- 1Department of Medical Laboratory, Shenzhen Longhua District Central Hospital, Shenzhen, China
- 2Xianning Medical College, Hubei University of Science and Technology, Xianning, China
Significant advancements have been made in comprehending the interactions between the microbiome and cancer. However, prevailing research predominantly directs its focus toward the gut microbiome, affording limited consideration to the interactions of intratumoral microbiota and tumors. Within the tumor microenvironment (TME), the intratumoral microbiome and its associated products wield regulatory influence, directing the modulation of cancer cell properties and impacting immune system functionality. However, to grasp a more profound insight into the intratumoral microbiota in cancer, further research into its underlying mechanisms is necessary. In this review, we delve into the intricate associations between intratumoral microbiota and cancer, with a specific focus on elucidating the significant contribution of intratumoral microbiota to the onset and advancement of cancer. Notably, we provide a detailed exploration of therapeutic advances facilitated by intratumoral microbiota, offering insights into recent developments in this burgeoning field.
1 Introduction
The presence of numerous microorganisms such as viruses, bacteria, fungi, and other microbes within the human body is vital for human health. These microorganisms exhibit colonization patterns in multiple anatomical sites, encompassing the oral cavity, skin, gastrointestinal tract, respiratory tract, and genitalia. Symbiotic interactions between humans and their microbiome are critical and contribute significantly to human health (1–3). Extensive inquiries into the human microbiome have illuminated variations in the microbial communities among individuals in a state of health and those experiencing pathological conditions. Moreover, the microbiome is closely linked to cancer by influencing the carcinogenesis process in the human body (4). The well-documented link between cancer and specific viruses, such as Epstein-Barr virus and human papillomavirus, underscores their potential to initiate oncogenic activation (5). Oncoviral infections have been shown to promote tumorigenesis by enabling the incorporation of oncogenes within the human genome structure (6, 7).
Research into host-microbial interactions has notably propelled the comprehension of intratumoral microbiota (8, 9). The advancement of detection technologies and enhanced comprehension of the TME have substantiated the presence of intratumoral bacteria. Tumor tissue presents a significantly reduced presence of microbial and fungal biomass when compared to the abundance observed in the gut environment (10, 11). Recent findings point to exclusive bacterial and fungal patterns characteristic of individual tumor types (12, 13). In comparison to normal tissues, tumor tissues manifested a heightened abundance of bacterial and fungal burdens. Remarkably, a substantial enrichment of multiple bacterial strains was observed specifically within tumor tissues. Intratumoral microbial components, distinguished in several tumor types, manifest meaningful correlations with the onset and advancement of cancer (14, 15). Recent studies underscore the fundamental importance of gut microbiota in governing the immune responses. Additionally, it has been demonstrated that the microbiota present within tumors can significantly shape the local immune responses in the TME, potentially affecting tumor progression (16). Within the TME, intratumoral microbiota conspicuously demonstrate anti-tumorigenic manifestations by orchestrating heightened antigen presentation, activating T and NK cells, executing proficient immunosurveillance, and synthesizing metabolites that suppress tumor progression. Conversely, pro-tumorigenic effects are characterized by elevated levels of reactive oxygen species (ROS), the emergence of driver mutations, the inactivation of T cells, and the induction of immunosuppression (3). The intratumoral microbiota manifests varied roles in anti-tumor immunity, with the potential to either enhance or suppress anti-tumor immune responses (17). Consequently, these roles have implications for the effectiveness of immunotherapy (16, 18). In recent years, there has been a surge in research interest delving into the intricate interplay between gut microbiota and the etiology as well as therapeutic responses in cancer. Nonetheless, increasing attention is being paid to intratumoral microbiota (3).
This review presents a thorough analysis of the burgeoning field of intratumoral microbiota research. We delve into its origins, the rich spectrum of its diversity, the intriguing links between intratumoral and gut microbiota, mechanistic involvement in tumorigenesis, and the exciting potential it holds for innovative tumor therapeutics. This review offers promising avenues for developing innovative therapeutic interventions leveraging intratumoral microbiota toward effective tumor management.
2 Intratumoral microbiota: unveiling their features
2.1 Origin of intratumoral microbiota
Despite the significant attention given to intratumoral microbiota, their origins have not been fully elucidated. Recent research has revealed that intratumoral microbiota may arise from distinct sources (Figure 1) (3, 11, 19, 20). The intratumoral microbiota may arise from breaches in mucosal barriers. Intratumoral microbiota is commonly found in cancers originating at mucosal sites, including colorectal, pancreatic, cervical, and lung cancer (21). These organs have externally exposed cavities, and the mucosal destruction that occurs during tumorigenesis can provide a pathway for microorganisms colonizing the mucosa to invade the tumor. Thus, the breach of mucosal barriers, with other factors, may lead to the colonization of microbiota in the TME and facilitate their complex interactions (16, 22). The identified representative bacteria within nasopharyngeal carcinoma tissues exhibit approximately 69% similarity in single-nucleotide variations to bacteria present in the nasopharyngeal microbiota. Subsequently, resemblances are observed with bacteria from the oral cavity (24.1%) and the gut (6.9%). These findings unequivocally establish the nasopharyngeal microbiota as the primary reservoir of intratumoral bacteria within nasopharyngeal carcinoma (23). Although there are abundant microbiomes in human mucosal organs, the idea that intratumoral microbiota can only come from the mucosal site through the mucosal barrier cannot explain all the intratumoral microbiota. A portion of intratumoral bacteria is rare within the mucosal organs of the corresponding tumors, while others are prevalent in non-mucosal origin tumors, such as breast cancer, suggesting other potential sources of intratumoral microbiota (11, 24). Therefore, additional investigation is necessary to clarify the mechanisms that facilitate microbial infiltration from mucosal organs into the TME.
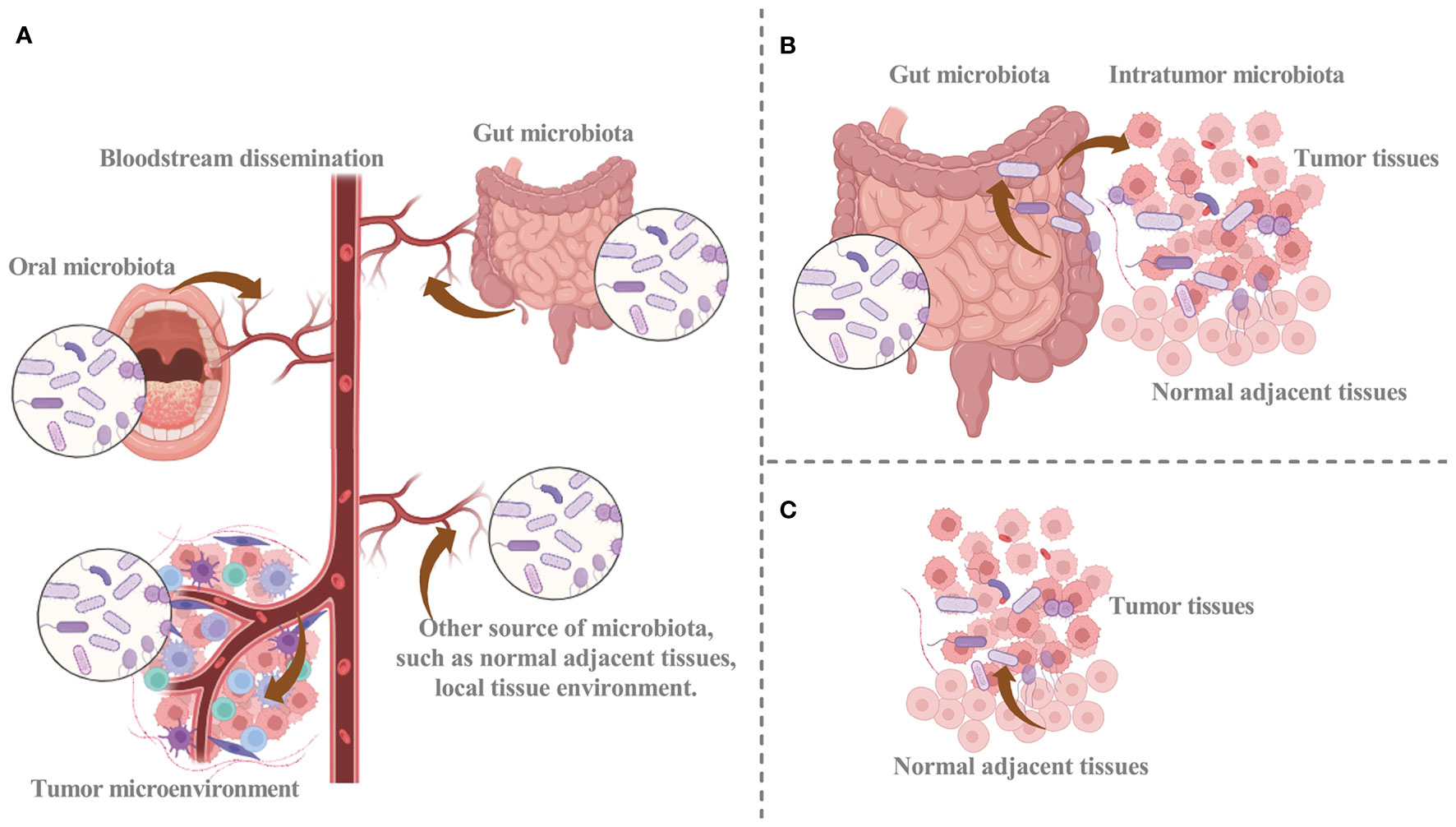
Figure 1 The potential sources of intratumoral microbiota. (A) Hematogenous spread facilitates the infiltration of intratumor microbes from oral, intestinal, and other sources into tumor sites. (B) Microbiota can disrupt the mucosal barrier and infiltrate tumor sites, and intratumoral microbiota of cancer may infiltrate tumor sites via the duct. (C) Normal adjacent tissue may provide a source for intratumor microbiota. Graphics created with BioRender.com.
The circulatory system represents another potential origin for intratumoral microbiota (3, 11). The chemotactic gradient of necrotic cell debris within a tumor is a mechanism that attracts microorganisms from different locations into the blood circulation. Malformed blood vessels provide a conducive setting for intratumoral microbiota to colonize the TME through hematogenous spread (9). Hematogenous spread facilitates the recruitment of microorganisms from various sites, including the oral cavity and intestines, to the tumor site, where they can colonize the tumor via infiltration through impaired blood vessels. The circulatory system, including blood, lymphatic fluid, and the internal passages of the alimentary tract, provides a plausible pathway for the transfer of microbiota. Considering the anatomical interconnectedness of the oral cavity, respiratory tract, and gastrointestinal tract, it is plausible that oral microbiota can easily migrate to these respective anatomical regions. When the oral microbiota undergoes ecological disruption, they may gain entry into the tumor and convert it into intratumoral microbiota (25).
