
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 02 February 2024
Sec. Comparative Immunology
Volume 15 - 2024 | https://doi.org/10.3389/fimmu.2024.1301877
Visceral white spot disease (VWND) caused by Pseudomonas plecoglossicida poses a major threat to the sustainable development of large yellow croaker (Larimichthys crocea) aquaculture. Genome-wide association analysis (GWAS) and RNA-seq research indicated that LcCD82a play an important role in resistance to visceral white spot disease in L. crocea, but the molecular mechanism of LcCD82a response to P. plecoglossicida infection is still unclear. In this study, we cloned and validated the Open Reading Frame (ORF) sequence of LcCD82a and explored the expression profile of LcCD82a in various tissues of L.crocea. In addition, two different transcript variants (LcCD82a-L and LcCD82a-S) of LcCD82a were identified that exhibit alternative splicing patterns after P. plecoglossicida infection, which may be closely related to the immune regulation during pathogenetic process of VWND. In order to explore the function of LcCD82a, we purified the recombinant protein of LcCD82a-L and LcCD82a-S. The bacterial agglutination and apoptosis function analysis showed that LcCD82a may involve in extracellular bacterial recognition, agglutination, and at the same time participate in the process of antigen presentation and induction of cell apoptosis. Collectively, our studies demonstrate that LcCD82a plays a crucial role in regulating apoptosis and antimicrobial immunity.
Large yellow croaker (Larimichthys crocea) is an important economic fish in China, and the huge market promotes the rapid development of the large yellow croaker farming industry (1). However, the highly intensive farming model has resulted in frequent occurrence of diseases including VWND caused by Pseudomonas plecoglossicida (2). Pseudomonas are a group of conditional pathogens, and the health status of the host and the surrounding environmental conditions (such as temperature) are the main factors affecting the pathogenicity of Pseudomonas (3). Pseudomonas plecoglossicida is a common pathogen in aquaculture, and can infect hosts such as large yellow croaker (Larimichthys crocea) (4), Plecoglossus altivelis (5), and Epinephelus coioides (6), causing visceral white-nodules disease (VWND). VWND caused by P. plecoglossicida has become a limiting factor for the healthy and sustainable development of aquaculture. Most studies on P. plecoglossicida have focused on the pathogenic mechanism and key virulence genes. Combination of transcriptomic and proteomic analysis indicated that the genes (such as PVDS2 and ICMF) involved in PVD synthesis and T6SS are involved in virulence of P. plecoglossicida (7). Dual RNA-seq in E. coioides revealed that secY is the key virulence gene for the pathogenicity of P. plecoglossicida, deepening the understanding of the pathogenic mechanism of P. plecoglossicida (8). Several pathological features for VWND can be observed, including white nodules in internal organs such as the spleen, liver and kidneys, as well as ulcers on the skin surface of infected L. crocea (9). Outbreaks of visceral white spot disease in cultured L. crocea are usually temperature-dependent (15 - 20°C is the optimal temperature), and the mortality rate of infection is extremely high (70 - 80%), which poses a major threat to the sustainability of the L. crocea aquaculture industry (3, 10). Studies have been conducted to investigate genes associated with P. plecoglossicida infection (11, 12), however the detailed pathogenesis of VWND and host resistance genes are unknown.
We have elucidated the genetic loci and the immune mechanisms of resistance to P. plecoglossicida in L. crocea by combined Genome-wide association analysis (GWAS) and RNA-seq analyses, and found that the CD82a gene and the p53 pathway play critical roles in VWDN resistance (9). CD82, also known as KAI1, belongs to the tetraspanin superfamily. Tetraspanins are cell-surface glycoproteins that are widely expressed in eukaryotic cell membranes and structurally comprise four hydrophobic membrane-spanning domains, which divide the protein molecule into a small extracellular loop (SEL), a large extracellular loop (LEL) with a CCG motif and two short intracellular termini (13, 14). Each member of tetraspanins assembles into multimeric complexes at the plasma membrane between or with other protein molecules such as integrins, adhesion molecules, and immunoglobulin superfamilies, thereby regulating various physiological processes such as cell differentiation, cell adhesion, apoptosis, viral infection, and immune responses (15–18).
CD82 has been demonstrated to have important roles in the immune system, including antigen (Ag) recognition and presentation, cell proliferation and apoptosis (19). Apoptosis is an essential component of normal physiological activity and is essential for the regulation of the cell cycle as well as for the growth, development and maintenance of homeostasis in the individual (20). In particular, apoptosis maintains tissue homeostasis and is therefore also involved in the development of some diseases and immune responses (21, 22). The widely reported role of CD82 is to inhibit the migration of epithelial cells, endothelial cells and the spread of cancer cells (23). Recently, it has been shown that CD82 also inhibits the migration of phagocytes, including neutrophils and dendritic cells, and promotes antigen presentation (24, 25). Antigen processing and presentation are complex processes that generate phagosomes, and previous studies have shown that CD82 is recruited to phagosomes during endocytosis of Gram-negative and Gram-positive bacteria (26). This indicates that CD82 is closely related to and plays an essential role in anti-bacterial infections.
Tetraspanin proteins have also been identified and characterized in fish and other lower vertebrates. A homologue of CD82, named Lja-CD82-like, exists in the lamprey Lampetra japonica and possesses highly conserved four transmembrane structural domains (27). The results showed that Lja-CD82-like was significantly expressed in immune tissues and that Lja-CD82-like was delivered in the cell membrane and cytoplasm after antigenic stimulation, suggesting that Lja-CD82-like may be involved in the immune defense process. Another gene family study in teleost fishes showed that each fish species possesses a different number of tetraspanin genes, and there was an intron gain during the evolution of the CD82 gene, which has a higher Ka/Ks value and a faster rate of change. Also, this study showed that CD82 expression level was differentially regulated in zebrafish, medaka, stickleback, and fugu under OP stress. This suggests that tetraspanin genes, including CD82 may be involved in regulating the immune response process in fish (28). However, the molecular characterization, expression pattern and immunologic function of CD82a in large yellow croaker are still unknown.
In this study, we performed cloning, recombinant expression of LcCD82a, explored the subcellular localization of LcCD82a, investigated the tissue distribution of LcCD82a and identified two different transcript variants, and initially characterized the function of LcCD82a in apoptosis and P. plecoglossicida infection. The results of this study will be helpful for understanding the molecular mechanism of LcCD82a in the immunoregulation of VWND in L. crocea.
This work was approved by the Animal Care and Use Committee at the College of Ocean and Earth Sciences, Xiamen University. All the methods used in this study were carried out following approved guidelines.
Healthy large yellow croaker purchased from Fufa Aquatic Breeding Company (Ningde, Fujian) were cultured temporarily in cement ponds (26 ± 0.2°C) for 15 days and then used for artificial infection experiments. During the temporary culture period, the fish were observed daily to ensure that the experimental fish were not infected with pathogens such as Cryptocaryon irritans and P. plecoglossicid. Feed at the same time every day and stop feeding one week before the start of the experiment. The experimental fish were randomly and equally distributed to two groups with intraperitoneal injection for P. plecoglossicid infection. Liver, spleen, gill and head kidney tissues were collected from 6 fish in the control group at 0 h followed by the same tissue samples as above at 72 h post-infection. All fish were anesthetized with tricaine methanesulfonate (MS-222; Sigma, St. Louis, MO, USA) at a concentration of 10 mg/L before sampling, and samples were snap-frozen in liquid nitrogen and stored at −80°C for subsequent RNA extraction and fluorescence quantification.
Total RNA was extracted from the collected tissues with TRIzol™ Reagent (Invitrogen), and cDNA was synthesized by reverse transcription with PrimeScript RT reagent Kit with gDNA Eraser Kit (TaKaRa). The open reading frame (ORF) sequence of LcCD82a was obtained from the L. crocea transcriptome data (accession number PRJNA836173), and then PCR amplification was performed with primers LcCD82a-verification-F/R (Table 1). The PCR products were gel-purified and cloned and then sent to Sangon for sequencing to confirm the correct sequence.
