
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol., 28 February 2024
Sec. Cancer Immunity and Immunotherapy
Volume 15 - 2024 | https://doi.org/10.3389/fimmu.2024.1295305
This article is part of the Research TopicTranscriptome and Single-Cell Sequencing Analyses to Classify Immune Subtypes, Uncover Novel Biomarkers, and Assess Immunotherapeutic Responses in CancerView all 16 articles
 Han Ding1,2†
Han Ding1,2† Jia-Cheng Xu3†
Jia-Cheng Xu3† Zhi-Guo Ding4†
Zhi-Guo Ding4† Lin-Feng Wu1,2†
Lin-Feng Wu1,2† Yan-Bo Liu1,2
Yan-Bo Liu1,2 Yi-Fei Zhang1,2
Yi-Fei Zhang1,2 Tian-Yin Chen1,2*‡
Tian-Yin Chen1,2*‡ Yi-Qun Zhang1,2*‡
Yi-Qun Zhang1,2*‡ Ping-Hong Zhou1,2*‡
Ping-Hong Zhou1,2*‡Introduction: Ubiquitination is a crucial biological mechanism in humans, essential for regulating vital biological processes, and has been recognized as a promising focus for cancer therapy. Our objective in this research was to discover potential enzymes associated with ubiquitination that may serve as therapeutic targets for individuals with esophageal carcinoma (ESCA).
Methods: To identify genes linked to the prognosis of ESCA, we examined mRNA sequencing data from patients with ESCA in the TCGA database. Further investigation into the role of the candidate gene in ESCA was conducted through bioinformatic analyses. Subsequently, we carried out biological assays to assess its impact on ESCA development.
Results: Through univariate Cox regression analysis, we identified Ubiquitin Conjugating Enzyme E2 B (UBE2B) as a potential gene associated with the prognosis of ESCA. UBE2B exhibited significant upregulation and was found to be correlated with survival outcomes in ESCA as well as other cancer types. Additionally, UBE2B was observed to be involved in various biological pathways linked to the development of ESCA, including TNF-a signaling via NF-κB, epithelial-mesenchymal transition, inflammatory response, and hypoxia. Moreover, immune-related pathways like B cell activation (GO: 0042113), B cell receptor signaling pathway (GO: 0050853) and B cell mediated immunity (GO:0019724) were also involved. It was found that high expression of UBE2B was correlated with the increase of several kinds of T cells (CD8 T cells, Th1 cells) and macrophages, while effector memory T cell (Tem) and Th17 cells decreased. Furthermore, UBE2B showed potential as a prognostic biomarker for ESCA, displaying high sensitivity and specificity. Notably, proliferation and migration in ESCA cells were effectively suppressed when the expression of UBE2B was knocked down.
Conclusions: To summarize, this study has made a discovery regarding the importance of gaining new insights into the role of UBE2B in ESCA. UBE2B might be an oncogene with good ability in predicting and diagnosing ESCA. Consequently, this discovery highlights the feasibility of targeting UBE2B as a viable approach for treating patients with ESCA.
Esophageal carcinoma (ESCA), which encompasses esophagus squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (ESAD), ranks eighth and poses a substantial threat to the global cancer burden (1). The global burden of ESCA was reflected in the high number of diagnoses in 2018, with over 572,000 patients being newly diagnosed, representing 3.2% of all newly diagnosed cancer cases worldwide (2). In spite of the disparities in the rates of occurrence among various geographical areas and ethnic backgrounds, there is a prevailing trend that indicates a progressive escalation in the incidence of ESCA (3–5). ESCA, a multifaceted ailment with intricate variations, is distinguished by numerous alterations in polymers and structural rearrangements (6, 7). The application of sequencing data analysis in the investigation of molecular targets relevant to the prognosis or development of ESCA has shown considerable promise.
Ubiquitination, a crucial protein degradation mechanism, serves as a vital component in human physiology. It plays a pivotal role in maintaining cellular homeostasis. Disturbed ubiquitination processes have been directly linked to numerous diseases, most notably cancer (8). Indeed, various studies have demonstrated the significant role of ubiquitination/deubiquitination processes in the development and progression of ESCA, ultimately contributing to its malignancy. Thus, directing attention towards molecules linked to the process of ubiquitination/deubiquitination presents a hopeful strategy for potentially efficacious treatment of ESCA. While previous research has predominantly concentrated on the impact of ubiquitin ligase enzyme (E3)/deubiquitination enzyme on the development of esophageal squamous cell carcinoma (ESCA) due to their remarkable substrate binding specificity, it is equally crucial to recognize the significance of ubiquitin-conjugating enzyme (E2) in the progression of cancer, including ESCA (9–11).
In a study that aimed to construct a competing endogenous RNA (ceRNA) network for esophageal adenocarcinoma (ESAD), Ubiquitin Conjugating Enzyme E2 B (UBE2B) was identified as a prognostic factor that could predict patient survival outcomes, implicating UBE2B as a potential marker for prognosis in ESAD (12). Therefore, attention has been drawn towards UBE2B, which has been observed to facilitate the progression of cancer in diverse types, including rectal, nasopharyngeal, and ovarian carcinoma (13–15). Although UBE2B was found to be related to overall survival (OS) in patients with ESCA based on mRNA sequencing data from The Cancer Genome Atlas Program (TCGA), its exact function and role in ESCA still need to be determined. Further studies are required to unravel the underlying mechanisms through which UBE2B impacts the progression of ESCA and to explore its potential as a therapeutic target for the disease.
By employing Cox regression analysis, we discerned a collection of ten genes linked to the OS of individuals diagnosed with ESCA, subsequently ordered by their respective p-values. Notably, UBE2B emerged as the sole gene related to ubiquitination within this grouping. Additionally, through bioinformatics analysis, distinctions were observed between ESCA patient groups categorized by varying levels of UBE2B expression. Ultimately, experimental findings substantiated the potential role of UBE2B within ESCA cells.
Among the entries in the TCGA database, there were a total of 185 ESCA patients for whom clinical information was available. Nonetheless, only 174 of these patients possessed mRNA sequencing data. Consequently, this study ultimately included 163 patient samples (including one duplicated sample) and 11 control samples. Prognostic data regarding these ESCA patients were procured from a previous investigation (16). Subsequent to the download, all mRNA sequencing data underwent normalization, resulting in transcripts per million (TPM) values. Prior to analysis, these TPM values were transformed to log2(TPM+1) for further exploration.
To explore the association between gene expression and prognosis in patients with ESCA, the cohort was divided into two groups based on the median expression value of each gene. An exploration into the association between the expression of each gene in these groups and the prognosis of ESCA patients was conducted via univariate Cox regression analyses. Subsequently, the obtained P-values were adjusted using the Benjamini and Hochberg (BH) method for correction. Protein-coding genes were then identified from the gene list, and the top ten genes were selected as candidate genes based on their ranking of P-values and hazard ratio (HR), with the most significant genes appearing at the top of the list and the least significant ones at the bottom.
We examined the expression of UBE2B in various cancer types using mRNA sequencing data obtained from the TCGA pan-cancer database. Similar to the previous method mentioned, the data underwent processing. Subsequently, the sequencing data was merged with the corresponding clinical data for each specific cancer type. The log-rank test was then employed to evaluate the prognostic significance of UBE2B in these cancer types.
The ESCA patients were divided into two groups, namely low-expression and high-expression, according to the median value of UBE2B expression. To identify genes that exhibited differential expression between these two groups, a differential gene expression analysis was performed using the DESeq2 R package (17). The selection criteria for identifying differently expressed genes (DEGs) were set as follows: | log2 fold change | ≥ 1 and an adjusted P value (adj. P) < 0.05.
Following the identification of DEGs based on the defined criteria, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were conducted. These analyses aimed to elucidate the underlying molecular mechanisms and functional implications of the DEGs (18, 19).
In addition to the GO and KEGG analyses, Gene Set Enrichment Analysis (GSEA) was utilized to investigate the relationship between UBE2B expression and pathways. This approach aims to identify and assess the enrichment of predefined gene sets or pathways based on their correlation with UBE2B expression levels. By employing GSEA, the potential associations between UBE2B expression and various pathways can be explored and analyzed (19, 20). The gene set used for GSEA was acquired from the MSigDB collections database, which can be found at the following link: https://www.gsea-msigdb.org/gsea/msigdb/collections.jsp. This database contains a wide range of curated gene sets, including pathways, molecular signatures, and functional annotations, which can be utilized in GSEA analyses to assess the enrichment of these gene sets in relation to UBE2B expression.
To investigate immune infiltration in two groups, Single-Sample Gene Set Enrichment Analysis (ssGSEA) was conducted. ssGSEA evaluates the enrichment of specific gene signatures in each sample based on their gene expression profiles. It assigns a relative enrichment score to each sample, reflecting the activity or abundance of specific biological pathways or cell types within the sample (21). In this study, the Spearman’s correlation test was utilized to assess the correlation between UBE2B expression and immune cell infiltration patterns in patients with ESCA (22). This analysis allows researchers to explore the potential involvement of UBE2B in regulating immune cell infiltration in ESCA and provides insights into the immunological aspects of ESCA.