Bacteria from adjacent normal tissues have been found in organs previously believed to lack microbial presence. Moreover, the bacterial composition within tumor tissues closely resembles that of adjacent normal tissues (3, 11, 26). The significant similarity of microbiota composition between tumor microbiota and normal adjacent tissue microbiota can be explained by the origin of normal adjacent tissue microbiota from TME. Within normal adjacent tissues, microorganisms from blood vessels or mucosal organs may infiltrate the TME stimulated by oxygen and chemotactic gradients (11). In addition, microorganisms in normal tissues may originate from the tumor site. Consequently, it is unclear whether normal adjacent tissues serve as a source of intratumoral microbiota, and further substantiation is necessary to elucidate this matter.
As knowledge of the origin and mechanisms of intratumoral microbiota grows, a more comprehensive understanding of intratumoral microbiota may assist in devising more potent therapeutic approaches. Exploring the various sources of intratumoral microbiota, analyzing their composition, and comparing them with the microbiome of other body sites may facilitate the identification of intratumoral microbiota. Furthermore, investigating the molecular mechanisms that underlie the infiltration of microorganisms into the TME is a compelling area of research.
2.2 Diversity of intratumoral microbiota
Given the possibility of multiple origins of intratumoral microbiota, it is plausible to suggest that the microbiome compositions of various cancer types are heterogeneous (15, 27). Within a variety of prevalent cancer types, there are distinct microbial signatures present in tissue and blood samples, each linked to a specific microbiota. Such microbial signatures have been utilized to differentiate healthy individuals from those with cancer, indicating that these signatures may have diagnostic potential (28). The utilization of a rigorous decontamination pipeline in analyzing The Cancer Genome Atlas (TCGA) database at the whole-genome and whole-transcriptome level has allowed for the discovery of unique microbial signatures present in both blood and tumor tissue that was specific to certain cancer types (15, 27). A recent pan-cancer study investigated the presence of cancer-associated fungi in 17,401 samples from 35 distinct cancer types. The findings indicate that fungal DNA and cells exhibit low abundance in several prevalent human cancers, with diverse community compositions across various cancer types. Distinct fungal species and corresponding cellular compositions were associated with specific types of cancer (15). Tumor microbial communities exhibit a predominance of bacteria, with a lower abundance of fungi. The composition of microbial communities in adjacent normal tissues is similar to that of tumor microbial communities. Some microorganisms have been identified in multiple types of tumors, although their abundance can differ depending on the specific cancer type (26).
Intratumoral bacteria possess some common characteristics. Their prevalence within cancerous tissues is significantly lower when compared to that of the gut, with qPCR and imaging quantification indicating that the bacterial presence is discernible in a fraction of cancer cells, varying from 0.1% to 10%. The microbial diversity is generally diminished in cancerous tissue as opposed to normal tissue, suggesting that tumors may foster a distinct milieu that selects for specific bacterial species. The majority of these bacteria are commensal organisms primarily inhabiting the intracellular compartment. The diverse bacterial ecosystems within cancer tissues could potentially contribute to multifunctional mechanisms when interacting with cancerous cells (14, 29).
The microbiota of colorectal cancer has been investigated, with some bacteria like Bacteroides fragilis, Escherichia coli, and Fusobacterium nucleatum frequently detected within tumor tissues. In addition, fungal species, such as Candida albicans, have been detected in some colorectal cancer samples (30–32). Helicobacter pylori, a bacterium responsible for chronic gastritis and peptic ulcers, is linked to the heightened risk of developing gastric cancer. Furthermore, some bacterial species like Streptococcus anginosus and Lactobacillus have been identified in some gastric cancer samples (33, 34). A pan-cancer analysis of the mycobiome across various anatomical locations revealed the presence of tumor-associated fungi and a significant abundance of Candida in gastrointestinal malignancies. Mycobiome communities in gastrointestinal tumors exhibit a high prevalence of Cyberlindnera jadinii, Saccharomyces cerevisiae, and Candida species. Blastomyces species are prevalent within pulmonary carcinomas, while Malassezia species are abundant within mammary tumors (13). Fusobacterium nucleatum, associated with colorectal tumors, also exhibited a higher prevalence in pancreatic and breast malignancies. Microbial compositions vary distinctly across different subtypes of tumors. For instance, multiple bacterial taxa exhibited distinct prevalence when comparing various subtypes of breast cancer, characterized by their human epidermal growth factor receptor 2 (HER2), estrogen receptor (ER), and progesterone receptor (PR) status. Granulicatella_Unknown species31 (species) and Dyadobacter (genus) exhibit enrichment in HER2+ breast cancer patients. Corynebacterium (genus) demonstrates enrichment in ER- breast cancer patients, while Actinomycetaceae (family), Sphingomonas_Unknown species124 (species), Streptophyta_Unknown genus116 (genus), Lautropia_Unknown species38 (species), and Actinomyces odontolyticus (species) manifest enrichment in ER+ breast cancer patients. Actinobacteria (class) displays enrichment in non-triple negative breast cancer. Conversely, Achromobacter denitrificans (species), Bacillus_Unknown species21 (species), Leptotrichia_Unknown species21 (species), Streptophyta_Unknown genus116 (genus), Nocardiopsaceae (family), and Achromobacter (genus) are enriched in triple-negative breast cancer. Moreover, breast tumors exhibited a heightened bacterial abundance in comparison to normal adjacent tissue (14).
2.3 The association between intratumoral and gut microbiota
The current research landscape is witnessing a surge in studies exploring the correlation between intratumoral and gut microbiota. Specific bacterial species within the gut microbiota have the potential to infiltrate the intestinal mucosa, enter the bloodstream, and inhabit neoplastic lesions, thus shaping the composition of the microbiota within tumors. The gut microbiota-tumor interplay has emerged as a critical factor influencing the onset and advancement of diverse forms of cancer. In glioma, intratumoral bacteria can originate not only from the gut microbiota but also from the oral cavity or adjacent brain tissue. Glioma-induced shifts in the local microenvironment, involving the disruption of the blood-brain barrier and immunosuppression, create conducive conditions for bacterial infiltration via either hematogenous or neuronal retrograde pathways. It is plausible that these bacteria existed in the brain tissue before tumorigenesis, with those adapting to the TME demonstrating growth throughout tumor development (35). Nevertheless, the precise mechanisms by which gut bacteria contribute to the intratumoral microbiota remain not completely elucidated and warrant emphasis (3).
The TME is subject to regulatory influences from both intratumoral and gut microbiota, involving modulation of immune responses and modification of cancer cell metabolism (16). Modulation of the TME is achievable through gut microbiota-mediated regulation of intestinal epithelial barrier components, resulting in the activation of lymphoid organs. The gut microbiota may mediate its impact on the TME via metabolites or the immune system, thereby potentially altering the activities of the microbiota within the tumor (24, 36).
Comparable to the gut microbiota, the intratumoral microbiota exhibits the potential to modulate host immune responses. The gut microbiota intricately shapes the effectiveness of immune checkpoint blockade and the ensuing immune responses against tumors (37). Diverse interactions among intratumoral microbiota can trigger unique immune responses, suggesting a potential interplay with gut microbiota (15). Further investigation is warranted to clarify the interplay between intratumoral and gut microbiota.
3 Mechanistic insights into tumorigenesis and intratumoral microbiota
Intratumoral bacteria can regulate cancer cell-intrinsic properties, such as mechanical stress, stem cell flexibility, epithelial-mesenchymal transition (EMT), and adhesion to endothelial cells, which can detrimentally impact the behavior of tumor cells in circulation. Intratumoral bacteria can regulate the extrinsic cancer milieu by releasing exosomes, thereby fostering metastasis, facilitating the breach of the vascular barriers for remote organ colonization, and contributing to the creation of a specialized premetastatic niche. Furthermore, they orchestrate the modulation of the adaptive and innate immune systems, ultimately dictating the resultant immune reaction (38). The intricate interplay between the intratumoral microbiota and cancer manifests in a multifaceted manner, exerting varied influences on cancer progression (Figure 2). These include promoting cancer growth and spread through increased mutagenesis, epigenetic modifications, modulation of oncogenes or oncogenic pathways, inflammation initiation, and immune response alteration (31, 38–40).
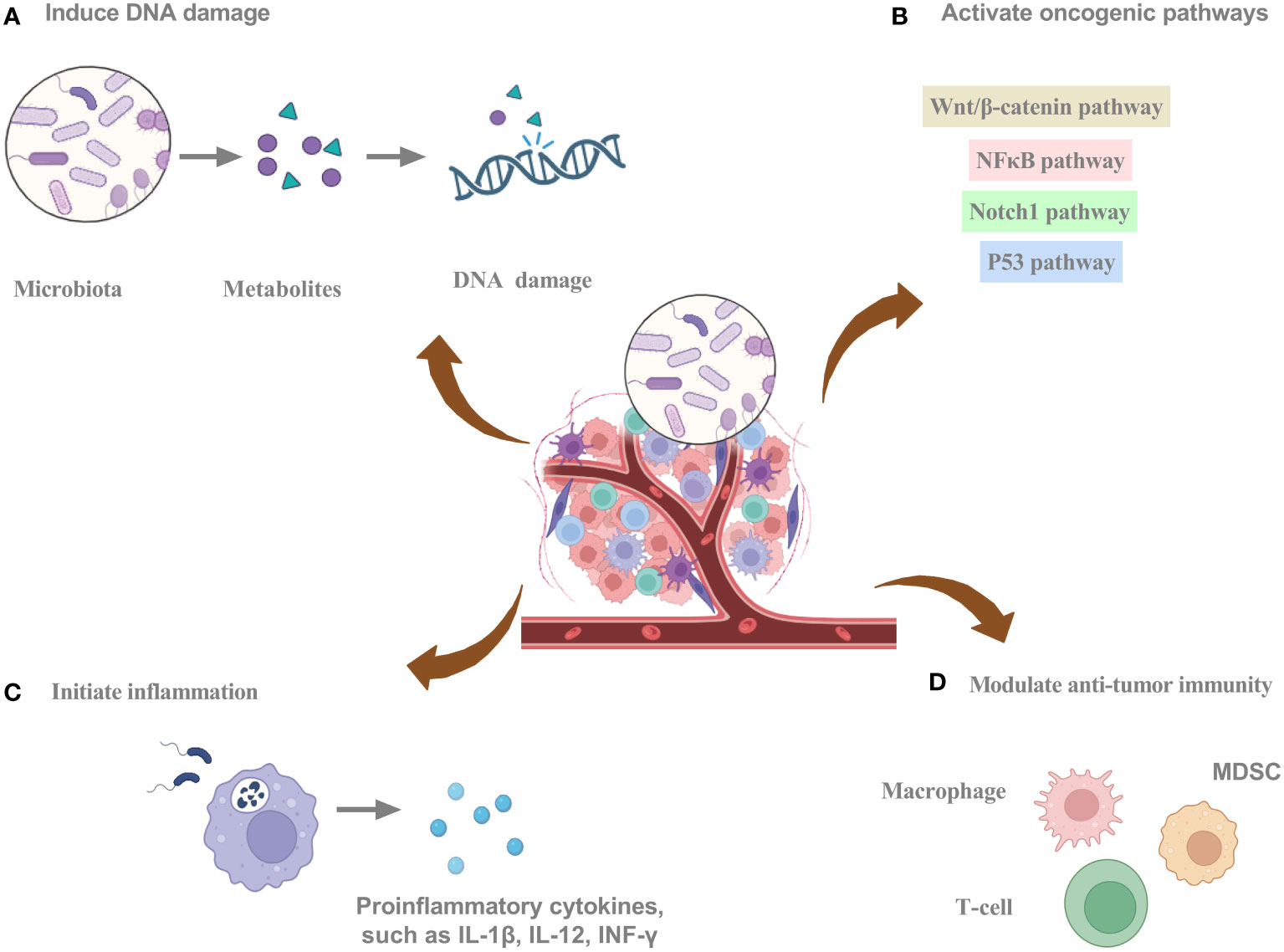
Figure 2 The potential mechanisms of intratumoral microbiota promoting tumorigenesis. (A) Intratumoral microbiota can secrete metabolites to induce DNA damage. (B) Intratumoral microbiota can activate oncogenic pathways. (C) Intratumoral microbiota can initiate inflammation. (D) Intratumoral microbiota can modulate anti-tumor immunity. Graphics created with BioRender.com.
