
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 06 February 2024
Sec. Nutritional Immunology
Volume 15 - 2024 | https://doi.org/10.3389/fimmu.2024.1299484
This article is part of the Research Topic Targeted Innovative Immunomodulation of Allergy and Inflammation by Natural Molecules and Probiotics View all 11 articles
 Nan Yang1*†
Nan Yang1*† Kamal Srivastava1†
Kamal Srivastava1† Yujuan Chen2
Yujuan Chen2 Hang Li3
Hang Li3 Anish Maskey4
Anish Maskey4 Patrick Yoo5
Patrick Yoo5 Xiaohong Liu6
Xiaohong Liu6 Raj K. Tiwari4
Raj K. Tiwari4 Jan Geliebter4
Jan Geliebter4 Anna Nowak-Wegrzyn7
Anna Nowak-Wegrzyn7 Jixun Zhan8
Jixun Zhan8 Xiu-Min Li4*
Xiu-Min Li4*Introduction: Peanut allergy is an immunoglobulin E (IgE) mediated food allergy. Rubia cordifolia L. (R. cordifolia), a Chinese herbal medicine, protects against peanut-induced anaphylaxis by suppressing IgE production in vivo. This study aims to identify IgE-inhibitory compounds from the water extract of R. cordifolia and investigate the underlying mechanisms using in vitro and in vivo models.
Methods: Compounds were isolated from R. cordifolia water extract and their bioactivity on IgE production was assessed using a human myeloma U266 cell line. The purified active compound, xanthopurpurin (XPP), was identified by LC-MS and NMR. Peanut-allergic C3H/HeJ mice were orally administered with or without XPP at 200µg or 400µg per mouse per day for 4 weeks. Serum peanut-specific IgE levels, symptom scores, body temperatures, and plasma histamine levels were measured at challenge. Cytokines in splenocyte cultures were determined by ELISA, and IgE + B cells were analyzed by flow cytometry. Acute and sub-chronic toxicity were evaluated. IL-4 promoter DNA methylation, RNA-Seq, and qPCR analysis were performed to determine the regulatory mechanisms of XPP.
Results: XPP significantly and dose-dependently suppressed the IgE production in U266 cells. XPP significantly reduced peanut-specific IgE (>80%, p <0.01), and plasma histamine levels and protected the mice against peanut-allergic reactions in both early and late treatment experiments (p < 0.05, n=9). XPP showed a strong protective effect even 5 weeks after discontinuing the treatment. XPP significantly reduced the IL-4 level without affecting IgG or IgA and IFN-γ production. Flow cytometry data showed that XPP reduced peripheral and bone marrow IgE + B cells compared to the untreated group. XPP increased IL-4 promoter methylation. RNA-Seq and RT-PCR experiments revealed that XPP regulated the gene expression of CCND1, DUSP4, SDC1, ETS1, PTPRC, and IL6R, which are related to plasma cell IgE production. All safety testing results were in the normal range.
Conclusions: XPP successfully protected peanut-allergic mice against peanut anaphylaxis by suppressing IgE production. XPP suppresses murine IgE-producing B cell numbers and inhibits IgE production and associated genes in human plasma cells. XPP may be a potential therapy for IgE-mediated food allergy.
Food allergy (FA) is an adverse immune response to food allergens. It can be life-threatening and the prevalence of FA has significantly increased over the past decades, affecting about 32 million people in the United States, of which 5.6 million are children under age 18 (1–9). Treatment options are extremely limited. Food avoidance and rescue medication after accidental exposure are the first lines of FA management. Peanut allergy (PNA) causes severe, and sometimes fatal reactions, often co-existing with other food allergies (1, 10–15). Food allergy is also increasing in other westernized and developing countries (2, 16). It negatively influences the quality of life for patients and their families. Unfortunately, there are currently limited treatment options for food allergies. Strict avoidance of allergens remains the standard treatment for the majority of food allergies, except for peanut (PN). Since 2020, an FDA approved drug (Palforzia) is available for peanut desensitization in children 4-17 years old (17). However, it is not approved for other age groups due to lack of efficacy, and requires a prolonged course of treatment with indefinite daily dosing (18, 19). Palforzia is associated with a high rate of adverse effects and relapse after treatment cessation (20). Moreover, real-world acceptance of Palforzia is reported to be low (21). Therefore, the need to develop treatments, particularly for milk and peanut (PN) allergies, is urgent and challenging.
Food allergies typically are IgE-mediated. IgE molecules binding to mast cells and basophils sensitize these cells. When IgE specifically binds to the epitopes of the food allergen, it activates mast cells and basophils to release histamine and many other mediators (22). These, in turn, elicit inflammation and immediate hypersensitivity (22). Since IgE plays a pivotal role in food allergies, the concept of decreasing IgE levels has been investigated as a therapeutic strategy for food allergy treatment. In 2003, the anti-IgE antibody, TNX901, was tested in a double-blind, randomized trial. The results showed that TNX-901 significantly increased the threshold of the sensitivity of patients to peanut (23, 24). In another study, omalizumab (Xolair), was also found to greatly reduce anaphylactic reactions during the oral immunotherapy (OIT) (25). These studies suggested potential for developing novel therapies to regulate IgE. However, the current form of anti-IgE therapy has limitations. It demands continuing bi-weekly or monthly injections to maintain effects, as this treatment neutralizes secreted IgE, but does not stop B-cell IgE production (26, 27). In addition, biologic therapies are very expensive. An alternative and cost-effective approach to inhibiting IgE production by B cells is highly desirable.
Previously, Lopez et al. screened 70 herbal medicines from traditional Chinese medicine (TCM) and found that Rubia cordifolia L. (R. cordifolia) inhibited IgE production in a dose-dependent manner in vitro (28). R. cordifolia also suppressed the PN-specific IgE levels and protected PN-allergic mice against anaphylactic reactions (28). In this study, we aimed to test the effect of a pure compound, xanthopurpurin (XPP), isolated from R. cordifolia, in a PN-allergic murine model and investigate underlying mechanisms. We, for the first time, demonstrate that XPP markedly reduced peanut-specific IgE, protected peanut-allergic mice from anaphylaxis, and reduced plasma histamine levels. Splenocytes and bone marrow IgE-producing B cells were significantly reduced, which was associated with reduction of IL-4 and epigenetic modulation of IL-4 gene promoter. RNA-Seq analysis of plasma cells revealed that XXP has the capacity to regulate genes associated with antibody production.
Water extract of R. cordifolia was purchased from E-Fong Herbs Inc. (South EI Monte, CA). Methanol, acetonitrile, and other chemicals were purchased from Fisher Scientifics (Pittsburgh, PA). Freshly ground, whole roasted PN (White Rose brand, NJ) were used to prepare the crude PN extract (CPE) as previously described (29, 30). Cholera toxin (CT) was purchased from List Biological Laboratories, Inc (Campbell, CA). The human myeloma cell line, U266 B cells, was purchased from American Type Culture Collection (Manassas, Virginia).
In order to isolate individual components from R. cordifolia, liquid-liquid extraction was used with different organic solvents. In brief, 200g of R. cordifolia water extract was first dissolved into 4L of DDH2O and extracted with 4L of dichloromethane for 24 hrs using a liquid-liquid extractor (Sigma-Aldrich, St. Louis, MO) at 100°C with a 2L heating mantle (EM2000/CX1, Barnstead International, Dubuque, IA). The dichloromethane extract of R. cordifolia was first dried using a rotary evaporator (Rotavapor R-210, BÜCHI, Switzerland), then redissolved into methanol and loaded onto Sephadex LH20 (Sigma-Aldrich, St. Louis, MO) column for further separation.
Liquid chromatography was performed using a Waters 2690 HPLC-PDA system (Waters, Milford, MA). R. cordifolia water extract (30 mg/mL) was first filtered through a 0.8μm Nalgene™ syringe filter (Thermofisher, Waltham, MA) and separated on a ZORBAX SB-C18 (4.6 × 150mm, 5µm) column (Agilent, Santa Clara, CA) on the HPLC system. The separation was carried out using mobile phase A (0.1% aqueous formic acid) and mobile phase B (acetonitrile with 0.1% formic acid) at a flow rate of 1mL/min. The separation gradient started at 2% of mobile phase B to 25% within 45 min, to 35% within 25 min, to 55% mobile phase B within 15 min, to 75% in 10 min and maintained at 75% mobile phase B for another 5 min. Data was collected and processed using Waters’ Empower 2 software. Mass spectra data was collected using Waters Alliance 2695 HPLC system coupled with Waters LCT premier TOF mass spectrometer in both positive mode and negative mode to characterize the molecular weight of unknown constituent. The parameters were set as: Capillary Voltage: 3200 v; Cone Voltage: 15 v; Aperture I: 25 v; Desolvation Temperature: 300 ˚C; Source Temperature: 110 ˚C; Desolvation gas: 500 L/h; Nebulizing gas: 40 L/h; Ionization mode: Electrospray; Positive Ion Acquisition Range: m/z 50-1000. The results were collected and analyzed by Empower and Masslynx software. 1H NMR (at 300 MHz) and 13C NMR (at 75 MHz) spectra were obtained on a JOEL instrument using DMSO-d6 as the solvent.
The human myeloma cell line (U266), purchased from ATCC (American Type Culture Collection; Rockville, MD), has been used to test different herb extracts and purified compounds for their IgE inhibitory effects (31). 2×105 cells/mL of cells were cultured at 37°C under 5% CO2 in complete media containing RPMI 1640 medium supplemented, 10% FBS, 1 mM sodium pyruvate, 1×10-5 M β-ME and 0.5% penicillin-streptomycin. XPP at 0, 2.5, 5, 10, 20 and 40 μg/mL were added at day 0. After 6 days, supernatants were harvested and IgE levels were measured by using an ELISA Kit (Mabtech Inc, OH). Briefly, samples and standards were added into prewashed 96-well plate coated with capture monoclonal antibodies (mAbs). After 2-hour incubation at room temperature, plate was washed 5 times and incubated with detection mAb for 1 hour. The plate was then washed for 5 times and incubated with streptavidin-HRP for 1hour at room temperature. The plate was then washed and developed with TMB substrate for 15 min. The optical density was measured using ELISA reader at 450nm. IgE concentrations were calculated based on the standard curve.
Six-week-old female C3H/HeJ mice were purchased from the Jackson Laboratory (Bar Harbor, ME). Mice were maintained on PN-free chow under specific pathogen-free conditions according to standard guidelines for the care and use of animals (32). The study protocol was approved by institutional animal care and use committee (IACUC) at Icahn Mount Sinai School of Medicine, New York (IACUC #LA11-00082). The institutional Public Health Service (PHS) animal welfare assurance number is D16-00069 (A3111-01).
