- 1Molecular Virology Lab, Atta-Ur-Rahman School of Applied Biosciences (ASAB), National University of Science & Technology (NUST), Islamabad, Pakistan
- 2Department of Community Dentistry, Islamabad Medical and Dental College (IMDC), Islamabad, Pakistan
- 3Institute of Dentistry, University of Eastern Finland (UEF), Kuopio, Finland
Many studies have investigated the antiviral activity of cytokines, including interleukin-6 (IL-6), interleukin-22 (IL-22), interleukin-32 gamma (IL-32γ), and interferon-lambda (IFN-λ) in diverse populations. This study aims to evaluate the role of these cytokines in inhibition of various human and animal viruses when administered exogenously. A comprehensive meta-analysis and systematic review were conducted on all the relevant studies from three databases. Standard mean differences (SMDs) of overall viral inhibition were used to generate the difference in the antiviral efficacy of these cytokines between control and experimental groups. A total of 4,618 abstracts for IL-6, 3,517 abstracts for IL-22, 2,160 abstracts for IL-32γ, and 1,026 abstracts for IFN-λ were identified, and 7, 4, 8, and 35 studies were included, respectively, for each cytokine. IFN-λ (SMD = 0.9540; 95% CI: 0.69–0.22) and IL-32γ (SMD = 0.459; 95% CI: 0.02–0.90) showed the highest influence followed by IL-6 (SMD = 0.456; CI: −0.04–0.95) and IL-22 (SMD = 0.244; 95% CI: −0.33–0.81). None of the cytokines represented heterogeneity (tau² > 0), but only IFN-λ indicated the funnel plot asymmetry (p = 0.0097). Results also indicated that IFN-λ and IL-32γ are more potent antivirals than IL-6 and IL-22. The collective findings of this study emphasize that exogenously administered pro-inflammatory cytokines, specifically IFN-λ and IL-32, exhibit a significant antiviral activity, thereby underscoring them as potent antiviral agents. Nonetheless, additional research is required to ascertain their clinical utility and potential for integration into combinatorial therapeutic regimens against viral infections.
Introduction
Viruses cause infections in human beings including adults and children. Behind most of the infections, the culprits are viruses and are far more frequent than bacterial, fungal, and other infections (1). On average, children suffer from viral respiratory infection around 10 times a year during early childhood (2). The spectrum of afflictions caused by viruses’ range from benign illnesses like common cold, flu (3), and warts (4) to severe diseases like acquired immune deficiency syndrome (5), coronavirus disease 2019 (COVID-19) (6), Influenza (7), and Ebola (8). Viruses are mainly composed of genetic information enclosed in proteins with keys to unlock the gateways of a specific or wide range of host cells. Researchers have so far identified approximately 270 species of viruses that are pathogenic to humans, but many are undiscovered (9).
In terms of their pathogenicity, viruses follow both acute and chronic courses. Manifestations of acute viral infections range from mild symptoms like cough, runny nose, and mild fever due to Rhinovirus, to severe morbidities like hemorrhagic fever, bleeding, persistent pain in the chest, shortness of breath, caused by dengue virus, and coronavirus 2. On the other hand, there are viruses capable of integrating themselves into the human genome and staying dormant, without showing any signs and symptoms for a while, leading to a chronic infection or carrier state. Example of these chronic viral pathogens include Epstein–Barr virus (10) and cytomegalovirus (CMV) (11). Whereas, some continue to express their genes at low concentrations and suddenly rise, e.g., human papillomavirus, hepatitis B virus (HBV), and hepatitis C virus (HCV) (12).
Antibiotics are no solutions to rescue human beings from viruses (13) although some antiviral drugs are available for the cure. Food and Drug Administration (FDA) has approved mostly small molecules and some large molecules such as oligonucleotides, interferons, and monoclonal antibodies against viral infections (14). Most of the antiviral drugs are inhibitors or base analogs to inhibit viral replication and gene expression (15). Despite the effectiveness of the approved antiviral drugs, there are several viral infections that cannot be cured with them, for example, influenza has developed the resistance against its PA inhibitor baloxavir marboxil (16), as well as ganciclovar resistance in transplant patients against CMV (17). Moreover, no or poor response, relapse, drug resistance mutation, narrow spectrum targets, and harmful side effects demand alternative strategies (18).
The immune system provides natural defense against viruses by activating innate (19) and adaptive (20) mechanisms, especially in eliminating acute viral infections, making them last for a week approximately (21). However, during chronic viral infections, the persistence of the virus results in desensitization and exhaustion of the immune system. The combination of antigen-presenting cells, phagocytes, and cytokines; the bridge between the cell-mediated and humoral immune system; and lymphocytes form an army to fight against the virus (22). Despite the powerful defense mechanism, viruses have developed different escape strategies by constantly evolving themselves via antigenic shift and drift.
Immunotherapy is an advanced field to accelerate the natural defense mechanism against viruses in which scientists have exploited different components of the immune system (23). Cytokine immunotherapy involves the exogenous intake of either cytokine expression vector or recombinant cytokines, either in vitro, in vivo, or both. These cytokines have been observed in inducing an effective antiviral state. Pro-inflammatory cytokines like type 1 interferons (IFNs), interleukin-1β (IL-1β), interleukin-6 (IL-6), interleukin-18 (IL-18), and tumor necrosis factor–α (TNF-α) can enhance viral neutralization, reduction in viral replication, and apoptosis of viral infected cells (24). The exact mechanism with most cytokines protects cells or a living system from viruses is yet to be fully understood. Growing evidence reports that they enhance antigen presentation by Major Histocompatibility Complex (MHC) molecules and stimulate cell-mediated immune cells such as natural killer cells, cytotoxic T cells, and CD8+ T cells. A direct antiviral state can also be achieved by some cytokines that enhance virus killing and virus-infected cell apoptosis (25).
There are insufficient data available on other pro-inflammatory cytokines that are used, but IFN-λ, IL-32γ, IL-6, and IL-22 have been extensively reviewed to determine their exogenous antiviral activity against different viruses. In order to obtain the valuable significance of such experimental projects, here, we performed a systematic review and meta-analysis of these studies. Pooling data from several studies to make a literature review sometimes leads to biasness of the subject. This meta-analysis presents a systematic and absolute conclusion on the antiviral effect of the mentioned cytokines through statistical analysis. As commercially available FDA-approved antiviral drugs are not effective against all viral diseases, the administration of these cytokines presents an excellent alternative medicine. The objective of this meta-analysis is to evaluate whether these selected cytokines can be either an alternative or combinatorial medicine.
Methods
Study design
This systematic review and meta-analysis was carried out in the light of Cochrane Handbook principles (26) and Preferred Reporting Items for Systematic Reviews and Meta-Analysis (27). Four separate sets of analyses were used to investigate the antiviral activity of each interleukin.
Study selection strategy
The search for the relevant studies began through major databases including Google Scholar, PubMed, and ScienceDirect. Only research papers from 1 January 2000 to 1 January 2023 were opted for and searched with the specific terms using advanced setting. The terms were decided with mutual agreement by all the authors as the chances were high to locate the required information (find below in inclusion criteria) in the articles mentioned with them. Separate searches were carried out using specific terms in the title of articles, “interleukin-28, interleukin-29, and interferon-lambda (as it is called with all the three names), and interleukin-22, interleukin 6, and interleukin-32” or their abbreviations “IL- 28, IL-29, and IFN-λ, and IL-22, IL-6, and IL-32” with “antiviral” or “virus” for each cytokine individually.
Three authors (Hina Kausar, Sarah Aqil, and R. Mudassir) were assigned to review all the titles, abstracts, and full texts to evaluate whether the selected articles meet the inclusion criteria for data extraction. In case of any disagreement about the article to be included or excluded, the corresponding author (Sobia Manzoor) reviewed the study and made the decision.
Inclusion criteria
Research articles from the year 2000 to 2023 were included to report recent progress in the relevant topic with experimental studies in vitro (primary and secondary cell cultures) or in vivo (animal models; mice, etc.) were included. The aim of the study could only be accomplished where the selected research must contain the experiments where cell culture(s) or an animal model were infected with any human virus and treated with any of the selected cytokines. Therefore, the inclusion of only in vitro or in vivo studies was crucial. Only the experimental studies utilizing exogenous IFN-λ, IL-6, IL-22, and IL-32γ (either recombinant proteins or expression vectors), treatment of pre– or post–human viral infections. Viral titers (quantity of viral DNA/RNA or/and viral proteins) must be mentioned as mean of the results with SEM (standard error of mean).
Exclusion criteria
Review articles, retrospective studies, and articles carrying endogenous antiviral effect of cytokines and animal and plant viruses were not included. The animal and plant systems are quite different from the human system in terms of the receptors that bind to viruses and also the responses that they generate during a viral infection; therefore, they should be separately reviewed and must not be included in this review. Articles following synergistic roles of selected cytokines with other compounds were also excluded. If other compounds are also used in combination with one of the selected cytokines to evaluate their antiviral potential, then it is difficult to report whether which one of these was truly responsible to generate that response. In addition, if the reduction in viral titer is due to their synergy, then the aim of the review is not justified, which is attributed to only the IFN-λ, IL-6, IL-22, and IL-32γ.