3.1 Induce DNA damage
Several bacterial species have evolved mechanisms to inflict DNA damage, which may instigate mutational events and ultimately promote carcinogenesis (41). Carcinogenic bacteria damage host DNA through a variety of mechanisms involving molecules, proteins, and metabolites. Fragile Bacteroidin exhibits the potential to cause DNA damage, thereby stimulating mutational events (3, 42). Single-cell RNA sequencing enables the identification of bacteria-associated host cells, their interactions, and the dysregulation of transcriptional pathways related to DNA damage repair, cell cycle, and the p53 signaling pathway (9). The production of colibactin by polyketide synthetase (pks)+ Escherichia coli can lead to DNA alkylation, provoking DNA damage and facilitating colorectal cancer progression (43). The pathogenic bacteria that adhere to the intestinal epithelium can induce episodes of diarrhea. The type 3 secretion system (T3SS) of these bacterial pathogens plays a crucial role in their interactions with intestinal epithelial cells, through which they can deliver genotoxin-UshA that damages the DNA of the host cells, contributing to the development of carcinogenesis (44). The involvement of microbes in instigating DNA damage through mutational processes is apparent. The mechanisms currently under consideration include Escherichia coli-mediated colibactin crosslinking, generating genotoxicity, and Helicobacter pylori-mediated aberrant cytidine expression. The exploration of mutational signatures through bioinformatics has opened the door to comprehending the processes underlying genomic alterations that drive oncogenesis. Microbes can elicit DNA damage that impacts the structure of the cancer genome, resulting in alterations to mutational spectra and mutational signatures (42). Additionally, the microbiota can convert numerous dietary metabolites into agents that damage DNA, and under conditions of dysbiosis, certain bacteria can produce toxins that cause DNA damage (3, 9, 45).
3.2 Activate carcinogenic pathways
Intratumor microbiota and their metabolites can influence signaling pathways that contribute to oncogenesis. Fusobacterium nucleatum has been implicated in the modulation of pathways and their associated molecules, exerting an influence on the landscape of pancreatic tumor development (46, 47). Through a Fap2-dependent pathway, Fusobacterium nucleatum engages with pancreatic cancer cells, inducing cytokine production. Through autocrine and paracrine pathways, cytokines stimulate cancer cell proliferation and enhance migration, ultimately propelling the evolution of the malignancy (47). Infections by bacteria lead to a substantial augmentation of signaling pathways, notably TNF, inflammatory responses, and hypoxia pathways. Furthermore, this fosters cancer cell progression through EMT and activation of the p53 pathway (9). Microbial metabolites can modulate signaling pathways such as transcription factor nuclear factor κB (NFκB) and Wnt/β‐catenin in tumor cells, thereby affecting tumor progression (3). In colorectal cancer, Fusobacterium nucleatum is recognized for its ability to trigger the initiation of the E-cadherin/β-catenin signaling cascades via FadA. This initiation eventuates in DNA damage, stimulation of cell growth, and augmentation of chk2 expression (48). CagA, a protein synthesized by Helicobacter pylori, can enter the host cell cytoplasm, triggering β-catenin signaling cascades, ultimately promoting the onset of gastric cancer (49). The involvement of Enterotoxigenic Bacteroides fragilis in breast cancer initiation is evident through both intraductal and intestinal colonization, emphasizing local and distant impacts. Elicitation of oncogenic effects by the Bacteroides fragilis toxin is potentially linked to the stimulation of the β-catenin and Notch1 signaling cascades (50).
3.3 Initiate inflammation
Chronic inflammation can elevate the likelihood of developing particular forms of cancer by activating inflammatory mediators and signaling cascades that promote tumor cell survival, proliferation, and invasion. Inflammatory mediators like ROS, cytokines, chemokines, and nitrogen species can facilitate tumor progression by fostering angiogenesis, elevating growth factor synthesis, and provoking the proliferation of cancerous cells (51, 52). Intratumoral bacteria can aggravate the inflammatory response, leading to the exacerbation of the disease (53). Intratumoral bacteria interacting with pattern recognition receptors (PRRs) can activate inflammatory pathways. Intratumoral bacteria may activate PRRs, leading to the secretion of cytokines and chemokines, the facilitation of angiogenesis, and immune cell recruitment (54, 55). An increased presence of Fusobacterium within tissues of head and neck squamous cell cancer has been linked to heightened inflammation and a less favorable prognosis. Moreover, complex interactions between competitive endogenous RNA networks and chromatin accessibility promote the development of microbiome-related inflammatory TME (56). Fusobacterium nucleatum can initiate the toll-like receptor 4 (TLR4)-mediated signaling cascade, which activates downstream signaling pathways and NFκB, leading to the induction of genes related to inflammation and the immune response (57). An elevated prevalence of Enterobacteriaceae is linked to heightened inflammatory activity, possibly attributed to their metabolizing inflammatory byproducts as an energy source (58). The secretion of virulence factors by Escherichia coli exacerbates the inflammatory response (59). The interplay between chronic inflammation and intratumoral bacteria requires further investigation.
3.4 Modulate anti-tumor immunity
Intratumoral microbiota can impact TME through several mechanisms, thus playing a role in tumorigenesis and cancer treatment (Table 1). Bacterial-induced modifications within the TME play a pivotal role in immunotherapy (69). Microbes within the TME elicit recognition by immune and cancer cells by presenting microbial antigens on their cell surfaces, stimulating an immune response and activating immune cells against the tumor (70). Moreover, some microbial antigens display structural resemblance to tumor antigens, activating immune cells that recognize these shared antigens. Consequently, the immune response triggered against microbial antigens can also target tumor cells expressing analogous antigens (71). In addition, some microbes in the TME can trigger immunogenic cell death, characterized by danger signal release and immune system activation, resulting in proinflammatory molecule secretion and tumor antigen presentation, facilitating an immune response against tumor cells (72). Furthermore, microbial component-mediated activation of PRRs boosts the immune response against tumors, eliciting the liberation of proinflammatory cytokines and heightened stimulation of immune cell activity (73, 74). Moreover, microbial-derived metabolites in the TME exert immunomodulatory effects by impacting immune cell behavior and remodeling the TME (75). Additionally, certain microbes in the TME can activate inhibitory checkpoints, diminish immune cell activity, and attenuate the anti-tumor immune response (72). Stimulated by intratumoral microbiota, the initiation of interleukin-17 production is triggered, fostering the infiltration of B cells into the complex microenvironment of tumor tissues. This intricately coordinated response emerges as a substantial factor in contributing to the progression of colon cancer. Within the milieu of colon cancer, polymorphonuclear neutrophils, recognized as highly abundant immune cells, have the potential to ameliorate microbial dysbiosis in colon cancer tissues. This is manifested by a decrease in tumor-associated Akkermansia and a concurrent increase in the prevalence of Proteobacteria (76). Within microsatellite instability-high colorectal cancers, the Fusobacterium nucleatum-enriched subset exhibits heightened tumor invasion. Furthermore, specific features within the immune microenvironment become evident, highlighting a significant reduction in FoxP3+ T cells spanning the entire tumor and a notable increase in the proportion of M2-polarized macrophages positioned within the tumor (77).
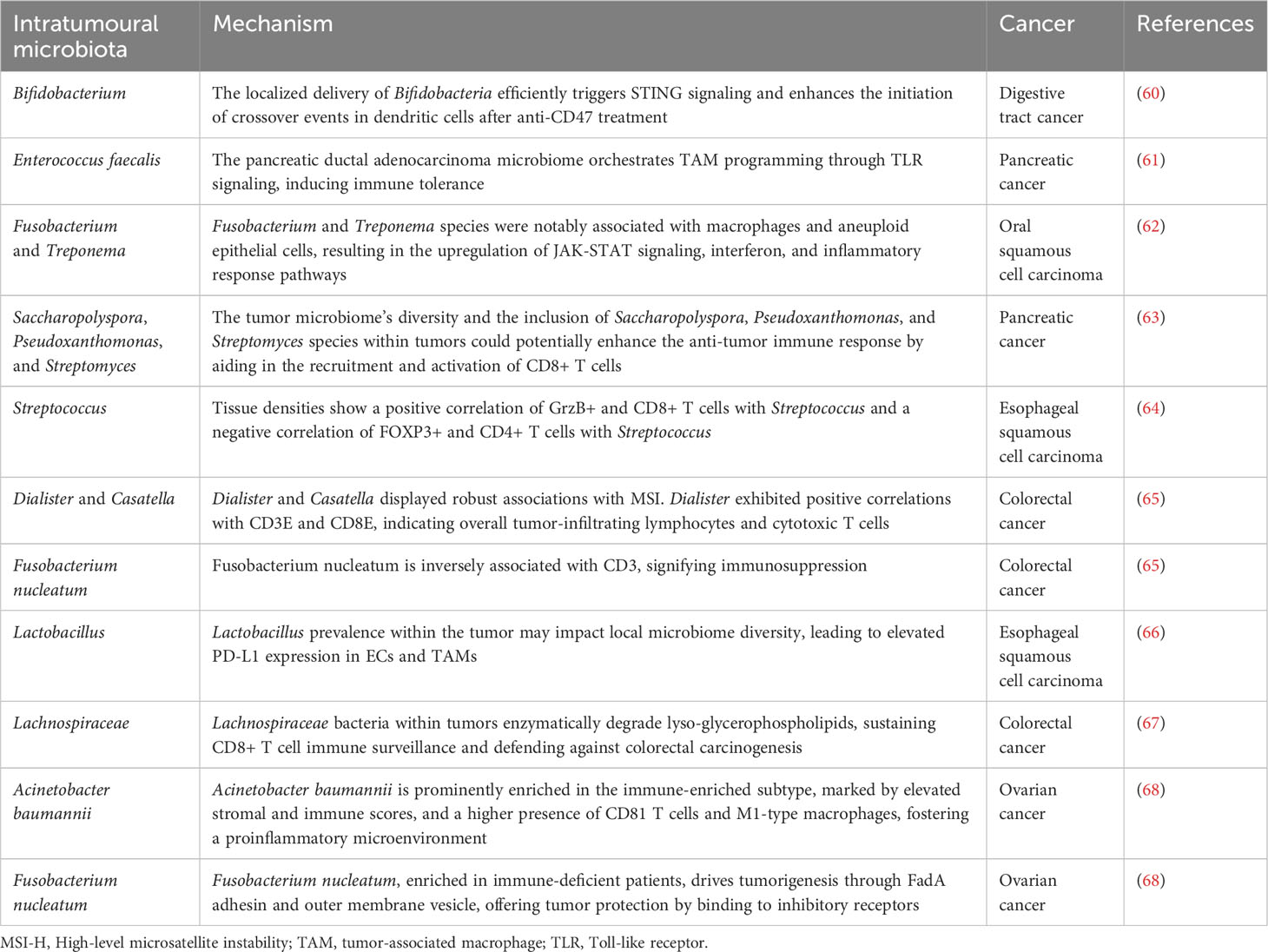
Table 1 Functional roles of intratumoral microbiota in the modulation of the tumor microenvironment.