As described previously (33), mice were systemically sensitized by intraperitoneal (i.p.) injection of 500 μg of crude PN extract with 2 mg of alum in PBS (Model #1). Sensitization was given in three weekly injections at weeks 0, 1 and 2. XPP treatment started at week 3, representing an early treatment protocol as PN allergy is not yet established at this time point. Treatment with XPP (400 μg/day dissolved in drinking water) was given once daily for a period of 8 weeks. Sham mice were orally given water daily. Naïve mice were used as controls which were not sensitized or treated. Post therapy oral challenge was given at week 18 with 200 mg of PN (Figure 1A).
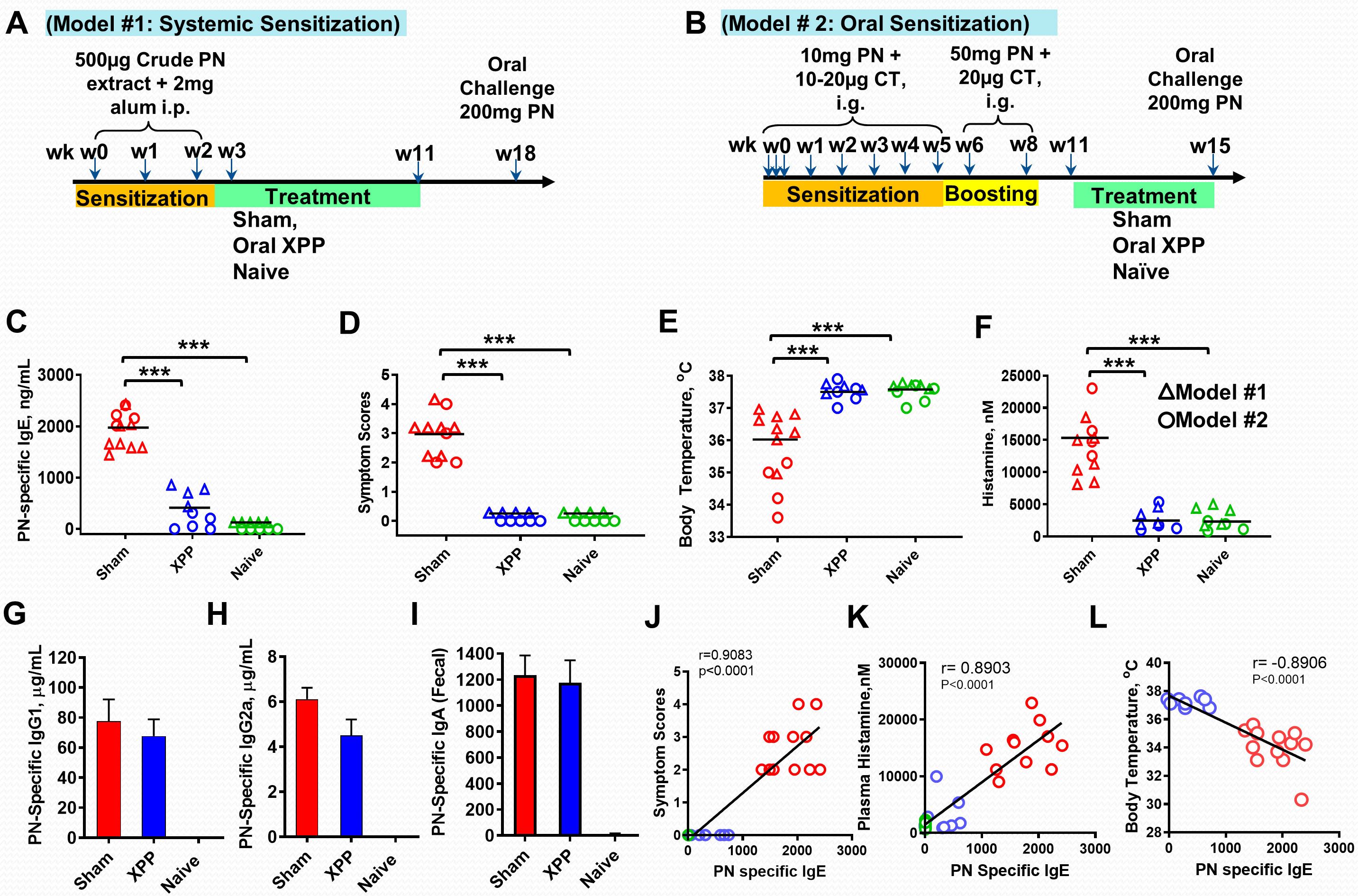
Figure 1 XPP prevented peanut-induced anaphylactic reactions in the systemic (triangle symbols) and Oral (circle symbols) sensitization protocols. (A) Experimental design for systemic i.p. PN sensitization; (B) Experimental design for oral PN sensitization; (C) Post-therapy PN-specific IgE in measure by ELISA; (D) Symptom Scores; (E) Body temperatures measured 30 minutes after PN challenge using a rectal probe; (F) Histamine levels in plasma obtained after PN challenge measured by ELISA; (G) Serum PN-specific IgG1 (Model #2); (H) Serum PN-specific IgG2a (Model #2); (I) Fecal PN-specific IgA (Model #2); (J) Correlation between PN specific IgE and Symptom score; (K) Correlation between Plasma histamine and PN specific IgE; (L). Correlation between Body Temperature and PN specific IgE; Pearson correlation coefficient (r) was calculated. Bars indicate group means. *** = P < 0.001 vs. Sham. N = 4~5 mice/group from 2 separate experiments.
Late treatment with XPP was tested in an oral sensitization model (Model #2). As described previously (34, 35), C3H/HeJ mice were intragastrically (i.g.) sensitized three times at week 0 with 10 mg of homogenized PN extract in 0.5 mL PBS containing 75 mg sodium bicarbonate, 10 µg of the mucosal adjuvant cholera toxin (List Laboratories, CA), and 16.5 µL (1.1 µL/g body weight) of 80 proof of Stolichnaya Vodka®. After the first week sensitization, mice were given the sensitization solution weekly as above with 20 µg CT for 5 weeks. 50 mg PN with 20 µg CT was given as the boosting dose at weeks 6 and 8 using the same gavage solution. Naïve mice were not sensitized. Oral XPP treatment (400 μg/day dissolved in drinking water) started at week 11 and continued through week 15. This treatment protocol represents late treatment as PN allergy is established and mice are reactive to challenge at this timepoint as shown by us previously (34). Sham group of mice received water daily.
Oral challenge with 200 mg PN was given at the end of week 15. Mice were fasted for 2 hrs prior to sensitization and challenge (Figure 1B).
Anaphylactic symptoms were evaluated 30-40 minutes following oral PN challenge as described previously (34, 35). Visually observed symptoms were scored utilizing the scoring system described previously (34): 0 - no symptoms; 1 - scratching and rubbing around the snout and head; 2 - puffiness around the eyes and snout, pilar erection, reduced activity, and/or decreased activity with increased respiratory rate; 3 - wheezing, labored respiration, cyanosis around the mouth and the tail; 4 - no activity after prodding, or tremor and convulsion; 5 - death. Cage identities were concealed during visual assessment of anaphylactic symptoms. Rectal temperatures were measured using a rectal probe (Harvard Apparatus, NJ).
PN specific-IgE in serum was measured as reported previously (34–36). Briefly, 96-well microtiter plates were coated with crude PN extract (sample wells) and rat anti-mouse IgE (reference wells) and held overnight at 4°C. After washing three times, plates were blocked for 2 hours at room temperature with 2% BSA-PBS. After three washes plates were incubated with diluted serum samples and purified mouse IgE antibody (in reference wells) overnight at 4°C. Biotinylated anti-IgE was added to the plates followed with the addition of avidin-peroxidase and ABTS substrate. For PN specific-IgG1 or IgG2a ELISAs, plates were coated with DNP-HSA in the reference wells and CPE in the sample wells. After the blocking, anti DNP-IgG1 or anti DNP-IgG2a were added to the reference wells, while samples were added into the sample wells. Plates were developed with biotinylated Rat anti-mouse IgG1 or IgG2a (BD Biosciences, CA) and ABTS (LGC, Searcare, MA). All plates were read using a spectrophotometer (Molecular Devices, San Jose, CA) using SoftMax software (Molecular Devices, San Jose, CA).
As previously performed (34, 35), blood samples were collected via sub-mandibular bleeding at 30 minutes after scoring and measurement of body temperature and plasma was isolated immediately after the blood collection. All samples were stored at -80°C for further usage. Histamine levels in the serum were measured using a commercial available ELISA kit (Fisher Scientific, Waltham, MA) as described by the manufacturer.
For acute toxicity analysis, naïve C3H/HeJ mice were fed with 4 mg of XPP per mouse, which is10 times of the daily dose. Mice were observed for 14 days. In the sub-chronic toxicity assay, naïve C3H/HeJ mice were fed with 5 times their daily therapeutic dose (2 mg/day) for 14 days. Control group mice (sham) were fed with water. Blood samples were collected at the end of each experiment. Blood urea nitrogen (BUN) and alanine aminotransferase (ALT) measurements for the evaluation of kidney and liver functions respectively and complete blood count (CBC) testing were performed by ALX laboratories, NY.
Mice were sacrificed at the end of each experiment. The splenocytes (SPCs) and mesenteric lymph node (MLN) cells of each mouse was collected as previously described (37). Cells (4 x106/well/mL) were cultured in 24 well plates in the presence or absence of CPE (200 µg/mL). After 72 hrs of incubation at 37°C under 5% CO2, supernatants were collected. Cytokine levels were measured using ELISA kits in triplicate according to the manufacturer’s instructions (BD Biosciences, San Jose, CA and R&D systems, Minneapolis, MN).
Cell viability was evaluated using a trypan blue exclusion as previously described (31). Briefly, a 10µL of cells suspension from each culture was mixed with equal volume of trypan blue dye. The mixture was loaded into a hemocytometer and cells were counted under a microscope. The percentage of viable cells was calculated as follows: Viable cells (%) = (total number of viable cells)/(total number of cells) × 100.
Single cell suspensions of spleen, bone marrow from two femurs and heparinized peripheral blood were isolated in AIM-V medium. Erythrocytes were lysed with red cell lysing buffer (Sigma-Aldrich, St. Louis, MO). For staining, cells were suspended in ice cold staining buffer (PBS including 0.5 mM EDTA, 0.05 mM Sodium Azide, 0.5% BSA). Surface staining was performed by incubating cells with unlabeled anti-IgE (to block membrane IgE), BV605 anti-B220, BV711-anti-CD3, anti-CD16/32 (Fc-block), all from BD Biosciences, CA). Live-dead discriminating dye (Live-Dead Aqua, Invitrogen, CA) was included. After incubation, cells were washed and suspended in fixation/permeabilization buffer for 15 mins. Cells were washed with permeabilization buffer and incubated with Fitc-anti IgE in permeabilization buffer for 30 mins in the dark on ice. After washing, cells were treated with Cytofix buffer for 15 mins for post-fixation and then washed 3 times with staining buffer and finally resuspended in 200 µL staining buffer for cell acquisition on LSRII flow cytometer (BD Bioscience, San Jose, CA). Flow cytometry analysis was performed using Flow Jo 5.4 software (Tree Star Inc., Ashland, Oregon) as follows. Singlet cells were selected on the basis of FSC-A/FSC-H profile and used to generate lymphocyte gate based on forward and side scatter properties (Supplementary Figure 1). Live cells were selected as cells negative for Live-Dead stain. From live cell gate, all IgE+ (Fitc-IgE) cells were gated and subsequently analyzed for IgE+ B cells (Fitc-IgE +; BV605-B220+ cells).