Transmembrane structural domains were analyzed using the SOSUI website (https://harrier.nagahama-ibio.ac.jp/sosui/). Alphafold (v. 2.2.2) was used for protein 3D structure prediction. The shear isoforms of LcCD82a were re-annotated using GMAP software, and the structures were visualized using R scripts. SnapGene was used for multiple sequence comparison of sequencing data.
In subsequent experiments (overexpression, flow cytometry), we explored the biological functions of LcCD82 using LYCK cell line. The LYCK cell line was a generous gift from the Fujian Academy of Agricultural Sciences. LYC cells were cultured in 199 Medium (M199, Gibco) containing 100U/ml penicillin-streptomycin and 10% fetal bovine serum (FBS, Gibco) at 28°C with 5% CO2. HEK-293T cells were cultured in DMEM Medium (Gibco) containing 100U/ml penicillin-streptomycin and 10% fetal bovine serum (FBS, Gibco) at 37°C with 5% CO2. In order to express LcCD82a, the ORF of LcCD82a was linked to the overexpression vector pCMV-flag to construct pCMV-flag-LcCD82a_L/S, which can express flag-tagged LcCD82a_L/S protein. In addition, we also used a vector with enhanced green fluorescent protein (EGFP) linked to LcCD82a_L/S, which can simultaneously express green fluorescent protein and LcCD82a_L/S protein, for exploring the subcellular localization of LcCD82a. To obtain recombinant proteins, LcCD82a-LEL was amplified by PCR using primers LcCD82a-LEL-F/R, ligated to pET32a vectors.
Cells were seeded into 96-well glass bottom plate and approximately 60% confluence at 28°C. Then, 100 ng of pCMV-EGFP-LcCD82a_L/S or pCMV-EGFP empty vector (control) were transfected into HEK-293T cells using Lipo 8000 Transfection Reagent (Beyotime) following the manufacturer’s protocol. After transfection 48 h, cells were washed with PBS, fixed with 4% paraformaldehyde for 15 min, treated with 0.1% ~ 0.2% Triton-X-100 for 5 min at room temperature, and then stained with DiD and Hoechst for 5 min at room temperature. Finally, the cells were washed five times with PBS and then observed under LSM780 (Carl Zeiss). Each experiment has three independent biological replicates and at least three technical replicates. Each group was repeated three times independently.
LYCK cells were seeded into a 6-well plate, and 2.5 µg of pCMV-myc-LcCD82a or pCMV-myc empty vector (control) were transfected into LYCK cells using Lipo 8000 Transfection Reagent (Beyotime) following the manufacturer’s protocol. The effect of overexpression was confirmed by qPT-PCR detection of mRNA expression level. Inactivated P. plecoglossicid (106 CFU/ml) was added to stimulate the cells after 8 h post-transfection. Cells that were not transfected were also subjected to the same treatment as a control. After 12 hours of treatment, the cells were washed with PBS, and the apoptosis was detected with Annexin V-FITC/PI Apoptosis Detection Kit (Beyotime) in a CytoFLEX (Beckman Coulter Life Science). Each experiment has three independent biological replicates and three technical replicates.
In order to obtain recombinant LcCD82a-LEL protein, use primers F and R to amplify the LcCD82a-LEL fragment with XhoI and EcoRI restriction sites, and insert it into pET-28a plasmid. Transform pET-28a-LcCD82a-LEL into E. coli, add IPTG (final concentration 0.5mM) when the transformant grows to OD600 = 0.6, and then continue to grow at 28°C for 4 h. Ultrasonic lysis of E. coli was performed on ice after replacing the bacterial culture solution with buffer (300 mM NaCl, 50 mM NaH2PO4, and 10 mM imidazole; pH 8.0). The supernatant was collected by centrifugation and filtered using a 0.22 μm acrodisc syringe filter, then recombinant proteins was eluted using elution buffer (300 mM NaCl, 50 mM NaH2PO4, and 250 mM imidazole; pH 8.0) after supernatant passing through a Ni-NTA column and dialyzed in PBS at 4°C to eliminate imidazole.
P. plecoglossicida and Staphylococcus aureus were amplified and cultured at 37°C to the exponential phase (OD600 = 0.6), centrifuged at 12000 g for 15 min, and the bacteria were resuspended in PBS to obtain a final bacterial suspension with OD600 = 0.2. Add 10 μl of bacterial suspension to 25 μl rLcCD82a-LEL dissolved in the same buffer as negative control and incubated at room temperature for 45 min, then observed under confocal microscope and electron microscope. Each experiment has three independent biological replicates and three technical replicates.
RNA was extracted from the collected tissues, and then, according to the protocol of PrimeScript™ RT reagent Kit (Takara, Japan), first-strand cDNA was synthesized using total RNA as a template. Expression patterns and temporal expression changes of LcCD82a in response to P. plecoglossicida infection were determined by qRT-PCR with primers LcCD82a-RT-F and LcCD82a-RT-R (Table 1). The reference gene is β-actin. The relative expression at each time point was analyzed using the 2-ΔΔCt method. Results were analyzed using one-way ANOVA and plotted using GraphPad Prism software. Each experiment has three independent biological replicates and three technical replicates.
Primers for LcCD82a were designed in the conserved regions of the two transcripts, PCR amplification and sequencing were performed using gill, liver, spleen and head kidney tissues sample as a template, and the results showed that transcript variants existed in gills and head kidney (Figure 1A), that is, there were two transcripts before infection, and only the transcript with a tetraspanin structure was present after infection. They were named LcCD82a-L and LcCD82a-S according to their length, the complete transcript length is 334 bp, and the variant is 262 bp. LcCD82a-S had a loss of the first exon (Figure 1B), and this caused a backward shift of the coding start site (Figures 1F, G), thereby changing the post-translational protein structure (Figures 1C–E). The visualization of the transmembrane structure, the reconstruction of the three-dimensional structure of the protein, and the prediction of the transmembrane region showed that LcCD82a-L has a classic four-transmembrane structure (two extracellular regions, three intracellular regions, and four transmembrane regions). The two extracellular regions were divided into LEL and SEL, but the intracellular segment and SEL structure of the N-terminal of LcCD82a-S protein were destroyed.