The microbiota may exert a significant impact on an immunosuppressive TME in pancreatic ductal adenocarcinoma (78). By translocating to the pancreas, the gut microbiome can initiate the formation of a TME exhibiting immunosuppressive, promoting tumorigenesis and metastatic spread, consequently impairing the potency of modulators targeting immune checkpoints (78). The increase of immune cells with immunosuppressive properties, such as myeloid-derived suppressor cells (MDSCs), regulatory T cells (Tregs), along with cytokines, obstruct TILs from penetrating the tumor site (78, 79). In oral cavity tumors, Fusobacterium nucleatum load exhibited a negative correlation with immune markers. Elevated Fusobacterium nucleatum levels were associated with decreased B lymphocytes, T helper lymphocytes, M2 macrophages, and fibroblasts. In tumors exhibiting a high load of Fusobacterium nucleatum, significant reductions were noted in the expressions of Toll-like receptor (TLR) 4 and OX40 ligand (TNFSF4). Significantly, TNFSF9 receptor (TNFRSF9) expression underwent a marked decrease, mirroring an escalation in its ligand (TNFSF9) expression with the mounting Fusobacterium nucleatum load. Simultaneously, there was a marked elevation in the levels of the pro-inflammatory cytokine IL-1ß (17). The presence of intratumoral microbiota has been identified as a pivotal factor in fostering an immunosuppressive TME by selectively recruiting specific immunosuppressive cellular populations, including Tregs, MDSCs, and TAMs. Consequently, this orchestrated recruitment acts as a deterrent to the efficacious infiltration of TILs (3, 17, 80). The depletion of CD4+ T cells of the Th1 subtype and CD8+ T cells with cytotoxic activity, accompanied by a shift towards Th2 T cells, as well as the shift of tumor-associated macrophages (TAMs) towards the M2 phenotype associated with immunosuppression, are associated with immune suppression and an unfavorable TME (78, 81, 82). The fibrogenic reprogramming of pancreatic ductal adenocarcinoma stellate cells results in a dense fibrotic stroma, impeding the penetration of therapeutic drugs and immune cells into the tumor locale. Furthermore, the activated pancreatic stellate cells recruit immunosuppressive cells, establishing a TME exhibiting immunosuppressive features, thus facilitating tumor growth and dampening effective immune reactions targeting tumors (78, 83).
Some microorganisms can interface with immune cells in the TME, potentially modulating their activity (11, 24). Fusobacterium nucleatum can impede the cytotoxicity exhibited by natural killer (NK) cells against tumors. Fusobacterium nuclei strains inhibit the cytotoxicity of NK cells by engaging with the Fap2 protein, leading to subsequent attachment to the inhibitory receptor TIGIT. Tumors exploit the Fap2 protein derived from Fusobacterium nucleatum to promote immune escape via TIGIT-mediated inhibition of immune cell function (84). Fusobacterium nucleatum can interact with carcinoembryonic antigen-related cell-adhesion molecule 1 (CEACAM1), thereby exerting an inhibitory effect on the function of T and NK cells (85). Commensal microbiota-mediated modulation of γδ T cell functionality impacts immune reactivity. Specifically, the microbiota elicits the activation of T cells, particularly those with the Vγ6+Vδ1+ phenotype, in lung cancer. These γδ T cells facilitate neutrophil penetration and stimulate the growth of tumor cells, thereby influencing the TME and tumor progression (86). Within colorectal carcinoma tissue, an inverse correlation has been observed between the prevalence of Fusobacterium nucleatum and the abundance of CD3+ T-cell count. A reduced CD3+ T-cell density can facilitate tumor progression by decreasing immune surveillance and impairing anti-tumor activity (87).
4 The potential of intratumoral microbiota for tumor therapy
Current research has established the considerable contribution of the microbiome to diverse aspects of cancer, such as oncogenesis, therapeutic response, and drug resistance (41). Strategic alteration of the gut microbiota holds promise for mitigation and management of cancer. However, the therapeutic potential of intratumoral microbiota warrants further investigation (22). Intratumoral microbiota may exert adverse or favorable effects on cancer therapy, depending on the underlying therapeutic mechanism (Figure 3; Table 2) (93). Two principal approaches for microbial-based treatments have progressed to the clinical stage. The first approach employs living or inactivated bacteria to stimulate an immune response via targeting specific antigens. The Bacillus Calmette-Guérin (BCG) vaccine, various bacterial vaccines, and the implementation of live, attenuated, double-deleted Listeria monocytogenes are notable examples of this strategy. The second strategy involves utilizing bacteria as carriers capable of the controlled release of immunostimulants, toxins, and other pharmaceutical agents. Engineered bacteria can elicit an anti-tumor response or serve as carriers for therapeutic applications. Through genetic modifications, engineered bacteria can release products or facilitate specific reactions that impede the progression of tumors. Furthermore, engineered bacteria can function as carriers for the targeted delivery of toxins, immunostimulants, or other therapeutic substances (11).
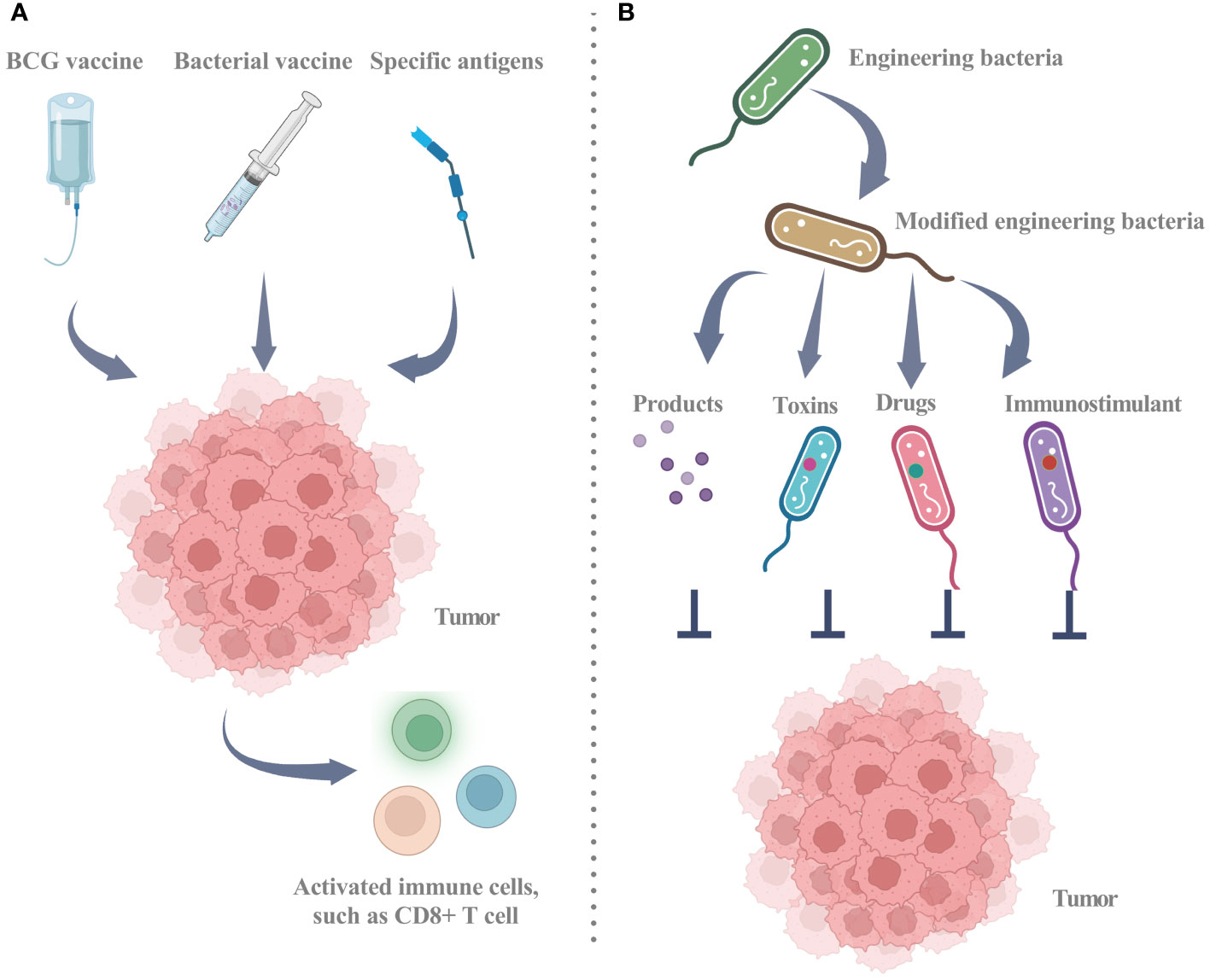
Figure 3 Utilizing intratumor microbiota for clinical treatment strategies. (A) Utilizing biological agents, such as the BCG vaccine and multiple bacterial vaccines, involves the use of either dead or living bacteria to recruit active immune cells, including CD8+ T cells, thereby triggering an anti-tumor immune response. The strategic utilization of specific antigens stands out as a pivotal mechanism to activate the immune system, fostering a heightened and robust CD8+ T cell response against cancer cells. (B) Engineered bacteria as a tool for tumor inhibition through the release of targeted products or reactions and as vehicles for delivering toxins, immunostimulants, or other drugs. Engineered bacteria can be programmatically designed to release targeted products or undergo specific reactions near tumor cells, encompassing toxins for direct cancer cell eradication, anti-angiogenic factors to impede vascular growth within tumors, or other agents impeding tumor progression. Engineered bacteria emerge as promising vehicles for the delivery of therapeutic agents, encompassing toxins, immunostimulants for immune response amplification against cancer, and conventional drugs. This targeted delivery system is designed to heighten treatment specificity and efficacy while mitigating potential harm to healthy tissues. BCG: Bacillus Calmette-Guérin. Graphics created with BioRender.com.