Genomic DNA was isolated from mouse splenocytes collected from orally sensitized mouse model#2 using Qiagen mini DNA/RNA extraction kit (Qiagen, Valencia, CA). Isolated DNA was bisulfite-modified using an Epitect plus DNA Bisulfite kit (Qiagen) following the manufacturer’s instructions. Bisulfite converted DNA was then amplified with the IL-4 primers listed below by PCR. The PCR products were subsequently purified and pyrosequenced using sequencing primers on a Pyromark Q24 system (Qiagen) as described previously (38). Primers for IL-4; Fwd: GTTTTTAAGGGGTTTTTATAGTAGGAAG; Rev-Biotin-AATTACCACTAACTCTCCTCCTACA. Sequencing (CpG -393) AGATTTTTTTGATATTATTTTGTT.
U266 cells were treated with XPP at 10 μg/mL. After 3 days of treatment, cells were collected and total RNA was isolated using Qiagen RNeasy MiniKits following the manufacturer’s instructions. The purity of total RNA was analyzed and the RNA-Seq was performed with an Illumina HiSeq2500 instrument at the Genomics Core facility at Icahn School of Medicine at Mount Sinai. Counts were converted to Log2 counts per million and normalized. A log2 fold change was calculated and the gene list with values ≥ 2 or ≤ -2 were uploaded to DAVID bioinformatic system for functional analysis.
RT-PCR analysis of genes related to Cell cycle, B cell differentiation, and IgE production was performed by QuantStudio 5 real-time PCR system (Thermo Fisher, Waltham, MA). For each PCR reaction, 12.5 μL maxima SYBR Green/ROX qPCR Master Mix 2x (Thermo Fisher, Waltham, MA) was mixed with 1.8 μL of 0.3 μM target primers and 300 ng of template DNA to make the total volume as 25 μL. The PCR procedure was set as 40 cycles at 25°C for 5 min, 42°C for 60 min and 70°C for 15 min. GAPDH was used as the housekeeping gene. The Delta Ct values for each gene were calculated by normalizing the Ct values with the housekeeping gene. The relative fold change in mRNA expression between different groups was calculated and expressed as 2−ΔΔCT. The primer sequence used are shown in Supplementary Table 1.
Data were analyzed using GraphPad Prism software (version 8.2.1, GraphPad Software. La Jolla, CA). Differences between two groups were analyzed using unpaired Student’s t-test. Differences between multiple groups were analyzed by One Way Analysis of Variance (One way ANOVA) followed by pair wise testing using Bonferroni’s adjustment. Pearson’s correlation coefficients were analyzed using GraphPad Prism software, and r values were generated. P values ≤0.05 were considered statistically significant.
R. cordifolia water extract was first extracted using dichloromethane, and further fractionated. Five fractions were collected and the fifth fraction (Fr.5) was found to have the highest anti-IgE effect at 10 µg/mL in human myeloma cell line (U266 cells) in vitro (Supplementary Figure 2). The major compound from this fraction was further collected and purified. The purity of the isolated compound was more than 95% as determined by analytical HPLC. The structure of this compound was identified using LC-MS (Figure 2A) and 1H and 13C NMR spectroscopy (Supplementary Table 2) (39). Mass spectra data showed a [M+H]+ ion peak of m/z 241 and [M-H]- ion peak of m/z 239 (Figure 2A). The molecular weight (MW) of this compound was then determined to be 240 g/mol. The 1H NMR data and the 13C NMR data of this compound was consistent with previously reported data of xanthopurpurin (Supplementary Table 2). Thus, based on the MS and NMR data, this compound was identified as xanthopurpurin (XPP).
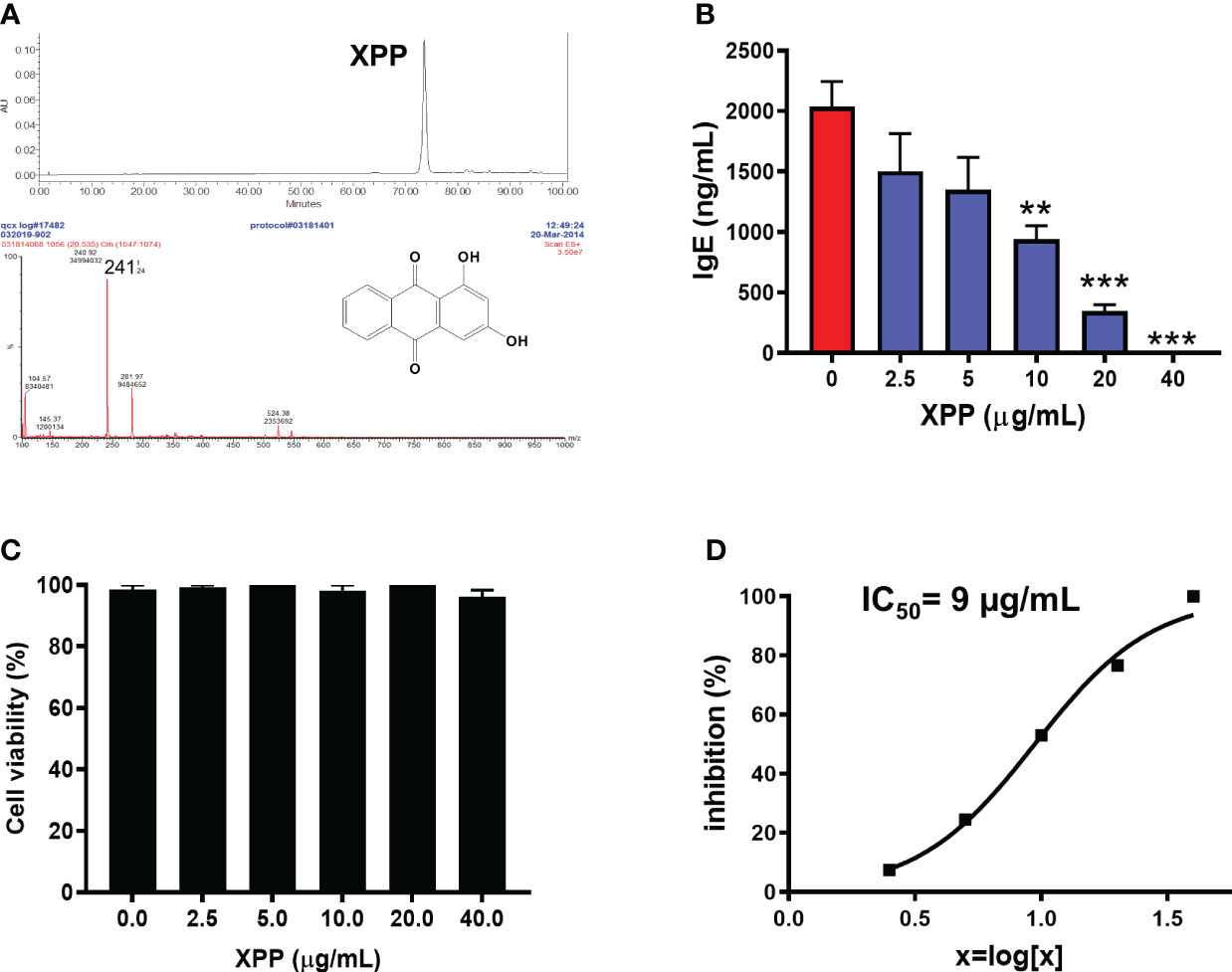
Figure 2 Characterization of xanthopurpurin (XPP) and the in vitro inhibitory effect of XPP on IgE by U266 cells. (A) HPLC chromatogram and Mass spectrum of XPP. The chemical structure of XPP was presented. (B) XPP dose-dependently inhibited IgE production by U266 cells (human IgE was measured by ELISA). (C) Cell viability assay of XPP on U266 cells by trypan blue exclusion. (D) IC50 values of XPP on IgE production in U266 cells. **p<0.01; ***p<0.001 vs. Untreated control (0 µg/mL). n=4-6.
We determined the biological effect of XPP on IgE production in vitro on U266 cells at various concentrations. XPP inhibited the IgE production by U266 cells in a dose-dependent manner (Figure 2B). Inhibition of IgE was first observed at XPP concentration of at 2.5 µg/mL, reaching statistically significant inhibition at 10 µg/mL (p < 0.01), and completely inhibition of IgE production was observed at 40 µg/mL. Cell viability was analyzed using trypan blue staining as previously published (31). No toxicity was observed at any concentration tested (Figure 2C). The IC50 value was calculated as 9 µg/mL (Figure 2D). To our knowledge, this is the first report that XPP inhibits IgE production. Notably, there was no cytotoxicity.
The in vitro data above prompted us to test whether XPP inhibits IgE production in vivo. We employed a well-established PN allergy model in which B and T cell responses and clinical reactions resemble human peanut- allergy. This model has been used by us and many other investigators (29, 36). We tested XPP effect in two separate experiments in which mice were either systemically sensitized with PN+ alum adjuvant (Model # 1, Figure 1A) or orally sensitized with PN + cholera toxin (Model #2, Figure 1B). Both sensitization protocols are well established (28, 29, 36, 40). To elicit anaphylaxis, all mice were challenged orally and XPP treatment was given i.g. as indicated (Figures 1A, B). PN-specific IgE levels at challenge in XPP-treated mice were significantly and markedly reduced compared to the sham-treated PNA mice in both models (75% reduction, p <0.001 vs. Sham, Figure 1C). Impressively, all XPP-treated mice were completely protected from anaphylaxis following PN challenge, evidenced by symptom scores of 0 (Figure 1D, p <0.001 vs. Sham), normal body temperatures (Figure 1E, p < 0.001 vs. Sham), and essentially normal plasma histamine levels (Figure 1F, p < 0.001). Protection in model #1 was observed 7 weeks after stopping therapy with significantly reduced symptom scores and PN-specific IgE levels and increased body temperatures, which all indicated a persistent effect (Supplementary Figure 3). Results in model #2 represent efficacy of late treatment with XPP as it was administered post-boosting, at a time when mice had established food allergy and were capable of anaphylaxis (34). Finally, XPP did not reduce PN-IgG1 (Figure 1G), PN-IgG2a (Figure 1H) or PN-IgA (Figure 1I) in the orally sensitized mouse model #2, which demonstrated its selectivity for IgE reduction. Furthermore, correlation analysis revealed that PN specific IgE showed significant positive correlation with symptom score and plasma histamine level (r = 0.9083, p < 0.0001; r = 0.8903, p < 0.0001, Figures 1J and K) and negatively correlated with body temperatures (r = -0.8906, p < 0.0001, Figure 1L).