After consolidating the clinical data and UBE2B expression levels in ESCA patients, we conducted Cox regression analyses, both univariate and multivariate. Factors with P<0.1 in the univariate Cox regression analysis were subjected into the multivariate Cox regression analysis, and factors with P<0.05 in the multivariate Cox regression analysis were finally identified as independent prognostic factors for ESCA patients. Subsequently, these independent prognostic factors were employed in the development of a nomogram. To assess the accuracy and performance of the developed prognostic signature, a validation plot was generated. Additionally, we evaluated the diagnostic ability of UBE2B in ESCA by employing the receiver operating characteristic (ROC) curve.
Two esophageal cancer cell lines, KYSE-150 and ECA-109, were cultured under specific conditions. RPMI 1640 medium was used to culture KYSE-150 cells, while ECA-109 cells were cultured in DMEM. Both media were supplemented with 10% fetal bovine serum (FBS) and penicillin/streptomycin solution (100 U/ml). Then, the cells were incubated in a humidified carbon dioxide incubator at a temperature of 37°C.
Once the cells reached 50% confluency in the 6-well culture plates, we proceeded with the RNA interference procedure following the instructions provided by the manufacturer of lipofectamine 2000 (Thermo Fisher, 11668019). After 48 hours, total RNA and proteins were collected to assess the effectiveness of UBE2B gene silencing using quantitative reverse transcription polymerase chain reaction (RT-qPCR) and Western Blot techniques. The small interference RNA (siRNA) targeting UBE2B (siUBE2B) utilized in this study was synthesized by Genepharm Technologies (Shanghai, China). Two specific sequences were employed: siUBE2B-1 (GCAGTTATATTTGGACCAGAA) and siUBE2B-2 (CGGGATTTCAAGCGGTTACAA).
To evaluate the viability of cells transfected with siUBE2B, we utilized the Cell Counting Kit-8 (CCK-8) reagent (Vazyme, A311-02-AA). Additionally, for the colony formation assay, UBE2B-silenced cells were seeded at a density of 1000 cells per well in 6-well culture plates and allowed to grow for approximately seven days. Subsequently, the cells were stained using a 0.1% (v/v) crystal violet solution.
For the migration assay, we used transwell chambers with a pore size of 8 mm (Corning,3422). The upper chamber was filled with 200 μl of serum-free media, while the lower chamber contained 500 μl of media supplemented with FBS (10% v/v). The cells were seeded in the upper chamber and allowed to migrate for 12 hours. Following this, the cells were fixed using a 4% (v/v) paraformaldehyde solution and subsequently stained with a 0.1% (v/v) crystal violet solution.
Total RNA extraction, reverse transcription, and real-time PCR were carried out using a commercial kit from Accurate Biotechnology (Changsha, Cat. No: AG21023, AG11706, and AG11718) following the manufacturer’s instructions. The primer sequences used for UBE2B and GAPDH were as follows: UBE2B Forward primer (5’-3’): TTGGACCAGAAGGGACACCT, UBE2B Reverse primer (5’-3’): CCTGGCTATTGGCTGGACTG; GAPDH Forward primer (5’-3’): GTCTCCTCTGACTTCAACAGCG, GAPDH Reverse primer (5’-3’): ACCACCCTGTTGCTGTAGCCAA.
Protein extraction and quantification were performed by commercial kit (Thermo Fisher Scientific; 78501 and 23227) following the kit’s instruction. Then, protein was uniformly loaded onto the SDS-PAGE gel (Shanghai Epizyme Biomedical Technology Co., Ltd; PG112). After about 90min, the protein was transferred to the nitrocellulose membrane (Millipore, HATF00010). Notably, the time of transference should be no more than 45 min. Subsequently, 5% non-fat milk was used to block the membrane at room temperature (1 hour). Finally, primary antibodies were employed to incubated the membrane at 4 °C overnight, and corresponding secondary antibodies were used to incubate at room temperature about 2 hours. The specific information about the antibodies were as the following: Anti-UBE2B (Abcam, ab128951, 1:1000), anti-GAPDH (Proteintech, 60004-1-Ig, 1:100000), secondary antibodies (ABclonal, AS014 and AS003,1:5000).
