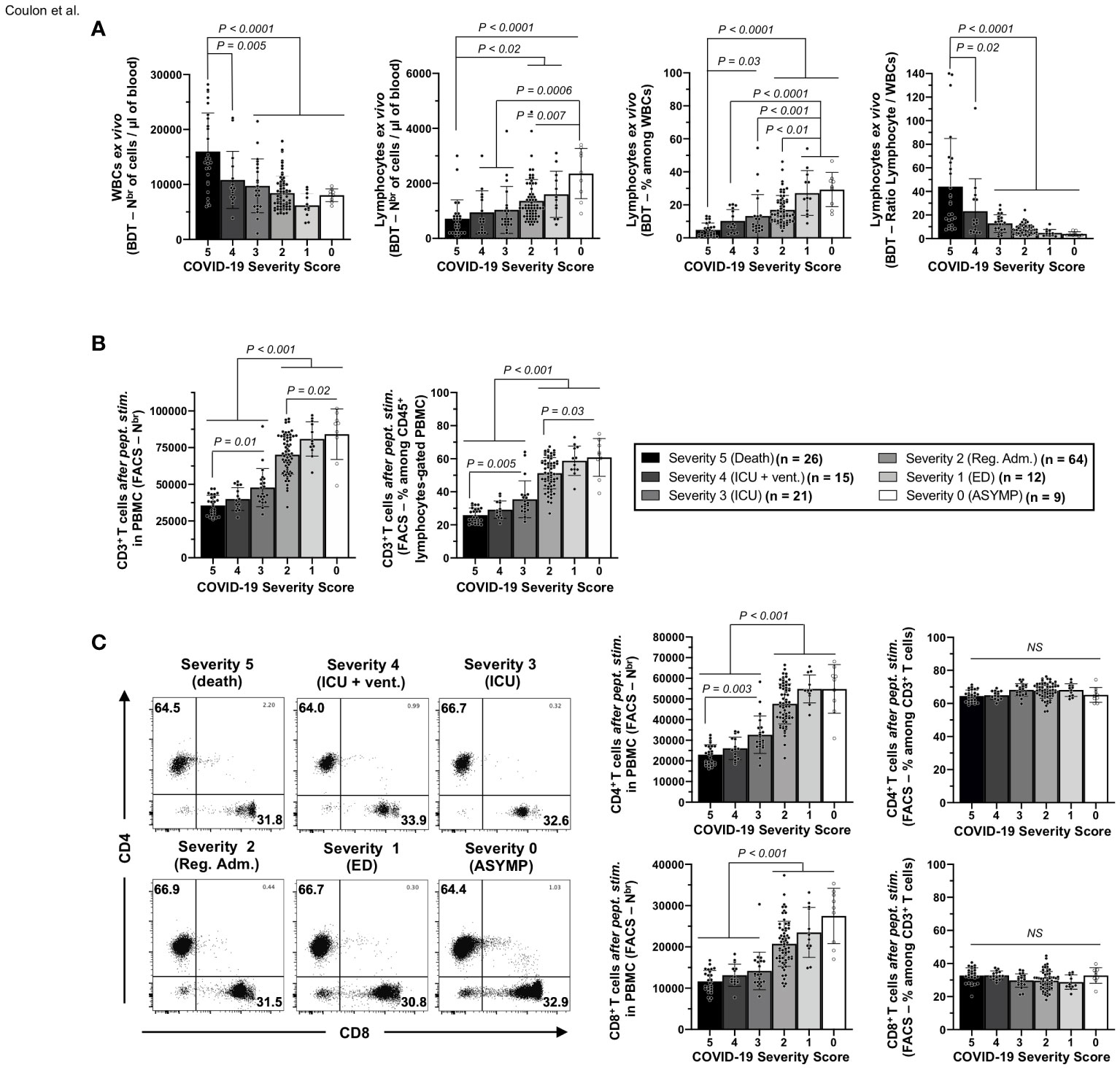
Figure 3 Frequencies of white blood cells, lymphocytes, and CD3+/CD4+/CD8+ T cells in the blood of unvaccinated COVID-19 patients with various degrees of disease severity. (A) numbers of white blood cells (WBCs) and total lymphocytes per µl of blood (left two panels) and percentages and ratios of total lymphocytes among WBCs (right two panels) measured ex vivo by blood differential test (BDT) in unvaccinated COVID-19 patients with various degrees of disease severity (n = 147). (B) Averages/means of numbers and frequencies of CD3+ T cells and (C) of total CD4+, and CD8+ T cells measured by flow cytometry from COVID-19 patients’ PBMCs with various severity scores after 72 h of stimulation with a pool of 16 CD4+ and 27 CD8+ CCCs/SARS-CoV-2 cross-reactive epitope peptides. The right panels show representative dot plots from patients with disease severity scores from 0 to 5. Data are expressed as the mean ±SD. Results are representative of two independent experiments and were considered statistically significant at p ≤ 0.05 (one-way ANOVA).
A significant increase in the numbers of WBCs was detected in unvaccinated COVID-19 patients with fatal outcomes, (i.e., patients with severity 5, ∼1.5- to ∼2.6-fold) when compared with all the remaining five groups of unvaccinated COVID-19 patients (i.e., patients with severity 0, 1, 2, 3, and 4; p ≤ 0.02, Figure 3A—left panel). However, significantly lower absolute numbers of total lymphocytes were detected in the blood of unvaccinated COVID-19 patients with fatal outcomes (i.e., patients with severity 5) compared to unvaccinated COVID-19 patients with mild disease (i.e., patients with severity 1 and 2: ∼1.9- to ∼2.3-fold decrease—p < 0.02) or to asymptomatic patients with no disease (i.e., patients with severity 0: ∼3.3-fold decrease—p < 0.0001) (Figure 3A—second panel from left). As a result, the more severe the disease, the lower the percentage of blood-derived lymphocytes within WBCs (Figure 3A—third panel from left), and the lower the ratio of lymphocyte/WBCs (Figure 3A—fourth panel from left).
Overall, these results indicate that unvaccinated severely ill COVID-19 patients and unvaccinated COVID-19 patients with fatal outcomes not only had a general leukocytosis but also lymphopenia, which developed as early as 4.8 days after reporting their first symptoms or their first PCR-positive test.
Furthermore, we found a significant CD3+ T-cell lymphopenia positively associated with the onset of severe disease in unvaccinated COVID-19 patients (Figure 3B). On average, the two groups of unvaccinated severely ill COVID-19 patients and unvaccinated COVID-19 patients with fatal outcomes (i.e., patients with severity 3, 4, and 5) had a ∼1.9-fold decrease in absolute number of CD3+ T cells compared to three groups of unvaccinated asymptomatic COVID-19 patients with low to no severe disease (i.e., patients with severity 0, 1, and 2, Figure 3B, p < 0.001). Similarly, the numbers of total CD4+ and CD8+ T cells within CD3+-gated cells were reduced early in the two groups of unvaccinated severely ill COVID-19 patients and unvaccinated COVID-19 patients with fatal outcomes (i.e., patients with severity 3, 4, and 5) compared to the three groups of unvaccinated asymptomatic COVID-19 patients with low to no severe disease (Figure 3C—left column graph).
Finally, we determined the frequencies of SARS-CoV-2-specific CD4+ and CD8+ T cells following a 72-h in vitro stimulation with individual CD4+ and CD8+ T epitope peptides (as illustrated in Supplementary Figure 2). We used tetramers specific to five highly conserved CCCs/SARS-CoV-2 cross-reactive DRB1*01:01-restricted CD4+ T-cell epitopes ORF1a1350–1365, S1–13, E26–40, M176–190, and ORF612–26 (Figure 4A) and five highly conserved CCCs/SARS-CoV-2 cross-reactive HLA-A*02:01-restricted CD8+ T-cell epitopes Orf1ab2210–2218, Orf1ab4283–4291, S976–984, S1220–1228, and ORF103–11 (Figure 4B).
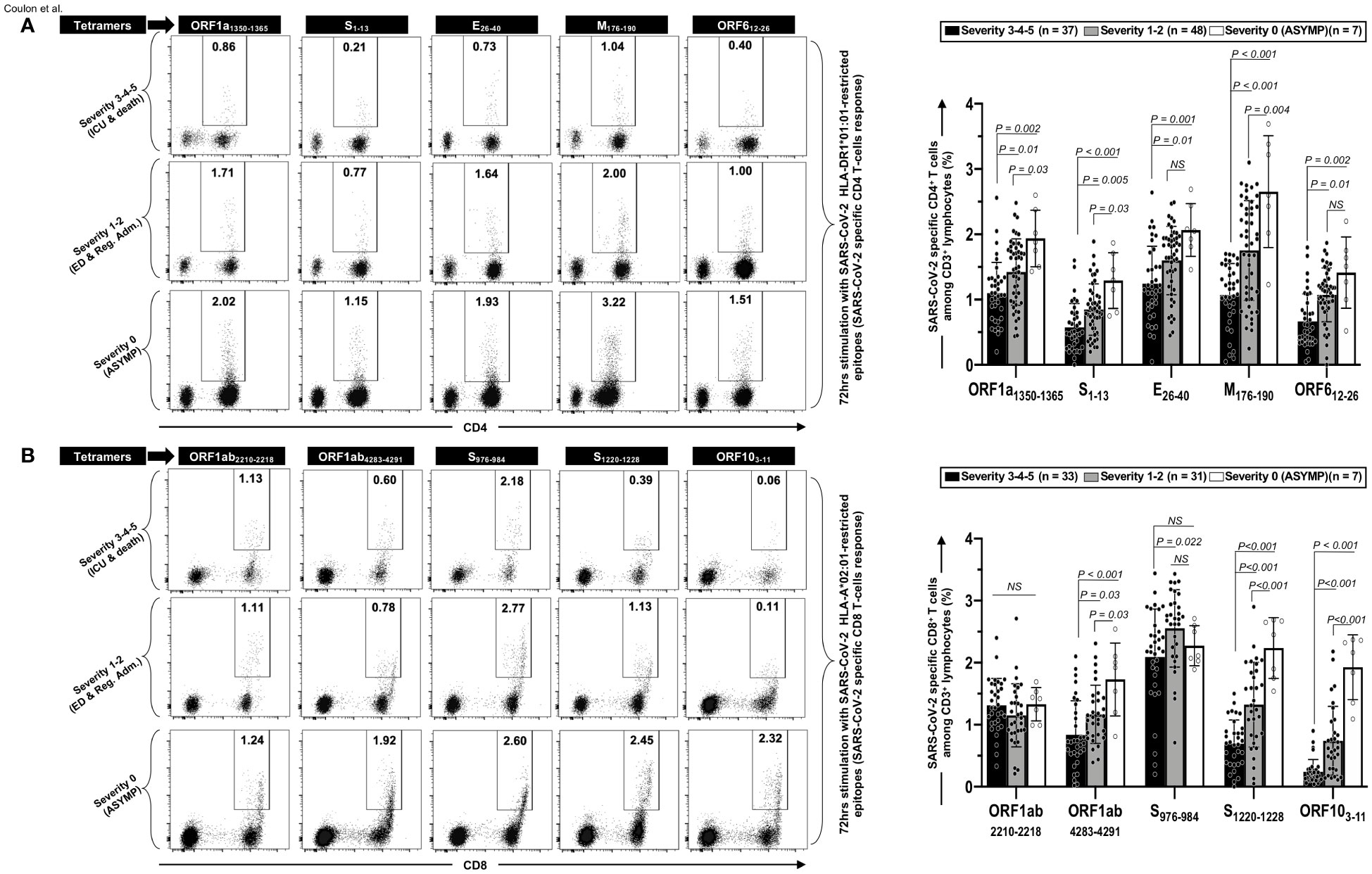
Figure 4 Frequencies of CCCs/SARS-CoV-2 cross-reactive CD4+ and CD8+ T cells in unvaccinated COVID-19 patients with various degrees of disease severity. PBMCs from HLA-DRB1*01:01-positive (n = 92) (A) or HLA-A*02:01-positive (n = 71) (B) unvaccinated COVID-19 patients with various degrees of disease severity were isolated and stimulated for 72 h with 10 μg/ml of indicated CCCs/SARS-CoV-2 cross-reactive CD4+ and CD8+ epitope peptides. The induced CD4+ and CD8+ T cells were then stained and analyzed by flow cytometry. The indicated epitope peptides were chosen among the CCCs/SARS-CoV-2 cross-reactive 16 CD4+ and 27 CD8+ epitope peptides based on tetramer availability. Panel (A) shows representative dot plots (left panels) and average frequencies of CCCs/SARS-CoV-2 cross-reactive CD4+ T cells (right panel) detected in three representatives COVID-19 patients, each falling into one of three groups of disease category: the unvaccinated asymptomatic COVID-19 patients (ASYMP, severity score 0), unvaccinated COVID-19 patients who developed mild to moderate disease (severity scores 1 and 2), and unvaccinated severely ill COVID-19 patients and unvaccinated patients with fatal COVID-19 outcomes (severity scores 3–5). Panel (B) shows representative dot plots (left panels) and average frequencies of CCCs/SARS-CoV-2 cross-reactive CD8+ T cells (right panel) detected in three representatives of COVID-19 patients and in panel (A). Data are expressed as the mean ± SD. Results are representative of two independent experiments and were considered statistically significant at p ≤ 0.05 (one-way ANOVA).