Given that XPP effectively suppressed IgE production and protected peanut allergic mice from anaphylaxis, it may have a potential for clinical use. It is important to understand its safety profile. Acute toxicity and sub-chronic toxicity experiments of XPP were performed. In the acute-toxicity assay, C3H/HeJ mice were fed with 10 times (4 mg/day) of the regular XPP treatment dose once and observed daily for 14 days. No deaths and abnormal behavior or diarrhea were observed (Table 1). In the sub-chronic toxicity assay, mice were fed 5 times (2 mg/day) of the regular treatment dose for 14 consecutive days. No deaths and abnormal behavior or diarrhea were observed either, and all mice appeared healthy. Mice were then sacrificed, and blood samples were obtained for chemical analysis by ALX laboratories (NY). Results showed that the serum ALT and BUN levels were similar to the control group and within the normal range (Table 1). The complete blood count (CBC) tests showed that the white blood cell, red blood cell, hemoglobin and platelet levels in the XPP treated group were also within the normal range and similar to the control group (Table 1). These results demonstrate that XPP has a high safety profile. These data are in line with the in vitro data that demonstrated no cytotoxicity of XPP or global immune suppression.
Since XPP significantly reduced the PN-specific IgE levels compared to sham-treated mice, we, therefore, addressed questions of whether the IgE-producing B cells numbers are modified by XPP treatment. IgE-producing B cells from the spleen and bone marrow (samples collected from orally sensitized mouse model #2) were evaluated using flow cytometry of cells stained for intracellular IgE (Figures 3A, B). The percentage of the IgE+B cells (over all IgE+ cells) was significantly reduced by XPP in both spleen and bone marrow cells (p < 0.01; p < 0.001, Figures 3C, D).
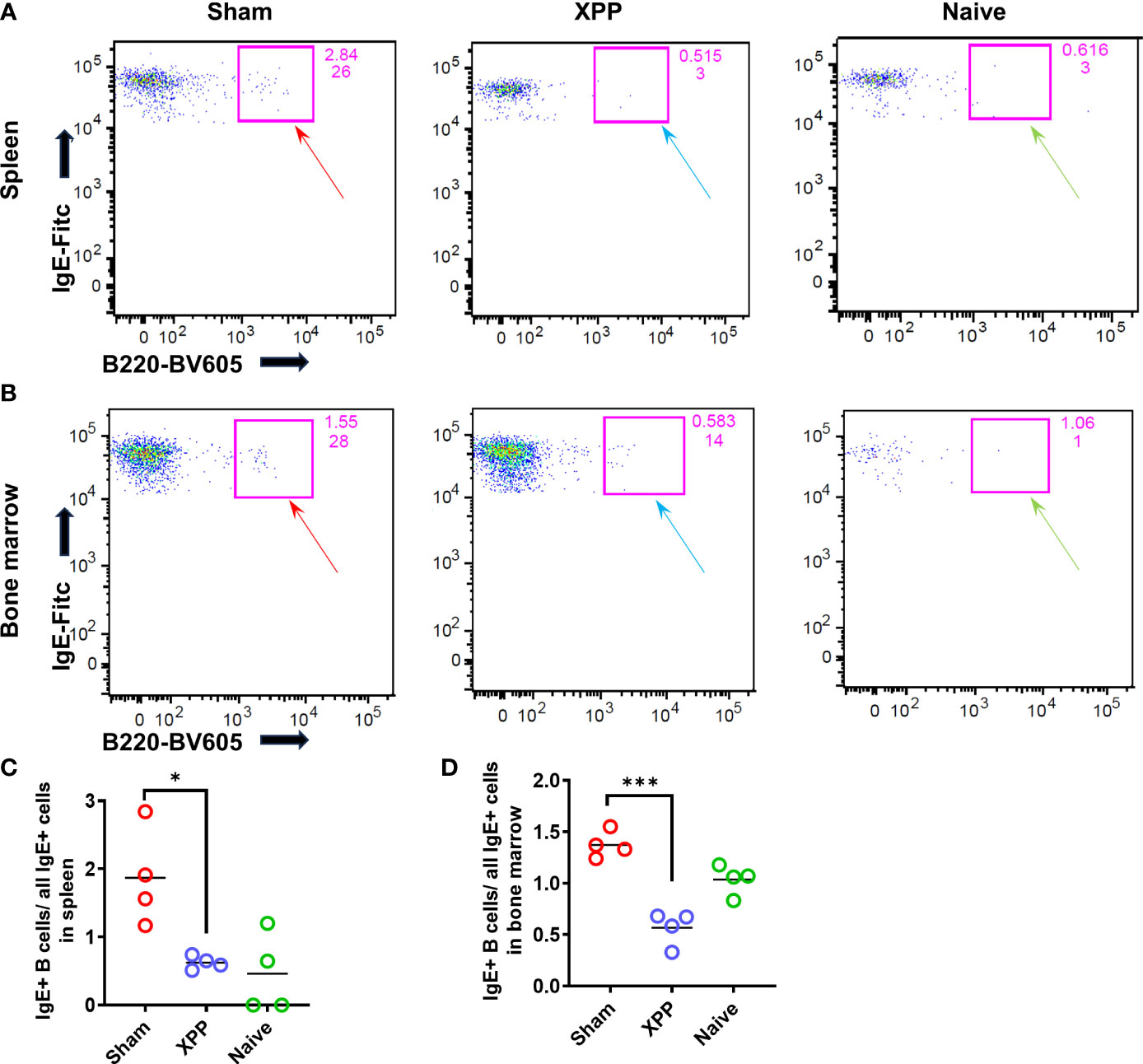
Figure 3 IgE+ B cells population in spleen and bone marrow samples. Splenocytes and bone marrow cells from oral sensitized model #2 were stained with antibodies to CD45R/B220 and IgE and analyzed by flow cytometry. (A) IgE+ B cells were found in the spleen of Sham, XPP treated, and naive mice; (B); IgE+ B cells were found in the bone marrow of Sham, XPP treated, and naive mice; (C) Percent of IgE+ B cells in spleen; (D) Percent of IgE+ B cells in bone marrow; *p ≤ 0.05; ***p < 0.001, n=4.
IL-4 is the key Th2 cytokine promoting IgE antibody switching and memory, while IFN- γ and IL-10 inhibit IL-4 production (41). We determined IL-4 production by splenocytes (SPCs) and MLN cells harvested from each group of mice from the orally sensitized mouse model #2 following the last challenge. XPP-treated mice showed significantly inhibited IL-4 level in both splenocyte and MLN cultures after the crude peanut extract (CPE) stimulation (Figures 4A, D). There were no significant differences in IL-10 (Figures 4B, E) or IFN-γ production (Figures 4C, F) in XPP-treated group in both the SPC or MLN cultures compared with the sham group. These data suggest that XPP may have a direct effect on IL-4 regulation.
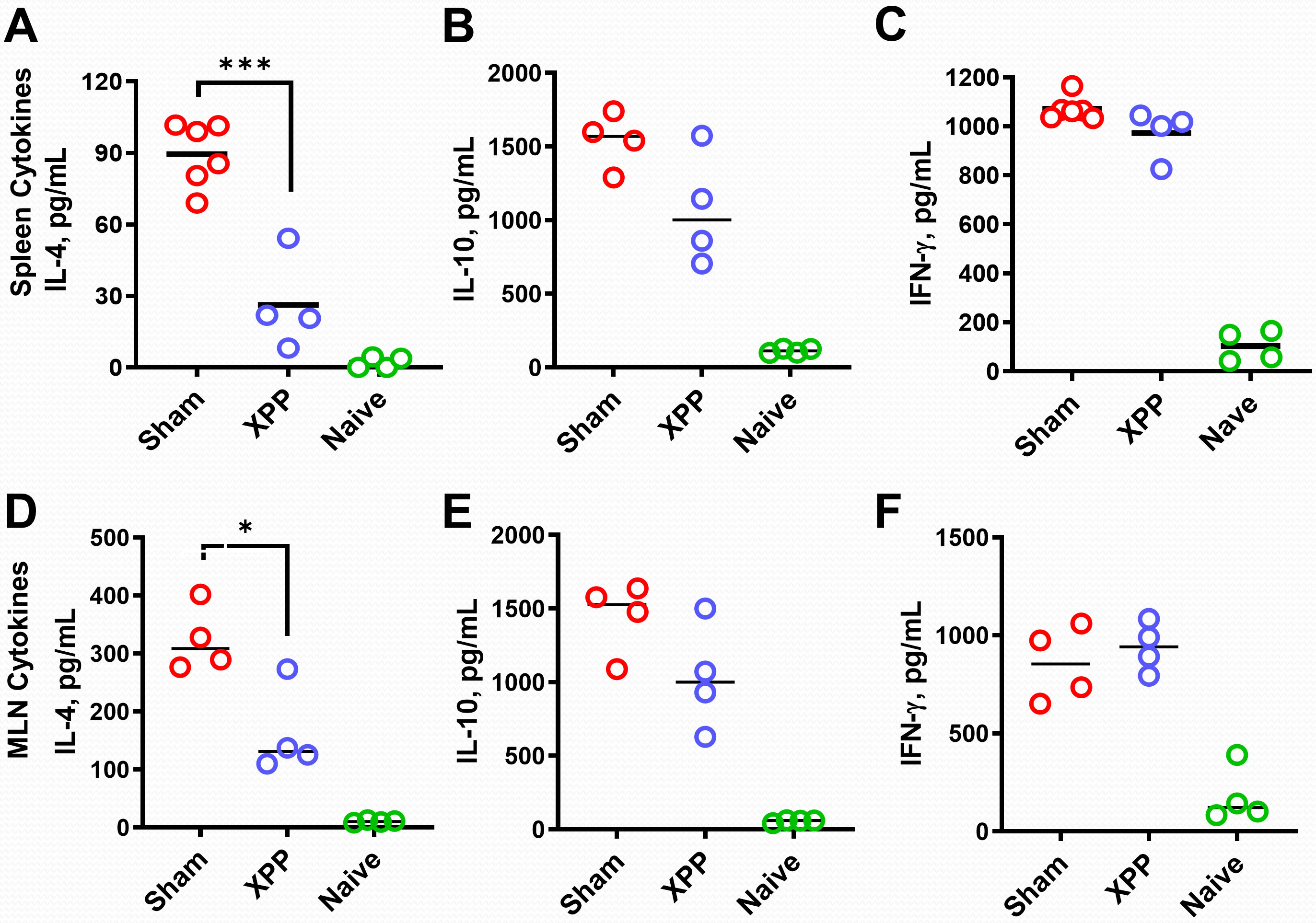
Figure 4 Effect of XPP on cytokine levels and IgE-producing B cell numbers. Splenocytes culture and the MLN cell cultures (from oral sensitized model #2) were stimulated with protein extracts of peanut (CPE). IL-4 (A), IL-10 (B) and IFN-γ (C) production in murine splenocytes; IL-4 (D), IL-10 (E) and IFN-γ (F) production in murine MLN were measured by ELISA. Bars indicate group means. N = 4-5 mice/group. *: p ≤0.05, ***: P<0.001 vs. Sham.