The quantitative data were represented as mean and median standard deviation (SD). At the same time, the qualitative data were presented as counts and percentage values. The count of colony formation and migration assays were calculated by ImageJ software. The results of biological assays were analyzed by t test, and p < 0.05 was regarded as statistically significant. All assays were repeated 3 times and showed the representative findings. The analyses were performed by R software and GraphPad Prism 8.0.
The flowchart of this study was shown in Figure 1. A sum of 601 genes encoding protein-coding related to the prognosis were detected (Supplementary Table S1). Out of these genes, UBE2B emerged as a noteworthy contender, ranking sixth due to its statistically significant low p-value as shown in Table 1, and fourth due to its considerably high hazard ratio as indicated in Table 2. The evidence strongly indicates the crucial involvement of UBE2B in the unfavorable prognosis of individuals with ESCA. It is worth mentioning that UBE2B stood out as the sole gene directly associated with ubiquitination, further enhancing its importance as a potential target of interest. Further investigations of UBE2B expression in unpaired samples of pan-cancer revealed diverse expression levels observed across different types of cancers. Figures 2A, B illustrated a notable increase in UBE2B expression in tumor samples obtained from various cancers, including CHOL, COAD, ESCA, KIRC, LIHC, and STAD. Similarly, in a paired pan-cancer analysis, UBE2B expression was found to be consistently high in the majority of tumor samples, including ESCA (Figures 2C, D). Furthermore, the expression of UBE2B was observed to be correlated with unfavorable prognosis in multiple tumor types, impacting both overall survival (OS) and disease-specific survival (DSS) (Figure 3).
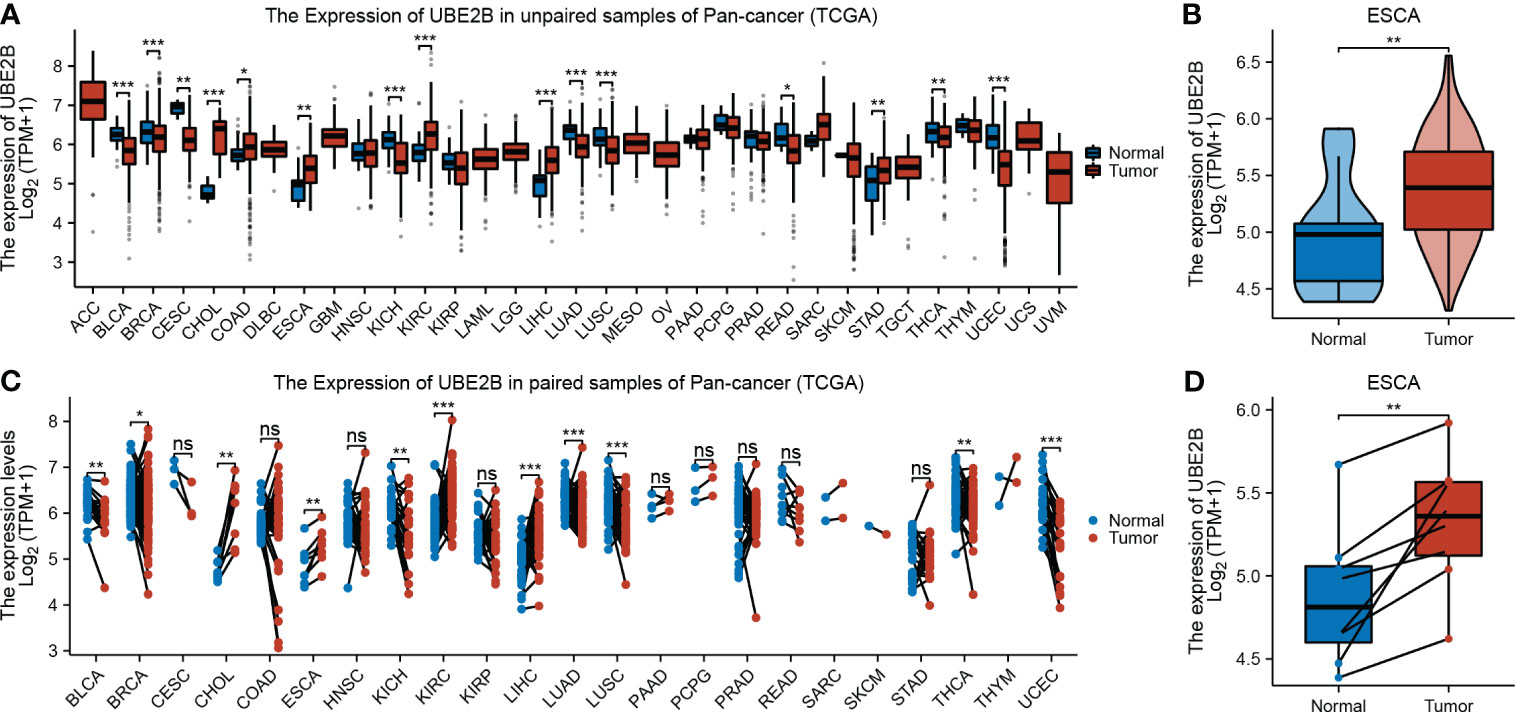
Figure 2 Expression of UBE2B in cancer. (A, B). Expression of UBE2B in unpaired samples of patients with Pan-cancer and ESCA (Esophageal Carcinoma). (C, D). Expression of UBE2B in paired samples of patients with Pan-cancer and ESCA (Esophageal Carcinoma). (*P < 0.05, **P < 0.01 and ***P < 0.001; ns, no significance)
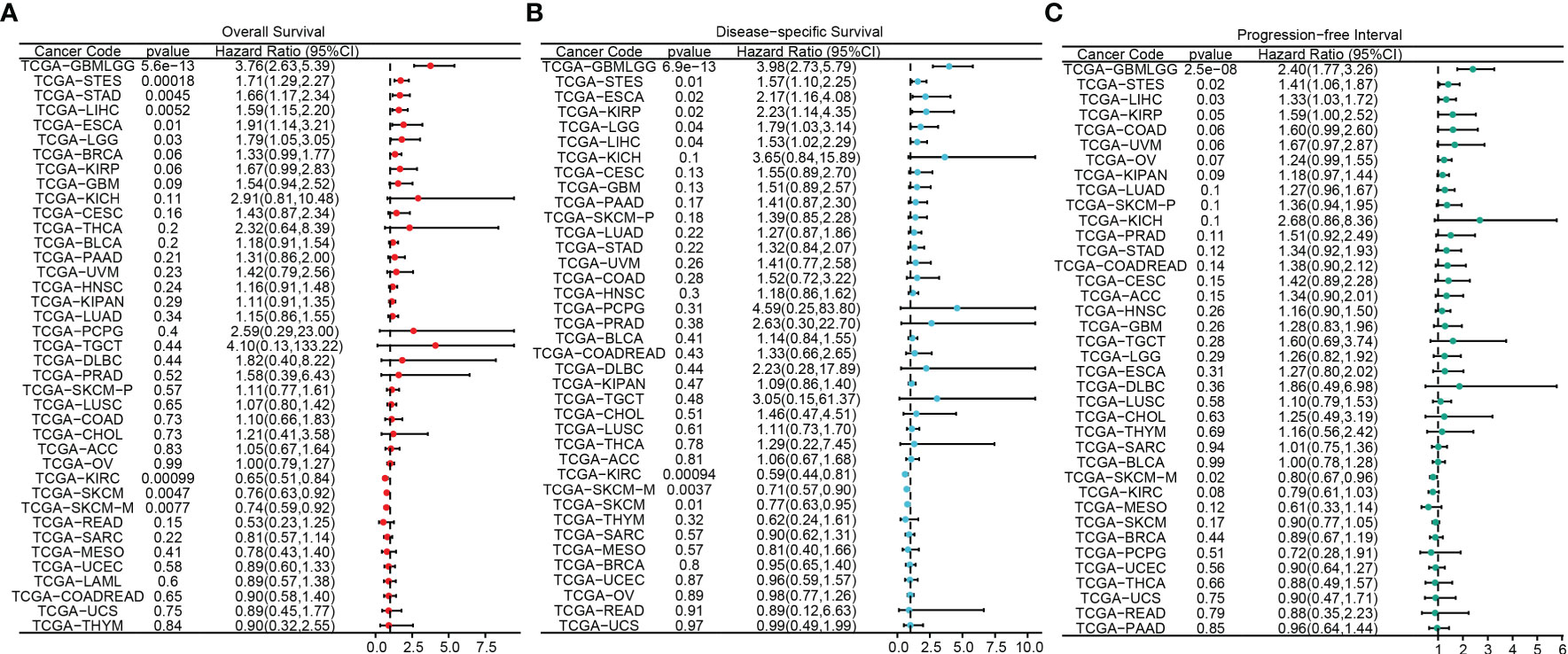
Figure 3 Association between UBE2B expression and prognosis in pan-cancers. (A) Overall survival analysis based on UBE2B expression. (B) Disease-specific survival analysis based on UBE2B expression. (C) Progression-free interval analysis based on UBE2B expression.
Table 3 presents the clinical characteristics of 162 ESCA patients. It encompasses several factors including age, gender, race, height, weight, BMI, smoking status, alcohol history, presence of Barrett’s esophagus, history of reflux, presence of columnar mucosa dysplasia, presence of columnar metaplasia, tumor central location, TNM stage, pathologic stage, histological type, histologic grade, primary therapy outcome, presence of residual tumor, and administration of radiation therapy. The chi-squared test results demonstrated that the clinical characteristics of ESCA patients in the low and high expression groups of UBE2B did not have a statistically significant impact on the expression of UBE2B. Therefore, these findings suggest the importance of conducting further analysis to investigate the independent role of UBE2B in ESCA. Further analysis revealed significant differences in UBE2B expression based on weight and histological type (Figures 4A, B). Specifically, high expression of UBE2B was associated with poorer survival outcomes, especially in cases of squamous cell carcinoma (Figures 4C, D).
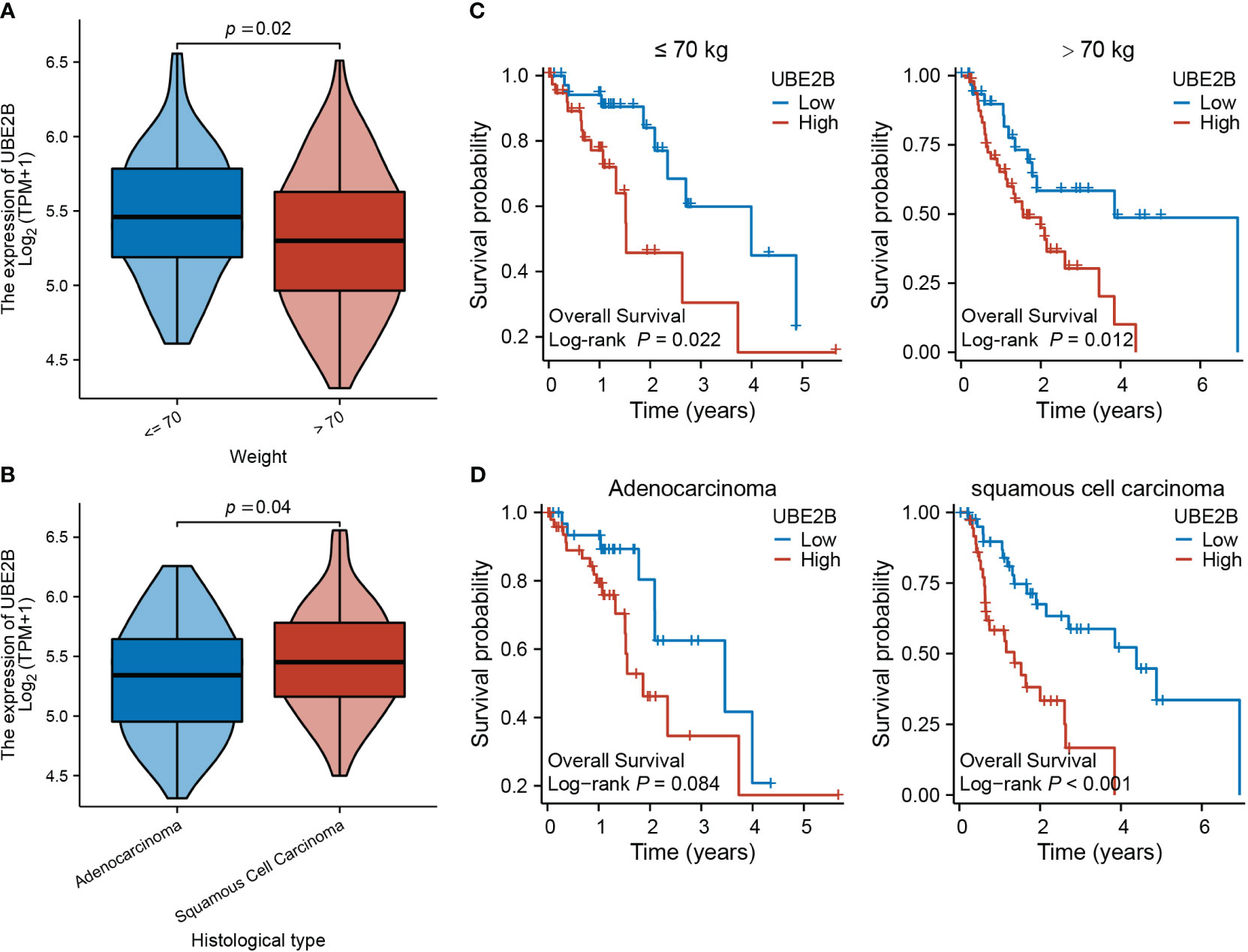
Figure 4 Expression of UBE2B in different clinical groups of patients with ESCA (Esophageal Carcinoma). (A, B) Differential UBE2B expression in different weight groups and histological types of ESCA patients. (C, D) The Kaplan-Meier (KM) plots showing the effect of UBE2B expression on patient survival indifferent weight and histological groups.
After applying the selection criteria, a total of 713 differentially expressed genes (DEGs) were identified in ESCA. These consisted of 488 up-regulated genes and 225 down-regulated genes, as depicted in Figure 5A. Interestingly, among these DEGs, three E3 ligases (TRIM23, PJA2, and AFF4) showed a positive correlation with UBE2B expression, suggesting potential interactions between these E3 ligases and UBE2B in the ubiquitination process (Figure 5B).
