
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Immunol. , 03 January 2024
Sec. Viral Immunology
Volume 14 - 2023 | https://doi.org/10.3389/fimmu.2023.1331981
This article is part of the Research Topic Immunotherapeutics Development against Hantaviruses View all 6 articles
 Zhiyong Wang
Zhiyong Wang Pei Wei*
Pei Wei*In this review article, we explore the characteristics of RNA viruses and their potential threats to humanity. We also provide a brief overview of the primary contemporary techniques used for the early detection of such viruses. After thoroughly analyzing the strengths and limitations of these methods, we highlight the importance of integrating nucleic acid testing with immunological assays in RNA virus detection. Although notable methodological differences between nucleic acid testing and immune assays pose challenges, the emerging single-molecule immunoassay-digital ELISA may be applied to technically integrate these techniques. We emphasize that the greatest value of digital ELISA is its extensive compatibility, which creates numerous opportunities for real-time, large-scale testing of RNA viruses. Furthermore, we describe the possible developmental trends of digital ELISA in various aspects, such as reaction carriers, identification elements, signal amplification, and data reading, thus revealing the remarkable potential of single-molecule digital ELISA in future RNA virus detection.
RNA viruses encompass a range of pathogens characterized by their use of RNA as a genetic material (1, 2). They include different types such as influenza virus (3, 4), Ebola virus (5, 6), and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (7, 8). Because of their high mutation rate and potential to cause severe diseases, they pose a significant threat to global public health. For this reason, they must be detected early to prevent disease transmission and facilitate effective treatments. Current detection methods include virus isolation and culture, viral sequencing, nucleic acid-based detection, and immunological-based detection (9, 10). Each method has unique advantages and challenges in terms of sensitivity, specificity, time efficiency, and resource requirements. Among them, nucleic acid- and immunological-based detection are the main methods for the early diagnosis of RNA viruses. Our viewpoint is that the integration of nucleic acid- and immunological-based approaches based on digital ELISA may be the future trend in RNA virus detection because it can minimize their respective limitations and maximize their advantages; thus, such integration can improve the accuracy and efficiency of RNA virus detection.
In this mini-review, we do not intend to provide an exhaustive survey of the field of digital detection because it has been extensively covered in the existing literature (11, 12). Instead, in the field of digital assay research, we focus on the detection of RNA viruses, specifically highlighting the convergence of nucleic acid testing techniques and immunoassays in the context of digital ELISA. We aim to offer an insightful yet succinct perspective on the revolutionary impact of digital ELISA technologies on the detection of RNA viruses.
RNA viruses, including double-stranded RNA viruses, single-stranded RNA viruses, and retroviruses, are a class of viruses that utilize RNA as their genetic material (1, 2). These viruses are characterized by their large size and short generation time; because their RNA-dependent RNA polymerase (RdRp) lacks a proofreading activity, they also have an extremely high mutation rate (13, 14). With a high mutation rate, RNA viruses in a single host can generate numerous genetic variations, which may result in viral phenotypes, including virulence and pathogenicity, thereby affecting the transmission and infection capacity of viruses (15, 16). They are capable of human-to-human transmission through various routes, including direct contact and inhalation; they can also be transmitted from humans to animals (17, 18). Because of the frequent recombination of genomes and the increased activity at the human–animal interface, RNA viruses are highly prone to causing new epidemics. In addition to their inherently high genetic diversity and rapid evolution, their transmission characteristics depend on their interaction with host immunity and the environment. The “survival path” of a newly emerging virus largely depends on the host’s immune response, and individuals with impaired adaptive immune responses more likely favor virus production and mutations (19, 20). During virus transmission, virus mutations may help evade previously existing immune responses and even change mechanisms that cross over with host cell pathways and are beneficial to infection; consequently, the probability of the emergence of new viral strains increases. This adaptive evolution, combined with the complexity of host immune responses, determines whether RNA viruses can cause localized, epidemic, or pandemic outbreaks (21, 22). In the past century, RNA viruses caused several major pandemics, posing significant threats to global health, and the COVID-19 pandemic in 2019 served as a stark reminder of this fact. Given the high frequency and widespread dissemination of RNA viruses, the detection and identification of these viruses are crucial for preventing potential pandemics and mitigating their societal impact. The clinical diagnosis of diseases caused by RNA virus infection primarily involves an integration of clinical symptoms, history of exposure to viruses, and virus detection. Because clinical manifestations and signs in infected patients are not entirely consistent, especially considering potential asymptomatic or mildly symptomatic infections, early viral detection is imperative for handling and controlling the large-scale transmission of RNA viruses.
Currently, detection or quantification methods for RNA viruses mainly include virus isolation and culture, nucleic acid-based detection methods, and immunological-based detection methods (9, 10). Among them, virus isolation and culture are the most reliable because they contribute to further analysis and characterization of the target virus (23, 24). However, most RNA viruses can be isolated only in laboratories with biosafety level 3 facilities. Additionally, virus isolation and culture are relatively time consuming, often requiring several days or even weeks (23, 24); therefore, they are unable to meet the demands for rapid diagnosis in most cases.