We found a significant decrease in the frequencies of CD4+ T cells specific to all the five highly conserved CCCs/SARS-CoV-2 cross-reactive DRB1*01:01-restricted epitopes in the three groups of unvaccinated severely ill COVID-19 and unvaccinated COVID-19 patients with fatal outcomes (i.e., patients with severity 3, 4, and 5) compared to the remaining three groups of unvaccinated COVID-19 patients with low to no severe disease (i.e., patients with severity 1, 2—p ≤ 0.01) and to unvaccinated asymptomatic COVID-19 patients (severity 0—p ≤ 0.002) (Figure 4A). Similarly, we found a significant decrease in the frequencies of CD8+ T cells specific to three out the five highly conserved CCCs/SARS-CoV-2 cross-reactive HLA-A*02:01-restricted CD8+ T-cell epitopes (Orf1ab4283–4291, S1220–1228, and ORF103–11) in the three groups of unvaccinated severely ill COVID-19 and unvaccinated COVID-19 patients with fatal outcomes (i.e., patients with severity 3, 4, and 5) compared to unvaccinated COVID-19 patients with low to no severe disease (i.e., patients with severity 1 and 2—p ≤ 0.03) and to unvaccinated asymptomatic COVID-19 patients (severity 0—p < 0.001) (Figure 4B). In contrast, similar frequencies of EBV BMLF-1280–288-specific CD8+ T cells were detected across the six groups of unvaccinated COVID-19 patients, regardless of disease severity, indicating that the decrease in the frequencies of T cells in severely ill COVID-19 patients specifically affected highly conserved and CCCs/SARS-CoV-2 cross-reactive T cells (Supplementary Figure 3A).
Taken together, our findings demonstrate that, compared to asymptomatic COVID-19 patients who presented with little to no disease, the severely ill patients and patients with fatal COVID-19 outcomes showed the following: (i) a broad and early lymphopenia (and leukocytosis), (ii) a decrease of bulk CD3+ T-cell lymphocytes number (equally affecting CD4+ and CD8+ T cells), and (iii) a reduction in CD4+ and CD8+ T cells specific to highly conserved CCCs/SARS-CoV-2 cross-reactive epitopes from structural, non-structural, and accessory protein antigens.
Unvaccinated severely ill COVID-19 patients present high frequencies of phenotypically and functionally exhausted CCCs/SARS-CoV-2 cross-reactive CD4+ and CD8+ T cells, detected both ex vivo and in vitro
We next compared the phenotype and function of CD4+ and CD8+ T cells specific to CCCs/SARS-CoV-2 cross-reactive epitopes in unvaccinated asymptomatic COVID-19 patients, with little to no disease, versus the unvaccinated severely ill COVID-19 patients and the unvaccinated COVID-19 patients with fatal outcomes.
Co-expression of four main exhaustion markers (PD-1, TIM3, TIGIT, and CTLA4) and two activation markers (AIMs) CD137 (4-1BB) and CD134 (OX40) were compared using FACS and tetramers specific to five highly conserved CCCs/SARS-CoV-2 cross-reactive DRB1*01:01-restricted CD4+ T-cell epitopes, ORF1a1350–1365, S1–13, E26–40, M176–190, and ORF612–26 both in vivo (Figure 5) and ex vitro (Supplementary Figure 6) and five highly conserved CCCs/SARS-CoV-2 cross-reactive HLA-A*02:01-restricted CD8+ T-cell epitopes, Orf1ab2210–2218, Orf1ab4283–4291, S976–984, S1220–1228, and ORF103–11 both in vitro (Figure 6) and ex vivo (Supplementary Figure 6).
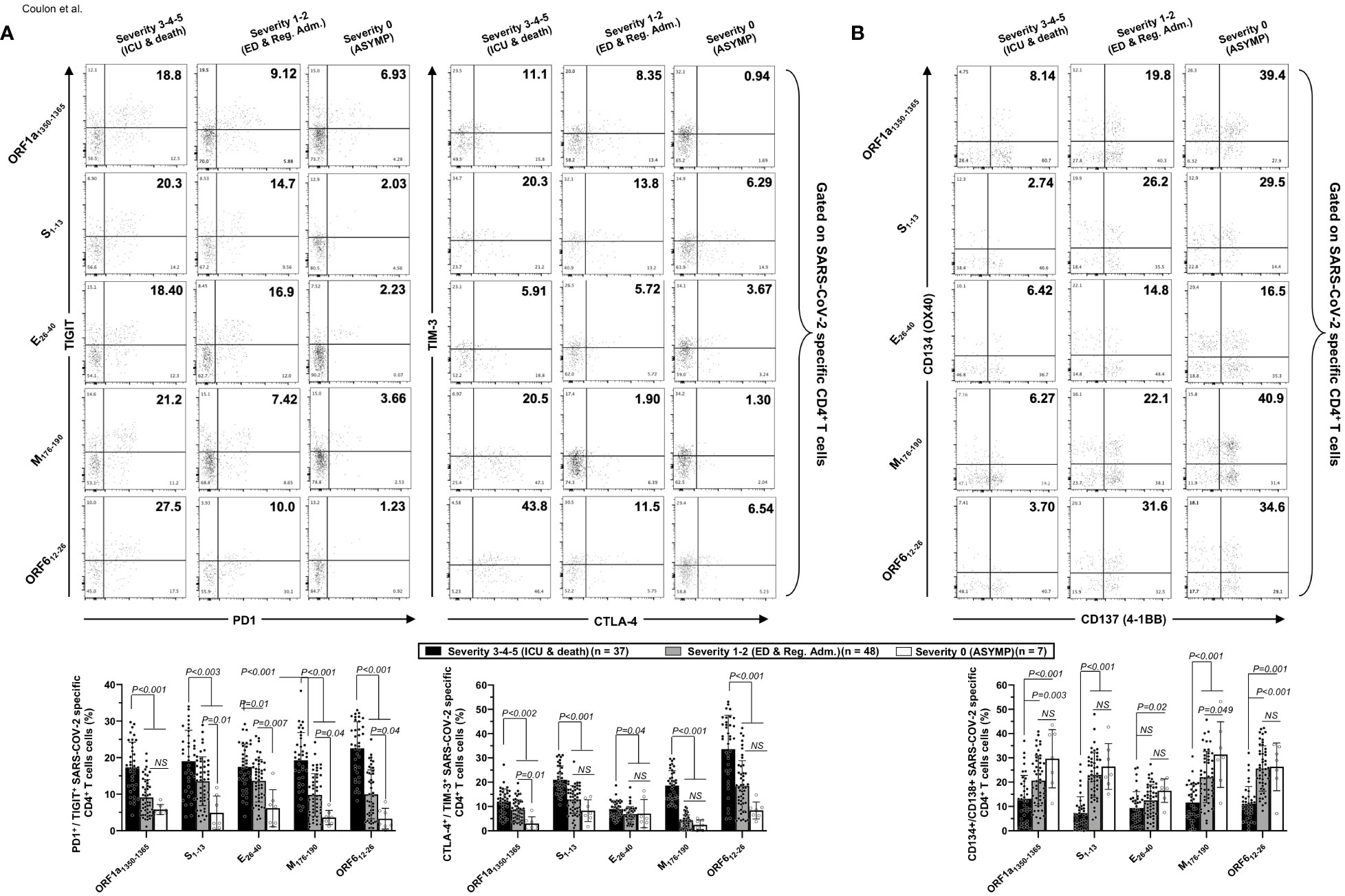
Figure 5 Co-expression of exhaustion and activation markers on CCCs/SARS-CoV-2 cross-reactive CD4+ T cells from unvaccinated COVID-19 patients with various degrees of disease severity. PBMCs from HLA-DRB1*01:01-positive unvaccinated COVID-19 patients with various degrees of disease severity were isolated and stimulated for 72 h with 10 μg/ml of five CCCs/SARS-CoV-2 cross-reactive CD4+ T-cell epitope peptides. The induced CD4+ T cells were then stained and analyzed by flow cytometry for the frequency of tetramer-specific CD4+ cells co-expressing exhaustion and activation markers. Panel (A) shows representative dot plots (upper panels) and average (lower panels) frequencies of CCCs/SARS-CoV-2 cross-reactive CD4+ T cells expressing exhaustion markers PD1/TIGIT and TIM-3/CTLA-4 detected in three representative groups of unvaccinated COVID-19 patients with various degrees of disease severity. Panel (B) shows representative dot plots (upper panels) and average (lower panels) frequencies of CCCs/SARS-CoV-2 cross-reactive CD4+ T cells expressing activation markers (AIMs) CD134/CD137 detected in three representative groups of unvaccinated COVID-19 patients with various degrees of disease severity. Results are representative of two independent experiments, and data are expressed as the mean ± SD and were considered statistically significant at p ≤ 0.05 calculated using one-way ANOVA.
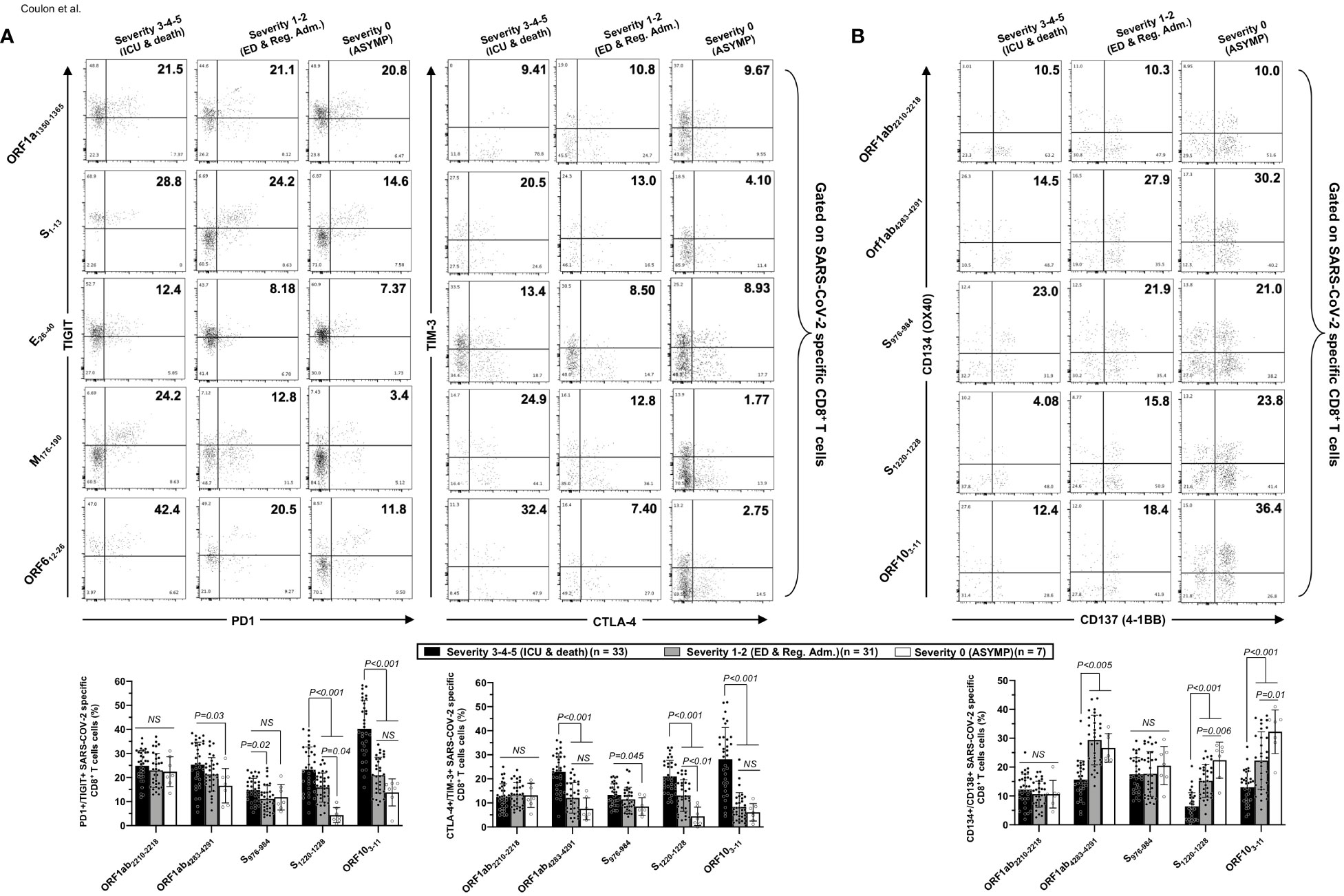
Figure 6 Co-expression of exhaustion and activation markers on CCCs/SARS-CoV-2 cross-reactive CD8+ T cells from unvaccinated COVID-19 patients with various degrees of disease severity. PBMCs from HLA-A*02:01-positive unvaccinated COVID-19 patients with various degrees of disease severity were isolated and stimulated for 72 h with 10 μg/ml of five CCCs/SARS-CoV-2 cross-reactive CD8+ T-cell epitope peptides. The induced CD8+ T cells were then stained and analyzed by flow cytometry for the frequency of tetramer-specific CD8+ cells co-expressing exhaustion and activation markers. Panel (A) shows representative dot plots (upper panels) and average frequencies of CCCs/SARS-CoV-2 cross-reactive CD8+ T cells (lower panel) expressing exhaustion markers PD1/TIGIT and TIM-3/CTLA-4 detected in three representative groups of unvaccinated COVID-19 patients with various degrees of disease severity. Panel (B) shows representative dot plots (upper panels) and average frequencies of CCCs/SARS-CoV-2 cross-reactive CD8+ T cells (lower panel) expressing activation markers (AIMs) CD134/CD137 detected in three representative groups of unvaccinated COVID-19 patients with various degrees of disease severity. Results are representative of two independent experiments, and data are expressed as the mean ± SD and were considered statistically significant at p ≤ 0.05 (one-way ANOVA).