Data extraction
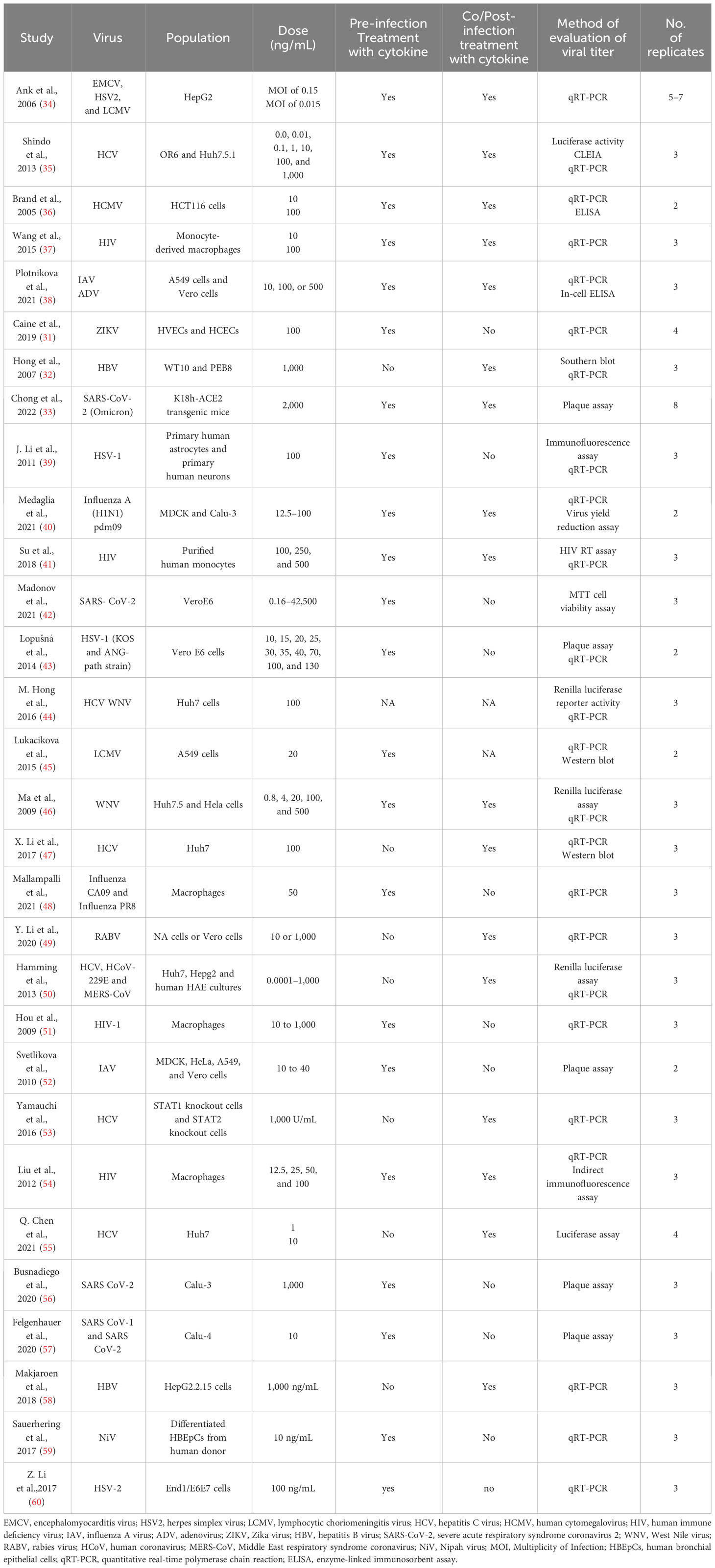
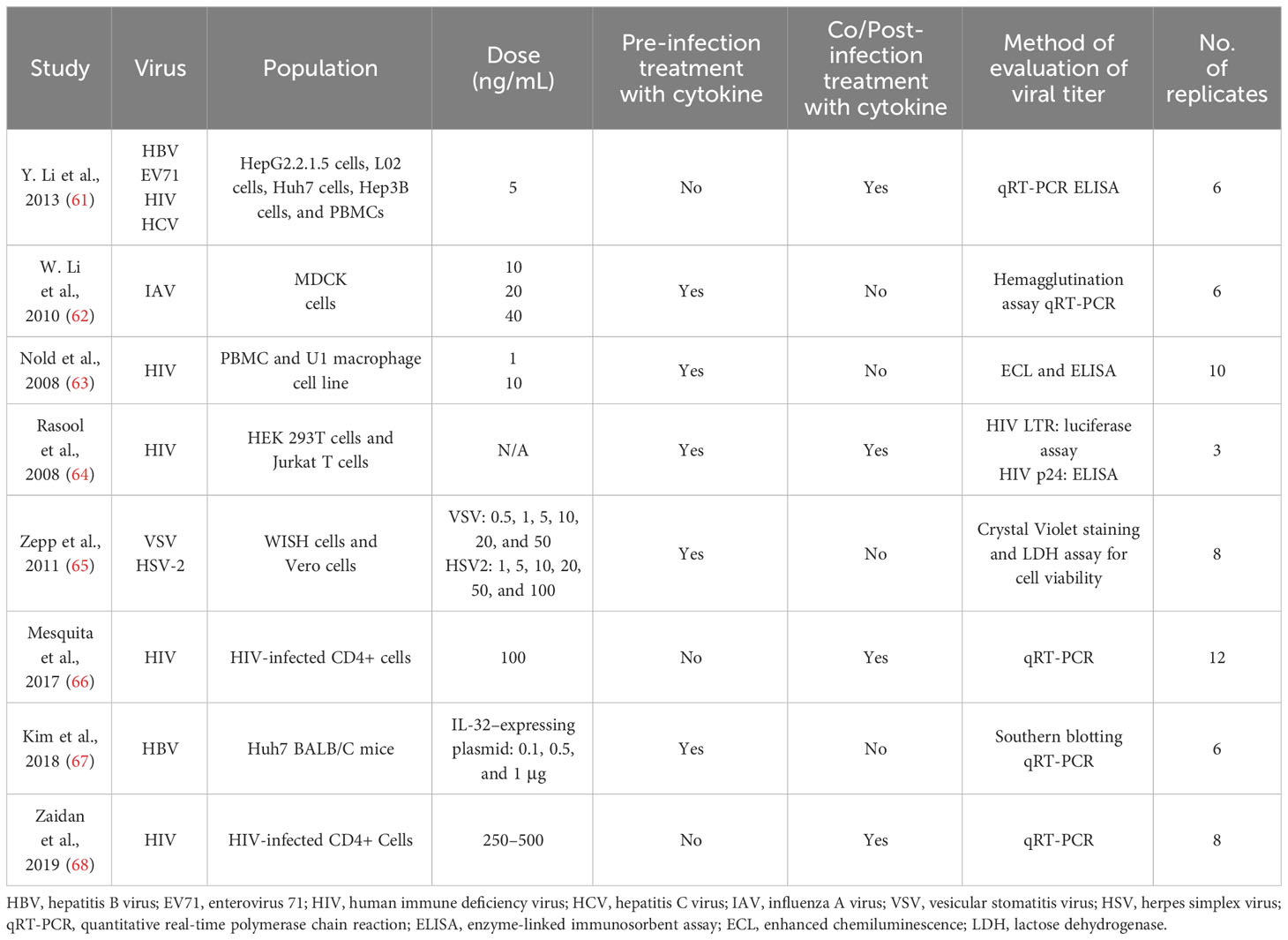
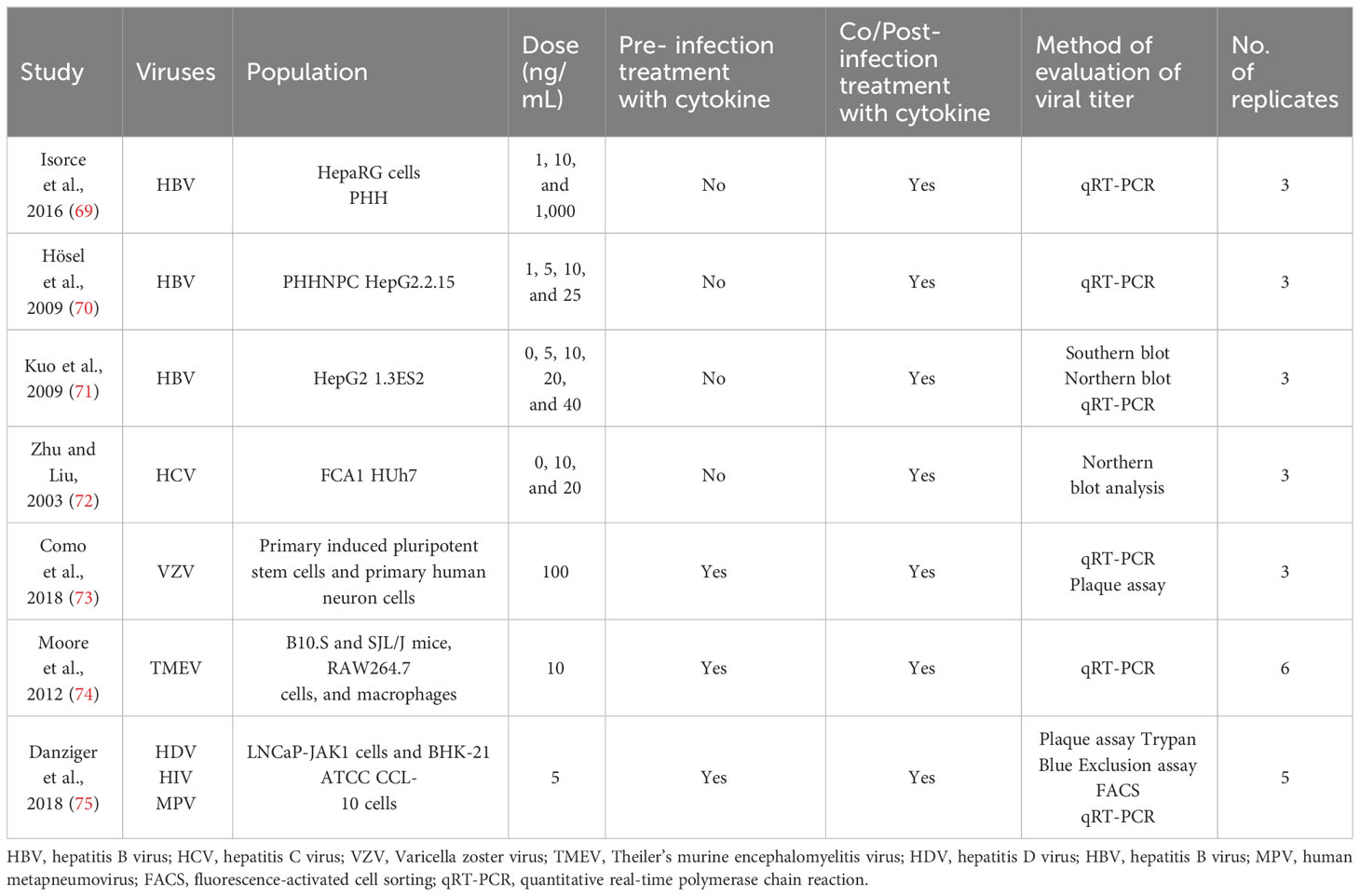
Articles that were present in duplication were assessed by Zotero to use it only once (28). Separate data sheets were created for all of the four cytokines. They included different columns, study name and year, name of viruses, name of cell lines/animal model, pre- or post-infection treatment with cytokines, cytokine dose, techniques for measuring viral titter, mean viral titers in the control (untreated), and cytokine-treated group and number of repetition of experiments. The mean viral titers were measured through the graphs present in the studies. Studies with multiple viruses were divided into separate rows. Standard deviation was calculated with the given mean and number of replicates.