Nucleic acid-based RNA virus detection methods include sequencing, such as next-generation sequencing (NGS) (25, 26), and nucleic acid amplification-based detection technologies, such as isothermal and non-isothermal nucleic acid amplification (27–30). Sequencing technologies utilize extensive reference databases to explore new mutations and evolution of isolated RNA virus strains; determine the mutation rate of viruses and other related recombinants; and evaluate potential contact tracing, virus evolution research, and molecular epidemiology (25, 26). However, sequencing analyses require highly skilled personnel to operate expensive instruments under strict laboratory conditions. They are time-consuming and relatively costly, severely limiting their widespread application in RNA virus detection (31). By comparison, detection technologies based on nucleic acid amplification feature high sensitivity, specificity, speed, quantifiable results, and automation; therefore, they are suitable for large-scale screening in the early stages of an epidemic and have become the most widely accepted RNA virus diagnostic method (25–28). However, they are also limited in terms of experimental conditions, equipment costs, personnel costs, and detection cycles. For instance, sample contamination caused by factors such as aerosols during nucleic acid amplification negatively affects detection accuracy (32). These limitations, combined with the inherent instability of RNA molecules, further restricts the application of such detection technologies in large-scale screening for early-stage RNA virus infections.
Immunological detection strategies utilize antibody–antigen reactions to detect target viral proteins (antigens) in samples or antibodies against viruses in serum (33, 34). An example of such strategies is enzyme-linked immunosorbent assay (ELISA) (35, 36). Immunoassay-based detection methods are cost effective, rapid, and simple, making them suitable for wide screening. However, their widespread application in early stages is restricted because of some inherent limitations. First, immunological detection has lower sensitivity than nucleic acid amplification techniques and may produce false-negative results in cases of low viral loads or limited serum antibody concentrations. Second, for viral antigen detection, the preparation and production of corresponding detecting antibodies require time, impeding the urgent demand for large-scale early screening. Third, the occurrence of cross-reactivity in antigen–antibody reactions may lead to false-positive results, thus limiting the accuracy of results to some extent (37).
Nucleic acid and immune detection techniques are the two main methods for the early screening of RNA virus infections, each with their own unique advantages and inherent limitations. As such, these methods should be integrated into a unified approach to form a more comprehensive and effective diagnostic system. Because of the fundamental methodological differences between nucleic acid testing and immune testing, they have a considerable gap. By comparison, ELISA exhibits strong compatibility, in addition to its numerous advantages, such as simplicity, practicality, speed, and reasonable cost; thus, it offers the potential for achieving technical integration between nucleic acid testing and immune testing.
Immunological assays have evolved over years of development, leveraging their exceptional compatibility to be integrated with various interdisciplinary fields; as a result, a series of noteworthy technological innovations has been developed. These advancements are evident in several aspects of immunological assays, including reaction carriers, recognition elements, signal amplification, and reading methods. Of particular importance is the fusion of single-molecule detection technology and immunological assays, resulting in single-molecule counting immunity, which is also known as digital ELISA (38, 39). This revolutionary technique has overcome the sensitivity limitations of traditional immunological assays, thus offering an extraordinary level of sensitivity and high resolution that macroscopic measurements have failed to achieve.
The pursuit of improving the detection limit of biomolecules has given rise to the trend of digital measurement. The term “digital” in computing refers to the binary code of 0 s and 1 s. Similarly, in detection systems, the switching of specific signals of biomolecules between discrete states (such as presence or absence, occurrence or non-occurrence, and binding or non-binding) is also a binary event. Unlike traditional bioassays typically conducted in a single reactor, in digital assays, individual target molecules are randomized into microchambers (11, 12). Once the signal separation is established between positive microchambers with a single target molecule and negative microchambers without the target molecule (reaction microchambers display binary discrete signals representing “1” or “0”), the absolute quantity of target molecules can be determined by counting positive reactors after digitizing signals with sufficient thresholds (11, 12). Figure 1 provides a schematic diagram comparing digital bioassays and traditional bioassays. In digital bioassays, the reaction in each microchamber displays a positive or negative signal, meeting the prerequisite conditions for a binomial trial. Therefore, digital bioassays should use the Poisson distribution, which is a limiting case of the binomial distribution (11, 12). Unlike the continuous signal recording noted in traditional ELISA, analysis is performed by recording discrete signals in digital ELISA. This strategic deviation circumvents the inaccuracies introduced by macro-level signal fluctuations, thereby providing an overwhelming advantage for detecting target molecules at low concentrations. In fact, for the same target molecule, the detection limit of digital assays is generally at least 1/1000 that of conventional assays (39, 40).