We detected significantly higher frequencies of phenotypically exhausted SARS-CoV-2-specific CD4+ T cells in unvaccinated symptomatic COVID-19 patients with high severity scores (i.e., patients with severity 3, 4, and 5) compared to unvaccinated asymptomatic COVID-19 patients (i.e., patients with severity 0) (Figure 5A—up to ∼6.9-fold increase for ORF612–26-specific PD-1+TIGIT+CD4+ T cells and up to ∼7.8-fold increase for M176–190-specific TIM-3+CTLA-4+CD4+ T cells). Similarly, there were significantly higher frequencies of phenotypically exhausted CD8+ T cells in unvaccinated severely ill COVID-19 and patients with fatal outcomes compared to unvaccinated asymptomatic COVID-19 patients (Figure 6A—up to ∼3.6-fold increase for S1220–1228-specific PD-1+TIGIT+CD8+ T cells and up to ∼4.6-fold increase for S1220–1228- and ORF103-11-specific TIM-3+CTLA-4+CD8+ T cells). Overall, except for Orf1ab2210–2218- and S976–984-specific-CD8+ T cells, the unvaccinated severely ill and fatal patients (i.e., patients with severity 3, 4, and 5) had significantly higher frequencies of exhausted CD8+ T cells co-expressing PD-1+TIGIT+ or TIM-3+CTLA-4 compared to unvaccinated asymptomatic COVID-19 patients with little to no disease (i.e., patients with severity 0, 1, and 2). The Orf1ab2210–2218- and S976–984-specific-CD8+ T cells did not demonstrate any significantly higher phenotypic exhaustion in unvaccinated severely ill COVID-19 patients. We confirmed ex vivo that the unvaccinated severely ill and fatal patients (i.e., patients with severity 3, 4, and 5) had significantly higher frequencies of exhausted CD8+ T cells co-expressing PD-1+TIGIT+ or TIM-3+CTLA-4 compared to unvaccinated asymptomatic COVID-19 patients with little to no disease (i.e., patients with severity 0, 1, and 2, p < 0.05, Supplementary Figure 6).
Accordingly, we also detected low frequencies of functional CD134+CD137+CD4+ T cells (Figure 5B) and low frequencies of functional CD134+CD137+CD8+ T cells (Figure 6B) in unvaccinated severely ill COVID-19 patients and unvaccinated patients with fatal COVID-19 outcomes. This applied to CD134+CD137+CD4+ T cells specific to CCCs/SARS-CoV-2 cross-reactive epitopes from all five structural and non-structural proteins and to CD134+CD137+CD8+ T cells specific to three out of five CCCs/SARS-CoV-2 cross-reactive epitopes from structural and non-structural proteins.
As expected, no differences were observed in phenotypic and functional exhaustion of EBV BMLF-1280–288-specific CD8+ T cells across the six groups of COVID-19 patients with various disease severities (Supplementary Figure 3B), suggesting that the exhaustion of CD4+ and CD8+ T cells in severely ill COVID-19 patients and to patients with fatal COVID-19 outcomes was specific to CCCs/SARS-CoV-2 cross-reactive epitopes.
Altogether, these results (i) indicate that phenotypic and functional exhaustion of CD4+ and CD8+ T cells, detected both ex vivo and in vitro, specific to highly conserved and CCCs/SARS-CoV-2 cross-reactive epitopes from both structural and non-structural antigens was associated with symptomatic and fatal infections in unvaccinated COVID-19 patients and (ii) suggest the importance of functional CCCs/SARS-CoV-2 cross-reactive CD4+ and CD8+ T cells, directed toward structural, non-structural, and accessory protein antigens, for protection against symptomatic and fatal infections in unvaccinated COVID-19 patients.
Higher rates of co-infection with alpha common cold coronavirus 229E present unvaccinated asymptomatic COVID-19 patients
We next compared the co-infection with each of the four main and seasonal α and β CCCs (i.e., α-CCC-NL63, α-CCC-229E, β-CCC-HKU1, and β-CCC-OC43) in a cohort of 85 unvaccinated COVID-19 patients divided into six groups based on the severity of COVID-19 symptoms, as above (i.e., patients with severity 5 to severity 0, Figures 7A, B). Using RT-PCR performed on nasopharyngeal swab samples, we found co-infection with the α-CCC species to be more common with significantly higher rates in the asymptomatic COVID-19 patients (i.e. unvaccinated naturally protected from severe symptoms) compared to severely ill COVID-19 patients and to unvaccinated patients with fatal outcomes (i.e., unvaccinated that were not naturally protected from severe symptoms) (Figure 7A—right panel; ∼2.6-fold increase in groups 1–2–3 versus groups 4–5–6 of disease severity; p = 0.0418 calculated with Fisher’s exact test). Co-infection with the CoV-229E α-CCC species was more common with significantly higher rates in the unvaccinated asymptomatic COVID-19 patients compared to unvaccinated severely ill COVID-19 patients and unvaccinated patients with fatal outcomes (Figure 7B, right panels: ∼4.2-fold increase between unvaccinated asymptomatic COVID-19 patients and unvaccinated severely ill COVID-19 patients (i.e., patients with severity of 4–5–6; p = 0.0223). However, there was no significant difference in the rates of co-infection with β-CCC species (nor with any of the four CCC species) across all six groups of COVID-19 patients with various severity symptoms (Figure 7A, central and left panels, and Figure 7B, left two panels).
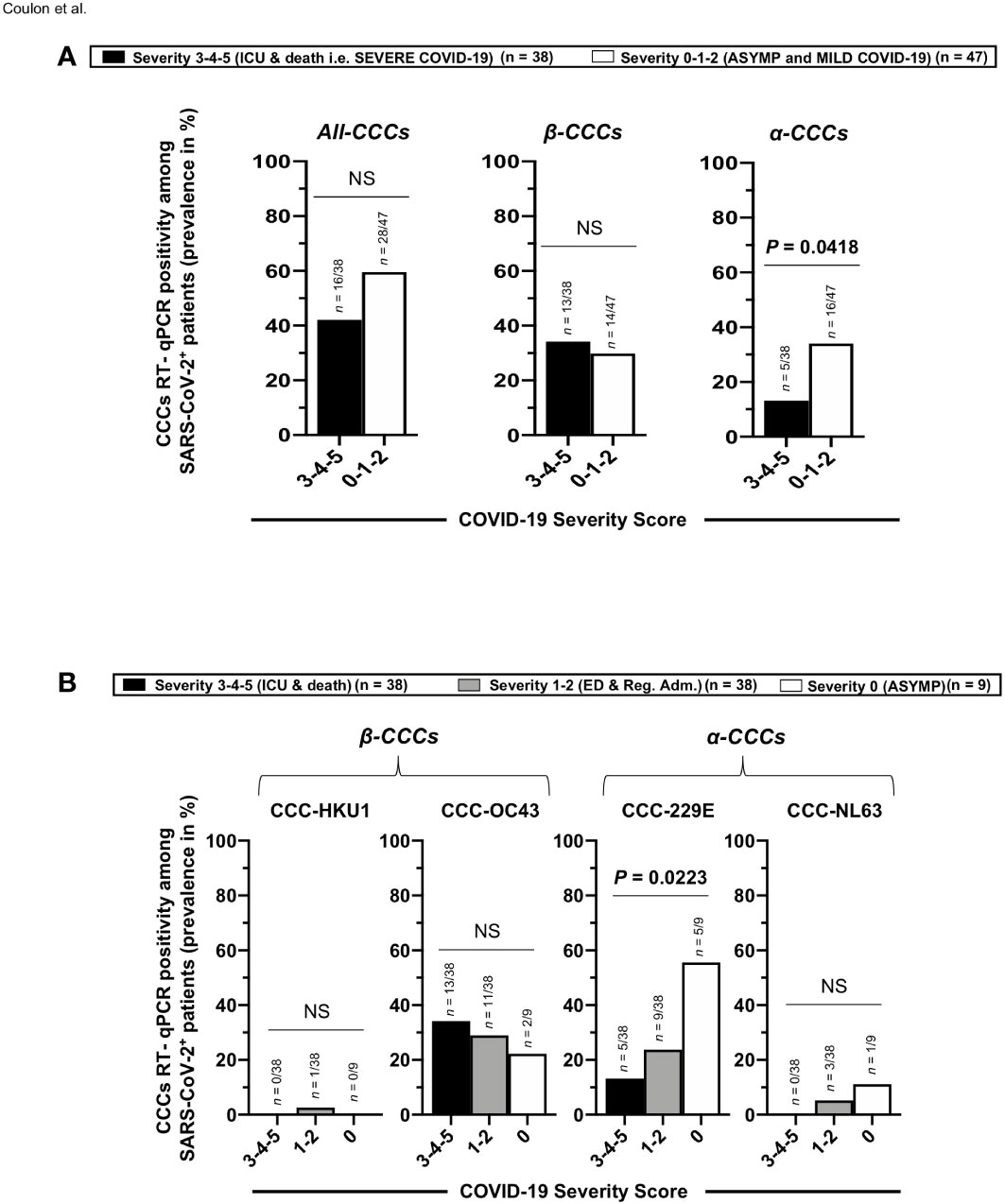
Figure 7 Rates (frequency) of co-infection with seasonal common cold coronavirus species α-CCC-NL63, α-CCC-229E, β-CCC HKU1, and β-CCC-OC43 in unvaccinated COVID-19 patients with various degrees of disease severity. Four major human common cold coronaviruses species, CCC-HKU1, CCC-OC43, CCC-229E, and CCC-NL63, were detected using RT-PCR in the nasopharyngeal swabs of COVID-19 patients (n = 85, first column) who developed various disease severity. Panel (A) shows all four α-CCCs and β-CCCs species (left panel), β-CCC species alone (middle panel), and α-CCC species alone (right panel), detected in unvaccinated severely ill COVID-19 patients and unvaccinated patients with fatal COVID-19 outcomes (severity scores 3–4–5) vs. unvaccinated COVID-19 patients who developed no, mild, and moderate disease (severity score 1–2–3). (B) The rate (%) of co-infection with each one of the four major species, CCC-HKU1, CCC-OC43, CCC-229E, and CCC-NL63, detected in unvaccinated severely ill COVID-19 patients and unvaccinated patients with fatal COVID-19 outcomes (severity scores 3–4–5), in unvaccinated COVID-19 patients who developed mild to moderate disease (severity score 1–2), and in unvaccinated asymptomatic COVID-19 patients (severity score 0). The p-values calculated using the Chi-squared test compare the rate (%) of co-infection with each CCC species between unvaccinated COVID-19 patients with various degrees of disease severity. Results are representative of two independent experiments, and data are expressed as the mean ± SD and were considered statistically significant at p ≤ 0.05 calculated using Fisher’s exact test.
As illustrated in Figure 8, these results indicate that (i) compared to severely ill COVID-19 patients and patients with fatal COVID-19 outcomes, the asymptomatic COVID-19 patients presented significantly higher rates of co-infection with the α-CCC species, and with the 229E of α-CCCs, in particular and (ii) suggest that co-infection with the α species of CCCs (particularly the 229E species of α-CCCs, but not the β species) was associated with the natural protection from symptomatic and fatal infections in unvaccinated COVID-19 patients with yet-to-be-determined mechanisms(s).