Statistical analysis
The analysis was carried out on Jamovi version 2.3.28 (29), using the standardized mean difference as the outcome measure (30). The statistical significance was measured through z-test. A random-effects model was fitted to the data. The amount of heterogeneity (i.e., tau2) was estimated using restricted maximum likelihood estimator (31). In addition, to estimate of tau2, the Q-test for heterogeneity (32) and the I2 statistics are reported. In case any amount of heterogeneity detected (i.e., tau2 > 0, regardless of the results of Q-test), a prediction interval for the true outcome was also provided. Studentized residuals and Cook’s distances were used to examine whether studies may be outliers and/or influential in the context of our specified model. Studies with studentized residual larger than 100 × (1 − 0.05/2 × k)th percentile of a standard normal distribution were considered potential outliers (i.e., using Bonferroni correction with two-sided alpha = 0.05 for K studies included in the meta-analysis). Studies with a Cook’s distance larger than the median plus six times the interquartile range of the Cook’s distances were considered to be influential. The rank correlation test and the regression test, using the standard error of the observed outcomes as the predictor, are used to check funnel plot asymmetry (33).
Results
The advanced search on each data base revealed 1,026 abstracts for IFN-λ, 2,160 for IL-32γ, 4,618 for IL-6, and 3,517 for IL-22, collectively. A total of 956 abstracts for IFN-λ, 2,150 for IL-32γ, 4,606 for IL-6, and 3,505 for IL-22 were excluded due to the absence of exogenous treatment of these cytokines to evaluate their antiviral potential. Most of the excluded articles were about innate antiviral response of cytokines, especially in the case of IL-6, and others did not include the information about viral quantification or number of antiviral experiments. Subsequently, a total of 36 full texts for IFN-λ, 2 for IL-32γ, 5 for IL-6, and 8 for IL-22 were excluded. The included studies contained the viral titers and number of experiments, following the inclusion criteria. The greatest number of studies found was 32 for IFN-λ, then 8 for IL-32γ, 7 for IL-6, and 4 for IL-22. The inclusion and exclusion breakdown has been shown in Figure 1.
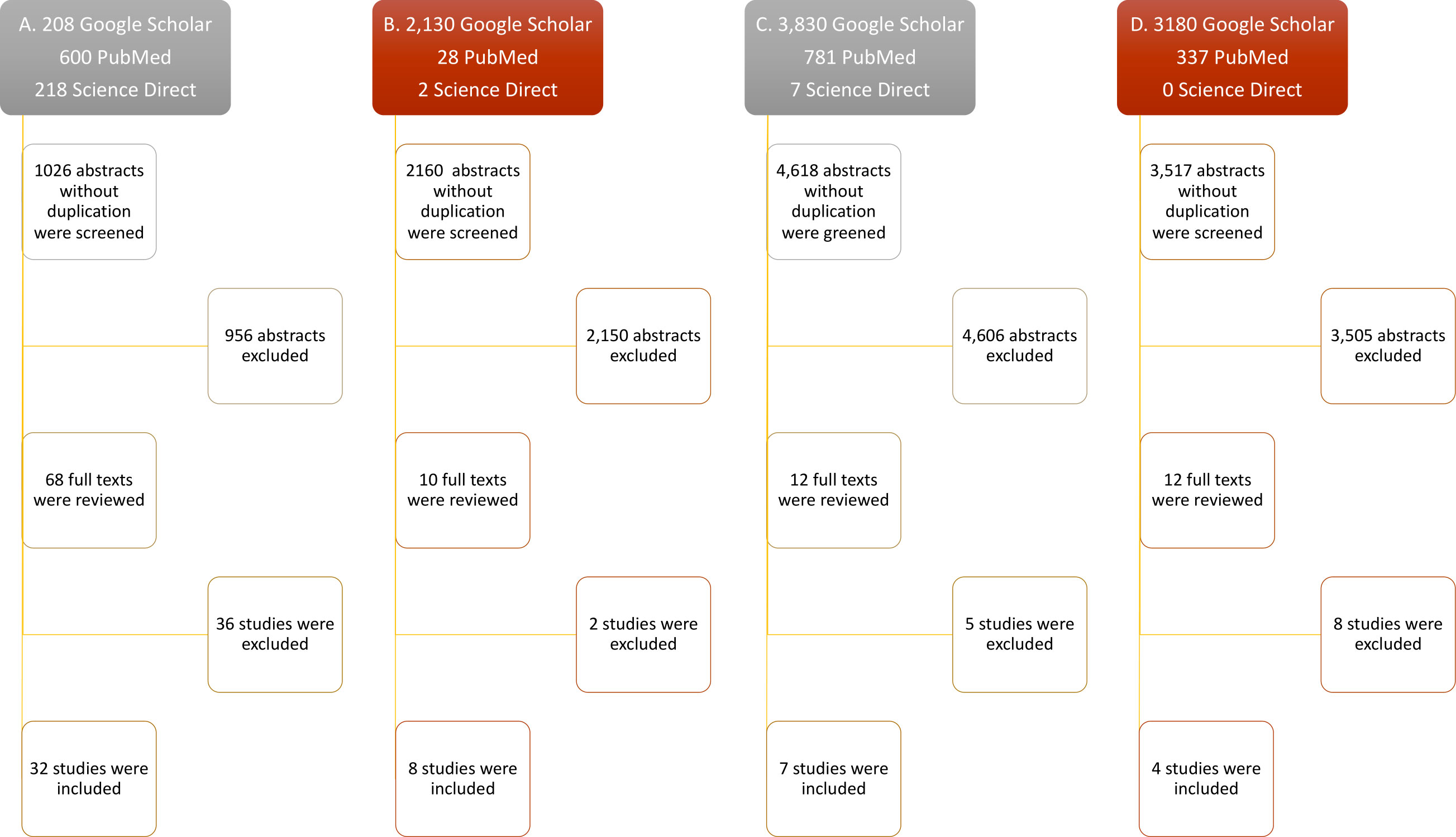
Figure 1 Flow diagram of study selection. Only research articles’ titles with the keywords (mentioned in methods) were selected for abstract reading. The relevant abstracts were included after duplication removal. Accessible full texts were screened and included. Full texts lacking exogenous cytokine treatment against viruses, mean viral titters with SEM, and number of replicates were excluded. (A–D) Study selection steps of IL-32, IFN-λ, IL-6, and IL-22, respectively.
Tables 1–4 represent systematic reviews of each of the cytokines with the extracted data that are required according to the inclusion criteria. Nearly all the most prevalent critical viral disease causative agents were included in the studies to observe the antiviral response of cytokines treatment on them. The cytokines were used either in the form of recombinant proteins, plasmids, or viral vectors expressing them. The experiments were conducted mostly on primary or secondary cell cultures and some on animal models. The treatments were either before or after viral infection, and, later, the viral titers were determined through various methods, out of which quantitative real-time polymerase chain reaction (qRT-PCR) being the most commonly used.
Standard mean differences of IFN-λ, IL-32γ, IL-6, and IL-22
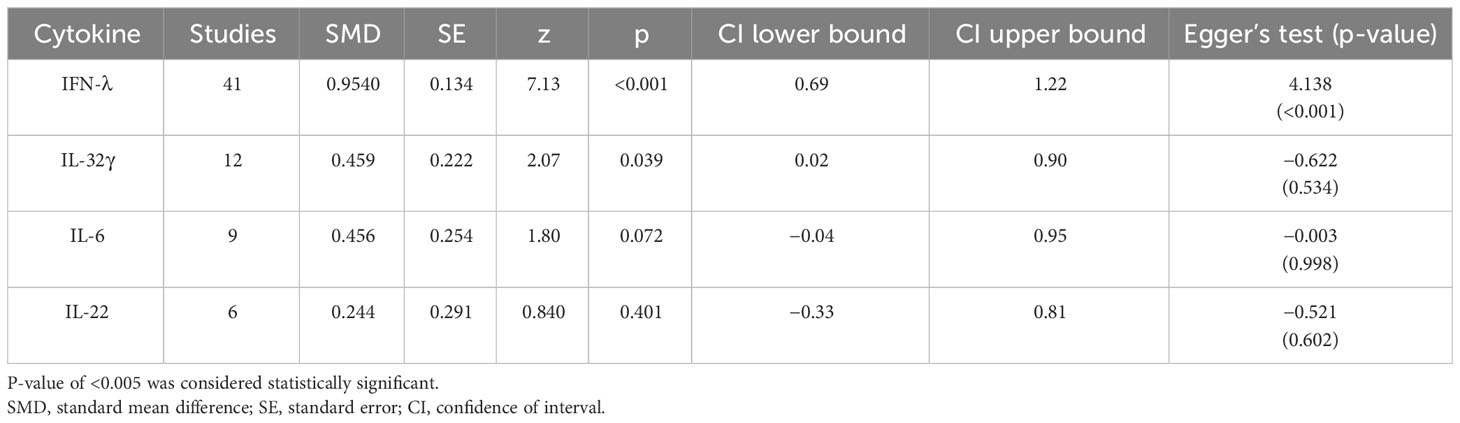
The analysis was carried out using the standardized mean difference as the outcome measure. A random-effects model was fitted to the data. Highest SMD was noted for IFN-λ with 0.9540, then IL-32γ with 0.459, IL-6 with 0.456, and IL-22 with 0.244 (Table 5). The average outcome differed significantly from zero for only IFN-λ and IL-32γ with z-score of 8.06 and 2.07 and p-values of <0.001 and 0.039, respectively, and, therefore, considered to be significant. The average outcome did not differ significantly from zero for IL-6 and IL-22 with z = 1.7978 and 0.8397, and p = 0.0722 and 0.4011, respectively.