Methylation status at the IL-4 promoter directly regulates IL-4 transcription; i.e. increased methylation suppresses IL-4 transcription whereas decreased methylation increases IL-4 transcription (42, 43). We therefore determined methylation levels at CpG-393 of IL-4 promoter as previously described (34). Percentages of CpG methylation for CpG-393 showed an increase in XPP-treated mice compared to sham (Figure 5A) suggesting that XPP treatment leads to a more closed IL-4 promoter. In all groups, methylation of IL-4 promoter CpG site showed robust significant inverse correlation with IL-4 production (Figure 5B, r = -0.6338, p < 0.05) and PN-specific IgE (Figure 5C, r = -0.6960, p < 0.0082). These data suggest that regulation of IL-4 promoter CpG methylation status may be a contributing mechanism of XPP suppression of IL-4 production.
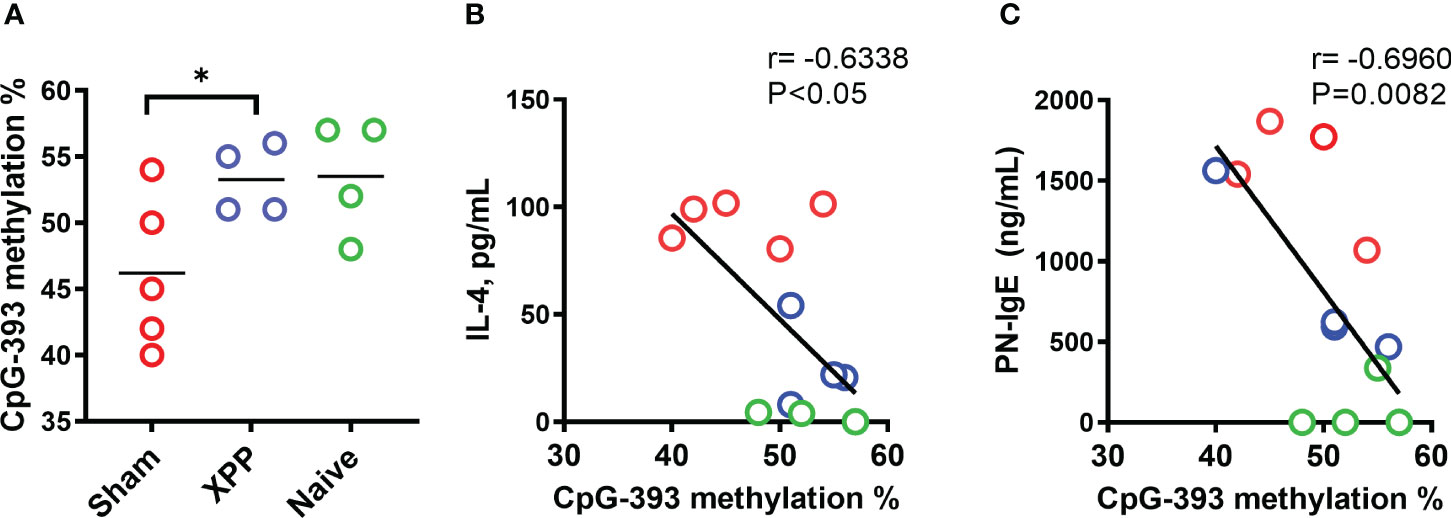
Figure 5 Percentage methylation of CpG sites in IL-4 promoter. Percentages of DNA methylation at the CpG-393 (A) site of IL-4 gene promoters in splenocytes collected from oral sensitized model #2 was assessed by pyrosequencing of bisulfite converted genomic DNA. Bars in A is group means. Pearson correlation between splenocyte IL-4 production vs. methylation percentage of CpG-393 (B) and PN-specific serum IgE vs. methylation percentage of CpG-393 (C) were calculated. N = 4-5 mice/group. * P<0.05.
RNA-Seq was used to evaluate the full gene-expression profiles of XPP-treated IgE producing human plasma cells. Overall, 4009 genes were upregulated, and 5094 genes were downregulated (Figure 6A). To further analyze the RNA-Seq results, pathway enrichment was investigated using DAVID. 53 related genes were matched to cell cycle process using David database. 65 related genes were matched to B cell differentiation- related genes, and 26 related genes were matched to IgE-related genes from Pubmed database (Figure 6B). The overlap graph showed the number of genes found in our RNA-Seq data using each method. Six pathways were involved in the regulation of XPP-treated IgE-producing plasma cells. These pathways are related to plasma B cells, IgE production, B cell differentiation, cell cycle, P53 pathway, and DNA replication. The heatmap for each pathway is shown in Figure 6C. As hundreds of genes were identified in the six enriched pathways, we further narrowed down candidate regulatory genes by filtering for differential expression using Log2C values > 2 or < -2 as cut-off values. Upregulated genes are shown in yellow whereas downregulated genes are shown in green (Figure 6D).
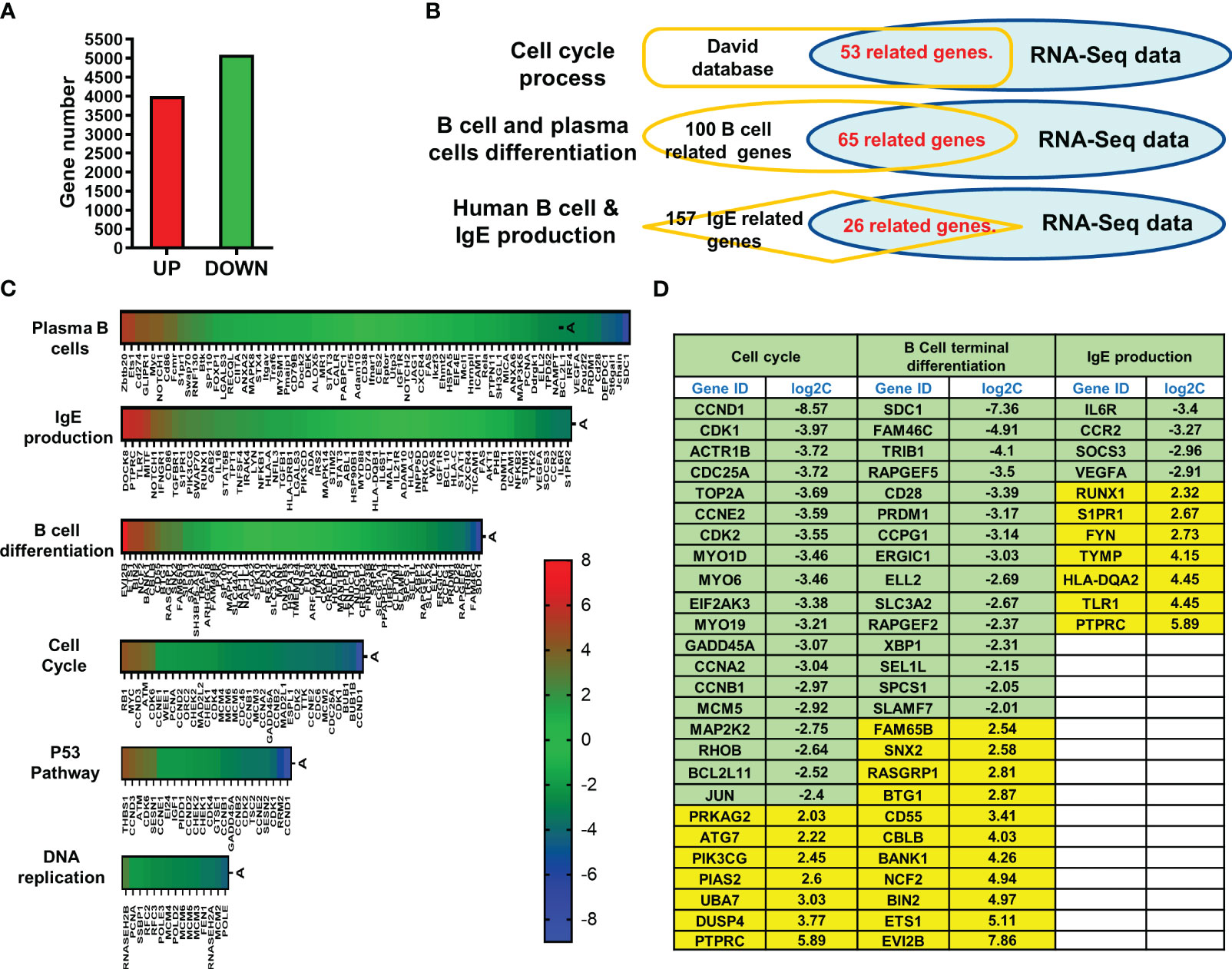
Figure 6 RNA sequencing of XPP treated U266 cells and pathway analysis. (A) Bar graph of total up-regulated a down-regulated genes; (B) RNA-Seq expression profile of most up- or down-regulated genes in cell cycle, B cell differentiation, and IgE production. (C) Heatmap of RNA-Seq results resulted to 6 pathways; (D) Table of candidate genes which have most significant changes. The threshold of log2C values was set as lower than -2.0 or higher than 2.0.
In order to further verify XPP mechanism of action at the molecular level, the expression levels of some of these target genes were validated by qRT-PCR. We chose the CCND1, DUSP4, and PTPARC from the cell cycle related genes, SCD1 and ETS1 from the B cell terminal differentiation gene list, and IL6R and PTPRC from the IgE production related gene list as the targets for RT-PCR validation. The RT-PCR confirmed that the CCND1, SCD1, and IL6R genes expression were significantly reduced in U266 cells with the treatment of XPP at the concentration of 20 μg/mL (Figures 7A–C, p < 0.001), while the DUSP4 and PTPRC gene expression was significantly increased by XPP at the same concentration (Figures 7D, F, p < 0.01). ETS1 gene expression also showed the upregulation trend with the increasing concentration of XPP, but no significant value was observed (Figure 7E). Overall, we were able to identify key genes associated with different pathways involving disease progression, and successfully validated some of these genes by qRT-PCR.