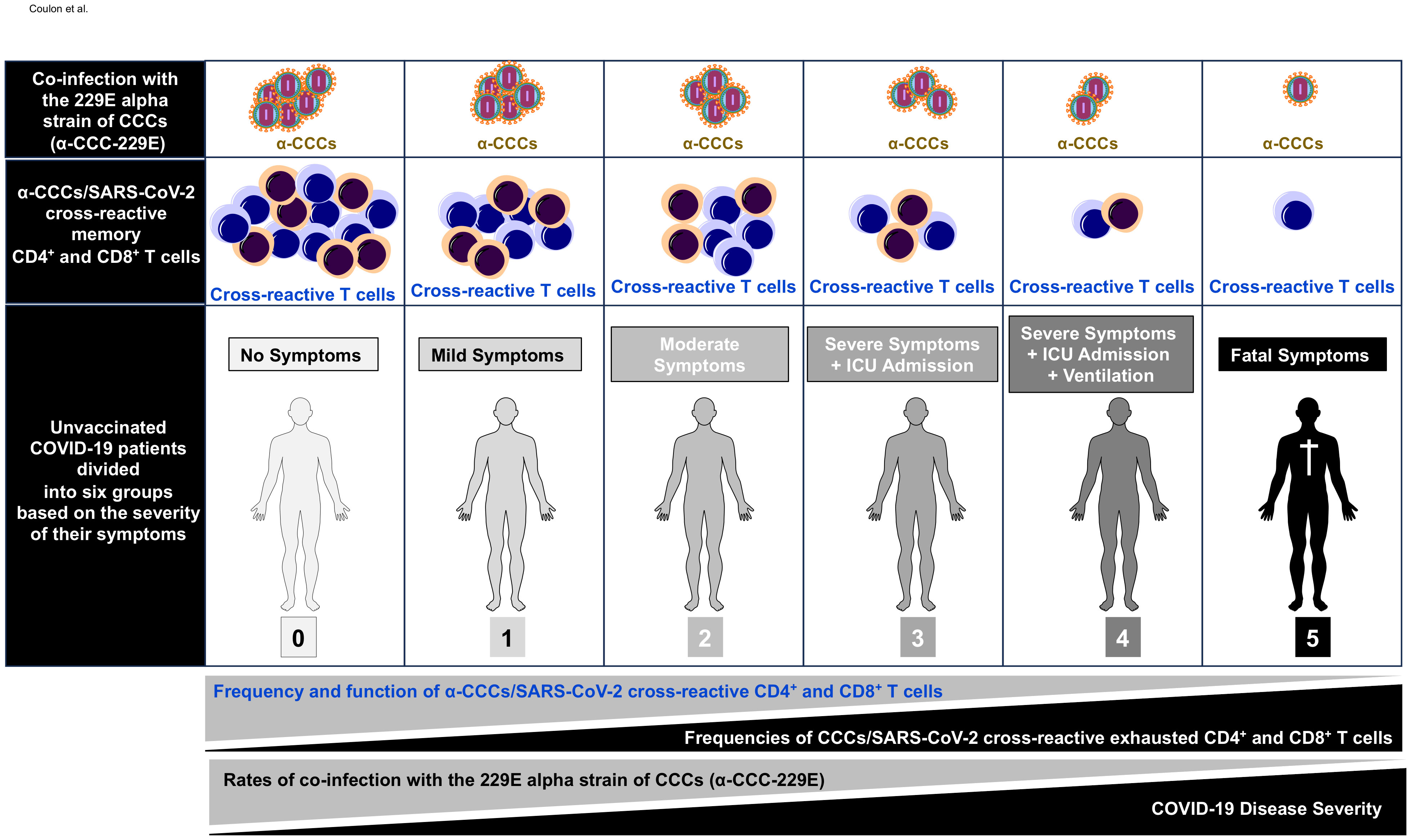
Figure 8 Illustration showing higher frequencies of common cold coronavirus/SARS-CoV-2 cross-reactive CD4+ and CD8+ T cells detected in unvaccinated asymptomatic COVID-19 patients is associated with higher rates of co-infection with alpha common cold coronavirus strain 229E (α-CCC-229E). The first row shows increasing copies of α-CCC-229E detected in unvaccinated asymptomatic COVID-19 patients compared to unvaccinated symptomatic COVID-19 patients. The middle row shows increasing numbers of common cold coronavirus/SARS-CoV-2 cross-reactive CD4+ and CD8+ memory T cells detected in unvaccinated asymptomatic COVID-19 patients compared to unvaccinated symptomatic COVID-19 patients. The bottom row shows symptoms detected in unvaccinated COVID-19 patients with symptoms increasing from severity 0 in asymptomatic COVID-19 patients (left) to severity 5 in COVID-19 patients with fatal COVID-19 outcomes (right) as detailed in Materials and methods.”.
High frequencies of α-CCCs/SARS-CoV-2 cross-reactive memory CD4+ and CD8+ T cells are associated with natural protection from symptomatic and fatal infections in unvaccinated COVID-19 patients
Next, we determined whether (i) the higher rates of co-infection with α-CCC species observed in the unvaccinated asymptomatic COVID-19 patients were associated with high frequencies of CCCs/SARS-CoV-2 cross-reactive CD4+ and CD8+ T cells detected in these asymptomatic COVID-19 groups and (ii) the high frequencies of α-CCCs/SARS-CoV-2 cross-reactive epitope-specific CD4+ and CD8+ T cells were associated with fewer symptoms observed in unvaccinated COVID-19 patients. To this end, we determined the percentage of unvaccinated asymptomatic COVID-19 patients, unvaccinated severely ill COVID-19, and unvaccinated patients with fatal outcomes who presented significant IFN-γ+CD4+ and IFN-γ+CD8+ T-cell responses (i.e., IFN-γ-ELISpot SFCs > 50) specific to α-CCCs/SARS-CoV-2 cross-reactive epitopes.
Significantly higher percentages of unvaccinated asymptomatic COVID-19 patients with significant IFN-γ+CD4+ and IFN-γ+CD8+ T-cell responses specific to α-CCCs/SARS-CoV-2 cross-reactive epitopes were observed (p < 0.001). Similarly, the α-CCCs/SARS-CoV-2 cross-reactive epitopes were strongly cross-recognized by IFN-γ+CD4+ T cells (SFCs>50) and CD8+ T cells from both unexposed pre-pandemic healthy individuals (UPPHI) and unvaccinated asymptomatic COVID-19 patients. In contrast, low frequencies of unvaccinated severely ill COVID-19 patients and unvaccinated patients with fatal outcomes significant IFN-γ+CD4+, and IFN-γ+CD8+ T-cell responses specific to α-CCCs/SARS-CoV-2 cross-reactive epitopes (p < 0.001) (Supplementary Table 4). We also found that unexposed pre-pandemic healthy individuals (UPPHI) who were never exposed to SARS-CoV-2 presented α-CCCs/SARS-CoV-2 cross-reactive IFN-γ+CD4+ and IFN-γ+CD8+ T cells specific to highly conserved SARS-CoV-2 epitopes (Supplementary Figure 7), confirming our report and others’ previous reports (1, 16, 21, 41, 43, 44).
Taken together, these results demonstrate that, compared to low proportions of severely ill COVID-19 patients and patients with fatal outcomes, significant proportions of both unvaccinated asymptomatic COVID-19 patients and unexposed pre-pandemic healthy individuals (UPPHI) presented significant α-CCCs/SARS-CoV-2 strong cross-reactive CD4+ and CD8+ T-cell responses. These findings suggest a crucial role of functional α-CCCs/SARS-CoV-2 cross-reactive memory CD4+ and CD8+ T cells, induced following previous α-CCC seasonal exposures, in protection against subsequent severe symptomatic SARS-CoV-2 infection, as illustrated in Figure 8.
Cross-reactive CD4+ and CD8+ T-cell epitopes from α-CCCs and SARS-CoV-2 that present high similarity and identity are associated with natural protection from symptomatic and fatal infections in unvaccinated COVID-19 patients
Using both the Multiple Sequences Alignments (MSA) and the Epitope Conservancy Tool (ECT) algorithms and software, we determined the identity (%id) and the similarity scores (Ss) of cross-reactive CD4+ and CD8+ T-cell epitopes, between the four major CCC species (α-hCCC-NL63, α-hCCC-229E, and β-hCCC-HKU1, β-hCCC-OC43), on the one hand, and SARS-CoV-2, on the other hand, as described in Materials and methods (58), (Table 2 and Supplementary Tables 1-3).
Of the 16 highly conserved CD4+ T-cell epitopes (Supplementary Figure 8), the ORF1ab5019–5033 epitope was highly conserved (%id ≥ 67%) and highly similar (SS ≥ 0.8) between SARS-CoV-2 and the two β-CCC species (β-CCC-HKU1 and β-CCC-OC43), while the ORF1ab6088–6102 epitope was highly conserved between SARS-CoV-2 and both β-CCC-HKU1 and α-CCC-NL63 species (Table 2 and Supplementary Tables 1-3). Five out of the 27 CD8+ T-cell epitopes (ORF1ab3013–3021, ORF1ab6749–6757, S958–966, E20–28, and M52–60) were highly conserved (% id ≥67%) and highly similar (SS ≥ 0.8) between SARS-CoV-2 and the α-CCCs and/or β-CCC species. Specifically, the ORF1ab3013–3021 CD8+ T-cell epitope was highly conserved between SARS-CoV-2 and the two β-CCC species (β-CCC-HKU1 and β-CCC-OC43); the ORF1ab6749–6757 epitope was highly conserved between SARS-CoV-2 and all the four CCC species; the S958–966 epitope was highly conserved between SARS-CoV-2, the two β-CCC species, and the α-CCC-NL63 species; the E20–28 epitope was highly conserved between SARS-CoV-2 and the β-CCC-HKU1 species; and the M52-60 epitope was highly conserved between SARS-CoV-2, the two β-CCC species (β-CCC-HKU1 and β-CCC-OC43) and the α-CCC-229E species (Supplementary Figure 9, Table 2, and Supplementary Tables 1-3). While the E20–28 epitope was conserved (%id = 67%) between SARS-CoV-2 and α-CCC-NL63 species, it did not present significant similarity (Table 2 and Supplementary Tables 1-3).
Next, we determined the corresponding NL63 peptide (SS = 0.76). While the S976–984 epitope was conserved between SARS-CoV-2 and three CCC species (%id = 67%), it did not present significant similarity with the corresponding CCC peptides [β-CCC-HKU1 (SS=0.78), β-CCC-OC43 (SS=0.78) and α-CCC-NL63 (SS = 0.73)]. Finally, while the S2–10 epitope was highly similar between SARS-CoV-2 and α-CCC-NL63 (SS = 0.82), it was not significantly identical (id% = 56%) (Table 2 and Supplementary Tables 1-3).
Next, we determined whether the CCCs/SARS-CoV-2-cross-reactive epitopes were cross-recognized preferentially by the CD4+ and CD8+ T cells from either unvaccinated asymptomatic COVID-19 patients, or unvaccinated severely ill COVID-19 patients and unvaccinated patients with fatal outcomes (Supplementary Table 4). No significant differences were detected when the slopes S of the SARS-CoV-2-specific CD4+ and CD8+ T-cell responses were applied towards epitopes that have no significant identity nor similarity to epitopes from the four CCCs. Significant differences were detected when the slopes S of the SARS-CoV-2-specific CD4+ and CD8+ T-cell responses were applied to epitopes that have significant identity and/or similarity to epitopes from at least one of the four CCCs (Supplementary Table 4). In contrast, SARS-CoV-2 CD4+ or CD8+ T cells cross-recognizing epitopes that are highly identical and similar exclusively in β-CCC species, but not in α-CCC species (i.e., epitopes ORF1ab5019-5033 and ORF1ab3013-3021), presented a significantly lower slope S (p = 0.04) (Supplementary Table 4). The ORF1ab5019–5033 and ORF1ab3013–3021 epitopes have slopes S close to 0 among all epitopes (Supplementary Table 4).
These data indicated that (i) CCCs/SARS-CoV-2-cross-reactive CD4+ or CD8+ T-cell epitopes that share high identity and similarity exclusively with the α-CCC species were cross-recognized mainly by CD4+ or CD8+ T cells from asymptomatic COVID -19 patients; (ii) in contrast, the CCCs/SARS-CoV-2-cross-reactive CD4+ or CD8+ T cell epitopes that share high identity and similarity exclusively with the β-CCC species were cross-recognized mainly by CD4+ or CD8+ T cells from severely ill symptomatic patients; and (iii) compared to severely ill COVID-19 patients and patients with fatal outcomes, the asymptomatic COVID-19 patients presented significantly higher frequencies of α-CCCs/SARS-CoV-2 cross-reactive CD4+ and CD8+ T cells. The findings suggest a crucial role of functional, poly-antigenic α-CCCs/SARS-CoV-2 cross-reactive memory CD4+ and CD8+ T cells, induced following previous α-CCC seasonal exposures, in protection against subsequent severe symptomatic SARS-CoV-2 infection.