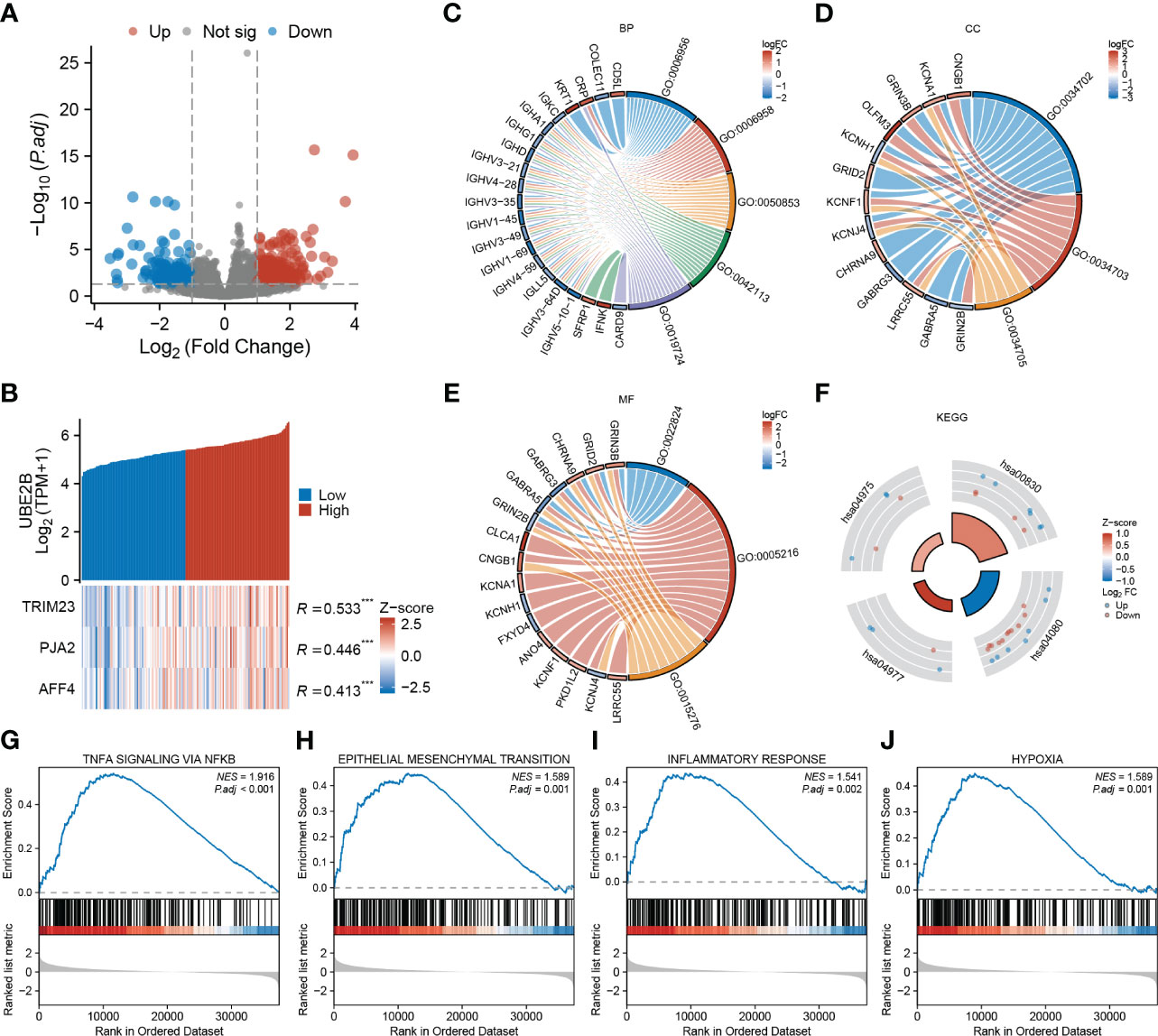
Figure 5 Functional enrichment analysis based on UBE2B expression in patients with ESCA (Esophageal Carcinoma). (A) Volcano plot depicting differentially expressed genes based on UBE2B expression. (B) Prediction of the E3 ligase that interacts with UBE2B. (C–F). Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses. (G–J). Gene Set Enrichment Analysis (GSEA) based on UBE2B expression.
Additionally, GO enrichment analyses unveiled that UBE2B expression was linked to various immune-related biological processes (BP), including complement activation (GO:0006956 and GO:0006958), B cell-mediated immunity (GO:0050853, GO:0042113, GO:0019724) and humoral immune response (GO:0006959) (Figure 5C). Additionally, ion channel-related cellular components (CC) including ion channel complex (GO:0034702), cation channel complex (GO:0034703) and potassium channel complex (GO:0034705), as well ion channel- related molecular functions (MF) such as transmitter-gated ion channel activity (GO:0022824), ion channel activity (GO:0005216) and ligand-gated ion channel activity (GO:0015276) were also enriched (Figures 5D, E). Furthermore, KEGG analyses indicated that UBE2B-associated DEGs might participate in some processes (Figure 5F) like fat digestion and absorption (hsa04975), retinol metabolism (hsa00830), neuroactive ligand-receptor interaction (hsa04080) and vitamin digestion and absorption (hsa04977). Utilizing gene sets from the MSigDB collection (h.all.v7.5.1. symbols [Hallmarks] (50)), several signaling pathways were found to be enriched, including TNF-a signaling via NF-κB, epithelial-mesenchymal transition (EMT), inflammatory response and hypoxia (Figures 5G–J). These results suggested that UBE2B could affect the development of ESCA by regulating the immune response, ion channel, and some biological pathways.
Furthermore, based on immune infiltration scores, the infiltration levels of CD8 T cells, macrophages, effector memory T cell (Tem), Th1 cells and Th17 cells were significantly different in low- and high-expression of UBE2B groups (Figure 6A). Additionally, the expression of UBE2B was found to be correlated with macrophages, Th1 cells, CD8 T cells, cytotoxic cells, B cells, Tem and Th17 cells (Figure 6B).
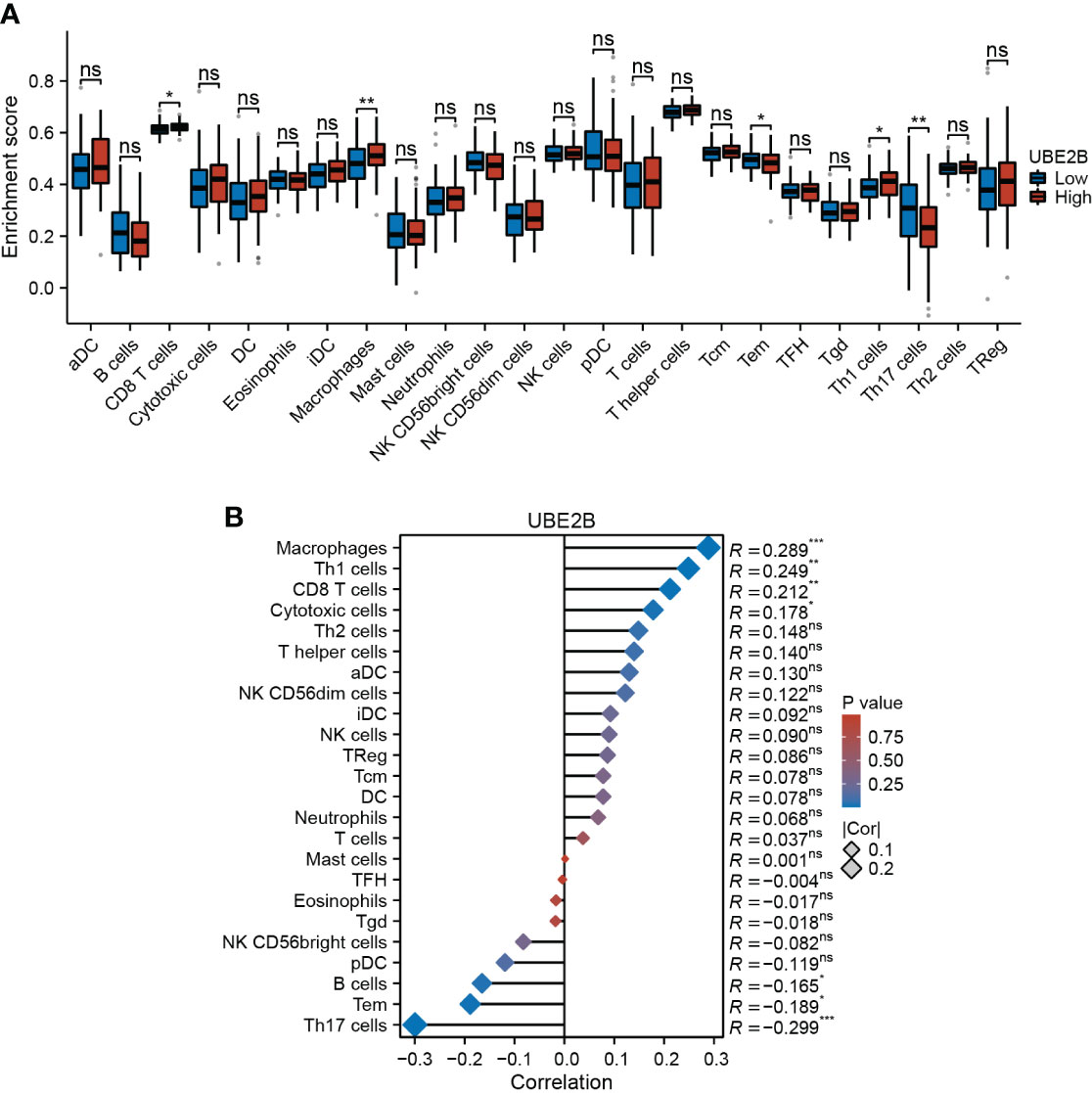
Figure 6 Immune infiltration in two UBE2B expression groups. (A) The different immune cells are in two groups. (B) Correlation analysis between immune cell types and UBE2B expression.