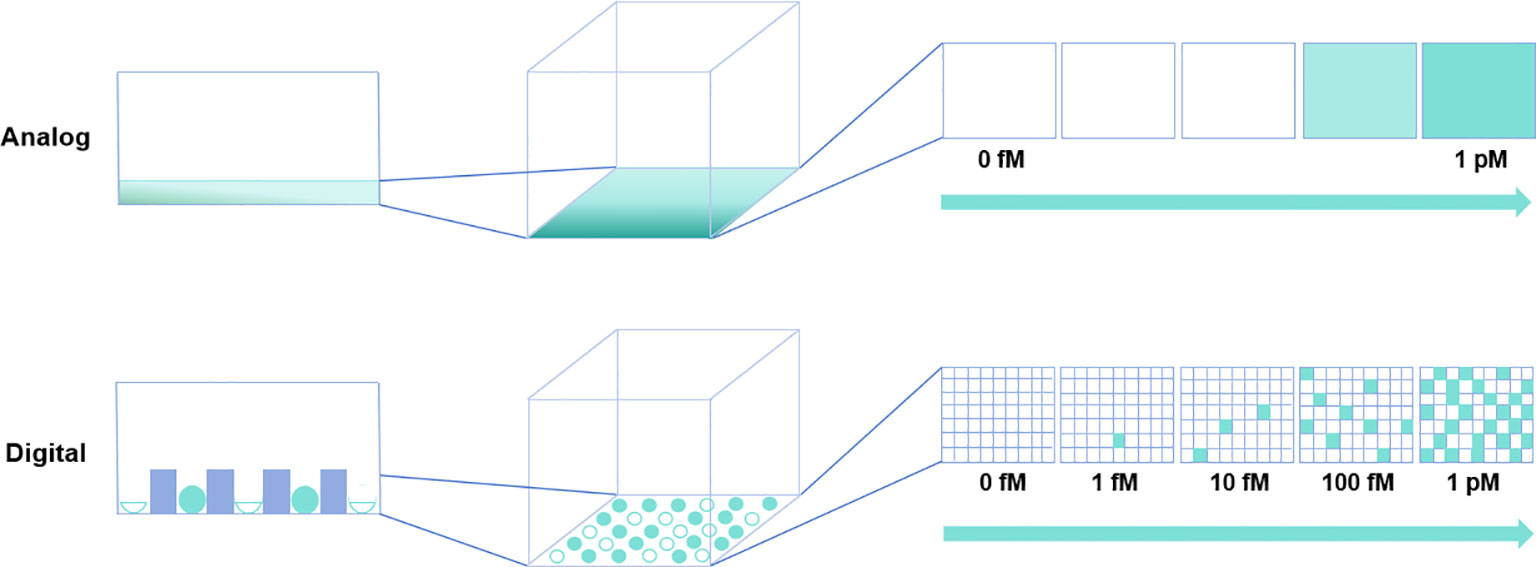
Figure 1 Comparative schematic of traditional versus digital bioassays. In the traditional (analog) assay depicted in the upper panel, rapid diffusion of reaction products is suboptimal for detecting highly diluted target molecules. Conversely, in the digital bioassay shown in the lower panel, reaction products are compartmentalized within microchambers, enabling detection at the single-molecule level.
Digital biodetection systems consist of three main components: microchamber reactors for microsegmentation, markers for studying or labeling target molecules, and substrates or chemical probes for providing detectable signals (11, 12). Depending on the type of signal amplification in the microreactor, digital assays for RNA viruses can be divided into two categories. The first category includes digital PCR and other digital nucleic acid amplification assays, which rely on the exponential amplification of viral RNA transcribed into cDNA. However, the non-linear enzyme-catalyzed reverse transcription process introduces quantitative bias (41, 42). Moreover, the contamination of samples during the nucleic acid amplification process greatly affects the accuracy of quantitative results (32). Therefore, digital detection methods based on non-reverse transcription and non-nucleic acid amplification can be more effective for the quantification of RNA viruses. Unlike the first category involving digital PCR, which relies on the amplification of the target molecule, the other category of digital detection of RNA viruses relies on the amplification of detection signals (such as enzymes and fluorescent probes); one such example is digital ELISA.
The principal methodology of digital ELISA involves a four-step process: the formation of immune complexes to capture target molecules (formation), spatial or temporal partition of these complexes to make them distinguishable and quantifiable (partition), recognition of signals generated by the target molecules through optical or electrical methods (recognition), and absolute quantification using the Poisson distribution (quantification) (12, 43). Advances in the development of RNA antibody mimetics (44, 45), nucleic acid aptamers (46, 47), and molecularly imprinted polymers (48, 49) have made it feasible to directly detect trace amounts of viral RNA using digital ELISA; this is a significant breakthrough. This capability implies that high-sensitivity RNA virus detection can be performed even without the need for a nucleic acid amplification step, thereby reducing the assay time and completely bypassing issues related to sample contamination that can occur during amplification. This ultimately reduces the stringent environmental controls required for nucleic acid assays and has profound implications for the real-time detection of RNA viruses. Figure 2 provides a schematic representation of the use of digital ELISA for the direct detection of viral RNA. Currently, successful commercial digital ELISA detection platforms, such as Simoa (50) and Erenna (its upgraded version is called SMCxPro) (51), are widely used for practical applications. With these commercial digital ELISA technology platforms, quantitative analysis of antibodies (IgG, IgM, and IgA) in the blood of SARS-CoV-2 infected individuals can be performed on the same day when the symptoms first appear and the nucleic acid test yields positive results, with a sample volume of less than 1 μL (52). Most excitingly, research utilizing the Simoa platform has demonstrated the capability to detect SARS-CoV-2 RNA in saliva, achieving a remarkable detection limit of 3.4 fM (53).
