
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 11 December 2023
Sec. Parasite Immunology
Volume 14 - 2023 | https://doi.org/10.3389/fimmu.2023.1303446
Introduction: Pre-erythrocytic malaria vaccines hold the promise of inducing sterile protection thereby preventing the morbidity and mortality associated with Plasmodium infection. The main surface antigen of P. falciparum sporozoites, i.e., the circumsporozoite protein (CSP), has been extensively explored as a target of such vaccines with significant success in recent years. Systematic adjuvant selection, refinements of the immunization regimen, and physical properties of the antigen may all contribute to the potential of increasing the efficacy of CSP-based vaccines. Protection appears to be dependent in large part on CSP antibodies. However due to a knowledge gap related to the exact correlates of immunity, there is a critical need to improve our ability to down select candidates preclinically before entering clinical trials including with controlled human malaria infections (CHMI).
Methods: We developed a novel multiplex competition assay based on well-characterized monoclonal antibodies (mAbs) that target crucial epitopes across the CSP molecule. This new tool assesses both, quality and epitope-specific concentrations of vaccine-induced antibodies by measuring their equivalency with a panel of well-characterized, CSP-epitope-specific mAbs.
Results: Applying this method to RTS,S-immune sera from a CHMI trial demonstrated a quantitative epitope-specificity profile of antibody responses that can differentiate between protected vs. nonprotected individuals. Aligning vaccine efficacy with quantitation of the epitope fine specificity results of this equivalency assay reveals the importance of epitope specificity.
Discussion: The newly developed serological equivalence assay will inform future vaccine design and possibly even adjuvant selection. This methodology can be adapted to other antigens and disease models, when a panel of relevant mAbs exists, and could offer a unique tool for comparing and down-selecting vaccine formulations.
Understanding immune correlates of protection can greatly facilitate the development of effective vaccines. Antibodies have been identified as correlates of protection for some vaccines [reviewed in (1)]. The Plasmodium falciparum sporozoite’s Circumsporozoite Protein (CSP) is the target of most pre-erythrocytic malaria vaccines. This includes the vaccine RTS,S/AS01E that is based on a CSP fragment, and the vaccine was recommended for widespread use among children in sub-Saharan Africa and in other regions with moderate to high P. falciparum malaria transmission. This recommendation was based on results from an ongoing pilot implementation program in more than 900,000 children in Ghana, Kenya and Malawi (2–5). In 2022, RTS,S/AS01E (Mosquirix) became the first malaria vaccine to receive prequalification approval from the WHO, which enables UNICEF to purchase the vaccine (6).
Additional CSP-based vaccine candidates have also shown promise in clinical studies, thus bringing the community closer to additional protective malaria vaccine(s) [(7), and Robben et al., manuscript in preparation]. Assessing vaccine efficacy through surrogate immunological parameters is hampered by the lack of confirmed immune correlates of protection mediated by CSP-specific immune responses. Antibody titers, as measured by enzyme linked immunosorbent assays (ELISA), alone have not been able to predict protection (8–10), requiring a multi-faceted assessment of antibody isotypes, functional activity, and avidity (9, 11). Various rodent models have been employed to assess and screen candidate vaccines, and some are undergoing interrogation for correlation with controlled human malaria infection model (CHMI) results (Locke et al., manuscript in preparation). However, the CHMI trial continues to be the best way to assess vaccine efficacy in early, reasonably sized clinical studies, therefore lengthy development time and great cost must be expended before a definitive clinical field efficacy readout can be obtained. The objective of the present study was to develop a serological competition assay, the CSP-based assay for serological quantification and equivalency (CBASQE), that assesses the equivalency of vaccine-induced antibodies in relation to well-characterized monoclonal antibodies against key epitopes of CSP. In this assay, the equivalence, i.e., the ability of CSP vaccine-induced antibodies to successfully compete with well characterized, relevant mAbs for binding to their epitopes, determines epitope specificity, implied avidity, and concentration of antibodies in sera from vaccinees. One of the key features of the assay – and difference from standard assays for evaluating serum samples - is its ability to evaluate both preclinical and clinical samples, unbiased by the use of different, species-specific secondary antibodies (Bolton et al. manuscript in preparation).
Our CBASQE assay was developed using an electro-chemiluminescence immune assay (ECLIA)-based multiplex platform that has unique qualifying features: (1) high sensitivity with exceptionally low inter- and intra-assay variability; (2) wide linear range over 4-5 logs; and (3) suitability for testing closely related antigens without cross reactivity due to antigenic similarity and competition; and (4) a proven suitability for samples from human malaria vaccinees (12–15).
The CBASQE assay was designed as a competition assay using well-defined monoclonal antibodies specific for crucial epitopes within CSP with most having demonstrated functional activity in preclinical models and/or the clinic. To this end, we selected the N-terminus specific mAb 5D5 (16, 17), the junctional region-specific (NPDP) mAb CIS43 (18, 19), the minor repeat-specific (NVDP) mAb L9 (18, 20, 21), the major repeat-specific (NPNA6) mAb 317 (19, 22–24), and the C-terminal mAbs 236 (16) and 369. These important epitopes are linear except for those in the C-terminus. Importantly, no steric inhibition between these mAbs, even with the two C-terminus-specific mAbs, is anticipated nor has it been observed (Bolton et al, manuscript in preparation) allowing for the independent quantitation of these regional responses. Plate antigen selection was based on the reported roles of the various regions of CSP during sporozoite migration to the liver and invasion of host cells: The N-terminus sequence plays a crucial role during the passage of the sporozoite to the liver (25, 26). CSP undergoes cleavage of the N-terminus right before infecting hepatocytes, a process during which the C-terminus is revealed, allowing it to bind to cellular receptors and mediate entry into the host cell (25). The junctional region and the repeat region are critical for successful invasion, at least for some of the Plasmodium species, particularly P. falciparum. The junction-specific mAb CIS43 and minor repeat-specific mAb L9 have recently been tested in clinical trials (NCT04206332 and NCT05019729) for their ability to protect against progression to blood-stage infections after intravenous delivery with subsequent sporozoite CHMI and demonstrated significant efficacy (27, 28). The major repeat region is represented by six NPNA repeats, which is long enough to measure binding of homodimerizing mAbs (19, 24, 29–32), but minimizes cross reactive binding by mAbs CIS43 and L9 (21, 33). The CBASQE assay was used to dissect repeat region-specific antibody responses to determine the importance of junction vs. minor- vs. major repeat-specific antibodies.
