
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 19 January 2024
Sec. Vaccines and Molecular Therapeutics
Volume 15 - 2024 | https://doi.org/10.3389/fimmu.2024.1339467
Mycobacterium tuberculosis (M.tb) effectively manipulates the host processes to establish the deadly respiratory disease, Tuberculosis (TB). M.tb has developed key mechanisms to disrupt the host cell health to combat immune responses and replicate efficaciously. M.tb antigens such as ESAT-6, 19kDa lipoprotein, Hip1, and Hsp70 destroy the integrity of cell organelles (Mitochondria, Endoplasmic Reticulum, Nucleus, Phagosomes) or delay innate/adaptive cell responses. This is followed by the induction of cellular stress responses in the host. Such cells can either undergo various cell death processes such as apoptosis or necrosis, or mount effective immune responses to clear the invading pathogen. Further, to combat the infection progression, the host secretes extracellular vesicles such as exosomes to initiate immune signaling. The exosomes can contain M.tb as well as host cell-derived peptides that can act as a double-edged sword in the immune signaling event. The host-symbiont microbiota produces various metabolites that are beneficial for maintaining healthy tissue microenvironment. In juxtaposition to the above-mentioned mechanisms, M.tb dysregulates the gut and respiratory microbiome to support its replication and dissemination process. The above-mentioned interconnected host cellular processes of Immunometabolism, Cellular stress, Host Microbiome, and Extracellular vesicles are less explored in the realm of exploration of novel Host-directed therapies for TB. Therefore, this review highlights the intertwined host cellular processes to control M.tb survival and showcases the important factors that can be targeted for designing efficacious therapy.
Mycobacterium tuberculosis (M.tb), the causative bacterium of the deadly disease Tuberculosis (TB) stands as one of humanity’s most enduring pathogens (1). The extensive co-evolutionary history has equipped M.tb with formidable arsenals to counteract host immune responses (2). The interplay between the host and pathogen represents a highly intricate and ever-evolving phenomenon that is pivotal in determining disease progression or the successful clearance of the invading pathogen (3). The dynamic relationship between M.tb and the host immune system operates at multiple levels, encompassing various cellular processes, such as immune signaling, metabolic responses, the management of various cellular stresses (including those affecting the endoplasmic reticulum, mitochondria, and DNA integrity), host-microbiome balance, the release of extracellular components like exosomes and cell-free nucleic acids, and many other regulatory factors (4, 5) (Figure 1). For instance, M.tb deploys multiple factors such as Hip1, ESAT-6, etc, to delay T-cell responses by targeting immune metabolism (6). The immune cells, in turn, maintain high ATP levels to synthesize and secrete pro-inflammatory cytokines to limit M.tb infection (7).
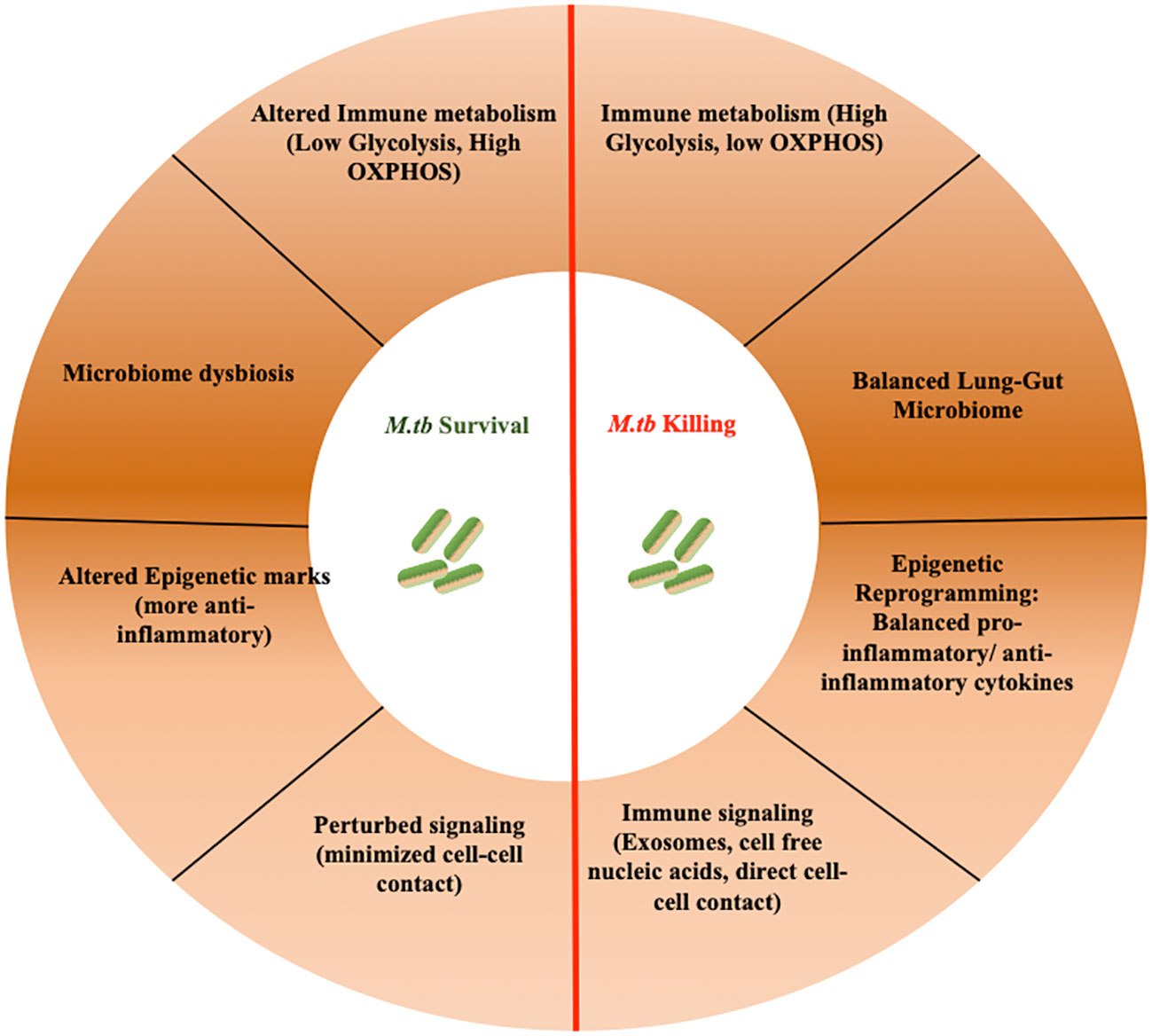
Figure 1 Over-view of various integrated host processes that governs TB progression. TB progression is influenced by an interplay of integrated host processes. These intricate mechanisms govern the pathogenesis of TB, balancing disease clearance and progression.
The source for the functioning of host immune cells depends on several metabolic pathways. There is a reciprocal relationship between the host and M.tb in terms of their metabolism. The presence of the host-microbiome adds complexity to the host immune system functioning (8). The microbiome-derived metabolic intermediates are involved in intricating host immune signaling pathways. Studies using the murine model have illustrated the differential expansion of T cell subsets in the presence of varying gut symbionts. For instance, the Th1 response dominates in mice colonized with Bacteroides fragilis (9) while the Th2 response is predominant in mice colonized with Clostridial strains (10). Antibiotic treatment such as Isoniazid/Pyrazinamide has been observed to alter the gut microbiome in TB murine model. The altered gut microbiome changes the metabolites in peripheral circulation which in turn has been found to alter the metabolism of alveolar macrophages. This results in defective microbicidal activity and enhanced susceptibility of macrophages to M.tb (11, 12).
Apart from impacting the host immune system, the metabolites can also have a direct anti-mycobacterial activity (13). For instance, Indole-3 propionic acid (IPA) generated from tryptophan (trp) metabolism by gut symbiont Clostridium sporogenes was found to showcase anti-mycobacterial properties in a murine model of TB (14). Thus the metabolites derived from the gut symbionts impact M.tb survival/clearance either through direct anti-mycobacterial activity or indirectly through the activation of host immune responses.
The translocation of metabolites and folded/unfolded proteins from infected cells initiates signaling processes for the immune cells. Host and M.tb derived proteins are transported through the exosomes. The contents of exosomes act as a double-edged sword. Exosomes have been found to promote protective Th1mediated immunity during TB in murine models (15). Despite positive immune activation by exosomes, few studies have elucidated the negative immune regulation during TB. In the later stages of macrophage infection by M.tb, a notable decrease in macrophage activation followed by upregulation of apoptotic and autophagic pathways has been studied (16, 17).
Understanding and decoding these complex cellular and microbial events will help us understand TB in a more effective way. It is noteworthy to realize that the host cellular processes mentioned above are intertwined and maintain the symbiotic relationship between the host and M.tb. In the contemporary scientific landscape, researchers are delving extensively into characterizing host-M.tb interactions that will aid in decoding M.tb pathogenesis (1, 18). This endeavour is essential for developing more effective immunotherapies that can enhance protection against active TB (ATB), latent TB (LTBI), and reactivation of the infection.
This review provides a concise overview of the lesser-explored host processes- Immunometabolism, Endoplasmic (ER) stress, Host microbiome, and Extracellular vesicles. These processes are few amongst the countless host processes such as transcriptional and translational reprograming, innate/adaptive cells activation, inflammatory mediators signaling, cellular metabolism, granuloma formation, cell death and survival processes etc, which are involved in the dynamic interaction between the host and M.tb. These crucial interactions offer potential targets for the development of innovative Host Directed Therapies aimed at combating TB and warding off the emergence of antibiotic-resistant strains.
M.tb interacts with numerous biomolecules and different cell types to establish the infection successfully. Upon aerosol inhalation, M.tb interacts with respiratory epithelial cells and persists for nearly 48 hours without replicating (19). The general notion in the early stage of M.tb infection implies that M.tb released from dying, necrotic cells interact higher with alveolar epithelial cells, irrespective of replication (20).These epithelial cells are well known to modulate the local immune microenvironment against M.tb as they secrete pro-inflammatory molecules that drive the myeloid cell activation (19). This is followed by the entry of M.tb through M cells to the underlying lymphoid layer and predominantly infecting the alveolar macrophages through pattern recognition receptors (PRRs). The PRRs recognize various pathogen-associated molecular patterns (PAMPs) present or secreted by M.tb Such molecules are mycolic acids, lipoarabinomannan (LAM), and secreted proteins such as ESAT-6, facilitating the activation of the MyD88 signaling pathway. This, in turn, induces an inflammatory response to control M.tb infection. Some studies reported that M.tb induces Irg1 expression via MyD88 and STING pathway, eventually controlling M.tb infection (21). It has been demonstrated that M.tb lipids such as phthiocerol dimycocerosate (PDIM) facilitate the fusion of M.tb with the membrane of macrophages and ensure successful entrance to the cell through the endocytic pathway (22). Within these macrophages, M.tb may either be degraded by lysosomal acidification or autophagy, or it may surpass these barriers to infect other immune cells such as dendritic cells, neutrophils, or myeloid cells (23). These infected tissue-resident alveolar macrophages initiate signaling cascades which is essential to synthesize anti-microbial peptides, pro-inflammatory cytokines, and prostaglandins to effectively clear M.tb. Macrophage pro-inflammatory phenotype is obtained by shifting predominantly to glycolysis, initiating transcriptional programs that supports acquisition of an anti-bacterial state (24). M.tb has adopted various mechanisms to subvert the lysosomal degradation within the macrophages. The cell wall organization of M.tb, especially mannose-capped lipoarabinomannan (manLAM) sanctions the survival within the phagosomes of the macrophages by preventing phosphorylation of phosphatidylinositol-3-phosphate (PI3P) and further prevents maturation of early phagosomes to late phagosomes (25, 26). Nonetheless, M.tb utilizes its type VII secretion system and secretes ESAT-6 to exit out of these macrophages and infect neutrophils and dendritic cells in the lung interstitium (27).
Recent pieces of evidence show that TB patients have a higher number of neutrophils which is associated with the failure of TB regimen, i.e., anti-tubercular therapy, and show higher lung pathology (28). Within neutrophils, M.tb replicates and is subjected to degradation by an arsenal of proteolytic enzymes such as elastase, proteinase 3, cathepsin G, and Reactive oxygen species (ROS) (29). SLAMF1 is one of the co-stimulatory proteins that governs autophagy in neutrophils, and projects M.tb degradation. However, SLAMF1 levels are downregulated in TB patients when compared to healthy controls, and thus unable to control M.tb progression (28). Neutrophils govern the adaptive immune response by directing dendritic cells (DCs) to present antigens to T cells (30). There are two classes of thoughts for the role of neutrophils in TB (31). Firstly, neutrophils are known to control the early stage of infection during pulmonary TB (32), whereas another study suggests that neutrophils are permissive to M.tb and help in disease progression during the granulomatous stage of infection, and cause tissue destruction (33). In later stages of infection, neutrophils expedite the growth of the bacteria and facilitate the dissemination of M.tb (33). It has been shown that neutrophils interact with T cells and macrophages within the granuloma and secrete immunosuppressive cytokines that downregulates T cell responses (34). The detailed role of neutrophils during different stages of M.tb infection is yet to be elucidated.