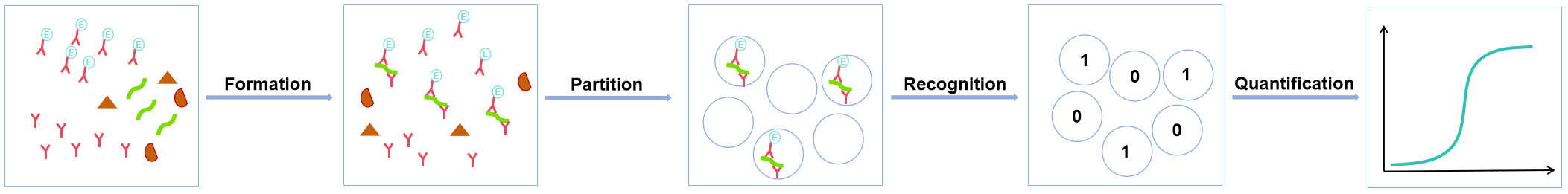
Figure 2 Schematic outline of the digital ELISA process for detecting viral RNA. Viral RNA is captured by specific antibodies (or nucleic acid aptamers or molecular polymers) to form an immune complex, which is then compartmentalized into discrete, countable units. Quantification of individual immunocomplex molecules is achieved by recognizing the signal generated by the marker molecules conjugated to the antibodies.
Currently, digital ELISA is undergoing rapid development; for example, the widespread application of droplet discretization method has achieved the full loading of immunocomplexes (54, 55). Scholars such as Akama proposed a soft discretization method using enzyme-catalyzed deposition, which is expected to avoid the dependence on complex droplet generation equipment (56). Other researchers developed a particle diffusion recognition method, which allows particles to exhibit characteristic Brownian motion in a limited space; in this way, uniform immunoanalysis can be digitized without washing (57, 58). Chen and others visualized single immunocomplexes by using microbubbles produced through hydrogen peroxide catalysis with platinum nanoparticles (59). Zhang and others used the enhancement effect of microsphere lenses to visualize single target labels successfully under a 20× objective (60). Furthermore, emerging evidence suggests that digital ELISA will make further breakthroughs in portability, readiness, simplicity, high throughput, and ultrasensitivity (61, 62).
Because of factors such as testing costs, efficiency, reagent stability, and maintenance costs, the large-scale application of digital ELISA for early RNA virus detection remains impractical. However, looking back at the entire history of diagnostic technology development, the improvement of any detection technique requires a long period of exploration. The significance of digital ELISA in RNA virus detection lies in its ability to integrate nucleic acid and immune detection, rather than just improving existing detection techniques. Equally important is the powerful compatibility of ELISA, which provides a wide range of options for further expanding RNA virus detection.
For example, in the optimization of reaction carriers, digital ELISA can be combined with paper-based biosensors (63, 64) to develop shorter, lower-cost, and field-deployable paper-based detection systems that meet the needs of large-scale, real-time RNA virus detection. With the development of low-cost data reading devices, such as desktop scanners or mobile cameras (65), RNA viruses can be detected early in resource-limited areas on a large scale. In terms of recognition element optimization, given that antibodies as recognition molecules have limitations, such as poor stability, high costs, and difficulty in large-scale production, digital ELISA can be combined with nucleic acid aptamers (46, 47) or molecularly imprinted polymers (48, 49) to eliminate dependence on antibodies. For RNA viruses, these alternatives further remove barriers to the integration of nucleic acid and immunoassays. In the aspect of signal amplification optimization, various nanomaterials, such as nanospheres (60, 66), nanopores (67), upconversion nanoparticles (68, 69), and quantum dots (70, 71), when combined with digital ELISA, can greatly enhance the amplification of collected signals. Additionally, the combination of CRISPR-Cas system with traditional ELISA can greatly enhance the detection sensitivity of the latter (72). Therefore, the effect produced by the combination of CRISPR-Cas system with digital ELISA is worth anticipating. The development of these methods ensures the sensitivity of digital ELISA, compensating for the loss of sensitivity because of avoiding nucleic acid amplification in the unification of nucleic acid and immunoassays. In terms of reading methods, neither fluorescence scanning after enzyme-catalyzed signal amplification in the Simoa detection system (50) nor device-dependent single-molecule signal scanning in the Erenna (SMCxPro) system (51) is suitable for early large-scale screening of RNA viruses. However, through explorations such as bright-field or dark-field imaging, digital ELISA is expected to achieve smartphone-based reading methods (65). Particularly, it can be applied to potentially integrate digital reading content with cutting-edge technologies, such as artificial intelligence (AI), machine learning (ML), the Internet, and the Internet of Things (IoT). By integrating AI and ML into diagnostics, we can achieve unprecedented analysis speed and accuracy. For example, these technologies can screen large amounts of patient data, identify patterns and trends that may indicate an outbreak, and even predict epidemics before they reach pandemic levels. In addition, the integration of digital ELISA with the Internet and IoT can revolutionize remote healthcare. Virus detection can be performed in real time, and data can be transmitted immediately to healthcare providers regardless of their geographical location. Thus, response times can be remarkably accelerated, and more targeted interventions can be implemented, especially in rural or underserved areas. However, these processes must be built on robust privacy and cybersecurity measures to protect sensitive health data.