Intratumoral bacteria have been implicated in altering tumor cell responsiveness to chemotherapy. Specific bacterial enzymes have been noted to mediate the metabolic conversion of gemcitabine into an inactive metabolite. The colonization of pancreatic tumors by Gammaproteobacteria has been correlated with their ability to degrade gemcitabine, which subsequently contributes to an enhanced chemoresistance of the tumor (94). In colon cancer, intratumoral Gammaproteobacteria facilitated resistance to gemcitabine through the synthesis of bacterial cytidine deaminase (CDDL) enzyme and was subsequently eradicated through the concurrent administration of ciprofloxacin (92). Analysis of taxonomic distributions revealed higher levels of Gammaproteobacteria in cholangiocarcinoma tumor tissues resistant to low-dose gemcitabine, low-dose cisplatin, and high-dose gemcitabine, while the abundance of Actinobacteria was lower in low-dose gemcitabine and high-dose gemcitabine resistant groups (95). The intratumoral presence of CDDL-expressing bacteria facilitates the metabolism of gemcitabine into 2’2-difluorodeoxyuridine (dFdU), thus preventing the inhibition of DNA replication within malignant cells. The reduction in bacterial-mediated resistance upon depletion of NupC, the transporter for bacterial nucleosides, in bacteria with active CDDL expression, indicates the involvement of NupC in the internalization of gemcitabine by the bacteria (96). Post neoadjuvant chemotherapy, a substantial augmentation of Pseudomonas within breast tumors was witnessed. Moreover, breast malignancies in individuals experiencing distant metastatic spread demonstrated an elevated prevalence of Staphylococcus and Brevundimonas (97). Variations in intratumoral microbiota signatures distinguish responders from non-responders to neoadjuvant chemoimmunotherapy (NACI) in patients with esophageal squamous cell carcinoma. Responders displayed heightened levels of tumor-resident Streptococcus, establishing a positive correlation with the increased infiltration of CD8+ T cells and GrzB+ T cells. Fecal microbial transplantation (FMT) from NACI responders restructured the intratumoral microbiota composition, resulting in Streptococcus enrichment in tumor tissues, increased infiltration of CD8+ T cells, and the promotion of positive results with anti-PD-1 therapy (64).
Intratumoral microbiota may exert both immunostimulatory and immunosuppressive effects on anti-tumor immunity, with the potential to promote the advancement of cancer by inducing processes such as heightened production of ROS, fostering an anti-inflammatory milieu, impairing T cell function, and instigating immunosuppressive responses (3). To elucidate the correlation between a specific intratumor microbial signature and the response to immunotherapy, a comparative analysis of metastatic melanomas was carried out. Examination of distinct microbial taxa profiles in patients, including immune checkpoint inhibitor responders (n=29) and non-responders (n=48), unveiled noteworthy distinctions. There were 18 high-abundance taxa and 28 low-abundance taxa among responders compared with non-responders. Notably, responders showed an increased abundance of Clostridium, whereas non-responders exhibited a higher Gardnerella vaginalis (14). The attenuated vaccine BCG, originating from Mycobacterium bovis, has been implemented in clinical therapies for bladder cancer (98). The efficacy of traditional cancer treatments, including radiation and chemotherapy, is diminished in areas with low oxygen levels. Clostridium novyi-NT can thrive in this oxygen-deprived environment, facilitating the destruction of hypoxic and necrotic regions within tumors. Clostridium novyi-NT bacteria can replicate and selectively target cancer cells. The production of toxins by these bacteria can inflict damage upon tumor cells and incite an immune response leading to the eradication of the tumor (99). In the phase I trial (NCT01924689) involving 24 individuals with solid neoplasms, the intratumoral administration of Clostridium novyi-NT initiated the activation of bacterial spores, leading to a 42% incidence of tumor mass breakdown. Among the evaluated cohort of 22 individuals, 41% exhibited a decline in injected tumor dimensions, and 86% showed a stable disease (100). Bifidobacterium fosters the effectiveness of anti-CD47 immunotherapy through its accumulation within the TME, mediated by interferon-dependent mechanisms and the activation of the Stimulator of interferon genes (STING) pathway (60). Following bacterial ablation, the pancreatic ductal adenocarcinoma TME underwent immunogenic reprogramming, characterized by diminished MDSCs and heightened M1 macrophage differentiation, facilitating the Th1 polarization in CD4+ T cells and stimulating the induction of CD8+ T-cell. Augmented PD-1 levels following bacterial ablation were associated with improved efficacy of immunotherapy. An abundant and distinct microbiome triggers the differentiation of suppressive monocytic cells in pancreatic cancer by selectively activating Toll-like receptors (TLRs), ultimately resulting in T-cell anergy (61).
Disruptions in the microbiota contribute to the accumulation of toxic metabolites and the persistence of inflammatory reactions, thus fostering cancer development and the evolution of treatment resistance (2). Remodeling intratumoral microbiota has emerged as a promising avenue for potential therapeutic strategies. Probiotics, antibiotics, and fecal microbiota transplantation are the prevailing techniques utilized for systemic microbiota, offering a feasible avenue for their application in targeting the intratumoral microbiota associated with cancer (31, 101).
5 Conclusions
Amidst the burgeoning interest in unraveling the relationship between gut microbiota and tumors, attention is now directed toward probing the effects of intratumoral microbiota on tumorigenesis and its implications for cancer treatment. Advances in techniques for analyzing the gut and tumor microbiome have enhanced the understanding of the microbiome’s impact on human health. Nevertheless, the exploration of intratumoral bacteria is still in its preliminary phase. Recent findings demonstrate the widespread occurrence of intratumor microbiota in various tumor types. The complexity and ambiguity of the host-intratumoral microbiota interplay necessitate future studies to improve the understanding of the intratumor microbiota in carcinogenesis. Intratumoral microbiota exerts immunomodulatory effects within the TME, influencing tumor outcomes by promoting inflammatory responses or regulating anti-tumor activity. Intratumoral microbiota exerts a significant influence on therapeutic effectiveness, offering novel avenues for cancer therapy, diagnostic and prognostic assessment, and potential therapeutic targets (Figure 4). In particular, the complex interactions among intratumoral microbiota, antitumor immunity, and therapeutic efficacy in tumors require further investigation. Comprehensive profiling of distinct intratumor microbiota holds promise for manipulating these bacterial communities to advance cancer treatment. Further research into the molecular mechanisms of intratumoral microbiota is also necessary. Targeting the intratumoral microbiota presents opportunities for potential universal therapies and synergistic combination approaches with approved chemotherapeutics and immunotherapies. Considering the significant impact of microbial metabolites, integrating microbiome and metabolome profiles may emerge as a pivotal approach for personalized therapies. Undoubtedly, the significance of intratumoral microbiota within tumor biology is poised to assume a pivotal role in forthcoming decades of carcinogenesis investigations.
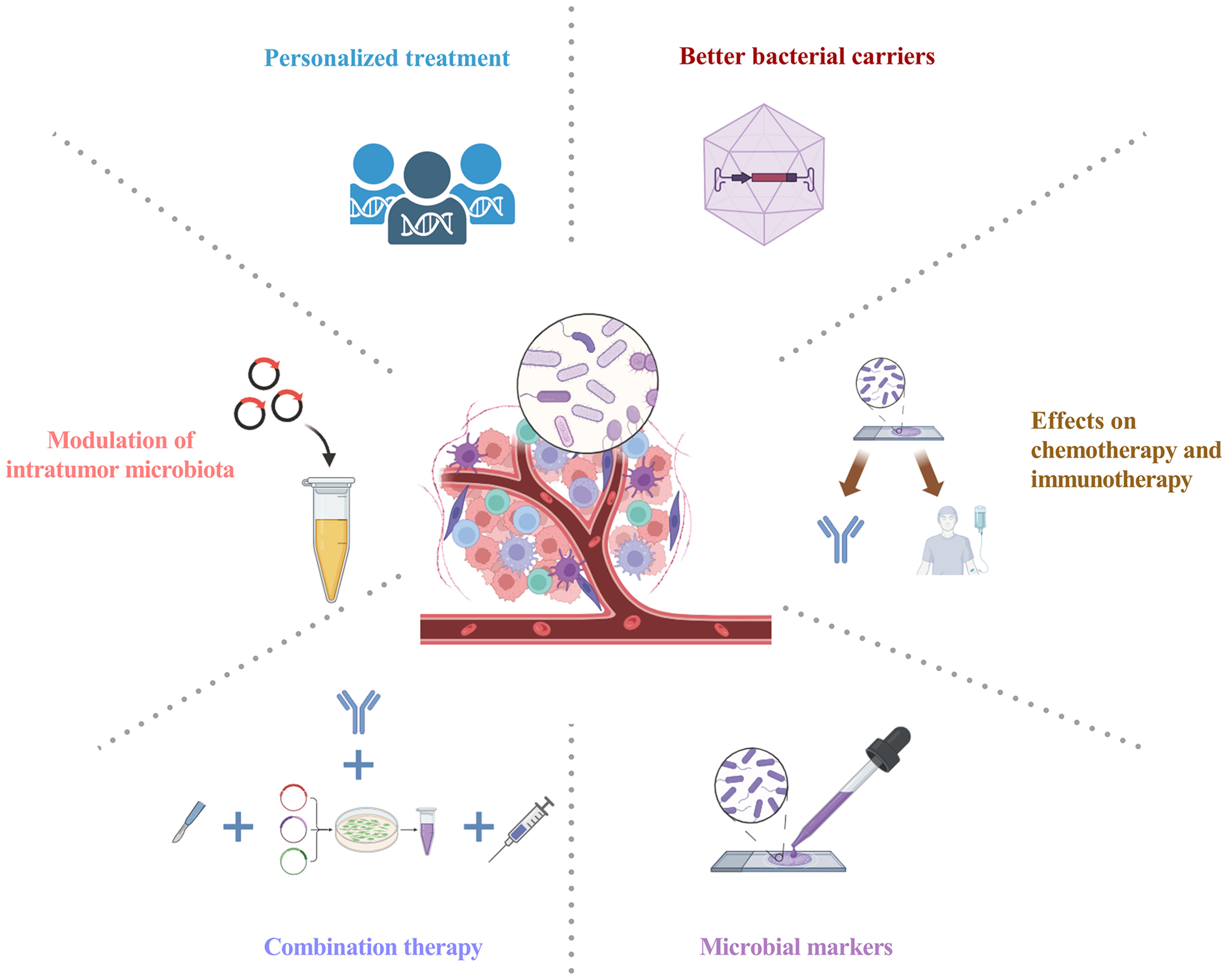
Figure 4 Intratumoral microbiota: prospects for clinical application. Personalized treatment: Integrating advanced sequencing techniques allows for the comprehensive analysis of intratumoral microbiota, shedding light on microbial-derived antigens and paving the way for personalized treatment modalities; Modulation of intratumor microbiota: Leveraging probiotics, antibiotics, and targeted interventions stands as a promising strategy for intratumoral microbiota modulation, aiming to reinstate a harmonized microbial community; Combination therapy: Combining antibiotics or bacterial therapies with other anti-tumor treatments, such as chemotherapy or immunotherapy, seeks to optimize cancer therapy by targeting both tumor cells and the intratumor microbiota; Better bacterial carriers: This innovative strategy maximizes specific bacterial attributes, utilizing advanced carriers for precise tumor therapeutics with strong targeting, lower infection risk, and superior payload efficiency; Microbial markers: Utilizing intratumoral microbiota for early cancer diagnosis, prognosis, and monitoring; Effects on chemotherapy and immunotherapy: Impact of intratumoral microbiota on chemotherapy and immunotherapy, evaluating efficacy, tolerability, and toxicity. Graphics created with BioRender.com.