The plate antigens representing the CSP C-terminus used in our assay span most of the non-repeat region of the RTS,S vaccine construct (CSP aa 272-389) and harbor many highly polymorphic amino acid positions presumably selected by immune pressure present in the field. A subset of these sequences was observed to be associated with allele-specific vaccine protection in a previous study (34). The role of C-terminal antibodies is disputed, especially when assessing the functional activity of mAbs in rodent models (35), but there is mounting evidence from studies of clinical samples suggesting antibodies with C-terminal specificity are indeed a component of protective responses following RTS,S/AS01 vaccination (15, 36–39). Furthermore, previous work from our laboratory has demonstrated that the breadth of C-terminal antibody responses is associated with protection (15). In that study, eight variant C-terminal peptides derived from a database of naturally occurring CSP sequences in an RTS,S/AS01E phase III ancillary genotyping study (34) were tested for reactivity with sera from malaria-naïve RTS,S vaccinees (15). Neafsey et al. had determined the Hamming distance of each of the variant peptides to the RTS,S vaccine’s 3D7 sequence at polymorphic amino acid positions (34). The results of this C-terminal breadth assay suggested the breadth of antibody reactivity is an indicator of a focused response against the conserved portion of the molecule. These responses to the conserved region are likely mediating functional activity. To account for variants present in the field, the assay described here includes the 3D7 sequence of the C-terminus that is represented in many CSP-based vaccines including RTS,S (40) and R21 (41). The H18 peptide represents mid-Hamming distant divergence [Hamming distance = 5], and the H50 represents the most divergent variant [Hamming distance = 10] from the 3D7 sequence.
To demonstrate the power of the novel CBASQE assay in discerning qualitative differences and assessing antibody equivalency, we tested a panel of clinical samples from an RTS,S trial (42) with known protective status of the vaccinees (15, 42). With these data, we established serological profiles of protective responses in humans induced by RTS,S vaccination and can assess potential contributing and hierarchical roles of the distinct epitopes in protection. These results emphasize the importance of dissecting minor/major repeat and C-terminal specific immune responses and their role in mediating protection. Systematic comparisons of CBASQE profiles of preclinical, Phase I/IIa and Phase IIb samples will be invaluable when assessing the contributions of region-specific antibodies to protection and guide future vaccine design by indicating the critical epitopes to be included in next-generation CSP-based vaccines designed to induce both high quantity and high-quality vaccine responses. Such an approach could result in greater durability than has been observed with current CSP-based vaccines.
Seven biotinylated PfCSP peptides spanning across the CSP molecule were produced (CS Bio, Menlo Park, CA), Atlantic Peptides Inc, Concord, NH)) and sequences are listed in Table 1. Human monoclonal antibodies (mAb 5D5, CIS43, L9, 317, 236, and 369) were tagged using the MSD Gold-Sulfo-Tag NHS- Ester Conjugation Pack (MesoScale Discovery (MSD) Inc, Gaithersburg, MD), per manufacturer’s instructions. The Sulfo-tagged mAbs were used as competitors, i.e., determining the strength of serum antibodies to compete with the epitope-specific mAbs. The fine specificities of these mAbs are listed in Table 1.
The use of serum samples was reviewed by the WRAIR Human Subjects Protection Branch (Protocol WRAIR#2142). Pre-immune and day of challenge sera (two weeks post third immunization from a previously conducted clinical trial (NCT00075049) (42) in which study participants vaccinated with RTS,S adjuvanted with AS01B or AS02A (n =18 protected subjects, n =18 non-protected subjects) were tested. The study enrolled both, male and female healthy U.S. residents ages 18-45 years with no travel history to malaria-endemic countries. Preliminary experiments did not show differences between the two adjuvant cohorts in the level of their antibody responses to the variant peptides (13) nor their avidity to repeat and C-terminal 3D7 peptide. A human CSP-immune serum pool (CSP-AV) and commercial human AB pooled serum were used as positive and negative assay controls, respectively.
The CBASQE assay is based on the Mesoscale platform and uses the 10-spot, U-PLEX format (MSD, Gaithersburg, MD) (Bolton et al, manuscript in preparation, graphic summary of assay in Figure 1). The U-plex format utilizes specific U-plex Linkers for targeted coating of specific spots within the wells of a U-plex plate. Previous work from our lab established that none of the ten available Linkers differ in their efficiency to bind to their specific spot (13).
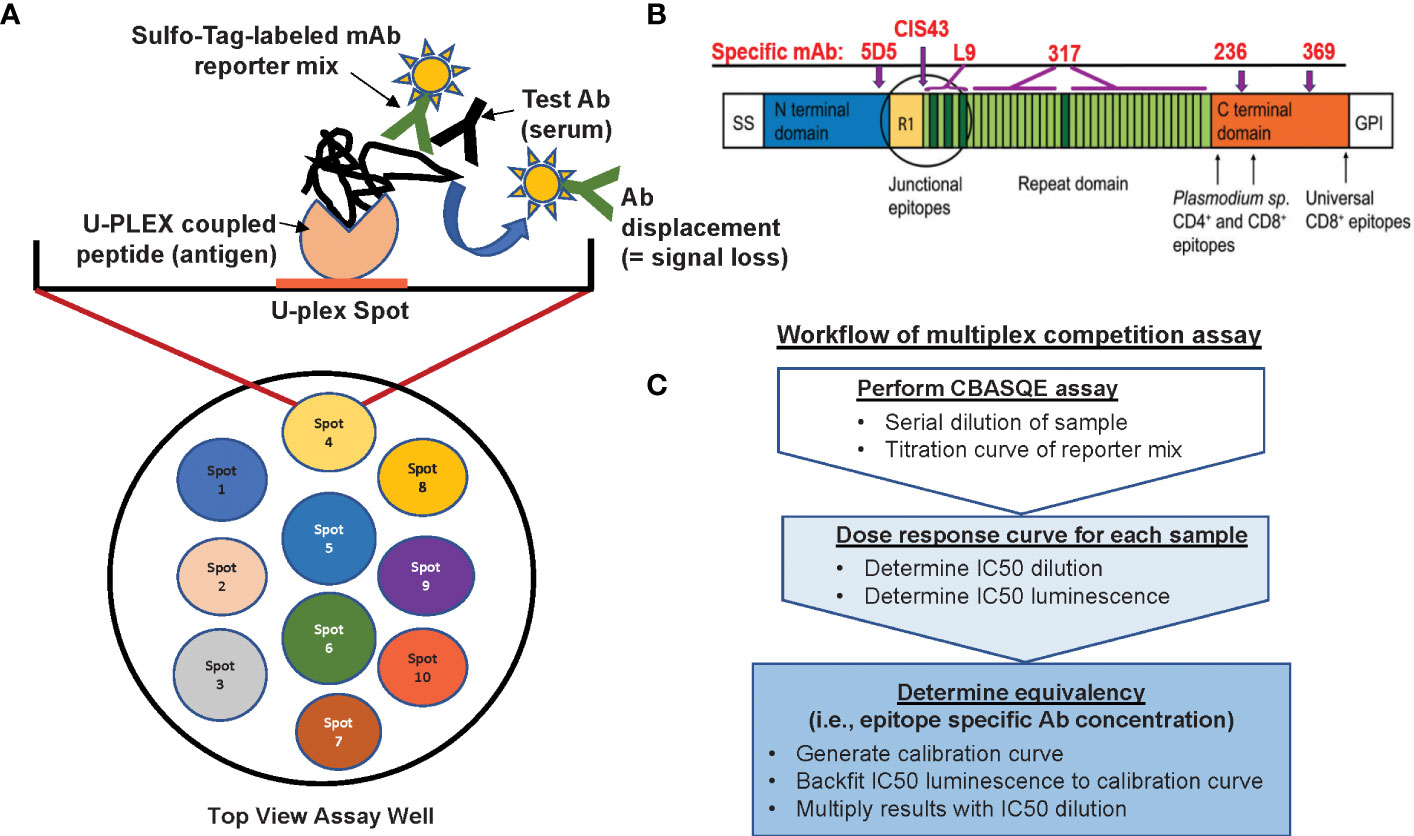
Figure 1 Overview of the CBASQE assay. (A) The 10-spot MSD plate accommodates up to ten different antigens per well (left column). Wells were coated with a mixture containing seven U-plex coupled peptides (spots 4-6 were not used). Serially diluted test serum was mixed with the Sulfo-tag labeled mAb reporter mix and added to the U-plex plates. Loss/reduction in luminescence is seen when serum antibodies successfully competed off the Sulfo-tag labeled reporter mix. (B) Schematic of the CSP protein visualizing the regions represented by the peptides used in this study and where epitope-specific mAbs are binding. (C) Analysis workflow for conducting the serological equivalence assay.