Although the development of digital ELISA faces numerous challenges, its potential advantages are remarkable. This technology may be used to technically integrate nucleic acid testing and immunoassay, thereby transforming the paradigm of RNA virus detection and improving speed, accuracy, and diagnostic range relative to those of current methods. The exceptional sensitivity and specificity of digital ELISA can enhance the early detection of viruses; consequently, therapeutic responses will be faster and more targeted, considerably mitigating the spread and severity of diseases. Addressing the high mutation rates of RNA viruses, digital ELISA has a rapid detection capability and can serve as an efficient and effective tool, thereby identifying and possibly preventing the spread of emergent and recurrent infections. Therefore, the advancement of digital ELISA technology promises to revolutionize RNA virus detection.
ZW: Writing – original draft. PW: Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Science and Technology Support Program of Guizhou Province ([2020]4Y191).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Markiewicz L, Drazkowska K, Sikorski PJ. Tricks and threats of RNA viruses–towards understanding the fate of viral RNA. RNA Biol (2021) 18(3):1–19. doi: 10.1080/15476286.2021.1875680
2. Alvarezmunoz S, Upeguiporras N, Gomez AP, Ramirez-Nieto G. Key factors that enable the pandemic potential of RNA viruses and inter-species transmission: A systematic review. Viruses (2021) 13(4):537. doi: 10.3390/v13040537
3. Pleschka S. Overview of influenza viruses. Curr Top Microbiol Immunol (2013) 370:1–20. doi: 10.1007/82_2012_272
4. Nuwarda RF, Alharbi AA, Kayser V. An overview of influenza viruses and vaccines. Vaccines (2021) 9(9):1032. doi: 10.3390/vaccines9091032
5. Furuyama W, Marzi A. Ebola virus: Pathogenesis and countermeasure development. Annu Rev Virol (2019) 6(1):435–58. doi: 10.1146/annurev-virology-092818-015708
6. Tomori O, Kolawole MO. Ebola virus disease: Current vaccine solutions. Curr Opin Immunol (2021) 71:27–33. doi: 10.1016/j.coi.2021.03.008
7. Hu B, Guo H, Zhou P, Shi ZL. Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol (2021) 19(3):141–54. doi: 10.1038/s41579-020-00459-7
8. Jackson CB, Farzan M, Chen B, Choe H. Mechanisms of SARS-CoV-2 entry into cells. Nat Rev Mol Cell Biol (2022) 23(1):3–20. doi: 10.1038/s41580-021-00418-x
9. Khan J, Asoom L, Khan M, Chakrabartty I, Dandoti S, Rudrapal M, et al. Evolution of RNA viruses from SARS to SARS-CoV-2 and diagnostic techniques for COVID-19: A review. Beni Suef Univ J Basic Appl Sci (2021) 10(1):60. doi: 10.1186/s43088-021-00150-7
10. Cella LN, Blackstock D, Yates MA, Mulchandani A, Chen W. Detection of RNA viruses: Current technologies and future perspectives. Crit Rev Eukaryot Gene Expr (2013) 23(2):125–37. doi: 10.1615/critreveukaryotgeneexpr.2013006974
11. Zhang Y, Noji H. Digital bioassays: Theory, applications, and perspectives. Anal Chem (2017) 89(1):92–101. doi: 10.1021/acs.analchem.6b04290
12. Noji H, Minagawa Y, Ueno H. Enzyme-based digital bioassay technology–key strategies and future perspectives. Lab Chip (2022) 22(17):3092–109. doi: 10.1039/d2lc00223j
13. Malpica JoséM, Fraile A, Moreno I, Obies CI, Drake JW, García-Arenal F. The rate and character of spontaneous mutation in an RNA virus. Genetics (2003) 162(4):1505–11. doi: 10.1093/genetics/162.4.1505
14. Ferris MT, Joyce P, Burch CL. High frequency of mutations that expand the host range of an RNA virus. Genetics (2007) 176(2):1013–22. doi: 10.1534/genetics.106.064634
15. Lam TY, Zhu H, Guan Y, Holmes EC. Genomic analysis of the emergence, evolution, and spread of human respiratory RNA viruses. Annu Rev Genomics Hum Genet (2016) 17(1):193. doi: 10.1146/annurev-genom-083115-022628