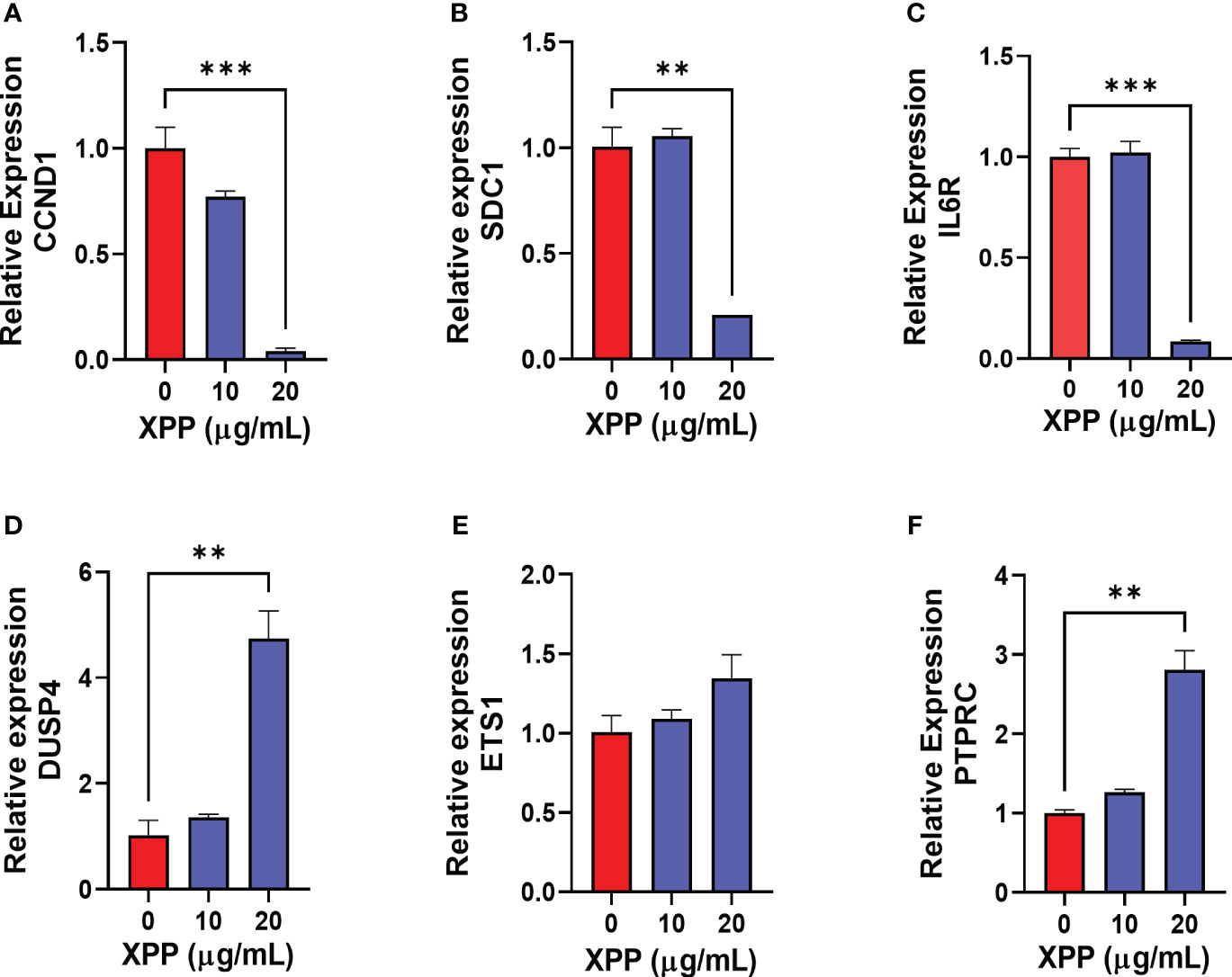
Figure 7 Relative gene expression of 6 down-or up-regulated genes in XPP treated U266 cells. The relative expression level of CCND1 (A), SDC1 (B), IL6R (C), DUSP4 (D), ETS1 (E), and PTPRC (F) were determined by comparing with GAPDH mRNA expression. **p<0.01 ***p<0.001.
Efficacious reduction of IgE for the treatment of food allergy remains elusive. Herbal medicines have been investigated for their inhibition effect on IgE production. We previously showed that the herbal formula FAHF-2 and its refined form showed significant IgE inhibition, both in vitro and in vivo (44–46). The active compound, berberine, was isolated from P. chinensis, which is one of the nine herbal constituents in FAHF-2 formula (31). We showed that berberine significantly inhibited the IgE production in vitro and modulated the ε-germline transcript expression through regulation of the phosphorylation of IƙBα, STAT-3 and T-bet (31). However, the bioavailability of berberine is poor. Oral berberine alone did not reduce PN-specific IgE levels and failed to protect PN-allergic mice from anaphylaxis, while the whole formula showed significant suppression of IgE levels (47).
Previously, our lab determined that the herbal medicine, R. cordifolia, inhibited the IgE production both in vitro and in vivo (28). R. cordifolia also reduced the anaphylactic symptoms in PN allergic mouse model (28). Historically, R. cordifolia is a widely used herbal medicine in Asia. Recent studies have shown that R. cordifolia roots have antibacterial, antioxidant and anti-inflammatory activities (28). R. cordifolia is enriched for anthraquinones, which have been isolated and identified, such as, alizarin (1,3-dihydroxy-2-ethoxymethyl-9,10 anthraquinone), purpurin (1,2,4-trihydroxyanthraquinone), mollugin (1-hydroxy-2-methy-9,10-anthraquinone) (48–50). We, for the first time, investigated the effect of xanthopurpurin on PN specific food allergy using murine models. Importantly, complete protection from anaphylaxis persisted for 7 weeks after stopping treatment which is longer than observed for OIT approaches in murine models (Supplementary Figure 3) (34). Protection was also observed when late treatment XPP was given after establishing PN allergy. Other compounds from R. cordifolia, such as purpurin and alizarin, inhibited IgE production only in vitro, but failed in vivo (Data not shown). Xanthopurpurin (XPP) significantly inhibited IgE production not only in vitro, but also in vivo, similar to the whole R. cordifolia herbal medicine (28). Thus, XPP has promise as a therapy for food allergy.
Food-induced anaphylaxis is an IgE-dependent type-I hypersensitivity reaction. Allergen-specific IgE produced by B cells is increased in response to allergen stimulation due to increased number of IgE-producing B cells. Regulation of IgE levels using anti-IgE molecules has been employed in many studies targeting various allergic diseases (26, 51). However, omalizumab can neutralize the free serum IgE (52), but less information about targeting to IgE production B cells and plasma cells. Compared with anti-IgE therapies, the XPP treatment not only inhibited IgE production, but it also suppressed the number of IgE producing B cells in our animal model. Downregulation of IgE-producing B cells provides evidence of a direct effect on IgE production.
Food allergy is associated with the imbalance of Th1and Th2 response. Th2 response was dramatically increased while Th1 response was diminished in food allergy (53). Th2 cytokines play important role in the pathogenesis of food allergy. IL-4 is pivotal for the activation of Th2 response and INF-γ is associated with the active Th1 response. To investigate the effect of XPP on PN allergic, we utilized two mouse models, systemic model and oral model, in this study. Both systemic model (i.p. sensitization) and oral model (i.g. sensitization) induced persistent peanut hypersensitivity. Systemic model using i.p. sensitization method in C3H/HeJ mice is most efficient, fast and simple to achieve, with high levels of PN sIgE and hypersensitivity. Oral model with i.g sensitization is more similar to the real food allergy scenario on human who was exposed to the PN allergen by food consumption. The oral model required to use the cholera toxin as the adjuvant, which is toxic, and need a longer time to introduce the allergic reactions. In this study, we first tested XPP on a fast established systemic sensitization model. Then, we investigated the effect of XPP on oral sensitization model which mimic the peanut allergy in reality. XPP treatment reduced IgE production, but did not show obvious effects on PN-sIgG1, -sIgG2a, and -sIgA levels (Figures 1G–I). IgG2a is Th1-associated immunoglobulin. Our data showed that the XPP treatment significantly suppressed the IL-4 production, but did not significantly inhibit the production of IL-10 and INF-γ. We also performed the in vitro experiment of XPP on human IgG producing myeloma cell line, ARH-77 (ATCC, Manassas, VA). XPP didn’t show significant inhibition on the IgG production. No toxicity was observed in this experiment (Supplementary Figure 4). All these data together suggest that XPP treatment selectively regulates Th2 response, without global immunosuppression. However, whether XPP has an effect on other IgG-producing myeloma cells lines, or on IgA- IgM- or IgD-isotype antibody producing myeloma cells are unknown, requiring further investigation.
Epigenetic mechanisms have been suggested in food allergy studies (54). Prior studies reported that allergic subjects showed significantly lower methylation level at IL-4 promoter region compared with health subjects (55). We found that XPP treatment increased DNA methylation status of CpG at site -393 compared with the sham group (p = 0.05). Decreased DNA methylation in Sham group is consistent with higher IL-4 gene expression, whereas higher DNA methylation in XPP-treated group implies decreased transcription of IL-4, which in turn reduces production of IL-4. Correlation data also confirmed that the increased DNA methylation at CpG-393 was negatively correlated with IL-4 expression and IgE production.
Additional mechanisms contributing to the regulatory effect of XPP on IgE production may involve targeting of genes related to other signal pathways. Our RNA-seq data of XPP-treatment on U266 cell identified 6 pathways/processes as being enriched, including Plasma/B cells and IgE production, among others. Significantly up or down regulated genes (log 2C value > 2 or < 2) were selected as candidates for the validation using qPCR. Top 3 down regulated and 3 upregulated genes showed CCND1, SDC1 and IL-6R were significantly reduced whereas DUSP4 and PTPRC expressions were significantly increased. The ETS1 gene showed mild non-significant increase at 20 μg/mL of XPP. Previous reports of these genes in the context of food allergy are scarce. CCND1 and IL-6R have been reported to be associated with asthma inflammation and high serum IgE in patients (56–58). IL-6 signaling directly related to the survival and maturation of B cells and Plasma cells (59, 60). SDC1 is a maker of long-lived plasma cells and myeloma pathogenesis (61, 62). DUSP4, on the other hand, was reported as a tumor suppressor (63, 64). DUSP4 expression reduced the expansion of antigen-specific B cells and the production of antibodies (65). While the direct immunoregulatory targets of XPP remain to be identified, our data suggests that the XPP treatment decreases Th2-immune responses and pathways related to IgE production.