To prepare the assay plates, each biotinylated peptide (Table 1), representing the N-terminus, junctional region, minor repeat, major repeat, and C-terminus, was diluted to 300 nM with the coating diluent (PBS with 0.5% BSA) based on our previous work (13). The peptides were then mixed and incubated at RT with their designated U-plex Linker provided in the MSD U-plex assay kit for 30 mins. MSD Stop Solution (MSD) was added to the peptides/Linker solution (U-PLEX coupled solution) and incubated at RT for another 30 min. After incubation, the individual U-PLEX-coupled solutions were combined into one tube, creating a multiplex coating solution. The 10-spot U-plex plates were then coated with the multiplex coating solution containing the various CSP peptides and incubated at RT for 1hr. After incubation, the U-plex plates were washed with 1x MSD Wash Buffer and used immediately.
During the incubation of the U-PLEX multiplex coating solution, samples were prepared. The reference sample was created by combining unlabeled (no Sulfo-tag) mAb containing, 4.5 ng/mL N-terminus specific mAb 5D5, 1.6 ng/mL junctional peptide-specific mAb CIS43, 14 ng/mL minor repeat-specific mAb L9, 14 ng/mL NPNA6-specific mAb 317, and 4.6 ng/mL mAb236 and 4.6 ng/mL mAb 369 for C-term mAbs. This reference sample was tested at eight, 3-fold dilutions (from neat-1:2,187). Test sera were diluted three-fold spanning four dilutions from 1:3 to 1:81.
In a 96 well plate (Corning, Glendale, AZ), the sera samples and the reference sample were combined with the Sulfo-tag reporter mix containing 25 ng/mL mAb 5D5, 25 ng/mL mAb CIS43, 60 ng/mL mAb L9, 25ng/mL mAb 317, 16 ng/mL mAb 236, and 32ng/mL mAb 369 at a constant concentration, then transferred to the antigen-coated U-plex plate, and incubated at RT for 1hr.
After incubation, the plates were washed three times with 1x MSD Wash Buffer. MSD Read Buffer B was added to each well and the plates were immediately read on the MESO QuickPlex SQ 120 (MSD), per the manufacturer’s instructions. Raw data are reported as mean luminescence signal (MLS).
Test sera were diluted three-fold (1:3 to 1:81) to generate a dose response curve based on the percentage inhibition. The dose response curve of each sample is entered into the Quest Graph™ IC50 Calculator (https://www.aatbio.com/tools/ic50-calculator). This calculator models an experimental data set based on a four-parameter logistic regression model and provides the dilution and the luminescence signal corresponding to the IC50. Testing eight different concentrations of the reference/reporter mix in each experiment was done to generate a standard curve for (a) the conversion of the IC50 into absolute concentrations of epitope-specific antibodies in the sample, and (b) the monitoring of the assay performance from experiment to experiment. The computational manipulations (i.e., generating the standard curve and calculating the antibody concentrations) were performed using the Standard Curve Analysis app (OriginLab Inc, Northampton, MA). For the conversion of the IC50 to a concentration of epitope-specific serum antibodies, the luminescence signal is backfitted to the standard curve. The calculated IC50 concentration is then multiplied with the dilution factor to determine the absolute concentration of epitope-specific antibody (equivalency) in the sample.
Univariate analysis between protected and unprotected subjects was performed to determine the correlation between MSD intensity (reactivity) to the various peptides and protection status. Prior to any t-test, we carried out a Shapiro-Wilks test to determine if the to-be-compared data points were normally distributed. If both were normally distributed (p < 0.05 by the Shapiro-Wilks test), we applied a two-sided Student’s t-test. If either distribution was not normally distributed, we applied the Wilcoxon signed-rank test. Correlations and correlation matrices (to determine the relationship between the various epitope-specificities) were calculated and plotted using R Studio (Version 2023.6.1 + 524). R scripts are available upon request.
Establishing the serological profile of malaria-naïve, RTS,S-vaccinated subjects and stratifying the results based on protective status revealed unique roles of repeat- and C-terminal antibodies in protection (Figure 2). Based on the vaccine design, we assume that the responses to the junctional sequences are due to the presence of NANP/NVDP in the junctional peptide (KQPADGNPDPNANPNVDP). Interestingly, the magnitude in reactivity to the minor repeat peptide was not statistically different between protected vs. non-protected individuals. In contrast, protected individuals had significantly stronger major repeat-specific (NPNA)6 and C-terminal antibody responses compared to non-protected individuals. Also noteworthy was the fact that C-terminal antibodies of protected vaccinees demonstrated more cross-reactivity to the H18 and H50 peptides, which agrees with prior observations (15). No responses to the N-terminus were detected confirming the specificity of the assay results since this region is not included in the RTS,S vaccine.