16. Xiao Y, Rouzine IM, Bianco S, Acevedo A, Goldstein EF, Farkov M, et al. RNA recombination enhances adaptability and is required for virus spread and virulence. Cell Host Microbe (2016) 19(4):493–503. doi: 10.1016/j.chom.2016.03.009
17. Woolhouse MEJ, Brierley L. Epidemiological characteristics of human-infective RNA viruses. Sci Data (2018) 5:180017. doi: 10.1038/sdata.2018.17
18. Woolhouse ME, Brierley L, McCaffery C, Lycett S. Assessing the epidemic potential of RNA and DNA viruses. Emerg Infect Dis (2016) 22(12):2037–44. doi: 10.3201/eid2212.160123
19. Banerjee S, Guedj J, Ribeiro RM, Moses M, Perelson AS. Estimating biologically relevant parameters under uncertainty for experimental within-host murine West Nile virus infection. J R Soc Interface (2016) 13(117):20160130. doi: 10.1098/rsif.2016.0130
20. Warger J, Gaudieri S. On the evolutionary trajectory of SARS-CoV-2: host immunity as a driver of adaptation in RNA viruses. Viruses (2022) 15(1):70. doi: 10.3390/v15010070
21. Enard D, Petrov DA. Ancient RNA virus epidemics through the lens of recent adaptation in human genomes. Philos Trans R Soc Lond B Biol Sci (2020) 375(1812):20190575. doi: 10.1098/rstb.2019.0575
22. Carrasco-Hernandez R, Jácome R, López Vidal Y, Ponce de León S. Are RNA viruses candidate agents for the next global pandemic? A review. ILAR J (2017) 58(3):343–58. doi: 10.1093/ilar/ilx026
24. Yuan X, Yang C, He Q, Chen J, Yu D, Li J, et al. Current and perspective diagnostic techniques for COVID-19. ACS Infect Dis (2020) 6(8):1998–2016. doi: 10.1021/acsinfecdis.0c00365
25. Mizutani T, Endoh D, Okamoto M, Shirato K, Shimizu H, Arita M, et al. Rapid genome sequencing of RNA viruses. Emerg Infect Dis (2007) 13(2):322–4. doi: 10.3201/eid1302.061032
26. Matranga CB, Gladden-Young A, Qu J, Winnicki S, Nosamiefan D, Levin JZ, et al. Unbiased deep sequencing of RNA viruses from clinical samples. J Vis Exp (2016) 113:54117. doi: 10.3791/54117
27. Qian J, Boswell SA, Chidley C, Lu ZX, Pettit ME, Gaudio BL, et al. An enhanced isothermal amplification assay for viral detection. Nat Commun (2020) 11(1):5920. doi: 10.1038/s41467-020-19258-y
28. Biyani R, Sharma K, Kojima K, Biyani M, Sharma V, Kumawat T, et al. Development of robust isothermal RNA amplification assay for lab-free testing of RNA viruses. Sci Rep (2021) 11(1):15997. doi: 10.1038/s41598-021-95411-x
29. Sule WF, Oluwayelu DO. Real-time RT-PCR for COVID-19 diagnosis: challenges and prospects. Pan Afr Med J (2020) 35:121. doi: 10.11604/pamj.supp.2020.35.24258
30. Fernández-Carballo BL, McBeth C, McGuiness I, Kalashnikov M, Baum C, Borrós S, et al. Continuous-flow, microfluidic, qRT-PCR system for RNA virus detection. Anal Bioanal Chem (2018) 410(1):33–43. doi: 10.1007/s00216-017-0689-8
31. Sikkema-Raddatz B, Johansson LF, de Boer EN, Almomani R, Boven LG, van den Berg MP, et al. Targeted next-generation sequencing can replace Sanger sequencing in clinical diagnostics. Hum Mutat (2013) 34(7):1035–42. doi: 10.1002/humu.22332
32. Kong XQ, Wang YJ, Fang ZX, Yang TC, Tong ML. False-positive results of SARS-CoV-2 RT-PCR in oropharyngeal swabs from vaccinators. Front Med (2022) 9:847407. doi: 10.3389/fmed.2022.847407
33. Trinh KTL, Do HDK, Lee NY. Recent advances in molecular and immunological diagnostic platform for virus detection: A review. Biosensors (2023) 13(4):490. doi: 10.3390/bios13040490
34. Ong DSY, Fragkou PC, Schweitzer VA, Chemaly RF, Moschopoulos CD, Skevaki C, et al. How to interpret and use COVID-19 serology and immunology tests. Clin Microbiol Infect (2021) 27(7):981–6. doi: 10.1016/j.cmi.2021.05.001
35. Gan SD, Patel KR. Enzyme immunoassay and enzyme-linked immunosorbent assay. J Invest Dermatol (2013) 133(9):1–3. doi: 10.1038/jid.2013.287
36. Hayrapetyan H, Tran T, Tellez-Corrales E, Madiraju C. Enzyme-linked immunosorbent assay: Types and applications. Methods Mol Biol (2023) 2612:1–17. doi: 10.1007/978-1-0716-2903-1_1