Author contributions
JW: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Resources, Visualization, Writing – original draft, Writing – review & editing. PZ: Conceptualization, Data curation, Formal analysis, Methodology, Project administration, Resources, Software, Writing – original draft. WM: Conceptualization, Supervision, Validation, Writing – review & editing. CZ: Funding acquisition, Supervision, Writing – review & editing, Writing – original draft.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This present study was supported in part by the National Natural Science Foundation of China (82270940), and the Science and Technology Project of Shenzhen of China (JCYJ2021032414261403).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
TME, tumor microenvironment; TCGA, The Cancer Genome Atlas; PR, Progesterone receptor; ER, Estrogen receptor; HER2, Human epidermal growth factor receptor 2; T3SS, Type 3 secretion system; EMT, Epithelial-mesenchymal transition; NFκB, Transcription factor nuclear factor κB; PRRs, Pattern recognition receptors; TLRs, Toll-like receptors; TLR4, Toll-like receptor 4; TNFSF4, Toll-like receptor (TLR) 4 and OX40 ligand; Tregs, Regulatory T cells; MDSCs, Myeloid-derived suppressor cells; TAMs, Tumor-associated macrophages; NK, Natural killer; CEACAM1, Carcinoembryonic antigen-related cell-adhesion molecule 1; BCG, Bacillus Calmette-Guérin; CDDL, Bacterial enzyme cytidine deaminase; dFdU, 2’2-difluorodeoxyuridine; ROS, Reactive oxygen species; STING, Stimulator of interferon genes; FMT, Fecal microbial transplantation; NACI, neoadjuvant chemoimmunotherapy.
References
1. Dekaboruah E, Suryavanshi MV, Chettri D, Verma AK. Human microbiome: an academic update on human body site specific surveillance and its possible role. Arch Microbiol (2020) 202:2147–67. doi: 10.1007/s00203-020-01931-x
2. Hou K, Wu ZX, Chen XY, Wang JQ, Zhang D, Xiao C, et al. Microbiota in health and diseases. Signal Transduct Target Ther (2022) 7:135. doi: 10.1038/s41392-022-00974-4
3. Yang L, Li A, Wang Y, Zhang Y. Intratumoral microbiota: roles in cancer initiation, development and therapeutic efficacy. Signal Transduct Target Ther (2023) 8:35. doi: 10.1038/s41392-022-01304-4
4. Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet (2012) 13:260–70. doi: 10.1038/nrg3182
5. Morales-Sanchez A, Fuentes-Panana EM. Human viruses and cancer. Viruses (2014) 6:4047–79. doi: 10.3390/v6104047
6. Azevedo MM, Pina-Vaz C, Baltazar F. Microbes and cancer: friends or faux? Int J Mol Sci (2020) 21:3115. doi: 10.3390/ijms21093115
7. Vyshenska D, Lam KC, Shulzhenko N, Morgun A. Interplay between viruses and bacterial microbiota in cancer development. Semin Immunol (2017) 32:14–24. doi: 10.1016/j.smim.2017.05.003
8. Contreras AV, Cocom-Chan B, Hernandez-Montes G, Portillo-Bobadilla T, Resendis-Antonio O. Host-microbiome interaction and cancer: potential application in precision medicine. Front Physiol (2016) 7:606. doi: 10.3389/fphys.2016.00606
9. Galeano Nino JL, Wu H, LaCourse KD, Kempchinsky AG, Baryiames A, Barber B, et al. Effect of the intratumoral microbiota on spatial and cellular heterogeneity in cancer. Nature (2022) 611:810–7. doi: 10.1038/s41586-022-05435-0
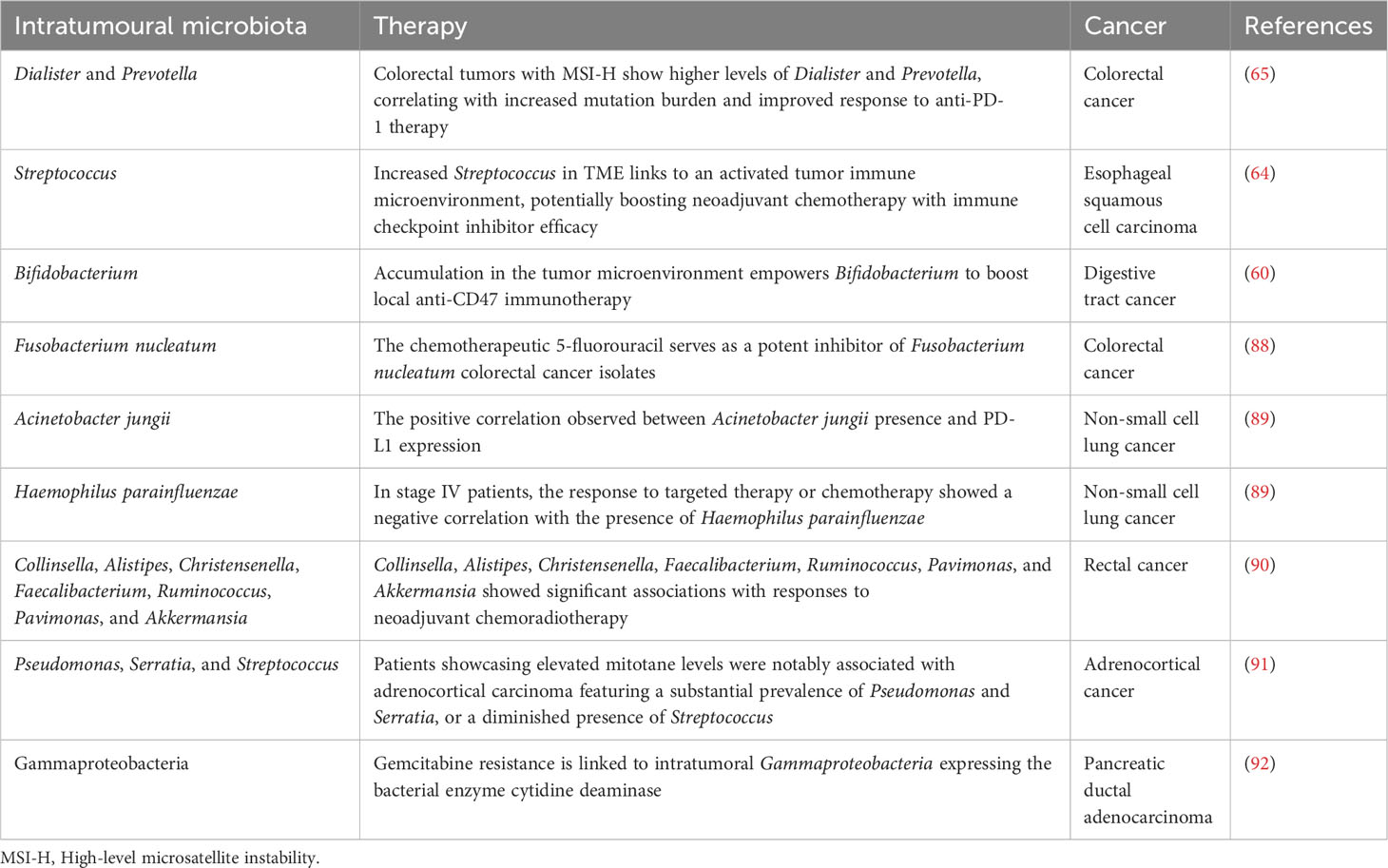
10. Chen Y, Wu FH, Wu PQ, Xing HY, Ma T. The role of the tumor microbiome in tumor development and its treatment. Front Immunol (2022) 13:935846. doi: 10.3389/fimmu.2022.935846
11. Xie Y, Xie F, Zhou X, Zhang L, Yang B, Huang J, et al. Microbiota in tumors: from understanding to application. Adv Sci (Weinh) (2022) 9:e2200470. doi: 10.1002/advs.202200470
12. Li X, Saxena D. The tumor mycobiome: A paradigm shift in cancer pathogenesis. Cell (2022) 185:3648–51. doi: 10.1016/j.cell.2022.09.013
13. Dohlman AB, Klug J, Mesko M, Gao IH, Lipkin SM, Shen X, et al. A pan-cancer mycobiome analysis reveals fungal involvement in gastrointestinal and lung tumors. Cell (2022) 185:3807–22.e12. doi: 10.1016/j.cell.2022.09.015
14. Nejman D, Livyatan I, Fuks G, Gavert N, Zwang Y, Geller LT, et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science (2020) 368:973–80. doi: 10.1126/science.aay9189
15. Narunsky-Haziza L, Sepich-Poore GD, Livyatan I, Asraf O, Martino C, Nejman D, et al. Pan-cancer analyses reveal cancer-type-specific fungal ecologies and bacteriome interactions. Cell (2022) 185:3789–806.e17. doi: 10.1016/j.cell.2022.09.005
16. Chen Y, Liu B, Wei Y, Kuang DM. Influence of gut and intratumoral microbiota on the immune microenvironment and anti-cancer therapy. Pharmacol Res (2021) 174:105966. doi: 10.1016/j.phrs.2021.105966
17. Wang M, Yu F, Li P. Intratumor microbiota in cancer pathogenesis and immunity: from mechanisms of action to therapeutic opportunities. Front Immunol (2023) 14:1269054. doi: 10.3389/fimmu.2023.1269054
18. Ferrari V, Rescigno M. The intratumoral microbiota: friend or foe? Trends Cancer (2023) 9:472–9. doi: 10.1016/j.trecan.2023.03.005
19. Gong Y, Huang X, Wang M, Liang X. Intratumor microbiota: a novel tumor component. J Cancer Res Clin Oncol (2023) 149:6675–91. doi: 10.1007/s00432-023-04576-7
20. Jiang Z, Zhang W, Zhang Z, Sha G, Wang D, Tang D. Intratumoral microbiota: A new force in diagnosing and treating pancreatic cancer. Cancer Lett (2023) 554:216031. doi: 10.1016/j.canlet.2022.216031
21. Xue C, Chu Q, Zheng Q, Yuan X, Su Y, Bao Z, et al. Current understanding of the intratumoral microbiome in various tumors. Cell Rep Med (2023) 4:100884. doi: 10.1016/j.xcrm.2022.100884
22. Sepich-Poore GD, Zitvogel L, Straussman R, Hasty J, Wargo JA, Knight R. The microbiome and human cancer. Science (2021) 371:eabc4552. doi: 10.1126/science.abc4552
23. Qiao H, Tan XR, Li H, Li JY, Chen XZ, Li YQ, et al. Association of intratumoral microbiota with prognosis in patients with nasopharyngeal carcinoma from 2 hospitals in China. JAMA Oncol (2022) 8:1301–9. doi: 10.1001/jamaoncol.2022.2810
24. Gao F, Yu B, Rao B, Sun Y, Yu J, Wang D, et al. The effect of the intratumoral microbiome on tumor occurrence, progression, prognosis and treatment. Front Immunol (2022) 13:1051987. doi: 10.3389/fimmu.2022.1051987
25. Zheng HH, Du CT, Yu C, Tang XY, Huang RL, Zhang YZ, et al. The relationship of tumor microbiome and oral bacteria and intestinal dysbiosis in canine mammary tumor. Int J Mol Sci (2022) 23:10928. doi: 10.3390/ijms231810928
26. Liang Y, Li Q, Liu Y, Guo Y, Li Q. Awareness of intratumoral bacteria and their potential application in cancer treatment. Discovery Oncol (2023) 14:57. doi: 10.1007/s12672-023-00670-x
27. Dohlman AB, Arguijo Mendoza D, Ding S, Gao M, Dressman H, Iliev ID, et al. The cancer microbiome atlas: a pan-cancer comparative analysis to distinguish tissue-resident microbiota from contaminants. Cell Host Microbe (2021) 29:281–98.e5. doi: 10.1016/j.chom.2020.12.001
28. Poore GD, Kopylova E, Zhu Q, Carpenter C, Fraraccio S, Wandro S, et al. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature (2020) 579:567–74. doi: 10.1038/s41586-020-2095-1
29. Fu A, Yao B, Dong T, Chen Y, Yao J, Liu Y, et al. Tumor-resident intracellular microbiota promotes metastatic colonization in breast cancer. Cell (2022) 185:1356–72.e26. doi: 10.1016/j.cell.2022.02.027
30. Rye MS, Garrett KL, Holt RA, Platell CF, McCoy MJ. Fusobacterium nucleatum and Bacteroides fragilis detection in colorectal tumours: Optimal target site and correlation with total bacterial load. PloS One (2022) 17:e0262416. doi: 10.1371/journal.pone.0262416
31. Liu J, Zhang Y. Intratumor microbiome in cancer progression: current developments, challenges and future trends. biomark Res (2022) 10:37. doi: 10.1186/s40364-022-00381-5
32. Zhang Y, Zhang L, Zheng S, Li M, Xu C, Jia D, et al. Fusobacterium nucleatum promotes colorectal cancer cells adhesion to endothelial cells and facilitates extravasation and metastasis by inducing ALPK1/NF-kappaB/ICAM1 axis. Gut Microbes (2022) 14:2038852. doi: 10.1080/19490976.2022.2038852