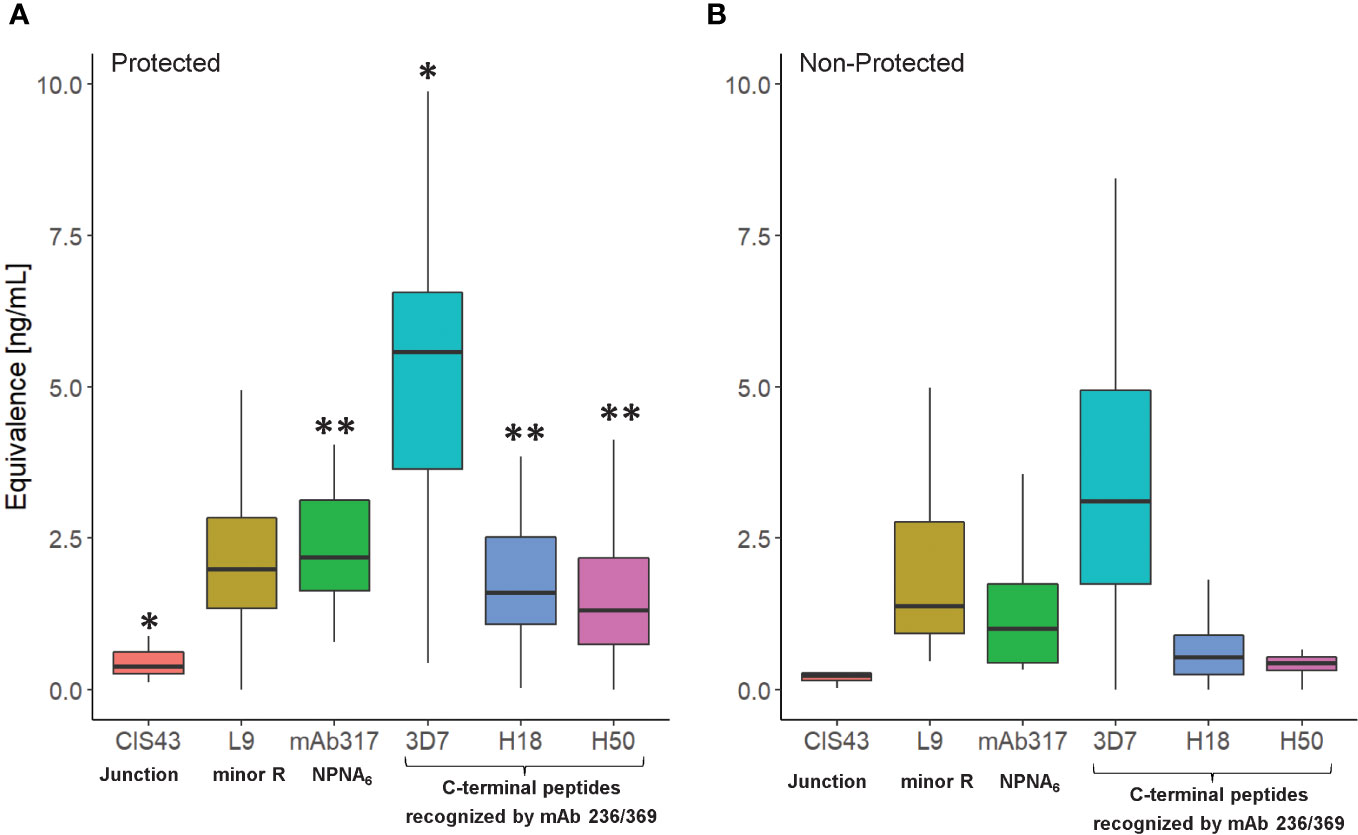
Figure 2 Serological profile of RTS,S vaccinees (NCT00075049) as determined in the CBASQE assay. Data expressed as ng/ml epitope-specific antibody concentration in sera of protected (n=18, (A)) or non-protected (n=18, (B)) RTS,S vaccinees. Equivalency was determined using mAbs CIS43, 317, 236, and 369. Results for N-terminus omitted since no reactivity of the sera was measured. Asterisks indicate statistical significance between protected vs. non-protected RTS,S vaccinees (two-sample T-test, *p< 0.05, **< 0.005).
Correlation matrices were generated to explore the interplay between the concentrations of epitope-specific antibodies in sera of protected vs. non-protected RTS,S vaccinees (Figure 3). In protected individuals, significant correlations were observed between the C-terminal specific antibodies (3D7, H18, H50) (Figure 3A). Weaker correlations were observed for repeat-specific antibodies with only mAbs L9 and CIS43 correlating significantly. Notably, the magnitude of responses to the C-terminal peptides correlated strongly with the magnitude to the junctional region (CIS43 epitope). Taken together, the data suggest that a broad immune response to various epitopes on CSP is associated with protection. One contributing factor to the stronger correlations in protected individuals can also be a higher quality, i.e., antibodies with higher affinity and/or avidity. High quality antibodies will be more successful in competing with the reporter mAbs than lower quality antibodies. In contrast, the breadth of responses to the various epitopes on CSP in non-protected individuals is limited (Figure 3B) to correlations between the C-terminal responses as well as between mAbs CIS43 and L9-epitope specific responses.
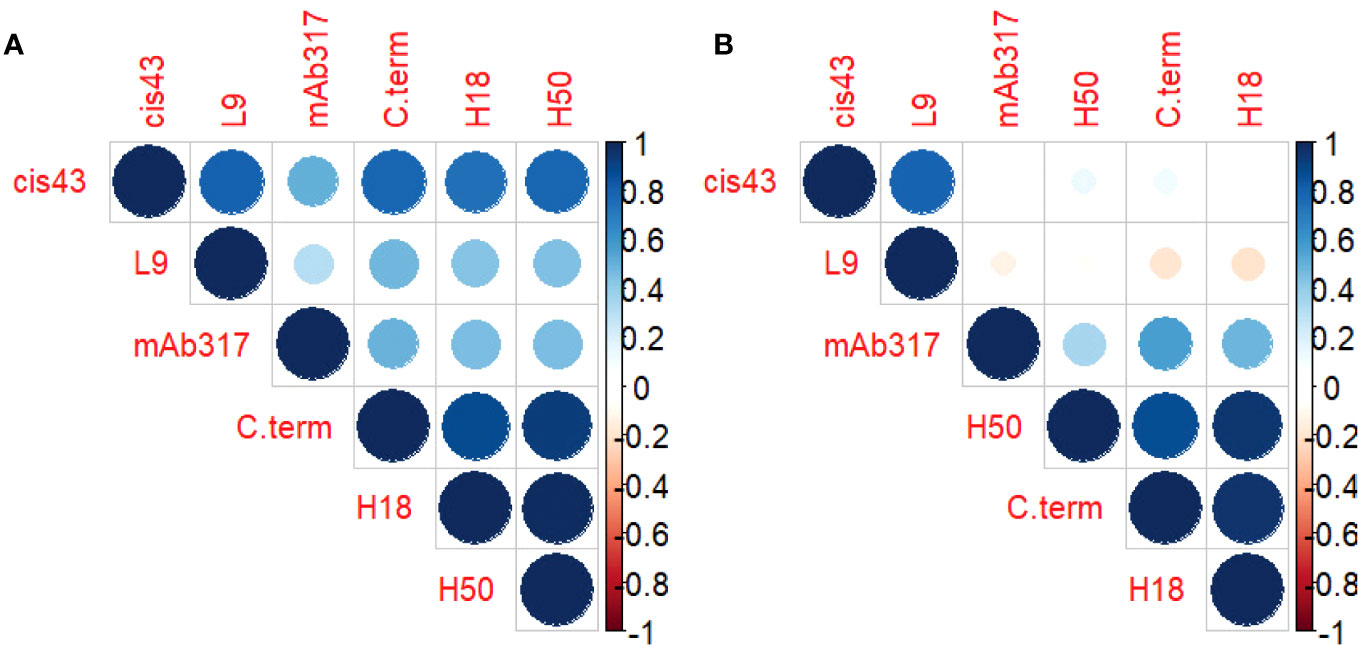
Figure 3 Antibody profile of protected vs. non-protected RTS,S immunized vaccinees to CSP regions. Correlation matrices indicate the relationship between the equivalencies of the antibody responses to the various CSP epitope specific mAbs. The color and size of the dots (scale below graphs) indicate the degree of correlation between the different CSP peptides (small to large indicating low to high correlation). Correlation matrices stratified by protective status ((A) = protected, (B) = non-protected individuals).
Next, we compared the CBASQE data with previously generated ELISA data assessing total IgG, using the same plate antigens for human RTS,S vaccinees (42) to determine the level of agreement between the readout methods. First, we determined the correlation between ELISA and CBASQE on the entire sample set stratified by protective status (Table 2).