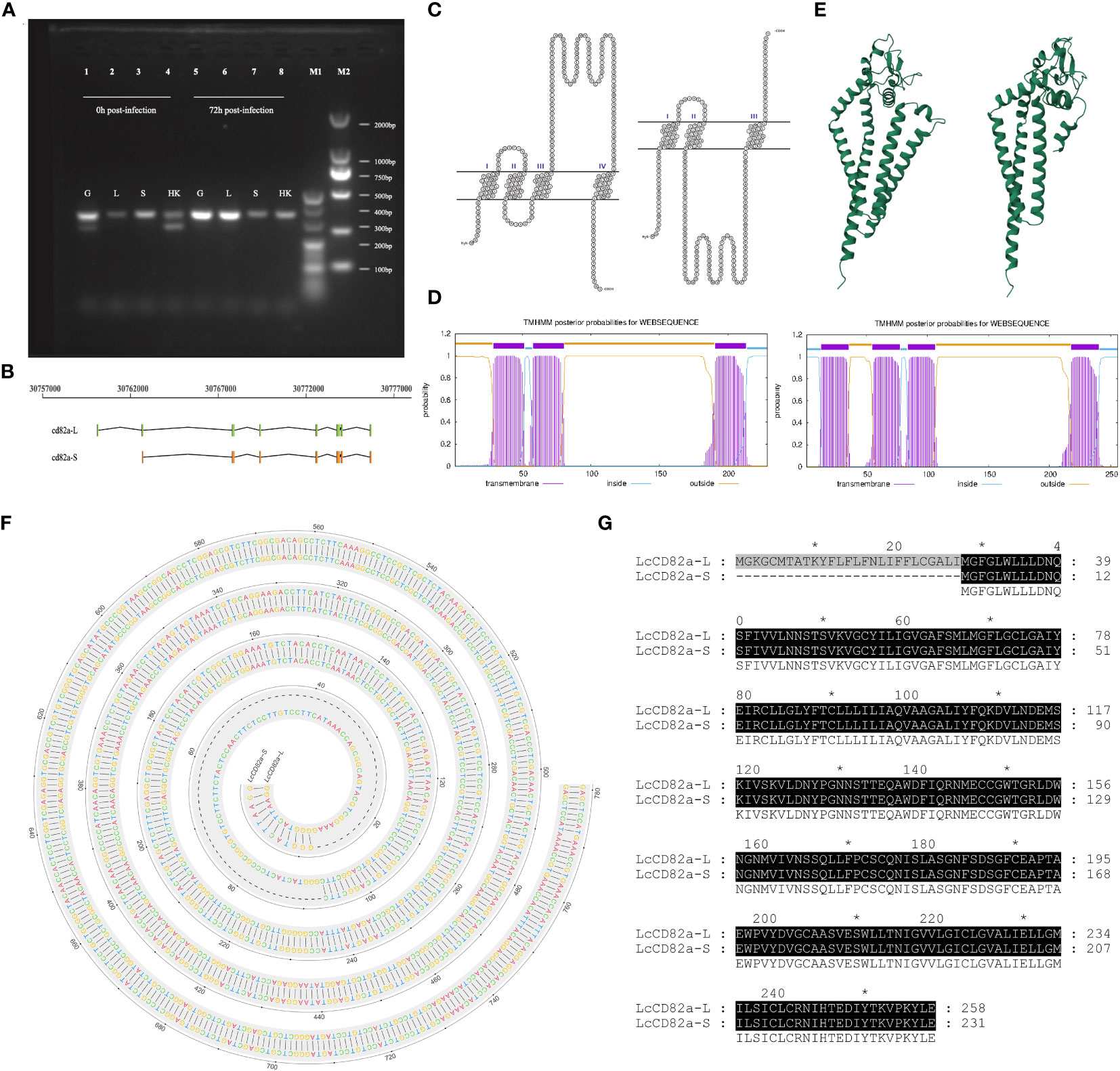
Figure 1 Structural prediction and splicing pattern analysis of transcript variants LcCD82a-L/S. (A) PCR verification of alternative splicing patterns. (B) Linear structures of two transcript variants. (C) Visualization of transmembrane structure of transcript variants. (D) Three-dimensional structural reconstruction of proteins encoded by transcript variants. (E) Prediction of transmembrane regions of proteins encoded by transcript variants. (F) DNA sequence alignment of CD82a-L and CD82a-S. The “-” indicates differences. (G) Amino acid sequence alignment of CD82a-L and CD82a-S. The results of the homology analysis showed 100% sequence homology between CD82a-L and CD82a-S, except for the first exon region (indicated by black markers in the figure, with darker colors indicating higher homology).
The bacteria in the fermentation broth were sonicated, the target protein was expressed in its soluble form, and the eluted sample was purer and at a higher concentration when the concentration of imidazole was 250 mM (Figure S1). The samples with high purity were mixed to obtain about 5 ml of high-purity LcCD82a-LEL protein, and the protein concentration was about 0.33 mg/ml.
To explore the differences in the expression levels of LcCD82a among different tissues, we examined it in 11 tissues of healthy L. crocea. Furthermore, the LcCD82a gene was quantified in immune tissues before and after infection, to explore whether the infection of P. plecoglossicida in immune tissues would cause the differential regulation of LcCD82a. The results showed that the LcCD82a gene was widely expressed in most tissues of L.crocea (Figure 2A), with the highest expression in brain (B), intestinal tract (I), and liver (L) tissues, kidney (K), head kidney (HK), and spleen (S) had the lowest expression. After infection with P. plecoglossicida, the expression of LcCD82a was down-regulated in spleen (S) and head kidney (HK) (Figure 2B), which were about one-fifth (26% and 20%) of the pre-infection level respectively. The expression levels in livers (L) and gills (G) were up-regulated, which were 2.32 times and 1.65 times that before infection, respectively, and the differences were significant (P < 0.05).
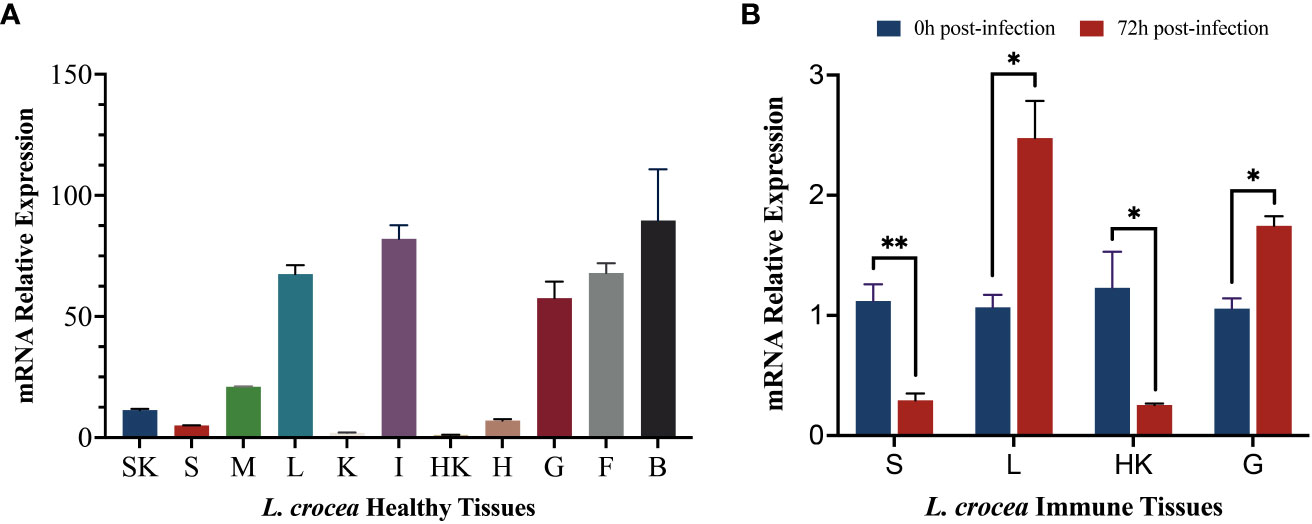
Figure 2 mRNA expression profiles of LcCD82a in healthy tissues and immune tissues. The data represents the log2 fold change of expression at different time points compared to the control group. (A) Relative mRNA expression levels of LcCD82a in various tissues. (B) The mRNA expression profiles of LcCD82a in immunized tissues at different time points after challenge. *p < 0.05, **p<0.01.
The cell lines were transfected with pCMV-LcCD82a-S-EGFP and pCMV-LcCD82a-S-EGFP two plasmids with green fluorescent markers for subcellular localization, and the empty plasmid (pCMV-C-EGFP) was used as a control group. The fusion proteins of rLcCD82a-L and rLcCD82a-S share the same distributional location in the cell, being strongly localized to the cell membrane, the nuclear membrane and various organelle membranes (Figure 3). This suggests that the structural differences between LcCD82a-L and LcCD82a-S do not affect the position of distribution in the cell, and that both proteins function on the biomembrane system. The green fluorescent protein of the control group was widely distributed in the nucleus, cytoplasm and cell membrane (Figure 3), indicating that the green fluorescent protein and the carrier plasmid did not affect the subcellular localization of the target protein.