Discussion
Characterizing the underlying T-cell mechanisms associated with protection against COVID-19 severity in unvaccinated asymptomatic patients is a challenging task today, since most individuals have received at least one dose of COVID-19 vaccine (39). Only 15.2% of adults in the United States are unvaccinated (37, 38). This study is one of the few to comprehensively characterize the cross-reactive memory CD4+ and CD8+ T cells in unvaccinated symptomatic and asymptomatic COVID-19 patients. We compared the antigen specificity, frequency, phenotype, and function of CCCs/SARS-CoV-2 cross-reactive memory CD4+ and CD8+ T cells, cross-recognizing genome-wide conserved epitopes in a cohort of 147 unvaccinated COVID-19 patients, divided into six groups based on the severity of their symptoms. The findings demonstrate several relationships between antigen-specific T-cell responses and disease outcome. Specifically, severely ill symptomatic COVID-19 patients who required admission to intensive care units (ICUs) and patients with fatal COVID-19 outcomes, versus unvaccinated asymptomatic COVID-19 patients, displayed significantly (i) higher rates of co-infection with the 229E alpha species of CCCs (α-CCC-229E); (ii) higher frequencies of α-CCCs/SARS-CoV-2 cross-reactive functional memory CD134+CD137+CD4+ and CD134+CD137+CD8+ T cells, directed toward conserved epitopes from structural, non-structural, and accessory SARS-CoV-2 proteins; and (iii) lower frequencies of CCCs/SARS-CoV-2 cross-reactive and exhausted PD-1+TIM3+TIGIT+CTLA4+CD4+ and PD-1+TIM3+TIGIT+CTLA4+CD8+ T cells. These observations (i) support a crucial role for functional, poly-antigenic α-CCCs/SARS-CoV-2 cross-reactive memory CD4+ and CD8+ T cells, induced following previous α-CCC seasonal exposures, in protection against subsequent severe symptomatic SARS-CoV-2 infection and (ii) provide critical insights into developing broadly protective, multi-antigen, CD4+ and CD8+ T-cell-based, universal pan-Coronavirus vaccines capable of conferring cross-species protection.
The present comprehensive study of cross-reactive SARS-CoV-2 epitope-specific CD4+ and CD8+ T cells suggests that pre-pandemic exposure to seasonal α-CCC species, but not to β-CCC species, may have conferred protection from symptomatic COVID-19 infections by an as-yet-to-be-determined mechanism(s). It is likely that pre-existing CCCs/SARS-CoV-2 cross-reactive memory CD4+ and CD8+ T cells, induced in UPPHI by seasonal α-CCC species, cross-recognized protective SARS-CoV-2 epitopes. These data are consistent with previous studies showing that high levels of CCCs immunity in convalescent patients are associated with improved survival in COVID-19 patients (60, 61).
In the present study, we detected pre-existing CCCs/SARS-CoV-2 cross-reactive memory CD4+ and CD8+ T cells specific to many conserved SARS-CoV-2 epitopes in UPPHI. These results extend previous reports on the presence of specific repertoires of protective clones of memory CD4+ and CD8+ T cells in UPPHI possibly primed by previous exposure to seasonal CCCs infections and the rapid recall of α-CCCs/SARS-CoV-2 cross-reactive memory CD4+ and CD8+ T cells (1, 21, 41, 43, 62–66). UPPHI likely have different repertoires of protective and pathogenic memory CD4+ and CD8+ T cells targeting cross-reactive CCCs/SARS-CoV-2 epitopes of structural, non-structural, and accessory protein antigens that are associated with different disease outcomes in COVID-19 patients (13, 67–69). Indeed, we discovered that concomitant SARS-CoV-2/β-CCCs species (i.e., β-CCCs-HKU1 and β-CCCs-OC43) co-infection correlated with a trend toward more severe COVID-19 disease, whereas SARS-CoV-2/α-CCCs species (i.e., α-CCCs-NL63 and mainly α-CCCs-229E) co-infection significantly correlated with less severe COVID-19 disease.
The positive correlation between functional α-CCCs/SARS-CoV-2 cross-reactive memory CD4+ and CD8+ T cells and better disease outcomes in asymptomatic COVID-19 patients supports the importance of developing CoV vaccines that cross-recognize functional α-CCCs/SARS-CoV-2 cross-reactive memory CD4+ and CD8+ T cells (70–72). Pre-existing T cells cross-recognizing conserved SARS-CoV-2 epitopes that cross-react with α-CCCs, but not β-CCCs, may be important in preventing severe COVID-19 symptoms. We are currently assessing whether candidate multi-epitope-based vaccines expressing the epitopes associated with good disease outcomes that cross-react with α-CCC species (in contrast to symptomatic epitopes that cross-react with β-CCC species) would confer cross-species protection in the HLA-A2/DR1/hACE2 triple transgenic mice.
While many SARS-CoV-2 epitopes present high identity and high similarity with the four α-CCCs and β-CCC species, they did not necessarily recall the strongest SARS-CoV-2 epitope-specific CD4+ and CD8+ T-cell responses in UPPHI. For example, the SARS-CoV-2 epitopes ORF1a1350–1365, S1–13, M176–190, and ORF612–26 recalled strong CD4+ T-cell responses in UPPHI but were not identical or similar with any epitopes from the four α-CCCs and β-CCC species. The same observation applies to the CD8+ epitopes ORF1ab1675–1683, S1000–1008, and S1220–1228. This suggests that the SARS-CoV-2-specific CD4+ and CD8+ T-cell responses in UPPHI may have been induced by other non-CCC pathogens, as has been reported by (73–76). Thus, in line with previous reports, we found that not all SARS-CoV-2 T-cell epitopes cross-reacted with CCC epitopes (41, 73–77). For instance, CMV T cells cross-react with SARS-CoV-2 T cells, despite low sequence homology between the two viruses, and this may contribute to the pre-existing immunity against SARS-CoV-2 (74). This is in agreement with our finding that eight of the 27 CD8+ T-cell epitopes (ORF1ab1675–1683, ORF1ab5470–5478, ORF1ab6749–6757, S2–10, S958–966, S1220–1228, E20–28, and E26–34) shared highly identical sequences (%id equal to 67%–78%) with epitopes from common human pathogens (EBV, Streptococcus pneumoniae, Bordetella pertussis, and Corynebacterium diphtheriae) and to widely distributed BCG and DTa/wP vaccines. Six of those also shared high similarity scores (SS≥0.8) with epitopes from EBV, S. pneumoniae, B. pertussis, and C. diphtheriae and widely distributed BCG and DTa/wP vaccines. CD8+ T cells specific to SARS-CoV-2 epitopes that share high identity and similarity with the DTwP vaccine (but not BCG vaccine) epitopes were significantly associated with asymptomatic COVID-19 infection. The most functional CD8+ T cells cross-recognized SARS-CoV-2 common epitopes that are highly similar and identical to epitopes from the DTwP vaccine. These findings are consistent with a previous study that described a correlation between DTwP vaccination and fewer COVID-19 deaths (59). Overall, our findings suggest that the pre-existing SARS-CoV-2-specific CD4+ and CD8+ T-cell responses in UPPHI may be the consequence of heterologous immunity induced by CCCs (31, 44, 75, 78–82), other pathogens (77), and widely administered vaccines (BCG, DTwP) (73–76).
The present comprehensive analysis demonstrates, both in vitro and ex vivo, that unvaccinated severely ill COVID-19 patients had higher frequencies of phenotypically and functionally exhausted CCCs/SARS-CoV-2 cross-reactive CD4+ and CD8+ T cells. In contrast, higher frequencies of functional CD4+ and CD8+ T cells specific to CCCs/SARS-CoV-2 epitopes were detected in unvaccinated asymptomatic COVID-19 patients. Although older COVID-19 patients tend to be more symptomatic compared to younger COVID-19 patients, the symptomatic COVID-19 patients tend to have less functional SARS-CoV-2-specific T cells, regardless of age. Similar results were obtained when age-matched symptomatic and asymptomatic COVID-19 patients were compared, suggesting that the frequency of functional SARS-CoV-2-specific T cells is age independent (data not shown). Besides CD134 and CD137 functional markers, we recently assessed the expression of additional activation and cytotoxicity markers by CCCs/SARS-CoV-2-cross-reactive CD4+ and CD8+ T cells from COVID-19 patients and healthy individuals (1). Higher frequencies of CCCs/SARS-CoV-2-cross-reactive functional memory CD8+ T cells were detected in both COVID-19 patients and healthy individuals. However, we have observed that COVID-19 patients and unexposed healthy individuals exhibited a different pattern of CD8+ T-cell immunodominance. Unlike for CD8+ T cells, higher frequencies of multifunctional CCCs/SARS-CoV-2-cross-reactive memory CD4+ T cells, expressing CD69, CD107a/b, and TNF-α were detected in COVID-19 patients compared to healthy individuals (1). However, the association of T-cell exhaustion with symptomatic and fatal COVID-19 infections in unvaccinated patients is currently being debated (6, 83). Reports using small cohorts of patients did not identify a link between higher expression of exhaustion markers and impaired function of SARS-CoV-2-specific CD4+ and CD8+ T cells in convalescent patients (84, 85). In contrast, our study used larger cohorts of COVID-19 patients with detailed clinical differentiation of symptomatic and asymptomatic patients. Our data are consistent with previous reports in which a broad T-cell exhaustion with impaired function was found in both the peripheral compartment (PBMCs), the lungs, and the brain of symptomatic patients (15, 86–88) and increased levels of PD-1 in severe cases compared to those in non-severe cases (83, 89).
Moreover, we extended those reports by characterizing the exhausted SARS-CoV-2-specific CD4+ and CD8+ T cells co-expressing multiple markers of exhaustion, TIM3, TIGIT, and CTLA4, besides PD-1. There is no consensus on a specific combination of inhibitory molecules of clusters of exhaustion markers to conclude phenotypic and functional exhaustion of epitope-specific CD4+ and CD8+ T cells. Overall, the major markers (or pathways) described as associated with CD4+ and CD8+ T-cell exhaustion include PD-1, TIGIT, CTLA-4, and TIM-3. While various combinations of these exhaustion have been used to demonstrate T-cell exhaustion, typically the PD-1 and TIM-3 combination is mainly used to demonstrate CD4+ and CD8+ T-cell exhaustion and dysfunction. In this study, we have used the combination of PD-1 and TIGIT exhaustion markers, on the one hand, and the combination of CTLA-4 and TIM-3 exhaustion markers, on the other hand, to demonstrate that increased frequencies of phenotypically exhausted SARS-CoV-2 epitope-specific CD4+ and CD8+ T cells are associated with severe COVID-19 disease, as previously reported in other systems (90–97). These findings suggest impaired functionality in SARS-CoV-2-specific CD4+ and CD8+ T cells, along with generally lower interferon-gamma (IFN-γ) and tumor necrosis factor-alpha (TNF-α) production, is associated with symptomatic and fatal Infections in unvaccinated COVID-19 patients. Our results also agree with previous reports highlighting that a prior “original antigenic sin” (OAS) potentially linked to prior exposure to seasonal CCCs might skew CCCs/SARS-CoV-2 cross-reactive CD4+ and CD8+ T cells toward an exhausted phenotype (98). Because severely ill patients preferentially developed higher frequencies of co-infection with β-CCC species and higher frequencies of pre-existing β-CCCs/SARS-CoV-2 cross-reactive memory CD4+ and CD8+ T cells, T-cell exhaustion may be related to prior exposure to seasonal β-species of CCCs.
The present study has comprehensively characterized CCCs/SARS-CoV-2 cross-reactive memory CD4+ and CD8+ T cells in blood samples from over 140 unvaccinated symptomatic and asymptomatic COVID-19 patients. However, there remain several gaps in our understanding. First, the study of CCCs/SARS-CoV-2 cross-reactive memory CD4+ and CD8+ T cells in unvaccinated symptomatic and asymptomatic COVID-19 patients has not been adjusted retrospectively to previous CCCs infections, due to the lack of pre-COVID-19 samples. At this point, the vast majority of adults in the United States have been infected and/or received at least one dose of the COVID-19 vaccine (37, 38); thus, going forward, characterizing pre-COVID-19 cross-reactive memory CD4+ and CD8+ T cells in unvaccinated COVID 19 patients will be very difficult (39). Second, the study did not follow up with the COVID-19 patients at later times points after convalescence; hence, the reported CCCs/SARS-CoV-2 cross-reactive memory CD4+ and CD8+ T cell characteristics are reflective of their status shortly after exposure to SARS-CoV-2 or during the symptomatic disease. Although we assessed SARS-CoV-2-specific CD4+ and CD8+ T cell responses at an early stage of the disease (blood sampled on average 5 days after the appearance of the first reported symptoms), the precise timing of the patient’s first exposure to SARS-CoV-2 is not known. Third, since the T-cell responses reported in this study were assessed in the peripheral blood, this may not reflect tissue-resident CD4+ and CD8+ T cells in the lungs and the brain. The reduced number of functional CCCs/SARS-CoV-2 cross-reactive memory CD4+ and CD8+ T cells detected in the peripheral blood of symptomatic COVID-19 patients may be due to T-cell redistribution to other organs, such as the lungs and the brain. The asymptomatic infections in unvaccinated COVID-19 patients might be attributed to homing and redistribution of high numbers of functional CCCs/SARS-CoV-2 cross-reactive CD4+ and CD8+ T cells into the lungs of unvaccinated asymptomatic COVID-19 patients, rather than in peripheral blood. In this context, we recently reported that high frequencies of functional lung-resident memory CD4+ and CD8+ T cells contributed to protection against COVID-19-like symptoms and death caused by SARS-CoV-2 infection in a mouse model (2). Thus, future studies should investigate tissue-resident CD4+ and CD8+ T cells in the lungs to determine whether their frequency and function correlate with protection from symptomatic and fatal infections in unvaccinated COVID-19 patients. Finally, while the study enrolled 600 patients overall, the study compared the antigen specificity, frequency, phenotype, and function of common cold coronaviruses (CCCs) and SARS-CoV-2 cross-reactive memory CD4+ and CD8+ T cells, targeting genome-wide conserved epitopes in a cohort of 147 unvaccinated COVID-19 patients screened for two HLA types, HLA-DRB1*01:01 and HLA-A*02:01. Thus, future studies are being conducted to assess T cells from other HLA types. Nevertheless, our results are consistent with the hypothesis that the early presence of high numbers of functional α-CCCs/SARS-CoV-2 cross-reactive CD4+ and CD8+ T cells targeting multiple antigens was associated with protection from symptomatic and fatal SARS-CoV-2 infections in unvaccinated COVID-19 patients (99).