Overall, the correlations between the data sets are strong when analyzing the data of all subjects (r ≥ 0.73) even though the CBASQE assay detects only selected epitopes across the CSP, i.e., epitopes of the respective mAbs used in the CBASQE assay. Stratifying the data based on protective status, however, revealed there is a qualitative difference between the serological profiles of protected vs. non-protected individuals. The degree of correlation was lower in non-protected individuals for both the repeat and the C-terminal region. This difference was visualized by generating scatterplots for each of the plate antigens (Figure 4).
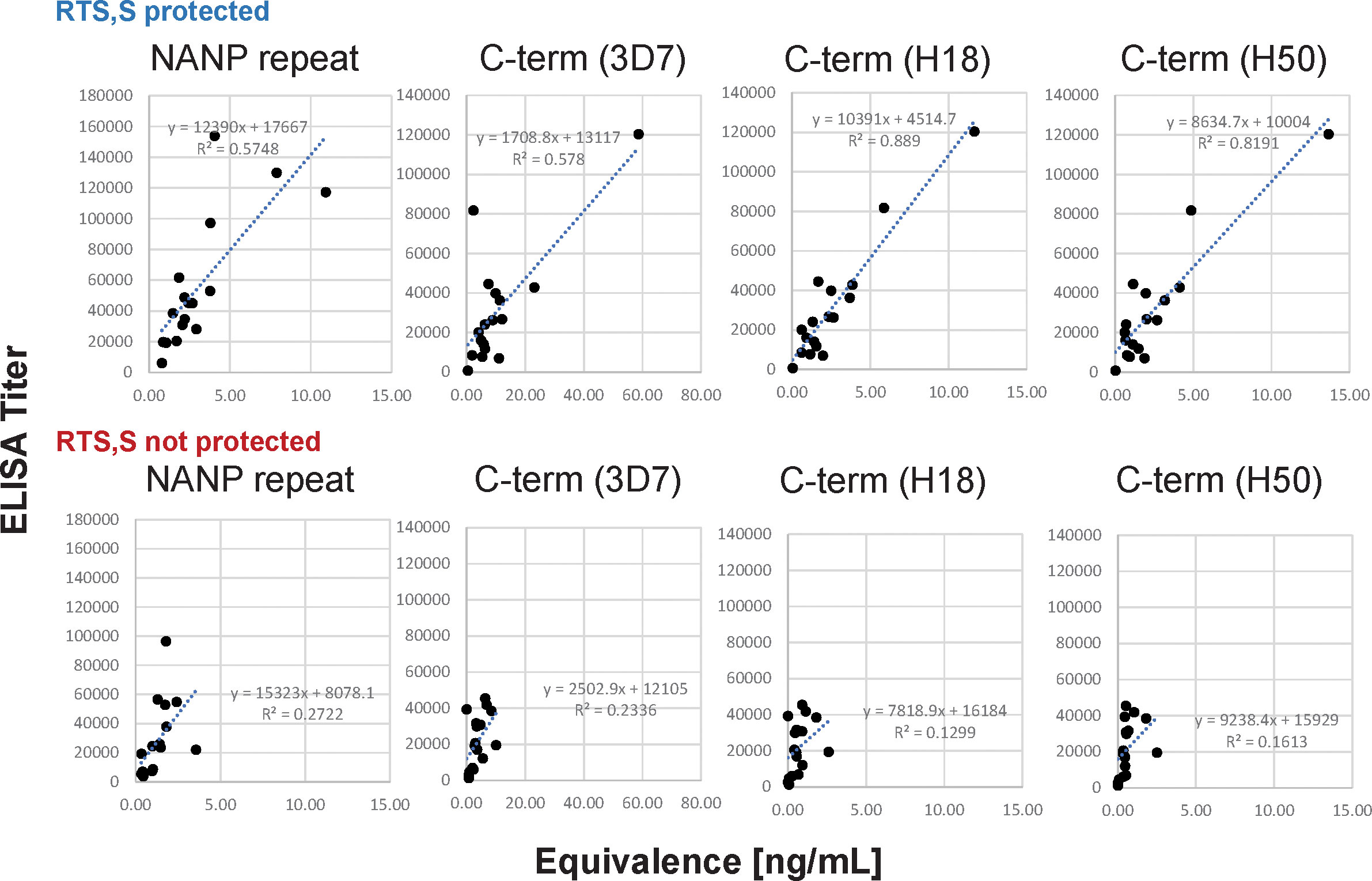
Figure 4 Agreement between ELISA- and CBASQE (Equivalence) results dependent on protective status. Log-transformed ELISA titers (y-axis) and Equivalence (x-axis) results for repeat-specific and C-terminal responses were stratified by protective status: responses of sera from protected (n=18, top panels) and non-protected individuals (n=18, bottom panels).
Despite decades of rational vaccine design for CSP-based candidates, there is still a major knowledge gap when it comes to immune mechanisms mediating protection against P. falciparum infection whether from naturally acquired or vaccine-induced immunity. Sporozoite CSP has been extensively studied as a major target of vaccine development but there is much still unknown regarding critical epitopes of the CSP molecule leading to highly active, durable vaccine responses. The newly developed CBASQE assay simultaneously establishes a serological profile of antigen specificities across the CSP vaccine antigen and measures the quality of the response by determining the concentration of antibodies with equivalence to functionally-active monoclonal antibodies. The CBASQE assay seeks to narrow this knowledge gap and inform next generation vaccine design. Our data demonstrate the following capabilities of the assay:
(a) Sensitive and highly robust assay platform enables the assessment of small volume samples: The high-throughput and high-dimensional assessment of sera is conducive to testing samples from large clinical trials which provide statistical sample sizes adequate for determining the functional role of specific epitopes.
(b) Establishing functional serological profiles of clinical samples (Figures 2, 3): This is invaluable as immunodominance may be different across species and may have an impact on the ability to induce antibodies that have functional activity either in vitro or in vivo. Conventional serological assays such as ELISA and bead-based flow cytometry (e.g., Luminex) used to determine the quality and quantity of antibodies rely on species-specific secondary antibodies. Therefore, direct inter-species comparisons are confounded by the use of different, species-specific secondary reagents. This also hampers inter-laboratory comparisons as different reagents such as different lots of the same secondary antibody can affect the overall results.
(c) Superiority of the CBASQE assay over established ELISA-based methods for assessing the magnitude of serological responses by using CSP-monoclonal, repeat-specific mAbs (43): Our novel assay can assess serological responses in a species-independent manner and report the results as mass concentration. The only serological assay reported to date measures the magnitude of antibody responses to the repeat region and also failed to detect correlation with protection (43).
(d) The CBASQE assay measures the absolute concentration of epitope-specific antibodies in serum/plasma samples and simultaneously infers the binding strength (avidity) of these antibodies by competition with potent mAbs: The distinct feature of the CBASQE assay becomes apparent when comparing the data with ELISA results (Figure 4). ELISA titers to the central repeat region have been reported as an immune correlate of protection (8, 44). The CBASQE assay shows a significantly higher concentration of major repeat-directed antibodies in protected subjects (Figure 2), but antibodies to the minor repeats were not significantly higher in that cohort. The correlation between ELISA and CBASQE results is strong in protected individuals (r ≥ 0.74) but lower for non-protected individuals suggesting more qualitative differences in the serological response. Qualitative differences between protected and non-protected vaccinees become even more apparent when comparing ELISA vs. CBASQE results for C-terminal antibody responses as further discussed below and when comparing the profile (ie., when establishing the breadth of the response to epitopes across the entire CSP molecule) (Figure 2).