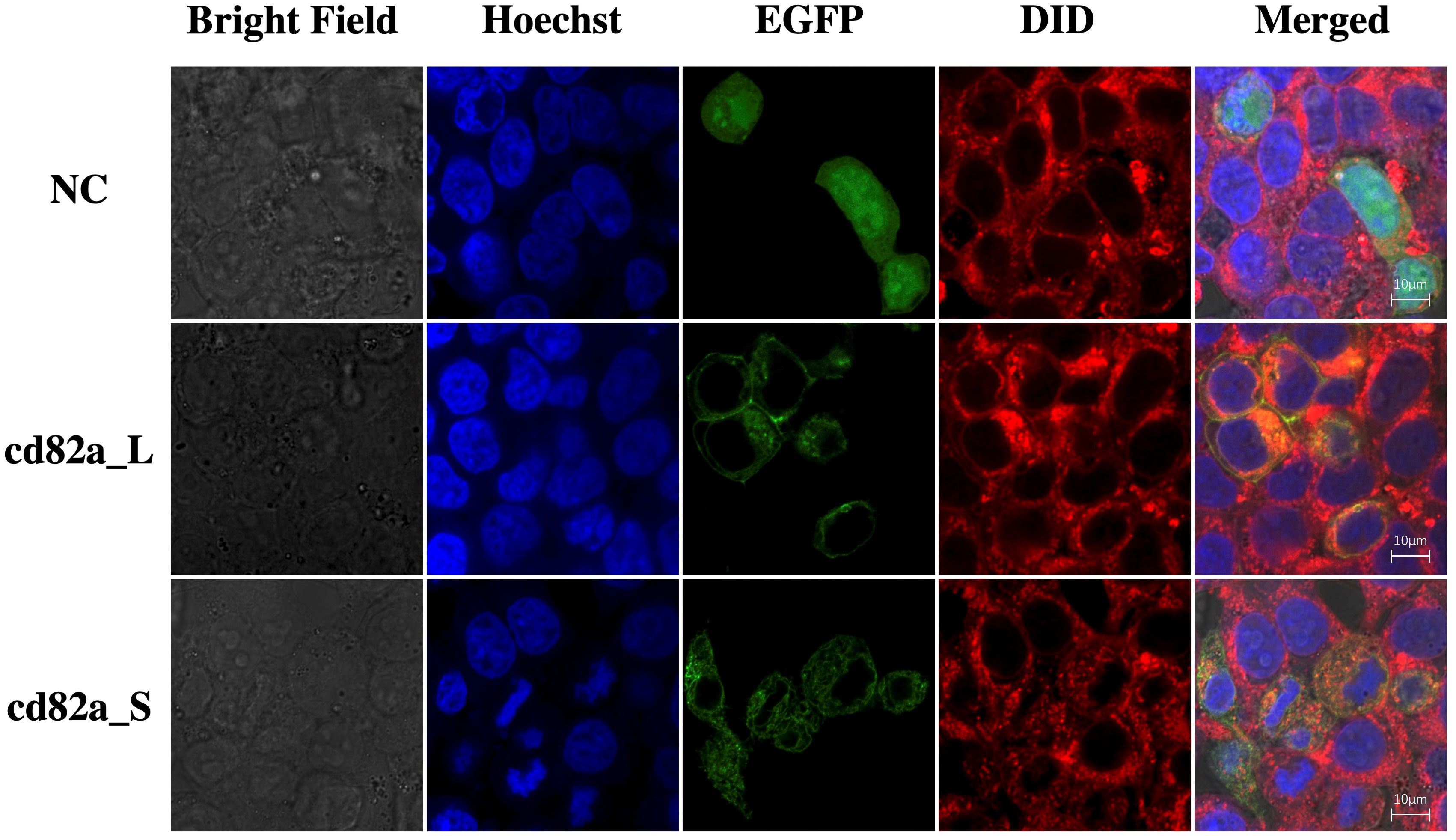
Figure 3 Subcellular localization of transcript variants LcCD82a-L/S. The first column is the cell morphology taken in bright field, the second column is the nuclear staining with Hoechst dye, the third column is the fusion protein of the target sequence and EGFP, the fourth column is the cell membrane staining with DID dye, and the last column is the result after merge.
In order to explore the molecular functions of LcCD82a, LcCD82a-L and LcCD82a-S were overexpressed in LYCK cells, which were stimulated with inactivated P. plecoglossicida, and then detected by flow cytometry. Figures 4A, B show the control groups. The percentage of normal cells in cells transfected with pCMV-LcCD82a-S was 17.0% (Figure 4D), which was lower than that in the control group (24.6%), and the apoptosis rate was 82.6%, of which 40.6% were early apoptotic cells and 42.0% were late apoptotic cells. The apoptosis rate after transfection of pCMV-LcCD82a-S was significantly higher than that of the control group (P < 0.05), but the percentage of late apoptotic cells was significantly lower than that of the control group (48.4%), mainly caused by the significantly higher percentage of early apoptotic cells than that of the control group (Figure 4C). The percentage of normal cells in cells transfected with pCMV-LcCD82a-L was 11.6% (Figure 4E), and the apoptosis rate was 87.9%, of which 18.1% were early apoptotic cells and 69.8% were late apoptotic cells. The apoptosis rate after transfection with pCMV-LcCD82a-L was higher than that of the control group (74.9%), but the number of early apoptotic cells was reduced, mainly contributed by the number of late apoptotic cells. Figure 4F demonstrates that the apoptosis rate of LYCK cells was significantly upregulated after overexpression of LcCD82a-L (P < 0.01).
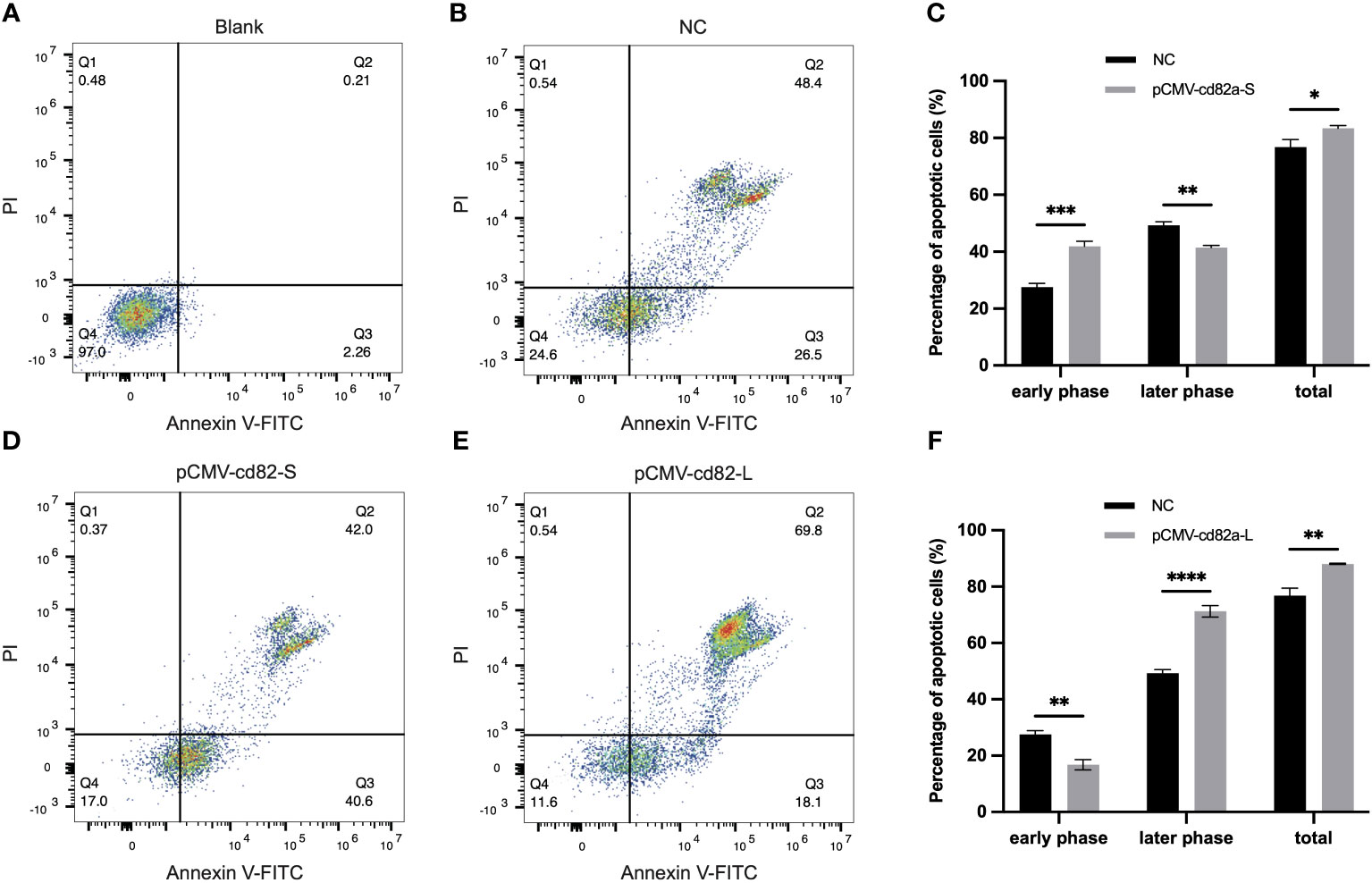
Figure 4 LcCD82a-L/S activates apoptosis of LYCK cells stimulated by P. plecoglossicida. (A, B) Blank control and negative control. (C, D) Changes in apoptotic rate of LYCK cells after LcCD82a-S overexpression followed by P. plecoglossicida challenge. *p < 0.05, **p < 0.01, ***p < 0.001. (E, F) Changes in apoptotic rate of LYCK cells after LcCD82a-L overexpression followed by P. plecoglossicida challenge. **p < 0.01, ***p < 0.001, ****p < 0.0001.