Forest plots and heterogeneity statistics of IFN-λ, IL-32γ, IL-6, and IL-22
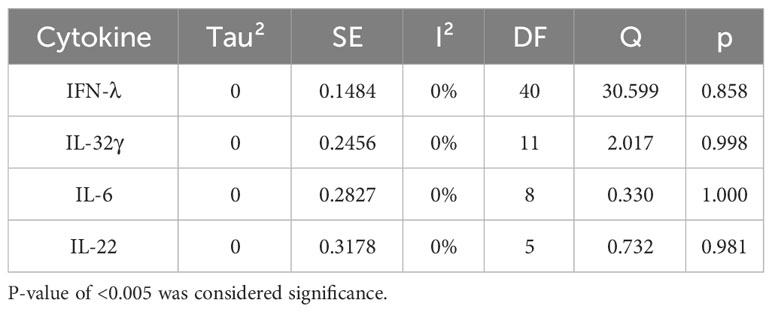
Forest plots revealing statistical heterogeneity of estimated effects of the treatment in all the included studies are shown in Figures 2–5. The heterogeneity tests scores are mentioned in Table 6 for all the four cytokines. None of four cytokines’ true outcomes appear to be heterogeneous (tau² < 0). The examination of the studentized residuals indicated no outlier data from any of the four cytokines. According to the Cook’s distances, none of the studies of IL-6 and IL-22 could be overly influential. However, study by Chong et al. (2022b) of IFN-λ and another study by Zaidan et al. (2019) of IL-32γ could be overly influential.
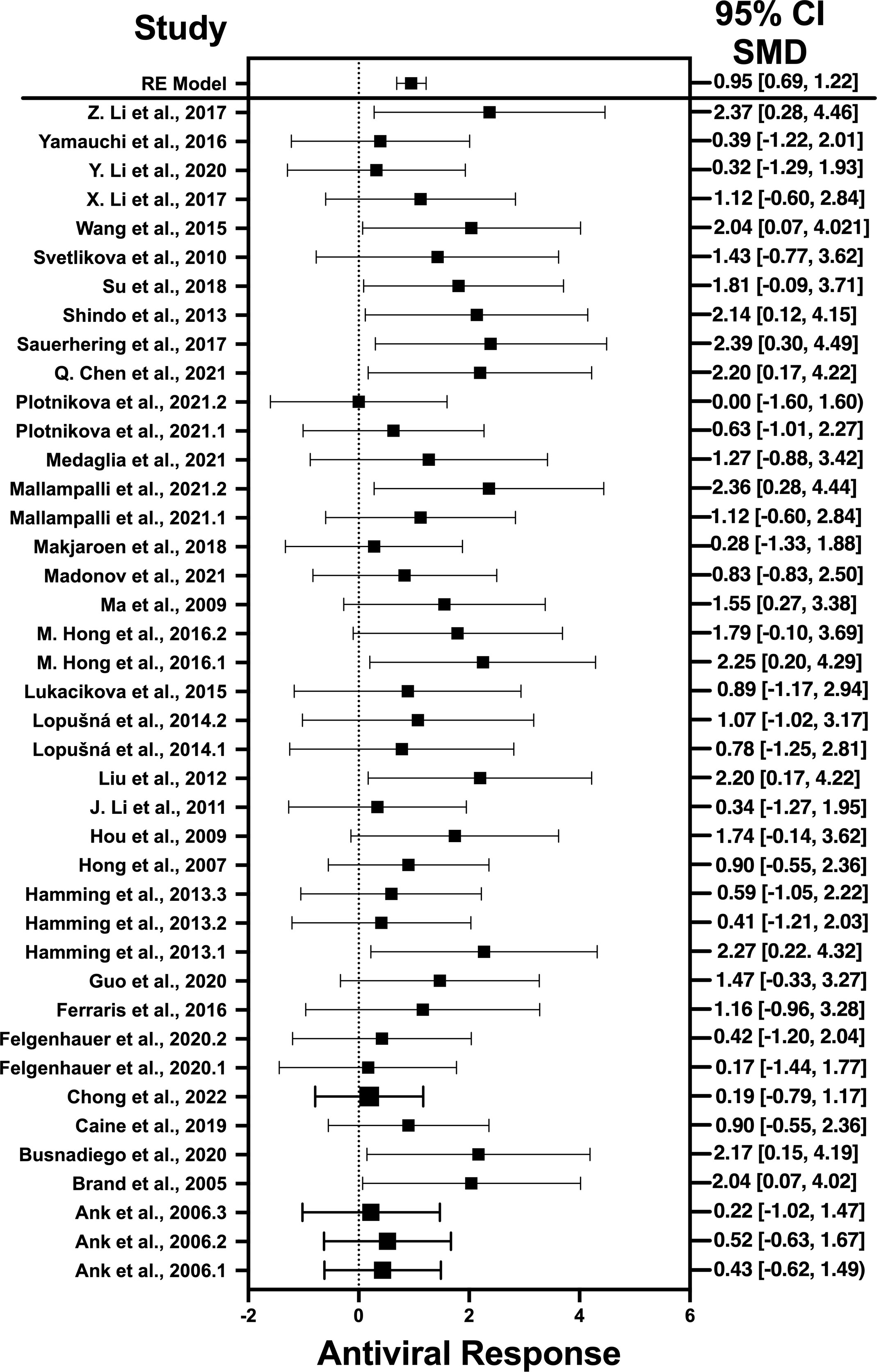
Figure 2 Forest plot illustrating antiviral response of IFN-λ according to the estimated effect sizes. The vertical line on the zero represents null hypothesis; the length of horizontal lines represents the total effect size of each study, starting with a lower limit and ending on an upper limit; and the squares are the mean differences. The antiviral effect IFN-λ is rated on a scale of 0 to 5. Square positioned above “0” is considered antiviral effect of IFN-λ, whereas a square lying below “0” will favor its proviral role.
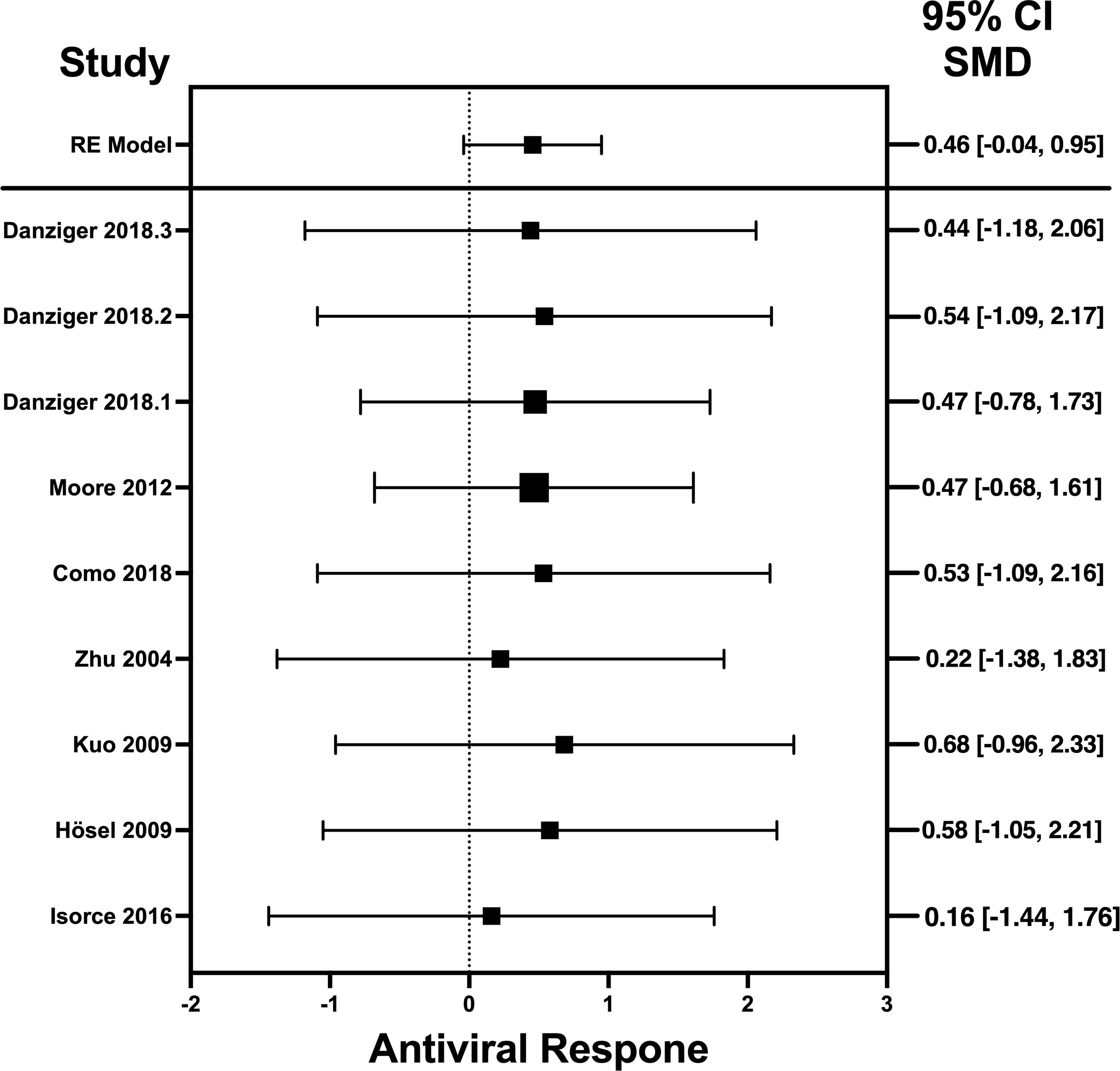
Figure 3 Forest plot illustrating antiviral response of IL-32γ according to the estimated effect sizes. The vertical line on the zero represents null hypothesis, the length of horizontal lines represents the total effect size of each study, starting with a lower limit and ending on an upper limit and the squares are the mean differences. The antiviral effect IL-32γ is rated on a scale of 0 to 5. Square positioned above “0” is considered antiviral effect of IL-32γ, whereas a square lying below “0” will favor its proviral role.