This report also confirms previous reports that (i) early and broad lymphopenia positively correlated with COVID-19 disease severity and mortality (86, 100–102); (ii) broad leukocytosis combined with T cell lymphopenia was present in severe COVID-19 patients and extended those findings by demonstrating that the observed T-cell lymphopenia was particularly prevalent for SARS-CoV-2-specific T cells (86, 100); and (iii) a significant age-dependent and comorbidity-associated susceptibility to COVID-19 disease, with patients over 60 years of age, and those with pre-existing diabetic and hypertension comorbidities being the most susceptible to severe COVID-19 disease (13, 20).
In conclusion, the present comprehensive analysis of specific and cross-reactive SARS-CoV-2 epitope-specific T cells reveals clear relationships between T-cell responses and disease outcomes in unvaccinated COVID-19 patients. Compared to severely ill COVID-19 patients and patients with fatal COVID-19 outcomes, the asymptomatic COVID-19 patients presented high rates of co-infection with the α-CCC species and more functional and less exhausted α-CCCs/SARS-CoV-2 cross-reactive memory CD4+ and CD8+ T cells, targeting structural, non-structural, and accessory proteins. The findings suggest functional, poly-antigenic α-CCCs/SARS-CoV-2 cross-reactive memory CD4+ and CD8+ T cells, induced following CCCs repetitive exposures, are contributing factors in reducing the severity of SARS-CoV-2 infection, as illustrated in Figure 8. Most of the >10 billion doses of first-generation COVID-19 vaccines are based on the Spike antigen alone (103, 104) and function mainly by inducing neutralizing antibodies (105). Because the Spike protein has undergone a substantial number of mutations with each successive viral variant, these first-generation subunit vaccines are susceptible to immune evasion by new variants and subvariants, such as XBB.1.5, EG.5 (Eris), and HV.1 sub-variants of Omicron (71, 72). To overcome this critical limitation, the next generation of COVID-19 vaccines should also target other highly conserved structural and non-structural SARS-CoV-2 antigens capable of inducing protection by cross-reactive CD4+ and CD8+ T cells (1, 106). Herein, the findings of this report provide a roadmap for developing next-generation α-CCCs/SARS-CoV-2 cross-reactive CD4+ and CD8+ T cell-based, multi-antigen, pan-Coronavirus vaccines capable of conferring cross-species protection.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The studies involving humans were approved by Between July 2020 to November 2023, 600 patients were enrolled at the University of California Irvine Medical Center with mild to severe COVID-19 under an approved Institutional Review Board–approved protocol (IRB#-2020-5779). Written informed consent was obtained from participants before inclusion. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
LB: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. PC: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft. SP: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Software, Validation, Visualization, Writing – original draft. ND: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Writing – original draft. RS: Data curation, Formal analysis, Methodology, Project administration, Writing – original draft, Conceptualization. LZ: Data curation, Methodology, Writing – original draft. DT: Data curation, Resources, Writing – original draft. RE: Data curation, Resources, Writing – original draft. CF: Data curation, Resources, Writing – original draft. SS: Resources, Writing – original draft. LH: Resources, Writing – original draft. AN: Writing – review & editing, Writing – original draft. BK: Resources, Writing – original draft. EB: Data curation, Methodology, Writing – original draft. HV: Data curation, Writing – original draft. DG: Conceptualization, Formal analysis, Writing – review & editing. TJ: Conceptualization, Writing – original draft. JU: Conceptualization, Formal analysis, Writing – review & editing, Writing – original draft.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. These studies were supported in part by Public Health Service Research grants AI158060, AI150091, AI143348, AI147499, AI143326, AI138764, AI124911, and AI110902 from the National Institutes of Allergy and Infectious Diseases (NIAID) to LB and by R43AI174383 to TechImmune, LLC. The funder TechImmune, LLC was not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication.
Acknowledgments
The authors would like to thank the NIH Tetramer Facility (Emory University, Atlanta, GA) for providing the Tetramers used in this study. We thank the UC Irvine Center for Clinical Research (CCR) and the Institute for Clinical and Translational Science (ICTS) for providing human blood samples and nasopharyngeal swab samples used in this study. A special thanks to Dr. Alessandro Ghigi and Dr. Kai Zheng for providing patients’ clinical information. We also thank those who contributed directly or indirectly to this COVID-19 project: Gavin S. Herbert, Dr. Steven A. Goldstein, Dr. Michael J. Stamos, Dr. Suzanne B. Sandmeyer, Jim Mazzo, Dr. Daniela Bota, Dr. Dan Forthal, Christine Dwight, Janice Briggs, Marge Brannon, Beverley Alberola, Jessica Sheldon, Rosie Magallon, and Andria Pontello.
Conflict of interest
Authors HV, DG, TJ and JU were employed by company TechImmune LLC. LB has an equity interest in TechImmune, LLC., a company that may potentially benefit from the research results and serves on the company’s Scientific Advisory Board. LB’s relationship with TechImmune, LLC., has been reviewed and approved by the University of California, Irvine by its conflict-of-interest policies.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1343716/full#supplementary-material
References
1. Prakash S, Srivastava R, Coulon PG, Dhanushkodi NR, Chentoufi AA, Tifrea DF, et al. Genome-wide B cell, CD4(+), and CD8(+) T cell epitopes that are highly conserved between human and animal coronaviruses, identified from SARS-CoV-2 as targets for preemptive pan-coronavirus vaccines. J Immunol. (2021) 206:2566–82. doi: 10.4049/jimmunol.2001438
2. Prakash S, Dhanushkodi NR, Zayou L, Ibraim IC, Quadiri A, Coulon PG, et al. Cross-protection induced by highly conserved human B, CD4 (+,) and CD8 (+) T cell epitopes-based coronavirus vaccine against severe infection, disease, and death caused by multiple SARS-CoV-2 variants of concern. Front Immunol. (2023) 15:1328905. doi: 10.1101/2023.05.24.541850
4. Harris E. Most COVID-19 deaths worldwide were among older people. JAMA. (2023) 329:704. doi: 10.1001/jama.2023.1554
5. Bahgat MM, Nadeem R, Nasraa MH, Amer K, Hassan WA, ELGarhy FM, et al. Proinflammatory cytokine profiles in both mild symptomatic and asymptomatic SARS-CoV-2-infected Egyptian individuals and a proposed relationship to post-COVID-19 sequela. Viral Immunol. (2023) 36:600–9. doi: 10.1089/vim.2023.0060
6. Wilk AJ, Rustagi A, Zhao NQ, Roque J, Martínez-Colón GJ, McKechnie JL, et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat Med. (2020) 26:1070–6. doi: 10.1038/s41591-020-0944-y
7. Baek MS, Lee MT, Kim WY, Choi JC, Jung SY. COVID-19-related outcomes in immunocompromised patients: A nationwide study in Korea. PLoS One. (2021) 16:e0257641. doi: 10.1371/journal.pone.0257641
8. Helleberg M, Niemann CU, Moestrup KS, Kirk O, Lebech AM, Lane C, et al. Persistent COVID-19 in an immunocompromised patient temporarily responsive to two courses of remdesivir therapy. J Infect Dis. (2020) 222:1103–7. doi: 10.1093/infdis/jiaa446
9. Tsang TK, Wang C, Fang VJ, Perera R, So HC, Ip DKM, et al. Reconstructing household transmission dynamics to estimate the infectiousness of asymptomatic influenza virus infections. Proc Natl Acad Sci U S A. (2023) 120:e2304750120. doi: 10.1073/pnas.2304750120
10. Bostanci A, Gazi U, Tosun O, Suer K, Unal Evren E, Evren H, et al. Long-COVID-19 in asymptomatic, non-hospitalized, and hospitalized populations: A cross-sectional study. J Clin Med. (2023) 12. doi: 10.3390/jcm12072613
11. Wherry EJ, Barouch DH. T cell immunity to COVID-19 vaccines. Science. (2022) 377:821–2. doi: 10.1126/science.add2897
12. Perez-Potti A, Lange J, Buggert M. Deciphering the ins and outs of SARS-CoV-2-specific T cells. Nat Immunol. (2021) 22:8–9. doi: 10.1038/s41590-020-00838-5
13. Peng Y, Mentzer AJ, Liu G, Yao X, Yin Z, Dong D, et al. Broad and strong memory CD4(+) and CD8(+) T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat Immunol. (2020) 21:1336–45. doi: 10.1038/s41590-020-0782-6
14. Schub D, Klemis V, Schneitler S, Mihm J, Lepper PM, Wilkens H, et al. High levels of SARS-CoV-2-specific T cells with restricted functionality in severe courses of COVID-19. JCI Insight. (2020) 5:e142167. doi: 10.1172/jci.insight.142167
15. Sette A, Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell. (2021) 184:861–80. doi: 10.1016/j.cell.2021.01.007
16. Grifoni A, Sidney J, Vita R, Peters B, Crotty S, Weiskopf D, et al. SARS-CoV-2 human T cell epitopes: Adaptive immune response against COVID-19. Cell Host Microbe. (2021) 29:1076–92. doi: 10.1016/j.chom.2021.05.010
17. Rydyznski Moderbacher C, Ramirez SI, Dan JM, Grifoni A, Hastie KM, Weiskopf D, et al. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell. (2020) 183:996–1012.e19. doi: 10.1016/j.cell.2020.09.038
18. Tan AT, Linster M, Tan CW, Le Bert N, Chia WN, Kunasegaran K, et al. Early induction of functional SARS-CoV-2-specific T cells associates with rapid viral clearance and mild disease in COVID-19 patients. Cell Rep. (2021) 34:108728. doi: 10.1016/j.celrep.2021.108728
19. Tarke A, Zhang Y, Methot N, Narowski TM, Phillips E, Mallal S, et al. Targets and cross-reactivity of human T cell recognition of common cold coronaviruses. Cell Rep Med. (2023) 4:101088. doi: 10.1016/j.xcrm.2023.101088
20. Brand I, Gilberg L, Bruger J, Gari M, Wieser A, Eser TM, et al. Broad T cell targeting of structural proteins after SARS-CoV-2 infection: high throughput assessment of T cell reactivity using an automated interferon gamma release assay. Front Immunol. (2021) 12:688436. doi: 10.3389/fimmu.2021.688436
21. Nelde A, Bilich T, Heitmann JS, Maringer Y, Salih HR, Roerden M, et al. SARS-CoV-2-derived peptides define heterologous and COVID-19-induced T cell recognition. Nat Immunol. (2021) 22:74–85. doi: 10.1038/s41590-020-00808-x
22. Bange EM, Han NA, Wileyto P, Kim JY, Gouma S, Robinson J, et al. CD8(+) T cells contribute to survival in patients with COVID-19 and hematologic cancer. Nat Med. (2021) 27:1280–9. doi: 10.1038/s41591-021-01386-7
23. Munoz-Fontela C, Dowling WE, Funnell SGP, Gsell PS, Riveros-Balta AX, Albrecht RA, et al. Animal models for COVID-19. Nature. (2020) 586:509–15. doi: 10.1038/s41586-020-2787-6