In this study, bacterial agglutination assays were performed using rLcCD82a-LEL recombinant protein, using S. aureus and P. plecoglossicida (Figure S2) as model pathogens, respectively. Fluorescent labeling was performed on both bacteria, which were photographed using fluorescence confocal microscopy (100×) and electron microscopy (50,000x), respectively. S. aureus and P. plecoglossicida without recombinant protein added were both evenly distributed in the field of view with regular, intact shapes. After adding rLcCD82a-LEL, bacterial agglutination was observed in both S. aureus and P. plecoglossicida (Figure 5). And under the electron microscope, it could be observed that the morphology of the bacteria changed, and some of them ruptured and became wrinkled after death.
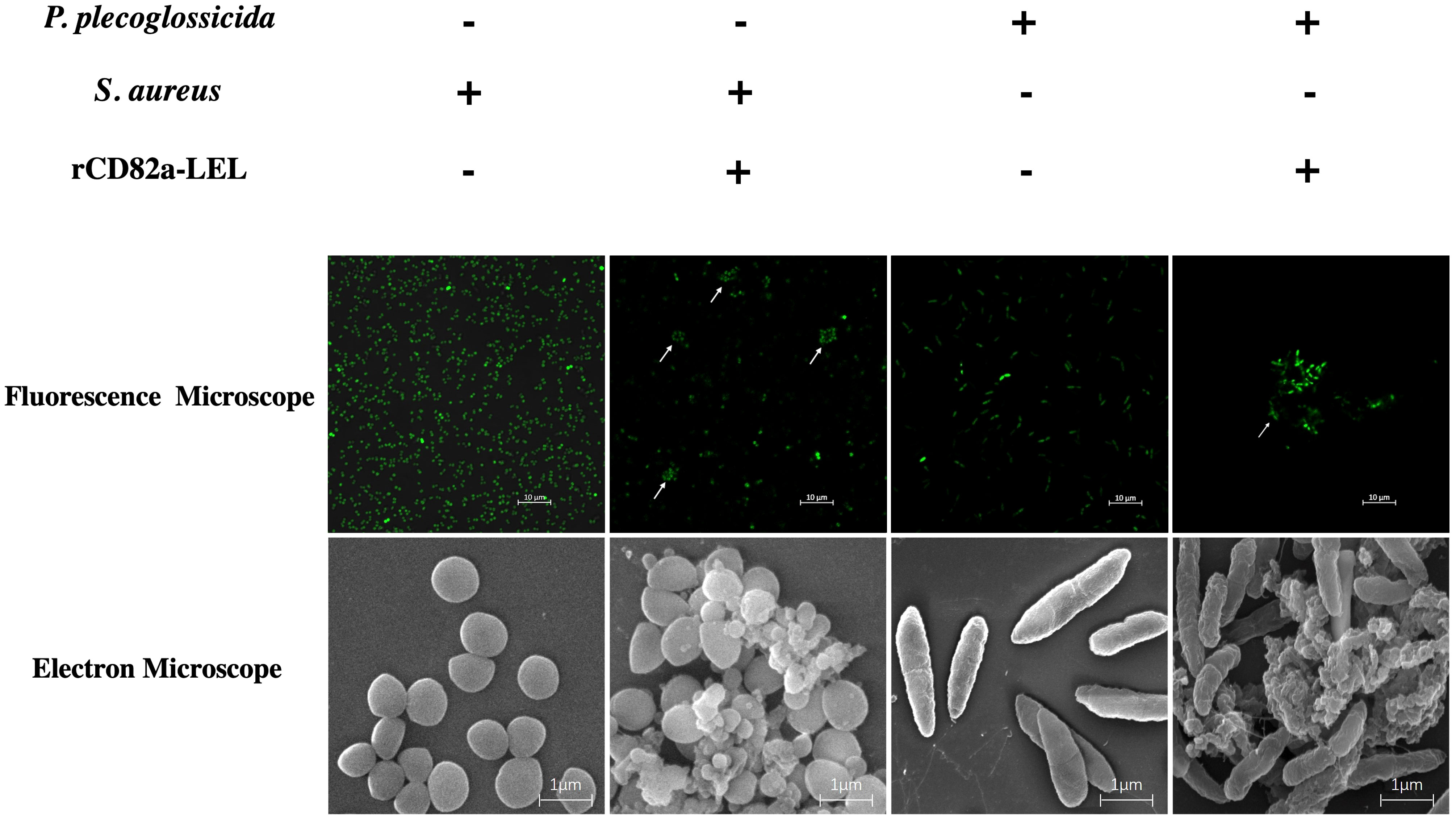
Figure 5 Agglutination activity of rLcCD82a-LEL against FITC-labeled Gram-positive bacteria (S. aureus) and Gram-negative bacteria (P. plecoglossicida). FITC-labeled S. aureus and P. plecoglossicida were co-incubated with rLcCD82a-LEL, and images were taken using an LSM 780 confocal microscope, with arrows showing bacterial agglutination.
As an important factor for maintaining normal physiological activities, CD82a is widely distributed in various tissues and is specifically expressed in different physiological and pathological states (29–33). Our results demonstrated that LcCD82a was expressed in various tissues of healthy L. crocea, with the highest expression level in the brain, intestine, and liver. Previous studies in Lampetra japonica have shown similar results, with CD82 mRNA detected in several tissues and highest in blood cells and liver. It was reported that CD82 play a significant role in the defense against pathogens such as bacteria (26). In this study, exploration of LcCD82a expression patterns in immune tissues during P. plecoglossicida infection revealed significant differential expression in all tested immune tissues during the peak infection period (upregulation in liver and gills, downregulation in spleen and head kidney), which was consistent with the pathogen distribution identified using immunohistochemistry (IHC) and confirming the presence of immune cell aggregation in tissues such as liver and gills of Pseudosciaena crocea (34, 35).