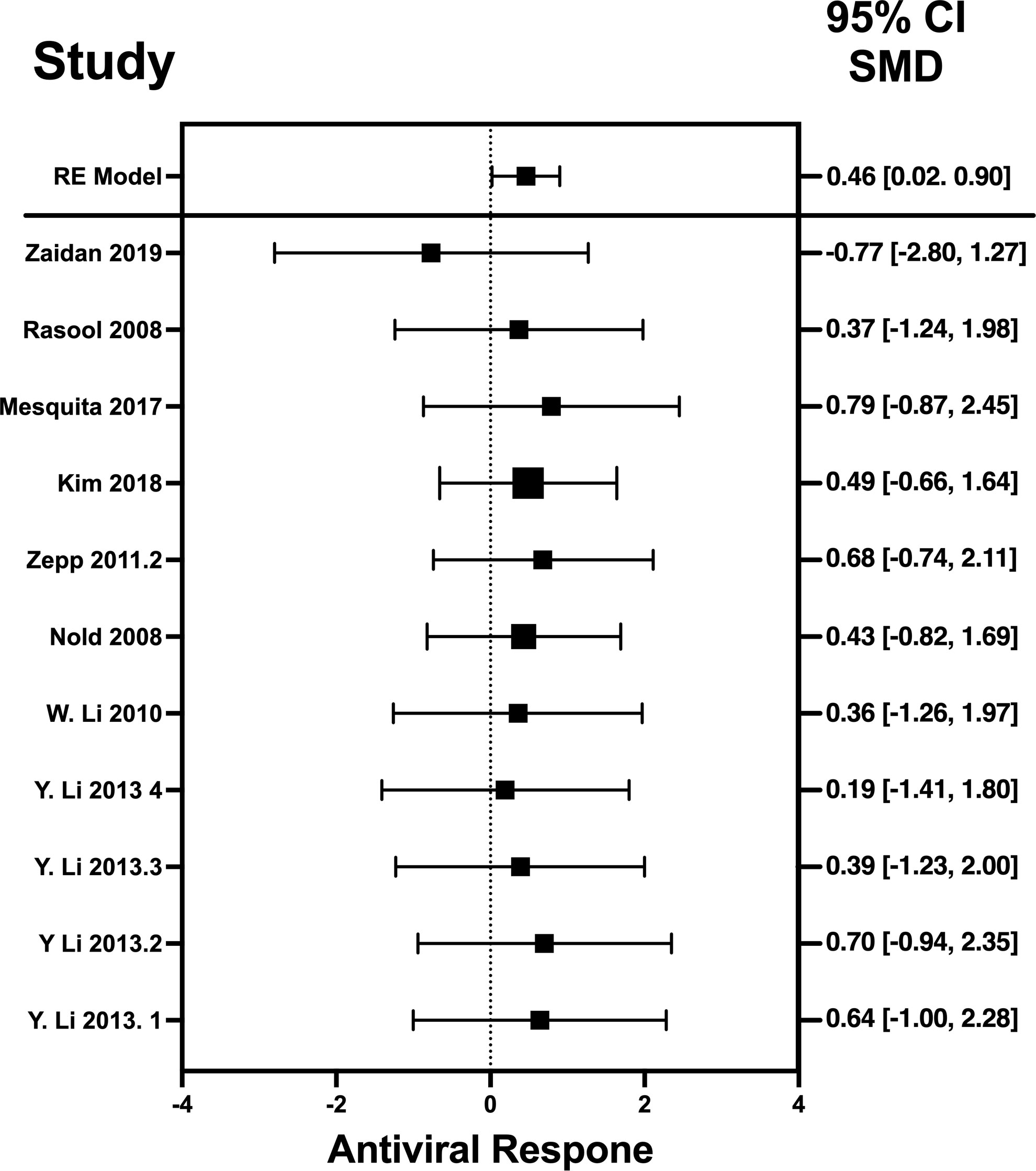
Figure 4 Forest plot illustrating antiviral response of IL-6 according to the estimated effect sizes. The vertical line on the zero represents null hypothesis; the length of horizontal lines represents the total effect size of each study, starting with a lower limit and ending on an upper limit; and the squares are the mean differences. The antiviral effect IL-6 is rated on a scale of 0 to 5. Square positioned above “0” is considered antiviral effect of IL-6, whereas a square lying below “0” will favor its proviral role. Square positioned above “0” is considered antiviral effect of IL-6, whereas a square lying below “0” will favor its proviral role.
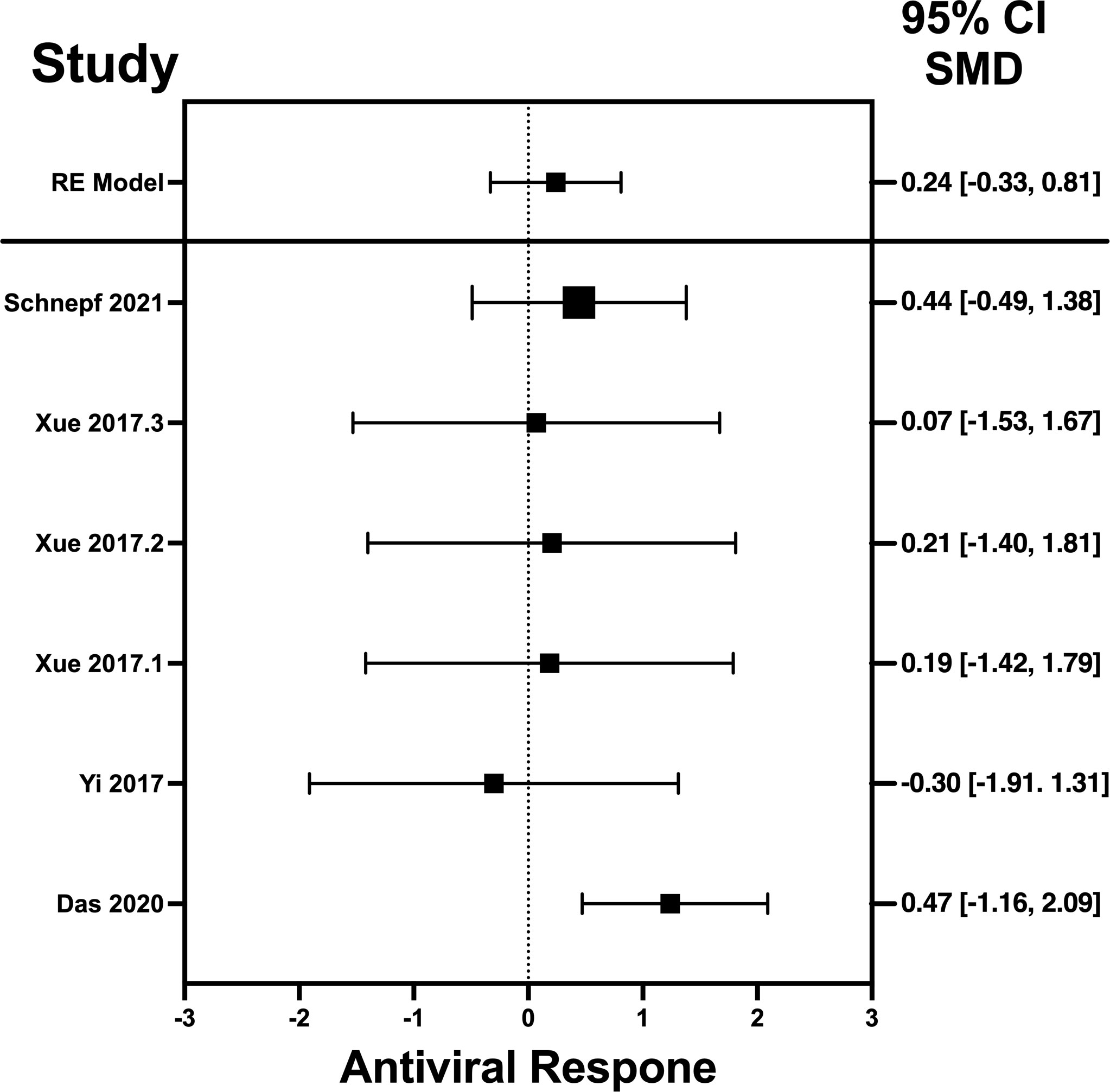
Figure 5 Forest plot illustrating antiviral response of IL-22 according to the estimated effect sizes. The vertical line on the zero represents null hypothesis; the length of horizontal lines represents the total effect size of each study, starting with a lower limit and ending on an upper limit; and the squares are the mean differences. The antiviral effect IL-22 is rated on a scale of 0 to 5. Square positioned above “0” is considered antiviral effect of IL-22, whereas a square lying below “0” will favor its proviral role. Square positioned above “0” is considered antiviral effect of IL-22, whereas a square lying below “0” will favor its proviral role.
Risk of publication bias
According to Egger’s linear regression test of IFN-λ, the funnel plot showed asymmetry with a p-value of 0.001. This may suggest that one or more studies included for IFN-λ carry risk of publication bias. Other than that, IL-32γ, IL-6, and IL-22 Egger’s test did not reveal any funnel plot asymmetry (p-values given in Table 5).
Discussion
This systematic review and meta-analysis was conducted to aggregate data from multiple studies, revealing that cytokines such as IL-6, IL-22, IL-32γ, and IFN-λ have antiviral potential when administered exogenously. These pro-inflammatory cytokines regulate immune system communication and serve as mediators in the defense against viral pathogens. In addition to their well-known role in inflammation and immune regulation, cytokines are considered to exhibit direct antiviral activity (79).
To our knowledge, this is the inaugural study that systematically reviews existing evidence and performs a comprehensive meta-analysis to evaluate pro-inflammatory cytokines as alternative therapeutic agents to conventional antiviral drugs. Although various interleukins like IL-1β, IL-18, and TNF-α have been shown to inhibit viral infections (24), IL-6, IL-22, IL-32γ, and IFN-λ were selected due to high number of studies focusing on them.
The SMD measured for all the selected cytokines revealed that it was larger for IFN-λ compared to IL-6, IL-22, and IL-32γ. This high SMD indicates that IFN-λ has a more substantial antiviral impact than the other three interleukins. Then, a high number of investigations (please refer to Table 1) have been conducted on the IFN-λ’s antiviral activity, and, now, it is in phase 2 clinical trials as well (80).
The heterogeneity of the cytokines studied in this systematic review and meta-analysis was represented through Forest plots. As suggested and depicted by the Forest plots, the lack of variability in the antiviral activity of any of the four cytokines studied in this systematic review and meta-analysis suggests that the observed antiviral effects of the cytokines were more likely attributable to their true antiviral effects of these cytokines.