24. Li Z, Wang Z, Dinh PC, Zhu D, Popowski KD, Lutz H, et al. Cell-mimicking nanodecoys neutralize SARS-CoV-2 and mitigate lung injury in a non-human primate model of COVID-19. Nat Nanotechnol. (2021) 16:942–51. doi: 10.1038/s41565-021-00923-2
25. Morita R, Kubota-Koketsu R, Lu X, Sasaki T, Nakayama EE, Liu YC, et al. COVID-19 relapse associated with SARS-CoV-2 evasion from CD4(+) T-cell recognition in an agammaglobulinemia patient. iScience. (2023) 26:106685. doi: 10.1016/j.isci.2023.106685
26. Speletas M, Raftopoulou S, Farmaki E, Gatselis N, Germanidis G, Mouchtouri VA, et al. and COVID-19: lessons from patients with agammaglobulinemia and the study of functional B-cell polymorphisms. J Investig Allergol Clin Immunol. (2021) 32:53–5. doi: 10.18176/jiaci
27. Soresina A, Moratto D, Chiarini M, Paolillo C, Baresi G, Foca E, et al. Two X-linked agammaglobulinemia patients develop pneumonia as COVID-19 manifestation but recover. Pediatr Allergy Immunol. (2020) 31:565–9. doi: 10.1111/pai.13263
28. Quinti I, Lougaris V, Milito C, Cinetto F, Pecoraro A, Mezzaroma I, et al. A possible role for B cells in COVID-19? Lesson from patients with agammaglobulinemia. J Allergy Clin Immunol. (2020) 146:211–13.e4. doi: 10.1016/j.jaci.2020.04.013
29. Sabatino JJ Jr., Mittl K, Rowles WM, McPolin K, Rajan JV, Laurie MT, et al. Multiple sclerosis therapies differentially affect SARS-CoV-2 vaccine-induced antibody and T cell immunity and function. JCI Insight. (2022) 7. doi: 10.1172/jci.insight.156978
30. Gao Y, Cai C, Wullimann D, Niessl J, Rivera-Ballesteros O, Chen P, et al. Immunodeficiency syndromes differentially impact the functional profile of SARS-CoV-2-specific T cells elicited by mRNA vaccination. Immunity. (2022) 55:1732–46.e5. doi: 10.1016/j.immuni.2022.07.005
31. Beretta A, Cranage M, Zipeto D. Is cross-reactive immunity triggering COVID-19 immunopathogenesis? Front Immunol. (2020) 11:567710. doi: 10.3389/fimmu.2020.567710
32. Saini SK, Hersby DS, Tamhane T, Povlsen HR, Amaya Hernandez SP, Nielsen M, et al. SARS-CoV-2 genome-wide T cell epitope mapping reveals immunodominance and substantial CD8(+) T cell activation in COVID-19 patients. Sci Immunol. (2021) 6. doi: 10.1126/sciimmunol.abf7550
33. Thieme CJ, Anft M, Paniskaki K, Blazquez-Navarro A, Doevelaar A, Seibert FS, et al. Robust T cell response toward spike, membrane, and nucleocapsid SARS-CoV-2 proteins is not associated with recovery in critical COVID-19 patients. Cell Rep Med. (2020) 1:100092. doi: 10.1016/j.xcrm.2020.100092
34. Huang P, Zuo Q, Li Y, Oduro PK, Tan F, Wang Y, et al. A vicious cycle: in severe and critically ill COVID-19 patients. Front Immunol. (2022) 13:930673. doi: 10.3389/fimmu.2022.930673
35. Tizazu AM, Mengist HM, Demeke G. Aging, inflammaging and immunosenescence as risk factors of severe COVID-19. Immun Ageing. (2022) 19:53. doi: 10.1186/s12979-022-00309-5
36. Wratil PR, Schmacke NA, Karakoc B, Dulovic A, Junker D, Becker M, et al. Evidence for increased SARS-CoV-2 susceptibility and COVID-19 severity related to pre-existing immunity to seasonal coronaviruses. Cell Rep. (2021) 37:110169. doi: 10.1016/j.celrep.2021.110169
37. Griggs EP, Mitchell PK, Lazariu V, Gaglani M, McEvoy C, Klein NP, et al. Clinical epidemiology and risk factors for critical outcomes among vaccinated and unvaccinated adults hospitalized with COVID-19-VISION Network, 10 States, June 2021-March 2023. Clin Infect Dis. (2023). doi: 10.1093/cid/ciad505
38. Johnson AG, Linde L, Payne AB, Ali AR, Aden V, Armstrong B, et al. Notes from the Field: Comparison of COVID-19 Mortality Rates Among Adults Aged ≥65 Years Who Were Unvaccinated and Those Who Received a Bivalent Booster Dose Within the Preceding 6 Months - 20 U.S. Jurisdictions, September 18, 2022-April 1, 2023. MMWR Morb Mortal Wkly Rep. (2023) 72(24):667–9. doi: 10.15585/mmwr.mm7224a6
39. Meng L, Masters NB, Lu PJ, Singleton JA, Kriss JL, Zhou T, et al. Cluster analysis of adults unvaccinated for COVID-19 based on behavioral and social factors, National Immunization Survey-Adult COVID Module, United States. Prev Med. (2023) 167:107415. doi: 10.1016/j.ypmed.2022.107415
40. Grifoni A, Weiskopf D, Ramirez SI, Mateus J, Dan JM, Moderbacher CR, et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. (2020) 181:1489–501.e15. doi: 10.1016/j.cell.2020.05.015
41. Tan CCS, Owen CJ, Tham CYL, Bertoletti A, van Dorp L, Balloux F. Pre-existing T cell-mediated cross-reactivity to SARS-CoV-2 cannot solely be explained by prior exposure to endemic human coronaviruses. Infect Genet Evol. (2021) 95:105075. doi: 10.1016/j.meegid.2021.105075
42. Sattler A, Angermair S, Stockmann H, Heim KM, Khadzhynov D, Treskatsch S, et al. SARS-CoV-2-specific T cell responses and correlations with COVID-19 patient predisposition. J Clin Invest. (2020) 130:6477–89. doi: 10.1172/JCI140965
43. Sekine T, Perez-Potti A, Rivera-Ballesteros O, Stralin K, Gorin JB, Olsson A, et al. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell. (2020) 183:158–68.e14. doi: 10.1016/j.cell.2020.08.017
44. Mateus J, Grifoni A, Tarke A, Sidney J, Ramirez SI, Dan JM, et al. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science. (2020) 370:89–94. doi: 10.1126/science.abd3871
45. Grifoni E, Vannucchi V, Valoriani A, Cei F, Lamanna R, Gelli AMG, et al. Interleukin-6 added to CALL score better predicts the prognosis of COVID-19 patients. Intern Med J. (2021) 51:146–7. doi: 10.1111/imj.14974
46. Diniz MO, Mitsi E, Swadling L, Rylance J, Johnson M, Goldblatt D, et al. Airway-resident T cells from unexposed individuals cross-recognize SARS-CoV-2. Nat Immunol. (2022) 23:1324–9. doi: 10.1038/s41590-022-01292-1
47. Le Bert N, Tan AT, Kunasegaran K, Tham CYL, Hafezi M, Chia A, et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. (2020) 584:457–62. doi: 10.1038/s41586-020-2550-z
48. Bacher P, Rosati E, Esser D, Martini GR, Saggau C, Schiminsky E, et al. Low-avidity CD4(+) T cell responses to SARS-CoV-2 in unexposed individuals and humans with severe COVID-19. Immunity. (2020) 53:1258–71.e5. doi: 10.1016/j.immuni.2020.11.016
49. Yu ED, Narowski TM, Wang E, Garrigan E, Mateus J, Frazier A, et al. Immunological memory to common cold coronaviruses assessed longitudinally over a three-year period pre-COVID19 pandemic. Cell Host Microbe. (2022) 30:1269–78.e4. doi: 10.1016/j.chom.2022.07.012
50. An P, Song P, Wang Y, Liu B. Asymptomatic patients with novel coronavirus disease (COVID-19). Balkan Med J. (2020). doi: 10.4274/balkanmedj
51. Olerup O, Zetterquist H. HLA-DRB1*01 subtyping by allele-specific PCR amplification: a sensitive, specific and rapid technique. Tissue Antigens. (1991) 37:197–204. doi: 10.1111/j.1399-0039.1991.tb01872.x
52. Gatz SA, Pohla H, Schendel DJ. A PCR-SSP method to specifically select HLA-A*0201 individuals for immunotherapeutic studies. Tissue Antigens. (2000) 55:532–47. doi: 10.1034/j.1399-0039.2000.550604.x
53. Herr W, Ranieri E, Gambotto A, Kierstead LS, Amoscato AA, Gesualdo L, et al. Identification of naturally processed and HLA-presented Epstein-Barr virus peptides recognized by CD4(+) or CD8(+) T lymphocytes from human blood. Proc Natl Acad Sci U.S.A. (1999) 96:12033–8. doi: 10.1073/pnas.96.21.12033
54. Dolton G, Tungatt K, Lloyd A, Bianchi V, Theaker SM, Trimby A, et al. More tricks with tetramers: a practical guide to staining T cells with peptide-MHC multimers. Immunology. (2015) 146:11–22. doi: 10.1111/imm.12499
55. Kuypers J, Martin ET, Heugel J, Wright N, Morrow R, Englund JA. Clinical disease in children associated with newly described coronavirus subtypes. Pediatrics. (2007) 119:e70–6. doi: 10.1542/peds.2006-1406
56. Gunson RN, Collins TC, Carman WF. Real-time RT-PCR detection of 12 respiratory viral infections in four triplex reactions. J Clin Virol. (2005) 33:341–4. doi: 10.1016/j.jcv.2004.11.025
57. Cui L-J, Zhang C, Zhang T, Lu R-J, Xie Z-D, Zhang L-L, et al. Human coronaviruses HCoV-NL63 and HCoV-HKU1 in hospitalized children with acute respiratory infections in Beijing, China. Adv Virol. (2011) 2011:129134–4. doi: 10.1155/2011/129134
58. Frankild S, De Boer RJ, Lund O, Nielsen M, Kesmir C. Amino acid similarity accounts for T cell cross-reactivity and for “holes” in the T cell repertoire. PLoS One. (2008) 3:e1831. doi: 10.1371/journal.pone.0001831
59. Reche PA. Potential cross-reactive immunity to SARS-CoV-2 from common human pathogens and vaccines. Front Immunol. (2020) 11:586984. doi: 10.3389/fimmu.2020.586984
60. Greenbaum U, Klein K, Martinez F, Song J, Thall PF, Ramdial JL, et al. High levels of common cold coronavirus antibodies in convalescent plasma are associated with improved survival in COVID-19 patients. Front Immunol. (2021) 12:675679. doi: 10.3389/fimmu.2021.675679
61. Abbasi J. COVID-19 and the common cold-preexisting coronavirus antibodies may hinder SARS-CoV-2 immunity. JAMA. (2022) 327:609–10. doi: 10.1001/jama.2022.0326
62. Pedersen J, Koumakpayi IH, Babuadze G, Baz M, Ndiaye O, Faye O, et al. Cross-reactive immunity against SARS-CoV-2 N protein in Central and West Africa precedes the COVID-19 pandemic. Sci Rep. (2022) 12:12962. doi: 10.1038/s41598-022-17241-9
63. Kundu R, Narean JS, Wang L, Fenn J, Pillay T, Fernandez ND, et al. Cross-reactive memory T cells associate with protection against SARS-CoV-2 infection in COVID-19 contacts. Nat Commun. (2022) 13:80. doi: 10.1038/s41467-021-27674-x
64. Jaago M, Rahni A, Pupina N, Pihlak A, Sadam H, Tuvikene J, et al. Differential patterns of cross-reactive antibody response against SARS-CoV-2 spike protein detected for chronically ill and healthy COVID-19 naive individuals. Sci Rep. (2022) 12:16817. doi: 10.1038/s41598-022-20849-6
65. Yin D, Han Z, Lang B, Li Y, Mai G, Chen H, et al. Effect of seasonal coronavirus immune imprinting on the immunogenicity of inactivated COVID-19 vaccination. Front Immunol. (2023) 14:1195533. doi: 10.3389/fimmu.2023.1195533
66. Lundgren A, Leach S, Axelsson H, Isakson P, Nystrom K, Scharf L, et al. Plasmablasts in previously immunologically naive COVID-19 patients express markers indicating mucosal homing and secrete antibodies cross-reacting with SARS-CoV-2 variants and other beta-coronaviruses. Clin Exp Immunol. (2023) 213:173–89. doi: 10.1093/cei/uxad044
67. Santos MMS, Pereira IJ, Cuboia N, Reis-Pardal J, Adriao D, Cardoso T, et al. Predictors of early and long-term mortality after ICU discharge in critically ill COVID-19 patients: A prospective cohort study. PLoS One. (2023) 18:e0293883. doi: 10.1371/journal.pone.0293883
68. Wang S, Zhu R, Zhang C, Guo Y, Lv M, Zhang C, et al. Effects of the pre-existing coronary heart disease on the prognosis of COVID-19 patients: A systematic review and meta-analysis. PLoS One. (2023) 18:e0292021. doi: 10.1371/journal.pone.0292021