It is conventionally known that innate immune cells provide the first line of defense against M.tb and commands adaptive immune cells to contain the bacteria within the hypoxic granulomas. This bridging is primarily done by antigen-presenting cells, mainly dendritic cells that are least phagocytic in nature. They phagocytose M.tb and degrade it to derive M.tb antigens. The infected DCs then migrate to the distal lymph nodes in response to CCL19/21 gradients to induce effective cell-mediated immunity (35). Once, enough antigens are generated, DCs downregulate their phagocytic activity and divert their energy status in antigen processing and presentation to T cells. However, accumulating shreds of evidence suggest that M.tb has employed various mechanisms to delay the ability of DCs to present antigens to T cells (26). Certain M.tb antigens such as serine hydrolase Hip1, ESAT-6, and hsp70 impair the maturation and migration of DCs and thereby prevent T cell proliferation (36–38). Another M.tb antigen 19kDa lipoprotein is known to downregulate the expression of MHC class II and HLA-DM on DCs, thereby delaying the T cell priming (39, 40). This poses barrier in restricting the infection, and hence M.tb disseminates to other organs such as the central nervous system, kidney, liver, pancreas, etc (41). As DCs present M.tb antigens to naïve CD4+ T cells via MHC II, the CD4+ T cells are activated and subsequently differentiated into Th1 type to secrete IFN-γ, TNF-α, and IL-2. These activated CD4+ T cells are crucial in containing the M.tb within granuloma, and further activating CD8+ T cells to enable its cytotoxic effector functions. CD4+ T cells act synergistically with CD8+ T cells to prevent the growth of M.tb (42). However, chronic exposure to M.tb antigens leads to T cell exhaustion in which T cells are functionally impaired. Such T cells express TIM-3 along with other co-inhibitory molecules such as PD-1, LAG-3, CTLA-4 which results in lower production of IL-2, IFN-γ, and other pro-inflammatory cytokines (43). It is also reported that CD4+ T cells are necessary to prevent CD8+ T cell exhaustion (42). Conversely, a recent report suggests that patients with Tuberculous Pleural Effusion (TPE) have higher numbers of inflammatory granzyme-secreting CD8+ T cells in the pleural space, possibly involved in disease progression (44). Single-cell RNA sequencing and TCR sequencing have revealed different subsets of T cells in different compartments of infected lungs in TPE patients (44). This is necessary to realize that the migration of activated T cells to the site of infection is primarily based on the expression of chemokines and their cognate receptors. CCL5, CCL19, and CCL21 are the only known chemokines that facilitate the migration of T cells from distal lymph nodes to the infected lungs (45, 46).
The collective efforts and responses of immune cells to defeat the so-called ‘Trojan horse’ are indispensable. Contrarywise, M.tb has evolved its arsenal of immune-escaping and immune-suppressive mechanisms to conquer the complex processes of the host (17). It is important to highlight that both host and M.tb are genetically complex and thus harbor various other pathways of interactions, which are yet to be investigated.
Various host processes collectively regulate disease pathogenesis, which are conquered by M.tb. Such processes involve DNA replication, transcriptional reprogramming, translation, cell death pathways, cell survival, metabolism, cell transport process, stress events such as ER stress, mitochondrion stress, etc. These processes integrate to control M.tb infection, and prevent tissue pathology. In this review, we aim to focus on Immunometabolism, ER stress, Microbiome and Exosome mediated immune signaling that modulates the host-M.tb interaction dynamics.
The process of mounting an antimicrobial response and inducing effector functions is a thermodynamically and energetically expensive process in immune cells (47). The cells employ bioenergetics to meet the energy demand according to their status. These needs are met by metabolic plasticity that serves to be crucial for macrophages and T cells to synthesize antimicrobial molecules and prompt effector functions (48). Conventionally, the integrated processes involving glycolysis, tricarboxylic acid cycle (TCA), oxidative phosphorylation (OXPHOS), and fatty acid metabolism supply the energy demands universally in all cell types at their quiescent stage (49, 50). However, metabolic plasticity is central in immune cells as these cells execute highly energy-intensive functions (51). This includes increased synthesis of antimicrobial peptides, pro-inflammatory and anti-inflammatory cytokines, chemokines and their cognate receptors, antigen processing and presentation, synthesis of immunoglobulins and exhibit cytolytic functions (52). It is crucial to highlight that immunometabolism is influenced by the stage of M.tb infection such as active state, re-infection and latent TB (granuloma) (53, 54). In this section, we highlighted the alterations between metabolic processes and host immune cells upon different stages of M.tb infection.
During active state, upon M.tb phagocytosis through toll-like receptors (TLRs), especially TLR2/4, the alveolar macrophages become activated in MyD88 dependent manner which leads to acquiring a pro-inflammatory ‘M1’ phenotype (55). ‘M1’ macrophages require a massive number of ATPs to synthesize pro-inflammatory cytokines such as IL-1β and antimicrobial peptides, therefore these macrophages reprogram the metabolic processes and shift to glycolysis to meet their energy requirements (56). Laura et al., have shown that M.tb induces a metabolic shift towards aerobic glycolysis and perturbs OXPHOS in alveolar macrophages (57). This results in the accumulation of TCA cycle products such as succinate that induces the expression of hypoxia-inducible factor 1α (HIF-1α) (53). HIF-1α is the master regulator of the glycolysis pathway that regulates the expression of glycolytic enzymes which further acts in a positive feedback manner to increase the glycolysis (58). Metabolic plasticity in macrophages has been extensively reviewed elsewhere (53, 59). It has been broadly studied that M.tb induces ‘Warburg effect’ by increasing the expression of glucose receptors that ultimately leads to enhanced uptake of glucose within the infected macrophages (60). This is accompanied by the increase in end- product of glycolysis, i.e., lactate secretion in extracellular spaces. It has been demonstrated that lactate secreted from the infected cells drive the negative regulation of glycolysis in the resting macrophages (61). The decrease in glycolysis is balanced by the increase in OXPHOS, thereby lactate act as immunomodulator by inhibiting the production of IL-1β and TNF. However, it is interesting to note that lactate rewires the metabolic responses in resting macrophages so that subsequent infection with M.tb is diminished (61). Conversely, a recent study suggests that enhanced glucose uptake or during Diabetes Mellitus in TB patients (TB-DM), the increased glucose levels negatively regulate HIF-1α, and thus mediates the survival of M.tb in the infected macrophages (62). Another study demonstrated that alveolar macrophages offer nutrient rich and permissive environment for M.tb and thus enables higher bacterial replication when compared with interstitial macrophages Further sequencing analysis revealed that alveolar macrophages have higher fatty acid oxidation that allows release of fatty acids and cholesterol available for uptake by M.tb (63). Furthermore, M.tb maintains low replicating stage, and sustain in extracellular compartment within the granuloma. This is likely due to low amounts for fatty acids and cholesterol, which are necessary for its replication (64).
Nonetheless, M.tb has evolved key mechanisms to drive the transition from M1 to M2 phenotype by ravaging mitochondrial integrity. M.tb secretes ESAT-6 that damages mitochondrion upon mTOR inhibition, destroys mitochondrial membrane and disrupts the membrane potential (65). This in turn interrupts the contacts between mitochondrion and phagolysosome, which is thought to be necessary for M.tb killing (10.1016/j.micinf.2015.06.003). Another defensive strategy employed by M.tb is delayed apoptosis and enhanced upregulation of OXPHOS in M.tb infected macrophages. Rv1813c is an effector M.tb protein that when overexpressed in macrophages, is deposited in mitochondrial matrix and prevents the release of cytochrome c (6). However, M.tb secretes various potent antigenic proteins that collectively promotes M.tb survival, but their collective role in altering macrophage metabolism is yet to be studied. Also, this has been known for ages that during latent TB, M.tb secretes ESAT-6 in granuloma during latent TB, and simultaneously, macrophages exhibit anti-inflammatory properties (66). However, least efforts have been made to visualize what happens at metabolic level that do not allow the macrophage cell death, even with loss of mitochondrial integrity. Metabolomic analysis of M.tb infected macrophages revealed higher levels of NAD+, glutathione and creatinine, and it has been reported that these metabolites collectively regulate anti-mycobacterial activity of macrophages as compared to uninfected macrophages (56). Additionally, M.tb upregulates glutathione metabolism in macrophages resulting in higher levels of glutamine, that supports M2 polarization (67). Recent studies have classified ratios of amino acids in plasma to whole blood as biomarkers with 84% sensitivity in detecting active TB patients. Amino acids such as leucine, asparagine, ornithine, arginine, tyrosine and serine are present in higher ratios in TB patients when compared to cured TB patients and healthy controls (68). These are probably secreted in the blood and are taken up by macrophages for Nitric oxide (NO) production. Conversely, one of the interesting studies suggested that administration of L-tyrosine curbs mycobacterial survival in TB granulomas, possible due to increased production of ROS (69). However, amino acid metabolism in host macrophages is yet to be elucidated with respect to TB progression.
Innate immune cells such as macrophages and DCs bridge the antigen encounter with naïve adaptive immune cells through antigen presentation. M.tb infected M1 macrophages secrete certain cytokines such as IL-6, IL-10, and IL-12 in the local environment where naïve T cells sense these cytokines, and enhance their glycolysis to prompt effector functions (70). Together with this, T cells need to proliferate and expand clonally which necessitates the rapid synthesis of proteins, lipids and DNA. These phenotypes are acquired by rapid nutrient uptake such as glucose, and other metabolites, and calcium mobilization. In accordance to T cell effector functions, ketolysis is an alternative to glycolytic pathway that drives CD8+ T cell cytolytic and cytokines functions such as IFN-γ and TNF-α (71). It is relevant to investigate ketolysis in TB progression and to delineate the possible protective mechanisms against TB.
In general, TCR interaction with co-stimulatory molecules such as CD80/86 induces mitochondrial biogenesis, that also enhances the upregulation of AMP kinase, and further supplements ATPs for rapid T cell proliferation and effector functions (72). A remarkable study demonstrated that mitochondrial matrix protein, Cyclophilin D (CypD) controls M.tb infection by controlling T cell responses, and maintains disease tolerance. In CypD deficient M.tb infected mice, there is higher infiltration of T cells in lungs, which leads to lung pathology. The study has established that CypD tightly controls aerobic glycolysis in T cells in ROS dependent manner that is known for inducing effector functions such as production of IFN-γ, TNF-α and IL2 (73). As discussed by Koeken et al., T cells and other myeloid cells are evolving in conjunction with M.tb to promote disease tolerance that eventually leads to lower immunopathology (74).
Nonetheless, chronic exposure of M.tb results in T cell dysfunction, primarily, CD8+ T cells. M.tb exposure stems metabolic reprogramming in T cells which subsequently ensues the expression of exhaustion markers such as PD-1, CTLA-4 (75). Through transcriptomic and experimental analysis, these observations are finely correlated with reduced glucose uptake by M.tb specific CD8+ T cells. The series of bioenergetics further demonstrated that chronic exposure of CD8+ T cells dampens mitochondrial health, declining OXPHOS, and reduced production of cytokines, which marks T cell exhaustion (75). However, CD4+ T cells are known to prevent CD8+ T cell exhaustion during M.tb infection by an unknown mechanism to produce more IFN-γ, TNF-α, granzymes and perforins (42). Based on these evidences, this could be speculated that CD4+ T cells modulate CD8+ T cells by preventing mitochondrial dysfunction, and more pronounced aerobic glycolysis. Nonetheless, these aforementioned speculations need experimental validations and analysis. Conventional to the metabolism in all immune cells, HIF1α is known in CD4+ T cells to upregulate glycolysis to produce ATP rapidly. This is paramount for release of cytokines such as IFN-γ that drives macrophage activation. Nevertheless, using conditional knockout mechanisms, Liu et al., have shown that deletion of negative regulator of HIF-1α, VHL, dampens the host’s ability to control M.tb infection, and hence lower accumulation of M.tb specific T cells in the lungs of these mice. This is explained by the fact that VHL is vital for stabilizing the appropriate extent of HIF1α in controlling CD4+ T cell response against M.tb. This further accompanies to prevent CD4+ T cell exhaustion during M.tb infection. Altogether, VHL is required for T cell activation, DNA replication and proliferation (76).
The above- mentioned reports are well described in the context of effector functions of T cells. Conversely, minimal efforts are put into identifying the metabolic reprogramming in stemming the memory responses of T cells, which are crucial for the basis of any therapeutic development. Our group has previously shown that M.tb induces higher expression of specific histone deacetylase (HDACs), such as Sirtuin 2 in CD4+ T cells that deacetylates NF-ĸB at lysine 310 which impairs macrophage- CD4+ T cells interactions to restrict M.tb growth (77). Recently, the study from our group has discovered that pharmacological inhibition of Sirtuin 2 by AGK2 induces glycolysis by modulating Wnt-β-catenin pathway, that subsequently enhances T-cell stem cell like memory (CD4+ Tscm) responses during M.tb infection. This is further complemented by adjunct therapy with BCG as well as DOTS therapy, that results in higher Tscm responses that further give rise to different memory pools and effector T cells (77, 78). Similarly, whole proteomics analysis of Purified protein derivative (PPD+) healthy individuals when treated with an immunomodulator, Berberine, showed upregulation of glycolytic pathway along with NOTCH3, that may possibly play role in prompting protective T cell effector function against TB (79).
In conclusion, it is imperative to note that delving into immunometabolism of the host during M.tb infection will help researchers refine the vaccine development. By incorporating insights into the metabolic requirements of immune cells during M.tb infection, researchers can design vaccines that enhance the overall efficacy of the immune response, potentially leading to more successful prevention and treatment strategies for TB.