Results revealed no outliers or overly influential studies in the IL-6 and IL-22 datasets, although the SMDs of the two studies, Zaidan et al. (2019) (76) and Chong et al. (2022) (33) deviated from the average SMDs of all studies in the IL-32γ and IFN-λ datasets, respectively. In fact, the study by Zaidan et al. (2019) (68) indicated a proviral effect of IL-32, suggesting that the effect of IL-32 on viral infection is complex. It is possible that IL-32 has different effects on different viruses or that its effects vary depending on the specific population that is studied. Egger’s linear regression test was done to estimate the publication bias (81). The publication bias is checked through the depiction of asymmetry of the funnel plot generated as part of output of Egger’s regression. Among all cytokines studied, only the funnel plot of IFN-λ was asymmetrical regardless of zero heterogeneity. This asymmetry indicates publication bias, and one of the major reasons is the suppression of the null findings by the authors (82). Another possible reason for this asymmetry could be the use of the random-effects model (83).
IL-6, IL-22, IL-32, and IFN-λ are now being considered as potential allies in the struggle against viral infections. IFN-λ, also known as type III interferon, has demonstrated notable action against a variety of viruses, including respiratory viruses like influenza and SARS-CoV-2 (84). Being a pro-inflammatory cytokine, IL-6 has been shown to have antiviral effects against several viral infections (HBV, HCV, VZV, etc.) and is essential for immune responses. IL-22, another pro-inflammatory and tissue-protective cytokine, has shown antiviral activity against several viruses, including hepatitis C and the human papillomavirus. IL-32 has demonstrated antiviral potential by preventing viral replication and modifying host immune responses (85). These cytokines promise as possible antiviral therapeutics, whether used alone or in conjunction with already available antiviral medications.
Although several antiviral drugs are commercially available, they often come with side effects. These side effects could be reversible or mild like flu-like symptoms or can significantly cause the neurotoxic effects in patients (86). There is multiple evidence of scientific research in which scientists are using the combination of antiviral drugs with cytokines for best results. Chudhary et al. (2023) used the multiple antiviral drugs against HCV and concluded pegylated IFN + Ribavirin to be more effective (87).
Results indicate that, among the selected pro-inflammatory cytokines, IFN-λ has shown the best antiviral activity. Among the selected studies, Sauerhering et al. (2017) (67) has the highest effect size of 2.39 (CI: 0.30–4.49). This indicates its high antiviral activity against the Nipah virus, whereas IL-22 presents the lowest antiviral activity among the selected viruses. This is because IL-22 does not produce a direct antiviral response by interacting with the viral genes, instead it initiates the underlying signaling pathways that recruits the IFN-λ or cooperates with IL-18 to induce the interferon-stimulated gene (ISG) production and, hence, provides the protective role (88). Another restricting element is that not every pro-inflammatory cytokine generates the adaptive immune response to induce the viral suppression; instead, they generate the innate immune response to combat the viral infection (89). That is why the results of IL-22 was different from the other selected cytokines as it acts as a key modulator of innate immune response and is not directly involved in the adaptive immunity (90). Similar trend is observed when IL-32γ induces the antiviral responses against influenza virus (70), vesicular stomatitis virus (VSV) (91), and human immunodeficiency virus (HIV) (72). It produce IFN-λ that, in turn, suppresses the viral infection via generation of immune modulators (75). IL-6 not only produces the immune response against viruses but also interacts with certain viral genes that are responsible for the viral entry (92) and viral replication (78).
On the other hand, IFN-λ has superior antiviral activity than IL-6, IL-22, and IL-32γ. This is attributed to the induction of specific antiviral genes as well as the upregulation of ISGs. IFN-λ has been shown through in vitro experiments to specifically upregulate the antiviral genes in the epithelial cells. This results in the unique antiviral transcript profile and a strong induction of ISGs like MX Dynamin Like GTPase 2 (MX2), ISG15, and Interferon-induced protein with tetratricopeptide repeats 3 (IFIT3), which all contribute to the strong antiviral activity (93). Furthermore, IFN-λ offers strong defense at anatomical barriers such as epithelial surfaces and exhibits the notable antiviral activity against specific virus like EMCV, LCMV, and HSV (94).
This comprehensive systematic review and meta-analysis provides valuable insights. On the other hand, it has some limitations too, including the paucity of studies involving exogenously administered cytokines and the restricted range of cytokines tested for antiviral effects. This paucity regarding the availability of ample literature can be a major possible reason for the biasness of the reported data for this study. Another limiting factor of this study could be the evaluation of very few cytokines for their antiviral effect. To address these limitations, scientists should prioritize research involving the administration of recombinant cytokines or their expression vectors to provide a robust understanding of their antiviral effect in in vitro and in vivo settings as well. In addition, more cytokines should be explored for their potential antiviral properties.
In conclusion, this meta-analysis provides significant evidence supporting the potent antiviral properties of exogenous cytokines, specifically IL-6, IL-22, IL-32γ, and IFN-λ, against human and animal viral infections. Overall, these findings highlight the potential therapeutic applications of four important cytokines in the development of novel antiviral strategies, thus warranting further investigation to elucidate their precise mechanisms of action and optimize treatment protocols.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
Author contributions
AS: Conceptualization, Data curation, Investigation, Methodology, Project administration, Software, Writing – original draft, Writing – review & editing. AQ: Conceptualization, Data curation, Investigation, Methodology, Project administration, Software, Writing – original draft, Writing – review & editing. SA: Data curation, Methodology, Writing – original draft. RM: Data curation, Methodology, Writing – original draft. HK: Data curation, Writing – original draft, Investigation. SM: Formal analysis, Supervision, Validation, Writing – review & editing. JA: Formal analysis, Validation, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Verhoef J, van Kessel K, Snippe H. Immune response in human pathology: infections caused by bacteria, viruses, fungi, and parasites. Nijkamp Parnhams Princ Immunopharmacol (2019), 165–78. doi: 10.1007/978-3-030-10811-3_10
2. Moriyama M, Hugentobler WJ, Iwasaki A. Seasonality of respiratory viral infections. Annu Rev Virol (2020) 7:83–101. doi: 10.1146/annurev-virology-012420-022445
3. Pappas DE. The common cold. Princ Pract Pediatr Infect Dis (2018), 199–202.e1. doi: 10.1016/B978-0-323-40181-4.00026-8
4. Bristow I. Paediatric cutaneous warts and verrucae: an update. Int J Environ Res Public Health (2022) 19:16400. doi: 10.3390/ijerph192416400
5. Schwetz TA, Fauci AS. The extended impact of human immunodeficiency virus/AIDS research. J Infect Dis (2019) 219:6–9. doi: 10.1093/infdis/jiy441
6. Wilder-Smith A. COVID-19 in comparison with other emerging viral diseases: risk of geographic spread via travel. Trop Dis Travel Med Vaccines (2021) 7:3. doi: 10.1186/s40794-020-00129-9
7. Moghadami M. A narrative review of influenza: A seasonal and pandemic disease. Iran J Med Sci (2017) 42:2–13.
8. Rojas M, Monsalve DM, Pacheco Y, Acosta-Ampudia Y, Ramírez-Santana C, Ansari AA, et al. Ebola virus disease: An emerging and re-emerging viral threat. J Autoimmun (2020) 106:102375. doi: 10.1016/j.jaut.2019.102375
9. Forni D, Cagliani R, Clerici M, Sironi M. Disease-causing human viruses: novelty and legacy. Trends Microbiol (2022) 30:1232–42. doi: 10.1016/j.tim.2022.07.002
10. Xu M, Zhang W-L, Zhu Q, Zhang S, Yao Yy, Xiang T, et al. Genome-wide profiling of Epstein-Barr virus integration by targeted sequencing in Epstein-Barr virus associated Malignancies. Theranostics (2019) 9:1115–24. doi: 10.7150/thno.29622
11. Mauch-Mücke K, Schön K, Paulus C, Nevels MM. Evidence for tethering of human cytomegalovirus genomes to host chromosomes. Front Cell Infect Microbiol (2020) 10:577428. doi: 10.3389/fcimb.2020.577428
12. Deigendesch N, Stenzel W. Acute and chronic viral infections. Handb Clin Neurol (2017) 145:227–43. doi: 10.1016/B978-0-12-802395-2.00017-1
13. Silva HM. Antibiotics against viruses: Brazilian doctors adrift. Infect Control Hosp Epidemiol (2022) 43:1992–3. doi: 10.1017/ice.2021.434
14. Tompa DR, Immanuel A, Srikanth S, Kadhirvel S. Trends and strategies to combat viral infections: A review on FDA approved antiviral drugs. Int J Biol Macromol (2021) 172:524–41. doi: 10.1016/j.ijbiomac.2021.01.076
15. Kausar S, Said Khan F, Ishaq Mujeeb Ur Rehman M, Akram M, Riaz M, Rasool G, et al. A review: Mechanism of action of antiviral drugs. Int J Immunopathol Pharmacol (2021) 35:20587384211002621. doi: 10.1177/20587384211002621
16. Takashita E. Influenza polymerase inhibitors: mechanisms of action and resistance. Cold Spring Harb Perspect Med (2021) 11:a038687. doi: 10.1101/cshperspect.a038687
17. Ross SA. 206 - cytomegalovirus. In: Long SS, editor. Pract. Pediatr. Infect. Dis, Sixth. Philadelphia: Elsevier (2023). p. 1092–1099.e2.