The results of univariate and multivariate Cox regression analyses identified several prognostic factors (Table 4), including pathologic N stage (HR: 6.675, 95% CI: 1.362 - 32.709, P=0.019), pathologic M stage (HR: 29.443, 95% CI: 1.842 - 470.748, P=0.017) and the expression of UBE2B (HR: 4.990, 95% CI: 1.101 - 22.623, P=0.037). Subsequently, a multivariate Cox regression analysis was performed based on these three prognostic factors. The results are illustrated in the forest plot (Figure 7A). Moreover, the ROC curve demonstrated that UBE2B exhibited good sensitivity and specificity in diagnosing ESCA (AUC: 0.749, 95%CI: 0.572-0.925), indicating that UBE2B could be a diagnostic biomarker for patients with ESCA (Figure 7B). Based on the results from Figure 7A, a nomogram was constructed to predict the 1-, 2- and 3-year survival probabilities of patients with ESCA and performance was assessed using calibration plots (Figures 7C, D). In addition, the time-dependent ROC curve further showed the capability of UBE2B in predicting the OS of patients with ESCA (Figure 7E). Furthermore, the log-rank test revealed that UBE2B was significantly correlated with the OS (P<0.001) and DSS (P=0.003) of patients with ESCA, but not progression-free interval (Figures 7F–H). These findings establish that UBE2B was an independent prognostic factor for patients with ESCA.
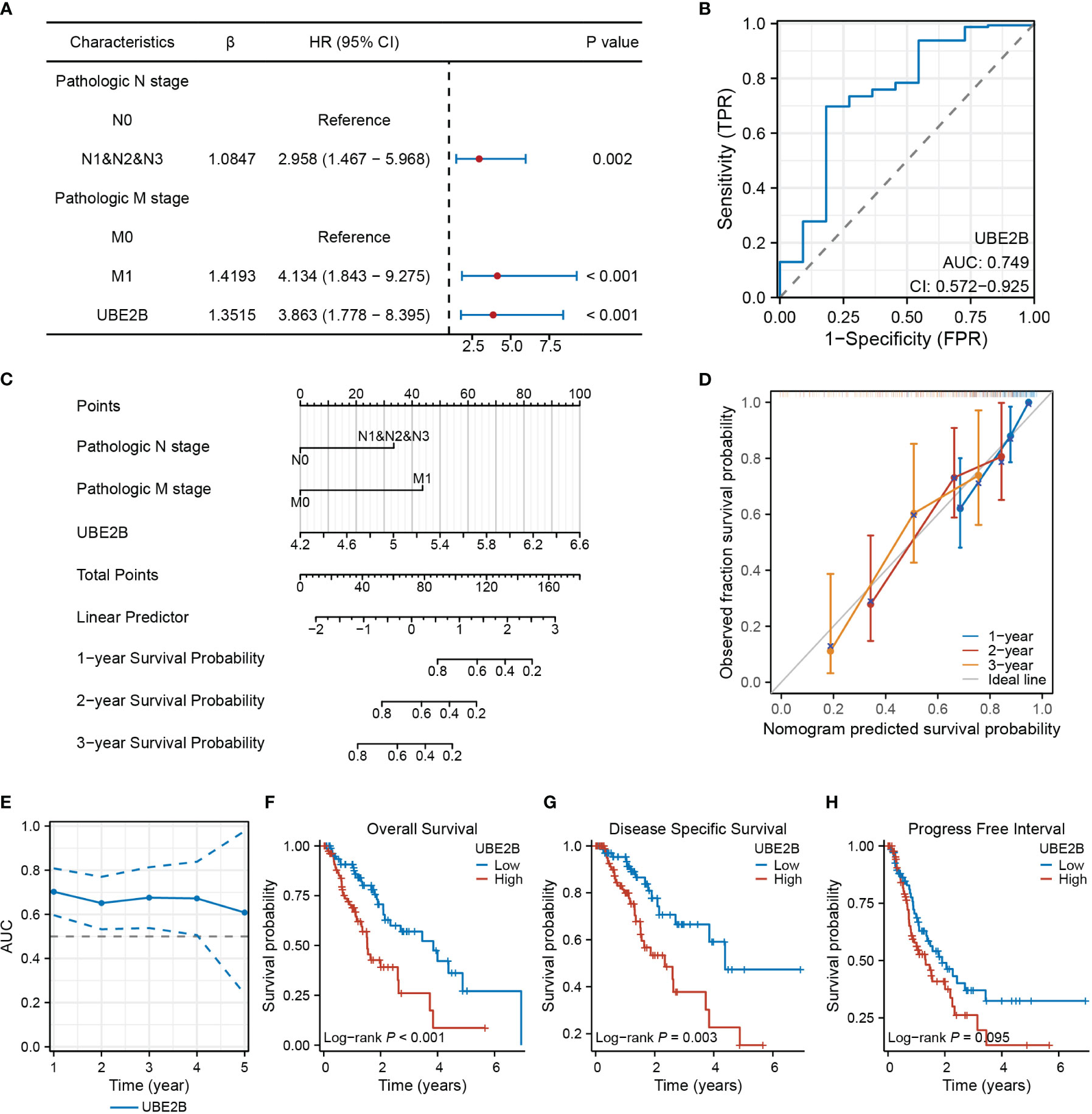
Figure 7 UBE2B as a biomarker in patients with ESCA (Esophageal Carcinoma). (A) Forest plot showing factors related to the prognosis of patients with ESCA (Esophageal Carcinoma). (B) Receiver Operating Characteristic (ROC) curve assessing the diagnostic ability of UBE2B in ESCA (Esophageal Carcinoma). (C) Nomogram for predicting the survival probability of patients with ESCA (Esophageal Carcinoma). (D) Calibration plot of the predictive performance of nomogram. (E) UBE2B evaluation in predicting the prognosis of patients with ESCA (Esophageal Carcinoma). (F-H). The Kaplan-Meier (KM) plots showing the effect of UBE2B expression on the survival outcomes of patients with ESCA (Esophageal Carcinoma), including overall survival, disease specific survival and progress free interval, respectively.
The RT-qPCR and western blot were used to detect the silencing efficacy of siUBE2B. It was found that the expression of UBE2B was suppressed after KYSE-150/ECA-109 was transfected with siUBE2B (Figure 8A). Subsequently, CCK-8 assay and colony formation assay revealed a significant inhibition in the proliferation of both the cell lines upon UBE2B knockdown (∗p < 0.05,∗∗p < 0.01, ∗∗∗p < 0.001; Figures 8B, 9A). Moreover, the UBE2B-knockdown cells demonstrated decreased migration ability (Figure 9B). These findings suggested that UBE2B could be an oncogene promoting cell proliferation and migration of ESCA cells.