Apart from modulating immunometabolism, M.tb induces ER stress as its survival strategy, and hijack the host immune responses (80). ER stress is caused due to imbalance between protein synthesis and protein folding. Consequently, unfolded proteins accumulate in the cell and induce ‘Unfolded Protein Response’ (UPR) (81). This UPR/ER stress is caused by M.tb during ATB and LTBI for retrieving nutrients as well as evading immune responses. Siemons et al., reported for the first time in 2010 that M.tb infection induces ER stress-related genes in granulomas in humans and mice. Immuno-histochemistry demonstrated that the markers of ER stress such as CHOP, eIF2α, Ire1α, and ATF3 have higher expression in the M.tb infected macrophages within human and mice granulomas (82). Thereafter, there has been a surge in identifying M.tb proteins that induce ER stress and the significance of ER stress during M.tb pathogenesis. There are couple of M.tb proteins identified till date that induce ER stress for survival, dissemination and nutrient uptake. Through over-expression studies, Choi et al., reported that ESAT-6 induces ROS in A549 epithelial cells by increasing intracellular Ca2+ concentration, that subsequently induces ER stress (83). Moreover, ER stress induction subsequently triggers apoptosis of the infected cells through activation of Caspase-12 that lies near ER compartment, hence modulates intracellular survival of M.tb (84). This study has demonstrated that initially higher expression of ER chaperones, BiP is essential for host cell survival, whereas later during M.tb infection, M.tb induces higher expression of CHOP pathway, that subsequently activates Caspase-12 to induce apoptosis (84). Conventionally, apoptotic bodies carrying M.tb undergo efferocytosis, and hence helps in reduction in M.tb burden. However, researchers delved deeper into identifying what provokes ER stress-induced apoptosis in M.tb infected cells. Further, they observed that another ER chaperone, Calreticulin is upregulated upon M.tb infection. Calreticulin forms complex with CXCR1 and TNFR and induces apoptosis, thus suppressing intracellular survival of M.tb (85). These are the cumulative host defense strategy for controlling M.tb infection. Nevertheless, M.tb has evolved to surpass the host defense strategy by targeting ER for deriving lipids and cholesterol for its own replication. Also, M.tb secretes CDP-diglyceride hydrolase (CdhM) that accumulates in ER, and damages its integrity, which induces cell death. This mechanism aids in M.tb dissemination to extracellular environment (86). Similarly, other M.tb proteins such as Rv0297 has PGRS domain, that enables the accumulation of CdhM in ER, and causes ER-induced apoptosis (87). The PE- PGRS domain containing proteins are antigenic in nature and are known for M.tb persistence during latent TB (88), hence are one of the potent targets for vaccine development.
It is remarkable to understand that ER stress results in halting in mRNA translation. This may ultimately ensue the accumulation of untranslated mRNA and other folded/unfolded proteins in a membrane less structure called stress granules (89). These stress granules are known to prevent ER mediated apoptosis in acute liver failure (90). This unexplored area of research in TB could serve as the another host defense strategy in preventing M.tb dissemination and thus controlling M.tb replication. This is important to realize that ER stress-induced apoptosis confers M.tb survival in later stages of granuloma and promotes dissemination (82). M.tb can counter host defense response at multitude level, hence it is noteworthy that researchers must dive in designing multi-epitopes vaccine (91). The multi epitope vaccines can target M.tb proteins that induce cellular stress and may target different arms of immune escape. For instance, our group has designed a peptide based multi-epitope vaccine (peptide-TLR agonist-liposome; PTL) harboring ESAT-6 and Ag85B. The intranasal delivery of PTL in conjunction with BCG, significantly improved the immune responses against murine model of TB (86). Despite numerous peptide- based multi-epitope vaccine in research pipeline against TB, limited research is available on the effects of these vaccines in combating cellular stress.
Nonetheless, this is a very less explored area of research to discover the role of ER stress during latency and reactivation. ER stress is potentially utilized by M.tb to persist within the host and evade from host killing mechanisms. Therefore, delineating the interaction between ER responses and M.tb is crucial in terms of designing better pharmacological therapies to limit M.tb progression.
The various organs in human are co-inhabited by a diverse symbiotic microbial community. The majority of these microbiome reside in the respiratory tract and the gastrointestinal tract (92). The microbiota niche performs diverse protective functions in the host such as regulation of metabolism, maintaining barrier integrity, immune activation, trophic functions, etc. throughout the body, thereby maintaining tissue homeostasis (93). Several respiratory diseases including TB is characterized with changes in host microbiome composition in lung and gut which dictates the course of the disease pathogenesis.
The upper respiratory tract acts as a gateway to the lower respiratory tract and to the final destination of M.tb colonization- the lung (94). Similarly, metabolites produced by the gut microbiome can reach the lungs through blood (95). Several studies have hypothesized that dysbiosis of the microbiota in the upper respiratory tract, lungs, and gut increases the chances of M.tb primary infection, reinfection, and reactivation of LTBI (96).
Microbiome colonization is initiated post birth in the mucosal sites. In healthy human lungs, resident microbiome belongs to the following phyla- Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria. Among these, the most prevalent genera have been identified as following- Prevotella, Streptococcus, Veillonella, Fusobacterium, and Haemophilus (97).
Recent literature suggests an alteration in the microbial community of the upper respiratory tract upon infection with TB. The microbial composition is speculated to have a role in the disease progression from healthy to ATB or LTBI. ATB shows enhancement in gram-positive and gram-negative species. Proteobacteria, Bacteroidata, and Bacillota are the most observed phylum while Balutia and Acinoetbacter are the dominant classes in ATB as compared to the Healthy cohort (HC). HCs were observed to have enrichment in the Lacnospiraceae family and the Faecalibacterium genus. Comparative analysis between the HC and LTBI cohort signified the presence of the Kosakonia genus in HC while the Faecalibacterium genus was enriched in LTBI (98).
Prospective cohort studies have indicated the role of the microbial community in predicting the susceptibility to TB. Notably, Healthy Household Contacts (HHCs) who did not acquire LTBI had an elevated relative abundance of Corynebacterium, Dolosigranulum, Cutibacterium, and Staphylococcus. Conversely, in HHCs with established LTBI, the abundance of Corynebacterium, Lawsonella, Paracoccus, Cutibacterium, Dolosigranulum, and Moraxella was notably higher. It is worth mentioning that the bacterial genera Dolosigranulum and Variovax were exclusively found in HC, leading to the conclusion that these genera are integral components of a healthy nasopharyngeal microbiome. Furthermore, in pre-LTBI patients, the absence of Dolosigranulum, coupled with the presence of Rothia and Megasphaera genera, has raised speculation regarding their potential involvement in modulating host immunity and increasing susceptibility to M.tb infection (99).
In a separate investigation led by Botero et al., noteworthy alterations were observed in the bacterial and fungal communities within the oropharynx of TB patients when compared to individuals in good health. The study unveiled a significant increase in the presence of the Ascomycota phylum in TB patients, indicative of fungal diversity shifts. Conversely, the genus Cryptococcus showed a notable decline in abundance within the oropharynx of TB patients. It is crucial to acknowledge that one of the key limitations of this study is the relatively small sample size, which may impact the generalizability of the findings (100).
These findings suggested a resemblance in nasopharyngeal species diversity and composition among HC, ATB, and LTBI groups, shedding light on the intricate interplay between upper respiratory microbiota and TB infection. These studies have however failed to conclusively show an association between bacterial dysbiosis in the nasopharyngeal tract and the chances of acquiring TB or progressing to LTBI.
Most of the studies focussing on studying lung microbiome diversity in TB rely on sputum samples for their downstream analysis (101). Sputum samples are spuriously contaminated with pharyngeal microbiota; thus, the result fails to be a true indicator of the lung microbiome (102). The investigation of the accurate picture of the lung microbiome is hindered by invasive techniques such as bronchioalveolar lavage fluid (BALF) collection and throat swabs (102). These techniques provide a better picture of the lung microbiome than the traditional sputum collection method.
In the pursuit of a deeper understanding of the lung microbiome, Xiao et al., conducted a study that harnessed BALF samples and employed metagenomics to explore microbial enrichment at the species level. Their findings revealed intriguing disparities between the HC and the Untreated Pulmonary TB Group (UTG). Specifically, the HC displayed enrichment in species like Prevotella melaninogenica and Ralstonia pickettii, while the UTG exhibited a heightened presence of S. aureus and Neisseria gonorrhoeae. These outcomes starkly contrast with those reported by Hu et al., in their BALF-based investigation (103).
Another pertinent study conducted by Pérez et al., shed light on differences in alveolar microbiota among patients with pulmonary TB. Their analysis of BALF samples uncovered a reduction in the Streptococcus genus, while concurrently identifying an increase in the abundance of Mycobacterium, Lactobacillus, and Acinetobacter (104). These findings underscore the complexity and variability in lung microbiota profiles associated with pulmonary TB, emphasizing the need for further research to elucidate the underlying factors and implications of these differences. These two studies provide crucial insights into the lung microbiome in TB patients but are severely limited by the patient sample size.
In a comprehensive study by Kang et al., the investigation unearthed distinct characteristics within the airway microbiota that set patients with M.tb infection apart from those without. Notably, a novel genus, Anoxybacillus, exhibited an increased presence in BALF samples. What sets this study apart is its robust patient sample size, which bolsters the need for further exploration to establish a definitive correlation between the prevalence of Anoxybacillus and TB (105).
Similarly, Zhou et al., research delivered significant findings, underscoring the prevalence of the Cupriavidus genus as dominant in TB patients, while Streptococcus emerged as the dominant genus in healthy subjects. Notably, their study was carried out utilizing BALF samples (106). These observations underscore the utility of BALF in uncovering crucial distinctions in lung microbiota between TB patients and healthy individuals, thus emphasizing the potential significance of these findings in understanding TB pathogenesis and treatment strategies. Despite the vast literature available, there are limited studies linking microbiome alteration to functional disease progression.
Xia et al., undertook a study with the objective of establishing a connection between changes in the microbial composition and variations in cytokine levels within BALF and their potential implications in the progression of TB (107). This investigation aimed to elucidate the interplay between microbial alterations and cytokine dynamics, shedding light on their role in the advancement of TB.
Meta-analysis serves as a valuable avenue for reinforcing scientific findings when the available sample size is constrained. These comprehensive studies amalgamate data derived from multiple investigations centered on a common research question, effectively surmounting the limitations posed by small sample sizes. Notably, a meta-analysis conducted by Hong et al., offered intriguing insights by proposing a co-abundance relationship between R. mucilaginosa and A. graevenitzii with M.tb within the lung microbiota of patients with ATB. It is important to acknowledge that this study is bounded by the number of source studies employed and the origin of the microbiota samples used to mimic the lung microbiome. Nonetheless, it introduces the intriguing possibility that the co-occurrence of specific bacterial communities may serve as a valuable indicator of disease progression and hold potential for predicting treatment outcomes (108).
Microbiome in healthy gut harbor around 50 different bacterial phyla with Bacteroidetes and the Firmicutes as the two dominant gut resident phyla (109). TB progression is marked by dysbiosis of resident gut microbiome (110). Several cohort studies undertaken on the Chinese population indicate differences in diversity, composition, and function of gut microbiota in HC vs TB patients. The studies suggested dysbiosis of the gut microbiome in TB patients with a lower abundance of short-chain fatty acid producers namely microbiome genera such as Lachnospiraceae and species - Haemophilus parainfluenzae, Roseburia inulinivorans, and Roseburia hominis. TB patients also show a notable increase in opportunistic pathogens of the genera Enterococcus and Rothia. The studies suggest dysregulated host metabolism such as decrease in the biosynthesis capacity of amino acids and fatty acids and altered carbohydrate metabolism patterns in the gut in TB patients as compared to HC (13). Butyrate, a short-chain fatty acid, is a key metabolic pathway that is downregulated in notably all the cohort studies involving active TB patients (111). Butyrate production is inversely linked with the severity of inflammatory and metabolic diseases (112).
Demographics, geographic characteristics, and antibiotic usage also impact microbial diversity. Cohort studies from Pakistani, Chinese and Indian populations show differential enrichment of gut microbial phyla in healthy and TB patients (113).
In the Chinese cohort phylum Actinobacteria increased while phylum Bacteroidetes diminished in healthy cohort while in TB patients Proteobacteria and Bacteroidates have been notablyenriched. Firmicutes which shows enrichment in the Pakistani cohort significantly diminished in the Chinese cohort. While Firmicutes and Actinobacteria showed relative enhancement in a cohort study in the Indian population (13).
These microbial signatures need further investigation as the majority of the study uses TB and HC which does not provide confirmatory results on how these signatures might differ from non-TB inflammatory diseases (113).
The secondary metabolites, immune signals and microbial remnant produced by gut/lung microbiome can affect the microbiota composition and abundance in a bidirectional fashion termed as the gut lung axis. The resident microbiome composition can dictate the disease progression. Recent murine studies have demonstrated that enriching the murine gut with Lactobacillus can alleviate asthma-like symptoms in mice (114).
Moreover, administration of IPA produced by gut Clostidia species have shown anti-tubercular activity in murine model by targeting trp and Short chain fatty acids (SCFA) biosynthesis in M.tb (115). It is worth noting that patients with ATB often exhibit increased regulatory T cells (Tregs). Interestingly, Faecal transplantation in murine model have shown to bear fruitful results in reducing T reg population and increasing protective cytokines such as IFN-γ/TNF-α (116).