18. Fung S, Choi HSJ, Gehring A, Janssen HLA. Getting to HBV cure: The promising paths forward. Hepatol Baltim Md (2022) 76:233–50. doi: 10.1002/hep.32314
19. Bai H, Si L, Jiang A, Belgur C, Zhai Y, Plebani R, et al. Mechanical control of innate immune responses against viral infection revealed in a human lung alveolus chip. Nat Commun (2022) 13:1928. doi: 10.1038/s41467-022-29562-4
20. Moss P. The T cell immune response against SARS-CoV-2. Nat Immunol (2022) 23:186–93. doi: 10.1038/s41590-021-01122-w
21. Marshall JS, Warrington R, Watson W, Kim HL. An introduction to immunology and immunopathology. Allergy Asthma Clin Immunol (2018) 14:49. doi: 10.1186/s13223-018-0278-1
22. Thompson MR, Kaminski JJ, Kurt-Jones EA, Fitzgerald KA. Pattern recognition receptors and the innate immune response to viral infection. Viruses (2011) 3:920–40. doi: 10.3390/v3060920
23. Wallis RS, O’Garra A, Sher A, Wack A. Host-directed immunotherapy of viral and bacterial infections: past, present and future. Nat Rev Immunol (2023) 23:121–33. doi: 10.1038/s41577-022-00734-z
24. Isorce N, Testoni B, Locatelli M, Fresquet J, Rivoire M, Luangsay S, et al. Antiviral activity of various interferons and pro-inflammatory cytokines in non-transformed cultured hepatocytes infected with hepatitis B virus. Antiviral Res (2016) 130:36–45. doi: 10.1016/j.antiviral.2016.03.008
25. Atallah-Yunes SA, Robertson MJ. Cytokine based immunotherapy for cancer and lymphoma: biology, challenges and future perspectives. Front Immunol (2022) 13:872010. doi: 10.3389/fimmu.2022.872010
26. Tanriver-Ayder E, Faes C, van de Casteele T, McCann SK, Macleod MR. Comparison of commonly used methods in random effects meta-analysis: application to preclinical data in drug discovery research. BMJ Open Sci (2021) 5:e100074. doi: 10.1136/bmjos-2020-100074
27. Almalik O, Zhan Z, van den Heuvel ER. Tests for publication bias are unreliable in case of heteroscedasticity. Contemp Clin Trials Commun (2021) 22:100781. doi: 10.1016/j.conctc.2021.100781
28. Francis G. Replication, statistical consistency, and publication bias. J Math Psychol (2013) 57:153–69. doi: 10.1016/j.jmp.2013.02.003
29. Huynh TLD, Wu J, Duong AT. Information Asymmetry and firm value: Is Vietnam different? J Econ Asymmetries (2020) 21:e00147. doi: 10.1016/j.jeca.2019.e00147
30. Lozhkov AA, Klotchenko SA, Ramsay ES, Moshkoff HD, Moshkoff DA, Vasin AV, et al. The key roles of interferon lambda in human molecular defense against respiratory viral infections. Pathogens (2020) 9:989. doi: 10.3390/pathogens9120989
31. Viechtbauer W. Conducting meta-analyses in R with the metafor package. J Stat Softw (2010) 36:1–48. doi: 10.18637/jss.v036.i03
32. Cochran WG. The combination of estimates from different experiments. Biometrics (1954) 10:101–29. doi: 10.2307/3001666
33. Nakagawa S, Lagisz M, Jennions MD, Koricheva J, Noble DWA, Parker TH, et al. Methods for testing publication bias in ecological and evolutionary meta-analyses. Methods Ecol Evol (2022) 13:4–21. doi: 10.1111/2041-210X.13724
34. Cumpston MS, McKenzie JE, Welch VA, Brennan SE. Strengthening systematic reviews in public health: guidance in the Cochrane Handbook for Systematic Reviews of Interventions, 2nd edition. J Public Health (2022) 44:e588–92. doi: 10.1093/pubmed/fdac036
35. Arya S, Kaji AH, Boermeester MA. PRISMA reporting guidelines for meta-analyses and systematic reviews. JAMA Surg (2021) 156:789–90. doi: 10.1001/jamasurg.2021.0546
36. McKeown S, Mir ZM. Considerations for conducting systematic reviews: evaluating the performance of different methods for de-duplicating references. Syst Rev (2021) 10:38. doi: 10.1186/s13643-021-01583-y
37. Şahi̇n M, Aybek E. Jamovi: an easy to use statistical software for the social scientists. Int J Assess Tools Educ (2020) 6:670–92. doi: 10.21449/ijate.661803
38. Murad MH, Wang Z, Chu H, Lin L. When continuous outcomes are measured using different scales: guide for meta-analysis and interpretation. BMJ (2019) 364:k4817. doi: 10.1136/bmj.k4817
39. Ank N, West H, Bartholdy C, Eriksson K, Thomsen AR, Paludan SR. Lambda interferon (IFN-λ), a type III IFN, is induced by viruses and IFNs and displays potent antiviral activity against select virus infections in vivo. J Virol (2006) 80:4501–9. doi: 10.1128/JVI.80.9.4501-4509.2006
40. Shindo H, Maekawa S, Komase K, Miura M, Kadokura M, Sueki R, et al. IL-28B (IFN-λ3) and IFN-α synergistically inhibit HCV replication. J Viral Hepat (2013) 20:281–9. doi: 10.1111/j.1365-2893.2012.01649.x
41. Brand S, Beigel F, Olszak T, Zitzmann K, Eichhorst ST, Otte J-M, et al. IL-28A and IL-29 mediate antiproliferative and antiviral signals in intestinal epithelial cells and murine CMV infection increases colonic IL-28A expression. Am J Physiol-Gastrointest Liver Physiol (2005) 289:G960–8. doi: 10.1152/ajpgi.00126.2005
42. Wang Y, Li J, Wang X, Zhou Y, Zhang T, Ho W. Comparison of antiviral activity of lambda-interferons against HIV replication in macrophages. J Interferon Cytokine Res (2015) 35:213–21. doi: 10.1089/jir.2014.0064
43. Plotnikova M, Lozhkov A, Romanovskaya-Romanko E, Baranovskaya I, Sergeeva M, Klotchenko S, et al. IFN-λ1 displays various levels of antiviral activity in vitro in a select panel of RNA viruses. Viruses (2021) 13:1602. doi: 10.3390/v13081602
44. Caine EA, Scheaffer SM, Arora N, Zaitsev K, Artyomov MN, Coyne CB, et al. Interferon lambda protects the female reproductive tract against Zika virus infection. Nat Commun (2019) 10:280. doi: 10.1038/s41467-018-07993-2
45. Hong S-H, Cho O, Kim K, Shin H-J, Kotenko SV, Park S. Effect of interferon-lambda on replication of hepatitis B virus in human hepatoma cells. Virus Res (2007) 126:245–9. doi: 10.1016/j.virusres.2007.03.006
46. Chong Z, Karl CE, Halfmann PJ, Kawaoka Y, Winkler ES, Keeler SP, et al. Nasally delivered interferon-λ protects mice against infection by SARS-CoV-2 variants including Omicron. Cell Rep (2022) 39:110799. doi: 10.1016/j.celrep.2022.110799
47. Li J, Hu S, Zhou L, Ye L, Wang X, Ho J, et al. Interferon lambda inhibits herpes simplex virus type I infection of human astrocytes and neurons. Glia (2011) 59:58–67. doi: 10.1002/glia.21076
48. Medaglia C, Zwygart AC-A, Silva PJ, Constant S, Huang S, Stellacci F, et al. Interferon lambda delays the emergence of influenza virus resistance to oseltamivir. Microorganisms (2021) 9:1196. doi: 10.3390/microorganisms9061196
49. Su Q-J, Wang X, Zhou R-H, Guo L, Liu H, Li J-L, et al. IFN-λ4 inhibits HIV infection of macrophages through signalling of IFN-λR1/IL-10R2 receptor complex. Scand J Immunol (2018) 88:e12717. doi: 10.1111/sji.12717
50. Madonov PG, Svyatchenko VA, Legostaev SS, Kikhtenko NA, Kotlyarova AA, Oleinik LA, et al. Evaluation of the anti-viral activity of human recombinant interferon lambda-1 against SARS-coV-2. Bull Exp Biol Med (2021) 172:53–6. doi: 10.1007/s10517-021-05330-0
51. Lopušná K, Režuchová I, Kabát P, Kúdelová M. Interferon lambda induces antiviral response to herpes simplex virus 1 infection. Acta Virol (2014) 58:325–32. doi: 10.4149/av_2014_03_325
52. Hong M, Schwerk J, Lim C, Kell A, Jarret A, Pangallo J, et al. Interferon lambda 4 expression is suppressed by the host during viral infection. J Exp Med (2016) 213:2539–52. doi: 10.1084/jem.20160437
53. Lukacikova L, Oveckova I, Betakova T, Laposova K, Polcicova K, Pastorekova S, et al. Antiviral effect of interferon lambda against lymphocytic choriomeningitis virus. J Interferon Cytokine Res (2015) 35:540–53. doi: 10.1089/jir.2014.0083
54. Ma D, Jiang D, Qing M, Weidner JM, Qu X, Guo H, et al. Antiviral effect of interferon lambda against West Nile virus. Antiviral Res (2009) 83:53–60. doi: 10.1016/j.antiviral.2009.03.006
55. Li X, Li Y, Fang S, Su J, Jiang J, Liang B, et al. Downregulation of autophagy-related gene ATG5 and GABARAP expression by IFN-λ1 contributes to its anti-HCV activity in human hepatoma cells. Antiviral Res (2017) 140:83–94. doi: 10.1016/j.antiviral.2017.01.016
56. Mallampalli RK, Adair J, Elhance A, Farkas D, Chafin L, Long ME, et al. Interferon lambda signaling in macrophages is necessary for the antiviral response to influenza. Front Immunol (2021) 12:735576. doi: 10.3389/fimmu.2021.735576
57. Li Y, Zhao L, Luo Z, Zhang Y, Lv L, Zhao J, et al. Interferon-λ Attenuates rabies virus infection by inducing interferon-stimulated genes and alleviating neurological inflammation. Viruses (2020) 12:405. doi: 10.3390/v12040405
58. Hamming OJ, Terczyńska-Dyla E, Vieyres G, Dijkman R, Jørgensen SE, Akhtar H, et al. Interferon lambda 4 signals via the IFNλ receptor to regulate antiviral activity against HCV and coronaviruses: Recombinant IFNλ4 is antiviral. EMBO J (2013) 32:3055–65. doi: 10.1038/emboj.2013.232
59. Hou W, Wang X, Ye L, Zhou L, Yang Z-Q, Riedel E, et al. Lambda interferon inhibits human immunodeficiency virus type 1 infection of macrophages. J Virol (2009) 83:3834–42. doi: 10.1128/JVI.01773-08