In conclusion, we, for the first time, demonstrate that xanthopurpurin, an active compound isolated from R. cordifolia, suppressed the IgE production in vivo and protected peanut allergic mice from anaphylactic reactions. Furthermore, xanthopurpurin also exhibits regulatory effects on IL-4 promoter DNA methylation, and on expression of several genes related to B cell survival and IgE production.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethical approval was not required for the studies on humans in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used. The animal study was approved by Institutional animal care and use committee (IACUC) at Icahn Mount Sinai School of Medicine, New York. The study was conducted in accordance with the local legislation and institutional requirements.
NY: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Supervision, Writing – original draft, Writing – review & editing. KS: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Validation, Writing – review & editing. YC: Data curation, Formal analysis, Investigation, Methodology, Writing – review & editing. HL: Data curation, Investigation, Methodology, Writing – review & editing. AM: Data curation, Investigation, Methodology, Writing – review & editing. PY: Data curation, Investigation, Methodology, Writing – review & editing. XL: Conceptualization, Writing – review & editing. RT: Writing – review & editing. JG: Writing – review & editing. AN: Writing – review & editing. JZ: Data curation, Writing – review & editing. X-ML: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was partially supported by Winston Wolkoff Fund, Natural IgE Inhibitor Allergy Product, and Integrative Medicine Study to X-ML.
We thank Henry Ehrlich for reading the manuscript. We thank Anna Sherbakova and her family for their initial support of this project.
X-ML received research support to her institution from the National Institutes of Health (NIH)/National Center for Complementary and Alternative Medicine (NCCAM) # 1P01 AT002644725-01 “Center for Chinese Herbal Therapy (CHT) for Asthma”, and grant #1R01AT001495-01A1 and 2R01 AT001495-05A2, NIH/NIAID R43AI148039, NIH/NIAID 1R21AI176061-01, NIH/NIAID 1R44AI177183-01, NIH/NIAID 1R41AI172572-01A1, Food Allergy Research and Education (FARE), Winston Wolkoff Integrative Medicine Fund for Allergies and Wellness, the Parker Foundation and Henan University of Chinese Medicine; received consultancy fees from FARE and Johnson & Johnson Pharmaceutical Research & Development, L.L.C. Bayer Global Health LLC; received royalties from UpToDate; received travel expenses from the NCCAM and FARE; share US patent US7820175B2 (FAHF-2), US10500169B2 (XPP), US10406191B2 (S. Flavescens), US10028985B2 (WL); US11351157B2 (nanoBBR): take compensation from her practice at Center for Integrative Health and Acupuncture PC; US Times Technology Inc is managed by her related party; is a member of General Nutraceutical Technology LLC and Health Freedom LLC. NY received research support from the National Institutes of Health (NIH)/National Center for Complementary and Alternative Medicine (NCCAM), NIH/NIAID R43AI148039, NIH/NIAID 1R21AI176061-01, NIH/NIAID 1R44AI177183-01, NIH/NIAID 1R41AI172572-01A1; shares US patent: US10500169B2 (XPP), US10406191B2 (S. Flavescens), US10028985B2 (WL); and is a member of General Nutraceutical Technology, LLC and Health Freedom LLC; receives a salary from General Nutraceutical Technology, LLC. KS shares US patent: US11351157B2 (nanoBBR).
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1299484/full#supplementary-material
1. Cianferoni A, Muraro A. Food-induced anaphylaxis. Immunol Allergy Clin North Am (2012) 32(1):165–95. doi: 10.1016/j.iac.2011.10.002
2. Gupta RS, Warren CM, Smith BM, Jiang J, Blumenstock JA, Davis MM, et al. Prevalence and severity of food allergies among US adults. JAMA Netw Open (2019) 2(1):e185630. doi: 10.1001/jamanetworkopen.2018.5630
3. Sasaki M, Koplin JJ, Dharmage SC, Field MJ, Sawyer SM, McWilliam V, et al. Prevalence of clinic-defined food allergy in early adolescence: The SchoolNuts study. J Allergy Clin Immunol (2018) 141(1):391–398.e4. doi: 10.1016/j.jaci.2017.05.041
4. Dunlop JH, Keet CA. Epidemiology of food allergy. Immunol Allergy Clin North Am (2018) 38(1):13–25. doi: 10.1016/j.iac.2017.09.002
5. Gupta RS, Springston EE, Warrier MR, Smith B, Kumar R, Pongracic J, et al. The prevalence, severity, and distribution of childhood food allergy in the United States. Pediatrics (2011) 128(1):e9–17. doi: 10.1542/peds.2011-0204
6. Bunyavanich S, Rifas-Shiman SL, Platts-Mills TA, Workman L, Sordillo JE, Gillman MW, et al. Peanut allergy prevalence among school-age children in a US cohort not selected for any disease. J Allergy Clin Immunol (2014) 134(3):753–5. doi: 10.1016/j.jaci.2014.05.050
7. Gupta R, Holdford D, Bilaver L, Dyer A, Holl JL, Meltzer D. The economic impact of childhood food allergy in the United States. JAMA Pediatr (2013) 167(11):1026–31. doi: 10.1001/jamapediatrics.2013.2376
8. Gupta R, Holdford D, Bilaver L, Dyer A, Meltzer D. The high economic burden of childhood food allergy in the United States. J Allergy Clin Immunol (2013) 131(2):AB223. doi: 10.1016/j.jaci.2012.12.1464
9. FARE. Food Allergy Research and Education. Facts and Statistics. Available at: https://www.foodallergy.org/resources/facts-and-statistics#:~:text=How%20Many%20People%20Have%20Food,roughly%20two%20in%20every%20classroom.
10. Savage J, Sicherer S, Wood R. The natural history of food allergy. J Allergy Clin Immunol Pract (2016) 4(2):196–203;quiz 204. doi: 10.1016/j.jaip.2015.11.024
11. Brough HA, Caubet JC, Mazon A, Haddad D, Bergmann MM, Wassenberg J, et al. Defining challenge-proven coexistent nut and sesame seed allergy: A prospective multicenter European study. J Allergy Clin Immunol (2020) 145(4):1231–9. doi: 10.1016/j.jaci.2019.09.036
12. Warren CM, Jiang J, Gupta RS. Epidemiology and burden of food allergy. Curr Allergy Asthma Rep (2020) 20(2):6. doi: 10.1007/s11882-020-0898-7
13. Wang J, Sampson HA. Food anaphylaxis. Clin Exp Allergy (2007) 37(5):651–60. doi: 10.1111/j.1365-2222.2007.02682.x
14. FDA Encourages Manufacturers to Clearly Declare All Uses of Sesame in Ingredient List on Food Labels (2020). Available at: https://www.fda.gov/news-events/press-announcements/fda-encourages-manufacturers-clearly-declare-all-uses-sesame-ingredient-list-food-labels.
15. Motosue MS, Bellolio MF, Van Houten HK, Shah ND, Campbell RL. National trends in emergency department visits and hospitalizations for food-induced anaphylaxis in US children. Pediatr Allergy Immunol (2018) 29(5):538–44. doi: 10.1111/pai.12908
16. Tang ML, Mullins RJ. Food allergy: is prevalence increasing? Intern Med J (2017) 47(3):256–61. doi: 10.1111/imj.13362
17. Smith SS, Hilas O. Peanut (Arachis hypogaea) allergen powder-dnfp: the first FDA-approved oral immunotherapy for desensitization of peanut allergy in children. J Pediatr Pharmacol Ther (2021) 26(7):669–74. doi: 10.5863/1551-6776-26.7.669
18. Vickery BP, Vereda A, Casale TB, Beyer K, du Toit G, Hourihane JO, et al. AR101 oral immunotherapy for peanut allergy. N Engl J Med (2018) 379(21):1991–2001. doi: 10.1056/NEJMoa1812856
19. Bird JA, Spergel JM, Jones SM, Rachid R, Assa'ad AH, Wang J, et al. Efficacy and safety of AR101 in oral immunotherapy for peanut allergy: results of ARC001, a randomized, double-blind, placebo-controlled phase 2 clinical trial. J Allergy Clin Immunol Pract (2018) 6(2):476–485.e3. doi: 10.1016/j.jaip.2017.09.016
20. Perkin MR. Palforzia for peanut allergy: Panacea or predicament. Clin Exp Allergy (2022) 52(6):729–31. doi: 10.1111/cea.14145
21. Mustafa SS, Patrawala S. Real world adoption of FDA-approved peanut oral immunotherapy with palforzia. J Allergy Clin Immunol (2021) 147(2):1. doi: 10.1016/j.jaci.2020.12.401
22. Li X-M. Alternative/Integrative Medical Approaches in Allergy and Sleep: Basic Principles and Clinical Practice, in Allergy and Sleep. Fishbein A, Sheldon SH, editors. Switzerland: Springer Nature (2019).
23. Leung DY, Shanahan WR, Li XM, Sampson HA. New approaches for the treatment of anaphylaxis. Novartis Found Symp (2004) 257:248–60;discussion 260-4,276-85. doi: 10.1002/0470861193.ch20
24. Leung DY, Sampson HA, Yunginger JW, Burks AW, Schneider LC, Wortel CH, et al. Effect of anti-IgE therapy in patients with peanut allergy. N Engl J Med (2003) 348(11):986–93. doi: 10.1056/NEJMoa022613
25. Umetsu DT, Rachid R, Schneider LC. Oral immunotherapy and anti-IgE antibody treatment for food allergy. World Allergy Organ J (2015) 8(1):20. doi: 10.1186/s40413-015-0070-3
26. Hendeles L, Sorkness CA. Anti-immunoglobulin E therapy with omalizumab for asthma. Ann Pharmacother (2007) 41(9):1397–410. doi: 10.1345/aph.1K005
27. Luger EO, Wegmann M, Achatz G, Worm M, Renz H, Radbruch A. Allergy for a lifetime? Allergol Int (2010) 59(1):1–8. doi: 10.2332/allergolint.10-RAI-0175
28. López-Expósito I, Castillo A, Yang N, Liang B, Li XM. Chinese herbal extracts of Rubia cordifolia and Dianthus superbus suppress IgE production and prevent peanut-induced anaphylaxis. Chin Med (2011) 6:35. doi: 10.1186/1749-8546-6-35
29. Li XM, Serebrisky D, Lee SY, Huang CK, Bardina L, Schofield BH, et al. A murine model of peanut anaphylaxis: T- and B-cell responses to a major peanut allergen mimic human responses. J Allergy Clin Immunol (2000) 106(1 Pt 1):150–8. doi: 10.1067/mai.2000.107395
30. Beyer K, Morrow E, Li XM, Bardina L, Bannon GA, Burks AW, et al. Effects of cooking methods on peanut allergenicity. J Allergy Clin Immunol (2001) 107(6):1077–81. doi: 10.1067/mai.2001.115480
31. Yang N, Wang J, Liu C, Song Y, Zhang S, Zi J, et al. Berberine and limonin suppress IgE production by human B cells and peripheral blood mononuclear cells from food-allergic patients. Ann Allergy Asthma Immunol (2014) 113(5):556–564.e4. doi: 10.1016/j.anai.2014.07.021
32. Institute of laboratory animal resources commission of life sciences NRC. In: Guide for the Care and Use of Laboratory Animals. Washington DC: National Academic Press.