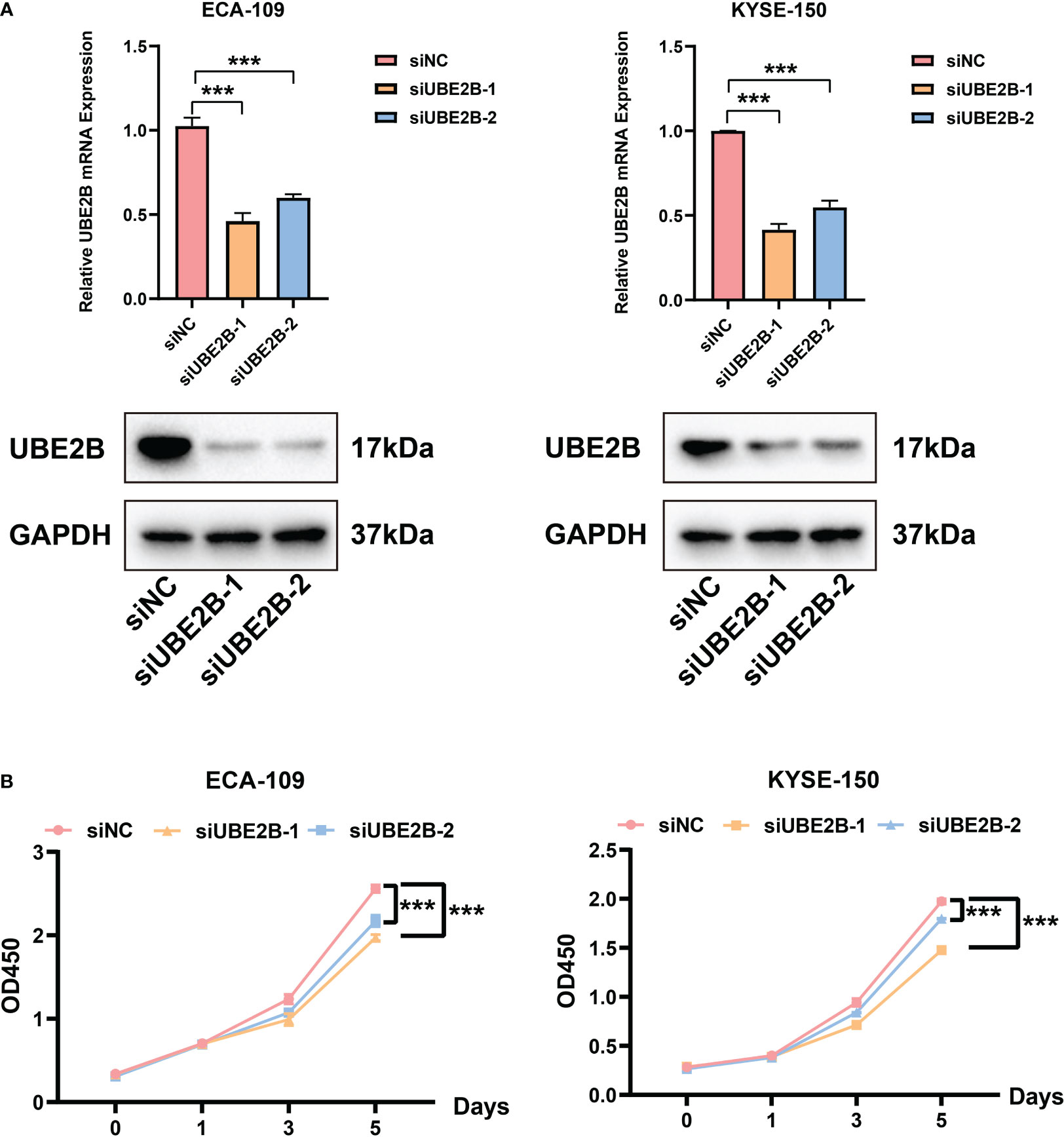
Figure 8 Silencing UBE2B suppressed the proliferation of ECA-109 and KYSE-150 cells. (A) Assessment of silencing efficacy of UBE2B. (B) CCK-8 assay to measure cell proliferation. (***P < 0.001).
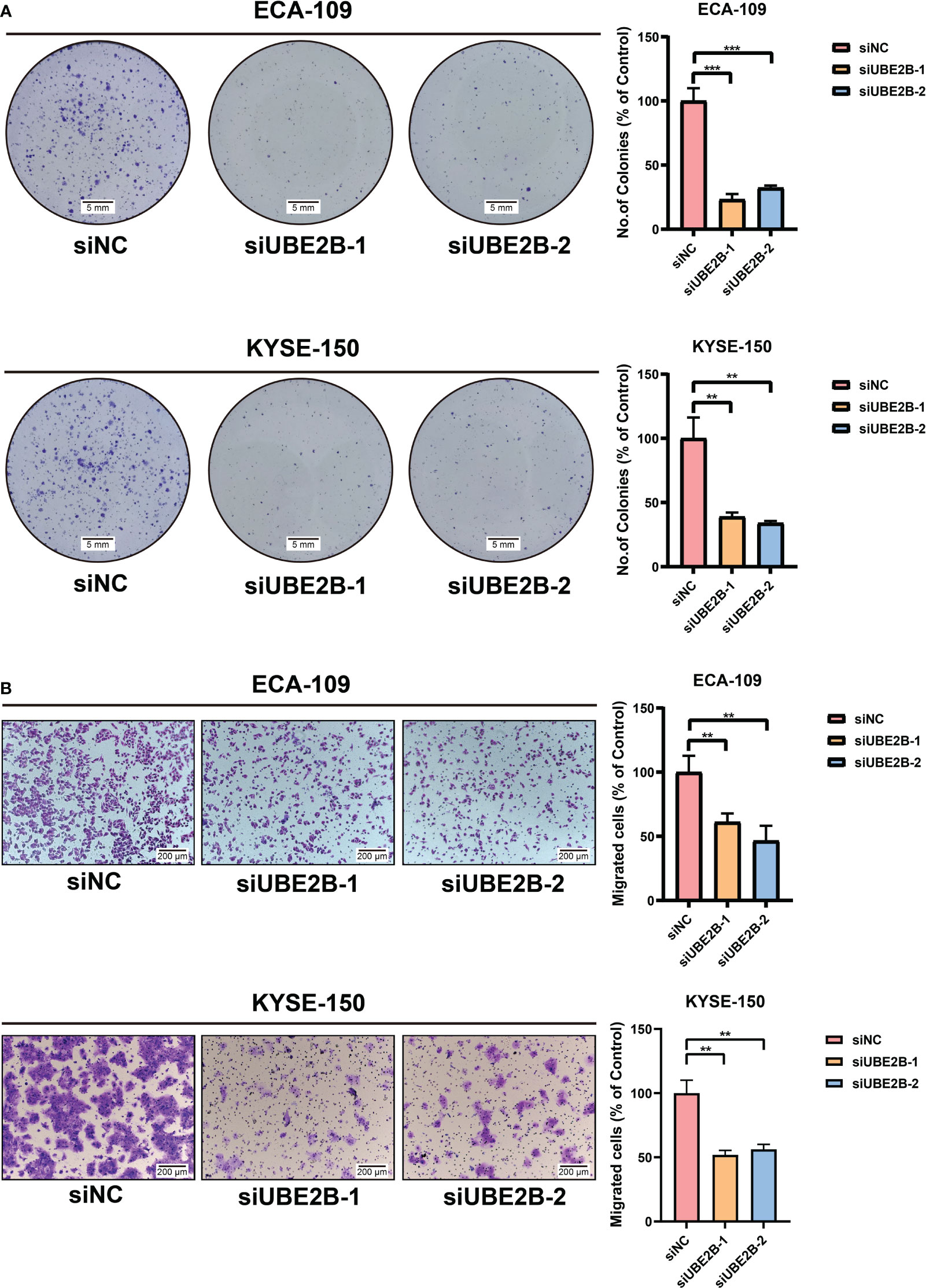
Figure 9 Silencing UBE2B inhibited the growth and migration of ECA-109 and KYSE-150 cells. (A) Colony formation assay to investigate cell growth ability. (B) Detection of cell migration using a migration assay. (**P < 0.01, ***P < 0.001).
Research has provided evidence that genes responsible for regulating ubiquitination enzymes are implicated in the development of different types of cancers, including ESCA. A prime illustration of this is the E3 ligase, such as NF168, which has been determined to intensify the aggressiveness of ESCA by bolstering the stability of STAT1 (23). Additionally, HECTD3 has been identified as a facilitator of ESCA cell growth and survival by inhibiting the activation of caspase-9 (24). Furthermore, in ESCA tissues, E2 enzymes like UBE2C are found to be significantly overexpressed, and their levels are associated with unfavorable survival outcomes for ESCA patients (25). Indeed, based on the identified roles of ubiquitination-related enzymes in ESCA initiation and progression, targeting these enzymes could potentially be a promising strategy for treating ESCA. In this study, the emphasis was on investigating the role of UBE2B in ESCA. UBE2B is a gene that is highly expressed in ESCA tissues and was found to have a significant correlation with the prognosis of ESCA patients, as indicated by its high P value and HR. However, few studies systematically analyzed the expression of UBE2B and its specific function in ESCA.
UBE2B, an evolutionarily conserved enzyme among eukaryotes, interacted with RAD18 to play a pivotal role in post-replicative DNA repair (26). In addition, UBE2B directly affects the tolerance and repair of DNA damage by its involvement in modifying chromatin through the ubiquitination of histone 2B, which leads to changes in chromatin structure and facilitates the recruitment of DNA repair factors (27–29). Studies reported that UBE2B acted as a positive regulator of the canonical Wnt signaling pathway by affecting the stability and activity of β-catenin, promoting cancer development (30). In addition to its involvement in the progression of oral squamous cancer, UBE2B also played a role in enhancing the cisplatin resistance of triple-negative breast cancer (31). Consequently, the activation of UBE2B leads to the enhancement of stem cell gene expression and promotion of malignant characteristics in ovarian cells, thereby contributing to chemotherapy resistance, metastasis, and recurrence in patients with ovarian cancer (32). As such, targeting UBE2B may present a promising therapeutic approach for the prevention and treatment of various cancers. This research demonstrates that UBE2B functions as an oncogene, driving the aggressive growth and migration of ESCA cells, while also serving as a significant prognostic marker for patients with ESCA. However, the specific E3 ligases that interact with UBE2B in ESCA and contribute to its role in tumor development are not yet fully understood. Further investigation is needed to better confirm and understand the molecular mechanisms underlying UBE2B-mediated ESCA development.