SCFA produced by gut microbiome can enter the circulatory pathway and reach various peripheral organs including bone marrow. Within the bone marrow SCFA can induce differentiation of progenitor macrophage and DC which can then migrate to the lung and thus modulate lung microenvironment (117) (Figure 2). Anaerobic microbiota in the lower airways produce SCFAs, and elevated serum SCFA levels have been linked to the induction of FoxP3-expressing Tregs in BALF lymphocytes. Pulmonary SCFA produced by anaerobic microbiota has also been associated with a decrease in IFN-γ levels in active TB (118, 119).
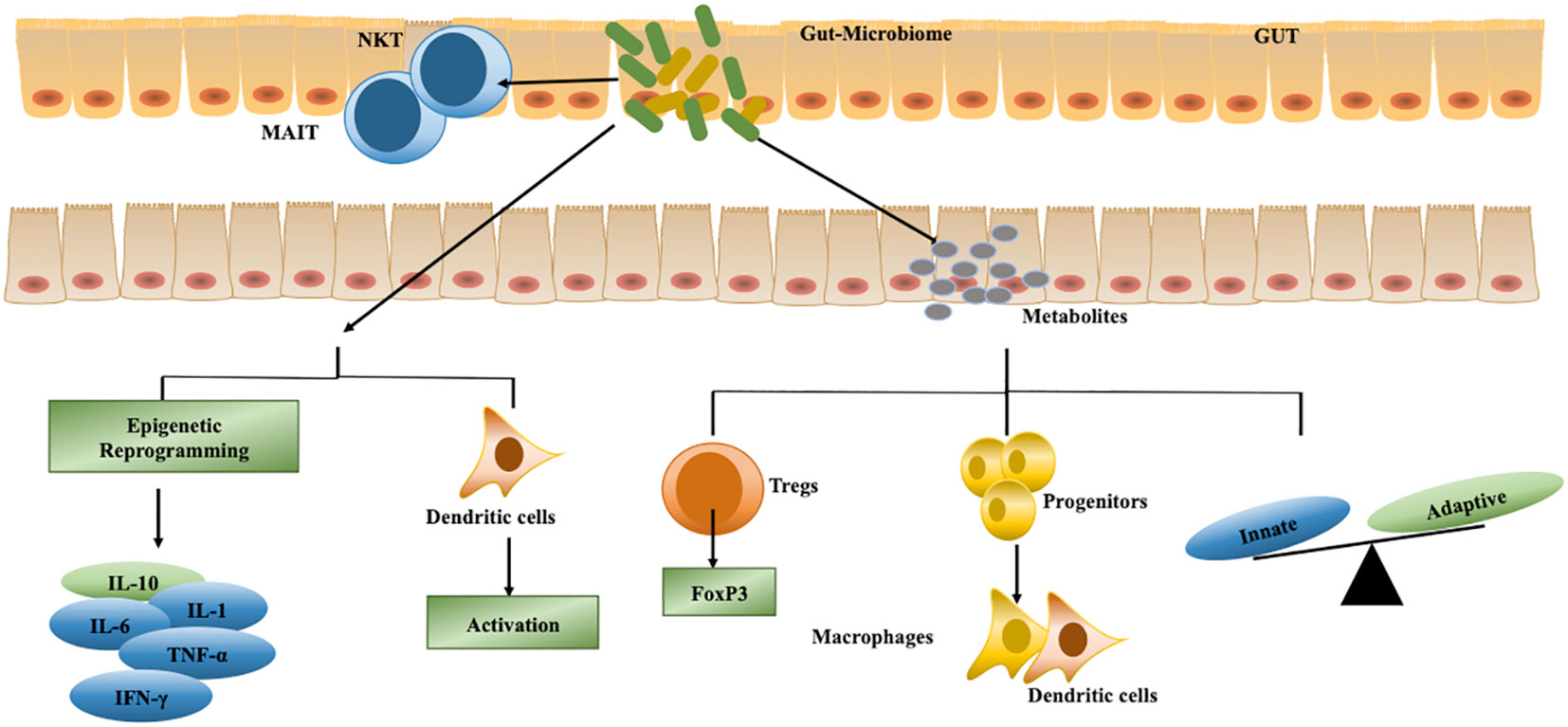
Figure 2 Microbiome controls various host cellular processes to regulate M.tb survival. Gut microbiota secretes various metabolites such as short-chain fatty acids (SCFA), indole propionic acid (IPA), flagellin, lipopolysaccharide (LPS), and peptidoglycan to regulate differentiation of immune cells that traverse to the lung for containing M.tb replication. These metabolites also regulate the balance of innate and adaptive immunity. Apart from protecting against TB, a few metabolites secreted by gut microbiota suppress immune responses against M.tb by enhancing Tregs function in the lung. Also, the microbiome in the respiratory tract can reprogram immune cells to secrete differential cytokines and induce dendritic cell activation to enhance antigen presentation to T cells.
Metabolites from the gut commensal microbiome have role in steering the abundance and function of non-conventional T cells such as Natural killer T cells (NKT) and Mucosal associated invariant T cells (MAIT). Studies using germ-free mice have shown the complete absence of MAIT cells, thus pointing towards a probable role of microbiota in affecting the abundance of MAIT cells. MAIT cells have been found to play important role in protection against TB (96).
Although there is limited research on the modulation of the gut-lung axis in TB, an intriguing study conducted by Negi et al., in a murine TB model highlights the role of dendritic cell activation by the gut microbiome of TB. This research suggests gut dysbiosis can lead to low mincle expression on lung DC which negatively regulates antimycobacterial T cell activation in the lung. The study suggests restoration of Lactobacillus as a possible measure to augment mincle based DC signaling (120).
Inflammation mediated by IFN-γ expression is crucial for controlling the initial stages of TB progression. Shen et al., revealed possible association of lncRNA-CGB produced by gut Bacteroides fragilis in positively regulating IFN-γ production by epigenetic reprogramming (121). The long-term anti TB treatment is known to induce significant alteration in microbial community. The altered microbiome can impact effective immune responses generation during the treatment (122). Limited literature is available that can explain the mechanism of lung microbiome mediated changes in the gut microbiome composition upon the initiation of colonization of lung by M.tb.
The mechanism linked to generation of optimal vaccine responses in human is a very complex phenomenon. Several intrinsic and extrinsic factors such as age, genetics, nutritional status, pre-existing immunity and comorbidities dictate the nature of generation of vaccine responses (123). Recent studies have demonstrated yet another intrinsic factor of microbiome that can influence vaccine immunogenicity (124). Antibiotics-induced gut microbiome dysbiosis has been linked to diminished immune response generation to vaccination in healthy adults (125). Optimal vaccine-induced antibody response has been linked to the prevalence of gut commensals in diseases such as bacterial diarrhea, influenza, polio, etc (126). Studies using germ free animal models have linked defective Th1/Th2 responses, reduced intraepithelial lymphocytes, reduced antibody generation and a dysregulation in Th17 cell responses to the host microbiome (127).
In the context of TB, study conducted by Nadeem et al., reported gut microbiome dysbiosis as important determinant in dictating BCG vaccine efficacy in TB. The altered microbial community had a decreased prevalence of Bifidobacterium, Lactobacillus and an increased prevalence of Bacteroides. The dysbiosis resulted in decreased activation and frequency of various T cells subsets including the memory subset. The dysbiosis also impacted the secretion of pro-inflammatory cytokines and overall hampered the successful clearance of M.tb in the murine model of TB (128).
A cohort study of western European population suggested the role of gut microbiome in altering BCG-induced specific and trained immune responses. The trained immune response against Staphylococcus aureus was negatively correlated with the abundance of Roseburia. Roseburia secreted metabolites affected the phenylalanine metabolism. The disturbed phenylalanine metabolism can dampen BCG-induced trained immunity. The abundance of Ruminococcus and Eggerthella lenta was correlated to be a positive indicator for M.tb control (129).
Actinobacterial abundance was correlated with positive T-cell response to BCG administration in a cohort study on Bangladeshi infants. The prevalence of Enterobacteriales, Pseudomonadales, and Clostridiales was demonstrated to be associated with neutrophilia and reduced vaccine efficacy (130).
Immunomodulatory molecules produced by microbiota such as flagellin, peptidoglycan, lipopolysaccharides, and SCFA acts as ligands to PRRs (Toll like receptors, NOD-2 receptors) and modulate innate and adaptive immune cells activation and metabolism. SCFA production has been linked to the generation of efficient B cell response through modulating B cell differentiation into antibody producing cells (131). Gut microbiome has also been shown to regulate antigen specific T cell responses. The microbiota has notable role in generation of type-1 interferon by plasmacytoid dendritic cells which can eventually signal conventional dendritic cells to prime T cell more efficiently (132). Microbial-encoded cross-reactive epitopes could also potentially modulate vaccine immune response (124). Administration of bacterial species in form of pro/prebiotics, or faecal transplant can be an important determinant towards optimizing microbiota composition. This optimization can prove to be beneficial therapeutic avenue to positively impact vaccine immunogenicity (133) Thus, understanding and harnessing the influence of the gut microbiome on vaccination responses could lead to more effective and targeted strategies for disease prevention.
Exosomes are extracellular vesicles in the size range of 30-150nm. Exosomes are secreted from cells by the process of exocytosis and mediate cellular signaling and communication. Exosomes present surface protein from the parental cell as well the target cell. This feature can help in predicting the dynamic cellular turnover and organ specific metastasis of cellular contents, thus overcoming the limitation of conventional blood-based biomarker. The content of exosomes is highly diverse consisting of soluble and membrane bound proteins, lipids, and nucleic acids (134). Overall, the exosomes can be an excellent blood-based biomarker for early disease diagnosis and predicting disease progression. Blood-based biomarkers have the potential to significantly enhance TB detection, particularly in cases involving children and elderly individuals who may not be able to produce sputum (135). This approach also mitigates the risk of TB transmission to healthcare workers during sputum collection. In the pursuit of point-of-care diagnostics, exosomes offer a promising platform for blood-based TB detection. Additionally, exosomes can serve as an excellent vaccine development and delivery system (136).
Exosomes are an important component in the cellular signaling cascade. Extracellular vesicles (EVs) derived from M.tb-infected cells are commonly found to express M.tb cell wall, cell membrane, respiration proteins and lipoproteins (137). A report from a paediatric cohort followed by a validation study in Non-Human Primate (NHP) model suggests differential expression of M.tb cell wall components in ATB vs LTBI cases. Downregulation of Lipoarabinomanan (LAM) and upregulation of lipoprotein LprG have been observed in NHPs with latent infection. ATB results in upregulation of LAM and LprG in serum EVs of NHP (138). Multiple reaction monitoring assays have identified M.tb secretory antigens such as Culture filtrate protein (CFP2) and Mtp32 peptides as potential biomarkers for active TB, while in individuals with LTBI, the SVF peptide from Glutamine synthetase (GlnA1) have been detected in approximately 82% of participants (139, 140).
Exosomes in the bloodstream carry small RNAs, including microRNAs (miRNAs), which are well-protected from nucleases and are under active investigation as essential blood-based biomarkers, particularly in cancer. In a pilot study, miR-484, miR-425, and miR-96 were found to be significantly upregulated in patients with active TB compared to HC. These altered miRNAs have functional relevance in metabolic disorders and insulin signaling, which is reflected in disrupted sugar metabolism in macrophages infected with M.tb (141).
Profiling miRNAs from serum exosomes in individuals with LTBI and ATB, in comparison to HC, revealed differential expression patterns among the groups. Specifically, LTBI patients exhibited unique miRNA profiles, including hsa-let-7e-5p, hsa-let-7d-5p, hsa-miR-450a-5p, and hsa-miR-140-5p. In contrast, ATB exhibited an enrichment of hsa-miR-1246, hsa-miR-2110, hsa-miR-370-3p, hsa-miR-28-3p, and hsa-miR-193b-5p. Several other miRNAs associated with the activation of phagocytosis (has-mir-142-3p), apoptosis (has-mir-29a), and inflammatory signaling pathways were upregulated in TB patients (142). In another report, ATB has been documented to be characterized by the enhanced expression of let-7i-5p, miR-148a-3p, miR-21-5p, and miR-423-5p. In contrast, LTBI has been associated with upregulation in the expression of miR-122-5p and miR-451a (143). Studies characterizing miRNA from patient serum exosomes are very limited and vary extensively between different cohorts. The studies also lack functional in-depth characterization of miRNA targets and downstream signaling events that might prove to be beneficial or detrimental to the host. Several factors such as host sex, age, basal immunity, pre-existing disease, exosome isolation protocols, etc contribute to variation in exosome characteristics and functions (144). These confounders need to be explored in-depth to utilize exosomes as a highly precise diagnostic tool.
In addition to M.tb peptides, exosomes also transport host peptides with altered regulation. Comparative proteomic studies using isobaric tags for relative and absolute quantification (iTRAQ) revealed deregulated host proteins in serum extracellular vesicles when comparing ATB, non-TB (respiratory diseases other than TB; NTB), and HC. Host proteins such as Kynurenine-oxoglutarate transaminase 3 (KYAT3), Alpha-1-Antitrypsin (SERPINA1), Haptoglobin (HP), and Apolipoprotein C-III (APOC3) are primarily involved in neutrophil degranulation, plasma heme scavenging, kynurenine metabolism, and lipid metabolism, respectively (145) and shown to exhibit deregulation in ATB. It is essential to note that while there have been numerous studies characterizing the content of exosomes in the context of TB, most of them have focused on in vitro and in vivo models. Only a limited number of studies have been validated in large patient populations and the functional implication of the altered metabolism in TB progression Additionally, many studies have primarily compared ATB to HC, neglecting the consideration of NTB and LTBI.