We characterized two LcCD82a gene transcript variants, LcCD82a-L and LcCD82a-S, with transcript differences primarily located in the first exon region. The shorter transcript variant LcCD82a-S lacks a portion of the first exon, resulting in the disruption of the N-terminal cytoplasmic domain and extracellular loop, leaving only the remaining three major transmembrane regions. Moreover, an analysis of differential splicing patterns in gills and head kidney tissues before and after infection indicated selective splicing of LcCD82a-S specifically before infection. CD82 has functions such as antigen presentation (19), and the head kidney is an important immune organ that can produce a variety of immune cells, and the gills are rich in blood and leukocyte (36). Therefore, the LcCD82a-L may be involved in the immune defense of organisms against bacterial infections, and the coding region of the first exon may be closely related to bacterial infections, antigen recognition and presentation.
Subcellular localization can explore the specific location of a certain protein or expression product distribution in cells, thus providing a research direction for understanding the mechanism of action of genes. We explored the subcellular localization of LcCD82a and found that both LcCD82a-L and LcCD82a-S were localized in the cytoplasm, nuclear membrane, organelle membrane and cell membrane. Previous evidence has shown that CD82 is a tetraspanin that is ubiquitously present in various biological membranes, such as the inner membrane of exosomes carrying major histocompatibility complex (MHC) class II molecules (37). The subcellular localization of LcCD82a provides strong evidence for its function. LcCD82a is a member of the tetraspanin family and plays an antigen-presenting function on biological membranes. Its localization in the nuclear membrane and cytoplasm is closely related to apoptosis.
Apoptosis plays an important role in maintaining normal tissue development and homeostasis, and dysregulation of apoptotic signaling can lead to various pathological conditions in organisms. Cell apoptosis is an important pathway limiting pathogen replication and dissemination, and early apoptosis following pathogen infection can contribute to disease control. Therefore, we overexpressed LcCD82a-L and LcCD82a-S in LYCK cells to investigate the effects of two different transcriptional variants of LcCD82a on apoptosis after stimulation by P. plecoglossicida. Our results showed that although the two transcript variants affected apoptosis in different ways and degrees, they both promoted apoptosis after pathogen infection in general. Previous research has shown that some viruses have genes that inhibit the apoptosis of infected cells and promote their own survival and proliferation. For example, the proteins encoded by the BHRF-1 gene of Epstein-Barr virus (EBV) (38) and the LMW5-HL gene of African swine fever virus (ASFV) (39) are similar to the BCL-2 protein sequence, so they also play the role of inhibiting apoptosis. In addition, the gene LMP-1 of EBV inhibits p53-triggered apoptosis during the incubation period of infection, creating favorable survival conditions for infected B cells (40). Consequently, LcCD82a promoted apoptosis of P. plecoglossicida-infected LYCK cells, which could clear the pathogen and inhibit its proliferation to a certain extent, thereby playing a positive role in the process of resisting P. plecoglossicida in L. crocea. In addition, cells are phagocytized by phagocytic cells after apoptosis, and the cell contents do not flow out without triggering inflammation, which is a relatively mild process (41). LcCD82a induces apoptosis in infected cells, which reduces the extent of damage in the diseased area and improves the survival rate of L. crocea as compared to inflammation caused by excessive immune response.
Cell apoptosis and antigen presentation processes are interdependent, with apoptotic cells or apoptotic bodies serving as the major source of cross-presentation of antigens (42). Phagocytosis is a primitive defense mechanism, and teleost M1 macrophages are an important component of teleost innate immunity, which can recognize and phagocytose invading pathogens (43–45). In addition, macrophages are also an important bridge connecting the innate immunity and adaptive immunity of teleost. Macrophages present intracellular pathogen-derived antigens to T cells, thereby triggering adaptive immunity (46–48). Antigen presenting cells (APCs) are a type of immune cells that can ingest, process and present antigens to T lymphocytes, and play an important role in immune response. CD82 and other tetraspanins were previously found in APCs (17, 49). To investigate the effect of CD82 on pathogen recognition and antigen presentation, we performed bacterial agglutination assays using rLcCD82a-LEL. The results showed that in the groups to which rLcCD82a-LEL was added, the bacteria all appeared to agglutinate and cluster. CD82 has been shown to be actively recruited to phagosomes containing fungi (Cryptococcus neoformans, Candida albicans and Aspergillus fumigatus), Gram-positive bacteria (Staphylococcus aureus), or Gram-negative bacteria (Escherichia coli). And CD82 overlaps with the endocytic pathway of lipopolysaccharide (LPS), suggesting that CD82 is related to the presentation of antigen-associated molecules (26). Moreover, previous studies have found that there are antimicrobial proteins and peptides that can agglutinate bacteria. Antimicrobial proteins in saliva cause bacteria in the mouth to agglutinate and act as the first barrier of defense against pathogens (50). Eosinophil cationic protein (ECP) is a secreted protein with strong antimicrobial effects, and it has been reported to have bacterial agglutination activity against Gram-negative bacteria in a concentration range close to the minimal inhibitory concentration, suggesting that agglutination of bacteria plays an essential role in the bactericidal process (51, 52). Based on the above evidence, we reckon that LcCD82a is involved in the phagocytosis and antigen presentation of P. plecoglossicida, and can agglutinate invading pathogens to inhibit and kill them.
The spatial and temporal expression patterns of LcCD82a in the kidney cell line and individual specimens of the fish were characterized. Two transcript variants, LcCD82a-L and LcCD82a-S, were identified with differential transcriptional patterns before and after infection with P. plecoglossicida, suggesting a potential association with post-infection immune regulation. Further investigation into the molecular functions of the transcript variants LcCD82a-L/S and the simulated peptide LcCD82-LEL revealed that LcCD82a plays a critical role as a membrane protein, involved in extracellular bacterial recognition and aggregation. These findings would be useful for understanding the molecular mechanism of VWND in large yellow croaker.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Ethical approval was not required for the studies on humans in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used. The animal study was approved by the Animal Care and Use Committee at the College of Ocean and Earth Sciences, Xiamen University. The study was conducted in accordance with the local legislation and institutional requirements.
YL: Data curation, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing. YB: Data curation, Methodology, Writing – review & editing. SC: Data curation, Validation, Writing – review & editing. FP: Supervision, Writing – review & editing. YL: Project administration, Writing – review & editing. HC: Resources, Writing – review & editing. ZZ: Resources, Writing – review & editing. PX: Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing. TZ: Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National key research and development program (2022YFD2401002), the National Sciences Fund for Distinguished Young Scholars (32225049), the National Natural Science Foundation of China (U21A20264), the Seed Industry Innovation and Industrialization Project of Fujian Province ( 2021FJSCZY01).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1301877/full#supplementary-material
Supplementary Figure 1 | The identification of purity of proteins collected from elution samples by SDS-PAGE. Lane 1: protein marker, lane 2: pET32a-LcCD82a-LEL bacterial liquid (not induced by IPTG), lane 3: pET32a-LcCD82a-LEL bacterial liquid (induced by IPTG), lane 4: Supernatant of pET32a-LcCD82a-LEL bacterial solution after sonication, lane 5: The remaining liquid after the supernatant of pET32a-LcCD82a-LEL bacterial solution (after sonication) passed through the NTA column, lane 6: Precipitation of pET32a-LcCD82a-LEL bacterial solution after sonication, lane 7: The liquid was eluted by 0 mM imidazole, lane 8: the liquid was eluted by 50 mM imidazole, lane 9: the liquid was eluted by 100 mM imidazole, lane 10: the liquid was eluted by 250 mM imidazole, lane 11: the liquid was eluted by 500 mM imidazole.