Bioinformatic analyses have revealed that UBE2B expression levels vary across different clinical groups, with a significant increase observed in overweight patients with ESCA. This finding is noteworthy as previous studies have established a correlation between high weight, increased ESCA risk, and adverse survival outcomes (3). Furthermore, our findings show that UBE2B expression levels are significantly elevated in patients with ESCC and are strongly correlated with poor prognosis in ESCC patients. However, no such association was observed in patients with esophageal adenocarcinoma ESAD. Additionally, UBE2B demonstrated promising potential as a predictive and diagnostic biomarker for ESCA, independently predicting patient outcomes. Despite establishing a relationship between UBE2B expression and clinical data in ESCA patients, it is important to note that further investigations are necessary to validate these results. This could involve larger patient cohorts, prospective studies, and functional experiments to elucidate the underlying correlation between UBE2B expression and clinical information of patients with ESCA.
The nuclear factor-κB (NF-κB) pathway plays a role in promoting proliferation and angiogenesis in multiple types of human cancers, among other signaling pathways involved in oncogenesis (33).. Resistance to chemotherapeutic drugs in ESCC is observed in the NF-κB pathway, which not only enhances proliferation, angiogenesis, and metastasis, but also contributes to the development of ESCC (34). In addition, tumor necrosis factor-alpha (TNF-α) has been implicated in the development of ESCC from precancerous to cancerous lesions (35). TNF-α activates the NF-κB pathway, leading to the initiation of inflammation, which in turn contributes to the progression of ESCC (36). By suppressing the body’s anti-tumor immune response, inflammatory reactions contribute significantly to the progression and spread of cancer. Moreover, these reactions have the potential to enhance cancer resistance through the activation of tumor stem cells (37). Thus, previous research has utilized inflammatory response biomarkers as individual factors to anticipate the survival prognosis of patients with ESCA (38, 39). On the other hand, inflammatory cytokines can instigate EMT, an essential mechanism that initiates invasion and metastasis of ESCA (40). Inhibiting ESCA progression has been regarded as an effective strategy by targeting EMT. Additionally, it has been observed that hypoxia disrupts the oxidative stress equilibrium in normal cells, resulting in therapeutic resistance and cancer progression (41). Hypoxia-related genes modulate cellular metabolism and molecular responses by regulating the EMT transcription factors/repressors and EMT-related signaling pathways (42, 43). In ESCC, HIF-2α has been found to promote the proliferation and migration of tumor cells by inducing the process of EMT through the activation of the NOTCH signaling pathway (44). In this study, it was intriguingly observed that these signaling pathways and biological processes that promote the aggressive behavior of tumor cells were enriched specifically in patients with ESCA who had high expression of UBE2B. Considering the involvement of UBE2B in driving the malignant progression of ESCA by modulating these pathways and processes, it is possible that UBE2B plays a role in the progression of ESCA and contributes to poor prognosis.
In ESCA, the number of tumor-infiltrating macrophages and T cells are higher than those in other solid tumors (45). Although it was found that high expression of UBA1 increased the level of CD8 T cells and macrophages, the level of Tem decreased. As a member of memory T cell, Tem have high levels of receptors to mediate strong immediate effector function to response the inflammation, which also participates in anti-tumor (46). Therefore, UBA1 might help tumor immune escape by suppressing Tem. Additionally, it was found that the expression of UBA1 was negatively correlated with the level of Th17 cells. Th17 cells were mainly reported in autoimmune diseases in previous studies, but now they are found in many cancer types like melanoma, ovarian cancer, and colorectal cancer (47). However, the role of Th17 cells in cancers seemed to be contradictory. One study compared the impact of IL-17 and Th17 cells on cancer patient survival, and found that high Th17 cell frequencies improved the survival outcomes, whereas high IL-17 contributed to poor prognosis (48). Therefore, whether UBA1 promote the development by inhibiting the number of Th17 cells in ESCA needs further researches.
In summary, this study aimed to elucidate the specific functions and mechanisms of UBE2B in ESCA development and progression, potentially identifying it as a novel therapeutic target or prognostic marker. By focusing on UBE2B, the researchers aimed to gain insights into its potential role in ESCA biology and its significance in patient outcomes.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
HD: Data curation, Formal analysis, Validation, Writing – original draft, Writing – review & editing. J-CX: Formal analysis, Visualization, Writing – review & editing. Z-GD: Validation, Writing – review & editing. L-FW: Software, Writing – review & editing. Y-BL: Visualization, Writing – review & editing. Y-FZ: Visualization, Writing – review & editing. T-YC: Conceptualization, Methodology, Writing – review & editing. Y-QZ: Conceptualization, Funding acquisition, Writing – review & editing. P-HZ: Conceptualization, Project administration, Supervision, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by the National Natural Science Foundation of China (No. 82172787).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1295305/full#supplementary-material
Supplementary Table 1 | The list of genes associated with the prognosis of patients with ESCA.
1. Sun Y, Zhang T, Wu W, Zhao D, Zhang N, Cui Y, et al. Risk factors associated with precancerous lesions of esophageal squamous cell carcinoma: a screening study in a high risk chinese population. J Canc. (2019) 10:3284–90. doi: 10.7150/jca.29979.
2. Thrift AP. Global burden and epidemiology of Barrett oesophagus and oesophageal cancer. Nat Rev Gastroenterol hepatol. (2021) 18:432–43. doi: 10.1038/s41575-021-00419-3.
3. Vingeliene S, Chan DSM, Vieira AR, Polemiti E, Stevens C, Abar L, et al. An update of the WCRF/AICR systematic literature review and meta-analysis on dietary and anthropometric factors and esophageal cancer risk. Ann Oncol Off J Eur Soc Med Oncol. (2017) 28:2409–19. doi: 10.1093/annonc/mdx338.
4. Lv H, He Z, Wang H, Du T, Pang Z. Differential expression of miR-21 and miR-75 in esophageal carcinoma patients and its clinical implication. Am J Trans Res. (2016) 8:3288–98.
5. Yang F, Wei K, Qin Z, Shao C, Shu Y, Shen H. Association between TNF-ɑ-308G/A polymorphism and esophageal cancer risk: An updated meta-analysis and trial sequential analysis. J Canc. (2019) 10:1086–96. doi: 10.7150/jca.29390.
6. Lordick F, Janjigian YY. Clinical impact of tumour biology in the management of gastroesophageal cancer. Nat Rev Clin Oncol. (2016) 13:348–60. doi: 10.1038/nrclinonc.2016.15.
7. Martincorena I, Fowler JC, Wabik A, Lawson ARJ, Abascal F, Hall MWJ, et al. Somatic mutant clones colonize the human esophagus with age. Sci (New York NY). (2018) 362:911–7. doi: 10.1126/science.aau3879.
8. Wloka C, Van Meervelt V, van Gelder D, Danda N, Jager N, Williams CP, et al. Label-free and real-time detection of protein ubiquitination with a biological nanopore. ACS nano. (2017) 11:4387–94. doi: 10.1021/acsnano.6b07760.
9. Zhou X, Li Y, Wang W, Wang S, Hou J, Zhang A, et al. Regulation of Hippo/YAP signaling and Esophageal Squamous Carcinoma progression by an E3 ubiquitin ligase PARK2. Theranostics. (2020) 10:9443–57. doi: 10.7150/thno.46078.
10. Guo X, Zhu R, Luo A, Zhou H, Ding F, Yang H, et al. EIF3H promotes aggressiveness of esophageal squamous cell carcinoma by modulating Snail stability. J Exp Clin Cancer Res CR. (2020) 39:175. doi: 10.1186/s13046-020-01678-9.
11. Wang Q, Wu L, Cao R, Gao J, Chai D, Qin Y, et al. Fbxo45 promotes the Malignant development of esophageal squamous cell carcinoma by targeting GGNBP2 for ubiquitination and degradation. Oncogene. (2022) 41:4795–807. doi: 10.1038/s41388-022-02468-7.
12. Wang Y, Liang N, Xue Z, Xue X. Identifying an eight-gene signature to optimize overall survival prediction of esophageal adenocarcinoma using bioinformatics analysis of ceRNA network. OncoTargets Ther. (2020) 13:13041–54. doi: 10.2147/OTT.S287084.
13. Zeng X, Zheng W, Sheng Y, Ma H. UBE2B promotes ovarian cancer growth via promoting RAD18 mediated ZMYM2 monoubiquitination and stabilization. Bioengineered. (2022) 13:8000–12. doi: 10.1080/21655979.2022.2048991.
14. Huang WL, Luo CW, Chou CL, Yang CC, Chen TJ, Li CF, et al. High expression of UBE2B as a poor prognosis factor in patients with rectal cancer following chemoradiotherapy. Anticancer Res. (2020) 40:6305–17. doi: 10.21873/anticanres.14651.
15. Kao WC, Hsu SH, Lin CL, Lin CY, Chen SW, Chen YX, et al. Role of high ubiquitin-conjugating enzyme E2 expression as a prognostic factor in nasopharyngeal carcinoma. Oncol letters. (2022) 23:194. doi: 10.3892/ol.
16. Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, et al. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. (2018) 173:400–16.e11. doi: 10.1016/j.cell.2018.02.052
17. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. (2014) 15:550. doi: 10.1186/s13059-014-0550-8.