Furthermore, the response to TB treatment in DOTS responders versus non-responders has not been extensively explored and validated in the context of differences in exosomal content. An open randomized controlled trial comparison of patient plasma exosome proteins suggested downregulation of Complement component binding protein 4 (C4BPA) and Calgranulin B (S100A9) might be involved in a positive host response to LTBI treatment. S100A9 which is a calcium binding protein, has been reported to be upregulated in chronic pulmonary obstructive disease and notably impairs innate immune cell polarization in obesity models (146, 147). C4BPA has a functional role in modulating host-M.tb interaction (148). Exosomes are being speculated to have a role in the development and pathogenesis of drug resistance phenotypes. Drug-resistant TB (DR-TB) has a dysregulated cholesterol metabolism in comparison to non-drug-resistant TB (NDR) patients. DR-TB patients show a decrease in the expression of various apolipoprotein family members such as APOA1, APOB, and APOC1. These apolipoproteins have been linked to the formation of foamy macrophages and reduction of cellular uptake of anti-TB drugs thus leading to failure of DR-TB treatment (149). Several pieces of research are being conducted to evaluate urine-based M.tb diagnostic assays which can be used as adjunct detection methods for smear and PCR-negative patients (150, 151).
One of the important limitations of blood-based circulating biomarkers is that they provide inconclusive detail about immune processes taking place in human tissues. Emerging studies suggest liquid biopsies can provide an alternative to the standard circulating biomarkers assays. Liquid biopsies are currently being used for prenatal testing, cancer monitoring, predicting vaccine efficacy, and investigating cases of graft rejection (152).
Liquid biopsies are been widely researched in the quest to improve the diagnosis of TB. Cf nucleic acids are primarily released by dying cells. Profiling of CfDNA coupled to epigenetic marks on cfDNA can provide information about mechanisms of cellular injury, death, systemic immune dynamics, and interaction of immune cells in tissues (152, 153). cfDNA in humans can have host as well as microbial origin. In a proof-of-concept study using CRISPR-Cas13, mycobacterial regions of IS6110 and IS1081 were detected in three separate active TB patient cohorts. Future studies can help elucidate the correlation of microbial cfDNA distribution in different states of TB progression (154). Monitoring DNA methylome in immune cell-derived cfDNA can prove to be of paramount importance in understanding immune responses and cellular exhaustion in inflammatory and chronic diseases (152).
In addition to the role as biomarkers, exosomes have the potential to be used for drug delivery systems due to their nano-lipid properties (155). Several in-vitro and in-vivo studies demonstrate the immune-activating properties of M.tb-infected innate immune cell-derived exosomes. M.tb antigen-loaded exosomes have demonstrated immune boosting properties which are critical in vaccine and adjuvant development for developing novel vaccine and immunomodulatory candidates (156, 157).
Exosomes carrying potent M.tb antigens hold a great promise to serve as a productive cell-free vaccine candidate to replace/boost BCG immunizations. Exosomes from M.tb-infected macrophages can direct the polarization of naïve macrophages. Additionally, exosomes derived from infected innate immune cells can effectively prime the adaptive T cells in the in-vivo model of TB (16). In yet another study on the murine model of TB, exosome vaccination has been found to prime as well as boost BCG immunization. Mice displayed a Th1-mediated immune response with limited Th2 response as compared to BCG-immunized mice. The purified exosomes used for vaccination showed notable enrichment with antigenic mycobacterial proteins (157).
EV derived from human neutrophils stimulated with M.tb results in a decrease in intracellular M.tb in the macrophages. The EVs derived from the in-vitro stimulated human neutrophils contains TLR ligands and co-stimulatory molecules required for innate and adaptive immune cell activation, respectively. The EVs induce autophagy to clear intracellular M.tb from the macrophages (158). Another research suggests the use of ubiquitin-tagged Ag85B-ESAT6 fusion protein-based exosome can be used as a novel vaccine candidate. This antigen-specific exosome-based delivery system has been observed to induce enhanced IFN-γ response by T cells and it enhances mycobacterial clearance (159).
Exosomes have also been tested as vaccine candidates for cancer in murine melanoma models. DC-derived exosomes were found to induce adaptive immune responses. The exosomes significantly augmented CD8+ T cell proliferation and differentiation into central memory cells (160). EVs have also been found to act as a superior adjuvant and elicit an effective cell-mediated immune response (161). In a study conducted on the murine model, exosomal adjuvant enhanced the IFN-γ secretion upon immunization with hepatitis B surface antigen. The role of exosomes as adjuvants during BCG immunization requires further research in TB (162). Exosomes function as a double-edged sword in the host-pathogen interaction. There are few studies suggesting exosomal contents can limit inflammation and delay mycobacterial clearance (16, 163).
Nevertheless, more research in the exciting field of exosomes is required to realize its full potential as a vaccine and adjuvant candidate to provide better protection against TB.
M.tb has developed diverse defensive strategies to counter host immune responses, targeting nearly every significant host process. Importantly, disruptions in any host cellular process intricately influence connected pathways, thereby regulating M.tb pathogenesis. For instance, M.tb causes a shift in metabolic pathways through impacting immunometabolism that can induce ER stress. The Unfolded protein-initiated ER stress leads to the inclusion of unfolded proteins, spliced mRNA, and metabolites within EV. The EVs secreted out of the infected cells alter immune signaling in the adjacent bystander immune cells. The immune mediators and the intermediatory metabolic pathway products can also affect the diversity and composition of host microbiome in the various organs. Cohort studies suggests the dysregulation of gut/respiratory microbiome is correlated to M.tb pathogenesis. The metabolites generated by host and the host harbored microbiome can modulate signaling events that affect M.tb growth, survival and interactions within the host. This is noteworthy that dysregulation of the microbiome also affects the metabolism of immune cells which impacts the energy status of the cell, and consequently alters immune responses against M.tb. Through this review, we have highlighted the less explored host pathways that prevail in M.tb pathogenesis. Currently, many researchers are investigating the effect of these integrated pathways in M.tb pathogenesis. Yet, extensive efforts are needed to understand the modulations in infected cells or activated immune cells that collectively affect M.tb pathogenesis. Despite having many vaccines and drug candidates in clinical pipelines, a well-designed approach to prevent TB and reduce tissue damage is still an urgent need to provide better protection against TB. In conclusion, it is necessary to design novel host directed therapies that can improve the protection against TB.
AG: Data curation, Investigation, Writing – original draft, Writing – review & editing. AV: Data curation, Investigation, Writing – original draft, Writing – review & editing. AB: Data curation, Formal Analysis, Investigation, Visualization, Writing – original draft, Writing – review & editing. VD: Conceptualization, Funding acquisition, Investigation, Resources, Supervision, Visualization, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. We would also like to acknowledge the funding support from Central Council of Research in Homeopathy, Ministry of Ayush, Government of India and Science and Engineering Research Board (SERB), Department of Science and Technology, Government of India and ICGEB, New Delhi, India. However the funders has no role in study design.
AG and AV are the recipient of Junior Research Fellowship from Department of Biotechnology (DBT), Government of India. AB is the recipient of DST-INSPIRE Faculty Fellowship, Department of Science and Technology, Government of India and HGK-IYBA Fellowship from Department of Biotechnology, Government of India and Women Excellence Award from Science and Engineering Research Board, Government of India. VD is also a recipient of DST-INSPIRE Faculty Fellowship, Department of Science and Technology, Government of India and an Early Career Research Award from Science and Engineering Research Board (SERB), Department of Science and Technology, Government of India.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Smith I. Mycobacterium tuberculosis pathogenesis and molecular determinants of virulence. Clin Microbiol Rev (2003) 16(3):463–96. doi: 10.1128/CMR.16.3.463-496.2003
2. Goldberg MF, Saini NK, Porcelli SA. Evasion of innate and adaptive immunity by mycobacterium tuberculosis. Microbiol Spectr (2014) 2(5). doi: 10.1128/microbiolspec.MGM2-0005-2013
3. Jo EK. Interplay between host and pathogen: immune defense and beyond. Exp Mol Med (2019) 51(12):1–3. doi: 10.1038/s12276-019-0281-8
4. Cooper AM. Cell-mediated immune responses in tuberculosis. Annu Rev Immunol (2009) 27:393–422. doi: 10.1146/annurev.immunol.021908.132703
5. Pal R, Bisht MK, Mukhopadhyay S. Secretory proteins of Mycobacterium tuberculosis and their roles in modulation of host immune responses: focus on therapeutic targets. FEBS J (2022) 289(14):4146–71. doi: 10.1111/febs.16369
6. Martin M, deVisch A, Boudehen YM, Barthe P, Gutierrez C, Turapov O, et al. A Mycobacterium tuberculosis Effector Targets Mitochondrion, Controls Energy Metabolism, and Limits Cytochrome c Exit. Microbiol Spectr (2023) 11(3):e0106623. doi: 10.1128/spectrum.01066-23
7. Soto-Heredero G, Gómez de Las Heras MM, Gabandé-Rodríguez E, Oller J, Mittelbrunn M. Glycolysis - a key player in the inflammatory response. FEBS J (2020) 287(16):3350–69. doi: 10.1111/febs.15327
8. Chandra P, Coullon H, Agarwal M, Goss CW, Philips JA. Macrophage global metabolomics identifies cholestenone as host/pathogen cometabolite present in human Mycobacterium tuberculosis infection. J Clin Invest (2022) 132(3):e152509. doi: 10.1172/JCI152509
9. Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell (2005) 122(1):107–18. doi: 10.1016/j.cell.2005.05.007
10. Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science (2011) 331(6015):337–41. doi: 10.1126/science.1198469
11. Khan N, Mendonca L, Dhariwal A, Fontes G, Menzies D, Xia J, et al. Intestinal dysbiosis compromises alveolar macrophage immunity to Mycobacterium tuberculosis. Mucosal Immunol (2019) 12(3):772–83. doi: 10.1038/s41385-019-0147-3