33. Kulis M, Li Y, Lane H, Pons L, Burks W. Single-tree nut immunotherapy attenuates allergic reactions in mice with hypersensitivity to multiple tree nuts. J Allergy Clin Immunol (2011) 127(1):81–8. doi: 10.1016/j.jaci.2010.09.014
34. Srivastava KD, Song Y, Yang N, Liu C, Goldberg IE, Nowak-Wegrzyn A. B-FAHF-2 plus oral immunotherapy (OIT) is safer and more effective than OIT alone in a murine model of concurrent peanut/tree nut allergy. Clin Exp Allergy (2017) 47(8):1038–49. doi: 10.1111/cea.12936
35. Srivastava KD, Siefert A, Fahmy TM, Caplan MJ, Li XM, Sampson HA. Investigation of peanut oral immunotherapy with CpG/peanut nanoparticles in a murine model of peanut allergy. J Allergy Clin Immunol (2016) 138(2):536–43.e4. doi: 10.1016/j.jaci.2014.12.1701
36. Srivastava KD, Bardina L, Sampson HA, Li XM. Efficacy and immunological actions of FAHF-2 in a murine model of multiple food allergies. Ann Allergy Asthma Immunol (2012) 108(5):351–358.e1. doi: 10.1016/j.anai.2012.03.008
37. Qu C, Srivastava K, Ko J, Zhang TF, Sampson HA, Li XM. Induction of tolerance after establishment of peanut allergy by the food allergy herbal formula-2 is associated with up-regulation of interferon-gamma. Clin Exp Allergy (2007) 37(6):846–55. doi: 10.1111/j.1365-2222.2007.02718.x
38. Song Y, Liu C, Hui Y, Srivastava K, Zhou Z, Chen J, et al. Maternal allergy increases susceptibility to offspring allergy in association with TH2-biased epigenetic alterations in a mouse model of peanut allergy. J Allergy Clin Immunol (2014) 134(6):1339–1345.e7. doi: 10.1016/j.jaci.2014.08.034
39. Berger Y, Castonguay A. The carbon-13 nuclear magnetic resonance spectra of anthraquinone, eight polyhydroxyanthraquinones and eight polymethoxyanthraquinones. Organic Magnetic Resonance (1978) 11(8):3. doi: 10.1002/mrc.1270110802
40. Brandt EB, Strait RT, Hershko D, Wang Q, Muntel EE, Scribner TA, et al. Mast cells are required for experimental oral allergen-induced diarrhea. J Clin Invest (2003) 112(11):1666–77. doi: 10.1172/JCI19785
41. Elser B, Lohoff M, Kock S, Giaisi M, Kirchhoff S, Krammer PH, et al. IFN-gamma represses IL-4 expression via IRF-1 and IRF-2. Immunity (2002) 17(6):703–12. doi: 10.1016/S1074-7613(02)00471-5
42. Wilson CB, Rowell E, Sekimata M. Epigenetic control of T-helper-cell differentiation. Nat Rev Immunol (2009) 9(2):91–105. doi: 10.1038/nri2487
43. Kwon NH, Kim JS, Lee JY, Oh MJ, Choi DC. DNA methylation and the expression of IL-4 and IFN-gamma promoter genes in patients with bronchial asthma. J Clin Immunol (2008) 28(2):139–46. doi: 10.1007/s10875-007-9148-1
44. Srivastava KD, Kattan JD, Zou ZM, Li JH, Zhang L, Wallenstein S, et al. The Chinese herbal medicine formula FAHF-2 completely blocks anaphylactic reactions in a murine model of peanut allergy. J Allergy Clin Immunol (2005) 115(1):171–8. doi: 10.1016/j.jaci.2004.10.003
45. Srivastava K, Yang N, Chen Y, Lopez-Exposito I, Song Y, Goldfarb J, et al. Efficacy, safety and immunological actions of butanol-extracted Food Allergy Herbal Formula-2 on peanut anaphylaxis. Clin Exp Allergy (2011) 41(4):582–91. doi: 10.1111/j.1365-2222.2010.03643.x
46. Yang N, Maskey AR, Srivastava K, Kim M, Wang Z, Musa I, et al. Inhibition of pathologic immunoglobulin E in food allergy by EBF-2 and active compound berberine associated with immunometabolism regulation. Front Immunol (2023) 14:1081121. doi: 10.3389/fimmu.2023.1081121
47. Yang N, Srivastava K, Song Y, Liu C, Cho S, Chen Y, et al. Berberine as a chemical and pharmacokinetic marker of the butanol-extracted Food Allergy Herbal Formula-2. Int Immunopharmacol (2017) 45:120–7. doi: 10.1016/j.intimp.2017.01.009
48. Itokawa H, Ibraheim ZZ, Qiao YF, Takeya K. Anthraquinones, naphthohydroquinones and naphthohydroquinone dimers from Rubia cordifolia and their cytotoxic activity. Chem Pharm Bull (Tokyo) (1993) 41(10):1869–72. doi: 10.1248/cpb.41.1869
49. Murthy HN, Joseph KS, Paek KY, Park SY. Anthraquinone production from cell and organ cultures of. Metabolites (2022) 13(1):39. doi: 10.3390/metabo13010039
50. Yang HS, Wang J, Guo C, Liu W, Chen YY, Wei JF, et al. Simultaneous determination of alizarin and rubimaillin in Rubia cordifolia by ultrasound-assisted ionic liquid-reversed phase liquid chromatography. Zhongguo Zhong Yao Za Zhi (2015) 40(13):2617–23.
51. Gauvreau GM, Arm JP, Boulet LP, Leigh R, Cockcroft DW, Davis BE, et al. Efficacy and safety of multiple doses of QGE031 (ligelizumab) versus omalizumab and placebo in inhibiting allergen-induced early asthmatic responses. J Allergy Clin Immunol (2016) 138(4):1051–9. doi: 10.1016/j.jaci.2016.02.027
52. Nyborg AC, Zacco A, Ettinger R, Jack Borrok M, Zhu J, Martin T, et al. Development of an antibody that neutralizes soluble IgE and eliminates IgE expressing B cells. Cell Mol Immunol (2016) 13(3):391–400. doi: 10.1038/cmi.2015.19
53. Romagnani S. Immunologic influences on allergy and the TH1/TH2 balance. J Allergy Clin Immunol (2004) 113(3):395–400. doi: 10.1016/j.jaci.2003.11.025
54. Cañas JA, Núñez R, Cruz-Amaya A, Gómez F, Torres MJ, Palomares F, et al. Epigenetics in food allergy and immunomodulation. Nutrients (2021) 13(12):4345. doi: 10.3390/nu13124345
55. Berni Canani R, Paparo L, Nocerino R, Cosenza L, Pezzella V, Di Costanzo M, et al. Differences in DNA methylation profile of Th1 and Th2 cytokine genes are associated with tolerance acquisition in children with IgE-mediated cow's milk allergy. Clin Epigenet (2015) 7:38. doi: 10.1186/s13148-015-0070-8
56. Li CH, Chiu KL, Hsia TC, Shen TC, Chen LH, Yu CC, et al. Significant association of cyclin D1 promoter genotypes with asthma susceptibility in Taiwan. In Vivo (2021) 35(4):2041–6. doi: 10.21873/invivo.12473
57. Thun GA, Imboden M, Berger W, Rochat T, Probst-Hensch NM. The association of a variant in the cell cycle control gene CCND1 and obesity on the development of asthma in the Swiss SAPALDIA study. J Asthma (2013) 50(2):147–54. doi: 10.3109/02770903.2012.757776
58. Wang Y, Hu H, Wu J, Zhao X, Zhen Y, Wang S, et al. The IL6R gene polymorphisms are associated with sIL-6R, IgE and lung function in Chinese patients with asthma. Gene (2016) 585(1):51–7. doi: 10.1016/j.gene.2016.03.026
59. Vazquez MI, Catalan-Dibene J, Zlotnik A. B cells responses and cytokine production are regulated by their immune microenvironment. Cytokine (2015) 74(2):318–26. doi: 10.1016/j.cyto.2015.02.007
60. Tiburzy B, Kulkarni U, Hauser AE, Abram M, Manz RA. Plasma cells in immunopathology: concepts and therapeutic strategies. Semin Immunopathol (2014) 36(3):277–88. doi: 10.1007/s00281-014-0426-8
61. Halliley JL, Tipton CM, Liesveld J, Rosenberg AF, Darce J, Gregoretti IV, et al. Long-lived plasma cells are contained within the CD19(-)CD38(hi)CD138(+) subset in human bone marrow. Immunity (2015) 43(1):132–45. doi: 10.1016/j.immuni.2015.06.016
62. Ren Z, Spaargaren M, Pals ST. Syndecan-1 and stromal heparan sulfate proteoglycans: key moderators of plasma cell biology and myeloma pathogenesis. Blood (2021) 137(13):1713–8. doi: 10.1182/blood.2020008188
63. Hanna A, Nixon MJ, Estrada MV, Sanchez V, Sheng Q, Opalenik SR, et al. Combined Dusp4 and p53 loss with Dbf4 amplification drives tumorigenesis via cell cycle restriction and replication stress escape in breast cancer. Breast Cancer Res (2022) 24(1):51. doi: 10.1186/s13058-022-01542-y
64. Kim GC, Lee CG, Verma R, Rudra D, Kim T, Kang K, et al. ETS1 suppresses tumorigenesis of human breast cancer via trans-activation of canonical tumor suppressor genes. Front Oncol (2020) 10:642. doi: 10.3389/fonc.2020.00642
Keywords: Rubia cordifolia L., food allergy, IgE, transcriptome, RNA-Seq
Citation: Yang N, Srivastava K, Chen Y, Li H, Maskey A, Yoo P, Liu X, Tiwari RK, Geliebter J, Nowak-Wegrzyn A, Zhan J and Li X-M (2024) Sustained silencing peanut allergy by xanthopurpurin is associated with suppression of peripheral and bone marrow IgE-producing B cell. Front. Immunol. 15:1299484. doi: 10.3389/fimmu.2024.1299484
Received: 22 September 2023; Accepted: 11 January 2024;
Published: 06 February 2024.
Edited by:
Ebenezer Satyaraj, Nestle Purina PetCare Company, United StatesReviewed by:
Michał Zarobkiewicz, Medical University of Lublin, PolandCopyright © 2024 Yang, Srivastava, Chen, Li, Maskey, Yoo, Liu, Tiwari, Geliebter, Nowak-Wegrzyn, Zhan and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiu-Min Li, WGl1TWluX0xpQG55bWMuZWR1; Nan Yang, bmFuLnlhbmdAZ250LXVzLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.