Supplementary Figure 2 | Electron microscopic photographs of P. plecoglossicida. Scale bars are 1 µm length in image.
1. Bai Y, Liu Y, Qu A, Wang J, Zhao J, Ke Q, et al. Dual RNA-seq reveals a host-pathogen interaction transcriptional regulation pattern between Cryptocaryon irritans and large yellow croaker (Larimichthys crocea). Aquaculture (2023) 565. doi: 10.1016/j.aquaculture.2022.739104
2. Li C, Wang S, Ren Q, He T, Chen X. An outbreak of visceral white nodules disease caused by Pseudomonas plecoglossicida at a water temperature of 12 degrees C in cultured large yellow croaker (Larimichthys crocea) in China. J Fish Dis (2020) 43(11):1353–61. doi: 10.1111/jfd.13206
3. Li C, Wang S, Ren Q, He T, Chen X. An outbreak of visceral white nodules disease caused by Pseudomonas plecoglossicida at a water temperature of 12°C in cultured large yellow croaker (Larimichthys crocea) in China. J Fish Dis (2020) 43(11):1353–61. doi: 10.1111/jfd.13206
4. Hu J, Zhang F, Xu X, Su Y, Qin Y, Ma Y, et al. Identification and virulence of the pathogen of white-spots disease in internal organs of Pseudosciaena Crocea. Oceanologia Et Limnologia Sin (2014) 45(2):409–17. doi: 10.11693/hyhz20140300078
5. Nishimori E, Kita-Tsukamoto K, Wakabayashi H. Pseudomonas plecoglossicida sp. nov., the causative agent of bacterial haemorrhagic ascites of ayu, Plecoglossus altivelis. Int J Syst Evol Microbiol (2000) 50 Pt 1:83–9. doi: 10.1099/00207713-50-1-83
6. Zhang B, Luo G, Zhao L, Huang L, Qin Y, Su Y, et al. Integration of RNAi and RNA-seq uncovers the immune responses of Epinephelus coioides to L321_RS19110 gene of Pseudomonas plecoglossicida. Fish Shellfish Immunol (2018) 81:121–9. doi: 10.1016/j.fsi.2018.06.051
7. Huang L, Liu W, Jiang Q, Zuo Y, Su Y, Zhao L, et al. Integration of transcriptomic and proteomic approaches reveals the temperature-dependent virulence of pseudomonas plecoglossicida. Front Cell Infect Microbiol (2018) 8:207. doi: 10.3389/fcimb.2018.00207
8. Luo G, Sun Y, Huang L, Su Y, Zhao L, Qin Y, et al. Time-resolved dual RNA-seq of tissue uncovers Pseudomonas plecoglossicida key virulence genes in host-pathogen interaction with Epinephelus coioides. Environ Microbiol (2020) 22(2):677–93. doi: 10.1111/1462-2920.14884
9. Bai Y, Qu A, Liu Y, Chen X, Wang J, Zhao J, et al. Integrative analysis of GWAS and transcriptome reveals p53 signaling pathway mediates resistance to visceral white-nodules disease in large yellow croaker. Fish Shellfish Immunol (2022) 130:350–8. doi: 10.1016/j.fsi.2022.09.033
10. Bai Y, Wang J, Zhao J, Ke Q, Qu A, Deng Y, et al. Genomic selection for visceral white-nodules diseases resistance in large yellow croaker. Aquaculture (2022) 559. doi: 10.1016/j.aquaculture.2022.738421
11. Wang L, Sun Y, Zhao L, Xu X, Huang L, Qin Y, et al. Dual RNA-seq uncovers the immune response of Larimichthys crocea to the secY gene of Pseudomonas plecoglossicida from the perspective of host-pathogen interactions. Fish Shellfish Immunol (2019) 93:949–57. doi: 10.1016/j.fsi.2019.08.040
12. Wang LY, Liu ZX, Zhao LM, Huang LX, Qin YX, Su YQ, et al. Dual RNA-seq provides novel insight into the roles of dksA from Pseudomonas plecoglossicida in pathogen-host interactions with large yellow croakers ( Larimichthys crocea). Zool Res (2020) 41(4):410–22. doi: 10.24272/j.issn.2095-8137.2020.048
13. Stipp CS, Kolesnikova TV, Hemler ME. Functional domains in tetraspanin proteins. Trends Biochem Sci (2003) 28(2):106–12. doi: 10.1016/S0968-0004(02)00014-2
14. Lu X, An L, Fan G, Zang L, Huang W, Li J, et al. EGFR signaling promotes nuclear translocation of plasma membrane protein TSPAN8 to enhance tumor progression via STAT3-mediated transcription. Cell Res (2022) 32(4):359–74. doi: 10.1038/s41422-022-00628-8
15. Huang Y, Zucker B, Zhang S, Elias S, Zhu Y, Chen H, et al. Migrasome formation is mediated by assembly of micron-scale tetraspanin macrodomains. Nat Cell Biol (2019) 21(8):991–1002. doi: 10.1038/s41556-019-0367-5
16. Yáñez-Mó M, Barreiro O, Gordon-Alonso M, Sala-Valdés M, Sánchez-Madrid F. Tetraspanin-enriched microdomains: a functional unit in cell plasma membranes. Trends Cell Biol (2009) 19(9):434–46. doi: 10.1016/j.tcb.2009.06.004
17. Levy S, Shoham T. The tetraspanin web modulates immune-signalling complexes. Nat Rev Immunol (2005) 5(2):136–48. doi: 10.1038/nri1548
18. Zhang S, Kodys K, Babcock GJ, Szabo G. CD81/CD9 tetraspanins aid plasmacytoid dendritic cells in recognition of hepatitis C virus-infected cells and induction of interferon-alpha. Hepatology (2013) 58(3):940–9. doi: 10.1002/hep.25827
19. Rocha-Perugini V, Sánchez-Madrid F, Martínez Del Hoyo G. Function and dynamics of tetraspanins during antigen recognition and immunological synapse formation. Front Immunol (2015) 6:653. doi: 10.3389/fimmu.2015.00653
20. King KL, Cidlowski JA. Cell cycle regulation and apoptosis. Annu Rev Physiol (1998) 60(1):601–17. doi: 10.1146/annurev.physiol.60.1.601
21. Favaloro B, Allocati N, Graziano V, Di Ilio C, De Laurenzi V. Role of apoptosis in disease. Aging (Albany NY) (2012) 4(5):330–49. doi: 10.18632/aging.100459
22. Schwartzman RA, Cidlowski JA. Apoptosis: the biochemistry and molecular biology of programmed cell death. Endocr Rev (1993) 14(2):133–51. doi: 10.1210/edrv-14-2-133
23. Zöller M. Tetraspanins: push and pull in suppressing and promoting metastasis. Nat Rev Cancer (2009) 9(1):40–55. doi: 10.1038/nrc2543
24. Jones EL, Wee JL, Demaria MC, Blakeley J, Ho PK, Vega-Ramos J, et al. Dendritic cell migration and antigen presentation are coordinated by the opposing functions of the tetraspanins CD82 and CD37. J Immunol (2016) 196(3):978–87. doi: 10.4049/jimmunol.1500357
25. McGowan ENS, Wong O, Jones E, Nguyen J, Wee J, Demaria MC, et al. Tetraspanin CD82 restrains phagocyte migration but supports macrophage activation. iScience (2022) 25(7):104520. doi: 10.1016/j.isci.2022.104520
26. Artavanis-Tsakonas K, Kasperkovitz PV, Papa E, Cardenas ML, Khan NS, van der Veen AG, et al. The tetraspanin CD82 is specifically recruited to fungal and bacterial phagosomes prior to acidification. Infect Immun (2011) 79(3):1098–106. doi: 10.1128/IAI.01135-10
27. Zhang X, Song X, Su P, Gou M, Wang H, Liu X, et al. Molecular cloning, expression pattern, and phylogenetic analysis of a tetraspanin CD82-like molecule in lamprey Lampetra japonica. Dev Genes Evol (2016) 226(2):87–98. doi: 10.1007/s00427-016-0530-y