12. Lee WJ, Hase K. Gut microbiota-generated metabolites in animal health and disease. Nat Chem Biol (2014) 10(6):416–24. doi: 10.1038/nchembio.1535
13. Eribo OA, du Plessis N, Ozturk M, Guler R, Walzl G, Chegou NN. The gut microbiome in tuberculosis susceptibility and treatment response: guilty or not guilty? Cell Mol Life Sci (2020) 77(8):1497–509. doi: 10.1007/s00018-019-03370-4
14. Negatu DA, Liu JJJ, Zimmerman M, Kaya F, Dartois V, Aldrich CC, et al. Whole-cell screen of fragment library identifies gut microbiota metabolite indole propionic acid as antitubercular. Antimicrob Agents Chemother (2018) 62(3):e01571–17. doi: 10.1128/AAC.01571-17
15. Smith VL, Cheng Y, Bryant BR, Schorey JS. Exosomes function in antigen presentation during an in vivo Mycobacterium tuberculosis infection. Sci Rep (2017) 7(1):43578. doi: 10.1038/srep43578
16. Singh PP, LeMaire C, Tan JC, Zeng E, Schorey JS. Exosomes released from M. tuberculosis infected cells can suppress IFN-γ mediated activation of naïve macrophages. PloS One (2011) 6(4):e18564. doi: 10.1371/journal.pone.0018564
17. Zhai W, Wu F, Zhang Y, Fu Y, Liu Z. The immune escape mechanisms of mycobacterium tuberculosis. Int J Mol Sci (2019) 20(2):340. doi: 10.3390/ijms20020340
18. Yang J, Zhang L, Qiao W, Luo Y. Mycobacterium tuberculosis: Pathogenesis and therapeutic targets. MedComm (2020) (2023) 4(5):e353. doi: 10.1002/mco2.353
19. Reuschl AK, Edwards MR, Parker R, Connell DW, Hoang L, Halliday A, et al. Innate activation of human primary epithelial cells broadens the host response to Mycobacterium tuberculosis in the airways. PloS Pathog (2017) 13(9):e1006577. doi: 10.1371/journal.ppat.1006577
20. Scordo JM, Knoell DL, Torrelles JB. Alveolar epithelial cells in mycobacterium tuberculosis infection: active players or innocent bystanders? J Innate Immun (2016) 8(1):3–14. doi: 10.1159/000439275
21. Bomfim CCB, Fisher L, Amaral EP, Mittereder L, McCann K, Correa AAS, et al. Mycobacterium tuberculosis induces irg1 in murine macrophages by a pathway involving both TLR-2 and STING/IFNAR signaling and requiring bacterial phagocytosis. Front Cell Infect Microbiol (2022) 12:862582. doi: 10.3389/fcimb.2022.862582
22. Augenstreich J, Haanappel E, Ferré G, Czaplicki G, Jolibois F, Destainville N, et al. The conical shape of DIM lipids promotes Mycobacterium tuberculosis infection of macrophages. Proc Natl Acad Sci U.S.A. (2019) 116(51):25649–58. doi: 10.1073/pnas.1910368116
23. Sia JK, Rengarajan J. Immunology of mycobacterium tuberculosis infections. Microbiol Spectr (2019) 7(4). doi: 10.1128/microbiolspec.GPP3-0022-2018
24. Kolliniati O, Ieronymaki E, Vergadi E, Tsatsanis C. Metabolic regulation of macrophage activation. J Innate Immun (2022) 14(1):51–68. doi: 10.1159/000516780
25. Deretic V, Singh S, Master S, Harris J, Roberts E, Kyei G, et al. Mycobacterium tuberculosis inhibition of phagolysosome biogenesis and autophagy as a host defence mechanism. Cell Microbiol (2006) 8(5):719–27. doi: 10.1111/j.1462-5822.2006.00705.x
26. Shaler CR, Horvath C, Lai R, Xing Z. Understanding delayed T-cell priming, lung recruitment, and airway luminal T-cell responses in host defense against pulmonary tuberculosis. Clin Dev Immunol (2012) 2012:628293. doi: 10.1155/2012/628293
27. Bussi C, Gutierrez MG. Mycobacterium tuberculosis infection of host cells in space and time. FEMS Microbiol Rev (2019) 43(4):341–61. doi: 10.1093/femsre/fuz006
28. Pellegrini JM, Sabbione F, Morelli MP, Tateosian NL, Castello FA, Amiano NO, et al. Neutrophil autophagy during human active tuberculosis is modulated by SLAMF1. Autophagy (2021) 17(9):2629–38. doi: 10.1080/15548627.2020.1825273
29. Hilda JN, Das S, Tripathy SP, Hanna LE. Role of neutrophils in tuberculosis: A bird’s eye view. Innate Immun (2020) 26(4):240–7. doi: 10.1177/1753425919881176
30. Li Y, Wang W, Yang F, Xu Y, Feng C, Zhao Y. The regulatory roles of neutrophils in adaptive immunity. Cell Commun Signal (2019) 17:147. doi: 10.1186/s12964-019-0471-y
31. Lowe DM, Redford PS, Wilkinson RJ, O’Garra A, Martineau AR. Neutrophils in tuberculosis: friend or foe? Trends Immunol (2012) 33(1):14–25. doi: 10.1016/j.it.2011.10.003
32. Martineau AR, Newton SM, Wilkinson KA, Kampmann B, Hall BM, Nawroly N, et al. Neutrophil-mediated innate immune resistance to mycobacteria. J Clin Invest (2007) 117(7):1988–94. doi: 10.1172/JCI31097
33. Hult C, Mattila JT, Gideon HP, Linderman JJ, Kirschner DE. Neutrophil dynamics affect mycobacterium tuberculosis granuloma outcomes and dissemination. Front Immunol (2021) 12:712457. doi: 10.3389/fimmu.2021.712457
34. Gideon HP, Phuah J, Junecko BA, Mattila JT. Neutrophils express pro- and anti-inflammatory cytokines in granulomas from Mycobacterium tuberculosis-infected cynomolgus macaques. Mucosal Immunol (2019) 12(6):1370–81. doi: 10.1038/s41385-019-0195-8
35. McCormick S, Shaler CR, Xing Z. Pulmonary mucosal dendritic cells in T-cell activation: implications for TB therapy. Expert Rev Respir Med (2011) 5(1):75–85. doi: 10.1586/ers.10.81
36. Madan-Lala R, Sia JK, King R, Adekambi T, Monin L, Khader SA, et al. Mycobacterium tuberculosis impairs dendritic cell functions through the serine hydrolase Hip1. J Immunol (2014) 192(9):4263–72. doi: 10.4049/jimmunol.1303185
37. Sreejit G, Ahmed A, Parveen N, Jha V, Valluri VL, Ghosh S, et al. The ESAT-6 protein of Mycobacterium tuberculosis interacts with beta-2-microglobulin (β2M) affecting antigen presentation function of macrophage. PloS Pathog (2014) 10(10):e1004446. doi: 10.1371/journal.ppat.1004446
38. Motta A, Schmitz C, Rodrigues L, Ribeiro F, Teixeira C, Detanico T, et al. Mycobacterium tuberculosis heat-shock protein 70 impairs maturation of dendritic cells from bone marrow precursors, induces interleukin-10 production and inhibits T-cell proliferation in vitro. Immunology (2007) 121(4):462–72. doi: 10.1111/j.1365-2567.2007.02564.x
39. Gehring AJ, Rojas RE, Canaday DH, Lakey DL, Harding CV, Boom WH. The Mycobacterium tuberculosis 19-kilodalton lipoprotein inhibits gamma interferon-regulated HLA-DR and Fc gamma R1 on human macrophages through Toll-like receptor 2. Infect Immun (2003) 71(8):4487–97. doi: 10.1128/IAI.71.8.4487-4497.2003
40. Pai RK, Convery M, Hamilton TA, Boom WH, Harding CV. Inhibition of IFN-gamma-induced class II transactivator expression by a 19-kDa lipoprotein from Mycobacterium tuberculosis: a potential mechanism for immune evasion. J Immunol (2003) 171(1):175–84. doi: 10.4049/jimmunol.171.1.175
41. Harding JS, Rayasam A, Schreiber HA, Fabry Z, Sandor M. Mycobacterium-infected dendritic cells disseminate granulomatous inflammation. Sci Rep (2015) 5(1):15248. doi: 10.1038/srep15248
42. Lu YJ, Barreira-Silva P, Boyce S, Powers J, Cavallo K, Behar SM. CD4 T cell help prevents CD8 T cell exhaustion and promotes control of Mycobacterium tuberculosis infection. Cell Rep (2021) 36(11):109696. doi: 10.1016/j.celrep.2021.109696
43. Jayaraman P, Jacques MK, Zhu C, Steblenko KM, Stowell BL, Madi A, et al. TIM3 Mediates T Cell Exhaustion during Mycobacterium tuberculosis Infection. PloS Pathog (2016) 12(3):e1005490. doi: 10.1371/journal.ppat.1005490
44. Cai Y, Wang Y, Shi C, Dai Y, Li F, Xu Y, et al. Single-cell immune profiling reveals functional diversity of T cells in tuberculous pleural effusion. J Exp Med (2022) 219(3):e20211777. doi: 10.1084/jem.20211777
45. Vesosky B, Rottinghaus EK, Stromberg P, Turner J, Beamer G. CCL5 participates in early protection against Mycobacterium tuberculosis. J Leukoc Biol (2010) 87(6):1153–65. doi: 10.1189/jlb.1109742
46. Khader SA, Rangel-Moreno J, Fountain JJ, Martino CA, Reiley WW, Pearl JE, et al. In a murine tuberculosis model, the absence of homeostatic chemokines delays granuloma formation and protective immunity. J Immunol (2009) 183(12):8004–14. doi: 10.4049/jimmunol.0901937
47. DiAngelo JR, Bland ML, Bambina S, Cherry S, Birnbaum MJ. The immune response attenuates growth and nutrient storage in Drosophila by reducing insulin signaling. Proc Natl Acad Sci USA (2009) 106(49):20853–8. doi: 10.1073/pnas.0906749106
48. Slack M, Wang T, Wang R. T cell metabolic reprogramming and plasticity. Mol Immunol (2015) 68(2 Pt C):507–12. doi: 10.1016/j.molimm.2015.07.036
49. Sun (孙 李 哲) L, Yang (杨晓 峰) X, Yuan (袁祖贻) Z, Wang (王 虹) H. Metabolic reprogramming in immune response and tissue inflammation. Arterioscler Thromb Vasc Biol (2020) 40(9):1990–2001. doi: 10.1161/ATVBAHA.120.314037
50. Zhang X, Zink F, Hezel F, Vogt J, Wachter U, Wepler M, et al. Metabolic substrate utilization in stress-induced immune cells. ICMx (2020) 8(S1):28. doi: 10.1186/s40635-020-00316-0
51. Hasan MN, Capuk O, Patel SM, Sun D. The role of metabolic plasticity of tumor-associated macrophages in shaping the tumor microenvironment immunity. Cancers (Basel) (2022) 14(14):3331. doi: 10.3390/cancers14143331
52. Ganeshan K, Chawla A. Metabolic regulation of immune responses. Annu Rev Immunol (2014) 32:609–34. doi: 10.1146/annurev-immunol-032713-120236
53. Howard NC, Khader SA. Immunometabolism during Mycobacterium tuberculosis Infection. Trends Microbiol (2020) 28(10):832–50. doi: 10.1016/j.tim.2020.04.010
54. Qualls JE, Murray PJ. Immunometabolism within the tuberculosis granuloma: amino acids, hypoxia, and cellular respiration. Semin Immunopathol (2016) 38(2):139–52. doi: 10.1007/s00281-015-0534-0
55. Liu CH, Liu H, Ge B. Innate immunity in tuberculosis: host defense vs pathogen evasion. Cell Mol Immunol (2017) 14(12):963–75. doi: 10.1038/cmi.2017.88
56. Vrieling F, Kostidis S, Spaink HP, Haks MC, Mayboroda OA, Ottenhoff THM, et al. Analyzing the impact of Mycobacterium tuberculosis infection on primary human macrophages by combined exploratory and targeted metabolomics. Sci Rep (2020) 10(1):7085. doi: 10.1038/s41598-020-62911-1
57. Gleeson LE, Sheedy FJ, Palsson-McDermott EM, Triglia D, O’Leary SM, O’Sullivan MP, et al. Cutting edge: mycobacterium tuberculosis induces aerobic glycolysis in human alveolar macrophages that is required for control of intracellular bacillary replication. J Immunol (2016) 196(6):2444–9. doi: 10.4049/jimmunol.1501612
58. Nagao A, Kobayashi M, Koyasu S, Chow CCT, Harada H. HIF-1-dependent reprogramming of glucose metabolic pathway of cancer cells and its therapeutic significance. Int J Mol Sci (2019) 20(2):238. doi: 10.3390/ijms20020238
59. Kumar R, Singh P, Kolloli A, Shi L, Bushkin Y, Tyagi S, et al. Immunometabolism of phagocytes during mycobacterium tuberculosis infection. Front Mol Biosci (2019) 6:105. doi: 10.3389/fmolb.2019.00105
60. Cumming BM, Pacl HT, Steyn AJC. Relevance of the warburg effect in tuberculosis for host-directed therapy. Front Cell Infection Microbiol (2020) 10:576596. doi: 10.3389/fcimb.2020.576596
61. Maoldomhnaigh C Ó, Cox DJ, Phelan JJ, Mitermite M, Murphy DM, Leisching G, et al. Lactate alters metabolism in human macrophages and improves their ability to kill mycobacterium tuberculosis. Front Immunol (2021) 12:663695. doi: 10.3389/fimmu.2021.663695