The overall agreement between ELISA and CBASQE results is only observed in protected RTS,S vaccinees (Table 2; Figure 4). One possible explanation is the impact of avidity on the CBASQE results. The assay measures epitope-specific antibodies that are capable of successfully competing with specific mAbs. Successful competition of test sera with the epitope specific mAbs is a function of antibody concentration and avidity/affinity. Low-avidity antibodies may not be able to compete with the high-affinity/avidity mAbs in the CBASQE assay but will generate titers in an ELISA assay. Previous work on data integration of serological parameters revealed antibody avidity as one of the hallmarks of a protective response (9).
One of the key differences between ELISA and CASQE assay is the need by ELISAs for secondary reagents that can bias the readout due to their isotype- and subclass-specificity. The CBASQE assay is isotype/subclass agnostic and, therefore, provides a global assessment of the serological response.
We would like to note that the CBASQE assay reports lower concentrations of antigen-specific antibodies in sera compared to quantitative ELISAs. This difference is the result of several factors: (i) most plate antigens present more than one epitope. For example, using the C-terminal peptide Pf16 (45) in ELISA assays results in the binding of a multitude of different antibody clones. In the case of the CBASQE assay, only the binding to mAb 236- and mAb 369-specific epitopes is measured. Therefore, reporting the data as such and distinguishing between a multi-epitope assessment (ELISA) vs. a mono- or oligo-epitope assessment (CBASQE) is imperative. Measuring epitope-specific responses rather than the global response to a plate antigen may reveal clear associations between epitope and protection. Another noteworthy aspect is the fact that CBASQE results do not only reflect the magnitude of the epitope specific response, but also include a qualitative assessment (avidity/affinity high enough to compete with the reference mAb).
This present study is the first of its kind in that it applies a competition multiplex assay to assess equivalence of serological responses to known benchmark mAbs (i.e., mAbs shown to target critical epitopes and/or mediate functional activity against the pathogen). Here, we used six mAbs specific for distinct epitopes across the CSP molecule. The limited number of mAbs could be seen as a limitation of the assay. When and if new epitopes involved in protection or immune escape are identified (e.g., Friedman-Klabanoff et al, manuscript in preparation), both the peptides representing the respective epitopes and the mAbs can be readily added to the assay. Another limitation of any serological assay seeking to determine correlation of the antibody responses with protection is the fact that the contribution of cellular responses to protection are de-emphasized, apart from the impact of helper T cells on the isotype profile of the antibody response.
The present study addresses the controversial role of antibodies to the CSP’s C-term by revealing that vaccine-induced C-term responses are associated with protection. We had previously reported on a multiplex assay for determining the breadth of C-terminal antibody responses (15). All of the samples for that study were obtained from malaria-naïve RTS,S vaccinees, and the cross-reactivity to other C-terminal variant peptides, defined as the breadth, correlated with protection. In that study, we concluded that the breadth of C-term binding is a surrogate marker for reactivity to conserved epitopes within the C-terminus. Here, we confirm, using a complementary approach, that breadth as measured by the concentration of antibodies that can cross-react with a subset of C-term variants represented by H18 and H50 is also associated with protection (Figures 2, 3). We acknowledge that this assessment was made with the same set of clinical samples. Future studies are planned to assess the generalizability of this observation in other RTS,S as well as other CSP-based vaccine candidate trials.
The objective of this study was to establish a new assay platform that addresses shortcomings of commonly-used readout methods for establishing serological profiles. We used a sample set from an RTS,S clinical trial where participants were challenged (CHMI) after immunization to validate the CBASQE approach. The next step is to apply this new assay to additional, larger CHMI sample sets to further support these findings, and to monitor the development and maturation of protective responses during the course of vaccination. Ultimately, the CBASQE assay will need to be applied to field samples to establish functional serological profiles in residents of malaria-endemic regions (46, 47) as compared to profiles induced by vaccination of malaria-naïve individuals and with various vaccine formulations.
Lastly, this concept of measuring the equivalence of antibody responses in comparison to mAbs with known therapeutic/protective activity or even sera from protected/immune individuals provides a high-throughput readout to decipher functionally relevant serological responses responsible for protection. These data are invaluable for next-generation vaccine design and down-selection of vaccine formulations.
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
The studies involving humans were approved by WRAIR internal review board/human subject protection branch reviewed and approved the original trial as well as this exempt human use protocol governing this study. The studies were conducted in accordance with the local legislation and institutional requirements. The human samples used in this study were acquired from primarily isolated as part of your previous study for which ethical approval was obtained. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and institutional requirements.
JB: Methodology, Writing – original draft, Investigation, Validation. RM: Investigation, Resources, Writing – review & editing, Visualization. EL: Resources, Writing – review & editing, Investigation, Visualization. JR: Resources, Writing – review & editing, Funding acquisition, Investigation. EB-L: Funding acquisition, Resources, Writing – review & editing, Conceptualization, Data curation, Formal Analysis, Methodology, Visualization, Writing – original draft, Supervision.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by PATH and the Military Infectious Disease Research Program (MIDRP).
EB-L and JR are government employees. Title 17 U.S.C. § 105 provides that “Copyright protection under this title is not available for any work of the United States Government, but the United States Government”. Title 17 U.S.C. § 101 defines US Government work as “work prepared by a military service member or employee of the US Government as part of that person’s official duties”. The Malaria-027 clinical trial (NCT00075049) was sponsored by the US Army Medical Research and Development Command in collaboration with Walter Reed Army Institute of Research (WRAIR) and GlaxoSmithKline Biologicals SA (GSK). GSK had the opportunity to review the publication. The authors are solely responsible for final content and interpretation. The authors would like to thank Dr. Lorraine Soisson and Dr. C. Richter King for continued scientific discussions and Ms. Tanisha Robinson for technical assistance.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Material has been reviewed by the Walter Reed Army Institute of Research. There is no objection to its presentation and/or publication. The opinions or assertions contained herein are the private views of the authors, and are not to be construed as official, or as reflecting the views of the Department of the Army or the Department of Defense. The investigators have adhered to the policies for protection of human subjects as prescribed in AR 70-25. This paper has been approved for public release with unlimited distribution.
1. Plotkin SA. Updates on immunologic correlates of vaccine-induced protection. Vaccine (2020) 38(9):2250–7. doi: 10.1016/j.vaccine.2019.10.046
2. Kelleher K, Drysdale C. Who Recommends Groundbreaking Malaria Vaccine for Children at Risk (2019). World Health Organization. Available at: https://www.who.int/news/item/06-10-2021 (Accessed March 16, 2023).