18. Walter W, Sánchez-Cabo F, Ricote M. GOplot: an R package for visually combining expression data with functional analysis. Bioinf (Oxford England). (2015) 31:2912–4. doi: 10.1093/bioinformatics/btv300.
19. Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics J Integr Biol. (2012) 16:284–7. doi: 10.1089/omi.2011.0118.
20. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci United States Am. (2005) 102:15545–50. doi: 10.1073/pnas.0506580102.
21. Yoshihara K, Shahmoradgoli M, Martínez E, Vegesna R, Kim H, Torres-Garcia W, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. (2013) 4:2612. doi: 10.1038/ncomms3612.
22. Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. (2013) 39:782–95. doi: 10.1016/j.immuni.2013.10.003.
23. Yu N, Xue M, Wang W, Xia D, Li Y, Zhou X, et al. RNF168 facilitates proliferation and invasion of esophageal carcinoma, possibly via stabilizing STAT1. J Cell Mol Med. (2019) 23:1553–61. doi: 10.1111/jcmm.14063.
24. Li Y, Wu X, Li L, Liu Y, Xu C, Su D, et al. The E3 ligase HECTD3 promotes esophageal squamous cell carcinoma (ESCC) growth and cell survival through targeting and inhibiting caspase-9 activation. Cancer letters. (2017) 404:44–52. doi: 10.1016/j.canlet.2017.07.004.
25. Li R, Pang XF, Huang ZG, Yang LH, Peng ZG, Ma J, et al. Overexpression of UBE2C in esophageal squamous cell carcinoma tissues and molecular analysis. BMC canc. (2021) 21:996. doi: 10.1186/s12885-021-08634-6.
26. Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. (2002) 419:135–41. doi: 10.1038/nature00991.
27. Robzyk K, Recht J, Osley MA. Rad6-dependent ubiquitination of histone H2B in yeast. Sci (New York NY). (2000) 287:501–4. doi: 10.1126/science.287.5452.501.
28. Kao CF, Hillyer C, Tsukuda T, Henry K, Berger S, Osley MA. Rad6 plays a role in transcriptional activation through ubiquitylation of histone H2B. Genes Dev. (2004) 18:184–95. doi: 10.1101/gad.1149604.
29. Gajan A, Martin CE, Kim S, Joshi M, Michelhaugh SK, Sloma I, et al. Alternative splicing of RAD6B and not RAD6A is selectively increased in melanoma: identification and functional characterization. Cells. (2019) 8(11):1375. doi: 10.3390/cells8111375.
30. Sarma A, Gajan A, Kim S, Gurdziel K, Mao G, Nangia-Makker P, et al. RAD6B loss disrupts expression of melanoma phenotype in part by inhibiting WNT/β-catenin signaling. Am J pathol. (2021) 191:368–84. doi: 10.1016/j.ajpath.2020.10.015.
31. Haynes B, Gajan A, Nangia-Makker P, Shekhar MP. RAD6B is a major mediator of triple negative breast cancer cisplatin resistance: Regulation of translesion synthesis/Fanconi anemia crosstalk and BRCA1 independence. Biochim Biophys Acta Mol Bas Dis. (2020) 1866:165561. doi: 10.1016/j.bbadis.2019.165561.
32. Somasagara RR, Spencer SM, Tripathi K, Clark DW, Mani C, Madeira da Silva L, et al. RAD6 promotes DNA repair and stem cell signaling in ovarian cancer and is a promising therapeutic target to prevent and treat acquired chemoresistance. Oncogene. (2017) 36:6680–90. doi: 10.1038/onc.2017.279.
33. Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. (2006) 441:431–6. doi: 10.1038/nature04870.
34. Chen Y, Wang D, Peng H, Chen X, Han X, Yu J, et al. Epigenetically upregulated oncoprotein PLCE1 drives esophageal carcinoma angiogenesis and proliferation via activating the PI-PLCϵ-NF-κB signaling pathway and VEGF-C/ Bcl-2 expression. Mol canc. (2019) 18:1. doi: 10.1186/s12943-018-0930-x.
35. Tselepis C, Perry I, Dawson C, Hardy R, Darnton SJ, McConkey C, et al. Tumour necrosis factor-alpha in Barrett's oesophagus: a potential novel mechanism of action. Oncogene. (2002) 21:6071–81. doi: 10.1038/sj.onc.1205731.
36. Webster JD, Vucic D. The balance of TNF mediated pathways regulates inflammatory cell death signaling in healthy and diseased tissues. Front Cell Dev Biol. (2020) 8:365. doi: 10.3389/fcell.2020.00365.
37. Aguilar-Cazares D, Chavez-Dominguez R, Marroquin-Muciño M, Perez-Medina M, Benito-Lopez JJ, Camarena A, et al. The systemic-level repercussions of cancer-associated inflammation mediators produced in the tumor microenvironment. Front Endocrinol. (2022) 13:929572. doi: 10.3389/fendo.2022.929572.
38. Chen CJ, Lee CT, Tsai YN, Tseng CM, Chen TH, Hsu MH, et al. Prognostic significance of systemic inflammatory response markers in patients with superficial esophageal squamous cell carcinomas. Sci Rep. (2022) 12:18241. doi: 10.1038/s41598-022-21974-y.
39. Hirahara N, Matsubara T, Kawahara D, Nakada S, Ishibashi S, Tajima Y. Prognostic significance of preoperative inflammatory response biomarkers in patients undergoing curative thoracoscopic esophagectomy for esophageal squamous cell carcinoma. Eur J Surg Oncol J Eur Soc Surg Oncol Br Assoc Surg Oncol. (2017) 43:493–501. doi: 10.1016/j.ejso.2016.11.018.
40. Zhang H, Qin G, Zhang C, Yang H, Liu J, Hu H, et al. TRAIL promotes epithelial-to-mesenchymal transition by inducing PD-L1 expression in esophageal squamous cell carcinomas. J Exp Clin Cancer Res CR. (2021) 40:209. doi: 10.1186/s13046-021-01972-0.
41. Jiang R, Hu J, Zhou H, Wei H, He S, Xiao J. A novel defined hypoxia-related gene signature for prognostic prediction of patients with ewing sarcoma. Front Genet. (2022) 13:908113. doi: 10.3389/fgene.2022.908113.
42. Imai T, Horiuchi A, Wang C, Oka K, Ohira S, Nikaido T, et al. Hypoxia attenuates the expression of E-cadherin via up-regulation of SNAIL in ovarian carcinoma cells. Am J pathol. (2003) 163:1437–47. doi: 10.1016/S0002-9440(10)63501-8.
43. Joseph JP, Harishankar MK, Pillai AA, Devi A. Hypoxia induced EMT: A review on the mechanism of tumor progression and metastasis in OSCC. Oral Oncol. (2018) 80:23–32. doi: 10.1016/j.oraloncology.2018.03.004.
44. Ji Z, Bao S, Li L, Wang D, Shi M, Liu X. Hypoxia-inducible factor-2α promotes EMT in esophageal squamous cell carcinoma through the Notch pathway. Adv Clin Exp Med Off Organ Wroclaw Med University. (2022) 31:795–805. doi: 10.17219/acem/147270.
45. Zheng Y, Chen Z, Han Y, Han L, Zou X, Zhou B, et al. Immune suppressive landscape in the human esophageal squamous cell carcinoma microenvironment. Nat Commun. (2020) 11:6268. doi: 10.1038/s41467-020-20019-0.
46. Liu Q, Sun Z, Chen L. Memory T cells: strategies for optimizing tumor immunotherapy. Protein Cell. (2020) 11:549–64. doi: 10.1007/s13238-020-00707-9.
47. Kryczek I, Banerjee M, Cheng P, Vatan L, Szeliga W, Wei S, et al. Phenotype, distribution, generation, and functional and clinical relevance of Th17 cells in the human tumor environments. Blood. (2009) 114:1141–9. doi: 10.1182/blood-2009-03-208249.
Keywords: bioinformatics, esophageal carcinoma, immune infiltration, prognostic biomarker, UBE2B
Citation: Ding H, Xu J-C, Ding Z-G, Wu L-F, Liu Y-B, Zhang Y-F, Chen T-Y, Zhang Y-Q and Zhou P-H (2024) Identification and validation of UBE2B as a prognostic biomarker promoting the development of esophageal carcinomas. Front. Immunol. 15:1295305. doi: 10.3389/fimmu.2024.1295305
Received: 16 September 2023; Accepted: 08 February 2024;
Published: 28 February 2024.
Edited by:
Jie Shen, Nanjing Drum Tower Hospital, ChinaCopyright © 2024 Ding, Xu, Ding, Wu, Liu, Zhang, Chen, Zhang and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tian-Yin Chen, Y2hlbi50aWFueWluQHpzLWhvc3BpdGFsLnNoLmNu; Yi-Qun Zhang, emhhbmcueWlxdW5AenMtaG9zcGl0YWwuc2guY24=; Ping-Hong Zhou, emhvdS5waW5naG9uZ0B6cy1ob3NwaXRhbC5zaC5jbg==
†These authors have contributed equally to this work and share first authorship
‡These authors have contributed equally to this work and share last authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.