62. Terán G, Li H, Catrina SB, Liu R, Brighenti S, Zheng X, et al. High glucose and carbonyl stress impair HIF-1-regulated responses and the control of mycobacterium tuberculosis in macrophages. mBio (2022) 13(5):e0108622. doi: 10.1128/mbio.01086-22
63. Huang L, Nazarova EV, Tan S, Liu Y, Russell DG. Growth of Mycobacterium tuberculosis in vivo segregates with host macrophage metabolism and ontogeny. J Exp Med (2018) 215(4):1135–52. doi: 10.1084/jem.20172020
64. Ehrt S, Schnappinger D, Rhee KY. Metabolic principles of persistence and pathogenicity in Mycobacterium tuberculosis. Nat Rev Microbiol (2018) 16(8):496–507. doi: 10.1038/s41579-018-0013-4
65. Pagán AJ, Lee LJ, Edwards-Hicks J, Moens CB, Tobin DM, Busch-Nentwich EM, et al. mTOR-regulated mitochondrial metabolism limits mycobacterium-induced cytotoxicity. Cell (2022) 185(20):3720–3738.e13. doi: 10.1016/j.cell.2022.08.018
66. Refai A, Gritli S, Barbouche MR, Essafi M. Mycobacterium tuberculosis virulent factor ESAT-6 drives macrophage differentiation toward the pro-inflammatory M1 phenotype and subsequently switches it to the anti-inflammatory M2 phenotype. Front Cell Infect Microbiol (2018) 8:327. doi: 10.3389/fcimb.2018.00327
67. Jha AK, Huang SCC, Sergushichev A, Lampropoulou V, Ivanova Y, Loginicheva E, et al. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity (2015) 42(3):419–30. doi: 10.1016/j.immuni.2015.02.005
68. Sun Y, Liao Y, Xiong N, He X, Zhang H, Chen X, et al. Amino acid profiling as a screening and prognostic biomarker in active tuberculosis patients. Clinica Chimica Acta (2023) 548:117523. doi: 10.1016/j.cca.2023.117523
69. Gao Y, Li J, Guo X, Guan L, Wang J, Huang X, et al. L-tyrosine limits mycobacterial survival in tuberculous granuloma. Pathogens (2023) 12(5):654. doi: 10.3390/pathogens12050654
70. Duan T, Du Y, Xing C, Wang HY, Wang RF. Toll-like receptor signaling and its role in cell-mediated immunity. Front Immunol (2022) 13:812774. doi: 10.3389/fimmu.2022.812774
71. Luda KM, Longo J, Kitchen-Goosen SM, Duimstra LR, Ma EH, Watson MJ, et al. Ketolysis drives CD8+ T cell effector function through effects on histone acetylation. Immunity (2023) 56(9):2021–35. doi: 10.1016/j.immuni.2023.07.002
72. Zhang L, Zhang W, Li Z, Lin S, Zheng T, Hao B, et al. Mitochondria dysfunction in CD8+ T cells as an important contributing factor for cancer development and a potential target for cancer treatment: a review. J Exp Clin Cancer Res (2022) 41(1):227. doi: 10.1186/s13046-022-02439-6
73. Tzelepis F, Blagih J, Khan N, Gillard J, Mendonca L, Roy DG, et al. Mitochondrial cyclophilin D regulates T cell metabolic responses and disease tolerance to tuberculosis. Sci Immunol (2018) 3(23):eaar4135. doi: 10.1126/sciimmunol.aar4135
74. Koeken VACM, van Crevel R, Netea MG. T cell metabolism has evolved to tolerate tuberculosis. Cell Metab (2018) 28(3):332–3. doi: 10.1016/j.cmet.2018.08.015
75. Russell SL, Lamprecht DA, Mandizvo T, Jones TT, Naidoo V, Addicott KW, et al. Compromised Metabolic Reprogramming Is an Early Indicator of CD8+ T Cell Dysfunction during Chronic Mycobacterium tuberculosis Infection. Cell Rep (2019) 29(11):3564–79. doi: 10.1016/j.celrep.2019.11.034
76. Liu R, Muliadi V, Mou W, Li H, Yuan J, Holmberg J, et al. HIF-1 stabilization in T cells hampers the control of Mycobacterium tuberculosis infection. Nat Commun (2022) 13(1):5093. doi: 10.1038/s41467-022-32639-9
77. Bhaskar A, Kumar S, Khan MZ, Singh A, Dwivedi VP, Nandicoori VK. Host sirtuin 2 as an immunotherapeutic target against tuberculosis. eLife (2020) 9:e55415. doi: 10.7554/eLife.55415
78. Bhaskar A, Pahuja I, Negi K, Verma A, Ghoshal A, Mathew B, et al. SIRT2 inhibition by AGK2 enhances mycobacteria-specific stem cell memory responses by modulating beta-catenin and glycolysis. iScience (2023) 26(5):106644. doi: 10.1016/j.isci.2023.106644
79. Pahuja I, Negi K, Kumari A, Agarwal M, Mukhopadhyay S, Mathew B, et al. Berberine governs NOTCH3/AKT signaling to enrich lung-resident memory T cells during tuberculosis. PloS Pathog (2023) 19(3):e1011165. doi: 10.1371/journal.ppat.1011165
80. Sharma N, Shariq M, Quadir N, Singh J, Sheikh JA, Hasnain SE, et al. Mycobacterium tuberculosis protein PE6 (Rv0335c), a novel TLR4 agonist, evokes an inflammatory response and modulates the cell death pathways in macrophages to enhance intracellular survival. Front Immunol (2021) 12:eelo. doi: 10.3389/fimmu.2021.696491
81. Lin JH, Walter P, Yen TSB. Endoplasmic reticulum stress in disease pathogenesis. Annu Rev Pathol (2008) 3:399–425. doi: 10.1146/annurev.pathmechdis.3.121806.151434
82. Seimon TA, Kim MJ, Blumenthal A, Koo J, Ehrt S, Wainwright H, et al. Induction of ER stress in macrophages of tuberculosis granulomas. PloS One (2010) 5(9):e12772. doi: 10.1371/journal.pone.0012772
83. Choi HH, Shin DM, Kang G, Kim KH, Park JB, Hur GM, et al. Endoplasmic reticulum stress response is involved in Mycobacterium tuberculosis protein ESAT-6-mediated apoptosis. FEBS Lett (2010) 584(11):2445–54. doi: 10.1016/j.febslet.2010.04.050
84. Lim YJ, Choi JA, Choi HH, Cho SN, Kim HJ, Jo EK, et al. Endoplasmic reticulum stress pathway-mediated apoptosis in macrophages contributes to the survival of Mycobacterium tuberculosis. PloS One (2011) 6(12):e28531. doi: 10.1371/journal.pone.0028531
85. Jo SH, Choi JA, Lim YJ, Lee J, Cho SN, Oh SM, et al. Calreticulin modulates the intracellular survival of mycobacteria by regulating ER-stress-mediated apoptosis. Oncotarget (2017) 8(35):58686–98. doi: 10.18632/oncotarget.17419
86. Xu P, Tang J, He ZG. Induction of endoplasmic reticulum stress by cdhM mediates apoptosis of macrophage during mycobacterium tuberculosis infection. Front Cell Infect Microbiol (2022) 12:877265. doi: 10.3389/fcimb.2022.877265
87. Grover S, Sharma T, Singh Y, Kohli S, Singh A, Manjunath P, et al. The PGRS Domain of Mycobacterium tuberculosis PE_PGRS Protein Rv0297 Is Involved in Endoplasmic Reticulum Stress-Mediated Apoptosis through Toll-Like Receptor 4. mBio (2018) 9(3):e01017-18. doi: 10.1128/mBio.01017-18
88. Campuzano J, Aguilar D, Arriaga K, León JC, Salas-Rangel LP, González-y-Merchand J, et al. The PGRS domain of Mycobacterium tuberculosis PE_PGRS Rv1759c antigen is an efficient subunit vaccine to prevent reactivation in a murine model of chronic tuberculosis. Vaccine (2007) 25(18):3722–9. doi: 10.1016/j.vaccine.2006.12.042
89. Child JR, Chen Q, Reid DW, Jagannathan S, Nicchitta CV. Recruitment of endoplasmic reticulum-targeted and cytosolic mRNAs into membrane-associated stress granules. RNA (2021) 27(10):1241–56. doi: 10.1261/rna.078858.121
90. Li WY, Yang F, Li X, Wang LW, Wang Y. Stress granules inhibit endoplasmic reticulum stress-mediated apoptosis during hypoxia-induced injury in acute liver failure. World J Gastroenterol (2023) 29(8):1315–29. doi: 10.3748/wjg.v29.i8.1315
91. Verma A, Ghoshal A, Dwivedi VP, Bhaskar A. Tuberculosis: The success tale of less explored dormant Mycobacterium tuberculosis. Front Cell Infect Microbiol (2022) 12:1079569. doi: 10.3389/fcimb.2022.1079569
92. Rastogi S, Mohanty S, Sharma S, Tripathi P. Possible role of gut microbes and host’s immune response in gut-lung homeostasis. Front Immunol (2022) 13:954339. doi: 10.3389/fimmu.2022.954339
93. Malard F, Dore J, Gaugler B, Mohty M. Introduction to host microbiome symbiosis in health and disease. Mucosal Immunol (2021) 14(3):547–54. doi: 10.1038/s41385-020-00365-4
94. Costantini C, Nunzi E, Romani L. From the nose to the lungs: the intricate journey of airborne pathogens amid commensal bacteria. Am J Physiology-Cell Physiol (2022) 323(4):C1036–43. doi: 10.1152/ajpcell.00287.2022
95. Haldar S, Jadhav SR, Gulati V, Beale DJ, Balkrishna A, Varshney A, et al. Unravelling the gut-lung axis: insights into microbiome interactions and Traditional Indian Medicine’s perspective on optimal health. FEMS Microbiol Ecol (2023) 99(10):fiad103. doi: 10.1093/femsec/fiad103
96. Comberiati P, Di Cicco M, Paravati F, Pelosi U, Di Gangi A, Arasi S, et al. The role of gut and lung microbiota in susceptibility to tuberculosis. Int J Environ Res Public Health (2021) 18(22):12220. doi: 10.3390/ijerph182212220
97. Shah T, Shah Z, Baloch Z, Cui X. The role of microbiota in respiratory health and diseases, particularly in tuberculosis. BioMed Pharmacother (2021) 143:112108. doi: 10.1016/j.biopha.2021.112108
98. Huang Y, Tang Jh, Cai Z, Qi Y, Jiang S, Ma Tt, et al. Alterations in the nasopharyngeal microbiota associated with active and latent tuberculosis. Tuberculosis (2022) 136:102231. doi: 10.1016/j.tube.2022.102231
99. Ruiz-Tagle C, Ugalde JA, Naves R, Araos R, García P, Balcells ME. Reduced microbial diversity of the nasopharyngeal microbiome in household contacts with latent tuberculosis infection. Sci Rep (2023) 13(1):7301. doi: 10.1038/s41598-023-34052-8
100. Botero LE, Delgado-Serrano L, Cepeda ML, Bustos JR, Anzola JM, Del Portillo P, et al. Respiratory tract clinical sample selection for microbiota analysis in patients with pulmonary tuberculosis. Microbiome (2014) 2(1):29. doi: 10.1186/2049-2618-2-29
101. Sala C, Benjak A, Goletti D, Banu S, Mazza-Stadler J, Jaton K, et al. Multicenter analysis of sputum microbiota in tuberculosis patients. PloS One (2020) 15(10):e0240250. doi: 10.1371/journal.pone.0240250
102. Dickson RP, Erb-Downward JR, Martinez FJ, Huffnagle GB. The microbiome and the respiratory tract. Annu Rev Physiol (2016) 78:481–504. doi: 10.1146/annurev-physiol-021115-105238
103. Hu Y, Cheng M, Liu B, Dong J, Sun L, Yang J, et al. Metagenomic analysis of the lung microbiome in pulmonary tuberculosis - a pilot study. Emerg Microbes Infect (2020) 9(1):1444–52. doi: 10.1080/22221751.2020.1783188
104. Vázquez-Pérez JA, Carrillo CO, Iñiguez-García MA, Romero-Espinoza I, Márquez-García JE, Falcón LI, et al. Alveolar microbiota profile in patients with human pulmonary tuberculosis and interstitial pneumonia. Microbial Pathogenesis (2020) 139:103851. doi: 10.1016/j.micpath.2019.103851
105. Hu Y, Kang Y, Liu X, Cheng M, Dong J, Sun L, et al. Distinct lung microbial community states in patients with pulmonary tuberculosis. Sci China Life Sci (2020) 63(10):1522–33. doi: 10.1007/s11427-019-1614-0
106. Zhou Y, Lin F, Cui Z, Zhang X, Hu C, Shen T, et al. Correlation between either cupriavidus or porphyromonas and primary pulmonary tuberculosis found by analysing the microbiota in patients’ Bronchoalveolar lavage fluid. Cardona PJ editor. PloS One (2015) 10(5):e0124194. doi: 10.1371/journal.pone.0124194
107. Xia X, Chen J, Cheng Y, Chen F, Lu H, Liu J, et al. Comparative analysis of the lung microbiota in patients with respiratory infections, tuberculosis, and lung cancer: A preliminary study. Front Cell Infect Microbiol (2022) 12:1024867. doi: 10.3389/fcimb.2022.1024867
108. Hong By, Paulson JN, Stine OC, Weinstock GM, Cervantes JL. Meta-analysis of the lung microbiota in pulmonary tuberculosis. Tuberculosis (2018) 109:102–8. doi: 10.1016/j.tube.2018.02.006
109. Bull MJ. Plummer NT. Part 1: the human gut microbiome in health and disease. Integr Med (Encinitas) (2014) 13(6):17–22.