3. Chandramohan D, Zongo I, Sagara I, Cairns M, Yerbanga RS, Diarra M, et al. Seasonal malaria vaccination with or without seasonal malaria chemoprevention. New Engl J Med (2021) 385(11):1005–17. doi: 10.1056/NEJMoa2026330
4. RTS SCTP. Efficacy and safety of rts,S/as01 malaria vaccine with or without a booster dose in infants and children in Africa: final results of a phase 3, individually randomised, controlled trial. Lancet (2015) 386(9988):31–45. doi: 10.1016/s0140-6736(15)60721-8
5. Cairns M, Barry A, Zongo I, Sagara I, Yerbanga SR, Diarra M, et al. The duration of protection against clinical malaria provided by the combination of seasonal rts,S/as01(E) vaccination and seasonal malaria chemoprevention versus either intervention given alone. BMC Med (2022) 20(1):352. doi: 10.1186/s12916-022-02536-5
6. WHO. World malaria report, 2022. Geneva/Switzerland: WHO (2022). Available at: https://www.who.int/teams/global-malaria-programme/reports/world-malaria-report-2022.
7. Datoo MS, Natama MH, Somé A, Traoré O, Rouamba T, Bellamy D, et al. Efficacy of a low-dose candidate malaria vaccine, R21 in adjuvant matrix-M, with seasonal administration to children in Burkina Faso: A randomised controlled trial. Lancet (2021) 397(10287):1809–18. doi: 10.1016/s0140-6736(21)00943-0
8. Chaudhury S, Ockenhouse CF, Regules JA, Dutta S, Wallqvist A, Jongert E, et al. The biological function of antibodies induced by the rts,S/as01 malaria vaccine candidate is determined by their fine specificity. Malar J (2016) 15:301. doi: 10.1186/s12936-016-1348-9
9. Chaudhury S, Regules JA, Darko CA, Dutta S, Wallqvist A, Waters NC, et al. Delayed fractional dose regimen of the rts,S/as01 malaria vaccine candidate enhances an igg4 response that inhibits serum opsonophagocytosis. Sci Rep (2017) 7(1):7998. doi: 10.1038/s41598-017-08526-5
10. Regules JA, Cicatelli SB, Bennett JW, Paolino KM, Twomey PS, Moon JE, et al. Fractional third and fourth dose of rts,S/as01 malaria candidate vaccine: A phase 2a controlled human malaria parasite infection and immunogenicity study. J Infect Dis (2016) 214(5):762–71. doi: 10.1093/infdis/jiw237
11. Young WC, Carpp LN, Chaudhury S, Regules JA, Bergmann-Leitner ES, Ockenhouse C, et al. Comprehensive data integration approach to assess immune responses and correlates of rts,S/as01-mediated protection from malaria infection in controlled human malaria infection trials. Front Big Data (2021) 4:672460. doi: 10.3389/fdata.2021.672460
12. Bolton JS, Chaudhury S, Dutta S, Gregory S, Locke E, Pierson T, et al. Comparison of elisa with electro-chemiluminescence technology for the qualitative and quantitative assessment of serological responses to vaccination. Malar J (2020) 19(1):159. doi: 10.1186/s12936-020-03225-5
13. Bolton J, Chaudhury S, MacGill RS, Early AM, King CR, Locke E, et al. Multiplex serological assay for establishing serological profiles of polymorphic, closely related peptide antigens. MethodsX (2021) 8:101345. doi: 10.1016/j.mex.2021.101345
14. Chaudhury S, Bolton JS, Eller LA, Robb M, Ake J, Ngauy V, et al. Assessing Prevalence and Transmission Rates of Malaria through Simultaneous Profiling of Antibody Responses against Plasmodium and Anopheles Antigens. J Clin Med (2022) 11(7):1839. doi: 10.3390/jcm11071839
15. Chaudhury S, MacGill RS, Early AM, Bolton JS, King CR, Locke E, et al. Breadth of humoral immune responses to the C-terminus of the circumsporozoite protein is associated with protective efficacy induced by the rts,S malaria vaccine. Vaccine (2021) 39(6):968–75. doi: 10.1016/j.vaccine.2020.12.055
16. Beutler N, Pholcharee T, Oyen D, Flores-Garcia Y, MacGill RS, Garcia E, et al. A novel CSP C-terminal epitope targeted by an antibody with protective activity against plasmodium falciparum. PloS Pathog (2022) 18(3):e1010409. doi: 10.1371/journal.ppat.1010409
17. Thai E, Costa G, Weyrich A, Murugan R, Oyen D, Flores-Garcia Y, et al. A high-affinity antibody against the csp N-terminal domain lacks plasmodium falciparum inhibitory activity. J Exp Med (2020) 217(11):e20200061. doi: 10.1084/jem.20200061
18. Wang LT, Pereira LS, Kiyuka PK, Schön A, Kisalu NK, Vistein R, et al. Protective effects of combining monoclonal antibodies and vaccines against the plasmodium falciparum circumsporozoite protein. PloS Pathog (2021) 17(12):e1010133. doi: 10.1371/journal.ppat.1010133
19. Pholcharee T, Oyen D, Torres JL, Flores-Garcia Y, Martin GM, González-Páez GE, et al. Diverse antibody responses to conserved structural motifs in plasmodium falciparum circumsporozoite protein. J Mol Biol (2020) 432(4):1048–63. doi: 10.1016/j.jmb.2019.12.029
20. Flores-Garcia Y, Wang LT, Park M, Asady B, Idris AH, Kisalu NK, et al. The P. Falciparum CSP repeat region contains three distinct epitopes required for protection by antibodies in vivo. PloS Pathog (2021) 17(11):e1010042. doi: 10.1371/journal.ppat.1010042
21. Wang LT, Pereira LS, Flores-Garcia Y, O’Connor J, Flynn BJ, Schön A, et al. A potent anti-malarial human monoclonal antibody targets circumsporozoite protein minor repeats and neutralizes sporozoites in the liver. Immunity (2020) 53(4):733–44.e8. doi: 10.1016/j.immuni.2020.08.014
22. Pholcharee T, Oyen D, Flores-Garcia Y, Gonzalez-Paez G, Han Z, Williams KL, et al. Structural and biophysical correlation of anti-nanp antibodies with in vivo protection against P. Falciparum. Nat Commun (2021) 12(1):1063. doi: 10.1038/s41467-021-21221-4
23. Oyen D, Torres JL, Wille-Reece U, Ockenhouse CF, Emerling D, Glanville J, et al. Structural basis for antibody recognition of the nanp repeats in plasmodium falciparum circumsporozoite protein. Proc Natl Acad Sci (2017) 114(48):E10438–E45. doi: 10.1073/pnas.1715812114
24. Martin GM, Torres JL, Pholcharee T, Oyen D, Flores-Garcia Y, Gibson G, et al. Affinity-matured homotypic interactions induce spectrum of pfcsp-antibody structures that influence protection from malaria infection. bioRxiv (2022), 2022.09.20.508747. doi: 10.1101/2022.09.20.508747
25. Coppi A, Natarajan R, Pradel G, Bennett BL, James ER, Roggero MA, et al. The malaria circumsporozoite protein has two functional domains, each with distinct roles as sporozoites journey from mosquito to mammalian host. J Exp Med (2011) 208(2):341–56. doi: 10.1084/jem.20101488