37. Terato K, Do C, Chang J, Waritani T. Preventing further misuse of the ELISA technique and misinterpretation of serological antibody assay data. Vaccine (2016) 34(39):4643–4. doi: 10.1016/j.vaccine.2016.08.007
38. Leirs K, Tewari Kumar P, Decrop D, Pérez-Ruiz E, Leblebici P, Van Kelst B, et al. Bioassay development for ultrasensitive detection of influenza a nucleoprotein using digital ELISA. Anal Chem (2016) 88(17):8450–8. doi: 10.1021/acs.analchem.6b00502
39. Hasegawa T, Shibayama S, Osumi Y, Sentsui H, Kato M. Quantitative performance of digital ELISA for the highly sensitive quantification of viral proteins and influenza virus. Anal Bioanal Chem (2023) 415(10):1897–904. doi: 10.1007/s00216-023-04600-2
40. O’Connell GC, Alder ML, Smothers CG, Still CH, Webel AR, Moore SM. Use of high-sensitivity digital ELISA improves the diagnostic performance of circulating brain-specific proteins for detection of traumatic brain injury during triage. Neurol Res (2020) 42(4):346–53. doi: 10.1080/01616412.2020.1726588
41. Sanders R, Mason DJ, Foy CA, Huggett JF. Evaluation of digital PCR for absolute RNA quantification. PloS One (2013) 8(9):e75296. doi: 10.1371/journal.pone.0075296
42. Ståhlberg A, Håkansson J, Xian X, Semb H, Kubista M. Properties of the reverse transcription reaction in mRNA quantification. Clin Chem (2004) 50(3):509–15. doi: 10.1373/clinchem.2003.026161
43. Basu AS. Digital assays part II: Digital protein and cell assays. SLAS Technol (2017) 22(4):387–405. doi: 10.1177/2472630317705681
44. Banna HA, Das NK, Ojha M, Koirala D. Advances in chaperone-assisted RNA crystallography using synthetic antibodies. BBA Adv (2023) 4:100101. doi: 10.1016/j.bbadva.2023.100101
45. Dey A, Prakash J, Das R, Shelar S, Saini A, Cherian S, et al. Development of a rapid and ultra-sensitive RNA : DNA hybrid immunocapture based biosensor for visual detection of SARS-CoV-2 RNA. PNAS Nexus (2023) 2(3):pgad031. doi: 10.1093/pnasnexus/pgad031
46. Li L, Xu S, Yan H, Li X, Yazd HS, Li X, et al. Nucleic acid aptamers for molecular diagnostics and therapeutics: advances and perspectives. Angew Chem Int Ed Engl (2021) 60(5):2221–31. doi: 10.1002/anie.202003563
47. Park KS. Nucleic acid aptamer-based methods for diagnosis of infections. Biosens Bioelectron (2018) 102:179–88. doi: 10.1016/j.bios.2017.11.028
48. Jamalipour Soufi G, Iravani S, Varma RS. Molecularly imprinted polymers for the detection of viruses: challenges and opportunities. Analyst (2021) 146(10):3087–100. doi: 10.1039/d1an00149c
49. Cui F, Zhou Z, Zhou HS. Molecularly imprinted polymers and surface imprinted polymers based electrochemical biosensor for infectious diseases. Sensors (2020) 20(4):996. doi: 10.3390/s20040996
50. Rissin D, Kan C, Campbell T, Howes S, Fournier D, Linan S, et al. Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nat Biotechnol (2010) 28(6):595–9. doi: 10.1038/nbt.1641
51. Fischer SK, Joyce A, Spengler M, Yang TY, Zhuang Y, Fjording MS, et al. Emerging technologies to increase ligand binding assay sensitivity. AAPS J (2015) 17(1):93–101. doi: 10.1208/s12248-014-9682-8
52. Norman M, Gilboa T, Ogata AF, Maley AM, Cohen L, Busch EL, et al. Ultrasensitive high-resolution profiling of early seroconversion in patients with COVID-19. Nat BioMed Eng (2020) 4(12):1180–7. doi: 10.1038/s41551-020-00611-x
53. Ter-Ovanesyan D, Gilboa T, Lazarovits R, Rosenthal A, Yu X, Li JZ, et al. Ultrasensitive measurement of both SARS-CoV-2 RNA and antibodies from saliva. Anal Chem (2021) 93(13):5365–70. doi: 10.1021/acs.analchem.1c00515
54. Liu C, Xu X, Li B, Situ B, Pan W, Hu Y, et al. Single-exosome-counting immunoassays for cancer diagnostics. Nano Lett (2018) 18(7):4226–32. doi: 10.1021/acs.nanolett.8b01184
55. Tian S, Zhang Z, Chen J, Du M, Li Z, Yang H, et al. Digital analysis with droplet-based microfluidic for the ultrasensitive detection of β-gal and AFP. Talanta (2018) 186:24–8. doi: 10.1016/j.talanta.2018.04.016