110. Barbosa-Amezcua M, Galeana-Cadena D, Alvarado-Peña N, Silva-Herzog E. The microbiome as part of the contemporary view of tuberculosis disease. Pathogens (2022) 11(5):584. doi: 10.3390/pathogens11050584
111. Pant A, Das B, Arimbasseri GA. Host microbiome in tuberculosis: disease, treatment, and immunity perspectives. Front Microbiol (2023) 14:1236348. doi: 10.3389/fmicb.2023.1236348
112. van Deuren T, Blaak EE, Canfora EE. Butyrate to combat obesity and obesity-associated metabolic disorders: Current status and future implications for therapeutic use. Obes Rev (2022) 23(10):e13498. doi: 10.1111/obr.13498
113. Khaliq A, Ravindran R, Afzal S, Jena PK, Akhtar MW, Ambreen A, et al. Gut microbiome dysbiosis and correlation with blood biomarkers in active-tuberculosis in endemic setting. PloS One (2021) 16(1):e0245534. doi: 10.1371/journal.pone.0245534
114. Fujimura KE, Lynch SV. Microbiota in allergy and asthma and the emerging relationship with the gut microbiome. Cell Host Microbe (2015) 17(5):592–602. doi: 10.1016/j.chom.2015.04.007
115. Negatu DA, Yamada Y, Xi Y, Go ML, Zimmerman M, Ganapathy U, et al. Gut microbiota metabolite indole propionic acid targets tryptophan biosynthesis in mycobacterium tuberculosis. mBio (2019) 10(2):e02781–18. doi: 10.1128/mBio.02781-18
116. Wang Z, Hua W, Li C, Chang H, Liu R, Ni Y, et al. Protective role of fecal microbiota transplantation on colitis and colitis-associated colon cancer in mice is associated with treg cells. Front Microbiol (2019) 10:2498. doi: 10.3389/fmicb.2019.02498
117. Liu X, Cheng Y, Zang D, Zhang M, Li X, Liu D, et al. The role of gut microbiota in lung cancer: from carcinogenesis to immunotherapy. Front Oncol (2021) 11:720842. doi: 10.3389/fonc.2021.720842
118. Naidoo CC, Nyawo GR, Wu BG, Walzl G, Warren RM, Segal LN, et al. The microbiome and tuberculosis: state of the art, potential applications, and defining the clinical research agenda. Lancet Respir Med (2019) 7(10):892–906. doi: 10.1016/S2213-2600(18)30501-0
119. Segal LN, Clemente JC, Li Y, Ruan C, Cao J, Danckers M, et al. Anaerobic bacterial fermentation products increase tuberculosis risk in antiretroviral-drug-treated HIV patients. Cell Host Microbe (2017) 21(4):530–7. doi: 10.1016/j.chom.2017.03.003
120. Negi S, Pahari S, Bashir H, Agrewala JN. Gut microbiota regulates mincle mediated activation of lung dendritic cells to protect against mycobacterium tuberculosis. Front Immunol (2019) 10:1142. doi: 10.3389/fimmu.2019.01142
121. Yang F, Yang Y, Chen L, Zhang Z, Liu L, Zhang C, et al. The gut microbiota mediates protective immunity against tuberculosis via modulation of lncRNA. Gut Microbes (2022) 14(1):2029997. doi: 10.1080/19490976.2022.2029997
122. Wipperman MF, Fitzgerald DW, Juste MAJ, Taur Y, Namasivayam S, Sher A, et al. Antibiotic treatment for Tuberculosis induces a profound dysbiosis of the microbiome that persists long after therapy is completed. Sci Rep (2017) 7(1):10767. doi: 10.1038/s41598-017-10346-6
123. Wiggins KB, Smith MA, Schultz-Cherry S. The nature of immune responses to influenza vaccination in high-risk populations. Viruses (2021) 13(6):1109. doi: 10.3390/v13061109
124. Lynn DJ, Benson SC, Lynn MA, Pulendran B. Modulation of immune responses to vaccination by the microbiota: implications and potential mechanisms. Nat Rev Immunol (2022) 22(1):33–46. doi: 10.1038/s41577-021-00554-7
125. Hagan T, Cortese M, Rouphael N, Boudreau C, Linde C, Maddur MS, et al. Antibiotics-driven gut microbiome perturbation alters immunity to vaccines in humans. Cell (2019) 178(6):1313–28. doi: 10.1016/j.cell.2019.08.010
126. Collins N, Belkaid Y. Do the microbiota influence vaccines and protective immunity to pathogens?: engaging our endogenous adjuvants. Cold Spring Harb Perspect Biol (2018) 10(2):a028860. doi: 10.1101/cshperspect.a028860
127. Zheng D, Liwinski T, Elinav E. Interaction between microbiota and immunity in health and disease. Cell Res (2020) 30(6):492–506. doi: 10.1038/s41422-020-0332-7
128. Nadeem S, Maurya SK, Das DK, Khan N, Agrewala JN. Gut dysbiosis thwarts the efficacy of vaccine against mycobacterium tuberculosis. Front Immunol (2020) 11:726. doi: 10.3389/fimmu.2020.00726
129. Stražar M, Mourits VP, Koeken VACM, De Bree LCJ, Moorlag SJCFM, Joosten LAB, et al. The influence of the gut microbiome on BCG-induced trained immunity. Genome Biol (2021) 22(1):275. doi: 10.1186/s13059-021-02482-0
130. Huda MN, Lewis Z, Kalanetra KM, Rashid M, Ahmad SM, Raqib R, et al. Stool microbiota and vaccine responses of infants. Pediatrics (2014) 134(2):e362–72. doi: 10.1542/peds.2013-3937
131. Qu S, Gao Y, Ma J, Yan Q. Microbiota-derived short-chain fatty acids functions in the biology of B lymphocytes: From differentiation to antibody formation. Biomedicine Pharmacotherapy (2023) 168:115773. doi: 10.1016/j.biopha.2023.115773
132. Schaupp L, Muth S, Rogell L, Kofoed-Branzk M, Melchior F, Lienenklaus S, et al. Microbiota-induced type I interferons instruct a poised basal state of dendritic cells. Cell (2020) 181(5):1080–96. doi: 10.1016/j.cell.2020.04.022
133. Zimmermann P. The immunological interplay between vaccination and the intestinal microbiota. NPJ Vaccines (2023) 8(1):24. doi: 10.1038/s41541-023-00627-9
134. Zhou B, Xu K, Zheng X, Chen T, Wang J, Song Y, et al. Application of exosomes as liquid biopsy in clinical diagnosis. Signal Transduct Target Ther (2020) 5(1):144. doi: 10.1038/s41392-020-00258-9
135. Goletti D, Petruccioli E, Joosten SA, Ottenhoff THM. Tuberculosis biomarkers: from diagnosis to protection. Infect Dis Rep (2016) 8(2):6568. doi: 10.4081/idr.2016.6568
136. Huda MN, Nurunnabi M. Potential application of exosomes in vaccine development and delivery. Pharm Res (2022) 39(11):2635–71. doi: 10.1007/s11095-021-03143-4
137. Rodriguez GM, Prados-Rosales R. Functions and importance of mycobacterial extracellular vesicles. Appl Microbiol Biotechnol (2016) 100(9):3887–92. doi: 10.1007/s00253-016-7484-x
138. Zheng W, LaCourse SM, Song B, Singh DK, Khanna M, Olivo J, et al. Diagnosis of paediatric tuberculosis by optically detecting two virulence factors on extracellular vesicles in blood samples. Nat BioMed Eng (2022) 6(8):979–91. doi: 10.1038/s41551-022-00922-1
139. Mehaffy C, Dobos KM, Nahid P, Kruh-Garcia NA. Second generation multiple reaction monitoring assays for enhanced detection of ultra-low abundance Mycobacterium tuberculosis peptides in human serum. Clin Proteom (2017) 14(1):21. doi: 10.1186/s12014-017-9156-y
140. Mehaffy C, Kruh-Garcia NA, Graham B, Jarlsberg LG, Willyerd CE, Borisov A, et al. Identification of mycobacterium tuberculosis peptides in serum extracellular vesicles from persons with latent tuberculosis infection. J Clin Microbiol (2020) 58(6):e00393–20. doi: 10.1128/JCM.00393-20
141. Alipoor SD, Tabarsi P, Varahram M, Movassaghi M, Dizaji MK, Folkerts G, et al. Serum exosomal miRNAs are associated with active pulmonary tuberculosis. Dis Markers (2019) 2019:1–9. doi: 10.1155/2019/1907426
142. Lyu L, Zhang X, Li C, Yang T, Wang J, Pan L, et al. Small RNA profiles of serum exosomes derived from individuals with latent and active tuberculosis. Front Microbiol (2019) 10:1174. doi: 10.3389/fmicb.2019.01174
143. Hashimoto S, Zhao H, Hayakawa M, Nakajima K, Taguchi Y h, Murakami Y. Developing a diagnostic method for latent tuberculosis infection using circulating miRNA. Transl Med Commun (2020) 5(1):25. doi: 10.1186/s41231-020-00078-7
144. Beetler DJ, Di Florio DN, Bruno KA, Ikezu T, March KL, Cooper LT, et al. Extracellular vesicles as personalized medicine. Mol Aspects Med (2023) 91:101155. doi: 10.1016/j.mam.2022.101155
145. Arya R, Dabral D, HMd F, Mazumdar H, SJ P, Deka T, et al. Serum small extracellular vesicles proteome of tuberculosis patients demonstrated deregulated immune response. Proteomics Clin Apps (2020) 14(1):1900062. doi: 10.1002/prca.201900062
146. Railwah C, Lora A, Zahid K, Goldenberg H, Campos M, Wyman A, et al. Cigarette smoke induction of S100A9 contributes to chronic obstructive pulmonary disease. Am J Physiol Lung Cell Mol Physiol (2020) 319(6):L1021–35. doi: 10.1152/ajplung.00207.2020
147. Franz S, Ertel A, Engel KM, Simon JC, Saalbach A. Overexpression of S100A9 in obesity impairs macrophage differentiation via TLR4-NFkB-signaling worsening inflammation and wound healing. Theranostics (2022) 12(4):1659–82. doi: 10.7150/thno.67174
148. Du Y, Xin H, Cao X, Liu Z, He Y, Zhang B, et al. Association between plasma exosomes S100A9/C4BPA and latent tuberculosis infection treatment: proteomic analysis based on a randomized controlled study. Front Microbiol (2022) 13:934716. doi: 10.3389/fmicb.2022.934716
149. Wu M, Yang Q, Yang C, Han J, Liu H, Qiao L, et al. Characteristics of plasma exosomes in drug-resistant tuberculosis patients. Tuberculosis (2023) 141:102359. doi: 10.1016/j.tube.2023.102359
150. Patel K, Nagel M, Wesolowski M, Dees S, Rivera-Milla E, Geldmacher C, et al. Evaluation of a urine-based rapid molecular diagnostic test with potential to be used at point-of-care for pulmonary tuberculosis. J Mol Diagnostics (2018) 20(2):215–24. doi: 10.1016/j.jmoldx.2017.11.005
151. Labugger I, Heyckendorf J, Dees S, Häussinger E, Herzmann C, Kohl TA, et al. Detection of transrenal DNA for the diagnosis of pulmonary tuberculosis and treatment monitoring. Infection (2017) 45(3):269–76. doi: 10.1007/s15010-016-0955-2
152. Fox-Fisher I, Shemer R, Dor Y. Epigenetic liquid biopsies: a novel putative biomarker in immunology and inflammation. Trends Immunol (2023) 44(5):356–64. doi: 10.1016/j.it.2023.03.005
153. Chang A, Loy CJ, Lenz JS, Steadman A, Andama A, Nhung NV, et al. Circulating cell-free RNA in blood as a host response biomarker for the detection of tuberculosis [Internet]. Infect Dis (except HIV/AIDS) (2023). doi: 10.1101/2023.01.11.23284433
154. Thakku SG, Lirette J, Murugesan K, Chen J, Theron G, Banaei N, et al. Genome-wide tiled detection of circulating Mycobacterium tuberculosis cell-free DNA using Cas13. Nat Commun (2023) 14(1):1803. doi: 10.1038/s41467-023-37183-8
155. Yadav D, Malviya R. Exploring potential of exosomes drug delivery system in the treatment of cancer: Advances and prospective. Med Drug Discovery (2023) 20:100163. doi: 10.1016/j.medidd.2023.100163
156. Sun YF, Pi J, Xu JF. Emerging role of exosomes in tuberculosis: from immunity regulations to vaccine and immunotherapy. Front Immunol (2021) 12:628973. doi: 10.3389/fimmu.2021.628973
157. Cheng Y, Schorey JS. Exosomes carrying mycobacterial antigens can protect mice against Mycobacterium tuberculosis infection. Eur J Immunol (2013) 43(12):3279–90. doi: 10.1002/eji.201343727
158. Alvarez-Jiménez VD, Leyva-Paredes K, García-Martínez M, Vázquez-Flores L, García-Paredes VG, Campillo-Navarro M, et al. Extracellular vesicles released from mycobacterium tuberculosis-infected neutrophils promote macrophage autophagy and decrease intracellular mycobacterial survival. Front Immunol (2018) 9:272. doi: 10.3389/fimmu.2018.00272
159. Cheng Y, Schorey JS. Targeting soluble proteins to exosomes using a ubiquitin tag. Biotech Bioengineering (2016) 113(6):1315–24. doi: 10.1002/bit.25884
160. Hao S, Bai O, Li F, Yuan J, Laferte S, Xiang J. Mature dendritic cells pulsed with exosomes stimulate efficient cytotoxic T-lymphocyte responses and antitumour immunity. Immunology (2007) 120(1):90–102. doi: 10.1111/j.1365-2567.2006.02483.x
161. Hao S, Liu Y, Yuan J, Zhang X, He T, Wu X, et al. Novel exosome-targeted CD4+ T cell vaccine counteracting CD4+25+ Regulatory T cell-mediated immune suppression and stimulating efficient central memory CD8+ CTL responses. J Immunol (2007) 179(5):2731–40. doi: 10.4049/jimmunol.179.5.2731
162. Jesus S, Soares E, Cruz MT, Borges O. Exosomes as adjuvants for the recombinant hepatitis B antigen: First report. Eur J Pharmaceutics Biopharmaceutics (2018) 133:1–11. doi: 10.1016/j.ejpb.2018.09.029
163. Juffermans NP, Dekkers PEP, Peppelenbosch MP, Speelman P, Van Deventer SJH, van der Poll T. Expression of the chemokine receptors CXCR1 and CXCR2 on granulocytes in human endotoxemia and tuberculosis: involvement of the p38 mitogen–activated protein kinase pathway. J Infect Dis (2000) 182(3):888–94. doi: 10.1086/315750
Keywords: Mycobacterium tuberculosis, immunometabolism, ER stress, microbiome, exosomes, cell-free DNA, biomarkers
Citation: Ghoshal A, Verma A, Bhaskar A and Dwivedi VP (2024) The uncharted territory of host-pathogen interaction in tuberculosis. Front. Immunol. 15:1339467. doi: 10.3389/fimmu.2024.1339467
Received: 17 November 2023; Accepted: 03 January 2024;
Published: 19 January 2024.
Edited by:
T. Mark Doherty, GlaxoSmithKline, BelgiumReviewed by:
Tousif Sultan, University of Alabama at Birmingham, United StatesCopyright © 2024 Ghoshal, Verma, Bhaskar and Dwivedi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ved Prakash Dwivedi, dmVkcHJha2FzaGJ0QGdtYWlsLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.