26. Aldrich C, Magini A, Emiliani C, Dottorini T, Bistoni F, Crisanti A, et al. Roles of the amino terminal region and repeat region of the plasmodium berghei circumsporozoite protein in parasite infectivity. PloS One (2012) 7(2):e32524. doi: 10.1371/journal.pone.0032524
27. Gaudinski MR, Berkowitz NM, Idris AH, Coates EE, Holman LA, Mendoza F, et al. A monoclonal antibody for malaria prevention. New Engl J Med (2021) 385(9):803–14. doi: 10.1056/NEJMoa2034031
28. Wu RL, Idris AH, Berkowitz NM, Happe M, Gaudinski MR, Buettner C, et al. Low-dose subcutaneous or intravenous monoclonal antibody to prevent malaria. New Engl J Med (2022) 387(5):397–407. doi: 10.1056/NEJMoa2203067
29. Oyen D, Torres JL, Cottrell CA, Richter King C, Wilson IA, Ward AB. Cryo-em structure of P. Falciparum circumsporozoite protein with a vaccine-elicited antibody is stabilized by somatically mutated inter-fab contacts. Sci Adv (2018) 4(10):eaau8529. doi: 10.1126/sciadv.aau8529
30. Pholcharee T, Oyen D, Flores-Garcia Y, Gonzalez-Paez G, Han Z, Williams KL, et al. Structural and biophysical correlation of anti-nanp antibodies with in vivo protection against P. Falciparum. Nat Commun (2021) 12(1):1063. doi: 10.1038/s41467-021-21221-4
31. Murugan R, Scally SW, Costa G, Mustafa G, Thai E, Decker T, et al. Evolution of protective human antibodies against plasmodium falciparum circumsporozoite protein repeat motifs. Nat Med (2020) 26(7):1135–45. doi: 10.1038/s41591-020-0881-9
32. Imkeller K, Scally SW, Bosch A, Martí GP, Costa G, Triller G, et al. Antihomotypic affinity maturation improves human B cell responses against a repetitive epitope. Science (2018) 360(6395):1358–62. doi: 10.1126/science.aar5304
33. Kisalu NK, Idris AH, Weidle C, Flores-Garcia Y, Flynn BJ, Sack BK, et al. A human monoclonal antibody prevents malaria infection by targeting a new site of vulnerability on the parasite. Nat Med (2018) 24(4):408–16. doi: 10.1038/nm.4512
34. Neafsey DE, Juraska M, Bedford T, Benkeser D, Valim C, Griggs A, et al. Genetic diversity and protective efficacy of the rts,S/as01 malaria vaccine. New Engl J Med (2015) 373(21):2025–37. doi: 10.1056/NEJMoa1505819
35. Scally SW, Murugan R, Bosch A, Triller G, Costa G, Mordmuller B, et al. Rare pfcsp C-terminal antibodies induced by live sporozoite vaccination are ineffective against malaria infection. J Exp Med (2018) 215(1):63–75. doi: 10.1084/jem.20170869
36. Ubillos I, Ayestaran A, Nhabomba AJ, Dosoo D, Vidal M, Jimenez A, et al. Baseline exposure, antibody subclass, and hepatitis B response differentially affect malaria protective immunity following rts,S/as01e vaccination in African children. BMC Med (2018) 16(1):197. doi: 10.1186/s12916-018-1186-4
37. Dobano C, Sanz H, Sorgho H, Dosoo D, Mpina M, Ubillos I, et al. Concentration and avidity of antibodies to different circumsporozoite epitopes correlate with rts,S/as01e malaria vaccine efficacy. Nat Commun (2019) 10(1):2174. doi: 10.1038/s41467-019-10195-z
38. Dennison SM, Reichartz M, Abraha M, Spreng RL, Wille-Reece U, Dutta S, et al. Magnitude, specificity, and avidity of sporozoite-specific antibodies associate with protection status and distinguish among rts,S/as01 dose regimens. Open Forum Infect Dis (2020) 8(2). doi: 10.1093/ofid/ofaa644
39. Suscovich TJ, Fallon JK, Das J, Demas AR, Crain J, Linde CH, et al. Mapping functional humoral correlates of protection against malaria challenge following rts,S/as01 vaccination. Sci Transl Med (2020) 12(553). doi: 10.1126/scitranslmed.abb4757
40. Laurens MB. Rts,S/as01 vaccine (Mosquirix): an overview. Hum Vaccin Immunother (2020) 16(3):480–9. doi: 10.1080/21645515.2019.1669415
41. Collins KA, Snaith R, Cottingham MG, Gilbert SC, Hill AVS. Enhancing protective immunity to malaria with a highly immunogenic virus-like particle vaccine. Sci Rep (2017) 7:46621. doi: 10.1038/srep46621
42. Kester KE, Cummings JF, Ofori-Anyinam O, Ockenhouse CF, Krzych U, Morris P, et al. Randomized, double-blind, phase 2a trial of falciparum malaria vaccines rts,S/as01b and rts,S/as02a in malaria-naive adults: safety, efficacy, and immunologic associates of protection. J Infect Dis (2009) 200:337–46. doi: 10.1086/600120
43. Radin K, Clement F, Jongert E, Sterckx YGJ, Ockenhouse C, Regules J, et al. A monoclonal antibody-based immunoassay to measure the antibody response against the repeat region of the circumsporozoite protein of plasmodium falciparum. Malaria J (2016) 15(1):543. doi: 10.1186/s12936-016-1596-8
44. White MT, Bejon P, Olotu A, Griffin JT, Riley EM, Kester KE, et al. The relationship between rts,S vaccine-induced antibodies, cd4(+) T cell responses and protection against plasmodium falciparum infection. PloS One (2013) 8(4):e61395. doi: 10.1371/journal.pone.0061395
45. Schwenk R, DeBot M, Porter M, Nikki J, Rein L, Spaccapelo R, et al. Igg2 Antibodies against a Clinical Grade Plasmodium Falciparum CSP Vaccine Antigen Associate with Protection against Transgenic Sporozoite Challenge in Mice. PloS One (2014) 9(10):e111020. doi: 10.1371/journal.pone.0111020
46. Triller G, Scally SW, Costa G, Pissarev M, Kreschel C, Bosch A, et al. Natural parasite exposure induces protective human anti-malarial antibodies. Immunity (2017) 47(6):1197–209.e10. doi: 10.1016/j.immuni.2017.11.007
Keywords: serology, circumsporozoite protein, antigen-specificity, equivalency, vaccine, protection, malaria
Citation: Bolton JS, MacGill RS, Locke E, Regules JA and Bergmann-Leitner ES (2023) Novel antibody competition binding assay identifies distinct serological profiles associated with protection. Front. Immunol. 14:1303446. doi: 10.3389/fimmu.2023.1303446
Received: 28 September 2023; Accepted: 21 November 2023;
Published: 11 December 2023.
Edited by:
José Roberto Mineo, Federal University of Uberlandia, BrazilReviewed by:
Toshihiro Horii, Osaka University, JapanCopyright © 2023 Bolton, MacGill, Locke, Regules and Bergmann-Leitner. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Elke S. Bergmann-Leitner, ZWxrZS5zLmJlcmdtYW5uLWxlaXRuZXIuY2l2QGhlYWx0aC5taWw=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.