33. Wroblewski LE, Peek RM Jr., Wilson KT. Helicobacter pylori and gastric cancer: factors that modulate disease risk. Clin Microbiol Rev (2010) 23:713–39. doi: 10.1128/CMR.00011-10
34. Wei Q, Zhang Q, Wu Y, Han S, Yin L, Zhang J, et al. Analysis of bacterial diversity and community structure in gastric juice of patients with advanced gastric cancer. Discovery Oncol (2023) 14:7. doi: 10.1007/s12672-023-00612-7
35. Liang J, Li T, Zhao J, Wang C, Sun H. Current understanding of the human microbiome in glioma. Front Oncol (2022) 12:781741. doi: 10.3389/fonc.2022.781741
36. Noguti J, Chan AA, Bandera B, Brislawn CJ, Protic M, Sim MS, et al. Both the intratumoral immune and microbial microenvironment are linked to recurrence in human colon cancer: results from a prospective, multicenter nodal ultrastaging trial. Oncotarget (2018) 9:23564–76. doi: 10.18632/oncotarget.25276
37. Khan MAW, Ologun G, Arora R, McQuade JL, Wargo JA. Gut microbiome modulates response to cancer immunotherapy. Dig Dis Sci (2020) 65:885–96. doi: 10.1007/s10620-020-06111-x
38. Fu A, Yao B, Dong T, Cai S. Emerging roles of intratumor microbiota in cancer metastasis. Trends Cell Biol (2022) 33:583–93. doi: 10.1016/j.tcb.2022.11.007
39. Wang G, He X, Wang Q. Intratumoral bacteria are an important "accomplice" in tumor development and metastasis. Biochim Biophys Acta Rev Cancer (2023) 1878:188846. doi: 10.1016/j.bbcan.2022.188846
40. Chen J, Li T, Liang J, Huang Q, Huang JD, Ke Y, et al. Current status of intratumour microbiome in cancer and engineered exogenous microbiota as a promising therapeutic strategy. BioMed Pharmacother (2022) 145:112443. doi: 10.1016/j.biopha.2021.112443
41. Bhatt AP, Redinbo MR, Bultman SJ. The role of the microbiome in cancer development and therapy. CA Cancer J Clin (2017) 67:326–44. doi: 10.3322/caac.21398
42. Barrett M, Hand CK, Shanahan F, Murphy T, O'Toole PW. Mutagenesis by microbe: the role of the microbiota in shaping the cancer genome. Trends Cancer (2020) 6:277–87. doi: 10.1016/j.trecan.2020.01.019
43. Pleguezuelos-Manzano C, Puschhof J, Rosendahl Huber A, van Hoeck A, Wood HM, Nomburg J, et al. Mutational signature in colorectal cancer caused by genotoxic pks(+) E. coli. Nat (2020) 580:269–73. doi: 10.1038/s41586-020-2080-8
44. Liu Y, Fu K, Wier EM, Lei Y, Hodgson A, Xu D, et al. Bacterial genotoxin accelerates transient infection-driven murine colon tumorigenesis. Cancer Discovery (2022) 12:236–49. doi: 10.1158/2159-8290.CD-21-0912
45. Rivas-Dominguez A, Pastor N, Martinez-Lopez L, Colon-Perez J, Bermudez B, Orta ML. The role of DNA damage response in dysbiosis-induced colorectal cancer. Cells (2021) 10:1934. doi: 10.3390/cells10081934
46. Bellotti R, Speth C, Adolph TE, Lass-Florl C, Effenberger M, Ofner D, et al. Micro- and mycobiota dysbiosis in pancreatic ductal adenocarcinoma development. Cancers (Basel) (2021) 13:3431. doi: 10.3390/cancers13143431
47. Udayasuryan B, Ahmad RN, Nguyen TTD, Umana A, Monet Roberts L, Sobol P, et al. Fusobacterium nucleatum induces proliferation and migration in pancreatic cancer cells through host autocrine and paracrine signaling. Sci Signal (2022) 15:eabn4948. doi: 10.1126/scisignal.abn4948
48. Guo P, Tian Z, Kong X, Yang L, Shan X, Dong B, et al. FadA promotes DNA damage and progression of Fusobacterium nucleatum-induced colorectal cancer through up-regulation of chk2. J Exp Clin Cancer Res (2020) 39:202. doi: 10.1186/s13046-020-01677-w
49. Song X, Xin N, Wang W, Zhao C. Wnt/beta-catenin, an oncogenic pathway targeted by H. pylori in gastric carcinogenesis. Oncotarget (2015) 6:35579–88. doi: 10.18632/oncotarget.5758
50. Parida S, Wu S, Siddharth S, Wang G, Muniraj N, Nagalingam A, et al. A procarcinogenic colon microbe promotes breast tumorigenesis and metastatic progression and concomitantly activates notch and beta-catenin axes. Cancer Discovery (2021) 11:1138–57. doi: 10.1158/2159-8290.CD-20-0537
51. Greten FR, Grivennikov SI. Inflammation and cancer: triggers, mechanisms, and consequences. Immunity (2019) 51:27–41. doi: 10.1016/j.immuni.2019.06.025
52. Zhao H, Wu L, Yan G, Chen Y, Zhou M, Wu Y, et al. Inflammation and tumor progression: signaling pathways and targeted intervention. Signal Transduct Target Ther (2021) 6:263. doi: 10.1038/s41392-021-00658-5
53. Armstrong H, Bording-Jorgensen M, Dijk S, Wine E. The complex interplay between chronic inflammation, the microbiome, and cancer: understanding disease progression and what we can do to prevent it. Cancers (Basel) (2018) 10:83. doi: 10.3390/cancers10030083
55. Villemin C, Six A, Neville BA, Lawley TD, Robinson MJ, Bakdash G. The heightened importance of the microbiome in cancer immunotherapy. Trends Immunol (2023) 44:44–59. doi: 10.1016/j.it.2022.11.002
56. Qiao H, Li H, Wen X, Tan X, Yang C, Liu N. Multi-omics integration reveals the crucial role of fusobacterium in the inflammatory immune microenvironment in head and neck squamous cell carcinoma. Microbiol Spectr (2022) 10:e0106822. doi: 10.1128/spectrum.01068-22
57. Yang Y, Weng W, Peng J, Hong L, Yang L, Toiyama Y, et al. Fusobacterium nucleatum increases proliferation of colorectal cancer cells and tumor development in mice by activating toll-like receptor 4 signaling to nuclear factor-kappaB, and up-regulating expression of microRNA-21. Gastroenterology (2017) 152:851–66.e24. doi: 10.1053/j.gastro.2016.11.018
58. He Y, Zhang Q, Yu X, Zhang S, Guo W. Overview of microbial profiles in human hepatocellular carcinoma and adjacent nontumor tissues. J Transl Med (2023) 21:68. doi: 10.1186/s12967-023-03938-6
59. Wang Y, Fu K. Genotoxins: the mechanistic links between escherichia coli and colorectal cancer. Cancers (Basel) (2023) 15:1152. doi: 10.3390/cancers15041152
60. Shi Y, Zheng W, Yang K, Harris KG, Ni K, Xue L, et al. Intratumoral accumulation of gut microbiota facilitates CD47-based immunotherapy via STING signaling. J Exp Med (2020) 217:e20192282. doi: 10.1084/jem.20192282
61. Pushalkar S, Hundeyin M, Daley D, Zambirinis CP, Kurz E, Mishra A, et al. The pancreatic cancer microbiome promotes oncogenesis by induction of innate and adaptive immune suppression. Cancer Discovery (2018) 8:403–16. doi: 10.1158/2159-8290.CD-17-1134
62. Bullman S. INVADEseq to study the intratumoural microbiota at host single-cell resolution. Nat Rev Cancer (2023) 23:189. doi: 10.1038/s41568-023-00553-x
63. Riquelme E, Zhang Y, Zhang L, Montiel M, Zoltan M, Dong W, et al. Tumor microbiome diversity and composition influence pancreatic cancer outcomes. Cell (2019) 178:795–806 e12. doi: 10.1016/j.cell.2019.07.008
64. Wu H, Leng X, Liu Q, Mao T, Jiang T, Liu Y, et al. Intratumoral microbiota composition regulates chemoimmunotherapy response in esophageal squamous cell carcinoma. Cancer Res (2023) 83:3131–44. doi: 10.1158/0008-5472.CAN-22-2593
65. Byrd DA, Fan W, Greathouse KL, Wu MC, Xie H, Wang X. The intratumor microbiome is associated with microsatellite instability. J Natl Cancer Inst (2023) 115:989–93. doi: 10.1093/jnci/djad083
66. Zhang S, Zhang S, Ma X, Zhan J, Pan C, Zhang H, et al. Intratumoral microbiome impacts immune infiltrates in tumor microenvironment and predicts prognosis in esophageal squamous cell carcinoma patients. Front Cell Infect Microbiol (2023) 13:1165790. doi: 10.3389/fcimb.2023.1165790
67. Zhang X, Yu D, Wu D, Gao X, Shao F, Zhao M, et al. Tissue-resident Lachnospiraceae family bacteria protect against colorectal carcinogenesis by promoting tumor immune surveillance. Cell Host Microbe (2023) 31:418–32.e8. doi: 10.1016/j.chom.2023.01.013
68. Sheng D, Yue K, Li H, Zhao L, Zhao G, Jin C, et al. The interaction between intratumoral microbiome and immunity is related to the prognosis of ovarian cancer. Microbiol Spectr (2023) 11:e0354922. doi: 10.1128/spectrum.03549-22