56. Akama K, Shirai K, Suzuki S. Droplet-free digital enzyme-linked immunosorbent assay based on a tyramide signal amplification aystem. Anal Chem (2016) 88(14):7123–9. doi: 10.1021/acs.analchem.6b01148
57. Akama K, Noji H. Multiplexed homogeneous digital immunoassay based on single-particle motion analysis. Lab Chip (2020) 20(12):2113–21. doi: 10.1039/d0lc00079e
58. Akama K, Noji H. Multiparameter single-particle motion analysis for homogeneous digital immunoassay. Analyst (2021) 146(4):1303–10. doi: 10.1039/d0an02056g
59. Chen H, Li Z, Zhang L, Sawaya P, Shi J, Wang P. Quantitation of femtomolar-level protein biomarkers using a simple microbubbling digital assay and bright-field smartphone imaging. Angew Chem Int Ed Engl (2019) 58(39):13922–8. doi: 10.1002/anie.201906856
60. Zhang Q, Li J, Pan X, Liu X, Gai H. Low-numerical aperture microscope objective boosted by liquidimmersed dielectric microspheres for quantum dotbased digital immunoassays. Anal Chem (2021) 93(38):12848–53. doi: 10.1021/acs.analchem.1c02709
61. Matson RS. ELISA-based biosensors. Methods Mol Biol (2023) 2612:225–38. doi: 10.1007/978-1-0716-2903-1_16
62. Curtin K, Fike BJ, Binkley B, Godary T, Li P. Recent advances in digital biosensing technology. Biosensors (2022) 12(9):673. doi: 10.3390/bios12090673
63. Lee D, Asmare N, Sarioglu AF. Paper-based multi-well depletion ELISA. Lab Chip (2023) 23(2):225–38. doi: 10.1039/d2lc00960a
64. Antiochia R. Paper-based biosensors: Frontiers in point-of-care detection of COVID-19 disease. Biosensors (2021) 11(4):110. doi: 10.1016/j.bios.2021.113485
65. Zhdanov A, Keefe J, Franco-Waite L, Konnaiyan KR, Pyayt A. Mobile phone based ELISA (MELISA). Biosens Bioelectron (2018) 103:138–42. doi: 10.1016/j.bios.2017.12.033
66. Zhang Q, Zhang X, Li J, Gai H. Nonstochastic protein counting analysis for precision biomarker detection: suppressing poisson noise at ultralow concentration. Anal Chem (2020) 92(1):654–8. doi: 10.1021/acs.analchem.9b04809
67. He L, Tessier DR, Briggs K, Tsangaris M, Charron M, McConnell EM, et al. Digital immunoassay for biomarker concentration quantification using solid-state nanopores. Nat Commun (2021) 12(1):5348. doi: 10.1038/s41467-021-25566-8
68. Farka Z, Mickert MJ, Hlaváček A, Skládal P, Gorris HH. Single molecule upconversion-linked immunosorbent assay with extended dynamic range for the sensitive detection of diagnostic biomarkers. Anal Chem (2017) 89(21):11825–30. doi: 10.1021/acs.analchem.7b03542
69. Mickert MJ, Farka Z, Kostiv U, Hlaváček A, Horák D, Skládal P, et al. Upconversion-linked immunosorbent assay measures subfemtomolar concentrations of prostate-specific antigen through single-molecule counting. Anal Chem (2019) 91(15):9435–41. doi: 10.1021/acs.analchem.9b02872
70. Liu X, Huang C, Zong C, Liang A, Wu Z, Zhang Y, et al. A singlemolecule homogeneous immunoassay by counting spatially”overlapping”two-color quantum dots with wide-field fluorescence microscopy. ACS Sens (2018) 3(12):2644–50. doi: 10.1021/acssensors.8b01092
71. Liu X, Sun Y, Lin X, Pan X, Wu Z, Gai H. Digital duplex homogeneous immunoassay by counting immuno complex labeled with quantum dots. Anal Chem (2021) 93(6):3089–95. doi: 10.1021/acs.analchem.0c04020
Keywords: RNA viruses, nucleic acid testing, immunoassays, digital ELISA, single molecule detection
Citation: Wang Z and Wei P (2024) Shifting the paradigm in RNA virus detection: integrating nucleic acid testing and immunoassays through single-molecule digital ELISA. Front. Immunol. 14:1331981. doi: 10.3389/fimmu.2023.1331981
Received: 02 November 2023; Accepted: 12 December 2023;
Published: 03 January 2024.
Edited by:
Adi Idris, Queensland University of Technology, AustraliaReviewed by:
Lucia Vojtech, University of Washington, United StatesCopyright © 2024 Wang and Wei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pei Wei, d2VpcGVpQHptdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.