69. Silva RCD, de Lima SC, Dos Santos Reis WPM, de Magalhaes JJF, Magalhaes RNO, Rathi B, et al. Comparison of DNA extraction methods for COVID-19 host genetics studies. PLoS One. (2023) 18:e0287551. doi: 10.1371/journal.pone.0287551
70. Kared H, Redd AD, Bloch EM, Bonny TS, Sumatoh H, Kairi F, et al. SARS-CoV-2-specific CD8+ T cell responses in convalescent COVID-19 individuals. J Clin Invest. (2021) 131. doi: 10.1172/JCI145476
71. Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, et al. Safety and efficacy of the BNT162b2 mRNA COVID-19 vaccine. New Engl J Med. (2020) 383:2603–15. doi: 10.1056/NEJMoa2034577
72. Baden LR, El Sahly HM, Essink B, Kotloff K, Frey S, Novak R, et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. New Engl J Med. (2021) 384:403–16. doi: 10.1056/NEJMoa2035389
73. Namuniina A, Muyanja ES, Biribawa VM, Okech BA, Ssemaganda A, Price MA, et al. Proportion of Ugandans with pre-pandemic SARS-CoV-2 cross-reactive CD4+ and CD8+ T-cell responses: A pilot study. PloS Glob Public Health. (2023) 3:e0001566. doi: 10.1371/journal.pgph.0001566
74. Pothast CR, Dijkland RC, Thaler M, Hagedoorn RS, Kester MGD, Wouters AK, et al. SARS-CoV-2-specific CD4(+) and CD8(+) T cell responses can originate from cross-reactive CMV-specific T cells. Elife 11. (2022). doi: 10.7554/eLife.82050
75. Schmidt KG, Nganou-Makamdop K, Tenbusch M, El Kenz B, Maier C, Lapuente D, et al. SARS-CoV-2-seronegative subjects target CTL epitopes in the SARS-CoV-2 nucleoprotein cross-reactive to common cold coronaviruses. Front Immunol. (2021) 12:627568. doi: 10.3389/fimmu.2021.627568
76. Steiner S, Sotzny F, Bauer S, Na IK, Schmueck-Henneresse M, Corman VM, et al. HCoV- and SARS-CoV-2 cross-reactive T cells in CVID patients. Front Immunol. (2020) 11:607918. doi: 10.3389/fimmu.2020.607918
77. Ogbe A, Kronsteiner B, Skelly DT, Pace M, Brown A, Adland E, et al. T cell assays differentiate clinical and subclinical SARS-CoV-2 infections from cross-reactive antiviral responses. Nat Commun. (2021) 12:2055. doi: 10.1101/2020.09.28.20202929
78. Murray SM, Ansari AM, Frater J, Klenerman P, Dunachie S, Barnes E, et al. The impact of pre-existing cross-reactive immunity on SARS-CoV-2 infection and vaccine responses. Nat Rev Immunol. (2023) 23:304–16. doi: 10.1038/s41577-022-00809-x
79. Becerra-Artiles A, Nanaware PP, Muneeruddin K, Weaver GC, Shaffer SA, Calvo-Calle JM, et al. Immunopeptidome profiling of human coronavirus OC43-infected cells identifies CD4 T-cell epitopes specific to seasonal coronaviruses or cross-reactive with SARS-CoV-2. PLoS Pathog. (2023) 19:e1011032. doi: 10.1371/journal.ppat.1011032
80. Klompus S, Leviatan S, Vogl T, Mazor RD, Kalka IN, Stoler-Barak L, et al. Cross-reactive antibodies against human coronaviruses and the animal coronavirome suggest diagnostics for future zoonotic spillovers. Sci Immunol 6. (2021). doi: 10.1126/sciimmunol.abe9950
81. Johansson AM, Malhotra U, Kim YG, Gomez R, Krist MP, Wald A, et al. Cross-reactive and mono-reactive SARS-CoV-2 CD4+ T cells in prepandemic and COVID-19 convalescent individuals. PLoS Pathog. (2021) 17:e1010203. doi: 10.1371/journal.ppat.1010203
82. Lipsitch M, Grad YH, Sette A, Crotty S. Cross-reactive memory T cells and herd immunity to SARS-CoV-2. Nat Rev Immunol. (2020) 20:709–13. doi: 10.1038/s41577-020-00460-4
83. Silva BSA, Pereira T, Minuzzi LG, Padilha CS, Figueiredo C, Olean-Oliveira T, et al. Mild to moderate post-COVID-19 alters markers of lymphocyte activation, exhaustion, and immunometabolic responses that can be partially associated by physical activity level- an observational sub-analysis fit- COVID study. Front Immunol. (2023) 14:1212745. doi: 10.3389/fimmu.2023.1212745
84. Mazzoni A, Salvati L, Maggi L, Annunziato F, Cosmi L. Hallmarks of immune response in COVID-19: Exploring dysregulation and exhaustion. Semin Immunol. (2021) 55:101508. doi: 10.1016/j.smim.2021.101508
85. Zhang JY, Wang XM, Xing X, Xu Z, Zhang C, Song JW, et al. Single-cell landscape of immunological responses in patients with COVID-19. Nat Immunol. (2020) 21:1107–18. doi: 10.1038/s41590-020-0762-x
86. Adamo S, Chevrier S, Cervia C, Zurbuchen Y, Raeber ME, Yang L, et al. Profound dysregulation of T cell homeostasis and function in patients with severe COVID-19. Allergy. (2021) 76:2866–81. doi: 10.1111/all.14866
87. Wang N, Vuerich M, Kalbasi A, Graham JJ, Csizmadia E, Manickas-Hill ZJ, et al. Limited TCR repertoire and ENTPD1 dysregulation mark late-stage COVID-19. iScience. (2021) 24:103205. doi: 10.1016/j.isci.2021.103205
88. Shahbaz S, Xu L, Sligl W, Osman M, Bozorgmehr N, Mashhouri S, et al. The quality of SARS-CoV-2-specific T cell functions differs in patients with mild/moderate versus severe disease, and T cells expressing coinhibitory receptors are highly activated. J Immunol. (2021) 207:1099–111. doi: 10.4049/jimmunol.2100446
89. Files JK, Boppana S, Perez MD, Sarkar S, Lowman KE, Qin K, et al. Sustained cellular immune dysregulation in individuals recovering from SARS-CoV-2 infection. J Clin Invest. (2021) 131. doi: 10.1172/JCI140491
90. Yin K, Peluso MJ, Luo X, Thomas R, Shin MG, Neidleman J, et al. Long COVID manifests with T cell dysregulation, inflammation and an uncoordinated adaptive immune response to SARS-CoV-2. Nat Immunol. (2024) 25:218–25. doi: 10.1038/s41590-023-01724-6
91. Paine SK, Choudhury P, Alam M, Bhattacharyya C, Pramanik S, Tripathi D, et al. Multi-faceted dysregulated immune response for COVID-19 infection explaining clinical heterogeneity. Cytokine. (2024) 174:156434. doi: 10.1016/j.cyto.2023.156434
92. Cezar R, Kundura L, Andre S, Lozano C, Vincent T, Muller L, et al. T4 apoptosis in the acute phase of SARS-CoV-2 infection predicts long COVID. Front Immunol. (2023) 14:1335352. doi: 10.3389/fimmu.2023.1335352
93. Chentoufi AA, Dhanushkodi NR, Srivastava R, Prakash S, Coulon PA, Zayou L, et al. Combinatorial herpes simplex vaccine strategies: from bedside to bench and back. Front Immunol. (2022) 13:849515. doi: 10.3389/fimmu.2022.849515
94. Srivastava R, Roy S, Coulon P, Vahed H, Parkash S, Dhanushkodi N, et al. Therapeutic mucosal vaccination of HSV-2 infected Guinea pigs with the ribonucleotide reductase 2 (RR2) protein boosts antiviral neutralizing antibodies and tissue-resident CD4+ and CD8+ TRM cells associated with protection against recurrent genital herpes. J Virol In Press. (2019). doi: 10.1128/JVI.02309-18
95. Srivastava R, Roy S, Coulon PG, Vahed H, Prakash S, Dhanushkodi N, et al. Therapeutic mucosal vaccination of herpes simplex virus 2-infected guinea pigs with ribonucleotide reductase 2 (rr2) protein boosts antiviral neutralizing antibodies and local tissue-resident cd4+ and cd8+ trm cells associated with protection against recurrent genital herpes. J Virol. (2019) 93(9):e02309-18. doi: 10.1128/JVI.02309-18
96. Roy S, Coulon PG, Srivastava R, Vahed H, Kim GJ, Walia SS, et al. Blockade of LAG-3 immune checkpoint combined with therapeutic vaccination restore the function of tissue-resident anti-viral CD8(+) T cells and protect against recurrent ocular herpes simplex infection and disease. Front Immunol. (2018) 9:2922. doi: 10.3389/fimmu.2018.02922
97. Allen SJ, Hamrah P, Gate D, Mott KR, Mantopoulos D, Zheng L, et al. The role of LAT in increased CD8+ T cell exhaustion in trigeminal ganglia of mice latently infected with herpes simplex virus 1. J Virol. (2011) 85:4184–97. doi: 10.1128/JVI.02290-10
98. Aydillo T, Rombauts A, Stadlbauer D, Aslam S, Abelenda-Alonso G, Escalera A, et al. Immunological imprinting of the antibody response in COVID-19 patients. Nat Commun. (2021) 12:3781. doi: 10.1038/s41467-021-23977-1
99. Bertoletti A, Le Bert N, Qui M, Tan AT. SARS-CoV-2-specific T cells in infection and vaccination. Cell Mol Immunol. (2021) 18:2307–12. doi: 10.1038/s41423-021-00743-3
100. Huang G, Kovalic AJ, Graber CJ. Prognostic value of leukocytosis and lymphopenia for coronavirus disease severity. Emerg Infect Dis. (2020) 26:1839–41. doi: 10.3201/eid2608.201160
101. Penaloza HF, Lee JS, Ray P. Neutrophils and lymphopenia, an unknown axis in severe COVID-19 disease. PLoS Pathog. (2021) 17:e1009850. doi: 10.1371/journal.ppat.1009850
102. Veiga-Parga T, Suryawanshi A, Mulik S, Gimenez F, Sharma S, Sparwasser T, et al. On the role of regulatory T cells during viral-induced inflammatory lesions. J Immunol. (2012) 189:5924–33. doi: 10.4049/jimmunol.1202322
103. Mengist HM, Kombe Kombe AJ, Mekonnen D, Abebaw A, Getachew M, Jin T. Mutations of SARS-CoV-2 spike protein: Implications on immune evasion and vaccine-induced immunity. Semin Immunol. (2021) 55:101533. doi: 10.1016/j.smim.2021.101533
104. Ahmad L. Implication of SARS-CoV-2 immune escape spike variants on secondary and vaccine breakthrough infections. Front Immunol. (2021) 12:742167. doi: 10.3389/fimmu.2021.742167
105. Sharma S, Vercruysse T, Sanchez-Felipe L, Kerstens W, Rasulova M, Bervoets L, et al. Updated vaccine protects against SARS-CoV-2 variants including Omicron (B.1.1.529) and prevents transmission in hamsters. Nat Commun. (2022) 13:6644. doi: 10.1038/s41467-022-34439-7
Keywords: SARS-CoV-2, symptomatic, asymptomatic, COVID-19, common cold coronavirus, CD4 + T cells, CD8 + T cells, exhaustion α-CCCs/SARS-CoV-2 cross-reactive T cells in asymptomatic COVID-19 infection
Citation: Coulon P-G, Prakash S, Dhanushkodi NR, Srivastava R, Zayou L, Tifrea DF, Edwards RA, Figueroa CJ, Schubl SD, Hsieh L, Nesburn AB, Kuppermann BD, Bahraoui E, Vahed H, Gil D, Jones TM, Ulmer JB and BenMohamed L (2024) High frequencies of alpha common cold coronavirus/SARS-CoV-2 cross-reactive functional CD4+ and CD8+ memory T cells are associated with protection from symptomatic and fatal SARS-CoV-2 infections in unvaccinated COVID-19 patients. Front. Immunol. 15:1343716. doi: 10.3389/fimmu.2024.1343716
Received: 24 November 2023; Accepted: 08 March 2024;
Published: 28 March 2024.
Edited by:
Pedro A. Reche, Complutense University of Madrid, SpainReviewed by:
Sarah Elizabeth Jackson, University of Cambridge, United KingdomRicardo Da Silva Antunes, La Jolla Institute for Immunology (LJI), United States
Copyright © 2024 Coulon, Prakash, Dhanushkodi, Srivastava, Zayou, Tifrea, Edwards, Figueroa, Schubl, Hsieh, Nesburn, Kuppermann, Bahraoui, Vahed, Gil, Jones, Ulmer and BenMohamed. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lbachir BenMohamed, TGJlbm1vaGFAdWNpLmVkdQ==
†These authors have contributed equally to this work
 Pierre-Gregoire Coulon
Pierre-Gregoire Coulon Swayam Prakash1†
Swayam Prakash1† Elmostafa Bahraoui
Elmostafa Bahraoui Trevor M. Jones
Trevor M. Jones Lbachir BenMohamed
Lbachir BenMohamed