28. Cao J, Tan X. Comparative analysis of the tetraspanin gene family in six teleost fishes. Fish Shellfish Immunol (2018) 82:432–41. doi: 10.1016/j.fsi.2018.08.048
29. Tagawa K, Arihiro K, Takeshima Y, Hiyama E, Yamasaki M, Inai K. Down-regulation of KAI1 messenger RNA expression is not associated with loss of heterozygosity of the KAI1 gene region in lung adenocarcinoma. Jpn J Cancer Res (1999) 90(9):970–6. doi: 10.1111/j.1349-7006.1999.tb00843.x
30. Dong JT, Suzuki H, Pin SS, Bova GS, Schalken JA, Isaacs WB, et al. Down-regulation of the KAI1 metastasis suppressor gene during the progression of human prostatic cancer infrequently involves gene mutation or allelic loss. Cancer Res (1996) 56(19):4387–90. doi: 10.1097/00002820-199610000-00009
31. Liu FS, Dong JT, Chen JT, Hsieh YT, Ho ES, Hung MJ. Frequent down-regulation and lack of mutation of the KAI1 metastasis suppressor gene in epithelial ovarian carcinoma. Gynecol Oncol (2000) 78(1):10–5. doi: 10.1006/gyno.2000.5801
32. Miyazaki T, Kato H, Shitara Y, Yoshikawa M, Tajima K, Masuda N, et al. Mutation and expression of the metastasis suppressor gene KAI1 in esophageal squamous cell carcinoma. Cancer (2000) 89(5):955–62. doi: 10.1002/1097-0142(20000901)89:5<955::AID-CNCR3>3.0.CO;2-Z
33. Jackson P, Millar D, Kingsley E, Yardley G, Ow K, Clark S, et al. Methylation of a CpG island within the promoter region of the KAI1 metastasis suppressor gene is not responsible for down-regulation of KAI1 expression in invasive cancers or cancer cell lines. Cancer Lett (2000) 157(2):169–76. doi: 10.1016/S0304-3835(00)00483-3
34. Chen Z, Shi H, Xie JJ. Dynamic Pathology and Pathogen Distribution of Pseudosciaena crocea Infected by Pseudomonas plecoglossicida. J Zhejiang Ocean University (Natural Science) (2018) 37(6):483–8. doi: 10.3969/j.issn.1008-830X.2018.06.002
35. Zhang DF, An SW, Zhou SM, Wang GL. Histopathology and ultrastructure of visceral white-spots in Pseudosciaena crocea. Prog IN FISHERY Sci (2017) 38(7):11–6. doi: 10.11758/yykxjz.20160616001
36. Press CM, Evensen Ø. The morphology of the immune system in teleost fishes. Fish Shellfish Immunol (1999) 9(4):309–18. doi: 10.1006/fsim.1998.0181
37. Escola JM, Kleijmeer MJ, Stoorvogel W, Griffith JM, Yoshie O, Geuze HJ. Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes. J Biol Chem (1998) 273(32):20121–7. doi: 10.1074/jbc.273.32.20121
38. Flanagan AM, Letai A. BH3 domains define selective inhibitory interactions with BHRF-1 and KSHV BCL-2. Cell Death Differ (2008) 15(3):580–8. doi: 10.1038/sj.cdd.4402292
39. Neilan JG, Lu Z, Afonso CL, Kutish GF, Sussman MD, Rock DL. An African swine fever virus gene with similarity to the proto-oncogene bcl-2 and the Epstein-Barr virus gene BHRF1. J Virol (1993) 67(7):4391–4. doi: 10.1128/jvi.67.7.4391-4394.1993
40. Okan I, Wang Y, Chen F, Hu LF, Imreh S, Klein G, et al. The EBV-encoded LMP1 protein inhibits p53-triggered apoptosis but not growth arrest. Oncogene (1995) 11(6):1027–31. doi: 10.1016/0167-4889(95)00078-7
41. Call JA, Eckhardt SG, Camidge DR. Targeted manipulation of apoptosis in cancer treatment. Lancet Oncol (2008) 9(10):1002–11. doi: 10.1016/S1470-2045(08)70209-2
42. Taylor G, Wyld S, Valarcher JF, Guzman E, Thom M, Widdison S, et al. Recombinant bovine respiratory syncytial virus with deletion of the SH gene induces increased apoptosis and pro-inflammatory cytokines in vitro, and is attenuated and induces protective immunity in calves. J Gen Virol (2014) 95(Pt 6):1244–54. doi: 10.1099/vir.0.064931-0
43. Zhou D, Huang C, Lin Z, Zhan S, Kong L, Fang C, et al. Macrophage polarization and function with emphasis on the evolving roles of coordinated regulation of cellular signaling pathways. Cell Signal (2014) 26(2):192–7. doi: 10.1016/j.cellsig.2013.11.004
44. Rieger AM, Hall BE, Barreda DR. Macrophage activation differentially modulates particle binding, phagocytosis and downstream antimicrobial mechanisms. Dev Comp Immunol (2010) 34(11):1144–59. doi: 10.1016/j.dci.2010.06.006
45. Janeway CA Jr. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb Symp Quant Biol (1989) 54 Pt 1:1–13. doi: 10.1101/SQB.1989.054.01.003
46. Sever L, Vo NT, Bols NC, Dixon B. Expression of tapasin in rainbow trout tissues and cell lines and up regulation in a monocyte/macrophage cell line (RTS11) by a viral mimic and viral infection. Dev Comp Immunol (2014) 44(1):86–93. doi: 10.1016/j.dci.2013.11.019
47. Sever L, Vo NT, Lumsden J, Bols NC, Dixon B. Induction of rainbow trout MH class I and accessory proteins by viral haemorrhagic septicaemia virus. Mol Immunol (2014) 59(2):154–62. doi: 10.1016/j.molimm.2014.02.001
48. Sever L, Vo NTK, Bols NC, Dixon B. Tapasin's protein interactions in the rainbow trout peptide-loading complex. Dev Comp Immunol (2018) 81:262–70. doi: 10.1016/j.dci.2017.12.015
49. Tarrant JM, Robb L, van Spriel AB, Wright MD. Tetraspanins: molecular organisers of the leukocyte surface. Trends Immunol (2003) 24(11):610–7. doi: 10.1016/j.it.2003.09.011
50. Van Nieuw Amerongen A, Bolscher JG, Veerman EC. Salivary proteins: protective and diagnostic value in cariology? Caries Res (2004) 38(3):247–53. doi: 10.1159/000077762
51. Pulido D, Moussaoui M, Andreu D, Nogues MV, Torrent M, Boix E. Antimicrobial action and cell agglutination by the eosinophil cationic protein are modulated by the cell wall lipopolysaccharide structure. Antimicrob Agents Chemother (2012) 56(5):2378–85. doi: 10.1128/AAC.06107-11
Keywords: Larimichthys crocea, Pseudomonas plecoglossicida, CD82a, apoptosis, antigen presentation
Citation: Liu Y, Bai Y, Chen S, Pu F, Li Y, Chi H, Zheng Z, Xu P and Zhou T (2024) Molecular characterization, expression pattern and immunologic function of CD82a in large yellow croaker (Larimichthys crocea). Front. Immunol. 15:1301877. doi: 10.3389/fimmu.2024.1301877
Received: 25 September 2023; Accepted: 17 January 2024;
Published: 02 February 2024.
Edited by:
Gregory Todd Pharr, Mississippi State University, United StatesCopyright © 2024 Liu, Bai, Chen, Pu, Li, Chi, Zheng, Xu and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tao Zhou, enRAeG11LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.