69. Tang Q, Peng X, Xu B, Zhou X, Chen J, Cheng L. Current status and future directions of bacteria-based immunotherapy. Front Immunol (2022) 13:911783. doi: 10.3389/fimmu.2022.911783
70. Fan JY, Huang Y, Li Y, Muluh TA, Fu SZ, Wu JB. Bacteria in cancer therapy: A new generation of weapons. Cancer Med (2022) 11:4457–68. doi: 10.1002/cam4.4799
71. Boesch M, Baty F, Rothschild SI, Tamm M, Joerger M, Fruh M, et al. Tumour neoantigen mimicry by microbial species in cancer immunotherapy. Br J Cancer (2021) 125:313–23. doi: 10.1038/s41416-021-01365-2
72. Ma J, Huang L, Hu D, Zeng S, Han Y, Shen H. The role of the tumor microbe microenvironment in the tumor immune microenvironment: bystander, activator, or inhibitor? J Exp Clin Cancer Res (2021) 40:327. doi: 10.1186/s13046-021-02128-w
73. Li D, Wu M. Pattern recognition receptors in health and diseases. Signal Transduct Target Ther (2021) 6:291. doi: 10.1038/s41392-021-00687-0
74. Rumpret M, von Richthofen HJ, Peperzak V, Meyaard L. Inhibitory pattern recognition receptors. J Exp Med (2022) 219:e20211463. doi: 10.1084/jem.20211463
75. Rossi T, Vergara D, Fanini F, Maffia M, Bravaccini S, Pirini F. Microbiota-derived metabolites in tumor progression and metastasis. Int J Mol Sci (2020) 21:2786. doi: 10.3390/ijms21165786
76. Triner D, Devenport SN, Ramakrishnan SK, Ma X, Frieler RA, Greenson JK, et al. Neutrophils restrict tumor-associated microbiota to reduce growth and invasion of colon tumors in mice. Gastroenterology (2019) 156:1467–82. doi: 10.1053/j.gastro.2018.12.003
77. Lee JA, Yoo SY, Oh HJ, Jeong S, Cho NY, Kang GH, et al. Differential immune microenvironmental features of microsatellite-unstable colorectal cancers according to Fusobacterium nucleatum status. Cancer Immunol Immunother (2021) 70:47–59. doi: 10.1007/s00262-020-02657-x
78. Panebianco C, Ciardiello D, Villani A, Maiorano BA, Latiano TP, Maiello E, et al. Insights into the role of gut and intratumor microbiota in pancreatic ductal adenocarcinoma as new key players in preventive, diagnostic and therapeutic perspective. Semin Cancer Biol (2022) 86:997–1007. doi: 10.1016/j.semcancer.2021.11.007
79. Chou WC, Rampanelli E, Li X, Ting JP. Impact of intracellular innate immune receptors on immunometabolism. Cell Mol Immunol (2022) 19:337–51. doi: 10.1038/s41423-021-00780-y
80. Labani-Motlagh A, Ashja-Mahdavi M, Loskog A. The tumor microenvironment: A milieu hindering and obstructing antitumor immune responses. Front Immunol (2020) 11:940. doi: 10.3389/fimmu.2020.00940
81. Boutilier AJ, Elsawa SF. Macrophage polarization states in the tumor microenvironment. Int J Mol Sci (2021) 22:6995. doi: 10.3390/ijms22136995
82. Wei F, Zhong S, Ma Z, Kong H, Medvec A, Ahmed R, et al. Strength of PD-1 signaling differentially affects T-cell effector functions. Proc Natl Acad Sci U.S.A. (2013) 110:E2480–9. doi: 10.1073/pnas.1305394110
83. Boulter L, Bullock E, Mabruk Z, Brunton VG. The fibrotic and immune microenvironments as targetable drivers of metastasis. Br J Cancer (2021) 124:27–36. doi: 10.1038/s41416-020-01172-1
84. Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M, et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity (2015) 42:344–55. doi: 10.1016/j.immuni.2015.01.010
85. Gur C, Maalouf N, Shhadeh A, Berhani O, Singer BB, Bachrach G, et al. Fusobacterium nucleatum supresses anti-tumor immunity by activating CEACAM1. Oncoimmunology (2019) 8:e1581531. doi: 10.1080/2162402X.2019.1581531
86. Jin C, Lagoudas GK, Zhao C, Bullman S, Bhutkar A, Hu B, et al. Commensal Microbiota Promote Lung Cancer Development via gammadelta T Cells. Cell (2019) 176:998–1013.e16. doi: 10.1016/j.cell.2018.12.040
87. Mima K, Sukawa Y, Nishihara R, Qian ZR, Yamauchi M, Inamura K, et al. Fusobacterium nucleatum and T cells in colorectal carcinoma. JAMA Oncol (2015) 1:653–61. doi: 10.1001/jamaoncol.2015.1377
88. LaCourse KD, Zepeda-Rivera M, Kempchinsky AG, Baryiames A, Minot SS, Johnston CD, et al. The cancer chemotherapeutic 5-fluorouracil is a potent Fusobacterium nucleatum inhibitor and its activity is modified by intratumoral microbiota. Cell Rep (2022) 41:111625. doi: 10.1016/j.celrep.2022.111625
89. Zhang M, Zhang Y, Sun Y, Wang S, Liang H, Han Y. Intratumoral microbiota impacts the first-line treatment efficacy and survival in non-small cell lung cancer patients free of lung infection. J Healthc Eng (2022) 2022:5466853. doi: 10.1155/2022/5466853
90. Sun L, Qu J, Ke X, Zhang Y, Xu H, Lv N, et al. Interaction between intratumoral microbiota and tumor mediates the response of neoadjuvant therapy for rectal cancer. Front Microbiol (2023) 14:1229888. doi: 10.3389/fmicb.2023.1229888
91. Cantini G, Niccolai E, Canu L, Di Gloria L, Baldi S, Propato AP, et al. Intratumour microbiota modulates adrenocortical cancer responsiveness to mitotane. Endocr Relat Cancer (2023) 30:e230094. doi: 10.1530/ERC-23-0094
92. Geller LT, Barzily-Rokni M, Danino T, Jonas OH, Shental N, Nejman D, et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science (2017) 357:1156–60. doi: 10.1126/science.aah5043
93. Cogdill AP, Gaudreau PO, Arora R, Gopalakrishnan V, Wargo JA. The impact of intratumoral and gastrointestinal microbiota on systemic cancer therapy. Trends Immunol (2018) 39:900–20. doi: 10.1016/j.it.2018.09.007
94. Sayin S, Rosener B, Li CG, Ho B, Ponomarova O, Ward DV, et al. Evolved bacterial resistance to the chemotherapy gemcitabine modulates its efficacy in co-cultured cancer cells. Elife (2023) 12:e83140. doi: 10.7554/eLife.83140
95. Sitthirak S, Suksawat M, Phetcharaburanin J, Wangwiwatsin A, Klanrit P, Namwat N, et al. Chemotherapeutic resistant cholangiocarcinoma displayed distinct intratumoral microbial composition and metabolic profiles. PeerJ (2022) 10:e13876. doi: 10.7717/peerj.13876
96. Geller LT, Straussman R. Intratumoral bacteria may elicit chemoresistance by metabolizing anticancer agents. Mol Cell Oncol (2018) 5:e1405139. doi: 10.1080/23723556.2017.1405139
97. Chiba A, Bawaneh A, Velazquez C, Clear KYJ, Wilson AS, Howard-McNatt M, et al. Neoadjuvant chemotherapy shifts breast tumor microbiota populations to regulate drug responsiveness and the development of metastasis. Mol Cancer Res (2020) 18:130–9. doi: 10.1158/1541-7786.MCR-19-0451
98. Cardillo F, Bonfim M, da Silva Vasconcelos Sousa P, Mengel J, Ribeiro Castello-Branco LR, Pinho RT. Bacillus calmette-guerin immunotherapy for cancer. Vaccines (Basel) (2021) 9:439. doi: 10.3390/vaccines9050439
99. Staedtke V, Roberts NJ, Bai RY, Zhou S. Clostridium novyi-NT in cancer therapy. Genes Dis (2016) 3:144–52. doi: 10.1016/j.gendis.2016.01.003
100. Janku F, Zhang HH, Pezeshki A, Goel S, Murthy R, Wang-Gillam A, et al. Intratumoral injection of clostridium novyi-NT spores in patients with treatment-refractory advanced solid tumors. Clin Cancer Res (2021) 27:96–106. doi: 10.1158/1078-0432.CCR-20-2065
Keywords: intratumoral microbiota, immunotherapy, cancer, treatment, tumor microenvironment
Citation: Wu J, Zhang P, Mei W and Zeng C (2024) Intratumoral microbiota: implications for cancer onset, progression, and therapy. Front. Immunol. 14:1301506. doi: 10.3389/fimmu.2023.1301506
Received: 25 September 2023; Accepted: 28 December 2023;
Published: 16 January 2024.
Edited by:
Nathella Pavan Kumar, National Institute of Research in Tuberculosis (ICMR), IndiaReviewed by:
Beatrice Omusiro Ondondo, University Hospitals of Leicester NHS Trust, United KingdomAbdullah Saeed, City of Hope National Medical Center, United States
Copyright © 2024 Wu, Zhang, Mei and Zeng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Changchun Zeng, emVuZ2NoY2hAZ2xtYy5lZHUuY24=
 Jinmei Wu1
Jinmei Wu1 Pengfei Zhang
Pengfei Zhang Changchun Zeng
Changchun Zeng