60. Svetlikova D, Kabat P, Ohradanova A, Pastorek J, Betakova T. Influenza A virus replication is inhibited in IFN-λ2 and IFN-λ3 transfected or stimulated cells. Antiviral Res (2010) 88:329–33. doi: 10.1016/j.antiviral.2010.10.005
61. Yamauchi S, Takeuchi K, Chihara K, Honjoh C, Kato Y, Yoshiki H, et al. STAT1 is essential for the inhibition of hepatitis C virus replication by interferon-λ but not by interferon-α. Sci Rep (2016) 6:38336. doi: 10.1038/srep38336
62. Liu M-Q, Zhou D-J, Wang X, Zhou W, Ye L, Li J-L, et al. IFN-λ3 inhibits HIV infection of macrophages through the JAK-STAT pathway. PloS One (2012) 7:e35902. doi: 10.1371/journal.pone.0035902
63. Chen Q, Coto-Llerena M, Suslov A, Teixeira RD, Fofana I, Nuciforo S, et al. Interferon lambda 4 impairs hepatitis C viral antigen presentation and attenuates T cell responses. Nat Commun (2021) 12:4882. doi: 10.1038/s41467-021-25218-x
64. Busnadiego I, Fernbach S, Pohl MO, Karakus U, Huber M, Trkola A, et al. Antiviral activity of type I, II, and III interferons counterbalances ACE2 inducibility and restricts SARS-coV-2. mBio (2020) 11:e01928–20. doi: 10.1128/mBio.01928-20
65. Felgenhauer U, Schoen A, Gad HH, Hartmann R, Schaubmar AR, Failing K, et al. Inhibition of SARS–CoV-2 by type I and type III interferons. J Biol Chem (2020) 295:13958–64. doi: 10.1074/jbc.AC120.013788
66. Makjaroen J, Somparn P, Hodge K, Poomipak W, Hirankarn N, Pisitkun T. Comprehensive proteomics identification of IFN-λ3-regulated antiviral proteins in HBV-transfected cells. Mol Cell Proteomics MCP (2018) 17:2197–215. doi: 10.1074/mcp.RA118.000735
67. Sauerhering L, Müller H, Behner L, Elvert M, Fehling SK, Strecker T, et al. Variability of interferon-λ induction and antiviral activity in Nipah virus infected differentiated human bronchial epithelial cells of two human donors. J Gen Virol (2017) 98:2447–53. doi: 10.1099/jgv.0.000934
68. Li Z, Lu X, Zhu Y, Cheng P, Liu S, Zhang Y, et al. Lambda-interferons inhibit herpes simplex virus type 2 replication in human cervical epithelial cells by activating the JAK/STAT pathway. Jpn J Infect Dis (2017) 70:416–22. doi: 10.7883/yoken.JJID.2016.465
69. Li Y, Xie J, Xu X, Liu L, Wan Y, Liu Y, et al. Inducible interleukin 32 (IL-32) exerts extensive antiviral function via selective stimulation of interferon λ1 (IFN-λ1) *. J Biol Chem (2013) 288:20927–41. doi: 10.1074/jbc.M112.440115
70. Li W, Sun W, Liu L, Yang F, Li Y, Chen Y, et al. IL-32: A Host Proinflammatory Factor against Influenza Viral Replication Is Upregulated by Aberrant Epigenetic Modifications during Influenza A Virus Infection. J Immunol (2010) 185:5056–65. doi: 10.4049/jimmunol.0902667
71. Nold MF, Nold-Petry CA, Pott GB, Zepp JA, Saavedra MT, Kim S-H, et al. Endogenous IL-32 controls cytokine and HIV-1 production1. J Immunol (2008) 181:557–65. doi: 10.4049/jimmunol.181.1.557
72. Rasool ST, Tang H, Wu J, Li W, Mukhtar MM, Zhang J, et al. Increased level of IL-32 during human immunodeficiency virus infection suppresses HIV replication. Immunol Lett (2008) 117:161–7. doi: 10.1016/j.imlet.2008.01.007
73. Zepp JA, Nold-Petry CA, Dinarello CA, Nold MF. Protection from RNA and DNA viruses by IL-32. J Immunol (2011) 186:4110–8. doi: 10.4049/jimmunol.1000081
74. Mesquita PMM, Preston-Hurlburt P, Keller MJ, Vudattu N, Espinoza L, Altrich M, et al. Role of interleukin 32 in human immunodeficiency virus reactivation and its link to human immunodeficiency virus–herpes simplex virus coinfection. J Infect Dis (2017) 215:614–22. doi: 10.1093/infdis/jiw612
75. Kim DH, Park E-S, Lee AR, Park S, Park YK, Ahn SH, et al. Intracellular interleukin-32γ mediates antiviral activity of cytokines against hepatitis B virus. Nat Commun (2018) 9:3284. doi: 10.1038/s41467-018-05782-5
76. Zaidan SM, Leyre L, Bunet R, Larouche-Anctil E, Turcotte I, Sylla M, et al. Upregulation of IL-32 isoforms in virologically-suppressed HIV-infected individuals: Potential role in persistent inflammation and transcription from stable HIV-1 reservoirs. J Acquir Immune Defic Syndr (20191999) 82:503–13. doi: 10.1097/QAI.0000000000002185
77. Hösel M, Quasdorff M, Wiegmann K, Webb D, Zedler U, Broxtermann M, et al. Not interferon, but interleukin-6 controls early gene expression in hepatitis B virus infection. Hepatol Baltim Md (2009) 50:1773–82. doi: 10.1002/hep.23226
78. Kuo T-M, Hu C, Chen Y-L, Hong M-H, Jeng K-S, Liang C-CT, et al. HBV replication is significantly reduced by IL-6. J BioMed Sci (2009) 16:41. doi: 10.1186/1423-0127-16-41
79. Zhu H, Liu C. Interleukin-1 inhibits hepatitis C virus subgenomic RNA replication by activation of extracellular regulated kinase pathway. J Virol (2003) 77:5493–8. doi: 10.1128/JVI.77.9.5493-5498.2003
80. Como CN, Pearce CM, Cohrs RJ, Baird NL. Interleukin-6 and type 1 interferons inhibit varicella zoster virus replication in human neurons. Virology (2018) 522:13–8. doi: 10.1016/j.virol.2018.06.013
81. Danziger O, Pupko T, Bacharach E, Ehrlich M. Interleukin-6 and Interferon-α Signaling via JAK1-STAT Differentially Regulate Oncolytic versus Cytoprotective Antiviral States. Front Immunol (2018) 9:94. doi: 10.3389/fimmu.2018.00094
82. Das S, St Croix C, Good M, Chen J, Zhao J, Hu S, et al. Interleukin-22 inhibits respiratory syncytial virus production by blocking virus-mediated subversion of cellular autophagy. iScience (2020) 23:101256. doi: 10.1016/j.isci.2020.101256
83. Yi P, Liang Y, Yuan DMK, Jie Z, Kwota Z, Chen Y, et al. A tightly regulated IL-22 response maintains immune functions and homeostasis in systemic viral infection. Sci Rep (2017) 7:3857. doi: 10.1038/s41598-017-04260-0
84. Xue M, Zhao J, Ying L, Fu F, Li L, Ma Y, et al. IL-22 suppresses the infection of porcine enteric coronaviruses and rotavirus by activating STAT3 signal pathway. Antiviral Res (2017) 142:68–75. doi: 10.1016/j.antiviral.2017.03.006
85. Schnepf D, Hernandez P, Mahlakõiv T, Crotta S, Sullender ME, Peterson ST, et al. Rotavirus susceptibility of antibiotic-treated mice ascribed to diminished expression of interleukin-22. PloS One (2021) 16:e0247738. doi: 10.1371/journal.pone.0247738
86. Björkström NK, Strunz B, Ljunggren H-G. Natural killer cells in antiviral immunity. Nat Rev Immunol (2022) 22:112–23. doi: 10.1038/s41577-021-00558-3
87. Feld JJ, Kandel C, Biondi MJ, Kozak RA, Zahoor MA, Lemieux C, et al. Peginterferon lambda for the treatment of outpatients with COVID-19: a phase 2, placebo-controlled randomised trial. Lancet Respir Med (2021) 9:498–510. doi: 10.1016/S2213-2600(20)30566-X
88. Gimeno Brias S, Stack G, Stacey MA, Redwood AJ, Humphreys IR. The role of IL-22 in viral infections: paradigms and paradoxes. Front Immunol (2016) 7. doi: 10.3389/fimmu.2016.00211
89. Rouse BT, Sehrawat S. Immunity and immunopathology to viruses: what decides the outcome? Nat Rev Immunol (2010) 10:514–26. doi: 10.1038/nri2802
90. Dudakov JA, Hanash AM, van den Brink MRM. Interleukin-22: immunobiology and pathology. Annu Rev Immunol (2015) 33:747–85. doi: 10.1146/annurev-immunol-032414-112123
91. Bae S, Kang D, Hong J, Chung B, Choi J, Jhun H, et al. Characterizing antiviral mechanism of interleukin-32 and a circulating soluble isoform in viral infection. Cytokine (2012) 58:79–86. doi: 10.1016/j.cyto.2011.12.024
92. Bouezzedine F, Fardel O, Gripon P. Interleukin 6 inhibits HBV entry through NTCP down regulation. Virology (2015) 481:34–42. doi: 10.1016/j.virol.2015.02.026
93. Martínez-Espinoza I, Guerrero-Plata A. Current landscape of IFN-λ: induction, inhibition, and potential clinical applications to treat respiratory viral infections. ImmunoHorizons (2023) 7:265–72. doi: 10.4049/immunohorizons.2200010
Keywords: interleukin-6, interleukin-22, interleukin-32 gamma, interferon-lambda, antiviral
Citation: Sattar AA, Qaiser A, Kausar H, Aqil S, Mudassar R, Manzoor S and Ashraf J (2024) The potential of IFN-λ, IL-32γ, IL-6, and IL-22 as safeguards against human viruses: a systematic review and a meta-analysis. Front. Immunol. 15:1303115. doi: 10.3389/fimmu.2024.1303115
Received: 27 September 2023; Accepted: 17 January 2024;
Published: 14 February 2024.
Edited by:
Ying Zhu, Wuhan University, ChinaReviewed by:
Sebastian Reyes-Cerpa, Major University, ChileYushun Wan, Chongqing Medical University, China
Copyright © 2024 Sattar, Qaiser, Kausar, Aqil, Mudassar, Manzoor and Ashraf. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sobia Manzoor, ZHIuc29iaWFtYW56b29yQGFzYWIubnVzdC5lZHUucGs=
†These authors have contributed equally to this work and share first authorship
‡These authors have contributed equally to this work
 Areej A. Sattar
Areej A. Sattar Ariba Qaiser
Ariba Qaiser Hina Kausar
Hina Kausar Sarah Aqil
Sarah Aqil Rida Mudassar
Rida Mudassar Sobia Manzoor
Sobia Manzoor Javed Ashraf2,3
Javed Ashraf2,3